- 1Centre of Biological Engineering, Laboratory of Research in Biofilms Rosário Oliveira (LIBRO), University of Minho, Braga, Portugal
- 2Institut für Medizinische Mikrobiologie, Virologie und Hygiene, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany
- 3Instituto de Investigação e Inovação em Saúde (i3S), Universidade do Porto, Porto, Portugal
- 4Instituto de Biologia Molecular e Celular (IBMC), Universidade do Porto, Porto, Portugal
- 5Instituto de Ciências Biomédicas de Abel Salazar, Universidade do Porto (ICBAS-UP), Porto, Portugal
Staphylococcus epidermidis is one of the most important commensal microorganisms of human skin and mucosae. However, this bacterial species is also the cause of severe infections in immunocompromised patients, specially associated with the utilization of indwelling medical devices, that often serve as a scaffold for biofilm formation. S. epidermidis strains are often multidrug resistant and its association with biofilm formation makes these infections hard to treat. Their remarkable ability to form biofilms is widely regarded as its major pathogenic determinant. Although a significant amount of knowledge on its biofilm formation mechanisms has been achieved, we still do not understand how the species survives when exposed to the host harsh environment during invasion. A previous RNA-seq study highlighted that iron-metabolism associated genes were the most up-regulated bacterial genes upon contact with human blood, which suggested that iron acquisition plays an important role in S. epidermidis biofilm development and escape from the host innate immune system. In this perspective article, we review the available literature on the role of iron metabolism on S. epidermidis pathogenesis and propose that exploiting its dependence on iron could be pursued as a viable therapeutic alternative.
Introduction
Health care-associated infections (HAIs) are a significant cause of morbidity and mortality worldwide and represent an increasing problem in modern medicine (Haque et al., 2018). More than 4 million patients are affected by HAIs every year in Europe, with an average prevalence rate of 7.1%, which accounts for an annual cost of approximately 7 billion (Allegranzi et al., 2011; European Centre for Disease Prevention and Control, 2018). In developing countries, the estimated prevalence rates of HAIs are even higher, ranging from 5.7% to 19.1% (pooled prevalence rate of 10.1%) (Pittet et al., 2008; Allegranzi et al., 2011). Patients admitted to intensive care units are particularly susceptible to these infections, not only due to their immunocompromised status, but also due to the extensive use of invasive procedures and devices (e.g., mechanical ventilators and catheters) (Vincent, 2003). Bloodstream infections, surgical site infections and urinary tract infections account for the majority of HAIs (Magill et al., 2014). According to the type of infection, the causative agents may vary, although staphylococci, and in particular S. epidermidis, play an important role in HAIs episodes (Zarb et al., 2012). The establishment of S. epidermidis as a successful nosocomial pathogen stems from its intrinsic ability to form biofilms on the surface of implantable medical devices, which is often accompanied by multidrug resistance (Águila-Arcos et al., 2017) and increased antibiotic tolerance (Dengler Haunreiter et al., 2019), rendering the treatment of HAIs extremely challenging. Therefore, a deeper understanding of their pathogenic mechanisms, particularly those associated with biofilm formation, is, more than ever before, pivotal in the identification of new antibacterial drug targets.
Bacterial Biofilms
Our perception of how bacterial growth takes place in the environment, particularly during infection, has changed over the years. If on one hand bacteria were initially thought to grow mostly under a planktonic mode of growth, as it occurs artificially in a liquid culture, it is currently well-established that forming a biofilm is the preferred mode of growth for most bacterial species (Flemming et al., 2016). The classic definition of a biofilm is a structured community of microorganisms adhered to each other and/or to a surface, which is often embedded in a self-produced matrix of extracellular polymeric substance (Donlan and Costerton, 2002). Despite its frequent association with infectious diseases, this mode of growth is also adopted by non-pathogenic bacteria in different locations of the human body, such as skin (Brandwein et al., 2016) or gastrointestinal tract (De Vos, 2015). Therefore, the ability to grow as a biofilm can be generally regarded as a way bacteria employ to cope with harsh environments (Flemming et al., 2016). In S. epidermidis biofilm assembly follows a basic stepwise process comprising distinct stages: primary attachment of cells to a surface, accumulation of cells in multiple layers and maturation of the biofilm structure, and detachment of cells from the biofilm and their dispersal (Fey and Olson, 2010).
S. epidermidis Biofilms in Infection
During infection, S. epidermidis face severe restriction in the availability of essential nutrients, a phenomenon usually referred to as “nutritional immunity” (Cassat and Skaar, 2013). Iron is one of those essential nutrients, which bacteria must acquire for their own cellular functions (Andrews et al., 2003). As a result, iron acquisition has been considered as a key process in bacterial pathogenicity (Skaar, 2010; Oliveira et al., 2021a). Interestingly, there has been increasing evidence suggesting that iron may also exert a modulatory effect over S. epidermidis biofilm formation (Oliveira et al., 2017), although the exact mechanisms are not completely understood. Nevertheless, the study of biofilms under iron-restricted conditions represents not only a closer approximation to the environmental conditions found in the human host, but it is also pivotal for a better comprehension of the molecular mechanisms behind biofilm formation in an infection scenario.
The matrix of S. epidermidis biofilms is a complex mixture of polysaccharides, proteins and nucleic acids. The polysaccharide intercellular adhesin (also known as poly-N-acetylglucosamine; PIA/PNAG), which biosynthesis is mediated by the products of the ica (intercellular adhesion) operon (Heilmann et al., 1996) was one of the first molecules found to be implicated in S. epidermidis biofilm accumulation (Ziebuhr et al., 1997). Since then, other factors mediating intercellular adhesion and biofilm accumulation have been identified such as the accumulation-associated protein (Aap) (Rohde et al., 2005), the extracellular matrix-binding protein (Embp) (Christner et al., 2010), or the small basic protein (Sbp) (Decker et al., 2015). The matrix of S. epidermidis biofilms has been shown to impede the penetration of antimicrobial molecules, phagocytic cells, reactive oxygen species, among others (Otto, 2012), which partly explains why S. epidermidis biofilm-associated infections are hard to eradicate or frequently relapse. Nevertheless, the understanding that the biofilm matrix acts solely as a physical barrier has been challenged over the years. In a study addressing biofilms formed by S. epidermidis, Staphylococcus aureus, Escherichia coli, and Klebsiella pneumoniae it was demonstrated that the ability of different classes of antibiotics to kill biofilm cells are independent of penetration (Singh et al., 2016). Another important issue about biofilm-associated infections is that biofilm cells employ different mechanisms to evade the host immune response. It was previously demonstrated that the diffusion of antibodies through S. epidermidis biofilms is not hindered by the biofilm matrix itself, but instead antibodies penetrate the matrix and bind to specific receptors within the matrix (i.e. PIA/PNAG), which reduce the available antibodies during opsonophagocytosis (Cerca et al., 2006). Inactivation of antimicrobial peptides (AMPs) and complement proteins has also been observed in S. epidermidis biofilms (Kristian et al., 2008). Moreover, biofilm-forming S. epidermidis strains were found to impair macrophage cell activation (Schommer et al., 2011). Another issue related with biofilms is the fact that cells adopting this mode of growth exhibit a decreased metabolic rate, which leads to lower efficiency of antibiotics whose action is dependent on actively growing cells (Cerca et al., 2005). Moreover, low pro-inflammatory properties have been attributed to dormant S. epidermidis biofilm cells (Cerca et al., 2011).
