
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 26 May 2022
Sec. Clinical Microbiology
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.774494
This article is part of the Research Topic Antimicrobial Resistance: a One Health Perspective View all 9 articles
The prevalence and transmission of mobile colistin resistance (mcr) genes have led to a severe threat to humans and animals. Escherichia fergusonii is an emerging pathogen which is closely related to a variety of diseases. However, the report of mcr genes harboring E. fergusonii is still rare. One study in Brazil reported the E. fergusonii isolates with IncHI2-type plasmids harboring mcr-1. A Chinese study reported two strains carrying mcr-1 gene with the same plasmid type IncI2. Here, we identified two strains of E. fergusonii carrying mcr-1 gene from farm environments with IncX4-type and IncI2-type plasmids, respectively. To our best knowledge, this is the first report about mcr-1 gene located on IncX4-type plasmid in E. fergusonii. We investigate the resistance mechanism of colistin-resistant Escherichia fergusonii strains 6S41-1 and 5ZF15-2-1 and elucidate the genetic context of plasmids carrying mcr-1 genes. In addition, we also investigated chromosomal mutations mediated colistin resistance in these two strains. Species identification was performed using MALDI-TOF MS and 16S rRNA gene sequencing. The detection of mcr-1 gene was determined by PCR and Sanger sequencing. S1-pulsed-field gel electrophoresis (PFGE), Southern blotting, antimicrobial susceptibility testing, conjugation experiments, complete genome sequencing, and core genome analysis were conducted to investigate the characteristics of isolates harboring mcr-1. The mcr-1 genes on two strains were both plasmids encoded and the typical IS26-parA-mcr-1-pap2 cassette was identified in p6S41-1 while a nikA-nikB-mcr-1 locus sites on the conjugative plasmid p5ZF15-2-1. In addition, Core genome analysis reveals that E. fergusonii 6S41-1 and 5ZF15-2-1 have close genetic relationships. The mcr-1 gene is located on conjugative IncI2-type plasmid p5ZF15-2-1, which provides support for its further transmission. In addition, there’s the possibility of mcr-1 spreading to humans through farm environments and thereby threatening public health. Therefore, continuous monitoring and investigations of mcr-1 among Enterobacteriaceae in farm environments are necessary to control the spread.
Escherichia fergusonii is a species with high genotypic and phenotypic similarity to Escherichia coli. DNA hybridization shows that it has 64% similarity with Escherichia coli-Shigella (Farmer et al., 1985a). It was once known as Enteric Group 10 until Farmer et al. proposed it as a new species within the genus Escherichia and family Enterobacteriaceae in 1985 (Farmer et al., 1985b). Since then, E. fergusonii has been considered as a significant emerging opportunistic pathogen both for animals and humans. E. fergusonii was initially isolated from blood samples of clinical patients (Farmer et al., 1985b). To this day, E. fergusonii has been widely isolated from animals, humans, and the environment. In specimens of animal origin, it has been isolated from the feces of goats, sheep, horses, cattle, pigs, chickens, turkeys, reindeer, and ostriches that display symptoms of salmon-like infection including diarrhea, meningitis, mastitis, abortion, and septicemia (Farmer et al., 1985b; Bain and Green, 1999; Herráez et al., 2005; Hariharan et al., 2007; Foster et al., 2010; Oh et al., 2012). While in clinical specimens, E. fergusonii is often isolated from blood, urine, feces, gallbladder fluid, spinal fluid, and abdominal wound samples of patients with septicemia, intestinal diseases, urinary tract infections, and pancreatic cancer (Farmer et al., 1985b; Funke et al., 1993). In addition, E. fergusonii was also isolated from food products during routine screening procedures and water samples (Fegan et al., 2006; Maifreni et al., 2013; Maheux et al., 2014).
Colistin is a group of polypeptide antibiotics produced by Paenibacillus polymyxa, which was first discovered in the 1940s and is considered to be the last line of defense against severe infection caused by pan-drug-resistant Gram-negative pathogens (Storm et al., 1977; Sun et al., 2018). For decades, colistin has been used in curative treatments and is widely regarded as a preventive drug in veterinary medicine (Skov and Monnet, 2016). However, the highly selective pressure caused by the extensive use of colistin in animals has led to the selection and the spread of colistin-resistant pathogens (Asai et al., 2005; Diarra et al., 2007). The resistance mechanism of Enterobacteriaceae to colistin could be mediated both by chromosomes and plasmids (Huang et al., 2020). For a long period, the resistance was thought to be caused by chromosomal mutations such as two-component systems and its regulators, or the overexpression of efflux pump proteins. None of these were transferable (Anyanwu et al., 2020). However, since the first discovery of mobile colistin resistance gene mcr-1 in E. coli from China (Liu et al., 2016), 10 different mcr genes and several variants have been identified across the globe which demonstrates a horizontal transfer mechanism for colistin resistance (Luo et al., 2017; Wang et al., 2020a). The emergence of mcr genes has aroused widespread attention and concern around the world. Although China banned colistin as a feed additive for animals in 2017 (Walsh and Wu, 2016), the problem of antibiotic resistance caused by mcr genes is still severe (Wang et al., 2020b).
Among the 10 different mcr genes, mcr-1 is most widespread and commonly detected in Enterobacteriaceae, especially Escherichia coli (Jeannot et al., 2017). However, the detection of mcr-1 gene in E. fergusonii is still rarely reported, which is worthy of our attention and further study. In addition, the multi-drug resistance and even pan-drug resistance pathogens caused by the coexistence of mcr-1 and other drug resistance genes make the option of anti-infective therapy in a dilemma and pose a serious threat to public health. Here, we identified two strains of E. fergusonii carrying mcr-1, which were isolated from farm soils and feces of a healthy pig, respectively, and performed core genome analysis by whole-genome sequencing data. Furthermore, we studied the drug resistance profile, plasmid characteristics, and chromosomal mutations to reveal the potential resistance mechanism of these two strains.
In a routine surveillance study of antimicrobial resistance of bacteria from farm environments, we collected samples from conventional farms since May 2019. All samples were collected using sterile cotton swabs and were kept in -20°C during transportation. And all samples were enriched within 72 h after sampling. The enriched samples were coated on MacConkey agar (OXOID, Hampshire, United Kingdom) plates containing 2 mg/L colistin for preliminary screening. Identification of species was performed using both matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (Bruker Daltonik GmbH, Bremen, Germany) and 16S rRNA gene sequencing. The mcr-1 gene was detected under PCR and Sanger sequencing, as described previously (Zheng et al., 2015).
The size and number of plasmids of E. fergusonii 6S41-1 and 5ZF15-2-1 were identified by the S1-PFGE. In addition, the location of mcr-1 gene was determined according to Southern blotting and hybridization with digoxigenin-labeled mcr-1 specific probe. The transferability of the plasmid carrying mcr-1 gene was verified by conjugation experiments using rifampicin-resistant E. coli 600 as the recipient strain according to the previous study (Zheng et al., 2017). The transconjugants were selected on Mueller-Hinton medium containing 200mg/L rifampicin and 1mg/L colistin. Then, the transconjugants were identified by MALDI-TOF/MS, and the mcr-1 gene was detected by PCR to confirm whether the plasmids were successfully transferred into the recipients.
