
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 25 November 2022
Sec. Parasite and Host
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.1072385
This article is part of the Research Topic Advances in Molecular Biology, Pathogenesis, Diagnosis, Vaccines, and Treatment of Diseases Caused by Apicomplexan Parasites View all 14 articles
 Xiao-Man Li1†
Xiao-Man Li1† Hong-Li Geng1†
Hong-Li Geng1† Yong-Jie Wei1
Yong-Jie Wei1 Wei-Lan Yan1
Wei-Lan Yan1 Jing Liu2*
Jing Liu2* Xin-Yu Wei3
Xin-Yu Wei3 Miao Zhang1
Miao Zhang1 Xiang-Yu Wang1
Xiang-Yu Wang1 Xiao-Xuan Zhang1
Xiao-Xuan Zhang1 Gang Liu1*
Gang Liu1*Intoduction: Cryptosporidiosis is a zoonotic disease caused by Cryptosporidium infection with the main symptom of diarrhea. The present study performed a metaanalysis to determine the global prevalence of Cryptosporidium in Equus animals.
Methods: Data collection was carried out using Chinese National Knowledge Infrastructure (CNKI), VIP Chinese journal database (VIP), WanFang Data, PubMed, and ScienceDirect databases, with 35 articles published before 2021 being included in this systematic analysis. This study analyzed the research data through subgroup analysis and univariate regression analysis to reveal the factors leading to high prevalence. We applied a random effects model (REM) to the metadata.
Results: The total prevalence rate of Cryptosporidium in Equus was estimated to be 7.59% from the selected articles. The prevalence of Cryptosporidium in female Equus was 2.60%. The prevalence of Cryptosporidium in Equus under 1-year-old was 11.06%, which was higher than that of Equus over 1-year-old (2.52%). In the experimental method groups, the positive rate detected by microscopy was the highest (10.52%). The highest Cryptosporidium prevalence was found in scale breeding Equus (7.86%). The horses had the lowest Cryptosporidium prevalence (7.32%) among host groups. C. muris was the most frequently detected genotype in the samples (53.55%). In the groups of geographical factors, the prevalence rate of Cryptosporidium in Equus was higher in regions with low altitude (6.88%), rainy (15.63%), humid (22.69%), and tropical climates (16.46%).
Discussion: The search strategy use of five databases might have caused the omission of some researches. This metaanalysis systematically presented the global prevalence and potential risk factors of Cryptosporidium infection in Equus. The farmers should strengthen the management of young and female Equus animals, improve water filtration systems, reduce stocking densities, and harmless treatment of livestock manure.
Cryptosporidium is a zoonotic coccidian parasite, which mainly parasitizes in the epithelial cells of the small intestine in vertebrates (Xue, 2021). The life cycle of Cryptosporidium in hosts comprises asexual and sexual phases, and finally, the oocysts were shed with faeces. The oocyst containing infective sporozoites can survive for months in moist conditions (Chalmers et al., 2019). The Cryptosporidium-infected patients usually display signs of diarrhea and abdominal pain, and other clinical features include nausea, vomiting, and low-grade fever. The occasional signs include myalgia, weakness, malaise, headache, and anorexia. The severity, persistence, and eventual outcome of infection generally depend on the characteristics of Cryptosporidium and host factors (Bouzid et al., 2013).
Cryptosporidium was examined in mucosal tissues of mice by Tyzzer in 1907 (Zhang and Jiang, 2001). Cryptosporidium was the first detection happened to be in immunodeficient Arabian horse foals in 1978 (Snyder et al., 1978). People didn’t pay attention to the disease at first. At the end of the nineteenth century, the events of death of AIDS patients who were infected with Cryptosporidium caused a full attention of cryptosporidiosis (Current et al., 1983). Cryptosporidium is worldwide distributed, and the infections of Cryptosporidium in organisms have been reported in more than 70 countries (Xue, 2021).
Equus animals are mainly distributed in Eurasia and Africa, roughly divided into three species: horse, donkey and zebra. Horses are now mainly used for entertainment or competition, donkeys are kept as companions or used as working or production animals, whereas mules are mainly used for working purposes (Jian, 2012). Because they are maintained in a close association with their owners and veterinary personnel, Equus animals are the important reservoirs for transmission of pathogens (such as Cryptosporidium hominis and Toxoplasma gondii) to humans and other animals (Alvarado-Esquivel et al., 2015; Jian et al., 2016). The cases of Equus cryptosporidiosis have been reported in many countries around the world, such as China, Italy, and United States. (Veronesi et al., 2010; Qi et al., 2015; Wagnerová et al., 2016).
At present, the systematic evaluation and analysis of Equus cryptosporidiosis are still absent. Therefore, a systematic review and meta-analysis were conducted to evaluate the prevalence of Equus cryptosporidiosis in the world. The collected information was used to discuss the factors affecting the infection of Cryptosporidium in Equus (Page et al., 2021).
To evaluate the prevalence of Cryptosporidium infections in Equus around the world, we performed a comprehensive review of literatures both in Chinese and English published from the beginning of the creation to October 1, 2021. The articles were derived from five databases, including CNKI, VIP Chinese Journal Database, Wanfang Data, PubMed, and ScienceDirect. In the three Chinese databases, the advanced search was carried out using “Equus (in Chinese)” and “Cryptosporidium (in Chinese)” as keywords. In Science Direct, the keywords “Cryptosporidium” and “Equus” were used for a search. We used MeSH terms “Cryptosporidium” and “Equine” and their entry terms, such as “Horse”, “Horse, Domestic”, “Domestic Horse”, “Domestic Horses”, “Horses, Domestic”, “Equus” “caballus”, and “Equus przewalskii” in PubMed. We used boolean operators “AND” to connect MeSH terms and “OR” to connect the entry terms. Finally, the search formula was “((Cryptosporidium) OR Cryptosporidiums) AND ((((((((Horse) OR Horse, Domestic) OR Domestic Horse) OR Horses, Domestic) OR Equus caballus) OR Equus przewalskii) OR Horse genus) “. Endnote (X9.2 version) was employed to organize the obtained article information. A protocol for the literature review was devised (Figure 1) in accordance with the PRISMA guidelines.
