
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 15 June 2022
Sec. Microbiome in Health and Disease
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.795343
This article is part of the Research Topic C. elegans Host-Microbiome Interactions: From Medical to Ecological and Evolutionary Model View all 8 articles
Gut bacteria are often described by the neutral term commensals. However, the more we learn about their interactions with hosts, the more apparent it becomes that gut commensals often contribute positively to host physiology and fitness. Whether hosts can prefer beneficial bacteria, and how they do so, is not clear. This is of particular interest in the case of the bacterivore C. elegans, which depends on bacteria as food source, but also as gut colonizers that contribute to its physiology, from development to immunity. It is further unclear to what extent worms living in their microbially-diverse habitats can sense and distinguish between beneficial bacteria, food, and pathogens. Focusing on Enterobacteriaceae and members of closely related families, we isolated gut bacteria from worms raised in compost microcosms, as well as bacteria from the respective environments and evaluated their contributions to host development. Most isolates, from worms or from the surrounding environment, promoted faster development compared to the non-colonizing E. coli food strain. Pantoea strains further showed differential contributions of gut isolates versus an environmental isolate. Characterizing bacterial ability to hinder pathogenic colonization with Pseudomonas aeruginosa, supported the trend of Pantoea gut commensals being beneficial, in contrast to the environmental strain. Interestingly, worms were attracted to the beneficial Pantoea strains, preferring them over non-beneficial bacteria, including the environmental Pantoea strain. While our understanding of the mechanisms underlying these host-microbe interactions are still rudimentary, the results suggest that hosts can sense and prefer beneficial commensals.
Bacterial community composition in plant and animal hosts are typically distinct from those in the surrounding environments (Backhed et al., 2005; Hacquard et al., 2015; Wippel et al., 2021). What determines this distinction, and which factors dominate the process of microbiota assembly is an active area of research and a topic of debate (Ley et al., 2008; Gilbert et al., 2018; Sieber et al., 2019). Environmental availability and external environmental factors, such as temperature, toxins and host diet are important (Spor et al., 2011; Smith et al., 2015; Callens et al., 2018; Simon et al., 2019). Equally important is host filtering, which is dependent on host genetics and is dominated by host immunity, internal nutrient availability and behavior (Hacquard et al., 2015; Berg et al., 2016a; 2019; Davenport et al., 2017; Goodrich et al., 2017), but is also affected by bacterial interactions, which can impact microbiota diversity and are known to affect host physiology and fitness (Lawley and Walker, 2013; Simon et al., 2019; Johnke et al., 2020).
The nematode C. elegans is well-suited for studying host-microbe interactions. Clonal populations and tight genetic control reduce inter-individual variation when examining gut microbiota composition, and a transparent body permits in vivo monitoring of bacterial colonization (Shapira, 2017). Over the past several years, work in this system has yielded important insights into the processes that shape the gut microbiota, in particular with respect to the role of host genes (Berg et al., 2019; Ortiz et al., 2021; Zhang et al., 2021). C. elegans feeds on bacteria from its environment, but specific bacteria resist digestion and persist in the worm gut giving rise to a characteristic gut microbiota shaped by environmental availability as well as by host genetics (Berg et al., 2016a; 2016b; Zhang et al., 2017; 2021). Work in the past several years has characterized the composition and diversity of the C. elegans gut microbiota, and common taxa were identified across worm populations isolated from different geographical locations (Berg et al., 2016a; Dirksen et al., 2016; Samuel et al., 2016). Bacteria of the Enterobacteriaceae family are among the most prevalent, and several species of this family were shown to have positive effects on host development (Berg et al., 2019; Dirksen et al., 2020) [although this may come with a trade-off for adult stress resistance (Slowinski et al., 2020)]. Additional families including Pseudomonadaceae, Xanthomondaceae, Comamonadaceae and Brucellaceae, contain members that have similar contributions to worm development (Watson et al., 2014; Samuel et al., 2016; Dirksen et al., 2020; Zhang et al., 2021).
Gut bacteria are often described as commensals, but the more they are studied, the clearer it becomes that many contribute positively to host functions, such as development, as described above, metabolism (Zimmermann et al., 2020), and often, to colonization resistance - preventing invasion by pathogens, but also by non-pathogenic colonizers that may perturb homeostasis. Such colonization resistance depends on competition for nutrients, inter-bacterial warfare mediated by secreted toxins, or activation of host immunity (Lawley and Walker, 2013; Sorbara and Pamer, 2019). In C. elegans, colonization resistance has been so far shown to depend on secretion of anti-microbial peptides (Dirksen et al., 2016; Kissoyan et al., 2019) and on immune priming (Montalvo-Katz et al., 2013; Berg et al., 2016b; Willis et al., 2021). While research focuses on beneficial commensals, not all environmentally available bacteria are necessarily beneficial. Whether hosts can distinguish between species or strains that are beneficial and those that are less so is unclear. Previous work with Enterobacter commensals isolated from C. elegans or from C. briggsae demonstrated that commensals of one host species could protect it from a subsequent exposure to a pathogen, but failed to protect the other host species, suggesting a functional adaptation between hosts and their cognate commensals, the basis for which is still unknown (Berg et al., 2016b). If host preference for certain bacteria existed, it could be mediated by differential bacterial abilities to colonize and persist in the worm gut, or alternatively, by host behavior, as previously described for C. elegans preference of food sources that better supported its growth (Shtonda and Avery, 2006; Kim and Flavell, 2020).
In this study, we set out to expand the repertoire of beneficial gut bacteria, focusing on members of the Enterobacteriaceae family and of the Enterobacterales order, to examine whether some taxa are more likely than others to colonize the worm gut, and to explore the worm’s ability to prefer beneficial ones. This analysis identified the genus Pantoea [part of the Erwiniaceae family, a recent splinter off the Enterobacteriaceae family (Adeolu et al., 2016)], of which gut isolates showed significant contributions to host fitness. Further analysis demonstrated that beneficial Pantoea strains were preferred as gut commensals due to a greater ability to colonize than a non-beneficial congeneric, but also through host attraction toward these bacteria.
All experiments were performed using worms of the Bristol N2 strain. Those were raised either on nematode growth medium (NGM) or on peptone-free medium (PFM). E. coli strain OP50 was used as the standard food to raise worms and as a reference point in comparisons with other bacteria. A GFP-expressing version of P. aeruginosa strain PA14 was used as a model pathogen. The N2, OP50 and PA14-GFP strains were originally obtained from the Caenorhabditis Genome Center (CGC).
