- 1College of Veterinary Medicine, Qingdao Agricultural University, Qingdao, China
- 2College of Life Sciences, Changchun Sci-Tech University, Shuangyang, China
- 3General Monitoring Station for Wildlife-Borne Infectious Diseases, State Forestry and Grass Administration, Shenyang, China
- 4Key Laboratory of Zoonosis Research, Ministry of Education, College of Veterinary Medicine, Jilin University, Changchun, China
- 5College of Animal Science and Veterinary Medicine, Heilongjiang Bayi Agricultural University, Daqing, China
Blastocystis is a protozoan that parasitizes the intestines. A number of hosts of Blastocystis have been found, including human and animals. However, there has been no research on the prevalence of Blastocystis in Tibetan antelope. Here, a molecular test was performed using 627 Tibetan antelope fecal samples collected on Tibet in China from 2019 to 2020. The result showed that 30 (4.8%) samples were Blastocystis positive. The highest prevalence of Blastocystis was in Shuanghu County (25/209, 12.0%), followed by Shenza County (2/103, 1.9%), Nyima County (3/182, 1.6%), and Baigoin County (0/133, 0.0%). In addition, logistic regression analysis showed that the gender, sampling year, and area of Tibetan antelope were risk factors for Blastocystis prevalence. Three subtypes (ST10, ST13, and ST14) of Blastocystis were found in Tibetan antelope through a subtype sequence analysis, and ST13 was identified to be the dominant subtype. This is the first investigation for the infection of Blastocystis in Tibetan antelope. Collectively, the data in this study have expanded the host range of Blastocystis and provided basic information for the distribution of Blastocystis subtypes, which could support the prevention of Blastocystis infection in wild animals.
Introduction
Blastocystis is a protozoan that parasitizes the intestines (Jiménez et al., 2019; Paik et al., 2019). It can infect a variety of hosts, such as mammals, amphibians, birds, and insects (Zhu et al., 2020). Blastocystis is transmitted through the fecal–oral route or water and food between susceptible hosts (Asghari et al., 2019; Deng et al., 2019). Hosts infected with Blastocystis could develop clinical signs, e.g., diarrhea, abdominal pain, and vomiting. Immunocompromised individuals are more susceptible to Blastocystis (Wang et al., 2018a; Paik et al., 2019).
Blastocystis was first isolated from animal feces in 1911 (Alexeieff, 1911). Since then, more and more animals and humans, such as cattle, deer, sheep, and goats, were identified to be the hosts of Blastocystis (Table 1). In China, Blastocystis was first reported in children in 1990 (Li et al., 1990). A large number of investigations regarding Blastocystis prevalence in different hosts were performed previously (Song et al., 2017; Zhao et al., 2017; Wang et al., 2018a; Wang et al., 2018b). So far, the infection of Blastocystis has been reported in many animals, including domestic and wild animals (Zhao et al., 2017; Wang et al., 2018a).
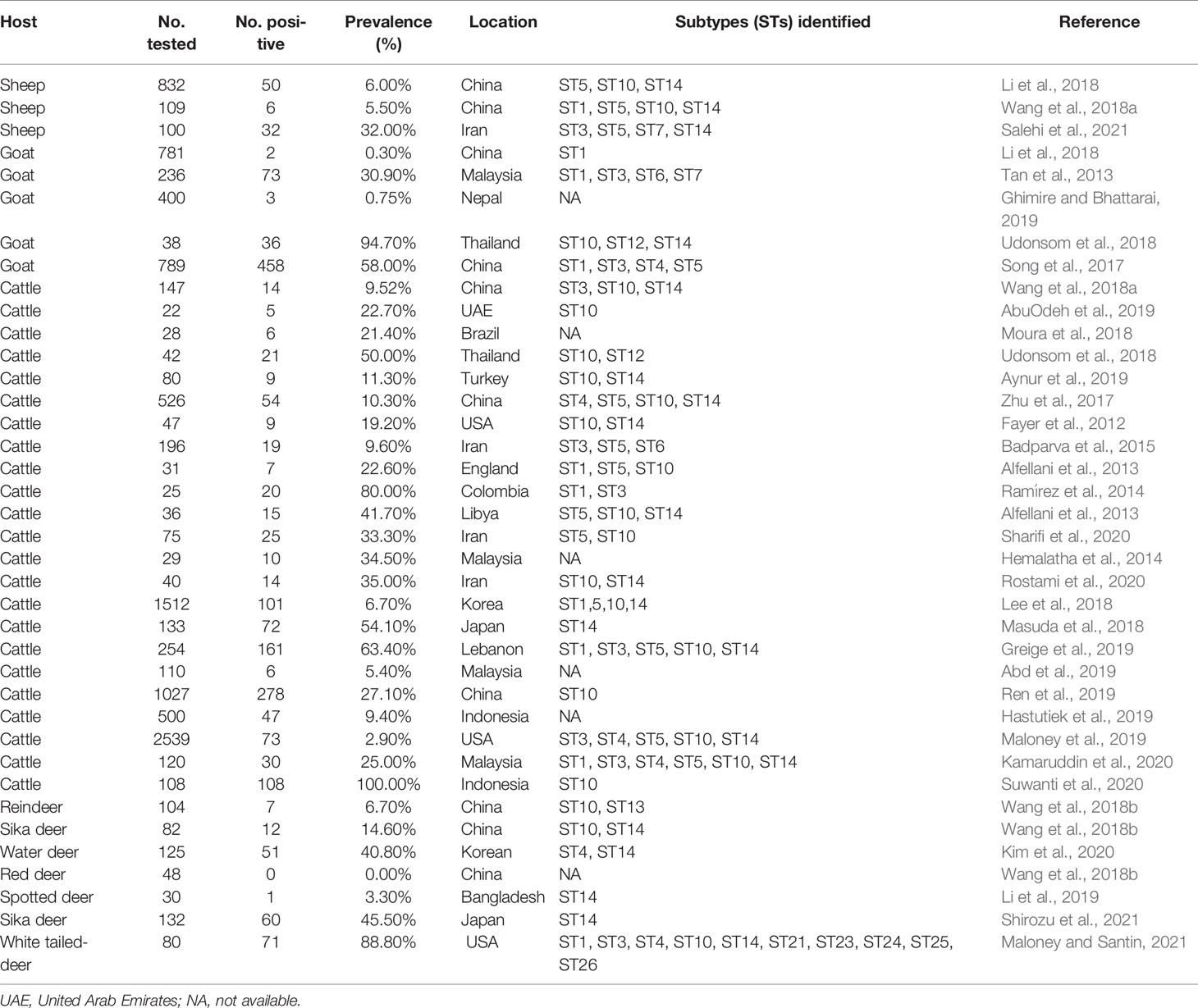
Table 1 Subtypes and prevalence of Blastocystis sp. detected from the ruminants worldwide (2010–2021).
