- 1Human Biology Division, Fred Hutchinson Cancer Research Center, Seattle, WA, United States
- 2Department of Obstetrics, Gynecology and Reproductive Sciences, University of California San Diego, San Diego, CA, United States
- 3Department of Pediatrics, Division of Infectious Diseases, Washington University in St. Louis School of Medicine, St. Louis, MO, United States
Recurrent urinary tract infections (rUTI) are a costly clinical problem affecting millions of women worldwide each year. The majority of rUTI cases are caused by uropathogenic Escherichia coli (UPEC). Data from humans and mouse models indicate that some instances of rUTI are caused by UPEC emerging from latent reservoirs in the bladder. Women with vaginal dysbiosis, typically characterized by high levels of Gardnerella and other anaerobes, are at increased risk of UTI. Multiple studies have detected Gardnerella in urine collected by transurethral catheterization (to limit vaginal contamination), suggesting that some women experience routine urinary tract exposures. We recently reported that inoculation of Gardnerella into the bladder triggers rUTI from UPEC bladder reservoirs in a mouse model. Here we performed whole bladder RNA-seq to identify host pathways involved in Gardnerella-induced rUTI. We identified a variety host pathways differentially expressed in whole bladders following Gardnerella exposure, such as pathways involved in inflammation/immunity and epithelial turnover. At the gene level, we identified upregulation of Immediate Early (IE) genes, which are induced in various cell types shortly following stimuli like infection and inflammation. One such upregulated IE gene was the orphan nuclear receptor Nur77 (aka Nr4a1). Pilot experiments in Nur77-/- mice suggest that Nur77 is necessary for Gardnerella exposure to trigger rUTI from UPEC reservoirs. These findings demonstrate that bladder gene expression can be impacted by short-lived exposures to urogenital bacteria and warrant future examination of responses in distinct cell types, such as with single cell transcriptomic technologies. The biological validation studies in Nur77-/- mice lay the groundwork for future studies investigating Nur77 and the Immediate Early response in rUTI.
Introduction
In humans, the urinary tract is the second most common site of infection, most frequently by uropathogenic Eschericia coli (UPEC) (Foxman, 2014). Approximately 1% of bladder infections (cystitis) progress to more serious kidney infections (pyelonephritis), and some of these become life-threatening systemic infections (Ki et al., 2004; Jolley et al., 2012). Recurrent urinary tract infections (rUTIs) are very common: 24% of women with an initial UTI will have rUTI within 6 months, and up to 70% will have rUTI within 1 year (Foxman et al., 2000; Foxman, 2014). Approximately 1% of all women (~35 million worldwide) have six or more rUTIs each year (Foxman et al., 2000; Foxman et al., 2002; Foxman, 2014). These women are often given prophylactic antibiotics, contributing to the problem of antibiotic resistance.
It is now appreciated that UPEC has an intracellular niche within the bladder. Multiple studies of UTI and rUTI in adults and children have identified intracellular UPEC, both in bladder epithelial cells shed in urine and in bladder biopsies (Elliott et al., 1985; Rosen et al., 2007; Robino et al., 2014; Liu et al., 2016; De Nisco et al., 2019). One study using confocal microscopy detected intracellular bacteria in 36.8% of samples from children with acute UTI. Notably, a medical chart review demonstrated that the children with intracellular bacteria were significantly more likely to have a history of rUTI (OR, 8.0; 95% CI, 2.3-27.4) (Robino et al., 2014). In C57BL/6 mice, UPEC can persist within bladder epithelial cells for months after inoculation, without bacteriuria (bacteria in urine) and despite antibiotic treatment (Mulvey et al., 2001; Schilling et al., 2002; Eto et al., 2006; Mysorekar and Hultgren, 2006). In patients, most rUTIs (as many as 82%) are caused by a UPEC strain identical to that of the previous infection (Ejrnaes et al., 2006; Czaja et al., 2009; Luo et al., 2012; Skjot-Rasmussen et al., 2013; Koljalg et al., 2014), even when appropriate antibiotic therapy is given. This could reflect reintroduction of UPEC to the bladder from the gut or vaginal reservoir, but is also consistent with the concept that rUTI can be caused by emergence of the initially infecting UPEC strain from an intracellular bladder reservoir (Mulvey et al., 2001; Hickling and Nitti, 2013; Glover et al., 2014). The potential for bladder reservoirs to seed rUTI has prompted preclinical investigations into strategies to eliminate bladder reservoirs as a means of rUTI. Gardnerella strain JCP8151B causes epithelial exfoliation in the vagina in a mouse model (Gilbert et al., 2013). We hypothesized that Gardnerella could likewise cause bladder epithelial (urothelial) exfoliation and thus be a trigger of UPEC rUTI from bladder reservoirs in women. This hypothesis was further supported by three clinical observations potentially linking Gardnerella with UTI. First, women with bacterial vaginosis, a vaginal dysbiosis in which the vagina is overpopulated by a polymicrobial mixture of bacteria including Gardnerella, have an up to 13-fold higher likelihood of UTI than those with Lactobacillus-dominated vaginal microbiotas (OR 2.21-13.75) (Harmanli et al., 2000; Hillebrand et al., 2002; Sharami et al., 2007; Sumati and Saritha, 2009; Amatya et al., 2013). Second, women with rUTI who received vaginal interventions that influence the vaginal microbiota (e.g., vaginal estrogen, probiotic intravaginal Lactobacillus crispatus) experienced fewer rUTIs than those who received placebo (Stapleton et al., 2011; Rahn et al., 2014; Sadahira et al., 2021). Third, multiple urinary microbiome studies have detected Gardnerella in urine collected directly from the bladder (via suprapubic aspiration or catheterization), suggesting that Gardnerella routinely gains access to the bladder in some women (Wolfe et al., 2012; Pearce et al., 2014; Pearce et al., 2015; Thomas-White et al., 2016; Gottschick et al., 2017). Intriguingly, Gardnerella can co-aggregate with UPEC in an in vitro biofilm assay (Castro et al., 2016), suggesting possible synergy between these bacterial species.
To test our hypothesis, we developed a mouse model to determine whether Gardnerella could cause exfoliation in the bladder and UPEC emergence from bladder reservoirs (Gilbert et al., 2017; O’Brien et al., 2020). We found that in mice with intracellular UPEC reservoirs in the bladder, two transurethral Gardnerella inoculations triggered UPEC emergence from reservoirs, leading to rUTI. Gardnerella exposure caused urothelial apoptosis and exfoliation, which likely enabled UPEC to emerge from reservoirs. Notably, Gardnerella did not stably colonize the bladder, demonstrating that a brief exposure was sufficient to elicit UTI pathogenesis. Here we sought to further characterize the effect of Gardnerella bladder exposures in order to identify host processes involved in UPEC rUTI that could serve as potential biomarkers or therapeutic targets. We performed RNA sequencing (RNA-seq) on bladders harboring intracellular UPEC reservoirs, with and without exposure to Gardnerella. We found that host gene expression changes following Gardnerella exposure were modest, in keeping with a bacterial exposure that does not result in stable colonization. However, gene set enrichment analyses revealed upregulation of many host pathways. Some of the pathways, such as apoptosis and inflammatory cytokines, corroborated the phenotypic results we previously reported in this model, and other pathways were generally related to UTI. Five genes in the Immediate Early response pathway were significantly upregulated, including Nur77 (aka Nr4a1). Notably, Nur77-/- mice were protected from Gardnerella-induced rUTI, confirming its role in this model.
Results
Mouse Model of Gardnerella Bladder Exposure
We performed RNA-seq on whole bladders to identify early host responses to Gardnerella bladder exposure that could contribute to UPEC rUTI. Figure 1 outlines the experimental timeline. First, we established UPEC bladder reservoirs by administering an initial intravesical inoculation (directly into the bladder through the urethra) of 107 colony-forming units (CFU) of the UPEC strain UTI89. Mice that cleared UPEC bacteriuria by 4 weeks post infection (wpi), as determined by weekly urine dilution plating and CFU enumeration, were inoculated intravesically with Gardnerella strain JCP8151B or vehicle (PBS) as a control. Our previous experiments demonstrated that Gardnerella is cleared by 12 hours (h), so we refer to this as an ‘exposure’ rather than an infection. One group of mice received a single exposure and bladders were collected 12 h later in order to examine early host responses to Gardnerella. These groups are labelled PBS-1 and Gard-1. Our previous studies demonstrated that two exposures to Gardnerella were required to cause urothelial exfoliation and to trigger UPEC rUTI. Therefore, to understand the host responses to Gardnerella that could ultimately lead to exfoliation and rUTI, another group of mice, labelled PBS-2 and Gard-2, received two exposures, 12 h apart, and bladders were collected 12 h after the second exposure (Gilbert et al., 2017; O’Brien et al., 2020). Consistent with our previous report that UPEC emergence from reservoirs usually occurs 24-72 h after Gardnerella exposure, only one mouse in the current study (in the Gard-1 group) had detectable reservoir emergence indicated by UPEC titers in urine collected immediately prior to sacrifice.