Microcosms were prepared with 5 gr oak knoll soil composted for two weeks either with chopped bananas or apples, cured of endogenous non-C. elegans nematodes and reconstituted with the original bacterial community, as previously described (Berg et al., 2016a). Initially germ-free L1 larvae were added to microcosms, raised at 20°C for four days, following which adults were harvested, washed intensively and surface sterilized, ascertaining removal of external bacteria by plating final wash solutions on lysogeny broth (LB) plates/no antibiotics (Berg et al., 2016a). Bacteria were released from worms by grinding the latter with a motorized pestle in a volume of 100 µl M9, pelleting debris, and plating bacteria from supernatants on Enterobacteriaceae/Enterobacterales-selective violet red bile glucose (VRBG) agar (Difco) or on non-selective rich media, tryptic soy (TS) or LB agar. Plates were incubated at 25°C for 48 hours, from which colonies were regrown, and frozen at -80°C till subsequent analyses. Bacteria were similarly cultured from the respective microcosm environments, following resuspension in M9 solution, vortexing to disrupt larger soil aggregates and plating supernatant after spinning down (one minute at 1800 rpm) soil particles.
Twenty bacterial clones were randomly selected from each microcosm environments. Approximately 160 strains were isolated in total, of which 69 (41 from soil, 28 from worms) were taxonomically classified at the genus level following Sanger sequencing of full-length 16S rRNA gene amplified using primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′).
The impact of individual bacterial strains on C. elegans growth rate was assessed by comparing the percentage of animals reaching larval L4 stage to that in animals raised on E. coli OP50. Each strain was tested in at least two independent experiments. Worm populations were raised from eggs obtained by bleaching gravid worms, or from starvation-synchronized L1 larvae (50-100 worms per plate), on lawns of bacterial isolates (cultured overnight in LB at 28°C, concentrated 10x and plated on NGM plates) for 40 hours at 25°C, at which point the percentage of L4 larvae was evaluated. Each plate was divided to quarters. Starting to the upper left of the quadrant, the stage of the first 25 worms observed was assessed.
L1 worms were grown at 25°C to early adulthood (36 hours) on PFM plates with lawns of different Pantoea strains (prepared as described above). Adult worms were washed off plates, washed twice with M9, transferred to slow killing plates seeded with PA14-GFP (Shapira and Tan, 2008), and further replenished with the respective Pantoea strains (50 ml 20-fold concentrated two-day culture). Following additional 40 to 48 hours at 25°C, 20-30 worms were picked off plates, washed twice with 1 mL M9 in an Eppendorf tube, paralyzed with 10mM levamisole and mounted on 2% to 4% agarose pads to be imaged using a Leica MZ16F stereoscope equipped with a QImaging MicroPublisher 5.0 camera. Images of worms of different experimental groups were all taken using identical exposure settings.
Quantification of fluorescence signal was achieved using a protocol implemented on the Fiji plugin within the ImageJ package v2.10/1.53c (Schindelin et al., 2012). Fluorescent signal per worm was calculated using Integrated Density values, following subtraction of background mean gray values and autofluorescence, and subsequently normalized for worm size.
To bring together results from different experiments, normalized fluorescent signal values per worm were additionally scaled by the median value in worms raised on OP50 of the respective experiment.
Normalized PA14-GFP signal values correlated well with the number of colonizing bacteria as evaluated by colony-forming unit (CFU) counts (Supplementary Figure 1).
Bacteria were killed by a one-hour incubation at 28°C with paraformaldehyde (PFA) at a final concentration of 0.5% as previously described (Beydoun et al., 2021). Dead bacteria were then washed five times, concentrated, OD adjusted and plated on the appropriate plates.
Adult worms raised at 20°C (72 hours) on PFM plates with lawns of the examined bacterial strains were rinsed off and washed as previously described with several extra washes (Dirksen et al., 2020). Three 10-worm samples were taken from each population and washed five times more with 1 mL M9/0.01% Triton. Each 10-worm sample was transferred in a 100 mL volume to tubes containing 10-15 zirconia beads and broken to release bacteria using a PowerLyser (30 sec, 4000 rpm). Lysates were serially diluted, plated on VRBG plates, and colonies appearing after 24 hours of incubation at 28°C were counted. Post-wash worm-free supernatant was plated in parallel to ascertain the lack of contaminating external bacteria.
Worms were raised to young adulthood on OP50 lawns on PFM plates and shifted following three M9 washes to the center of test plates equidistantly spotted (5 uL volume, 1.5 cm from center) with tested strains at similar densities. In pairwise comparisons, plates contained two spots of each strain; worms at each spot were counted following 3 hours at 20°C, calculating a choice index (CI) as # Worms on strain A - #Worms on strain B/Total # of scored worms (Abada et al., 2009). In multiple strain plates, worms were offered a choice of each of the Pantoea strains as well as of OP50, and the percentage in each spot was evaluated after 3 hours.
Statistical evaluation was performed in R (v 3.6.3). Plots were generated using the ggplot2 R package (Villanueva and Chen, 2019).
To explore C. elegans preferences of bacteria from its environment, and bacterial contributions to host fitness, we isolated bacteria from worms raised in compost microcosms, as well as from their immediate environments. Microcosms were made of local soil composted with produce to emulate environments from which C. elegans has been previously isolated (Berg et al., 2016a). Bacteria were isolated by culturing on VRBG selective medium (see Methods), to focus on bacteria of the Enterobacteriaceae family/Enterobacterales order, which were previously shown to be a significant component of the worm gut microbiota and have diverse effects on host fitness (Berg et al., 2016a; 2016b; Samuel et al., 2016; Zhang et al., 2017; Slowinski et al., 2020). Culturing on rich media plates further supplemented isolation efforts with non-Enterobacterales isolates. Sixty-nine of the isolates were characterized by full-length 16S (identified at genus-level resolution, although not always unambiguously) and were employed in subsequent analysis (Figure 1A; Supplementary Table S1, Accession numbers OK487509-OK487574). Among the different genera, some were represented mostly (or exclusively) by environmental isolates (Erwinia, Pseudomonas, Rahnella), while others were represented primarily by gut isolates (Kluyvera and Raoultella). This may suggest preferred niches for the respective genera. However, it should be noted that bacteria of the Pseudomonas genus (which also grew on VRBG with a different colony morphology than Enterobacterales), here not found in worm guts, were previously isolated from worms (Montalvo-Katz et al., 2013; Schulenburg and Felix, 2017; Zimmermann et al., 2020).
