- 1Health Science Center, Shenzhen University, Shenzhen, Guangdong, China
- 2Key Laboratory of Livestock Infectious Diseases in Northeast China, Ministry of Education, Key Laboratory of Zoonosis, Shenyang Agricultural University, Shenyang, China
- 3Research and Development Center, Panjin Guanghe Crab Industry Co., Ltd., Panjin, China
Background: Enterobacter bugandensis is an emerging human pathogen in which multidrug resistant strains have been continuously isolated from various environments. Thus, this organism possesses the potential to pose challenges in human healthcare. However, the mechanisms, especially the efflux pumps, responsible for the multidrug resistance in E. bugandensis remain to be well elucidated.
Methods: The Enterobacter strain CMCC(B) 45301 was specifically identified using whole genome sequencing. The specific CMCC(B) 45301 homologues of the TolC dependent efflux-pump genes characterized in Escherichia coli were identified. The tolC deletion mutant in CMCC(B) 45301 was constructed and subjected to susceptibility tests using 26 different antimicrobial agents, along with the wild type strain. The synergistic effects combining the Bacillus crude extract (BCE) and several other TolC-affected compounds against CMCC(B) 45301 were assayed.
Results: We reclassified the Enterobacter CMCC(B) 45301 strain from species cloacae to bugandensis, on the basis of its whole genome sequence. We found that the CMCC(B) 45301 TolC, AcrAB, AcrD, AcrEF, MdtABC, EmrAB, and MacAB exhibit high similarity with their respective homologues in E. coli and Enterobacter cloacae. Our results for the susceptibility tests revealed that lacking tolC causes 4- to 256-fold decrease in the minimal inhibitory concentrations of piperacillin, gentamicin, kanamycin, tetracycline, norfloxacin, ciprofloxacin, chloramphenicol, and erythromycin against CMCC(B) 45301. In addition, the inhibition zones formed by cefuroxime, cefoperazone, amikacin, streptomycin, minocycline, doxycycline, levofloxacin, florfenicol, trimethoprim-sulfamethoxazole, azithromycin, lincomycin, and clindamycin for the tolC mutant were larger or more obvious than that for the parent. Our data suggested the important role played by TolC in CMCC(B) 45301 susceptibility to common antibiotic families covering ß-lactam, aminoglycoside, tetracycline, fluoroquinolone, phenicol, folate pathway antagonist, macrolide, and lincosamide. Deletion for tolC also increased the susceptibility of CMCC(B) 45301 to berberine hydrochloride and BCE, two natural product-based agents. Finally, we found that erythromycin, norfloxacin, and ciprofloxacin can potentiate the antibacterial activity of BCE against CMCC(B) 45301.
Discussion: The present study elaborated the comprehensive TolC effect on the antimicrobial susceptibility profile in E. bugandensis, which might contribute to the development of more therapeutic options against this nosocomial pathogen.
Introduction
Enterobacter spp. are gram-negative bacteria inhabiting a wide range of environmental niches with the human gastrointestinal tract being the most noticeable one (Mezzatesta et al., 2012). Some members of genus Enterobacter are well-known nosocomial pathogens, capable of causing infections in the blood streams, lungs, urinary tracts, and peritoneum of immunocompromised individuals, especially those accepting intensive cares in hospitals (Chow et al., 1991; Fernandez-Baca et al., 2001; Davin-Regli and Pages, 2015; Gravey et al., 2020). In the past decades, clinical isolates possessing multidrug resistance resulted from production of extended spectrum ß-lactamases (ESBLs) and carbapenemases, overexpression of AmpC, reduction in membrane permeability, and overexpression of efflux pumps are frequently isolated in Enterobacter, which poses a tremendous challenge to human healthcare, world-widely (Sanders and Sanders, 1997; Hilty et al., 2013; Chavda et al., 2016; Telke et al., 2017; Liu et al., 2021). Enterobacter bugandensis was identified as a novel species by Doijad and colleagues (Doijad et al., 2016), and subsequently expanded as the identification of strains isolated from the international space station (ISS), various nosocomial settings, and vegetables (Singh et al., 2018; Falgenhauer et al., 2019; Matteoli et al., 2020; Moon et al., 2021). As known, conventional typing methods based on 16S rDNA sequencing and phenotypic assays are not adequate for accurate identification of species within genus Enterobacter (Annavajhala et al., 2019). Recently, as the growing use of whole genome sequencing, the resolution of molecular typing of bacterial strains has been elevated to the genome level (Federhen et al., 2016; Ciufo et al., 2018). This facilitated the reclassification of some strains from Enterobacter cloacae to E. bugandensis, which was based on the average nucleotide identity (ANI) as compared their respective genomes against known E. bugandensis strains (Ciufo et al., 2018; Matteoli et al., 2020). Notably, the first identified E. bugandensis strains were neonatal-blood isolates, indicating their potential causing severe systemic infections in human beings (Doijad et al., 2016). The pathogenicity of E. bugandensis was evidenced by the results that E. bugandensis EB-247 establishes faster infections in Galleria mellonella and yields a higher survival rate grown in human serum, compared with E. cloacae ATCC 13047, a known pathogenic strain of Enterobacter (Pati et al., 2018). Thus, E. bugandensis has been considered as the most pathogenic species within genus Enterobacter (Pati et al., 2018; Matteoli et al., 2020). Besides, the multidrug resistant (MDR) strains of E. bugandensis have been continuously isolated (Doijad et al., 2016; Moon et al., 2021). Taken together, reclassification or specific identification of the E. bugandensis strains is exceedingly required, in order to provide references important for precise reaction combating this organism and effective genetic engineering for better understanding of this species.
Efflux pumps are important mechanisms for the emergence of multidrug resistance in gram-negative bacteria and comprise classes of RND, MF, ABC, MATE, SMR, and PACE, see reference Nikaido and Zgurskaya, 1999; Li et al., 2015 and Nishino et al., 2021 for reviews. The RND transporters are tripartite efflux pumps, among which AcrAB-TolC is the major one responsible for expelling many antimicrobial agents in Enterobacter (Perez et al., 2007; Guerin et al., 2016). It has been described that substrates of AcrAB-TolC encompass almost all the commonly-used antibiotic families, namely ß-lactam inhibitor, aminoglycoside, tetracycline, macrolide, lincosamide, fluoroquinolone, folate pathway antagonist, and phenicol, in some Enterobacter strains (Perez et al., 2007; Guerin et al., 2016; Liu et al., 2018; Gravey et al., 2020). Alarmingly, AcrAB-TolC can confer, at least in part, the increased resistance to some effective pharmaceutical options against MDR strains, like the combination of ß-lactam/ß-lactamase inhibitor and the last-line drugs, carbapenems, polymyxins, and tigecycline, in Enterobacter spp. (Perez et al., 2007; Telke et al., 2017; Huang et al., 2019; Gravey et al., 2020; Senchyna et al., 2021). Efflux of antimicrobial natural products can also be performed by AcrAB-TolC in Enterobacter strains (Kuete et al., 2010). Furthermore, Enterobacter strains are able to gain spontaneous mutations in the acrB gene or genes encoding regulators of acrAB and tolC, for their adaptations to antibiotic stresses (Telke et al., 2017; Gravey et al., 2020; Senchyna et al., 2021). As an outer-membrane channel, TolC can form efflux pumps with some other RND-type transporters, MF transporters, and ABC transporters, overexpression of which is capable of restoring the resistance to one or more antibiotic agents in bacterial strains devoid of AcrB (Kobayashi et al., 2001; Nagakubo et al., 2002; Nishino et al., 2003; Li et al., 2015; Yousefian et al., 2021; Nishino et al., 2021). Thus, studies regarding the TolC-involved efflux pumps in gram-negative bacteria including Enterobacter spp. are imperative for developing novel therapeutic strategies, for instance, the combination of efflux-pumped antibiotic/efflux-pump inhibitor, against the MDR strains (Morita et al., 2016; Li et al., 2017; Grimsey et al., 2020; Li et al., 2021b).
In E. bugandensis, some isolates are tested positive for the resistance/reduced susceptibility to cephalosporins, imipenem, aminoglycosides, fluoroquinolones, and polymyxin B (Doijad et al., 2016; Moon et al., 2021; Sarangi et al., 2022). Studies revealed that the plasmid-borne resistance genes and the chromosomally integrated gene encoding imipenemase IMI-1 might contribute to the multidrug resistance in E. bugandensis EB-247 and the imipenem resistance in E. bugandensis S68-1, respectively (Pati et al., 2018; Moon et al., 2021). However, investigations of the multidrug resistance/susceptibility concerning efflux pumps in E. bugandensis are still lacking. To our knowledge, a comprehensive study about the correlation between TolC or TolC-related efflux pumps and multidrug resistance/susceptibility in E. bugandensis has not been reported. Here, we constructed a clean deletion mutant of tolC in the E. bugandensis CMCC(B) 45301 strain, and tested the changes in its susceptibility to a range of 26 antimicrobial agents, including antibiotics belonging to different families and natural products, as compared with the parent strain. In addition, certain combinations of the TolC-affected antibiotic and natural product were assessed for their synergistic effects against the wild-type strain, for exploring novel antibacterial options combating E. bugandensis. The present study mainly aimed to elucidate the effect of TolC on the antimicrobial susceptibility profile in E. bugandensis, in advance providing information assisting actions against this bacterium in the future.
Results
Whole genome sequencing-based reclassification of E. cloacae CMCC(B) 45301
In this study, we used E. cloacae CMCC(B) 45301, a strain collected in China National Center for Medical Culture Collections (CMCC; http://www.cmccb.org.cn/cmccbnew/) and initially identified as E. cloacae, as the experimental material. To obtain the comprehensive genetic information of this strain, we sequenced and assembled its complete genome using the combination of Pacbio and Illumina platforms, see materials and methods. Our results showed that the circular chromosome DNA of CMCC(B) 45301 (GenBank accession No. CP097255) possesses a size of 4,631,472 base pairs (bp) and a G+C content of 56.13%. In addition, strain CMCC(B) 45301 harbors an 81,691-bp plasmid (GenBank accession No. CP097254) with a G+C content of 47.35%. The chromosome and plasmid sequences contain a total of 4,499 genes annotated via the NCBI Prokaryotic Genome Annotation Pipeline (PGAP). Importantly, according to the result of the quality-control test that compared the genome sequence of CMCC(B) 45301 against the type-strain genomes throughout GenBank as previously described (Federhen et al., 2016), strain CMCC(B) 45301 should be identified more specifically as an E. bugandensis organism, given the 98.8% ANI comparing its genomic assembly (GenBank accession No. GCA_023374275.1) with the best-matching type-strain assembly (GenBank accession No. GCA_019046905.1) of E. bugandensis (Ciufo et al., 2018). Thus, the original E. cloacae CMCC(B) 45301 strain was reidentified and designated as E. bugandensis CMCC(B) 45301 (EBU45301).
