
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 10 February 2022
Sec. Bacteria and Host
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.748461
This article is part of the Research TopicListeria: Environmental Adaption, Infection and Host InteractionsView all 7 articles
 Zhiting Ling1,2,3,4†
Zhiting Ling1,2,3,4† Dan Zhao1,2,3,4†
Dan Zhao1,2,3,4† Xinyu Xie1,2,3,4
Xinyu Xie1,2,3,4 Hao Yao1,2,3,4
Hao Yao1,2,3,4 Yuting Wang1,2,3,4
Yuting Wang1,2,3,4 Suwei Kong1,2,3,4
Suwei Kong1,2,3,4 Xiang Chen1,2,3,4
Xiang Chen1,2,3,4 Zhiming Pan1,2,3,4
Zhiming Pan1,2,3,4 Xin’an Jiao1,2,3,4*
Xin’an Jiao1,2,3,4* Yuelan Yin1,2,3,4*
Yuelan Yin1,2,3,4*The internalin family proteins, which carry the leucine repeat region structural motif, play diverse roles in Listeria monocytogenes (Lm) infection and pathogenesis. Although Internalin F, encoded by inlF, was identified more than 20 years ago, its role in the Lm anti-inflammatory response remains unknown. Lm serotype 4b isolates are associated with the majority of listeriosis outbreaks, but the function of InlF in these strains is not fully understood. In this study, we aimed to elucidate the role of inlF in modulating the inflammatory response and pathogenesis of the 4b strain Lm NTSN. Strikingly, although inlF was highly expressed at the transcriptional level during infection of five non-phagocytic cell types, it was not involved in adherence or invasion. Conversely, inlF did contributed to Lm adhesion and invasion of macrophages, and dramatically suppressed the expression of pro-inflammatory cytokines interleukin (IL)-1β and tumor necrosis factor (TNF-α). Consistent with the in vitro results, during Lm infection mice, inlF significantly inhibited the expression of IL-1β and IL-6 in the spleen, as well as IL-1β, IL-6, and TNF-α in the liver. More importantly, inlF contributed to Lm colonization in the spleen, liver, and ileum during the early stage of mouse infection via intragastric administration, inducing severe inflammatory injury and histopathologic changes in the late stage. To our knowledge, this is the first report to demonstrate that inlF mediates the inhibition of the pro-inflammatory response and contributes to the colonization and survival of Lm during the early stage of infection in mice. Our research partly explains the high pathogenicity of serovar 4b strains and will lead to new insights into the pathogenesis and immune evasion of Lm.
Listeria monocytogenes (Lm) is a ubiquitous foodborne pathogen with worldwide distribution, comprising of 14 serotypes that, are determined by the serological reaction between specific surface carbohydrate (O-) antigens and flagella (H-) antigens (Yin et al., 2019). Of the 14 serovars identified in the Lm species, serovars 1/2a, 1/2b, and 4b strains are responsible for over 95% isolations from clinical cases of human listeriosis, among which, 4b strains alone account for 49% of Listeria-related foodborne diseases (Goulet et al., 2006). Chen et al. reported that the risk factor of 4b strains causing listeriosis was 100-fold higher than that of 1/2a and 1/2c strains (Chen et al., 2007). Thus far, invasive 4b strains are responsible for economic losses in the animal husbandry industry and threaten human health. Therefore, unveiling the interaction between invasive 4b strain and host can better understand the pathogenic mechanism and contribute to control Listeria infection. Whereas, previous studies have mostly dealt with the identification and functional characterization of bacterial cell surface-associated virulence proteins in the Lm serovar group 1/2.
Surface proteins retained by the bacterial cell wall play a critical role in Lm infection and pathogenesis. Of particular importance are the internalin family proteins, which contain leucine-rich repeats (LRRs) that mediate entry into non-phagocytic cells, allowing membrane translocation and systemic infection. Two well-known internalin proteins InlA and InlB are required for bacterial adhesion and internalization. InlA interaction with its human host receptor E-cadherin mediates the crossing of the intestinal barrier (Sahu et al., 2007). InlB triggers bacterial entry by interacting with the hepatocyte growth factor receptor (Met), gC1q-R, and proteoglycans (Jonquières et al., 2001), thus contributing to Lm colonization in the liver and spleen (Lecuit et al., 2004). InlC is involved in the adhesion and invasion of microvascular endothelial cells as well as in the weakening of the innate immune response. InlJ is exclusively expressed in vivo during animal infection, where it interacts with mucin secreted by intestinal epithelial cells and contributes to the virulence of Lm (Sabet et al., 2005; Sabet et al., 2008). InlH is not involved in adhesion and invasion, but can interfere with the host immune response and contribute to immune evasion (Schubert et al., 2001). InlK protein can interact specifically with the fornix protein (MVP) in the cytoplasm to mediate bacterial autophagy escape in a manner that does not cross-react with ActA actin polymerization (Dortet et al., 2011). InlP is a newly discovered, secreted internalin protein that is important for listeriosis and has strong tropism for the placenta (Faralla et al., 2016). Recently, increasing attention has been paid to the integral role of InlF in regulating a series of processes related to infection. Although many members of the internalin family of virulence factors have been discovered, their functional elucidation is only the tip of an iceberg, with the mechanisms of Lm infection and host immune evasion still poorly understood.
Once Lm has colonized the gastrointestinal tract, immediate immune responses are essential for the clearance of these bacteria. An innate immune response is required to defend against early Lm infection ahead of the T-cell-mediated immune responses mounted to eradicate the intracellular bacteria (Unanue, 1997; Pamer, 2004; Stavru et al., 2011). Cytokines play a crucial role in the host’s activation of the innate immune system (Zharkova et al., 2017). It mainly includes the pro-inflammatory cytokine tumor necrosis factor (TNF)-α, which has an important role in inducing the T helper type 1 (Th1) immune response against cellular pathogens (Allie et al., 2013; Romagnoli et al., 2017). Interleukin (IL)-6 is a key cytokine in the control of inflammation, promoting T -cell differentiation, regulating the cellular immune response, and clearing intracellular Lm (Hoge et al., 2013). Recent studies have highlighted multiple strategies by which Lm evades innate defenses. For instance, N-deacetylation of the peptidoglycan in the bacterial cell wall helps Lm to avoid recognition by Toll-like receptor 2 and NOD-like receptors NOD1 and NOD2, thus escaping via manipulation of the host’s autophagic machinery (Boneca et al., 2007). Another survival strategy involves the Lm virulence factor listeriolysin O, which uses a mitochondrial sequence NOD-like receptor NLRX1 to hijack the host cell’s mitotic homeostasis system (Zhang Y. et al., 2019). Although there are couples of reports about inlF, its role in the inflammatory response has not yet been reported, further efforts are required to bridge this knowledge gap.
In this study, we investigated the role of inlF in the infection and pathogenesis of Lm serovar 4b strain NTSN, which was isolated from an ovine outbreak. We compared the transcriptional expression characteristics inlF and other internalin genes, examined the adhesion and invasion abilities of inlF in the wild type, deletion and complemented strain in five non-phagocytic cells and macrophages, and used in vitro and in vivo models to unravel the role of inlF in the modulation of the host innate immune response. To our knowledge, these results represent the first report of inlF-mediated interference with the innate immune response and contribution to Lm survival during the early stage of infection.
We isolated the virulent Lm strain NTSN from a case of ovine listeriosis. The Escherichia coli DH5α strain and pGEM-T and pAULA vectors were obtained from the Jiangsu Key Laboratory of Zoonosis, and the shuttle vector pKSV7 was kindly donated by Professor Zhu Guoqiang (Yangzhou University, China). The cell lines used in this study are listed in Supplementary Table 1 and were propagated in Dulbecco’s modified Eagle medium (Gibco, USA) supplemented with 10% fetal bovine serum (CLARK, USA).