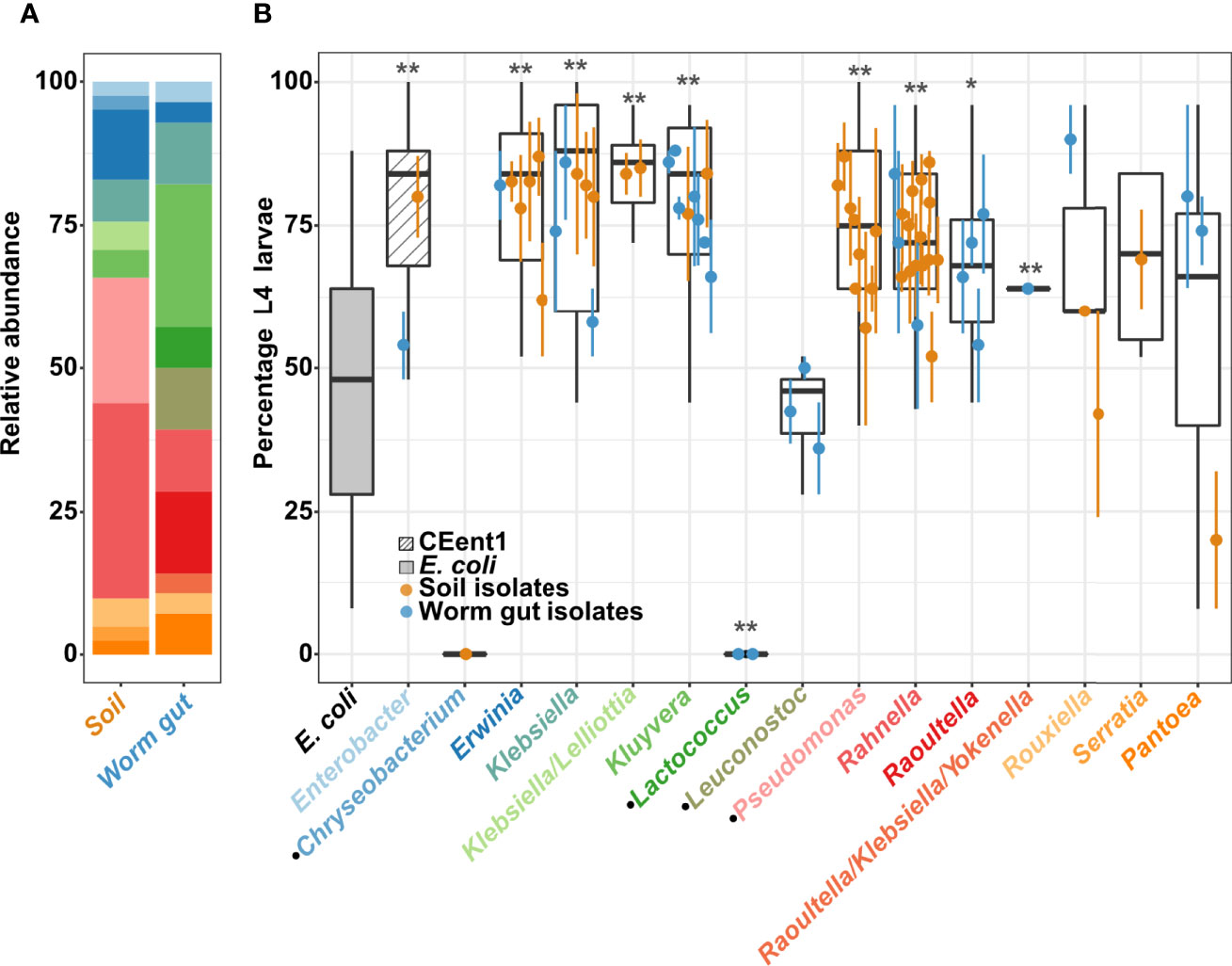
Figure 1 Effects of environmental and worm gut isolates on C. elegans development. (A) Bacterial isolates distribution. Relative abundance of genera recovered from soil microcosms and from worm guts, identified based on full length 16S Sanger sequencing and colored as in panel B x-axis (see also Supplementary Table 1). (B) Percentage of L4 larvae in worm populations raised from eggs for 40 hours at 25°C on environmental or gut bacterial isolates, as designated, grouped at the genus level; N=25/strain/experiment, dots represent averages (± SE) of 2 independent experiments. E. coli strain OP50 and Enterobacter hormaechei strain CEent1, in the gray and the hatched box, respectively, represent values from 41 experiments and provide reference for comparisons. Dotted lines represent the mean value for development on OP50, mean + SD and mean + 2 SD’s. Boxes extend from first to third quartiles, with black lines representing the median. *p < 0.05; **p < 0.001, in comparison to E. coli (Student’s t-test, corrected for multiple testing using Bonferroni). Black dots at x-axis labels indicate non-Enterobacteriales isolates.
Initial analysis examined the effects of individual bacterial isolates on host development, quantifying the percentage of worms reaching the last larval stage (L4) after 40 hours. Overall, most isolates (either gut or soil microcosm strains) of the examined genera promoted significantly faster development than the standard food E. coli strain OP50, and similar to the previously described Enterobacter hormaechaei CEent1 gut commensal (Berg et al., 2016b; Dirksen et al., 2020). In contrast, raising worms on most non-Enterobacterales genera, i.e., Chryseobacterium, Leuconostoc and Lactococcus, demonstrated slower or similarly-paced development, compared to OP50, in agreement with previous reports (Dirksen et al., 2016; Samuel et al., 2016; Dirksen et al., 2020).
Interestingly, among members of the Erwiniaceae genus Pantoea (a recent splinter off Enterobacteriaceae), gut isolates promoted significantly faster development than an environmental isolate (Figure 1B and Supplementary Figure 2). Isolates of the Rouxiella genus showed a similar distinction between gut and soil isolates, but not as significant as with Pantoea isolates. Pantoea bacteria, together with other Enterobacterales genera, are known to be part of the C. elegans microbiota (Berg et al., 2016a), and a previously identified Pantoea gut isolate, BIGb0393, which is part of the CeMBio community, has been shown to promote fast development in C. elegans despite not being a prolific colonizer (Dirksen et al., 2020). Our screen further suggests that a subset of Pantoea bacteria contributes to fast development in C. elegans.