To date, approximately 29 proposed Blastocystis subtypes have been identified in a large number of literatures (Ma et al., 2020). ST1-9 and ST12 subtypes were identified in humans, while ST10-17 and ST21-28 subtypes were detected in animals (Stensvold and Clark, 2016; Ning et al., 2020; Hublin et al., 2021). Of note, some subtypes were identified in both humans and animals, such as ST1, ST3, and ST5 subtypes (Song et al., 2017; Wang et al., 2018a). ST4 was found in deer (Wang et al., 2018b; Kim et al., 2020; Shirozu et al., 2021), and ST12 was found in yaks in the plateau area (Ren et al., 2019).
Tibetan antelope (Pantholops hodgsonii) belongs to genus Pantholops, family Bovidae, order Cetartiodactyla according to the IUCN Red List in 2016 (IUCN SSC Antelope Specialist Group, 2016). Tibetan antelope is one of the most rare and endangered wild animals. There are approximately 100,000 to 150,000 Tibetan antelope in India and China (IUCN SSC Antelope Specialist Group 2016). Tibetan antelope can carry various pathogens, such as Mycoplasma capricolum subspecies, capripneumoniae (Mccp) (Yu et al., 2012), and Escherichia coli (Bai et al., 2016).
However, the existing data indicate that sheep may carry several potential Blastocystis subtypes, including ST1, ST3, ST4, ST5, ST6, and ST7 (Tan et al., 2013; Song et al., 2017; Wang et al., 2018a; Salehi et al., 2021). So far, studies on the prevalence and subtype diversity of Blastocystis in Tibetan antelope are unknown and the relevant public health impact is still unclear. This study provides important information on the diversity of Blastocystis subtypes in Tibetan antelope, and would help determine the role of Tibetan antelope in the transmission of Blastocystis to humans and other animals.
Materials and Methods
Specimen Collection
From August 2019 to September 2020, the feces of 627 wild Tibetan antelope was collected in four areas in Tibet (Table 2 and Figure 1). This study randomly observed Tibetan antelope in the field. Fresh fecal samples were put into a PE glove immediately after defecation onto the ground, and then were placed into ice boxes and transported to the laboratory. This study was approved by the Ethics Committee of Jilin University. Appropriate permission was obtained from the General Monitoring Station for Wildlife-Borne Infectious Diseases, State Forestry and Grass Administration.
DNA Extraction and PCR Amplification
Genomic DNA was extracted using the E.Z.N.A.® Stool DNA Kit (Omega Biotek Inc., Norcross, GA, USA) according to the manufacturer’s instructions and stored at −20°C until PCR amplification. SSU rRNA gene was the target for PCR analysis using primers RD5 (5′-ATCTGGTTGATCCTGCCAGT-3′) and BhRDr (5′-GAGCTTTTTAACTGCAACAACG-3′) as described previously to amplify an approximately 600-bp region (Scicluna et al., 2006). Positive and negative controls were included in each test. PCR products were observed using UV light after electrophoresis at a 1.5% agarose gel containing ethidium bromide.
Sequence and Phylogenetic Analyses
The Blastocystis-positive PCR products were sent to Sangon Biotech Company (Shanghai, China) for sequencing. The sequence accuracy was confirmed by bidirectional sequencing. The sequencing was re-performed if variation was present in the previous result. Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/blast/) was employed to compare consensus sequences with the similar sequences on GenBank. The subtypes of Blastocyst isolates were determined through the online platform PubMLST (https://pubmlst.org/bigsdb?db=pubmlst_blastocystis_seqdef). The obtained sequences were aligned using ClustalX 1.83 program. The alignment was trimmed using the trimAI v1.2 software (http://trimal.cgenomics.org/downloads) (Li et al., 2019). All positions with gaps were eliminated, and 104 unambiguously aligned sites were used for phylogenetic inference. The maximum likelihood (ML) method (Kimura two-parameter model) was employed to reconstruct phylogenetic trees by using MEGA X. Representative nucleotide sequences were submitted to GenBank under accession numbers: MZ444657–MZ444662.
Statistical Analysis
The variation of Blastocystis prevalence (y) in Tibetan antelope on the basis of sampling year (x1), gender (x2), and collecting region (x3) was analyzed with χ2 test using SAS version 9.4 (SAS Institute Inc., USA). In the multivariable regression analysis, each of the variables was independently contained in the binary Logit model. The best model was judged by Fisher’s scoring algorithm. All tests were two-sided, and the results were considered statistically significant when p < 0.05. To explore the association between Blastocystis prevalence and the investigated factors, the odds ratios (ORs) and their 95% confidence intervals (95% CIs) were calculated.