Iron and Its Biological Importance
Considering that a large proportion of nosocomial infections is associated with biofilm formation and that S. epidermidis relies on iron acquisition to survive in the host environment, it is noteworthy to explore the link between these two bacterial processes. Iron belongs to the subfamily of transition elements and is one of the most abundant metals on Earth (Frey and Reed, 2012). It is a key nutrient for almost all living organisms, including bacteria, with very few exceptions (Archibald, 1983; Troxell et al., 2012), since it participates in essential biochemical processes, such as electron transfer and catalysis (Hudson et al., 2005). In nature, most iron exists under the form of two oxidative states: ferrous (Fe2+) and ferric (Fe3+) iron. Under aqueous, aerobic environments, Fe2+ is spontaneously oxidized to Fe3+, leading to the formation of ferric hydroxide (Morgan and Lahav, 2007). Additionally, the solubility of ferric hydroxide under neutral pH conditions usually found in the human body is extremely low (Schwertmann, 1991). To overcome this low solubility issue, superior organisms produce proteins (e.g. transferrin and ferritin) that are able to bind Fe3+ and maintain it stable while making it simultaneously available for biochemical processes (Brock, 1989).
Of note, the adult human body contains approximately 3-5 g of iron (Zhang and Enns, 2009). Even though this represents a large quantity, the levels of free ferric ion available in the body are kept to a minimum (~10-24 M) (Raymond et al., 2003; Waldvogel-Abramowski et al., 2014). Therefore, most of iron is complexed as Fe2+ in several proteins, such as metalloproteins. In these proteins, iron is mostly found in the form of heme prosthetic groups (Liu et al., 2014). Hemoglobin, a well-known metalloprotein present in erythroid precursors and mature erythrocytes, represents the major iron reservoir in humans (∼65%). The remaining iron is stored in hepatocytes, bound to ferritin, and within macrophages (Meynard et al., 2014). A small proportion can be found in muscles within myoglobin, or as part of other cellular iron-containing proteins (Soares and Hamza, 2016). Another fraction of the iron is present in the so-called labile iron pool, which consists of redox-active iron ions (both Fe2+ and Fe3+) bound to a variety of low affinity ligands (Kakhlon and Cabantchik, 2002).
Bacterial Iron Acquisition Systems
During infection, S. epidermidis faces very harsh conditions, particularly iron restriction, once it reaches the bloodstream (Cassat and Skaar, 2013). The tiny amount of iron residing extracellularly is mostly bound by high affinity iron-binding proteins. This ensures that the concentration of free iron in body fluids and tissues can be as low as 10-24 M (Raymond et al., 2003). This is an extremely low level to support bacterial proliferation, as microorganisms typically require iron concentrations of approximately 10-6 M for growth (Miethke, 2013). Surprisingly, there is a lack of comprehensive studies available to date on the iron acquisition mechanisms in S. epidermidis, despite being a major source of bloodstream infections (Kleinschmidt et al., 2015). With very few exceptions (Archibald, 1983; Troxell et al., 2012), most pathogens rely on their ability to scavenge several biologically essential metals, including iron, for their survival, both in vitro and in vivo (Takase et al., 2000; Raymond et al., 2003; Dale et al., 2004; Beasley et al., 2009). ATP-binding cassette (ABC) transporters are among the most common bacterial iron acquisition systems (Cartron et al., 2006), which allow the uptake of iron bound either to host-derived proteins (e.g., transferrin) (Taylor and Heinrichs, 2002) or bacterial-derived siderophores (Beasley et al., 2009; Brillet et al., 2012) and hemophores (Rossi et al., 2001).
Siderophore-mediated iron uptake is a widely spread strategy among bacteria to survive in iron-restricted environments. While no siderophore has been described in S. epidermidis so far, there are findings suggesting that this species is able to produce at least one siderophore (Oliveira et al., 2017; Oliveira et al., 2021b). Siderophores are a class of small (usually less than 1 kDa), potent iron-chelating organic molecules with high affinity for Fe3+ (Wilson et al., 2016). Siderophores generally form hexadentate, octahedral, complexes with ferric ions in a 1:1 ratio of siderophore to iron (Hider and Kong, 2010). These molecules are synthesized intracellularly and secreted into the environment as iron-free compounds (Hider and Kong, 2010). Once Fe3+-siderophore complexes are formed, their transport across the bacterial cell membrane to the cytoplasm takes place through a myriad of uptake systems, particularly ABC transporters (Chu et al., 2010). The subsequent release of iron from high affinity siderophores may follow two different mechanisms: (i) enzymatic reduction of siderophore-bound Fe3+ to Fe2+ (Matzanke et al., 2004; Ganne et al., 2017) or (ii) enzyme-catalyzed siderophore hydrolysis (Lin et al., 2005). By the time bacterial iron requirements are met, the transcription of genes encoding iron transport systems is downregulated through the action of a repressor protein called Fur (Ferric uptake regulator) (Troxell and Hassan, 2013).
Regulation of Iron Acquisition Systems
The expression of different bacterial virulence factors, particularly those associated with iron acquisition, is triggered by a decreased intracellular iron content (Porcheron and Dozois, 2015). The regulatory protein Fur is key in a conserved mechanism across bacteria responsible for the regulation of transcriptional responses to iron deprivation, and is now recognized as the canonical global iron-responsive regulator in bacteria (Troxell and Hassan, 2013). In general, when the intracellular iron content surpasses the level required for proper cellular function, there is an association of one Fe2+ ion with two Fur monomers. In its dimeric form, Fur is able to bind a conserved 19-bp DNA motif within the operator region of target genes, designated as Fur box, which blocks RNA polymerase and ultimately leads to repression of gene transcription (Fillat, 2014). Once the intracellular iron levels become depleted, the Fe2+ ion dissociates from the Fur dimer, the Fur box becomes unoccupied, and transcription of target genes is resumed (Fillat, 2014). Current knowledge about the involvement of iron and Fur as transcription regulators in S. epidermidis is limited but in S. aureus, Fur has been shown to regulate the transcription of iron acquisition (Hazmanian et al., 2003; Torres et al., 2006; Beasley et al., 2009) and other virulence-related genes (Torres et al., 2010), and was implicated in biofilm formation (Johnson et al., 2005).
Interplay between iron and biofilm formation in S. epidermidis
As part of the normal microflora of human skin and mucosae (Grice et al., 2009), S. epidermidis, and to a lesser extent S. aureus, are frequent sources of biofilm infections associated with the use of indwelling medical devices (e.g., catheter systems, prosthetic joints, and a range of other polymer and metal implants) (McCann et al., 2008; Hogan et al., 2015; Lourtet-Hascoët et al., 2016) and are also implicated in more serious medical conditions (e.g., sepsis) (Kleinschmidt et al., 2015; Tong et al., 2015). Staphylococcal biofilms have been the focus of intensive research, and the role of iron in this process has been explored. However, the regulatory role of iron in S. epidermidis biofilms is not well understood, mostly due to the considerable lack of studies. The first study dates back to early 90s (Deighton and Borland, 1993) and demonstrated that most strains displayed an enhanced biofilm formation ability under conditions of iron limitation, although this phenotype was strain-dependent and, in some cases, only becomes apparent after a prolonged incubation period (48 hours). During the first 24 hours, this stimulatory effect was only evident for strains classified as weak or moderate biofilm producers, while iron limitation produced an inhibitory effect for strong biofilm producers. A renewed interest in this field was raised by the astonishing finding that catecholamine inotropic drugs, which are frequently administered in intensive-care units, significantly promote biofilm formation by S. epidermidis through sequestration of iron from transferrin (Neal et al., 2001; Lyte et al., 2003). The importance of iron for S. epidermidis biofilms is underscored by the fact that biofilm cells upregulate the transcription of genes involved in iron acquisition upon contact with human blood (França et al., 2014). In another study, it was demonstrated that the ability of different strains, including the reference strain S. epidermidis ATCC 35984 (RP62A), to form biofilms is strongly inhibited under iron-limiting conditions due to a delayed growth rate, reduced cell viability and impaired PIA/PNAG production (Oliveira et al., 2017). Iron excess (1 mM) produces a mildly detrimental effect in biofilm formation that is not related with PIA/PNAG (Oliveira et al., 2017). Siderophore biosynthesis or iron/siderophore transport systems, although not fundamental for planktonic growth under iron starvation, are absolutely required for biofilm formation under these conditions (Oliveira et al., 2021b).