To expand examination of additional fitness measures that may be affected by Pantoea isolates, we tested their ability to prevent pathogenic colonization by the virulent Pseudomonas aeruginosa PA14. We found that worms raised on the three Pantoea strains recovered from the C. elegans gut (the previously described BIGb0393 and the two identified in this study (V8 and T16)) were more resistant to subsequent colonization by PA14-GFP than worms raised on E. coli, and additionally, more resistant than worms raised on a Pantoea strain isolated from the worm microcosm environment (T14) (Figures 2A, B). Subsequent analyses showed that gut Pantoea were better at colonizing the worm gut than the environmental strain (Figure 2C), similar to previous reports for BIGb0393 (Dirksen et al., 2020). However, better colonization, and presumably competition, did not seem to be at the basis of Pantoea’s ability to prevent infection, since CFU analysis of worms following PA14-GFP colonization (after 44 hours of exposure) identified only a few remaining Pantoea in the gut, which was not correlated with the extent of PA14-GFP colonization (Figure 2D), and since dead Pantoea were as capable of preventing PA14 colonization as live ones (Figure 2B). Together, these results suggest that Pantoea strains help prevent PA14 colonization but not through competition.
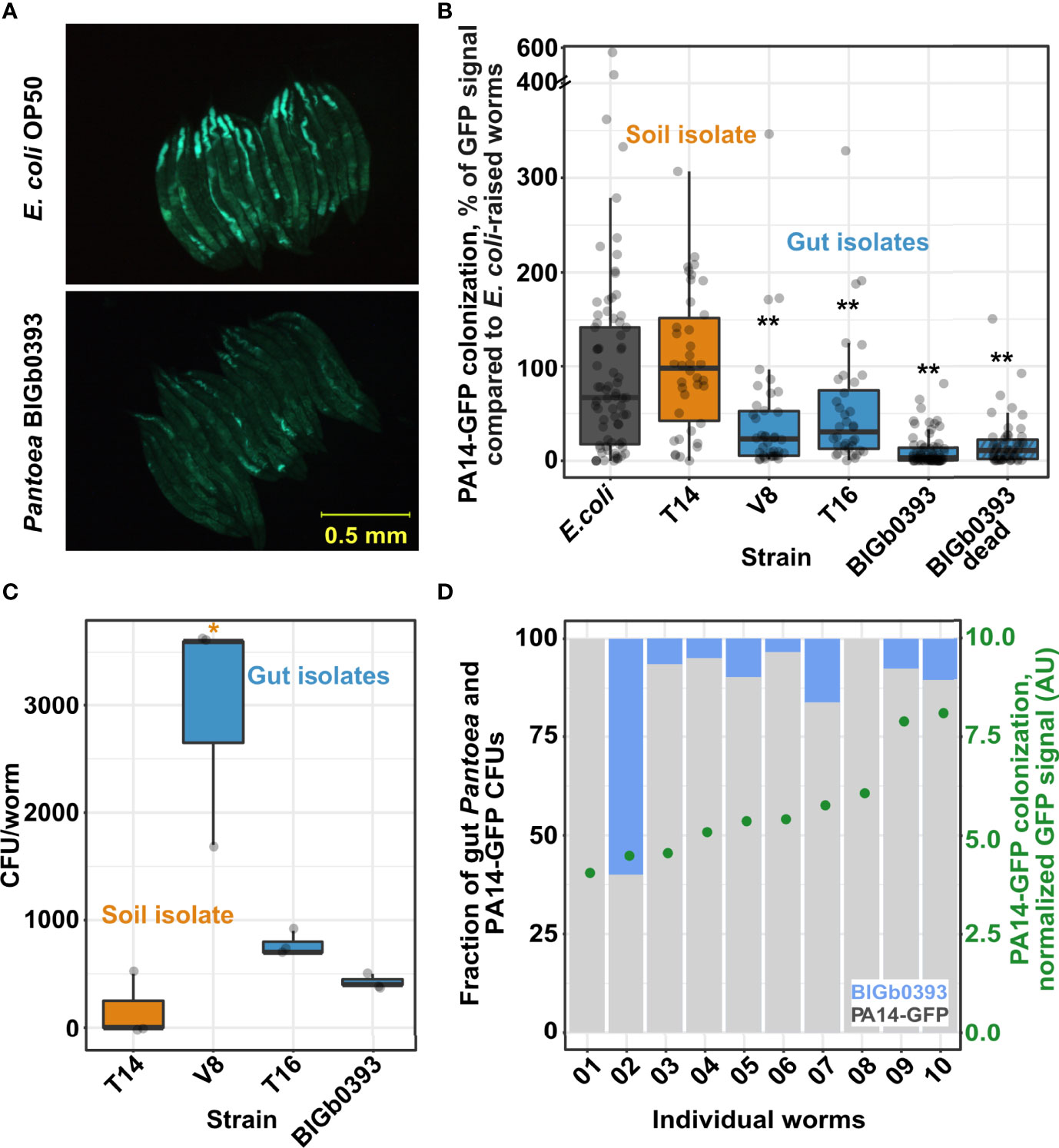
Figure 2 Gut isolates but not an environmental Pantoea strain provide protection from pathogenic colonization by Pseudomonas aeruginosa PA14. (A) Worm colonization by PA14-GFP following 43-45 hours of exposure. (B) Quantification of PA14-GFP fluorescent signal in images as in A; N=36-75 worms from two or three independent experiments. (C) Worm colonization by individual Pantoea strains. CFU counts were estimated in worms raised for 72 hours starting at the L1 stage. Shown are measurements performed in triplicate (N=10 worms each). B and (C) * and **, indicate p < 0.05 and p < 0.001, respectively (Student’s t-test). (D) Gut colonization in individual worms raised on BIGb0393 and shifted for 44 hours to P. aeruginosa PA14-GFP. No correlation is observed between Pantoea colonization (left y-axis, based on CFU counts), and PA14-GFP colonization [right y-axis, fluorescent signal, arbitrary units (AU)].