The antimicrobial susceptibility profiles of 6S41-1 and 5ZF15-2-1, and their corresponding transconjugants, were determined using the agar dilution method and broth microdilution method. The results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) standards (https://clsi.org), except colistin and tigecycline which are interpreted according to EUCAST clinical breakpoints (https://www.eucast.org/). E. coli ATCC 25922 was used as a quality control.
Genomic DNA was extracted by using a Bacterial DNA Kit (QIAGEN, Hilden, Germany). The harvested DNA was detected by the agarose gel electrophoresis and quantified by Qubit® 2.0 Fluorometer (Thermo Scientific). Then the DNA was sequenced both on the Illumina NovaSeq 6000 (Illumina, San Diego, CA, United States) and Oxford Nanopore platforms (Oxford Nanopore Technologies, Oxford, United Kingdom) to obtain short-read data and long-read data, respectively. The raw llumina reads were assembled using SPAdes3.10.0, and then the sequencing results were hybrid assembled with Unicycler v0.4.7 to get the complete genome sequence (Wick et al., 2017). The bacterial genomes were annotated using Prokka. Additionally, the acquired antimicrobial resistance genes and replicon type of plasmid were determined using online tools (http://www.genomicepidemiology.org/), while the transposon and IS elements were identified using the ISFinder database (http://www-is.biotoul.fr/). The virulence genes of isolates were identified using VFDB. Finally, the circular image of multiple plasmids comparisons was plotted by the BLAST Ring Image Generator (BRIG) (Alikhan et al., 2011). The comparison figure of the genetic environment surrounding the mcr-1 gene was generated by Easyfig 2.2.3 (Sullivan et al., 2011).
Based on WGS data, amino acid sequences of isolates 6S41-1 and 5ZF15-2-1 were compared with reference strain E. fergusonii RHB19-C05. PROVEAN (http://provean.jcvi.org/index.php) was used to predict whether the amino acid substitutions in two-component systems, AcrAB-TolC pump system and its regulators, affect protein function. Additionally, SMART analysis (http://smart.embl-heidelberg.de/) was performed to determine the corresponding domain architectures.
Genome sequences for 114 strains of E. fergusonii were downloaded from the NCBI database, assembly section (Table S4). According to Wu et al. (Wu et al., 2021), the inclusion of confounding strains may introduce important biases. Thus, we performed average nucleotide identity (ANI) analysis using pyani (https://github.com/widdowquinn/pyani). Then these genomes, plus E. fergusonii 6S41-1 and 5ZF15-2-1, were performed phylogenetic analyses using Roary (Page et al., 2015), a tool that builds rapid large-scale prokaryote pan genomes and identifies core genes. Next, a maximum likelihood phylogenetic tree was generated by MEGA X using core genes. And the visualization and modification were performed by iTOL (https://itol.embl.de/).
In total, we collected 80 soil samples and 534 fecal samples from six randomly selected farms in Jiaxing, Zhejiang Province. Among them, two distinct colistin-resistant E. fergusonii isolates were identified and named 6S41-1 and 5ZF15-2-1, respectively. In more detail, strains 6S41-1 and 5ZF15-2-1were isolated from the soils and the feces of a healthy pig in two different farms, respectively. The single colonies selected from the selective mediums were identified as E. fergusonii using MALDI-TOF/MS and 16S rRNA, and the mcr-1 gene was confirmed by PCR and sequencing.
A total of 17 antibiotics were included in this study (Table 1). The AST revealed that isolate 6S41-1 exhibited resistance to colistin, gentamicin, and chloramphenicol but was shown to be susceptible to tigecycline, amoxicillin-clavulanate, piperacillin/tazobactam, ceftazidime, ceftriaxone, cefepime, cefotaxime, imipenem, meropenem, trimethoprim/sulfamethoxazole, amikacin, and aztreonam. In addition, the minimum inhibitory concentration (MIC) values of ciprofloxacin and levofloxacin for 6S41-1 were determined as intermediate. Strain 5ZF15-2-1 was sensitive to almost all antibiotics except colistin, with MIC value of 4 mg/L. Moreover, the transconjugants 5ZF15-2-1-E. coli 600 showed the same antibiotic resistance profile to 5ZF15-2-1 but was intermediate to colistin.
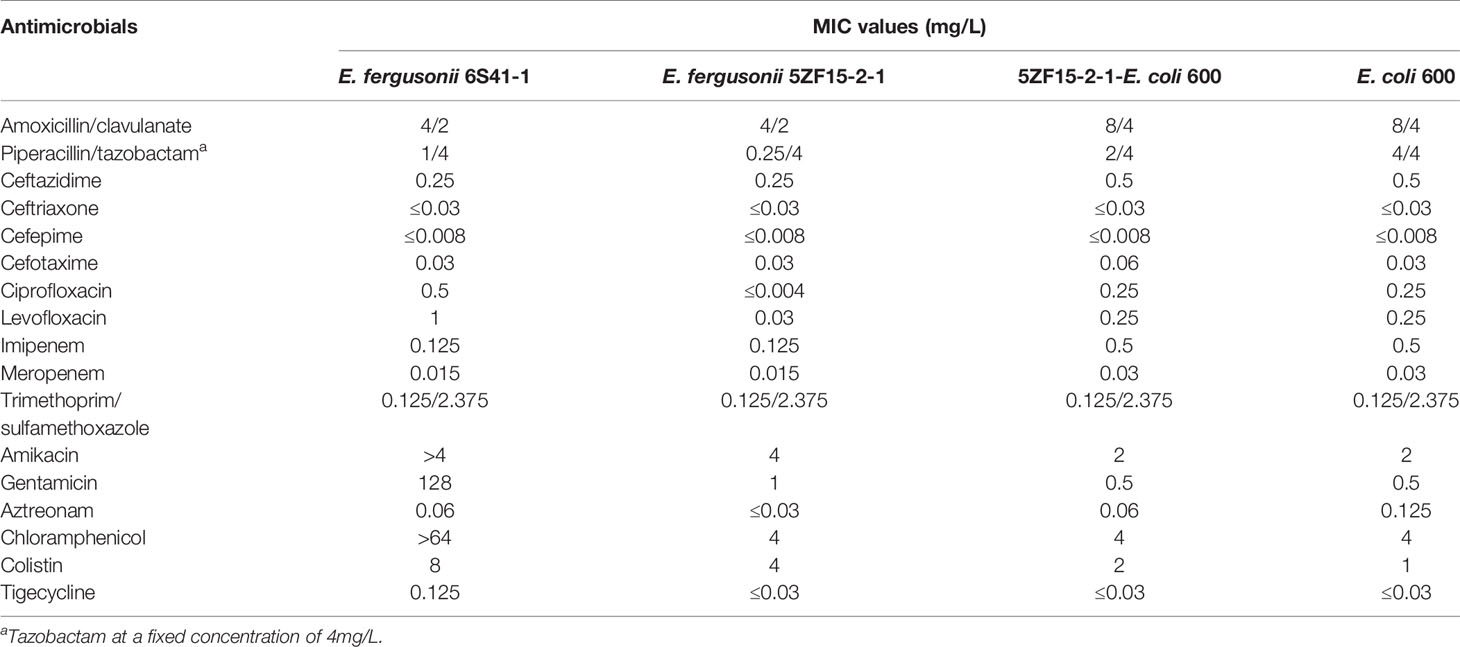
Table 1 MIC values of antimicrobials for E. fergusonii 6S41-1, 5ZF15-2-1, transconjugant 5ZF15-2-1-E. coli 600 and recipient strain E. coli 600.
