- 1Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia
- 2State Research Center of Dermatovenerology and Cosmetology, Russian Ministry of Health, Moscow, Russia
Comparative whole-genome analysis was performed for Neisseria gonorrhoeae isolates belonging to the Neisseria gonorrhoeae multiantigen sequence typing (NG-MAST) types predominant worldwide — 225, 1407, 2400, 2992, and 4186 — and to genogroup 807, the most common genogroup in the Russian Federation. Here, for the first time, the complete genomes of 25 N. gonorrhoeae isolates from genogroup 807 were obtained. For NG-MAST types 225, 1407, 2400, 2992, and 4186, genomes from the Pathogenwatch database were used. The phylogenetic network constructed for 150 genomes showed that the clustering according to NG-MAST type corresponded to the clustering according to genome. Comparisons of genomes of the six sequence types revealed 8-20 genes specific to each sequence type, including the loci for phase variations and genetic components of the gonococcal genetic island (GGI). NG-MAST type 2992 and 4186 isolates either lacked the GGI or carried critical mutations in genes essential for DNA secretion. In all analyzed genogroup 807 isolates, substitution of the essential atlA gene with the eppA gene was found, accompanied by a change in the traG allele, replacement of the ych gene with ych1, and the absence of the exp1 gene, which is likely to result in loss of GGI functionality. For the NG-MAST type 225, 1407 and 2400 isolates, no premature stop codons or reading frameshifts were found in the genes essential for GGI function. A relationship between isolate susceptibility to ciprofloxacin, penicillin, tetracycline and the presence of lesions in GGI genes necessary for DNA secretion was established. The N. gonorrhoeae evolutionary pathways, which allow a particular sequence type to maintain long-term predominance in the population, may include changes in genes responsible for adhesion and virulence, changes in the GGI structure, preservation of genes carrying drug resistance determinants, and changes in genes associated with host adaptation or encoding enzymes of biochemical pathways.
Introduction
Gonococcal infection caused by Neisseria gonorrhoeae is one of the most common sexually transmitted diseases. N. gonorrhoeae can quickly acquire resistance to antimicrobial drugs used for the treatment (Unemo and Shafer, 2014; Unemo and Jensen, 2017; Unemo et al., 2019). The development of resistance to third-generation cephalosporins (ceftriaxone and cefixime), the modern drugs of choice for the treatment of gonorrhea worldwide, poses a great danger, since gonorrhea may become an incurable disease (Unemo, 2015; Tacconelli et al., 2018).
The Neisseria gonorrhoeae multiantigen sequence typing (NG-MAST) scheme is a traditional tool for studying the molecular epidemiology of gonococcal infection (ECDC report, 2018). This method allows one to perform two related challenging endeavors: first, to isolate a meaningful number of genetic variants [sequence types (STs)] within the N. gonorrhoeae species and, on this basis, to analyze the transmission routes of gonococcal infection (Martin et al., 2004); second, to identify and control the spread of the most epidemiologically dangerous clones with multiple resistance to antimicrobial drugs (Chisholm et al., 2013).
New approaches to the study of N. gonorrhoeae molecular epidemiology are based on the use of whole-genome sequencing (WGS) technologies, which allow simultaneous evaluation of both the set of genes characterizing the origin and transfer of the analyzed clinical isolate (De Silva et al., 2016) and the set of genetic determinants of antibiotic resistance (Harrison et al., 2016). In fact, WGS allows typing problems to be solved at a higher level than does the NG-MAST technique, directly linking the origin of the analyzed clinical isolates with their antibiotic resistance (Harrison et al., 2020). WGS data have been successfully used to identify the determinants of resistance in multiresistant isolates from Europe (Jacobsson et al., 2016; Abrams and Trees, 2017; Harris et al., 2018; Ryan et al., 2018) and the USA (Grad et al., 2016); to study phylogenetic relationships, population structure and molecular epidemiology (Demczuk et al., 2015; Ezewudo et al., 2015; Grad et al., 2016; Abrams and Trees, 2017; Ryan et al., 2018; Harrison et al., 2020); and to predict the level of resistance to various drugs (Eyre et al., 2017; Golparian et al., 2018).
Horizontal gene transfer is an important driving force of bacterial evolution. The development of genetic diversity can lead to the accumulation of genes and alleles that help bacteria survive by responding to selection pressures, for example, by acquiring antibiotic resistance genes, virulence/pathogenic factors, genes that contribute to evasion of the host immune response, etc. (Dubnau, 1999; Hamilton and Dillard, 2006). For N. gonorrhoeae, a key mode of chromosomal DNA transfer is most likely a transformation that occurs frequently and efficiently owing to the natural competence of this bacterium (Hamilton et al., 2005; Hamilton and Dillard, 2006). The type IV secretion system (T4SS) allows the bacterial cell to produce and secrete single-stranded DNA (ssDNA), which can then be specifically recognized by pili on recipient cells via DNA uptake sequences (DUS) sequences and recombined into the genome (Hamilton and Dillard, 2006). The T4SS is encoded by genes located on a gonococcal genetic island (GGI) of approximately 59 kb in length (Hamilton et al., 2005; Callaghan et al., 2017; Callaghan et al., 2021). A GGI is present in the genome of ~80% of N. gonorrhoeae isolates. Like all genetic islands, it is a mobile element and itself was once acquired by horizontal transfer (Rotman and Seifert, 2014). The site-specific recombination system XerCD is responsible for its mobility, cutting the flanking difA and difB sites and inserting the island (Dillard and Seifert, 2001; Hamilton et al., 2005; Ramsey et al., 2011; Harrison et al., 2016; Callaghan et al., 2017; Callaghan et al., 2021). As has been shown experimentally, among the 66 GGI genes, only 21 are essential for the function of the system, and 2/3 of the essential genes are tra genes, which are homologous to the genes encoding T4SS of the F-plasmid of E. coli (Hamilton et al., 2005; Pachulec et al., 2014; Callaghan et al., 2017).
Analysis of the PubMLST (https://pubmlst.org) and Pathogenwatch (https://pathogen.watch/genomes/all?organismId=485) databases showed that the most common STs worldwide are NG-MAST types 225, 1407, 2400, 2992 and 4186. NG-MAST 1407 is predominant in many European countries and causes anxiety due to its multidrug resistance, including resistance to third-generation cephalosporins (ECDC report, 2013; Unemo and Shafer, 2014; Unemo and Jensen, 2017; Unemo et al., 2019; Młynarczyk-Bonikowska et al., 2020). Our phylogenetic analysis of Russian isolates in previous works showed that the Russian population of N. gonorrhoeae differs from the European population. Isolates of NG-MAST type 1407 were found only sporadically; isolates of NG-MAST types 225, 2400, 2992, and 4186 were also rare (Kandinov et al., 2020; Shaskolskiy B. et al., 2020; Shaskolskiy B. L. et al., 2020b). The most abundant genogroup in the Russian Federation was the G807 genogroup, accounting for more than 20% of all samples and including the most common NG-MAST types in the Russian Federation, i.e., NG-MAST types 807, 228, 1544, 9570, 9576, and 5941 (Shaskolskiy B. et al., 2020). Notably, ST 807 is rare in Europe: only 5 isolates of this type were identified among 1189 European Centre for Disease Prevention and Control (ECDC) isolates (ECDC report, 2013).
The goal of this work was to perform a comparative whole-genome analysis of N. gonorrhoeae isolates of NG-MAST types predominant worldwide and in the Russian Federation that could identify genes and strongly differing alleles specific to each of the STs and identify phylogenetic relationships for the predominant STs. An important part of the work was analysis of the GGI, which is responsible for horizontal gene transfer, and consideration of the relationship between antimicrobial susceptibility and structural changes in the GGI.
Materials and Methods
WGS of the Isolates From the Russian Federation
For WGS, 25 previously analyzed clinical N. gonorrhoeae isolates were selected (Kubanov et al., 2019; Shaskolskiy et al., 2019; Kandinov et al., 2020) (Table 1). The isolates were assigned to ST 807 and to STs 228, 5941, 9570, and 9576 (all belonging to genogroup 807), in which the combined sequences of the porB and tbpB gene fragments used for NG-MAST typing differed by no more than one nucleotide.
