- Department of Comparative Immunobiology, Zoological Institute, Christian-Albrechts University, Kiel, Germany
In comparison with the standard monoxenic maintenance in the laboratory, rearing the nematode Caenorhabditis elegans on its natural microbiota improves its fitness and immunity against pathogens. Although C. elegans is known to exhibit choice behavior and pathogen avoidance behavior, little is known about whether C. elegans actively chooses its (beneficial) microbiota and whether the microbiota influences worm behavior. We examined eleven natural C. elegans isolates in a multiple-choice experiment for their choice behavior toward four natural microbiota bacteria and found that microbiota choice varied among C. elegans isolates. The natural C. elegans isolate MY2079 changed its choice behavior toward microbiota isolate Ochrobactrum vermis MYb71 in both multiple-choice and binary-choice experiments, in particular on proliferating bacteria: O. vermis MYb71 was chosen less than other microbiota bacteria or OP50, but only after preconditioning with MYb71. Examining escape behavior and worm fitness on MYb71, we ruled out pathogenicity of MYb71 and consequently learned pathogen avoidance behavior as the main driver of the behavioral change toward MYb71. The change in behavior of C. elegans MY2079 toward microbiota bacterium MYb71 demonstrates how the microbiota influences the worm’s choice. These results might give a baseline for future research on host–microbiota interaction in the C. elegans model.
Introduction
The nematode Caenorhabditis elegans has been extensively used as a research model and is usually kept in monoxenic culture with the Escherichia coli strain OP50 as food. This is in stark contrast to C. elegans natural environment (e.g., rotting fruits, decomposing plant matter, and compost) where the nematode lives in close association with a huge variety of microbes (Félix and Braendle, 2010; Petersen et al., 2015; Dirksen et al., 2016). In the past years, research on the natural C. elegans microbiota revealed that the species-rich bacterial community of C. elegans is distinct from that of the corresponding natural environment and is dominated by Enterobacteriaceae and members of the genera Pseudomonas, Stenotrophomonas, Ochrobactrum, and Sphingomonas (Berg et al., 2016a; Dirksen et al., 2016; Samuel et al., 2016). Various microbiota genera are able to colonize and persist in the worm’s intestine and show beneficial effects on population growth and resistance against abiotic and biotic stress (Berg et al., 2016b; Dirksen et al., 2016; Samuel et al., 2016; Zimmermann et al., 2020; Pees et al., 2021). Protective effects of Pseudomonas, Bacillus, and Enterobacter strains against pathogen infection have been characterized (Montalvo-Katz et al., 2013; Berg et al., 2016b; Kissoyan et al., 2019). Furthermore, assessment of the inducible transcriptome response revealed influences of two Ochrobactrum strains on C. elegans dietary response, development, fertility, immunity, and energy metabolism indicating that further microbiota-mediated host functions are conceivable (Yang et al., 2019).
The influence of microbiota on vertebrate and invertebrate host behavior has been repeatedly demonstrated, for example, on olfactory behavior (Fischer et al., 2017; Qiao et al., 2019), microbial preference, and foraging decisions in Drosophila melanogaster (Wong et al., 2017) and social preference and depressive-like behavior in mice (Arentsen et al., 2015; Luo et al., 2018). In particular, natural C. elegans isolates show different behavioral responses depending on genotype, time of isolation from the wild, and sampling site (Volkers et al., 2013; Petersen et al., 2015). These behavioral responses are often regulated by the presence of food bacteria and can influence feeding, egg laying, and movement of the worm (Horvitz et al., 1982; Avery and Horvitz, 1990). Certain bacteria and small molecules can alter C. elegans chemotactic responses (Bargmann and Horvitz, 1991; Grewal and Wright, 1992; Shtonda and Avery, 2006). Differentiation between benign and harmful bacteria is particularly important under natural conditions in which worms face a multitude of microorganisms. Some natural C. elegans populations exhibit a preference for naturally coexisting bacterial species (Volkers et al., 2013) indicating that choice behavior may be involved in the identification and acquisition of symbionts, commensals, and microbiota (O’Donnell et al., 2020). However, information on the influence of the microbiota on C. elegans behavior is still limited.
The aim of this study was hence to identify and characterize the potential effects of the microbiota on the behavior of C. elegans. We focused on a trait of likely relevance under natural conditions, namely, choice behavior. We demonstrated that natural C. elegans strains differ in their choice behavior toward their microbiota bacteria. Particularly, preconditioning of natural C. elegans isolate MY2079 with proliferating Ochrobactrum vermis MYb71 resulted in a behavioral change upon a second exposure. Accordingly, colonizing MYb71 is able to alter C. elegans preference for the same bacterial strain. Our results contribute to the understanding of C. elegans behavior to its natural gut microbiota.
Materials and Methods
Nematode and Bacterial Strains
We used eleven natural C. elegans isolates (Supplementary Table S1A) isolated at two North German locations between 2002 and 2012 (Haber, 2005; Petersen et al., 2014) and the C. elegans laboratory strain N2. The natural isolates were chosen based on genotypic differences at the microsatellite level that translated into phenotypic variation (Petersen et al., 2015). All strains were maintained following standard procedures (Stiernagle, 2006).
Six bacterial strains were used as representatives for different genera of C. elegans native microbiota able to colonize the worm’s intestine (Supplementary Table S1B) (Dirksen et al., 2016; Dirksen et al., 2020; Zimmermann et al., 2020; Pees et al., 2021): the Gram-negative bacteria Acinetobacter guillouiae MYb10, Pseudomonas fluorescens MYb11, Ochrobactrum anthropi MYb49, O. vermis MYb71, and Ochrobactrum pseudogrignonense MYb237 and the Gram-positive bacterium Rhodococcus erythropolis MYb53 (Dirksen et al., 2016). Microbiota isolates were thawed freshly, cultured for 2 days on tryptic soy agar (TSA) at 25°C, and subsequently grown in tryptic soy broth (TSB) in a shaking incubator for 42 h at 28°C (MYb10, MYb49, MYb53, MYb71) or 16 h (MYb11). E. coli OP50 was cultured in TSB for 16 h at 37°C. For the experiments, all bacteria were adjusted to OD60010 in their supernatant if not stated otherwise.
RNA Isolation
Bacterial lawns (OD60010) were grown for 24 h at 25°C on 9-cm nematode growth medium (NGM) plates. The lawns were scraped off, pooled from five plates, and pelleted at 4,000 g at 4°C for 15 min, and 100 µl of bacterial pellet was resuspended in 1 ml of RNAmagic (Bio-Budget Technology GmbH, Krefeld, Germany). Aliquots of 800 µl were frozen in liquid nitrogen, thawed at 45°C and frozen in three repeated cycles, and stored at −80°C until RNA isolation. Total RNA was isolated using MACHEREY-NAGEL (Düren, Germany) NucleoSpin Kit using the protocol for bacterial cells.
Microbiota Choice Experiments
Choice behavior was examined using either multiple-choice experiments offering five bacterial strains simultaneously or binary-choice experiments with only two bacterial strains. C. elegans strains were either unconditioned or preconditioned with a specific microbiota bacterium. Worms touching a bacterial spot were scored, and all other worms were not taken into account. The position of the bacteria across assay plates was randomized, and all experiments were performed without current knowledge of worm and bacterial strain identity to avoid any observer bias.
