- 1Shaanxi Clinical Research Center for Oral Diseases, National Clinical Research Center for Oral Diseases, State Key Laboratory of Military Stomatology, Department of Oral Medicine, School of Stomatology, The Fourth Military Medical University, Xi’an, China
- 2Shaanxi International Joint Research Center for Oral Diseases, State Key Laboratory of Military Stomatology, Department of Histology and Pathology, School of Stomatology, The Fourth Military Medical University, Xi’an, China
Granulomatous inflammation (GI) diseases are a group of chronic inflammation disorders characterized by focal collections of multinucleated giant cells, epithelioid cells and macrophages, with or without necrosis. GI diseases are closely related to microbes, especially virulent intracellular bacterial infections are important factors in the progression of these diseases. They employ a range of strategies to survive the stresses imposed upon them and persist in host cells, becoming the initiator of the fighting. Microbe-host communication is essential to maintain functions of a healthy host, so defense capacity of hosts is another influence factor, which is thought to combine to determine the result of the fighting. With the development of gene research technology, many human genetic loci were identified to be involved in GI diseases susceptibility, providing more insights into and knowledge about GI diseases. The current review aims to provide an update on the most recent progress in the identification and characterization of bacteria in GI diseases in a variety of organ systems and clinical conditions, and examine the invasion and escape mechanisms of pathogens that have been demonstrated in previous studies, we also review the existing data on the predictive factors of the host, mainly on genetic findings. These strategies may improve our understanding of the mechanisms underlying GI diseases, and open new avenues for the study of the associated conditions in the future.
Introduction
Granulomatous inflammation (GI) is a special type of chronic inflammation, which is characterized by focal collections of multinucleated giant cells, epithelioid cells and macrophages in response to a persistent inflammatory stimulus (Williams and Williams, 1983). GI is a protective response to encapsulate foreign material, and is found throughout both vertebrate and invertebrate species. A wide range of stimuli can result in GI, including microbes, parasites and fungi (Shah et al., 2017), some viruses have also been described in association with GI diseases.
Among these stimuli, microbes are the important triggers for the development of GI. In recent decades, an increasing body of studies implicated microbes and their alterations in many GI diseases, i.e., tuberculosis (TB), leprae, Crohn’s disease (CD) and Buruli ulcer (BU). Evidence of Mycobacterium, Nocardia, Salmonella, etc., involved in GI pathogenesis has been found. In some GI diseases, clear proof-of-concept of causality is still lacking, but the possibility of an infectious origin has been postulated.
The complexity of GI pathogenesis, involving multiple distinct elements, pathogens maybe necessary but not always sufficient, especially for the GI diseases initiated by less virulent bacteria or resident bacteria, A previous study showed that a sibling of a CD patient who develops the diseases is at 13 to 36-fold higher risk compared with that of the general population (Ahmad et al., 2001), therefore the genetic factors of hosts have long been considered another major contributor to GIs. With widely applied genome-wide approaches, many genetic loci have been identified to be involved in GI disease susceptibility, moreover some unexpected overlap in genetic architecture between different GI diseases have been revealed, which further emphasizes the genetic role in the pathogenesis of GI diseases, although other predisposing conditions of the host, (including diet and healthy status), and environmental risk factors may also play a role (Alcais et al., 2009; Fox et al., 2016).
Here, we will focus on the bacterial involvement in GIs, parasites, fungi, virus and non-infectious agents will not be discussed, and provide an overview of the recent advances in both bacteriology and genetics that impact the development of GI diseases.
Microbes Associated With Granulomatous Inflammation
Actinobacteria
The members of Actinobacteria are by far the most common causes of GI diseases (Table 1). TB, caused by infection with Mycobacterium tuberculosis is a classic and ancient GI disease, and remains one of the leading causes of morbidity and mortality globally (Lawn and Zumla, 2011). Leprosy is a GI disease caused by M. leprae, another member of Mycobacterium spp. that mainly affects skin and peripheral nerves (Kaplan and Cohn, 1986). Other GI diseases verified by scientific research as having an association with Mycobacterium spp. include BU (Mac et al., 1948; Ruf et al., 2011), a chronic necrotizing GI skin disease; sarcoidosis, a multisystem GI diseases (Esteves et al., 2016); CD, a GI disease of the gastrointestinal tract (Kirkwood et al., 2009); malakoplakia, a rare GI disease that affects a wide variety of organs (Remond et al., 1994; Mitchell and Dugas, 2019); and Takayasu arteritis, a rare large-vessel GI vasculitis (Mwipatayi et al., 2005; Soto et al., 2012).
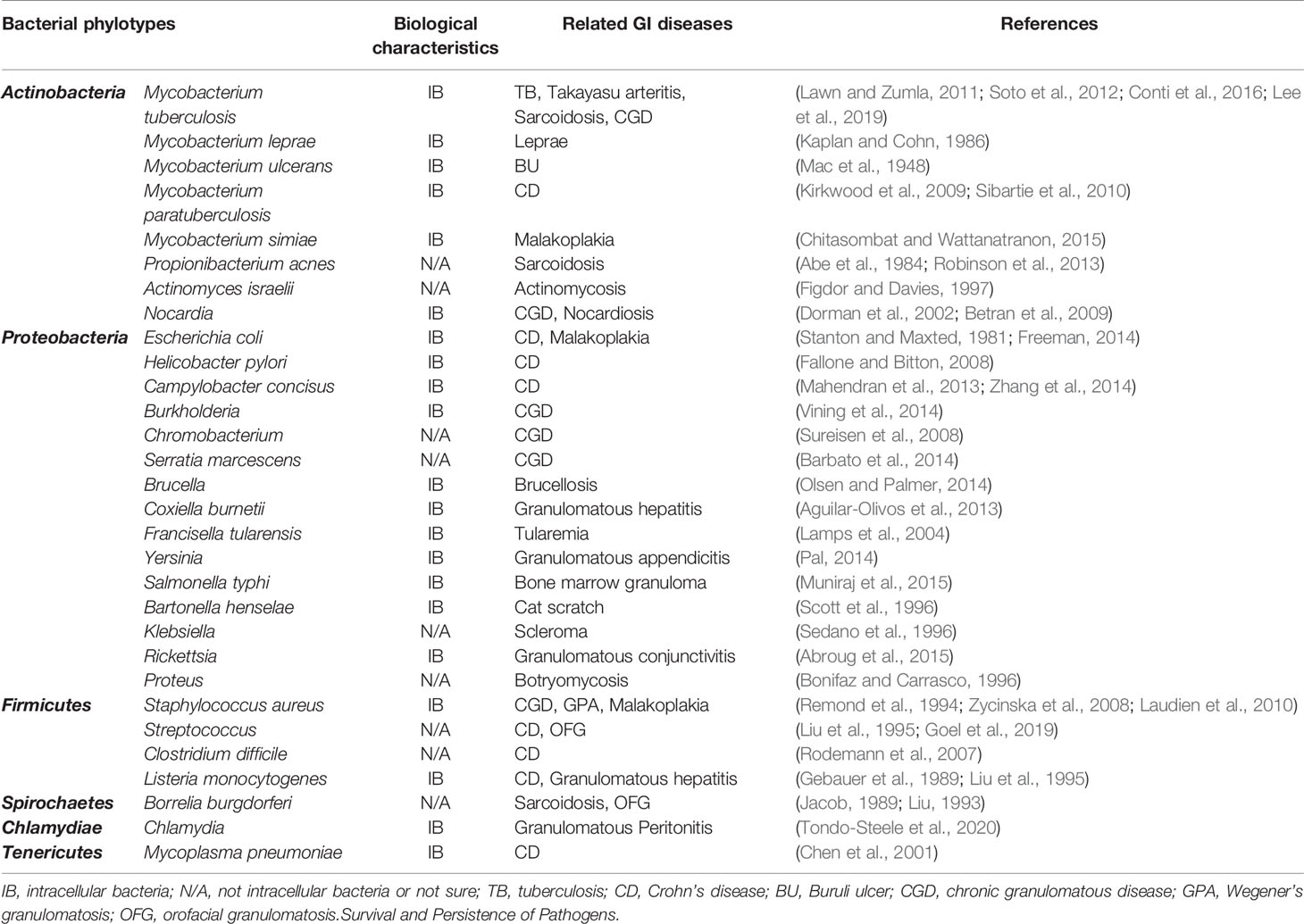
Table 1 Overview of the bacterial phylotypes involved in GI diseases and their biological characteristics.
Mycobacterium spp. are aerobic rod-shaped, obligate intracellular bacteria enriched in lipids with long-chain mycolic acids in cell envelope (Daffe and Etienne, 1999; Rahlwes et al., 2019), and there are many common biological characteristics among them, therefore, scientists presumed that they may have evolved from a common ancestor (Mostowy and Behr, 2005; Stinear et al., 2007; Roltgen et al., 2012; Boritsch et al., 2014). Mycobacterium spp. can exert pathogenic capacity in a direct way, for example 6-kDa early secreted antigenic target (ESAT-6) secretion system 1 (ESX-1), possess membrane-lysing activity, and induce host cell necrosis and spread of bacteria to adjacent cells (Van Der Wel et al., 2007; Smith et al., 2008; Welin et al., 2011); Mycolic acid, hallmarks of Mycobacterium spp. serves as a special barrier critical for many of the disease-inducing and physiological aspects of Mycobacterium (Forrellad et al., 2013). Also, Mycobacterium spp. can exert pathogenic capacity in an indirect way, e.g. they release a mannose-containing glycoconjugate, which can impair the ability of macrophages to kill other phagocytosed pathogens (Mpofu et al., 2007; Friswell et al., 2010), the extensive sequence homology between human stress proteins and Mycobacterium may cause cross-reactivity against vascular peptides that mimic the antigens of M. tuberculosis, which might be the etiological factor of Takayasu arteritis (Castillo-Martinez and Amezcua-Guerra, 2012).
In addition to Mycobacterium spp., other numbers of Actinobacteria, such as Propionibacterium, Actinomyces and Nocardia have been also reported in GI diseases (Table 1). More specific strategies they employ will be discussed below.
Proteobacteria
Escherichia coli (E. coli) is the most typical and the best-characterized species of the Proteobacteria phylum in GI, mainly in CD. In contrast to pathogenic Mycobacterium species, E. coli is a resident bacterium in normal intestinal flora, where it plays an important role in maintaining normal intestinal homeostasis. However, over the years, many studies have implicated E. coli as a provocative factor for the development of CD (Freeman, 2014), although a direct causal relationship between E.coli and CD has not been established in humans. E.coli is also a commonly cultured microorganism alone or along with other pathogens in malakoplakia lesions (Stanton and Maxted, 1981).
E. coli usually colonizes different epithelial surfaces with genetic and phenotypic diversities. However, upon infection E.coli from the outer loose mucin layer can penetrate and interact with the epithelial lining and deeper layers, showing unique adherent and invasive properties, so that it was named adherent invasive E. coli (AIEC). Studies showed that AIEC adhere to intestinal epithelial cells through carcinoembryonic antigen-related cell adhesion molecule 6 receptors (CEACAM6 receptors), enter cells via a macropinocytosis-like process, lyse the endocytic vacuole (Boudeau et al., 1999), survive and replicate within both epithelial cells and macrophages (Mann and Saeed, 2012), which may promote granuloma formation (Glasser et al., 2001; Barnich et al., 2007; Carvalho et al., 2009; Matricon et al., 2010).
In addition to E. coli, there are other Proteobacteria species that have been reported to be associated with GI diseases, including Gammaproteobacteria members Francisella, Coxiella, Yersinia, Salmonella, Proteus and Serratia; Alphaproteobacteria members, Brucella, Bartonella, and Rickettsia; Betaproteobacteria members, Burkholderia, Klebsiella and chromobacterium; Epsilonproteobacteria members, Helicobacter and Campylobacter (Table 1).
Firmicutes
Of the phylotypes of the Firmicutes phylum, Staphylococcus aureus, Streptococcus, Clostridium and Listeria were reported to be involved in GI diseases (Table 1). They are all Gram-positive pathogens, and widely distributed in nature. S. aureus infection is the signature complication of chronic granulomatous disease (CGD), which is a genetic immune disease caused by the deficiency of the phagocyte transmembrane nicotinamide adenine dinucleotide (NADPH) oxidase (Buvelot et al., 2017). S. aureus was also occasionally isolated from some malakoplakia patients (Remond et al., 1994). In search of a specific pathogenic bacterial agent for Wegener’s granulomatosis (also known as granulomatosis with polyangiitis, GPA), it was found that chronic nasal carriage of S. aureus is approximately three times higher in GPA patients than in healthy control (Stegeman et al., 1994).
Streptococcus and Listeria were identified immunochemically in giant cells, macrophages and lymph nodes of CD patients (Liu et al., 1995). In a recent study of orofacial granulomatosis (OFG), which is considered to be close related to CD, the increased abundance of the Streptococcus was found in saliva of OFG patients (Goel et al., 2019).
Despite clinical evidence suggesting that these members of Firmicutes may be implicated in the pathophysiology of GI diseases, laboratory investigation of the possible mechanisms by which they are involved in GI is relatively limited. As Streptococcal immunoreactivity was also found in some normal tissue, Liu et al. concluded that Streptococcus more than likely acted as secondary rather than primary agents associated with CD (Liu et al., 1995). However, by virtue of that they possess the ability to produce putative virulence determinants, evade from phagocytes killing, they as mediators of GI are still attractive targets for further investigation.
Spirochaetes
Spirochaetes, a group of gram-negative bacteria, have long and spiral cell bodies, endoflagella that reside in the periplasmic space and flagella-dependent motility that sets them apart from other bacteria (Johnson, 1977; Harwood and Canale-Parola, 1984). Taxonomically, the phylum Spirochaetes is classified into the Spirochaetaceae, Leptospiraceae, Brevinemataceae and Brachyspiraceae families. They are very heterogeneous, with cell dimensions varying from diameters of 0.09μm to lengths of 500μm. The morphologies of the cell body and endoflagella greatly differ among species. Compared with that of other bacteria, the scientific understating of the physiology and molecular biology of spirochetes remains very limited. Flagella and motility are known virulence factors of pathogenic Spirochaetes, and are related to invasion and adhesion (Josenhans and Suerbaum, 2002; Haiko and Westerlund-Wikstrom, 2013).
The major means of spirochetes motility is swimming. They are attracted to areas of higher viscosity and exhibit a viscosity-dependent increase in swimming speed (Petrino and Doetsch, 1978), which may assist the accumulation of Spirochaetes in the mucus layer in vivo. In addition, some Spirochaetes species can move on a solid surface through twitching motility (Wall and Kaiser, 1999). The spirochaetal movements over host cell surfaces have been shown to be related to the severity of the symptoms caused by this microorganism.
The hypothesis that Spirochaetes is a possible pathogen for sarcoidosis was first documented in 1989 in epidemiological studies (Jacob, 1989), but was not substantiated further (Arcangeli et al., 1994). There are also a few studies on the role of Spirochaetes in the etiology of OFG. The detection of antibodies against Spirochaetes in as many as 77.8% of patients with OFG has been reported (Liu, 1993; Liu et al., 1994). However, in some other studies, Spirochaetes were not detected (Muellegger et al., 2000), so currently, a definitive relationship between Spirochaetes and the pathogenesis of GI diseases is still debated, more studies are needed to illuminate the possible association.
In addition to the above pathogens, Chlamydiae and Tenericutes may be involved in GIs, and listed in Table 1.
In the pathogenic bacteria known to be associated with GI diseases, the vast majority are intracellular bacteria (e.g., M. tuberculosis, M. leprae, L. monocytogenes and S. typhimurium) (Table 1). E. coli and S. aureus were considered extracellular pathogens for many years. However, recent growing evidence has demonstrated that they have the potential to invade and persist within host cells (Foster et al., 2014), including long-lived phagocytes (DCs and macrophages), as well as nonprofessional phagocytes (e.g., endothelial and epithelial cells, osteoblasts, and fibroblasts (Sinha et al., 1999). Compared to extracellular bacteria, intracellular pathogens are adapted to life within phagocytes, and employ a range of strategies to create a niche within host cells and survive the stresses imposed upon them, playing an underappreciated role in GI development (Figure 1). Various mechanisms by which GI pathogens survive and persist in host cells are discussed below.