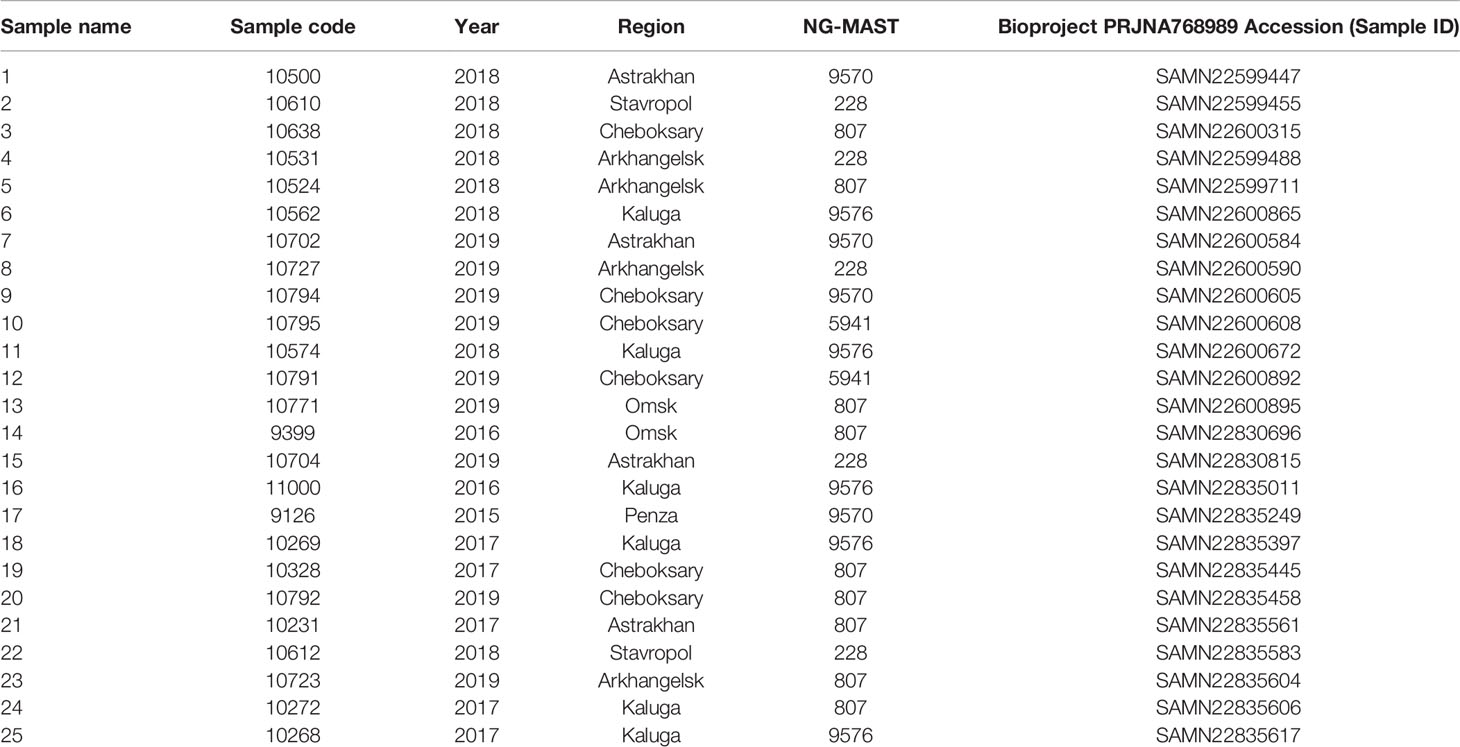
Table 1 Clinical isolates of N. gonorrhoeae from the Russian Federation used for whole-genome sequencing.
Isolates were grown aseptically on separate chocolate agar plates at 37°C in the presence of 5% CO2. Genomic DNA was isolated from overnight cultures of gonococcal cells using a Monarch Genomic DNA Purification Kit (New England Biolabs, UK). The obtained DNA preparations were additionally purified using AMPure XP magnetic beads (Beckman, USA). Final DNA concentrations were measured using a NanoDrop 2000 spectrophotometer and a Qubit 4 Fluorometer (both Thermo Scientific, USA) and ranged from 20-100 ng/μL.
WGS was performed on two platforms: FLO-MIN110 R9 and R10 flow cells in a MinION device (Oxford Nanopore Technologies, UK) and the MiniSeq system (Illumina, USA). For sequencing in the MinION device, a library of DNA fragments was prepared using Oxford Nanopore and New England Biolabs reagent kits according to the native barcoding genomic DNA protocol (with EXP-NBD104, EXP-NBD114, and SQK-LSK109). According to the manufacturer’s protocol, damaged DNA ends were repaired, and subsequent bar coding and ligation of the adapters were carried out. The final library (5-50 fmol) was loaded onto the flow cell.
DNA libraries for sequencing on the Illumina platform were prepared using the DNA fragmentation method followed by PCR and indexing according to the Nextera XT DNA Library (Illumina) protocol. After purification of the libraries, their sizes and concentrations were checked using an automated TapeStation 4150 capillary electrophoresis platform (Agilent, USA). The final library was normalized and denatured according to the instructions for the MiniSeq instrument. The density of clusters averaged 170-250 K/mm2 in all runs. The data output averaged 10.5 gigabytes (GB) out of the 12 GB theoretically possible with this system.
Fifteen N. gonorrhoeae isolates were sequenced in parallel on the Oxford Nanopore and Illumina platforms, and 10 isolates were sequenced only on the Illumina platform. In total, ~1 GB of raw data in fastQ format was collected for each sample from each platform. Hybrid assembly of de novo genomes after sequencing on two platforms (15 genomes) was carried out using the Unicycler program (https://github.com/rrwick/Unicycler). The genomes of the remaining 10 isolates were assembled using SPAdes v3.13.1 (https://github.com/ablab/spades). Quality trimming and adapter clipping were carried out with Trimmomatic v0.39 (https://github.com/usadellab/Trimmomatic). All sequences were uploaded to GenBank, Bioproject PRJNA768989, under the assigned accession numbers (Table 1).
Selection of N. gonorrhoeae Genomes From the Database
To compare genomes and identify genes specific to one ST compared with another, we used the genomes of isolates of the most common STs worldwide available in the Pathogenwatch database (https://pathogen.watch/genomes/all?organismId=485): NG-MAST types 225, 1407, 2400, 2992 and 4186; 25 genomes of each ST; 125 genomes in total. Samples from the database were randomly selected as follows: for each analyzed ST, a list of genomes in the database for the years 2004-2017 was compiled, a set of 25 random unrepeated numbers ranging from 1 to the length of the list was obtained, and genomes were selected in accordance with the set of random numbers. Selected samples are listed in Table S1 of the Supplementary Material.
Construction of the Phylogenetic Network
The selected genomes of N. gonorrhoeae isolates were processed using the Prokka 1.14.6 program (https://github.com/tseemann/prokka) to obtain gff files with annotated genomes. The gff files were then analyzed by rapid large-scale prokaryote pangenome analysis using Roary software (Page et al., 2015). After Roary processing, core genomes containing aligned sequences of concatenated orthologs were obtained. Based on the obtained core genome sequences of isolates belonging to six STs, a phylogenetic network characterizing the relationships between different NG-MAST types was constructed using the SplitsTree 4.17.1 program (Huson and Bryant, 2006).
Identification of Genes and Allelic Variants Specific to Each ST
Upon processing the gff files generated with Prokka 1.14.6 using Roary software, a distribution table of genes in the genomes of all 150 analyzed samples (25 samples x 6 STs), consisting of 2856 genes, was compiled. To clarify the function of the identified genes and to divide them into genes and allelic variants, we carried out annotation using pubMLST. To identify genes specific to the isolates of each analyzed ST, the frequencies of the genes in each of the six STs were calculated. First, genes that were identical across all STs, i.e., genes present in at least 80% of the genomes of all STs were removed. Furthermore, by conducting a pairwise comparison of the gene lists for two compared STs, we determined the number of genes present in 80% (in 20 out of 25 genomes) or more of the genomes of isolates belonging to one ST but were absent from or found in no more than 28% (in 7 out of 25 genomes) of the genomes of isolates belonging to another ST. This selection process yielded a list of 329 genes, which are shown in Table S2 of the Supplementary Material. All specific genes identified by Prokka 1.14.6 were annotated with the MPI Bioinformatics Toolkit (https://toolkit.tuebingen.mpg.de) (ProtBLAST, HHpred) (Gabler et al., 2020) to refine gene functions.
Structural Analysis of GGIs
A sample comprising 150 isolates, i.e., sequences of 125 isolates belonging to STs 225, 1407, 2400, 2992 and 4186, was obtained from the Pathogenwatch database, and the sequences of the 25 Russian isolates of genogroup 807, that we obtained in this work, were analyzed using PubMLST (https://pubmlst.org, section “Single sequence query”) according to the selected typing scheme (“Gonococcal genetic island”). If there was no complete coincidence of the analyzed sequence with the gene sequence in the PubMLST database, for example, if a sequence contained a mutation or stop codon, or the allele was not contained in the database, this sequence was analyzed separately with the choice of a specific locus, for example, locus “NEIS2273(traG)”, “NEIS2275(ych)”, etc., and further checked in the BioEdit program (Ibis Biosciences, Carlsbad, CA) for the presence of mutations, stop codons, or frameshifts.
Comparison of Drug Resistance of NG-MAST 225, 807, 1407, 2400, 2992, and 4186 Isolates
The data on resistance to azithromycin, cefixime, ceftriaxone, ciprofloxacin, penicillin, and tetracycline for N. gonorrhoeae isolates belonging to ST 225 (81 isolates), 1407 (570 isolates), 2400 (269 isolates), 2992 (674 isolates), and 4186 (333 isolates) were obtained from the Pathogenwatch database. For comparison, we used the characteristics of 137 previously studied Russian isolates (Kubanov et al., 2019; Shaskolskiy et al., 2019; Kandinov et al., 2020) belonging to genogroup 807. Isolates with comparisons of their antibiotic susceptibility and their characteristics are listed in Table S3 of the Supplementary Material.
Results
Phylogenetic Network of the Relationship Between Isolates of the Studied NG-MAST Types
To evaluate the genetic diversity of the six NG-MAST types, a phylogenetic network was constructed based on the genomic data of the corresponding isolates (Figure 1). As seen in the figure, the clustering of isolates according to NG-MAST type corresponded to clustering according to the core genomes of N. gonorrhoeae STs. The genetic diversity within each ST differed. The greatest diversity was observed for STs 2992 and 2400; the lowest, for ST 4186.
