- 1Key Laboratory of Shaanxi Province for Craniofacial Precision Medicine Research, College of Stomatology, Xi’an Jiaotong University, Xi’an, China
- 2Clinical Research Center of Shaanxi Province for Dental and Maxillofacial Diseases, Center of Oral Public Health, College of Stomatology, Xi’an Jiaotong University, Xi’an, China
- 3State Key Laboratory of Oral Diseases, Department of Preventive Dentistry, West China Hospital of Stomatology, Sichuan University, Chengdu, China
Dental caries is one of the most prevalent and costly biofilm-associated infectious diseases worldwide. Streptococcus mutans (S. mutans) is well recognized as the major causative factor of dental caries due to its acidogenicity, aciduricity and extracellular polymeric substances (EPSs) synthesis ability. The EPSs have been considered as a virulent factor of cariogenic biofilm, which enhance biofilms resistance to antimicrobial agents and virulence compared with planktonic bacterial cells. The traditional anti-caries therapies, such as chlorhexidine and antibiotics are characterized by side-effects and drug resistance. With the development of computer technology, several novel approaches are being used to synthesize or discover antimicrobial agents. In this mini review, we summarized the novel antimicrobial agents targeting the S. mutans biofilms discovery through computer technology. Drug repurposing of small molecules expands the original medical indications and lowers drug development costs and risks. The computer-aided drug design (CADD) has been used for identifying compounds with optimal interactions with the target via silico screening and computational methods. The synthetic antimicrobial peptides (AMPs) based on the rational design, computational design or high-throughput screening have shown increased selectivity for both single- and multi-species biofilms. These methods provide potential therapeutic agents to promote targeted control of the oral microbial biofilms in the near future.
Introduction
The human oral cavity has over 700 bacterial species, harboring the second most diverse microbiome in the body (Pathak et al., 2021). Dental caries is one of the prominent chronic diseases worldwide. S. mutans is considered as the most significant contributor to dental caries due to its cariogenic ability of acid production, acid tolerance and biofilm formation (Pitts et al., 2017; Cui et al., 2019). The biofilm is a structured community that consists of a wide range of microbial species embedded in a self-organized matrix of extracellular polymeric substances (EPSs) (Flemming et al., 2016; Jakubovics et al., 2021). The EPSs not only provide a scaffold for biofilm maturation but also enhance the biofilm tolerance to antimicrobial agents (Jakubovics et al., 2021). The traditional antimicrobial agents, such as the broad-spectrum antimicrobials, pose the challenges for maintaining commensal bacteria (Huang et al., 2021). Though chlorhexidine has a strong bactericidal ability (Coelho et al., 2017), the side effects such as tooth staining, unpleasant taste, mouth irritation, and disturbing the homeostasis of the oral microbiome have limited its usage (Brookes et al., 2020). A larger number of evidence showed that natural products and their derivatives exhibited inhibitory activities against S. mutans growth (Sparks et al., 2017). However, the identification and isolation of active compounds from plants are complex and time-consuming. Moreover, the emergence of antibiotic resistance has necessitated the search for novel antibacterial agents target specific oral bacterial pathogens and inhibit EPSs formation (Coelho et al., 2017; Xia et al., 2019).
The ideal anti-biofilm approach is to facilitate the biofilm dispersion, inhibit the reproduction and metabolism of pathogens, as well as avoid the emergence of antibiotic-resistant bacteria and not disturb the homeostasis of the oral microbiome (Kanwar et al., 2017; Simon et al., 2019). Based on FDA-approved drugs database and small molecule database screening, the drug repurposing has advantages of lower toxicity and faster clinical transition (Wishart et al., 2006; Cui et al., 2019). And the rapid advance of computer technology makes it possible to investigate the biomolecular interactions and design/redesign chemical molecules, which enhance a better understanding the mechanism of inhibition and reveal essential structural properties at the molecular level. The antimicrobial peptides (AMPs) have been considered as potential drug candidates (Bin Hafeez et al., 2021), but it is challenging to improve the specificity of AMPs against S. mutans (Mai et al., 2011). Computational tools enable exploration of previously unexplored regions of AMP sequence space which may yield synthetic peptides with enhanced biological function (Torres et al., 2021). This review introduced emerging computer approaches including drugs repurposing of small-molecule compounds, computer-aided drug design (CADD), and new synthetic antimicrobial peptides (Figure 1).
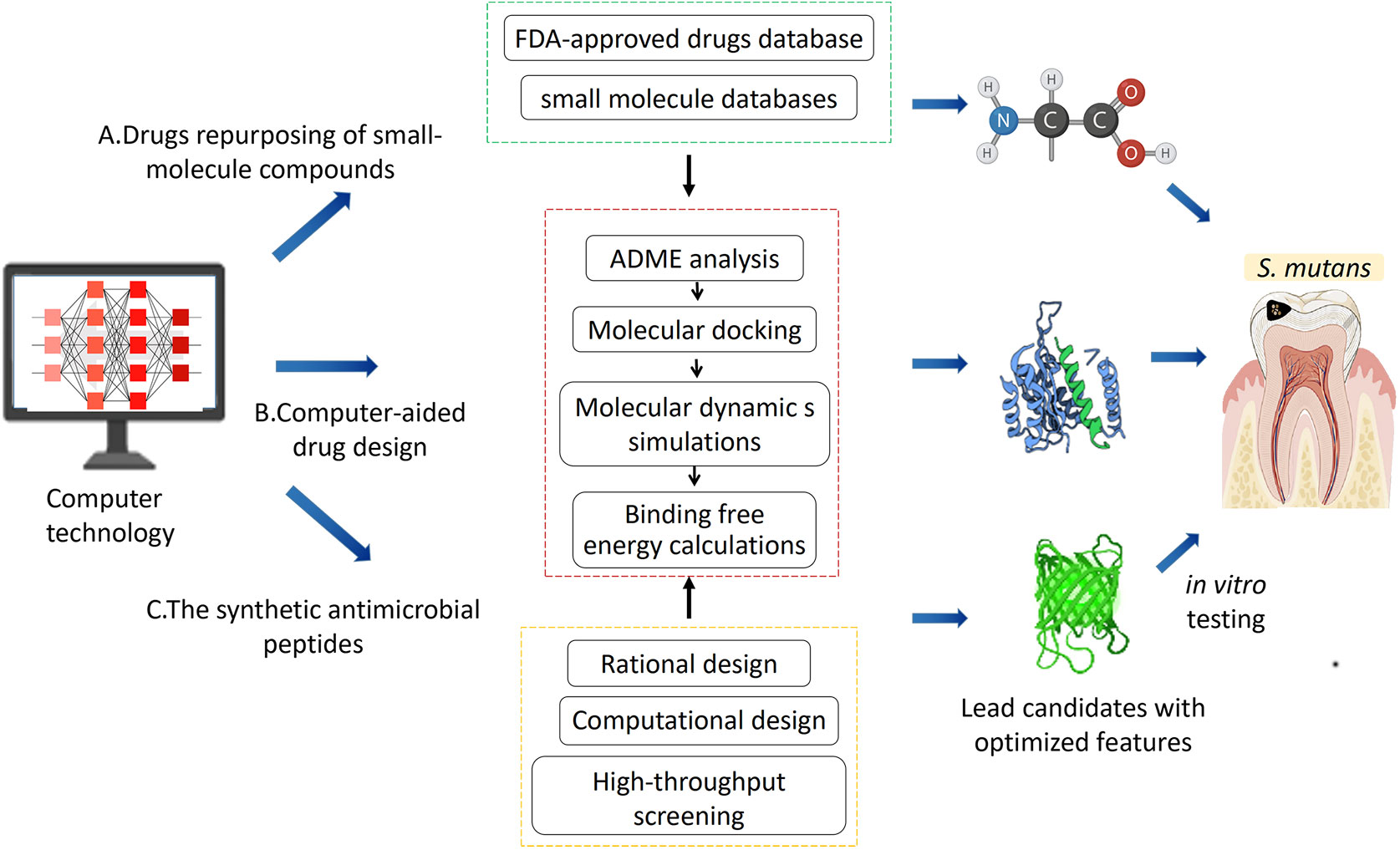
Figure 1 The computer technologies employed to discover novel antimicrobial agents including: (A) drug repurposing of small-molecule compounds; (B) computer-aided drug design; (C) the synthetic antimicrobial peptides. Then in vitro testing targeting the S.mutans was conducted to select the lead candidates with optimal features.
