- 1College of Life Science, Changchun Sci-Tech University, Shuangyang, China
- 2College of Veterinary Medicine, Qingdao Agricultural University, Qingdao, China
- 3State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China
- 4Center of Prevention and Control Biological Disaster, State Forestry and Grassland Administration, Shenyang, China
- 5Veterinary Department, Muyuan Foods Co., Ltd., Nanyang, China
- 6College of Animal Science and Veterinary Medicine, Heilongjiang Bayi Agricultural University, Daqing, China
Enterocytozoon (E.) bieneusi and Cryptosporidium spp. are the most important zoonotic enteric pathogens associated with diarrheal diseases in animals and humans. However, it is still not known whether E. bieneusi and Cryptosporidium spp. are carried by wild rodents in Shanxi, Guangxi, Zhejiang, Shandong, and Inner Mongolia, China. In the present study, a total of 536 feces samples were collected from Rattus (R.) norvegicus, Mus musculus, Spermophilus (S.) dauricus, and Lasiopodomys brandti in six provinces of China, and were detected by PCR amplification of the SSU rRNA gene of Cryptosporidium spp. and ITS gene of E. bieneusi from June 2017 to November 2020. Among 536 wild rodents, 62 (11.6%) and 18 (3.4%) samples were detected as E. bieneusi- and Cryptosporidium spp.-positive, respectively. Differential prevalence rates of E. bieneusi and Cryptosporidium spp. were found in different regions. E. bieneusi was more prevalent in R. norvegicus, whereas Cryptosporidium spp. was more frequently identified in S. dauricus. Sequence analysis indicated that three known Cryptosporidium species/genotypes (Cryptosporidium viatorum, Cryptosporidium felis, and Cryptosporidium sp. rat genotype II/III) and two uncertain Cryptosporidium species (Cryptosporidium sp. novel1 and Cryptosporidium sp. novel2) were present in the investigated wild rodents. Meanwhile, 5 known E. bieneusi genotypes (XJP-II, EbpC, EbpA, D, and NCF7) and 11 novel E. bieneusi genotypes (ZJR1 to ZJR7, GXM1, HLJC1, HLJC2, and SDR1) were also observed. This is the first report for existence of E. bieneusi and Cryptosporidium spp. in wild rodents in Shanxi, Guangxi, Zhejiang, and Shandong, China. The present study also demonstrated the existence of E. bieneusi and Cryptosporidium spp. in S. dauricus worldwide for the first time. This study not only provided the basic data for the distribution of E. bieneusi and Cryptosporidium genotypes/species, but also expanded the host range of the two parasites. Moreover, the zoonotic E. bieneusi and Cryptosporidium species/genotypes were identified in the present study, suggesting wild rodents are a potential source of human infections.
Introduction
The rodents are one of the largest families of mammals. Wild rodents (e.g., wild rats) are the most widely distributed worldwide. They can shed many pathogens (e.g., Enterocytozoon (E.) bieneusi and Cryptosporidium spp.) into the environment due to living in an open environment, thus becoming potential sources for transmission of pathogens to other animals (Deng et al., 2016; García-Livia et al., 2020; Gui et al., 2020). In addition, the rodents have a closed relationship with humans. Thus, many pathogens, including E. bieneusi and Cryptosporidium spp., might be transmitted from rodents to humans. (García-Livia et al., 2020; Gui et al., 2020; Zhao et al., 2020).
E. bieneusi and Cryptosporidium spp. are the common zoonotic enteric pathogens responsible for a majority of parasitic diarrhea diseases worldwide (Qi et al., 2015; Zhang X. et al., 2018; Zhao et al., 2018; Wang S. N. et al., 2020). Both of them can infect humans and a wide variety of animals (e.g., rodents) (Wang et al., 2013; Zhao et al., 2018; Li and Xiao, 2020; Wang S. N. et al., 2020) mainly through water-borne and food-borne routes (Wang et al., 2013; Zhao et al., 2018). In general, healthy people infected with both pathogens are asymptomatic or manifest symptoms of self-limiting diarrhea. However, the infection of E. bieneusi and Cryptosporidium spp. in immunocompromised individuals may cause chronic or life-threatening diarrheas (Wang et al., 2013; Sutthikornchai et al., 2021). Owing to their significance in public health, Cryptosporidium spp. and E. bieneusi have been put into Category B Priority Pathogen list by the National Institute of Allergy and Infectious Diseases (NIAID) (NIAID, 2018). Moreover, E. bieneusi is also listed on the Environmental Protection Agency (EPA) microbial contaminant candidate list of concern for waterborne transmission (Didier et al., 2009).
E. bieneusi is consist of more than 500 genotypes, which are classified into 11 groups based on the sequences of the internal transcribed spacer (ITS) region of the rRNA gene (Santin, 2015; Zhang Y. et al., 2018; Zhao et al., 2018; Li W. et al., 2019; Wang S. N. et al., 2020; Abarca et al., 2021). Group 1, identified as zoonotic, is responsible for a vast majority of human infections (Wang S. N. et al., 2020). Groups 2-11 are mainly composed of host-specific or host-adapted genotypes (Guo et al., 2014; Wang S. N. et al., 2020). To date, a total of 36 ITS genotypes of E. bieneusi have been found in rodent species and 15 (Type IV, BEB6, EbpA, EbpC, C, D, H, CZ3, S6, Peru6, Nig7, Peru8, Peru11, Peru16, and PigITS5) were considered as zoonotic genotypes (Danišová et al., 2015; Cama et al., 2007; Sak et al., 2011; Guo et al., 2014; Perec-Matysiak et al., 2015; Qi et al., 2015; Roellig et al., 2015; Deng et al., 2016).
Cryptosporidium spp. contains more than 100 species/genotypes based on the sequence of the small subunit (SSU) rRNA gene (Feng et al., 2018; Holubová et al., 2019). To date, 38 of them have been identified in humans, whereas only C. hominis and C. parvum were frequently found in humans (Essid et al., 2018; Krumkamp et al., 2021), and the remaining genotypes/species were occasionally observed in humans. Rodents are one of the most important reservoirs of Cryptosporidium spp. More than 30 Cryptosporidium species/genotypes have been identified in rodent species (Zhang X. et al., 2018). Among them, at least ten Cryptosporidium species (including C. parvum, C. andersoni, C. muris, C. wrairi, C. tyzzeri, C. scrofarum, C. ubiquitum, C. hominis, C. suis, and C. meleagridis) and more than 20 Cryptosporidium genotypes, such as ground squirrel genotypes (I-III), rat genotypes (I-IV), deer mouse genotypes (I-IV), chipmunk genotypes II, vole genotype, and mouse genotypes (II, III), have been identified in humans (Bajer et al., 2002; Nakai et al., 2004; Feng et al., 2007; Foo et al., 2007; Kimura et al., 2007; Kvác et al., 2008; Lv et al., 2009; Paparini et al., 2012; Backhans et al., 2013; Murakoshi et al., 2013; Ng-Hublin et al., 2013; Song et al., 2015; Stenger B. et al., 2015; Stenger B. L. et al., 2015; Zhao et al., 2015; Gholipoury et al., 2016; Saki et al., 2016; Danišová et al., 2017; Wang S. N. et al., 2020).
In view of such severe situations, it is essential to investigate the prevalence of E. bieneusi and Cryptosporidium spp. in different rodent species and identify their species/genotypes. However, information regarding Cryptosporidium spp. infection in rodents was limited in China, which was only reported in Microtus fuscus (Qinghai vole) and Ochotona curzoniae (wild plateau pika) in Qinghai (Zhang X. et al., 2018), brown rats (Rattus norvegicus) in Heilongjiang (Li et al., 2016), bamboo rats in Sichuan (Liu et al., 2015), pet chinchillas in Beijing, Henan and Guizhou (Qi et al., 2015), commensal rodents in Henan and Fujian (Zhao et al., 2015), brown rats in Heilongjiang (Zhao et al., 2018), wild, laboratory, and pet rodents in Beijing, Henan, Fujian and Sichuan (Lv et al., 2009), bamboo rats in Guangdong, Hunan, Guangxi, Jiangxi and Hainan (Wei et al., 2019; Li et al., 2020a; Li et al., 2020b), Asian house rats, brown rats, Edward’s long-tailed rats and muridae in Hainan (Zhao et al., 2019). In China, E. bieneusi in rodents has been only reported in Heilongjiang (Zhao et al., 2018), Beijing (Qi et al., 2015), Henan (Qi et al., 2015; Wang J. et al., 2020), Guizhou (Qi et al., 2015), Sichuan (Deng et al., 2016), Shandong (Wang J. et al., 2020), Guangdong (Wang et al., 2019), Hunan (Wang et al., 2019; Gui et al., 2020), Jiangxi (Wang et al., 2019), Chongqing (Wang et al., 2019), Guangxi (Wang et al., 2019), and Hainan (Zhao et al., 2020).
However, it is still not known whether E. bieneusi and Cryptosporidium spp. are carried by wild rodents in Shanxi, Guangxi, Zhejiang, Shandong, and Inner Mongolia, China. Thus, the present study was performed to estimate the prevalence and genotypes of E. bieneusi and Cryptosporidium spp. in wild rodents by the molecular biological method.