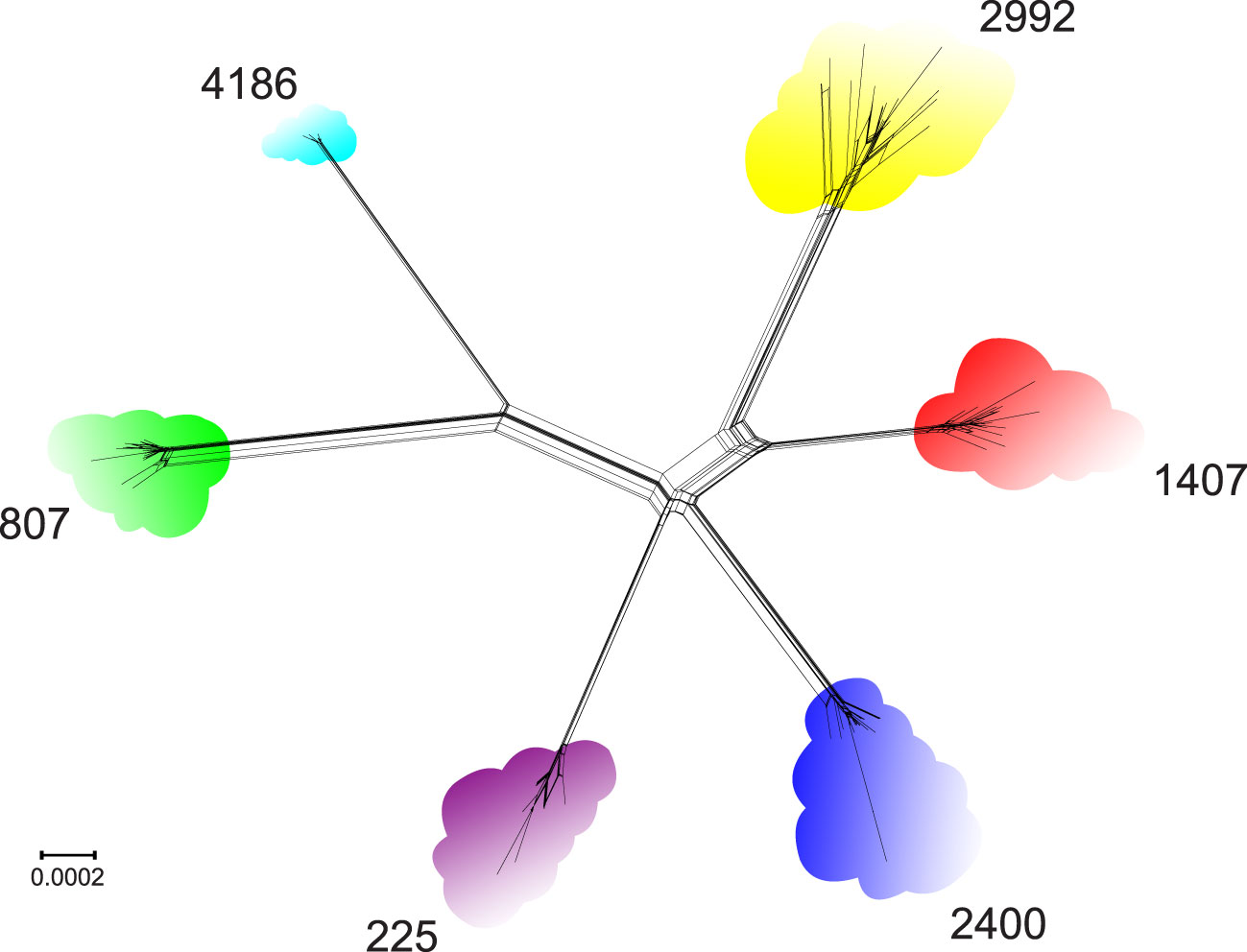
Figure 1 Core genome phylogenetic network of the relationships between N. gonorrhoeae genomes of isolates belonging to STs 225, 807, 1407, 2400, 2992 and 4186.
Structural Comparison of Genetic Gonococcal Islands in Isolates of STs 225, 807, 1407, 2400, 2992, and 4186
Detailed sequence analysis of the genes that compose the GGI in isolates of the six STs (25 isolates per ST) revealed significant differences in the structures of gonococcal islands. All identified sequence alterations are described in Table 2, and the structures of the GGIs in STs 807 and 1407 with the indicated mutations are shown in Figure 2. Both in Table 2 and in Figure 2, GGI genes essential for DNA secretion (Harrison et al., 2016) are highlighted in bold. Allele numbers are given according to the PubMLST nomenclature. Notably, alleles of genes with identified mutations in the GGI genes were not present in the current version of the PubMLST database.
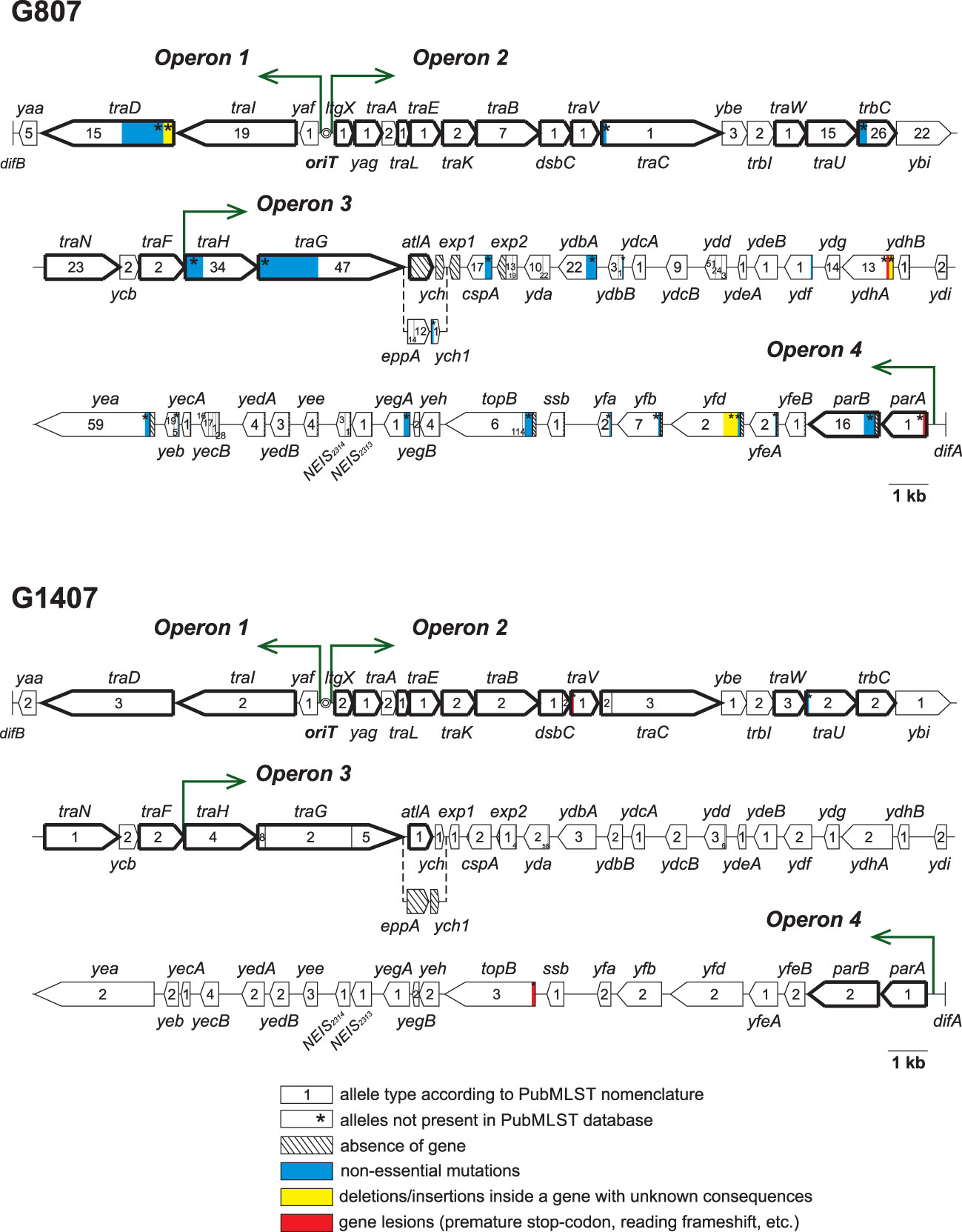
Figure 2 GGI structure of ST 807 and ST 1407 isolates, with definition of genes, alleles and all identified mutations. GGI genes essential for DNA secretion are highlighted in bold.
Not all studied isolates contained a GGI. Most isolates belonging to STs 225, 807, 1407, 2400, and 4186 (24-25 of 25) contained a GGI; however, in isolates of ST 2992, a GGI was present in only 4 of 25 samples.
In the isolates of ST 225, all GGI genes were intact (non-mutated), indicating that they were functioning properly. An exception was the putative helicase gene yea, in which the 1861C→T substitution was found in five samples, leading to the formation of a premature stop codon (Table 2). However, the yea gene is nonessential for GGI function (Harrison et al., 2016).
In the ST 807 isolates, significant changes were revealed in genes both essential and nonessential for GGI function. The identified key change was the replacement of the essential atlA gene with the eppA gene, which occurred in all 25 isolates and was accompanied by the replacement of the ych gene with the ych1 gene and the absence of the exp1 gene. In some isolates of ST 807, changes in traG alleles also occurred, i.e., mutation of NEIS2273(traG) allele 47 or replacement of allele 47 with allele 49, accompanied by mutation (Table 2). These changes can result in the loss of the T4SS DNA secretion ability, as, for example, was shown by Kohler et al. (2013) and Pachulec et al. (2014). Interestingly, although the function of the proteins encoded by the atlA (autolysin A, peptidoglycan transglycosylase), and eppA (endopeptidase, M23 metallopeptidase) genes is cleavage of the peptidoglycan during the formation of a pore in the bacterial cell wall for installing the T4SS, the Epp protein cannot replace AtlA, for an unknown reason (Kohler et al., 2013).
