- 1The Second Affiliated Hospital of Dalian Medical University, Liaoning, China
- 2Department of Bacteriology, Capital Institute of Pediatrics, Beijing, China
- 3Department of Daily Clinic, Seventh Medical Center of People’s Liberation Army (PLA) General Hospital, Beijing, China
Klebsiella pneumoniae carbapenemase genes (blaKPC) play an important role in carbapenem-resistant Enterobacteriaceae in China. A rapid detection method for blaKPC genes and investigations into the molecular characteristics of blaKPC positive Klebsiella pneumoniae were necessary. In this study, an easy and rapid recombinase aided amplification assay (RAA) for blaKPC was established. This protocol could be completed at 39°C in 15–20 min. The sensitivity of this assay was determined as 48 copies per reaction, and the specificity was 100%. The blaKPC RAA method could be used for clinical diagnosis and epidemiological investigation. Among 801 fecal samples from inpatients, 34 blaKPC positive isolates were identified from each sample, of which 23 isolates were K. pneumoniae. ST11 with blaKPC-2 was the most prevalent type. All these strains were multidrug resistant and carried various virulence genes. Fecal carriage of blaKPC positive carbapenem-resistant K.pneumoniae poses significant challenges for public health control.
Introduction
Carbapenem antibiotics have once been regarded as the last line of defense for the treatment of resistant Enterobacteriaceae bacteria. In recent years, with the extensive application of carbapenem antibiotics, carbapenem-resistant Enterobacteriaceae (CRE) have shown a steadily increasing trend (Xu et al., 2015). Klebsiella pneumoniae carbapenemase (KPC) is one of the significant carbapenemases that belong to class A of the Ambler classification system, and they are capable of hydrolyzing carbapenems and β-lactamase. Rectal colonization or carriage of KPC serves as an important reservoir for dissemination of resistant bacteria (Borer et al., 2012). This could result in both horizontal and vertical spreading between and within species. Therefore, a rapid and simple detection method is integral to the control of KPC dissemination during clinical diagnosis and treatment.
At present, molecular detection methods for blaKPC are slow and laborious. Recombinase aided amplification assay (RAA) is a novel isothermal amplification technology that uses specific enzymes and proteins for DNA amplification at 39°C within 15–20 min. In the RAA assay, the three major proteins including single strand DNA binding protein, recombinase UvsX (anneals the primers to the template DNA), and DNA polymerase (for amplification and extension) make the reaction system rapid and specific. Recently, RAA has been successfully used to detect various microbial pathogens (Zhang et al., 2017; Qin et al., 2021), but it has not been used to detect resistance genes.
In this study, we established an RAA detection method to accurately identify the blaKPC gene. In addition, as KPC positive species in fecal samples are frequently encountered, approximately 90%–100% in K. pneumoniae (Kpn) (Liu et al., 2019; Pan et al., 2019; Xu et al., 2020), we further analyzed the characteristics of KPC positive K. pneumoniae including multilocus sequence typing (MLST), serotype, mucus phenotype, and virulence genes. Our results provided a rapid clinical screening test to improve the control of K. pneumoniae.
Materials and Methods
Clinical Samples, Strain Collection, and DNA Extraction
A total of 801 nonduplicate stool specimens for routine analysis were randomly collected from 434 adult inpatients in a general hospital and 367 pediatric inpatients in a pediatrics hospital in Beijing, China from December, 2020 to February, 2021. The two hospitals were both tertiary comprehensive hospitals. Clinical information for these isolates was simultaneously collected.
The collected specimens were processed in two parts. A 200 mg sample was taken from each fecal sample for total DNA extraction, according to the instructions of the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The remaining specimens were cultured on MacConkey Agar plates. The isolated strains were verified using a VITEK-2 compact system (bioMerieux, Marcy l’Etoile, France) and 16s-rRNA Sequencing (Sangon Biotech Co., Ltd., Shanghai, China).
Primers and Probe Design for RAA
A total of 87 blaKPC genes were present in the NCBI website, all these genes were downloaded for the primer design. The conserved regions of 87 blaKPC genes were aligned and contrasted using BioEdit Sequence Alignment Editor 7.2.1 software. The primers and probe were manually designed within the highly conserved regions (Figure 1), according to the principles of RAA primer and probe design. NCBI Primer-BLAST was used to check and ensure the specificity of the primers and probe. Oligo7 software was used to carefully analyze dimer formation among themselves (self-dimers) and the formation of secondary structures (hairpins). The primers and probe were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China).
