- 1State Key Laboratory of Oral Diseases and National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu, China
- 2Department of Cariology and Endodontics, West China Hospital of Stomatology, Sichuan University, Chengdu, China
- 3Stomatology Hospital, School of Stomatology, Zhejiang University School of Medicine, Hangzhou, China
- 4Clinical Research Center for Oral Diseases of Zhejiang Province, Key Laboratory of Oral Biomedical Research of Zhejiang Province, Cancer Center of Zhejiang University, Hangzhou, China
- 5Basic and Translation Sciences, Penn Dental Medicine, University of Pennsylvania, Philadelphia, PA, United States
Taste receptors, originally identified in taste buds, function as the periphery receptors for taste stimuli and play an important role in food choice. Cohort studies have revealed that single nucleotide polymorphisms of taste receptors such as T1R1, T1R2, T2R38 are associated with susceptibility to oral diseases like dental caries. Recent studies have demonstrated the wide expression of taste receptors in various tissues, including intestinal epithelia, respiratory tract, and gingiva, with an emerging role of participating in the interaction between mucosa surface and microorganisms via monitoring a wide range of metabolites. On the one hand, individuals with different oral microbiomes exhibited varied taste sensitivity, suggesting a potential impact of the oral microbiota composition on taste receptor function. On the other hand, animal studies and in vitro studies have uncovered that a variety of oral cells expressing taste receptors such as gingival solitary chemosensory cells, gingival epithelial cells (GECs), and gingival fibroblasts can detect bacterial signals through bitter taste receptors to trigger host innate immune responses, thus regulating oral microbial homeostasis. This review focuses on how taste receptors, particularly bitter and sweet taste receptors, mediate the oral microbiota-host interaction as well as impact the occurrence and development of oral diseases. Further studies delineating the role of taste receptors in mediating oral microbiota-host interaction will advance our knowledge in oral ecological homeostasis establishment, providing a novel paradigm and treatment target for the better management of dental infectious diseases.
Introduction
Taste is triggered by signals from the oral sensory structures known as taste buds, which are stimulated by tastants and then conveyed to the central gustatory nervous system (Small, 2012; von Molitor et al., 2021). Various tastes including sweet, bitter, umami, sour, and salt are mainly perceived by specific taste receptors located at taste buds, which help the host to discriminate between nutrients and poisonous and harmful substances (Margolskee, 1993; Roper, 2013). General health may be impaired by dysfunctional or diseased states of taste receptors. Past studies have well illustrated that gustatory sensitivity is influenced by single nucleotide polymorphisms (SNPs) of taste receptors and leads to different food preferences, which contribute to varied oral microbiota and disease susceptibility in the host (Chamoun et al., 2018b).
Recent studies have found that taste receptors are not only distributed in cells within taste buds, but are also expressed on a variety of cell types both orally and extra-orally, such as tuft cells, airway smooth muscle cells, macrophages, and so on (Deshpande et al., 2010; Sbarbati et al., 2010; Grassin-Delyle et al., 2019). Meanwhile, studies on taste receptors both on and off the taste buds support the view that taste receptors play an essential role as chemoreceptors in diverse non-gustatory physiological and pathological processes (Lee et al., 2014; Schneider et al., 2019; Gopallawa et al., 2021). Taking the oral taste receptors as an example, on the one hand, taste receptors detect various metabolites and other toxins derived from the oral microbiota, contributing to the establishment of the host immune response and the maintenance of homeostasis (Gil et al., 2015; Zheng et al., 2019; Medapati et al., 2021b). On the other hand, the perceptive capacity of taste receptors is in turn shaped by the oral microbiota (Solemdal et al., 2012; Zhu et al., 2021). Studies in other tissues have yielded similar findings (Tizzano et al., 2010; Deckmann and Kummer, 2016). The investigation of how taste receptors are involved in the interaction between host and oral microbiota will facilitate a thorough understanding of the chemosensory function of taste receptors, which is of great significance for the elucidation of the association between taste receptors and oral diseases. Accordingly, the role of taste receptors in regulating oral health and disease states will be discussed in this review, with a focus on the impact of genotypic and phenotypic changes in receptors on the composition of the oral microbiota, as well as the key role of taste receptors mediating innate immune responses in oral microbiota-host interaction.
Molecular Mechanisms of Taste Signal Transduction
Taste receptors are chemosensory receptors that exist in both taste buds and extra-gustatory tissues (Chandrashekar et al., 2006; Carey and Lee, 2019). Five widely accepted and fundamental tastes (sweet, umami, salt, sour, bitter) are initiated from these taste receptors (Lindemann, 2001). In recent studies, kokumi and fat are likely to be potential candidates for new basic tastes (Khan et al., 2019; Rhyu et al., 2020; Hichami et al., 2021). The receptors, including multiple members of the G protein-coupled receptor (GPCR) superfamily and some ionic channels, put different specificity to stimuli (Nuemket et al., 2017; Teng et al., 2019; Ahmad and Dalziel, 2020). In practical terms, sweet means consuming carbohydrates as vital energy source, salt indicates ingestion of sodium, and umami promotes detecting amino acids, which are respectively indispensable for energy metabolism, ionic homeostasis and building proteins (Yamaguchi and Ninomiya, 2000; Dias et al., 2012; Laffitte et al., 2014). Sour, which perceives rotten food, and bitter, which implicate a variety of toxic substances such as alkaloids and cyanogenic glycosides, are revolting tastes (Glendinning, 1994; Huang et al., 2008). Therefore, sour and bitter tastes promote the establishment of early warning against the intake of underlying toxins.
Bitter, sweet and umami taste signals are supposed to converge on a common intracellular signaling transduction in the Type II cells. Bitter, sweet, umami tastes are activated by GPCRs, which are seven-transmembrane proteins of two main classes. GPCRs that detect sweet and umami stimuli are named as taste receptor family 1 member (T1R) (Zhao et al., 2003), and those that transduce bitter compounds are named as taste receptor family 2 member (T2R) (Chandrashekar et al., 2000). When activated by corresponding stimuli, G protein coupled to these taste receptors is resolved into Gβγ subunit and Gα subunit including Gα-gustducin, Gα14 and Gαi (McLaughlin et al., 1992; Huang et al., 1999; Tizzano et al., 2008). With further hydrolysis of Gβγ, Gβ3 and Gγ13 can be formed, which stimulates phospholipase Cβ2 (PLCB2) to increase intracellular Ca2+ levels (Zhang et al., 2007). Gα subunit is considered to influence cyclic adenosine monophosphate (cAMP) signaling (Clapp et al., 2008). When bitter and umami receptors are activated by tastants, Gα subunit diminishes intracellular cAMP levels, which inhibits activity of protein kinase A. As a result, it weakens the inhibition of protein kinase A on the PLCB2-IP3 pathway, and further promotes the release of Ca2+ in the endoplasmic reticulum. When sweet receptors are activated by tastants, Gα subunit increases intracellular cAMP levels, which enhances the activity of protein kinase A and inhibits K+ channels, thereby promoting extracellular Ca2+ influx. At last, elevated intracellular Ca2+ levels promote the opening of transient receptor potential cation channel subfamily M member 5 (TRPM5), which is an ion channel that results in membrane depolarization and causes action potential followed by the release of ATP (Figure 1) (Iwata et al., 2014).
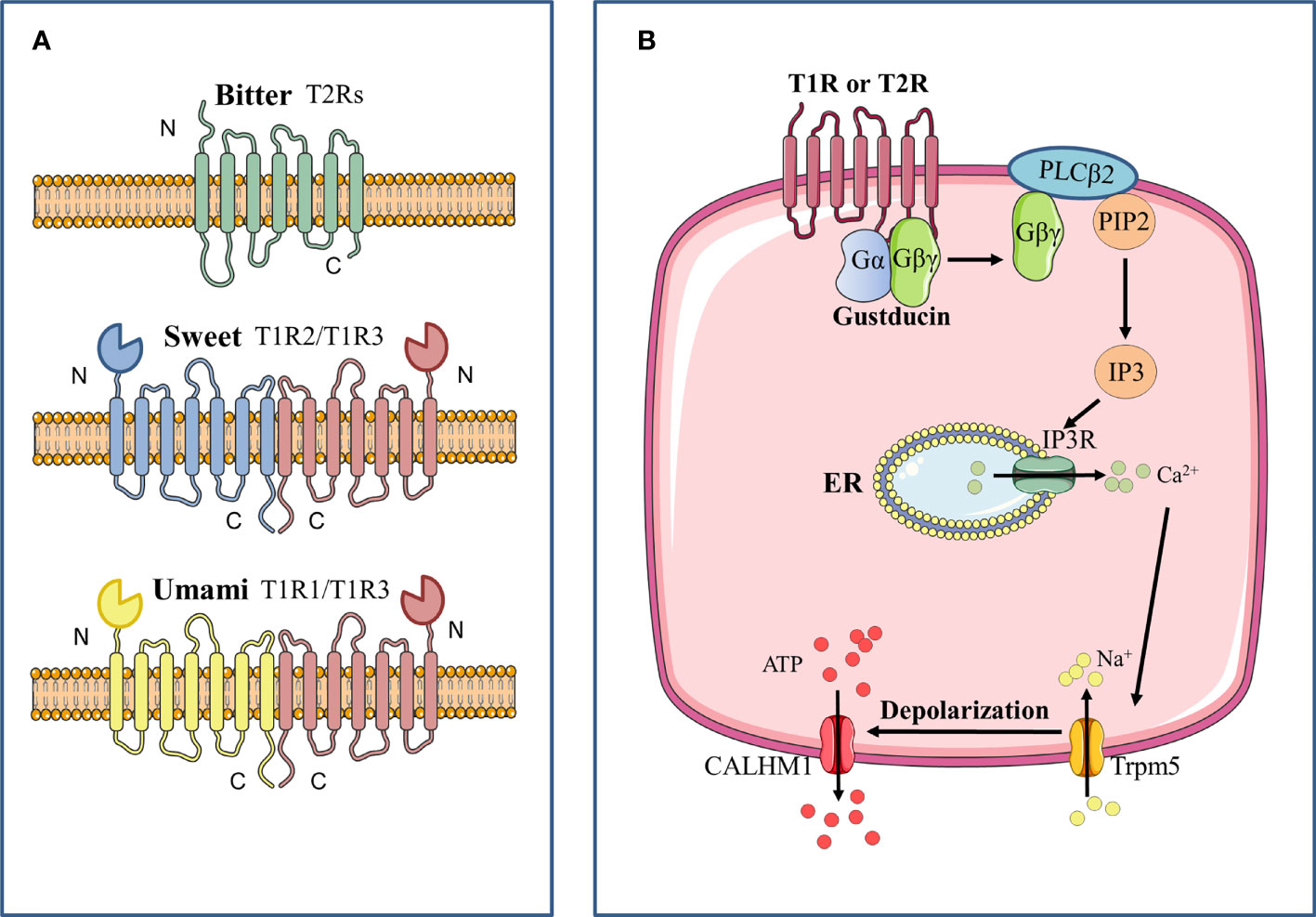
Figure 1 Signal transduction pathway of bitter, sweet, and umami GPCRs. (A) Bitter, sweet and umami receptors are all G-protein coupled-receptors. Bitter receptors are composed of T2Rs, while sweet (T1R2/T1R3) and umami (T1R2/T1R3) receptors are composed of T1Rs, which are characterized by a large N terminal domain that forms a Venus flytrap structure. (B) After stimulation of the taste receptor, the downstream Gβγ complex is mobilized, which then activates phospholipase C isoform β2 (PLCβ2) to induce the production of inositol 1,4,5-trisphosphate (IP3). IP3 then activates the IP3 receptor (IP3R), an intracellular ion channel that allows the release of Ca2+ from the endoplasmic reticulum (ER), resulting in an increase in intracellular Ca2+. The complex of transient receptor potential cation channel subfamily M member 5 (TRPM5) is then activated and triggers the inward Na+ diffusion. The depolarization causes activation of the complex of calcium homeostasis modulator 1 (CALHM1) channels, thus resulting in the release of ATP as the neurotransmitter.