As part of the eligibility for inclusion, titles that suggested the topic Cryptosporidium in Equus were selected. The abstracts from the selected reference titles were reviewed by two independent reviewers to determine if the studies met the inclusion criteria and, if so, the entire articles were reviewed in full. The inclusion criteria for the systematic review and meta-analysis were as follows: (1) the object of research was Equus; (2) the diseased individuals were positive for Cryptosporidium in the research; (3) the research contained clear information, including the number of sick individuals and the population, the number of positive samples, the location of the test, and the location of sampling; (4) the article should contain a full text; (5) the research must be designed for a cross-sectional extension. Articles that do not meet these criteria were excluded. Unpublished reports, comments and copies were also excluded.
The extracted data included article title, first author, publication year, detection method, breeding environment, breeding method, genotypes, sampling year, article quality, detailed geographic and climatic factors, total number, number of positives, age and gender of the research object, geographic location (latitude and longitude), altitude, relative humidity, annual average temperature, annual precipitation, climate, and detection method type. The meteorological data of the years involved were from the China Meteorological Data Service Center (CMDC, http://data.cma.cn/) and national centers of environmental information (https://www.ncei.noaa.gov/maps/monthly/), such as temperature, rainfall, longitude, latitude, humidity and altitude. The database was established by using Microsoft Excel (version 16.32). Two authors independently extracted and recorded data from each selected study. The differences derived from reviewers or uncertainty about the qualifications of the research were further assessed by another author of this paper. The grouping method is based on previous studies (Wei et al., 2021a; Wei et al., 2021b).
The quality of the included studies was evaluated according to the criteria based on the recommended grading evaluation, formulation, and the Grading of Recommendations Assessment, Development and Evaluation (GRADE) (Xie and Machado, 2021). The scoring criteria were as follows: (1) whether a clear detection method was employed; (2) whether the sampling animal was clear; (3) whether three or more influencing factors were included; (4) the sample size was greater than enough; and (5) whether the sampling year was clear. Therefore, the studies could be scored between 0 and 5 points.
The meta package in R software version 4.0.3 (“R core team, R: A language and environment for statistical computing” R core team 2018) was employed to analyze the data in this study (Zeng et al., 2020). Before performing the meta-analysis, we tested five transformation methods to bring the data closer to a Gaussian distribution, namely no transformation (PRAW), logarithmic transformation (PLN), logit transform (PLOGIT), arcsine transform (PAS), and double arcsine transform (PFT) (Table 1). The conversion rate was based on a Shapiro-Wilk normal test. The W-value close to 1 and the P-value greater than 0.05 was close to the Gaussian distribution criterion. The heterogeneity between studies was calculated by Cochran-Q, I2 statistics, and χ2 test, the P-value < 0.05 and I2 = 50% was used to define the degree of heterogeneity with a statistical significance. According to the heterogeneity of the included articles, a random effect model was selected for analysis (Ni et al., 2020). The forest plots were used for a comprehensive analysis. The funnel plot and Egger’s test were used to evaluate publication bias of studies. When there is publication bias in the included articles, the funnel plot is asymmetric, and the distribution is skewed (Egger et al., 1997). The Egger’s test is expected to have a regression intercept of 0 in the absence of bias (Lin and Chu, 2018). The stability of a study was evaluated by the trim and fill analysis and sensitivity analysis (Wang et al., 2021).
To further study the potential sources of heterogeneity, the individual and multivariate model factors were analyzed to determine factors that affected heterogeneity. The factors included sampling surveys (before 2008 vs. others), diagnostic methods (molecular diagnostics vs. other methods), age ≤ 1year vs. age >1year, genotype (C. parvum vs. others), gender (female vs. male), feeding method (cage-free vs. scale breeding), host (horse vs. others), country (China vs. others), the quality level of publications (5 points vs. others), longitude (<50° vs. others), latitude (<30° vs. others), annual average rainfall (< 500 mm vs. ≥ 500 mm), annual average temperature (< 10°C vs. ≥ 10°C), annual average humidity (40%-50% vs. others), altitude (<50 m vs. others), and climate (temperate climate vs. others)
In this study, 1,182 related articles were identified after searching five databases, and 67 articles were selected after the initial screening and removal of duplicates. According to the selection criteria described in section, an additional 32 indeterminate articles were excluded after checking the full text. Finally, 35 articles were selected for the meta-analysis. (Figure 1; Table 2).
The included articles involved 13 countries. In the 35 studies, the total number of samples was 9,817, and the number of positives was 816 (Table 3). According to our quality criteria, 11 articles were considered to be 5 points, 21 were of medium-quality (three or four points), and the remaining 3 articles were deemed to be of low-quality (zero to two points; Table 3).
In the included studies, the forest plot showed the degree of heterogeneity of all data (χ2 = 0.0267, I2 = 96.0%, P < 0.01) (Figure 2). According to the funnel chart, the distribution of points was observed to be incompletely symmetrical, which might be due to a publication bias (0.6344) or a small sample bias (Figure 3). Six supplementary studies were found in trim and fill analysis, which changed the aggregate estimate (Figure 4). The Egger’s test was used to assess the potential publication bias in the analysis with a P-value (0.7398) greater than 0.05, thus indicating that no publication bias was present in the data (Figure 5). The sensitivity test showed that the reorganized data were not significantly affected after excluding any study, and the results were consistent (Figure 6), which verified the rationality and reliability of this analysis. Figure 7 is a map of Cryptosporidium prevalence in Equus worldwide. A chord diagram shows the relationship between the prevalence of Cryptosporidium in Equus species and epidemiological variables (stripe width indicates prevalence) (Figure 8). C. parvum genotype had a larger proportion in North America in the geographic area grouping. Genotype C. hominis accounted for a large proportion of donkey in the host group. Genotype C. andersoni accounted for more than 1- year-old in the age group.