Six-week-old female C57BL/6 and BALB/c mice were purchased from the Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Animal experiments were conducted following the guidelines laid down for the welfare and ethics of experimental animals. All animals were kept at the animal biosafety facilities and the study was conducted in accordance with procedures approved by the Institutional Animal Ethics Committee of Yangzhou University.
Total RNA from mouse tissues or bacterial cultures was extracted using an RNeasy Plus Mini Kit (Qiagen, Germany). The mRNA in mouse liver or spleen was collected using an RNA prep pure animal tissues Extraction Kit (Tiangen, China). Synthesis of cDNA was performed with a PrimeScript™ RT reagent kit protocol (Takara, China) using 1 µg total RNA. Primers used for quantitative real-time PCR (qRT-PCR) are listed in Supplementary Table 3. The qRT-PCR operation program followed the recommended thermal cycling conditions for the FastStart Universal SYBR Green Master reaction mix (Rox, Germany). To objectively reflect differences in gene expression, gyrb and gapdh were selected as internal reference genes. The 2ΔΔCt (ΔCt = Ctobjective gene − Ctreference gene) method was used to calculate the relative changes in expression.
To achieve homologous recombination, a plasmid carrying the inlF gene flanking regions inlFa and inlFb (pGEM-T-inlFab) was constructed by amplifying inlFa and inlFb by PCR using primers (inlFa-F/inlFa-R and inlFb-F/inlb-R) designed from the Lm NTSN genome sequence. Oligonucleotide primer sequences are provided in Supplementary Table 4. A plasmid carrying inlFab-(pKSV7-inlFab) was positively identified, introduced into a competent culture of Lm NTSN by electroporation in accordance with a previously described protocol (Park and Stewart, 1990), and cultured in an incubator at 30°C. Positive clones were screened by inlFW1/inlFW2 and the obtained mutant was named NTSNΔinlF.
To obtain the complemented strain NTSNΔinlF::inlF, a plasmid carrying inlFcd-(pAULA-inlFcd) was constructed using primers inlFc-F/inlFc-R and inlFd-F/inld-R as described above, then electroporated into NTSNΔinlF competent cells. Positive clones were screened by inlFW3/inlFW4 on brain heart infusion (BHI) plates containing 5 mg/L erythromycin (Sigma, USA) at 42°C.
All constructed vectors were verified by sequencing. The cDNA isolated from Lm NTSN, NTSNΔinlF and NTSNΔinlF::inlF was used as template for RT-PCR with primers inlF1/inlF2 to determine the deletion and reversion of inlF.
Bacterial from exponentially growing cultures of Lm NTSN, NTSN Δ inlF and NTSNΔ inlF::inlF were harvested and the optical density at 600 nm (OD600) was measured and adjusted to 0.05. Three parallel replicates were set for each strain. The OD600 value of each flask was measured at 1.5-h intervals.
Lm NTSN and NTSNΔinlF were scraped from plates using an inoculating loop and transferred into 5 ml of 0.45% normal saline. The bacterial turbidity was controlled at ∼1.0 with a nephelometer and biochemical characteristics were identified using a VITEK® 2 GN card (Biomérieux, France).
To compare the invasive capacities of Lm NTSN, NTSNΔinlF and NTSNΔinlF::inlF, five non-phagocytic cell lines (Caco-2 human intestinal epithelial cells, HepG2 human hepatocytes, Hela human cervical cancer cells, BRL3A murine hepatocytes, and NIH3T3 murine fibroblasts) and the RAW264.7 macrophage cell line passaged under stabilized growth conditions were seeded into 24-well cell culture plates and cultured for 12–16 h to form monolayers with ~90% confluence. Lm NTSN, NTSNΔinlF and NTSNΔinlF::inlF cultures were transferred to 10 ml fresh BHI broth (1:50 ratio) and grown at 37°C to an OD600 value of 0.8. Cell monolayers (5 × 105 cells) were infected with bacteria at a multiplicity of infection (MOI) of 20 for 1 h, after which the medium was removed and the cells were washed twice with phosphate-buffered saline (PBS). This was followed by the addition of 1 ml DMEM medium containing 100 μg/ml gentamicin sulfate, and incubation for 1 h to assess invasion ability (Yin et al., 2019). Cells were subsequently washed twice with PBS, 0.2% Triton X-100 was added and incubated for 9 min at room temperature to induce cell lysis. Serial dilutions of lysates in PBS were plated onto agar for 20 h of incubation at 37°C before determination of bacterial numbers. The bacterial invasion rate was calculated with the following formula: (number of bacteria internalized/number of initial bacteria in wells) × 100%.
Six-week-old female C57BL/6 mice were used to estimate organ colonization properties following intra-gastric inoculation with 1 × 109 colony-forming units (CFU) of Lm. Mice were given 15 mg/300 µl CaCO3 by gavage 30 min before the bacterial inoculation to neutralize gastric acid. Lm NTSN, NTSNΔinlF and NTSNΔinlF::inlF (n = 7/group) were evaluated at 6, 24 and 72 h post infection by determining bacterial loads in the ileum, spleen, and liver of mice. The ileum samples were treated as follows: a 2-centimeter-length was taken and washed with 5 ml PBS by syringe to remove the luminal contents (Yin et al., 2019). The ileum was homogenized, and serial dilutions of the organ homogenates were plated onto improved oxford medium (BD Diagnostics, USA) foundation plates. Serial dilutions of livers and spleens homogenates were plated onto BHI plate.
For the determination of inflammatory cytokines, 6-week-old C57BL/6 female mice were randomly divided into three groups and inoculated abdominally with Lm NTSN, NTSNΔinlF or NTSNΔinlF::inlF (100 μl) at a dose of 2 × 105 CFU. The spleen and liver of each mouse were harvested at 6 h post infection and used to extract total RNA. The transcriptional expression of cytokines IL-6, IL-10, TNF-α, and IL-1β was determined by qRT-PCR performed according to the specific steps described in Analysis of Gene Expression at the Transcriptional Level above.
Six-week-old BALB/c mice were dissected at 72 h post-intragastric inoculation with Lm at a dose of 1 × 109 CFU. Following the bacterial load assessment, samples of the spleen, liver, ileum and brain were separated and fixed in 12% formaldehyde to prepare histopathologic sections. The fixed sections were subjected to hematoxylin and eosin staining at WuXi AppTec Co., Ltd.
To understand the expression features of inlF, inlA, and inlB during Lm infection of multiple cell types (Supplementary Table 1), the mRNA transcriptional levels of each were determined and compared. Interestingly, the expression of inlF at the transcriptional level was higher than that of inlA and inlB (p <0.0001) during Lm infection of five non-phagocytic cell types, and was higher than inlB (p <0.01) during Lm invasion of RAW264.7 cells (Figures 1A–F). These results indicated that inlF may play an important role in the process of Lm invasion of various types of cells.
