
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 14 January 2022
Sec. Bacteria and Host
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.830180
This article is part of the Research TopicObligate Intracellular Bacteria: Evasion and Adaptative Tactics Shaping the Host-Pathogen InterfaceView all 11 articles
Ehrlichia chaffeensis is an obligatory intracellular bacterium that causes human monocytic ehrlichiosis, an emerging, potentially fatal tick-borne infectious disease. The bacterium enters human cells via the binding of its unique outer-membrane invasin EtpE to the cognate receptor DNase X on the host-cell plasma membrane; this triggers actin polymerization and filopodia formation at the site of E. chaffeensis binding, and blocks activation of phagocyte NADPH oxidase that catalyzes the generation of microbicidal reactive oxygen species. Subsequently, the bacterium replicates by hijacking/dysregulating host-cell functions using Type IV secretion effectors. For example, the Ehrlichia translocated factor (Etf)-1 enters mitochondria and inhibits mitochondria-mediated apoptosis of host cells. Etf-1 also induces autophagy mediated by the small GTPase RAB5, the result being the liberation of catabolites for proliferation inside host cells. Moreover, Etf-2 competes with the RAB5 GTPase-activating protein, for binding to RAB5-GTP on the surface of E. chaffeensis inclusions, which blocks GTP hydrolysis and consequently prevents the fusion of inclusions with host-cell lysosomes. Etf-3 binds ferritin light chain to induce ferritinophagy to obtain intracellular iron. To enable E. chaffeensis to rapidly adapt to the host environment and proliferate, the bacterium must acquire host membrane cholesterol and glycerophospholipids for the purpose of producing large amounts of its own membrane. Future studies on the arsenal of unique Ehrlichia molecules and their interplay with host-cell components will undoubtedly advance our understanding of the molecular mechanisms of obligatory intracellular infection and may identify hitherto unrecognized signaling pathways of human hosts. Such data could be exploited for development of treatment and control measures for ehrlichiosis as well as other ailments that potentially could involve the same host-cell signaling pathways that are appropriated by E. chaffeensis.
Ehrlichia chaffeensis is a tick-borne Gram-negative obligatory intracellular bacterium of the family Anaplasmataceae in the order Rickettsiales. Infection causes severe flu-like febrile disease called human monocytic ehrlichiosis (HME), which is often accompanied by hematologic abnormalities and signs similar to those of hepatitis (Dawson et al., 1991; Paddock and Childs, 2003). Tick-borne diseases are on the rise (Biggs et al., 2016; Madison-Antenucci et al., 2020; Alkishe et al., 2021). Discovered in 1986 (Maeda et al., 1987), HME is currently among the most prevalent life-threatening tick-borne zoonoses (Adams et al., 2017). HME diagnosis is challenging, as early signs and symptoms are indistinct or mimic other illnesses. No HME vaccine exists, and the only effective therapy is the broad-spectrum antibiotic doxycycline. However, treatment is often delayed (or even not initiated) owing to misdiagnosis or comorbidity with an unrelated underlying illness or injury, stress, immunosuppression, and/or co-infection with other tick-borne pathogens, which collectively can lead to severe complications or death, with a mortality rate of 1–5% among different populations (Paddock and Childs, 2003). Ehrlichia spp. also can negatively impact livestock agroeconomics and working and companion animals, as the various species and strains of Ehrlichia can cause severe and potentially fatal diseases in animals (Rikihisa, 1991).
Ehrlichia chaffeensis replicates within monocytes and macrophages, which are primary immune cells that recognize pathogen-associated molecular patterns (PAMPs) to unleash potent innate antimicrobial defenses. As a survival strategy, E. chaffeensis has lost genes encoding major PAMPs such as lipopolysaccharide, peptidoglycan, flagella, and common pili (Lin and Rikihisa, 2003). It has a single small (1.18 Mbp) circular genome that lacks most genes for amino-acid biosynthesis and intermediary metabolism (Dunning Hotopp et al., 2006); consequently, the bacterium depends on host cells for these molecules. Molecular and cellular research on E. chaffeensis has revealed unique mechanisms that mediate its parasitism. Foremost is the E. chaffeensis invasin EtpE (entry-triggering protein of Ehrlichia), which binds its cognate host-cell surface receptor DNase X, thereby inducing its internalization. This occurs without eliciting host-derived signals that normally would activate the phagocyte NADPH oxidase 2 (NOX2) complex, that catalyzes the production of microbicidal reactive oxygen species (ROS) from molecular oxygen (Mohan Kumar et al., 2013; Teymournejad et al., 2017). Once internalized, the bacterium replicates within a membrane-bound compartment (inclusion); this is also secluded from components of the NOX2 complex (Lin and Rikihisa, 2007).
Ehrlichia chaffeensis inclusions rapidly fuse with host-cell early endosomes, thereby acquiring early-endosome markers including the small GTPase RAB5 and its effectors such as early endosome antigen 1, VPS34, and Rabankyrin-5. This facilitates subsequent fusion with other early endosomes that contain transferrin receptor (TfR). Exogenous iron-loaded transferrin (Tf) enters into inclusions through the TfR-Tf endosome recycling pathway (Barnewall et al., 1997). Recent studies revealed that the inclusions have features of the early amphisome, which is the vesicular compartment formed by fusion of early endosomes with early autophagosomes, as ATG5, but not LC3 or ATG14L, was also found in inclusions (Lin et al., 2016). This review primarily focuses on recent findings pertaining to invasin, Type IV secretion system (T4SS) effectors, and host-cell membrane lipids that are acquired by E. chaffeensis. Readers are referred to several informative reviews for discussion of other aspects of E. chaffeensis (Paddock and Childs, 2003; Rikihisa, 2010; McBride and Walker, 2011; Rikihisa, 2011; Rikihisa, 2015; McClure et al., 2017; Byerly et al., 2021).
As an obligatory intracellular bacterium, E. chaffeensis cannot survive without entry into permissive host cells. To enter host monocytes and macrophages, E. chaffeensis uses the C-terminus of its unique outer-membrane protein, EtpE-C, to directly bind the host-cell DNase X (DNASE1like1), a cell-surface glycosylphosphatidylinositol-anchored receptor that senses extracellular DNA (Mohan Kumar et al., 2013) (Figure 1). Actin polymerization is not required for E. chaffeensis binding to host cells but is necessary for entry, and thus entry can be inhibited by cytochalasin D (Mohan Kumar et al., 2015).
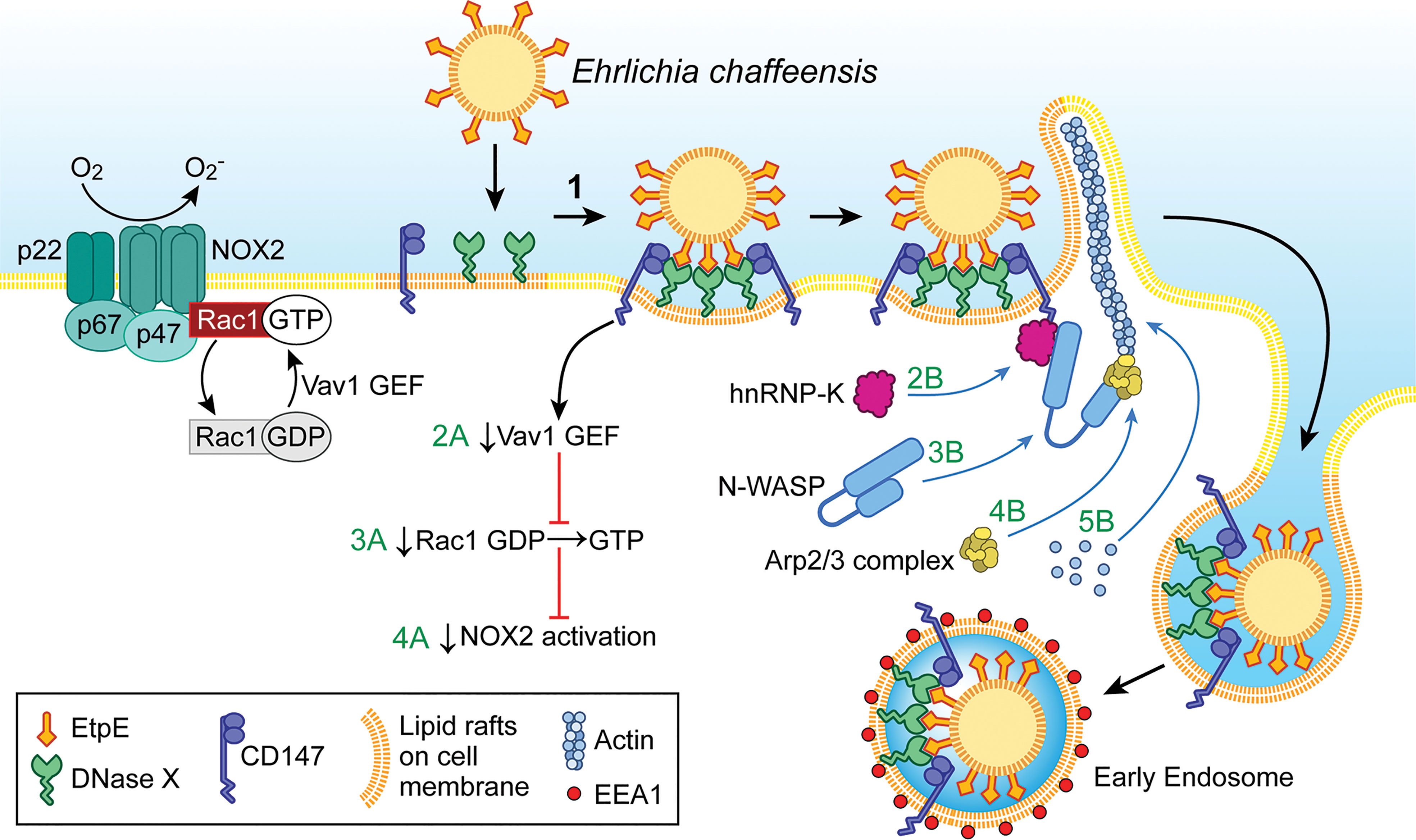
Figure 1 Ehrlichia entry is coupled with blockade of the activation of the phagocyte NADPH oxidase (NOX2) complex. Extracellular E. chaffeensis uses the C-terminal region of its surface protein EtpE to bind DNase X on the host-cell surface. The consequent lateral redistribution of DNase X within dynamic lipid rafts brings CD147 into association with the EtpE–DNase X complex (1). CD147 relays the signal to downregulate Vav1 GEF (guanine-nucleotide exchange factor; 2A) that prevents Rac1 activation (3A) and consequently prevents activation of the NOX2 complex (4A). CD147 also recruits hnRNP-K to bind N-WASP, leading to activation of N-WASP (conformational change) (2B and 3B). Activated N-WASP binds the Arp2/3 actin-nucleation complex (4B), leading to spatiotemporal actin polymerization and filopodia formation to internalize E. chaffeensis into endosomes (5B). The drawing was modified from (Mohan Kumar et al., 2015), copyright 2015 ASM.