Drugs repurposing of small-molecule compounds
Drug repurposing is a method for discovering new uses of the original drugs beyond the scope of the medical indications. Compared to new drug discovery and development, it offers the advantages of lowering drug development cost and risk as existing drugs have already gone through clinical development stages (Ashburn and Thor, 2004). Small molecule compounds, with a molecular weight <1000 Da, have shown good antimicrobial activity, good stability and low toxicity (Xie et al., 2017; Roman, 2021). Recently, a number of comprehensive small molecule databases have emerged including DrugBank (Wishart et al., 2006), ChEMBL (Bento et al., 2014), PharmGKB (Hewett et al., 2002), Zinc (Williams, 2008), and PubChem (Wang et al., 2012), which have improved the chances of success by enabling the pre-selection of active compounds to test in vitro. In silico prediction of interactions between drugs and target proteins provides a convenient method to predict the new drug–target interactions (DTIs) (Wang and Zeng, 2013). Molecular docking, ligand-based and network-based approaches have been commonly used in virtually screening for a large number of compounds against a target protein (Ekins et al., 2007; Wang and Zeng, 2013). Moreover, Li (Li et al., 2011) have obtained a high enrichment of true positives predictions by using known interaction docking, consensus scoring, and specificity criteria.
Nitro group is critical for the anti–infective activity of nitro-based antimicrobial drugs, including nitroimidazoles, nitrothiazoles, and nitrofurans (Stover et al., 2000; Torreele et al., 2010). Based on the antimicrobial activity of nitrofurans against S.mutans, Zhang (Zhang et al., 2019) synthesized a novel water–soluble hybrid of indolin-2-one and nitrofuran ZY354, which exhibited low cytotoxicity and remarkable antimicrobial activity against S.mutans in multi-species biofilms. Toremifene, an FDA-approved drug for treating breast cancer, has also shown good inhibitory effect on the growth of S. mutans (Gerits et al., 2017a). Kuang (Kuang et al., 2020) identified a natural anticancer compound napabucasin (NAP) showed antimicrobial activity against oral streptococci. Using NAP as a lead compound, Xiao (Lyu et al., 2021) redesigned and synthesized a novel molecule LCG-N25. LCG-N25 exhibited stronger antimicrobial activity toward S.mutans with lower cytotoxicity.
Vitamin C and vitamin D are essential nutrients to human health. Interestingly, increased evidence showed that salivary vitamin C and serum vitamin D levels were associated with the occurrence of dental caries (Syed et al., 2019). Vitamin C has been reported to inhibit the synthesis of EPSs via inhibition of the quorum sensing and other stationary phase regulatory mechanisms (Pandit et al., 2017). Moreover, there was a concentration-dependent inhibitory effect of vitamin C on S. mutans growth and biofilm formation (Eydou et al., 2020). Vitamin D plays a key role in tooth mineralization, and it can lead to the “rachitic tooth” if the levels are unregulated (Foster et al., 2014). Saputo (Saputo et al., 2018) had screened FDA-approved drugs to identify old drugs with new therapeutic effects against S. mutans, and identified the vitamin D derivative doxercalciferol could interfere with S. mutans wall synthesis. Doxercalciferol exhibited synergistic activity in combination with bacitracin and possessed lytic activity against S. mutans through a bacitracin resistance mechanism of MbrABCD. Ferumoxytol was an FDA approved nanoparticle to treat iron deficiency (Schwenk, 2010). Liu (Liu et al., 2018) proved that ferumoxytol could disrupt intractable oral biofilms and prevent dental caries via intrinsic peroxidase-like activity.
Other drugs, such as dihydrofolate reductase inhibitor, trimetrexate analogues (Zhang et al., 2015), antiasthmatic drug zafirlukast (Gerits et al., 2017b), antifungal azoles lotrimazole and econazole (Qiu et al., 2017), efflux inhibitors reserpine (Zeng et al., 2017) have also been shown as potential inhibitors against S. mutans growth (Cui et al., 2019). Dipeptidyl peptidase (DPP IV) is a well-known therapeutic target in Type II diabetes. Anti-human DPP IV drugs saxagliptin can affect S. mutans growth (De et al., 2016; De et al., 2018).
Although drug repositioning has advantages such as lower cost, shorter development timelines and higher safety, the side effects and adverse reactions are yet to be solved (Yang et al., 2021). Firstly, the cytotoxicity of the novel molecules, particularly for the synthetic molecules should be comprehensively evaluated before clinical translation. In addition, drug resistance by oral bacteria still requires long-term evaluation both in vitro and in vivo models (Saputo et al., 2018). And the indications of reused drugs are narrow compared to antibiotics due to their original effects.
Computer-aided drug design (CADD)
Rapid developments in computer technology makes it possible to investigate biomolecular interactions at the molecular level and design/redesign new chemical molecules through computer-aided drug design (CADD) (Dixon et al., 2006). High research costs and significant decrease in the number of new drug approvals have made commercial pharmaceutical companies hesitant to spend on drug discovery research to some extent (Kitchen et al., 2004). CADD provides information about the bioactive parts of compounds virtually and allow rapid examination of the synthesis processes in a resource-efficient, more reliable, and cost-effective way without actually manufacturing them (Kitchen et al., 2004; Thomford et al., 2018; Xu et al., 2022a). Structure-based virtual screening (SBVS) and ligand-based virtual screening (LBVS) are two main strategies commonly applied in computer-aided drug discovery (Khan et al., 2019). SBVS depends on the structure of the target and interactions with the ligands, while LBVS relies on the central similarity-property principle which indicates that similar molecules should exhibit similar properties (Ekins et al., 2007). The chemical similarity calculations are the core of LBVS (Willett, 2003). The methods of LBVS include similarity and substructure searching, quantitative structure-activity relationships (QSAR), and pharmacophore and 3D shape matching (Lavecchia and Di Giovanni, 2013). On the other hand, SBVS employs the 3D structure of the biological target to dock the candidate molecules and ranks them based on their predicted binding affinity or complementarity to the binding site (Lavecchia and Di Giovanni, 2013). SBVS and LBVS significantly minimize the complexity of finding potential therapeutic compounds against the pathogenic bacterial (Tan et al., 2008). Carmen identified ALS-31 as a small molecule inhibitor of S. mutans superoxide dismutase (SOD) by LBVS and SBVS, which inhibited planktonic growth and biofilm formation of S. mutans (Cerchia et al., 2022). Pushkaran and his colleagues (Pushkaran et al., 2019) computationally evaluated repurposing of an FDA approved drug Diosmin (DIO) using structure-based drug design method. They identified Diosmin (DIO) targeting the active site residues of L, D-transpeptidase (Ldt) enzymes which involved in Mycobacterium tuberculosis (M. bt) cell wall biosynthesis.
