
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 07 February 2022
Sec. Molecular Bacterial Pathogenesis
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.755545
This article is part of the Research TopicAntimicrobial Resistance Dissemination and Horizontal Gene TransferView all 5 articles
 Zhihai Liu1,2,3†
Zhihai Liu1,2,3† Ke Wang1,4,5†
Ke Wang1,4,5† Yaru Zhang6
Yaru Zhang6 Lining Xia4
Lining Xia4 Li Zhao1
Li Zhao1 Changmei Guo1,4
Changmei Guo1,4 Xudong Liu1
Xudong Liu1 Liting Qin7
Liting Qin7 Zhihui Hao2*
Zhihui Hao2*The objective of this study was to understand the diversity characteristics of ESBL-producing Escherichia coli (ESBL-EC) in chicken, pig, and cattle. A high prevalence of ESBL-EC (260/344) was observed in all food animals with prevalence rates of 78.6% (110/140) for chicken, 70.7% (58/82) for cattle, and 75.4% (92/122) for swine. However, the resistance rates presented significant differences in different animal origin ESBL-EC, where resistance to CTX, GEN, IMP, NEO, and OFL was the highest in chicken ESBL-EC, then in cattle, and the lowest in swine. Seriously, most ESBL-EC harbor multidrug resistance to antibiotics (MDR, ≥3 antibiotic categories), and the MDR rates of ESBL-EC were the highest in chicken (98.18%), followed by swine (93.48%), and the lowest in cow (58.62%), while the same trend also was observed in MDR of ≥5 antibiotic categories. This high prevalence and resistance can be partly interpreted by the high carriage rates of the β-lactamases CTX-M (n = 89), OXA (n = 59), SHV (n = 7), and TEM (n = 259). A significant difference of β-lactamase genes also presented in different animal species isolates, where the chicken origin ESBL-EC possessed higher carriage rates of almost all genes tested than cattle and swine. Notably, eight chicken origin ESBL-EC carried transferable plasmid-mediated blaNDM-1 or blaNDM-5, especially, of which four ESBL-EC also contained the colistin resistance gene mcr-1, as confirmed by genomic analysis. More interestingly, two deletion events with a 500-bp deletion in ΔISAba125 and a 180-bp deletion in dsbC were observed in three blaNDM-5 IncX3 plasmids, which, as far as we know, is the first discovery. This showed the instability and horizontal transfer of blaNDM genetic context, suggesting that blaNDM is evolving to “pack light” to facilitate rapid and stable horizontal transfer. Sequence types (STs) and PFGE showed diversity patterns. The most prevalent STs were ST48 (n = 5), ST189 (n = 5), ST206 (n = 4), ST6396 (n = 3), ST10 (n = 3), and ST155 (n = 3), where ST48 ESBL-EC originated from three food animal species. The STs of all blaNDM-positive ESBL-EC were attributed to three STs, namely, ST6396 (n = 2), ST206 (n = 2), and ST189 (n = 4), where ST189 was also the unique type for four mcr-1-carrying ESBL-EC. In conclusion, we suggest that the three animal species ESBL-EC show similar high prevalence, diversity in isolate lineages, and significant discrepancies in antibiotic resistance and resistance genes. This suggests that monitoring and anti-infection of different food animal origin ESBL-EC need different designs, which deserves more attention and further surveillance.
Antimicrobial resistance (AMR) is a serious threat to public health. The annual human deaths will rise from the current 0.7 million to 10 million with an estimated $100 trillion in economic losses by 2050 due to AMR (O’Neill, 2016; Friedrich, 2019). The extensive use of antimicrobials in humans, animals, and the environment has generated populations of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae, carbapenem-resistant Enterobacteriaceae (CRE), vancomycin-resistant enterococci (VRE), methicillin-resistant Staphylococcus aureus (MRSA), and so on across the globe (Okpara et al., 2018), in which ESBL-producing Enterobacteriaceae and CRE, such as NDM-producing Enterobacteriaceae, are the most prevalent in animals (Wang et al., 2017). Animals, especially food animals, are not only the reservoirs of antibiotic resistance genes (ARGs) and antibiotic resistance bacteria (ARB) but also the generators of novel resistance mechanisms and genes. Recently, novel mechanisms and genes were first revealed in food animal bacteria, such as mobile colistin resistance (mcr-1) in pig (Liu et al., 2016) and plasmid-mediated tigecycline resistance gene tet(X) in pig and chicken (He et al., 2019; Sun et al., 2019). In our previous work, a series of novel variant genes were also firstly reported in food animal origin isolates, such as novel carbapenemase-encoding genes blaNDM-17 (Liu et al., 2017), blaNDM-20 (Liu et al., 2018), and blaVIM-48 (Liu et al., 2019); novel mcr variants mcr-3 (Yin et al., 2017); and plasmid-mediated high-level tigecycline resistance genes tet(X3) and tet(X4) (He et al., 2019). Currently, the important ARGs in animals are bla, mcr, cfr, and tet genes and their variants (de Alcântara Rodrigues et al., 2020; Ling et al., 2020), where bla (blaKPC, blaNDM, blaVIM, and blaOXA-48/23), mcr, and tet gene-mediated AMR are closely associated to the last resort antibiotics in the human clinic (Guerra et al., 2014). Worryingly, all those ARB and ARGs can transfer to human through the consumption of products of animal origin (de Alcântara Rodrigues et al., 2020). Accordingly, AMR and ARGs in food animals need more attention.
Among numerous resistance mechanisms, the expression of ESBL is the most prevalent and increasingly common and has become the basic resistance mechanism that persists over other ARGs (Bush and Bradford, 2020). For example, blaESBLs genes were observed in almost all CRE or mcr-positive pathogens (Bush and Bradford, 2020; Ling et al., 2020). ESBLs can hydrolyze β-lactam antibiotics including the penicillins and cephalosporins, which is still a common clinical difficulty in fighting infection. The most prevalent and typical ESBLs are the blaTEM, blaSHV, and blaCTX-M genes which have evolved into dozens of subtypes with increasing enzyme activities by amino acid substitutions. These β-lactamases have been isolated from humans (Pitout and Laupland, 2008; Musicha et al., 2017; Wittekamp et al., 2018), the environment (Runcharoen et al., 2017), and animals in many countries including China (White et al., 2001) and represent an emerging general public health threat. In particular, animals and farm environments are important reservoirs of ESBL-producing bacteria (Seiffert et al., 2013; Kim et al., 2019). A high prevalence of ESBL producers along with a high level of diversity in ESBL genes has been reported in broilers and poultry production chains (Dame-Korevaar et al., 2019). In addition, animal wastes carrying ESBLs lead to biological contamination and accumulation in animal food, vegetables, water, and soil (Caltagirone et al., 2017; Dorado-García et al., 2018; Sen and Sarkar, 2018). So, ESBL genes can spread from animals to humans and can be directly transmitted to farmers (Kola et al., 2012; Dohmen et al., 2015). Seriously, the coexistence of other resistance genes, such as fosA, sul, blaNDM, and mcr, is frequent in ESBL-producing pathogens, resulting in multidrug resistance (Wang et al., 2017). Consequently, this will compromise the effectiveness of β-lactams for the treatment of disease in both people and animals, which will ultimately aggravate financial burden on society.
Antimicrobial resistance is derived from antibiotic selection and exposure, and dissemination of ESBL-producing bacteria is associated with the heavy use of cephalosporins (Allcock et al., 2017). Globally, 63,151 tons of antibiotics (all classes) were used to treat livestock in 2010, and it is predicted to reach 105,596 tons by 2030 (Van Boeckel et al., 2015). Given the global estimates of antimicrobial consumption based on species-specific coefficients of antimicrobial consumption per population correction unit (PCU), cattle, chicken, and pig are the primary consumers of antibiotics and the highest use is reported in pig (Van Boeckel et al., 2015), where a significant difference in antimicrobial consumption is estimated which is generally lower (45 mg/PCU) for cattle than for chicken (148 mg/PCU) and pig (172 mg/PCU). Studies have shown that resistance correlates with antibiotic consumption (Mather et al., 2012). Accordingly, the resistance patterns should follow the usage pattern. Withal, the discrepancy in their physiology, species, and growth environment may also affect AMR. All those may influence the prevalence of drug-resistant bacteria.
So, in China, as the largest consumer of veterinary drugs, in which most are β-lactams used to treat livestock (Van Boeckel et al., 2015), a comprehensive study is necessary to better understand the prevalence characteristics of ESBL-producing Escherichia coli (ESBL-EC) in animals. The aim of this study was to characterize the diversity characteristics of ESBL-producing E. coli and the prevalence characteristics and the prevalence of blaNDM, mcr, and blaESBLs from the different food animals, such as chicken, pig, and cattle.
The fecal samples (n = 344) in this study were collected from livestock farms in Guangdong, Shandong, Xinjiang, and Heilongjiang provinces during 2015–2019. Samples were taken from chicken, swine, and cattle feces. These farms had records of β-lactam antibiotic usage for preventing and treating bacterial infections (data not shown). Immediately upon receipt of samples by the laboratory, they were plated on MacConkey agar plates (Luqiao, Beijing, China) and incubated at 37°C for 16–18 h. Red colonies were selected and enriched by cultivation in 2 ml Mueller–Hinton broth (Luqiao) and plated on methylene blue agar (Luqiao) plates containing 2 µg/ml of cefotaxime. Presumptive ESBL-producing E. coli were collected and genomic DNA was extracted using a Fast Pure Bacteria DNA Isolation Mini Kit (Vazyme Biotech, Nanjing, China). Bacterial species were confirmed using 16S rDNA PCR amplification and sequencing as previously described (Kim et al., 2010).
The broth microdilution method was used to determine the minimal inhibitory concentrations (MIC) for the following antibiotic categories: i) carbapenems: imipenem (IMP); ii) penicillins: amoxicillin (AMX); iii) cephalosporins: cephalexin (CN), cefotaxime (CTX), and cefepime (FEP); iv) tetracyclines: tetracycline (TET) and tigecycline (TGC); v) aminoglycosides: gentamicin (GEN) and neomycin (NEO); vi) aminocyclitol: spectinomycin (SPT); vii) colistin (CST); and viii) quinolones: ofloxacin (OFL) (Hubei Widely Chemical Technology Co., Ltd.). Escherichia coli ATCC 25922 was used as the quality control strain in MIC testing. All procedures and test interpretations followed the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2021). Multidrug-resistant (MDR) isolates were defined as possessing resistance to three or more antibiotic categories (Magiorakos et al., 2012).
All presumptive ESBL-producing isolates were screened by PCR for the β-lactamase genes blaSHV, blaTEM, and blaCTX-M and carbapenemase-encoding genes blaNDM, blaIMP, blaKPC, blaVIM, blaOXA, blaAIM, blaBIC, blaDIM, blaGIM, blaSIM, and blaSPM (Poirel et al., 2011; Casella et al., 2018), and each sample was tested at least twice. The genes blaCTX-M, blaNDM, and blaOXA were subtyped by PCR and sequencing as previously described (Poirel et al., 2011), and blaNDM subtypes were further confirmed by whole genome sequencing (WGS). PCR primers and conditions for PCR amplification are listed in Table 1. PCR amplicons were separated by electrophoresis through 1.5% agarose gels, stained with EtBr, and visualized under UV light. Amplicons were sequenced to confirm identity and analyzed using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast/). Escherichia coli CCD1 as a blaNDM-positive control in our previous report (Liu et al., 2018) and E. coli ATCC 25922, as the negative control, were used to assay the feasibility of the PCR test to exclude adverse factors.
