
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 16 November 2022
Sec. Microbiome in Health and Disease
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.1057668
 Damian Muszyński1
Damian Muszyński1 Anna Kudra1
Anna Kudra1 Bartosz Kamil Sobocki2
Bartosz Kamil Sobocki2 Marcin Folwarski3
Marcin Folwarski3 Ermanno Vitale4
Ermanno Vitale4 Veronica Filetti4
Veronica Filetti4 Wojciech Dudzic5
Wojciech Dudzic5 Karolina Kaźmierczak-Siedlecka6*
Karolina Kaźmierczak-Siedlecka6* Karol Połom6
Karol Połom6There is an urgent need to search for new screening methods that allow early detection of esophageal cancer and thus achieve better clinical outcomes. Nowadays, it is known that the esophagus is not a sterile part of the gastrointestinal tract. It is colonized with various microorganisms therefore a “healthy” esophageal microbiome exists. The dysbiotic changes of esophageal microbiome can lead to the development of esophageal diseases including esophageal cancer. There is a strong consensus in the literature that the intestinal microbiome may be involved in esophageal carcinogenesis. Recently, emphasis has also been placed on the relationship between the oral microbiome and the occurrence of esophageal cancer. According to recent studies, some of the bacteria present in the oral cavity, such as Tannerella forsythia, Streptococcus anginosus, Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, and Fusobacterium nucleatum may contribute to the development of this cancer. Moreover, the oral microbiome of patients with esophageal cancer differs significantly from that of healthy individuals. This opens new insights into the search for a microbiome-associated marker for early identification of patients at high risk for developing this cancer.
For years, esophageal cancer has been considered as one of the most common cancers worldwide with a reported 604,100 new cases in 2020 (Uhlenhopp et al., 2020; Sung et al., 2021). A common symptom of esophageal cancer is dysphagia (Yuen et al., 2019), which leads to low amount of food intake and consequently contributes to the development of disease-related malnutrition. According to some data, an estimated 79% of these patients are malnourished (Jordan et al., 2018). The etiology of esophageal cancer is complicated and involves several factors, which are shown in Figure 1 (Huang and Yu, 2018; Uhlenhopp et al., 2020).
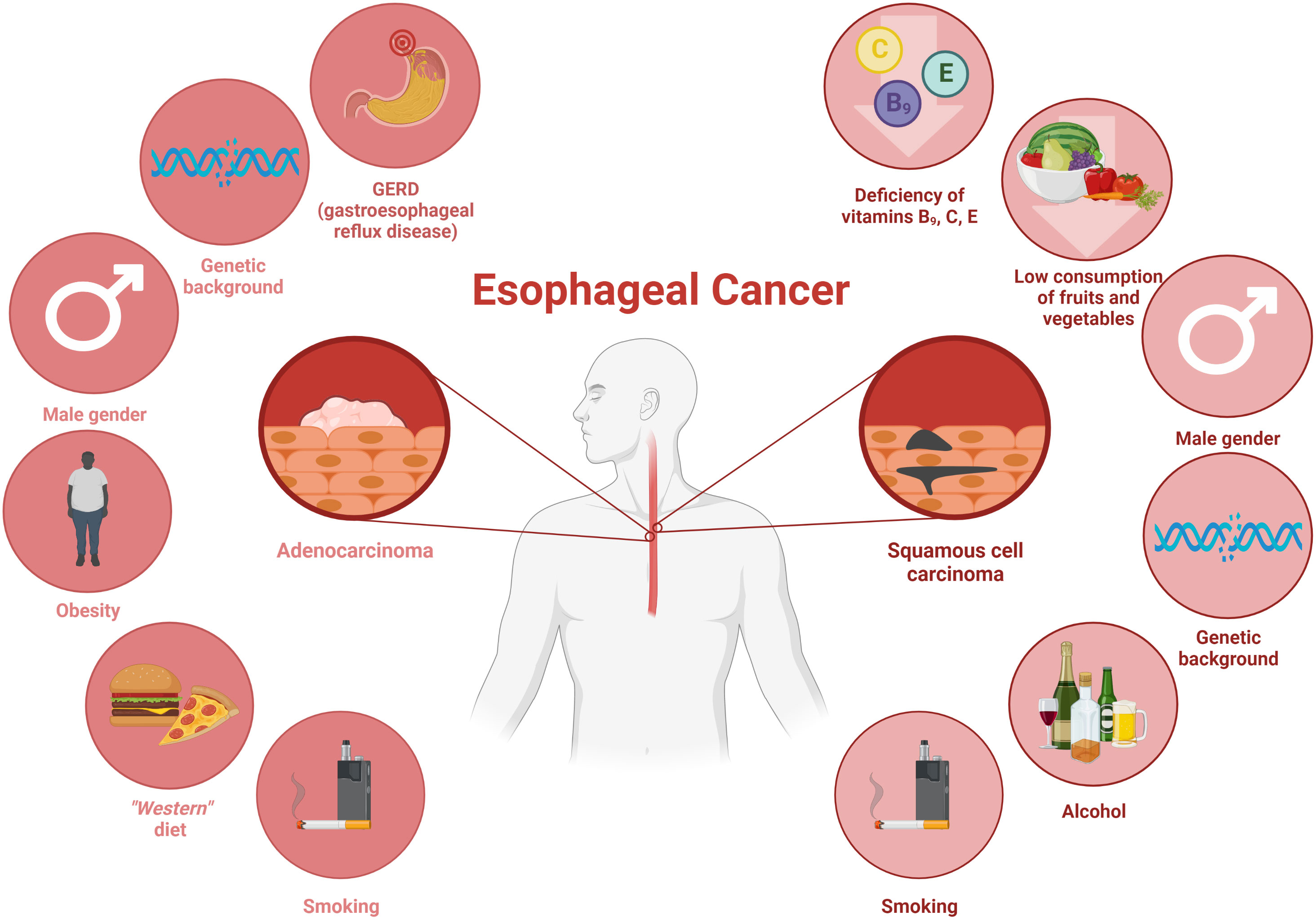
Figure 1 The main risk factors contributing to the development of esophageal cancers: adenocarcinoma and squamous cell carcinoma. Own elaboration based on the literature (Parent et al., 2000; Domper Arnal et al., 2015; Clin et al., 2017; Coleman et al., 2018; Huang and Yu, 2018). This figure was created with Biorender.com.
