
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 14 December 2021
Sec. Clinical Microbiology
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.763687
This article is part of the Research TopicCOVID-19: New Variants and Host DemographyView all 15 articles
 Hardeep Singh Tuli1*
Hardeep Singh Tuli1* Katrin Sak2
Katrin Sak2 Poonam Aggarwal3
Poonam Aggarwal3 Ashif Iqubal4
Ashif Iqubal4 Sushil K. Upadhaya1
Sushil K. Upadhaya1 Jagjit Kaur5
Jagjit Kaur5 Ginpreet Kaur6
Ginpreet Kaur6 Diwakar Aggarwal1
Diwakar Aggarwal1Within almost the last 2 years, the world has been shaken by the coronavirus disease 2019 (COVID-19) pandemic, which has affected the lives of all people. With nearly 4.92 million deaths by October 19, 2021, and serious health damages in millions of people, COVID-19 has been the most serious global challenge after the Second World War. Besides lost lives and long-term health problems, devastating impact on economics, education, and culture will probably leave a lasting impression on the future. Therefore, the actual extent of losses will become obvious only after years. Moreover, despite the availability of different vaccines and vaccination programs, it is still impossible to forecast what the next steps of the virus are or how near we are to the end of the pandemic. In this article, the route of molecular evolution of the coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is thoroughly compiled, highlighting the changes that the virus has undergone during the last 2 years and discussing the approaches that the medical community has undertaken in the fight against virus-induced damages.
From the end of 2019, life has been greatly affected by the coronavirus disease 2019 (COVID-19) all over the world. Based on the data from Worldometers, this pandemic has afflicted more than 241.97 million human lives and has claimed nearly 4.92 million lives around the globe during the last 1.5 years (https://www.worldometers.info/coronavirus/; data from October 19, 2021). At that, the elderly people and those with underlying cardiovascular, respiratory, and metabolic disorders have been found to be especially vulnerable by severe course of the disease, causing bilateral pneumonia, acute respiratory distress syndrome (ARDS), failure of multiple organs (including, but not limited to, the brain, heart, liver, and kidneys), or even mortality (Abdullahi et al., 2020; Li et al., 2021). In addition to the direct health damages, devastating impact on education, culture, economics, and general public welfare proceeding from the strict restrictions in social contacts established for the disease prevention cannot be underestimated (Sood et al., 2020).
COVID-19 is caused by an infection with the single-stranded RNA virus with positive polarity, i.e., severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that transmits mainly via respiratory droplets, aerosols, and fomites (Abdullahi et al., 2020; Kawabata et al., 2020; Li et al., 2021; Mallah et al., 2021). Coronaviruses consist of enveloped virus particles with 80–120 nm of diameter; they have typically spherical or pleomorphic structure with spike-like projections of glycoproteins on surface, giving them a crown-like appearance under electron microscopy (Tuli et al., 2021). The initial reservoir of SARS-CoV-2 is hypothesized to be bats transmitting the virus particles to human beings (Tuli et al., 2021). Within the time of the pandemic course, SARS-CoV-2 virus has been in a continuous molecular evolution, displaying genetic diversity and mutations with varied degrees of transmission and virulence (Abdullahi et al., 2020; Deimel et al., 2021). Such mutations can help virus particles to escape the immune system and/or replicate more efficiently once it has entered the host organism, making the virus more infectious and pathogenic (Adedokun et al., 2021; Hossain et al., 2021). The impact of viral changes on the COVID-19 pandemic has been apparent in the disease outbreaks occurring disproportionately in different parts of the world (Abdullahi et al., 2020; Fraser, 2020; Vudathaneni et al., 2021). Therefore, the virus variants are designated by the geographical regions where the mutations have emerged, including the UK (B.1.1.7), Brazilian (B.1.1.248), and South African (1.351) strains, among others (Hossain et al., 2021). Furthermore, as mutations in the virus genome can change also the susceptibility of the virus to both clinically used drugs and vaccines, concerns have been arisen about the efficacy of current preventive and therapeutic interventions for stopping the pandemic (Chiam et al., 2021; Hossain et al., 2021; Matta et al., 2021; Robinson et al., 2021).
In this state-of-the-art review article, molecular characteristics of the currently emerged variants of SARS-CoV-2 are under discussion, analyzing their infectivity, morbidity, and mortality potential, as well as susceptibility to the current intervention measures applied for achieving control over the pandemic.
Mutations originate as a result of viral replication during circulation. Despite being an RNA virus, coronaviruses undergo fewer mutations because of their strong proofread mechanism. Moreover, the fate of mutations is determined by the natural selection, meaning that those favored with respect to viral better survival will increase in frequency, and those that reduce viral fitness tend to be eliminated from the population of circulating viruses. However, mutations can also happen due to chance events. Therefore, the interplay of natural selection and chance events leads to virus evolution.
The SARS-CoV-2 virus has been mutated over time, resulting in different genetic variations in the population of circulating viral strains over the course of the COVID-19 pandemic. The evolution of SARS-CoV-2 suggests strong purifying selection and modest divergence; one of the most closely related strain of SARS-CoV-2 is “RaTG13” found in a bat sample from Yunnan Province, China, in 2013. RaTG13 (horseshoe bat, Rhinolophus affinis) shows 96% similarity to SARS-CoV-215. Though RaTG13 is closely related to SARS-CoV-2, there is a significant level of variation in sequence similarity across the genomes of these two viruses, ranging between 93.1% and 99.6% (Zhou et al., 2020). However, comparisons with other coronavirus strains suggest complex recombination events during its evolution. Various recombinations were detected across the genome majorly in ORF1a and in the region marking the N-terminus of the S protein (Li et al., 2003; Li, 2016; Hoffmann et al., 2020; Wan et al., 2020). S protein binds to angiotensin-converting enzyme 2 (ACE2) receptors and mediates viral entry into the human cells. One such mutation, D614G, arises as a result of single-nucleotide polymorphism (SNP) and results in amino acid change from an aspartate [D] to a glycine [G] at residue 614, increasing the efficiency of viral entry into the human cells (Isabel et al., 2020; Korber et al., 2020).
The D614G mutation in the spike glycoprotein of SARS-CoV-2 was significantly detected for the first time in early March 2020 and has spread globally across multiple geographic regions over the next month (Korber et al., 2020). However, various sequencing studies have already identified the D614G mutation in viruses in China in late January, which dispersed globally. Similarly, the population genetics analysis of more than 25,000 sequences from the United Kingdom also found that viruses with 614G are more transmissible and affect larger phylogenetic clusters (Volz et al., 2021). Even parallel studies in animal models also indicate that 614G viruses are more transmissible. As a result of more favored mutation, this strain has now become a dominant global strain (Hou et al., 2020; Plante et al., 2021).
Apart from its evolution in humans, there is evidence of cross-specific transmission in other animals like mink, which can even lead to emergence of potentially dangerous recombinant SARS-CoV-2 strains. Outbreaks of SARS-CoV-2 on mink farms in the Netherlands and Denmark that started in late spring and early summer 2020 demonstrated human-to-mink, mink-to-mink, and mink-to-human transmissions (European Centre for Disease Prevention and Control, 2020, Oude Munnink et al., 2021). In early November 2020, 214 cases of mink-associated human COVID-19 were reported. These cases where Y453F mutation in the receptor binding domain of spike might be responsible for increased binding affinity for ACE2 in mink. Eleven patients from the Danish outbreak had a cluster 5 variant having three additional mutations in spike (del69_70, I692V, and M1229I). An investigation of human serum samples in nine patients showed a significant reduction in neutralization activity against cluster 5 viruses (mean, 3.58-fold; range, 0–13.5). Therefore, continued evolution and adaptation of SARS-CoV-2 in an animal reservoir resulted in novel SARS-CoV-2 from mink to humans and other mammals.
Another lineage B.1.1.7 (also called 501Y.V1) was identified in southeastern England (Rambaut et al., 2021) and became one of the variants of the highest concern. This variant has already highly evolved, having 17 lineage-defining mutations even prior to its detection in early September. Seven of these mutations were in the spike proteins only that later formed the basis for the vaccine in the United Kingdom. This variant was found to be 56% more transmissible and was responsible for approximately 28% of cases of SARS-CoV-2 infection in England within 1 month (Davies et al., 2020). Unlike D614G, which could be because of chance events, B.1.1.7 (Alpha variant) strongly seems to have arisen as a result of natural selection. It came into existence after outcompeting already circulating widespread SARS-CoV variants.
Most of the mutations in B.1.1.7 lineage include mutations in the spike glycoprotein, N501Y in the receptor binding domain, deletion 69_70, and P681H in the furin cleavage site, which could probably influence ACE2 binding and viral replication. Specifically, the 501Y spike variants were predicted to have an increased affinity for human ACE2, and another variant, also with an N501Y mutation, was spreading fast in South Africa (Beta variant—B.a351, B.1.351.2, and B.1.353.3). Immunogenic effects of these mutations are currently not clear. Similarly, the Gamma variant (P.1) was emerged in the Amazon city of Manaus in December 2020 and has led to a surge in cases in Brazil (Buss et al., 2021).
Recently, the Delta variant (B.1.617.2, AY.1, and AY.2) having multiple mutations originated in India is of major concern (Centers for Disease Control and Prevention (CDC), 2021; Public Health England, 2021). This variant is the highest transmissible variant and hence favored by evolution. Therefore, different mutants originated in different geographical areas as a combinatorial result of selective advantage or chance mutation. Variants having mutations in spike to increase transmissibility could quickly outcompete and replace other circulating variants. Moreover, widespread infection among humans is now posing a huge threat to other mammals that usually interact with human populations and worsen the severity of disease by creating more dangerous recombinant SARS-CoV-2 strains. It would be important to consider the epidemiological, genetic, and functional studies of different variants and come up with a strong strategy to stop its transmission across the species.