Our results suggested that the characterized beneficial Pantoea were better able to colonize the worm intestine, which is likely what made them gut commensals. However, behavioral mechanisms were also shown to affect worm interactions with bacteria. Worms are known to avoid pathogenic bacteria (Pradel et al., 2007) or show attraction to food bacteria that better support growth (Shtonda and Avery, 2006). We therefore investigated whether worms preferred specific Pantoea strains. Both multiple-choice and pairwise choice assays revealed significant preference of the beneficial Pantoea gut isolates over the environmental isolate or over control OP50 (Figures 3A, B). These results indicate that behavioral mechanisms further promote preference of beneficial commensals over a similar yet non-beneficial congeneric.
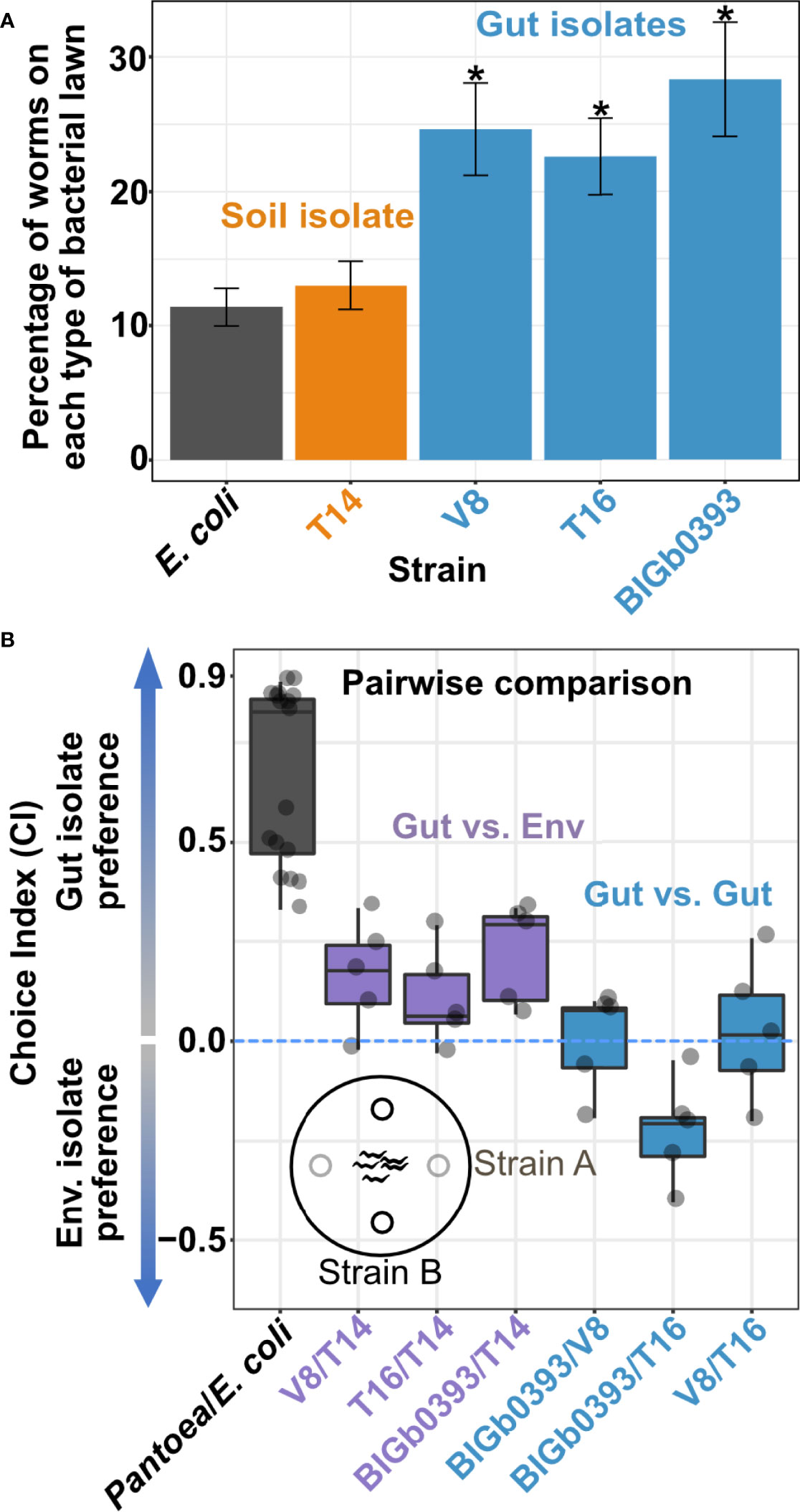
Figure 3 C. elegans shows preference towards beneficial Pantoea. (A) Multi-strain plates. Shown are percentages of young adult worms localized to spots of the designated strains after a three-hour incubation at 25°C. Shown are averages ± SEM for 10 plates (N=36-164 worms each). *p < 0.01 (Student’s t-test). (B) Pairwise preference comparisons. Shown are choice index values of young adult worms exposed to designated strains for 3 hours. Each dot represents an independent pairwise comparison between the designated Pantoea strains (N=21-123 worms per comparison), with boxes defining the 25 to 75 percentile values and lines representing the median value.
In exploring the adaptive capacities of environmentally-acquired commensals for C. elegans fitness, we identified members of the genus Pantoea as multi-purpose beneficial commensals. Gut Pantoea promoted fast development and further hindered pathogenic colonization. While the number of strains available for examination was rather low, it appears that benefits were limited to Pantoea isolated from the worm gut and were missing from a congeneric soil isolate. Gut Pantoea were better at colonizing the worm, and in addition, worms seemed to prefer them over the less beneficial soil Pantoea or the E. coli food bacteria. This behavior adds to previous reports of worms preferring bacteria that support their growth and development (Shtonda and Avery, 2006), by expanding such preference to bacteria that are beneficial beyond nutrition.
C. elegans behavioral food preferences are linked to its neuronal responses to external bacterial cues (e.g., bacterial odors), helping to pre-emptively avoid pathogenic bacteria and promoting interactions with beneficial ones. In addition, C. elegans can exhibit learned behaviors within one to three hours after sampling different food sources, where in some cases, consumption of preferred bacteria has been correlated with increased lifespan and, to a lesser extent, with increased growth rates (Troemel et al., 1997; Law et al., 2004; Shtonda and Avery, 2006; Sowa et al., 2015; Worthy et al., 2018). The capability of C. elegans to prefer beneficial microbes may be comparable to that observed in other eukaryotic models. In Drosophila melanogaster, for example, olfactory signals synthesized by gut-associated microbes were shown to modify host foraging behavior, egg laying, and nutritional as well as microbial preferences (Fischer et al., 2017; Wong et al., 2017). Unlike the microbial signals in the drosophila model, the molecular basis for Pantoea preference by C. elegans is yet unknown. However, an intriguing parallel in the effects that bacteria exerts on their host is presented by a recently described Providencia gut commensal, which was shown to promote worm growth and was able to attract worms by modulating host neurotransmission (Samuel et al., 2016; O’Donnell et al., 2020).