According to the data obtained from the selected articles, the global combined prevalence of Equus Cryptosporidium infection was 7.59% (95% CI: 4.86-10.87) (Table 3). Before 2008, the prevalence of Cryptosporidium in Equus was 10.77% (95% CI: 3.92-20.32), which was significantly higher than that in other time periods (P < 0.01). The highest positive rate of Equus Cryptosporidium was 17.91% (95% CI: 9.73-27.92) in Oceania. Among the detection methods, microscopic examination had the highest rate of 10.52% (95% CI: 4.99-17.19). The prevalence of Cryptosporidium in Equus ≤ 1-year-old was 11.06% (95% CI: 6.13-17.22), which was significantly higher than in other age groups (P < 0.01). The highest prevalence of Cryptosporidium in female Equus was 2.60% (95% CI: 0.78-5.44). The highest prevalence of Cryptosporidium among the rearing method groups was 7.86% (95% CI: 4.39-12.22) for collectivized breeding. Among the host groups, mules had the highest rate of 20.00% (95% CI: 10.20-32.09). The highest prevalence of Equus Cryptosporidium was 19.58% (95% CI: 6.03–38.43) in Italy, which was significantly higher than that in other countries (Table 4). The highest genotype prevalence of Cryptosporidium in Equus was C. muris (53.55%; 95% CI: 11.65-92.51), followed by C. hominis, with a rate of 43.94% (95% CI: 23.11–65.95) (Table 5).
Among the analyzed geographical factors, the positive rate of Cryptosporidium in Equus at northern latitude < 30° was the highest (8.77%; 95% CI: 2.88-17.44). The positive rate of Cryptosporidium in Equus with longitude > 100° was the highest (9.03%, 95% CI: 3.15-17.53). The information for other geographical latitude subgroup analyses included precipitation range (≥ 500 mm; 15.63%; 95% CI: 8.60-24.28), temperature range (≥ 10°C; 8.04%, 95% CI: 3.30-14.60), humidity range (70%-80%; 22.69%; 95% CI: 13.76-33.10) and altitude range (< 50 m; 6.88%; 95% CI: 2.70-12.78). According to the analysis of climatic factors, we found that tropical climate had the highest positive rate of Equus Cryptosporidium (16.46%; 95% CI: 0.48-48.11; Table 6).
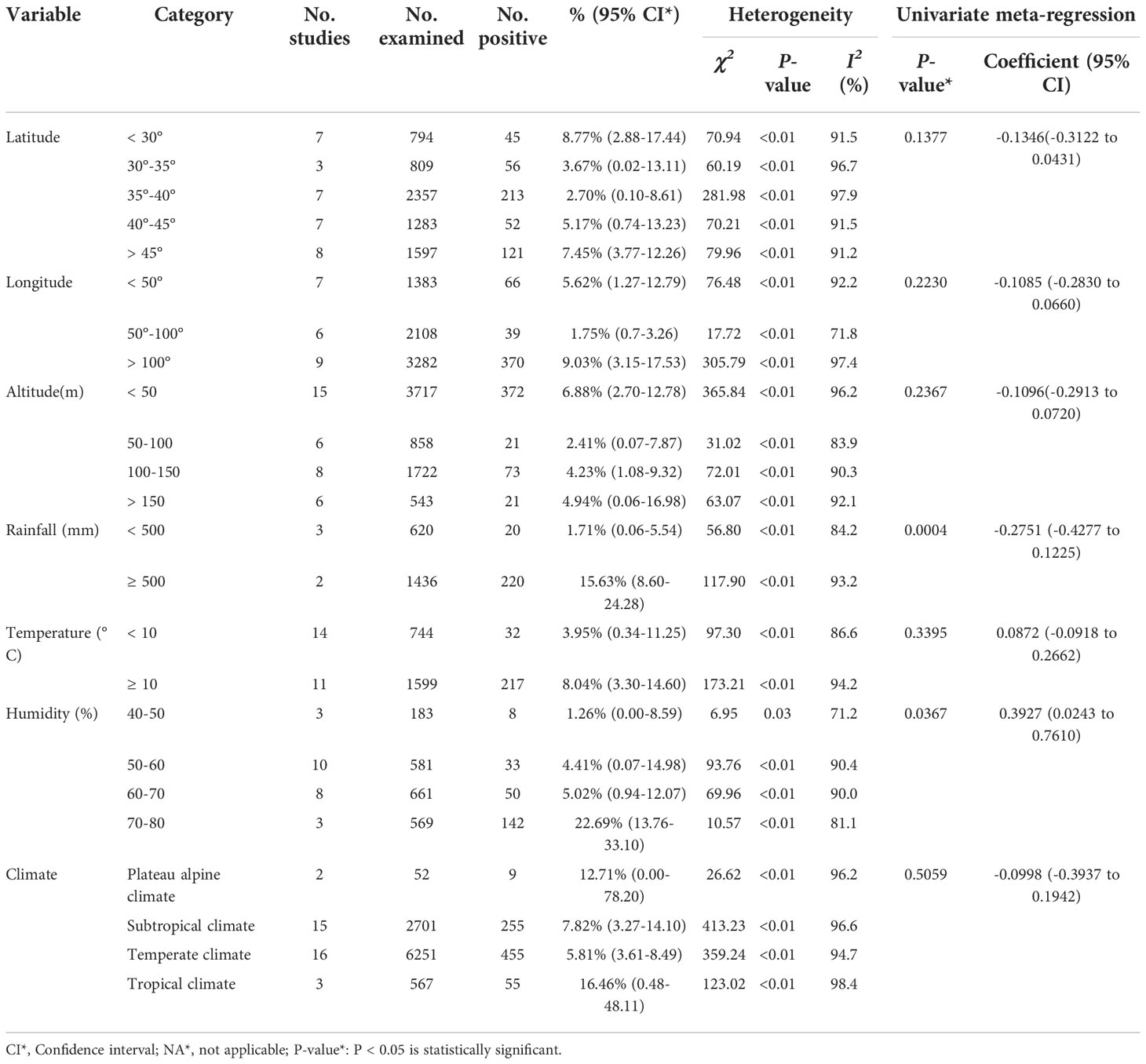
Table 6 A subgroup analysis of the prevalence of Cryptosporidium in equine genus by geographical location, climate and other variables.
