- 1College of Life Sciences, Henan Agricultural University, Zhengzhou, China
- 2Clinical Lab, Henan Provincial Chest Hospital, Zhengzhou, China
Protein palmitoylation—a lipid modification in which one or more cysteine thiols on a substrate protein are modified to form a thioester with a palmitoyl group—is a significant post-translational biological process. This process regulates the trafficking, subcellular localization, and stability of different proteins in cells. Since palmitoylation participates in various biological processes, it is related to the occurrence and development of multiple diseases. It has been well evidenced that the proteins whose functions are palmitoylation-dependent or directly involved in key proteins’ palmitoylation/depalmitoylation cycle may be a potential source of novel therapeutic drugs for the related diseases. Many researchers have reported palmitoylation of proteins, which are crucial for host-virus interactions during viral infection. Quite a few explorations have focused on figuring out whether targeting the acylation of viral or host proteins might be a strategy to combat viral diseases. All these remarkable achievements in protein palmitoylation have been made to technological advances. This paper gives an overview of protein palmitoylation modification during viral infection and the methods for palmitoylated protein detection. Future challenges and potential developments are proposed.
1 Introduction
Protein lipidation is an important post-translational modification in which lipid moieties are covalently attached to proteins. This process increases the hydrophobicity of proteins, thereby resulting in changes in protein conformation, stability, membrane association, localization, and binding properties. Palmitoylation refers to the covalent modification of proteins with palmitoyl groups. S-palmitoylation of proteins, which is one of the most common forms of palmitoylation, occurs when saturated C16 fatty acids covalently attach to the side chains of cysteine residues through unstable thioester-bonds (Fukata and Fukata, 2010; Blanc et al., 2013; Hentschel et al., 2016; Resh, 2016; Zaballa and van der Goot, 2018). Studies have shown the existence of more than 40 different molecular types of free fatty acids, including a variety of long-chain/short-chain, saturated/unsaturated acids, that can participate in the esterification of proteins (Quehenberger et al., 2010). Palmitoylation of proteins is a reversible dynamic process, and depalmitoylation occurs when, under certain conditions, the thioester bond is hydrolyzed, and the palmitate is separated from the cysteine residue. This kind of reversible modification is present in all eukaryotes, including mammals, plants (Hemsley, 2017), and parasites (Blanc et al., 2013; Brown et al., 2017; Corvi and Turowski, 2019); it regulates trafficking (Linder and Deschenes, 2007; Daniotti et al., 2017; Ernst et al., 2018; Tortosa and Hoogenraad, 2018; Sun et al., 2020), localization (Gamage et al., 2017; Sada et al., 2019; Nakamura et al., 2020), stability (Gok et al., 2020; Wang et al., 2020), interaction (Zaballa and van der Goot, 2018), and signal transduction of proteins (Zheng et al., 2012; Globa and Bamji, 2017; Zingler et al., 2019).
Furthermore, palmitoylation modification is associated with various kinds of diseases, such as neurological diseases (Alzheimer’s disease, Huntington’s disease, and schizophrenia) (Sanders and Hayden, 2015; Cho and Park, 2016), infectious diseases, and even cancer (Blanc, Blaskovic et al., 2013; Hornemann, 2015; Resh, 2017). In recent years, considerable reports have shown that the glycoproteins of almost all of the enveloped viruses have at least one S-acylated site (Gadalla and Veit, 2020). Also, the role played by palmitoylated viral proteins in the infection process has been discussed from different angles (Veit, 2012; Veit et al., 2013; Sobocinska et al., 2017). Therefore, it is important to develop reliable and practical detection methods to analyze the palmitoylation modification of proteins. Researchers have utilized the latest technologies and have made considerable progress in exploring the pathogenesis of diseases related to palmitoylation (Brigidi and Bamji, 2013; Gu and Robinson, 2016; Gao and Hannoush, 2018; Peng and Hang, 2019). However, in the last decade, only a few research teams have systematically reviewed the detection methods of palmitoylation (Lanyon-Hogg et al., 2017; Gao and Hannoush, 2018; Zaballa and van der Goot, 2018; Lu and Fang, 2020). This paper reviewed the palmitoylation modification and the different analytical methods to detect palmitoylated proteins. The palmitoylation of proteins related to viral infection was emphasized, and future research prospects were reviewed.
2 Protein Palmitoylation/Depalmitoylation Cycle
2.1 S-Palmitoylation
S-palmitoylation is a post-translational modification of proteins resulting from the covalent attachment of the palmitoyl chain to a cysteine residue(s) of proteins through a reversible thioester bond. This unique reversible process can act as a molecular switch, just like phosphorylation or ubiquitination (Blaskovic et al., 2013). Due to the obvious heterogeneity with other fatty acids linked to cysteine residues, the fatty acylation process is generally known as “thioacylation” or “S-acylation” rather than palmitoylation (Linder and Deschenes, 2007). Although “S-palmitoylation” implies that cysteine residues are only modified by 16:0 fatty acids, in the cell, they can also be modified by long-chain fatty acids with different degrees of unsaturation (Thinon et al., 2016; Greaves et al., 2017). S-palmitoylation of some proteins (e.g., cytosolic proteins) helps them acquire hydrophobic anchors, resulting in an improved association with membrane, trafficking, and localization. In many cases, palmitoylation of key cellular transmembrane proteins also occurs, including adhesion molecules, tetraspanins, G-protein-coupled receptors, receptor ligands, and viral glycoproteins (Blaskovic et al., 2013). In theory, palmitates should be added to the N-terminal of cysteine residues—known as N-palmitoylation—to form a more stable and irreversible structure. However, N-palmitoylation is rare in natural viral proteins, and its biological function has not been explored so far. Therefore, unless specified, palmitoylation of proteins usually refers to S-palmitoylation at the cysteine site.
Since the identification of the first protein to have undergone S-palmitoylation, nearly a thousand eukaryotic proteins have been identified as the substrates of palmitoylation (Blanc et al., 2015; Blanc et al., 2019). Some experts in this field have reviewed the roles of S-palmitoylation in synaptic plasticity, membrane trafficking, and physiological processes, such as metabolism, cell death, cell polarity and migration, cancer, and innate immunity (Chamberlain and Shipston, 2015; Daniotti et al., 2017; Zareba-Koziol et al., 2018); therefore, this part will not be discussed here.
2.2 Palmitoylation/Depalmitoylation: A Dynamic Reversible Cycle
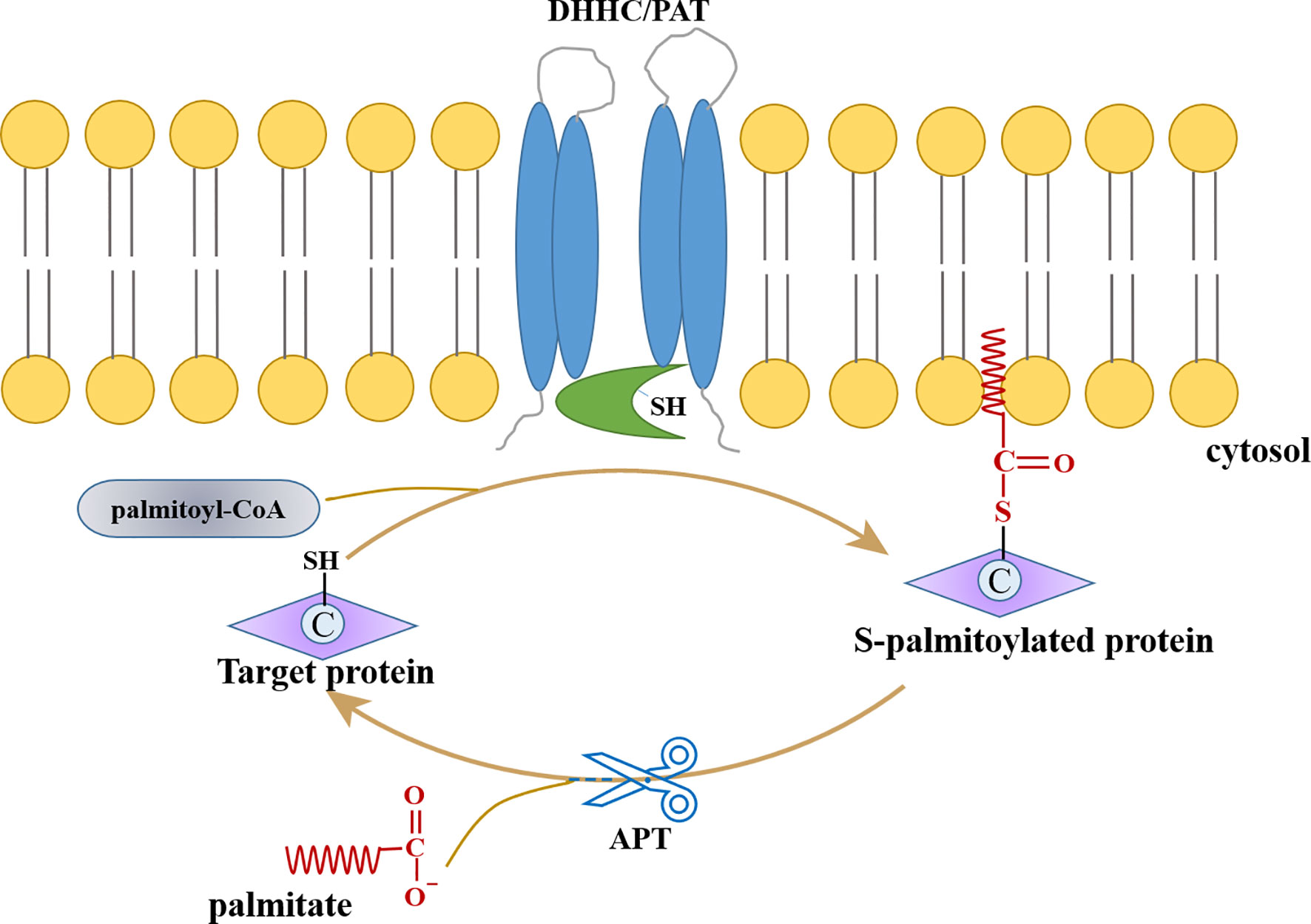
S-palmitoylation is a reversible and dynamic modification; palmitoyl acyltransferases (PATs) catalyze the addition of palmitic acid, while the removal of palmitic acid is mediated by acyl protein thioesterases (APTs) (Figure 1). Since PATs contain Zn2+ binding domains and conserved “Asp-His-His-Cys” (DHHC) motifs, they are considered to be part of the zDHHC group of enzymes (Mitchell et al., 2006; Gottlieb and Linder, 2017). The first discovered palmitoyl transferases are Erf2p and Erf4p of Saccharomyces cerevisiae and are identified as Ras protein acyltransferases (Ras PAT). Erf2p, containing conserved DHHC-cysteine-rich domain, forms a complex with Erf4p to function as the effectors of Ras protein in the endoplasmic reticulum (ER) membrane (Lobo et al., 2002; Roth et al., 2002). Since then, twenty-three human DHHC proteins have been identified or predicted to form a protein superfamily.
The zDHHC enzymes, usually present in Golgi membrane or ER, catalyzes the palmitoylation process of substrate proteins; this is a two-step process, where palmitate is transferred onto the thiol group of cysteine from cytosolic palmitoyl-CoA by PAT. First, the Asp and His act as a proton shuttle, thereby converting the Cys residue of the zDHHC enzyme into a thiolate nucleophile. In the second step, the carbonyl carbon of the palmityl-CoA thioester is attacked by Cys thiolate, resulting in instantaneous auto-acylation of cysteine residues in its DHHC motif. This facilitates the PAT-mediated transfer of the palmitoyl residue to the substrate protein (Stix et al., 2020) (Figure 1). The mutational analysis has revealed that the first histidine of the DHHC motif is especially important during the second step (Mitchell et al., 2010; Jennings and Linder, 2012; Tsai et al., 2014; Gonzalez Montoro et al., 2015). Recent research by Rana et al. revealed the crystal structures of two DHHC palmitoyl transferases leading to an understanding that the active site resides at the membrane-cytosol interface. Based on this, it was concluded that the thioester-exchange reaction is catalyzed by DHHC enzymes and that the membrane-proximal cysteines are potential sites for palmitoylation (Rana et al., 2018; Rana et al., 2019; Stix et al., 2020).
Compared to palmitoylation, i.e., its reverse reaction, depalmitoylation, is poorly understood; and, only seven APTs have been identified to catalyze depalmitoylation. APT (commonly referred to as palmitoyl protein thioesterase or PPT) catalyzes the hydrolysis of thioesters to dissolve and replace the substrate protein on the membrane. Structural studies show that two depalmitoylases (APT1 and APT2), which were identified earlier (Duncan and Gilman, 1998; Toyoda et al., 1999), contain convergent acyl binding channels, indicating that factors other than acyl chain recognition also mediate the selection of substrate (Devedjiev et al., 2000). Recombinant PPT1 enzyme from baculovirus expression system acts as a powerful depalmitoylase for H-Ras protein (Camp and Hofmann, 1993). The mammalian α/β hydrolase-domain containing protein (ABHD) recently became the novel candidate for regulating lipid metabolism. ABHD17A and its two isoforms, ABHD17B and ABHD17C, were identified as depalmitoylases targeting Ras-family GTPases and synaptic proteins in neurons (Lin and Conibear, 2015). Moreover, another depalmitoylase, ABHD10, is known to act on S-palmitoylated peroxiredoxin-5 protein and regulates redox homeostasis in mitochondria (Cao et al., 2019).
The reversibility of protein palmitoylation modification suggests a subtle mechanism, which avoids imbalance. To date, quite a few reports have demonstrated the relationship between the defects of palmitoylation/depalmitoylation process and human diseases, including Huntington’s disease (Sanders and Hayden, 2015), schizophrenia (Ota et al., 2013), Alzheimer’s disease (Mizumaru et al., 2009; Vetrivel et al., 2009), Goltz syndrome (Biechele et al., 2011; Doubravska et al., 2011), and other diseases (Yeste-Velasco et al., 2015; Ko and Dixon, 2018) related to PATs. However, it is still unclear how the cycle works in detail, especially, which PAT participates or maintains this dynamic equilibrium of the target proteins involved in the above diseases.
