- 1Department of Pathogen Biology and Immunology, Kunming Medical University, Kunming, China
- 2Department of Internal Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL, United States
Drug resistance in Plasmodium vivax may pose a challenge to malaria elimination. Previous studies have found that P. vivax has a decreased sensitivity to antimalarial drugs in some areas of the Greater Mekong Sub-region. This study aims to investigate the ex vivo drug susceptibilities of P. vivax isolates from the China–Myanmar border and genetic variations of resistance-related genes. A total of 46 P. vivax clinical isolates were assessed for ex vivo susceptibility to seven antimalarial drugs using the schizont maturation assay. The medians of IC50 (half-maximum inhibitory concentrations) for chloroquine, artesunate, and dihydroartemisinin from 46 parasite isolates were 96.48, 1.95, and 1.63 nM, respectively, while the medians of IC50 values for piperaquine, pyronaridine, mefloquine, and quinine from 39 parasite isolates were 19.60, 15.53, 16.38, and 26.04 nM, respectively. Sequence polymorphisms in pvmdr1 (P. vivax multidrug resistance-1), pvmrp1 (P. vivax multidrug resistance protein 1), pvdhfr (P. vivax dihydrofolate reductase), and pvdhps (P. vivax dihydropteroate synthase) were determined by PCR and sequencing. Pvmdr1 had 13 non-synonymous substitutions, of which, T908S and T958M were fixed, G698S (97.8%) and F1076L (93.5%) were highly prevalent, and other substitutions had relatively low prevalences. Pvmrp1 had three non-synonymous substitutions, with Y1393D being fixed, G1419A approaching fixation (97.8%), and V1478I being rare (2.2%). Several pvdhfr and pvdhps mutations were relatively frequent in the studied parasite population. The pvmdr1 G698S substitution was associated with a reduced sensitivity to chloroquine, artesunate, and dihydroartemisinin. This study suggests the possible emergence of P. vivax isolates resistant to certain antimalarial drugs at the China–Myanmar border, which demands continuous surveillance for drug resistance.
Introduction
Plasmodium vivax is the most widely distributed malaria parasite, which can also cause life-threatening diseases. P. vivax was responsible for 6.4 million cases globally in 2019, with nearly 51.7% of the cases in Southeast Asia (WHO, 2020). Countries within the Greater Mekong Sub-region (GMS) are pursuing malaria elimination by 2030 (WHO, 2015). As the GMS is progressing toward eliminating P. falciparum by 2025, the proportion of malaria cases caused by P. vivax infections has increased steadily. Myanmar carries the heaviest malaria burden, with malaria transmission concentrated along its international borders (Geng et al., 2019). In most vivax-endemic areas, the combination of chloroquine (CQ) and primaquine remains the first-line treatment for uncomplicated P. vivax, but CQ resistance has been an increasing concern. Although artemisinin-based combination therapies (ACTs) as a unified treatment of P. falciparum and P. vivax infection may have significant advantages in areas where both species coexist (Douglas et al., 2010), ACT treatment of vivax malaria is commonly deployed in areas such as Indonesia, where P. vivax CQ resistance was evident (Baird, 2011).
CQ was first produced in 1934 and soon proved to be one of the most successful and important antimalarial drugs (Wellems and Plowe, 2001). However, the extensive use of CQ in the following decades eventually led to drug resistance. P. falciparum developed resistance in various areas since the 1950s, but drug-resistant P. vivax was not reported until the 1980s in Indonesia and Papua New Guinea (Rieckmann et al., 1989; Baird et al., 1991). To date, CQ-resistant (CQR) P. vivax has been detected in more than ten countries (Price et al., 2014). As early as 1993, CQR P. vivax cases were reported in Myanmar (Myat Phone et al., 1993; Marlar et al., 1995; Guthmann et al., 2008). In recent years, clinical failures after CQ treatment, indicating RI or even RII type resistance as defined by the WHO criteria (WHO, 1967), have been reported in many regions of Myanmar (Yuan et al., 2015; Htun et al., 2017; Xu et al., 2020). At the Thai–Myanmar border, 8% of P. vivax patients developed recurrent parasitemia within 28 days after CQ treatment (Chu et al., 2018). In addition, although the recurrence rate of P. vivax infection within 28 days after CQ treatment in other parts of the GMS such as Vietnam (3.5%) and western Thailand (8%) was still low (Rijken et al., 2011; Amaratunga et al., 2014a), CQR P. vivax isolates were confirmed in these areas (Rijken et al., 2011; Thanh et al., 2015). Therefore, the CQR parasites have become a focus of attention in the GMS, especially since this parasite is targeted for elimination.