Multiple-Choice Experiment
For the multiple-choice experiments, four microbiota bacteria and OP50 were adjusted to OD60010 in phosphate-buffered saline (PBS), and 25 µl of each bacterium and a PBS control were pipetted equidistantly in a circle onto 9-cm peptone-free NGM (PFM) plates or NGM plates and left to dry for 2 h. Approximately 100 synchronized C. elegans in the fourth larval stage (L4) were pipetted to the center of the plate. The number of worms residing on each bacterium was determined after 2 or 24 h. We calculated the mean proportion of worms per bacterium = number of worms on a bacterium/total number of worms on all bacteria per plate.
Preconditioning of Caenorhabditis elegans
C. elegans was grown on OP50 until the L4 stage, washed three times in sterile M9 buffer, and pipetted to preconditioning plates freshly inoculated with one bacterium in OD60010. Preconditioning was performed with live and heat-killed (80°C for 40 min) MYb71 and OP50, with OP50 lawns spiked with 21 ng of total RNA of MYb71 or OP50, or with the odor of viable MYb71 or OP50. For the latter, L4 worms were placed on OP50 lawns with either 500 µl MYb71 or OP50 on a thin NGM layer in the lid of the plate. Lining the edge of the plate with 1% palmitic acid in ethanol kept the worms on the OP50 lawn. After 24 h, the preconditioned adults were washed repeatedly in M9 buffer and subsequently applied to binary-choice assay plates.
Binary-Choice Assay
Approximately 70 naïve L4 worms or preconditioned adults were pipetted centrally between two opposing 30-µl bacterial spots on 6-cm NGM plates. The number of worms residing on each bacterium was determined after 2 and/or 24 h. We calculated a choice index = (number of worms on the bacterium opposing the microbiota bacterium − number of worms on the microbiota bacterium)/(total number of worms on both bacteria). To test for bacterial signals produced upon contact with worms, the initially placed worms were removed after 24 h, and naïve adult worms (from OP50) were pipetted centrally onto the same plates. The choice index for the newly introduced worms was determined after 2 h (26 h total time).
Bacterial Lawn Leaving Experiment
Synchronized C. elegans MY2079 L4 worms were preconditioned on MYb71 or OP50 for 24 h. Adults were washed repeatedly in M9 buffer, and approximately 50 adults were pipetted onto NGM plates inoculated with 50 µl of either MYb71 or OP50. The number of worms on and outside the bacterial lawn was scored after 2 h. We calculated the proportion of escaped worms = number of worms outside the lawn/total number of worms on the plate.
Brood Size Assay
NGM and PFM plates were inoculated daily with 50 µl MYb71 or OP50 at OD60010 adjusted in PBS and stored at 20°C for 24 h. Initially, one L4 worm was picked onto the plate and transferred daily to a new plate. We scored the hatched offspring of each worm until the worm stopped laying eggs. Data of worms that died or escaped from the plate before the end of the egg-laying period were excluded.
Statistics
Statistical calculations were performed with R studio software (version 1.3.1093) and can be found in Supplementary Table S2. Graphs were produced with R Studio (version 1.3.1093) and edited with Inkscape (version 1.0.1).
Results
Microbiota Bacteria Influence the Choice Behavior of Natural Caenorhabditis elegans Isolate MY2079
N2 can be considered as a laboratory animal, domesticated to a monoxenic environment with only E. coli OP50 as a food source. Natural C. elegans isolates can differ in their behavioral response to bacterial cues (Petersen et al., 2015; Pradhan et al., 2019). Since C. elegans microbiota was originally extracted from natural C. elegans isolates, we selected eleven genotypically divergent natural C. elegans isolates (Petersen et al., 2015) and the laboratory strain N2 to screen their choice behavior toward four microbiota isolates from different genera and OP50 as control (Figure 1A).
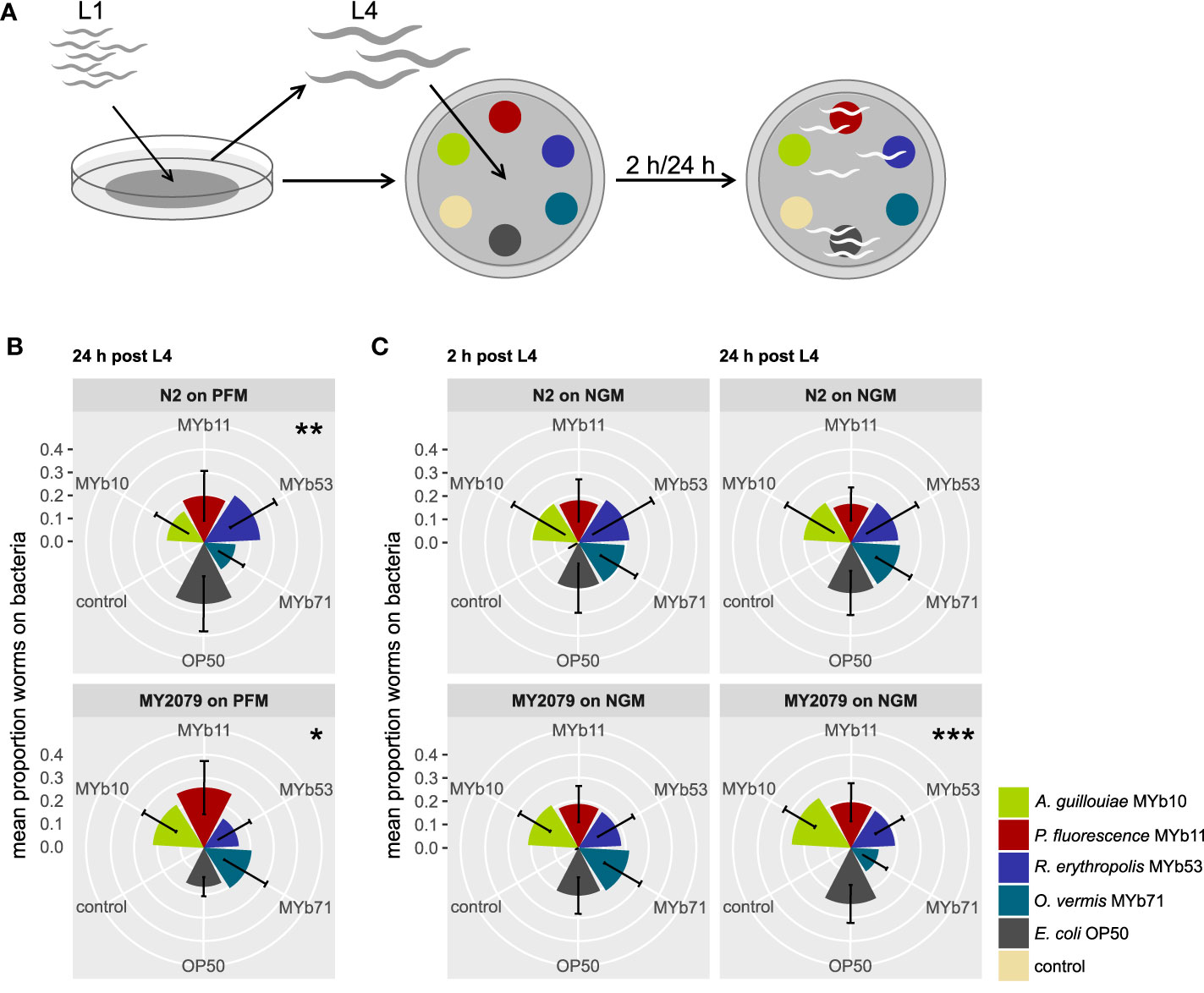
Figure 1 Natural Caenorhabditis elegans isolate MY2079 alters its choice behavior on proliferating microbiota after 24h. (A) Synchronized C. elegans were grown on OP50 until L4 stage and then used in multiple-choice experiments. The choice behavior of C. elegans N2 and MY2079 was evaluated toward a subset of (B) non-proliferating microbiota on peptone-free medium (PFM) after 24 h and (C) proliferating microbiota on nematode growth medium (NGM) after 2 and 24 h post L4. Shown are means ± SD of n = 15 (with ~100 worms per n), Kruskal–Wallis (Kruskal and Wallis, 1952) with false discovery rate correction for multiple testing (Benjamini and Hochberg, 1995); asterisks indicate difference within the given treatment. Significance is designated to the following scale: ***p < 0.001, **p < 0.01, *p < 0.05.