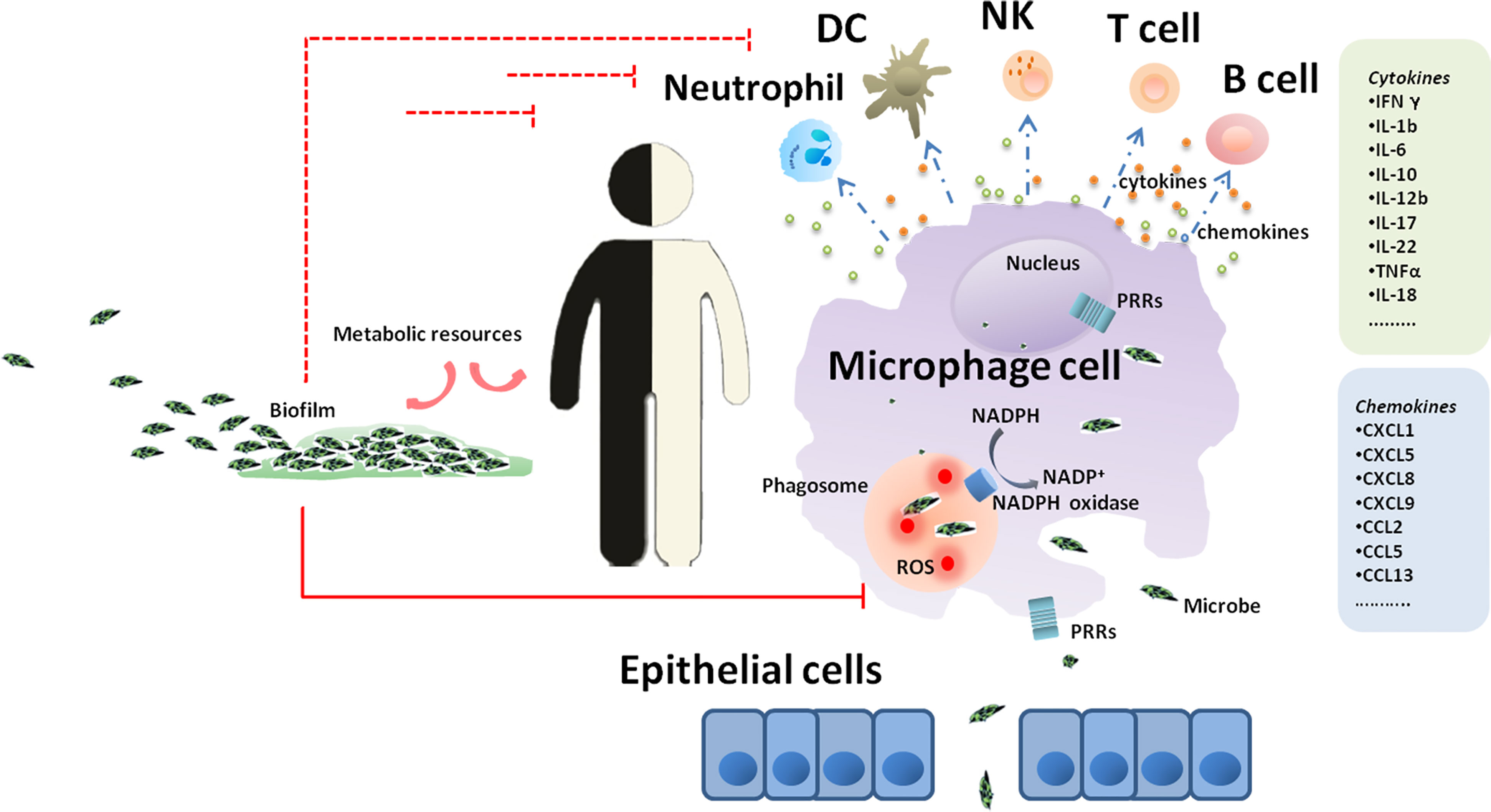
Figure 1 A journey of GI microbes in the host. Epithelium is the gateway for the introduction of GI microbes inside the host and subsequent progression of GI diseases. Once microbes get through the gateway, the process of phagocytosis is initiated, and macrophages are the first line of defense. In macrophages, invading microbes are engulfed by phagocytosis into phagosomal structures and encounter a complex and dynamic range of host defenses. The recognition of bacterial antigens by a range of pattern recognition receptors expressed by macrophages triggers a signaling cascade leading to the recruitment of a diverse cell type complement including neutrophils, dendritic cells, natural killer cells, T lymphocytes and B lymphocytes.
Adherence and Biofilm Formation
The capacity of long-term colonization of cell or tissue surfaces is the key characteristic of pathogens to cause infection. To initiate infection, the pathogens adhere to the host cells or tissues, which confer on them the potential to further invade host cells (Boudeau et al., 1999). For instance, the adhesions present in Mycobacterium spp. surface not only mediate the attachment of bacteria to the surfaces of tissue, but also are capable of sustaining their survival (De Lima et al., 2009; Silva et al., 2013); S. aureus cells express binding proteins that promote attachment to host proteins (e.g., fibronectin and laminin), and damaged tissue where the underlying layers exposed (Bisognano et al., 2004).
Once the bacteria attach to the surface of the cell or tissue, they initiate the synthesis of the extracellular matrix, which is composed of glycopeptides and other molecules, and then form fully developed biofilms (Zambrano and Kolter, 2005), unless some intracellular bacteria invade the host cells without the need of creating a biofilm. The biofilms provide a permeability barrier and stable community for the pathogens, host immune defenses are often inefficacious against bacteria growing in it (Costerton et al., 1999). The protection offered by biofilms makes a large proportion of bacterial biomass possible, which is another pathogenic trait. Furthermore, the high cell density in biofilms facilitates horizontal gene transfer between bacteria (Sorensen et al., 2005), playing a vital role in resistance to host defenses.
Evasion of Host Immunity
The encounter between the host and pathogens leads to a multifaceted and complex immune response. The different cell lines of innate and adaptive immunity come into play at different times in the battle, composing an immune surveillance network that restricts infection. However, GI pathogens have a vast arsenal of defenses against the host immune system, and can evade it on many levels.
Escaping Immune surveillance
As the first line of defense during infections, macrophages are not only the primary targets of intracellular bacteria to be abused, but also the center of infection, which play decisive roles in host responses to intracellular bacteria through a myriad of defense strategies (Figure 1). The recognition of bacterial antigens by a range of pattern recognition receptors (PRRs) expressed by macrophages triggers a signaling cascade leading to the expression of cytokines, chemokines and peptides and to the activation and recruitment of a diverse cell type complement including neutrophils, natural killer (NK) cells and dendritic cells (DCs) (Ehlers and Schaible, 2012). PRR activation also initiates the expression of costimulatory molecules on the surface of macrophages and DCs, which is important for the onset of adaptive immunity (Nunes-Alves et al., 2014).
GI pathogens escape the immune surveillance in different ways. First, the pathogens subvert PRR recognition, and consequently avoid PRR activation (Baxt et al., 2013). For instance, E. coli invade bladder epithelial cells in a type 1 pilus-dependent way, thus shunning TLR4-mediated exocytic processes (Anderson et al., 2004). Second, the pathogens express some proteins that directly or indirectly impair immune reactions or even cause host cell death, thereby facilitating pathogen survival (Derrick and Morris, 2007). For instance, S. aureus expresses staphylococcal complement inhibitor, SCIN, which blocks opsonization by C3b and prevents C3a activation, which is important for mast cell degranulation (Rooijakkers et al., 2009). More recently, increasing attention has been directed at the emerging evidence showing that the host ubiquitin system is targeted by GI pathogens for immune evasion (Penn et al., 2018). Third, bacterial effectors were found to enter the nucleus of infected cell to destroy host nuclear processes, disturbing in host gene transcription and DNA replication or repair (Bierne and Cossart, 2012; Sharma et al., 2015).
Anti-Phagocytosis
After invasion, the pathogens are engulfed by phagocytosis into phagosomal structures that will fuse with endosomes, eventually with lysosomes, where the engulfed bacteria are destroyed (Turner et al., 2017). Intracellular bacteria can survive and acquire antiphagocytic capabilities. For instance, M. tuberculosis has an antiphagocytic capsule, which can limit and control the interaction of the bacterium with macrophages (Stokes et al., 2004); capsular polysaccharide (CP) and protein A (SpA) expressed by S. aureus (Thakker et al., 1998), stringent starvation protein A and the macrophage growth locus protein MglA secreted by F. tularensis (Bell et al., 2010; Bent et al., 2013), acid-tolerant proteins in Mycobacteria can help them to avoid phagocytic killing (Sundaramurthy et al., 2017); M. tuberculosis can even reduce the formation of phagosomes through increasing the expression of miR-142-3p and decreasing the actin binding protein N-Wasp in macrophages (Bettencourt et al., 2013); additionally, type VII secretion systems expressed in Mycobacterium spp. and alpha toxin expressed by S. aureus can help them to evade from phagocytes and subvert host defenses (Van Der Wel et al., 2007; Pang et al., 2010).
Autophagy is an essential process in which cytoplasmic constituents are engulfed by double-membrane autophagosomes, and finally degraded in vacuoles or lysosomes (Nakamura and Yoshimori, 2017). Autophagy was initially identified as a nonselective degradation to maintain cellular homeostasis. Recent studies have clearly shown that autophagy is also involved in the host defense against intracellular pathogen infection (Yoshimori and Amano, 2009; Manzanillo et al., 2013). In the phagocytes, intracellular pathogen is ubiquitinated and recognized by autophagy receptors, then is trapped in autophagosomes. Improving autophagy has been shown to promote bacterial killing (Gutierrez et al., 2004; Wang et al., 2013). Reversely, the autophagy process can be modulated by intracellular bacteria. M. tuberculosis and M. Leprae were all shown to dampen autophagy in human cells as an immune escape mechanism (Silva et al., 2017; Siregar et al., 2022). Similarly, infection with AIEC reduces the autophagy response in host cells by reducing the expression of proteins required for autophagy, whereby pathogens acquire enhanced intracellular survival (Palmela et al., 2018). S. aureus can escape autophagic degradation by blocking autophagy flux, inhibit the fusion of autophagosomes with lysosomes and increasing the pH in autolysosomes, also can utilize autophagy for its own intracellular survivial (Cai et al., 2020).
Anti-Oxidation
The pathogens engulfed by phagocytosis encounter a dynamic range of host defenses, including reactive oxygen, acidification, nitrogen intermediates, antimicrobial peptides (Stallings and Glickman, 2010) and environmental stringencies (Houben et al., 2012), of which the process known as an oxidative burst is crucial for the clearance of pathogens (Gan et al., 2008; Silva, 2010). Reactive oxygen species (ROS), which result from the activation of NADPH oxidase and generate O−2, are highly toxic to bacteria. ROS production can initiate many oxidative reactions, either directly destroying protein, DNA, and lipids or indirectly damaging nucleic acids through oxidation of the nucleotide pool (Van Acker and Coenye, 2017). In addition, ROS can trigger inflammatory signaling cascades via genomic expression of proinflammatory regulators, transcription factors and protein kinase pathways, leading to an overactivated immune system (Choudhury and Macnee, 2017).
GI associated pathogens generally exert strong antioxidant activity. As the leading pathogen in GI diseases, M. tuberculosis has developed complex mechanisms to survive high oxygen stress (OS) burden in the host. Not only do the mycolic acids form a physical barrier to counter host-generated exogenous OS (Portevin et al., 2014), but they also secret a specific protein, namely, enhanced intracellular survivial (Eis) protein, which can sense ROS and respond in a counteractive manner (Awuh and Flo, 2017). As well as, M. tuberculosis generates and secretes antioxidant enzymes (e.g., superoxide dismutases and glutathione peroxidase), helping them to persist in an abnormal redox environment. Additionally, M. tuberculosis possesses the ability to repair and remove oxidative damaged proteins (Jaeger, 2007). Antioxidative system highly effective in protecting intracellular bacteria against ROS stress was discovered in other GI pathogens as well, it was shown that Coxiella burnetii produces an acid phosphatase with inhibitory effects on free radical release from phagocytes NADPH oxidase (Hill and Samuel, 2011), and Francisella tularensis synthesizes factors which inhibit or disrupt NADPH oxidase activity, conducing bacterial colonization and virulence (Mccaffrey et al., 2010).
Interference in Immunoresponses
In the context of an infection with GI pathogens, multiple cytokines (both pro- and anti-inflammatory cytokines) implicate in controlling or promoting pathogenesis, which are important for the fate of GI pathogens. It was shown that GI pathogens can manipulate cytokine responses to their advantage. For example, Mycobacteria increase secretion of IL-10, the anti-inflammatory cytokine by macrophage, maintaining the macrophages in a resting state (Mcclean and Tobin, 2016); induce the expression of SH2 domain-containing protein (CISH) and host Suppressor of Cytokine Signaling 1 (SOCS1), thus dampening pro-inflammatory responses (Duncan et al., 2017). Also, mycobacterial infection results in the expansion of regulatory cells. All these findings are helpful for understanding the biology of microbes associated with GI.
Metabolic Regulation
For the intracellular bacteria, a suitable substance supply, including nutrients, ions and carbon influences their behavior and plays a crucial role in their persisting long enough within host cells. However, the substances needed for the metabolism of intracellular bacteria are found only in the infected host cells. Therefore for this purpose, intracellular bacteria reprogram their metabolism to contend with the limited metabolic resources of the host. They rely on various or similar host-derived carbon sources to replicate, e.g. L. monocytogenes residing within the host cell cytosol uses host glycerol and G6P as sources of carbon and energy (Chatterjee et al., 2006; Joseph et al., 2006), Brucella induces the upregulation of glucose uptake and glycolysis to support their growth and survival in the intracellular niche (Czyz et al., 2017). Some bacteria, e.g. M. tuberculosis acquire nutrients via multiple metabolic pathways (Bloch and Segal, 1956; Mehrotra et al., 2014).
Metal ions, including iron, copper, manganese and zinc, as components of metalloproteins or as structural elements for enzymes are required in many biological processes. During infection, the host restricts the availability of essential metals from invading pathogens. Meanwhile, the toxicity of metals such as zinc and copper can be used as a host defense mechanism to facilitate bacterial killing (Porcheron et al., 2013). Studies have shown that pathogens evolve sophisticated systems to control the transportation of these metals to ensure their physiological needs while countering metal toxicity, e.g. M. tuberculosis and E. coli are able to acquire soluble iron from host iron proteins through siderophores (Chakraborty et al., 2012; Liu et al., 2014), ferroportin expressed in the membrane of M. tuberculosis-containing phagosomes can provide intraphagosomal iron that favors the pathogen (Van Zandt et al., 2008); S. typhimurium induces the formation of estrogen-related receptor-γ, which triggers hepcidin expression and iron retention in macrophage cells (Kim et al., 2014). In contrast, M. tuberculosis synthesizes mycobacterial copper transport protein B (MctB), ATPase CtpV (Hodgkinson and Petris, 2012; Neyrolles et al., 2013) and p-type ATPase (Neyrolles et al., 2013), which enable them to resist metal intoxication within the phagosomes of macrophages.
In addition to the above strategies used in survival and persistence of GI pathogens, antimicrobial resistance (AMR) is also closely related to intracellular survival of the pathogen upon antimicrobial agents, which lead to treatment failure. Under extreme nutrient-limiting conditions or during latency, GI pathogens obtain additional survival advantages by slowing growth, reducing respiration rate, etc., increase their tolerance against stresses imposed upon them (Wayne and Hayes, 1996; Leistikow et al., 2010). Associated discoveries will not be included in this review.
Predisposing Factors in Hosts
Microbial Recognition Defect
PRRs of the host identify microbial pathogens and form the foundation of the innate immune system. PRRs include Toll-like receptors (TLRs), retinoic acid inducible gene-I(RIG-I)-like receptors (RLRs), C-type lectin receptors, oligo-adenylate synthetase (OAS)-like receptors, absent in melanoma 2 (AIM2)-like receptors and nucleotide-binding oligomerization (NOD)-like receptors (NLRs) (Thompson et al., 2011). Based on N-terminal domain, NLRs are divided into four subfamilies, NLRA, NLRB, NLRC, and NLRP, which have a common domain organization with a central NOD. Different bacterial molecules have been characterized as ligands or stimulators of PRRs (Takeuchi and Akira, 2010).
Defects in PRR signaling contribute to the pathogenesis of different GIs. Several single-nucleotide polymorphisms (SNPs) in TLR1 have been recognized to be associated with susceptibility or resistance to leprosy and leprosy reactions (Johnson et al., 2007; Marques Cde et al., 2013). Genetic variants of TLR1, 2, 4, and 6 were shown to be involved in the activation of immune cells during the development of CD, sarcoidosis and leprae (Rock et al., 1998; Arbour et al., 2000; Ferguson et al., 2007; Chen et al., 2010; Marques Cde et al., 2013).
NOD2 in the cytoplasm recognizes bacterial peptidoglycan in the cell walls of Gram-negative and Gram-positive bacteria, leads to NF-κB activation and production of IL-6, IL-1b, TNF-α, IL-8 and α-defensins (Negroni et al., 2018). Also it interacts with autophagy-related proteins to help destroy intracellular pathogens (Naser et al., 2012). NOD2 was one of the first PPRs identified to be a strongly associated genetic risk factor for CD (Ogura et al., 2001). Likewise, polymorphisms in the NOD2 gene region were found to be associated with leprosy reactions (Berrington et al., 2010). In addition, polymorphisms in the NLRP1, NLRP3 and NLRP6 were linked with CD and leprosy susceptibility as well (Villani et al., 2009; Pontillo et al., 2013).
