- 1Global and Tropical Health Division, Menzies School of Health Research, Charles Darwin University, Darwin, N T, Australia
- 2Infectious Diseases Society Kota Kinabalu Sabah – Menzies School of Health Research Clinical Research Unit, Kota Kinabalu, Sabah, Malaysia
- 3Clinical Research Centre, Queen Elizabeth Hospital, Ministry of Health, Kota Kinabalu, Sabah, Malaysia
- 4Department of Medicine, Queen Elizabeth Hospital II, Kota Kinabalu, Sabah, Malaysia
- 5Faculty of Medicine and Health Sciences, Universiti Malaysia Sabah, Kota Kinabalu, Sabah, Malaysia
- 6Clinical Malaria, Queensland Institute of Medical Research (QIMR) Berghofer Medical Research Institute, Brisbane, QLD, Australia
- 7Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, United Kingdom
- 8Malaria and Fever, Foundation for Innovative New Diagnostics (FIND), Geneva, Switzerland
Background: Plasmodium knowlesi causes zoonotic malaria across Southeast Asia. First-line diagnostic microscopy cannot reliably differentiate P. knowlesi from other human malaria species. Rapid diagnostic tests (RDTs) designed for P. falciparum and P. vivax are used routinely in P. knowlesi co-endemic areas despite potential cross-reactivity for species-specific antibody targets.
Methods: Ten RDTs were evaluated: nine to detect clinical P. knowlesi infections from Malaysia, and nine assessing limit of detection (LoD) for P. knowlesi (PkA1-H.1) and P. falciparum (Pf3D7) cultures. Targets included Plasmodium-genus parasite lactate dehydrogenase (pan-pLDH) and P. vivax (Pv)-pLDH.
Results: Samples were collected prior to antimalarial treatment from 127 patients with microscopy-positive PCR-confirmed P. knowlesi mono-infections. Median parasitaemia was 788/µL (IQR 247-5,565/µL). Pan-pLDH sensitivities ranged from 50.6% (95% CI 39.6–61.5) (SD BIOLINE) to 87.0% (95% CI 75.1–94.6) (First Response® and CareStart™ PAN) compared to reference PCR. Pv-pLDH RDTs detected P. knowlesi with up to 92.0% (95% CI 84.3-96.7%) sensitivity (Biocredit™). For parasite counts ≥200/µL, pan-pLDH (Standard Q) and Pv-pLDH RDTs exceeded 95% sensitivity. Specificity of RDTs against 26 PCR-confirmed negative controls was 100%. Sensitivity of six highest performing RDTs were not significantly different when comparing samples taken before and after (median 3 hours) antimalarial treatment. Parasite ring stages were present in 30% of pre-treatment samples, with ring stage proportions (mean 1.9%) demonstrating inverse correlation with test positivity of Biocredit™ and two CareStart™ RDTs.
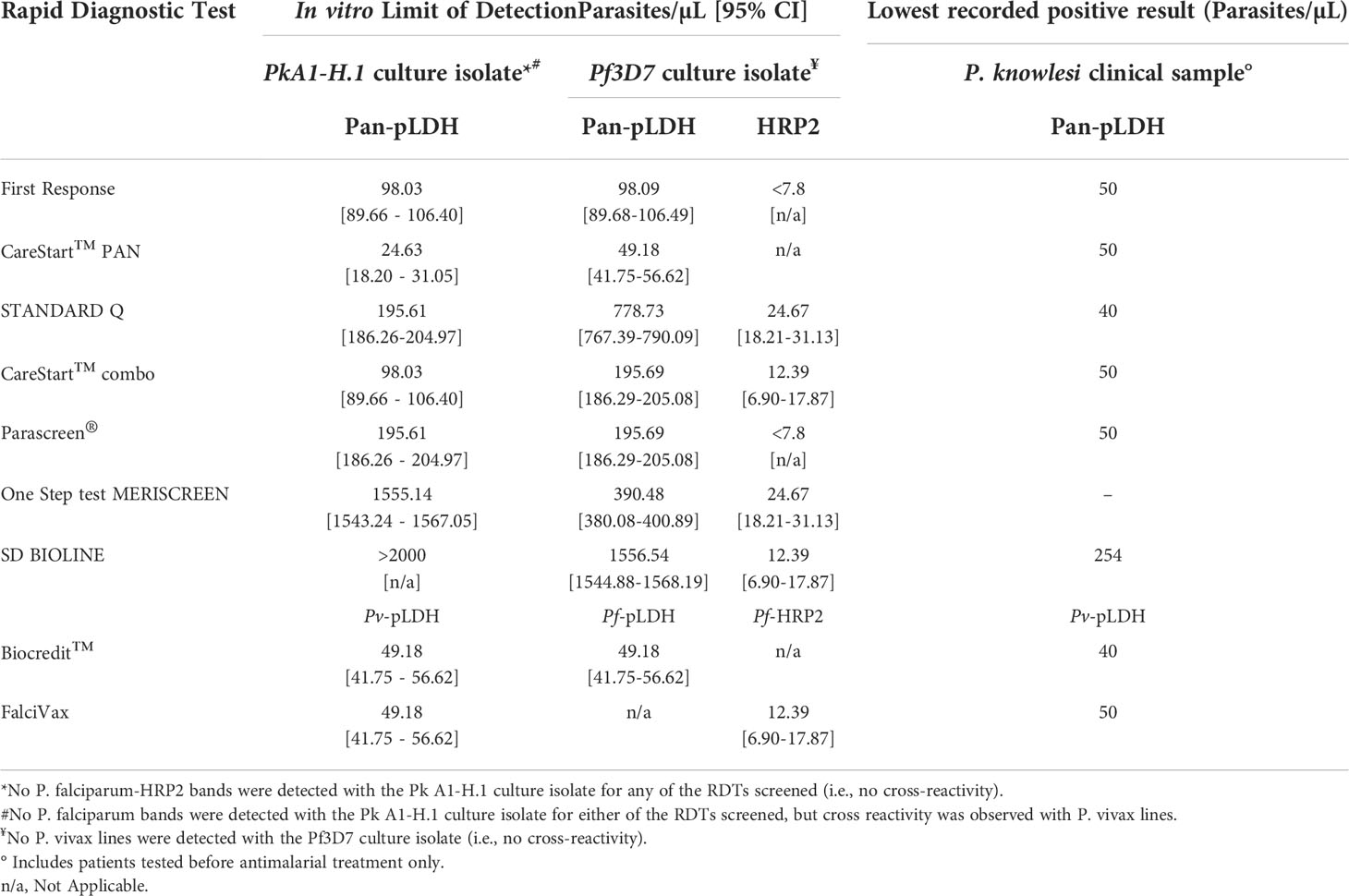
For cultured P. knowlesi, CareStart™ PAN demonstrated the lowest LoD at 25 parasites/µL; LoDs of other pan-pLDH ranged from 98 to >2000 parasites/µL. Pv-pLDH LoD for P. knowlesi was 49 parasites/µL. No false-positive results were observed in either P. falciparum-pLDH or histidine-rich-protein-2 channels.
Conclusion: Selected RDTs demonstrate sufficient performance for detection of major human malaria species including P. knowlesi in co-endemic areas where microscopy is not available, particularly for higher parasite counts, although cannot reliably differentiate among non-falciparum malaria.
Introduction
The emergence of the simian malaria parasite Plasmodium knowlesi, known to infect humans across most of Southeast Asia, poses significant challenges to regional malaria control programs and World Health Organization (WHO) human malaria elimination progress (World Health Organization, 2020). In Malaysia, a substantial increase in P. knowlesi cases has been reported since 2008 (Chin et al., 2020; Cooper et al., 2020), and P. knowlesi is now almost the sole cause of malaria with over 2600 documented cases in 2020 (World Health Organization, 2021). Other countries with a higher burden of human malaria, such as Indonesia (Coutrier et al., 2018), India (Tyagi et al., 2013), Cambodia (Khim et al., 2011), Myanmar (Jiang et al., 2010), and Thailand (Putaporntip et al., 2009) have also reported P. knowlesi cases using molecular methods, with these cases initially being misidentified as other species using routine light microscopy. Due to similarities in morphology between P. knowlesi and other endemic Plasmodium species (Lee et al., 2009), there are major limitations to microscopy as the primary method of diagnosis in endemic areas, with regional prevalence likely underestimated (Barber et al., 2013b; Grigg et al., 2021). Sensitive and specific malaria diagnostic tools are vital to ensure timely diagnosis, particularly in areas of high prevalence (World Health Organization, 2011) where misidentification of P. knowlesi as other Plasmodium species can lead to delayed or inappropriate treatment and increased risk of fatal outcome (Rajahram et al., 2019).
The utilization of antibody panels for human malaria has enabled the design of antigen-capture rapid diagnostic tests (RDTs) as an alternative to microscopy (Moody, 2002). Although no RDT has been designed for specific detection of P. knowlesi, the current list of WHO-prequalified in vitro diagnostics (IVD) for malaria includes seven tests that can detect the genus-conserved parasite lactate dehydrogenase (pLDH) antigen (World Health Organization, 2022b). However, the performance of these RDTs for detecting P. knowlesi infections has not been investigated. Moreover, in the last round of the WHO-FIND Malaria RDT Evaluation Programme published in 2018, composite test positivity was considerably increased compared to earlier rounds, with the best-performing pLDH-based tests for P. falciparum and P. vivax detection approaching 100% even at low parasite counts of 200/µL (World Health Organization, 2018). McCutchan et al. previously detailed a panel of monoclonal antibodies targeting multiple epitopes against LDH isoforms of the major human Plasmodium species in addition to P. knowlesi. Three epitopes with nominal specific binding capacity to LDH from P. vivax and P. falciparum were found to cross-react with P. knowlesi LDH, although not with LDH from other simian malaria species (McCutchan et al., 2008). However, previous studies evaluating the utility of a small number of P. vivax and P. falciparum-pLDH based RDTs from previous WHO RDT testing rounds demonstrated poor sensitivity for the detection of clinical P. knowlesi infections (Barber et al., 2013a; Foster et al., 2014; Grigg et al., 2014).