Colonization resistance conferred by gut bacteria can be due to direct antagonism of invaders (i.e., resource competition or metabolic warfare) or through induction of host immune responses (Kamada et al., 2013; Montalvo-Katz et al., 2013; Iatsenko et al., 2014; Granato et al., 2019). While the basis of Pantoea’s ability to promote such resistance is not known, other Pantoea species have been reported to produce antimicrobial molecules, effective, among others, against Pseudomonas aeruginosa (Williams and Stavrinides, 2020). In the example we identified, hindering of pathogen colonization was likely not due to competition, as BIGb0393 conferred protection from PA14 colonization even when dead. Another alternative is immune priming, which was previously shown to provide protection from PA14 infection through activation of the p38 MAPK pathway (Montalvo-Katz et al., 2013). Other studies have shown that immune priming can be conferred by heat-killed bacteria (Lee et al., 2015; Yuen and Ausubel, 2018), suggesting that immune activation may be triggered by recognition of heat-labile microbe-associated molecular patterns (MAMPs), in line with previous studies (Twumasi-Boateng and Shapira, 2012). Lastly, it is also possible that the nutritional value of Pantoea, similar in live bacteria as in dead ones, makes for healthier worms, which can then better withstand infection. However, generally better health is more likely to contribute to long-term tolerance of infection and survival than on the initial stages of colonization, which are dependent on specific immune pathways.
The expansion in the reported abilities of Pantoea strains to accelerate development and contribute to host fitness present them as important members of the worm gut microbiota. The difference in the effects of, and host affinity to, some members of this taxa (gut isolates) compared to others (the soil isolate), distinguishes this genus from other examined genera. This, together with what appears to be mutual adaptations, including better ability of beneficial Pantoea to colonize the worm gut (compared to a non-beneficial congeneric) and host preference of its cognate commensals, suggest long-standing interactions between the worm host and its Pantoea gut commensals. How these interactions have evolved and how tightly they link the fitness of the two partners, remains to be further studied. Furthermore, research into C. elegans interactions with additional Pantoea strains is required to ascertain the paradigm proposed here and to provide further support.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
RC, OMPC, and MS contributed to the conception of the study and the preparation of the manuscript. MM conducted soil microcosm experiments, isolated bacteria and analyzed their effects on host development. OMPC and RC lead colonization experiments with pathogens. OMPC conducted preference assays. OMPC, RC, and BP contributed to designing colonization experiments, data acquisition, image quantification, and data analysis and interpretation. VN contributed to valuable experiments and results that were omitted in the final version. All authors contributed to the article and approved the submitted version.
Work described in this manuscript was supported by NIH grants R01OD024780 and R01AG061302.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MP declared a shared affiliation with the authors to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Andrew Ly for sequencing bacterial strains.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.795343/full#supplementary-material
Abada, E. A., Sung, H., Dwivedi, M., Park, B. J., Lee, S. K., Ahnn, J. (2009). C. Elegans Behavior of Preference Choice on Bacterial Food. Mol. Cells 28, 209–213. doi: 10.1007/s10059-009-0124-x
Adeolu, M., Alnajar, S., Naushad, S., S. Gupta, R. (2016). Genome-Based Phylogeny and Taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales Ord. Nov. Divided Into the Families Enterobacteriaceae, Erwiniaceae Fam. Nov., Pectobacteriaceae Fam. Nov., Yersiniaceae Fam. Nov., Hafniaceae Fam. Nov., Morganellaceae Fam. Nov., and Budviciaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. doi: 10.1099/ijsem.0.001485
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., Gordon, J. I. (2005). Host-Bacterial Mutualism in the Human Intestine. Science 307, 1915–1920. doi: 10.1126/science.1104816
Berg, M., Monnin, D., Cho, J., Nelson, L., Crits-Christoph, A., Shapira, M. (2019). Tgfβ/BMP Immune Signaling Affects Abundance and Function of C. Elegans Gut Commensals. Nat. Commun. 10, 604. doi: 10.1038/s41467-019-08379-8
Berg, M., Stenuit, B., Ho, J., Wang, A., Parke, C., Knight, M., et al. (2016a). Assembly of the Caenorhabditis Elegans Gut Microbiota From Diverse Soil Microbial Environments. ISME J. 10, 1998–2009. doi: 10.1038/ismej.2015.253
Berg, M., Zhou, X. Y., Shapira, M. (2016b). Host-Specific Functional Significance of Caenorhabditis Gut Commensals. Front. Microbiol. 7, 1622. doi: 10.3389/fmicb.2016.01622
Beydoun, S., Choi, H. S., Dela-Cruz, G., Kruempel, J., Huang, S., Bazopoulou, D., et al. (2021). An Alternative Food Source for Metabolism and Longevity Studies in Caenorhabditis Elegans. Commun. Biol. 4, 258. doi: 10.1038/s42003-021-01764-4
