- 1Department of Parasitology, National Institute of Infectious Diseases, Shinjuku-ku, Tokyo, Japan
- 2Department of Infectious Diseases, Tokai University School of Medicine, Isehara, Kanagawa, Japan
- 3Department of Medicine, University of Virginia, Charlottesville, VA, United States
- 4Graduate School of Medicine, The University of Tokyo, Bunkyo, Tokyo, Japan
Entamoeba histolytica is the causative agent of amoebic dysentery and liver abscess in humans. The parasitic lifestyle and the virulence of the protist require elaborate biological processes, including vesicular traffic and stress management against a variety of reactive oxygen and nitrogen species produced by the host immune response. Although the mechanisms for intracellular traffic of representative virulence factors have been investigated at molecular levels, it remains poorly understood whether and how intracellular traffic is involved in the defense against reactive oxygen and nitrogen species. Here, we demonstrate that EhArfX2, one of the Arf family of GTPases known to be involved in the regulation of vesicular traffic, was identified by comparative transcriptomic analysis of two isogenic strains: an animal-passaged highly virulent HM-1:IMSS Cl6 and in vitro maintained attenuated avirulent strain. EhArfX2 was identified as one of the most highly upregulated genes in the highly virulent strain. EhArfX2 was localized to small vesicle-like structures and largely colocalized with the marker for the trans-Golgi network SNARE, EhYkt6, but neither with the endoplasmic reticulum (ER)-resident chaperon, EhBip, nor the cis-Golgi SNARE, EhSed5, and Golgi-luminal galactosyl transferase, EhGalT. Expression of the dominant-active mutant form of EhArfX2 caused an increase in the number of lysosomes, while expression of the dominant-negative mutant led to a defect in lysosome formation and cysteine protease transport to lysosomes. Expression of the dominant-negative mutant in the virulent E. histolytica strain caused a reduction of the size of liver abscesses in a hamster model. This defect in liver abscess formation was likely at least partially attributed to reduced resistance to nitrosative, but not oxidative stress in vitro. These results showed that the EhArfX2-mediated traffic is necessary for the nitrosative stress response and virulence in the host.
Introduction
Entamoeba histolytica is the etiological agent of human amoebiasis, which presents clinical manifestations including colitis, dysentery, and hepatic liver abscess (Stanley, 2003; Santi-Rocca et al., 2009). Invasion of the intestinal mucosa by the trophozoites leads to colitis and dysentery. In 5%–10% of the symptomatic intestinal infections, the trophozoites are disseminated through the portal vein to other organs including the liver. The survival of trophozoites in the tissues and subsequent formation of niche for parasitism require mechanisms for inactivation of or evasion from the hostile host environments including the immune response.
Multiple cellular mechanisms such as motility, adherence, and cytolysis are coordinately involved in the pathogenesis of E. histolytica (Faust and Guillen, 2012). Adherence to the host cells is primarily mediated by 260-kDa Gal/GalNAc lectin on the plasma membrane, which is a heterodimer of 170-kDa heavy and 35/31-kDa light subunits (Petri et al., 2002). The lysine- and glutamic acid-rich protein (KERP1) was initially discovered by the proteome analysis of brush-border binding proteins (Perdomo et al., 2016). KERP1 is implicated in the virulence because KERP1 is involved in the binding to epithelial cells, and its gene expression is upregulated during liver abscess development (Santi-Rocca et al., 2008) and shown to be necessary for the progression of liver abscess (Seigneur et al., 2005). The traffic of KERP1 remains largely elusive because it was shown to be transported extracellularly via the non-classical endoplasmic reticulum (ER)–Golgi-independent pathway (Perdomo et al., 2016). Cysteine proteases (CPs) and amoebapores (APs) have been shown to be essential for tissue invasion and abscess progression (Thibeaux et al., 2012). Among 50 CP genes that are encoded in the genome of E. histolytica, only three, CP-A1, CP-A2, and CP-A5, are highly expressed in steady state and account for 90% of the CP activity in trophozoite lysates (Irmer et al., 2009). APs are ~100 amino acid (aa) small peptides containing the “saposin-like protein” (SAPLIP) domain (Nowak et al., 2004). CPs and APs are stored in the lysosomes of trophozoites (Saito-Nakano et al., 2004) and are secreted in a constitutive and contact-dependent manner with host cells, respectively (Leippe et al., 1995). Anti-sense repression of CP-A5 (Ankri et al., 1999) or AP-A (Zhang et al., 2004) caused decreased abscess size, suggesting that both factors are essential for abscess formation.
Membrane traffic is mediated by mechanisms that are highly conserved among eukaryotes. It is controlled by two major families of small GTPases and their accessory molecules (Hutagalung and Novick, 2011). The Arf and Rab families of GTPases are involved in vesicular budding and fusion, respectively, and their functions are regulated according to the status of the bound guanine nucleotides (GTP-bound active or GDP-bound inactive state) (Hutagalung and Novick, 2011; Jackson and Bouvet, 2014). The attachment of vesicles to the target membrane is promoted by the membrane-targeted Rab-regulated tethering complex. Subsequently, membrane fusion finally occurs by the action of soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptors (SNAREs) and their associated regulatory components (Ungermann and Langosch, 2005). The Entamoeba genome apparently contains more than 100 Rab genes. The number is comparable to or sometimes higher than those in multicellular eukaryotes (Saito-Nakano et al., 2005; Nakada-Tsukui et al., 2010). Due to their complexity and heterogeneity, only nine E. histolytica Rab proteins have been analyzed for their functions and shown to be involved in pathogenesis at least in vitro. EhRab5, EhRab7A, EhRab8A, EhRabA, and EhRabB were shown to be involved in phagocytosis and trogocytosis (Saito-Nakano et al., 2004; Nakada-Tsukui et al., 2005; Welter and Temesvari, 2009; Juarez-Hernandez et al., 2013; Hanadate et al., 2016). EhRab7A, EhRab7B, EhRab7D, and EhRab11B were shown to play a role in the intracellular CP transport and maturation (Nakada-Tsukui et al., 2005; Mitra et al., 2007; Saito-Nakano et al., 2007; Saito-Nakano et al., 2021). EhRab21 was demonstrated to be associated with attachment actin cytoskeleton reorganization (Emmanuel et al., 2015). EhRabB is the only Rab protein that was implicated in the virulence in vivo and in vitro. It was shown that expression of the dominant-negative mutant of EhRabB caused a reduction of phagocytosis in vitro and reduced the size of liver abscesses in the hamster model (Juarez-Hernandez et al., 2013). While biological roles of the representative Rabs have been elucidated, those of Arfs remain largely elusive.
In the present study, we describe the identification and characterization of one member of Arf GTPase family, EhArfX2. We identified EhArfX2 as one of the highly upregulated genes in the hamster liver-passed virulent strain when compared to the isogenic laboratory-maintained attenuated strain. We further demonstrated that EhArfX2 and the EhArfX2-mediated traffic pathway are involved in the defense against nitrosative stress, which is known to occur during liver abscess formation. We have further demonstrated that EhArfX2 is localized to the trans-Golgi network and involved in trafficking from the trans-Golgi network to lysosomes. Finally, we demonstrated the in vivo role of EhArfA2; expression of dominant-negative mutant EhArfX2 caused a decrease in the formation of liver abscesses in hamsters, which was likely attributable to decreased resistance to nitrosative stress.
Materials and Methods
Ethics Statement
The animal procedures were approved by the Institutional Animal Care and Use Committee (No. 211075) and conducted at the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited National Institute of Infectious Diseases, Japan.
Cell Culture
Trophozoites of the E. histolytica strain-attenuated HM-1:IMSS Cl6 (a-HM1) were cultured axenically at 35°C in BI-S-33 medium, as previously described (Diamond et al., 1972; Diamond et al., 1978). Liver-passed virulent HM-1 (v-HM1) was cultured in YIMDHA-S medium supplemented with live Crithidia fasciculata at 35°C (Kobayashi et al., 2005).
Transcriptome Analysis
The Affymetrix (Santa Clara, CA, USA) custom array containing probe sets for 9,435 open reading frames (ORFs) from the E. histolytica genome database was used (Gilchrist et al., 2006). Synthesis of cDNA, hybridization, washing, staining, scanning of the arrays, and data acquisition (Gene Chip scanner 3000I, Affymetrix) were performed according to standard Affymetrix protocols (Gilchrist et al., 2006). Differentially expressed genes were identified by the following criteria: a fold change of at least 2-fold upregulated or downregulated in the virulent strain (v-HM1) as compared to the attenuated strain (a-HM1), a raw signal intensity of >10, a standard deviation of triplicates of the normalized signal intensity of <0.5, and a p-value of Student’s t-test of <0.05.
Quantitative Real-Time PCR
The mRNA expression of EhArf and EhSar genes was analyzed by quantitative real-time PCR analyses, essentially as previously described (Gilchrist et al., 2006; Saito-Nakano et al., 2007), with some modifications (Applied Biosystems 7500, Foster City, CA, USA). RNA polymerase II served as an internal control (GenBank accession number XP_649091). The primers used are listed in Supplementary Table S1. The following parameters were used: an initial denaturation step at 95°C for 9 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 65°C for 1 min. A final step at 95°C for 9 s, 60°C for 9 s, and 95°C for 9 s was used to remove primer dimers.
Collection of EhArf GTPases and EhSNARE Orthologs
The E. histolytica genome database was searched using S. cerevisiae and Homo sapiens Sar, Arf, and Arl proteins as queries by BLASTP. The E. histolytica genome database was also searched by BLASTP using EhSar1a, EhArfX1, and EhArfX2. All the possible Entamoeba Sar/Arf/Arl protein sequences were reexamined with BLASTP analysis using individual amoebic protein as an inquiry sequence against the human database at the National Center of Biotechnology Information (NCBI) as reported previously (Saito-Nakano et al., 2005; Nakada-Tsukui et al., 2010).
Phylogenetic Analysis
Multiple alignment of sequences of EhArf GTPases was obtained using MUSCLE (Edgar, 2004) and was further corrected by manual inspection. With 22 sequences, 152 unambiguously aligned aa sites were used for the below analysis, corresponding to residues 5–10, 14–92, 94–113, 115–130, and 145–175 of the EhArfX2 sequence. Phylogenetic analyses by the maximum likelihood (ML), neighbor-joining (NJ), and maximum parsimony (MP) methods were performed using PROML, PROTDIST + NEIGHBOR, and PROTPARS programs, respectively, in the PHYLIP package (http://evolution.gs.washington.edu/phylip.html). To analyze the ML and NJ methods with the JTT model considering heterogeneous substitution rates across sites (the JTT + G model), the Γ-shape parameter of the discrete Γ-distribution with four categories that approximated site rates was estimated using the CODEML program in PAML package (Yang, 1997) and the ML best tree with homogeneous substitution rates across sites. Bootstrap analysis for each of the three methods was performed by the ML (the JTT + G model), NJ (the JTT + G model), or MP methods to the resampled data sets produced by the SEQBOOT programs in the PHYLIP package. One thousand resamplings were performed for all analyses. A consensus tree was generated using the CONSENSE program, in the PHYLIP package, based on the bootstrap analysis of the ML method. The AU test in the CONSEL program was used for statistical comparisons among the alternative trees (Shimodaira, 2002). The alternative trees for the test were prepared by grafting EhArfX2 branch to all branches of the backbone tree.
Plasmid Construction
To construct plasmids expressing C-terminus 3HA-tagged or 3myc-tagged EhArfX2, the EhArfX2 coding region was PCR amplified from cDNA synthesized from a-HM1 and inserted in the BglII site of pEhExHA or pKT-3M vector, respectively (Saito-Nakano et al., 2004; Nakada-Tsukui et al., 2005). Plasmids expressing EhArfX2Q68L and EhArfX2T28N mutants were constructed by PCR-mediated mutagenesis using a PrimeSTAR Mutagenesis Basal Kit (Takara, Japan). To construct plasmids expressing green fluorescence protein (GFP)-fusion protein, fragments coding EhGalT, EhSed5, and EhYkt6 were amplified from cDNA and inserted in the pEhExGFP vector (Watanabe et al., 2020), which was removed myc-tagged sequence from a previously reported myc-GFP fusion-expressing vector, pKT-MG (Saito-Nakano et al., 2004). The necessary information for plasmid construction is shown in Supplementary Table S1. To generate a vector for double-expressing CP-A5-HA and 3myc-tagged EhArfX2, a 1.7-kb fragment containing the 3myc-EhArfX2 coding region-flanked 5’ upstream and 3’ downstream untranslated regions (UTRs) was cloned into the SpeI site of CP-A5-HA-expressing plasmid (Sato et al., 2006a) as described previously (Saito-Nakano et al., 2004). Oligonucleotide sequences used for the plasmid construction were listed in Supplementary Table S1.
Amoeba Transformants
Plasmids were introduced into the a-HM1 and v-HM1 strains by lipofection as previously described (Nozaki et al., 1999). Transformant amoebae were selected by the addition of 1 µg/ml Geneticin after 24-h transfection and gradually increased to 6 µg/ml for 2 weeks.
Antibodies
Anti-Bip (Hanadate et al., 2016), anti-Vps26 (Nakada-Tsukui et al., 2005), anti-CPBF1 (Furukawa et al., 2012), and anti-CS1 (Okada et al., 2006) antibodies were previously described, respectively. Anti-GFP, anti-HA, and anti-Myc antibodies were purchased as follows: rabbit anti-GFP, A11122 (Molecular Probes, Invitrogen, Carlsbad, CA, USA); mouse anti-GFP, 11814460001 (Roche Diagnostics, Indianapolis, IN, USA); mouse anti-HA, clone 16B12, and rabbit anti-Myc, clone A-14 (Santa Cruz Biotech, Santa Cruz, CA, USA).
Indirect Immunofluorescence Analysis
An indirect immunofluorescence assay was conducted essentially as previously described (Saito-Nakano et al., 2004). Trophozoites were attached to 8-mm round wells on a glass slide, fixed with 3.7% paraformaldehyde, permeabilized with 0.1% Tx100, and reacted with antibodies. Alexa 488- and Alexa 568-conjugated immunoglobulin G (IgG; Molecular Probes) were used as secondary antibodies. Acidic compartments of trophozoites were stained with 500-fold dilution of LysoTracker Red DND-99 (Invitrogen) for 12 h. Images were acquired using LSM510 or LSM780 confocal laser scanning microscope (Zeiss, Germany). Colocalization analyses were processed using Fiji software (Schindelin et al., 2012).