Autophagy Defect
Autophagy is an important intracellular process by which invading pathogens are degraded inside the lysosomes (Deretic et al., 2013). The defect in autophagy has been shown to result in excess production of cytokine (Van De Veerdonk and Dinarello, 2014), several autophagy-related genes have been identified that predispose individuals to a higher GI diseases risk. For example, ATG16L1 is expressed in T cells, antigen-presenting cells and intestinal Paneth cells (Cadwell et al., 2008). It interacts with IRGM and NOD2 to form a molecular complex to regulate autophagy responses to microbial invading (Chauhan et al., 2015). It was shown that knocking down ATG16L1 reduces the ability of cells to capture bacteria and abrogates autophagy of S. typhimurium in host cells, which may promote the onset of CD (Kuballa et al., 2008; Schultz et al., 2017). Mutations in autophagy-related gene PTPN2 not only lead to defective autophagosome formation, but also promote T cell differentiation into Th1 and Th17 types (Spalinger et al., 2015; Spalinger et al., 2018). Two other autophagy-related genes, LRRK2 and MUC19 were also reported to be associated with CD risk (Barrett et al., 2008; Tong et al., 2010). PRKN/PARK2 was identified as a genetic susceptibility factor for leprosy and BU(Table 2), and was shown to play a role in the degradation of intracellular Salmonella, Mycobacteria and Listeria (Manzanillo et al., 2013). CGD is an inherited GI disease caused by a defect in the production of reactive oxygen species, but both mouse studies and human studies have shown that defective autophagy is involved in its pathogenesis similarly (Van De Veerdonk and Dinarello, 2014), although the exact mechanisms are not yet clear.
Oxidation Defect
NADPH oxidase, an enzyme mainly contained in the plasma membrane of macrophages and neutrophils, represents an important defense mechanism in microbial killing (Minakami and Sumimotoa, 2006). The functional NADPH oxidase complex is composed of 5 subunits. The genes encoding the five subunits of the NADPH oxidase enzyme are CYBA, CYBB, NCF1, NCF2 and NCF4. Molecular defects in any one of these genes can result in CGD, which is characterized by the impaired production of ROS and failure to eliminate pathogens and tissue granuloma formation (Idh et al., 2017).
Notably, nearly 50% of CGD patients develop an inflammatory bowel disease that resembles CD (Marks et al., 2009). Consistently, a recent study identified missense mutations in CYBB, CYBA, NCF1, NCF2 and NCF4 in some patients with CD (Denson et al., 2018). In (1990), Nielsen et al. reported that alveolar macrophages from patients with sarcoidosis showed a weak oxidative burst response in vitro stimulation, which is thought to be involved in the pathology of pulmonary sarcoidosis (Nielsen et al., 1990). However, a genetic polymorphism investigation failed to find a significant association of polymorphisms in CYBB, CYBA, NCF1, NCF2, NCF4 that led to increased susceptibility to sarcoidosis (Lee et al., 2006). In (2017), Werner et al. generated a mouse model of GI using a strain of P. acnes isolated from a patient with sarcoidosis, and showed that a deficiency in CYBB is linked with increased granuloma formation in the lung (Werner et al., 2017).
In addition to NADPH, some gene coding enzymes related to oxide metabolism are also involved. For instance, SOD2 encodes superoxide dismutase 2, a homotetrameric mitochondrial enzyme that converts superoxide derivatives of oxidative phosphorylation into hydrogen peroxide and diatomic oxygen (Wiener et al., 2007). A family-based analysis revealed that SOD2 is a risk gene conferring susceptibility to leprosy (Ramos et al., 2016). With the development of gene research technology, more potential risk gene may be discovered in the future.
Dysregulated Immunoresponses
Resistance to infection involves a set of interrelated defenses. If the recruited and activated macrophages are unable to remove invading pathogens efficiently, a further immune response may be triggered, which works to control the pathogen.
Antigen presentation is important for the initiation of adaptive immune responses. Due to the crucial role of human leukocyte antigens (HLAs) in antigen presentation and immunomodulation, the SNPs of major histocomatibility complex (MHC) locus have been investigated intensively, which enhances our understanding of the underlying mechanisms. Studies have shown that HLA alleles and haplotypes are involved genetic factors controlling susceptibility to GI diseases, including OFG (Gibson and Wray, 2000), CD (Orchard et al., 2002), leprosy (Jarduli et al., 2013), TB (Sveinbjornsson et al., 2016) and sarcoidosis (Design of a Case Control Etiologic Study of Sarcoidosis (ACCESS). ACCESS Research Group, 1999). Certain HLA alleles, e.g. HLA-DPB1*1701 allele and DPB1*2:01 allele have been recognized as risk factor for GI diseases in humans (Richeldi et al., 1993).
Outside SNPs of MHC, the genes associated with the development, proliferation, apoptosis, migration of immune cells, and antibody production may affect the clinical phenotype and behavior of GI disease (Table 2), Not only that, there is crosstalk between different genes, for instance, NOD2 activation triggers autophagy of immune cells with the participation of ATG16L1, and deficiency in ATG16L1 heightens cytokine production via NOD (Sorbara et al., 2013), the patients with high-risk NOD2 or ATG16L1 variants exhibit impaired MHCII antigen presentation (Cooney et al., 2010). The absence of effective crosstalk may lead to altered inflammation, increasing susceptibility to development of GIs further.
Interestingly, many polymorphisms are not disease specific, some unexpected overlap in genetic architecture between different GI diseases have been revealed (Table 2), for example, CD loci were also markedly enriched in genes involved in leprosy. Generally, in most cases a negligible number of single mutations have not been found that cause GI diseases, and many of the associated genes are thought to combine to produce a predisposition, although their exact contribution to GI disease have not been fully evaluated. In addition, some other genes were selected as positional and functional candidates for association studies, but their function is not yet known (Medeiros et al., 2016; Nakauchi et al., 2016).
Epigenetic Modifications
Extensive research into genetic predispositions that increase the susceptibility to GIs was performed though, in some investigations researchers still failed to uncover specific functional genes that are associated with the susceptibility to GIs, thus epigenetics arouses increasing attention. Epigenetics refers to changes in the activity, expression or function of genes that are not mediated by DNA sequence, mechanisms of epigenetics include DNA methylation, histone modifications, and non-coding RNA (Kitazawa et al., 2022). Data showed that epigenetic modifications, resulting from interactions between the host and exposome potentiate host susceptibility to GIs. This notion is compelling given that epigenetic alternations have been linked to bacterial infectious diseases (Compare et al., 2011). Consistently, an increased risk of developing CD among people migrating from low- to high-incidence regions of CD provided important epidemiological information to support the pathogenic role of epigenetic changes (Benchimol et al., 2015).
Studies have demonstrated some key metabolites of the bacteria, such as mycobacterial lipoprotein (Pennini et al., 2006), mannosylated lipoarabinomannan (ManLam) and Eis protein (Kim et al., 2012)serve as regulators of the host cellular transcriptional machinery, participating in epigenetic processes associated with GI (Miro-Blanch and Yanes, 2019); a variety of short-chain fatty acids (SCFAs) produced by bacteria can epigenetically regulate the immune response (Olza et al., 2017); microRNA was shown to be participated in the immune response to some GI pathogens (Kalla et al., 2015); global methylation analysis of peripheral blood mononuclear cells (PBMCs) from TB patients revealed that DNA methylation signatures may regulate certain immune responses in vivo (Chen et al., 2017). These findings offer new insights into the pathogenesis of GI diseases.
Other Cells Involved
Recently, eosinophils and platelets in GI diseases were gradually recognized (Requena and Fernandez-Figueras, 2007; Lugo-Villarino and Neyrolles, 2014; Furuta et al., 2019). They all contain major granule proteins and a large amount of cytokines and chemokines (Requena and Fernandez-Figueras, 2007; Deppermann and Kubes, 2016), but their potential role in GI diseases was rarely addressed.
After a long period of dominance by classic immune cells, somatic cells, such as epithelial cells and fibroblasts have been discovered to be additional actors in the maintenance of the immunological integrity of the human body. The epithelium acts as a barrier, separating the bacteria from the immune cells, altered physical epithelial barrier function, a thinner mucus layer have been identified as risk factors for GIs (Hui et al., 2011; Turpin et al., 2018).
GI occurs in all tissue sites, however varies considerably in their degree of complexity, physical size and organization. The structural composition plays a role as a primary host-defense mechanism for containing bacteria, also provides a shelter for pathogens, the cause of the specific microarchitecture and inflammatory status of GI deserves further study.
Other Predisposing Conditions
In addition to affecting people with inherent factors, as stated above, GI diseases usually affect patients with other predisposing conditions, e.g., lactation, psychological stress, pregnancy, intercurrent infections, puberty, vaccination or various environmental stimuli (Kahawita and Lockwood, 2008). Immunocompromised individuals, including those with leukemia, lymphoma, diabetes and uncontrolled HIV infection, and patients who are taking immune-suppressing biologics prescribed for common immune-mediated diseases and cancers are at risk for GI diseases (Robson, 2000; Doherty et al., 2008; Agarwal, 2011), which makes the situation more complex and changeable.
Conclusions
GI diseases present challenges to scientific inquiries and clinical managements. The existing literature suggests that the unstable balance between bacterial virulence and host immunity determines the pathological features of the infections related to these diseases, however, the interconnecting mechanisms still remain largely elusive. Recently, microbial dysbiosis in the commensal community has received great attention in GI research, especially for the resident bacteria associated GIs. Taking advantage of high-throughput data on genetic and microbiome may open up a new avenue for GI research.
Author Contributions
XW: Writing-Original draft preparation. YL: Conceptualization, supervision and validation. All authors contributed to the article and approved the submitted version.
Funding
This work is supported by grants from the National Clinical Research Center for Oral Disease of China (LCA202008) and the State Key Laboratory of Military Stomatology (2018ZB01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
GI, granulomatous inflammation; TB, tuberculosis; CD, Crohn’s disease; BU, Buruli ulcer; GPA, granulomatosis with polyangiitis; OFG, orofacial granulomatosis; CGD, chronic granulomatous disease; IB, intracellular bacteria; NADPH, transmembrane nicotinamide adenine dinucleotide oxidase; AMR, antimicrobial resistance; PRRs, pattern recognition receptors; TLRs, Toll-like receptors; NKs, natural killer cells; DCs, dendritic cells; ROS, Reactive oxygen species; SNPs, single-nucleotide polymorphisms; NLRs, nucleotide-binding oligomerization (NOD)-like receptors; HLAs, human leukocyte antigens; MHC, major histocomatibility complex.
References
Abe, C., Iwai, K., Mikami, R., Hosoda, Y. (1984). Frequent Isolation of Propionibacterium Acnes From Sarcoidosis Lymph Nodes. Zentralbl. Bakteriol. Mikrobiol. Hyg. A. 256, 541–547. doi: 10.1016/s0174-3031(84)80032-3
Abroug, N., Khairallah-Ksiaa, I., Kahloun, R., Khochtali, S., Zaouali, S., Khairallah, M. (2015). Parinaud's Oculoglandular Syndrome Revealing Subclinical Rickettsia Conorii Infection. Int. Ophthalmol. 35, 717–719. doi: 10.1007/s10792-015-0094-2
Adams, L. A., Moller, M., Nebel, A., Schreiber, S., van der Merwe, L., Van Helden, P. D., et al. (2011). Polymorphisms in MC3R Promoter and CTSZ 3'UTR Are Associated With Tuberculosis Susceptibility. Eur. J. Hum. Genet. 19, 676–681. doi: 10.1038/ejhg.2011.1
Adrianto, I., Lin, C. P., Hale, J. J., Levin, A. M., Datta, I., Parker, R., et al. (2012). Genome-Wide Association Study of African and European Americans Implicates Multiple Shared and Ethnic Specific Loci in Sarcoidosis Susceptibility. PLoS One 7, e43907. doi: 10.1371/journal.pone.0043907
Agarwal, S. K. (2011). Biologic Agents in Rheumatoid Arthritis: An Update for Managed Care Professionals. J. Manag. Care Pharm. 17, S14–S18. doi: 10.18553/jmcp.2011.17.s9-b.S14
Aguilar-Olivos, N., Manzano-Robleda, M. D. C., Gutierrez-Grobe, Y., Chable-Montero, F., Albores-Saavedra, J., Lopez-Mendez, E. (2013). Granulomatous Hepatitis Caused by Q Fever: A Differential Diagnosis of Fever of Unknown Origin. Ann. Hepatol. 12, 138–141. doi: 10.1016/S1665-2681(19)31396-1
Ahmad, T., Satsangi, J., Mcgovern, D., Bunce, M., Jewell, D. P. (2001). Review Article: The Genetics of Inflammatory Bowel Disease. Aliment. Pharmacol. Ther. 15, 731–748. doi: 10.1046/j.1365-2036.2001.00981.x
Alcais, A., Abel, L., Casanova, J. L. (2009). Human Genetics of Infectious Diseases: Between Proof of Principle and Paradigm. J. Clin. Invest. 119, 2506–2514. doi: 10.1172/JCI38111
Alter, A., De Leseleuc, L., Van Thuc, N., Thai, V. H., Huong, N. T., Ba, N. N., et al. (2010). Genetic and Functional Analysis of Common MRC1 Exon 7 Polymorphisms in Leprosy Susceptibility. Hum. Genet. 127, 337–348. doi: 10.1007/s00439-009-0775-x
Alter, A., Huong, N. T., Singh, M., Orlova, M., Van Thuc, N., Katoch, K., et al. (2011). Human Leukocyte Antigen Class I Region Single-Nucleotide Polymorphisms Are Associated With Leprosy Susceptibility in Vietnam and India. J. Infect. Dis. 203, 1274–1281. doi: 10.1093/infdis/jir024
Anderson, G. G., Dodson, K. W., Hooton, T. M., Hultgren, S. J. (2004). Intracellular Bacterial Communities of Uropathogenic Escherichia Coli in Urinary Tract Pathogenesis. Trends Microbiol. 12, 424–430. doi: 10.1016/j.tim.2004.07.005
Arbour, N. C., Lorenz, E., Schutte, B. C., Zabner, J., Kline, J. N., Jones, M., et al. (2000). TLR4 Mutations Are Associated With Endotoxin Hyporesponsiveness in Humans. Nat. Genet. 25, 187–191. doi: 10.1038/76048
Arcangeli, G., Calabro, S., Cisno, F., Zambotto, F. M., Drigo, R., Ferraresso, A. (1994). Determination of Antibodies to Borrelia Burgdorferi in Sarcoidosis. Sarcoidosis 11, 32–33.
(1999). Design of a Case Control Etiologic Study of Sarcoidosis (ACCESS). ACCESS Research Group. J. Clin. Epidemiol. 52, 1173–1186.
