- 1Intrahospital Infections Laboratory, National Centre for Microbiology, Instituto de Salud Carlos III (ISCIII), Majadahonda, Spain
- 2Departamento de Biotecnología Microbiana y de Plantas, Centro de Investigaciones Biológicas Margarita Salas (CSIC), Madrid, Spain
- 3Centro de Investigación Biomédica en Red de Enfermedades Respiratorias (CIBERES), Madrid, Spain
Bacteriophages (phages) are viruses that infect bacteria. They are the most abundant biological entity on Earth (current estimates suggest there to be perhaps 1031 particles) and are found nearly everywhere. Temperate phages can integrate into the chromosome of their host, and prophages have been found in abundance in sequenced bacterial genomes. Prophages may modulate the virulence of their host in different ways, e.g., by the secretion of phage-encoded toxins or by mediating bacterial infectivity. Some 70% of Streptococcus pneumoniae (the pneumococcus)—a frequent cause of otitis media, pneumonia, bacteremia and meningitis—isolates harbor one or more prophages. In the present study, over 4000 S. pneumoniae genomes were examined for the presence of prophages, and nearly 90% were found to contain at least one prophage, either defective (47%) or present in full (43%). More than 7000 complete putative integrases, either of the tyrosine (6243) or serine (957) families, and 1210 full-sized endolysins (among them 1180 enzymes corresponding to 318 amino acid-long N-acetylmuramoyl-L-alanine amidases [LytAPPH]) were found. Based on their integration site, 26 different pneumococcal prophage groups were documented. Prophages coding for tRNAs, putative virulence factors and different methyltransferases were also detected. The members of one group of diverse prophages (PPH090) were found to integrate into the 3’ end of the host lytASpn gene encoding the major S. pneumoniae autolysin without disrupting it. The great similarity of the lytASpnand lytAPPH genes (85–92% identity) allowed them to recombine, via an apparent integrase-independent mechanism, to produce different DNA rearrangements within the pneumococcal chromosome. This study provides a complete dataset that can be used to further analyze pneumococcal prophages, their evolutionary relationships, and their role in the pathogenesis of pneumococcal disease.
Introduction
Bacteriophages (phages) are viruses that infect bacteria. They are the most abundant biological entities on Earth — current estimates suggest there to be close to 1031 phage particles (Mushegian, 2020) — and can be found nearly everywhere. Temperate bacteriophages infect and kill bacteria to release phage progeny, but on occasion they may integrate into the host genome via site‐specific recombination events, and replicate vertically. Integrated phages are stably maintained in the chromosome (i.e., as a prophage) in a state known as lysogeny (Lwoff, 1953). Campbell (1962) was the first to propose a model for the integration of λ prophage into the bacterial chromosome. This model consists of two phases: (1) the circularization of the linear phage DNA molecule injected into the cell, and (2) the linear insertion of the phage DNA into the bacterial chromosome via the activity of a specific integrase (Int) that catalyzes the site-specific recombination of the phage attachment site (attP) and bacterial attachment site (attB). These two sequences generally share a short stretch of identical bases (the core sequence) where this site-specific recombination occurs. After recombination, the phage genome is left integrated into the bacterial chromosome, flanked by two duplicated hybrid att sites: attL and attR. Under certain circumstances (prophage induction), the prophage becomes excised from the bacterial chromosome and viral replication begins via the lytic cycle.
Endolysins are phage-encoded enzymes capable of hydrolyzing the bacterial cell wall; they are synthesized at the end of the lytic cycle to allow the release of phage progeny. In this era of global increase in antibacterial resistance, endolysins are being tested as an alternative (or complement) to the use of antibiotics (for recent reviews see Vázquez et al., 2018; Fernández et al., 2021; Murray et al., 2021). It is well documented that, while integrated into the host genome, genes encoding Ints and endolysins are located at either end of Streptococcus prophages (Canchaya et al., 2003).
Prophages are often found in sequenced bacterial genomes. Indeed, nearly half of bacterial genomes appear to contain at least one prophage. The minimum doubling time is the trait most strongly correlated with lysogeny, followed by genome size and, interestingly, pathogenicity (Touchon et al., 2016).
Streptococcus pneumoniae (the pneumococcus) is a major human pathogen and a frequent cause of non-invasive diseases such as otitis, conjunctivitis and pneumonia, but also life-threatening invasive sepsis, bacteremic pneumonia, and meningitis. Pneumococcal pneumonia ranks first in terms of associated mortality among all lower respiratory tract diseases, and is responsible for more than one million deaths every year (GBD 2016 Lower Respiratory Infections Collaborators, 2018). That prophages may contribute to bacterial virulence is well established, particularly in Pseudomonas aeruginosa, Salmonella enterica, Escherichia coli, Vibrio cholerae, Staphylococcus spp., and Clostridium spp. (Schroven et al., 2021). Prophages can alter the phenotype of their hosts at different levels, e.g., by causing them to secrete toxins, by modifying the bacterial envelope, and/or impacting bacterial infectivity and bacterial cell regulation.
High-throughput next generation sequencing techniques have now provided numerous de novo sequenced and assembled bacterial genomes, and over 8000 pneumococcal genomes are currently included in the NCBI Reference Sequence Database (RefSeq) database. However, while first reported in 1977 and suggested to be widespread (Bernheimer, 1977; Ramirez et al., 1999), pneumococcal prophages (PPHs) have remained relatively unexamined — that is until recently (García et al., 2005). Based on comparisons of complete prophages and their predicted encoded proteins, PPHs have been grouped into three major groups with only a small number of prophages falling outside these classes (Romero et al., 2009a; Romero et al., 2009b). More recently, pairwise comparisons of prophage sequences showed four major prophage clusters and one single prophage (Brueggemann et al., 2017). Phages belonging to the same phylogenetic group share high sequence similarity in their packaging, morphology, and lysis modules, and are typically associated with one or two main integrase types. Besides, incomplete pneumococcal prophages can be highly conserved over long periods of time and clustered into five major groups that differed from those of intact PPHs (Rezaei Javan et al., 2019).
In 2017, a pan-genome-wide association study identified PPHs as being associated with reduced pneumococcal carriage duration, although this was attributed more to the disruption caused by the integration of the phage genome and the genetic transformation competence system than to any property of the prophage itself (Lees et al., 2017). Other data have shown that a role for prophages in pneumococcal virulence and patient mortality is linked to PblA and/or PblB, two prophage-encoded proteins known to be involved in enhanced platelet activation together with higher formation of platelet-monocyte complexes (Tunjungputri et al., 2017). Prophages may naturally enter the lytic cycle, or be induced to do so via exposure to fluoroquinolones (López et al., 2014) (with a concomitant increase in PblA/PblB expression). A recent study involving patients with invasive pneumococcal disease determined that the 30-day mortality of pneumococcal meningitis was 11% in pblB-positive patients compared to <1% in pblB-negative patients, although the authors recognize that this finding does not prove causality (Cremers et al., 2019). It has also recently been suggested that incomplete PPHs and certain prophage genes may be involved in pneumococcal pathogenesis (Rezaei Javan et al., 2019). In addition, transcriptomic analyses have shown that a defective PPH may serve as a switch that controls the expression of a bacterial gene (ychF) located immediately downstream of the prophage int gene that is involved in nasopharyngeal colonization (Chen et al., 2019). A review is available on the recent advances in the genomic and functional characterization of pneumococcal temperate phages, and their contribution to pneumococcal pathogenesis and genome evolution (Garriss and Henriques-Normark, 2020).
In the present study, the genomic sequences of over 4000 S. pneumoniae isolates from diverse lineages were examined to obtain the broadest possible picture of PPH diversity. The precise integration sites and the att core sequences of most of them were elucidated and used to identify the families to which a number of previously reported but incompletely studied PPHs belong. In addition, recombination between prophage and host lytA genes is shown to be an important source of chromosomal rearrangement in S. pneumoniae.
Materials and Methods
Compilation of the Pneumococcal Genome Dataset
This study was performed using a dataset mined from the National Center for Biotechnology Information (NCBI) database (available at https://www.ncbi.nlm.nih.gov/genome/?term=Streptococcus+pneumoniae). The latter contains whole-genome sequences (assembled or otherwise) for more than 8500 pneumococcal genomes (last accessed January 30, 2021). Sequence types (STs) (Enright and Spratt, 1998; Jolley et al., 2018) of the strains included in the dataset were determined on the basis of whole genome sequencing data (Larsen et al., 2012).
Search for Pneumococcal Prophages
To detect PPHs, the dataset was searched, using the BLAST platform (Johnson et al., 2008), for homologs of int- and lytAPPH-like genes encoding (respectively) Ints and endolysins [with respect to the latter, only N-acetylmuramoyl-L-alanine amidases (NAM-amidases; EC 3.5.1.28) of the Amidase_2 family (Pfam database identifier: PF01510) (Morales et al., 2010) are currently known]. The corresponding genes/proteins of previously characterized PPHs, such as MM1 (Obregón et al., 2003a), фSpn_OXC, фSpn_6, and фSpn_18 (Romero et al., 2009a), and others (Càmara et al., 2018). The endolysins encoded by various virulent pneumococcal phages (see below) were used as query sequences. The original and newly found genes/proteins with a coverage ≥ 90% and ≥ 50% identity were used in searches in an iterative manner.
Bioinformatic Analyses
Sequence comparison and alignments were performed using the BLAST platform and/or Clustal Omega package (Sievers and Higgins, 2021) running at the European Bioinformatics Institute (EMBL-EBI) website (https://www.ebi.ac.uk). Potential tRNAs genes were identified using ARAGORN (Laslett and Canback, 2004) and tRNAscan-SE (Schattner et al., 2005) software. Detections were recorded only when both programs coincided in the results returned. The codon usage of S. pneumoniae D39 was obtained from the corresponding database (Nakamura et al., 2000). Protein domains were preliminarily identified using CDD/SPARCLE (Lu et al., 2020). PHASTER (running at https://phaster.ca/) was also employed to identify phage-homologous regions in bacterial genomes (Arndt et al., 2016). To ascertain equivalent detection of protein datasets, open reading frames were identified and translated from prophage sequences using Prodigal v2.6.3 (Hyatt et al., 2010). Prophage proteomes were then compared all against all through the calculation of the weighted gene repertoire relatedness (wGRR) (Pfeifer et al., 2021). Best bi-directional hits were found from hits detected by the ‘easy-search’ workflow of MMseqs2 (Mirdita et al., 2019) with e-value < 10−4, identity ≥ 35%, coverage ≥ 50% thresholds. Agglomerative hierarchical clustering from the resulting wGRR matrix was carried out by the linkage tool of the scipy.cluster.hierarchy python library using the ‘ward’ method on euclidean distances.
Results
Temperate Bacteriophages Are Abundant in S. pneumoniae
The dataset produced contained 4003 strains, including 126 whose genomes were sequenced to complete or near-complete (chromosome) assembly level and deposited in the RefSeq database (O'Leary et al., 2016) (last accessed, October 11, 2020) (labeled in green in Table S1). The strains included in the dataset represent 447 different STs. Moreover, up to 50% of the strains belonged to one of 35 (out of a possible 43) Pneumococcal Molecular Epidemiology Network (PMEN) clones [including single and double locus variants (McGee et al., 2001)]. PMEN clones are resistant to one or more antibiotics in wide clinical use and dominate the population of antibiotic-resistant pneumococci. Globally susceptible clones known to be important in disease are also included in the PMEN clone dataset (https://www.pneumogen.net/pmen/, last accessed September 15, 2020). Further, 2557 strains in the dataset were assigned to one of the 169 Global Pneumococcal Sequence Clusters (GPSCs) and, among these strains, 1096 could be classified as belonging to one of the 35 dominant GPSCs (Gladstone et al., 2019). From a total of 46 serogroups (numbered 1–48) described to date (numbers 26 and 30 are not in use) (Lund and Henrichsen, 1978), 3573 strains belonged to one of 40 serogroups. In addition, the dataset included 4 non-encapsulated laboratory mutants and 426 non-typeable (NT) isolates (Table S1).
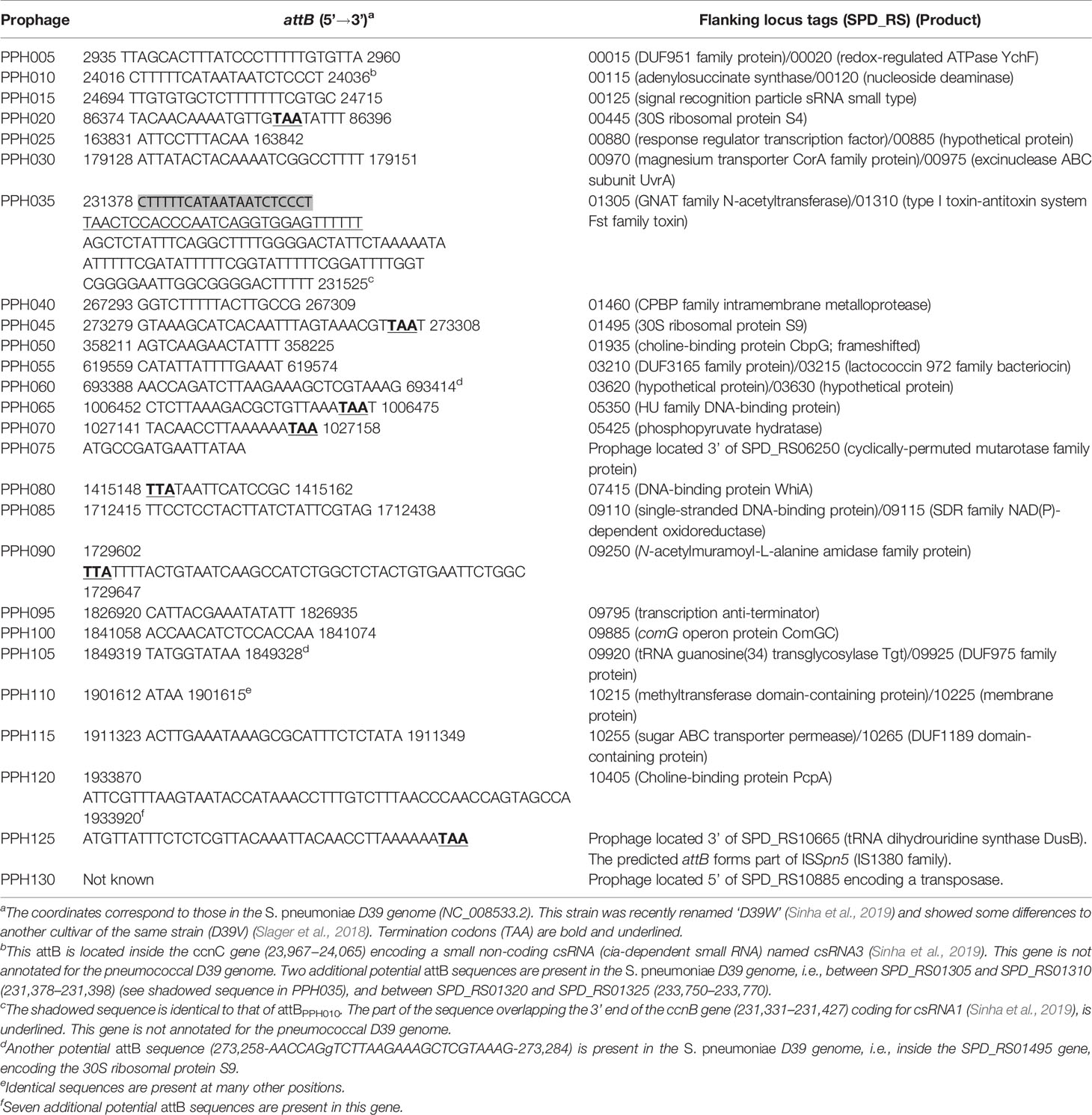
In agreement with data reported in previous studies (Ramirez et al., 1999; Brueggemann et al., 2017), PPHs were seen to be widely distributed across different S. pneumoniae isolates. Indeed, only 434 strains in the dataset (10.8%) (highlighted with a red background in Table S1) appeared to lack temperate phages. In addition, among the 126 RefSeq strains with complete (or near complete) genomes (see above), only 26 (20.6%) lacked any discernible prophage. On the basis of their Ints, more than 7000 putative PPHs (including putatively full-length and partial prophage sequences) were found in the dataset. Visual inspection of bacterial genes flanking the prophages revealed up to 26 different integration sites (Table 1). PPHs were clustered into 26 groups [named from PPH005 to PPH130 (in steps of 5)] according to their purported insertion sites, and arranged in order using the genome of the non-lysogenic S. pneumoniae D39 (Acc. No. NC_008533.2) as a reference. It is worth noting that many more complete Ints (7234) than endolysins (1210) could be identified. In addition, the proportion of partial sequences was much higher for the endolysin genes than the Ints genes. Thus, 42% of the endolysins were found incomplete (887 incomplete out of 2097 total proteins), whereas only 7% of the Ints were apparently incomplete or partly deleted (548 out of a total of 7782) (Table 1). This might be because endolysin-coding genes are among those known to be difficult to assemble from short read data due to the presence of choline-binding motif clusters (Croucher et al., 2017).