We defined choice behavior as present if worms preferred at least one of the offered bacteria over the others. In an initial screen, we identified general significant choice behavior after 24 h in one natural C. elegans isolate, MY2079 (p = 0.022; Supplementary Figure S1), and a trend in eight natural C. elegans isolates (p < 0.063). We confirmed the results of the screen for MY2079 with an additional multiple-choice experiment using a higher replication (n = 15), in which not only MY2079 (p = 0.012) but this time also N2 (p = 0.008) showed choice behavior after 24 h. Yet the choice behavior between MY2079 and N2 differed significantly (p < 0.001; chi-squared test) (Figure 1B).
In its ephemeral natural habitats, C. elegans lives a boom-and-bust life cycle (Frézal and Félix, 2015), because as the availability of plant material varies, so do the growth conditions and the availability of food organisms. Hence, we wondered whether the availability of nutrients for bacterial growth influences the choice behavior toward microbiota. We examined the effect of bacterial proliferation on the choice behavior of C. elegans N2 and the natural isolate MY2079 in a multiple-choice experiment on NGM, which supports bacterial proliferation. C. elegans MY2079 showed choice behavior after 24 h (p < 0.001) but not after 2 h, whereas C. elegans N2 showed generally no choice behavior on NGM (Figure 1C). Further, the choice behavior between C. elegans MY2079 and N2 on NGM differed significantly after 2 h (p < 0.001; chi-squared test) and 24 h (p < 0.001; chi-squared test) (Figure 1C).
These results indicate that the choice behavior of N2 and MY2079 differs and that C. elegans MY2079 shows choice behavior particularly in the presence of proliferating microbiota.
Natural Caenorhabditis elegans Isolate MY2079 Avoids Ochrobactrum vermis MYb71 Over Time
We noticed in the multiple-choice experiment on proliferating bacteria that MY2079 changed its choice behavior toward MYb71 and OP50 over time and eventually avoided MYb71 in favor of OP50 (Figure 1C). Hence, we wondered whether the avoidance behavior toward MYb71 is due to repulsion from MYb71 or increased attraction toward one of the other bacteria. We performed a binary-choice experiment with MYb71 opposing one test bacterial strain at a time (Figure 2A). C. elegans N2 did not change its choice behavior over time and chose MYb71 and the opposing test bacteria equally. Similarly, C. elegans MY2079 chose MYb10, MYb11, MYb53, and OP50 equally to MYb71 at 2 h (Figure 2B). However, after 24 h, MY2079 preferred all other bacteria over MYb71 (p = 0.028). These results suggest that the change in choice behavior toward MYb71 over time is specific for C. elegans MY2079 and is caused by avoidance of proliferating MYb71.
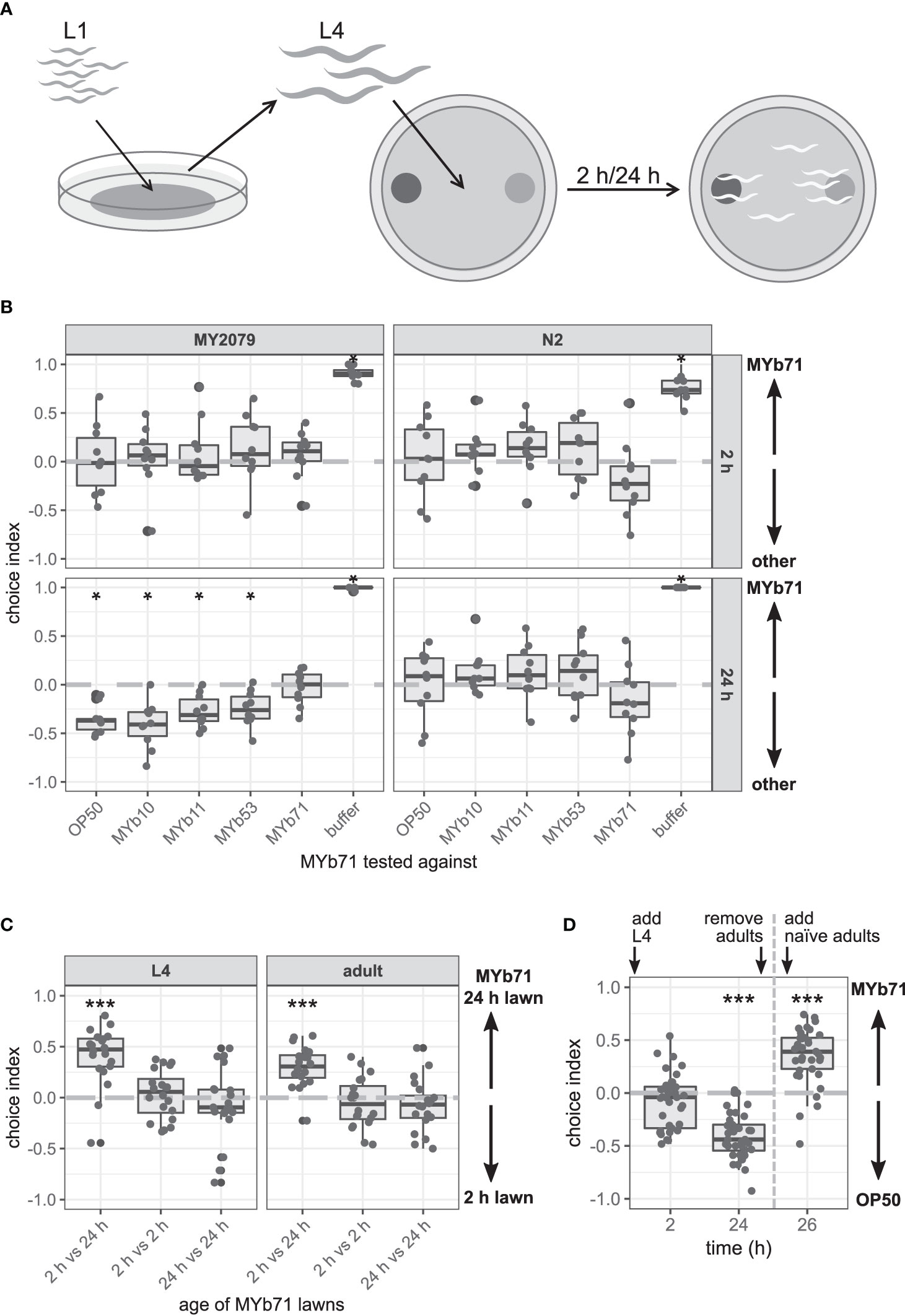
Figure 2 The late microbiota avoidance response is specific to MYb71, independent of MYb71 lawn age, and is not shown in naïve MY2079. (A) Synchronized Caenorhabditis elegans were grown on OP50 until L4 stage and then used in binary-choice experiments using MYb71 (dark gray) vs. a test bacterium (light gray). The choice index was determined at 2 and 24 h post L4. (B) The choice behavior of C. elegans N2 and MY2079 toward MYb71 was tested against individual microbiota isolates or OP50 (n = 10, with ~70 worms per n). (C) Choice behavior of MY2079 L4 larvae and adults toward a “new” (2 h) and “old” (24 h) lawn (n = 20, with ~70 worms per n). (D) MYb71 and OP50 on choice plates were exposed to MY2079 worms, and the choice behavior of MY2079 was evaluated after 2 and 24 h. After removal of the initially placed worms, the choice behavior of naïve adults was evaluated after 2 h on the same plates (n = 34-40, with ~70 worms per n). A negative choice index indicates a choice of (B) OP50, MYb10, MYb11, MYb53, or a buffer control; or (C) the 2 h lawn; or (D) OP50. A positive choice index indicates a choice of (B, D) MYb71 or (C) the 24 h MYb71 lawn. A choice index of 0 indicates the equal choice of bacteria on both sides. Wilcoxon signed-rank test (Wilcoxon, 1945) with false discovery rate (FDR) correction for multiple testing was applied for comparing the choice indices to 0. Data are presented in boxplots; each gray dot represents one replicate. Significance is designated to the following scale: ***p < 0.001, *p < 0.05.