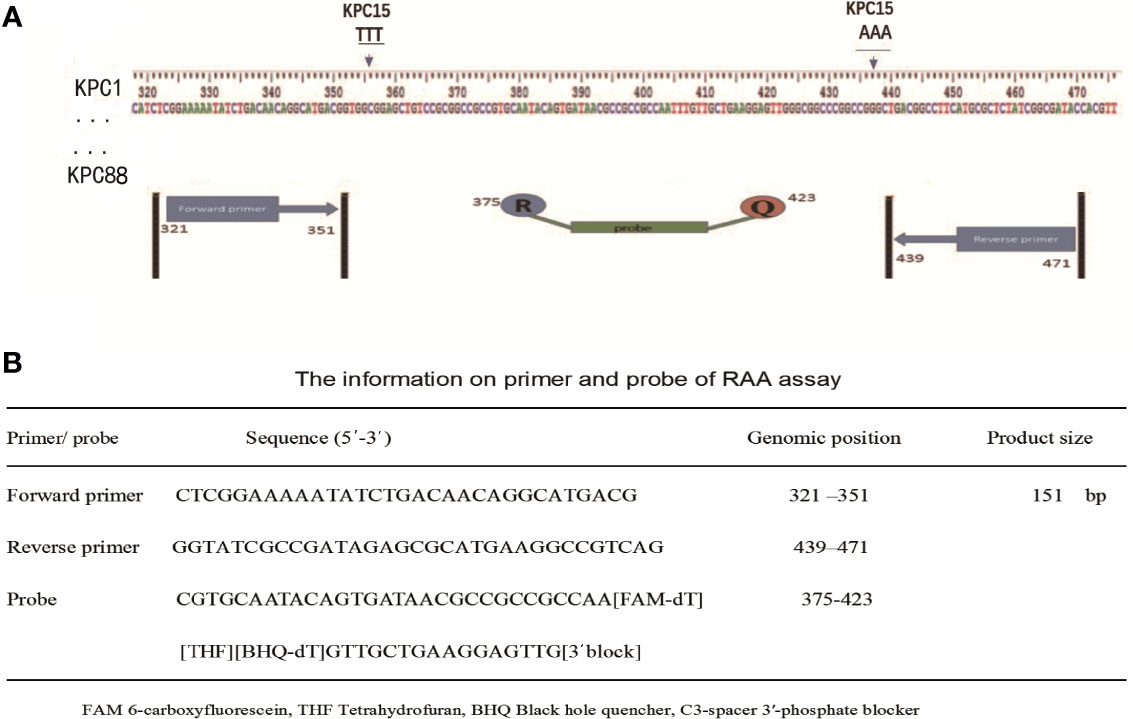
Figure 1 Selection of specific regions and primer positions. (A) A total of 87 blaKPC genes (blaKPC-1 to blaKPC-82 and blaKPC-84 to blaKPC-88) were released in the NCBI website, and blaKPC83 was not released. The primers and probe sets were designed within the highly conserved regions of 87 blaKPC genes (320-472 bp region) by avoiding two mutation regions of blaKPC-15. (B) The primer and probe sequences used for RAA assays.
RAA Assay
The full-length blaKPC-2 gene (Kpn ATCC1705) was PCR-amplified and cloned into vector pUC57 (Tiangen Biotech Co., Ltd, Beijing, China). A dilution series with the recombinant plasmids (ranging from 107 to 100 copies/μl) was prepared to evaluate the sensitivity of the RAA assay.
The RAA amplification assays were performed in a 50 μL reaction volume using a commercial RAA kit (Qitian Biological Co., Ltd., Jiangsu, China). The reaction mixture contained 2 μL of extracted DNA template, 25 μL of reaction buffer, 15.7 μL of DNase-free water, 2.1 μL of primer F, 2.1 μL of primer R, 0.6 μL of probe, and 2.5 μL of magnesium acetate. The tubes with the reaction mixture containing the RAA enzyme mix in a lyophilized form, were placed into a B6100 Oscillation mixer (QT-RAA-B6100, Qitian Bio-Tech Co. Ltd., Jiangsu, China) and incubated for 4 min, mixed briefly, centrifuged, and finally transferred to a fluorescence detector (QT-RAA-1620, Qitian Bio-Tech Co. Ltd., Jiangsu, China) to be measured for 10–20 min at 39°C. A positive control (blaKPC positive plasmid) and negative control (double-distilled water) were included in each run.
Analytical Sensitivity, Specificity, and Reproducibility of the RAA Assay
Analytical sensitivity of the RAA assay was carried out using a serial diluted recombinant plasmid ranging from 107 to 100 copies/μL. The specificity assay was evaluated by the testing of Kpn ATCC1705 (carrying blaKPC-2), Acinetobacter baumannii (carrying blaKPC-3), Pseudomonas aeruginosa (carrying blaKPC-2), and another 12 bacterial samples without blaKPC genes (PCR verified, data not shown), as follows: Kpn ATCC2146 (carrying blaNDM-1), K. oxytoca (carrying blaIMP-4), Escherichia coli (carrying blaOXA-48), Shigella sonnei, Salmonella enteritidis, Proteus mirabilis, Yersinia enterocolitica, Campylobacter jejuni, Citrobacter freundii, Burkholderia cepacia, Haemophilus influenzae and Morganella morganii. All the above mentioned clinical strains were originally preserved at the Department of Bacteriology of Capital Institute of Pediatrics. In addition, six replicates were performed on six different days to validate the reproducibility of the RAA assay.
PCR Detection and Sequencing
There was currently no commercial kit for blaKPC detection, therefore standard PCR was used for the reference. A 50 μL reaction volume containing the following components was used for all the PCRs: 25 μL of PCR Master Mix reagent (Tiangen Biotech Co., Ltd, Beijing, China), 19 μL of double-distilled water, 2 μL of 10 μM KPC-F primer (5′-ATGTCACTGTATCGCCGTCT-3′) and KPC-R primer (5′-TTACTGCCCGTTGACGC-3′), and 2 μL of DNA template. PCR products were sequenced at Sangon Biotech Co., Ltd. (Shanghai, China) and compared to the NCBI database (http://blast.ncbi.nlm.nih.gov).
Detection and Evaluation of the RAA Assay With Clinical Samples
All the blaKPC-positive samples and the randomly selected blaKPC-negative samples that were detected by standard PCR from 801 samples were used to validate the performance of the established RAA method. The results of the RAA assay were compared with those from a standard PCR assay.
Molecular Typing Analyses of the KPC Positive Kpn Isolates
MLST and KL serotyping were conducted to phylogenically classify the KPC positive Kpn. Seven housekeeping genes of Kpn were amplified and sequenced based on protocols as described (Diancourt et al., 2005). The wzi gene for each strain was amplified with specific primers as reported previously (Brisse et al., 2013). Sequence types (ST) and KL-genotypes were identified using the online database at the Pasteur Institute Multilocus Sequence Typing website for Kpn (https://bigsdb.pasteur.fr).