Awuh, J. A., Flo, T. H. (2017). Molecular Basis of Mycobacterial Survival in Macrophages. Cell Mol. Life Sci. 74, 1625–1648. doi: 10.1007/s00018-016-2422-8
Barbato, M., Ragusa, G., Civitelli, F., Marcheggiano, A., Di Nardo, G., Iacobini, M., et al. (2014). Chronic Granulomatous Disease Mimicking Early-Onset Crohn's Disease With Cutaneous Manifestations. BMC Pediatr. 14, 156. doi: 10.1186/1471-2431-14-156
Barnich, N., Carvalho, F. A., Glasser, A. L., Darcha, C., Jantscheff, P., Allez, M., et al. (2007). CEACAM6 Acts as a Receptor for Adherent-Invasive E. Coli, Supporting Ileal Mucosa Colonization in Crohn Disease. J. Clin. Invest. 117, 1566–1574. doi: 10.1172/JCI30504
Barrett, J. C., Hansoul, S., Nicolae, D. L., Cho, J. H., Duerr, R. H., Rioux, J. D., et al. (2008). Genome-Wide Association Defines More Than 30 Distinct Susceptibility Loci for Crohn's Disease. Nat. Genet. 40, 955–962. doi: 10.1038/ng.175
Baxt, L. A., Garza-Mayers, A. C., Goldberg, M. B. (2013). Bacterial Subversion of Host Innate Immune Pathways. Science 340, 697–701. doi: 10.1126/science.1235771
Bell, B. L., Mohapatra, N. P., Gunn, J. S. (2010). Regulation of Virulence Gene Transcripts by the Francisella Novicida Orphan Response Regulator PmrA: Role of Phosphorylation and Evidence of MglA/SspA Interaction. Infect. Immun. 78, 2189–2198. doi: 10.1128/IAI.00021-10
Benchimol, E. I., Mack, D. R., Guttmann, A., Nguyen, G. C., To, T., Mojaverian, N., et al. (2015). Inflammatory Bowel Disease in Immigrants to Canada and Their Children: A Population-Based Cohort Study. Am. J. Gastroenterol. 110, 553–563. doi: 10.1038/ajg.2015.52
Bent, Z. W., Brazel, D. M., Tran-Gyamfi, M. B., Hamblin, R. Y., Vandernoot, V. A., Branda, S. S. (2013). Use of a Capture-Based Pathogen Transcript Enrichment Strategy for RNA-Seq Analysis of the Francisella Tularensis LVS Transcriptome During Infection of Murine Macrophages. PLoS One 8, e77834. doi: 10.1371/journal.pone.0077834
Berrington, W. R., Macdonald, M., Khadge, S., Sapkota, B. R., Janer, M., Hagge, D. A., et al. (2010). Common Polymorphisms in the NOD2 Gene Region are Associated With Leprosy and Its Reactive States. J. Infect. Dis. 201, 1422–1435. doi: 10.1086/651559
Betran, A., Rezusta, A., Lezcano, M. A., Villuendas, M. C., Revillo, M. J., Boiron, P., et al. (2009). First Spanish Case of Nocardiosis Caused by Nocardia Takedensis. J. Clin. Microbiol. 47, 1918–1919. doi: 10.1128/JCM.00090-09
Bettencourt, P., Marion, S., Pires, D., Santos, L. F., Lastrucci, C., Carmo, N., et al. (2013). Actin-Binding Protein Regulation by microRNAs as a Novel Microbial Strategy to Modulate Phagocytosis by Host Cells: The Case of N-Wasp and miR-142-3p. Front. Cell Infect. Microbiol. 3, 19. doi: 10.3389/fcimb.2013.00019
Bhanothu, V., Lakshmi, V., Theophilus, J. P., Rozati, R., Badhini, P., Vijayalaxmi, B. (2015). Investigation of Toll-Like Receptor-2 (2258g/A) and Interferon Gamma (+874t/A) Gene Polymorphisms Among Infertile Women With Female Genital Tuberculosis. PLoS One 10, e0130273. doi: 10.1371/journal.pone.0130273
Bhattacharyya, C., Majumder, P. P., Pandit, B. (2019). An Exome Wide Association Study of Pulmonary Tuberculosis Patients and Their Asymptomatic Household Contacts. Infect. Genet. Evol. 71, 76–81. doi: 10.1016/j.meegid.2019.03.006
Bibert, S., Bratschi, M. W., Aboagye, S. Y., Collinet, E., Scherr, N., Yeboah-Manu, D., et al. (2017). Susceptibility to Mycobacterium Ulcerans Disease (Buruli Ulcer) Is Associated With IFNG and iNOS Gene Polymorphisms. Front. Microbiol. 8 1903. doi: 10.3389/fmicb.2017.01903
Bierne, H., Cossart, P. (2012). When Bacteria Target the Nucleus: The Emerging Family of Nucleomodulins. Cell Microbiol. 14, 622–633. doi: 10.1111/j.1462-5822.2012.01758.x
Bisognano, C., Kelley, W. L., Estoppey, T., Francois, P., Schrenzel, J., Li, D., et al. (2004). A recA-LexA-Dependent Pathway Mediates Ciprofloxacin-Induced Fibronectin Binding in Staphylococcus Aureus. J. Biol. Chem. 279, 9064–9071. doi: 10.1074/jbc.M309836200
Bloch, H., Segal, W. (1956). Biochemical Differentiation of Mycobacterium Tuberculosis Grown In Vivo and In Vitro. J. Bacteriol. 72, 132–141. doi: 10.1128/jb.72.2.132-141.1956
Boisson-Dupuis, S., Ramirez-Alejo, N., Li, Z., Patin, E., Rao, G., Kerner, G., et al. (2018). Tuberculosis and Impaired IL-23-Dependent IFN-Gamma Immunity in Humans Homozygous for a Common TYK2 Missense Variant. Sci. Immunol. 3 (30), eaau8714. doi: 10.1016/s0895-4356(99)00142-0
Bonifaz, A., Carrasco, E. (1996). Botryomycosis. Int. J. Dermatol. 35, 381–388. doi: 10.1111/j.1365-4362.1996.tb03015.x
Boritsch, E. C., Supply, P., Honore, N., Seemann, T., Stinear, T. P., Brosch, R. (2014). A Glimpse Into the Past and Predictions for the Future: The Molecular Evolution of the Tuberculosis Agent. Mol. Microbiol. 93, 835–852. doi: 10.1111/mmi.12720
Boudeau, J., Glasser, A. L., Masseret, E., Joly, B., Darfeuille-Michaud, A. (1999). Invasive Ability of an Escherichia Coli Strain Isolated From the Ileal Mucosa of a Patient With Crohn's Disease. Infect. Immun. 67, 4499–4509. doi: 10.1128/IAI.67.9.4499-4509.1999
Braun, K., Wolfe, J., Kiazyk, S., Kaushal Sharma, M. (2015). Evaluation of Host Genetics on Outcome of Tuberculosis Infection Due to Differences in Killer Immunoglobulin-Like Receptor Gene Frequencies and Haplotypes. BMC Genet. 16, 63. doi: 10.1186/s12863-015-0224-x
Bulat-Kardum, L. J., Etokebe, G. E., Lederer, P., Balen, S., Dembic, Z. (2015). Genetic Polymorphisms in the Toll-Like Receptor 10, Interleukin (IL)17A and IL17F Genes Differently Affect the Risk for Tuberculosis in Croatian Population. Scand. J. Immunol. 82, 63–69. doi: 10.1111/sji.12300
Buvelot, H., Posfay-Barbe, K. M., Linder, P., Schrenzel, J., Krause, K. H. (2017). Staphylococcus Aureus, Phagocyte NADPH Oxidase and Chronic Granulomatous Disease. FEMS Microbiol. Rev. 41, 139–157. doi: 10.1093/femsre/fuw042
Cadwell, K., Liu, J. Y., Brown, S. L., Miyoshi, H., Loh, J., Lennerz, J. K., et al. (2008). A Key Role for Autophagy and the Autophagy Gene Atg16l1 in Mouse and Human Intestinal Paneth Cells. Nature 456, 259–263. doi: 10.1038/nature07416
Cadwell, K., Patel, K. K., Maloney, N. S., Liu, T. C., Ng, A. C., Storer, C. E., et al. (2010). Virus-Plus-Susceptibility Gene Interaction Determines Crohn's Disease Gene Atg16L1 Phenotypes in Intestine. Cell 141, 1135–1145. doi: 10.1016/j.cell.2010.05.009
Cai, J., Li, J., Zhou, Y., Wang, J., Cui, L., Meng, X., et al. (2020). Staphylococcus Aureus Facilitates Its Survival in Bovine Macrophages by Blocking Autophagic Flux. J. Cell Mol. Med. 24, 3460–3468. doi: 10.1111/jcmm.15027
Cao, Y., Schmitz, J. L., Yang, J., Hogan, S. L., Bunch, D., Hu, Y., et al. (2011). DRB1*15 Allele Is a Risk Factor for PR3-ANCA Disease in African Americans. J. Am. Soc. Nephrol. 22, 1161–1167. doi: 10.1681/ASN.2010101058
Capela, C., Dossou, A. D., Silva-Gomes, R., Sopoh, G. E., Makoutode, M., Menino, J. F., et al. (2016). Genetic Variation in Autophagy-Related Genes Influences the Risk and Phenotype of Buruli Ulcer. PLoS Negl. Trop. Dis. 10, e0004671. doi: 10.1371/journal.pntd.0004671
Cardoso, C. C., Pereira, A. C., De Sales Marques, C., Moraes, M. O. (2011). Leprosy Susceptibility: Genetic Variations Regulate Innate and Adaptive Immunity, and Disease Outcome. Future Microbiol. 6, 533–549. doi: 10.2217/fmb.11.39
Carr, E. J., Niederer, H. A., Williams, J., Harper, L., Watts, R. A., Lyons, P. A., et al. (2009). Confirmation of the Genetic Association of CTLA4 and PTPN22 With ANCA-Associated Vasculitis. BMC Med. Genet. 10, 121. doi: 10.1186/1471-2350-10-121
Carvalho, F. A., Barnich, N., Sivignon, A., Darcha, C., Chan, C. H., Stanners, C. P., et al. (2009). Crohn's Disease Adherent-Invasive Escherichia Coli Colonize and Induce Strong Gut Inflammation in Transgenic Mice Expressing Human CEACAM. J. Exp. Med. 206, 2179–2189. doi: 10.1084/jem.20090741
Castillo-Martinez, D., Amezcua-Guerra, L. M. (2012). Self-Reactivity Against Stress-Induced Cell Molecules: The Missing Link Between Takayasu's Arteritis and Tuberculosis? Med. Hypotheses 78, 485–488. doi: 10.1016/j.mehy.2012.01.012
Cervino, A. C., Lakiss, S., Sow, O., Bellamy, R., Beyers, N., Hoal-Van Helden, E., et al. (2002). Fine Mapping of a Putative Tuberculosis-Susceptibility Locus on Chromosome 15q11-13 in African Families. Hum. Mol. Genet. 11, 1599–1603. doi: 10.1093/hmg/11.14.1599
Chakraborty, S., Kaur, S., Guha, S., Batra, S. K. (2012). The Multifaceted Roles of Neutrophil Gelatinase Associated Lipocalin (NGAL) in Inflammation and Cancer. Biochim. Biophys. Acta 1826, 129–169. doi: 10.1016/j.bbcan.2012.03.008
Chatterjee, S. S., Hossain, H., Otten, S., Kuenne, C., Kuchmina, K., Machata, S., et al. (2006). Intracellular Gene Expression Profile of Listeria Monocytogenes. Infect. Immun. 74, 1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006
Chauhan, S., Mandell, M. A., Deretic, V. (2015). IRGM Governs the Core Autophagy Machinery to Conduct Antimicrobial Defense. Mol. Cell 58, 507–521. doi: 10.1016/j.molcel.2015.03.020
Chen, Y. C., Chao, T. Y., Leung, S. Y., Chen, C. J., Wu, C. C., Fang, W. F., et al. (2017). Histone H3K14 Hypoacetylation and H3K27 Hypermethylation Along With HDAC1 Up-Regulation and KDM6B Down-Regulation are Associated With Active Pulmonary Tuberculosis Disease. Am. J. Transl. Res. 9, 1943–1955.
Chen, W., Li, D., Paulus, B., Wilson, I., Chadwick, V. S. (2001). High Prevalence of Mycoplasma Pneumoniae in Intestinal Mucosal Biopsies From Patients With Inflammatory Bowel Disease and Controls. Dig. Dis. Sci. 46, 2529–2535. doi: 10.1023/A:1012352626117
Chen, E. S., Song, Z., Willett, M. H., Heine, S., Yung, R. C., Liu, M. C., et al. (2010). Serum Amyloid A Regulates Granulomatous Inflammation in Sarcoidosis Through Toll-Like Receptor-2. Am. J. Respir. Crit. Care Med. 181, 360–373. doi: 10.1164/rccm.200905-0696OC
Chitasombat, M. N., Wattanatranon, D. (2015). Disseminated Mycobacterium Simiae With Pelvic Malakoplakia in an AIDS Patient. Clin. Med. Insights Case Rep. 8, 89–91. doi: 10.4137/CCRep.S31751
Choudhury, G., Macnee, W. (2017). Role of Inflammation and Oxidative Stress in the Pathology of Ageing in COPD: Potential Therapeutic Interventions. COPD 14, 122–135. doi: 10.1080/15412555.2016.1214948
Christophi, G. P., Caza, T., Curtiss, C., Gumber, D., Massa, P. T., Landas, S. K. (2014). Gene Expression Profiles in Granuloma Tissue Reveal Novel Diagnostic Markers in Sarcoidosis. Exp. Mol. Pathol. 96, 393–399. doi: 10.1016/j.yexmp.2014.04.006
Cleynen, I., Gonzalez, J. R., Figueroa, C., Franke, A., Mcgovern, D., Bortlik, M., et al. (2013). Genetic Factors Conferring an Increased Susceptibility to Develop Crohn's Disease Also Influence Disease Phenotype: Results From the IBDchip European Project. Gut 62, 1556–1565. doi: 10.1136/gutjnl-2011-300777
Compare, D., Rocco, A., Liguori, E., D'armiento, F. P., Persico, G., Masone, S., et al. (2011). Global DNA Hypomethylation Is an Early Event in Helicobacter Pylori-Related Gastric Carcinogenesis. J. Clin. Pathol. 64, 677–682. doi: 10.1136/jcp.2010.087858
Conti, F., Lugo-Reyes, S. O., Blancas Galicia, L., He, J., Aksu, G., Borges De Oliveira, E., Jr., et al. (2016). Mycobacterial Disease in Patients With Chronic Granulomatous Disease: A Retrospective Analysis of 71 Cases. J. Allergy Clin. Immunol. 138, 241–248 e243. doi: 10.1016/j.jaci.2015.11.041
Cooney, R., Baker, J., Brain, O., Danis, B., Pichulik, T., Allan, P., et al. (2010). NOD2 Stimulation Induces Autophagy in Dendritic Cells Influencing Bacterial Handling and Antigen Presentation. Nat. Med. 16, 90–97. doi: 10.1038/nm.2069
Costerton, J. W., Stewart, P. S., Greenberg, E. P. (1999). Bacterial Biofilms: A Common Cause of Persistent Infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Curtis, J., Luo, Y., Zenner, H. L., Cuchet-Lourenco, D., Wu, C., Lo, K., et al. (2015). Susceptibility to Tuberculosis Is Associated With Variants in the ASAP1 Gene Encoding a Regulator of Dendritic Cell Migration. Nat. Genet. 47, 523–527. doi: 10.1038/ng.3248
Czyz, D. M., Willett, J. W., Crosson, S. (2017). Brucella Abortus Induces a Warburg Shift in Host Metabolism That Is Linked to Enhanced Intracellular Survival of the Pathogen. J. Bacteriol. 199 (15), e00227-17. doi: 10.1128/JB.00227-17
Daffe, M., Etienne, G. (1999). The Capsule of Mycobacterium Tuberculosis and Its Implications for Pathogenicity. Tuber. Lung Dis. 79, 153–169. doi: 10.1054/tuld.1998.0200
Davoudi, S., Chang, V. S., Navarro-Gomez, D., Stanwyck, L. K., Sevgi, D. D., Papavasileiou, E., et al. (2018). Association of Genetic Variants in RAB23 and ANXA11 With Uveitis in Sarcoidosis. Mol. Vis. 24, 59–74.
De Lima, C. S., Marques, M. A., Debrie, A. S., Almeida, E. C., Silva, C. A., Brennan, P. J., et al. (2009). Heparin-Binding Hemagglutinin (HBHA) of Mycobacterium Leprae is Expressed During Infection and Enhances Bacterial Adherence to Epithelial Cells. FEMS Microbiol. Lett. 292, 162–169. doi: 10.1111/j.1574-6968.2009.01488.x
Denson, L. A., Jurickova, I., Karns, R., Shaw, K. A., Cutler, D. J., Okou, D. T., et al. (2018). Clinical and Genomic Correlates of Neutrophil Reactive Oxygen Species Production in Pediatric Patients With Crohn's Disease. Gastroenterology 154, 2097–2110. doi: 10.1053/j.gastro.2018.02.016
Deppermann, C., Kubes, P. (2016). Platelets and Infection. Semin. Immunol. 28, 536–545. doi: 10.1016/j.smim.2016.10.005
Deretic, V., Saitoh, T., Akira, S. (2013). Autophagy in Infection, Inflammation and Immunity. Nat. Rev. Immunol. 13, 722–737. doi: 10.1038/nri3532
Derrick, S. C., Morris, S. L. (2007). The ESAT6 Protein of Mycobacterium Tuberculosis Induces Apoptosis of Macrophages by Activating Caspase Expression. Cell Microbiol. 9, 1547–1555. doi: 10.1111/j.1462-5822.2007.00892.x
Doherty, S. D., Van Voorhees, A., Lebwohl, M. G., Korman, N. J., Young, M. S., Hsu, S., et al. (2008). National Psoriasis Foundation Consensus Statement on Screening for Latent Tuberculosis Infection in Patients With Psoriasis Treated With Systemic and Biologic Agents. J. Am. Acad. Dermatol. 59, 209–217. doi: 10.1016/j.jaad.2008.03.023
Dorman, S. E., Guide, S. V., Conville, P. S., Decarlo, E. S., Malech, H. L., Gallin, J. I., et al. (2002). Nocardia Infection in Chronic Granulomatous Disease. Clin. Infect. Dis. 35, 390–394. doi: 10.1086/341416
Du, J., Han, J., Li, X., Zhang, Y., Li, H., Yang, S. (2015). StIL-17 Gene Polymorphisms in the Development of Pulmonary Tuberculosis. Int. J. Clin. Exp. Pathol. 8, 3225–3229.