Results
Prevalence of Blastocystis sp.
In the present study, 30 out of 627 Tibetan antelope feces were identified to be Blastocystis positive (Figure 2). The infection rate of Blastocystis in Tibetan antelope in 2019 (0.6%, 2/322) was lower than that (9.2%, 28/305) in 2020. The prevalence of Blastocystis in different investigated counties ranged from 0% to 12% (Table 3). Blastocystis were detected in all three counties, except for Baigoin County. The highest prevalence of Blastocystis was in Shuanghu County (25/209, 12.0%), followed by Shenza County (2/103, 1.9%) and Nyima County (3/182, 1.6%; Table 3). The infection rate of Blastocystis in Tibetan antelope in females (2.7%, 8/300) was significantly lower than that (6.7%, 22/327) in males (p = 0.017).
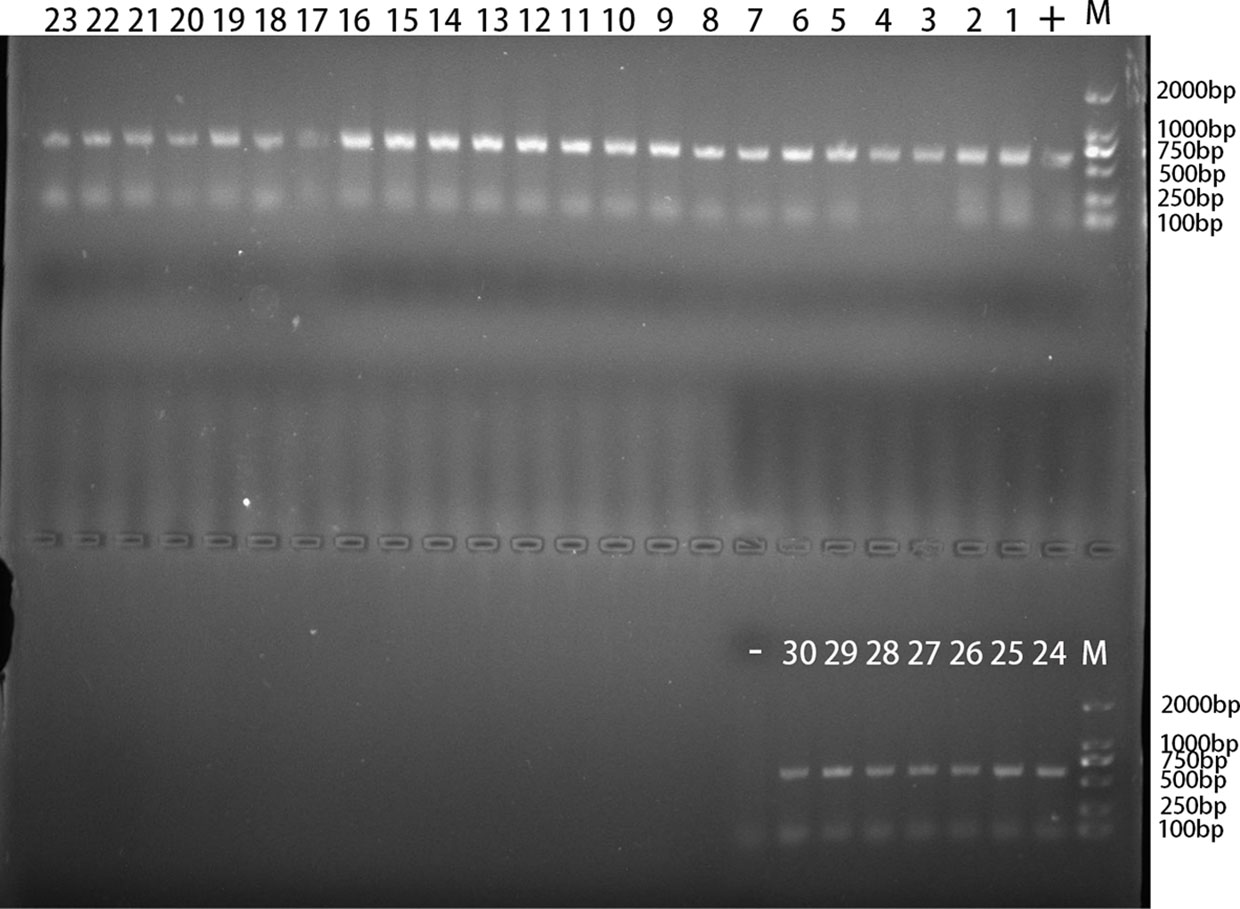
Figure 2 The PCR amplification result of Blastocystis rRNA gene. “1–30”: 30 positive samples of Blastocystis in TA25, TA26, TA72, TA100, TA103, TA105, TA107, TA108, TA111, TA118, TA124, TA128, TA130, TA131, TA147, TA148, TA156, TA157, TA169, TA175, TA214, TA216, TA228, TA230, TA232, TA235, TA238, TA252, TA291, and TA307; “-”: negative control; “+”: positive control; “M”: DL2000 Marker.