Antimicrobial Susceptibility Testing of the KPC Positive Kpn
Antimicrobial susceptibility testing was performed using the VITEK2 compact system. The minimal inhibitory concentration (MIC) values for co-trimoxazole, aztreonam, ampicillin/sulbactam, piperacillin/tazobactam, cefazolin, cefuroxime, ceftazidime, cefoperazone/sulbactam, cefotaxime, cefepime, gentamicin, amikacin, ciprofloxacin, levofloxacin, tetracycline, imipenem, and meropenem were determined. Escherichia coli ATCC25922 and Pseudomonas aeruginosa ATCC27853 were used for quality control. The results were recorded, according to the 2020 Clinical Laboratory Standards Institute’s threshold.
Hypermucoviscosity Phenotype, Growth Curve, and Virulence Genes Assay of the KPC Positive Kpn
The string test of strains was performed to determine the hypermucoviscosity phenotype, as previously described (Shon et al., 2013). Growth curve assays for a randomly selected isolate of each type of Kpn were conducted to assess the in vitro fitness of strains for growth kinetics analysis. Experiments were repeated in triplicate. The growth curve results were calculated by statistical analysis and the average absorbance values were used for drawing. Various virulence genes involved in invasion infection and colonization (rmpA2, rmpA, magA, fimH, wabG, uge, allS, mrkD, aerobactin, wcaG, kfuBC, iucA, iroN, ybtA, ureA, iucB, entB, and iroB) were amplified, which were related to capsular polysaccharide synthesis, fimbriae synthesis, iron acquisition system, urease, and lipopolysaccharide. Carrier rates of virulence factors grouped by the differing MLST and KL-type were documented.
Statistical Analysis
Statistical analyses of the kappa and P-values of the RAA and standard PCR assays (with sequencing), were calculated with SPSS 21.0 (IBM, Armonk, NY, USA). The trials were performed in triplicate.
Results
Primers and Probe Design of the RAA Assay
As shown in Figure 1A, the RAA primers and probe for the blaKPC gene were manually designed within the conserved regions of the 87 blaKPC types, and alignment among the different blaKPC types was presented. Sequences for two optimal primers and one probe were listed in Figure 1B.
Analytical Sensitivity and Specificity of the RAA Assay
The sensitivity of the RAA assay for blaKPC was shown in Figure 2A. An increase in the fluorescence signal was observed from 102 to 107 copies/uL. The detection limit of the RAA assay was 48 copies per reaction while standard PCR was 480 copies per reaction. RAA is more sensitive when compared with standard PCR.
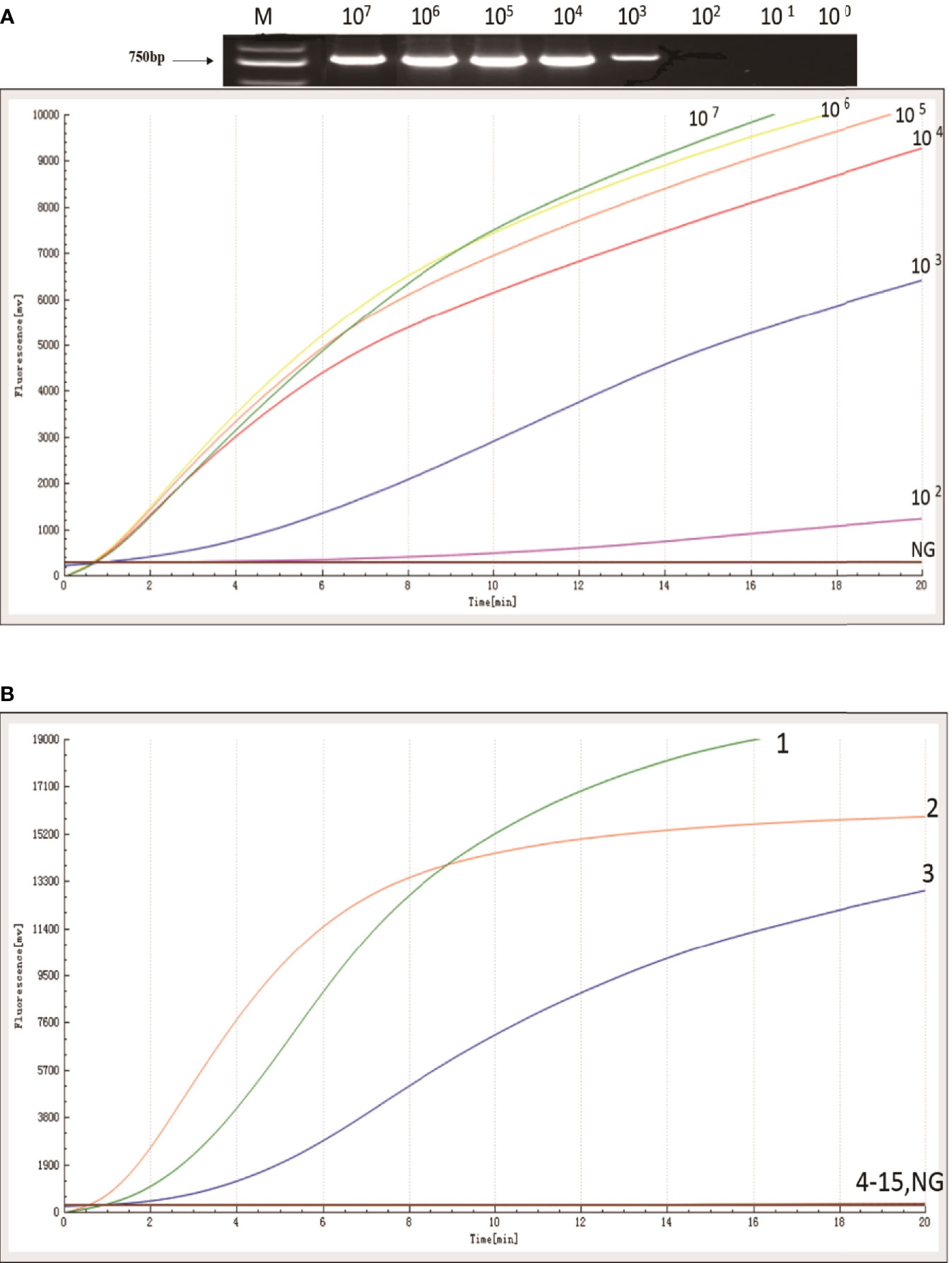
Figure 2 Sensitivity and specificity of RAA assay. (A) An increase in the fluorescence signal was observed from 1× 107 to 1 × 102 copies/reaction. (B) Kpn ATCC1705, Acinetobacter baumannii, and Pseudomonas aeruginosa produced amplification signals (1, 2, and 3), while the other KPC negative bacterial strains (4-15) and negative control were negative.