The relatedness of ESBL-EC was assessed using pulsed field gel electrophoresis (PFGE) as previously described (Tenover et al., 1995). Here, 45 typical isolates were selected mainly based on source, resistance genes, and resistance (MIC). Normally, only one representative strain was selected among isolates possessing similar MIC or resistance genes, while the final sample size takes into account the number of different sources. They included swine (n = 10), cattle (n = 9), and chicken (n = 26). Briefly, agarose-embedded DNA was digested with Xba1 (Takara, Beijing, China) for 3 h at 37°C. DNA fragments were separated by electrophoresis in 0.5× TBE buffer at 14°C for 18 h using a CHEF-DR III electrophoresis system (Bio-Rad, Hercules, CA, USA) with pulse times of 2.2–54.2 s. Escherichia coli strain H9812 was used as standard for DNA size measurements. The gels were stained with EtBr and processed as above. PFGE results were analyzed using BioNumerics software v3.0 (Applied Maths Kortrijk, Belgium). All isolates successfully typed using PFGE were subjected to multilocus sequence typing (MLST) using the following loci: recA, adk, fumC, icd, mdh, purA, and gyrB (Kim et al., 2015). These PCR products were sequenced and the data were interpreted using the MLST database (http://enterobase.warwick.ac.uk/species/ecoli/allele_st_search) (Zhou et al., 2020). Cluster analysis of ST (sequence type) was performed using the BioNumerics software.
Transferability of blaNDM from animal blaNDM-positive E. coli isolates was assayed by conjugation using the azide-resistant strain E. coli J53 as the recipient (Liu et al., 2018). In brief, equal volumes of donor and recipient strains were mixed and filtered onto 0.45 µm filters that were then placed on Mueller–Hinton agar plates containing 100 µg/ml sodium azide and 1 µg/ml meropenem (Liu et al., 2019). At the same time, 10-fold dilutions were plated to determine transfer frequencies and presumptive transconjugants were screened using PCR (see above).
The blaNDM-positive E. coli were further characterized by WGS using DNA extracted as described above (Wang et al., 2017). The NEXT Ultra DNA Library Prep kit (New England Biolabs, Beverly, MA, USA) was used to establish gene libraries of 150 bp with paired ends and sequenced using the Illumina HiSeq 2500 system at Bionova Biotech (Beijing, China). The SPAdes algorithm v.3.10.0 (http://cab.spbu.ru/software/spades/) was used to assemble raw data, and the assembled sequences were further analyzed using workflows obtained from the bacterial analysis pipeline (https://cge.cbs.dtu.dk/services/cge) in CGE (Center for Genomic Epidemiology) services.
In our study, we identified 260 ESBL-EC isolates from the 344 fecal samples, including 110 (78.6%, 110/140) from chicken, 58 (70.7%, 58/82) from cattle, and 92 (75.4%, 92/122) from swine. Our presumptive ESBL isolates were also examined for the three most common β-lactamase genes, in which the blaTEM (99.62%, n = 259) was dominant and obviously higher than blaSHV (2.69%, n = 7) and blaCTX-M (34.2%, n = 89). Meanwhile, the carriage rate of blaCTX-M in chicken was 80.9%, higher than that in swine (7.6%) and cattle (0%). Similarly, the carriage rate of blaTEM and blaSHV genes from chicken was higher than those from swine and cattle. Comparatively, the genes blaTEM, blaSHV, and blaCTX-M from chicken were more prevalent than from swine and cattle. The carbapenemase-encoding genes in our sample population were represented by the blaNDM gene, only found in chicken (n = 8), and by the blaOXA gene in chicken, cattle, and swine at 33.6%, 25.9%, and 7.6%, respectively. Overall, blaTEM-1 was the predominant β-lactamase gene among the animal species screened in this study, and the blaCTX-M-55 gene was the major ESBL gene found in chicken.
Allele subtyping analysis indicated that 89 isolates harboring the blaCTX-M gene were present in our ESBL-EC population with 7 known and 1 CTX-M-untypable. These were grouped as CTX-M-55 (77%), CTX-M-15 (5.6%), CTX-M-14 (4.45%), CTX-M-3 (4.45%), CTX-M-123 (1.1%), CTX-M-98 (2.2%), and CTX-M-65 (4.45%). We found only seven blaCTX-M-positive E. coli from swine. The CTX-M-55 variant was present in isolates from swine (n = 6) and chicken (n = 63), while only the CTX-M-55 and CTX-M-untypable subtypes were observed in swine.
When we examined the isolates from chicken, we found a diversity of β-lactamase. For example, the TEM subtype was TEM-1B, while the OXA series were represented by OXA-2 (n = 5) and OXA-10 (n = 54) variants. In the group of seven SHV subtypes, only one (SHV-12) was present and the others were untypable and tentatively named SHV-like. Importantly, eight isolates carried the blaNDM gene with seven blaNDM-5 and a single blaNDM-1 isolate, in which four isolates also contained blaNDM-5 and mcr-1. All eight were collected from chicken. The other β-lactamase-related ARGs we examined for this study were absent in all isolates. In addition, ARG coexistence of resistance genes was common and OXA-10/TEM (n = 18), OXA-10/CTX-M/TEM (n = 32), and CTX-M/TEM (n = 48) were the primary groupings. The coexistence schemes of most genes were TEM/CTX-M/OXA-10/NDM (n = 4) and OXA-2/OXA-10/CTX-M/TEM (n = 1) (Figure 1).
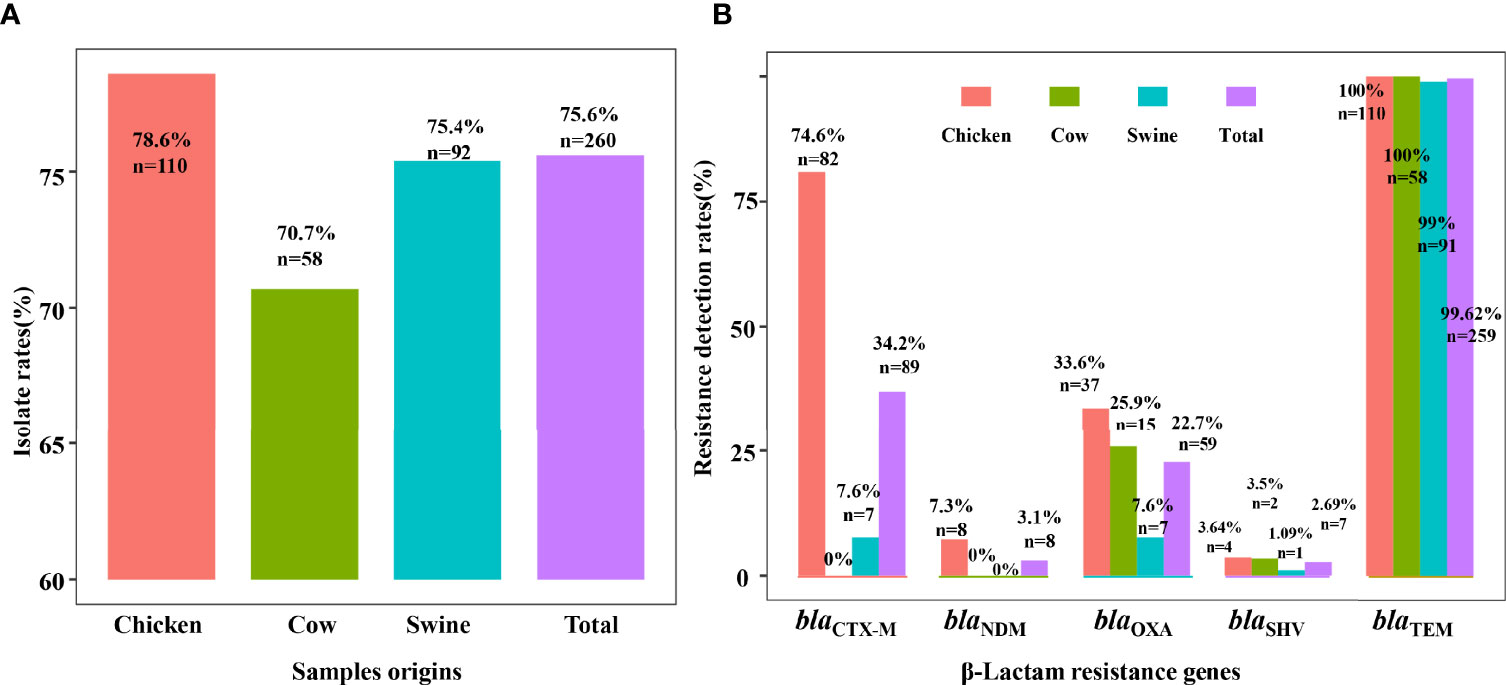
Figure 1 The prevalence of ESBL-EC and β-lactam resistance genes. Detection rates of ESBL-EC are shown in (A) and the carriage rates of β-lactam resistance genes are shown in (B). The bars showed the detection rates of resistant isolates or carriage rates of resistance genes in ESBL-EC among different food animals. In total, 260 isolates were detected from 344 samples, including 110 from chicken (red), 58 from cow (green), and 92 from swine (blue).
Overall, the prevalence of drug-resistant isolates from chicken was higher than that from cattle and swine. All ESBL-EC were resistant to cephalexin and most were tigecycline susceptible. Resistance to CTX, GEN, IMP, NEO, and OFL was the highest in chicken, then in cattle, and the lowest in swine. Noticeably, more than 50% ESBL-EC from any animal origin were also resistant to colistin. Specifically, ESBL-EC from chicken exhibited diverse MDR patterns that included resistance to all tested antibiotics. The individual resistance rates for all antibiotics in isolates from chicken were at least 38% and the rates for seven antibiotics were >70% except for tigecycline (3.6%). The resistance rates for ESBL-EC from cattle were <51.7% (except for LEX). Similarly, ESBL-EC from swine also showed susceptibility to IMP and TGC and low resistance rates to FEP (1.1%) (Figure 2A).
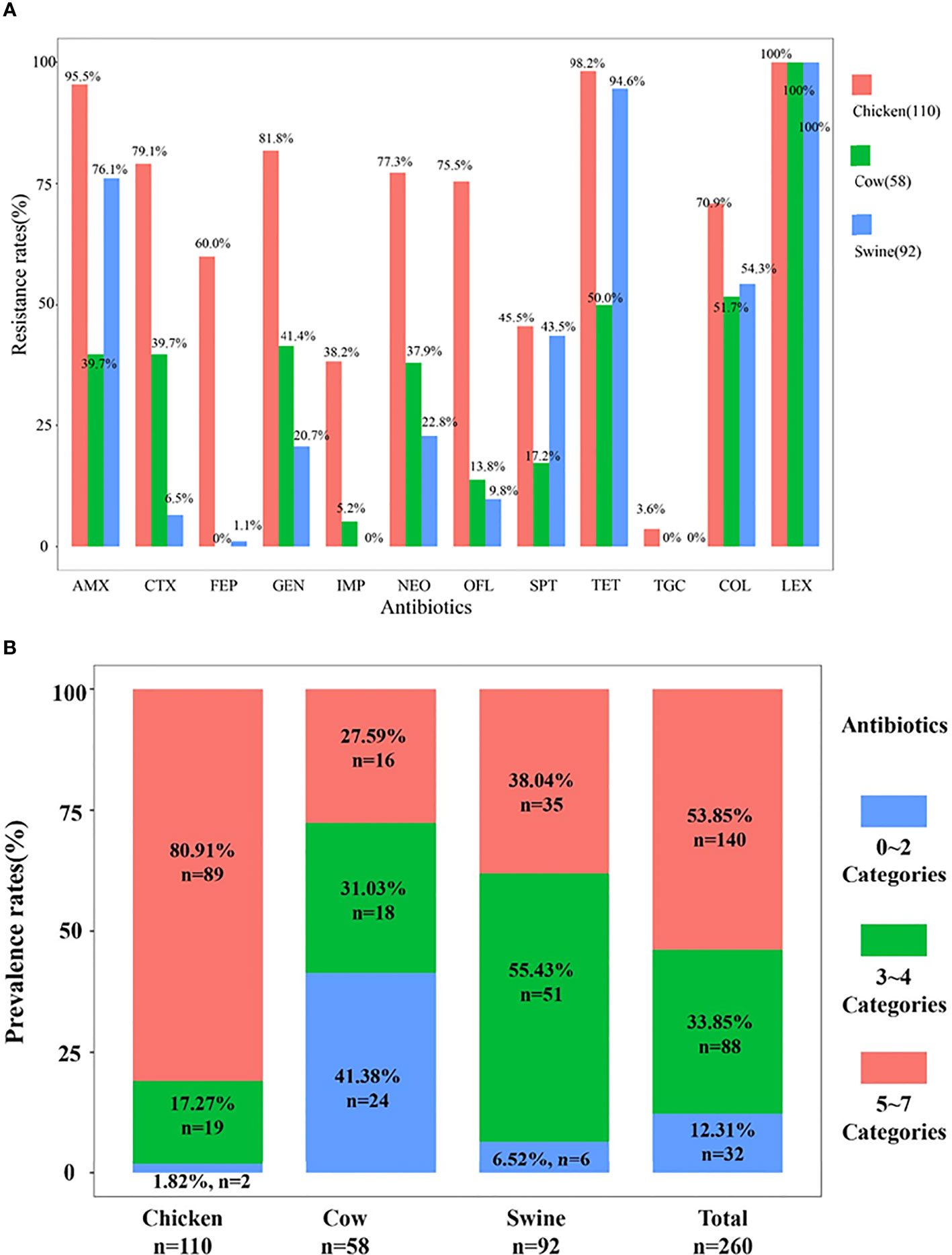
Figure 2 The resistance rates and multidrug resistance rates. The resistance rates of 260 isolates are shown in (A). The prevalence rates of multidrug resistance among 260 ESBL-EC are shown in (B).