Identification of the TolC dependent efflux-pump genes in EBU45301
As known, TolC can form efflux pumps with the RND-type transporters, AcrAB, AcrAD, AcrEF, MdtEF, and MdtABC, the MF-type transporters, EmrAB and EmrKY, and the ABC-type transporter MacAB in Escherichia coli (Nishino et al., 2003). Recently, effects of acrD, acrEF, mdtABC, and some other RND efflux-pump genes on the resistance/susceptibility to various antibiotics in the E. cloacae ATCC 13047 strain (ECL13047) were characterized, describing a landscape of TolC-dependent RND efflux pumps in E. cloacae highly similar to that in E. coli (Guerin et al., 2016). To locate the respective EBU45301 homologues of the TolC dependent efflux-pump genes in E. coli K-12 MG1655 (GenBank accession No. U00096.3), BLAST was performed comparing each of these E. coli genes against the whole genome of EBU45301, and the locus whose product displayed distinguishable-high identities/positives with that of the corresponding E. coli gene was considered as a specific homologue. The identified EBU45301 efflux-pump genes were also subjected to comparisons with their respective homologues in ECL13047 (GenBank accession No. NC_014121), for further confirmation. Our results revealed that specific homologues of tolC, acrAB, acrD, acrEF, mdtABC, emrAB, and macAB are encoded in both EBU45301 and ECL13047, and yet such homologues for mdtEF and emrKY can be found throughout the genome of neither of these strains (Table 1). EBU45301 harbors genes encoding for some other RND efflux-pump components, whose specific homologues are either present or absent in ECL13047 (Tables S1, S2), exhibiting potential in interaction with TolC (Guerin et al., 2016). In addition, several MF efflux pump proteins showing partial homology with the respective EmrAB- and EmrKY-TolC components were identified in EBU45301 (Table S3), indicating that there are EmrAB/EmrKY-like efflux pumps involving TolC in this bacterium.
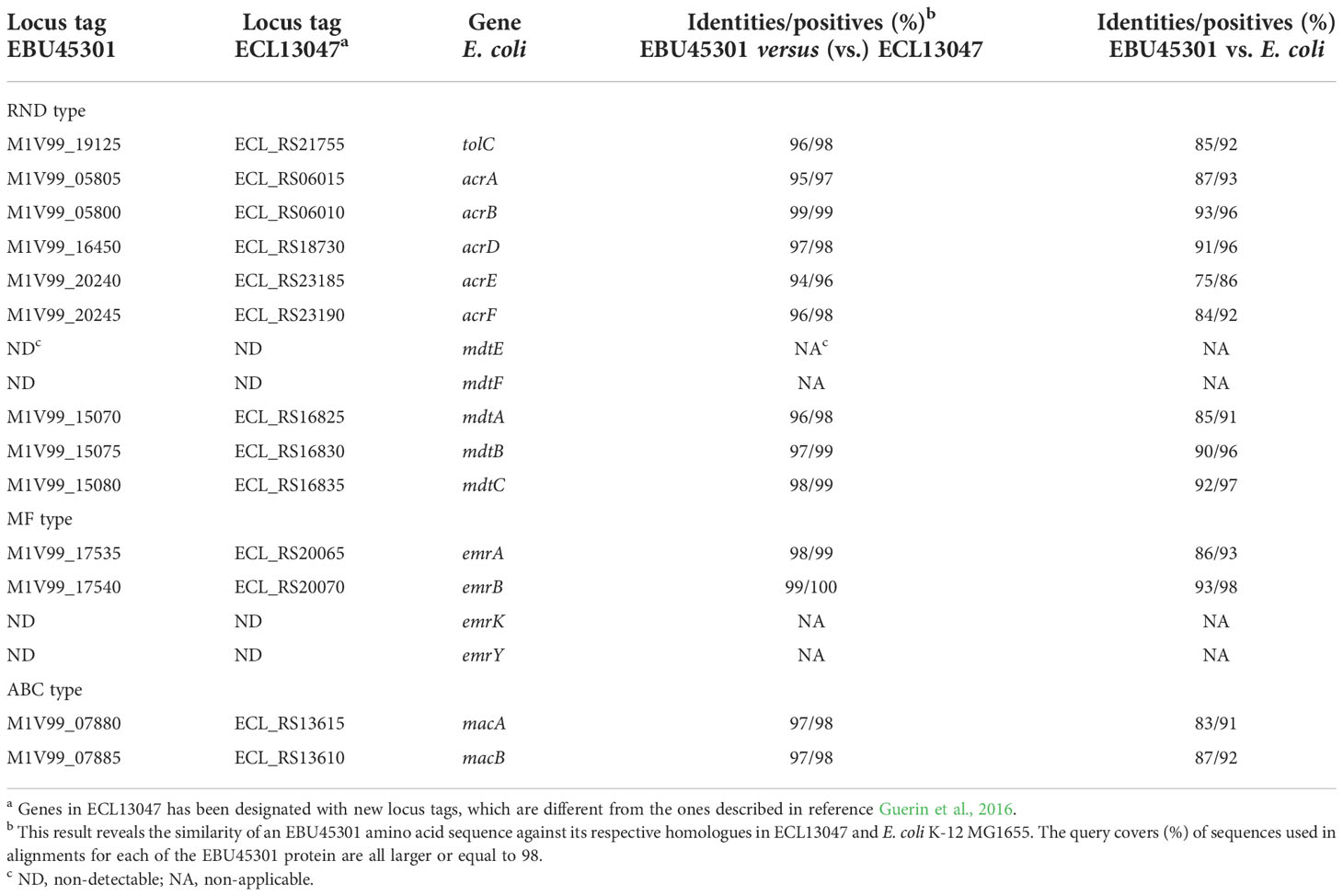
Table 1 TolC related efflux-pump genes in EBU45301 located based on their homologies with the respective sequences in E. cloacae and E. coli.
We also compared the tolC gene of EBU45301 with that of the type or recommended reference (by the NCBI Genome database) strains representing the species belonging to genus Enterobacter. A maximum-likelihood tree was constructed based on tolC, and it is shown that EBU45301 is clustered with E. bugandensis EB-247 in the same clade closely related to Enterobacter chuandaensis 090028 and Enterobacter sichuanensis WCHECL1597 (Figure 1A). Furthermore, the TolC amino-acid sequences from EBU45301, E. bugandensis EB-247, Enterobacter chuandaensis 090028, Enterobacter sichuanensis WCHECL1597, Enterobacter cloacae ATCC 13047, Enterobacter asburiae JCM 6051, Enterobacter hormaechei ATCC 49162, and E. coli K-12 MG1655 were aligned for a comparison, to investigate the homology of EBU45301 TolC with its respective homologues in the closely related species and species in which AcrAB-TolC has been reported as a powerful MDR mechanism (Li et al., 2015; Guerin et al., 2016; Telke et al., 2017; Gravey et al., 2020). The alignment result revealed that EBU45301 TolC is highly homologous with the other TolC sequences (Figure 1B), suggesting its function transporting multiple compounds. Taken together, we hypothesized that TolC plays a magnificent role in mediating the antimicrobial susceptibility profile of EBU45301.
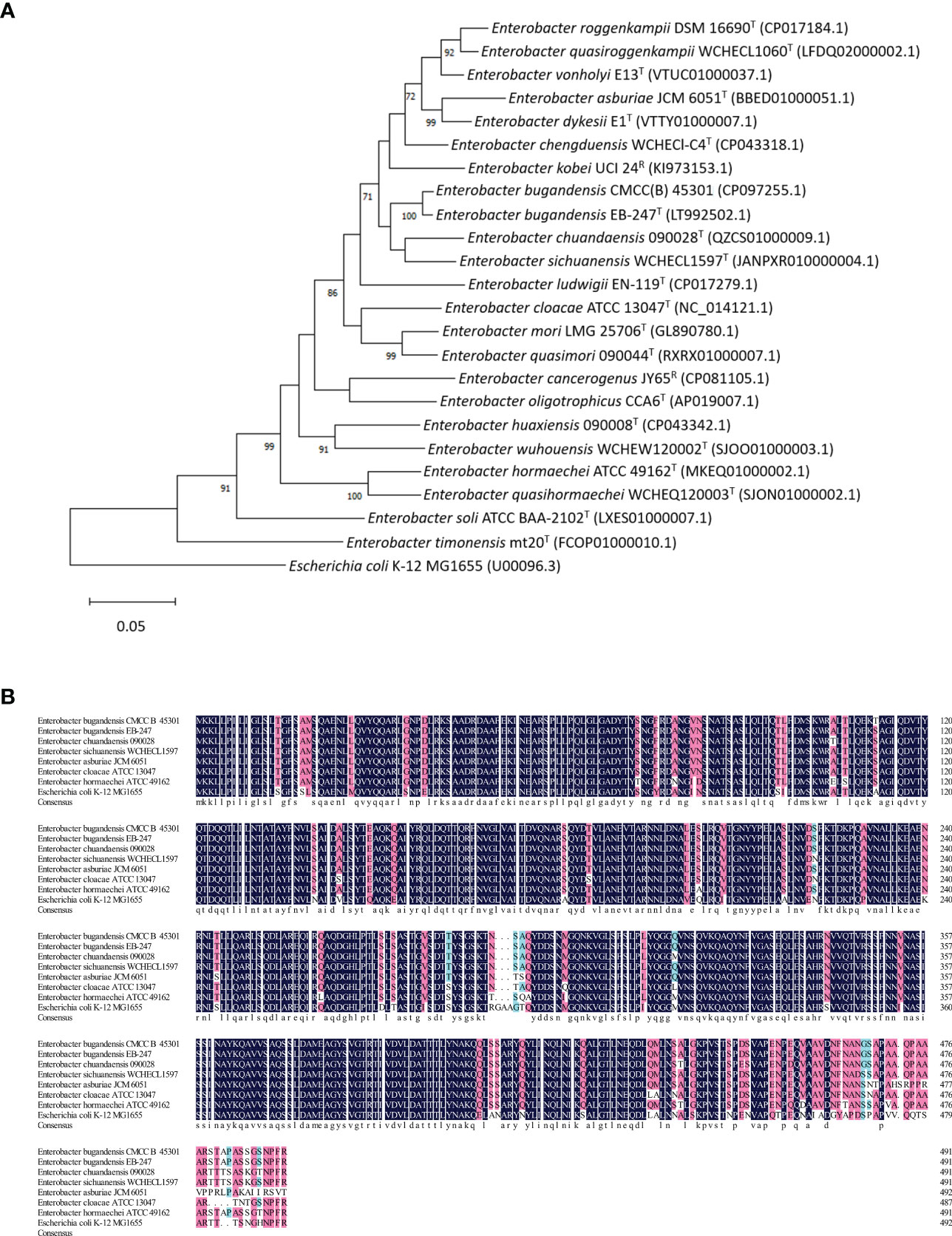
Figure 1 Comparative analyses of EBU45301 and the type/reference strains representing Enterobacter species based on the tolC gene. (A) A maximum-likelihood tree was constructed based on the tolC gene sequences retrieved from the genomes of the Enterobacter strains as indicated. The Genbank accession No. for each of the used genomic sequence was shown in parentheses. The shown values on certain branch nodes were bootstrap values >70%, based on 500 replications. The bar represents 0.05 substitutions per site. E. coli K-12 MG1655 was used to root the tree. T, type; R, reference. (B) An alignment of the amino acid sequences of TolC from EBU45301 and several other Enterobacter strains (as indicated) was performed. The homology levels were shaded as follows: ≥50%, light blue; ≥70%, pink; =100%, dark blue. The consensus sequence was shown at the bottom of this alignment in lower case.
