- 1Program in Vector-Borne Diseases, Department of Veterinary Microbiology and Pathology, College of Veterinary Medicine, Washington State University, Pullman, WA, United States
- 2Veterinary Pest Genetic Research Unit, USDA-ARS, Kerrville, TX, United States
- 3Animal Disease Research Unit, USDA-ARS, Pullman, WA, United States
Bovine babesiosis caused by Babesia bigemina and Babesia bovis is an economically important disease that affects cattle worldwide. Both B. bigemina and B. bovis are transovarially transmitted by Rhipicephalus ticks. However, little is known regarding parasite gene expression during infection of the tick vector or mammalian host, which has limited the development of effective control strategies to alleviate the losses to the cattle industry. To understand Babesia gene regulation during tick and mammalian host infection, we performed high throughput RNA-sequencing using samples collected from calves and Rhipicephalus microplus ticks infected with B. bigemina. We evaluated gene expression between B. bigemina blood-stages and kinetes and compared them with previous B. bovis RNA-seq data. The results revealed similar patterns of gene regulation between these two tick-borne transovarially transmitted Babesia parasites. Like B. bovis, the transcription of several B. bigemina genes in kinetes exceeded a 1,000-fold change while a few of these genes had a >20,000-fold increase. To identify genes that may have important roles in B. bigemina and B. bovis transovarial transmission, we searched for genes upregulated in B. bigemina kinetes in the genomic datasets of B. bovis and non-transovarially transmitted parasites, Theileria spp. and Babesia microti. Using this approach, we identify genes that may be potential markers for transovarial transmission by B. bigemina and B. bovis. The findings presented herein demonstrate common Babesia genes linked to infection of the vector or mammalian host and may contribute to elucidating strategies used by the parasite to complete their life cycle.
Introduction
Bovine babesiosis is a tick-borne disease of worldwide impact caused by Apicomplexan intracellular parasites of the genus Babesia. The four species of Babesia responsible for most of the clinical cases in cattle are Babesia bigemina, Babesia bovis, Babesia divergens, and Babesia major (Bock et al., 2004). The first two are found in several countries in tropical and subtropical regions, including Central and South America, Australia, Asia, Africa, and Southern Europe with high prevalence of 22% for B. bigemina and 20% for B. bovis (Jacob et al., 2020).
The severity of bovine babesiosis can vary according to several factors, including age, genetics, and status of the immune system, among other characteristics, including the relative virulence of the Babesia strains involved (Bock et al., 2004; Schnittger et al., 2012). The most common manifestations of acute cases include fever, hemolytic anemia, jaundice, weakness, and hemoglobinuria while chronic infections are usually asymptomatic (Bock et al., 2004; Schnittger et al., 2012). Another symptom of acute B. bovis infection is the manifestation of neurological signs (ataxia, lethargy, inappetence, incoordination, teeth grinding, involuntary movements of the legs) which are related to sequestration of infected erythrocytes in capillaries of different tissues, including the brain. However, this manifestation does not happen during B. bigemina infections (reviewed by (Gallego-Lopez et al., 2019). In acute infections, the maximum percentage of infected erythrocytes is ~10% in B. bigemina and less than 1% in B. bovis (Chaudhry et al., 2010), but these estimates may be biased by massive sequestration of B. bovis infected erythrocytes in capillaries. Disease triggered by B. bigemina infection causes mortality rates of up to 30% in untreated animals, whereas infections by B. bovis may have much higher mortality rates (between 70-80%) (Jacob et al., 2020).
Both B. bigemina and B. bovis are transmitted to cattle by Rhipicephalus ticks (Bock et al., 2004), the definitive host of Babesia. The vertebrate host infection starts when sporozoites are injected along with tick saliva by the infected nymph (B. bigemina) or larva (B. bovis) (Suarez and Noh, 2011). Invasion occurs after several random collisions between parasite and erythrocytes which facilitates orientation of the anterior Babesia apical complex. After a tight junction between sporozoites and the erythrocyte cell membrane is established, proteins are released from the secretory organelles, and the sporozoite actively penetrates the erythrocyte. Internalized parasites differentiate into trophozoites which reproduce asexually into merozoites. Mature merozoites egress from the infected erythrocyte, causing their destruction, and invade other erythrocytes, where the cycle of asexual multiplication is perpetuated (Mosqueda et al., 2002; Jalovecka et al., 2018). If the animal survives acute disease, they become chronically infected for months to years (Johnston et al., 1978; Chung et al., 2017). When ticks ingest the parasites during blood meal acquisition, gamogony begins with the formation of gametocytes in the tick midgut lumen. Gametocytes undergo metamorphosis resulting in haploid gamete formation (Jalovecka et al., 2018). Babesia fertilization is induced by contact between male and female gametes and results in the formation of a diploid zygote, which invades tick gut epithelial cells. Inside the intestinal cells, zygotes undergo meiotic division resulting in the formation of a single cell kinete. Kinetes are released to the tick hemolymph and migrate throughout the whole tick body, including ovaries which results in vertical transmission to the tick offspring. In the infected larvae, Babesia infects the salivary glands and undergoes sporogony to become sporozoites. These infectious forms are continuously released to the blood of the vertebrate host during larval and nymphal feeding (Jalovecka et al., 2018).
All these Babesia stages and transitions are required to complete the parasite life cycle and are characterized by specific gene expression of proteins involved in distinct cell biological processes and molecular mechanisms, with different signaling pathways. However, our current knowledge regarding these events is limited (Jalovecka et al., 2018; Suarez et al., 2019; Ueti et al., 2020; Ueti et al., 2021). This knowledge gap precludes the understanding of host-tick-Babesia interaction, constraining the discovery of novel drug therapies and identification of new targets for vaccine development (Suarez et al., 2019).
In this study, we identified differentially expressed genes in both B. bigemina kinetes and blood-stages and compared our results with a recently published B. bovis transcriptome data set (Ueti et al., 2020; Ueti et al., 2021). The findings presented herein demonstrate common Babesia genes linked to tick vector and mammalian host infection that may contribute to elucidating strategies used by the parasite to complete their biological life cycle.
Materials and methods
Ethics statement
All animal procedures were carried out in accordance with the Animal Welfare Act and Regulations (United States Department of Agriculture). The study was conducted under Animal use protocol #IACUC 2018-16, approved by the Institutional Animal Care and Use Committee of the University of Idaho, Moscow, ID, USA and adhered to the principles and standards put forth in the Guide for the Care and Use of Laboratory Animals (The National Academy of Sciences) and the Guide for Care and Use of Agricultural Animals in Agricultural Research and Teaching (The American Dairy Science Association, the American Society of Animal Science, and the Poultry Science Association).
Cattle, Rhipicephalus microplus and Babesia bigemina
Three splenectomized naïve Holstein calves (C46527, C1437 and C46541) approximately 4 months old and confirmed to be Babesia-free by competitive enzyme-linked immunosorbent assay and nested PCR (Goff et al., 2006; Suarez et al., 2012) were used for feeding R. microplus, the La Minita strain. Approximately 40,000 larvae were applied under a cloth patch on the dorsal region for acquisition of B. bigemina parasites. Fourteen days after tick application, when approximately 1% of the ticks had molted to unfed adults, ~107 B. bigemina infected erythrocytes, the Mexico strain, were inoculated intravenously into each calf to synchronize female tick acquisition feeding with an ascending B. bigemina parasitemia (Bohaliga et al., 2019). The presence of B. bigemina in the blood was determined by Giemsa-stained blood smear.
Isolation of Babesia bigemina blood-stages and kinetes
For obtaining B. bigemina blood-stages, blood of each infected calf was collected individually immediately before euthanasia in sterile flasks with glass beads and shaken to prevent coagulation. Defibrinated red blood cells (RBCs) were washed with Puck’s saline G solution, placed in culture flasks, and incubated with 5% CO2 to increase the percentage of parasitized RBCs, as described (Levy and Ristic, 1980; Ueti et al., 2021). After 3-5 days of in vitro incubation, the culture media was removed by centrifugation at 2800 ×g for 3 min, cells were suspended in TRIzol® reagent (Life technologies, Carlsbad, CA), and stored at -80 °C until used.
For obtaining kinetes, replete female ticks that fed on calves infected with B. bigemina during an ascending parasitemia were collected daily beginning at 6 days post-infection. Ticks were cleaned and rinsed in warm running water, distributed in 24-well plates, placed in containers with saturated KNO3 solution, and incubated at 26 °C at 92% relative humidity for 6 days to allow accumulation of B. bigemina kinetes in the tick’s hemolymph as previously described (Howell et al., 2007; Bohaliga et al., 2019). To identify ticks infected with B. bigemina, a distal leg segment was removed, a drop of exuding hemolymph was collected, put on a slide and Giemsa stained to examine for the presence of kinetes by light microscopy. Babesia bigemina kinetes were collected from ticks with >50 kinetes/hemolymph as previously described (Howell et al., 2007). The hemolymph samples were collected, pooled, concentrated by centrifugation at 4000 ×g for 2 min (Bohaliga et al., 2019), suspended in TRIzol reagent, and stored at -80 °C until used.
RNA extraction and quality control
Parasite samples were suspended in TRIzol Reagent for total RNA extraction. Total RNA was extracted individually from paired blood-stage and kinete samples and treated with TURBO DNase (ThermoFisher Scientific) according to the manufacturer’s instructions to eliminate contaminating genomic DNA. RNA isolation was accomplished using the RNA Cleanup and Concentrator kit (RNA-5 and RNA-25) (Zymo Research, Irvine, CA) according to the manufacturer’s instructions. RNA concentration was measured by Nanodrop spectrophotometer (ThermoFisher Scientific, ND 1000) and tested for residual DNA by PCR targeting the CCp1 gene (Bastos et al., 2013). RNA quality was assessed by Nanodrop absorbances at 260/280 nm and 260/230 nm and Agilent Bioanalyzer Nano RNA chip (Agilent Technologies, Santa Clara, CA, USA), following the manufacturer’s recommendations. The RNA integrity number values for blood-stages were 9.5, 10 and 9.3 and kinetes were 9.7, 7.9 and 8.8. Purified RNA samples were stored at -80 °C until used.
RNA-seq library preparation and sequencing
The RNA-seq libraries were constructed using the Illumina TruSeq mRNA stranded protocols (Illumina San Diego, CA, USA) following manufacturer’s recommendation. Briefly, from the total RNA, messenger RNA (mRNA) was enriched using poly-A selection, mRNA was chemically fragmented and denatured before transcription into first-stranded cDNA followed by double-stranded cDNA. Double-stranded cDNA was end-repaired and polyadenylated. Illumina dual index adapters were ligated to the cDNA samples followed by 9 cycles of PCR amplification. The libraries were quantified, normalized, and then multiplexed as previously described (Ueti et al., 2021). The multiplexed samples were sequenced on three lanes of a HiSeq 2500 (Illumina). Approximately 60 million total reads per sample were obtained. FASTQ files were generated with pipelines for the removal of adapter sequences from the 3’ ends of reads.
Differential expression analysis was performed at the Fred Hutchinson Cancer Institute (Seattle, WA, USA). The combined RNA-seq run resulted in six separate fastq files representing each library, containing sequenced paired reads and quality control data. The HiSeq Control software HCS 2.2.58 version and Real Time Analysis software RTA 1.18.64 version, supplied by Illumina Inc, were used. All raw and processed RNA sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE214115.
Identification and analysis of differentially expressed genes
Differential gene expression analysis between kinetes and blood-stages was performed as follows. Low quality reads were filtered before alignment to the B. bigemina BOND reference genome (NCBIRefSeq GCF_000981445.1) using STAR v2.5.2a (2-pass mapping) (Dobin et al., 2013). Counts were generated from alignments for each gene using the Subread feature of Counts v1.6.0 (https://spark.apache.org/docs/1.6.0/programming-guide.html). Genes without at least 1 read per million mapped reads across all three samples within a group were removed, data was normalized using the Trimmed Mean of M-Values (TMM method) and analyzed for differential expression significance testing using the generalized linear model likelihood ratio test (GLM LRT) method in edgeR v3.20.9 (https://bioconductor.stastistik.tu-dortmund.de/packages/3.6/bioc/html/edgeR.html) (Robinson et al., 2010; McCarthy et al., 2012; Chen et al., 2016). The false discovery rate (FDR) method was employed to correct for multiple testing and genes were termed significantly differentially expressed if their log Fold Change (logFC) value was greater than or equal to 1 and the FDR set to 5%. The p-values were adjusted for multiple testing according to FDR correction (Benjamini and Hochberg, 1995).
In silico analysis of genes in tick-borne pathogens
Orthologs of B. bigemina proteins were identified for B. bovis, Babesia ovata and B. divergens using the PiroplasmaDB resource (https://piroplasmadb.org/piro/app). Locus tags for B. bigemina genes were searched and the synteny genes were identified. Using the syntenic sequences track of the genome browser allowed the identification of orthologs among related Apicomplexan species.
Genes upregulated in B. bigemina kinetes was translated to proteins and used as queries to select potentially homologous sequences from B. bovis (T2Bo) [taxid:484906], Theileria spp. [taxid:5873] (that include Theileria annulata, Theileria parva, Theileria orientalis and Theileria equi), and B. microti [taxid:5868] based on the results of the protein BLAST platform (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against the non-redundant protein sequences database. Sequences identified by BLASTp analyses were extracted using a custom script, and all results were manually inspected.