Siderophores and their role in bacterial pathogenesis
The perception of bacterial iron acquisition systems, particularly siderophores, as virulence factors is derived from the observation that their inactivation results in a measurable loss of virulence (Meyer et al., 1996; Rabsch et al., 2003; Montañez et al., 2005; Beasley et al., 2011; Reddy et al., 2013; Runci et al., 2019). One of the host mechanisms to counteract bacterial iron acquisition is the secretion of lipocalin 2 (Lcn2) by cells of the innate immune system, blocking the action of some siderophores, such as enterobactin (Flo et al., 2004). Bacteria overcome this issue through the production of diverse, functionally redundant siderophores, some of them being resistant to the action of Lcn2 (also referred to as “stealth siderophores”) (Fischbach et al., 2006; Bachman et al., 2011). This, together with the implication of siderophores in roles other than iron sequestration, such as the modulation of host cellular pathways (Holden and Bachman, 2015), seems to provide an adaptive advantage over the host immune response and contributes to bacterial pathogenesis. Assigning this kind of roles across different siderophores is difficult not only due to their structural and chemical diversity, but also because their biosynthesis may follow different pathways even in the most closely related species. Staphylococci are a paradigmatic example: S. aureus produces two different siderophores (staphyloferrins A and B) (Beasley et al., 2009; Cotton et al., 2009), S. epidermidis synthesizes staphyloferrin A only (Oliveira et al., 2021b), and S. lugdunensis hijacks siderophores from other staphylococci instead of producing them (Brozyna et al., 2014) (Figure 1A). Another striking difference regarding iron acquisition among staphylococci is that S. aureus and S. lugdunensis have a system dedicated to heme-bound iron acquisition, called iron-regulated surface determinant (Isd) (Torres et al., 2006; Heilbronner et al., 2011), which is absent in S. epidermidis. Therefore, the absence of the Isd system, in combination with the production of a single siderophore, underscores the relevance of siderophore-mediated iron acquisition in S. epidermidis.
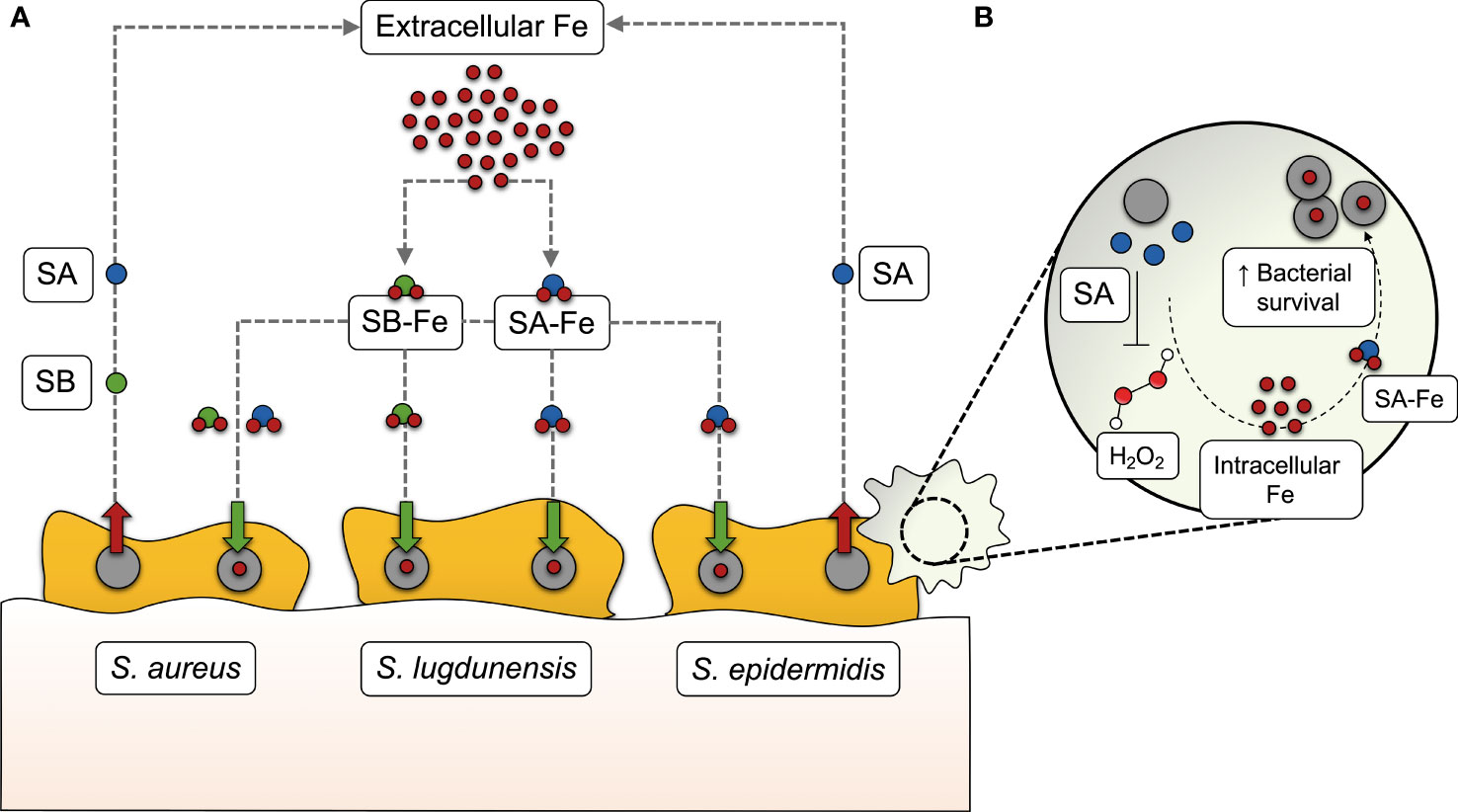
Figure 1 Siderophore-mediated iron acquisition in staphylococci. (A) While S. aureus produces two different siderophores (staphyloferrin A, SA, and staphyloferrin B, SB), S. epidermidis synthesizes SA only, and S. lugdunensis hijacks SA and SB from these two staphylococci species instead of producing them. After being secreted into the extracellular medium, SA and SB bind to extracellular iron (Fe) and make their way back to the cell as siderophore-iron complexes (SA-Fe and SB-Fe), providing the iron levels required for several bacterial processes, including biofilm formation. (B) In S. epidermidis, SA-mediated iron acquisition has recently been found to contribute to bacterial survival in human macrophages, by withstanding the action of certain reactive oxygen species, such as hydrogen peroxide (H2O2).