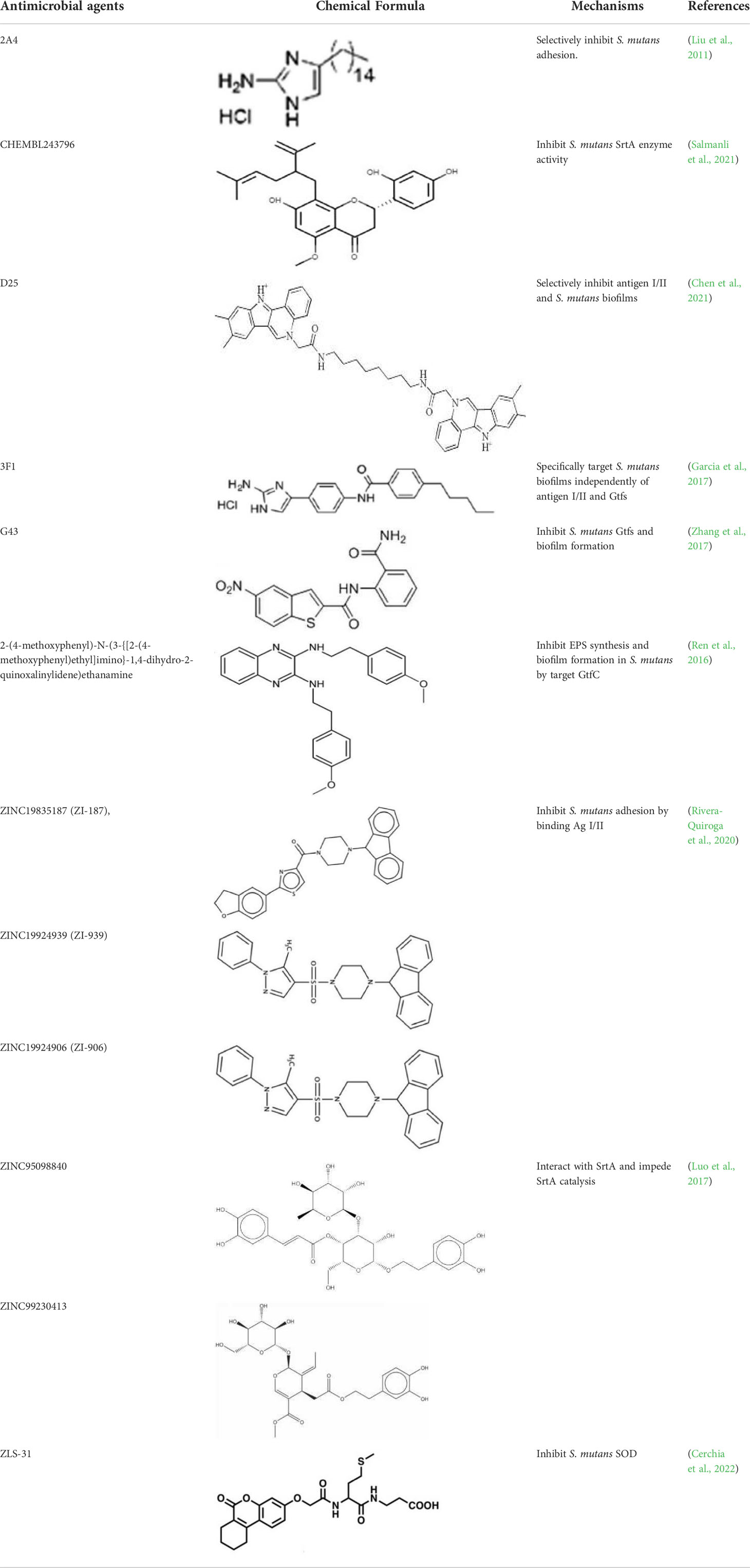
The process of CADD usually includes the following steps. First, various compounds aimed at the target enzyme undergo high-throughput screening. Then an absorption, distribution, metabolism and excretion (ADME) analysis is performed to determine the pharmacokinetic properties of these screened compounds. Next, molecular docking is applied to investigate the mechanism of interaction between these compounds and the target enzyme structure at the molecular level. Finally, molecular dynamic (MD) simulations and binding free energy calculations are performed to analyze the structure stability (Kitchen et al., 2004; Thomford et al., 2018). Key virulence factors such as antigens I/II, Gtfs and SrtA are usually exploited as the targets for computer-aided drug design against S. mutans (Table 1).
Antigens I/II
The antigen I/II (Ag I/II) family of adhesins are widely distributed on the cell surface of many streptococci, which also involved in S. mutans adhesion to the tooth surface and the bacterial co-aggregation (Matsumoto-Nakano, 2018; Yang et al., 2018). Through virtual searching for inhibitors based on Ag I/II protein structures, Rivera (Rivera-Quiroga et al., 2020) found three molecules ZINC19835187 (ZI-187), ZINC19924939 (ZI-939) and ZINC19924906 (ZI-906) inhibited about 90% adhesion of S. mutans. S. mutans cell-surface-localized adhesin P1, is an amyloid-forming protein (Tang et al., 2016). The interactions between C123 (C-terminal segment) and P1 contribute to biofilm-related events such as amyloid fibrils formation, suggesting that C3 would serve as a promising anti-amyloid target (Rivière et al., 2020). Chen (Chen et al., 2021) selected small molecules targeting C3 through structure-based virtual screening, and found that D25 selectively inhibited amyloid fibrils and S. mutans biofilms but had little influence on biofilms formed by S. gordonii and S. sanguinis.
Sortase A
Sortase A (SrtA), one of the membrane-associated sortase enzymes, is responsible for anchoring of numerous virulence-associated surface proteins, including FruA, GbpC, Pac, WapA and Dex, and thus contributes to the biofilm formation of S. mutans (Luo et al., 2017). Although small molecules and natural products including trans-chalcone and flavonoid compounds exhibit effective inhibition against SrtA (Hu et al., 2013; Huang et al., 2014; Singh et al., 2014; Panche et al., 2016). The multi-drug resistance and side-effects make the discovery of new inhibitors for SrtA necessary (Luo et al., 2017). After high-throughput screening, CHEMBL243796 (kurarinone) was found to have especially good inhibitory activity against S. mutans SrtA (Salmanli et al., 2021). Luo (Luo et al., 2017) revealed that several similar compounds including acteoside (ZINC95098840) and oleuropein (ZINC98230413), with good affinities and appropriate pharmacokinetic parameters, were potential inhibitors to impede the catalysis of SrtA.
Glucosyltransferases
The glucosyltransferases (Gtfs) of S. mutans play essential roles in the etiology and pathogenesis of dental caries. The EPSs, mainly synthesized by Gtfs, provide binding sites that promote accumulation of microorganisms on the tooth surface and further establishment of pathogenic biofilms (Koo et al., 2010). Based on SBVS method, Zhang (Zhang et al., 2017) found two small-molecule compounds #G16 and #G43 specifically inhibited Gtfs and S. mutans biofilm formation. The compound #G43 showed great inhibitory effect on the activity of GtfB and GtfC than #G16. Ren (Ren et al., 2016) screened approximately 150,000 compounds from commercially available databases and identified a quinoxaline derivative, 2-(4-methoxyphenyl)-N-(3-{[2-(4-methoxyphenyl)ethyl]imino}-1,4-dihydro-2-quinoxalinylidene) ethanamine as a potential GtfC inhibitor. The computational techniques in drug design have improved the development and optimization of active compounds (Ejalonibu et al., 2021). Liu (Liu et al., 2011) screened a focused small-molecule library and found eight active compounds inhibit S. mutans production of Ag I/II and Gtf, shareing similar structural of 2-aminoimidazole (2-AI), which helped to design the derivatives of marine natural products to inhibit both Gram-positive and Gram-negative bacteria.