3 Protein Palmitoylation Modification During Viral Infection
3.1 Palmitoylation of Viral Proteins
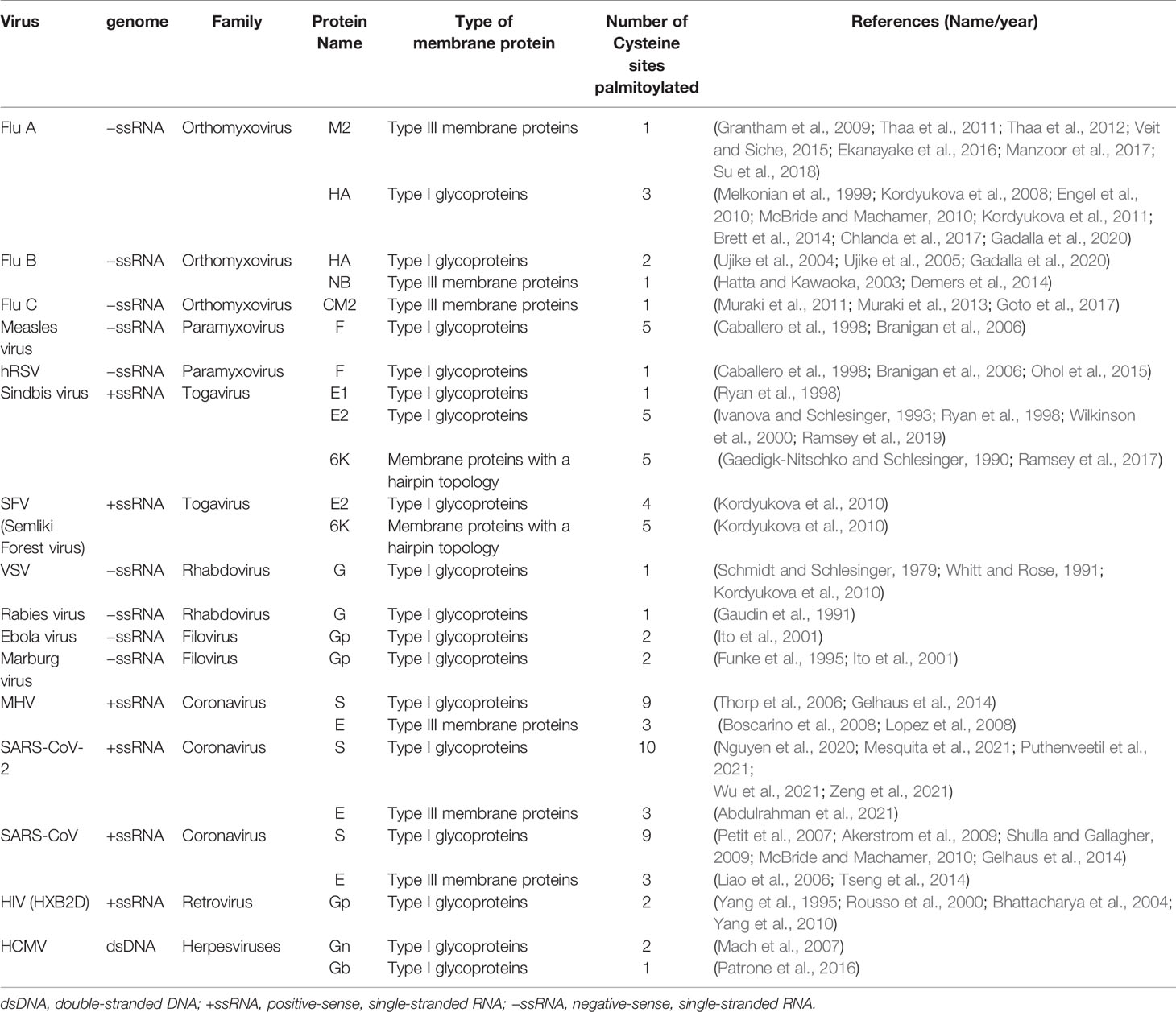
Many cell-and-animal-based studies have reported the multiple roles of protein modifications in viral infectious diseases. Palmitoylation of viral proteins, which are involved in virus assembly or pathogenesis, has been found in most of the eukaryotic permissive cell types, including yeast, insect, plant, and vertebrate cells (Veit, 2012; Blanc et al., 2013; Lin et al., 2017). In Table 1, we have summarized the viral proteins, especially structural proteins of virus envelope that undergo palmitoylation during viral infections leading to serious diseases in humans or animals. Also, we have provided select examples illustrating key findings and the important roles of palmitoylated viral proteins during viral infections.
Since the identification of palmitoylated glycoproteins of Sindbis virus and Vesicular stomatitis virus in 1979 (Schmidt et al., 1979; Schmidt and Schlesinger, 1979), many other palmitoylated proteins of viruses have been reported, such as hemagglutinin (HA) and proton channel M2 of influenza virus, glycoproteins of filoviruses and retrovirus (including HIV), fusion protein (F) of measles virus, the S-protein of coronavirus (CoV) (Veit, 2012; Kordyukova et al., 2019). It is noteworthy that the palmitoylation sites of SARS-CoV-2 spike protein have recently been identified and proved to be essential for SARS-CoV-2 fusion with the host cell (Mesquita et al., 2021; Puthenveetil et al., 2021). Palmitoylation modification of proteins may facilitate trafficking of glycoproteins on viral membranes, thereby promoting assembly and budding of progeny virions on infected epithelial cells (Veit et al., 2013; Demers et al., 2014). Moreover, the palmitoylation of non-structural viral proteins, such as Chikungunya virus (CHIKV) nsP1 and hepatitis C virus (HCV) NS2, are crucial to the successful infection (Utt et al., 2019; Wu et al., 2019; Zhang et al., 2019; Bakhache et al., 2020). In mammalian cells, CHIKV replication complex anchors on the plasma membrane. Palmitoylation of the cysteine residue at the C-terminus of CHIKV nsP1 enhances its interaction with the lipid bilayer, which is critical for targeting cholesterol-rich lipid rafts and viral genome replication (Utt et al., 2019; Zhang et al., 2019). The subcellular localization of HCV NS2 is regulated through palmitoylation, which also promotes the replication of HCV RNA. Therefore, palmitoylation of HCV NS2 could be utilized to inhibit HCV RNA replication and viral assembly, providing a novel alternative strategy effective against HCV infection (Wu et al., 2019).
Besides the above-mentioned mammalian viruses, in the viruses infecting aquatic animals and plants, palmitoylation of proteins plays an important role in viral replication, protein localization, and other related functions. A recent study reported that exogenous palmitic acid promotes red-spotted grouper nervous necrosis virus (RGNNV) infection, and interfering with the palmitoylation and phospholipid synthesis significantly inhibits RGNNV replication (Huang et al., 2020). In plant cells, the S-palmitoylation of Mungbean yellow mosaic virus (MYMV) AC4 allows its accumulation on the plasma membrane, thereby counteracting the host defense mechanism by suppressing the post-transcriptional gene silencing (PTGS) (Carluccio et al., 2018).
In fact, another important function of palmitoylation is to target modified proteins to lipid rafts (Diaz-Rohrer et al., 2014; Stepanek et al., 2014). These rafts provide the membrane platform for the entry, assembly, and budding of different viruses. It is speculated that the specific inhibition of acyltransferases that dramatically inhibits viral replication will not affect the palmitoylation of intracellular proteins because the transportation of fatty acids could be mediated by other members of the zDHHC family (Veit and Siche, 2015; Gadalla and Veit, 2020). Therefore, some researchers proposed that inhibition of certain protein palmitoylation may be a promising strategy for treating related diseases (Chavda et al., 2014; Lanyon-Hogg et al., 2017; Lin et al., 2017; Chen et al., 2018). The zDHHC protein family and APTs could be utilized as key modifiers of protein palmitoylation cycles, making them potential drug targets, which means ZDHHCs targeting viral proteins need to be identified (Gadalla and Veit, 2020). However, although palmitoylation of viral proteins had already been discovered 43 years ago (Schmidt and Schlesinger, 1979), the identification of PATs responsible for S-acylation of viral proteins remained a major gap till several zDHHCs catalyzing the S-acylation of proteins of influenza viruses and SARS-CoV-2 were found. Using CRISPR/Cas9 knockout and siRNA screening, DHHC2, 8, 15, and 20 were found to be involved in S-acylation of HA and M2 of IAV and human influenza viruses (Gadalla et al., 2020). DHHC20, -8, and -9 were verified to play certain roles in SARS-CoV-2 spike protein(S) and enveloped protein (Abdulrahman et al., 2021; Mesquita et al., 2021; Puthenveetil et al., 2021). However, conflicting results regarding the palmitoylation sites and palmitoylation enzymes of SARS-CoV-2 spike protein were obtained by different researchers (Mesquita et al., 2021; Puthenveetil et al., 2021; Zeng et al., 2021; Li et al., 2022). Li et al. found the cysteines of C-terminal and N‐terminal C15 of the spike protein were palmitoylated (Li et al., 2022), while Wu et al. reported that N-terminal C15 mutant did not affect the S-acylation of the spike (Wu, Zhang et al., 2021). Using computational, lipidomic, and biochemical approaches, Mesquita et al. deciphered the roles of zDHHC20 and zDHHC9 in the palmitoylation of the C terminal cysteines of spike protein in in vivo and in vitro conditions (Mesquita et al., 2021). The reasons for the conflicting results may be the use of different viruses and cell models or different methods. In addition no specific molecule drug for inhibiting a certain zDHHC substrate interaction has been developed so far (Abdulrahman et al., 2021). Looking forward, we believe that the discovery and identification of enzymes and their specific inhibitors acting on the palmitoylation cycle of viral proteins would be a new challenge and almost certainly would become one of the most interesting topics to investigate. Further biochemical and structural analysis of different zDHHCs and a series of different viral and cellular substrates are needed to understand the mechanism of acylation reactions (Gadalla and Veit, 2020).
3.2 Palmitoylation of Host Cell Proteins
Besides the expression of immune-related genes, viral infection can trigger a series of immune responses in host cells, including the modification of related proteins. However, so far, few such proteins have been reported. Furthermore, the palmitoylated host proteins that have been reported during viral infection are associated with interferon (IFN) pathways, e.g., stimulator of interferon genes (STING) and interferon-induced transmembrane (IFITM).
STING, a central signal component of DNA sensing pathways in cells, is a class of small molecular proteins located in the endoplasmic reticulum (Ishikawa et al., 2009; Sun et al., 2013). It is the optimal inducer of type I IFN in response to invading viruses or bacteria (Ishikawa and Barber, 2008; Hansen et al., 2014; Christensen et al., 2016; Holm et al., 2016). Excessive activation of the STING signaling pathway gives rise to abnormal responses of the innate immune system, leading to a series of autoinflammatory diseases (Hansen et al., 2019). Researchers have found that STING activation depends on its palmitoylation in the Golgi complex (Ishikawa and Barber, 2008). A non-specific inhibitor for protein palmitoylation, 2-bromopalmitate (2-BP), can inhibit palmitoylation of STING and eliminate type I IFN response (Mukai et al., 2016). Another important finding is the identification of a class of highly effective and selective antagonists blocking STING palmitoylation by covalently binding to the predicted transmembrane Cys91 (Mukai et al., 2016). Experimental evidence showed the antagonists, and their derivatives can down-regulate STING-mediated inflammatory cytokine expression (Haag et al., 2018). Together, these data imply the potential of STING-palmitoylation-targeted therapy in the treatment of autoinflammatory diseases.
Another broad-spectrum antiviral protein family known as IFITMs has recently emerged as promising palmitoylation targets (Brass et al., 2009; Yount et al., 2010; Yount et al., 2012; Diamond and Farzan, 2013; McMichael et al., 2017). The cysteine palmitoylation of IFITMs plays an important role in their antiviral activities because this modification may help in the regulation of IFITMs localization (Hach et al., 2013). Besides, localization in the vesicle membrane allows IFITM to directly fuse with the specific virus particles. This fusion complex is then trafficked to the lysosome, which relies on the palmitoylation of its three conserved cysteines at 70,71, and 105 sites (Spence et al., 2019). Furthermore, preventing the fusion of the viral membrane with the cells or the fusion among the infected cells could be the main antiviral mechanism of IFITM (Buchrieser et al., 2019; Zani et al., 2019). A very interesting study showed that placental damage and abortion caused by the virus-induced upregulation of type I IFN during pregnancy is associated with the palmitoylation of IFITM3. This can be caused by the mutation in three cysteine residues of IFITM3, which suppresses the inhibition of cell fusion (Buchrieser et al., 2019). In one of our studies, we confirmed that resistance to the Japanese encephalitis virus (JEV) infection requires the S-palmitoylation modification of IFITM in swine (Xu et al., 2020). Therefore, the discovery of activators or inhibitors targeting the palmitoylation/depalmitoylation cycle of IFITM would be a promising subject for antiviral drugs or immunomodifiers of the diseases caused by viral infections or immune disorders.
4 Detection Methods for Palmitoylation Modifications
The researchers have developed different approaches to detect S-palmitoylation, which has effectively contributed to the identification and functional research. This section summarizes the principles and brief procedures of these methods and discusses their major limitations. According to the different targets of analysis in the S-palmitoyl group, the identification methods can be divided roughly into two categories, the palmitate-targeted and the cysteine-targeted (also called lipid-and cysteine-centric). All these techniques are label-based, except the technique of gas chromatography-mass spectrometry (GC-MS) and MALDI-TOF MS, which is a relatively direct way. When palmitic acid is the target of analysis, it is usually labeled with an analog of palmitic acid or a probe, and then the labeled proteins are enriched by affinity purification or observed with optical instruments. However, such methods cannot be applied to body fluids or tissue samples of in vivo experiments because it requires the metabolic activity of the radioactive label or chemically modified palmitic acid analogs. Besides, when palmitic acid analog is labeled on the target protein, other shorter, longer, or unsaturated fatty acid modifications cannot be analyzed except for palmitoylated modifications. In the cysteine-centric method, biotin or molecular mass labels are utilized to detect palmitoylation proteins by affinity enrichment purification or western blotting. Because this method does not require metabolic labeling in live cells, it can be used to analyze not only samples of cells but also tissues and body fluids. The general operating procedures of the commonly used strategies mentioned above are described separately.