The 12 antibiotics we tested in our study were from nine categories, and we found that MDR isolates were prevalent at rates of 87.69% (n = 228), where 88 isolates were resistant to 3–4 categories, while 140 isolates were resistant to 5–7 categories. MDR isolates from chicken and swine were the most prevalent (98.18%, n = 108; 93.48%, n = 84) and followed by cattle (>50%). The MDR isolates from chicken were primarily resistant to 5–7 antibiotic categories (81.9%, n = 89), and this rate was significantly higher than those for swine and cattle. Chicken also possessed a significant number of isolates that possessed resistance to 3–4 categories (17.27%, n = 19). In contrast, the cattle isolates displayed MDR rates to 3–4 and 5–7 categories that were almost equal. However, in swine MDR isolates, resistance to 3–4 categories (55.43%) greatly exceeded those in the 5–7 categories (Figure 2B). Therefore, different animal origin ESBL-EC demonstrated different MDR patterns.
Relationships of ESBL-EC isolates were analyzed using PFGE and MLST. Consequently, 37 isolates among 45 typical ESBL-EC were successfully identified by PFGE. These isolates could be divided into three major lineages containing 30 branches. Significantly, four different origin ESBL-EC isolates, consisting of two from chicken and one each from swine and cattle, showed the same PFGE pattern. Additionally, two small groups of chicken and swine isolates presented the same PFGE maps. Meanwhile, there were a few consistent PFGE patterns derived from the same animal species, such as w49 and w48 from swine and w161 and w160 from cattle (Figure 3). In general, the pattern of PFGE showed diversity but did not reveal dominant clone types.
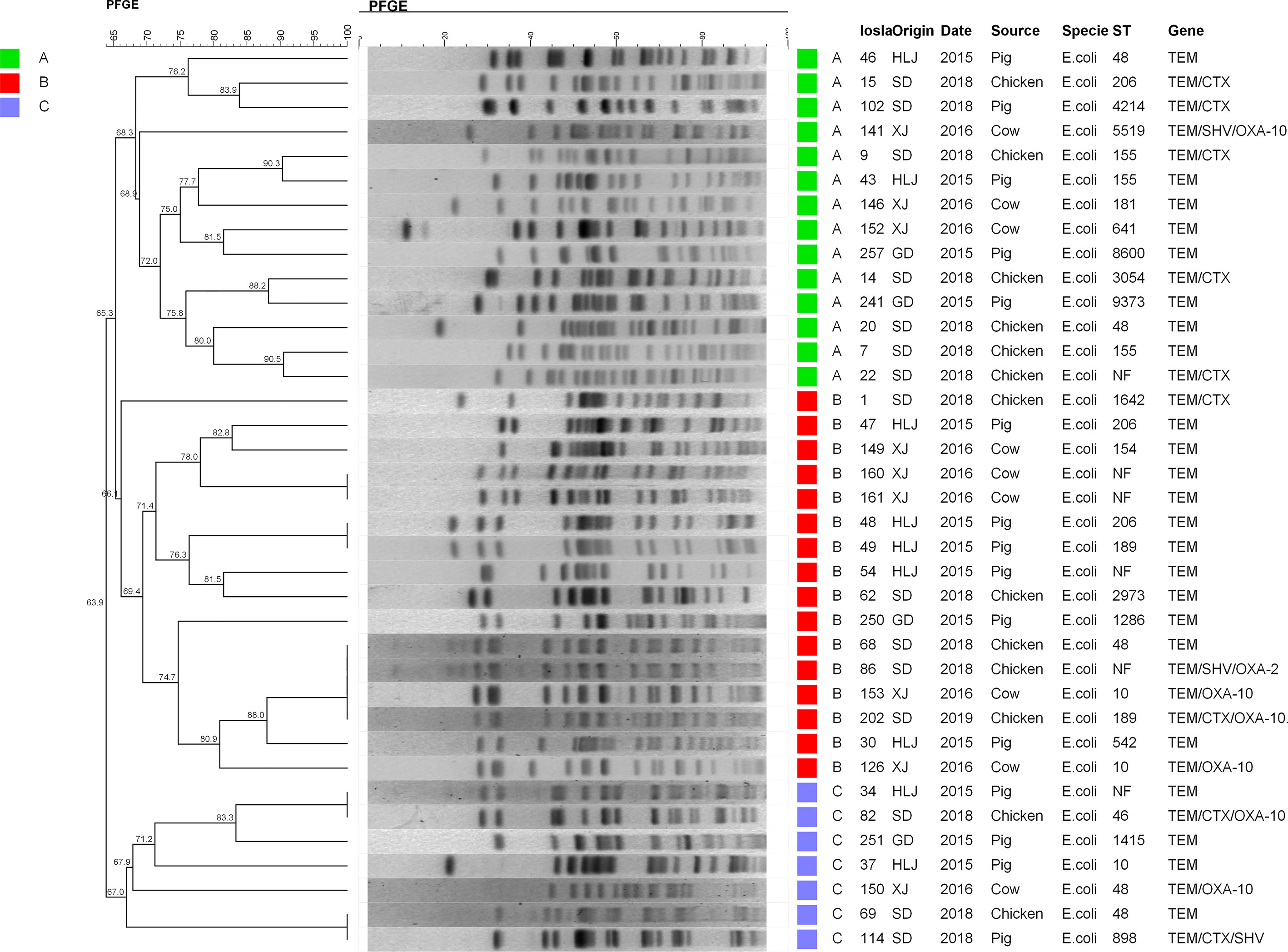
Figure 3 PFGE patterns of 37 ESBL-EC. All strains were mainly divided into three lineages: A (green), B (red), and C (blue). From left to right, PFGE results and phylogenetic analysis, strain number, collection place, collection time, source of strain, species, ST typing, and drug resistance genes carried are represented. HLJ, Heilongjiang; SD, Shandong; XJ, Xinjiang; GD, Guangdong.
The MLST assay was successful for 45 isolates, where 39 STs were identified and 6 isolates were not defined using the MLST database. The large clusters were ST48 (n = 5), ST189 (n = 5), ST206 (n = 4), ST6396 (n = 3), ST10 (n = 3), and ST155 (n = 3). The ST48 was present in three chicken isolates and one each from cattle and swine isolates. The five ST189 isolates consisted of one from swine and four from chicken. Interestingly, the four ST189 ESBL-EC from chicken showed coexistence of blaNDM-5 and mcr-1, and ST189 is the unique ST type for mcr-1 clones in our study (Table 2). The STs of the remaining blaNDM-positive ESBL-EC were attributed to two STs, namely, ST6396 (n = 2) and ST206 (n = 2). All five ST48 isolates generated five diverse PFGE patterns and similar results occurred for ST189, ST206, ST10, and ST155 (Figure 3). Noteworthy, while three ST155 showed slightly different PFGE patterns, they still exhibited high similarity >70% (Figures 3, 4). These data indicated that our ESBL-EC possessed diversity in PFGE and ST classifications and the same clonal type was commonly represented in all animal species analyzed in this study.

Table 2 Information for the eight blaNDM-positive Escherichia coli isolates identified in the current study.
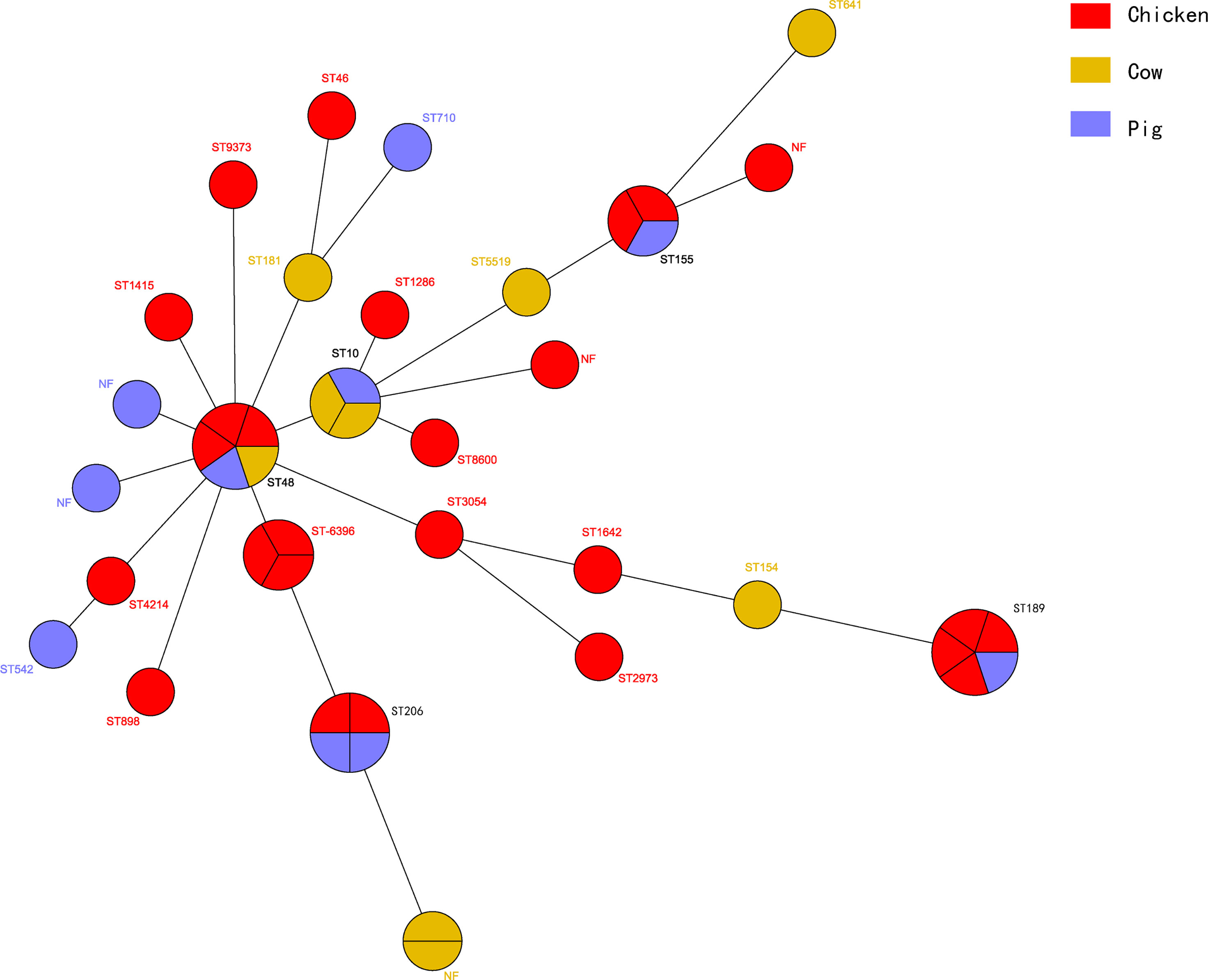
Figure 4 Phylogenetic analysis of STs among 45 ESBL-EC isolates. The clones from chicken (n = 26) were colored red, while those of cow (n = 9) yellow and those of pig (n = 10) blue.