EtpE-C-induced actin polymerization is dependent on DNase X as well as activation of the actin nucleation–promoting factor neuronal Wiskott–Aldrich Syndrome protein(N-WASP) (Mohan Kumar et al., 2015). The N-WASP inhibitor wiskostatin or overexpression of the WA domain of N-WASP, which exerts a dominant-negative effect on N-WASP, inhibits actin polymerization and E. chaffeensis entry (Mohan Kumar et al., 2015). How does EtpE-C binding to DNase X on the macrophage surface activate cytoplasmic signaling? EtpE-C binding to DNase X on the macrophage surface recruits three factors, namely the type I transmembrane glycoprotein CD147 (basigin/extracellular matrix metalloproteinase inducer), cytoplasmic heterogeneous nuclear ribonucleoprotein K (hnRNP-K), and N-WASP, to facilitate actin polymerization at the site of E. chaffeensis binding (Mohan Kumar et al., 2015) (Figure 1). CD147 is the key molecule to relay signals started with DNase X membrane receptor to the inside of the cell. Thus, the extracellular bone marrow–derived macrophages from CD147flox/flox-Lyz2-Cre mice, in which Cre expression (driven by the Lyz2 promoter) is used for myeloid cell–specific CD147 knockout, are significantly less susceptible to E. chaffeensis infection (Teymournejad and Rikihisa, 2020). hnRNP-K binds N-WASP and activates the Arp2/3 complex to nucleate actin polymerization in vitro (Yoo et al., 2006), and the intracellular nanoantibody clone #47 (iAB-47), which binds and confines hnRNP-K to the nucleus (Inoue et al., 2007), potently blocks E. chaffeensis entry (Mohan Kumar et al., 2015). Entry requires host-cell energy but not Ehrlichia energy, as demonstrated by the fact that latex beads coated with recombinant EtpE-C could bind DNase X and enter phagocytes as well as non-phagocytic cells that are permissive to E. chaffeensis infection (Mohan Kumar et al., 2013).
Phagocytes, such as monocytes and neutrophils, produce powerful NADPH oxidase (NOX2 complex), a multicomponent enzyme composed of a membrane-bound heterodimeric cytochrome b558 component (gp91phox [NOX2] and p22phox), three cytoplasmic subunits (p67phox, p47phox, and p40phox), and the small GTPase Rac1 or Rac2 (Panday et al., 2015). In resting phagocytes, these NOX2 components are dissociated and hence the enzyme is inactive. Phagocyte-activating agents such as phorbol myristate acetate (PMA), invading pathogens, or an N-formyl peptide can induce rapid assembly of all NOX2 components into a holoenzyme that catalyzes the production of superoxide anion from molecular oxygen (Debeurme et al., 2010). is secreted extracellularly and into the lumen of phagosomes and serves as starting material for the production of microbicidal reactive oxygen species (ROS), including hydrogen peroxide (H2O2), oxidized halogens, hydroxyl radicals, and singlet oxygen (Gabig and Babior, 1981). Paradoxically, E. chaffeensis isolated from host cells is quite sensitive to ROS, and infectivity decreases rapidly when bacteria are exposed to ROS (Lin and Rikihisa, 2007). In fact, the E. chaffeensis genome lacks genes encoding enzymes that facilitate ROS detoxification, free-radical scavenging, repair of ROS-induced damage, and the oxidative stress response (Dunning Hotopp et al., 2006; Lin and Rikihisa, 2007). How, then, does E. chaffeensis prevent or the overcome ROS assault by host macrophages? Remarkably, unlike most other bacteria, E. chaffeensis does not induce ROS production in human monocytes and rapidly blocks generation induced by PMA (Lin and Rikihisa, 2007). This inhibition is specific to monocytes, as E. chaffeensis cannot block ROS production by PMA-stimulated neutrophils, and a host cell-surface protein is required (Lin and Rikihisa, 2007). This surface protein was later revealed to be DNase X, as inhibition of NOX2-complex activation could be initiated by the binding of EtpE-C to DNase X (Teymournejad et al., 2017) (Figure 1). Thus, DNase X–mediated entry and ROS blockade are coupled to ensure E. chaffeensis survival during entry. However, neutrophils do not express DNase X (Teymournejad et al., 2017), which is likely the primary reason why E. chaffeensis neither infects neutrophils nor blocks activation of the NOX2 complex. The binding of E. chaffeensis or of recombinant EtpE-C–coated beads to DNase X can trigger activation of N-WASP (Mohan Kumar et al., 2015). However, N-WASP activation is not involved in the blockade of ROS production initiated by E. chaffeensis or EtpE-C binding to DNase X (Teymournejad et al., 2017). Rac GTPases act as binary switches for the activation of NOX2 (Seifert et al., 1986; Roberts et al., 1999; Zhao et al., 2003; Bokoch and Zhao, 2006). Two Rac isoforms exist, namely Rac1 and Rac2, and Rac2 is the predominant isoform in human neutrophils, whereas Rac1 predominates in monocytes, the latter accounting for 90% of cellular Rac (Zhao et al., 2003). For Rac activation, GTP-for-GDP exchange is facilitated by a membrane-localized, Rac-specific guanine-nucleotide exchange factor (GEF) (Bokoch et al., 1994), and Rac becomes inactivated upon GTP hydrolysis catalyzed by a GTPase-activating protein specific for Rac (Geiszt et al., 2001). Vav1 is a hemopoiesis-specific Rho/Rac guanine-nucleotide exchange factor that plays a prominent role in adhesion-mediated suppression of ROS generation in phagocytes (Zhao et al., 2003). Engagement of EtpE-C with DNase X triggers CD147-dependent suppression of the PMA-induced activation of Vav1 (Teymournejad and Rikihisa, 2020) (Figure 1). Consequently, E. chaffeensis and EtpE-C, upon binding DNase X, block Rac1 activation (Teymournejad and Rikihisa, 2020) (Figure 1). Actin polymerization led by Rac/wave activation is a well-known mechanism for the entry of several intracellular bacteria including Listeria, Yersinia, Salmonella, and Chlamydia into non-phagocytes (Alrutz et al., 2001; Carabeo et al., 2004; Bosse et al., 2007; Humphreys et al., 2013). However, E. chaffeensis does not utilize this mode of entry (Mohan Kumar et al., 2015) to colonize phagocytes, as Rac-dependent actin polymerization and entry would activate phagocyte NOX2 as well.
Immunization of mice and dogs with recombinant EtpE-C significantly inhibits E. chaffeensis infection via intraperitoneal or infected-tick challenge (Mohan Kumar et al., 2013; Budachetri et al., 2020). Thus, EtpE-C could be included in a candidate vaccine to counter tick-transmitted ehrlichiosis.
The T4SS can transfer bacterial proteins or nucleoprotein complexes across the membrane of eukaryotic cells (Alvarez-Martinez and Christie, 2009). The T4SS has several ancestral lineages including the archetype virB/virD system of Agrobacterium tumefaciens and the dot/icm system of Legionella pneumophila, sometimes referred to as T4aSS and T4bSS, respectively (Alvarez-Martinez and Christie, 2009). All members of the order Rickettsiales, which includes E. chaffeensis, have T4aSS (Gillespie et al., 2010). T4SS functions through its effectors/substrates. To date, three T4SS effectors have been experimentally demonstrated, namely Ehrlichia translocated factor (Etf)-1, -2, and -3 (Liu et al., 2012; Lin et al., 2016; Yan et al., 2018; Yan et al., 2021).