Sulfadoxine-pyrimethamine (SP) has a long history of combating malaria due to its safety, good tolerance, and long-lasting activity (Müller and Hyde, 2013). In China, as a component of the two combination prophylactic regimens, SP was widely used for malaria prophylaxis between the mid-1960s and early 1990s (Huang et al., 2014). Under this scenario, all malaria parasite species should be under a selection pressure by antifolate drugs. In addition, although SP has rarely been used to treat vivax malaria, the prevalence of mixed P. falciparum/P. vivax infections in the region (Li et al., 2014) suggests that P. vivax might also be under selection by antifolate drugs used to treat P. falciparum infections. A decade ago, P. falciparum resistance to artemisinin family drugs was detected first in Cambodia (Noedl et al., 2008; Amaratunga et al., 2014b) and then in other countries of the GMS (Holmgren et al., 2007; Dondorp et al., 2009; Ashley et al., 2014). P. falciparum has also developed resistance to ACT partner drugs such as mefloquine (MFQ) and piperaquine (PPQ) (Price et al., 2004; Nawaz et al., 2009; Cui et al., 2012). Cross-sectional surveys in the Thai–Myanmar border area, Cambodia and Vietnam detected 1% of infections as mixed P. falciparum/P. vivax infections (Imwong et al., 2015). This rate reached 14% among febrile patients at the China–Myanmar border. Furthermore, misdiagnosis of malaria cases is also common; a recent survey of acute malaria cases showed that as much as 20% of P. vivax cases were misdiagnosed in the field (Geng et al., 2019). These studies highlight the inevitable collateral selection of P. vivax by the commonly used antimalarial drugs used to treat P. falciparum.
The underlying mechanisms of antimalarial drug resistance in P. vivax are less well understood than those in P. falciparum. P. vivax CQR mechanism appears different from that for P. falciparum. Mutations in the P. falciparum chloroquine resistance transporter (pfcrt) are the main determinants of CQ resistance (Fidock et al., 2000), but studies failed to identify an association between CQ resistance and mutations in the pfcrt ortholog, P. vivax chloroquine resistance transporter-o (pvcrt-o) (Nomura et al., 2001). The pfmdr1 (P. falciparum multidrug resistance 1) gene is often used to monitor the resistance of P. falciparum to 4-aminoquinolines and MFQ (Duraisingh et al., 2000; Djimdé et al., 2001; Price et al., 2004; Happi et al., 2006). Its ortholog in P. vivax, pvmdr1, may also play a role in CQ resistance. Some identified an association of the pvmdr1 Y976F substitution with CQR P. vivax in vitro (Suwanarusk et al., 2007), but others did not find such an association (Sá et al., 2005; Barnadas et al., 2008; Gomes et al., 2016). The P. falciparum multidrug resistance protein 1 (pfmrp1) gene, encoding an ABC transporter transmembrane protein, is a potential multidrug-resistance candidate in P. falciparum with mutations associated with a reduced susceptibility to CQ (Veiga et al., 2011; Gupta et al., 2014). Therefore, although several genes are suspected to be associated with CQ resistance in P. vivax, there are no definitive molecular markers for monitoring P. vivax CQ resistance. Mutations in dihydrofolate reductase (pvdhfr) and dihydropteroate synthase (pvdhps) have been associated with the altered clinical response to SP (de Pécoulas et al., 1998; Eldin de Pécoulas et al., 1998; Imwong et al., 2001; Tjitra et al., 2002; Imwong et al., 2003; Korsinczky et al., 2004). To track the ex vivo drug susceptibilities of P. vivax parasites in northeast Myanmar, we collected clinical samples and profiled their ex vivo susceptibilities to seven antimalarial drugs, which are the first-line drugs for treating vivax or falciparum malaria in the GMS. Furthermore, we surveyed the polymorphisms of candidate resistance markers pvmdr1, pvmrp1, pvdhfr, and pvdhps in these isolates.