The synthetic antimicrobial peptides
Antimicrobial peptides (AMPs) are small bioactive proteins that comprise a part of the body’s first line to inactivate pathogens (Magana et al., 2020). AMPs can inhibit the growth of bacteria, disrupt bacterial cell membrane structure and compete with bacteria for adhesion (Jenssen et al., 2006; Guaní-Guerra et al., 2010; Magana et al., 2020). However, natural antimicrobial peptides generally have limitations, such as a short half-life, unstable in the variable oral environment, and might lead to bacterial resistance (Torres et al., 2019; Niyonsaba et al., 2020). The synthetic AMPs has the advantages including slower emergence of resistance, increased selectivity, and decreased cytotoxicity toward healthy cells (Sullivan et al., 2011; Dalzini et al., 2016; Pletzer et al., 2016). The artificial antimicrobial peptides also have favorable pharmacokinetics and desirable stability (Niu et al., 2021a; Niu et al., 2021b). The synthesized antimicrobial peptide SET-M33D, an isomeric form with D amino acids, can kill multi-resistant pathogens, including Gram-positive S. aureus, S. saprophyticus, and various Gram-negative Enterobacteriaceae with high efficacy and low toxicity (Brunetti et al., 2020). The synthesized antimicrobial peptide GH12 induced low toxicity in human gingival fibroblasts and significantly reduce the cariogenic properties of S. mutans by decreasing the lactic acid production and water-insoluble EPS synthesis (Wang et al., 2018). GH12 could make S. sanguinis and S. gordonii expand their ecological advantages by promoting hydrogen peroxide production, shifting the microbial composition to a more balanced one (Jiang et al., 2018; Jiang et al., 2020).
The methods of AMPs synthesis or discovery can be grouped into three approaches: rational design, computational design and high-throughput screening (Torres et al., 2019; Lei et al., 2021).
Rational design
The fusion of targeting and killing peptides is a common rational design strategy (Guo et al., 2015). A new class of pathogen-selective molecules, called selectively or specifically targeted antimicrobial peptides (STAMPs) was constructed (Eckert et al., 2006b; Huo et al., 2018). STAMP contains a pathogen-specific target peptide and antimicrobial peptide and/or connecting regions can selectively kill bacteria (Huo et al., 2018). S. mutans quorum-sensing (QS) system regulates the gene expression, bacteriocin production and biological behavior partially through competence stimulating peptide (CSP) (Huo et al., 2018). CSP serves as a STAMP targeting domain to mediate S. mutans-specific delivery of the antimicrobial peptide domain (Eckert et al., 2006a). When fused with broad antimicrobial peptide G2 at either the C terminus or N terminus, the CSP-derived STAMP C16G2, M8G2 and C16-33 showed robust, specific activity against S. mutans grown in planktonic cultures and biofilms in both single and multi-species biofilm states (Eckert et al., 2006a; Sullivan et al., 2011; De La Fuente-Nunez, 2019). The STAMPs C16G2, M8G2, C16-33, and M8-33 can target S. mutans without disturbing noncariogenic oral streptococci, indicating that they can maintain a normal ecological balance (Tan et al., 2008). Li (Li et al., 2010) synthesized S. mutans-specific targeting peptide 2_1G2, a derivative in which the S. mutans-specific CSPC16 targeting domain was replaced with peptide 2_1. The STAMP 2_1G2 could lead to protective biofilms formation with the ability to prevent secondary surface colonization by cariogenic S. mutans. A series of STAMPs C8H, C11H, C12H, C13H, and C14H were synthesized, and their selective antibacterial activity against S. mutans on single species and multi-species biofilms were studied, and a total of 21 protein spots were downregulated after C11H treatment.
Computational design
Advances in computational-resources have facilitated the discovery and synthesis of novel AMPs (Porto et al., 2017; De La Fuente-Nunez, 2019), including structure-activity relationships (SAR) study (Abdel Monaim et al., 2018), neural networks (Müller et al., 2018), deep learning (Hamid and Friedberg, 2019). Since understanding the role and importance of each amino acid residue in a given sequence is fundamental for programming peptide, quantitative structure–activity relationship studies (QSAR) have been used to describe amino acid residues and their features (Torres et al., 2021). The approaches applied for the identification of AMPs from databases, including local alignments, regular expressions (REGEX), activity prediction by machine learning (Porto et al., 2017). By using supervised machine learning and a genetic algorithm, Boone (Boone et al., 2021) found a peptide active against S. epidermidis, with an improved ease of synthesis. Yazici (Yazici et al., 2016) analyzed the structures and predicted ternary conformations of the engineered chimeric peptides through computational modeling methods, which exhibited antimicrobial activity against S. mutans, S. epidermidis, and E. coli.
High-throughput screening
High-throughput screening of peptides is also an effective way. The main advantage of large screens is the higher probability of obtaining hits, which helping to select and optimize complex molecular descriptors, and obtain a more precise definition of simple molecular descriptors (Torres et al., 2021). Xie have developed an effective high-throughput screening system for designing and screening peptides that acted selectively on microbial membranes (Xie et al., 2006). However, the reported number of molecules needed to achieve conclusive SAR studies is higher than that required for structure-based screening approaches (Torres et al., 2021).
Conclusion and future prospects
Traditional anti-caries agents, such as fluoride, chlorhexidine, and antibiotics, are characterized by side-effects and low selectivity, which may destroy the homeostasis of oral microbiome. More attention is being directed to find alternative agents to control biofilm-related diseases. Drug repurposing of small molecule compounds lower drug development cost and risk. However, it has come to a consensus that preventing or treating dental biofilms are particularly challenging with small-molecule drugs due to low solubility, brief topical-exposure regimens, salivary clearance, and limited drug diffusion into EPS biofilm matrix (Sims et al., 2020; Osman et al., 2022). Nanoparticle carriers have shown good ability to increase small- molecule drug penetration to biofilms, improve drug stability, and enhance drug bioavailability (Sims et al., 2020; Zhang et al., 2021), which may be potential candidates for clinical translation (Xu et al., 2022b). CADD provides information about the bioactive parts of compounds virtually and allow rapid examination of the synthesis processes. Using friendly and publicly accessible web-servers may be the future direction of prediction methods and computational tools development (Porto et al., 2018). And one of the limitations of computer-guided methods is the need for standardized and reliable biological data as input for designing process. The new synthesized antimicrobial peptide showed increased selectivity and decreased cytotoxicity, but AMPs perform poorly in the oral cavity due to low target specificity in solution, anionic protein adsorption and the diluting effects of saliva (Ramburrun et al., 2021). Designing hydroxyapatite-binding antimicrobial peptide (HBAMP), or incorporation into liquid crystalline systems (LCS) may be effective in the delivery of peptides, more strategies are still required to improve AMPs physiological in vitro and in vivo stability.
Moreover, it is noteworthy that current studies have mainly focused on in vitro or animal studies using single-species biofilms, which would limit the clinical translation of these approaches. The mechanism of small-molecule compounds’ inhibitory effect on the biofilm is still unclear. The multi-species microbial, saliva, and enzyme make the oral cavity environment complex, affecting the efficacy of the novel agents in vivo. Combining these new methods, such as computational tools and algorithms, may be an effective way to synthesize novel antimicrobial agents. Further studies are also necessary to evaluate the antimicrobial activities, bioactivity, and biocompatibility of the novel drug more comprehensively and find effective drug delivery systems.
Author contributions
BZ contributed to original draft preparation and writing. MZ and JT contributed to review and editing. LL and RH contributed to revise the manuscript. All authors contributed to writing and reviewing the manuscript.