Sour taste receptors are located in a subset of taste receptor cells, the Type III cells, on the tongue and palate epithelium (Chandrashekar et al., 2006). Recent studies have demonstrated that sour taste is initiated by a proton-selective channel Otopetrin-1(OTOP1), which is a 24 transmembrane protein (Tu et al., 2018; Zhang et al., 2019). Up to date, OTOP1 and other otopetrin family members are supposed to be the only voltage-insensitive and proton-selective ionic channels (Teng et al., 2019), which may explain how Type III cells are activated by small changes in proton levels and tolerate much more massive changes in other ion levels when in food intake. The increasing intracellular proton levels mediated by OTOP1, on one hand, proximately change the membrane potential (Zhang et al., 2019), on the other hand, subsequently block KIR2.1, a member of inward-rectifier K+ channels, to generate membrane depolarization (Ye et al., 2016). However, how OTOP1 gates and passes through protons and whether there are other elements in sour taste transduction remain to be solved in the future.
Salty taste in most mammals is divided into two pathways: appetitive salt taste and high-salt taste. Animals and humans adapt easily to appetitive salty taste in a low concentration (<100 mM) (Heck et al., 1984). This appetite is likely to meet the host requirement of Na+ to maintain an ionic homeostasis. By comparison, high-salt taste (>300 mM) is significantly aversive (Oka et al., 2013), which indicates a reflex that protects individuals against hypernatremia and dehydration. Appetitive salt taste is activated by epithelial sodium channels (ENaC), a heterotrimer consisted of α, β and γ subunits (Canessa et al., 1994), in some taste cells within fungiform papillae (Nomura et al., 2020). With the access of Na+ through ENaC, it causes a membrane depolarization driving action potentials. Notably, there is no change in intracellular Ca2+ levels with the generation of action potentials in the taste cells. ATP is released by calcium homeostasis modulator 1 (CALHM1) and CALHM3 subunits which are switched on under depolarization. Some studies have indicated that activation of s high-salt taste is associated with some types of bitter and sour taste cells in the foliate papillae and circumvallate papillae (Kretz et al., 1999; Lewandowski et al., 2016). However, there are no specific receptors and well-defined salty transduction signaling that have been demonstrated to function with high-salt taste.
Genetic Variation of Taste Receptors Correlated With Oral Diseases
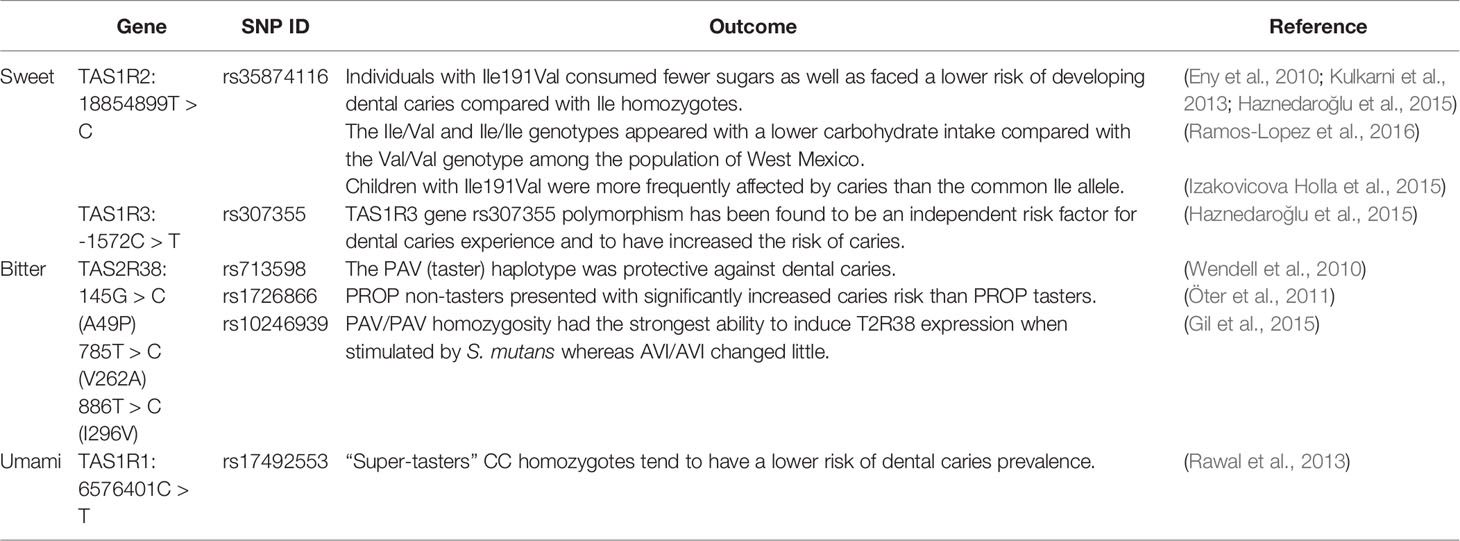
Varied dietary patterns among individuals are influenced by differences in taste perception abilities, thus shaping different oral health status. Genetic variations in certain genes associated with sweet, bitter, umami, salt, and sour tastes have been shown to correlate with taste function in varying degrees, and are thought to be potentially relevant affectors of oral disease susceptibility (Table 1) (Chamoun et al., 2018b).
Sweet Taste Receptors
The perception of sweet taste is mediated by a heterodimer receptor composed of T1R2 and T1R3 together (Li et al., 2002). Various kinds of sweet taste compounds are detected by the T1R2/T1R3 sweet taste receptor, such as natural sugars, nonnutritive sweeteners, sweet-tasting proteins, and so on (Chandrashekar et al., 2006). The activation of the sweet taste receptor initiates intracellular signal transduction, mediating the regulation of the production and secretion of physiologically significant hormones and proteins like insulin and glucagon-like peptide-1 (GLP-1) (Kojima and Nakagawa, 2011; Kyriazis et al., 2012). Moreover, the sweet taste is a source of hedonic liking and greatly influences people’s dietary choices (Jayasinghe et al., 2017). Disruption or loss of sweet taste receptor function would cause a wide range of health problems including metabolic disorders and oral diseases (Murovets et al., 2015; Smith et al., 2016).
Specifically, differences in the capacity of sweet taste receptors are associated with their genetic variation (Garcia-Bailo et al., 2009). SNPs of T1R2 and T1R3 determine sweet taste perception thresholds and the degree of sweet food preference and consumption (Jayasinghe et al., 2017). Eny et al. pointed out Ile191Val (rs35874116) variations in T1R2 were associated with different sugar intake, as Val allele carriers consumed fewer sugars compared with the Ile homozygotes (Eny et al., 2010). A recent study conducted in a Mexican population reported that a higher carbohydrate intake, as well as the risk of hypertriglyceridemia (HTG), was found in the Val/Val genotype individuals versus the Ile/Val and Ile/Ile genotypes (Ramos-Lopez et al., 2016). In addition, the studies of T1R3 revealed the association between SNPs and sweet taste sensitivity as well (Fushan et al., 2009; Murovets et al., 2020).
As aforementioned, SNPs of sweet taste receptors lead to differences in dietary sugar intake (Garcia-Bailo et al., 2009). The ingested carbohydrates are metabolized to produce acid in the oral biofilm made up of microorganisms that adhere to the tooth surface (Bradshaw and Lynch, 2013). When the pH of the tooth surface falls below 5.5, minerals are lost from the tooth surface faster than remineralization, which ultimately leads to dental caries. Put simply, sweet taste receptor gene polymorphisms shape the risk of caries by altering sweet food preferences. Studies conducted by Kulkarni et al. and Haznedaroglu et al. concluded that individuals who hold an Ile191Val polymorphism consumed fewer sugars with a lower risk of developing dental caries, while Ile homozygotes whose carbohydrate intake are higher showed more frequent high-risk caries experience (>8 caries) (Kulkarni et al., 2013; Haznedaroğlu et al., 2015). These conclusions are consistent with most of the other studies (Eny et al., 2010; Ramos-Lopez et al., 2016). However, one study reported an opposite finding that children who carried the Val allele were more frequently affected by caries than the common Ile allele (Izakovicova Holla et al., 2015). One possible explanation is that differences in race, environment, and food culture among populations are also involved in the effect of sweet taste receptor SNPs on dietary choices as well as the risk of caries. Another possibility is that the different genotypes of taste receptors not merely determine the sensitivity of taste perception, but are also involved in the oral microbiota-host interaction through other mechanisms.
Additionally, the increased intake of sugar caused by T1R2/T1R3 SNPs also modifies the oral microecological environment, leading to the alteration of the composition of oral microflora. A recent study revealed that children with low sensitivity to sweet taste presented with a higher incidence of dental caries mainly due to their more frequent consumption of sweet food, who also appeared to contain an increased presence of cariogenic Streptococcus mutans (S. mutans) (Jurczak et al., 2020). Altered microbial diversity occurs after a sugar-rich diet, which greatly increases the presence of cariogenic species, mediating the further development of caries (Tanner et al., 2018).
