
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 27 October 2022
Sec. Bacteria and Host
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.1009901
This article is part of the Research Topic Roles of Non-coding RNAs in Infectious Diseases - Vol II View all 6 articles
 Kusuma Sai Davuluri
Kusuma Sai Davuluri Devendra S. Chauhan*
Devendra S. Chauhan*Despite more than a decade of active study, tuberculosis (TB) remains a serious health concern across the world, and it is still the biggest cause of mortality in the human population. Pathogenic bacteria recognize host-induced responses and adapt to those hostile circumstances. This high level of adaptability necessitates a strong regulation of bacterial metabolic characteristics. Furthermore, the immune reponse of the host virulence factors such as host invasion, colonization, and survival must be properly coordinated by the pathogen. This can only be accomplished by close synchronization of gene expression. Understanding the molecular characteristics of mycobacterial pathogenesis in order to discover therapies that prevent or resolve illness relies on the bacterial capacity to adjust its metabolism and replication in response to various environmental cues as necessary. An extensive literature details the transcriptional alterations of host in response to in vitro environmental stressors, macrophage infection, and human illness. Various studies have recently revealed the finding of several microRNAs (miRNAs) that are believed to play an important role in the regulatory networks responsible for adaptability and virulence in Mycobacterium tuberculosis. We highlighted the growing data on the existence and quantity of several forms of miRNAs in the pathogenesis of M. tuberculosis, considered their possible relevance to disease etiology, and discussed how the miRNA-based signaling pathways regulate bacterial virulence factors.
Non-coding RNAs are single-stranded transcripts that regulate the mRNA (coding gene) expression by degrading them. microRNAs (miRNAs) are small molecules of non-coding RNA that contain 17–25 nucleotides and modulate gene expression (Esteller, 2011). From the past few decades, there is great progress in miRNA research, which is believed to be important in regulating various pathological processes (Aguilar et al., 2019). Over 20 years ago, the first miRNA discovery made a mark in a molecular biology new era. Over 2,000 miRNAs have been identified in humans, and it is thought that they collectively regulate one-third of the genes in the genome. miRNAs have been linked to a variety of human diseases and are being researched for use as clinical diagnostic and therapeutic targets through biogenesis, involving the multitude of mechanisms that inactive miRNA converts into mature miRNA (Taneja and Dutta, 2019). Disruption in the maturation process of miRNA, for example, miR-146a irregular expression and impaired miRNA regulatory mechanisms, leads to neoplasia, ischemic heart disease, neurodegenerative diseases, etc. Yang et al., 2019 (Chen et al., 2019). miRNA formation process occurs in the nucleus by RNA polymerase II. Initially, the primary transcript with hair pin structure that encodes miRNA sequences is regulated by RNA polymerase II transcription factors, epigenetic and histone modifiers. Primary miRNA goes through maturation processes by cropping the loop end of pri-miRNA (pre-miRNA) in the nucleus. Later, the resulting product is exported to the cytoplasm by exportin-5 for further maturation steps. RNase, Dicer crops the loop end one more time resulting in the small RNA duplex (Bogerd et al., 2014). Only six nucleotides that match are required to obtain functional miRNA (Bartel, 2009). Recent research studies reveal the genesis and role of miRNA in regulating several bacterial pathogenesis-associated signaling pathways (Zhang et al., 2017b; Stutz et al., 2018). miRNAs can be reliable in therapeutic settings. These factors became important in screening the diseases with high specificity, sensitivity, and accessibility (Walker and Harland., 2009 Ostrik et al., 2021). Northern blotting, microarray analysis, and quantitative polymerase chain reaction (qPCR) are traditional methods for miRNA detection. To improve the sensitivity and selectivity of miRNA detection, new technology methods always rely on signal amplification strategies, such as nanoparticle-based amplification, isothermal exponential amplification, rolling circle amplification, hybridization chain reaction, and combinations of these (Ye et al., 2019). Our literature review reveals the role of miRNAs as modulators of signaling pathways in tuberculosis (TB). miRNAs act as genetic switches that make them regulators of cellular signaling pathways. We can predict the targets of miRNA easily nowadays through the discovery of high-throughput genomic screening methods. Understanding the role of miRNA in signaling pathways might lead to novel therapies. New kinase inhibitors are being studied to treat many diseases by detailed understanding of the role of miRNA in regulating the kinase cascade pathways. We will indeed be able to create new therapeutical platforms, such as locked genomic technology, for synthesizing and providing efficient RNA-based chemotherapeutic agents. miRNA expression patterns differ in active TB, latent tuberculosis infection (LTBI), and healthy individuals (Sabir et al., 2018). miRNA synthesis mainly influences the action of various immune cells (Chandan et al., 2020). We summarize some of them and discuss their benefits and drawbacks for improving miRNA detection design. Research studies found significant variations in miRNA patterns that help in identifying LTBI and long-term TB infection. Compared to LTBI, active tuberculosis shows upregulation of miR-194-5p, miR-21, miR-29c-3, miR-150-5p, miR-365a-3p, miR-223-3p, miR-451a-5p, miR-44-5p, and miR-144-3p (Wang et al., 2011). Innate immune response: miR-146a (IRAK)-1/ (TRAF)-6], miR-9 (NF-κB1), miR-125b (ERK)1 (Zhou et al., 2010), miR-26-5p (KLF4), miR-132-3p [(TLR)]; Regulation of inflammation: miR-21-5p (TLR4), miR-146a-5a (TRAF-6), miR-20b-5p (NLRP3), miR-223-3p (NFIA), miR-27b-3p (Bag2), miR-99b-5p [(TNF)-α and TNF receptor superfamily (TNFRSF)-4], miR-125-5p (TNF-α), miR-142-3pN (Wasp), miR-144(IFN-γ and TNF-α), miR-27a (IRAK-4); Autophagy: miR-155 (Rheb), miR-27a (Cacna2d3), miR-889 (TWEAK), miR-106a (ULK1, ATG7, ATG16L1) (Yang J et al., 2019), miR-125 (DRAM2), miR-142-3p (ATG16L1) (Yang Y et al., 2019), miR-17 (ATG7), miR-144-3p (ATG4a), miR-20a (ATG7/ATG16L1) Cui et al., 2022a, miR-23a-5p (TLR2/MyD88/NF-κB), miR-26a (KLF4); Apoptosis: miR-27a, miR-96 (FOXO3) (Guttilla and White, 2009), miR-20a-5p (JNK)2, miR-27b (Bag2), miR-21 [PI3K/Akt NF-κB], Let-7e (Caspase-3), miR-29a (Caspase-7) (Pattnaik et al., 2022). In this review, we focused on the six main signaling pathways involved in the major pathogenic mechanisms such as autophagy, inflammation, and apoptosis. miRNAs that show strong research evidence of regulation according to the target scan and miRbase software were discussed. Finding out the function of various miRNAs in the regulation of various pathogenic signaling pathways may lead to identifying new therapeutic targets. Inactive mRNA undergoes splicing/processing to convert into mature mRNA. Mature mRNA then transported from nucleus to cytosol where it is translated as shown in Figure 1.
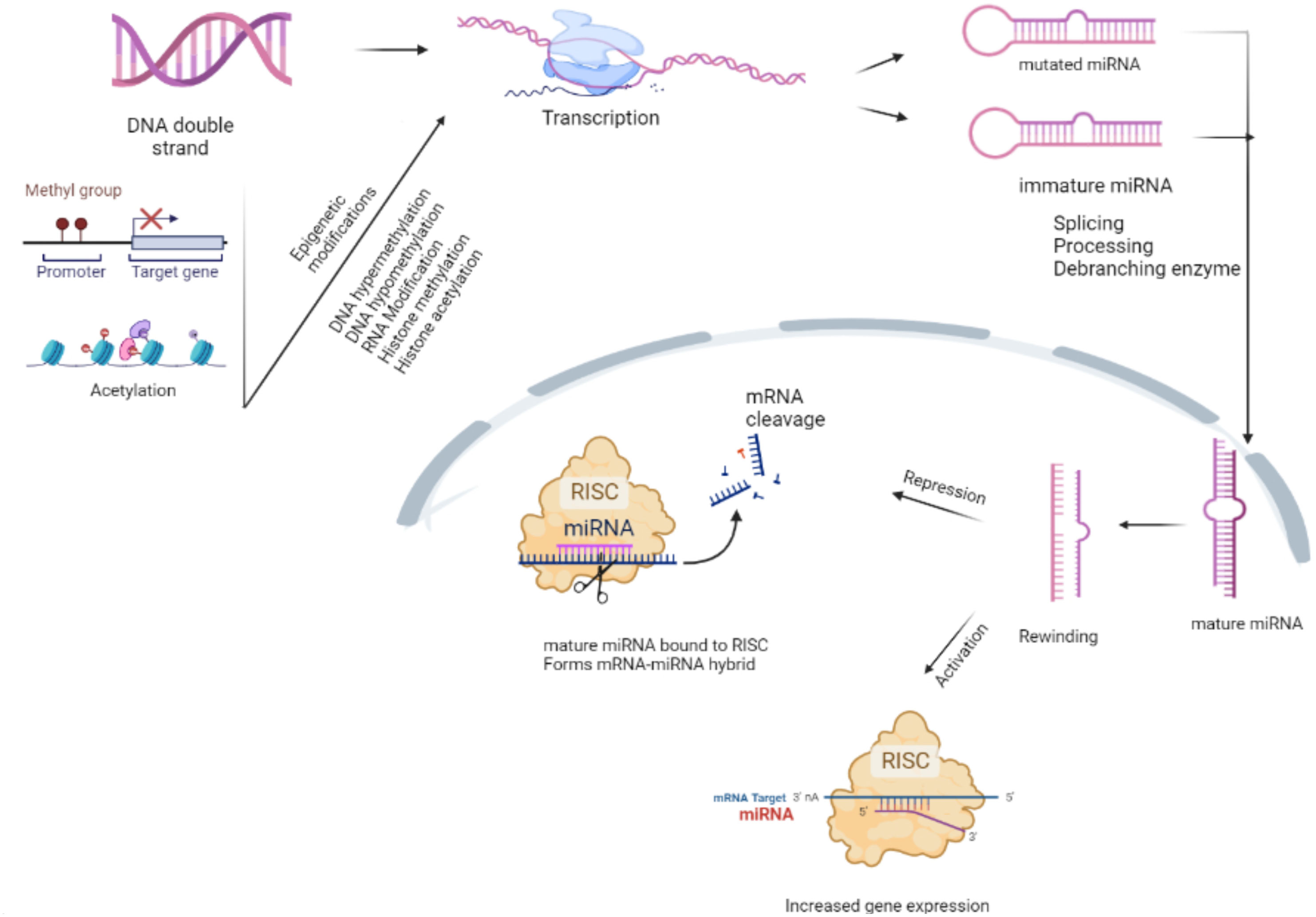
Figure 1 Maturation of microRNA (miRNA). RNA polymerase II or III modifies the primary miRNA into the cap structure through polyadenylation. The Drosha complex crops the miRNA into a hairpin-shaped pre-miRNA during the initial processing of pri-miRNA in the nucleus. Immature miRNA is exported to the cytoplasm by the exportin-5/Ran-GTP complex for Dicer processing. One of the miRNA duplex strands forms miRNA-RISC, which engages on the target mRNA to mediate gene silencing via translational repression or mRNA degradation/deadenylation (detailed in Figure 1). The epigenetic modifications during the transcription process render the miRNA unable to carry out its normal functions. Messenger ribonucleic acid (mRNA); micro ribonucleic acid (miRNA); deoxyribo nucleic acid (DNA); RNA-induced silencing complex (RISAC).
miRNA expression patterns in patients with active TB were shown to be distinct from those of individuals with LTBI or healthy controls (Fu et al., 2011; Harapan et al., 2013; De Araujo et al., 2019). miRNA synthesis may influence the activation of natural killer cells, macrophages, dendritic cells, and T cells (Xu et al., 2019). To avoid the immune system, Mycobacterium tuberculosis may either enhance or inhibit miRNA expression. TNF-α and interferon (IFN)-γ are the host cytokines associated with autophagy during bacterial infection. Myeloid cells triggered by TLR signaling have been demonstrated to be negatively affected by higher levels of miRNA-146a-5p, miR-21-5p, miR-155, miR-199b, and miR132-5p (Kim et al., 2017). The overexpression of miR-27a-5p and miR-33 in M. tuberculosis-infected cells inhibits the creation of autophagosomes and the killing of M. tuberculosis by macrophages (Liu et al., 2018). M. tuberculosis-infected macrophages overexpress miRNAs that target IFN-γ and TNF-α, which suppress the immunological response against M. tuberculosis (Chakrabarty et al., 2019). As an additional line of defense against intracellular infections, host miRNAs such as miR-325-3p and miR-20b-5p impact cell death and inflammasome activation (Lou et al., 2017; Fu et al., 2020). Host innate and adaptive immune systems, such as miR-155-5p and let-7f, both have a role in the activation of miRNAs during M. tuberculosis infection, which is necessary for the clearance of pathogens (Rothchild et al., 2016). In cohorts that comprised persons with LTBI, active TB, and healthy controls, researchers have studied miRNA expression profiles in serum/plasma or blood cells (Lyu et al., 2019). Compared to LTBI, the expression of five miRNAs was higher in TB patient PBMCs (miR-365a-3p, miR-223-3p, miR-451a-5p, miR-44-5p, and miR-144-3p), with target predictions pointing to a possible role in TB patients’ hematopoiesis. According to another research, active TB patients had an improved expression of miR-194-5p and other miRNAs, including miR-21, miR-29c-3, and miR-150-5p. Upregulation of miR-29a-3p was shown to be a helpful prospective biomarker for qRT-PCR-based differentiation among active TB and LTBI (Zhang et al., 2014; Kanniappan et al., 2017). Individuals infected with HIV and those who were not exhibited similar levels of miRNAs.