Investigation of the TolC effect on the susceptibility of EBU45301 to different antimicrobial agents
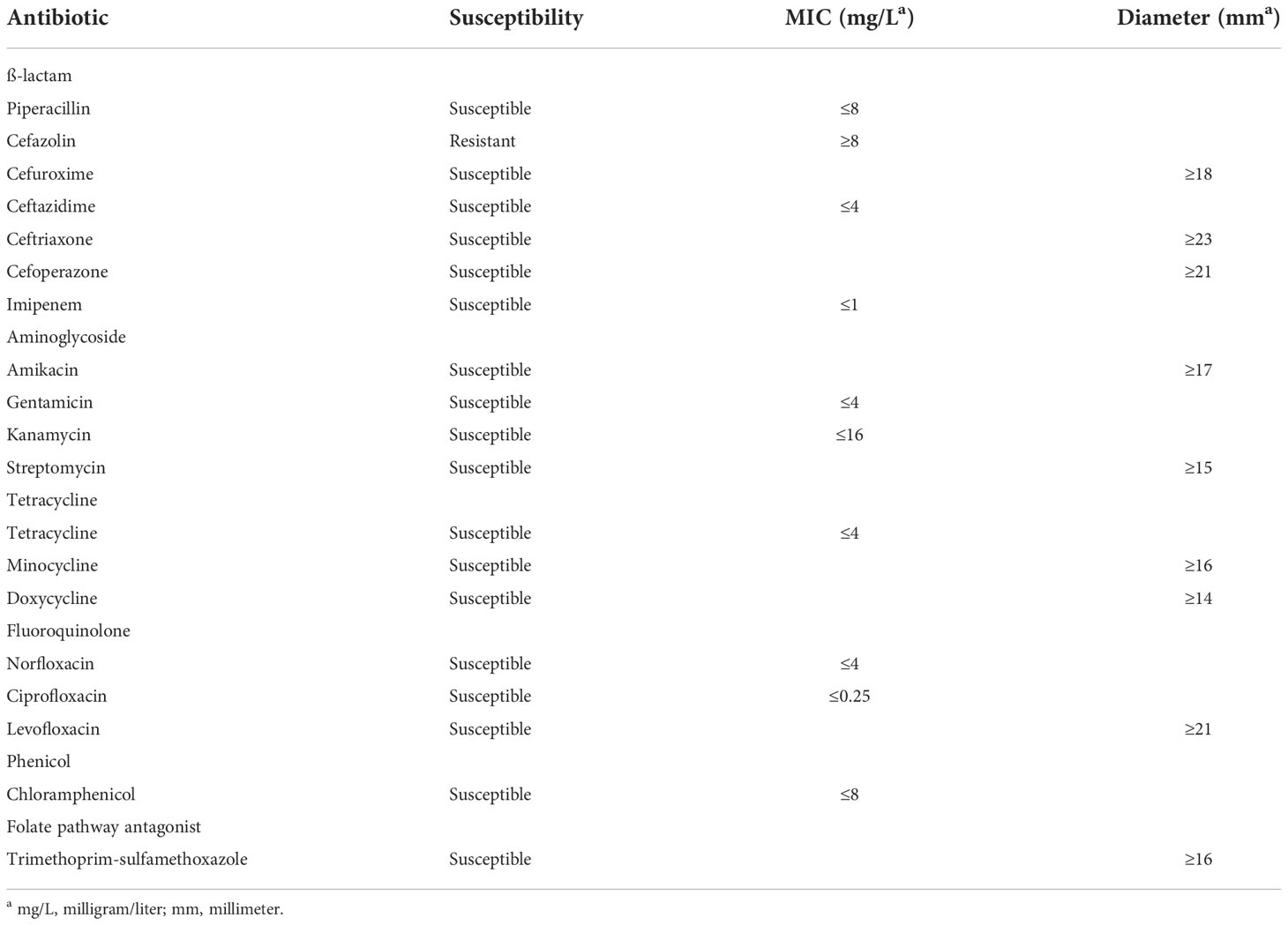
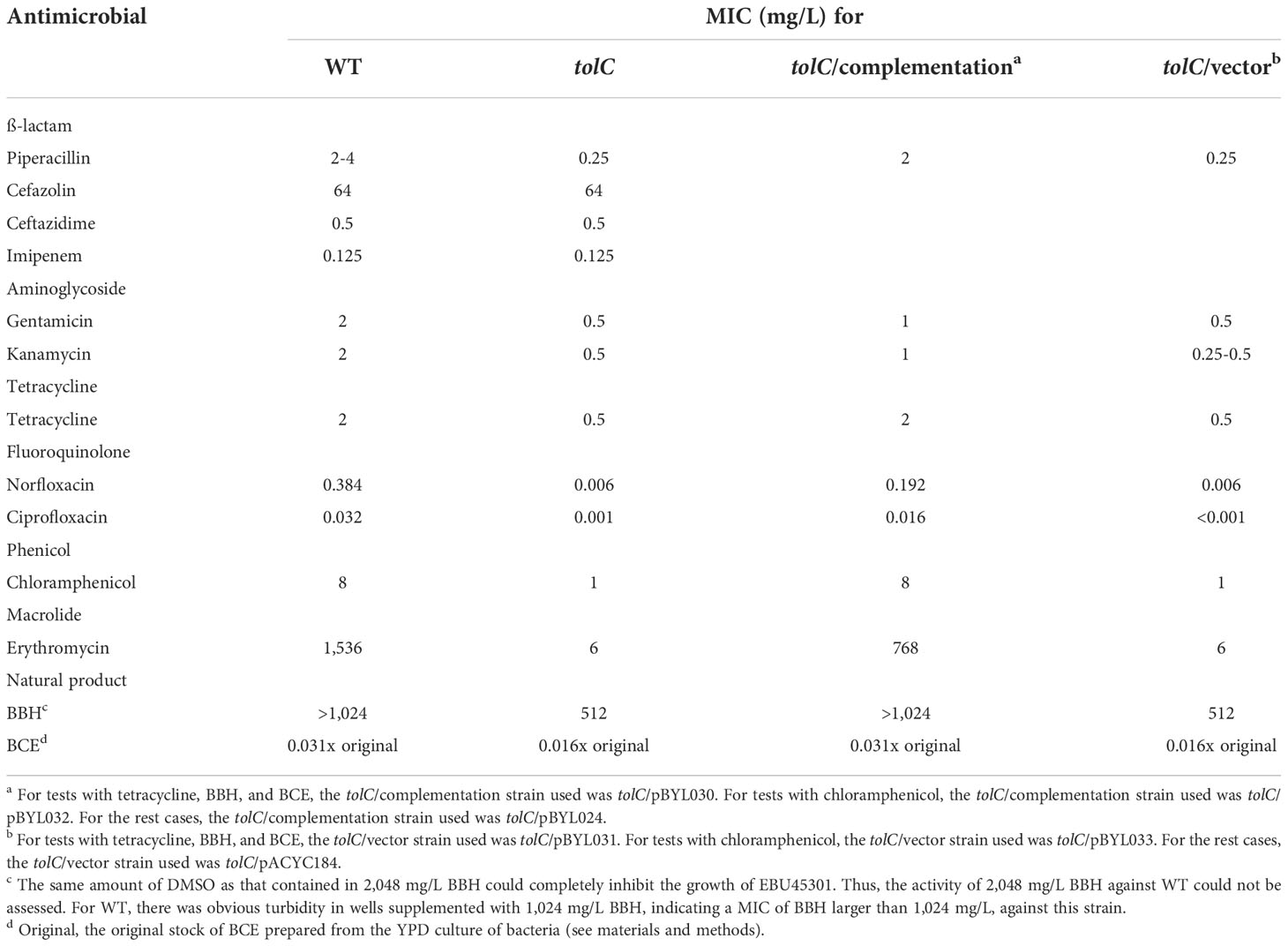
To study the role of TolC in EBU45301 physiology, we constructed a clean deletion mutant for tolC via the pRE112 system (Edwards et al., 1998). It has been described that the TolC-dependent efflux pumps affect the susceptibility of E. coli and some Enterobacter species to a broad spectrum of antibiotics (see Table S4) and natural products like isobavachalcone and Diospyrone (Kuete et al., 2010). Here, we assayed the susceptibility of the tolC mutant (tolC) and the wild-type strain (WT) to a range of 26 antimicrobial agents, covering the extensively-applied antibiotic families, ß-lactam, aminoglycoside, tetracycline, fluoroquinolone, macrolide, lincosamide, phenicol, and folate pathway antagonist, and two other agents, berberine hydrochloride (BBH) and the Bacillus crude extract (BCE), see materials and methods for details.
Our results showed that the wild-type EBU45301 is intrinsically resistant to cefazolin and susceptible to piperacillin, cefuroxime, ceftazidime, ceftriaxone, cefoperazone, imipenem, amikacin, gentamicin, kanamycin, streptomycin, tetracycline, minocycline, doxycycline, norfloxacin, ciprofloxacin, levofloxacin, chloramphenicol, and trimethoprim-sulfamethoxazole (Tables 2–4), according to the respective breakpoints recommended in the performance standards provided by CLSI, 2022. We also found that the tolC deletion causes the increased susceptibility of EBU45301 to all the tested antimicrobial agents, except for cefazolin, ceftazidime, ceftriaxone, and imipenem (Tables 3, 4). The minimal inhibitory concentration (MIC) of piperacillin for the tolC mutant was 8-16 times smaller than that for the parent (Table 3), in accordance with the disk-diffusion results for piperacillin (Table 4 and Figure S1). In terms of drugs classified as cephalosporins, the tolC deletion rendered EBU45301 more sensitive to cefuroxime and cefoperazone, different from that for cefazolin, ceftazidime, and ceftriaxone (Tables 3, 4, and Figure S1). In addition, loss of tolC resulted in a 4-fold decrease in the MICs of gentamicin, kanamycin, and tetracycline against EBU45301 (Table 3), which was consistent with the slightly larger inhibition zones for the tolC mutant than WT, revealed in the disk-diffusion tests for these drugs (Table 4 and Figure S1). Similarly, in the cases of the other two aminoglycosides, amikacin and streptomycin, the effect of TolC was observed but not very obvious (Table 4 and Figure S1). However, not like that for tetracycline, strain tolC appeared to be much more sensitive to minocycline and doxycycline, also belonging to family tetracycline, than WT, which might be due to the weaker activity of these two antibiotics against the wild-type EBU45301 (Table 4 and Figure S1). The TolC effect was found remarkable on the tested fluoroquinolones. The respective fold changes in MIC for norfloxacin and ciprofloxacin were 64 and 32, comparing tolC with WT (Table 3). These phenotypes in susceptibility observed using the broth-microdilution method were confirmed via the well-diffusion method, for norfloxacin and ciprofloxacin (Table 4 and Figure S1). Moreover, the inhibition zone using levofloxacin for strain tolC was obviously larger than strain WT, suggesting a TolC effect on levofloxacin efflux as strong as that for norfloxacin or ciprofloxacin (Table 4 and Figure S1). Moreover, compared with the wild type, the tolC mutant exhibited defects in resisting chloramphenicol and florfenicol, which were determined as the 8-fold decrease in MIC (Table 3) and the much larger inhibition zone as shown (Table 4 and Figure S2), respectively. Trimethoprim-sulfamethoxazole, a combination of folate pathway antagonists, showed higher activity combating tolC than WT, assayed through the disk-diffusion tests (Table 4 and Figure S2). We also tested the TolC effect on EBU45301 susceptibility to antibiotics that are not recommended for cases of Enterobacteriaceae, such as macrolides and lincosamides. Larger and clearer inhibition zones were observed for the tolC mutant than the WT strain, when the susceptibility to erythromycin, azithromycin, and clindamycin was tested (Table 4 and Figure S2). In assays using lincomycin, there were areas with fewer bacteria instead of clear zones around the disks for tolC, and yet none of these were found for WT (Table 4 and Figure S2). Note that there is a 256-fold decrease in the MIC of erythromycin against tolC, compared with WT (Table 3), indicating that the efflux of erythromycin is tightly related to TolC in EBU45301.