The contribution of staphyloferrins in staphylococcal pathogenicity has been evidenced by the reduced virulence of siderophore-deficient S. aureus in different murine infection models (Dale et al., 2004; Nakaminami et al., 2017). This may be partly explained by the protective effect of staphyloferrin A to cells residing in host phagocytic cells. Deletion of S. epidermidis sfaABCD, which mediates siderophore biosynthesis, leads to compromised fitness in the macrophage intracellular milieu and increased susceptibility to reactive oxygen species (Oliveira et al., 2021b) (Figure 1B).
Exploiting the bacterial dependence on iron for therapeutic purposes
The importance of iron and siderophores for biofilm formation and virulence across different nosocomial pathogens offers an opportunity for the development of new strategies to tackle biofilm-associated infections. Bacterial iron uptake systems, particularly siderophore and heme transport systems, can be exploited as gateways for the delivery of so-called “trojan horse” compounds into the bacterial cytoplasm. One possible approach is to use the heme synthetic analog gallium-protoporphyrin IX (GaPP) to facilitate the delivery of gallium into biofilms. GaPP has demonstrated remarkable antibiofilm activity against S. aureus, which is enhanced in combination with other compounds, such as gallium nitrate, iron chelating agents and conventional antibiotics (Chang et al., 2016; Richter et al., 2016; Richter et al., 2017a; Richter et al., 2017b; Choi et al., 2019a; Choi et al., 2019b). The use of GaPP is encouraged by the safety it demonstrated across cytotoxicity studies in different human cell lines, primary human cells and mice (Chang et al., 2016), although further studies are required on this matter. Nevertheless, care must be taken when selecting carrier molecules for the bacterial uptake of gallium, which should be based on the target species. For instance, despite the efficacy of GaPP against staphylococci, complexation of gallium with their native siderophore staphyloferrin A results in poor antimicrobial activity, raising some concerns about the suitability of native siderophores as carriers of gallium-based compounds (Kelson et al., 2013). The “trojan horse” strategy may also be employed for improved delivery of conventional antibiotics to pathogens through their conjugation with siderophores. Surprisingly, while there is evidence about the promising antibacterial activity of this group of compounds against Gram-positive and Gram-negative pathogens (Ji et al., 2012; Wencewicz et al., 2013; Ito et al., 2016; Ito-Horiyama et al., 2016), its efficacy against biofilms, in which antibiotic penetration is particularly dampened, remains unknown and is worth further investigation.
Iron acquisition-related molecules are also regarded as suitable target candidates for vaccine development since they display a good degree of conservation and their expression is readily induced as soon as pathogens invade the host and face nutritional immunity (Sheldon and Heinrichs, 2012). The Syntiron/Sanofi Pasteur consortium recently started preclinical trials on a multivalent vaccine based on four iron-regulated lipoproteins for the prevention of S. aureus skin and soft tissue infection (Syntiron, Sanofi Pasteur, 2018), although no detailed information has been made publicly available.
Conclusions and Perspectives
S. epidermidis biofilm-associated infections are an increasing issue worldwide and have posed huge challenges to healthcare professionals. In the years to come, the global spread of multidrug-resistant lineages (Lee et al., 2018) may render the treatment of S. epidermidis biofilm-associated infections extremely difficult. Given this likely scenario, there is an urgent need to identify alternative bacterial targets for the development of novel anti-infective strategies. During the last years, we have been witnessing a renewed interest in bacterial iron acquisition mechanisms, mostly due to very promising findings underscoring the complex regulatory role of iron in biofilm formation, as well as the major role of siderophores in the virulence of several pathogens, including S. epidermidis. Consequently, this has put a spotlight on iron acquisition-related processes and brought a new hope for the development of a much-needed new generation of therapeutic strategies against life-threatening nosocomial infections. Nevertheless, there is still a long road ahead, as we still need to achieve a deeper understanding of the different biological roles that siderophores may assume in the pathogenicity of biofilm-associated infections and, most importantly, of the whole range of consequences of inhibiting iron acquisition in bacteria.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author Contributions
NC and FO drafted the first version of the manuscript. HR and MV reviewed and edited the first version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
FO is supported by the Fundação para a Ciência e a Tecnologia research project with reference PTDC/BIA-MOL/29553/2017, under the scope of COMPETE2020 (POCI-01-0145-FEDER-029553).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Águila-Arcos, S., Álvarez-Rodríguez, I., Garaiyurrebaso, O., Garbisu, C., Grohmann, E., Alkorta, I. (2017). Biofilm-Forming Clinical Staphylococcus Isolates Harbor Horizontal Transfer and Antibiotic Resistance Genes. Front. Microbiol. 8, 2018. doi: 10.3389/fmicb.2017.02018
Allegranzi, B., Nejad, S. B., Combescure, C., Graafmans, W., Attar, H., Donaldson, L, et al. (2011). Report on the Burden of Endemic Health Care-Associated Infection Worldwide (Geneva 27, Switzerland:WHO Library Cataloguing-in-Publication Data).