Material and Methods
Ethical Approvals, Study Site, and Parasite Isolates
This study was approved by the institutional review boards of Kunming Medical University and Pennsylvania State University. Signed informed consent/assent forms were obtained from all patients and guardians in the case of minors. P. vivax clinical isolates were obtained from P. vivax patients aged seven years and older who attended the malaria clinics located at the Laiza town, Kachin State, Myanmar, in May and July of 2016. Patients with clinical symptoms of malaria (e.g., rigor, fever, headache, malaise, muscle pains) were diagnosed by a microscopic examination of Giemsa-stained blood smears. Parasites were staged based on morphological characteristics described earlier (Russell et al., 2003). Only patients with >0.5% parasitemia and >70% ring stage on presentation and without taking antimalarial medicine were recruited. Patients younger than 7 years, anemic patients (hemoglobin level <7 g/dl), pregnant and lactating women, and those who took antimalarial drugs in the preceding two weeks were excluded. Three (for patients aged 7–17 years) or five milliliters (for patients aged ≥18 years) of blood were drawn from each patient by venipuncture into a heparin-coated tube, transported to the nearby laboratory in a 37°C thermos, and used for ex vivo drug assays within four hours of collection.
Ex Vivo Drug Assay
CQ, MFQ, quinine (QN), and pyronaridine phosphate (PND) were obtained from Sigma. PPQ was obtained from Chongqing Kangle Pharmaceutical Co., Ltd (Chongqing, China). Dihydroartemisinin (DHA) and artesunate (AS) were obtained from Kunming Pharmaceutical Corp. (Kunming, China). The stock solutions and serial dilutions were made according to the previous description (Li et al., 2020). Each drug concentration was tested in triplicate. The testing plates were dried inside a biosafety hood, sealed, and stored at 4°C for use within two weeks. The ex vivo drug assay, i.e., the schizont maturation assay, was performed as described earlier (Li et al., 2017; Li et al., 2020). Two milliliters of P. vivax-infected blood were centrifuged at 1,000 rpm for 10 min, and the cell pellet was resuspended with two volumes of RPMI 1640. Leukocytes were removed using an NWF filter (Zhi Xing Bio, Bengbu, China) (Tao et al., 2011). Then, 400 μL of the packed erythrocytes were resuspended in 19.6 mL of culture medium containing McCoy’s 5A (11.9 g/L, Sigma, St. Louis, MO, USA), HEPES (25 ml/L, Sigma), gentamicin (5 mg/L, Jinan Limin Pharmaceutical Co., Ltd, Jinan, China), 7.5% NaHCO3 (2.1 g/L, Sigma), and 25% AB+ serum from malaria-naive donors to make a 2% hematocrit. The red blood cell (RBC) suspension (100 μL/well) was dispensed into the wells of a Costar 96-well flat-bottom microtiter plate (Sigma), which was pre-dosed with antimalarial drugs. The plates were then incubated in a candle jar at 37°C for 26–36 h depending on the stage of the parasites. Twelve hours later, parasite development in the drug-free wells was monitored every 1–4 h. When more than 40% of the parasites in the control wells developed to the mature schizont stage, all wells were collected to prepare thick and thin films (Lu et al., 2012). Blood films were fixed with methanol, stained with 10% Giemsa (Sigma) solution for 30 min, and examined microscopically under oil immersion. Schizonts with at least three well-defined chromatin dots were counted. The number of schizonts per 200 asexual parasites at each drug concentration was determined and normalized with the control well.