Subcellular Fractionation
The subcellular fractionation was performed as reported previously with some modifications (Saito-Nakano et al., 2007; Hanadate et al., 2016). Approximately 3 × 105 amoeba cells were washed with cold phosphate buffered saline (PBS) containing 2% glucose, resuspended in homogenization buffer (250 mM sucrose, 50 mM Tris, pH 7.5, 50 mM NaCl, 0.1 mg/ml of E-64), and homogenized on ice with 30 strokes with a Dounce homogenizer. After unbroken cells were removed by centrifugation at 400 × g for 2 min, the supernatant was centrifuged at 13,000 × g at 4°C for 10 min to obtain the pellet (p13) and supernatant (s13) fractions. The s13 fraction was further separated by centrifugation at 100,000 × g at 4°C for 1 h to obtain soluble (s100) fractions. These fractions were subjected to immunoblot analyses with anti-HA, anti-CPBF1, or anti-CS1 antibodies.
Cysteine Protease Assay
CP activity in the culture supernatant and parasite lysates was assayed as previously described (Saito-Nakano et al., 2007). CP activity was measured using z-Arg-Arg-7-amino-4-trifluoromethylcoumarin substrate as described (Leippe et al., 1995).
Phagocytosis Assay
Phagocytosis of erythrocytes was described previously (Saito-Nakano et al., 2004). Amoeba trophozoites were attached to 8-mm round wells on a glass slide and then incubated with 1 × 107 erythrocytes/ml in biosate, yeast extract, iron-serum (BIS) medium under the indicated incubation times at 37°C. Cells were fixed with paraformaldehyde, ingested erythrocytes were stained with diaminobenzidine, and EhArfX2-HA mutant-expressing trophozoites were stained with anti-HA antibody. The number of ingested erythrocytes in EhArfX2-HA-expressing trophozoites was counted under the microscope.
Hamster Liver Abscess Model
Approximately 4 × 105 v-HM1 were injected into the left lobe of the liver of 5-week-old Syrian hamsters. Animals were sacrificed 6 days post infection, and the liver and abscess were dissected and weighed separately (Sato et al., 2006b).
Assays for Sensitivity to Nitric Oxide and H2O2
EhArfX2 transformants established in the v-HM1 strain were seeded in 96-well plates at 1 × 104 cells/well in the presence of the indicated concentration of drugs with BIS medium. After 18-h culture, the medium was removed and the viability of attached trophozoites was measured by the addition of 10% WST-1 reagent (Roche) in Opti-MEM medium for 20 min at 37°C (Penuliar et al., 2015). The absorbance at 450 nm was measured using a DTX880 Multimode Detector (Beckman Coulter, Brea, CA, USA). Experiments were repeated three times with triplicate replicates per experiment.
Results
Comparative Transcriptomic Analysis of Animal-Passaged Virulent and Culture-Attenuated HM-1 Strains Led to Identification of Upregulated and Downregulated E. histolytica Genes
To identity genes that are involved in virulence in vivo, we exploited comparative transcriptome analysis using a custom-made microarray containing probe sets for all ORFs from the Entamoeba genome database, which we previously designed and used in a number of studies (Gilchrist et al., 2006; Husain et al., 2011; Penuliar et al., 2012; Penuliar et al., 2015; Jeelani et al., 2017). Gene expression profiles were compared between two isogenic strains with the identical genetic background but maintained in different ways: one that had been passaged through the hamster liver (virulent HM-1:IMSS Cl6 strain, v-HM1 hereinafter) and one that had been continuously cultured in vitro and was no longer capable of producing abscesses in hamsters (a-HM1) (see Figure 4C, described below). The transcriptomic comparison of v-HM1 and a-HM1 identified 59 genes that were differentially expressed (>4-fold, q-value <0.05) with the significance of gene expression was determined using the significance analysis of microarray statistical program (https://tibshirani.su.domains/SAM/) (Gilchrist et al., 2006). Among them, 30 genes were upregulated (Table 1) and 29 were downregulated in v-HM1 (Supplementary Table S2). Among the 30 upregulated genes, three genes appear to be associated with cellular functions related to membrane traffic based on annotation: two Arf GTPases (EHI_189960, EHI_073480) and ATG4 cysteine peptidase (EHI_055660). Neither Rab GTPase nor SNARE was identified as upregulated or downregulated genes. In this study, we describe our analysis of the Arf GTPases that we identified by the comparative transcriptomic analysis. Characterization of other upregulated and downregulated genes will be reported elsewhere.
Verification of Specific Overexpression of EhArfX2 in a Virulent Strain of E. histolytica
The E. histolytica genome contains genes encoding 14 members of the Sar/Arf family of GTPases based on the E. histolytica genome database at AmoebaDB (http://amoebadb.org/amoeba/, version 54 released on September 8, 2021). Phylogenetic analysis revealed none of the E. histolytica Sar/Arf family GTPases has close kinship with known human and yeast counterparts at statistically supported levels (bootstrap values >70 by at least one of the following analyses: ML, DM, and MP) (Figure 1A). Thus, assignment of orthologs is not possible. We alternatively assigned E. histolytica Arf genes with “X” as a prefix, as all genes appear to be lineage specific. E. histolytica Arfs were numerically designated in a descending order of the percentage identity to human Arf1 with the “X” prefix (Supplementary Table S3). In a case where two E. histolytica Arf proteins show >99% identity, they are considered to be allelic isotypes and designated with alphabets in lowercase, e.g., EhArfX1a and EhArfX1b. EhArfX5 and EhArfX6 form monophyly and are positioned closely to human and yeast Arl1, although not statistically supported. EhArfX7–EhArfX9 also form monophyly and represent an E. histolytica-specific clade (Figure 1A). Three allelic isotypes of EhSar1a–EhSar1c form a well-supported monophylic clade with >99% identity and show high similarity with human and yeast Sar1 (Supplementary Table S3). EhArfX1a/b, X2, and X3 appear to form monophyly, but the tree topology was not statistically supported.
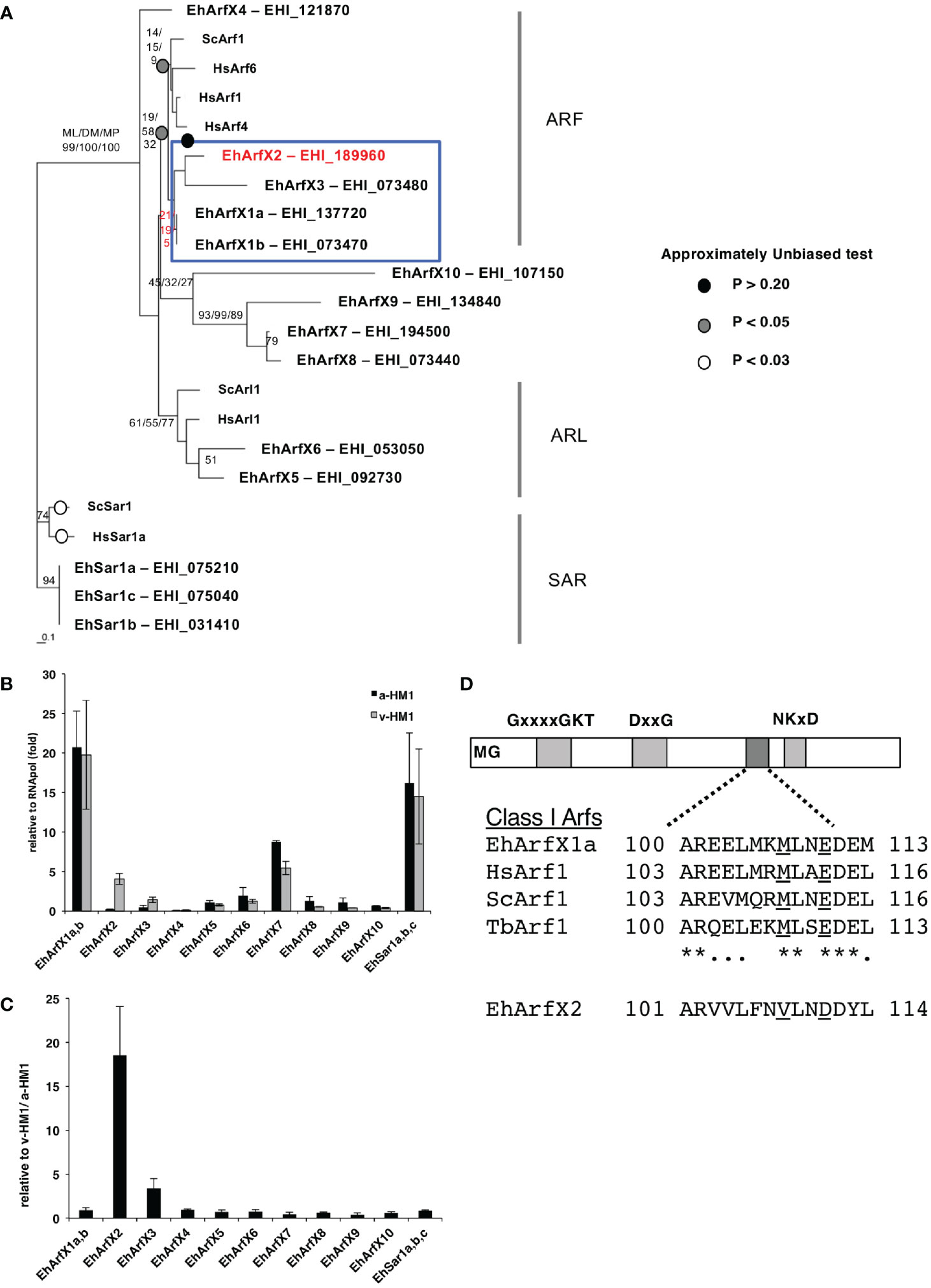
Figure 1 Phylogenetic position, mRNA expression, and structure of E. histolytica Arf proteins. (A) The consensus maximum likelihood (ML) tree of 14 members of the Sar/Arf family of GTPases from E. histolytica and representative members from yeast and human. Bootstrap probabilities by the ML method are attached to the internal branches. Branches with less than 50% bootstrap support are unmarked. For the node of interest, bootstrap values determined by the distance matrix (DM) and maximum parsimony (MP) methods are also shown. Circles highlighted by white, gray, and black indicate the branches in which p-value by the AU test is more than 0.05. The branches in which p-value is less than 0.01 are unmarked. (B) Relative abundance of mRNA of 14 E. histolytica Sar/Arf genes. Quantitative real-time PCR was performed using cDNA synthesized with total RNA from a-HM1 and v-HM1 as templates. As EhArfX1a (EHI_137720) and EhArfX1b (EHI_073470) shared 99.8% nucleotide identity, and three EhSar1 genes (EHI_075210, EHI_075040, EHI_031410) shared 99.8% nucleotide identities, it was impossible to distinguish each isotype. Bars indicate standard deviation (n = 3). (C) Normalized mRNA expression levels of 14 E. histolytica Sar/Arf genes in v-HM1 relative to a-HM1. Bars indicate standard deviation (n = 3). (D) Schematic diagram of class I Arf proteins. EhArfX2 lacks the Golgi-targeting motif that is observed in EhArfX1 and Arf1 from other organisms. The consensus MxxE motif is underlined. *conserved amino acids.
The mRNA levels of EhArfX2 and EhArfX3 in a-HM-1 were relatively low when compared to other members of 14 Sar/Arf GTPase family genes, as suggested by our microarray analysis and further validated by quantitative real-time PCR (Figure 1B). In v-HM1, mRNA levels were higher compared to a-HM-1: 18 ± 5.5-fold for EhArfX2 and 3.4 ± 1.1-fold for EhArfX3, respectively (Figure 1C). In the subsequent study, we focused on the biological and pathogenic roles of EhArfX2.
Structural Features of E. histolytica EhArfX2
Among the Arf subfamily of GTPases, the function of Arf1, which is primarily localized to the Golgi apparatus in humans, yeasts, Trypanosoma brucei, and Arabidopsis thaliana (Teal et al., 1994; Price et al., 2007; Matheson et al., 2008), has been well studied. It was shown that the Golgi targeting of Arf1 is regulated via interaction of the MxxE motif present in the second (DxxG) and third (NKxD) GTP-binding consensus sequences (Honda et al., 2005) (Figure 1D). Although EhArfX1a does not show high similarity to Arf1 based on protein alignment and phylogenetic analysis (Figure 1A), the MxxE motif is well conserved in EhArfX1a and EhArfX1b (Figure 1D). EhArfX1a and EhArfX1b may be the functional orthologs of mammalian Arf1. In contrast, EhArfX2 does not possess the MxxE motif, and subcellular localization and function of EhArfX2 could not be presumed in silico.
EhArfX2 Is Colocalized With the E. histolytica Ortholog of Trans-Golgi SNARE EhYkt6 but Not With Either Cis-Golgi or Endoplasmic Reticulum Markers
The Arf family of GTPases is known to be localized to the Golgi apparatus, the trans-Golgi network, endosomes, and the plasma membrane in other organisms (Jackson and Bouvet, 2014). We examined the subcellular localization of EhArfX2 using ER and the Golgi markers (Makiuchi and Nozaki, 2014). Indirect immunofluorescence assay using the transformant expressing GFP fused with the signal peptide (SP) from Gal/GalNAc lectin at the N-terminus and ER retention signal (KDEL) at the C-terminus (SP-GFP-KDEL) (Teixeira and Huston, 2008) with antiserum against E. histolytica Bip, the ER luminal chaperon (EHI_199890) (Munro and Pelham, 1986; Hanadate et al., 2016), revealed colocalization with high Pearson’s correlation coefficient (R = 0.67; Supplementary Figure S1A). While indirect immunofluorescence assay using the transformant expressing EhArfX2 fused with the HA tag at the C-terminus (EhArfX2-HA), anti-HA and anti-Bip antibodies, revealed different staining patterns (R = 0.14; Figure 2A). Next, an E. histolytica ortholog of Golgi-luminal galactosyl transferase (EhGalT), which has the N-terminal SP but lacks a transmembrane domain, was used for the Golgi luminal marker [EHI_000660, 45% aa identity to Strongylocentrotus purpuratus (sea urchin) galactosyl transferase, e-value = 3e-6]. GalT presumably, by analogy, catalyzes a transfer of galactose, N-acetylgalactosamine, and N-acetylglucosamine in the Golgi lumen (Magnelli et al., 2008). HA-tagged EhGalT, detected with anti-HA antibody, showed dot-like structures that were well associated with ER stained by anti-Bip antibody (R = 0.74; Supplementary Figure S1B). The high-resolution 3D image revealed that the dot-like EhGalT signals were associated with the continuous Bip signal (Supplementary Figure S1C). The distribution of EhArfX2-HA detected with anti-HA antibody differed from the pattern of EhGalT-GFP detected with anti-GFP antibody (R = 0.15; Figure 2B).