Materials and Methods
Specimen Collection
A total of 536 feces samples were collected from four rodent species from Daqing City in Heilongjiang (n = 41; 39 S. dauricus, 2 R. norvegicus), Taigu County in Shanxi (n = 53, R. norvegicus), Nanning City in Guangxi (n = 74, M. musculus), Weihai City in Shandong (n = 227, R. norvegicus), Jiaxing City in Zhejiang (n = 119, R. norvegicus) and Xilingol League in Inner Mongolia (n = 22, L. brandti), China from June 2017 to November 2020. These rodents were captured by trapping method. The rodents had been euthanized by CO2 inhalation, and then the fresh feces sample (approximately 500 mg) was collected directly from the intestinal and rectal content of each rodent, and then was placed into ice boxes and sent to the laboratory. Information regarding sampling time, region, and species was recorded. This study was approved by the Ethics Committee of Qingdao Agricultural University.
DNA Extraction and PCR Amplification
Genomic DNA was extracted from fecal sample of approximately 200 mg using the E.Z.N.A.® Stool DNA Kit (Omega Biotek Inc., Norcross, GA, USA) according to the manufacturer’s instructions, and then was stored at -20°C prior to PCR. The prevalence and genotypes of E. bieneusi were identified by PCR amplification of the ITS gene according to the previous description (Zhao et al., 2018). Cryptosporidium spp. in the fecal samples was confirmed by PCR amplification of the SSU rRNA gene according to the previous report (Zhao et al., 2018). The positive and negative controls were included in each test. The secondary PCR products were observed using UV light after an electrophoretic analysis at a 1.5% agarose gel containing ethidium bromide.
Sequence and Phylogenetic Analyses
The positive PCR specimens were sent to Sangon Biotech Company (Shanghai, China) for sequencing. A new PCR product should be sequenced if previously produced sequences had single nucleotide substitutions, insertions or deletions. The nucleotide sequences were aligned and analyzed with reference sequences by using the Clustal X 1.83 program and Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/), in order to determine the species/genotypes of Cryptosporidium spp. and E. bieneusi. The phylogenetic trees were reconstructed with Mega 5.0 using neighbor-joining (NJ) method under Kimura 2-parameter model (1,000 replicates). All nucleotide sequences were deposited in GenBank with accession numbers MT647749 – MT647806 and OK117929 – OK117932 for E. bieneusi, and MT561508 – MT561533 for Cryptosporidium spp.
Statistical Analysis
The statistical analysis for the prevalence of E. bieneusi and Cryptosporidium in wild rodents from different region, season, sampling year, and species were performed by using χ2 test in SAS version 9.1 (SAS Institute, Cary, NC, USA). The results were considered to be statistically significant when P < 0.05. Odds ratios (ORs) and their 95% confidence intervals (95% CIs) were also calculated to compare the magnitude of various risk factors for E. bieneusi and Cryptosporidium prevalence.
Results
Prevalence of Cryptosporidium spp. and E. bieneusi
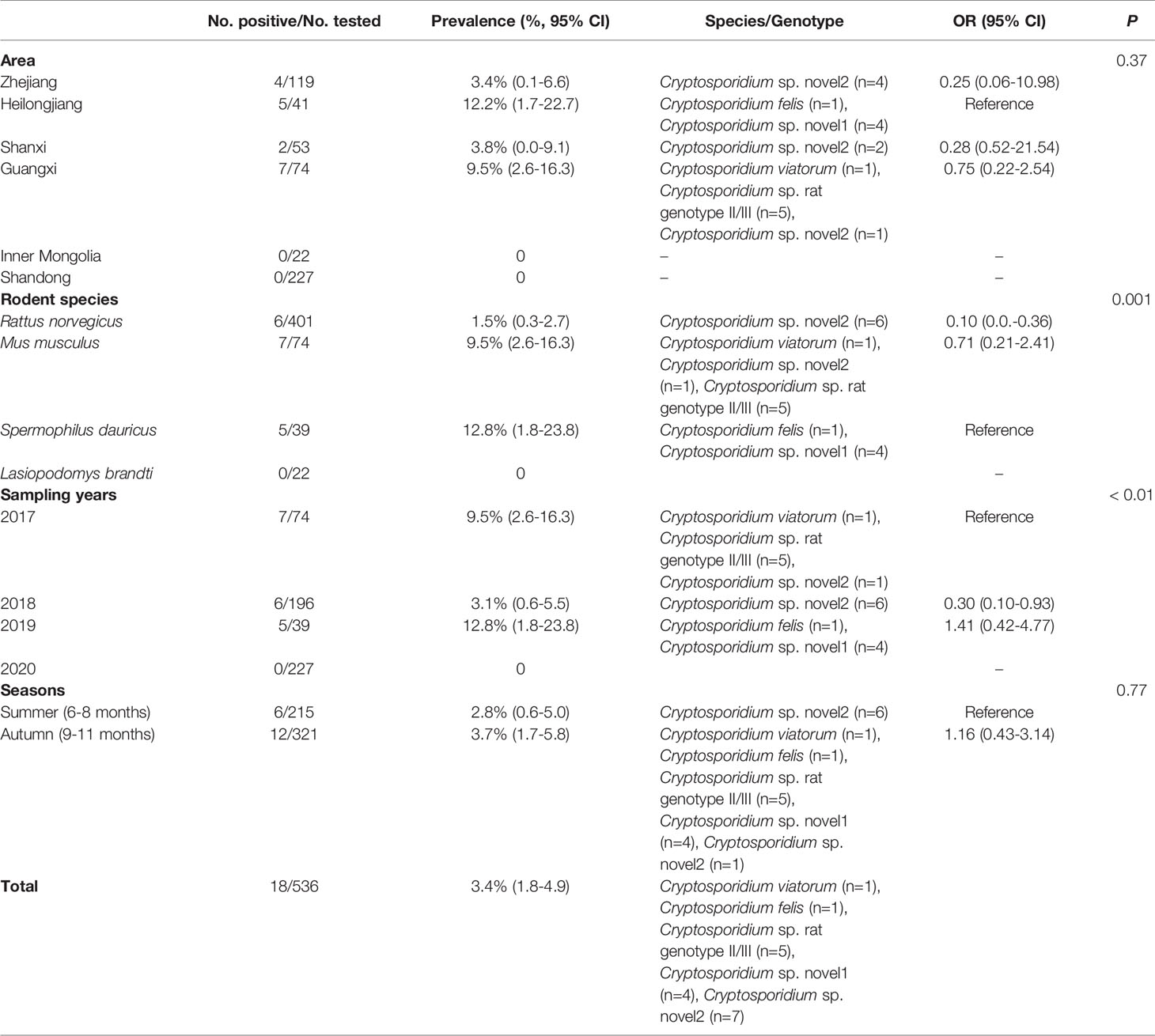
In the present study, 18 out of 536 (3.4%) fecal samples were identified as Cryptosporidium-positive (Table 1). The prevalence rates of Cryptosporidium in different species of rodents were 15% (6/401) in R. norvegicus, 9.5% (7/74) in M. musculus, 12.8% (5/39) in S. dauricus, and 0% (0/22) in L. brandti (Table 1). Moreover, the prevalence of Cryptosporidium in different regions ranged from 0% in Inner Mongolia (0/22) and Shandong (0/227) to 12.2% in Heilongjiang (5/41) (Table 1). Furthermore, the prevalence in different collection years ranged from 0% to 12.8% (Table 1). The prevalence of Cryptosporidium in rodent feces collected in autumn (3.7%, 12/321) was slightly higher than that in summer (2.8%, 6/215) (Table 1).