Within Southeast Asia, there is large variance in the use of microscopy or RDTs as the first-line malaria diagnostic tool, in addition to the specific type of RDTs targeting different antigens such as pLDH or P. falciparum-specific histidine-rich protein 2 (HRP2) commercially sourced between endemic countries. For example, Malaysia uses microscopy as the first-line detection tool, whereas a variety of different malaria RDTs are primarily used within certain areas of other countries such as Indonesia (World Health Organization, 2020). At a regional level, India is the biggest consumer of RDTs as part of their national malaria control strategy due to the prevalence of both P. falciparum and P. vivax (World Health Organization, 2019; Kojom Foko et al., 2021). Although parasitological diagnosis using RDTs is recommended by the WHO where microscopy is unavailable, many countries still face logistical and cost difficulties in providing access to RDTs in remote areas, compounded by the limited analytical sensitivity of pLDH-based RDTs, with cases potentially treated or referred to tertiary centers based on clinical suspicion (Abba et al., 2014; Betty et al., 2015; Jimenez et al., 2017; Yerlikaya et al., 2018; Cunningham et al., 2019).
In this study we evaluate the utility of commercially-available RDTs for detecting clinical P. knowlesi infections from an endemic setting. We also estimate the limit of detection of RDTs using dilution series of P. knowlesi and P. falciparum laboratory cultures (Hussin et al., 2020).
Methods
Rapid diagnostic tests selection
The study included all seven WHO-prequalified malaria RDTs that target pan-pLDH (World Health Organization, 2022b). Based on the WHO round 8 malaria RDT product testing results available at the time of the study design (World Health Organization, 2018), two additional RDTs were selected according to the best panel detection score for both P. falciparum and P. vivax at parasite count thresholds of 200 and 2000 parasites/µL. RDTs with a lower false positivity rate were then preferentially included. Subject to source availability from manufacturer, the final ten RDTs were selected (Table 1). Eight RDTs were evaluated on both clinical P. knowlesi samples and laboratory cultured P. knowlesi and P. falciparum isolates. One RDT (careUS™) was tested only on clinical samples, and another (Meriscreen OneStep™) on only laboratory samples.
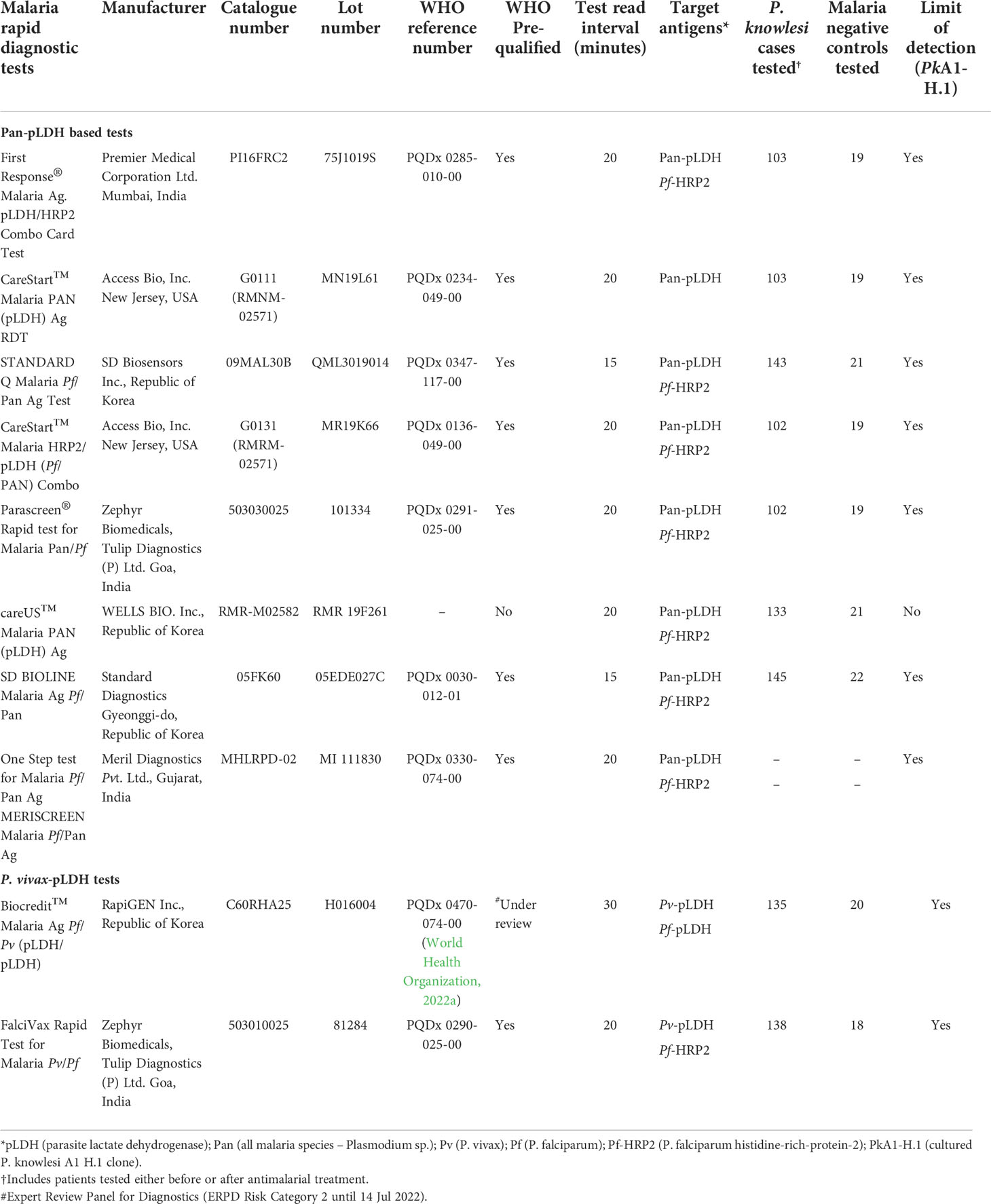
Table 1 Product information, target antigens and clinical data for the ten rapid diagnostic tests evaluated for P. knowlesi detection.
Study sites, subjects, and ethical approval
Patients were enrolled from Ranau District Hospital in Sabah, Malaysia, a secondary referral center servicing the administrative district (area of 3609 km2), accepting referrals from 16 primary health clinics, including all cases of malaria for in-patient management. Patient blood samples were collected and demographic and clinical data were recorded on standardized case record forms as part of an ongoing prospective observational malaria study. Patients presenting to the hospital study site were included if they were positive for P. knowlesi by microscopy, were more than 1 year of age, and provided appropriate written informed consent. Malaria-negative individuals presenting with or without a fever at the same healthcare facility were also prospectively enrolled as controls. The study was approved by the Medical Research and Ethics Committee of the Ministry of Health, Malaysia (NMRR-10-584-6684) and Menzies School of Health Research, Australia (HREC 10-1431).
Blood sample procedures
Venous whole blood samples were collected in EDTA vacutainers prior to antimalarial treatment where possible. All patients had thick and thin blood films prepared from pre-treatment whole blood for verification of parasitaemia by microscopy. Microscopic quantification of P. knowlesi parasitaemia (parasites per microlitre) was conducted by an experienced research microscopist equivalent to WHO Level 1 competency; parasitaemia was calculated from the number of parasites per 200 white blood cells on thick blood film, multiplied by the individual patient’s total white cell count (Ba et al., 2015; World Health Organization et al., 2015) obtained from routine hospital laboratory flow cytometry. Specific blood volumes specified in manufacturers’ instructions were used to evaluate the RDTs. The remaining EDTA whole blood samples were stored at -80°C for subsequent malaria species confirmation by reference real time PCR.
Plasmodium species confirmation by reference real-time PCR
All microscopically diagnosed malaria cases and microscopy-negative controls were tested by PCR for P. knowlesi, P. falciparum, P. vivax, P. malariae and P. ovale spp. In brief, genomic DNA was extracted from 200 μL of whole blood using QIAamp DNA Blood Mini Kits (Cat. No.: 51106; QIAGEN) according to the manufacturer’s manual, with a final elution volume of 200 μL. PCR detection of Plasmodium species was performed by laboratory research members blinded to the microscopy results. A real-time PCR assay, QuantiFast™ targeting the 18S SSU rRNA gene was conducted once for each sample in accordance with the manufacturers’ and Sabah Public Health Laboratory protocols, using the Bio-Rad CFX96 Touch™ PCR machine (Bio-Rad, USA). QuantiFast™ real-time PCR was carried out over two separate reactions, with the first reaction for Plasmodium genus screening, followed by subsequent P. knowlesi and other human Plasmodium species-specific detection if positive (Nuin et al., 2020).
RDTs conducted on clinical P. knowlesi samples
The nine available antigen-based RDTs were transported and stored with appropriate conditions and temperature monitoring. All RDTs were conducted according to the manufacturers’ instructions at Ranau District Hospital in Sabah. Test results were independently interpreted by two readers, with a positive result documented when both the result and control test strip lines were deemed to be present. In the event of a result discrepancy, a third individual blinded to the initial results performed a tie-break reading. The intensity of each test band on the RDT was graded visually on a scale from 0 to 4 according to a standard color chart provided by FIND, with 0 indicating no line observed and 4 indicating the highest intensity line observed. A threshold score of ≥ 1 was used to indicate a confirmed parasite signal. Due to staggered enrolments and differences in timing of procurement, RDTs were not all obtained simultaneously and tested on exact sample sets. Key clinical and demographic parameters were tested across samples used for individual RDTs to ensure no confounding group differences.
Limit of detection for in vitro P. knowlesi and P. falciparum
Cultures of P. knowlesi (PkA1-H.1) and P. falciparum (Pf3D7) parasites were maintained in vitro as described previously (van Schalkwyk et al., 2019). Parasite dilutions for RDT testing were performed in whole blood obtained from either the UK National Health Service Blood and Transplant (NHSBT) or Cambridge Bioscience. Negative controls (whole blood without Plasmodium parasites) and positive controls (whole blood containing P. falciparum parasites at 2,000 parasites/µL) were supplied by FIND, the global alliance for diagnostics (FIND, 2022). For RDT testing, P. knowlesi or P. falciparum cultured parasites were diluted two-fold in whole blood from 2,000 parasites/µL. All controls and test samples were applied on RDT cassettes according to the manufacturer’s instructions. Experiments were performed in duplicate at the London School of Hygiene & Tropical Medicine, UK, and repeated on four separate occasions for P. knowlesi and between two to four times for P. falciparum. Signals were assessed and classified visually according to the same RDT color chart supplied by FIND for the clinical evaluation. The threshold used to indicate a confirmed parasite signal present was a consistent faint line with a score of ≥ 1. The careUS™ pan-pLDH RDT was not included in the LoD evaluation due to procurement difficulties from the manufacturer at the time of testing.