Etf-1, -2, and -3 directly bind the E. chaffeensis T4SS coupling ATPase VirD4 and are then transferred from the bacterium into the host-cell cytoplasm by crossing three membranes (inner and outer Ehrlichia membranes, and inclusion membrane) (Liu et al., 2012; Lin et al., 2016; Yan et al., 2018; Yan et al., 2021). Each Etf is required for E. chaffeensis infection, as downregulation of any Etf gene by electroporation of E. chaffeensis with an individual Etf-specific antisense peptide nucleic acid significantly reduces the expression of the corresponding mRNA and hence the bacteria’s ability to infect host cells (Sharma et al., 2017; Yan et al., 2018; Yan et al., 2021). This type of inhibition could be trans-complemented by ectopic expression of the corresponding GFP-coupled Etf in host cells, underscoring the critical roles of the three T4SS effectors in E. chaffeensis replication (Sharma et al., 2017; Yan et al., 2018; Yan et al., 2021). Characteristics of the three T4SS effectors of E. chaffeensis is listed in Table 1.
E. chaffeensis inhibits host-cell apoptosis to maximize bacterial proliferation inside host cells (Liu et al., 2011; Liu et al., 2012). Etf-1 is highly upregulated during early exponential growth of E. chaffeensis in human monocytes (Liu et al., 2012). In Etf-1–transfected mammalian cells, Etf-1 was found to localize to mitochondria and inhibit apoptosis induced by the treatment with etoposide (Liu et al., 2012) (Figure 2). Moreover, in similar experiments with yeast, Etf-1 also localized to mitochondria and inhibited apoptosis induced by heterologous expression of human Bax (Liu et al., 2012). The N-terminal 24 amino-acid residues of Etf-1, especially residue K23, play a critical role in mitochondrial targeting of Etf-1, as deletion mutation of this residue significantly decreased Etf-1 localization to mitochondria (Zhang et al., 2021). The mitochondrial matrix protein manganese superoxide dismutase (MnSOD) maintains a basal level of ROS in cells by scavenging and is essential for maintaining aerobic life (Holley et al., 2012). The MnSOD level was found to increase in E. chaffeensis–infected cells or Etf-1–transfected cells, and the amount of ROS in infected or Etf-1-transfected cells was significantly lower than that in uninfected or control plasmid–transfected cells (Liu et al., 2012; Yan et al., 2021). These data suggest that, by upregulating mitochondrial MnSOD, Etf-1 serves as an antioxidant to prevent ROS-induced cellular damage and apoptosis to allow intracellular infection (Liu et al., 2012).
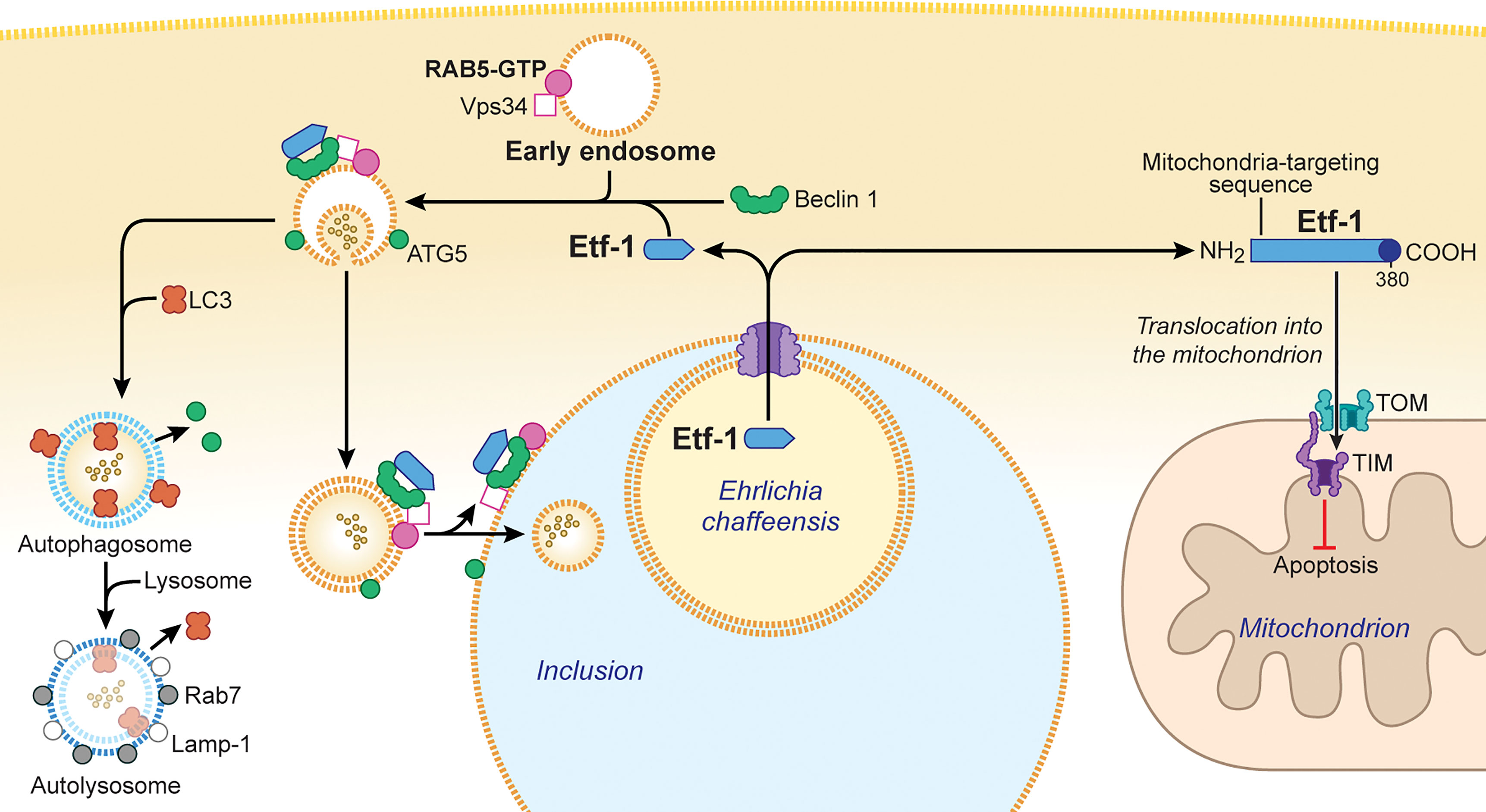
Figure 2 Ehrlichia chaffeensis Etf-1 localizes to mitochondria to block apoptosis of host cells. Alternatively, Etf-1 binds to the Belin 1–VPS34–RAB5-GTP complex and induces RAB5-regulated autophagy. (Right) Etf-1 is depicted in blue with the putative T4SS signal depicted in dark blue. Etf-1 has a mitochondria-targeting presequence and localizes to mitochondria. Mitochondria-localized Etf-1 blocks apoptosis of eukaryotic host cells by preventing loss of mitochondrial membrane potential. TOM: transporter outer membrane complex; TIM: transporter inner membrane complex. (Left) Etf-1 binds the Beclin 1–VPS34–RAB5-GTP complex and induces RAB5-regualted autophagy. Etf-1 autophagosomes are recruited to E. chaffeensis inclusions and deliver captured host cytoplasmic contents. If not recruited to inclusions, Etf-1 autophagosomes mature to autolysosomes, in which captured substrates are degraded and catabolites are released to the cytoplasmic to promote bacterial proliferation. The drawing was modified from (Rikihisa, 2019), copyright 2017 Taylor & Francis, and from (Rikihisa, 2017), copyright 2017 Springer.
To verify the functions of intracellular Etf-1 and investigate the possibility that Etf-1 could be used as a therapeutic target, Etf-1−specific nanobodies were developed by immunizing a llama (Zhang et al., 2021). One particular nanobody could form a stable complex with Etf-1 and thereby block the mitochondrial localization of Etf-1 (Zhang et al., 2021). Intracellular expression of this anti−Etf-1 nanobody inhibited three activities of Etf-1 and E. chaffeensis: upregulation of mitochondrial MnSOD, reduction of intracellular ROS, and inhibition of apoptosis (Zhang et al., 2021). Conjugation of this nanobody to cyclized cell-permeable peptide 12 facilitated effective entrance into mammalian cells, where it abrogated the blockade of apoptosis caused by E. chaffeensis and inhibited infection by E. chaffeensis in cultured cells and in a mouse model of severe combined immunodeficiency (Zhang et al., 2021). Thus, in principle, intracellular nanobodies that interfere with T4SS effector functions could be developed as research tools as well as therapeutic agents.
