
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 13 December 2021
Sec. Bacteria and Host
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.761335
This article is part of the Research TopicListeria: Environmental Adaption, Infection and Host InteractionsView all 7 articles
 Changyong Cheng†
Changyong Cheng† Feng Liu†
Feng Liu† Haobo Jin
Haobo Jin Xiangfei Xu
Xiangfei Xu Jiali Xu
Jiali Xu Simin Deng
Simin Deng Jing Xia
Jing Xia Yue Han
Yue Han Lei Lei
Lei Lei Xian Zhang
Xian Zhang Houhui Song*
Houhui Song*Listeria monocytogenes is more heat-resistant than most other non-spore-forming foodborne pathogens, posing a severe threat to food safety and human health, particularly during chilled food processing. The DegU orphan response regulator is known to control heat resistance in L. monocytogenes; however, the underlying regulatory mechanism is poorly understood. Here, we show that DegU contributes to L. monocytogenes exponential growth under mild heat-shock stress. We further demonstrate that DegU directly senses heat stress through autoregulation and upregulates the hrcA-grpE-dnaK-dnaJ operon, leading to increased production of heat-shock proteins. We also show that DegU can directly regulate the expression of the hrcA-grpE-dnaK-dnaJ operon. In conclusion, our results shed light on the regulatory mechanisms underlying how DegU directly activates the hrcA-grpE-dnaK-dnaJ operon, thereby regulating heat resistance in L. monocytogenes.
Listeria monocytogenes is a facultative intracellular Gram-positive bacterium that causes listeriosis, which is associated with a mortality rate of 20%-30% in immunocompromised individuals (de Noordhout et al., 2014; Pasechnek et al., 2020). This bacterial pathogen is widespread in the environment and can survive under a wide range of stress conditions, such as low and high temperatures, dryness, low pH, and high salinity, which allow it to persist in food manufacturing sites for several years (Ming and Daeschel, 1993; Keto-Timonen et al., 2007; Camargo et al., 2017). L. monocytogenes can reportedly grow under temperatures as high as 45°C and is more heat resistant than many other non-spore-forming foodborne pathogens (Lin et al., 2012; Pontinen et al., 2017; Ballom et al., 2020). Although the general heat stress properties of L. monocytogenes have been described (Nair et al., 2000; van der Veen et al., 2007; Soni et al., 2011), further investigations are required to reveal the response mechanisms triggered by heat stress in the pathogen.
Pathogenic bacteria rely on two-component systems (TCSs) to sense chemical and physical changes in the environment and respond accordingly (Lemmer et al., 2020; Salvail and Groisman, 2020). Canonical TCSs comprise a signal-sensing histidine kinase and a cytoplasmic response regulator. In these systems, the histidine kinase undergoes autophosphorylation under certain stimuli and activates a cognate response regulator via the transfer of the phosphoryl group (Murret-Labarthe et al., 2020; Rapun-Araiz et al., 2020; Tiwari et al., 2020). Genome-wide analysis has revealed that L. monocytogenes encodes 15 paired two-component systems and one orphan response regulator (Williams et al., 2005).
The DegS/DegU two-component system of B. subtilis was one of the first described in Gram-positive bacteria (Tokunaga et al., 1994; Mader et al., 2002; Cairns et al., 2015); however, unlike B. subtilis, L. monocytogenes expresses only DegU, the cognate response regulator, and not DegS, the sensor histidine kinase. Hence, DegU is considered an orphan response regulator in this bacterium (Mauder et al., 2008). Previous studies have shown that DegU is a pleiotropic regulator in L. monocytogenes, and is required for biofilm formation, chemotaxis, motility, and growth at high temperatures (Gueriri et al., 2008a; Gueriri et al., 2008b; Mauder et al., 2008). Although DegU has been reported to play a crucial role in resistance to heat stress in L. monocytogenes, how DegU allows it to respond rapidly to rising temperatures is unknown. Here, the principal objective of this study was to investigate the mechanism underlying the DegU-mediated heat-stress tolerance in this bacterium.
L. monocytogenes strain EGD-e was used as the reference strain. All Escherichia coli strains were grown at 37°C in LB medium (Oxoid Ltd, Basingstoke, United Kingdom), and all L. monocytogenes strains were grown at 37°C in BHI broth (Oxoid Ltd). E. coli DH5α was used for transformation, and E. coli BL21 was used for protein expression. When needed, the following antibiotics were added to E. coli or L. monocytogenes cultures: 100 µg/mL ampicillin, 10 µg/mL chloramphenicol, or 50 µg/mL kanamycin. Primers were purchased from Tsingke (Tsingke Biotechnology Co., Ltd, Hangzhou, China), and the sequences are shown in Table S1.
The ΔdegU mutant was constructed by a two-step allelic exchange procedure using the pKSV7 shuttle plasmid as previously described (Cheng et al., 2021). The degU complementation strains were generated using the integrative plasmid pIMK2 as previously described (Zhang et al., 2020). The targeted degU gene was cloned into pIMK2 via a one-step cloning method and then electroporated into competent L. monocytogenes cells. The mutant and complement strains were verified by PCR and DNA sequencing.
A degU-lacZ fusions containing the promoter region of degU and the lacZ gene, was cloned into the Sac I and BamH I sites of the plasmid pIMK2. Then, the recombinant plasmid was introduced into the wild-type EGD-e and the ΔdegU mutant strain. All strains were grown overnight in BHI broth at 37°C, diluted 1:1,000 in fresh BHI broth, and grown at 43°C to an OD600 of 0.6. The collected culture was assayed for β-Galactosidase activity using a β-galactosidase (β-GAL) Activity Assay Kit(Micromethod; Sangon Biotech) according to the manufacturer’s specification.
The wild-type, ΔdegU mutant, and complement strains were grown in BHI broth overnight, diluted 1:1,000 in fresh BHI broth, and incubated at 43°C to an OD600 of 0.6. Total bacterial RNA was extracted using the Bacteria Total RNA Isolation Kit (Sangon Biotech, China) and reverse-transcribed into cDNA using reverse transcriptase from TOYOBO. According to the manufacturer’s specifications, real-time qPCR was performed using an Mx3000P qPCR system (Stratagene-Agilent) and SYBR qPCR Mix (TOYOBO). All expression results were normalized relative to the housekeeping gene rpoB. Relative transcription levels were determined using the 2−ΔΔCt method. RT-PCR was performed as previously described (Liu et al., 2018). The PCR products were electrophoresed on a 1% agarose gel and photographed using a SmartView Pro 2400 system (Major Science, USA).
