- Department of Molecular Immunology, Ruhr-University Bochum, Bochum, Germany
Infections caused by Gram-negative pathogens pose a major health burden. Both respiratory and gastrointestinal infections are commonly associated with these pathogens. With the increase in antimicrobial resistance (AMR) over the last decades, bacterial infections may soon become the threat they have been before the discovery of antibiotics. Many Gram-negative pathogens encode virulence-associated Type III and Type IV secretion systems, which they use to inject bacterial effector proteins across bacterial and host cell membranes into the host cell cytosol, where they subvert host cell functions in favor of bacterial replication and survival. These secretion systems are essential for the pathogens to cause disease, and secretion system mutants are commonly avirulent in infection models. Hence, these structures present attractive targets for anti-virulence therapies. Here, we review previously and recently identified inhibitors of virulence-associated bacterial secretions systems and discuss their potential as therapeutics.
Introduction
The development and spread of antimicrobial resistance (AMR) are among the biggest challenges for global health (Jee et al., 2018; Zhang et al., 2022). With the microbiota serving as a reservoir for resistance genes that can be passed on to pathogens via horizontal gene transfer, multi-resistant pathogens emerge in ever greater numbers (Kent et al., 2020; Hofer, 2022). As antibiotics target bacterial growth and survival mechanisms, this leads to an increase in selective pressure which will only aid the development of new resistance mutations (Martinez and Baquero, 2000; Perry and Wright, 2014). Furthermore, there is an increase in the prescription of last-resort antibiotics (Osei Sekyere, 2016; Klein et al., 2018). Their increased use will lead to further spread of resistance and limit treatment options for bacterial infections. In 2019, an estimated 1.2 million deaths globally were associated with antibiotic-resistant or multi-resistant bacterial infections (Antimicrobial Resistance, 2022). For Europe alone, this attributed to a cost of 1.5 billion Euros in healthcare-associated expenses and productivity loss (Anderson et al., 2019). Furthermore, only 17 new antimicrobials have been approved for use in the past decade (Chahine et al., 2022).
Due to the increase in AMR and the speed at which resistances to new antibiotics arise, research has shifted some of its focus to anti-virulence strategies, which interfere with virulence properties such as toxin production, quorum sensing or adhesion, thus preventing pathogenesis. Here, one of the aims is to identify so-called pathoblockers, compounds that target bacterial virulence factors without affecting the growth or survival of either the pathogen or the microbiota. This strategy renders bacteria avirulent but does not kill them, limiting survival pressure. Moreover, avirulent bacteria either cannot colonize or may be recognized and cleared by the immune system (Keyser et al., 2008; Duncan et al., 2012; Mühlen and Dersch, 2016; Hotinger et al., 2021). Furthermore, many virulence factors at which pathoblockers are targeted are exposed on the bacterial cell surface, bypassing the expulsion by bacterial efflux pumps (Smith and Romesberg, 2007).
Bacterial virulence factors include toxins, adhesins, invasins, secretion systems, and translocated proteins (effectors) essential for adherence, colonization, invasion, and disease establishment (Rasko and Sperandio, 2010; Mühlen and Dersch, 2016; Fleitas Martinez et al., 2019). Eleven different bacterial secretion systems have been described to date (Abrusci et al., 2014; Green and Mecsas, 2016; Grossman et al., 2021; Palmer et al., 2021). Type III and Type IV secretion systems (T3SSs and T4SSs, respectively) are commonly found in Gram-negative pathogenic bacteria and are associated with bacterial pathogenesis in eukaryotic hosts (Coburn et al., 2007; Keyser et al., 2008; Baron, 2010). They translocate effector proteins from the bacteria into the host cell cytosol. The genes encoding for T3- and T4SSs are mainly found in pathogenic bacteria and are usually located within mobile genetic elements associated with virulence, such as pathogenicity islands or plasmids. T3SSs are found in more than 25 species of Gram-negative pathogens, including pathogenic E. coli, Salmonella, Chlamydia, and Pseudomonas (Cornelis, 2006; Coburn et al., 2007; Cornelis, 2010). T4SSs are associated with virulence in pathogens such as Legionella, Neisseria, Helicobacter, and Coxiella (Cascales and Christie, 2003). Virulence-associated T3- and T4- secretion systems can also be found in plant pathogens including Pseudomonas syringae, Agrobacterium tumefaciens and Xanthomonas spp. (Hueck, 1998; Cascales and Christie, 2003; Mansfield et al., 2012). However, the focus of this review will be on human pathogenic bacteria. Both, T3- and T4SSs span the inner and outer bacterial membrane and protrude from the bacterial surface. Upon contact with a host cell, a pore is formed in the host cell membrane at the tip of the secretion system, which creates a connection from the pathogen to the host cell, enabling it to translocate effector proteins directly into the host cell cytosol, where they subvert cell signaling pathways in favor of bacterial survival and persistence (Dean, 2011; Green and Mecsas, 2016; Bienvenu et al., 2021).
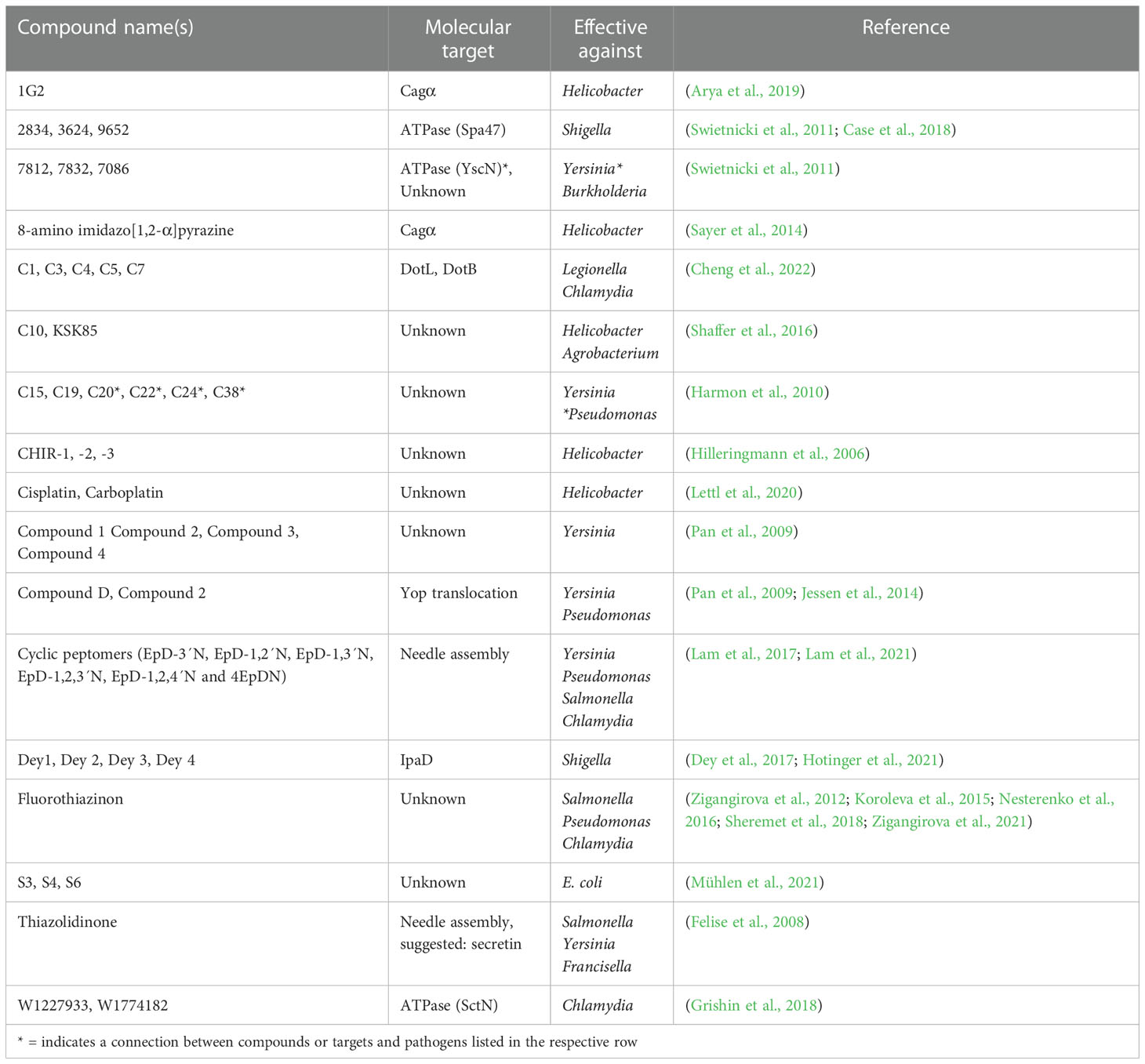
T3SSs encoded by Gram-negative pathogens play a central role in virulence (Coburn et al., 2007; Cornelis, 2010), making them promising targets for pathoblockers (Keyser et al., 2008; Duncan et al., 2012; Beckham and Roe, 2014; Mühlen and Dersch, 2016; Boudaher and Shaffer, 2019; Hotinger et al., 2021). They are evolutionarily related and therefore highly similar to flagellar T3SSs and consist of more than 20 proteins (Hueck, 1998; Cornelis, 2010; Abby and Rocha, 2012; Deng et al., 2017). T3SSs can be divided into cytosolic components, the basal body, and the needle. The cytosolic components include an ATPase required for unfolding effector proteins and powering their translocation. The cytosolic components are located underneath the basal body, which consists of an inner and an outer ring integrated into the inner and outer bacterial membrane, respectively. The needle complex consists of a base that is integrated into the bacterial membrane and a needle-shaped extension that protrudes from the bacterial cell (Figure 1A). In some instances, such as the T3SSs of pathogenic E. coli, the needle is followed by a filament, which extends from the former. The needle or filament ends in the tip complex, which is involved in sensing eukaryotic cells. Once in contact with a host cell, the proteins that make up the translocon are translocated through the needle and inserted into the host cell membrane where they form a pore (Dey et al., 2019). As the structures and functions of the secretion systems show high conservation between the different strains of bacteria, so do their synthesis and assembly (Notti and Stebbins, 2016; Condry and Nilles, 2017; Deng et al., 2017; Horna and Ruiz, 2021; Jenkins et al., 2022).
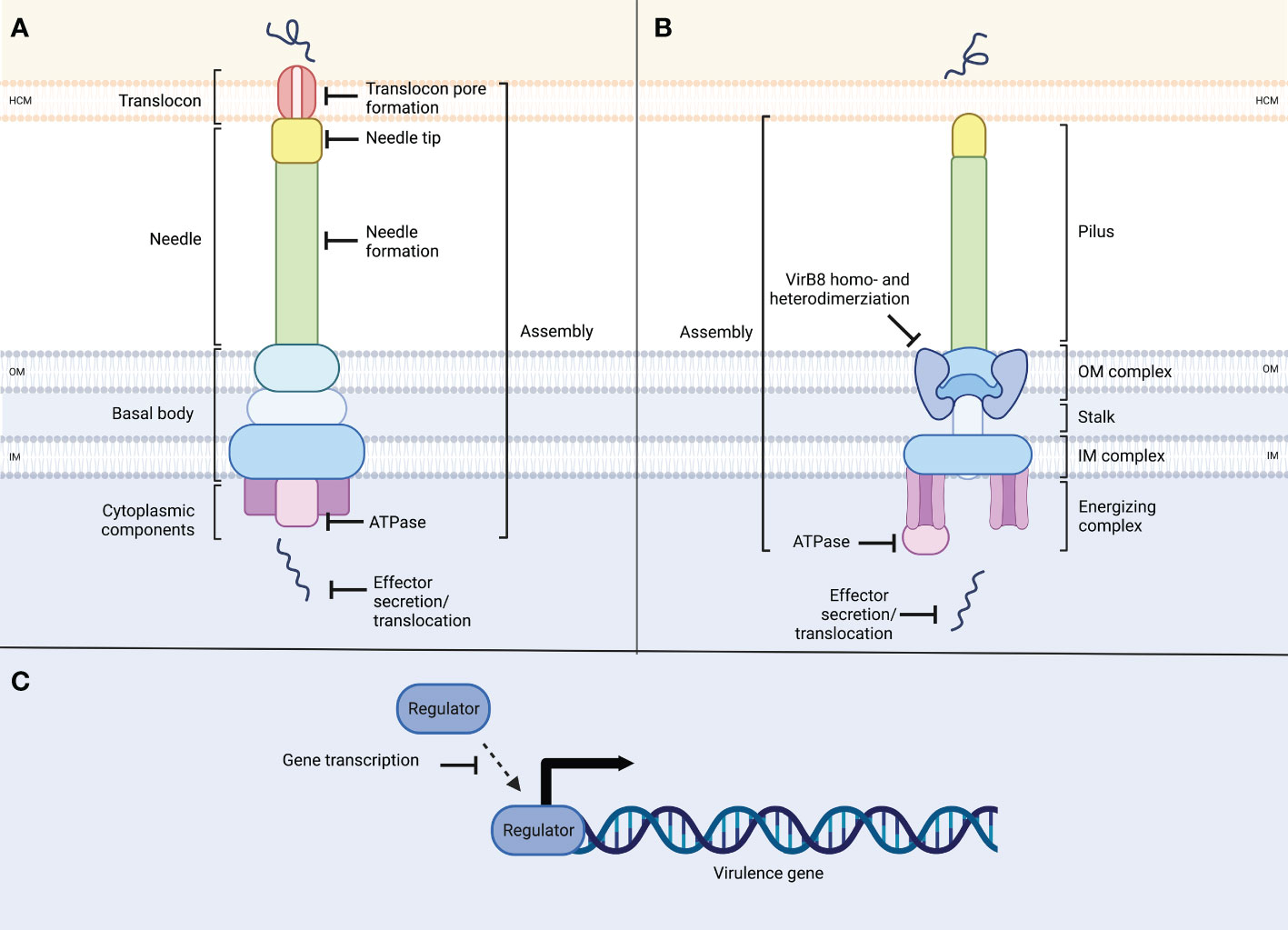
Figure 1 Schematic overview of the structure of type III and type IV secretion systems and potential inhibitor targets. (A) T3SSs consist of ring-shaped structures which form the basal body, spanning the inner (IM) and outer membrane (OM) of the bacterium. Cytoplasmic components, including the ATPase complex power needle assembly as well as the translocation of effector proteins. The needle ends in the needle tip, and the translocon forms a pore within the host cell membrane (HCM) through which unfolded effector proteins are translocated into the host cell. T3SSs target sites include secretion system assembly, needle or needle tip formation, translocon pore formation, ATPase function or effector protein secretion/translocation. (B) T4SSs consists of ring-like structures in the outer and inner membranes, which are connected through the stalk. At the inner membrane complex, an ATPase complex is located, providing energy for assembly and protein translocation. The pilus connects to the host cell membrane through which unfolded effector proteins are secreted into the host cell. Targets in T4SSs are secretion system assembly, VirB8 dimerization, ATPase function and effector secretion. (C) Additionally, inhibition can target virulence regulators, interfering with virulence gene or secretion system transcription.