Autophagy is the process by which eukaryotic cells routinely degrade cellular components to ensure homeostasis and is considered a part of the innate immune response that clears a variety of intracellular pathogens (Levine et al., 2011; Deretic, 2012). However, intracellular replication of E. chaffeensis is enhanced by the autophagy inducer rapamycin and inhibited by the autophagy inhibitor 3-methyl adenine (Lin et al., 2016). Use of Spautin-1 (a cell-permeable inhibitor of the autophagy regulator Beclin 1), Beclin 1 small interfering RNA, or mouse bone marrow–derived macrophages from atg5flox/flox-Lyz2-Cre mice (in which Lyz2 promoter–driven Cre expression is used for myeloid cell–specific Atg5 knockout) demonstrated that autophagy not only enhances ehrlichial infection but also is required for E. chaffeensis replication (Lin et al., 2016). In fact, E. chaffeensis induces a unique type of cellular autophagy to recycle host-cell catabolites for use during its replication (Lin et al., 2016). E. chaffeensis–induced autophagy is independent of the general cellular ubiquitination pathways as well as the canonical autophagy pathway involving MTOR, ULK1, and AMPK (Lin et al., 2016). Etf-1 binds Beclin 1 and VPS34 and activates the class III PtdIns3K (phosphatidylinositol 3-kinase) complex, which is an essential component and master regulator of autophagy initiation (Figure 2), but this Etf-1 complex does not recruit the endoplasmic reticulum resident ATG14L, unlike Ats-1 of Anaplasma phagocytophilum, that also binds Beclin 1 and VPS34 and induces autophagy (Niu et al., 2010). Rather, the Etf-1-Beclin 1 complex recruits RAB5-GTP (Lin et al., 2016) (Figure 2). This type of autophagy is referred to as “RAB5-regulated autophagy” (Ravikumar et al., 2008), as constitutively active RAB5 induces autophagy by binding to the RAB5 effector VPS34, which binds Beclin 1 and hence the class III PtdIns3K complex. Expansion of a polyglutamine tract within the Huntingtin protein due to the mutation causes its accumulation and aggregation in the cytoplasm, leading to the neurodegenerative genetic disorder Huntington’s disease (Raspe et al., 2009). The mutant Huntingtin protein, is poorly degraded in proteasomes but can be degraded via RAB5-regulated autophagy (Ravikumar et al., 2008). Etf-1–induced RAB5-regulated autophagy was found to clear an aggregation-prone mutant Huntingtin protein in a class III PtdIns3K–dependent manner (Lin et al., 2016).
During the exponential growth stage of E. chaffeensis, the concentrations of free/cytoplasmic L-glutamine and L-glutamate in infected human monocytes increase substantially, making them available for ehrlichial growth (Lin et al., 2016). Indeed, host cell–preincorporated radioactive L-glutamine could be readily taken up by E. chaffeensis in an autophagy-dependent manner, and the human cytoplasmic autophagy cargo protein GAPDH could be delivered into E. chaffeensis inclusions as well (Lin et al., 2016). In addition to several early-endosome markers, Etf-1 and the early autophagosome marker ATG5 (but not LC3) are present on the membrane of E. chaffeensis inclusions (Barnewall et al., 1997; Mott et al., 1999; Lin et al., 2016) (Figure 2), and thus the inclusions can be considered as large amphisomes formed by fusion of early endosomes and early autophagosomes. Etf-1–induced autophagy releases host-cell small-molecule catabolites into the host cytoplasm to provide nutrients (e.g., amino acids) to E. chaffeensis. Furthermore, Etf-1–induced autophagy creates a host cytoplasmic space for E. chaffeensis to grow without lysing host cells.
How are the two competing functions of Etf-1 distributed within E. chaffeensis–infected cells? The translocase of the outer membrane of mitochondria (TOM) complex is the main pore for the import of nuclear-encoded proteins into mitochondria, and mitochondrial membrane potential is required for import (Pfanner and Truscott, 2002). The majority of Etf-1 targets mitochondria during the early stage of infection when mitochondrial membrane potential is maximal. As infection progresses, Etf-1 is diverted to autophagosomes as mitochondria begin to lose membrane potential (Wurm et al., 2011). This suggests that host-cell physiologic conditions during infection influence the distribution of Etf-1 between mitochondria and autophagosomes, consequently affecting E. chaffeensis growth.
Although Etf-1 interacts with RAB5-GTP via Beclin 1 and localizes to E. chaffeensis inclusions, inhibition of lysosome fusion with inclusions by keeping RAB5 on inclusions, requires another T4SS effector, Etf-2, because Etf-1-GFP vesicles mature to autolysosomes (Lin et al., 2016).
Ehrlichia chaffeensis sequesters the regulator of endosomal traffic, RAB5, on its membrane-bound inclusions to avoid being routed to host-cell phagolysosomes (Barnewall et al., 1997; Mott et al., 1999). How is RAB5 sequestered on the ehrlichial inclusion membrane? The answer is via its association with Etf-2. Etf-2 directly binds RAB5-GTP on the membrane of early endosomes and of E. chaffeensis–containing inclusions (Yan et al., 2018) (Figure 3). A yeast two-hybrid assay and a microscale thermophoresis assay revealed that Etf-2 binds tightly to RAB5-GTP but not RAB5-GDP (Yan et al., 2018). This is because Etf-2 contains two conserved motifs of RAB GAP Tre2-Bub2-Cdc16 domain, namely an Arg finger and a Gln finger, although it lacks RAB5-specific GAP activity (Yan et al., 2018). Thus, Etf-2 binding to RAB5-GTP blocks RAB5-GTP engagement with RABGAP5 (Figure 3), and consequently RAB5-GTP hydrolysis is delayed on E. chaffeensis inclusions (Yan et al., 2018).
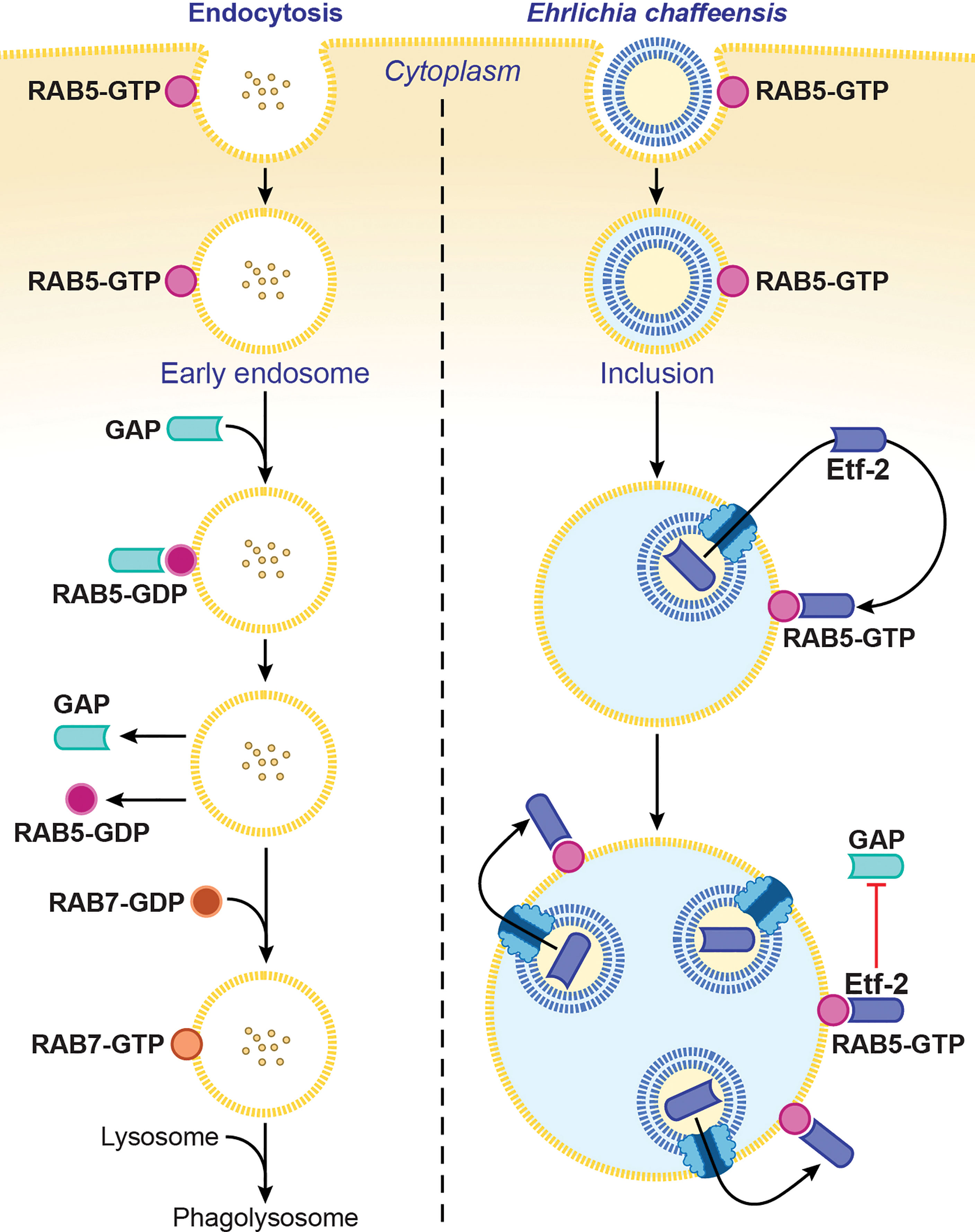
Figure 3 Ehrlichia chaffeensis Etf-2 binds RAB5-GTP and blocks RABGAP5 from acting on RAB5. RAB5-GTP hydrolysis by the RAB5-specific GAP is required for endosome maturation and lysosomal fusion (left). Etf-2 is responsible for blocking lysosomal fusion with E. chaffeensis inclusions by localizing to E. chaffeensis inclusions via binding to RAB5-GTP and competitively blocking RABGAP5 from acting on RAB5 on the inclusion surface (right). The drawing is from (Yan et al., 2018), copyright 2018 PNAS.