Bacterial Cell Density and Worm Developmental Stage are not Responsible for Late Microbiota Avoidance Response
What might be responsible for the change in the choice behavior of MY2079 toward MYb71 over time – a phenomenon that we as of now call “late microbiota avoidance response”? In the course of the choice experiment, two main factors changed: 1) bacterial proliferation led to an increase in cell density and therefore higher concentration of bacterial products after 24 compared with 2 h. 2) At the same time, the worms developed from the L4 stage to the adult stage. We therefore compared the attraction of C. elegans MY2079 toward MYb71 in OD60010 and OD60030 (directly adjusted from a liquid culture) and toward MYb71 lawns grown on a plate for 2 or 24 h. The latter was done for L4 larvae and adults to examine developmental influences. Both L4 larvae (p < 0.001) and adults (p < 0.001) chose the 24 h MYb71 lawn over the 2 h lawn (Figure 2C). Similarly, L4 larvae of N2 (p = 0.003) and MY2079 (p = 0.014) chose MYb71 at OD60030 over OD60010 (Supplementary Figure S2).
These findings indicate that C. elegans prefers a higher MYb71 cell density in direct comparison with a lower MYb71 cell density independent of the developmental stage. However, in the presence of an alternative bacterial lawn, the MYb71 lawn after 24 h is less attractive (Figure 2B). Therefore, an increased cell density in the course of the experiment did not explain the late microbiota avoidance response toward MYb71 in the previous experiments.
The Late Microbiota Avoidance Response Is Independent of Worm-Exposed Ochrobactrum vermis MYb71
Bacterial communities are able to react to their environment, including being grazed on by bacterivores. Polynucleobacter asymbioticus, for example, changes its transcriptional profile upon grazing by chrysophytes concerning transcriptional, translational, and stress responses (Beisser et al., 2019). These transcriptional differences in bacteria and resulting changes in the availability of bacterial products might be perceived by C. elegans and consequently influence its behavior. To test the potential production of worm-repelling compounds after worm exposure, we exposed MYb71 to C. elegans in a binary-choice experiment, removed the worms after 24 h, and exposed naïve worms to the same bacteria. As seen before, the initially placed C. elegans showed the late microbiota avoidance response toward MYb71 (p < 0.001; Figure 2D). Interestingly, naïve worms did not avoid the 24-h-old, worm-exposed MYb71 but preferred this lawn over OP50 instead (p < 0.001).
These findings suggest that the production of a repellent bacterial product triggered by exposure to worms is rather unlikely.
The Late Microbiota Avoidance Response Can Be Induced by Preconditioning Worms With MYb71 and Is Specific to MYb71
As we did not observe a late microbiota avoidance response in naïve worms but in worms that have likely fed on MYb71, we hypothesized that worms had to be exposed to MYb71 to show the late microbiota avoidance response. Therefore, we preconditioned worms with MYb71 or OP50, i.e., exposed L4 worms for 24 h to either MYb71 or OP50, and subsequently performed a binary-choice experiment scoring behavior only after 2 h. Interestingly, we found the late microbiota avoidance response in preconditioned worms as in previous assays after 24 h, whereas worms preconditioned with OP50 did not show the late microbiota avoidance response (p < 0.001; Figure 3A and Supplementary Figures S3A, B). This indicates that preconditioning with MYb71 is enough to induce the late microbiota avoidance response.
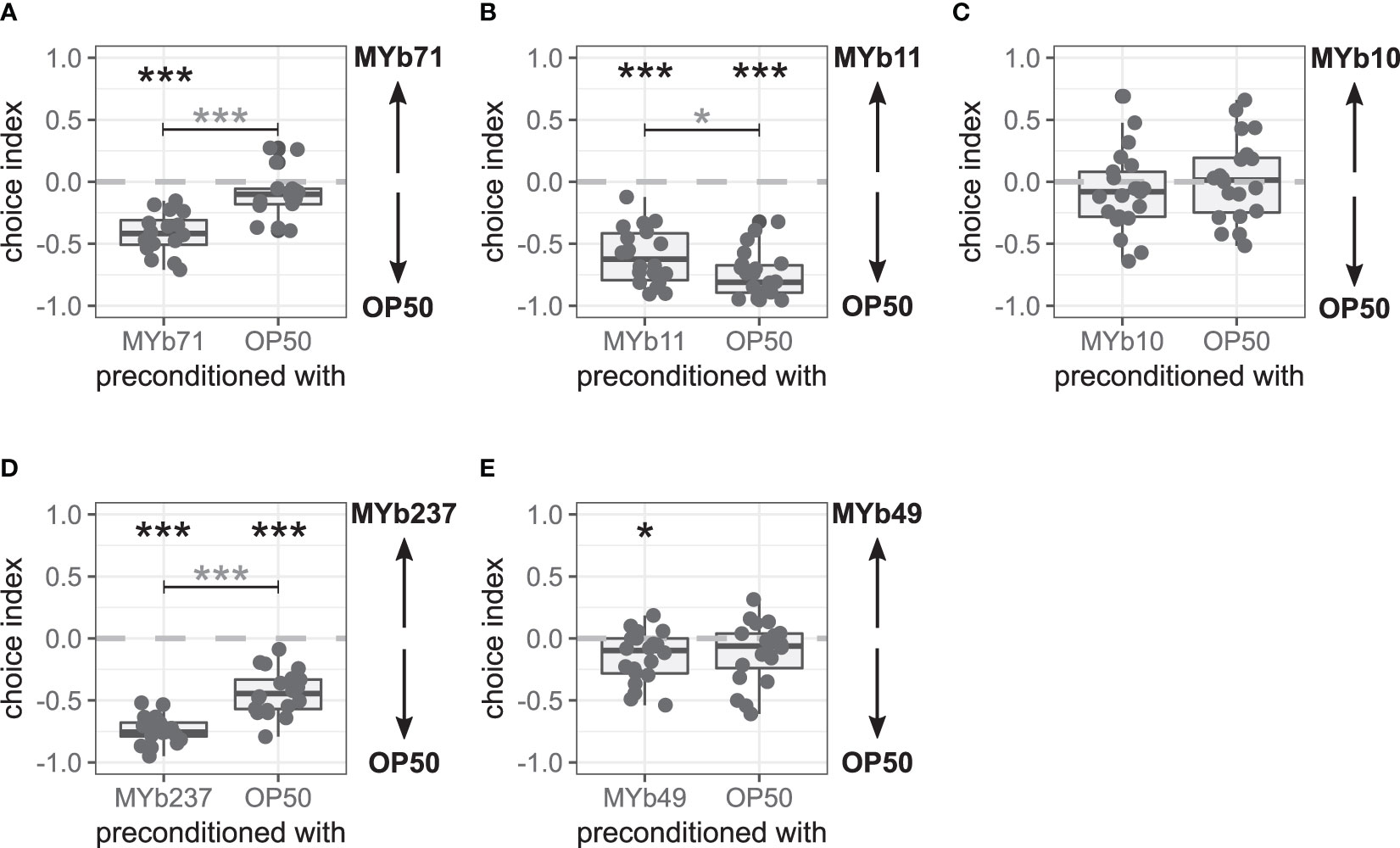
Figure 3 Late microbiota avoidance response of Caenorhabditis elegans after bacterial preconditioning is species-specific. Choice behavior of C. elegans MY2079 2 h after preconditioning with (A) OP50 or microbiota Ochrobactrum vermis MYb71 (n = 16, with ~70 worms per n), (B) Pseudomonas lurida MYb11 (n = 22, with ~70 worms per n), (C) Acinetobacter guillouiae MYb10 (n = 22, with ~70 worms per n), (D) Ochrobactrum pseudogrignonense MYb237 (n = 20, with ~70 worms per n), or (E) Ochrobactrum anthropi MYb49 (n = 20, with ~70 worms per n). Additional runs are shown in Supplementary Figure S3. Wilcoxon signed-rank test with false discovery rate (FDR) correction for multiple testing was applied for comparing the choice indices to 0. Data are presented in boxplots; each gray dot represents one replicate. A negative choice index indicates a choice of OP50, a positive choice index indicates a choice of the microbiota, and a choice index of 0 indicates the equal choice of both bacteria. Asterisks indicate a difference to a choice index of 0 (black) or a difference between preconditioning treatments (gray). Significance is designated to the following scale: ***p < 0.001, *p < 0.05.