Statistical analysis
The primary analysis compared the diagnostic accuracy of each RDT to detect microscopy-positive, PCR-confirmed P. knowlesi clinical infections collected prior to antimalarial treatment. Diagnostic accuracy was evaluated using the number of true positive, false negative, false positive and true negative results to calculate sensitivity and specificity (McNeil et al., 1975). Exact binomial confidence intervals (CI) of 95% for each diagnostic metric were calculated and reported. Diagnostic positive and negative predictive values incorporated the estimated prevalence of malaria, calculated from the proportion of febrile patients with a positive microscopy result presenting to the district hospital site during the study period (Safari et al., 2015). Independent comparisons between the sensitivity of RDT test components were conducted using Fisher’s exact test for: a) parasitaemia of above or below 200 parasites/µL, and b) samples collected before versus after administration of antimalarial treatment. For the latter, logistic regression was used to further compare the sensitivity of RDT antigen targets when controlling for median time in hours post treatment. Group differences across RDTs compared log normalized age of participants and parasite count distribution using one-way ANOVA, and sex using Chi-square test. Interobserver agreement between RDT line intensity readers was measured using the kappa coefficient (McHugh, 2012). Spearman’s correlation test was used to assess the association between line intensity gradings and parasitaemia as well as correlation between proportion of early trophozoite (ring) stages to RDT test positivity. The limit of detection (LoD) measurements with 95% confidence intervals were estimated using logistic regression analysis in R (version 4.0.5, the R Foundation for Statistical Computing, 2020). All other statistical analyses were carried out using STATA v16 (TX, USA).
Results
Baseline demographics
From July 2020 to August 2021, 1,698 febrile patients presented to the hospital site and were screened by microscopy, with 508 identified as P. knowlesi infections. Of these, 492 were subsequently confirmed on PCR, resulting in a background malaria prevalence of 28.9% (95% CI 26.8 - 31.1%) in this cohort. A total of 202 microscopically determined P. knowlesi clinical cases and 26 malaria-negative controls who were able to be approached by the research team and gave appropriate consent were prospectively enrolled for RDT testing.
Of the P. knowlesi-positive samples with RDTs tested, 127 (63%) were collected from patients prior to anti-malarial treatment and were included in the primary analysis. The median age of patients with P. knowlesi malaria was 34 years (range 4 – 87 years). PCR-confirmed negative controls had a median age of 34 years (range 12 – 78 years) and included 21 (81%) febrile patients. Most malaria patients and controls were male (79% and 69%, respectively). There were no statistically significant differences in age or sex between malaria cases and negative controls across the RDTs conducted.
The geometric mean parasite count of P. knowlesi infected patients was 734 parasites/µL (95% CI 563 – 956), with a range of 18 – 41,883 parasites/µL. Of 202 P. knowlesi enrolled patients, 55 (27%) had a parasite count of less than 200/µL, which included 26 (47%) with samples collected prior to treatment. Three patients were classified with severe P. knowlesi malaria using WHO research criteria (World Health Organization, 2014). There were no statistically significant differences in the distribution of parasite counts used for individual RDT testing.
Sensitivity of pan-pLDH based RDTs for P. knowlesi detection
The target pan-pLDH antigen made up the major component of seven RDTs tested using clinical isolates (Table 1). The First Response® and CareStart™ PAN RDTs contained the highest performing pan-pLDH tests to detect clinical P. knowlesi infections, with a sensitivity of 87.0% (95% CI 75.1 - 94.6) and lowest detected parasitaemia of 50/µL for both (Table 2). The poorest performing pan-pLDH based RDT was the SD Bioline with a sensitivity of 50.6% (95% CI 39.6 - 61.5), and a parasitemia threshold for positivity of 254 parasites/µL. The median parasitaemia of patients with a false-negative pan-pLDH result was 260 parasites/µL (IQR 125 – 737). False-negative results were reported at high parasite counts for the SD Bioline (8,315 parasites/µL) and careUS™ (6,341 parasites/µL) RDTs.
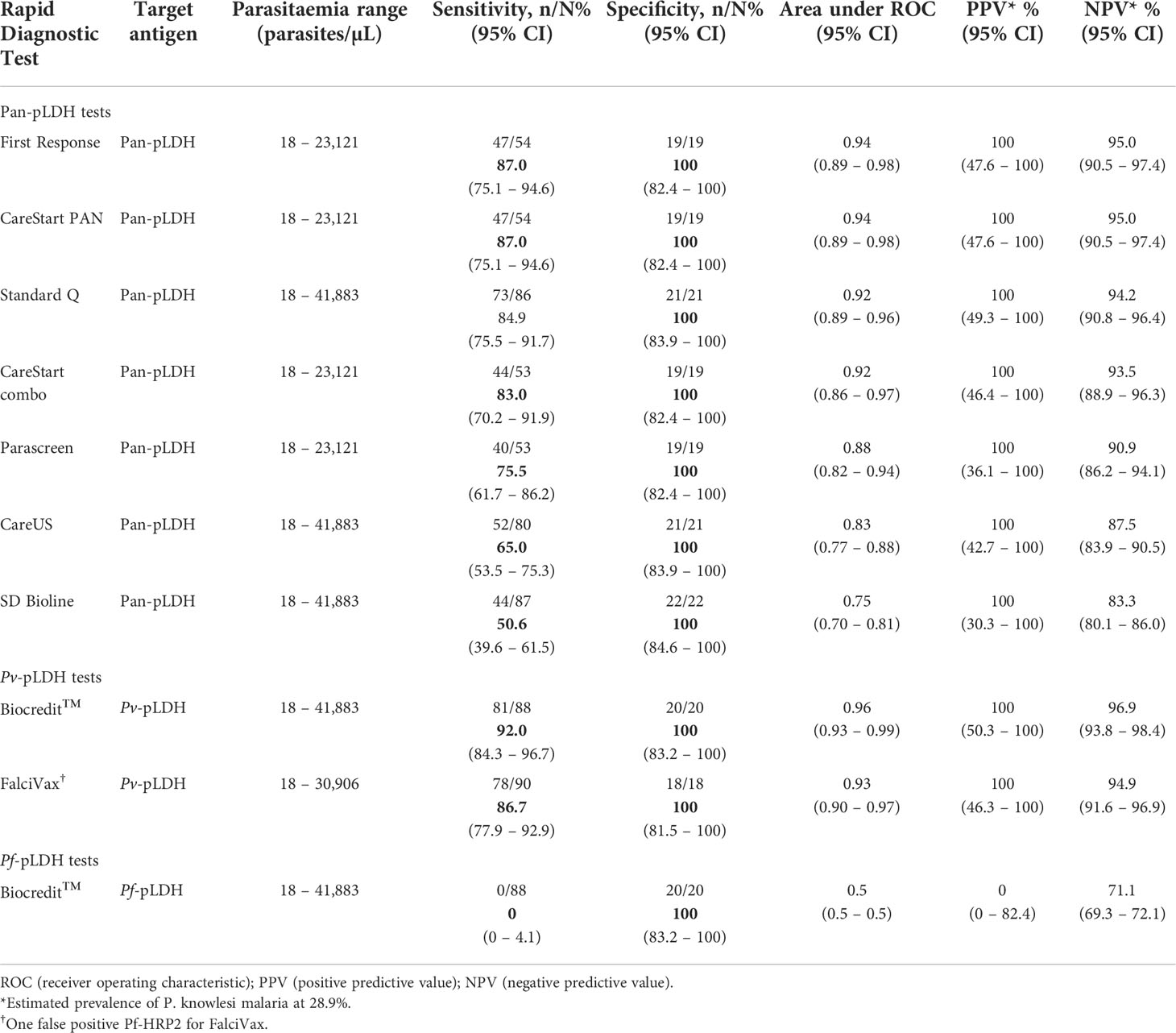
Table 2 Performance of RDTs in detecting P. knowlesi clinical infections (prior to antimalarial treatment).
Cross-reactivity and sensitivity of P. vivax-pLDH to detect P. knowlesi
The Pv-pLDH target antigen of the Biocredit™ RDT demonstrated a sensitivity of 92.0% (95% CI 84.3 - 96.7), the highest for detecting P. knowlesi among all RDTs tested. The Biocredit™ RDT also had the lowest recorded parasite counts of 32 parasites/µL and 40 parasites/µL for clinical samples tested either before or after antimalarial treatment, respectively (Table 3). The Pv-pLDH test component of the FalciVax RDT also cross-reacted with P. knowlesi, albeit with a slightly lower sensitivity of 86.7% (95% CI 77.9 - 92.9) and lowest detected clinical parasitaemia of 50 parasites/µL. The median parasitaemia of patients with a false-negative Pv-pLDH result was 83 parasites/µL (IQR 41 – 203), with the highest false-negative result for Biocredit™ recorded at 251 parasites/µL and for FalciVax at 422 parasites/µL.
Cross-reactivity of P. falciparum-specific antibodies with P. knowlesi
Amongst the RDTs tested, only the Biocredit™ RDT included a Pf-pLDH component for which no cross-reactivity was observed for any of the P. knowlesi clinical samples tested. Of the seven RDTs which contain a Pf-HRP2 component, a single false-positive result was recorded for the FalciVax RDT at a P. knowlesi count of 31 parasites/µL (Table 2). Six other false positive Pf-HRP2 results were observed when the following RDTs were tested on samples collected after antimalarial treatment: Parascreen® (150/µL), First Response (148/µL and 1,924/µL) and FalciVax (49/µL, 162/µL and 608/µL).
Sensitivity of RDT detection at different levels of P. knowlesi parasitaemia
Of P. knowlesi-infected patients whose samples were collected prior to treatment, 26 (20%) had a parasite count less than 200/µL, with a median parasitaemia of 88 parasites/µL (IQR 50 – 133) (Supplementary Table 1). Consistent with the overall data, the best performing test in detecting samples with low parasitaemia was Pv-pLDH from the Biocredit™ RDT with a sensitivity of 73.7% (95% CI 48.8 to 90.9%), followed by the FalciVax Pv-pLDH test at 50%. The pan-pLDH tests performed poorly at these low parasite counts, with the SD Bioline unable to detect a single P. knowlesi-positive sample and both the CareStart™ PAN and First Response® tests demonstrating a low sensitivity of 42.9% (95% CI 9.9 - 81.6%); Figure 1.