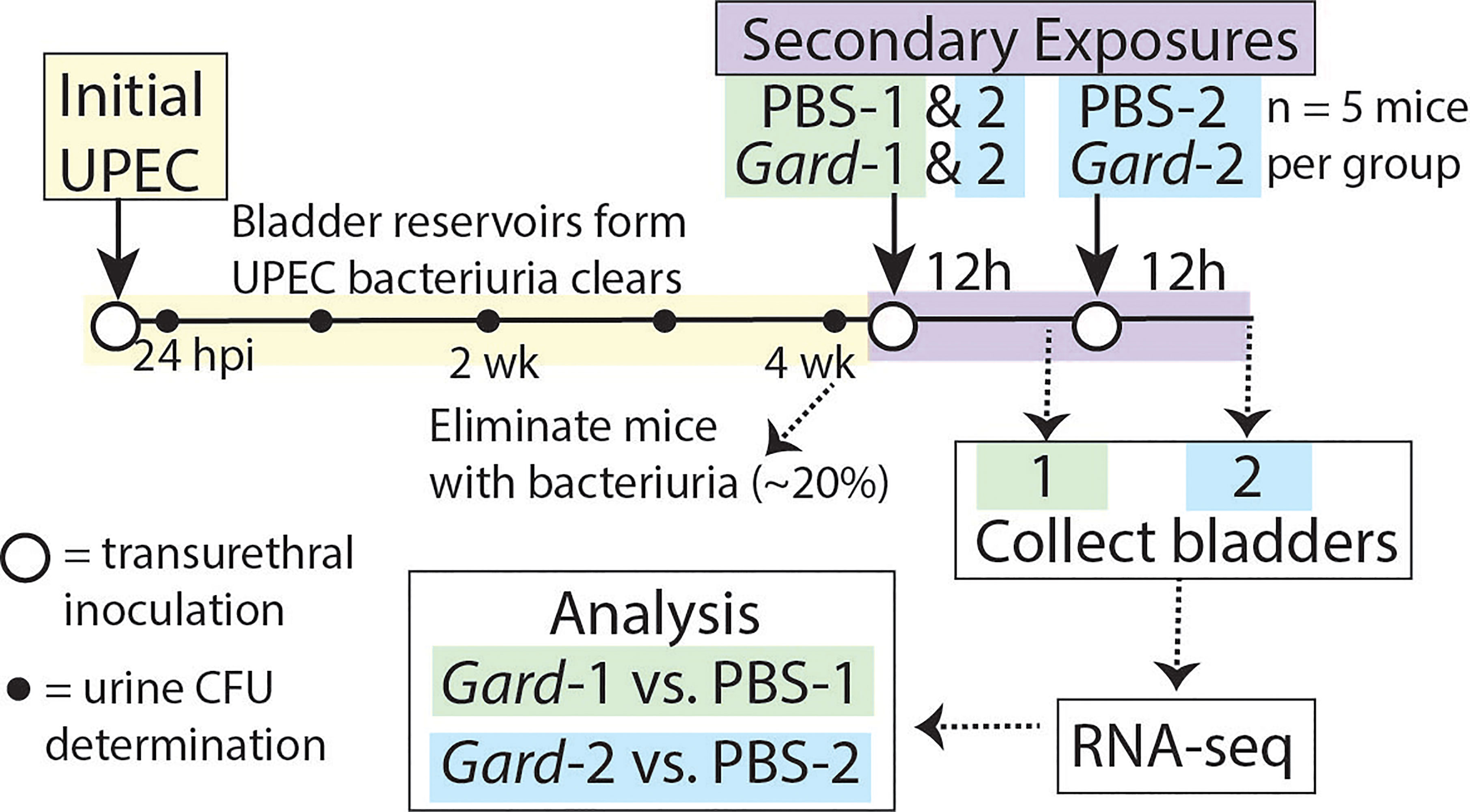
Figure 1 Mouse model schematic for RNA-seq experiments. The time line of the model depicts the ‘Initial UPEC’ reservoir formation phase in yellow and the ‘Secondary Exposure’ phase in purple. Female C57BL/6 mice were inoculated transurethrally (open circles) with UPEC and bacteriuria was monitored weekly (closed circles). Mice that cleared UPEC bacteriuria were inoculated transurethrally with either PBS as a control or Gardnerella. Twelve hours later Exposure Group 1 was sacrificed while Exposure Group 2 received an additional transurethral inoculation and was sacrificed twelve hours (h) later. For the bladder titer experiments described in Figures 4, 5 the second exposure occurred one week later instead of 12 h later and bladders were collected 72 h after the second exposure.
RNA was extracted from bladders from each experimental group (5 mice per group) and used for RNA-seq to assess host gene expression changes as a result of Gardnerella exposures. A total of 640,624,040 RNA-seq reads were generated. Of these, 457,166,961 could be aligned to the Mus musculus reference genome. Further details of the RNA-seq reads are found in Supplementary Table 1. Principal component analysis and multi-dimensional scatter plots did not reveal obvious clustering of any of the individual exposure groups (not shown).
Gene Set Enrichment Analysis Shows Pathways Related to Urothelial Turnover and Inflammation
We determined which gene ontology (GO) terms were differentially expressed following each Gardnerella exposure relative to PBS controls from the same time points (i.e. PBS-1 vs. Gard-1 and PBS-2 vs. Gard-2). Four GO molecular functions and 54 GO biological processes were significantly down-regulated (FDR adjusted P < 0.05, log2FC > 2) following the first Gardnerella exposure (Figure 2A). The majority of the affected pathways were related to host immune and inflammatory processes (Supplementary Tables 2, 3). More substantial differences were seen following the second Gardnerella exposure (Figure 2A): 215 GO molecular functions and 1,488 GO biological processes were significantly up-regulated (FDR adjusted P < 0.05, log2FC > 2). In addition to immune and inflammatory processes, many GO functions were related to urothelial integrity and turnover (Supplementary Tables 4, 5). Also, 33 GO terms related to apoptosis were increased, which is consistent with our previous observation of increased cleaved Casp-3 staining and TUNEL-positive urothelial cells following two Gardnerella exposures (Gilbert et al., 2017).
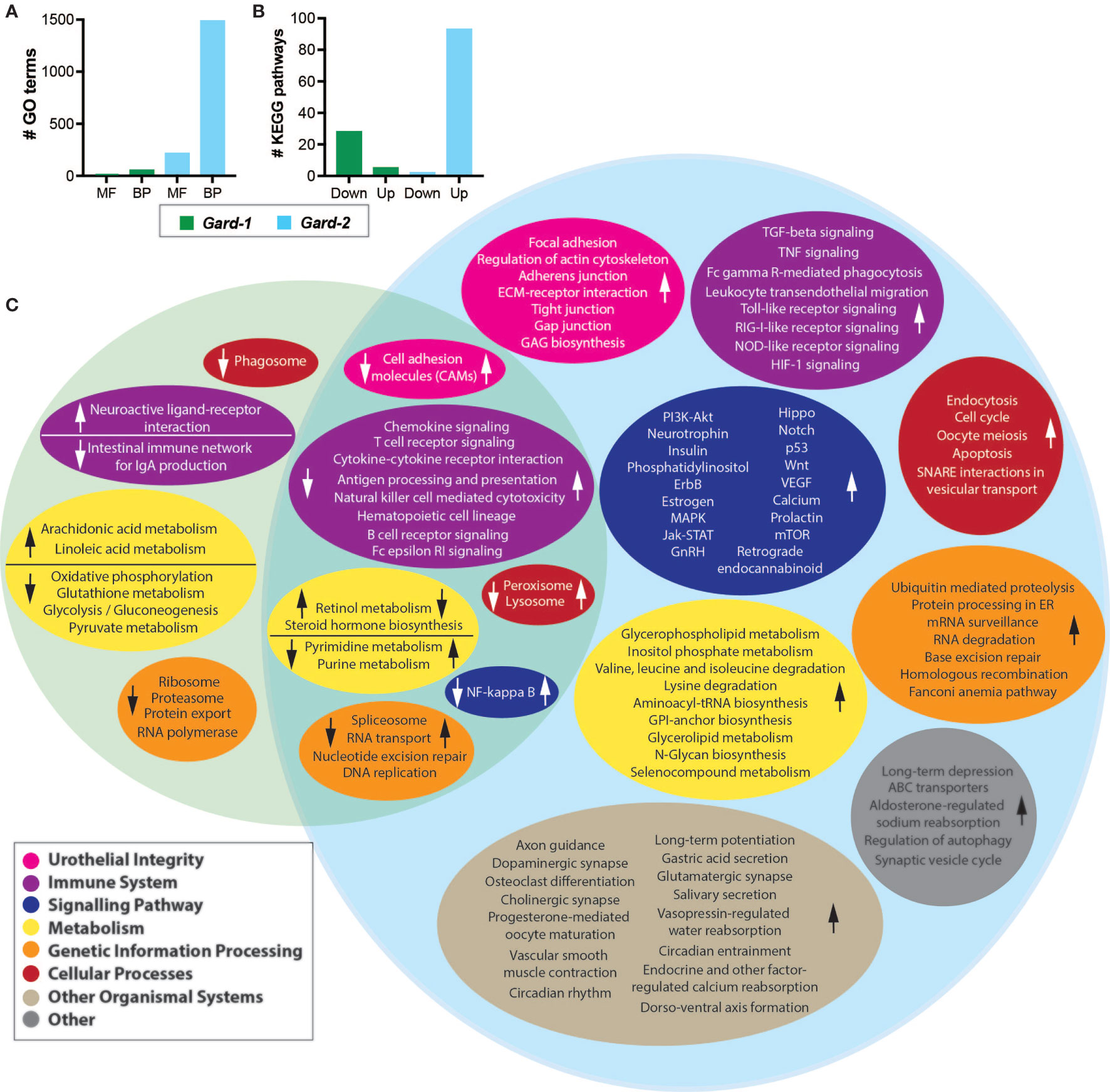
Figure 2 Differential expression analysis. Shown are the host gene expression pathways that were differentially expressed following each Gardnerella (Gard) exposure. (A) The indicates number of Gene Ontology (GO) terms were significantly up- or down-regulated. MF, molecular functions; BP, biological processes. (B) The indicated number of KEGG pathways were significantly up- or down-regulated following Gardnerella exposure. (C) The Venn diagram displays KEGG pathways that were significantly different between PBS-1 and Gard-1 (left, green circle) or PBS-2 and Gard-2 (right, blue circle). Pathways with similar functions are organized into color-coded ovals as indicated in the figure legend. The direction of the change in the Gard group relative to the PBS group is indicated by arrows in each colored oval. The overlapping section of the Venn diagram indicates pathways impacted in both comparisons, with the arrows on the left in each colored oval indicating the direction of the change for Gard-1 and the arrow on the right indicating the direction of the change for Gard-2.