Accumulation of mutations within the genome is the primary driving force in viral evolution within an endemic setting (Dan et al., 2020; Baden et al., 2021). This inherent feature often leads to altered virulence, infectivity and transmissibility, and antigenic shifts to escape host immunity, which might compromise the efficacy of vaccines and antiviral drugs (Upadhyay et al., 2021; Yadav et al., 2021a). The SARS-CoV-2 as RNA virus lacks mismatch repair mechanism and replication accompanied by a high mutation rate (Domingo and Holland, 1997). Therefore, the mutations of the coronavirus are commonsensical and predictable, which leads to several rapidly spreading variants (Table 1). At present, emergence of fast-spreading three SARS-CoV-2 variants (B.1.1.7, B.1.351, and B.1.1.28.1) due to rapid mutations in ACE2 became dominant strains all around the world, causing concern on prevention and treatment of COVID-19 (Krammer, 2020; Callaway, 2021; Zhou and Wang, 2021). The morphological and physiological assessments of the P.1 or B.1.1.28.1 variant of SARS-CoV-2 from Brazil reflected less resistance to antibodies produced from natural infection or vaccination compared with other parallel variants B.1.351 from South Africa, and B.1.1.7 from the United Kingdom (Faria et al., 2021). It is noteworthy that P.1, B.1.1.7, and B.1.351 have accrued multiple mutations in the NTD (N-terminal domain) and can be neutralized by a monoclonal antibody, mAb 222 (Cerutti et al., 2021; Dejnirattisai et al., 2021). In addition, these mutated residues also have the potential to modulate vaccine-induced antibody responses (Supasa et al., 2021; Zhou et al., 2021). The three central variants by analyzing 160 sequences claimed that B-type viruses (with substitution, NS8_L84S) were common in East Asia, whereas A-type (ancestral lineage) and C-type (NS3_G251V variant) viruses were prevalent in Europe and North America (Forster et al., 2020). Along with other co-evolving mutations, NSP12_P323L and S_D614G probably provide variants with an evolutionary advantage over their ancestral types, allowing them to survive and circulate in this densely populated region (Becerra-Flores and Cardozo, 2020; Islam et al., 2021). Thus, the recent emergence of a number of variants of concern (VOCs) has led to design of new vaccines that will be able to protect against the emerging viral variants.
The comprehensive analysis of whole-genome sequences of 837 Indian SARS-CoV-2 strains revealed the occurrence of 33 different mutations, 18 of which were unique to India (Tang et al., 2020; Sarkar et al., 2021b). The second SARS-CoV-2 epidemic wave in India began around March 2021, and just weeks after, it became the dominant lineage by superseding the previous lineages (Kar et al., 2021; Salvatore et al., 2021). Almost all new cases of COVID-19 are the Delta variant (B.1.617.2) with augmented cases, but the rate of growth is slower than that of the Alpha variant (O’Dowd, 2021). The data showed the even at the higher risk of hospitalization for patients with the Delta variant compared with the Alpha variant (B.1.1.7), two doses of vaccine gave a high degree (90%) of protection (Shrotri et al., 2021; Stowe et al., 2021; Williams et al., 2021). The identification and spread of various dreading variants including B.1.1.7, B.1.351, and P.1 in India led to global VOCs (Alai et al., 2021). The Kappa and Delta variant lineages of SARS-CoV-2 were first detected in December 2020 in India (Cherian et al., 2021). Rapidly between January and February 2021, the Delta (B.1.617.2) variant became dominant in Maharashtra and was marked as a VOC in early May by the WHO (2021b). Therefore, it is imperative that currently known variants of COVID-19 and new variants should be carefully considered in the design of an effective vaccine.
The case fatality rate (CFR) in COVID-19 seems to be elevated than that of in seasonal influenza, whereas both diseases principally have an effect on older adults above 65 years of age with infirmity (Dan et al., 2020; Yadav et al., 2021b). The augmented fatality rate of COVID-19 could be because of variations in underlying comorbidities of patients, pathogenicity of the causative agent SARS-CoV-2, immunity of population, and responses of host to the infection (Jha et al., 2020; Upadhyay et al., 2020a; Upadhyay et al., 2020b). It has been reported that the COVID-19 patients were more frequently obese and suffered from diabetes, hypertension, and dyslipidemia than influenza patients; on the contrary, the influenza patients often had cardiac failure, chronic respiratory disease, cirrhosis, and anemia (Piroth et al., 2021). Patients admitted to care centers with new variant of SARS-CoV-2 more frequently experienced acute respiratory failure, pulmonary embolism, septic shock, or hemorrhagic stroke, but less frequently developed myocardial infarction or atrial fibrillation (Dan et al., 2020). In-hospital mortality was comparatively multifold higher in patients with COVID-19 than conventional influenza patients (16.9% vs. 5.8%, respectively), with a relative risk of death of 2.9 (West et al., 2021b). Quantitatively, there was less pediatric patients (<18 years) for COVID-19 than influenza among the patients admitted in the hospital, but a bigger proportion of patients younger than 5 years required intensive care unit (ICU) support to COVID-19 than influenza (Piroth et al., 2021). As per the report, in-hospital mortality of adolescents (11–17 years) was manyfold higher for COVID-19 than for influenza as well. Thus, the effect of the SARS-CoV-2 variant is tremendous for all sex and age groups of the human population but was supposed to be the most common challenging health risk factor to immunocompromised septuagenarians and octogenarians (Figure 1).
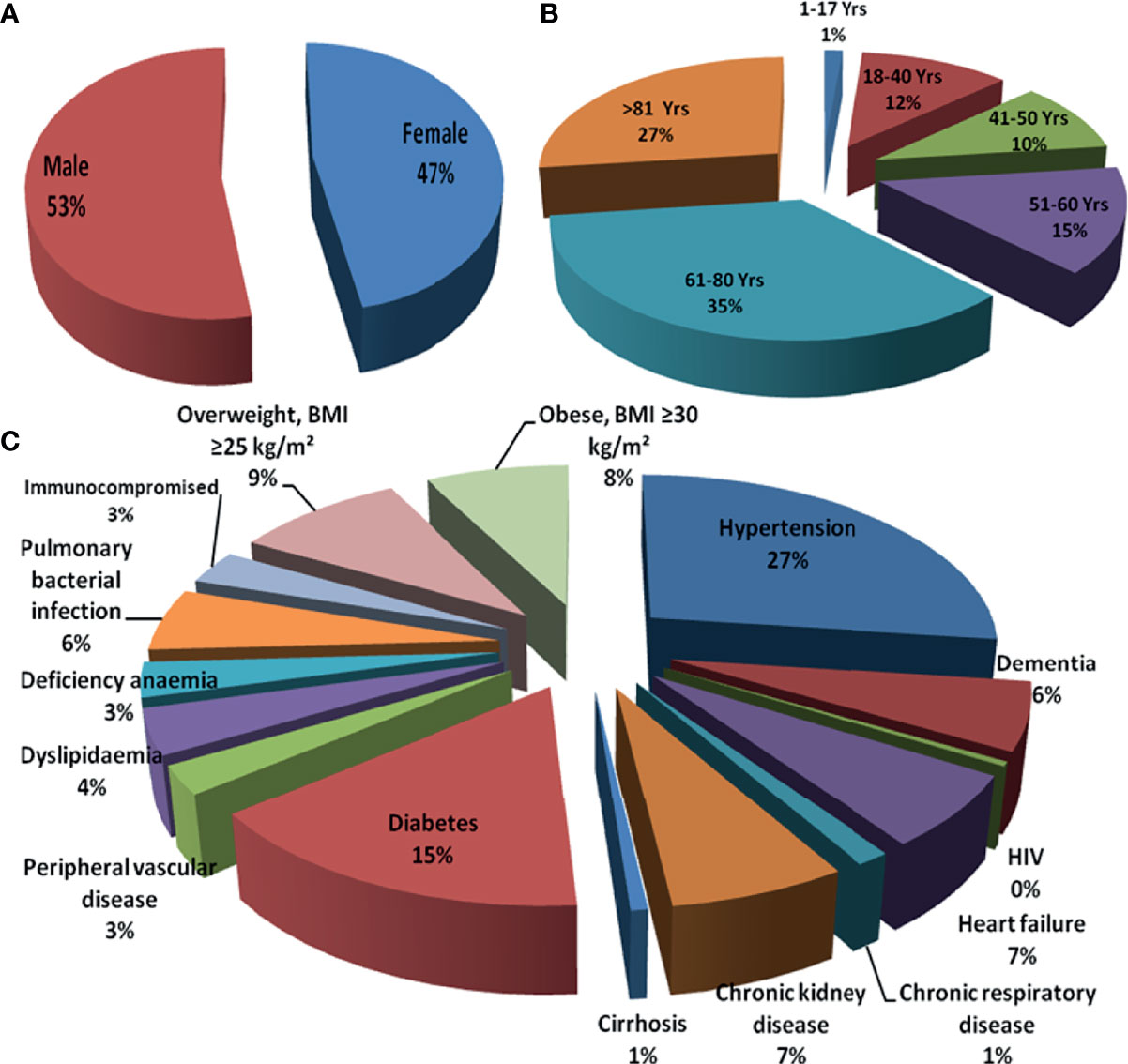
Figure 1 Risk of infectivity and morbidity among COVID-19 patients: (A) sex based, (B) age based, and (C) comorbidities. COVID-19, coronavirus disease 2019.