The genomic features of E. fergusonii 6S41-1 and 5ZF15-2-1 are displayed in Table 2. E. fergusonii 6S41-1 genome consists of a 4,724,978 bp circular chromosome with an average G+C content of 49.8% and six plasmids. The size and average G+C content of the plasmid conferring mcr-1 was 46,439 bp and 44.4%, respectively. The genome of 5ZF15-2-1 consists of a chromosome of 4,934,492 bp and four plasmids The plasmid types of these two strains that do not carry mcr-1 gene are shown in Table S1. A screening for acquired resistance determinants found that no resistance gene was encoded on the chromosome, and only one resistance gene, mcr-1, was encoded on the plasmid p5ZF15-2-1. Moreover, plasmid p6S41-1 encoding acquired resistance genes both mcr-1 and qnrS1, the latter mediating resistance to ciprofloxacin (Table S2). Virulome analysis showed that the majority of virulence genes are encoded on chromosomes in both strains only cseA which encoded adhesin protein sited on a IncFII type plasmid of E. fergusonii 6S41-1 (Table S1). In particular, the virulence profiles of these two strains were almost identical. For instance, they both contained virulence factors encoded secretions systems (tssALMJ, clpV/tssH and hcp2/tssD2), transcriptional regulators (rcsB), and outer membrane proteins (ompA).
S1-PFGE and Southern blot confirmed that 6S41-1 isolate contained a ~46 kb plasmid harboring mcr-1 gene (Figure 1). The WGS results demonstrated that the mcr-1-encoding plasmid, designated as p6S41-1, was an IncX4-type plasmid with the size of 46,394 bp and contained 135 protein-coding genes with a GC content of 44.4% (Table 2). We tried to transfer plasmid p6S41-1 to the recipient strain E. coli 600. However, in vitro conjugation experiments were unsuccessful. We also tried to transfer plasmid extracted from isolate, and repeated transformation methods failed to move the plasmid to recipient E. coli DH5α cells. p6S41-1 contains a collection of genes involved in segregation, stability, replication, and conjugative transfer of the plasmid (dnaJ, pir, parA, hns, topB and virB4,8,9,11), which together constructed the basic backbone of the plasmid. The genetic context of mcr-1 was characterized by an IS26 element upstream of the mcr-1-pap2 element, and similar structures could be seen in E. coli plasmid pMFDS1318.1 (accession number: MK875282), E. coli plasmid pWI2-mcr (accession number: LT838201), and E. coli plasmid pICBEC7Pmcr (accession number: CP017246) (Figure 2A).
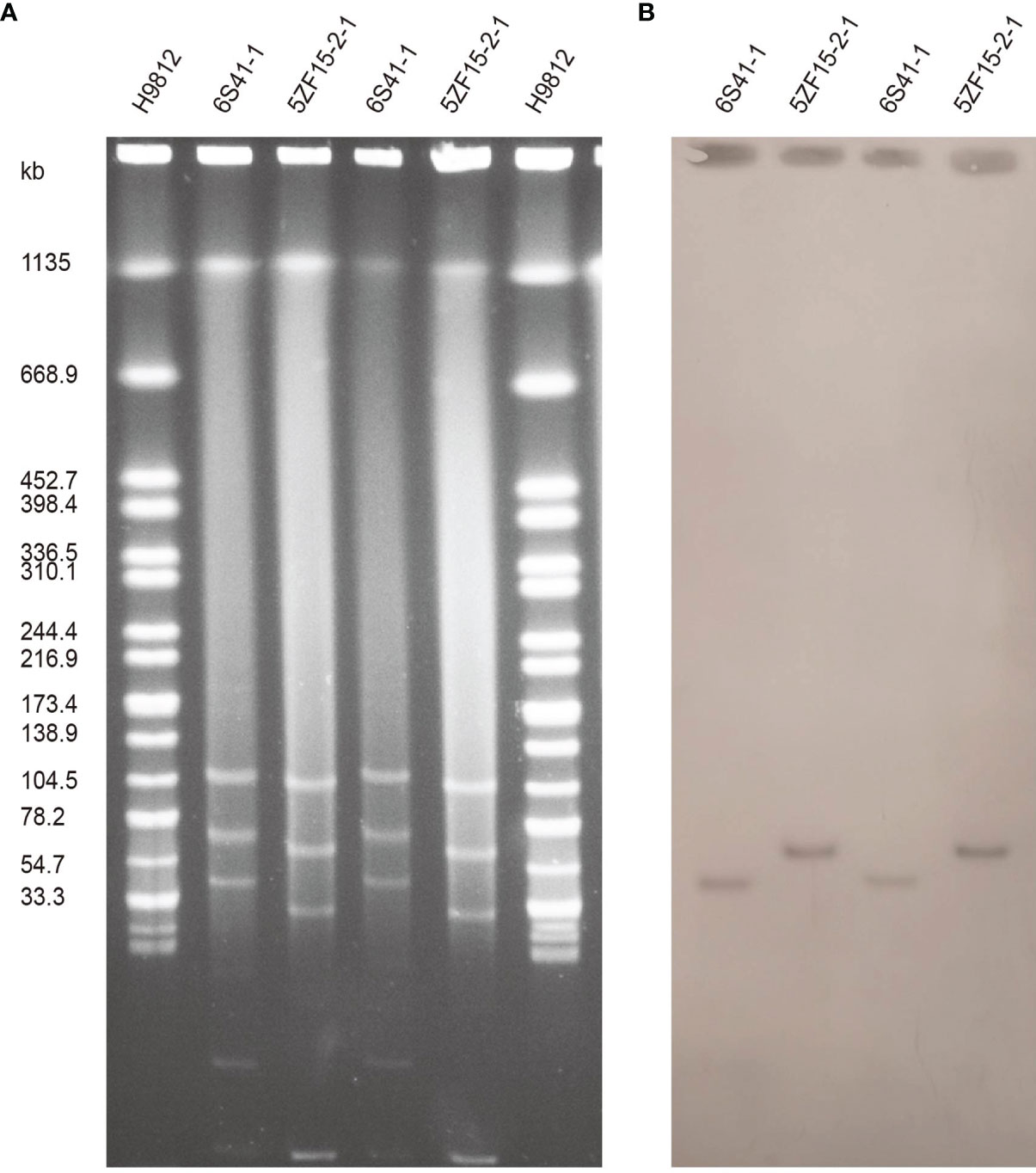
Figure 1 Plasmid profiles of E. fergusonii 6S41-1 and 5ZF15-2-1. (A) Plasmid size determination by S1-PFGE, with Salmonella enterica serotype Braenderup H9812 as the size marker. (B) Southern blotting hybridization with an mcr-1-specific probe.