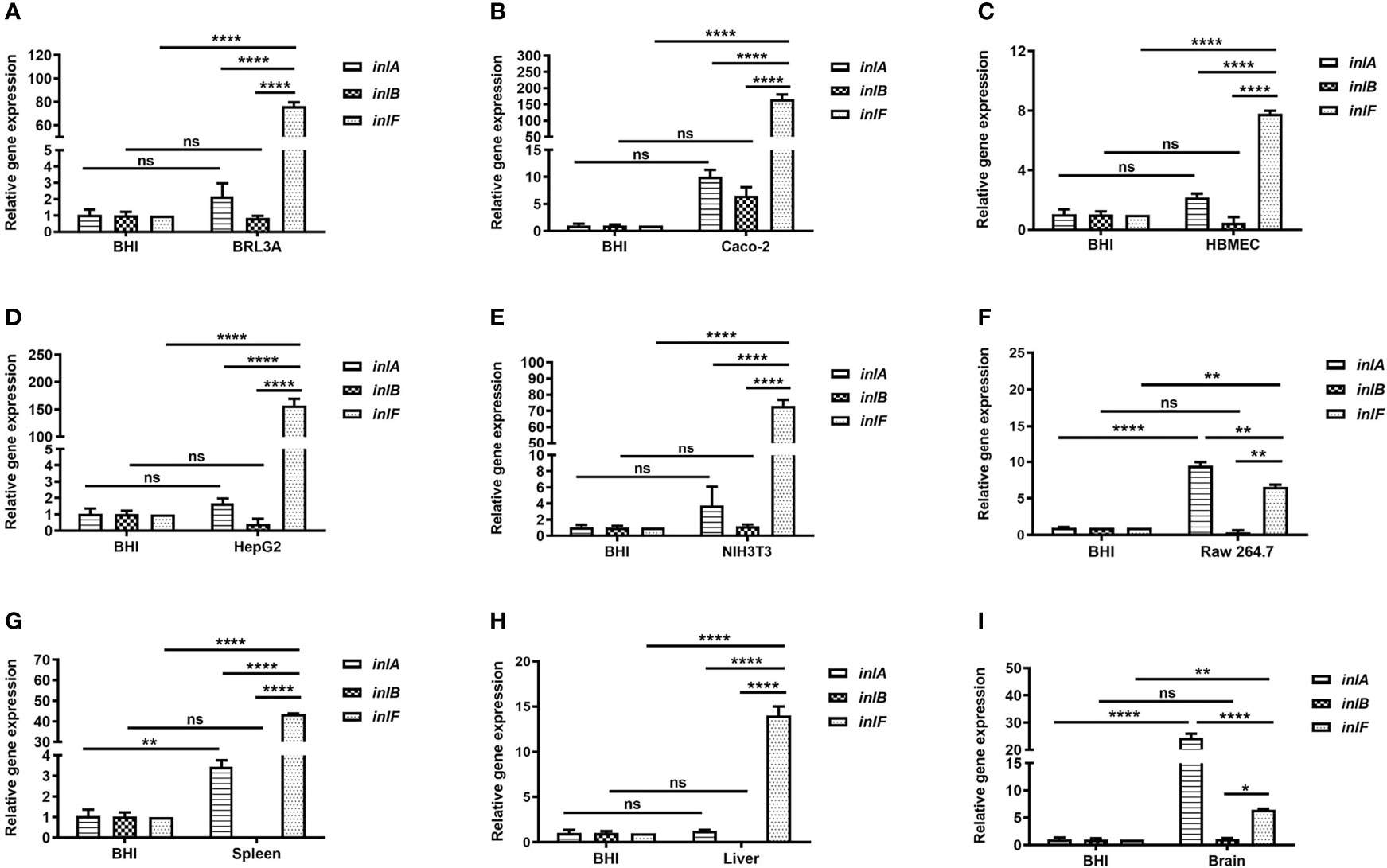
Figure 1 Analysis of the transcriptional expression of internalin genes. Internalins production during Lm NTSN invasion of BRL3A (A), Caco-2 (B), HBMEC (C), HepG2 (D), NIH3T3 (E) cells was determined by qRT-PCR. The detailed description of these cells is shown in Supplementary Table 1. Internalins expression in Lm NTSN infection of mice spleen, liver and brain is shown in (F-H), respectively. In the experiment, six-week-old BALB/c mice were intragastrically infused at a dose of 1 × 109 CFU. After 72 h, RNA was extracted from the liver, spleen and brain. BHI refers to each internalin gene’s expression under the in vitro culture condition. Error bars represented SEM, n = 3 independent experiments. Statistical analysis was carried out by Tukey’s multiple comparisons test: *P < 0.05, **P < 0.01, ****P < 0.0001, ns, no significance.
Lm crosses the intestinal barrier for dissemination to the organs by the blood circulation, and can also breach the blood–brain or placental barriers (Zheng and Xun, 2013). To further evaluate the role of internalins in the process of Lm infection in vivo, the expression of inlF, inlA, and inlB in the spleen, liver and brain of infected mice was measured using RT-PCR. The expression of inlF in the three organs was dramatically upregulated compared with Lm cultured in BHI, and its transcriptional level was significantly higher than those of inlA and inlB (p <0.0001) in liver and spleen (Figures 1G, H). In brain tissues, the transcriptional level of inlF was significantly higher than that of inlB (p <0.05), but significantly lower than that of inlA (p <0.0001) (Figure 1I). These differences in expression of inlF, inlA, and inlB suggested that they may play different roles in Lm invasion of different organs, with a possible involvement of inlF in the liver, spleen, and brain.
The equivalent comparison between mutant and wild strain can provide a breakthrough for the interpretation of experimental phenomena. To decipher the role of inlF in Lm serovar 4b infection, we compared the biological and immunological characteristics of an inlF deletion strain, the parental strain and the complemented strain using in vitro and in vivo models. We constructed a deletion mutant strain (NTSNΔinlF) and its reverse strain (NTSNΔinlF::inlF), with verification of the genomic knockout of inlF shown by RT-PCR with no DNA amplification of products with the length of 184 bp (Supplementary Figure 1A). The deletion of inlF was further confirmed by Western blotting showing the absence of an 88kDa-band in the component of the cell wall (Supplementary Figure 1B). Taken together, these results confirmed the successful construction of the NTSNΔinlF mutant strain via homologous recombination technology.
The growth features of the parental strain, inlF-deficient and reverse strains were compared by measuring growth curves. All three Lm strains cultured at 37°C exhibited similar growth trends, with logarithmic growth within 6 h, followed by entry into a stationary phase (Supplementary Figure 2). The growth curves verified that the deletion of inlF did not affect the growth of Lm in culture. The biochemical characteristics of each strain were determined by an automatic microbial identification instrument used to evaluate a potential effect of inlF deletion on Listeria metabolism. The results suggested that inlF was not involved in Lm NTSN physiologic or biochemical reactions of Lm NTSN (Supplementary Table 2).
Testing the bacterial infection properties of a variety of different cell types is a widely used approach that offers convenience, good repeatability and a short experimental cycle. To evaluate the role of inlF in virulence, we examined the adhesion and invasion abilities of Lm NTSN, NTSNΔinlF, and NTSNΔinlF::inlF in five non-phagocytic cell types and macrophages RAW264.7 (Supplementary Table 1). The results showed that inlF was not required for Listeria adhesion or invasion of five non-phagocytic cell types in vitro (Figures 2A–E). By contrast, the deletion of inlF significantly decreased the Lm adhesion and invasion capability in RAW264.7 (p <0.05) (Figure 2F). These results indicated that inlF may contribute to Lm infection via interaction with macrophages.