The degU gene was amplified from genomic DNA of L. monocytogenes EGD-e with the primer pair PdegU-F and PdegU-R and cloned into the Nde I and Xho I sites of the expression vector pET30a. Then, the recombinant plasmid pET30a-degU was transformed into E. coli BL21 to express His6-DegU protein. The expression was induced by isopropyl-β-D-thiogalactopyranoside (0.5 mM) at 16°C for 5 h and purified by Ni-nitrilotriacetic acid (Ni-NTA) resin affinity chromatography. The purified proteins were confirmed by running the SDS-PAGE and stored at -80°C until use.
The recombinant protein DegU was obtained and phosphorylated according to previously described procedures (Goodman et al., 2020). DNA probes were purified with a Gel Extraction Kit (TIANGEN, China) and were labeled using the Biotin Labeling Kit for the EMSA (Beyotime, China). EMSA was performed with the Chemiluminescent EMSA Kit (Beyotime). Samples were analyzed by 4% non-denaturing polyacrylamide gel electrophoresis in 0.5 × TBE buffer. The gel was then transferred to a nylon membrane (Beyotime) followed by UV crosslinking. The bands were detected using the Chemiluminescent EMSA Kit (Beyotime).
DNase I footprinting experiments were carried out as previously described (Li et al., 2018). To prepare the fluorescent FAM-labeled probes, the promoter region of hrcA was PCR amplified using a 2× KOD One PCR Master Mix (TOYOBO) from the plasmid T-hcrA using primers containing 6-FAM at the 5′ end. The labeled probes (300 ng) were then mixed with purified DegU in a 40-µL reaction volume at 25°C for 30 min. Subsequently, 0.015 units of DNase I (Promega) and the reaction buffer were added, followed by incubation for 1 min at 37°C. The reaction was terminated by adding 140 µL of DNase I stop solution. Digested DNA samples were extracted with phenol-chloroform, and pellets containing DNA were resuspended in 30 µL of water. The results were analyzed using Peak Scanner software v1.0 (Applied Biosystems).
Data were analyzed using GraphPad Prism version 5.0 (GraphPad Software, La Jolla, CA, USA) using two-tailed Student’s t-tests and are presented as means ± SD.
To verify whether DegU plays an important role in the heat tolerance of L. monocytogenes, we generated the in-frame deletion mutant strain ΔdegU and the complemented strain CΔdegU. When exposed to heat stress (43°C), the mutant strain ΔdegU showed a significant growth defect on BHI agar plates compared with the wild-type EGD-e and CΔdegU strains (Figure 1A). The promoter activity of PdegU-lacZ in the WT and ΔdegU strains had no obvious change under heat stress (Figure S3), which indicated that DegU protein level was not elevated under heat stress. Previous studies have demonstrated that the Pta-AckA pathway plays a role in DegU protein phosphorylation (Gueriri et al., 2008a). The RT-qPCR results showed that the mRNA level of the pta and ackA genes, which are responsible for DegU activation, were significantly elevated under heat stress in the WT strain (Figure 1B). Collectively, these results strongly indicated that DegU contributes to the heat resistance of L. monocytogenes.
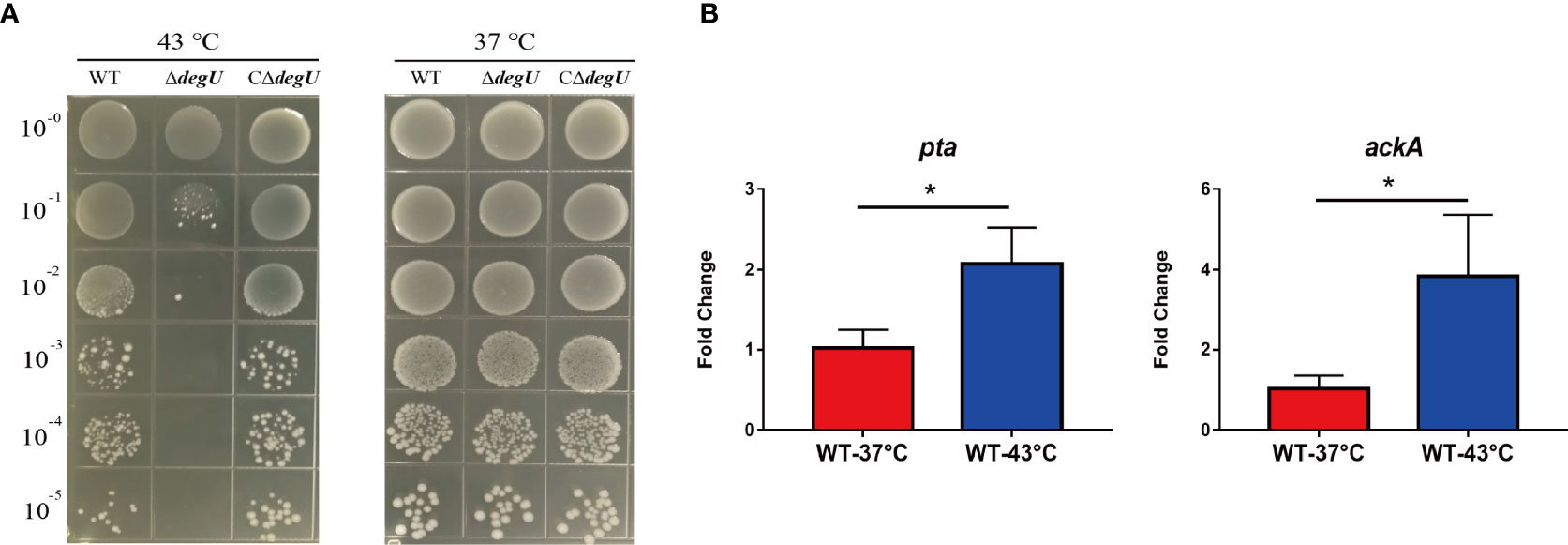
Figure 1 DegU modulates Listeria monocytogenes responses to heat stress. (A) Bacterial growth under heat stress. The wild-type (WT) EGD-e and the ΔdegU mutant L. monocytogenes were spotted on BHI agar plates using 10-fold serial dilutions and incubated at 37°C and 43°C. (B) The relative expression levels of the pta and ackA genes in response to heat stress. WT L. monocytogenes EGD-e cells were grown at 43°C. Data are presented as the means ± S.D. of three independent experiments. *p < 0.05.