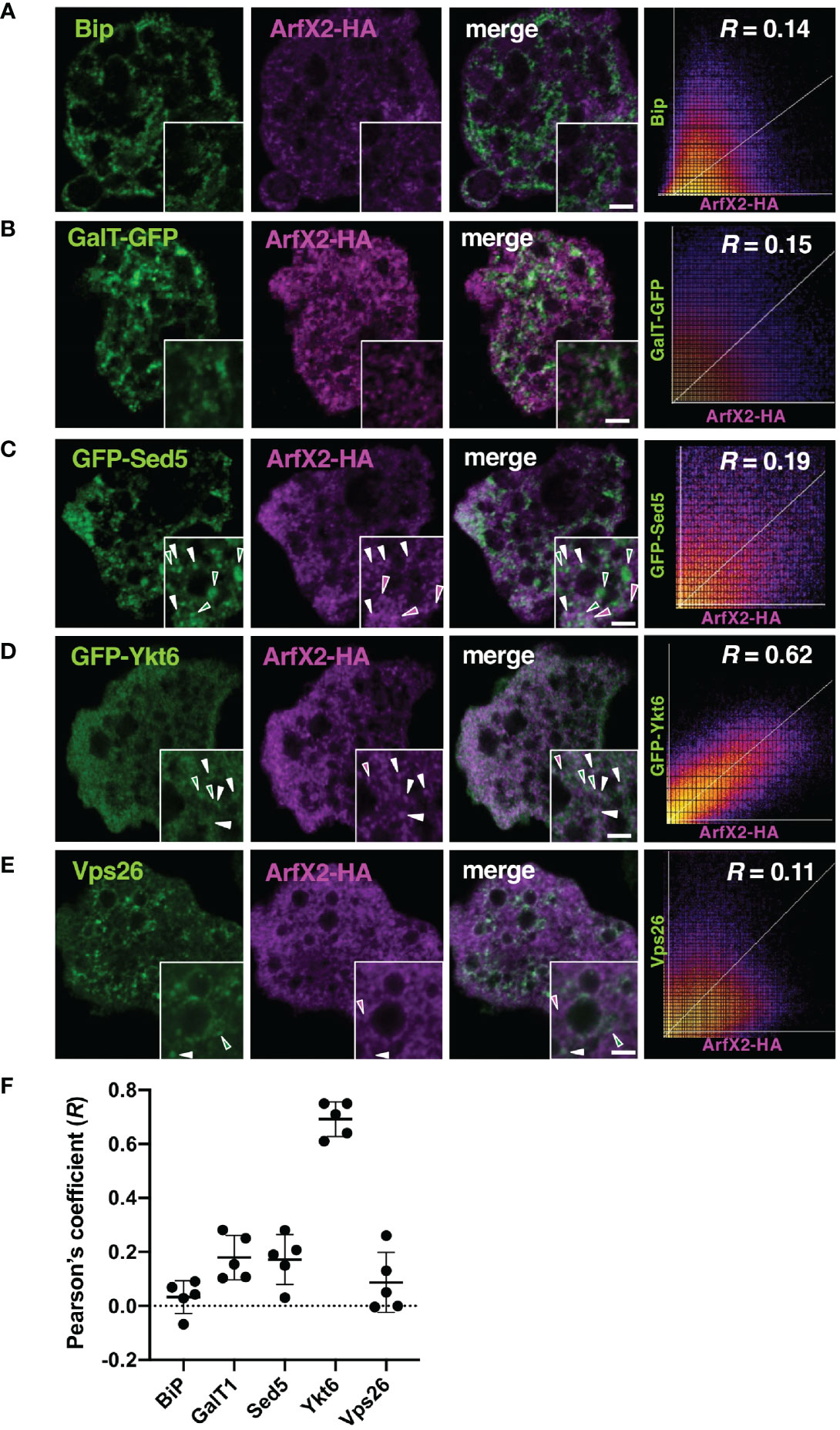
Figure 2 Subcellular localization of EhArfX2-HA and other organelle markers. EhArfX2-HA-expressing cells were stained with anti-HA antibodies (magenta) and anti-Bip (A) or anti-Vps26 antibodies (E), respectively (green). EhArfX2-HA and EhGalT-GFP double-expressing (B), EhArfX2-HA and GFP-EhSed5 double-expressing (C), or EhArfX2-HA and GFP-EhYkt6 double-expressing (D) cells were stained with anti-GFP and anti-HA antibodies. Colocalized vesicular signals are indicated with white arrowheads. Scatter plots depict colocalization analysis. R = Pearson’s correlation coefficient. Thick bars, 2 µm. (F) The R values obtained from panels (A–E) are plotted, and average (bold bars) and standard deviation (thin bars) are indicated (n = 5).
We further attempted to localize EhArfX2 by examining if EhArfX2 is colocalized with some organelle-specific SNAREs, which are known to mediate membrane fusion together with Rab GTPases (Hong, 2005). In membrane fusion, the coiled-coil SNARE motif of R-SNAREs on the acceptor vesicle membrane (previously called v-SNARE) makes specific interaction with the SNARE motif of Q-SNARE (or called t-SNARE) on the donor membrane to form a cognate complex. The SNARE motif is ubiquitous and evolutionally conserved throughout eukaryotic lineages (Hong, 2005; Kloepper et al., 2007). We identified 25 SNARE, 16 Q-SNARE, and nine R-SNARE genes in the E. histolytica genome (http://amoebadb.org/amoeba/, version 54 released on September 8, 2021) by the genome wide survey using the conserved coiled-coil SNARE motif domain, followed by phylogenetic analysis (Supplementary Figures S2A, S3A). The description of the entire repertoire of SNAREs will be reported elsewhere. Among them, two SNAREs were used in this study as cis-Golgi and trans-Golgi markers. EHI_181290 is an E. histolytica ortholog of cis-Golgi SNARE Sed5 from S. cerevisiae and A. thaliana (also annotated as syntaxin-5 in human). The mutual overall aa identity of the Sed5 orthologs among organisms such as yeasts, plants, and mammals is low (31%–37%, e-value = 8e-33–2e-48); similarly, it is also low between E. histolytica Sed5 ortholog, EhSed5 (EHI_181290), and orthologs from these organisms (e.g., 28% aa identity to human syntaxin-5, e-value = 3e-18) (Supplementary Figure S2A). Despite the low level of aa conservation, all known Sed5 orthologs are known to localize to the cis-Golgi membrane, warranting validation as the cis-Golgi marker (Shestakova et al., 2007; Chatre et al., 2009). Three Golgi-associated signatures that were identified in Sed5 from other organisms are well conserved in EhSed5: the short stretch of aa close to the N terminus (RDRTxEF) (Yamaguchi et al., 2002), the coiled-coil SNARE domain of ~60 aa close to the C terminus, and the short (15 aa) C-terminal transmembrane segment (Supplementary Figure S2B). It was established that the RDRTxEF motif is required for the SNARE complex assembly and functions in ER–Golgi fusion (Yamaguchi et al., 2002). Compared to the plasma membrane-localized syntaxins, which typically contain a relatively long (the average of 23 aa) transmembrane segment, the transmembrane region of Sed5 and Syn5 from yeast and human, respectively, is shorter and rich in phenylalanine (Supplementary Table S4) (Bretscher and Munro, 1993; Banfield et al., 1994). On the other hand, EHI_052110 is an apparent E. histolytica ortholog of trans-Golgi SNARE Ykt6 (37% aa identity to human Ykt6, e-value = 3e-23) (Tai et al., 2004; Chen et al., 2005) and used as a trans-Golgi marker (Supplementary Figure S3A). Unlike other SNARE members, Ykt6 lacks a transmembrane domain but possesses a conserved C-terminal dicysteine motif, which is modified by farnesylation and geranylgeranylation (Sakata et al., 2021) (Supplementary Figure S3B). EhYkt6 showed good colocalization with EhArfX2-HA (R = 0.62) (Supplementary Figure S4A), whereas EhSed5 did not (R = 0.19) (Figures 2C, D). The subcellular localization of EhSed5, Bip, and EhYkt6 was clearly distinct (R < 0.1; Supplementary Figures S4B, C). The localization of EhArfX2 was also different from the EhVps26-positive endosomal-like compartment (Nakada-Tsukui et al., 2005) (Figure 2E). These results indicate that EhArfX2 showed the highest colocalization with EhYkt6 among ER and the Golgi markers (Figure 2F) and localizes to the trans-Golgi or its equivalent compartment in E. histolytica.
EhArfX2 Is Involved in Lysosomal Trafficking and Biogenesis
We further investigated the trafficking pathway mediated by EhArfX2, the amoeba transformants that expressed either a constitutively active GTP-bound or an inactive GDP-bound form of EhArfX2 mutant, which corresponds to human Arf1Q71L or Arf1T31N, respectively (Dascher and Balch, 1994; Zhang et al., 1994). We took advantage of an approach of dominant-negative mutant because gene repression by small antisense RNA-mediated transcriptional gene silencing (Morf et al., 2013; Nakada-Tsukui et al., 2018) was not feasible based on the fact that EhArfX2 gene was not expressed in a-HM1, and neither in G3 strain, which is commonly used for gene silencing (Figure 1B). We assumed that EhArfX2 may be involved in trafficking from the trans-Golgi to lysosomes. The dominant-active EhArfX2Q68L and dominant-negative EhArfX2T28N mutants fused with the HA-tag on the C-terminus were constitutively expressed (Figure 3A). This episomal plasmid-driven expression caused up to 10–58-fold increase in the sum (i.e., intrinsic plus exogenous) of the steady-state EhArfX2 mRNA in a-HM-1 (Supplementary Figure S5), and the sum level of the EhArfX2 transcripts was comparable to the level of EhArfX2 mRNA in v-HM1 (Figure 1C). Indirect immunofluorescence assay using anti-HA antibody showed that both HA-tagged EhArfX2Q68L and EhArfX2T28N mutant proteins were distributed to small vesicles and the cytosol, similar to that of wild-type EhArfX2-HA (Figure 3A). The expression of EhArfX2 mutants affected the number of lysosomes visualized with LysoTracker Red. The number of lysosomes per section was increased in EhArfX2wt (4.8 ± 0.4 lysosomes per section, p < 0.05 compared to mock control) and EhArfX2Q68L (6.0 ± 0.7, p < 0.005) compared to mock control (4.0 ± 0.5). On the contrary, the number of lysosomes was decreased by the expression of EhArfX2T28N (2.7 ± 0.3 lysosomes per section, p < 0.01) (Figure 3B). Subcellular fractionation of cell lysates followed by immunoblot analysis showed that the amount of EhArfX2WT-HA in the cell lysate and the soluble fraction (s100) were much higher than those of EhArfX2Q68L and EhArfX2T28N-HA (Figure 3C). A fraction of EhArfX2WT-HA, EhArfX2Q68L-HA, and EhArfX2T28N-HA was also distributed to the membrane-associated fractions (p13 and p100). A majority of EhArfX2Q68L-HA protein (68% ± 4%) was associated with the membrane (p13 and p100 fractions), while 35% ± 6.7% EhArfX2wt-HA and, unexpectedly, 48% ± 16% EhArfX2T28N-HA were partitioned to the membrane-associated fractions (Figure 3C). These data are consistent with a premise that EhArfX2 cycles between small vesicular membranes and the cytosol in a nucleotide-dependent manner and is involved in lysosome biogenesis.
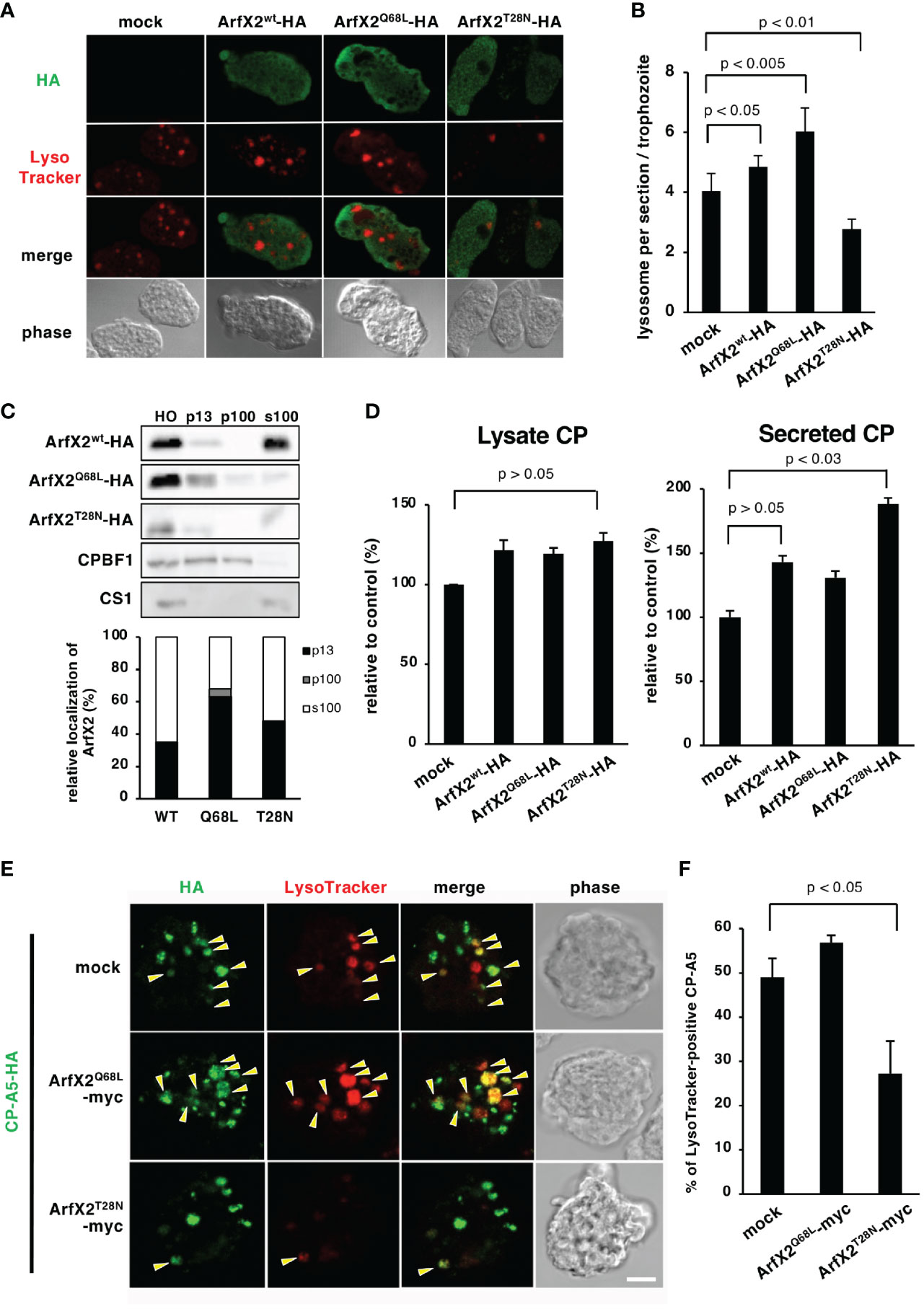
Figure 3 Effects of expression of EhArfX2 wild type and mutants on lysosome trafficking. (A) Indirect immunofluorescence assay of EhArfX2WT-HA, EhArfX2Q68L-HA, and EhArfX2T28N-HA. EhArfX2 and lysosomes were stained with anti-HA (green) and LysoTracker Red (red), respectively. (B) Number of lysosomes in EhArfX2WT-HA-, EhArfX2Q68L-HA-, and EhArfX2T28N-HA-expressing transformant cells. Lysosomes were visualized with LysoTracker Red, and the number of lysosomes per confocal section was counted in independent 30 trophozoites. (C) Subcellular fractionation of EhArfX2WT-HA, EhArfX2Q68L-HA, and EhArfX2T28N-HA. Homogenate (HO) was separated into low-speed pellet (13,000 × g, p13), high-speed pellet (100,000 × g, p100), and soluble (100,000 × g, s100) fractions, respectively (upper panel). EhArfX2-HA was probed with anti-HA antibody. Antibodies against CP-binding protein family 1 (CPBF1) and cysteine synthase 1 (CS1) were used as a marker of membrane and cytosol fractions, respectively. Quantification of EhArfX2-HA in the p13, p100, and s100 fractions by chemiluminescence measurement (lower graph). (D) CP activity in the culture medium (left) and cell lysates (right) in the control (mock), EhArfX2wt-HA-, EhArfX2Q68L-HA-, and EhArfX2T28N-HA-expressing cells. (E) Localization of HA-tagged CP-A5 in EhArfX2Q68L-myc- and EhArfX2T28N-myc-expressing cells. CP-A5-HA and lysosomes were stained with anti-HA antibody (green) and LysoTracker Red (red), respectively. LysoTracker-positive CP-A5-HA signals are indicated by yellow arrowheads. Scale bar, 10 µm. (F) Relative colocalization of CP-A5-HA and LysoTracker. The number of LysoTracker and CP-A5-HA double-positive compartments and the total number of CP-A5-HA-positive compartments in the cells were counted (n = 30 cells). The percentages of the double-positive compartments per total number of CP-A5-HA-positive compartments are shown. Significance was evaluated by the Student’s t-test.
