- 1Department of Otolaryngology, University Of California San Diego, La Jolla, CA, United States
- 2Department of Otorhinolaryngology - Head & Neck Surgery, Inje University Busan Paik Hospital, Busan, South Korea
- 3Research Section, Veterans Administration (VA) San Diego Healthcare System, La Jolla, CA, United States
Background: Pathogens of otitis media (OM) induce inflammatory responses in the middle ear (ME), characterized by mucosal hyperplasia, leukocyte infiltration, and inflammatory mediators, including arachidonic acid metabolites. We studied the role of the eicosanoid leukotriene B4 (LTB4) in OM.
Methods: Expression of LTB4-related genes was evaluated by gene array and single-cell RNA-Seq in MEs infected with nontypeable Haemophilus influenzae (NTHi). An inhibitor of LTB4 receptor 1 (i.e. U75302) was also used to block LTB4 responses.
Results: ME expression of LTB4-related genes was observed by gene arrays and scRNA-Seq. However, not all genes involved in LTB4 generation occurred in any one specific cell type. Moreover, LTB4 receptor inhibition significantly reduced mucosal hyperplasia and virtually eliminated leukocyte infiltration.
Conclusions: ME expression of LTB4-related genes suggest a functional role in OM disease. The fact that LTB4-generation is spread across different cell types is consistent with a transcellular pathway of eicosanoid biosynthesis involving cell-to-cell signaling as well as transfer of biosynthetic intermediates between cells. The dramatic reduction in ME leukocyte infiltration caused by U75302 indicates that LTB4 plays a major role in ME inflammatory cell recruitment, acting via the LTB4R1 receptor. Given that there are many other chemotactic factors that occur in the ME during OM, the ability of LTB4 to activate leukocytes and stimulate their extravasation may explain the effects of inhibition. Reduction in mucosal hyperplasia due to U75302 administration may be secondary to the reduction in leukocytes since LTB4R1 is not expressed by mucosal epithelial or stromal cells. The results suggest that LTB4 receptor antagonists could be useful in treating OM.
Introduction
Otitis media (OM) is the most prevalent disease of childhood (Casselbrant et al., 1993). More than 90% of children experience at least one episode of OM before the age of five. While the majority of acute OM is self-limiting and resolves within a few days even without treatment, 10-15% of children suffer from chronic or recurrent OM (Teele et al., 1989; Kaur et al., 2017). OM peaks between the ages of 6 months and two years (Pichichero, 2016), which is a critical period for the development of speech and language.
In developed countries, OM causes significant health and financial burdens. In the US, OM causes more pediatrician visits, antibiotic prescriptions, and surgeries than any other condition for children under 5 years of age (Rovers et al., 2008), at a cost estimated at $5 billion per year (Rosenfeld and Bluestone, 2003; Klein, 2000; Tong et al., 2018). Hearing loss due to chronic OM has been linked to delays in speech (Friel-Patti and Finitzo, 1993), language acquisition (Klausen et al., 2000), deficits in learning (Williams and Jacobs, 2009), and disorders of central auditory processing (Klein et al., 1988). However, in parts of the developing world, OM is a much more serious public health problem. The WHO estimates that undertreated OM, leading to chronic suppurative OM (CSOM), leads to 30,000 deaths per year due to meningitis. WHO also estimates that CSOM is responsible for half of the world’s burden of handicapping hearing loss, more than 275 million cases (WHO, 2004; Leach et al., 2020; WHO, 2020). It has even been suggested that CSOM represents a neglected tropical disease (Li et al., 2015).
OM is a multifactorial disease, with contributions from Eustachian tube dysfunction, prior upper respiratory viral infection, genetics, and environmental factors (Schilder et al., 2016). However, these factors often lead to a final common pathway of bacterial infection, primarily by nontypeable Haemophilus influenzae (NTHi), Streptococcus pneumoniae and/or Moraxcela cattharalis, with some viral co-infection (Ruohola et al., 2006). While streptococcal vaccines have reduced ME infections by covered strains, OM is an opportunistic infection and the incidence of ME infection by other species and streptococcal strains has increased (Casey et al., 2010). The result is that OM incidence has declined only modestly due to vaccinations (Vojtek et al., 2017).
Despite many years of research, treatments for OM have seen little improvement. Antibiotics remain the most common therapy for OM. Although antibiotics have been shown to be of little benefit for children over two, antibiotic prescriptions remain very common due largely to parental demand for treatment (McGrath et al., 2013). Pressure equalization tubes are recommended for chronic/recurrent OM, but they require surgery and general anesthesia for children, and can lead to scarring of the tympanic membrane (Yaman et al., 2010). Clearly, new therapies are desirable. Recent research on OM has illuminated pathways that contribute to OM pathogenesis and recovery (Kurabi et al., 2016). These pathways offer potential opportunities for alternative interventions.
The invading pathogens that are the hallmark of OM trigger innate and, eventually, adaptive immunity. Acute, uncomplicated OM typically resolves in a few days even without treatment (Rosenfeld and Kay, 2003). Because this is too soon for adaptive immune activation, it is clear that innate immunity is the default process for OM resolution. Many studies in animals and patients (see Kurabi et al., 2016 for review) have confirmed that innate immune receptors and effectors contribute to OM resolution. However, innate immune activation of inflammation also appears to be responsible for many of the pathogenic features of OM, including fluid infiltration of the ME producing serous OM, mucus generation leading to glue ear, and mucosal hyperplasia (Mittal et al., 2014). Understanding the roles of innate immune processes in OM may lead to the development of novel therapies.
A major component of inflammation is the recruitment of pro-inflammatory leukocytes, which invade the ME in large numbers in response to infection (Lim and Birck, 1971). Key among these pro-inflammatory cells are polymorphonuclear cells (PMNs) and monocytes (Qvarnberg et al., 1984), which enter the ME from the circulation in response to chemotactic factors. These can be secreted by macrophages that reside in the ME, or by non-immune ME cell types (Ryan et al., 2020). Monocytes that enter the ME typically differentiate into macrophages. PMNs and macrophages phagocytose and kill pathogens, but they also secrete pro-inflammatory substances that can harm middle ear tissues. Moreover, because of their relatively short life span in tissue (Colotta et al., 1992), dying PMNs release toxic substances such as the reactive oxygen species resident in their numerous intracellular granules as well as cellular debris that can impede clearance of ME fluid. The resultant inflammation induces fluid extravasation into the ME and hyperplasia of the ME mucosa, along with mucosal secretion of mucus and other factors.
Inflammation also induces arachidonic acid (AA) metabolism (Hanna and Hafez, 2018). As shown in Figure 1, the cytosolic enzyme phospholipase A2 group IV-A (PLA2G4A) converts cell membrane lipids into AA. AA can be metabolized by cyclo-oxygenases into prostaglandins, or by arachidonate-5-lipoxygenase (ALOX5) into the eicosanoid leukotriene A4 (LTA4). The latter process requires ALOX5 activating protein (ALOX5AP), which anchors ALOX5 to the membrane and plays an essential role in the transfer of AA to ALOX5. LTA4 in turn is converted by ALOX12 or ALOX15 into lipoxins. Alternatively, it can be converted by leukotriene A4 hydrolase (LTA4H) into leukotriene B4 (LTB4). The biological effects of leukotrienes and lipoxins often oppose each other, with LTB4 generally being pro-inflammatory and lipoxins being anti-inflammatory. LTB4 is a major chemoattractant of inflammatory leukocytes. LTB4 signals primarily via two LTB4 receptors, LTB4R1 and LTB4R2, which are expressed by leukocytes. LTB4R1 is a high-affinity receptor specific for LTB4, while LTB4R2 has a lower affinity and can also respond to other eicosanoids (Sheppe and Edelmann, 2021). Alternative, eicosanoid pathways lead to the production of other leukotrienes or lipoxygenases, while the cyclo-oxygenase pathway can generate lipoxygenases, prostaglandins, and thromboxane.
