
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Cell. Infect. Microbiol., 24 November 2022
Sec. Clinical Microbiology
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.1083501
This article is part of the Research TopicDeveloping Therapeutics for Antimicrobial Resistant PathogensView all 12 articles
 Raja Veerapandian1*
Raja Veerapandian1* Parveez Ahamed Abdul Azees2
Parveez Ahamed Abdul Azees2 Thiruselvam Viswanathan3
Thiruselvam Viswanathan3 Bennett Tochukwu Amaechi2*
Bennett Tochukwu Amaechi2* Govindsamy Vediyappan4*
Govindsamy Vediyappan4*Editorial on the Research Topic
Developing therapeutics for antimicrobial resistant pathogens
The World Health Organization (WHO) declared antimicrobial resistance (AMR) as one of the top 10 global public health issues with an alarming note that the world is running out of antibiotics (WHO, 2017). It is estimated that by 2050 the total healthcare cost for AMR may increase up to US$100 trillion and cause up to10 million deaths each year (de Kraker et al., 2016). A recent study provided the most comprehensive analysis of the burden of AMR to date and highlights that its magnitude on human health is potentially much larger than the major human diseases (Murray et al., 2022). AMR is already a multi-dimensional complex challenge, and the influence of COVID-19 pandemic even worsen the situation. Thus, there is need for safe and effective drugs against AMR pathogens. Given the complexity of AMR and its impact on humans, animals, and the environment, we need multiple approaches to overcome these obstacles (Figure 1). As a step towards finding solutions for AMR threat, our Research Topic brings a collection of 11 manuscripts with 8 research and 3 review articles exploring different agents against AMR pathogens.
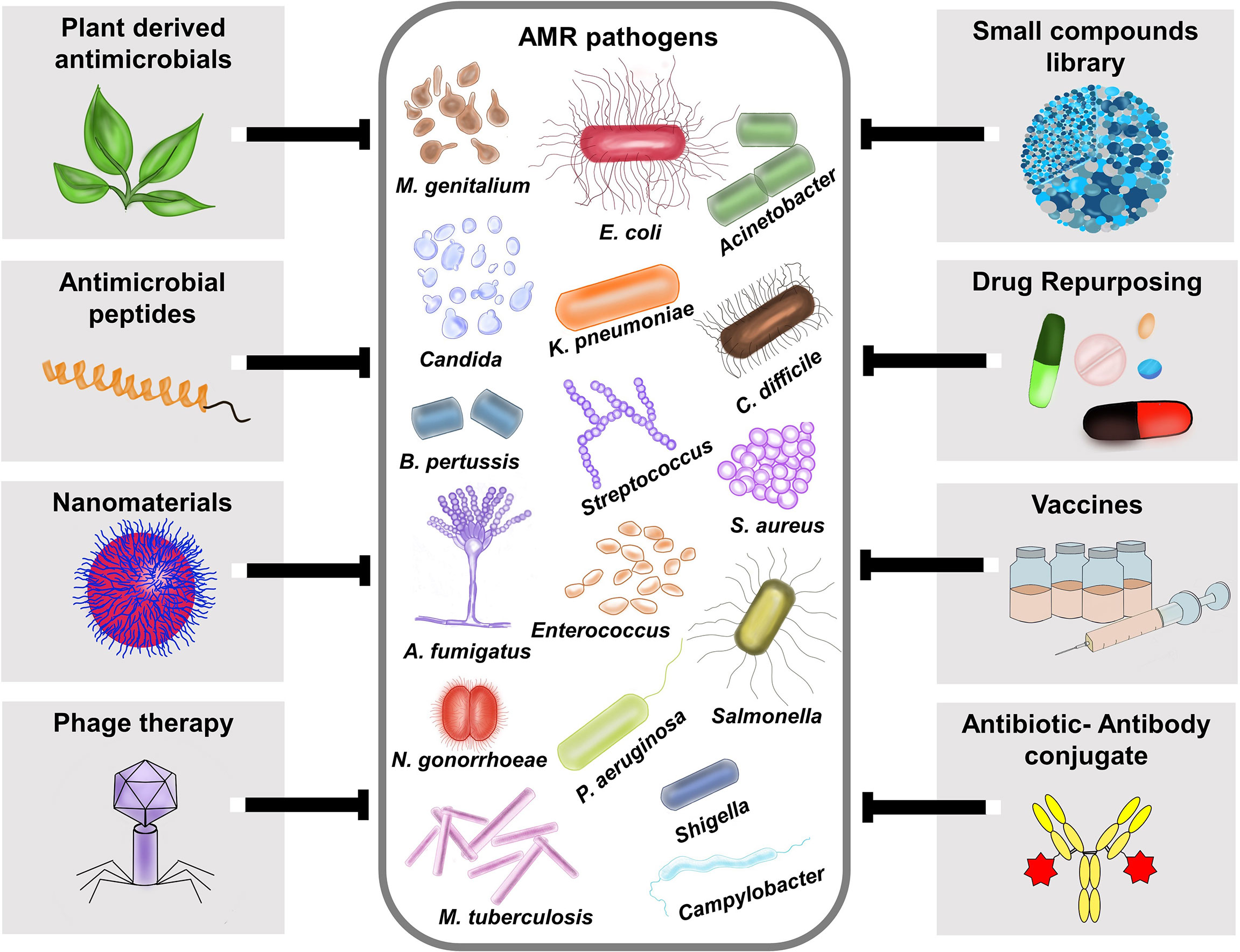
Figure 1 Different therapeutic strategies for combating antimicrobial resistant pathogens. AMR pathogens depicted in the figure is based on the Centers for Disease Control and Prevention (CDC), 2019 antibiotic resistance threats report, which categorized the AMR pathogens into urgent threats, serious threats, concerning threats and watch list https://www.cdc.gov/drugresistance/biggest-threats.html. Various therapeutic approaches like plant- derived antimicrobials, antimicrobial peptides (AMPs), Nanomaterials, Phage therapy, small compound library screening, Drug repurposing, Vaccines, and Antibiotic-Antibody conjugates (AAC) are currently available to fight against AMR pathogens.