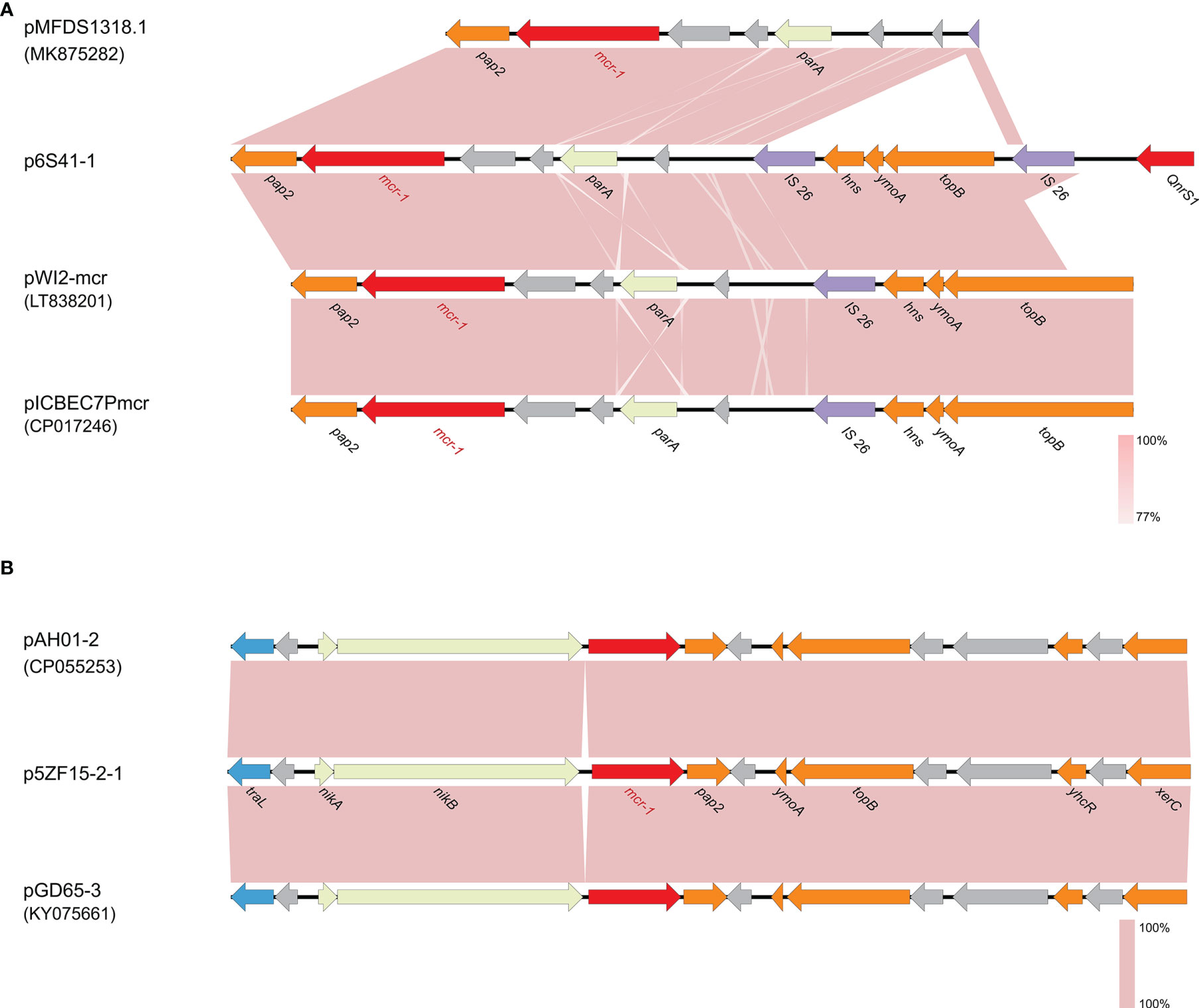
Figure 2 The genetic context of mcr-1 gene on p6S41-1 and p5ZF15-2-1. (A) Comparison of genes surrounding mcr-1 on p6S41-1, pMFDS1318.1 (accession number: MK875282), pWI2-mcr (accession number: LT838201) and pICBEC7Pmcr (accession number: CP017246). (B) Comparison of genes surrounding mcr-1 on p5ZF15-2-1, pAH01-2 (accession number: CP055253) and pGD65-3 (accession number: KY075661). Open reading frames (ORFs) are shown as arrows and indicated according to their putative functions. Purple indicates genes related to mobile elements, red indicates genes related to drug resistance, light yellow indicates genes related to plasmid stability, blue indicates genes involved in conjugation and orange represents other functional genes. Hypothetical protein encoded genes are colored by grey. Regions with a high degree of homology are indicated by pink shading.
The plasmid harboring mcr-1 of E. fergusonii 5ZF15-2-1 was 61,228 bp in length with 75 CDSs and a GC content of 42.4% (Table 2). In silico analysis indicated that plasmid p5ZF15-2-1 belongs to IncI2. Conjugation experiments revealed that the transmission of mcr-1 from E. fergusonii 5ZF15-2-1 to E. Coli 600 was successful, which was consistent with the antimicrobial susceptibility profile of the transconjugant (Table 1). Annotation revealed that mcr-1 was the only resistance gene on plasmid p5ZF15-2-1, which carries genes coding for replication initiation and conjugative transfer assembly proteins, plasmid stability proteins, and other functional proteins. Exploration of the genetic context surrounding mcr-1 showed that the genes traL, nikA, and nikB were present in the upstream region, and the genes pap2, ymoA, topB, yhcR, and xerC were present in the downstream region. As shown in Figure 2B, the sequence encompassing the traL-hp-nikA-nikB-mcr-1-pap2-hp-ymoA-topB-hp-hp-yhcR-hp-xerC region in this study showed a 100% nucleotide identity with the corresponding region of the IncI2 plasmids pAH01-2 (accession number: CP055253) and pGD65-3 (accession number: KY075661) which both E. coli strains and isolated from Anhui and Guangzhou, respectively. Further, plasmid p5ZF15-2-1 carried genes encoding pilus and conjugative transfer proteins pilU, pilT, pilQ, pilO, which provided evidence for transmission of the plasmid.
By the search of the nr/nt database, plasmid p6S41-1 in our study was closely related to E. coli pWI2-mcr (accession number: LT838201), S. enterica pSH16G1509 (accession number: MK477615), E. coli pMCR-13EC-C962A (accession number: KX555452), and E. coli pMFDS1318.1 (accession number: MK875282), from the same incompatibility. Further, plasmid p5ZF15-2-1 exhibiting 99% nucleotide identity and 99%-100% query coverages to pM-64-826 (accession number: MT773675), pAH01-2 (accession number: CP055253), pVE362 (accession number: AP018355), and pXDL-MCR-1.18 (accession number: CP043036). As shown in Figure 3, the backbone sequences of these plasmids were almost identical.