Currently, the strategy to treat the COVID-19 infection comprises social distancing and vaccination. However, with the sharp rise in the cases and variable symptoms, various pharmacotherapies were explored for enhancing viral clearance and other symptomatic relief (Rahman and Idid, 2021). Until now, no specific drug for the treatment and management of COVID-19 has been developed. Hence, the focus has been shifted towards drug repurposing, which is time saving, is an accepted approach, and has an unmet need of time (Stasi et al., 2020). At present, many of the existing drugs have been repurposed and tested in preclinical and clinical trials (Table 2). However, with the advancement and better understanding of pathophysiology and clinical presentation among patients, it was noticed that the clinical efficacy of these drugs depends on timing of use, disease stage, and dose regimen (Iqubal et al., 2021a). Antiviral drugs are important when used during the early stage, as they inhibit viral entry and replication (Şimşek Yavuz and Ünal, 2020). Among antiviral drugs, remdesivir is one of the extensively used drugs. Initially, the in vitro study has shown antiviral potential against COVID-19. Later on, the US Food and Drug Administration (FDA) approved this drug to shorten the recovery time in adults and children (below the age of 12) (Young et al., 2021). However, the outcome of the WHO SOLIDARITY trial that involved 11,330 patients across 40 countries showed a non-significant effect on reducing mortality, duration of hospitalization, and need of a mechanical ventilator (Horby et al., 2020). Lopinavir/ritonavir is a combination therapy for HIV, and it was proposed to be an effective therapy for COVID-19 (Cao et al., 2020). Ivermectin is an approved antiparasitic drug (Caly et al., 2020). Initially, the in vitro study showed that ivermectin significantly inhibited the replication; but based on the outcome of a double-blinded randomized trial, no clinical efficacy of lopinavir/ritonavir and ivermectin among COVID-19-infected patients were found (López-Medina et al., 2021). These drugs are not in use now. Hydroxychloroquine and chloroquine were also claimed to be promising therapeutic modality against COVID-19 infection, but the outcome of the randomized trial showed a non-significant effect against symptomatic relief among COVID-19 patients (Mitjà et al., 2021).
Use of corticosteroids and immunotherapy is preferred during cytokine storms or at the hyperinflammatory stage, and inappropriate use of these drugs often results in fetal immunogenic reactions (Esmaeilzadeh and Elahi, 2021; Rabaan et al., 2021).
Based on various clinical findings, corticosteroids were reported to be effective against cytokine storm and hyperinflated lungs (Hassan et al., 2020; Shang et al., 2020). The outcome of the landmark RECOVERY trial that involved confirmed patients of COVID-19 showed that the use of dexamethasone resulted in reduced mortality and need of mechanical ventilators or oxygen supply (Hamilton et al., 2021). Based on this trial, dexamethasone was approved among critically ill patients, either alone or in combination with remdesivir (Vetter et al., 2020; Mehta et al., 2021). Interferon-β-1a, a cytokine, exhibits an immunogenic response against viral infection (Yuen et al., 2020). Previously, interferon-β-1a showed clinical ineffectiveness against ARDS but exhibited a positive response among the patients of COVID-19 (Bosi et al., 2020; Kali et al., 2021; Tortajada et al., 2021). Interferon-β-1a, when used during the early stage of infection, reduced the duration of hospitalization and mortality rate (Davoudi-Monfared et al., 2020). However, recent findings have shown that interferon-β-1a is ineffective against Alpha (B.1.1.7), Beta (B.1.351), Gamma (P1), and Delta (B.1.617.2) strains (Davoudi-Monfared et al., 2020). Currently, interferon-β-1a is not recommended for treating COVID-19 patients (Davoudi-Monfared et al., 2020). Similar to interferon-β-1a, anakinra (interleukin-1 antagonist) was found to be effective in reducing mortality during the initial investigation, but recent findings have shown its ineffectiveness against B.1.1.7; B.1.351, and P.1 variants and, hence, are not recommended to treat COVID-19-infected patients (Huet et al., 2020). Tocilizumab (IL-6 receptor antibody) and sarilumab as well as siltuximab (IL-receptor antagonist) are effective during hyperinflammatory state; and hence, they were explored for possible protective effects in COVID-19 infection (Michot et al., 2020). Some clinical trials, such as REMAP and RECOVERY, showed the benefit of using tocilizumab, sarilumab, and siltuximab, which reduced mortality and showed a better safety profile among infected patients (Michot et al., 2020; Gordon et al., 2021). Janus kinase (JAK) inhibitors (baricitinib, ruxolitinib, and tofacitinib) are well-known drugs approved for rheumatoid arthritis and other inflammatory conditions (Stebbing et al., 2020). Baricitinib is considered as one of the potential drug candidates against COVID-19 infection (Saber-Ayad et al., 2021). This drug acts by inhibiting viral endocytosis in the in vitro study and inhibits the altered hyperinflammatory signaling pathway (Richardson et al., 2020). In ACTT-2 trial, when baricitinib was used in combination with remdesivir, it showed superior clinical efficacy in reducing ARDS and mortality rate as compared with baricitinib alone (Kalil et al., 2021). Currently, the combination of baricitinib and remdesivir is approved by the US FDA for the treatment of COVID-19 infection (Kalil et al., 2021). Apart from JAK inhibitor, Bruton’s tyrosine kinase inhibitors such as rilzabrutinib, ibrutinib, and acalabrutinib are currently approved by the US FDA for the treatment of hematological malignancy (Table 2) (Rezaei et al., 2020; Benner and Carson, 2021; Rada et al., 2021). These drugs act as an inhibitor macrophage activation, which is a rate-limiting step during cytokine storm (Roschewski et al., 2020). Therefore, these drugs are hypothesized to be a future therapeutic candidate against COVID-19 infection (Figure 2). More recently, anti-SARS-CoV-2-neutralizing antibodies such as casirivimab, bamlanivimab, imdevimab, etesevimab, and sotrovimab were approved by the US FDA for the treatment of non-hospitalized patients with a confirmed report of COVID-19 infection (Mahase, 2021; Verderese et al., 2021; Weinreich et al., 2021).
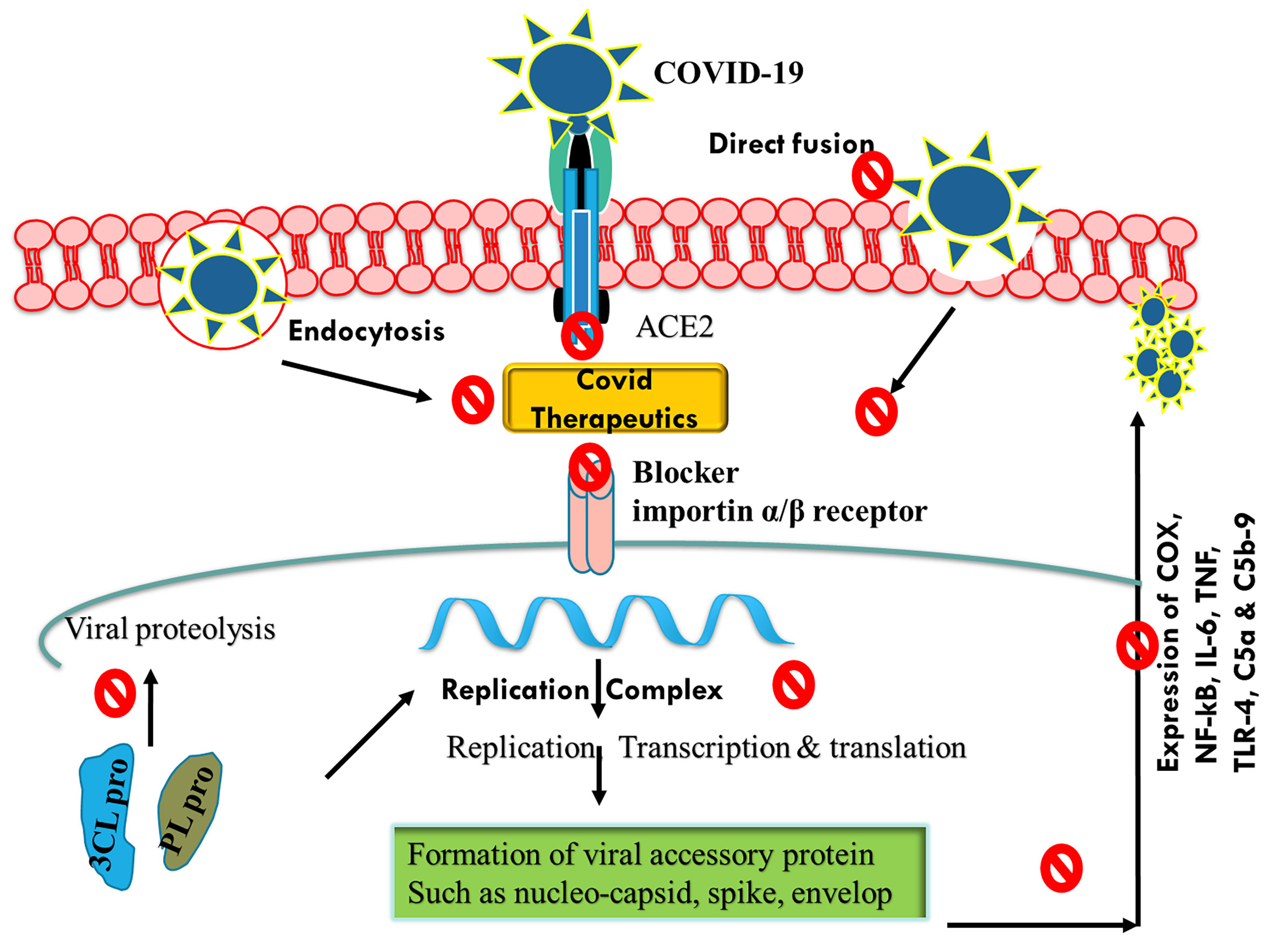
Figure 2 Schematic representation of mechanisms of action of COVID-19 therapeutics by inhibiting endocytosis, ACE2 receptor, and viral replication. COVID-19, coronavirus disease 2019; ACE2, angiotensin-converting enzyme 2.