Cryptosporidium is a waterborne pathogen that infects livestock, poultry, and companion animals, thus posing a great threat to public health (Gao et al., 2021). Many large Cryptosporidium outbreaks were caused by contamination of water sources with animal feces (Rodríguez et al., 2012). Cryptosporidiosis can cause slow growth of sick animals, extreme weight loss, decreased resistance, and huge losses to the animal husbandry (Han et al., 2020). In this study, a publication bias was observed according to the Egger’s test. Pass I2 statistics results, the prevalence of Cryptosporidium in Equus species in the world was highly heterogeneous in the eligible studies, which may be caused by differences in detection methods, age, gender, geographic factors, and countries.
The prevalence of Cryptosporidium in female Equus was identified to be higher than that in male Equus. This is probably due to a weaker body resistance of female Equus than males, especially after giving birth, and more susceptible to Cryptosporidium infection (Chen et al., 2013). The mare is considered to be a carrier of Cryptosporidium, and the young animals could be infected with Cryptosporidium by sharing a pasture or barn with the foals (Qi and Zhang, 2018). The eggs of Cryptosporidium in infected female Equus have no obvious shedding phenomenon, but the amount of shedding during the perinatal period increases (Skerrett and Holland, 2001). This significantly increases the probability of young animals being infected with Cryptosporidium.
In this study, the prevalence of Cryptosporidium in Equus of ≤ 1 year was higher than that of Equus > 1 year, which is basically in line with the previous reports (Langkjaer et al., 2007; Wang, 2020). The maternal antibodies in the young animals will lose protective effects in 2-6 months, resulting in a decreased resistance of the young animals (Lu et al., 2008). In addition, the weaning stress response in young animals will lead to changes at hormone levels and immune function, thus causing immune system suppression, and thereby increasing the possibility of being infected with Cryptosporidium (Ren et al., 2018). Therefore, the breeding of dams and cubs needs to be strengthened to improve the resistance of animals, and the dams can be isolated during the breeding period.
Poor sanitary conditions increase the infection risk of Cryptosporidium in animals and expand the spread of Cryptosporidium species (Gao et al., 2021). Our study of the rearing group showed that mules and donkeys had higher rates of Cryptosporidium infection than horses. The living environment of mules and donkeys is relatively complicated, and they usually suffer from poor harness, lack of veterinary care, improper nutrition, and low status and value, in spite of their usefulness (Davis, 2019). Meanwhile, the feces cannot be cleaned in time, which is more likely to cause the transmission and infection of Cryptosporidium in animals (Jian, 2012). An analysis of the breeding environment also showed that the prevalence of Cryptosporidium was higher in Equus than that farmed on scale breeding. The high stocking densities of large-scale farming might result in high rates of Cryptosporidium infection in high-density captive equines. This may be due to the facts that more oocysts are scattered in high-density pens and the transmission speed is fast (Wang et al., 2008). Therefore, we recommend reducing stocking density and enhancing the animal welfare of donkeys and mules.
The investigation of sampling year in the selected articles showed that the prevalence of Cryptosporidium in Equus animals before 2012 was higher. In 2008, a worldwide economic crisis occurred and started to recover until 2010 (Zhang, 2018). The sluggish world economy may make people slack in Equus breeding, which may be one of the reasons for the high prevalence of Cryptosporidium. After 2013, the world economy gradually recovered, and many countries began to pay more attention to the animal husbandry. The gradually increased animal welfare may contributed to the lower prevalence of Cryptosporidium in Equus.
In the continent and country groups, the highest positive rate of Cryptosporidium was found in Oceania and the lowest was in Africa. Among them, New Zealand in Oceania had the highest positive rate, and Algeria in Africa had the lowest positive rate. New Zealand has a temperate maritime climate, with a warm and humid climate throughout the year, and its long coastline and abundant water resources make it more suitable for Cryptosporidium to survive. The frequent rainfall may lead to the transmission of Cryptosporidium in animal feces from fields to surface waters, thus resulting in a higher prevalence of Cryptosporidium in this genus of horses (Sterk et al., 2016). In contrast, the Algerian coast has a Mediterranean climate, with high temperature and rainy winter, while the central and southern parts have a savanna and tropical desert climate, which is dry and rainy, with cold winter and hot summer. The increase of temperature will reduce the life activity of the worms on the land surface (Moriarty et al., 2012). Therefore, the prevalence of Cryptosporidium in Equus in Africa is low.
In the geographical groups, the positive rate of Cryptosporidium was the highest in regions with longitude > 100° and latitude < 30°. These two factors are mainly concentrated in the central and southern parts of China. The central and southern parts of China belong to the temperate monsoon climate and the subtropical monsoon climate. The rainfall types of these two climates are high temperature and rainy in summer, low temperature and less rain in winter (Wang, 2009). Meanwhile, the positive rate of Cryptosporidium in Equus that live in areas with humidity 70%-80%, rainfall > 500 mm, temperature ≥ 10°C, and altitude < 50 m was higher than that in other regions. High rainfall will increase the pollution of water sources, and Cryptosporidium is mainly transmitted through water sources, thereby increasing the prevalence of Cryptosporidium (Young et al., 2015). As altitude decreases, the temperature gradually increases (Zhu et al., 2013). Suitable temperature and hot and humid weather are the reasons for an increase in the prevalence of Cryptosporidium. This is in line with previous reports (Burton et al., 2010; Lv et al., 2021).
The genotypes of Cryptosporidium in infected Equus were analyzed. The main genotypes of Equus Cryptosporidium were C. parvum, horse genotype, C. andersoni, C. muris, C. hominis, and C. cuniculus (Wang, 2015; Wagnerová et al., 2016; Deng, 2018; Zhang, 2021). Among them, C. parvum, C. andersoni, and C. hominis are the genotypes that infect humans. In this study, the positive rates of C. muris and C. hominis genotypes were higher. Therefore, strengthening the feeding, management, and control of Equus Cryptosporidium are of great importance to the public safety of humans and animals.