Andrews, S. C., Robinson, A. K., Rodríguez-Quiñones, F. (2003). Bacterial Iron Homeostasis. FEMS Microbiol. Rev. 27 (2–3), 215–237. doi: 10.1016/S0168-6445(03)00055-X
Archibald, F. (1983). Lactobacillus plantarum, an Organism Not Requiring Iron. FEMS Microbiol. Lett. 19 (1), 29–32. doi: 10.1111/j.1574-6968.1983.tb00504.x
Bachman, M. A., Oyler, J. E., Burns, S. H., Caza, M., Lépine, F., Dozois, C. M, et al. (2011). Klebsiella pneumoniae Yersiniabactin Promotes Respiratory Tract Infection Through Evasion of Lipocalin 2. Infect. Immun 79 (8), 3309–16. doi: 10.1128/IAI.05114-11
Beasley, F. C., Marolda, C. L., Cheung, J., Buac, S., Heinrichs, D. E. (2011). Staphylococcus aureus Transporters Hts, Sir, and Sst Capture Iron Liberated From Human Transferrin by Staphyloferrin A, Staphyloferrin B, and Catecholamine Stress Hormones, Respectively, and Contribute to Virulence. Infect. Immun. 79 (6), 2345–2355. doi: 10.1128/IAI.00117-11
Beasley, F. C., Vinés, E. D., Grigg, J. C., Zheng, Q., Liu, S., Lajoie, G. A, et al. (2009). Characterization of Staphyloferrin A Biosynthetic and Transport Mutants in Staphylococcus aureus. Mol. Microbiol. 72 (4), 947–963. doi: 10.1111/j.1365-2958.2009.06698.x
Brandwein, M., Steinberg, D., Meshner, S. (2016). Microbial Biofilms and the Human Skin Microbiome. NPJ Biofilms. Microbiom. 2, 3. doi: 10.1038/s41522-016-0004-z
Brillet, K., Ruffenach, F., Adams, H., Journet, L., Gasser, V., Hoegy, F, et al. (2012). An ABC Transporter With Two Periplasmic Binding Proteins Involved in Iron Acquisition in Pseudomonas aeruginosa. ACS Chem. Biol 7 (12), 2036–45. doi: 10.1021/cb300330v
Brock, J. H. (1989). Iron-Binding Proteins. Acta Paediatri. Scand. Supplement. 361, 31–43. doi: 10.1111/apa.1989.78.s361.31
Brozyna, J. R., Sheldon, J. R., Heinrichs, D. E. (2014). Growth Promotion of the Opportunistic Human Pathogen, Staphylococcus lugdunensis, by Heme, Hemoglobin, and Coculture With Staphylococcus aureus. MicrobiologyOpen 3 (2), 182–195. doi: 10.1002/mbo3.162
Cartron, M. L., Maddocks, S., Gillingham, P., Craven, C. J., Andrews, S. C. (2006). Feo - Transport of Ferrous Iron Into Bacteria. BioMetals 19 (2), 143–57. doi: 10.1007/s10534-006-0003-2
Cassat, J. E., Skaar, E. P. (2013). Iron in Infection and Immunity. Cell Host Microbe 13 (5), 509–519. doi: 10.1016/j.chom.2013.04.010
Cerca, F., Andrade, F., França, Â., Andrade, E. B., Ribeiro, A., Almeida, A. A, et al. (2011). Staphylococcus epidermidis Biofilms With Higher Proportions of Dormant Bacteria Induce a Lower Activation of Murine Macrophages. J. Med. Microbiol. 60, 1717–1724. doi: 10.1099/jmm.0.031922-0
Cerca, N., Jefferson, K. K., Oliveira, R., Pier, G. B., Azeredo, J. (2006). Comparative Antibody-Mediated Phagocytosis of Staphylococcus epidermidis Cells Grown in a Biofilm or in the Planktonic State. Infect. Immun. 74 (8), 4849–4855. doi: 10.1128/IAI.00230-06
Cerca, N., Martins, S., Cerca, F., Jefferson, K. K., Pier, G. B., Oliveira, R, et al. (2005). Comparative Assessment of Antibiotic Susceptibility of Coagulase-Negative Staphylococci in Biofilm Versus Planktonic Culture as Assessed by Bacterial Enumeration or Rapid XTT Colorimetry. J. Antimicrobial. Chemother. 56 (2), 331–336. doi: 10.1093/jac/dki217
Chang, D., Garcia, R. A., Akers, K. S., Mende, K., Murray, C. K., Wenke, J. C, et al. (2016). Activity of Gallium Meso-and Protoporphyrin Ix Against Biofilms of Multidrug-Resistant Acinetobacter baumannii Isolates. Pharmaceuticals 9 (1), 16. doi: 10.3390/ph9010016
Choi, S. R., Britigan, B. E., Narayanasamy, P. (2019a). Iron/heme Metabolism-Targeted Gallium(III) Nanoparticles are Active Against Extracellular and Intracellular Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrobial. Agents Chemother 63 (4), e02643–18. doi: 10.1128/AAC.02643-18
Choi, S. R., Britigan, B. E., Narayanasamy, P. (2019b). Dual Inhibition of Klebsiella pneumoniae and Pseudomonas aeruginosa Iron Metabolism Using Gallium Porphyrin and Gallium Nitrate. ACS Infect. Dis 5 (9), 1559–1569. doi: 10.1021/acsinfecdis.9b00100
Christner, M., Franke, G. C., Schommer, N. N., Wendt, U., Wegert, K., Pehle, P, et al. (2010). The Giant Extracellular Matrix-Binding Protein of Staphylococcus epidermidis Mediates Biofilm Accumulation and Attachment to Fibronectin. Mol. Microbiol. 75 (1), 187–207. doi: 10.1111/j.1365-2958.2009.06981.x
Chu, B. C., Garcia-Herrero, A., Johanson, T. H., Krewulak, K. D., Lau, C. K., Peacock, R. S, et al. (2010). Siderophore Uptake in Bacteria and the Battle for Iron With the Host; a Bird’s Eye View. BioMetals 23 (4), 601–611. doi: 10.1007/s10534-010-9361-x
Cotton, J. L., Tao, J., Balibar, C. J. (2009). Identification and Characterization of the Staphylococcus aureus Gene Cluster Coding for Staphyloferrin a. Biochemistry 48 (5), 1025–1035. doi: 10.1021/bi801844c
Dale, S. E., Doherty-Kirby, A., Lajoie, G., Heinrichs, D. E. (2004). Role of Siderophore Biosynthesis in Virulence of Staphylococcus aureus: Identification and Characterization of Genes Involved in Production of a Siderophore. Infect. Immun. 72 (1), 29–37. doi: 10.1128/IAI.72.1.29-37.2004
Decker, R., Burdelski, C., Zobiak, M., Büttner, H., Franke, G., Christner, M, et al. (2015). An 18 kDa Scaffold Protein Is Critical for Staphylococcus epidermidis Biofilm Formation. PloS Pathog. 11 (3), e1004735. doi: 10.1371/journal.ppat.1004735
Deighton, M., Borland, R. (1993). Regulation of Slime Production in Staphylococcus epidermidis By Iron Limitation. Infect. Immun. 61 (10), 4473–4479. doi: 10.1128/iai.61.10.4473-4479.1993
Dengler Haunreiter, V., Boumasmoud, M., Häffner, N., Wipfli, D., Leimer, N., Rachmühl, C, et al. (2019). In-Host Evolution of Staphylococcus epidermidis in a Pacemaker-Associated Endocarditis Resulting in Increased Antibiotic Tolerance. Nat. Commun. 10 (1), 1149. doi: 10.1038/s41467-019-09053-9
De Vos, W. M. (2015). Microbial Biofilms and the Human Intestinal Microbiome. NPJ Biofilms. Microbiom. 1, 15005. doi: 10.1038/npjbiofilms.2015.5
Donlan, R. M., Costerton, J. W. (2002). Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 15 (2), 167–193. doi: 10.1128/CMR.15.2.167-193.2002
European Centre for Disease Prevention and Control. (2018) Annual Epidemiological Report for 2016: Healthcare-Associated Infections in Intensive Care Units. Available at: https://ecdc.europa.eu/sites/portal/files/documents/AER_for_2016-HAI_0.pdf.