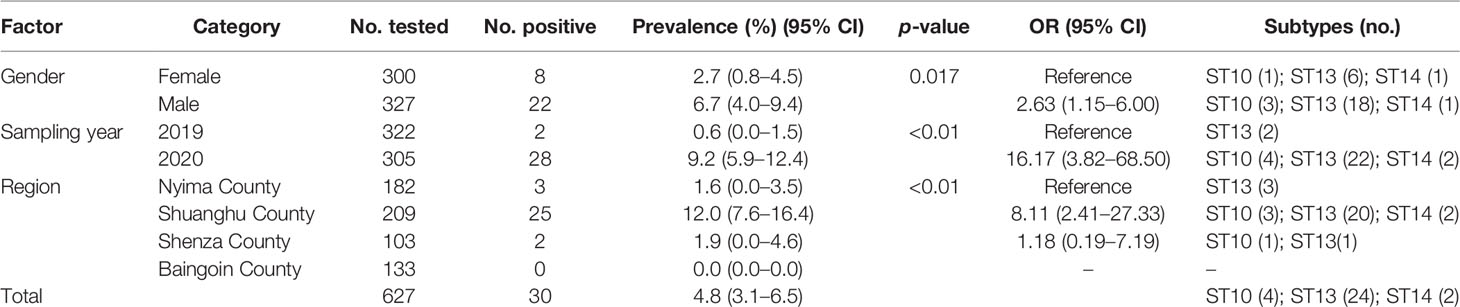
Table 3 Occurrence and subtype distribution of Blastocystis sp. in tibetan antelope (Pantholops hodgsonii).
Risk Factors of Blastocystis sp.
To expose gender, sampling year, and collecting region of Tibetan antelope, and Blastocystis prevalence, univariate analysis was also conducted in the present study (Table 3). A Fisher’s scoring method-based positive stepwise logistic regression analysis was performed to estimate the influence of multiple variables on Blastocystis infection. Only one variable was found to have effects on the Blastocystis infection in the final model, as described by the equation: y = 1.3138x3 + 0.7097. Collecting region had a positive impact on the risk of Blastocystis infection with the OR of 3.720 (95% CI 2.061–6.715). Nyima County (1.6%, 95% CI 0.0–3.5) was considered to have lower prevalence than Shuanghu County (OR = 8.11, 95% CI 2.41–27.33) and Shenza County (OR = 1.18, 95% CI 0.19–7.19) (Table 3).
Distribution and Phylogenetic Analysis of Blastocystis Subtypes
Three Blastocystis subtypes (ST10, ST13, and ST14) were detected in this study. Among them, the ST13 subtype was found in 24 individuals and was widely distributed in different gender subgroups, sampling years, and collecting regions. In the sampling years, all three Blastocystis subtypes appeared in Tibetan antelope in 2020, and only ST13 was found in the Tibetan antelope in 2019 (Table 3). In the gender subgroups, although all of the three subtypes appeared in the Tibetan antelope, the infection of ST10 and ST13 in males was higher than that in females. Tibetan antelope in Shuanghu County was found to be infected with three Blastocystis subtypes in the regional subgroups. However, only the ST13 subtype was found in the Tibetan antelope in Nyima County (Table 3).
The six representative sequences in this study and 49 sequences on GenBank were used to construct a phylogenetic tree. According to the phylogenetic tree analysis, the sequences of the three subtypes (ST10, ST13, and ST14) obtained from this study were clustered with their reference subtypes (Figure 3). The sequence of ST10 isolate has 99% homology with that of ST10 isolated from sika deer (MK930358) and sheep (MW850529). The sequence identified as ST13 in this study has a high degree of homology (99%) with the sequence identified in white Kangaroo (MT672637) and reindeer (MH325366). The ST14 sequence has 98% homology with the known reference sequence identified in sheep (MF186707).
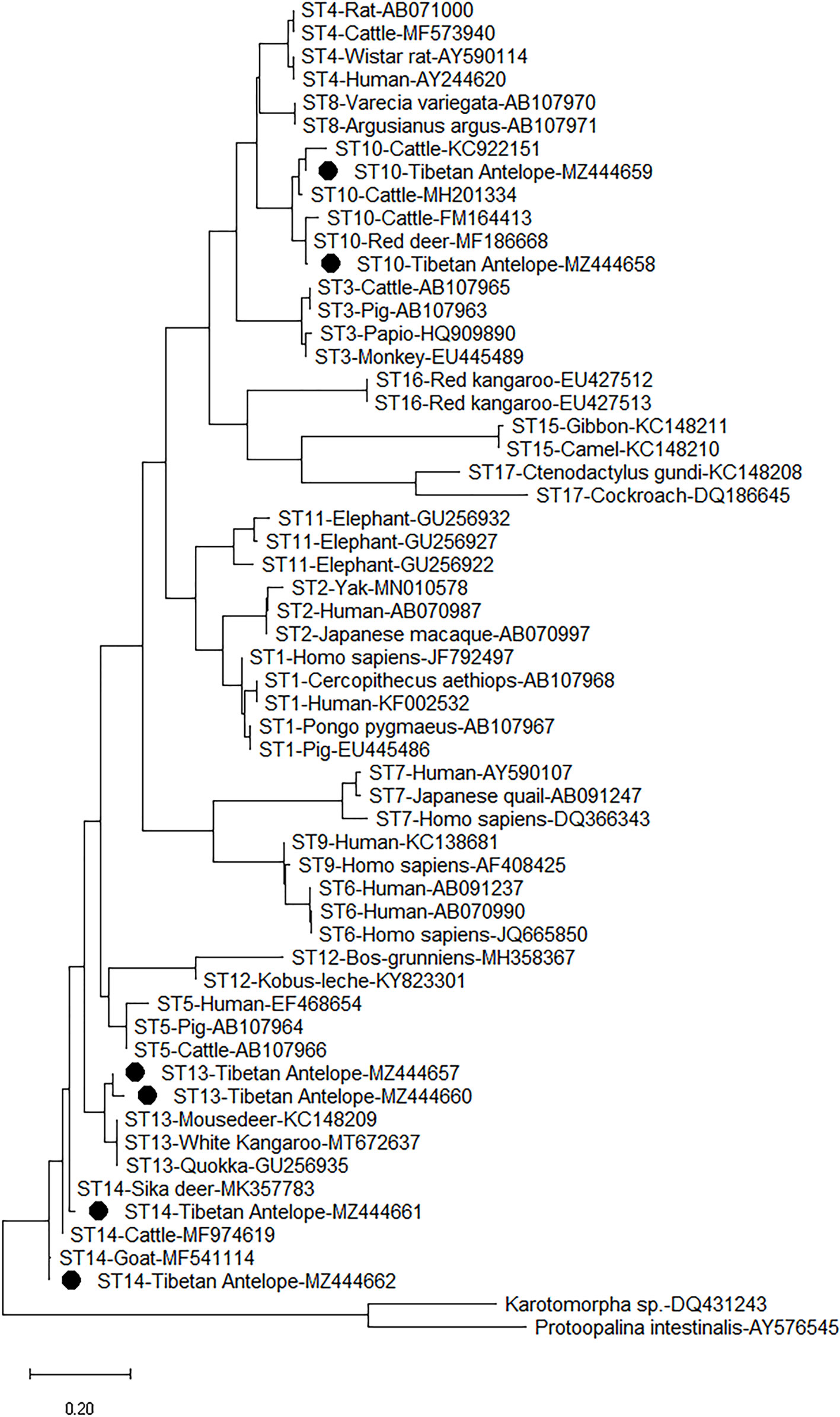
Figure 3 Phylogenetic analyses of Blastocystis using (ML) method (Kimura two-parameter model). Bootstrap values below 50% from 1,000 replicates are not shown. Blastocystis isolates identified in the present study are indicated by solid circles.