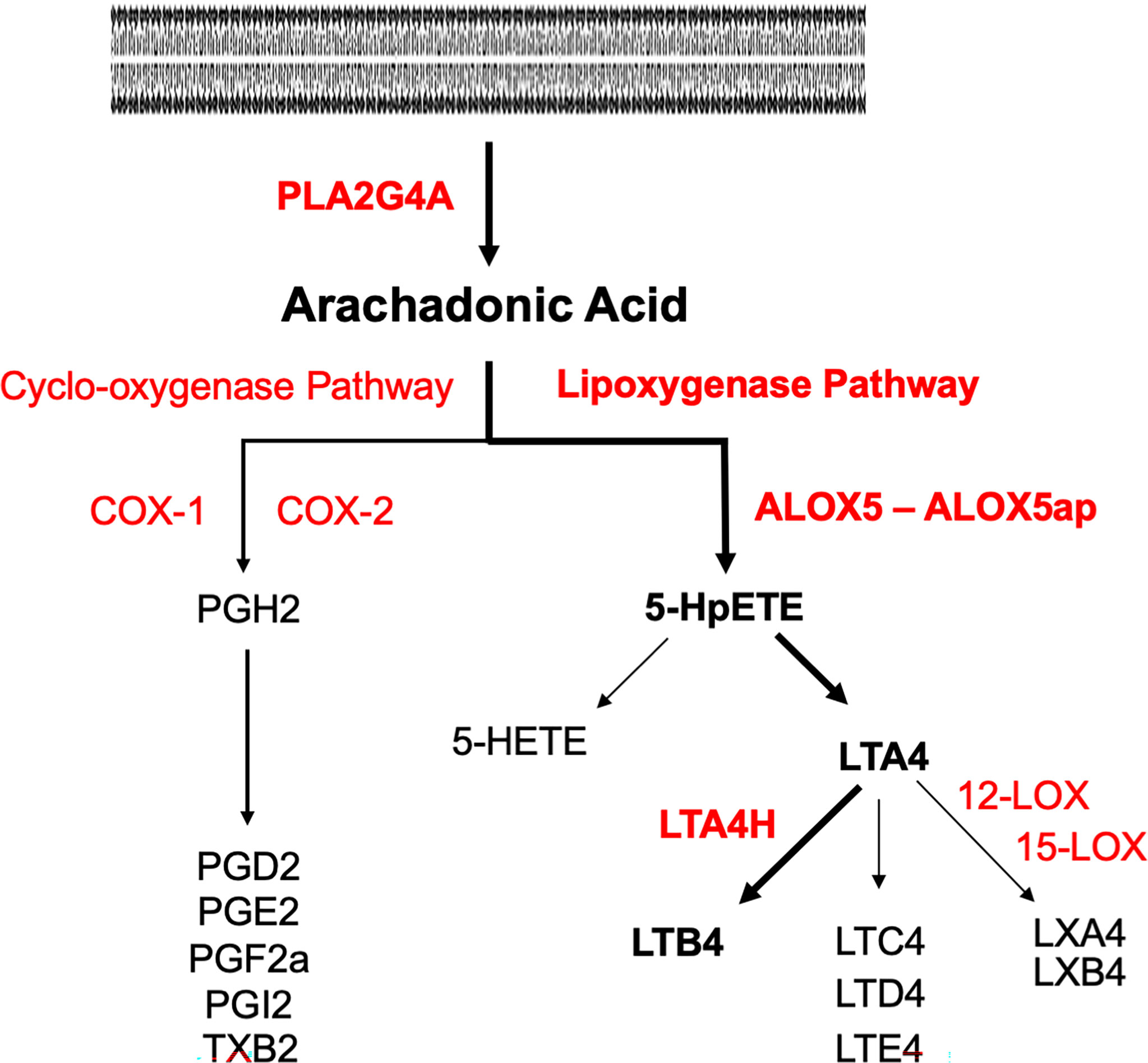
Figure 1 Pathways of arachidonic acid metabolism. Enzymes are in red, metabolites are in black. The pathway to LTB4 via the lipoxygenase pathway is delineated in bold. Phospholipase A2 (PLA2G4A) generates AA from cell membranes. Lipoxygenase A5 (ALOX5) and its activating protein (ALOX5AP) convert AA to 5HpETE, which then partitions between reduction to the 5-HETE and conversion to leukotriene A4 (LTA4). LTA4 hydrolase (LTA4H) converts LTA4 to LTB4.
The role of LTB4 in OM is largely unexplored. The purpose of the present study was to evaluate the expression of genes associated with LTB4 generation as well as those encoding its receptors, during an acute episode of OM. Gene expression data are used because LTB4 is a lipid moiety with a half-life in minutes, and therefore difficult to quantify. To explore the functional role of LTB4 in OM, inhibition of the LTB4 receptor LTB4R1 was employed.
Methods
Animals
Mice used for gene array analysis were 60-90 day old C57BL/6:CB F1 hybrids, while those for single-cell RNA-Seq were C57BL/6 (Jackson Labs, Bar Harbor, ME, USA). Sprague-Dawley rats (Harlan Sprague Dawley, Indianapolis, IN, USA) were used for inhibitor studies. All experiments were performed to National Institutes of Health guidelines and approved by the VA San Diego Medical Center IACUC.
OM Generation
Mice or rats were deeply anesthetized with rodent cocktail (ketamine 50 mg/kg, xylazine 1 mg/kg, acepromazine 5 mg/kg in 50 µl, i.p.) and the ME bulla surgically exposed. A small opening was created in each bulla and Haemophilus influenzae strain 3655 (nontypeable, biotype II; NTHi), was infected in 5 μl PBS for mice and 50 μl for rats, at a titer of 104.
Gene Array
Gene arrays were used to provide quantitative information on gene expression levels within the ME, since mRNA is extracted in a uniform manner from all cells in a tissue. Forty mice per time point were inoculated in the ME bilaterally with NTHi. Uninoculated animals served as controls. Mucosal tissue and exudate were harvested from 20 mice at each of the following intervals: 0 hours (0h, no treatment), 3h, 6h, 24h, 2 days (2d), 3d, 5d and 7d after inoculation, and pooled. The tissue was homogenized in TRIzol (Life Technologies, Carlsbad, CA) and total RNA extracted, reverse transcribed, amplified and transcribed in vitro to generate biotinylated cRNA probes that were hybridized to 2 Affymetrix (Agilent Technologies, Santa Clara, CA) MU430 2.0 microarrays, according to the manufacturer’s protocol. This procedure was duplicated for each time point to obtain a second, independent biological replicate. Thus each data point represents 2 separate samples consisting of 20 mice each, and 4 Affymetrix arrays. Specific genes were assessed at individual time points using Genespring GX 7.3 (Agilent Technologies, Santa Clara, CA). To identify changes in gene expression, the data were first analyzed using a variance modeling approach. The raw MAS5 expression values were imported into the VAMPIRE software without prior normalization. This program uses a Bayesian approach to identify significantly altered genes (Hsiao et al., 2005). Hybridization to probes for genes involved in AA metabolism and signaling were assessed at individual time points for difference from control (uninfected) samples, after Bonferonni correction for multiple tests and using ANOVA in Genespring GX 7.3 (Agilent Technologies, Santa Clara, CA). Additional details of methods are provided in our previous publication (Hernandez et al., 2015) in which the genes evaluated here were not included.