Funding
This work was funded by the Health Research Fund of Shaanxi Province (NO. 2021E020) and Interdisciplinary Innovation Projects of West China Hospital of Stomatology (RD-03-202103).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdel Monaim, S., Jad, Y. E., El-Faham, A., de la Torre, B. G., Albericio, F. (2018). Teixobactin as a scaffold for unlimited new antimicrobial peptides: SAR study. Bioorg. Med. Chem. 26 (10), 2788–2796. doi: 10.1016/j.bmc.2017.09.040
Ashburn, T. T., Thor, K. B. (2004). Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discovery 3 (8), 673–683. doi: 10.1038/nrd1468
Bento, A. P., Gaulton, A., Hersey, A., Bellis, L. J., Chambers, J., Davies, M., et al. (2014). The ChEMBL bioactivity database: an update. Nucleic Acids Res. 42 (Database issue), D1083–D1090. doi: 10.1093/nar/gkt1031
Bin Hafeez, A., Jiang, X., Bergen, P. J., Zhu, Y. (2021). Antimicrobial peptides: An update on classifications and databases. Int. J. Mol. Sci. 22 (21), 11691–11742. doi: 10.3390/ijms222111691
Boone, K., Wisdom, C., Camarda, K., Spencer, P., Tamerler, C. (2021). Combining genetic algorithm with machine learning strategies for designing potent antimicrobial peptides. BMC Bioinf. 22 (1), 239–255. doi: 10.1186/s12859-021-04156-x
Brookes, Z. L. S., Bescos, R., Belfield, L. A., Ali, K., Roberts, A. (2020). Current uses of chlorhexidine for management of oral disease: a narrative review. J. Dent. 103, 103497–103505. doi: 10.1016/j.jdent.2020.103497
Brunetti, J., Carnicelli, V., Ponzi, A., Di Giulio, A., Lizzi, A. R., Cristiano, L., et al. (2020). Antibacterial and anti-inflammatory activity of an antimicrobial peptide synthesized with d amino acids. Antibiot. (Basel) 9 (12), 840–856. doi: 10.3390/antibiotics9120840
Cerchia, C., Roscetto, E., Nasso, R., Catania, M. R., De Vendittis, E., Lavecchia, A., et al. (2022). In silico identification of novel inhibitors targeting the homodimeric interface of superoxide dismutase from the dental pathogen Streptococcus mutans. Antioxid. (Basel) 11 (4), 785–803. doi: 10.3390/antiox11040785
Chen, Y., Cui, G., Cui, Y., Chen, D., Lin, H. (2021). Small molecule targeting amyloid fibrils inhibits Streptococcus mutans biofilm formation. AMB Express 11 (1), 171–184. doi: 10.1186/s13568-021-01333-2
Coelho, A., Paula, A. B. P., Carrilho, T. M. P., Da Silva, M., Botelho, M., Carrilho, E. (2017). Chlorhexidine mouthwash as an anticaries agent: A systematic review. Quintessence Int. 48 (7), 585–591. doi: 10.3290/j.qi.a38353
Cui, T., Luo, W., Xu, L., Yang, B., Zhao, W., Cang, H. (2019). Progress of antimicrobial discovery against the major cariogenic pathogen Streptococcus mutans. Curr. Issues Mol. Biol. 32, 601–644. doi: 10.21775/cimb.032.601
Dalzini, A., Bergamini, C., Biondi, B., De Zotti, M., Panighel, G., Fato, R., et al. (2016). The rational search for selective anticancer derivatives of the peptide trichogin GA IV: a multi-technique biophysical approach. Sci. Rep. 6, 24000–24013. doi: 10.1038/srep24000
De La Fuente-Nunez, C. (2019). Toward autonomous antibiotic discovery. mSystems 4 (3), e00151–e00119. doi: 10.1128/mSystems.00151-19
De, A., Lupidi, G., Petrelli, D., Vitali, L. A. (2016). Molecular cloning and biochemical characterization of xaa-pro dipeptidyl-peptidase from Streptococcus mutans and its inhibition by anti-human DPP IV drugs. FEMS Microbiol. Lett. 363 (9), 066–072. doi: 10.1093/femsle/fnw066
De, A., Pompilio, A., Francis, J., Sutcliffe, I. C., Black, G. W., Lupidi, G., et al. (2018). Antidiabetic "gliptins" affect biofilm formation by Streptococcus mutans. Microbiol. Res. 209, 79–85. doi: 10.1016/j.micres.2018.02.005
Dixon, S. L., Smondyrev, A. M., Rao, S. N. (2006). PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chem. Biol. Drug Des. 67 (5), 370–372. doi: 10.1111/j.1747-0285.2006.00384.x
Eckert, R., He, J., Yarbrough, D. K., Qi, F., Anderson, M. H., Shi, W. (2006a). Targeted killing of Streptococcus mutans by a pheromone-guided "smart" antimicrobial peptide. Antimicrob. Agents Chemother. 50 (11), 3651–3657. doi: 10.1128/aac.00622-06
Eckert, R., Qi, F., Yarbrough, D. K., He, J., Anderson, M. H., Shi, W. (2006b). Adding selectivity to antimicrobial peptides: rational design of a multidomain peptide against Pseudomonas spp. Antimicrob. Agents Chemother. 50 (4), 1480–1488. doi: 10.1128/aac.50.4.1480-1488.2006
Ejalonibu, M. A., Ogundare, S. A., Elrashedy, A. A., Ejalonibu, M. A., Lawal, M. M., Mhlongo, N. N., et al. (2021). Drug discovery for Mycobacterium tuberculosis using structure-based computer-aided drug design approach. Int. J. Mol. Sci. 22 (24), 13259–13297. doi: 10.3390/ijms222413259
Ekins, S., Mestres, J., Testa, B. (2007). In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling. Br. J. Pharmacol. 152 (1), 9–20. doi: 10.1038/sj.bjp.0707305
Eydou, Z., Jad, B. N., Elsayed, Z., Ismail, A., Magaogao, M., Hossain, A. (2020). Investigation on the effect of vitamin c on growth & biofilm-forming potential of Streptococcus mutans isolated from patients with dental caries. BMC Microbiol. 20 (1), 231–241. doi: 10.1186/s12866-020-01914-4
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14 (9), 563–575. doi: 10.1038/nrmicro.2016.94
Foster, B. L., Nociti, F. H., Jr., Somerman, M. J. (2014). The rachitic tooth. Endocr. Rev. 35 (1), 1–34. doi: 10.1210/er.2013-1009
Garcia, S. S., Blackledge, M. S., Michalek, S., Su, L., Ptacek, T., Eipers, P., et al. (2017). Targeting of Streptococcus mutans biofilms by a novel small molecule prevents dental caries and preserves the oral microbiome. J. Dent. Res. 96 (7), 807–814. doi: 10.1177/0022034517698096
Gerits, E., Defraine, V., Vandamme, K., De Cremer, K., De Brucker, K., Thevissen, K., et al. (2017a). Repurposing toremifene for treatment of oral bacterial infections. Antimicrob. Agents Chemother. 61 (3), e01846–e01816. doi: 10.1128/aac.01846-16
Gerits, E., van der Massen, I., Vandamme, K., De Cremer, K., De Brucker, K., Thevissen, K., et al. (2017b). In vitro activity of the antiasthmatic drug zafirlukast against the oral pathogens Porphyromonas gingivalis and Streptococcus mutans. FEMS Microbiol. Lett. 364 (2), 005–011. doi: 10.1093/femsle/fnx005
Guaní-Guerra, E., Santos-Mendoza, T., Lugo-Reyes, S. O., Terán, L. M. (2010). Antimicrobial peptides: general overview and clinical implications in human health and disease. Clin. Immunol. 135 (1), 1–11. doi: 10.1016/j.clim.2009.12.004