The included articles mainly employed three types of detection methods, including molecular detection, immunological detection, and microscopic detection. Molecular examination is mainly based on PCR. Immunological tests include IFAT, DAF, and ELISA. Among them, the positive rate detected by IFAT technology is the highest, followed by microscopy and PCR. The microscopic detection of Cryptosporidium is easily operated and has a low cost but may lead to misdiagnosis and increase the probability of false positives (Robinson and Chalmers, 2020a). For immunological detection, although ELISA has the advantages of high sensitivity and specificity, it cannot perform species identification (Mittal et al., 2014). IFAT is prone to false positive, thus resulting in a high positive rate (Robinson et al., 2020b). The PCR detection method have high sensitivity and specificity than that of microscopy, has become the optimal method for Cryptosporidium detection (Zhang et al., 2017). It is recommended to use PCR method to detect Cryptosporidium in epidemiological investigations.
These results reflect the prevalence of Cryptosporidium in Equus to a certain extent around the world. In this meta-analysis, the reasons for losing points in some studies are: (1) less than 3 risk factors; (2) limited samples; (3) unclear sampling year; and (4) absence of specific sampling location. It is recommended that researchers should take more samples and explore more influencing factors to provide more data that support for the prevention and treatment of Cryptosporidium infection in Equus.
The advantages of this study are rigorous method and lots of risk factors. The publication bias was tested by using funnel chart and more accurate Egger’s test. The subgroup analysis and regression analysis correctly identified the influence of heterogeneity on the results. The combination of multiple data analysis methods made our findings reasonable. A comprehensive analysis of subgroups can replenish the previous articles and provide more complete data for follow-up research. However, this study also has some limitations. First, we searched in five databases, and the search strategy might have caused the omission of some researches. Second, the data in some subgroups, such as Italy in the city subgroup, are covered only by one article, which can lead to unstable outcomes.
A systematic review and meta-analysis of 35 articles provided a comprehensive overview of the global epidemiology of Equus Cryptosporidium. The results showed that cryptosporidiosis was widespread in Equus, and cryptosporidiosis was prone to occur in the areas with warm climate. Equus under 1 year were more susceptible to Cryptosporidium infection. In view of the high incidence of cryptosporidiosis, correct prevention and control measures should be taken in time for specific age groups and regions. During the breeding process, good hygiene habits should be developed, regional prevention and control should be strengthened, and water pollution should be minimized.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethical review and approval was not required for the animal study.
X-ML and H-LG: Data curation, Methodology, Supervision, Writing-review and editing. W-LY and X-XZ: Writing-review and editing. Y-JW, Xin-YW: Data curation, Resources, Software. MZ and Xia-YW: Data curation, Methodology, Visualization. JL and GL: Conceptualization, Supervision, Funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by the Research Foundation for Distinguished Scholars of Qingdao Agricultural University (grant no. 665-1120044 and 665-1122009).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alvarado-Esquivel, C., Alvarado-Esquivel, D., Dubey, J. P. (2015). Prevalence of toxoplasma gondii antibodies in domestic donkeys (Equus asinus) in durango, mexico slaughtered for human consumption. BMC Vet. Res. 11, 6. doi: 10.1186/s12917-015-0325-9
Bouzid, M., Hunter, P. R., Chalmers, R. M., Tyler, K. M. (2013). Cryptosporidium pathogenicity and virulence. Clin. Microbiol. Rev. 26 (1), 115–134. doi: 10.1128/CMR.00076-12
Burton, A. J., Nydam, D. V., Dearen, T. K., Mitchell, K., Bowman, D. D., Xiao, L. (2010). The prevalence of Cryptosporidium, and identification of the Cryptosporidium horse genotype in foals in new York state. Vet. Parasitol. 174 (1-2), 139–144. doi: 10.1016/j.vetpar.2010.08.019
Caffara, M., Piva, S., Pallaver, F., Iacono, E., Galuppi, R. (2013). Molecular characterization of Cryptosporidium spp. from foals in Italy. Vet. J. 198 (2), 531–533. doi: 10.1016/j.tvjl.2013.09.004
Chalmers, R. M., Davies, A. P., Tyler, K. (2019). Cryptosporidium. Microbiol. (Reading) 165 (5), 500–502. doi: 10.1099/mic.0.000764
Chen, J., Shen, K. F., Ren, H. X. (2013). Investigation of goats Cryptosporidium infection in some breeding farms of chongqing. Chin. J. Vet. Med. 48 (12), 15–17.
Couso-Pérez, S., de Limia, F. B., Ares-Mazás, E., Gómez-Couso, H. (2020). First report of zoonotic Cryptosporidium parvum GP60 subtypes IIaA15G2R1 and IIaA16G3R1 in wild ponies from the northern Iberian Peninsula. Parasitol. Res. 119 (1), 249–254. doi: 10.1007/s00436-019-06529-x
Current, W. L., Reese, N. C., Ernst, J. V., Bailey, W. S., Heyman, M. B., Weinstein, W. M. (1983). Human cryptosporidiosis in immunocompetent and immunodeficient persons. studies of an outbreak and experimental transmission. N Engl. J. Med. 308 (21), 1252–1257. doi: 10.1056/NEJM198305263082102
Davis, E. (2019). Donkey and mule welfare. Vet. Clin. North Am. Equine Pract. 35 (3), 481–491. doi: 10.1016/j.cveq.2019.08.005
De Souza, P. N., Bomfim, T. C., Huber, F., Abboud, L. C., Gomes, ,. R. S. (2009). Natural infection by Cryptosporidium sp., Giardia sp. and Eimeria leuckarti in three groups of equines with different handlings in Rio de Janeiro, Brazil. Vet. Parasitol. 160 (3-4), 327–333. doi: 10.1016/j.vetpar.2008.10.103
Deng, L. (2018). Phylogenetic and multilocus genotypes of Cryptosporidium and Enterocytozoon bieneusi in horses. Sichuan Agric. Univ 2, 1–63.