The isolates of ST 1407 showed a small number of changes in both essential and nonessential GGI genes, and all the changes were observed in only one or two of the 25 samples. In the ST 2400 samples, all GGI genes were intact and appeared to be functioning.
Only a small fraction of the ST 2992 samples carried the GGI. One sample with the GGI harbored substitution of the atlA gene with the eppA gene, which could lead to loss of the functional activity of the GGI.
The isolates of ST 4186 carried several mutations in essential and nonessential GGI genes. The most important change was the presence of a stop codon in the important traI gene, encoding the relaxase, in all 24 GGI-harboring samples; thus, none of these isolates were likely to be able to secrete ssDNA.
Thus, although 126 (84%) of the studied isolates in the sample carried a GGI in their genome, only 75 of them (59.5%) were potentially capable of secreting ssDNA.
Antimicrobial Resistance of N. gonorrhoeae Isolates of Different STs and Its Association With GGI Structure
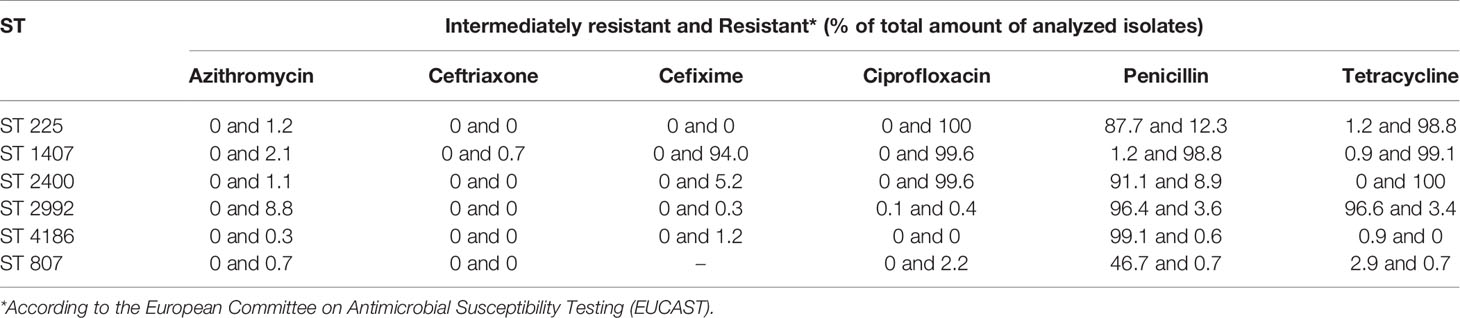
Next, we investigated the susceptibility of isolates of different STs to antimicrobial drugs, including antimicrobials currently used (azithromycin, ceftriaxone, and cefixime) and those previously used (ciprofloxacin, penicillin, and tetracycline) to treat gonococcal infections. Data on the drug susceptibility of ST 225, 1407, 2400, 2992, and 4186 isolates obtained from the Pathogenwatch database and data on the Russian isolates of ST 807 were used, for 2064 isolates in total. The ratio of drug-resistant isolates differed across the STs (Table 3; initial data are provided in Table S3 of the Supplementary Material).
Most ST 807 and ST 4186 isolates were susceptible to antimicrobials except for penicillin. No data on the resistance of ST 807 isolates to cefixime were available since this drug is almost never used for the treatment of gonococcal infection in the Russian Federation. The most resistant isolates were those of ST 1407, almost all of which were resistant to cefixime, ciprofloxacin, penicillin, and tetracycline. Isolates of this ST, which is widespread worldwide, carry multiple drug resistance determinants, in particular, nucleotide substitutions in the porB gene encoding the porin protein that impede the entry of antibiotics into the cell and a deletion in the promoter region of the pump regulator gene mtrR that leads to increased efflux of antibiotics; moreover, most isolates of ST 1407 carry a mosaic allele of the penicillin-binding protein gene penA (Unemo and Shafer, 2014; Unemo and Jensen, 2017; Unemo et al., 2019).
Thus, phylogenetically different STs (Figure 1) also differ in antibiotic resistance profiles (Table 3), which can be explained not only by the specifics of antimicrobial drug use in the countries where the isolates were collected but also by differences in evolutionary and adaptation pathways of the considered STs that allow them to survive and occupy a prominent position in the gonococcal population.
Assuming that the GGI structural regularities found for the studied STs were preserved, we compared the drug susceptibility of isolates of different STs with the presence of a “properly functioning” GGI, i.e., a GGI without lesions in the essential genes (Figure 3). As seen in the figure, isolates of STs 225, 1407, and 2400 with a “properly functioning” island without obvious lesions/critical changes in essential genes (premature stop codons or reading frameshifts) demonstrated increased resistance to ciprofloxacin, penicillin, and tetracycline. Isolates of STs 1407 and 2400 were also resistant to cefixime. On the other hand, most isolates carrying gene lesions or not containing a GGI, e.g., isolates of STs 807, 2992 and 4186, did not show an increase in resistance to these antibiotics. However, it should be mentioned that the proportion of isolates resistant to azithromycin was higher for the isolates without “properly functioning” GGI (8.8% azithromycin-resistant isolates for ST 2992).
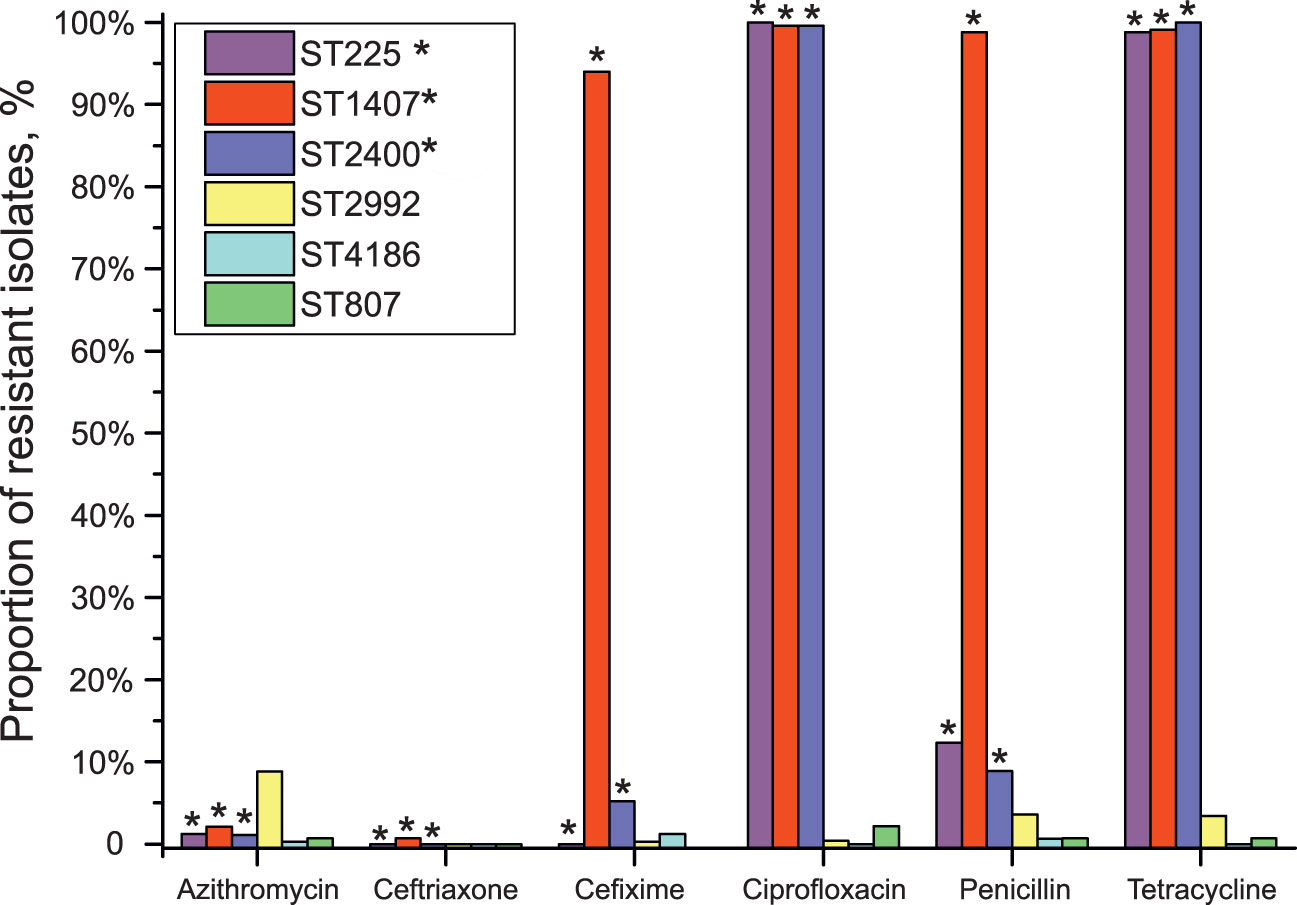
Figure 3 Association between GGI function and the resistance to azithromycin, ceftriaxone, cefixime, ciprofloxacin, penicillin, and tetracycline for isolates belonging to STs 225, 807, 1407, 2400, 2992 and 4186. No data on the resistance of ST 807 isolates to cefixime are available. STs carrying a GGI without lesions in genes essential for GGI function (“properly functioning GGI”) are marked with asterisks.