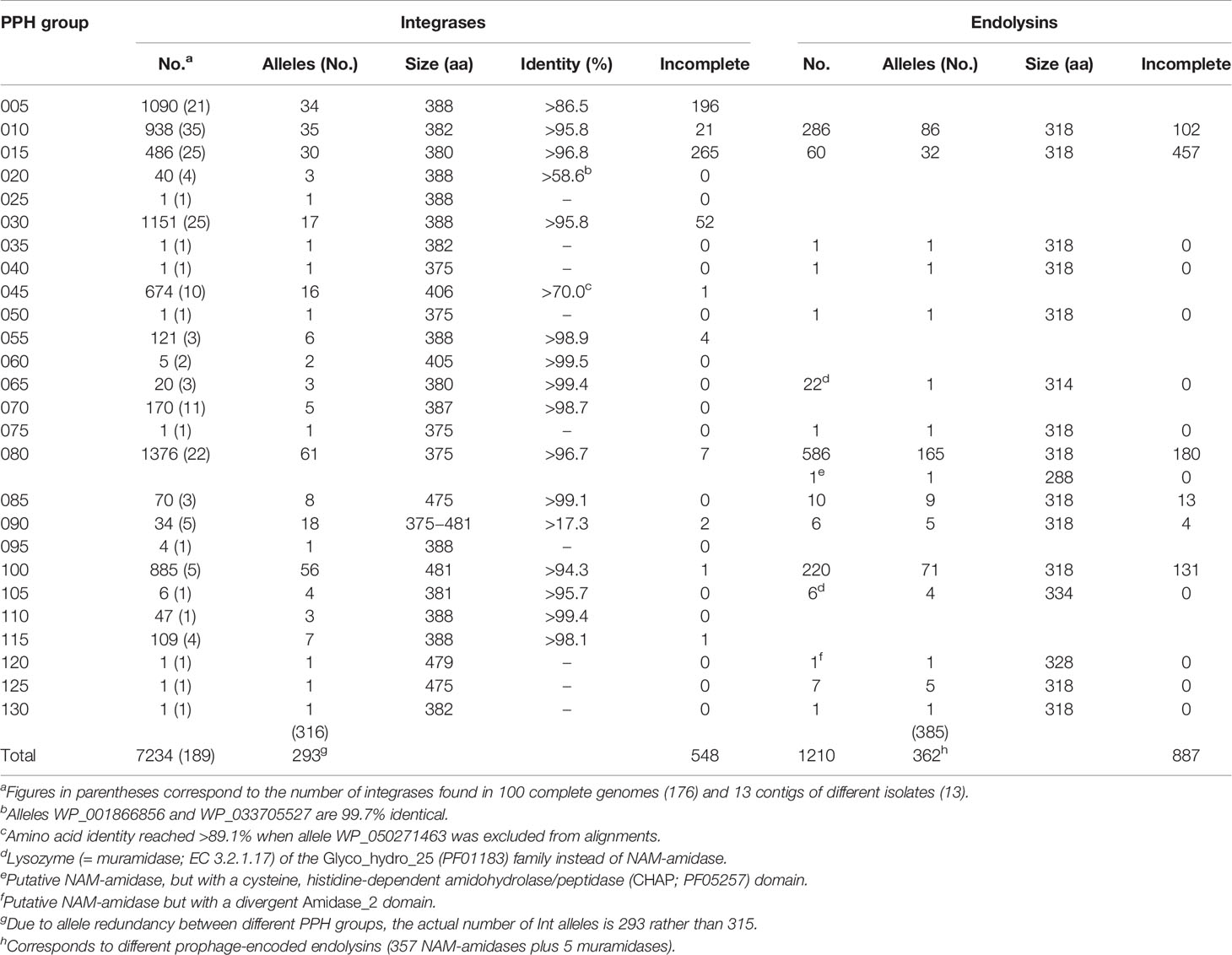
Table 1 Number and distribution of integrases and endolysins in the pneumococcal prophages (PPHs) analyzed.
In sharp contrast to the moderate number of endolysins compared to Ints in the current dataset, the number of different PPH endolysin alleles (362) was similar to that of the Int alleles (293). This strongly suggests that the PPH endolysin-coding gene shows great genetic variability. Nevertheless, 11 PPH groups, namely 005, 020, 025, 030, 045, 055, 060, 070, 095, 110, and 115, appeared to completely lack an endolysin gene and thus were understood to represent incomplete (or defective) prophages. Taking into account the number of full Ints, these incomplete prophages correspond to nearly half (47%) of the total PPHs.
PPH Integrases
On the basis of amino acid (aa) sequence similarities and catalytic residues, site-specific Ints (recombinases) can be classified into two major families: the tyrosine or serine families (Groth and Calos, 2004). These have different structures and functional mechanisms indicating that they have evolved separately. Thus, serine Ints are usually larger than tyrosine Ints, and the aa residues important for catalysis and structure, as well as their locations, can be very different. For example, the N-terminal domain of tyrosine Ints is involved in binding the arm-type sites of attP and the C-terminal domains involved in catalysis, whereas in serine Ints the opposite is true (Groth and Calos, 2004; Van Duyne and Rutherford, 2013).
Table S2 shows the complete catalog of the Ints found in this study. Excluding the prophages of the PPH090 group, which will be analyzed separately, only three Ints were found in more than one PPH group. These corresponded to the most frequent type of each group, i.e., WP_00876735 (in PPH010 and PPH130), WP_00266847 (in PPH040, PPH075, and PPH080), and WP_000704678 (in PPH055, PPH095, and PPH110). This observation agrees with the common idea that strong (albeit not absolute) specificity exists between prophage Ints and attB/attP sequences. For example, it has long been recognized that Int-mediated recombination occurs between attP of phage λ and the secondary attachment sites in the host genome, although the frequency is low (Shimada et al., 1975). In the present work, sequence identities between Ints were usually >90% within each PPH group. The PPHs contained either tyrosine or serine Ints, although PPHs with tyrosine Ints were 6.5 times more common (6243) than the latter (957) in the dataset (the Ints of PPH090 were not taken into account) (Table 2). Serine Ints (groups PPH085, PPH100, PPH120, and PPH125) were much more similar to each other than were the tyrosine Ints (Table 3). Among the tyrosine Ints, those from groups PPH010/PPH130 and PPH035 diverged greatly from the rest.
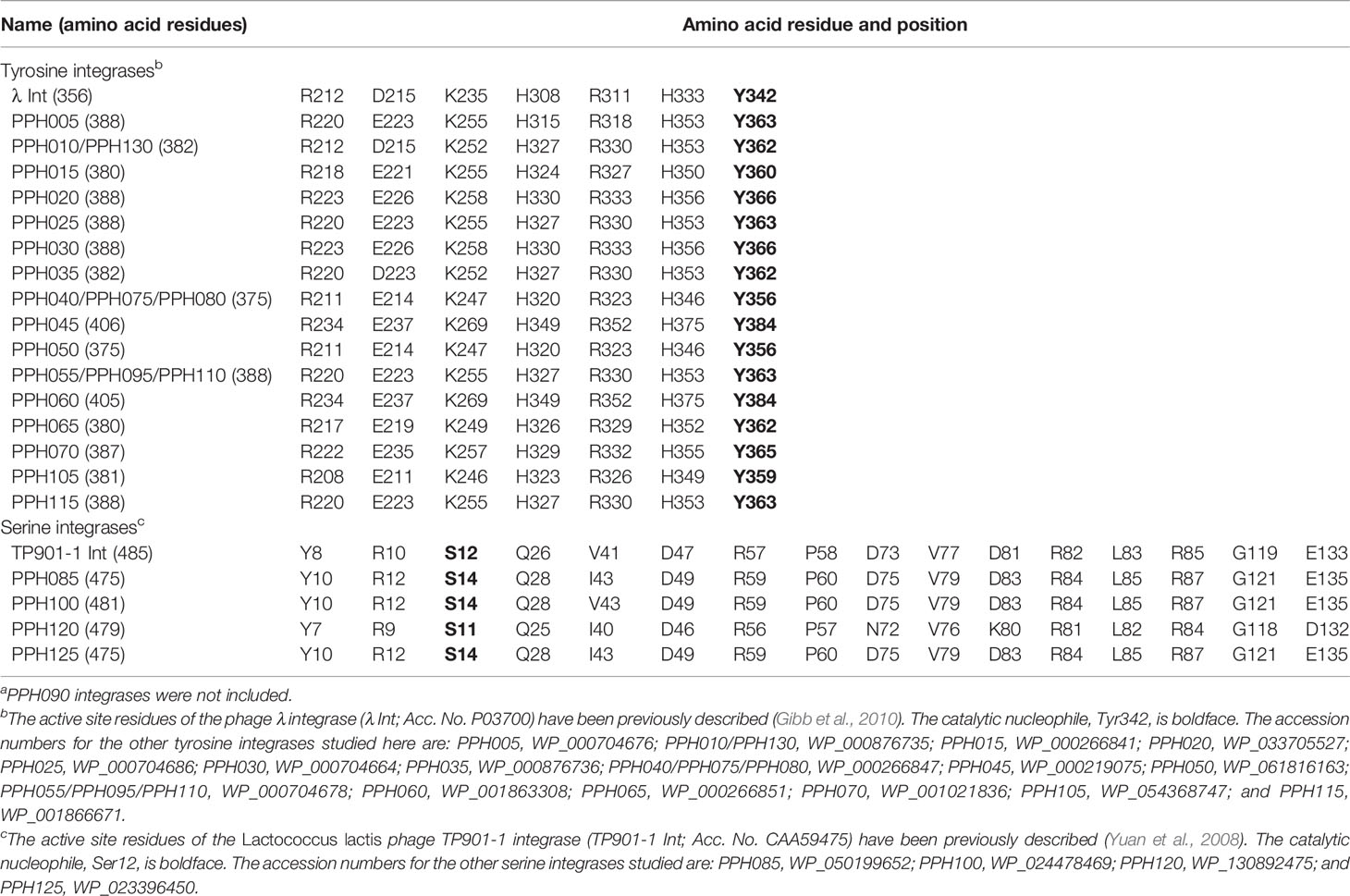
Table 2 Active site residues of PPH tyrosine and serine recombinasesa.
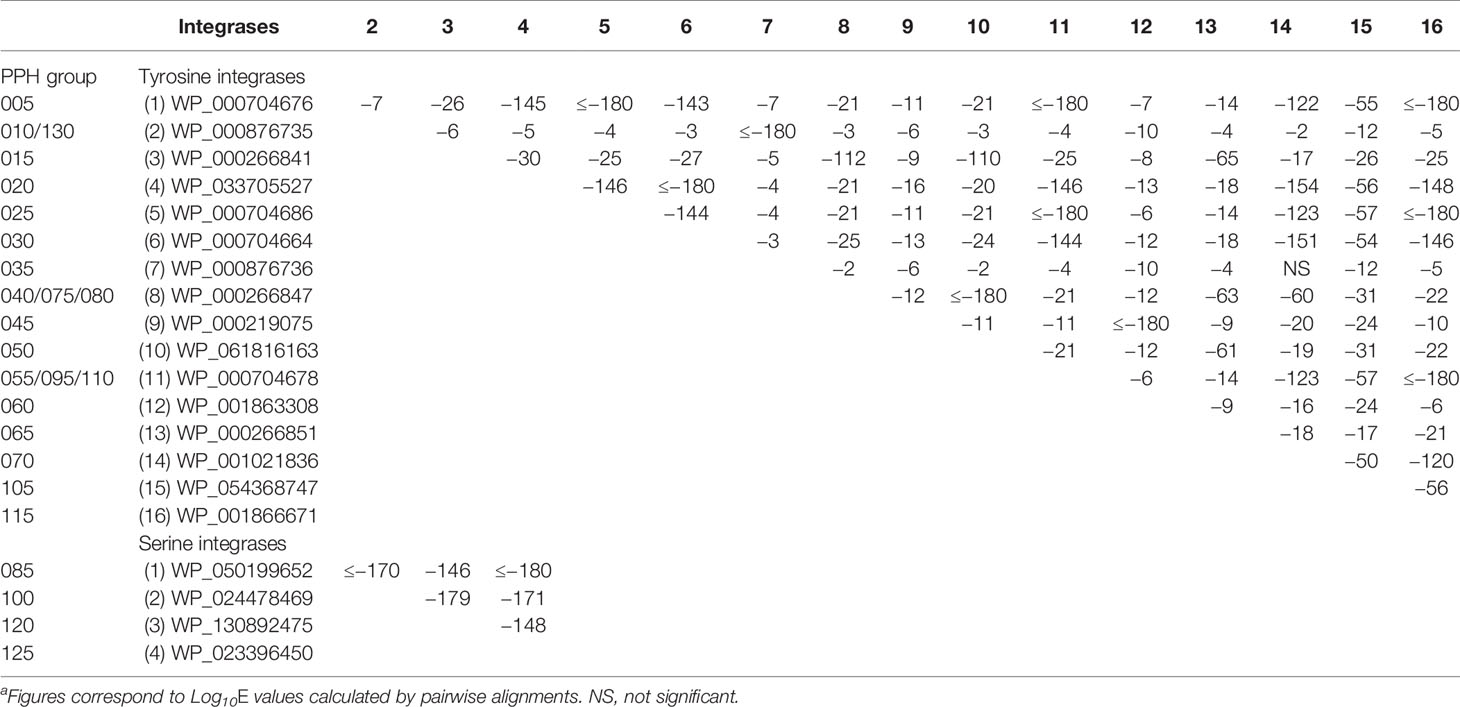
Table 3 Sequence similarities among tyrosine and serine integrases of PPHs a.
PPH Endolysins
Endolysins encoded by virulent (lytic) phages infecting S. pneumoniae have been extensively reviewed (López and García, 2004; García et al., 2005; Galán-Bartual et al., 2015; Maestro and Sanz, 2016; Vázquez et al., 2018). Briefly, three biochemically and structurally different peptidoglycan hydrolases of phage origin have been described so far. Two of them are choline-binding proteins: the NAM-amidase Pal (YP_004306947; 296 aa), with an Amidase_5 domain (PF05382), and the Cpl-1 lysozyme (=muramidase) (NP_044837; 339 aa) with a Glyco_hydro_25 (PF01183) domain. Both enzymes contain a C-terminal cell wall-binding domain composed of six choline-binding repeats (Choline_bind_1; PF01473). Cpl-7 (YP_009623604; 342 aa) is a choline-independent muramidase that differs from Cpl-1 in the C-terminal domain responsible for peptidoglycan binding (CW_7 repeats; PF08230). Pal is encoded by phage Dp-1, whereas Cpl-1 and Cpl-7 are the endolysins of phages Cp-1 and Cp-7, respectively. The endolysin from a recently isolated virulent podovirus very similar to Cp-1 (SOCP) (Ouennane et al., 2015) (NP_044837; 339 aa), is a muramidase 100% identical to Cpl-1. Similarly, the NAM-amidase (AQY55407; 295 aa) of the MS1 pneumococcal siphovirus (Kot et al., 2017; Silva et al., 2020) is 81% identical (89% similar) to Pal.
Taking into account that only a few virulent S. pneumoniae-infecting phages have been currently isolated, the diversity of their endolysins contrasts with the lytic enzymes encoded by PPHs. The latter are 318 aa-long, belong to the Amidase_2 family (PF01510) of NAM-amidases, and contain six Choline_bind_1 repeats in the C-terminal domain (Morales et al., 2010; Morales et al., 2015). Remarkably, in addition to having the same length (957 bp), the known phage genes coding for these endolysins (lytAPPH) are closely related (85–92% identity) to lytASpn, which encodes the major pneumococcal autolysin —also a NAM-amidase of the Amidase_2 family (Morales et al., 2010). LytASpn is a well-known virulence factor that plays a role(s) during different steps of infection (Canvin et al., 1995; Ramos-Sevillano et al., 2015; Ramos-Sevillano et al., 2016; Corsini et al., 2021). Taking into account the strong similarities between bacterial and phage NAM-amidases, the latter may also be important in pneumococcal pathogenesis.
The current notion of an exclusive presence for lytA-like genes among PPHs could not be fully confirmed in the present study. As already mentioned, 1210 full-length endolysins were identified in the dataset (Table 1 and Table S3). Among those, 29 proteins (≈2.5%) did not correspond to NAM-amidases of the Amidase_2 family. There were 28 proteins homologous to the Cpl-1 lysozyme, with one containing a cysteine, histidine-dependent amidohydrolase/peptidase (CHAP) domain (PF05257) instead of an Amidase_2 domain at the N-terminal moiety. The lysozymes were endolysins of PPH065 (22 identical proteins; WP_000739159) and PPH105 (6 proteins; 4 alleles: WP_054365577, WP_054368721, WP_054380492, and WP_054392100), which are 314 aa- and 334 aa-long respectively. Notably, all strains harboring PPH065 are members of GPSC19 and have serotype 22F (the only exception is strain 2245STDY6178828 which belongs to serotype 42) (see below).
The main differences between Cpl-1 and the PPH lysozymes detected in the present study are located in the linker region connecting the N- and the C-terminal domains. Thus, in Cpl-1, the linker (189-DDEEDDKPKTA-199) (Hermoso et al., 2003) is longer than (and different to) that of the newly discovered endolysins (188-DDEEAKAK-195). Moreover, WP_000739159 lacks the fourth Choline_bind_1 repeat that forms part of the C-terminal domain of the enzyme, and which is responsible for binding the enzyme to the cell wall. A detailed analysis of other PPHs (156 additional genomes, either complete or not) revealed four additional examples of 334 aa-long Cpl-1 homologs, specifically those encoded by prophages IPP16, IPP25, IPP27, and by a nameless prophage harbored by S. pneumoniae strain R34-3131 (Table S4). The latter prophage (which belongs to the PPH105 family) was already present in the dataset (Table S1).