Fey, P. D., Olson, M. E. (2010). Current Concepts in Biofilm Formation of Staphylococcus epidermidis. Future Microbiol. 5 (6), 917–933. doi: 10.2217/fmb.10.56
Fillat, M. F. (2014). The FUR (Ferric Uptake Regulator) Superfamily: Diversity and Versatility of Key Transcriptional Regulators. Arch. Biochem. Biophys. 546, 41–52. doi: 10.1016/j.abb.2014.01.029
Fischbach, M. A., Lin, H., Zhou, L., Yu, Y., Abergel, R. J., Liu, D. R, et al. (2006). The Pathogen-Associated iroA Gene Cluster Mediates Bacterial Evasion of Lipocalin 2. Proc Natl Acad Sci USA 103 (44), 16502–16507. doi: 10.1073/pnas.0604636103
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., Kjelleberg, S. (2016). Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 14 (9), 563–575. doi: 10.1038/nrmicro.2016.94
Flo, T. H., Smith, K. D., Sato, S., Rodriguez, D. J., Holmes, M. A., Strong, R. K, et al. (2004). Lipocalin 2 Mediates an Innate Immune Response to Bacterial Infection by Sequestrating Iron. Nature 432 (7019), 917–21. doi: 10.1038/nature03104
França, A., Carvalhais, V., Maira-Litrán, T., Vilanova, M., Cerca, N., Pier, G. (2014). Alterations in the Staphylococcus epidermidis Biofilm Transcriptome Following Interaction With Whole Human Blood. Pathog. Dis. 70 (3), 444–448. doi: 10.1111/2049-632X.12130
Frey, P. A., Reed, G. H. (2012). The Ubiquity of Iron. ACS Chem. Biol. 7 (9), 1477–1481. doi: 10.1021/cb300323q
Ganne, G., Brillet, K., Basta, B., Roche, B., Hoegy, F., Gasser, V, et al. (2017). Iron Release From the Siderophore Pyoverdine in Pseudomonas aeruginosa Involves Three New Actors: FpvC, FpvG, and FpvH. ACS Chem. Biol. 12 (4), 1056–1065. doi: 10.1021/acschembio.6b01077
Grice, E. A., Kong, H. H., Conlan, S., Deming, C. B., Davis, J., Young, A. C, et al. (2009). Topographical and Temporal Diversity of the Human Skin Microbiome. Science 324 (5931), 1190–1192. doi: 10.1126/science.1171700
Haque, M., Sartelli, M., McKimm, J., Bakar, M. A. (2018). Health Care-Associated Infections – An Overview. Infect. Drug Resist. 11, 2321–2333. doi: 10.2147/IDR.S177247
Hazmanian, S. K., Skaar, E. P., Gaspar, A. H., Humayun, M., Gornicki, P., Jelenska, J, et al. (2003). Passage of Heme-Iron Across the Envelope of Staphylococcus aureus. Science 299 (5608), 906–909. doi: 10.1126/science.1081147
Heilbronner, S., Holden, M. T. G., van Tonder, A., Geoghegan, J. A., Foster, T. J., Parkhill, J, et al. (2011). Genome Sequence of Staphylococcus lugdunensis N920143 Allows Identification of Putative Colonization and Virulence Factors. FEMS Microbiol. Lett. 322 (1), 60–67. doi: 10.1111/j.1574-6968.2011.02339.x
Heilmann, C., Schweitzer, O., Gerke, C., Vanittanakom, N., Mack, D., Götz, F. (1996). Molecular Basis of Intercellular Adhesion in the Biofilm-Forming Staphylococcus epidermidis. Mol. Microbiol. 20 (5), 1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x
Hider, R. C., Kong, X. (2010). Chemistry and Biology of Siderophores. Natural Product. Rep. 27 (5), 637–657. doi: 10.1039/b906679a
Hogan, S., Stevens, N. T., Humphreys, H., Gara, J. P. O., Neill, E. O. (2015). Current and Future Approaches to the Prevention and Treatment of Staphylococcal Medical Device-Related Infections. Curr. Pharm. Design. 21 (1), 100–113. doi: 10.2174/1381612820666140905123900
Holden, V. I., Bachman, M. A. (2015). Diverging Roles of Bacterial Siderophores During Infection. Metallomics 7 (6), 986–995. doi: 10.1039/C4MT00333K
Hudson, J. M., Heffron, K., Kotlyar, V., Sher, Y., Maklashina, E., Cecchini, G, et al. (2005). Electron Transfer and Catalytic Control by the Iron-Sulfur Clusters in a Respiratory Enzyme, E. Coli Fumarate Reductase. J. Am. Chem. Soc. 127 (19), 6977–6989. doi: 10.1021/ja043404q
Ito-Horiyama, T., Ishii, Y., Ito, A., Sato, T., Nakamura, R., Fukuhara, N, et al. (2016). Stability of Novel Siderophore Cephalosporin S-649266 Against Clinically Relevant Carbapenemases. Antimicrobial. Agents Chemother 60 (7), 4384–6. doi: 10.1128/AAC.03098-15
Ito, A., Kohira, N., Bouchillon, S. K., West, J., Rittenhouse, S., Sader, H. S, et al. (2016). In Vitro Antimicrobial Activity of S-649266, a Catechol-Substituted Siderophore Cephalosporin, When Tested Against Non-Fermenting Gram-Negative Bacteria. J. Antimicrobial. Chemother 71 (3), 670–7. doi: 10.1093/jac/dkv402
Ji, C., Miller, P. A., Miller, M. J. (2012). Iron Transport-Mediated Drug Delivery: Practical Syntheses and In Vitro Antibacterial Studies of Tris-Catecholate Siderophore-Aminopenicillin Conjugates Reveals Selectively Potent Antipseudomonal Activity. J. Am. Chem. Soc 134 (24), 9898–901. doi: 10.1021/ja303446w
Johnson, M., Cockayne, A., Williams, P. H., Morrissey, J. A. (2005). Iron-Responsive Regulation of Biofilm Formation in Staphylococcus aureus Involves Fur-Dependent and Fur-Independent Mechanisms. J. Bacteriol. 187 (23), 8211–8215. doi: 10.1128/JB.187.23.8211-8215.2005
Kakhlon, O., Cabantchik, Z. I. (2002). The Labile Iron Pool: Characterization, Measurement, and Participation in Cellular Processes. Free Radical Biol. Med. 33 (8), 1037–1046. doi: 10.1016/S0891-5849(02)01006-7
Kelson, A. B., Carnevali, M., Truong-Le, V. (2013). Gallium-Based Anti-Infectives: Targeting Microbial Iron-Uptake Mechanisms. Curr. Opin. Pharmacol 13 (5), 707–16. doi: 10.1016/j.coph.2013.07.001
Kleinschmidt, S., Huygens, F., Faoagali, J., Rathnayake, I. U., Hafner, L. M. (2015). Staphylococcus epidermidis as a Cause of Bacteremia. Future Microbiol. 10 (11), 1859–1879. doi: 10.2217/fmb.15.98
Kristian, S. A., Birkenstock, T. A., Sauder, U., Mack, D., Götz, F., Landmann, R. (2008). Biofilm Formation Induces C3a Release and Protects Staphylococcus epidermidis From IgG and Complement Deposition and From Neutrophil-Dependent Killing. J. Infect. Dis. 197 (7), 1028–1035. doi: 10.1086/528992
Lee, J. Y. H., Monk, I. R., Gonçalves da Silva, A., Seemann, T., Chua, K. Y. L., Kearns, A, et al. (2018). Global Spread of Three Multidrug-Resistant Lineages of Staphylococcus epidermidis. Nat. Microbiol. 3 (10), 1175–1185. doi: 10.1038/s41564-018-0230-7
