
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Cell. Infect. Microbiol., 14 February 2023
Sec. Bacteria and Host
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.1075768
This article is a correction to:
Genetic Inactivation of Chlamydia trachomatis Inclusion Membrane Protein CT228 Alters MYPT1 Recruitment, Extrusion Production, and Longevity of Infection
 Jennifer H. Shaw1*
Jennifer H. Shaw1* Charlotte E. Key1
Charlotte E. Key1 Timothy A. Snider2
Timothy A. Snider2 Prakash Sah3
Prakash Sah3 Edward I. Shaw3
Edward I. Shaw3 Derek J. Fisher4
Derek J. Fisher4 Erika I. Lutter3*
Erika I. Lutter3*By Shaw JH, Key CE, Snider TA, Sah P, Shaw EI, Fisher DJ and Lutter EI (2018) 8:415. doi: 10.3389/fcimb.2018.00415
In the published article, there was an error in Figure 3A; panel MLC2 as published: Images for MLCK were duplicated in place of MLC2. The corrected Figure 3A; panel for MLC2 and its caption appear below.
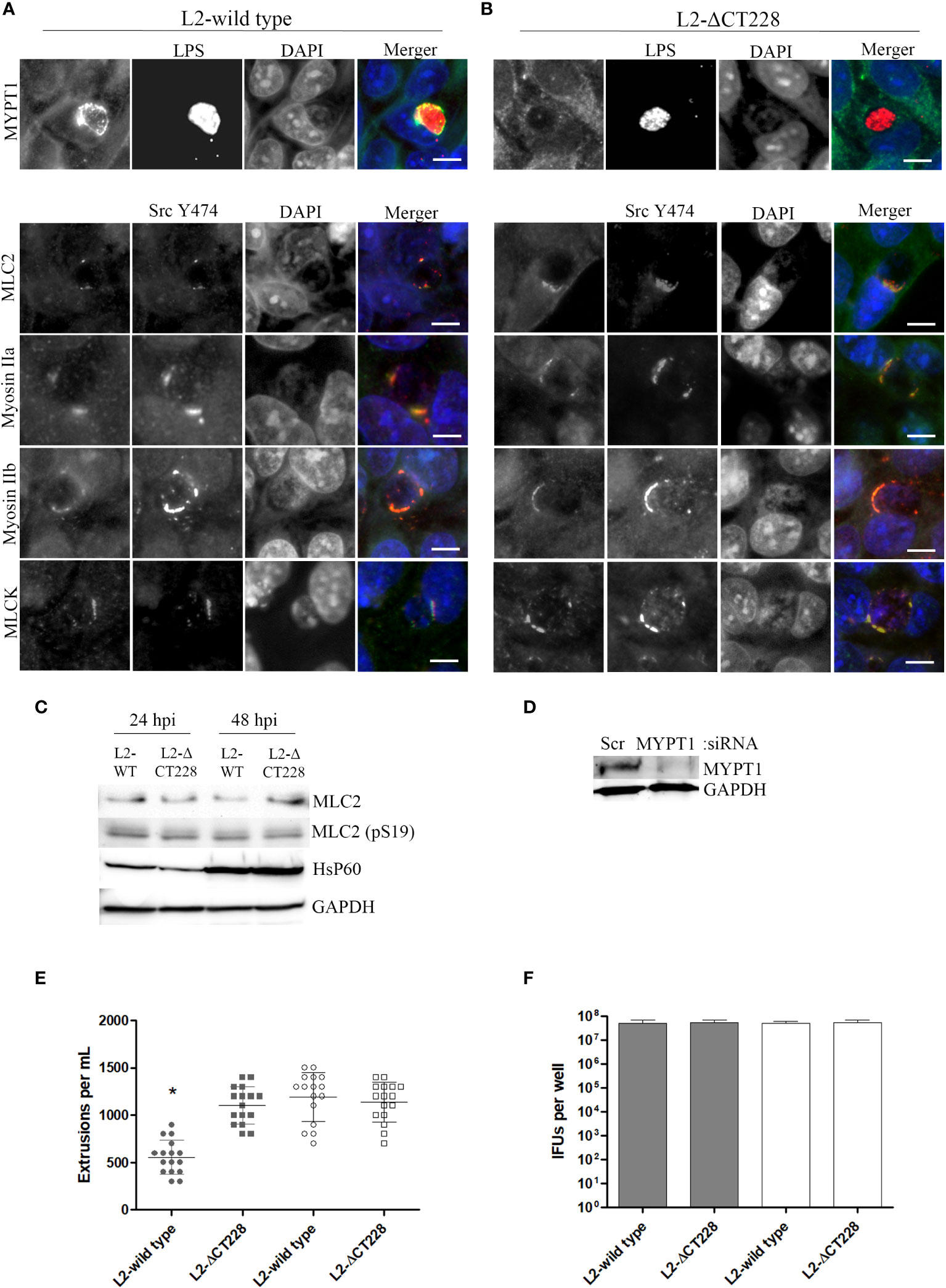
Figure 3 Recruitment of MYPT1 and Myosin phosphatase pathway components and extrusion production by C. trachomatis L2-wild type and L2-1CT228. HeLa cell monolayers were infected at a MOI of ∼0.5 with L2-wild type and L2-1CT228 for 18 h (in technical triplicate). Cells were fixed and stained with primary antibodies to MYPT1, Chlamydia LPS, MLC2 (pS19), Src Y474, MLCK (pY471), non-muscle Myosin IIa and IIb followed by fluorescent secondary antibodies. Experiments were repeated on three separate occasions and representative images were selected. (A, B) Top panel shows individual and merged images of MYPT1 recruitment (green) and Chlamydia LPS staining (red) in both the L2-wild type and L2-1CT228. Lower panel of individual and merged images show MLC2 (pS19), MLCK (pY471), and