Deng, L., Li, W., Zhong, Z., Gong, C., Cao, X., Song, Y., et al. (2017). Occurrence and Genetic Characteristics of Cryptosporidium hominis and Cryptosporidium andersoni in Horses from Southwestern China. J. Eukaryot Microbiol. 64 (5), 716–720. doi: 10.1111/jeu.12399
Egger, M., Davey, S. G., Schneider, M., Minder, C. (1997). Bias in meta-analysis detected by a simple, graphical test. BMJ. 315 (7109), 629–634. doi: 10.1136/bmj.315.7109.629
Galuppi, R., Piva, S., Castagnetti, C., Iacono, E., Tanel, S., Pallaver, F., et al. (2015). Epidemiological survey on Cryptosporidium in an Equine Perinatology Unit. Vet. Parasitol. 210 (1-2), 10–18. doi: 10.1016/j.vetpar.2015.03.021
Gao, H. H., Wang, J. D., Kang, X. D., Wang, P. Z., Wu, X. Q., Li, Y. K. (2021). Prevalence and risk factors of Cryptosporidium in sheep and goat of China. Chin. J. Vet. Sci. 41 (2), 401–406.
Grinberg, A., Pomroy, W. E., Carslake, H. B., Shi, Y., Gibson, I. R., Drayton, ,. B. M. (2009). A study of neonatal cryptosporidiosis of foals in New Zealand. N Z Vet. J. 57 (5), 284–289. doi: 10.1080/00480169.2009.58622
Guo, P. F., Chen, T. T., Tsaihong, J. C., Ho, G. D., Cheng, P. C., Tseng, Y. C., et al. (2014). Prevalence and species identification of Cryptosporidium from fecal samples of horses in Taiwan. Southeast Asian J. Trop. Med. Public Health 45 (1), 6–12.
Han, M. N., Ma, L., Zhou, L. L., Ren, M., Wang, D., Lin, Q. (2020). Cryptosporidiosis and its prevalence in yak. Prog. Vet. Med. 41 (1), 10812.
Hijjawi, N., Mukbel, R., Yang, R., Ryan, U. (2016). Genetic characterization of Cryptosporidium in animal and human isolates from Jordan. Vet. Parasitol. 228, 116–120. doi: 10.1016/j.vetpar.2016.08.015
Inácio, S. V., de Brito, R. L., Zucatto, A. S., Coelho, W. M., de Aquino, M. C., Aguirre, ,. A., et al. (2012). Cryptosporidium spp. infection in mares and foals of the northwest region of São Paulo State, Brazil. Rev. Bras. Parasitol. Vet. 21 (4), 355–358. doi: 10.1590/s1984-29612012005000003
Inácio, S. V., Widmer, G., de Brito, R. L., Zucatto, A. S., de Aquino, M. C., Oliveira, B. C., et al. (2017). First description of Cryptosporidium hominis GP60 genotype IkA20G1 and Cryptosporidium parvum GP60 genotypes IIaA18G3R1 and IIaA15G2R1 in foals in Brazil. Vet. Parasitol. 233, 48–51. doi: 10.1016/j.vetpar.2016.11.021
Jian, F. (2012). Molecular epidemiology of Cryptosporidium and Giardia of equine in some regions of China. Henan Agric. Univ 7, 1–9.
Jian, F., Liu, A., Wang, R., Zhang, S., Qi, M., Zhao, W., et al. (2016). Common occurrence of Cryptosporidium hominis in horses and donkeys. Infect. Genet. Evol. 43, 261–266. doi: 10.1016/j.meegid.2016.06.004
Johnson, E., Atwill, E. R., Filkins, M. E., Kalush, J. (1997). The prevalence of shedding of Cryptosporidium and Giardia spp. based on a single fecal sample collection from each of 91 horses used for backcountry recreation. J. Vet. Diagn. Invest. 9 (1), 56–60. doi: 10.1177/104063879700900110
Kostopoulou, D., Casaert, S., Tzanidakis, N., van Doorn, D., Demeler, J., von Samson-Himmelstjerna, G., et al. (2015). The occurrence and genetic characterization of Cryptosporidium and Giardia species in foals in Belgium, The Netherlands, Germany and Greece. Vet. Parasitol. 211 (3-4), 170–174. doi: 10.1016/j.vetpar.2015.04.018
Laatamna, A. E., Wagnerová, P., Sak, B., Květoňová, D., Aissi, M., Rost, M., et al. (2013). Equine cryptosporidial infection associated with Cryptosporidium hedgehog genotype in Algeria. Vet. Parasitol. 197 (1-2), 350–353. doi: 10.1016/j.vetpar.2013.04.041
Laatamna, A. E., Wagnerová, P., Sak, B., Květoňová, D., Xiao, L., Rost, M., et al. (2015). Microsporidia and Cryptosporidium in horses and donkeys in Algeria: detection of a novel Cryptosporidium hominis subtype family (Ik) in a horse. Vet. Parasitol. 208 (3-4), 135–142. doi: 10.1016/j.vetpar.2015.01.007
Langkjaer, R. B., Vigre, H., Enemark, H. L., Maddox-Hyttel, C. (2007). Molecular and phylogenetic characterization of Cryptosporidium and Giardia from pigs and cattle in Denmark. Parasitology. 134 (3), 339. doi: 10.1017/S0031182006001533
Li, F., Su, J., Chahan, B., Guo, Q., Wang, T., Yu, Z., et al. (2019). Different distribution of Cryptosporidium species between horses and donkeys. Infect. Genet. Evol. 75, 103954. doi: 10.1016/j.meegid.2019.103954
Lin, L., Chu, H. (2018). Quantifying publication bias in meta-analysis. Biometrics. 74 (3), 785–794. doi: 10.1111/biom
Liu, A., Zhang, J., Zhao, J., Zhao, W., Wang, R., Zhang, L. (2015). The first report of Cryptosporidium andersoni in horses with diarrhea and multilocus subtype analysis. Parasit Vectors. 8, 483. doi: 10.1186/s13071-015-1102-0
Lu, Q. B., Qiu., S. X., Ru, B. R. (2008). Epidemiological investigation of cryptosporidiosis in dairy calves in some prefectures of henan province. Vet. Sci. China 2008 (3), 261–267.