Berberine is known as a plant-isolated natural product exhibiting antibacterial activity against nosocomial pathogens like Acinetobacter baumannii and Pseudomonas aeruginosa. Also, berberine (hydrochloride) was considered as a substrate of the AcrB-homologous transporters in A. baumannii and P. aeruginosa (Morita et al., 2016; Li et al., 2021b). Here, we assessed the activity of BBH against the EBU45301 strains, and found that there is an at least 2-fold decrease in the MIC of BBH for tolC, compared with WT (Table 3). This result indicated that BBH displays intrinsic antibacterial activity for EBU45301 and can interact with one or more TolC-related efflux pumps in this bacterium.
It has been well characterized that Bacillus strains are able to synthesize various secondary metabolites, including fengycin, iturin, surfactin, difficidin, bacilysin, bacillibactin, bacillaene, amylocyclicin, subtilosin A, sublancin, and macrolactin, that possess antibacterial activity (Wilson et al., 1987; Scholz et al., 2014; Cavera et al., 2015; Liu et al., 2019; de Souza Freitas et al., 2020; Lv et al., 2020; Li et al., 2021a; Chakraborty et al., 2022; Erega et al., 2022; Zhou et al., 2022). There is a recent study showing that the transportation of bacilysin and bacillaene can be affected by RND-type transporters in Campylobacter jejuni (Erega et al., 2022). In the present study, we used the crude extract from strain GHZJ-1, which is a laboratory stock initially identified as Bacillus sp. and found exhibiting anti-yeast activity (data not shown), as one antimicrobial agent used for the susceptibility tests with the EBU45301 strains, see materials and methods for details. Obviously, comparing with the commercial antibiotic products, BCE used here was a crude agent and its exact concentration was difficult to determine. Thus, we first tested the MIC of BCE prepared from different batches with the EBU45301 strains. Our data revealed that BCE displays antibacterial activity against EBU45301 and that its MICs against WT and tolC are 0.031x original (BCE stock) and 0.016x original, respectively, no matter which batch of the BCE product was used (Table 3). Moreover, using the well-diffusion method, BCE formed larger inhibition zones for tolC than that for WT (10.4 mm vs. 7.2 mm; Table 4), and the variations among different batches were highly small (Figure S2), indicating that the antibacterial activity of BCE against EBU45301 is fairly stable. These results suggested the existence of the TolC pump-transported antibacterial metabolite(s) within BCE, like that observed in C. jejuni (Erega et al., 2022). Undoubtedly, identification of these specific metabolite(s) warrants more experiments.
The increased susceptibility to antimicrobial agents in the tolC mutant was complemented via introduction of the wild-type tolC (Tables 3, 4, Figures S1, Figure S2), which confirmed the TolC effect on the susceptibility profile in this organism. Note that some of the complementation performed using plasmid pBYL024 is not exactly complete. This is likely due to the slight growth defect in strains harboring the pACYC184-based plasmids (Figure S3), which was not observed for strains carrying the plasmids (pBYL030-033) whose introduction was facilitated via integration into the chromosome (data not shown).
Synergistic effects combining BCE and other TolC-affected antibiotics
The altered susceptibility to BCE observed in the tolC mutant suggests that there are substances capable of traveling through one or more TolC-mediated efflux pumps in this extract (Tables 3, 4). Thus, BCE can be considered as a competitor of some other TolC-affected antibiotics for the capacity of efflux pumps regarding TolC. Besides, BCE is likely to contain lipopeptides, for instance, surfactin, that are able to change the cell-wall permeability and therefore potentiate the activity of other antibiotics against gram-negative bacteria (Liu et al., 2019). Taken together, we hypothesized that combinations of BCE and some TolC-affected antibiotics can confer synergistic effects against EBU45301. Here, we selected erythromycin, norfloxacin, and ciprofloxacin, whose MICs were decreased the most in EBU45301 strains devoid of tolC (Table 3), as the agents respectively used in association with BCE, for synergistic assays. As shown in Table 5, weak synergistic effects were observed for combinations of BCE/erythromycin: in the presence of 0.5x MIC BCE, the MIC of erythromycin against EBU45301 WT was 384 mg/L (0.25x MIC), and vice versa. However, such synergistic effects were not observed for combinations of BCE/norfloxacin or ciprofloxacin (Table 5). It has been characterized that the checkerboard method is unable to fully test synergistic effects due to its twofold dilution feature (Grimsey et al., 2020). Thus, we also performed synergistic assays using the well-diffusion method, as previously described (Grimsey et al., 2020). Our results showed that in the presence of 12 mg/L (0.008x MIC) erythromycin, the antibacterial activity of BCE against WT is increased, compared with that in the absence of antibiotics (9.9 mm vs. 7.4 mm; Figure 2). This increased phenotype in BCE activity was enhanced in the presence of 24 mg/L (0.016x MIC) erythromycin (11.6 mm vs. 7.4; Figure 2). Similarly, the BCE activity against WT was elevated in the presence of 0.006 mg/L (0.016x MIC) norfloxacin (9.9 mm vs. 7.4 mm) or 0.5 µg/L (0.016x MIC) ciprofloxacin (8.8 mm vs. 7.4 mm), and yet the existence of neither of these antibiotics at 0.008x MIC (0.003 mg/L or 0.25 µg/L) could potentiate the BCE activity (Figure 2). It appeared that 0.016x MIC norfloxacin increases the BCE activity slightly more than 0.016x MIC ciprofloxacin (Figure 2). These results revealed that at sub-MICs, erythromycin, norfloxacin, and ciprofloxacin can all enhance the BCE activity inhibiting EBU45301 with erythromycin being the most efficient. Our findings also supported that BCE used in this study can serve as a resource developing novel antibacterial agents or antibiotic adjuvants against E. bugandensis. Of course, analysis and isolation of the active substances within BCE is inevitable in future studies.
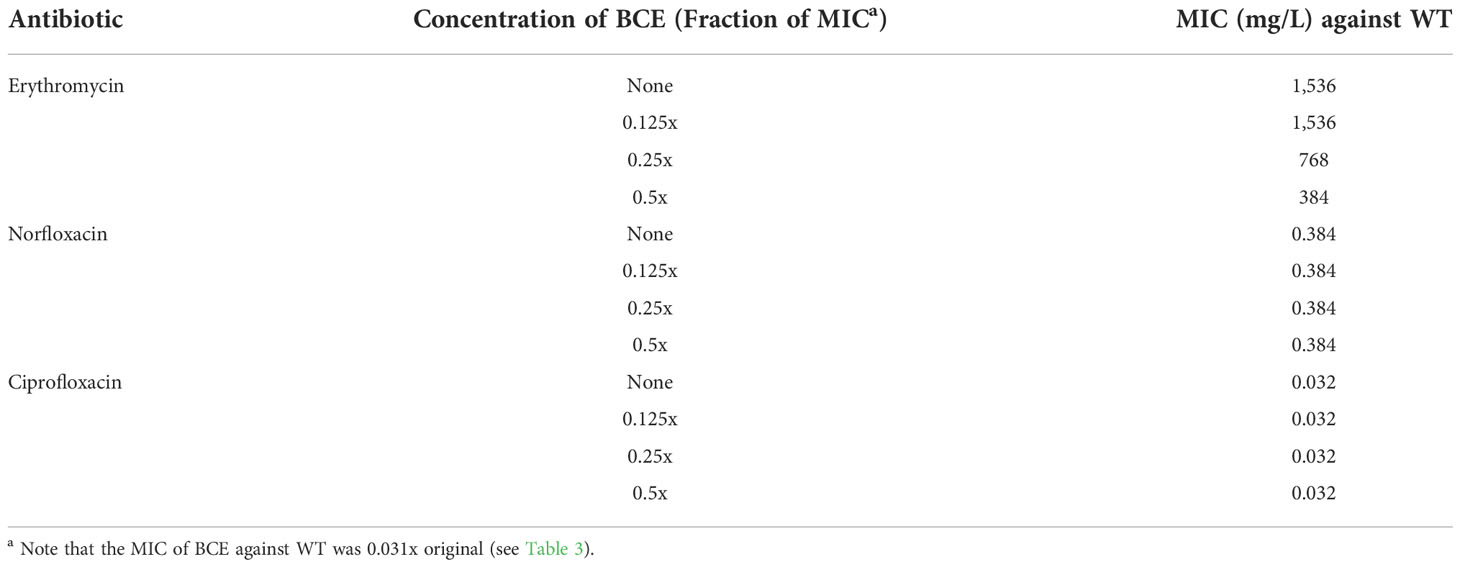
Table 5 Synergistic tests using BCE in combination with certain TolC-affected antibiotics (checkerboard method).