Validation of RNA-seq data using RT-qPCR
Three hundred nanograms of the total RNA extracted from each preparation of parasite were treated with DNase I (Invitrogen, USA) to eliminate contaminating genomic DNA, and reverse transcribed (RT) using a SuperScript III kit. The resulting cDNA was used as a template in RT-qPCR with the SsoFast™ EvaGreen® Supermix (BioRad, CA, USA). Gene expression was quantified on a CFX96™ Real-Time PCR Detection System (Bio-RAD). The amplification reaction was performed in a final volume of 20 µL using 0.8 µL of 10 µM of each primer set, 6.9 µL of nuclease-free water, 1.5 µL of a cDNA as template and 10 µL of SsoFast EvaGreen Supermix. The qPCR conditions consisted of an initial denaturation cycle at 95 °C for 3 min, followed by 40 cycles at 95 °C for 30 sec for denaturation, 60 °C for 30 sec for annealing, and 72 °C for 30 sec for extension. The specific primers (Supplementary Table 1) were designed using Primer3 software (Rozen and Skaletsky, 2000) and synthesized by Eurofins Genomics (Louisville, KY, USA). The specificity of qPCR was assessed by melting curve analysis. To confirm the nucleotide sequence of the specific DNA fragments for each designed primer, all amplicons were sequenced (data not shown). CFX Manager Software (Bio-Rad) was used to obtain quantification cycle (Cq) values (Bustin et al., 2009) and analyze expression data. The transcription level was calculated as a relative expression using 2-ΔΔCq method (Livak and Schmittgen, 2001) to calculate the relative expression of the target gene after normalization to two reference genes, Mitogen-activated protein kinase (MAPK) and Proteosome A (ProtA) (Supplementary Table 1). These reference genes were selected from the RNA-seq data, as there was no variation in expression between the two groups evaluated (kinetes and blood-stages).
Results and discussion
RNA-seq analysis: Differential gene expression
Approximately 120 million reads mapped to B. bigemina sequences were obtained from the analysis, of which 92 million corresponded to blood-stages, and 27 million to kinetes (Supplementary Table 2). Low rRNA rates were obtained (Supplementary Table 2), indicating the successful enrichment of mRNA from all blood and tick samples. Principal component analysis of the RNA-seq data resulted in separation of the biological replicates of the same condition into two distinct groups corresponding to blood-stages and kinetes (Figure 1A). The first principal component explained 90% of the variation between groups and the second principal component showed that blood-stage transcripts were more similar to each other as compared to kinete transcripts (Figure 1A). This pattern was observed in a previous B. bovis blood-stage and kinete RNA-seq study (Ueti et al., 2021) and may be associated with individual variability of samples due to the presence of tick hemocyte mRNA.
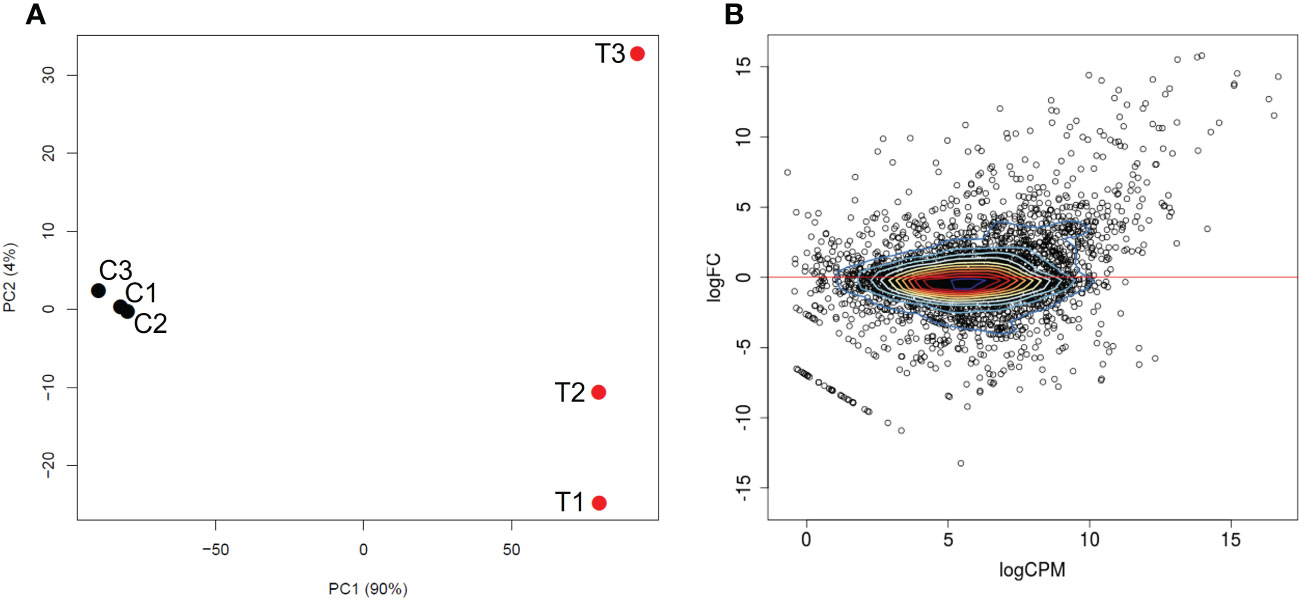
Figure 1 Differential transcriptional expression by B. bigemina blood-stages versus kinetes. (A) PCA plot between B. bigemina stages from blood (C1, C2, C3) and versus kinetes (T1, T2, T3). C indicates samples from calves and T indicates samples from ticks. (B) A MA plot shows the relationship of the log2 fold-change (FC) of B. bigemina blood-stages and kinetes versus the log2 normalized mean expression counts per million (CPM).
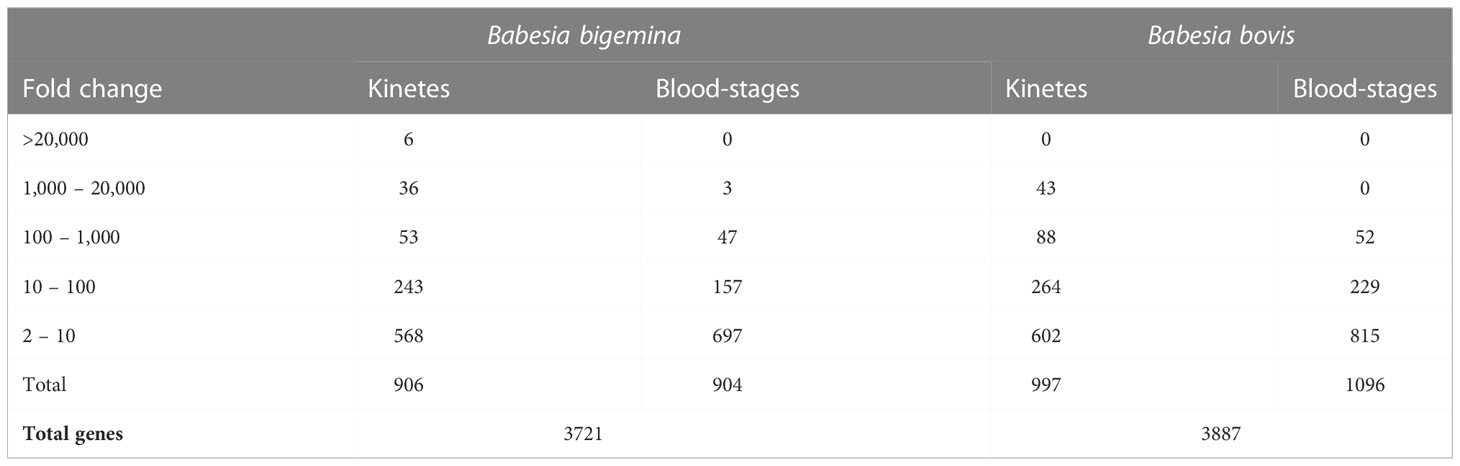
A total of 3,721 genes were identified in both Babesia stages (Figure 1B, Table 1). Supplemental Data 1 provides a list of all differentially expressed genes (DEGs) found between B. bigemina kinetes and blood-stages. Among the DEGs, 906 genes were upregulated in kinetes, and 904 by blood-stages (Table 1). Interestingly, the magnitude of gene expression was greater in the kinete, reaching over ~56,000-fold increase as compared to the blood-stages which had only an ~9,600-fold increase. For comparison, the magnitude of expression in the B. bovis RNA-seq was >19,000-fold in kinetes and <300-fold increase in blood-stages (Ueti et al., 2021). Table 1 shows the comparative gene expression in B. bigemina and B. bovis.
In B. bigemina kinetes, 42 genes had a fold change ranging from ~1,000 to 56,767, while in blood-stages, only 3 genes were upregulated in this range (Table 1). These kinete upregulated genes may be linked to the importance of these genes in biological functions related to invasion and survival of the parasite within the host tick. However, it can be speculated that upregulated genes may not be associated with protein expression as previously described (Lindner et al., 2019). A study in Plasmodium sporozoites analyzed highly abundant transcripts and correlated them with no or low amounts of detected protein, evidencing translational repression (Lindner et al., 2019). To understand more completely the biological importance that a given gene may have, studies at both the proteomic and functional levels must be also included. Furthermore, it is important to point out that gene products from less abundant transcripts may also be essential for parasite function.
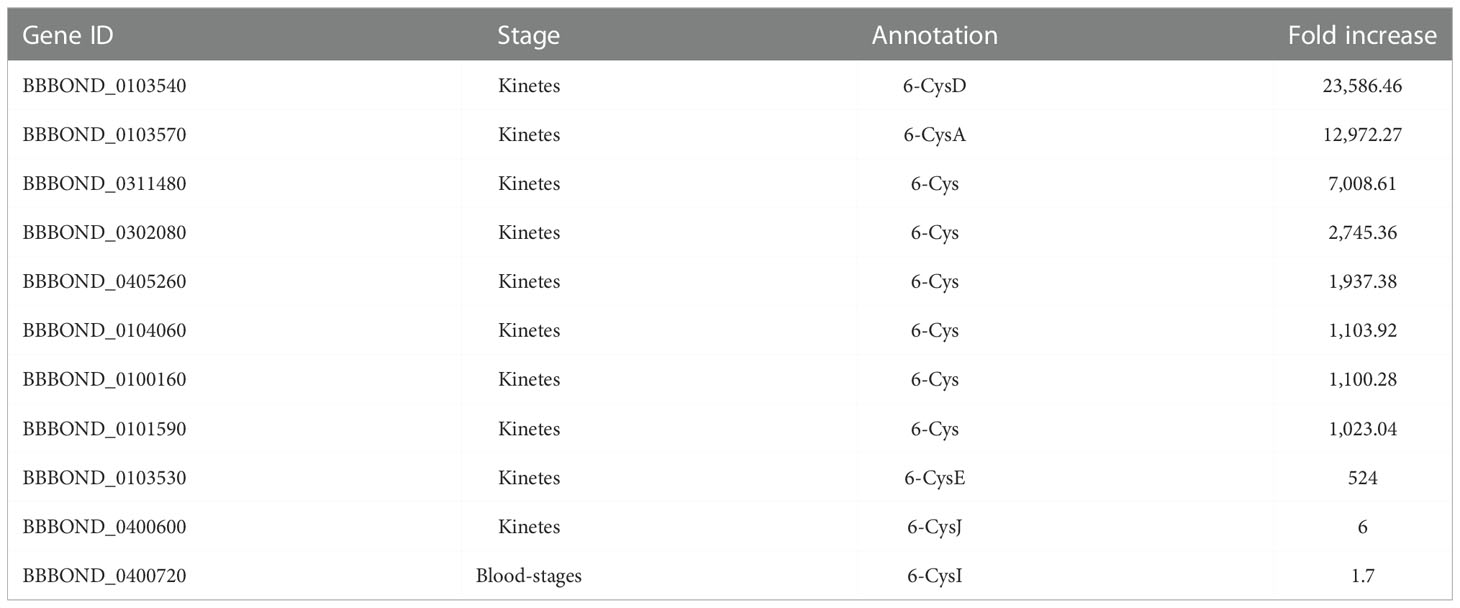
In B. bigemina, highly expressed genes found in kinetes (over 1,000-fold increase) included kinete specific protein (KSP), rhomboid 4 (ROM4), membrane and RNA-binding protein, thrombospondin-related anonymous protein (TRAP), 6-cysteine (6-cys), inner membrane complex family protein, vesicle coat complex membrane and Der1-like family (Der-1) (Table 2). Orthologs of these genes were found in B. bovis, B. ovata and B. divergens (Table 2). Some highly expressed genes in B. bigemina kinetes were similar to B. bovis kinetes (Table 2), suggesting that these genes may play important roles during Babesia development within tick hemolymph including the invasion of the ovary and egg mass and may serve as potential candidate targets for the development of transmission blocking vaccines or drugs (Bohaliga et al., 2019). Gene expression by species is categorized in Table 2.