The specificity of the RAA assay for blaKPC was confirmed by testing three blaKPC-positive samples and another 12 blaKPC-negative samples. In contrast with the other blaKPC-negative bacterial samples and the distilled water control (4 to 15, in Figure 2B), only Kpn ATCC1705, Acinetobacter baumannii, and Pseudomonas aeruginosa produced amplification signals (1, 2, and 3, in Figure 2B); meanwhile, no cross-reactivity was observed for other types of carbapenemase gene. The RAA assay for the detection of blaKPC was specific (100%).
Evaluation of the RAA Assay Using Clinical Samples
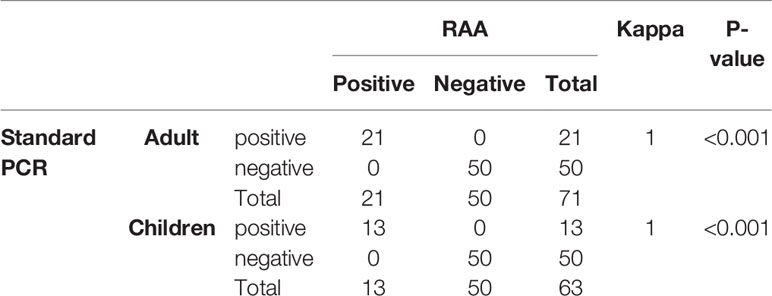
Initially, during the study period, 801 stool samples were used for screening blaKPC genes by standard PCR, of which 34 (4.2%, 34/801) samples were positive for blaKPC. Subsequently, all 34 blaKPC-positive samples and 100 randomly selected blaKPC-negative samples were used for RAA validation. The results of the RAA assay were in complete agreement with those obtained with standard PCR. No false-positives were found between the RAA assay and the standard PCR, as shown in Table 1.
Clinical Characteristics of blaKPC-Positive Strains, MLST, and KL-Serotype
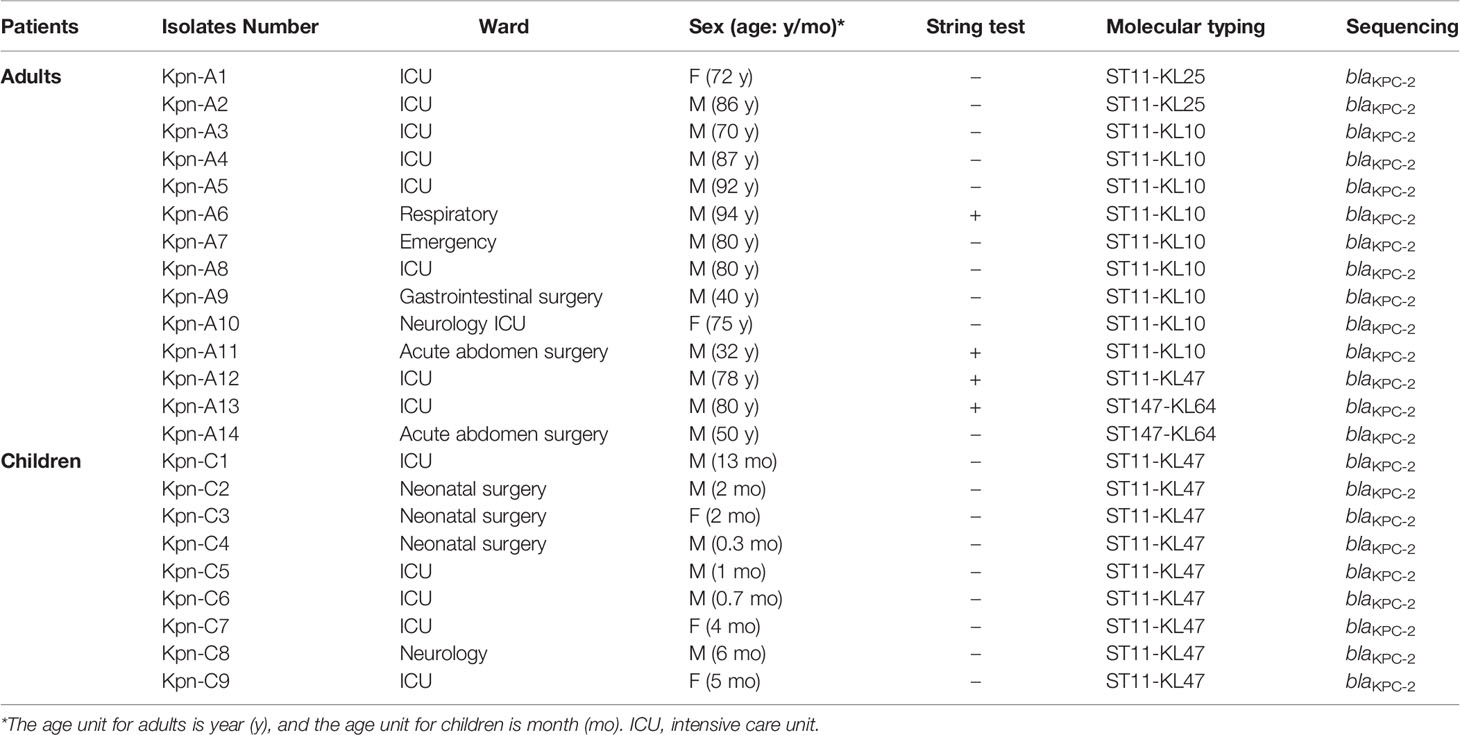
The carbapenem resistant strains were isolated from all the 34 blaKPC-positive samples that were detected by standard PCR and RAA. Of them, 23 Kpn (67.6%, 23/34) were identified, which referred to 2.9% (23/801) in fecal samples. All the blaKPC-positive Kpn strains carried blaKPC-2. Characteristics, including age, gender, ward distribution, and sequencing results for the 23 cases were shown in Table 2. Overall, most of the blaKPC-positive Kpn cases (20/23) came from ICU wards and surgical wards.