DNA Amplification and Sequencing
QiAamp DNA Mini Kit (Qiagen, Hilden, Germany) was used to extract parasite DNA from 200 μL of whole blood. Single-species P. vivax infections were confirmed by nested PCR targeting the 18S rRNA gene (Snounou et al., 1993). Then, P. vivax merozoite surface protein- 3α (pvmsp-3α) was genotyped using a PCR and restriction fragment length polymorphism (RFLP) protocol to confirm monoclonal P. vivax infections (Bruce et al., 1999). Multiple molecular markers, including pvmdr1, pvmrp1, pvdhps, and pvdhfr, which are related to drug resistance in P. vivax, were amplified and sequenced. The full-length pvmdr1 (4,395 bp) was amplified by PCR using Phanta Max Super-Fidelity DNA polymerase P505d (Nanjing Vazyme Biotech, China) containing high-fidelity Pfu DNA polymerase with primers pvmdr1F (5′- CAGCAGACACCATTTAAGG-3′) and pvmdr1R (5′- CCGTTTGTTGATTAGTTGC-3′). Fragments of pvmrp1 (497 bp), pvdhps (767 bp), and pvdhfr (632 bp), which covered potential drug resistance-associated mutations, were amplified by nested PCR using primers reported from previous studies (Mint Lekweiry et al., 2012; Chehuan et al., 2013) (Supplementary Table S2). All reactions were performed in 25 μL containing 12.5 μL of 2× Phanta Max Buffer, 10 mM of each dNTP mix, 0.5 μM of each primer, 0.5 U of Super-Fidelity DNA polymerase P505d, and 1 μL (5–10 ng) of genomic DNA. The PCR cycling parameters were as follows: initial denaturation at 95°C for 3 min, followed by 35 cycles at 95°C for 15 sec, 58°C for 15 sec, 72°C for 3 min, and then a final extension at 72°C for 5 min. The PCR products were resolved on a 1.2% agarose gel, and the sizes of the PCR products were determined using LD2000 or LD5000 DNA ladder (TaKaRa, Japan). Amplification products were sequenced on both strands by Sangon Biotech Co. Ltd. (Shanghai, China). Sequences were assembled using the SeqMan program of the Lasergene software (DNASTAR, Madison, WI, USA). Individual sequences were aligned to the Salvador I strain reference sequences retrieved from the GenBank (XM_001613678 for pvmdr1, XP_001612680 for pvmrp1, XP_001617209 for pvdhps, and XP_001615082 for pvdhfr) to determine the presence of single nucleotide polymorphisms in the respective genes. Nucleotide sequences were translated into amino acid sequences to examine mutant codons using BioEdit (version 7.2.5).
Data Analysis