As it was mentioned above, there are two types of esophageal cancer (Figure 1), namely esophageal squamous cell carcinoma (ESCC, also called OSCC) and esophageal adenocarcinoma (OAC) (Smyth et al., 2017; Short et al., 2017). ESCC is common in East Asia, East Africa, Southern Africa, and Southern Europe, while OAC is more common in developed countries (Huang and Yu, 2018). Over the past four decades, OAC has been found to dominate well ahead of OSCC in terms of incidence (Smyth et al., 2017). The development of esophageal cancer depends on the types of esophageal cancer, due to the fact that they differ in both their biological and anatomical aspects (Smyth et al., 2017). OSCC has similar features to squamous cell carcinoma of the head or neck (Huang and Yu, 2018). In contrast, its precursor in the form of Barrett’s esophagus plays a key role in the development of OAC (Huang and Yu, 2018).
Overall, the above risk factors for esophageal cancer are well described in the literature in contrast to the gut microbiota which, may also play a role in carcinogenesis according to recent published studies (Chen C et al., 2021; Zhou et al., 2021). Therefore, in this review, we briefly discussed the role of the gut microbiota in esophageal carcinogenesis and its alterations in esophageal cancer patients. Then, the relationship between oral microbiota and esophageal cancer development is reviewed based on the recent studies in this field.
The esophageal microbiome is not well understood, however it is known that the esophagus is not a sterile part of the gastrointestinal tract (Laserna-Mendieta et al., 2021). Rapid food passage is observed in the esophagus, which probably limits the presence of microbes. Nevertheless, the pH in healthy individuals is quite stable (around 7) providing a good condition for a variety of microbes (Di Pilato et al., 2016). Analysis of the microbiome revealed that the esophagus is resided with some microorganisms (Figure 2). Notably, upper, middle as well as lower part of the esophagus have a similar composition of the microbiome. Overall, the esophageal microbiome consists of 6 phyla, such as Firmicutes, Bacteroides, Actinobacteria, Proteobacteria, Fusobacteria, and TM7 (Lv et al., 2019). Among the Gram-positive bacteria a diverse microbial population is observed. In particular, the Streptococcus genus is most abundant in the esophagus of healthy individuals (Corning et al., 2018). In addition, Veillonella and Prevotella also occur in the esophagus (Laserna-Mendieta et al., 2021). In esophageal diseases the microbiome is altered (Lv et al., 2019). The imbalanced changes of esophageal microbiome can be used as a marker for the detection of esophageal diseases (Okereke et al., 2019).
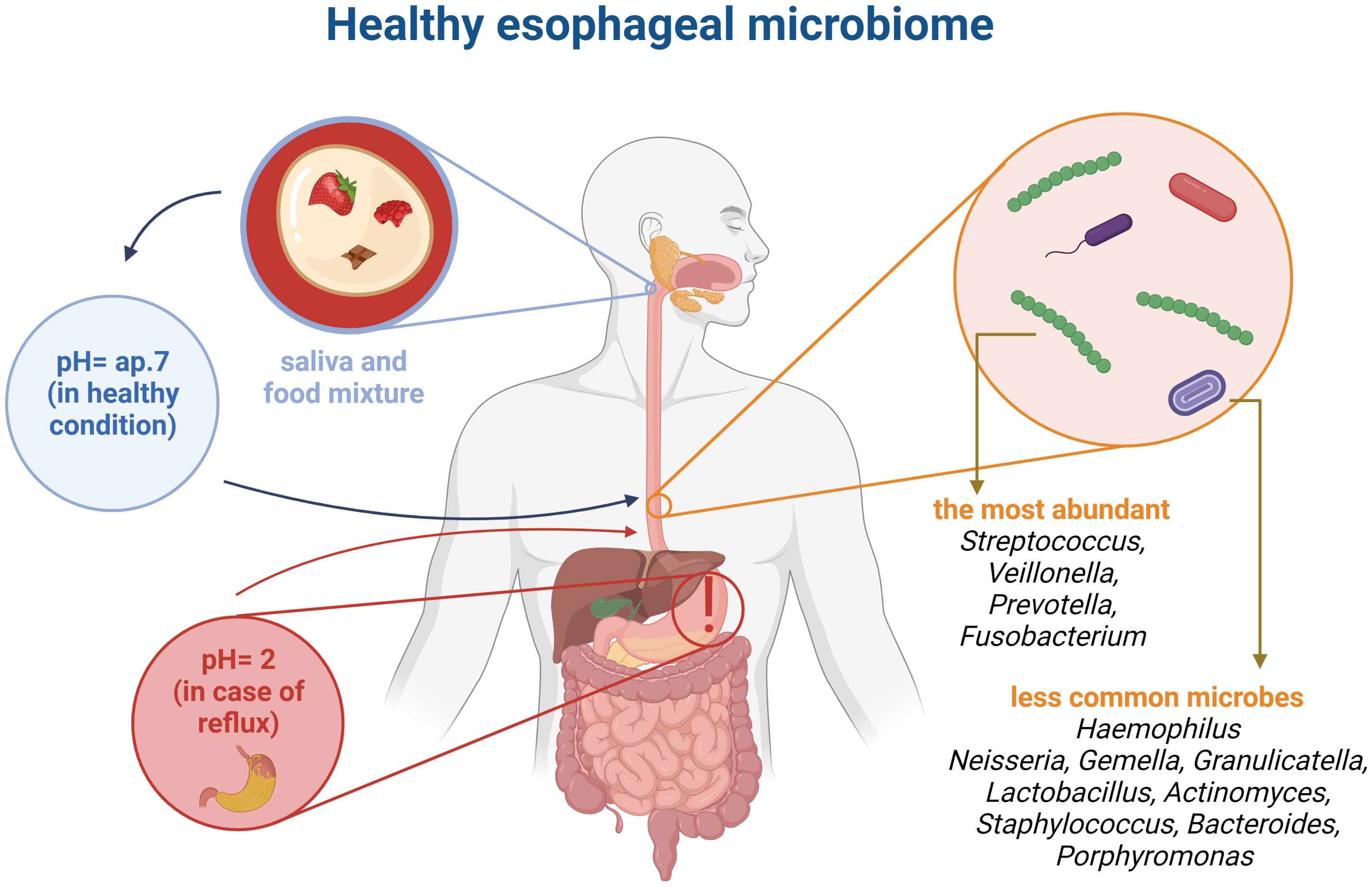
Figure 2 Comparison pointer value pH in healthy organism and with a reflux. Healthy esophageal microbiome based on literature (Di Pilato et al., 2016; Corning et al., 2018; Lv et al., 2019; Laserna-Mendieta et al., 2021). This figure was created using Biorender.com.