MLST analysis revealed that ST11 was the predominant ST (91.3%, 21/23), followed by ST147 (8.7%, 2/23). We further investigated the KL serotypes of these strains: KL47 (43.5%, 10/23), KL10 (15.6%, 9/23), and KL25 (8.7%, 2/23). There were three KL serotypes in ST11; ST11-KL47 were the most common type (10/21), followed by ST11-KL10 (9/21), ST11-KL25 (2/21), and ST147-KL64 (2/21) (Table 2).
Interestingly, a male ICU patient (Kpn-A13 in Table 2) diagnosed with colon cancer, had a sputum culture result that was carbapenem-resistant Kpn, following clinical laboratory tests. We isolated the blaKPC-positive Kpn stain from his stool sample and sputum sample, and they shared the same genotype (ST147-KL64).
Antimicrobial Susceptibility of blaKPC-Positive Kpn Isolates
Antimicrobial-susceptibility testing revealed that all blaKPC-positive Kpn isolates were resistant to Carbapenem antibiotics (Imipenem and Meropenem, 100%) and Beta-lactam antibiotics (aztreonam, piperacillin/tazobactam, cefazolin, ceftazidime, cefoperazone/sulbactam, cefepime; 100%) (Supplementary Figure S1); furthermore, they were also multidrug resistant. However, 95.7% of these isolates were susceptible to Tigecycline (22/23) and 52.2% retained susceptibility to Sulfonamides antibiotics (12/23).
Hypermucoviscosity Phenotype, Growth Curve, and Virulence Genes of blaKPC-Positive Kpn Isolates
The string test showed that four strains were hypermucoviscous (Table 2), which belonged to ST11(75%) and ST147 (25%), respectively. The blaKPC-positive strains were in a logarithmic growth phase for 1–8 hours, reached a maximum at ~8 hours, and then maintained a relatively stable state (Figure 3A). There was no statistically significant difference in growth of blaKPC-positive and blaKPC-negative strains (P > 0.05).
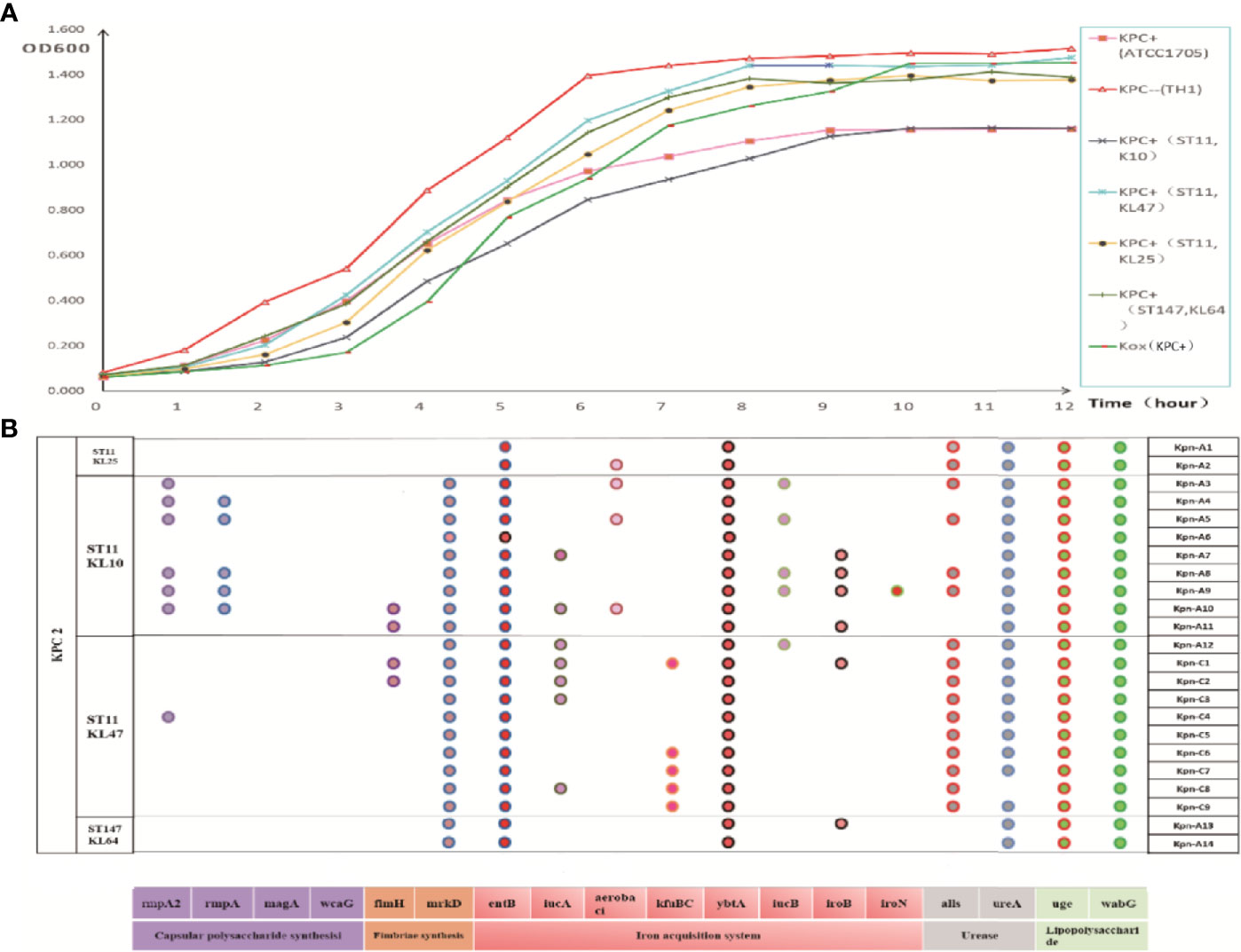
Figure 3 (A) Growth curve of each group of strains. The data exhibited are the average absorbance values of three independent experiments. (B) Distribution of virulence gene in strains. The presence of virulence genes in a specific genome is indicated by different colored circles. Virulence genes and their roles are shown at the bottom.