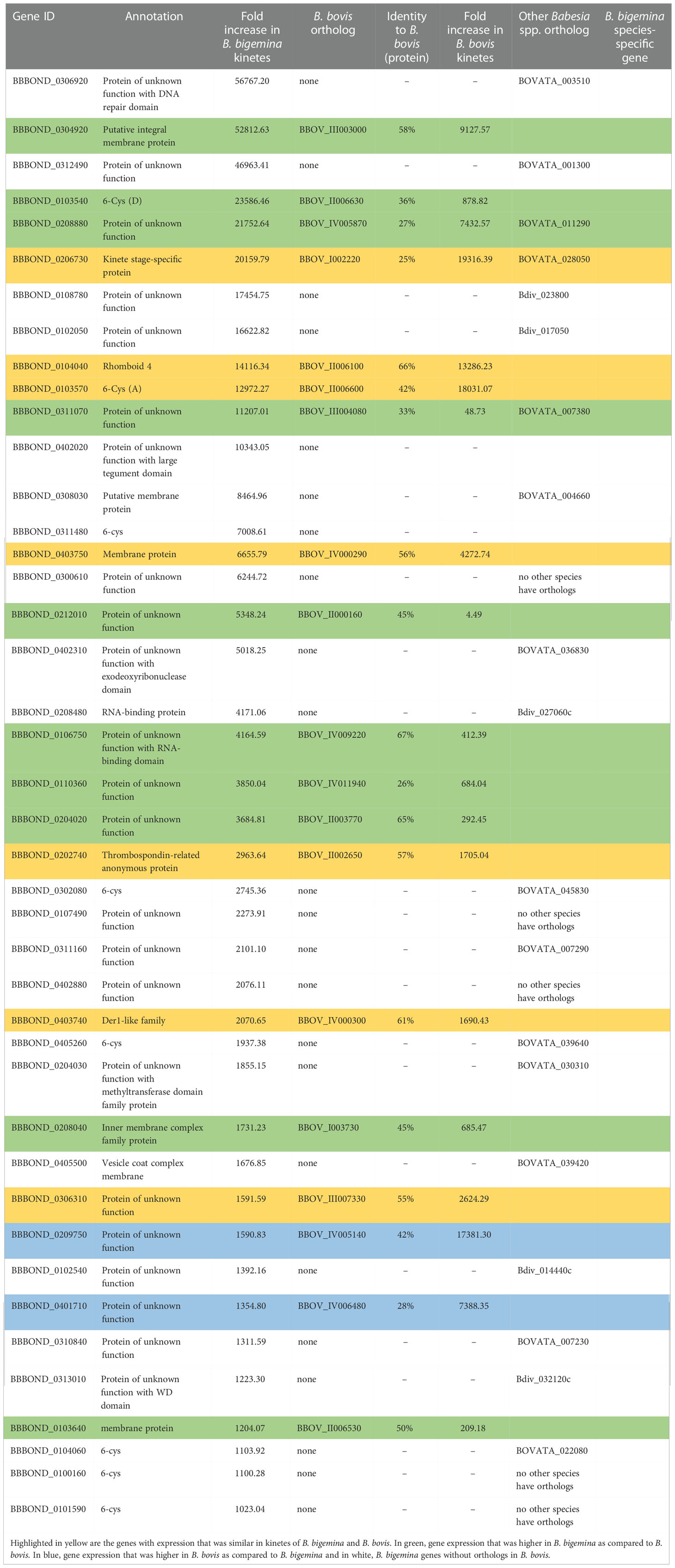
Table 2 Differential expression of genes highly expressed in B. bigemina kinetes. Orthologs of these genes in B. bovis (BBOV), B. ovata (BOVATA) and B. divergens (Bdiv) were found by synteny using PiroplasmaDB.
In B. bigemina blood-stages, 43 genes with increased expression (>120-fold change) were identified (Table 3). In addition to many proteins of unknown function, two ribosome-binding proteins and one probable fructokinase were found in B. bigemina blood-stages (Table 3). The functions of the gene products of most of the upregulated genes have not yet been characterized and may have biological importance related to invasion, gliding motility, and egress (González et al., 2019). Expression of two B. bigemina blood-stage genes exceeded that of B. bovis orthologs (Table 3).
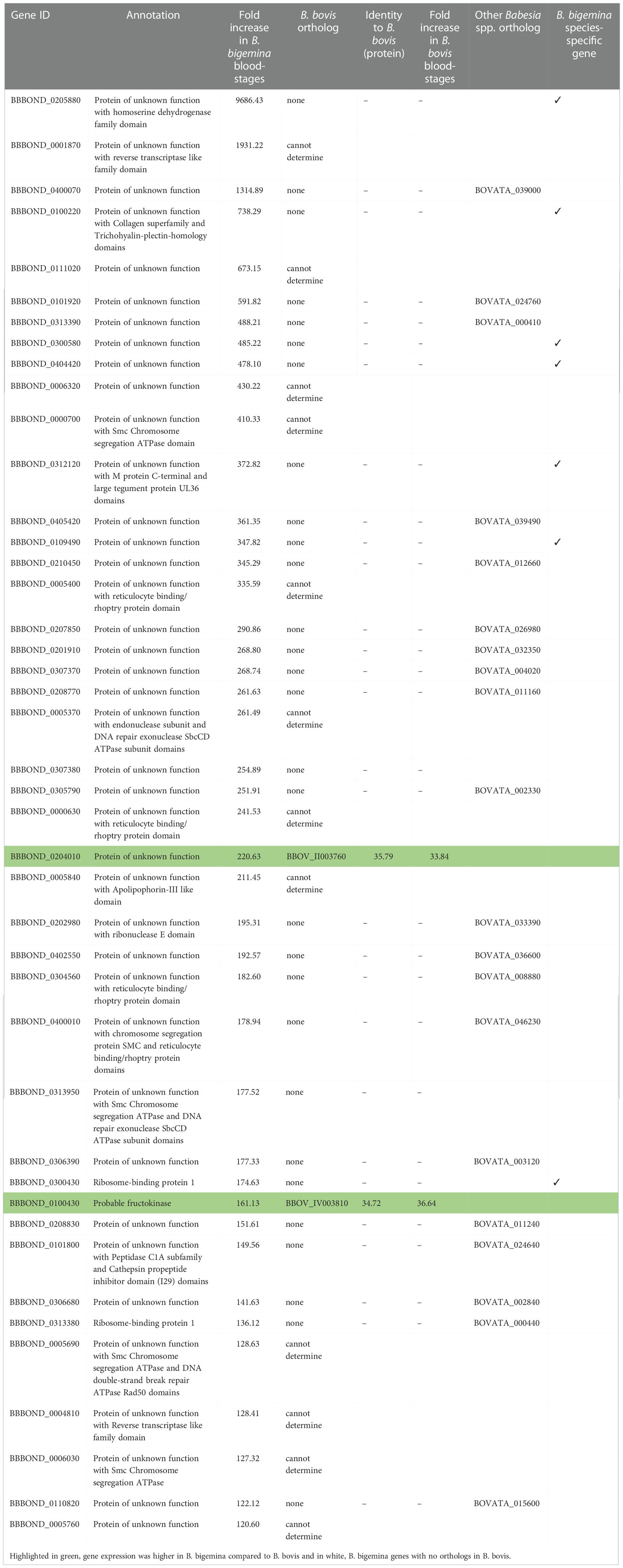
Table 3 Differential expression of genes with over 120-fold increase in B. bigemina blood-stages. Orthologs of these genes were found in B. bovis (BBOV) and B. ovata (BOVATA) by synteny using PiroplasmaDB.
It is interesting to note that of the 42 highly expressed genes in B. bigemina kinetes, 20 had orthologs in B. bovis. However, this was not the case for blood-stages, as only two of the 43 genes upregulated by >120-fold had orthologs in B. bovis (Tables 2 and 3). In the RNA-seq of B. bovis, differentially expressed kinete genes with levels >1,000-fold change were KSP, ROM family proteins, membrane proteins, TRAP, 6-cys, protein kinases, Der-1, GCC2/GCC3 protein, and AP2 families (Ueti et al., 2020; Ueti et al., 2021). Importantly, of the genes that exceeded >1,000-fold change, 34 genes were unique for Babesia when compared to the bovine genome by BLASTp (data not shown).
The list of B. bovis genes expressed by blood-stages (>120-fold increase) is quite distinct from those genes expressed by B. bigemina during the same life cycle phase in the mammalian host. In B. bovis blood-stages, small open reading frame (SmORF), variant erythrocyte surface antigen (VES), merozoite surface antigen (MSA), spherical body protein (SBP), membrane proteins, ROM family proteins, rhoptry-associated protein 1 (RAP-1) family protein and others were upregulated (Ueti et al., 2020; Ueti et al., 2021). B. bovis infected RBC (iRBC), but not B. bigemina iRBC, can adhere to vascular endothelial cells and accumulate in the host microvasculature, a process that is apparently mediated by VES proteins. Babesia bovis changes the protein and lipid composition of the iRBC membrane, modifying its architecture resulting in increased cellular adhesiveness and high antigenic variability (Suarez et al., 2019). The VES gene family is also responsible for this antigenic variation, and this phenomenon, combined with epithelial cell cytoadhesion by iRBCs, allows B. bovis to evade the vertebrate host immune response. The B. bovis genome contains 133 members of VES (Allred et al., 2000; Eichenberger et al., 2017; Ueti et al., 2021). However, the role of VES proteins in B. bigemina is unknown and understanding its mechanisms may be a key to potential vaccine candidates (Suarez et al., 2019). (Jackson et al., 2014) described some VES genes in B. bigemina BOND, JG29, Puerto Rico, and BbiS3P strains, however, in our RNA-seq experiments no differential expression of VES genes was found in either kinetes or blood-stages. In addition, the SmORF genes may assist in capillary sequestration (Suarez et al., 2019). However, SmORF genes are present only in B. bovis, and not in B. bigemina (Suarez et al., 2019). Altogether, these differences in gene expression may contribute to increased virulence and pathogenicity of B. bovis when compared to B. bigemina.
Synteny analysis using the Piroplasma genome database showed that nine genes classified as protein of unknown function, three 6-cys and one ribosome-binding protein 1 had no orthologs in B. ovata and B. divergens, suggesting that they are B. bigemina-specific genes (Tables 2 and 3).
Babesia bigemina differentially expressed genes
HAP2/GCS1 domain protein
The fusion of male and female gametes to generate a zygote is an important step in the life cycle of many organisms and has been considered a target for transmission blocking vaccines. The gene HAP2/GCS1 (hapless 2/generative cell specific 1) was identified in plants, Plasmodium, and other organisms (Johnson et al., 2004; Mori et al., 2006; Hirai et al., 2008) as important keys in gamete fertilization. In bovine Babesia, HAP2 was found to be essential for the development of sexual forms in hap2 KO parasites and expressed on the surface of B. bovis gametes (Hussein et al., 2017). Interestingly, anti-HAP2 antibodies blocked B. bigemina zygote formation (Camacho-Nuez et al., 2017). The expression of HAP2-GCS1 was increased 20-fold in B. bigemina kinetes (BBBOND_0307410). This gene was also increased in B. bovis kinetes (BBOV_III006770, 258-fold) (Ueti et al., 2021), but 13 times more than B. bigemina. HAP2 is a gamete fusion protein and may be is involved in attachment of kinetes to ovary epithelial cells (Ueti et al., 2021). However, further investigation is needed to test if HAP2 protein is expressed on the surface of Babesia kinetes.
CCp family
The CCp gene family is related to the development of sexual stages in Apicomplexa organisms, including Plasmodium, Theileria, and Babesia (Pradel et al., 2004; Becker et al., 2010; Bastos et al., 2013). In the B. bigemina RNA-seq, three members of the CCp family, named CCp1, CCp2 and CCp3 (BBBOND_0307800, BBBOND_0203950 and, BBBOND_0312400, respectively), containing LCCL (Limulus coagulation factor C) motifs, were upregulated in blood-stages, which ranged from 1.8 to 8.9-fold. The same pattern was observed in B. bovis (BBOV_III006360, BBOV_II003700, and BBOV_III008930) with blood-stage expression varying from 2.2 to 8.8-fold (Ueti et al., 2021). In a previous study, the gene expression and proteins of CCp1-3 was proven in Puerto Rico strain B. bigemina induced sexual stages (Bastos et al., 2013). A recent study showed that in addition to the described CCp1-3, two novel non-canonical members of the CCp gene family, named CCp5 and FNPA, were also found in B. bovis. Both genes are highly conserved among geographically distinct B. bovis strains (Ozubek et al., 2022). Babesia bovis FNPA protein has a predicted transmembrane domain, and this fact characterizes it as a possible target of a transmission-blocking vaccine against B. bovis (Ozubek et al., 2022). In the B. bigemina and B. bovis blood-stages RNA-seqs, upregulation of CCp5 (BBBOND_0306080, 2.8-fold and BBOV_III007200, 2-fold) and FNPA (BBBOND_0206110, 3.4-fold and BBOV_I002720, 2.8-fold increase) (Ueti et al., 2021) were found.