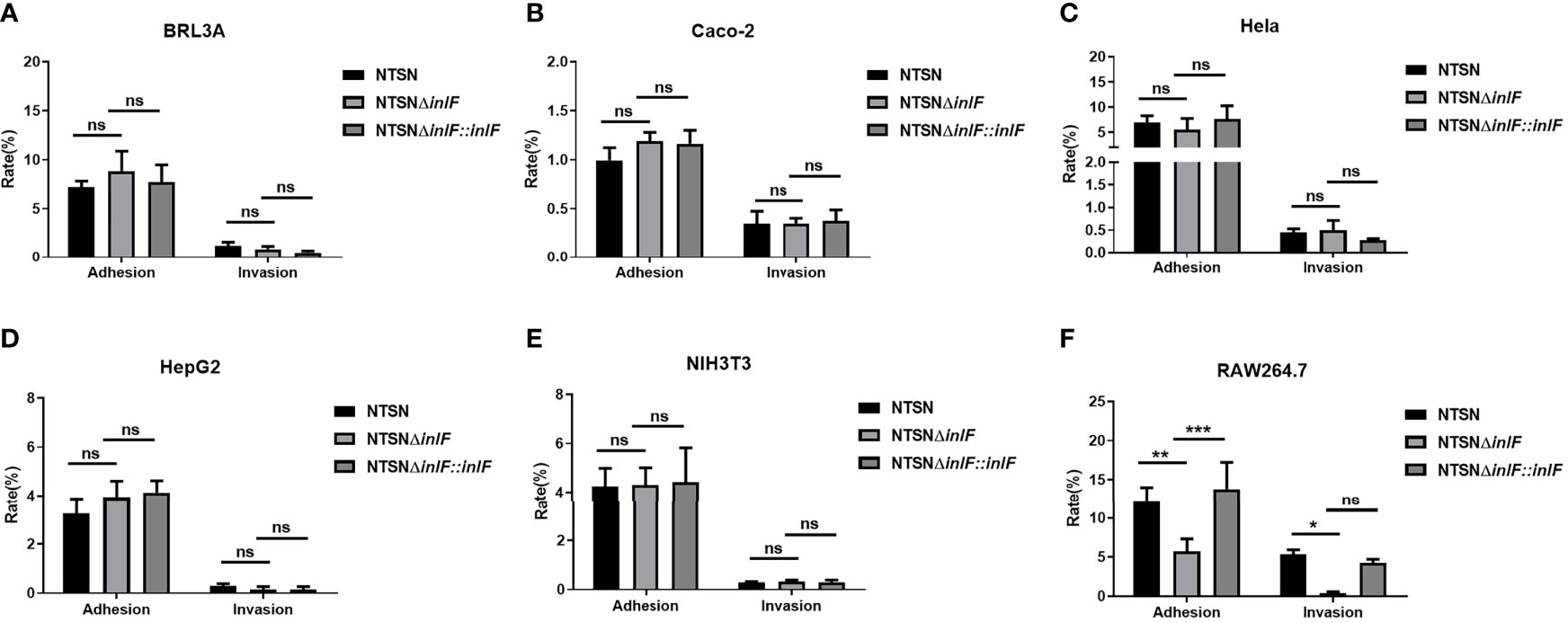
Figure 2 Comparative invasive capacities of Lm NTSN NTSNΔinlF and NTSNΔinlF::inlF. Six types of cells (Supplementary Table 1) were infected (MOI = 20) with Lm NTSN, NTSNΔinlF, and NTSNΔinlF::inlF, respectively. The percentage of intracellular bacterium was calculated after 1h of bacteria invasion. Shows are the adhesion and invasion of Lm to 5 types of non-phagocytes (A–E), to macrophage RAW264.7 is shown in (F). Among them, the PBS negative control group is represented by horizontal line bar, NTSN is represented by black and white check bar and the mutant is represented by black dot bar. Error bars represented SEM, n = 3 independent experiments. Statistical analysis was carried out by Tukey’s multiple comparisons test: *P < 0.05, **P < 0.01, ***P < 0.001, ns, no significance.
RAW264.7, a line of mouse peritoneal macrophages, is a commonly used inflammatory cell model. The interaction between Lm and macrophages RAW264.7 was assessed by examining the expression of inflammatory cytokines IL-10, IL-6, IL-1β, and TNF-α. While there were no statistical differences in the transcriptional expression of IL-10 or IL-6 between Lm NTSNΔinlF and the wild parental and reversion strains (p >0.05), the expressions of TNF-α (p <0.01) and IL-1β (p <0.01) were significantly higher in the mutant strain (Figure 3). These results indicated that the deletion of inlF triggered a strong upregulation in the expression of pro-inflammatory factors, and that inlF may act as a modulator of the anti-inflammatory response during Lm infection in vitro.
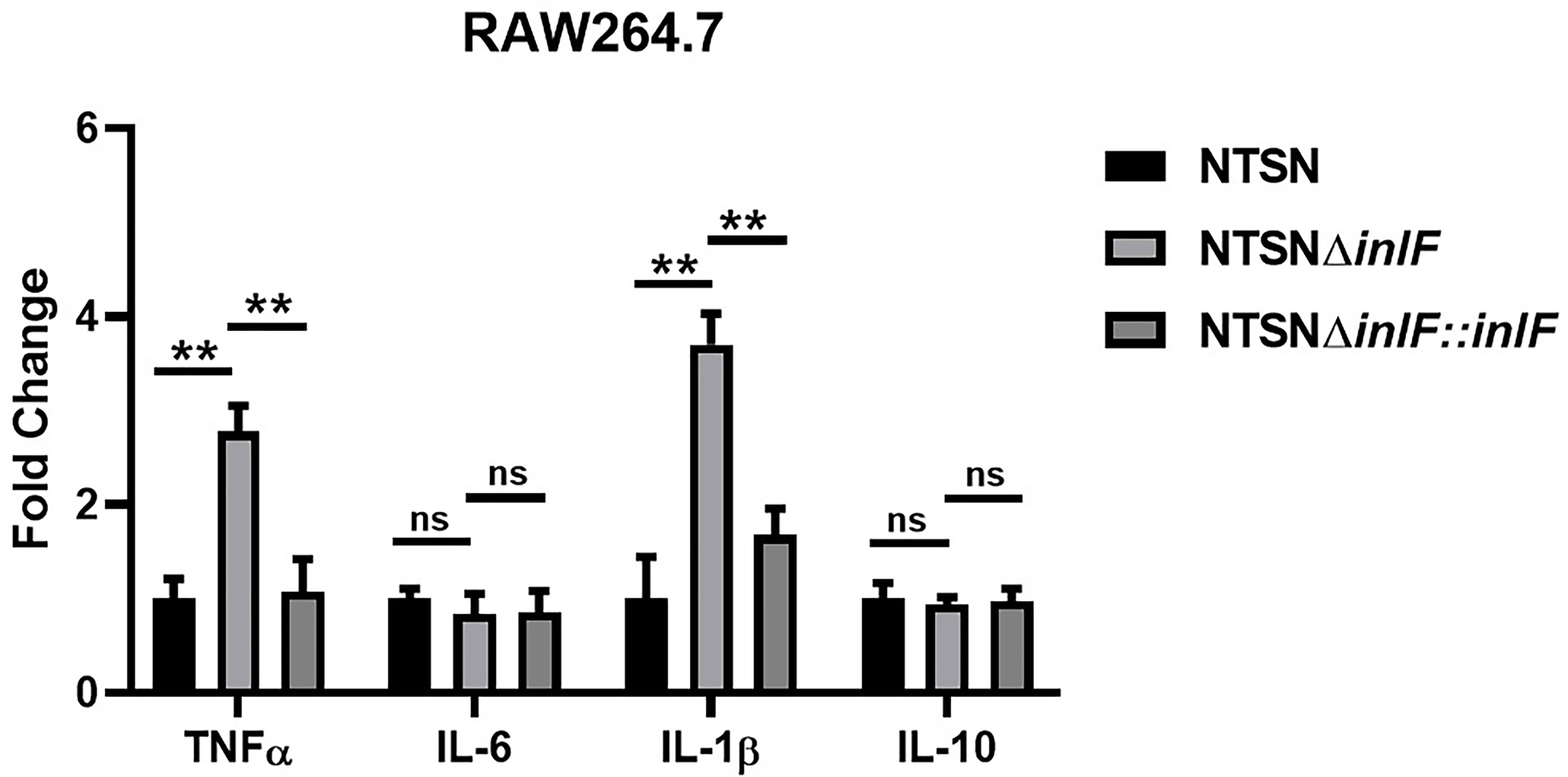
Figure 3 InlF impairs the proinflammatory response in vitro. RAW264.7 cells were infected (MOI = 20) with Lm NTSN NTSNΔinlF and NTSNΔinlF::inlF. The determination was performed 1h of bacteria invasion. Among them, the PBS negative control group is represented by horizontal line bar, NTSN is represented by black and white check bar and the mutant is represented by black dot bar. Values are mean with error bars; n = 3 independent experiments. Statistical analysis was carried out by Tukey’s multiple comparisons test: **P < 0.01, ns, no significance.