The CHAP domain-containing endolysin (WP_057595562; 288 aa) found in the genome of S. pneumoniae strain SMRU392 (serotype 35F) is encoded by one of the PPH080 group of phages, and is identical to that of prophage 33888. The sequence of the latter was recently reported in an independent study (van Tonder et al., 2019) (Table S4). Since the host strain of prophage 33888 was the same as in the present study (SMRU392), and the Ints were also identical (WP_ 050256063), both prophages are probably the same. The WP_057595562 PPH endolysin is 63% identical (77% similar) to Skl, a proven NAM-amidase (WP_033686260; 288 aa) of an unnamed temperate phage of Streptococcus mitis SK137 (Llull et al., 2006). The complete genomic sequence of, presumably, the same S. mitis prophage (now designated as Javan331; MK448732) has recently been reported (Rezaei Javan et al., 2019). Remarkably, the existence of putative CHAP-endolysins encoded by those prophages was not mentioned in any study from other laboratories.
Among the 1181 endolysins with a predicted Amidase_2 domain, the presence of one atypical protein was noted. This protein (WP_130892444; PPH120) is a 328 aa-long, putative NAM-amidase with a divergent Amidase_2 domain. It is an endolysin encoded by the 36.7 kb-long prophage inserted into PPH010_4, giving rise to PPH120 (see below). Searches for proteins very similar (≥87% identity) to WP_13089244 revealed several endolysins of the same length encoded by prophages of three members of the Mitis group streptococci, i.e., Streptococcus pseudopneumoniae, S. mitis, and Streptococcus oralis. Specifically, these prophages are 277_SPSE, 289_SPSE, and 380_SPSE (WP_049511163) from S. pseudopneumoniae (Roach et al., 2015), S. mitis strains SK564 (WP_000238871) (Kilian et al., 2014) and DD22 (WP_061864892) (Denapaite et al., 2016), S. oralis U-o11 (Javan367; QBX17588) (Rezaei Javan et al., 2019), and S. oralis subsp. tigurinus 859 (WP_084868230) (Diene et al., 2016). Sequence alignments also revealed the variant domain of WP_130892444 to be remarkably similar to that of the Amidase_2 domain of LysGH15, the endolysin encoded by a myovirus phage (GH15) that infects Staphylococcus aureus (Gu et al., 2011). This lysin possesses a modular structure containing an N-terminal CHAP domain, a central Amidase_2 domain, and a C-terminal SH3_5 (PF08460) bacterial-binding domain. Elucidation of the crystal structure of the Amidase_2 domain of LysGH15 (Gu et al., 2014) revealed the aa residues involved in Zn2+ binding (H214, H324, and C332), catalysis (E282, and T330), as well as other important residues (W263, and N75), to be conserved in WP_130892444 at comparable positions (Figure S1). In sharp contrast, some of the equivalent residues in the NAM-amidase of S. pneumoniae TIGR4 (Mellroth et al., 2014; Li et al., 2015)—namely H26 and D149, as zinc ligands, and the catalytic residue H147—differed from those of WP_130892444 (and LysGH15) but were fully conserved among the widespread 318 aa-long NAM-amidases (Figure S1).
Insights Into the PPH Genomes and Identification of Their Attachment Sites
Only four att core sequences for PPHs were reported in studies published up until 2009 (Gindreau et al., 2000; Obregón et al., 2003b; Romero et al., 2009a), and additional core sequences have only been seldom reported in more recent papers (Càmara et al., 2018; Garriss and Henriques-Normark, 2020). To the best of our knowledge, only seven different attB core sequences have been previously described (Table S5). Remarkably, genome examination of many PPHs currently deposited in public databases (131 out of 158; last accessed, January 30, 2021) allow for no precise identification of the attP core site because most of the reported phage genomes only include the DNA region running from the first nucleotide of the int gene to the last one of the endolysin-coding gene (even though the definition was that of ‘complete genome’). Further, they lack the corresponding intermediate sequence where attP ought to be located (Table S4).
To gain insight into the sequences and locations of the integration sites of the PPHs described in the present study, a detailed analysis of all the PPHs harbored by 126 strains with near complete (chromosome assembly level) genomes was performed. The strains included in this subset represent 88 different STs, 27 PMEN clones, 66 GPSCs, and 27 different serotypes. In addition, the dataset included one non-encapsulated laboratory mutant and four NT isolates (Table S6). As mentioned above, 100 out of 126 strains (79.4%) were lysogenic and fulfilled the above assembly requirements in the original dataset. Fifty one isolates harbored only one PPH per genome whereas 49 were polylysogenic, and one of them (strain GPSC72) contained up to five different prophages (Table S6). Taking advantage of previously determined locations of int genes, the genomes of these strains were examined in detail and the sequences corresponding to the attL and attR sites of 176 prophages belonging to 20 different PPH groups recorded. The sequences and locations for the remaining six groups (PPH035, PPH065, PPH090, PPH095, PPH105, and PPH110) were estimated using the partial genomes (either at the contig or scaffold assembly level) of nine additional strains included in the dataset. Prophage genomes were defined as the DNA sequence running from the first nucleotide of attL to the last one preceding attR. The attL sequences (obviously identical or near identical to their corresponding attR within each PPH group) were used as queries to map the corresponding attB sites on the S. pneumoniae D39 genome (Table 4). With the exception of PPH130, the core sequence of 24 different attB could be determined. As mentioned above, seven of them had been already reported, namely, PPH005, PPH010, PPH015, PPH035, PPH080, PPH085, and PPH100 (compare the data of Table 4 with those of Table S5). With the exception of PPH075, PPH125 and PPH130, in which attB sites appear to be located inside insertion sequences (ISs), the precise chromosomal location of the different attB sites could be established. Although many PPH insertion sites are located in intergenic spaces (something typical among prophages), six core attachment sites mapped within the 3’ region of genes annotated for the D39 genome, and included the termination codon (TAA) of the gene where the prophage was integrated (Table 4). In addition, attBPPH035 was found to partially overlap (but apparently not interrupt) the 3’ part of ccnB encoding csRNA2, one of the five small non-coding csRNAs (cia-dependent small RNAs) that form part of the two-component regulatory system CiaRH (Sinha et al., 2019). It should be underlined that ccnB is not annotated for the S. pneumoniae D39 genome. On the contrary, seven prophage groups potentially interrupt gene translation: 1) the core integration site of PPH010 viruses (attBPPH010), previously reported to lie between SPD_RS00115 and SPD_RS00120 (Romero et al., 2009a) but actually located in ccnC coding for csRNA3 (Furi et al., 2019); 2) PPH015, which integrates into SPD_RS00125 encoding the signal recognition particle sRNA involved in membrane protein targeting (Steinberg et al., 2018); 3) PPH040, which integrates into SPD_RS01460, potentially encoding an intramembrane metalloprotease of the CAAX proteases and bacteriocin-processing enzymes (CPBP) family (Pei et al., 2011); 4) PPH050, inserted into SPD_RS01935 encoding a frameshifted choline-binding protein (CbpG) in D39 (Frolet et al., 2010); 5) a gene (SPD_RS09795) encoding UlaR [a transcriptional activator of the ula operon in the presence of ascorbic acid (Afzal et al., 2015)], which may be inactivated by the insertion of bacteriophages of the PPH095 group; 6) PPH100, which integrates into SPD_RS09885 encoding ComGC, the major subunit of the competence pilus (Laurenceau et al., 2015); 7) and bacteriophages of the PPH120 group, which disrupt SPD_RS10405, i.e., the gene encoding for the PcpA choline-binding protein (Sánchez-Beato et al., 1998) (now an important component of several new protein-based, pneumococcal vaccines currently under evaluation) (Masomian et al., 2020).
Table S6 attempts to organize the different PPHs in a manner that takes into account their wide genetic diversity. For this, a concept of ‘equivalent PPH genomes’ was followed, i.e., genomes that overlapped for ≥90% of their total length and showed ≥90% sequence identity were considered ‘equivalent’, and only one of them was used in further comparisons. Figure S2 shows a comparison of the different PPH genomes. PPH095 and PPH110 are not depicted since, although they inserted into two different attBs, these PPHs were equivalent to PPH055 (see below). Due to the evident sequence variability of prophage genomes, pairwise nucleotide alignments were initially performed only among PPHs of the same group. Several main conclusions were then drawn: 1) int genes were present in every prophage found, but this was not the case for endolysin-coding genes; 2) two major PPH groups were recognized, one with genome lengths ranging from ≈10–20 kb on one side (e.g., PPH005 and PPH030 groups), and the other from ≈35–45 kb on the other side (e.g., PPH010 and PPH080 groups); 3) major differences can still be seen among the members of a PPH group (e.g., PPH015, PPH020, PPH045 and PPH090 show large differences); 5) several PPHs with genomes of 51–92 kb appear to result from recombination events between two different prophages, e.g., i) PPH075 is the result of the insertion of an unknown defective prophage (12,227 bp) into a PPH085_5 equivalent. This defective prophage is flanked by a 33 bp-long sequence (5’-CCCTAGACTTGAAATAAAGCGCATTTCTCTATA-3’) located at positions 32,873–32,905 and 45,100–45,132 of the PPH075 genome; a near-identical sequence (1,911,317-TCCTAGACTTGAAATAAAGCGCATTTCTCTATA-1,911,349) may contain part of the promoter region of SPD_RS10265 (malA) in the D39 genome (Nieto et al., 1997); ii) PPH080_11 appears to result from the integration of a PPH010-like prophage into a PPH080_3 equivalent; as expected, the insertion of the PPH010-like phage occurs in the sequence 5’-CTTTTTCATAATAATCTCCCT-3’ at positions 2069–2089 and 32,251–32,271 of the PPH080_11 genome; iii) the insertion and rearrangement of a PPH010_4-equivalent into a previously unknown prophage gives rise to PPH120, possibly with the assistance of two ISs (ISSpn5 and IS1167) and the addition of a gene cluster encoding several tRNAs. Sequence comparison revealed this prophage to be very similar (82–85% query coverage and >95% nucleotide sequence identity) to two Streptococcus anginosus prophages, namely Javan83 (Rezaei Javan et al., 2019) and SA01 (van der Kamp et al., 2020); interestingly, the latter two prophages appear to harbor no tRNA genes. In addition, both encode an endolysin 76% identical (88% similar) to Cpl-7 (see above), whereas, as already discussed, the new component of PPH120 codes for a peculiar 328 aa-long NAM-amidase of the Amidase_2 family of proteins; iv) a tandem insertion of two near identical PPHs (PPH080_1A and PPH080_1B) gives rise to the 92 kb-long hybrid prophage PPH080_1AB.
Among the various prophage identification tools, we used the PHASTER program to analyze the 109 genomes included in Table S6 (186 PPH). Up to 263 putative PPHs were predicted; 40 of them were designated as ‘intact’ by the program and actually corresponded to real prophages (Table S6). The predicted limits of the PPHs did match the real coordinates in only few cases (e.g., PPH030 from strain SP64 was almost correctly located (real: 177,043–189,925; predicted: 177,043–189,926). Unfortunately, in most instances, this was not the case. For example, in strain GPSC47 (NZ_LR216060), PPH010_9 is located between coordinates 3152 and 36,649 whereas PHASTER predicted the range 1771–42,683). Of note, the existence of PPHs of the groups 010, 015, 080, and 110 was well predicted, whereas that of those belonging to groups 020, 045 and 115 were not (Table S7). In our hands, the sensitivity of PHASTER (0.74) was similar to that previously reported (Lopes de Sousa et al., 2018; Reis-Cunha et al., 2019).
Pairwise nucleotide alignments were then performed among genomes belonging to different PPH groups. In addition to equivalent PPHs (see above), a second level of similarity was allowed, i.e., those prophages with an overlap of between 80 and 89% of their genomes and showing nucleotide identities of ≥90%. Figure 1A shows ‘equivalent’ and ‘very similar’ genomes on white or gray backgrounds respectively. It should be underlined that equivalent prophages were distributed between different PPH groups. Thus, in addition to the equivalency between the defective prophages PPH055, PPH095, PPH110 and PPH115_2 partly mentioned above, other equivalent, putatively complete prophages of different groups were seen: 1) PPH010_2, PPH035 and PPH130; 2) PPH015_1, PPH080_8 and PPH090_3; and 3) PPH040 and PPH080_3. As expected, most of the ‘very similar’ category of prophages corresponded to different members of the same group, as exemplified by members of the PPH010 group such as PPH010_7–PPH010_10 and PPH010_12–PPH010_14 (Figure 1A). Pairwise comparison of 80 PPH sequences and a dendrogram depicted seven major prophage clusters, and three singletons (Figure 1B). Four of the clusters matched those described by Brueggemann et al. (2017) and one more to the defective prophages mentioned by Rezaei Javan et al. (2019) that were recognized as separate entities from full-length PPHs. The remaining PPHs (either clustered or not) correspond to previously undescribed prophages.
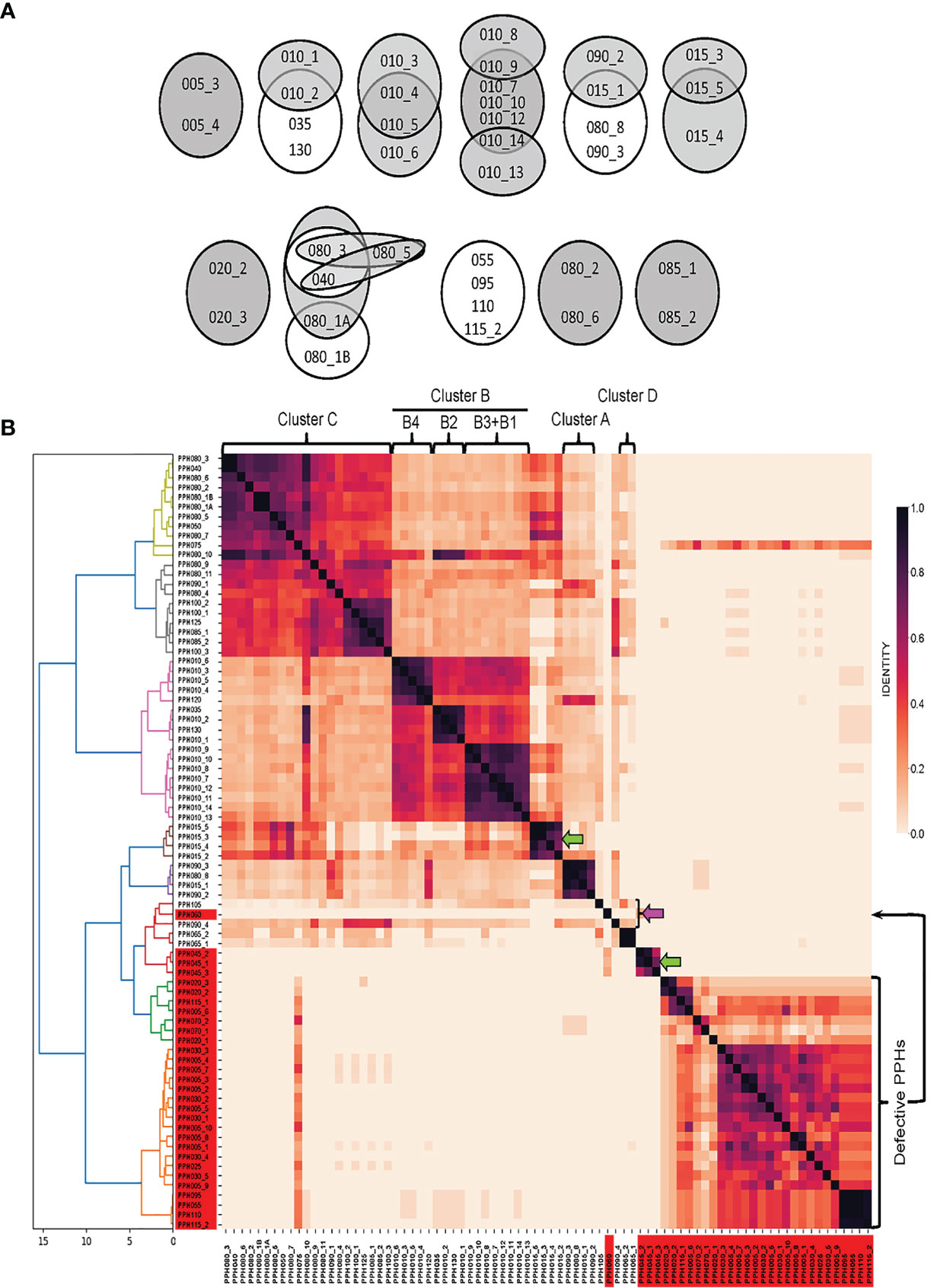
Figure 1 Sequence similarities between PPHs genomes. (A) Venn diagrams showing equivalent or very similar pneumococcal prophages. Two PPH genomes were considered ‘equivalent’ when they overlapped for ≥90% of their length and showed ≥90% sequence identity (shown on a white background). When two genomes overlapped between 80 and 89% of their length, and nucleotide identities were ≥90%, the corresponding PPHs were considered as ‘very similar’. They are shown on a gray background. (B). wGRR similarity heatmap depicting the amino acid identity shared between pairs of prophage sequences (either full-length or defective prophages). Different colors were applied to clusters showing distance thresholds > 4. For agglomerative hierarchical clustering, distances are shown in the X axis. The dendrogram and heatmap python tools of the scipy.cluster.hierarchy library were used to depict final agglomerative hierarchical clustering and heatmap plots, respectively. Green and pink arrows point to new clusters or singletons respectively. The correspondence between PPHs and the cluster organization of pneumococcal prophages previously reported (Brueggemann et al., 2017; Rezaei Javan et al., 2019) is indicated at the upper part of this panel. See Table S4 for a proposed correlation between previously described pneumococcal prophages and PPH groups.