KEGG Pathway Analysis Shows a Dynamic Response to Gardnerella Exposure
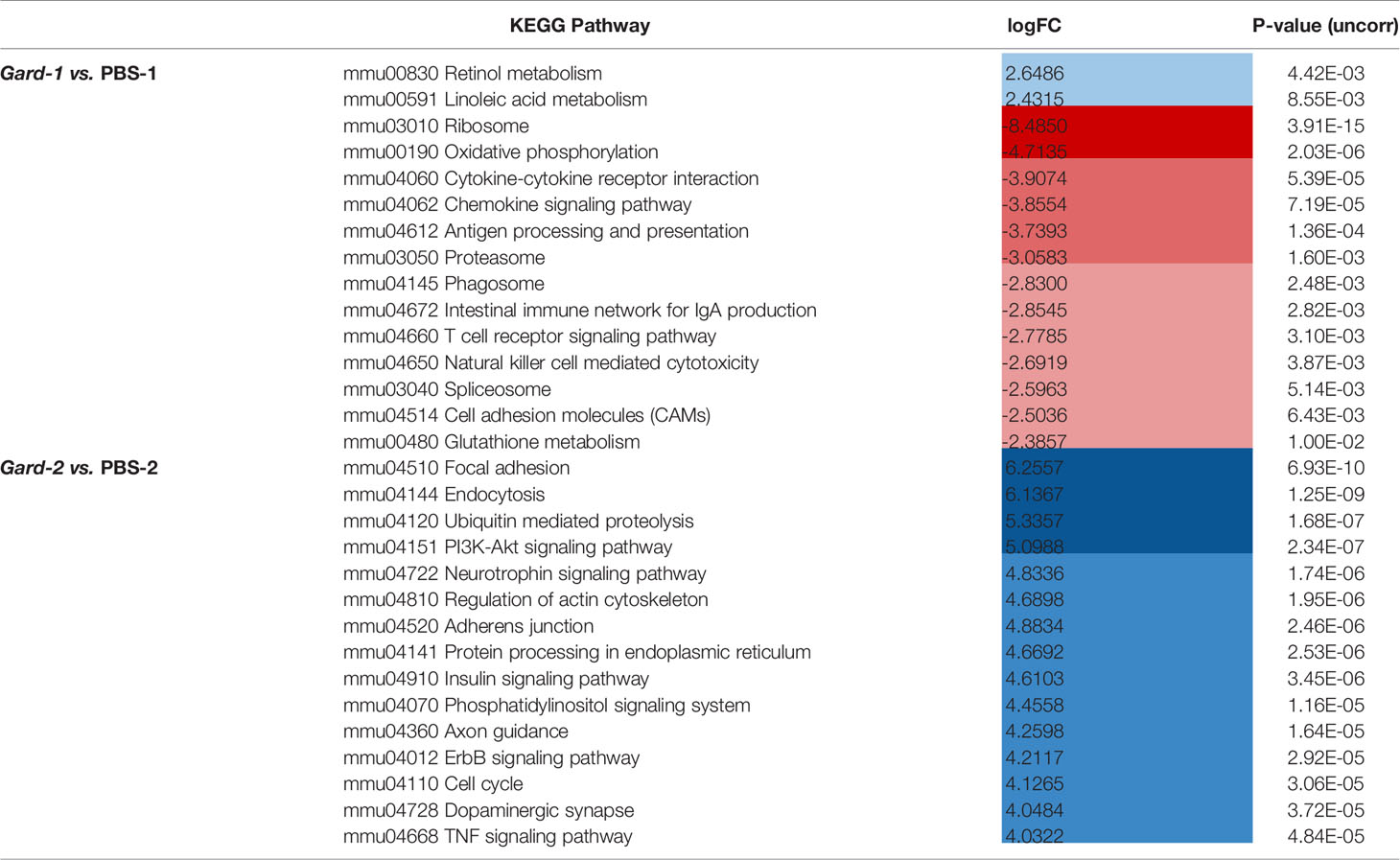
We performed a similar gene set analysis (comparing PBS-1 vs. Gard-1 and PBS-2 vs. Gard-2) using the KEGG Pathway database. Echoing the results from the GO term analysis, more substantial changes occurred following two Gardnerella exposures. Compared to one Gardnerella exposure, two exposures resulted in a greater number of pathways affected and greater with greater fold changes (Figures 2B, C and Table 1). Our previous studies showed that two exposures are necessary to elicit significant urothelial exfoliation and rUTI, which is consistent with greater changes to gene expression in the bladder after the second Gardnerella exposure.
The pathways that were activated following one Gardnerella exposure were primarily related to metabolism, while the pathways with decreased expression included several related to the immune system and signaling (Supplementary Table 6). In contrast to one Gardnerella exposure, where more pathways were downregulated than upregulated, after two Gardnerella exposures all but two of the affected pathways were upregulated (Supplementary Table 7). Consistent with our previous observation of urothelial exfoliation at this time point, several affected pathways were related to epithelial integrity and renewal, such as focal adhesion, cell junctions (adherens, tight, gap), actin cytoskeleton regulation, and Wnt signaling. Many of the upregulated pathways were related to mucosal immune responses, as would be expected following a bacterial exposure. In addition to pathways specific for certain immune cell types, such as T cells, NK cells, and B cells, changes were observed for immune processes like phagocytosis, antigen processing and presentation and cytokine/chemokine signaling. Several signaling pathways related to inflammation and immunity were also affected, including PI3K-Akt, NF-κB, Jak-STAT and Hippo.
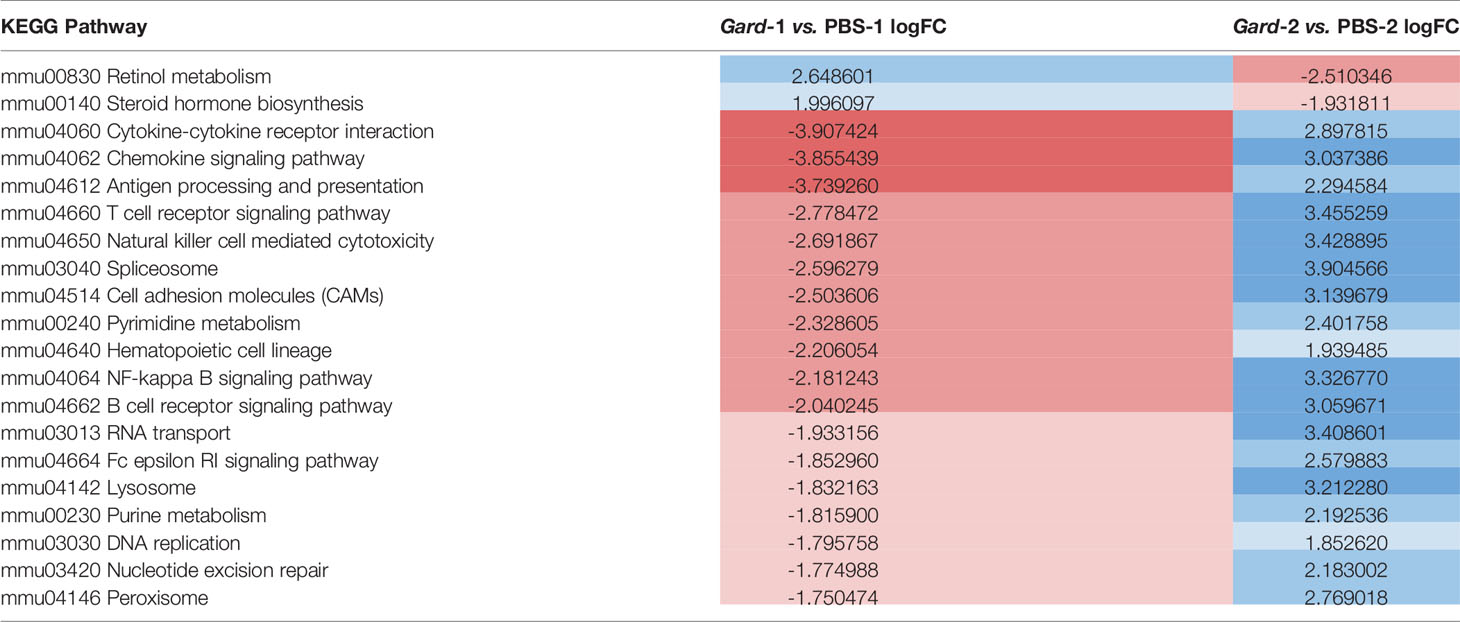
When considering the temporal dynamics of the pathway changes, we found that 20 pathways were significantly different in expression at both time points, but for each of these the change was in opposite directions (Figure 2C arrows; Table 2). For example, pathways relating to retinol metabolism and steroid hormone biosynthesis were upregulated after the first exposure but downregulated after the second. In contrast, inflammatory pathways like cytokine-cytokine receptor interaction, chemokine signaling, and antigen processing and presentation were downregulated after the first exposure and upregulated after the second. These results demonstrate that Gardnerella elicits a dynamic transcriptional response in the bladder.
Differential Gene Expression Suggests a Role for the Immediate Early Response Pathway
It is not atypical for gene set enrichment analyses to uncover more effects than can be detected when assessing significantly differentially expressed individual genes, especially in biologically complex samples like whole organ homogenates. Considering the complexity of the model, which depends on the outcomes of the initial UPEC reservoir formation phase and on the duration of time Gardnerella is maintained in the bladder after each inoculation, it was not entirely surprising that few significant differences were detectable when the data were analyzed at the gene level, comparing PBS-1 vs. Gard-1 and PBS-2 vs. Gard-2. Five genes were significantly increased (FDR adjusted P < 0.05, log2FC > 1.6) following the first Gardnerella exposure (Figure 3 and Supplemental Table 8), and no genes withstood FDR correction after the second Gardnerella exposure. Notably, all of the upregulated genes (Atf3, Fosb, Nur77, Nurr1 and Arc) belong to the class of Immediate Early (IE) response genes that are rapidly co-induced in multiple cell types in response to external stimuli, such as infection and inflammatory signals (Wilson et al., 2010; Bahrami and Drablos, 2016; Crean and Murphy, 2021). This suggests that the presence of these genes in the dataset was not random or artifactual, but reflects a coordinated rapid response to Gardnerella exposure. The fact that these IE genes were increased at the first time point, but not at the second, is consistent with previous reports demonstrating rapid induction and then return to baseline of IE genes (Pei et al., 2005; Tan et al., 2020).
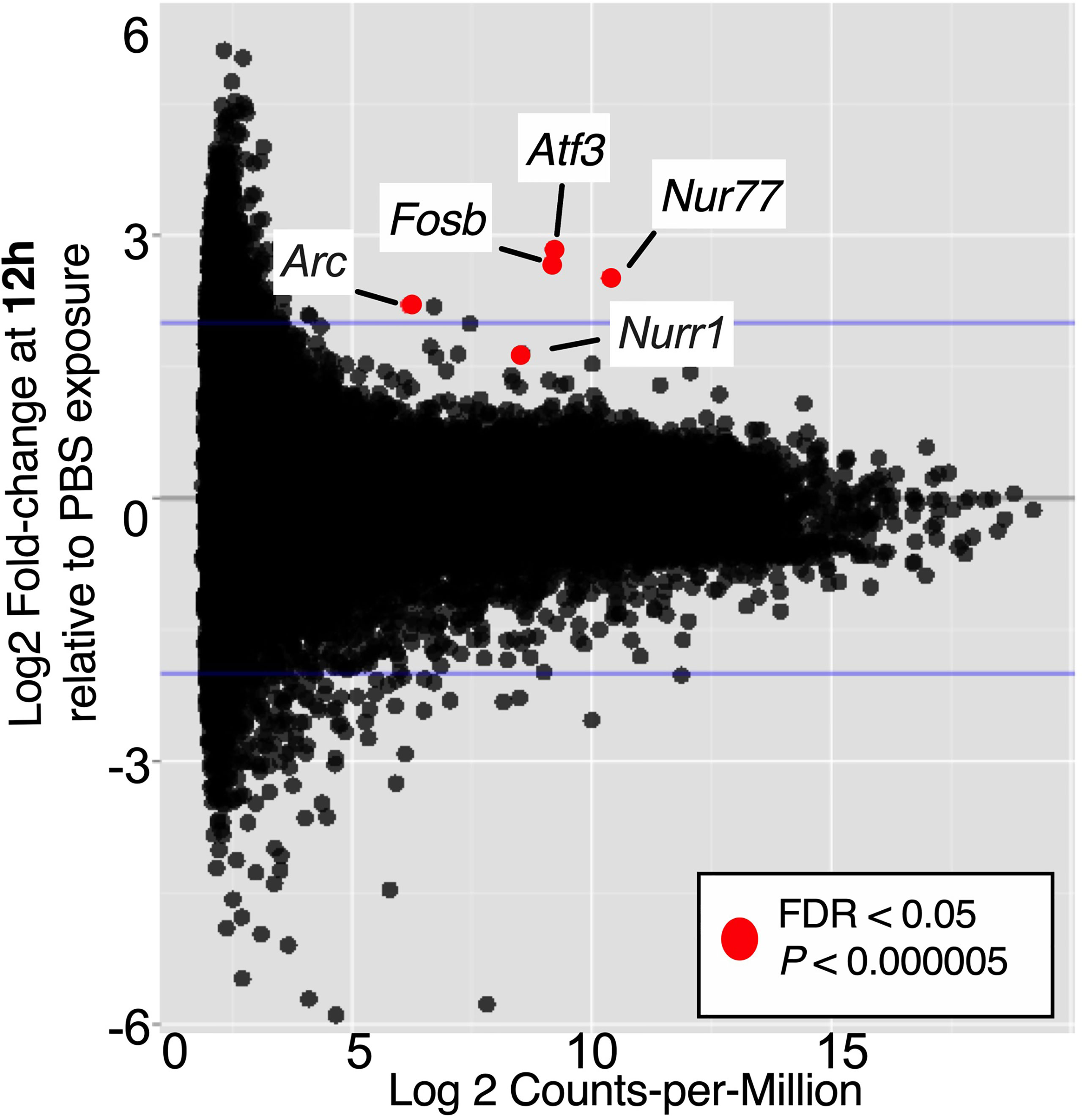
Figure 3 Immediate Early genes upregulated in bladders 12 h after one Gardnerella exposure. The volcano plot indicates genes that had significantly higher expression in Gard-1 compared to PBS-1 bladders. FDR, false discovery rate correction for multiple comparisons.