Bitter Taste Receptors
Human detects numerous different bitter tastes through 25 types of T2Rs, while T2R38 plays the most vital role that genetically controls the perception capacity of bitter taste (Kim et al., 2003; Dong et al., 2009). Bitter taste receptors often work in conjunction with sweet taste receptors to influence our diet. Simply as people with high sweetness sensitivity reduce their intake of sweet food, people with high bitter sensitivity often tend to avoid bitter food. However, reduced intake of bitter foods such as antioxidant-rich vegetables and fruits may lead to a higher risk of cardiovascular disease or cancer (Basson et al., 2005; Roura et al., 2016).
The bitter taste sensation of agonists PTC and PROP is mediated by their combination with T2R38 (Kim et al., 2003; Bufe et al., 2005). Three SNPs of T2R38 caused by different combinations of amino acid substitutions respectively at positions 49 (Pro49Ala, rs713598), 262 (Ala262Val, rs1726866), and 296 (Val296Ile, rs10246939) mainly regulated the diverse senses of bitter taste (Kim et al., 2003). Homozygous for the PAV haplotype are characterized by a stronger perception of bitterness than the average person and are called “super-tasters”, while those homozygous for the AVI haplotype manifest as “non-tasters” whose sense of bitter taste are less sensitive. The performance of heterozygotes is intermediate (Bartoshuk et al., 1994).
Various studies have confirmed that some undesirable diet may be attributed to the “supertaster” phenotype and have further linked the effects of bitter taste receptor SNPs leading to food preferences to health-disease transformation. Wendell et al. reported that the PAV (taster) haplotype is proven to be protective against dental caries (Wendell et al., 2010). The result of a subsequent study by Öter et al. confirmed this surprising view, as PROP non-tasters presented with significantly increased caries risk than PROP tasters (Öter et al., 2011). Interestingly, the capacity of T2R38 expression stimulated by S. mutans exhibits a polarity. PAV/PAV homozygosity had the strongest ability to induce T2R38 expression whereas AVI/AVI changed little (Gil et al., 2015). However, the theory that taste receptor mediates food selection fails to explain this phenomenon, as “super-tasters” have a stronger perception of bitterness and tend to consume less healthful bitter foods, yet they have a lower risk of caries and a lower abundance of pathogenic bacteria. This suggests the presence of pathogenic factors independent of taste receptor-mediated food selection. Further studies are needed to elucidate the pathogenesis involved.
In addition, several bitter taste receptors other than T2R38 have recently been shown to be as well involved in interactions with the oral flora and induction of disease. A recent study uncovered the novel role of bitter taste receptor T2R14 in detecting S. mutans and mediating innate immune defense in GECs (Medapati et al., 2021b). Moreover, CA6 gene polymorphisms were associated with S. mutans colonization, oral microbiota composition and the risk of dental caries, which were linked to bitter taste and smell perception (Esberg et al., 2019).
Other Taste Receptors
Umami taste is initiated and enhanced through the heterodimer T1R1/T1R3 (Zhao et al., 2003). The primary umami taste substance is free l-glutamate, a compound that we are familiar with its another form called monosodium glutamate (MSG) (Ikeda, 2002). Two SNPs caused the substitutions of amino acid in the T1R1 respectively at positions 110 (Ala110Val, rs41278020) and 372 (Ala372Thr, rs34160967) are associated with sensitivity in umami taste (Raliou et al., 2009). An intronic SNP (rs17492553) in T1R1 is related to taste intensity in sweet, bitter, salty and sour (Rawal et al., 2013). CC homozygotes appeared to possess stronger taste intensity as “super-tasters” than that in TT homozygotes from rs17492553 groups. Similar to the trend in rs17492553 that in rs34160967, GG groups are stronger in overall taste intensity than that in AA/AG groups (Rawal et al., 2013). A recent study conducted in a Brazilian pre-adolescent population has shown that rs17492553 in T1R1 was relevant to dental caries prevalence. “Super-tasters” CC homozygotes tend to have a lower risk of dental caries prevalence. The impact of SNPs in other taste receptors such as ENaC and OTOP1 on taste perception and oral disease susceptibility has remained obscure.
Regulation of Taste Receptors on Oral Microbiota-Host Interaction
Since its discovery, the associations between taste receptors and oral diseases have been extensively studied. The prevailing notion is that the SNPs of taste receptors enable the host to perceive various tastes differently, which influenced dietary choices and host oral microbiome, regulating the host’s metabolism along with internal environmental homeostasis and ultimately mediating the reciprocal transition between health and disease states. Meanwhile, as we noted previously, food selection conducted by taste receptors SNPs fails to explain the full range of disease events. Several recent studies have focused on revealing how the taste receptor acts as a chemoreceptor to engage in signal transduction and immune responses in the oral cavity, which provides fresh insights into the role that taste receptors play in oral diseases. The latest relevant advancements are presented and summarized below, providing fresh perspectives on how taste receptors interact with oral microbiota to manage oral health status.
Taste-Shaped Diet Impacts Host Metabolism and Microbial Homeostasis
As previously mentioned, the differential perceptive capacity of taste receptors due to SNPs influences individual food preferences. Recent studies have confirmed that taste preferences for sweet, bitter, sour and salt are affected genetically in part and have an impact on food and beverage choices (Chamoun et al., 2018a; Eriksson et al., 2019; Chamoun et al., 2021). Ample research evidence suggests that diet is one of the most important factors influencing human health and that different dietary patterns greatly regulate host metabolism and microbial homeostasis. For example, a high-fat diet in healthy adults leads to altered gut microbiota, including higher levels of Alistipes and Bacteroides species and a decrease in Faecalibacterium species, as well as increased faecal metabolites p-resol and indole, which are implicated in a higher risk of cardiovascular and metabolic disturbances (Wan et al., 2019). Some researchers have also suggested that microbiota can in turn alter dietary patterns by manipulating host taste perception (Cattaneo et al., 2019b). Overall, taste perception could be involved in host-microbiota interaction by influencing dietary patterns.
Further, impairment or loss of taste function poses a systemic and multilayered risk to people, rather than solely oral diseases. As a consequence of altered taste perception, many foods may be averted owing to the difficulty in appreciating the diet, thus shifting people towards unhealthy eating habits and potentially leading to serious consequences (Chamoun et al., 2018b). For instance, an increase in salt taste recognition threshold may lead people to consume more salt to improve food palatability, thereby elevating their risk of cardiovascular diseases (Pilic and Mavrommatis, 2018; Tapanee et al., 2021). Notably, a national health survey in the US reported that participants who met the recommendations of the Healthy Eating Index (HEI-2015) scores were at a lower risk of developing untreated coronal caries than those who did not conform to the recommendations (Kaye et al., 2020). In addition, shifted taste perception has also been proven to be correlated with depression and anxiety (Esposito et al., 2021).
In addition, taste receptors are associated with the regulation of body metabolism and internal environmental homeostasis. Several studies have revealed the secretory function of taste receptors in the gastrointestinal tract. For example, TAS2Rs regulate intestinal anion secretion to protect the host from ingesting dietary toxins (Glendinning, 1994). Smith et al. proved that intestinal sweet taste receptors modulate gut hormone responses and glucose absorption, as glucose-dependent stimulation of intestinal T1R2/T1R3 chemo-sensors on L-cells enhances glucose absorption through GLP-2-mediated activation of GLUT2 transporter in enterocytes (Smith et al., 2018). Serrano et al. subsequently found that T1R2 gene variant Ile191Val causes a partial loss of function and results in reduced glucose excursions during an OGTT, which is independent of the variations in beta-cell function or insulin sensitivity (Serrano et al., 2021). Additionally, bitter taste receptors have also been revealed of the function to evoke airway smooth muscle relaxation via localized calcium flux, predicting the potential role in counteracting asthmatic bronchoconstriction (Deshpande et al., 2010). Recent studies have also found that older carriers of variant rs236514 (A) of the KCNJ2 gene, which is located in Type III sour-sensing taste cells and affects sour perception thresholds, have a higher preference for acid and lower energy intake, along with an increased risk of mild-to-severe cognitive impairment (Chamoun et al., 2018b; Ferraris et al., 2021a; Ferraris et al., 2021b).
Microbiota Modulate the Perceptive Capacity of Taste Receptors
Significant relationships between oral microbiota composition and taste sensitivity were found recently. A study conducted by Solemdal et al. in acutely hospitalized elderly showed reduced taste ability in patients with poor oral hygiene such as caries activity, among whom patients with high Lactobacillus growth had the most significant impairment in sour taste (Solemdal et al., 2012). This result may be in relation to the adaptation of sour taste perception due to acid produced by bacteria and the increased sour taste threshold. Cattaneo et al. revealed a significant difference among subjects with varied responsiveness to PROP in terms of the relative abundance of some taxa (Cattaneo et al., 2019a). Subjects characterized by greater PROP responsiveness showed overrepresentation mainly in five bacterial genera, including Actinomyces, Oribacterium, Solobacterium, Catonella and Campylobacter. Their further study also found that specific bacterial taxa mainly composed of Clostridiales and Bacteroidales may influence host sensitivity to salt and acid, as these taxa were negatively correlated with perceived taste thresholds for salt and acid (Cattaneo et al., 2019b). Similarly, Feng et al. reported that elevated proportions of Actinomyces and Firmicutes in the saliva were associated with reduced taste sensitivity, whereas increased taste sensitivity resulted from higher proportions of Bacteroides in the tongue film (Feng et al., 2018). Besides, a study by Besnard et al. demonstrated that the low-fat tasters showed greater oral bacterial diversity and a high Bacteroides/Lactobacillus ratio which significantly promotes host inflammation compared to the high-fatty tasters (Besnard et al., 2020). Impairment of oral fat perception may form a feedback loop with such a pro-inflammatory bacterial community composition, as obesity-induced inflammation has been shown to damage taste buds in mice and reduce fatty taste sensitivity in human (Jilani et al., 2017; Kaufman et al., 2018). Our latest study as well illustrated that bacterial lipopolysaccharide-induced alternative splicing of the mouse sweet taste receptor T1R2, thus contributing to a significant upregulation of its non-functional isoform expression and inhibition of sweet taste perception (Zhu et al., 2021). It can be hypothesized that these impaired or non-functional isoforms of taste receptors would similarly lack the ability to detect and mediate the elimination of pathogens.
