- 1Research Center of Avian Disease, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 3Key Laboratory of Animal Disease and Human Health of Sichuan Province, Sichuan Agricultural University, Chengdu, China
microRNAs (miRNAs), non-coding RNAs about 22 nt long, regulate the post-transcription expression of genes to influence many cellular processes. The expression of host miRNAs is affected by virus invasion, which also affects virus replication. Increasing evidence has demonstrated that miRNA influences RNA virus multiplication by binding directly to the RNA virus genome. Here, the knowledge relating to miRNAs’ relationships between host miRNAs and RNA viruses are discussed.
Introduction
The discovery of microRNA (miRNA) began with Caenorhabditis elegans in the 1990s, in which lin-4, a small non-coding RNA (ncRNA), was discovered (Wightman et al., 1993; Moss et al., 1997). Later, scientists found that lin-14 and let-7 from C. elegans were able to bind to the 3' untranslated region (3’UTR) of post-transcriptional products, thus regulating gene expression in order to control the development of C. elegans (Hong et al., 2000; Reinhart et al., 2000). Since then, scientists have located miRNA in a variety of organisms, ranging from plants to animals, and most of the miRNAs were evolutionarily conserved. After miRbase (http://www.mirbase.org/) had been established, it became possible to search the database for miRNA sequences from different species.
In eukaryotic organisms, miRNAs regulate the post-transcriptional expression of host and virus genes by degrading mRNA and inhibiting protein translation. Some scholars suggest that this mechanism may have originally evolved from a small RNA-mediated antiviral defense system that now coordinates various developmental processes and is strongly linked to cellular homeostasis (Aguado and TenOever, 2018). Because of this, miRNA expression is also susceptible to disease occurrence via virus invasion and other mechanisms. Furthermore, the expression of miRNAs in the organism changes following viral infection, suggesting that they may be involved in the virus infection process itself or in the innate immunity of the organism as modulators of cellular gene post-transcriptional expression (Zhang et al., 2016; Pu et al., 2019; Diallo et al., 2021). In addition to regulating the expression of intracellular host genes, miRNA can also influence the replication and pathogenesis of RNA viruses by binding directly to the RNA virus genome (Chen et al., 2011; Trobaugh et al., 2014; Trobaugh and Klimstra, 2017). Moreover, some special host miRNAs bind to RNA within RNA viruses and affect the spatial structure of this RNA to influence viral replication (Li et al., 2013). The role of miRNAs on RNA virus genomes has the potential to treat viruses; therefore, the roles of miRNAs on RNA viral genomes are both summarized and discussed.
Interactions Between miRNAs and Viruses
In plants, worms, and insects, RNA interference (RNAi) is an ancient, dominating, and strong antiviral defense mechanism, whereas RNAi plays an antiviral role only in undifferentiated embryonic stem cells in vertebrates (Maillard et al., 2019). The antiviral effects in vertebrates depends heavily on interferon (IFN)-mediated antiviral responses, such as type I IFN and the IFN-stimulated genes (ISGs) (Mazewski et al., 2020). Even so, the vertebrate RNAi system is still vital for the body, mainly relying on tissue- and cell-specific miRNAs to bind mRNA, thus inhibiting its translation or reducing its stability relating to the regulation of cellular protein expression during the growth, development, reproduction, and death of organisms (Maillard et al., 2013). Furthermore, the expression of miRNAs is susceptible to transcriptional efficiency, epigenetic, intracellular homeostasis, and extracellular environmental influences, leading to time-specific and tissue-specific miRNA expression (Catalanotto et al., 2016). There is evidence that the invasion of microorganisms, such as bacteria and viruses, can lead to the variable miRNA expression profiles observed in cells (Trobaugh and Klimstra, 2017).
As for the virus, we know that many experimental results have shown that a virus infection can alter the miRNA expression profiles of host cells (Skalsky and Cullen, 2010; Fu et al., 2019; Seong et al., 2020). Due to the fact that differing changes with an intracellular miRNA expression profile are caused by different viruses, we can use the specific expression of some specific miRNAs in cells as specific biomarkers of viral infection (Atherton et al., 2019; Biswas et al., 2019). Understanding these changes following virus pathogenesis helps us to learn more about the characteristics of virus infection in different hosts or tissues and to seek meaningful miRNAs within the changing miRNA expression profiles (Ninomiya et al., 2016). These changes in miRNA expression profiles could be a strategy used by RNA viruses when escaping from the host cell immune system through the fine-tuning of miRNA on intracellular protein expression, or it could be an immune reaction, meaning that the body perceives the invasion of RNA viruses and fine-tunes immune-related genes through the miRNA system to coordinate the whole immune response. Since miRNA can bind to mRNA and regulate its translation, it is reasonable to suggest that the positive RNA from RNA viruses, which carry translatable genetic information, could be targeted and regulated by the host miRNAs (Zheng et al., 2018; Li et al., 2019; Chen et al., 2021). Up until now, there have been numerous experiments conducted to try and confirm this hypothesis. In addition, studies have found that some viruses actually produce miRNAs during an infection in order to manipulate host genes or RNA genes, thus helping self-replication, mostly noted in DNA viruses. For example, as a herpesvirus, Epstein-Barr Virus (EBV) was found to encode miRNA (Cai et al., 2006; Amoroso et al., 2011). Most herpesviruses are believed to encode miRNAs based on their genome size and the number of proteins they encode (Amoroso et al., 2011; Grundhoff and Sullivan, 2011). To date, the role of miRNAs encoded by RNA viruses, as opposed to those encoded by DNA viruses with distinct characteristics, remains controversial. So far, miRNAs have been identified in several RNA virus families, including Retroviridae, Orthomyxoviridae, Flaviviridae, Filoviridae, and Coronaviridae (Grundhoff and Sullivan, 2011; Nanbo et al., 2021). In summary, both the virus and the host can regulate the viral or host genes and thus control the virus proliferation process by encoding miRNAs.
The Process of miRNA Biogenesis
As shown in Figure 1, the miRNA genes are transcribed for primary miRNA (pri-miRNA). This occurs mainly via polymerase II and partly by polymerase III, which includes thousands of nucleotides and contains at least a hairpin structure (Ha and Kim, 2014). MiRNAs can be transcribed for the intron clusters of pre-mRNA, independent gene units, or long noncoding RNA (Stavast and Erkeland, 2019). Next, a “microprocessor” complex, mainly composed of the RNase III enzyme Drosha and DiGeorge critical region 8 (DGCR8) (named DGCR8 in mammals and Pasha in other animals), cuts the pri-miRNA to form stem-loop (S-L) precursor miRNAs (pre-miRNA) 60–90 nt long in the nucleus (Lee et al., 2003); then, the pre-miRNA is transferred into the cytoplasm using the export receptor, exportin-5 (Ohrt et al., 2006). The complex composed of Dicer (which belongs to the RNase III family) and transactivation-responsive RNA-binding protein (TRBP) cleaves the pre-miRNA apical loop within the cytoplasm to form double-stranded miRNAs about 22 nt in length (Bennasser et al., 2011; Kim et al., 2016). After the miRNA duplex has been transferred to the RNA-induced silencing complex, loading complex (RLC), which is mainly constituted by Argonaute (AGO) protein, and other proteins (involving translin-associated factor X, TRANSLIN and heat shock protein 90 in addition to others), the RNA duplex structure is unwound, and the passenger strand is ejected (Ye et al., 2011; Sahu et al., 2014). Different AGO proteins have distinct functions and cleavage activities that are probably determined by the PIWI domain of the AGO protein (Tang, 2005), and based on the cleavage activity of AGO, RNA-induced silencing complex (RISC) is divided into non-cleaving RISC and cleaving RISC. Subsequently, the miRNA-RISC (miRISC) complex recruits downstream factors, such as glycine–tryptophan protein of 182 kDa (GW182), including the trinucleotide repeat-containing gene 6A–6C (TNRC6A–TNRC6C), to mediate gene silencing associated with RNAi (Ding and Han, 2007; Fabian and Sonenberg, 2012).