We further wondered whether the choice behavior changed due to general intestinal colonization with microbiota and examined the choice behavior after preconditioning with two other representatives of colonizing microbiota, P. lurida MYb11 and A. guillouiae MYb10. C. elegans MY2079 chose OP50 over MYb11 independent of the preconditioning (p < 0.001; Figure 3B and Supplementary Figures S3C, D). However, worms preconditioned with MYb11 chose MYb11 slightly more compared with worms preconditioned with OP50 (p = 0.014). Preconditioning with MYb10 had no effect on choice behavior, and MYb10 and OP50 were always chosen equally (Figure 3C and Supplementary Figures S3E, F).
We used two additional Ochrobactrum microbiota isolates, O. pseudogrignonense MYb237 and O. anthropi MYb49, to test for a genus-specific preconditioning effect on choice behavior. C. elegans MY2079 chose OP50 over MYb237 independent of the preconditioning (p < 0.001), but OP50 was even more preferred in worms preconditioned with MYb237 (p < 0.001; Figure 3D and Supplementary Figure S3G). Preconditioning with MYb49 led to the choice of OP50 over MYb49 (p = 0.014), whereas worms preconditioned with OP50 chose OP50 and MYb49 equally in two out of three runs (Figure 3E and Supplementary Figures S3H, I). Overall, preconditioning with MYb49 compared with preconditioning with OP50 had no effect on the choice behavior.
These results support that preconditioning with colonizing microbiota can influence C. elegans choice behavior. However, neither colonization with microbiota nor preconditioning with bacteria of the genus Ochrobactrum resulted in a late avoidance response of the microbiota, in which avoidance occurred only after preconditioning with the test bacterium. Hence, the late microbiota avoidance response appears to be specific for O. vermis MYb71.
The Late Microbiota Avoidance Response Is Independent of Bacterial RNA but Dependent on MYb71 Colonization of the Worm
Pathogenic bacteria, bacteria with insufficient nutritional value, or bacteria that negatively affect fitness in any other way can be avoided by C. elegans in order to reduce their uptake (Schulenburg and Müller, 2004; Meisel and Kim, 2014). Since we observed avoidance of MYb71, we evaluated whether the late microbiota avoidance response is linked to potential pathogenicity of MYb71. We found neither any lawn leaving behavior from MYb71 (Supplementary Figure S4A) nor a detrimental effect of MYb71 on the worm’s fitness (Supplementary Figure S4B). These observations suggest that MYb71 does not have a pathogenic effect on C. elegans.
Even without obvious pathogenicity of MYb71 to C. elegans, the question still remains whether C. elegans displays learned avoidance behavior mediated by bacterial products or whether gut colonization is necessary to elicit the worm’s late microbiota avoidance response.
Recently, the learned pathogen avoidance behavior of C. elegans against Pseudomonas aeruginosa PA14 has been demonstrated to depend on a single non-coding RNA derived from PA14 (Kaletsky et al., 2020; Moore et al., 2021). Hence, we tested the influence of preconditioning with total RNA of MYb71 or OP50 on choice behavior and found that worms from both treatments chose OP50 over MYb71 in a binary choice assay (p = 0.044, Figure 4A).
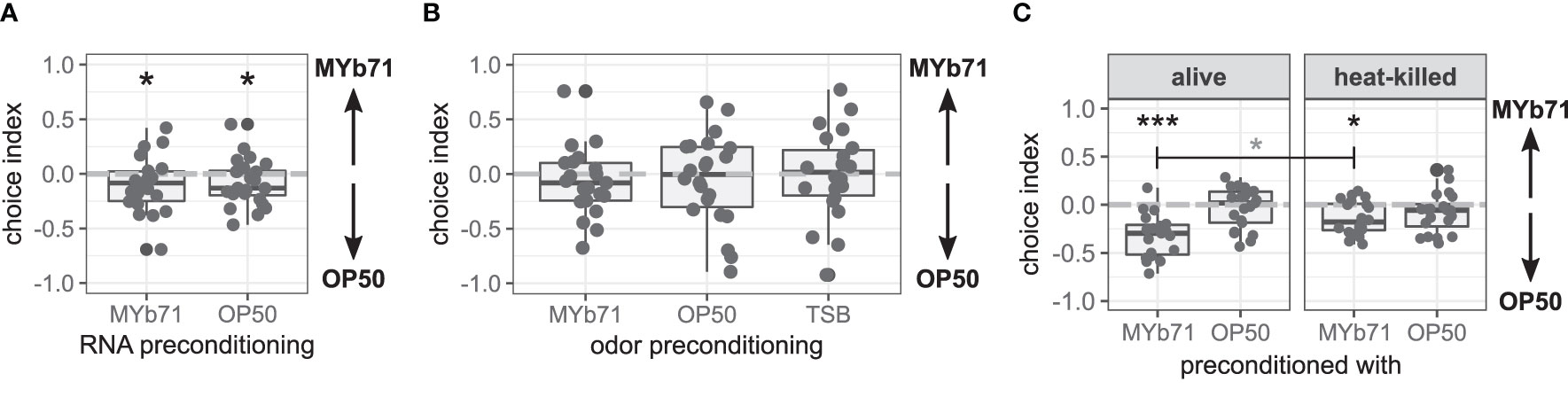
Figure 4 Late microbiota avoidance response is independent of total RNA of MYb71 or MYb71 odor but is more pronounced with viable MYb71. (A) Choice behavior after preconditioning with RNA of MYb71 or OP50 applied to viable OP50 (n = 24, with ~70 worms per n); or (B) after preconditioning with the odor of MYb71, OP50, or TSB (n = 22, with ~70 worms per n); or (C) after preconditioning with alive or heat-killed MYb71 or OP50 (n = 20, with ~70 worms per n). A negative choice index indicates the choice of OP50, a positive choice index indicates the choice of MYb71, and a choice index of 0 indicates the equal choice of both bacteria. Wilcoxon signed-rank test, false discovery rate (FDR)-corrected. Data are presented in boxplots; each gray dot represents one replicate. Asterisks indicate a difference to a choice index of 0 (black) or difference between treatments (gray). Significance is designated to the following scale: ***p < 0.001, *p < 0.05.