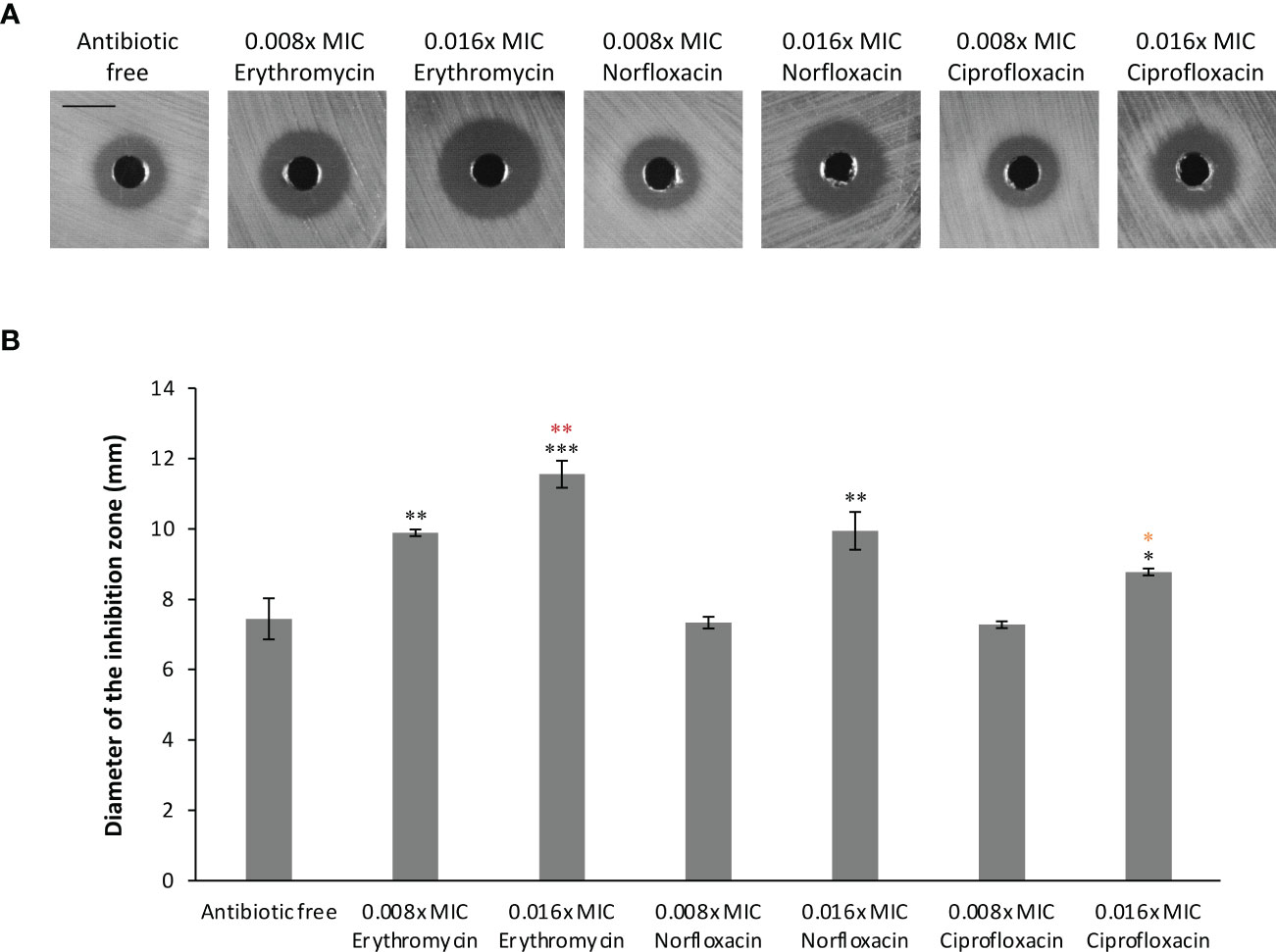
Figure 2 Synergistic effects combining BCE and erythromycin, norfloxacin, or ciprofloxacin against EBU45301. The susceptibility of WT to BCE was tested in the presence of different concentrations (fraction of MIC) of erythromycin, norfloxacin, or ciprofloxacin as indicated, using the well-diffusion method (see materials and methods). The inhibition zones formed by different BCE/antibiotic combinations against WT (A) were observed and determined for diameters (B). The shown images are a representative of three independent experiments. All the black bars represent 10 mm. The diameter of inhibition zones was calculated as average ± SD of three independent experiments. * (black), p ≤ 0.05; ** (black), p ≤ 0.01; *** (black), p ≤ 0.001 comparing the inhibition-zone diameter with that for BCE against WT, in the absence of antibiotic. ** (red), p ≤ 0.01 comparing the inhibition-zone diameter for BCE in combination with 0.016x MIC erythromycin with that for BCE in combination with 0.008x MIC erythromycin. * (orange), p ≤ 0.05 comparing the inhibition-zone diameter for BCE in combination with 0.016x MIC ciprofloxacin with that for BCE in combination with 0.016x MIC norfloxacin.
Discussion
E. bugandensis is an emerging pathogen in which MDR strains are frequently isolated, which poses serious problems in their treatments during nosocomial infections (Doijad et al., 2016; Pati et al., 2018; Singh et al., 2018; Falgenhauer et al., 2019; Matteoli et al., 2020; Moon et al., 2021). Genomic analyses have demonstrated that many efflux pump genes are included in genomes of E. bugandensis strains (Matteoli et al., 2020), and yet a comprehensive description of the relationship between efflux pumps and antibiotic susceptibility/resistance in this species has not been reported. Here, we studied the effect of TolC, an outer-membrane component of different efflux pumps, on the susceptibility to a range of 26 antimicrobial agents in a newly reclassified E. bugandensis strain, CMCC(B) 45301 (EBU45301). We also investigated whether particular combinations of the TolC-affected natural product/antibiotic exhibited synergistic effects. The goal of this study is to elucidate the physiological role of TolC in the antimicrobial susceptibility profile of EBU45301, which might contribute to the development and application of new therapeutic options against (MDR) E. bugandensis.
In the present study, we sequenced the complete genome and plasmid harbored in the Enterobacter strain CMCC(B) 45301 (GenBank accession No. CP097254-CP097255), and reidentified this organism from E. cloacae to E. bugandensis, on the basis of the ANI of its genomic assembly against the best-matching type-strain assembly of E. bugandensis (Ciufo et al., 2018). This provides a full-detail reference of the EBU45301 genome, which is still scarce within species bugandensis. Using this reference sequence, we identified the specific EBU45301 homologues of the respective tolC dependent efflux-pump genes characterized in E. coli and E. cloacae (Table 1). In addition, some putative efflux-pump genes whose products display homology with components involved in the known TolC-related efflux pumps were located in the EBU45301 genome (Tables S1-3). The specific homologues for some of these genes were not found in the genome of E. cloacae ATCC 13047 (Tables S2, Table S3), indicating that they are not part of the core resistome in genus Enterobacter. All these comparative results suggested a significant role of TolC in EBU45301. Intriguingly, we noticed that genes of M1V99_14575, M1V99_14580 (Table S2), M1V99_14525, M1V99_14530, and M1V99_14535 (Table S3) are included within a ~90,000 bp sequence (Position 2864027-2954705) in the EBU45301 genome that is not highly identical to any sequences but one within the genome of E. ludwigii strain UW5 (Position 3536652-3627328), BLASTing it against the NCBI database. Moreover, several hypothetical MF transporters are encoded in this big-chunk DNA fragment, suggesting a unique armory of MDR mechanisms in EBU45301. More research regarding this fragment may provide insights into areas of how MDR genes are transferred horizontally among environmental strains.
We constructed a deletion mutant for tolC and subjected it to antimicrobial susceptibility tests, along with the wild type. It is found that, following the guidelines of breakpoints recommended for Enterobacteriaceae, the wild-type EBU45301 is resistant to cefazolin, weakly susceptible to streptomycin, doxycycline, minocycline, and chloramphenicol, susceptible to piperacillin, cefuroxime, ceftazidime, ceftriaxone, cefoperazone, imipenem, amikacin, gentamicin, kanamycin, tetracycline, norfloxacin, ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole (Tables 2-4). Furthermore, the tolC deletion resulted in the increased susceptibility to all the assayed antimicrobial agents, except cefazolin, ceftazidime, ceftriaxone, and imipenem, in EBU45301 (Tables 2-4). However, it has been described that efflux pumps regarding TolC are likely to be capable of transporting cefazolin, ceftazidime, ceftriaxone, and imipenem in E. coli or certain Enterobacter species (Chang et al., 2007; Perez et al., 2007; Moosavian et al., 2021). The phenotype unresponsive to the tolC deletion for these four ß-lactams in EBU45301 indicated that the deletion phenotype is not obvious enough, as the overexpression phenotype, or that even similar efflux pumps identified in closely-related species vary in their specific substrates (Perez et al., 2007; Chang et al., 2007; Guerin et al., 2016). Despite of this, the effect of TolC-involved efflux pumps on other tested antibiotics belonging to ß-lactam, aminoglycoside, tetracycline, fluoroquinolone, phenicol, macrolide, lincosamide, and folate pathway antagonist observed in EBU45301 is similar to that found in E. coli or Enterobacter, as listed in Table S4. Moreover, the tolC strain exhibited increased susceptibility to the natural product-based agents, BBH and BCE (Tables 3, 4), in EBU45301, which to our best knowledge has not been characterized for Enterobacter.
Berberine or BBH can be used as an efflux-pump inhibitor in association with some efflux-pumped antibiotics against MDR A. baumannii and P. aeruginosa (Morita et al., 2016; Li et al., 2021b). However, there was no synergistic effects detected combining BBH and the respective TolC-affected antibiotics, erythromycin, norfloxacin, ciprofloxacin, chloramphenicol, and tetracycline, in EBU45301 (data not shown). This might be due to the just-constitutive expression of the efflux pump genes in EBU45301, different from the strains previously used for testing the activity of certain combinations of efflux-pumped antibiotic/efflux-pump inhibitor, which are either spontaneously or artificially MDR (Morita et al., 2016; Grimsey et al., 2020; Li et al., 2021b). Therefore, in our future studies, synergistic tests using BBH and other TolC-affected antibiotics will be performed with artificial MDR strains, such as the ramR mutant, in EBU45301. Given the fact that BCE includes substances TolC efflux pump-transported, we also assayed the synergistic effects using BCE in combination with erythromycin, norfloxacin, or ciprofloxacin, three antibiotics the susceptibility to which was affected the most in tolC. Our results demonstrated that erythromycin, norfloxacin, and ciprofloxacin can potentiate the activity of BCE against EBU45301, at their sub-MICs (Table 5 and Figure 2). Erythromycin exhibited the highest ability to potentiate the BCE activity against EBU45301, and norfloxacin seemed to be more efficient as an adjuvant for BCE than ciprofloxacin (Figure 2). It appeared that the more an antibiotic is affected by TolC, the better a synergistic compound for BCE against EBU45301 it is. To elucidate this, more experiments are required. In terms of BCE, we surely can consider it as a reservoir isolating for active substances as the antibacterial agents or antibiotic adjuvants against E. bugandensis in the future. However, we still have a long way to go. First, whole genome sequencing of the Bacillus strain used in this study is warranted for typing it to a specific species and providing a genetic reference including clusters encoding different secondary metabolites. Secondly, antibacterial substances, especially those with the efflux-pumped features, like bacilysin and bacillaene in C. jejuni (Erega et al., 2022), and the antibiotic-adjuvant features, like surfactin in E. coli and subtilosin A in Gardnerella vaginalis (Cavera et al., 2015; Liu et al., 2019), are required to be identified and isolated for more specific studies. Finally, experiments about genetic modification or incubation conditions promoting the production of useful metabolites of our Bacillus strain are desired, for a more efficient application of this organism.
In summary, the present study revealed that TolC is involved in the efflux of a broad range of antimicrobial agents encompassing antibiotic families of ß-lactam, aminoglycoside, tetracycline, fluoroquinolone, phenicol, macrolide, lincosamide, and folate pathway antagonist, and natural product-based agents, like BBH and BCE, in EBU45301. In addition, certain combinations of the TolC-affected agents, namely BCE and erythromycin, norfloxacin, or ciprofloxacin, displayed synergistic effects combating EBU45301. This investigation provided a comprehensive description of the TolC effect on multidrug susceptibility/resistance in E. bugandensis, which can be used as a reference for the future emergence of MDR isolates belonging to this species and as research endorsing the development of more therapeutic strategies against this opportunistic pathogen.
Materials and methods
Bacterial strains and growth conditions
All the bacterial strains used in the present study are listed in Table 6. Except where indicated, the E. coli and EBU45301 strains were grown in LB broth or agar (Solarbio, Beijing, China) at 37°C aerobically. If required, 12.5 mg/L chloramphenicol (Solarbio) or 10 mg/L tetracycline (Solarbio), was used for the E. coli and EBU45301 strains.