As shown in Figure 3B, blaKPC-positive Kpn strains contained multiple virulence genes including entB (100%, iron acquisition system), uge (100%, lipopolysaccharide), ybtA (100%, iron acquisition system), wabG (100%, lipopolysaccharide), mrkD (91%, fimbriac synthesis), alls (70%, urease), and ureA (96%, urease). ST11-KL10 strains harbored almost all the virulence genes. ST11 strains (48%, 183/378) carried more virulence genes than ST147 strains (36%, 13/36).
Discussion
The blaKPC gene is a carbapenem resistance gene that is located in mobile transposon Tn4401, which facilitates its transfer between plasmids, species, patients, and different countries (Munoz and Quinn, 2009). The KPC epidemic is problematic for global health. Therefore, identifying and screening blaKPC genes in clinical settings would be extremely helpful.
In the present study, we established an RAA assay for the detection of the blaKPC gene. The primers and a probe were designed in the conserved region of all 87 existing blaKPC genes, thus allowing the amplification of all these genes. This assay, therefore, had good specificity. The sensitivity of the blaKPC RAA assay was superior to standard PCR and reached the same level as other nucleic acid detections for blaKPC, such as real-time PCR (5–40 copies/reaction) (Chen et al., 2011; Subirats et al., 2017) and loop-mediated isothermal amplification assay (LAMP) (40-105CFU/mL) (Nakano et al., 2015; Srisrattakarn et al., 2017). The agreement between the RAA and standard PCR assays was 100%, which suggested that the RAA assay was of sufficient quality to be used for clinical blaKPC screening.
RAA assay was superior to LAMP or real-time PCR assays, in terms of the simplicity, rapidity and cost (Shen et al, 2019). Real-time PCR requires varying cycling temperatures under rigorous conditions, while RAA assay only need a constant 39°C simple device. In addition to unsophisticated laboratory setting, RAA assay could be completed in 15-20 min, while real-time PCR requires 2-3 hours, which represents a significant reduction of the turnaround time. Real-time PCR is difficult to perform in laboratories with limited funds, poor equipment, and a shortage of technicians. As regards the cost of assay, each test for LAMP assay costs 5-6 $. the RAA cost for one sample is about half of that by LAMP. The high cost of LAMP assays restricts its use in clinical settings.
In the present study, the most common blaKPC genotype (blaKPC-2) and the fecal carriage rate were consistent with the other studies (Zhao et al., 2014; Liu et al., 2019). Also, we found blaKPC positive patients mainly in intensive care units (ICU) and surgical wards, where patients received more invasive treatment and numerous antibiotics (including carbapenem), which may lead to the spreading of blaKPC. Some reports showed that carbapenem treatments led to the emergence of carbapenem-resistant K. pneumoniae (CR-Kpn) in patients’ gastrointestinal tracts, which subsequently caused infections in other body sites (Zhang et al., 2018). Fecal colonization with CRE is a risk factor for bacterial translocation resulting in subsequent endogenous infections, which can also be seen from our research. One male patient hospitalized for colon cancer, gradually developed pneumonia. The same blaKPC-positive Kpn strains (ST147-KL64) were detected from his stool and sputum samples. This implied that CR-Kpn can colonize the gastrointestinal tract and cause disease under special conditions. Long-time asymptomatic carriage of CR-Kpn in the intestines of adults and children have recently attracted more attention (Lübbert et al., 2014; Ghaith et al., 2019). In this study, most strains carrying blaKPC were Kpn (67.6%). This was consistent with other studies (Liu et al., 2019). It is therefore very important for us to study the molecular characteristics of these strains.
In the present study, ST11 with blaKPC2 was the most prevalent type in blaKPC positive Kpn strains, which was consistent with the other studies (Liu et al., 2019; Pan et al., 2019; Chen et al., 2020). To further investigate the detailed difference in ST11, serotypes were tested and three KL types (KL25, KL47, KL10) were found. Type KL-47 has been reported in other studies in China (Zheng et al., 2020). In addition to ST11, we also isolated two ST147 with KL64 type Kpn strains. This was consistent with another Italian study where they found ST147 with KL64 blaKPC positive Kpn from rectal swab specimens (Arena et al., 2020). Diverse KL types of Kpn strains determine different degrees of detection by the innate immune system involving immune clearance (Doorduijn et al., 2016).
Results of the present study illustrated that only 17.4% (4/23) of strains were hypermucoviscous, which is consistent with other studies (10.9%, 6/55) (Zhang et al., 2020). In the current study, these blaKPC-positive Kpn contained multiple virulence genes associated with synthesis or regulation related factors for infection, including capsular polysaccharide synthesis, fimbriae synthesis, iron acquisition system, urease, and lipopolysaccharide. These strains will definitely have an impact and be problematic in the control and prevention of mutual transmissions.