Nur77 Is Necessary for Gardnerella-Induced UPEC rUTI
The RNA-seq gene-level data suggested that IE genes could play a role in Gardnerella-induced UPEC rUTI. The orphan nuclear receptor Nur77 (also called Nr4a1) is expressed early in the IE pathway, acting as a transcription factor for other IE genes including Atf3 (Yoon et al., 2011; Gao et al., 2016). We used Nur77-/- mice (whole-body) to investigate whether RNA-seq findings could translate to rUTI outcomes. We first established latent UPEC reservoirs in wild type C57BL/6 mice and age-matched Nur77-/- mice on the C57BL/6 background (Lee et al., 1995). Since the role of Nur77 during UTI has not previously been studied, we examined whether the absence of Nur77 impacted UPEC titers during initial infection (yellow bar on Figure 1). There was no significant difference in the overall level of acute UPEC bacteriuria at 24 hours post infection (hpi) during the initial infection between the mouse strains (Figure 4A), although the proportion of mice with low titers (<104 CFU/mL) was significantly greater in Nur77-/- compared to WT mice (0% WT, 14% Nur77-/-; Fisher’s exact P < 0.05). In a subset of animals, we examined UPEC titers during the initial infection phase in bladder tissue following clearance of bacteriuria at 2 wpi (a measure of intracellular reservoirs) and there was no significant difference between WT and Nur77-/- mice (Figure 4B). The proportion of mice that had chronic bacteriuria that persisted at > 104 CFU/mL urine at all time points during the initial infection phase out to 4 wpi was indistinguishable between the two groups (2/39 WT, 2/67 Nur77-/-) (Figure 4C). Together these data demonstrate that Nur77 is not essential for initial UPEC UTI in naïve mice.
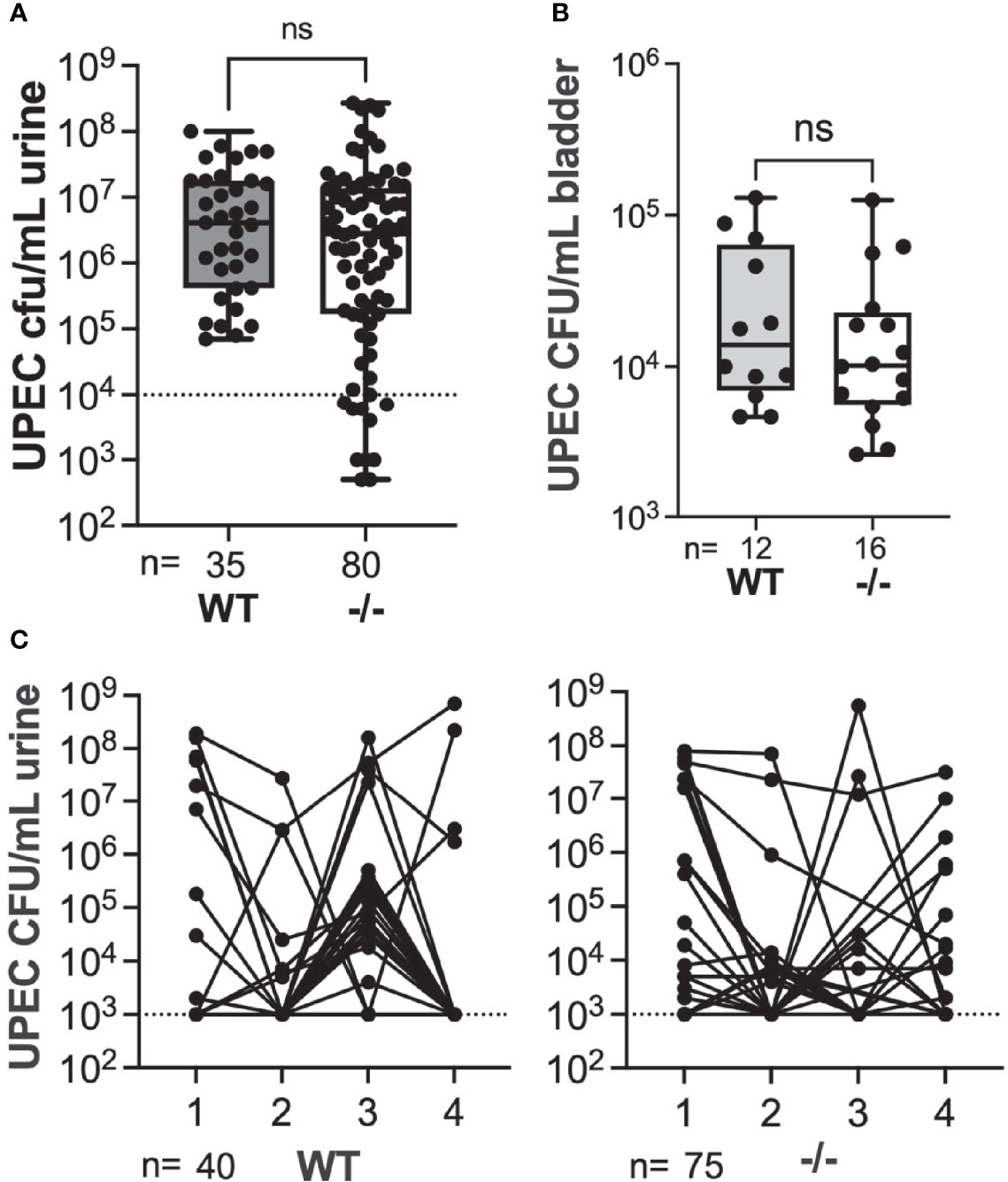
Figure 4 Absence of Nur77 does not affect acute or chronic UPEC bacteriuria or bladder reservoir titers. (A) Acute UPEC bacteriuria 24 hpi following initial infection in wild type (WT) mice and mice globally lacking Nur77 (-/-). (B) UPEC titers in bladders collected 2 weeks after initial UPEC infection. All mice had cleared UPEC bacteriuria prior to bladder analysis, thus titers represent intracellular reservoirs. (C) UPEC weekly bacteriuria titers following initial UPEC infection. Each dot represents data from an individual mouse. ns, not significant.
Next, we took mice that had cleared initial UPEC bacteriuria by 4 wpi and gave them two secondary exposures, 1 week apart, to Gardnerella or PBS. We examined bacteriuria during the subsequent 72 hr after the second exposure as we described previously (Gilbert et al., 2017; O’Brien et al., 2020). We used this exposure model (rather than two exposures 12 h apart, as in the RNA-seq experiment) because it has a higher overall rate of rUTI and because the effects of Gardnerella exposure are also evident in the bladder UPEC titers at the experimental endpoint. Similar to our previous studies (Gilbert et al., 2017), the rate of spontaneous emergence in WT PBS control animals was 11% (Figure 5A). Although knockout mice appeared to have a somewhat higher baseline of emergence compared to WT, there was no statistically significant difference in rUTI incidence between WT and Nur77-/- mice exposed to PBS (11% WT versus 26% Nur77-/-; P = 0.247, Fisher’s exact test). Consistent with our previous results, Gardnerella exposure increased the incidence of UPEC rUTI in WT mice by approximately 5-fold (56% Gard vs. 11% PBS in WT mice; P < 0.01). Conversely, the incidence of rUTI was indistinguishable between the Gardnerella and PBS exposure groups in Nur77-/- mice 32% Gard vs. 29% PBS, Figure 5A). In line with our previous findings, Gardnerella-exposed WT mice had lower bladder UPEC burdens at the experimental endpoint (Figure 5B). This is presumably due to egress of UPEC from the tissue concordant with development of bacteriuria. In contrast, in Nur77-/- mice, bladder titers were no different between mice exposed to Gardnerella and those exposed to PBS, which is consistent with the urine titer data. Taken together, these results corroborate the RNA-seq data and point to Nur77 as a necessary host factor for Gardnerella-induced UPEC rUTI from bladder reservoirs.