4.1 Mass Spectrometry
4.1.1 Gas Chromatography–Coupled Mass Spectrometry
GC-MS is a relatively direct and reproducible method and has been widely used for analyzing hundreds of metabolites in biological fluids or tissue samples (Zeki et al., 2020). The method mainly comprises the following steps: 1) preparation of purified protein samples; 2) washing the samples to remove non-covalently coupled lipids; 3) hydrogenation with hydroxylamine for cleavage of thioester bonds and simultaneous ethyl esterification with anhydrous formic acid and ethanol; 4) liquid phase extraction to separate free fatty acids; and 5) the detection of the exact lipid fraction with GC-MS (Sorek and Yalovsky, 2010; Sorek et al., 2013). Based on GC-MS analysis, the purified recombinant Rho-related GTPase ROP6 (Thomas et al., 1992) and calcium sensor protein CBL1 (Uchida and Stadtman, 1992) of Arabidopsis have been identified as C-16 palmitic acid and C-18 stearic acid-modified proteins, respectively (Batistic et al., 2008; Sorek et al., 2011; Sorek et al., 2013). Given the range of protein concentration and volume of samples required for GC-MS analysis, S-palmitoylation can be detected for a purified protein sample with a concentration as low as 1 µg (Li et al., 2011). However, to a certain extent, the application of GC-MS in the identification of palmitoylated proteins is limited because of its need for purified protein samples. Since most of the palmitoylated proteins are membrane-localized and have strong hydrophobicity, their purification is a difficult process.
4.1.2 MALDI-TOF Mass Spectrometry
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) is the only method that allows detecting directly the exact cysteine binding of fatty acid and the type of fatty acid attached to the S-acylated hydrophobic molecules (Webster and Oxley, 2012). In addition, this approach could be applied to the proteins that are difficult purify from protein mixtures. Besides the identification of palmitoylation, the MALDI-TOF MS-based approach revealed lipidation modifications of several cellular proteins and the heterogeneous fatty acylation of neuromodulin GAP-43 and GNAI (Liang et al., 2002) (Nuskova et al., 2021).
Further, by this method, the differential S-acylation of many viral proteins was confirmed (Serebryakova et al., 2006; Kordyukova et al., 2008; Kordyukova et al., 2010; Mineev et al., 2013; Vitale et al., 2013; Brett et al., 2014). Therefore, MALDI-TOF MS is an approach that uniquely detects hydrophobic viral lipopeptides, which assist in finding therapeutic targets to prevent viruses.
In contrast to MALDI-MS, which is perfect to identify the exact type of fatty acid attached to the exact S-acylation site, the LC-MS/MS based methods are adopted mainly in combination with other approaches, for example, ssABE (Collins et al., 2017; Woodley and Collins, 2019) and PalmPISC (Yang et al., 2010; Dowal et al., 2011) are all combined with acyl-biotin exchange (ABE) method, which will be mentioned below.
4.2 Radioactive Palmitic Acid Metabolic Labeling
Radioactive labeling has been a relatively straightforward approach because the palmitoyl labeled with radioisotope 3H or 14C could be developed by autoradiography. Following metabolic labeling of the samples, the protein can be easily immunoprecipitated and analyzed using SDS-PAGE; the radiolabeled target protein can be visualized by autoradiography (Peseckis et al., 1993; Swarthout et al., 2005; Zou et al., 2011; Tsai et al., 2014). The main steps of this approach include: 1) incubating the cultured cells in the serum-free medium; 2) labeling cells with radiolabeled 3H-palmitic acid; 3) washing cells to remove unbound 3H-palmitic acid; 4) immunoprecipitation and analysis of the labeled proteins by SDS-PAGE, and 5) visualization of the 3H emission signal by X-ray film exposure. This method was utilized to identify the palmitoylation of influenza A glycoprotein hemagglutinin (HA) and hepatitis E virus (HEV) ORF3 protein (Gouttenoire et al., 2018; Gadalla et al., 2020). However, since this method is less sensitive, tedious, time-consuming, and hazardous (because of radioactive material), it has not been used very often in recent years (Lu and Fang, 2020).
4.3 Click Chemistry
Click chemistry provides an alternative way to analyze S-palmitoylated proteins. By means of Huisgen’s copper (I)-catalyzed azide-alkyne cycloaddition reaction, target proteins are labeled with terminal azide or alkynyl groups, purified by affinity chromatography, and identified with mass spectrometry or gel-based visualization (Speers and Cravatt, 2004; Sobocinska et al., 2017). The commercially available alkyne fatty acid analog, 17-octadecynoic acid (17-ODYA), is commonly used as a non-radioactive derivative of palmitic acid. This is because it can enter into palmitoylated endogenous sites by metabolic pathways of the cellular palmitoylation mechanism. In addition, based on the biological orthogonal method (this allows multiple methods of labeling to be used in the same biosystems), dual-click chemistry pulse-chase scheme has been developed. For example, both 17-ODYA and methionine surrogate L-azidohomoalanine (L–AHA) are used for detecting dynamic palmitoylation modification and monitoring the turnover rate of proteins (Lin and Conibear, 2015). Besides, the click chemistry-based method can be used to study the subcellular localization of palmitoylated proteins through in situ probe combination techniques (Gao and Hannoush, 2014; Jiang et al., 2018; Stypulkowski et al., 2018). Also, if combined with mass spectrometry-based proteomics, this kind of non-radioactive labeling of alkynyl fatty acids is far more sensitive and safety, making it easier to analyze the palmitoylated protein globally and dynamically (Charron et al., 2009; Martin and Cravatt, 2009; Yount et al., 2010; Martin et al., 2011). However, under in situ environment, analogs of palmitic acid may hinder the normal metabolism and other processes because of the complexity of eukaryotic cell metabolism; for example, N-, and O-palmitoylation and N-myristoylation sites of proteins could reduce the efficiency and accuracy of detection methods (Jones et al., 2012; Gao and Hannoush, 2014; Wright et al., 2014).
4.4 Acyl-Biotin Exchange (ABE) and Acyl-Resin Assisted Capture (acyl-RAC)
These two methods are cysteine-centric and involve non-radioactive labeling, and to some extent, simplify the identification and quantification of palmitoylated proteins. These methods include several key steps: completely blocking the free sulfhydryl group, using hydroxylamine to hydrolyze all thioester bonds, and capturing the newly released free sulfhydryl group with a pyridyl-disulfide bond biotin conjugate (Forrester et al., 2011; Guo et al., 2014) to form biotin linked by disulfide bonds (ABE), or directly capturing them by pyridyl disulfide bond resin (Acyl-RAC). For the detection of S-palmitoylation, the peptides labeled with biotin are enriched with streptavidin resin, and then subjected to mass spectrometry.
Since ABE and acyl-RAC are easier, convenient, and timesaving, by far, hundreds of S-palmitoylated proteins have been identified, and their S-palmitoylated sites have been annotated (Linder et al., 1993; Mumby and Muntz, 1995; Forrester et al., 2011; Zhou et al., 2019). However, there are certain disadvantages of ABE, including the high background caused by captured non-S-palmitoylated proteins and the fact that the type of lipid attached to the protein cannot be accurately identified and needs further analysis. Furthermore, these indirect detection methods cannot accurately quantify the palmitoylation sites of endogenous proteins. By adding 2,2′-dithiodipyridine, Zhou et al. developed a low-background ABE (LB-ABE) method based on the blocking of residual free cysteine residues before the biotin-HPDP reaction and identified thousands of candidate S-palmitoylated proteins (Zhou et al., 2019).
4.5 Acyl-PEGyl Exchange Gel-Shift (APEGS) Method
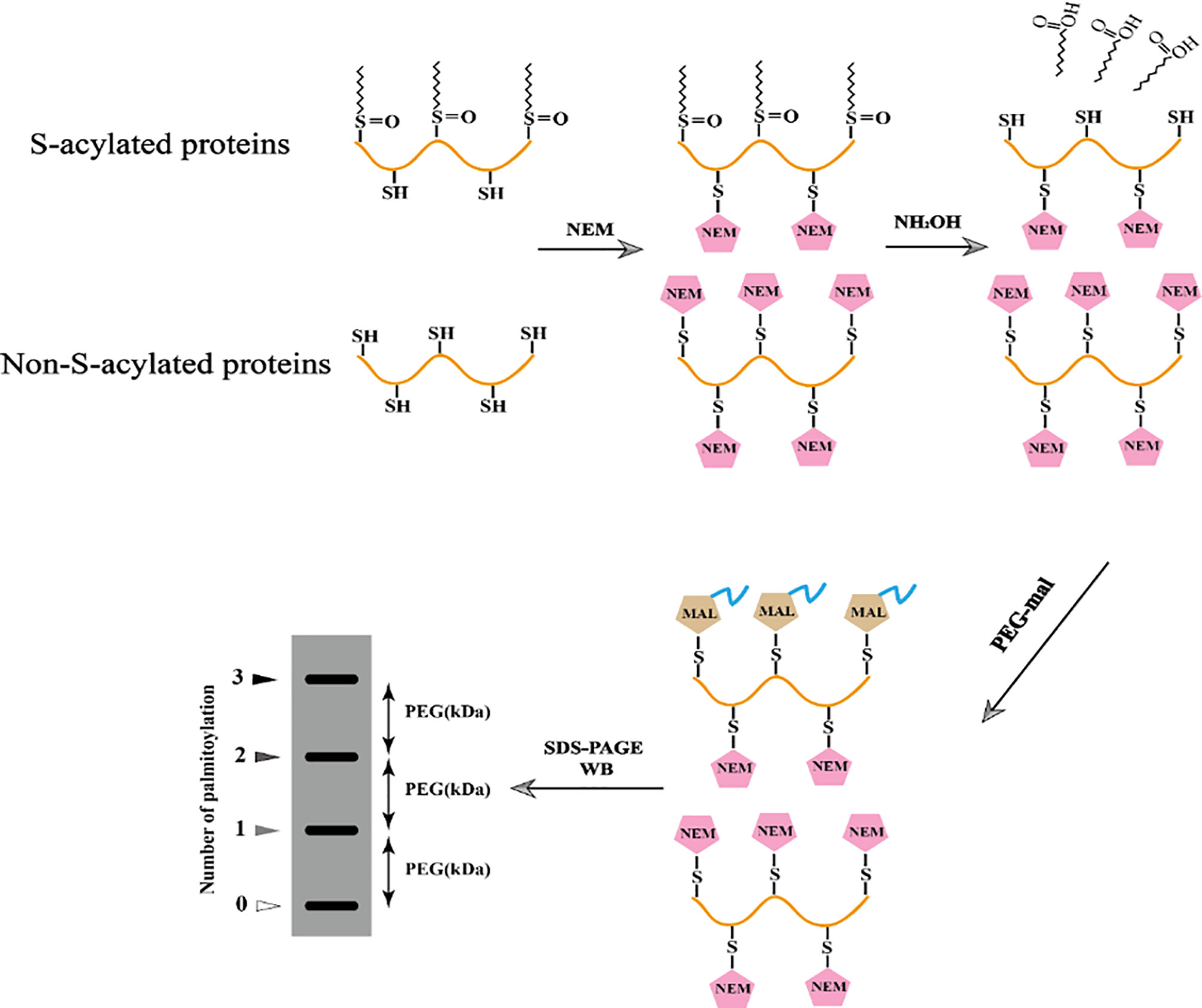
In 2016, a novel method based on Acyl-PEGyl exchange gel-shift (APEGS) was reported, in which the palmitoylated proteins were labeled with a fixed mass label, such as 2 kDa, 5 kDa, or 10 kDa methoxy polyethylene glycol maleimide (MPEG-mal) and detected by western blotting (Percher et al., 2016) (Figure 2).
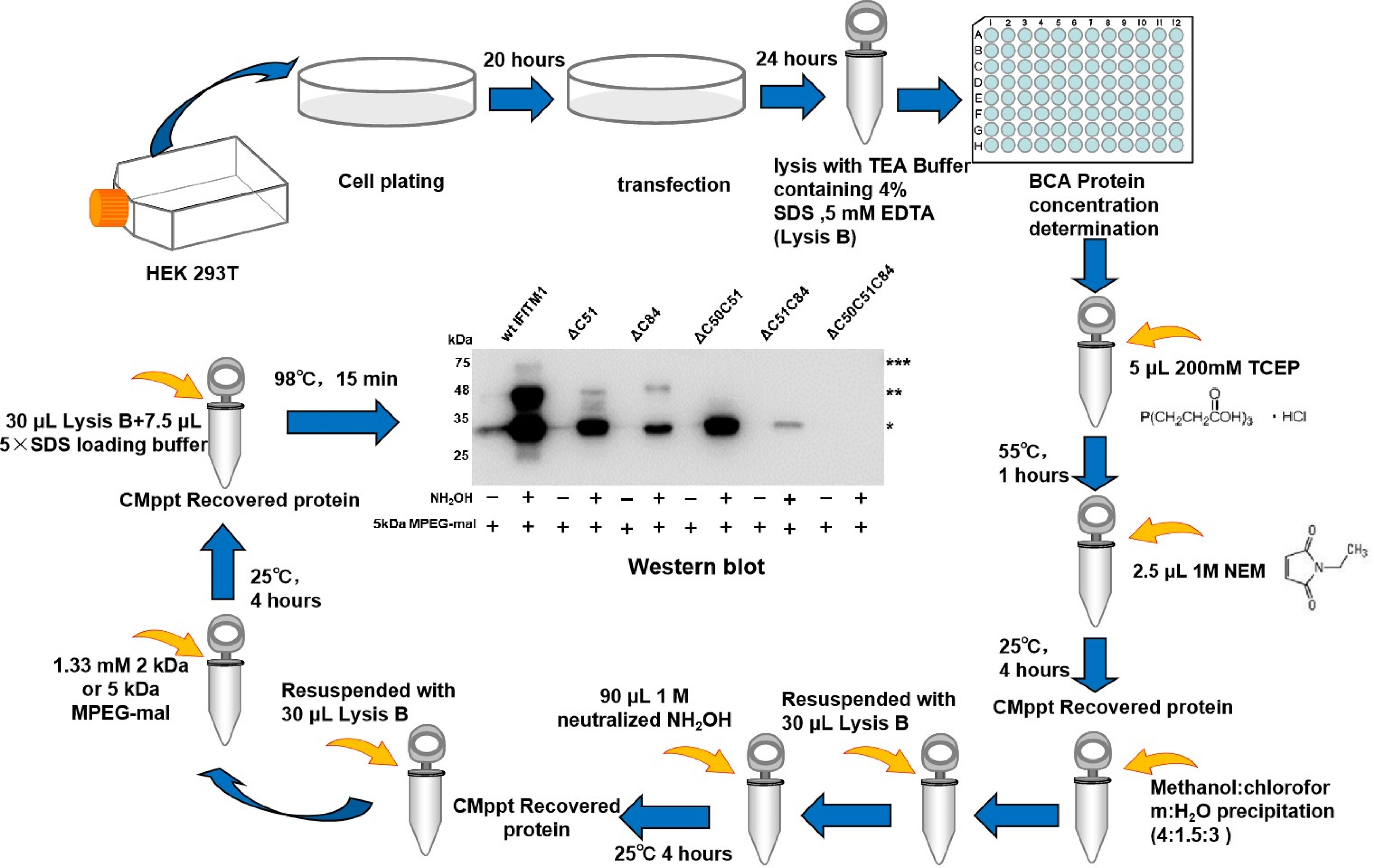
First, Tris (2-carboxyethyl) phosphine (TCEP) is used to reduce all disulfide bonds in cell lysate proteins. All non-palmitoylated cysteines or free hydroxyl groups in the protein sequence are blocked with N-ethylmaleimide (NEM). After activation with hydroxylamine hydrochloride (NH2OH), the cysteine sites where palmitoylation occurs are reduced to a free sulfhydryl state. Next, these free sulfhydryl groups are replaced by the newly added mass label, MPEG-mal, and finally, the palmitoylation is detected by western blotting. Due to its strong sensitivity and high specificity, this novel technology can determine the degree of palmitoylation and identify the number of palmitoylation sites of proteins with the help of gray-scale analysis software, such as the frequently used ones: Image J, Image-Pro Plus, and Quantity One. Based on the procedures of Takashi Kanadome (Kanadome et al., 2019) and Avital Perchera (Percher et al., 2016), we successfully identified three S-palmitoylation sites of swine IFITM1 and also analyzed the characteristics of each palmitoylation site by mathematical calculations of gray-scale analysis software (Xu et al., 2020) (Figure 3).