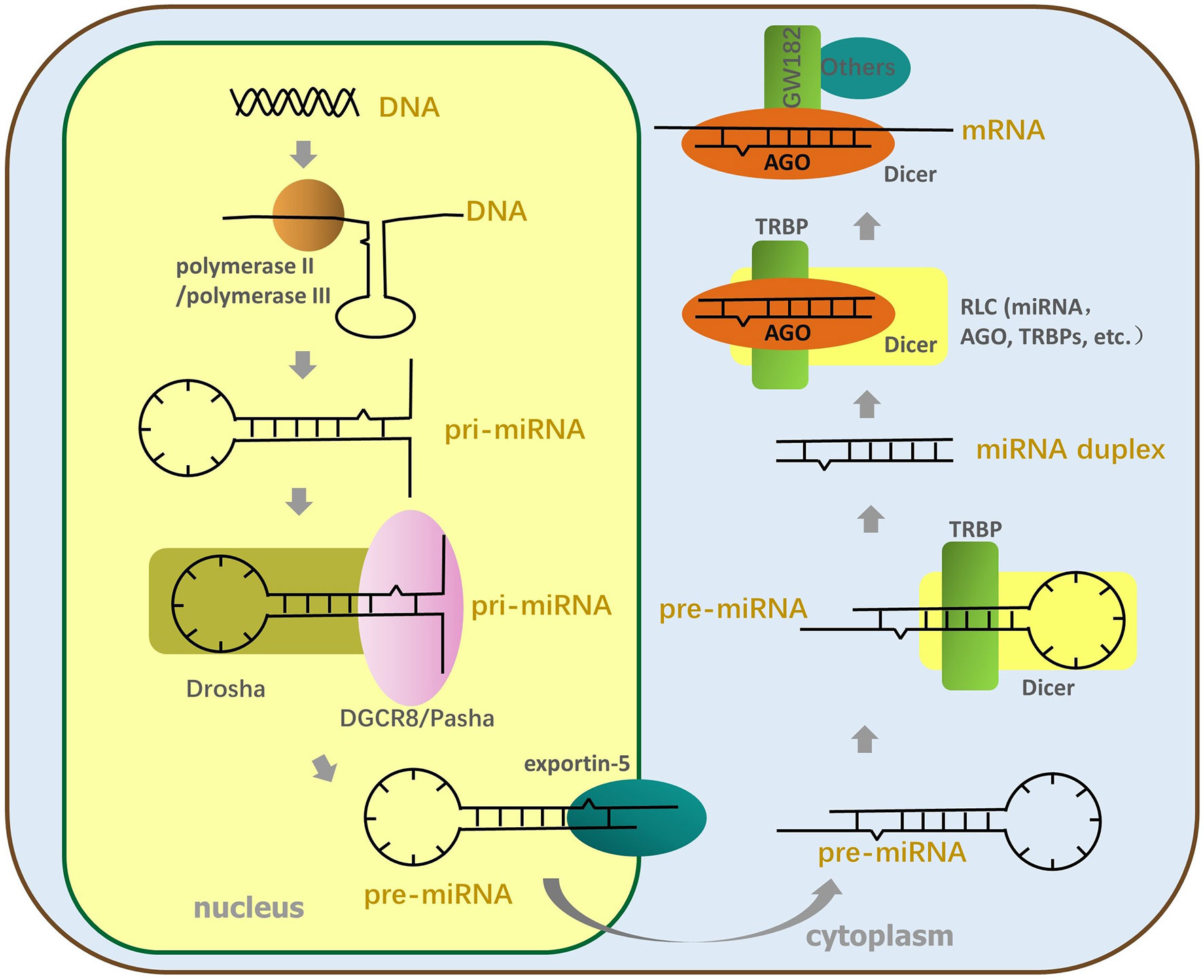
Figure 1 The process of miRNA formation. The miRNA genes are transcribed for pri-miRNA by polymerase II or polymerase III; then, Drosha and DGCR8 split the pri-miRNA to form SL pre-miRNA, transferred to the cytoplasm by the export receptor, exportin-5. Next, the TL element of pre-miRNA is cut off by Dicer and TRBP to produce miRNA duplex. The miRNA duplex is transferred to RLC constituted by AGO, Dicer, TRBP, and so on and is then unfastened twice. The end, mature single-stranded miRNA enters RISC, and AGO of RISC recruits downstream factors to perform RNA interference.
miRNAs Regulate Gene Expression
In general, the seed sequence (the 2-7 nucleotide sequences of the 5’ end of mature single-stranded miRNA) helps the RISC complex target transcripts through the principle of base complementation (Chendrimada et al., 2007; Eichhorn et al., 2014). But, in fact, the other bases on some miRNAs can also help the RISC complex to locate mRNA (Lim et al., 2005; Zheng et al., 2013; Agarwal et al., 2015; Zhang Y. et al., 2017; Jiang et al., 2020). After the formation of miRNA into miRISC complexes, the methodologies of the miRNAs mainly depend on the AGO proteins of the miRISC and the degree of complementarity between the targeted mRNA sequence and the core sequence of miRNA (Figure 2) (Tang, 2005; Loeb et al., 2012). When the transcript target and core sequence of the miRNA are perfectly complementary for cleaving miRISC, miRISC performs the slicing of the mRNA and efficiently degrades the mRNA (Tang, 2005; Ipsaro and Joshua-Tor, 2015). But, in animals, the transcript targets rarely provide complete complementary sequences to the core sequence of the miRNA, and AGO proteins do not all have cleavage activity, which precludes mRNA cleavage by AGO proteins (Ipsaro and Joshua-Tor, 2015). Owing to the fact that AGO proteins are insufficient alone to mediate silencing, miRISC uses the GW182 protein as a platform to recruit other proteins (Takimoto et al., 2009; Kobayashi and Tomari, 2016; Sheu-Gruttadauria and MacRae, 2018). The GW182 proteins have two special structures: an amino-terminal AGO-binding domain (ABD), which binds to AGO, and a silencing domain (SD), which interacts with silencing effectors such as cytoplasmic poly(A)-binding protein (PABP), the cytoplasmic deadenylase complexes PAN2-PAN3, and CCR4 NOT (Guo et al., 2010; Braun et al., 2013). Some scholars hold the view that the interaction of these proteins with GW182 proteins interferes with the PABPC1-eIF4G interaction, which results in a decline in translational efficiency and distorts the conformation of transfer RNAs, making the 5’ cap and poly(A) tail of mRNA more readily available to utilize to degrade mRNA (Elkayam et al., 2017; Niaz and Hussain, 2018). In contrast, there is another view that the interactions between PABPC and GW182 catalyze the miRNA-mediated deadenylation of target transcripts (Meijer et al., 2013; Niaz and Hussain, 2018).
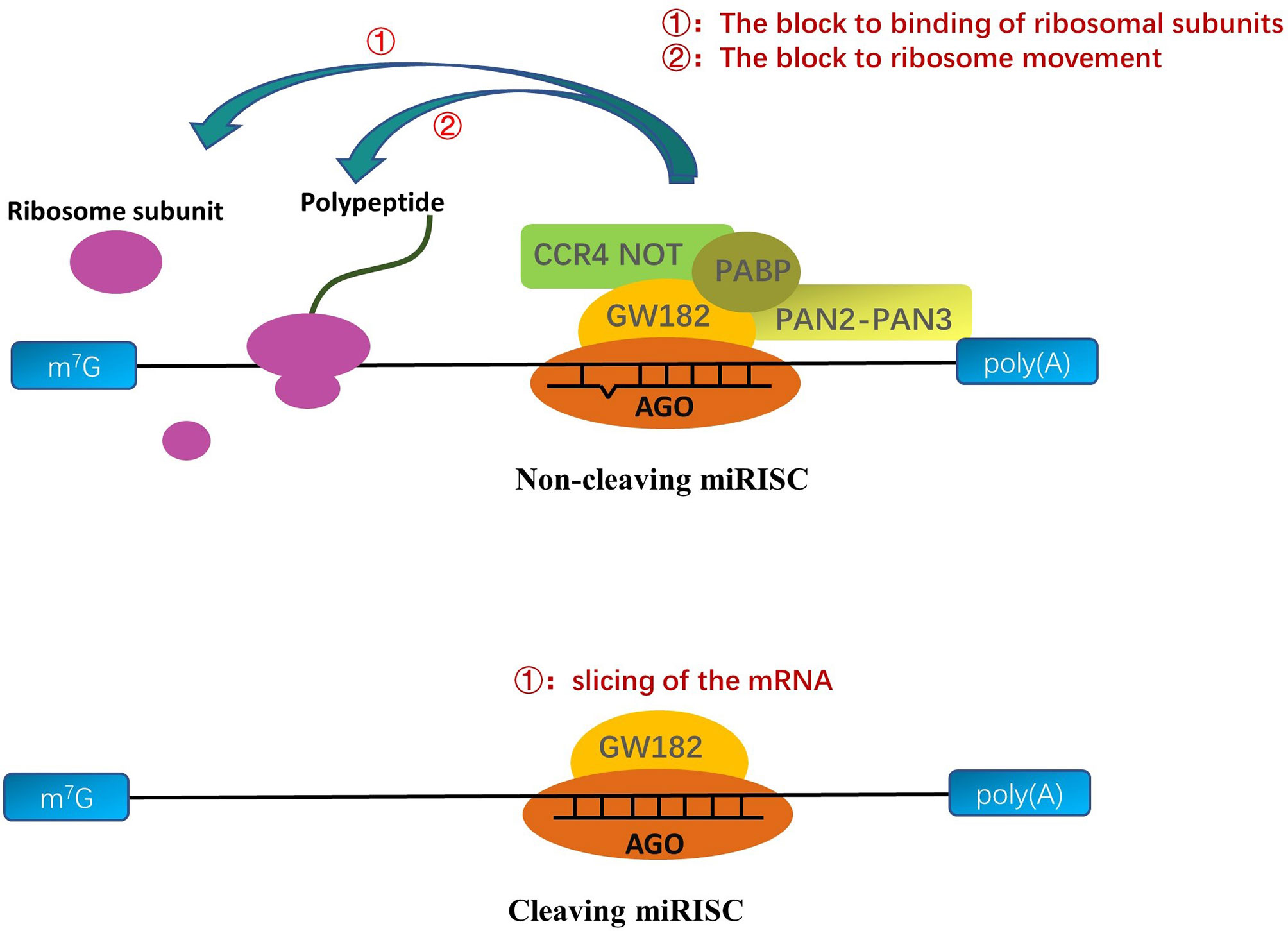
Figure 2 A molecular understanding of miRNA-mediated gene silencing. When the miRNA sequences are partly complementary to the targeted RNA sequences, the AGO protein binds to GW182; then, the complex recruits PABP, CCR4 NOT, and PAN2-PAN3 to interferences’ ribosome movement on the mRNA/viral RNA (vRNA) or prevents the binding of ribosomal large and small subunits to inhibit translation. When the miRNA sequences are perfect complementary to the targeted RNA sequences, cleaving-miRISC composed of miRNA and AGO protein cuts and degrades the targeted transfer RNA to inhibit translation.