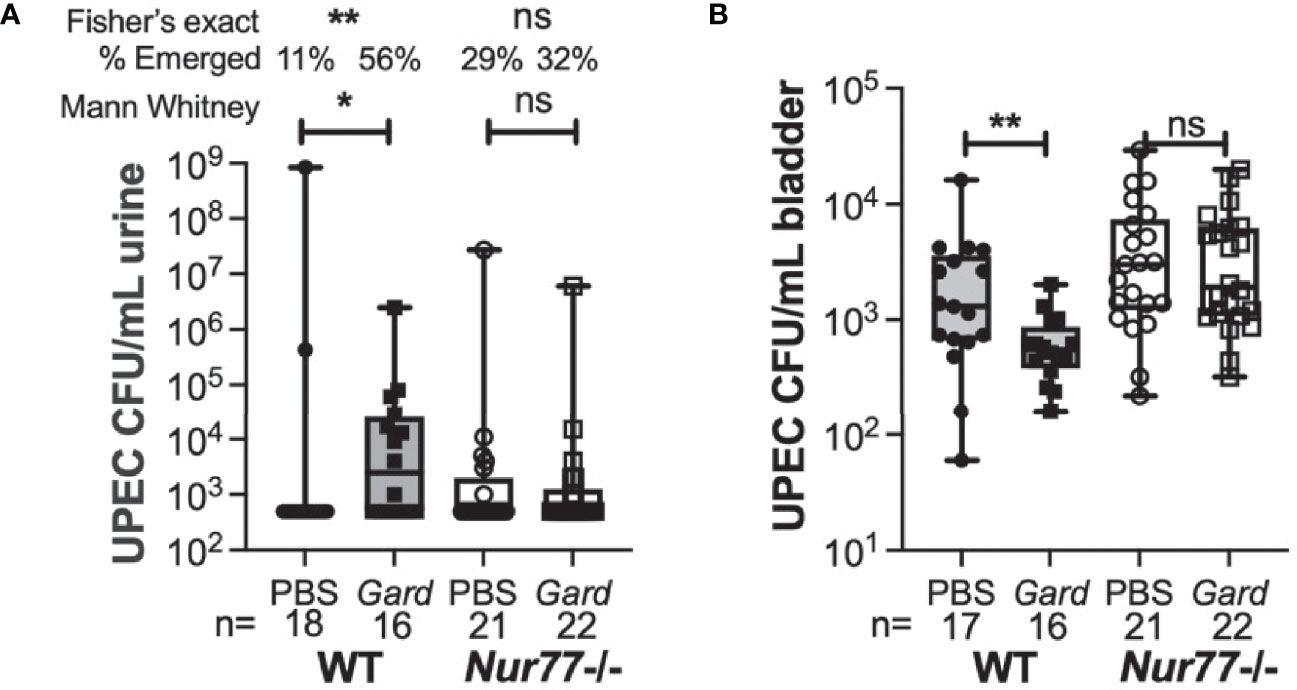
Figure 5 Mice lacking Nur77 are protected from Gardnerella-induced UPEC reservoir emergence. Mice harboring bladder UPEC reservoirs were exposed to PBS or Gardnerella twice at a one-week interval. Urines were collected daily for three days after the second exposure and mice were euthanized after three days. (A) Shown are the highest UPEC urine titers of each individual mouse. (B) Bladder UPEC titers are shown. The number of mice in each group is given at the bottom of the graphs. *P < 0.05; **P < 0.01.
Discussion
Millions of women suffer from recurrent UPEC UTI (Foxman et al., 2000; Foxman, 2014), most commonly caused by the same strain that caused the initial infection (Ejrnaes et al., 2006; Czaja et al., 2009; Skjot-Rasmussen et al., 2013; Koljalg et al., 2014). At least some instances of same-strain rUTI are likely due to UPEC emergence from protected bladder intracellular reservoirs (Mulvey et al., 2001; Hickling and Nitti, 2013; Glover et al., 2014). We and others have demonstrated that UPEC can persist intracellularly in the mouse bladder for weeks or months, and can emerge and elicit rUTI after bladder exfoliation is experimentally induced (Mulvey et al., 2001; Schilling et al., 2002; Eto et al., 2006; Mysorekar and Hultgren, 2006). In our model, we use two Gardnerella exposures as the trigger for bladder exfoliation and rUTI (Gilbert et al., 2017; O’Brien et al., 2020). In the present study we demonstrate that Gardnerella exposures in mice containing UPEC reservoirs affected the expression of host pathways related to urothelial exfoliation and regeneration, mucosal inflammation and immunity, and other processes. Immediate Early (IE) genes were upregulated following one Gardnerella exposure. The orphan nuclear receptor Nur77 is a key IE gene (Yoon et al., 2011; Gao et al., 2016), and we found that whole-body Nur77 knockout mice were protected from recurrent UPEC UTI following Gardnerella exposure – that is to say, the rate of rUTI was not increased in Nur77-/- mice exposed to Gardnerella compared to those mice exposed to PBS. Thus, the IE response may contribute to Gardnerella-induced recurrent UPEC UTI.
Gardnerella has frequently been isolated as the most abundant organism in “urinary microbiome” studies, including those using expanded culture techniques to detect live bacteria (Wolfe et al., 2012; Pearce et al., 2014; Hilt et al., 2014; Pearce et al., 2015; Karstens et al., 2016; Wu et al., 2017). Importantly, these studies collected urine by catheterization or suprapubic needle aspiration to limit vaginal contamination of urine specimens, supporting the conclusion that Gardnerella can indeed be found within the bladder. In a recent longitudinal culturomics study of healthy women, Gardnerella was often present at high relative abundance in urine sample pairs collected from the same woman at different time points (Ksiezarek et al., 2021). Another large study found hospital inpatients with G. vaginalis in their urine were more likely to have a history of rUTI than patients in whom G. vaginalis was not detected (Josephson et al., 1988). Although these clinical studies make it clear that Gardnerella can be found in the bladder and urine, it is not yet known whether Gardnerella stably colonizes the urinary tract, vs. being repeatedly re-introduced into the bladder by hygienic and/or sexual behaviors. We have shown that Gardnerella does not stably colonize the mouse bladder and is cleared within 12 h of exposure, but nonetheless triggers exfoliation and UPEC rUTI (Gilbert et al., 2017). Here we detected significant changes in expression of several pathways even relatively early after a single Gardnerella exposure; more substantial changes occurred after two Gardnerella exposures as indicated by significant changes in GO terms and KEGG pathways. These findings further support a model in which repeated introduction of Gardnerella into the bladder (which may occur in women after sexual activity) can drive host responses and rUTI phenotypes. Thus, our study provides further support for “covert pathogenesis,” the notion that a brief exposure to a microbe (here, Gardnerella) can affect the host enough to drive disease attributed to another microbe (here, E. coli) (Gilbert and Lewis, 2019).
Several pathways identified in this RNA-seq analysis are corroborated by phenotypic observations that we previously reported in this mouse exposure model. As would be expected following bacterial exposure, here we observed changes in many host pathways and processes related to the mucosal immune response. The conclusion that Gardnerella exposures trigger an inflammatory response in bladders harboring UPEC reservoirs is further supported by our previous finding of higher levels of IL-12, IFN-g, and RANTES in bladder homogenates (Gilbert et al., 2017). Interestingly, pathways related to T and B cells were upregulated after the second Gardnerella exposure. This observation could implicate adaptive immunity generated by the initial UPEC infection as a contributor to UPEC reservoir emergence. A previous study showed that an adaptive immune response is necessary to clear a challenge UPEC infection that was introduced by a second UPEC inoculation two weeks after a primary UPEC infection in C57BL/6 mice (Mora-Bau et al., 2015). Other studies have demonstrated that bladder inflammation differs between first and second UPEC exposures and that severe bladder inflammation can impact UTI outcomes (O’Brien et al., 2018; Yu et al., 2019). More work is needed to understand bladder immune responses to Gardnerella exposure and UPEC reservoir emergence. The RNA-seq pathway analysis also implicated urothelial integrity and turnover, as well as apoptosis. Our previous studies corroborate these findings, as we have shown that our model strain, Gardnerella JCP8151B (also used in the present study), elicits both vaginal (Gilbert et al., 2013; Gilbert et al., 2019) and urothelial exfoliation (Gilbert et al., 2017). In the same urinary tract exposure model used here, mice exposed to Gardnerella displayed evidence of apoptosis in the urothelium. Compared to mice exposed to PBS, those exposed to Gardnerella had increased urothelial TUNEL staining and cleaved Caspase-3 12 h after two Gardnerella exposures (Gilbert et al., 2017). Thus, the RNA-seq findings of pathways related to inflammation and urothelial integrity echo the biological phenotypes we previously reported in the mouse model and the findings from our whole organ analysis support the relevance of RNA-seq methods for identifying biologically relevant host responses to Gardnerella bladder exposures.
The model and data presented here have some limitations. Although whole bladder RNA-seq is an established method for investigating host response during UTI (O’Brien et al., 2016; Yu et al., 2019), transcriptional changes cannot be attributed to specific cell types, but rather reflect changes that occurred at the organ level. As well, using the whole organ for bulk RNA-seq may mask gene expression changes that only occur in a small subset of cells, which could be one explanation for the relatively modest number of genes for which expression changes after Gardnerella exposure were statistically significant following FDR correction. Nonetheless, the RNA-seq data corroborated previously reported phenotypic findings in this model (apoptosis, inflammatory cytokines) and identified a host gene (Nur77) that we subsequently found to be necessary for Gardnerella-induced rUTI. These findings support the utility of RNA-seq methods for assessing host responses to even transient bladder microbial exposures and warrant future single-cell transcriptomic studies to specifically examine urothelial, immune, and other bladder cell responses.
The model presented here involves multiple exposures to two different microbes that could independently or synergistically stimulate host responses over time. While this likely reflects the situation occurring in women, it does present some challenges for interpretation. A caveat of the current dataset is that it cannot distinguish changes that required a second Gardnerella exposure from changes that resulted from the first exposure, but required additional time to become apparent. However, since we previously reported that a single Gardnerella exposure was rapidly cleared and did not cause urothelial exfoliation or UPEC rUTI, it is very likely that most of the pathway upregulation observed here was due to the second exposure, but this should be determined in future studies. Our finding that host gene expression pathways were primarily downregulated after one Gardnerella exposure and upregulated after two exposures could suggest that the second time point reflects a host genetic signature of UPEC reservoir reactivation. Additionally, since all of the mice used in the present study harbored UPEC reservoirs (because of our interest in rUTI), we cannot distinguish changes that were due specifically and solely to Gardnerella from changes that resulted from “re-awakening” of UPEC reservoirs. Relatively little is understood with respect to how UPEC emerge from quiescent intracellular reservoirs and what host processes are involved. Furthermore, UPEC infection has a lasting effect on the bladder mucosa that is likely to also impact how the bladder responds to subsequent exposures. Consistent with this idea, we previously reported that the bladder cytokine changes caused by Gardnerella were distinct between naive mice and those with UPEC reservoirs (Gilbert et al., 2017). Future studies aimed at distinguishing between these various possibilities could examine the bladder transcriptome following Gardnerella exposure in naive mice and following induction of UPEC rUTI from reservoirs by other means (e.g. protamine sulfate (Mysorekar and Hultgren, 2006) or chitosan (Blango et al., 2014)).