Many heat-shock-related genes have been previously identified, including hrcA, grpE, dnaK dnaJ, htpX, and groEL (Diamant and Goloubinoff, 1998; Arsene et al., 2000; van der Veen and Abee, 2010). In this study, we examined the link between DegU and the transcript levels of these genes using RT-qPCR. Under heat stress, the transcript levels of the hrcA, grpE, dnaK, and dnaJ genes were significantly lower in the ΔdegU mutant strain than in the wild-type EGD-e and complemented strains (Figure 2A), and the htpX and groEL genes were not changed (data not show). These four genes are adjacent to the L. monocytogenes chromosome (Figure 2B). To verify if they are co-transcribed as a polycistronic mRNA under heat stress, we performed RT-PCR across the hrcA-grpE, grpE-dnaK, and dnaK-dnaJ junctions. The results confirmed that the hrcA, grpE, dnaK, and dnaJ genes comprise an operon (Figure 2B). However, it should be recognized that RT-PCR is not sufficient to conclude that these genes constitute a complete operon, which may be short overlapping transcripts generated from internal promoters and terminators. These findings indicated that DegU is essential for the transcriptional regulation of the hrcA-grpE-dnaK-dnaJ operon.
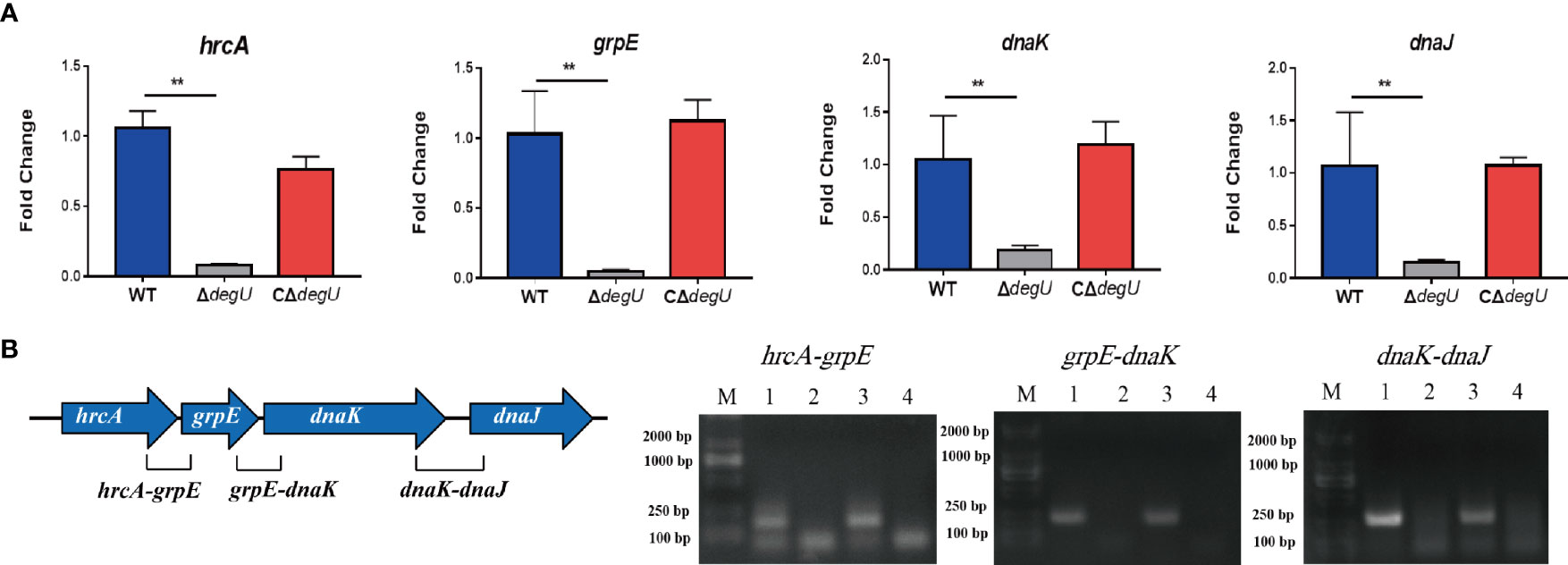
Figure 2 hrcA-dnaJ transcription is activated by DegU. (A) The relative mRNA levels of hrcA, grpE, dnaK, and dnaJ as determined by RT-qPCR in WT, ΔdegU, and CΔdegU strains at 43°C. (B) Schematic diagram showing the gene order. Co-expression results confirmed that the hrcA, grpE, dnaK, and dnaJ genes form an operon under heat-stress conditions. Lane 1, cDNA; lane 2, total RNA; lane 3, genomic DNA; and lane 4, no-template control. Data are presented as the means ± S.D. of three independent experiments. **p < 0.01.
To further investigate the mechanism involved in the DegU-mediated regulation of the hrcA-grpE-dnaK-dnaJ operon, we determined the binding site for DegU in the promoter regions of hrcA and dnaJ using EMSA, with the degU and groES promoters respectively serving as a positive or negative control for DegU binding. Marked band shifts were observed with the hrcA promoter region but not with that of dnaJ (Figure 3A). These EMSA data showed that DegU bound to the promoter region of hrcA but not to that of dnaJ. To map the precise binding sequence of DegU, a DNase I footprinting assay was performed using FAM-labeled probes. As shown in Figure 3B, a 56-bp sequence (3′-AACCGCACTATTTGACCTATTTTGACCAAACAATCCTACTTTAGTCTGAAATCGAG-5′) appeared to be protected from DNase I digestion by DegU binding. To identify the minimum sequence required for DegU binding, the fragment of the hrcA promoter region used for EMSA was divided into segments so that the specific binding site was confined within the remaining 50 bp (Figure 3C).