Single-Cell RNA-Seq
Gene arrays generated from bulk tissue cannot identify which cells are expressing a given gene. We used single-cell RNA-Seq (scRNA-Seq) to provide precise, cell-level data. However, because the isolation of different cell types will vary in efficiency depending upon fragility and strength of bonding to other cells, this method is less able to determine overall ME levels of gene expression. Groups of six C57BL/6 mice each were untreated as controls. Additional groups were inoculated in the ME with NTHi and mucosal tissue and exudate harvested 24 hours later, when the gene array data indicated maximum regulation of genes related to LTB4. The pooled tissue for each sample was digested with thermolysin (0.5 mg/ml, Sigma-Aldrich, #T7902) followed by FACSMax cell dissociation solution (Genlantis, #T200100), and triturated into single cells. Dissociated cells were diluted to 700 cells/μL. Three replicates were performed to obtain independent biological samples of control MEs, while four replicates were performed 24 hours after NTHi inoculation. Single-cell libraries were prepared using the Chromium Controller (10X Genomics, Pleasanton, CA) according to the manufacturers’ instructions. The libraries were sequenced on an Illumina HiSeq 2500 (Illumina, San Diego) and yielded approximately 200 million reads per sample.
Bar-coded reads were demultiplexed using Cellranger 2.0.2 (10X Genomics) and mkfastq in conjunction with bcl2fastq 2.17.1.14 (Illumina) and aligned to a murine reference genome mm10 (Ensembl 93) provided by 10X Genomics. Reads were filtered to remove short reads and reference genome mismatches to improve library quality, quantified and subjected to principal component analysis (PCA) clustering. For the three samples from normal MEs, the average number of analyzed cells/sample was 2,257, and the number of genes detected/sample averaged 17,323. For the four samples at 24 hours the number of cells/sample averaged 3,978 and genes detected averaged 17,413. Identification of cells in each cluster was based on the following marker genes: epithelial cells, high expression of Epcam and Krt18; ciliated epithelial cells, Epcam and Hydin; immature epithelial cells, low Epcam and Krt18; stromal cells, Col1a2; vascular endothelial cells, Egfl7 and Flt4; lymphatic endothelial cells Egfl7 and Flt1; pericytes, Rgs5; monocytes, Csf1r; lymphocytes, Ptprcap; and melanocytes, Mlana. After infection, PMNs were identified by expression of Il1f9 and Stfa2l1 and RBCs by Hba-a1. Graph-based and K-means analysis of gene expression was then performed for genes related to LTB4 generation and sensing. Expression of genes by ME cells was visualized using 10X Genomics cLoupe. A cluster plot for each gene was generated using log2 data, for optimal visualization of expressing cells. Violin plots of expression levels for each cluster were log-normalized for optimal comparison across the cluster cell population. Additional details of methods are available in our previous publication on normal ME scRNA-Seq (Ryan et al., 2020), in which no genes related to AA metabolism were reported. The gene data is available at the Science Data Bank (https://www.scidb.cn/en) under doi:10.11922/sciencedb.01353.
Inhibitor Studies
Because of the small size (5 μL) of the mouse ME, inhibitor studies were performed in rats. Rats and mice are closely related and have highly similar responses to ME NTHi inoculation (Melhus et al., 1994; Melhus and Ryan, 2003). For the inhibitor study, 4 rat MEs per treatment were inoculated with NTHi as above. Control rats received NTHi alone in PBS plus 0.1% DMSO. Inhibitor groups received NTHi with different doses of the specific BLT1 inhibitor U75302 [(6-(6-(3R-hydroxy-1E,5Z-undecadien-1-yl)-2-pyridinyl)-1,5S-hexanediol; refs; Cayman Chemical Company, Ann Arbor, MI, USA] plus DMSO. Three U75302 dosages were used: 1.5 μM, 15 μM, 150 μM. 150 μM U75302 only into the ME served as an additional control. Rats were sacrificed for ME harvest 48 hours after inoculation.
Histology
Rats were sacrificed by decapitation under deep anesthesia. ME bullae were extracted, post-fixed with 4% PFA overnight, and decalcified in an 8% EDTA/4% PFA solution over a 14-day period. The bullae were embedded in paraffin and sectioned at 7 μm. Sections were stained with hematoxylin-eosin and mounted. As described previously (Leichtle et al., 2010), from four sections at the midpoint of each ME, mucosal thickness was measured at six standard locations and averaged to obtain a measure for that ME. From the same sections, the total area of the ME lumen and the percent of the lumen area occupied by leukocytes were digitally determined using NIH Image software. A 300X image of the ME infiltrate was captured for each section, and the number of neutrophils and macrophages quantified to capture the proportions of each cell type in the infiltrate. Leukocyte data were not normally distributed and were analyzed using the Kruskal-Wallace nonparametric ANOVA.
Results
Gene Array
As assessed by cRNA probe hybridization to gene arrays, several genes involved in LTB4 production and sensing were significantly upregulated following NTHi inoculation of the ME (Figure 2A). The Pla2g4a gene was only slightly (1.5-fold) upregulated during OM. The Alox5 gene increased expression 3-fold at 1 day, while Alox5ap increased expression 5-fold at 3 days after inoculation. The Lta4h gene for the LTB4 generating LTA4 hydrolase enzyme increased 2-fold at 1 day. Most dramatically, the LTB4 receptor gene Ltb4r1 increased by 170-fold at 1 day after NTHi inoculation of the ME, while Ltb4r2 was not significantly regulated. Some other enzymes of AA metabolism were also significantly regulated, especially Cox2 but also Alox12 and Alox15 (Figure 2B). Detailed data on fold change ranges and statistics are presented in the Supplementary Table.
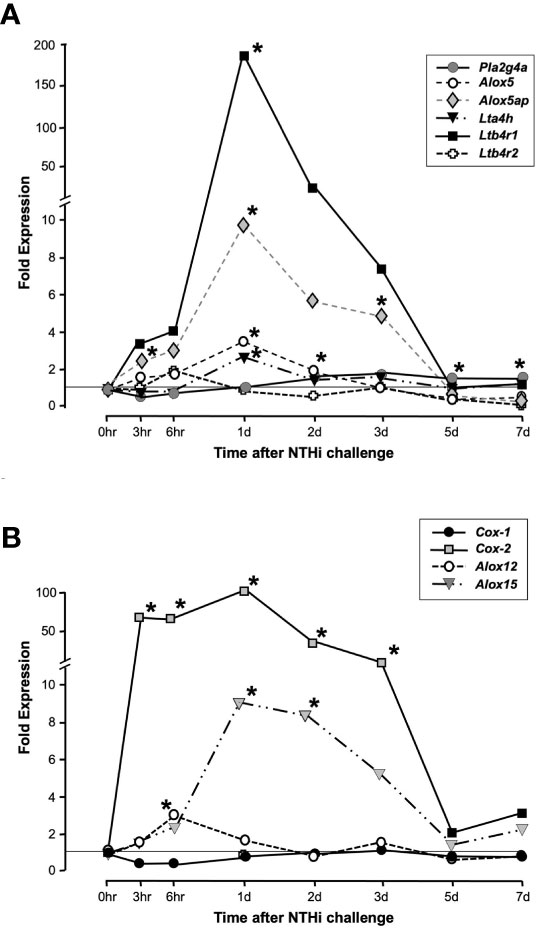
Figure 2 Expression of genes involved in LTB4 generation and sensing during a complete episode of acute NTHi-induced OM in the mouse, from initiation to recovery, evaluated by gene arrays. Panel (A) shows AA Lipoxygenase pathway related genes. Ltb4r1 is very strongly and significantly upregulated during OM, with lesser but significant upregulation of Alox5, Alox5ap and Lta4h. Ltb4r2 was not significantly regulated. Panel (B) shows related enzymes of AA metabolism. *and gray color = p <.05, ANOVA. Detailed data on fold change ranges and p values are presented in the Supplementary Table.