It was observed that miR-1246, miR-2110, miR370-3p, miR-28-3p, and miR-193b-5p were overexpressed in active TB, whereas miR-3675-5p was downregulated (Duffy et al., 2018). There was no validity testing done on the patients in the second cohort. Pathological Biomarkers for Tuberculosis Progression and Therapy Response researchers want to find predictive miRNA signatures for LTBI-to-TB progression and anti-TB medication response. According on published data rather than a screening in the lab, these miRNAs were selected. TB patients who received successful TB treatment were shown to have lower levels of other miRNAs than those who did not react to treatment (Lyu et al., 2019). While the concept and therapeutic regimen were the same, the screening method was different in a Chinese study. Compared to miR-148b-3p, miR-92a-3p, and miR-21-5p, miR-125a-5p was elevated in this case (Zhao et al., 2013; Duffy et al., 2018). Due to discrepancies in results, standardization of screening techniques is needed to provide more accurate results. In patients with active TB, LTBI, and isoniazide-treated LTBI, researchers found three miRNAs (let-7a-5p, a small nucleolar RNA miR-196b-5p, and SNORD104) as highly sensitive classifiers to distinguish TB from non-TB group members using insignificant RNA sequencing (RNA-seq) of whole blood (Barry et al., 2018).
Regardless of the prevalence of HIV-1 coinfection, small RNA levels in plasma dropped dramatically before and after therapy. Although miR-29a-3p, SNORD61, miR-17-3p, and miR-133a levels were reduced among persons who reacted to medicine compared to those who did not, no single miRNA or combination of small RNAs was shown to be a significant predictor of successful TB therapy (Wang et al., 2018). To investigate whether there was a similar profile of differential miRNA expression across trials from patients with active TB and healthy controls, samples from patients with active TB were compared to those of healthy controls. Since the past decade or so, researchers have used this method to find miRNA markers in serum/plasma and blood cells. miRNAs described employing broad-spectrum unbiased procedures such as small RNA-seq will likely provide new accurate results than researchers who just concentrate on a few possible miRNAs. Because only a few miRNAs are available in the signature revealed by two or more studies, there is a lack of consistency in the outcomes of such screenings. An array of patient demographics and different types of RNA-seq and microarrays may be at fault. It is thought that these miRNAs play an essential role in TB pathogenesis by decreasing the host’s innate and acquired immune response to intracellular infections, both directly and indirectly. Anti-inflammatory miRNAs, such as miR-21-5p and miR-146a-5p, may also be used to discriminate among active TB and LTBI or an otherwise wholesome condition (Spinelli et al., 2013). As a putative biomarker of active TB, the M. tuberculosis inducing miR-155-5p, which is overexpressed in sufferers, plays a vital part in host defense (Etna et al., 2018). Role of different miRNAs in the pathogenesis of tuberculosis is tabulated in Table 1.
AMP-activated protein kinase (AMPK) is a crucial metabolic sensor that responds to alternations in the cellular AMP/ATP ratio following the activation of catabolic energy production. The AMPK pathway is also activated in response to various bacterial infections and inflammation. Various bacterial antigens activate AMPK signaling cascades associated with host response modulation that can either increase or decrease pathogen survival (Grahame Hardie, 2016; Prantner et al., 2017). Previous research has revealed a wide range of AMPK pathway functions, including the regulation of host signaling and participation in significant events. mTOR kinase phosphorylation was more activated in macrophages than AMPK in a time-dependent manner following M. tuberculosis infection (Yang et al., 2014). There is evidence that cytosolic M. tuberculosis colocalizes with p62 and LC3, which are autophagic machinery components. Watson et al. 2012 found it in only 30% of total M. tuberculosis phagosomes, providing evidence that the majority of intracellular M. tuberculosis could inhibit xenophagy activation because of TFEB nuclear translocation downregulation. Activation of mTOR during. M. tuberculosis infection raises the levels of miRNA-33 and miRNA-33a (Oneyama et al., 2011). miR-124 reduces cell proliferation through G1 phase cell cycle arrest (Gong et al., 2016; He et al., 2020). Furthermore, overexpression of miR-124 overexpression reduces both cell growth and glucose consumption in cells (Zhao et al., 2017), which is consistent with AMPK downregulation. Increased levels of miR-101a and miR-199a decrease AMPK signaling (Liu et al., 2016b) (Li et al., 2020), and miR-101a can both directly and indirectly inactivate AMPK (Li et al., 2018; Liu et al., 2018). miRNAs that were downregulated during hypoxia are also involved in AMPK signaling, which plays an important role in reducing oxidative stress, autophagy, and apoptosis during hypoxia (Li et al., 2016; Tran et al., 2017; da Cruz et al., 2018; Sun et al., 2018; Zhu et al., 2018; Zhao T. et al., 2020). Starvation, genotoxic stress, hypoxia, ER stress, and reactive oxygen species (ROS) all activate signaling pathways that either initiate or regulate autophagy cascades. AMPK-mTORC1 regulates autophagy by integrating multiple stimuli and pathways into a signal for the ULK complex, which is the starting point for autophagy. Several miRNAs have been identified as regulators of AMPK-mTORC1. miRNAs acts as both positive and negative regulators of the gene expression. Upregulated miRNAs mainly target the signaling pathways associated with pathogenesis during the infection. The miRNAs targeting the various pathways are shown in the Figure 2.
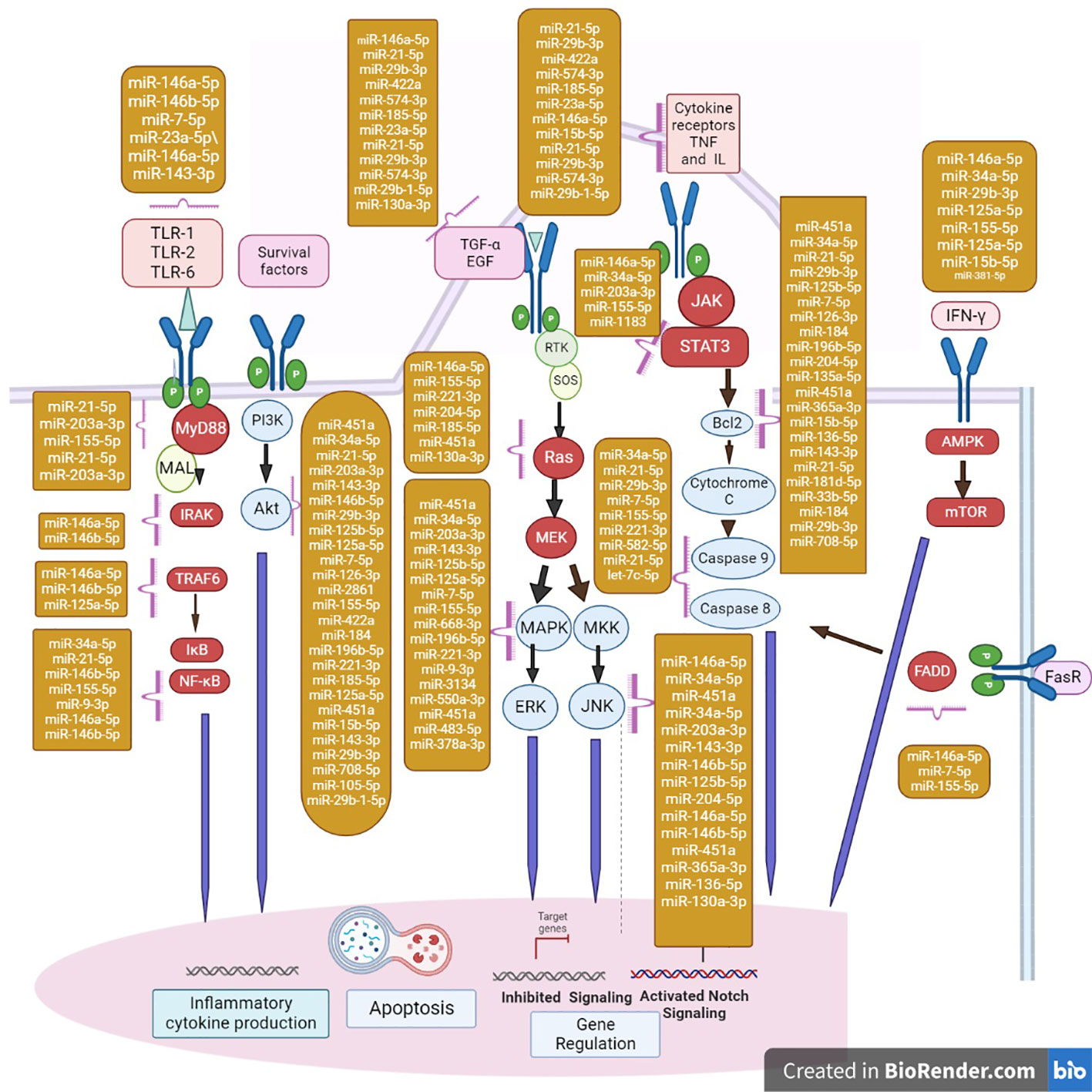
Figure 2 Major miRNAs that regulate apoptosis effectors are shown in the diagram in the yellow box. FasR, Fas Receptor; FADD, Fas-associated death domain protein. miRNAs regulate the major cascades of autophagy. The action of miRNAs involved in the regulation of key members of autophagy cascades; repression/activation of mRNA are shown in the nucleus. mTORC1 induces and regulates the autophagy by miRNAs. AMPK-mTORC1 lies at the heart of regulation of autophagy by integrating numerous stimuli and pathways into a signal for the starting point of autophagy. In addition, ER stress and ROS regulate autophagy independently of the AMPK-mTORC1 pathway. In TNF-α-induced necroptosis, the engagement of TNFR1 recruits Complex I (composed of TRADD, TRAF2). This complex promotes the NF-κB activation and promotes cell survival and inhibits apoptosis. When growth factor receptors are activated, the class I PI3K complex and a small GTPase, Ras, are activated, which activate the PI3K-PKD1-AKT and Ras-Raf-1-MEK1/2-ERK1/2 pathways, respectively. Both AKT and ERK1/2 phosphorylate and inhibit tuberous sclerosis complex, thereby stabilizing Ras homolog, which activates mTORC1, resulting in autophagy inhibition. JNK1-mediated Bcl-2 phosphorylation reduces the binding activity of Bcl-2 and Bcl-xL to initiate autophagy, which promotes cell survival. TLR signaling and miRNAs form a complex network. TLR recruits adaptor proteins and activates downstream signaling cascades that activate the NF-κB signaling pathway and the MAPK signaling pathway in response to specific microbial recognition. This activation causes inflammatory mediators and miRNA genes to be expressed.