Other Features of Pneumococcal Prophages
Many prophages from both Gram-negative and Gram-positive bacteria integrate into tRNA genes (Williams, 2002) although this is not the case for PPHs (see above). Nevertheless, sequence analysis revealed the presence of potential tRNA genes in some PPH genomes (Table S8). Of note, PPH120 has nine tRNA genes located at the right end of the prophage, whereas other PPHs have these genes located closer to the middle. A search of PPH genomes reported elsewhere revealed that five previously sequenced phages (namely, IP8, IP16, IP25, IP26, and IP27) also harbor a tRNA-Ser gene, a feature not mentioned in previous reports (Table S8).
PblB may be an important virulence determinant in some PPHs. PblB has been described as a phage minor tail protein (a putative anti-receptor) that behaves as an adhesin mediating the galactose-specific adhesion activity of pneumococci to platelets and human lung epithelial cells in vitro. It is also required for nasopharyngeal and lung colonization in a mouse model of infection (Hsieh et al., 2015). The sequence diversity of PblB is notable (Brueggemann et al., 2017). Amino acid sequence alignments in the PPH dataset fully confirmed this (as shown diagramatically in Figure S3), even among PPHs of the same group (e.g., in PPH010_7, in which the length of PblB was 2134 aa in strain GPSC97 but 3256 aa in GPSC25). It is also reported that the PblB encoded by the temperate phage SM1 infecting S. mitis (1062 aa) (Bensing et al., 2001) is very different to the protein (P15; 1987 aa) previously analyzed in the S. pneumoniae NTUH-P15 strain (Hsieh et al., 2015). Moreover, these two proteins differ notably from most of the PblB proteins found in the present study, particularly in their middle regions where different combinations of five different (albeit related) sequence repeats were found (Figure S3). PblB-like proteins were encoded by some of the phages belonging to the PPH010, PPH035, PPH040, PPH080, PPH085, and PPH100 groups. Willner et al. showed that the S. mitis prophage SM1 can be induced by commonly ingested substances and that the oral cavity is indeed a reservoir of pblA and pblB genes (Willner et al., 2011).
First described in the Gram-negative bacterium Dichelobacter nodosus (an ovine footrot pathogen), virus-associated protein E (VapEDno) has been proposed involved in virulence (Billington et al., 1996). Proteins similar to VapE have also been reported encoded by prophages of other microorganisms such as S. aureus (Lindsay et al., 1998), Pseudomonas putida (Canchaya et al., 2003), Vibrio parahaemolyticus (Seguritan et al., 2003), Staphylococcus epidermidis (Chen et al., 2013) and Elizabethkingia anophelis (Peng et al., 2020), although direct evidence of the involvement of VapE in virulence was provided in neither of these studies. In S. pneumoniae, Romero et al. (2009a) first noted that several prophages encoded proteins similar (about 30% identity; E value = 10–34) to VapEDno (also designated as virE). More recently, several gene products from defective prophages have been shown similar to a Streptococcus suis protein named VapESsu (Rezaei Javan et al., 2019), a protein apparently unrelated in primary structure to VapEDno but which has been shown involved in virulence in a mouse model (Ji et al., 2016). The translated nucleotide dataset of PPHs was compared to both VapEDno and VapESsu and using an E value cutoff of 10–20, two sets of putative virulence-associated proteins were found (Table S9).
It is well known that bacteria have developed defense mechanisms to help them survive phage attacks. These mechanisms may interfere with the adsorption of the phage onto the cell surface, with phage genome injection, or target the foreign nucleic acid after its injection, etc. (for recent reviews see Labrie et al., 2010; Koonin et al., 2017; Rostøl and Marraffini, 2019). Phages, in turn, have evolved mechanisms to overcome restriction-modification (R-M) systems. One of these is based on the presence in phage genomes (both virulent and temperate) of orphan methyltransferase (Mtase) genes (i.e., without concomitant endonuclease-coding genes) (Murphy et al., 2013). There are three functional classes of type II DNA Mtases that function by transferring a methyl group from S-adenosyl-L-methionine to a target nucleotide base, forming either N-6-methyladenine (class I), N-4-methylcytosine (class II), or C-5-methylcytosine (class III). Our laboratory previously reported the existence of putative Mtases-encoding genes in two S. pneumoniae temperate phages, namely MM1 (Obregón et al., 2003a) and VO1 (Obregón et al., 2003b). Taking the Mtases of these two phages as query sequences, the complete set of PPH genomes was screened. Interestingly, more than 30 putative orphan Mtases of either the MM1 or VO1 class were detected. None of them was encoded by defective PPHs (Table S10).
PPH090, a Group of Prophages That Integrate Into the lytASpn Gene via an Int-Independent Mechanism
The proposed core sequence of attBPPH090 (46 bp-long) mapped to the 3’ end of the lytASpn gene overlapping its termination codon (Table 4). Consequently, phage integration should not disturb lytA translation. In sharp contrast with other PPHs, the most striking feature of prophages of the PPH090 group is that there is no preference for a particular Int (or group of Ints) (Table S2). Specifically, different PPHs of the PPH090 group encode Ints of the tyrosine (12 entries) or serine (6 entries) families, some of which are identical to those of other prophage groups: 1) tyrosine Ints [PPH010 (WP_000876726, WP_000876733, WP_000876735, and WP_050199954), PPH015 (WP_023396066, WP_044813689, WP_050127433, W_050203801, and WP_050221901), PPH080 (WP_000266839, WP_000266847, and WP_000266854)]; and 2) serine Ints [PPH100 (WP_024478469, WP_044812715, WP_050203932, WP_050226267, WP_050230313, and WP_050974546)]. These data strongly suggest that PPH integration into lytASpn does not require a specific interaction between attB, attP, and the Int, and that some other factors may be involved—possibly recombination events between lytASpn and lytAPPH (see below).
The four PPH090 genomes shown in Figure S2 encode, respectively, one of three different Ints of the tyrosine family (PPH090_1–PPH090_3) and one Int of the serine family (PPH090_4). Among them, the genome of the NT strain SMRU257 (master record NZ_CKNY01000000) appears to be unique since, in addition to the PPH090_2 genome (39,182 bp; contig NZ_CKNY01000002), it contains a 39,182 bp-long sequence (contig NZ_CKNY01000010) inserted into the ffs gene (corresponding to SPD_RS00125 in S. pneumoniae D39) (Table 4). Moreover, this prophage harbors all the characteristics of a full member of the PPH015 group, including the near identical flanking attL and attR core sequences (5’-TTGTGTGCTCTTTTTTCGTGC-3’). Remarkably, PPH090_2 and the ‘new’ PPH015 only differ at two nucleotide positions, i.e., 918 (G) and 951 (T) of the 957 bp-long lytAPPH and lytAPPH* genes (the asterisk indicates that the phage and bacterial lytA genes were probably chimeric [a hybrid] because they contain sequences of both phage and bacterial origin) (Figure S4).
The hypothesis that lytAPPH recombines with the lytASpn gene and integrates into the pneumococcal genome was tested by making a detailed analysis of both the lytA genes and the DNA regions located between int and lytASpn*, as well as downstream of lytAPPH*, in the integrated state (Figure 2A). Figure 2B shows the appropriateness of the hypothesis since, immediately after the termination codon of lytAPPH*, the sequence was syntenic, i.e. shares the same genetic order, with the DNA region located 3’ of lytASpnin a non-lysogenic strain. In fact, previous studies had already provided different kinds of evidence on the existence of recombination between these genes (Romero et al., 1990; Whatmore and Dowson, 1999; Morales et al., 2010). It has also been reported that lytASpn and plyA (the gene encoding the main pneumolysin, a cholesterol-dependent cytolysin) form part of a pathogenicity island in S. pneumoniae, characterized by the presence of direct repeats (ISs), phage-related genes, and/or genes potentially encoding virulence factors and mobility proteins (Ints, transposases). These findings also suggest that the plyA–lytA island is a recombination hotspot. At least eight genomic arrangements of this island have been recognized (Morales et al., 2015). During the present study, an additional arrangement that appears to be exclusive to some NT pneumococci was found (Figure 2B). It is worth mentioning that a larger protein (3628 aa residues; WP_142367134) of unknown function is encoded within the plyA–lytA island in the pneumococcal strain SMRU257.
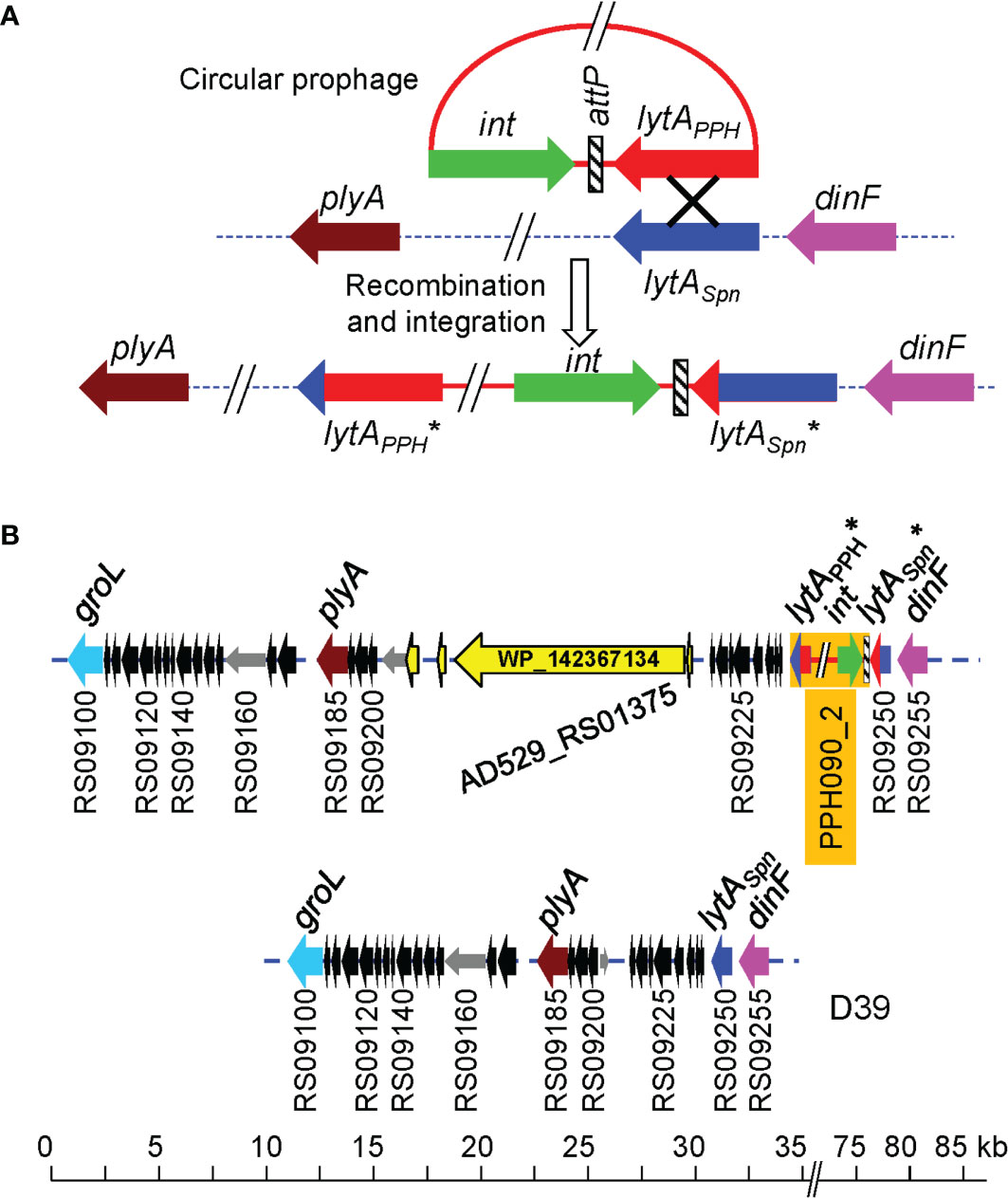
Figure 2 A proposal for phage integration into the S. pneumoniae genome after the recombination of lytASpn and lytAPPH. (A) The circular phage genome (red) and the pneumococcal chromosome (dotted line) are shown. Genes are drawn as arrows indicating the direction of transcription. An ‘X’ indicates the recombination event. The pneumococcal genes plyA and dinF that flank the lytASpn gene are shown for reference (Morales et al., 2015). The asterisk (*) indicates that the phage and bacterial lytA genes were probably chimeric because they contain sequences of both phage and bacterial origin. (B) Novel genomic arrangement of this plyA—lytA island in the pneumococcal strain SMRU257. The location of a gene (AD529_RS01375) encoding a large protein (3628 aa residues; WP_142367134) is shown. For comparison, the corresponding DNA region of the control D39 strain is shown at the bottom.
The DNA region located immediately 3’ of the corresponding lytAPPH* gene of each of the four PPH090 was aligned with a 2 kb-long region located immediately downstream of the lytASpn gene of the non-lysogenic S. pneumoniae D39 strain (NC_008533.2; complement [1,727,602–1,729,601; the position of the last nucleotide of lytASpn is 1,729,602]). In agreement with the hypothesis formulated above, the four PPH090 genomes showed >96.5% nucleotide identity with the D39 genome in that region (data not shown). The PPH090 regions that run from the 3’ end of int to the 3’ of lytASpn* after integration, were also aligned (Figure 3). This region was 548 bp-long in PPH090_4 (serine Int family), and shorter (305–306 bp) in prophages with tyrosine Int-coding genes. The DNA regions corresponding to the two putative PPH015 prophages (PPH090_2 and PPH090_3) were very similar (97.7%; 298 identical nucleotides in a 305 nt overlap). The presence of a 51 bp-long sequence (5’-ATAGAAAGGAAACTTTCTAAATTGTTCTTTCACCGCAGGCTTAGGCTTGCG-3’) located immediately after the termination codon of the lytASpn* gene and found to be conserved in all the PPHs studied (indicated by orange rectangles in Figure 3) should be noted. Among the 126 pneumococcal strains with either complete or near complete genomes, this 51 bp-long signature was absent from the 26 non-lysogenic strains (as expected), but it was found immediately downstream of the 3’ end of the lytASpn gene among two lysogenic strains: 4041STDY6836166 (Acc. No. NZ_LS483451; positions 1,865,445–1,865,394) and GPSC72 (Acc. No. NZ_LR216049; positions 1,745,355–1,745,304). In these strains, the 51 bp-long sequence is part of a ≈700 bp-long fragment that is ≥95% identical to another fragment located 3’ of several lytAPPH genes, particularly those within the PPH010 group (unpublished observations). This finding indicates that those strains were once lysogenic, but that at some point, most of the prophage was lost. Interestingly, a putative transcriptional terminator is located close to the 3’ end of the lytAPPH (and lytASpn) genes and may function in a manner similar to that of the transcriptional terminator of the lytASpn gene previously determined (Díaz and García, 1990) (Figure S5).
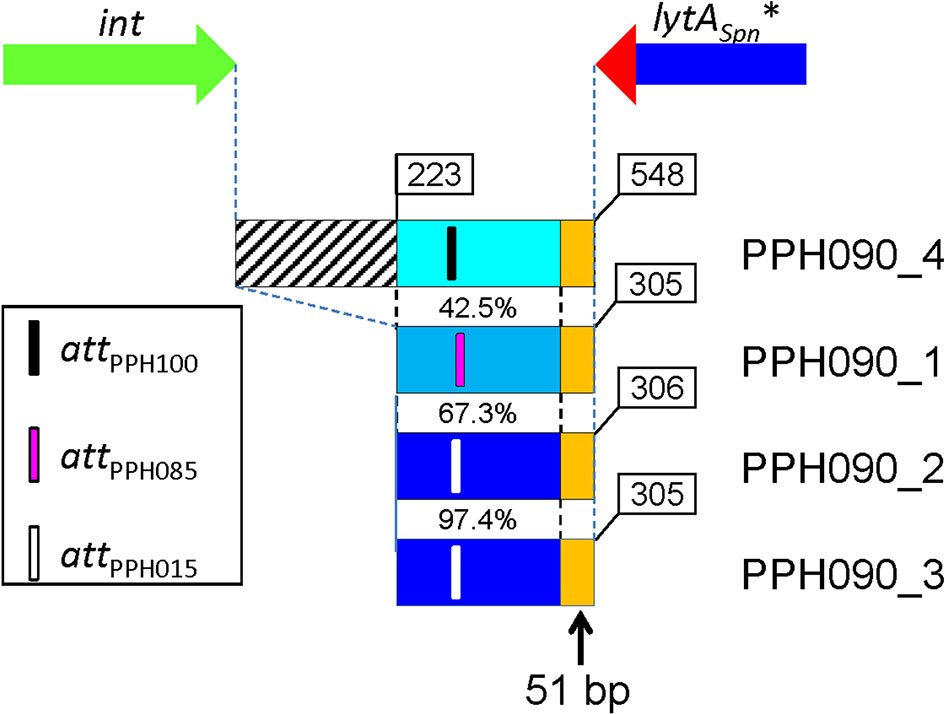
Figure 3 Organization of the DNA region located between the 3’ ends of the int and lytASpn* genes. Genes are not drawn to scale. Four prophages of the PPH090 family were examined. The intergenic region is longer in PPH090_4 (belonging to the serine Int family) than in the other PPHs with tyrosine Ints. The percentage nucleotide identities are indicated. The approximate positions of the attP core sequences are shown. Two of the prophages belong to the PPH015 group and the corresponding whole regions are 97.4% identical. Orange rectangles represent the 51 bp-long perfectly conserved sequence located immediately 3’ from lytAPPH and lytASpn*.