Ehrlichia is an obligate aerobe that requires the electron transport chain, thus iron, because its glycolytic pathway is incomplete and it lacks ATP-ADP translocase, unlike Rickettsia and Chlamydia (Dunning Hotopp et al., 2006). Ehrlichia chaffeensis lacks the siderophore biosynthesis pathway and Fe3+ uptake regulator (Dunning Hotopp et al., 2006). Instead, Ehrlichia acquires iron from the host-cell labile cellular iron pool, and pretreating human monocytes with deferoxamine, a membrane-permeable chelator of this iron pool, blocks E. chaffeensis infection (Barnewall and Rikihisa, 1994). Ehrlichia enhances host-cell iron uptake via upregulating TfR mRNA (Barnewall et al., 1999) and acquires iron from holoTf, as E. chaffeensis endosomes intersect with TfR-recycling endosomes and are slightly acidic—enough to release iron from holoTf (Barnewall et al., 1997). In fact, treatment of macrophages with interferon-γ downregulates TfR mRNA and almost completely inhibits Ehrlichia infection, and addition of holoTf abrogates this inhibition (Barnewall and Rikihisa, 1994). However, TfR mRNA levels return to basal level after 24 h post-infection, when bacterial exponential growth begins (Barnewall et al., 1999); at that time, treatment with interferon-γ can no longer inhibit infection (Barnewall and Rikihisa, 1994). How, then, does exponentially growing Ehrlichia acquire iron from host cells? The answer is Etf-3, which binds directly and tightly to ferritin (the primary eukaryotic cytoplasmic iron storage protein) and thereby induces ferritinophagy, a selective form of autophagy by recruiting NCOA4 (nuclear receptor coactivator 4), a cargo receptor that mediates ferritinophagy, and LC3, an autophagosome biogenesis protein (Yan et al., 2021) (Figure 4). Etf-3–induced ferritinophagy causes ferritin degradation and significantly increases the labile cellular iron pool, which can feed E. chaffeensis (Figure 4). Indeed, an increase in cellular ferritin by adding ferric ammonium citrate to the culture medium, or overexpression of Etf-3 or NCOA4, enhances E. chaffeensis proliferation, whereas knockdown of Etf-3 in Ehrlichia via transfection with a plasmid encoding an Etf-3 antisense peptide nucleic acid inhibits Ehrlichia proliferation (Yan et al., 2021).
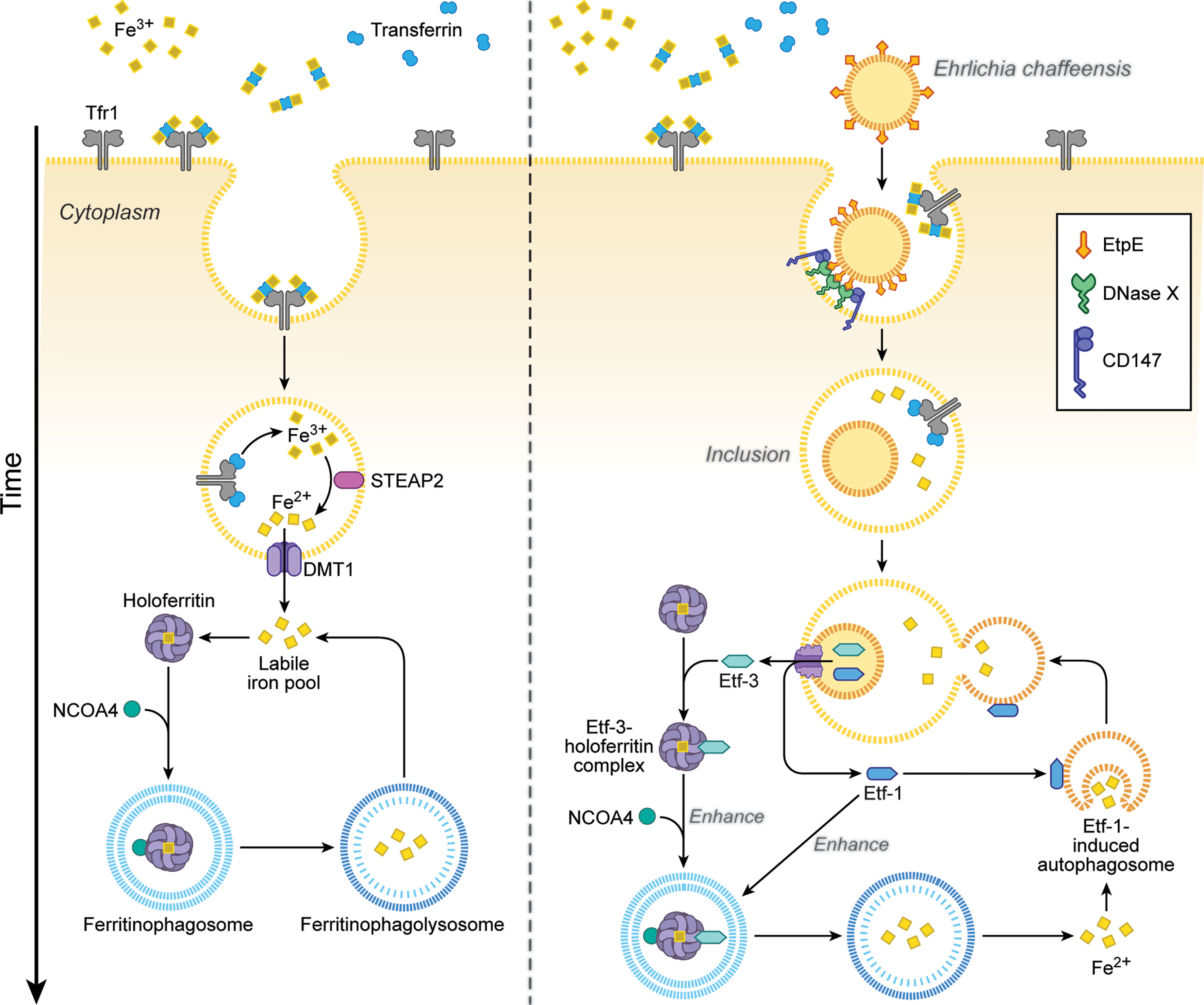
Figure 4 Ehrlichia chaffeensis Etf-3 binds ferritin light chain to induce ferritinophagy to increase the labile-iron pool for acquisition of iron by E. chaffeensis. Iron homeostasis is tightly regulated in host cells to maintain the labile cellular iron pool (left). Etf-3 directly binds ferritin via ferritin light chain and induces ferritinophagy to increase the labile cellular iron pool, thereby providing Fe2+ for E. chaffeensis proliferation. Etf-1–induced autophagy synergizes with Etf-3 to deliver extra Fe2+ to Ehrlichia inclusions (right). Tf: transferrin, which binds Fe3+ and transports it into cells. TfR: transferrin receptor, which binds and delivers iron-saturated transferrin via endocytosis. STEAP2: Six-transmembrane epithelial antigen of prostate-2, a metalloreductase that reduces Fe3+ to Fe2+. DMT1: Divalent metal transporter 1 that transports Fe2+ from endosomes to the cytoplasm. NCOA4: Nuclear receptor coactivator 4, a cargo receptor that mediates ferritinophagy. The drawing is from (Yan et al., 2021), copyright 2021 PNAS.
Excessive ferritinophagy induces the generation of toxic ROS, which could presumably kill both Ehrlichia and host cells. During Ehrlichia proliferation, however, there is concomitant upregulation of Ehrlichia Fe-superoxide dismutase, the gene that is co-regulated with the Ehrlichia T4SS operon, and increase in mitochondrial MnSOD in response to the co-secreted Etf-1 (Yan et al., 2021). Consequently, despite enhanced ferritinophagy, cellular ROS levels are reduced in Ehrlichia-infected cells compared with uninfected cells (Yan et al., 2021). Thus, Ehrlichia robs host-cell iron sequestered in ferritin without killing the host cell.