In recent years, new studies have developed our knowledge of the relationship between alterations in the gut microbiota and esophageal carcinogenesis. It has been suggested that this relationship may be crucial in both tumor formation and development (Zhou et al., 2021). Previous studies have confirmed that viruses such as human papillomavirus (HPV) and Epstein-Barr virus, as well as alterations in intestinal bacteria may be involved in esophageal carcinogenesis (Meng et al., 2018). A close association has been noted- high rates of HPV are found in areas that also have high rates of ESCC (Xu et al., 2015). Carcinogenesis can be stimulated by the microbiota in several ways. It has an impact on the immune response. Using a mouse model of Barret’s esophagus, Münch et al. showed that changes in the gut microbiota induced by a high-fat diet led to increased levels of proinflammatory cytokines and immune cells and subsequently to a pro-tumor immune phenotype. Importantly, this study demonstrates the necessary role of the gut microbiota in the transmission of dietary influences via inflammatory mechanisms (Münch et al., 2019). A similar mechanism involving the gut microbiota has also been described for a high-fructose diet (Proaño-Vasco et al., 2021). In addition, the study by Lei et al. proved that an adequate composition of the microbiota (in this case, through Saccharomyces boulardii supplementation) prevents unfavorable immune responses (Li B et al., 2022). Another study by Wu et al. showed that the gut microbiota plays a mainly protective role and that its perturbations and the decrease in its level lead to an increased inflammatory response and a shortened survival time. In light of these studies, shaping the composition of the gut microbiota may be a useful approach in order to achieve an effective and specific antitumor immune response. The very important part of treatment is adequate supplementation. For instance, riboflavin deficiency leads to oxidative DNA damage and DNA double-strand breaks (Pan et al., 2020). For this reason, new compounds that should be administered to patients with from esophageal cancer need to be better characterized and discovered.
Another potential mechanism linking the microbiota to esophageal carcinogenesis is overexpression of inducible nitric oxide synthase (iNOS) (Ledda et al., 2019; Gillespie et al., 2021). Park et al. indicated that Gram-negative bacteria expressing lipopolysaccharide could increase iNOS expression and thereby decrease esophageal sphincter relaxation, leading to GERD associated with an increased risk of esophageal cancer (Park et al., 2002). The Warburg effect is one of the best known mechanisms involved in carcinogenesis. Studies by Zhou et al. and Deshpande et al. found that lactate-producing bacterial species are significantly increased in Barrett’s esophagus, gastroesophageal reflux disease (GERD), or esophageal adenocarcinoma. Lactate produced by bacteria may be one of the factors that actively support the transition to anaerobic metabolism and stimulate the growth of esophageal cancer (Deshpande et al., 2018; Zhou et al., 2020). The gut microbiota may influence OAC by upregulating of COX-2 and activating Toll-like receptors (Grover et al., 2021). Some driver bacteria can also produce genotoxins that can stimulate cell proliferation and mutation (Sheflin et al., 2014).
The gut microbiota can also secrete some metabolites. One of the examples is polyamines, which promote tumour progression. Another substance of interest is butyrate which generally has tumour-inhibitory properties; however, at low concentrations, it can also stimulate cancer progression (Buda et al., 2003; Clarke et al., 2008; Donohoe et al., 2012; Belcheva et al., 2014; Donohoe et al., 2014).
Although some evidence has now been assembled, the relationship between the gut microbiota and esophageal adenocarcinoma appears to be poorly studied. Future studies should focus on finding possible link mechanisms.
The association between the gut microbiota and gut microbiota-derived metabolites and many diseases, including cancers, has been studied many times in in vitro and in vivo studies (Mohseni and Fu, 2020). The gut microbiome was investigated in study by Deng et al. involving 23 patients with esophageal cancer and 23 matched healthy individuals (Deng et al., 2021). The gut microbiome was analysed from fresh stool samples by 16S rRNA gene sequencing. Considering the strain, patients with esophageal cancer were found to have significantly higher levels of Firmicutes and Actinobacteria and lower levels of Bacteroidetes compared to healthy individuals. The authors have reported that the abundance of bacteria, producing short-chain fatty acids (SCFAs) is reduced in patients with esophageal cancer while the amount of lipopolysaccharide-producing bacteria is increased (Deng et al., 2021). It should be emphasized that SCFAs pool plays an important role as it has anti-inflammatory effects and increases the integrity of the intestinal barrier, among others. It is also worth noting that butyrate itself is produced by anaerobes in the distal parts of the digestive system, which may indicate its important role in the development of neoplasms in this system, including in the case of esophageal cancer (Guan et al., 2021). Similarly, in another study in ESCC patients (n=18), it was observed that the amount of Bacteroidetes, Fusobacteria and Spirochaetes is decreased (Yang et al., 2021). Additionally, the diversity of intestinal microbiota is also reduced in these patients.
Other studies showed that there is a relationship between intestinal microbiome and esophageal cancer. Analysis of the feces of patients with esophageal cancer (n=40) in comparison with the feces of healthy peoples (n=147) showed that the microbiome of the two groups is different. In this study, the fecal microbiota was additionally analysed in gastric cancer (n=46) and colorectal cancer (n=44). The differences between cancer patients and control subjects were related to the number of Bacteroides fragilis, Escherichia coli, Akkermansia muciniphila, Clostridium hathewayi and Alistipes finegoldii in the faeces, while in cancer patients their amount was increased (Li N et al., 2021). In the case of the control group, the increased number of bacteria was especially Faecalibacterium prausnitzii, Roseburia faecis, Clostridium clostridioforme, and also Bifidobacterium adolescent (Li N et al., 2021).
High-fat diet (HFD) negatively affects the gut microbiome and the bile acid composition. The changes in bile acids composition induced by HFD may contribute to the development of Barrett’s esophagus and esophageal cancer, as shown in a study in mice (Tong et al., 2021). Overall, obesity is related to higher risk of conditions/disorders, such GERD, Barrett’s esophagus as well as OAC (Lagergren, 2011). Distribution of fat in abdominal part plays a significant role in this context (Lagergren, 2011). It should be also mentioned that OAC, which is associated with obesity, may have a different carcinogenic pathway (Schlottmann et al., 2020).