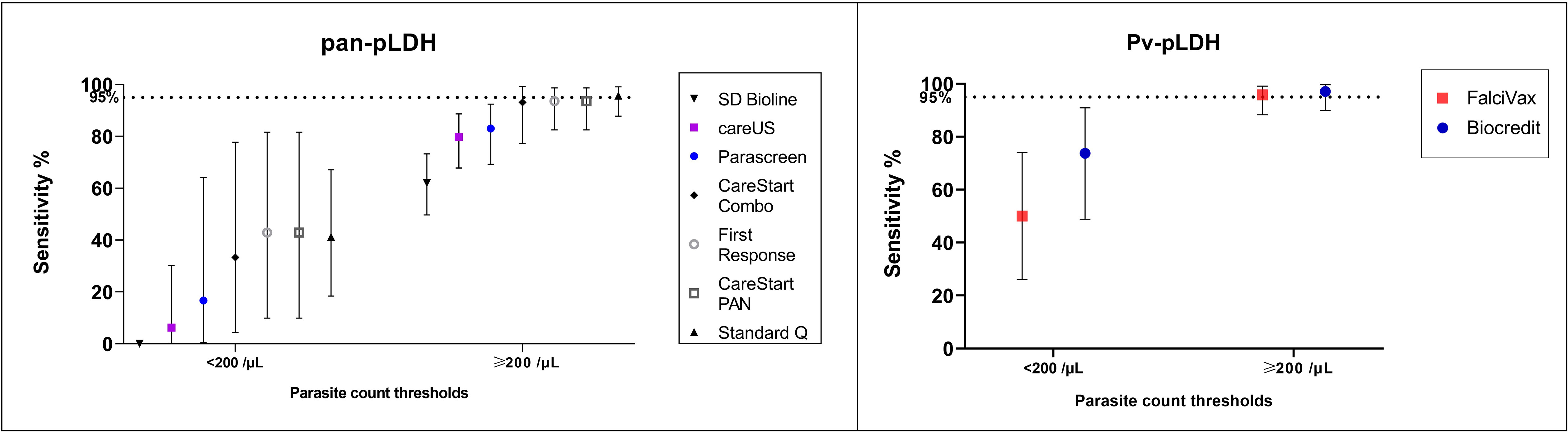
Figure 1 Sensitivity (and 95% confidence intervals) of (left) pan-pLDH and (right) Pv-LDH in detecting P. knowlesi for parasite count groups of <200/µL and ≥200/µL.
The parasite count group of 200 parasites/µL and above comprised of 101 (80%) P. knowlesi-infected participants, with a median parasitaemia of 1,245/µL (IQR 489 – 6,224). Both pan-pLDH and Pv-pLDH targets demonstrated excellent utility for P. knowlesi detection, with three RDTs exceeding the predefined sensitivity threshold of 95% (Diagnostics, 2011) at which performance is considered sufficient for clinical use (Figure 1). These RDTs included the Pv-pLDH component of the Biocredit™ and FalciVax RDTs with sensitivities of 97.1% (95% CI 89.9 - 99.6%) and 95.8% (95% CI 88.3 – 99.1%) respectively, in addition to the pan-pLDH of Standard Q at 95.7% sensitivity (95% CI 87.8 – 99.1%). Comparable performance was seen for the pan-pLDH of the CareStart™ PAN and First Response® RDTs with a sensitivity of 93.6% for both. Performance further improved for the 74 (58.3%) P. knowlesi malaria cases with parasitaemia of more than 500 parasites/µL, with the same five highest performing RDTs all recording 100% sensitivity. The SD Bioline pan-pLDH test also performed poorest in this parasite count group with a sensitivity of 62.0%.
Proportion of ring stages affect test positivity
Microscopic evaluation of parasite life-stages were carried out on the 127 clinical samples with P. knowlesi infections collected prior to antimalarial treatment. The mean proportions of early trophozoite (rings), late trophozoite and schizont asexual life-stages in a single infection were 1.9% (95% CI 0.8 – 2.9), 92.8% (95% CI 90.0 – 95.6) and 5.3% (95% CI 2.7 – 8.0) respectively. Asynchronous life-stages were found in 52% (66/127) of P. knowlesi infections, with synchronous late trophozoites and schizonts present in 46% and 2% respectively. The proportion of ring stages inversely correlate with test positivity for CareStart™ combo, CareStart™ PAN and Biocredit™ RDTs (Spearman’s rho < -0.32; p<0.002); Supplementary Table 2. Logistic regression controlling for parasitaemia demonstrated that the probability of a pan-pLDH or Pv-pLDH positive test outcome decreases by 0.9% (95% CI 0.8 – 1.0) for CareStart™ combo (p=0.044) and 0.8% (95% CI 0.7 – 1.0) for Biocredit™ (p=0.016) with each percentage increase in mean ring stage proportion. In comparison, in vitro cultures were all asynchronous with geometric mean proportions of rings and late trophozoites of 59.1% (95%CI 49.7 - 70.4) and 37.6% (95%CI 28.8 - 49.0) respectively.
Specificity against malaria-negative samples
All pan-pLDH and Pv-pLDH RDT tests exhibited 100% specificity for malaria diagnosis, as demonstrated by the lack of response for all malaria-negative controls in individually tested RDTs. Two false-positives were recorded for Pf-HRP2 antibody, one each by First Response® and Parascreen® respectively (Table 2).
Performance of RDTs to detect sub-microscopic P. knowlesi infections
Five patients excluded from the primary analysis were initially enrolled as microscopy-negative febrile controls; however, subsequent reference PCR detected P. knowlesi. Sample DNA was re-extracted and PCR repeated for confirmation, and research microscopy cross-checked 500 slide fields to confirm the infection was below both routine and research standard microscopic limit of detection. A single submicroscopic P. knowlesi infection was positive on a pan-pLDH test (CareStart™ PAN) and the Pv-pLDH based RDTs (FalciVax and Biocredit™). The other 4 submicroscopic P. knowlesi infections were negative on all RDTs. Inclusion of these submicroscopic P. knowlesi infections in the primary analysis would not have resulted in statistically significant differences in reported RDT sensitivities.
RDT detection for P. knowlesi in samples collected before versus after anti-malarial treatment
RDTs were evaluated independently on 75 P. knowlesi-infected patients after administration of weight-based antimalarial treatment with oral artemether-lumefantrine, including 6.5% of whom initially received at least 1 dose of intravenous artesunate. RDTs were conducted a median of 3.3 hours (IQR 1.7 - 7.1 hours) after antimalarial treatment, with no statistically significant difference in time among the evaluated RDTs. Of the seven RDTs with pan-pLDH as the target antigen, two had significantly higher sensitivities when tested on pre- versus post-treatment samples: SD Bioline (50.6% vs 29.3% respectively; p = 0.01) and Parascreen® (75.5% vs 55.1%; p = 0.03) (Figure 2). Diagnostic sensitivity for all primary target antigens when categorized to parasite count groups of below and above 200/µL was not otherwise affected by whether samples were taken before or after administration of anti-malarial treatment (Supplementary Table 3).
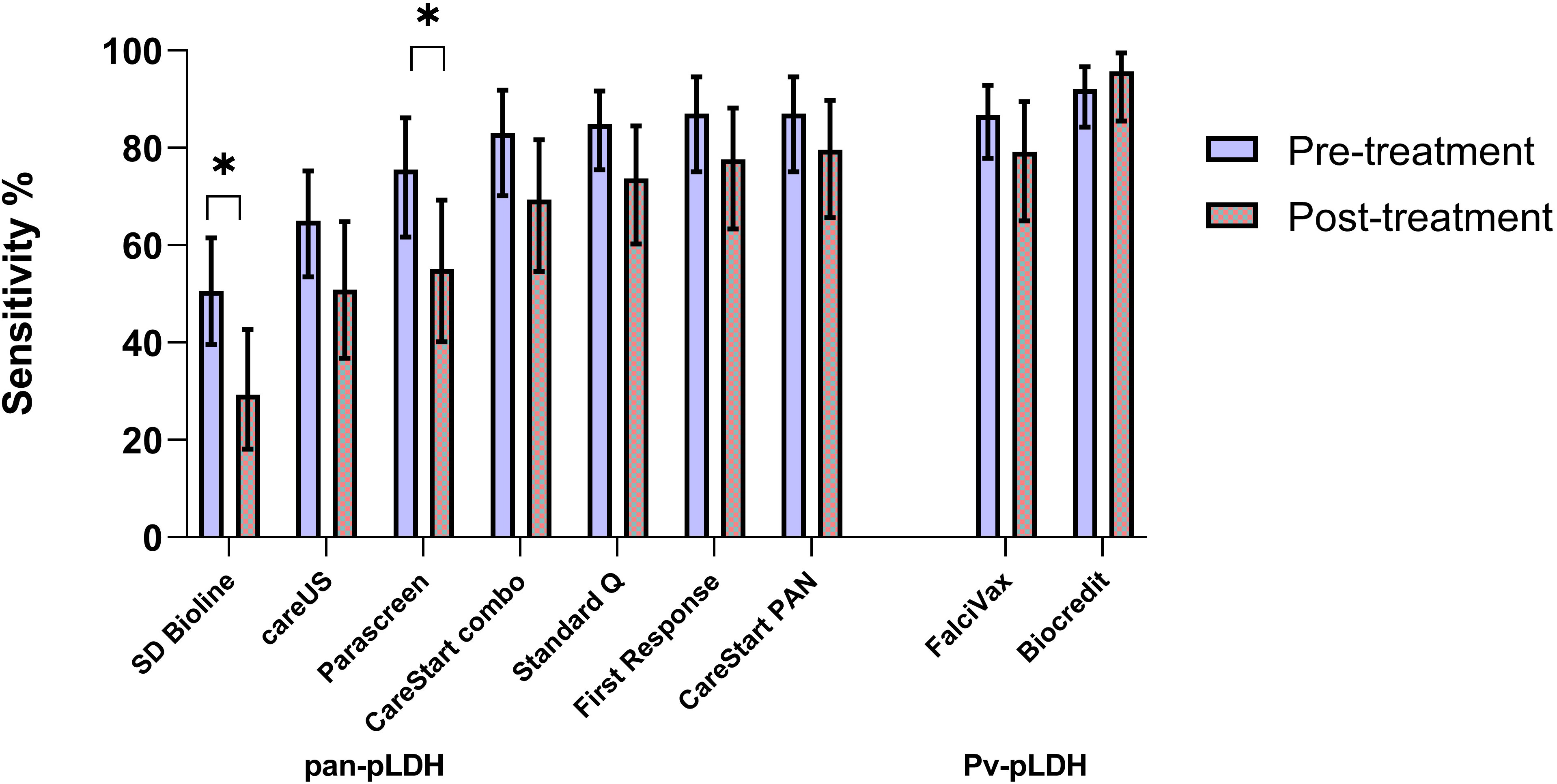
Figure 2 Comparative sensitivity of RDT targets (pre versus post antimalarial treatment) for P. knowlesi including 95% confidence intervals (*) indicate p < 0.05.