Macrophages are widely distributed in various tissues and function in homeostasis (Zhang X. M. et al., 2019). To understand the role of inlF in mediating the Lm defense against host immune responses, we measured the transcriptional expression of inflammatory factors IL-10, IL-6, IL-1β, and TNF-α in the spleen and liver of infected mice. Strikingly, infection with NTSNΔinlF triggered significantly higher production of cytokines IL-6 (p <0.05) and IL-1β (p <0.05) in the spleen; there was no obvious difference in TNF-α elicited by three Lm strains (p >0.05) (Figure 4A). Consistently, the transcriptional expression of IL-6 (p <0.01) and IL-1β (p <0.001) was also significantly higher in the liver of mice infected with NTSNΔinlF. Importantly, following the infection of NTSNΔinlF, there was significantly higher expression of TNF-α in the liver (p <0.001) (Figure 4B). The above data suggested that inlF played a crucial role in modulating the suppression of the pro-inflammatory response in vivo.
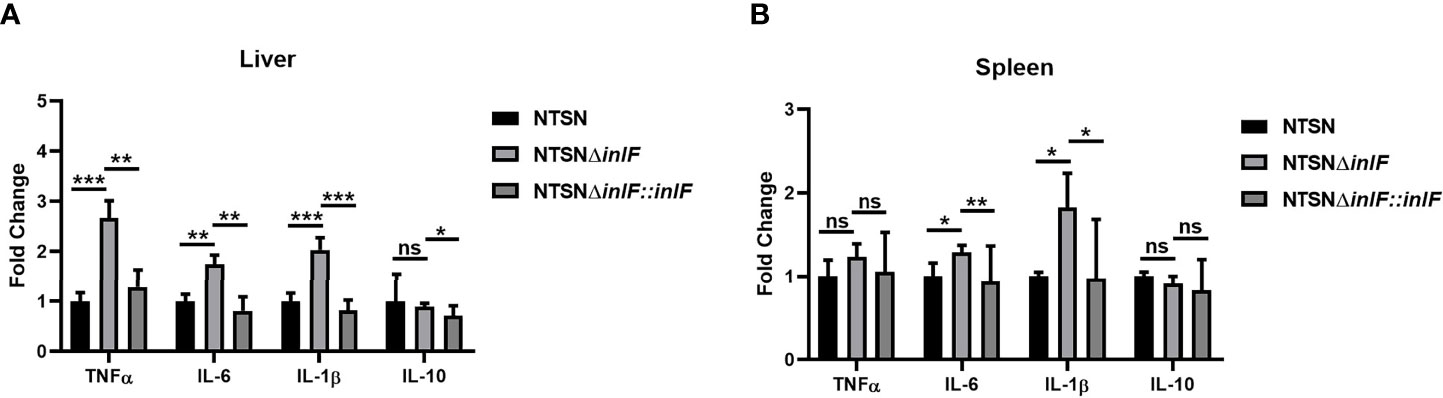
Figure 4 InlF modulates the inflammatory process in vivo. Mice were inoculated intraperitoneally with Lm NTSN NTSNΔinlF and NTSNΔinlF::inlF, the levels of cytokines in the spleen and liver were measured at 6 h post-infection. qRT-PCR detected the expression of inflammatory cytokines TNF-α, IL-6, IL-1β, and IL-10 in mice liver (A) and spleen (B). Among them, the PBS negative control group is represented by horizontal line bar, NTSN is represented by black and white check bar and the mutant is represented by black dot bar. Values are mean with error bars; n = 3 independent experiments. Statistical analysis was carried out by Tukey’s multiple comparisons test: *P < 0.05, **P < 0.01, ***P < 0.001, ns, no significance.
As a facultative intracellular parasite, Lm adheres to and invades intestinal cells with the contribution of internalin proteins, which enable it to break through the intestinal barrier. We compared the colonization abilities of the parental strain and the inlF deletion strain following the inoculation of mice via the intragastric route. Six hours after inoculation, the bacterial loads in the spleen (p <0.01), liver (p <0.01), and ileum (p<<0.05) were significantly lower in mice infected with NTSNΔinlF compared with the parental and reversion strains (Figure 5A). At later time points after infection, 24 h (Figure 5B) and 72 h (Figure 5C), there were no statistical differences between strains in the bacterial loads in the liver, spleen, and ileum. Taken together, these results indicated that inlF was involved in Lm colonization in the early stage of infection.
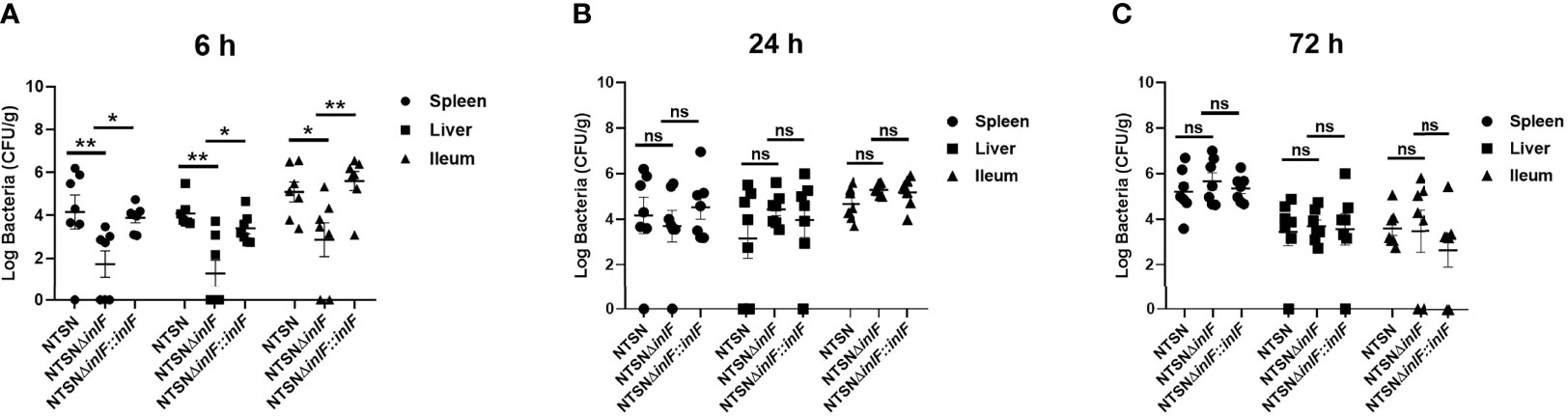
Figure 5 InlF contributes to Lm infection during murine listeriosis. Lm (1 × 109 CFU) were intragastrically inoculated to mice (n = 7). Animals were euthanized 6 h (A), 24 h (B), and 72 h (C) after infection. The spleen, liver and ileum were recovered, homogenized and plated on BHI or MOX plates. The numbers of bacterium able to colonize the organs are shown. Among them, the PBS negative control group is represented by horizontal line bar, NTSN is represented by black and white check bar and the mutant is represented by black dot bar. Values are mean ± SEM; n = 3 independent experiments. Statistical analysis was carried out by Tukey’s multiple comparisons test: *P < 0.05, **P < 0.01, ns, no significance.