4.6 Other Supporting Strategies
Identification of palmitoylation sites by the above-mentioned methods, whether in vivo or in vitro, is usually time-consuming and labor-intensive. According to the characteristics of amino acids sequences and the structures of the palmitoylated proteins, researchers developed the software or websites to predict the possible palmitoylation sites for proteins. The in silico-based prediction platforms, such as CSS-Palm, IFS-Palm, WAP-Palm, PalmPred, SwissPalm database, and others, can narrow down the possible palmitoylation sites and can therefore guide further experimental designs. Here, we highlight two of them. The clustering and scoring strategy for palmitoylation sites prediction (CSS-Palm) system was implemented by Zhou et al., who developed a free accessible web server (Zhou et al., 2006; Ren et al., 2008). Since then, with CSS-Palm, many potential palmitoylated proteins of cells have been predicted, including PD-L1 (Zhou et al., 2019; Wang et al., 2020), MC1R (Chen et al., 2017), and differential S-acylation of enveloped viruses (Kordyukova et al., 2019). In 2021, a novel predictor, graphic presentation system for the prediction of S-palmitoylation (GPS-palm) was developed. This platform showed great improvement in the general prediction of S-palmitoylation sites compared with other existing tools (Ning et al., 2021). GPS-palm also provides two species-specific predictors for predicting human-and mouse-specific sites produced by the same research group would be useful tools for researchers of the related fields.
To confirm the amino acid sites of palmitoylation or to investigate the function or localization of palmitoylated proteins, mutagenesis is utilized as the standard and common approach. In this method, the potentially palmitoylated cysteine residue is replaced by serine or alanine. Because of its similar structure to cysteine, serine substitution can maintain the properties of putative palmitoylated protein; however, this substitution might cause side-chain defects and false positivity due to the higher hydrophilicity of hydroxyl groups.
5 Conclusion/Perspective
In the last few years, considerable headway has been made in the field of protein S-palmitoylation, including the increased number of the identified S-palmitoylated proteins, the drastically improved detecting methods, the developed labels, and some promising inhibitors. The scientific community has reached a consensus on the significance of palmitoylation in the occurrence and progression of certain diseases. Studies on protein palmitoylation have gained importance after the realization of the harmful effects caused by a variety of viruses. Quite a few researchers believe that palmitoylation-related proteins or enzymes are likely to be a breakthrough point in the treatment of major diseases in the future. Based on APEGS and pulse-chase protocol (click chemistry), a team of Harvard Medical School found that fatty acid and zDHHC19-mediated palmitoylation are key factors for STAT3 signal regulation (Niu et al., 2019), which provided important evidence of the relationship between palmitoylation and inflammation or cancer. The studies mentioned in this review indicate that palmitoylation affects the crucial function of proteins by regulating the interactions of proteins. Therefore, it is predicable that potential therapeutic targets for related diseases would be found with further studies on protein palmitoylation. Recently, an inhibitor of epithelial growth factor receptor (EGFR), DHHC20 antagonist, has been discovered and may develop into a drug candidate for treating patients with KRAS mutation tumors (Kharbanda et al., 2020).
Despite the recent progress in studies related to the palmitoylation of proteins associated with viral infections, there are many questions to be answered, for example, how the palmitoylation of virial proteins regulates viral replication dynamically. The following findings and speculations may provide important references for researchers in this field. The palmitoylation of influenza virus HA may serve as a raft-targeting signal, which can recruit HA to the plasma membrane and form a large raft structure (Leser and Lamb, 2005; Hess et al., 2007); thus, it provides a platform for the assembly and budding of the virions (Gerl et al., 2012). S-palmitoylation of influenza virus M2 may facilitate the shedding of virus particles from the plasma membrane and their budding (Schroeder et al., 2005). Efficient replication of CHIKV depends on the palmitoylation of two membrane-associated loops of nsP1. This is because the acylation leads to a hydrophobic state and helps in the electrostatic interaction of the protein with the inner leaflet of the plasma membrane; thus, enhancing viral replication (Zhang et al., 2021). A recent study reported that the palmitoylation of coronavirus proteins and the ensuing formation of complex lipid membranes were crucial to virus replication and assembly (Tanner and Alfieri, 2021), which undoubtedly provides a clue for digging into potential application of palmitoylated proteins, especially during the COVID-19 pandemic. Besides, we think that in-depth exploration of the following directions would contribute to understanding protein palmitoylation at a higher level:
i. The exploration of the relationship between the intracellular molecular mechanisms or pathways of virus entry or replication and the palmitoylated proteins involved in order to discover the target whose palmitoylation/depalmitoylation cycle should be regulated.
ii. The identification of enzymes catalyzing the cycle of protein palmitoylation and screening of proteins that interact with palmitoylated proteins so that they could be utilized as targets to regulate the palmitoylation cycle. For example, till now, only zDHHC family of palmitoylases have been identified (Mitchell et al., 2006). Also, there are very few newly confirmed depalmitoylases, except for ABHD17 and ABHD10 (Lin and Conibear, 2015; Cao et al., 2019). APTs and PPTs that catalyze the cycle of protein palmitoylation in other mammalian species have not been reported yet.
iii. High-throughput methods for unbiased and efficient identification of palmitoylated proteins involved in the whole process of virus-host interactions should be improved. Due to the strong hydrophobicity of palmitoylated proteins, many challenges exist in the detection and analysis of such proteins. Moreover, this kind of modification usually occurs in low or medium abundance proteins, which makes the identification disjointed as no specific antibody can be used (Yang et al., 2010). All these bottlenecks suggest that more reliable and effective methods should be developed.
iv. The discovery of specific inhibitors of the enzymes participating in protein palmitoylation. Although 2-bromopalmitate had been recognized as an inhibitor of protein palmitoylation, it has widely been recognized to exert off-target effects by inhibiting other enzymes involved in lipid metabolism and is also cytotoxic to cells (Pedro et al., 2013). A new inhibitor of DHHC, cyano-myracrylamide (CMA), was reported recently, which has a broad-spectrum inhibitory effect on DHHC family members like 2-bromopalmitate but with less toxicity and off-target effects (Azizi et al., 2021).
v. Analysis of the structures of enzymes and viral proteins involved in the palmitoylation/depalmitoylation cycle would be helpful for a better understanding of the subtle ways of host-virus interactions and designing new inhibitors or drugs.
This review gives a brief overview of protein palmitoylation and provides some theoretical foundation for further research on palmitoylation and the biological function of target proteins that are involved in various diseases, including viral infections. We hope more researchers will demonstrate their interesting findings based on varying functions of the palmitoylated proteins in the future.
Author Contributions
JX designed research. JX and XL wrote the paper. XL and ZX draw the picture. LS contributed the table. LS, WL, and AL modified the paper. All authors contributed to the article and approved the submitted version.
Funding
This project was financially supported by the National Natural Science Fund (U1804108), the Foundation of Henan Educational Committee (19A230005), and the Preferred Foundation for Returned Scholar from Overseas of Ministry of Human Resources and Social Security of China (Study abroad personnel and expert service center of Henan Province) (2017–02).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Zihan Li and SageSCI for their contribution in modifying the figures and polishing the manuscript.
References
Abdulrahman, D. A., Meng, X., Veit, M. (2021). S-Acylation of Proteins of Coronavirus and Influenza Virus: Conservation of Acylation Sites in Animal Viruses and DHHC Acyltransferases in Their Animal Reservoirs. Pathogens 10 (6), 669. doi: 10.3390/pathogens10060669
Akerstrom, S., Gunalan, V., Keng, C. T., Tan, Y. J., Mirazimi, A. (2009). Dual Effect of Nitric Oxide on SARS-CoV Replication: Viral RNA Production and Palmitoylation of the S Protein Are Affected. Virology 395 (1), 1–9. doi: 10.1016/j.virol.2009.09.007
Azizi, S. A., Lan, T., Delalande, C., Kathayat, R. S., Banales Mejia, F., Qin, A., et al. (2021). Development of an Acrylamide-Based Inhibitor of Protein S-Acylation. ACS Chem. Biol. 16 (8), 1546–1556. doi: 10.1021/acschembio.1c00405
Bakhache, W., Neyret, A., Bernard, E., Merits, A., Briant, L. (2020). Palmitoylated Cysteines in Chikungunya Virus Nsp1 Are Critical for Targeting to Cholesterol-Rich Plasma Membrane Microdomains With Functional Consequences for Viral Genome Replication. J. Virol. 94 (10), e02183–19. doi: 10.1128/JVI.02183-19
Batistic, O., Sorek, N., Schultke, S., Yalovsky, S., Kudla, J. (2008). Fatty Acyl Modification Determines the Localization and Plasma Membrane Targeting of CBL/CIPK Ca2+ Signaling Complexes in Arabidopsis. Plant Cell 20 (5), 1346–1362. doi: 10.1105/tpc.108.058123
Bhattacharya, J., Peters, P. J., Clapham, P. R. (2004). Human Immunodeficiency Virus Type 1 Envelope Glycoproteins That Lack Cytoplasmic Domain Cysteines: Impact on Association With Membrane Lipid Rafts and Incorporation Onto Budding Virus Particles. J. Virol. 78 (10), 5500–5506. doi: 10.1128/jvi.78.10.5500-5506.2004
Biechele, S., Cox, B. J., Rossant, J. (2011). Porcupine Homolog is Required for Canonical Wnt Signaling and Gastrulation in Mouse Embryos. Dev. Biol. 355 (2), 275–285. doi: 10.1016/j.ydbio.2011.04.029
Blanc, M., Blaskovic, S., van der Goot, F. G. (2013). Palmitoylation, Pathogens and Their Host. Biochem. Soc. Trans. 41 (1), 84–88. doi: 10.1042/BST20120337
Blanc, M., David, F., Abrami, L., Migliozzi, D., Armand, F., Burgi, J., et al. (2015). SwissPalm: Protein Palmitoylation Database. F1000Res 4, 261. doi: 10.12688/f1000research.6464.1
Blanc, M., David, F. P. A., van der Goot, F. G. (2019). SwissPalm 2: Protein S-Palmitoylation Database. Methods Mol. Biol. 2009, 203–214. doi: 10.1007/978-1-4939-9532-5_16
Blaskovic, S., Blanc, M., van der Goot, F. G. (2013). What Does S-Palmitoylation do to Membrane Proteins? FEBS J. 280 (12), 2766–2774. doi: 10.1111/febs.12263
Boscarino, J. A., Logan, H. L., Lacny, J. J., Gallagher, T. M. (2008). Envelope Protein Palmitoylations Are Crucial for Murine Coronavirus Assembly. J. Virol. 82 (6), 2989–2999. doi: 10.1128/JVI.01906-07