Single-Cell Gene Expression
ScRNA-Seq was used to map the expression of LTB4 synthesis and sensing genes to uninfected ME cells, and cells harvested at 24 hours after NTHi inoculation when expression of most genes peaked (Figure 2A). PCA generated 10-15 clusters for each condition, which corresponded to different ME cell types. Some cell types, such as epithelial cells, consisted of multiple PCA clusters that corresponded to recognized subtypes. Other cell types, such as PMNs, consisted of several clusters not easily defined. After identification of the cell types in each cluster, genes related to LTB4 generation and sensing were visualized using the 10X Genomics cLoupe function.
Figure 3 presents gene expression by cells in the uninfected ME. The expression by cell types is illustrated in the cLoupe PCA projection of Figure 3A, which shows the results in a representative control sample. As can be seen in the figure, Pla2g4a mRNA was expressed by a small number of most ME cell types, with the exception of ciliated epithelial cells, where most cells were positive, and endothelial cells, melanocytes and lymphocytes, which were essentially negative. Alox5 was observed in small subsets of monocytes and lymphocytes. Alox5ap was expressed in most monocytes and approximately half of the stromal cells, as well as a few epithelial cells and lymphocytes. Lta4h was observed in small subsets of all ME cell types, while Ltb4r1 was prevalent only in subsets of monocytes and lymphocytes. Ltb4r2 was not expressed by any ME cells. Figure 3B shows violin plots of gene expression by PCA cluster, from the same control sample. The plots provide a relative quantitative analysis of gene expression. They confirm that only a small number of cells expressed LTB4-related genes at high levels, with the majority of cells not expressing. The exceptions were monocytes and stromal cells, of which many cells expressed Alox5ap at appreciable levels.
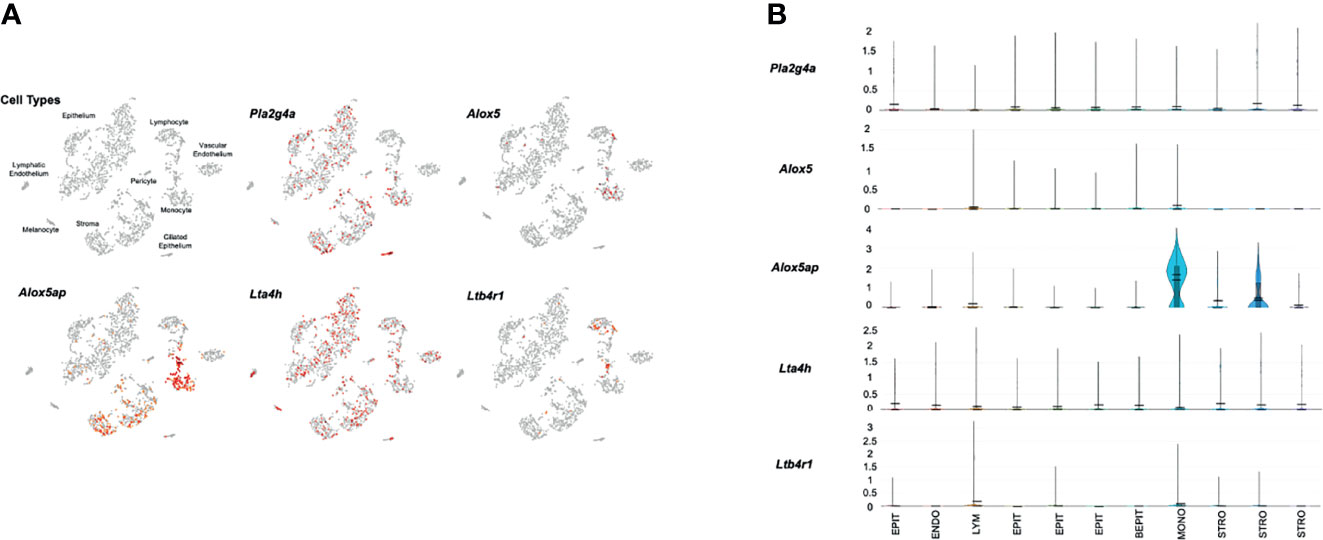
Figure 3 Expression of LTB4-related genes by individual ME cell types, before inoculation of the murine ME with NTHi. For each sample, twelve MEs were pooled and digested into a single-cell suspension for single-cell RNA-Seq. Panel (A) represents 2,858 cells recovered from the normal ME, clustered by principal component analysis, and identified by recognized cell-type marker genes. Expression of each gene by cell is indicated in red. Panel (B) presents violin plots for individual PCA clusters, labeled by cell type, with some cell types represented by multiple clusters. The width of each plot represents the frequency of cells expressing the gene at that log normalized level. The upper horizontal line in each plot represents the cluster mean, and the lower line the median. The height of the vertical line represents the highest expressing cell.
Figure 4 presents results from a representative ME sample collected 24 hours after NTHi infection. All of the cell types observed in control MEs were present, except for melanocytes. In addition, a new category of immature epithelial cells was present, and large numbers of neutrophils and a few erythrocytes had infiltrated the ME. In agreement with our array data, substantially more expression of LTB4-related genes was observed 1 day after infection. Pla2g4a mRNA was expressed in virtually all monocytes and variable subsets of other cell types. Alox5 mRNA was observed primarily in a subset of infiltrating neutrophils, and a few monocytes. Alox5ap was expressed by almost all monocytes, the majority of neutrophils and small subsets of other cell types. Lta4h was present in most epithelial cells and monocytes, but also by small subsets of other cell types. Ltb4r1 was produced exclusively by monocytes and a subset of neutrophils. Ltb4r2 expression again was not observed. The violin plots of Figure 4B confirm the distribution of LTB4-related gene expression by cell type, and provide relative quantitative expression levels which can be compared to those for the cell types in Figure 3B. Each of the genes was differentially expressed at a significant level (p<.05, ANOVA) between control and infected MEs, for the cell types with major differences in violin plots. PMNs, which were not present in the control sample, could not be thus analyzed.