Similar to T3SSs, T4SSs are functionally and structurally diverse (Cascales and Christie, 2003; Fronzes et al., 2009; Bhatty et al., 2013; Low et al., 2014). They are found in Gram-negative and Gram-positive bacteria and can be separated into minimized and expanded T4SSs (Grohmann et al., 2018; Costa et al., 2021; Liu et al., 2022; Sheedlo et al., 2022). They consist of three large groups: bacterial conjugation systems, DNA uptake and release systems, and effector translocators (Fronzes et al., 2009; Wallden et al., 2010; Sgro et al., 2019). The latter subgroup includes the virulence-associated T4SSs used by bacterial pathogens to translocate effector proteins or protein-DNA complexes into host cells to promote their proliferation and survival inside host cells (Cascales and Christie, 2003). The T4SS itself consists of the inner membrane complex spanning the bacterial inner membrane and the outer membrane core complex, which are connected by a stalk. Cytoplasmic components of the inner membrane complex include energy-generating ATPases. A pilus extends from the outer membrane core complex into the extracellular space and enhances protein and nucleic acid transport to the target cells (Figure 1B). Several components are conserved among the T4SSs from different pathogens. However, there are also species-specific features, including variations in the number of components and differences in the size and symmetries of the complexes (Low et al., 2014). Thus far, the structures of four T4SS have been identified, including the Vir, Cag, and Dot/Icm T4SS families (Cascales and Christie, 2003; Vincent et al., 2006; Wallden et al., 2010; Sgro et al., 2019; Sheedlo et al., 2022). The Vir secretion system belongs to the minimized T4SSs. First described in Helicobacter pylori, it has also been identified in Brucella and Bartonella (Cascales and Christie, 2003). The Vir T4SS can further be found in the plant pathogen A. tumefaciens (Cascales and Christie, 2003). It is composed of 12 subunits, which are VirB1 to VirB11 and VirD4 (Low et al., 2014). A key component of the Vir T4SS is the protein VirB8. To fulfill its function in system assembly, it dimerizes with either VirB8 or VirB10, forming the inner membrane channel of the T4SS together with VirB6 (Paschos et al., 2011; Sivanesan and Baron, 2011). Furthermore, the complex consists of the pilus, which is composed of a major and minor subunit, and the outer membrane complex. Three ATPases (VirB4, VirB11, and VirD4) assist in the assembly of this system (Savvides et al., 2003; Low et al., 2014). Expanded T4SSs are found in the human pathogens H. pylori, Legionella pneumophila, and Coxiella burnetii (Cascales and Christie, 2003; Nagai and Kubori, 2011). First described in H. pylori which translocates the oncogenic effector protein CagA, this family of secretion systems is called the Cag T4SS family (Segal et al., 1998; Fischer, 2011; Cover et al., 2020). The Cag T4SS is a multiprotein complex composed of 20 different proteins. Like the Vir T4SS, it spans the inner and the outer membrane and contains three putative ATPases, essential for CagA secretion (Fischer, 2011; Chung et al., 2019; Hu et al., 2019). The Dot/Icm system of L. pneumophila is one of the best-studied T4SSs. It is composed of at least 27 components that assemble into different subcomplexes, including the outer membrane complex, a periplasmic ring, and the inner membrane complex. A channel at the center of the stalk connects the outer and inner membrane complex. Cytosolic ATPases (DotL, DotB, DotO) provide energy for protein translocation through the T4SS (Chetrit et al., 2018; Ghosal et al., 2019).
The virulence-associated T3SS and T4SSs have been at the center of many high-throughput screens in an attempt to identify inhibitors that block the pathogenicity mediated by translocated effector proteins (Baron, 2010; Duncan et al., 2012; Mühlen and Dersch, 2016; Boudaher and Shaffer, 2019; Pendergrass and May, 2019; Hotinger et al., 2021). Many studies used known secretion system-induced virulence phenotypes, such as secretion system-mediated hemolysis, translocation of effector proteins by effector-reporter fusion proteins, effector secretion, or effector-mediated outcomes such as NF-κB induction using reporter gene fusions as the basis of their screens (Beckham and Roe, 2014; Fasciano et al., 2019). Particularly in recent years, there has been a shift toward structure-based computational predictions. Furthermore, several previously identified inhibitors have been modified in attempts to increase their potency, permeability, or target specificity. As the T3SSs of Gram-negative pathogens show very high architectural homology, the aim is to improve inhibitory compounds to identify novel substances with low toxicity, a broad target spectrum, and high selectivity for T3SS-positive pathogens. While many studies have aimed at developing or identifying inhibitors targeting the T3SS of bacteria, fewer have focused on pathogens using T4SSs (Boudaher and Shaffer, 2019). This is unfortunate because bacterial pathogens expressing T4SSs also cause a diverse range of severe diseases. In the case of H. pylori, a link to the development of gastric cancer has even been proven (Ishaq and Nunn, 2015; Norwood et al., 2022). Furthermore, as T4SSs were shown to translocate not only effector proteins but also DNA, inhibiting these systems may also aid in reducing the transfer and spread of acquired resistance genes within a community (Juhas et al., 2008; Boudaher and Shaffer, 2019). While no natural inhibitors were found to date that target T4SSs, several chemical compounds have been identified (Boudaher and Shaffer, 2019).
Several processes present themselves as potential targets for anti-virulence strategies: synthesis of needle components, assembly of the secretion system, interaction with the host cell, and secretion/translocation of the substrates (Figure 1). Due to the high similarity between each family of secretion systems, inhibitors can likely be found that target not only one but also several different pathogens at once. Furthermore, as only pathogenic bacteria express these secretion systems, non-pathogenic bacteria will not be targeted. Additionally, as secretion system inhibitors are selected for lack of impact on bacterial survival, the selective pressure to develop resistance should remain low.
Salicylidene acylhydrazides
Salicylidene acylhydrazides (SAHs) are a group of potent chemical inhibitors active against the T3- and T4SSs of several pathogens, including Salmonella, Pseudomonas, Chlamydia, and Brucella. They were among the earliest identified secretion system inhibitors and remain the best-studied compound family to date (Table 1) (Duncan et al., 2012; Beckham and Roe, 2014; Mühlen and Dersch, 2020).
SAHs and their derivatives were first described as inhibitors of the Yersinia T3SS by Kauppi et al. (2003). The authors used a transcriptional fusion of the yopE gene (encoding the effector protein YopE) and luciferase and identified 30 molecules that significantly reduced luciferase expression. Compounds 2 to 4 decreased luciferase activity without or with only minor effects on bacterial growth. Furthermore, when grown in medium containing these compounds, Yop secretion was decreased in a dose-dependent manner. While being the most potent, Compound 2, an SAH, also affected bacterial motility. Compound 2, later renamed INP0007, was further investigated and shown to specifically block Yop secretion and translocation by Yersinia without adverse effects on bacterial or cell viability (Kauppi et al., 2003; Nordfelth et al., 2005).
Hudson et al. assessed the SAHs INP0007 and INP0403 as inhibitors of the Salmonella T3SS-1 (SPI-1). Both compounds inhibited effector secretion and SPI-1-mediated hemolysis in a dose-dependent manner. In vitro invasion assays with pre-incubated bacteria further showed a significant decrease in the number of intracellular bacteria in the presence of INP0007 (Hudson et al., 2007). Moreover, Negrea et al. tested nine SAHs (INP0007, INP0010, INP0400, INP0401, INP0402, INP0403, INP0404, INP0405, and INP0406) in Salmonella infections. All compounds reduced SPI-1-mediated invasion and effector secretion, and INP0400 and INP0007 also inhibited intracellular replication. All compounds, except INP0010, also inhibited the secretion of a SipB-β-lactamase fusion protein. In INP0010-treated cells, the fusion protein was expressed but not secreted. Furthermore, transcriptional reporter fusions suggested that this compound interfered with the transcription of SPI-1. INP0404 or INP0405 were shown to affect Salmonella motility (Negrea et al., 2007). Interestingly, INP0403 was later shown to reduce the transcription of genes involved in T3-secretion and iron acquisition and that its inhibitory activity was reversed when iron was supplied exogenously (Layton et al., 2010).
As the T3SSs of Yersinia and Chlamydia are highly conserved (Hsia et al., 1997), Wolf et al. investigated a potential role for INP0007 in inhibiting the T3SS of obligate intracellular Chlamydia (Wolf et al., 2006). Here, treatment with INP0007 inhibited C. trachomatis development in a dose-dependent manner, while bacterial viability was unaffected. Moreover, the bacteria remained metabolically active but could not divide, and their differentiation from reticulate to elementary bodies was inhibited. Addition of the inhibitor after infection blocked the conversion from established reticulate bodies back to elementary bodies in in vitro infection experiments. These effects were reversible when the inhibitor was removed (Nordfelth et al., 2005).
Additional well-characterized SAHs targeting Chlamydia are INP0010, INP0400, and INP0341. They were demonstrated to inhibit the C. pneumoniae developmental cycle (INP0010) and intracellular replication of C. pneumoniae and C. trachomatis in vitro (Muschiol et al., 2006; Bailey et al., 2007; Slepenkin et al., 2007). Treating epithelial cells with INP0010 significantly reduced intracellular levels of the effector proteins IncB and IncC. Transcription of incB was increased upon treatment, while incC transcription remained unaltered, indicating that INP0010 affected effector translocation but not gene expression (Bailey et al., 2007). While treatment with INP0400 at the time of infection partially blocked the entry of elementary C. trachomatis bodies into host cells, treatment with INP0400 during the mid-cycle reduced translocation of the effector protein IncA (Muschiol et al., 2006). INP0341, on the other hand, completely inhibited the expression of the T3SS chaperone LcrH-1 and the structural protein CopB in C. trachomatis during the mid- and late developmental stages (Slepenkin et al., 2003; Slepenkin et al., 2007). Consequently, downregulation of incA but not incC and incG could be observed. It was suggested that INP0341 works by sequestering iron within the host cell, inhibiting Chlamydia development within the host (Slepenkin et al., 2007). Further in vitro studies showed that this inhibitor affected neither essential members of the vaginal microbiome, nor the integrity of the vaginal mucosa, and showed only mild cytotoxicity. It was also observed that it could protect from vaginal Chlamydia infection in a mouse model (Slepenkin et al., 2011; Pedersen et al., 2014). After the initial in vivo experiments, the inhibitor was improved and formulated as a vaginal gel suitable for use in humans (Pedersen et al., 2014). Ur-Rehman et al. later investigated a library of SAHs for their effect on C. pneumoniae and C. trachomatis (Ur-Rehman et al., 2012) and chose ME0177 and ME0192 for further investigation due to their low toxicity. ME0192, the more potent compound, showed potent activity against intracellular chlamydial growth in vitro. However, its short half-life (< 1h) limits its application to topical treatment. Promisingly, the inhibitory effects of ME0192 were unaffected in the presence of vaginal simulant fluid or healthy vaginal microbiota. Hence, the effect of ME0192 was subsequently assessed in vivo by topical application before, during, and five days after C. trachomatis infection. Here, a significant reduction in the number of colonized mice and bacterial burden could be observed. ME0177, on the other hand, had a concentration-dependent effect in vivo, which lasted even after its removal, making it a promising compound for systemic treatment (Ur-Rehman et al., 2012).
INP0341 also inhibited the expression and secretion of the Pseudomonas exotoxin ExoS and P. aeruginosa-mediated cytotoxicity in vitro. In in vivo infection with P. aeruginosa, topical treatment of infected mice could delay mortality although it was unable to prevent the systemic spread of infection (Uusitalo et al., 2017). Subsequent research investigated its inhibitory effect on P. aeruginosa infections of human corneal epithelial cells and in a murine keratitis model (Sharma et al., 2020). These studies confirmed that INP0341 inhibited P. aeruginosa-mediated cytotoxicity without affecting bacterial viability. Additionally, in vivo experiments showed that topical application of INP0341 to corneal scratches could inhibit bacterial growth and facilitate bacterial clearance (Sharma et al., 2020).
The SAHs INP0400, INP0402, INP0404, and INP0405 were also investigated in Shigella infections, where they were shown to inhibit T3SS-mediated effector secretion. Moreover, the ability of Shigella to invade epithelial cells and cause apoptosis in macrophages was impaired. The most substantial effect could be observed when the compounds were added to the bacterial growth cultures. Electron microscopy analysis of inhibitor-treated bacteria demonstrated that these compounds interfered with the assembly of the Shigella T3SS needle (Veenendaal et al., 2009).