Branigan, P. J., Day, N. D., Liu, C., Gutshall, L. L., Melero, J. A., Sarisky, R. T., et al. (2006). The Cytoplasmic Domain of the F Protein of Human Respiratory Syncytial Virus Is Not Required for Cell Fusion. J. Gen. Virol. 87 (Pt 2), 395–398. doi: 10.1099/vir.0.81481-0
Brass, A. L., Huang, I. C., Benita, Y., John, S. P., Krishnan, M. N., Feeley, E. M., et al. (2009). The IFITM Proteins Mediate Cellular Resistance to Influenza A H1N1 Virus, West Nile Virus, and Dengue Virus. Cell 139 (7), 1243–1254. doi: 10.1016/j.cell.2009.12.017
Brett, K., Kordyukova, L. V., Serebryakova, M. V., Mintaev, R. R., Alexeevski, A. V., Veit, M. (2014). Site-Specific S-Acylation of Influenza Virus Hemagglutinin: The Location of the Acylation Site Relative to the Membrane Border is the Decisive Factor for Attachment of Stearate. J. Biol. Chem. 289 (50), 34978–34989. doi: 10.1074/jbc.M114.586180
Brigidi, G. S., Bamji, S. X. (2013). Detection of Protein Palmitoylation in Cultured Hippocampal Neurons by Immunoprecipitation and Acyl-Biotin Exchange (ABE). J. Vis. Exp. 72, 50031. doi: 10.3791/50031
Brown, R. W., Sharma, A. I., Engman, D. M. (2017). Dynamic Protein S-Palmitoylation Mediates Parasite Life Cycle Progression and Diverse Mechanisms of Virulence. Crit. Rev. Biochem. Mol. Biol. 52 (2), 145–162. doi: 10.1080/10409238.2017.1287161
Buchrieser, J., Degrelle, S. A., Couderc, T., Nevers, Q., Disson, O., Manet, C., et al. (2019). IFITM Proteins Inhibit Placental Syncytiotrophoblast Formation and Promote Fetal Demise. Science 365 (6449), 176–180. doi: 10.1126/science.aaw7733
Caballero, M., Carabana, J., Ortego, J., Fernandez-Munoz, R., Celma, M. L. (1998). Measles Virus Fusion Protein is Palmitoylated on Transmembrane-Intracytoplasmic Cysteine Residues Which Participate in Cell Fusion. J. Virol. 72 (10), 8198–8204. doi: 10.1128/JVI.72.10.8198-8204.1998
Camp, L. A., Hofmann, S. L. (1993). Purification and Properties of a Palmitoyl-Protein Thioesterase That Cleaves Palmitate From H-Ras. J. Biol. Chem. 268 (30), 22566–22574. doi: 10.1016/S0021-9258(18)41567-0
Cao, Y., Qiu, T., Kathayat, R. S., Azizi, S. A., Thorne, A. K., Ahn, D., et al. (2019). ABHD10 is an S-Depalmitoylase Affecting Redox Homeostasis Through Peroxiredoxin-5. Nat. Chem. Biol. 15 (12), 1232–1240. doi: 10.1038/s41589-019-0399-y
Carluccio, A. V., Prigigallo, M. I., Rosas-Diaz, T., Lozano-Duran, R., Stavolone, L. (2018). S-Acylation Mediates Mungbean Yellow Mosaic Virus AC4 Localization to the Plasma Membrane and in Turns Gene Silencing Suppression. PloS Pathog. 14 (8), e1007207. doi: 10.1371/journal.ppat.1007207
Chamberlain, L. H., Shipston, M. J. (2015). The Physiology of Protein S-Acylation. Physiol. Rev. 95 (2), 341–376. doi: 10.1152/physrev.00032.2014
Charron, G., Zhang, M. M., Yount, J. S., Wilson, J., Raghavan, A. S., Shamir, E., et al. (2009). Robust Fluorescent Detection of Protein Fatty-Acylation With Chemical Reporters. J. Am. Chem. Soc. 131 (13), 4967–4975. doi: 10.1021/ja810122f
Chavda, B., Arnott, J. A., Planey, S. L. (2014). Targeting Protein Palmitoylation: Selective Inhibitors and Implications in Disease. Expert Opin. Drug Discovery 9 (9), 1005–1019. doi: 10.1517/17460441.2014.933802
Chen, B., Sun, Y., Niu, J., Jarugumilli, G. K., Wu, X. (2018). Protein Lipidation in Cell Signaling and Diseases: Function, Regulation, and Therapeutic Opportunities. Cell Chem. Biol. 25 (7), 817–831. doi: 10.1016/j.chembiol.2018.05.003
Chen, S., Zhu, B., Yin, C., Liu, W., Han, C., Chen, B., et al. (2017). Palmitoylation-Dependent Activation of MC1R Prevents Melanomagenesis. Nature 549 (7672), 399–403. doi: 10.1038/nature23887
Chlanda, P., Mekhedov, E., Waters, H., Sodt, A., Schwartz, C., Nair, V., et al. (2017). Palmitoylation Contributes to Membrane Curvature in Influenza A Virus Assembly and Hemagglutinin-Mediated Membrane Fusion. J. Virol. 91 (21), e00947–17. doi: 10.1128/JVI.00947-17
Cho, E., Park, M. (2016). Palmitoylation in Alzheimer’s Disease and Other Neurodegenerative Diseases. Pharmacol. Res. 111, 133–151. doi: 10.1016/j.phrs.2016.06.008
Christensen, M. H., Jensen, S. B., Miettinen, J. J., Luecke, S., Prabakaran, T., Reinert, L. S., et al. (2016). HSV-1 ICP27 Targets the TBK1-Activated STING Signalsome to Inhibit Virus-Induced Type I IFN Expression. EMBO J. 35 (13), 1385–1399. doi: 10.15252/embj.201593458
Collins, M. O., Woodley, K. T., Choudhary, J. S. (2017). Global, Site-Specific Analysis of Neuronal Protein S-Acylation. Sci. Rep. 7 (1), 4683. doi: 10.1038/s41598-017-04580-1
Corvi, M. M., Turowski, V. R. (2019). Palmitoylation in Apicomplexan Parasites: From Established Regulatory Roles to Putative New Functions. Mol. Biochem. Parasitol. 230, 16–23. doi: 10.1016/j.molbiopara.2019.04.001
Daniotti, J. L., Pedro, M. P., Valdez Taubas, J. (2017). The Role of S-Acylation in Protein Trafficking. Traffic 18 (11), 699–710. doi: 10.1111/tra.12510
Demers, A., Ran, Z., Deng, Q., Wang, D., Edman, B., Lu, W., et al. (2014). Palmitoylation is Required for Intracellular Trafficking of Influenza B Virus NB Protein and Efficient Influenza B Virus Growth In Vitro. J. Gen. Virol. 95 (Pt 6), 1211–1220. doi: 10.1099/vir.0.063511-0
Devedjiev, Y., Dauter, Z., Kuznetsov, S. R., Jones, T. L., Derewenda, Z. S. (2000). Crystal Structure of the Human Acyl Protein Thioesterase I From a Single X-Ray Data Set to 1.5 a. Structure 8 (11), 1137–1146. doi: 10.1016/s0969-2126(00)00529-3
Diamond, M. S., Farzan, M. (2013). The Broad-Spectrum Antiviral Functions of IFIT and IFITM Proteins. Nat. Rev. Immunol. 13 (1), 46–57. doi: 10.1038/nri3344
Diaz-Rohrer, B. B., Levental, K. R., Simons, K., Levental, I. (2014). Membrane Raft Association is a Determinant of Plasma Membrane Localization. Proc. Natl. Acad. Sci. U.S.A. 111 (23), 8500–8505. doi: 10.1073/pnas.1404582111
Doubravska, L., Krausova, M., Gradl, D., Vojtechova, M., Tumova, L., Lukas, J., et al. (2011). Fatty Acid Modification of Wnt1 and Wnt3a at Serine is Prerequisite for Lipidation at Cysteine and is Essential for Wnt Signalling. Cell. Signal. 23 (5), 837–848. doi: 10.1016/j.cellsig.2011.01.007
Dowal, L., Yang, W., Freeman, M. R., Steen, H., Flaumenhaft, R. (2011). Proteomic Analysis of Palmitoylated Platelet Proteins. Blood 118 (13), e62–e73. doi: 10.1182/blood-2011-05-353078
Duncan, J. A., Gilman, A. G. (1998). A Cytoplasmic Acyl-Protein Thioesterase That Removes Palmitate From G Protein Alpha Subunits and P21(RAS). J. Biol. Chem. 273 (25), 15830–15837. doi: 10.1074/jbc.273.25.15830
Ekanayake, E. V., Fu, R., Cross, T. A. (2016). Structural Influences: Cholesterol, Drug, and Proton Binding to Full-Length Influenza A M2 Protein. Biophys. J. 110 (6), 1391–1399. doi: 10.1016/j.bpj.2015.11.3529
Engel, S., Scolari, S., Thaa, B., Krebs, N., Korte, T., Herrmann, A., et al. (2010). FLIM-FRET and FRAP Reveal Association of Influenza Virus Haemagglutinin With Membrane Rafts. Biochem. J. 425 (3), 567–573. doi: 10.1042/BJ20091388
Ernst, A. M., Syed, S. A., Zaki, O., Bottanelli, F., Zheng, H., Hacke, M., et al. (2018). S-Palmitoylation Sorts Membrane Cargo for Anterograde Transport in the Golgi. Dev. Cell 47 (4), 479–493 e477. doi: 10.1016/j.devcel.2018.10.024
Forrester, M. T., Hess, D. T., Thompson, J. W., Hultman, R., Moseley, M. A., Stamler, J. S., et al. (2011). Site-Specific Analysis of Protein S-Acylation by Resin-Assisted Capture. J. Lipid Res. 52 (2), 393–398. doi: 10.1194/jlr.D011106
Fukata, Y., Fukata, M. (2010). Protein Palmitoylation in Neuronal Development and Synaptic Plasticity. Nat. Rev. Neurosci. 11 (3), 161–175. doi: 10.1038/nrn2788
Funke, C., Becker, S., Dartsch, H., Klenk, H. D., Muhlberger, E. (1995). Acylation of the Marburg Virus Glycoprotein. Virology 208 (1), 289–297. doi: 10.1006/viro.1995.1151
Gadalla, M. R., Abrami, L., van der Goot, F. G., Veit, M. (2020). Hemagglutinin of Influenza A, But Not of Influenza B and C Viruses is Acylated by ZDHHC2, 8, 15 and 20. Biochem. J. 477 (1), 285–303. doi: 10.1042/BCJ20190752
Gadalla, M. R., Veit, M. (2020). Toward the Identification of ZDHHC Enzymes Required for Palmitoylation of Viral Protein as Potential Drug Targets. Expert Opin. Drug Discovery 15 (2), 159–177. doi: 10.1080/17460441.2020.1696306
Gaedigk-Nitschko, K., Ding, M. X., Levy, M. A., Schlesinger, M. J. (1990). Site-Directed Mutations in the Sindbis Virus 6K Protein Reveal Sites for Fatty Acylation and the Underacylated Protein Affects Virus Release and Virion Structure. Virology 175 (1), 282–291. doi: 10.1016/0042-6822(90)90210-i
Gaedigk-Nitschko, K., Schlesinger, M. J. (1990). The Sindbis Virus 6K Protein can be Detected in Virions and Is Acylated With Fatty Acids. Virology 175 (1), 274–281. doi: 10.1016/0042-6822(90)90209-a
Gamage, D. G., Leikina, E., Quinn, M. E., Ratinov, A., Chernomordik, L. V., Millay, D. P. (2017). Insights Into the Localization and Function of Myomaker During Myoblast Fusion. J. Biol. Chem. 292 (42), 17272–17289. doi: 10.1074/jbc.M117.811372
Gao, X., Hannoush, R. N. (2014a). Method for Cellular Imaging of Palmitoylated Proteins With Clickable Probes and Proximity Ligation Applied to Hedgehog, Tubulin, and Ras. J. Am. Chem. Soc. 136 (12), 4544–4550. doi: 10.1021/ja410068g
Gao, X., Hannoush, R. N. (2014b). Single-Cell Imaging of Wnt Palmitoylation by the Acyltransferase Porcupine. Nat. Chem. Biol. 10 (1), 61–68. doi: 10.1038/nchembio.1392
Gao, X., Hannoush, R. N. (2018). A Decade of Click Chemistry in Protein Palmitoylation: Impact on Discovery and New Biology. Cell Chem. Biol. 25 (3), 236–246. doi: 10.1016/j.chembiol.2017.12.002
Gaudin, Y., Tuffereau, C., Benmansour, A., Flamand, A. (1991). Fatty Acylation of Rabies Virus Proteins. Virology 184 (1), 441–444. doi: 10.1016/0042-6822(91)90866-a
Gelhaus, S., Thaa, B., Eschke, K., Veit, M., Schwegmann-Wessels, C. (2014). Palmitoylation of the Alphacoronavirus TGEV Spike Protein S is Essential for Incorporation Into Virus-Like Particles But Dispensable for S-M Interaction. Virology 464, 397–405. doi: 10.1016/j.virol.2014.07.035
Gerl, M. J., Sampaio, J. L., Urban, S., Kalvodova, L., Verbavatz, J. M., Binnington, B., et al. (2012). Quantitative Analysis of the Lipidomes of the Influenza Virus Envelope and MDCK Cell Apical Membrane. J. Cell Biol. 196 (2), 213–221. doi: 10.1083/jcb.201108175
Globa, A. K., Bamji, S. X. (2017). Protein Palmitoylation in the Development and Plasticity of Neuronal Connections. Curr. Opin. Neurobiol. 45, 210–220. doi: 10.1016/j.conb.2017.02.016
Gok, C., Plain, F., Robertson, A. D., Howie, J., Baillie, G. S., Fraser, N. J., et al. (2020). Dynamic Palmitoylation of the Sodium-Calcium Exchanger Modulates Its Structure, Affinity for Lipid-Ordered Domains, and Inhibition by XIP. Cell Rep. 31 (10), 107697. doi: 10.1016/j.celrep.2020.107697
Gonzalez Montoro, A., Chumpen Ramirez, S., Valdez Taubas, J. (2015). The Canonical DHHC Motif Is Not Absolutely Required for the Activity of the Yeast S-Acyltransferases Swf1 and Pfa4. J. Biol. Chem. 290 (37), 22448–22459. doi: 10.1074/jbc.M115.651356
Goto, T., Shimotai, Y., Matsuzaki, Y., Muraki, Y., Sho, R., Sugawara, K., et al. (2017). Effect of Phosphorylation of CM2 Protein on Influenza C Virus Replication. J. Virol. 91 (22). doi: 10.1128/JVI.00773-17
Gottlieb, C. D., Linder, M. E. (2017). Structure and Function of DHHC Protein S-Acyltransferases. Biochem. Soc. Trans. 45 (4), 923–928. doi: 10.1042/BST20160304