Considering the importance and significance of blaNDM, the blaNDM-positive ESBL-EC were further characterized. Transconjugation tests were employed to confirm the transferability of the blaNDM gene and all were mobilized to strain J53 (Table 3). All the transconjugants displayed similar resistance profiles and conferred only a slightly lower resistance to carbapenems relative to the parental isolates. This was further evidence that blaNDM was successfully transferred from our ESBL-EC isolates, suggesting that all blaNDM may be located in mobile plasmids or conjugative transposons.
We subjected eight isolates to WGS analysis (Table 2), in which seven clean genomic datasets were obtained, where the clean data excluded impure and low-quality data, such as adaptor, barcode, low-quality reads, compared with the raw data, and one was excluded for the analysis due to poor data quality. Subsequently, the genomics were analyzed by CGE services, and the results are shown in Table 2. The results showed that these included three STs (ST189 n = 4, ST6396 and ST206 n = 2 each), where the ST of E. coli w206 (ST206) was performed by PCR. The coexisting resistance genes mainly included blaNDM, blaCTX-M, blaOXA, fosA, mph(A), aph(4)-Ia, aac(3)-Iva, aph(3’)-Ia, aac(6’)Ib-cr, and mcr-1, notable among which was the coexistence of blaNDM and mcr-1. All ST189 blaNDM isolates contained similar ARGs, virulence genes, and plasmid types, and similar results were also observed in the two ST6396 isolates (Table 2). For exploring genetic environments and plasmid characteristics of blaNDM-5, all contigs harboring blaNDM-5 were extracted and annotated. Consequently, seven contigs were successfully obtained, consisting of four contigs with 45,539 bp and three contigs with 42,928 bp. These contigs shared homology >99% with many blaNDM-5 plasmid sequences in the NCBI database, such as i) pEC135 (MH347484.1) from a human E. coli isolate in China, ii) pJN11NDM5 (MN092230) from a human E. coli in China, iii) pGSH8M-2 from wastewater in Tokyo Bay, and iv) pHNAH699 (MH286952) from a chicken E. coli isolate in China. These plasmids were identified as IncX3 type based on typical characteristics of replication, partitioning, plasmid maintenance, transcriptional activation, and conjugation genes, as shown in Figure 5. Although the complete plasmid sequence was not successfully assembled, this did not prevent the seven plasmids from being identified as IncX3 type because of the typical characteristics for plasmid type harbored in these contigs. Because the remaining isolate (E. coli w206) genome was not successfully assembled due to low quality, it was difficult to extract a blaNDM-harboring contig and achieve plasmid typing. Given this analysis and successful transconjugation results, it can be concluded that all blaNDM-5 were located in transferable IncX3 type plasmids.
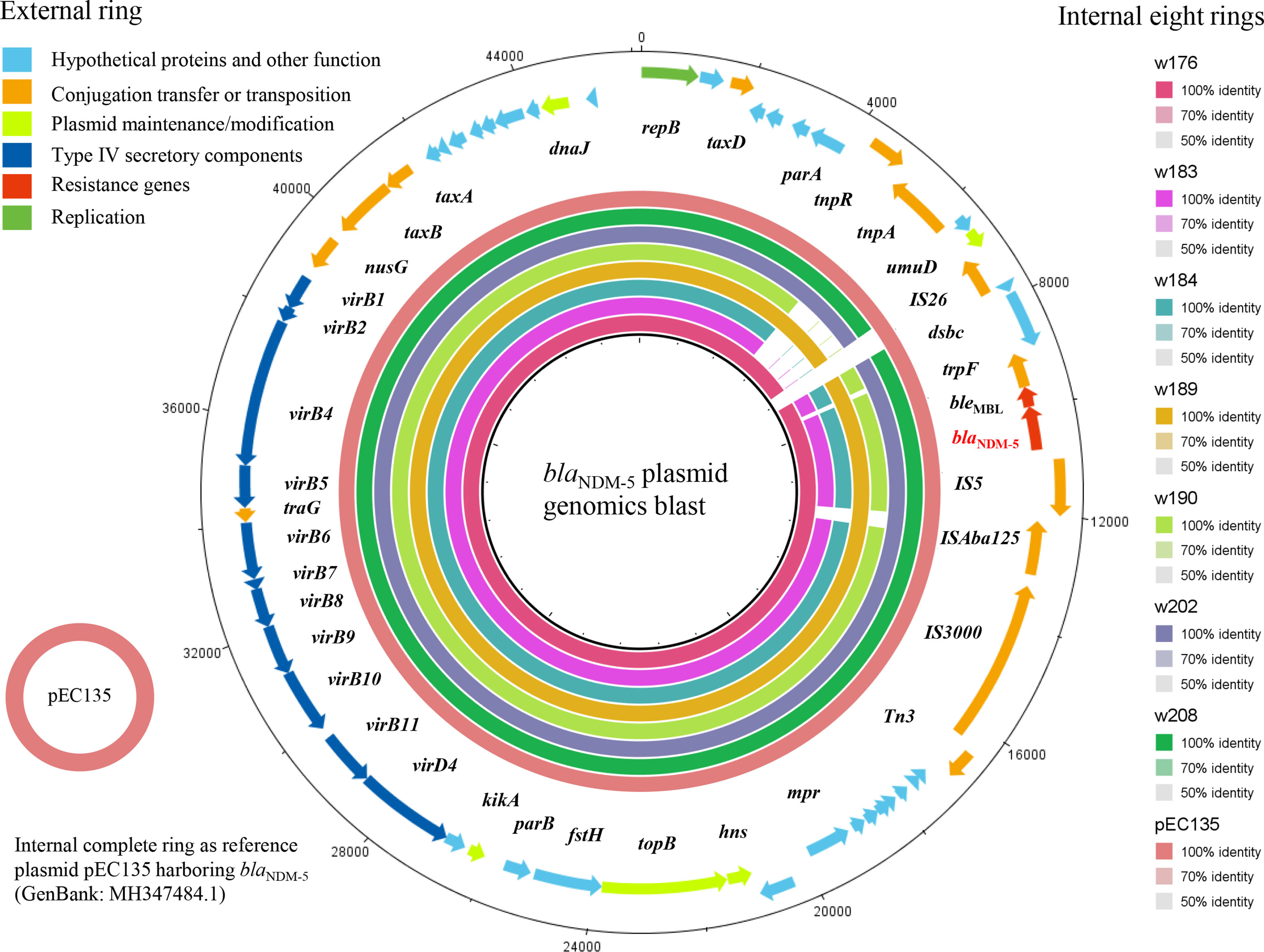
Figure 5 The blast and annotation of seven blaNDM-harboring plasmid genomes (constructed by BRIG). The eight small external rings represent different blaNDM-harboring plasmids and are shown in different colors, which are w176, w183, w184, w189, w190, w202, and w208 and the reference plasmid pEC135, from inside to outside. The external ring represents the annotation of plasmid pEC135 (GenBank accession no. MF347484). Genes are color-coded depending on functional annotations.
To decipher the genetic context of blaNDM-5, a genomics Blast and annotation were performed on the seven contigs using pEC135 as a reference sequence (Figure 5). The results are exhibited in Figure 6, where blaNDM-5 was flanked in a typical and common genetic structure ΔISAba125-IS5-blaNDM-ble-trpF-dsbC-IS26, as shown in Figure 6A. No other resistance genes were observed in this context except for blaNDM and bleMBL, a bleomycin resistance gene. The results of Blast showed a high identity in all seven genomes; however, a few segment gaps in three contigs (w183, 184, and w190) were observed in umuD, dsbC, and ΔISAba125 regions relative to the other four genomes, which exhibit deletions in dsbC (180-bp deletion) and ΔISAba125 (500-bp deletion). These deletions were internal to the extracted contigs rather than being generated by misassembly, suggesting that the ΔISAba125 was unstable. Moreover, the genetic environments of mcr-1 were dissected in four isolates and four genome contigs carrying the mcr-1 were successfully extracted from them, which were small segments with sizes of 8,246 bp (w176) and 8,308 bp (w189, w202, and w208). The Blast results indicated that they shared 100% identity and the same genetic environment, which is a common sequence in the NCBI database. mcr-1 was flanked in the upstream region by pap2 and downstream by several hypothetical protein encoding genes, as shown in Figure 6B.
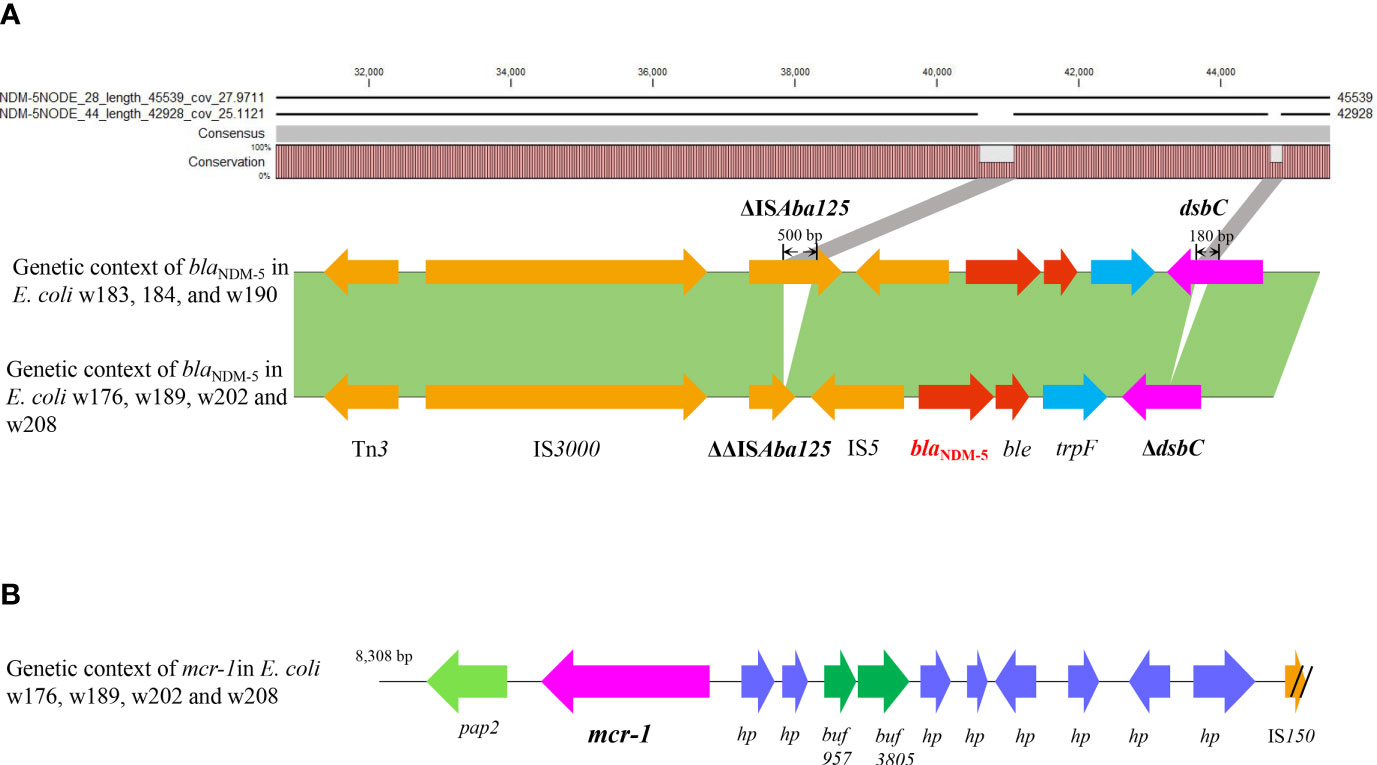
Figure 6 The analysis of blaNDM and mcr-1 genetic environments. Genetic context of blaNDM-5 (red) and the analysis of deletion fragments in ΔISAba125 and dsbC by Blast are shown (A). Genetic organization of mcr-1 (purple) is shown in (B).