Guo, L., Mclean, J. S., Yang, Y., Eckert, R., Kaplan, C. W., Kyme, P., et al. (2015). Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 112 (24), 7569–7574. doi: 10.1073/pnas.1506207112
Hamid, M. N., Friedberg, I. (2019). Identifying antimicrobial peptides using word embedding with deep recurrent neural networks. Bioinformatics 35 (12), 2009–2016. doi: 10.1093/bioinformatics/bty937
Hewett, M., Oliver, D. E., Rubin, D. L., Easton, K. L., Stuart, J. M., Altman, R. B., et al. (2002). PharmGKB: the pharmacogenetics knowledge base. Nucleic Acids Res. 30 (1), 163–165. doi: 10.1093/nar/30.1.163
Huang, P., Hu, P., Zhou, S. Y., Li, Q., Chen, W. M. (2014). Morin inhibits sortase a and subsequent biofilm formation in Streptococcus mutans. Curr. Microbiol. 68 (1), 47–52. doi: 10.1007/s00284-013-0439-x
Huang, Y., Liu, Y., Shah, S., Kim, D., Simon-Soro, A., Ito, T., et al. (2021). Precision targeting of bacterial pathogen via bi-functional nanozyme activated by biofilm microenvironment. Biomaterials 268, 120581–120605. doi: 10.1016/j.biomaterials.2020.120581
Hu, P., Huang, P., Chen, W. M. (2013). Curcumin inhibits the sortase a activity of the Streptococcus mutans UA159. Appl. Biochem. Biotechnol. 171 (2), 396–402. doi: 10.1007/s12010-013-0378-9
Huo, L., Huang, X., Ling, J., Liu, H., Liu, J. (2018). Selective activities of STAMPs against Streptococcus mutans. Exp. Ther. Med. 15 (2), 1886–1893. doi: 10.3892/etm.2017.5631
Jakubovics, N. S., Goodman, S. D., Mashburn-Warren, L., Stafford, G. P., Cieplik, F. (2021). The dental plaque biofilm matrix. Periodontol. 86 (1), 32–56. doi: 10.1111/prd.12361
Jenssen, H., Hamill, P., Hancock, R. E. (2006). Peptide antimicrobial agents. Clin. Microbiol. Rev. 19 (3), 491–511. doi: 10.1128/cmr.00056-05
Jiang, W., Wang, Y., Luo, J., Chen, X., Zeng, Y., Li, X., et al. (2020). Antimicrobial peptide GH12 prevents dental caries by regulating dental plaque microbiota. Appl. Environ. Microbiol. 86 (14), e00527–e00520. doi: 10.1128/aem.00527-20
Jiang, W., Wang, Y., Luo, J., Li, X., Zhou, X., Li, W., et al. (2018). Effects of antimicrobial peptide GH12 on the cariogenic properties and composition of a cariogenic multispecies biofilm. Appl. Environ. Microbiol. 84 (24), e01423–e01418. doi: 10.1128/aem.01423-18
Kanwar, I., Sah, A. K., Suresh, P. K. (2017). Biofilm-mediated antibiotic-resistant oral bacterial infections: mechanism and combat strategies. Curr. Pharm. Des. 23 (14), 2084–2095. doi: 10.2174/1381612822666161124154549
Khan, S. U., Ahemad, N., Chuah, L. H., Naidu, R., Htar, T. T. (2019). Sequential ligand- and structure-based virtual screening approach for the identification of potential G protein-coupled estrogen receptor-1 (GPER-1) modulators. RSC Adv. 9 (5), 2525–2538. doi: 10.1039/c8ra09318k
Kitchen, D. B., Decornez, H., Furr, J. R., Bajorath, J. (2004). Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discovery 3 (11), 935–949. doi: 10.1038/nrd1549
Koo, H., Xiao, J., Klein, M. I., Jeon, J. G. (2010). Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 192 (12), 3024–3032. doi: 10.1128/jb.01649-09
Kuang, X., Yang, T., Zhang, C., Peng, X., Ju, Y., Li, C., et al. (2020). Repurposing napabucasin as an antimicrobial agent against oral streptococcal biofilms. BioMed. Res. Int. 2020, 8379526–8379534. doi: 10.1155/2020/8379526
Lavecchia, A., Di Giovanni, C. (2013). Virtual screening strategies in drug discovery: a critical review. Curr. Med. Chem. 20 (23), 2839–2860. doi: 10.2174/09298673113209990001
Lei, M., Jayaraman, A., Van Deventer, J. A., Lee, K. (2021). Engineering selectively targeting antimicrobial peptides. Annu. Rev. BioMed. Eng. 23, 339–357. doi: 10.1146/annurev-bioeng-010220-095711
Li, Y. Y., An, J., Jones, S. J. (2011). A computational approach to finding novel targets for existing drugs. PloS Comput. Biol. 7 (9), e1002139. doi: 10.1371/journal.pcbi.1002139
Li, L. N., Guo, L. H., Lux, R., Eckert, R., Yarbrough, D., He, J., et al. (2010). Targeted antimicrobial therapy against Streptococcus mutans establishes protective non-cariogenic oral biofilms and reduces subsequent infection. Int. J. Oral. Sci. 2 (2), 66–73. doi: 10.4248/ijos10024
Liu, Y., Naha, P. C., Hwang, G., Kim, D., Huang, Y., Simon-Soro, A., et al. (2018). Topical ferumoxytol nanoparticles disrupt biofilms and prevent tooth decay in vivo via intrinsic catalytic activity. Nat. Commun. 9 (1), 2920–2931. doi: 10.1038/s41467-018-05342-x
Liu, C., Worthington, R. J., Melander, C., Wu, H. (2011). A new small molecule specifically inhibits the cariogenic bacterium Streptococcus mutans in multispecies biofilms. Antimicrob. Agents Chemother. 55 (6), 2679–2687. doi: 10.1128/aac.01496-10
Luo, H., Liang, D. F., Bao, M. Y., Sun, R., Li, Y. Y., Li, J. Z., et al. (2017). In silico identification of potential inhibitors targeting Streptococcus mutans sortase a. Int. J. Oral. Sci. 9 (1), 53–62. doi: 10.1038/ijos.2016.58
Lyu, X., Li, C., Zhang, J., Wang, L., Jiang, Q., Shui, Y., et al. (2021). A novel small molecule, LCG-N25, inhibits oral streptococcal biofilm. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.654692
Magana, M., Pushpanathan, M., Santos, A. L., Leanse, L., Fernandez, M., Ioannidis, A., et al. (2020). The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 20 (9), e216–e230. doi: 10.1016/s1473-3099(20)30327-3
Mai, J., Tian, X. L., Gallant, J. W., Merkley, N., Biswas, Z., Syvitski, R., et al. (2011). A novel target-specific, salt-resistant antimicrobial peptide against the cariogenic pathogen Streptococcus mutans. Antimicrob. Agents Chemother. 55 (11), 5205–5213. doi: 10.1128/aac.05175-11
Matsumoto-Nakano, M. (2018). Role of Streptococcus mutans surface proteins for biofilm formation. Jpn. Dent. Sci. Rev. 54 (1), 22–29. doi: 10.1016/j.jdsr.2017.08.002
Müller, A. T., Hiss, J. A., Schneider, G. (2018). Recurrent neural network model for constructive peptide design. J. Chem. Inf. Model. 58 (2), 472–479. doi: 10.1021/acs.jcim.7b00414
Niu, J. Y., Yin, I. X., Mei, M. L., Wu, W. K. K., Li, Q. L., Chu, C. H. (2021a). The multifaceted roles of antimicrobial peptides in oral diseases. Mol. Oral. Microbiol. 36 (3), 159–171. doi: 10.1111/omi.12333
Niu, J. Y., Yin, I. X., Wu, W. K. K., Li, Q. L., Mei, M. L., Chu, C. H. (2021b). Antimicrobial peptides for the prevention and treatment of dental caries: A concise review. Arch. Oral. Biol. 122, 105022–105033. doi: 10.1016/j.archoralbio.2020.105022