Lv, X. Q., Qin, S. Y., Lyu, C., Leng, X., Zhang, J. F., Gong, Q. L. (2021). A systematic review and meta-analysis of Cryptosporidium prevalence in deer worldwide. Microb. Pathog. 157, 105009. doi: 10.1016/j.micpath.2021.105009
Majewska, A. C., Solarczyk, P., Tamang, L., Graczyk, ,. T. K. (2004). Equine Cryptosporidium parvum infections in western Poland. Parasitol. Res. 93 (4), 274–278. doi: 10.1007/s00436-004-1111-y
Majewska, A. C., Werner, A., Sulima, P., Luty, T. (1999). Survey on equine cryptosporidiosis in Poland and the possibility of zoonotic transmission. Ann. Agric. Environ. Med. 6 (2), 161–165.
Mittal, S., Sharma, M., Chaudhary, U., Yadav, A. (2014). Comparison of ELISA and microscopy for detection of Cryptosporidium in stool. J. Clin. Diagn. Res. 8 (11), DC07–DC08. doi: 10.7860/JCDR/2014/9713.5088
Moriarty, E. M., Weaver, L., Sinton, L. W., Gilpin, B. (2012). Survival of Escherichia coli, Enterococci and Campylobacter jejuni in Canada goose faeces on pasture. Zoonoses Public Health 59 (7), 490–497. doi: 10.1111/zph.12014
Ni, H. B., Gong, Q. L., Zhao, Q., Li, X. Y., Zhang, X. X. (2020). Prevalence of Haemophilus parasuis "Glaesserella parasuis" in pigs in China: A systematic review and meta-analysis. Prev. Vet. Med. 182, 105083. doi: 10.1016/j.prevetmed.2020.105083
Olson, M. E., Thorlakson, C. L., Deselliers, L., Morck, D. W., McAllister, ,. T. A. (1997). Giardia and Cryptosporidium in Canadian farm animals. Vet. Parasitol. 68 (4), 375–381. doi: 10.1016/s0304-4017(96)01072-2
Page, M. J., McKenzie, J. E., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 74 (9), 790–799. doi: 10.1136/bmj.n71
Perrucci, S., Buggiani, C., Sgorbini, M., Cerchiai, I., Otranto, D., Traversa, D. (2011). Cryptosporidium parvum infection in a mare and her foal with foal heat diarrhoea. Vet. Parasitol. 182 (2-4), 333–336. doi: 10.1016/j.vetpar.2011.05.051
Qi, M., Zhang, L. X. (2018). Advance of research on Cryptosporidium and cryptosporidiosis of equine. Chin. J. Zoonoses. 34 (3), 267–271.
Qi, M., Zhou, H., Wang, H., Wang, R., Xiao, L., Arrowood, M. J., et al. (2015). Molecular identification of cryptosporidium spp. and Giardia duodenalis in grazing horses from xinjiang, China. Vet. Parasitol. 209 (3-4), 169–172. doi: 10.1016/j.vetpar.2015.02.030
Raue, K., Heuer, L., Böhm, C., Wolken, S., Epe, C., Strube, C. (2017). 10-year parasitological examination result to 2012) of faecal samples from horses, ruminants, pigs, dogs, cats, rabbits and hedgehogs. Parasitol. Res. 116 (12), 3315–3330. doi: 10.1007/s00436-017-5646-0
Ren, G. J., Wang, X. T., Jing, S., Song, J. K., Zhao, G. H. (2018). Infection status of cryptosporidium spp.in post-weaned qinchuan calves in partial regions of shanxi province. Chin. J. Vet. Sci. 38 (7), 1355–1358.
Robinson, G., Chalmers, R. M. (2020a). Cryptosporidium diagnostic assays: Microscopy. Methods Mol. Biol. 2052, 1–10. doi: 10.1007/978-1-4939-9748-0_1
Robinson, G., Elwin, K., Chalmers, R. M. (2020b). Cryptosporidium diagnostic assays: Molecular detection. Methods Mol. Biol. 2052, 11–22. doi: 10.1007/978-1-4939-9748-0_2
Rodríguez, D. C., Pino, N., Peñuela, G. (2012). Microbiological quality indicators in waters of dairy farms: Detection of pathogens by PCR in real time. Sci. Total Environ. 427-8, 314–318. doi: 10.1016/j.scitotenv.2012.03.052
Skerrett, H. E., Holland, C. V. (2001). Asymptomatic shedding of Cryptosporidium oocysts by red deer hinds and calves. Vet. Parasitol. 94 (4), 239–246. doi: 10.1016/s0304-4017(00)00405-2
Snyder, S. P., England, J. J., McChesney, A. E. (1978). Cryptosporidiosis in immunodeficient arabian foals. Vet. Pathol. 15 (1), 12–17. doi: 10.1177/030098587801500102
Song, Y., Li, W., Zhong, Z. J., Peng, J. L., Xie, N., Huang, X. M. (2017). Investigation and identification of Cryptosporidium from animals in Guiyang Zoo. Vet. Sci. China 47 (3), 295–300. doi: 10.16656/j.issn.1673-4696.2017.03.004
Sterk, A., Schijven, J., de, Roda, Husman, A. M., de, Nijs, T. (2016). Effect of climate change on runoff of Campylobacter and Cryptosporidium from land to surface water. Water Res. 95, 90–102. doi: 10.1016/j.watres.2016.03.005
Veronesi, F., Passamonti, F., Cacciò, S., Diaferia, M., Piergili, Fioretti, D. (2010). Epidemiological survey on equine Cryptosporidium and Giardia infections in Italy and molecular characterization of isolates. Zoonoses Public Health 57 (7-8), 510–517. doi: 10.1111/j.1863-2378.2009.01261.x
Wagnerová, P., Sak, B., McEvoy, J., Rost, M., Sherwood, D., Holcomb, K., et al. (2016). Cryptosporidium parvum and Enterocytozoon bieneusi in American mustangs and chincoteague ponies. Exp. Parasitol. 162, 24–27. doi: 10.1016/j.exppara.2015.12.004
Wang, B. J. (2009). Use comparative methods to fully grasp climate knowledge. Test questions Res. 36), 38–46.