Whole genome sequencing of EBU45301
In this study, the whole genome sequencing of EBU45301 was accomplished using a combination of the Pacbio and Illumina platforms. For sequencing performed by the Pacbio platform, the total DNA isolated from EBU45301 was subjected to construction of the 10Kb (kilo base) SMRTbell library, using the SMRTbell™ Template kit version 1.0. The constructed library was tested using Qubit and Agilent 2100 for its quality and fragment-size, respectively, followed by the sequencing conducted via PacBio Sequel. For sequencing performed by the Illumina platform, the total DNA was randomly broken into fragments of ~350 bp using the Covaris shearing instrument, and a library of these fragments was constructed according to the manufacturer’s instructions of the NEBNext®Ultra™ DNA Library Prep Kit for Illumina (NEB, MA, USA). Similarly, the Illumina library was subjected to quality tests by Qubit and fragment-size checking by Agilent 2100, which was followed by the sequencing conducted via Illumina NovaSeq PE150. The original sequencing data were filtered for the elimination of low-quality reads and adaptors, and the obtained clean data were assembled using the software SMRT Link version 5.0.1, as previously described (Ardui et al., 2018; Reiner et al., 2018).
Construction of phylogenic tree based on the tolC gene sequences
The tolC gene sequences were retrieved from the genomic sequences of the type or recommended reference strains representing their species in Enterobacter, as indicated in Figure 1A. These tolC sequences were aligned by MUSCLE (Codons) in MEGA version 11, the result of which was subsequently saved and used for phylogenic-tree construction via the method of maximum-likelihood. The bootstrap values were determined based on 500 replications.
Alignment of the amino acid sequences of TolC from different Enterobacter strains
The alignment of the nucleotide sequences of tolC described above (for the phylogenic tree) was translated into amino acid sequences using the translating function for alignments in MEGA version 11. The resulted alignment of the amino acid sequences was saved as a FASTA file. This file was opened, edited for display settings, and finally exported as a figure using the software DNAMAN version 10.
Construction of strains for the tolC deletion and complementation
To construct a clean deletion for tolC in EBU45301, a pRE112-based system was used as previously described (Edwards et al., 1998). Generally, the upstream and downstream of the EBU45301 tolC gene were PCR amplified using the primer pairs tolCUP-F: 5’-GAG CTC CCA GAT AGC TCA ACA CCG GT-3’/tolCUP-R: 5’-GGC GTG ATA ACA CTC TTG CAT TCC TTG TTG TGA AG-3’ and tolCDOWN-F: 5’-CAA CAA GGA ATG CAA GAG TGT TAT CAC GCC CTC TC-3’/tolCDOWN-R: 5’-GGT ACC CAT CAT GTA ACC TGC CAT TAA T-3’, respectively. These two DNA fragments were subsequently spliced by overlap extension (SOE) PCR (Thornton, 2016), to obtain the DNA product of ΔtolC, flanked by the built-in sequences of Sac I and Kpn I. The ΔtolC fragment was subcloned into pRE112 between Sac I and Kpn I, resulting in plasmid pBYL009. pBYL009 was then transformed into E. coli S17-1 λpir, and the resultant strain was used along with the wild-type EBU45301 for conjugation performed at 30°C. The conjugants of EBU45301 were selected as chloramphenicol-resistant (Cmr) and cefazolin-resistant (Czr) colonies in LB plates supplemented with 20 mg/L cefazolin (Apexbio, TX, USA) and 12.5 mg/L chloramphenicol. These conjugants were subsequently streaked on LB plates supplemented with 6% (w/v) sucrose (Solarbio) for separated single colonies, randomly picking and patching of which were conducted simultaneously on LB plates supplemented with or without chloramphenicol. The sucrose-resistant but chloramphenicol-sensitive colonies were further subjected to PCR tests using the primer pair tolCUP-F/tolCDOWN-R, and isolates of the tolC mutant were screened out as the ones from which a ~1100 bp product was amplified by this primer pair.
To complement the tolC deletion, fragment of the wild-type tolC gene was PCR amplified using the primer pair tolC-F1: 5’-CCC GTC CTG TGG ATC CTC GCC CTC TTC GAT CAT CC-3’/tolC-R1: 5’-CCG GCG TAG AGG ATC CAT CAT GTA ACC TGC CAT TAA T-3’. In this way, the tolC fragment was flanked by the sequences (underlined) homologous with those in plasmid pACYC184. This tolC fragment and the linear pACYC184, digested at site BamH I, were ligated according to the manufacturer’s instructions of the ClonExpress Ultra One Step Cloning Kit (Vazyme Biotech Co., Ltd, Nanjing, China), and the resulted plasmid was designated as pBYL024. The tolC mutant was subsequently transformed with pBYL024, and the tolC/pBYL024 strains were isolated as Cmr colonies. In the meanwhile, the tolC mutant was transformed with pACYC184, and the Cmr transformants were isolated and used as a control strain for the empty vector. Given that the tetracycline-resistant (Tcr) feature of pACYC184 is not suitable for the susceptibility tests using antibiotics like tetracycline and doxycycline, the fragment including gene tolC and its upstream and downstream was inserted in pRE112, to obtain an alternative complementing plasmid. Briefly, the tolC fragment was PCR amplified using the primer pair tolC-F2: 5’-TAT CGC ATG CGG TAC CTC GCC CTC TTC GAT CAT CC-3’/tolC-R2: 5’-TTC TTC TAG AGG TAC CAT CAT GTA ACC TGC CAT TAA T-3’. The sequences homologous with those in pRE112 have been underlined. This fragment was then subcloned in pRE112 at site Kpn I, in the same manner as that described for pBYL024, and the resulted plasmid was designated as pBYL030. As shown in Figure S4, the suicide plasmid pBYL030 is introduced and integrated into the genome of the tolC mutant, via the homologous recombination during the conjugation of S17-1 λpir/pBYL030 and tolC. The tolC/pBYL030 strain used in this study was selected as Cmr and Czr colonies in which the homologous recombination was occurred within the upstream of tolC (Figure S4). To check this, PCR tests using the primer pair tolC-CF:5’-AGG GCG GTC AGG TAA ACT CT-3’/tolC-CR1: 5’-GCG TGT TAC GGT GAA AAC CT-3’ were performed, as described in Figure S4. The vector-control strain, tolC/pBYL031, was constructed in the same manner as that described for tolC/pBYL030, except the different primer pair used for cloning of the upstream of tolC (tolCUP; Figure S5). This primer pair was tolC-F2/tolC-R3: 5’-TTC TTC TAG AGG TAC TTG CAT TCC TTG TTG TGA AG-3’. In addition, to perform complementation suitable for the susceptibility tests using chloramphenicol, the other alternative complementing plasmid, pBYL032, and its vector-control, pBYL033, were obtained, in the same manner as that described for pBYL030 and pBYL031, except that pDMS197 was used instead of pRE112 for cloning. Strain tolC/pBYL032 and tolC/pBYL033 were selected as Tcr and Czr colonies, via the same pipelines constructing tolC/pBYL030 and tolC/pBYL031 (Figures S4, S5). To check the desirable tolC/pBYL032 isolates, the primer pair used for PCR was tolC-CF/tolC-CR2: 5’-GAC AGC ATC GCC AGT CAC TA-3’.
Initial identification of the Bacillus sp. GHZJ-1 strain
The bacterial strain GHZJ-1 was isolated from a waste-water pond possessed by the Panjin Guanghe Crab Industry Co., Ltd and exhibited anti-yeast activity in other studies of our group (data not shown). To identify this bacterium, the partial region of its 16S rDNA gene was PCR amplified using the universal primer pair 27F/1492R. The PCR products were subjected to sanger sequencing, and the 16S rDNA sequence of GHZJ-1 was subsequently assembled (GenBank accession No. OP316901). BLASTing this sequence against the NCBI database, its homologues in strains identified as Bacillus amyloliquefaciens and Bacillus velezensis were shown with the highest score (bits), 100% identities and 100% query cover. Thus, GHZJ-1 was initially identified as a Bacillus strain.
Preparation of BCE
To prepare the crude extract of Bacillus sp. GHZJ-1, the strain was grown in YPD broth (Solarbio) at 28°C with aeration for 72 hours (h). Subsequently, the YPD culture of GHZJ-1 was subjected to 1.5-h sonication (every 15-second sonication process was followed by a 15-second pause) and centrifugation at 4°C, to obtain the supernatant. This supernatant was further sterilized via the 0.22-micrometer filters (Sangon Biotech, Shanghai, China) and finally used as the BCE agent (original) in susceptibility tests against EBU45301. In this manner, three independent batches of BCE were prepared for the susceptibility tests. The BCE stocks were preserved at -80°C and thawed on ice before use.
Antimicrobial susceptibility test
In this study, the MICs of piperacillin, cefazolin, ceftazidime, imipenem, gentamicin, kanamycin, tetracycline, norfloxacin, ciprofloxacin, chloramphenicol, and erythromycin were determined using the method of broth microdilution, according to the standards for the dilution methods provided by CLSI, 2018a. Briefly, the tested antibiotics at desired concentrations were prepared using cation-adjusted Mueller-Hinton broth (CAMHB; Solarbio) in a twofold-dilution manner and aliquoted in 96-well microtiter plates. The final cell density of the inoculum for each well was 5x 105 CFU/milliliter (mL). The microtiter plates were incubated at 37°C for 18 h in an ambient air incubator, and the MIC of antibiotics was measured as their lowest concentration that completely inhibit bacteria growth (CLSI, 2018a). The concentrations used for the tested antibiotics ranged as follows: piperacillin (Solarbio), 64-0.031 mg/L; cefazolin (Apexbio), 512-0.5 mg/L; ceftazidime (Solarbio), 64-0.031 mg/L; imipenem (Solarbio), 16-0.008 mg/L; gentamicin (Solarbio), 32-0.016 mg/L; kanamycin (Solarbio), 32-0.016 mg/L; tetracycline (Solarbio), 32-0.016 mg/L; norfloxacin (Solarbio), 6.144-0.0015 mg/L; ciprofloxacin (Solarbio), 0.512-0.00025 mg/L; chloramphenicol (Solarbio), 128-0.06 mg/L; erythromycin (Solarbio), 6,144-0.75 mg/L. Three independent experiments were performed to determine the MICs of these antibiotics, and in every independent experiment, the tests for each concentration used were conducted in triplicates. The disk-diffusion tests were performed according to the standards for the methods of disk diffusion provided by CLSI, 2018b, using the antibiotic disks (BKMAM, Hunan, China) as follows: piperacillin, 100 micrograms (µg); cefuroxime, 30 µg; ceftriaxone, 30 µg; cefoperazone, 75 µg; amikacin, 30 µg; gentamicin, 10 µg; kanamycin, 30 µg; streptomycin, 10 µg; tetracycline, 30 µg; minocycline, 30 µg; doxycycline, 30 µg; levofloxacin, 5 µg; florfenicol, 30 µg; trimethoprim-sulfamethoxazole, 1.25-23.75 µg; erythromycin, 15 µg; azithromycin, 15 µg; lincomycin, 2 µg; clindamycin, 2 µg. Briefly, the tested bacteria were seeded in the Mueller-Hinton agar (MHA; Solarbio) plates via evenly coating the plates with streaks of the inoculum (0.5 McFarland standard). The antibiotic disks were then placed properly in the inoculated MHA plates, which was followed by 18-h incubation at 37°C of these plates in an ambient air incubator (CLSI, 2018b). The well-diffusion tests were performed for ciprofloxacin and norfloxacin. Basically, the MHA plates were evenly coated with bacterial cultures, as that described for the disk-diffusion tests (CLSI, 2018b). Afterwards, cylinder wells with 6 mm-diameter base sides were drilled in the inoculated agar with a height of approximately 3 mm. 75 microliters (µL) of 0.16 mg/L ciprofloxacin or 1.92 mg/L norfloxacin (both diluted in CAMHB) was aliquoted into each well for the tests. Finally, all the treated agar plates were incubated at 37°C for 18 hours in an ambient air incubator, to observe the inhibition zones. 75 µL CAMHB was used as the negative control for each well. Diameter of the inhibition zone for well-diffusion tests was determined as the value subtracting the well diameter (6 mm) from the whole diameter for the inhibition circle. For each tested antibiotic in disk-diffusion and well-diffusion assays, diameter of the inhibition zone was measured as average ± SD based on three independent experiments. The comparative analyses of data were performed using student’s t tests. The susceptibility/resistance of EBU45301 to piperacillin, cefazolin, ceftazidime, imipenem, gentamicin, kanamycin, tetracycline, norfloxacin, ciprofloxacin, and chloramphenicol was determined according to the MIC-based breakpoints provided by CLSI, 2022. The susceptibility/resistance of EBU45301 to cefuroxime, ceftriaxone, cefoperazone, amikacin, streptomycin, minocycline, doxycycline, levofloxacin, and trimethoprim-sulfamethoxazole was determined according to the breakpoints based on the inhibition-zone diameters, provided by CLSI, 2022. For the susceptibility tests the readings of which were used for susceptibility/resistance determination, the E. coli ATCC 25922 strain was used as a quality-control strain (CLSI, 2022).