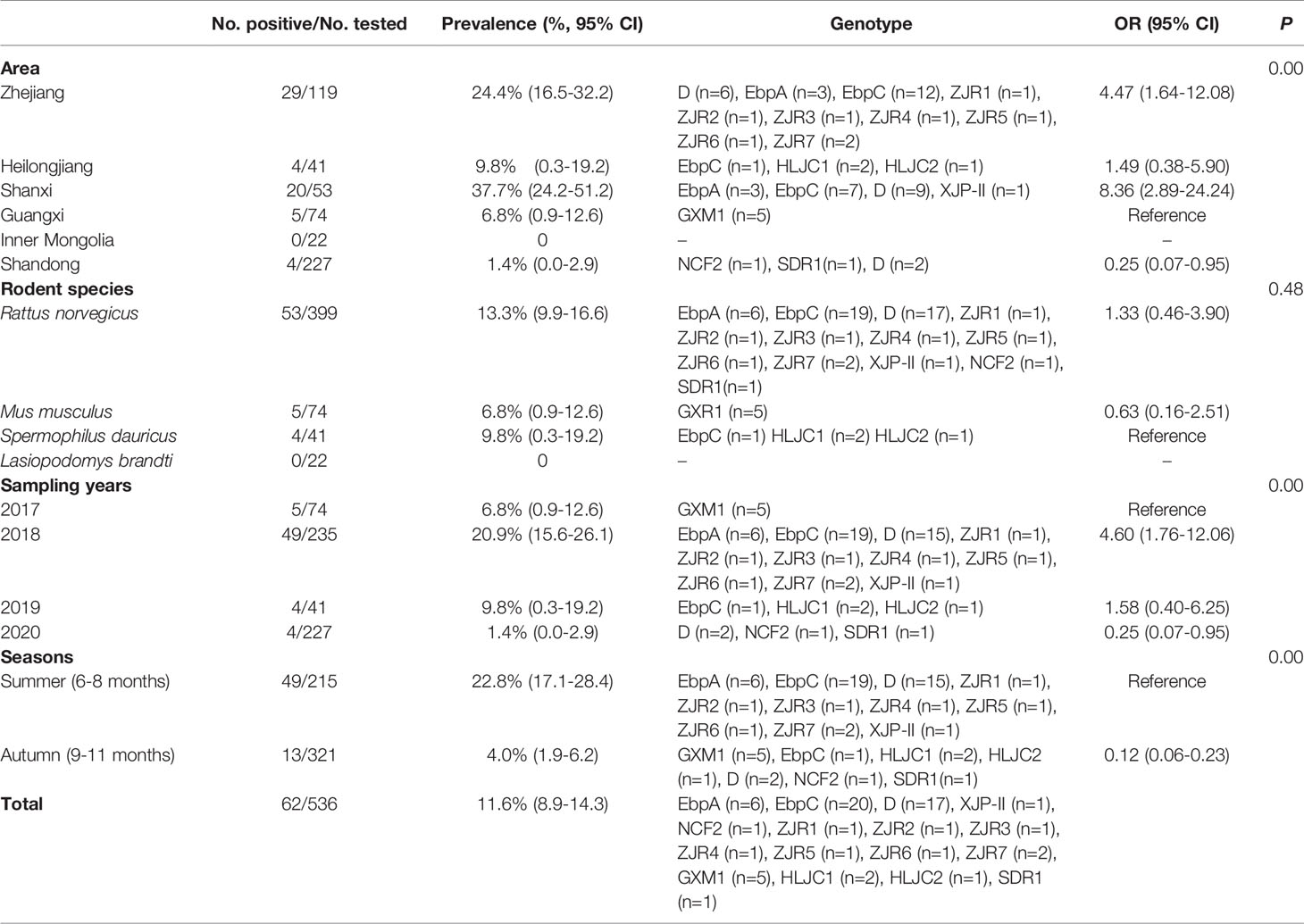
Among 536 rodents, 62 samples (11.6%) were detected to be E. bieneusi-positive in three rodent species, with 13.3% (53/399) in R. norvegicus, 6.8% (5/74) in M. musculus, and 9.8% (4/41) in S. dauricus (Table 2). The highest prevalence of E. bieneusi was found in Shanxi (37.7%, 20/53), and followed by Zhejiang (24.4%, 29/119), Heilongjiang (9.8%, 4/41), Guangxi (6.8%, 5/74), and Shandong (1.4%, 4/227) (Table 2). The prevalence of E. bieneusi was 6.8% (5/74), 20.9%, (49/235) 9.8% (4/41), and 1.4% (4/227) in rodents collected in 2017, 2018, 2019, and 2020, respectively (Table 2). The prevalence of E. bieneusi in rodents was 22.8% in summer (49/215) and 4.0% in autumn (13/321), respectively (Table 2).
E. bieneusi and Cryptosporidium spp. coinfection was found in three wild rodents in this study. All of them were R. norvegicus collected in 2018. Two were collected from Zhejiang Province, and the remaining one was collected from Shanxi Province.
Distribution of Cryptosporidium spp. and E. bieneusi
Cryptosporidium sp. rat genotype II/III, Cryptosporidium felis, and Cryptosporidium viatorum were identified in the investigated rodents through the analysis of SSU rRNA gene of Cryptosporidium. Furthermore, two Cryptosporidium genotypes with uncertain species status were observed (Figure 1 and Table 1). Cryptosporidium sp. novel1 and C. felis were found in S. dauricus in Heilongjiang. C. viatorum and Cryptosporidium sp. rat genotype II/III were only identified in M. musculus in Guangxi. Cryptosporidium sp. novel2 was found in three provinces Zhejiang (R. norvegicus), Shanxi (R. norvegicus), and Guangxi (M. musculus) (Table 1).
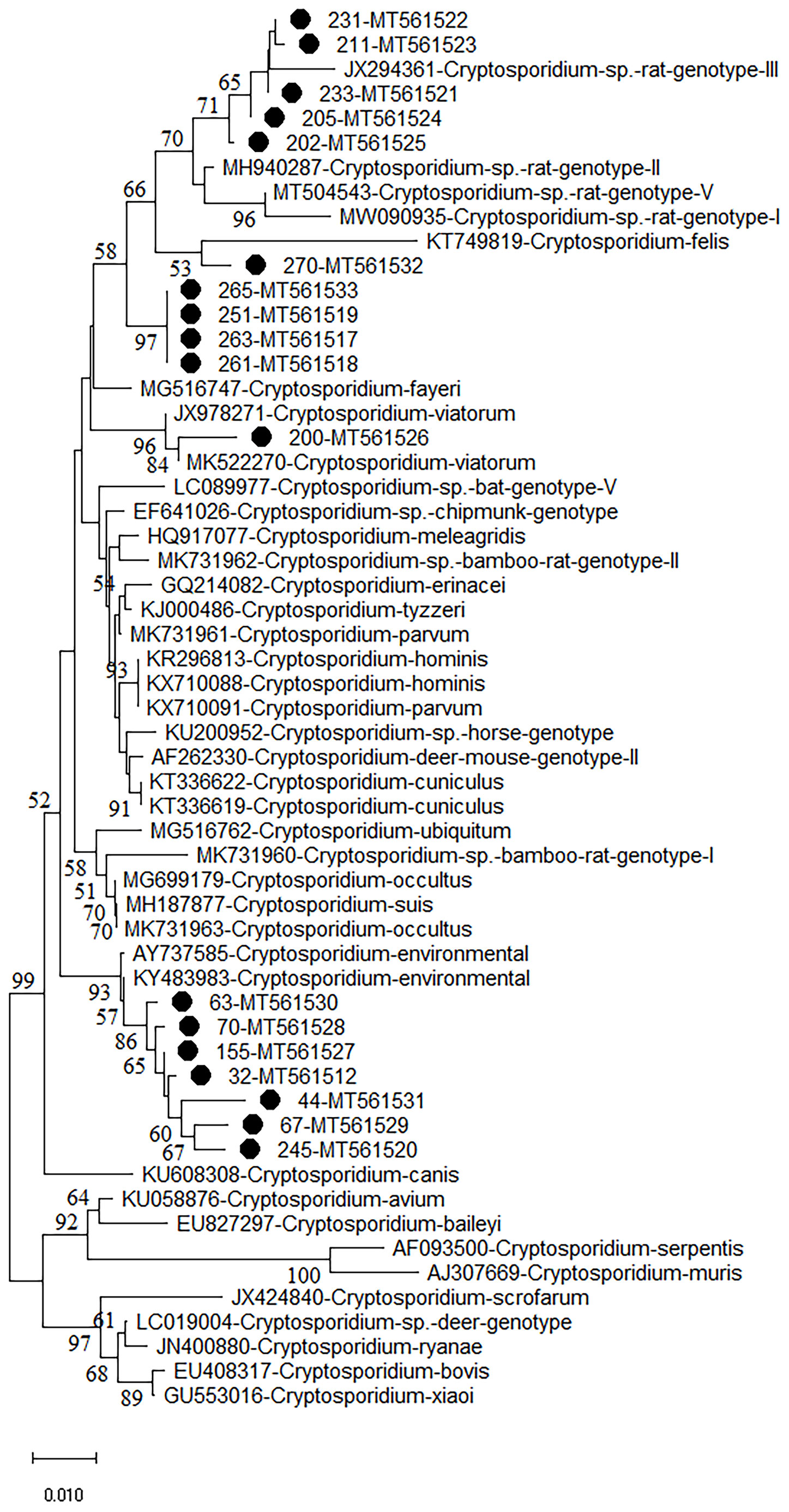
Figure 1 Phylogenetic analyses of SSU rRNA gene of Cryptosporidium spp. using neighbor-joining (NJ) method (Kimura 2-parameter model, 1,000 replicates). Bootstrap values below 50% are not shown. Cryptosporidium isolates identified in the present study are pointed out by solid circles.
A total of 16 E. bieneusi genotypes were identified in this study, including 5 known genotypes (XJP-II, EbpC, EbpA, D, and NCF7) and 11 novel genotypes (ZIR1 to ZJR7, GXM1, HLJC1, HLJC2, and SDR1) (Figure 2 and Table 2). Among them, genotype D was found in R. norvegicus in Zhejiang, Shanxi, and Shandong. EbpA was only found in R. norvegicus in Zhejiang and Shanxi, whereas EbpC was identified in Zhejiang (R. norvegicus), Shanxi (R. norvegicus), and Heilongjiang (S. dauricus). Moreover, NCF2 (R. norvegicus in Shandong), XJP-II (R. norvegicus in Shanxi), ZIR1 to ZJR7 (R. norvegicus in Zhejiang), GXM1 (M. musculus in Guangxi), HLJC1 (S. dauricus in Heilongjiang), HLJC2 (S. dauricus in Heilongjiang), and SDR1 (R. norvegicus in Shandong) were only found in one province (Table 2).