There are several possible interpretations of the perceptive changes in taste receptors owing to the growth of specific oral bacteria. One possibility lies in the ability of oral bacteria to modulate the food preferences of the host. Microbes in the gastrointestinal tract have shown a potential role in manipulating host diet by regulating chemoreceptor expression (van de Wouw et al., 2017). This is in line with the recent study which indicated that higher relative abundance of oral Clostridia is associated with increased total energy, fat and protein intake as well as reduced fiber consumption, while Proteobacteria phylum and Prevotella genus exhibited the reverse correlation (Cattaneo et al., 2019b). Another plausible explanation rests on the fact that bacterial metabolites of the compounds can activate or modulate host taste perception. For instance, host sensitivity to sucrose is correlated with the sucrose catabolism of different oral bacteria in vivo (Gardner et al., 2020; Sedghi et al., 2021). Some specific intraoral bacteria like Porphyromonas gingivalis (P. gingivalis) are capable of utilizing salivary glutamates so as to modulate their concentration, ultimately affecting the taste perception (Takahashi, 2015). Consistent findings were observed in investigations that examined the impacts of different oral microbiota on the metabolism and aroma perception of cysteine conjugates as well as glycosides (Starkenmann et al., 2008; Parker et al., 2020). Furthermore, oral microbiota also modulates host chemosensation by altering taste receptor density, which is attributed to the ability of certain microorganisms to mediate the secretion of inflammatory factors by host cells and trigger inflammation. On the one hand, the immune response can diminish taste perception through damaging taste buds (Wang et al., 2009; Cohn et al., 2010). On the other hand, several studies have also demonstrated that taste receptors themselves have varying degrees of responsiveness to microorganisms and are directly engaged in the host immune regulation (Gil et al., 2015; Zheng et al., 2019). We elaborate below on the latest progress in studying the participation of taste receptors in the host immune response.
Taste Receptors Implicated in the Host Immune Response to Microorganisms
As previously stated, taste receptors act as chemoreceptors that not only sense chemicals known as tastants to initiate taste signals, but also recognize microorganisms such as bacteria and activate downstream signaling cascades, thereby contributing to innate host defense and the maintenance of microbial homeostasis (Lee et al., 2014; Gil et al., 2015; Jaggupilli et al., 2018). In the past, great progress has been made on the involvement of taste receptors in extraoral organs such as airways and gastrointestinal tract in host innate immunity and microbial regulation, while the role of taste receptors in the oral cavity in mediating microbiota-host interaction is minimally known. A number of recent studies have yielded new light on oral taste receptors that detect bacterial signals and subsequently coordinate immune responses (Figure 2).
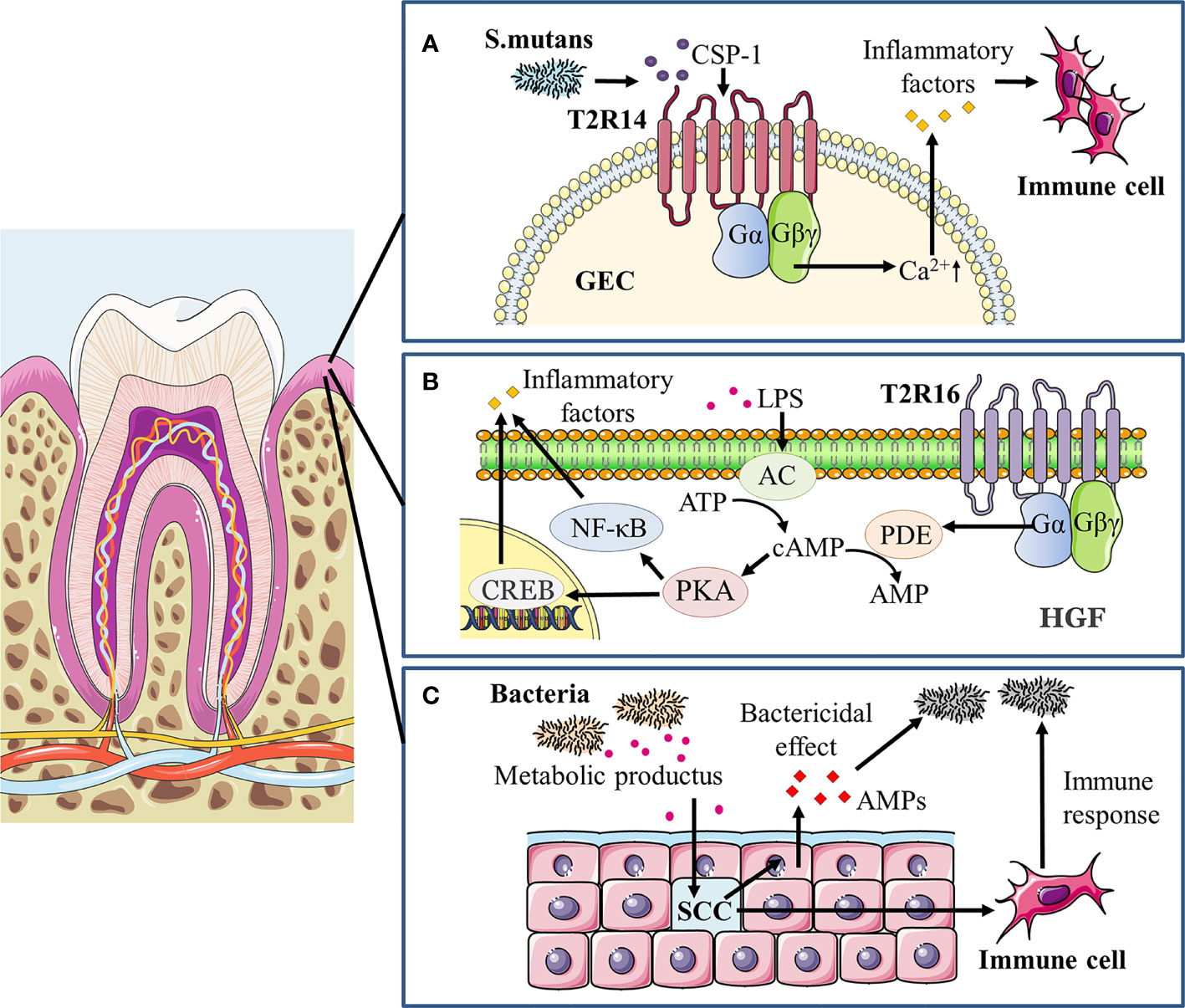
Figure 2 Taste receptors modulate oral immune response to microorganisms. (A) In gingiva epithelial cells (GECs) treated with S. mutans competence stimulating peptide-1 (CSP-1), a vigorous increase in intracellular calcium mobilization and secretion of cytokines/chemokines including IL-6, IL-8 and TNF-α occurred primarily through the T2R14-Gβγ-PLCβ pathway, recruiting immune cells and mediating the immune response to pathogens. (B) LPS can induce a significant dose- and time-dependent increase in adenylate cyclase (AC) activity and thereafter elevates intracellular cAMP levels, positively affecting NF-κB activity through its major effector protein kinase A (PKA) and stimulating inflammatory responses. In addition, the cAMP/PKA/cAMP response element binding protein (CREB) signaling pathway may promote the LPS-induced release of pro-inflammatory cytokines, including IL-6, IL-33, and TNF-α. In human gingival fibroblasts (HGFs), agonist-stimulated T2R16 mobilizes downstream α-gustducin to activate the phosphodiesterase (PDE) that hydrolyzes cAMP, thereby reducing intracellular cAMP levels and alleviating the LPS-induced inflammation as well as tissue injury. (C) Taste receptors of gingival solitary chemosensory cells (gSCCs) activated by bacterial metabolites would stimulate epithelial cells to release antimicrobial peptides (AMPs) as a direct bactericidal effect and also recruit immune cells to modulate the oral immune response.
Taste receptors have been detected to be expressed on a variety of tissues and cells, and are known to be engaged in host immunity (Workman et al., 2015; Bloxham et al., 2020; Welcome, 2020). Expression of bitter receptors is observed in immune cells such as monocytes and neutrophils, which aids in the recognition of bacterial quorum sensing molecules (QSMs) and facilitates the innate immune response to disease (Maurer et al., 2015). T2Rs (mainly T2R38 and T2R14) in airway epithelial cells specifically bind to the Pseudomonas aeruginosa derived QSMs acyl homoserine lactones (AHLs) and initiates the host immune response, activating chemotaxis of immune cells and promoting the secretion of antimicrobial peptides (AMPs) as well as inflammatory factors (Hariri et al., 2017; Freund et al., 2018; Jaggupilli et al., 2018). In addition, the mechanism of T2R participation in the immune response has been further revealed by the work of Gopallawa et al. in macrophages (Gopallawa et al., 2021). Bitter taste metabolite from bacteria stimulates bitter taste receptors and activates calcium signaling, leading to activation of nitric oxide synthase (NOS) isoforms with nitric oxide (NO) production. NO may upregulate phagocytosis of macrophages via several mechanisms (Jun et al., 1996). Simultaneously, bacterial co-stimulation of macrophages and T2Rs also enhanced phagocytosis by reducing cAMP when cAMP levels were elevated from baseline. Interestingly, sweet taste may regulate innate immunity in reverse to bitter taste signals. Lee et al. demonstrated that T1R2/3 receptors activated by sugars or bacterial d-amino acids could inhibit T2R-dependent calcium signaling and downstream AMP secretion of adjoining epithelial cells, diminishing the immune response to microbial bitter products (Lee et al., 2014; Lee et al., 2017). Furthermore, T2Rs activation can also induce anti-inflammatory effects as antagonizing lipopolysaccharide (LPS)-provoked production of inflammatory mediators in human peripheral mononuclear blood cells and lung macrophages (Tran et al., 2018; Grassin-Delyle et al., 2019). The anti-inflammatory activity of T2Rs was likewise associated with SNPs, as the functional PAV haplotype T2R38 receptor exhibited significant inhibition of TNF-α release after agonist stimulation compared to the non-functional AVI/AVI diplotype (Tran et al., 2018).