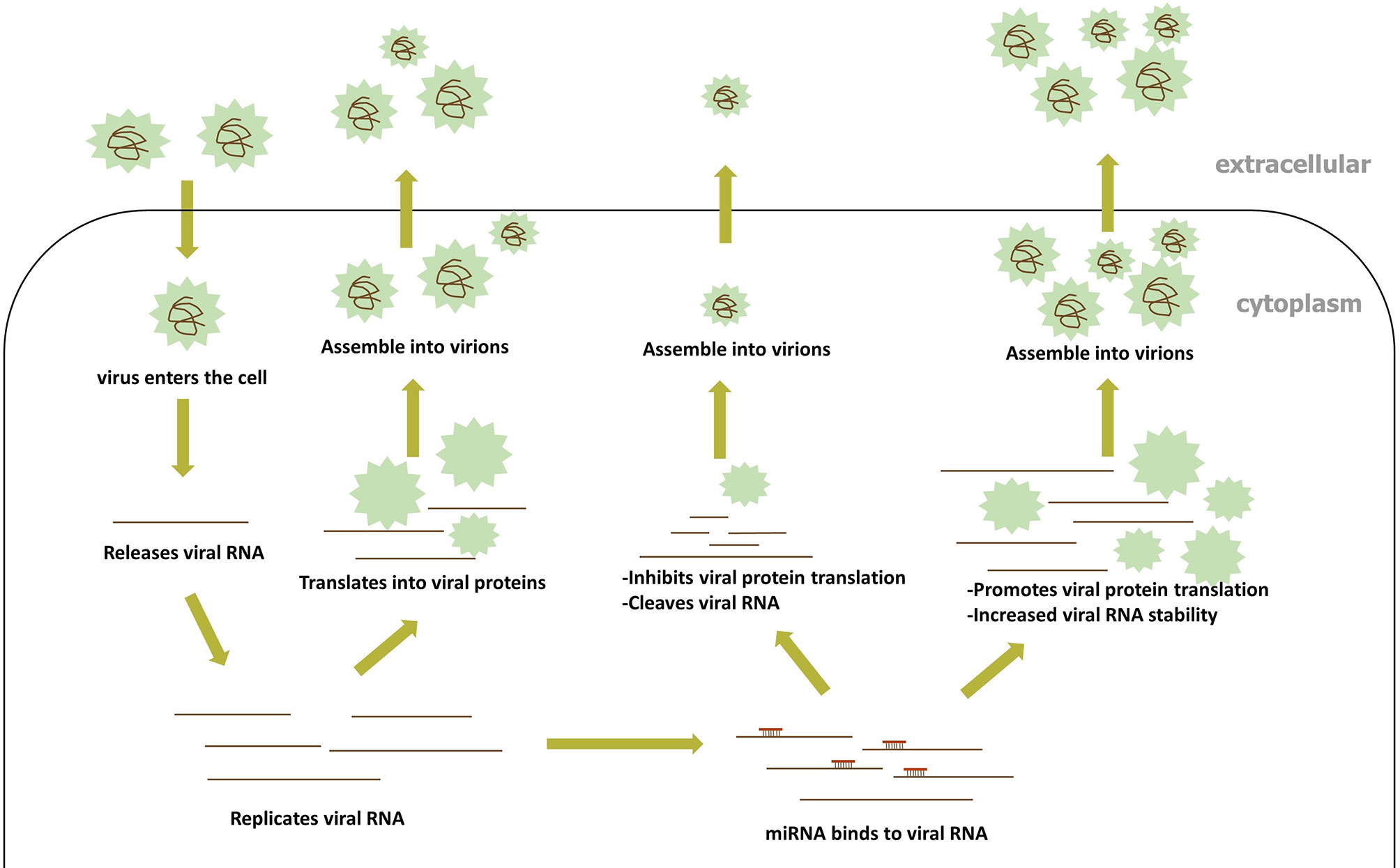
Accumulating evidence demonstrates that host miRNAs participate in the replication and pathogenesis of the viruses by binding directly to the RNA of many RNA viruses. During RNA virus infection, miRNA can then target RNA virus genes in a manner similar to the way they target host genes (Teterina et al., 2014; Bakre et al., 2017). In general, the 5’UTR and 3’UTR of the RNA virus gene have the most natural binding sites to miRNA, but evidence accumulated toward proving the presence of miRNA-binding sites in the RNA virus protein open reading frame (ORF). A positive-strand RNA virus genome can bind directly to the host’s miRNAs in the same way as host mRNA binding, while the positive-strand intermediate of dsRNA and negative RNA virus work in a similar fashion. The direct targeting of RNA viruses by miRNA encompasses two methods of modulating virus replication (Figure 3): (1) the inhibition of viral RNA translation reduces viral replication, and (2) changes in the stability of the viral RNA secondary structure; both methods are discussed in detail below.
Translation Inhibition by the Host miRNA Binding to the RNA Within the RNA Virus
Many relevant examples have proved that miRNA inhibits viral proliferation and replication by targeting viral RNA and gene silencing. For example, the infectious bursal disease virus (IBDV), a member of the family Birnaviridae, is targeted by gga-miR-21 that downregulates the expression of IBDV VP1 at the translational level rather than the mRNA level, which therefore represses IBDV replication (Wang et al., 2013). Similarly, the virulent newcastle disease virus (NDV) is a negative-strand RNA virus and is translated into viral protein after NDV has been transcribed for a positive-sense RNA upon entry into host cells. gga-miR-1603 and gga-miR-1794 (gga, chicken) were confirmed to target two highly conserved regions of the L gene of NDV, to inhibit the expression of the L protein at both the protein and RNA levels, thus suppressing NDV replication (Chen et al., 2021). Both of these examples are based on the RNA-silencing effects of miRNA in inhibiting protein translation within RNA viruses, and some more interesting examples of miRNA and RNA viruses are mentioned below.
Previously, only the single regulation of host miRNA on the host genes or viral genes was focused on. With further research, miRNA was later found to be able to simultaneously target both the host and virus gene to limit virus proliferation. A few examples to reference this procedure is described below. Studies have shown that gga-miR-130b in DF-1 cells can simultaneously inhibit viral replication by targeting the IBDV segment A and enhance the expression of type interferon by targeting the intracellular cytokine signal suppressor 5 (SOCS5) (Fu et al., 2017). The genomic segment B of IBDV binds to the gga-miR-454 of DF-1 to inhibit virus replication, while cellular cytokine signal suppressor 6 (SOCS6) is inhibited by gga-miR-454 to boost the immune response (Fu et al., 2018). The miR-324-5p directly targets the H5N1 virus PB1 gene and the cellular CUEDC2 gene, the negative regulator of the interferon pathway, to inhibit the gene expression of both genes, thus enhancing innate immunity (Kumar et al., 2018). In these cases, on the one hand, the host miRNA directly blocks the expression of the viral proteins by targeting viral genes, and on the other hand, increases the host immune response by inhibiting the expression of negative immune regulators in the host, in order to concomitantly limit viral proliferation in vivo. Another example is the porcine epidemic diarrhea virus (PEDV), which belongs to the genus Alphacoronavirus of the Coronavirus family, a single-stranded, positive-sense RNA virus. It was confirmed that miR-221-5p acted as a dose-dependent inhibitor for PEDV, by directly targeting the 3’UTR of the PEDV genomic RNA, and stimulating NF-κB signaling via p65 nuclear translocation, to upregulate IFN-β and related ISGs (Zheng et al., 2018). Due to the mechanism used by miRNAs when binding to the RNA described, and the examples above, miRNAs can simultaneously target both viral and host genes, cocreating an environment that affects viral replication.