The host component against pathogenic organisms is NF-κB, which is a regulator of cell pro-inflammatory responses (Zhang Q et al., 2017a). NF-κB has been linked to the emergence of chronic inflammation and bacterial infections. Recent studies have reported that several miRNAs have been associated with the inflammatory responses by regulating the M. tuberculosis replication and induced pathogenesis by targeting the TRAF-6 signaling pathways (Cui et al., 2018). The detailed mechanisms have to be illustrated further. TRAF-6 belongs to the TNF receptor protein family, acts as an important regulator in many cellular pathways and regulates signal transduction of the TNF receptor superfamily (Chen et al., 2020). TRAF-6 also acts as a link among IRAK-1/IRAK and NF-κB/IB kinase signaling pathways in response to pro-inflammatory cytokines by binding with TGF-β-activated kinase-1 (TAK1) and supporting IκB kinase phosphorylation, ubiquitination, and deterioration after the stimulation of many innate immunity-associated genes (Li H. et al., 2018). During the M. tuberculosis infection, upregulation of miRNA-125a in macrophages depends on TLR4 signaling by targeting TRAF-6 and modulating NF-κB (Cui J et al., 2022a; 2022b). This mechanism leads to the attenuation of the immune response and enhances the survival of mycobacteria. The NF-κB pathway is associated with bacteria–host interactions (Westermann, 2018). By identifying the PI3K-AKT-mTOR signaling pathway (PTEN), miR-26b tends to promote the LPS-induced NF-κB signaling pathway and enhances the expression of pro-inflammatory factors (Huang et al., 2012). Studies showed that genetic disruption of the p50 subunit of NF-κB restricts the M. tuberculosis infection. Pharmacologic regulation of NF-κB activation decreases the viability of intracellular mycobacteria (Liu et al., 2016a). NF-κB inhibition increases the apoptosis of macrophages and autophagy, which is the established defense mechanism (Bai et al., 2013). Inactivation of NF-κB downregulates the expression of PTEN that regulates cellular activities that may be crucial for pathogen resistance. PTEN signaling regulates infection by affecting various intracellular mycobacterial pathogens (Fang et al., 2016). PTEN deficiency renders susceptibility to infection in multiple cells infected with mycoplasma and Mycobacterium. PTEN’s lipid phosphatase activity is critical for infection tolerance. Mycobacterium infectious disease activates Akt phosphorylation, and suppression of Akt or PI3K activity regulates cellular infection (Huang et al., 2012). M. tuberculosis-infected macrophages secrete cytokines, showing an effective defense mechanism against the pathogen. NF-κB and mitogen-activated protein kinase (MAPK) signaling pathways regulate the expression of various cytokines (Gañán-Gómez et al., 2014). Cytokines such as TNF-α, IL-6, and IL-1β are potent mediators showing immune response against the M. tuberculosis bacilli (Cui et al., 2021). Targeting the cytokines and their regulatory pathways restricts the host immune response. For example, M. tuberculosis virulence protein PtpA arrests the NF-κB and JNK signaling pathways (Rothchild et al., 2016). Treatment of cells with early secreted antigenic target-6 (ESAT-6) prevents TLR-associated NF-κB activation (Yang et al., 2015). TAK is a serine/threonine protein kinase associated with the activation of NF-κB pathway. Studies show that miRNAs regulate TAK expression to promote chemoresistance. Upregulation of miR-143 attenuates the function of TAK. miR-146a and miR-26b also target the TAK to promote apoptosis and are associated with the NF-κB pathway inhibition. miR-143 and miR-146a inhibit the NF-κB signaling pathway (Chen Y et al., 2016; Chen et al., 2019).
During the M. tuberculosis infection, TLR2-deficient animals were more susceptible than control mice, but TLR2- and TLR4-deficient mice were as vulnerable as control mice in a low-dose M. tuberculosis challenge (Ju et al., 2018). Pattern recognition receptors (PRRs) expressed on leukocytes activate phagocytosis and host defense mechanisms through the promotion of signaling cascades. TLRs and mannose receptors associated with PRRs play a critical role in immune response and detect the pathogen-derived molecules. Most of the mycobacterial antigens act as agonists for TLRs (Liu Y. et al., 2016). Inoculation of BCG was believed to be dependent on TLR2 and TLR4. Most of the mycobacterial proteins and lipids are associated with the TLR-dependent signaling cascades. TLR regulates hundreds of the host genes that are associated with signaling and acts against microbial antigens, so studying about TLR molecular mechanisms has great importance (Shariq et al., 2021). TLR signaling was negatively affected by higher levels of miRNA-146a-5p, miR-21-5p, miR-99b-5p, and miR132-5p 9 (Wu et al., 2012). The overexpression of miR-27a-5p and miR-33 in M. tuberculosis-infected cells inhibits the creation of autophagosomes and the killing of M. tuberculosis by macrophages (Marcinowski et al., 2012). As an additional line of defense against intracellular infections, host miRNAs such as miR-325-3p and miR-20b-5p impact cell death and inflammasome activation (Kumar et al., 2015). miRNAs associated with the host innate and adaptive immune systems, such as miR-155-5p and let-7f, have a role in the activation of signaling pathways during M. tuberculosis infection, which is necessary for the clearance of pathogens (Iwai et al., 2015; Li et al., 2016). Although miR-29a-3p, SNORD61, miR-17-3p, and miR-133a levels were reduced among people who reacted to medicine compared to those who did not, no single miRNA or combination of small RNAs was shown to be a significant predictor of successful TB therapy (Dersch et al., 2017). M. tuberculosis-mediated TLR2/1 signaling increases the expression of the vitamin D receptor and the vitamin D hydroxylase, resulting in enhanced production of antimicrobial peptides (Lv et al., 2017). TLR4 may contribute to M. tuberculosis resistance; however, no agreement has been achieved at this point. TLR4 has a protective role in adaptive immunity against pulmonary TB in vivo; the non-functional TLR4 causes high mortality and increased bacterial burden in the lungs. miR-146a-5p, miR-21-5p, miR-99b-5p, and miR-132-5p are highly expressed in TB patients and adversely regulate host signaling cytokines in myeloid cells triggered by TLR signaling, promoting M. tuberculosis survival (He et al., 2018). Other miRNAs that are upregulated in M. tuberculosis-infected macrophages, such as miR-27a-5p, miR-33, miR-125-5p, and miR-144-5p, inhibit autophagy formation and M. tuberculosis killing by macrophages. Both miR-29a-3p and miR-125-5p are upregulated in infected macrophages and directly target IFN and TNF, thereby reducing the immune reaction to intracellular M. tuberculosis (Stepanov et al., 2015). Cell necrosis and inflammasome formation are two other mechanisms of defensive strategy against intracellular pathogens that are controlled by M. tuberculosis-induced host miRNAs such as miR-325-3p and miR-20b-5p. However, some miRNAs that are influenced during M. tuberculosis infection, such as miR-155-5p and let-7f, play a crucial role in the activation of host innate and adaptive immunity, as well as microbial clearance (Sinigaglia et al., 2020).
Ligand identification activates the TIR-containing adaptor receptor MyD88, which further binds to IRAK-1 and IRAK-4 (Li et al., 2013; Gu et al., 2017). IRAK has a destruction domain and a serine/threonine kinase domain, and there are four members of the IRAK family: IRAK-1, IRAK-2, IRAK-M, and IRAK-4 (Li et al., 2002). Studies have revealed that IRAK-4 functions upstream of IRAK-1 in the TLR complex (Zhang et al., 2019). Mutations in the IRAK-4 gene have been linked to a higher sensitivity to bacterial infection in patients with Mendelian susceptibility to mycobacterial disease, and M. tuberculosis-infected patients are resistant to TLR ligands (Cui et al., 2018). Moreover, NF-κB essential modulator (NEMO) and IRAK-4 were revealed to be important in IL-12 formation and increased IFN-γ production in humans and mice, which would be vital to creating protective immune responses against mycobacterial infection (Wu et al., 2019). In response to TLR stimulation, IRAK-4 associates with IRAK-1, and the emergence of a dominant negative form of IRAK-4 negative regulator IRAK-1 activation (Li S et al., 2002). IRAK-deficient mice secrete more cytokines in response to TLR ligands (Lomaga et al., 1999). IRAK-M is the negative regulator of the TLR signaling that shows the important role of this protein in suppressing mycobacteria-induced inflammasome activation and TLR signaling pathways. The MAPK pathways are triggered by primary stimulations such as mycobacterial products or whole mycobacteria, resulting in the stimulation of transcription factors such as NF-κB and activator protein-1 (AP-1) (Wang et al., 2001). miRNAs regulating the IRAK signaling pathway were shown in Table 2.
Death ligands such as Fas ligand (FasL) bind to death receptors in the FasR receptor. Following this interaction, the death-inducing signaling complex (DISC) is formed, which includes the Fas-associated death domain-containing protein (FADD) and procaspase-8/10. RNAi-mediated FADD knockdown in cells reduced NF-κB signaling. Exogenous FADD expression prevented NF-κB signaling (Dockrell, 2003). FADD loss-of-function mutations in the death effector inhibited the caspase-8 and NF-κB activation that promotes apoptosis. Caspase-8 deficiency inhibited TNF-related apoptosis-inducing ligand (TRAIL)-induced NF-κB activation. These findings reveal a mechanism for TRAIL-induced NF-κB activation that involves the TRAIL receptors DD, FADD, and caspase-8. These proteins play an important role in apoptosis signaling and are the mediators of non-apoptotic CD95 signaling during T-cell proliferation (Welz et al., 2011). FADD-deficient T cells show reduced proliferation, implying that FADD plays an important role in proliferation signaling. FADD can be regulated transcriptionally by miR-155 (Wang et al., 2011) or miR-128a. miR-128a ectopic expression conferred Fas resistance in cells by directly targeting FADD, but antagonizing miR-128a function made cells susceptible to Fas-mediated apoptosis (Yamada et al., 2014).
Following the binding of activated calcium ions to cAMP, they activate small Ras-like GTPases like Ras-proximate-1 (Rap1), which is primarily involved in cell adhesion and junction formation during cell proliferation. cAMP is also known to increase ERK1/2 phosphorylation via ROS-dependent activation of Ras. Through the negative feedback regulation, miR-146 plays an important role in the control of TLRs and cytokine signaling (Brooks et al., 2014). miRNAs have the ability to regulate the levels of molecules by being involved in the negative feedback of PRR-induced signaling (Mao et al., 2005). miR-124 was discovered to be a negative regulator of inflammation by targeting several pathways, including signal transducer and activator of transcription (STAT) and TLRs (O’Shea et al., 2013). miR-124 inhibits intestinal inflammation by attenuating the production of IL-6 and TNF-α via targeting STAT3, a major factor in inflammatory response, and acetylcholinesterase, a negative regulator of the cholinergic anti-inflammatory signal. Sun et al. (2018) reported that miR-124 inhibits STAT3 to reduce IL-6 production and TNF-α-converting enzyme to inhibit TNF-α release in response to LPS. Lower levels of miR-124 and higher levels of STAT3 promote inflammation and disease pathogenesis. miR-124 expression is increased in pulmonary TB patients. miR-124 negatively regulates multiple TLR signaling components, including TLR6, MyD88, TNF-α, and TRAF6, implying an underlying negative feedback loop between miR-124 and TLR signaling to prevent excessive inflammation (Wang S. et al., 2018). In both calves and humans, a decrease in miR-124 expression contributes to high proliferation and pulmonary inflammation.
MyD88 is an intracellular molecule connected to IRAK and TLRs to transduce signals. MyD88 activates MAPK, PI3K, NF-κB, and IRAK following the initiation of the signal cascade. MyD88 deficiency impairs the macrophage response to bacterial antigens and makes the individual susceptible to infection. However, macrophages activate antibacterial immunity through MyD88-independent mechanisms (Cervantes, 2017). MyD88 deficiency improved resistance to polymicrobial sepsis, indicating that both MyD88-dependent and MyD88-independent antibacterial mechanisms exist. Many studies that showed the regulation of individual genes in macrophages by subcellular microbial products through the TLR/MyD88 signal transduction pathway have been conducted (Huang et al., 2019). However, there appears to be no study showing the role of MyD88 in macrophage activation showing antimicrobial activity. Three unexpected findings emerged in MyD88-deficient mice, implying that the current understanding of macrophage activation needs to be revised. Macrophages undergo active self-priming activation, which is dependent on MyD88. MyD88 is not involved in the IFN-γ signaling pathway; however, the expression of many genes in macrophages in response to IFN-γ is mostly dependent on MyD88. The majority of transcriptional responses of macrophages against M. tuberculosis do not require MyD88. This suggests that TLRs are not the primary receptors for recognizing M. tuberculosis or that TLR-dependent responses are mediated by MyD88-independent signaling pathways (O’Connell et al., 2010; Sharbati et al., 2011). miR-155 modulates the production of inflammatory mediators in response to microbial stimuli by negatively regulating the expression of an important TAK1- and TRAF6-binding protein 2 (TAB2) (Ceppi et al., 2009). Additionally, miR-146a inhibits TLR signaling, thereby inhibiting the production of inflammatory mediators (Taganov et al., 2006; Chen et al., 2007). When PAMPs are recognized, TLR signaling is activated, which leads to the transcriptional activation of genes encoding pro-inflammatory mediators following the activation of antigen-specific adaptive immune response via a MyD88-dependent or -independent pathway (Medzhitov et al., 1998). Various signalling pathways are regulated by more than one miRNAs. The miRNAs that regulate the various signalling pathway during the tuberculosis infection are shown in the Table 2.