Figure 2 Phylogenetic analyses of ITS gene of Enterocytozoon bieneusi using neighbor-joining (NJ) method (Kimura 2-parameter model, 1,000 replicates). Bootstrap values below 50% are not shown. E. bieneusi isolates identified in the present study are pointed out by solid circles.
Phylogenetic Relationships of Cryptosporidium spp. and E. bieneusi
The phylogenetic analysis of various Cryptosporidium species/genotypes showed two uncertain species status and three known species/genotypes (Figure 1). The sequences of Cryptosporidium sp. novel2, including seven Cryptosporidium spp. sequences (isolates 32, 44, 63, 67, 70, 155, and 245), were clustered with Cryptosporidium spp. sequences identified from environmental samples (Figure 1). Five sequences (isolates 202, 205, 211, 231, and 233) were clustered with Cryptosporidium sp. rat genotype II/III in a same clade (Figure 1). Sequences of isolates 251, 261, 263, and 265 (Cryptosporidium sp. novel1) were grouped into a novel separate clade (Figure 1). Sequences of isolates 270 and 200 were clustered with that of C. felis and C. viatorum in a same clade, respectively (Figure 1).
The Neighbor-Joining analysis for sequences of E. bieneusi species/genotypes obtained in this study revealed that 5 known genotypes and 11 novel genotypes (Figure 2). Fourteen genotypes (5 known genotypes and 9 novel genotypes) were divided into Group 1, with ZJR7, SDR1, and D in 1a, EbpC, ZJR5, and ZJR1 in 1d, HLJC1, ZJR4, EbpA, XJP-II, and ZJR3 in 1e, NCF2 in 1b, GXM1 in 1i, and ZJR6 in 1j (Figure 2). Furthermore, HLJC2 was grouped in Group 2, and ZJR2 was classified into Group 10 (Figure 2).
Discussion
In this study, the total prevalence of Cryptosporidium spp. was 3.4% (18/536) in four rodent species (R. norvegicus, M. musculus, L. brandti, and S. dauricus), which was consistent with previous reports showing the prevalence rates ranged from 0.8% to 80.0% in a variety of rat species (Feng, 2010; Mirzaghavami et al., 2016; Wei et al., 2019), e.g., 1.5-38.0% in brown rats, 8.0-31.4% in mice, and 0.8-73.0% in voles (Feng, 2010; Wei et al., 2019; Ježková et al., 2021). The present study found that the prevalence rates of Cryptosporidium spp. in R. norvegicus, M. musculus, L. brandti, and S. dauricus were 1.5% (6/401), 9.5% (7/74), 0% (0/22), and 12.8% (5/39), respectively with statistical significance (P < 0.05). There was a 0.10- (OR = 0.10, 95% CI 0.0-0.36) and 0.71- (OR = 0.71, 95% CI 0.21-2.41) fold increase of Cryptosporidium spp. infection risk in R. norvegicus (1.5%, 95% CI 0.3-2.7), M. musculus (9.5%, 95% CI 2.6-16.3) compared with that in S. dauricus (12.8%, 95% CI 1.8-23.8). Furthermore, the prevalence of E. bieneusi in rodents varied in different countries, e.g., 87.5% in Peru (Cama et al., 2007), 28.6-42.9% in Poland (Perec-Matysiak et al., 2015), 1.1% in Slovakia (Danišová et al., 2015), 20.0-100% in USA (Roellig et al., 2015). In the present study, the overall E. bieneusi prevalence was 11.6% (62/536), with 13.3% (53/399) in R. norvegicus, 6.8% (5/74) in M. musculus, 9.8% (4/41) in S. dauricus, and 0% (0/22) in L. brandti. In China, E. bieneusi infection has also been reported in many rodent species, such as Bamboo rat (5.1%, 22/435; 15.4%, 18/117) (Wang et al., 2019; Zhao et al., 2020), Brown rat (7.9%, 19/242; 14.3%, 8/58) (Zhao et al., 2018; Zhao et al., 2020), Chinchilla (3.6%, 5/140) (Qi et al., 2015), Indo-Chinese forest rat (9.3%, 5/54) (Zhao et al., 2020), Asiatic brush-tailed porcupine (7.5%, 7/93) (Zhao et al., 2020), Bower’s white-toothed rat (31.6%, 37/117) (Zhao et al., 2020), Edward's long-tailed rat (7.9%, 3/38) (Zhao et al., 2020), Chipmunk (17.6%, 49/279 (Deng et al., 2018), Asian house rat (23.1%, 31/134) (Zhao et al., 2020), Chinese white-bellied rat (18.2% 6/33) (Zhao et al., 2020), Lesser rice-feld rat (36.4%, 16/44) (Zhao et al., 2020). Coinfection (n = 3) of E. bieneusi and Cryptosporidium spp. was also found in the present study. Different susceptibility of different rodent species, different sampling time and sample size, animal age, and animal welfare could affect the prevalence of Cryptosporidium spp. and E. bieneusi in different rodent species in different regions.
Although Cryptosporidium spp. in rodent feces collected in summer (6/215, 2.8%, 95% CI 0.6-5.0) has a slightly lower prevalence than those collected in autumn (12/321, 3.7%, 95% CI 1.7-5.8), the difference was not significant statistically (P = 0.77) (Table 1). Moreover, the temperature and humidity in summer (49/215, 22.8%, 95% CI 17.1-28.4) may be more suitable for the survival of E. bieneusi oocysts than in autumn (13/321, 4.0%, 95% CI 1.9-6.2), the infection risk of E. bieneusi had 0.12-fold increase (OR = 0.12, 95% CI 0.06-0.23) in rodent feces collected in autumn (4.0%, 95% CI 1.9-6.2) than that in summer (22.8%, 95% CI 17.1-28.4) in the investigated rodents (Table 2). The investigated rodents were more active in the summer temperature, which might be the other reason for these rodents to be infection and transmission increase. Other ecological factors such as climate, food resources, breeding, physical activity, etc, which might affect the accuracy of prevalence of the two pathogens, should also be investigated in the further study.
More than 30 Cryptosporidium species/genotypes have been identified in rodents. However, only five species/genotypes were identified in this study, including C. viatorum, C. felis, Cryptosporidium sp. rat genotype II/III, Cryptosporidium sp. novel1, and Cryptosporidium sp. novel2. Among them, Cryptosporidium sp. rat genotype II/III, previously reported in rodents (García-Livia et al., 2020; Ježková et al., 2021), was also identified in this study, which was further confirmed that Cryptosporidium sp. rat genotype II/III was one of the prevalent Cryptosporidium genotypes in rodents. Moreover, two uncertain species of Cryptosporidium (Cryptosporidium sp. novel1 and novel2) were also identified in this study. Cryptosporidium sp. novel1 (isolates 251, 261, 263, and 265) was grouped into a new separate clade. Cryptosporidium sp. novel2 (isolates 32, 44, 63, 67, 70, 155, and 245), grouped with Cryptosporidium environmental. The results indicate two new genotypes/species that have clustered a branch in phylogenetic analysis with environmental isolates of Cryptosporidium spp. One of the reasons that in environmental samples, it is difficult to determine the species and genotype is the simultaneous contamination of several species and genotypes in samples that after sequencing cannot detect a known species or genotype. Unfortunately, other genes such as COWP and HSP70 of the uncertain species have also not been successfully amplified. Thus, the investigation should be continue performed to further confirm whether presence of the two uncertain species of Cryptosporidium in wild rodents. C. viatorum, has been identified in humans (Insulander et al., 2013; Lebbad et al., 2013; Adamu et al., 2014; Ayinmode et al., 2014; De Lucio et al., 2016; Sanchez et al., 2017; Ukwah et al., 2017; Sannella et al., 2019). C. viatorum was first found in travellers who returned to the United Kingdom from the Indian subcontinent, with clinical signs of diarrhea, fever, headache, abdominal pain, nausea, vomiting, and marked weight loss (Elwin et al., 2012). So far, C. viatorum has been documented in the following countries: Bangladesh, Ethiopia, Barbados, Kenya, Colombia, Nigeria, Pakistan, Guatemala, India, and Nepal (Insulander et al., 2013; Lebbad et al., 2013; Adamu et al., 2014; Ayinmode et al., 2014; De Lucio et al., 2016; Sanchez et al., 2017; Ukwah et al., 2017; Sannella et al., 2019). Besides, C. viatorum was also found in China, such as Hainan Province (Leopoldamys edwardsi), Guangdong Province (Berylms bowersi), and Chongqing City (Leopoldamys edwardsi) in China, and in Australia (Rattus lutreolus) (Koehler et al., 2018; Chen et al., 2019; Zhao et al., 2019). C. felis has been widely reported in cats (Jiang et al., 2020), in addition to patients with HIV/AIDS in Peru, Ethiopia, Nigeria, Jamaica, and Portugal (Cama et al., 2003; Jiang et al., 2020). In this study, C. viatorum and C. felis were found in M. musculus and S. dauricus, which was worth for further research, e.g., whether wild rodents are potentially important reservoirs for C. viatorum and C. felis transmission to humans. More importantly, this is the first study showing existence of Cryptosporidium spp. in S. dauricus, which has expanded the host ranges of Cryptosporidium.