Histologic analysis was performed on tissue sections of mice infected with Lm NTSN or NTSNΔinlF 72 h post infection. Compared with a PBS control, NTSN infection led to severe lesions in multiple tissues; spleens exhibited local lesions, necrosis and neutrophil infiltration, some hepatocytes appeared fatty with granular degeneration; there was erosion of the lamina propria of the intestinal mucosa; and the nuclei of nuclei in cells that comprise the intestinal glands of the lower ileum were fragmented or dissolved. Lm NTSNΔinlF caused mild pathologic changes, including proliferation of macrophages in the spleen, neutrophil infiltration, and increased phagocytes in the connective tissue of the liver hilar region. Additionally, there were no obvious pathologic changes in the brains of either of the infected groups, which might be explained by the relatively short infection time (Figure 6). These observations result indicated that inlF was critical for Lm virulence and contributed to infection and pathogenesis in vivo in mice.
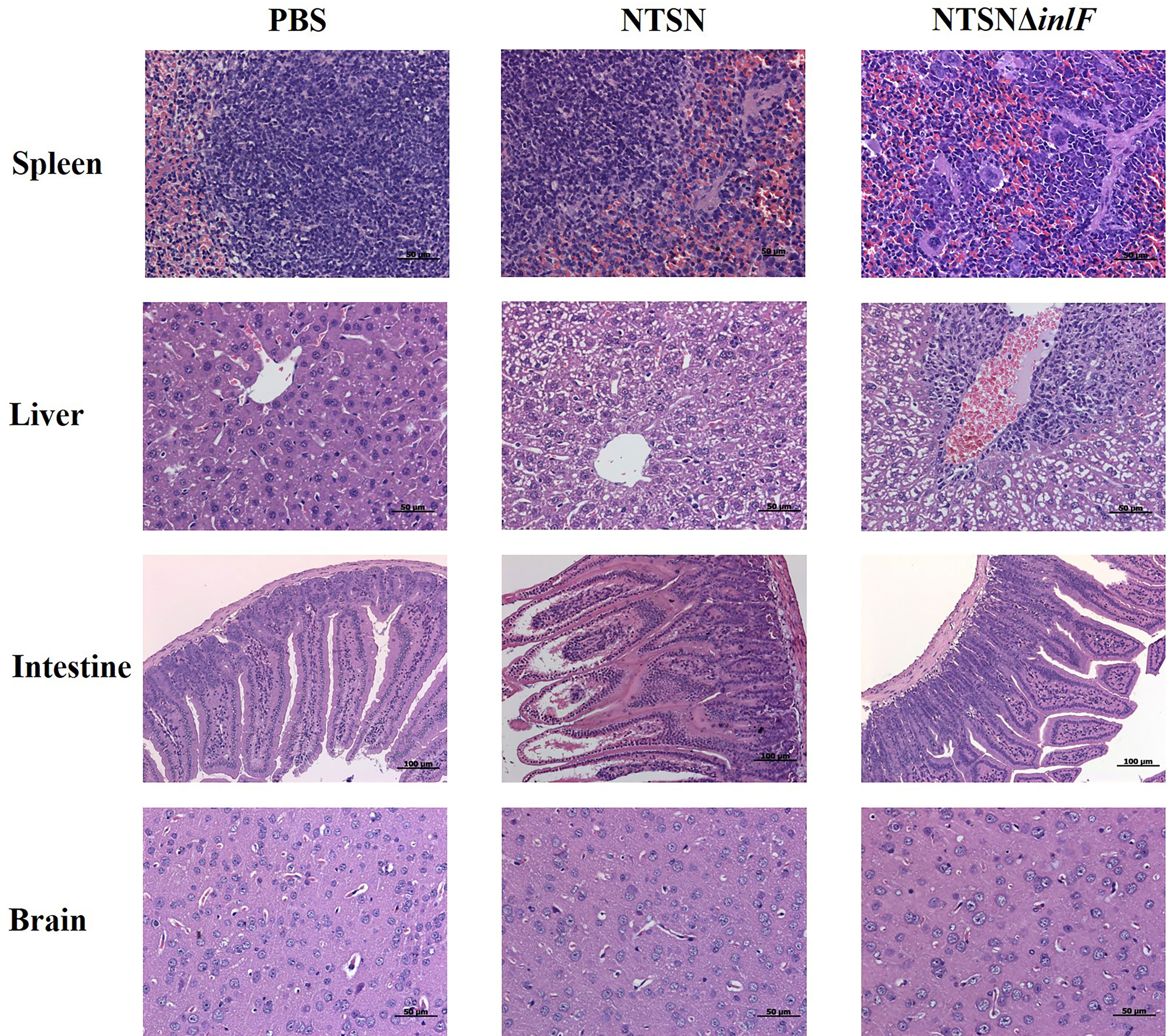
Figure 6 Histopathological section of infected mice organs. Six-week-old female mice were randomly assigned to 3 groups as follows to receive either NTSN or NTSNΔinlF combined with PBS as a control. Experiments were performed at 72 h post-infection. Sections of the spleens, livers, ileum, and brains are shown at the same magnification.
Pathogenic Lm has the ability to invade various cell types, and encodes internalin proteins with specific roles in bacterial infection and immunity. Since the first internalin protein (InlA) was identified and functionalized in 1991, 25 internalins containing the LRRs have been successively identified in the Lm strain EGD-e. Surface protein InlF, also a member of the internalin family, was identified in the Lm strain EGD as early as 1997; however, based on in vitro and in vivo experiments, Dramsi et al. and Senay et al. verified that it did not play a role in the adhesion and invasion of non-phagocytic cells, and was not essential for infecting mice (Dramsi et al., 1997; Senay et al., 2020). Kirchner and colleagues investigated the impact of the ROCK pathway on Lm serovar 1/2a 10403S infection of mammalian cells and found that inhibition of ROCK activity contributed to InlF-mediated invasion and virulence (Kirchner and Higgins, 2008). A recent study verified that inlF from intravenous inoculation with Lm 10403S was able to coordinate with host cell-surface vimentin and contribute to Lm invasion of the brain (Ghosh et al., 2018). Previous research on InlF was mainly conducted on evolutionary lineageII strains; however, there is only 78% homology in the amino acid sequence of InlF protein between lineage I and lineageII strains (Supplementary Figure 3). Furthermore, because the majority of invasive 4b strains in human outbreaks of listeriosis belonged to lineage I, the contribution of InlF in mediating infection and pathogenesis is not fully understood.
To obtain a more comprehensive understanding the roles of inlF in Lm serovar 4b infection, we compared its transcriptional expression with inlA and inlB during infection of five non-phagocytic cell types and macrophages RAW264.7 and also analyzed its expression in the liver, spleen and brain of infected mice. Unexpectedly, we found that inlF was significantly upregulated not only in non-phagocytic cells and macrophages in vitro, but also in all three organs in vivo. These results suggested that inlF played an important role in the interaction between Lm and various types of host cells. The dramatically enhanced colonization capability in the ileum during the early stage of infection in mice, and following subsequent severe pathologic damage to the liver, spleen, and ileum, strongly supports a role for inlF in Lm virulence and systemic infection. Authors of previous studies of inlF using Lm 4b serovar JF5203 concluded that the lack of inlF had no significant effect on infection or growth (Rupp et al., 2017). Senay et al. constructed an inlF-deficient 4b strain to assess its role in the infection of multiple cell types and found, that inlF was not required for hyper-invasiveness (Senay et al., 2020). By contrast, our infection experiments in mice have demonstrated a role of inlF in the virulence and pathogenesis of a serovar 4b strain.