To assess the susceptibility to BBH in the EBU45301 strains, 18 mg/mL BBH was prepared using DMSO as the solvent. The MICs of BBH against the tested strains were determined using the broth-microdilution method as described above. In the meanwhile, the antibacterial activity of DMSO at the concentrations same as that contained in the serially diluted BBH was tested independently, to rule out the interference of DMSO in inhibition of EBU45301 growth. The concentrations of BBH used for the tests ranged from 2,048 mg/L to 4 mg/L.
The susceptibility of the EBU45301 strains to BCE was tested using both the broth-microdilution and well-diffusion method, as described above. To determine the MIC of BCE against the EBU45301 strains, the concentrations ranged from 0.5x original (BCE) to 0.001x original (BCE) were prepared by performing serial twofold dilutions with CAMHB and used for the tests. In the meanwhile, the antibacterial activity of YPD broth at the concentrations same as that contained in the serially diluted BCE was tested independently, to rule out the interference of YPD broth in inhibition of EBU45301 growth. Using the same batch of BCE (three batches in total), two independent experiments were performed for each of the EBU45301 strain, and in every independent experiment, the tests for each concentration used were conducted in triplicates. In the well-diffusion assays, 75 µL BCE (original) was aliquoted into each well, and 75 µL YPD broth was used as the negative control for each well. Three independent experiments were performed with each one using BCE from different batches for comparisons. To determine the diameter of the inhibition zones formed by BCE, the average reading for the three BCE batches used in the same independent experiment was calculated first, and the final value of diameter was determined as average ± SD based on the average readings for the three independent experiments. The comparative analyses of data were performed using student’s t tests.
Synergistic assay
The synergistic assays testing BCE in association with erythromycin, norfloxacin, or ciprofloxacin against the EBU45301 WT were performed using both the checkerboard method and the well-diffusion method. The checkerboard arrangements for the tested combinations were in the same manner as previously described (MacNair et al., 2018). In the checkerboard assays, the fractional inhibitory concentrations of BCE, erythromycin, norfloxacin, and ciprofloxacin against WT were determined using the broth-microdilution protocols for these four antibiotics, respectively, as described above. To better assay the synergistic effects combining BCE and erythromycin, norfloxacin, or ciprofloxacin against WT, the well-diffusion protocol described for testing the antibacterial activity of BCE was used. The inhibition zones formed by BCE in the seeded MHA plates supplemented with 12 mg/L erythromycin, 24 mg/L erythromycin, 0.003 mg/L norfloxacin, 0.006 mg/L norfloxacin, 0.25 µg/L ciprofloxacin, 0.5 µg/L ciprofloxacin, or none antibiotics were observed and determined for diameters, as described above. The comparative analyses of data were performed using student’s t tests.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
BL and JZ conducted the experiments. BL, JZ, and XL analyzed the data. BL and XL drafted the manuscript on the basis of the analyzed data. BL and XL designed the whole project. All authors contributed to the article and approved the submitted version.
Funding
XL was supported by the Liaoning Province “The Open Competition Mechanism to Select the Best Candidates” Project, with the Grant/Award Number 2021JH1/10400040.
Acknowledgments
We appreciate the generosity of Panjin Guanghe Crab Industry Co., Ltd by providing their waste-water pond as the resource for isolation of the strains used in our studies.
Conflict of interest
Author XL is employed by Panjin Guanghe Crab Industry Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1036933/full#supplementary-material
References
Annavajhala, M. K., Gomez-Simmonds, A., Uhlemann, A. C. (2019). Multidrug-resistant enterobacter cloacae complex emerging as a global, diversifying threat. Front. Microbiol. 10, 44. doi: 10.3389/fmicb.2019.00044
Ardui, S., Ameur, A., Vermeesch, J. R., Hestand, M. S. (2018). Single molecule real-time (Smrt) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 46, 2159–2168. doi: 10.1093/nar/gky066
Cavera, V. L., Volski, A., Chikindas, M. L. (2015). The natural antimicrobial subtilosin a synergizes with lauramide arginine ethyl ester (Lae), epsilon-Poly-L-Lysine (Polylysine), clindamycin phosphate and metronidazole, against the vaginal pathogen gardnerella vaginalis. Probiotics Antimicrob. Proteins 7, 164–171. doi: 10.1007/s12602-014-9183-1
Chakraborty, K., Kizhakkekalam, V. K., Joy, M., Chakraborty, R. D. (2022). Bacillibactin class of siderophore antibiotics from a marine symbiotic bacillus as promising antibacterial agents. Appl. Microbiol. Biotechnol. 106, 329–340. doi: 10.1007/s00253-021-11632-0
Chang, A. C., Cohen, S. N. (1978). Construction and characterization of amplifiable multicopy dna cloning vehicles derived from the P15a cryptic miniplasmid. J. Bacteriol. 134, 1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978
Chang, T. M., Lu, P. L., Li, H. H., Chang, C. Y., Chen, T. C., Chang, L. L. (2007). Characterization of fluoroquinolone resistance mechanisms and their correlation with the degree of resistance to clinically used fluoroquinolones among escherichia coli isolates. J. Chemother. 19, 488–494. doi: 10.1179/joc.2007.19.5.488
Chavda, K. D., Chen, L., Fouts, D. E., Sutton, G., Brinkac, L., Jenkins, S. G., et al. (2016). Comprehensive genome analysis of carbapenemase-producing enterobacter spp.: New insights into phylogeny, population structure, and resistance mechanisms. Mbio 7 (6), e02093–16. doi: 10.1128/mBio.02093-16
Chow, J. W., Fine, M. J., Shlaes, D. M., Quinn, J. P., Hooper, D. C., Johnson, M. P., et al. (1991). Enterobacter bacteremia: Clinical features and emergence of antibiotic resistance during therapy. Ann. Intern. Med. 115, 585–590. doi: 10.7326/0003-4819-115-8-585
Ciufo, S., Kannan, S., Sharma, S., Badretdin, A., Clark, K., Turner, S., et al. (2018). Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes At the ncbi. Int. J. Syst. Evol. Microbiol. 68, 2386–2392. doi: 10.1099/ijsem.0.002809
CLSI (2018a). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically CLSI standard M07. 11th ed (Wayne, PA: CLSI).
CLSI (2018b). Performance standards for antimicrobial disk susceptibility tests CLSI standard M02. 13th ed (Wayne, PA: CLSI).
CLSI (2022). Performance standards for antimicrobial susceptibility testing CLSI supplement M100. 32nd ed (Wayne, PA: CLSI).
Davin-Regli, A., Pages, J. M. (2015). Enterobacter aerogenes and enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 6, 392. doi: 10.3389/fmicb.2015.00392
de Souza Freitas, F., de Assis Lage, T. C., Ayupe, B. A. L., de Paula Siqueira, T., De Barros, M., Totola, M. R. (2020). Bacillus subtilis Tr47ii as a source of bioactive lipopeptides against gram-negative pathogens causing nosocomial infections. 3 Biotech. 10, 474. doi: 10.1007/s13205-020-02459-z
Doijad, S., Imirzalioglu, C., Yao, Y., Pati, N. B., Falgenhauer, L., Hain, T., et al. (2016). Enterobacter bugandensis sp. nov., isolated from neonatal blood. Int. J. Syst. Evol. Microbiol. 66, 968–974. doi: 10.1099/ijsem.0.000821
Edwards, R. A., Keller, L. H., Schifferli, D. M. (1998). Improved allelic exchange vectors and their use to analyze 987p fimbria gene expression. Gene 207, 149–157. doi: 10.1016/S0378-1119(97)00619-7
Erega, A., Stefanic, P., Danevcic, T., Smole Mozina, S., Mandic Mulec, I. (2022). Impact of bacillus subtilis antibiotic bacilysin and campylobacter jejuni efflux pumps on pathogen survival in mixed biofilms. Microbiol. Spectr. 10 (4), e0215622. doi: 10.1128/spectrum.02156-22
Falgenhauer, J., Imirzalioglu, C., Falgenhauer, L., Yao, Y., Hauri, A. M., Erath, B., et al. (2019). Whole-genome sequences of clinical enterobacter bugandensis isolates from Germany. Microbiol. Resour. Announc. 8 (29), e00465–19. doi: 10.1128/MRA.00465-19
Federhen, S., Rosello-Mora, R., Klenk, H. P., Tindall, B. J., Konstantinidis, K. T., Whitman, W. B., et al. (2016). Meeting report: Genbank microbial genomic taxonomy workshop (12–13 may 2015). Stand Genom. Sci. 11, 15. doi: 10.1186/s40793-016-0134-1
Fernandez-Baca, V., Ballesteros, F., Hervas, J. A., Villalon, P., Dominguez, M. A., Benedi, V. J., et al. (2001). Molecular epidemiological typing of enterobacter cloacae isolates from a neonatal intensive care unit: Three-year prospective study. J. Hosp. Infect. 49, 173–182. doi: 10.1053/jhin.2001.1053
Gravey, F., Cattoir, V., Ethuin, F., Fabre, L., Beyrouthy, R., Bonnet, R., et al. (2020). Ramr deletion in an enterobacter hormaechei isolate as a consequence of therapeutic failure of key antibiotics in a long-term hospitalized patient. Antimicrob. Agents Chemother. 64 (10), e00962–20. doi: 10.1128/AAC.00962-20
Grimsey, E. M., Fais, C., Marshall, R. L., Ricci, V., Ciusa, M. L., Stone, J. W., et al. (2020). Chlorpromazine and amitriptyline are substrates and inhibitors of the acrb multidrug efflux pump. Mbio 11 (3), e00465–20. doi: 10.1128/mBio.00465-20
Guerin, F., Lallement, C., Isnard, C., Dhalluin, A., Cattoir, V., Giard, J. C. (2016). Landscape of resistance-Nodulation-Cell division (Rnd)-type efflux pumps in enterobacter cloacae complex. Antimicrob. Agents Chemother. 60, 2373–2382. doi: 10.1128/AAC.02840-15
Hilty, M., Sendi, P., Seiffert, S. N., Droz, S., Perreten, V., Hujer, A. M., et al. (2013). Characterisation and clinical features of enterobacter cloacae bloodstream infections occurring At a tertiary care university hospital in Switzerland: Is cefepime adequate therapy? Int. J. Antimicrob. Agents 41, 236–249. doi: 10.1016/j.ijantimicag.2012.10.022
Huang, L., Feng, Y., Zong, Z. (2019). Heterogeneous resistance to colistin in enterobacter cloacae complex due to a new small transmembrane protein. J. Antimicrob. Chemother. 74, 2551–2558. doi: 10.1093/jac/dkz236
Kobayashi, N., Nishino, K., Yamaguchi, A. (2001). Novel macrolide-specific abc-type efflux transporter in escherichia coli. J. Bacteriol. 183, 5639–5644. doi: 10.1128/JB.183.19.5639-5644.2001
Kuete, V., Ngameni, B., Tangmouo, J. G., Bolla, J. M., Alibert-Franco, S., Ngadjui, B. T., et al. (2010). Efflux pumps are involved in the defense of gram-negative bacteria against the natural products isobavachalcone and diospyrone. Antimicrob. Agents Chemother. 54, 1749–1752. doi: 10.1128/AAC.01533-09
Li, J., Chen, J., Yang, G., Tao, L. (2021a). Sublancin protects against methicillin-resistant staphylococcus aureus infection by the combined modulation of innate immune response and microbiota. Peptides 141, 170533. doi: 10.1016/j.peptides.2021.170533
Li, Y., Huang, J., Li, L., Liu, L. (2017). Synergistic activity of berberine with azithromycin against pseudomonas aeruginosa isolated from patients with cystic fibrosis of lung In vitro and In vivo. Cell Physiol. Biochem. 42, 1657–1669. doi: 10.1159/000479411
Li, X. Z., Plesiat, P., Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/CMR.00117-14
Li, X., Song, Y., Wang, L., Kang, G., Wang, P., Yin, H., et al. (2021b). A potential combination therapy of berberine hydrochloride with antibiotics against multidrug-resistant acinetobacter baumannii. Front. Cell Infect. Microbiol. 11, 660431. doi: 10.3389/fcimb.2021.660431
Liu, S., Fang, R., Zhang, Y., Chen, L., Huang, N., Yu, K., et al. (2021). Characterization of resistance mechanisms of enterobacter cloacae complex Co-resistant to carbapenem and colistin. BMC Microbiol. 21, 208. doi: 10.1186/s12866-021-02250-x
Liu, J., Wang, X., Shi, W., Qian, Z., Wang, Y. (2019). Sensitization of avian pathogenic escherichia coli to amoxicillin In vitro and In vivo in the presence of surfactin. PloS One 14, E0222413. doi: 10.1371/journal.pone.0222413
Liu, L., Yu, J., Tang, M., Liu, J. (2018). Mechanisms of resistance in clinical isolates of enterobacter cloacae that are less susceptible to cefepime than to ceftazidime. Ann. Clin. Lab. Sci. 48, 355–362.