At present, more than 400 genotypes of E. bieneusi have been identified, most of which exhibit host specificity (Santín and Fayer, 2011; Wang S. N. et al., 2020). At least 48 genotypes of E. bieneusi infect both human and animals, bringing zoonoses risks (Li and Xiao, 2019). Through phylogenetic analysis, these genotypes were divided into at least 11 groups, e.g., Group 1 to Group 11 (Zhao et al., 2018; Wang S. N. et al., 2020). To date, some genotypes were found in rodents, of which 15 genotypes (CZ3, Peru6, BEB6, C, D, EbpA, EbpC, H, Peru8, Peru11, Peru16, PigITS5, S6, IV, and Nig7) were reported to infect human. In China, EbpA, EbpC, CHY1, N, D, Peru11, S7, SCC-2, PGP, Peru6, J, PigEBITS7, BR1 and BR2, Type IV, Peru8, ESH02, CHG5, HNR-I to HNR-VII, K, CQR-1, CQR-2, CQR-3, GDR-1, GDR-2, GDR-3. SCC-1, SCC-3, SCC-4, CHY1, Nig7 CHG9, ChG14, BEB6, CHG2, SC02, CE01 and CE02 genotypes were reported in rodents (Feng et al., 2009; Zhao et al., 2018; Wang et al., 2019; Li J. et al., 2020; Wang J. et al., 2020; Zhao et al., 2020). However, only 5 known genotypes (XJP-II, EbpC, EbpA, D, and NCF7) and 11 novel genotypes (ZIR1 to ZJR7, GXM1, HLJC1, HLJC2, and SDR1) were identified in the present study. Among them, 14 genotypes were clustered into a highly-supported monophyletic clade (Group 1), indicating that these genotypes are human-pathogenic types and may cause infection between humans and rodents, thus becoming a public health significance. This was the first record of E. bieneusi in S. dauricus. Eleven novel genotypes (ZIR1 to ZJR7, GXM1, HLJC1, HLJC2, and SDR1) were recorded in rodents for the first time. Of which, ZJR1, ZJR3, ZJR4, ZJR5, ZJR6, ZJR7, SDR1, HLJC1, and GXM1 were grouped into Group 1 (Figure 2), thus suggesting that rodents (R. norvegicus, M. musculus, and S. dauricus) may play an important role in the transmission of E. bieneusi between rodents and humans. Genotype XJP-II was previously found in pigs in Xinjiang (Li D. F. et al., 2019b), and NCF2 was also identified in farmed foxes (Vulpes lagopus) (Zhang et al., 2016; Ma et al., 2020) and raccoon dogs (Nyctereutes procyonoides) (Xu et al., 2016) in China, Kangaroo in Australia (Zhang Y. et al., 2018). Genotypes EbpC, EbpA, and D were frequently found in humans and a broad range of animals (Wang et al., 2013; Liu et al., 2017; Qi et al., 2018; Zhang X. X. et al., 2018; Zou et al., 2018; Wang H. et al., 2020; Wang Y. et al., 2020; Yu et al., 2020). The results showed that natural transmission of E. bieneusi among rodents, humans and many other animals may occur. More importantly, the three ITS genotypes were also found in water in China, which should be paid more attention to prevent the water-borne transmission of E. bieneusi (Hu et al., 2014).
Collectively, the present study firstly demonstrated that existence of Cryptosporidium spp. (3.4%, 18/536) and E. bieneusi (11.6%, 62/536) in rodents in Shanxi, Guangxi, and Zhejiang, China. Three known Cryptosporidium species/genotypes (C. viatorum, C. felis, and Cryptosporidium sp. rat genotype II/III), two uncertain Cryptosporidium species/genotypes (Cryptosporidium sp. novel1 and Cryptosporidium sp. novel2), 5 known E. bieneusi genotypes (XJP-II, EbpC, EbpA, D, and NCF7) and 11 novel E. bieneusi genotypes (ZJR1 to ZJR7, GXM1, HLJC1, HLJC2, and SDR1) were identified in the investigated rodents, suggesting rodents can act as a potential source of human and animal infections. E. bieneusi was more prevalent in R. norvegicus, whereas Cryptosporidium spp. was more frequently identified in S. dauricus. The present study also demonstrated that S. dauricus was the host of E. bieneusi and Cryptosporidium spp. for the first time. This study expanded the host range of these two parasites, which not only provided basic data for distribution of E. bieneusi and Cryptosporidium genotypes/species, but also provided foundation data for the prevention and control of E. bieneusi and Cryptosporidium spp. in China.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MT647749-MT647806, OK117929-OK117932, MT561508-MT561533.
Ethics Statement
This study was approved by the Ethics Committee of Qingdao Agricultural University.
Author Contributions
QZ, Y-CW, and H-TS conceived and designed the study and critically revised the manuscript. H-BN, S-YQ, DY, Z-HF, Z-HG, H-XW, H-YQ, and NX collected the samples. Z-YS, MZ, and Y-ZS performed the experiments. H-BN, Y-ZS, and S-YQ analyzed the data and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China, the National Innovation and Entrepreneurship Training Program for College Students of Shandong Province (202110435018), the State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (SKLVEB2019KFKT012), the Wild Animal Disease Monitoring and Early Warning System Maintenance Project (2130211), and the Research Foundation for Distinguished Scholars of Qingdao Agricultural University (665-1120046).
Conflict of Interest
Author Y-CW is employed by Veterinary Department, Muyuan Foods Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abarca, N., Santín, M., Ortega, S., Maloney, J. G., George, N. S., Molokin, A., et al. (2021). Molecular Detection and Characterization of Blastocystis Sp. and Enterocytozoon Bieneusi in Cattle in Northern Spain. Vet. Sci. 8, 191. doi: 10.3390/vetsci8090191
Adamu, H., Petros, B., Zhang, G., Kassa, H., Amer, S., Ye, J., et al. (2014). Distribution and Clinical Manifestations of Cryptosporidium Species and Subtypes in HIV/AIDS Patients in Ethiopia. PloS Negl. Trop. Dis. 8 (4), e2831. doi: 10.1371/journal.pntd.0002831
Ayinmode, A. B., Zhang, H., Dada-Adegbola, H. O., Xiao, L. (2014). Cryptosporidium Hominis Subtypes and Enterocytozoon Bieneusi Genotypes in HIV-Infected Persons in Ibadan, Nigeria. Zoonoses Public Health 61 (4), 297–303. doi: 10.1111/zph.12072
Backhans, A., Jacobson, M., Hansson, I., Lebbad, M., Lambertz, S. T., Gammelgård, E., et al. (2013). Occurrence of Pathogens in Wild Rodents Caught on Swedish Pig and Chicken Farms. Epidemiol. Infect. 141 (9), 1885–1891. doi: 10.1017/S0950268812002609
Bajer, A., Bednarska, M., Pawelczyk, A., Behnke, J. M., Gilbert, F. S., Sinski, E. (2002). Prevalence and Abundance of Cryptosporidium Parvum and Giardia Spp. In Wild Rural Rodents From the Mazury Lake District Region of Poland. Parasitology 125 (Pt 1), 21–34. doi: 10.1017/s0031182002001865
Cama, V. A., Bern, C., Sulaiman, I. M., Gilman, R. H., Ticona, E., Vivar, A., et al. (2003). Cryptosporidium Species and Genotypes in HIV-Positive Patients in Lima, Peru. J. Eukaryot Microbiol. 50 (Suppl), 531–533. doi: 10.1111/j.1550-7408.2003.tb00620.x
Cama, V. A., Pearson, J., Cabrera, L., Pacheco, L., Gilman, R., Meyer, S., et al. (2007). Transmission of Enterocytozoon Bieneusi Between a Child and Guinea Pigs. J. Clin. Microbiol. 45 (8), 2708–2710. doi: 10.1128/JCM.00725-07