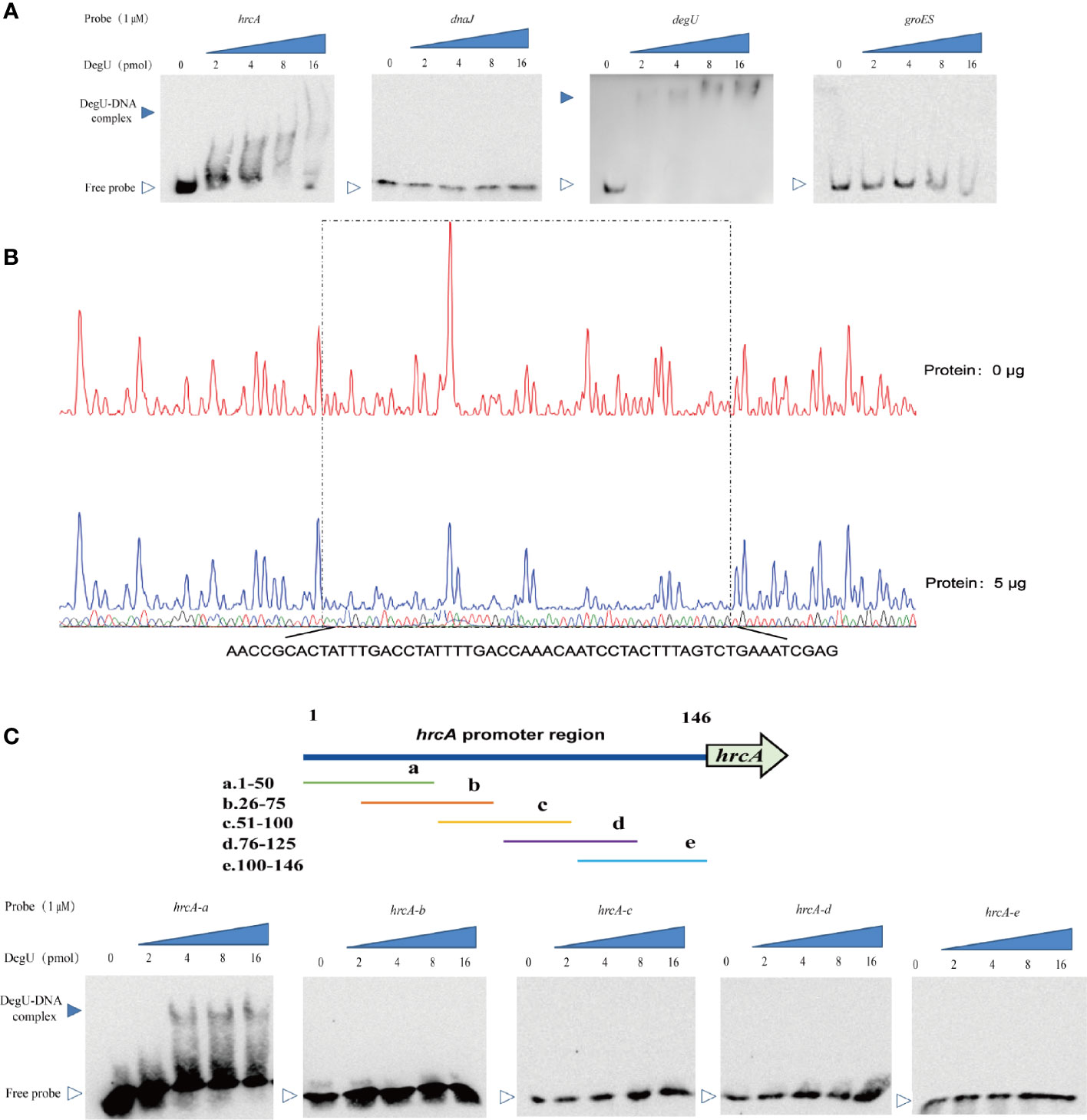
Figure 3 Identification of DegU binding sites in the hrcA promoter. (A) Electrophoretic mobility shift assay (EMSA) analysis of the direct binding of DegU-P to the hrcA, dnaJ, degU (positive control), and groES (negative control) promoter regions. (B) DNase I footprinting assay analysis of DegU-P binding to the hrcA promoter. FAM-labeled hrcA probes (300 ng) were used for the binding reactions in the absence (upper panel) or presence of 5 µg (lower panel) of DegU. The nucleotide sequences protected by DegU binding are shown below the electropherogram. (C) Electrophoretic mobility shift assay (EMSA) analysis of the binding of DegU-P to various truncated forms of the hrcA promoter. A total of six probes (left panel) were used to identify the DegU binding site in the hrcA promoter region by EMSA (right panel). Each experiment was performed three times, and the representative results were shown.
According to previous studies, the TSS (designated as +1) of hrcA was found to be located 45 bp upstream of its start codon and designated as C (Figure 4) (Wurtzel et al., 2012). In addition, analysis of the hrcA promoter region revealed a putative −10 AATTTACCA box and a putative −35 AGTCAA box respectively located at 8 bp and 31 bp downstream of the TSS (Figure 4). Furthermore, the specific DegU binding sequence was mapped from 52 to 101 bp upstream from the hrcA TSS (Figure 4).