Discussion
The overall infection rate of Blastocystis in Tibetan antelope was 4.8% (30/627) in Tibet, which was lower than the prevalence of 5.5% (6/109) and 6.0% (50/832) identified in sheep (Wang et al., 2018a; Li et al., 2018) in China and 19.3% (9/150) and 32.0% (32/100) in sheep in Iran (Rostami et al., 2020; Salehi et al., 2021). The difference in the prevalence of Blastocystis may be related to the living environment and geographical factors of different countries (Tan, 2008). The infection rate of Blastocystis in different species is different. For example, the prevalence of Tibetan antelope in this study and sheep, goats, and cattle in other studies were 4.8% (30/627), 0.75% (3/400) (Ghimire and Bhattarai, 2019), and 14.43% (72/500) (Hastutiek et al., 2019), respectively. The results showed that the prevalence of Blastocystis might be related to the sensitivity of animals to Blastocystis. Therefore, future research should collect more samples to better understand the population characteristics of Blastocystis in Tibetan antelope.
The average temperature in Baingoin County is −17.1°C annually, which is much lower than that of other counties. The survival of Blastocystis may be affected by the low-temperature environment in Baingoin County. Blastocystis might survive in warm and humid environment (Sari et al., 2021). This is probably the reason why the infection rate of Blastocystis in Baingoin County (0.0%, 0/133) was significantly lower than that of the other three counties.
Previous studies have shown that the infection rate of Blastocystis in males (4.8%, 25/517) was higher than that in females (3.1%, 9/291) in cattle (Lee et al., 2018) and in sambar (males: 38.2%, 21/55 vs. females: 23.3%, 7/30) (Kim et al., 2020). This study also found that the infection rate of Blastocystis (6.7%, 22/327) was higher in males than in females (2.7%, 8/300) of Tibetan antelope. This may be due to the fact that males have a wider range of activities than females, and have a relatively higher chance for contacting with cysts.
At present, 29 proposed Blastocystis subtypes have been identified (Maloney and Santin, 2021). Among them, ST1, ST3, ST5, ST10, and ST14 subtypes were detected in sheep and goats (Song et al., 2017; Li et al., 2018; Wang et al., 2018a), among which ST10 and ST14 were the most common subtypes (Fayer et al., 2012; Zhao et al., 2017; Hublin et al., 2021). ST10 and ST14 were also detected in Tibetan antelope. However, it is worth noting that ST10 (n = 4) and ST14 (n = 2) were not common in the samples of Tibetan antelope in this study. On the contrary, ST13 (n = 24) represented the infection trend of Blastocystis in Tibetan antelope. ST13 subtype is a relatively rare subtype. ST13 has been detected in deer, flying squirrels, kangaroo, monkeys, and other animals (Parkar et al., 2010; Alfellani et al., 2013; Wang et al., 2018b; Li et al., 2019; Xiao et al., 2019). Compared with domestic animals, ST13 may be more common in wild animals. Therefore, the follow-up research should focus on the distribution of Blastocystis genotypes in wild animals.
In summary, this is the first report of Blastocystis infection in Tibetan antelope in Tibet, China. The total prevalence of Blastocystis was 4.11% (30/627). Moreover, ST10, ST13, and ST14 subtypes were found in Tibetan antelope, among which ST13 was the dominant subtype. These results not only expanded the knowledge of hosts of Blastocystis, but also provided data for further studies on the distribution of Blastocystis subtypes in Tibetan antelope, and also provided data supporting for the prevention of Blastocystis infection in wild animals.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee of Jilin University.
Author Contributions
H-BN and JJ conceived and designed the study and critically revised the manuscript. S-YQ, H-TS, J-HZ, Z-JW, and TM collected the samples. H-LG, Y-ZS, Y-GL, and N-YX performed the experiments. H-LG and Y-ZS analyzed the data and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Research Foundation for Distinguished Scholars of Qingdao Agricultural University (665-1120046).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Chuang Lyu (Shandong New Hope Liuhe Group Co., Ltd. and Qingdao Jiazhi Biotechnology Co., Ltd., Qingdao, China) for critically revising the manuscript.
References
Abd, R. N. A., Yusof, A. M., Mohammad, M. (2019). Identification of Blastocystis Sp. Infection Form Cattle, Goat and Sheep Isolated From Farms in Pahang, Malaysia. Int. J. Allied Health Sci. 3 (3), 810–810.