Studies in the oral cavity have expanded the role of taste receptors, especially bitter taste receptors, in the recognition of oral pathogenic microorganisms and modulation of oral microbial homeostasis. Gil et al. showed that the T2R38 gene polymorphism in the oral cavity determined the degree of innate immune response induced by oral bacteria (Gil et al., 2015). T2R38 mRNA expression in GECs carrying PAV/PAV genotype increased significantly when stimulated with S. mutans, while AVI/AVI genotype showed little change. Human β-defensin-2 (hBD-2) and IL-1α secretion after the stimulation of S. mutans was decreased in PAV/PAV cell line after the T2R38 knockdown. The result agrees with Wendell et al. who reported that the PAV haplotype of T2R38 exhibited a protective effect against caries in primary dentition, whereas the AVI haplotype posed a higher risk of suffering from caries (Wendell et al., 2010). In the same study by Gil et al., an opposite result was found for stimulation from P. gingivalis or Fusobacterium nucleatum, as the expression of T2R38 was significantly upregulated in the GECs of AVI/AVI genotype, while no significant changes were observed in PAV/PAV or PAV/AVI genotype (Gil et al., 2015). This suggests that different pathogenic bacteria may act through different signaling pathways to stimulate taste receptors and elicit subsequent immune responses in the organism. The study by Medapati and his colleagues then uncovered an emerging role of T2R14 in recognizing oral S. mutans with activation of innate immune response in GECs (Medapati et al., 2021b). T2R14 in GECs mediates calcium signaling and secretion of pro-inflammatory cytokines upon recognition of competence stimulating peptide-1 (CSP-1) secreted by S. mutans, as well as attracting differentiated HL-60 immune cells (dHL-60). A subsequent study by Medapati et al. further pointed out that T2R14 may mediate the internalization of Staphylococcus aureus (S. aureus) in GECs through activating p21-activated kinase 1 associated actin and F-actin, and enhanced hBD-2 secretion that inhibits S. aureus (Medapati et al., 2021a). However, the knockdown of T2R14 alone does not affect the level of the internalization of S. mutans and the secretion of hBD-2 in GECs. It is interesting to observe that T2R14-dependent inhibition of S. aureus occurred when GECs were treated with CSP-1 from S. mutans. This hints that bitter receptor-mediated bactericidal effects may be one of the important mechanisms by which S. mutans overcome competition with commensal bacteria in the oral cavity. Of note, study evidence also suggests that bitter taste receptors may similarly assume a repressive role against inflammation development in the oral cavity. Our recent work unveiled that the bitter agonist salicin decreased LPS-induced cAMP accumulation in human gingival fibroblasts in a T2R16-dependent manner, and subsequently suppressed the expression of inflammatory cytokines along with NF-κB activation (Zhou et al., 2021). The simultaneous function of T2Rs in mediating the secretion of antimicrobial substances and preventing excessive inflammatory responses prompted its potential use as a target for the treatment of periodontitis.
Impairment of taste receptors, which are crucial for the host to detect bacteria and mediate regulation oral homeostasis, can lead to overgrowth of pathogenic microorganisms and disease progression. Our study on gingival solitary chemosensory cells (gSCCs) showed that knockout of taste signaling molecules increases bacterial load as well as pathogenicity, ultimately exacerbating periodontitis (Zheng et al., 2019). GSCCs are found in the mouse gingival junctional epithelium, utilizing bitter taste receptors and coupled taste transduction elements to detect bacterial components and modulate immune responses. α-gustducin-null (Gnat3-/-) mice which knocked out one of the taste signaling receptors altered the oral microbiome, resulting in greater alveolar bone loss. Meanwhile, topical treatment of bitter denatonium benzoate in wildtype mice could alleviate periodontitis by upregulating the expression of antimicrobial peptides, which was abolished in Gnat3-/- mice. This is coherent with past studies in nasal SCCs (Tizzano et al., 2010; Lee et al., 2014). One plausible hypothesis is that the lack of taste signaling in gSCCs causes insufficient antimicrobial peptides secretion and dysbiosis of microbiota, increasing the risk of periodontitis. These outcomes suggest that taste signaling plays a key role in host oral immune modulation.
Conclusion
There is now substantial evidence to support that multiple taste receptors are expressed on different cells in the oral cavity to detect bacterial metabolites and distinguish pathogenic from commensal bacteria, playing a critical role in host regulation of oral microbial homeostasis as well as oral health (Zheng et al., 2019). For instance, recent studies in GECs have shown that the expressed bitter taste receptors T2R14 and T2R38 detect various bacterial QSMs or metabolites through different pathways, with the secretion of AMPs to remove pathogenic bacteria (Gil et al., 2015; Medapati et al., 2021b). Functional alterations in taste receptors can also occur as a result of oral microbiota and disease (Zhu et al., 2021). Targeting taste receptors may be a promising alternative therapy to treat oral diseases caused by specific pathogens without antibiotics.
In parallel, numerous studies have described the influence of host genetic polymorphisms on susceptibility to oral diseases (Piekoszewska-Ziętek et al., 2017; Kozak et al., 2020). Similarly, taste receptor SNPs not only determine the distinct taste perception capacities and thus affect food preferences, but also shape the recognition of microbial metabolites, leading to different levels of immune defense in response to specific oral microbes (Chisini et al., 2021). Taste receptor genotypic and phenotypic variations may have potential implications in predicting susceptibility to oral disease and the efficacy of therapy, thereby facilitating the development of personalized treatment based on individual receptor genotypes.
However, current research on the non-gustatory perceptive functions of taste receptors in the oral cavity is still limited, with many crucial questions awaiting further explanations. To name a few, are sour or salt taste receptors in oral likewise engaged in the detection and response to bacterial metabolites? Are there any differences in the transduction pathways between gustatory perception and non-gustatory perception functions? Furthermore, the present understanding of the links and mechanisms between the taste receptor SNPs and oral microbiota as well as diseases is still far from enough. We look forward to further studies on taste receptors to reveal their key role as chemoreceptors in host-pathogen interactions both in oral and systemic settings.
Author Contributions
Conceptualization: XX, XZ, and MT. Writing original draft: HD, JL, JZ, ZZ. Writing, review, and editing: XP, XDZ, XZ, XX, and MT. funding acquisition: XX and XZ. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81900995 to XZ, 81771099 to XX), the China Postdoctoral Science Foundation (2020M673266 to XZ), and the Research funding for talents developing, West China Hospital of Stomatology Sichuan University (RCDWJS2020-11 to XZ); the National Institute of Dental and Craniofacial Research (NIDCR) (R01DC028979 to MT), and National Institute on Deafness and Other Communication Disorders (NIDCD) (R01DC016598 to MT).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Figures were produced with the assistance of Servier Medical Art (https://smart.servier.com/).
References
Ahmad, R., Dalziel, J. E. (2020). G Protein-Coupled Receptors in Taste Physiology and Pharmacology. Front. Pharmacol. 11. doi: 10.3389/fphar.2020.587664
Bartoshuk, L. M., Duffy, V. B., Miller, I. J. (1994). PTC/PROP Tasting: Anatomy, Psychophysics, and Sex Effects. Physiol. Behav. 56 (6), 1165–1171. doi: 10.1016/0031-9384(94)90361-1
Basson, M. D., Bartoshuk, L. M., Dichello, S. Z., Panzini, L., Weiffenbach, J. M., Duffy, V. B. (2005). Association Between 6-N-Propylthiouracil (PROP) Bitterness and Colonic Neoplasms. Digestive Dis. Sci. 50 (3), 483–489. doi: 10.1007/s10620-005-2462-7
Besnard, P., Christensen, J. E., Bernard, A., Simoneau-Robin, I., Collet, X., Verges, B., et al. (2020). Identification of an Oral Microbiota Signature Associated With an Impaired Orosensory Perception of Lipids in Insulin-Resistant Patients. Acta Diabetol. 57 (12), 1445–1451. doi: 10.1007/s00592-020-01567-9
Bloxham, C. J., Foster, S. R., Thomas, W. G. (2020). A Bitter Taste in Your Heart. Front. Physiol. 11. doi: 10.3389/fphys.2020.00431
Bradshaw, D. J., Lynch, R. J. M. (2013). Diet and the Microbial Aetiology of Dental Caries: New Paradigms. Int. Dental J. 63 Suppl 2, 64–72. doi: 10.1111/idj.12082