Chen, Y. W., Zheng, W. B., Zhang, N. Z., Gui, B. Z., Lv, Q. Y., Yan, J. Q., et al. (2019). Identification of Cryptosporidium Viatorum XVa Subtype Family in Two Wild Rat Species in China. Parasit. Vectors 12 (1), 502. doi: 10.1186/s13071-019-3763-6
Danišová, O., Valenčáková, A., Stanko, M., Luptáková, L., Hasajová, A. (2015). First Report of Enterocytozoon Bieneusi and Encephalitozoon Intestinalis Infection of Wild Mice in Slovakia. Ann. Agric. Environ. Med. 22 (2), 251–252. doi: 10.5604/12321966.1152075
Danišová, O., Valenčáková, A., Stanko, M., Luptáková, L., Hatalová, E., Čanády, A. (2017). Rodents as a Reservoir of Infection Caused by Multiple Zoonotic Species/Genotypes of C. Parvum, C. Hominis, C. Suis, C. Scrofarum, and the First Evidence of Cryptosporidium Muskrat Genotypes I and II of Rodents in Europe. Acta Trop. 172, 29–35. doi: 10.1016/j.actatropica.2017.04.013
De Lucio, A., Amor-Aramendia, A., Bailo, B., Saugar, J. M., Anegagrie, M., Arroyo, A., et al. (2016). Prevalence and Genetic Diversity of Giardia Duodenalis and Cryptosporidium Spp. Among School Children in a Rural Area of the Amhara Region, North-West Ethiopia. PloS One 11 (7), e0159992. doi: 10.1371/journal.pone.0159992
Deng, L., Li, W., Yu, X., Gong, C., Liu, X., Zhong, Z., et al. (2016). First Report of the Human-Pathogenic Enterocytozoon Bieneusi From Red-Bellied Tree Squirrels (Callosciurus Erythraeus) in Sichuan, China. PloS One 11 (9), e0163605. doi: 10.1371/journal.pone.0163605
Deng, L., Li, W., Zhong, Z., Chai, Y., Yang, L., Zheng, H., et al. (2018). Molecular Characterization and New Genotypes of Enterocytozoon Bieneusi in Pet Chipmunks (Eutamias Asiaticus) in Sichuan Province. China BMC Microbiol. 18 (1), 37. doi: 10.1186/s12866-018-1175-y
Didier, E. S., Weiss, L. M., Cali, A., Marciano-Cabral, F. (2009). Overview of the Presentations on Microsporidia and Free-Living Amebae at the 10th International Workshops on Opportunistic Protists. Eukaryot Cell. 8 (4), 441–445. doi: 10.1128/EC.00302-08
Elwin, K., Hadfield, S. J., Robinson, G., Crouch, N. D., Chalmers, R. M. (2012). Cryptosporidium Viatorum N. Sp. (Apicomplexa: Cryptosporidiidae) Among Travellers Returning to Great Britain From the Indian Subcontinen-2011. Int. J. Parasitol. 42 (7), 675–682. doi: 10.1016/j.ijpara.2012.04.016
Essid, R., Menotti, J., Hanen, C., Aoun, K., Bouratbine, A. (2018). Genetic Diversity of Cryptosporidium Isolates From Human Populations in an Urban Area of Northern Tunisia. Infect. Genet. Evol. 58, 237–242. doi: 10.1016/j.meegid.2018.01.004
Feng, Y. (2010). Cryptosporidium in Wild Placental Mammals. Exp. Parasitol. 124 (1), 128–137. doi: 10.1016/j.exppara.2008.11.005
Feng, Y., Alderisio, K. A., Yang, W., Blancero, L. A., Kuhne, W. G., Nadareski, C. A., et al. (2007). Cryptosporidium Genotypes in Wildlife From a New York Watershed. Appl. Environ. Microbiol. 73 (20), 6475–6483. doi: 10.1128/AEM.01034-07
Feng, X., Reddy, V. K., Mayanja-Kizza, H., Weiss, L. M., Marton, L. J., Tzipori, S. (2009). Therapeutic Evaluation of Polyamine Analogue Drug Candidates Against Enterocytozoon Bieneusi in a SCID Mouse Model. Antimicrob. Agents Chemother. 53 (6), 2417–2423. doi: 10.1128/AAC.01113-08
Feng, Y., Ryan, U. M., Xiao, L. (2018). Genetic Diversity and Population Structure of Cryptosporidium. Trends Parasitol. 34 (11), 997–1011. doi: 10.1016/j.pt.2018.07.009
Foo, C., Farrell, J., Boxell, A., Robertson, I., Ryan, U. M. (2007). Novel Cryptosporidium Genotype in Wild Australian Mice (Mus Domesticus). Appl. Environ. Microbiol. 73 (23), 7693–7696. doi: 10.1128/AEM.00848-07
García-Livia, K., Martín-Alonso, A., Foronda, P. (2020). Diversity of Cryptosporidium Spp. In Wild Rodents From the Canary Islands, Spain. Parasit. Vectors 13 (1), 445. doi: 10.1186/s13071-020-04330-9
Gholipoury, M., Rezai, H. R., Namroodi, S., Arab Khazaeli, F. (2016). Zoonotic and Non-Zoonotic Parasites of Wild Rodents in Turkman Sahra, Northeastern Iran. Iran J. Parasitol. 11 (3), 350–357.
Gui, B. Z., Zou, Y., Chen, Y. W., Li, F., Jin, Y. C., Liu, M. T., et al. (2020). Novel Genotypes and Multilocus Genotypes of Enterocytozoon Bieneusi in Two Wild Rat Species in China: Potential for Zoonotic Transmission. Parasitol. Res. 119 (1), 283–290. doi: 10.1007/s00436-019-06491-8
Guo, Y., Alderisio, K. A., Yang, W., Cama, V., Feng, Y., Xiao, L. (2014). Host Specificity and Source of Enterocytozoon Bieneusi Genotypes in a Drinking Source Watershed. Appl. Environ. Microbiol. 80 (1), 218–225. doi: 10.1128/AEM.02997-13
Holubová, N., Zikmundová, V., Limpouchová, Z., Sak, B., Konečný, R., Hlásková, L., et al. (2019). Cryptosporidium Proventriculi Sp. N. (Apicomplexa: Cryptosporidiidae) in Psittaciformes Birds. Eur. J. Protistol. 69, 70–87. doi: 10.1016/j.ejop.2019.03.001
Hu, Y., Feng, Y., Huang, C., Xiao, L. (2014). Occurrence, Source, and Human Infection Potential of Cryptosporidium and Enterocytozoon Bieneusi in Drinking Source Water in Shanghai, China, During a Pig Carcass Disposal Incident. Environ. Sci. Technol. 48 (24), 14219–14227. doi: 10.1021/es504464t
Insulander, M., Silverlas, C., Lebbad, M., Karlsson, L., Mattsson, J. G., Svenungsson, B. (2013). Molecular Epidemiology and Clinical Manifestations of Human Cryptosporidiosis in Sweden. Epidemiol. Infect. 141 (5), 1009–1020. doi: 10.1017/S0950268812001665
Ježková, J., Prediger, J., Holubová, N., Sak, B., Konečný, R., Feng, Y., et al. (2021). Cryptosporidium Ratti N. Sp. (Apicomplexa: Cryptosporidiidae) and Genetic Diversity of Cryptosporidium Spp. In Brown Rats (Rattus Norvegicus) in the Czech Republic. Parasitology 148 (1), 84–97. doi: 10.1017/S0031182020001833
Jiang, W., Roellig, D. M., Lebbad, M., Beser, J., Troell, K., Guo, Y., et al. (2020). Subtype Distribution of Zoonotic Pathogen Cryptosporidium Felis in Humans and Animals in Several Countries. Emerg. Microbes Infect. 9 (1), 2446–2454. doi: 10.1080/22221751.2020.1840312
Kimura, A., Edagawa, A., Okada, K., Takimoto, A., Yonesho, S., Karanis, P. (2007). Detection and Genotyping of Cryptosporidium From Brown Rats (Rattus Norvegicus) Captured in an Urban Area of Japan. Parasitol. Res. 100 (6), 1417–1420. doi: 10.1007/s00436-007-0488-9
Koehler, A. V., Wang, T., Haydon, S. R., Gasser, R. B. (2018). Cryptosporidium Viatorum From the Native Australian Swamp Rat Rattus Lutreolus - An Emerging Zoonotic Pathogen? Int. J. Parasitol. Parasit. Wildl. 7 (1), 18–26. doi: 10.1016/j.ijppaw.2018.01.004
Krumkamp, R., Aldrich, C., Maiga-Ascofare, O., Mbwana, J., Rakotozandrindrainy, N., Borrmann, S., et al. (2021). Transmission of Cryptosporidium Species Among Human and Animal Local Contact Networks in Sub-Saharan Africa: A Multicountry Study. Clin. Infect. Dis. 72 (8), 1358–1366. doi: 10.1093/cid/ciaa223
Kvác, M., Hofmannová, L., Bertolino, S., Wauters, L., Tosi, G., Modrý, D. (2008). Natural Infection With Two Genotypes of Cryptosporidium in Red Squirrels (Sciurus Vulgaris) in Italy. Folia Parasitol. (Praha) 55 (2), 95–99. doi: 10.14411/fp.2008.012