Wang, H. (2015). Molecular epidemiology of three intestinal protozoas on livestock in henan province. China Agric. Univ 8, 1–106.
Wagnerová, P., Sak, B., McEvoy, J., Rost, M., Matysiak, A. P., Ježková, J., et al. (2015). Genetic diversity of Cryptosporidium spp. including novel identification of the Cryptosporidium muris and Cryptosporidium tyzzeri in horses in the Czech Republic and Poland. Parasitol. Res. 114 (4), 1619–1624. doi: 10.1007/s00436-015-4353-y
Wang, W. (2020). Investigation of intestinal parasites infection in donkey in parts of xinjiang [D]. Tarim Univ.
Wang, Y. L., Cui, B., Jian, F. C., Zhang, L. X., Ning, C. S., Dong, H. P., et al. (2008). Epidemiological investigation of ovine cryptosporidiosis in henan province. Vet. Sci. China. 2), 160–164.
Wang, W., Gong, Q. L., Zeng, A., Li, M. H., Zhao, Q., Ni, H. B. (2021). Prevalence of Cryptosporidium in pigs in China: A systematic review and meta-analysis. Transbound Emerg. Dis. 68 (3), 1400–1413. doi: 10.1111/tbed.13806
Wei, X. Y., Gao, Y., Lv, C., Wang, W., Chen, Y., Zhao, Q., et al. (2021a). The global prevalence and risk factors of Toxoplasma gondii among foxes: A systematic review and meta-analysis. Microb. Pathog. 150, 104699. doi: 10.1016/j.micpath.2020.104699
Wei, X. Y., Gong, Q. L., Zeng, A., Wang, W., Wang, Q., Zhang, X. X. (2021b). Seroprevalence and risk factors of Toxoplasma gondii infection in goats in China from 2010 to 2020: A systematic review and meta-analysis. Prev. Vet. Med. 186, 105230. doi: 10.1016/j.prevetmed.2020.105230
Wei, Z. L. (2020). Investigation of intestinal parasites infection in racehorses in parts of China. Tarim Univ. 11, 1–52. doi: 10.27708/d.cnki.gtlmd.2020.000054
Xiao, L., Herd, ,. R. P. (1994). Epidemiology of equine Cryptosporidium and Giardia infections. Equine Vet. J. 26 (1), 14–17. doi: 10.1111/j.2042-3306.1994.tb04323.x
Xie, C. X., Machado, G. C. (2021). Clinimetrics: Grading of recommendations, assessment, development and evaluation (GRADE). J. Physiother. 67 (1), 66. doi: 10.1016/j.jphys.2020.07.003
Xue, N. Y. (2021). Molecular epidemiological investigation and genotype analysis of four kinds of intestinal protozoa in longjiang wagyu cattle in heilongjiang province. Heilongjiang Bayi Agric. Univ 9, 1–74.
Young, I., Smith, B. A., Fazil, A. (2015). A systematic review and meta-analysis of the effects of extreme weather events and other weather-related variables on Cryptosporidium and Giardia in fresh surface waters. J. Water Health 13 (1), 1–17. doi: 10.2166/wh.2014.079
Zeng, A., Gong, Q. L., Wang, Q., Wang, C. R., Zhang, X. X. (2020). The global seroprevalence of toxoplasma gondii in deer from 1978 to 2019: A systematic review and meta-analysis. Acta Trop. 208, 105529. doi: 10.1016/j.actatropica.2020.105529
Zhang, B. L. (2018). Main trends of contemporary world economic development. Forum Leadership Sci. 2018 (8), 65–77.
Zhang, Q. Y. (2021). Dynamic investigation of Cryptosporidium infection in foals in parts of xinjiang. Tarim Univ 8, 1–50.
Zhang, Q., Zhang, Z., Ai, S., Wang, X., Zhang, R., Duan, Z. (2019). Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis from animal sources in the Qinghai-Tibetan Plateau Area (QTPA) in China. Comp. Immunol. Microbiol. Infect. Dis. 67, 101346. doi: 10.1016/j.cimid.2019.101346
Zhang, L. X., Jiang, J. S. (2001). Research progress of Cryptosporidium and cryptosporidiosis. Acta Parasitol. Et Med. Entomologica Sinica. 2001 (3), 184–192.
Zhang, Y. X., Zhou, M. J., Yu, Z. J., Chen, S. Y., Wang, L. Y., Yu, Y. L. (2017). Investigation on infection of Cryptosporidium in domestic dogs of Beijing. Prog. Vet. Med. 38 (6), 120–123.
Zhou, H. (2015). A Survey of Intestinal Parasites in Donkeys of animal-exchange market in Jiaxiang County, Shandong and Multilocus Sequence Typing of Cryptosporidium Donkey Genotype. Henan Agric. Univ. 7, 1–59.
Keywords: Cryptosporidium, Equus, meta-analysis, prevalence, zoonotic diseases
Citation: Li X-M, Geng H-L, Wei Y-J, Yan W-L, Liu J, Wei X-Y, Zhang M, Wang X-Y, Zhang X-X and Liu G (2022) Global prevalence and risk factors of Cryptosporidium infection in Equus: A systematic review and meta-analysis. Front. Cell. Infect. Microbiol. 12:1072385. doi: 10.3389/fcimb.2022.1072385
Received: 17 October 2022; Accepted: 08 November 2022;
Published: 25 November 2022.
Edited by:
Mingming Liu, Hubei University of Arts and Science, ChinaCopyright © 2022 Li, Geng, Wei, Yan, Liu, Wei, Zhang, Wang, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Liu, bGl1bGlhbmc5OTgzQDE2My5jb20=; Gang Liu, Z2FuZ2xpdUBxYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.