In the course of a previously published study, it was found that mutations in miRNA targets within some RNA viruses could render the functions of miRNAs targeting RNA viruses ineffective. These phenomena probably occur because RNA polymerases lack the same collation activity as DNA polymerases and miRNA sequences are highly conserved, thus allowing RNA viruses to mutate and fleetly escape from the host’s miRNA inhibition. For instance, as a member of the Orthomyxoviridae family, the influenza A virus (IAV) is a negative single-stranded RNA virus and has multiple subtype strains. A study verified that hsa-miR- 1307-3p, as a novel potent suppressor, could bind directly to the NS1 RNA of IAV to suppress NS1 expression and influenza virus replication, but a new mutation has been identified in the NS1 gene, which is targeted by hsa-miR- 1307-3p, of more than 100 strain types represented by A(H1N1)pdm09 (a subtype strain of IAV), which invalidated the inhibition of miRNA (Bavagnoli et al., 2019). Analogously, ssc-miR-204 and ssc-miR-4331 only exhibited an inhibitory effect on SIV-H1N1/2009 (a subtype strain of IAV) replication due to mutations in the peer sequences of other strains (Zhang S. et al., 2017). Many similar examples have led to the viewpoint that mutations may represent a means by which RNA viruses evade the direct binding inhibition effects of the host miRNAs. To avoid the influence of gene mutation on the RNA virus protein expression is critical as such mutations are synonymous in many similar findings under natural conditions. Another example, enterovirus 71 (EV71) of the family Picornaviridae, a single-strand positive-sense RNA virus, could have its VP1 and VP3 protein levels regulated by hsa-miR-296-5p to inhibit virus infection (Zheng et al., 2013). Whereas, by aligning the sequences of hsa-miR-296-5p binding to the genome in each EV71 subtype, some strains were found to have synonymous mutations in the same location, research confirmed that these viruses could escape the suppression of hsa-miR-296-5p (Zheng et al., 2013). However, many RNA viruses are still inhibited by the host miRNA. The randomness and uncertainty of mutations may make correct mutations less efficient. Moreover, the short-term inhibition of miRNAs on virus replication may not hurt or hinder the entire process of virus proliferation, so not all RNA viruses need to escape the effects of miRNAs. Naturally, miRNA-binding sites in RNA viruses that create no positive effects on viral replication are more likely to have been deleted in vivo than miRNA-binding sites that have positive effects on viral replication.
Given the evidence pointing toward the importance of binding sites, scholars studied the highly conserved miRNA-binding sites in RNA viruses. It was found that the antiviral activities of miRNAs caused by direct targeting RNA viruses may be used by the virus to evade immunity, prolong incubation periods, or even increase the virulence of the virus during evolution. Under some conditions, the downregulation of virus replication induced by miRNA may in fact be necessary for the persistence of viral infection (Mahajan et al., 2009). Studies have shown that the eastern equine encephalitis virus (EEEV) utilized the inhibitory effect of miRNAs to prevent the virus from being detected by the immune system prematurely, resulting in the enhanced neurovirulence of the virus. EEEV, a single-stranded positive-sense RNA mosquito-borne alphavirus, encodes four strongly conserved miRNA-binding sites for has-miR-142-3p in myeloid-lineage cells (Trobaugh et al., 2014). The hematopoietic cell-specific miR-142-3p restricts EEEV replication to determine virus tropism and to suppress innate immune responses in the myeloid cell, which is conducive to microbial infection and exacerbates EEEV neurovirulence (Trobaugh et al., 2014). The presence of the miRNA-binding sequences is necessary for efficient EEEV replication in mosquitoes, so it is legitimate to assume that these sites are essential for virus transmission (Trobaugh et al., 2019a). For EEEV, the number of miR-142-3p-binding sites is dominant in the suppression of EEEV replication. Even though the miRNA inhibits EEEV expression, the inhibitory effect of miRNA on EEEV is very important for both prolonging the incubation period and increasing virulence during the whole virus proliferation process in vivo. Besides this, after human immunodeficiency virus type 1 (HIV-1)- infected resting primary CD4+ T cells, the significantly enriched miRNAs including, miR-28, miR-150, miR-223, and miR-382 potently inhibited HIV-1 production by targeting the 3’UTR of the HIV-1 RNA and related host proteins (Huang et al., 2007). The inhibition of related host proteins including Tat and Rev, which are key factors for transcription and translation in viral RNA, could further strengthen the viral incubation period to help the virus multiply (Huang et al., 2007). Additionally, studies have shown that miR-326, miR-196b, and miR-1290 have similar effects (Houzet et al., 2012; Wang et al., 2015). The chikungunya virus (CHIKV) is a positive-sense single-stranded RNA virus in the Alphavirus genus, transmitted by Aedes aegypti (Ae. aegypti) mosquitoes. Ae. aegypti miR-2944b-5p can bind to the 3’UTR of CHIKV and reduce CHIKV replication, meanwhile regulating the cellular target, vps-13 (vacuolar protein sorting) (Dubey et al., 2019). CHIKV may be using miR-2944b-5p, alongside its target vps-13, to maintain the cellular integrity of the mitochondrial membrane potential, therefore helping the CHIKV survive in mosquito cells (Dubey et al., 2019). This miRNA can inhibit excessive proliferation of CHIKV in the intermediate host, and maintain the titer of the virus in the intermediate host together with the genes of the intermediate host in vivo, which also contributes toward the transmission of the virus. In general, the highly conserved miRNA-binding sites in RNA viruses have positive implications for virus survival.
miRNA Influence Upon the Spatial Structure of Viral RNA
It is already known that RNA has a certain spatial structure under natural conditions. This did lead to the question as to whether the binding of host miRNA to viral RNA could alter viral RNA to enable interactions with other proteins by changing the spatial structure of viral RNA once the RNA virus enters the host cell to begin replication or translation. There have been some specific examples that have helped elucidate this matter. The hepatitis C virus (HCV), a single-stranded RNA virus with the Flaviviridae family, only has an ORF and requires liver-specific miR-122 interactions with the sequences from the HCV RNA 5’UTR to maintain a high viral RNA abundance in the liver (Jopling, 2008). The 5’UTR and 3’UTR of the HCV RNA have four and three SL structures, respectively, and the ORF region encodes four structural proteins and six non-structural proteins (Jopling, 2008). Two miR-122-binding sites in the 5’UTR, which are separated by a highly conserved 14-nucleotide sequence, are occupied by miR-122 to synergistically give HCV RNA higher stability rather than inhibiting viral replication through RNAi-related functions (Jopling, 2008). Some studies have suggested that efficient HCV replication require the annealing binding of miR-122 to these two sites, which then forms a trimolecular RNA structure that is essential for efficient virus proliferation (Amador-Cañizares et al., 2018; Chahal et al., 2019). However, some scholars believe that any small RNA binding to the miR-122-binding sites on the HCV 5’UTR can promote the HCV life cycle (Kunden et al., 2020). Furthermore, some researchers have found that the insertion of the viral miR-122-binding site into the 3’NCR of a reporter mRNA leads to the downregulation of mRNA expression, which implies that the location of the miRNA-binding site may dictate its effects on gene regulation (Jopling, 2008; Jopling et al., 2008). Further research has shown that miR-122 binds to the 5’NTR of the HCV gene, to protect uncapped HCV RNA genes by defending the degradation caused by 5’ exonuclease Xrn1 and Xrn2, whereas miR-122 negatively regulates the expression of the reporter mRNA by binding to the 3’NCR of the mRNA in a general manner (Li et al., 2013; Sedano and Sarnow, 2014). This interaction promotes HCV RNA accumulation by stabilizing viral RNA, leading to changes in the secondary structure of the viral genome, stimulating the generation of the canonical HCV IRES RNA structure (Schult et al., 2018; Chahal et al., 2019; Kunden et al., 2020). The multiple effects of miR-122 on HCV proliferation are based on the changes observed to the HCV secondary structure after miRNA has bound to viral RNA. Some similar examples are also described in other systems. When the dengue virus (DENV) infects human cells in vitro, endogenous miR-548g-3p can bind to the conserved stem-loop promoter (SLA) (a promoter for DENV RNA transcription) in the 5’NTR of the DENV genome to suppress DENV intracellular replication (Wen et al., 2015). Based on this study, it can be inferred that the combination of miR-548g-3p and the targeted site of DENV, the natural SL structure of the promoter element needed to regulate the binding efficiency of DENV promoter and RNA-dependent viral RNA polymerase may be destroyed (Yu et al., 2008; Wen et al., 2015). Besides this example, the bovine viral diarrhea virus is also targeted by miR-17 and let-7 through binding to S2 and S1 present in the virus genome 3’-NTR, respectively (Scheel et al., 2016; Kokkonos et al., 2020). AGO2 and miR-17 binding were essential for viral replication, whereas let-7 binding increased virus translation.