Recombination Between Prophage and Bacterial lytA Genes Is a Source of Chromosomal Rearrangements in S. pneumoniae
PPH090-like prophages appear to be made via recombination of the lytAPPH and lytASpn genes after the prophage is spontaneously excised from the host genome. However, recombination may also take place during the lysogenic (integrated) state of the prophage. In this instance, however, lytA recombination may result not in the emergence of a new PPH090 prophage but in chromosomal rearrangements. To gain insight into the recombination events that appear to occur between lytASpn and lytAPPH, the corresponding genes from the four PPH090 reported here were aligned and the position of nucleotides conserved in lytASpn* and lytAPPH* —but not in both groups— were annotated (Figure 4A). A search for polymorphisms at those positions was then performed in all the lytASpn (325) and lytAPPH (356) alleles of the dataset (Figure 4B). Few polymorphic positions appeared between positions 1 and 100 (approximately), but beyond there the average polymorphism reached 7 and 14% in the tested positions of lytASpn* and lytAPPH* respectively. This suggests that recombination events (if any) are more frequent in the 3’ than the 5’ moiety of the gene. The possible significance of the notable polymorphism observed at three nucleotide positions (14 and 17 in lytAPPH, and 282 in lytASpn) was not further analyzed, but the importance of the polymorphism at position 951 (very close to the 3’ end of lytA which is located at position 957) mentioned above was studied in detail. As shown in Figure 4B, searching among the 325 lytASpn alleles in the resulting data showed 84 of them (154 strains) to have a ‘T’ instead of an ‘A’ nucleotide at position 951. Table S11 summarizes a systematic analysis of this region among a subset of 51 of those 84 lytASpn alleles (111 strains). The results indicate the existence of recombination events that produced the translocation of several DNA fragments from their original position (taking the non-lysogenic S. pneumoniae D39 genome as a reference) to a region located immediately downstream of the lytASpn* gene (Figure 5). Five major rearrangement events were recorded and, in agreement with the genes found, the direct participation of PPH010 (SPD_RS00120), PPH015 (SPD_RS00130), PPH080 (SPD_RS07410), PPH085 (SPD_RS09110), or PPH100 (SPD_RS09880) was deduced. By comparing these arrangements with those of the different PPHs described in this study, the bacterial and phage lytA genes were seen to represent inverted repeats in arrangements A and B, and individual direct repeats in arrangements C, D, and E. A recombination event in cases A or B would likely produce a chromosomal inversion (Table S11). In addition, since the region between the lytA genes (the spacer) is asymmetrical around the origin of replication, a chromosomal inversion may induce changes in the relative positions of the origin and terminus of replication, as previously noted (Achaz et al., 2003). In arrangements C, D and E, a recombination event produced between repeat sequences in the same orientation on the same chromosome could result in a translocation (Hughes, 2000). A close examination of the region located immediately downstream of lytASpn* (SPD_RS09250; depicted as gray squares in Figure 5) in 14 different strains revealed complex organizations of different length that contained prophage fragments or even putative full length prophages. For example, strain SMRU51 harbored a prophage equivalent to PPH035 (Figure 6). Of note, an additional recombination event in strain SMRU1319 (arrangement D2) produced an inversion of the genes located upstream of SPD_RS09115 and downstream of SPD_RS09110 (Table S12).
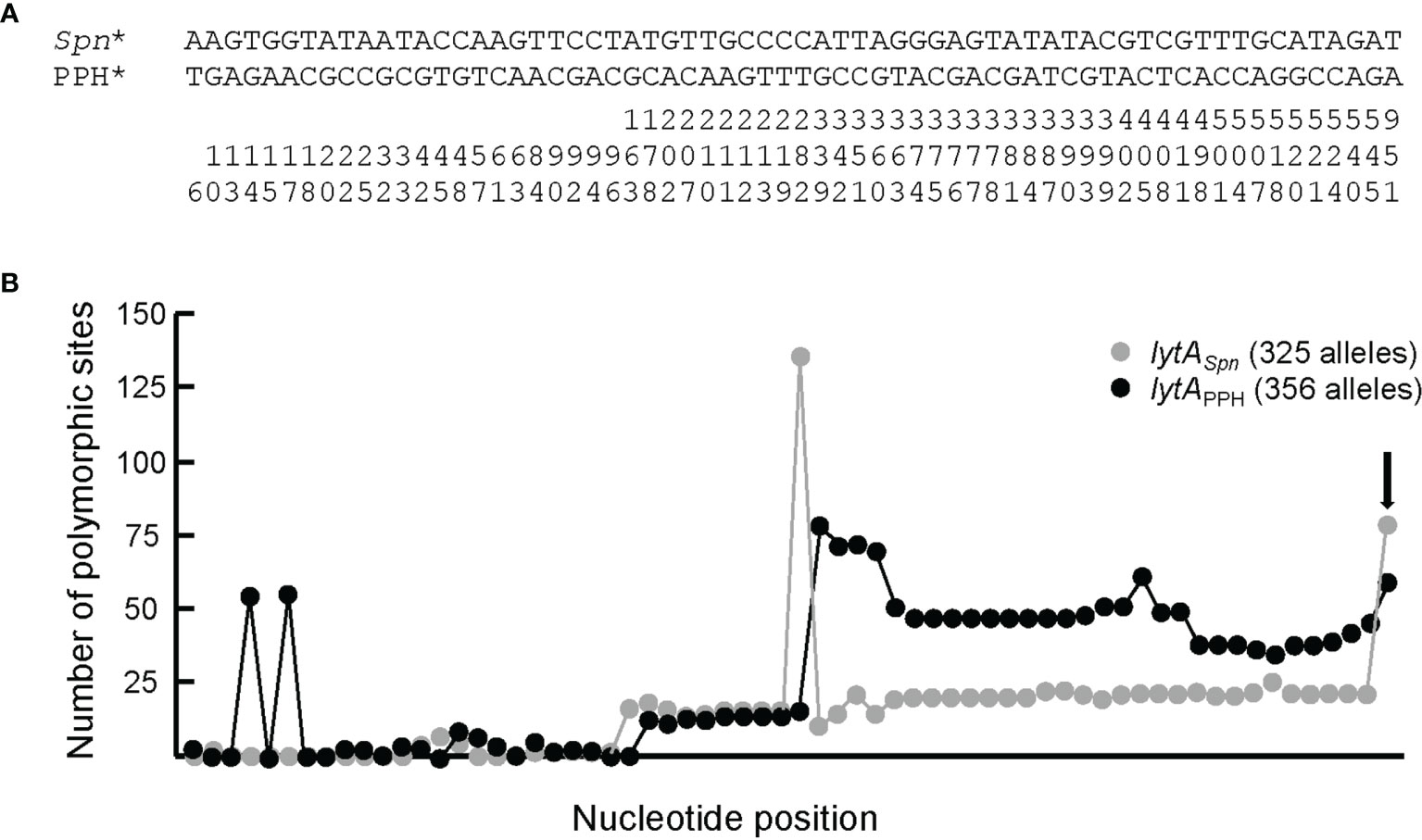
Figure 4 Sequence signatures of lytASpn* and lytAPPH_090* alleles. (A) Bacterial and prophage lytA genes of strains harboring PPH090 group phages were aligned and the positions where the bacterial (on one side) and phage (on the other) genes diverged were marked. For simplicity, the highly polymorphic region characteristic of the lytA gene (positions 421–480) (Morales et al., 2010) was not included in the alignment. The nucleotide positions (taking 1 as the first nucleotide of the ATG initiation codon) should be read vertically. (B) Distribution of polymorphic sites in bacterial and prophage alleles according to the alignment shown in panel (A) The vertical arrow points to the data at the polymorphic position 951. The total length of the lytA gene is 957 bp.
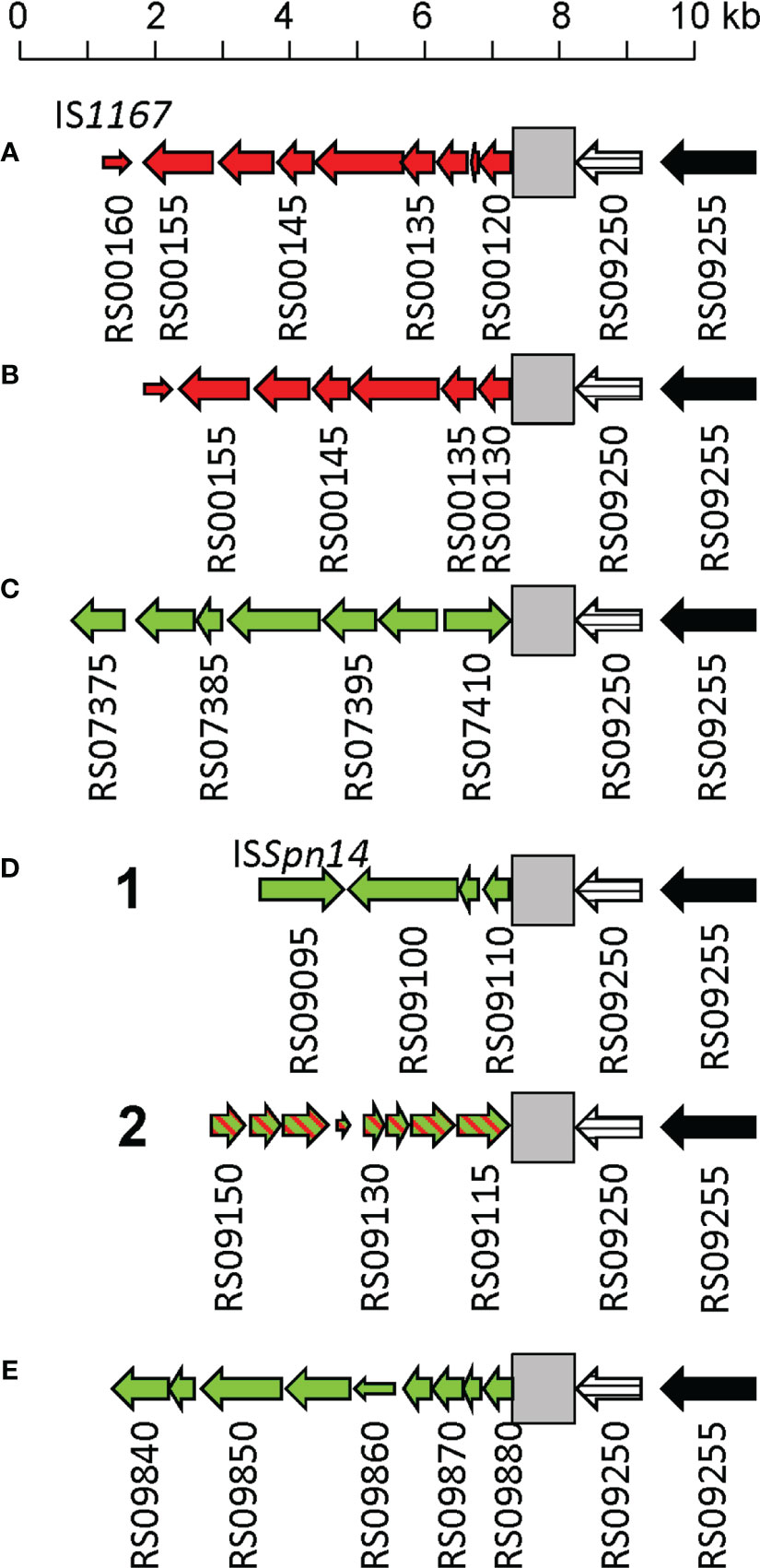
Figure 5 Schematic representation of several DNA rearrangements in the 3’ region of lytASpn*. Genes are represented as arrows indicating the direction of transcription and are numbered as those of the S. pneumoniae D39 strain (SPD_). Narrow open arrows indicate frameshifted open reading frames. The lytASpn gene (SPD_RS09250) is represented by a striped arrow. Panels (A–E) respectively show the rearrangements of genes located in the vicinity of SPD_RS00120 (coding for a nucleoside deaminase), SPD_RS00125 (ffs encoding the signal recognition particle sRNA), SPD_RS07415 (whiA), SPD_RS09110 (coding for a single-stranded DNA-binding protein), or within SPD_RS09885 (encoding ComGC, the major subunit of the competence pilus). Green and red arrows respectively indicate genes translocated or inverted with respect to their position in S. pneumoniae D39. In arrangement D2, an additional recombination event caused the inversion of several genes (indicated by hatched arrows). A gray square indicates the region located 3’ of lytASpn shown in detail in Figure 6.
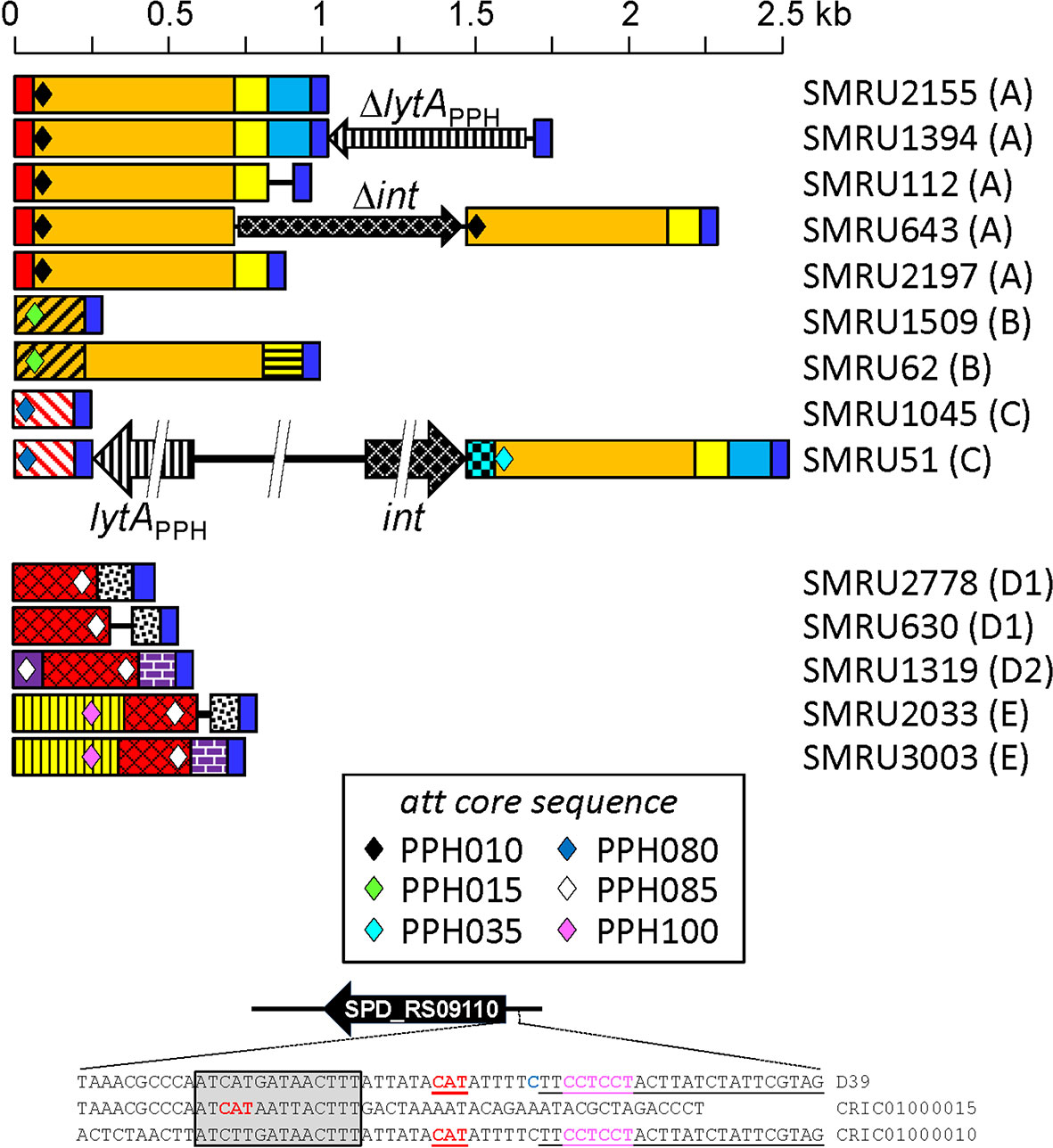
Figure 6 Diagram of the region located immediately downstream of the termination codon of lytASpn* in 14 pneumococcal strains with differently rearranged genomes. The region corresponds to that indicated with a gray square in Figure 4. DNA regions sharing ≥95% nucleotide identity are shown with the same color and shadowing. The type of arrangement (A to E) is indicated in parentheses to the right of the name of each strain. The different core attachment sites are shown as diamonds. The deep blue rectangles represent the conserved 51 bp-long sequence mentioned in the text. The red rectangles correspond to the reverse complement of positions 24037–24121 of the S. pneumoniae D39 genome (between SPD_RS00120 encoding a nucleoside deaminase and SPD_RS00115 coding for an adenylosuccinate synthase). The purple rectangle corresponds to positions 1,712,485–1,712,386 (between SPD_RS09110 and SPD_RS09115 in the D39 chromosome). For additional information see Tables S6 and S8. At the bottom, the nucleotide sequence surrounding the initiation codon of SPD_RS09110 in strains D39 and SMRU1319 (arrangement D2) is shown. The correct initiation codon is shown in red, bold font, and underlined. Another in-frame initiation codon is not underlined and apparently lacks a potential ribosome-binding site (RBS) (Acc. No. CRIC01000015). The predicted RBS of SPD_RS09110 is shown in pink lettering. The 14 bp-long repeat potentially responsible for a chromosomal inversion in strain SMRU1319 is inserted in a gray box. The att core sequence of PPH085 is underlined.