The Bcl-2 family members that promote and prevent apoptosis are controlled differently by M. tuberculosis. The prototypical antiapoptotic protein Bcl-2 has homologs in the Bcl-2 family (Klingler et al., 1997). The antiapoptotic family member bfl-1 is upregulated in macrophages during the Mycobacterium bovis BCG infection (Perskvist et al., 2002). An antiapoptotic gene called Bcl-xL was upregulated during M. tuberculosis infection after Bcl-2 was downregulated (Mogga et al., 2002). Infection with M. tuberculosis causes neutrophils to produce more of the proapoptotic family protein Bcl-2-associated X-protein (Bax) and less of the antiapoptotic family protein Bcl-xL (Harris and Thompson, 2000). Bcl-2 was upregulated and Bax was downregulated in animal models of TB. By reducing neutrophil levels and increasing B-cell levels, a number of miRNAs have been linked to the regulation of the apoptotic pathway (van Rensburg et al., 2018). miR-365, which is highly expressed in cells, directly targets the proapoptotic protein Bax, and these interactions are linked to drug resistance in pancreatic cancer cells. By inhibiting Bax expression, miR-125b conferred drug resistance in breast cancer cells (Zhou et al., 2010). By specifically targeting Bcl-xL and inducing apoptosis, miR-491 reduces the viability of cells. Treatment with miR-491 prevents tumor growth in naive mice in vivo (Nakano et al., 2010). Downregulation of miR-133a has been linked to tumor development and prognosis. Restoration of miR-133a inhibits cell division and triggers apoptosis. Bcl-xL regulation by miR-608 has also been demonstrated (Zhang et al., 2014). The expression of Bcl-2 was discovered to be inversely correlated with miR-15a and miR-16-1 (Cimmino et al., 2005). These two miRNAs directly inhibit Bcl-2 at the posttranscriptional level, according to a subsequent study, and also cause apoptosis (Zhang Y. et al., 2014). Bcl-2 protein signaling was elevated when miR-204 was downregulated. MiR-148a and miR-24-2c also directly inhibit Bcl-2 expression (Srivastava et al., 2011; Zhang et al., 2011). Apoptosis is regulated by the endogenous miR-23a/b and miR-27a/b inhibitors of apoptotic peptidase-activating factor (Apaf)-1 expression. It has been demonstrated that miR-133 and miR-24a directly repress caspase-9 to regulate cell fate (Xu et al., 2007; Walker and Harland, 2009; Ji et al., 2013; Chen et al., 2014).
Caspases play an important role in classical apoptosis (Kroemer and Martin., 2005). Caspase activation is not necessarily important in all types of apoptosis. Apoptosis can be triggered by the extrinsic pathway and the intrinsic pathway involving ligation of cell surface death receptors through respective ligands and regulating the Bcl-2 family of pro- and antiapoptotic proteins, respectively. Suppression of caspase activity is cytoprotective when cells are stimulated to undergo apoptosis via death receptor ligation (Maquarre et al., 2005). Caspases, on the other hand, are terminal effectors of the mitochondrial pathway, and this type of cell death is mostly caspase independent (Jäättelä, 2001; Hudson et al., 2013). Few studies suggest that apoptotic cell death can occur in the absence of caspases or in the presence of both caspase and non-caspase protease activity (Sacconi et al., 2012). TNF-α activates caspases and can initiate several cell death pathways, including lysosomal permeabilization mediated by cathepsin B release, which activates the mitochondrial apoptosis pathway (Guicciardi et al., 2000). Although these caspase-independent pathways have not been fully characterized, calpains and serine proteases have been implicated as cell death mediators (Zhang L. et al., 2012; Guo et al., 2013). Overexpression of miR-337-3p and miR-17-5p/miR-132-3p/-212-3p, respectively, can regulate executioner caspase-3 and caspase-7. Furthermore, miRNA overexpression, particularly miR-337-3p, reduces TRAIL cytotoxicity.
MAPK-regulated ERK, JNK, and p38 groups alter gene expression. MAPK signaling pathways are activated during mycobacterial infection and are related to mycobacterial pathogenesis (Song et al., 2003; Pasquinelli et al., 2013; Guo et al., 2019). The p38 MAPK pathway is associated with mycobacteria-induced IL-10, and other cytokines such as TNF-α/IL-4/IFN-γ are produced (Bachstetter and Van Eldik, 2010; de Souza et al., 2014). TNF-α expression in human macrophages is increased by ERK1/2 signaling (Surewicz et al., 2004). MAPK signaling pathways are involved in the regulation of antimycobacterial pathways such as phagosome acidification, apoptosis, and antigen presentation via MHC class II expression. Previous research indicates that the p38 MAPK pathway could serve as a means for mycobacteria to be suppressed. Suppression of p38 MAPK activity increases phagosome acidification. Inactivation of the p38 MAPK pathway causes an increase in phagosome acidification and a significant increase in monocytes’ ability to kill mycobacteria (Klug et al., 2011). Synthesis of TNF in human macrophages is inhibited by lipomannan from virulent M. tuberculosis, but not by avirulent Myocobacterium smegmatis. This variation in response is due to TB and lipomannan induces the stimulation causing TNF mRNA transcripts to destabilize following the reduced expression of TNF protein. Mycobacterium smegmatis Lipomannan increases MAPK-activated protein kinase 2 (MK2) phosphorylation, which is important for maintaining TNF mRNA stability by contributing miRNAs. miR-125b binds to the 3’ UTR region of TNF mRNA and destabilizes the transcript, whereas miR-155 increases TNF production by increasing TNF mRNA half-life and reducing the expression of SHIP1, which is the negative regulator of the PI3K/Akt pathway (Rajaram et al., 2011). Signaling via ERK1/2 and p38 inhibits a well-known mycobacterial TLR2 agonist. Thus, the p38 MAPK and ERK1/2 pathways regulate macrophage antimicrobial function and antigen presentation by infected macrophages, potentially contributing to host immune evasion (Liu P et al., 2016; Liu P et al., 2016; Hölscher et al., 2020). TNF mRNA is stabilized by activated MK2 (Campbell et al., 2014). Non-phosphorylated TTP Tristetraprolin binds to the Adenine/Guanine rich elements ARE region of target mRNAs and causes rapid degradation via a variety of mechanisms (Chen et al., 2013). During the Mycobacterium infection, TLR2-dependent MAPK p38 and the PI3K/Akt pathway stimulates an increase in TNF mRNA expression. Mycobacteria cause the activation and expression of MK2, miR125b, and miR-155 to differ. TNF expression in mycobacteria-infected macrophages is significantly influenced by MAPK p38 and Akt activation. miR-125b inhibits TNF production by targeting the 3′ UTR of the TNF transcript. It also increases the stability of B-Ras2, an inhibitor of NF-κB signaling in human macrophages, lowering the inflammatory response (Niu et al., 2018). TNF production is regulated by hsa-miR-155, which targets the inositol phosphatase for degradation via its 3′ UTR interaction (Yang et al., 2015). Mycobacteria cause differential expression of miRNAs, which are involved in mRNA signal transduction in human macrophages (Schifano et al., 2017). The JNK signaling pathway is important in many biological processes, including embryogenesis. These kinases regulate the expression of host genes involved with apoptotic cell death pathways and carcinogenesis, thereby controlling the functions of neurons and the immune system. Several miRNAs and long noncoding RNAs (lncRNAs) are functionally related to JNKs (Ghafouri-Fard et al., 2021). miR-138 targets mixed-lineage kinase-3 (MLK3), an important component of the JNK/mitogen-activated kinase pathway. miR-138 upregulation diminished proapoptosis factors and apoptosis rate. Upregulation of miR-138 decreased the expression of JNK, phosphorylated JNK (p-JNK), c-jun, p38 MAPK, p-p38 MAPK, iNOS, and COX-2 (Ghafouri-Fard et al., 2021). Low concentrations of MLK3 proteins and inhibition of the JNK/MAPK signaling pathways provide protection. miRNA-363-3p transcriptional regulation is mediated by DNA methylation. The dual-specificity phosphatase 10 targets miRNA-363-3p, and its inhibition promotes JNK phosphorylation. The miRNA-363-3p/DUSP10/JNK axis was linked to the inhibition of homologous recombination and DNA repair pathways. An innovative therapy is thought to be the miRNA-363-3p/DUSP10/JNK axis (Zhou et al., 2022). miR-517a controls oxidative stress. miR-517a suppression enhances cleaved caspase-3 expression, Bax/Bcl-2 ratio, ROS and MDA levels, and cell apoptosis while decreasing ERK1/2 phosphorylation, T-AOC levels, SOD activity, cell proliferation, and mitochondrial membrane potential. Lower levels of miR-517a result in the inactivation of the JNK signaling pathway. As a result, melanoma cells experience increased oxidative stress (Tofannin et al., 2011). miR-221 demonstrates the impact of cyclin-dependent kinase inhibitor on the occurrence and progression of cell cycle progression (Sun et al., 2011). miRNA-31 identifies the cell division cycle protein 42 and forms a negative feedback chain for JNK inactivation upon the formation of miR-31/Cdc42/phosphorylated MLK3 (p-MLK3). miR-31 and p-JNK were found in high concentrations in the liver tissues of Drug induced lung injury patients with various causes. miR-31 can inhibit the overactivation of the ROS/JNK/mitochondrial diseased death loop in Acetaminophen-induced DILI hepatocytes, suggesting a new therapeutic potential for JNK overactivation-based liver injury. The triggering of apoptosis was mediated. Lower levels of miR-517a result in the inactivation of the JNK signaling pathway. As a result, melanoma cells experience increased oxidative stress (Tofannin et al., 2011). miR-221 demonstrates the impact of cyclin-dependent kinase inhibitor on the occurrence and progression of cell cycle progression (Sun et al., 2011). miRNA-31 identifies the cell division cycle protein 42 and forms a negative feedback chain for JNK inactivation upon the formation of miR-31/Cdc42/p-MLK3. miR-31 and p-JNK were found in high concentrations in the liver tissues of DILI patients with various causes. miR-31 can inhibit the overactivation of the ROS/JNK/mitochondrial diseased death loop in APAP-induced DILI hepatocytes, suggesting a new therapeutic potential for JNK overactivation-based liver injury. The triggering of apoptosis was mediated. Apoptosis was induced by activating the JNK pathway and using a JNK specific inhibitor, which was found to entirely inhibit miR-10b-induced apoptosis.
In this review, we narrated the upregulation and downregulation of miRNAs that target the components of six signaling pathways activated in TB infection. After a careful review of the bibliography, we observed the upregulation and downregulation of miRNAs, playing an important role in the pathways. Most of the signaling pathways remain active during the TB infection. We mentioned the important roles of pathways and their regulating miRNAs. These miRNAs can be considered as therapeutic targets. Therefore, targeting their expression can modulate the activity of signaling pathways. We reviewed the importance of miRNAs, posttranscriptional regulators that control mRNA stability in signaling pathways. miRNAs can multiply and regulate cellular outcomes in response to various extracellular signals by acting as genetic switches or fine-tuners. Signaling networks, on the other hand, control the stability, biogenesis, and abundance of miRNAs over time by regulating layers of the miRNA biogenesis pathway. The detailed study of the miRNAs regulating the immune-associated pathways is useful for the development of miRNA mimetic/inhibitor molecules. Immune effects induced by miRNA drugs are currently the major challenges of miRNA therapeutics.
KD conceptualized the idea, analyzed the data and wrote the manuscript. DC helped in improving the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.

Aguilar, C., Mano, M., Eulalio, A. (2019). MicroRNAs at the host-bacteria interface: Host defense or bacterial offense. Trends Microbiol. 27 (3), 206–218. doi: 10.1016/j.tim.2018.10.011
Bachstetter, A. D., Van Eldik, L. J. (2010). The p38 MAP kinase family as regulators of proinflammatory cytokine production in degenerative diseases of the CNS. Aging Dis. 1 (3), 199–211.