EhArfX2 Is Involved in Cysteine Protease Transport and Maturation
CPs were shown to be involved in a variety of biological processes in E. histolytica including degradation of mucin and extracellular matrix (Moncada et al., 2006), target cell cytolysis (Thibeaux et al., 2012), inactivation of immune anaphylatoxins, immune evasion, and implicated in tissue invasion in the colon and the liver (Reed et al., 1995; Ankri et al., 1999; Moncada et al., 2006; Thibeaux et al., 2012; St-Pierre et al., 2017). It was shown that CPs are stored in lysosomes and constitutively secreted to the culture medium (Leippe et al., 1995; Saito-Nakano et al., 2004; Sato et al., 2006a). We examined whether overexpression of EhArfX2WT or mutants affects the lysosome biogenesis and lysosomal functions, more specifically, intracellular storage and secretion of CPs. EhArfX2T28N-expressing cells, which showed a defect in lysosome formation, showed 1.8 ± 0.2-fold increased secretion of CP to the medium when compared to the mock transformant cells (Figure 3D, right). The secretion of CP also appeared to be increased, although statistically not proven to be significant, in EhArfX2WT- and EhArfX2Q68L-expressing transformants (Figure 3D, right). On the contrary, the total amount of CP activity in lysates was not changed (Figure 3D, left). Increased secretion of CP in EhArfX2T28N cells was not caused by the increase in the transcription levels of CP-A2 and CP-A5 mRNA because the transcript levels of CP-A2 and CP-A5 mRNA were comparable among the strains expressing EhArfX2 WT or mutants and mock strain (Supplementary Figure S5).
Interference of CP trafficking to lysosomes and subsequent missecretion of CPs by EhArfX2T28N expression was further examined by the visualization of intracellular CPs. One of the major CPs implicated in pathogenesis, CP-A5, was tagged with HA at the C-terminus (Sato et al., 2006a) and co-expressed with either EhArfX2Q68L or EhArfX2T28N tagged with myc (Figure 3E). EhArfX2 with the C-terminal myc tag showed indistinguishable distribution from HA-tagged EhArfX2; EhArfX2-HA was also localized to small vesicular and cytosolic patterns (Supplementary Figure S6). CP-A5-HA was predominantly localized to lysosomes, stained with LysoTracker Red, in EhArfX2Q68L-expressing cells, as previously shown in the parental line (Sato et al., 2006a) (Figure 3E, mock). In EhArfX2T28N-expressing cells, 27% ± 7.3% of CP-A5-HA-positive compartments were stained with LysoTracker Red (Figures 3E, F). In contrast, in mock and EhArfX2Q68L cells, 48% ± 4% and 56% ± 1% of CP-A5-positive compartments were LysoTracker Red positive, respectively (Figure 3F). These data indicate that expression of EhArfX2T28N inhibits the intracellular transport of CP-A5 to lysosomes. In order to directly show that the traffic of lysosomal enzymes is regulated by EhArfX2, we monitored traffic and activity of CPs, which are the representative lysosomal enzymes and also the virulence-associated factor of the parasite. Gelatin substrate gel assay showed that the secretion of CPs to the medium increased in EhArfX2WT-HA, EhArfX2Q68L-HA, and EhArfX2T28N-HA strains (Supplementary Figure S7A). The intensity of the band corresponding to EhCP-A1 and the top band of two bands corresponding to EhCP-A2 appear to be increased in EhArfX2WT-HA, EhArfX2Q68L-HA, and EhArfX2T28N-HA strains. Note that EhCP-A2 was previously shown to localize to lysosomes (Saito-Nakano et al., 2007). Furthermore, destruction of the Chinese hamster ovary (CHO) monolayer by EhArfX2T28N-HA strain also increased compared to mock control strain (Supplementary Figure S7B). Furthermore, the increased cytopathic activity was canceled by CP inhibitor E64, validating that the observed cytopathic effect was attributable to CP activity. The gelatin substrate gel electrophoresis (Tillack et al., 2006) using culture supernatant of EhArfX2T28N-HA revealed increased secretion of EhCP-A2 (Supplementary Figure S7A), which is localized to the amoebic lysosomes (Saito-Nakano et al., 2007). Increased secretion of CP in EhArfX2T28N-HA was verified by the disruption of monolayers of cultured mammalian cells, such as CHO cells in vitro, and the in vitro virulence was abolished by the addition of CP inhibitor E64 (Supplementary Figure S7B). These results further support the premise that expression of EhArfX2T28N inhibited transport of CPs to lysosomes from the trans-Golgi network and caused subsequent mistargeting of CPs to the culture medium. These results indicate that the transport of lysosomal enzymes including CPs is regulated by EhArfX2 and also suggest that EhArfX2T28N expression may influence the pathogenesis of the parasite through CPs.
Expression of EhArfX2T28N Causes a Reduction in Abscess Formation
To further investigate the causal connection between EhArfX2 expression and pathogenic activities, we next evaluated in vitro cell growth (Figure 4A) and ingestion of erythrocytes via trogocytosis or phagocytosis (Ralston et al., 2014; Somlata et al., 2017) (Figure 4B). The population doubling time of EhArfX2WT-, EhArfX2Q68L-, and EhArfX2T28N-expressing lines was unchanged when compared with the mock control (Figure 4A). In addition, erythrophagocytosis activity was also comparable among the strains (Figure 4B). We next evaluated the effect of EhArfX2WT and the mutants on in vivo virulence, e.g., liver abscess formation, using the hamster liver abscess model. Based on the disturbed traffic of CPs observed in EhArfX2T28N-expressing transformant line (Figure 3D), it was expected that EhArfX2T28N-expressing lines may show a change in the ability of liver abscess formation. The transformant lines expressing EhArfX2WT or dominant-negative EhArfX2T28N were established using v-HM1 as a parental line and then injected into the hamster liver (Figure 4C). It should be noted that none of the transformants derived from a-HM1 was capable of forming liver abscesses in hamsters. Overexpression of EhArfX2WT-HA in v-HM1 did not affect the apparent in vivo virulence, i.e., liver abscess formation, when compared to mock control strain. Parental (non-transformant) v-HM1, mock transformant, and EhArfX2WT-HA-expressing transformant strains formed abscesses that occupied 39.5% ± 6.7%, 36% ± 8.7%, and 32.9% ± 3.4% of the whole liver weight, respectively. In contrast, EhArfX2T28N-HA-expressing strain formed significantly smaller abscesses, accounting for 15.4% ± 3.8% of the whole liver weight. These results indicate that expression of EhArfX2T28N, which caused impairment of lysosome biogenesis and regulation of CP trafficking, also resulted in a reduction in the ability of liver abscess formation despite comparable rate of growth and erythrophagocytosis in vitro. Overexpression of EhArfX2WT-HA upon v-HM1 genetic background did not have an additive effect on liver abscess formation.
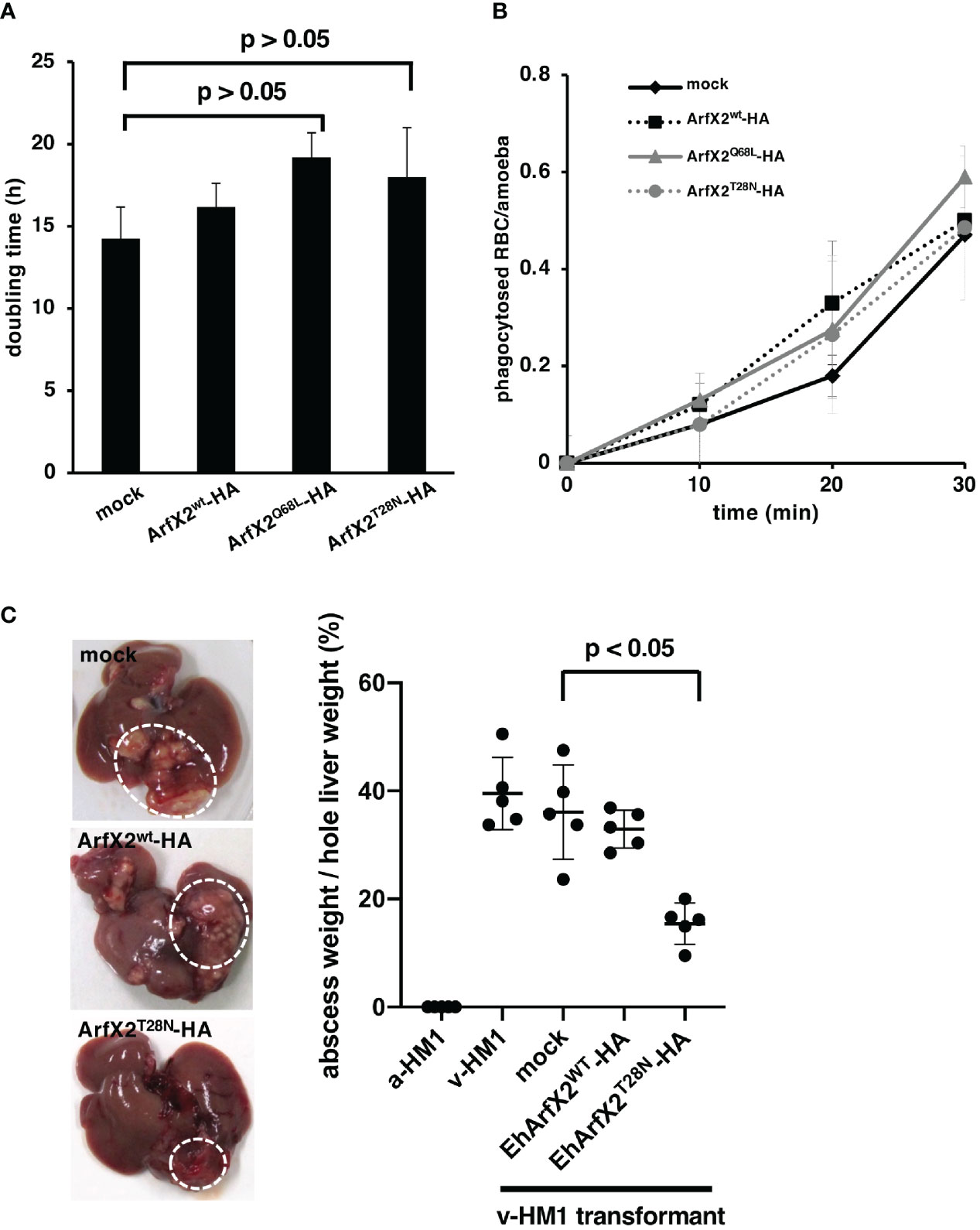
Figure 4 Effects of EhArfX2WT-HA, EhArfX2Q68L-HA, and EhArfX2T28N-HA expression on growth, erythrophagocytosis, and liver abscess formation. (A) The doubling time of EhArfX2WT-HA, EhArfX2Q68L-HA, EhArfX2T28N-HA, and mock control cells (n = 3). (B) Efficiency of phagocytosis of human erythrocytes. Amoeba transformant cells were incubated with human erythrocytes at the indicated times. Cells were fixed with paraformaldehyde, and the number of ingested erythrocytes were counted under the microscope (n = 30). (C) Amoebic liver abscess formation caused by infection with a-HM1, v-HM1, v-HM1 transformants expressing EhArfX2WT-HA, EhArfX2T28N-HA, and mock control cells. Six days after inoculation, hamsters were sacrificed and whole liver and separated liver abscesses were weighted (n = 5). Statistical significance was evaluated by the Student’s t-test.
EhArfX2T28N Caused a Defect in Nitrosative Stress Response
We speculated that EhArfX2T28N-expressing transformant has an additional defect other than oversecretion of CPs that explains less capacity for liver abscess formation. To this end, we hypothesized if EhArfX2T28N-expressing transformant has decreased resistance to environmental stresses because, during liver abscess formation, trophozoites are exposed to oxidative or nitrosative stresses, which are mainly produced by activated macrophages (Lin and Chadee, 1992; Dodson et al., 2013; Jeelani and Nozaki, 2016). The viability of EhArfX2T28N-HA- or EhArfX2WT-HA-expressing transformant that had been created from v-HM1 was evaluated under the exposure of nitrosative [sodium nitroprusside (SNP)] (Santi-Rocca et al., 2012) or oxidative (H2O2 and paraquat) stresses (Husain et al., 2012). Interestingly, EhArfX2T28N-HA-expressing line showed increased sensitivity to SNP when compared to EhArfX2WT-HA-expressing or mock transformant (Figure 5A). In contrast, sensitivity to H2O2 and paraquat was comparable to that of EhArfX2 WT-expressing and mock lines (Figures 5B, C). These results indicate that EhArfX2 is involved in the anti-nitrosative stress response.