Since the identification of the first plasmid-coded ESBL Enterobacteriaceae in the 1960s (Urbánek et al., 2007), these strains have spread globally and are a major threat to public health. In Asia, the prevalence rates of ESBL-EC are 20%~70% (Chong et al., 2018). Especially in China, the frequency of ESBL-EC is 60%~70% (Jean et al., 2016) and has been steadily increasing. Studies from 23 centers in 16 cities in China revealed that the presence rates of ESBL-EC increased from 36.1% to 68.1% among 3,074 E. coli over the last 10 years (P < 0.001) (Yang et al., 2013). A similar epidemiology has been seen in other Asian countries. Data from the Study for Monitoring Antimicrobial Resistance Trends (SMART) reported that the prevalence rate of ESBL-EC, in 2007, was 79% in India, 50.8% in Thailand, 34.4% in Vietnam, 17.8% in Hong Kong, 33.3% in South Korea and Singapore, 17.0% in the Philippines, 12.7% in Taiwan, and 55% in China (Hawser et al., 2009). By 2012, these rates had increased to 54.2% in Thailand, 28.3% in Hong Kong, 19.0% in the Philippines, 31.8% in Taiwan, and 68.8% in China (Jean et al., 2016). Recently, a global surveillance demonstrated that the rates of ESBL-EC were ~15% in Europe, ~10% in North America (Chong et al., 2018), 54%~71% (2011) in Latin America, and <15% in Africa (most countries) (Jones et al., 2013). ESBL-EC were also omnipresent, which can spread in animal food, seafood (Sanjit Singh et al., 2017), fruits (Randall et al., 2017), vegetables (Reuland et al., 2014; Randall et al., 2017), food animals (Carattoli, 2008; Day et al., 2019), wild animals (Yang et al., 2019), the environment, and even in other media, such as boots, air, insects, and transports (Müller et al., 2016; Dame-Korevaar et al., 2019). These data indicated that there is a high probability that ESBL-EC distribute to humans through food animals (Schmithausen et al., 2015; Dame-Korevaar et al., 2019). It is assessed that about 60% of human pathogens come from animals by diverse delivery media (Woolhouse, 2006). There is a serious and significantly high prevalence rate of ESBL-EC in animals relative to humans. For instance, a London study claimed that the percentage of ESBL-EC in humans was 17% (678) among 3,995 samples, but that in food animals was 65% (104) (Day et al., 2019). Chicken, cattle, and swine are the major food animals that play important roles in human health and food security, which is a global concern. Undoubtedly, the dissemination of ESBL-EC in animals is a significant risk to public safety, so effective strategies to monitor ESBL-EC prevalence should be implemented in food animals.
The emergence and prevalence of ESBL-EC are related to antibiotic usage (Ghafourian et al., 2015). However, the use of antibiotics for the treatment of infections in humans and animals is almost irreplaceable. Currently, the global annual consumption of antibiotics is between 100,000 and 200,000 tons, including human and veterinary medicine (Wang et al., 2010). Global consumption of antimicrobials in animals will increase from 63,151 in 2010 to 105,596 tons in 2030 (Van Boeckel et al., 2015), where China, USA, Brazil, India, and Germany are the largest consumers. In China, 23% of antibiotics are used for food animals at levels of 45, 148, and 172 mg/kg for cattle, chicken, and swine, respectively (Van Boeckel et al., 2015). Of all the antibiotics, the β-lactams are the most commonly used for animals, including penicillins and cephalosporins (Wei et al., 2011), both of which are associated with ESBL-EC. Accordingly, we believe, with the irreplaceable use of antibiotics in the future, the emergence and prevalence of ESBL-EC will be more serious in food animals in the absence of effective monitors.
The differences in the types and amounts of veterinary antibiotics used in different animals may result in different rates of emergence and prevalence of ESBL-EC. In support of this, our study showed that resistance rates presented a significant difference in different animal origin ESBL-EC, in which the resistance to CTX, GEN, IMP, NEO, and OFL was the highest in chicken ESBL-EC, then in cattle, and the lowest in swine. A similar difference in multidrug resistance was also being observed. Meanwhile, a high prevalence of ESBL-EC was also observed, in which that of chicken (78.6%) was higher than those of cattle (70.7%) and swine (75.4%). A recent survey showed a rapid rise of chicken origin ESBL-EC from 23.8%, 25.7%, 41.2%, 44.9%, 49.6%, 50.0% to 57.0% during 2008~2014, respectively (Wu et al., 2018), but our data from chicken was even higher (78.6%). The ESBL-EC prevalence in chicken is considered to be becoming widespread and more serious than other food animals. A recent report in South Korea revealed the presence rates of ESBL-EC in chicken are the highest up to 94.1% (Song et al., 2020), followed by 69.5% in pig, while only 7.0% in cattle, where a more significant difference was observed relative to our data. The trend in prevalence rates of ESBL-EC may also reflect resistance conditions. We found that chicken origin ESBL-EC were highly resistant to all antibiotics tested in this study, compared with pig and cattle, which was similar to a previous study (Ho et al., 2011). Besides, the lower rates for pig and cattle seem to be related to antibiotic type. For example, in our results, cattle isolates exhibited high resistance rates to CTX, GEN, IMP, NEO, and OFL relative to swine, while the converse was true for AMX, SPT, TET, FEP, and COL.
The prevalence of ESBL-EC can be partly interpreted by blaESBL genes, which are the root occurrence of ESBL-EC. In our research, the prevalence, resistance, and blaESBL carriage rates of ESBL-EC present a similar trend in different food animals, where those in chicken were the highest and most serious. So, the difference of resistance in different food animals should be attributed to blaESBL genes. In detail, blaTEM was present in almost all isolates, where TEM was commonly the foundation for the other β-lactamases (Pimenta et al., 2014), such as CTX-M (Bush, 2018). Over the past 20 years, TEM and SHV have been the primary β-lactamases found in ESBL-EC isolates (Zhang et al., 2014). Interestingly, the rate for SHV was low, so TEM might be the facilitator of ESBL resistance in the isolates in this study. The CTX-M type is currently the most prevalent β-lactamase globally (Zhang et al., 2014), of which CTX-M-15 and CTX-M-14 predominate in both animals and humans (Bevan et al., 2017). However, only five CTX-M-15 and four CTX-M-14 were detected, while CTX-M-55 (n = 69) was the predominant CTX-M prevalent subtype in food animals in our study, which agrees with the report by Pimenta et al. (2014). In China, CTX-M-55 was rare before 2009 (Zhuo et al., 2009); however, it is now the predominant ESBL and is widespread. Recently, Rao et al. described 186 CTX-M-55-expressing isolates from food animals and 47 isolates in humans (Rao et al., 2014; Zhang et al., 2014). Although CTX-M-65 is also a major CTX-M subtype in China, we only found four ESBL-EC producing CTX-M-65. As CTX-M-1 and CTX-M-14 were the dominant subtypes in other countries, CTX-M-55 spread mainly in China (Seiffert et al., 2013). This has been evidenced by recent reports in China; for instance, Fu et al. revealed 32 (84.21%) blaCTX-M-55 from 38 blaCTX-M from humans in Shanghai (Fu et al., 2020), Zhang et al. claimed CTX-M-55 (12/27, 44.44%) was the most prevalent ESBL type from foodborne animals (Zhang et al., 2019), and Jiang et al. showed the gene blaCTX-M-55 (31/64) was the predominant blaCTX-M subtype, followed by blaCTX-M-14 (18/64) and blaCTX-M-65 (14/64) from chicken in their study (Jiang et al., 2017).
More significantly, although the use of carbapenem antibiotics is banned in feed for food animals, eight NDM-producing ESBL-EC were found in 45 carbapenem-resistant isolates from chicken, consisting of seven NDM-5 and one NDM-1. As the major type of carbapenemase, NDM can impair the efficacy of almost all β-lactams (except aztreonam) and the therapeutic options are limited mostly to polymyxins and tigecycline, so acquiring additional NDM gene resistance is worrisome. Previous reports revealed that a significantly higher incidence of sepsis and bloodstream infection was caused by NDM-1 (Chatterjee et al., 2016). Since blaNDM-1 was first identified in New Delhi, India, in 2008 (Yong et al., 2009), more than 38 subtypes of NDM have been deposited in GenBank. So far, the presence of NDM has been reported in at least 55 countries and regions and in more than 60 species of bacteria (Wu et al., 2019). In our previous survey, 161 blaNDM-positive CRE (carbapenem-resistant Enterobacteriaceae) were identified among 736 food animal samples, including 84 NDM-5 (Wang et al., 2017). Currently, NDM-5 has become the predominant NDM variant among CRE from chicken (Wang et al., 2017), which is supported by our study, while NDM-1 is the predominant global NDM variant (Nordmann et al., 2011). Besides, there are many carbapenem-resistant ESBL-EC without known carbapenem resistance genes in our study, which need further exploration. Meanwhile, all transconjugation of blaNDM-5 was successfully performed, confirming the transferability of blaNDM-5 and suggesting blaNDM location on plasmids. Subsequently, seven blaNDM-5 harboring genomic contigs were identified as IncX3 plasmid sequences. All these affirmed that the seven blaNDM-5 genes were harbored on mobile IncX3 plasmids which likely caused the rapid spread of blaNDM in China (Ma et al., 2020). The low fitness cost of blaNDM-5-carrying IncX3 plasmid may demonstrate why it can become the predominantly transferred plasmid (Ma et al., 2020). There is no doubt that eight blaNDM-5 harboring ESBL-EC may transfer to human by the food chain, resulting in a potential threat to public health.
The genetic environment showed that all blaNDM-5 genes were flanked by the same genetic structure ΔISAba125-IS5-blaNDM-5-bleMBL-trpF-dsbC-IS26, which is a common blaNDM genetic environment (Liu et al., 2020). Although it was regarded as being responsible for the horizontal transfer and integration of blaNDM-5 (Liu et al., 2020), further evidence is still needed to support this view. Notably, the genomes blast revealed that the dsbC and ΔISAba125 regions of three blaNDM-5 plasmid genomes contained deletions, with 500-bp absent from ΔISAba125 and 180 bp deleted in dsbC, compared with the other four isolates. This is a significant and powerful evidence that ΔISAba125 and dsbC were unstable, and ever in flux, and indicated that blaNDM as a exogenous gene was recombined into the IncX3 plasmid by insertion or transposition based on IS structure. The blaNDM is originally from Acinetobacter spp. and harbored in complete transposition structure Tn125, including a complete ISAba125 on each end (Bontron et al., 2016). Subsequently, the Tn125 composite transposon slightly changed due to each ISAba125 element by the part integration of IS26, forming an intermediate composite transposon Tn125-IS26 which is adjacent to ΔISAba125 and IS26 at both termini (Weber et al., 2019). With either terminal ISAba125 completely replaced by IS26, a new composite structure ΔISAba125-IS26 is generated, and IS26 becomes the predominant element responsible for blaNDM transfer (Liu et al., 2020). However, analysis of the travel history of genetic organizations of blaNDM indicates that it currently occurs mainly in IncN/A/C type plasmids rather than IncX3 (Kopotsa et al., 2020). All these still warn us that the genetic organizations of blaNDM are being simplified. Now, a novel evolutionary trajectory of blaNDM genetic environment evolution is taking place in IncX3 plasmids in our study, where ΔISAba125 is truncated by deletion of 500-bp sequences without IS26 insertion and forms a shorter ΔΔISAba125, which, as far as we know, is the first report. Unexpectedly, a 180-bp segment was also lost in the dsbC gene downstream of the blaNDM gene, but dsbC is a gene encoding an oxidoreductase rather than a transposable element. Nevertheless, it seems to suggest instability and shortening of the blaNDM genetic locus. Accordingly, the occurrence of segment deletions in the ISAba125 region and even the dsbC gene gives us reason to hypothesize that blaNDM loci are evolving to “pack light” to facilitate rapid and stable transfer.