RDT line intensity and interobserver agreement
All control channels for all RDTs were graded 4 and there were no invalid results recorded. Line intensity gradings positively correlated with increasing P. knowlesi parasitaemia, including for both pan-pLDH and Pv-pLDH target antigens (Spearman’s rho >0.73; p<0.001) (Figure 3). Reporting of binary results for the primary pan-pLDH and Pv-pLDH antigens for all nine RDTs demonstrated excellent interobserver agreement with kappa values of >0.90. For each individual RDT, overall interobserver agreement was assessed for line intensity grades 1-4, indicative of independently reproducible readings for each colour intensity. Pan-pLDH-based tests recorded kappa values ranging from minimal (κ=0.40; SD Bioline) to moderate (κ=0.77; First Response®) agreement, whereas the Pv-pLDH lines gave weak kappa values of 0.50 (Biocredit™) and 0.57 (FalciVax).
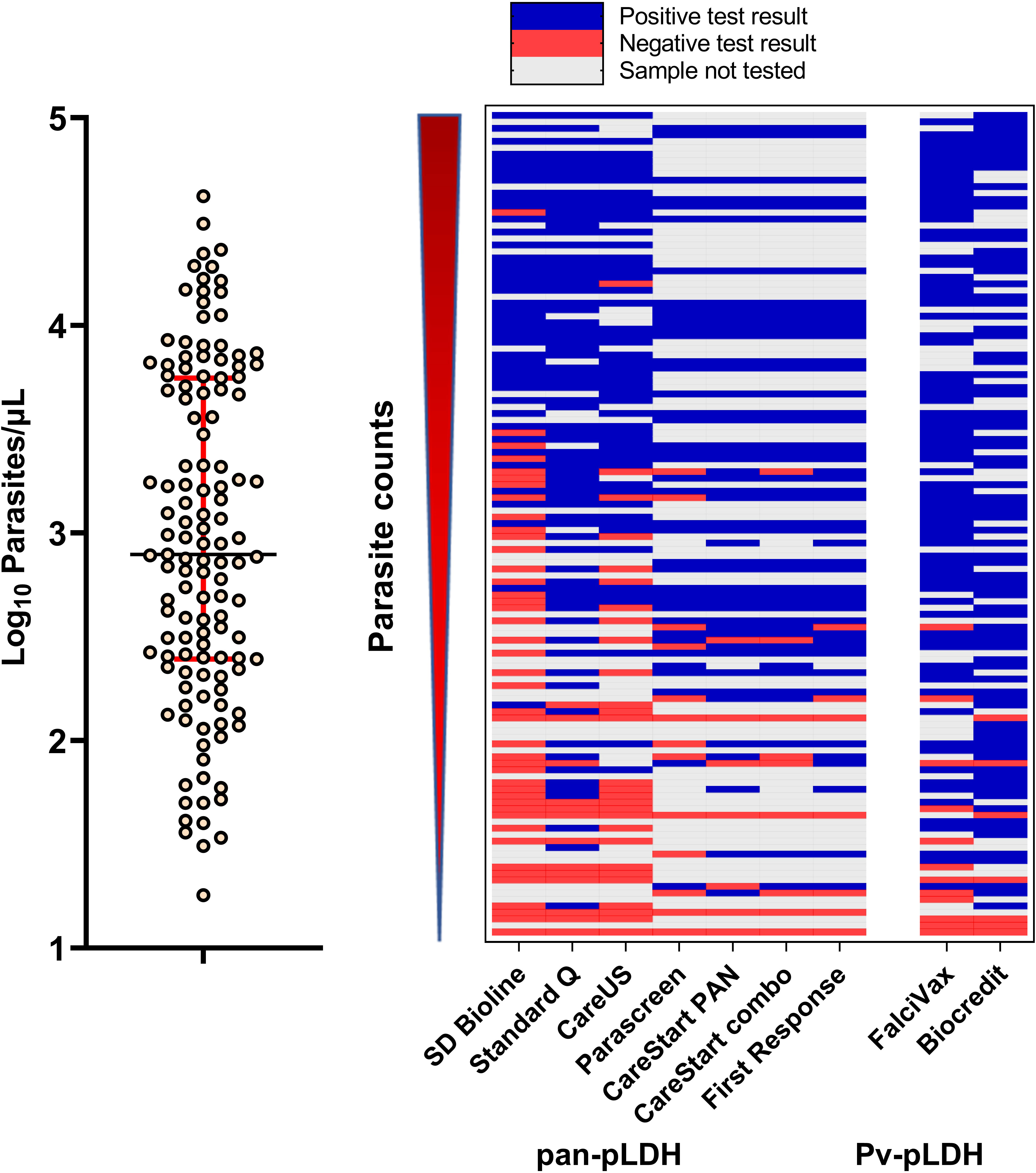
Figure 3 (Left) Distribution of parasite counts of clinical samples tested. (Right) Overview of tested samples with increasing parasite counts and corresponding outcomes in the nine RDTs tested for detection of clinical P. knowlesi.
Detection limits for in vitro P. knowlesi and P. falciparum cultures
P. knowlesi and P. falciparum parasites cultured in vitro were detected by all pan-pLDH channels of the RDTs tested. For the detection of PkA1-H.1 via the pan-pLDH channel, the most sensitive RDT was the CareStart™ PAN with a LoD of 25 parasites/µL, followed by CareStart™ combo and First Response® with a LoD of 98 parasites/µL for both (Table 3). The least sensitive RDTs were the Meriscreen One Step Test (LoD of 1,555 parasites/µL) and SD Bioline, with the latter not able to detect PkA1-H.1 even at the highest parasite concentration tested of 2,000 parasites/µL. Similar to the PkA1-H.1 dilutions, the pan-pLDH channel of CareStart™ PAN recorded the highest sensitivity for the Pf3D7 cultures (LoD of 49 parasites/µL), whereas SD Bioline was the least sensitive (LoD of 1,556 parasites/µL). All RDTs with HRP2 channels were able to detect Pf3D7 with LoD values of >25 parasites/µL.
Culture-derived PkA1-H.1 were also detected by the Pv-pLDH channels of Biocredit™ and FalciVax®. No cross-reactivity occurred in the corresponding P. falciparum-pLDH or HRP2 channels, similar to test results for clinical samples. The LoD of cultured parasites in the Pv-pLDH channels were determined to be 49 parasites/µL for both RDTs, also consistent with the lowest parasitaemia detected from clinical samples (Table 3). In comparison, culture-derived Pf3D7 was detected by P. falciparum channels of both the Biocredit™ (LoD 49 parasites/µL) and FalciVax (LoD 12 parasites/µL) RDTs, with no cross-reactivity occurring in the Pv-pLDH channels.
Discussion
Rapid parasitological confirmation of malaria is a key pillar of effective clinical management recommended by the WHO in all endemic settings (World Health Organization, 2015). Extending this rationale to include zoonotic malaria requires the availability of accurate point-of-care detection methods for P. knowlesi. Given the lack of routine microscopy available in many areas of Southeast Asia, the need to develop and validate RDTs for P. knowlesi diagnosis was identified as a key research priority in 2017 by the P. knowlesi WHO Evidence Review Group (Abeysinghe, 2017). The most sensitive antibody for P. knowlesi detection in this study was the anti-Pv-pLDH component of the Biocredit™ RDT at 92%, much higher than that seen in previously published RDT evaluations encompassing earlier iterations of Pv-pLDH antibodies (Barber et al., 2013a; Foster et al., 2014; Grigg et al., 2014). Among the RDTs with pan-pLDH as the primary antibody target, CareStart™ PAN and First Response® were the most sensitive for detecting P. knowlesi clinical samples at 87%. No RDTs met the overall 95% sensitivity threshold deemed sufficient by the WHO to replace microscopy as the preferred first-line diagnostic tool (World Health Organization, 2000). However, performance was excellent for parasite counts of more than 200/μL, with both Pv-pLDH based RDTs (Biocredit™ and Falcivax®) and one pan-pLDH based RDT (Standard Q) demonstrating sensitivity above 95%, increasing further to 100% for parasite counts more than 500/μL. These RDTs may have reasonable utility in areas where microscopy is not routinely available, although in endemic regions such as Sabah, Malaysia, symptomatic P. knowlesi infections commonly occur at low parasite densities, with previous large studies reporting around 12% of patients having a parasite count less than 200/μL (Grigg et al., 2018a).
The performance of WHO pre-qualified/Round 8 pre-validated RDTs used for detection of P. knowlesi in this study were markedly improved from previously evaluated RDTs selected from earlier WHO testing rounds, using similar pLDH targets with commercially undisclosed epitopes. Previous studies aiming to detect low-level P. knowlesi clinical monoinfections in Sabah using pan-pLDH (OptiMAL-IT) and VOM (non-P. falciparum)-pLDH (CareStart™) RDTs demonstrated poor sensitivities of less than 42% (Grigg et al., 2014). Other pan-pLDH (First Response®, [Cat. No : II6FRC30]) and pan-Plasmodium aldolase (ParaHIT) RDTs also demonstrated insufficient sensitivities of 74% and 23%, respectively (Barber et al., 2013a). A separate study in Sarawak, Malaysia, reported comparably low sensitivity values: 71% for pan-pLDH (OptiMAL-IT), 40% for pan-pLDH (Paramax-3) and 29% for aldolase (BinaxNOW®) (Foster et al., 2014). Test results with pLDH-based assays more broadly, including for P. falciparum detection, have been shown to have variable performance (World Health Organization, 2000; Park et al., 2020) and are influenced by several factors including binding affinity of the target protein, parasite density in blood samples and the prozone effect (Wongsrichanalai et al., 2007; Gillet et al., 2011; Bosco et al., 2020). This reflects differing characteristics of the antibodies used by each manufacturer in these proprietary test platforms.