The E. chaffeensis cell membrane is cholesterol-rich (Lin and Rikihisa, 2003), but the bacterium cannot synthesize cholesterol and partially lacks genes for glycerophospholipid biosynthesis (Lin et al., 2020). As small Gram-negative bacteria, Ehrlichia spp. require abundant membrane lipids for rapid intracellular proliferation. Thus, E. chaffeensis must acquire these membrane lipids from host cells. Furthermore, by incorporating eukaryotic lipids such as phosphatidylcholine and cholesterol, E. chaffeensis mimics the eukaryotic plasma membrane and, by doing so, adapts to the cellular environment of the host. Indeed, exogenous 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD)-phosphatidylcholine, Bodipy-phosphatidylethanolamine, and Bodipy (TopFluor)-cholesterol are rapidly trafficked to ehrlichia inclusions in infected cells (Figure 5). DiI (3,3’-dioctadecylindocarbocyanine)-prelabeled host-cell membranes are unidirectionally trafficked to Ehrlichia inclusions and the bacterial membrane (Figure 5), but DiI-prelabeled Ehrlichia membranes are not reversibly trafficked to host-cell membranes (Lin et al., 2020). The trafficking of host-cell membranes to Ehrlichia inclusions is dependent on both the host endocytic and autophagic pathways as well as bacterial protein synthesis, as the respective inhibitors block the trafficking of DiI-labeled host membranes to Ehrlichia as well as infection (Lin et al., 2020). Cryosections of infected cells show numerous membranous vesicles inside Ehrlichia inclusions as well as multivesicular bodies docked on the inclusion surface, both of which can be labeled by GFP-tagged 2×FYVE protein that binds to phosphatidylinositol 3-phosphate, a product of PtdIns3K activity (Lin et al., 2020). Focused ion-beam scanning electron microscopy of infected cells has validated the existence of numerous membranous structures inside bacterial inclusions (Lin et al., 2020). These results support the notion that Ehrlichia inclusions are amphisomes formed through fusion of early endosomes, multivesicular bodies, and early autophagosomes induced by Etf-1, and they provide the host-cell membrane glycerophospholipids and cholesterol necessary for bacterial proliferation.
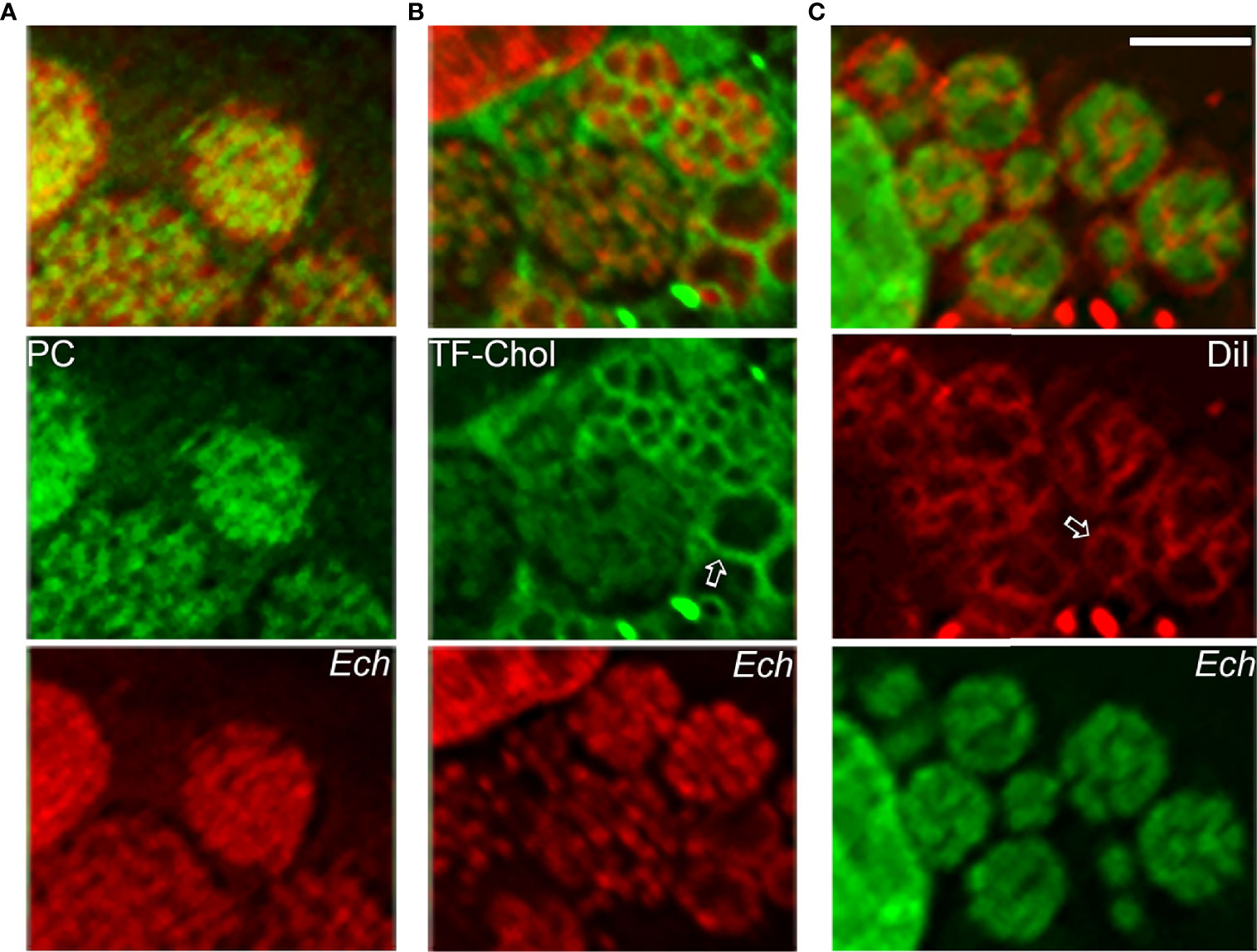
Figure 5 Host-cell membrane lipids (cholesterol and glycerophospholipids) are trafficked to the membrane of E. chaffeensis and its inclusions. NBD-phosphatidylcholine (A), TopFluor-cholesterol (B), or DiI (C) is added to E. chaffeensis–infected RF/6A cells to monitor the intracellular distribution of the fluorescent lipids. DNAs of bacteria and host are stained with Hoechst 33342 (pseudocolored green and red, respectively). Note labeling of densely packed intraluminal E. chaffeensis (Ech, small cocci stained with Hoechst 33342) with the three fluorescent lipids (A–C) and labeling of the bacterial inclusion membrane (open arrows) and intraluminal membranes with TopFluor-cholesterol (B) and DiI (C). Bar: 3 µm. The drawing was modified from (Lin et al., 2020), copyright 2020 PNAS.
Ehrlichia species propagate via perpetual transmission between ticks and mammalian hosts and can proliferate in each of these two distinct environments. Owing to multiple technical limitations, little is known about the bacterial components that enable Ehrlichia to thrive throughout this lifecycle. Recently, Himer1 transposon mutagenesis was successfully applied to E. chaffeensis as well as other Ehrlichia species, and functional knockout mutants have been cloned (Cheng et al., 2013; Bekebrede et al., 2020). Moreover, the application of targeted mutagenesis techniques to Rickettsia and Ehrlichia species is on the horizon (McClure et al., 2017). Combined with advanced analysis of functional genes in ticks along with molecular and cellular techniques to manipulate ticks, it is expected that Ehrlichia Himer1 transposon insertional mutant libraries will facilitate this line of investigation. Further experimental discoveries of bacterial factors and their functions during the natural life cycle of Ehrlichia—in which humans are merely accidental hosts—are expected to reveal the remarkable molecular evolution of these tick-borne pathogens and inform the development of effective therapeutic strategies and preventative measures for diseases caused by Ehrlichia species.
The author confirms being the sole contributor of this review and has approved it for publication.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The author thanks previous and current laboratory members, and T. Vojt for help in preparing the figures. Some of the studies from the author’s laboratory reported in this review were supported by grants (R01AI121124, R21AI146736 and R01AI151065) from the National Institutes of Health and CDMRP Award W81XWH-17-1-0519 from the Department of Defense.