ST and PFGE diversity were observed in the major ESBL-EC in our study, and shared STs and PFGE patterns were exhibited. The presence of similar ST and PFGE types indicated that it is possible for ESBL-EC to transfer among the different food animals. Presently, ST131 has become a major ST across the globe for ESBL-EC (Marie-Hélène et al., 2014), especially those carrying blaCTX-M. There is widespread ST131 in both humans and animals (Belas et al., 2019), especially in chicken, swine, and cattle (Duggett et al., 2021). However, there were no ST131 isolates identified in our study, while a distribution of types was noted including ST48 (n = 5), ST189 (n = 5), ST206 (n = 4), ST6396 (n = 3), ST10 (n = 3), and ST155 (n = 3). The same five ST189 isolates included one from swine and four from chicken. Notably, four ST189 from chicken harbored the same resistance genes, namely, blaCTX-M-65, blaOXA-10, blaNDM-5, and mcr-1. ST189 E. coli mainly spread in poultry and is thought to have spread from food animals to human (Kim et al., 2015; Hasan et al., 2016). Previously, ST189 ESBL-EC was reported to be associated with blaCTX-M (Hasan et al., 2016), in agreement with our study. Recently, a series of reports revealed mcr-1 was found in ST189 E. coli from food animals and human (Zając et al., 2019; Yamaguchi et al., 2020), suggesting that ST189 ESBL-EC have become the reservoirs, even and preferred clones of mcr genes, as we also found. However, blaNDM or variants were hardly reported in ST189 E. coli, indicating that our study revealed novel findings on blaNDM in ST189 ESBL-EC. The occurrence of ST206 E. coli was mainly in poultry farms, which often carried the blaESBLs. Recently, Ayeni et al. reported that 35 (68.6%) ST206 E. coli harbored blaESBLs in poultry in Nigeria (Ayeni et al., 2020), and CTX-M-27 was deemed to be almost exclusively found in ST206 isolates (Ayeni et al., 2020). ST206 E. coli was often identified in carbapenemase-producing CRE or colistin-resistant isolates from animal, environment, and human, often harboring blaNDM or mcr-1, sometimes even with coexisting blaNDM and/or mcr-1 genes. ST206 E. coli harboring blaNDM or/and mcr-1 have been found in acute diarrhea patients or outpatients (Zheng et al., 2018), retail vegetables (Luo et al., 2017), pig, and water samples (Li et al., 2019). This was also observed in our study, where two ST206 E. coli were derived from food animals. All these suggested that mcr and/or blaNDM-carrying ST206 E. coli can transfer to human by the food production chain (Kawamura et al., 2017; Li et al., 2019). On the other hand, ST48 ESBL-EC was present in three animal species, but these displayed different PFGE patterns, suggesting that they were different strains. ST48 was reported in human and animal isolates as associated with ESBL lactamases, especially the CTX-M type (Chen et al., 2019; Nüesch-Inderbinen et al., 2020), but we did not find the latter in any of our ST48 isolates. ST10 is considered to be the second most prevalent clone after ST131 in ESBL-EC worldwide and is mainly spread among livestock and poultry (Aibinu et al., 2012), such as swine, chicken, and cattle (Pietsch et al., 2017). This was supported by our research, as two ST10 ESBL-EC were from cattle and the other one was from swine. ST10 with blaCTX-M was not found in our study as the three ST10 ESBL-EC did not carry this gene. Similarly, ST155 was previously detected in CTX-M-1-producing E. coli strains, while no blaCTX-M was observed in our three ST155 ESBL-EC. Currently, the major prevalent source of ST155 ESBL-EC is chicken and its environments. Like ST155, ST6396 is also a niche prevalent clone, but detected in human and animals (Zhao et al., 2021). In general, shared ST clones will become more frequent in food animals. In other words, the diversity of ESBL-EC would be more common in any food animal in the future.
In our study, ESBL-EC showed high prevalence and diverse isolate lineages among chicken, swine, and cattle, together with a significant difference in prevalence rates of blaESBL genes and resistance determinants. All these indicate that farm animals have become reservoirs of ESBL-EC and facilitators of blaESBL gene transfers and which can also show genetic environment change. Meanwhile, the coexistence of blaNDM and/or mcr has emerged in ESBL-EC among food animals, posing further threats to human health. Accordingly, it is time for more effective measures to monitor the prevalence of ESBL-EC in different food animals, reducing the risk of transmission to humans.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject accession no. PRJNA784017 including JAJNDN000000000, JAJNDO000000000, JAJNDP000000000, JAJNDQ000000000, JAJNDR000000000, 572 JAJNDS000000000, JAJNDT000000000 (https://www.ncbi.nlm.nih.gov/genbank/).
The animal study was reviewed and approved by Qingdao Agricultural University Animal Experiment Committee. Written informed consent was obtained from the owners for the participation of their animals in this study.
ZH and ZL are responsible for the study design. KW, ZL, LX, LQ, LZ, CG, and XL assisted in the data collection. KW and ZL interpreted the data. ZL and YZ completed the written report. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Plan (No. 2016YFD0501007); National Natural Science Foundation of China (No. 32172911); 100 foreign experts and teams plan (No. WST2017010); Shandong Provincial Natural Science Foundation, China (No. ZR2020QC199); and China Postdoctoral Science Foundation (No. 2021M701105).
Author YZ was employed by Shandong New Hope Liuhe Group Ltd. Author KW was employed by company Autobio Labtec Instruments Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.755545/full#supplementary-material
Aibinu, I., Odugbemi, T., Koenig, W., Ghebremedhin, B. (2012). Sequence Type ST131 and ST10 Complex (ST617) Predominant Among CTX-M-15-Producing Escherichia Coli Isolates From Nigeria. Clin. Microbiol. Infect. 18 (3), E49–E51. doi: 10.1111/j.1469-0691.2011.03730.x
Allcock, S., Young, E. H., Holmes, M., Gurdasani, D., Dougan, G., Sandhu, M. S., et al. (2017). Antimicrobial Resistance in Human Populations: Challenges and Opportunities. Glob. Health Epidemiol. Genom. 2, 135–137. doi: 10.1017/gheg.2017.4
Ayeni, F. A., Falgenhauer, J., Schmiedel, J., Schwengers, O., Chakraborty, T., Falgenhauer, L. (2020). Detection of blaCTX-M-27-Encoding Escherichia Coli ST206 in Nigerian Poultry Stocks. J. Antimicrob. Chemoth. 75 (10), 3070–3072. doi: 10.1093/jac/dkaa293
Belas, A., Marques, C., Aboim, C., Pomba, C. (2019). Emergence of Escherichia Coli ST131 H 30/H 30-Rx Subclones in Companion Animals. J. Antimicrob. Chemoth. 74, 266–269. doi: 10.1093/jac/dky381
Bevan, E. R., Jones, A. M., Hawkey, P. M. (2017). Global Epidemiology of CTX-M β-Lactamases: Temporal and Geographical Shifts in Genotype. J. Antimicrob. Chemoth. 72 (8), 2145–2155. doi: 10.1093/jac/dkx146
Bontron, S., Nordmann, P., Poirel, L. (2016). Transposition of Tn125 Encoding the NDM-1 Carbapenemase in Acinetobacter Baumannii. Antimicrob. Agents Chemother. 60 (12), 7245–7251. doi: 10.1128/AAC.01755-16
Bush, K. (2018). Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 62, e01076–e01018. doi: 10.1128/AAC.01076-18
Bush, K., Bradford, P. A. (2020). Epidemiology of Beta-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 33 (2), e19–e47. doi: 10.1128/CMR.00047-19
Caltagirone, M., Nucleo, E., Spalla, M., Zara, F., Novazzi, F., Marchetti, V. M., et al. (2017). Occurrence of Extended Spectrum β-Lactamases, KPC-Type, and Mcr-1.2-Producing Enterobacteriaceae From Wells, River Water, and Wastewater Treatment Plants in Oltrepò Pavese Area, Northern Italy. Front. Microbiol. 8, 2232. doi: 10.3389/fmicb.2017.02232
Carattoli, A. (2008). Animal Reservoirs for Extended Spectrum β-Lactamase Producers. Clin. Microbiol. Infect. 14, 117–123. doi: 10.1111/j.1469-0691.2007.01851.x
Casella, T., Haenni, M., Madela, N. K., Andrade, L. K. D., Pradela, L. K., Andrade, L. N. D., et al. (2018). Extended-Spectrum Cephalosporin-Resistant Escherichia Coli Isolated From Chickens and Chicken Meat in Brazil is Associated With Rare and Complex Resistance Plasmids and Pandemic St Lineages. J. Antimicrob. Chemoth. 73 (12), 3293–3297. doi: 10.1093/jac/dky335
Chatterjee, S., Datta, S., Roy, S., Ramanan, L., Saha, A., Viswanathan, R., et al. (2016). Carbapenem Resistance in Acinetobacter Baumannii and Other Acinetobacter Spp. Causing Neonatal Sepsis: Focus on NDM-1 and its Linkage to Isaba125. Front. Microbiol. 7:1126. doi: 10.3389/fmicb.2016.01126
Chen, Y., Liu, Z., Zhang, Y., Zhang, Z., Lei, L., Xia, Z. (2019). Increasing Prevalence of ESBL-Producing Multidrug Resistance Escherichia Coli From Diseased Pets in Beijing, China From 2012 to 2017. Front. Microbiol. 10, 2852. doi: 10.3389/fmicb.2019.02852
Chong, Y., Shimoda, S., Shimono, N. (2018). Current Epidemiology, Genetic Evolution and Clinical Impact of Extended-Spectrum β-Lactamase-Producing Escherichia Coli and Klebsiella Pneumoniae. Infect. Genet. Evol. 61, 185–188. doi: 10.1016/j.meegid.2018.04.005
Clinical and laboratory standards institute (CLSI) (2021). Performance Standards for Antimicrobial Susceptibility Testing. 31th (Wayne, PA: Clinical and Laboratory Standards Institute). Available at: https://clsi.org/standards/products/microbiology/documents/m100/. M100.