GCC2/GCC3 domain containing protein family
The GCC2/GCC3 domain containing proteins belong to the cysteine repeat modular proteins (CRMPs) family, which is an important specific protein of apicomplexan parasites. In Plasmodium, it was postulated that all four CRMP are required for host cell interaction. Analysis of P. berghei sporozoites lacking PCRMP1 – 4, showed they were incapable of invading the mosquito salivary gland. In addition, mutant sporozoites lacking PCRMP3 or 4 did not proliferate inside hepatocytes (Douradinha et al., 2011). In Plasmodium GCC2/GCC3 are differentially expressed in pairs during the parasite life cycle. This was also found to be true for B. bovis where different GCC2/GCC3 pairs were upregulated based on life stage. In B. bovis kinetes, BBOV_III011730, 761-fold and BBOV_IV006250, 1,082-fold, were upregulated while BBOV_III011740, 5.6-fold and BBOV_IV006260, 14-fold were upregulated in blood-stages (Ueti et al., 2021). In contrast, in the B. bigemina RNA-seq, three kinete genes were up-regulated (BBBOND_0208310 and BBBOND_0208320 (both 98-fold) and BBBOND_0404220 (137-fold)). In B. bovis, GCC2/GCC3 genes are arranged in tandem, however, the BOND annotation indicated an intervening gene between BBBOND_0208300 and BBBOND_0208320. In silico and sequencing analysis of RT-PCR amplicons using forward and reverse primers spanning BBBOND_0208310 and BBBOND_0208320, respectively, suggested that there was not an intervening gene, and that the gene model may need to be updated (Supplementary Figure 1). Expression of GCC2/GCC3 proteins in B. bigemina blood-stages did not follow the paired upregulation of B. bovis as only a single protein, BBBOND_0404210 (1.8-fold), was upregulated, and another, BBBOND_0208300, had no difference in expression.
The AP2 family
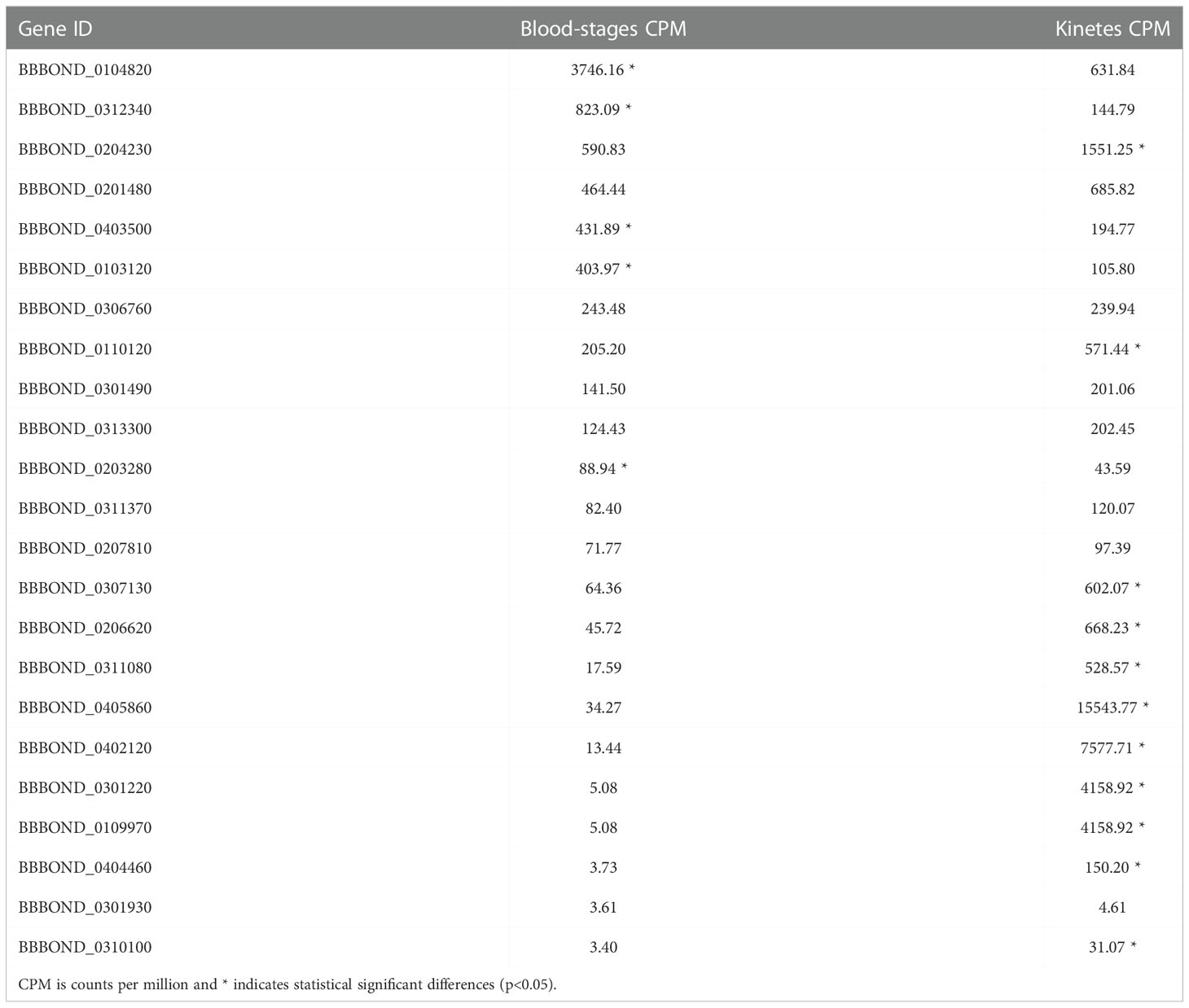
The Apetala 2 (AP2) family are nuclear transcription factors and contain conserved structural motifs that are directly involved in DNA binding (Alzan et al., 2016). AP2 encompasses the main transcription factors specific to Apicomplexa (Balaji et al., 2005). These genes are associated with transcriptional control of other genes involved in parasite development (Suarez et al., 2019). Several described B. bigemina genes encoding proteins containing AP2 domains (Alzan et al., 2016) and found in B. bovis (Ueti et al., 2021) were differentially expressed in B. bigemina blood-stage and kinetes. Table 4 identifies the B. bigemina AP2 genes that had orthologs in B. bovis, although their architectures may show considerable variability (28 - 74% identity). In B. bigemina, there are a total of 23 AP2 genes that were identified. Eleven were upregulated in B. bigemina kinetes and five in the blood-stages, while seven were not different between groups (Table 4), suggesting that they could be differentially expressed by forms at other points of the biological cycle, such as gametes or sporozoites. The B. bigemina AP2 gene-family expression profile suggests the highest expression level for BBBOND_0104820 in blood-stage parasites (Table 5). Its ortholog BBOV_II005480 was also the highest expressed AP2 in B. bovis blood-stages (Ueti et al., 2021). However, in kinetes, the correlation between B. bigemina and B. bovis was not maintained. Interestingly, B. bigemina BBBOND_0312340 and BBBOND_0403500 AP2 genes were upregulated in blood-stages but their orthologs in B. bovis (BBOV_III008870 and BBOV_IV000500) were upregulated in kinetes. Further investigation of the functions of the AP2 genes in these two Babesia are necessary to understand why the pattern of expression of these two AP2 genes is distinct between B. bigemina and B. bovis. This differential expression may be linked to differences in virulence or the needs of the parasite at different stages in its life cycle (in different hosts). Babesia bigemina kinetes had a higher magnitude of gene expression when compared to B. bovis (Table 1) and perhaps differences in AP2 genes contribute to the transcriptional control of other genes required by kinetes for survival/development/interaction in the invertebrate host.
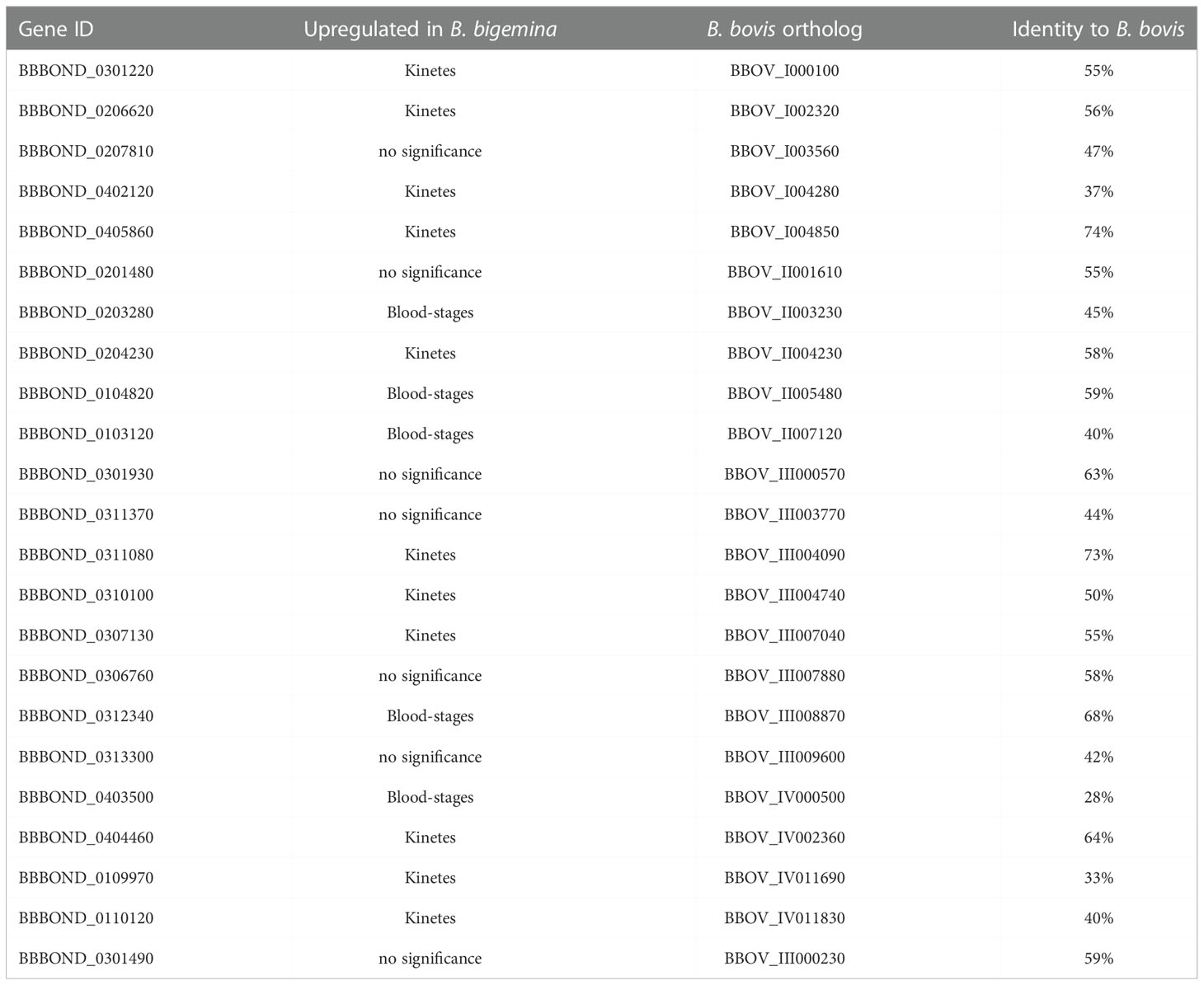
Table 4 AP2 gene family expressed in kinetes and blood-stages in B. bigemina and their respective orthologs in B. bovis.
6-cys gene family
The 6-cys gene family was originally identified in Plasmodium and plays important roles in gamete stages and oocyte formation. Members of the 6-cys family are considered strong candidates for vaccine development (van Dijk et al., 2010; Alzan et al., 2017). These proteins are defined by domains that contain 6 positionally conserved cysteines, but arrangements of 4, 5 or 7 cysteine residues can also be found in Babesia (Alzan et al., 2016). In addition, most 6-cys proteins have signal peptides, suggesting these proteins are likely to be expressed on the surface of the parasite, or secreted (Alzan et al., 2016). In B. bovis, this family is composed of ten genes (A-J) that are related to the development of gametes in the R. microplus midgut (Alzan et al., 2016). Babesia bovis RNA-seq revealed that all ten 6-cys genes were upregulated in kinetes (Ueti et al., 2021). In fact, four members of 6-cys (C, A, B and E) (BBOV_II006620, BBOV_II006600, BBOV_II006610 and BBOV_II006640) were highly expressed in kinetes (19,316, 18,031, 14,454 and 1,731-fold increase respectively) (Ueti et al., 2021). In B. bigemina, some 6-Cys family members unique to B. bigemina or shared with a B. ovata ortholog were highly upregulated in kinetes (Table 2), suggesting that they may play a role during tick infection. The 6-Cys family is comprised of 11 members with ten upregulated in kinetes and a single member upregulated in blood-stages (Table 6). In Plasmodium vivax, three 6-Cys family proteins were expressed in the blood-stage and might be involved in merozoite invasion activity (Wang et al., 2014).
ABC transporter
The ATP binding cassette (ABC) transporter genes are present in all prokaryotes and eukaryotes, including Apicomplexan parasites. This family of genes codes for different proteins which translocate amino acids, ions, peptides, proteins, cholesterol, metabolites, and toxins across extra- and intracellular membranes and contribute to drug resistance through decreased drug uptake into the cell or efflux (which expel toxins and drugs out of the cell) (El-Awady et al., 2016). Two putative ABC transporters were upregulated in B. bigemina kinetes (BBBOND_0403540 and BBBOND_0207340, both over 11-fold), and five members were upregulated in the blood-stage (BBBOND_0309830, BBBOND_0105520, BBBOND_0100850, BBBOND_0313150 and BBBOND_0300960) by 1.65 to 3-fold. In a B. bovis RNA-seq, ABC transporters were also upregulated in kinetes (BBOV_III006450, BBOV_III011170 and BBOV_I003180) and in blood-stages (BBOV_IV008140, BBOV_III005570, BBOV_I000600) (Ueti et al., 2021). The expression varied between 3.5 to 85.8-fold change. ABC transporter proteins are immunogenic and have been used as antigens in the formulation of vaccines against bacteria (Riquelme-Neira et al., 2013; Murphy et al., 2016). In protozoan parasites, like P. falciparum and Leishmania sp., the ABC transporters are targets of antiprotozoal drugs (Klokouzas et al., 2003; El-Awady et al., 2016).