Lm is a facultative intracellular pathogen that can escape from phagocytic vesicles into the host cytoplasm, where it regulates the innate and adaptive immune responses, and decreases host resistance to systemic infection (Stockinger et al., 2004; Opitz et al., 2006; Boneca et al., 2007). Multiple Listeria-specific genes are involved in the interference with the innate immune response that occurs during Lm–phagocytic cell interaction. PdgA and OatA, which are associated with the synthesis of peptidoglycan for the bacterial cell wall, mediate Lm evasion from the innate immune response (Boneca et al., 2007; Aubry et al., 2011), while the internalin family protein InlC dampens the nuclear factor (NF)-кB signaling pathway by targeting the inhibitor of NF-кB, IкB kinase (Gouin et al., 2010). InlH from the serovar 1/2a EGD-e strain has been reported to mediate IL-6 secretion during murine listeriosis. InlF protein contains 13 LRR domains and an LPXTG motif, marking it structurally similar to InlH (Bierne et al., 2007). Whether inlF also contribute to evasion of the host immune response remains unclear and deserves further elucidation.
To investigate the potential role of inlF in mediating Lm interference with the innate immune response, we constructed an inlF-deficient and reverse Lm strain, compared it with the parental strain for effects on cytokines during infection of macrophages and mice. Interestingly, we found that inlF did significantly inhibit the production of pro-inflammatory cytokines TNF-α and IL-1β, enhancing the survival of Lm in macrophages in vitro. In vivo, inlF strongly suppressed the production of TNF-α, IL-6, and IL-1β in the liver, as well as IL-6 and IL-1β in the spleen. As the effector cytokines, IL-6, TNFα, and IL-1β are clearly required for initiation of the Th1-type immune response and for direct bacterial killing (Allie et al., 2013; Romagnoli et al., 2017). The suppression of TNF-α and IL-1β production undoubtedly promoted the ability of Lm NTSN to evade the direct bacteria-killing of macrophages, thus contributing to its survival during the early stage of infection. The inhibitory effect of inlF on IL-6 in vitro and in vivo was consistent with the known involvement of inlH in immune invasion; however, the obvious difference in invasion ability between inlF and inlH in RAW264.7 cells suggested that inlF may play a more important role in immune evasion (Personnic et al., 2010). Pro-inflammatory cytokines (e.g., IL-6, TNFα) and chemokines (e.g., CXCL2) are upregulated by the activation of the NF-κB pathway upon the host sensing the presence of cytosolic Lm via target receptors (D'Orazio, 2019). We deduced that inlF may be involved in blocking the NF-κB pathway, thereby downregulating the inflammatory response and contributing to Lm infection and colonization of the host.
Taken together, our results suggest that inlF from a serovar 4b strain of Lm plays a crucial role in modulating the innate immune response through interference with the production of multiple pro-inflammatory cytokines, thereby contributing to survival in macrophages and colonization in the early stage of infection. This study has verified that inlF as an important virulence factor involved in immune evasion and pathogenesis.
The original contributions presented in the study are included in the article/Supplementary Matreial. Further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by the Institutional Animal Ethics Committee of Yangzhou University.
YY and XJ designed the experiments. ZL and DZ performed the experiments and analyzed the results. YW and SK were involved in animal experiments. XX and HY participated in experiments related to immune escape. ZL and YY wrote the manuscript. XC and ZP provided a lot of guidance in the design and writing process. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (Nos. 2017YFC1601201), the National Natural Science Foundation of China (No. 31472193), the Jiangsu agricultural science and technology independent innovation funds (CX(21)1004), the Key Research and Development Program (Modern Agriculture) Project of Jiangsu Province (No. BE2017341), the Jiangsu Agricultural Science and Technology Independent Innovation Funds [No. CX(21)1049], and the Priority Academic Development Program of Jiangsu Higher Education Institutions (PAPD).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.748461/full#supplementary-material
Allie, N., Grivennikov, S. I., Keeton, R., Hsu, N. J., Bourigault, M. L., Court, N., et al. (2013). Prominent Role for T Cell-Derived Tumour Necrosis Factor for Sustained Control of Mycobacterium Tuberculosis Infection. Sci. Rep. 3, 1809. doi: 10.1038/srep01809
Aubry, C., Goulard, C., Nahori, M. A., Cayet, N., Decalf, J., Sachse, M., et al. (2011). OatA, A Peptidoglycan O-Acetyltransferase Involved in Listeria Monocytogenes Immune Escape, Is Critical for Virulence. J. Infect. Dis. 204 (5), 731–740. doi: 10.1093/infdis/jir396
Bierne, H., Sabet, C., Personnic, N., Cossart, P. (2007). Internalins: A Complex Family of Leucine-Rich Repeat-Containing Proteins in Listeria Monocytogenes. Microbes Infect. 9 (10), 1156–1166. doi: 10.1016/j.micinf.2007.05.003
Boneca, I. G., Dussurget, O., Cabanes, D., Nahori, M. A., Sousa, S., Girardin, S. E., et al. (2007). A Critical Role for Peptidoglycan N-Deacetylation in Listeria Evasion From the Host Innate Immune System. Proc. Natl. Acad. Sci. U. S. A. 104 (3), 997–1002. doi: 10.1073/pnas.0609672104
Chen, Y., Zhang, W., Knabel, S. J. (2007). Multi-Virulence-Locus Sequence Typing Identifies Single Nucleotide Polymorphisms Which Differentiate Epidemic Clones and Outbreak Strains of Listeria Monocytogenes. Clin. Microbiol. 45 (3), 835–846. doi: 10.1128/JCM.01575-06
D'Orazio, S. E. F. (2019). Innate and Adaptive Immune Responses During Listeria Monocytogenes Infection. Microbiol. Spectr. 7, G3–G0065. doi: 10.1128/microbiolspec.GPP3-0065-2019
Dortet, L., Mostowy, S., Louaka, A. S., Gouin, E., Nahori, M. A., Wiemer, E. A. (2011). Recruitment of the Major Vault Protein by InlK: A Listeria Monocytogenes Strategy to Avoid Autophagy. PLoS Pathog. 7 (8), e1002168. doi: 10.1371/journal.ppat.1002168
Dramsi, S., Dehoux, P., Lebrun, M., Goossens, P. L., Cossart, P. (1997). Identification of Four New Members of the Internalin Multigene Family of Listeria Monocytogenes EGD. Infect. Immun. 65 (5), 1615–1625. doi: 10.1128/iai.65.5.1615-1625.1997
Faralla, C., Rizzuto, G. A., Lowe, D. E., Kim, B., Cooke, C., Shiow, L. R. (2016). InlP, A New Virulence Factor With Strong Placental Tropism. Infect. Immun. 84 (12), 3584–3596. doi: 10.1128/IAI.00625-16
Ghosh, P., Halvorsen, E. M., Ammendolia, D. A., Mor-Vaknin, N., O'Riordan, M. X. D., Higgins, D. E., et al. (2018). Invasion of the Brain by Listeria Monocytogenes Is Mediated by InlF and Host Cell Vimentin. Mol. Microbiol. 9 (1), e00160–e00118. doi: 10.1128/mBio.00160-18