This study confirms the cross-reactivity of P. knowlesi LDH antigens with Pv-pLDH antibodies used in two commercially available RDTs tested (Biocredit™ and FalciVax®). P. knowlesi LDH has previously been shown to have high sequence identity (>96%) with the P. vivax ortholog (Singh et al., 2012), however the separate LDH isoforms exhibit both shared and unique surface residues associated with differential epitope binding of targeted antibodies (McCutchan et al., 2008). With both RDTs demonstrating high test positivity for P. knowlesi despite not being developed or previously validated for this purpose, specificity for P. vivax detection in co-endemic areas is likely to be significantly lower than manufacturers report. In areas where both species are endemic, misidentification could potentially lead to inappropriate clinical management, such as chloroquine for unsuspected severe knowlesi malaria (Rajahram et al., 2012), or unnecessary liver-stage treatment with primaquine for presumed P. vivax (Barber et al., 2021). However, in areas where P. vivax has been successfully eliminated, the cross-reactivity seen in Pv-LDH channels of both these RDTs could be used to accurately detect P. knowlesi infections and initiate appropriate antimalarial treatment. RDTs may also be potentially used to confirm P. knowlesi infections in patients diagnosed with less reliable microscopy and given antimalarial treatment at peripheral health clinics prior to early referral hospital transfer. Additionally, RDTs may have a future role in hospital inpatient treatment monitoring to confirm parasite clearance and allow hospital discharge, for which current Malaysian Ministry of Health guidelines stipulate two consecutive negative microscopy results (Ministry of Health Malaysia, 2014).
Exploiting differences in the degree of cross-reactivity with pan-pLDH and P. vivax or P. falciparum-pLDH has also been hypothesized as a potential strategy to distinguish separate Plasmodium species. A recent study assessed quantified Pv-LDH cross-reactivity against both recombinant pLDH proteins and laboratory cultured isolates from P. knowlesi, in addition to the related simian species P. cynomolgi (Barney et al., 2021). P. knowlesi-pLDH recombinant protein cross-reacted with both the pan-pLDH and Pv-pLDH antibodies; however, the P. cynomolgi-pLDH protein only reacted with the latter. Genetically, P. cynomolgi is also closely related to P. vivax and likely shares high sequence identity with orthologous Pv-pLDH targets. In the current study, P. knowlesi parasites from both clinical and laboratory-cultured samples were also detected by all pan-pLDH and Pv-pLDH components tested, meaning they are likely unable to differentiate P. vivax. The quantified ratio of pan- to Pv-pLDH responses could provide additional information if this approach was used during the development of the RDTs (Kho et al., 2022), however this requires further investigation. With the exception of CareStart™ PAN, in vitro LoD values from the pan-pLDH based RDTs were generally higher than the lowest detected parasitaemia of the clinical samples. The presumed larger pLDH-to-parasite ratio in clinical whole blood samples could be occurring due to either higher proportions of late parasite stages with greater pLDH production compared to cultured samples or hypothesized elevated pLDH secreted into the peripheral circulation by hidden extravascular parasites, as seen in the spleen in P. vivax (Kho et al., 2021a; Kho et al., 2021b). LDH expression levels may also differ between parasite species and needs to be considered as a potential source of variability in inter-species sensitivity, in addition to primary sequence variation.
The single Pf-pLDH based test (Biocredit™) evaluated in this study did not demonstrate any cross-reactivity with P. knowlesi, despite a Pf -pLDH containing RDT in a previous study demonstrating low sensitivities of up to 29% (Grigg et al., 2014). This is consistent with previous modelling of pLDH isoforms which also demonstrated both shared and unique residues between P. falciparum and P. knowlesi associated with specific epitope antibody binding or lack thereof (McCutchan et al., 2008). For public health surveillance purposes including measuring progress towards elimination of P. falciparum, RDTs used in co-endemic areas would ideally need to differentiate between P. falciparum and P. knowlesi, given similarities in the microscopic morphology of their early ring-stages (Lee et al., 2009; Barber et al., 2013b). Correct identification of P. falciparum also allows appropriate transmission-blocking administration of single-dose primaquine, not required for P. knowlesi due to the gametocidal effect of current blood-stage treatments in this species (Grigg et al., 2016; Grigg et al., 2018b). In this context, the Pf-HRP2 antigen remains the preferred test component to differentiate non-falciparum malaria in areas such as Malaysia where hrp2/3 gene deletions have not been reported (Grigg et al., 2014). When compared to previous studies of diagnostic accuracy for P. knowlesi which also reported false-positive results for Pf-HRP2 channels (Foster et al., 2014; Grigg et al., 2014), the proportion seen for P. falciparum was lower in the RDTs evaluated in this study. Additionally, no bands were observed for any RDT in the Pf-HRP2 channel with the cultured PkA1-H.1, further confirming the high specificity of this test for P. falciparum parasites.
A novel aspect of this study is the systematic evaluation of RDT detection for submicroscopic P. knowlesi infections. A single positive result on the same sample for the three highest performing RDTs demonstrated their potential ability to detect at least a small proportion (20%) of these extremely low-level infections. The small number of patients with submicroscopic infections included in this subgroup analysis limits meaningful interpretation, although the true sensitivity is likely to remain low. These findings are consistent with the lowest reported limit of detection of 25 parasites/µL demonstrated with the CareStart™ PAN RDT, marginally below the microscopic detection threshold of approximately 40 parasites/µL (World Health Organization, 2015). Lower median parasite counts for symptomatic P. knowlesi infections compared to P. falciparum and P. vivax are well-established, indicating a lower fever threshold (Grigg et al., 2018a). Submicroscopic P. knowlesi infections have been widely reported across Southeast Asia (Anstey et al., 2021), including in Malaysia (Fornace et al., 2016; Siner et al., 2017; Grignard et al., 2019; Noordin et al., 2020), however the majority of these were asymptomatic. Further study is required to understand the role of submicroscopic and asymptomatic infections in P. knowlesi transmission, including what proportion resolve spontaneously and whether they may facilitate onwards transmission to human or macaque hosts.
Limitations of the study included a lack of P. vivax and P. falciparum clinical infections in the primary analysis due to near elimination of these Plasmodium species in Malaysia, which would have enabled evaluation of diagnostic specificity across malaria species. Ideally the same P. knowlesi infections evaluated in the primary analysis would be conducted on all RDTs, however due to staggered timing of RDT procurement and manufacturer expiry dates this was not possible. Potential performance bias related to differences in P. knowlesi infections is minimized by similar parasite count distributions between RDTs tested, and the large number of clinical P. knowlesi infections included overall. Other methodological strengths include the careful selection of the top-performing pan- and Pv-pLDH based RDTs from WHO prequalification testing, including those known to be used commonly as the first-line point-of-care diagnostic tool in other P. knowlesi endemic countries such as Indonesia. These key antibody targets based on hypothesised cross-reactivity with P. knowlesi were evaluated with robust quality assurance practices and inter-reader variability to improve reliability of findings.
Current efforts to develop novel malaria RDTs or modify those currently available to improve specificity for P. knowlesi and other clinically relevant simian malaria species face considerable obstacles (Yerlikaya et al., 2018) as manufacturers of existing commercial malaria RDTs do not often disclose which pLDH epitopes are bound by their proprietary antibodies (Grigg et al., 2021). Current malaria RDT development is appropriately focused on developing highly-sensitive tests in order to facilitate elimination of P. falciparum and P. vivax, due to the increasingly recognized role that low-level asymptomatic infections have on sustaining transmission (Stresman et al., 2020). However, simian malaria species such as P. knowlesi and P. cynomolgi are not routinely included as part of the RDT development or validation process. The inclusion of reliable P. knowlesi-specific antigen targets in RDTs with satisfactory sensitivity would play an important role in malaria case management, allow accurate assessment of progress towards WHO elimination goals for P. falciparum and P. vivax, and provide a vital epidemiological tool for understanding regional zoonotic malaria transmission and the true burden of disease, especially in resource-constrained areas.
Conclusion
The findings of this study support utilising RDTs for initial rapid detection, but not identification, of knowlesi malaria in areas of higher endemism where access to microscopy is limited or unavailable. Improvement in the performance of pLDH-based RDTs for P. knowlesi detection was apparent with high sensitivities recorded particularly for parasite counts above 200/µL. When used as a single parasitological diagnostic test, malaria RDTs should incorporate a Pf-HRP2 component for non-falciparum species differentiation. The development of newer generation malaria RDTs to be deployed in P. knowlesi endemic areas, including through the WHO RDT product testing process, should include validation against P. knowlesi and other zoonotic Plasmodium species capable of human infection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Medical Research and Ethics Committee of the Ministry of Health, Malaysia and Menzies School of Health Research, Australia. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
MG, DS, SY, NA, CS, SK, BB, SD, GR and TW conceptualised, designed the study, critically revised the manuscript and were involved in general oversight of study. Patient enrolments were conducted by AT and SS, laboratory work were carried out by AT, SS, DS and MI, and AT, SY, DS and MG were involved in data analysis and manuscript preparation and revisions. All authors approved the submitted version of the manuscript.
Funding
This work was supported by the Australian Centre of Research Excellence in Malaria Elimination, the National Health and Medical Research Council, Australia (Grant Numbers 1037304 and 1045156, fellowships to NMA [1042072], MJG [1138860]), the National Institutes of Health, USA (R01AI160457-01 and R01AI160457-02) and Malaysian Ministry of Health (grant number BP00500/117/1002) awarded to GSR. AFT is supported via a Malaysia Australia Colombo Plan Commemoration (MACC) and Australian Government Research Training Program (RTP) Scholarship at Charles Darwin University. MJG is supported by the Australian Centre for International Agricultural Research, Australian Government (LS-2019-116). The analytical performance analysis part of this work was conducted with funds from the Bill & Melinda Gates Foundation (grant no. OPP1172683).
Acknowledgments
We thank the study participants, the IDSKKS malaria research team (Maizatul Farina binti Abd Mutalib and Noorazela binti Mohamed Yassin), Ranau District Hospital director and clinical/laboratory staff, Dr. Fiona Binzini of Ranau Public Health District (PKD Ranau), Dr. Dyang Ruksuna Md Ruhul, Dr. Wan Mohamad Haziman Bin Wan Taib and Dr. Mohd Fakhrurizal Bin Matdiris as well as the Sabah State Public Health Laboratory. We would like to thank the Director General of Health Malaysia for the permission to publish this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MN declared a past co-authorship with the author TW to the handling editor.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1023219/full#supplementary-material
References
Abba, K., Kirkham, A. J., Olliaro, P. L., Deeks, J. J., Donegan, S., Garner, P., et al. (2014). Rapid diagnostic tests for diagnosing uncomplicated non-falciparum or Plasmodium vivax malaria in endemic countries. Cochrane Database Systematic Rev. 12), 1465–1858.