Our RNA-seq data implicated the nuclear receptor Nur77 (aka Nr4a1) as an early responder to Gardnerella exposure. Nur77 regulates myriad cellular processes, that intersect with the UPEC UTI pathogenic cascade and could influence rUTI outcomes. For example, Nur77 regulates apoptosis in multiple tissue types (Rajpal et al., 2003; Herring et al., 2019), and we previously found that Gardnerella induced bladder exfoliation via apoptosis (Gilbert et al., 2017). Whether or not Nur77 drives exfoliation in the Gardnerella-exposed urothelium remains to be determined. Nur77 also modulates inflammation (Rodriguez-Calvo et al., 2017) and has been specifically implicated in T cell responses (Liebmann et al., 2018) and Ly6C- monocytes (Hanna et al., 2011). Nur77 modulated inflammatory responses to E. coli in the lung during pneumonia in mice (Cui et al., 2019), but the role of Nur77 in mediating bladder responses to UPEC has not been examined. In the present study we did not conduct an in-depth characterization of acute UPEC UTI in Nur77-/- mice. However, the first phase of our rUTI model entails UPEC infection and monitoring of urine bacterial titers over time during the initial infection phase, prior to secondary exposures. We observed no differences between wild type and Nur77-/- mice in the initial UPEC infection phase of the model. As well, we found that Nur77-/- mice did harbor stable bladder UPEC reservoirs. Thus, Nur77 may be dispensable for initial UPEC bladder infection in C57BL/6 mice, though we did not assess chronic bladder or kidney infection or other mouse UTI phenotypes. Strikingly, we found that Nur77-/- mice were protected from Gardnerella-induced recurrent UPEC UTI, as evidenced by no difference in the incidence of post-exposure bacteriuria or remaining bladder reservoir titers between Gardnerella-exposed vs. PBS-exposed Nur77-/- mice. In this pilot study, the wild type and Nur77-/- mice were not littermates. Future studies will directly compare Nur77-/- mice to Nur77+/- and wild type littermate controls to validate our rUTI findings. Nur77 is a druggable target, with several ligands being explored for treatment of diseases such as cancer, metabolic disorders, hyperinflammatory responses and endometriosis (Wu and Chen, 2018; Mohankumar et al., 2020). If our findings translate to rUTI in humans, this could open up a therapeutic avenue that is much needed in the current climate of increasing antibiotic resistance (Tamadonfar et al., 2019). Future studies will more closely examine the mechanism(s) for Nur77-mediated response to Gardnerella in the bladder, will investigate whether IE responses impact UTI phenotypes in other mouse models, and will test whether Nur77 could also play a role in host response to Gardnerella in the vagina.
Materials and Methods
Ethics Statement
Mouse experiments were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals. The Institutional Animal Care and Use Committee (IACUC) of Washington University School of Medicine approved the protocol (Protocol Number: 20170081).
Bacterial Strains and Growth Conditions
Uropathogenic E. coli strain UTI89, harboring a kanamycin resistance cassette (Wright et al., 2005), was grown aerobically at 37°C in static liquid culture in Lysogeny Broth (LB) medium, or on LB agar plates with 25 μg/ml kanamycin. Gardnerella strain JCP8151B (Lewis et al., 2013), historically regarded as G. vaginalis and recently referred to as G. piotii (Hill et al., 2019), was grown anaerobically at 37°C in shaking liquid culture in NYCIII medium, or on NYCIII agar plates with 1 mg/ml streptomycin. Mouse inocula were prepared as previously described (O’Brien et al., 2020).
Mice
Six- to seven-week-old female C57BL/6 mice (“wild type”) were obtained from Charles River (Fredericks facility). Mice globally deficient in Nur77 (a.k.a. Nr4a1) were obtained from Jackson Laboratories (B6;129S2-Nr4a1tm1Jmi/J, catalog #006187). Mice were given a regular chow diet in a specific pathogen-free facility with a 12 h light/12 h dark cycle at Washington University School of Medicine. Mice were allowed to acclimate to the facility after transport for 1 week prior to experiments.
Mouse Urinary Tract Inoculation Experiments for RNA-Seq
Experiments were performed essentially as described previously (Gilbert et al., 2017; O’Brien et al., 2020). Briefly, mice were anesthetized with isoflurane and then inoculated transurethrally with 50 μL prepared 1 x 107 CFU UPEC inoculum. Urine was collected at 24 hpi, and weekly thereafter for 4 weeks. Mice that no longer had detectable UTI89 in urine at 4 weeks, reflecting resolution of the initial bladder lumen infection, were then inoculated transurethrally with 50 uL prepared inoculum of 1 x 108 CFU Gardnerella strain JCP8151B (10 mice) or PBS (10 mice). Twelve hours later, five mice from each exposure group (Gard-1 and PBS-1) were sacrificed and their bladders were collected aseptically and flash frozen in liquid nitrogen for future RNA isolation. The remaining five mice per group received a second transurethral inoculation with JCP8151B or PBS and were sacrificed another 12 h later to collect bladders for RNA isolation (Gard-2 and PBS-2).
Library Preparation and Sequencing
Bladders were homogenized and RNA was extracted using the RNeasy Plus Mini kit (Qiagen). Libraries were prepared from each bladder individually with 10 ng of total RNA and RNA integrity was determined using an Agilent Bioanalyzer, with a Bioanalyzer RIN score greater than 8.0 obtained for all samples. ds-cDNA was prepared using the SMARTer Ultra Low RNA kit for Illumina Sequencing (Takara-Clontech) per the manufacturer’s protocol. cDNA was fragmented using a Covaris E220 sonicator using peak incident power 18, duty factor 20%, cycles/burst 50, time 120 seconds to yield an average size of 200 base pairs (bp). cDNA was then blunt ended, had an A base added to the 3’ ends, and then had Illumina sequencing adapters ligated to the ends. Ligated fragments were then amplified for 12 cycles using primers incorporating unique index tags. Fragments were multiplexed with 5-6 samples per lane and were sequenced on an Illumina HiSeq 2500 using single end 50 bp reads to target 30M per sample.
RNA-Seq Data Acquisition, Quality Control, and Processing
RNA-seq reads from the twenty individual libraries (5 mice per exposure group) were demultiplexed using a custom demultiplexing script written in Python and then aligned to the Ensembl GRCm38.76 (Mus musculus) assembly with STAR version 2.0.4b. Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount version 1.4.5. Transcript counts were produced by Sailfish version 0.6.3. Sequencing performance was assessed for total number of aligned reads, total number of uniquely aligned reads, genes and transcripts detected, ribosomal fraction, known junction saturation and read distribution over known gene models with RSeQC version 2.3. All gene-level and transcript counts were then imported into the R/Bioconductor package EdgeR and TMM-normalized to adjust for differences in library size. Genes or transcripts not expressed in any sample were excluded from further analysis. Performance of the samples was assessed with a Spearman correlation matrix and multi-dimensional scaling plots. Generalized linear models with robust dispersion estimates were created to test for gene/transcript level differential expression. The fits of the trended and tagwise dispersion estimates were then plotted to confirm proper fit of the observed mean to variance relationship where the tagwise dispersions are equivalent to the biological coefficients of variation of each gene. Differentially expressed genes and transcripts (comparing PBS-1 vs. Gard-1 and PBS-2 vs. Gard-2) were then filtered for FDR adjusted P values less than or equal to 0.05. For each EdgeR contrast, global perturbations in known Gene Ontology (GO) terms and KEGG pathways were detected using the R/Bioconductor package GAGE to test for changes in expression of the reported log2 fold-changes reported by edgeR in each term versus the background log2 fold-changes of all genes found outside the respective term. The R/Bioconductor package heatmap3 was used to display heatmaps across groups of samples for each GO term with a Benjamini-Hochberg false-discovery rate adjusted P value less than or equal to 0.05.
Recurrent UTI Experiments in Wild Type and Nur77 -/- Mice
Mice were anesthetized with isoflurane and then inoculated transurethrally with 50 μL prepared UPEC inoculum. Urine was collected at 24 hpi, and weekly thereafter for 4 weeks. A subset of mice were sacrificed at 2 weeks to compare bladder reservoir titers. Mice that no longer had detectable UTI89 in urine at 4 weeks, reflecting resolution of the initial bladder lumen infection, were used for recurrent UTI experiments. The groups were frequency matched based upon the time course of clearance of UPEC urine titers during the initial infection (O’Brien et al., 2020). Mice were given two bladder exposures of PBS or Gardnerella, 1 week apart (transurethral inoculations prepared as in experiments described above). Urine was collected at 24, 48 and 72 h after the second exposure and titers were enumerated by serial dilution and plating on selective media (LB+kanamycin to detect UPEC; NYCIII+streptomycin to detect Gardnerella). At 72 h, mice were humanely sacrificed via cervical dislocation under isofluorane anaesthesia and bladders and kidneys were aseptically harvested. Homogenates were prepared in 1 mL sterile PBS and plated on appropriate selective media. Bacterial burden in each sample was calculated as CFU/bladder. Samples with no colonies were plotted at one-half of the limit of detection.
Data Availability Statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE186800 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186800).
Ethics Statement
The animal study was reviewed and approved by The Institutional Animal Care and Use Committee (IACUC) of Washington University School of Medicine
Author Contributions
NG and VO’B performed experiments. NG, VO’B, and AL analyzed the data. NG and VO’B drafted the manuscript. All authors provided funding to support the research. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Institutes of Health NIAID [R01 AI114635 to AL and R21 AI152049 to AL and NG] and NIDDK [R21 DK092586 to AL and K01 DK110225 to NG], by the National Science Foundation [Graduate Research Fellowship to VO’B #DGE–1143954], by the American Heart Association [Postdoctoral Fellowship to NMG] and by the Center for Women’s Infectious Disease Research at Washington University School of Medicine in St. Louis [Pilot Research Grant to NG]. Some of the animal studies were performed in a facility supported by the NCRR [C06 RR015502]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Eric Tycksen at the Genome Technology Access Center (GTAC) for performing the RNA-seq data analysis and for helpful responses to our questions throughout the duration of the project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.788229/full#supplementary-material
References
Amatya, R., et al. (2013). Urinary Tract Infection in Vaginitis: A Condition Often Overlooked. Nepal. Med. Coll. J. 15 (1), 65–67.