Tree et al. on the other hand discovered that the SAHs INP0010, INP0031, INP0401, and INP0403 did not affect the T3SS of enterohemorrhagic E. coli (EHEC) directly but acted via regulators, such as the locus of enterocyte effacement (LEE) regulator ler (Tree et al., 2009). For the most part, the mechanisms of action of the SAH inhibitors remain unclear. Later, new classes of SAHs were created by Zambelloni et al., using INP0031 as a scaffold. The authors developed eight novel compounds by incorporating the imine moiety into two different hydrazine-containing heterocycles. Two of these, RCZ12 and RCZ20, displayed potent EHEC T3SS inhibition. In vitro pull-down assays identified the coiled-coil domain 1 of the effector protein and translocon component EspD as the binding site of these inhibitors (Zambelloni et al., 2017).
The suppression of T3SSs of different pathogens by SAHs suggests that they may affect a conserved factor, with direct effects on T3S, the T3SS, or regulatory proteins being discussed (Tree et al., 2009; Veenendaal et al., 2009; Wang et al., 2011). For INP0031, Wang et al. identified three potential target proteins, WrbA, Tpx, and FolX, which they confirmed in different bacterial pathogens, including E. coli, P. aeruginosa and Shigella. All of these proteins are known to contribute to the expression of virulence factors, including T3SSs and flagella (Wang et al., 2011) and Tpx and WrbA have also been attributed an important role in motility activation (Nachin et al., 2005). SAHs may, therefore, increase the activity of Tpx and WrbA and enhance their repressive effects on the T3SS. As the tested inhibitors did not affect expression levels of Tpx and WrbA, the authors proposed a direct interaction of the inhibitor with these proteins, resulting in their activation or stabilization. SAHs may further bind to several proteins important for cellular stress responses, which, in turn, impair the transcription, expression, and assembly of the T3SS (Wang et al., 2011).
B8I-1 to -5 are salicylidene acylhydrazides with structural similarities to broad-spectrum inhibitors of T3SSs (Smith et al., 2012). Interestingly, they were identified to target VirB8 of the Vir T4SS of Brucella abortus and did not show broad-spectrum activity against other types of T4SSs, suggesting high target specificity (Paschos et al., 2011). B8I-2 inhibited T4SS assembly by binding within a surface groove near the dimerization site in VirB8, thus preventing VirB8 dimerization by inhibiting conformational changes. Moreover, it reduced the levels of several VirB proteins, suggesting that inhibitor binding destabilized the VirB8 protein (Smith et al., 2012).
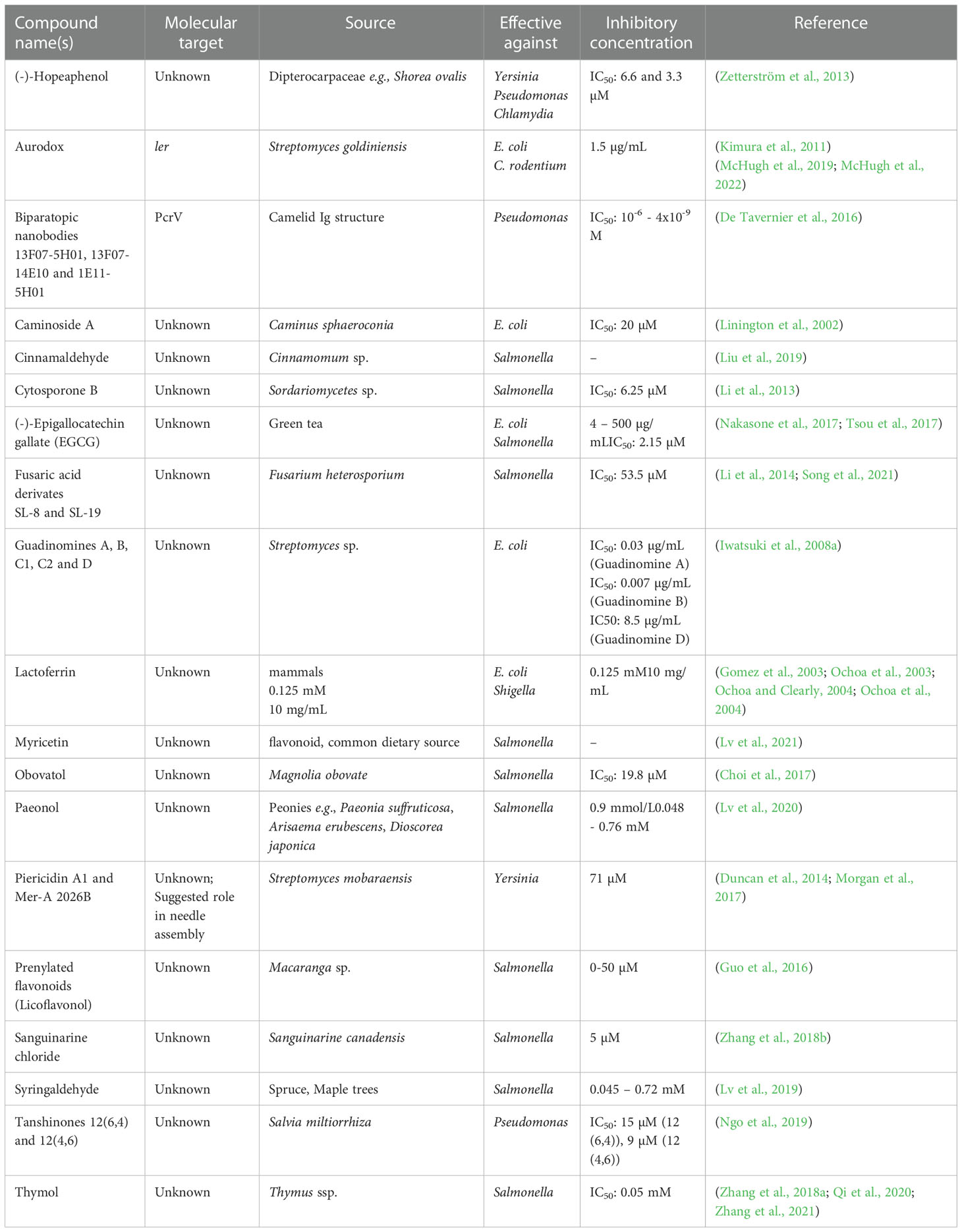
Other chemical secretion system inhibitors
Many high-throughput screens aimed at identifying secretion system inhibitors have used commercially-available chemical compound libraries. Some of these libraries also included well-characterized pharmacological substances. Various chemical substances have been identified as secretion system inhibitors in this way (Table 2).
Chemical compounds interfering with T3SSs
Felise et al. screened for small molecules inhibiting the Salmonella T3SS using different effector proteins fused to a phospholipase. In this screen, the authors obtained seven hits that reduced the phospholipase activity and secretion of effector proteins. However, only thiazolidinone did not affect the production of transcriptional regulators of the T3SS machinery. This compound inhibited the formation of needle complexes and thus blocked the T3SSs in Yersinia and Francisella. Additionally, it blocked the T2SS, but not the function of flagella in Francisella, suggesting the targeting of secretin, the only protein homolog shared between T2- and T3SSs (Felise et al., 2008).
In an early study, Pan et al. screened a chemical compound library for substances that reversed bacterial growth inhibition, which occurs in Y. pestis under T3 secretion conditions. Four inhibitory compounds were identified, designated Compound 1-4. All of these compounds showed a concentration-dependent inhibition of Yop effector secretion. Interestingly, Compounds 1 and 3 inhibited the secretion of Yops to varying degrees. Compounds 1, 3, and 4 only inhibited T3S-mediated cytotoxicity, while Compound 3 also inhibited the translocation of a YopE-β-lactamase fusion protein. Compounds 1 and 3 further inhibited T3-mediated secretion of the effector protein Tir from enteropathogenic E. coli (EPEC) (Pan et al., 2009). Jessen et al. tested Compound 2 and its isomer Compound D for their effect on Y. pestis (Jessen et al., 2014). They confirmed that Compound D also inhibited Yop secretion and additionally observed the inhibition of effector protein secretion by P. aeruginosa and Y. pseudotuberculosis. While the mechanisms remain unknown, it was suggested that Compound D inhibits Yop translocation via interaction with the translocon component YopD. Compound 2 appears to employ a different mechanism as, unlike Compound D, it was also active in the presence of the Yersinia Yop secretion inhibitor calcium. Unfortunately, both compounds were toxic to eukaryotic cells, and their modification will be necessary to improve safety (Jessen et al., 2014).
Six additional chemical compounds were identified, which block the translocation of a Y. pseudotuberculosis YopB-β-lactamase fusion protein into eukaryotic cells. In the study, C15, C19, C20, C22, C24, and C38 inhibited the YopB translocation without affecting the expression of T3SS components or effector proteins, formation of the T3SS, or the secretion of effectors. C20 was further shown to reduce the binding of Yersinia to host cells. Interestingly, all compounds except C15 and C19 also inhibited ExoS secretion from P. aeruginosa, as shown by the reduction of ExoS-mediated cell death. Therefore, the molecular targets of these compounds appear to be conserved between Y. pseudotuberculosis and P. aeruginosa (Harmon et al., 2010).
Zigangirova et al. developed a new small-molecule inhibitor, Fluorothiazinon (FT, previously called CL-55) (Zigangirova et al., 2012). FT was one of the first T3SS inhibitors which showed potential for therapeutic treatment of Salmonella infections (Nesterenko et al., 2016; Zigangirova et al., 2021). It was characterized by low toxicity, high stability, and therapeutic efficacy in animal models (Sheremet et al., 2018). FT inhibited the secretion of early Salmonella SPI-1 effectors without affecting intracellular bacterial growth (Zigangirova et al., 2021), while suppressing bacterial growth within macrophages. This suggested that FT inhibited not only SPI-1 but also Salmonella T3SS SPI-2, which is essential for intracellular survival and systemic spread (Zigangirova et al., 2021). In a mouse model of acute Salmonella infection, treatment with FT decreased the bacterial load and increased survival rates (Nesterenko et al., 2016; Sheremet et al., 2018). Further investigation in a murine model of systemic Salmonella infection showed that FT could suppress systemic spread when used as either therapeutic (seven days after infection) or preventative treatment (four days before infection) (Zigangirova et al., 2021). In a P. aeruginosa in vivo airway infection model, FT inhibited the secretion of the effector proteins ExoT and ExoY in a dose-dependent manner. Moreover, it was shown that the treatment could reduce tissue damage and prevent systemic spread and mortality (Sheremet et al., 2018). A phase II trial is currently underway, which includes FT as an active ingredient in the treatment of patients with urinary infections caused by P. aeruginosa (Zigangirova et al., 2021). FT was further shown to block the T3SS-mediated virulence of C. trachomatis in vitro. Upon treatment, the effector protein IncA was absent from chlamydial inclusions, whereas its transcription in bacteria was unimpaired, suggesting that FT affects the T3SS and not bacterial metabolism (Zigangirova et al., 2012). In a subsequent study, the inhibitory effect of FT was further studied in C. trachomatis-infected mice. Here, the number of bacteria in vaginal excretions was reduced upon infection, and a decrease in chlamydial DNA was observed in ovaries and uteri (Koroleva et al., 2015).
Schwochert et al. designed a library of synthetic cyclic peptomers (Schwochert et al., 2016) inspired by phepropeptin D produced by Streptomyces (Sekizawa et al., 2001), which Lam et al. used to screen for inhibitors of NF-κB activation in response to Yersinia infection. They showed that EpD-3’N, EpD-1,2N, EpD-1,3’N, EpD-1,2,3’N, and EpD-1,2,4’N strongly inhibited effector secretion by Yersinia and P. aeruginosa without affecting bacterial growth and with only low cytotoxicity in mammalian cells (Lam et al., 2017). The authors then developed isomers based on the cyclic peptomer EpD-1,2N. They observed that 4EpDN was more potent than its parental compound and that it inhibited the T3SSs of both, Yersinia and S. Typhimurium without affecting the transcription of T3SS genes. In addition, 4EpDN reduced the number of T3SS needles on the surface of Y. pseudotuberculosis, suggesting that it may specifically prevent needle assembly in a variety of pathogens (Lam et al., 2021). Because the compound reduced the localization of the inner ring protein YscD to the Yersinia inner cell membrane, it has been suggested that 4EpDN may interact with the lumen of the T3SS needle, affecting the secretion of large cargo proteins more than that of small cargo. Alternatively, it may disrupt the assembly of the T3SS needle subunit, thereby resulting in non-functional needles and impairing effector translocation (Lam et al., 2021).
Other small molecules inhibiting needle formation were identified by Dey et al., who performed a structure-based compound screen to identify molecules binding to the effector protein IpaD of Shigella (Dey et al., 2017). The authors found four small molecules with scaffolds based on quinoline, pyrrolidine aniline, hydroxyindole, and morpholinoaniline, later named Dey 1-4, respectively (Hotinger et al., 2021). After binding of the compounds to IpaD, conformational changes were induced, inhibiting translocon assembly by a yet unknown mechanism (Dey et al., 2017).
The effector protein Tir is essential for intimate attachment to enterocytes by EPEC and EHEC. Mühlen et al. recently described the inhibitory substances S3, S4, and S6, which reduced the translocation of a Tir-β−lactamase fusion protein, as well as Tir-mediated attaching and effacing (AE) lesion formation and effector-dependent cell detachment. None of the compounds affected the expression of T3SS components EspA, EspB, or EspD or that of the effector protein Tir. The three identified compounds belong to different chemical classes and varied in their ability to inhibit T3SS-mediated hemolysis, suggesting that they have different modes of action. As neither of the compounds induced the expression of Shiga toxin in a Shiga toxin reporter strain, they may be promising candidates for treating EHEC infections if developed further (Mühlen et al., 2021).
Chemical inhibitors targeting T4SSs
While T3SSs have been the focus of many inhibitor screens, few studies have aimed to identify substances, which can interfere with T4SS-mediated virulence. In 2016, Shaffer et al. identified two closely related small molecules, called C10 and KSK85, which impaired delivery of the H. pylori T4-effector CagA and peptidoglycan into host cells. KSK85 inhibited the formation of Cag secretion system-associated pili, while C10 impaired protein transport. Neither inhibitor showed any adverse effects on the viability of gastric epithelial cells or bacteria (Shaffer et al., 2016). A distinctive feature of these compounds was their activity against other T4SSs, such as the Vir T4SS of A. tumefaciens (Shaffer et al., 2016).