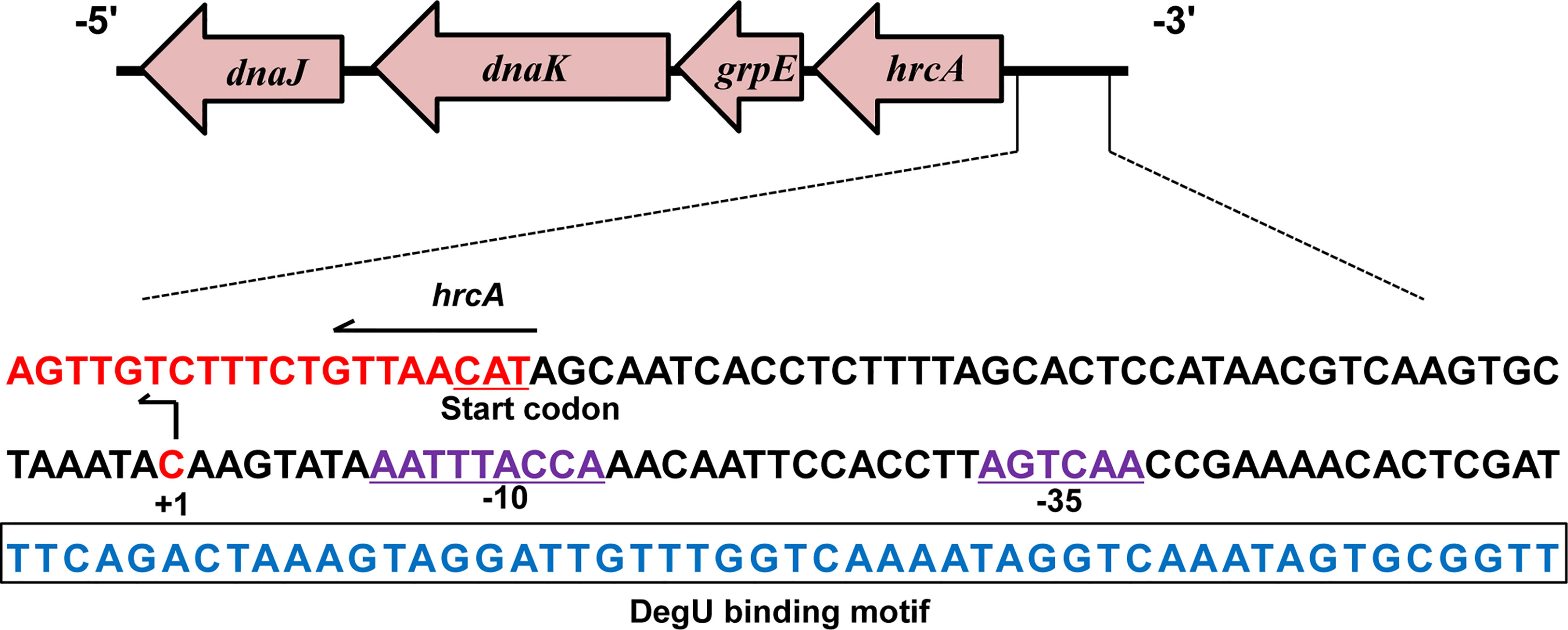
Figure 4 Characterizing the DegU Binding Site. DNA sequences of the hrcA promoter region. The DegU-binding site is shown in blue nucleotides boxed in black. The -35 and -10 regions are underlined and shown in purple. The TSS is denoted by +1 (bent arrow). The start codon of hrcA is shown in red. In panels A, experiment was performed at least three times and representative result is shown.
The DegS/DegU system is a pleiotropic TCS of B. subtilis involved in controlling many biological processes, such as chemotaxis, motility, and degradative enzyme production (Gupta and Rao, 2014). In L. monocytogenes, DegU is considered an orphan response regulator as this bacterium lacks DegS, the sensor histidine kinase (Gueriri et al., 2008a). Studies have previously shown that DegU is required for L. monocytogenes to grow in RPMI 1640 synthetic medium and BHI broth at 44°C (Gueriri et al., 2008b). In our study, experimental mutation of the degU gene in this pathogen inhibited its growth on BHI agar at 43°C, suggesting that DegU plays a crucial role in heat resistance in L. monocytogenes.
In L. monocytogenes LM1009, the deletion of the pta and ackA genes completely blocked acetyl phosphate synthesis, suggesting that Pta and AckA are essential for the synthesis of acetyl phosphate which plays an important role in modulating DegU activity in L. monocytogenes (Gueriri et al., 2008a). So, the mRNA level of the pta and ackA genes elevated under heat stress could enhance DegU activity. That the degU mutant could not respond to heat stress suggested that DegU is essential for inducing the transcription of heat-shock proteins in L. monocytogenes. To elucidate the underlying regulatory mechanisms, several differentially expressed heat-shock-related genes were selected for RT-qPCR analysis of the transcriptional changes induced by heat stress (Kornitzer et al., 1991; Hanawa et al., 2000; Cardoso et al., 2010; Somolinos et al., 2010). The results suggested that heat-shock-related genes such as hrcA, grpE, dnaK, and dnaJ, well-known to play important roles in response to heat shock, were under the control of DegU in L. monocytogenes.
As the fact is that the class I heat-shock response is activated under heat shock and is essential for prokaryotic cells surviving in environmental stresses. Previous studies have shown that HrcA is a transcription repressor for the class I heat-shock response; GrpE, DnaJ, and DnaK are the class I heat-shock response chaperone proteins (van der Veen and Abee, 2010). DnaK can bind denatured proteins and assists the refolding of denatured polypeptides into active proteins (Hartl, 1996; Pierpaoli et al., 1997). DnaJ and GrpE can increase the rate of protein folding and release from DnaK through the transfer of non-native proteins to DnaK (Liberek et al., 1991).
As previously reported, the hrcA-grpE-dnaK-dnaJ operon can be transcribed from various sites (Hanawa et al., 2000). Sequence analysis led to the identification of the promoter sequence and two transcriptional initiation sites, one upstream of hrcA and the other upstream of dnaJ, which corresponded to the independent expression of the dnaJ gene (Hanawa et al., 2000). Interestingly, RT-PCR analysis showed that hrcA, grpE, dnaK, and dnaJ were co-transcribed as a single transcript from the transcriptional initiation site of the hrcA gene under heat stress conditions. EMSA and DNase I footprinting indicated that DegU directly interacted with a 50-bp sequence in the hrcA promoter region but did not bind to the dnaJ promoter.
In conclusion, for the first time, we have revealed the regulatory mechanisms associated with the orphan response regulator DegU in the heat resistance of L. monocytogenes. The findings indicated that DegU contributes to regulating the expression of heat-shock-related genes via a complicated regulatory network involving the hrcA-grpE-dnaK-dnaJ operon (Figure 5). Many stress proteins are known to be essential for the survival of L. monocytogenes, both in the external environment and inside the host (Hu et al., 2007; Seifart Gomes et al., 2011; Zhang et al., 2013; Curtis et al., 2017). However, further research is needed to better understand the mechanisms underlying the signal transduction pathways employed by L. monocytogenes during environmental adaptation and host infection.