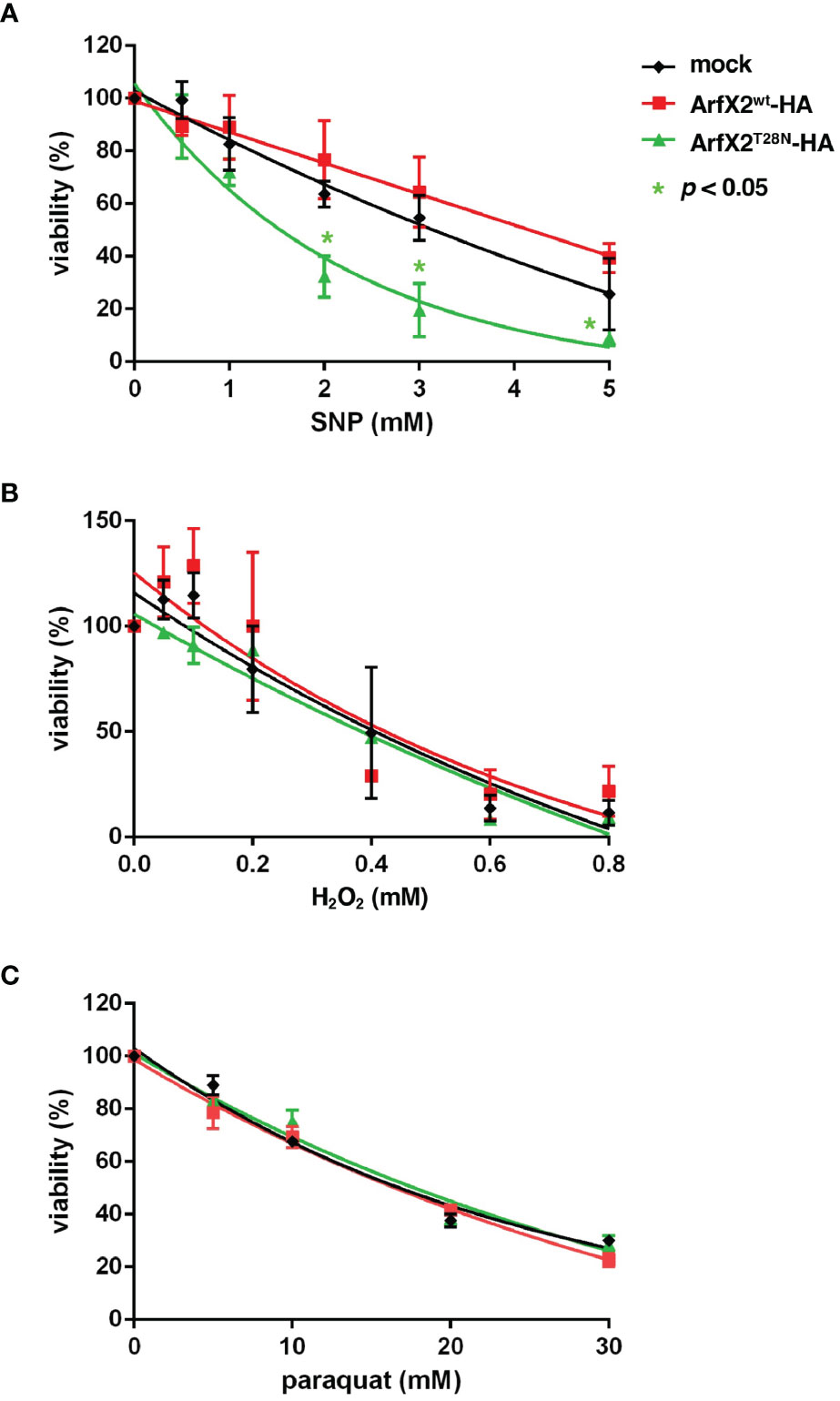
Figure 5 In vitro sensitivity to oxidative and nitrative stress of EhArfX2WT-HA and EhArfX2T28N-HA expression. Viability of trophozoites of EhArfX2WT-HA, EhArfX2T28N-HA, and mock transformants after 18-h treatment with varying concentrations of sodium nitroprusside (SNP) (A), H2O2 (B), or paraquat (C) is shown.
Discussion
EhArfX2 Is Involved in Cysteine Protease Transport, Nitrosative Stress Response, and Amoebic Liver Abscess Formation
In this study, we have demonstrated for the first time that Arf GTPase plays important roles in intracellular trafficking of the major virulence-associated factor (CPs) and the defense against nitrosative stresses and is thus involved in liver abscess formation in animals. This is the first report on the involvement of Arf GTPases in the pathogenesis of E. histolytica and on the engagement of Arf GTPases in anti-nitrosative stress responses. It has been well established that Rab-regulated membrane trafficking (Nakada-Tsukui et al., 2005; Mitra et al., 2007; Saito-Nakano et al., 2007; Hanadate et al., 2016; Saito-Nakano et al., 2021), calcium and phosphoinositide signaling (Bharadwaj et al., 2017; Somlata et al., 2017; Nakada-Tsukui et al., 2019; Sharma et al., 2019; Babuta et al., 2020; Watanabe et al., 2020), and cytoskeletal regulation (Sahoo et al., 2004; Aslam et al., 2012; Mansuri et al., 2014; Babuta et al., 2018) play important roles in host cell recognition and ingestion (phago- and trogocytosis), secretion of virulence factors, and degradation of ingested materials in the lysosomes, trogosomes, and phagosomes. Thus, this study has underpinned Arf GTPase as a new player that regulates vesicular trafficking involved in the traffic of virulence factors, stress response, and thus in vivo virulence. The implications of this study should provide insights into multiple roles of membrane trafficking in diverse cellular functions in E. histolytica.
Discovery of EhArfX2 as a Key Determinant of In Vivo Virulence by Comparative Transcriptomic Analysis of Virulent and Avirulent Isogenic E. histolytica Strains
It was striking that EhArfX2 gene was exclusively identified among >100 Rab, 14 Arf/Sar, and other traffic-related genes including SNAREs and coatomer proteins (COPs) by transcriptomic comparisons of liver-passed v-HM1 and in vitro culture-attenuated a-HM1. These data underpin EhArfX2 to be a key determinant in liver abscess formation. Two studies of comparative transcriptomic analysis of attenuated and animal-passaged virulent strains with an identical genetic background were previously conducted (Santi-Rocca et al., 2008; Meyer et al., 2016). EhArfX2 was identified in neither of the studies. In the study by Santi-Rocca et al. (2008), 33 genes were found to be upregulated (>2-fold) in the animal-passed virulent HM-1 when compared to the attenuated avirulent strain. One of the genes encoded a parasite surface-associated protein, lysine and glutamic acid rich protein 1 (KERP1), and was further investigated, showing that KERP1 was involved in the adherence to the host cell and liver abscess development (Santi-Rocca et al., 2008). Meyer et al. (2016) identified 30 genes including tyrosine kinase and Myb family proteins as upregulated genes by comparing two isogenic clones: non-virulent HM-1:IMSS-A and virulent HM-1:IMSS-B clones. Among 30 genes, it was shown that gene silencing of one hypothetical protein EHI_127670 caused a decrease in CP secretion in vitro and the liver pathology in vivo when expression was induced in HM-1:IMSS-B (Meyer et al., 2016). In our study, the original strain we used was clonal and the genome was sequenced (Kawano-Sugaya et al., 2020), and we obtained virulent and attenuated lines derived from this clonal strain of HM-1:IMSS Cl6 for transcriptomic comparisons. The fact that EhArfX2 was not identified in the two previous studies (Santi-Rocca et al., 2008; Meyer et al., 2016) suggests the presence of multiple mechanisms for attenuation and virulence because no universal gene expression change, either upregulation or downregulation of a gene or a set of genes, was identified in three independent studies (Table 1 and Supplementary Table S2). Note that genes encoding so-called classic virulence factors, such as Gal/GalNAc lectin, CPs, and amoebapores, were not differentially expressed, similar to the observations in two previous reports (Santi-Rocca et al., 2008; Meyer et al., 2016). The uniqueness and significance of the present study are that a novel EhArfX2 GTPase was identified to be involved in liver abscess development and nitrosative stress resistance (Figure 5).
EhArfX2 Is Involved in Lysosome Trafficking From the Trans-Golgi Network and Lysosome Biogenesis
Arf/Sar GTPases are the key regulators of eukaryotic cell organization and membrane traffic together with Rab GTPases (Jackson and Bouvet, 2014). We identified 11 genes encoding EhArfs and three genes encoding EhSar1 in the E. histolytica genome (Figure 1A). N-terminal glycine residue, responsible for an N-terminal myristoylation and the insertion into the lipid bilayer, is conserved in EhArfX2, but the MxxE motif characteristic for Golgi-resident Arf1 is not present (Figure 1D). It has been shown that in human, the binding of ER-residing Arf1 to a membrane-localized SNARE in the cis-Golgi targeting is necessary for the ER–to–cis-Golgi traffic (Honda et al., 2005). We have demonstrated the trans-Golgi localization of EhArfX2 (Figure 2F), utilizing Golgi-resident luminal EhGalT and two SNAREs. The presence of Golgi-localized glycosylation enzymes was previously suggested by the bioinformatic approach in Entamoeba (Magnelli et al., 2008). In this study, we have shown that the small dot-like localization of one of the representative glycosylation enzymes from E. histolytica, EhGalT, responsible for the transfer of Gal, GalNAc, and GlcNAc to proteins, was closely associated with the ER (Supplementary Figure S1C), suggesting that amoebic glycosylation occurs at the region adjacent to the ER, similar to the case of the Golgi in plants, where the Golgi stacks are located in the vicinity of the ER exit site (Ito et al., 2018). EhGalT was not colocalized with EhArfX2 (Figure 2B), indicating that EhArfX2 does not localize to the EhGalT-positive Golgi membrane. SNAREs are essential for the fusion of transport vesicles with an acceptor membrane (Hong, 2005). The mechanisms of action and the subcellular localization of all SNAREs have been established in human, Arabidopsis, and Saccharomyces (Gupta and Brent Heath, 2002; Uemura et al., 2004; Hong, 2005; Kloepper et al., 2007). Phylogenetic analysis of 16 Q-SNAREs and nine R-SNAREs from E. histolytica predicted EhSed5 and EhYkt6 being associated with cis- and trans-Golgi, respectively (Supplementary Figures S2A, S3A). The short (15 aa) transmembrane region in EhSed5 well agrees with the premise of EhSed5 being a Golgi membrane resident (Supplementary Table S4). The Golgi-residing transmembrane proteins are typically short and likely due to the fact that the cholesterol and its analogs are less abundant in the Golgi apparatus than in the plasma membrane in humans (Bretscher and Munro, 1993). It is known that the transmembrane region of Sed5 orthologs is rich in phenylalanine, but its significance is not fully understood (Bretscher and Munro, 1993). Double prenylation of Ykt6 members at the C-terminus is essential for the membrane targeting of Ykt6 and the correct sorting of lysosomal hydrolases, cathepsin D and β-hexosaminidase, at the trans-Golgi in humans (Sakata et al., 2021) (Supplementary Figure S3B). It was shown that mutations of the C-terminus cysteine residues caused the missorting of lysosomal hydrolases at the trans-Golgi, leading to their missecretion into the extracellular space (Sakata et al., 2021). Taken together, these data imply the functional relevance of EhArfX2 and EhYkt6 in the trafficking from the trans-Golgi to the lysosomes in E. histolytica (Figures 2D, 3D). Clearly distinct localization of EhSed5 and EhYkt6 is consistent with the premise that the Golgi membrane is functionally divided into cis- and trans-cisternae in E. histolytica (Supplementary Figure S4C), although the Golgi morphology was previously described as vacuole-like (Mazzuco et al., 1997).
Dominant-Negative Effect of EhArfX2T28N
It was shown in human cells that Arf1Q71L constitutively binds to GTP and inhibits the ER–to–Golgi and intra-Golgi transport via its dominant active effects (Dascher and Balch, 1994; Donaldson and Jackson, 2011). Expression of a GDP-bound inactive form of human Arf1 (Arf1T31N) failed to recruit coat protein I (COPI) coatomer to the membrane and inhibited the pre-Golgi transport (Dascher and Balch, 1994; Beck et al., 2009). Thus, the COPI uncoating reaction depends on GTP hydrolysis by Arf1, and both Arf mutants prevent the traffic from the Golgi (Tanigawa et al., 1993). In the present study, expression of either EhArfX2Q68L or EhArfX2T28N mutant showed positive or negative effects on lysosome biogenesis, respectively (Figure 3A). However, neither showed a strong dominant effect on cell growth and erythrophagocytosis activity (Figures 4A, B). Thus, the role of the GTPase cycle of EhArfX2 may differ from that of human Arf1, and the GTP-bound state may be required for the EhArfX2 function. The fact that lysosomal traffic was inhibited by the expression of EhArfX2T28N mutant (Figures 3D, E) supported the hypothesis that GTP-bound state of EhArfX2 is required for the lysosomal fusion (Figure 3A). EhArfX2T28N displayed reduced lysosome formation and abnormal accumulation of CP in the pre-lysosomal non-acidic compartment (Figures 3D, E). It remains to be investigated how CPs are transported from the trans-Golgi to lysosomes in an EhArfX2-dependent fashion.
Localization of overexpressed EhArfX2WT-HA to the cytosol and EhArfX2T28N-HA to the membrane fraction, demonstrated by subcellular fractionation and immunoblot analyses (Figure 3C), was unexpected because it was previously shown in human cells that wild-type Arf1–Arf5 were, when overexpressed, mainly localized to the Golgi and the amount of wild-type Arf1–Arf5 distributed to the cytosol also increased. Furthermore, inactive GDP-bound Arf1–Arf5 mutants in humans were also shown to be accumulated in the cytosol and formed a complex with Arf guanine nucleotide exchange factors (Arf GEFs) but failed to be activated at the Golgi membrane (Cohen and Donaldson, 2010). However, it has been recently shown that Arf-GDP on the Golgi membrane can recruit Arf GEF via an unidentified protease-sensitive receptor (Quilty et al., 2018). Arf is released from the Golgi membrane once GTP is hydrolyzed. It was speculated that the limited amount of the receptor controls the constitutive level of Arf-GTP on the Golgi membrane (Quilty et al., 2018). Identification of Arf GEF, responsible for GTP loading on EhArfX2, and an Arf GEF receptor on the trans-Golgi membrane may further clarify the detailed trafficking pathway from the trans-Golgi to lysosomes in E. histolytica. It is worth noting that seven candidates for Arf GEF, Sec7, are present in the E. histolytica genome (Pipaliya et al., 2019).