Duncan, S. A., Baganizi, D. R., Sahu, R., Singh, S. R., Dennis, V. A. (2017). SOCS Proteins as Regulators of Inflammatory Responses Induced by Bacterial Infections: A Review. Front. Microbiol. 8 2431. doi: 10.3389/fmicb.2017.02431
Ehlers, S., Schaible, U. E. (2012). The Granuloma in Tuberculosis: Dynamics of a Host-Pathogen Collusion. Front. Immunol. 3, 411. doi: 10.3389/fimmu.2012.00411
Esteves, T., Aparicio, G., Garcia-Patos, V. (2016). Is There Any Association Between Sarcoidosis and Infectious Agents?: A Systematic Review and Meta-Analysis. BMC Pulm. Med. 16, 165. doi: 10.1186/s12890-016-0332-z
Fallone, C. A., Bitton, A. (2008). Is IBD Caused by a Helicobacter Pylori Infection? Inflammation Bowel. Dis. 14 Suppl 2, S37–S38. doi: 10.1002/ibd.20552
Ferguson, L. R., Shelling, A. N., Browning, B. L., Huebner, C., Petermann, I. (2007). Genes, Diet and Inflammatory Bowel Disease. Mutat. Res. 622, 70–83. doi: 10.1016/j.mrfmmm.2007.05.011
Festen, E. A., Goyette, P., Green, T., Boucher, G., Beauchamp, C., Trynka, G., et al. (2011). A Meta-Analysis of Genome-Wide Association Scans Identifies IL18RAP, PTPN2, TAGAP, and PUS10 as Shared Risk Loci for Crohn's Disease and Celiac Disease. PLoS Genet. 7, e1001283. doi: 10.1371/journal.pgen.1001283
Figdor, D., Davies, J. (1997). Cell Surface Structures of Actinomyces Israelii. Aust. Dent. J. 42, 125–128. doi: 10.1111/j.1834-7819.1997.tb00109.x
Fischer, A., Ellinghaus, D., Nutsua, M., Hofmann, S., Montgomery, C. G., Iannuzzi, M. C., et al. (2015). Identification of Immune-Relevant Factors Conferring Sarcoidosis Genetic Risk. Am. J. Respir. Crit. Care Med. 192, 727–736. doi: 10.1164/rccm.201503-0418OC
Fischer, A., Schmid, B., Ellinghaus, D., Nothnagel, M., Gaede, K. I., Schurmann, M., et al. (2012). A Novel Sarcoidosis Risk Locus for Europeans on Chromosome 11q13.1. Am. J. Respir. Crit. Care Med. 186, 877–885. doi: 10.1164/rccm.201204-0708OC
Forrellad, M. A., Klepp, L. I., Gioffre, A., Sabio Y Garcia, J., Morbidoni, H. R., de la Paz Santangelo, M., et al. (2013). Virulence Factors of the Mycobacterium Tuberculosis Complex. Virulence 4, 3–66. doi: 10.4161/viru.22329
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., Hook, M. (2014). Adhesion, Invasion and Evasion: The Many Functions of the Surface Proteins of Staphylococcus Aureus. Nat. Rev. Microbiol. 12, 49–62. doi: 10.1038/nrmicro3161
Fox, G. J., Orlova, M., Schurr, E. (2016). Tuberculosis in Newborns: The Lessons of the "Lubeck Disaster" (1929-1933). PLoS Pathog. 12, e1005271. doi: 10.1371/journal.ppat.1005271
Franke, A., Fischer, A., Nothnagel, M., Becker, C., Grabe, N., Till, A., et al. (2008). Genome-Wide Association Analysis in Sarcoidosis and Crohn's Disease Unravels a Common Susceptibility Locus on 10p12.2. Gastroenterology 135, 1207–1215. doi: 10.1053/j.gastro.2008.07.017
Freeman, H. J. (2014). Natural History and Long-Term Clinical Course of Crohn's Disease. World J. Gastroenterol. 20, 31–36. doi: 10.3748/wjg.v20.i1.31
Friswell, M., Campbell, B., Rhodes, J. (2010). The Role of Bacteria in the Pathogenesis of Inflammatory Bowel Disease. Gut. Liver. 4, 295–306. doi: 10.5009/gnl.2010.4.3.295
Furuta, S., Iwamoto, T., Nakajima, H. (2019). Update on Eosinophilic Granulomatosis With Polyangiitis. Allergol. Int. 68, 430–436. doi: 10.1016/j.alit.2019.06.004
Gan, H., Lee, J., Ren, F., Chen, M., Kornfeld, H., Remold, H. G. (2008). Mycobacterium Tuberculosis Blocks Crosslinking of Annexin-1 and Apoptotic Envelope Formation on Infected Macrophages to Maintain Virulence. Nat. Immunol. 9, 1189–1197. doi: 10.1038/ni.1654
Garman, L., Pezant, N., Pastori, A., Savoy, K. A., Li, C., Levin, A. M., et al. (2020). Genome-Wide Association Study of Ocular Sarcoidosis Confirms HLA Associations and Implicates Barrier Function and Autoimmunity in African Americans. Ocul. Immunol. Inflamm. (2), 244–249. doi: 10.1080/09273948.2019.1705985
Gebauer, K., Hall, J. C., Donlon, J. B., Herrmann, R., Rofe, S., Platell, C. (1989). Hepatic Involvement in Listeriosis. Aust. N. Z. J. Med. 19, 486–487. doi: 10.1111/j.1445-5994.1989.tb00316.x
Geluk, A., Ottenhoff, T. H. (2006). HLA and Leprosy in the Pre and Postgenomic Eras. Hum. Immunol. 67, 439–445. doi: 10.1016/j.humimm.2006.03.009
Gibson, J., Wray, D. (2000). Human Leucocyte Antigen Typing in Orofacial Granulomatosis. Br. J. Dermatol. 143, 1119–1121. doi: 10.1046/j.1365-2133.2000.03877.x
Glasser, A. L., Boudeau, J., Barnich, N., Perruchot, M. H., Colombel, J. F., Darfeuille-Michaud, A. (2001). Adherent Invasive Escherichia Coli Strains From Patients With Crohn's Disease Survive and Replicate Within Macrophages Without Inducing Host Cell Death. Infect. Immun. 69, 5529–5537. doi: 10.1128/IAI.69.9.5529-5537.2001
Goel, R. M., Prosdocimi, E. M., Amar, A., Omar, Y., Escudier, M. P., Sanderson, J. D., et al. (2019). Streptococcus Salivarius: A Potential Salivary Biomarker for Orofacial Granulomatosis and Crohn's Disease? Inflammation Bowel. Dis. 25, 1367–1374. doi: 10.1093/ibd/izz022
Grant, A. V., Cobat, A., Van Thuc, N., Orlova, M., Huong, N. T., Gaschignard, J., et al. (2014). CUBN and NEBL Common Variants in the Chromosome 10p13 Linkage Region are Associated With Multibacillary Leprosy in Vietnam. Hum. Genet. 133, 883–893. doi: 10.1007/s00439-014-1430-8
Grant, A. V., El Baghdadi, J., Sabri, A., El Azbaoui, S., Alaoui-Tahiri, K., Abderrahmani Rhorfi, I., et al. (2013). Age-Dependent Association Between Pulmonary Tuberculosis and Common TOX Variants in the 8q12-13 Linkage Region. Am. J. Hum. Genet. 92, 407–414. doi: 10.1016/j.ajhg.2013.01.013
Grant, A. V., Sabri, A., Abid, A., Abderrahmani Rhorfi, I., Benkirane, M., Souhi, H., et al. (2016). A Genome-Wide Association Study of Pulmonary Tuberculosis in Morocco. Hum. Genet. 135, 299–307. doi: 10.1007/s00439-016-1633-2
Gutierrez, M. G., Master, S. S., Singh, S. B., Taylor, G. A., Colombo, M. I., Deretic, V. (2004). Autophagy Is a Defense Mechanism Inhibiting BCG and Mycobacterium Tuberculosis Survival in Infected Macrophages. Cell 119, 753–766. doi: 10.1016/j.cell.2004.11.038
Haiko, J., Westerlund-Wikstrom, B. (2013). The Role of the Bacterial Flagellum in Adhesion and Virulence. Biol. (Basel). 2, 1242–1267. doi: 10.3390/biology2041242
Harwood, C. S., Canale-Parola, E. (1984). Ecology of Spirochetes. Annu. Rev. Microbiol. 38, 161–192. doi: 10.1146/annurev.mi.38.100184.001113
Hatta, M., Ratnawati, Tanaka, M., Ito, J., Shirakawa, T., Kawabata, M. (2010). NRAMP1/SLC11A1 Gene Polymorphisms and Host Susceptibility to Mycobacterium Tuberculosis and M. Leprae in South Sulawesi, Indonesia. Southeast. Asian J. Trop. Med. Public Health 41, 386–394.
Hijikata, M., Matsushita, I., Le Hang, N. T., Thuong, P. H., Tam, D. B., Maeda, S., et al. (2016). Influence of the Polymorphism of the DUSP14 Gene on the Expression of Immune-Related Genes and Development of Pulmonary Tuberculosis. Genes Immun. 17, 207–212. doi: 10.1038/gene.2016.11
Hill, J., Samuel, J. E. (2011). Coxiella Burnetii Acid Phosphatase Inhibits the Release of Reactive Oxygen Intermediates in Polymorphonuclear Leukocytes. Infect. Immun. 79, 414–420. doi: 10.1128/IAI.01011-10
Hodgkinson, V., Petris, M. J. (2012). Copper Homeostasis at the Host-Pathogen Interface. J. Biol. Chem. 287, 13549–13555. doi: 10.1074/jbc.R111.316406
Houben, E. N., Bestebroer, J., Ummels, R., Wilson, L., Piersma, S. R., Jimenez, C. R., et al. (2012). Composition of the Type VII Secretion System Membrane Complex. Mol. Microbiol. 86, 472–484. doi: 10.1111/j.1365-2958.2012.08206.x
Hu, Q., Hua, H., Zhou, L., Zou, X. (2020). Association Between Interleukin-8 -251a/T Polymorphism and the Risk of Tuberculosis: A Meta-Analysis. J. Int. Med. Res. 48, 300060520917877. doi: 10.1177/0300060520917877
Hui, Y., Wohlers, J., Podschun, R., Hedderich, J., Lamprecht, P., Ambrosch, P., et al. (2011). Antimicrobial Peptides in Nasal Secretion and Mucosa With Respect to S. Aureus Colonisation in Wegener s Granulomatosis. Clin. Exp. Rheumatol. 29, S49–S56.
Hulpke, S., Tomioka, M., Kremmer, E., Ueda, K., Abele, R., Tampe, R. (2012). Direct Evidence That the N-Terminal Extensions of the TAP Complex Act as Autonomous Interaction Scaffolds for the Assembly of the MHC I Peptide-Loading Complex. Cell Mol. Life Sci. 69, 3317–3327. doi: 10.1007/s00018-012-1005-6
Husmann, C. A., Holle, J. U., Moosig, F., Mueller, S., Wilde, B., Cohen Tervaert, J. W., et al. (2014). Genetics of Toll Like Receptor 9 in ANCA Associated Vasculitides. Ann. Rheum. Dis. 73, 890–896. doi: 10.1136/annrheumdis-2012-202803
Idh, J., Andersson, B., Lerm, M., Raffetseder, J., Eklund, D., Woksepp, H., et al. (2017). Reduced Susceptibility of Clinical Strains of Mycobacterium Tuberculosis to Reactive Nitrogen Species Promotes Survival in Activated Macrophages. PLoS One 12, e0181221. doi: 10.1371/journal.pone.0181221
Jacob, F. (1989). Could Borrelia Burgdorferi be a Causal Agent of Sarcoidosis? Med. Hypotheses 30, 241–243. doi: 10.1016/0306-9877(89)90032-7
Jaeger, T. (2007). Peroxiredoxin Systems in Mycobacteria. Subcell. Biochem. 44, 207–217. doi: 10.1007/978-1-4020-6051-9_9
Jafari, M., Nasiri, M. R., Sanaei, R., Anoosheh, S., Farnia, P., Sepanjnia, A., et al. (2016). The NRAMP1, VDR, TNF-Alpha, ICAM1, TLR2 and TLR4 Gene Polymorphisms in Iranian Patients With Pulmonary Tuberculosis: A Case-Control Study. Infect. Genet. Evol. 39, 92–98. doi: 10.1016/j.meegid.2016.01.013
Jagiello, P., Aries, P., Arning, L., Wagenleiter, S. E., Csernok, E., Hellmich, B., et al. (2005). The PTPN22 620w Allele Is a Risk Factor for Wegener's Granulomatosis. Arthritis Rheum. 52, 4039–4043. doi: 10.1002/art.21487
Jarduli, L. R., Sell, A. M., Reis, P. G., Sippert, E. A., Ayo, C. M., Mazini, P. S., et al. (2013). Role of HLA, KIR, MICA, and Cytokines Genes in Leprosy. BioMed. Res. Int. 2013, 989837. doi: 10.1155/2013/989837
Jiao, L., Song, J., Ding, L., Liu, T., Wu, T., Zhang, J., et al. (2020). A Novel Genetic Variation in NCF2, the Core Component of NADPH Oxidase, Contributes to the Susceptibility of Tuberculosis in Western Chinese Han Population. DNA Cell Biol. 39, 57–62. doi: 10.1089/dna.2019.5082
Johnson, R. C. (1977). The Spirochetes. Annu. Rev. Microbiol. 31, 89–106. doi: 10.1146/annurev.mi.31.100177.000513
Johnson, C. M., Lyle, E. A., Omueti, K. O., Stepensky, V. A., Yegin, O., Alpsoy, E., et al. (2007). Cutting Edge: A Common Polymorphism Impairs Cell Surface Trafficking and Functional Responses of TLR1 But Protects Against Leprosy. J. Immunol. 178, 7520–7524. doi: 10.4049/jimmunol.178.12.7520
Josenhans, C., Suerbaum, S. (2002). The Role of Motility as a Virulence Factor in Bacteria. Int. J. Med. Microbiol. 291, 605–614. doi: 10.1078/1438-4221-00173
Joseph, B., Przybilla, K., Stuhler, C., Schauer, K., Slaghuis, J., Fuchs, T. M., et al. (2006). Identification of Listeria Monocytogenes Genes Contributing to Intracellular Replication by Expression Profiling and Mutant Screening. J. Bacteriol. 188, 556–568. doi: 10.1128/JB.188.2.556-568.2006
Kahawita, I. P., Lockwood, D. N. (2008). Towards Understanding the Pathology of Erythema Nodosum Leprosum. Trans. R. Soc. Trop. Med. Hyg. 102, 329–337. doi: 10.1016/j.trstmh.2008.01.004
Kalla, R., Ventham, N. T., Kennedy, N. A. (2015). MicroRNAs: New Players in Inflammatory Bowel Disease. Gut 64 (3), 504–517. doi: 10.1136/gutjnl-2014-307891
Kanazawa, N., Okafuji, I., Kambe, N., Nishikomori, R., Nakata-Hizume, M., Nagai, S., et al. (2005). Early-Onset Sarcoidosis and CARD15 Mutations With Constitutive Nuclear factor-kappaB Activation: Common Genetic Etiology With Blau Syndrome. Blood 105, 1195–1197. doi: 10.1182/blood-2004-07-2972
Kaplan, G., Cohn, Z. A. (1986). Regulation of Cell-Mediated Immunity in Lepromatous Leprosy. Lepr. Rev. 57 Suppl 2, 199–202. doi: 10.5935/0305-7518.19860072
Kim, K. H., An, D. R., Song, J., Yoon, J. Y., Kim, H. S., Yoon, H. J., et al. (2012). Mycobacterium Tuberculosis Eis Protein Initiates Suppression of Host Immune Responses by Acetylation of DUSP16/MKP-7. Proc. Natl. Acad. Sci. U.S.A. 109, 7729–7734. doi: 10.1073/pnas.1120251109
Kim, H. S., Choi, D., Lim, L. L., Allada, G., Smith, J. R., Austin, C. R., et al. (2011). Association of Interleukin 23 Receptor Gene With Sarcoidosis. Dis. Markers 31, 17–24. doi: 10.1155/2011/185106
Kim, D. K., Jeong, J. H., Lee, J. M., Kim, K. S., Park, S. H., Kim, Y. D., et al. (2014). Inverse Agonist of Estrogen-Related Receptor Gamma Controls Salmonella Typhimurium Infection by Modulating Host Iron Homeostasis. Nat. Med. 20, 419–424. doi: 10.1038/nm.3483
Kirkwood, C. D., Wagner, J., Boniface, K., Vaughan, J., Michalski, W. P., Catto-Smith, A. G., et al. (2009). Mycobacterium Avium Subspecies Paratuberculosis in Children With Early-Onset Crohn's Disease. Inflammation Bowel. Dis. 15, 1643–1655. doi: 10.1002/ibd.20967
Kitazawa, S., Ohno, T., Haraguchi, R., Kitazawa, R. (2022). Histochemistry, Cytochemistry and Epigenetics. Acta Histochem. Cytochem. 55, 1–7. doi: 10.1267/ahc.21-00095
Kordjazy, N., Haj-Mirzaian, A., Rohani, M. M., Gelfand, E. W., Rezaei, N., Abdolghaffari, A. H. (2018). Role of Toll-Like Receptors in Inflammatory Bowel Disease. Pharmacol. Res. 129, 204–215. doi: 10.1016/j.phrs.2017.11.017
Kouhpayeh, H. R., Taheri, M., Baziboroon, M., Naderi, M., Bahari, G., Hashemi, M. (2016). CCL5 Rs2107538 Polymorphism Increased the Risk of Tuberculosis in a Sample of Iranian Population. Prague. Med. Rep. 117, 90–97. doi: 10.14712/23362936.2016.9
Kuballa, P., Huett, A., Rioux, J. D., Daly, M. J., Xavier, R. J. (2008). Impaired Autophagy of an Intracellular Pathogen Induced by a Crohn's Disease Associated ATG16L1 Variant. PLoS One 3, e3391. doi: 10.1371/journal.pone.0003391
Lahiri, A., Hedl, M., Yan, J., Abraham, C. (2017). Human LACC1 Increases Innate Receptor-Induced Responses and a LACC1 Disease-Risk Variant Modulates These Outcomes. Nat. Commun. 8, 15614. doi: 10.1038/ncomms15614
Lamps, L. W., Havens, J. M., Sjostedt, A., Page, D. L., Scott, M. A. (2004). Histologic and Molecular Diagnosis of Tularemia: A Potential Bioterrorism Agent Endemic to North America. Mod. Pathol. 17, 489–495. doi: 10.1038/modpathol.3800087
Lareau, C. A., Adrianto, I., Levin, A. M., Iannuzzi, M. C., Rybicki, B. A., Montgomery, C. G. (2015). Fine Mapping of Chromosome 15q25 Implicates ZNF592 in Neurosarcoidosis Patients. Ann. Clin. Transl. Neurol. 2, 972–977. doi: 10.1002/acn3.229
Laudien, M., Gadola, S. D., Podschun, R., Hedderich, J., Paulsen, J., Reinhold-Keller, E., et al. (2010). Nasal Carriage of Staphylococcus Aureus and Endonasal Activity in Wegener s Granulomatosis as Compared to Rheumatoid Arthritis and Chronic Rhinosinusitis With Nasal Polyps. Clin. Exp. Rheumatol. 28, 51–55.