Grouping of STs according to the presence/absence of a “properly functioning” GGI (a group containing STs 225, 1407 and 2400 and a group containing STs 807, 2992 and 4186) and analysis with the χ-square test revealed differences (p <0.001) in susceptibility to cefixime, ciprofloxacin, penicillin and tetracycline between isolates carrying a GGI without lesions in essential genes and isolates either without a GGI or carrying a GGI with lesions in essential genes. Importantly, we considered the association of resistance not with the presence of a GGI itself but with the presence of a “properly functioning” GGI without lesions in genes essential for its function.
Genomic Comparison of Isolates Belonging to Different STs and Identification of Genes Specific to Each ST
Analysis and annotation of genes in the genomes of 150 N. gonorrhoeae isolates belonging to STs 225, 807, 1407, 2400, 2992, and 4186 using the Prokka program showed the presence of 2856 genes in the pangenome. Annotation with pubMLST allowed us to separate genes and allelic variants and to clarify gene functions. After analysis with the pubMLST program, 2149 genes remained in the pangenome. Among these 2149 genes, 1799 were present in at least 80% of the genomes, 164 were present in more than 15% but less than 80% of the genomes, and 184 were present in less than 15% of the genomes.
To identify genes specific to a given ST, we performed a pairwise comparison of ST genomes using the following parameters: the number of genes (determined using the Prokka program) present in at least 80% of the genomes of isolates belonging to one ST and either were not detected at all or were detected in no more than 28% of the genomes of isolates belonging to another ST (minor components). The combined pairwise comparisons of the genomes of the six STs are shown in Figure 4 as Venn diagrams. For example, comparison of the genomes of ST 807 isolates with the genomes of other ST isolates (Figure 4B) showed the following:
- 13 genes were absent or were minor components in isolates of ST 225 but were present in isolates of the five other STs;
- 10 genes were absent or were minor components in isolates of ST 4186 but were present in isolates of the five other STs;
- 66 genes were absent or were minor components in isolates of ST 2992 but were present in isolates of the five other STs;
- 9 genes were absent or were minor components in isolates of ST 2400 but were present in isolates of the five other STs;
- 7 genes were absent or were minor components in isolates of ST 1407 but were present in isolates of the five other STs;
- 20 genes were specific to ST 807 but were minor components in isolates of the five other STs.
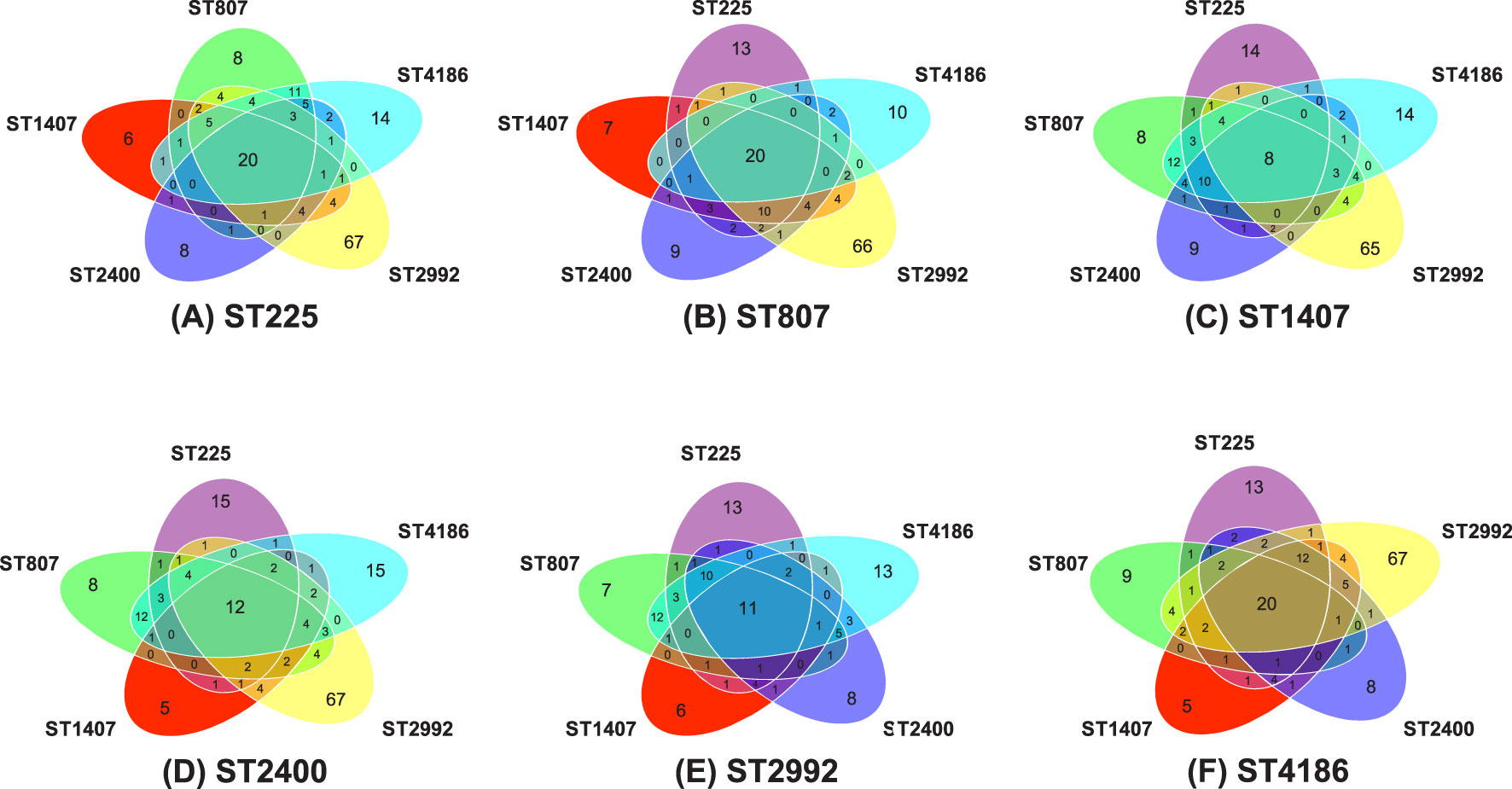
Figure 4 Venn diagrams characterizing pairwise comparisons of the genomes of isolates belonging to one ST with the genomes of isolates belonging to the five other STs. Reference STs: ST 225 (A), ST 807 (B), ST 1407 (C), ST 2400 (D), ST 2992 (E) and ST 4186 (F). The numbers on the diagrams indicate the numbers of genes present in at least 80% of the reference ST isolates and not detected or present in no more than 28% of isolates belonging to another ST. The center of each diagram shows the number of genes that are specific to the indicated ST but are minor components in the five other STs.
As a result of 30 pairwise comparisons of the genomes of the six STs, 329 genes specific to one ST but minor components in another ST were identified. All 329 genes are listed in Table S2 of the Supplementary Material. Intersections of the lists of pairwise comparisons revealed genes specific to a particular sequence type, 8-20 genes for each sequence type (Table 4). Thus, Table 4 presents a list of genes specific to one ST but not specific to any of the other five STs and the functions of the proteins encoded by these genes that were revealed by annotation. The list of genes includes the specific alleles of the tbpB gene encoding transferrin binding protein B (TbpB), which determine an NG-MAST type and have been identified for all STs.

Table 4 Loci and gene alleles specific to the analyzed STs and the functions of the proteins encoded by these genes.
For all studied STs, variable alleles of genes encoding pili proteins (PilE, PilH, PilX, PilJ, etc.) were identified as specific alleles; these genes are highly variable in N. gonorrhoeae. As seen in Table 4, the genes specific to each ST also include genes encoding hypothetical proteins and proteins whose functions have not been established.
For ST 225, the specific genetic loci included genes encoding the transmembrane transport protein xanthine/uracil permease XanQ; the putative microbial toxin AbiEii; which induces cell death during phage infection; type IV toxin-antitoxin system (Dy et al., 2014); a MafB family polymorphic toxin involved in interbacterial competition (Jamet and Nassif, 2015; Arenas et al., 2020); the type I restriction-modification system protein NgoAV; and a protein of the sel1 repeat family associated with upregulation of a set of genes negatively regulated by ferric uptake regulator (Fur) and downregulation of a set of genes positively regulated by Fur (Li et al., 2017).
The genes specific to ST 807 included a gene encoding the multiple adhesin family protein MafA, whose expression products are involved in virulence, adhesion and transcytosis in pathogenic Neisseria and are required for the function of the polymorphic toxin MafB (Arenas et al., 2020); a gene encoding a protein of the ComF family and required for transformation (Tribble et al., 2012); an M48 family metallopeptidase gene; a gene encoding a protein of the thermonuclease family belonging to the replication, recombination and repair system; a gene encoding the modification methylase ModA (restriction-modification system); which is involved in the control of phase variations in representatives of the Neisseria genus (Gawthorne et al., 2012); and a gene encoding an AAA family ATPase that may act as a toxin for a type IV toxin-antitoxin resistance system (Yamamoto et al., 2009). We also identified a number of specific genes that constituted the GGI, in particular, traG, eppA, and ych1, in which alterations or lesions have occurred (premature stop codons or reading frameshifts).
For ST 1407, the specific genes included a type XXXIV mosaic allele of the penA gene encoding the PBP 2 protein, whose presence reduces the susceptibility of N. gonorrhoeae to β-lactam drugs, and genes encoding the opacity protein and the MafB family polymorphic toxin HNH endonuclease (Jamet and Nassif, 2015; Arenas et al., 2020).
The list of genes specific to ST 2400 included a gene for an acetyltransferase involved in lipid A biosynthesis (Bartling and Raetz, 2009); a gene encoding an efflux pump protein of the major facilitator superfamily (MFS) that carry out transfer of small molecules across cell membranes (Bosshart and Fotiadis, 2019); a gene encoding the lytic transglycosylase RlpA, which exhibits unusual specificity for naked glycans and is necessary for cell division and the correct biogenesis of the bacterial cell wall (Jorgenson et al., 2014); and a gene encoding the phosphoshikimate 1-carboxyvinyltransferase enzyme of the HAD hydrolase family.