Gouttenoire, J., Pollan, A., Abrami, L., Oechslin, N., Mauron, J., Matter, M., et al. (2018). Palmitoylation Mediates Membrane Association of Hepatitis E Virus ORF3 Protein and is Required for Infectious Particle Secretion. PloS Pathog. 14 (12), e1007471. doi: 10.1371/journal.ppat.1007471
Grantham, M. L., Wu, W. H., Lalime, E. N., Lorenzo, M. E., Klein, S. L., Pekosz, A. (2009). Palmitoylation of the Influenza A Virus M2 Protein Is Not Required for Virus Replication In Vitro But Contributes to Virus Virulence. J. Virol. 83 (17), 8655–8661. doi: 10.1128/JVI.01129-09
Greaves, J., Munro, K. R., Davidson, S. C., Riviere, M., Wojno, J., Smith, T. K., et al. (2017). Molecular Basis of Fatty Acid Selectivity in the zDHHC Family of S-Acyltransferases Revealed by Click Chemistry. Proc. Natl. Acad. Sci. U.S.A. 114 (8), E1365–E1374. doi: 10.1073/pnas.1612254114
Guo, J., Gaffrey, M. J., Su, D., Liu, T., Camp, D. G., 2nd, Smith, R. D., et al. (2014). Resin-Assisted Enrichment of Thiols as a General Strategy for Proteomic Profiling of Cysteine-Based Reversible Modifications. Nat. Protoc. 9 (1), 64–75. doi: 10.1038/nprot.2013.161
Gu, L., Robinson, R. A. (2016). Proteomic Approaches to Quantify Cysteine Reversible Modifications in Aging and Neurodegenerative Diseases. Proteomics Clin. Appl. 10 (12), 1159–1177. doi: 10.1002/prca.201600015
Haag, S. M., Gulen, M. F., Reymond, L., Gibelin, A., Abrami, L., Decout, A., et al. (2018). Targeting STING With Covalent Small-Molecule Inhibitors. Nature 559 (7713), 269–273. doi: 10.1038/s41586-018-0287-8
Hach, J. C., McMichael, T., Chesarino, N. M., Yount, J. S. (2013). Palmitoylation on Conserved and Nonconserved Cysteines of Murine IFITM1 Regulates its Stability and Anti-Influenza A Virus Activity. J. Virol. 87 (17), 9923–9927. doi: 10.1128/JVI.00621-13
Hansen, A. L., Mukai, K., Schopfer, F. J., Taguchi, T., Holm, C. K. (2019). STING Palmitoylation as a Therapeutic Target. Cell Mol. Immunol. 16 (3), 236–241. doi: 10.1038/s41423-019-0205-5
Hansen, K., Prabakaran, T., Laustsen, A., Jorgensen, S. E., Rahbaek, S. H., Jensen, S. B., et al. (2014). Listeria Monocytogenes Induces IFNbeta Expression Through an IFI16-, cGAS- and STING-Dependent Pathway. EMBO J. 33 (15), 1654–1666. doi: 10.15252/embj.201488029
Hatta, M., Kawaoka, Y. (2003). The NB Protein of Influenza B Virus Is Not Necessary for Virus Replication In Vitro. J. Virol. 77 (10), 6050–6054. doi: 10.1128/jvi.77.10.6050-6054.2003
Hemsley, P. A. (2017). An Outlook on Protein S-Acylation in Plants: What are the Next Steps? J. Exp. Bot. 68 (12), 3155–3164. doi: 10.1093/jxb/erw497
Hentschel, A., Zahedi, R. P., Ahrends, R. (2016). Protein Lipid Modifications–More Than Just a Greasy Ballast. Proteomics 16 (5), 759–782. doi: 10.1002/pmic.201500353
Hess, S. T., Gould, T. J., Gudheti, M. V., Maas, S. A., Mills, K. D., Zimmerberg, J. (2007). Dynamic Clustered Distribution of Hemagglutinin Resolved at 40 Nm in Living Cell Membranes Discriminates Between Raft Theories. Proc. Natl. Acad. Sci. U.S.A. 104 (44), 17370–17375. doi: 10.1073/pnas.0708066104
Holm, C. K., Rahbek, S. H., Gad, H. H., Bak, R. O., Jakobsen, M. R., Jiang, Z., et al. (2016). Influenza A Virus Targets a cGAS-Independent STING Pathway That Controls Enveloped RNA Viruses. Nat. Commun. 7, 10680. doi: 10.1038/ncomms10680
Hornemann, T. (2015). Palmitoylation and Depalmitoylation Defects. J. Inherit. Metab. Dis. 38 (1), 179–186. doi: 10.1007/s10545-014-9753-0
Huang, Y., Zhang, Y., Zheng, J., Wang, L., Qin, Q., Huang, X. (2020). Metabolic Profiles of Fish Nodavirus Infection In Vitro: RGNNV Induced and Exploited Cellular Fatty Acid Synthesis for Virus Infection. Cell Microbiol. 22 (9), e13216. doi: 10.1111/cmi.13216
Ishikawa, H., Barber, G. N. (2008). STING Is an Endoplasmic Reticulum Adaptor That Facilitates Innate Immune Signalling. Nature 455 (7213), 674–678. doi: 10.1038/nature07317
Ishikawa, H., Ma, Z., Barber, G. N. (2009). STING Regulates Intracellular DNA-Mediated, Type I Interferon-Dependent Innate Immunity. Nature 461 (7265), 788–792. doi: 10.1038/nature08476
Ito, H., Watanabe, S., Takada, A., Kawaoka, Y. (2001). Ebola Virus Glycoprotein: Proteolytic Processing, Acylation, Cell Tropism, and Detection of Neutralizing Antibodies. J. Virol. 75 (3), 1576–1580. doi: 10.1128/JVI.75.3.1576-1580.2001
Ivanova, L., Schlesinger, M. J. (1993). Site-Directed Mutations in the Sindbis Virus E2 Glycoprotein Identify Palmitoylation Sites and Affect Virus Budding. J. Virol. 67 (5), 2546–2551. doi: 10.1128/JVI.67.5.2546-2551.1993
Jennings, B. C., Linder, M. E. (2012). DHHC Protein S-Acyltransferases Use Similar Ping-Pong Kinetic Mechanisms But Display Different Acyl-CoA Specificities. J. Biol. Chem. 287 (10), 7236–7245. doi: 10.1074/jbc.M111.337246
Jiang, H., Zhang, X., Chen, X., Aramsangtienchai, P., Tong, Z., Lin, H. (2018). Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies. Chem. Rev. 118 (3), 919–988. doi: 10.1021/acs.chemrev.6b00750
Jones, M. L., Collins, M. O., Goulding, D., Choudhary, J. S., Rayner, J. C. (2012). Analysis of Protein Palmitoylation Reveals a Pervasive Role in Plasmodium Development and Pathogenesis. Cell Host Microbe 12 (2), 246–258. doi: 10.1016/j.chom.2012.06.005
Kanadome, T., Yokoi, N., Fukata, Y., Fukata, M. (2019). Systematic Screening of Depalmitoylating Enzymes and Evaluation of Their Activities by the Acyl-PEGyl Exchange Gel-Shift (APEGS) Assay. Methods Mol. Biol. 2009, 83–98. doi: 10.1007/978-1-4939-9532-5_7
Kharbanda, A., Walter, D. M., Gudiel, A. A., Schek, N., Feldser, D. M., Witze, E. S. (2020). Blocking EGFR Palmitoylation Suppresses PI3K Signaling and Mutant KRAS Lung Tumorigenesis. Sci. Signal 13 (621), eaax2364. doi: 10.1126/scisignal.aax2364
Ko, P. J., Dixon, S. J. (2018). Protein Palmitoylation and Cancer. EMBO Rep. 19 (10), e46666. doi: 10.15252/embr.201846666
Kordyukova, L. V., Serebryakova, M. V., Baratova, L. A., Veit, M. (2008). S Acylation of the Hemagglutinin of Influenza Viruses: Mass Spectrometry Reveals Site-Specific Attachment of Stearic Acid to a Transmembrane Cysteine. J. Virol. 82 (18), 9288–9292. doi: 10.1128/JVI.00704-08
Kordyukova, L. V., Serebryakova, M. V., Baratova, L. A., Veit, M. (2010). Site-Specific Attachment of Palmitate or Stearate to Cytoplasmic Versus Transmembrane Cysteines Is a Common Feature of Viral Spike Proteins. Virology 398 (1), 49–56. doi: 10.1016/j.virol.2009.11.039
Kordyukova, L. V., Serebryakova, M. V., Khrustalev, V. V., Veit, M. (2019). Differential S-Acylation of Enveloped Viruses. Protein Pept. Lett. 26 (8), 588–600. doi: 10.2174/0929866526666190603082521
Kordyukova, L. V., Serebryakova, M. V., Polyansky, A. A., Kropotkina, E. A., Alexeevski, A. V., Veit, M., et al. (2011). Linker and/or Transmembrane Regions of Influenza A/Group-1, A/Group-2, and Type B Virus Hemagglutinins Are Packed Differently Within Trimers. Biochim. Biophys. Acta 1808 (7), 1843–1854. doi: 10.1016/j.bbamem.2011.03.005
Lanyon-Hogg, T., Faronato, M., Serwa, R. A., Tate, E. W. (2017). Dynamic Protein Acylation: New Substrates, Mechanisms, and Drug Targets. Trends Biochem. Sci. 42 (7), 566–581. doi: 10.1016/j.tibs.2017.04.004
Leser, G. P., Lamb, R. A. (2005). Influenza Virus Assembly and Budding in Raft-Derived Microdomains: A Quantitative Analysis of the Surface Distribution of HA, NA and M2 Proteins. Virology 342 (2), 215–227. doi: 10.1016/j.virol.2005.09.049
Liang, X., Lu, Y., Neubert, T. A., Resh, M. D. (2002). Mass Spectrometric Analysis of GAP-43/Neuromodulin Reveals the Presence of a Variety of Fatty Acylated Species. J. Biol. Chem. 277 (36), 33032–33040. doi: 10.1074/jbc.M204607200
Liao, Y., Yuan, Q., Torres, J., Tam, J. P., Liu, D. X. (2006). Biochemical and Functional Characterization of the Membrane Association and Membrane Permeabilizing Activity of the Severe Acute Respiratory Syndrome Coronavirus Envelope Protein. Virology 349 (2), 264–275. doi: 10.1016/j.virol.2006.01.028
Li, L., Dong, L., Xia, L., Li, T., Zhong, H. (2011). Chemical and Genetic Probes for Analysis of Protein Palmitoylation. J. Chromatogr. B Analyt. Technol. BioMed. Life Sci. 879 (17-18), 1316–1324. doi: 10.1016/j.jchromb.2010.11.018
Li, D., Liu, Y., Lu, Y., Gao, S., Zhang, L. (2022). Palmitoylation of SARS-CoV-2 S Protein Is Critical for S-Mediated Syncytia Formation and Virus Entry. J. Med. Virol. 94 (1), 342–348. doi: 10.1002/jmv.27339
Lin, D. T., Conibear, E. (2015). ABHD17 Proteins are Novel Protein Depalmitoylases That Regulate N-Ras Palmitate Turnover and Subcellular Localization. Elife 4, e11306. doi: 10.7554/eLife.11306
Lin, D. T. S., Davis, N. G., Conibear, E. (2017). Targeting the Ras Palmitoylation/Depalmitoylation Cycle in Cancer. Biochem. Soc. Trans. 45 (4), 913–921. doi: 10.1042/BST20160303
Linder, M. E., Deschenes, R. J. (2007). Palmitoylation: Policing Protein Stability and Traffic. Nat. Rev. Mol. Cell Biol. 8 (1), 74–84. doi: 10.1038/nrm2084
Linder, M. E., Middleton, P., Hepler, J. R., Taussig, R., Gilman, A. G., Mumby, S. M. (1993). Lipid Modifications of G Proteins: Alpha Subunits are Palmitoylated. Proc. Natl. Acad. Sci. U.S.A. 90 (8), 3675–3679. doi: 10.1073/pnas.90.8.3675
Lobo, S., Greentree, W. K., Linder, M. E., Deschenes, R. J. (2002). Identification of a Ras Palmitoyltransferase in Saccharomyces Cerevisiae. J. Biol. Chem. 277 (43), 41268–41273. doi: 10.1074/jbc.M206573200
Lopez, L. A., Riffle, A. J., Pike, S. L., Gardner, D., Hogue, B. G. (2008). Importance of Conserved Cysteine Residues in the Coronavirus Envelope Protein. J. Virol. 82 (6), 3000–3010. doi: 10.1128/JVI.01914-07
Lu, H., Fang, C. (2020). Methodology for Detecting Protein Palmitoylation. Adv. Exp. Med. Biol. 1248, 425–430. doi: 10.1007/978-981-15-3266-5_17
Mach, M., Osinski, K., Kropff, B., Schloetzer-Schrehardt, U., Krzyzaniak, M., Britt, W. (2007). The Carboxy-Terminal Domain of Glycoprotein N of Human Cytomegalovirus Is Required for Virion Morphogenesis. J. Virol. 81 (10), 5212–5224. doi: 10.1128/JVI.01463-06
Manzoor, R., Igarashi, M., Takada, A. (2017). Influenza A Virus M2 Protein: Roles From Ingress to Egress. Int. J. Mol. Sci. 18 (12), 2649. doi: 10.3390/ijms18122649
Martin, B. R., Cravatt, B. F. (2009). Large-Scale Profiling of Protein Palmitoylation in Mammalian Cells. Nat. Methods 6 (2), 135–138. doi: 10.1038/nmeth.1293
Martin, B. R., Wang, C., Adibekian, A., Tully, S. E., Cravatt, B. F. (2011). Global Profiling of Dynamic Protein Palmitoylation. Nat. Methods 9 (1), 84–89. doi: 10.1038/nmeth.1769
McBride, C. E., Machamer, C. E. (2010). Palmitoylation of SARS-CoV S Protein is Necessary for Partitioning Into Detergent-Resistant Membranes and Cell-Cell Fusion But Not Interaction With M Protein. Virology 405 (1), 139–148. doi: 10.1016/j.virol.2010.05.031
McMichael, T. M., Zhang, L., Chemudupati, M., Hach, J. C., Kenney, A. D., Hang, H. C., et al. (2017). The Palmitoyltransferase ZDHHC20 Enhances Interferon-Induced Transmembrane Protein 3 (IFITM3) Palmitoylation and Antiviral Activity. J. Biol. Chem. 292 (52), 21517–21526. doi: 10.1074/jbc.M117.800482
Melkonian, K. A., Ostermeyer, A. G., Chen, J. Z., Roth, M. G., Brown, D. A. (1999). Role of Lipid Modifications in Targeting Proteins to Detergent-Resistant Membrane Rafts. Many Raft Proteins are Acylated, While Few are Prenylated. J. Biol. Chem. 274 (6), 3910–3917. doi: 10.1074/jbc.274.6.3910
Mesquita, F. S., Abrami, L., Sergeeva, O., Turelli, P., Qing, E., Kunz, B., et al. (2021). S-Acylation Controls SARS-CoV-2 Membrane Lipid Organization and Enhances Infectivity. Dev. Cell 56 (20), 2790–2807 e2798. doi: 10.1016/j.devcel.2021.09.016