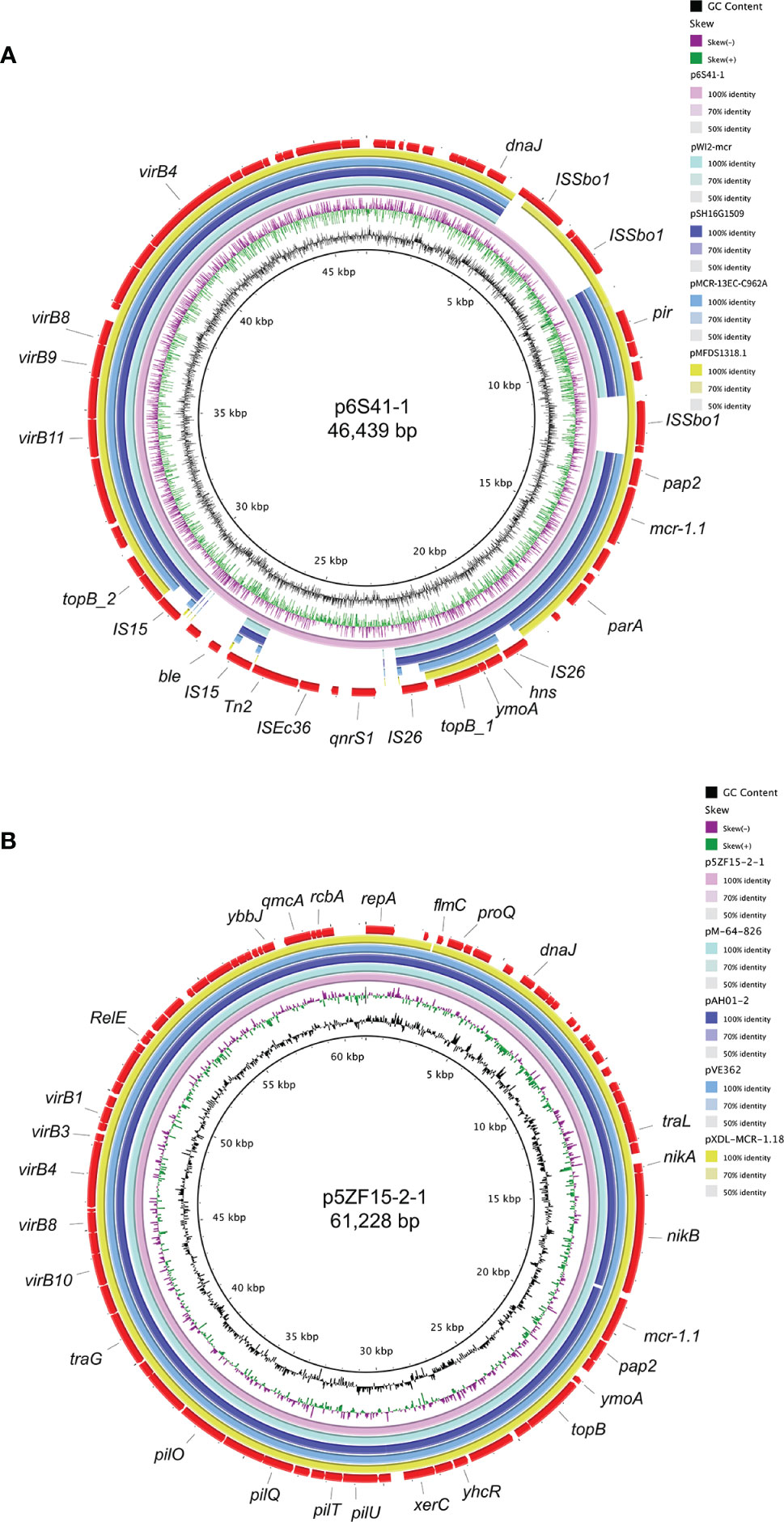
Figure 3 The genetic features of plasmid p6S41-1 and p5ZF15-2-1. (A) Circular comparison between mcr-1 bearing IncX4 plasmids p6S41-1 in this study and four similar IncX4 plasmids in NCBI nr/nt database. p6S41-1 was used as the reference in the outermost ring. (B) Circular comparison between mcr-1 bearing IncI2 plasmids p5ZF15-2-1 in this study and four similar IncI2 plasmids in NCBI nr/nt database. p5ZF15-2-1 was used as the reference in the outermost ring.
The results of amino acid sequences comparison show that amino acid mutations occurred in PmrA, PhoQ, and MgrB (Table S3). Among them, the substitution T299I in PhoQ was predicted to affect protein functions according to PROVEAN. In more detail, it might have affected phosphate transfer for it is located on His Kinase A (phosphoacceptor) domain which is the key element in two-component signal transduction systems according to SMART analysis. In accordance with the results of AST, the MIC of colistin for transconjugants 5ZF15-2-1-E. coli 600 was lower than that of 5ZF15-2-1 which indicated that isolate 5ZF15-2-1 may acquire colistin resistance via chromosomal mutation on PhoQ besides mcr-1 gene.
According to the result of ANI analysis, one strain (SAMEA5771506, Figure S1) was removed in subsequential analyses. The result of phylogenetic analysis of 115 core genomes (Figure 4) shows strains 6S41-1 (BioSample: SAMN20239770) and 5ZF15-2-1 (BioSample: SAMN20243900) in this study exhibited close phylogenetic relationships. In addition, the two strains are far from the other nine strains from China (SAMN20033693, SAMN14604091, SAMN10145472, SAMN08534259, SAMN07682644, SAMN07682645, SAMN07682646, SAMN08534257, SAMN10531910) but closely related to (SAMN15148372, SAMN15148373, SAMN15148374, SAMN15148375, SAMN15148376) from the United Kingdom. The five strains from the United Kingdom were all isolated from sheep feces on a farm in 2017. Further, we conducted drug resistance gene analysis on 113 strains from the database via Resfinder and found that seven strains carried mcr-1. Interestingly, six of seven strains were isolated from China and came from a variety of sources including sludge, anus swab, and feces, which indicates China may be a main reservoir of mcr-1-bearing E. fergusonii (Figure 4). In terms of the phylogenetic tree, the nine strains (including two in this study) from China could be divided into four clusters and are distantly related.

Figure 4 The maximum likelihood phylogenetic tree based on the core genome sequences of 115 E. fergusonii strains. E. fergusonii 6S41-1 (BioSample: SAMN20239770) and 5ZF15-2-1 (BioSample: SAMN20243900) are indicated in red. The other mcr-1-bearing strains were marked by dark red. The three circles around the phylogenetic tree indicate the location (inner circle), collection date and source (outer circle) of these strains.
Enterobacteriaceae is one of the leading causes of both nosocomial and community-acquired infections. And genus Escherichia is the most important one among it. E. fergusonii, as the emerging pathogen of genus Escherichia in recent years, is increasing in prevalence and spread among humans and animals worldwide (Glover et al., 2017; Adesina et al., 2019). It was reported that infections caused by E. fergusonii have occurred in many countries (Savini et al., 2008; Lai et al., 2011). In addition, according to recent reports, the resistance of E. fergusonii to antibiotics is increasingly observed. For example, E. fergusonii has been found harboring β-lactams genes and carbapenemase-resistant genes, which contribute significantly to the growing clinical drug selection burden (Savini et al., 2008; Lagacé-Wiens et al., 2010). However, only a few studies have reported mcr-1 gene in E. fergusonii (da Silva Pontes et al., 2020), and the phylogenetic analysis of E. fergusonii is still lacking. Therefore, we conducted core genome analysis of E. fergusonii 6S41-1 and 5ZF15-2-1 by whole-genome sequencing data. In addition, we also elucidated the molecular characteristics and genetic context of plasmids carrying mcr-1 gene.