Statistical analyses were performed using GraphPad Prism6 (GraphPad Software, Inc, San Diego, CA, USA). Geometric means of the ex vivo IC50s (half-maximum inhibitory concentrations) were calculated by fitting the drug response data to a sigmoid curve. Medians and interquartile ranges (IQRs) were calculated because the data were not normally distributed. IC50s between groups were compared by using the Mann-Whitney U test and the Wilcoxon matched-pairs signed-rank test. Correlations between IC50s of drugs were determined using Spearman’s test in the R package.
Results
Ex Vivo Susceptibilities of Parasite Isolates to Antimalarial Drugs
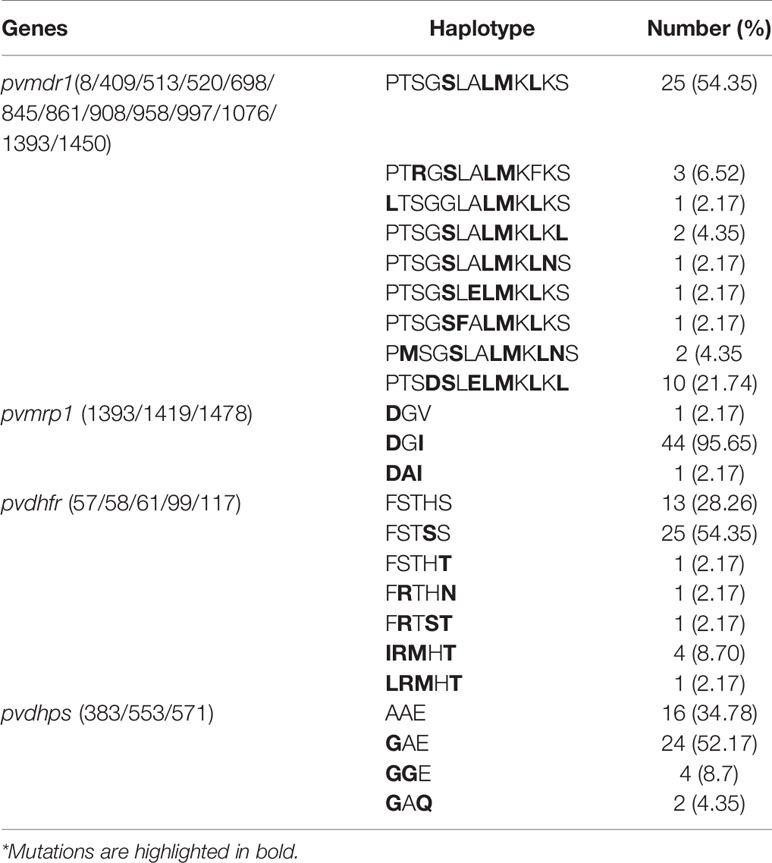
Parasite samples were obtained from 97 P. vivax patients fitting the recruitment criteria and were used to determine susceptibilities to seven antimalarial drugs. Among them, 57 isolates successfully developed to mature schizonts. Genotyping by RFLP analysis of pvmsp3α determined that 46 were monoclonal P. vivax infections. All 46 isolates were successfully assayed for CQ, AS, and DHA. Since seven samples were not sufficient for assaying all drugs, 39 isolates were assayed for PPQ, PND, MFQ, and QN (Table 1 and Figure 1A). For CQ, the median ex vivo IC50 was 96.48 nM (range: 36.5–303.4 nM), with 4.4% (2/46) isolates having IC50 values above 220 nM. For the artemisinin derivatives AS and DHA, the median IC50 was 1.95 (range: 0.4–11.8 nM) and 1.63 (range: 0.3–10.2 nM), respectively. For PND, the median IC50 was 15.5 nM (range: 4.5–38.8 nM). For MFQ, the median IC50 was 16.4 nM (range: 5.8–48.5 nM). For PPQ, the median IC50 was 19.6 nM (range: 8.5–52.8 nM). Isolates had a median ex vivo IC50 value of 26.0 nM (range: 7.2–149.2 nM) to QN. Compared to the ex vivo assays performed in 2012–2015, we found a rising trend of IC50 for PND only (Supplementary Table 3). To detect whether the susceptibilities to different drugs were correlated, Spearman’s correlation analysis was performed between IC50s of pairs of the drugs. There is no correlation between the IC50 values of the seven antimalarial drugs of the parasite isolates in the study, except for a weak correlation between DHA and CQ (Figure 1B, r = 0.34, P < 0.05).
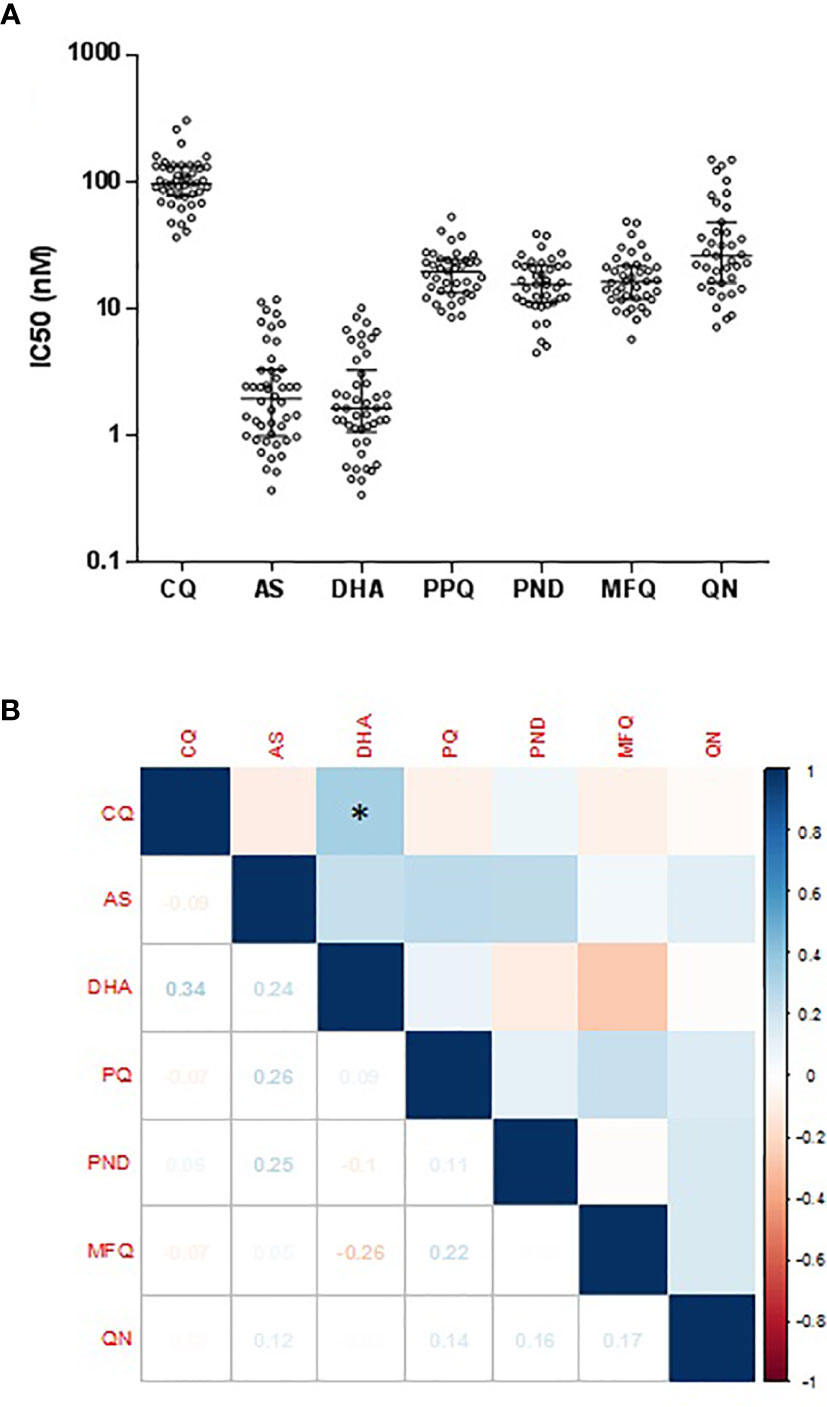
Figure 1 IC50 values of each antimalarial drug among P. vivax isolates P. vivax isolates for each antimalarial drug. (A) Dot plots of ex vivo susceptibilities of P. vivax isolates to seven antimalarial drugs. Comparison of ex vivo IC50 values (in nM) to seven antimalarial drugs. (B) Correlations between IC50s of P. vivax isolates to seven antimalarial drugs. Used Spearman’s test to analyze the correlation between IC50s Spearman’s correlation analysis between IC50 values of parasite isolates to seven antimalarial drugs. The values of correlation coefficient based on the right side of the color scale. The values of the correlation coefficient were based on the color scale shown on the right. *indicates a significant correlation.