Bai, X., Feldman, N. E., Chmura, K., Ovrutsky, A. R., Su, W. L., Griffin, L., et al. (2013). Inhibition of nuclear factor-kappa b activation decreases survival of mycobacterium tuberculosis in human macrophages. PloS One 8 (4), e61925. doi: 10.1371/journal.pone.0061925
Barry, S. E., Ellis, M., Yang, Y., Guan, G., Wang, X., Britton, W. J., et al. (2018). Identification of a plasma microRNA profile in untreated pulmonary tuberculosis patients that is modulated by anti-mycobacterial therapy. J. Infect. 77, 341–348. doi: 10.1016/j.jinf.2018.03.006
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 (2), 215–233. doi: 10.1016/j.cell.2009.01.002
Bogerd, H. P., Whisnant, A. W., Kennedy, E. M., Flores, O., Cullen, B. R. (2014). Derivation and characterization of dicer- and microRNA-deficient human cells. RNA 20 (6), 923–937. doi: 10.1261/rna.044545.114
Brooks, A. J., Dai, W., O’Mara, M. L., Abankwa, D, Chhabra, Y, Pelekanos, RA, et al. (2014). Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 344 (6185), 1249783. doi: 10.1126/science.1249783
Campbell, RM, Anderson, BD, Brooks, NA, Brooks, HB, Chan, EM, De Dios, A, et al (2014). Characterization of LY2228820 dimesylate, a potent and selective inhibitor of p38 MAPK with antitumor activity. Mol Cancer Ther. 13(2), 364–74. doi: 10.1158/1535-7163.MCT-13-0513
Ceppi, M, Pereira, PM, Dunand-Sauthier, I, Barras, E, Reith, W, Santos, MA, et al (2009). MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci U S A. 106(8), 2735–40. doi: 10.1073/pnas.0811073106
Cervantes, J. L. (2017). MyD88 in mycobacterium tuberculosis infection. Med. Microbiol. Immunol. 206 (3), 187–193. doi: 10.1007/s00430-017-0495-0
Chakrabarty, S., Kumar, A., Raviprasad, K., Mallya, S., Satyamoorthy, K., Chawla, K. (2019). Host and MTB genome encoded miRNA markers for diagnosis of tuberculosis. Tuberculosis 116, 37–43. doi: 10.1016/j.tube.2019.04.002
Chandan, K., Gupta, M., Sarwat, M. (2020). Role of host and pathogen-derived MicroRNAs in immune regulation during infectious and inflammatory diseases. Front. Immunol. 10, 3081. doi: 10.3389/fimmu.2019.03081
Chen, XM, Splinter, PL, O'Hara, SP, LaRusso, NF (2007). A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem 282: 28929–28938.
Chen, L., Dong, R., Lu, Y., Zhou, Y., Li, K., Zhang, Z., et al. (2019). MicroRNA-146a protects against cognitive decline induced by surgical trauma by suppressing hippocampal neuroinflammation in mice. Brain Behav. Immun. 78, 188–201. doi: 10.1016/j.bbi.2019.01.020
Chen, Y. C., Lee, C. P., Hsiao, C. C., Hsu, P. Y., Wang, T. Y., Wu, C. C., et al. (2020). MicroRNA-23a-3p down-regulation in active pulmonary tuberculosis patients with high bacterial burden inhibits mononuclear cell function and phagocytosis through TLR4/TNF-α/TGF-β1/IL-10 signaling via targeting IRF1/SP1. Int. J. Mol. Sci. 21 (22), 8587. doi: 10.3390/ijms21228587
Chen, Q., Xu, J., Li, L., Li, H., Mao, S., Zhang, F., et al. (2014). MicroRNA-23a/b and microRNA-27a/b suppress apaf-1 protein and alleviate hypoxia-induced neuronal apoptosis. Cell Death Disease. 5, e1132. doi: 10.1038/cddis.2014.92
Chen, Y., Min, L., Ren, C., Xu, X, Yang, J., Sun, X., et al (2017). miRNA-148a serves as a prognostic factor and suppresses migration and invasion through Wnt1 in non-small cell lung cancer. PLoS One. 12 (2), e0171751. doi: 10.1371/journal.pone.0171751
Chen, Y., Zeng, Z., Shen, X., Wu, Z., Dong, Y., Cheng, J. C. (2016). MicroRNA-146a-5p negatively regulates pro-inflammatory cytokine secretion and cell activation in lipopolysaccharide stimulated human hepatic stellate cells through inhibition of toll-like receptor 4 signaling pathways. Int. J. Mol. Sci. 17 (7), 1076. doi: 10.3390/ijms17071076
Chen, Y. L., Jiang, Y. W., Su, Y. L., Lee, S. C., Chang, M. S., Chang, C. J., et al (2013). Transcriptional regulation of tristetraprolin by NF-κB signaling in LPS-stimulated macrophages. Mol Biol Rep. 40 (4), 2867–2877. doi: 10.1007/s11033-012-2302-8
Cimmino, A., Calin, G. A., Fabbri, M., Iorio, M. V., Ferracin, M., Shimizu, M., et al. (2005). miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U.S.A. 102, 13944–13949.
Cui, J., Gu, L., Zhong, L., Liu, X., Sun, Y., Xu, T. (2022a). microRNA-20-1 and microRNA-101a suppress the NF-κB-Mediated inflammation production by targeting TRAF6 in miiuy croaker. Infect. Immun. 90 (1), e0058521. doi: 10.1128/IAI.00585-21
Cui, J., Li, Z., Cui, K., Gao, Y., Zhang, B., Niu, J., et al. (2021). MicroRNA-20a-3p regulates the host immune response to facilitate the mycobacterium tuberculosis infection by targeting IKKβ/NF-κB pathway. Int. Immunopharmacol. 91, 107286. doi: 10.1016/j.intimp.2020.107286
Cui, Y., Wang, Y., Zhao, D., Feng, X., Zhang, L., Liu, C. (2018). Loganin prevents BV-2 microglia cells from Aβ1-42 -induced inflammation via regulating TLR4/TRAF6/NF-κB axis. Cell Biol. Int. 42 (12), 1632–1642. doi: 10.1002/cbin.11060
Cui, J., Zheng, W., Sun, Y., Xu, T. (2022b). Inducible MicroRNA-132 inhibits the production of inflammatory cytokines by targeting TRAF6, TAK1, and TAB1 in teleost fish. Infect. Immun. 90 (5), e0012022. doi: 10.1128/iai.00120-22
da Cruz, R. S., Carney, E. J., Clarke, J., Cao, H., Cruz, M. I., Benitez, C., et al (2018). Paternal malnutrition programs breast cancer risk and tumor metabolism in offspring. Breast Cancer Res. 20, 99. doi: 10.1186/s13058-018-1034-7
De Araujo, L. S., Ribeiro-Alves, M., Leal-Calvo, T., Leung, J., Durán, V., Samir, M., et al. (2019). Reprogramming of small noncoding RNA populations in peripheral blood reveals host biomarkers for latent and active mycobacterium tuberculosis infection. mBio 10, e01037-19. doi: 10.1128/mBio.01037-19
Dersch, P., Khan, M. A., Mühlen, S., Görke, B. (2017). Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front. Microbiol. 8, 803. doi: 10.3389/fmicb.2017.00803
de Souza, A. P., Vale, V. L., Silva Mda, C., Araújo, I. B., Trindade, S. C., de Moura-Costa, L. F., et al. (2014). MAPK involvement in cytokine production in response to corynebacterium pseudotuberculosis infection. BMC Microbiol., 14:230. doi: 10.1186/s12866-014-0230-6
Dockrell, D. (2003). The multiple role of FAS ligand in the pathogenesis of infectious disease. Clin. Microbiol. Infect. 9, 766–779. doi: 10.1046/j.1469-0691.2003.00669.x
Duffy, F. J., Thompson, E., Downing, K., Suliman, S., Mayanja-Kizza, H., Boom, W. H., et al. (2018). A serum circulating miRNA signature for short-term risk of progression to active tuberculosis among household contacts. Front. Immunol. 9, 661. doi: 10.3389/fimmu.2018.00661
Esteller, M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12 (12), 861–874. doi: 10.1038/nrg3074
Etna, M. P., Sinigaglia, A., Grassi, A., Giacomini, E., Romagnoli, A., Pardini, M., et al. (2018). Mycobacterium tuberculosis-induced miR-155 subverts autophagy by targeting ATG3 in human dendritic cells. PloS Pathog. 14, e1006790. doi: 10.1371/journal.ppat.1006790
Fang, Y., Chen, H., Hu, Y., Li, Q., Hu, Z., Ma, T., et al. (2016). Burkholderia pseudomallei-derived miR-3473 enhances NF-κB via targeting TRAF3 and is associated with different inflammatory responses compared to burkholderia thailandensis in murine macrophages. BMC Microbiol. 16 (1), 283. doi: 10.1186/s12866-016-0901-6
Foghsgaard, L., Wissing, D., Mauch, D., Lademann, U., Bastholm, L., Boes, M., et al. (2001). Cathepsin b acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J. Cell Biol. 153, 999–1010. doi: 10.1083/jcb.153.5.999
Fu, B., Xue, W., Zhang, H., Zhang, R., Feldman, K., Zhao, Q., et al. (2020). MicroRNA-325-3p facilitates immune escape of mycobacterium tuberculosis through targeting LNX1 via NEK6 accumulation to promote anti-apoptotic STAT3 signaling. mBio 11 (3), e00557-20. doi: 10.1128/mBio.00557-20
Fu, Y., Yi, Z., Wu, X., Li, J., Xu, F. (2011). Circulating microRNAs in patients with active pulmonary tuberculosis. J. Clin. Microbiol. 49 (12), 4246–4251. doi: 10.1128/JCM.05459-11
Fu, X., Zeng, L., Liu, Z., Ke, X., Lei, L., Li, G. (2016). MicroRNA-206 regulates the secretion of inflammatory cytokines and MMP9 expression by targeting TIMP3 in mycobacterium tuberculosis-infected THP-1 human macrophages. Biochem. Biophys. Res. Commun. 477 (2), 167–173. doi: 10.1016/j.bbrc.2016.06.038
Gañán-Gómez, I., Wei, Y., Yang, H., Pierce, S., Bueso-Ramos, C., Calin, G., et al. (2014). Overexpression of miR-125a in myelodysplastic syndrome CD34+ cells modulates NF-κB activation and enhances erythroid differentiation arrest. PloS One 9 (4), e93404. doi: 10.1371/journal.pone.0093404
Ghafouri-Fard, S., Abak, A., Mohaqiq, M., Shoorei, H., Taheri, M. (2021). Interaction between non-coding RNAs and JNK in human disorders. BioMed. Pharmacother. 138, 111497. doi: 10.1016/j.biopha.2021.111497
Gong, X., Wang, H., Ye, Y., Shu, Y., Deng, Y., He, X., et al. (2016). miR-124 regulates cell apoptosis and autophagy in dopaminergic neurons and protects them by regulating AMPK/mTOR pathway in parkinson's disease. Am. J. Transl. Res. 8, 2127–2137. doi: 10.1016/j.ymthe.2022.06.003
Grahame Hardie, D. (2016). Regulation of AMP-activated protein kinase by natural and synthetic activators. Acta Pharm. Sin. B. 6 (1), 1–19. doi: 10.1016/j.apsb.2015.06.002
Gu, X., Gao, Y., Mu, D. G., Fu, E. Q. (2017). MiR-23a-5p modulates mycobacterial survival and autophagy during mycobacterium tuberculosis infection through TLR2/MyD88/NF-κB pathway by targeting TLR2. Exp. Cell Res. 354 (2), 71–77. doi: 10.1016/j.yexcr.2017.03.039
Guicciardi, M. E., Deussing, J., Miyoshi, H., Bronk, S. F., Svingen, P. A., Peters, C., et al. (2000). Cathepsin b contributes to TNF-α-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Investig. 106, 1127–1137. doi: 10.1172/JCI9914
Guo, Q., Bi, J., Li, M., Ge, W., Xu, Y., Fan, W., et al. (2019). ESX secretion-associated protein c from Mycobacterium tuberculosis induces macrophage activation through the toll-like receptor-4/Mitogen-Activated protein kinase signaling pathway. Front. Cell Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00158
Guo, Q., Zhang, J., Li, J., Zou, L., Zhang, J., Xie, Z., et al. (2013). Forced miR-146a expression causes autoimmune lymphoproliferative syndrome in mice via downregulation of fas in germinal center b cells. Blood 121, 4875–4883. doi: 10.1182/blood-2012-08-452425
Guttilla, I. K., White, B. A. (2009). Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 284 (35), 23204–23216. doi: 10.1074/jbc.M109.031427