Lebbad, M., Beser, J., Insulander, M., Karlsson, L., Mattsson, J. G., Svenungsson, B., et al. (2013). Unusual Cryptosporidiosis Cases in Swedish Patients: Extended Molecular Characterization of Cryptosporidium Viatorum and Cryptosporidium Chipmunk Genotype I. Parasitology 140 (14), 1735–1740. doi: 10.1017/S003118201300084X
Li, W., Feng, Y., Santin, M. (2019). Host Specificity of Enterocytozoon Bieneusi and Public Health Implications. Trends Parasitol. 35, 436–451. doi: 10.1016/j.pt.2019.04.004
Li, J., Jiang, Y., Wang, W., Chao, L., Jia, Y., Yuan, Y., et al. (2020). Molecular Identification and Genotyping of Enterocytozoon Bieneusi in Experimental Rats in China. Exp. Parasitol. 210, 107850. doi: 10.1016/j.exppara.2020.107850
Li, Q., Li, L., Tao, W., Jiang, Y., Wan, Q., Lin, Y., et al. (2016). Molecular Investigation of Cryptosporidium in Small Caged Pets in Northeast China: Host Specificity and Zoonotic Implications. Parasitol. Res. 115 (7), 2905–2911. doi: 10.1007/s00436-016-5076-4
Liu, H., Jiang, Z., Yuan, Z., Yin, J., Wang, Z., Yu, B., et al. (2017). Infection by and Genotype Characteristics of Enterocytozoon Bieneusi in HIV/AIDS Patients From Guangxi Zhuang Autonomous Region, China. BMC Infect. Dis. 17 (1), 684. doi: 10.1186/s12879-017-2787-9
Liu, X., Zhou, X., Zhong, Z., Zuo, Z., Shi, J., Wang, Y., et al. (2015). Occurrence of Novel and Rare Subtype Families of Cryptosporidium in Bamboo Rats (Rhizomys Sinensis) in China. Vet. Parasitol. 207 (1-2), 144–148. doi: 10.1016/j.vetpar.2014.11.009
Li, W., Xiao, L. (2019). Multilocus Sequence Typing and Population Genetic Analysis of Enterocytozoon Bieneusi: Host Specificity and Its Impacts on Public Pealth. Front. Genet. 10:307. doi: 10.3389/fgene.2019.00307
Li, W., Xiao, L. (2020). Ecological and Public Health Significance of Enterocytozoon Bieneusi. One Health 12, 100209. doi: 10.1016/j.onehlt.2020.100209
Li, F., Zhang, Z., Hu, S., Zhao, W., Zhao, J., Kváč, M., et al. (2020b). Common Occurrence of Divergent Cryptosporidium Species and Cryptosporidium Parvum Subtypes in Farmed Bamboo Rats (Rhizomys Sinensis). Parasit. Vectors 13 (1), 149. doi: 10.1186/s13071-020-04021-5
Li, D. F., Zhang, Y., Jiang, Y. X., Xing, J. M., Tao, D. Y., Zhao, A. Y., et al. (2019). Genotyping and Zoonotic Potential of Enterocytozoon Bieneusi in Pigs in Xinjiang, China. Front. Microbiol. 10:2401. doi: 10.3389/fmicb.2019.02401
Li, F., Zhao, W., Zhang, C., Guo, Y., Li, N., Xiao, L., et al. (2020a). Cryptosporidium Species and C. Parvum Subtypes in Farmed Bamboo Rats. Pathogens 9 (12):1018. doi: 10.3390/pathogens9121018
Lv, C., Zhang, L., Wang, R., Jian, F., Zhang, S., Ning, C., et al. (2009). Cryptosporidium Spp. In Wild, Laboratory, and Pet Rodents in China: Prevalence and Molecular Characterization. Appl. Environ. Microbiol. 75 (24), 7692–7699. doi: 10.1128/AEM.01386-09
Ma, Y. Y., Zou, Y., Ma, Y. T., Nie, L. B., Xie, S. C., Cong, W., et al. (2020). Molecular Detection and Genotype Distribution of Enterocytozoon Bieneusi in Farmed Silver Foxes (Vulpes Vulpes) and Arctic Foxes (Vulpes Lagopus) in Shandong Province, Eastern China. Parasitol. Res. 119 (1), 321–326. doi: 10.1007/s00436-019-06538-w
Mirzaghavami, M., Sadraei, J., Forouzandeh, M. (2016). Detection of Cryptosporidium Spp. In Free Ranging Animals of Tehran, Iran. J. Parasit. Dis. 40 (4), 1528–1531. doi: 10.1007/s12639-015-0720-y
Murakoshi, F., Fukuda, Y., Matsubara, R., Kato, Y., Sato, R., Sasaki, T., et al. (2013). Detection and Genotyping of Cryptosporidium Spp. In Large Japanese Field Mice, Apodemus Speciosus. Vet. Parasitol. 196 (1-2), 184–188. doi: 10.1016/j.vetpar.2013.02.011
Nakai, Y., Hikosaka, K., Sato, M., Sasaki, T., Kaneta, Y., Okazaki, N. (2004). Detection of Cryptosporidium Muris Type Oocysts From Beef Cattle in a Farm and From Domestic and Wild Animals in and Around the Farm. J. Vet. Med. Sci. 66 (8), 983–984. doi: 10.1292/jvms.66.983
Ng-Hublin, J. S., Singleton, G. R., Ryan, U. (2013). Molecular Characterization of Cryptosporidium Spp. From Wild Rats and Mice From Rural Communities in the Philippines. Infect. Genet. Evol. 16, 5–12. doi: 10.1016/j.meegid.2013.01.011
NIAID Emerging Infectious Diseases/Pathogens. Available at: https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens (Accessed 7 May 2018).
Paparini, A., Jackson, B., Ward, S., Young, S., Ryan, U. M. (2012). Multiple Cryptosporidium Genotypes Detected in Wild Black Rats (Rattus Rattus) From Northern Australia. Exp. Parasitol. 131 (4), 404–412. doi: 10.1016/j.exppara.2012.05.009
Perec-Matysiak, A., Buńkowska-Gawlik, K., Kváč, M., Sak, B., Hildebrand, J., Leśniańska, K. (2015). Diversity of Enterocytozoon Bieneusi Genotypes Among Small Rodents in Southwestern Poland. Vet. Parasitol. 214 (3-4), 242–246. doi: 10.1016/j.vetpar.2015.10.018
Qi, M., Li, J., Zhao, A., Cui, Z., Wei, Z., Jing, B., et al. (2018). Host Specificity of Enterocytozoon Bieneusi Genotypes in Bactrian Camels (Camelus Bactrianus) in China. Parasit. Vectors 11 (1), 219. doi: 10.1186/s13071-018-2793-9
Qi, M., Luo, N., Wang, H., Yu, F., Wang, R., Huang, J., et al. (2015). Zoonotic Cryptosporidium Spp. And Enterocytozoon Bieneusi in Pet Chinchillas (Chinchilla Lanigera) in China. Parasitol. Int. 64 (5), 339–341. doi: 10.1016/j.parint.2015.05.007
Roellig, D. M., Salzer, J. S., Carroll, D. S., Ritter, J. M., Drew, C., Gallardo-Romero, N., et al. (2015). Identification of Giardia Duodenalis and Enterocytozoon Bieneusi in an Epizoological Investigation of a Laboratory Colony of Prairie Dogs, Cynomys Ludovicianus. Vet. Parasitol. 210 (1-2), 91–97. doi: 10.1016/j.vetpar.2015.03.022
Saki, J., Foroutan-Rad, M., Asadpouri, R. (2016). Molecular Characterization of Cryptosporidium Spp. In Wild Rodents of Southwestern Iran Using 18S rRNA Gene Nested-PCR-RFLP and Sequencing Techniques. J. Trop. Med. 2016:6834206. doi: 10.1155/2016/6834206
Sak, B., Kváč, M., Květoňová, D., Albrecht, T., Piálek, J. (2011). The First Report on Natural Enterocytozoon Bieneusi and Encephalitozoon Spp. Infections in Wild East-European House Mice (Mus Musculus Musculus) and West-European House Mice (M. M. Domesticus) in a Hybrid Zone Across the Czech Republic-Germany Border. Vet. Parasitol. 178 (3-4), 246–250. doi: 10.1016/j.vetpar.2010.12.044
Sanchez, A., Munoz, M., Gomez, N., Tabares, J., Segura, L., Salazar, A., et al. (2017). Molecular Epidemiology of Giardia, Blastocystis and Cryptosporidium Among Indigenous Children From the Colombian Amazon Basin. Front. Microbiol. 8, 248. doi: 10.3389/fmicb.2017.00248
Sannella, A. R., Suputtamongkol, Y., Wongsawat, E., Caccio, S. M. (2019). A Retrospective Molecular Study of Cryptosporidium Species and Genotypes in HIV-Infected Patients From Thailand. Parasit. Vectors 12 (1), 91. doi: 10.1186/s13071-019-3348-4
Santin, M. (2015). “Enterocytozoon Bieneusi,” in Biology of Foodborne Parasites. Eds. Xiao, L., Ryan, U., Feng, Y. (Boca Raton, FL, USA: CRC Press), 149–174.