Mineev, K. S., Lyukmanova, E. N., Krabben, L., Serebryakova, M. V., Shulepko, M. A., Arseniev, A. S., et al. (2013). Structural Investigation of Influenza Virus Hemagglutinin Membrane-Anchoring Peptide. Protein Eng. Des. Sel. 26 (9), 547–552. doi: 10.1093/protein/gzt034
Mitchell, D. A., Mitchell, G., Ling, Y., Budde, C., Deschenes, R. J. (2010). Mutational Analysis of Saccharomyces Cerevisiae Erf2 Reveals a Two-Step Reaction Mechanism for Protein Palmitoylation by DHHC Enzymes. J. Biol. Chem. 285 (49), 38104–38114. doi: 10.1074/jbc.M110.169102
Mitchell, D. A., Vasudevan, A., Linder, M. E., Deschenes, R. J. (2006). Protein Palmitoylation by a Family of DHHC Protein S-Acyltransferases. J. Lipid Res. 47 (6), 1118–1127. doi: 10.1194/jlr.R600007-JLR200
Mizumaru, C., Saito, Y., Ishikawa, T., Yoshida, T., Yamamoto, T., Nakaya, T., et al. (2009). Suppression of APP-Containing Vesicle Trafficking and Production of Beta-Amyloid by AID/DHHC-12 Protein. J. Neurochem. 111 (5), 1213–1224. doi: 10.1111/j.1471-4159.2009.06399.x
Mukai, K., Konno, H., Akiba, T., Uemura, T., Waguri, S., Kobayashi, T., et al. (2016). Activation of STING Requires Palmitoylation at the Golgi. Nat. Commun. 7, 11932. doi: 10.1038/ncomms11932
Mumby, S. M., Muntz, K. H. (1995). Receptor Regulation of G Protein Palmitoylation. Biochem. Soc. Trans. 23 (1), 156–160. doi: 10.1042/bst0230156
Muraki, Y., Okuwa, T., Furukawa, T., Matsuzaki, Y., Sugawara, K., Himeda, T., et al. (2011). Palmitoylation of CM2 Is Dispensable to Influenza C Virus Replication. Virus Res. 157 (1), 99–105. doi: 10.1016/j.virusres.2011.02.013
Muraki, Y., Okuwa, T., Himeda, T., Hongo, S., Ohara, Y. (2013). Effect of Cysteine Mutations in the Extracellular Domain of CM2 on the Influenza C Virus Replication. PloS One 8 (4), e60510. doi: 10.1371/journal.pone.0060510
Nakamura, A., Oki, C., Sawada, S., Yoshii, T., Kuwata, K., Rudd, A. K., et al. (2020). Designer Palmitoylation Motif-Based Self-Localizing Ligand for Sustained Control of Protein Localization in Living Cells and Caenorhabditis Elegans. ACS Chem. Biol. 15 (4), 837–843. doi: 10.1021/acschembio.0c00014
Nguyen, H. T., Zhang, S., Wang, Q., Anang, S., Wang, J., Ding, H., et al. (2020). Spike Glycoprotein and Host Cell Determinants of SARS-CoV-2 Entry and Cytopathic Effects. J. Virol. 95 (5), e02304–20. doi: 10.1128/JVI.02304-20
Ning, W., Jiang, P., Guo, Y., Wang, C., Tan, X., Zhang, W., et al. (2021). GPS-Palm: A Deep Learning-Based Graphic Presentation System for the Prediction of S-Palmitoylation Sites in Proteins. Brief Bioinform. 22 (2), 1836–1847. doi: 10.1093/bib/bbaa038
Niu, J., Sun, Y., Chen, B., Zheng, B., Jarugumilli, G. K., Walker, S. R., et al. (2019). Fatty Acids and Cancer-Amplified ZDHHC19 Promote STAT3 Activation Through S-Palmitoylation. Nature 573 (7772), 139–143. doi: 10.1038/s41586-019-1511-x
Nuskova, H., Serebryakova, M. V., Ferrer-Caelles, A., Sachsenheimer, T., Luchtenborg, C., Miller, A. K., et al. (2021). Stearic Acid Blunts Growth-Factor Signaling via Oleoylation of GNAI Proteins. Nat. Commun. 12 (1), 4590. doi: 10.1038/s41467-021-24844-9
Ohol, Y. M., Wang, Z., Kemble, G., Duke, G. (2015). Direct Inhibition of Cellular Fatty Acid Synthase Impairs Replication of Respiratory Syncytial Virus and Other Respiratory Viruses. PloS One 10 (12), e0144648. doi: 10.1371/journal.pone.0144648
Ota, V. K., Gadelha, A., Assuncao, I. B., Santoro, M. L., Christofolini, D. M., Bellucco, F. T., et al. (2013). ZDHHC8 Gene may Play a Role in Cortical Volumes of Patients With Schizophrenia. Schizophr. Res. 145 (1-3), 33–35. doi: 10.1016/j.schres.2013.01.011
Patrone, M., Coroadinha, A. S., Teixeira, A. P., Alves, P. M. (2016). Palmitoylation Strengthens Cholesterol-Dependent Multimerization and Fusion Activity of Human Cytomegalovirus Glycoprotein B (Gb). J. Biol. Chem. 291 (9), 4711–4722. doi: 10.1074/jbc.M115.682252
Pedro, M. P., Vilcaes, A. A., Tomatis, V. M., Oliveira, R. G., Gomez, G. A., Daniotti, J. L. (2013). 2-Bromopalmitate Reduces Protein Deacylation by Inhibition of Acyl-Protein Thioesterase Enzymatic Activities. PloS One 8 (10), e75232. doi: 10.1371/journal.pone.0075232
Peng, T., Hang, H. C. (2019). Chemical Proteomic Profiling of Protein Fatty-Acylation in Microbial Pathogens. Curr. Top. Microbiol. Immunol. 420, 93–110. doi: 10.1007/82_2018_126
Percher, A., Ramakrishnan, S., Thinon, E., Yuan, X., Yount, J. S., Hang, H. C. (2016). Mass-Tag Labeling Reveals Site-Specific and Endogenous Levels of Protein S-Fatty Acylation. Proc. Natl. Acad. Sci. U.S.A. 113 (16), 4302–4307. doi: 10.1073/pnas.1602244113
Peseckis, S. M., Deichaite, I., Resh, M. D. (1993). Iodinated Fatty Acids as Probes for Myristate Processing and Function. Incorporation Into Pp60v-Src. J. Biol. Chem. 268 (7), 5107–5114. doi: 10.1016/S0021-9258(18)53508-0
Petit, C. M., Chouljenko, V. N., Iyer, A., Colgrove, R., Farzan, M., Knipe, D. M., et al. (2007). Palmitoylation of the Cysteine-Rich Endodomain of the SARS-Coronavirus Spike Glycoprotein is Important for Spike-Mediated Cell Fusion. Virology 360 (2), 264–274. doi: 10.1016/j.virol.2006.10.034
Puthenveetil, R., Lun, C. M., Murphy, R. E., Healy, L. B., Vilmen, G., Christenson, E. T., et al. (2021). S-Acylation of SARS-CoV-2 Spike Protein: Mechanistic Dissection, In Vitro Reconstitution and Role in Viral Infectivity. J. Biol. Chem. 297 (4), 101112. doi: 10.1016/j.jbc.2021.101112
Quehenberger, O., Armando, A. M., Brown, A. H., Milne, S. B., Myers, D. S., Merrill, A. H., et al. (2010). Lipidomics Reveals a Remarkable Diversity of Lipids in Human Plasma. J. Lipid Res. 51 (11), 3299–3305. doi: 10.1194/jlr.M009449
Ramsey, J., Chavez, M., Mukhopadhyay, S. (2019). Domains of the TF Protein Important in Regulating its Own Palmitoylation. Virology 531, 31–39. doi: 10.1016/j.virol.2019.02.016
Ramsey, J., Renzi, E. C., Arnold, R. J., Trinidad, J. C., Mukhopadhyay, S. (2017). Palmitoylation of Sindbis Virus TF Protein Regulates Its Plasma Membrane Localization and Subsequent Incorporation Into Virions. J. Virol. 91 (3), e02000–16. doi: 10.1128/JVI.02000-16
Rana, M. S., Kumar, P., Lee, C. J., Verardi, R., Rajashankar, K. R., Banerjee, A. (2018). Fatty Acyl Recognition and Transfer by an Integral Membrane S-Acyltransferase. Science 359 (6372), eaao6326. doi: 10.1126/science.aao6326
Rana, M. S., Lee, C. J., Banerjee, A. (2019). The Molecular Mechanism of DHHC Protein Acyltransferases. Biochem. Soc. Trans. 47 (1), 157–167. doi: 10.1042/BST20180429
Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y., Yao, X. (2008). CSS-Palm 2.0: An Updated Software for Palmitoylation Sites Prediction. Protein Eng. Des. Sel. 21 (11), 639–644. doi: 10.1093/protein/gzn039
Resh, M. D. (2016). Fatty Acylation of Proteins: The Long and the Short of it. Prog. Lipid Res. 63, 120–131. doi: 10.1016/j.plipres.2016.05.002
Resh, M. D. (2017). Palmitoylation of Proteins in Cancer. Biochem. Soc. Trans. 45 (2), 409–416. doi: 10.1042/BST20160233
Roth, A. F., Feng, Y., Chen, L., Davis, N. G. (2002). The Yeast DHHC Cysteine-Rich Domain Protein Akr1p is a Palmitoyl Transferase. J. Cell Biol. 159 (1), 23–28. doi: 10.1083/jcb.200206120
Rousso, I., Mixon, M. B., Chen, B. K., Kim, P. S. (2000). Palmitoylation of the HIV-1 Envelope Glycoprotein is Critical for Viral Infectivity. Proc. Natl. Acad. Sci. U.S.A. 97 (25), 13523–13525. doi: 10.1073/pnas.240459697
Ryan, C., Ivanova, L., Schlesinger, M. J. (1998). Effects of Site-Directed Mutations of Transmembrane Cysteines in Sindbis Virus E1 and E2 Glycoproteins on Palmitylation and Virus Replication. Virology 249 (1), 62–67. doi: 10.1006/viro.1998.9281
Sada, R., Kimura, H., Fukata, Y., Fukata, M., Yamamoto, H., Kikuchi, A. (2019). Dynamic Palmitoylation Controls the Microdomain Localization of the DKK1 Receptors CKAP4 and LRP6. Sci. Signal 12 (608), eaat9519. doi: 10.1126/scisignal.aat9519
Sanders, S. S., Hayden, M. R. (2015). Aberrant Palmitoylation in Huntington Disease. Biochem. Soc. Trans. 43 (2), 205–210. doi: 10.1042/BST20140242
Schmidt, M. F., Bracha, M., Schlesinger, M. J. (1979). Evidence for Covalent Attachment of Fatty Acids to Sindbis Virus Glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 76 (4), 1687–1691. doi: 10.1073/pnas.76.4.1687
Schmidt, M. F., Schlesinger, M. J. (1979). Fatty Acid Binding to Vesicular Stomatitis Virus Glycoprotein: A New Type of Post-Translational Modification of the Viral Glycoprotein. Cell 17 (4), 813–819. doi: 10.1016/0092-8674(79)90321-0
Schroeder, C., Heider, H., Moncke-Buchner, E., Lin, T. I. (2005). The Influenza Virus Ion Channel and Maturation Cofactor M2 Is a Cholesterol-Binding Protein. Eur. Biophys. J. 34 (1), 52–66. doi: 10.1007/s00249-004-0424-1
Serebryakova, M. V., Kordyukova, L. V., Baratova, L. A., Markushin, S. G. (2006). Mass Spectrometric Sequencing and Acylation Character Analysis of C-Terminal Anchoring Segment From Influenza A Hemagglutinin. Eur. J. Mass Spectrom. (Chichester) 12 (1), 51–62. doi: 10.1255/ejms.792
Shulla, A., Gallagher, T. (2009). Role of Spike Protein Endodomains in Regulating Coronavirus Entry. J. Biol. Chem. 284 (47), 32725–32734. doi: 10.1074/jbc.M109.043547
Sobocinska, J., Roszczenko-Jasinska, P., Ciesielska, A., Kwiatkowska, K. (2017). Protein Palmitoylation and Its Role in Bacterial and Viral Infections. Front. Immunol. 8, 2003. doi: 10.3389/fimmu.2017.02003
Sorek, N., Akerman, A., Yalovsky, S. (2013). Analysis of Protein Prenylation and S-Acylation Using Gas Chromatography-Coupled Mass Spectrometry. Methods Mol. Biol. 1043, 121–134. doi: 10.1007/978-1-62703-532-3_13
Sorek, N., Gutman, O., Bar, E., Abu-Abied, M., Feng, X., Running, M. P., et al. (2011). Differential Effects of Prenylation and s-Acylation on Type I and II ROPS Membrane Interaction and Function. Plant Physiol. 155 (2), 706–720. doi: 10.1104/pp.110.166850
Sorek, N., Yalovsky, S. (2010). Analysis of Protein S-Acylation by Gas Chromatography-Coupled Mass Spectrometry Using Purified Proteins. Nat. Protoc. 5 (5), 834–840. doi: 10.1038/nprot.2010.33
Speers, A. E., Cravatt, B. F. (2004). Profiling Enzyme Activities In Vivo Using Click Chemistry Methods. Chem. Biol. 11 (4), 535–546. doi: 10.1016/j.chembiol.2004.03.012
Spence, J. S., He, R., Hoffmann, H. H., Das, T., Thinon, E., Rice, C. M., et al. (2019). IFITM3 Directly Engages and Shuttles Incoming Virus Particles to Lysosomes. Nat. Chem. Biol. 15 (3), 259–268. doi: 10.1038/s41589-018-0213-2
Stepanek, O., Draber, P., Horejsi, V. (2014). Palmitoylated Transmembrane Adaptor Proteins in Leukocyte Signaling. Cell Signal 26 (5), 895–902. doi: 10.1016/j.cellsig.2014.01.007
Stix, R., Lee, C. J., Faraldo-Gomez, J. D., Banerjee, A. (2020). Structure and Mechanism of DHHC Protein Acyltransferases. J. Mol. Biol. 432 (18), 4983–4998. doi: 10.1016/j.jmb.2020.05.023
Stypulkowski, E., Asangani, I. A., Witze, E. S. (2018). The Depalmitoylase APT1 Directs the Asymmetric Partitioning of Notch and Wnt Signaling During Cell Division. Sci. Signal 11 (511), eaam8705. doi: 10.1126/scisignal.aam8705
Sun, C., Wang, P., Dong, W., Liu, H., Sun, J., Zhao, L. (2020). LncRNA PVT1 Promotes Exosome Secretion Through YKT6, RAB7, and VAMP3 in Pancreatic Cancer. Aging (Albany NY) 12 (11), 10427–10440. doi: 10.18632/aging.103268
Sun, L., Wu, J., Du, F., Chen, X., Chen, Z. J. (2013). Cyclic GMP-AMP Synthase is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 339 (6121), 786–791. doi: 10.1126/science.1232458