Cruz-Lopez et al. have reviewed the efficacy of recently approved antibiotics, β-lactam inhibitors, especially those in advanced clinical phases of development, for treating multi-drug resistant (MDR) infections caused by Gram-negative bacteria. The authors also convey the message that these new drugs should be used wisely. Liao et al., in their review discussed the recent advances in antivirulence drug strategy against Pseudomonas aeruginosa, an opportunistic nosocomial MDR pathogen, and elaborates the uses of various drug candidates like small molecules derived from chemical libraries, repurposed drugs, and natural compounds, supplemented by several engineered antibodies, and alginate oligomers against specific virulence mechanisms (e.g. T3SS, QS, LPS, etc.).
Plants are the major sources of bioactive products used medicinally since ancient times against infectious diseases (Holzmeyer et al., 2020; Veerapandian et al., 2020). More than 200,000 natural products have been discovered from plants while there are abundant bioactive products yet to be discovered (Hartmann, 2007). Suganya et al. presented a broad and noteworthy review article addressing the use of conventional and complex phytochemicals against MDR bacteria. They have listed the potential bioactive phytochemicals that are effective against MDR pathogens.
Diseases caused by fungi in humans are difficult to manage where mostly they affect individuals with serious underlying illness and compromised host defense (Köhler et al., 2014). Fungi respond to antifungals immediately and treatment failure is common due to antifungal resistance development (Fisher et al., 2018; Fisher et al., 2022). Since fungi are eukaryotes, it is challenging to develop effective antifungals without host cell toxicity. Jothi et al. reported bacterial quorum-sensing molecule (QSM) as promising natural inhibitors against Candida albicans virulence, after virtual screening of bacterial QSMs against modeled fungal targets such as Cyr1 and Ras1, which are involved in hyphal growth regulatory pathways. They have validated the hyphal inhibitory effects of the identified compounds through in silico analysis and in vitro experiments. Recently, many studies have targeted medicinal plants in finding effective drugs against dual species biofilms (Veerapandian and Vediyappan, 2019). A study by Qian et al. has reported the antifungal and antibiofilm efficacy of paeonol, a major phenolic component of Moutan cortex Radicis, the root cortex of Paeonia suffruticosa Andrews. Interestingly, this compound has strong antifungal and antibiofilm activities against C. albicans and Cryptococcus neoformans both as mono- or dual-species. Furthermore, RNA-Seq analysis revealed that the C. albicans biofilm was inhibited by downregulating the expression of genes involved in filamentation, adhesion, and growth-related genes. Whereas C. neoformans biofilm was inhibited by upregulation of ergosterol biosynthesis-related genes.