In this study, all blaKPC-positive Kpn isolates were resistant to carbapenems and beta-lactam antibiotics, which revealed that blaKPC genotype matched the phenotype. However, most of them were susceptible to tigecycline (95.7%), and some of them retained susceptibility to sulfonamides (52.2%). The CR-Kpn infection therapeutic options are limited to tigecycline, colistin, polymyxin B, etc. Effective surveillance and strict infection control strategies should be implemented to prevent the dissemination of CR-Kpn.
There were some limitations in this study, like the small number of cases and the short duration for a retrospective study.
Conclusion
In conclusion, the RAA assay established in this study had a high specificity and sensitivity and provides a simple and reliable method for blaKPC detection, which is suitable for application in primary laboratories or clinical practice. In blaKPC-positive Kpn strains from hospitalized patients’ stool samples, ST11 carrying blaKPC-2 was the most prevalent type. These strains were multidrug resistant and contained various virulence genes. Effective surveillance and control measures are urgently required to control the transmission of blaKPC-positive strains.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
JY, GX, and RL designed the study. WZ, YF, HZ, CY, JF, LG, and JC performed the experiments. RZ, SL, SD, NL, WX, and JH analyzed the results. WZ and GX wrote the article. JY, GX, and RL revised it. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by Public service development and reform pilot project of Beijing Medical Research Institute (BMR2019-11), and Research Foundation of Capital Institute of Pediatrics (GZ-2021-06 and CXYJ-2021-04). National Natural Science Foundation for Key Programs of China Grants (82130065), National Natural Science Foundation of China (31370093 82002191 and 32170201) and FENG foundation (FFBR 202103) to JY
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge Jiangsu Qitian Bio-Tech Co. Ltd. for providing the RAA detection Kit.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.746325/full#supplementary-material
References
Arena, F., Di, ,. P. V., Vannetti, F., Fabbri, L., Antonelli, A., Coppi, M., et al. (2020). Population Structure of KPC Carbapenemase-Producing Klebsiella Pneumoniae in a Long-Term Acute-Care Rehabilitation Facility: Identification of a New Lineage of Clonal Group 101, Associated With Local Hyperendemicity. Microb. Genom. 6 (1), e000308. doi: 10.1099/mgen.0.000308
Borer, A., Odes, L. S., Eskira, S., Nativ, R., Riesenberg, K., Riven, L. I., et al. (2012). Risk Factors for Developing Clinical Infection With Carbapenem-Resistant Klebsiella Pneumoniae in Hospital Patients Initially Only Colonized With Carbapenem-Resistant K Pneumoniae. Am. J. Infect. Control. 40 (5), 421–425. doi: 10.1016/j.ajic.20
Brisse, S., Passet, V., Haugaard, A. B., Babosan, A., Kassis-Chikhani, N., Struve, C., et al. (2013). Wzi Gene Sequencing, a Rapid Method for Determination of Capsular Type for Klebsiella Strains. J. Clin. Microbiol. 51 (12), 4073–4078. doi: 10.1128/JCM.01924-13
Chen, X., Liu, Q., Liu, W. E., Yan, Q. (2020). Risk Factors for Subsequential Carbapenem-Resistant Klebsiella Pneumoniae Clinical Infection Among Rectal Carriers With Carbapenem-Resistant Klebsiella Pneumoniae. Infect. Drug Resist. 13, 1299–1305. doi: 10.2147/IDR.S247101
Chen, L., Mediavilla, J. R., Endimiani, A., Rosenthal, M. E., Zhao, Y., Bonomo, R. A., et al. (2011). Multiplex Real-Time PCR Assay for Detection and Classification of Klebsiella Pneumoniae Carbapenemase Gene (Bla KPC) Variants. J. Clin. Microbiol. 49 (2), 579–585. doi: 10.1128/JCM.01588-10
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A., Brisse, S. (2005). Multilocus Sequence Typing of Klebsiella Pneumoniae Nosocomial Isolates. J. Clin. Microbiol. 43 (8), 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005
Doorduijn, D. J., Rooijakkers, S. H., van Schaik, W., Bardoel, B. W. (2016). Complement Resistance Mechanisms of Klebsiella Pneumoniae. Immunobiology 221 (10), 1102–1109. doi: 10.1016/j.imbio.2016.06.01
Ghaith, D. M., Mohamed, Z. K., Farahat, M. G., Aboulkasem, S. W., Mohamed, H. O. (2019). Colonization of Intestinal Microbiota with Carbapenemase-Producing Enterobacteriaceae in Paediatric Intensive Care Units in Cairo, Egypt. Arab. J. Gastroenterol. 20 (1), 19–22. doi: 10.1016/j.ajg.2019.01.002
Liu, Q., Liu, L., Li, Y., Chen, X., Yan, Q., Liu, W. E. (2019). Fecal Carriage and Epidemiology of Carbapenem-Resistant Enterobacteriaceae Among Hospitalized Patients in a University Hospital. Infect. Drug Resist. 12, 3935–3942. doi: 10.2147/IDR.S233795
Lübbert, C., Lippmann, N., Busch, T., Kaisers, U. X., Ducomble, T., Eckmanns, T., et al. (2014). Long-term Carriage of Klebsiella pneumoniae Carbapenemase-2-Producing K Pneumoniae after a Large Single-Center Outbreak in Germany. Am. J. Infect. Cont. 42 (4), 376–380. doi: 10.1016/j.ajic.2013.12.001