Bufe, B., Breslin, P. A. S., Kuhn, C., Reed, D. R., Tharp, C. D., Slack, J. P., et al. (2005). The Molecular Basis of Individual Differences in Phenylthiocarbamide and Propylthiouracil Bitterness Perception. Curr. Biol.: CB 15 (4), 322–327. doi: 10.1016/j.cub.2005.01.047
Canessa, C. M., Schild, L., Buell, G., Thorens, B., Gautschi, I., Horisberger, J. D., et al. (1994). Amiloride-Sensitive Epithelial Na+ Channel is Made of Three Homologous Subunits. Nature 367 (6462), 463–467. doi: 10.1038/367463a0
Carey, R. M., Lee, R. J. (2019). Taste Receptors in Upper Airway Innate Immunity. Nutrients 11 (9), 2017. doi: 10.3390/nu11092017
Cattaneo, C., Gargari, G., Koirala, R., Laureati, M., Riso, P., Guglielmetti, S., et al. (2019a). New Insights Into the Relationship Between Taste Perception and Oral Microbiota Composition. Sci. Rep. 9 (1), 3549. doi: 10.1038/s41598-019-40374-3
Cattaneo, C., Riso, P., Laureati, M., Gargari, G., Pagliarini, E. (2019b). Exploring Associations Between Interindividual Differences in Taste Perception, Oral Microbiota Composition, and Reported Food Intake. Nutrients 11 (5), 1167. doi: 10.3390/nu11051167
Chamoun, E., Carroll, N. A., Duizer, L. M., Qi, W., Feng, Z., Darlington, G., et al. (2018a). The Relationship Between Single Nucleotide Polymorphisms in Taste Receptor Genes, Taste Function and Dietary Intake in Preschool-Aged Children and Adults in the Guelph Family Health Study. Nutrients 10 (8), 990. doi: 10.3390/nu10080990
Chamoun, E., Liu, A. S., Duizer, L. M., Feng, Z., Darlington, G., Duncan, A. M., et al. (2021). Single Nucleotide Polymorphisms in Sweet, Fat, Umami, Salt, Bitter and Sour Taste Receptor Genes are Associated With Gustatory Function and Taste Preferences in Young Adults. Nutr. Res. (New York N.Y.) 85, 40–46. doi: 10.1016/j.nutres.2020.12.007
Chamoun, E., Mutch, D. M., Allen-Vercoe, E., Buchholz, A. C., Duncan, A. M., Spriet, L. L., et al. (2018b). A Review of the Associations Between Single Nucleotide Polymorphisms in Taste Receptors, Eating Behaviors, and Health. Crit. Rev. Food Sci. Nutr. 58 (2), 194–207. doi: 10.1080/10408398.2016.1152229
Chandrashekar, J., Hoon, M. A., Ryba, N. J. P., Zuker, C. S. (2006). The Receptors and Cells for Mammalian Taste. Nature 444 (7117), 288–294. doi: 10.1038/nature05401
Chandrashekar, J., Mueller, K. L., Hoon, M. A., Adler, E., Feng, L., Guo, W., et al. (2000). T2Rs Function as Bitter Taste Receptors. Cell 100 (6), 703–711. doi: 10.1016/S0092-8674(00)80706-0
Chisini, L. A., Cademartori, M. G., Conde, M. C. M., Costa, F. D. S., Salvi, L. C., Tovo-Rodrigues, L., et al. (2021). Single Nucleotide Polymorphisms of Taste Genes and Caries: A Systematic Review and Meta-Analysis. Acta Odontol. Scand. 79 (2), 147–155. doi: 10.1080/00016357.2020.1832253
Clapp, T. R., Trubey, K. R., Vandenbeuch, A., Stone, L. M., Margolskee, R. F., Chaudhari, N., et al. (2008). Tonic Activity of Galpha-Gustducin Regulates Taste Cell Responsivity. FEBS Lett. 582 (27), 3783–3787. doi: 10.1016/j.febslet.2008.10.007
Cohn, Z. J., Kim, A., Huang, L., Brand, J., Wang, H. (2010). Lipopolysaccharide-Induced Inflammation Attenuates Taste Progenitor Cell Proliferation and Shortens the Life Span of Taste Bud Cells. BMC Neurosci. 11, 72. doi: 10.1186/1471-2202-11-72
Deckmann, K., Kummer, W. (2016). Chemosensory Epithelial Cells in the Urethra: Sentinels of the Urinary Tract. Histochem. Cell Biol. 146 (6), 673–683. doi: 10.1007/s00418-016-1504-x
Deshpande, D. A., Wang, W. C. H., McIlmoyle, E. L., Robinett, K. S., Schillinger, R. M., An, S. S., et al. (2010). Bitter Taste Receptors on Airway Smooth Muscle Bronchodilate by Localized Calcium Signaling and Reverse Obstruction. Nat. Med. 16 (11), 1299–1304. doi: 10.1038/nm.2237
Dias, A. G., Rousseau, D., Duizer, L., Cockburn, M., Chiu, W., Nielsen, D., et al. (2012). Genetic Variation in Putative Salt Taste Receptors and Salt Taste Perception in Humans. Chem. Senses 38 (2), 137–145. doi: 10.1093/chemse/bjs090
Dong, D., Jones, G., Zhang, S. (2009). Dynamic Evolution of Bitter Taste Receptor Genes in Vertebrates. BMC Evol. Biol. 9, 12. doi: 10.1186/1471-2148-9-12
Eny, K. M., Wolever, T. M., Corey, P. N., El-Sohemy, A. (2010). Genetic Variation in TAS1R2 (Ile191Val) is Associated With Consumption of Sugars in Overweight and Obese Individuals in 2 Distinct Populations. Am. J. Clin. Nutr. 92 (6), 1501–1510. doi: 10.3945/ajcn.2010.29836
Eriksson, L., Esberg, A., Haworth, S., Holgerson, P. L., Johansson, I. (2019). Allelic Variation in Taste Genes Is Associated With Taste and Diet Preferences and Dental Caries. Nutrients 11 (7), 1491. doi: 10.3390/nu11071491
Esberg, A., Haworth, S., Brunius, C., Lif Holgerson, P., Johansson, I. (2019). Carbonic Anhydrase 6 Gene Variation Influences Oral Microbiota Composition and Caries Risk in Swedish Adolescents. Sci. Rep. 9 (1), 452–452. doi: 10.1038/s41598-018-36832-z
Esposito, C. M., Ceresa, A., Buoli, M. (2021). The Association Between Personality Traits and Dietary Choices: A Systematic Review. Adv. Nutr. (Bethesda Md.) 12 (4), 1149–1159. doi: 10.1093/advances/nmaa166
Feng, Y., Licandro, H., Martin, C., Septier, C., Zhao, M., Neyraud, E., et al. (2018). The Associations Between Biochemical and Microbiological Variables and Taste Differ in Whole Saliva and in the Film Lining the Tongue. BioMed. Res. Int. 2018, 2838052. doi: 10.1155/2018/2838052
Ferraris, C., Turner, A., Scarlett, C., Veysey, M., Lucock, M., Bucher, T., et al. (2021a). Association Between Sour Taste SNP -Rs236514, Diet Quality and Mild Cognitive Impairment in an Elderly Cohort. Nutrients 13 (3), 719. doi: 10.3390/nu13030719
Ferraris, C., Turner, A., Scarlett, C. J., Veysey, M., Lucock, M., Bucher, T., et al. (2021b). Sour Taste SNP -Rs236514 and Differences in Nutrient Intakes and Metabolic Health Markers in the Elderly. Front. Nutr. 8. doi: 10.3389/fnut.2021.701588
Freund, J. R., Mansfield, C. J., Doghramji, L. J., Adappa, N. D., Palmer, J. N., Kennedy, D. W., et al. (2018). Activation of Airway Epithelial Bitter Taste Receptors by Quinolones Modulates Calcium, Cyclic-AMP, and Nitric Oxide Signaling. J. Biol. Chem. 293 (25), 9824–9840. doi: 10.1074/jbc.RA117.001005
Fushan, A. A., Simons, C. T., Slack, J. P., Manichaikul, A., Drayna, D. (2009). Allelic Polymorphism Within the TAS1R3 Promoter is Associated With Human Taste Sensitivity to Sucrose. Curr. Biol.: CB 19 (15), 1288–1293. doi: 10.1016/j.cub.2009.06.015
Garcia-Bailo, B., Toguri, C., Eny, K. M., El-Sohemy, A. (2009). Genetic Variation in Taste and its Influence on Food Selection. Omics: J. Integr. Biol. 13 (1), 69–80. doi: 10.1089/omi.2008.0031
Gardner, A., So, P. W., Carpenter, G. H. (2020). Intraoral Microbial Metabolism and Association With Host Taste Perception. J. Dental Res. 99 (6), 739–745. doi: 10.1177/0022034520917142
Gil, S., Coldwell, S., Drury, J. L., Arroyo, F., Phi, T., Saadat, S., et al. (2015). Genotype-Specific Regulation of Oral Innate Immunity by T2R38 Taste Receptor. Mol. Immunol. 68 (2 Pt C), 663–670. doi: 10.1016/j.molimm.2015.10.012
Glendinning, J. I. (1994). Is the Bitter Rejection Response Always Adaptive? Physiol. Behav. 56 (6), 1217–1227. doi: 10.1016/0031-9384(94)90369-7
Gopallawa, I., Freund, J. R., Lee, R. J. (2021). Bitter Taste Receptors Stimulate Phagocytosis in Human Macrophages Through Calcium, Nitric Oxide, and Cyclic-GMP Signaling. Cell. Mol. Life Sci.: CMLS 78 (1), 271–286. doi: 10.1007/s00018-020-03494-y
Grassin-Delyle, S., Salvator, H., Mantov, N., Abrial, C., Brollo, M., Faisy, C., et al. (2019). Bitter Taste Receptors (TAS2Rs) in Human Lung Macrophages: Receptor Expression and Inhibitory Effects of TAS2R Agonists. Front. Physiol. 10. doi: 10.3389/fphys.2019.01267
Hariri, B. M., McMahon, D. B., Chen, B., Freund, J. R., Mansfield, C. J., Doghramji, L. J., et al. (2017). Flavones Modulate Respiratory Epithelial Innate Immunity: Anti-Inflammatory Effects and Activation of the T2R14 Receptor. J. Biol. Chem. 292 (20), 8484–8497. doi: 10.1074/jbc.M116.771949
Haznedaroğlu, E., Koldemir-Gündüz, M., Bakır-Coşkun, N., Bozkuş, H. M., Çağatay, P., Süsleyici-Duman, B., et al. (2015). Association of Sweet Taste Receptor Gene Polymorphisms With Dental Caries Experience in School Children. Caries Res. 49 (3), 275–281. doi: 10.1159/000381426
Heck, G. L., Mierson, S., DeSimone, J. A. (1984). Salt Taste Transduction Occurs Through an Amiloride-Sensitive Sodium Transport Pathway. Science (New York N.Y.) 223 (4634), 403–405. doi: 10.1126/science.6691151
Hichami, A., Khan, A. S., Khan, N. A. (2021). Cellular and Molecular Mechanisms of Fat Taste Perception. Handb. Exp. Pharmacol. 6, 1–24. doi: 10.1007/164_2021_437
Huang, Y. A., Maruyama, Y., Stimac, R., Roper, S. D. (2008). Presynaptic (Type III) Cells in Mouse Taste Buds Sense Sour (Acid) Taste. J. Physiol. 586 (12), 2903–2912. doi: 10.1113/jphysiol.2008.151233