Gouin, E., Adib, C. M., Balestrino, D., Nahori, M. A., Villiers, V., Cossart, P., et al. (2010). The Listeria Monocytogenes InlC Protein Interferes With Innate Immune Responses by Targeting the Iκb Kinase Subunit Ikkα. Proc. Natl. Acad. Sci. U. S. A. 107 (40), 17333–17338. doi: 10.1073/pnas.1007765107
Goulet, V., Jacquet, C., Martin, P., Vaillant, V., Laurent, E., de Valk, H. (2006). Surveillance of Human Listeriosis in France, 2001-2003. Euro. Surveill. 11 (6), 79–81. doi: US1195165A
Hoge, J., Yan, I., Janner, N., Schumacher, V., Chalaris, A., Steinmetz, O. M., et al. (2013). IL-6 Controls the Innate Immune Response Against Listeria Monocytogenes via Classical IL-6 Signaling. J. Immunol. 190, 703–711. doi: 10.4049/jimmunol.1201044
Jonquières, R., Pizarro-Cerdá, J., Cossart, P. (2001). Synergy Between the N-And C-Terminal Domains of InlB for Efficient Invasion of Non-Phagocytic Cells by Listeria Monocytogenes. Mol. Microbiol. 42 (4), 955–965. doi: 10.1046/j.1365-2958.2001.02704.x
Kirchner, M., Higgins, D. E. (2008). Inhibition of ROCK Activity Allows InlF-Mediated Invasion and Increased Virulence of Listeria Monocytogenes. Mol. Microbiol. 68 (3), 749–767. doi: 10.1111/j.1365-2958.2008.06188.x
Lecuit, M., Nelson, D. M., Smith, S. D., Khun, H., Huerre, M., Vacher-Lavenu, M. C. (2004). Targeting and Crossing of the Human Maternofetal Barrier by Listeria Monocytogenes: Role of Internalin Interaction With Trophoblast E-Cadherin. Proc. Natl. Acad. Sci. 101 (16), 6152–6157. doi: 10.1073/pnas.0401434101
Opitz, B., Puschel, A., Beermann, W., Hocke, A. C., Forster, S., Schmeck, B., et al. (2006). Listeria Monocytogenes Activated P38 MAPK and Induced IL-8 Secretion in a Nucleotide-Binding Oligomerization Domain 1-Dependent Manner in Endothelial Cells. J. Immunol. 176, 484–490. doi: 10.4049/jimmunol.176.1.484
Pamer, E. G. (2004). Immune Responses to Listeria Monocytogenes. Nat. Rev. Immunol. 4, 812–823. doi: 10.1038/nri1461
Park, S. F., Stewart, G. S. (1990). High-Efficiency Transformation of Listeria Monocytogenes by Electroporation of Penicillin-Treated Cells. Gene 94 (1), 129–132. doi: 10.1016/0378-1119(90)90479-b
Personnic, N., Bruck, S., Nahori, M. A., Toledo-Arana, A., Nikitas, G., Lecuit, M., et al. (2010). The Stress-Induced Virulence Protein InlH Controls Interleukin-6 Production During Murine Listeriosis. Infect. Immun. 78 (5), 1979–1989. doi: 10.1128/IAI.01096-09
Romagnoli, P. A., Fu, H. H., Qiu, Z., Khairallah, C., Pham, Q. M., Puddington, L., et al. (2017). Differentiation of Distinct Long-Lived Memory CD4 T Cells in Intestinal Tissues After Oral Listeria Monocytogenes Infection. Mucosal Immunol. 10, 520–530. doi: 10.1038/mi.2016.66
Rupp, S., Bärtschi, M., Frey, J., Oevermann, A. (2017). Hyperinvasiveness and Increased Intercellular Spread of Listeria Monocytogenes Sequence Type 1 are Independent of Listeriolysin S, Internalin F and Internalin J1. J. Med. Microbiol. 66 (7), 1053–1062. doi: 10.1099/jmm.0.000529
Sabet, C., Lecuit, M., Cabanes, D., Cossart, P., Bierne, H. (2005). LPXTG Protein InlJ, a Newly Identified Internalin Involved in Listeria Monocytogenes Virulence. Infect. Immun. 73 (10), 6912–6922. doi: 10.1128/IAI.73.10.6912-6922.2005
Sabet, C., Toledo-Arana, A., Personnic, N., Lecuit, M., Dubrac, S., Poupel, O. (2008). The Listeria Monocytogenes Virulence Factor InlJ Is Specifically Expressed In Vivo and Behaves as an Adhesin. Infect. Immun. 76 (4), 1368–1378. doi: 10.1128/IAI.01519-07
Sahu, S. C., Gaines, D. W., Williams, K. M., Raybourne, R. B.. (2007). A Synthetic Polypeptide Based on Human E-Cadherin Inhibits Invasion of Human Intestinal and Liver Cell Lines by Listeria Monocytogenes. J. Med. Microbiol. 56 (8), 1011–1016. doi: 10.1099/jmm.0.47194-0
Schubert, W. D., Göbel, G., Diepholz, M., Darji, A., Kloer, D., Hain, T. (2001). Internalins From the Human Pathogen Listeria Monocytogenes Combine Three Distinct Folds Into a Contiguous Internalin Domain. J. Mol. Biol. 312 (4), 783–794. doi: 10.1006/jmbi.2001.4989
Senay, T. E., Ferrell, J. L., Garrett, F. G., Albrecht, T. M., Cho, J., D'Orazio, S. E. F., et al. (2020). Neurotropic Lineage III Strains of Listeria Monocytogenes Disseminate to the Brain Without Reaching High Titer in the Blood. mSphere 5 (5), e00871–e00820. doi: 10.1128/mSphere.00871-20
Stavru, F., Archambaud, C., Cossart, P. (2011). Cell Biology and Immunology of Listeria Monocytogenes Infections: Novel Insights. Immunol. Rev. 240, 160–184. doi: 10.1111/j.1600-065X.2010.00993.x
Stockinger, S., Reutterer, B., Schaljo, B., Schellack, C., Brunner, S., Materna, T., et al. (2004). IFN Regulatory Factor 3-Dependent Induction of Type I IFNs by Intracellular Bacteria Is Mediated by a TLR- and Nod2-Independent Mechanism. J. Immunol. 173, 7416–7425. doi: 10.4049/jimmunol.173.12.7416
Unanue, E. R. (1997). Studies in Listeriosis Show the Strong Symbiosis Between the Innate Cellular System and the T-Cell Response. Immunol. Rev. 158, 11–25. doi: 10.1111/j.1600-065x.1997.tb00988.x
Yin, Y., Yao, H., Doijad, S., Kong, S., Shen, Y., Cai, X., et al. (2019). A Hybrid Sub-Lineage of Listeria Monocytogenes Comprising Hypervirulent Isolates. Nat. Commun. 10, 4283. doi: 10.1038/s41467-019-12072-1
Zhang, X. M., Chen, D. G., Li, S. C., Zhu, B., Li, Z. J. (2019). Embryonic Origin and Subclonal Evolution of Tumor-Associated Macrophages Imply Preventive Care for Cancer. Cells 10 (4), 903. doi: 10.3390/cells10040903
Zhang, Y., Yao, Y., Qiu, X., Wang, G., Hu, Z., Chen, S., et al. (2019). Listeria Hijacks Host Mitophagy Through a Novel Mitophagy Receptor to Evade Killing. Nat. Immunol. 20 (4), 433–446. doi: 10.1038/s41590-019-0324-2
Zharkova, O., Celhar, T., Cravens, P. D., Satterthwaite, A. B., Fairhurst, A. M., Davis, L. S. (2017). Pathways Leading to an Immunological Disease: Systemic Lupus Erythematosus. Rheumatology 56 (suppl_1), i55–i66. doi: 10.1093/rheumatology/kew427
Keywords: Listeria monocytogenes, inlF, serovar 4b, immune evasion, inflammatory injury, colonization
Citation: Ling Z, Zhao D, Xie X, Yao H, Wang Y, Kong S, Chen X, Pan Z, Jiao X and Yin Y (2022) inlF Enhances Listeria monocytogenes Early-Stage Infection by Inhibiting the Inflammatory Response. Front. Cell. Infect. Microbiol. 11:748461. doi: 10.3389/fcimb.2021.748461
Received: 28 July 2021; Accepted: 09 November 2021;
Published: 10 February 2022.
Edited by:
Houhui Song, Zhejiang Agriculture & Forestry University, ChinaReviewed by:
Chun Fang, Yangtze University, ChinaCopyright © 2022 Ling, Zhao, Xie, Yao, Wang, Kong, Chen, Pan, Jiao and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin’an Jiao, amlhb0B5enUuZWR1LmNu; Yuelan Yin, eXlsYW5AeXp1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.