Santín, M., Fayer, R. (2011). Microsporidiosis: Enterocytozoon Bieneusi in Domesticated and Wild Animals. Res. Vet. Sci. 90 (3), 363–371. doi: 10.1016/j.rvsc.2010.07.014
Song, J., Kim, C. Y., Chang, S. N., Abdelkader, T. S., Han, J., Kim, T. H., et al. (2015). Detection and Molecular Characterization of Cryptosporidium Spp. From Wild Rodents and Insectivores in South Korea. Korean J. Parasitol. 53 (6), 737–743. doi: 10.3347/kjp.2015.53.6.737
Stenger, B. L., Clark, M. E., Kváč, M., Khan, E., Giddings, C. W., Dyer, N. W., et al. (2015). Highly Divergent 18S rRNA Gene Paralogs in a Cryptosporidium Genotype From Eastern Chipmunks (Tamias Striatus). Infect. Genet. Evol. 32, 113–123. doi: 10.1016/j.meegid.2015.03.003
Stenger, B., Clark, M. E., Kváč, M., Khan, E., Giddings, C. W., Prediger, J., et al. (2015). North American Tree Squirrels and Ground Squirrels With Overlapping Ranges Host Different Cryptosporidium Species and Genotypes. Infect. Genet. Evol. 36, 287–293. doi: 10.1016/j.meegid.2015.10.002
Sutthikornchai, C., Popruk, S., Mahittikorn, A., Arthan, D., Soonthornworasiri, N., Paratthakonkun, C., et al. (2021). Molecular Detection of Cryptosporidium Spp., Giardia Duodenalis, and Enterocytozoon Bieneusi in School Children at the Thai-Myanmar Border. Parasitol. Res. 120 (8), 2887–2895. doi: 10.1007/s00436-021-07242-4
Ukwah, B. N., Ezeonu, I. M., Ezeonu, C. T., Roellig, D., Xiao, L. (2017). Cryptosporidium Species and Subtypes in Diarrheal Children and HIV-Infected Persons in Ebonyi and Nsukka, Nigeria. J. Infect. Dev. Ctries. 11 (2), 173–179. doi: 10.3855/jidc.8034
Wang, H., Lin, X., Sun, Y., Qi, N., Lv, M., Xiao, W., et al. (2020). Occurrence, Risk Factors and Genotypes of Enterocytozoon Bieneusi in Dogs and Cats in Guangzhou, Southern China: High Genotype Diversity and Zoonotic Concern. BMC Vet. Res. 16 (1), 201. doi: 10.1186/s12917-020-02421-4
Wang, H., Liu, Q., Jiang, X., Zhang, Y., Zhao, A., Cui, Z., et al. (2019). Dominance of Zoonotic Genotype D of Enterocytozoon Bieneusi in Bamboo Rats (Rhizomys Sinensis). Infect. Genet. Evol. 73, 113–118. doi: 10.1016/j.meegid.2019.04.025
Wang, J., Lv, C., Zhao, D., Zhu, R., Li, C., Qian, W. (2020). First Detection and Genotyping of Enterocytozoon Bieneusi in Pet Fancy Rats (Rattus Norvegicus) and Guinea Pigs (Cavia Porcellus) in China. Parasite 27, 21. doi: 10.1051/parasite/2020019
Wang, S. N., Sun, Y., Zhou, H. H., Lu, G., Qi, M., Liu, W. S., et al. (2020). Prevalence and Genotypic Identification of Cryptosporidium Spp. And Enterocytozoon Bieneusi in Captive Asiatic Black Bears (Ursus Thibetanus) in Heilongjiang and Fujian Provinces of China. BMC Vet. Res. 16 (1), 84. doi: 10.1186/s12917-020-02292-9
Wang, Y., Zhang, K., Zhang, Y., Wang, K., Gazizova, A., Wang, L., et al. (2020). First Detection of Enterocytozoon Bieneusi in Whooper Swans (Cygnus Cygnus) in China. Parasit. Vectors 13 (1), 5. doi: 10.1186/s13071-020-3884-y
Wang, L., Zhang, H., Zhao, X., Zhang, L., Zhang, G., Guo, M., et al. (2013). Zoonotic Cryptosporidium Species and Enterocytozoon Bieneusi Genotypes in HIV-Positive Patients on Antiretroviral Therapy. J. Clin. Microbiol. 51 (2), 557–563. doi: 10.1128/JCM.02758-12
Wei, Z., Liu, Q., Zhao, W., Jiang, X., Zhang, Y., Zhao, A., et al. (2019). Prevalence and Diversity of Cryptosporidium Spp. In Bamboo Rats (Rhizomys Sinensis) in South Central China. Int. J. Parasitol. Parasit. Wildl. 9, 312–316. doi: 10.1016/j.ijppaw.2019.06.010
Xu, C., Ma, X., Zhang, H., Zhang, X. X., Zhao, J. P., Ba, H. X., et al. (2016). Prevalence, Risk Factors and Molecular Characterization of Enterocytozoon Bieneusi in Raccoon Dogs (Nyctereutes Procyonoides) in Five Provinces of Northern China. Acta Trop. 161, 68–72. doi: 10.1016/j.actatropica.2016.05.015
Yu, F., Cao, Y., Wang, H., Liu, Q., Zhao, A., Qi, M., et al. (2020). Host-Adaptation of the Rare Enterocytozoon Bieneusi Genotype CHN4 in Myocastor Coypus (Rodentia: Echimyidae) in China. Parasit. Vectors 13 (1), 578. doi: 10.1186/s13071-020-04436-0
Zhang, X. X., Cong, W., Lou, Z. L., Ma, J. G., Zheng, W. B., Yao, Q. X., et al. (2016). Prevalence, Risk Factors and Multilocus Genotyping of Enterocytozoon Bieneusi in Farmed Foxes (Vulpes Lagopus), Northern China. Parasit. Vectors 9, 72. doi: 10.1186/s13071-016-1356-1
Zhang, X. X., Jiang, R. L., Ma, J. G., Xu, C., Zhao, Q., Hou, G., et al. (2018). Enterocytozoon Bieneusi in Minks (Neovison Vison) in Northern China: A Public Health Concern. Front. Microbiol. 9, 1221. doi: 10.3389/fmicb.2018.01221
Zhang, X., Jian, Y., Li, X., Ma, L., Karanis, G., Karanis, P. (2018). The First Report of Cryptosporidium Spp. In Microtus Fuscus (Qinghai Vole) and Ochotona Curzoniae (Wild Plateau Pika) in the Qinghai-Tibetan Plateau Area, China. Parasitol. Res. 117 (5), 1401–1407. doi: 10.1007/s00436-018-5827-5
Zhang, Y., Koehler, A. V., Wang, T., Haydon, S. R., Gasser, R. B. (2018). New Operational Taxonomic Units of Enterocytozoon in Three Marsupial Species. Parasit. Vectors 11 (1), 371. doi: 10.1186/s13071-018-2954-x
Zhao, W., Wang, J., Ren, G., Yang, Z., Yang, F., Zhang, W., et al. (2018). Molecular Characterizations of Cryptosporidium Spp. And Enterocytozoon Bieneusi in Brown Rats (Rattus Norvegicus) From Heilongjiang Province, China. Parasit. Vectors 11 (1), 313. doi: 10.1186/s13071-018-2892-7
Zhao, Z., Wang, R., Zhao, W., Qi, M., Zhao, J., Zhang, L., et al. (2015). Genotyping and Subtyping of Giardia and Cryptosporidium Isolates From Commensal Rodents in China. Parasitology 142 (6), 800–806. doi: 10.1017/S0031182014001929
Zhao, W., Zhou, H., Huang, Y., Xu, L., Rao, L., Wang, S., et al. (2019). Cryptosporidium Spp. In Wild Rats (Rattus Spp.) From the Hainan Province, China: Molecular Detection, Species/Genotype Identification and Implications for Public Health. Int. J. Parasitol. Parasit. Wildl. 9, 317–321. doi: 10.1016/j.ijppaw.2019.03.017
Zhao, W., Zhou, H., Yang, L., Ma, T., Zhou, J., Liu, H., et al. (2020). Prevalence, Genetic Diversity and Implications for Public Health of Enterocytozoon Bieneusi in Various Rodents From Hainan Province, China. Parasit. Vectors 13 (1), 438. doi: 10.1186/s13071-020-04314-9
Keywords: Cryptosporidium spp., Enterocytozoon bieneusi, prevalence, genotyping, wild rats, China
Citation: Ni H-B, Sun Y-Z, Qin S-Y, Wang Y-C, Zhao Q, Sun Z-Y, Zhang M, Yang D, Feng Z-H, Guan Z-H, Qiu H-Y, Wang H-X, Xue N-Y and Sun H-T (2021) Molecular Detection of Cryptosporidium spp. and Enterocytozoon bieneusi Infection in Wild Rodents From Six Provinces in China. Front. Cell. Infect. Microbiol. 11:783508. doi: 10.3389/fcimb.2021.783508
Received: 26 September 2021; Accepted: 02 November 2021;
Published: 25 November 2021.
Edited by:
Wei Cong, Shandong University, Weihai, ChinaReviewed by:
Wen-Bin Zheng, Shanxi Agricultural University, ChinaKatarzyna Buńkowska-Gawlik, University of Wrocław, Poland
Jianhai Yin, National Institute of Parasitic Diseases, China
Majid Pirestani, Tarbiat Modares University, Iran
Copyright © 2021 Ni, Sun, Qin, Wang, Zhao, Sun, Zhang, Yang, Feng, Guan, Qiu, Wang, Xue and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-Chun Wang, MzgxNzcwNzIyQHFxLmNvbQ==; Quan Zhao, emhhb3F1YW4wODI1QDE2My5jb20=; He-Ting Sun, eGlhb2ZlbmdzaHRAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Hong-Bo Ni
Hong-Bo Ni Yu-Zhe Sun2†
Yu-Zhe Sun2† Si-Yuan Qin
Si-Yuan Qin Quan Zhao
Quan Zhao Nian-Yu Xue
Nian-Yu Xue