Abeysinghe, R. (2017). Outcomes from the evidence review group on Plasmodium knowlesi. Present. Malar. Policy Advis. Commun. Meet 1, 22–24. Available at: http://iris.wpro.who.int/handle/10665.1/13663
Anstey, N. M., Grigg, M. J., Rajahram, G. S., Cooper, D. J., William, T., Kho, S., et al. (2021). “Chapter one - knowlesi malaria: Human risk factors, clinical spectrum, and pathophysiology,” in Advances in parasitology. Ed. Drakeley, C. (Cambridge, Massachusetts, USA: Academic Press), 1–43.
Ba, E. H., Baird, J. K., Barnwell, J., Bell, D., Carter, J., Dhorda, M., et al. (2015). Microscopy for the detection, identification and quantification of malaria parasites on stained thick and thin blood films in research settings (version 1.0): procedure: methods manual. 1, 21–23. Available at: https://apps.who.int/iris/handle/10665/163782
Barber, B. E., Grigg, M. J., Cooper, D. J., van Schalkwyk, D. A., William, T., Rajahram, G. S., et al. (2021). “Chapter two - clinical management of plasmodium knowlesi malaria,” in Advances in parasitology. Ed. Drakeley, C. (Cambridge, Massachusetts, USA: Academic Press), 45–76.
Barber, B. E., William, T., Grigg, M. J., Piera, K., Yeo, T. W., Anstey, N. M. (2013a). Evaluation of the sensitivity of a pLDH-based and an aldolase-based rapid diagnostic test for diagnosis of uncomplicated and severe malaria caused by PCR-confirmed Plasmodium knowlesi, Plasmodium falciparum, and Plasmodium vivax. J. Clin. Microbiol. 51 (4), 1118–1123. doi: 10.1128/JCM.03285-12
Barber, B. E., William, T., Grigg, M. J., Yeo, T. W., Anstey, N. M. (2013b). Limitations of microscopy to differentiate Plasmodium species in a region co-endemic for Plasmodium falciparum, Plasmodium vivax and Plasmodium knowlesi. Malar J. 12, 8. doi: 10.1186/1475-2875-12-8
Barney, B., Velasco, M., Cooper, C., Rashid, A., Kyle, D., Moon, R., et al. (2021). Diagnostic characteristics of lactate dehydrogenase on a multiplex assay for malaria detection including the zoonotic parasite Plasmodium knowlesi. Am. J. Trop. Med. Hygiene 106, 275–282. doi: 10.4269/ajtmh.21-0532
Betty, R., Niniek Lely, P., Rukmini, R., Widodo, J. P. (2015). Analysis of implementation the policy on malaria elimination in Indonesia. Buletin Penelitian Sistem Kesehatan 18 (3), 277–284. doi: 10.22435/hsr.v18i3.4549.277-284
Bosco, A. B., Nankabirwa, J. I., Yeka, A., Nsobya, S., Gresty, K., Anderson, K., et al. (2020). Limitations of rapid diagnostic tests in malaria surveys in areas with varied transmission intensity in Uganda 2017-2019: Implications for selection and use of HRP2 RDTs. PloS One 15 (12), e0244457. doi: 10.1371/journal.pone.0244457
Chin, A. Z., Maluda, M. C. M., Jelip, J., Jeffree, M. S. B., Culleton, R., Ahmed, K. (2020). Malaria elimination in Malaysia and the rising threat of Plasmodium knowlesi. J. Physiol. Anthropol 39 (1), 36–45. doi: 10.1186/s40101-020-00247-5
Cooper, D. J., Rajahram, G. S., William, T., Jelip, J., Mohammad, R., Benedict, J., et al. (2020). Plasmodium knowlesi malaria in sabah, malaysi–2017: Ongoing increase in incidence despite near-elimination of the human-only Plasmodium species. Clin. Infect. Dis. 70 (3), 361–367. doi: 10.1093/cid/ciz237
Coutrier, F. N., Tirta, Y. K., Cotter, C., Zarlinda, I., Gonzalez, I. J., Schwartz, A., et al. (2018). Laboratory challenges of Plasmodium species identification in aceh province, Indonesia, a malaria elimination setting with newly knowlesi. PloS Negl. Trop. Dis. 12 (11), e0006924. doi: 10.1371/journal.pntd.0006924
Cunningham, J., Jones, S., Gatton, M. L., Barnwell, J. W., Cheng, Q., Chiodini, P. L., et al. (2019). A review of the WHO malaria rapid diagnostic test product testing programm –2018): performance, procurement and policy. Malaria J. 18 (1), 1–15. doi: 10.1186/s12936-019-3028-z
Diagnostics (2011). A research agenda for malaria eradication: Diagnoses and diagnostics. PloS Med. 8 (1), e1000396. doi: 10.1371/journal.pmed.1000396
FIND (2022) Malaria specimen bank (Geneva, Switzerland). Available at: https://www.finddx.org/biobank-services/specimen-bank/specimens-mal/ (Accessed 09-09-2022).
Fornace, K. M., Nuin, N. A., Betson, M., Grigg, M. J., William, T., Anstey, N. M., et al. (2016). Asymptomatic and submicroscopic carriage of Plasmodium knowlesi malaria in household and community members of clinical cases in sabah, Malaysia. J. Infect. Dis. 213 (5), 784–787. doi: 10.1093/infdis/jiv475
Foster, D., Cox-Singh, J., Mohamad, D. S., Krishna, S., Chin, P. P., Singh, B. (2014). Evaluation of three rapid diagnostic tests for the detection of human infections with Plasmodium knowlesi. Malar J. 13, 60. doi: 10.1186/1475-2875-13-60
Gillet, P., Scheirlinck, A., Stokx, J., De Weggheleire, A., Chaúque, H. S., Canhanga, O. D., et al. (2011). Prozone in malaria rapid diagnostics tests: how many cases are missed? Malaria J. 10 (1), 166. doi: 10.1186/1475-2875-10-166
Grigg, M. J., Lubis, I. N., Tetteh, K. K. A., Barber, B. E., William, T., Rajahram, G. S., et al. (2021). “Chapter three - plasmodium knowlesi detection methods for human infections–diagnosis and surveillance,” in Advances in parasitology. Ed. Drakeley, C. (Cambridge, Massachusetts, USA: Academic Press), 77–130.
Grigg, M. J., William, T., Barber, B. E., Parameswaran, U., Bird, E., Piera, K., et al. (2014). Combining parasite lactate dehydrogenase-based and histidine-rich protein 2-based rapid tests to improve specificity for diagnosis of malaria due to Plasmodium knowlesi and other Plasmodium species in sabah, Malaysia. J. Clin. Microbiol. 52 (6), 2053–2060. doi: 10.1128/JCM.00181-14
Grigg, M. J., William, T., Barber, B. E., Rajahram, G. S., Menon, J., Schimann, E., et al. (2018a). Age-related clinical spectrum of Plasmodium knowlesi malaria and predictors of severity. Clin. Infect. Dis. 67 (3), 350–359. doi: 10.1093/cid/ciy065
Grigg, M. J., William, T., Barber, B. E., Rajahram, G. S., Menon, J., Schimann, E., et al. (2018b). Artemether-lumefantrine versus chloroquine for the treatment of uncomplicated Plasmodium knowlesi malaria: An open-label randomized controlled trial CAN KNOW. Clin. Infect. Dis. 66 (2), 229–236. doi: 10.1093/cid/cix779
Grigg, M. J., William, T., Menon, J., Dhanaraj, P., Barber, B. E., Wilkes, C. S., et al. (2016). Artesunate-mefloquine versus chloroquine for treatment of uncomplicated Plasmodium knowlesi malaria in Malaysia (ACT KNOW): an open-label, randomised controlled trial. Lancet Infect. Dis. 16 (2), 180–188. doi: 10.1016/S1473-3099(15)00415-6
Grignard, L., Shah, S., Chua, T. H., William, T., Drakeley, C. J., Fornace, K. M. (2019). Natural human infections with Plasmodium cynomolgi and other malaria species in an elimination setting in sabah, Malaysia. J. Infect. Dis. 220 (12), 1946–1949. doi: 10.1093/infdis/jiz397
Hussin, N., Lim, Y. A., Goh, P. P., William, T., Jelip, J., Mudin, R. N. (2020). Updates on malaria incidence and profile in Malaysia from 2013 to 2017. Malar J. 19 (1), 55. doi: 10.1186/s12936-020-3135-x
Jiang, N., Chang, Q., Sun, X., Lu, H., Yin, J., Zhang, Z., et al. (2010). Co-Infections with Plasmodium knowlesi and other malaria parasites, Myanmar. Emerging Infect. Dis. 16 (9), 1476–1478. doi: 10.3201/eid1609.100339
Jimenez, A., Rees-Channer, R. R., Perera, R., Gamboa, D., Chiodini, P. L., González, I. J., et al. (2017). Analytical sensitivity of current best-in-class malaria rapid diagnostic tests. Malaria J. 16 (1), 1–9. doi: 10.1186/s12936-017-1780-5
Khim, N., Siv, S., Kim, S., Mueller, T., Fleischmann, E., Singh, B., et al. (2011). Plasmodium knowlesiInfection in humans, cambodi –2010. Emerging Infect. Dis. 17 (10), 1900–1902. doi: 10.3201/eid1710.110355
Kho, S., Anstey, N. M., Barber, B. E., Piera, K. A., William, T., Kenangalem, E., et al. (2022). Diagnostic performance of a 5-plex immunoassay against species-specific and pan Plasmodium antigens in regions co-endemic for p. falciparum, p. vivax, p. knowlesi, p. malariae and p. ovale. Sci. Rep. 12, 7286–7295. doi: 10.1038/s41598-022-11042-w
Kho, S., Qotrunnada, L., Leonardo, L., Andries, B., Wardani, P. A. I., Fricot, A., et al. (2021a). Evaluation of splenic accumulation and colocalization of immature reticulocytes and Plasmodium vivax in asymptomatic malaria: A prospective human splenectomy study. PloS Med. 18 (5), e1003632. doi: 10.1371/journal.pmed.1003632
Kho, S., Qotrunnada, L., Leonardo, L., Andries, B., Wardani, P. A. I., Fricot, A., et al. (2021b). Hidden biomass of intact malaria parasites in the human spleen. New Engl. J. Med. 384 (21), 2067–2069. doi: 10.1056/nejmc2023884
Kojom Foko, L. P., Pande, V., Singh, V. (2021). Field performances of rapid diagnostic tests detecting human Plasmodium species: A systematic review and meta-analysis in India 1990-2020. Diagnostics (Basel Switzerland) 11 (4), 590. doi: 10.3390/diagnostics11040590
Lee, K. S., Cox-Singh, J., Singh, B. (2009). Morphological features and differential counts of Plasmodium knowlesi parasites in naturally acquired human infections. Malar J. 8, 73. doi: 10.1186/1475-2875-8-73
McCutchan, T. F., Piper, R. C., Makler, M. T. (2008). Use of malaria rapid diagnostic test to IdentifyPlasmodium knowlesiInfection. Emerging Infect. Dis. 14 (11), 1750–1752. doi: 10.3201/eid1411.080840
McHugh, M. L. (2012). Interrater reliability: the kappa statistic. Biochemia Med. 22 (3), 276–282. doi: 10.11613/BM.2012.031
McNeil, B. J., Keeler, E., Adelstein, S. J. (1975). Primer on certain elements of medical decision making. New Engl. J. Med. 293 (5), 211–215. doi: 10.1056/NEJM197507312930501
Ministry of Health Malaysia. (2014). “Manangement guideline of malaria in malaysia”. 1st ed. (Putrajaya: Vector Borne Disease Sector, Disease Control Division, Ministry of Health, Malaysia).