Adams, D. A., Thomas, K. R., Jajosky, R. A., Foster, L., Baroi, G., Sharp, P., et al. (2017). Summary of Notifiable Infectious Diseases and Conditions - United States, 2015. MMWR Morb Mortal Wkly Rep. 64, 1–143. doi: 10.15585/mmwr.mm6453a1
Alkishe, A., Raghavan, R. K., Peterson, A. T. (2021). Likely Geographic Distributional Shifts Among Medically Important Tick Species and Tick-Associated Diseases Under Climate Change in North America: A Review. Insects 12, 1–25. doi: 10.3390/insects12030225
Alrutz, M. A., Srivastava, A., Wong, K. W., D’Souza-Schorey, C., Tang, M., Ch’Ng, L. E., et al. (2001). Efficient Uptake of Yersinia Pseudotuberculosis via Integrin Receptors Involves a Rac1-Arp 2/3 Pathway That Bypasses N-WASP Function. Mol. Microbiol. 42, 689–703. doi: 10.1046/j.1365-2958.2001.02676.x
Alvarez-Martinez, C. E., Christie, P. J. (2009). Biological Diversity of Prokaryotic Type IV Secretion Systems. Microbiol. Mol. Biol. Rev. 73, 775–808. doi: 10.1128/MMBR.00023-09
Barnewall, R. E., Ohashi, N., Rikihisa, Y. (1999). Ehrlichia chaffeensis and E. sennetsu, But Not the Human Granulocytic Ehrlichiosis Agent, Colocalize With Transferrin Receptor and Up-Regulate Transferrin Receptor mRNA by Activating Iron-Responsive Protein 1. Infect. Immun. 67, 2258–2265. doi: 10.1128/IAI.67.5.2258-2265.1999
Barnewall, R. E., Rikihisa, Y. (1994). Abrogation of Gamma Interferon-Induced Inhibition of Ehrlichia Chaffeensis Infection in Human Monocytes With Iron-Transferrin. Infect. Immun. 62, 4804–4810. doi: 10.1128/iai.62.11.4804-4810.1994
Barnewall, R. E., Rikihisa, Y., Lee, E. H. (1997). Ehrlichia chaffeensis Inclusions Are Early Endosomes Which Selectively Accumulate Transferrin Receptor. Infect. Immun. 65, 1455–1461. doi: 10.1128/iai.65.4.1455-1461.1997
Bekebrede, H., Lin, M., Teymournejad, O., Rikihisa, Y. (2020). Discovery of In Vivo Virulence Genes of Obligatory Intracellular Bacteria by Random Mutagenesis. Front. Cell Infect. Microbiol. 10, 2. doi: 10.3389/fcimb.2020.00002
Biggs, H. M., Behravesh, C. B., Bradley, K. K., Dahlgren, F. S., Drexler, N. A., Dumler, J. S., et al. (2016). Diagnosis and Management of Tickborne Rickettsial Diseases: Rocky Mountain Spotted Fever and Other Spotted Fever Group Rickettsioses, Ehrlichioses, and Anaplasmosis - United States. MMWR Recomm Rep. 65, 1–44. doi: 10.15585/mmwr.rr6502a1
Bokoch, G. M., Bohl, B. P., Chuang, T. H. (1994). Guanine Nucleotide Exchange Regulates Membrane Translocation of Rac/Rho GTP-Binding Proteins. J. Biol. Chem. 269, 31674–31679. doi: 10.1016/S0021-9258(18)31748-4
Bokoch, G. M., Zhao, T. (2006). Regulation of the Phagocyte NADPH Oxidase by Rac GTPase. Antioxid Redox Signal 8, 1533–1548. doi: 10.1089/ars.2006.8.1533
Bosse, T., Ehinger, J., Czuchra, A., Benesch, S., Steffen, A., Wu, X., et al. (2007). Cdc42 and Phosphoinositide 3-Kinase Drive Rac-Mediated Actin Polymerization Downstream of C-Met in Distinct and Common Pathways. Mol. Cell Biol. 27, 6615–6628. doi: 10.1128/MCB.00367-07
Budachetri, K., Teymournejad, O., Lin, M., Yan, Q., Mestres-Villanueva, M., Brock, G. N., et al. (2020). An Entry-Triggering Protein of Ehrlichia Is a New Vaccine Candidate Against Tick-Borne Human Monocytic Ehrlichiosis. mBio 11, 1–13. doi: 10.1128/mBio.00895-20
Byerly, C. D., Patterson, L. L., McBride, J. W. (2021). Ehrlichia TRP Effectors: Moonlighting, Mimicry and Infection. Pathog. Dis. 79, 1–16. doi: 10.1093/femspd/ftab026
Carabeo, R. A., Grieshaber, S. S., Hasenkrug, A., Dooley, C., Hackstadt, T. (2004). Requirement for the Rac GTPase in Chlamydia Trachomatis Invasion of Non-Phagocytic Cells. Traffic 5, 418–425. doi: 10.1111/j.1398-9219.2004.00184.x
Cheng, C., Nair, A. D., Indukuri, V. V., Gong, S., Felsheim, R. F., Jaworski, D., et al. (2013). Targeted and Random Mutagenesis of Ehrlichia Chaffeensis for the Identification of Genes Required for In Vivo Infection. PloS Pathog. 9, e1003171. doi: 10.1371/journal.ppat.1003171
Dawson, J. E., Anderson, B. E., Fishbein, D. B., Sanchez, J. L., Goldsmith, C. S., Wilson, K. H., et al. (1991). Isolation and Characterization of an Ehrlichia sp. From a Patient Diagnosed With Human Ehrlichiosis. J. Clin. Microbiol. 29, 2741–2745. doi: 10.1128/jcm.29.12.2741-2745.1991
Debeurme, F., Picciocchi, A., Dagher, M. C., Grunwald, D., Beaumel, S., Fieschi, F., et al. (2010). Regulation of NADPH Oxidase Activity in Phagocytes: Relationship Between FAD/NADPH Binding and Oxidase Complex Assembly. J. Biol. Chem. 285, 33197–33208. doi: 10.1074/jbc.M110.151555
Deretic, V. (2012). Autophagy: An Emerging Immunological Paradigm. J. Immunol. 189, 15–20. doi: 10.4049/jimmunol.1102108
Dunning Hotopp, J. C., Lin, M., Madupu, R., Crabtree, J., Angiuoli, S. V., Eisen, J., et al. (2006). Comparative Genomics of Emerging Human Ehrlichiosis Agents. PloS Genet. 2, e21. doi: 10.1371/journal.pgen.0020021
Gabig, T. G., Babior, B. M. (1981). The Killing of Pathogens by Phagocytes. Annu. Rev. Med. 32, 313–326. doi: 10.1146/annurev.me.32.020181.001525
Geiszt, M., Dagher, M. C., Molnar, G., Havasi, A., Faure, J., Paclet, M. H., et al. (2001). Characterization of Membrane-Localized and Cytosolic Rac-GTPase-Activating Proteins in Human Neutrophil Granulocytes: Contribution to the Regulation of NADPH Oxidase. Biochem. J. 355, 851–858. doi: 10.1042/bj3550851
Gillespie, J. J., Brayton, K. A., Williams, K. P., Diaz, M. A., Brown, W. C., Azad, A. F., et al. (2010). Phylogenomics Reveals a Diverse Rickettsiales Type IV Secretion System. Infect. Immun. 78, 1809–1823. doi: 10.1128/IAI.01384-09
Holley, A. K., Dhar, S. K., Xu, Y., St Clair, D. K. (2012). Manganese Superoxide Dismutase: Beyond Life and Death. Amino Acids 42, 139–158. doi: 10.1007/s00726-010-0600-9
Humphreys, D., Davidson, A. C., Hume, P. J., Makin, L. E., Koronakis, V. (2013). Arf6 Coordinates Actin Assembly Through the WAVE Complex, a Mechanism Usurped by Salmonella to Invade Host Cells. Proc. Natl. Acad. Sci. U. S. A. 110, 16880–16885. doi: 10.1073/pnas.1311680110
Inoue, A., Sawata, S. Y., Taira, K., Wadhwa, R. (2007). Loss-Of-Function Screening by Randomized Intracellular Antibodies: Identification of hnRNP-K as a Potential Target for Metastasis. Proc. Natl. Acad. Sci. U. S. A. 104, 8983–8988. doi: 10.1073/pnas.0607595104
Levine, B., Mizushima, N., Virgin, H. W. (2011). Autophagy in Immunity and Inflammation. Nature 469, 323–335. doi: 10.1038/nature09782
Lin, M., Grandinetti, G., Hartnell, L. M., Bliss, D., Subramaniam, S., Rikihisa, Y. (2020). Host Membrane Lipids Are Trafficked to Membranes of Intravacuolar Bacterium Ehrlichia Chaffeensis. Proc. Natl. Acad. Sci. U. S. A. 117, 8032–8043. doi: 10.1073/pnas.1921619117
Lin, M., Liu, H., Xiong, Q., Niu, H., Cheng, Z., Yamamoto, A., et al. (2016). Ehrlichia Secretes Etf-1 to Induce Autophagy and Capture Nutrients for Its Growth Through RAB5 and Class III Phosphatidylinositol 3-Kinase. Autophagy 12, 2145–2166. doi: 10.1080/15548627.2016.1217369
Lin, M., Rikihisa, Y. (2003). Ehrlichia chaffeensis and Anaplasma phagocytophilum Lack Genes for Lipid A Biosynthesis and Incorporate Cholesterol for Their Survival. Infect. Immun. 71, 5324–5331. doi: 10.1128/IAI.71.9.5324-5331.2003
Lin, M., Rikihisa, Y. (2007). Degradation of P22phox and Inhibition of Superoxide Generation by Ehrlichia chaffeensis in Human Monocytes. Cell Microbiol. 9, 861–874. doi: 10.1111/j.1462-5822.2006.00835.x
Liu, H., Bao, W., Lin, M., Niu, H., Rikihisa, Y. (2012). Ehrlichia Type IV Secretion Effector ECH0825 Is Translocated to Mitochondria and Curbs ROS and Apoptosis by Upregulating Host MnSOD. Cell Microbiol. 14, 1037–1050. doi: 10.1111/j.1462-5822.2012.01775.x