Bahrami, S., Drablos, F. (2016). Gene Regulation in the Immediate-Early Response Process. Adv. Biol. Regul. 62, 37–49. doi: 10.1016/j.jbior.2016.05.001
Blango, M. G., Ott, E. M., Erman, A., Veranic, P., Mulvey, M. A. (2014). Forced Resurgence and Targeting of Intracellular Uropathogenic Escherichia Coli Reservoirs. PLoS One 9 (3), e93327. doi: 10.1371/journal.pone.0093327
Castro, J., Machado, D., Cerca, N. (2016). Escherichia Coli and Enterococcus Faecalis are Able to Incorporate and Enhance a Pre-Formed Gardnerella Vaginalis Biofilm. Pathog. Dis. 74 (3), ftw007. doi: 10.1093/femspd/ftw007
Crean, D., Murphy, E. P. (2021). Targeting NR4A Nuclear Receptors to Control Stromal Cell Inflammation, Metabolism, Angiogenesis, and Tumorigenesis. Front. Cell Dev. Biol. 9, 589770. doi: 10.3389/fcell.2021.589770
Cui, P., Wu, S., Xu, X., Ye, H., Hou, J., Liu, X., et al. (2019). Deficiency of the Transcription Factor NR4A1 Enhances Bacterial Clearance and Prevents Lung Injury During Escherichia Coli Pneumonia. Shock 51 (6), 787–794. doi: 10.1097/SHK.0000000000001184
Czaja, C. A., Stamm, W. E., Stapleton, A. E., Roberts, P. L., Hawn, T. R., Scholes, D., et al. (2009). Prospective Cohort Study of Microbial and Inflammatory Events Immediately Preceding Escherichia Coli Recurrent Urinary Tract Infection in Women. J. Infect. Dis. 200 (4), 528–536. doi: 10.1086/600385
De Nisco, N. J., Neugent, M., Mull, J., Chen, L., Kuprasertkul, A., de Souza Santos, M., et al. (2019). Direct Detection of Tissue-Resident Bacteria and Chronic Inflammation in the Bladder Wall of Postmenopausal Women With Recurrent Urinary Tract Infection. J. Mol. Biol. 431 (21), 4368–4379. doi: 10.1016/j.jmb.2019.04.008
Ejrnaes, K., Sandvang, D., Lundgren, B., Ferry, S., Holm, S., Monsen, T., et al. (2006). Pulsed-Field Gel Electrophoresis Typing of Escherichia Coli Strains From Samples Collected Before and After Pivmecillinam or Placebo Treatment of Uncomplicated Community-Acquired Urinary Tract Infection in Women. J. Clin. Microbiol. 44 (5), 1776–1781. doi: 10.1128/JCM.44.5.1776-1781.2006
Elliott, T. S., Reed, L., Slack, R. C., Bishop, M. C. (1985). Bacteriology and Ultrastructure of the Bladder in Patients With Urinary Tract Infections. J. Infect. 11 (3), 191–199. doi: 10.1016/S0163-4453(85)92997-4
Eto, D. S., Sundsbak, J. L., Mulvey, M. A. (2006). Actin-Gated Intracellular Growth and Resurgence of Uropathogenic Escherichia Coli. Cell Microbiol. 8 (4), 704–717. doi: 10.1111/j.1462-5822.2006.00691.x
Foxman, B. (2014). Urinary Tract Infection Syndromes: Occurrence, Recurrence, Bacteriology, Risk Factors, and Disease Burden. Infect. Dis. Clin. North Am. 28 (1), 1–13. doi: 10.1016/j.idc.2013.09.003
Foxman, B., Gillespie, B., Koopman, J., Zhang, L., Palin, K., Tallman, P., et al. (2000). Risk Factors for Second Urinary Tract Infection Among College Women. Am. J. Epidemiol. 151 (12), 1194–1205. doi: 10.1093/oxfordjournals.aje.a010170
Foxman, B., Manning, S. D., Tallman, P., Bauer, R., Zhang, L., Koopman, J. S., et al. (2002). Uropathogenic Escherichia Coli Are More Likely Than Commensal E. Coli to be Shared Between Heterosexual Sex Partners. Am. J. Epidemiol. 156 (12), 1133–1140. doi: 10.1093/aje/kwf159
Gao, H., Chen, Z., Fu, Y., Yang, X., Weng, R., Wang, R., et al. (2016). Nur77 Exacerbates PC12 Cellular Injury In Vitro by Aggravating Mitochondrial Impairment and Endoplasmic Reticulum Stress. Sci. Rep. 6, 34403. doi: 10.1038/srep34403
Gilbert, N. M., Lewis, W. G., Li, G., Sojka, D. K., Lubin, J. B., Lewis, A. L., et al. (2019). Gardnerella Vaginalis and Prevotella Bivia Trigger Distinct and Overlapping Phenotypes in a Mouse Model of Bacterial Vaginosis. J. Infect. Dis. 220 (7), 1099–1108. doi: 10.1093/infdis/jiy704
Gilbert, N. M., Lewis, A. L. (2019). Covert Pathogenesis: Transient Exposures to Microbes as Triggers of Disease. PLoS Pathog. 15 (3), e1007586. doi: 10.1371/journal.ppat.1007586
Gilbert, N. M., Lewis, W. G., Lewis, A. L. (2013). Clinical Features of Bacterial Vaginosis in a Murine Model of Vaginal Infection With Gardnerella Vaginalis. PLoS One 8 (3), e59539. doi: 10.1371/journal.pone.0059539
Gilbert, N. M., O’Brien, V. P., Lewis, A. L. (2017). Transient Microbiota Exposures Activate Dormant Escherichia Coli Infection in the Bladder and Drive Severe Outcomes of Recurrent Disease. PLoS Pathog. 13 (3), e1006238. doi: 10.1371/journal.ppat.1006238
Glover, M., Moreira, CG., Sperandio, V., Zimmern, P. (2014). Recurrent Urinary Tract Infections in Healthy and Nonpregnant Women. Urol. Sci. 25 (1), 1–8. doi: 10.1016/j.urols.2013.11.007
Gottschick, C., Deng, Z. L., Vital, M., Masur, C., Abels, C., Pieper, D. H., et al. (2017). The Urinary Microbiota of Men and Women and Its Changes in Women During Bacterial Vaginosis and Antibiotic Treatment. Microbiome 5 (1), 99. doi: 10.1186/s40168-017-0305-3
Hanna, R. N., Carlin, L. M., Hubbeling, H. G., Nackiewicz, D., Green, A. M., Punt, J. A., et al. (2011). The Transcription Factor NR4A1 (Nur77) Controls Bone Marrow Differentiation and the Survival of Ly6C- Monocytes. Nat. Immunol. 12 (8), 778–785. doi: 10.1038/ni.2063
Harmanli, O. H., Cheng, G. Y., Nyirjesy, P., Chatwani, A., Gaughan, J. P. (2000). Urinary Tract Infections in Women With Bacterial Vaginosis. Obstet. Gynecol. 95 (5), 710–712. doi: 10.1016/s0029-7844(99)00632-8
Herring, J. A., Elison, W. S., Tessem, J. S. (2019). Function of Nr4a Orphan Nuclear Receptors in Proliferation, Apoptosis and Fuel Utilization Across Tissues. Cells 8 (11), 1373. doi: 10.3390/cells8111373
Hickling, D. R., Nitti, V. W. (2013). Management of Recurrent Urinary Tract Infections in Healthy Adult Women. Rev. Urol. 15 (2), 41–48.
Hill, J. E., Albert, A. Y. K., V.R. Group (2019). Resolution and Cooccurrence Patterns of Gardnerella Leopoldii, G. Swidsinskii, G. Piotii and G. Vaginalis Within the Vaginal Microbiome. Infect. Immun. 87 (12), e00532–19. doi: 10.1128/IAI.00532-19
Hillebrand, L., Harmanli, O. H., Whiteman, V., Khandelwal, M. (2002). Urinary Tract Infections in Pregnant Women With Bacterial Vaginosis. Am. J. Obstet. Gynecol. 186 (5), 916–917. doi: 10.1067/mob.2002.123987
Hilt, E. E., McKinley, K., Pearce, M. M., Rosenfeld, A. B., Zilliox, M. J., Mueller, E. R., et al. (2014). Urine Is Not Sterile: Use of Enhanced Urine Culture Techniques to Detect Resident Bacterial Flora in the Adult Female Bladder. J. Clin. Microbiol. 52 (3), 871–876. doi: 10.1128/JCM.02876-13
Jolley, J. A., Kim, S., Wing, D. A. (2012). Acute Pyelonephritis and Associated Complications During Pregnancy in 2006 in US Hospitals. J. Matern. Fetal. Neonatal. Med. 25 (12), 2494–2498. doi: 10.3109/14767058.2012.704441
Josephson, S., Thomason, J., Sturino, K., Zabransky, R., Williams, J. (1988). Gardnerella Vaginalis in the Urinary Tract: Incidence and Significance in a Hospital Population. Obstet. Gynecol. 71 (2), 245–250.