Harapan, H., Fitra, F., Ichsan, I., Mulyadi, M., Miotto, P., NA, H., et al. (2013). The roles of microRNAs on tuberculosis infection: meaning or myth? Tuberculosis (Edinb) 93 (6), 596–605. doi: 10.1016/j.tube.2013.08.004
Harris, M. H., Thompson, C. B. (2000). The role of the bcl-2 family in the regulation of outer mitochondrial membrane permeability. Cell Death Differ 7, 1182. doi: 10.1038/sj.cdd.4400781
He, Y., Yang, Y., Liao, Y., Xu, J., Liu, L., Li, C., et al. (2020). miR-140-3p inhibits cutaneous melanoma progression by disrupting AKT/p70S6K and JNK pathways through ABHD2. Mol. Ther. Oncolytics 17, 83–93. doi: 10.1016/j.omto.2020.03.009
He, X., Zheng, Y., Liu, S., Shi, S., Liu, Y., He, Y., et al. (2018). MiR-146a protects small intestine against ischemia/reperfusion injury by down-regulating TLR4/TRAF6/NF-κB pathway. J. Cell Physiol. 233 (3), 2476–2488. doi: 10.1002/jcp.26124
Hölscher, C., Gräb, J., Hölscher, A., Müller, AL, Schäfer, SC, Rybniker, J. (2020). Chemical p38 MAP kinase inhibition constrains tissue inflammation and improves antibiotic activity in. Mycobacterium tuberculosis-infected mice. Sci. Rep. 10(1), 13629. doi: 10.1038/s41598-020-70184-x
Huang, X., Qiao, F., Xue, P. (2019). The protective role of microRNA-140-5p in synovial injury of rats with knee osteoarthritis via inactivating the TLR4/Myd88/NF-κB signaling pathway. Cell Cycle 18 (18), 2344–2358. doi: 10.1080/15384101.2019.1647025
Huang, G., Redelman-Sidi, G., Rosen, N., Glickman, M. S., Jiang, X. (2012). Inhibition of mycobacterial infection by the tumor suppressor PTEN. J. Biol. Chem. 287 (27), 23196–23202. doi: 10.1074/jbc.M112.351940
Hudson, R., Yi, M., Esposito, D., Glynn, S., Starks, A., Yang, Y., et al. (2013). MicroRNA-106b-25 cluster expression is associated with early disease recurrence and targets caspase-7 and focal adhesion in human prostate cancer. Oncogene 32, 4139–4147. doi: 10.1038/onc.2012.424
Iwai, H., Funatogawa, K., Matsumura, K., Kato-Miyazawa, M., Kirikae, F., Kiga, K., et al. (2015). MicroRNA-155 knockout mice are susceptible to mycobacterium tuberculosis infection. Tuberculosis (Edinb). 95 (3), 246–250. doi: 10.1016/j.tube.2015.03.006
Jäättelä, M. L. M. (2001). Four deaths and a funeral: from caspases to alternative mechanisms. Nat. Rev. Mol. Cell Biol. 2, 589–598. doi: 10.1038/35085008
Ji, F., Zhang, H., Wang, Y., Li, M., Xu, W., Kang, Y., et al. (2013). MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting bcl-xL and mcl-1. Bone 56, 220–226. doi: 10.1016/j.bone.2013.05.020
Ju, M., Liu, B., He, H., Gu, Z., Liu, Y., Su, Y., et al. (2018). MicroRNA-27a alleviates LPS-induced acute lung injury in mice via inhibiting inflammation and apoptosis through modulating TLR4/MyD88/NF-κB pathway. Cell Cycle 17 (16), 2001–2018. doi: 10.1080/15384101.2018.1509635
Kanniappan, P., Ahmed, S. A., Rajasekaram, G., Marimuthu, C., Ch'ng, E. S., Lee, L. P., et al. (2017). RNomic identification and evaluation of npcTB_6715, a non-protein-coding RNA gene as a potential biomarker for the detection of mycobacterium tuberculosis. J. Cell Mol. Med. 21 (10), 2276–2283. doi: 10.1111/jcmm.13148
Kim, J. K., Kim, T. S., Basu, J., Jo, E. K. (2017). MicroRNA in innate immunity and autophagy during mycobacterial infection. Cell Microbiol. 19 (1). doi: 10.1111/cmi.12687
Kim, J. K., Yuk, J. M., Kim, S. Y., Kim, T. S., Jin, H. S., Yang, C. S., et al. (2015). MicroRNA-125a inhibits autophagy activation and antimicrobial responses during mycobacterial infection. J. Immunol. 194 (11), 5355–5365. doi: 10.4049/jimmunol.1402557
Klingler, K., Tchou-Wong, K. M., Brandli, O., Aston, C., Kim, R., Chi, C., et al. (1997). Effects of mycobacteria on regulation of apoptosis in mononuclear phagocytes. Infect. Immun. 65, 5272. doi: 10.1128/iai.65.12.5272-5278.1997
Klug, K., Ehlers, S., Uhlig, S., Reiling, N. (2011). Mitogen-activated protein kinases p38 and ERK1/2 regulated control of mycobacterium avium replication in primary murine macrophages is independent of tumor necrosis factor-α and interleukin-10. Innate Immun. 17 (5), 470–485. doi: 10.1177/1753425910377799
Kong, L., Sun, M., Jiang, Z., Li, L., Lu, B. (2018). MicroRNA-194 inhibits lipopolysaccharide-induced inflammatory response in nucleus pulposus cells of the intervertebral disc by targeting TNF receptor-associated factor 6 (TRAF6). Med. Sci. Monit. 24, 3056–3067. doi: 10.12659/MSM.907280
Kroemer, G., Martin, S. J. (2005). Caspase-independent cell death. Nat. Med. 11, 725–730. doi: 10.1038/nm1263
Kumar, M., Sahu, S. K., Kumar, R., Subuddhi, A., Maji, R. K., Jana, K., et al. (2015). MicroRNA let-7 modulates the immune response to mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway. Cell Host Microbe 17 (3), 345–356. doi: 10.1016/j.chom.2015.01.007
Lang, Q., Ling, C. (2012). MiR-124 suppresses cell proliferation in hepatocellular carcinoma by targeting PIK3CA. Biochem. Biophys. Res. Commun. 426, 247–252. doi: 10.1016/j.bbrc.2012.08.075
Liang, S., Song, Z., Wu, Y., Gao, Y., Gao, M., Liu, F., et al. (2018). MicroRNA-27b modulates inflammatory response and apoptosis during Mycobacterium tuberculosis infection. J. Immunol. 200 (10), 3506–3518. doi: 10.4049/jimmunol.1701448
Li, X., Huang, S., Yu, T., Liang, G., Liu, H., Pu, D., et al. (2019). MiR-140 modulates the inflammatory responses of mycobacterium tuberculosis-infected macrophages by targeting TRAF6. J. Cell Mol. Med. 23 (8), 5642–5653. doi: 10.1111/jcmm.14472
Li, J., Li, J., Wei, T., Li, J. (2016). Down-regulation of microRNA-137 improves high glucose-induced oxidative stress injury in human umbilical vein endothelial cells by up-regulation of AMPKalpha1. Cell. Physiol. Biochem. 39, 847–859. doi: 10.1159/000447795
Li, Z, Qu, Z, Wang, Y, Qin, M, Zhang, H (2020). miR-101-3p sensitizes non-small cell lung cancer cells to irradiation. Open Med (Wars). 15(1), 413–423. doi: 10.1515/med-2020-0044
Li, S., Strelow, A., Fontana, E.J., Wesche, H. (2002). IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc. Natl. Acad. Sci. USA. 99, 5567–5572
Liu, H. Y. (2016). Down-regulation of miR-144 after mycobacterium tuberculosis infection promotes inflammatory factor secretion from macrophages through the Tpl2/ERK pathway. Cell Mol. Biol. (Noisy-le-grand). 62 (2), 87–93. doi: 10.14715/cmb/2016.62.2.15
Liu, F., Chen, J., Wang, P., Li, H., Zhou, Y., Liu, H., et al. (2018). MicroRNA-27a controls the intracellular survival of mycobacterium tuberculosis by regulating calcium-associated autophagy. Nat. Commun. 9 (1), 4295. doi: 10.1038/s41467-018-06836-4
Liu, Y., Li, J. Y., Chen, S. T., Huang, H. R., Cai, H. (2016a). The rLrp of mycobacterium tuberculosis inhibits proinflammatory cytokine production and downregulates APC function in mouse macrophages via a TLR2-mediated PI3K/Akt pathway activation-dependent mechanism. Cell Mol. Immunol. 13 (6), 729–746. doi: 10.1038/cmi.2015.58
Liu, P., Ye, F., Xie, X., Li, X., Tang, H., Li, S., et al. (2016b). Mir-101-3p is a key regulator of tumor metabolism in triple negative breast cancer targeting AMPK. Oncotarget 7, 35188–35198. doi: 10.18632/oncotarget.9072
Li, M., Wang, J., Fang, Y., Gong, S., Li, M., Wu, M., et al. (2016). microRNA-146a promotes mycobacterial survival in macrophages through suppressing nitric oxide production. Sci. Rep. 6, 23351. doi: 10.1038/srep23351
Li, Y., Wang, S., Gao, X., Zhao, Y., Li, Y., Yang, B., et al. (2018). Octreotide alleviates autophagy by up-regulation of microRNA-101 in intestinal epithelial cell line caco-2. Cell. Physiol. Biochem. 49, 1352–1363. doi: 10.1159/000493413
Li, H., Wang, Y., Song, Y. (2018). MicroRNA-26b inhibits the immune response to mycobacterium tuberculosis (M.tb) infection in THP-1 cells via targeting TGFβ-activated kinase-1 (TAK1), a promoter of the NF-κB pathway. Int. J. Clin. Exp. Pathol. 11 (3), 1218–1227.
Li, S., Yue, Y., Xu, W., Xiong, S. (2013). MicroRNA-146a represses mycobacteria-induced inflammatory response and facilitates bacterial replication via targeting IRAK-1 and TRAF-6. PloS One 8 (12), e81438. doi: 10.1371/journal.pone.0081438
Li, W. T., Zhang, Q. (2019). MicroRNA-708-5p regulates mycobacterial vitality and the secretion of inflammatory factors in mycobacterium tuberculosis-infected macrophages by targeting TLR4. Eur. Rev. Med. Pharmacol. Sci. 23 (18), 8028–8038. doi: 10.26355/eurrev_201909_19019
Lou, J., Wang, Y., Zhang, Z., Qiu, W. (2017). MiR-20b inhibits mycobacterium tuberculosis induced inflammation in the lung of mice through targeting NLRP3. Exp. Cell Res. 358 (2), 120–128. doi: 10.1016/j.yexcr.2017.06.007
Lu, H., Buchan, R. J., Cook, S. A. (2010). MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc. Res. 86, 410–420. doi: 10.1093/cvr/cvq010
Lv, J., He, X., Wang, H., Wang, Z., Kelly, G. T., Wang, X., et al. (2017). TLR4-NOX2 axis regulates the phagocytosis and killing of mycobacterium tuberculosis by macrophages. BMC Pulm Med. 17 (1), 194. doi: 10.1186/s12890-017-0517-0
Lyu, L., Zhang, X., Li, C., Yang, T., Wang, J., Pan, L., et al. (2019). Small RNA profiles of serum exosomes derived from individuals with latent and active tuberculosis. Front. Microbiol. 10, 1174. doi: 10.3389/fmicb.2019.01174
Ma, C., Li, Y., Li, M., Deng, G., Wu, X., Zeng, J., et al. (2014). microRNA-124 negatively regulates TLR signaling in alveolar macrophages in response to mycobacterial infection. Mol. Immunol. 62 (1), 150–158. doi: 10.1016/j.molimm.2014.06.014
Mao, X., Ren, Z., Parker, G. N., Sondermann, H, Pastorello, MA, Wang, W, et al. (2005). Structural bases of unphosphorylated STAT1 association and receptor binding. Mol. Cell. 17 (6), 761–771. doi: 10.1016/j.molcel.2005.02.021
Maquarre, E., Artus, C., Gadhoum, Z., Jasmin, C., Smadja-Joffe, F., Robert-Lezenes, J. (2005). CD44 ligation induces apoptosis via caspase- and serine protease-dependent pathways in acute promyelocytic leukemia cells. Leukemia 19, 2296–2303. doi: 10.1038/sj.leu.2403944
Marcinowski, L., Tanguy, M., Krmpotic, A., Rädle, B., Lisnić, V. J., Tuddenham, L., et al. (2012). Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PloS Pathog. 8 (2), e1002510. doi: 10.1371/journal.ppat.1002510
Medzhitov, R, Preston-Hurlburt, P, Kopp, E. (1998). MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell. 2(2), 253–258. doi: 10.1016/s1097-2765(00)80136-7
Mogga, S., Mustafa, T., Sviland, L., Nilsen, R. (2002). Increased bcl-2 and reduced bax expression in infected macrophages in slowly progressive primary murine mycobacterium tuberculosis infection. Scand. J. Immunol. 56, 383. doi: 10.1046/j.1365-3083.2002.01140.x