The following specific genes were identified for ST 2992: a gene encoding the phage transcriptional regulator of the “helix-turn-helix” motif, a gene encoding the DNA-binding transcriptional regulator protein, a gene encoding subunit C of aspartyl/glutamyl-tRNA amidotransferase, and a gene encoding the permease subunit of the RND-type MtrD efflux pump associated with the resistance to penicillins, tetracyclines, macrolides (azithtomycin) and cephalosporins (Shafer et al., 2016).
For ST 4186, the specific genes included the gene encoding the maltose phosphorylase MapA, which is involved in maltose metabolism; a gene encoding an Smr/MutS family protein involved in DNA repair (Fukui and Kuramitsu, 2011); a gene encoding the heat shock protein GrpE; a gene encoding an M48 family metallopeptidase; and the specific allele 142 of the traG gene, a GGI component.
Genomic Comparison of ST 807 and ST 1407
The pairwise comparison of the genomes of ST 807, which is the most widespread ST in the Russian Federation, and the genomes of ST 1407, which is otherwise the most common ST worldwide but not in the Russian Federation, was particularly interesting. This comparison revealed 53 loci for ST 807 and 62 specific loci for ST 1407, including pseudogenes and genes encoding hypothetical proteins. First, notably, the ST 807 isolates carried a type I nonmosaic allele of the penA gene, while the ST 1407 isolates carried a type XXXIV mosaic allele associated with resistance to third-generation cephalosporins.
As shown above, the structure of the GGI differed between ST 807 and ST 1407 (Figure 2). In both STs, an allele of the traG gene specific to that type was found. However, unlike the ST 807 genome, the ST 1407 genome contained the atlA gene instead of the eppA gene, which may contribute to the pathogenesis of gonococcal infection (Kohler et al., 2013; Pachulec et al., 2014).
The genomes of both STs were characterized by specific genes and alleles encoding proteins that are components of pili and proteins involved in pilus biogenesis (pilS, pilE, pilV, pilI, pilJ, pilX, pilH, pilV); unique alleles of genes encoding pathogenicity factors, including the opacity protein OpaA (which interacts with receptors of the CEACAM family); and specific alleles of genes encoding proteins of the ComF family that are important for the uptake of exogenous DNA by naturally competent bacteria. The genomes of both STs contained specific proteins responsible for the initiation of replication; a characteristic ATP-dependent chaperone, ClpB; a 7-carboxy-7-deazaguanine synthase, QueE; a metallopeptidase of the M48 family; and a protein of the thermonuclease family.
Proteins that were present in most ST 1407 samples but were absent from or minor components in the ST 807 samples included an HNH endonuclease of the MafB family, an Eco29kI family restrictase, a DNA (cytosine-5)-methyltransferase and type II toxin-antitoxin system VapC family toxin, and proteins of the transport and efflux systems, including a HlyD family efflux transporter protein and a peptidase domain-containing ABC transporter.
Proteins that were present in most ST 807 samples but were absent from or minor components in the ST 1407 samples included MafA adhesins; immunity 8 family protein, a toxin belonging to the most common family of immunity proteins whose C-terminal domain is characterized by significant variability due to recombination with genes encoding other toxins (Zhang et al., 2012); enzymes in biochemical pathways, including glycosyl transferases (protein glycosylation system); a Zn-dependent alcohol dehydrogenase; and a restriction endonuclease of the NgoFVII family.
Discussion
In this work, the complete genomes of 25 Russian isolates belonging to ST 807 and closely related STs were obtained for the first time. Genogroup G807 has been predominant in the Russian Federation over the past 10 years, and its notable feature is its susceptibility to antimicrobial drugs (Kubanov et al., 2019; Kandinov et al., 2020; Shaskolskiy et al., 2020a). The genomes of N. gonorrhoeae isolates belonging to the NG-MAST types that are the most widespread worldwide (ST 225, 1407, 2400, 2992, and 4186) and in the Russian Federation (ST 807) were compared. The constructed phylogenetic network for 25 core genomes of each ST showed that the clustering of the isolates according to NG-MAST type corresponded to the clustering according to the core genome, consistent with the results of Harrison et al. (2020). Thus, the NG-MAST scheme provides information about and correctly describes the phylogeny of N. gonorrhoeae isolates as revealed by WGS. Database analysis showed that phylogenetically different STs also differed in the profiles of resistance to the drugs that are currently and were previously used for gonorrhea treatment.
The genomes of N. gonorrhoeae isolates of different STs were compared to identify genes that distinguish one sequence type from the others. To compare genomes, the following criteria were chosen: genes and gene alleles present in at least 80% (that is, in 20 out of 25 genomes under study) of the genomes of isolates belonging to one ST and either were absent from or were present in no more than 28% (that is, in 7 out of 25 genomes under study) of the genomes of isolates belonging to another ST. For comparison, in the work of Golparian et al. (2020), when the entire Danish gonococcal population for the period from 1928 to 2013 was analyzed, the corresponding criteria of ≥ 99% and less than 15% were applied. In our work, we analyzed a much smaller population containing only individual STs; moreover, we sought to avoid information loss due to errors in the sequencing results.
The differences in the genomes of N. gonorrhoeae isolates belonging to different NG-MAST types included differences in a large set of genes and gene alleles: the genetic lines of the studied STs contained from 8 to 20 different genes specific to each of these types. The functions of specific genes differed, with the exception of the genes encoding pili proteins, which were found in all STs; this similarity is predictable given the pathogenic role of pili and their phase variations. Analysis of the set of genes that distinguished individual NG-MAST types showed the presence of genes associated with host adaptation. In particular, ST 225 was characterized by the presence of MafB, a toxin for the defense against other bacteria, as well as a protein of the Sel1 repeat family, which plays an important role in iron metabolism. For type 807, the mafA gene, whose expression products are involved in virulence, adhesion and transcytosis in pathogenic Neisseria and are necessary for the function of MafB (Arenas et al., 2020), was notable.
Analysis of genes specific for each ST indicated the presence of two genes contributing to the antibiotic resistance of N. gonorrhoeae: a mosaic allele of penA gene in ST 1407, that is strongly associated with resistance to cephalosporins, and an mtrD gene of multidrug efflux transporter in ST 2992 associated with the resistance to penicillins, tetracyclines, macrolides (azithtomycin) and cephalosporins. The presence of specific mtrD gene in isolates of ST 2992 may contribute to the increased resistance to azithromycin (Figure 3 and Table 3).
Harrison et al., on the basis of WGS data for 3750 N. gonorrhoeae isolates, obtained core genomes and identified the most conserved and differing genomic loci (Harrison et al., 2020). The most conserved loci were NEIS0415 (ribosome biogenesis GTP-binding protein YsxC) and NEIS2686 (hypothetical protein). The less conserved loci included those associated with pilin biosynthesis (NEIS0827), cell division (NEIS0116 and NEIS0128), iron acquisition (NEIS0338), and MafA proteins in the toxin-antitoxin system. Several genes that we identified as specific coincided with the genes described earlier (Harrison et al., 2020), for example, the gene encoding a type IV biogenesis protein (NEIS0827) in ST 225 and the mafA gene in ST 807 (see Table 4).
A detailed study of the GGI gene sequences showed significant differences in the GGI structure between isolates belonging to different STs. Isolates of ST 807 and ST 4186 harbored mutations in the GGI genes, which play an essential role in DNA secretion, leading to loss of functionality. In all isolates of ST 807, the atlA gene, which is critically important for GGI function, was replaced with the eppA gene, accompanied alteration of the traG allele, replacement of the ych gene with the ych1 gene, and loss of the exp1 gene. Only a small proportion of ST 2992 isolates (4 of 25) carried GGI genes.
The isolates of STs 225, 1407, and 2400 lacked significant lesions in the essential GGI genes, i.e., these isolates are predicted to secrete DNA. The proportion of isolates resistant to ciprofloxacin, penicillin, and tetracycline among isolates with STs DNA secretion capability was significantly higher than that among isolates of STs without a GGI or with a GGI harboring lesions in essential genes. Thus, our results indicated the presence of a relationship between the decrease in the susceptibility of isolates to ciprofloxacin, penicillin, and tetracycline and the absence of lesions in the GGI genes necessary for DNA secretion.
The association between the presence of a GGI and genotype-predicted resistance to numerous antibiotics, including cefixime, ciprofloxacin, and penicillin, was noted earlier by Harrison et al, 2016. However, as our results indicate, simply the presence of a GGI, which is found in ~80% of N. gonorrhoeae isolates, does not guarantee its proper function for ssDNA secretion, and lesions/changes in the key genes that could interfere with the entire T4SS system must be considered.
Thus, WGS indicated genomic differences between isolates belonging to the most common NG-MAST types. The evolutionary pathways of N. gonorrhoeae, which allow a particular NG-MAST type to maintain long-term predominance in the population for many years, may include changes in genes responsible for adhesion and virulence, changes in the GGI structure, preservation of genes that carry the determinants of drug resistance, and changes in genes associated with host adaptation and genes encoding enzymes of biochemical pathways. In-depth analysis of the significance of such differences in the ongoing evolution of N. gonorrhoeae is undoubtedly required. However, the results obtained in this work for the comparison of this limited number of genomes (25 genomes of each of the six STs) provide information on the presence of genes specific to each ST, which can be used to create in vivo and in vitro models of the pathogenesis of gonococcal infection.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Ethical approval/written informed consent was not required for the study of animals/human participants in accordance with the local legislation and institutional requirements.