AbuOdeh, R., Ezzedine, S., Madkour, M., Stensvold, C. R., Samie, A., Nasrallah, G., et al. (2019). Molecular Subtyping of Blastocystis From Diverse Animals in the United Arab Emirates. Protist 170 (5), 125679. doi: 10.1016/j.protis.2019.125679
Alexeieff, A. (1911). Sur La Nature Des Formations Dites “Kystes De Trichomonas Intestinalis”. Comptes Rendus Des. Seances la Sociiti Biologie 71, 296298.
Alfellani, M. A., Taner-Mulla, D., Jacob, A. S., Imeede, C. A., Yoshikawa, H., Stensvold, C. R., et al. (2013). Genetic Diversity of Blastocystis in Livestock and Zoo Animals. Protist 164, 497–509. doi: 10.1016/j.protis.2013.05.003
Asghari, A., Sadraei, J., Pirestani, M., Mohammadpour, I. (2019). First Molecular Identification and Subtype Distribution of Blastocystis Sp. Isolated From Hooded Crows (Corvus Cornix) and Pigeons (Columba Livia) in Tehran Province, Iran. Comp. Immunol. Microbiol. Infect. Dis. 62, 25–30. doi: 10.1016/j.cimid.2018.11.013
Aynur, Z. E., Güçlü, Ö., Yıldız, İ., Aynur, H., Ertabaklar, H., Bozdoğan, B., et al. (2019). Molecular Characterization of Blastocystis in Cattle in Turkey. Parasitol Res. 118 (3), 1055–1059. doi: 10.1007/s00436-019-06243-8
Badparva, E., Sadraee, J., Kheirandish, F. (2015). Genetic Diversity of Blastocystis Isolated From Cattle in Khorramabad, Iran. Jundishapur J. Microbiol. 8 (3), e14810. doi: 10.5812/jjm.14810
Bai, X., Hu, B., Xu, Y., Sun, H., Zhao, A., Ba, P., et al. (2016). Molecular and Phylogenetic Characterization of Non-O157 Shiga Toxin-Producing Escherichia Coli Strains in China. Front. Cell Infect. Microbiol. 6, 143. doi: 10.3389/fcimb.2016.00143
Deng, L., Chai, Y., Zhou, Z., Liu, H., Zhong, Z., Hu, Y., et al. (2019). Epidemiology of Blastocystis Sp. Infection in China: A Systematic Review. Parasite 26, 41. doi: 10.1051/parasite/2019042
Fayer, R., Santin, M., Macarisin, D. (2012). Detection of Concurrent Infection of Dairy Cattle With Blastocystis, Cryptosporidium, Giardia, and Enterocytozoon by Molecular and Microscopic Methods. Parasitol Res. 111 (3), 1349–1355. doi: 10.1007/s00436-012-2971-1
Ghimire, T. R., Bhattarai, N. (2019). A Survey of Gastrointestinal Parasites of Goats in a Goat Market in Kathmandu, Nepal. J. Parasit. Dis. 43 (4), 686–695. doi: 10.1007/s12639-019-01148-w
Greige, S., El Safadi, D., Khaled, S., Gantois, N., Baydoun, M., Chemaly, M., et al. (2019). First Report on the Prevalence and Subtype Distribution of Blastocystis Sp. In Dairy Cattle in Lebanon and Assessment of Zoonotic Transmission. Acta Trop. 194, 23–29. doi: 10.1016/j.actatropica.2019.02.013
Hastutiek, P., Yuniarti, W. M., Djaeri, M., Lastuti, N. D. R., Suprihati, E., Suwanti, L. T. (2019). Prevalence and Diversity of Gastrointestinal Protozoa in Madura Cattle at Bangkalan Regency, East Java, Indonesia. Vet. World 12 (2), 198–204. doi: 10.14202/vetworld.2019.198-204
Hemalatha, C., Chandrawathani, P., Suresh Kumar, G., Premaalatha, S., Geethamalar, B., Lily Rozita, M. H., et al. (2014). The Diagnosis of Blastocystis Sp. Form Animals-an Emerging Zoonosis. Malaysian J. Vet. Res. 5, 15–21.
Hublin, J. S. Y., Maloney, J. G., Santin, M. (2021). Blastocystis in Domesticated and Wild Mammals and Birds. Res. Vet. Sci. 135, 260–282. doi: 10.1016/j.rvsc.2020.09.031
Jiménez, P. A., Jaimes, J. E., Ramírez, J. D. (2019). A Summary of Blastocystis Subtypes in North and South America. Parasit. Vectors 12 (1), 376. doi: 10.1186/s13071-019-3641-2
Kamaruddin, S. K., Mat, Y. A., Mohammad, M. (2020). Prevalence and Subtype Distribution of Blastocystis Sp. In Cattle From Pahang, Malaysia. Trop. Biomed. 37 (1), 127–141.
Kim, K. T., Noh, G., Lee, H., Kim, S. H., Jeong, H., Kim, Y., et al. (2020). Genetic Diversity and Zoonotic Potential of Blastocystis in Korean Water Deer, Hydropotes Inermis Argyropus. Pathogens 9 (11), 955. doi: 10.3390/pathogens9110955
Lee, H., Lee, S. H., Seo, M. G., Kim, H. Y., Kim, J. W., Lee, Y. R., et al. (2018). Occurrence and Genetic Diversity of Blastocystis in Korean Cattle. Vet. Parasitol. 258, 70–73. doi: 10.1016/j.vetpar.2018.06.010
Li, J., Karim, M. R., Li, D., Rahaman Sumon, S. M. M., Siddiki, S. H. M. F., Rume, F. I., et al. (2019). Molecular Characterization of Blastocystis Sp. In Captive Wildlife in Bangladesh National Zoo: Non-Human Primates With High Prevalence and Zoonotic Significance. Int. J. Parasitol Parasites Wildl. 10, 314–320. doi: 10.1016/j.ijppaw.2019.11.003
Li, W. C., Wang, K., Gu, Y. (2018). Occurrence of Blastocystis Sp. And Pentatrichomonas Hominis in Sheep and Goats in China. Parasit. Vectors 11 (1), 93. doi: 10.1186/s13071-018-2671-5
Li, X. M., Zhou, H., Zhong, D. M., He, J. G. (1990). Diagnosis and Treatment of Two Cases of Blastocystis Hominis. Chin. J. Pediatr. 28, 8.