COVID-19 vaccines play a critical role in helping the countries to overcome the challenging pandemic that they are currently grappling with. It is believed that the severity of the pandemic will gradually reduce as the herd immunity is achieved. However, there may be factors that make it difficult to achieve herd immunity such as receiving only one dose of the vaccine for which two doses are required, denial to get vaccinated, and shortage of the vaccines. Therefore, it is very important to mass vaccinate the population completely if we want to win the battle over the pandemic (Chen and Lu, 2021). A public–private partnership was initiated by the US government to speed up development, approval, and distribution of the COVID-19 vaccines (Corey et al., 2020). Most of the COVID-19 vaccines have spike glycoprotein of SARS-CoV-2 as their basis. The commonly used vaccines are as follows: BNT162b2 (Pfizer-BioNTech) (Polack et al., 2020), ChAdOx1 nCOV19 (Oxford-AstraZeneca) (Voysey et al., 2021), NVX-CoV2373 (Novavax) (Keech et al., 2020), mRNA-1273 (NIAID-Moderna) (Baden et al., 2021), and Ad26COV2S (Janssen) (Sadoff et al., 2021). There are several preprints, peer-reviewed publications, press releases, policy documents, and public regulatory documents that demonstrate the efficacy and safety of these vaccines (Keech et al., 2020; Polack et al., 2020; Baden et al., 2021; Voysey et al., 2021). A study was conducted to study the efficacy of BNT162b2 vaccine (Dagan et al., 2021) during the mass vaccination in Israel. The participants were followed up 7 days after the second dose, and it was found that the vaccine has an efficacy of 94% for symptomatic COVID-19 participants, 92% for people with severe COVID-19, 92% for people with documented infection, and 87% for the people admitted in the hospitals. It was also concluded that the effectiveness of the vaccine was lower in people who suffer from various coexisting medical conditions like hypertension and obesity than in healthy individuals. Similar results were found in England for adults aged 70 years and over, indicating that the BNT162b2 vaccine showed 85%–90% efficacy after the second dose (Lopez Bernal et al., 2021). The risk of being admitted to hospitals was reduced by 44% in the vaccinated people, whereas the risk of death was reduced by 51%. They also studied the efficacy of ChAdOx1-S vaccine and found out that a single dose was 60%–75% effective in people with symptomatic COVID-19 and that the risks of hospital admission were reduced up to 80% in the vaccinated people. Various vaccines are being manufactured and distributed across the globe (Table 3) to control the pandemic. Figure 3 summarizes the mechanisms of action of investigated anti-COVID-19 vaccines. These vaccines have helped in reducing the number of COVID-19 cases; however, the efficacy may vary in different studies. In the earlier phases of vaccination, it was found that the people receiving the vaccination were more prone to COVID-19 infection, which encouraged people to defer the vaccines. However, it was found that the infection occurred when people travelled to infected region or encountered COVID-19-positive patients, and the risk of infection was higher in the first 3 days of vaccination. This period was before the incubation of vaccine occurred, which rules out the odds of vaccination.
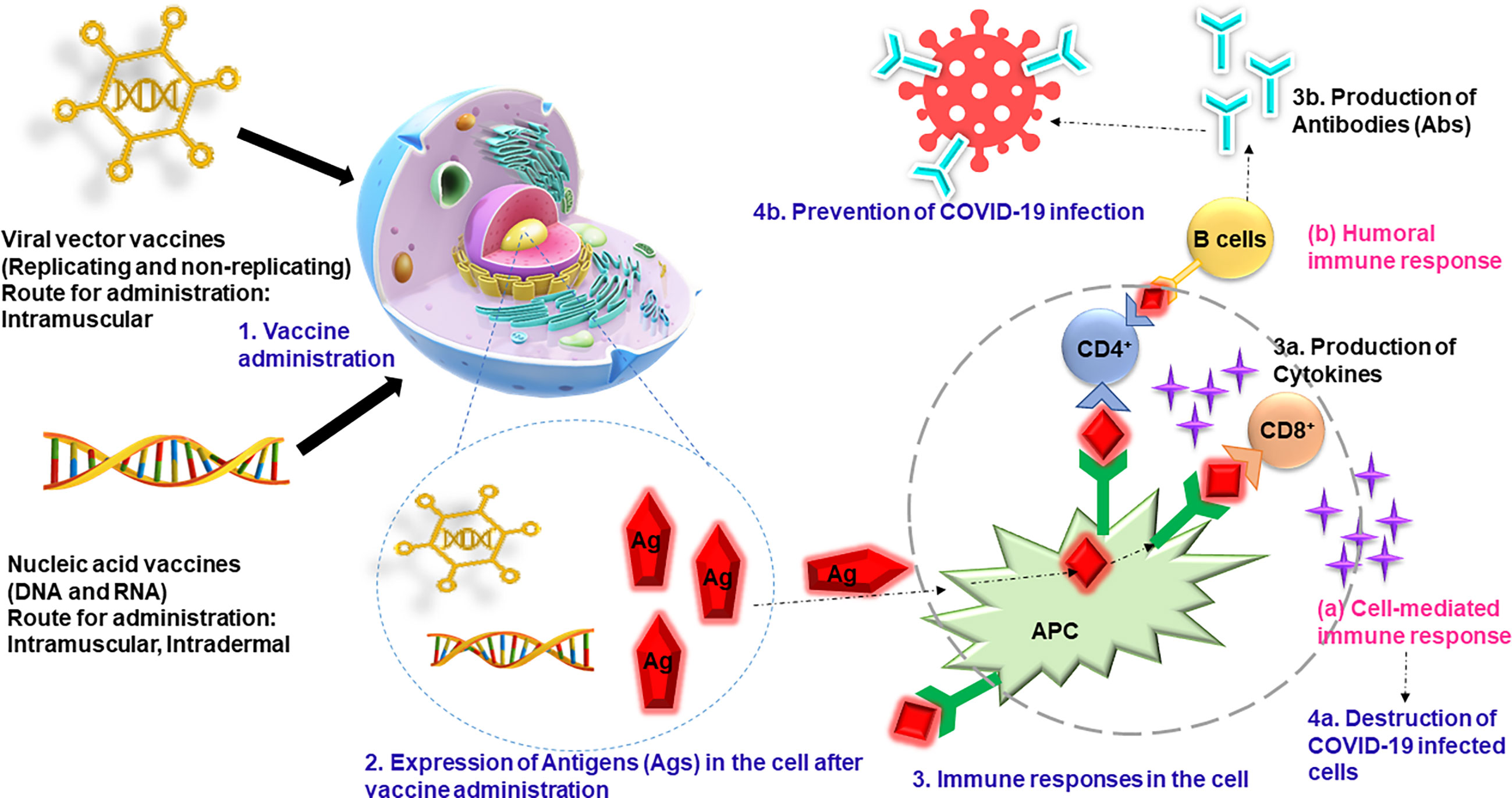
Figure 3 The vaccines (viral vector and nucleic acid vaccines) are administered through intramuscular or intradermal routes, and antigen expression is initiated in the cells. The B cells and T cells generate the humoral immune response and cell-mediated immune response, respectively. The cell-mediated immune response produces the cytokines that kill the infected cells, and humoral cells produce antibodies that prevent the COVID-19 infection. COVID-19, coronavirus disease 2019.
Most of these vaccines were manufactured against the original strain of SARS-CoV-2, and since then, the virus has mutated several times. It is crucial to develop a wide-spectrum vaccine that is effective against the various strains of SARS-CoV-2. In addition, for controlling the COVID-19, it is very important that the global population may be vaccinated completely. It is the duty of the officials to build trust among the public and encourage them to get vaccinated. The eradication of this disease is only possible when the herd immunity is achieved by vaccinating the people globally.
Considering the rapid molecular evolution of SARS-CoV-2 virus from its emergence to the present moment, continuous surveillance is required to identify novel mutations with potential ability to bypass current measures for controlling COVID-19. In the near future, readiness to react to such changes in virus genome is probably unavoidable. Rapid ongoing vaccination with continuously improved and updated vaccines or even vaccine cocktails is obviously the only human-controlled proactive way to impede the pandemic. Taking into consideration the fact that increased transmission can enhance the probability of further mutations (Matta et al., 2021), quick vaccination of the most active (younger) age groups seems to be the best strategy for preventing the appearance of novel hazardous mutations. On the other hand, the possibility of emergence of a mutant virus variant with high prevalence (high transmissibility) but low virulence cannot be avoided, overriding the spread of the current high-lethality strains and changing the fatal disease course to be much milder, thereby ensuring the “friendly” coexistence of virus and humankind in the future. Which of these scenarios will come true is just the question of time; still, it is clear that the lessons that this pandemic has taught to humankind are absolutely unique and tremendous.
Within already nearly the last 2 years, humankind of the 21st century has undergone unexpectedly complicated challenges related to the COVID-19 pandemic, from total social isolation to different mass-vaccination campaigns. However, despite biotechnological prosperity and ultrafast preparation of vaccines, we still cannot look to the future with peace of mind, as the virus is circulating among populations even after the use of current vaccines, and we have no means to forecast the virulence and lethality of potentially developing novel strains. Therefore, our location within this pandemic can be decided only retrospectively, and it remains to be hoped that after 5 years we will estimate today’s position as the end of the pandemic.
HT performed the literature survey and data extraction. KS contributed in the introduction and conclusion. PA contributed in the molecular evolution. AI contributed in the therapeutic section. SU contributed in the geographic distribution section. JK contributed in the vaccination section. GK and DA contributed in final proofing and editing. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdullahi, I. N., Emeribe, A. U., Ajayi, O. A., Oderinde, B. S., Amadu, D. O., Osuji, A. I. (2020). Implications of SARS-CoV-2 Genetic Diversity and Mutations on Pathogenicity of the COVID-19 and Biomedical Interventions. J. Taibah Univ. Med. Sci. 15 (4), 258–264. doi: 10.1016/j.jtumed.2020.06.005
Adedokun, K. A., Olarinmoye, A. O., Olayemi, L. O., Shehu, M. R., Mustapha, J. O., Kamorudeen, R. T., et al (2021). Addressing the Global Surge of COVID-19 Cases: Insights From Diagnostics, Improved Treatment Strategies, Vaccine Development and Application. J. Clin. Transl. Res. 7 (2), 127–139. doi: 10.18053/jctres.07.202102.00
Alai, S., Gujar, N., Joshi, M., Gautam, M., Gairola, S. (2021). Pan-India Novel Coronavirus SARS-CoV-2 Genomics and Global Diversity Analysis in Spike Protein. Heliyon 7 (3):e06564. doi: 10.1016/j.heliyon.2021.e06564
Baden, L. R., El Sahly, H. M., Essink, B., Kotloff, K., Frey, S., Novak, R., et al (2021). Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 384 (5), 403–416. doi: 10.1056/NEJMoa2035389
Becerra-Flores, M., Cardozo, T. (2020). SARS-CoV-2 Viral Spike G614 Mutation Exhibits Higher Case Fatality Rate. Int. J. Clin. Pract. 74 (8), e13525. doi: 10.1111/ijcp.13525
Benner, B., Carson, W. E. (2021). Observations on the Use of Bruton's Tyrosine Kinase Inhibitors in SAR-CoV-2 and Cancer. J. Hematol. Oncol. 14 (1), 15. doi: 10.1186/s13045-020-00999-8
Biotech, B. (2021). Bharat Biotech Announces Phase 3 Results of COVAXIN: Indias First COVID-19 Vaccine Demonstrates Interim Clinical Efficacy of 81% (Hyderabad, TS), 3. Available at: https://www.bharatbiotech.com/images/press/covaxin-phase3-efficacy-results.pdf.