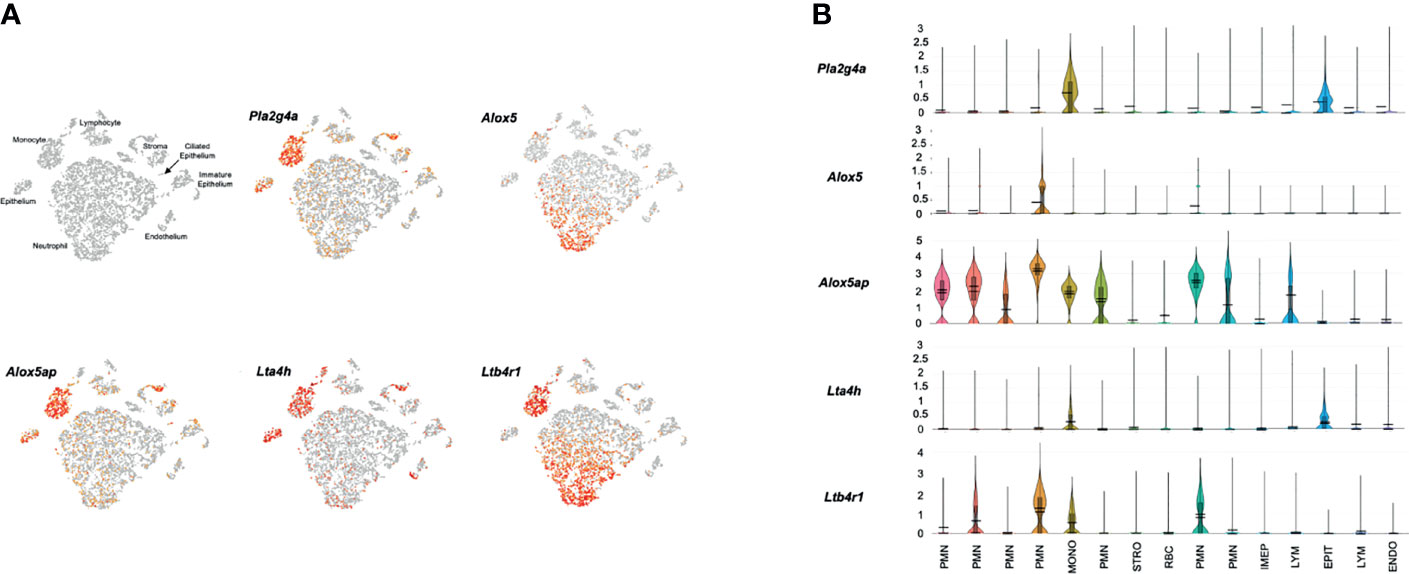
Figure 4 Expression of LTB4-related genes by individual ME cell types, 24 hours after inoculation of the murine ME with NTHi. (A) PCA clusters of 9,604 cells, with cell types identified and cells expressing each gene indicted in red. (B) Violin plots demonstrating cell type expression patterns as well as the much higher level expression of LTB4-related genes.
Mucosal Hyperplasia
Figures 5 shows representative examples of ME histology for control MEs, before infection, for MEs 48 hours after inoculation with NTHi alone, for MEs after 48 hours after inoculation with NTHi plus one of three dosages of U75302, and in MEs 72 hours after injection of the highest dosage of U75302 alone. Quantitative analysis of mucosal thickness is provided in Figure 6A. ME infection with NTHi substantially increased mucosal thickness when compared to control MEs. All three dosages of U75302 significantly reduced mucosal thickness (p <.05; ANOVA). Thickness after injection of U75302 alone was not significantly different from control.
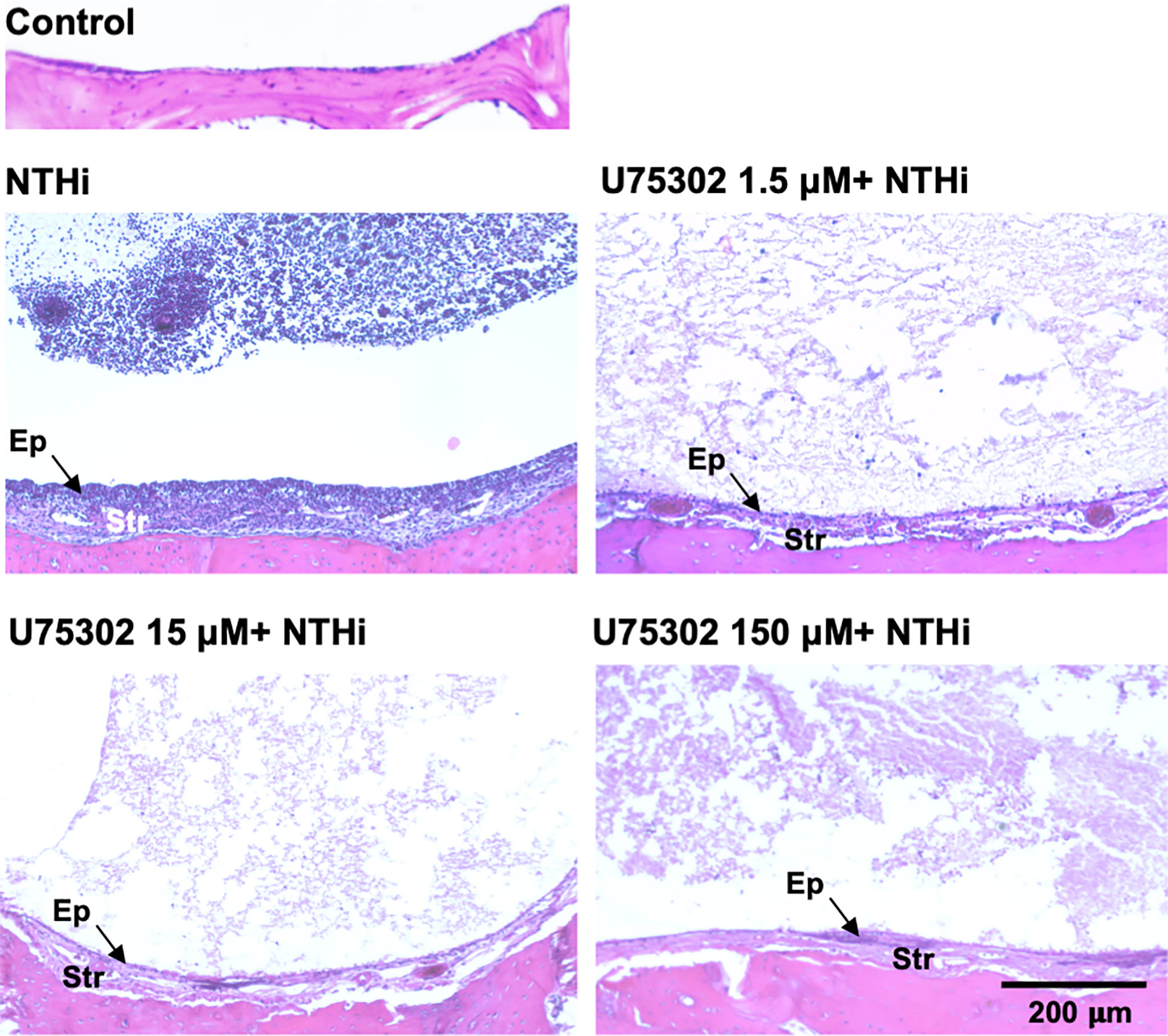
Figure 5 The MEs of a control, untreated mouse and of mice 2 days after infection with NTHi, or NTHi plus different doses of the specific LTB4R1 inhibitor U75302. At all doses, hyperplasia of both the mucosal epithelium (Ep) and stromal (Str) was reduced. In addition, infiltration of the ME lumen by leukocytes was virtually eliminated. A luminal precipitate was observed in all treated ears, which presumably represents an acellular exudate.