Nakano, H., Miyazawa, T., Kinoshita, K., Yamada, Y., Yoshida, T. (2010). Functional screening identifies a microRNA, miR-491 that induces apoptosis by targeting bcl-XL in colorectal cancer cells. Int. J. Cancer. 127, 1072–1080. doi: 10.1002/ijc.25143
Nara, K., Kawashima, N., Noda, S., Fujii, M., Hashimoto, K., Tazawa, K., et al. (2019). Anti-inflammatory roles of microRNA 21 in lipopolysaccharide-stimulated human dental pulp cells. J. Cell Physiol. 234 (11), 21331–21341. doi: 10.1002/jcp.28737
Niu, W., Sun, B., Li, M., Cui, J., Huang, J., Zhang, L. (2018). TLR-4/microRNA-125a/NF-κB signaling modulates the immune response to mycobacterium tuberculosis infection. Cell Cycle 17 (15), 1931–1945. doi: 10.1080/15384101.2018.1509636
Oneyama, C., Ikeda, J., Okuzaki, D., Suzuki, K., Kanou, T., Shintani, Y., et al. (2011). MicroRNA-mediated downregulation of mTOR/FGFR3 controls tumor growth induced by src-related oncogenic pathways. Oncogene 30, 3489–3501. doi: 10.1038/onc.2011.63
O'Connell, RM, Kahn, D, Gibson, WS, Round, JL, Scholz, RL, Chaudhuri, AA, et al (2010). MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 33(4), 607–19. doi: 10.1016/j.immuni.2010.09.009
O’Shea, J. J., Holland, S. M., Staudt, L. M. (2013). JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl. J. Med. 368 (2), 161–170. doi: 10.1056/NEJMra1202117
Ostrik, A. A., Azhikina, T. L., Salina, E. G. (2021). Small noncoding RNAs and their role in the pathogenesis of mycobacterium tuberculosis infection. Biochem. (Mosc). 86 (Suppl 1), S109–S119. doi: 10.1134/S000629792114008X
Ouimet, M., Koster, S., Sakowski, E., Ramkhelawon, B., van Solingen, C., Oldebeken, S., et al. (2016). Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 17 (6), 677–686. doi: 10.1038/ni.3434
Pasquinelli, V., Rovetta, A. I., Alvarez, I. B., Jurado, J. O., Musella, R. M., Palmero, D. J., et al. (2013). Phosphorylation of mitogen-activated protein kinases contributes to interferon γ production in response to mycobacterium tuberculosis. J. Infect. Dis. 207 (2), 340–350. doi: 10.1093/infdis/jis672
Pattnaik, B., Patnaik, N., Mittal, S., Mohan, A., Agrawal, A., Guleria, R., et al. (2022). Micro RNAs as potential biomarkers in tuberculosis: A systematic review. Noncoding RNA Res. 7 (1), 16–26. doi: 10.1016/j.ncrna.2021.12.005
Perskvist, N., Long, M., Stendahl, O., Zheng, L. (2002). Mycobacterium tuberculosis promotes apoptosis in human neutrophils by activating caspase-3 and altering expression of Bax/Bcl-xL via an oxygen-dependent pathway. J. Immunol. 168, 6358. doi: 10.4049/jimmunol.168.12.6358
Prantner, D., Perkins, D. J., Vogel, S. N. (2017). AMP-activated kinase (AMPK) promotes innate immunity and antiviral defense through modulation of stimulator of interferon genes (STING) signaling. J. Biol. Chem. 292 (1), 292–304. doi: 10.1074/jbc.M116.763268
Qin, J., Zhou, J., Teng, L., Han, Y. (2018). MicroRNA-10b promotes apoptosis via JNK pathway in clear cell renal cell carcinoma. Nephron 139 (2), 172–180. doi: 10.1159/000486017
Rajaram, M. V., Ni, B., Morris, J. D., Brooks, M. N., Carlson, T. K., Bakthavachalu, B., et al. (2011). Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc. Natl. Acad. Sci. U S A. 108 (42), 17408–17413. doi: 10.1073/pnas.1112660108
Ren, R., Chen, S. D., Fan, J., Zhang, G., Li, J. B. (2018). miRNA-138 regulates MLK3/JNK/MAPK pathway to protect BV-2 cells from H2O2-induced apoptosis. Bratisl Lek Listy. 119 (5), 284–288. doi: 10.4149/BLL_2018_053
Rothchild, A. C., Sissons, J. R., Shafiani, S., Plaisier, C., Min, D., Mai, D., et al. (2016). MiR-155-regulated molecular network orchestrates cell fate in the innate and adaptive immune response to mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U S A. 113 (41), E6172–E6181. doi: 10.1073/pnas.1608255113
Sabir, N., Hussain, T., Shah, S. Z. A., Peramo, A., Zhao, D., Zhou, X. (2018). miRNAs in tuberculosis: New avenues for diagnosis and host-directed therapy. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00602
Sacconi, A., Biagioni, F., Canu, V., Mori, F., Di Benedetto, A., Lorenzon, L., et al. (2012). miR-204 targets bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Disease. 3, e423. doi: 10.1038/cddis.2012.160
Schifano, JM, Corcoran, K, Kelkar, H, Dittmer, DP. (2017). Expression of the Antisense-to-Latency Transcript Long Noncoding RNA in Kaposi's Sarcoma-Associated Herpesvirus. J Virol. 91(4), e01698-16. doi: 10.1128/JVI.01698-16
Sharbati, J, Lewin, A, Kutz-Lohroff, B, Kamal, E, Einspanier, R, Sharbati, S. (2011). Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS One. 6(5), e20258. doi: 10.1371/journal.pone.0020258
Shariq, M., Quadir, N., Sharma, N., Singh, J., Sheikh, J. A., Khubaib, M., et al. (2021). Mycobacterium tuberculosis RipA dampens TLR4-mediated host protective response using a multi-pronged approach involving autophagy, apoptosis, metabolic repurposing, and immune modulation. Front. Immunol. 12, 636644. doi: 10.3389/fimmu.2021.636644
Sharma, N., Verma, R., Kumawat, K. L., Basu, A., Singh, S. K. (2015). miR-146a suppresses cellular immune response during Japanese encephalitis virus JaOArS982 strain infection in human microglial cells. J. Neuroinflammation. 12, 30. doi: 10.1186/s12974-015-0249-0
Shi, G., Mao, G., Xie, K., Wu, D., Wang, W. (2018). MiR-1178 regulates mycobacterial survival and inflammatory responses in mycobacterium tuberculosis-infected macrophages partly via TLR4. J. Cell Biochem. 119 (9), 7449–7457. doi: 10.1002/jcb.27054
Sinigaglia, A., Peta, E., Riccetti, S., Venkateswaran, S., Manganelli, R., Barzon, L. (2020). Tuberculosis-associated MicroRNAs: From pathogenesis to disease biomarkers. Cells 9 (10), 2160. doi: 10.3390/cells9102160
Song, C. H., Lee, J. S., Lee, S. H., Lim, K., Kim, H. J., Park, J. K., et al. (2003). Role of mitogen-activated protein kinase pathways in the production of tumor necrosis factor-alpha, interleukin-10, and monocyte chemotactic protein-1 by mycobacterium tuberculosis H37Rv-infected human monocytes. J. Clin. Immunol. 23 (3), 194–201. doi: 10.1023/a:1023309928879
Spinelli, S. V., Diaz, A., D’Attilio, L., Marchesini, M. M., Bogue, C., Bay, M. L., et al. (2013). Altered microRNA expression levels in mononuclear cells of patients with pulmonary and pleural tuberculosis and their relation with components of the immune response. Mol. Immunol. 53, 265–269. doi: 10.1016/j.molimm.2012.08.008
Srivastava, N., Manvati, S., Srivastava, A., Pal, R., Kalaiarasan, P., Chattopadhyay, S., et al. (2011). miR-24-2 controls H2AFX expression regardless of gene copy number alteration and induces apoptosis by targeting antiapoptotic gene BCL-2: a potential for therapeutic intervention. Breast Cancer Res. 13, R39. doi: 10.1186/bcr2861
Stepanov, G. A., Filippova, J. A., Komissarov, A. B., Kuligina, E. V., Richter, V. A., Semenov, D. V. (2015). Regulatory role of small nucleolar RNAs in human diseases. BioMed. Res. Int. 2015, 206849. doi: 10.1155/2015/206849
Stutz, M. D., Clark, M. P., Doerflinger, M., Pellegrini, M. (2018). Mycobacterium tuberculosis: Rewiring host cell signaling to promote infection. J. Leukoc. Biol. 103 (2), 259–268. doi: 10.1002/JLB.4MR0717-277R
Sun, T., Li, M.-Y., Li, P.-F., Cao, J.-M. (2018). MicroRNAs in cardiac autophagy: small molecules and big role. Cells 7, 104. doi: 10.3390/cells7080104
Sun, K, Wang, W, Zeng, JJ, Wu, CT, Lei, ST, Li, GX, et al (2011). MicroRNA-221 inhibits CDKN1C/p57 expression in human colorectal carcinoma. Acta Pharmacol Sin. 32(3), 375–84. doi: 10.1038/aps.2010.206
Surewicz, K, Aung, H, Kanost, RA, Jones, L, Hejal, R, Toossi, Z, et al (2004). The differential interaction of p38 MAP kinase and tumor necrosis factor-alpha in human alveolar macrophages and monocytes induced by Mycobacterium tuberculois. Cell Immunol. 228(1), 34–41. doi: 10.1016/j.cellimm.2004.03.007
Taganov, KD, Boldin, MP, Chang, KJ, Baltimore, D (2006). NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 103(33), 12481–6. doi: 10.1073/pnas.0605298103
Taneja, S., Dutta, T. (2019). On a stake-out: Mycobacterial small RNA identification and regulation. Noncoding RNA Res. 4 (3), 86–95. doi: 10.1016/j.ncrna.2019.05.001
Toffanin, S, Hoshida, Y, Lachenmayer, A, Villanueva, A, Cabellos, L, Minguez, B, et al (2011). MicroRNA-based classification of hepatocellular carcinoma and oncogenic role of miR-517a. Gastroenterology. 140(5), 1618–1628. doi: 10.1053/j.gastro.2011.02.009
Tran, M., Lee, S.-M., Shin, D.-J., Wang, L. (2017). Loss of miR-141/200c ameliorates hepatic steatosis and inflammation by reprogramming multiple signaling pathways in NASH. JCI Insight 2 2(21), e96094. doi: 10.1172/jci.insight.96094
Vandenabeele, P., Orrenius, S., Zhivotovsky, B. (2005). Serine proteases and calpains fulfill important supporting roles in the apoptotic tragedy of the cellular opera. Cell Death Differ. 12, 1219–1224. doi: 10.1038/sj.cdd.4401719
Van Rensburg, I. C., Du Toit, L., Walzl, G., Du Plessis, N., Loxton, A. G. (2018). Decreased neutrophil–associated miRNA and increased b-cell associated miRNA expression during tuberculosis. Gene 655, 35–41. doi: 10.1016/j.gene.2018.02.052
Walker, J. C., Harland, R. M. (2009). microRNA-24a is required to repress apoptosis in the developing neural retina. Genes Dev. 23, 1046–1051. doi: 10.1101/gad.1777709
Wang, Y., Chen, C., Xu, X. D., Li, H., Cheng, M. H., Liu, J., et al. (2020). Levels of miR-125a-5p are altered in mycobacterium avium-infected macrophages and associate with the triggering of an autophagic response. Microbes Infect. 22 (1), 31–39. doi: 10.1016/j.micinf.2019.07.002
Wang, J., Cui, Z., Liu, L., Zhang, S., Zhang, Y., Zhang, Y., et al. (2019). MiR-146a mimic attenuates murine allergic rhinitis by downregulating TLR4/TRAF6/NF-κB pathway. Immunotherapy 11 (13), 1095–1105. doi: 10.2217/imt-2019-0047
Wang, S., Wu, G., Han, Y., Song, P., Chen, J., Wu, Y., et al. (2018). miR-124 regulates STAT3-mediated cell proliferation, migration and apoptosis in bladder cancer. Oncol. Lett. 16 (5), 5875–5881. doi: 10.3892/ol.2018.9341
Wang, C., Yang, S., Liu, C. M., Jiang, T. T., Chen, Z. L., Tu, H. H., et al. (2018). Screening and identification of four serum miRNAs as novel potential biomarkers for cured pulmonary tuberculosis. Tuberculosis 108, 26–34. doi: 10.1016/j.tube.2017.08.010
Wang, C., Yang, S., Sun, G., Tang, X., Lu, S., Neyrolles, O., et al. (2011). Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PloS One 6 (10), e25832. doi: 10.1371/journal.pone.0025832