Maloney, J. G., Jang, Y., Molokin, A., George, N. S., Santin, M. (2021). Wide Genetic Diversity of Blastocystis in White-Tailed Deer (Odocoileus Virginianus) From Maryland, USA. Microorganisms 9 (6), 1343. doi: 10.3390/microorganisms9061343
Maloney, J. G., Lombard, J. E., Urie, N. J., Shivley, C. B., Santin, M. (2019). Zoonotic and Genetically Diverse Subtypes of Blastocystis in US Pre-Weaned Dairy Heifer Calves. Parasitol Res. 118 (2), 575–582. doi: 10.1007/s00436-018-6149-3
Maloney, J. G., Santin, M. (2021). Mind the Gap: New Full-Length Sequences of Blastocystis Subtypes Generated via Oxford Nanopore Minion Sequencing Allow for Comparisons Between Full-Length and Partial Sequences of the Small Subunit of the Ribosomal RNA Gene. Microorganisms 9 (5), 997. doi: 10.3390/microorganisms9050997
Ma, L., Qiao, H., Wang, H., Li, S., Zhai, P., Huang, J., et al. (2020). Molecular Prevalence and Subtypes of Blastocystis Sp. In Primates in Northern China. Transbound Emerg. Dis. 67 (6), 2789–2796. doi: 10.1111/tbed.13644
Masuda, A., Sumiyoshi, T., Ohtaki, T., Matsumoto, J. (2018). Prevalence and Molecular Subtyping of Blastocystis From Dairy Cattle in Kanagawa, Japan. Parasitol Int. 67 (6), 702–705. doi: 10.1016/j.parint.2018.07.005
Moura, R. G. F., Oliveira-Silva, M. B., Pedrosa, A. L., Nascentes, G. A. N., Cabrine-Santos, M. (2018). Occurrence of Blastocystis Spp. In Domestic Animals in Triângulo Mineiro Area of Brazil. Rev. Soc. Bras. Med. Trop. 51 (2), 240–243. doi: 10.1590/0037-8682-0484-2016
Ning, C. Q., Hu, Z. H., Chen, J. H., Ai, L., Tian, L. G. (2020). Epidemiology of Blastocystis Infection From 1990 to 2019 in China. Infect. Dis. Poverty 9 (1), 168. doi: 10.1186/s40249-020-00779-z
Paik, S., Jung, B. Y., Lee, H., Hwang, M. H., Han, J. E., Rhee, M. H., et al. (2019). Molecular Detection and Subtyping of Blastocystis in Korean Pigs. Korean J. Parasitol. 57 (5), 525–529. doi: 10.3347/kjp.2019.57.5.525
Parkar, U., Traub, R. J., Vitali, S., Elliot, A., Levecke, B., Robertson, I., et al. (2010). Molecular Characterization of Blastocystis Isolates From Zoo Animals and Their Animal-Keepers. Vet. Parasitol. 169 (1-2), 8–17. doi: 10.1016/j.vetpar.2009.12.032
Ramírez, J. D., Sánchez, L. V., Bautista, D. C., Corredor, A. F., Flórez, A. C., Stensvold, C. R. (2014). Blastocystis Subtypes Detected in Humans and Animals From Colombia. Infect. Genet. Evol. 22, 223–228. doi: 10.1016/j.meegid.2013.07.020
Ren, M., Song, J. K., Yang, F., Zou, M., Wang, P. X., Wang, D., et al. (2019). First Genotyping of Blastocystis in Yaks From Qinghai Province, Northwestern China. Parasit. Vectors 12 (1), 171. doi: 10.1186/s13071-019-3436-5
Rostami, M., Fasihi-Harandi, M., Shafiei, R., Aspatwar, A., Derakhshan, F. K., Raeghi, S. (2020). Genetic Diversity Analysis of Blastocystis Subtypes and Their Distribution Among the Domestic Animals and Pigeons in Northwest of Iran. Infect. Genet. Evol. 86:104591. doi: 10.1016/j.meegid.2020.104591
Salehi, R., Rostami, A., Mirjalali, H., Stensvold, C. R., Haghighi, A. (2021). Genetic Characterization of Blastocystis From Poultry, Livestock Animals and Humans in the Southwest Region of Iran-Zoonotic Implications. Transbound Emerg. Dis. doi: 10.1111/tbed.14078
Sari, I. P., Audindra, S., Zhafira, A. S., Rahma, A. A., Wahdini, S. (2021). Nutritional Status of School-Aged Children With Intestinal Parasite Infection in South Jakarta, Indonesia. Open Access Macedonian J. Med. Sci. 9 (E), 95–100. doi: 10.3889/oamjms.2021.5711
Scicluna, S. M., Tawari, B., Clark, C. G. (2006). DNA Barcoding of Blastocystis. Protist 157 (1), 77–85. doi: 10.1016/j.protis.2005.12.001
Sharifi, Y., Abbasi, F., Shahabi, S., Zaraei, A., Mikaeili, F., Sarkari, B. (2020). Comparative Genotyping of Blastocystis Infecting Cattle and Human in the South of Iran. Comp. Immunol. Microbiol. Infect. Dis. 72, 101529. doi: 10.1016/j.cimid.2020.101529