These examples indicate the importance of the miRNA binding site location for targeting the virus RNA. If the binding site is located in the folding of the secondary structure of RNA, SL structure, or the binding site of some proteins, the corresponding miRNA may have an important influence on the expression or function of this genome (Gerresheim et al., 2017). Interestingly, the unconventional relationship between miRNA and viral RNA has also been found in some other RNA viruses, which most or all enhancing the stability of the viral RNA to positively regulate RNA virus replication.
Conclusions
Eukaryotic miRNAs participate in the regulation of most physiological activities within cells, including RNA virus infection. RNA viruses use RNA as a gene carrier, and miRNAs bind to RNA in a base complementary way, which allows some miRNAs to directly target the RNA within RNA viruses. Most of this binding occurs in the ORF or 3’UTR of the RNA virus RNA, whereas miRNA works with RNA-binding proteins to inhibit RNA viral replication. In addition, some cellular miRNAs are able to bind to RNA viral RNA to avoid the cleavage of the related enzymes or to stabilize the spatial structure of the RNA to positively regulate RNA virus replication. However, as there is a lack of any proofreading function, RNA viruses can escape from miRNA through gene mutation in the face of the miRNA’s direct targeted inhibition. From the above-cited publications, which include many examples, on the one hand, RNA viruses utilize some specific cellular miRNAs to create an intracellular environment conducive to the transmission of viral infection. Yet, on the other hand, it is commonplace that most miRNAs that directly or indirectly strongly inhibit RNA virus replication are manipulated by the RNA virus to reduce expression, or are shunned by RNA viruses with genetic mutations. Moreover, miRNAs in eukaryotes are highly conserved and are not easily mutated, which makes it difficult for miRNAs to be dominant antiviral factors against RNA viruses in nature.
Research has shown that it is feasible for researchers to insert the binding sequences of intracellular specific miRNA into the RNA virus genes in order to prepare miRNA vaccines according to the inhibition of targeted RNAs by some miRNAs (Tsetsarkin et al., 2015). The insertion of single or multiple copies of the brain-expressed miRNA target sequence in the 3’NTR of the genome of the neurotropic chimeric tick-borne encephalitis virus/dengue virus 4 (TBEV/DEN4) flavivirus could reduce the neurotoxicity of the virus (Heiss et al., 2012). However, the artificial RNA virus can also escape from miRNA to regain its neurotoxicity by accumulating gene mutations or via deletion (Heiss et al., 2012). This phenomenon can be alleviated by increasing the copy number of inserted sequences and setting appropriate spacing. Similarly, the deletion of the miR-142-3p-binding sites in the EEEV 3’ UTR can lead to efficient EEEV infection of myeloid cells, resulting in a decrease in the virulence of the virus, but the accumulation of genetic mutations may restore virus virulence during serial passages of the virus (Trobaugh et al., 2019b). How to stabilize the artificial sequence while also inhibiting the virulence of the virus needs further research. Host miRNA is hijacked by invading RNA viruses and participates in the pathogenic process of viruses, or miRNAs inhibit RNA virus replication by targeting their RNA, which makes it reasonable for miRNA to be a potential new therapeutic target for RNA viral therapy. There have been attempts to develop antiviral drugs based on miRNA studies. In the laboratory, artificial miRNAs are designed to target the conserved genomic regions of the West Nile virus NS5 and NS2A or consensus sequence of 3’NTR of JEV, and all show effective suppression of the virus in vitro (Sharma et al., 2018; Karothia et al., 2020). Scientists have tried to use similar RNAi technology to treat some viral diseases in clinical trials, such as the inhibition of HBV replication in chronic HBV infection (Wooddell et al., 2017; van den Berg et al., 2020), treating Ebola virus patients to improve survival and recovery rates (Thi et al., 2015) and targeting HIV-1 replication for cell transplant therapy (Scarborough and Gatignol, 2017). It is not only miRNAs in animals that can limit the influence of RNA viruses, as miRNAs in plants have also been found to inhibit the replication of certain viruses. MiR-2911 from honeysuckle (a traditional Chinese medicine) can suppress IAVs with a broad spectrum by directly targeting the IAV RNA, and the let-7a from this plant can inhibit the replication of DENV by targeting its NS1 sequence (Zhou et al., 2015; Lee et al., 2017).
Intracellular miRNA, as a link of the intracellular regulatory network, can simultaneously target multiple cellular mRNAs and the RNA of the invading RNA virus. Cellular miRNAs, the host mRNA, and RNA viruses are closely related and interact with each other, affecting the whole body. This connection makes miRNAs a potential breakthrough in the future to tackle the current ravages of many RNA viruses. However, in order to be an antiviral drug, miRNA requires comprehensive consideration for the variable of cell homeostasis and other physiological conditions caused by changes in the miRNA. Moreover, a large number of convenient, cheap, and accurate synthesis methods of artificial miRNA still need further study. At present, miRNA as a new approach against RNA viruses is still far from being realized, and a breakthrough in the field is expected.
Author Contributions
LL was responsible for the collection and analysis of materials and organizing and writing the manuscript. RJ, AC, and MW gave the main modification reviews and suggestions. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31872475), Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (CARS-SVDIP), and China Agricultural Research System (CARS-42-17).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the team of “Research Center of Avian Disease” and the National Natural Science Foundation of China, and they provide a good platform and financial support. The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn/) for the expert linguistic services provided.
References
Agarwal, V., Bell, G. W., Nam, J. W., Bartel, D. P. (2015). Predicting Effective microRNA Target Sites in Mammalian mRNAs. Elife 4, e05005. doi: 10.7554/eLife.05005
Aguado, L. C., TenOever, B. (2018). RNA Virus Building blocks-miRNAs Not Included. PloS Pathog. 14 (5), e1006963. doi: 10.1371/journal.ppat.1006963
Amador-Cañizares, Y., Panigrahi, M., Huys, A., Kunden, R. D., Adams, H. M., Schinold, M. J., et al. (2018). miR-122, Small RNA Annealing and Sequence Mutations Alter the Predicted Structure of the Hepatitis C Virus 5’ UTR RNA to Stabilize and Promote Viral RNA Accumulation. Nucleic Acids Res. 46 (18), 9776–9792. doi: 10.1093/nar/gky662