Su, W. C., Yu, W. Y., Huang, S. H., Lai, M. M. C. (2018). Ubiquitination of the Cytoplasmic Domain of Influenza A Virus M2 Protein Is Crucial for Production of Infectious Virus Particles. J. Virol. 92 (4), e01972–17. doi: 10.1128/JVI.01972-17
Swarthout, J. T., Lobo, S., Farh, L., Croke, M. R., Greentree, W. K., Deschenes, R. J., et al. (2005). DHHC9 and GCP16 Constitute a Human Protein Fatty Acyltransferase With Specificity for H- and N-Ras. J. Biol. Chem. 280 (35), 31141–31148. doi: 10.1074/jbc.M504113200
Tanner, J. E., Alfieri, C. (2021). The Fatty Acid Lipid Metabolism Nexus in COVID-19. Viruses 13 (1), 90. doi: 10.3390/v13010090
Thaa, B., Levental, I., Herrmann, A., Veit, M. (2011). Intrinsic Membrane Association of the Cytoplasmic Tail of Influenza Virus M2 Protein and Lateral Membrane Sorting Regulated by Cholesterol Binding and Palmitoylation. Biochem. J. 437 (3), 389–397. doi: 10.1042/BJ20110706
Thaa, B., Tielesch, C., Moller, L., Schmitt, A. O., Wolff, T., Bannert, N., et al. (2012). Growth of Influenza A Virus Is Not Impeded by Simultaneous Removal of the Cholesterol-Binding and Acylation Sites in the M2 Protein. J. Gen. Virol. 93 (Pt 2), 282–292. doi: 10.1099/vir.0.038554-0
Thinon, E., Percher, A., Hang, H. C. (2016). Bioorthogonal Chemical Reporters for Monitoring Unsaturated Fatty-Acylated Proteins. Chembiochem 17 (19), 1800–1803. doi: 10.1002/cbic.201600213
Thomas, D. W., van Kuijk, F. J., Stephens, R. J. (1992). Quantitative Determination of Hydroxy Fatty Acids as an Indicator of In Vivo Lipid Peroxidation: Oxidation Products of Arachidonic and Docosapentaenoic Acids in Rat Liver After Exposure to Carbon Tetrachloride. Anal. Biochem. 206 (2), 353–358. doi: 10.1016/0003-2697(92)90377-j
Thorp, E. B., Boscarino, J. A., Logan, H. L., Goletz, J. T., Gallagher, T. M. (2006). Palmitoylations on Murine Coronavirus Spike Proteins are Essential for Virion Assembly and Infectivity. J. Virol. 80 (3), 1280–1289. doi: 10.1128/JVI.80.3.1280-1289.2006
Tortosa, E., Hoogenraad, C. C. (2018). Polarized Trafficking: The Palmitoylation Cycle Distributes Cytoplasmic Proteins to Distinct Neuronal Compartments. Curr. Opin. Cell Biol. 50, 64–71. doi: 10.1016/j.ceb.2018.02.004
Toyoda, T., Sugimoto, H., Yamashita, S. (1999). Sequence, Expression in Escherichia Coli, and Characterization of Lysophospholipase II. Biochim. Biophys. Acta 1437 (2), 182–193. doi: 10.1016/s1388-1981(99)00007-4
Tsai, F. D., Wynne, J. P., Ahearn, I. M., Philips, M. R. (2014). Metabolic Labeling of Ras With Tritiated Palmitate to Monitor Palmitoylation and Depalmitoylation. Methods Mol. Biol. 1120, 33–41. doi: 10.1007/978-1-62703-791-4_3
Tseng, Y. T., Wang, S. M., Huang, K. J., Wang, C. T. (2014). SARS-CoV Envelope Protein Palmitoylation or Nucleocapid Association is Not Required for Promoting Virus-Like Particle Production. J. BioMed. Sci. 21, 34. doi: 10.1186/1423-0127-21-34
Uchida, K., Stadtman, E. R. (1992). Selective Cleavage of Thioether Linkage in Proteins Modified With 4-Hydroxynonenal. Proc. Natl. Acad. Sci. U.S.A. 89 (12), 5611–5615. doi: 10.1073/pnas.89.12.5611
Ujike, M., Nakajima, K., Nobusawa, E. (2004). “Influence of Acylation Sites of Influenza B Virus Hemagglutinin on Fusion Pore Formation and Dilation. J. Virol. 78 (21), 11536–11543. doi: 10.1128/JVI.78.21.11536-11543.2004
Ujike, M., Nakajima, K., Nobusawa, E. (2005). Influence of Additional Acylation Site(s) of Influenza B Virus Hemagglutinin on Syncytium Formation. Microbiol. Immunol. 49 (4), 355–359. doi: 10.1111/j.1348-0421.2005.tb03740.x
Utt, A., Rausalu, K., Jakobson, M., Mannik, A., Alphey, L., Fragkoudis, R., et al. (2019). Design and Use of Chikungunya Virus Replication Templates Utilizing Mammalian and Mosquito RNA Polymerase I-Mediated Transcription. J. Virol. 93 (18), e00794–19. doi: 10.1128/JVI.00794-19
Veit, M. (2012). Palmitoylation of Virus Proteins. Biol. Cell 104 (9), 493–515. doi: 10.1111/boc.201200006
Veit, M., Serebryakova, M. V., Kordyukova, L. V. (2013). Palmitoylation of Influenza Virus Proteins. Biochem. Soc. Trans. 41 (1), 50–55. doi: 10.1042/BST20120210
Veit, M., Siche, S. (2015). S-Acylation of Influenza Virus Proteins: Are Enzymes for Fatty Acid Attachment Promising Drug Targets? Vaccine 33 (49), 7002–7007. doi: 10.1016/j.vaccine.2015.08.095
Vetrivel, K. S., Meckler, X., Chen, Y., Nguyen, P. D., Seidah, N. G., Vassar, R., et al. (2009). Alzheimer Disease A Beta Production in the Absence of S-Palmitoylation-Dependent Targeting of BACE1 to Lipid Rafts. J. Biol. Chem. 284 (6), 3793–3803. doi: 10.1074/jbc.M808920200
Vitale, R., Roine, E., Bamford, D. H., Corcelli, A. (2013). Lipid Fingerprints of Intact Viruses by MALDI-TOF/mass Spectrometry. Biochim. Biophys. Acta 1831 (4), 872–879. doi: 10.1016/j.bbalip.2013.01.011
Wang, X., Qian, P., Cui, H., Yao, L., Yuan, J. (2020). A Protein Palmitoylation Cascade Regulates Microtubule Cytoskeleton Integrity in Plasmodium. EMBO J. 39 (13), e104168. doi: 10.15252/embj.2019104168
Webster, J., Oxley, D. (2012). Protein Identification by MALDI-TOF Mass Spectrometry. Methods Mol. Biol. 800, 227–240. doi: 10.1007/978-1-61779-349-3_15
Whitt, M. A., Rose, J. K. (1991). Fatty Acid Acylation is Not Required for Membrane Fusion Activity or Glycoprotein Assembly Into VSV Virions. Virology 185 (2), 875–878. doi: 10.1016/0042-6822(91)90563-q
Wilkinson, T. A., Yin, J., Pidgeon, C., Post, C. B. (2000). Alkylation of Cysteine-Containing Peptides to Mimic Palmitoylation. J. Pept. Res. 55 (2), 140–147. doi: 10.1034/j.1399-3011.2000.00164.x
Woodley, K. T., Collins, M. O. (2019). Quantitative Analysis of Protein S-Acylation Site Dynamics Using Site-Specific Acyl-Biotin Exchange (ssABE). Methods Mol. Biol. 1977, 71–82. doi: 10.1007/978-1-4939-9232-4_6
Wright, M. H., Clough, B., Rackham, M. D., Rangachari, K., Brannigan, J. A., Grainger, M., et al. (2014). Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach. Nat. Chem. 6 (2), 112–121. doi: 10.1038/nchem.1830
Wu, M. J., Shanmugam, S., Welsch, C., Yi, M. (2019). Palmitoylation of Hepatitis C Virus NS2 Regulates Its Subcellular Localization and NS2-NS3 Autocleavage. J. Virol. 94 (1), e00906–19. doi: 10.1128/JVI.00906-19
Wu, Z., Zhang, Z., Wang, X., Zhang, J., Ren, C., Li, Y., et al. (2021). Palmitoylation of SARS-CoV-2 S Protein is Essential for Viral Infectivity. Signal Transduct. Target. Ther. 6 (1), 231. doi: 10.1038/s41392-021-00651-y
Xu, Z., Li, X., Xue, J., Shen, L., Zheng, W., Yin, S., et al. (2020). S-Palmitoylation of Swine Interferon-Inducible Transmembrane Protein is Essential for its Anti-JEV Activity. Virology 548, 82–92. doi: 10.1016/j.virol.2020.06.004
Yang, P., Ai, L. S., Huang, S. C., Li, H. F., Chan, W. E., Chang, C. W., et al. (2010). The Cytoplasmic Domain of Human Immunodeficiency Virus Type 1 Transmembrane Protein Gp41 Harbors Lipid Raft Association Determinants. J. Virol. 84 (1), 59–75. doi: 10.1128/JVI.00899-09
Yang, W., Di Vizio, D., Kirchner, M., Steen, H., Freeman, M. R. (2010). Proteome Scale Characterization of Human S-Acylated Proteins in Lipid Raft-Enriched and non-Raft Membranes. Mol. Cell Proteomics 9 (1), 54–70. doi: 10.1074/mcp.M800448-MCP200
Yang, C., Spies, C. P., Compans, R. W. (1995). The Human and Simian Immunodeficiency Virus Envelope Glycoprotein Transmembrane Subunits are Palmitoylated. Proc. Natl. Acad. Sci. U.S.A. 92 (21), 9871–9875. doi: 10.1073/pnas.92.21.9871
Yeste-Velasco, M., Linder, M. E., Lu, Y. J. (2015). Protein S-Palmitoylation and Cancer. Biochim. Biophys. Acta 1856 (1), 107–120. doi: 10.1016/j.bbcan.2015.06.004
Yount, J. S., Karssemeijer, R. A., Hang, H. C. (2012). S-Palmitoylation and Ubiquitination Differentially Regulate Interferon-Induced Transmembrane Protein 3 (IFITM3)-Mediated Resistance to Influenza Virus. J. Biol. Chem. 287 (23), 19631–19641. doi: 10.1074/jbc.M112.362095
Yount, J. S., Moltedo, B., Yang, Y. Y., Charron, G., Moran, T. M., Lopez, C. B., et al. (2010). Palmitoylome Profiling Reveals S-Palmitoylation-Dependent Antiviral Activity of IFITM3. Nat. Chem. Biol. 6 (8), 610–614. doi: 10.1038/nchembio.405
Zaballa, M. E., van der Goot, F. G. (2018). The Molecular Era of Protein S-Acylation: Spotlight on Structure, Mechanisms, and Dynamics. Crit. Rev. Biochem. Mol. Biol. 53 (4), 420–451. doi: 10.1080/10409238.2018.1488804
Zani, A., Zhang, L., McMichael, T. M., Kenney, A. D., Chemudupati, M., Kwiek, J. J., et al. (2019). Interferon-Induced Transmembrane Proteins Inhibit Cell Fusion Mediated by Trophoblast Syncytins. J. Biol. Chem. 294 (52), 19844–19851. doi: 10.1074/jbc.AC119.010611
Zareba-Koziol, M., Figiel, I., Bartkowiak-Kaczmarek, A., Wlodarczyk, J. (2018). Insights Into Protein S-Palmitoylation in Synaptic Plasticity and Neurological Disorders: Potential and Limitations of Methods for Detection and Analysis. Front. Mol. Neurosci. 11. doi: 10.3389/fnmol.2018.00175
Zeki, O. C., Eylem, C. C., Recber, T., Kir, S., Nemutlu, E. (2020). Integration of GC-MS and LC-MS for Untargeted Metabolomics Profiling. J. Pharm. BioMed. Anal. 190, 113509. doi: 10.1016/j.jpba.2020.113509
Zeng, X. T., Yu, X. T., Cheng, W. (2021). The Interactions of ZDHHC5/GOLGA7 With SARS-CoV-2 Spike (S) Protein and Their Effects on S Protein’s Subcellular Localization, Palmitoylation and Pseudovirus Entry. Virol. J. 18 (1), 257. doi: 10.1186/s12985-021-01722-w
Zhang, K., Law, Y. S., Law, M. C. Y., Tan, Y. B., Wirawan, M., Luo, D. (2021). Structural Insights Into Viral RNA Capping and Plasma Membrane Targeting by Chikungunya Virus non-Structural Protein 1. Cell Host Microbe. 29 (5), 757–64.e3. doi: 10.1016/j.chom.2021.02.018
Zhang, N., Zhao, H., Zhang, L. (2019). Fatty Acid Synthase Promotes the Palmitoylation of Chikungunya Virus Nsp1. J. Virol. 93 (3), e01747–18. doi: 10.1128/JVI.01747-18
Zheng, H., Pearsall, E. A., Hurst, D. P., Zhang, Y., Chu, J., Zhou, Y., et al. (2012). Palmitoylation and Membrane Cholesterol Stabilize Mu-Opioid Receptor Homodimerization and G Protein Coupling. BMC Cell Biol. 13, 6. doi: 10.1186/1471-2121-13-6
Zhou, B., Wang, Y., Yan, Y., Mariscal, J., Di Vizio, D., Freeman, M. R., et al. (2019). Low-Background Acyl-Biotinyl Exchange Largely Eliminates the Coisolation of Non-S-Acylated Proteins and Enables Deep S-Acylproteomic Analysis. Anal. Chem. 91 (15), 9858–9866. doi: 10.1021/acs.analchem.9b01520
Zhou, F., Xue, Y., Yao, X., Xu, Y. (2006). CSS-Palm: Palmitoylation Site Prediction With a Clustering and Scoring Strategy (CSS). Bioinformatics 22 (7), 894–896. doi: 10.1093/bioinformatics/btl013
Zingler, P., Sarchen, V., Glatter, T., Caning, L., Saggau, C., Kathayat, R. S., et al. (2019). Palmitoylation is Required for TNF-R1 Signaling. Cell Commun. Signal 17 (1), 90. doi: 10.1186/s12964-019-0405-8
Keywords: post-translational modification, S-palmitoylation, viral infection, virus-host interaction, detection methods
Citation: Li X, Shen L, Xu Z, Liu W, Li A and Xu J (2022) Protein Palmitoylation Modification During Viral Infection and Detection Methods of Palmitoylated Proteins. Front. Cell. Infect. Microbiol. 12:821596. doi: 10.3389/fcimb.2022.821596
Received: 24 November 2021; Accepted: 12 January 2022;
Published: 27 January 2022.
Edited by:
Qing Deng, Purdue University, United StatesReviewed by:
Larisa Kordyukova, Lomonosov Moscow State University, RussiaMark Oliver Collins, The University of Sheffield, United Kingdom
Copyright © 2022 Li, Shen, Xu, Liu, Li and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Xu, eHVqdW5AaGVuYXUuZWR1LmNu; eHVqdW4wODI4QDEyNi5jb20=
 Xiaoling Li
Xiaoling Li Lingyi Shen1
Lingyi Shen1 Wei Liu
Wei Liu Jun Xu
Jun Xu