An interesting research article from Ramalingam et al. has identified a potential compound from marine actinomycete Nocardiopsis exhalans, isolated from the mucus of Scleractinia coral Acropora formosa. The purified bioactive fraction of cell-free culture of N. exhalans was identified as N-(2-hydroxyphenyl)-2-phenazinamine (NHP), which showed potent biofilm inhibitory activity against Escherichia coli, P. aeruginosa, and Staphylococcus aureus. Combination antibiotic therapy is found to be superior and may directly or indirectly block the development of antibiotic resistance (Vipin et al., 2020). In this direction, She et al. reported the SPR741, a polymyxin B derivative that has a strong antimicrobial activity against extensively drug-resistant and pandrug-resistant Klebsiella pneumoniae when used in combination with macrolide antibiotics (erythromycin and clarithromycin). Yu et al. conducted a meta-analysis of clinical trials comparing novel carbapenem–β-lactamase inhibitor combinations with comparators to assess the clinical and microbiological responses, mortality, and adverse events (AEs). They concluded that imipenem–cilastatin/relebactam (ICR) and meropenem–vaborbactam (MEV) provided better outcome and could be applied as antimicrobial armamentarium against complicated infections. Song and Han have evaluated the Pharmacokinetic/pharmacodynamic exposure of vancomycin, at three infusion modes, intermittent infusion, continuous infusion, and optimal two-step infusion, for MRSA infections in critically ill patients, and reported the optimal two-step infusion mode to be most effective among them.
Antimicrobial peptides (AMPs) are one of the promising alternative solutions for pathogenic microbes that are difficult to treat with conventional antibiotics. AMPs are made up of short amino acid residues, which are present in all species of life (Parthasarathy et al., 2021). To date, more than 3000 AMPs have been reported, out of which >70% show antimicrobial activities (Wang et al., 2016). Wang et al. have reported a potential truncated peptide, named Spgillcin177–189 exerting antibacterial activity against several MDR bacterial pathogens. The authors have derived this peptide from the mud crab Scylla paramamosain. They showed that the peptide could affect the membrane permeability and cause the accumulation of intracellular reactive oxygen species in the pathogens. In addition, it has strong anti-biofilm activity against S. aureus and P. aeruginosa. Finally, an interesting study by Chang et al. reported gurmarin, a medicinal plant-derived cyclic peptide that blocks S. aureus biofilm formation in vitro and in vivo (a rat-implant biofilm model). This can be further developed for therapeutic uses. Using biochemical and electron microscopic methods, the authors demonstrated that exposure of the peptide to S. aureus diminishes the synthesis of N-acetylglucosamine polysaccharide during biofilm growth. The transcriptomic analysis further shows the upregulation of genes involved in oxidoreductases and the downregulation of genes involved in transferases and hydrolases. The cyclic peptides are known to have diverse bioactivity and are resistant to host proteolytic activity.
Overall, we believe that the articles presented in this collection contribute new knowledge on the discovery and development of antimicrobial agents and constitute a step toward finding potential therapeutics against AMR pathogens.
All authors listed here made substantial, direct, and intellectual contribution to managing the Research Topic, writing this editorial and approved it for publication.
The K-INBRE postdoctoral support to RV and the Innovative Research Award from Johnson Cancer Research Center, KSU to GV are kindly acknowledged.