Amoroso, R., Fitzsimmons, L., Thomas, W. A., Kelly, G. L., Rowe, M., Bell, A. I. (2011). Quantitative Studies of Epstein-Barr Virus-Encoded microRNAs Provide Novel Insights Into Their Regulation. J. Virol. 85 (2), 996–1010. doi: 10.1128/jvi.01528-10
Atherton, L. J., Jorquera, P. A., Bakre, A. A., Tripp, R. A. (2019). Determining Immune and miRNA Biomarkers Related to Respiratory Syncytial Virus (RSV) Vaccine Types. Front. Immunol. 10. doi: 10.3389/fimmu.2019.02323
Bakre, A. A., Shim, B. S., Tripp, R. A. (2017). MicroRNA-134 Regulates Poliovirus Replication by IRES Targeting. Sci. Rep. 7 (1), 12664. doi: 10.1038/s41598-017-12860-z
Bavagnoli, L., Campanini, G., Forte, M., Ceccotti, G., Percivalle, E., Bione, S., et al. (2019). Identification of a Novel Antiviral Micro-RNA Targeting the NS1 Protein of the H1N1 Pandemic Human Influenza Virus and a Corresponding Viral Escape Mutation. Antiviral Res. 171, 104593. doi: 10.1016/j.antiviral.2019.104593
Bennasser, Y., Chable-Bessia, C., Triboulet, R., Gibbings, D., Gwizdek, C., Dargemont, C., et al. (2011). Competition for XPO5 Binding Between Dicer mRNA, pre-miRNA and Viral RNA Regulates Human Dicer Levels. Nat. Struct. Mol. Biol. 18 (3), 323–327. doi: 10.1038/nsmb.1987
Biswas, S., Haleyurgirisetty, M., Lee, S., Hewlett, I., Devadas, K. (2019). Development and Validation of Plasma miRNA Biomarker Signature Panel for the Detection of Early HIV-1 Infection. EBioMedicine 43, 307–316. doi: 10.1016/j.ebiom.2019.04.023
Braun, J. E., Huntzinger, E., Izaurralde, E. (2013). The Role of GW182 Proteins in miRNA-Mediated Gene Silencing. Adv. Exp. Med. Biol. 768, 147–163. doi: 10.1007/978-1-4614-5107-5_9
Cai, X., Schäfer, A., Lu, S., Bilello, J. P., Desrosiers, R. C., Edwards, R., et al. (2006). Epstein-Barr Virus microRNAs are Evolutionarily Conserved and Differentially Expressed. PloS Pathog. 2 (3), e23. doi: 10.1371/journal.ppat.0020023
Catalanotto, C., Cogoni, C., Zardo, G. (2016). MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 17 (10), 1712. doi: 10.3390/ijms17101712
Chahal, J., Gebert, L. F. R., Gan, H. H., Camacho, E., Gunsalus, K. C., MacRae, I. J., et al. (2019). miR-122 and Ago Interactions With the HCV Genome Alter the Structure of the Viral 5’ Terminus. Nucleic Acids Res. 47 (10), 5307–5324. doi: 10.1093/nar/gkz194
Chendrimada, T. P., Finn, K. J., Ji, X., Baillat, D., Gregory, R. I., Liebhaber, S. A., et al. (2007). MicroRNA Silencing Through RISC Recruitment of Eif6. Nature 447 (7146), 823–828. doi: 10.1038/nature05841
Chen, Y., Shen, A., Rider, P. J., Yu, Y., Wu, K., Mu, Y., et al. (2011). A Liver-Specific microRNA Binds to a Highly Conserved RNA Sequence of Hepatitis B Virus and Negatively Regulates Viral Gene Expression and Replication. FASEB J. 25 (12), 4511–4521. doi: 10.1096/fj.11-187781
Chen, Y., Zhu, S., Hu, J., Hu, Z., Liu, X., Wang, X., et al. (2021). gga-miR-1603 and gga-miR-1794 Directly Target Viral L Gene and Function as a Broad-Spectrum Antiviral Factor Against NDV Replication. Virulence 12 (1), 45–56. doi: 10.1080/21505594.2020.1864136
Diallo, I., Ho, J., Laffont, B., Laugier, J., Benmoussa, A., Lambert, M., et al. (2021). Altered microRNA Transcriptome in Cultured Human Liver Cells Upon Infection With Ebola Virus. Int. J. Mol. Sci. 22 (7), 3792. doi: 10.3390/ijms22073792
Ding, L., Han, M. (2007). GW182 Family Proteins are Crucial for microRNA-Mediated Gene Silencing. Trends Cell Biol. 17 (8), 411–416. doi: 10.1016/j.tcb.2007.06.003
Dubey, S. K., Shrinet, J., Sunil, S. (2019). Aedes Aegypti microRNA, miR-2944b-5p Interacts With 3’UTR of Chikungunya Virus and Cellular Target Vps-13 to Regulate Viral Replication. PloS Negl. Trop. Dis. 13 (6), e0007429. doi: 10.1371/journal.pntd.0007429
Eichhorn, S. W., Guo, H., McGeary, S. E., Rodriguez-Mias, R. A., Shin, C., Baek, D., et al. (2014). mRNA Destabilization is the Dominant Effect of Mammalian microRNAs by the Time Substantial Repression Ensues. Mol. Cell 56 (1), 104–115. doi: 10.1016/j.molcel.2014.08.028
Elkayam, E., Faehnle, C. R., Morales, M., Sun, J., Li, H., Joshua-Tor, L. (2017). Multivalent Recruitment of Human Argonaute by GW182. Mol. Cell 67 (4), 646–658.e643. doi: 10.1016/j.molcel.2017.07.007
Fabian, M. R., Sonenberg, N. (2012). The Mechanics of miRNA-Mediated Gene Silencing: A Look Under the Hood of miRISC. Nat. Struct. Mol. Biol. 19 (6), 586–593. doi: 10.1038/nsmb.2296
Fu, M., Wang, B., Chen, X., He, Z., Wang, Y., Li, X., et al. (2017). MicroRNA gga-miR-130b Suppresses Infectious Bursal Disease Virus Replication via Targeting of the Viral Genome and Cellular Suppressors of Cytokine Signaling 5. J. Virol. 92 (1), e01646–e01617. doi: 10.1128/jvi.01646-17
Fu, M., Wang, B., Chen, X., He, Z., Wang, Y., Li, X., et al. (2018). gga-miR-454 Suppresses Infectious Bursal Disease Virus (IBDV) Replication via Directly Targeting IBDV Genomic Segment B and Cellular Suppressors of Cytokine Signaling 6 (Socs6). Virus Res. 252, 29–40. doi: 10.1016/j.virusres.2018.05.015
Fu, Y., Zhang, L., Zhang, R., Xu, S., Wang, H., Jin, Y., et al. (2019). Enterovirus 71 Suppresses miR-17-92 Cluster Through Up-Regulating Methylation of the miRNA Promoter. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00625
Gerresheim, G. K., Dünnes, N., Nieder-Röhrmann, A., Shalamova, L. A., Fricke, M., Hofacker, I., et al. (2017). microRNA-122 Target Sites in the Hepatitis C Virus RNA NS5B Coding Region and 3’ Untranslated Region: Function in Replication and Influence of RNA Secondary Structure. Cell Mol. Life Sci. 74 (4), 747–760. doi: 10.1007/s00018-016-2377-9
Grundhoff, A., Sullivan, C. S. (2011). Virus-Encoded microRNAs. Virology 411 (2), 325–343. doi: 10.1016/j.virol.2011.01.002
Guo, H., Ingolia, N. T., Weissman, J. S., Bartel, D. P. (2010). Mammalian microRNAs Predominantly Act to Decrease Target mRNA Levels. Nature 466 (7308), 835–840. doi: 10.1038/nature09267
Ha, M., Kim, V. N. (2014). Regulation of microRNA Biogenesis. Nat. Rev. Mol. Cell Biol. 15 (8), 509–524. doi: 10.1038/nrm3838
Heiss, B. L., Maximova, O. A., Thach, D. C., Speicher, J. M., Pletnev, A. G. (2012). MicroRNA Targeting of Neurotropic Flavivirus: Effective Control of Virus Escape and Reversion to Neurovirulent Phenotype. J. Virol. 86 (10), 5647–5659. doi: 10.1128/jvi.07125-11
Hong, Y., Lee, R. C., Ambros, V. (2000). Structure and Function Analysis of LIN-14, A Temporal Regulator of Postembryonic Developmental Events in Caenorhabditis Elegans. Mol. Cell Biol. 20 (6), 2285–2295. doi: 10.1128/mcb.20.6.2285-2295.2000
Houzet, L., Klase, Z., Yeung, M. L., Wu, A., Le, S. Y., Quiñones, M., et al. (2012). The Extent of Sequence Complementarity Correlates With the Potency of Cellular miRNA-Mediated Restriction of HIV-1. Nucleic Acids Res. 40 (22), 11684–11696. doi: 10.1093/nar/gks912
Huang, J., Wang, F., Argyris, E., Chen, K., Liang, Z., Tian, H., et al. (2007). Cellular microRNAs Contribute to HIV-1 Latency in Resting Primary CD4+ T Lymphocytes. Nat. Med. 13 (10), 1241–1247. doi: 10.1038/nm1639
Ipsaro, J. J., Joshua-Tor, L. (2015). From Guide to Target: Molecular Insights Into Eukaryotic RNA-Interference Machinery. Nat. Struct. Mol. Biol. 22 (1), 20–28. doi: 10.1038/nsmb.2931