Lawn, S. D., Zumla, A. I. (2011). Tuberculosis. Lancet 378, 57–72. doi: 10.1016/S0140-6736(10)62173-3
Lee, S. W., Chuang, T. Y., Huang, H. H., Liu, C. W., Kao, Y. H., Wu, L. S. (2016). VDR and VDBP Genes Polymorphisms Associated With Susceptibility to Tuberculosis in a Han Taiwanese Population. J. Microbiol. Immunol. Infect. 49, 783–787. doi: 10.1016/j.jmii.2015.12.008
Lee, H., Eom, M., Kim, S. H., Wang, H. Y., Choi, E. H. (2019). Identification of Mycobacterium Tuberculosis and Non-Tuberculous Mycobacteria From Cutaneous Sarcoidosis Lesions by Reverse Blot Hybridization Assay. J. Dermatol. 46, 917–921. doi: 10.1111/1346-8138.15042
Lee, P. L., West, C., Crain, K., Wang, L. (2006). Genetic Polymorphisms and Susceptibility to Lung Disease. J. Negat. Result. BioMed. 5, 5. doi: 10.1186/1477-5751-5-5
Leistikow, R. L., Morton, R. A., Bartek, I. L., Frimpong, I., Wagner, K., Voskuil, M. I. (2010). The Mycobacterium Tuberculosis DosR Regulon Assists in Metabolic Homeostasis and Enables Rapid Recovery From Nonrespiring Dormancy. J. Bacteriol. 192, 1662–1670. doi: 10.1128/JB.00926-09
Levin, A. M., Iannuzzi, M. C., Montgomery, C. G., Trudeau, S., Datta, I., Adrianto, I., et al. (2014). Admixture Fine-Mapping in African Americans Implicates XAF1 as a Possible Sarcoidosis Risk Gene. PLoS One 9, e92646. doi: 10.1371/journal.pone.0092646
Li, M., Jiao, L., Lyu, M., Song, J., Bai, H., Zhang, C., et al. (2020). Association of IL27 and STAT3 Genetic Polymorphism on the Susceptibility of Tuberculosis in Western Chinese Han Population. Infect. Genet. Evol. 83, 104324. doi: 10.1016/j.meegid.2020.104324
Li, Y., Pabst, S., Kubisch, C., Grohe, C., Wollnik, B. (2010). First Independent Replication Study Confirms the Strong Genetic Association of ANXA11 With Sarcoidosis. Thorax 65, 939–940. doi: 10.1136/thx.2010.138743
Liu, H. G. (1993). [A Study on the Relationship Between Cheilitis Granulomatosa and Melkersson-Rosenthal Syndrome]. Zhonghua. Kou. Qiang. Yi. Xue. Za. Zhi. 28, 323–324, 383.
Liu, Z., Reba, S., Chen, W. D., Porwal, S. K., Boom, W. H., Petersen, R. B., et al. (2014). Regulation of Mammalian Siderophore 2,5-DHBA in the Innate Immune Response to Infection. J. Exp. Med. 211, 1197–1213. doi: 10.1084/jem.20132629
Liu, Y., Van Kruiningen, H. J., West, A. B., Cartun, R. W., Cortot, A., Colombel, J. F. (1995). Immunocytochemical Evidence of Listeria, Escherichia Coli, and Streptococcus Antigens in Crohn's Disease. Gastroenterology 108, 1396–1404. doi: 10.1016/0016-5085(95)90687-8
Liu, H., Wang, Z., Li, Y., Yu, G., Fu, X., Wang, C., et al. (2017). Genome-Wide Analysis of Protein-Coding Variants in Leprosy. J. Invest. Dermatol. 137, 2544–2551. doi: 10.1016/j.jid.2017.08.004
Liu, Q., Wu, S., Xue, M., Sandford, A. J., Wu, J., Wang, Y., et al. (2016). Heterozygote Advantage of the Rs3794624 Polymorphism in CYBA for Resistance to Tuberculosis in Two Chinese Populations. Sci. Rep. 6, 38213. doi: 10.1038/srep38213
Liu, H. G., Zhang, L. F., Wu, Q. G. (1994). [Histopathological Findings of 59 Cases of Cheilitis Granulomatosa]. Zhonghua. Kou. Qiang. Yi. Xue. Za. Zhi. 29198–200, 254.
Lugo-Villarino, G., Neyrolles, O. (2014). Of Clots and Granulomas: Platelets are New Players in Immunity to Tuberculosis. J. Infect. Dis. 210, 1687–1690. doi: 10.1093/infdis/jiu356
Lyons, P. A., Rayner, T. F., Trivedi, S., Holle, J. U., Watts, R. A., Jayne, D. R., et al. (2012). Genetically Distinct Subsets Within ANCA-Associated Vasculitis. N. Engl. J. Med. 367, 214–223. doi: 10.1056/NEJMoa1108735
Mac, C. P., Tolhurst, J. C., et al. (1948). A New Mycobacterial Infection in Man. J. Pathol. Bacteriol. 60, 93–122. doi: 10.1002/path.1700600111
Mahasirimongkol, S., Yanai, H., Mushiroda, T., Promphittayarat, W., Wattanapokayakit, S., Phromjai, J., et al. (2012). Genome-Wide Association Studies of Tuberculosis in Asians Identify Distinct at-Risk Locus for Young Tuberculosis. J. Hum. Genet. 57, 363–367. doi: 10.1038/jhg.2012.35
Mahendran, V., Tan, Y. S., Riordan, S. M., Grimm, M. C., Day, A. S., Lemberg, D. A., et al. (2013). The Prevalence and Polymorphisms of Zonula Occluden Toxin Gene in Multiple Campylobacter Concisus Strains Isolated From Saliva of Patients With Inflammatory Bowel Disease and Controls. PLoS One 8, e75525. doi: 10.1371/journal.pone.0075525
Malkova, A., Starshinova, A., Zinchenko, Y., Basantsova, N., Mayevskaya, V., Yablonskiy, P., et al. (2020). The Opposite Effect of Human Leukocyte Antigen Genotypes in Sarcoidosis and Tuberculosis: A Narrative Review of the Literature. ERJ. Open Res. 6 (3), 00155–2020. doi: 10.1183/23120541.00155-2020
Mann, E. A., Saeed, S. A. (2012). Gastrointestinal Infection as a Trigger for Inflammatory Bowel Disease. Curr. Opin. Gastroenterol. 28, 24–29. doi: 10.1097/MOG.0b013e32834c453e
Manzanillo, P. S., Ayres, J. S., Watson, R. O., Collins, A. C., Souza, G., Rae, C. S., et al. (2013). The Ubiquitin Ligase Parkin Mediates Resistance to Intracellular Pathogens. Nature 501, 512–516. doi: 10.1038/nature12566
Marks, D. J., Miyagi, K., Rahman, F. Z., Novelli, M., Bloom, S. L., Segal, A. W. (2009). Inflammatory Bowel Disease in CGD Reproduces the Clinicopathological Features of Crohn's Disease. Am. J. Gastroenterol. 104, 117–124. doi: 10.1038/ajg.2008.72
Marques Cde, S., Brito-De-Souza, V. N., Guerreiro, L. T., Martins, J. H., Amaral, E. P., Cardoso, C. C., et al. (2013). Toll-Like Receptor 1 N248S Single-Nucleotide Polymorphism Is Associated With Leprosy Risk and Regulates Immune Activation During Mycobacterial Infection. J. Infect. Dis. 208, 120–129. doi: 10.1093/infdis/jit133
Matricon, J., Barnich, N., Ardid, D. (2010). Immunopathogenesis of Inflammatory Bowel Disease. Self. Nonself. 1, 299–309. doi: 10.4161/self.1.4.13560
Mccaffrey, R. L., Schwartz, J. T., Lindemann, S. R., Moreland, J. G., Buchan, B. W., Jones, B. D., et al. (2010). Multiple Mechanisms of NADPH Oxidase Inhibition by Type A and Type B Francisella Tularensis. J. Leukoc. Biol. 88, 791–805. doi: 10.1189/jlb.1209811
Mcclean, C. M., Tobin, D. M. (2016). Macrophage Form, Function, and Phenotype in Mycobacterial Infection: Lessons From Tuberculosis and Other Diseases. Pathog. Dis. 74 (7), ftw068. doi: 10.1093/femspd/ftw068
Mcdougal, K. E., Fallin, M. D., Moller, D. R., Song, Z., Cutler, D. J., Steiner, L. L., et al. (2009). Variation in the Lymphotoxin-Alpha/Tumor Necrosis Factor Locus Modifies Risk of Erythema Nodosum in Sarcoidosis. J. Invest. Dermatol. 129, 1921–1926. doi: 10.1038/jid.2008.456
Medeiros, P., Da Silva, W. L., De Oliveira Gimenez, B. B., Vallezi, K. B., Moraes, M. O., De Souza, V. N. B., et al. (2016). The GATA3 Gene Is Involved in Leprosy Susceptibility in Brazilian Patients. Infect. Genet. Evol. 39, 194–200. doi: 10.1016/j.meegid.2016.01.015
Mehrotra, P., Jamwal, S. V., Saquib, N., Sinha, N., Siddiqui, Z., Manivel, V., et al. (2014). Pathogenicity of Mycobacterium Tuberculosis Is Expressed by Regulating Metabolic Thresholds of the Host Macrophage. PLoS Pathog. 10, e1004265. doi: 10.1371/journal.ppat.1004265
Minakami, R., Sumimotoa, H. (2006). Phagocytosis-Coupled Activation of the Superoxide-Producing Phagocyte Oxidase, a Member of the NADPH Oxidase (Nox) Family. Int. J. Hematol. 84, 193–198. doi: 10.1532/IJH97.06133
Mira, M. T., Alcais, A., Nguyen, V. T., Moraes, M. O., Di Flumeri, C., Vu, H. T., et al. (2004). Susceptibility to Leprosy Is Associated With PARK2 and PACRG. Nature 427, 636–640. doi: 10.1038/nature02326
Miro-Blanch, J., Yanes, O. (2019). Epigenetic Regulation at the Interplay Between Gut Microbiota and Host Metabolism. Front. Genet. 10, 638. doi: 10.3389/fgene.2019.00638
Mitchell, A., Dugas, A. (2019). Malakoplakia of the Colon Following Renal Transplantation in a 73 Year Old Woman: Report of a Case Presenting as Intestinal Perforation. Diagn. Pathol. 14, 22. doi: 10.1186/s13000-019-0799-z
Mostowy, S., Behr, M. A. (2005). The Origin and Evolution of Mycobacterium Tuberculosis. Clin. Chest. Med. 26, 207–216, v-vi. doi: 10.1016/j.ccm.2005.02.004
Mpofu, C. M., Campbell, B. J., Subramanian, S., Marshall-Clarke, S., Hart, C. A., Cross, A., et al. (2007). Microbial Mannan Inhibits Bacterial Killing by Macrophages: A Possible Pathogenic Mechanism for Crohn's Disease. Gastroenterology 133, 1487–1498. doi: 10.1053/j.gastro.2007.08.004
Muellegger, R. R., Weger, W., Zoechling, N., Kaddu, S., Soyer, H. P., El Shabrawi-Caelen, L., et al. (2000). Granulomatous Cheilitis and Borrelia Burgdorferi: Polymerase Chain Reaction and Serologic Studies in a Retrospective Case Series of 12 Patients. Arch. Dermatol. 136, 1502–1506. doi: 10.1001/archderm.136.12.1502
Muniraj, K., Padhi, S., Phansalkar, M., Sivakumar, P., Varghese, R. G., Kanungo, R. (2015). Bone Marrow Granuloma in Typhoid Fever: A Morphological Approach and Literature Review. Case Rep. Infect. Dis. 2015, 628028. doi: 10.1155/2015/628028
Mwipatayi, B. P., Jeffery, P. C., Beningfield, S. J., Matley, P. J., Naidoo, N. G., Kalla, A. A., et al. (2005). Takayasu Arteritis: Clinical Features and Management: Report of 272 Cases. ANZ. J. Surg. 75, 110–117. doi: 10.1111/j.1445-2197.2005.03312.x
Naderi, M., Hashemi, M., Abedipour, F., Bahari, G., Rezaei, M., Taheri, M. (2016). Evaluation of Interferon-Induced Transmembrane Protein-3 (IFITM3) Rs7478728 and Rs3888188 Polymorphisms and the Risk of Pulmonary Tuberculosis. BioMed. Rep. 5, 634–638. doi: 10.3892/br.2016.763
Nakamura, S., Yoshimori, T. (2017). New Insights Into Autophagosome-Lysosome Fusion. J. Cell Sci. 130, 1209–1216. doi: 10.1242/jcs.196352
Nakauchi, A., Wong, J. H., Mahasirimongkol, S., Yanai, H., Yuliwulandari, R., Mabuchi, A., et al. (2016). Identification of ITPA on Chromosome 20 as a Susceptibility Gene for Young-Onset Tuberculosis. Hum. Genome Var. 3, 15067. doi: 10.1038/hgv.2015.67
Naser, S. A., Arce, M., Khaja, A., Fernandez, M., Naser, N., Elwasila, S., et al. (2012). Role of ATG16L, NOD2 and IL23R in Crohn's Disease Pathogenesis. World J. Gastroenterol. 18, 412–424. doi: 10.3748/wjg.v18.i5.412
Neela, V. S., Suryadevara, N. C., Shinde, V. G., Pydi, S. S., Jain, S., Jonnalagada, S., et al. (2015). Association of Taq I, Fok I and Apa I Polymorphisms in Vitamin D Receptor (VDR) Gene With Leprosy. Hum. Immunol. 76, 402–405. doi: 10.1016/j.humimm.2015.04.002
Negroni, A., Pierdomenico, M., Cucchiara, S., Stronati, L. (2018). NOD2 and Inflammation: Current Insights. J. Inflammation Res. 11, 49–60. doi: 10.2147/JIR.S137606
Neyrolles, O., Mintz, E., Catty, P. (2013). Zinc and Copper Toxicity in Host Defense Against Pathogens: Mycobacterium Tuberculosis as a Model Example of an Emerging Paradigm. Front. Cell Infect. Microbiol. 3, 89. doi: 10.3389/fcimb.2013.00089
Nielsen, H., Frederiksen, J., Sherson, D., Milman, N. (1990). Comparison of Alveolar Macrophage and Blood Monocyte Oxidative Burst Response in Pulmonary Sarcoidosis. APMIS 98, 401–406. doi: 10.1111/j.1699-0463.1990.tb01050.x
Nonghanphithak, D., Reechaipichitkul, W., Namwat, W., Lulitanond, V., Naranbhai, V., Faksri, K. (2016). Genetic Polymorphisms of CCL2 Associated With Susceptibility to Latent Tuberculous Infection in Thailand. Int. J. Tuberc. Lung Dis. 20, 1242–1248. doi: 10.5588/ijtld.16.0017
Nunes-Alves, C., Booty, M. G., Carpenter, S. M., Jayaraman, P., Rothchild, A. C., Behar, S. M. (2014). In Search of a New Paradigm for Protective Immunity to TB. Nat. Rev. Microbiol. 12, 289–299. doi: 10.1038/nrmicro3230
Ogura, Y., Bonen, D. K., Inohara, N., Nicolae, D. L., Chen, F. F., Ramos, R., et al. (2001). A Frameshift Mutation in NOD2 Associated With Susceptibility to Crohn's Disease. Nature 411, 603–606. doi: 10.1038/35079114
Olsen, S. C., Palmer, M. V. (2014). Advancement of Knowledge of Brucella Over the Past 50 Years. Vet. Pathol. 51, 1076–1089. doi: 10.1177/0300985814540545
Olza, J., Aranceta-Bartrina, J., Gonzalez-Gross, M., Ortega, R. M., Serra-Majem, L., Varela-Moreiras, G., et al. (2017). Reported Dietary Intake and Food Sources of Zinc, Selenium, and Vitamins A, E and C in the Spanish Population: Findings From the ANIBES Study. Nutrients 9 (7), 697. doi: 10.3390/nu9070697
Orchard, T. R., Chua, C. N., Ahmad, T., Cheng, H., Welsh, K. I., Jewell, D. P. (2002). Uveitis and Erythema Nodosum in Inflammatory Bowel Disease: Clinical Features and the Role of HLA Genes. Gastroenterology 123, 714–718. doi: 10.1053/gast.2002.35396
Orchard, T. R., Thiyagaraja, S., Welsh, K. I., Wordsworth, B. P., Hill Gaston, J. S., Jewell, D. P. (2000). Clinical Phenotype Is Related to HLA Genotype in the Peripheral Arthropathies of Inflammatory Bowel Disease. Gastroenterology 118, 274–278. doi: 10.1016/S0016-5085(00)70209-5
Pal, K. (2014). Granulomatous Appendicitis in Children: A Single Institutional Experience. Afr. J. Paediatr. Surg. 11, 26–31. doi: 10.4103/0189-6725.129207
Palmela, C., Chevarin, C., Xu, Z., Torres, J., Sevrin, G., Hirten, R., et al. (2018). Adherent-Invasive Escherichia Coli in Inflammatory Bowel Disease. Gut 67, 574–587. doi: 10.1136/gutjnl-2017-314903