Mysoin IIa and IIb (green) co-localizing with active Src Y474 kinase (red) in microdomains at the periphery of inclusions in both L2-wild type and L2-1CT228. Scale bar, 10µm. (C) Total protein from L2-wild type and L2-1CT228 infected HeLa cells at 24 and 48 h post-infection were assessed for MLC2, MLC2 (pS19), HsP60, and GAPDH levels by western blot analysis. (D) HeLa cells were treated with either Scramble (Scr) or MYPT1 siRNA for 48 h prior to infection with L2-wild type and L2-1CT228. Protein samples were assessed for MYPT1 and GAPDH levels by western blot. (E) Extrusions collected and (F) IFUs were assessed for L2 wild-type and L2-1CT228 at 48 h post-infection in either Scramble (symbols and solid bars) or MYPT1 (open symbols and white bars) siRNA treated HeLa cells. *p < 0.0001.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: Chlamydia, extrusion, lymphogranuloma venereum, L2 serovar, sexually transmitted infection, urogenital infection, CT228, mouse infection
Citation: Shaw JH, Key CE, Snider TA, Sah P, Shaw EI, Fisher DJ and Lutter EI (2023) Corrigendum: Genetic inactivation of Chlamydia trachomatis inclusion membrane protein CT228 alters MYPT1 recruitment, extrusion production, and longevity of infection. Front. Cell. Infect. Microbiol. 12:1075768. doi: 10.3389/fcimb.2022.1075768
Received: 20 October 2022; Accepted: 25 October 2022;
Published: 14 February 2023.
Edited and Reviewed by:
Rey Carabeo, University of Nebraska Medical Center, United StatesCopyright © 2023 Shaw, Key, Snider, Sah, Shaw, Fisher and Lutter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer H. Shaw, amVubmlmZXIuaC5zaGF3QG9rc3RhdGUuZWR1; Erika I. Lutter, ZXJpa2EubHV0dGVyQG9rc3RhdGUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.