Oral microbiota imbalance can be associated with the development of digestive cancer. Recently in 2021, a systematic review found that there is a difference in the composition of the oral microbiota between patients with digestive cancer and controls subjects (Reitano et al., 2021). Therefore, it is suggested that the oral microbiome may influence the occurrence of esophageal cancer (Peters et al., 2017). Recently, a study by Kawasaki et al. investigated, the relationship between the oral microbiota and esophageal cancer (Kawasaki et al., 2021). In this study, 61 patients with esophageal cancer and 62 matched individuals (without cancer) participated. Samples of both unstimulated saliva and subgingival plaque were collected for analysis of the oral microbiome. The results of this study suggest that bacteria such as Tannerella forsythia and Streptococcus anginosus (from dental plaque) and Aggregatibacter actinomycetemcomitans (from unstimulated saliva) may be associated with higher risk of esophageal cancer (Kawasaki et al., 2021). Notably, virulence factors of A. actinomycetemcomitans include leukotoxin and cytotoxic distension toxin. There are seven different serotypes of this bacterium, i.e., serotypes from a to g. Especially, A. actinomycetemcomitans serotype b causes an aggressive form of periodontitis. An important virulence factor could be a genomic cagE, which is also a kind of marker (Johansson et al., 2019). The study by Peters et al. also found that Tannerella forsythia was associated with a higher risk of EAC whereas Porphyromonas gingivalis was associated with a higher risk of ESCC (Peters et al., 2017). In addition, the authors reported that lower levels of Neisseira as well as Streptococcus pneumoniae may be associated with a lower risk of EAC. The oral microbiome was analysed by 16S rRNA gene sequencing (Peters et al., 2017). T. forsythia is known to be a periodontal pathogen. Its virulence is possible thanks to the O-glycan structures, present in the S-layer of this bacterium, which probably play a crucial role in the development of infection (Chinthamani et al., 2021).
Fusobacterium nucleatum is known to cause periodontal disease, but may also be associated with tumour development (Yamamura et al., 2016). Rapid disease development is possible thanks to specific virulence factors of F. nucleatum, such as adhesin, which allows strong adhesion to host cells (Han, 2015). Recently, it was shown that tissues from esophageal cancer contained significantly more F. nucleatum DNA compared with normal esophageal mucosa (p = 0.021). The analysis was performed by qPCR. It has also been reported that F. nucleatum is associated with shorter survival in these tissues of esophageal cancer (Yamamura et al., 2016). Another pathogen that may influence carcinogenesis is P. gingivalis, which mediates cell transformation in many cancers (Sobocki et al., 2022). Previously, it has been confirmed that the presence of P. gingivalis in the esophageal mucosa is a negative prognostic factor that promotes theprognosis of esophageal cancer (Gao et al., 2016). Therefore, eradication of P. gingivalis may be considered as a potential treatment option. However, some studies have attempted to investigate the potential mechanism of action. Cell line - based experiments by Liang et al. indicated the role of the miR-194/GRHL3/PTEN/Akt axis. In the group of patients with P. gingivalis infection, there were significant differences such as: up-regulation of miR-194 and Akt, down -regulation of GRLH3 (direct target of miR-194) and PTEN compared to patients with negative status. Together, these changes enhanced the pro-proliferative and pro-migratory phenotype of esophageal tumour (Liang et al., 2020). On the other hand, Chen et al. pointed out the mechanism based on the increased production of IL-6 induced by P. gingivalis followed by promoted epithelial-mesenhymal transition and recruitment of myeloid – derived suppressor cells (Chen MF et al., 2021). The review by Malinowski et al. indicated other potential mechanisms such as pro-inflammatory IL-1β cytokine production and secretion of gingipain K by P. gingivalis which causes degradation of immunoglobulins and the complement system. In addition, infection was associated with significant upregulation of MMP-2 and GLUT transporters (Malinowski et al., 2019). However, these hypotheses should be carefully validated in the future.
Apart from the effects of the above mentioned pathogens, other experiments showed different possibilities of the influence of the oral microbiota. A study on about oral microbiota in patients with esophageal cancer was conducted by Hezi Li et al. (Li H et al., 2021). In this study, microbiota analysis was performed by sequencing the 16S rRNA of V3-V4 gene regions. A variety of microbes were examined and the results showed differences in the esophageal microbiota between healthy individuals and patients with esophageal cancer. At the phylum level, patients with OSCC had a reduced amount of Proteobacteria (17.0% vs 20.1% in healthy controls), however slightly higher levels of Bacteroidetes (25.3% vs 24.9%) and Firmicutes (34% vs 31.1%). At the genus level, the difference between OSCC patients and healthy control subjects is seen in Streptococcus (17.3% vs 14.5%), Prevotella (8.6% vs 8.5%) and Neisseria (8.1% vs 10.7%) (Li H et al., 2021).
In another study, Liu et al. investigated the association between the oral microbiome and the risk of malignant esophageal lesions (Liu et al., 2020). The microbiome was studied by sequencing 16s RNA genes. The results suggest that the oral microbiome, its composition, and its content play an important role the esophageal cancer, and thus may be a biomarker that can be considered in early detection. A similar study in a similar region of the world was performed on 39 patients with esophageal cancer and 51 volunteers as a control group (Zhao et al., 2020). The oral microbiome was analysed by 16S rDNA gene sequencing. The study also revealed differences in the oral microbiome between healthy individuals and patients with esophageal cancer. Notably, in the case of patients with esophageal cancer, increased numbers of Firmicutes, Negativicutes, Selenomonadales, Prevotellaceae, Prevotella, and Veillonellaceae were found in patients with esophageal cancer. The percentage of taxa, such as Proteobacteria, Betaproteobacteria, Neisseriales, Neisseriaceae, and Neisseria was reduced. In conclusion the significant difference in the oral microbiome between healthy individuals and those with esophageal cancer suggests that the oral microbiome may be used as a biomarker for the prediction of esophageal cancer in the future. Nevertheless, it should be mentioned that the establishment of a microbial biomarker is associated with several general problems, such as the dependence of the microbiome on ethnicity as well as geographic regions, and many others. In addition Zhang et al, have noted that although changes may have occurred in the oral microbiome of esophageal cancer patients they have observed inconsistencies in research over the years. This is particularly true for Streptococcus, which are widely distributed in both the mouth and esophagus. Consistency is disrupted by geographic locations or inconsistent laboratory methods for isolation and quantification.
The relevant conclusions about the changes of oral microbiota composition and levels from esophageal precancerous lesions to ESCC were drawn by Li et al. study (Li Z et al., 2021). The authors compared the saliva samples by PICRUSt2 analysis and showed that there are significant differences in terms of nitrate oxidoreductase alpha and beta subunits, and nitrate reductase gamma subunit activities between saliva of healthy subjects and patients with ESCC (Li Z et al., 2021). The role of these secondary metabolites produced by oral microbiota should be further investigated and their changes ought to be confirmed. Moreover, the authors proved that alpha diversity in the saliva decreased parallelly to disease progression (Li Z et al., 2021). They also identified common bacterial biomarkers in the group of patients with cancer: Bosea, Solobacterium, Gemella, and Peptostreptococcus and high – grade dysplasia: Lactobacillus. Considering that saliva samples are relatively easy to collect material, oral microbiota and for instance small RNAs (Li K et al., 2022), may be faster and efficient method of screening and diagnosis in the future and should be further investigated. The consistent results were also obtained in Wang et al. study which proved that alpha and beta diversity were significantly lower whereas the variability was higher comparing saliva of patients with ESCC to healthy subjects group (Wang et al., 2019). In addition, this study showed that high risk of esophageal cancer may be linked to both Actinomyces and Atopobium presence in the saliva (Wang et al., 2019). Although the investigated group is limited (20 patients with ESCC vs 21 healthy subjects), the conclusions are promising and should be tested in more numerous cohorts in the future.