EhArfX2 Is Involved in Anti-Nitrosative Stress Response During Liver Abscess Formation
E. histolytica CPs have been implicated for liver pathology because antisense inhibition of expression of CP-A5 reduced activity on formation of liver abscess in hamsters (Ankri et al., 1999). Thus, we expected that expression of EhArfX2T28N, which caused increased secretion of CP (Figure 3D), may also result in more pronounced hepatic abscesses in vivo. However, the results were counterintuitive: EhArfX2T28N expression increased the in vitro CHO monolayer destruction (Supplementary Figure S7A), while it reduced the size of liver abscesses (Figure 4C). It is conceivable that in vivo virulence requires a tight regulation of the processing, posttranslational modifications (PTMs), and trafficking of EhCP-A5 at the trans-Golgi and lysosomes. However, the tight regulation of CP trafficking was presumably affected by overexpression of EhArfX2T28N-HA, leading to dysregulation and missecretion of the immature non-functional (or less active) CPs. Alternatively, unidentified factors, which are involved in, e.g., anti-nitrosative defense, are less efficiently transported in EhArfX2T28N strain, leading to the reduction of in vivo virulence. The major virulent factor, EhCP-A5, is predicted to be glycosylated at Asn272 (Bruchhaus et al., 2003), and N-linked glycosylation on EhCP-A5 was verified by its inhibition by tunicamycin (Nakada-Tsukui et al., 2012). Thus, our findings, together with the implications by previous studies, suggest that the correct processing (i.e., a cleavage of the SP and the pro domain) and posttranslational modifications, including N-glycosylation, in the ER, the trans-Golgi, and lysosomes are impaired in EhArfX2T28N-expressing strain, and CP was missecreted into the extracellular milieu. It should be noted that the in vitro CP assay used in the present study (Figure 3D and Supplementary Figure S7A) does not distinguish precursor unprocessed (or partially processed) CPs and fully matured CPs because precursor CPs are also expected to be self-activated in vitro. Further investigations are needed to assess the pathological role of EhCP-A5 in tissue invasion by utilizing ex vivo model (Thibeaux et al., 2012). It was worth noting that it was shown that gene silencing of one hypothetical protein EHI_127670 caused a decrease in CP secretion in vitro and the liver pathology in vivo when the gene was overexpressed in the apparently highly virulent liver-passaged line, HM-1:IMSS B (Meyer et al., 2016). The underlying reason for this apparent discrepancy from our observation remains elusive. These findings indicate that the essentiality of EhArfX2 for the in vivo survival and the liver abscess is not attributable to its involvement in traffic and regulated secretion of CPs but likely management of stress in the hostile host environment during liver abscess formation. The importance of EhArfX2 on cell survival was supported, indirectly, by the repeated failure of gene silencing of EhArfX2 (Morf et al., 2013; Nakada-Tsukui et al., 2018) in v-HM1 strain (data not shown). We have shown that EhArfX2 is involved in the defense against nitrosative, but not oxidative, stresses. To date, the defense mechanisms of anti-nitrosative stress, which is mainly caused by nitric oxide (NO) from macrophages, are not clearly understood. During development of amoebic liver abscess, exposure of NO from macrophages primarily causes nitrosylation of cysteine residues in parasite proteins (Lin and Chadee, 1992), inactivation of key metabolic enzymes, and ER fragmentation (Santi-Rocca et al., 2012). More than 100 proteins including CPs, alcohol dehydrogenase (ADH), and lectin heavy subunit were previously reported to be S-nitrosylated (Siman-Tov and Ankri, 2003; Hertz et al., 2014), including two Rab GTPases (EHI_108610 EhRab1A and EHI_177520 EhRabX11) and one Sar1 GTPase (EHI_075040 EhSar1c) (Hertz et al., 2014), indicating that the intracellular traffic between the ER to the Golgi is likely affected by NO.
In our comparative transcriptomic analysis, we also found that one of the Atg4 orthologs was upregulated in liver-passed v-HM1 (Table 1), suggesting the involvement of autophagy and possible cross talk with EhArfX2. Atg4 cleaves the ubiquitin-like Atg8 near the C-terminus glycine, allowing the conjugation of Atg8 with phosphatidylethanolamine (PE) to generate membrane-bound Atg8-PE for subsequent autophagosome formation (Kirisako et al., 2000; Kabeya et al., 2004). At the initial step of autophagosome formation, two different GTPases are required: TGN localized Arl1 and Ypt6 in S. cerevisiae (Yang and Rosenwald, 2016), which may partially explain a possible link between autophagy and post-TGN traffic and between autophagy and EhArfX2 in stress response. However, the specific role of EhAtg4 in trans-Golgi to lysosome traffic remains elusive. In mammalian cells, it has been demonstrated that damaged proteins by oxidative stress are degraded by the autophagy–lysosomal pathway under mild oxidative conditions (Dodson et al., 2013) because 26S proteasome is sensitive to oxidative stress (Ishii et al., 2005). Furthermore, it has been also demonstrated that the oxidative stress induces autophagy in mammalian cells (Scherz-Shouval et al., 2007). It has been also shown that under starvation conditions, cells produce reactive oxygen species (ROS), specifically H2O2, and inhibit Atg4 (Kirisako et al., 2000). Although it is not known if nitrosative stress also affects autophagy, damaged proteins by oxidative and nitrosative stresses may be degraded by lysosomal and autophagic degradation systems during host tissue invasion and liver abscess development.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/, EhArfX1a (BR001420), EhArfX1b (BR001421), EhArfX2 (BR001422), EhArfX3 (BR001423), EhArfX4 (BR001424), EhArfX5 (BR001425), EhArfX6 (BR001426), EhArfX7 (BR001427), EhArfX8 (BR001428), EhArfX9 (BR001429), EhArfX10 (BR001430), EhSar1a (BR001431), EhSar1b (BR001432), EhSar1c (BR001433), EhSed5 (BR001434), EhSyn1A (BR001435), EhSyn1B (BR001436), EhSyn1C (BR001437), EhSynA (BR001438), EhSynB (BR001439), EhSynC (BR001440), EhSynD (BR001441), EhSynE (BR001442), EhSynF (BR001443), EhSynG (BR001444), EhSynH (BR001445), EhSynI (BR001446), EhSynJ (BR001447), EhVti11 (BR001448), EhVti12 (BR001449), EhYkt6 (BR001450), EhVampB (BR001451), EhVampD1 (BR001452), EhVampD2 (BR001453), EhVampD3 (BR001454), EhVampE (BR001455), EhVampF (BR001456), EhVampG (BR001457), EhVampH (BR001458).
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (No. 211075) and conducted at the AAALAC-accredited National Institute of Infectious Diseases, Japan.
Author Contributions
Conceptualization: YS-N, MT, WP, and TN. Investigation: YS-N, TM, and MT. Analyzed the data: YS-N, TM, and MT. Contributed analysis tools: CG and WP. Funding acquisition: YS-N and TN. Writing—original draft: YS-N, TM, MT, and TN. Writing—review and editing: YS-N, TM, MT, CG, WP, and TN. All authors contributed to the article and approved the submitted version.
Funding
This research was partially funded by Grants-in-Aid for Scientific Research (C) (JP19K07531 to YS-N) and Grant-in-Aid for Scientific Research (B) (JP18H0265, JP21H02723) to TN from the Japan Society for the Promotion of Science, Grant for Research on Emerging and Re-emerging Infectious Diseases from Japan Agency for Medical Research and Development (AMED, JP20fk0108138 and JP20fk0108139 to YS-N and TN), and a Grant for Science and Technology Research Partnership for Sustainable Development (SATREPS) from AMED and Japan International Cooperation Agency (JICA) (JP20jm0110022) to TN.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Seiki Kobayashi and Eiko Nakasone for technical assistance, Christopher D. Huston for providing the GFP-KDEL plasmid, and Kumiko Nakada-Tsukui, Ghulam Jeelani, and all members of our laboratory for valuable discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.794152/full#supplementary-material
References
Ankri, S., Stolarsky, T., Bracha, R., Padilla-Vaca, F., Mirelman, D. (1999). Antisense Inhibition of Expression of Cysteine Proteinases Affects Entamoeba histolytica-Induced Formation of Liver Abscess in Hamsters. Infect. Immun. 67 (1), 421–422. doi: 10.1128/IAI.67.1.421-422.1999
Aslam, S., Bhattacharya, S., Bhattacharya, A. (2012). The Calmodulin-Like Calcium Binding Protein EhCaBP3 of Entamoeba histolytica Regulates Phagocytosis and is Involved in Actin Dynamics. PLoS Pathog. 8 (12), e1003055. doi: 10.1371/journal.ppat.1003055
Babuta, M., Bhattacharya, S., Bhattacharya, A. (2020). Entamoeba histolytica and Pathogenesis: A Calcium Connection. PLoS Pathog. 16 (5), e1008214. doi: 10.1371/journal.ppat.1008214
Babuta, M., Kumar, S., Gourinath, S., Bhattacharya, S., Bhattacharya, A. (2018). Calcium-Binding Protein EhCaBP3 is Recruited to the Phagocytic Complex of Entamoeba histolytica by Interacting With Arp2/3 Complex Subunit 2. Cell Microbiol. 20 (12), e12942. doi: 10.1111/cmi.12942
Banfield, D. K., Lewis, M. J., Rabouille, C., Warren, G., Pelham, H. R. (1994). Localization of Sed5, a Putative Vesicle Targeting Molecule, to the Cis-Golgi Network Involves Both its Transmembrane and Cytoplasmic Domains. J. Cell Biol. 127 (2), 357–371. doi: 10.1083/jcb.127.2.357
Beck, R., Rawet, M., Wieland, F. T., Cassel, D. (2009). The COPI System: Molecular Mechanisms and Function. FEBS Lett. 583 (17), 2701–2709. doi: 10.1016/j.febslet.2009.07.032
Bharadwaj, R., Arya, R., Shahid Mansuri, M., Bhattacharya, S., Bhattacharya, A. (2017). EhRho1 Regulates Plasma Membrane Blebbing Through PI3 Kinase in Entamoeba histolytica. Cell Microbiol. 19 (10), e12751. doi: 10.1111/cmi.12751
Bretscher, M. S., Munro, S. (1993). Cholesterol and the Golgi Apparatus. Science 261 (5126), 1280–1281. doi: 10.1126/science.8362242
Bruchhaus, I., Loftus, B. J., Hall, N., Tannich, E. (2003). The Intestinal Protozoan Parasite Entamoeba histolytica Contains 20 Cysteine Protease Genes, of Which Only a Small Subset is Expressed During In Vitro Cultivation. Eukaryot. Cell 2 (3), 501–509. doi: 10.1128/EC.2.3.501-509.2003
Chatre, L., Wattelet-Boyer, V., Melser, S., Maneta-Peyret, L., Brandizzi, F., Moreau, P. (2009). A Novel Di-Acidic Motif Facilitates ER Export of the Syntaxin SYP31. J. Exp. Bot. 60 (11), 3157–3165. doi: 10.1093/jxb/erp155
Chen, Y., Shin, Y. K., Bassham, D. C. (2005). YKT6 is a Core Constituent of Membrane Fusion Machineries at the Arabidopsis Trans-Golgi Network. J. Mol. Biol. 350 (1), 92–101. doi: 10.1016/j.jmb.2005.04.061
Cohen, L. A., Donaldson, J. G. (2010). Analysis of Arf GTP-Binding Protein Function in Cells. Curr. Protoc. Cell Biol. Chapter 3, Unit 14 12 11–17. doi: 10.1002/0471143030.cb1412s48
Dascher, C., Balch, W. E. (1994). Dominant Inhibitory Mutants of ARF1 Block Endoplasmic Reticulum to Golgi Transport and Trigger Disassembly of the Golgi Apparatus. J. Biol. Chem. 269 (2), 1437–1448. doi: 10.1016/S0021-9258(17)42277-0
Diamond, L. S., Harlow, D. R., Cunnick, C. C. (1978). A New Medium for the Axenic Cultivation of Entamoeba histolytica and Other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72 (4), 431–432. doi: 10.1016/0035-9203(78)90144-x
Diamond, L. S., Mattern, C. F., Bartgis, I. L. (1972). Viruses of Entamoeba Histolytica. I. Identification of Transmissible Virus-Like Agents. J. Virol. 9 (2), 326–341. doi: 10.1128/JVI.9.2.326-341.1972
Dodson, M., Darley-Usmar, V., Zhang, J. (2013). Cellular Metabolic and Autophagic Pathways: Traffic Control by Redox Signaling. Free Radic. Biol. Med. 63, 207–221. doi: 10.1016/j.freeradbiomed.2013.05.014
Donaldson, J. G., Jackson, C. L. (2011). ARF Family G Proteins and Their Regulators: Roles in Membrane Transport, Development and Disease. Nat. Rev. Mol. Cell Biol. 12 (6), 362–375. doi: 10.1038/nrm3117
Edgar, R. C. (2004). MUSCLE: Multiple Sequence Alignment With High Accuracy and High Throughput. Nucleic Acids Res. 32 (5), 1792–1797. doi: 10.1093/nar/gkh340
Emmanuel, M., Nakano, Y. S., Nozaki, T., Datta, S. (2015). Small GTPase Rab21 Mediates Fibronectin Induced Actin Reorganization in Entamoeba histolytica: Implications in Pathogen Invasion. PLoS Pathog. 11 (3), e1004666. doi: 10.1371/journal.ppat.1004666
Faust, D. M., Guillen, N. (2012). Virulence and Virulence Factors in Entamoeba histolytica, the Agent of Human Amoebiasis. Microbes Infect. 14 (15), 1428–1441. doi: 10.1016/j.micinf.2012.05.013