Lin, H., Fischbach, M. A., Liu, D. R., Walsh, C. T. (2005). In Vitro Characterization of Salmochelin and Enterobactin Trilactone Hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 127 (31), 11075–11084. doi: 10.1021/ja0522027
Liu, J., Chakraborty, S., Hosseinzadeh, P., Yu, Y., Tian, S., Petrik, I, et al. (2014). Metalloproteins Containing Cytochrome, Iron–Sulfur, or Copper Redox Centers. Chem. Rev. 114 (8), 4366–4469. doi: 10.1021/cr400479b
Lourtet-Hascoët, J., Bicart-See, A., Félicé, M. P., Giordano, G., Bonnet, E. (2016). Staphylococcus lugdunensis, a Serious Pathogen in Periprosthetic Joint Infections: Comparison to Staphylococcus aureus and Staphylococcus epidermidis. Int. J. Infect. Dis 51:56–61. doi: 10.1016/j.ijid.2016.08.007
Lyte, M., Freestone, P. P. E., Neal, C. P., Olson, B. A., Haigh, R. D., Bayston, R, et al. (2003). Stimulation of Staphylococcus epidermidis Growth and Biofilm Formation by Catecholamine Inotropes. Lancet [Internet] 361 (9352), 130–135. doi: 10.1016/S0140-6736(03)12231-3
Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A, et al. (2014). Multistate Point-Prevalence Survey of Health Care–Associated Infections. New Engl. J. Med. 370 (13), 1198–1208. doi: 10.1056/NEJMoa1306801
Matzanke, B. F., Anemüller, S., Schünemann, V., Trautwein, A. X., Hantke, K. (2004). FhuF, Part of a Siderophore-Reductase System. Biochemistry 43 (5), 1386–1392. doi: 10.1021/bi0357661
McCann, M. T., Gilmore, B. F., Gorman, S. P. (2008). Staphylococcus epidermidis Device-Related Infections: Pathogenesis and Clinical Management. J. Pharm. Pharmacol. 60 (12), 1551–1571. doi: 10.1211/jpp/60.12.0001
Meyer, J. M., Neely, A., Stintzi, A., Georges, C., Holder, I. A. (1996). Pyoverdin is Essential for Virulence of Pseudomonas aeruginosa. Infect. Immun. 64 (2), 518–523. doi: 10.1128/iai.64.2.518-523.1996
Meynard, D., Babitt, J. L., Lin, H. Y. (2014). The Liver: Conductor of Systemic Iron Balance. Blood 123 (2), 168–176. doi: 10.1182/blood-2013-06-427757
Miethke, M. (2013). Molecular Strategies of Microbial Iron Assimilation: From High-Affinity Complexes to Cofactor Assembly Systems. Metallomics 5 (1), 15–28. doi: 10.1039/C2MT20193C
Montañez, G. E., Neely, M. N., Eichenbaum, Z. (2005). The Streptococcal Iron Uptake (Siu) Transporter is Required for Iron Uptake and Virulence in a Zebrafish Infection Model. Microbiology 151 (Pt 11), 3749–3757. doi: 10.1099/mic.0.28075-0
Morgan, B., Lahav, O. (2007). The Effect of pH on the Kinetics of Spontaneous Fe(II) Oxidation by O2 in Aqueous Solution - Basic Principles and a Simple Heuristic Description. Chemosphere 68 (11), 2080–4. doi: 10.1016/j.chemosphere.2007.02.015
Nakaminami, H., Chen, C., Truong-Bolduc, Q. C., Kim, E. S., Wang, Y., Hooper, D. C. (2017). Efflux Transporter of Siderophore Staphyloferrin A in Staphylococcus aureus Contributes to Bacterial Fitness in Abscesses and Epithelial Cells. Infect. Immun. 85 (8), e00358–e00317. doi: 10.1128/IAI.00358-17
Neal, C. P., Freestone, P. P. E., Maggs, A. F., Haigh, R. D., Williams, P. H., Lyte, M. (2001). Catecholamine Inotropes as Growth Factors for Staphylococcus epidermidis and Other Coagulase-Negative Staphylococci. FEMS Microbiol. Lett. 194 (2), 163–169. doi: 10.1111/j.1574-6968.2001.tb09463.x
Oliveira, F., França, Â., Cerca, N. (2017). Staphylococcus epidermidis is Largely Dependent on Iron Availability to Form Biofilms. Int. J. Med. Microbiol. 307 (8), 552–563. doi: 10.1016/j.ijmm.2017.08.009
Oliveira, F., Lima, T., Correia, A., Silva, A. M., Soares, C., Morais, S, et al. (2021b). Siderophore-Mediated Iron Acquisition Plays a Critical Role in Biofilm Formation and Survival of Staphylococcus epidermidis Within the Host. Front. Med. 8, 799227. doi: 10.3389/fmed.2021.799227
Oliveira, F., Rohde, H., Vilanova, M., Cerca, N. (2021a). The Emerging Role of Iron Acquisition in Biofilm-Associated Infections. Trends Microbiol. 29 (9), 772–775. doi: 10.1016/j.tim.2021.02.009
Otto, M. (2012). Molecular Basis of Staphylococcus epidermidis Infections. Semin. Immunopathol 34 (2), 201–14. doi: 10.1007/s00281-011-0296-2
Pittet, D., Allegranzi, B., Storr, J., Nejad, S. B., Dziekan, G., Leotsakos, A, et al. (2008). Infection Control as a Major World Health Organization Priority for Developing Countries. J. Hosp. Infect. 68 (4), 285–292. doi: 10.1016/j.jhin.2007.12.013
Porcheron, G., Dozois, C. M. (2015). Interplay Between Iron Homeostasis and Virulence: Fur and RyhB as Major Regulators of Bacterial Pathogenicity. Vet. Microbiol. 179 (1–2), 2–14. doi: 10.1016/j.vetmic.2015.03.024
Rabsch, W., Methner, U., Voigt, W., Tschäpe, H., Reissbrodt, R., Williams, P. H. (2003). Role of Receptor Proteins for Enterobactin and 2,3-Dihydroxybenzoylserine in Virulence of Salmonella Enterica. Infect. Immun. 71 (12), 6953–6956. doi: 10.1128/IAI.71.12.6953-6961.2003
Raymond, K. N., Dertz, E. A., Kim, S. S. (2003). Enterobactin: An Archetype for Microbial Iron Transport. Proc. Natl. Acad. Sci. 100 (7), 3584–3588. doi: 10.1073/pnas.0630018100
Reddy, P. V., Puri, R. V., Chauhan, P., Kar, R., Rohilla, A., Khera, A, et al. (2013). Disruption of Mycobactin Biosynthesis Leads to Attenuation of Mycobacterium tuberculosis for Growth and Virulence. J. Infect. Dis. 208 (8), 1255–1265. doi: 10.1093/infdis/jit250
Richter, K., Ramezanpour, M., Thomas, N., Prestidge, C. A., Wormald, P. J., Vreugde, S. (2016). Mind “De GaPP”: In Vitro Efficacy of Deferiprone and Gallium-Protoporphyrin Against Staphylococcus aureus Biofilms. Int. Forum Allergy Rhinol 6 (7), 737–43. doi: 10.1002/alr.21735
Richter, K., Thomas, N., Claeys, J., McGuane, J., Prestidge, C. A., Coenye, T, et al. (2017b). A Topical Hydrogel With Deferiprone and Gallium-Protoporphyrin Targets Bacterial Iron Metabolism and has Antibiofilm Activity. Antimicrobial. Agents Chemother 61 (6), e00481–17. doi: 10.1128/AAC.00481-17
Richter, K., Thomas, N., Zhang, G., Prestidge, C. A., Coenye, T., Wormald, P. J, et al. (2017a). Deferiprone and Gallium-Protoporphyrin Have the Capacity to Potentiate the Activity of Antibiotics in Staphylococcus aureus Small Colony Variants. Front. Cell. Infect. Microbiol 7, 280. doi: 10.3389/fcimb.2017.00280