Colistin was reconsidered as a valid therapeutic option with the increasing carbapenem-resistant bacteria (Bialvaei and Samadi Kafil, 2015). But the emergence of mobile colistin resistance gene mcr-1 threatens public health and aroused concern over the world. To date, mcr-1 has been found in more than 50 countries and regions across six continents and has been observed in 15 bacterial species, including Acinetobacter baumannii, Enterobacter cloacae, E. coli, Klebsiella aerogenes, and so on (Xiaomin et al., 2020). In this study, we identified two strains of E. fergusonii carrying mcr-1 and performed core genome analysis. The results showed that E. fergusonii 6S41-1 and 5ZF15-2-1 have close genetic relationships with strains (SAMN15148375, SAMN15148373, SAMN15148372, SAMN15148376, SAMN15148376) for they all belong to the same major cluster and all isolated from farm environment. On the other hand, E. fergusonii has been isolated from different sources and has been detected in different countries and at different times. The main source and country are feces and the United Kingdom, respectively.
In this study, the typical IS26-parA-mcr-1-pap2 cassette was identified in plasmid p6S41-1, which is similar to other IncX4 plasmids (Figure 2A) (Du et al., 2020). In addition, p6S41-1 did not have an ISApl1 insertion sequence upstream of the mcr-1 gene, which is in accordance with other studies (Sun et al., 2017). Generally, ISApl1 played a pivotal role in the mobilization of mcr-1 (Snesrud et al., 2017). However, ISApl1 in front of mcr-1 was absent in all IncX4-type plasmids (Sun et al., 2017). Reports provided evidence that ISApl1 is involved in the initial transposition of the mcr-1 element and then is lost for the stability of mcr-1 on IncX4 plasmids (Li et al., 2017; Snesrud et al., 2017). In similarity to some other known IncI2-type plasmids (Wang et al., 2017), ISApl1 was also absent on plasmid p5ZF15-2-1. And a nikA-nikB-mcr-1 locus sites on p5ZF15-2-1. The same structure was also present in studies by Zheng et al. and Wang et al. (Wang et al., 2017; Zheng et al., 2019). Wang et al. speculated these IncI2 plasmids have contributed to the spread of mcr-1 genes among different species via horizontal means by diversified conjugation-aided mechanisms without the assistance of the ISApl1 gene (Wang et al., 2017). This hypothesis was consistent with the results of conjugation assay in this study. Interestingly, the chromosomal mutations T299I in PhoQ of E. fergusonii 6S41-1 was predicted to affect protein function which might contribute to colistin resistance besides mcr-1 gene. This suggests that multiple drug resistance mechanisms co-exist in E. fergusonii 6S41-1. In addition, the pap2 gene, which can be found in both these two plasmids, was confirmed as necessary along with mcr-1 to reduce colistin susceptibility (Choi et al., 2020).
Different replicons have been found carrying mcr-1 since the first report in 2016 (Liu et al., 2016), including IncI2, IncX3, IncX4, IncF, IncFII, IncH1, IncHI1, IncHI2, IncP, and IncY (Hu et al., 2021). Most plasmids carrying mcr-1 proved to be transferable, with IncX4 and IncI2 as dominant types among them (Xiaomin et al., 2020). To date, IncX4 and IncI2 plasmids carrying mcr-1 have been identified in different species of Enterobacteriaceae, including K. pneumoniae, Salmonella, E. coli, C. braakii, and C. sakazakii, which come from various sources, including humans, food animals, and animal production environments (Zhou et al., 2017; Wang et al., 2018; Zheng et al., 2019; Xiaomin et al., 2020). A previous study reported the E. fergusonii isolates with IncHI2-type plasmids harboring mcr-1 in Brazil (da Silva Pontes et al., 2020). That same year, a Chinese study reported two strains carrying mcr-1 gene isolated from caecal contents of chickens with the same plasmid type IncI2 (Li et al., 2020). In this study, we isolated two strains of E. fergusonii carrying mcr-1 which the plasmids types were IncX4 and IncI2, respectively. To our best knowledge, this is the first report about mcr-1 gene located on IncX4-type plasmid in E. fergusonii. It demonstrated the further spread of mcr-1 with different replicons among different species of Enterobacteriaceae, indicating the need to pay more attention to it. Of note, these two plasmids, both carrying genes encoding type IV secretion systems, could increase bacterial competitiveness and may contribute to the fitness of the antibiotic-resistant bacteria.
According to the “One Health” concept, the environment plays a vital role in safeguarding public health. And environmental AMR was identified as six emerging issues of greatest concern by the United Nations Environment Programme (UNEP) (Anyanwu et al., 2020). According to current reports, mcr-1 bearing isolates have been detected in various farm environments, including E. coli strains isolated from dairy cow feces in China with mcr-1 genes encoded on IncI2-type plasmid and chromosomes respectively, mcr-1 bearing E. coli strains isolated from farm soils in China in 2017 with various of plasmids types and MLST types, and mcr-1-positive strains from manure, swabs, and flies in German swine farms (Guenther et al., 2017; Zheng et al., 2017; Zheng et al., 2019). This indicates that farm environments are important reservoirs for the storage and spread of mcr-1. This result is most likely due to the fact that colistin is used as a feed additive to promote animal growth. In this study, E. fergusonii isolates 6S41-1 and 5ZF15-2-1 carrying mcr-1 from farm soils and pig feces imply the extended dissemination of mcr-1 among different species of Enterobacteriaceae in farm environments, indicating the urgent need for surveillance and further study of mcr-1 gene in farm environments.
In summary, we identified two E. fergusonii isolates harboring mcr-1 and described the complete sequence by whole-genome sequencing. The mcr-1 genes located on IncX4 and IncI2 plasmids spread widely among species of Enterobacteriaceae worldwide. Not only that, the mcr-1 gene located on IncI2-type plasmid p5ZF15-2-1 has the ability of horizontal transfer, which provides support for its further transmission. For mcr-1 gene can spread via food chains, the source of two strains highlights the possibility that mcr-1 may spread to humans through farm environments and thereby threaten public health. Therefore, continuous monitoring and investigations of mcr-1 in farm environments are necessary to control its spread.
The datasets presented in this study can be found in online repositories: https://www.ncbi.nlm.nih.gov/ (accession numbers CP079884-CP079890).