Discussion
According to the data presented in this and previous studies, prophages are very common in S. pneumoniae: up to 90% of pneumococcal isolates may harbor temperate phages, although nearly half of them (in the present work 47%) appear to be defective and lack the genes involved in morphogenesis and lysis. These can be considered ‘defective’ (incomplete or remnant) PPHs that may be in a state of mutational decay (Casjens, 2003). It should be remembered that genome decay does not take place homogeneously. Previous reports have shown that, in incomplete prophages, there is an enrichment in int genes, whereas genes involved in phage lytic functions are preferentially lost (Bobay et al., 2014; Khan et al., 2020). Of note, some defective prophages are capable of excision from the bacterial chromosome (Chen et al., 2019). Sometimes, defective prophages are also designated as ‘cryptic prophages’ (Wang and Wood, 2016), although in theory this term should include only fully functional prophages that have never been induced to follow a lytic cycle (Casjens, 2003). Recently, incomplete streptococcal prophages were termed ‘satellite’ prophages (Rezaei Javan et al., 2019), although the characteristic dependence of true satellite prophages upon proteins produced by a helper prophage for packaging and progeny phage release was not demonstrated. A defective prophage (possibly a PPH005_1 relative) has been reported as capable of excising from the chromosome, but, as expected, it does not seem to enter the lytic cycle (Chen et al., 2019).
The present work fully confirms that the vast majority of full length PPHs harbor lytAPPH, a gene homologous to lytASpn, encoding a 318-aa long NAM-amidase (Amidase_2 family) endolysin. This clearly indicates the enhancement of phage and host fitness. A small (but significant) number of PPH endolysins (28), however, corresponded to Cpl-1-like lysozymes, and only one to a CHAP-containing enzyme. The lysozymes were the hallmark endolysins of the PPH065 and PPH105 groups. Notably, all strains harboring PPH065 were members of GPSC19 with serotype 22F (the only exception being strain 2245STDY6178828, which belongs to serotype 42). Serotype 22F is an emerging serotype (Tin Tin Htar et al., 2019) that is not included in the current 13-valent pneumococcal conjugate vaccine, although it has been incorporated into the next generation of 15- and 20-valent pneumococcal conjugate vaccines currently under evaluation (Masomian et al., 2020). All these isolates belong to clonal complex 433, which is predominant in Spain (Sempere et al., 2020). In addition, strains harboring PPH105 are of serotype 35B (sequence type 558): a single-locus variant of the PMEN Utah35B-24 clone, also an emerging, multidrug-resistant serotype associated with non-vaccine serotypes (Andam et al., 2017). It has been shown that pediatric isolates of serotypes 22F form better biofilms than adult isolates (Sempere et al., 2020) and that serotype 35B pneumococci are good biofilm formers (Domenech et al., 2015). Whether this phenotype is directly related to the presence of prophages, as has been proposed (Loeffler and Fischetti, 2006; Carrolo et al., 2010; Harvey et al., 2016), remains to be determined. Previous experiments have shown that the Cpl-1 endolysin is very effective in destroying in vitro formed biofilms either of S. pneumoniae, or of the type strains of S. pseudopneumoniae and S. oralis (Domenech et al., 2011). In addition, a recent study has shown that ClyJ-3 (an engineered chimera with a very short linker) shows improved thermal stability and has greater bactericidal activity against pneumococci, as well as reduced cytotoxicity, than does its parental enzyme (Yang et al., 2020). Since the Cpl-1-like lysozymes found in the present work also have linkers shorter than that of Cpl-1, detailed in vitro and in vivo studies of the newly discovered PPH lysozymes are warranted.
Using a subset of the database formed by complete or near complete pneumococcal genomes, up to 24 different attB sites were detected. Taking into account the capacity of some prophages to insert at different sites, the existence of 20 different PPH groups was disclosed. Various prophages integrate into genes encoding csRNAs. In S. pneumoniae, these non-coding sRNAs show a high degree of similarity to each other, and have been shown to affect pneumococcal physiology pleiotropically, e.g., they are involved in β-lactam resistance, and regulate natural competence development. The mechanism of action of csRNAs appears to be additive (Brantl and Brückner, 2014). Previous studies have also reported that prophage insertion into SPD_RS09885 interrupted competence development (Croucher et al., 2011; Croucher et al., 2014; Croucher et al., 2016) and it was found that having an intact comGC (also named comYC) gene was significantly associated with increased carriage duration (Lees et al., 2017).
A tandem prophage insertion (PPH080_1AB) was also found. This is usually interpreted as the integration of two phages that use the same att (in this case, 5’-TTATAATTCATCCGC-3’). The differences between the parental prophages in PPH080_1B involve a tandem duplication of a cluster of nine genes in PPH080_1A (marked with different colors in Figure S2). Further, some rare events related to PPH insertion were also noted. PPH075, PPH080_3, and PPH120 contained attB sequences, which would allow another phage to be integrated into their genomes, forming a prophage-in-prophage configuration and allowing the duplication of prophages possibly encoding medically or biologically important genes. To the best of our knowledge, this phenomenon has only previously been described in Shiga toxin-encoding prophages in E. coli (Nakamura et al., 2021).
Although first described in phage T4 in 1968 (Weiss et al., 1968), the role of tRNA genes in phages has been subject to debate (Bailly-Bechet et al., 2007). Viruses, which depend on the host protein expression machinery, have evolved various strategies to optimize translation, either by adapting their host codon usage to that of the host or encoding their own tRNAs. It has been noted that most phages contain only one or two tRNA genes, while a few may contain ≥20 (Bailly-Bechet et al., 2007). In general, temperate phages harbor fewer tRNA genes than do virulent phages. It has been suggested that tRNA acquisition may contribute to greater virulence (Bailly-Bechet et al., 2007). A more recent study of different mycobacteriophage clusters suggests that the benefits of having tRNA genes may be associated with either a better growth in their hosts (larger burst size and shorter latency) or the ability to infect more hosts (Delesalle et al., 2016). Although an increasing number of prophages are described that carry tRNA genes (Canchaya et al., 2004), their presence among Streptococcus prophages appears to be uncommon. In a recent analysis of 13,200 viral sequences (Morgado and Vicente, 2019), only four streptococcal phages were found to harbor tRNA genes; three were virulent [SMP from S. suis (Acc. No. EF116926), SP-QS1 from S. pneumoniae (Acc. No. HE962497), and P5652 from Streptococcus thermophilus (Acc. No. KY705261)], and only one was temperate [ΦARI0746 from S. pneumoniae (Acc. No. KT337365)].
All the VapESsu-like proteins were encoded by defective prophages (with the exception of PPH08_12), as previously reported (Rezaei Javan et al., 2019). Both defective and complete prophages coded for VapEDno homologs. Experimental evidence exists that the VapESsu homolog of the defective prophage Javan757 (Acc. No. WP_000434359) is involved in virulence. Using a sepsis model, it was reported that mice infected with the wild-type pneumococcal strain had significantly greater blood and spleen bacterial counts than the isogenic ΔvapE mutant (Rezaei Javan et al., 2019). In addition, it was observed that the reduced virulence of the mutant could be explained by a significant growth delay when cultivated in serum. Further, vapE was upregulated only when the pneumococcal strain grew under planktonic conditions, analogous to bacteremic growth. Whether the other VapEDno homologs described here also play a significant role in pneumococcal pathogenicity warrants further investigation.
S. pneumoniae does not contain an endogenous CRISPR/Cas system; this is consistent with interference with natural transformation and thereby lateral gene transfer crucial for pneumococcal host adaptation (Bikard et al., 2012) (https://crispr.i2bc.paris-saclay.fr/). Among the defense mechanisms developed by bacteria to combat phage attack, R-M systems are pervasive. Their general role is to spot foreign DNA via base modifications and to defend the host DNA from restriction enzymes via Mtase activity. Recently, however, more functions for R-M systems have been described (Vasu and Nagaraja, 2013). Their activities are due to several heterogeneous proteins classified into at least four groups. Most S. pneumoniae isolates express either the DpnI or DpnII R-M system, although a DpnIII system has also been reported (Eutsey et al., 2015). Interestingly, the virulent pneumophage Cp-1 cannot be restricted in S. pneumoniae because it does not contain the corresponding target sequence (5’-GATC-3’) in its genome (Martín et al., 1996). The virulent pneumophage Dp-1 probably defends against host-induced DNA restriction by incorporating modified bases into it (García et al., 2005). This is consistent with a more recent report showing that resistance to Dp-1 in S. pneumoniae results from mutations in a single gene (SPD_RS05930) coding for a type IV restriction endonuclease (Leprohon et al., 2015). It is noteworthy that type IV restriction systems differ from other types in that the Mtase and endonuclease activities are combined in a single enzyme that requires base modification to act (Loenen and Raleigh, 2014). A phase-variable type I R-M system has also been identified in various strains of S. pneumoniae. This system operates as an epigenetic switch that regulates gene expression, virulence, and phase variation (opaque versus transparent phenotype) in pneumococci (Li and Zhang, 2019). Moreover, the importance that phase-variable type I R-M systems have in the multifunctional defense against prophage SpSL1 infection in S. pneumoniae has been demonstrated (Furi et al., 2019).
The present study provides clear evidence of recombination events between pneumococcal and phage lytA homologs. Recombination is apparently independent of the phage Int and is facilitated by the noticeable sequence similarity between the phage and host genes (≥85% identity). Although uncommon, a similar process has been reported to occur between the thyP3 and the thyA genes encoding the thymidylate synthase of the temperate bacteriophage φ3T and that of the Bacillus subtilis host (Tucker, 1969; Stroynowski, 1981; Stout et al., 1998; Fox et al., 1999). In this case, the nucleotide sequence identity reaches 96% (Kenny et al., 1985; Tam and Borriss, 1995). Unfortunately, whether recombination between the B. subtilis and the phage φ3T genes also causes genome rearrangements, as is the case of the PPH090 group, is unknown.
The present results provide a comprehensive view of the lysogenic state of phages in S. pneumoniae. As in most phage genomes currently under study, the majority of the PPH genes play uncharacterized roles. There is increasing evidence that although bacteriophages do not infect eukaryotic cells, they do interact with innate immune cells via Toll-like receptors (which appears to be particularly true for temperate bacteriophages) (Cieślik et al., 2021; Podlacha et al., 2021; Popescu et al., 2021), but the phage components involved in this are virtually unknown.
Finally, the consequences of genome rearrangements involving lytA genes in bacteria and phage physiology deserve to be further studied.
Author’s Note
This work is dedicated to our mentor and friend, Concepción Ronda, who fostered the research on pneumococcal bacteriophages in our laboratory.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Authors Contributions
EG designed the study. AM-G and EG conducted the analyses and wrote the article. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The help of Carmen Ascaso, who granted the infrastructure and peaceful environment required for this study to be completed, is greatly acknowledged. We thank P. García (CIB, CSIC) for carefully revising the article and suggestions. This research was supported by grants MPY 509/19 from the Instituto de Salud Carlos III (ISCIII) and SAF2017-88664-R from the Spanish Ministerio de Economía, Industria y Competitividad (MEICOM). The Centro de Investigación Biomédica en Red de Enfermedades Respiratorias (CIBERES) is an initiative of the Instituto de Salud Carlos III (ISCIII). AM-G is the recipient of a Miguel Servet contract by the ISCIII.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.775402/full#supplementary-material
Abbreviations
aa, amino acid(s); CHAP, a cysteine, histidine-dependent amidohydrolase/peptidase domain; csRNA, cia-dependent small RNA; GPSCs, Global Pneumococcal Sequence Clusters; Int, integrase; IS, insertion sequence; lytASpn, gene coding for the major pneumococcal autolysin; lytAPPH, phage gene encoding a NAM-amidase of the Amidase_2 family; NAM-amidase, N-acetylmuramoyl-L-alanine amidase; NCBI, National Center for Biotechnology Information; Mtase, DNA methyltransferase; NT, non-typeable; PMEN, Pneumococcal Molecular Epidemiology Network; PPH, pneumococcal prophage; RefSeq, NCBI Reference Sequence Dabatase; R-M, restriction-modification; ST, sequence type.
References
Achaz, G., Coissac, E., Netter, P., Rocha, E. P. C. (2003). Associations Between Inverted Repeats and the Structural Evolution of Bacterial Genomes. Genetics 164, 1279–1289. doi: 10.1093/genetics/164.4.1279
Afzal, M., Shafeeq, S., Henriques-Normark, B., Kuipers, O. P. (2015). UlaR Activates Expression of the Ula Operon in Streptococcus Pneumoniae in the Presence of Ascorbic Acid. Microbiology 161, 41–49. doi: 10.1099/mic.0.083899-0
Andam, C. P., Worby, C. J., Gierke, R., McGee, L., Pilishvili, T., Hanage, W. P. (2017). Penicillin Resistance of Nonvaccine Type Pneumococcus Before and After PCV13 Introduction, United States. Emerg. Infect. Dis. 23, 1012–1015. doi: 10.3201/eid2306.161331
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al (2016). PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Bailly-Bechet, M., Vergassola, M., Rocha, E. (2007). Causes for the Intriguing Presence of tRNAs in Phages. Genome Res. 17, 1486–1495. doi: 10.1101/gr.6649807
Bensing, B. A., Siboo, I. R., Sullam, P. M. (2001). Proteins PblA and PblB of Streptococcus Mitis, Which Promote Binding to Human Platelets, are Encoded Within a Lysogenic Bacteriophage. Infect. Immun. 69, 6186–6192. doi: 10.1128/IAI.69.10.6186-6192.2001
Bernheimer, H. P. (1977). Lysogeny in Pneumococci Freshly Isolated From Man. Science 195, 66–68. doi: 10.1126/science.12565
Bikard, D., Hatoum-Aslan, A., Mucida, D., Marraffini, L. A. (2012). CRISPR Interference can Prevent Natural Transformation and Virulence Acquisition During In Vivo Bacterial Infection. Cell Host Microbe 12, 177–186. doi: 10.1016/j.chom.2012.06.003
Billington, S. J., Johnston, J. L., Rood, J. I. (1996). Virulence Regions and Virulence Factors of the Ovine Footrot Pathogen, Dichelobacter Nodosus. FEMS Microbiol. Lett. 145, 147–156. doi: 10.1111/j.1574-6968.1996.tb08570.x