Iron transporter
Iron receptors and transporters are associated with the pathogen’s virulence and immunogenicity and are usually located on the cell surface, making them good targets for drugs or vaccines (Glanfield et al., 2007). Iron-sulfur clusters (Fe-S) are small inorganic metal cofactors, and proteins that contain Fe-S clusters have functions that include electron transport, regulation of gene expression, substrate binding and activation, radical generation, and DNA repair (Wachnowsky et al., 2018). An important antiparasitic drug, artemisinin, has iron-dependent activity, and the artemisinin drug family is a potent first-line antimalarial drugs (Sibmooh et al., 2001; Olliaro and Taylor, 2004). Artesunate, a derivative of artemisin, effectively inhibited growth of cultures of B. bovis and B. gibsoni (Goo et al., 2013). The role of iron uptake and metabolism in Babesia should be better studied for new potential therapeutic targets. Babesia bigemina kinetes express an Fe-S cluster biosynthesis domain gene (BBBOND_0307200, 57-fold increase), Fe-S cluster assembly protein NFU-like (BBBOND_0211120, 2-fold) and CDGSH Fe-S domain (BBBOND_0202830, 2-fold). In a B. bovis RNA-seq, Fe-S cluster assembly scaffold family protein (BBOV_III007290, 2-fold and BBOV_IV004050, 3-fold) were found in blood-stages and kinetes (Ueti et al., 2021).
Virulence genes
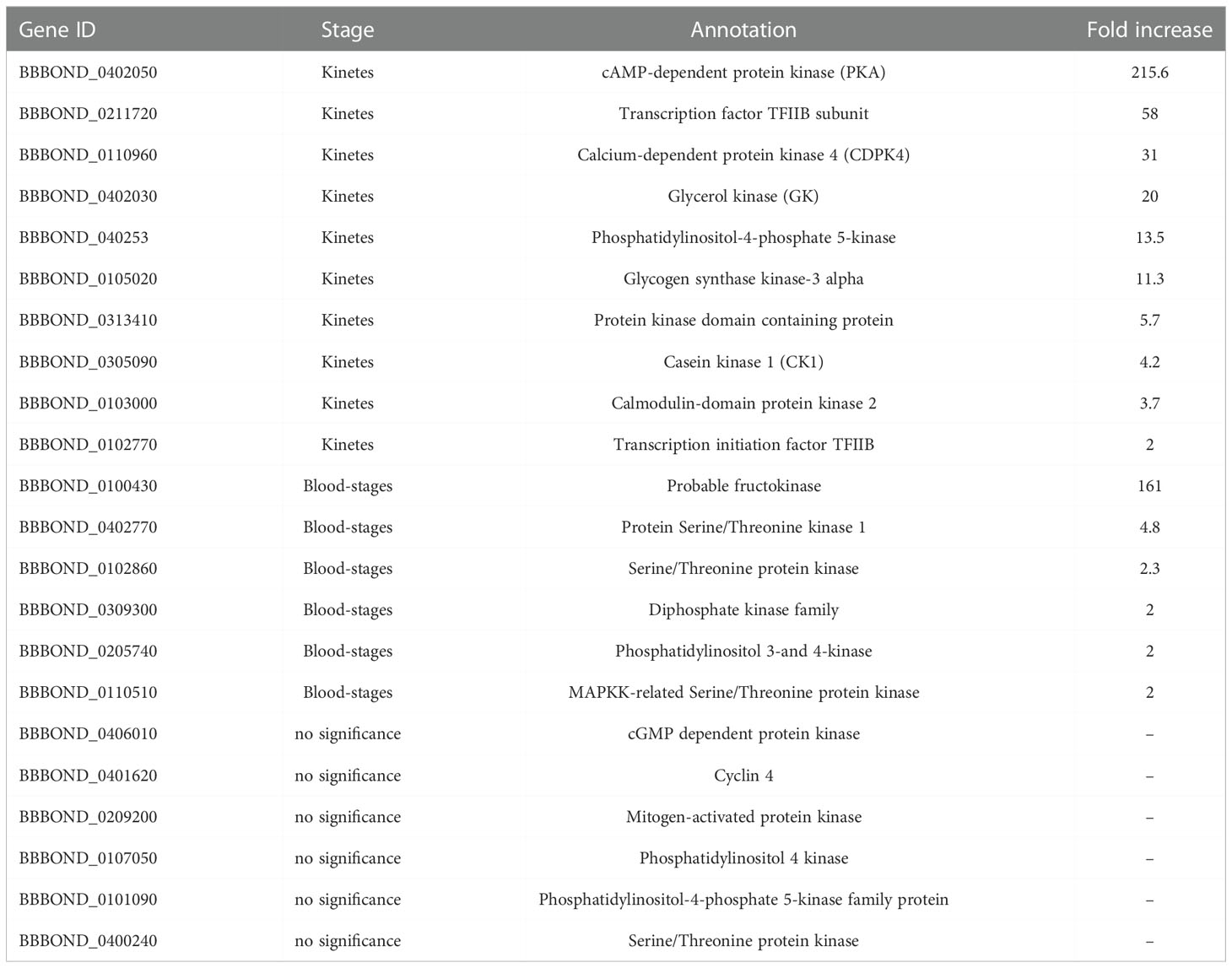
Sixteen virulence genes described and analyzed in the virulome of Mexican B. bigemina strain (Sachman-Ruiz et al., 2021) were found to be upregulated in kinetes and blood-stages (Table 7) in this RNA-seq. Most virulence genes are protein kinases (PKA), which is a family of enzymes that modifies other proteins by adding phosphates. The phosphorylation by PK has been reported to regulate important cellular processes such as transcription, translation, protein synthesis, cell cycle, and apoptosis in Apicomplexa parasites (Kato et al., 2012). Use of staurosporine, an inhibitor of serine/threonine protein kinase, causes the inhibition of host cell invasion or growth of several Apicomplexa species (Plasmodium, Trypanosoma, and Leishmania) (Bork et al., 2006). Furthermore, it led to the complete clearing of B. bovis parasitemia and caused a significant increase in the percentage of extracellular merozoites, probably due to inhibition of erythrocyte invasion by the parasite (Bork et al., 2006).
In humans, the transcription factor TFIIB is phosphorylated in vivo at serine 65 and this phosphorylation is important for the transcription process (Wang et al., 2010). Unfortunately, no study reports the function of TFIIB in Apicomplexa parasites. However, in B. bigemina, TFIIB was upregulated in kinetes (Table 7) and in B. bovis (BBOV_II007500, 2.2-fold) (Ueti et al., 2021).
In P. falciparum gametocytes, whole genome microarray analysis identified glycerol kinase (GK) as the second most highly upregulated gene, and its function is linked to the phosphorylation of host-glycerol (Schnick et al., 2009). GK deletion from P. falciparum had no effect on asexual parasite growth, gametocyte development or exflagellation, suggesting that these life cycle stages do not utilize host-derived glycerol as a carbon source (Schnick et al., 2009). GK was upregulated in both B. bigemina (Table 7) and B. bovis kinetes (BBOV_I004170, 11-fold) (Ueti et al., 2021).
The virulence factor casein kinase I members are serine/threonine protein kinases, highly conserved in eukaryotes (protozoan to humans) and involved in various cellular processes such invasion, protein trafficking, transcription, cell cycle regulation, apoptosis, phosphorylation of upstream kinases, protein-protein interactions, and others (Rachidi et al., 2021). Some studies in Plasmodium, Toxoplasma and Leishmania evaluated the role of this protein and suggest CK1 was an important vaccine candidate, but to date, drug screening has only been studied in Leishmania (Rachidi et al., 2021). In Leishmania, CK1 has functions on host immune system, with phosphorylation in human C3a (complement system) and inducing immune evasion (Lamotte et al., 2017). In addition, CK1 inhibitors have been shown to block the growth of extracellular Leishmania in in vitro culture (Allocco et al., 2006). In Babesia, CK1 was upregulated in kinetes of B. bigemina (Table 7) and B. bovis (BBOV_III002900, 21.3-fold). Further studies should assess whether the protein encoded by this gene expressed in Babesia kinetes interacts with the tick’s innate immune system, leading to avoidance of the complement-like system and, thus, permitting success in infecting tick tissues.
Additionally, another six virulence genes cited in (Sachman-Ruiz et al., 2021) were identified, however, there was no difference between groups (Table 7). These virulence genes may be expressed in other phases of the biological cycle of B. bigemina and consequently, not evaluated in this dataset.
Babesia bigemina genes related to host cell invasion and egress molecular processes
The mechanism of cell invasion of the Babesia-related apicomplexan parasites, including Toxoplasma and Plasmodium, involves initial pathogen attachment to molecules on the cell membrane of the target cell, followed by reorientation, leaving its apical pole facing the cell surface before the parasite actively penetrates the host cell. This active penetration mechanism utilizes an actin/myosin-based motor complex. Some specific structural characteristics of Apicomplexan parasites are related to their motility/migration and invasion/egress of host cells (Suarez et al., 2019). The parasites have an outer plasmatic membrane (PM) and an inner plasmatic complex (IMC), and the space between the two membranes forms the molecular complex (glideosome) that allows the parasite to move. Another important structure involved in the invading process of Apicomplexan parasites is an apical complex that includes several organelles such as rhoptries, micronemes and conoids. However, cell invasion by Babesia parasites does not involve conoids (Suarez et al., 2019).
Gliding motility
Babesia bigemina kinete and blood-stage RNA-seq revealed 39 genes putatively related to the components of the molecular invasion/egress processes that were upregulated between 1.6- to 14,116.3-fold increase in kinetes and 1.7- to 56.7-fold change in blood-stages (Table 8). Of the components that make up the glideosome, actin, myosin A and gliding-associated protein 45 (GAP45) were upregulated in B. bigemina kinetes (Table 8). A previous B. bovis RNA-seq also found actin (BBOV_IV009790, and BBOV_I000300, with 16.6- and 12.6-fold increase in kinetes and BBOV_I004010, BBOV_IV003760, BBOV_IV005480, BBOV_III010900, with 2.2- to 3.7-fold increase in blood-stage), myosin A (BBOV_I003490, 9.8-fold) and GAP45 (BBOV_II005470, 2.3-fold increase) were increased (Ueti et al., 2021). Babesia bigemina actin, myosin A, and GAP45 have high identity with their B. bovis orthologs (63 to 98%) (Table 8). Babesia bigemina myosin B and myosin light chain (MLC) genes were found to be upregulated in blood-stages (Table 8), suggesting that the movement in B. bigemina could be governed by alternative myosin motors, similar to B. divergens and P. falciparum (Hernández et al., 2017; González et al., 2019). In the B. bovis RNA-seq, myosin B expression (BBOV_IV012030) was not significantly different between stages (Ueti et al., 2021).
Microneme
Host cell invasion/egress is largely regulated by the secretion of proteins from specialized organelles, such as micronemes. Many studies have identified the most common post-translational modification to be phosphorylation by protein kinases (PK) and protein phosphatases (PP). Calcium-dependent phosphorylation, driven by a unique family of calcium-dependent protein kinases (CDPKs), plays an essential role in regulation of the protein secretion and increase in the parasite’s cytosolic calcium concentration (Frénal and Soldati-Favre, 2013; Yang and Arrizabalaga, 2017). Calcium-dependent proteins are important for motility, cell invasion, and egress of Apicomplexa parasites (Chakraborty et al., 2017; Dubois and Soldati-Favre, 2019). In B. bigemina kinetes, CDPK4 and CDPK were upregulated (Table 7, Table 8). In B. bovis kinetes, the expression of CDPK4 (BBOV_IV003210, 76-fold) and CDPK (BBOV_II007640, 3-fold) were also upregulated (Ueti et al., 2021). Protein phosphatases are characterized based on target amino acid: serine/threonine, tyrosine, or dual specificity (acting on the three residues) (Yang and Arrizabalaga, 2017). In B. bigemina, the targets of all three PP identified (PP, PP2A and PP2C) were serine/threonine (Table 8). In addition to cell motility, PP2A is also involved in other biological processes such as proliferation, apoptotic cell death, and cell cycle control (Lillo et al., 2014). In B. bovis kinetes, PP2C (BBOV_I000710, 7.7-fold) was upregulated (Ueti et al., 2021). In B. bigemina kinetes, a single PPKL serine/threonine phosphatase was found (Table 8), corroborating with Yang and Arrizabalaga (2017) who found that four apicomplexan parasites (Cryptosporidium parvum, B. bovis, P. falciparum, and T. gondii) have only one PPKL.