Callens, M., Watanabe, H., Kato, Y., Miura, J., Decaestecker, E. (2018). Microbiota Inoculum Composition Affects Holobiont Assembly and Host Growth in Daphnia. Microbiome 6, 56. doi: 10.1186/s40168-018-0444-1
Davenport, E. R., Sanders, J. G., Song, S. J., Amato, K. R., Clark, A. G., Knight, R. (2017). The Human Microbiome in Evolution. BMC Biol. 15, 127. doi: 10.1186/s12915-017-0454-7
Dirksen, P., Assié, A., Zimmermann, J., Zhang, F., Tietje, A.-M., Marsh, S. A., et al. (2020). CeMbio - The Caenorhabditis Elegans Microbiome Resource. G3 (Bethesda Md.) 10, 3025–3039. doi: 10.1534/g3.120.401309
Dirksen, P., Marsh, S. A., Braker, I., Heitland, N., Wagner, S., Nakad, R., et al. (2016). The Native Microbiome of the Nematode Caenorhabditis Elegans: Gateway to a New Host-Microbiome Model. BMC Biol. 14, 38. doi: 10.1186/s12915-016-0258-1
Fischer, C. N., Trautman, E. P., Crawford, J. M., Stabb, E. V., Handelsman, J., Broderick, N. A. (2017). Metabolite Exchange Between Microbiome Members Produces Compounds That Influence Drosophila Behavior. Elife 6, e18855. doi: 10.7554/eLife.18855
Gilbert, J. A., Blaser, M. J., Caporaso, J. G., Jansson, J. K., Lynch, S., Knight, R. (2018). Current Understanding of the Human Microbiome. Nat. Med. 24, 392–400. doi: 10.1038/nm.4517
Goodrich, J. K., Davenport, E. R., Clark, A. G., Ley, R. E. (2017). The Relationship Between the Human Genome and Microbiome Comes Into View. Annu. Rev. Genet. 51, 413–433. doi: 10.1146/annurev-genet-110711-155532
Granato, E. T., Meiller-Legrand, T. A., Foster, K. R. (2019). The Evolution and Ecology of Bacterial Warfare. Curr. Biol. 29, R521–R537. doi: 10.1016/j.cub.2019.04.024
Hacquard, S., Garrido-Oter, R., González, A., Spaepen, S., Ackermann, G., Lebeis, S., et al. (2015). Microbiota and Host Nutrition Across Plant and Animal Kingdoms. Cell Host Microbe 17, 603–616. doi: 10.1016/j.chom.2015.04.009
Iatsenko, I., Yim, J. J., Schroeder, F. C., Sommer, R. J. (2014). B. Subtilis GS67 Protects C. Elegans From Gram-Positive Pathogens via Fengycin-Mediated Microbial Antagonism. Curr. Biol. 24, 2720–2727. doi: 10.1016/j.cub.2014.09.055
Johnke, J., Dirksen, P., Schulenburg, H. (2020). Community Assembly of the Native C. Elegans Microbiome is Influenced by Time, Substrate and Individual Bacterial Taxa. Enviro Microbiol. 22, 1265–1279. doi: 10.1111/1462-2920.14932
Kamada, N., Chen, G. Y., Inohara, N., Núñez, G. (2013). Control of Pathogens and Pathobionts by the Gut Microbiota. Nat. Immunol. 14, 685–690. doi: 10.1038/ni.2608
Kim, D. H., Flavell, S. W. (2020). Host-Microbe Interactions and the Behavior of Caenorhabditis Elegans. J. Neurogenet 34, 500–509. doi: 10.1080/01677063.2020.1802724
Kissoyan, K. A. B., Drechsler, M., Stange, E.-L., Zimmermann, J., Kaleta, C., Bode, H. B., et al. (2019). Natural C. Elegans Microbiota Protects Against Infection via Production of a Cyclic Lipopeptide of the Viscosin Group. Curr. Biol. 29, 1030–1037.e5. doi: 10.1016/j.cub.2019.01.050
Lawley, T. D., Walker, A. W. (2013). Intestinal Colonization Resistance. Immunology 138, 1–11. doi: 10.1111/j.1365-2567.2012.03616.x
Law, E., Nuttley, W. M., van der Kooy, D. (2004). Contextual Taste Cues Modulate Olfactory Learning in C. Elegans by an Occasion-Setting Mechanism. Curr. Biol. 14, 1303–1308. doi: 10.1016/j.cub.2004.06.066
Lee, J., Choe, J., Kim, J., Oh, S., Park, S., Kim, S., et al. (2015). Heat-Killed Lactobacillus Spp. Cells Enhance Survivals of Caenorhabditis Elegans Against Salmonella and Yersinia Infections. Lett. Appl. Microbiol. 61, 523–530. doi: 10.1111/lam.12478
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R., Gordon, J. I. (2008). Worlds Within Worlds: Evolution of the Vertebrate Gut Microbiota. Nat. Rev. Microbiol. 6, 776–788. doi: 10.1038/nrmicro1978
Montalvo-Katz, S., Huang, H., Appel, M. D., Berg, M., Shapira, M. (2013). Association With Soil Bacteria Enhances P38-Dependent Infection Resistance in Caenorhabditis Elegans. Infect. Immun. 81, 514–520. doi: 10.1128/IAI.00653-12
O’Donnell, M. P., Fox, B. W., Chao, P.-H., Schroeder, F. C., Sengupta, P. (2020). A Neurotransmitter Produced by Gut Bacteria Modulates Host Sensory Behaviour. Nature 583, 415–420. doi: 10.1038/s41586-020-2395-5
Ortiz, A., Vega, N. M., Ratzke, C., Gore, J. (2021). Interspecies Bacterial Competition Regulates Community Assembly in the C. Elegans Intestine. ISME J. 15, 2131–2145. doi: 10.1038/s41396-021-00910-4
Pradel, E., Zhang, Y., Pujol, N., Matsuyama, T., Bargmann, C. I., Ewbank, J. J. (2007). Detection and Avoidance of a Natural Product From the Pathogenic Bacterium Serratia Marcescens by Caenorhabditis Elegans. Proc. Natl. Acad. Sci. U.S.A. 104, 2295–2300. doi: 10.1073/pnas.0610281104
Samuel, B. S., Rowedder, H., Braendle, C., Félix, M.-A., Ruvkun, G. (2016). Caenorhabditis Elegans Responses to Bacteria From its Natural Habitats. Proc. Natl. Acad. Sci. U.S.A. 113, E3941. doi: 10.1073/pnas.1607183113
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schulenburg, H., Felix, M. A. (2017). The Natural Biotic Environment of Caenorhabditis Elegans. Genetics 206, 55–86. doi: 10.1534/genetics.116.195511
Shapira, M. (2017). Host–microbiota Interactions in Caenorhabditis Elegans and Their Significance. Curr. Opin. Microbiol. 38, 142–147. doi: 10.1016/j.mib.2017.05.012