Lv, J., Da, R., Cheng, Y., Tuo, X., Wei, J., Jiang, K., et al. (2020). Mechanism of antibacterial activity of bacillus amyloliquefaciens c-1 lipopeptide toward anaerobic clostridium difficile. BioMed. Res. Int. 2020, 3104613. doi: 10.1155/2020/3104613
MacNair, C. R., Stokes, J. M., Carfrae, L. A., Fiebig-Comyn, A. A., Coombes, B. K., Mulvey, M. R., et al. (2018). Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 9, 458. doi: 10.1038/s41467-018-02875-z
Matteoli, F. P., Passarelli-Araujo, H., Pedrosa-Silva, F., Olivares, F. L., Venancio, T. M. (2020). Population structure and pangenome analysis of enterobacter bugandensis uncover the presence of blactx-M-55, blandm-5 and blaimi-1, along with sophisticated iron acquisition strategies. Genomics 112, 1182–1191. doi: 10.1016/j.ygeno.2019.07.003
Mezzatesta, M. L., Gona, F., Stefani, S. (2012). Enterobacter cloacae complex: Clinical impact and emerging antibiotic resistance. Future Microbiol. 7, 887–902. doi: 10.2217/fmb.12.61
Moon, S. H., Udaondo, Z., Li, X., Yang, X., Jun, S. R., Huang, E. (2021). Isolation and characterisation of carbapenemase-producing and polymyxin b-resistant enterobacter bugandensis from a vegetable. J. Glob. Antimicrob. Resist. 26, 264–265. doi: 10.1016/j.jgar.2021.07.013
Moosavian, M., Sima, M. K., Khosravi, N. A., Montazeri, E. A. (2021). Detection of oqxab efflux pumps, a multidrug-resistant agent in bacterial infection in patients referring to teaching hospitals in ahvaz, southwest of Iran. Int. J. Microbiol. 2021, 2145176. doi: 10.1155/2021/2145176
Morita, Y., Nakashima, K., Nishino, K., Kotani, K., Tomida, J., Inoue, M., et al. (2016). Berberine is a novel type efflux inhibitor which attenuates the mexxy-mediated aminoglycoside resistance in pseudomonas aeruginosa. Front. Microbiol. 7, 1223. doi: 10.3389/fmicb.2016.01223
Nagakubo, S., Nishino, K., Hirata, T., Yamaguchi, A. (2002). The putative response regulator baer stimulates multidrug resistance of escherichia coli Via a novel multidrug exporter system, mdtabc. J. Bacteriol. 184, 4161–4167. doi: 10.1128/JB.184.15.4161-4167.2002
Nikaido, H., Zgurskaya, H. I. (1999). Antibiotic efflux mechanisms. Curr. Opin. Infect. Dis. 12, 529–536. doi: 10.1097/00001432-199912000-00001
Nishino, K., Yamada, J., Hirakawa, H., Hirata, T., Yamaguchi, A. (2003). Roles of tolc-dependent multidrug transporters of escherichia coli in resistance to beta-lactams. Antimicrob. Agents Chemother. 47, 3030–3033. doi: 10.1128/AAC.47.9.3030-3033.2003
Nishino, K., Yamasaki, S., Nakashima, R., Zwama, M., Hayashi-Nishino, M. (2021). Function and inhibitory mechanisms of multidrug efflux pumps. Front. Microbiol. 12, 737288. doi: 10.3389/fmicb.2021.737288
Pati, N. B., Doijad, S. P., Schultze, T., Mannala, G. K., Yao, Y., Jaiswal, S., et al. (2018). Enterobacter bugandensis: A novel enterobacterial species associated with severe clinical infection. Sci. Rep. 8, 5392. doi: 10.1038/s41598-018-23069-z
Perez, A., Canle, D., Latasa, C., Poza, M., Beceiro, A., Tomas Mdel, M., et al. (2007). Cloning, nucleotide sequencing, and analysis of the acrab-tolc efflux pump of enterobacter cloacae and determination of its involvement in antibiotic resistance in a clinical isolate. Antimicrob. Agents Chemother. 51, 3247–3253. doi: 10.1128/AAC.00072-07
Reiner, J., Pisani, L., Qiao, W., Singh, R., Yang, Y., Shi, L., et al. (2018). Cytogenomic identification and long-read single molecule real-time (Smrt) sequencing of a bardet-biedl syndrome 9 (Bbs9) deletion. NPJ Genom. Med. 3, 3. doi: 10.1038/s41525-017-0042-3
Sanders, W. E., Jr., Sanders, C. C. (1997). Enterobacter spp.: Pathogens poised to flourish At the turn of the century. Clin. Microbiol. Rev. 10, 220–241. doi: 10.1128/CMR.10.2.220
Sarangi, J., Matsuo, N., Nonogaki, R., Hayashi, M., Kawamura, K., Suzuki, M., et al. (2022). Molecular epidemiology of enterobacter cloacae complex isolates with reduced carbapenem susceptibility recovered by blood culture. Jpn J. Infect. Dis. 75, 41–48. doi: 10.7883/yoken.JJID.2021.141
Scholz, R., Vater, J., Budiharjo, A., Wang, Z., He, Y., Dietel, K., et al. (2014). Amylocyclicin, a novel circular bacteriocin produced by bacillus amyloliquefaciens Fzb42. J. Bacteriol. 196, 1842–1852. doi: 10.1128/JB.01474-14
Senchyna, F., Tamburini, F. B., Murugesan, K., Watz, N., Bhatt, A. S., Banaei, N. (2021). Comparative genomics of enterobacter cloacae complex before and after acquired clinical resistance to ceftazidime-avibactam. Diagn. Microbiol. Infect. Dis. 101, 115511. doi: 10.1016/j.diagmicrobio.2021.115511
Singh, N. K., Bezdan, D., Checinska Sielaff, A., Wheeler, K., Mason, C. E., Venkateswaran, K. (2018). Multi-drug resistant enterobacter bugandensis species isolated from the international space station and comparative genomic analyses with human pathogenic strains. BMC Microbiol. 18, 175. doi: 10.1186/s12866-018-1325-2
Telke, A. A., Olaitan, A. O., Morand, S., Rolain, J. M. (2017). Soxrs induces colistin hetero-resistance in enterobacter asburiae and enterobacter cloacae by regulating the acrab-tolc efflux pump. J. Antimicrob. Chemother. 72, 2715–2721. doi: 10.1093/jac/dkx215
Thornton, J. A. (2016). Splicing by overlap extension pcr to obtain hybrid dna products. Methods Mol. Biol. 1373, 43–49. doi: 10.1007/7651_2014_182
Wilson, K. E., Flor, J. E., Schwartz, R. E., Joshua, H., Smith, J. L., Pelak, B. A., et al. (1987). Difficidin and oxydifficidin: Novel broad spectrum antibacterial antibiotics produced by bacillus subtilis. ii. isolation and physico-chemical characterization. J. Antibiot. (Tokyo) 40, 1682–1691. doi: 10.7164/antibiotics.40.1682
Yousefian, N., Ornik-Cha, A., Poussard, S., Decossas, M., Berbon, M., Daury, L., et al. (2021). Structural characterization of the emrab-tolc efflux complex from e. coli. Biochim. Biophys. Acta Biomembr. 1863, 183488. doi: 10.1016/j.bbamem.2020.183488
Keywords: TolC, efflux pumps, antimicrobial agents, susceptibility, synergistic effects, Enterobacter bugandensis
Citation: Li B, Zhang J and Li X (2022) A comprehensive description of the TolC effect on the antimicrobial susceptibility profile in Enterobacter bugandensis. Front. Cell. Infect. Microbiol. 12:1036933. doi: 10.3389/fcimb.2022.1036933
Received: 05 September 2022; Accepted: 24 November 2022;
Published: 09 December 2022.
Edited by:
Alessandra Oliva, Department of Public Health and Infectious Diseases, Sapienza University of Rome, ItalyReviewed by:
Tieli Zhou, First Affiliated Hospital of Wenzhou Medical University, ChinaAlberto Antonelli, University of Florence, Italy
Copyright © 2022 Li, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingyu Li, YnlsaTE5ODhAZm94bWFpbC5jb20=; Xiaodong Li, bGl4aWFvZG9uZ0BzeWF1LmVkdS5jbg==
 Bingyu Li
Bingyu Li Ji Zhang
Ji Zhang Xiaodong Li2,3*
Xiaodong Li2,3*