Furukawa, A., Nakada-Tsukui, K., Nozaki, T. (2012). Novel Transmembrane Receptor Involved in Phagosome Transport of Lysozymes and Beta-Hexosaminidase in the Enteric Protozoan Entamoeba histolytica. PLoS Pathog. 8 (2), e1002539. doi: 10.1371/journal.ppat.1002539
Gilchrist, C. A., Houpt, E., Trapaidze, N., Fei, Z., Crasta, O., Asgharpour, A., et al (2006). Impact of Intestinal Colonization and Invasion on the Entamoeba histolytica Transcriptome. Mol. Biochem. Parasitol. 147 (2), 163–176. doi: 10.1016/j.molbiopara.2006.02.007
Gupta, G. D., Brent Heath, I. (2002). Predicting the Distribution, Conservation, and Functions of SNAREs and Related Proteins in Fungi. Fungal Genet. Biol. 36 (1), 1–21. doi: 10.1016/S1087-1845(02)00017-8
Hanadate, Y., Saito-Nakano, Y., Nakada-Tsukui, K., Nozaki, T. (2016). Endoplasmic Reticulum-Resident Rab8A GTPase is Involved in Phagocytosis in the Protozoan Parasite Entamoeba histolytica. Cell Microbiol. 18 (10), 1358–1373. doi: 10.1111/cmi.12570
Hertz, R., Ben Lulu, S., Shahi, P., Trebicz-Geffen, M., Benhar, M., Ankri, S. (2014). Proteomic Identification of S-Nitrosylated Proteins in the Parasite Entamoeba histolytica by Resin-Assisted Capture: Insights Into the Regulation of the Gal/GalNAc Lectin by Nitric Oxide. PLoS One 9 (3), e91518. doi: 10.1371/journal.pone.0091518
Honda, A., Al-Awar, O. S., Hay, J. C., Donaldson, J. G. (2005). Targeting of Arf-1 to the Early Golgi by Membrin, an ER-Golgi SNARE. J. Cell Biol. 168 (7), 1039–1051. doi: 10.1083/jcb.200409138
Hong, W. (2005). SNAREs and Traffic. Biochim. Biophys. Acta 1744 (3), 493–517. doi: 10.1016/j.bbamcr.2005.03.014
Husain, A., Jeelani, G., Sato, D., Nozaki, T. (2011). Global Analysis of Gene Expression in Response to L-Cysteine Deprivation in the Anaerobic Protozoan Parasite Entamoeba histolytica. BMC Genomics 12, 275. doi: 10.1186/1471-2164-12-275
Husain, A., Sato, D., Jeelani, G., Soga, T., Nozaki, T. (2012). Dramatic Increase in Glycerol Biosynthesis Upon Oxidative Stress in the Anaerobic Protozoan Parasite Entamoeba histolytica. PLoS Negl. Trop. Dis. 6 (9), e1831. doi: 10.1371/journal.pntd.0001831
Hutagalung, A. H., Novick, P. J. (2011). Role of Rab GTPases in Membrane Traffic and Cell Physiology. Physiol. Rev. 91 (1), 119–149. doi: 10.1152/physrev.00059.2009
Irmer, H., Tillack, M., Biller, L., Handal, G., Leippe, M., Roeder, T., et al (2009). Major Cysteine Peptidases of Entamoeba histolytica are Required for Aggregation and Digestion of Erythrocytes But are Dispensable for Phagocytosis and Cytopathogenicity. Mol. Microbiol. 72 (3), 658–667. doi: 10.1111/j.1365-2958.2009.06672.x
Ishii, T., Sakurai, T., Usami, H., Uchida, K. (2005). Oxidative Modification of Proteasome: Identification of an Oxidation-Sensitive Subunit in 26 S Proteasome. Biochemistry 44 (42), 13893–13901. doi: 10.1021/bi051336u
Ito, Y., Uemura, T., Nakano, A. (2018). The Golgi Entry Core Compartment Functions as a COPII-Independent Scaffold for ER-To-Golgi Transport in Plant Cells. J. Cell Sci. 131 (2), cs203893. doi: 10.1242/jcs.203893
Jackson, C. L., Bouvet, S. (2014). Arfs at a Glance. J. Cell Sci. 127 (Pt 19), 4103–4109. doi: 10.1242/jcs.144899
Jeelani, G., Nozaki, T. (2016). Entamoeba Thiol-Based Redox Metabolism: A Potential Target for Drug Development. Mol. Biochem. Parasitol. 206 (1-2), 39–45. doi: 10.1016/j.molbiopara.2016.01.004
Jeelani, G., Sato, D., Soga, T., Nozaki, T. (2017). Genetic, Metabolomic and Transcriptomic Analyses of the De Novo L-Cysteine Biosynthetic Pathway in the Enteric Protozoan Parasite Entamoeba histolytica. Sci. Rep. 7 (1), 15649. doi: 10.1038/s41598-017-15923-3
Juarez-Hernandez, L. J., Garcia-Perez, R. M., Salas-Casas, A., Garcia-Rivera, G., Orozco, E., Rodriguez, M. A. (2013). Entamoeba histolytica: The Over Expression of a Mutated EhRabB Protein Produces a Decrease of In Vitro and In Vivo Virulence. Exp. Parasitol. 133 (3), 339–345. doi: 10.1016/j.exppara.2012.12.008
Kabeya, Y., Mizushima, N., Yamamoto, A., Oshitani-Okamoto, S., Ohsumi, Y., Yoshimori, T. (2004). LC3, GABARAP and GATE16 Localize to Autophagosomal Membrane Depending on Form-II Formation. J. Cell Sci. 117 (Pt 13), 2805–2812. doi: 10.1242/jcs.01131
Kawano-Sugaya, T., Izumiyama, S., Yanagawa, Y., Saito-Nakano, Y., Watanabe, K., Kobayashi, S., et al (2020). Near-Chromosome Level Genome Assembly Reveals Ploidy Diversity and Plasticity in the Intestinal Protozoan Parasite Entamoeba histolytica. BMC Genomics 21 (1), 813. doi: 10.1186/s12864-020-07167-9
Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., et al (2000). The Reversible Modification Regulates the Membrane-Binding State of Apg8/Aut7 Essential for Autophagy and the Cytoplasm to Vacuole Targeting Pathway. J. Cell Biol. 151 (2), 263–276. doi: 10.1083/jcb.151.2.263
Kloepper, T. H., Kienle, C. N., Fasshauer, D. (2007). An Elaborate Classification of SNARE Proteins Sheds Light on the Conservation of the Eukaryotic Endomembrane System. Mol. Biol. Cell 18 (9), 3463–3471. doi: 10.1091/mbc.e07-03-0193
Kobayashi, S., Imai, E., Haghighi, A., Khalifa, S. A., Tachibana, H. (2005). And Takeuchi, TAxenic Cultivation of Entamoeba Dispar in Newly Designed Yeast Extract-Iron-Gluconic Acid-Dihydroxyacetone-Serum Medium. J. Parasitol. 91 (1), 1–4. doi: 10.1645/GE-3386
Leippe, M., Sievertsen, H. J., Tannich, E., Horstmann, R. D. (1995). Spontaneous Release of Cysteine Proteinases But Not of Pore-Forming Peptides by Viable Entamoeba histolytica. Parasitology 111 ( Pt 5), 569–574. doi: 10.1017/s0031182000077040
Lin, J. Y., Chadee, K. (1992). Macrophage Cytotoxicity Against Entamoeba histolytica Trophozoites is Mediated by Nitric Oxide From L-Arginine. J. Immunol. 148 (12), 3999–4005.
Magnelli, P., Cipollo, J. F., Ratner, D. M., Cui, J., Kelleher, D., Gilmore, R., et al (2008). Unique Asn-Linked Oligosaccharides of the Human Pathogen Entamoeba histolytica. J. Biol. Chem. 283 (26), 18355–18364. doi: 10.1074/jbc.M800725200
Makiuchi, T., Nozaki, T. (2014). Highly Divergent Mitochondrion-Related Organelles in Anaerobic Parasitic Protozoa. Biochimie 100, 3–17. doi: 10.1016/j.biochi.2013.11.018
Mansuri, M. S., Bhattacharya, S., Bhattacharya, A. (2014). A Novel Alpha Kinase EhAK1 Phosphorylates Actin and Regulates Phagocytosis in Entamoeba histolytica. PLoS Pathog. 10 (10), e1004411. doi: 10.1371/journal.ppat.1004411
Matheson, L. A., Suri, S. S., Hanton, S. L., Chatre, L., Brandizzi, F. (2008). Correct Targeting of Plant ARF GTPases Relies on Distinct Protein Domains. Traffic 9 (1), 103–120. doi: 10.1111/j.1600-0854.2007.00671.x
Mazzuco, A., Benchimol, M., De Souza, W. (1997). Endoplasmic Reticulum and Golgi-Like Elements in Entamoeba. Micron 28 (3), 241–247. doi: 10.1016/s0968-4328(97)00024-3
Meyer, M., Fehling, H., Matthiesen, J., Lorenzen, S., Schuldt, K., Bernin, H., et al (2016). Overexpression of Differentially Expressed Genes Identified in non-Pathogenic and Pathogenic Entamoeba histolytica Clones Allow Identification of New Pathogenicity Factors Involved in Amoebic Liver Abscess Formation. PLoS Pathog. 12 (8), e1005853. doi: 10.1371/journal.ppat.1005853
Mitra, B. N., Saito-Nakano, Y., Nakada-Tsukui, K., Sato, D., Nozaki, T. (2007). Rab11B Small GTPase Regulates Secretion of Cysteine Proteases in the Enteric Protozoan Parasite Entamoeba histolytica. Cell Microbiol. 9 (9), 2112–2125. doi: 10.1111/j.1462-5822.2007.00941.x
Moncada, D., Keller, K., Ankri, S., Mirelman, D., Chadee, K. (2006). Antisense Inhibition of Entamoeba histolytica Cysteine Proteases Inhibits Colonic Mucus Degradation. Gastroenterology 130 (3), 721–730. doi: 10.1053/j.gastro.2005.11.012
Morf, L., Pearson, R. J., Wang, A. S., Singh, U. (2013). Robust Gene Silencing Mediated by Antisense Small RNAs in the Pathogenic Protist Entamoeba histolytica. Nucleic Acids Res. 41 (20), 9424–9437. doi: 10.1093/nar/gkt717
Munro, S., Pelham, H. R. (1986). An Hsp70-Like Protein in the ER: Identity With the 78 Kd Glucose-Regulated Protein and Immunoglobulin Heavy Chain Binding Protein. Cell 46 (2), 291–300. doi: 10.1016/0092-8674(86)90746-4
Nakada-Tsukui, K., Saito-Nakano, Y., Ali, V., Nozaki, T. (2005). A Retromerlike Complex is a Novel Rab7 Effector That is Involved in the Transport of the Virulence Factor Cysteine Protease in the Enteric Protozoan Parasite Entamoeba histolytica. Mol. Biol. Cell 16 (11), 5294–5303. doi: 10.1091/mbc.e05-04-0283
Nakada-Tsukui, K., Saito-Nakano, Y., Husain, A., Nozaki, T. (2010). Conservation and Function of Rab Small GTPases in Entamoeba: Annotation of E. Invadens Rab and its Use for the Understanding of Entamoeba Biology. Exp. Parasitol. 126 (3), 337–347. doi: 10.1016/j.exppara.2010.04.014
Nakada-Tsukui, K., Sekizuka, T., Sato-Ebine, E., Escueta-de Cadiz, A., Ji, D. D., Tomii, K., et al (2018). AIG1 Affects In Vitro and In Vivo Virulence in Clinical Isolates of Entamoeba histolytica. PLoS Pathog. 14 (3), e1006882. doi: 10.1371/journal.ppat.1006882
Nakada-Tsukui, K., Tsuboi, K., Furukawa, A., Yamada, Y., Nozaki, T. (2012). A Novel Class of Cysteine Protease Receptors That Mediate Lysosomal Transport. Cell Microbiol. 14 (8), 1299–1317. doi: 10.1111/j.1462-5822.2012.01800.x
Nakada-Tsukui, K., Watanabe, N., Maehama, T., Nozaki, T. (2019). Phosphatidylinositol Kinases and Phosphatases in Entamoeba histolytica. Front. Cell Infect. Microbiol. 9:150. doi: 10.3389/fcimb.2019.00150
Nowak, N., Lotter, H., Tannich, E., Bruchhaus, I. (2004). Resistance of Entamoeba histolytica to the Cysteine Proteinase Inhibitor E64 is Associated With Secretion of Pro-Enzymes and Reduced Pathogenicity. J. Biol. Chem. 279 (37), 38260–38266. doi: 10.1074/jbc.M405308200
Nozaki, T., Asai, T., Sanchez, L. B., Kobayashi, S., Nakazawa, M., Takeuchi, T. (1999). Characterization of the Gene Encoding Serine Acetyltransferase, a Regulated Enzyme of Cysteine Biosynthesis From the Protist Parasites Entamoeba histolytica and Entamoeba Dispar. Regulation and Possible Function of the Cysteine Biosynthetic Pathway in Entamoeba. J. Biol. Chem. 274 (45), 32445–32452. doi: 10.1074/jbc.274.45.32445
Okada, M., Huston, C. D., Oue, M., Mann, B. J., Petri, W. A., Jr., Kita, K., et al (2006). Kinetics and Strain Variation of Phagosome Proteins of Entamoeba histolytica by Proteomic Analysis. Mol. Biochem. Parasitol. 145 (2), 171–183. doi: 10.1016/j.molbiopara.2005.10.001
Penuliar, G. M., Furukawa, A., Nakada-Tsukui, K., Husain, A., Sato, D., Nozaki, T. (2012). Transcriptional and Functional Analysis of Trifluoromethionine Resistance in Entamoeba histolytica. J. Antimicrob. Chemother. 67 (2), 375–386. doi: 10.1093/jac/dkr484
Penuliar, G. M., Nakada-Tsukui, K., Nozaki, T. (2015). Phenotypic and Transcriptional Profiling in Entamoeba histolytica Reveal Costs to Fitness and Adaptive Responses Associated With Metronidazole Resistance. Front. Microbiol. 6:354. doi: 10.3389/fmicb.2015.00354
Perdomo, D., Manich, M., Syan, S., Olivo-Marin, J. C., Dufour, A. C., Guillen, N. (2016). Intracellular Traffic of the Lysine and Glutamic Acid Rich Protein KERP1 Reveals Features of Endomembrane Organization in Entamoeba histolytica. Cell Microbiol. 18 (8), 1134–1152. doi: 10.1111/cmi.12576
Petri, W. A., Jr., Haque, R., Mann, B. J. (2002). The Bittersweet Interface of Parasite and Host: Lectin-Carbohydrate Interactions During Human Invasion by the Parasite Entamoeba histolytica. Annu. Rev. Microbiol. 56, 39–64. doi: 10.1146/annurev.micro.56.012302.160959
Pipaliya, S. V., Schlacht, A., Klinger, C. M., Kahn, R. A., Dacks, J. (2019). Ancient Complement and Lineage-Specific Evolution of the Sec7 ARF GEF Proteins in Eukaryotes. Mol. Biol. Cell 30 (15), 1846–1863. doi: 10.1091/mbc.E19-01-0073
Price, H. P., Stark, M., Smith, D. F. (2007). Trypanosoma Brucei ARF1 Plays a Central Role in Endocytosis and Golgi-Lysosome Trafficking. Mol. Biol. Cell 18 (3), 864–873. doi: 10.1091/mbc.e06-08-0736
Quilty, D., Chan, C. J., Yurkiw, K., Bain, A., Babolmorad, G., Melancon, P. (2018). The Arf-GDP-Regulated Recruitment of GBF1 to Golgi Membranes Requires Domains HDS1 and HDS2 and a Golgi-Localized Protein Receptor. J. Cell Sci. 132 (4), jcs208199. doi: 10.1242/jcs.208199
Ralston, K. S., Solga, M. D., Mackey-Lawrence, N. M., Somlata, Bhattacharya, A., Petri, W. A., Jr. (2014). Trogocytosis by Entamoeba histolytica Contributes to Cell Killing and Tissue Invasion. Nature 508 (7497), 526–530. doi: 10.1038/nature13242
Reed, S. L., Ember, J. A., Herdman, D. S., DiScipio, R. G., Hugli, T. E., Gigli, I. (1995). The Extracellular Neutral Cysteine Proteinase of Entamoeba histolytica Degrades Anaphylatoxins C3a and C5a. J. Immunol. 155 (1), 266–274.