Wang, J., Yang, K., Zhou, L., Wu, M., Wu, Y., Zhu, M., et al. (2013). MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting rheb. PloS Pathog. 9, e1003697. doi: 10.1371/journal.ppat.1003697
Wang, H. Q., Yu, X. D., Liu, Z. H., Cheng, X., Samartzis, D., Jia, L. T., et al. (2011). Deregulated miR-155 promotes fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J. pathology. 225, 232–242. doi: 10.1002/path.2931
Wang, J.G., Miyazu, M., Matsushita, E., Sokabe, M., Naruse, K.. (2001). Uni-axial cyclic stretch induces focal adhesion kinase (FAK) tyrosine phosphorylation followed by mitogen-activated protein kinase (MAPK) activation. Biochem. Biophys. Res.Comm. 288, 356–361
Watson, R. O., Manzanillo, P. S., Cox, J. S. (2012). Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150, 803–815. doi: 10.1016/j.cell.2012.06.040
Welz, P. S., Wullaert, A., Vlantis, K., Kandylis, V., Fernandez-Majad, V., Ermalaeva, M., et al. (2011). FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Natur 477, 330–335. doi: 10.1038/nature10273
Westermann, A. J. (2018). Regulatory RNAs in virulence and host-microbe interactions. Microbiol. Spectr. 6 (4). doi: 10.1128/microbiolspec.RWR-0002-2017
Woo, S. J., Kim, Y., Jung, H., Lee, J. J., Hong, J. Y. (2022). MicroRNA 148a suppresses tuberculous fibrosis by targeting NOX4 and POLDIP2. Int. J. Mol. Sci. 23 (6), 2999. doi: 10.3390/ijms23062999
Wu, Z., Lu, H., Sheng, J., Li, L. (2012). Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and bcl-2. FEBS Lett. 586 (16), 2459–2467. doi: 10.1016/j.febslet.2012.06.004
Wu, J., Niu, P., Zhao, Y., Cheng, Y., Chen, W., Lin, L., et al. (2019). Impact of miR-223-3p and miR-2909 on inflammatory factors IL-6, IL-1ß, and TNF-α, and the TLR4/TLR2/NF-κB/STAT3 signaling pathway induced by lipopolysaccharide in human adipose stem cells. PloS One 14 (2), e0212063. doi: 10.1371/journal.pone.0212063
Wu, M, Song, D, Li, H, Yang, Y, Ma, X, Deng, S, et al. (2019). Negative regulators of STAT3 signaling pathway in cancers. Cancer Manag Res. 11, 4957–4969. doi: 10.2147/CMAR.S206175
Xiang, J., Chao, D. T., Korsmeyer, S. J. (1996). BAX-induced cell death may not require interleukin 1β-converting enzyme-like proteases. Proc. Natl. Acad. Sci. U.S.A. 93, 14559–14563. doi: 10.1073/pnas.93.25.14559
Xu, S. J., Hu, H. T., Li, H. L., Chang, S. (2019). The role of miRNAs in immune cell development, immune cell activation, and tumor immunity: With a focus on macrophages and natural killer cells. Cells 8 (10), 1140. doi: 10.3390/cells8101140
Xu, C., Lu, Y., Pan, Z., Chu, W., Luo, X., Lin, H., et al. (2007). The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J. Cell Sci. 120, 3045–3052. doi: 10.1242/jcs.010728
Yamada, N., Noguchi, S., Kumazaki, M., Shinohara, H., Miki, K., Naoe, T., et al. (2014). Epigenetic regulation of microRNA-128a expression contributes to the apoptosis-resistance of human T-cell leukaemia jurkat cells by modulating expression of fas-associated protein with death domain (FADD). Biochim. Biophys. Acta (BBA)-Molecular Cell Res. 1843, 590–602. doi: 10.1016/j.bbamcr.2013.11.022
Yang, J., Chen, Y., Jiang, K., Yang, Y., Zhao, G., Guo, S., et al. (2019). MicroRNA-106a provides negative feedback regulation in lipopolysaccharide-induced inflammation by targeting TLR4. Int. J. Biol. Sci. 15 (11), 2308–2319. doi: 10.7150/ijbs.33432
Yang, C. S., Kim, J. J., Lee, H. M., Jin, H. S., Lee, S. H., Park, J. H., et al. (2014). The AMPK-PPARGC1A pathway is required for antimicrobial host defense through activation of autophagy. Autophagy. 10 (5), 785–802. doi: 10.4161/auto.28072
Yang, S., Li, F., Jia, S., Zhang, K., Jiang, W., Shang, Y., et al. (2015). Early secreted antigen ESAT-6 of mycobacterium tuberculosis promotes apoptosis of macrophages via targeting the microRNA155-SOCS1 interaction. Cell Physiol. Biochem. 35 (4), 1276–1288. doi: 10.1159/000373950
Yang, Y., Yang, C., Guo, Y. F., Liu, P., Guo, S., Yang, J., et al. (2019). MiR-142a-3p alleviates escherichia coli derived lipopolysaccharide-induced acute lung injury by targeting TAB2. Microb. Pathog. 136, 103721. doi: 10.1016/j.micpath.2019.103721
Yang, C., Yang, C., Huang, Z., Zhang, J., Chen, N., Guo, Y., et al. (2022). Reduced expression of MiR-125a-5p aggravates LPS-induced experimental acute kidney injury pathology by targeting TRAF6. Life Sci. 288, 119657. doi: 10.1016/j.lfs.2021.119657
Yang, C., Yan, Z., Hu, F., Wei, W, Sun, Z, Xu, W. (2020). Silencing of microRNA-517a induces oxidative stress injury in melanoma cells via inactivation of the JNK signaling pathway by upregulating CDKN1C. Cancer Cell Int. 20, 32. doi: 10.1186/s12935-019-1064-y
Ye, J., Xu, M., Tian, X., Cai, S., Zeng, S. (2019). Research advances in the detection of miRNA. J. Pharm. Anal. 9 (4), 217–226. doi: 10.1016/j.jpha.2019.05.004
Ying, J, Yu, X, Ma, C, Zhang, Y, Dong, J. (2017). MicroRNA-363-3p is downregulated in hepatocellular carcinoma and inhibits tumorigenesis by directly targeting specificity protein 1. Mol Med Rep. 16(2), 1603–1611. doi: 10.3892/mmr.2017.6759
Zhang, L., Dong, L., Li, Y., Hong, Z., Wei, W. (2012). The microRNA miR-181c controls microglia-mediated neuronal apoptosis by suppressing tumor necrosis factor. J. neuroinflammation. 9, 211. doi: 10.1186/1742-2094-9-211
Zhang, X., Guo, Y., Xu, X., Tang, T., Sun, L., Wang, H., et al. (2019). miR-146a promotes borna disease virus 1 replication through IRAK1/TRAF6/NF-κB signaling pathway. Virus Res. 271, 197671. doi: 10.1016/j.virusres.2019.197671
Zhang, H., Li, Y., Huang, Q., Ren, X., Hu, H., Sheng, H., et al. (2011). MiR-148a promotes apoptosis by targeting bcl-2 in colorectal cancer. Cell Death Differentiation. 18, 1702–1710. doi: 10.1038/cdd.2011.28
Zhang, Y., Schiff, D., Park, D., Abounader, R. (2014). MicroRNA-608 and MicroRNA-34a regulate chordoma malignancy by targeting EGFR, bcl-xL and MET. PloS One 9, e91546. doi: 10.1371/journal.pone.0091546
Zhang, Q., Jiang, X., He, W., Wei, K., Sun, J., Qin, X., et al. (2017a). MCL plays an anti-inflammatory role in Mycobacterium tuberculosis-induced immune response by inhibiting NF-κB and NLRP3 inflammasome activation. Mediators Inflamm. 2017, 2432904. doi: 10.1155/2017/2432904
Zhang, Q., Sun, J., Wang, Y., He, W., Wang, L., Zheng, Y., et al. (2017b). Antimycobacterial and anti-inflammatory mechanisms of baicalin via induced autophagy in macrophages infected with mycobacterium tuberculosis. Front. Microbiol. 8, 2142. doi: 10.3389/fmicb.2017.02142
Zhang, H., Sun, Z., Wei, W., Liu, Z., Fleming, J., Zhang, S., et al. (2014). Identification of serum microRNA biomarkers for tuberculosis using RNA-seq. PloS One 9, e88909. doi: 10.1371/journal.pone.0088909
Zhang, Z. M., Zhang, A. R., Xu, M., Lou, J., Qiu, W. Q. (2017). TLR-4/miRNA-32-5p/FSTL1 signaling regulates mycobacterial survival and inflammatory responses in mycobacterium tuberculosis-infected macrophages. Exp. Cell Res. 352 (2), 313–321. doi: 10.1016/j.yexcr.2017.02.025
Zhang, C, Jiang, H, Wang, P, Liu, H, Sun, X (2017). Transcription factor NF-kappa B represses ANT1 transcription and leads to mitochondrial dysfunctions. Sci Rep. 7, 44708. doi: 10.1038/srep44708
Zhao, Z., Hao, J., Li, X., Chen, Y., Qi, X. (2019). MiR-21-5p regulates mycobacterial survival and inflammatory responses by targeting bcl-2 and TLR4 in mycobacterium tuberculosis-infected macrophages. FEBS Lett. 593 (12), 1326–1335. doi: 10.1002/1873-3468.13438
Zhao, X., Lu, C., Chu, W., Zhang, B., Zhen, Q., Wang, R., et al. (2017). MicroRNA-124 suppresses proliferation and glycolysis in non-small cell lung cancer cells by targeting AKT-GLUT1/HKII. Tumour Biol. 39, 1010428317706215. doi: 10.1177/1010428317706215
Zhao, T., Qiu, Z., Gao, Y. (2020). MiR-137-3p exacerbates the ischemia-reperfusion injured cardiomyocyte apoptosis by targeting KLF15. Naunyn Schmiedebergs Arch. Pharmacol. 393, 1013–1024. doi: 10.1007/s00210-019-01728-w
Zhao, N., Wang, R., Zhou, L., Zhu, Y., Gong, J., Zhuang, S.-M. (2014). MicroRNA-26b suppresses the NF-κB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Mol. cancer. 13, 35. doi: 10.1186/1476-4598-13-35
Zhao, L., Xue, M., Zhang, L., Guo, B, Qin, Y, Jiang, Q, et al. (2020). MicroRNA-4268 inhibits cell proliferation via AKT/JNK signalling pathways by targeting Rab6B in human gastric cancer. Cancer Gene Ther. 27(6), 461–472. doi: 10.1038/s41417-019-0118-6
Zhao, G., Zhang, J.-G., Liu, Y., Qin, Q., Wang, B., Tian, K., et al. (2013). MiR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKα1. Mol. Cancer Ther. 12, 83–93. doi: 10.1158/1535-7163.MCT-12-0534-T
Zheng, D., Zang, Y., Xu, H., Wang, Y., Cao, X., Wang, T., et al. (2019). MicroRNA-214 promotes the calcification of human aortic valve interstitial cells through the acceleration of inflammatory reactions with activated MyD88/NF-κB signaling. Clin. Res. Cardiol. 108 (6), 691–702. doi: 10.1007/s00392-018-1398-9
Zhou, M., Liu, Z., Zhao, Y., Ding, Y., Liu, H., Xi, Y., et al. (2010). MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem. 285, 21496–21507. doi: 10.1074/jbc.M109.083337
Zhou, W., Xu, Y., Zhang, J., Zhang, P., Yao, Z., Yan, Z., et al (2022). MiRNA-363-3p/DUSP10/JNK axis mediates chemoresistance by enhancing DNA damage repair in diffuse large B-cell lymphoma. Leukemia 36 (7), 1861–1869. doi: 10.1038/s41375-022-01565-6
Zhu, W., Lei, J., Bai, X., Wang, R., Ye, Y., Bao, J. (2018). MicroRNA-503 regulates hypoxia-induced cardiomyocytes apoptosis through PI3K/Akt pathway by targeting IGF-1R. Biochem. Biophys. Res. Commun. 506, 1026–1031. doi: 10.1016/j.bbrc.2018.10.160
Zhu, T., Liu, H., Su, L., Xiong, X., Wang, J., Xiao, Y., et al. (2021). MicroRNA-18b-5p downregulation favors Mycobacterium tuberculosis clearance in macrophages via HIF-1α by promoting an inflammatory response. ACS Infect. Dis. 7 (4), 800–810. doi: 10.1021/acsinfecdis.0c00650
Keywords: miRNA, tuberculosis, pathogenesis, signaling pathways, anti-TB treatment
Citation: Davuluri KS and Chauhan DS (2022) microRNAs associated with the pathogenesis and their role in regulating various signaling pathways during Mycobacterium tuberculosis infection. Front. Cell. Infect. Microbiol. 12:1009901. doi: 10.3389/fcimb.2022.1009901
Received: 02 August 2022; Accepted: 03 October 2022;
Published: 27 October 2022.
Edited by:
Bikash R. Giri, Tongji University, ChinaReviewed by:
Md. Aejazur Rahman, Africa Health Research Institute (AHRI), South AfricaCopyright © 2022 Davuluri and Chauhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Devendra S. Chauhan, NjA0ZGtzQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.