Summarizing all above mentioned data/facts, it can be concluded that the link between alterations of oral microbiota and development of esophageal cancer exists. The summary of above mentioned aspects is presented on Figure 3.
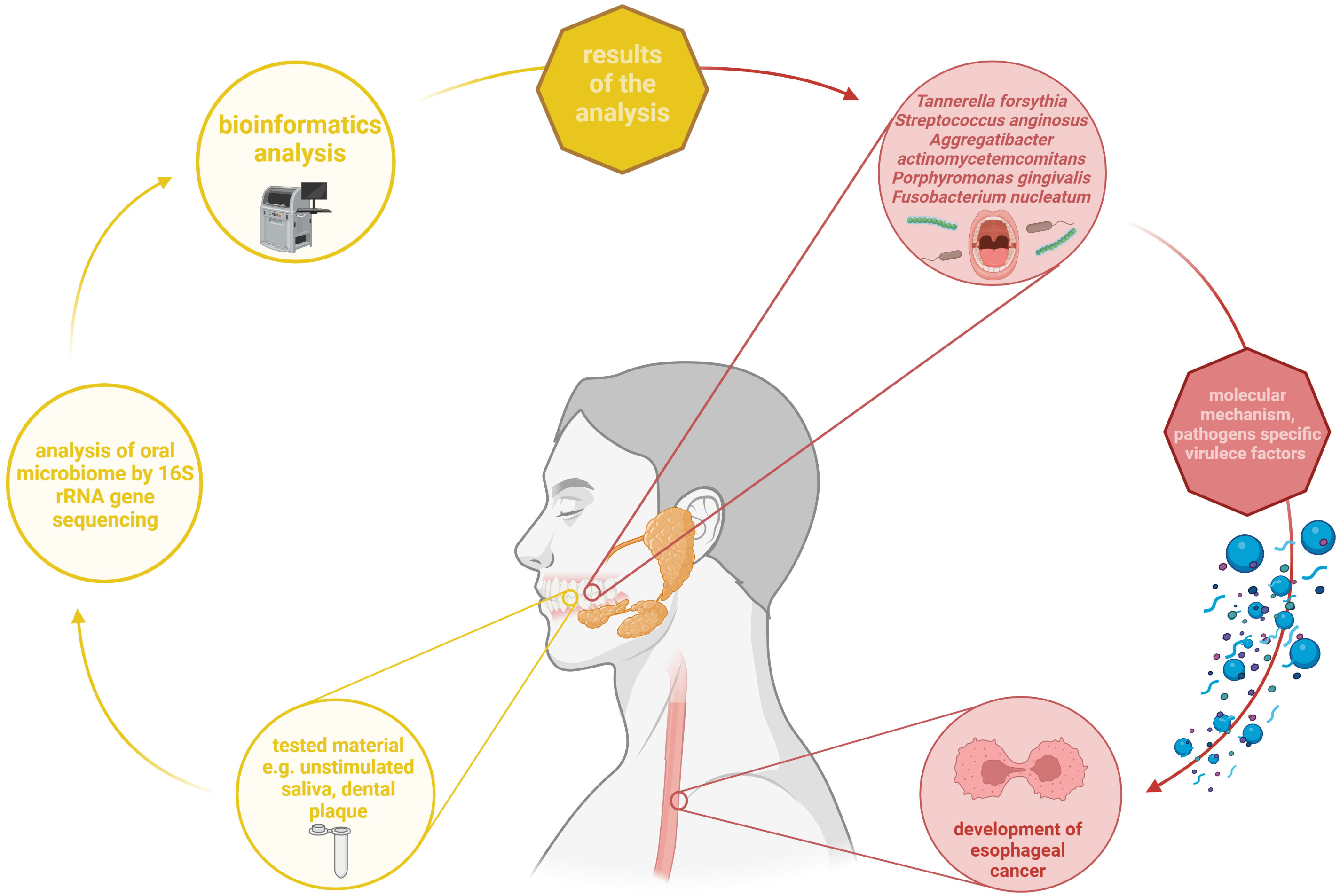
Figure 3 Summary of the relationship between changes in the oral microbiota and development of esophageal cancer. Based on up-to-date studies, some of bacteria such as T. forsythia, S. anginosus, A. actinomycetemcomitans, P. gingivalis, and F. nucleatum may contribute to esophageal cancer occurrence. They are characterized by presenting specific virulence factors that allow them to participate in this process. In most reviewed studies, oral microbiome was analysed from dental plaque and unstimulated saliva was analysed using 16S rRNA gene sequencing. Own elaboration based on the literature (Yamamura et al., 2016; Johansson et al., 2019; Chinthamani et al., 2021). This figure was created using Biorender.com.
There is observed increasing insight into the usage of probiotics in gastrointestinal cancers. Probiotics can be administered as supportive treatment of esophageal cancer to, among others, improve the nutritional status and functioning of immune system. As it was previously mentioned, it is estimated that 79% of esophageal cancer patients are malnourished (Jordan et al., 2018). Recently, in study protocol Liu et al. have reported that they will analyse the impact of probiotics on both nutritional status as well as gastrointestinal complications in patients with esophageal cancer in postoperative period (Liu et al., 2021). The impact of probiotics on nutritional status was analysed in Kaźmierczak-Siedlecka et al. double-blind, randomized and placebo-controlled study regarding cancer patients (also with esophageal cancer) (Kaźmierczak-Siedlecka et al., 2020). In this study probiotic strain Lactobacillus plantarum 299v was given per 4 weeks. It was observed that the level of albumin was significantly higher in group receiving probiotics (p=0.032). The level of albumin is one of the laboratory parameters used to assess the nutritional status besides prealbumin, total protein, and total lymphocyte count (Kaźmierczak-Siedlecka et al., 2020). Another probiotic Lactobacillus rhamnosus (PTCC 1637) may also be promising in case of esophageal cancer, what has been confirmed in Hashemi-Khah et al. study (Hashemi-Khah et al., 2022). Despite the fact that overall Lactobacillus spp. and Bifidobacterium spp. are the most commonly used probiotics (Kaźmierczak-Siedlecka et al., 2021), the other probiotics can also be considered. Nevertheless, specialists should pay attention to probiotic strains and their related properties.