Author Contributions
BS designed and directed the project and performed bioinformatic studies. DK, IK, SG, and BS carried out WGS and analyzed WGS results. DK analyzed the structures of gonococcal genetic islands. AK supervised N. gonorrhoeae sample collection. VS and DD carried out NG-MAST typing and preparation of samples for WGS. ED carried out gene annotation and wrote the manuscript. DG designed and supervised the project and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Russian Science Foundation, grant number 17-75-20039 (WGS and analysis of genomes) and by the Ministry of Science and Higher Education of the Russian Federation to the EIMB Center for Precision Genome Editing and Genetic Technologies for Biomedicine under the Federal Research Program for Genetic Technologies Development for 2019-27, agreement number 075-15-2019-1660 (analysis of drug resistance of N. gonorrhoeae isolates). Collection and susceptibility testing of N. gonorrhoeae clinical isolates were performed according to the Ministry of Health of the Russian Federation assignment number 056-03-2021-124.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.831336/full#supplementary-material
References
Abrams, A. J., Trees, D. L. (2017). Genomic Sequencing of Neisseria gonorrhoeae to Respond to the Urgent Threat of Antimicrobial-Resistant Gonorrhea. Pathog. Dis. 75 (4), 10.1093/femspd/ftx041. doi: 10.1093/femspd/ftx041
Arenas, J., Catón, L., van den Hoeven, T., de Maat, V., Cruz Herrero, J., Tommassen, J. (2020). The Outer-Membrane Protein MafA of Neisseria meningitidis Constitutes a Novel Protein Secretion Pathway Specific for the Fratricide Protein MafB. Virulence 11 (1), 1701–1715. doi: 10.1080/21505594.2020.1851940
Bartling, C. M., Raetz, C. R. (2009). Crystal Structure and Acyl Chain Selectivity of Escherichia coli LpxD, the N-Acyltransferase of Lipid A Biosynthesis. Biochemistry 48 (36), 8672–8683. doi: 10.1021/bi901025v
Bosshart, P. D., Fotiadis, D. (2019). Secondary Active Transporters. Subcell Biochem. 92, 275–299. doi: 10.1007/978-3-030-18768-2_9
Callaghan, M. M., Heilers, J. H., van der Does, C., Dillard, J. P. (2017). Secretion of Chromosomal DNA by the Neisseria gonorrhoeae Type IV Secretion System. Curr. Top. Microbiol. Immunol. 413, 323–345. doi: 10.1007/978-3-319-75241-9_13
Callaghan, M. M., Klimowicz, A. K., Shockey, A. C., Kane, J., Pepperell, C. S., Dillard, J. P. (2021). Transcriptional and Translational Responsiveness of the Neisseria gonorrhoeae Type IV Secretion System to Conditions of Host Infections. Infect. Immun. 89 (12), e0051921. doi: 10.1128/IAI.00519-21
Chisholm, S. A., Unemo, M., Quaye, N., Johansson, E., Cole, M. J., Ison, C. A., et al. (2013). Molecular Epidemiological Typing Within the European Gonococcal Antimicrobial Resistance Surveillance Programme Reveals Predominance of a Multidrug-Resistant Clone. Euro Surveill. 18 (3), 20358. doi: 10.2807/ese.18.03.20358-en
Demczuk, W., Lynch, T., Martin, I., Van Domselaar, G., Graham, M., Bharat, A., et al. (2015). Whole-Genome Phylogenomic Heterogeneity of Neisseria gonorrhoeae Isolates With Decreased Cephalosporin Susceptibility Collected in Canada Between 1989 and 2013. J. Clin. Microbiol. 53 (1), 191–200. doi: 10.1128/JCM.02589-14
De Silva, D., Peters, J., Cole, K., Cole, M. J., Cresswell, F., Dean, G., et al. (2016). Whole-Genome Sequencing to Determine Transmission of Neisseria gonorrhoeae: An Observational Study. Lancet Infect. Dis. 16 (11), 1295–1303. doi: 10.1016/S1473-3099(16)30157-8
Dillard, J. P., Seifert, H. S. (2001). A Variable Genetic Island Specific for Neisseria gonorrhoeae Is Involved in Providing DNA for Natural Transformation and Is Found More Often in Disseminated Infection Isolates. Mol. Microbiol. 41 (1), 263–277. doi: 10.1046/j.1365-2958.2001.02520.x
Dubnau, D. (1999). DNA Uptake in Bacteria. Annu. Rev. Microbiol. 53, 217–244. doi: 10.1146/annurev.micro.53.1.217
Dy, R. L., Przybilski, R., Semeijn, K., Salmond, G. P., Fineran, P. C. (2014). A Widespread Bacteriophage Abortive Infection System Functions Through a Type IV Toxin-Antitoxin Mechanism. Nucleic Acids Res. 42 (7), 4590–4605. doi: 10.1093/nar/gkt1419
European Centre for Disease Prevention and Control (2018). Molecular Typing of Neisseria gonorrhoeae – A Study of 2013 Isolates (Stockholm: ECDC). Available at: https://www.ecdc.europa.eu/sites/portal/files/documents/Molecular-typing-N-gonorrhoeae-web.pdf.
Eyre, D. W., De Silva, D., Cole, K., Peters, J., Cole, M. J., Grad, Y. H., et al. (2017). WGS to Predict Antibiotic MICs for Neisseria gonorrhoeae. J. Antimicrob. Chemother. 72, 1937–1947. doi: 10.1093/jac/dkx067
Ezewudo, M. N., Joseph, S. J., Castillo-Ramirez, S., Dean, D., Del Rio, C., Didelot, X., et al. (2015). Population Structure of Neisseria gonorrhoeae Based on Whole Genome Data and Its Relationship With Antibiotic Resistance. Peer J. 3, e806. doi: 10.7717/peerj.806
Fukui, K., Kuramitsu, S. (2011). Structure and Function of the Small MutS-Related Domain. Mol. Biol. Int. 2011, 691735. doi: 10.4061/2011/691735
Gabler, F., Nam, S. Z., Till, S., Mirdita, M., Steinegger, M., Söding, J., et al. (2020). Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinf. 72 (1), e108. doi: 10.1002/cpbi.108
Gawthorne, J. A., Beatson, S. A., Srikhanta, Y. N., Fox, K. L., Jennings, M. P. (2012). Origin of the Diversity in DNA Recognition Domains in Phasevarion Associated modA Genes of Pathogenic Neisseria and Haemophilus influenzae. PloS One 7 (3), e32337. doi: 10.1371/journal.pone.0032337
Golparian, D., Donà, V., Sánchez-Busó, L., Foerster, S., Harris, S., Endimiani, A., et al. (2018). Antimicrobial Resistance Prediction and Phylogenetic Analysis of Neisseria gonorrhoeae Isolates Using the Oxford Nanopore MinION Sequencer. Sci. Rep. 8, 17596. doi: 10.1038/s41598-018-35750-4
Golparian, D., Harris, S. R., Sánchez-Busó, L., Hoffmann, S., Shafer, W. M., Bentley, S. D., et al. (2020). Genomic Evolution of Neisseria gonorrhoeae Since the Preantibiotic Era, (1928-2013): Antimicrobial Use/Misuse Selects for Resistance and Drives Evolution. BMC Genomics 21 (1), 116. doi: 10.1186/s12864-020-6511-6
Grad, Y. H., Harris, S. R., Kirkcaldy, R. D., Green, A. G., Marks, D. S., Bentley, S. D., et al. (2016). Genomic Epidemiology of Gonococcal Resistance to Extended-Spectrum Cephalosporins, Macrolides, and Fluoroquinolones in the United States 2000-2013. J. Infect. Dis. 214 (10), 1579–1587. doi: 10.1093/infdis/jiw420
Hamilton, H. L., Dillard, J. P. (2006). Natural Transformation of Neisseria gonorrhoeae: From DNA Donation to Homologous Recombination. Mol. Microbiol. 59 (2), 376–385. doi: 10.1111/j.1365-2958.2005.04964.x
Hamilton, H. L., Domínguez, N. M., Schwartz, K. J., Hackett, K. T., Dillard, J. P. (2005). Neisseria gonorrhoeae Secretes Chromosomal DNA via a Novel Type IV Secretion System. Mol. Microbiol. 55 (6), 1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x
Harris, S. R., Cole, M. J., Spiteri, G., Sánchez-Busó, L., Golparian, D., Jacobsson, S., et al. (2018). Public Health Surveillance of Multidrug-Resistant Clones of Neisseria gonorrhoeae in Europe: A Genomic Survey. Lancet Infect. Dis. 18, 758–768. doi: 10.1016/S1473-3099(18)30225-1
Harrison, O. B., Cehovin, A., Skett, J., Jolley, K. A., Massari, P., Genco, C. A., et al. (2020). Neisseria gonorrhoeae Population Genomics: Use of the Gonococcal Core Genome to Improve Surveillance of Antimicrobial Resistance. J. Infect. Dis. 222 (11), 1816–1825. doi: 10.1093/infdis/jiaa002
Harrison, O. B., Clemence, M., Dillard, J. P., Tang, C. M., Trees, D., Grad, Y. H., et al. (2016). Genomic Analyses of Neisseria gonorrhoeae Reveal an Association of the Gonococcal Genetic Island With Antimicrobial Resistance. J. Infect. 73 (6), 578–587. doi: 10.1016/j.jinf.2016.08.010