Jiang, H., Bai, L., Ji, L., Bai, Z., Su, J., Qin, T., et al. (2020). Degradation of MicroRNA miR-466d-3p by Japanese Encephalitis Virus NS3 Facilitates Viral Replication and Interleukin-1β Expression. J. Virol. 94 (15), e00294–e00220. doi: 10.1128/jvi.00294-20
Jopling, C. L. (2008). Regulation of Hepatitis C Virus by microRNA-122. Biochem. Soc. Trans. 36 (Pt 6), 1220–1223. doi: 10.1042/bst0361220
Jopling, C. L., Schütz, S., Sarnow, P. (2008). Position-Dependent Function for a Tandem microRNA miR-122-Binding Site Located in the Hepatitis C Virus RNA Genome. Cell Host Microbe 4 (1), 77–85. doi: 10.1016/j.chom.2008.05.013
Karothia, D., Kumar Dash, P., Parida, M., Bhagyawant, S. S., Kumar, J. S. (2020). Vector Derived Artificial miRNA Mediated Inhibition of West Nile Virus Replication and Protein Expression. Gene 729, 144300. doi: 10.1016/j.gene.2019.144300
Kim, Y. K., Kim, B., Kim, V. N. (2016). Re-Evaluation of the Roles of DROSHA, Export in 5, and DICER in microRNA Biogenesis. Proc. Natl. Acad. Sci. U.S.A. 113 (13), E1881–E1889. doi: 10.1073/pnas.1602532113
Kobayashi, H., Tomari, Y. (2016). RISC Assembly: Coordination Between Small RNAs and Argonaute Proteins. Biochim. Biophys. Acta 1859 (1), 71–81. doi: 10.1016/j.bbagrm.2015.08.007
Kokkonos, K. G., Fossat, N., Nielsen, L., Holm, C., Hepkema, W. M., Bukh, J., et al. (2020). Evolutionary Selection of Pestivirus Variants With Altered or No microRNA Dependency. Nucleic Acids Res. 48 (10), 5555–5571. doi: 10.1093/nar/gkaa300
Kumar, A., Kumar, A., Ingle, H., Kumar, S., Mishra, R., Verma, M. K., et al. (2018). MicroRNA hsa-miR-324-5p Suppresses H5N1 Virus Replication by Targeting the Viral PB1 and Host Cuedc2. J. Virol. 92 (19), e01057–e01018. doi: 10.1128/jvi.01057-18
Kunden, R. D., Khan, J. Q., Ghezelbash, S., Wilson, J. A. (2020). The Role of the Liver-Specific microRNA, miRNA-122 in the HCV Replication Cycle. Int. J. Mol. Sci. 21 (16), 5677. doi: 10.3390/ijms21165677
Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., et al. (2003). The Nuclear RNase III Drosha Initiates microRNA Processing. Nature 425 (6956), 415–419. doi: 10.1038/nature01957
Lee, Y. R., Yeh, S. F., Ruan, X. M., Zhang, H., Hsu, S. D., Huang, H. D., et al. (2017). Honeysuckle Aqueous Extract and Induced Let-7a Suppress Dengue Virus Type 2 Replication and Pathogenesis. J. Ethnopharmacol. 198, 109–121. doi: 10.1016/j.jep.2016.12.049
Li, Y., Masaki, T., Yamane, D., McGivern, D. R., Lemon, S. M. (2013). Competing and Noncompeting Activities of miR-122 and the 5’ Exonuclease Xrn1 in Regulation of Hepatitis C Virus Replication. Proc. Natl. Acad. Sci. U.S.A. 110 (5), 1881–1886. doi: 10.1073/pnas.1213515110
Lim, L. P., Lau, N. C., Garrett-Engele, P., Grimson, A., Schelter, J. M., Castle, J., et al. (2005). Microarray Analysis Shows That Some microRNAs Downregulate Large Numbers of Target mRNAs. Nature 433 (7027), 769–773. doi: 10.1038/nature03315
Li, Y., Wang, L., Rivera-Serrano, E. E., Chen, X., Lemon, S. M. (2019). TNRC6 Proteins Modulate Hepatitis C Virus Replication by Spatially Regulating the Binding of miR-122/Ago2 Complexes to Viral RNA. Nucleic Acids Res. 47 (12), 6411–6424. doi: 10.1093/nar/gkz278
Loeb, G. B., Khan, A. A., Canner, D., Hiatt, J. B., Shendure, J., Darnell, R. B., et al. (2012). Transcriptome-Wide miR-155 Binding Map Reveals Widespread Noncanonical microRNA Targeting. Mol. Cell 48 (5), 760–770. doi: 10.1016/j.molcel.2012.10.002
Mahajan, V. S., Drake, A., Chen, J. (2009). Virus-Specific Host miRNAs: Antiviral Defenses or Promoters of Persistent Infection? Trends Immunol. 30 (1), 1–7. doi: 10.1016/j.it.2008.08.009
Maillard, P. V., Ciaudo, C., Marchais, A., Li, Y., Jay, F., Ding, S. W., et al. (2013). Antiviral RNA Interference in Mammalian Cells. Science 342 (6155), 235–238. doi: 10.1126/science.1241930
Maillard, P. V., van der Veen, A. G., Poirier, E. Z., Reis e Sousa, C. (2019). Slicing and Dicing Viruses: Antiviral RNA Interference in Mammals. EMBO J. 38 (8), e100941. doi: 10.15252/embj.2018100941
Mazewski, C., Perez, R. E., Fish, E. N., Platanias, L. C. (2020). Type I Interferon (IFN)-Regulated Activation of Canonical and Non-Canonical Signaling Pathways. Front. Immunol. 11. doi: 10.3389/fimmu.2020.606456
Meijer, H. A., Kong, Y. W., Lu, W. T., Wilczynska, A., Spriggs, R. V., Robinson, S. W., et al. (2013). Translational Repression and Eif4a2 Activity are Critical for microRNA-Mediated Gene Regulation. Science 340 (6128), 82–85. doi: 10.1126/science.1231197
Moss, E. G., Lee, R. C., Ambros, V. (1997). The Cold Shock Domain Protein LIN-28 Controls Developmental Timing in C. Elegans and is Regulated by the Lin-4 RNA. Cell 88 (5), 637–646. doi: 10.1016/s0092-8674(00)81906-6
Nanbo, A., Furuyama, W., Lin, Z. (2021). RNA Virus-Encoded miRNAs: Current Insights and Future Challenges. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.679210
Niaz, S., Hussain, M. U. (2018). Role of GW182 Protein in the Cell. Int. J. Biochem. Cell Biol. 101, 29–38. doi: 10.1016/j.biocel.2018.05.009
Ninomiya, M., Kondo, Y., Kimura, O., Funayama, R., Nagashima, T., Kogure, T., et al. (2016). The Expression of miR-125b-5p is Increased in the Serum of Patients With Chronic Hepatitis B Infection and Inhibits the Detection of Hepatitis B Virus Surface Antigen. J. Viral Hepat. 23 (5), 330–339. doi: 10.1111/jvh.12522
Ohrt, T., Merkle, D., Birkenfeld, K., Echeverri, C. J., Schwille, P. (2006). In Situ Fluorescence Analysis Demonstrates Active siRNA Exclusion From the Nucleus by Exportin 5. Nucleic Acids Res. 34 (5), 1369–1380. doi: 10.1093/nar/gkl001
Pu, M., Chen, J., Tao, Z., Miao, L., Qi, X., Wang, Y., et al. (2019). Regulatory Network of miRNA on its Target: Coordination Between Transcriptional and Post-Transcriptional Regulation of Gene Expression. Cell Mol. Life Sci. 76 (3), 441–451. doi: 10.1007/s00018-018-2940-7
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., et al. (2000). The 21-Nucleotide Let-7 RNA Regulates Developmental Timing in Caenorhabditis Elegans. Nature 403 (6772), 901–906. doi: 10.1038/35002607
Sahu, S., Philip, F., Scarlata, S. (2014). Hydrolysis Rates of Different Small Interfering RNAs (siRNAs) by the RNA Silencing Promoter Complex, C3PO, Determines Their Regulation by Phospholipase Cβ. J. Biol. Chem. 289 (8), 5134–5144. doi: 10.1074/jbc.M113.531467
Scarborough, R. J., Gatignol, A. (2017). RNA Interference Therapies for an HIV-1 Functional Cure. Viruses 10 (1), 8. doi: 10.3390/v10010008
Scheel, T. K., Luna, J. M., Liniger, M., Nishiuchi, E., Rozen-Gagnon, K., Shlomai, A., et al. (2016). A Broad RNA Virus Survey Reveals Both miRNA Dependence and Functional Sequestration. Cell Host Microbe 19 (3), 409–423. doi: 10.1016/j.chom.2016.02.007
Schult, P., Roth, H., Adams, R. L., Mas, C., Imbert, L., Orlik, C., et al. (2018). microRNA-122 Amplifies Hepatitis C Virus Translation by Shaping the Structure of the Internal Ribosomal Entry Site. Nat. Commun. 9 (1), 2613. doi: 10.1038/s41467-018-05053-3
Sedano, C. D., Sarnow, P. (2014). Hepatitis C Virus Subverts Liver-Specific miR-122 to Protect the Viral Genome From Exoribonuclease Xrn2. Cell Host Microbe 16 (2), 257–264. doi: 10.1016/j.chom.2014.07.006