Changes that occur during the development of esophageal neoplasms affect not only the esophageal microbiota, but also other elements of the human body, including the oral cavity microbiome and the intestinal microbiome. The increasing amount of F. nucleatum, S. anginosus or A. actinomycetemcomitans, among others, during the development of the disease may serve as biomarkers in the future. However, further research is needed in this direction. Increased risk of esophageal cancer may be caused by improper diet or hygiene, leading to a proliferation of bacteria responsible not only for the development of esophageal cancer, but also for other diseases, including gingivitis or circulatory disorders. Last, but not least it should be mentioned that pathogens present in oral cavity such as F. nucleatum and P. ginvivalis, seem to have a strong influence on progression and final outcome. Therefore, new studies should be conducted to investigate the impact of possible eradication as a therapeutic option on survival and other prognostic parameters. Determining the most influential and discovering new prognostic microbes in the oral cavity should definitely be a goal for further studies.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Belcheva, A., Irrazabal, T., Robertson, S. J., Streutker, C., Maughan, H., Rubino, S., et al. (2014). Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 158, 288–299. doi: 10.1016/j.cell.2014.04.051
Buda, A., Qualtrough, D., Jepson, M. A., Martines, D., Paraskeva, C., Pignatelli, M. (2003). Butyrate downregulates alpha2beta1 integrin: a possible role in the induction of apoptosis in colorectal cancer cell lines. Gut 52, 729–734. doi: 10.1136/gut.52.5.729
Chen, C., Chen, L., Lin, L., Jin, D., Du, Y., Lyu, J. (2021). Research progress on gut microbiota in patients with gastric cancer, esophageal cancer, and small intestine cancer. Appl. Microbiol. Biotechnol. 105, 4415–4425. doi: 10.1007/s00253-021-11358-z
Chen, M. F., Lu, M. S., Hsieh, C. C., Chen, W. C. (2021). Porphyromonas gingivalis promotes tumor progression in esophageal squamous cell carcinoma. Cell. Oncol. Dordr. 44, 373–384. doi: 10.1007/s13402-020-00573-x
Chinthamani, S., Settem, P. R., Honma, K., Nakajima, T., Sharma, A. (2021). Purification of tannerella forsythia surface-layer (S-layer) proteins. Methods Mol. Biol. Clifton N.J. 2210, 135–142. doi: 10.1007/978-1-0716-0939-2_13
Clarke, J. M., Topping, D. L., Bird, A. R., Young, G. P., Cobiac, L. (2008). Effects of high-amylose maize starch and butyrylated high-amylose maize starch on azoxymethane-induced intestinal cancer in rats. Carcinogenesis 29, 2190–2194. doi: 10.1093/carcin/bgn192
Clin, B., Thaon, I., Boulanger, M., Brochard, P., Chamming’s, S., Gislard, A., et al. (2017). Cancer of the esophagus and asbestos exposure. Am. J. Ind. Med. 60, 968–975. doi: 10.1002/ajim.22769
Coleman, H. G., Xie, S. H., Lagergren, J. (2018). The epidemiology of esophageal adenocarcinoma. Gastroenterology 154, 390–405. doi: 10.1053/j.gastro.2017.07.046
Corning, B., Copland, A. P., Frye, J. W. (2018). The esophageal microbiome in health and disease. Curr. Gastroenterol. Rep. 20, 39. doi: 10.1007/s11894-018-0642-9
Deng, Y., Tang, D., Hou, P., Shen, W., Li, H., Wang, T., et al. (2021). Dysbiosis of gut microbiota in patients with esophageal cancer. Microb. Pathog. 150, 104709. doi: 10.1016/j.micpath.2020.104709
Deshpande, N. P., Riordan, S. M., Castaño-Rodríguez, N., Wilkins, M. R., Kaakoush, N. O. (2018). Signatures within the esophageal microbiome are associated with host genetics, age, and disease. Microbiome 6, 227. doi: 10.1186/s40168-018-0611-4
Di Pilato, V., Freschi, G., Ringressi, M. N., Pallecchi, L., Rossolini, G. M., Bechi, P. (2016). The esophageal microbiota in health and disease. Ann. N. Y. Acad. Sci. 1381, 21–33. doi: 10.1111/nyas.13127
Domper Arnal, M. J., Ferrández Arenas, Á., Lanas Arbeloa, Á. (2015). Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J. Gastroenterol. W.J.G. 21, 7933–7943. doi: 10.3748/wjg.v21.i26.7933
Donohoe, D. R., Collins, L. B., Wali, A., Bigler, R., Sun, W., Bultman, S. J. (2012). The warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell. 48, 612–626. doi: 10.1016/j.molcel.2012.08.033
Donohoe, D. R., Holley, D., Collins, L. B., Montgomery, S. A., Whitmore, A. C., Hillhouse, A., et al. (2014). A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discovery 4, 1387–1397. doi: 10.1158/2159-8290.cd-14-0501
Gao, S., Li, S., Ma, Z., Liang, S., Shan, T., Zhang, M., et al. (2016). Presence of porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect. Agent Cancer 11, 3. doi: 10.1186/s13027-016-0049-x
Gillespie, M. R., Rai, V., Agrawal, S., Nandipati, K. C. (2021). The role of microbiota in the pathogenesis of esophageal adenocarcinoma. Biology 10, 697. doi: 10.3390/biology10080697
Grover, K., Gregory, S., Gibbs, J. F., Emenaker, N. J. (2021). A discussion of the gut microbiome’s development, determinants, and dysbiosis in cancers of the esophagus and stomach. J. Gastrointest. Oncol. 12, 290–300. doi: 10.21037/jgo-2019-gi-07
Guan, X., Li, W., Meng, H. (2021). A double-edged sword: Role of butyrate in the oral cavity and the gut. Mol. Oral. Microbiol. 36, 121–131. doi: 10.1111/omi.12322
Han, Y. W. (2015). Fusobacterium nucleatum: a commensal-turned pathogen. Curr. Opin. Microbiol. 23, 141–147. doi: 10.1016/j.mib.2014.11.013