Moody, A. (2002). Rapid diagnostic tests for malaria parasites. Clin. Microbiol. Rev. 15 (1), 66–78. doi: 10.1128/cmr.15.1.66-78.2002
Noordin, N. R., Lee, P. Y., Mohd Bukhari, F. D., Fong, M. Y., Abdul Hamid, M. H., Jelip, J., et al. (2020). Prevalence of asymptomatic and/or low-density malaria infection among high-risk groups in peninsular Malaysia. Am. J. Trop. Med. Hygiene 103 (3), 1107–1110. doi: 10.4269/ajtmh.20-0268
Nuin, N. A., Tan, A. F., Lew, Y. L., Piera, K. A., William, T., Rajahram, G. S., et al. (2020). Comparative evaluation of two commercial real-time PCR kits (QuantiFast and abTES) for the detection of Plasmodium knowlesi and other Plasmodium species in sabah, Malaysia. Malar J. 19 (1), 306. doi: 10.1186/s12936-020-03379-2
Park, S. H., Jegal, S., Ahn, S. K., Jung, H., Lee, J., Na, B. K., et al. (2020). Diagnostic performance of three rapid diagnostic test kits for malaria parasite Plasmodium falciparum. Korean J. Parasitol. 58 (2), 147–152. doi: 10.3347/kjp.2020.58.2.147
Putaporntip, C., Hongsrimuang, T., Seethamchai, S., Kobasa, T., Limkittikul, K., Cui, L., et al. (2009). Differential prevalence of Plasmodium infections and cryptic Plasmodium knowlesi malaria in humans in Thailand. J. Infect. Dis. 199 (8), 1143–1150. doi: 10.1086/597414
Rajahram, G. S., Barber, B. E., William, T., Menon, J., Anstey, N. M., Yeo, T. W. (2012). Deaths due to Plasmodium knowlesi malaria in sabah, Malaysia: association with reporting as Plasmodium malariae and delayed parenteral artesunate. Malar J. 11, 284. doi: 10.1186/1475-2875-11-284
Rajahram, G. S., Cooper, D. J., William, T., Grigg, M. J., Anstey, N. M., Barber, B. E. (2019). Deaths from Plasmodium knowlesi malaria: Case series and systematic review. Clin. Infect. Dis. 69 (10), 1703–1711. doi: 10.1093/cid/ciz011
Safari, S., Baratloo, A., Elfil, M., Negida, A. (2015). Evidence based emergency medicine part 2: Positive and negative predictive values of diagnostic tests. Emergency (Tehran Iran) 3 (3), 87–88. doi: 10.22037/aaem.v3i3.274
Siner, A., Liew, S.-T., Kadir, K. A., Mohamad, D. S. A., Thomas, F. K., Zulkarnaen, M., et al. (2017). Absence of plasmodium inui and plasmodium cynomolgi, but detection of plasmodium knowlesi and plasmodium vivax infections in asymptomatic humans in the betong division of Sarawak, Malaysian Borneo. Malaria J. 16 (1), 417–428. doi: 10.1186/s12936-017-2064-9
Singh, V., Kaushal, D. C., Rathaur, S., Kumar, N., Kaushal, N. A. (2012). Cloning, overexpression, purification and characterization of Plasmodium knowlesi lactate dehydrogenase. Protein Expression Purification 84 (2), 195–203. doi: 10.1016/j.pep.2012.05.008
Stresman, G., Sepúlveda, N., Fornace, K., Grignard, L., Mwesigwa, J., Achan, J., et al. (2020). Association between the proportion of Plasmodium falciparum and Plasmodium vivax infections detected by passive surveillance and the magnitude of the asymptomatic reservoir in the community: a pooled analysis of paired health facility and community data. Lancet Infect. Dis. 20 (8), 953–963. doi: 10.1016/s1473-3099(20)30059-1
Tyagi, R. K., Das, M. K., Singh, S. S., Sharma, Y. D. (2013). Discordance in drug resistance-associated mutation patterns in marker genes of Plasmodium falciparum and Plasmodium knowlesi during coinfections. J. Antimicrob Chemother 68 (5), 1081–1088. doi: 10.1093/jac/dks508
van Schalkwyk, D. A., Blasco, B., Nuñez, R. D., Liew, J. W., Amir, A., Lau, Y. L., et al. (2019). Plasmodium knowlesi exhibits distinct in vitro drug susceptibility profiles from those of Plasmodium falciparum. Int. J. Parasitol: Drugs Drug Resistance 9, 93–99. doi: 10.1016/j.ijpddr.2019.02.004
Wongsrichanalai, C., Barcus, M. J., Muth, S., Sutamihardja, A., Wernsdorfer, W. H. (2007). A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT). Am. J. Trop. Med. Hyg 77 (6 Suppl), 119–127. doi: 10.4269/ajtmh.2007.77.119
World Health Organization (2000). New perspectives: malaria diagnosis, report of a joint WHO/USAID: informal consultation held on 25–27 October 1999 Vol. 1999 (Geneva, Switzerland: World Health Organization), 4–48.
World Health Organization (2011). Malaria rapid diagnostic test performance: results of WHO product testing of malaria RDTs: round 3 (Geneva, Switzerland: World Health Organization), Vol. 2010-2011).
World Health Organization (2014). Severe malaria. Trop. Med. Int. Health 19, 7–131. doi: 10.1111/tmi.12313_2
World Health Organization (2015). Global technical strategy for malaria 2016-2030 (Geneva, Switzerland: World Health Organization).
World Health Organization (2018). Malaria rapid diagnostic test performance: summary results of WHO product testing of malaria RDTs: round 1-8 (2008–2018) (Geneva: World Health Organization).
World Health Organization (2019). World malaria report Vol. 2019 (Geneva: World Health Organization).
World Health Organization (2020). “World malaria report 2020: 20 years of global progress and challenges,” in World malaria report 2020: 20 years of global progress and challenges). (Geneva, Switzerland: World Health Organization)
World Health Organization (2021). World malaria report 2021. (Geneva, Switzerland: World Health Organization)
World Health Organization (2022a) In vitro diagnostics under assessment. Available at: https://extranet.who.int/pqweb/vitro-diagnostics/products-under-assessment (Accessed 21 March 2022).
World Health Organization (2022b) Prequalified in vitro diagnostics (Geneva, Switzerland). Available at: https://extranet.who.int/pqweb/vitro-diagnostics/vitro-diagnostics-lists (Accessed 14 Apr 2022).
World Health Organization, Research, U.N.U.W.B.W.S.P.f., Training in Tropical, D. (2015). Microscopy for the detection, identification and quantification of malaria parasites on stained thick and thin blood films in research settings (version 1.0): procedure: methods manual (Geneva: World Health Organization).
Keywords: malaria, Plasmodium knowlesi, rapid diagnostic test, diagnostic performance, point-of-care, Malaysia, parasite lactate dehydrogenase
Citation: Tan AF, Sakam SSb, Rajahram G S, William T, Abd Rachman Isnadi M F, Daim S, Barber B E, Kho S, Sutherland CJ, Anstey NM, Yerlikaya S, van Schalkwyk DA and Grigg MJ (2022) Diagnostic accuracy and limit of detection of ten malaria parasite lactate dehydrogenase-based rapid tests for Plasmodium knowlesi and P. falciparum. Front. Cell. Infect. Microbiol. 12:1023219. doi: 10.3389/fcimb.2022.1023219
Received: 19 August 2022; Accepted: 26 September 2022;
Published: 17 October 2022.
Edited by:
Kamal El Bissati, The University of Chicago, United StatesReviewed by:
Myat Htut Nyunt, Department of Medical Research, Myanmar;Kwame Kumi Asare, University of Cape Coast, GhanaCopyright © 2022 Tan, Sakam, Rajahram, William, Abd Rachman Isnadi, Daim, Barber, Kho, Sutherland, Anstey, Yerlikaya, van Schalkwyk and Grigg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angelica F. Tan, YW5nZWxpY2FmaW9uYS50YW5AbWVuemllcy5lZHUuYXU=; Matthew J. Grigg, bWF0dGhldy5ncmlnZ0BtZW56aWVzLmVkdS5hdQ==
†These authors contributed equally to this work
 Angelica F. Tan
Angelica F. Tan Sitti Saimah binti Sakam2
Sitti Saimah binti Sakam2 Mohammad Faruq Abd Rachman Isnadi
Mohammad Faruq Abd Rachman Isnadi Colin J. Sutherland
Colin J. Sutherland Nicholas M. Anstey
Nicholas M. Anstey Donelly A. van Schalkwyk
Donelly A. van Schalkwyk Matthew J. Grigg
Matthew J. Grigg