Bosi, E., Bosi, C., Rovere Querini, P., Mancini, N., Calori, G., Ruggeri, A., et al (2020). Interferon β-1a (IFNβ-1a) in COVID-19 Patients (INTERCOP): Study Protocol for a Randomized Controlled Trial. Trials 21 (1), 939. doi: 10.1186/s13063-020-04864-4
Brüssow, H. (2021). COVID-19: Emergence and Mutational Diversification of SARS-CoV-2. Microb. Biotechnol. 14 (3), 756–768. doi: 10.1111/1751-7915.13800
Buss, L. F., Prete, C. A., Jr., Abrahim, C., Mendrone, A., Salomon, T., de Almeida-Neto, C., et al (2021). Three-Quarters Attack Rate of SARS-CoV-2 in the Brazilian Amazon During a Largely Unmitigated Epidemic. Science (New York N.Y.) 371 (6526), 288–292. doi: 10.1126/science.abe9728
Callaway, E. (2021). Fast-Spreading COVID Variant can Elude Immune Responses. Nature 589, 500–501. doi: 10.1038/d41586-021-00121-z
Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., Wagstaff, K. M. (2020). The FDA-Approved Drug Ivermectin Inhibits the Replication of SARS-CoV-2 In Vitro. Antiviral Res. 178, 104787. doi: 10.1016/j.antiviral.2020.104787
Cao, B., Wang, Y., Wen, D., Liu, W., Wang, J., Fan, G., et al (2020). A Trial of Lopinavir-Ritonavir in Adults Hospitalized With Severe Covid-19. N. Engl. J. Med. 382 (19), 1787–1799. doi: 10.1056/NEJMoa2001282
Cavagna, L., Seminari, E., Zanframundo, G., Gregorini, M., Di Matteo, A., Rampino, T., et al (2020). Calcineurin Inhibitor-Based Immunosuppression and COVID-19: Results From a Multidisciplinary Cohort of Patients in Northern Italy. Microorganisms 8 (7), 977. doi: 10.3390/microorganisms8070977
CDC (2021). COVID-19: SARS-CoV-2 Variant Classifications and Definitions. Department of Health and Human Services (USA: CDC Atlanta, GA). Available at: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html.
Cerutti, G., Guo, Y., Zhou, T., Gorman, J., Lee, M., Rapp, M., et al (2021). Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite. Cell Host Microbe 29 (5), 819–833.e7. doi: 10.1101/2021.01.10.426120
Chen, J., Lu, H. (2021). New Challenges to Fighting COVID-19: Virus Variants, Potential Vaccines, and Development of Antivirals. Biosci. Trends 15 (2), 126–128. doi: 10.5582/bst.2021.01092
Cherian, S., Potdar, V., Jadhav, S., Yadav, P., Gupta, N., Das, M., et al (2021). Convergent Evolution of SARS-CoV-2 Spike Mutations, L452R, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms 9 (7), 1542. doi: 10.1101/2021.04.22.440932
Chiam, T., Subedi, K., Chen, D., Best, E., Bianco, F. B., Dobler, G., et al (2021). Hospital Length of Stay Among COVID-19-Positive Patients. J. Clin. Transl. Res. 7 (3), 377–385. doi: 10.1016/j.chom.2021.03.005
Chudik, A., Pesaran, M. H., Rebucci, A. (2021). COVID-19 Time-Varying Reproduction Numbers Worldwide: An Empirical Analysis of Mandatory and Voluntary Social Distancing. Cambridge, MA: (NBER Work. Pap. Ser). Available at: https://www.nber.org/system/files/working_papers/w28629/w28629.pdf.
Corey, L., Mascola, J. R., Fauci, A. S., Collins, F. S. (2020). A Strategic Approach to COVID-19 Vaccine R&D. Science (New York N.Y.) 368 (6494), 948–950. doi: 10.1126/science.abc5312
Dagan, N., Barda, N., Kepten, E., Miron, O., Perchik, S., Katz, M. A., et al (2021). BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 384 (15), 1412–1423. doi: 10.1056/NEJMoa2101765
Dan, S., Pant, M., Upadhyay, S. K. (2020). The Case Fatality Rate in Covid-19 Patients With Cardiovascular Disease: Global Health Challenge and Paradigm in the Current Pandemic. Curr. Pharmacol. Rep. 1–10. doi: 10.1007/s40495-020-00239-0
Davies, N. G., Barnard, R. C., Jarvis, C. I., Kucharski, A. J., Munday, J., Pearson, C. A. B., et al (2020). Estimated Transmissibility and Severity of Novel SARS-CoV-2 Variant of Concern 202012/01 in England. Science 372 (6538), eabg3055. doi: 10.1126/science.abg3055
Davoudi-Monfared, E., Rahmani, H., Khalili, H., Hajiabdolbaghi, M., Salehi, M., Abbasian, L., et al (2020). A Randomized Clinical Trial of the Efficacy and Safety of Interferon β-1a in Treatment of Severe COVID-19. Antimicrob. Agents Chemother. 64 (9), e01061–e01020. doi: 10.1128/AAC.01061-20
Deimel, L. P., Li, Z., Ranasinghe, C. (2021). Interleukin-13 as a Target to Alleviate Severe Coronavirus Disease 2019 and Restore Lung Homeostasis. J. Clin. Transl. Res. 7 (1), 116–120. doi: 10.18053/jctres.07.202101.0
Dejnirattisai, W., Zhou, D., Supasa, P., Liu, C., Mentzer, A. J., Ginn, H. M., et al (2021). Antibody Evasion by the P.1 Strain of SARS-CoV-2. Cell 184 (11), 2939–2954:e9. doi: 10.1016/j.cell.2021.03.055
Domingo, E., Holland, J. J. (1997). RNA Virus Mutations and Fitness for Survival. Annu. Rev. Microbiol. 51, 151–178. doi: 10.1146/annurev.micro.51.1.151
Esmaeilzadeh, A., Elahi, R. (2021). Immunobiology and Immunotherapy of COVID-19: A Clinically Updated Overview. J. Cell Physiol. 236 (4), 2519–2543. doi: 10.1002/jcp.30076
European Centre for Disease Prevention and Control (2020). Detection of New SARS-CoV-2 Variants Related to Mink (Stockholm: ECDC). Available at: https://www.ecdc.europa.eu/sites/default/files/documents/RRASARS-CoV-2-in-mink-12-nov-2020.pdf.
Faria, N. R., Claro, I. M., Candido, D., Moyses, Franco, L. A., Andrade, P. S., et al (2021) Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in Manaus: Preliminary Findings. SARS-CoV-2 Coronavirus / Ncov-2019 Genomic Epidemiology. Available at: https://virological.org/t/genomiccharacterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminaryfindings/586.
Forster, P., Forster, L., Renfrew, C., Forster, M. (2020). Phylogenetic Network Analysis of SARS-CoV-2 Genomes. Proc. Natl. Acad. Sci. U.S.A. 117 (17), 9241–9243. doi: 10.1073/pnas.2004999117
Fraser, B. (2020). COVID-19 Strains Remote Regions of Peru. Lancet 395 (10238), 1684. doi: 10.1016/S0140-6736(20)31236-8
Giannini, S., Passeri, G., Tripepi, G., Sella, S., Fusaro, M., Arcidiacono, G., et al (2021). Effectiveness of In-Hospital Cholecalciferol Use on Clinical Outcomes in Comorbid COVID-19 Patients: A Hypothesis-Generating Study. Nutrients 13 (1), 219. doi: 10.3390/nu13010219
Gordon, A. C., Mouncey, P. R., Al-Beidh, F., Rowan, K. M., Nichol, A. D., Arabi, Y. M., et al (2021). Interleukin-6 Receptor Antagonists in Critically Ill Patients With Covid-19. N. Engl. J. Med. 384 (16), 1491–1502. doi: 10.1056/NEJMoa2100433
Gottlieb, R. L., Nirula, A., Chen, P., Boscia, J., Heller, B., Morris, J., et al (2021). Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA 325 (7), 632–644. doi: 10.1001/jama.2021.0202
Hamilton, F. W., Lee, T., Arnold, D. T., Lilford, R., Hemming, K. (2021). Is Convalescent Plasma Futile in COVID-19? A Bayesian Re-Analysis of the RECOVERY Randomized Controlled Trial. Int. J. Infect. Dis. 109, 114–117. doi: 10.1016/j.ijid.2021.06.034
Haseltine, W. A. (2021). A New Variant in the Philippines. Philippines: Forbes. Available at: https://www.forbes.com/sites/williamhaseltine/2021/03/18/third-generation-covid-19-variant-described-in-the-philippines/?sh=617be2b973ca.