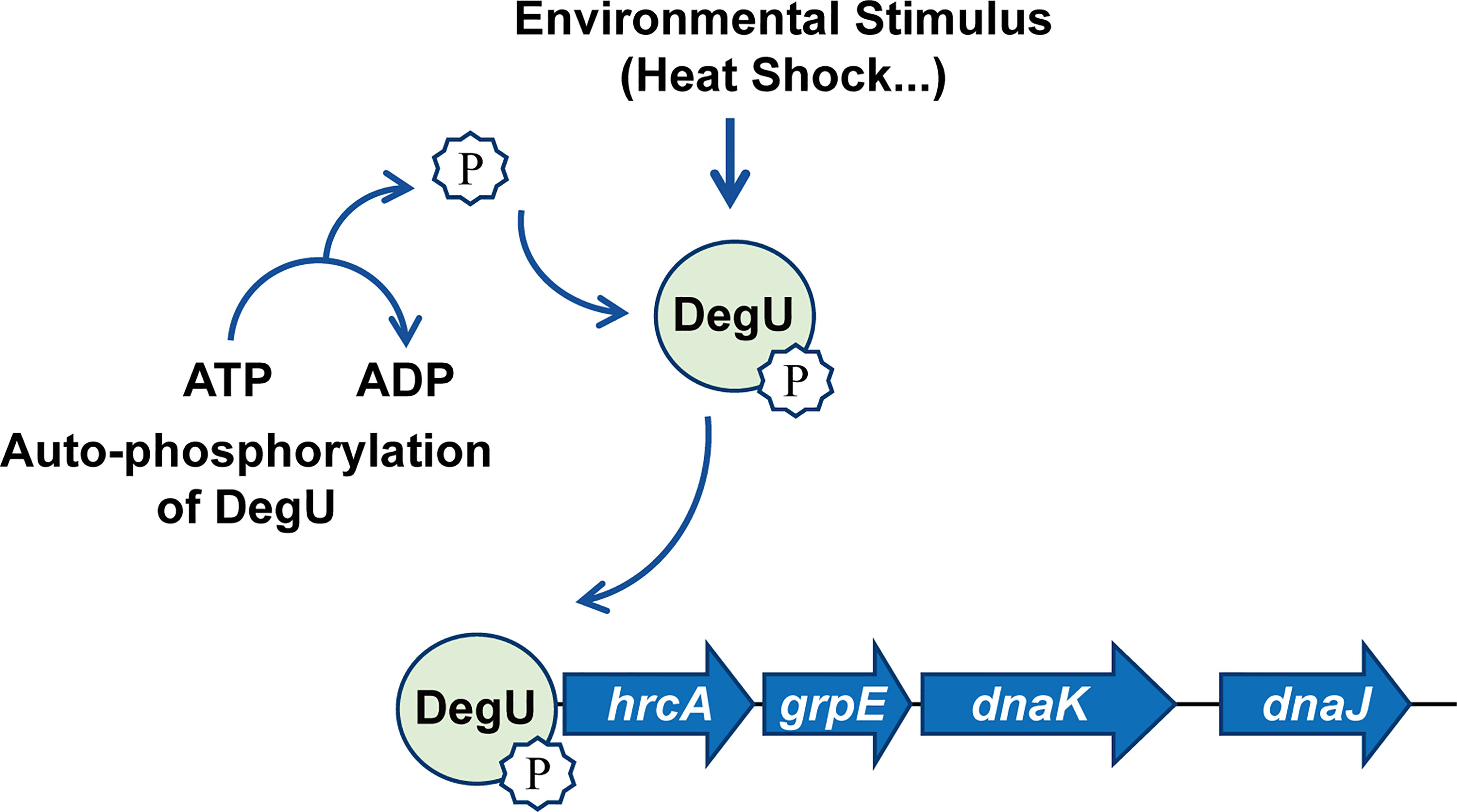
Figure 5 Model illustrating that DegU mediates heat resistance in Listeria monocytogenes. Under heat stress, DegU responds by undergoing autophosphorylation and directly activating the transcription of the hrcA heat-shock operon.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
CC and HS conceived and designed the experiments. CC, FL, HJ, XX, JXu, YH, and SD performed the experiments. CC, FL, JXi, YH, LL, and XZ analyzed the data. CC, HS and FL wrote the paper. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31872620, 31770040, 31972648, 32172849, and 32002358), the Fundamental Research Funds for the Provincial Universities of Zhejiang (2020KJ004), and the Natural Science Foundation of Zhejiang Province (LZ19C180001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.761335/full#supplementary-material
Arsene, F., Tomoyasu, T., Bukau, B. (2000). The Heat Shock Response of Escherichia Coli. Int. J. Food Microbiol. 55 (1-3), 3–9. doi: 10.1016/s0168-1605(00)00206-3
Ballom, K. F., Tsai, H. C., Taylor, M., Tang, J., Zhu, M. J. (2020). Stability of Listeria Monocytogenes in Non-Fat Dry Milk Powder During Isothermal Treatment and Storage. Food Microbiol. 87, 103376. doi: 10.1016/j.fm.2019.103376
Cairns, L. S., Martyn, J. E., Bromley, K., Stanley-Wall, N. R. (2015). An Alternate Route to Phosphorylating Degu of Bacillus Subtilis Using Acetyl Phosphate. BMC Microbiol. 15, 78. doi: 10.1186/s12866-015-0410-z
Camargo, A. C., Woodward, J. J., Call, D. R., Nero, L. A. (2017). Listeria Monocytogenes in Food-Processing Facilities, Food Contamination, and Human Listeriosis: The Brazilian Scenario. Foodborne Pathog. Dis. 14 (11), 623–636. doi: 10.1089/fpd.2016.2274
Cardoso, K., Gandra, R. F., Wisniewski, E. S., Osaku, C. A., Kadowaki, M. K., Felipach-Neto, V., et al. (2010). Dnak and Groel Are Induced in Response to Antibiotic and Heat Shock in Acinetobacter Baumannii. J. Med. Microbiol. 59 (Pt 9), 1061–1068. doi: 10.1099/jmm.0.020339-0
Cheng, C., Han, X., Xu, J., Sun, J., Li, K., Han, Y., et al. (2021). Yjbh Mediates the Oxidative Stress Response and Infection by Regulating Spxa1 and the Phosphoenolpyruvate-Carbohydrate Phosphotransferase System (PTS) in Listeria Monocytogenes. Gut Microbes 13 (1), 1–19. doi: 10.1080/19490976.2021.1884517
Curtis, T. D., Takeuchi, I., Gram, L., Knudsen, G. M. (2017). The Influence of the Toxin/Antitoxin Mazef on Growth and Survival of Listeria Monocytogenes Under Stress. Toxins (Basel) 9 (1), 31. doi: 10.3390/toxins9010031
de Noordhout, C. M., Devleesschauwer, B., Angulo, F. J., Verbeke, G., Haagsma, J., Kirk, M., et al. (2014). The Global Burden of Listeriosis: A Systematic Review and Meta-Analysis. Lancet Infect. Dis. 14 (11), 1073–1082. doi: 10.1016/S1473-3099(14)70870-9
Diamant, S., Goloubinoff, P. (1998). Temperature-Controlled Activity of Dnak-Dnaj-Grpe Chaperones: Protein-Folding Arrest and Recovery During and After Heat Shock Depends on the Substrate Protein and the Grpe Concentration. Biochemistry 37 (27), 9688–9694. doi: 10.1021/bi980338u
Goodman, K. N., Powers, M. J., Crofts, A. A., Trent, M. S., Hendrixson, D. R. (2020). Campylobacter Jejuni Bumsr Directs a Response to Butyrate via Sensor Phosphatase Activity to Impact Transcription and Colonization. Proc. Natl. Acad. Sci. U. S. A. 117 (21), 11715–11726. doi: 10.1073/pnas.1922719117
Gueriri, I., Bay, S., Dubrac, S., Cyncynatus, C., Msadek, T. (2008a). The Pta-Acka Pathway Controlling Acetyl Phosphate Levels and the Phosphorylation State of the Degu Orphan Response Regulator Both Play a Role in Regulating Listeria Monocytogenes Motility and Chemotaxis. Mol. Microbiol. 70 (6), 1342–1357. doi: 10.1111/j.1365-2958.2008.06496.x
Gueriri, I., Cyncynatus, C., Dubrac, S., Arana, A. T., Dussurget, O., Msadek, T. (2008b). The Degu Orphan Response Regulator of Listeria Monocytogenes Autorepresses Its Own Synthesis and Is Required for Bacterial Motility, Virulence and Biofilm Formation. Microbiol. (Reading) 154 (Pt 8), 2251–2264. doi: 10.1099/mic.0.2008/017590-0
Gupta, M., Rao, K. K. (2014). Phosphorylation of Degu Is Essential for Activation of Amye Expression in Bacillus Subtilis. J. Biosci. 39 (5), 747–752. doi: 10.1007/s12038-014-9481-5