Pang, Y. Y., Schwartz, J., Thoendel, M., Ackermann, L. W., Horswill, A. R., Nauseef, W. M. (2010). Agr-Dependent Interactions of Staphylococcus Aureus USA300 With Human Polymorphonuclear Neutrophils. J. Innate. Immun. 2, 546–559. doi: 10.1159/000319855
Pennini, M. E., Pai, R. K., Schultz, D. C., Boom, W. H., Harding, C. V. (2006). Mycobacterium Tuberculosis 19-kDa Lipoprotein Inhibits IFN-Gamma-Induced Chromatin Remodeling of MHC2TA by TLR2 and MAPK Signaling. J. Immunol. 176, 4323–4330. doi: 10.4049/jimmunol.176.7.4323
Penn, B. H., Netter, Z., Johnson, J. R., Von Dollen, J., Jang, G. M., Johnson, T., et al. (2018). An Mtb-Human Protein-Protein Interaction Map Identifies a Switch Between Host Antiviral and Antibacterial Responses. Mol. Cell 71 637–648, e635. doi: 10.1016/j.molcel.2018.07.010
Petit-Bertron, A. F., Pedron, T., Gross, U., Coppee, J. Y., Sansonetti, P. J., Cavaillon, J. M., et al. (2005). Adherence Modifies the Regulation of Gene Expression Induced by Interleukin-10. Cytokine 29, 1–12. doi: 10.1016/j.cyto.2004.09.001
Petrino, M. G., Doetsch, R. N. (1978). 'Viscotaxis', a New Behavioural Response of Leptospira Interrogans (Biflexa) Strain B16. J. Gen. Microbiol. 109, 113–117. doi: 10.1099/00221287-109-1-113
Pontillo, A., Laurentino, W., Crovella, S., Pereira, A. C. (2013). NLRP1 Haplotypes Associated With Leprosy in Brazilian Patients. Infect. Genet. Evol. 19, 274–279. doi: 10.1016/j.meegid.2013.06.006
Porcheron, G., Garenaux, A., Proulx, J., Sabri, M., Dozois, C. M. (2013). Iron, Copper, Zinc, and Manganese Transport and Regulation in Pathogenic Enterobacteria: Correlations Between Strains, Site of Infection and the Relative Importance of the Different Metal Transport Systems for Virulence. Front. Cell Infect. Microbiol. 3, 90. doi: 10.3389/fcimb.2013.00090
Portevin, D., Sukumar, S., Coscolla, M., Shui, G., Li, B., Guan, X. L., et al. (2014). Lipidomics and Genomics of Mycobacterium Tuberculosis Reveal Lineage-Specific Trends in Mycolic Acid Biosynthesis. Microbiologyopen 3, 823–835. doi: 10.1002/mbo3.193
Qrafli, M., Amar, Y., Bourkadi, J., Ben Amor, J., Iraki, G., Bakri, Y., et al. (2014). The CYP7A1 Gene Rs3808607 Variant Is Associated With Susceptibility of Tuberculosis in Moroccan Population. Pan. Afr. Med. J. 18, 1. doi: 10.11604/pamj.2014.18.1.3397
Rahlwes, K. C., Sparks, I. L., Morita, Y. S. (2019). Cell Walls and Membranes of Actinobacteria. Subcell. Biochem. 92, 417–469. doi: 10.1007/978-3-030-18768-2_13
Ramos, G. B., Salomao, H., Francio, A. S., Fava, V. M., Werneck, R. I., Mira, M. T. (2016). Association Analysis Suggests SOD2 as a Newly Identified Candidate Gene Associated With Leprosy Susceptibility. J. Infect. Dis. 214, 475–478. doi: 10.1093/infdis/jiw170
Reischl, S., Troger, J., Jesinghaus, M., Kasajima, A., Wilhelm, D. F., Friess, H., et al. (2020). Annexin A1 Expression Capacity as a Determinant for Disease Severity in Crohn's Disease. Dig. Dis. 38, 398–407. doi: 10.1159/000505910
Remond, B., Dompmartin, A., Moreau, A., Esnault, P., Thomas, A., Mandard, J. C., et al. (1994). Cutaneous Malacoplakia. Int. J. Dermatol. 33, 538–542. doi: 10.1111/j.1365-4362.1994.tb02888.x
Requena, L., Fernandez-Figueras, M. T. (2007). Subcutaneous Granuloma Annulare. Semin. Cutan. Med. Surg. 26, 96–99. doi: 10.1016/j.sder.2007.02.006
Richeldi, L., Sorrentino, R., Saltini, C. (1993). HLA-DPB1 Glutamate 69: A Genetic Marker of Beryllium Disease. Science 262, 242–244. doi: 10.1126/science.8105536
Robinson, L. A., Smith, P., Sengupta, D. J., Prentice, J. L., Sandin, R. L. (2013). Molecular Analysis of Sarcoidosis Lymph Nodes for Microorganisms: A Case-Control Study With Clinical Correlates. BMJ Open 3, e004065. doi: 10.1136/bmjopen-2013-004065
Robson, C. D. (2000). Imaging of Granulomatous Lesions of the Neck in Children. Radiol. Clin. North Am. 38, 969–977. doi: 10.1016/S0033-8389(05)70215-3
Rock, F. L., Hardiman, G., Timans, J. C., Kastelein, R. A., Bazan, J. F. (1998). A Family of Human Receptors Structurally Related to Drosophila Toll. Proc. Natl. Acad. Sci. U.S.A. 95, 588–593. doi: 10.1073/pnas.95.2.588
Rodemann, J. F., Dubberke, E. R., Reske, K. A., Seo, D. H., Stone, C. D. (2007). Incidence of Clostridium Difficile Infection in Inflammatory Bowel Disease. Clin. Gastroenterol. Hepatol. 5, 339–344. doi: 10.1016/j.cgh.2006.12.027
Roltgen, K., Stinear, T. P., Pluschke, G. (2012). The Genome, Evolution and Diversity of Mycobacterium Ulcerans. Infect. Genet. Evol. 12, 522–529. doi: 10.1016/j.meegid.2012.01.018
Rooijakkers, S. H., Wu, J., Ruyken, M., Van Domselaar, R., Planken, K. L., Tzekou, A., et al. (2009). Structural and Functional Implications of the Alternative Complement Pathway C3 Convertase Stabilized by a Staphylococcal Inhibitor. Nat. Immunol. 10, 721–727. doi: 10.1038/ni.1756
Ruf, M. T., Sopoh, G. E., Brun, L. V., Dossou, A. D., Barogui, Y. T., Johnson, R. C., et al. (2011). Histopathological Changes and Clinical Responses of Buruli Ulcer Plaque Lesions During Chemotherapy: A Role for Surgical Removal of Necrotic Tissue? PLoS Negl. Trop. Dis. 5, e1334. doi: 10.1371/journal.pntd.0001334
Salie, M., Daya, M., Lucas, L. A., Warren, R. M., van der Spuy, G. D., Van Helden, P. D., et al. (2015). Association of Toll-Like Receptors With Susceptibility to Tuberculosis Suggests Sex-Specific Effects of TLR8 Polymorphisms. Infect. Genet. Evol. 34, 221–229. doi: 10.1016/j.meegid.2015.07.004
Schultz, B. M., Paduro, C. A., Salazar, G. A., Salazar-Echegarai, F. J., Sebastian, V. P., Riedel, C. A., et al. (2017). A Potential Role of Salmonella Infection in the Onset of Inflammatory Bowel Diseases. Front. Immunol. 8, 191. doi: 10.3389/fimmu.2017.00191
Schuring, R. P., Hamann, L., Faber, W. R., Pahan, D., Richardus, J. H., Schumann, R. R., et al. (2009). Polymorphism N248S in the Human Toll-Like Receptor 1 Gene Is Related to Leprosy and Leprosy Reactions. J. Infect. Dis. 199, 1816–1819. doi: 10.1086/599121
Scott, M. A., Mccurley, T. L., Vnencak-Jones, C. L., Hager, C., Mccoy, J. A., Anderson, B., et al. (1996). Cat Scratch Disease: Detection of Bartonella Henselae DNA in Archival Biopsies From Patients With Clinically, Serologically, and Histologically Defined Disease. Am. J. Pathol. 149, 2161–2167.
Sedano, H. O., Carlos, R., Koutlas, I. G. (1996). Respiratory Scleroma: A Clinicopathologic and Ultrastructural Study. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. Endod. 81, 665–671. doi: 10.1016/S1079-2104(96)80072-2
Seshadri, C., Thuong, N. T., Mai, N. T., Bang, N. D., Chau, T. T., Lewinsohn, D. M., et al. (2017). A Polymorphism in Human MR1 Is Associated With mRNA Expression and Susceptibility to Tuberculosis. Genes Immun. 18, 8–14. doi: 10.1038/gene.2016.41
Shah, K. K., Pritt, B. S., Alexander, M. P. (2017). Histopathologic Review of Granulomatous Inflammation. J. Clin. Tuberc. Other. Mycobact. Dis. 7, 1–12. doi: 10.1016/j.jctube.2017.02.001
Sharma, G., Upadhyay, S., Srilalitha, M., Nandicoori, V. K., Khosla, S. (2015). The Interaction of Mycobacterial Protein Rv2966c With Host Chromatin Is Mediated Through non-CpG Methylation and Histone H3/H4 Binding. Nucleic Acids Res. 43, 3922–3937. doi: 10.1093/nar/gkv261
Sibartie, S., Scully, P., Keohane, J., O'neill, S., O'mahony, J., O'hanlon, D., et al. (2010). Mycobacterium Avium Subsp. Paratuberculosis (MAP) as a Modifying Factor in Crohn's Disease. Inflammation Bowel. Dis. 16, 296–304. doi: 10.1002/ibd.21052
Silva, M. T. (2010). When Two is Better Than One: Macrophages and Neutrophils Work in Concert in Innate Immunity as Complementary and Cooperative Partners of a Myeloid Phagocyte System. J. Leukoc. Biol. 87, 93–106. doi: 10.1189/jlb.0809549
Silva, B. J., Barbosa, M. G., Andrade, P. R., Ferreira, H., Nery, J. A., Corte-Real, S., et al. (2017). Autophagy Is an Innate Mechanism Associated With Leprosy Polarization. PLoS Pathog. 13, e1006103. doi: 10.1371/journal.ppat.1006103
Silva, C. A., Danelishvili, L., Mcnamara, M., Berredo-Pinho, M., Bildfell, R., Biet, F., et al. (2013). Interaction of Mycobacterium Leprae With Human Airway Epithelial Cells: Adherence, Entry, Survival, and Identification of Potential Adhesins by Surface Proteome Analysis. Infect. Immun. 81, 2645–2659. doi: 10.1128/IAI.00147-13
Sinha, B., Francois, P. P., Nusse, O., Foti, M., Hartford, O. M., Vaudaux, P., et al. (1999). Fibronectin-Binding Protein Acts as Staphylococcus Aureus Invasin via Fibronectin Bridging to Integrin Alpha5beta1. Cell Microbiol. 1, 101–117. doi: 10.1046/j.1462-5822.1999.00011.x
Siregar, T., Prombutara, P., Kanjanasirirat, P., Kunkaew, N., Tubsuwan, A., Boonmee, A., et al. (2022). The Autophagy-Resistant Mycobacterium Tuberculosis Beijing Strain Upregulates KatG to Evade Starvation-Induced Autophagic Restriction. Pathog. Dis. 80 (1), ftac004. doi: 10.1093/femspd/ftac004
Smith, J., Manoranjan, J., Pan, M., Bohsali, A., Xu, J., Liu, J., et al. (2008). Evidence for Pore Formation in Host Cell Membranes by ESX-1-Secreted ESAT-6 and Its Role in Mycobacterium Marinum Escape From the Vacuole. Infect. Immun. 76, 5478–5487. doi: 10.1128/IAI.00614-08
Sobota, R. S., Stein, C. M., Kodaman, N., Scheinfeldt, L. B., Maro, I., Wieland-Alter, W., et al. (2016). A Locus at 5q33.3 Confers Resistance to Tuberculosis in Highly Susceptible Individuals. Am. J. Hum. Genet. 98, 514–524. doi: 10.1016/j.ajhg.2016.01.015
Song, G. G., Kim, J. H., Lee, Y. H. (2014). Associations Between TNF-Alpha -308 a/G and Lymphotoxin-Alpha +252 a/G Polymorphisms and Susceptibility to Sarcoidosis: A Meta-Analysis. Mol. Biol. Rep. 41, 259–267. doi: 10.1007/s11033-013-2859-x
Sorbara, M. T., Ellison, L. K., Ramjeet, M., Travassos, L. H., Jones, N. L., Girardin, S. E., et al. (2013). The Protein ATG16L1 Suppresses Inflammatory Cytokines Induced by the Intracellular Sensors Nod1 and Nod2 in an Autophagy-Independent Manner. Immunity 39, 858–873. doi: 10.1016/j.immuni.2013.10.013
Sorensen, S. J., Bailey, M., Hansen, L. H., Kroer, N., Wuertz, S. (2005). Studying Plasmid Horizontal Transfer in Situ: A Critical Review. Nat. Rev. Microbiol. 3, 700–710. doi: 10.1038/nrmicro1232
Soto, M. E., Del Carmen Avila-Casado, M., Huesca-Gomez, C., Alarcon, G. V., Castrejon, V., Soto, V., et al. (2012). Detection of IS6110 and HupB Gene Sequences of Mycobacterium Tuberculosis and Bovis in the Aortic Tissue of Patients With Takayasu's Arteritis. BMC Infect. Dis. 12, 194. doi: 10.1186/1471-2334-12-194
Spalinger, M. R., Kasper, S., Chassard, C., Raselli, T., Frey-Wagner, I., Gottier, C., et al. (2015). PTPN2 Controls Differentiation of CD4(+) T Cells and Limits Intestinal Inflammation and Intestinal Dysbiosis. Mucosal Immunol. 8, 918–929. doi: 10.1038/mi.2014.122
Spalinger, M. R., Manzini, R., Hering, L., Riggs, J. B., Gottier, C., Lang, S., et al. (2018). PTPN2 Regulates Inflammasome Activation and Controls Onset of Intestinal Inflammation and Colon Cancer. Cell Rep. 22, 1835–1848. doi: 10.1016/j.celrep.2018.01.052
Stallings, C. L., Glickman, M. S. (2010). Is Mycobacterium Tuberculosis Stressed Out? A Critical Assessment of the Genetic Evidence. Microbes Infect. 12, 1091–1101. doi: 10.1016/j.micinf.2010.07.014
Stanton, M. J., Maxted, W. (1981). Malacoplakia: A Study of the Literature and Current Concepts of Pathogenesis, Diagnosis and Treatment. J. Urol. 125, 139–146. doi: 10.1016/S0022-5347(17)54940-X
Stegeman, C. A., Tervaert, J. W., Sluiter, W. J., Manson, W. L., De Jong, P. E., Kallenberg, C. G. (1994). Association of Chronic Nasal Carriage of Staphylococcus Aureus and Higher Relapse Rates in Wegener Granulomatosis. Ann. Intern. Med. 120, 12–17. doi: 10.7326/0003-4819-120-1-199401010-00003
Stienstra, Y., van der Werf, T. S., Oosterom, E., Nolte, I. M., van der Graaf, W. T., Etuaful, S., et al. (2006). Susceptibility to Buruli Ulcer Is Associated With the SLC11A1 (NRAMP1) D543N Polymorphism. Genes Immun. 7, 185–189. doi: 10.1038/sj.gene.6364281
Stinear, T. P., Seemann, T., Pidot, S., Frigui, W., Reysset, G., Garnier, T., et al. (2007). Reductive Evolution and Niche Adaptation Inferred From the Genome of Mycobacterium Ulcerans, the Causative Agent of Buruli Ulcer. Genome Res. 17, 192–200. doi: 10.1101/gr.5942807
Stokes, R. W., Norris-Jones, R., Brooks, D. E., Beveridge, T. J., Doxsee, D., Thorson, L. M. (2004). The Glycan-Rich Outer Layer of the Cell Wall of Mycobacterium Tuberculosis Acts as an Antiphagocytic Capsule Limiting the Association of the Bacterium With Macrophages. Infect. Immun. 72, 5676–5686. doi: 10.1128/IAI.72.10.5676-5686.2004
Sundaramurthy, V., Korf, H., Singla, A., Scherr, N., Nguyen, L., Ferrari, G., et al. (2017). Survival of Mycobacterium Tuberculosis and Mycobacterium Bovis BCG in Lysosomes In Vivo. Microbes Infect. 19, 515–526. doi: 10.1016/j.micinf.2017.06.008
Sureisen, M., Choon, S. K., Tai, C. C. (2008). Recurrent Chromobacterium Violaceum Infection in a Patient With Chronic Granulomatous Disease. Med. J. Malays. 63, 346–347.