The authors would like to thank Mahalakshmi Vijayaraghavan, Texas Tech University Health Sciences Center El Paso and Shruthi Vijayaraghavan for their technical support in generating Figure 1. We also thank all the authors of the articles and peer reviewers who contributed to this research article collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Centers for Disease Control and Prevention (2019) 2019 AR threats report. Available at: https://www.cdc.gov/drugresistance/biggest-threats.html (Accessed October 17, 2022).
de Kraker, M. E., Stewardson, A. J., Harbarth, S. (2016). Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 13 (11), e1002184. doi: 10.1371/journal.pmed.1002184
Fisher, M. C., Alastruey-Izquierdo, A., Berman, J., Bicanic, T., Bignell, E. M., Bowyer, P., et al. (2022). Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 20 (9), 557–571. doi: 10.1038/s41579-022-00720-1
Fisher, M. C., Hawkins, N. J., Sanglard, D., Gurr, S. J. (2018). Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science. 360 (6390), 739–742. doi: 10.1126/science.aap7999
Hartmann, T. (2007). From waste products to ecochemicals: fifty years research of plant secondary metabolism. Phytochemistry. 68 (22-24), 2831–2846. doi: 10.1016/j.phytochem.2007.09.017
Holzmeyer, L., Hartig, A. K., Franke, K., Brandt, W., Muellner-Riehl, A. N., Wessjohann, L. A., et al. (2020). Evaluation of plant sources for antiinfective lead compound discovery by correlating phylogenetic, spatial, and bioactivity data. Proc. Natl. Acad. Sci. U.S.A. 117 (22), 12444–12451. doi: 10.1073/pnas.1915277117
Köhler, J. R., Casadevall, A., Perfect, J. (2014). The spectrum of fungi that infects humans. Cold Spring Harb. Perspect. Med. 5 (1), a019273. doi: 10.1101/cshperspect.a019273
Murray, C. J. L., Ikuta, K. S., Sharara, F., Swetschinski, L., Aguilar, G. R., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399 (10325), 629–655. doi: 10.1016/S0140-6736(21)02724-0
Parthasarathy, A., Borrego, E. J., Savka, M. A., Dobson, R. C. J., Hudson, A. O. (2021). Amino acid-derived defense metabolites from plants: A potential source to facilitate novel antimicrobial development. J. Biol. Chem. 296, 100438. doi: 10.1016/j.jbc.2021.100438
Veerapandian, R., Paudyal, A., Chang, A., Vediyappan, G. (2020). Separation of bioactive small molecules, peptides from natural sources and proteins from microbes by preparative isoelectric focusing (IEF) method. J. Vis. Exp. 160), 10. doi: 10.3791/61101
Veerapandian, R., Vediyappan, G. (2019). Gymnemic acids inhibit adhesive nanofibrillar mediated Streptococcus gordonii-candida albicans mono-species and dual-species biofilms. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02328
Vipin, C., Saptami, K., Fida, F., Mujeeburahiman, M., Rao, S. S., Athmika, et al. (2020). Potential synergistic activity of quercetin with antibiotics against multidrug-resistant clinical strains of Pseudomonas aeruginosa. PLoS One 15 (11), e0241304. doi: 10.1371/journal.pone.0241304
Wang, G., Li, X., Wang, Z. (2016). APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44 (D1), D1087–D1093. doi: 10.1093/nar/gkv1278
World Health Organization (2017) The world is running out of antibiotics, WHO report confirms. Available at: https://www.who.int/news/item/20-09-2017-the-world-is.e-running-out-of-antibiotics-who-report-confirms (Accessed October 17, 2022).
Keywords: drugs, anti-virulence drugs, anti-biofilm drugs, repurposed drugs, antibiotic- antibody combination therapy, nanotechnology, in silico drug design
Citation: Veerapandian R, Abdul Azees PA, Viswanathan T, Amaechi BT and Vediyappan G (2022) Editorial: Developing therapeutics for antimicrobial resistant pathogens. Front. Cell. Infect. Microbiol. 12:1083501. doi: 10.3389/fcimb.2022.1083501
Received: 29 October 2022; Accepted: 14 November 2022;
Published: 24 November 2022.
Edited and Reviewed by:
Max MAURIN, Université Grenoble Alpes, FranceCopyright © 2022 Veerapandian, Abdul Azees, Viswanathan, Amaechi and Vediyappan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raja Veerapandian, ci52ZWVyYXBhbmRpYW5AdHR1aHNjLmVkdQ==; Bennett Tochukwu Amaechi, YW1hZWNoaUB1dGhzY3NhLmVkdQ==; Govindsamy Vediyappan, Z3ZlZGl5YXBAa3N1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.