Niyonsaba, F., Song, P., Yue, H., Sutthammikorn, N., Umehara, Y., Okumura, K., et al. (2020). Antimicrobial peptide derived from insulin-like growth factor-binding protein 5 activates mast cells via mas-related G protein-coupled receptor X2. Allergy 75 (1), 203–207. doi: 10.1111/all.13975
Niyonsaba, F., Song, P., Yue, H., Sutthammikorn, N., Umehara, Y., Okumura, K., et al. (2020). Antimicrobial peptide derived from insulin-like growth factor-binding protein 5 activates mast cells via mas-related G protein-coupled receptor X2. Allergy 75 (1), 203–207. doi: 10.1111/all.13975
Osman, N., Devnarain, N., Omolo, C. A., Fasiku, V., Jaglal, Y, Govender, T. (2016). Surface modification of nano-drug delivery systems for enhancing antibiotic delivery and activity. Wiley Interdiscip Rev Nanomed Nanobiotechnol, 14 (1), e1758–e61. doi: 10.1002/wnan.1758
Pandit, S., Ravikumar, V., Abdel-Haleem, A. M., Derouiche, A., Mokkapati, V., Sihlbom, C., et al. (2017). Low concentrations of vitamin c reduce the synthesis of extracellular polymers and destabilize bacterial biofilms. Front. Microbiol. 8(2599). doi: 10.3389/fmicb.2017.02599
Pathak, J. L., Yan, Y., Zhang, Q., Wang, L., Ge, L. (2021). The role of oral microbiome in respiratory health and diseases. Respir. Med. 185, 106475–106482. doi: 10.1016/j.rmed.2021.106475
Pitts, N. B., Zero, D. T., Marsh, P. D., Ekstrand, K., Weintraub, J. A., Ramos-Gomez, F., et al. (2017). Dental caries. Nat. Rev. Dis. Primers 3, 17030–17045. doi: 10.1038/nrdp.2017.30
Pletzer, D., Coleman, S. R., Hancock, R. E. (2016). Anti-biofilm peptides as a new weapon in antimicrobial warfare. Curr. Opin. Microbiol. 33, 35–40. doi: 10.1016/j.mib.2016.05.016
Porto, W. F., Fensterseifer, I. C. M., Ribeiro, S. M., Franco, O. L. (2018). Joker: An algorithm to insert patterns into sequences for designing antimicrobial peptides. Biochim. Biophys. Acta Gen. Subj. 1862 (9), 2043–2052. doi: 10.1016/j.bbagen.2018.06.011
Porto, W., Pires, A., Franco, O. (2017). Computational tools for exploring sequence databases as a resource for antimicrobial peptides. Biotechnol. Adv. 35 (3), 337–349. doi: 10.1016/j.biotechadv.2017.02.001
Pushkaran, A. C., Vinod, V., Vanuopadath, M., Nair, S. S., Nair, S. V., Vasudevan, A. K., et al. (2019). Combination of repurposed drug diosmin with amoxicillin-clavulanic acid causes synergistic inhibition of mycobacterial growth. Sci. Rep. 9 (1), 6800–6813. doi: 10.1038/s41598-019-43201-x
Qiu, W., Ren, B., Dai, H., Zhang, L., Zhang, Q., Zhou, X., et al. (2017). Clotrimazole and econazole inhibit Streptococcus mutans biofilm and virulence in vitro. Arch. Oral. Biol. 73, 113–120. doi: 10.1016/j.archoralbio.2016.10.011
Ramburrun, P., Pringle, N. A., Dube, A., Adam, R. Z., D'souza, S., Aucamp, M. (2021). Recent advances in the development of antimicrobial and antifouling biocompatible materials for dental applications. Mater. (Basel) 14 (12), 3167–3198. doi: 10.3390/ma14123167
Ren, Z., Cui, T., Zeng, J., Chen, L., Zhang, W., Xu, X., et al. (2016). Molecule targeting glucosyltransferase inhibits Streptococcus mutans biofilm formation and virulence. Antimicrob. Agents Chemother. 60 (1), 126–135. doi: 10.1128/aac.00919-15
Rivera-Quiroga, R. E., Cardona, N., Padilla, L., Rivera, W., Rocha-Roa, C., Diaz De Rienzo, M. A., et al. (2020). In silico selection and in vitro evaluation of new molecules that inhibit the adhesion of Streptococcus mutants through antigen I/II. Int. J. Mol. Sci. 22 (1), 377–393. doi: 10.3390/ijms22010377
Rivière, G., Peng, E. Q., Brotgandel, A., Andring, J. T., Lakshmanan, R. V., Agbandje-Mckenna, M., et al. (2020). Characterization of an intermolecular quaternary interaction between discrete segments of the Streptococcus mutans adhesin P1 by NMR spectroscopy. FEBS J. 287 (12), 2597–2611. doi: 10.1111/febs.15158
Roman, B. I. (2021). The expanding role of chemistry in optimizing proteins for human health applications. J. Med. Chem. 64 (11), 7179–7188. doi: 10.1021/acs.jmedchem.1c00294
Salmanli, M., Tatar Yilmaz, G., Tuzuner, T. (2021). Investigation of the antimicrobial activities of various antimicrobial agents on Streptococcus mutans sortase a through computer-aided drug design (CADD) approaches. Comput. Methods Programs BioMed. 212, 106454–106461. doi: 10.1016/j.cmpb.2021.106454
Saputo, S., Faustoferri, R. C., Quivey, R. G., Jr. (2018). Vitamin D compounds are bactericidal against Streptococcus mutans and target the bacitracin-associated efflux system. Antimicrob. Agents Chemother. 62 (1), e01675–e01617. doi: 10.1128/aac.01675-17
Schwenk, M. H. (2010). Ferumoxytol: a new intravenous iron preparation for the treatment of iron deficiency anemia in patients with chronic kidney disease. Pharmacotherapy 30 (1), 70–79. doi: 10.1592/phco.30.1.70
Simon, G., Bérubé, C., Voyer, N., Grenier, D. (2019). Anti-biofilm and anti-adherence properties of novel cyclic dipeptides against oral pathogens. Bioorg. Med. Chem. 27 (12), 2323–2331. doi: 10.1016/j.bmc.2018.11.042
Sims, K. R., Jr., Maceren, J. P., Liu, Y., Rocha, G. R., Koo, H., Benoit, D. S. W. (2020). Dual antibacterial drug-loaded nanoparticles synergistically improve treatment of Streptococcus mutans biofilms. Acta Biomater. 115, 418–431. doi: 10.1016/j.actbio.2020.08.032
Singh, P., Anand, A., Kumar, V. (2014). Recent developments in biological activities of chalcones: a mini review. Eur. J. Med. Chem. 85, 758–777. doi: 10.1016/j.ejmech.2014.08.033
Sparks, T. C., Hahn, D. R., Garizi, N. V. (2017). Natural products, their derivatives, mimics and synthetic equivalents: role in agrochemical discovery. Pest Manag. Sci. 73 (4), 700–715. doi: 10.1002/ps.4458
Stover, C. K., Warrener, P., Vandevanter, D. R., Sherman, D. R., Arain, T. M., Langhorne, M. H., et al. (2000). A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405 (6789), 962–966. doi: 10.1038/35016103
Sullivan, R., Santarpia, P., Lavender, S., Gittins, E., Liu, Z., Anderson, M. H., et al. (2011). Clinical efficacy of a specifically targeted antimicrobial peptide mouth rinse: targeted elimination of Streptococcus mutans and prevention of demineralization. Caries Res. 45 (5), 415–428. doi: 10.1159/000330510
Syed, S., Yassin, S. M., Dawasaz, A. A., Amanullah, M., Alshahrani, I., Togoo, R. A. (2019). Salivary 1,5-anhydroglucitol and vitamin levels in relation to caries risk in children. BioMed. Res. Int. 2019, 4503450–4503456. doi: 10.1155/2019/4503450