Huson, D. H., Bryant, D. (2006). Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 23 (2), 254–267. doi: 10.1093/molbev/msj030
Jacobsson, S., Golparian, D., Cole, M., Spiteri, G., Martin, I., Bergheim, T., et al. (2016). WGS Analysis and Molecular Resistance Mechanisms of Azithromycin-Resistant (MIC >2 mg/L) Neisseria gonorrhoeae Isolates in Europe From 2009 to 2014. J. Antimicrob. Chemother. 71 (11), 3109–3116. doi: 10.1093/jac/dkw279
Jamet, A., Nassif, X. (2015). Characterization of the Maf Family of Polymorphic Toxins in Pathogenic Neisseria Species. Microb. Cell 2 (3), 88–90. doi: 10.15698/mic2015.03.194
Jorgenson, M. A., Chen, Y., Yahashiri, A., Popham, D. L., Weiss, D. S. (2014). The Bacterial Septal Ring Protein RlpA Is a Lytic Transglycosylase That Contributes to Rod Shape and Daughter Cell Separation in Pseudomonas aeruginosa. Mol. Microbiol. 93 (1), 113–128. doi: 10.1111/mmi.12643
Kandinov, I., Dementieva, E., Kravtsov, D., Chestkov, A., Kubanov, A., Solomka, V., et al. (2020). Molecular Typing of Neisseria gonorrhoeae Clinical Isolates in Russia 2018-2019: A Link Between penA Alleles and NG-MAST Types. Pathogens 9 (11), 941. doi: 10.3390/pathogens9110941
Kohler, P. L., Chan, Y. A., Hackett, K. T., Turner, N., Hamilton, H. L., Cloud-Hansen, K. A., et al. (2013). Mating Pair Formation Homologue TraG Is a Variable Membrane Protein Essential for Contact-Independent Type IV Secretion of Chromosomal DNA by Neisseria gonorrhoeae. J. Bacteriol. 195 (8), 1666–1679. doi: 10.1128/JB.02098-12
Kubanov, A., Solomka, V., Plakhova, X., Chestkov, A., Petrova, N., Shaskolskiy, B., et al. (2019). Summary and Trends of the Russian Gonococcal Antimicrobial Surveillance Programme 2005-2016. J. Clin. Microbiol. 5 (6), e02024–e02018. doi: 10.1128/JCM.02024-18
Li, M. S., Langford, P. R., Kroll, J. S. (2017). Inactivation of NMB0419, Encoding a Sel1-Like Repeat (SLR) Protein, in Neisseria meningitidis Is Associated With Differential Expression of Genes Belonging to the Fur Regulon and Reduced Intraepithelial Replication. Infect. Immun. 85 (5), e00574–e00516. doi: 10.1128/IAI.00574-16
Martin, I. M., Ison, C. A., Aanensen, D. M., Fenton, K. A., Spratt, B. G. (2004). Rapid Sequence-Based Identification of Gonococcal Transmission Clusters in a Large Metropolitan Area. J. Infect. Dis. 189 (8), 1497–1505. doi: 10.1086/383047
Młynarczyk-Bonikowska, B., Majewska, A., Malejczyk, M., Młynarczyk, G., Majewski, S. (2020). Multiresistant Neisseria gonorrhoeae: A New Threat in Second Decade of the XXI Century. Med. Microbiol. Immunol. 209 (2), 95–108. doi: 10.1007/s00430-019-00651-4
Pachulec, E., Siewering, K., Bender, T., Heller, E. M., Salgado-Pabon, W., Schmoller, S. K., et al. (2014). Functional Analysis of the Gonococcal Genetic Island of Neisseria gonorrhoeae. PloS One 9 (10), e109613. doi: 10.1371/journal.pone.0109613
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 31 (22), 3691–3693. doi: 10.1093/bioinformatics/btv421
Ramsey, M. E., Woodhams, K. L., Dillard, J. P. (2011). The Gonococcal Genetic Island and Type IV Secretion in the Pathogenic Neisseria. Front. Microbiol. 2, 61. doi: 10.3389/fmicb.2011.00061
Rotman, E., Seifert, H. S. (2014). The Genetics of Neisseria Species. Annu. Rev. Genet. 48, 405–431. doi: 10.1146/annurev-genet-120213-092007
Ryan, L., Golparian, D., Fennelly, N., Rose, L., Walsh, P., Lawlor, B., et al. (2018). Antimicrobial Resistance and Molecular Epidemiology Using Whole-Genome Sequencing of Neisseria gonorrhoeae in Irelan-2016: Focus on Extended-Spectrum Cephalosporins and Azithromycin. Eur. J. Clin. Microbiol. Infect. Dis. 37 (9), 1661–1672. doi: 10.1007/s10096-018-3296-5
Shafer, W. M., Yu, E. W., Rouquette-Loughlin, C., Golparian, D., Jerse, A. E., Unemo, M. (2016). “Efflux Pumps in Neisseria gonorrhoeae: Contributions to Antimicrobial Resistance and Virulence,” in Efflux-Mediated Antimicrobial Resistance in Bacteria. Mechanisms, Regulation and Clinical Implications. Eds. Li, X.-Z., Elkins, C. A., Zgurskaya, H. I. (Switzerland: Springer International Publishing, Adis), 439–470.
Shaskolskiy, B., Dementieva, E., Kandinov, I., Chestkov, A., Kubanov, A., Deryabin, D., et al. (2020a). Genetic Diversity of Neisseria gonorrhoeae Multi-Antigen Sequence Types in Russia and Europe. Int. J. Infect. Dis. 93, 1–8. doi: 10.1016/j.ijid.2020.01.020
Shaskolskiy, B., Dementieva, E., Kandinov, I., Filippova, M., Petrova, N., Plakhova, X., et al. (2019). Resistance of Neisseria gonorrhoeae Isolates to Beta-Lactam Antibiotics (Benzylpenicillin and Ceftriaxone) in Russia 2015-2017. PloS One 14 (7), e0220339. doi: 10.1371/journal.pone.0220339
Shaskolskiy, B. L., Kandinov, I. D., Chestkov, A. V., Solomka, V. S., Kubanov, A. A., Deryabin, D. G., et al. (2020b). Comparative Phylogenetic Analysis of Neisseria gonorrhoeae Clinical Isolates in Russia, European Union, and Japan. Bull. RSMU 1, 5–13. doi: 10.24075/brsmu.2020.009
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 18 (3), 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tribble, G. D., Rigney, T. W., Dao, D. H., Wong, C. T., Kerr, J. E., Taylor, B. E., et al. (2012). Natural Competence Is a Major Mechanism for Horizontal DNA Transfer in the Oral Pathogen Porphyromonas gingivalis. mBio 3 (1), e00231–e00211. doi: 10.1128/mBio.00231-11
Unemo, M. (2015). Current and Future Antimicrobial Treatment of Gonorrhoea - the Rapidly Evolving Neisseria gonorrhoeae Continues to Challenge. BMC Infect. Dis. 15, 364. doi: 10.1186/s12879-015-1029-2
Unemo, M., Jensen, J. S. (2017). Antimicrobial-Resistant Sexually Transmitted Infections: Gonorrhoea and Mycoplasma genitalium. Nat. Rev. Urol. 14 (3), 139–152. doi: 10.1038/nrurol.2016.268
Unemo, M., Seifert, H. S., Hook, E. W., 3rd, Hawkes, S., Ndowa, F., Dillon, J. R. (2019). Gonorrhoea. Nat. Rev. Dis. Primers 5 (1), 79. doi: 10.1038/s41572-019-0128-6
Unemo, M., Shafer, W. M. (2014). Antimicrobial Resistance in Neisseria gonorrhoeae in the 21st Century: Past, Evolution, and Future. Clin. Microbiol. Rev. 27 (3), 587–613. doi: 10.1128/CMR.00010-14
Yamamoto, S., Kiyokawa, K., Tanaka, K., Moriguchi, K., Suzuki, K. (2009). Novel Toxin-Antitoxin System Composed of Serine Protease and AAA-ATPase Homologues Determines the High Level of Stability and Incompatibility of the Tumor-Inducing Plasmid pTiC58. J. Bacteriol. 191 (14), 4656–4566. doi: 10.1128/JB.00124-09
Keywords: Neisseria gonorrhoeae, whole-genome sequencing, NG-MAST, gonococcal genetic island, antimicrobial resistance
Citation: Shaskolskiy B, Kravtsov D, Kandinov I, Gorshkova S, Kubanov A, Solomka V, Deryabin D, Dementieva E and Gryadunov D (2022) Comparative Whole-Genome Analysis of Neisseria gonorrhoeae Isolates Revealed Changes in the Gonococcal Genetic Island and Specific Genes as a Link to Antimicrobial Resistance. Front. Cell. Infect. Microbiol. 12:831336. doi: 10.3389/fcimb.2022.831336
Received: 08 December 2021; Accepted: 28 January 2022;
Published: 18 February 2022.
Edited by:
Nathan Weyand, Ohio University, United StatesReviewed by:
Jennifer L. Edwards, The Ohio State University, United StatesJoseph P. Dillard, University of Wisconsin-Madison, United States
Copyright © 2022 Shaskolskiy, Kravtsov, Kandinov, Gorshkova, Kubanov, Solomka, Deryabin, Dementieva and Gryadunov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Boris Shaskolskiy, Yi5zaGFza29sc2tpeUBiaW9jaGlwLnJ1
 Boris Shaskolskiy
Boris Shaskolskiy Dmitry Kravtsov
Dmitry Kravtsov Ilya Kandinov
Ilya Kandinov Sofya Gorshkova
Sofya Gorshkova Alexey Kubanov2
Alexey Kubanov2 Dmitry Deryabin
Dmitry Deryabin Ekaterina Dementieva
Ekaterina Dementieva Dmitry Gryadunov
Dmitry Gryadunov