Lettl et al. screened a library of pharmaceutically-active compounds for their ability to inhibit CagA-β-lactamase translocation by H. pylori. They identified two potent platinum coordination complexes, cisplatin and carboplatin, which are known for their anti-tumorigenic functions (Jamieson and Lippard, 1999). These compounds were further investigated by measuring CagA translocation into eukaryotic cells upon H. pylori infection. Both compounds reduced translocation and, hence, the intracellular phosphorylation of CagA. The viability of H. pylori was not affected. Additionally, the levels of IL-8, usually increased upon infection, were reduced by cisplatin treatment in a dose-dependent manner as was the bacterial adherence to host cells in the presence of cisplatin. Interestingly, once H. pylori infection was established, the adherence could not be resolved. However, the translocation of CagA was still reduced, suggesting that cisplatin can rapidly interfere with and inhibit effector translocation (Lettl et al., 2020).
Using a high-throughput screen to identify compounds that interfere with the translocation of effector proteins from Legionella pneumophila into macrophages, Cheng et al. identified five compounds (C1, C3, C4, C5, and C7), which showed high efficiency in different assays that require a functional T4SS. These included β-lactamase-LidA translocation and intracellular replication in macrophages. While C4 was shown to reduce the levels of LidA-reporter protein translocation by L. pneumophila but not by Coxiella burnetii, compounds C1, C3, C5, and C7 reduced effector translocation from both bacteria. Additionally, none of the compounds showed any adverse effects on the growth of L. pneumophila or C. burnetii in culture. As the five compounds vary in molecular structure and chemical properties, the authors suggested they may have diverse targets (Cheng et al., 2022).
Chemical inhibitors of secretion system-powering ATPases
Secretion system-associated ATPases are required for powering the secretion of needle/pilus components and effector proteins into host cells. Swietnicki et al. used a structure-based screen for chemical compounds which could bind the Yersinia T3SS ATPase YscN. They identified the compounds 7812, 7832, and 7086. These compounds were confirmed to potently inhibit YscN in an in vitro ATPase assay and subsequently abolished the secretion of YopE into host cells upon infection. Furthermore, these compounds also inhibited the ATPase BsaS of the Burkholderia mallei T3SS, which shows sequence homology to YscN. Because the identified compounds were not or only slightly toxic to mammalian cells, they do not appear to interfere with the function of eukaryotic ATPases (Swietnicki et al., 2011).
In the H. pylori Cag T4SSs, the ATPase Cagα powers complex assembly, DNA unwinding and translocation (Hilleringmann et al., 2006). Hilleringmann et al. identified three inhibitors of Cagα ATPase activity, CHIR-1, -2, and -3. Due to its potency, CHIR-1 was used for further characterization. It inhibited CagA secretion by H. pylori in a dose-dependent manner, completely blocking its translocation after pre-incubation, suggesting it may interfere with the early stages of T4SS assembly. Additionally, pre-incubation of H. pylori with CHIR-1 before intragastric infection of mice decreased the number of infected animals as well as their colonization (Hilleringmann et al., 2006).
Sayer et al. later tested 8-amino imidazo[1,2-α]pyrazine derivates as inhibitors of Cagα. To characterize the inhibitory effects of these compounds, they purified Cagα from E. coli and tested it in an in vitro ATPase assay. The authors observed a partial inhibition of ATPase activity in the presence of Compounds 11 and 32. A prediction of the docking site of Compound 11 suggested that the inhibitor binds within the catalytic site (Sayer et al., 2014). However, the solubility of the compound was low and needed modification. To improve their selectivity for VirB11 ATPases, the authors later added a peptide sequence to the previously identified substances in an attempt to disrupt peptide-peptide interactions. Here, the inhibitory effect was increased in in vitro ATPase assays when the compound was linked to a PEG-carbamate linker (Sayer et al., 2021).
Further work focusing on Cagα used a fragment-based screening approach for molecules that bound to or stabilized the ATPase in an in vitro ATPase assay. The authors identified four fragments, of which 1G2 was the most potent. The binding of 1G2 to the ATPase led to alterations in Cagα conformation and appeared to interfere with Cagα hexamer formation. The inhibition appears to be partial since treatment of bacteria with 1G2 reduced CagA-induced IL-8 production in cells upon infection, but intracellular phosphorylation of CagA within the host cells remained unaffected (Arya et al., 2019).
Also using in silico docking simulations, Case et al. identified three inhibitors of the T3SS ATPase Spa47 of Shigella. These compounds (2834, 3624, and 9652) showed decreased ATPase activity in ATPase assays and suppressed effector protein secretion without significant effects on bacterial growth or cell viability. The study further indicated that the inhibitors did not act by disrupting Spa47 oligomers or by preventing T3SS formation (Case et al., 2018).
SctN is the ATPase powering the T3SS of C. trachomatis (Beeckman et al., 2008). Using structure-based virtual screening against modeled SctN, two compounds, W1227933 and W1774182, were identified, which bind to SctN, blocking the translocation of the effector protein IncA (Grishin et al., 2018). A significant decrease was observed in the number of intracellular C. trachomatis inclusions when cells were treated with W1227933 and W1774182 at the time of infection. Intracellular chlamydial survival was completely abolished at high concentrations. Both compounds showed low to medium concentration-dependent cytotoxicity in eukaryotic cells. As the exact structure of SctN is still unknown, Grishin et al. used homologous ATPases, such as the E. coli T3SS ATPase EscN and the Salmonella enterica flagellar ATPase FliI for modeling, suggesting that these ATPases may also be targeted (Grishin et al., 2018).
Natural compounds inhibiting virulence-associated secretion systems
While most antibiotics were derived from natural sources, most screens for secretion system inhibitors have used commercially-available chemical compound libraries. Lately, however, there has been a shift back to assessing the potential of natural compounds in targeting bacterial infections and a number of promising substances have been identified (Table 3).
Natural inhibitors isolated from animal sources
The glycolipid caminoside A was found in extracts of the marine sponge Caminus sphaeroconia and was the first T3SS inhibitor ever described. It decreased the secretion of the EPEC translocon component EspB into the supernatant without affecting bacterial growth or the secretion of other proteins (Linington et al., 2002).
One of the best-studied compounds is lactoferrin, which is widely represented in various body fluids such as milk, tears, and saliva and, thus, cytotoxicity is highly unlikely. Besides its bacteriostatic effect due to iron sequestration, it also decreased the virulence of several pathogens, including Salmonella, Shigella, and E. coli, by targeting their T3SS (Ochoa et al., 2003; Ochoa et al., 2004; Bessler et al., 2006; Ochoa and Cleary, 2009; Mosquito et al., 2010). Lactoferrin induced the degradation of the Shigella translocon proteins IpaB and IpaC (Gomez et al., 2003) and the E. coli tip and translocon proteins EspA, EspB, and EspD (Ochoa et al., 2004). Interestingly, it functions via two distinct mechanisms. By binding LPS, protein-protein interactions were disrupted, resulting in the instability of virulence proteins. Additionally, it directly degraded T3SS components through its intrinsic serine protease activity.
In addition, antibodies derived from camelids, so-called nanobodies, can also be used to inhibit the T3SS. De Tavernier et al. screened a large library of bivalent and biparatopic nanobodies for the functional inhibition of PcrV, which is an integral part of the P. aeruginosa T3SS, as it forms the needle tip (Goure et al., 2004; Goure et al., 2005; Sato and Frank, 2011). This screen yielded a group of nanobodies (13F07-5H01, 13F07-14E10, and 1E11-5H01) which inhibited P. aeruginosa-mediated cell death. In vivo mouse models of an acute, lethal P. aeruginosa lung infection showed 100% survival when the nanobodies were used as prophylactic treatment. 13F07-5H01 significantly improved the survival of infected mice under therapeutic conditions, when treatment started as late as 18 hours post-infection (De Tavernier et al., 2016).
Secretions system inhibitors derived from bacteria and fungi
In 2008, Iwatsuki et al. identified six inhibitory compounds produced in Streptomyces sp.: guadinomines A, B, C1, C2, and D, and guadinomic acid. All of these compounds exhibited dose-dependent inhibition of T3SS-mediated hemolysis by EPEC without any cytotoxic or antimicrobial effects (Iwatsuki et al., 2008a; Iwatsuki et al., 2008b).
Another T3SS inhibitor isolated from Streptomyces is the kirromycin derivative aurodox (Kimura et al., 2011). This linear polyketide compound is highly similar to kirromycin, from which it differs only in the methylation of its pyridone moiety (McHugh et al., 2022). Aurodox was first identified in a screen for inhibitors of T3SS-mediated hemolysis in EPEC and was subsequently shown to decrease the secretion of effector proteins EspB, EspF, and Map without affecting bacterial growth. Furthermore, aurodox protected mice from succumbing to a lethal dose of Citrobacter rodentium, the mouse homolog of EPEC (Kimura et al., 2011). Later, aurodox was shown to inhibit the T3SS and, hence, virulence in EHEC and EPEC by downregulating the transcription of the virulence master regulator, ler. As aurodox did not induce the expression of Shiga toxin in EHEC, the compound may be a promising treatment option for EHEC infections (McHugh et al., 2019).
Another well-studied natural compound is cytosporone B (Csn-B), which is produced by fungi of the class Sordariomycetes. Li et al. screened a library of cytosporone B analogs and identified eight compounds (Csn-B and seven analogs) as inhibitors of S. Typhimurium SPI-1. Four compounds inhibited the secretion of SPI-1 effector proteins, and three of these also interfered with Salmonella invasion into eukaryotic cells (Li et al., 2013). Csn-B was the most potent compound, inhibiting effector protein secretion in a dose-dependent manner without affecting the flagellar T3SS or adverse effects on bacterial cells. Interestingly, overexpression of the T3SS regulator HilD could counteract Csn-B inhibition, suggesting that the compound interferes with the HilD regulatory cascade. Indeed, Csn-B upregulated the transcription of hha and hns encoding Hha and H-NS, which bind to promoters of SPI-1 regulatory genes, including hilD to repress their transcription (Ellermeier et al., 2005; Olekhnovich and Kadner, 2007; Li et al., 2013).
In 2014, Duncan et al. reported the discovery and initial characterization of two new Streptomyces-derived T3SS-inhibitors, Piericidin A1 and its derivative Mer-A 2026B. In their screen, the authors showed that both compounds prevented T3SS-dependent activation of NF-κB by Y. pseudotuberculosis effector proteins. Both Piericidin A1 and Mer-A 2026B inhibited the translocation of YopM into eukaryotic cells in a dose-dependent manner. Furthermore, neither compound interfered with bacterial growth or showed toxicity when added to eukaryotic cells, suggesting inhibition of an early stage, such as T3SS needle assembly (Duncan et al., 2014) instead of blocking bacteria-host cell interactions. In a later study, Morgan et al. confirmed this hypothesis by observing a reduced number of needle complexes at the bacterial surface combined with a decreased T3SS formation and effector translocation in Piercidin A1-treated Yersinia (Morgan et al., 2017).
Li et al. initially identified the mycotoxin fusaric acid (FA) as an inhibitor of the Salmonella T3SS. FA inhibited the secretion of SPI-1 effectors and bacterial invasion into host cells without disrupting cell growth or cytotoxicity (Li et al., 2014). Later, Song et al. designed twenty-two diphenylsulfane derivatives based on FA. Among these, SL-8 and SL-19 possessed strong activities against the T3SS and bacterial invasion and showed an improved potency compared to FA. The inhibitory effect on SPI-1 was independent of regulatory genes and the assembly of the T3SS needle complex. Thus, the authors proposed that SL-8 and SL-19 influence the formation of the SicA/InvF-effector complex or other related proteins, which may act as classical transcriptional activators (Song et al., 2021).
Plant-based pathoblockers
(-)-Hopeaphenol is a plant-derived inhibitor that targets the T3SSs of different pathogens, including Y. pseudotuberculosis and P. aeruginosa (Zetterström et al., 2013). It is highly efficient in blocking the expression of the Yersinia effector protein YopE. It further interfered with the translocation of ExoS from P. aeruginosa at higher concentrations. Moreover, (-)-hopeaphenol reduced entry and subsequent intracellular growth of C. trachomatis. Interestingly, Y. pseudotuberculosis grown in the presence of (-)-hopeaphenol did not assemble T3SSs when transferred to T3SS-inducing conditions, suggesting an irreversible inhibitory effect. It was suggested that (-)-hopeaphenol interacts with T3SS components at the bacterial surface, as its molecular weight and size imply a low membrane permeability (Zetterström et al., 2013). As (-)-hopeaphenol did not affect cell viability and growth, it may be a promising drug against multiple Gram-negative pathogens. Unfortunately, (-)-hopeaphenol is produced by plants of the Anisoptera spp. which are endangered, making the compound impossible to access and synthesize for further studies (International Union for Conservation of Nature (IUCN) Red List, 2019).
Tanshinones are natural herbal compounds isolated from the plant Salvia miltiorrhiza. The compounds were shown to block the interaction of the P. aeruginosa needle protein PscF with its chaperones PscE and PscG. Hence, they prevented needle formation and subsequent effector translocation of ExoS into infected macrophages. Additionally, a dose-dependent inhibition of intracellular bacterial survival was observed without cytotoxic effects for the host cell. Furthermore, mice were challenged with lethal doses of P. aeruginosa in a model of acute pneumonia to investigate the effect of tanshinones in vivo. There, the compounds reduced the secretion of bacterial virulence factors and prevented mice from dying when the inhibitors were applied at the time of infection and every 12 h afterwards (Quinaud et al., 2005; Abrusci et al., 2014; Feng et al., 2019). Another research group found two other tanshinone deviates, Clioquniol and 3-APPA1, by target-based screening, which the authors assembled into a series of larger hybrids. Here, two potent inhibitors were identified, 12(6,4) and 12(4,6). Like other tanshinones, they interfered with needle assembly by interacting with PscG and PscE. These compounds strongly inhibited the secretion of effector proteins ExoS and ExoT, while the secretion of PopD was dose-dependent. Both inhibitors were non-cytotoxic in eukaryotic cells and did not affect bacterial viability. In vivo experiments performed in Galleria mellonella showed that these tanshinone derivatives significantly improve larval survival upon P. aeruginosa infection (Ngo et al., 2019).