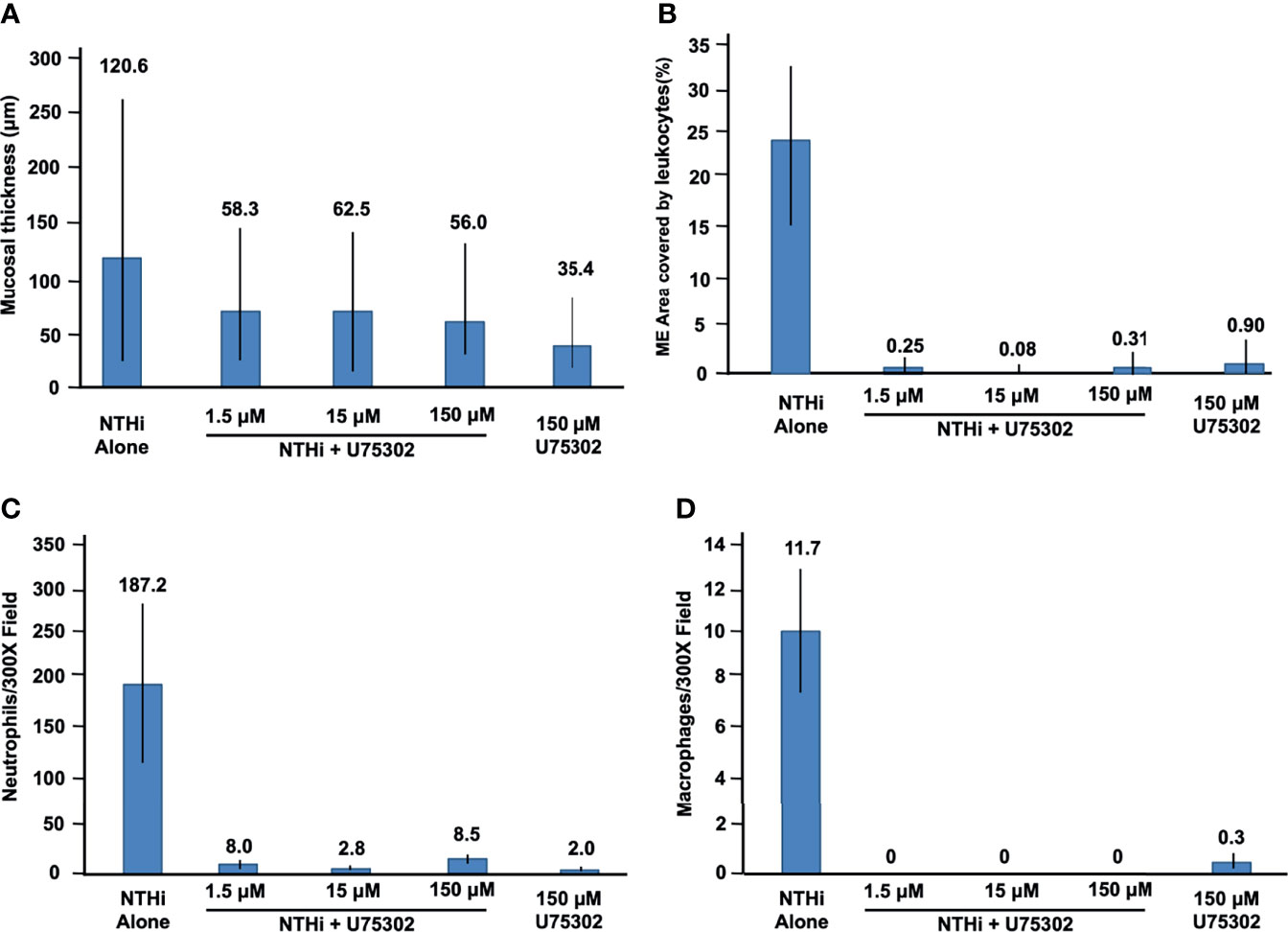
Figure 6 Quantitative evaluation of the effects of U75302 on OM. (A) ME mucosal thickness (mean, range: max and min). (B) Percent of the ME lumen occupied by leukocytes. (C) Number of neutrophils counted in high-power images of ME exudate. (D) Number of macrophages counted in high-power images of ME exudate.
Leukocyte Infiltration of the ME
Figure 5 also illustrates the leukocyte response of the ME to NTHI inoculation, with and without the LTB4R1 inhibitor U75302. It is clear from Figure 5 that leukocyte infiltration of the ME lumen was dramatically reduced by all three dosages of inhibitor. This is confirmed by the quantitative analysis presented in Figure 6B (p <.001; Kruskal Wallis ANOVA). However, the presence of substantial amounts of precipitate in the ME suggests that LTB4 inhibition did not reduce infiltration of the ME by fluid from the vasculature. U75302 alone did not induce significant cellular or fluid infiltration of the ME.
Leukocyte Cell Types in the ME
Infection by NTHi typically produces ME infiltration by large numbers of PMNs, and a lower number of monocytes/macrophages. This was also observed in the control animals of the present study. Figures 6C, D represent each cell type as counted in 300X micrographs of infiltrate in the ME lumen. Not surprisingly given the data shown in Figure 6B, treatment with U75302 virtually eliminated both cell types from the ME (p <.001; Kruskal Wallis ANOVA).
Discussion
Significant increases in the expression of genes related to LTB4 generation and receptor sensing after ME inoculation with NTHi provide evidence that this AA metabolite could play a role in OM. In particular, the dramatic rise in Ltb4r1 mRNA observed early in OM suggests that either ME cells generated large amounts of this receptor, or that cells expressing these receptors entered the ME in large numbers. Conversely there were no significant changes in the expression of Ltb4r2 mRNA. Genes involved in the generation of LTB4 also peaked at 24 hours, with Alox5ap also showing a significant increase earlier. Infection also increased the expression of other genes of the AA metabolic pathway, in both the cyclo-oxygenase and lipoxygenase pathways. These data suggest that other AA metabolites participate in OM, and their production has been documented in experimental OM (Goldie et al., 1993). However, responses to any of these would not be expected to be inhibited by U75302.
Single-cell RNA-Seq confirms and validates the expression of mRNA encoding components of the LTB4 synthetic pathway, and its specific receptor LTB4R1. Interestingly, all of the synthetic components were not present in any one ME cell type, but rather were distributed across a variety of cells. This is consistent with other cellular systems, where eicosanoids are often the result of intercellular interactions involving cell-to-cell signaling and the transfer of biosynthetic intermediates, such as LTA4, between cells (e.g. Fradin et al., 1989). This process is known as the transcellular pathway of eicosanoid biosynthesis (Sala et al., 2010). It requires one cell to synthesize and release a component of the biosynthetic pathway, and another cell to take up that intermediate and process it into a final active molecule. In the case of eicosanoids, this is aided by their lipid solubility.
This intercellular process appears to occur in the ME as well. From the pattern of Pla2g4a expression, it can be inferred that most AA is produced by ME epithelial cells, with a contribution from some resident leukocytes. The enzymes that produce LTB4 are primarily expressed by leukocytes, but the production of its precursor, LTA4H, is mostly by epithelial and stromal cells. The exchange of AA and LTA4H between epithelial and leukocytic cell types, and potentially signaling between the cell classes, appears to be necessary for the ME production of LTB4. The expression of LTB4R1 only in leukocytes was expected. The absence of cells expressing mRNA encoding the specific LTB4 receptor LTB4R2 indicates that inhibition of LTB4R1 would completely block LTB4 signaling in the ME.
The final step in LTB4 production involves the conversion of LTA4 to LTB4 by LTA4H (see Figure 1). Lta4h mRNA is highly expressed in epithelial cells and monocytes, and some stromal cells, vascular endothelial cells, and PMNs. These cells are likely to be the primary sources of direct LTB4 production. However, PMNs produce the most ALOX5 and are likely the primary source of the precursor molecules 5-HpETE and LTA4.