Sveinbjornsson, G., Gudbjartsson, D. F., Halldorsson, B. V., Kristinsson, K. G., Gottfredsson, M., Barrett, J. C., et al. (2016). HLA Class II Sequence Variants Influence Tuberculosis Risk in Populations of European Ancestry. Nat. Genet. 48, 318–322. doi: 10.1038/ng.3498
Takeuchi, O., Akira, S. (2010). Pattern Recognition Receptors and Inflammation. Cell 140, 805–820. doi: 10.1016/j.cell.2010.01.022
Thakker, M., Park, J. S., Carey, V., Lee, J. C. (1998). Staphylococcus Aureus Serotype 5 Capsular Polysaccharide Is Antiphagocytic and Enhances Bacterial Virulence in a Murine Bacteremia Model. Infect. Immun. 66, 5183–5189. doi: 10.1128/IAI.66.11.5183-5189.1998
Thompson, M. R., Kaminski, J. J., Kurt-Jones, E. A., Fitzgerald, K. A. (2011). Pattern Recognition Receptors and the Innate Immune Response to Viral Infection. Viruses 3, 920–940. doi: 10.3390/v3060920
Thuong, N. T., Dunstan, S. J., Chau, T. T., Thorsson, V., Simmons, C. P., Quyen, N. T., et al. (2008). Identification of Tuberculosis Susceptibility Genes With Human Macrophage Gene Expression Profiles. PLoS Pathog. 4, e1000229. doi: 10.1371/journal.ppat.1000229
Tondo-Steele, K., Rath, K., Niemeier, L. (2020). Granulomatous Peritonitis Secondary to Chlamydia Trachomatis: A Case Report. Gynecol. Oncol. Rep. 32, 100558. doi: 10.1016/j.gore.2020.100558
Tong, Y., Yamaguchi, H., Giaime, E., Boyle, S., Kopan, R., Kelleher, R. J. (2010). Loss of Leucine-Rich Repeat Kinase 2 Causes Impairment of Protein Degradation Pathways, Accumulation of Alpha-Synuclein, and Apoptotic Cell Death in Aged Mice. Proc. Natl. Acad. Sci. U.S.A. 107, 9879–9884. doi: 10.1073/pnas.1004676107
Turner, R. D., Chiu, C., Churchyard, G. J., Esmail, H., Lewinsohn, D. M., Gandhi, N. R., et al. (2017). Tuberculosis Infectiousness and Host Susceptibility. J. Infect. Dis. 216, S636–S643. doi: 10.1093/infdis/jix361
Turpin, W., Goethel, A., Bedrani, L., Croitoru Mdcm, K. (2018). Determinants of IBD Heritability: Genes, Bugs, and More. Inflammation Bowel. Dis. 24, 1133–1148. doi: 10.1093/ibd/izy085
Valentonyte, R., Hampe, J., Huse, K., Rosenstiel, P., Albrecht, M., Stenzel, A., et al. (2005). Sarcoidosis Is Associated With a Truncating Splice Site Mutation in BTNL2. Nat. Genet. 37, 357–364. doi: 10.1038/ng1519
Van Acker, H., Coenye, T. (2017). The Role of Reactive Oxygen Species in Antibiotic-Mediated Killing of Bacteria. Trends Microbiol. 25, 456–466. doi: 10.1016/j.tim.2016.12.008
Van Der Wel, N., Hava, D., Houben, D., Fluitsma, D., Van Zon, M., Pierson, J., et al. (2007). M. Tuberculosis and M. Leprae Translocate From the Phagolysosome to the Cytosol in Myeloid Cells. Cell 129, 1287–1298. doi: 10.1016/j.cell.2007.05.059
Van De Veerdonk, F. L., Dinarello, C. A. (2014). Deficient Autophagy Unravels the ROS Paradox in Chronic Granulomatous Disease. Autophagy 10, 1141–1142. doi: 10.4161/auto.28638
Van Limbergen, J., Wilson, D. C., Satsangi, J. (2009). The Genetics of Crohn's Disease. Annu. Rev. Genomics Hum. Genet. 10, 89–116. doi: 10.1146/annurev-genom-082908-150013
Van Zandt, K. E., Sow, F. B., Florence, W. C., Zwilling, B. S., Satoskar, A. R., Schlesinger, L. S., et al. (2008). The Iron Export Protein Ferroportin 1 is Differentially Expressed in Mouse Macrophage Populations and is Present in the Mycobacterial-Containing Phagosome. J. Leukoc. Biol. 84, 689–700. doi: 10.1189/jlb.1107781
Villani, A. C., Lemire, M., Fortin, G., Louis, E., Silverberg, M. S., Collette, C., et al. (2009). Common Variants in the NLRP3 Region Contribute to Crohn's Disease Susceptibility. Nat. Genet. 41, 71–76. doi: 10.1038/ng.285
Vining, M., Sharma, N., Guill, M. (2014). Atypical Presentation of Chronic Granulomatous Disease With Burkholderia Cepacia. BMJ Case Rep. 2014, bcr2013201524. doi: 10.1136/bcr-2013-201524
Wall, D., Kaiser, D. (1999). Type IV Pili and Cell Motility. Mol. Microbiol. 32, 1–10. doi: 10.1046/j.1365-2958.1999.01339.x
Wang, D., Fan, Y., Malhi, M., Bi, R., Wu, Y., Xu, M., et al. (2018). Missense Variants in HIF1A and LACC1 Contribute to Leprosy Risk in Han Chinese. Am. J. Hum. Genet. 102, 794–805. doi: 10.1016/j.ajhg.2018.03.006
Wang, J., Yang, K., Zhou, L., Minhaowu, Wu, Y., Zhu, M., et al. (2013). MicroRNA-155 Promotes Autophagy to Eliminate Intracellular Mycobacteria by Targeting Rheb. PLoS Pathog. 9, e1003697. doi: 10.1371/journal.ppat.1003697
Wayne, L. G., Hayes, L. G. (1996). An In Vitro Model for Sequential Study of Shiftdown of Mycobacterium Tuberculosis Through Two Stages of Nonreplicating Persistence. Infect. Immun. 64, 2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996
Welin, A., Eklund, D., Stendahl, O., Lerm, M. (2011). Human Macrophages Infected With a High Burden of ESAT-6-Expressing M. Tuberculosis Undergo Caspase-1- and Cathepsin B-Independent Necrosis. PLoS One 6, e20302. doi: 10.1371/journal.pone.0020302
Werner, J. L., Escolero, S. G., Hewlett, J. T., Mak, T. N., Williams, B. P., Eishi, Y., et al. (2017). Induction of Pulmonary Granuloma Formation by Propionibacterium Acnes Is Regulated by MyD88 and Nox2. Am. J. Respir. Cell Mol. Biol. 56, 121–130. doi: 10.1165/rcmb.2016-0035OC
Wieczorek, S., Hoffjan, S., Chan, A., Rey, L., Harper, L., Fricke, H., et al. (2009). Novel Association of the CD226 (DNAM-1) Gly307Ser Polymorphism in Wegener's Granulomatosis and Confirmation for Multiple Sclerosis in German Patients. Genes Immun. 10, 591–595. doi: 10.1038/gene.2009.44
Wieczorek, S., Holle, J. U., Muller, S., Fricke, H., Gross, W. L., Epplen, J. T. (2010). A Functionally Relevant IRF5 Haplotype Is Associated With Reduced Risk to Wegener's Granulomatosis. J. Mol. Med. (Berl.) 88, 413–421. doi: 10.1007/s00109-009-0580-y
Wiener, H. W., Perry, R. T., Chen, Z., Harrell, L. E., Go, R. C. (2007). A Polymorphism in SOD2 Is Associated With Development of Alzheimer's Disease. Genes Brain Behav. 6, 770–775. doi: 10.1111/j.1601-183X.2007.00308.x
Willcocks, L. C., Lyons, P. A., Clatworthy, M. R., Robinson, J. I., Yang, W., Newland, S. A., et al. (2008). Copy Number of FCGR3B, Which Is Associated With Systemic Lupus Erythematosus, Correlates With Protein Expression and Immune Complex Uptake. J. Exp. Med. 205, 1573–1582. doi: 10.1084/jem.20072413
Williams, G. T., Williams, W. J. (1983). Granulomatous Inflammation–A Review. J. Clin. Pathol. 36, 723–733. doi: 10.1136/jcp.36.7.723
Xie, G., Roshandel, D., Sherva, R., Monach, P. A., Lu, E. Y., Kung, T., et al. (2013). Association of Granulomatosis With Polyangiitis (Wegener's) With HLA-DPB1*04 and SEMA6A Gene Variants: Evidence From Genome-Wide Analysis. Arthritis Rheum. 65, 2457–2468. doi: 10.1002/art.38036
Xu, W. D., Xie, Q. B., Zhao, Y., Liu, Y. (2015). Association of Interleukin-23 Receptor Gene Polymorphisms With Susceptibility to Crohn's Disease: A Meta-Analysis. Sci. Rep. 5, 18584. doi: 10.1038/srep18584
Yamamoto, M., Yoshizaki, K., Kishimoto, T., Ito, H. (2000). IL-6 is Required for the Development of Th1 Cell-Mediated Murine Colitis. J. Immunol. 164, 4878–4882. doi: 10.4049/jimmunol.164.9.4878
Yi, Y. X., Han, J. B., Zhao, L., Fang, Y., Zhang, Y. F., Zhou, G. Y. (2015). Tumor Necrosis Factor Alpha Gene Polymorphism Contributes to Pulmonary Tuberculosis Susceptibility: Evidence From a Meta-Analysis. Int. J. Clin. Exp. Med. 8, 20690–20700.
Yi, L., Zhang, K., Mo, Y., Zhen, G., Zhao, J. (2015). The Association Between CD209 Gene Polymorphisms and Pulmonary Tuberculosis Susceptibility: A Meta-Analysis. Int. J. Clin. Exp. Pathol. 8, 12437–12445.
Yoshimori, T., Amano, A. (2009). Group a Streptococcus: A Loser in the Battle With Autophagy. Curr. Top. Microbiol. Immunol. 335, 217–226. doi: 10.1007/978-3-642-00302-8_10
Yuan, L., Ke, Z., Ma, J., Guo, Y., Li, Y. (2016). IRGM Gene Polymorphisms and Haplotypes Associate With Susceptibility of Pulmonary Tuberculosis in Chinese Hubei Han Population. Tuberculo. (Edinb.) 96, 58–64. doi: 10.1016/j.tube.2015.10.014
Zambrano, M. M., Kolter, R. (2005). Mycobacterial Biofilms: A Greasy Way to Hold it Together. Cell 123, 762–764. doi: 10.1016/j.cell.2005.11.011
Zhang, X., Cheng, Y., Zhang, Q., Wang, X., Lin, Y., Yang, C., et al. (2019). Meta-Analysis Identifies Major Histocompatiblity Complex Loci in or Near HLA-DRB1, HLA-DQA1, HLA-C as Associated With Leprosy in Chinese Han Population. J. Invest. Dermatol. 139, 957–960. doi: 10.1016/j.jid.2018.09.029
Zhang, F. R., Huang, W., Chen, S. M., Sun, L. D., Liu, H., Li, Y., et al. (2009). Genomewide Association Study of Leprosy. N. Engl. J. Med. 361, 2609–2618. doi: 10.1056/NEJMoa0903753
Zhang, J., Jiao, L., Bai, H., Wu, Q., Wu, T., Liu, T., et al. (2020). A Notch4 Missense Mutation is Associated With Susceptibility to Tuberculosis in Chinese Population. Infect. Genet. Evol. 78, 104145. doi: 10.1016/j.meegid.2019.104145
Zhang, L., Lee, H., Grimm, M. C., Riordan, S. M., Day, A. S., Lemberg, D. A. (2014). Campylobacter Concisus and Inflammatory Bowel Disease. World J. Gastroenterol. 20, 1259–1267. doi: 10.3748/wjg.v20.i5.1259
Zhang, F., Liu, H., Chen, S., Low, H., Sun, L., Cui, Y., et al. (2011). Identification of Two New Loci at IL23R and RAB32 That Influence Susceptibility to Leprosy. Nat. Genet. 43, 1247–1251. doi: 10.1038/ng.973
Zhang, S., Wang, X. B., Han, Y. D., Wang, C., Zhou, Y., Zheng, F. (2017). Certain Polymorphisms in SP110 Gene Confer Susceptibility to Tuberculosis: A Comprehensive Review and Updated Meta-Analysis. Yonsei. Med. J. 58, 165–173. doi: 10.3349/ymj.2017.58.1.165
Zhao, Y., Bu, H., Hong, K., Yin, H., Zou, Y. L., Geng, S. J., et al. (2015). Genetic Polymorphisms of CCL1 Rs2072069 G/A and TLR2 Rs3804099 T/C in Pulmonary or Meningeal Tuberculosis Patients. Int. J. Clin. Exp. Pathol. 8, 12608–12620.
Keywords: bacteria, granulomatous inflammation diseases, predisposing factors, immune response, gene, intracellular bacteria
Citation: Wang X and Liu Y (2022) Offense and Defense in Granulomatous Inflammation Disease. Front. Cell. Infect. Microbiol. 12:797749. doi: 10.3389/fcimb.2022.797749
Received: 19 October 2021; Accepted: 30 May 2022;
Published: 29 June 2022.
Edited by:
Antje Mueller, University of Lübeck, GermanyReviewed by:
Shibali Das, Washington University in St. Louis, United StatesSumanta Naik, Washington University in St. Louis, United States
Copyright © 2022 Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan Liu, bGl1eXVhbkBmbW11LmVkdS5jbg==
 Xinwen Wang
Xinwen Wang