In 2016, Guo et al. tested twenty prenylated flavonoids (1-20), a unique class of naturally occurring flavonoids isolated from trees of the Macaranga species, for their inhibitory activity against effector secretion by SPI-1. Among the tested flavonoids, licoflavonol exhibited the most substantial inhibitory effect on the secretion of SPI-1-associated effector proteins from Salmonella Typhimurium. Furthermore, it did not affect bacterial growth or secretion of the flagellar protein FliC (Guo et al., 2016).
(-)-Epigallocatechin gallate (EGCG), is a catechin most abundant in green tea. After infection of EGCG-treated epithelial cells with EPEC or EHEC, the bacterial adherence to cells was significantly reduced (Nakasone et al., 2017). Similarly, upon Salmonella or Yersinia infection of EGCG-treated epithelial cells, invasion was inhibited without affecting bacterial growth (Tsou et al., 2017).
In 2017, obovatol, derived from the Magnolia obovate plant, was shown to potently inhibit the T3SS of Salmonella Typhimurium (Choi et al., 2017). It blocked bacterial motility, mRNA expression, and effector protein secretion without inhibiting bacterial growth. Furthermore, obovatol was able to block Salmonella-induced hemolysis (Choi et al., 2017).
Another plant-derived T3SS inhibitor is sanguinarine chloride, isolated from the bloodroot plant Sanguinaria canadensis (Babich et al., 1996). In the 1970s and 80s, it was studied as a promising gingivitis treatment due to its anti-inflammatory properties. However, its high cytotoxicity mainly limits its therapeutic use to chemotherapy (Mondal et al., 2019). Sanguinarine chloride potently inhibited SipA-β−lactamase fusion protein translocation into epithelial cells and reduced the invasion of Salmonella. Additionally, it reduced the expression of the effector proteins SipA and SipB. However, sanguinarine chloride decreased the level of HilA, suggesting inhibition of HilA-induced virulence genes, as overexpression of HilA could compensate for these inhibitory effects (Zhang et al., 2018b).
Cinnamaldehyde is the active compound of cinnamon oil which can be extracted from a plant of the Cinnamomum genus. It has been described to exhibit anti-bacterial, anti-inflammatory, and anti-oxidative functions (Zhu et al., 2017). The compound reduced the expression of Salmonella SPI-1 effector proteins by downregulating the transcription of virulence regulator genes, including sipA and sipB (Liu et al., 2019). Hence, it is unsurprising that cinnamaldehyde blocks the translocation of several SPI-1-associated effector proteins. Furthermore, cinnamaldehyde-treated mice infected with S. Typhimurium showed decreased colonization and increased intestinal barrier functions compared to untreated animals.
Syringaldehyde can be found in the wood of spruce and maple trees and also acted as an effective inhibitor of SPI-1. Treatment with syringaldehyde potently inhibited the expression of the effector proteins SipA, SipB, and SipC, blocking bacterial invasion and subsequent cellular damage during infection of epithelial cells with Salmonella Typhimurium without affecting bacterial growth. The inhibitory effect of syringaldehyde was also confirmed in vivo. Here, S. Typhimurium-infected mice showed reduced bacterial loads, cecal damage, and systemic inflammation, resulting in an overall decrease in mortality (Lv et al., 2019).
The essential oil thymol derives from plants belonging to the Thymus genus. Thymol inhibited S. Typhimurium invasion into mammalian cells and protected mice from infection (Zhang et al., 2018a). Proteomic analysis of thymol-treated cells revealed the differential regulation of 144 proteins involved in metabolism and cellular structure. This suggests an antimicrobial role of thymol in altering membrane permeability, antioxidant response, and virulence of S. Typhimurium (Qi et al., 2020). A follow-up study by Zhang et al. revealed that thymol directly interacted with SipA, inducing conformational changes in the protein, making SipA susceptible to degradation by the Lon protease (Zhang et al., 2021).
The phenolic compound paeonol, found in peonies, was recently identified as an effective Salmonella SPI-1 inhibitor. It blocked the translocation of SipA into host cells without affecting bacterial growth. Moreover, paeonol-treated mice infected with S. Typhimurium showed decreased invasion of the bacteria into tissues, hence reducing tissue damage (Lv et al., 2020). Mechanistic studies revealed the transcriptional downregulation of the SPI-1 regulator hilA, resulting in the inhibition of effector protein expression (Lv et al., 2020).
Lastly, myricetin, a flavonoid with antioxidant properties that can be found in vegetables, fruits, and red wine (Lv et al., 2021) was recently discovered to prevent S. Typhimurium invasion into host cells, decreasing cell damage. Moreover, it blocked the translocation of the effector proteins SipA and SipB. Further analysis showed that it interfered with the regulatory network of SPI-1-related genes, leading to a significant decrease in the expression of crucial effector proteins, ultimately inhibiting T3SS-mediated virulence. In vivo, it was shown that myricetin treatment prevented pathological damage and death in infected mice (Lv et al., 2021).
Recent advances and the challenges for translation into the clinic
For over a decade, numerous high-throughput screens for pathoblockers have been conducted. In contrast to screening approaches for antibiotics, initial screens for secretion system inhibitors focused on commercially-available and sometimes overlapping chemical compound libraries (Duncan et al., 2012; Beckham and Roe, 2014; Boudaher and Shaffer, 2019; Hotinger et al., 2021). However, in recent years more and more natural compounds have been discovered which inhibit virulence-associated secretion systems in bacterial pathogens (Pendergrass and May, 2019). Unfortunately, the molecular targets of many pathoblockers identified over the years remain unknown, and no further research has been conducted to elucidate them. However, determining the targets of pathoblockers would enable their structural improvement (e.g., by chemical modification), potentially improving their efficacy or cell permeability and limiting cytotoxicity as well as potential cross-reactivity with eukaryotic proteins or processes (Miethke et al., 2021).
Several secretion system inhibitors have now been tested in mouse models of infection for their ability to inhibit virulence in vivo with encouraging results. However, the progression of promising compounds into clinical trials is hampered by difficulties in evaluating compound efficacy. While the therapeutic concentrations of antimicrobials targeting bacterial survival mechanisms are evaluated based on their minimal inhibitory concentration (MIC), this cannot be used as a guideline in the case of anti-infectives. Here, treatment does not lead to a measurable phenotype. Therefore, animal models are essential to determine the approximate therapeutic compound concentrations required for assessment in human trials.
Also, there is the question of bioavailability as well as the method of application. Topical treatments may be used for infections with Chlamydia or skin infections by P. aeruginosa. For other infections, oral administration is suggested for gastrointestinal infection with H. pylori, S. enterica, Shigella spp., and pathogenic E. coli, as well as enteropathogenic Yersinia species. For this, the compound has to be stable during its passage through the gastrointestinal tract and needs to be released at the site of infection. Pathoblockers targeting lung pathogens may need to be administered either as aerosols or by systemic application, posing yet other challenges.
Moreover, the potential of the different compounds as prophylactic versus therapeutic treatment must be considered. Most pathogens downregulate the expression of virulence genes including the energy-costly secretion systems after successful colonization of their target tissues (Heithoff et al., 1997; Heroven and Dersch, 2014; Connolly et al., 2018; Platenkamp and Mellies, 2018; Wang et al., 2019). Salmonella switches off the expression of SPI-1 upon invasion of host cells and, in turn, expresses SPI-2 once inside the host cell in a tightly regulated process to allow its survival and replication within the host (Lober et al., 2006; Singer et al., 2014; Lou et al., 2019). Yet other pathogens, such as C. burnetii, only express their secretion system once conditions inside host cells are favorable (Newton et al., 2020), escaping the recognition by secretion system inhibitors outside cells and limiting therapeutic treatment options.
Conclusion
With AMR on the rise and the number of last-resort antibiotics dwindling by the day, the focus has to shift towards intervention therapies that render pathogens avirulent but do not affect their viability to circumvent resistance development. With expression, assembly, and effector translocation of secretion systems being a metabolically-costly endeavor for the bacteria, interference with this process should not cause the (rapid) development of resistance. Future studies will have to determine the function and targets of identified inhibitors to modify the structures to make them either more or less specific. As secretion systems are highly similar in structure, it has surprised many that most pathoblockers appear specific to the pathogen they were tested in and showed little to no overlap between species. This suggests that these inhibitors were not targeting the virulence-associated secretion systems but other proteins found in the pathogen, specific for regulating the expression/formation of secretion systems. Researchers have attempted to circumvent these issues by structure-guided inhibitor synthesis. However, this approach, just as those approaches trying to identify inhibitors using overexpressed secreted proteins, has the drawback of providing no evidence of whether the identified molecules are functional in a cell-based setting. Furthermore, depending on the proposed target, pathoblockers have to be cell-permeable to reach their targets if these are not exposed on the bacterial cell surface. Several obligate or facultative intracellular bacteria including Coxiella, Shigella, and Chlamydia will also require the inhibitory compounds to be membrane-permeable in order to reach the bacteria and/or their potential targets. Membrane crossing compounds, do, however, pose the risk of affecting other intracellular host cell processes. Furthermore, due to the evolutionary relationship between bacterial flagella and T3SSs, compounds targeting the T3SS may always also affect bacterial motility. While in itself not affecting bacterial viability and, therefore, not inducing selective pressure, targeting flagella and hence motility will also affect motile members of the intestinal microbiota, which may result in the loss of these bacteria and may, in turn, suppress colonization resistance.
There is still a long road ahead, but the drastic increase in AMR and the limited number of approved antibiotics over the last decade call for more intense efforts to identify novel lead compounds available for anti-virulence approaches and to further develop these compounds in order to pave their way into the clinic.
Author contributions
All authors have made a substantial, direct, and intellectual contribution to this work and approved it for publication. All authors contributed to the article and approved the submitted version.
Funding
AKR was supported by the DFG (SCHM1586/3-2).
Acknowledgments
We thank Ingo Schmitz for his support. We acknowledge support by the Open Access Publication Funds of the Ruhr-Universität Bochum. All Figures were created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abby, S. S., Rocha, E. P. (2012). The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 8 (9), e1002983. doi: 10.1371/journal.pgen.1002983
Abrusci, P., McDowell, M. A., Lea, S. M., Johnson, S. (2014). Building a secreting nanomachine: a structural overview of the T3SS. Curr. Opin. Struct. Biol. 25, 111–117. doi: 10.1016/j.sbi.2013.11.001
Anderson, M., Clift, C., Schulze, K., Sagan, A., Nahrgang, S., Ait Ouakrim, D., et al. (2019). Averting the AMR crisis: What are the avenues for policy action for countries in Europe? (Copenhagen (Denmark).