JG and BZ conceived and designed the experiments. RL, SL, JQ, and HG collected samples and performed the experiments. HX and XG analyzed the data. RL wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by research grants from Henan Science and Technology Department (No. 192102310059), the National Key Research and Development Program of China (No.2016YFD0501105), the National Natural Science Foundation of China (No. 82072314), and Henan Province Medical Science and Technology Research Project Joint Construction Project (No. LHGJ20190232).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.774494/full#supplementary-material
Adesina, T., Nwinyi, O., De, N., Akinnola, O., Omonigbehin, E. (2019). First Detection of Carbapenem-Resistant Escherichia Fergusonii Strains Harbouring Beta-Lactamase Genes From Clinical Samples. Pathogens 8, E164. doi: 10.3390/pathogens8040164
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402
Anyanwu, M. U., Jaja, I. F., Nwobi, O. C. (2020). Occurrence and Characteristics of Mobile Colistin Resistance (Mcr) Gene-Containing Isolates From the Environment: A Review. Int. J. Environ. Res. Public Health 17, E1028. doi: 10.3390/ijerph17031028
Asai, T., Kojima, A., Harada, K., Ishihara, K., Takahashi, T., Tamura, Y. (2005). Correlation Between the Usage Volume of Veterinary Therapeutic Antimicrobials and Resistance in Escherichia Coli Isolated From the Feces of Food-Producing Animals in Japan. Jpn J. Infect. Dis. 58, 369–372. doi: 10.1097/01.qai.0000184861.26733.8c
Bain, M. S., Green, C. C. (1999). Isolation of Escherichia Fergusonii in Cases Clinically Suggestive of Salmonellosis. Vet. Rec 144, 511. doi: 10.1111/j.1740-8261.1999.tb00368.x
Bialvaei, A. Z., Samadi Kafil, H. (2015). Colistin, Mechanisms and Prevalence of Resistance. Curr. Med. Res. Opin. 31, 707–721. doi: 10.1185/03007995.2015.1018989
Choi, Y., Lee, J.-Y., Lim, S.-K., Ko, K. S. (2020). Intact Pap2 Downstream of Mcr-1 Appears to be Required for Colistin Resistance. Diagn. Microbiol. Infect. Dis. 97, 114997. doi: 10.1016/j.diagmicrobio.2020.114997
da Silva Pontes, L., Pimenta, R., Silveira, M. C., Tavares-Teixeira, C. B., Ferreira Pereira, N., da Conceiçāo Neto, O. C., et al. (2020). Escherichia Fergusonii Harboring IncHI2 Plasmid Containing Mcr-1 Gene: A Novel Reservoir for Colistin Resistance in Brazil. Microb. Drug Resist. 27, 721–725. doi: 10.1089/mdr.2020.0041
Diarra, M. S., Silversides, F. G., Diarrassouba, F., Pritchard, J., Masson, L., Brousseau, R., et al. (2007). Impact of Feed Supplementation With Antimicrobial Agents on Growth Performance of Broiler Chickens, Clostridium Perfringens and Enterococcus Counts, and Antibiotic Resistance Phenotypes and Distribution of Antimicrobial Resistance Determinants in Escherichia Coli Isolates. Appl. Environ. Microbiol. 73, 6566–6576. doi: 10.1128/AEM.01086-07
Du, C., Feng, Y., Wang, G., Zhang, Z., Hu, H., Yu, Y., et al. (2020). Co-Occurrence of the Mcr-1.1 and Mcr-3.7 Genes in a Multidrug-Resistant Escherichia Coli Isolate From China. Infect. Drug Resist. 13, 3649–3655. doi: 10.2147/IDR.S268787
Farmer, J. J., Davis, B. R., Hickman-Brenner, F. W., McWhorter, A., Huntley-Carter, G. P., Asbury, M. A., et al. (1985a). Biochemical Identification of New Species and Biogroups of Enterobacteriaceae Isolated From Clinical Specimens. J. Clin. Microbiol. 21, 46–76. doi: 10.1128/jcm.21.1.46-76.1985
Farmer, J. J., Fanning, G. R., Davis, B. R., O’Hara, C. M., Riddle, C., Hickman-Brenner, F. W., et al. (1985b). Escherichia Fergusonii and Enterobacter Taylorae, Two New Species of Enterobacteriaceae Isolated From Clinical Specimens. J. Clin. Microbiol. 21, 77–81. doi: 10.1128/JCM.21.1.77-81.1985
Fegan, N., Barlow, R. S., Gobius, K. S. (2006). Escherichia Coli O157 Somatic Antigen is Present in an Isolate of E. Fergusonii. Curr. Microbiol. 52, 482–486. doi: 10.1007/s00284-005-0447-6
Foster, G., Evans, J., Tryland, M., Hollamby, S., MacArthur, I., Gordon, E., et al. (2010). Use of Citrate Adonitol Agar as a Selective Medium for the Isolation of Escherichia Fergusonii From a Captive Reindeer Herd. Vet. Microbiol. 144, 484–486. doi: 10.1016/j.vetmic.2010.01.014
Funke, G., Hany, A., Altwegg, M. (1993). Isolation of Escherichia Fergusonii From Four Different Sites in a Patient With Pancreatic Carcinoma and Cholangiosepsis. J. Clin. Microbiol. 31, 2201–2203. doi: 10.1128/JCM.31.8.2201-2203.1993
Glover, B., Wentzel, J., Jenkins, A., Van Vuuren, M. (2017). The First Report of Escherichia Fergusonii Isolated From Non-Human Primates, in Africa. One Health 3, 70–75. doi: 10.1016/j.onehlt.2017.05.001
Guenther, S., Falgenhauer, L., Semmler, T., Imirzalioglu, C., Chakraborty, T., Roesler, U., et al. (2017). Environmental Emission of Multiresistant Escherichia Coli Carrying the Colistin Resistance Gene Mcr-1 From German Swine Farms. J. Antimicrob. Chemother. 72, 1289–1292. doi: 10.1093/jac/dkw585
Hariharan, H., López, A., Conboy, G., Coles, M., Muirhead, T. (2007). Isolation of Escherichia Fergusonii From the Feces and Internal Organs of a Goat With Diarrhea. Can. Vet. J. 48, 630–631. doi: 10.1080/03079450701344738
Herráez, P., Rodríguez, A. F., Espinosa de los Monteros, A., Acosta, A. B., Jaber, J. R., Castellano, J., et al. (2005). Fibrino-Necrotic Typhlitis Caused by Escherichia Fergusonii in Ostriches (Struthio Camelus). Avian Dis. 49, 167–169. doi: 10.1637/7221-061104r
Huang, J., Li, C., Song, J., Velkov, T., Wang, L., Zhu, Y., et al. (2020). Regulating Polymyxin Resistance in Gram-Negative Bacteria: Roles of Two-Component Systems PhoPQ and PmrAB. Future Microbiol. 15, 445–459. doi: 10.2217/fmb-2019-0322
Hu, Y., Nguyen, S. V., Wang, W., Gan, X., Dong, Y., Liu, C., et al. (2021). Antimicrobial Resistance and Genomic Characterization of Two Mcr-1-Harboring Foodborne Salmonella Isolates Recovered in China 2016. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.636284
Jeannot, K., Bolard, A., Plésiat, P. (2017). Resistance to Polymyxins in Gram-Negative Organisms. Int. J. Antimicrob. Agents 49, 526–535. doi: 10.1016/j.ijantimicag.2016.11.029
Lagacé-Wiens, P. R. S., Baudry, P. J., Pang, P., Hammond, G. (2010). First Description of an Extended-Spectrum-Beta-Lactamase-Producing Multidrug-Resistant Escherichia Fergusonii Strain in a Patient With Cystitis. J. Clin. Microbiol. 48, 2301–2302. doi: 10.1128/JCM.00364-10
Lai, C.-C., Cheng, A., Huang, Y.-T., Chung, K.-P., Lee, M.-R., Liao, C.-H., et al. (2011). Escherichia Fergusonii Bacteremia in a Diabetic Patient With Pancreatic Cancer. J. Clin. Microbiol. 49, 4001–4002. doi: 10.1128/JCM.05355-11