Hashemi-Khah, M. S., Arbab-Soleimani, N., Forghanifard, M. M., Gholami, O., Taheri, S., Amoueian, S. (2022). An In vivo study of lactobacillus rhamnosus (PTCC 1637) as a new therapeutic candidate in esophageal cancer. BioMed. Res. Int., 7607470. doi: 10.1155/2022/7607470
Huang, F. L., Yu, S. J. (2018). Esophageal cancer: Risk factors, genetic association, and treatment. Asian J. Surg. 41, 210–215. doi: 10.1016/j.asjsur.2016.10.005
Johansson, A., Claesson, R., Höglund Åberg, C., Haubek, D., Lindholm, M., Jasim, S., et al. (2019). Genetic profiling of aggregatibacter actinomycetemcomitans serotype b isolated from periodontitis patients living in Sweden. Pathog. Basel. Switz. 8, 153. doi: 10.3390/pathogens8030153
Jordan, T., Mastnak, D. M., Palamar, N., Kozjek, N. R. (2018). Nutritional therapy for patients with esophageal cancer. Nutr. Cancer. 70, 23–29. doi: 10.1080/01635581.2017.1374417
Kawasaki, M., Ikeda, Y., Ikeda, E., Takahashi, M., Tanaka, D., Nakajima, Y., et al. (2021). Oral infectious bacteria in dental plaque and saliva as risk factors in patients with esophageal cancer. Cancer 127, 512–519. doi: 10.1002/cncr.33316
Kaźmierczak-Siedlecka, K., Folwarski, M., Ruszkowski, J., Skonieczna-Żydecka, K., Szafrański, W., Makarewicz, W. (2020). Effects of 4 weeks of lactobacillus plantarum 299v supplementation on nutritional status, enteral nutrition tolerance, and quality of life in cancer patients receiving home enteral nutrition - a double-blind, randomized, and placebo-controlled trial. Eur. Rev. Med. Pharmacol. Sci. 24, 9684–9694. doi: 10.26355/eurrev_202009_23059
Kaźmierczak-Siedlecka, K., Roviello, G., Catalano, M., Polom, K. (2021). Gut microbiota modulation in the context of immune-related aspects of lactobacillus spp. Bifidobacterium spp. Gastrointestinal Cancers. Nutrients 13, 2674. doi: 10.3390/nu13082674
Lagergren, J. (2011). Influence of obesity on the risk of esophageal disorders. Nat. Rev. Gastroenterol. Hepatol. 8, 340–347. doi: 10.1038/nrgastro.2011.73
Laserna-Mendieta, E. J., FitzGerald, J. A., Arias-Gonzalez, L., Ollala, J. M., Bernardo, D., Claesson, M. J., et al. (2021). Esophageal microbiome in active eosinophilic esophagitis and changes induced by different therapies. Sci. Rep. 11, 7113. doi: 10.1038/s41598-021-86464-z
Ledda, C., Cannizzaro, E., Lovreglio, P., Vitale, E., Stufano, A., Montana, A., et al. (2019). Exposure to toxic heavy metals can influence homocysteine metabolism? Antioxid. Basel Switz. 9, 30. doi: 10.3390/antiox9010030
Liang, G., Wang, H., Shi, H., Zhu, M., An, J., Qi, Y., et al. (2020). Porphyromonas gingivalis promotes the proliferation and migration of esophageal squamous cell carcinoma through the miR-194/GRHL3/PTEN/Akt axis. ACS Infect. Dis. 6, 871–881. doi: 10.1021/acsinfecdis.0c00007
Li, N., Bai, C., Zhao, L., Sun, Z., Ge, Y., Li, X. (2021). The relationship between gut microbiome features and chemotherapy response in gastrointestinal cancer. Front. Oncol. 11. doi: 10.3389/fonc.2021.781697
Li, Z., Dou, L., Zhang, Y., He, S., Zhao, D., Hao, C., et al. (2021). Characterization of the oral and esophageal microbiota in esophageal precancerous lesions and squamous cell carcinoma. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.714162
Li, K., Lin, Y., Luo, Y., Xiong, X., Wang, L., Durante, K., et al. (2022). A signature of saliva-derived exosomal small RNAs as predicting biomarker for esophageal carcinoma: a multicenter prospective study. Mol. Cancer. 18, 21. doi: 10.1186/s12943-022-01499-8
Li, H., Luo, Z., Zhang, H., Huang, N., Li, D., Luo, C., et al. (2021). Characteristics of oral microbiota in patients with esophageal cancer in China. BioMed. Res. Int. 2021, 2259093. doi: 10.1155/2021/2259093
Liu, F., Liu, M., Liu, Y., Guo, C., Zhou, Y., Li, F., et al. (2020). Oral microbiome and risk of malignant esophageal lesions in a high-risk area of China: A nested case-control study. Chin. J. Cancer Res. Chung-Kuo Yen Cheng Yen Chiu. 32, 742–754. doi: 10.21147/j.issn.1000-9604.2020.06.07
Liu, C., Yang, J., Dong, W., Yuan, J. (2021). Effects of probiotics on gastrointestinal complications and nutritional status of postoperative patients with esophageal cancer. Med. (Baltimore). 100, 25138. doi: 10.1097/MD.0000000000025138
Li, B., Zhang, H., Shi, L., Li, R., Luo, Y., Deng, Y., et al. (2022). Saccharomyces boulardii alleviates DSS-induced intestinal barrier dysfunction and inflammation in humanized mice. Food Funct. 13, 102–112. doi: 10.1039/d1fo02752b
Lv, J., Guo, L., Liu, J. J., Zhao, H. P., Zhang, J., Wang, J. H. (2019). Alteration of the esophageal microbiota in barrett’s esophagus and esophageal adenocarcinoma. World J. Gastroenterol. 25, 2149–2161. doi: 10.3748/wjg.v25.i18.2149
Malinowski, B., Węsierska, A., Zalewska, K., Sokołowska, M. M., Bursiewicz, W., Socha, M., et al. (2019). The role of tannerella forsythia and porphyromonas gingivalis in pathogenesis of esophageal cancer. Infect. Agent Cancer 14, 3. doi: 10.1186/s13027-019-0220-2
Meng, C., Bai, C., Brown, T. D., Hood, L. E., Tian, Q. (2018). Human gut microbiota and gastrointestinal cancer. Genomics Proteomics Bioinf. 16, 33–49. doi: 10.1016/j.gpb.2017.06.002
Mohseni, A. H., Fu, X. (2020). Gut microbiota-derived metabolites and colorectal cancer: New insights and updates. Microb. Pathog. 149, 104569. doi: 10.1016/j.micpath.2020.104569
Münch, N. S., Fang, H. Y., Ingermann., J., Maurer, H. C., Anand, A., Kellner, V., et al. (2019). High-fat diet accelerates carcinogenesis in a mouse model of barrett’s esophagus via interleukin 8 and alterations to the gut microbiome. Gastroenterology 157, 492–506. doi: 10.1053/j.gastro.2019.04.013