Shirozu, T., Morishita, Y. K., Koketsu, M., Fukumoto, S. (2021). Molecular Detection of Blastocystis Sp. Subtype 14 in the Yezo Sika Deer (Cervus Nippon Yesoensis) in Hokkaido, Japan. Vet. Parasito. 100585, 2405–9390. doi: 10.1016/j.vprsr.2021.100585
Song, J. K., Yin, Y. L., Yuan, Y. J., Tang, H., Ren, G. J., Zhang, H. J., et al. (2017). First Genotyping of Blastocystis Sp. In Dairy, Meat, and Cashmere Goats in Northwestern China. Acta Trop. 176, 277–282. doi: 10.1016/j.actatropica
Stensvold, C. R., Clark, C. G. (2016). Current Status of Blastocystis: A Personal View. Parasitol Int. 65 (6 Pt B), 763–771. doi: 10.1016/j.parint.2016.05.015
Suwanti, L. T., Susana, Y., Hastutiek, P., Suprihati, E., Lastuti, N. D. R. (2020). Blastocystis Spp. Subtype 10 Infected Beef Cattle in Kamal and Socah, Bangkalan, Madura, Indonesia. Vet. World 13 (2), 231–237. doi: 10.14202/vetworld.2020.231-237
Tan, K. S. (2008). New Insights on Classification, Identification, and Clinical Relevance of Blastocystis Spp. Clin. Microbiol. Rev. 21 (4), 639–665. doi: 10.1128/CMR.00022-08
Tan, T. C., Tan, P. C., Sharma, R., Sugnaseelan, S., Suresh, K. G. (2013). Genetic Diversity of Caprine Blastocystis From Peninsular Malaysia. Parasitol Res. 112 (1), 85–89. doi: 10.1007/s00436-012-3107-3
Udonsom, R., Prasertbun, R., Mahittikorn, A., Mori, H., Changbunjong, T., Komalamisra, C., et al. (2018). Blastocystis Infection and Subtype Distribution in Humans, Cattle, Goats, and Pigs in Central and Western Thailand. Infect. Genet. Evol. 65, 107–111. doi: 10.1016/j.meegid.2018.07.007
Wang, J., Gong, B., Liu, X., Zhao, W., Bu, T., Zhang, W., et al. (2018b). Distribution and Genetic Diversity of Blastocystis Subtypes in Various Mammal and Bird Species in Northeastern China. Parasit. Vectors 11 (1), 522. doi: 10.1186/s13071-018-3106-z
Wang, J., Gong, B., Yang, F., Zhang, W., Zheng, Y., Liu, A. (2018a). Subtype Distribution and Genetic Characterizations of Blastocystis in Pigs, Cattle, Sheep and Goats in Northeastern China’s Heilongjiang Province. Infect. Genet. Evol. 57, 171–176. doi: 10.1016/j.meegid.2017.11.026
Xiao, X., Zhou, S. H., Jiang, N., Tian, D. Z., Zhou, Z. M., Zhang, M., et al. (2019). First Record of Leptospira and Blastocystis Infections in Captive Flying Squirrels (Trogopterus Xanthipes) From Enshi County, China. Acta Trop. 197, 105065. doi: 10.1016/j.actatropica.2019.105065
Yu, Z., Wang, T., Sun, H., Xia, Z., Zhang, K., Chu, D., et al. (2012). Contagious Caprine Pleuropneumonia in Endangered Tibetan Antelope, Chin. Emerg. Infect. Dis. 19 (12), 2051–2053. doi: 10.3201/eid1912.130067
Zhao, G. H., Hu, X. F., Liu, T. L., Hu, R. S., Yu, Z. Q., Yang, W. B., et al. (2017). Molecular Characterization of Blastocystis Sp. In Captive Wild Animals in Qinling Mountains. Parasitol Res. 116 (8), 2327–2333. doi: 10.1007/s00436-017-5506-y
Zhu, W., Tao, W., Gong, B., Yang, H., Li, Y., Song, M., et al. (2017). First Report of Blastocystis Infections in Cattle in China. Vet. Parasitol. 246, 38–42. doi: 10.1016/j.vetpar.2017.09
Keywords: Blastocystis, prevalence, subtypes, Tibetan antelope (Pantholops hodgsonii), PCR
Citation: Geng H-L, Sun Y-Z, Jiang J, Sun H-T, Li Y-G, Qin S-Y, Wang Z-J, Ma T, Zhu J-H, Xue N-Y and Ni H-B (2021) The Presence of Blastocystis in Tibetan Antelope (Pantholops hodgsonii). Front. Cell. Infect. Microbiol. 11:747952. doi: 10.3389/fcimb.2021.747952
Received: 27 July 2021; Accepted: 09 September 2021;
Published: 29 September 2021.
Edited by:
Guo-Hua Liu, Hunan Agricultural University, ChinaReviewed by:
Md Robiul Karim, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshZahra Babaei, Kerman University of Medical Sciences, Iran
Meysam Sharifdini, Guilan University of Medical Sciences, Iran
Copyright © 2021 Geng, Sun, Jiang, Sun, Li, Qin, Wang, Ma, Zhu, Xue and Ni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Jiang, amlhbmdqaW5neGlhb3lhb0AxNjMuY29t; Hong-Bo Ni, aG9uZ2JvbmlAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Hong-Li Geng1†
Hong-Li Geng1† Si-Yuan Qin
Si-Yuan Qin Nian-Yu Xue
Nian-Yu Xue Hong-Bo Ni
Hong-Bo Ni