Dame-Korevaar, A., Fischer, E. A., van der Goot, J., Stegeman, A., Mevius, D. (2019). Transmission Routes of ESBL/pAmpc Producing Bacteria in the Broiler Production Pyramid, a Literature Review. Prev. Vet. Med. 162, 136–150. doi: 10.1016/j.prevetmed.2018.12.002
Day, M. J., Hopkins, K. L., Wareham, D. W., Toleman, M. A., Elviss, N., Randall, L., et al. (2019). Extended-Spectrum β-Lactamase-Producing Escherichia Coli in Human-Derived and Food Chain-Derived Samples From England, Wales, and Scotland: An Epidemiological Surveillance and Typing Study. Lancet Infect. Dis. 19, 1325–1335. doi: 10.1016/S1473-3099(19)30273-7
de Alcântara Rodrigues, I., Ferrari, R. G., Panzenhagen, P., Mano, S. B., Conte-Junior, C. A. (2020). Antimicrobial Resistance Genes in Bacteria From Animal-Based Foods. Adv. Appl. Microbiol. 112, 143–183. doi: 10.1016/bs.aambs.2020.03.001
Dohmen, W., Bonten, M. J., Bos, M. E., van Marm, S., Scharringa, J., Wagenaar, J. A., et al. (2015). Carriage of Extended-Spectrum β-Lactamases in Pig Farmers is Associated With Occurrence in Pigs. Clin. Microbiol. Infect. 21 (10), 917–923. doi: 10.1016/j.cmi.2015.05.032
Dorado-García, A., Smid, J. H., Pelt, V. W., Mjm, B., Veldman, K. T. (2018). Molecular Relatedness of Esbl/Ampc-Producing Escherichia Coli From Humans, Animals, Food and the Environment: A Pooled Analysis. J. Antimicrob. Chemoth. 73 (2), 339–347. doi: 10.1093/jac/dkx397
Duggett, N., Ellington, M. J., Hopkins, K. L., Ellaby, N., Randall, L., Lemma, F., et al. (2021). Detection in Livestock of the Human Pandemic Escherichia Coli ST131 Fimh30(R) Clone Carrying blaCTX-M-27. J. Antimicrob. Chemother. 76 (1), 263–265. doi: 10.1093/jac/dkaa407
Friedrich, M. J. (2019). Un: All-Out Effort Needed to Combat Antimicrobial Resistance. Jama 321 (23), 2273. doi: 10.1001/jama.2019.7977
Fu, Y., Xu, X., Zhang, L., Xiong, Z., Ma, Y., Wei, Y., et al. (2020). Fourth Generation Cephalosporin Resistance Among Salmonella Enterica Serovar Enteritidis Isolates in Shanghai, China Conferred by blaCTX-M-55 Harboring Plasmids. Front. Microbiol. 11, 910. doi: 10.3389/fmicb.2020.00910
Ghafourian, S., Sadeghifard, N., Soheili, S., Sekawi, Z. (2015). Extended Spectrum Beta-Lactamases: Definition, Classification and Epidemiology. Curr. Issues Mol. Biol. 17 (1), 11–22. doi: 10.21775/cimb.017.011
Guerra, B., Fischer, J., Helmuth, R. (2014). An Emerging Public Health Problem: Acquired Carbapenemase-Producing Microorganisms are Present in Food-Producing Animals, Their Environment, Companion Animals and Wild Birds. Vet. Microbiol. 171, 290–297. doi: 10.1016/j.vetmic.2014.02.001
Hasan, B., Laurell, K., Rakib, M. M., Ahlstedt, E., Hernandez, J., Caceres, M., et al. (2016). Fecal Carriage of Extended-Spectrum Beta-Lactamases in Healthy Humans, Poultry, and Wild Birds in León, Nicaragua-a Shared Pool of blaCTX-M Genes and Possible Interspecies Clonal Spread of Extended-Spectrum Beta-Lactamases-Producing Escherichia Coli. Microb. Drug Resist. 22 (8), 682–687. doi: 10.1089/mdr.2015.0323
Hawser, S. P., Bouchillon, S. K., Hoban, D. J., Badal, R. E., Hsueh, P., Paterson, D. L. (2009). Emergence of High Levels of Extended-Spectrum-β-Lactamase-Producing Gram-Negative Bacilli in the Asia-Pacific Region: Data From the Study for Monitoring Antimicrobial Resistance Trends (SMART) Program 2007. Antimicrob. Agents Chemother. 53, 3280–3284. doi: 10.1128/AAC.00426-09
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of Plasmid-Mediated High-Level Tigecycline Resistance Genes in Animals and Humans. Nat. Microbiol. 4 (9), 1450–1456. doi: 10.1038/s41564-019-0445-2
Ho, P., Chow, K. H., Lai, E. L., Lo, W., Yeung, M. K., Chan, J., et al. (2011). Extensive Dissemination of CTX-M-Producing Escherichia Coli With Multidrug Resistance to ‘Critically Important’antibiotics Among Food Animals in Hong Kong 2008–2010. J. Antimicrob. Chemoth. 66 (4), 765–768. doi: 10.1093/jac/dkq539
Jean, S., Coombs, G., Ling, T., Balaji, V., Rodrigues, C., Mikamo, H., et al. (2016). Epidemiology and Antimicrobial Susceptibility Profiles of Pathogens Causing Urinary Tract Infections in the Asia-Pacific Region: Results From the Study for Monitoring Antimicrobial Resistance Trends (SMART), 2010–2013. Int. J. Antimicrob. Agents 47, 328–334. doi: 10.1016/j.ijantimicag.2016.01.008
Jiang, W., Men, S., Kong, L., Ma, S., Yang, Y., Wang, Y., et al. (2017). Prevalence of Plasmid-Mediated Fosfomycin Resistance Gene Fosa3 Among CTX-M-Producing Escherichia Coli Isolates From Chickens in China. Foodborne Pathog. Dis. 14 (4), 210–218. doi: 10.1089/fpd.2016.2230
Jones, R. N., Guzman-Blanco, M., Gales, A. C., Gallegos, B., Castro, A. L. L., Martino, M. D. V., et al. (2013). Susceptibility Rates in Latin American Nations: Report From a Regional Resistance Surveillance Progra). Braz. J. Infect. Dis. 17, 672–681. doi: 10.1016/j.bjid.2013.07.002
Kawamura, K., Nagano, N., Suzuki, M., Wachino, J., Kimura, K., Arakawa, Y. (2017). ESBL-Producing Escherichia Coli and its Rapid Rise Among Healthy People. Food Saf. (Tokyo). 5 (4), 122–150. doi: 10.14252/foodsafetyfscj.2017011
Kim, J., Guk, J., Mun, S., An, J., Song, H., Kim, J., et al. (2019). Metagenomic Analysis of Isolation Methods of a Targeted Microbe, Campylobacter Jejuni, From Chicken Feces With High Microbial Contamination. Microbiome 7 (1), 67. doi: 10.1186/s40168-019-0680-z
Kim, T., Kim, Y., Kim, S., Lee, J., Park, C., Kim, H. (2010). Identification and Distribution of Bacillus Species in Doenjang by Whole-Cell Protein Patterns and 16s rRNA Gene Sequence Analysis. J. Microbiol. Biotechn. 20 (8), 1210–1214. doi: 10.4014/jmb.1002.02008
Kim, S., Sung, J. Y., Choi, S. (2015). Molecular Characterization of Escherichia Coli Isolates From Humans and Chickens in the Chungcheong Area Using MLST Analysis. Korean J. Clin. Lab. Sci. 47 (2), 71–77. doi: 10.15324/kjcls.2015.47.2.71
Kola, A., Kohler, C., Pfeifer, Y., Schwab, F., Kühn, K., Schulz, K., et al. (2012). High Prevalence of Extended-Spectrum-β-Lactamase-Producing Enterobacteriaceae in Organic and Conventional Retail Chicken Meat, Germany. J. Antimicrob. Chemother. 146 (1-2), 2631–2634. doi: 10.1093/jac/dks295
Kopotsa, K., Mbelle, N. M., Sekyere, J. O. (2020). Epigenomics, Genomics, Resistome, Mobilome, Virulome and Evolutionary Phylogenomics of Carbapenem-Resistant Klebsiella Pneumoniae Clinical Strains. Microbial. Genomics 6 (12), mgen000474. doi: 10.1099/mgen.0.000474
Ling, Z., Yin, W., Shen, Z., Wang, Y., Shen, J., Walsh, T. R. (2020). Epidemiology of Mobile Colistin Resistance Genes Mcr-1 to Mcr-9. J. Antimicrob. Chemoth. 75 (11), 3087–3095. doi: 10.1093/jac/dkaa205
Liu, Z., Li, J., Wang, X., Liu, D., Ke, Y., Wang, Y., et al. (2018). Novel Variant of New Delhi Metallo-β-Lactamase, NDM-20, in Escherichia Coli. Front. Microbiol. 9, 248. doi: 10.3389/fmicb.2018.00248
Liu, Z., Wang, Y., Walsh, T. R., Liu, D., Shen, Z., Zhang, R., et al. (2017). Plasmid-Mediated Novel blaNDM-17 Gene Encoding a Carbapenemase With Enhanced Activity in a Sequence Type 48 Escherichia Coli Strain. Antimicrob. Agents Chemother. 61 (5), e2216–e2233. doi: 10.1128/AAC.02233-16
Liu, Y., Wang, Y., Walsh, T. R., Yi, L., Zhang, R., Spencer, J., et al. (2016). Emergence of Plasmid-Mediated Colistin Resistance Mechanism Mcr-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 16 (2), 161–168. doi: 10.1016/S1473-3099(15)00424-7
Liu, Z., Xiao, X., Liu, Y., Li, R., Wang, Z. (2020). Recombination of NDM-5-Producing Plasmids Mediated by IS26 Among Escherichia Coli. Int. J. Antimicrob. Ag. 55 (1), 105815. doi: 10.1016/j.ijantimicag.2019.09.019
Liu, Z., Zhang, R., Li, W., Yang, L., Liu, D., Wang, S., et al. (2019). Amino Acid Changes at the Vim-48 C-Terminus Result in Increased Carbapenem Resistance, Enzyme Activity and Protein Stability. J. Antimicrob. Chemoth. 74 (4), 885–893. doi: 10.1093/jac/dky536
Li, R., Zhang, P., Yang, X., Wang, Z., Fanning, S., Wang, J., et al. (2019). Identification of a Novel Hybrid Plasmid Coproducing Mcr-1 and Mcr-3 Variant From an Escherichia Coli Strain. J. Antimicrob. Chemoth. 74 (6), 1517–1520. doi: 10.1093/jac/dkz058
Luo, J., Yao, X., Lv, L., Doi, Y., Huang, X., Huang, S., et al. (2017). Emergence of Mcr-1 in Raoultella Ornithinolytica and Escherichia Coli Isolates From Retail Vegetables in China. Antimicrob. Agents Chemother. 61 (10), e1117–e1139. doi: 10.1128/AAC.01139-17
Ma, T., Fu, J., Xie, N., Ma, S., Lei, L., Zhai, W., et al. (2020). Fitness Cost of blaNDM-5-Carrying P3r-IncX3 Plasmids in Wild-Type NDM-Free Enterobacteriaceae. Microorganisms 8 (3), 377. doi: 10.3390/microorganisms8030377
Magiorakos, A., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 18 (3), 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Marie-Hélène, N. C., Xavier, B., Jean-Yves, M. (2014). Escherichia Coli ST131, an Intriguing Clonal Group. Clin. Microbiol. Rev. 27 (3), 543–574. doi: 10.1128/CMR.00125-13
Mather, A. E., Matthews, L., Mellor, D. J., Reeve, R., Denwood, M. J., Boerlin, P., et al. (2012). An Ecological Approach to Assessing the Epidemiology of Antimicrobial Resistance in Animal and Human Populations. Proc. Biol. Sci. 279, 1630–1639. doi: 10.1098/rspb.2011.1975
Müller, A., Stephan, R., Nüesch-Inderbinen, M. (2016). Distribution of Virulence Factors in ESBL-Producing Escherichia Coli Isolated From the Environment, Livestock, Food and Humans. Sci. Total Environ. 541, 667–672. doi: 10.1016/j.scitotenv.2015.09.135
Musicha, P., Cornick, J. E., Bar-Zeev, N., French, N., Masesa, C., Denis, B., et al. (2017). Trends in Antimicrobial Resistance in Bloodstream Infection Isolates at a Large Urban Hospital in Malaw-2016): A Surveillance Study. Lancet Infect. Dis. 17 (10), 1042. doi: 10.1016/S1473-3099(17)30394-8
Nordmann, P., Poirel, L., Walsh, T. R., Livermore, D. M. (2011). The Emerging NDM Carbapenemases. Trends Microbiol. 19 (12), 588–595. doi: 10.1016/j.tim.2011.09.005
Nüesch-Inderbinen, M., Kindle, P., Baschera, M., Liljander, A., Jores, J., Corman, V. M., et al. (2020). Antimicrobial Resistant and Extended-Spectrum ß-Lactamase (ESBL) Producing Escherichia Coli Isolated From Fecal Samples of African Dromedary Camels. Sci. Afr. 7, e274. doi: 10.1016/j.sciaf.2020.e00274
O’Neill, J. (2016). Review on Antimicrobial Resistance: Tackling Drug-Resistant Infections Globally: Final Report and Recommendations (Government of the United Kingdom: London, United Kingdom). Available at: https://apo.org.au/node/63983.