The learned pathogen avoidance response against PA14 is further mediated by bacteria-derived odors (Zhang et al., 2005). Here, worms preconditioned with MYb71 odor chose MYb71 and OP50 equally independent of the bacteria used for odor preconditioning (Figure 4B).
To answer the question of whether C. elegans needs to be colonized by MYb71, we tested the late microbiota avoidance response of worms preconditioned with either heat-killed bacteria or viable bacteria. Both worms preconditioned with viable (p < 0.001) and heat-killed MYb71 (p = 0.011) chose OP50 over MYb71 (Figure 4C). However, worms preconditioned with viable MYb71 resided more often on OP50 lawns than worms preconditioned with heat-killed MYb71 (p = 0.022). Worms from OP50 chose MYb71 and OP50 equally, independent of the preconditioning.
These results indicate that the late microbiota avoidance response is not caused by total bacterial RNA nor bacteria-derived odors but primarily by the worm’s colonization with viable MYb71.
Discussion
In the present study, we examined the choice behavior of the bacterivore nematode C. elegans toward its natural microbiota. We found that the natural C. elegans isolate MY2079 avoided specifically the microbiota isolate O. vermis MYb71 during a second exposure.
We observed the microbiota avoidance response of C. elegans MY2079 toward O. vermis MYb71 only either over time or during a second encounter. This behavior is reminiscent of the learned avoidance behavior in C. elegans against the pathogenic P. aeruginosa PA14 (Zhang et al., 2005). C. elegans trained for at least 4 h on PA14 lawns avoids PA14 during a second encounter more than untrained animals. On the host side, a complex interaction of TGFβ signaling (Zhang and Zhang, 2012; Meisel et al., 2014; Moore et al., 2019; Singh and Aballay, 2019), serotonin signaling (Zhang et al., 2005), and RNAi and piRNA signaling (Kaletsky et al., 2020) mediates the learned avoidance behavior. On the microbe side, bacterial secondary metabolites (Meisel et al., 2014) and non-coding RNAs (Kaletsky et al., 2020) trigger the learned behavioral response – in the case of non-coding RNA even across multiple worm generations (Kaletsky et al., 2020). Interestingly, so far only pathogenic bacteria such as PA14, Serratia marcescens ATCC 13880 (Zhang et al., 2005; Zhang and Zhang, 2012), or Enterococcus faecalis (Filipowicz et al., 2021) were known to elicit the learned avoidance behavior. Ochrobactrum, however, has been repeatedly isolated from wild C. elegans and has positive or neutral effects on C. elegans life-history traits such as population growth (Zimmermann et al., 2020), animal growth rates, and body size (Dirksen et al., 2020; Zhang et al., 2021). Ochrobactrum is therefore considered to be part of the natural, benign C. elegans microbiome (Berg et al., 2016a; Dirksen et al., 2016; Dirksen et al., 2020). The absence of lawn leaving behavior and lack of negative effects on brood size of C. elegans MY2079 presented in our study together with the mentioned published data support the benign nature of MYb71. Moreover, we excluded the small RNA-mediated mechanism of learned pathogen avoidance behavior (Kaletsky et al., 2020). Our finding is, hence, the first demonstration of an altered avoidance response after the first encounter as a reaction to a benign microbiota member. To explore to what extent such an avoidance response might benefit C. elegans MY2079 would be an interesting future direction.
Gut colonization with certain bacteria has been shown to alter C. elegans behavior in the context of microbiome bacteria: gut-colonizing Providencia bacteria manipulate host behavior by the production of the neuromodulator tyramine. Consequently, these bacteria are preferentially selected in food choice assays if C. elegans is colonized by Providencia (O’Donnell et al., 2020). Comparing preconditioning with heat-killed and viable MYb71, we showed that the avoidance was reduced when MYb71 was metabolically inactive. Therefore, bacteria-derived products could play a role in our observed late microbiota avoidance response. Further, O. vermis MYb71 colonizes and persists very efficiently in C. elegans (Dirksen et al., 2016; Zimmermann et al., 2020; Pees et al., 2021), providing the basis for host–microbe communication. Colonization by viable MYb71 is important for the late microbiota avoidance response because the worms showed no late avoidance response after exposure to MYb71 odor and reduced avoidance after exposure to heat-killed MYb71. As host response, MY2079 may synthesize substances responsible for the late microbiota avoidance response only when its gut is colonized by MY71. Moreover, the colonization with MYb71 might lead to a gut distention provoking the late microbiota avoidance response. The learned pathogen avoidance response against E. faecalis is caused by its accumulation in the anterior worm gut mediated by two transient receptor potential melastatin channels – even with an attenuated pathogen (Filipowicz et al., 2021).
Bacterial colonization of the C. elegans gut alone is certainly not the only trigger for the late microbiota avoidance response, as none of the other tested colonizing microbiota members phenocopied the response identified toward MYb71. The late microbiota avoidance response exhibits a very specific host–microbe interaction whose exchanged information and communicational signals remain to be discovered.
On the host side, the late microbiota avoidance response was specific for one natural C. elegans isolate, MY2079. In our initial multiple-choice screen, neither laboratory-adapted wild-type N2 nor the other 10 tested natural isolates showed significant choice behavior, i.e., did not prefer one of the offered bacterial spots over the others. However, the results of the multiple-choice screen are based on a relatively small number of replicates and many multiple comparisons. For the follow-up experiments, the number of replicates was greatly increased, and the number of comparisons was reduced, which then even led to significant choice behavior in N2, yet only on non-proliferating bacteria. The differences in choice behavior among the wild isolates exemplify the phenotypic diversity in C. elegans wild isolates as previously described (Petersen et al., 2015). The more profound differences in the behavioral response between N2 and MY2079 toward MYb71, however, can be attributed to the adaptation of N2 to laboratory conditions. Decades of maintenance on solely E. coli OP50 made the ability to choose specific microbiota bacteria increasingly redundant. A mutation in the G protein-coupled neuropeptide receptor gene npr-1, which leads to a solitary, non-bordering phenotype in N2 compared with a clumping and bordering phenotype in natural C. elegans isolates (de Bono and Bargmann, 1998; Reddy et al., 2009; Weber et al., 2010; Andersen et al., 2014), is only one example of the adaption of N2 to the laboratory.