Perforin-like proteins
Perforins form pores in the PM of the invertebrate or mammalian host cells facilitating the parasite’s egress process or assisting in traversing epithelial barriers during tissue passage (Sassmannshausen et al., 2020). In a recent study, 38 members of the perforin-like protein (PLP) gene family were identified in B. bovis, B. bigemina, B. ovata, B. divergens, B. microti and B. canis (Paoletta et al., 2021). However, unlike the data reported by Paoletta et al. (2021), during the B. bovis genome re-annotation process, it was established that gene BBOV_III000410 was split in two (BBOV_III000410 and BBOV_III000412), totaling seven PLPs identified in B. bovis (Ueti et al., 2021). Also, by bioinformatics tools, 60% of the Babesia PLP gene family have a signal peptide, suggesting that these proteins are secreted or reside in the lumen of the organelles (Paoletta et al., 2021). In B. bigemina RNA-seq, all PLPs2-8 genes were differentially upregulated in kinetes, with a 42- to 258-fold increase, and only PLP1 in B. bigemina blood-stages with a 1.65-fold increase (Table 8). As previously described, PLPs were upregulated in B. bovis kinetes with 10 to 870-fold increase, and only one PLP in blood-stages with 62-fold increase (Ueti et al., 2021). In addition, four more genes containing membrane attack complex/perforin (MACPF) domains (BBOV_II006750, BBOV_III000412, BBOV_III000414 with 17 to 654-fold increase) were identified in B. bovis kinetes (Ueti et al., 2021).
Bridging proteins
For binding of the actin-myosin motor to occur, bridging proteins are required. Two bridging proteins have been identified in Apicomplexa (Boucher and Bosch, 2015). Both bridging proteins were found in B. bigemina fructose-1,6-bisphosphate aldolase, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (BBBOND_0403010 and BBBOND_0202520) (Table 8). However, the change of GAPDH expression was not significant between blood-stages and kinetes. In B. bovis, the expression of orthologous aldolase BBOV_IV000790 was not significant in kinetes or blood-stages, and GAPDH BBOV_II002540 had a 3.7-fold increase in blood-stages (Ueti et al., 2021).
Thrombospondin-related anonymous proteins
A previously study demonstrated that TRAP proteins were associated with parasite invasion (Gaffar et al., 2004). In B. bigemina, we identified three TRAP genes. TRAP1 was overexpressed in kinetes whereas two TRAPs were upregulated in B. bigemina blood-stages (Table 8). In B. bovis, a TRAP-family protein was found on the apical side of merozoites (blood-stage) and is involved in host cell invasion (Gaffar et al., 2004). In a previous B. bovis RNA-seq, TRAP1 (BBOV_II002650) was also found to be upregulated in kinetes with 1,750-fold increase, while in blood-stages, TRAP2 (BBOV_II002890), TRAP3 (BBOV_II002630), and TRAP4 (BBOV_II002870) by 11-, 4- and 1.5-fold change, respectively (Ueti et al., 2021). A previous study demonstrated that TRAP1 protein was expressed only by B. bovis kinetes. In contrast, TRAP2, 3, and 4 proteins were expressed on the surface of B. bovis blood and sexual stages (Masterson et al., 2022).
Aspartyl protease 3
The secretory protein aspartyl protease 3 (ASP3) is associated with the rhoptry discharge during invasion of T. gondii and with the lysis of the host cell membrane during egress (Dogga et al., 2017). An anti-malarial compound (49c) based on a hydroxyethylamine scaffold interrupts the lytic cycle of T. gondii tachyzoites by targeting ASP3, and blocks invasion, egress and rhoptry discharge (Dogga et al., 2017). In B. bigemina, two ASPs were differentially upregulated in kinetes and blood-stages (Table 8). In B. bovis RNA-seq, ASPs (BBOV_IV007890, 35-fold and BBOV_IV009660, 9.8-fold) were found in kinetes, and in blood-stages (BBOV_IV010360, 21.8-fold and BBOV_III003510, 2.6-fold).
ROM proteases
ROM proteases contribute to processing of adhesins involved in attachment, invasion, intracellular replication, phagocytosis, and immune evasion, placing them at the vertex of host–parasite interactions (Sibley, 2013). ROM may be involved in releasing the parasite into the parasitophorous vacuole and RBC invasion by Babesia (Li et al., 2012; Suarez et al., 2019). In B. bovis, the parasitophorous membrane dissolves in a few minutes after merozoite invasion into the RBC (Asada et al., 2012). In P. falciparum, ROM4 function is related to cleavage of the adhesins of the cell surface during the invasion of RBC (O’Donnell et al., 2006). Babesia bigemina and B. bovis have three to five ROM4, and one ROM6, ROM7 and ROM8 (Gallenti et al., 2021; Gallenti et al., 2022). In B. bigemina RNA-seq, kinetes and blood-stages expressed genes encoding ROM proteases (Table 8). In addition, ROM6 (BBBOND_0208010) was also found in the RNA-seq of B. bigemina, however, it was not significant in the groups analyzed. In B. bovis RNA-seq, all ROM4 proteins were found in blood-stages (BBOV_II005940 [ROM4.4], BBOV_II005930 [ROM4.3], and BBOV_II005950 [ROM4.5], 150-, 14- and 6-fold change) and in kinetes (BBOV_II006100 [ROM4.1] and BBOV_II006070 [ROM4.2], 13,286.23 and 3,000-fold). In addition, ROM6 (BBOV_I003700, 1.54-fold in kinetes), ROM7 (BBOV_III000530, 2.67-fold in blood-stages) and ROM8 (BBOV_IV005790, 1.5-fold in blood-stages) were also upregulated (Ueti et al., 2021).
Rhoptry-associated proteins
Rhoptry-associated proteins are conserved in several species of Babesia, play an important functional role in parasite invasion and have been studied as a component in a vaccine against bovine babesiosis (Brown and Palmer, 1999). Monoclonal antibodies against B. bigemina and B. bovis RAP-1 inhibit parasite invasion (Figueroa and Buening, 1991; McElwain et al., 1991; Mosqueda et al., 2002). Like B. bovis (BBOV_IV009870 and BBOV_IV009860, with 122- and 35-fold increase, respectively), B. bigemina RAP-1 has been observed in blood-stages parasites (Table 8). In fact, there are important differences in the structure and organization of RAP-1 genes among these two parasites. While B. bovis only contains two identical RAP-1 genes, B. bigemina contains genes encoding for four distinct RAP-1a genes, five identical RAP-1b genes and a single copy RAP-1c gene (Suarez et al., 2003). Previous expression analysis performed on cultured blood-stages of the parasites revealed that only RAP-1a genes express RAP1a proteins, but the RAP-1b and RAP-1c genes generate low levels of transcripts, with no significant amounts of detectable proteins (Suarez et al., 2003). A RAP-1 related antigen (RRA) gene contributes to erythrocyte invasion and/or egression by B. bovis, and overall, to the survival of the parasite during infection (Suarez et al., 2011) and was found in B. bigemina (Table 8) and in B. bovis blood-stages (BBOV_IV010280, 18-fold increase) (Ueti et al., 2021).
Glycoprotein 45
The 45 kilodaltons glycoprotein (GP-45) is exposed on the surface of the B. bigemina merozoite, is highly immunogenic and it is believed to play a role in the invasion of erythrocytes (Mercado-Uriostegui et al., 2022). Nucleotide and amino acids sequences of GP-45 of 14 field isolates and strains revealed high percentage of similarity. In addition, sera from cattle naturally infected with B. bigemina contain antibodies that bind to specific peptides of GP-45 (Mercado-Uriostegui et al., 2022). In blood-stages of B. bigemina, the gene corresponding to GP-45 was upregulated (Table 8). Babesia bigemina GP-45 appears to be present as a single-copy gene (Fisher et al., 2001). In addition, GP-45 lacks introns and, unlike the tandem arrangement of B. bovis MSA-2 genes, GP45 is flanked by unrelated ORFs, similar to B. bovis MSA-1 (Suarez et al., 2000; Fisher et al., 2001). In B. bovis, all variable MSA (MSA1, MSA-2a1, MSA-2a2, MSA-2b and MSA-2c, with 86- to 170-fold) were found in blood-stages (Ueti et al., 2021).
Kinete-specific protein
The KSP gene that has been implicated to be important in transovarial transmission (Johnson et al., 2017; Bohaliga et al., 2019) was highly transcribed and specifically expressed in B. bigemina kinetes (Table 2, Table 8). This gene is homologous to the B. bovis KSP protein, sharing 25% amino acid identity. The fold change of both B. bigemina and B. bovis KSPs was similar (Table 2). In the B. bovis genome, a GPI anchorage site was identified in KSP, suggesting that the protein may be anchored on the parasite’s surface, and may play a role in invading tick ovary epithelial cells (Rodriguez et al., 2014; Johnson et al., 2017). So far, it is not known if other protozoan parasites share this protein (Johnson et al., 2017; Bohaliga et al., 2019). In fact, this gene does not appear in Theileria spp. or B. microti (Supplementary Table 3), perhaps because these parasites are not transovarially transmitted to the tick’s offspring (Jalovecka et al., 2018).
Genes upregulated by members of Babesia sensu stricto not found in Theileria spp. and Babesia microti
Babesia sensu stricto, such as B. bigemina and B. bovis, refers to protozoan parasites that are transovarially transmitted by the tick vectors. In contrast, Babesia sensu lato, such as B. microti, or Theileria are not transovarially transmitted by the vector (Schreeg et al., 2016; Jalovecka et al., 2018; Jalovecka et al., 2019). Discovering key molecules involved in Babesia sensu stricto transmission may lead to the development of interfering strategies to block the biological transmission of these parasites (Suarez et al., 2019). To identify genes unique to Babesia sensu stricto that are potentially involved in vertical transmission, all 906 genes upregulated in kinetes of B. bigemina were compared by BLASTp with B. bovis, Theileria spp. and B. microti genomes available in NCBI dataset. Of the genes upregulated in B. bigemina and B. bovis kinetes, 44 were not found in Theileria spp. and B. microti genomes (Supplementary Table 3). The genes listed in Supplementary Table 3 include KSP, membrane proteins, and proteins of unknown function. Some of these genes may encode proteins that have a function in kinete invasion into ovaries and transovarial transmission.
Validation of transcription levels by qPCR
To validate the RNA seq, four genes from kinete (TRAP1, ROM4, Der-1, and KSP) and four blood-stage genes (SBP4, CCp2, PFruc, and Ribok) were selected based on their magnitude of transcription. The similar profile of two reference genes (MAPK and ProtA) demonstrated a consistent pattern of the differential gene expression between kinete and blood-stage parasites (Supplementary Figure 2).
Conclusion
Omic studies that include cattle Babesia focus on analyzing genes of Babesia strains with distinct virulence, or between free merozoites versus intraerythrocytic stages (Pedroni et al., 2013; González et al., 2019). In contrast, our study compared genes differentially expressed by two different stages of the developmental cycle of B. bigemina: kinetes in the tick vector and blood-stages in bovine RBC. This approach revealed potential essential genes involved in parasite development within the tick and mammalian hosts.
Knowledge and understanding of the changes in gene expression patterns that occur as a result of mammalian-tick transitions is crucial for identifying candidates for development of efficient control strategies for bovine babesiosis. The results herein revealed important genetic markers to develop strategies to control the spread of bovine babesiosis caused by B. bigemina and B. bovis. Specifically, genes identified herein that are upregulated in both B. bigemina and B. bovis kinete could be used to determine critical genes or proteins to prevent tick infection and, consequently disrupt the transmission of the pathogen via tick vectors.
In this study, we provided a robust panel of expressed genes for B. bigemina by two distinct developmental stages and revealed potential markers of transovarial transmission. However, this study is based on differential expression gene analysis, and proteomic and functional studies will also be needed to strengthen our understanding of the biology of these parasites. We propose that the results generated in this study can serve as a basis for other research associated with the improvement of diagnostics, drugs, and effective vaccine development against bovine babesiosis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of the University of Idaho, Moscow, ID, USA.
Author contributions
MU and WJ conceived and supervised the project. JC-P drafted manuscript. JC-P, WJ, LK, and NT prepared figures and tables. KR, WJ and MU performed the experiments. JC-P, PS, WJ, LK, and MU analyzed the data. WJ, LK, CS, PS, HM, NT, KB and MU wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the United States Department of Agriculture - Agricultural Research Service (Project #2090-32000-040-000-D and #3094-22320-001-000-D) and National Institute of Food and Agriculture-Agriculture and Food Research Initiative Competitive Grant (Project #2016-67015-24968).