Okpara, E. O., Ojo, O. E., Awoyomi, O. J., Dipeolu, M. A., Oyekunle, M. A., Schwarz, S. (2018). Antimicrobial Usage and Presence of Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae in Animal-Rearing Households of Selected Rural and Peri-Urban Communities. Vet. Microbiol. 218, 31–39. doi: 10.1016/j.vetmic.2018.03.013
Pietsch, M., Eller, C., Wendt, C., Holfelder, M., Falgenhauer, L., Fruth, A., et al. (2017). Molecular Characterisation of Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia Coli Isolates From Hospital and Ambulatory Patients in Germany. Vet. Microbiol. 200, 130–137. doi: 10.1016/j.vetmic.2015.11.028
Pimenta, A. C., Fernandes, R., Moreira, I. S. (2014). Evolution of Drug Resistance: Insight on TEM β-Lactamases Structure and Activity and β-Lactam Antibiotics. Mini. Rev. Med. Chem. 14, 111–122. doi: 10.2174/1389557514666140123145809
Pitout, J., Laupland, K. (2008). Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae: An Emerging Public-Health Concern. Lancet Infect. Dis. 8 (3), 159–166. doi: 10.1016/S1473-3099(08)70041-0
Poirel, L., Walsh, T. R., Cuvillier, V., Nordmann, P. (2011). Multiplex PCR for Detection of Acquired Carbapenemase Genes. Diagn. Micr. Infect. Dis. 70 (1), 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Randall, L. P., Lodge, M. P., Elviss, N. C., Lemma, F. L., Hopkins, K. L., Teale, C. J., et al. (2017). Evaluation of Meat, Fruit and Vegetables From Retail Stores in Five United Kingdom Regions as Sources of Extended-Spectrum Beta-Lactamase (ESBL)-Producing and Carbapenem-Resistant Escherichia Coli. Int. J. Food Microbiol. 241, 283–290. doi: 10.1016/j.ijfoodmicro.2016.10.036
Rao, L., Lv, L., Zeng, Z., Chen, S., He, D., Chen, X., et al. (2014). Increasing Prevalence of Extended-Spectrum Cephalosporin-Resistant Escherichia Coli in Food Animals and the Diversity of CTX-M Genotypes During 2003–2012. Vet. Microbiol. 172 (3-4), 534–541. doi: 10.1016/j.vetmic.2014.06.013
Reuland, E. A., Al Naiemi, N., Raadsen, S. A., Savelkoul, P., Kluytmans, J., Vandenbroucke-Grauls, C. (2014). Prevalence of ESBL-Producing Enterobacteriaceae in Raw Vegetables. Eur. J. Clin. Microbiol. 33 (10), 1843–1846. doi: 10.1007/s10096-014-2142-7
Runcharoen, C., Raven, K. E., Reuter, S., Kallonen, T., Paksanont, S., Thammachote, J., et al. (2017). Whole Genome Sequencing of ESBL-Producing Escherichia Coli Isolated From Patients, Farm Waste and Canals in Thailand. Genome Med. 9 (1), 81. doi: 10.1186/s13073-017-0471-8
Sanjit Singh, A., Lekshmi, M., Prakasan, S., Nayak, B., Kumar, S. (2017). Multiple Antibiotic-Resistant, Extended Spectrum-β-Lactamase (ESBL)-Producing Enterobacteria in Fresh Seafood. Microorganisms 5, 53. doi: 10.3390/microorganisms5030053
Schmithausen, R. M., Schulze-Geisthoevel, S. V., Stemmer, F., El-Jade, M., Reif, M., Hack, S., et al. (2015). Analysis of Transmission of MRSA and ESBL-EC Among Pigs and Farm Personnel. PloS One 10 (9), e138173. doi: 10.1371/journal.pone.0138173
Seiffert, S. N., Hilty, M., Perreten, V., Endimiani, A. (2013). Extended-Spectrum Cephalosporin-Resistant Gram-Negative Organisms in Livestock: An Emerging Problem for Human Health? Drug Resist. Update 16 (1-2), 22–45. doi: 10.1016/j.drup.2012.12.001
Sen, S., Sarkar, K. (2018). Screening for ESBL Producing Bacterial Isolates of Agricultural Soil and Profiling for Multidrug Resistance. Ann. Agrar. Sci. 16 (3), 272–280. doi: 10.1016/j.aasci.2018.04.005
Song, J., Oh, S., Kim, J., Park, S., Shin, J. (2020). Clinically Relevant Extended-Spectrum Beta-Lactamase-Producing Escherichia Coli Isolates From Food Animals in South Korea. Front. Microbiol. 11, 604. doi: 10.3389/fmicb.2020.00604
Sun, J., Chen, C., Cui, C., Zhang, Y., Liu, X., Cui, Z., et al. (2019). Plasmid-Encoded Tet (X) Genes That Confer High-Level Tigecycline Resistance in Escherichia Coli. Nat. Microbiol. 4 (9), 1457–1464. doi: 10.1038/s41564-019-0496-4
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting Chromosomal DNA Restriction Patterns Produced by Pulsed-Field Gel Electrophoresis: Criteria for Bacterial Strain Typing. J. Clin. Microbiol. 33, 2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995
Urbánek, K., Kolář, M., Lovečková, Y., Strojil, J., Šantavá, L. (2007). Influence of Third-Generation Cephalosporin Utilization on the Occurrence of ESBL-Positive Kebsiella Pneumoniae Strains. J. Clin. Pharm. Ther. 32 (4), 403–408. doi: 10.1111/j.1365-2710.2007.00836.x
Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., et al. (2015). Global Trends in Antimicrobial Use in Food Animals. Proc. Natl. Acad. Sci. U. S. A. 112 (18), 5649–5654. doi: 10.1073/pnas.1503141112
Wang, Q., Mo, C., Li, Y., Gao, P., Tai, Y., Zhang, Y., et al. (2010). Determination of Four Fluoroquinolone Antibiotics in Tap Water in Guangzhou and Macao. Environ. Pollut. 158 (7), 2350–2358. doi: 10.1016/j.envpol.2010.03.019
Wang, Y., Zhang, R., Li, J., Wu, Z., Yin, W., Schwarz, S., et al. (2017). Comprehensive Resistome Analysis Reveals the Prevalence of NDM and Mcr-1 in Chinese Poultry Production. Nat. Microbiol. 2 (4), 1–7. doi: 10.1038/nmicrobiol.2016.260
Weber, R. E., Pietsch, M., Frühauf, A., Pfeifer, Y., Martin, M., Luft, D., et al. (2019). IS26-Mediated Transfer of blaNDM-1 as the Main Route of Resistance Transmission During a Polyclonal, Multispecies Outbreak in a German Hospital. Front. Microbiol. 10, 2817. doi: 10.3389/fmicb.2019.02817
Wei, R., Ge, F., Huang, S., Chen, M., Wang, R. (2011). Occurrence of Veterinary Antibiotics in Animal Wastewater and Surface Water Around Farms in Jiangsu Province, China. Chemosphere 82 (10), 1408–1414. doi: 10.1016/j.chemosphere.2010.11.067
White, D. G., Zhao, S., Sudler, R., Ayers, S., Friedman, S., Chen, S., et al. (2001). The Isolation of Antibiotic-Resistant Salmonella From Retail Ground Meats. N. Engl. J. Med. 345 (16), 1147–1154. doi: 10.1056/NEJMoa010315
Wittekamp, B. H., Plantinga, N. L., Cooper, B. S., Lopez-Contreras, J., Coll, P., Mancebo, J., et al. (2018). Decontamination Strategies and Bloodstream Infections With Antibiotic-Resistant Microorganisms in Ventilated Patients: A Randomized Clinical Trial. JAMA 320 (20), 2087–2098. doi: 10.1001/jama.2018.13765
Woolhouse, M. E. (2006). Where do Emerging Pathogens Come From? Microbe 1 (11), 511. doi: 10.1128/microbe.1.511.1
Wu, W., Feng, Y., Tang, G., Qiao, F., McNally, A., Zong, Z. (2019). NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 32 (2), e00115–e00118. doi: 10.1128/CMR.00115-18
Wu, C., Wang, Y., Shi, X., Wang, S., Ren, H., Shen, Z., et al. (2018). Rapid Rise of the ESBL and Mcr-1 Genes in Escherichia Coli of Chicken Origin in China 2008–2014. Emerg. Microbes Infect. 7, 1–10. doi: 10.1038/s41426-018-0033-1
Yamaguchi, T., Kawahara, R., Hamamoto, K., Hirai, I., Khong, D. T., Nguyen, T. N., et al. (2020). High Prevalence of Colistin-Resistant Escherichia Coli With Chromosomally Carried Mcr-1 in Healthy Residents in Vietnam. msphere 5 (2), e117–e120. doi: 10.1128/mSphere.00117-20
Yang, H., Rehman, M. U., Zhang, S., Yang, J., Li, Y., Gao, J., et al. (2019). High Prevalence of CTX-M Belonging to ST410 and ST889 Among ESBL Producing E. Coli Isolates From Waterfowl Birds in China’s Tropical Island, Hainan. Acta Trop. 194, 30–35. doi: 10.1016/j.actatropica.2019.03.008
Yang, Q., Zhang, H., Wang, Y., Xu, Y., Chen, M., Badal, R. E., et al. (2013). A 10 Years Surveillance for Antimicrobial Susceptibility of Escherichia Coli and Klebsiella Pneumoniae in Community- and Hospital-Associated Intra-Abdominal Infections in China. J. Med. Microbiol. 62, 1343–1349. doi: 10.1099/jmm.0.059816-0
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel Plasmid-Mediated Colistin Resistance Gene Mcr-3 in Escherichia Coli. mBio 8 (3), e517–e543. doi: 10.1128/mBio.00543-17
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a New Metallo-β-Lactamase Gene, blaNDM-1, and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella Pneumoniae Sequence Type 14 From India. Antimicrob. Agents Chemother. 53 (12), 5046–5054. doi: 10.1128/AAC.00774-09
Zając, M., Sztromwasser, P., Bortolaia, V., Leekitcharoenphon, P., Cavaco, L. M., Zitek-Barszcz, A., et al. (2019). Occurrence and Characterization of Mcr-1-Positive Escherichia Coli Isolated From Food-Producing Animals in Poland 2011-2016. Front. Microbiol. 10:1753. doi: 10.3389/fmicb.2019.01753
Zhang, C., Ding, X., Lin, X., Sun, R., Lu, Y., Cai, R., et al. (2019). The Emergence of Chromosomally Located blaCTX-M-55 in Salmonella From Foodborne Animals in China. Front. Microbiol. 10, 1268. doi: 10.3389/fmicb.2019.01268
Zhang, J., Zheng, B., Zhao, L., Wei, Z., Ji, J., Li, L., et al. (2014). Nationwide High Prevalence of CTX-M and an Increase of CTX-M-55 in Escherichia Coli Isolated From Patients With Community-Onset Infections in Chinese County Hospitals. BMC. Infect. Dis. 14 (1), 659. doi: 10.1186/s12879-014-0659-0
Zhao, Q., Berglund, B., Zou, H., Zhou, Z., Xia, H., Zhao, L., et al. (2021). Dissemination of blaNDM-5via IncX3 Plasmids in Carbapenem-Resistant Enterobacteriaceae Among Humans and in the Environment in an Intensive Vegetable Cultivation Area in Eastern China. Environ. Pollut. 273, 116370. doi: 10.1016/j.envpol.2020.116370
Zheng, B., Lv, T., Xu, H., Yu, X., Chen, Y., Li, J., et al. (2018). Discovery and Characterisation of an Escherichia Coli ST206 Strain Producing NDM-5 and MCR-1 From a Patient With Acute Diarrhoea in China. Int. J. Antimicrob. Ag. 51 (2), 273–275. doi: 10.1016/j.ijantimicag.2017.09.005
Zhou, Z., Alikhan, N., Mohamed, K., Fan, Y., Achtman, M., Brown, D., et al. (2020). The Enterobase User’s Guide, With Case Studies on Salmonella Transmissions, Yersinia Pestis Phylogeny, and Escherichia Core Genomic Diversity. Genome Res. 30 (1), 138–152. doi: 10.1101/gr.251678.119
Keywords: multidrug resistance, food animal, ESBL, Escherichia coli, NDM-5
Citation: Liu Z, Wang K, Zhang Y, Xia L, Zhao L, Guo C, Liu X, Qin L and Hao Z (2022) High Prevalence and Diversity Characteristics of blaNDM, mcr, and blaESBLs Harboring Multidrug-Resistant Escherichia coli From Chicken, Pig, and Cattle in China. Front. Cell. Infect. Microbiol. 11:755545. doi: 10.3389/fcimb.2021.755545
Received: 09 August 2021; Accepted: 13 December 2021;
Published: 07 February 2022.
Edited by:
Mingyu Wang, Shandong University, ChinaReviewed by:
Graciela Castro Escarpulli, Instituto Politécnico Nacional de México (IPN), MexicoCopyright © 2022 Liu, Wang, Zhang, Xia, Zhao, Guo, Liu, Qin and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihui Hao, aGFvemhpaHVpQGNhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.