The virtual elimination of leukocytic infiltration of the ME caused by the LTB4R1 inhibitor U75302 indicates that LTB4 activation of this receptor is required for the recruitment of PMNs and monocytes/macrophages to the tympanic cavity. Given that several other chemoattractants that target these two cell types are also expressed in the ME (Hernandez et al., 2015; Dennifel et al., 2017; Hur et al., 2021), the dominant role of LTB4 was unexpected. It seems likely that LTB4’s role in the activation of leukocytes, which is required for extravasation, is responsible for its ME effects since this could disable recruitment by all chemoattractants.
The results of our animal study have potential clinical implications. A prior study found that LTB4 is present in ME fluid from children, and that high levels are associated with treatment failure (Chonmaitree et al., 1996). The authors concluded that LTB4 may play role in delaying acute OM recovery and recurrence. They suggested that more effective treatment of OM would require the combined use of antibiotics that reduce inflammatory leukocyte function. Another study from the same group found that treatment with steroids did not decrease LTB4 in the ME fluid of children (McCormick et al., 2003). Our results indicate that LTB4 receptor antagonists, or perhaps natural pro-resolution compounds that oppose the effects of LTB4, such as resolvin E1 (Arita et al., 2005), may be useful in treating OM.
Data Availability Statement
The data presented in the study are deposited in the Science Data Bank repository, accession number 31253.11.sciencedb.01353 and can be viewed at https://www.scidb.cn/en/s/BJJBBv.
Ethics Statement
The animal study was reviewed and approved by VA Health IACUC.
Author Contributions
KH and KP performed experiments and data collection. AR, KH, and AK discussed the study idea, analyzed the data and generated the figures. KH wrote the initial manuscript draft. All authors contributed to final manuscript.
Funding
Supported by grants DC006279, DC000129, and DC012595 from the NIH/NIDCD and BX001205 from the VA.
Conflict of Interest
AR is a co-founder of Otonomy Inc., serves as a member of the Scientific Advisory Board, and holds an equity position in the company. The UCSD Committee on Conflict of Interest has approved this relationship. Otonomy, Inc. played no part in the research reported here.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.768815/full#supplementary-material
References
Arita, M., Yoshida, M., Hong, S., Tjonahen, E., Glickman, J. N., Petasis, N. A., et al. (2005). Resolvin E1, an Endogenous Lipid Mediator Derived From Omega-3 Eicosapentaenoic Acid, Protects Against 2,4,6-Trinitrobenzene Sulfonic Acid-Induced Colitis. Proc. Natl. Acad. Sci. U.S.A. 102, 7671–7676. doi: 10.1073/pnas.0409271102
Casey, J. R., Adlowitz, D., Pichichero, M. E. (2010). New Patterns in the Otopathogens Causing Acute Otitis Media Six to Eight Years After Introduction of Pneumococcal Conjugate Vaccine. Ped. Inf. Dis. J. 29, 304–309. doi: 10.1097/INF.0b013e3181c1bc48
Casselbrant, M. L., Mandel, E., Rockette, H., Bluestone, C. (1993). “Incidence of Otitis Media and Bacteriology of Acute Otitis Media During the First Two Years of Life,” in Recent Advances in Otitis Media. Eds. Lim, D., Bluestone, C., Klein, J., Nelson, J., Ogra, P. (Toronto: Decker), pp7–p11.
Chonmaitree, T., Patel, J. A., Sim, T., Garofalo, R., Uchida, T., Sim, T., et al. (1996). Role of Leukotriene B4 and Interleukin-8 in Acute Bacterial and Viral Otitis Media. Ann. Otol. Rhinol. Laryngol. 105, 968–974. doi: 10.1177/000348949610501207
Colotta, F., Re, F., Polentarutti, N., Sozzani, S., Mantovani, A. (1992). Modulation of Granulocyte Survival and Programmed Cell Death by Cytokines and Bacterial Products. Blood 80, 2012–2020. doi: 10.1182/blood.V80.8.2012.2012
Deniffel, D., Nuyen, B., Pak, K., Suzukawa, K., Hung, J., Wasserman, S. I. (2017). Otitis Media and Nasopharyngeal Colonization in CCL3-/- Mice. Infect Immun 85, e00148–17. doi: 10.1128/IAI.00148-17
Fradin, A., Zirrolli, J. A., Maclouf, J., Vausbinder, L., Henson, P. M., Murphy, R. C. (1989). Platelet-Activating Factor and Leukotriene Biosynthesis in Whole Blood. A Model for the Study of Transcellular Arachidonate Metabolism. J. Immunol. 143, 3680–3685.
Friel-Patti, S., Finitzo, T. (1993). “Speech-Language Learning in the First 3 Years in Children With Tympanostomy Tubes for Recurrent or Persistent OM,” in Recent Advances in Otitis Media. Eds. Lim, D., Bluestone, C., Klein, J., Nelson, J., Ogra, P. (Toronto:Decker), 523–552.
Goldie, P., Jung, T. T., Hellström, S. (1993). Arachidonic Acid Metabolites in Experimental Otitis Media and Effects of Anti-Inflammatory Drugs. Ann. Otol. Rhinol. Laryngol. 102, 954–960. doi: 10.1177/000348949310201208
Hanna, V. S., Hafez, E. A. A. (2018). Synopsis of Arachidonic Acid Metabolism: A Review. J. Adv. Res. 11, 23–32. doi: 10.1016/j.jare.2018.03.005
Hernandez, M., Leichtle, A., Pak, K., Webster, N. J., Wasserman, S. I., Ryan, A. F. (2015). The Transcriptome of a Complete Episode of Acute Otitis Media. BMC Genomics 16, 259. doi: 10.1186/s12864-015-1475-7
Hsiao, A., Ideker, T., Olefsky, J. M., Subramaniam, S. (2005). VAMPIRE Microarray Suite: A Web-Based Platform for the Interpretation of Gene Expression Data. Nucleic Acids Res 33, W627–W632. doi: 10.1093/nar/gki443
Hur, D. G., Kurabi, A., Lim, H. W., Spriggs, M., Pak, K., Ryan, A. F. (2021). Macrophage Depletion in CCR2-/- Mice Delays Bacterial Clearance and Enhances Neutrophil Infiltration in an Acute Otitis Media Model. J Infect Dis 223, 333–341. doi: 10.1093/infdis/jiaa353
Kaur, R., Morris, M., Pichichero, M. E. (2017). Epidemiology of Acute Otitis Media in the Post Pneumococcal Conjugate Vaccine Era. Pediatrics 140, e20170181. doi: 10.1542/peds.2017-0181
Klausen, O., Møller, P., Holmefjord, A., Reisaeter, S., Asbjørnsen, A. (2000). Lasting Effects Of OM With Effusion on Language Skills and Listening Performance. Acta Otolaryngol. Suppl. 543, 73–76. doi: 10.1080/000164800454026
Klein, J. O. (2000). The Burden of Otitis Media. Vaccine 19 Suppl 1, S2–S8. doi: 10.1016/s0264-410x(00)00271-1
Klein, J., Chase, C., Teele, D., Menyuk, P., Rosner, B., Allen, C., et al. (1988). “OM and the Development of Speech, Language and Cognitive Abilities at 7 Years of Age,” in Recent Advances in Otitis Media. Ed. Lim, D. J. (Toronto: Decker) pp 396–397.