Hassan, M. E., Hasan, H. M., Sridharan, K., Elkady, A., ElSeirafi, M. M. (2020). Dexamethasone in Severe COVID-19 Infection: A Case Series. Respir. Med. Case Rep. 31, 101205. doi: 10.1016/j.rmcr.2020.101205
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181 (2), 271–280.e8. doi: 10.1016/j.cell.2020.02.052
Horby, P., Mafham, M., Linsell, L., Bell, J. L., Staplin, N., Emberson, J. R., et al (2020). Effect of Hydroxychloroquine in Hospitalized Patients With Covid-19. N. Engl. J. Med. 383 (21), 2030–2040. doi: 10.1056/NEJMoa2022926
Hossain, M. K., Hassanzadeganroudsari, M., Apostolopoulos, V. (2021). The Emergence of New Strains of SARS-CoV-2. What Does it Mean for COVID-19 Vaccines? Expert Rev. Vaccines 1, 4. doi: 10.1080/14760584.2021.1915140
Hou, Y. J., Chiba, S., Halfmann, P., Ehre, C., Kuroda, M., Dinnon, K. H., et al (2020). SARS-CoV-2 D614G Variant Exhibits Efficient Replication Ex Vivo and Transmission In Vivo. Science (New York N.Y.) 370 (6523), 1464–1468. doi: 10.1126/science.abe8499
Huet, T., Beaussier, H., Voisin, O., Jouveshomme, S., Dauriat, G., Lazareth, I., et al (2020). Anakinra for Severe Forms of COVID-19: A Cohort Study. Lancet Rheumatol. 2 (7), e393–e400. doi: 10.1016/S2665-9913(20)30164-8
Iqubal, A., Hoda, F., Najmi, A. K., Haque, S. E. (2021a). Macrophage Activation and Cytokine Release Syndrome in COVID-19: Current Updates and Analysis of Repurposed and Investigational Anti-Cytokine Drugs. Drug Res. 71 (4), 173–179. doi: 10.1055/a-1291-7692
Iqubal, A., Iqubal, M. K., Ahmed, M., Haque, S. E. (2021b). Natural Products, a Potential Therapeutic Modality in Management and Treatment of Ncov-19 Infection: Preclinical and Clinical Based Evidence. Curr. Pharm. Des. 27 (9), 1153–1169. doi: 10.2174/1381612827999210111190855
Iqubal, A., Iqubal, M. K., Hoda, F., Najmi, A. K., Haque, S. E. (2021c). COVID-19 and Cardiovascular Complications: An Update From the Underlying Mechanism to Consequences and Possible Clinical Intervention. Expert Rev. Anti Infect. Ther. 19(9), 1083–1092. doi: 10.1080/14787210.2021.1893692
Isabel, S., Graña-Miraglia, L., Gutierrez, J. M., Bundalovic-Torma, C., Groves, H. E., Isabel, M. R. (2020). Evolutionary and Structural Analyses of SARS-CoV-2 D614G Spike Protein Mutation Now Documented Worldwide. Sci. Rep. 10 (1), 14031. doi: 10.1038/s41598-020-70827-z
Islam, O. K., Al-Emran, H. M., Hasan, M. S., Anwar, A., Jahid, M., Hossain, M. A. (2021). Emergence of European and North American Mutant Variants of SARS-CoV-2 in South-East Asia. Transbound Emerg. Dis. 68 (2), 824–832. doi: 10.1111/tbed.13748
Jha, D., Sharma, V., Sharma, J. K., Kumar, S., Sharma, V., Kamboj, P., et al (2020). Plausible State-Specific Plans and Recommendations to Avert COVID-19 Community Transmission. Bull. Pure Appl. Sci. (Zool.) 39A (2), 447–454. doi: 10.5958/2320-3188.2020.00051.0
Kali, S. K., Dröge, P., Murugan, P. (2021). Interferon β, an Enhancer of the Innate Immune Response Against SARS-CoV-2 Infection. Microb. Pathog. 158, 105105. doi: 10.1016/j.micpath.2021.105105
Kalil, A. C., Patterson, T. F., Mehta, A. K., Tomashek, K. M., Wolfe, C. R., Ghazaryan, V., et al (2021). Baricitinib Plus Remdesivir for Hospitalized Adults With Covid-19. N. Engl. J. Med. 384 (9), 795–807. doi: 10.1056/NEJMoa2031994
Kar, S. K., Ransing, R., Arafat, S., Menon, V. (2021). Second Wave of COVID-19 Pandemic in India: Barriers to Effective Governmental Response. EClinicalMedicine 36, 100915. doi: 10.1016/j.eclinm.2021.100915
Kawabata, H., Okazaki, Y., Watanabe, K., Inoue, T., Yamaguchi, K., Ueda, Y., et al (2020). A Box-Shaped Shielding Device for Reducing the Risk of COVID-19 Droplet Infection During Gastrointestinal Endoscopic Procedures. J. Clin. Transl. Res. 6 (6), 236–240. doi: 10.18053/jctres.06.202006.00
Keech, C., Albert, G., Cho, I., Robertson, A., Reed, P., Neal, S., et al (2020). Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 383 (24), 2320–2332. doi: 10.1056/NEJMoa2026920
Korber, B., Fischer, W. M., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., et al (2020). Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 182 (4), 812–827.e19. doi: 10.1016/j.cell.2020.06.043
Krammer, F. (2020). SARS-CoV-2 Vaccines in Development. Nature 586, 516–527. doi: 10.1038/s41586-020-2798-3
Laurence, J., Mulvey, J. J., Seshadri, M., Racanelli, A., Harp, J., Schenck, E. J., et al (2020). Anti-Complement C5 Therapy With Eculizumab in Three Cases of Critical COVID-19. Clin. Immunol. (Orlando Fla.) 219, 108555. doi: 10.1016/j.clim.2020.108555
Leung, K., Shum, M. H., Leung, G. M., Lam, T. T., Wu, J. T. (2021). Early Transmissibility Assessment of the N501Y Mutant Strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 26 (1), 2002106. doi: 10.2807/1560-7917.ES.2020.26.1.2002106
Li, F. (2016). Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 3 (1), 237–261. doi: 10.1146/annurev-virology-110615-042301
Li, T., Huang, T., Guo, C., Wang, A., Shi, X., Mo, X., et al (2021). Genomic Variation, Origin Tracing, and Vaccine Development of SARS-CoV-2: A Systematic Review. Innovation (N Y) 2 (2), 100116. doi: 10.1016/j.xinn.2021.100116
Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., et al (2003). Angiotensin-Converting Enzyme 2 is a Functional Receptor for the SARS Coronavirus. Nature 426 (6965), 450–454. doi: 10.1038/nature02145
Lipcsey, M., Persson, B., Eriksson, O., Blom, A. M., Fromell, K., Hultström, M., et al (2021). The Outcome of Critically Ill COVID-19 Patients is Linked to Thromboinflammation Dominated by the Kallikrein/Kinin System. Front. Immunol. 12, 627579. doi: 10.3389/fimmu.2021.627579
Lopez Bernal, J., Andrews, N., Gower, C., Robertson, C., Stowe, J., Tessier, E., et al (2021). Effectiveness of the Pfizer-BioNTech and Oxford-AstraZeneca Vaccines on Covid-19 Related Symptoms, Hospital Admissions, and Mortality in Older Adults in England: Test Negative Case-Control Study. BMJ (Clinical Res. Ed.) 373, n1088. doi: 10.1136/bmj.n1088
López-Medina, E., López, P., Hurtado, I. C., Dávalos, D. M., Ramirez, O., Martínez, E., et al (2021). Effect of Ivermectin on Time to Resolution of Symptoms Among Adults With Mild COVID-19: A Randomized Clinical Trial. JAMA 325 (14), 1426–1435. doi: 10.1001/jama.2021.3071
Mahase, E. (2021). Covid-19: FDA Authorises Neutralising Antibody Bamlanivimab for Non-Admitted Patients. BMJ (Clinical Res. ed.) 371:m4362. doi: 10.1136/bmj.m4362
Mallah, S. I., Ghorab, O. K., Al-Salmi, S., Abdellatif, O. S., Tharmaratnam, T., Iskandar, M. A., et al (2021). COVID-19: Breaking Down a Global Health Crisis. Ann. Clin. Microbiol. Antimicrob. 20 (1), 35. doi: 10.1186/s12941-021-00438-7
Matta, S., Rajpal, S., Chopra, K. K., Arora, V. K. (2021). Covid-19 Vaccines and New Mutant Strains Impacting the Pandemic. Indian J. Tuberc. 68 (2), 171–173. doi: 10.1016/j.ijtb.2021.03.010
Mehta, H. B., An, H., Andersen, K. M., Mansour, O., Madhira, V., Rashidi, E. S., et al (2021). Use of Hydroxychloroquine, Remdesivir, and Dexamethasone Among Adults Hospitalized With COVID-19 in the United States : A Retrospective Cohort Study. Ann. Intern. Med. 174(10), 1395–1403. doi: 10.7326/M21-0857
Merah, F., Lydia, L. M., Allam, I., Djidjik, R. (2021). Stratégies Vaccinales Contre Le SARS CoV-2. Rev. Algerienne Allergol. 6, 8–22 https://www.asjp.cerist.dz/en/PresentationRevue/588.