Huang, L., Shanker, Y. G., Dubauskaite, J., Zheng, J. Z., Yan, W., Rosenzweig, S., et al. (1999). Ggamma13 Colocalizes With Gustducin in Taste Receptor Cells and Mediates IP3 Responses to Bitter Denatonium. Nat. Neurosci. 2 (12), 1055–1062. doi: 10.1038/15981
Iwata, S., Yoshida, R., Ninomiya, Y. (2014). Taste Transductions in Taste Receptor Cells: Basic Tastes and Moreover. Curr. Pharm. Design 20 (16), 2684–2692. doi: 10.2174/13816128113199990575
Izakovicova Holla, L., Borilova Linhartova, P., Lucanova, S., Kastovsky, J., Musilova, K., Bartosova, M., et al. (2015). GLUT2 and TAS1R2 Polymorphisms and Susceptibility to Dental Caries. Caries Res. 49 (4), 417–424. doi: 10.1159/000430958
Jaggupilli, A., Singh, N., Jesus, V. C. D., Duan, K., Chelikani, P. (2018). Characterization of the Binding Sites for Bacterial Acyl Homoserine Lactones (AHLs) on Human Bitter Taste Receptors (T2Rs). ACS Infect. Dis. 4 (7), 1146–1156. doi: 10.1021/acsinfecdis.8b00094
Jayasinghe, S. N., Kruger, R., Walsh, D. C. I., Cao, G., Rivers, S., Richter, M., et al. (2017). Is Sweet Taste Perception Associated With Sweet Food Liking and Intake? Nutrients 9 (7), 750. doi: 10.3390/nu9070750
Jilani, H., Ahrens, W., Buchecker, K., Russo, P., Hebestreit, A. (2017). Association Between the Number of Fungiform Papillae on the Tip of the Tongue and Sensory Taste Perception in Children. Food Nutr. Res. 61 (1), 1348865. doi: 10.1080/16546628.2017.1348865
Jun, C. D., Han, M. K., Kim, U. H., Chung, H. T. (1996). Nitric Oxide Induces ADP-Ribosylation of Actin in Murine Macrophages: Association With the Inhibition of Pseudopodia Formation, Phagocytic Activity, and Adherence on a Laminin Substratum. Cell. Immunol. 174 (1), 25–34. doi: 10.1006/cimm.1996.0290
Jurczak, A., Jamka-Kasprzyk, M., Bębenek, Z., Staszczyk, M., Jagielski, P., Kościelniak, D., et al. (2020). Differences in Sweet Taste Perception and Its Association With the Cariogenic Profile in Preschool Children With Caries. Nutrients 12 (9), 2592. doi: 10.3390/nu12092592
Kaufman, A., Choo, E., Koh, A., Dando, R. (2018). Inflammation Arising From Obesity Reduces Taste Bud Abundance and Inhibits Renewal. PloS Biol. 16 (3), e2001959. doi: 10.1371/journal.pbio.2001959
Kaye, E. A., Sohn, W., Garcia, R. I. (2020). The Healthy Eating Index and Coronal Dental Caries in US Adults: National Health and Nutrition Examination Survey 2011-2014. J. Am. Dental Assoc. (1939) 151 (2), 78–86. doi: 10.1016/j.adaj.2019.09.009
Khan, A. S., Murtaza, B., Hichami, A., Khan, N. A. (2019). A Cross-Talk Between Fat and Bitter Taste Modalities. Biochimie 159, 3–8. doi: 10.1016/j.biochi.2018.06.013
Kim, U.-K., Jorgenson, E., Coon, H., Leppert, M., Risch, N., Drayna, D. (2003). Positional Cloning of the Human Quantitative Trait Locus Underlying Taste Sensitivity to Phenylthiocarbamide. Science (New York N.Y.) 299 (5610), 1221–1225. doi: 10.1126/science.1080190
Kojima, I., Nakagawa, Y. (2011). The Role of the Sweet Taste Receptor in Enteroendocrine Cells and Pancreatic β-Cells. Diabetes Metab. J. 35 (5), 451–457. doi: 10.4093/dmj.2011.35.5.451
Kozak, M., Dabrowska-Zamojcin, E., Mazurek-Mochol, M., Pawlik, A. (2020). Cytokines and Their Genetic Polymorphisms Related to Periodontal Disease. J. Clin. Med. 9 (12), 4045. doi: 10.3390/jcm9124045
Kretz, O., Barbry, P., Bock, R., Lindemann, B. (1999). Differential Expression of RNA and Protein of the Three Pore-Forming Subunits of the Amiloride-Sensitive Epithelial Sodium Channel in Taste Buds of the Rat. J. Histochem. Cytochem. 47 (1), 51–64. doi: 10.1177/002215549904700106
Kulkarni, G. V., Chng, T., Eny, K. M., Nielsen, D., Wessman, C., El-Sohemy, A. (2013). Association of GLUT2 and TAS1R2 Genotypes With Risk for Dental Caries. Caries Res. 47 (3), 219–225. doi: 10.1159/000345652
Kyriazis, G. A., Soundarapandian, M. M., Tyrberg, B. (2012). Sweet Taste Receptor Signaling in Beta Cells Mediates Fructose-Induced Potentiation of Glucose-Stimulated Insulin Secretion. Proc. Natl. Acad. Sci. U. S. A. 109 (8), E524–E532. doi: 10.1073/pnas.1115183109
Laffitte, A., Neiers, F., Briand, L. (2014). Functional Roles of the Sweet Taste Receptor in Oral and Extraoral Tissues. Curr. Opin. Clin. Nutr. Metab. Care 17 (4), 379–385. doi: 10.1097/MCO.0000000000000058
Lee, R. J., Hariri, B. M., McMahon, D. B., Chen, B., Doghramji, L., Adappa, N. D., et al. (2017). Bacterial D-Amino Acids Suppress Sinonasal Innate Immunity Through Sweet Taste Receptors in Solitary Chemosensory Cells. Sci. Signaling 10 (495), eaam7703. doi: 10.1126/scisignal.aam7703
Lee, R. J., Kofonow, J. M., Rosen, P. L., Siebert, A. P., Chen, B., Doghramji, L., et al. (2014). Bitter and Sweet Taste Receptors Regulate Human Upper Respiratory Innate Immunity. J. Clin. Invest. 124 (3), 1393–1405. doi: 10.1172/JCI72094
Lewandowski, B. C., Sukumaran, S. K., Margolskee, R. F., Bachmanov, A. A. (2016). Amiloride-Insensitive Salt Taste Is Mediated by Two Populations of Type III Taste Cells With Distinct Transduction Mechanisms. J. Neurosci. 36 (6), 1942–1953. doi: 10.1523/JNEUROSCI.2947-15.2016
Lindemann, B. (2001). Receptors and Transduction in Taste. Nature 413 (6852), 219–225. doi: 10.1038/35093032
Li, X., Staszewski, L., Xu, H., Durick, K., Zoller, M., Adler, E. (2002). Human Receptors for Sweet and Umami Taste. Proc. Natl. Acad. Sci. U. S. A. 99 (7), 4692–4696. doi: 10.1073/pnas.072090199
Margolskee, R. F. (1993). The Molecular Biology of Taste Transduction. BioEssays: News Rev. Mol. Cell. Dev. Biol. 15 (10), 645–650. doi: 10.1002/bies.950151003
Maurer, S., Wabnitz, G. H., Kahle, N. A., Stegmaier, S., Prior, B., Giese, T., et al. (2015). Tasting Pseudomonas Aeruginosa Biofilms: Human Neutrophils Express the Bitter Receptor T2R38 as Sensor for the Quorum Sensing Molecule N-(3-Oxododecanoyl)-L-Homoserine Lactone. Front. Immunol. 6. doi: 10.3389/fimmu.2015.00369
McLaughlin, S. K., McKinnon, P. J., Margolskee, R. F. (1992). Gustducin is a Taste-Cell-Specific G Protein Closely Related to the Transducins. Nature 357 (6379), 563–569. doi: 10.1038/357563a0
Medapati, M. R., Bhagirath, A. Y., Singh, N., Schroth, R. J., Bhullar, R. P., Duan, K., et al. (2021a). Bitter Taste Receptor T2R14 Modulates Gram-Positive Bacterial Internalization and Survival in Gingival Epithelial Cells. Int. J. Mol. Sci. 22 (18), 9920. doi: 10.3390/ijms22189920
Medapati, M. R., Singh, N., Bhagirath, A. Y., Duan, K., Triggs-Raine, B., Batista, E. L., et al. (2021b). Bitter Taste Receptor T2R14 Detects Quorum Sensing Molecules From Cariogenic Streptococcus Mutans and Mediates Innate Immune Responses in Gingival Epithelial Cells. FASEB J. 35 (3), e21375. doi: 10.1096/fj.202000208R
Murovets, V. O., Bachmanov, A. A., Zolotarev, V. A. (2015). Impaired Glucose Metabolism in Mice Lacking the Tas1r3 Taste Receptor Gene. PLoS One 10 (6), e0130997. doi: 10.1371/journal.pone.0130997
Murovets, V. O., Lukina, E. A., Sozontov, E. A., Andreeva, J. V., Khropycheva, R. P., Zolotarev, V. A. (2020). Allelic Variation of the Tas1r3 Taste Receptor Gene Affects Sweet Taste Responsiveness and Metabolism of Glucose in F1 Mouse Hybrids. PLoS One 15 (7), e0235913. doi: 10.1371/journal.pone.0235913
Nomura, K., Nakanishi, M., Ishidate, F., Iwata, K., Taruno, A. (2020). All-Electrical Ca-Independent Signal Transduction Mediates Attractive Sodium Taste in Taste Buds. Neuron 106 (5), 816–829.e6. doi: 10.1016/j.neuron.2020.03.006
Nuemket, N., Yasui, N., Kusakabe, Y., Nomura, Y., Atsumi, N., Akiyama, S., et al. (2017). Structural Basis for Perception of Diverse Chemical Substances by T1r Taste Receptors. Nat. Commun. 8, 15530. doi: 10.1038/ncomms15530
Oka, Y., Butnaru, M., von Buchholtz, L., Ryba, N. J. P., Zuker, C. S. (2013). High Salt Recruits Aversive Taste Pathways. Nature 494 (7438), 472–475. doi: 10.1038/nature11905
Öter, B., Ulukapı, I., Ulukapı, H., Topçuoğlu, N., Cıldır, S. (2011). The Relation Between 6-N-Propylthiouracil Sensitivity and Caries Activity in Schoolchildren. Caries Res. 45 (6), 556–560. doi: 10.1159/000332432
Parker, M., Onetto, C., Hixson, J., Bilogrevic, E., Schueth, L., Pisaniello, L., et al. (2020). Factors Contributing to Interindividual Variation in Retronasal Odor Perception From Aroma Glycosides: The Role of Odorant Sensory Detection Threshold, Oral Microbiota, and Hydrolysis in Saliva. J. Agric. Food Chem. 68 (38), 10299–10309. doi: 10.1021/acs.jafc.9b05450
Piekoszewska-Ziętek, P., Turska-Szybka, A., Olczak-Kowalczyk, D. (2017). Single Nucleotide Polymorphism in the Aetiology of Caries: Systematic Literature Review. Caries Res. 51 (4), 425–435. doi: 10.1159/000476075
Pilic, L., Mavrommatis, Y. (2018). Genetic Predisposition to Salt-Sensitive Normotension and its Effects on Salt Taste Perception and Intake. Br. J. Nutr. 120 (7), 721–731. doi: 10.1017/S0007114518002027
Raliou, M., Wiencis, A., Pillias, A. M., Planchais, A., Eloit, C., Boucher, Y., et al. (2009). Nonsynonymous Single Nucleotide Polymorphisms in Human Tas1r1, Tas1r3, and Mglur1 and Individual Taste Sensitivity to Glutamate. Am. J. Clin. Nutr. 90 (3), 789S–799S. doi: 10.3945/ajcn.2009.27462P