Okereke, I., Hamilton, C., Wenholz, A., Jala, V., Giang, T., Reynolds, S., et al. (2019). Associations of the microbiome and esophageal disease. J. Thorac. Dis. 11, 1588–1593. doi: 10.21037/jtd.2019.05.82
Pan, F., Xu, X., Zhang, L. L., Luo, H. J., Chen, Y., Long, L., et al. (2020). Dietary riboflavin deficiency induces genomic instability of esophageal squamous cells that is associated with gut microbiota dysbiosis in rats. Food Funct. 11, 10070–10083. doi: 10.1039/d0fo90058c
Parent, M. E., Siemiatycki, J., Fritschi, L. (2000). Workplace exposures and oesophageal cancer. Occup. Environ. Med. 57, 325–334. doi: 10.1136/oem.57.5.325
Park, H., Clark, E., Cullen, J. J., Koland, J. G., Kim, M. S., Conklin, J. L. (2002). Expression of inducible nitric oxide synthase in the lower esophageal sphincter of the endotoxemic opossum. J. Gastroenterol. 37, 1000–1004. doi: 10.1007/s005350200169
Peters, B. A., Wu, J., Pei, Z., Yang, L., Purdue, M. P., Freedman, N. D., et al. (2017). Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 77, 6777–6787. doi: 10.1158/0008-5472.can-17-1296
Proaño-Vasco, A., Baumeister, T., Metwaly, A., Reitmeier, S., Kleigrewe, K., Meng, C., et al. (2021). High-fructose diet alters intestinal microbial profile and correlates with early tumorigenesis in a mouse model of barrett’s esophagus. Microorganisms 9, 2432. doi: 10.3390/microorganisms9122432
Reitano, E., de’Angelis, N., Gavriilidis, P., Gaiani, F., Memeo, R., Inchingolo, R., et al. (2021). Oral bacterial microbiota in digestive cancer patients: A systematic review. Microorganisms 9, 2585. doi: 10.3390/microorganisms9122585
Schlottmann, F., Dreifuss, N. H., Patti, M. G. (2020). Obesity and esophageal cancer: GERD, barrett´s esophagus, and molecular carcinogenic pathways. Expert Rev. Gastroenterol. Hepatol. 14, 425–433. doi: 10.1080/17474124.2020.1764348
Sheflin, A. M., Whitney, A. K., Weir, T. L. (2014). Cancer-promoting effects of microbial dysbiosis. Curr. Oncol. Rep. 16, 406. doi: 10.1007/s11912-014-0406-0
Smyth, E. C., Lagergren, J., Fitzgerald, R. C., Lordick, F., Shah, M. A., Lagergren, P., et al. (2017). Oesophageal cancer. Nat. Rev. Dis. Primer. 3, 17048. doi: 10.1038/nrdp.2017.48
Sobocki, B. K., Basset, C. A., Bruhn-Olszewska, B., Olszewski, P., Szot, O., Kaźmierczak-Siedlecka, K., et al. (2022). Molecular mechanisms leading from periodontal disease to cancer. Int. J. Mol. Sci. 23, 970. doi: 10.3390/Fijms23020970
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. C.A. Cancer J. Clin. 71, 209–249. doi: 10.3322/caac.21660
Tong, Y., Gao, H., Qi, Q., Liu, X., Li, J., Gao, J., et al. (2021). High fat diet, gut microbiome and gastrointestinal cancer. Theranostics 11, 5889–5910. doi: 10.7150/thno.56157
Uhlenhopp, D. J., Then, E. O., Sunkara, T., Gaduputi, V. (2020). Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin. J. Gastroenterol. 13, 1010–1021. doi: 10.1007/s12328-020-01237-x
Wang, Q., Rao, Y., Guo, X., Liu, N., Liu, S., Wen, P., et al. (2019). Oral microbiome in patients with oesophageal squamous cell carcinoma. Sci. Rep. 13, 19055. doi: 10.1038/s41598-019-55667-w
Xu, W., Liu, Z., Bao, Q., Qian, Z. (2015). Viruses, other pathogenic microorganisms and esophageal cancer. Gastrointest. Tumors 2, 2–13. doi: 10.1159/000380897
Yamamura, K., Baba, Y., Nakagawa, S., Mima, K., Miyake, K., Nakamura, K., et al. (2016). Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin. Cancer Res. Off J. Am. Assoc. Cancer Res. 22, 5574–5581. doi: 10.1158/1078-0432.ccr-16-1786
Yang, W., Chen, C. H., Jia, M., Xing, X., Gao, L., Tsai, H. T., et al. (2021). Tumor-associated microbiota in esophageal squamous cell carcinoma. Front. Cell Dev. Biol. 9. doi: 10.3389/fcell.2021.641270
Yuen, M. T. Y., Tsang, R. K., Wong, I. Y. H., Chan, D. K. K., Chan, F. S. Y., Law, S. Y. K. (2019). Long-term pharyngeal dysphagia after esophagectomy for esophageal cancer-an investigation using videofluoroscopic swallow studies. Dis. Esophagus Off J. Int. Soc Dis. Esophagus 32. doi: 10.1093/dote/doy068
Zhao, Q., Yang, T., Yan, Y., Zhang, Y., Li, Z., Wang, Y., et al. (2020). Alterations of oral microbiota in Chinese patients with esophageal cancer. Front. Cell Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.541144
Zhou, J., Shrestha, P., Qiu, Z., Harman, D. G., Teoh, W. C., Al-Sohaily, S., et al. (2020). Distinct microbiota dysbiosis in patients with non-erosive reflux disease and esophageal adenocarcinoma. J. Clin. Med. 9, 2162. doi: 10.3390/jcm9072162
Keywords: esophageal cancer, esophageal microbiome, gut microbiome, oral microbiome, 16S rRNA gene sequencing
Citation: Muszyński D, Kudra A, Sobocki BK, Folwarski M, Vitale E, Filetti V, Dudzic W, Kaźmierczak-Siedlecka K and Połom K (2022) Esophageal cancer and bacterial part of gut microbiota – A multidisciplinary point of view. Front. Cell. Infect. Microbiol. 12:1057668. doi: 10.3389/fcimb.2022.1057668
Received: 29 September 2022; Accepted: 31 October 2022;
Published: 16 November 2022.
Edited by:
Pallavi Singh, Northern Illinois University, United StatesReviewed by:
Jay Sharma, UC San Diego Health, University of California, San Diego, United StatesCopyright © 2022 Muszyński, Kudra, Sobocki, Folwarski, Vitale, Filetti, Dudzic, Kaźmierczak-Siedlecka and Połom. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karolina Kaźmierczak-Siedlecka, bGVva2FkaWFAZ3VtZWQuZWR1LnBs
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.