Hanawa, T., Kai, M., Kamiya, S., Yamamoto, T. (2000). Cloning, Sequencing, and Transcriptional Analysis of the Dnak Heat Shock Operon of Listeria Monocytogenes. Cell Stress Chaperones 5 (1), 21–29. doi: 10.1379/1466-1268(2000)005<0021:CSATAO>2.0.CO;2
Hartl, F. U. (1996). Molecular Chaperones in Cellular Protein Folding. Nature 381 (6583), 571–579. doi: 10.1038/381571a0
Hu, Y., Oliver, H. F., Raengpradub, S., Palmer, M. E., Orsi, R. H., Wiedmann, M., et al. (2007). Transcriptomic and Phenotypic Analyses Suggest a Network Between the Transcriptional Regulators Hrca and Sigmab in Listeria Monocytogenes. Appl. Environ. Microbiol. 73 (24), 7981–7991. doi: 10.1128/AEM.01281-07
Keto-Timonen, R., Tolvanen, R., Lunden, J., Korkeala, H. (2007). An 8-Year Surveillance of the Diversity and Persistence of Listeria Monocytogenes in a Chilled Food Processing Plant Analyzed by Amplified Fragment Length Polymorphism. J. Food Prot 70 (8), 1866–1873. doi: 10.4315/0362-028x-70.8.1866
Kornitzer, D., Teff, D., Altuvia, S., Oppenheim, A. B. (1991). Isolation, Characterization, and Sequence of an Escherichia Coli Heat Shock Gene, Htpx. J. Bacteriol 173 (9), 2944–2953. doi: 10.1128/jb.173.9.2944-2953.1991
Lemmer, K. C., Alberge, F., Myers, K. S., Dohnalkova, A. C., Schaub, R. E., Lenz, J. D., et al. (2020). The Ntryx Two-Component System Regulates the Bacterial Cell Envelope. mBio 11 (3), e00957–20 . doi: 10.1128/mBio.00957-20
Liberek, K., Skowyra, D., Zylicz, M., Johnson, C., Georgopoulos, C. (1991). The Escherichia Coli Dnak Chaperone, the 70-Kda Heat Shock Protein Eukaryotic Equivalent, Changes Conformation Upon ATP Hydrolysis, Thus Triggering Its Dissociation From a Bound Target Protein. J. Biol. Chem. 266 (22), 14491–14496. doi: 10.1016/S0021-9258(18)98713-2
Li, H., Liu, F., Peng, W., Yan, K., Zhao, H., Liu, T., et al. (2018). The Cpxa/Cpxr Two-Component System Affects Biofilm Formation and Virulence in Actinobacillus Pleuropneumoniae. Front. Cell Infect. Microbiol. 8, 72. doi: 10.3389/fcimb.2018.00072
Lin, M. H., Chiang, M. L., Pan, C. L., Chou, C. C. (2012). Heat Shock and Cold Shock Treatments Affect the Survival of Listeria Monocytogenes and Salmonella Typhimurium Exposed to Disinfectants. J. Food Prot 75 (4), 695–700. doi: 10.4315/0362-028X.JFP-11-419
Liu, F., Peng, W., Liu, T., Zhao, H., Yan, K., Yuan, F., et al. (2018). Biological Role of Actinobacillus Pleuropneumoniae Type IV Pilus Proteins Encoded by the Apf and Pil Operons. Vet. Microbiol. 224, 17–22. doi: 10.1016/j.vetmic.2018.08.006
Mader, U., Antelmann, H., Buder, T., Dahl, M. K., Hecker, M., Homuth, G. (2002). Bacillus Subtilis Functional Genomics: Genome-Wide Analysis of the Degs-Degu Regulon by Transcriptomics and Proteomics. Mol. Genet. Genomics 268 (4), 455–467. doi: 10.1007/s00438-002-0774-2
Mauder, N., Williams, T., Fritsch, F., Kuhn, M., Beier, D. (2008). Response Regulator Degu of Listeria Monocytogenes Controls Temperature-Responsive Flagellar Gene Expression in Its Unphosphorylated State. J. Bacteriol 190 (13), 4777–4781. doi: 10.1128/JB.00258-08
Ming, X., Daeschel, M. A. (1993). Nisin Resistance of Foodborne Bacteria and the Specific Resistance Responses of Listeria Monocytogenes Scott a. J. Food Prot 56 (11), 944–948. doi: 10.4315/0362-028X-56.11.944
Murret-Labarthe, C., Kerhoas, M., Dufresne, K., Daigle, F. (2020). New Roles for Two-Component System Response Regulators of Salmonella Enterica Serovar Typhi During Host Cell Interactions. Microorganisms 8 (5), 722. doi: 10.3390/microorganisms8050722
Nair, S., Derre, I., Msadek, T., Gaillot, O., Berche, P. (2000). Ctsr Controls Class III Heat Shock Gene Expression in the Human Pathogen Listeria Monocytogenes. Mol. Microbiol. 35 (4), 800–811. doi: 10.1046/j.1365-2958.2000.01752.x
Pasechnek, A., Rabinovich, L., Stadnyuk, O., Azulay, G., Mioduser, J., Argov, T., et al. (2020). Active Lysogeny in Listeria Monocytogenes Is a Bacteria-Phage Adaptive Response in the Mammalian Environment. Cell Rep. 32 (4), 107956. doi: 10.1016/j.celrep.2020.107956
Pierpaoli, E. V., Sandmeier, E., Baici, A., Schonfeld, H. J., Gisler, S., Christen, P. (1997). The Power Stroke of the Dnak/Dnaj/Grpe Molecular Chaperone System. J. Mol. Biol. 269 (5), 757–768. doi: 10.1006/jmbi.1997.1072