Ramos-Lopez, O., Panduro, A., Martinez-Lopez, E., Roman, S. (2016). Sweet Taste Receptor TAS1R2 Polymorphism (Val191Val) Is Associated With a Higher Carbohydrate Intake and Hypertriglyceridemia Among the Population of West Mexico. Nutrients 8 (2), 101. doi: 10.3390/nu8020101
Rawal, S., Hayes, J. E., Wallace, M. R., Bartoshuk, L. M., Duffy, V. B. (2013). Do Polymorphisms in the TAS1R1 Gene Contribute to Broader Differences in Human Taste Intensity? Chem. Senses 38 (8), 719–728. doi: 10.1093/chemse/bjt040
Rhyu, M.-R., Song, A.-Y., Kim, E.-Y., Son, H.-J., Kim, Y., Mummalaneni, S., et al. (2020). Kokumi Taste Active Peptides Modulate Salt and Umami Taste. Nutrients 12 (4), 1198. doi: 10.3390/nu12041198
Roper, S. D. (2013). Taste Buds as Peripheral Chemosensory Processors. Semin. Cell Dev. Biol. 24 (1), 71–79. doi: 10.1016/j.semcdb.2012.12.002
Roura, E., Foster, S., Winklebach, A., Navarro, M., Thomas, W., Campbell, K., et al. (2016). Taste and Hypertension in Humans: Targeting Cardiovascular Disease. Curr. Pharm. Design 22 (15), 2290–2305. doi: 10.2174/1381612822666160216151545
Sbarbati, A., Bramanti, P., Benati, D., Merigo, F. (2010). The Diffuse Chemosensory System: Exploring the Iceberg Toward the Definition of Functional Roles. Prog. Neurobiol. 91 (1), 77–89. doi: 10.1016/j.pneurobio.2010.01.010
Schneider, C., O’Leary, C. E., Locksley, R. M. (2019). Regulation of Immune Responses by Tuft Cells. Nat. Rev. Immunol. 19 (9), 584–593. doi: 10.1038/s41577-019-0176-x
Sedghi, L., DiMassa, V., Harrington, A., Lynch, S. V., Kapila, Y. L. (2021). The Oral Microbiome: Role of Key Organisms and Complex Networks in Oral Health and Disease. Periodontol. 2000 87 (1), 107–131. doi: 10.1111/prd.12393
Serrano, J., Seflova, J., Park, J., Pribadi, M., Sanematsu, K., Shigemura, N., et al. (2021). The Ile191Val is a Partial Loss-of-Function Variant of the TAS1R2 Sweet-Taste Receptor and is Associated With Reduced Glucose Excursions in Humans. Mol. Metab. 54, 101339. doi: 10.1016/j.molmet.2021.101339
Small, D. M. (2012). Flavor is in the Brain. Physiol. Behav. 107 (4), 540–552. doi: 10.1016/j.physbeh.2012.04.011
Smith, K. R., Hussain, T., Karimian Azari, E., Steiner, J. L., Ayala, J. E., Pratley, R. E., et al. (2016). Disruption of the Sugar-Sensing Receptor T1R2 Attenuates Metabolic Derangements Associated With Diet-Induced Obesity. Am. J. Physiol. Endocrinol. Metab. 310 (8), E688–E698. doi: 10.1152/ajpendo.00484.2015
Smith, K., Karimian Azari, E., LaMoia, T. E., Hussain, T., Vargova, V., Karolyi, K., et al. (2018). T1R2 Receptor-Mediated Glucose Sensing in the Upper Intestine Potentiates Glucose Absorption Through Activation of Local Regulatory Pathways. Mol. Metab. 17, 98–111. doi: 10.1016/j.molmet.2018.08.009
Solemdal, K., Sandvik, L., Willumsen, T., Mowe, M., Hummel, T. (2012). The Impact of Oral Health on Taste Ability in Acutely Hospitalized Elderly. PLoS One 7 (5), e36557. doi: 10.1371/journal.pone.0036557
Starkenmann, C., Le Calvé, B., Niclass, Y., Cayeux, I., Beccucci, S., Troccaz, M. (2008). Olfactory Perception of Cysteine-S-Conjugates From Fruits and Vegetables. J. Agric. Food Chem. 56 (20), 9575–9580. doi: 10.1021/jf801873h
Takahashi, N. (2015). Oral Microbiome Metabolism: From “Who Are They?” to “What Are They Doing?” J. Dental Res. 94 (12), 1628–1637. doi: 10.1177/0022034515606045
Tanner, A. C. R., Kressirer, C. A., Rothmiller, S., Johansson, I., Chalmers, N. I. (2018). The Caries Microbiome: Implications for Reversing Dysbiosis. Adv. Dental Res. 29 (1), 78–85. doi: 10.1177/0022034517736496
Tapanee, P., Tidwell, D. K., Schilling, M. W., Peterson, D. G., Tolar-Peterson, T. (2021). Genetic Variation in Taste Receptor Genes (1, 1) and Its Correlation With the Perception of Saltiness in Normotensive and Hypertensive Adults. Int. J. Hypertension 2021, 5559831. doi: 10.1155/2021/5559831
Teng, B., Wilson, C. E., Tu, Y.-H., Joshi, N. R., Kinnamon, S. C., Liman, E. R. (2019). Cellular and Neural Responses to Sour Stimuli Require the Proton Channel Otop1. Curr. Biol.: CB 29 (21), 3647–3656.e5. doi: 10.1016/j.cub.2019.08.077
Tizzano, M., Dvoryanchikov, G., Barrows, J. K., Kim, S., Chaudhari, N., Finger, T. E. (2008). Expression of Galpha14 in Sweet-Transducing Taste Cells of the Posterior Tongue. BMC Neurosci. 9, 110. doi: 10.1186/1471-2202-9-110
Tizzano, M., Gulbransen, B. D., Vandenbeuch, A., Clapp, T. R., Herman, J. P., Sibhatu, H. M., et al. (2010). Nasal Chemosensory Cells Use Bitter Taste Signaling to Detect Irritants and Bacterial Signals. Proc. Natl. Acad. Sci. U. S. A. 107 (7), 3210–3215. doi: 10.1073/pnas.0911934107
Tran, H. T. T., Herz, C., Ruf, P., Stetter, R., Lamy, E. (2018). Human T2R38 Bitter Taste Receptor Expression in Resting and Activated Lymphocytes. Front. Immunol. 9. doi: 10.3389/fimmu.2018.02949
Tu, Y.-H., Cooper, A. J., Teng, B., Chang, R. B., Artiga, D. J., Turner, H. N., et al. (2018). An Evolutionarily Conserved Gene Family Encodes Proton-Selective Ion Channels. Science (New York N.Y.) 359 (6379), 1047–1050. doi: 10.1126/science.aao3264
van de Wouw, M., Schellekens, H., Dinan, T. G., Cryan, J. F. (2017). Microbiota-Gut-Brain Axis: Modulator of Host Metabolism and Appetite. J. Nutr. 147 (5), 727–745. doi: 10.3945/jn.116.240481
von Molitor, E., Riedel, K., Krohn, M., Hafner, M., Rudolf, R., Cesetti, T. (2021). Sweet Taste Is Complex: Signaling Cascades and Circuits Involved in Sweet Sensation. Front. Hum. Neurosci. 15. doi: 10.3389/fnhum.2021.667709
Wang, H., Zhou, M., Brand, J., Huang, L. (2009). Inflammation and Taste Disorders: Mechanisms in Taste Buds. Ann. New York Acad. Sci. 1170, 596–603. doi: 10.1111/j.1749-6632.2009.04480.x
Wan, Y., Wang, F., Yuan, J., Li, J., Jiang, D., Zhang, J., et al. (2019). Effects of Dietary Fat on Gut Microbiota and Faecal Metabolites, and Their Relationship With Cardiometabolic Risk Factors: A 6-Month Randomised Controlled-Feeding Trial. Gut 68 (8), 1417–1429. doi: 10.1136/gutjnl-2018-317609
Welcome, M. O. (2020). The Bitterness of Genitourinary Infections: Properties, Ligands of Genitourinary Bitter Taste Receptors and Mechanisms Linking Taste Sensing to Inflammatory Processes in the Genitourinary Tract. Eur. J. Obstetr. Gynecol. Reprod. Biol. 247, 101–110. doi: 10.1016/j.ejogrb.2020.02.015
Wendell, S., Wang, X., Brown, M., Cooper, M. E., DeSensi, R. S., Weyant, R. J., et al. (2010). Taste Genes Associated With Dental Caries. J. Dental Res. 89 (11), 1198–1202. doi: 10.1177/0022034510381502
Workman, A. D., Palmer, J. N., Adappa, N. D., Cohen, N. A. (2015). The Role of Bitter and Sweet Taste Receptors in Upper Airway Immunity. Curr. Allergy Asthma Rep. 15 (12), 72. doi: 10.1007/s11882-015-0571-8
Yamaguchi, S., Ninomiya, K. (2000). Umami and Food Palatability. J. Nutr. 130 (4), 921S–926S. doi: 10.1093/jn/130.4.921S
Ye, W., Chang, R. B., Bushman, J. D., Tu, Y.-H., Mulhall, E. M., Wilson, C. E., et al. (2016). The K+ Channel KIR2.1 Functions in Tandem With Proton Influx to Mediate Sour Taste Transduction. Proc. Natl. Acad. Sci. U. S. A. 113 (2), E229–E238. doi: 10.1073/pnas.1514282112
Zhang, J., Jin, H., Zhang, W., Ding, C., O’Keeffe, S., Ye, M., et al. (2019). Sour Sensing From the Tongue to the Brain. Cell 179 (2), 392–402.e15. doi: 10.1016/j.cell.2019.08.031
Zhang, Z., Zhao, Z., Margolskee, R., Liman, E. (2007). The Transduction Channel TRPM5 is Gated by Intracellular Calcium in Taste Cells. J. Neurosci. 27 (21), 5777–5786. doi: 10.1523/JNEUROSCI.4973-06.2007
Zhao, G. Q., Zhang, Y., Hoon, M. A., Chandrashekar, J., Erlenbach, I., Ryba, N. J., et al. (2003). The Receptors for Mammalian Sweet and Umami Taste. Cell 115 (3), 255–266. doi: 10.1016/s0092-8674(03)00844-4
Zheng, X., Tizzano, M., Redding, K., He, J., Peng, X., Jiang, P., et al. (2019). Gingival Solitary Chemosensory Cells are Immune Sentinels for Periodontitis. Nat. Commun. 10 (1), 4496. doi: 10.1038/s41467-019-12505-x
Zhou, Z., Xi, R., Liu, J., Peng, X., Zhao, L., Zhou, X., et al. (2021). TAS2R16 Activation Suppresses LPS-Induced Cytokine Expression in Human Gingival Fibroblasts. Front. Immunol. 12. doi: 10.3389/fimmu.2021.726546
Keywords: taste receptor, oral microbiota, innate immunity, periodontitis, dental caries, diet
Citation: Dong H, Liu J, Zhu J, Zhou Z, Tizzano M, Peng X, Zhou X, Xu X and Zheng X (2022) Oral Microbiota-Host Interaction Mediated by Taste Receptors. Front. Cell. Infect. Microbiol. 12:802504. doi: 10.3389/fcimb.2022.802504
Received: 26 October 2021; Accepted: 07 March 2022;
Published: 29 March 2022.
Edited by:
Kangmin Duan, University of Manitoba, CanadaReviewed by:
Manoj Reddy Medapati, University of Manitoba, CanadaNisha Singh, University of Manitoba, Canada
Copyright © 2022 Dong, Liu, Zhu, Zhou, Tizzano, Peng, Zhou, Xu and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Zheng, enh6ZGdAMTI2LmNvbQ==; Xin Xu, eGluLnh1QHNjdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Hao Dong
Hao Dong