Bobay, L.-M., Touchon, M., Rocha, E. P. C. (2014). Pervasive Domestication of Defective Prophages by Bacteria. Proc. Natl. Acad. Sci. U.S.A. 111, 12127–12132. doi: 10.1073/pnas.1405336111
Brantl, S., Brückner, R. (2014). Small Regulatory RNAs From Low-GC Gram-Positive Bacteria. RNA Biol. 11, 443–456. doi: 10.4161/rna.28036
Brueggemann, A. B., Harrold, C. L., Javan, R. R., van Tonder, A. J., McDonnell, A. J., Edwards, B. A. (2017). Pneumococcal Prophages are Diverse, But Not Without Structure or History. Sci. Rep. 7, 42976. doi: 10.1038/srep42976
Càmara, J., Cubero, M., Martín-Galiano, A. J., García, E., Grau, I., Nielsen, J. B., et al (2018). Evolution of the β-Lactam-Resistant Streptococcus Pneumoniae PMEN3 Clone Over a 30-Year Period in Barcelona, Spain. J. Antimicrob. Chemother. 73, 2941–2951. doi: 10.1093/jac/dky305
Canchaya, C., Fournous, G., Brüssow, H. (2004). The Impact of Prophages on Bacterial Chromosomes. Mol. Microbiol. 53, 9–18. doi: 10.1111/j.1365-2958.2004.04113.x
Canchaya, C., Proux, C., Fournous, G., Bruttin, A., Brüssow, H. (2003). Prophage Genomics. Microbiol. Mol. Biol. Rev. 67, 238–276. doi: 10.1128/mmbr.67.2.238-276.2003
Canvin, J. R., Marvin, A. P., Sivakumaran, M., Paton, J. C., Boulnois, G. J., Andrew, P. W., et al (1995). The Role of Pneumolysin and Autolysin in the Pathology of Pneumonia and Septicemia in Mice Infected With a Type 2 Pneumococcus. J. Infect. Dis. 172, 119–123. doi: 10.1093/infdis/172.1.119
Carrolo, M., Frias, M. J., Pinto, F. R., Melo-Cristino, J., Ramirez, M. (2010). Prophage Spontaneous Activation Promotes DNA Release Enhancing Biofilm Formation in Streptococcus Pneumoniae. PLoS One 5, e15678. doi: 10.1371/journal.pone.0015678
Casjens, S. (2003). Prophages and Bacterial Genomics: What Have We Learned So Far? Mol. Microbiol. 49, 277–300. doi: 10.1046/j.1365-2958.2003.03580.x
Chen, H.-J., Chang, Y.-C., Tsai, J.-C., Hung, W.-C., Lin, Y.-T., You, S.-J., et al (2013). New Structure of Phage-Related Islands Carrying fusB and a Virulence Gene in Fusidic Acid-Resistant Staphylococcus Epidermidis. Antimicrob. Agents Chemother. 57, 5737–5739. doi: 10.1128/AAC.01433-13
Chen, Y.-Y., Wang, J.-T., Lin, T.-L., Gong, Y.-N., Li, T.-H., Huang, Y.-Y., et al (2019). Prophage Excision in Streptococcus Pneumoniae Serotype 19A ST320 Promote Colonization: Insight Into its Evolution From the Ancestral Clone Taiwan 19F-14 (ST236). Front. Microbiol. 10, 205. doi: 10.3389/fmicb.2019.00205
Cieślik, M., Bagińska, N., Jończyk-Matysiak, E., Węgrzyn, A., Węgrzyn, G., Górski, A. (2021). Temperate Bacteriophages—The Powerful Indirect Modulators of Eukaryotic Cells and Immune Functions. Viruses 13, 1013. doi: 10.3390/v13061013
Corsini, B., Aguinagalde, L., Ruiz, S., Domenech, M., Yuste, J. (2021). Vaccination With LytA, LytC, or Pce of Streptococcus Pneumoniae Protects Against Sepsis by Inducing IgGs That Activate the Complement System. Vaccines 9, 186. doi: 10.3390/vaccines9020186
Cremers, A. J. H., Mobegi, F. M., van der Gaast-de Jongh, C., van Weert, M., van Opzeeland, F. J., Vehkala, M., et al (2019). The Contribution of Genetic Variation of Streptococcus Pneumoniae to the Clinical Manifestation of Invasive Pneumococcal Disease. Clin. Infect. Dis. 68, 61–69. doi: 10.1093/cid/ciy417
Croucher, N. J., Campo, J. J., Leb, T. Q., Liang, X., Bentley, S. D., Hanage, W. P., et al (2017). Diverse Evolutionary Patterns of Pneumococcal Antigens Identified by Pangenome-Wide Immunological Screening. Proc. Natl. Acad. Sci. U.S.A. 114, E357–E366. doi: 10.1073/pnas.1613937114
Croucher, N. J., Hanage, W. P., Harris, S. R., McGee, L., van der Linden, M., de Lencastre, H., et al (2014). Variable Recombination Dynamics During the Emergence, Transmission and 'Disarming' of a Multidrug-Resistant Pneumococcal Clone. BMC Biol. 12, 49. doi: 10.1186/1741-7007-12-49
Croucher, N. J., Harris, S. R., Fraser, C., Quail, M. A., Burton, J., van der Linden, M., et al (2011). Rapid Pneumococcal Evolution in Response to Clinical Interventions. Science 331, 430–434. doi: 10.1126/science.1198545
Croucher, N. J., Mostowy, R., Wymant, C., Turner, P., Bentley, S. D., Fraser, C. (2016). Horizontal DNA Transfer Mechanisms of Bacteria as Weapons of Intragenomic Conflict. PLoS Biol. 14, e1002394. doi: 10.1371/journal.pbio.1002394
Delesalle, V. A., Tanke, N. T., Vill, A. C., Krukonis, G. P. (2016). Testing Hypotheses for the Presence of tRNA Genes in Mycobacteriophage Genomes. Bacteriophage 6, e1219441. doi: 10.1080/21597081.2016.1219441
Denapaite, D., Rieger, M., Köndgen, S., Brückner, R., Ochigava, I., Kappeler, P., et al (2016). Highly Variable Streptococcus Oralis Strains are Common Among Viridans Streptococci Isolated From Primates. mSphere 1, e00041–e00015. doi: 10.1128/mSphere.00041-15
Díaz, E., García, J. L. (1990). Characterization of the Transcription Unit Encoding the Major Pneumococcal Autolysin. Gene 90, 157–162. doi: 10.1016/0378-1119(90)90454-y
Diene, S. M., François, P., Zbinden, A., Entenza, J. M., Resch, G. (2016). Comparative Genomics Analysis of Streptococcus Tigurinus Strains Identifies Genetic Elements Specifically and Uniquely Present in Highly Virulent Strains. PLoS One 11, e0160554. doi: 10.1371/journal.pone.0160554
Domenech, M., Damián, D., Ardanuy, C., Liñares, J., Fenoll, A., García, E. (2015). Emerging, non-PCV13 Serotypes 11A and 35B of Streptococcus Pneumoniae Show High Potential for Biofilm Formation In Vitro. PLoS One 10, e0125636. doi: 10.1371/journal.pone.0125636
Domenech, M., García, E., Moscoso, M. (2011). In Vitro Destruction of Streptococcus Pneumoniae Biofilms With Bacterial and Phage Peptidoglycan Hydrolases. Antimicrob. Agents Chemother. 55, 4144–4148. doi: 10.1128/aac.00492-11
Enright, M. C., Spratt, B. G. (1998). A Multilocus Sequence Typing Scheme for Streptococcus Pneumoniae: Identification of Clones Associated With Serious Invasive Disease. Microbiology 144, 3049–3060. doi: 10.1099/00221287-144-11-3049
Eutsey, R. A., Powell, E., Dordel, J., Salter, S. J., Clark, T. A., Korlach, J., et al (2015). Genetic Stabilization of the Drug-Resistant PMEN1 Pneumococcus Lineage by its Distinctive DpnIII Restriction-Modification System. mBio 6, e00173. doi: 10.1128/mBio.00173-15
Fernández, L., Cima-Cabal, M. D., Duarte, A. C., Rodríguez, A., García-Suárez, M., García, P. (2021). Gram-Positive Pneumonia: Possibilities Offered by Phage Therapy. Antibiotics (Basel) 10, 1000. doi: 10.3390/antibiotics10081000
Fox, K. M., Maley, F., Garibian, A., Changchien, L. M., Van Roey, P. (1999). Crystal Structure of Thymidylate Synthase A From Bacillus Subtilis. Protein Sci. 8, 538–544. doi: 10.1110/ps.8.3.538
Frolet, C., Beniazza, M., Roux, L., Gallet, B., Noirclerc-Savoye, M., Vernet, T., et al (2010). New Adhesin Functions of Surface-Exposed Pneumococcal Proteins. BMC Microbiol. 10, 190. doi: 10.1186/1471-2180-10-190
Furi, L., Crawford, L. A., Rangel-Pineros, G., Manso, A. S., De Ste Croix, M., Haigh, R. D., et al (2019). Methylation Warfare: Interaction of Pneumococcal Bacteriophages With Their Host. J. Bacteriol. 201, e00370–e00319. doi: 10.1128/jb.00370-19
Galán-Bartual, S., Pérez-Dorado, I., García, P., Hermoso, J. A. (2015). “Structure and Function of Choline-Binding Proteins,” in Streptococcus Pneumoniae. Molecular Mechanisms of Host-Pathogen Interactions. Eds. Brown, J., Hammerschimdt, S., Orihuela, C. (Amsterdam: Academic Press), 207–230. doi: 10.1016/b978-0-12-410530-0.00011-9
García, P., García, J. L., López, R., García, E. (2005). “Pneumococcal Phages,” in Phages: Their Role in Bacterial Pathogenesis and Biotechnology. Eds. Waldor, M. K., Friedman, D. L. I., Adhya, S. (Washington, D.C: ASM Press), 335–361. doi: 10.1128/9781555816506.ch17
Garriss, G., Henriques-Normark, B. (2020). Lysogeny in Streptococcus Pneumoniae. Microorganisms 8, 1546. doi: 10.3390/microorganisms8101546
GBD 2016 Lower Respiratory Infections Collaborators (2018). Estimates of the Global, Regional, and National Morbidity, Mortality, and Aetiologies of Lower Respiratory Infections in 195 Countries 1990–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 18, 1191–1210. doi: 10.1016/S1473-3099(18)30310-4
Gibb, B., Gupta, K., Ghosh, K., Sharp, R., Chen, J., Van Duyne, G. D. (2010). Requirements for Catalysis in the Cre Recombinase Active Site. Nucleic Acids Res. 38, 5817–5832. doi: 10.1093/nar/gkq384
Gindreau, E., López, R., García, P. (2000). MM1, a Temperate Bacteriophage of the 23F Spanish/USA Multiresistant Epidemic Clone of Streptococcus Pneumoniae: Structural Analysis of the Site-Specific Integration System. J. Virol. 74, 7803–7813. doi: 10.1128/jvi.74.17.7803-7813.2000
Gladstone, R. A., Lo, S. W., Lees, J. A., Croucher, N. J., van Tonder, A. J., Corander, J., et al (2019). International Genomic Definition of Pneumococcal Lineages, to Contextualise Disease, Antibiotic Resistance and Vaccine Impact. EBioMedicine 43, 338–346. doi: 10.1016/j.ebiom.2019.04.021
Groth, A. C., Calos, M. P. (2004). Phage Integrases: Biology and Applications. J. Mol. Biol. 335, 667–678. doi: 10.1016/j.jmb.2003.09.082
Gu, J., Feng, Y., Feng, X., Sun, C., Lei, L., Ding, W., et al (2014). Structural and Biochemical Characterization Reveals LysGH15 as an Unprecedented "EF-Hand-Like" Calcium-Binding Phage Lysin. PLoS Pathog. 10, e1004109. doi: 10.1371/journal.ppat.1004109
Gu, J., Xu, W., Lei, L., Huang, J., Feng, X., Sun, C., et al (2011). LysGH15, a Novel Bacteriophage Lysin, Protects a Murine Bacteremia Model Efficiently Against Lethal Methicillin-Resistant Staphylococcus Aureus Infection. J. Clin. Microbiol. 49, 111–117. doi: 10.1128/JCM.01144-10
Harvey, R. M., Trappetti, C., Mahdi, L. K., Wang, H., McAllister, L. J., Scalvini, A., et al (2016). The Variable Region of Pneumococcal Pathogenicity Island 1 is Responsible for Unusually High Virulence of a Serotype 1 Isolate. Infect. Immun. 84, 822–832. doi: 10.1128/IAI.01454-15
Hermoso, J. A., Monterroso, B., Albert, A., Galán, B., Ahrazem, O., García, P., et al (2003). Structural Basis for Selective Recognition of Pneumococcal Cell Wall by Modular Endolysin From Phage Cp-1. Structure 11, 1239–1249. doi: 10.1016/j.str.2003.09.005
Hsieh, Y.-C., Lin, T.-L., Lin, C.-M., Wang, J.-T. (2015). Identification of PblB Mediating Galactose-Specific Adhesion in a Successful Streptococcus Pneumoniae Clone. Sci. Rep. 5, 12265. doi: 10.1038/srep12265
Hughes, D. (2000). Evaluating Genome Dynamics: The Constraints on Rearrangements Within Bacterial Genomes. Genome Biol. 1, REVIEWS0006. doi: 10.1186/gb-2000-1-6-reviews0006
Hyatt, D., Chen, G.-L., LoCascio, P. F., Land, M. L., Larimer, F. W., Hauser, L. J. (2010). Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinformatics 11, 119. doi: 10.1186/1471-2105-11-119
Ji, X., Sun, Y., Liu, J., Zhu, L., Guo, X., Lang, X., et al (2016). A Novel Virulence-Associated Protein, VapE, in Streptococcus Suis Serotype 2. Mol. Med. Rep. 13, 2871–2877. doi: 10.3892/mmr.2016.4818
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., Madden, T. L. (2008). NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 36, W5–W9. doi: 10.1093/nar/gkn201
Jolley, K. A., Bray, J. E., Maiden, M. C. J. (2018). Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.org Website and Their Applications. Wellcome Open Res. 3, 124. doi: 10.12688/wellcomeopenres.14826.1
Kenny, E., Atkinson, T., Hartley, B. S. (1985). Nucleotide Sequence of the Thymidylate Synthetase Gene (Thyp3) From the Bacillus Subtilis Phage φ3T. Gene 34, 335–342. doi: 10.1016/0378-1119(85)90142-8
Khan, A., Burmeister, A. R., Wahl, L. M. (2020). Evolution Along the Parasitism-Mutualism Continuum Determines the Genetic Repertoire of Prophages. PLoS Comput. Biol. 16, e1008482. doi: 10.1371/journal.pcbi.1008482
Kilian, M., Riley, D. R., Jensen, A., Brüggemann, H., Tettelin, H. (2014). Parallel Evolution of Streptococcus Pneumoniae and Streptococcus Mitis to Pathogenic and Mutualistic Lifestyles. mBio 5, e01490–14. doi: 10.1128/mBio.01490-14
Koonin, E. V., Makarova, K. S., Wolf, Y. I. (2017). Evolutionary Genomics of Defense Systems in Archaea and Bacteria. Annu. Rev. Microbiol. 71, 233–261. doi: 10.1146/annurev-micro-090816-093830
Kot, W., Sabri, M., Gingras, H., Ouellette, M., Tremblay, D. M., Moineau, S. (2017). Complete Genome Sequence of Streptococcus Pneumoniae Virulent Phage MS1. Genome Announc. 5, e00333–17. doi: 10.1128/genomeA.00333-17
Labrie, S. J., Samson, J. E., Moineau, S. (2010). Bacteriophage Resistance Mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al (2012). Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Laslett, D., Canback, B. (2004). ARAGORN, a Program to Detect tRNA Genes and tmRNA Genes in Nucleotide Sequences. Nucleic Acids Res. 32, 11–16. doi: 10.1093/nar/gkh152
Laurenceau, R., Krasteva, P. V., Diallo, A., Ouarti, S., Duchateau, M., Malosse, C., et al (2015). Conserved Streptococcus Pneumoniae Spirosomes Suggest a Single Type of Transformation Pilus in Competence. PLoS Pathog. 11, e1004835. doi: 10.1371/journal.ppat.1004835
Lees, J. A., Croucher, N. J., Goldblatt, D., Nosten, F., Parkhill, J., Turner, C., et al (2017). Genome-Wide Identification of Lineage and Locus Specific Variation Associated With Pneumococcal Carriage Duration. eLife 6, e26255. doi: 10.7554/eLife.26255
Leprohon, P., Gingras, H., Ouennane, S., Moineau, S., Ouellette, M. (2015). A Genomic Approach to Understand Interactions Between Streptococcus Pneumoniae and its Bacteriophages. BMC Genomics 16, 972. doi: 10.1186/s12864-015-2134-8
Li, Q., Cheng, W., Morlot, C., Bai, X.-H., Jiang, Y.-L., Wang, W., et al (2015). Full-Length Structure of the Major Autolysin LytA. Acta Crystallogr. D Biol. Crystallogr. 71, 1373–1381. doi: 10.1107/S1399004715007403
Lindsay, J. A., Ruzin, A., Ross, H. F., Kurepina, N., Novick, R. P. (1998). The Gene for Toxic Shock Toxin is Carried by a Family of Mobile Pathogenicity Islands in Staphylococcus Aureus. Mol. Microbiol. 29, 527–543. doi: 10.1046/j.1365-2958.1998.00947.x
Li, J., Zhang, J. R. (2019). Phase Variation of Streptococcus Pneumoniae. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.GPP3-0005-2018
Llull, D., López, R., García, E. (2006). Skl, a Novel Choline-Binding N-Acetylmuramoyl-L-Alanine Amidase of Streptococcus Mitis SK137 Containing a CHAP Domain. FEBS Lett. 580, 1959–1964. doi: 10.1016/j.febslet.2006.02.060
Loeffler, J. M., Fischetti, V. A. (2006). Lysogeny of Streptococcus Pneumoniae With MM1 Phage: Improved Adherence and Other Phenotypic Changes. Infect. Immun. 74, 4486–4495. doi: 10.1128/IAI.00020-06
Loenen, W. A. M., Raleigh, E. A. (2014). The Other Face of Restriction: Modification-Dependent Enzymes. Nucleic Acids Res. 42, 56–69. doi: 10.1093/nar/gkt747
Lopes de Sousa, A., Maués, D., Lobato, A., Franco, E. F., Pinheiro, K., Araújo, F., et al (2018). PhageWeb – Web Interface for Rapid Identification and Characterization of Prophages in Bacterial Genomes. Front. Genet. 9, 644. doi: 10.3389/fgene.2018.00644
López, E., Domenech, A., Ferrándiz, M.-J., Frias, M. J., Ardanuy, C., Ramirez, M., et al (2014). Induction of Prophages by Fluoroquinolones in Streptococcus Pneumoniae: Implications for Emergence of Resistance in Genetically-Related Clones. PLoS One 9, e94358. doi: 10.1371/journal.pone.0094358
López, R., García, E. (2004). Recent Trends on the Molecular Biology of Pneumococcal Capsules, Lytic Enzymes, and Bacteriophage. FEMS Microbiol. Rev. 28, 553–580. doi: 10.1016/j.femsre.2004.05.002