Tang, W., Bhatt, A., Smith, A. N., Crowley, P. J., Brady, L. J., Long, J. R. (2016). Specific binding of a naturally occurring amyloidogenic fragment of Streptococcus mutans adhesin P1 to intact P1 on the cell surface characterized by solid state NMR spectroscopy. J. Biomol. NMR 64 (2), 153–164. doi: 10.1007/s10858-016-0017-1
Tan, L., Geppert, H., Sisay, M. T., Gütschow, M., Bajorath, J. (2008). Integrating structure- and ligand-based virtual screening: comparison of individual, parallel, and fused molecular docking and similarity search calculations on multiple targets. ChemMedChem 3 (10), 1566–1571. doi: 10.1002/cmdc.200800129
Thomford, N. E., Senthebane, D. A., Rowe, A., Munro, D., Seele, P., Maroyi, A., et al. (2018). Natural products for drug discovery in the 21st century: innovations for novel drug discovery. Int. J. Mol. Sci. 19 (6), 1578–1606. doi: 10.3390/ijms19061578
Torreele, E., Bourdin Trunz, B., Tweats, D., Kaiser, M., Brun, R., Mazué, G., et al. (2010). Fexinidazole–a new oral nitroimidazole drug candidate entering clinical development for the treatment of sleeping sickness. PloS Negl. Trop. Dis. 4 (12), e923–e937. doi: 10.1371/journal.pntd.0000923
Torres, M. D. T., Cao, J., Franco, O. L., Lu, T. K., de la Fuente-Nunez, C. (2021). Synthetic biology and computer-based frameworks for antimicrobial peptide discovery. ACS Nano 15 (2), 2143–2164. doi: 10.1021/acsnano.0c09509
Torres, M. D. T., Sothiselvam, S., Lu, T. K., de la Fuente-Nunez, C. (2019). Peptide design principles for antimicrobial applications. J. Mol. Biol. 431 (18), 3547–3567. doi: 10.1016/j.jmb.2018.12.015
Wang, Y., Wang, X., Jiang, W., Wang, K., Luo, J., Li, W., et al. (2018). Antimicrobial peptide GH12 suppresses cariogenic virulence factors of Streptococcus mutans. J. Oral. Microbiol. 10 (1), 1442089–1442099. doi: 10.1080/20002297.2018.1442089
Wang, Y., Xiao, J., Suzek, T. O., Zhang, J., Wang, J., Zhou, Z., et al. (2012). PubChem's bioAssay database. Nucleic Acids Res. 40, D400–D412. doi: 10.1093/nar/gkr1132
Wang, Y., Zeng, J. (2013). Predicting drug-target interactions using restricted Boltzmann machines. Bioinformatics 29 (13), i126–i134. doi: 10.1093/bioinformatics/btt234
Willett, P. (2003). Similarity-based approaches to virtual screening. Biochem. Soc. Trans. 31 (Pt 3), 603–606. doi: 10.1042/bst0310603
Williams, A. J. (2008). A perspective of publicly accessible/open-access chemistry databases. Drug Discovery Today 13 (11-12), 495–501. doi: 10.1016/j.drudis.2008.03.017
Wishart, D. S., Knox, C., Guo, A. C., Shrivastava, S., Hassanali, M., Stothard, P., et al. (2006). DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672. doi: 10.1093/nar/gkj067
Xia, M. Y., Xie, Y., Yu, C. H., Chen, G. Y., Li, Y. H., Zhang, T., et al. (2019). Graphene-based nanomaterials: the promising active agents for antibiotics-independent antibacterial applications. J. Control Release 307, 16–31. doi: 10.1016/j.jconrel.2019.06.011
Xie, X., Fu, Y., Liu, J. (2017). Chemical reprogramming and transdifferentiation. Curr. Opin. Genet. Dev. 46, 104–113. doi: 10.1016/j.gde.2017.07.003
Xie, Q., Matsunaga, S., Wen, Z., Niimi, S., Kumano, M., Sakakibara, Y., et al. (2006). In vitro system for high-throughput screening of random peptide libraries for antimicrobial peptides that recognize bacterial membranes. J. Pept. Sci. 12 (10), 643–652. doi: 10.1002/psc.774
Xu, Y., Huang, H., Wu, M., Tian, Y., Wan, Q., Shi, B., et al. (2022a). Rapid additive manufacturing of a superlight obturator for Large oronasal fistula in pediatric patient. Laryngoscope, 1–6. doi: 10.1002/lary.30352
Xu, Y., Xu, Y., Zhang, W., Li, M., Wendel, H. P., Geis-Gerstorfer, J., et al. (2022b). Biodegradable zn-Cu-Fe alloy as a promising material for craniomaxillofacial implants: An in vitro investigation into degradation behavior, cytotoxicity, and hemocompatibility. Front. Chem. 10, 860040–860056. doi: 10.3389/fchem.2022.860040
Yang, C., Scoffield, J., Wu, R., Deivanayagam, C., Zou, J., Wu, H. (2018). Antigen I/II mediates interactions between Streptococcus mutans and Candida albicans. Mol. Oral. Microbiol. 33 (4), 283–291. doi: 10.1111/omi.12223
Yang, S., Zhang, J., Yang, R., Xu, X. (2021). Small molecule compounds, a novel strategy against Streptococcus mutans. Pathogens 10 (12), 1540–1555. doi: 10.3390/pathogens10121540
Yazici, H., O'neill, M. B., Kacar, T., Wilson, B. R., Oren, E. E., Sarikaya, M., et al. (2016). Engineered chimeric peptides as antimicrobial surface coating agents toward infection-free implants. ACS Appl. Mater. Interfaces 8 (8), 5070–5081. doi: 10.1021/acsami.5b03697
Zeng, H., Liu, J., Ling, J. (2017). Efflux inhibitor suppresses Streptococcus mutans virulence properties. FEMS Microbiol. Lett. 364 (7), 033–041. doi: 10.1093/femsle/fnx033
Zhang, C., Kuang, X., Zhou, Y., Peng, X., Guo, Q., Yang, T., et al. (2019). A novel small molecule, ZY354, iInhibits dental caries-associated oral biofilms. Antimicrob. Agents Chemother. 63 (5), e02414–e02418. doi: 10.1128/aac.02414-18
Zhang, Q., Nguyen, T., Mcmichael, M., Velu, S. E., Zou, J., Zhou, X., et al. (2015). New small-molecule inhibitors of dihydrofolate reductase inhibit Streptococcus mutans. Int. J. Antimicrob. Agents 46 (2), 174–182. doi: 10.1016/j.ijantimicag.2015.03.015
Zhang, Q., Nijampatnam, B., Hua, Z., Nguyen, T., Zou, J., Cai, X., et al. (2017). Structure-based discovery of small molecule inhibitors of cariogenic virulence. Sci. Rep. 7 (1), 5974–5983. doi: 10.1038/s41598-017-06168-1
Keywords: extracellular polymeric substances, drug repurposing, computer-aided drug design, synthetic antimicrobial peptides, computer technology
Citation: Zhang B, Zhao M, Tian J, Lei L and Huang R (2022) Novel antimicrobial agents targeting the Streptococcus mutans biofilms discovery through computer technology. Front. Cell. Infect. Microbiol. 12:1065235. doi: 10.3389/fcimb.2022.1065235
Received: 09 October 2022; Accepted: 16 November 2022;
Published: 01 December 2022.
Edited by:
Keke Zhang, Wenzhou Medical University, ChinaReviewed by:
Cui Tao, Northwestern Polytechnical University, ChinaShu Deng, Boston University, United States
Zhili Zhao, Central South University, China
Copyright © 2022 Zhang, Zhao, Tian, Lei and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Lei, bGVpbGVpQHNjdS5lZHUuY24=; Ruizhe Huang, aHVhbmdyemhAbWFpbC54anR1LmVkdS5jbg==
 Bin Zhang
Bin Zhang Min Zhao
Min Zhao Jiangang Tian
Jiangang Tian Lei Lei
Lei Lei Ruizhe Huang
Ruizhe Huang