Rohde, H., Burdelski, C., Bartscht, K., Hussain, M., Buck, F., Horstkotte, M. A, et al. (2005). Induction of Staphylococcus epidermidis Biofilm Formation via Proteolytic Processing of the Accumulation-Associated Protein by Staphylococcal and Host Proteases. Mol. Microbiol. 55 (6), 1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x
Rossi, M. S., Fetherston, J. D., Létoffé, S., Carniel, E., Perry, R. D., Ghigo, J. M. (2001). Identification and Characterization of the Hemophore-Dependent Heme Acquisition System of Yersinia pestis. Infect. Immun 69 (11), 6707–6717. doi: 10.1128/IAI.69.11.6707-6717.2001
Runci, F., Gentile, V., Frangipani, E., Rampioni, G., Leoni, L., Lucidi, M, et al. (2019). Contribution of Active Iron Uptake to Acinetobacter baumannii Pathogenicity. Infect. Immun. 87 (4), e00755. doi: 10.1128/IAI.00755-18
Schommer, N. N., Christner, M., Hentschke, M., Ruckdeschel, K., Aepfelbacher, M., Rohde, H. (2011). Staphylococcus epidermidis Uses Distinct Mechanisms of Biofilm Formation To Interfere With Phagocytosis and Activation of Mouse Macrophage-Like Cells J774A.1. Infect. Immun. 79 (6), 2267–2276. doi: 10.1128/IAI.01142-10
Sheldon, J. R., Heinrichs, D. E. (2012). The Iron-Regulated Staphylococcal Lipoproteins. Front. Cell. Infect. Microbiol. 2(2235-2988 (2235-2988 (Electronic), 41. doi: 10.3389/fcimb.2012.00041
Singh, R., Sahore, S., Kaur, P., Rani, A., Ray, P. (2016). Penetration Barrier Contributes to Bacterial Biofilm-Associated Resistance Against Only Select Antibiotics, and Exhibits Genus-, Strain- and Antibiotic-Specific Differences. Pathog. Dis. 74 (6), ftw056. doi: 10.1093/femspd/ftw056
Skaar, E. P. (2010). The Battle for Iron Between Bacterial Pathogens and Their Vertebrate Hosts. PloS Pathog. 6 (8), e1000949. doi: 10.1371/journal.ppat.1000949
Soares, M. P., Hamza, I. (2016). Macrophages and Iron Metabolism. Immunity 44 (3), 492–504. doi: 10.1016/j.immuni.2016.02.016
Syntiron, Sanofi Pasteur. (2018) Research Programme: Staphylococcus aureus Vaccine - Syntiron/sanofi Pasteur. Available at: https://adisinsight.springer.com/drugs/800031401.
Takase, H., Nitanai, H., Hoshino, K., Otani, T. (2000). Impact of Siderophore Production on Pseudomonas aeruginosa Infections in Immunosuppressed Mice. Infect. Immun. 68 (4), 1834–1839. doi: 10.1128/IAI.68.4.1834-1839.2000
Taylor, J. M., Heinrichs, D. E. (2002). Transferrin Binding in Staphylococcus aureus: Involvement of a Cell Wall-Anchored Protein. Mol. Microbiol 43 (6), 1603–1614. doi: 10.1046/j.1365-2958.2002.02850.x
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., Fowler, V. G. (2015). Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev 28 (3), 603–661. doi: 10.1128/CMR.00134-14
Torres, V. J., Attia, A. S., Mason, W. J., Hood, M. I., Corbin, B. D., Beasley, F. C, et al. (2010). Staphylococcus aureus Fur Regulates the Expression of Virulence Factors That Contribute to the Pathogenesis of Pneumonia. Infect. Immun. 78 (4), 1618–1628. doi: 10.1128/IAI.01423-09
Torres, V. J., Pishchany, G., Humayun, M., Schneewind, O., Skaar, E. P. (2006). Staphylococcus aureus IsdB is a Hemoglobin Receptor Required for Heme Iron Utilization. J. Bacteriol. 188 (24), 8421–8429. doi: 10.1128/JB.01335-06
Troxell, B., Hassan, H. M. (2013). Transcriptional Regulation by Ferric Uptake Regulator (Fur) in Pathogenic Bacteria. Front. Cell. Infect. Microbiol. 3, 59. doi: 10.3389/fcimb.2013.00059
Troxell, B., Xu, H., Yang, X. F. (2012). Borrelia burgdorferi, a Pathogen That Lacks Iron, Encodes Manganese-Dependent Superoxide Dismutase Essential for Resistance to Streptonigrin. J. Biol. Chem. 287 (23), 19284–19293. doi: 10.1074/jbc.M112.344903
Vincent, J. L. (2003). Nosocomial Infections in Adult Intensive-Care Units. Lancet 361, 2068–2077. doi: 10.1016/S0140-6736(03)13644-6
Waldvogel-Abramowski, S., Waeber, G., Gassner, C., Buser, A., Frey, B. M., Favrat, B, et al. (2014). Physiology of Iron Metabolism. Transfusion. Med. Hemother. 41 (3), 213–221. doi: 10.1159/000362888
Wencewicz, T. A., Long, T. E., Möllmann, U., Miller, M. J. (2013). Trihydroxamate Siderophore-Fluoroquinolone Conjugates are Selective Sideromycin Antibiotics That Target Staphylococcus aureus. Bioconjugate. Chem. 24 (3), 473–486. doi: 10.1021/bc300610f
Wilson, B. R., Bogdan, A. R., Miyazawa, M., Hashimoto, K., Tsuji, Y. (2016). Siderophores in Iron Metabolism: From Mechanism to Therapy Potential. Trends Mol. Med. 22 (12), 1077–1090. doi: 10.1016/j.molmed.2016.10.005
Zarb, P., Coignard, B., Griskeviciene, J., Muller, A., Vankerckhoven, V., Weist, K, et al. (2012). The European Centre for Disease Prevention and Control (ECDC) Pilot Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use. Eurosurveillance 17 (46), 20316. doi: 10.2807/ese.17.46.20316-en
Zhang, A. S., Enns, C. A. (2009). Iron Homeostasis: Recently Identified Proteins Provide Insight Into Novel Control Mechanisms. J. Biol. Chem. 284 (2), 711–715. doi: 10.1074/jbc.R800017200
Ziebuhr, W., Heilmann, C., Götz, F., Meyer, P., Wilms, K., Straube, E, et al. (1997). Detection of the Intercellular Adhesion Gene Cluster (Ica) and Phase Variation in Staphylococcus epidermidis Blood Culture Strains and Mucosal Isolates. Infect. Immun. 65 (3), 890–896. doi: 10.1128/iai.65.3.890-896.1997
Keywords: iron acquisition systems, regulation of iron acquisition, Siderophores, S. epidermidis biofilms, role of iron in infection
Citation: Oliveira F, Rohde H, Vilanova M and Cerca N (2021) Fighting Staphylococcus epidermidis Biofilm-Associated Infections: Can Iron Be the Key to Success? Front. Cell. Infect. Microbiol. 11:798563. doi: 10.3389/fcimb.2021.798563
Received: 20 October 2021; Accepted: 11 November 2021;
Published: 30 November 2021.
Edited by:
Brendan Gilmore, Queen’s University Belfast, United KingdomReviewed by:
Pietro Speziale, University of Pavia, ItalyChristian Johann Lerche, Rigshospitalet, Denmark
Copyright © 2021 Oliveira, Rohde, Vilanova and Cerca. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nuno Cerca, bnVub2NlcmNhQGNlYi51bWluaG8ucHQ=
 Fernando Oliveira
Fernando Oliveira Holger Rohde
Holger Rohde Manuel Vilanova
Manuel Vilanova Nuno Cerca
Nuno Cerca