Kurabi, A., Pak, K., Ryan, A. F., Wasserman, S. I. (2016). Innate Immunity: Orchestrating Inflammation and Resolution of Otitis Media. Curr. Allergy Asthma Rep. 16, 6. doi: 10.1007/s11882-015-0585-2
Leach, A. J., Homøe, P., Chidziva, C., Gunasekera, H., Kong, K., Bhutta, M. F., et al. (2020). Panel 6: Otitis Media and Associated Hearing Loss Among Disadvantaged Populations and Low to Middle-Income Countries. Int. J. Pediatr. Otorhinolaryngol. 130 (Suppl 1):109857. doi: 10.1016/j.ijporl.2019.109857
Leichtle, A., Hernandez, M., Ebmeyer, J., Yamasaki, K., Lai, Y., Radek, K., et al. (2010). CC Chemokine Ligand 3 Overcomes the Bacteriocidal and Phagocytic Defect of Macrophages and Hastens Recovery From Experimental Otitis Media in TNF-/- Mice. J. Immunol. 184, 3087–3097. doi: 10.4049/jimmunol.0901167
Li, M. G., Hotez, P. J., Vrabec, J. T., Donovan, D. T. (2015). Is Chronic Suppurative Otitis Media a Neglected Tropical Disease? PloS Negl. Trop. Dis. 9, e0003485. doi: 10.1371/journal.pntd.0003761
Lim, D. J., Birck, H. (1971). Ultrastructural Pathology of the Middle Ear Mucosa in Serous Otitis Media. Ann. Otol. Rhinol. Laryngol. 8, , 838–, 853. doi: 10.1177/000348947108000611
McCormick, D. P., Saeed, K., Uchida, T., Baldwin, C. D., Deskin, R., Lett-Brown, M. A., et al. (2003). Middle Ear Fluid Histamine and Leukotriene B4 in Acute Otitis Media: Effect of Antihistamine or Corticosteroid Treatment. Int. J. Pediatr. Otorhinolaryngol. 67, 221–230. doi: 10.1016/s0165-5876(02)00372-5
McGrath, L., Becker-Dreps, S., Pate, V., Brookhart, M. (2013). Trends in Antibiotic Treatment of Acute Otitis Media and Treatment Failure in Children 2000-2011. PloS One 8, e81210. doi: 10.1371/journal.pone.0081210
Melhus, A., Hermansson, A., Prellner, K. (1994). Nontypeable and Encapsulated Haemophilus Influenzae Yield Different Clinical Courses of Experimental Otitis Media. Acta Otolaryngol. 114, 289–294. doi: 10.3109/00016489409126058
Melhus, A., Ryan, A. F. (2003). A Mouse Model for Acute Otitis Media. APMIS 111, 989–994. doi: 10.1034/j.1600-0463.2003.1111012.x
Mittal, R., Kodiyan, J., Gerring, R., Mathee, K., Li, J. D., Grati, M., et al. (2014). Role of Innate Immunity in the Pathogenesis of Otitis Media. Int. J. Infect. Dis. 29, 259–267. doi: 10.1016/j.ijid.2014.10.015
Pichichero, M. E. (2016). Ten-Year Study of Acute Otitis Media in Rochester, NY. Pedi Infects Dis. J. 35, 1027–1032. doi: 10.1097/INF.0000000000001216
Qvarnberg, Y., Holopainen, E., Palva, T. (1984). Aspiration Cytology in Acute Otitis Media. Acta Otolaryngol. 97, 443–449. doi: 10.3109/00016488409132919
Rosenfeld, R. M., Kay, D. (2003). Natural History of Untreated Otitis Media. Laryngoscope 113, 1645–1657. doi: 10.1097/00005537-200310000-00004
Rovers, M. M. (2008). The Burden of Otitis Media. Vaccine 26, Suppl 7, G2–G4. doi: 10.1016/j.vaccine.2008.11.005
Ruohola, A., Meurman, O., Nikkari, S., Skottman, T., Salmi, A., Waris, M., et al. (2006). Microbiology of Acute Otitis Media in Children With Tympanostomy Tubes: Prevalences of Bacteria and Viruses. Clin. Infect. Dis. 43, 1417–1422. doi: 10.1086/509332
Ryan, A. F., Nasamran, C. A., Pak, K., Draf, C., Fisch, K. M., Webster, N., et al. (2020). Single-Cell Transcriptomes Reveal a Complex Cellular Landscape in the Middle Ear and Differential Capacities for Acute Response to Infection. Front. Genet. 11:358. doi: 10.3389/fgene.2020.00358
Sala, A., Folco, G., Murphy, R. C. (2010). Transcellular Biosynthesis of Eicosanoids. Pharmacol. Rep. 62, 503–510. doi: 10.1016/s1734-1140(10)70306-6
Schilder, A. G. M., Chonmaitree, T., Cripps, A. W., Rosenfeld, R. M., Casselbrant, M. L., Haggard, M. P., et al. (2016). Otitis Media. Nat. Rev. Dis. Prim 2, 16063. doi: 10.1038/nrdp.2016.63
Sheppe, A. E. F., Edelmann, M. J. (2021). Roles of Eicosanoids in Regulating Inflammation and Neutrophil Migration as an Innate Host Response to Bacterial Infections. Infect. Immun., 89 00095–21. doi: 10.1128/IAI.00095-21
Teele, D. W., Klein, J. O., Rosner, B. (1989). Epidemiology of Otitis Media During the First Seven Years of Life in Children in Greater Boston: A Prospective, Cohort Study. J. Infect. Dis. 160, 83–94. doi: 10.1093/infdis/160.1.83
Tong, S., Amand, C., Kieffer, A., Kyaw, M. H. (2018). Trends in Healthcare Utilization and Costs Associated With Acute Otitis Media in the United States During 2008-2014. BMC Health Serv. Res. 18, 318–318. doi: 10.1186/s12913-018-3139-1
Vojtek, I., Nordgren, M., Hoet, B. (2017). Impact of Pneumococcal Conjugate Vaccines on Otitis Media: A Review of Measurement and Interpretation Challenges. Int J Pediatr Otorhinolaryngol 100, 174–82. doi: 10.1016/j.ijporl.2017.07.009
World Health Organization (WHO). (2004). Chronic Suppurative Otitis Media: Burden of Illness and Management Options. (Ed. Acuin, J.) Available at: http://www.who.int/pbd/deafness/activities/hearing_care/otitis_media.pdf.
World Health Organization (WHO). (2020). Deafness and Hearing Loss: Key Facts. Available at: https://www.who.int/en/news-room/fact-sheets/detail/deafness-and-hearing-loss.
Williams, C. J., Jacobs, A. M. (2009). The Impact of Otitis Media on Cognitive and Educational Outcomes. Med. J. Aust. 191 (suppl 9), S69–S72. doi: 10.5694/j.1326-5377.2009.tb02931.x
Keywords: leukocytes, otitis media (OM), leukotriene B4 (LTB4), mucosa, non-typeable Haemophilus influenzae (NTHi), arachidonic acid (AA), inflammation
Citation: Heo KW, Pak K, Kurabi A and Ryan AF (2021) Leukotriene B4 Is a Major Determinant of Leukocyte Recruitment During Otitis Media. Front. Cell. Infect. Microbiol. 11:768815. doi: 10.3389/fcimb.2021.768815
Received: 01 September 2021; Accepted: 30 November 2021;
Published: 22 December 2021.
Edited by:
Robyn Marsh, Charles Darwin University, AustraliaReviewed by:
Colin D. Bingle, The University of Sheffield, United KingdomLee-Ann H. Allen, University of Missouri, United States
Copyright © 2021 Heo, Pak, Kurabi and Ryan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arwa Kurabi, YWt1cmFiaUB1Y3NkLmVkdQ==
 Kyung Wook Heo
Kyung Wook Heo Kwang Pak
Kwang Pak Arwa Kurabi
Arwa Kurabi Allen F. Ryan
Allen F. Ryan