Pontinen, A., Aalto-Araneda, M., Lindstrom, M., Korkeala, H. (2017). Heat Resistance Mediated by Plm58 Plasmid-Borne Clpl in Listeria Monocytogenes. mSphere 2 (6), e00364–17 . doi: 10.1128/mSphere.00364-17
Rapun-Araiz, B., Haag, A. F., De Cesare, V., Gil, C., Dorado-Morales, P., Penades, J. R., et al. (2020). Systematic Reconstruction of the Complete Two-Component Sensorial Network in Staphylococcus Aureus. mSystems 5 (4), e00511–20. doi: 10.1128/mSystems.00511-20
Salvail, H., Groisman, E. A. (2020). The Phosphorelay Bara/Sira Activates the Non-Cognate Regulator Rcsb in Salmonella Enterica. PloS Genet. 16 (5), e1008722. doi: 10.1371/journal.pgen.1008722
Seifart Gomes, C., Izar, B., Pazan, F., Mohamed, W., Mraheil, M. A., Mukherjee, K., et al. (2011). Universal Stress Proteins Are Important for Oxidative and Acid Stress Resistance and Growth of Listeria Monocytogenes EGD-E In Vitro and In Vivo. PloS One 6 (9), e24965. doi: 10.1371/journal.pone.0024965
Somolinos, M., Espina, L., Pagan, R., Garcia, D. (2010). Sigb Absence Decreased Listeria Monocytogenes EGD-E Heat Resistance But Not Its Pulsed Electric Fields Resistance. Int. J. Food Microbiol. 141 (1-2), 32–38. doi: 10.1016/j.ijfoodmicro.2010.04.023
Soni, K. A., Nannapaneni, R., Tasara, T. (2011). The Contribution of Transcriptomic and Proteomic Analysis in Elucidating Stress Adaptation Responses of Listeria Monocytogenes. Foodborne Pathog. Dis. 8 (8), 843–852. doi: 10.1089/fpd.2010.0746
Tiwari, N., Lopez-Redondo, M., Miguel-Romero, L., Kulhankova, K., Cahill, M. P., Tran, P. M., et al. (2020). The Srrab Two-Component System Regulates Staphylococcus Aureus Pathogenicity Through Redox Sensitive Cysteines. Proc. Natl. Acad. Sci. U. S. A. 117 (20), 10989–10999. doi: 10.1073/pnas.1921307117
Tokunaga, T., Rashid, M. H., Kuroda, A., Sekiguchi, J. (1994). Effect of Degs-Degu Mutations on the Expression of Sigd, Encoding an Alternative Sigma Factor, and Autolysin Operon of Bacillus Subtilis. J. Bacteriol 176 (16), 5177–5180. doi: 10.1128/jb.176.16.5177-5180.1994
van der Veen, S., Abee, T. (2010). Hrca and Dnak Are Important for Static and Continuous-Flow Biofilm Formation and Disinfectant Resistance in Listeria Monocytogenes. Microbiol. (Reading) 156 (Pt 12), 3782–3790. doi: 10.1099/mic.0.043000-0
van der Veen, S., Hain, T., Wouters, J. A., Hossain, H., de Vos, W. M., Abee, T., et al. (2007). The Heat-Shock Response of Listeria Monocytogenes Comprises Genes Involved in Heat Shock, Cell Division, Cell Wall Synthesis, and the SOS Response. Microbiol. (Reading) 153 (Pt 10), 3593–3607. doi: 10.1099/mic.0.2007/006361-0
Williams, T., Bauer, S., Beier, D., Kuhn, M. (2005). Construction and Characterization of Listeria Monocytogenes Mutants With in-Frame Deletions in the Response Regulator Genes Identified in the Genome Sequence. Infect. Immun. 73 (5), 3152–3159. doi: 10.1128/IAI.73.5.3152-3159.2005
Wurtzel, O., Sesto, N., Mellin, J. R., Karunker, I., Edelheit, S., Becavin, C., et al. (2012). Comparative Transcriptomics of Pathogenic and Non-Pathogenic Listeria Species. Mol. Syst. Biol. 8, 853.
Zhang, X., Guan, C., Hang, Y., Liu, F., Sun, J., Yu, H., et al. (2020). An M29 Aminopeptidase From Listeria Monocytogenes Contributes to In Vitro Bacterial Growth But Not to Intracellular Infection. Microorganisms 8 (1), 110. doi: 10.3390/microorganisms8010110
Keywords: Listeria monocytogenes, orphan response regulator, DegU, heat resistance, heat-shock proteins
Citation: Cheng C, Liu F, Jin H, Xu X, Xu J, Deng S, Xia J, Han Y, Lei L, Zhang X and Song H (2021) The DegU Orphan Response Regulator Contributes to Heat Stress Resistance in Listeria monocytogenes. Front. Cell. Infect. Microbiol. 11:761335. doi: 10.3389/fcimb.2021.761335
Received: 19 August 2021; Accepted: 25 November 2021;
Published: 13 December 2021.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Jörgen Johansson, Umeå University, SwedenCopyright © 2021 Cheng, Liu, Jin, Xu, Xu, Deng, Xia, Han, Lei, Zhang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Houhui Song, c29uZ2hoQHphZnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.