Acknowledgments
We would like to acknowledge David Herndon for handling and shipping samples, receiving, and storing data files, and Sara Davis, Nicholas Durfee, Paul Lacy, Jacob Laughery, Ralph Horn, Kathy Mason, James Allison, and Megan Jacks for excellent technical and animal care support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1093338/full#supplementary-material
References
Allocco, J. J., Donald, R., Zhong, T., Lee, A., Tang, Y. S., Hendrickson, R. C., et al. (2006). Inhibitors of casein kinase 1 block the growth of leishmania major promastigotes In vitro. Int. J. Parasitol. 36, 1249–1259. doi: 10.1016/j.ijpara.2006.06.013
Allred, D. R., Carlton, J. M., Satcher, R. L., Long, J. A., Brown, W. C., Patterson, P. E., et al. (2000). The ves multigene family of B. Bovis encodes components of rapid antigenic variation at the infected erythrocyte surface. Mol. Cell 5, 153–162. doi: 10.1016/S1097-2765(00)80411-6
Alzan, H. F., Lau, A. O., Knowles, D. P., Herndon, D. R., Ueti, M. W., Scoles, G. A., et al. (2016). Expression of 6-cys gene superfamily defines babesia bovis sexual stage development within rhipicephalus microplus. PloS One 11, e0163791. doi: 10.1371/journal.pone.0163791
Alzan, H. F., Silva, M. G., Davis, W. C., Herndon, D. R., Schneider, D. A., Suarez, C. E. (2017). Geno- and phenotypic characteristics of a transfected babesia bovis 6-Cys-E knockout clonal line. Parasit. Vectors 10, 214. doi: 10.1186/s13071-017-2143-3
Asada, M., Goto, Y., Yahata, K., Yokoyama, N., Kawai, S., Inoue, N., et al. (2012). Gliding motility of babesia bovis merozoites visualized by time-lapse video microscopy. PloS One 7, e35227. doi: 10.1371/journal.pone.0035227
Balaji, S., Babu, M. M., Lyer, L. M., Aravind, L. (2005). Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33, 3994–4006. doi: 10.1093/nar/gki709
Bastos, R. G., Suarez, C. E., Laughery, J. M., Johnson, W. C., Ueti, M. W., Knowles, D. P. (2013). Differential expression of three members of the multidomain adhesion ccp family in babesia bigemina, babesia bovis and theileria equi. PloS One 8, e67765. doi: 10.1371/journal.pone.0067765
Becker, C. A., Malandrin, L., Depoix, D., Larcher, T., David, P. H., Chauvin, A., et al. (2010). Identification of three ccp genes in babesia divergens: Novel markers for sexual stages parasites. Mol. Biochem. Parasitol. 174, 36–43. doi: 10.1016/j.molbiopara.2010.06.011
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc.: Ser. B. (Methodol.) 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bock, R., Jackson, L., De Vos, A., Jorgensen, W. (2004). Babesiosis of cattle. Parasitology 129 Suppl, S247–S269. doi: 10.1017/S0031182004005190
Bohaliga, G. A. R., Johnson, W. C., Taus, N. S., Hussein, H. E., Bastos, R. G., Suarez, C. E., et al. (2019). Identification of proteins expressed by babesia bigemina kinetes. Parasit. Vectors 12, 271. doi: 10.1186/s13071-019-3531-7
Bork, S., Das, S., Okubo, K., Yokoyama, N., Igarashi, I. (2006). Effects of protein kinase inhibitors on the In vitro growth of babesia bovis. Parasitology 132, 775–779. doi: 10.1017/S0031182006009917
Boucher, L. E., Bosch, J. (2015). The apicomplexan glideosome and adhesins - structures and function. J. Struct. Biol. 190, 93–114. doi: 10.1016/j.jsb.2015.02.008
Brown, W. C., Palmer, G. H. (1999). Designing blood-stage vaccines against babesia bovis and b. Bigemina. Parasitol. Today 15, 275–281. doi: 10.1016/S0169-4758(99)01471-4
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time pcr experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Camacho-Nuez, M., Hernández-Silva, D. J., Castañeda-Ortiz, E. J., Paredes-Martínez, M. E., Rocha-Martínez, M. K., Alvarez-Sánchez, M. E., et al. (2017). Hap2, a novel gene in babesia bigemina is expressed in tick stages, and specific antibodies block zygote formation. Parasit. Vectors 10, 568. doi: 10.1186/s13071-017-2510-0
Chakraborty, S., Roy, S., Mistry, H. U., Murthy, S., George, N., Bhandari, V., et al. (2017). Potential sabotage of host cell physiology by apicomplexan parasites for their survival benefits. Front. Immunol. 8, 1261. doi: 10.3389/fimmu.2017.01261
Chaudhry, Z. I., Suleman, M., Younus, M., Aslim, A. S. (2010). Molecular detection of babesia bigemina and babesia bovis in crossbred carrier cattle through pcr. Pakistan J. of Zool. 42, 201–204.
Chen, Y., Lun, A. T., Smyth, G. K. (2016). From reads to genes to pathways: Differential expression analysis of RNA-seq experiments using rsubread and the edger quasi-likelihood pipeline. F1000res 5, 1438. doi: 10.12688/f1000research.8987.2
Chung, C. J., Suarez, C. E., Bandaranayaka-Mudiyanselage, C. L., Bandaranayaka-Mudiyanselage, C. B., Rzepka, J., Heiniger, T. J., et al. (2017). A novel modified-indirect ELISA based on spherical body protein 4 for detecting antibody during acute and long-term infections with diverse babesia bovis strains. Parasit. Vectors 10, 77. doi: 10.1186/s13071-017-2016-9
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Dogga, S. K., Mukherjee, B., Jacot, D., Kockmann, T., Molino, L., Hammoudi, P. M., et al. (2017). A druggable secretory protein maturase of toxoplasma essential for invasion and egress. Elife 6:e27480. doi: 10.7554/eLife.27480.049
Douradinha, B., Augustijn, K. D., Moore, S. G., Ramesar, J., Mota, M. M., Waters, A. P., et al. (2011). Plasmodium cysteine repeat modular proteins 3 and 4 are essential for malaria parasite transmission from the mosquito to the host. Malar. J. 10, 71. doi: 10.1186/1475-2875-10-71
Dubois, D. J., Soldati-Favre, D. (2019). Biogenesis and secretion of micronemes in toxoplasma gondii. Cell Microbiol. 21, e13018. doi: 10.1111/cmi.13018
Eichenberger, R. M., Ramakrishnan, C., Russo, G., Deplazes, P., Hehl, A. B. (2017). Genome-wide analysis of gene expression and protein secretion of babesia canis during virulent infection identifies potential pathogenicity factors. Sci. Rep. 7, 3357. doi: 10.1038/s41598-017-03445-x
El-Awady, R., Saleh, E., Hashim, A., Soliman, N., Dallah, A., Elrasheed, A., et al. (2016). The role of eukaryotic and prokaryotic ABC transporter family in failure of chemotherapy. Front. Pharmacol. 7, 535. doi: 10.3389/fphar.2016.00535
Figueroa, J. V., Buening, G. M. (1991). In vitro inhibition of multiplication of babesia bigemina by using monoclonal antibodies. J. Clin. Microbiol. 29, 997–1003. doi: 10.1128/jcm.29.5.997-1003.1991
Fisher, T. G., Mcelwain, T. F., Palmer, G. H. (2001). Molecular basis for variable expression of merozoite surface antigen Gp45 among American isolates of babesia bigemina. Infect. Immun. 69, 3782–3790. doi: 10.1128/IAI.69.6.3782-3790.2001
Frénal, K., Soldati-Favre, D. (2013). The glideosome, a unique machinery that assists the apicomplexa in gliding into host cells. Med. Sci. (Paris) 29, 515–522. doi: 10.1051/medsci/2013295015
Gaffar, F. R., Yatsuda, A. P., Franssen, F. F., De Vries, E. (2004). A babesia bovis merozoite protein with a domain architecture highly similar to the thrombospondin-related anonymous protein (TRAP) present in plasmodium sporozoites. Mol. Biochem. Parasitol. 136, 25–34. doi: 10.1016/j.molbiopara.2004.02.006
Gallego-Lopez, G. M., Cooke, B. M., Suarez, C. E. (2019). Interplay between attenuation- and virulence-factors of babesia bovis and their contribution to the establishment of persistent infections in cattle. Pathogens 8, 97. doi: 10.3390/pathogens8030097
Gallenti, R., Hussein, H. E., Alzan, H. F., Suarez, C. E., Ueti, M., Asurmendi, S., et al. (2022). Unraveling the complexity of the rhomboid serine protease 4 family of babesia bovis using bioinformatics and experimental studies. Pathogens 11, 344. doi: 10.3390/pathogens11030344
Gallenti, R., Poklepovich, T., Florin-Christensen, M., Schnittger, L. (2021). The repertoire of serine rhomboid proteases of piroplasmids of importance to animal and human health. Int. J. Parasitol. 51, 455–462. doi: 10.1016/j.ijpara.2020.10.010
Glanfield, A., Mcmanus, D. P., Anderson, G. J., Jones, M. K. (2007). Pumping iron: A potential target for novel therapeutics against schistosomes. Trends Parasitol. 23, 583–588. doi: 10.1016/j.pt.2007.08.018
Goff, W. L., Molloy, J. B., Johnson, W. C., Suarez, C. E., Pino, I., Rhalem, A., et al. (2006). Validation of a competitive enzyme-linked immunosorbent assay for detection of antibodies against babesia bovis. Clin. Vaccine Immunol. 13, 1212–1216. doi: 10.1128/CVI.00196-06
González, L. M., Estrada, K., Grande, R., Jiménez-Jacinto, V., Vega-Alvarado, L., Sevilla, E., et al. (2019). Comparative and functional genomics of the protozoan parasite babesia divergens highlighting the invasion and egress processes. PloS Negl. Trop. Dis. 13, e0007680. doi: 10.1371/journal.pntd.0007680
Goo, Y. K., Ueno, A., Terkawi, M. A., Aboge, G. O., Junya, Y., Igarashi, M., et al. (2013). Actin polymerization mediated by babesia gibsoni aldolase is required for parasite invasion. Exp. Parasitol. 135, 42–49. doi: 10.1016/j.exppara.2013.06.002
Hernández, P. C., Morales, L., Castellanos, I. C., Wasserman, M., Chaparro-Olaya, J. (2017). Myosin b of plasmodium falciparum (Pfmyob): In silico prediction of its three-dimensional structure and its possible interaction with MTIP. Parasitol. Res. 116, 1373–1382. doi: 10.1007/s00436-017-5417-y
Hirai, M., Arai, M., Mori, T., Miyagishima, S. Y., Kawai, S., Kita, K., et al. (2008). Male Fertility of malaria parasites is determined by Gcs1, a plant-type reproduction factor. Curr. Biol. 18, 607–613. doi: 10.1016/j.cub.2008.03.045
Howell, J. M., Ueti, M. W., Palmer, G. H., Scoles, G. A., Knowles, D. P. (2007). Transovarial transmission efficiency of babesia bovis tick stages acquired by rhipicephalus (Boophilus) microplus during acute infection. J. Clin. Microbiol. 45, 426–431. doi: 10.1128/JCM.01757-06
Hussein, H. E., Bastos, R. G., Schneider, D. A., Johnson, W. C., Adham, F. K., Davis, W. C., et al. (2017). The babesia bovis Hap2 gene is not required for blood stage replication, but expressed upon In vitro sexual stage induction. PloS Negl. Trop. Dis. 11, e0005965. doi: 10.1371/journal.pntd.0005965
Jackson, A. P., Otto, T. D., Darby, A., Ramaprasad, A., Xia, D., Echaide, I. E., et al. (2014). The evolutionary dynamics of variant antigen genes in babesia reveal a history of genomic innovation underlying host-parasite interaction. Nucleic Acids Res. 42, 7113–7131. doi: 10.1093/nar/gku322
Jacob, S. S., Sengupta, P. P., Paramanandham, K., Suresh, K. P., Chamuah, J. K., Rudramurthy, G. R., et al. (2020). Bovine babesiosis: An insight into the global perspective on the disease distribution by systematic review and meta-analysis. Vet. Parasitol. 283, 109136. doi: 10.1016/j.vetpar.2020.109136
Jalovecka, M., Hajdusek, O., Sojka, D., Kopacek, P., Malandrin, L. (2018). The complexity of piroplasms life cycles. Front. Cell Infect. Microbiol. 8, 248. doi: 10.3389/fcimb.2018.00248
Jalovecka, M., Sojka, D., Ascencio, M., Schnittger, L. (2019). Babesia life cycle - when phylogeny meets biology. Trends Parasitol. 35, 356–368. doi: 10.1016/j.pt.2019.01.007
Johnson, W. C., Taus, N. S., Reif, K. E., Bohaliga, G. A., Kappmeyer, L. S., Ueti, M. W. (2017). Analysis of stage-specific protein expression during babesia bovis development within female rhipicephalus microplus. J. Proteome Res. 16, 1327–1338. doi: 10.1021/acs.jproteome.6b00947
Johnson, M. A., Von Besser, K., Zhou, Q., Smith, E., Aux, G., Patton, D., et al. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168, 971–982. doi: 10.1534/genetics.104.029447
Johnston, L. A., Leatch, G., Jones, P. N. (1978). The duration of latent infection and functional immunity in droughtmaster and Hereford cattle following natural infection with babesia Argentina and babesia bigemina. Aust. Vet. J. 54, 14–18. doi: 10.1111/j.1751-0813.1978.tb00262.x
Kato, K., Sugi, T., Iwanaga, T. (2012). Roles of apicomplexan protein kinases At each life cycle stage. Parasitol. Int. 61, 224–234. doi: 10.1016/j.parint.2011.12.002
Klokouzas, A., Shahi, S., Hladky, S. B., Barrand, M. A., Van Veen, H. W. (2003). ABC Transporters and drug resistance in parasitic Protozoa. Int. J. Antimicrob. Agents 22, 301–317. doi: 10.1016/S0924-8579(03)00210-3
Lamotte, S., Späth, G. F., Rachidi, N., Prina, E. (2017). The enemy within: Targeting host-parasite interaction for antileishmanial drug discovery. PloS Negl. Trop. Dis. 11, E0005480. doi: 10.1371/journal.pntd.0005480
Levy, M. G., Ristic, M. (1980). Babesia bovis: Continuous cultivation in a microaerophilous stationary phase culture. Science 207, 1218–1220. doi: 10.1126/science.7355284
Li, H., Child, M. A., Bogyo, M. (2012). Proteases as regulators of pathogenesis: Examples from the apicomplexa. Biochim. Biophys. Acta 1824, 177–185. doi: 10.1016/j.bbapap.2011.06.002
Lillo, C., Kataya, A. R., Heidari, B., Creighton, M. T., Nemie-Feyissa, D., Ginbot, Z., et al. (2014). Protein phosphatases PP2A, PP4 and PP6: Mediators and regulators in development and responses to environmental cues. Plant Cell Environ. 37, 2631–2648. doi: 10.1111/pce.12364
Lindner, S. E., Swearingen, K. E., Shears, M. J., Walker, M. P., Vrana, E. N., Hart, K. J., et al. (2019). Transcriptomics and proteomics reveal two waves of translational repression during the maturation of malaria parasite sporozoites. Nat. Commun. 10, 4964. doi: 10.1038/s41467-019-12936-6
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Masterson, H. E., Taus, N. S., Johnson, W. C., Kappmeyer, L., Capelli-Peixoto, J., Hussein, H. E., et al. (2022). Thrombospondin-related anonymous protein (TRAP) family expression by babesia bovis life stages within the mammalian host and tick vector. Microorganisms 10, 2173. doi: 10.3390/microorganisms10112173
McCarthy, D. J., Chen, Y., Smyth, G. K. (2012). Differential expression analysis of multifactor rna-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
McElwain, T. F., Perryman, L. E., Musoke, A. J., Mcguire, T. C. (1991). Molecular characterization and immunogenicity of neutralization-sensitive babesia bigemina merozoite surface proteins. Mol. Biochem. Parasitol. 47, 213–222. doi: 10.1016/0166-6851(91)90181-5
Mercado-Uriostegui, M. A., Castro-Sánchez, L. A., Batiha, G. E., Valdez-Espinoza, U. M., Falcón-Neri, A., Ramos-Aragon, J. A., et al. (2022). The gp-45 protein, a highly variable antigen from babesia bigemina, contains conserved b-cell epitopes in geographically distant isolates. Pathogens 11, 591. doi: 10.3390/pathogens11050591
Mori, T., Kuroiwa, H., Higashiyama, T., Kuroiwa, T. (2006). Generative cell specific 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8, 64–71. doi: 10.1038/ncb1345
Mosqueda, J., Mcelwain, T. F., Stiller, D., Palmer, G. H. (2002). Babesia bovis merozoite surface antigen 1 and rhoptry-associated protein 1 are expressed in sporozoites, and specific antibodies inhibit sporozoite attachment to erythrocytes. Infect. Immun. 70, 1599–1603. doi: 10.1128/IAI.70.3.1599-1603.2002
Murphy, T. F., Brauer, A. L., Johnson, A., Kirkham, C. (2016). ATP-binding cassette (ABC) transporters of the human respiratory tract pathogen, moraxella catarrhalis: Role in virulence. PloS One 11, e0158689. doi: 10.1371/journal.pone.0158689
O’Donnell, R. A., Hackett, F., Howell, S. A., Treeck, M., Struck, N., Krnajski, Z., et al. (2006). Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J. Cell Biol. 174, 1023–1033. doi: 10.1083/jcb.200604136
Olliaro, P. L., Taylor, W. R. (2004). Developing artemisinin based drug combinations for the treatment of drug resistant falciparum malaria: A review. J. Postgrad. Med. 50, 40–44.