In conclusion, we found a novel avoidance response of the natural C. elegans isolate MY2079 specifically toward the microbiota isolate O. vermis MYb71. The exact factors of this inter-kingdom communication remain to be determined. Our findings, however, might initiate further research on how the behavior of C. elegans is influenced by its natural gut microbiota.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
CP: conceptualization, data curation, formal analysis, investigation, methodology, visualization, supervision of CMC, writing – original draft preparation, and writing – review and editing. BP: conceptualization, data curation, formal analysis, investigation, methodology, writing – original draft preparation, and writing – review and editing. CMC: investigation and methodology. ML: resources and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
The work was funded by the German Science Foundation within the Collaborative Research Center CRC 1182 on Origin and Function of Metaorganisms (project A1.3 to ML and project A1.1 funding for CP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Heidrun Ließegang for technical support, Daniel Schütz for help with statistical analyses, and Katja Dierking for her advice on the manuscript. We thank the Caenorhabditis Genetics Center (CGC), which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440), for originally providing the standard C. elegans laboratory strain N2.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.775634/full#supplementary-material
References
Andersen, E. C., Bloom, J. S., Gerke, J. P., Kruglyak, L. (2014). A Variant in the Neuropeptide Receptor Npr-1 Is a Major Determinant of Caenorhabditis elegans Growth and Physiology. PLoS Genet. 10, e1004156. doi: 10.1371/journal.pgen.1004156
Arentsen, T., Raith, H., Qian, Y., Forssberg, H., Heijtz, R. D. (2015). Host Microbiota Modulates Development of Social Preference in Mice. Microb. Ecol. Health Dis. 26. doi: 10.3402/mehd.v26.29719
Avery, L., Horvitz, H. R. (1990). Effects of Starvation and Neuroactive Drugs on Feeding in Caenorhabditis elegans. J. Exp. Zool. 253, 263–270. doi: 10.1002/jez.1402530305
Bargmann, C. I., Horvitz, H. R. (1991). Chemosensory Neurons With Overlapping Functions Direct Chemotaxis to Multiple Chemicals in C. elegans. Neuron 7, 729–742. doi: 10.1016/0896-6273(91)90276-6
Beisser, D., Bock, C., Hahn, M. W., Vos, M., Sures, B., Rahmann, S., et al. (2019). Interaction-Specific Changes in the Transcriptome of Polynucleobacter Asymbioticus Caused by Varying Protistan Communities. Front. Microbiol. 10, 1498. doi: 10.3389/fmicb.2019.01498
Benjamini, Y., Hochberg, Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc: Ser. B (Methodological) 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Berg, M., Stenuit, B., Ho, J., Wang, A., Parke, C., Knight, M., et al. (2016a). Assembly of the Caenorhabditis elegans Gut Microbiota From Diverse Soil Microbial Environments. ISME J. 10, 1998–2009. doi: 10.1038/ismej.2015.253
Berg, M., Zhou, X. Y., Shapira, M. (2016b). Host-Specific Functional Significance of Caenorhabditis Gut Commensals. Front. Microbiol. 7, 1622. doi: 10.3389/fmicb.2016.01622
de Bono, M., Bargmann, C. I. (1998). Natural Variation in a Neuropeptide Y Receptor Homolog Modifies Social Behavior and Food Response in C. elegans. Cell 94, 679–689. doi: 10.1016/S0092-8674(00)81609-8
Dirksen, P., Assié, A., Zimmermann, J., Zhang, F., Tietje, A.-M., Marsh, S. A., et al. (2020). CeMbio - The Caenorhabditis elegans Microbiome Resource. G3 Genes|Genomes|Genetics 10, 3025–3039. doi: 10.1534/g3.120.401309
Dirksen, P., Marsh, S. A., Braker, I., Heitland, N., Wagner, S., Nakad, R., et al. (2016). The Native Microbiome of the Nematode Caenorhabditis elegans: Gateway to a New Host-Microbiome Model. BMC Biol. 14. doi: 10.1186/s12915-016-0258-1
Félix, M.-A., Braendle, C. (2010). The Natural History of Caenorhabditis elegans. Curr. Biol. 20, R965–R969. doi: 10.1016/j.cub.2010.09.050
Filipowicz, A., Lalsiamthara, J., Aballay, A. (2021). TRPM Channels Mediate Learned Pathogen Avoidance Following Intestinal Distention. eLife 10, e65935. doi: 10.7554/eLife.65935
Fischer, C. N., Trautman, E. P., Crawford, J. M., Stabb, E. V., Handelsman, J., Broderick, N. A. (2017). Metabolite Exchange Between Microbiome Members Produces Compounds That Influence Drosophila Behavior. eLife 6. doi: 10.7554/eLife.18855
Frézal, L., Félix, M.-A. (2015). C. elegans Outside the Petri Dish. eLife 4, e05849. doi: 10.7554/eLife.05849
Grewal, P. S., Wright, D. J. (1992). Migration of Caenorhabditis elegans (Nematoda: Rhabditidae) Larvae Towards Bacteria and the Nature of the Bacterial Stimulus. Fundam. Appl. Nematol. 15, 8.
Haber, M. (2005). Evolutionary History of Caenorhabditis elegans Inferred From Microsatellites: Evidence for Spatial and Temporal Genetic Differentiation and the Occurrence of Outbreeding. Mol. Biol. Evol. 22, 160–173. doi: 10.1093/molbev/msh264
Horvitz, H., Chalfie, M., Trent, C., Sulston, J., Evans, P. (1982). Serotonin and Octopamine in the Nematode Caenorhabditis elegans. Science 216, 1012–1014. doi: 10.1126/science.6805073
Kaletsky, R., Moore, R. S., Vrla, G. D., Parsons, L. R., Gitai, Z., Murphy, C. T. (2020). Caenorhabditis elegans Interprets Bacterial Non-Coding RNAs to Learn Pathogenic Avoidance. Nature 586, 445–451. doi: 10.1038/s41586-020-2699-5
Kissoyan, K. A. B., Drechsler, M., Stange, E.-L., Zimmermann, J., Kaleta, C., Bode, H. B., et al. (2019). Natural C. elegans Microbiota Protects Against Infection via Production of a Cyclic Lipopeptide of the Viscosin Group. Curr. Biol. 29, 1–8. doi: 10.1016/j.cub.2019.01.050
Kruskal, W. H., Wallis, W. A. (1952). Use of Ranks in One-Criterion Variance Analysis. J Am Stat Assoc 47, 583–621. doi: 10.1080/01621459.1952.10483441
Luo, Y., Zeng, B., Zeng, L., Du, X., Li, B., Huo, R., et al. (2018). Gut Microbiota Regulates Mouse Behaviors Through Glucocorticoid Receptor Pathway Genes in the Hippocampus. Trans. Psychiatry 8. doi: 10.1038/s41398-018-0240-5
Meisel, J. D., Kim, D. H. (2014). Behavioral Avoidance of Pathogenic Bacteria by Caenorhabditis elegans. Trends Immunol. 35, 465–470. doi: 10.1016/j.it.2014.08.008