Li, X.-P., Sun, R.-Y., Song, J.-Q., Fang, L.-X., Zhang, R.-M., Lian, X.-L., et al. (2020). Within-Host Heterogeneity and Flexibility of Mcr-1 Transmission in Chicken Gut. Int. J. Antimicrob. Agents 55, 105806. doi: 10.1016/j.ijantimicag.2019.09.010
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Li, R., Xie, M., Zhang, J., Yang, Z., Liu, L., Liu, X., et al. (2017). Genetic Characterization of Mcr-1-Bearing Plasmids to Depict Molecular Mechanisms Underlying Dissemination of the Colistin Resistance Determinant. J. Antimicrob. Chemother. 72, 393–401. doi: 10.1093/jac/dkw411
Luo, Q., Yu, W., Zhou, K., Guo, L., Shen, P., Lu, H., et al. (2017). Molecular Epidemiology and Colistin Resistant Mechanism of Mcr-Positive and Mcr-Negative Clinical Isolated Escherichia Coli. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02262
Maheux, A. F., Boudreau, D. K., Bergeron, M. G., Rodriguez, M. J. (2014). Characterization of Escherichia Fergusonii and Escherichia Albertii Isolated From Water. J. Appl. Microbiol. 117, 597–609. doi: 10.1111/jam.12551
Maifreni, M., Frigo, F., Bartolomeoli, I., Innocente, N., Biasutti, M., Marino, M. (2013). Identification of the Enterobacteriaceae in Montasio Cheese and Assessment of Their Amino Acid Decarboxylase Activity. J. Dairy Res. 80, 122–127. doi: 10.1017/S002202991200074X
Oh, J.-Y., Kang, M.-S., An, B.-K., Shin, E.-G., Kim, M.-J., Kwon, J.-H., et al. (2012). Isolation and Epidemiological Characterization of Heat-Labile Enterotoxin-Producing Escherichia Fergusonii From Healthy Chickens. Vet. Microbiol. 160, 170–175. doi: 10.1016/j.vetmic.2012.05.020
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Savini, V., Catavitello, C., Talia, M., Manna, A., Pompetti, F., Favaro, M., et al. (2008). Multidrug-Resistant Escherichia Fergusonii: A Case of Acute Cystitis. J. Clin. Microbiol. 46, 1551–1552. doi: 10.1128/JCM.01210-07
Skov, R. L., Monnet, D. L. (2016). Plasmid-Mediated Colistin Resistance (Mcr-1 Gene): Three Months Later, the Story Unfolds. Euro Surveill 21, 30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155
Snesrud, E., Ong, A. C., Corey, B., Kwak, Y. I., Clifford, R., Gleeson, T., et al. (2017). Analysis of Serial Isolates of Mcr-1-Positive Escherichia Coli Reveals a Highly Active ISApl1 Transposon. Antimicrob. Agents Chemother. 61, e00056–e00017. doi: 10.1128/AAC.00056-17
Storm, D. R., Rosenthal, K. S., Swanson, P. E. (1977). Polymyxin and Related Peptide Antibiotics. Annu. Rev. Biochem. 46, 723–763. doi: 10.1146/annurev.bi.46.070177.003451
Sullivan, M. J., Petty, N. K., Beatson, S. A. (2011). Easyfig: A Genome Comparison Visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, J., Fang, L.-X., Wu, Z., Deng, H., Yang, R.-S., Li, X.-P., et al. (2017). Genetic Analysis of the IncX4 Plasmids: Implications for a Unique Pattern in the Mcr-1 Acquisition. Sci. Rep. 7, 424. doi: 10.1038/s41598-017-00095-x
Sun, J., Zhang, H., Liu, Y.-H., Feng, Y. (2018). Towards Understanding MCR-Like Colistin Resistance. Trends Microbiol. 26, 794–808. doi: 10.1016/j.tim.2018.02.006
Walsh, T. R., Wu, Y. (2016). China Bans Colistin as a Feed Additive for Animals. Lancet Infect. Dis. 16, 1102–1103. doi: 10.1016/S1473-3099(16)30329-2
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., Zong, Z. (2020a). Identification of Novel Mobile Colistin Resistance Gene Mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, J., Li, X., Li, J., Hurley, D., Bai, X., Yu, Z., et al. (2017). Complete Genetic Analysis of a Salmonella Enterica Serovar Indiana Isolate Accompanying Four Plasmids Carrying Mcr-1, ESBL and Other Resistance Genes in China. Vet. Microbiol. 210, 142–146. doi: 10.1016/j.vetmic.2017.08.024
Wang, X., Wang, Y., Wang, Y., Zhang, S., Shen, Z., Wang, S. (2018). Emergence of the Colistin Resistance Gene Mcr-1 and its Variant in Several Uncommon Species of Enterobacteriaceae From Commercial Poultry Farm Surrounding Environments. Vet. Microbiol. 219, 161–164. doi: 10.1016/j.vetmic.2018.04.002
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020b). Changes in Colistin Resistance and Mcr-1 Abundance in Escherichia Coli of Animal and Human Origins Following the Ban of Colistin-Positive Additives in China: An Epidemiological Comparative Study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving Bacterial Genome Assemblies From Short and Long Sequencing Reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, H., Wang, D., Gao, F. (2021). Toward a High-Quality Pan-Genome Landscape of Bacillus Subtilis by Removal of Confounding Strains. Brief Bioinform. 22, 1951–1971. doi: 10.1093/bib/bbaa013
Xiaomin, S., Yiming, L., Yuying, Y., Zhangqi, S., Yongning, W., Shaolin, W. (2020). Global Impact of Mcr-1-Positive Enterobacteriaceae Bacteria on “One Health.” Crit. Rev. Microbiol. 46, 565–577. doi: 10.1080/1040841X.2020.1812510
Zheng, B., Feng, C., Xu, H., Yu, X., Guo, L., Jiang, X., et al. (2019). Detection and Characterization of ESBL-Producing Escherichia Coli Expressing Mcr-1 From Dairy Cows in China. J. Antimicrob. Chemother. 74, 321–325. doi: 10.1093/jac/dky446
Zheng, B., Huang, C., Xu, H., Guo, L., Zhang, J., Wang, X., et al. (2017). Occurrence and Genomic Characterization of ESBL-Producing, MCR-1-Harboring Escherichia Coli in Farming Soil. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02510
Zheng, B., Zhang, J., Ji, J., Fang, Y., Shen, P., Ying, C., et al. (2015). Emergence of Raoultella Ornithinolytica Coproducing IMP-4 and KPC-2 Carbapenemases in China. Antimicrob. Agents Chemother. 59, 7086–7089. doi: 10.1128/AAC.01363-15
Keywords: IncX4, IncI2, farm environments, whole-genome sequencing, core genome analysis
Citation: Liu R, Xu H, Guo X, Liu S, Qiao J, Ge H, Zheng B and Gou J (2022) Genomic Characterization of Two Escherichia fergusonii Isolates Harboring mcr-1 Gene From Farm Environment. Front. Cell. Infect. Microbiol. 12:774494. doi: 10.3389/fcimb.2022.774494
Received: 22 September 2021; Accepted: 26 April 2022;
Published: 26 May 2022.
Edited by:
Rodrigo Cayô, Federal University of São Paulo, BrazilReviewed by:
Dr. Subhasree Roy, National Institute of Cholera and Enteric Diseases (ICMR), IndiaCopyright © 2022 Liu, Xu, Guo, Liu, Qiao, Ge, Zheng and Gou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianjun Gou, amlhbmp1bmdAenp1LmVkdS5jbg==; Beiwen Zheng, emhlbmdid0B6anUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.