Liu, Y., Zhang, Z., Jiang, Y., Zhang, L., Popov, V. L., Zhang, J., et al. (2011). Obligate Intracellular Bacterium Ehrlichia Inhibiting Mitochondrial Activity. Microbes Infect. 13, 232–238. doi: 10.1016/j.micinf.2010.10.021
Madison-Antenucci, S., Kramer, L. D., Gebhardt, L. L., Kauffman, E. (2020). Emerging Tick-Borne Diseases. Clin. Microbiol. Rev. 33, 1–18. doi: 10.1128/CMR.00083-18
Maeda, K., Markowitz, N., Hawley, R. C., Ristic, M., Cox, D., McDade, J. E. (1987). Human Infection With Ehrlichia canis, A Leukocytic Rickettsia. N. Engl. J. Med. 316, 853–856. doi: 10.1056/NEJM198704023161406
McBride, J. W., Walker, D. H. (2011). Molecular and Cellular Pathobiology of Ehrlichia Infection: Targets for New Therapeutics and Immunomodulation Strategies. Expert Rev. Mol. Med. 13, e3. doi: 10.1017/S1462399410001730
McClure, E. E., Chavez, A. S. O., Shaw, D. K., Carlyon, J. A., Ganta, R. R., Noh, S. M., et al. (2017). Engineering of Obligate Intracellular Bacteria: Progress, Challenges and Paradigms. Nat. Rev. Microbiol. 15, 544–558. doi: 10.1038/nrmicro.2017.59
Mohan Kumar, D., Lin, M., Xiong, Q., Webber, M. J., Kural, C., Rikihisa, Y. (2015). EtpE Binding to DNase X Induces Ehrlichial Entry via CD147 and hnRNP-K Recruitment, Followed by Mobilization of N-WASP and Actin. MBio 6, 1–15. doi: 10.1128/mBio.01541-15
Mohan Kumar, D., Yamaguchi, M., Miura, K., Lin, M., Los, M., Coy, J. F., et al. (2013). Ehrlichia chaffeensis Uses Its Surface Protein EtpE to Bind GPI-Anchored Protein DNase X and Trigger Entry Into Mammalian Cells. PloS Pathog. 9, e1003666. doi: 10.1371/journal.ppat.1003666
Mott, J., Barnewall, R. E., Rikihisa, Y. (1999). Human Granulocytic Ehrlichiosis Agent and Ehrlichia chaffeensis Reside in Different Cytoplasmic Compartments in HL-60 Cells. Infect. Immun. 67, 1368–1378. doi: 10.1128/IAI.67.3.1368-1378.1999
Niu, H., Kozjak-Pavlovic, V., Rudel, T., Rikihisa, Y. (2010). Anaplasma phagocytophilum Ats-1 Is Imported Into Host Cell Mitochondria and Interferes With Apoptosis Induction. PloS Pathog. 6, e1000774. doi: 10.1371/journal.ppat.1000774
Paddock, C. D., Childs, J. E. (2003). Ehrlichia chaffeensis: A Prototypical Emerging Pathogen. Clin. Microbiol. Rev. 16, 37–64. doi: 10.1128/CMR.16.1.37-64.2003
Panday, A., Sahoo, M. K., Osorio, D., Batra, S. (2015). NADPH Oxidases: An Overview From Structure to Innate Immunity-Associated Pathologies. Cell Mol. Immunol. 12, 5–23. doi: 10.1038/cmi.2014.89
Pfanner, N., Truscott, K. N. (2002). Powering Mitochondrial Protein Import. Nat. Struct. Biol. 9, 234–236. doi: 10.1038/nsb0402-234
Raspe, M., Gillis, J., Krol, H., Krom, S., Bosch, K., van Veen, H., et al. (2009). Mimicking Proteasomal Release of Polyglutamine Peptides Initiates Aggregation and Toxicity. J. Cell Sci. 122, 3262–3271. doi: 10.1242/jcs.045567
Ravikumar, B., Imarisio, S., Sarkar, S., O’Kane, C. J., Rubinsztein, D. C. (2008). Rab5 Modulates Aggregation and Toxicity of Mutant Huntingtin Through Macroautophagy in Cell and Fly Models of Huntington Disease. J. Cell Sci. 121, 1649–1660. doi: 10.1242/jcs.025726
Rikihisa, Y. (1991). The Tribe Ehrlichieae and Ehrlichial Diseases. Clin. Microbiol. Rev. 4, 286–308. doi: 10.1128/CMR.4.3.286
Rikihisa, Y. (2010). Anaplasma phagocytophilum and Ehrlichia chaffeensis: Subversive Manipulators of Host Cells. Nat. Rev. Microbiol. 8, 328–339. doi: 10.1038/nrmicro2318
Rikihisa, Y. (2011). Mechanisms of Obligatory Intracellular Infection With Anaplasma Phagocytophilum. Clin. Microbiol. Rev. 24, 469–489. doi: 10.1128/CMR.00064-10
Rikihisa, Y. (2015). Molecular Pathogenesis of Ehrlichia chaffeensis Infection. Annu. Rev. Microbiol. 69, 283–304. doi: 10.1146/annurev-micro-091014-104411
Rikihisa, Y. (2017). Role and Function of the Type IV Secretion System in Anaplasma and Ehrlichia Species. Curr. Top. Microbiol. Immunol. 413, 297–321. doi: 10.1007/978-3-319-75241-9_12
Rikihisa, Y. (2019). Subversion of RAB5-Regulated Autophagy by the Intracellular Pathogen Ehrlichia chaffeensis. Small GTPases 10, 343–349. doi: 10.1080/21541248.2017.1332506
Roberts, A. W., Kim, C., Zhen, L., Lowe, J. B., Kapur, R., Petryniak, B., et al. (1999). Deficiency of the Hematopoietic Cell-Specific Rho Family GTPase Rac2 Is Characterized by Abnormalities in Neutrophil Function and Host Defense. Immunity 10, 183–196. doi: 10.1016/S1074-7613(00)80019-9
Seifert, R., Rosenthal, W., Schultz, G. (1986). Guanine Nucleotides Stimulate NADPH Oxidase in Membranes of Human Neutrophils. FEBS Lett. 205, 161–165. doi: 10.1016/0014-5793(86)80886-9
Sharma, P., Teymournejad, O., Rikihisa, Y. (2017). Peptide Nucleic Acid Knockdown and Intra-Host Cell Complementation of Ehrlichia Type IV Secretion System Effector. Front. Cell Infect. Microbiol. 7, 228. doi: 10.3389/fcimb.2017.00228
Teymournejad, O., Lin, M., Rikihisa, Y. (2017). Ehrlichia Chaffeensis and Its Invasin EtpE Block Reactive Oxygen Species Generation by Macrophages in a DNase X-Dependent Manner. MBio 8. doi: 10.1128/mBio.01551-17
Teymournejad, O., Rikihisa, Y. (2020). Ehrlichia chaffeensis Uses an Invasin To Suppress Reactive Oxygen Species Generation by Macrophages via CD147-Dependent Inhibition of Vav1 To Block Rac1 Activation. mBio 11, 1–14. doi: 10.1128/mBio.00267-20
Wurm, C. A., Neumann, D., Lauterbach, M. A., Harke, B., Egner, A., Hell, S. W., et al. (2011). Nanoscale Distribution of Mitochondrial Import Receptor Tom20 Is Adjusted to Cellular Conditions and Exhibits an Inner-Cellular Gradient. Proc. Natl. Acad. Sci. U. S. A. 108, 13546–13551. doi: 10.1073/pnas.1107553108
Yan, Q., Lin, M., Huang, W., Teymournejad, O., Johnson, J. M., Hays, F. A., et al. (2018). Ehrlichia Type IV Secretion System Effector Etf-2 Binds to Active RAB5 and Delays Endosome Maturation. Proc. Natl. Acad. Sci. U. S. A. 115, E8977–E8986. doi: 10.1073/pnas.1806904115
Yan, Q., Zhang, W., Lin, M., Teymournejad, O., Budachetri, K., Lakritz, J., et al. (2021). Iron Robbery by Intracellular Pathogen via Bacterial Effector-Induced Ferritinophagy. Proc. Natl. Acad. Sci. U. S. A. 118, e2026598118. doi: 10.1073/pnas.2026598118
Yoo, Y., Wu, X., Egile, C., Li, R., Guan, J. L. (2006). Interaction of N-WASP With hnRNPK and Its Role in Filopodia Formation and Cell Spreading. J. Biol. Chem. 281, 15352–15360. doi: 10.1074/jbc.M511825200
Zhang, W., Lin, M., Yan, Q., Budachetri, K., Hou, L., Sahni, A., et al. (2021). An Intracellular Nanobody Targeting T4SS Effector Inhibits Ehrlichia Infection. Proc. Natl. Acad. Sci. U. S. A. 118, e2024102118. doi: 10.1073/pnas.2024102118
Zhao, T., Benard, V., Bohl, B. P., Bokoch, G. M. (2003). The Molecular Basis for Adhesion-Mediated Suppression of Reactive Oxygen Species Generation by Human Neutrophils. J. Clin. Invest. 112, 1732–1740. doi: 10.1172/JCI19108
Keywords: Ehrlichia chaffeensis, invasin, ROS, type IV secretion effector, RAB5, autophagy, ferritinophagy, membrane cholesterol
Citation: Rikihisa Y (2022) The “Biological Weapons” of Ehrlichia chaffeensis: Novel Molecules and Mechanisms to Subjugate Host Cells. Front. Cell. Infect. Microbiol. 11:830180. doi: 10.3389/fcimb.2021.830180
Received: 06 December 2021; Accepted: 20 December 2021;
Published: 14 January 2022.
Edited by:
Daniel E. Voth, University of Arkansas for Medical Sciences, United StatesReviewed by:
Damien F. Meyer, CIRAD, UMR ASTRE CIRAD/INRAe, FranceCopyright © 2022 Rikihisa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasuko Rikihisa, cmlraWhpc2EuMUBvc3UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.