Antimicrobial Resistance, C. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399 (10325), 629–655. doi: 10.1016/S0140-6736(21)02724-0
Arya, T., Oudouhou, F., Casu, B., Bessette, B., Sygusch, J., Baron, C. (2019). Fragment-based screening identifies inhibitors of ATPase activity and of hexamer formation of Cagalpha from the Helicobacter pylori type IV secretion system. Sci. Rep. 9 (1), 6474. doi: 10.1038/s41598-019-42876-6
Babich, H., Zuckerbraun, H. L., Barber, I. B., Babich, S. B., Borenfreund, E. (1996). Cytotoxicity of sanguinarine chloride to cultured human cells from oral tissue. Pharmacol. Toxicol. 78 (6), 397–403. doi: 10.1111/j.1600-0773.1996.tb00225.x
Bailey, L., Gylfe, A., Sundin, C., Muschiol, S., Elofsson, M., Nordstrom, P., et al. (2007). Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. FEBS Lett. 581 (4), 587–595. doi: 10.1016/j.febslet.2007.01.013
Baron, C. (2010). Antivirulence drugs to target bacterial secretion systems. Curr. Opin. Microbiol. 13 (1), 100–105. doi: 10.1016/j.mib.2009.12.003
Beckham, K. S., Roe, A. J. (2014). From screen to target: insights and approaches for the development of anti-virulence compounds. Front. Cell Infect. Microbiol. 4. doi: 10.3389/fcimb.2014.00139
Beeckman, D. S., Geens, T., Timmermans, J. P., Van Oostveldt, P., Vanrompay, D. C. (2008). Identification and characterization of a type III secretion system in Chlamydophila psittaci. Vet. Res. 39 (3), 27. doi: 10.1051/vetres:2008002
Bessler, H. C., de Oliveira, I. R., Giugliano, L. G. (2006). Human milk glycoproteins inhibit the adherence of Salmonella Typhimurium to HeLa cells. Microbiol. Immunol. 50 (11), 877–882. doi: 10.1111/j.1348-0421.2006.tb03863.x
Bhatty, M., Laverde Gomez, J. A., Christie, P. J. (2013). The expanding bacterial type IV secretion lexicon. Res. Microbiol. 164 (6), 620–639. doi: 10.1016/j.resmic.2013.03.012
Bienvenu, A., Martinez, E., Bonazzi, M. (2021). Undercover agents of infection: The stealth strategies of T4SS-equipped bacterial pathogens. Toxins (Basel) 13 (10). doi: 10.3390/toxins13100713
Boudaher, E., Shaffer, C. L. (2019). Inhibiting bacterial secretion systems in the fight against antibiotic resistance. Medchemcomm 10 (5), 682–692. doi: 10.1039/c9md00076c
Cascales, E., Christie, P. J. (2003). The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1 (2), 137–149. doi: 10.1038/nrmicro753
Case, H. B., Mattock, D. S., Dickenson, N. E. (2018). Shutting down Shigella secretion: Characterizing small molecule type three secretion system ATPase inhibitors. Biochemistry 57 (50), 6906–6916. doi: 10.1021/acs.biochem.8b01077
Chahine, E. B., Dougherty, J. A., Thornby, K. A., Guirguis, E. H. (2022). Antibiotic approvals in the last decade: Are we keeping up with resistance? Ann. Pharmacother. 56 (4), 441–462. doi: 10.1177/10600280211031390
Cheng, E., Dorjsuren, D., Lehman, S., Larson, C. L., Titus, S. A., Sun, H., et al. (2022). A comprehensive phenotypic screening strategy to identify modulators of cargo translocation by the bacterial type IVB secretion system. mBio 13 (2), e0024022. doi: 10.1128/mbio.00240-22
Chetrit, D., Hu, B., Christie, P. J., Roy, C. R., Liu, J. (2018). A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nat. Microbiol. 3 (6), 678–686. doi: 10.1038/s41564-018-0165-z
Choi, W. S., Lee, T. H., Son, S. J., Kim, T. G., Kwon, B. M., Son, H. U., et al. (2017). Inhibitory effect of obovatol from Magnolia obovata on the Salmonella type III secretion system. J. Antibiot (Tokyo) 70 (11), 1065–1069. doi: 10.1038/ja.2017.98
Chung, J. M., Sheedlo, M. J., Campbell, A. M., Sawhney, N., Frick-Cheng, A. E., Lacy, D. B., et al. (2019). Structure of the Helicobacter pylori cag type IV secretion system. Elife 8. doi: 10.7554/eLife.47644
Coburn, B., Sekirov, I., Finlay, B. B. (2007). Type III secretion systems and disease. Clin. Microbiol. Rev. 20 (4), 535–549. doi: 10.1128/CMR.00013-07
Condry, D. L., Nilles, M. L. (2017). Introduction to type III secretion systems. Methods Mol. Biol. 1531, 1–10. doi: 10.1007/978-1-4939-6649-3_1
Connolly, J. P. R., Slater, S. L., O'Boyle, N., Goldstone, R. J., Crepin, V. F., Ruano-Gallego, D., et al. (2018). Host-associated niche metabolism controls enteric infection through fine-tuning the regulation of type 3 secretion. Nat. Commun. 9 (1), 4187. doi: 10.1038/s41467-018-06701-4
Cornelis, G. R. (2006). The type III secretion injectisome. Nat. Rev. Microbiol. 4 (11), 811–825. doi: 10.1038/nrmicro1526
Cornelis, G. R. (2010). The type III secretion injectisome, a complex nanomachine for intracellular 'toxin' delivery. Biol. Chem. 391 (7), 745–751. doi: 10.1515/BC.2010.079
Costa, T. R. D., Harb, L., Khara, P., Zeng, L., Hu, B., Christie, P. J. (2021). Type IV secretion systems: Advances in structure, function, and activation. Mol. Microbiol. 115 (3), 436–452. doi: 10.1111/mmi.14670
Cover, T. L., Lacy, D. B., Ohi, M. D. (2020). The Helicobacter pylori cag type IV secretion system. Trends Microbiol. 28 (8), 682–695. doi: 10.1016/j.tim.2020.02.004
Dean, P. (2011). Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 35 (6), 1100–1125. doi: 10.1111/j.1574-6976.2011.00271.x
Deng, W., Marshall, N. C., Rowland, J. L., McCoy, J. M., Worrall, L. J., Santos, A. S., et al. (2017). Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15 (6), 323–337. doi: 10.1038/nrmicro.2017.20
De Tavernier, E., Detalle, L., Morizzo, E., Roobrouck, A., De Taeye, S., Rieger, M., et al. (2016). High throughput combinatorial formatting of PcrV nanobodies for efficient potency improvement. J. Biol. Chem. 291 (29), 15243–15255. doi: 10.1074/jbc.M115.684241
Dey, S., Anbanandam, A., Mumford, B. E., De Guzman, R. N. (2017). Characterization of small-molecule scaffolds that bind to the Shigella type III secretion system protein IpaD. ChemMedChem 12 (18), 1534–1541. doi: 10.1002/cmdc.201700348
Dey, S., Chakravarty, A., Guha Biswas, P., De Guzman, R. N. (2019). The type III secretion system needle, tip, and translocon. Protein Sci. 28 (9), 1582–1593. doi: 10.1002/pro.3682
Duncan, M. C., Linington, R. G., Auerbuch, V. (2012). Chemical inhibitors of the type three secretion system: disarming bacterial pathogens. Antimicrob. Agents Chemother. 56 (11), 5433–5441. doi: 10.1128/AAC.00975-12
Duncan, M. C., Wong, W. R., Dupzyk, A. J., Bray, W. M., Linington, R. G., Auerbuch, V. (2014). An NF-κB-based high-throughput screen identifies piericidins as inhibitors of the Yersinia pseudotuberculosis type III secretion system. Antimicrob. Agents Chemother. 58 (2), 1118–1126. doi: 10.1128/AAC.02025-13
Ellermeier, C. D., Ellermeier, J. R., Slauch, J. M. (2005). HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57 (3), 691–705. doi: 10.1111/j.1365-2958.2005.04737.x
Fasciano, A. C., Shaban, L., Mecsas, J. (2019). Promises and challenges of the type three secretion system injectisome as an antivirulence target. EcoSal Plus 8 (2). doi: 10.1128/ecosalplus.ESP-0032-2018
Felise, H. B., Nguyen, H. V., Pfuetzner, R. A., Barry, K. C., Jackson, S. R., Blanc, M. P., et al. (2008). An inhibitor of gram-negative bacterial virulence protein secretion. Cell Host Microbe 4 (4), 325–336. doi: 10.1016/j.chom.2008.08.001
Feng, C., Huang, Y., He, W., Cheng, X., Liu, H., Huang, Y., et al. (2019). Tanshinones: First-in-Class inhibitors of the biogenesis of the type 3 secretion system needle of Pseudomonas aeruginosa for antibiotic therapy. ACS Cent Sci. 5 (7), 1278–1288. doi: 10.1021/acscentsci.9b00452
Fischer, W. (2011). Assembly and molecular mode of action of the Helicobacter pylori cag type IV secretion apparatus. FEBS J. 278 (8), 1203–1212. doi: 10.1111/j.1742-4658.2011.08036.x
Fleitas Martinez, O., Cardoso, M. H., Ribeiro, S. M., Franco, O. L. (2019). Recent advances in anti-virulence therapeutic strategies with a focus on dismantling bacterial membrane microdomains, toxin neutralization, quorum-sensing interference and biofilm inhibition. Front. Cell Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00074
Fronzes, R., Christie, P. J., Waksman, G. (2009). The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7 (10), 703–714. doi: 10.1038/nrmicro2218
Ghosal, D., Jeong, K. C., Chang, Y. W., Gyore, J., Teng, L., Gardner, A., et al. (2019). Molecular architecture, polar targeting and biogenesis of the Legionella Dot/Icm T4SS. Nat. Microbiol. 4 (7), 1173–1182. doi: 10.1038/s41564-019-0427-4
Gomez, H. F., Ochoa, T. J., Carlin, L. G., Cleary, T. G. (2003). Human lactoferrin impairs virulence of Shigella flexneri. J. Infect. Dis. 187 (1), 87–95. doi: 10.1086/345875
Goure, J., Broz, P., Attree, O., Cornelis, G. R., Attree, I. (2005). Protective anti-V antibodies inhibit Pseudomonas and Yersinia translocon assembly within host membranes. J. Infect. Dis. 192 (2), 218–225. doi: 10.1086/430932
Goure, J., Pastor, A., Faudry, E., Chabert, J., Dessen, A., Attree, I. (2004). The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 72 (8), 4741–4750. doi: 10.1128/IAI.72.8.4741-4750.2004
Green, E. R., Mecsas, J. (2016). Bacterial secretion systems: An overview. Microbiol. Spectr. 4 (1). doi: 10.1128/microbiolspec.VMBF-0012-2015
Grishin, A. V., Luyksaar, S. I., Kapotina, L. N., Kirsanov, D. D., Zayakin, E. S., Karyagina, A. S., et al. (2018). Identification of chlamydial T3SS inhibitors through virtual screening against T3SS ATPase. Chem. Biol. Drug Des. 91 (3), 717–727. doi: 10.1111/cbdd.13130
Grohmann, E., Christie, P. J., Waksman, G., Backert, S. (2018). Type IV secretion in gram-negative and gram-positive bacteria. Mol. Microbiol. 107 (4), 455–471. doi: 10.1111/mmi.13896
Grossman, A. S., Mauer, T. J., Forest, K. T., Goodrich-Blair, H. (2021). A widespread bacterial secretion system with diverse substrates. mBio 12 (4), e0195621. doi: 10.1128/mBio.01956-21
Guo, Z., Li, X., Li, J., Yang, X., Zhou, Y., Lu, C., et al. (2016). Licoflavonol is an inhibitor of the type three secretion system of Salmonella enterica serovar Typhimurium. Biochem. Biophys. Res. Commun. 477 (4), 998–1004. doi: 10.1016/j.bbrc.2016.07.018
Harmon, D. E., Davis, A. J., Castillo, C., Mecsas, J. (2010). Identification and characterization of small-molecule inhibitors of yop translocation in Yersinia pseudotuberculosis. Antimicrob. Agents Chemother. 54 (8), 3241–3254. doi: 10.1128/AAC.00364-10
Heithoff, D. M., Conner, C. P., Hanna, P. C., Julio, S. M., Hentschel, U., Mahan, M. J. (1997). Bacterial infection as assessed by in vivo gene expression. Proc. Natl. Acad. Sci. U.S.A. 94 (3), 934–939. doi: 10.1073/pnas.94.3.934
Heroven, A. K., Dersch, P. (2014). Coregulation of host-adapted metabolism and virulence by pathogenic Yersiniae. Front. Cell Infect. Microbiol. 4. doi: 10.3389/fcimb.2014.00146
Hilleringmann, M., Pansegrau, W., Doyle, M., Kaufman, S., MacKichan, M. L., Gianfaldoni, C., et al. (2006). Inhibitors of Helicobacter pylori ATPase cagα block CagA transport and cag virulence. Microbiol. (Reading) 152 (Pt 10), 2919–2930. doi: 10.1099/mic.0.28984-0
Hofer, U. (2022). Mobilizing resistance genes in the human gut. Nat. Rev. Microbiol. 20 (6), 317. doi: 10.1038/s41579-022-00728-7
Horna, G., Ruiz, J. (2021). Type 3 secretion system of Pseudomonas aeruginosa. Microbiol. Res. 246, 126719. doi: 10.1016/j.micres.2021.126719
Hotinger, J. A., Pendergrass, H. A., May, A. E. (2021). Molecular targets and strategies for inhibition of the bacterial type III secretion system (T3SS); inhibitors directly binding to T3SS components. Biomolecules 11 (2). doi: 10.3390/biom11020316
Hsia, R. C., Pannekoek, Y., Ingerowski, E., Bavoil, P. M. (1997). Type III secretion genes identify a putative virulence locus of Chlamydia. Mol. Microbiol. 25 (2), 351–359. doi: 10.1046/j.1365-2958.1997.4701834.x
Hudson, D. L., Layton, A. N., Field, T. R., Bowen, A. J., Wolf-Watz, H., Elofsson, M., et al. (2007). Inhibition of type III secretion in Salmonella enterica serovar Typhimurium by small-molecule inhibitors. Antimicrob. Agents Chemother. 51 (7), 2631–2635. doi: 10.1128/AAC.01492-06
Hueck, C. J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62 (2), 379–433. doi: 10.1128/MMBR.62.2.379-433.1998
Hu, B., Khara, P., Song, L., Lin, A. S., Frick-Cheng, A. E., Harvey, M. L., et al. (2019). In situ molecular architecture of the Helicobacter pylori cag type IV secretion system. mBio 10 (3). doi: 10.1128/mBio.00849-19
International Union for Conservation of Nature (IUCN) Red List. [(accessed on 5September 2019)]; Available online: https://www.iucnredlist.org/search?query=anisoptera&searchType=species)
Ishaq, S., Nunn, L. (2015). Helicobacter pylori and gastric cancer: a state of the art review. Gastroenterol. Hepatol. Bed Bench 8 (Suppl 1), S6–S14.