Michot, J. M., Albiges, L., Chaput, N., Saada, V., Pommeret, F., Griscelli, F., et al (2020). Tocilizumab, an Anti-IL-6 Receptor Antibody, to Treat COVID-19-Related Respiratory Failure: A Case Report. Ann. Oncol. 31 (7), 961–964. doi: 10.1016/j.annonc.2020.03.300
Mitjà, O., Corbacho-Monné, M., Ubals, M., Alemany, A., Suñer, C., Tebé, C., et al (2021). A Cluster-Randomized Trial of Hydroxychloroquine for Prevention of Covid-19. N. Engl. J. Med. 384 (5), 417–427. doi: 10.1056/NEJMoa2021801
O’Dowd, A. (2021). Covid-19: Cases of Delta Variant Rise by 79%, But Rate of Growth Slows. BMJ 373:n1596. doi: 10.1136/bmj.n1596
Oldenburg, C. E., Doan, T. (2020). Azithromycin for Severe COVID-19. Lancet (London England) 396 (10256), 936–937. doi: 10.1016/S0140-6736(20)31863-8
Omarjee, L., Janin, A., Perrot, F., Laviolle, B., Meilhac, O., Mahe, G. (2020). Targeting T-Cell Senescence and Cytokine Storm With Rapamycin to Prevent Severe Progression in COVID-19. Clin. Immunol. 216:108464. doi: 10.1016/j.clim.2020.108464
Oude Munnink, B. B., Sikkema, R. S., Nieuwenhuijse, D. F., Molenaar, R. J., Munger, E., Molenkamp, R., et al (2021). Transmission of SARS-CoV-2 on Mink Farms Between Humans and Mink and Back to Humans. Science (New York N.Y.) 371 (6525), 172–177. doi: 10.1126/science.abe5901
Pang, J., Xu, F., Aondio, G., Li, Y., Fumagalli, A., Lu, M. (2021). Efficacy and Tolerability of Bevacizumab in Patients With Severe Covid-19. Nat. Commun. 12 (1), 814. doi: 10.1038/s41467-021-21085-8
Pater, A. A., Bosmeny, M. S., Barkay, C. L., Ovington, K. N., Chilamkurthy, R., Parasrampuria, M., et al (2021). Emergence and Evolution of a Prevalent New SARS-CoV-2 Variant in the United States. BioRxiv. doi: 10.1101/2021.01.11.426287
Peshimam, G. N., Farooq, U. (2021). CanSinoBIO's COVID-19 Vaccine 65.7% Effective in Global Trials, Pakistan Official Says Islamabad: Reuters. Available at: https://www.reuters.com/article/us-health-coronavirus-vaccine-pakistan-idUSKBN2A81N0.
Piroth, L., Cottenet, J., Mariet, A. S., Bonniaud, P., Blot, M., Tubert-Bitter, P., et al (2021). Comparison of the Characteristics, Morbidity, and Mortality of COVID-19 and Seasonal Influenza: A Nationwide, Population-Based Retrospective Cohort Study. Lancet Respir. Med. 9 (3), 251–259. doi: 10.1016/S2213-2600(20)30527-0
Plante, J. A., Liu, Y., Liu, J., Xia, H., Johnson, B. A., Lokugamage, K. G., et al (2021). Spike Mutation D614G Alters SARS-CoV-2 Fitness. Nature 592 (7852), 116–121. doi: 10.1038/s41586-020-2895-3
Polack, F. P., Thomas, S. J., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., et al (2020). Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 383 (27), 2603–2615. doi: 10.1056/NEJMoa2034577
Public Health England (2021). SARS-CoV-2 Variants of Concern and Variants Under Investigation in England—technical Briefing 17 (London, United Kingdom: Public Health England). Available at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/997418/Variants_of_Concern_VOC_Technical_Briefing_17.pdfpdficonexternalicon.
Rabaan, A. A., Al-Ahmed, S. H., Muhammad, J., Khan, A., Sule, A. A., Tirupathi, R., et al (2021). Role of Inflammatory Cytokines in COVID-19 Patients: A Review on Molecular Mechanisms, Immune Functions, Immunopathology and Immunomodulatory Drugs to Counter Cytokine Storm. Vaccines 9 (5):436. doi: 10.3390/vaccines9050436
Rada, M., Qusairy, Z., Massip-Salcedo, M., Macip, S. (2021). Relevance of the Bruton Tyrosine Kinase as a Target for COVID-19 Therapy Mol. Cancer. Res. 19, 4, 549–554. doi: 10.1158/1541-7786.MCR-20-0814
Rahman, M. T., Idid, S. Z. (2021). Can Zn Be a Critical Element in COVID-19 Treatment? Biol. Trace Elem. Res. 199 (2), 550–558. doi: 10.1007/s12011-020-02194-9
Rambaut, A., Loman, N., Pybus, O., Barclay, W., Barrett, J., Carabelli, A., et al (2021) Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel Set of Spike Mutations. Available at: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
Rezaei, M., Babamahmoodi, A., Marjani, M. (2020). Bruton's Tyrosine Kinase: A Promising Target for the Treatment of COVID-19. Tanaffos 19 (2), 85–88 https://pubmed.ncbi.nlm.nih.gov/33262793/.
Richardson, P., Griffin, I., Tucker, C., Smith, D., Oechsle, O., Phelan, A., et al (2020). Baricitinib as Potential Treatment for 2019-Ncov Acute Respiratory Disease. Lancet (London England) 395 (10223), e30–e31. doi: 10.1016/S0140-6736(20)30304-4
Robinson, J., Banerjee, I., Leclézio, A., Sathianm, B. (2021). COVID-19 and Mutations a Threat Level Assessment. Nepal J. Epidemiol. 11 (1), 983–987. doi: 10.3126/nje.v11i1.35659
Roschewski, M., Lionakis, M. S., Sharman, J. P., Roswarski, J., Goy, A., Monticelli, M. A., et al (2020). Inhibition of Bruton Tyrosine Kinase in Patients With Severe COVID-19. Sci. Immunol. 5 (48), eabd0110. doi: 10.1126/sciimmunol.abd0110
Roxby, P. (2020). Russian Covid Vaccine Shows Encouraging Results Moscow:(BBC News). Available at: https://www.bbc.com/news/health-54905330.
Şimşek Yavuz, S., Ünal, S. (2020). Antiviral Treatment of COVID-19. Turk. J. Med. Sci. 50 (SI-1), 611–619. doi: 10.3906/sag-2004-145
Saber-Ayad, M., Hammoudeh, S., Abu-Gharbieh, E., Hamoudi, R., Tarazi, H., Al-Tel, T. H., et al (2021). Current Status of Baricitinib as a Repurposed Therapy for COVID-19. Pharmaceuticals (Basel Switzerland) 14 (7):680. doi: 10.3390/ph14070680
Sabino, E. C., Buss, L. F., Carvalho, M., Prete, C. A., Jr., Crispim, M., Fraiji, N. A., et al (2021). Resurgence of COVID-19 in Manaus, Brazil, Despite High Seroprevalence. Lancet (London England) 397 (10273), 452–455. doi: 10.1016/S0140-6736(21)00183-5
Sadoff, J., Le Gars, M., Shukarev, G., Heerwegh, D., Truyers, C., de Groot, A. M., et al (2021). Interim Results of a Phase 1-2a Trial of Ad26.COV2.S Covid-19 Vaccine. N. Engl. J. Med. 384 (19), 1824–1835. doi: 10.1056/NEJMoa2034201
Salvatore, M., Bhattacharyya, R., Purkayastha, S., Zimmermann, L., Ray, D., Hazra, A., et al (2021). Resurgence of SARS-CoV-2 in India: Potential Role of the B.1.617.2 (Delta) Variant and Delayed Interventions. MedRxiv. doi: 10.1101/2021.06.23.21259405
Sarkar, R., Mitra, S., Chandra, P., Saha, P., Banerjee, A., Dutta, S., et al (2021b). Comprehensive Analysis of Genomic Diversity of SARS-CoV-2 in Different Geographic Regions of India: An Endeavour to Classify Indian SARS-CoV-2 Strains on the Basis of Co-Existing Mutations. Arch. Virol. 166 (3), 801–812. doi: 10.1007/s00705-020-04911-0
Sarkar, R., Saha, R., Mallick, P., Sharma, R., Kaur, A., Dutta, S., et al (2021a). Emergence of a New SARS-CoV-2 Variant From GR Clade With a Novel S Glycoprotein Mutation V1230L in West Bengal, India. MedRxiv. doi: 10.1101/2021.05.24.21257705
Shang, L., Zhao, J., Hu, Y., Du, R., Cao, B. (2020). On the Use of Corticosteroids for 2019-nCoV Pneumonia. Lancet (London England) 395 (10225), 683–684. doi: 10.1016/S0140-6736(20)30361-5
Shrotri, M., Krutikov, M., Palmer, T., Giddings, R., Azmi, B., Subbarao, S., et al (2021). Vaccine Effectiveness of the First Dose of ChAdOx1 Ncov-19 and BNT162b2 Against SARS-CoV-2 Infection in Residents of Long-Term Care Facilities (VIVALDI Study). Lancet Infect. Dis. 21(11), 1529–1538. doi: 10.1101/2021
Sood, S., Aggarwal, V., Aggarwal, D., Upadhyay, S. K., Sak, K., Tuli, H. S., et al (2020). COVID-19 Pandemic: From Molecular Biology, Pathogenesis, Detection, and Treatment to Global Societal Impact. Curr. Pharmacol. Rep. 1, 16. doi: 10.1007/s40495-020-00229-2
Stasi, C., Fallani, S., Voller, F., Silvestri, C. (2020). Treatment for COVID-19: An Overview. Eur. J. Pharmacol. 889, 173644. doi: 10.1016/j.ejphar.2020.173644
Stebbing, J., Phelan, A., Griffin, I., Tucker, C., Oechsle, O., Smith, D., et al (2020). COVID-19: Combining Antiviral and Anti-Inflammatory Treatments. Lancet Infect. Dis. 20 (4), 400–402. doi: 10.1016/S1473-3099(20)30132-8
Lopez Bernal, J., Andrews, N., Gower, C., Gallagher, E., Simmons, L., Thelwall, R., et al (2021). Effectiveness of COVID-19 Vaccines Against Hospital Admission With the Delta (B.1.617.2) Variant N Engl, J Med 385(7):585–94 . doi: 10.1056/NEJMoa2108891
Supasa, P., Zhou, D., Dejnirattisai, W., Liu, C., Mentzer, A. J., Ginn, H. M., et al (2021). Reduced Neutralization of SARS-CoV-2 B.1.1.7 Variant by Convalescent and Vaccine Sera. Cell 184 (8), 2201–2211:e7. doi: 10.1016/j.cell.2021.02.033
Tang, X., Wu, C., Li, X., Song, Y., Yao, X., Wu, X., et al (2020). On the Origin and Continuing Evolution of SARS-CoV-2. Natl. Sci. Rev. 7 (6), 1012–1023. doi: 10.1093/nsr/nwaa036
Tardif, J. C., Bouabdallaoui, N., L’Allier, P. L., Gaudet, D., Shah, B., Pillinger, M. H., et al (2021). Efficacy of Colchicine in non-Hospitalized Patients With COVID-19. Medrxiv. doi: 10.1101/2021.01.26.21250494v1
Tegally, H., Wilkinson, E., Lessells, R. J., Giandhari, J., Pillay, S., Msomi, N., et al (2021). Sixteen Novel Lineages of SARS-CoV-2 in South Africa. Nat. Med. 27 (3), 440–446. doi: 10.1038/s41591-021-01255-3
Tomkins-Tinch, C. H., Silbert, J., DeRuff, K. C., Siddle, K. J., Gladden-Young, A., Bronson, A., et al (2021) Detection of the Recurrent Substitution Q677H in the Spike Protein of SARS-CoV-2 in Cases Descended From the Lineage B.1.429. Available at: https://virological.org/c/novel-2019-coronavirus/ncov-2019-genomic-epidemiology/36.