Meisel, J. D., Panda, O., Mahanti, P., Schroeder, F. C., Kim, D. H. (2014). Chemosensation of Bacterial Secondary Metabolites Modulates Neuroendocrine Signaling and Behavior of C. elegans. Cell 159, 267–280. doi: 10.1016/j.cell.2014.09.011
Montalvo-Katz, S., Huang, H., Appel, M. D., Berg, M., Shapira, M. (2013). Association With Soil Bacteria Enhances p38-Dependent Infection Resistance in Caenorhabditis elegans. Infect. Immun. 81, 514–520. doi: 10.1128/IAI.00653-12
Moore, R. S., Kaletsky, R., Murphy, C. T. (2019). Piwi/PRG-1 Argonaute and TGF-β Mediate Transgenerational Learned Pathogenic Avoidance. Cell 177, 1827–1841.e12. doi: 10.1016/j.cell.2019.05.024
Moore, R. S., Kaletsky, R., Murphy, C. T. (2021). Protocol for Transgenerational Learned Pathogen Avoidance Behavior Assays in Caenorhabditis elegans. STAR Protoc. 2, 100384. doi: 10.1016/j.xpro.2021.100384
O’Donnell, M. P., Fox, B. W., Chao, P.-H., Schroeder, F. C., Sengupta, P. (2020). A Neurotransmitter Produced by Gut Bacteria Modulates Host Sensory Behaviour. Nature 583, 415–420. doi: 10.1038/s41586-020-2395-5
Pees, B., Johnke, J., Möhl, M., Hamerich, I. K., Leippe, M., Petersen, C. (2021). Microbes to-go: Slugs as Source for Caenorhabditis elegans Microbiota Acquisition. Environ. Microbiol. 23 (11), 6721–6733. doi: 10.1111/1462-2920.15730
Petersen, C., Dirksen, P., Prahl, S., Strathmann, E., Schulenburg, H. (2014). The Prevalence of Caenorhabditis elegans Across 1.5 Years in Selected North German Locations: The Importance of Substrate Type, Abiotic Parameters, and Caenorhabditis Competitors. BMC Ecol. 14, 4. doi: 10.1186/1472-6785-14-4
Petersen, C., Saebelfeld, M., Barbosa, C., Pees, B., Hermann, R. J., Schalkowski, R., et al. (2015). Ten Years of Life in Compost: Temporal and Spatial Variation of North German Caenorhabditis elegans Populations. Ecol. Evol. 5, 3250–3263. doi: 10.1002/ece3.1605
Pradhan, S., Quilez, S., Homer, K., Hendricks, M. (2019). Environmental Programming of Adult Foraging Behavior in C. elegans. Curr. Biol. 29, 2867–2879.e4. doi: 10.1016/j.cub.2019.07.045
Qiao, H., Keesey, I. W., Hansson, B. S., Knaden, M. (2019). Gut Microbiota Affects Development and Olfactory Behavior in Drosophila melanogaster. J. Exp. Biol. 222, jeb192500. doi: 10.1242/jeb.192500
Reddy, K. C., Andersen, E. C., Kruglyak, L., Kim, D. H. (2009). A Polymorphism in npr-1 Is a Behavioral Determinant of Pathogen Susceptibility in C. elegans. Science 323 (5912), 382–384. doi: 10.1126/science.1166527
Samuel, B. S., Rowedder, H., Braendle, C., Félix, M.-A., Ruvkun, G. (2016). Caenorhabditis elegans Responses to Bacteria From Its Natural Habitats. Proc. Natl. Acad. Sci. 113, E3941–E3949. doi: 10.1073/pnas.1607183113
Schulenburg, H., Müller, S. (2004). Natural Variation in the Response of Caenorhabditis elegans Towards Bacillus thuringiensis. Parasitology 128, 433–443. doi: 10.1017/S003118200300461X
Shtonda, B. B., Avery, L. (2006). Dietary Choice Behavior in Caenorhabditis elegans. J. Exp. Biol. 209, 89–102. doi: 10.1242/jeb.01955
Singh, J., Aballay, A. (2019). Intestinal Infection Regulates Behavior and Learning via Neuroendocrine Signaling. eLife 8, e50033. doi: 10.7554/eLife.50033
Stiernagle, T. (2006). Maintenance of C. elegans," in The C. elegans Research Community, WormBook. ed. WormBook. doi: 10.1895/wormbook.1.101.1
Volkers, R. J., Snoek, L., Hubar, C. J., van, H., Coopman, R., Chen, W., et al. (2013). Gene-Environment and Protein-Degradation Signatures Characterize Genomic and Phenotypic Diversity in Wild Caenorhabditis elegans Populations. BMC Biol. 11, 93. doi: 10.1186/1741-7007-11-93
Weber, K. P., De, S., Kozarewa, I., Turner, D. J., Babu, M. M., de Bono, M. (2010). Whole Genome Sequencing Highlights Genetic Changes Associated With Laboratory Domestication of C. elegans. PLoS One 5, e13922. doi: 10.1371/journal.pone.0013922
Wilcoxon, F. (1945). Individual Comparisons by Ranking Methods. Biom. Bull. 1, 80–83. doi: 10.2307/3001968
Wong, A. C.-N., Wang, Q.-P., Morimoto, J., Senior, A. M., Lihoreau, M., Neely, G. G., et al. (2017). Gut Microbiota Modifies Olfactory-Guided Microbial Preferences and Foraging Decisions in Drosophila. Curr. Biol. 27, 2397–2404.e4. doi: 10.1016/j.cub.2017.07.022
Yang, W., Petersen, C., Pees, B., Zimmermann, J., Waschina, S., Dirksen, P., et al. (2019). The Inducible Response of the Nematode Caenorhabditis elegans to Members of Its Natural Microbiota Across Development and Adult Life. Front. Microbiol. 10, 1793. doi: 10.3389/fmicb.2019.01793
Zhang, Y., Lu, H., Bargmann, C. I. (2005). Pathogenic Bacteria Induce Aversive Olfactory Learning in Caenorhabditis elegans. Nature 438, 179–184. doi: 10.1038/nature04216
Zhang, F., Weckhorst, J. L., Assié, A., Hosea, C., Ayoub, C. A., Khodakova, A. S., et al. (2021). Natural Genetic Variation Drives Microbiome Selection in the Caenorhabditis elegans Gut. Curr. Biol. 31, 2603–2618.e9. doi: 10.1016/j.cub.2021.04.046
Zhang, X., Zhang, Y. (2012). DBL-1, a TGF-β, Is Essential for Caenorhabditis elegans Aversive Olfactory Learning. PNAS 109, 17081–17086. doi: 10.1073/pnas.1205982109
Keywords: microbiota, C. elegans natural isolate MY2079, choice behavior, avoidance response, Ochrobactrum vermis MYb71
Citation: Petersen C, Pees B, Martínez Christophersen C and Leippe M (2021) Preconditioning With Natural Microbiota Strain Ochrobactrum vermis MYb71 Influences Caenorhabditis elegans Behavior. Front. Cell. Infect. Microbiol. 11:775634. doi: 10.3389/fcimb.2021.775634
Received: 14 September 2021; Accepted: 23 November 2021;
Published: 17 December 2021.
Edited by:
Michael A. Herman, University of Nebraska-Lincoln, United StatesReviewed by:
Yonghong Liu, Chinese Academy of Sciences, ChinaNatalia V. Kirienko, Rice University, United States
Roberto Ricardo Grau, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina
Copyright © 2021 Petersen, Pees, Martínez Christophersen and Leippe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carola Petersen, Y3BldGVyc2VuQHpvb2xvZ2llLnVuaS1raWVsLmRl
†Present address: Barbara Pees, Department of Integrative Biology, UC Berkeley, Berkeley, CA, United States
 Carola Petersen
Carola Petersen Barbara Pees†
Barbara Pees† Matthias Leippe
Matthias Leippe