Karstens, L., Asquith, M., Davin, S., Stauffer, P., Fair, D., Gregory, W. T., et al. (2016). Does the Urinary Microbiome Play a Role in Urgency Urinary Incontinence and Its Severity? Front. Cell Infect. Microbiol. 6, 78. doi: 10.3389/fcimb.2016.00078
Ki, M., Park, T., Choi, B., Foxman, B. (2004). The Epidemiology of Acute Pyelonephritis in South Korea, 1997-1999. Am. J. Epidemiol. 160 (10), 985–993. doi: 10.1093/aje/kwh308
Koljalg, S., Truusalu, K., Stsepetova, J., Pai, K., Vainumae, I., Sepp, E., et al. (2014). The Escherichia Coli Phylogenetic Group B2 With Integrons Prevails in Childhood Recurrent Urinary Tract Infections. APMIS 122 (5), 452–458. doi: 10.1111/apm.12167
Ksiezarek, M., Ugarcina-Perovic, S., Rocha, J., Grosso, F., Peixe, L. (2021). Long-Term Stability of the Urogenital Microbiota of Asymptomatic European Women. BMC Microbiol. 21 (1), 64. doi: 10.1186/s12866-021-02123-3
Lee, S. L., Wesselschmidt, R. L., Linette, G. P., Kanagawa, O., Russell, J. H., Milbrandt, J., et al. (1995). Unimpaired Thymic and Peripheral T Cell Death in Mice Lacking the Nuclear Receptor NGFI-B (Nur77). Science 269 (5223), 532–535. doi: 10.1126/science.7624775
Lewis, W. G., Robinson, L. S., Gilbert, N. M., Perry, J. C., Lewis, A. L. (2013). Degradation, Foraging, and Depletion of Mucus Sialoglycans by the Vagina-Adapted Actinobacterium Gardnerella Vaginalis. J. Biol. Chem. 288 (17), 12067–12079. doi: 10.1074/jbc.M113.453654
Liebmann, M., Hucke, S., Koch, K., Eschborn, M., Ghelman, J., Chasan, A. I., et al. (2018). Nur77 Serves as a Molecular Brake of the Metabolic Switch During T Cell Activation to Restrict Autoimmunity. Proc. Natl. Acad. Sci. U. S. A. 115 (34), E8017–E8026. doi: 10.1073/pnas.1721049115
Liu, S. C., Han, X. M., Shi, M., Pang, Z. L. (2016). Persistence of Uropathogenic Escherichia Coli in the Bladders of Female Patients With Sterile Urine After Antibiotic Therapies. J. Huazhong. Univ. Sci. Technolog. Med. Sci. 36 (5), 710–715. doi: 10.1007/s11596-016-1649-9
Luo, Y., Ma, Y., Zhao, Q., Wang, L., Guo, L., Ye, L., et al. (2012). Similarity and Divergence of Phylogenies, Antimicrobial Susceptibilities, and Virulence Factor Profiles of Escherichia Coli Isolates Causing Recurrent Urinary Tract Infections That Persist or Result From Reinfection. J. Clin. Microbiol. 50 (12), 4002–4007. doi: 10.1128/JCM.02086-12
Mohankumar, K., Li, X., Sung, N., Cho, Y. J., Han, S. J., Safe, S. (2020). Bis-Indole-Derived Nuclear Receptor 4a1 (NR4A1, Nur77) Ligands as Inhibitors of Endometriosis. Endocrinology 161 (4), bqaa027. doi: 10.1210/endocr/bqaa027
Mora-Bau, G., Platt, A. M., van Rooijen, N., Randolph, G. J., Albert, M. L., Ingersoll, M. A. (2015). Macrophages Subvert Adaptive Immunity to Urinary Tract Infection. PLoS Pathog. 11 (7), e1005044. doi: 10.1371/journal.ppat.1005044
Mulvey, M. A., Schilling, J. D., Hultgren, S. J. (2001). Establishment of a Persistent Escherichia Coli Reservoir During the Acute Phase of a Bladder Infection. Infect. Immun. 69 (7), 4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001
Mysorekar, I. U., Hultgren, S. J. (2006). Mechanisms of Uropathogenic Escherichia Coli Persistence and Eradication From the Urinary Tract. Proc. Natl. Acad. Sci. U. S. A. 103 (38), 14170–14175. doi: 10.1073/pnas.0602136103
O’Brien, V. P., Hannan, T. J., Yu, L., Livny, J., Roberson, E. D., Schwartz, D. J., et al. (2016). A Mucosal Imprint Left by Prior Escherichia Coli Bladder Infection Sensitizes to Recurrent Disease. Nat. Microbiol. 2, 16196. doi: 10.1038/nmicrobiol.2016.196
O’Brien, V. P., Dorsey, D. A., Hannan, T. J., Hultgren, S. J. (2018). Host Restriction of Escherichia Coli Recurrent Urinary Tract Infection Occurs in a Bacterial Strain-Specific Manner. PLoS Pathog. 14 (12), e1007457. doi: 10.1371/journal.ppat.1007457
O’Brien, V. P., Joens, M. S., Lewis, A. L., Gilbert, N. M. (2020). Recurrent Escherichia Coli Urinary Tract Infection Triggered by Gardnerella Vaginalis Bladder Exposure in Mice. J. Vis. Exp. 166, 10.3791/61967. doi: 10.7554/eLife.46677
Pearce, M. M., Hilt, E. E., Rosenfeld, A. B., Zilliox, M. J., Thomas-White, K., Fok, C., et al. (2014). The Female Urinary Microbiome: A Comparison of Women With and Without Urgency Urinary Incontinence. MBio 5 (4), e01283–e01214. doi: 10.1128/mBio.01283-14
Pearce, M. M., Zilliox, M. J., Rosenfeld, A. B., Thomas-White, K. J., Richter, H. E., Nager, C. W., et al. (2015). The Female Urinary Microbiome in Urgency Urinary Incontinence. Am. J. Obstet. Gynecol. 213 (3), 347 e1–347 11. doi: 10.1016/j.ajog.2015.07.009
Pei, L., Castrillo, A., Chen, M., Hoffmann, A., Tontonoz, P. (2005). Induction of NR4A Orphan Nuclear Receptor Expression in Macrophages in Response to Inflammatory Stimuli. J. Biol. Chem. 280 (32), 29256–29262. doi: 10.1074/jbc.M502606200
Rahn, D. D., Carberry, C., Sanses, T. V., Mamik, M. M., Ward, R. M., Meriwether, K. V., et al. (2014). Vaginal Estrogen for Genitourinary Syndrome of Menopause: A Systematic Review. Obstet. Gynecol. 124 (6), 1147–1156. doi: 10.1097/AOG.0000000000000526
Rajpal, A., Cho, Y. A., Yelent, B., Koza-Taylor, P. H., Li, D., Chen, E., et al. (2003). Transcriptional Activation of Known and Novel Apoptotic Pathways by Nur77 Orphan Steroid Receptor. EMBO J. 22 (24), 6526–6536. doi: 10.1093/emboj/cdg620
Robino, L., Scavone, P., Araujo, L., Algorta, G., Zunino, P., Pirez, M. C., et al. (2014). Intracellular Bacteria in the Pathogenesis of Escherichia Coli Urinary Tract Infection in Children. Clin. Infect. Dis. 59 (11), e158–e164. doi: 10.1093/cid/ciu634
Rodriguez-Calvo, R., Tajes, M., Vazquez-Carrera, M. (2017). The NR4A Subfamily of Nuclear Receptors: Potential New Therapeutic Targets for the Treatment of Inflammatory Diseases. Expert Opin. Ther. Targets 21 (3), 291–304. doi: 10.1080/14728222.2017.1279146
Rosen, D. A., Scavone, P., Araujo, L., Algorta, G., Zunino, P., Pirez, M. C., et al. (2007). Detection of Intracellular Bacterial Communities in Human Urinary Tract Infection. PLoS Med. 4 (12), e329. doi: 10.1371/journal.pmed.0040329
Sadahira, T., Wada, K., Araki, M., Mitsuhata, R., Yamamoto, M., Maruyama, Y., et al. (2021). Efficacy of Lactobacillus Vaginal Suppositories for the Prevention of Recurrent Cystitis: A Phase II Clinical Trial. Int. J. Urol. 28 (10), 1026–1031. doi: 10.1111/iju.14636
Schilling, J. D., Lorenz, R. G., Hultgren, S. J. (2002). Effect of Trimethoprim-Sulfamethoxazole on Recurrent Bacteriuria and Bacterial Persistence in Mice Infected With Uropathogenic Escherichia Coli. Infect. Immun. 70 (12), 7042–7049. doi: 10.1128/IAI.70.12.7042-7049.2002
Sharami, S. H., Afrakhteh, M., Shakiba, M. (2007). Urinary Tract Infections in Pregnant Women With Bacterial Vaginosis. J. Obstet. Gynaecol. 27 (3), 252–254. doi: 10.1080/01443610701194846
Skjot-Rasmussen, L., Olsen, S. S., Jakobsen, L., Ejrnaes, K., Scheutz, F., Lundgren, B., et al. (2013). Escherichia Coli Clonal Group A Causing Bacteraemia of Urinary Tract Origin. Clin. Microbiol. Infect. 19 (7), 656–661. doi: 10.1111/j.1469-0691.2012.03961.x
Stapleton, A. E., Au-Yeung, M., Hooton, T. M., Fredricks, D. N., Roberts, P. L., Czaja, C. A., et al. (2011). Randomized, Placebo-Controlled Phase 2 Trial of a Lactobacillus Crispatus Probiotic Given Intravaginally for Prevention of Recurrent Urinary Tract Infection. Clin. Infect. Dis. 52 (10), 1212–1217. doi: 10.1093/cid/cir183
Sumati, A. H., Saritha, N. K. (2009). Association of Urinary Tract Infection in Women With Bacterial Vaginosis. J. Glob. Infect. Dis. 1 (2), 151–152. doi: 10.4103/0974-777X.56254
Tamadonfar, K. O., Omattage, N. S., Spaulding, C. N., Hultgren, S. J. (2019). Reaching the End of the Line: Urinary Tract Infections. Microbiol. Spectr. 7 (3). doi: 10.1128/9781683670261.ch6
Tan, C., Hiwa, R., Mueller, J. L., Vykunta, V., Hibiya, K., Noviski, M., et al. (2020). NR4A Nuclear Receptors Restrain B Cell Responses to Antigen When Second Signals Are Absent or Limiting. Nat. Immunol. 21 (10), 1267–1279. doi: 10.1038/s41590-020-0765-7
Thomas-White, K. J., Hilt, E. E., Fok, C., Pearce, M. M., Mueller, E. R., Kliethermes, S., et al. (2016). Incontinence Medication Response Relates to the Female Urinary Microbiota. Int. Urogynecol. J. 27 (5), 723–733. doi: 10.1007/s00192-015-2847-x
Wilson, A. J., Chueh, A. C., Togel, L., Corner, G. A., Ahmed, N., Goel, S., et al. (2010). Apoptotic Sensitivity of Colon Cancer Cells to Histone Deacetylase Inhibitors Is Mediated by an Sp1/Sp3-Activated Transcriptional Program Involving Immediate-Early Gene Induction. Cancer Res. 70 (2), 609–620. doi: 10.1158/0008-5472.CAN-09-2327
Wolfe, A. J., Toh, E., Shibata, N., Rong, R., Kenton, K., Fitzgerald, M., et al. (2012). Evidence of Uncultivated Bacteria in the Adult Female Bladder. J. Clin. Microbiol. 50 (4), 1376–1383. doi: 10.1128/JCM.05852-11
Wright, K. J., Seed, P. C., Hultgren, S. J. (2005). Uropathogenic Escherichia Coli Flagella Aid in Efficient Urinary Tract Colonization. Infect. Immun. 73 (11), 7657–7668. doi: 10.1128/IAI.73.11.7657-7668.2005
Wu, L., Chen, L. (2018). Characteristics of Nur77 and Its Ligands as Potential Anticancer Compounds (Review). Mol. Med. Rep. 18 (6), 4793–4801. doi: 10.3892/mmr.2018.9515
Wu, P., Chen, Y., Zhao, J., Zhang, G., Chen, J., Wang, J., et al. (2017). Urinary Microbiome and Psychological Factors in Women With Overactive Bladder. Front. Cell Infect. Microbiol. 7, 488. doi: 10.3389/fcimb.2017.00488
Yoon, K., Lee, S. O., Cho, S. D., Kim, K., Khan, S., Safe, S., et al. (2011). Activation of Nuclear TR3 (NR4A1) by a Diindolylmethane Analog Induces Apoptosis and Proapoptotic Genes in Pancreatic Cancer Cells and Tumors. Carcinogenesis 32 (6), 836–842. doi: 10.1093/carcin/bgr040
Keywords: urinary tract infection (UTI), bacterial vaginosis (BV), Nur77, immediate early gene expression, bladder, urinary microbiome, orphan nuclear receptor 4A1 (NR4A1), RNA-seq - RNA sequencing
Citation: O’Brien VP, Lewis AL and Gilbert NM (2021) Bladder Exposure to Gardnerella Activates Host Pathways Necessary for Escherichia coli Recurrent UTI. Front. Cell. Infect. Microbiol. 11:788229. doi: 10.3389/fcimb.2021.788229
Received: 01 October 2021; Accepted: 12 November 2021;
Published: 06 December 2021.
Edited by:
Justin Merritt, Oregon Health and Science University, United StatesReviewed by:
Nuno Cerca, University of Minho, PortugalSheryl S. Justice, Nationwide Children’s Hospital, United States
Copyright © 2021 O’Brien, Lewis and Gilbert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicole M. Gilbert, Z2lsYmVydEB3dXN0bC5lZHU=
 Valerie P. O’Brien1
Valerie P. O’Brien1 Amanda L. Lewis
Amanda L. Lewis Nicole M. Gilbert
Nicole M. Gilbert