Iwatsuki, M., Uchida, R., Yoshijima, H., Ui, H., Shiomi, K., Kim, Y. P., et al. (2008a). Guadinomines, type III secretion system inhibitors, produced by Streptomyces sp. K01-0509. II: physico-chemical properties and structure elucidation. J. Antibiot (Tokyo) 61 (4), 230–236. doi: 10.1038/ja.2008.33
Iwatsuki, M., Uchida, R., Yoshijima, H., Ui, H., Shiomi, K., Matsumoto, A., et al. (2008b). Guadinomines, type III secretion system inhibitors, produced by Streptomyces sp. K01-0509. I: taxonomy, fermentation, isolation and biological properties. J. Antibiot (Tokyo) 61 (4), 222–229. doi: 10.1038/ja.2008.32
Jamieson, E. R., Lippard, S. J. (1999). Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev. 99 (9), 2467–2498. doi: 10.1021/cr980421n
Jee, Y., Carlson, J., Rafai, E., Musonda, K., Huong, T. T. G., Daza, P., et al. (2018). Antimicrobial resistance: a threat to global health. Lancet Infect. Dis. 18 (9), 939–940. doi: 10.1016/S1473-3099(18)30471-7
Jenkins, J., Worrall, L. J., Strynadka, N. C. J. (2022). Recent structural advances towards understanding of the bacterial type III secretion injectisome. Trends Biochem. Sci. 47 (9), 795–809. doi: 10.1016/j.tibs.2022.04.013
Jessen, D. L., Bradley, D. S., Nilles, M. L. (2014). A type III secretion system inhibitor targets YopD while revealing differential regulation of secretion in calcium-blind mutants of Yersinia pestis. Antimicrob. Agents Chemother. 58 (2), 839–850. doi: 10.1128/AAC.01170-13
Juhas, M., Crook, D. W., Hood, D. W. (2008). Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol. 10 (12), 2377–2386. doi: 10.1111/j.1462-5822.2008.01187.x
Kauppi, A. M., Nordfelth, R., Uvell, H., Wolf-Watz, H., Elofsson, M. (2003). Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10 (3), 241–249. doi: 10.1016/s1074-5521(03)00046-2
Kent, A. G., Vill, A. C., Shi, Q., Satlin, M. J., Brito, I. L. (2020). Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-c. Nat. Commun. 11 (1), 4379. doi: 10.1038/s41467-020-18164-7
Keyser, P., Elofsson, M., Rosell, S., Wolf-Watz, H. (2008). Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against gram-negative bacteria. J. Intern. Med. 264 (1), 17–29. doi: 10.1111/j.1365-2796.2008.01941.x
Kimura, K., Iwatsuki, M., Nagai, T., Matsumoto, A., Takahashi, Y., Shiomi, K., et al. (2011). A small-molecule inhibitor of the bacterial type III secretion system protects against in vivo infection with Citrobacter rodentium. J. Antibiot (Tokyo) 64 (2), 197–203. doi: 10.1038/ja.2010.155
Klein, E. Y., Van Boeckel, T. P., Martinez, E. M., Pant, S., Gandra, S., Levin, S. A., et al. (2018). Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. U.S.A. 115 (15), E3463–E3470. doi: 10.1073/pnas.1717295115
Koroleva, E. A., Kobets, N. V., Zayakin, E. S., Luyksaar, S. I., Shabalina, L. A., Zigangirova, N. A. (2015). Small molecule inhibitor of type three secretion suppresses acute and chronic Chlamydia trachomatis infection in a novel urogenital chlamydia model. BioMed. Res. Int. 2015, 484853. doi: 10.1155/2015/484853
Lam, H. N., Lau, T., Lentz, A., Sherry, J., Cabrera-Cortez, A., Hug, K., et al. (2021). Developing cyclic peptomers as broad-spectrum type III secretion system inhibitors in gram-negative bacteria. Antimicrob. Agents Chemother. 65 (7), e0169020. doi: 10.1128/AAC.01690-20
Lam, H., Schwochert, J., Lao, Y., Lau, T., Lloyd, C., Luu, J., et al. (2017). Synthetic cyclic peptomers as type III secretion system inhibitors. Antimicrob. Agents Chemother. 61 (9). doi: 10.1128/AAC.00060-17
Layton, A. N., Hudson, D. L., Thompson, A., Hinton, J. C., Stevens, J. M., Galyov, E. E., et al. (2010). Salicylidene acylhydrazide-mediated inhibition of type III secretion system-1 in Salmonella enterica serovar Typhimurium is associated with iron restriction and can be reversed by free iron. FEMS Microbiol. Lett. 302 (2), 114–122. doi: 10.1111/j.1574-6968.2009.01847.x
Lettl, C., Schindele, F., Testolin, G., Bar, A., Rehm, T., Bronstrup, M., et al. (2020). Inhibition of type IV secretion activity and growth of Helicobacter pylori by cisplatin and other platinum complexes. Front. Cell Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.602958
Li, J., Lv, C., Sun, W., Li, Z., Han, X., Li, Y., et al. (2013). Cytosporone b, an inhibitor of the type III secretion system of Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 57 (5), 2191–2198. doi: 10.1128/AAC.02421-12
Linington, R. G., Robertson, M., Gauthier, A., Finlay, B. B., van Soest, R., Andersen, R. J. (2002). Caminoside a, an antimicrobial glycolipid isolated from the marine sponge Caminus sphaeroconia. Org Lett. 4 (23), 4089–4092. doi: 10.1021/ol0268337
Li, J., Sun, W., Guo, Z., Lu, C., Shen, Y. (2014). Fusaric acid modulates type three secretion system of Salmonella enterica serovar Typhimurium. Biochem. Biophys. Res. Commun. 449 (4), 455–459. doi: 10.1016/j.bbrc.2014.05.044
Liu, X., Khara, P., Baker, M. L., Christie, P. J., Hu, B. (2022). Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids. Nat. Commun. 13 (1), 379. doi: 10.1038/s41467-022-28058-5
Liu, Y., Zhang, Y., Zhou, Y., Wang, T., Deng, X., Chu, X., et al. (2019). Cinnamaldehyde inhibits type three secretion system in Salmonella enterica serovar Typhimurium by affecting the expression of key effector proteins. Vet. Microbiol. 239, 108463. doi: 10.1016/j.vetmic.2019.108463
Lober, S., Jackel, D., Kaiser, N., Hensel, M. (2006). Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int. J. Med. Microbiol. 296 (7), 435–447. doi: 10.1016/j.ijmm.2006.05.001
Lou, L., Zhang, P., Piao, R., Wang, Y. (2019). Salmonella pathogenicity island 1 (SPI-1) and its complex regulatory network. Front. Cell Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00270
Low, H. H., Gubellini, F., Rivera-Calzada, A., Braun, N., Connery, S., Dujeancourt, A., et al. (2014). Structure of a type IV secretion system. Nature 508 (7497), 550–553. doi: 10.1038/nature13081
Lv, Q., Chu, X., Yao, X., Ma, K., Zhang, Y., Deng, X. (2019). Inhibition of the type III secretion system by syringaldehyde protects mice from Salmonella enterica serovar Typhimurium. J. Cell Mol. Med. 23 (7), 4679–4688. doi: 10.1111/jcmm.14354
Lv, Q., Li, S., Wei, H., Wen, Z., Wang, Y., Tang, T., et al. (2020). Identification of the natural product paeonol derived from peony bark as an inhibitor of the Salmonella enterica serovar Typhimurium type III secretion system. Appl. Microbiol. Biotechnol. 104 (4), 1673–1682. doi: 10.1007/s00253-019-10290-7
Lv, Q., Lv, Y., Dou, X., Wassy, S. L., Jia, G., Wei, L., et al. (2021). Myricetin inhibits the type III secretion system of Salmonella enterica serovar Typhimurium by downregulating the Salmonella pathogenic island I gene regulatory pathway. Microb. Pathog. 150, 104695. doi: 10.1016/j.micpath.2020.104695
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13 (6), 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Martinez, J. L., Baquero, F. (2000). Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44 (7), 1771–1777. doi: 10.1128/AAC.44.7.1771-1777.2000
McHugh, R. E., Munnoch, J. T., Braes, R. E., McKean, I. J. W., Giard, J., Taladriz-Sender, A., et al. (2022). Biosynthesis of Aurodox, a type III secretion system inhibitor from Streptomyces goldiniensis. Appl. Environ. Microbiol. 88 (15), e0069222. doi: 10.1128/aem.00692-22
McHugh, R. E., O'Boyle, N., Connolly, J. P. R., Hoskisson, P. A., Roe, A. J. (2019). Characterization of the mode of action of Aurodox, a type III secretion system inhibitor from Streptomyces goldiniensis. Infect. Immun. 87 (2). doi: 10.1128/IAI.00595-18
Miethke, M., Pieroni, M., Weber, T., Bronstrup, M., Hammann, P., Halby, L., et al. (2021). Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 5 (10), 726–749. doi: 10.1038/s41570-021-00313-1
Mondal, A., Gandhi, A., Fimognari, C., Atanasov, A. G., Bishayee, A. (2019). Alkaloids for cancer prevention and therapy: Current progress and future perspectives. Eur. J. Pharmacol. 858, 172472. doi: 10.1016/j.ejphar.2019.172472
Morgan, J. M., Duncan, M. C., Johnson, K. S., Diepold, A., Lam, H., Dupzyk, A. J., et al. (2017). Piericidin A1 blocks Yersinia Ysc type III secretion system needle assembly. mSphere 2 (1). doi: 10.1128/mSphere.00030-17
Mosquito, S., Ochoa, T. J., Cok, J., Cleary, T. G. (2010). Effect of bovine lactoferrin in Salmonella ser. Typhimurium infection in mice. Biometals 23 (3), 515–521. doi: 10.1007/s10534-010-9325-1
Mühlen, S., Dersch, P. (2016). Anti-virulence strategies to target bacterial infections. Curr. Top. Microbiol. Immunol. 398, 147–183. doi: 10.1007/82_2015_490
Mühlen, S., Dersch, P. (2020). Treatment strategies for infections with shiga toxin-producing Escherichia coli. Front. Cell Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.00169
Mühlen, S., Zapol'skii, V. A., Bilitewski, U., Dersch, P. (2021). Identification of translocation inhibitors targeting the type III secretion system of enteropathogenic Escherichia coli. Antimicrob. Agents Chemother. doi: 10.1128/AAC.00958-21
Muschiol, S., Bailey, L., Gylfe, A., Sundin, C., Hultenby, K., Bergstrom, S., et al. (2006). A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U.S.A. 103 (39), 14566–14571. doi: 10.1073/pnas.0606412103
Nachin, L., Nannmark, U., Nystrom, T. (2005). Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol 187 (18), 6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005
Nagai, H., Kubori, T. (2011). Type IVB secretion systems of Legionella and other gram-negative bacteria. Front. Microbiol. 2. doi: 10.3389/fmicb.2011.00136
Nakasone, N., Higa, N., Toma, C., Ogura, Y., Suzuki, T., Yamashiro, T. (2017). Epigallocatechin gallate inhibits the type III secretion system of gram-negative enteropathogenic bacteria under model conditions. FEMS Microbiol. Lett. 364 (13). doi: 10.1093/femsle/fnx111
Negrea, A., Bjur, E., Ygberg, S. E., Elofsson, M., Wolf-Watz, H., Rhen, M. (2007). Salicylidene acylhydrazides that affect type III protein secretion in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 51 (8), 2867–2876. doi: 10.1128/AAC.00223-07
Nesterenko, L. N., Zigangirova, N. A., Zayakin, E. S., Luyksaar, S. I., Kobets, N. V., Balunets, D. V., et al. (2016). A small-molecule compound belonging to a class of 2,4-disubstituted 1,3,4-thiadiazine-5-ones suppresses Salmonella infection in vivo. J. Antibiot (Tokyo) 69 (6), 422–427. doi: 10.1038/ja.2015.131
Newton, P., Thomas, D. R., Reed, S. C. O., Lau, N., Xu, B., Ong, S. Y., et al. (2020). Lysosomal degradation products induce Coxiella burnetii virulence. Proc. Natl. Acad. Sci. U.S.A. 117 (12), 6801–6810. doi: 10.1073/pnas.1921344117
Ngo, T. D., Ple, S., Thomas, A., Barette, C., Fortune, A., Bouzidi, Y., et al. (2019). Chimeric protein-protein interface inhibitors allow efficient inhibition of type III secretion machinery and Pseudomonas aeruginosa virulence. ACS Infect. Dis. 5 (11), 1843–1854. doi: 10.1021/acsinfecdis.9b00154
Nordfelth, R., Kauppi, A. M., Norberg, H. A., Wolf-Watz, H., Elofsson, M. (2005). Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73 (5), 3104–3114. doi: 10.1128/IAI.73.5.3104-3114.2005
Norwood, D. A., Montalvan, E. E., Dominguez, R. L., Morgan, D. R. (2022). Gastric cancer: Emerging trends in prevention, diagnosis, and treatment. Gastroenterol. Clin. North Am. 51 (3), 501–518. doi: 10.1016/j.gtc.2022.05.001
Notti, R. Q., Stebbins, C. E. (2016). The structure and function of type III secretion systems. Microbiol. Spectr. 4 (1). doi: 10.1128/microbiolspec.VMBF-0004-2015
Ochoa, T. J., Clearly, T. G. (2004). Lactoferrin disruption of bacterial type III secretion systems. Biometals 17 (3), 257–260. doi: 10.1023/b:biom.0000027701.12965.d4
Ochoa, T. J., Cleary, T. G. (2009). Effect of lactoferrin on enteric pathogens. Biochimie 91 (1), 30–34. doi: 10.1016/j.biochi.2008.04.006
Ochoa, T. J., Noguera-Obenza, M., Cleary, T. G. (2004). Lactoferrin blocks the initial host cell attachment mechanism of enteropathogenic E. coli (EPEC). Adv. Exp. Med. Biol. 554, 463–466. doi: 10.1007/978-1-4757-4242-8_65
Ochoa, T. J., Noguera-Obenza, M., Ebel, F., Guzman, C. A., Gomez, H. F., Cleary, T. G. (2003). Lactoferrin impairs type III secretory system function in enteropathogenic Escherichia coli. Infect. Immun. 71 (9), 5149–5155. doi: 10.1128/IAI.71.9.5149-5155.2003
Olekhnovich, I. N., Kadner, R. J. (2007). Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J. Bacteriol 189 (19), 6882–6890. doi: 10.1128/JB.00905-07