Munoz, P. L. S., Quinn, J. P. (2009). The Spread of Klebsiella Pneumoniae Carbapenemases: A Tale of Strains, Plasmids, and Transposons. Clin. Infect. Dis. 49 (11), 1739–1741. doi: 10.1086/648078
Nakano, R., Nakano, A., Ishii, Y., Ubagai, T., Kikuchi, U. T., Kikuchi, H., et al. (2015). Rapid Detection of the Klebsiella Pneumoniae Carbapenemase (KPC) Gene by Loop-Mediated Isothermal Amplification (LAMP). J. Infect. Chemother. 21 (3), 202–206. doi: 10.1016/j.jiac.2014.11.010
Pan, F., Tian, D., Wang, B., Zhao, W., Qin, H., Zhang, T. (2019). Fecal Carriage and Molecular Epidemiology of Carbapenem-Resistant Enterobacteriaceae From Outpatient Children in Shanghai. BMC Infect. Dis. 19 (1), 678. doi: 10.1186/s12879-019-4298-3
Qin, Z., Xue, L., Cai, W., Gao, J., Jiang, Y., Yang, J., et al. (2021). Development of a Recombinase-Aided Amplification Assay for Rapid Detection of Human Norovirus GII.4. BMC Infect. Dis. 21 (1), 248. doi: 10.1186/s12879-021-05942-x
Shen, X. X., Qiu, F. Z., Shen, L. P., Yan, T. F., Zhao, M. C., Qi, J. J., et al. (2019). A Rapid and Sensitive Recombinase Aided Amplification Assay to Detect Hepatitis B Virus Without DNA Extraction. BMC Infect. Dis. 19 (1), 229. doi: 10.1186/s12879-019-3814-9
Shon, A. S., Bajwa, R. P., Russo, T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A New and Dangerous Breed. Virulence 4 (2), 107–118. doi: 10.4161/viru.22718
Srisrattakarn, A., Lulitanond, A., Wilailuckana, C., Charoensri, N., Wonglakorn, L., Saenjamla, P., et al. (2017). Rapid and Simple Identification of Carbapenemase Genes, Blandm, Blaoxa-48, Blavim, Blaimp-14 and Blakpc Groups, in Gram-Negative Bacilli by in-House Loop-Mediated Isothermal Amplification With Hydroxynaphthol Blue Dye. World J. Microbiol. Biotechnol. 33 (7), 130. doi: 10.1007/s11274-017-2295-5
Subirats, J., Royo, E., Balcázar, J. L., Borrego, C. M. (2017). Real-Time PCR Assays for the Detection and Quantification of Carbapenemase Genes (Bla KPC, Bla NDM, and Bla OXA-48) in Environmental Samples. Environ. Sci. Pollut. Res. Int. 24 (7), 6710–6714. doi: 10.1007/s11356-017-8426-6
Xu, Y., Gu, B., Huang, M., Liu, H., Xu, T., Xia, W., et al. (2015). Epidemiology of Carbapenem Resistant Enterobacteriaceae (CRE) During 2000-2012 in Asia. J. Thorac. Dis. 7 (3), 376–385. doi: 10.3978/j.issn.2072-1439.2014.12.33
Xu, Q., Pan, F., Sun, Y., Wang, C., Shi, Y., Zhang, T., et al. (2020). Fecal Carriage and Molecular Epidemiology of Carbapenem-Resistant Enterobacteriaceae From Inpatient Children in a Pediatric Hospital of Shanghai. Infect. Drug Resist. 13, 4405–4415. doi: 10.2147/IDR.S27554
Zhang, R., Dong, N., Huang, Y., Zhou, H., Xie, M., Chan, E. W., et al. (2018). Evolution of Tigecycline- and Colistin-Resistant CRKP (Carbapenem-Resistant Klebsiella Pneumoniae) In Vivo and Its Persistence in the GI Tract. Emerg. Microbes Infect. 7 (1), 127. doi: 10.1038/s41426-018-0129-7
Zhang, X., Guo, L., Ma, R., Cong, L., Wu, Z., Wei, Y., et al. (2017). Rapid Detection of Salmonella With Recombinase Aided Amplification. J. Microbiol. Methods 139, 202–204. doi: 10.1016/j.mimet.2017.06.011
Zhang, Y., Jin, L., Ouyang, P., Wang, Q., Wang, R., Wang, J., et al. (2020). Evolution of Hypervirulence in Carbapenem-Resistant Klebsiella Pneumoniae in China: A Multicentre, Molecular Epidemiological Analysis. J. Antimicrob. Chemother. 75 (2), 327–336.
Zhao, Z. C., Xu, X. H., Liu, M. B., Wu, J., Lin, J., Li, B. (2014). Fecal Carriage of Carbapenem-Resistant Enterobacteriaceae in a Chinese University Hospital. Am. J. Infect. Control. 42 (5), e61–e64. doi: 10.1016/j.ajic.2014.01.024
Keywords: recombinase-aided amplification, rapid detection, blaKPC, characteristic, Klebsiella pneumoniae
Citation: Zhang W, Feng Y, Zhao H, Yan C, Feng J, Gan L, Cui J, Liu S, Zhang R, Du S, Li N, Xu W, Han J, Li R, Xue G and Yuan J (2021) A Recombinase Aided Amplification Assay for Rapid Detection of the Klebsiella pneumoniae Carbapenemase Gene and Its Characteristics in Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 11:746325. doi: 10.3389/fcimb.2021.746325
Received: 23 July 2021; Accepted: 24 August 2021;
Published: 20 September 2021.
Edited by:
Penghua Wang, University of Connecticut Health Center, United StatesReviewed by:
Xuejun Ma, Chinese Center For Disease Control and Prevention, ChinaSun Ying, Capital Medical University, China
Copyright © 2021 Zhang, Feng, Zhao, Yan, Feng, Gan, Cui, Liu, Zhang, Du, Li, Xu, Han, Li, Xue and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Yuan, eXVhbmppbmc2MjE2QDE2My5jb20=; Guanhua Xue, Z3Vhbmh1YXh1ZUAxNjMuY29t; Rongkuan Li, ZGFsaWFubHJrQDEyNi5jb20=
†These authors have contributed equally to this work
 Weiwei Zhang
Weiwei Zhang Yanling Feng
Yanling Feng Hanqing Zhao2
Hanqing Zhao2 Chao Yan
Chao Yan Jinghua Cui
Jinghua Cui Juqiang Han
Juqiang Han Guanhua Xue
Guanhua Xue Jing Yuan
Jing Yuan