Tortajada, C., Añón, S., Ortiz, M. M., Andreu-Ballester, J. C., Flores, J. (2021). Interferon β-1b for Patients With Moderate to Severe COVID-19 in the Inflammatory Phase of the Disease. J. Med. Virol. 93 (7), 4102–4107. doi: 10.1002/jmv.26976
Tuli, H. S., Sood, S., Kaur, J., Kumar, P., Seth, P., Punia, S., et al (2021). Mechanistic Insight Into Anti-COVID-19 Drugs: Recent Trends and Advancements. 3 Biotech 11, 110. doi: 10.1007/s13205-021-02644-8
Upadhyay, S. K., Dan, S., Girdhar, M., Rastogi, K. (2021). Recent Advancement in SARS-CoV-2 Diagnosis, Treatment, and Vaccine Formulation: A New Paradigm of Nanotechnology in Strategic Combating of COVID-19 Pandemic. Curr. Pharmacol. Rep., 1–14. doi: 10.1007/s40495-021-00250-z
Upadhyay, S. K., Singh, R., Babita, Kumar, G., Singh, G. (2020b). The Outbreak and Challenges of Novel Coronavirus (COVID-19): The Global Pandemic Emergency of Early 2K20 and Indian Scenario. Int. J. Biol. Pharm. Allied Sci. 9 (5), 1173–1199. doi: 10.31032/IJBPAS/2020/9.5.5126
Upadhyay, S. K., Singh, R., Singh, M., Kumar, V., Yadav, M., Aggarwal, D. (2020a). COVID-19 in Republic of India: A Report on Situation and Precautionary Strategies to Global Pandemic. Bull. Environ. Pharmacol. Life Sci. 9 (6), 39–48 http://bepls.com/bepls_may2020/7.pdf.
Urwyler, P., Moser, S., Charitos, P., Heijnen, I., Rudin, M., Sommer, G., et al (2020). Treatment of COVID-19 With Conestat Alfa, a Regulator of the Complement, Contact Activation and Kallikrein-Kinin System. Front. Immunol. 11:2072. doi: 10.3389/fimmu.2020.02072
Verderese, J. P., Stepanova, M., Lam, B., Racila, A., Kolacevski, A., Allen, D., et al (2021). Neutralizing Monoclonal Antibody Treatment Reduces Hospitalization for Mild and Moderate COVID-19: A Real-World Experience. Clin. Infect. Dis. ciab579. doi: 10.1093/cid/ciab579
Vetter, P., Kaiser, L., Calmy, A., Agoritsas, T., Huttnerm, A. (2020). Dexamethasone and Remdesivir: Finding Method in the COVID-19 Madness. Lancet Microbe 1 (8), e309–e310. doi: 10.1016/S2666-5247(20)30173-7
Volz, E., Hill, V., McCrone, J. T., Price, A., Jorgensen, D., O'Toole, Á., et al (2021). Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 184 (1), 64–75.e11. doi: 10.1016/j.cell.2020.11.020
Voysey, M., Clemens, S., Madhi, S. A., Weckx, L. Y., Folegatti, P. M., Aley, P. K., et al (2021). Safety and Efficacy of the ChAdOx1 Ncov-19 Vaccine (AZD1222) Against SARS-CoV-2: An Interim Analysis of Four Randomised Controlled Trials in Brazil, South Africa, and the UK. Lancet (London England) 397 (10269), 99–111. doi: 10.1016/S0140-6736(20)32661-1
Vudathaneni, V. K. P., Nadella, S. B., Lanke, R. B., Boyapati, R. (2021). Coronavirus Disease and Cardiovascular Disease: A Literature Review. J. Clin. Transl. Res. 7 (2), 156–162. doi: 10.18053/jctres.07.202102.00
Wan, Y., Shang, J., Graham, R., Baric, R. S., Li, F. (2020). Receptor Recognition by the Novel Coronavirus From Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 94 (7), e00127–e00120. doi: 10.1128/JVI.00127-20
Weinreich, D. M., Sivapalasingam, S., Norton, T., Ali, S., Gao, H., Bhore, R., et al (2021). REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients With Covid-19. N. Engl. J. Med. 384 (3), 238–251. doi: 10.1056/NEJMoa2035002
West, A. P., Barnes, C. O., Yang, Z., Bjorkman, P. J. (2021a). SARS-CoV-2 Lineage B.1.526 Emerging in the New York Region Detected by Software Utility Created to Query the Spike Mutational Landscape. BioRxiv. doi: 10.1101/2021.02.14.431043
West, A. P., Jr., Wertheim, J. O., Wang, J. C., Vasylyeva, T. I., Havens, J. L., Chowdhury, M. A., et al (2021b). Detection and Characterization of the SARS-CoV-2 Lineage B.1.526 in New York. Nat. Commun. 12 (1), 4886. doi: 10.1038/s41467-021-25168-4
WHO (2021a) Tracking SARS-CoV-2 Variants. Available at: https://www.who.int/activities/tracking-SARS-CoV-2-variants.
WHO (2021b) Weekly Epidemiological Update on COVID-19. Available at: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19—11- may-2021.
Williams, S. V., Vusirikala, A., Ladhani, S. N., Fernandez, R. O. E., Iyanger, N., Aiano, F., et al (2021). An Outbreak Caused by the SARS-CoV-2 Delta (B.1.617.2) Variant in a Care Home After Partial Vaccination With a Single Dose of the COVID-19 Vaccine Vaxzevria, London, England, April 2021. Euro Surveill. 26 (27), 2100626. doi: 10.2807/1560-7917.ES.2021.26.27.2100626
Yadav, R., Dhiman, U., Parihar, R. D., Upadhyay, S. K. (2021a). Virtual Screening of Potential Drug Molecules Against Covid-19 Targets: A Drug Repurposing Approach. Lett. Appl. NanoBioScience 11 (1), 2965–2980. doi: 10.33263/LIANBS111.29652980
Yadav, R., Parihar, R. D., Dhiman, U., Dhamija, P., Upadhyay, S. K., Imran, M., et al (2021b). Docking of FDA Approved Drugs Targeting NSP-16, N-Protein and Main Protease of SARS-CoV-2 as Dual Inhibitors. Biointerface Res. Appl. Chem. 11 (3), 9848–9861. doi: 10.33263/BRIAC113.98489861
Yan, Y., Pang, Y., Lyu, Z., Wang, R., Wu, X., You, C., et al (2021). The COVID-19 Vaccines: Recent Development, Challenges and Prospects. Vaccines 9 (4), 349. doi: 10.3390/vaccines9040349
Young, B., Tan, T. T., Leo, Y. S. (2021). The Place for Remdesivir in COVID-19 Treatment. Lancet Infect. Dis. 21 (1), 20–21. doi: 10.1016/S1473-3099(20)30911-7
Yuen, C. K., Lam, J. Y., Wong, W. M., Mak, L. F., Wang, X., Chu, H., et al (2020). SARS-CoV-2 Nsp13, Nsp14, Nsp15 and Orf6 Function as Potent Interferon Antagonists. Emerg. Microbes Infect. 9 (1), 1418–1428. doi: 10.1080/22221751.2020.1780953
Zhang, W., Davis, B. D., Chen, S. S., Sincuir Martinez, J. M., Plummer, J. T., Vail, E. (2021). Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA 325 (13), 1324–1326. doi: 10.1001/jama.2021.1612
Zhou, D., Dejnirattisai, W., Supasa, P., Liu, C., Mentzer, A. J., Ginn, H. M., et al (2021). Evidence of Escape of SARS-CoV-2 Variant B.1.351 From Natural and Vaccine-Induced Sera. Cell 184 (9), 2348–2361.e6. doi: 10.1016/j.cell.2021.02.037
Zhou, W., Wang, W. (2021). Fast-Spreading SARS-CoV-2 Variants: Challenges to and New Design Strategies of COVID-19 Vaccines. Signal Transduct. Target. Ther. 6, 226. doi: 10.1038/s41392-021-00644-x
Keywords: COVID pandemic, variants, molecular evolution, therapeutics, vaccination
Citation: Tuli HS, Sak K, Aggarwal P, Iqubal A, Upadhaya SK, Kaur J, Kaur G and Aggarwal D (2021) Molecular Evolution of Severe Acute Respiratory Syndrome Coronavirus 2: Hazardous and More Hazardous Strains Behind the Coronavirus Disease 2019 Pandemic and Their Targeting by Drugs and Vaccines. Front. Cell. Infect. Microbiol. 11:763687. doi: 10.3389/fcimb.2021.763687
Received: 24 August 2021; Accepted: 19 October 2021;
Published: 14 December 2021.
Edited by:
Rodolfo García-Contreras, National Autonomous University of Mexico, MexicoReviewed by:
Talha Bin Emran, Begum Gulchemonara Trust University, BangladeshCopyright © 2021 Tuli, Sak, Aggarwal, Iqubal, Upadhaya, Kaur, Kaur and Aggarwal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hardeep Singh Tuli, aGFyZGVlcC5iaW90ZWNoQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.