Osei Sekyere, J. (2016). Current state of resistance to antibiotics of last-resort in south Africa: A review from a public health perspective. Front. Public Health 4. doi: 10.3389/fpubh.2016.00209
Palmer, T., Finney, A. J., Saha, C. K., Atkinson, G. C., Sargent, F. (2021). A holin/peptidoglycan hydrolase-dependent protein secretion system. Mol. Microbiol. 115 (3), 345–355. doi: 10.1111/mmi.14599
Pan, N. J., Brady, M. J., Leong, J. M., Goguen, J. D. (2009). Targeting type III secretion in Yersinia pestis. Antimicrob. Agents Chemother. 53 (2), 385–392. doi: 10.1128/AAC.00670-08
Paschos, A., den Hartigh, A., Smith, M. A., Atluri, V. L., Sivanesan, D., Tsolis, R. M., et al. (2011). An in vivo high-throughput screening approach targeting the type IV secretion system component VirB8 identified inhibitors of Brucella abortus 2308 proliferation. Infect. Immun. 79 (3), 1033–1043. doi: 10.1128/IAI.00993-10
Pedersen, C., Slepenkin, A., Andersson, S. B., Fagerberg, J. H., Bergstrom, C. A., Peterson, E. M. (2014). Formulation of the microbicide INP0341 for in vivo protection against a vaginal challenge by Chlamydia trachomatis. PLoS One 9 (10), e110918. doi: 10.1371/journal.pone.0110918
Pendergrass, H. A., May, A. E. (2019). Natural product type III secretion system inhibitors. Antibiotics (Basel) 8 (4). doi: 10.3390/antibiotics8040162
Perry, J. A., Wright, G. D. (2014). Forces shaping the antibiotic resistome. Bioessays 36 (12), 1179–1184. doi: 10.1002/bies.201400128
Platenkamp, A., Mellies, J. L. (2018). Environment controls LEE regulation in enteropathogenic Escherichia coli. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01694
Qi, Y., Zhao, W., Wang, T., Pei, F., Yue, M., Li, F., et al. (2020). Proteomic analysis of the antimicrobial effects of sublethal concentrations of thymol on Salmonella enterica serovar Typhimurium. Appl. Microbiol. Biotechnol. 104 (8), 3493–3505. doi: 10.1007/s00253-020-10390-9
Quinaud, M., Chabert, J., Faudry, E., Neumann, E., Lemaire, D., Pastor, A., et al. (2005). The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J. Biol. Chem. 280 (43), 36293–36300. doi: 10.1074/jbc.M508089200
Rasko, D. A., Sperandio, V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discovery 9 (2), 117–128. doi: 10.1038/nrd3013
Sato, H., Frank, D. W. (2011). Multi-functional characteristics of the Pseudomonas aeruginosa type III needle-tip protein, PcrV; comparison to orthologs in other gram-negative bacteria. Front. Microbiol. 2. doi: 10.3389/fmicb.2011.00142
Savvides, S. N., Yeo, H. J., Beck, M. R., Blaesing, F., Lurz, R., Lanka, E., et al. (2003). VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 22 (9), 1969–1980. doi: 10.1093/emboj/cdg223
Sayer, J. R., Wallden, K., Koss, H., Allan, H., Daviter, T., Gane, P. J., et al. (2021). Design, synthesis, and evaluation of peptide-imidazo[1,2-a]pyrazine bioconjugates as potential bivalent inhibitors of the VirB11 ATPase HP0525. J. Pept. Sci. 27 (10), e3353. doi: 10.1002/psc.3353
Sayer, J. R., Wallden, K., Pesnot, T., Campbell, F., Gane, P. J., Simone, M., et al. (2014). 2- and 3-substituted imidazo[1,2-a]pyrazines as inhibitors of bacterial type IV secretion. Bioorg Med. Chem. 22 (22), 6459–6470. doi: 10.1016/j.bmc.2014.09.036
Schwochert, J., Lao, Y., Pye, C. R., Naylor, M. R., Desai, P. V., Gonzalez Valcarcel, I. C., et al. (2016). Stereochemistry balances cell permeability and solubility in the naturally derived phepropeptin cyclic peptides. ACS Med. Chem. Lett. 7 (8), 757–761. doi: 10.1021/acsmedchemlett.6b00100
Segal, G., Purcell, M., Shuman, H. A. (1998). Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U.S.A. 95 (4), 1669–1674. doi: 10.1073/pnas.95.4.1669
Sekizawa, R., Momose, I., Kinoshita, N., Naganawa, H., Hamada, M., Muraoka, Y., et al. (2001). Isolation and structural determination of phepropeptins a, b, c, and d, new proteasome inhibitors, produced by Streptomyces sp. J. Antibiot (Tokyo) 54 (11), 874–881. doi: 10.7164/antibiotics.54.874
Sgro, G. G., Oka, G. U., Souza, D. P., Cenens, W., Bayer-Santos, E., Matsuyama, B. Y., et al. (2019). Bacteria-killing type IV secretion systems. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01078
Shaffer, C. L., Good, J. A., Kumar, S., Krishnan, K. S., Gaddy, J. A., Loh, J. T., et al. (2016). Peptidomimetic small molecules disrupt type IV secretion system activity in diverse bacterial pathogens. mBio 7 (2), e00221–e00216. doi: 10.1128/mBio.00221-16
Sharma, P., Elofsson, M., Roy, S. (2020). Attenuation of Pseudomonas aeruginosa infection by INP0341, a salicylidene acylhydrazide, in a murine model of keratitis. Virulence 11 (1), 795–804. doi: 10.1080/21505594.2020.1776979
Sheedlo, M. J., Ohi, M. D., Lacy, D. B., Cover, T. L. (2022). Molecular architecture of bacterial type IV secretion systems. PLoS Pathog. 18 (8), e1010720. doi: 10.1371/journal.ppat.1010720
Sheremet, A. B., Zigangirova, N. A., Zayakin, E. S., Luyksaar, S. I., Kapotina, L. N., Nesterenko, L. N., et al. (2018). Small molecule inhibitor of type three secretion system belonging to a class 2,4-disubstituted-4H-[1,3,4]-thiadiazine-5-ones improves survival and decreases bacterial loads in an airway Pseudomonas aeruginosa infection in mice. BioMed. Res. Int. 2018, 5810767. doi: 10.1155/2018/5810767
Singer, H. M., Kuhne, C., Deditius, J. A., Hughes, K. T., Erhardt, M. (2014). The Salmonella Spi1 virulence regulatory protein HilD directly activates transcription of the flagellar master operon flhDC. J. Bacteriol 196 (7), 1448–1457. doi: 10.1128/JB.01438-13
Sivanesan, D., Baron, C. (2011). The dimer interface of Agrobacterium tumefaciens VirB8 is important for type IV secretion system function, stability, and association of VirB2 with the core complex. J. Bacteriol 193 (9), 2097–2106. doi: 10.1128/JB.00907-10
Slepenkin, A., Chu, H., Elofsson, M., Keyser, P., Peterson, E. M. (2011). Protection of mice from a Chlamydia trachomatis vaginal infection using a salicylidene acylhydrazide, a potential microbicide. J. Infect. Dis. 204 (9), 1313–1320. doi: 10.1093/infdis/jir552
Slepenkin, A., Enquist, P. A., Hagglund, U., de la Maza, L. M., Elofsson, M., Peterson, E. M. (2007). Reversal of the antichlamydial activity of putative type III secretion inhibitors by iron. Infect. Immun. 75 (7), 3478–3489. doi: 10.1128/IAI.00023-07
Slepenkin, A., Motin, V., de la Maza, L. M., Peterson, E. M. (2003). Temporal expression of type III secretion genes of Chlamydia pneumoniae. Infect. Immun. 71 (5), 2555–2562. doi: 10.1128/IAI.71.5.2555-2562.2003
Smith, M. A., Coincon, M., Paschos, A., Jolicoeur, B., Lavallee, P., Sygusch, J., et al. (2012). Identification of the binding site of Brucella VirB8 interaction inhibitors. Chem. Biol. 19 (8), 1041–1048. doi: 10.1016/j.chembiol.2012.07.007
Smith, P. A., Romesberg, F. E. (2007). Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat. Chem. Biol. 3 (9), 549–556. doi: 10.1038/nchembio.2007.27
Song, Y., Xu, G., Li, C., Li, Z., Lu, C., Shen, Y. (2021). Structural optimization of natural product fusaric acid to discover novel T3SS inhibitors of Salmonella. Biochem. Biophys. Res. Commun. 582, 72–76. doi: 10.1016/j.bbrc.2021.10.035
Swietnicki, W., Carmany, D., Retford, M., Guelta, M., Dorsey, R., Bozue, J., et al. (2011). Identification of small-molecule inhibitors of Yersinia pestis type III secretion system YscN ATPase. PLoS One 6 (5), e19716. doi: 10.1371/journal.pone.0019716
Tree, J. J., Wang, D., McInally, C., Mahajan, A., Layton, A., Houghton, I., et al. (2009). Characterization of the effects of salicylidene acylhydrazide compounds on type III secretion in Escherichia coli O157:H7. Infect. Immun. 77 (10), 4209–4220. doi: 10.1128/IAI.00562-09
Tsou, L. K., Yount, J. S., Hang, H. C. (2017). Epigallocatechin-3-gallate inhibits bacterial virulence and invasion of host cells. Bioorg Med. Chem. 25 (11), 2883–2887. doi: 10.1016/j.bmc.2017.03.023
Ur-Rehman, T., Slepenkin, A., Chu, H., Blomgren, A., Dahlgren, M. K., Zetterstrom, C. E., et al. (2012). Pre-clinical pharmacokinetics and anti-chlamydial activity of salicylidene acylhydrazide inhibitors of bacterial type III secretion. J. Antibiot (Tokyo) 65 (8), 397–404. doi: 10.1038/ja.2012.43
Uusitalo, P., Hagglund, U., Rhoos, E., Scherman Norberg, H., Elofsson, M., Sundin, C. (2017). The salicylidene acylhydrazide INP0341 attenuates Pseudomonas aeruginosa virulence in vitro and in vivo. J. Antibiot (Tokyo) 70 (9), 937–943. doi: 10.1038/ja.2017.64
Veenendaal, A. K., Sundin, C., Blocker, A. J. (2009). Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J. Bacteriol 191 (2), 563–570. doi: 10.1128/JB.01004-08
Vincent, C. D., Friedman, J. R., Jeong, K. C., Buford, E. C., Miller, J. L., Vogel, J. P. (2006). Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 62 (5), 1278–1291. doi: 10.1111/j.1365-2958.2006.05446.x
Wallden, K., Rivera-Calzada, A., Waksman, G. (2010). Type IV secretion systems: versatility and diversity in function. Cell Microbiol. 12 (9), 1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x
Wang, C., Ye, F., Chang, C., Liu, X., Wang, J., Wang, J., et al. (2019). Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals. Proc. Natl. Acad. Sci. U.S.A. 116 (44), 22331–22340. doi: 10.1073/pnas.1903695116
Wang, D., Zetterström, C. E., Gabrielsen, M., Beckham, K. S., Tree, J. J., Macdonald, S. E., et al. (2011). Identification of bacterial target proteins for the salicylidene acylhydrazide class of virulence-blocking compounds. J. Biol. Chem. 286 (34), 29922–29931. doi: 10.1074/jbc.M111.233858
Wolf, K., Betts, H. J., Chellas-Gery, B., Hower, S., Linton, C. N., Fields, K. A. (2006). Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61 (6), 1543–1555. doi: 10.1111/j.1365-2958.2006.05347.x
Zambelloni, R., Connolly, J. P. R., Huerta Uribe, A., Burgess, K., Marquez, R., Roe, A. J. (2017). Novel compounds targeting the enterohemorrhagic Escherichia coli type three secretion system reveal insights into mechanisms of secretion inhibition. Mol. Microbiol. 105 (4), 606–619. doi: 10.1111/mmi.13719
Zetterström, C. E., Hasselgren, J., Salin, O., Davis, R. A., Quinn, R. J., Sundin, C., et al. (2013). The resveratrol tetramer (-)-hopeaphenol inhibits type III secretion in the gram-negative pathogens Yersinia pseudotuberculosis and Pseudomonas aeruginosa. PLoS One 8 (12), e81969. doi: 10.1371/journal.pone.0081969
Zhang, Y., Liu, Y., Luo, J., Jie, J., Deng, X., Song, L. (2021). The herbal compound thymol targets multiple Salmonella Typhimurium virulence factors for lon protease degradation. Front. Pharmacol. 12. doi: 10.3389/fphar.2021.674955
Zhang, Y., Liu, Y., Qiu, J., Luo, Z. Q., Deng, X. (2018a). The herbal compound thymol protects mice from lethal infection by Salmonella Typhimurium. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01022
Zhang, Y., Liu, Y., Wang, T., Deng, X., Chu, X. (2018b). Natural compound sanguinarine chloride targets the type III secretion system of Salmonella enterica serovar Typhimurium. Biochem. Biophys. Rep. 14, 149–154. doi: 10.1016/j.bbrep.2018.04.011
Zhang, Z., Zhang, Q., Wang, T., Xu, N., Lu, T., Hong, W., et al. (2022). Assessment of global health risk of antibiotic resistance genes. Nat. Commun. 13 (1), 1553. doi: 10.1038/s41467-022-29283-8
Zigangirova, N. A., Nesterenko, L. N., Sheremet, A. B., Soloveva, A. V., Luyksaar, S. I., Zayakin, E. S., et al. (2021). Fluorothiazinon, a small-molecular inhibitor of T3SS, suppresses Salmonella oral infection in mice. J. Antibiot (Tokyo) 74 (4), 244–254. doi: 10.1038/s41429-020-00396-w
Keywords: T3SS, T4SS, bacterial pathogenesis, anti-virulence, pathoblocker
Citation: Blasey N, Rehrmann D, Riebisch AK and Mühlen S (2023) Targeting bacterial pathogenesis by inhibiting virulence-associated Type III and Type IV secretion systems. Front. Cell. Infect. Microbiol. 12:1065561. doi: 10.3389/fcimb.2022.1065561
Received: 09 October 2022; Accepted: 19 December 2022;
Published: 10 January 2023.
Edited by:
Abhinay Sharma, Hebrew University of Jerusalem, IsraelReviewed by:
Jenny-Lee Thomassin, University of Saskatchewan, CanadaAnastasia D. Gazi, Institut Pasteur, France
Copyright © 2023 Blasey, Rehrmann, Riebisch and Mühlen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sabrina Mühlen, c2FicmluYS5tdWVobGVuQHJ1aHItdW5pLWJvY2h1bS5kZQ==
†These authors have contributed equally to this work
‡ORCID: Sabrina Mühlen, orcid.org/0000-0001-7474-705X
Anna Katharina, orcid.org/0000-0003-4496-7881
 Nadja Blasey†
Nadja Blasey† Sabrina Mühlen
Sabrina Mühlen