Seong, R. K., Lee, J. K., Cho, G. J., Kumar, M., Shin, O. S. (2020). mRNA and miRNA Profiling of Zika Virus-Infected Human Umbilical Cord Mesenchymal Stem Cells Identifies miR-142-5p as an Antiviral Factor. Emerg. Microbes Infect. 9 (1), 2061–2075. doi: 10.1080/22221751.2020.1821581
Sharma, H., Tripathi, A., Kumari, B., Vrati, S., Banerjee, A. (2018). Artificial MicroRNA-Mediated Inhibition of Japanese Encephalitis Virus Replication in Neuronal Cells. Nucleic Acid Ther. 28 (6), 357–365. doi: 10.1089/nat.2018.0743
Sheu-Gruttadauria, J., MacRae, I. J. (2018). Phase Transitions in the Assembly and Function of Human miRISC. Cell 173 (4), 946–957.e916. doi: 10.1016/j.cell.2018.02.051
Skalsky, R. L., Cullen, B. R. (2010). Viruses, microRNAs, and Host Interactions. Annu. Rev. Microbiol. 64, 123–141. doi: 10.1146/annurev.micro.112408.134243
Stavast, C. J., Erkeland, S. J. (2019). The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 8 (11), 1465. doi: 10.3390/cells8111465
Takimoto, K., Wakiyama, M., Yokoyama, S. (2009). Mammalian GW182 Contains Multiple Argonaute-Binding Sites and Functions in microRNA-Mediated Translational Repression. Rna 15 (6), 1078–1089. doi: 10.1261/rna.1363109
Tang, G. (2005). siRNA and miRNA: An Insight Into RISCs. Trends Biochem. Sci. 30 (2), 106–114. doi: 10.1016/j.tibs.2004.12.007
Teterina, N. L., Liu, G., Maximova, O. A., Pletnev, A. G. (2014). Silencing of Neurotropic Flavivirus Replication in the Central Nervous System by Combining Multiple microRNA Target Insertions in Two Distinct Viral Genome Regions. Virology 456–457, 247–258. doi: 10.1016/j.virol.2014.04.001
Thi, E. P., Mire, C. E., Lee, A. C., Geisbert, J. B., Zhou, J. Z., Agans, K. N., et al. (2015). Lipid Nanoparticle siRNA Treatment of Ebola-Virus-Makona-Infected Nonhuman Primates. Nature 521 (7552), 362–365. doi: 10.1038/nature14442
Trobaugh, D. W., Gardner, C. L., Sun, C., Haddow, A. D., Wang, E., Chapnik, E., et al. (2014). RNA Viruses can Hijack Vertebrate microRNAs to Suppress Innate Immunity. Nature 506 (7487), 245–248. doi: 10.1038/nature12869
Trobaugh, D. W., Klimstra, W. B. (2017). MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Mol. Med. 23 (1), 80–93. doi: 10.1016/j.molmed.2016.11.003
Trobaugh, D. W., Sun, C., Bhalla, N., Gardner, C. L., Dunn, M. D., Klimstra, W. B. (2019a). Cooperativity Between the 3’ Untranslated Region microRNA Binding Sites is Critical for the Virulence of Eastern Equine Encephalitis Virus. PloS Pathog. 15 (10), e1007867. doi: 10.1371/journal.ppat.1007867
Trobaugh, D. W., Sun, C., Dunn, M. D., Reed, D. S., Klimstra, W. B. (2019b). Rational Design of a Live-Attenuated Eastern Equine Encephalitis Virus Vaccine Through Informed Mutation of Virulence Determinants. PloS Pathog. 15 (2), e1007584. doi: 10.1371/journal.ppat.1007584
Tsetsarkin, K. A., Liu, G., Kenney, H., Bustos-Arriaga, J., Hanson, C. T., Whitehead, S. S., et al. (2015). Dual miRNA Targeting Restricts Host Range and Attenuates Neurovirulence of Flaviviruses. PloS Pathog. 11 (4), e1004852. doi: 10.1371/journal.ppat.1004852
van den Berg, F., Limani, S. W., Mnyandu, N., Maepa, M. B., Ely, A., Arbuthnot, P. (2020). Advances With RNAi-Based Therapy for Hepatitis B Virus Infection. Viruses 12 (8), 851. doi: 10.3390/v12080851
Wang, Y. S., Ouyang, W., Pan, Q. X., Wang, X. L., Xia, X. X., Bi, Z. W., et al. (2013). Overexpression of microRNA gga-miR-21 in Chicken Fibroblasts Suppresses Replication of Infectious Bursal Disease Virus Through Inhibiting VP1 Translation. Antiviral Res. 100 (1), 196–201. doi: 10.1016/j.antiviral.2013.08.001
Wang, P., Qu, X., Zhou, X., Shen, Y., Ji, H., Fu, Z., et al. (2015). Two Cellular microRNAs, miR-196b and miR-1290, Contribute to HIV-1 Latency. Virology 486, 228–238. doi: 10.1016/j.virol.2015.09.016
Wen, W., He, Z., Jing, Q., Hu, Y., Lin, C., Zhou, R., et al. (2015). Cellular microRNA-miR-548g-3p Modulates the Replication of Dengue Virus. J. Infect. 70 (6), 631–640. doi: 10.1016/j.jinf.2014.12.001
Wightman, B., Ha, I., Ruvkun, G. (1993). Posttranscriptional Regulation of the Heterochronic Gene Lin-14 by Lin-4 Mediates Temporal Pattern Formation in C. Elegans. Cell 75 (5), 855–862. doi: 10.1016/0092-8674(93)90530-4
Wooddell, C. I., Yuen, M. F., Chan, H. L., Gish, R. G., Locarnini, S. A., Chavez, D., et al. (2017). RNAi-Based Treatment of Chronically Infected Patients and Chimpanzees Reveals That Integrated Hepatitis B Virus DNA is a Source of HBsAg. Sci. Transl. Med. 9 (409). doi: 10.1126/scitranslmed.aan0241
Ye, X., Huang, N., Liu, Y., Paroo, Z., Huerta, C., Li, P., et al. (2011). Structure of C3PO and Mechanism of Human RISC Activation. Nat. Struct. Mol. Biol. 18 (6), 650–657. doi: 10.1038/nsmb.2032
Yu, L., Nomaguchi, M., Padmanabhan, R., Markoff, L. (2008). Specific Requirements for Elements of the 5’ and 3’ Terminal Regions in Flavivirus RNA Synthesis and Viral Replication. Virology 374 (1), 170–185. doi: 10.1016/j.virol.2007.12.035
Zhang, Q., Huang, C., Yang, Q., Gao, L., Liu, H. C., Tang, J., et al. (2016). MicroRNA-30c Modulates Type I IFN Responses To Facilitate Porcine Reproductive and Respiratory Syndrome Virus Infection by Targeting Jak1. J. Immunol. 196 (5), 2272–2282. doi: 10.4049/jimmunol.1502006
Zhang, S., Wang, R., Su, H., Wang, B., Sizhu, S., Lei, Z., et al. (2017). Sus Scrofa miR-204 and miR-4331 Negatively Regulate Swine H1N1/2009 Influenza A Virus Replication by Targeting Viral HA and NS, Respectively. Int. J. Mol. Sci. 18 (4), 749. doi: 10.3390/ijms18040749
Zhang, Y., Zhang, Y., Xiao, L., Yu, T. X., Li, J. Z., Rao, J. N., et al. (2017). Cooperative Repression of Insulin-Like Growth Factor Type 2 Receptor Translation by MicroRNA 195 and RNA-Binding Protein Cugbp1. Mol. Cell Biol. 37 (19), e00225–e00217. doi: 10.1128/mcb.00225-17
Zheng, Z., Ke, X., Wang, M., He, S., Li, Q., Zheng, C., et al. (2013). Human microRNA hsa-miR-296-5p Suppresses Enterovirus 71 Replication by Targeting the Viral Genome. J. Virol. 87 (10), 5645–5656. doi: 10.1128/jvi.02655-12
Zheng, H., Xu, L., Liu, Y., Li, C., Zhang, L., Wang, T., et al. (2018). MicroRNA-221-5p Inhibits Porcine Epidemic Diarrhea Virus Replication by Targeting Genomic Viral RNA and Activating the NF-κB Pathway. Int. J. Mol. Sci. 19 (11), 3381. doi: 10.3390/ijms19113381
Keywords: miRNA, RNA virus, RISC complex, argonaute2, flavivirus
Citation: Lei L, Cheng A, Wang M and Jia R (2022) The Influence of Host miRNA Binding to RNA Within RNA Viruses on Virus Multiplication. Front. Cell. Infect. Microbiol. 12:802149. doi: 10.3389/fcimb.2022.802149
Received: 26 October 2021; Accepted: 14 March 2022;
Published: 21 April 2022.
Edited by:
Guofeng Cheng, Tongji University, ChinaReviewed by:
Bruno Sargueil, UMR8015 Laboratoire de cristallographie et RMN biologiques, FranceZengjun Lu, Lanzhou Veterinary Research Institute (CAS), China
Copyright © 2022 Lei, Cheng, Wang and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renyong Jia, amlhcnlAc2ljYXUuZWR1LmNu
 Lin Lei1,2,3
Lin Lei1,2,3 Mingshu Wang
Mingshu Wang Renyong Jia
Renyong Jia