Sahoo, N., Labruyere, E., Bhattacharya, S., Sen, P., Guillen, N., Bhattacharya, A. (2004). Calcium Binding Protein 1 of the Protozoan Parasite Entamoeba histolytica Interacts With Actin and is Involved in Cytoskeleton Dynamics. J. Cell Sci. 117 (Pt 16), 3625–3634. doi: 10.1242/jcs.01198
Saito-Nakano, Y., Loftus, B. J., Hall, N., Nozaki, T. (2005). The Diversity of Rab GTPases in Entamoeba histolytica. Exp. Parasitol. 110 (3), 244–252. doi: 10.1016/j.exppara.2005.02.021
Saito-Nakano, Y., Mitra, B. N., Nakada-Tsukui, K., Sato, D., Nozaki, T. (2007). Two Rab7 Isotypes, EhRab7A and EhRab7B, Play Distinct Roles in Biogenesis of Lysosomes and Phagosomes in the Enteric Protozoan Parasite Entamoeba histolytica. Cell Microbiol. 9 (7), 1796–1808. doi: 10.1111/j.1462-5822.2007.00915.x
Saito-Nakano, Y., Wahyuni, R., Nakada-Tsukui, K., Tomii, K., Nozaki, T. (2021). Rab7D Small GTPase is Involved in Phago-, Trogocytosis and Cytoskeletal Reorganization in the Enteric Protozoan Entamoeba histolytica. Cell Microbiol. 23 (1), e13267. doi: 10.1111/cmi.13267
Saito-Nakano, Y., Yasuda, T., Nakada-Tsukui, K., Leippe, M., Nozaki, T. (2004). Rab5-Associated Vacuoles Play a Unique Role in Phagocytosis of the Enteric Protozoan Parasite Entamoeba histolytica. J. Biol. Chem. 279 (47), 49497–49507. doi: 10.1074/jbc.M403987200
Sakata, N., Shirakawa, R., Goto, K., Trinh, D. A., Horiuchi, H. (2021). Double Prenylation of SNARE Protein Ykt6 is Required for Lysosomal Hydrolase Trafficking. J. Biochem. 169 (3), 363–370. doi: 10.1093/jb/mvaa111
Santi-Rocca, J., Rigothier, M. C., Guillen, N. (2009). Host-Microbe Interactions and Defense Mechanisms in the Development of Amoebic Liver Abscesses. Clin. Microbiol. Rev. 22 (1), 65–75, Table of Contents. doi: 10.1128/CMR.00029-08
Santi-Rocca, J., Smith, S., Weber, C., Pineda, E., Hon, C. C., Saavedra, E., et al (2012). Endoplasmic Reticulum Stress-Sensing Mechanism is Activated in Entamoeba histolytica Upon Treatment With Nitric Oxide. PLoS One 7 (2), e31777. doi: 10.1371/journal.pone.0031777
Santi-Rocca, J., Weber, C., Guigon, G., Sismeiro, O., Coppee, J. Y., Guillen, N. (2008). The Lysine- and Glutamic Acid-Rich Protein KERP1 Plays a Role in Entamoeba histolytica Liver Abscess Pathogenesis. Cell Microbiol. 10 (1), 202–217. doi: 10.1111/j.1462-5822.2007.01030.x
Sato, D., Nakada-Tsukui, K., Okada, M., Nozaki, T. (2006a). Two Cysteine Protease Inhibitors, EhICP1 and 2, Localized in Distinct Compartments, Negatively Regulate Secretion in Entamoeba histolytica. FEBS Lett. 580 (22), 5306–5312. doi: 10.1016/j.febslet.2006.08.081
Sato, D., Yamagata, W., Kamei, K., Nozaki, T., Harada, S. (2006b). Expression, Purification and Crystallization of L-Methionine Gamma-Lyase 2 From Entamoeba histolytica. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62 (Pt 10), 1034–1036. doi: 10.1107/S1744309106036694
Scherz-Shouval, R., Shvets, E., Fass, E., Shorer, H., Gil, L., Elazar, Z. (2007). Reactive Oxygen Species are Essential for Autophagy and Specifically Regulate the Activity of Atg4. EMBO J. 26 (7), 1749–1760. doi: 10.1038/sj.emboj.7601623
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al (2012). Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 9 (7), 676–682. doi: 10.1038/nmeth.2019
Seigneur, M., Mounier, J., Prevost, M. C., Guillen, N. (2005). A Lysine- and Glutamic Acid-Rich Protein, KERP1, From Entamoeba histolytica Binds to Human Enterocytes. Cell Microbiol. 7 (4), 569–579. doi: 10.1111/j.1462-5822.2005.00487.x
Sharma, S., Agarwal, S., Bharadwaj, R., Somlata, Bhattacharya, S., Bhattacharya, A. (2019). Novel Regulatory Roles of PtdIns(4,5)P2 Generating Enzyme EhPIPKI in Actin Dynamics and Phagocytosis of Entamoeba histolytica. Cell Microbiol. 21 (10), e13087. doi: 10.1111/cmi.13087
Shestakova, A., Suvorova, E., Pavliv, O., Khaidakova, G., Lupashin, V. (2007). Interaction of the Conserved Oligomeric Golgi Complex With T-SNARE Syntaxin5a/Sed5 Enhances Intra-Golgi SNARE Complex Stability. J. Cell Biol. 179 (6), 1179–1192. doi: 10.1083/jcb.200705145
Shimodaira, H. (2002). An Approximately Unbiased Test of Phylogenetic Tree Selection. Syst. Biol. 51 (3), 492–508. doi: 10.1080/10635150290069913
Siman-Tov, R., Ankri, S. (2003). Nitric Oxide Inhibits Cysteine Proteinases and Alcohol Dehydrogenase 2 of Entamoeba histolytica. Parasitol. Res. 89 (2), 146–149. doi: 10.1007/s00436-002-0716-2
Somlata, Nakada-Tsukui, K., Nozaki, T. (2017). AGC Family Kinase 1 Participates in Trogocytosis But Not in Phagocytosis in Entamoeba histolytica. Nat. Commun. 8 (1), 101. doi: 10.1038/s41467-017-00199-y
Stanley, S. L., Jr. (2003). Amoebiasis. Lancet 361 (9362), 1025–1034. doi: 10.1016/S0140-6736(03)12830-9
St-Pierre, J., Moreau, F., Cornick, S., Quach, J., Begum, S., Aracely Fernandez, L., et al (2017). The Macrophage Cytoskeleton Acts as a Contact Sensor Upon Interaction With Entamoeba histolytica to Trigger IL-1beta Secretion. PLoS Pathog. 13 (8), e1006592. doi: 10.1371/journal.ppat.1006592
Tai, G., Lu, L., Wang, T. L., Tang, B. L., Goud, B., Johannes, L., et al (2004). Participation of the Syntaxin 5/Ykt6/GS28/GS15 SNARE Complex in Transport From the Early/Recycling Endosome to the Trans-Golgi Network. Mol. Biol. Cell 15 (9), 4011–4022. doi: 10.1091/mbc.e03-12-0876
Tanigawa, G., Orci, L., Amherdt, M., Ravazzola, M., Helms, J. B., Rothman, J. E. (1993). Hydrolysis of Bound GTP by ARF Protein Triggers Uncoating of Golgi-Derived COP-Coated Vesicles. J. Cell Biol. 123 (6 Pt 1), 1365–1371. doi: 10.1083/jcb.123.6.1365
Teal, S. B., Hsu, V. W., Peters, P. J., Klausner, R. D., Donaldson, J. G. (1994). An Activating Mutation in ARF1 Stabilizes Coatomer Binding to Golgi Membranes. J. Biol. Chem. 269 (5), 3135–3138. doi: 10.1016/S0021-9258(17)41837-0
Teixeira, J. E., Huston, C. D. (2008). Evidence of a Continuous Endoplasmic Reticulum in the Protozoan Parasite Entamoeba histolytica. Eukaryot. Cell 7 (7), 1222–1226. doi: 10.1128/EC.00007-08
Thibeaux, R., Dufour, A., Roux, P., Bernier, M., Baglin, A. C., Frileux, P., et al (2012). Newly Visualized Fibrillar Collagen Scaffolds Dictate Entamoeba histolytica Invasion Route in the Human Colon. Cell Microbiol. 14 (5), 609–621. doi: 10.1111/j.1462-5822.2012.01752.x
Tillack, M., Nowak, N., Lotter, H., Bracha, R., Mirelman, D., Tannich, E., et al (2006). Increased Expression of the Major Cysteine Proteinases by Stable Episomal Transfection Underlines the Important Role of EhCP5 for the Pathogenicity of Entamoeba histolytica. Mol. Biochem. Parasitol. 149 (1), 58–64. doi: 10.1016/j.molbiopara.2006.04.009
Uemura, T., Ueda, T., Ohniwa, R. L., Nakano, A., Takeyasu, K., Sato, M. H. (2004). Systematic Analysis of SNARE Molecules in Arabidopsis: Dissection of the Post-Golgi Network in Plant Cells. Cell Struct. Funct. 29 (2), 49–65. doi: 10.1247/csf.29.49
Ungermann, C., Langosch, D. (2005). Functions of SNAREs in Intracellular Membrane Fusion and Lipid Bilayer Mixing. J. Cell Sci. 118 (Pt 17), 3819–3828. doi: 10.1242/jcs.02561
Watanabe, N., Nakada-Tsukui, K., Nozaki, T. (2020). Two Isotypes of Phosphatidylinositol 3-Phosphate-Binding Sorting Nexins Play Distinct Roles in Trogocytosis in Entamoeba histolytica. Cell Microbiol. 22 (3), e13144. doi: 10.1111/cmi.13144
Welter, B. H., Temesvari, L. A. (2009). Overexpression of a Mutant Form of EhRabA, a Unique Rab GTPase of Entamoeba histolytica, Alters Endoplasmic Reticulum Morphology and Localization of the Gal/GalNAc Adherence Lectin. Eukaryot. Cell 8 (7), 1014–1026. doi: 10.1128/EC.00030-09
Yamaguchi, T., Dulubova, I., Min, S. W., Chen, X., Rizo, J., Sudhof, T. C. (2002). Sly1 Binds to Golgi and ER Syntaxins via a Conserved N-Terminal Peptide Motif. Dev. Cell 2 (3), 295–305. doi: 10.1016/s1534-5807(02)00125-9
Yang, S., Rosenwald, A. G. (2016). Autophagy in Saccharomyces cerevisiae Requires the Monomeric GTP-Binding Proteins, Arl1 and Ypt6. Autophagy 12 (10), 1721–1737. doi: 10.1080/15548627.2016.1196316
Yang, Z. (1997). PAML: A Program Package for Phylogenetic Analysis by Maximum Likelihood. Comput. Appl. Biosci. 13 (5), 555–556. doi: 10.1093/bioinformatics/13.5.555
Zhang, C. J., Rosenwald, A. G., Willingham, M. C., Skuntz, S., Clark, J., Kahn, R. A. (1994). Expression of a Dominant Allele of Human ARF1 Inhibits Membrane Traffic In Vivo. J. Cell Biol. 124 (3), 289–300. doi: 10.1083/jcb.124.3.289
Zhang, X., Zhang, Z., Alexander, D., Bracha, R., Mirelman, D., Stanley, S. L., Jr. (2004). Expression of Amoebapores is Required for Full Expression of Entamoeba histolytica Virulence in Amebic Liver Abscess But is Not Necessary for the Induction of Inflammation or Tissue Damage in Amebic Colitis. Infect. Immun. 72 (2), 678–683. doi: 10.1128/IAI.72.2.678-683.2004
Keywords: Entamoeba histolytica, liver abscess, membrane traffic, Arf GTPase, stress resistance
Citation: Saito-Nakano Y, Makiuchi T, Tochikura M, Gilchrist CA, Petri WA Jr and Nozaki T (2021) ArfX2 GTPase Regulates Trafficking From the Trans-Golgi to Lysosomes and Is Necessary for Liver Abscess Formation in the Protozoan Parasite Entamoeba histolytica. Front. Cell. Infect. Microbiol. 11:794152. doi: 10.3389/fcimb.2021.794152
Received: 13 October 2021; Accepted: 24 November 2021;
Published: 17 December 2021.
Edited by:
Marcel I. Ramirez, Carlos Chagas Institute (ICC), BrazilReviewed by:
Bruno Goud, Centre National de la Recherche Scientifique (CNRS), FrancePatricia Talamás-Rohana, Centro de Investigaciones y Estudios Avanzados, Instituto Politécnico Nacional de México (CINVESTAV), Mexico
Esther Orozco, Centro de Investigaciones y Estudios Avanzados, Instituto Politécnico Nacional de México (CINVESTAV), Mexico
Copyright © 2021 Saito-Nakano, Makiuchi, Tochikura, Gilchrist, Petri and Nozaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoyoshi Nozaki, bm96YWtpQG0udS10b2t5by5hYy5qcA==
†Present address: Mami Tochikura, Quality Management, BioMerieux Japan Ltd., Minato-ku, Tokyo, Japan
 Yumiko Saito-Nakano
Yumiko Saito-Nakano Takashi Makiuchi2
Takashi Makiuchi2 Carol A. Gilchrist
Carol A. Gilchrist William A. Petri Jr
William A. Petri Jr Tomoyoshi Nozaki
Tomoyoshi Nozaki