Lund, E., Henrichsen, J. (1978). Laboratory Diagnosis, Serology and Epidemiology of Streptococcus Pneumoniae. Methods Microbiol. 12, 241–262. doi: 10.1016/S0580-9517(08)70365-9
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al (2020). CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Maestro, B., Sanz, J. M. (2016). Choline Binding Proteins From Streptococcus Pneumoniae: A Dual Role as Enzybiotics and Targets for the Design of New Antimicrobials. Antibiotics (Basel) 5, 21. doi: 10.3390/antibiotics5020021
Martín, A. C., López, R., García, P. (1996). Analysis of the Complete Nucleotide Sequence and Functional Organization of the Genome of Streptococcus Pneumoniae Bacteriophage Cp-1. J. Virol. 70, 3678–3687. doi: 10.1128/JVI.70.6.3678-3687.1996
Masomian, M., Ahmad, Z., Gew, L. T., Poh, C. L. (2020). Development of Next Generation Streptococcus Pneumoniae Vaccines Conferring Broad Protection. Vaccines 8, 132. doi: 10.3390/vaccines8010132
McGee, L., McDougal, L., Zhou, J., Spratt, B. G., Tenover, F. C., George, R., et al (2001). Nomenclature of Major Antimicrobial-Resistant Clones of Streptococcus Pneumoniae Defined by the Pneumococcal Molecular Epidemiology Network. J. Clin. Microbiol. 39, 2565–2571. doi: 10.1128/JCM.39.7.2565-2571.2001
Mellroth, P., Sandalova, T., Kikhney, A., Vilaplana, F., Hesek, D., Lee, M., et al (2014). Structural and Functional Insights Into Peptidoglycan Access for the Lytic Amidase LytA of Streptococcus Pneumoniae. mBio 5, e01120–e01113. doi: 10.1128/mBio.01120-13
Mirdita, M., Steinegger, M., Söding, J. (2019). MMseqs2 Desktop and Local Web Server App for Fast, Interactive Sequence Searches. Bioinformatics 35, 2856–2858. doi: 10.1093/bioinformatics/bty1057
Morales, M., García, P., de la Campa, A. G., Liñares, J., Ardanuy, C., García, E. (2010). Evidence of Localized Prophage-Host Recombination in the lytA Gene Encoding the Major Pneumococcal Autolysin. J. Bacteriol. 192, 2624–2632. doi: 10.1128/JB.01501-09
Morales, M., Martín-Galiano, A. J., Domenech, M., García, E. (2015). Insights Into the Evolutionary Relationships of LytA Autolysin and Ply Pneumolysin-Like Genes in Streptococcus Pneumoniae and Related Streptococci. Genome Biol. Evol. 7, 2747–2761. doi: 10.1093/gbe/evv178
Morgado, S., Vicente, A. C. (2019). Global in-Silico Scenario of tRNA Genes and Their Organization in Virus Genomes. Viruses 11, 180. doi: 10.3390/v11020180
Murphy, J., Mahony, J., Ainsworth, S., Nauta, A., van Sinderen, D. (2013). Bacteriophage Orphan DNA Methyltransferases: Insights From Their Bacterial Origin, Function, and Occurrence. Appl. Environ. Microbiol. 79, 7547–7555. doi: 10.1128/aem.02229-13
Murray, E., Draper, L. A., Ross, R. P., Hill, C. (2021). The Advantages and Challenges of Using Endolysins in a Clinical Setting. Viruses 13, 680. doi: 10.3390/v13040680
Mushegian, A. R. (2020). Are There 1031 Virus Particles on Earth, or More, or Fewer? J. Bacteriol. 202, e00052–20. doi: 10.1128/jb.00052-20
Nakamura, Y., Gojobori, T., Ikemura, T. (2000). Codon Usage Tabulated From International DNA Sequence Databases: Status for the Year 2000. Nucleic Acids Res. 28, 292. doi: 10.1093/nar/28.1.292
Nakamura, K., Ogura, Y., Gotoh, Y., Hayashi, T. (2021). Prophages Integrating Into Prophages: A Mechanism to Accumulate Type III Secretion Effector Genes and Duplicate Shiga Toxin-Encoding Prophages in Escherichia Coli. PLoS Pathog. 17, e1009073. doi: 10.1371/journal.ppat.1009073
Nieto, C., Espinosa, M., Puyet, A. (1997). The Maltose/Maltodextrin Regulon of Streptococcus Pneumoniae. Differential Promoter Regulation by the Transcriptional Repressor MalR. J. Biol. Chem. 272, 30860–30865. doi: 10.1074/jbc.272.49.30860
O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al (2016). Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Obregón, V., García, J. L., García, E., López, R., García, P. (2003a). Genome Organization and Molecular Analysis of the Temperate Bacteriophage MM1 of Streptococcus Pneumoniae. J. Bacteriol. 185, 2362–2368. doi: 10.1128/jb.185.7.2362-2368.2003
Obregón, V., García, P., López, R., García, J. L. (2003b). VO1, a Temperate Bacteriophage of the Type 19A Multiresistant Epidemic 8249 Strain of Streptococcus Pneumoniae: Analysis of Variability of Lytic and Putative C5 Methyltransferase Genes. Microb. Drug Resist. 9, 7–15. doi: 10.1089/107662903764736292
Ouennane, S., Leprohon, P., Moineau, S. (2015). Diverse Virulent Pneumophages Infect Streptococcus Mitis. PLoS One 10, e0118807. doi: 10.1371/journal.pone.0118807
Pei, J., Mitchell, D. A., Dixon, J. E., Grishin, N. V. (2011). Expansion of Type II CAAX Proteases Reveals Evolutionary Origin of γ-Secretase Subunit APH-1. J. Mol. Biol. 410, 18–26. doi: 10.1016/j.jmb.2011.04.066
Peng, S.-Y., Chen, L.-K., Wu, W.-J., Paramita, P., Yang, P.-W., Li, Y.-Z., et al (2020). Isolation and Characterization of a New Phage Infecting Elizabethkingia Anophelis and Evaluation of its Therapeutic Efficacy In Vitro and In Vivo. Front. Microbiol. 11, 728. doi: 10.3389/fmicb.2020.00728
Pfeifer, E., Moura de Sousa, J. A., Touchon, M., Rocha, E. P. C. (2021). Bacteria Have Numerous Distinctive Groups of Phage-Plasmids With Conserved Phage and Variable Plasmid Gene Repertoires. Nucleic Acids Res. 49, 2655–2673. doi: 10.1093/nar/gkab064
Podlacha, M., Grabowski, Ł., Kosznik-Kawśnicka, K., Zdrojewska, K., Stasiłojć, M., Węgrzyn, G., et al (2021). Interactions of Bacteriophages With Animal and Human Organisms—Safety Issues in the Light of Phage Therapy. Int. J. Mol. Sci. 22, 8937. doi: 10.3390/ijms22168937
Popescu, M., Van Belleghem, J. D., Khosravi, A., Bollyky, P. L. (2021). Bacteriophages and the Immune System. Annu. Rev. Virol. 8, 67–87. doi: 10.1146/annurev-virology-091919-074551
Ramirez, M., Severina, E., Tomasz, A. (1999). A High Incidence of Prophage Carriage Among Natural Isolates of Streptococcus Pneumoniae. J. Bacteriol. 181, 3618–3625. doi: 10.1128/JB.181.12.3618-3625.1999
Ramos-Sevillano, E., Urzainqui, A., Campuzano, S., Moscoso, M., González-Camacho, F., Domenech, M., et al (2015). Pleiotropic Effects of the Cell Wall Amidase LytA on Streptococcus Pneumoniae Sensitivity to the Host Immune Response. Infect. Immun. 83, 591–603. doi: 10.1128/iai.02811-14
Ramos-Sevillano, E., Urzainqui, A., de Andrés, B., González-Tajuelo, R., Domenech, M., González-Camacho, F., et al (2016). PSGL-1 on Leukocytes is a Critical Component of the Host Immune Response Against Invasive Pneumococcal Disease. PLoS Pathog. 12, e1005500. doi: 10.1371/journal.ppat.1005500
Reis-Cunha, J. L., Bartholomeu, D. C., Manson, A. L., Earl, A. M., Cerqueira, G. C. (2019). ProphET, Prophage Estimation Tool: A Stand-Alone Prophage Sequence Prediction Tool With Self-Updating Reference Database. PLoS One 14, e0223364. doi: 10.1371/journal.pone.0223364
Rezaei Javan, R., Ramos-Sevillano, E., Akter, A., Brown, J., Brueggemann, A. B. (2019). Prophages and Satellite Prophages are Widespread in Streptococcus and may Play a Role in Pneumococcal Pathogenesis. Nat. Commun. 10, 4852. doi: 10.1038/s41467-019-12825-y
Roach, D. J., Burton, J. N., Lee, C., Stackhouse, B., Butler-Wu, S. M., Cookson, B. T., et al (2015). A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota. PLoS Genet. 11, e1005413. doi: 10.1371/journal.pgen.1005413
Romero, P., Croucher, N. J., Hiller, N. L., Hu, F. Z., Ehrlich, G. D., Bentley, S. D., et al (2009a). Comparative Genomic Analysis of Ten Streptococcus Pneumoniae Temperate Bacteriophages. J. Bacteriol. 191, 4854–4862. doi: 10.1128/jb.01272-08
Romero, P., García, E., Mitchell, T. J. (2009b). Development of a Prophage Typing System and Analysis of Prophage Carriage in Streptococcus Pneumoniae. Appl. Environ. Microbiol. 75, 1642–1649. doi: 10.1128/AEM.02155-08
Romero, A., Lopez, R., Garcia, P. (1990). Sequence of the Streptococcus Pneumoniae Bacteriophage HB-3 Amidase Reveals High Homology With the Major Host Autolysin. J. Bacteriol. 172, 5064–5070. doi: 10.1128/jb.172.9.5064-5070.1990
Rostøl, J. T., Marraffini, L. (2019). (Ph)ighting Phages: How Bacteria Resist Their Parasites. Cell Host Microbe 25, 184–194. doi: 10.1016/j.chom.2019.01.009
Sánchez-Beato, A. R., López, R., García, J. L. (1998). Molecular Characterization of PcpA: A Novel Choline-Binding Protein of Streptococcus Pneumoniae. FEMS Microbiol. Lett. 164, 207–214. doi: 10.1016/s0378-1097(98)00206-7
Schattner, P., Brooks, A. N., Lowe, T. M. (2005). The tRNAscan-SE, Snoscan and snoGPS Web Servers for the Detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689. doi: 10.1093/nar/gki366
Schroven, K., Aertsen, A., Lavigne, R. (2021). Bacteriophages as Drivers of Bacterial Virulence and Their Potential for Biotechnological Exploitation. FEMS Microbiol. Rev. 45, fuaa041. doi: 10.1093/femsre/fuaa041
Seguritan, V., Feng, I. W., Rohwer, F., Swift, M., Segall, A. M. (2003). Genome Sequences of Two Closely Related Vibrio Parahaemolyticus Phages, VP16T and VP16C. J. Bacteriol. 185, 6434–6447. doi: 10.1128/JB.185.21.6434-6447.2003
Sempere, J., de Miguel, S., González-Camacho, F., Yuste, J., Domenech, M. (2020). Clinical Relevance and Molecular Pathogenesis of the Emerging Serotypes 22F and 33F of Streptococcus Pneumoniae in Spain. Front. Microbiol. 11, 309. doi: 10.3389/fmicb.2020.00309
Shimada, K., Weisberg, R. A., Gottesman, M. E. (1975). Prophage Lambda at Unusual Chromosomal Locations. III. The Components of the Secondary Attachment Sites. J. Mol. Biol. 93, 415–429. doi: 10.1016/0022-2836(75)90237-5
Sievers, F., Higgins, D. G. (2021). The Clustal Omega Multiple Alignment Package. Methods Mol. Biol. 2231, 3–16. doi: 10.1007/978-1-0716-1036-7_1
Silva, M. D., Oliveira, H., Faustino, A., Sillankorva, S. (2020). Characterization of MSlys, the Endolysin of Streptococcus Pneumoniae Phage MS1. Biotechnol. Rep. (Amst.) 28, e00547. doi: 10.1016/j.btre.2020.e00547
Sinha, D., Zimmer, K., Cameron, T. A., Rusch, D. B., Winkler, M. E., De Lay, N. R. (2019). Redefining the Small Regulatory RNA Transcriptome in Streptococcus Pneumoniae Serotype 2 Strain D39. J. Bacteriol. 201, e00764–18. doi: 10.1128/JB.00764-18
Slager, J., Aprianto, R., Veening, J.-W. (2018). Deep Genome Annotation of the Opportunistic Human Pathogen Streptococcus Pneumoniae D39. Nucleic Acids Res. 46, 9971–9989. doi: 10.1093/nar/gky725
Steinberg, R., Knüpffer, L., Origi, A., Asti, R., Koch, H.-G. (2018). Co-Translational Protein Targeting in Bacteria. FEMS Microbiol. Lett. 365, fny095. doi: 10.1093/femsle/fny095
Stout, T. J., Schellenberger, U., Santi, D. V., Stroud, R. M. (1998). Crystal Structures of a Unique Thermal-Stable Thymidylate Synthase From Bacillus Subtilis. Biochemistry 37, 14736–14747. doi: 10.1021/bi981270l
Stroynowski, I. T. (1981). Integration of the Bacteriophage Ф3T-Coded Thymidylate Synthetase Gene Into the Bacillus Subtilis Chromosome. J. Bacteriol. 148, 101–108. doi: 10.1128/jb.148.1.101-108.1981
Tam, N. H., Borriss, R. (1995). The thyA Gene From Bacillus Subtilis Exhibits Similarity With the Phage Ф3T Thymidylate Synthase Gene. Microbiology 141, 291–297. doi: 10.1099/13500872-141-2-291
Tin Tin Htar, M., Morato Martínez, J., Theilacker, C., Schmitt, H.-J., Swerdlow, D. (2019). Serotype Evolution in Western Europe: Perspectives on Invasive Pneumococcal Diseases (IPD). Expert Rev. Vaccines 18, 1145–1155. doi: 10.1080/14760584.2019.1688149
Touchon, M., Bernheim, A., Rocha, E. P. (2016). Genetic and Life-History Traits Associated With the Distribution of Prophages in Bacteria. ISME J. 10, 2744–2754. doi: 10.1038/ismej.2016.47
Tucker, R. G. (1969). Acquisition of Thymidylate Synthetase Activity by a Thymine-Requiring Mutant of Bacillus Subtilis Following Infection by the Temperate Phage φ3. J. Gen. Virol. 4, 489–504. doi: 10.1099/0022-1317-4-4-489
Tunjungputri, R. N., Mobegi, F. M., Cremers, A. J., van der Gaast-de Jongh, C. E., Ferwerda, G., Meis, J. F., et al (2017). Phage-Derived Protein Induces Increased Platelet Activation and is Associated With Mortality in Patients With Invasive Pneumococcal Disease. mBio 8, e01984–16. doi: 10.1128/mBio.01984-16
van der Kamp, I., Draper, L. A., Smith, M. K., Buttimer, C., Ross, R. P., Hill, C. (2020). A New Phage Lysin Isolated From the Oral Microbiome Targeting Streptococcus Pneumoniae. Pharmaceuticals (Basel) 13, 478. doi: 10.3390/ph13120478
Van Duyne, G. D., Rutherford, K. (2013). Large Serine Recombinase Domain Structure and Attachment Site Binding. Crit. Rev. Biochem. Mol. Biol. 48, 476–491. doi: 10.3109/10409238.2013.831807
van Tonder, A. J., Bray, J. E., Jolley, K. A., Jansen van Rensburg, M., Quirk, S. J., Haraldsson, G., et al (2019). Genomic Analyses of >3,100 Nasopharyngeal Pneumococci Revealed Significant Differences Between Pneumococci Recovered in Four Different Geographical Regions. Front. Microbiol. 10, 317. doi: 10.3389/fmicb.2019.00317
Vasu, K., Nagaraja, V. (2013). Diverse Functions of Restriction-Modification Systems in Addition to Cellular Defense. Microbiol. Mol. Biol. Rev. 77, 53–72. doi: 10.1128/MMBR.00044-12
Vázquez, R., García, E., García, P. (2018). Phage Lysins for Fighting Bacterial Respiratory Infections: A New Generation of Antimicrobials. Front. Immunol. 9, 2252. doi: 10.3389/fimmu.2018.02252
Wang, X., Wood, T. K. (2016). Cryptic Prophages as Targets for Drug Development. Drug Resist. Updat. 27, 30–38. doi: 10.1016/j.drup.2016.06.001
Weiss, S. B., Hsu, W.-T., Foft, J. W., Scherberg, N. H. (1968). Transfer RNA Coded by the T4 Bacteriophage Genome. Proc. Natl. Acad. Sci. U.S.A. 61, 114–121. doi: 10.1073/pnas.61.1.114
Whatmore, A. M., Dowson, C. G. (1999). The Autolysin-Encoding Gene (Lyta) of Streptococcus Pneumoniae Displays Restricted Allelic Variation Despite Localized Recombination Events With Genes of Pneumococcal Bacteriophage Encoding Cell Wall Lytic Enzymes. Infect. Immun. 67, 4551–4556. doi: 10.1128/iai.67.9.4551-4556.1999
Williams, K. P. (2002). Integration Sites for Genetic Elements in Prokaryotic tRNA and tmRNA Genes: Sublocation Preference of Integrase Subfamilies. Nucleic Acids Res. 30, 866–875. doi: 10.1093/nar/30.4.866
Willner, D., Furlan, M., Schmieder, R., Grasis, J. A., Pride, D. T., Relman, D. A., et al (2011). Metagenomic Detection of Phage-Encoded Platelet-Binding Factors in the Human Oral Cavity. Proc. Natl. Acad. Sci. U.S.A. 108 Suppl 1, 4547–4553. doi: 10.1073/pnas.1000089107
Yang, H., Luo, D., Etobayeva, I., Li, X., Gong, Y., Wang, S., et al (2020). Linker Editing of Pneumococcal Lysin ClyJ Conveys Improved Bactericidal Activity. Antimicrob. Agents Chemother. 64, e01610–19. doi: 10.1128/aac.01610-19
Keywords: Streptococcus pneumoniae, prophage, integrase, endolysin, lytic enzymes, tRNAs, virulence factors, genomic rearrangements
Citation: Martín-Galiano AJ and García E (2021) Streptococcus pneumoniae: a Plethora of Temperate Bacteriophages With a Role in Host Genome Rearrangement. Front. Cell. Infect. Microbiol. 11:775402. doi: 10.3389/fcimb.2021.775402
Received: 13 September 2021; Accepted: 29 October 2021;
Published: 18 November 2021.
Edited by:
Xueqing Wu, Zhejiang University, ChinaReviewed by:
Jeremy Brown, University College London, United KingdomJorge Moura de Sousa, Institut Pasteur, France
Yujiro Hirose, Osaka University, Japan
Copyright © 2021 Martín-Galiano and García. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ernesto García, ZS5nYXJjaWFAY2liLmNzaWMuZXM=
 Antonio J. Martín-Galiano
Antonio J. Martín-Galiano Ernesto García
Ernesto García