Shapira, M., Tan, M. W. (2008). Genetic Analysis of Caenorhabditis Elegans Innate Immunity. Methods Mol. Biol. 415, 429–442. doi: 10.1007/978-1-59745-570-1_25
Shtonda, B. B., Avery, L. (2006). Dietary Choice Behavior in Caenorhabditis Elegans. J. Exp. Biol. 209, 89–102. doi: 10.1242/jeb.01955
Sieber, M., Pita, L., Weiland-Brauer, N., Dirksen, P., Wang, J., Mortzfeld, B., et al. (2019). Neutrality in the Metaorganism. PloS Biol. 17, e3000298. doi: 10.1371/journal.pbio.3000298
Simon, J.-C., Marchesi, J. R., Mougel, C., Selosse, M.-A. (2019). Host-Microbiota Interactions: From Holobiont Theory to Analysis. Microbiome 7, 5. doi: 10.1186/s40168-019-0619-4
Slowinski, S., Ramirez, I., Narayan, V., Somayaji, M., Para, M., Pi, S., et al. (2020). Interactions With a Complex Microbiota Mediate a Trade-Off Between the Host Development Rate and Heat Stress Resistance. Microorganisms 8, 1781. doi: 10.3390/microorganisms8111781
Smith, C. C. R., Snowberg, L. K., Gregory Caporaso, J., Knight, R., Bolnick, D. I. (2015). Dietary Input of Microbes and Host Genetic Variation Shape Among-Population Differences in Stickleback Gut Microbiota. ISME J. 9, 2515–2526. doi: 10.1038/ismej.2015.64
Sorbara, M. T., Pamer, E. G. (2019). Interbacterial Mechanisms of Colonization Resistance and the Strategies Pathogens Use to Overcome Them. Mucosal Immunol. 12, 1–9. doi: 10.1038/s41385-018-0053-0
Sowa, J. N., Mutlu, A. S., Xia, F., Wang, M. C. (2015). Olfaction Modulates Reproductive Plasticity Through Neuroendocrine Signaling in Caenorhabditis Elegans. Curr. Biol. 25, 2284–2289. doi: 10.1016/j.cub.2015.07.023
Spor, A., Koren, O., Ley, R. (2011). Unravelling the Effects of the Environment and Host Genotype on the Gut Microbiome. Nat. Rev. Microbiol. 9, 279–290. doi: 10.1038/nrmicro2540
Troemel, E. R., Kimmel, B. E., Bargmann, C. I. (1997). Reprogramming Chemotaxis Responses: Sensory Neurons Define Olfactory Preferences in C. Elegans. Cell 91, 161–169. doi: 10.1016/S0092-8674(00)80399-2
Twumasi-Boateng, K., Shapira, M. (2012). Dissociation of Immune Responses From Pathogen Colonization Supports Pattern Recognition in C. Elegans. PloS One 7, e35400–e35400. doi: 10.1371/journal.pone.0035400
Villanueva, R. A. M., Chen, Z. J. (2019). Ggplot2: Elegant Graphics for Data Analysis, 2nd Edition. Measurement 17, 160–167. doi: 10.1080/15366367.2019.1565254
Watson, E., MacNeil, L. T., Ritter, A. D., Yilmaz, L. S., Rosebrock, A. P., Caudy, A. A., et al. (2014). Interspecies Systems Biology Uncovers Metabolites Affecting C. Elegans Gene Expression and Life History Traits. Cell 156, 759–770. doi: 10.1016/j.cell.2014.01.047
Williams, A. N., Stavrinides, J. (2020). Pantoea Natural Product 3 Is Encoded by an Eight-Gene Biosynthetic Gene Cluster and Exhibits Antimicrobial Activity Against Multi-Drug Resistant Acinetobacter Baumannii and Pseudomonas Aeruginosa. Microbiol. Res. 234, 126412. doi: 10.1016/j.micres.2020.126412
Willis, A. R., Sukhdeo, R., Reinke, A. W. (2021). Remembering Your Enemies: Mechanisms of Within-Generation and Multigenerational Immune Priming in Caenorhabditis Elegans. FEBS J. 288, 1759–1770. doi: 10.1111/febs.15509
Wippel, K., Tao, K., Niu, Y., Zgadzaj, R., Kiel, N., Guan, R., et al. (2021). Host Preference and Invasiveness of Commensal Bacteria in the Lotus and Arabidopsis Root Microbiota. Nat. Microbiol. 6, 1150–1162. doi: 10.1038/s41564-021-00941-9
Wong, A. C.-N., Wang, Q.-P., Morimoto, J., Senior, A. M., Lihoreau, M., Neely, G. G., et al. (2017). Gut Microbiota Modifies Olfactory-Guided Microbial Preferences and Foraging Decisions in Drosophila. Curr. Biol. 27, 2397–2404.e4. doi: 10.1016/j.cub.2017.07.022
Worthy, S. E., Haynes, L., Chambers, M., Bethune, D., Kan, E., Chung, K., et al. (2018). Identification of Attractive Odorants Released by Preferred Bacterial Food Found in the Natural Habitats of C. Elegans. PloS One 13, e0201158–e0201158. doi: 10.1371/journal.pone.0201158
Yuen, G. J., Ausubel, F. M. (2018). Both Live and Dead Enterococci Activate Caenorhabditis Elegans Host Defense via Immune and Stress Pathways. Virulence 9, 683–699. doi: 10.1080/21505594.2018.1438025
Zhang, F., Berg, M., Dierking, K., Félix, M.-A., Shapira, M., Samuel, B. S., et al. (2017). Caenorhabditis Elegans as a Model for Microbiome Research. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00485
Zhang, F., Weckhorst, J. L., Assié, A., Hosea, C., Ayoub, C. A., Khodakova, A. S., et al. (2021). Natural Genetic Variation Drives Microbiome Selection in the Caenorhabditis Elegans Gut. Curr. Biol. 31, 2603–2618.e9. doi: 10.1016/j.cub.2021.04.046
Keywords: C. elegans, microbiota, Enterobacteriaceae, Pantoea, commensal, preference
Citation: Pérez-Carrascal OM, Choi R, Massot M, Pees B, Narayan V and Shapira M (2022) Host Preference of Beneficial Commensals in a Microbially-Diverse Environment. Front. Cell. Infect. Microbiol. 12:795343. doi: 10.3389/fcimb.2022.795343
Received: 14 October 2021; Accepted: 24 May 2022;
Published: 15 June 2022.
Edited by:
Nic Vega, Emory University, United StatesReviewed by:
M. Fernanda Palominos, University of California, Berkeley, United StatesCopyright © 2022 Pérez-Carrascal, Choi, Massot, Pees, Narayan and Shapira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Shapira, bXNoYXBpcmFAYmVya2VsZXkuZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.