Ozubek, S., Alzan, H. F., Bastos, R. G., Laughery, J. M., Suarez, C. E. (2022). Identification of CCP5 and FNPA as novel non-canonical members of the CCp protein family in babesia bovis. Front. Vet. Sci. 9, 833183. doi: 10.3389/fvets.2022.833183
Paoletta, M. S., Laughery, J. M., Arias, L. S. L., Ortiz, J. M. J., Montenegro, V. N., Petrigh, R., et al. (2021). The key to egress? babesia bovis perforin-like protein 1 (Plp1) with hemolytic capacity is required for blood stage replication and is involved in the exit of the parasite from the host cell. Int. J. Parasitol. 51, 643–658. doi: 10.1016/j.ijpara.2020.12.010
Pedroni, M. J., Sondgeroth, K. S., Gallego-Lopez, G. M., Echaide, I., Lau, A. O. (2013). Comparative transcriptome analysis of geographically distinct virulent and attenuated babesia bovis strains reveals similar gene expression changes through attenuation. BMC Genomics 14, 763. doi: 10.1186/1471-2164-14-763
Pradel, G., Hayton, K., Aravind, L., Iyer, L. M., Abrahamsen, M. S., Bonawitz, A., et al. (2004). A multidomain adhesion protein family expressed in plasmodium falciparum is essential for transmission to the mosquito. J. Exp. Med. 199, 1533–1544. doi: 10.1084/jem.20031274
Rachidi, N., Knippschild, U., Späth, G. F. (2021). Dangerous duplicity: The dual functions of casein kinase 1 in parasite biology and host subversion. Front. Cell Infect. Microbiol. 11, 655700. doi: 10.3389/fcimb.2021.655700
Riquelme-Neira, R., Retamal-Díaz, A., Acuña, F., Riquelme, P., Rivera, A., Sáez, D., et al. (2013). Protective effect of a dna vaccine containing an open reading frame with homology to an abc-type transporter present in the genomic island 3 of brucella abortus in Balb/C mice. Vaccine 31, 3663–3667. doi: 10.1016/j.vaccine.2013.06.013
Robinson, M. D., Mccarthy, D. J., Smyth, G. K. (2010). Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rodriguez, A. E., Florin-Christensen, M., Flores, D. A., Echaide, I., Suarez, C. E., Schnittger, L. (2014). The glycosylphosphatidylinositol-anchored protein repertoire of babesia bovis and its significance for erythrocyte invasion. Ticks Tick Borne Dis. 5, 343–348. doi: 10.1016/j.ttbdis.2013.12.011
Rozen, S., Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. doi: 10.1385/1-59259-192-2:365
Sachman-Ruiz, B., Lozano, L., Lira, J. J., Martínez, G., Rojas, C., Álvarez, J. A., et al. (2021). A comparative genomic study of attenuated and virulent strains of babesia bigemina. Pathogens 10, 318. doi: 10.3390/pathogens10030318
Sassmannshausen, J., Pradel, G., Bennink, S. (2020). Perforin-like proteins of apicomplexan parasites. Front. Cell Infect. Microbiol. 10, 578883. doi: 10.3389/fcimb.2020.578883
Schnick, C., Polley, S. D., Fivelman, Q. L., Ranford-Cartwright, L. C., Wilkinson, S. R., Brannigan, J. A., et al. (2009). Structure and non-essential function of glycerol kinase in plasmodium falciparum blood stages. Mol. Microbiol. 71, 533–545. doi: 10.1111/j.1365-2958.2008.06544.x
Schnittger, L., Rodriguez, A. E., Florin-Christensen, M., Morrison, D. A. (2012). Babesia: A world emerging. Infect. Genet. Evol. 12, 1788–1809. doi: 10.1016/j.meegid.2012.07.004
Schreeg, M. E., Marr, H. S., Tarigo, J. L., Cohn, L. A., Bird, D. M., Scholl, E. H., et al. (2016). Mitochondrial genome sequences and structures aid in the resolution of piroplasmida phylogeny. PloS One 11, e0165702. doi: 10.1371/journal.pone.0165702
Sibley, L. D. (2013). The roles of intramembrane proteases in protozoan parasites. Biochim. Biophys. Acta 1828, 2908–2915. doi: 10.1016/j.bbamem.2013.04.017
Sibmooh, N., Udomsangpetch, R., Kujoa, A., Chantharaksri, U., Mankhetkorn, S. (2001). Redox reaction of artemisinin with ferrous and ferric ions in aqueous buffer. Chem. Pharm. Bull. (Tokyo) 49, 1541–1546. doi: 10.1248/cpb.49.1541
Suarez, C. E., Alzan, H. F., Silva, M. G., Rathinasamy, V., Poole, W. A., Cooke, B. M. (2019). Unravelling the cellular and molecular pathogenesis of bovine babesiosis: Is the sky the limit? Int. J. Parasitol. 49, 183–197. doi: 10.1016/j.ijpara.2018.11.002
Suarez, C. E., Florin-Christensen, M., Hines, S. A., Palmer, G. H., Brown, W. C., Mcelwain, T. F. (2000). Characterization of allelic variation in the babesia bovis merozoite surface antigen 1 (MSA-1) locus and identification of a cross-reactive inhibition-sensitive msa-1 epitope. Infect. Immun. 68, 6865–6870. doi: 10.1128/IAI.68.12.6865-6870.2000
Suarez, C. E., Laughery, J. M., Bastos, R. G., Johnson, W. C., Norimine, J., Asenzo, G., et al. (2011). A novel neutralization sensitive and subdominant RAP-1-Related antigen (RRA) is expressed by babesia bovis merozoites. Parasitology 138, 809–818. doi: 10.1017/S0031182011000321
Suarez, C. E., Laughery, J. M., Schneider, D. A., Sondgeroth, K. S., Mcelwain, T. F. (2012). Acute and persistent infection by a transfected Mo7 strain of babesia bovis. Mol. Biochem. Parasitol. 185, 52–57. doi: 10.1016/j.molbiopara.2012.05.003
Suarez, C. E., Noh, S. (2011). Emerging perspectives in the research of bovine babesiosis and anaplasmosis. Vet. Parasitol. 180, 109–125. doi: 10.1016/j.vetpar.2011.05.032
Suarez, C. E., Palmer, G. H., Florin-Christensen, M., Hines, S. A., Hötzel, I., Mcelwain, T. F. (2003). Organization, transcription, and expression of rhoptry associated protein genes in the babesia bigemina rap-1 locus. Mol. Biochem. Parasitol. 127, 101–112. doi: 10.1016/S0166-6851(02)00311-0
Ueti, M. W., Johnson, W. C., Kappmeyer, L. S., Herndon, D. R., Mousel, M. R., Reif, K. E., et al. (2020). Transcriptome dataset of babesia bovis life stages within vertebrate and invertebrate hosts. Data Brief 33, 106533. doi: 10.1016/j.dib.2020.106533
Ueti, M. W., Johnson, W. C., Kappmeyer, L. S., Herndon, D. R., Mousel, M. R., Reif, K. E., et al. (2021). Comparative analysis of gene expression between babesia bovis blood stages and kinetes allowed by improved genome annotation. Int. J. Parasitol. 51, 123–136. doi: 10.1016/j.ijpara.2020.08.006
van Dijk, M. R., Van Schaijk, B. C., Khan, S. M., Van Dooren, M. W., Ramesar, J., Kaczanowski, S., et al. (2010). Three members of the 6-cys protein family of plasmodium play a role in gamete fertility. PloS Pathog. 6, e1000853. doi: 10.1371/journal.ppat.1000853
Wachnowsky, C., Fidai, I., Cowan, J. A. (2018). Iron-sulfur cluster biosynthesis and trafficking - impact on human disease conditions. Metallomics 10, 9–29. doi: 10.1039/C7MT00180K
Wang, Y., Fairley, J. A., Roberts, S. G. (2010). Phosphorylation of TFIIB links transcription initiation and termination. Curr. Biol. 20, 548–553. doi: 10.1016/j.cub.2010.01.052
Wang, Y., Ma, A., Chen, S. B., Yang, Y. C., Chen, J. H., Yin, M. B. (2014). Genetic diversity and natural selection of three blood-stage 6-cys proteins in plasmodium vivax populations from the China-Myanmar endemic border. Infect. Genet. Evol. 28, 167–174. doi: 10.1016/j.meegid.2014.09.026
Keywords: Bovine babesiosis, B. bigemina, B. bovis, RNA-seq, blood-stages, kinetes, differential gene expression
Citation: Capelli-Peixoto J, Saelao P, Johnson WC, Kappmeyer L, Reif KE, Masterson HE, Taus NS, Suarez CE, Brayton KA and Ueti MW (2022) Comparison of high throughput RNA sequences between Babesia bigemina and Babesia bovis revealed consistent differential gene expression that is required for the Babesia life cycle in the vertebrate and invertebrate hosts. Front. Cell. Infect. Microbiol. 12:1093338. doi: 10.3389/fcimb.2022.1093338
Received: 08 November 2022; Accepted: 05 December 2022;
Published: 19 December 2022.
Edited by:
Shahid Karim, University of Southern Mississippi, United StatesReviewed by:
Ben J. Mans, Agricultural Research Council, South AfricaMelina Garcia Guizzo, National Institute of Allergy and Infectious Diseases (NIH), United States
Copyright © 2022 Capelli-Peixoto, Saelao, Johnson, Kappmeyer, Reif, Masterson, Taus, Suarez, Brayton and Ueti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Janaina Capelli-Peixoto, ai5jYXBlbGxpcGVpeG90b0B3c3UuZWR1
†Present address: Kathryn E. Reif, Department of Diagnostic Medicine/Pathobiology, College of Veterinary Medicine, Kansas State University, Manhattan, KS, United States
 Janaina Capelli-Peixoto
Janaina Capelli-Peixoto Perot Saelao
Perot Saelao Wendell C. Johnson3
Wendell C. Johnson3 Lowell Kappmeyer
Lowell Kappmeyer Kathryn E. Reif
Kathryn E. Reif Naomi S. Taus
Naomi S. Taus Carlos E. Suarez
Carlos E. Suarez Kelly A. Brayton
Kelly A. Brayton Massaro W. Ueti
Massaro W. Ueti