Polymorphisms in pvmdr1, pvmrp1, pvdhfr, and pvdhps Genes
The pvmdr1, pvmrp1, pcdhfr, and pvdhps genes were successfully sequenced in 46 P. vivax isolates. Compared with the Sal-I sequence, 13 non-synonymous substitutions (P8L, T409M, S513R, G520D, G698S, L845F, A861E, M908L, T958M, K997R, F1076L, K1393N, and S1450L) were identified in the pvmdr1 gene (Table 2). Two substitutions M908L and T958M were fixed, while G698S was present in 97.8% of the parasite population. The two substitutions Y976F and F1076L, which might be associated with CQ resistance (Brega et al., 2005), had completely different prevalences: Y976F was not detected, whereas F1076L was highly prevalent (93.5%). A total of nine haplotypes pvmdr1 were identified (Table 3). The most prevalent haplotype PTSGSLALMKLKS (54.4%) carried four substitutions, followed by PTSDSLELMKLKL (21.7%) with seven substitutions.

Table 2 The prevalence of pvmdr1, pvmrp1, pvdhfr and pvdhps genes substitutions in 46 parasite isolates and association of the mutations with altered ex vivo drug susceptibilities.
For part of the pvmrp1 gene (497 bp), compared with the Sal-I sequence, three non-synonymous substitutions (Y1393D, G1419A, and V1478I) were identified (Table 2). The Y1393D substitution was fixed, and G1419A was highly prevalent (97.8%). The most prevalent haplotype was DGI (95.7%) (Table 3).
Mutations in pvdhfr at codons 57, 58, 61, 99, and 117 were detected in 10.9, 15.2, 10.9, 56.5, and 17.4% of the isolates, respectively. No mutations were found at positions 13 or 173 (Table 2). The most prevalent haplotype FSTSS (54.4%) carried one mutation, which was followed by the wild type (28.3%) (Table 3). Three different tandem repeat variations were found in the pvdhfr gene. Type 1 was identical to the Sal-I reference sequence, type 2 had the H99S mutation, and Type 3 carried a deletion of six amino acids at positions 98–103 (THGGDN) (Supplementary Figure S2). Type II was the most common at 56.5% (26/46), followed by Type III at 28.3% (13/46). Of note, Type 3 carried only the S117N mutation. Compared with pvdhfr, pvdhps showed a relatively lower prevalence of mutation genotypes. A553G and E571Q were present at low frequencies (Table 2), A383G was detected in more than half of the isolates. The most prevalent haplotype GAE (52.17%) carried one mutation, which was followed by the wild type (34.8%) (Table 3).
Correlation Between pvmdr1 and pvmrp1 Gene Polymorphisms and Ex Vivo Drug Susceptibilities
Of the 13 non-synonymous pvmdr1 SNPs (P8L, T409M, S513R, G520D, G698S, L845F, A861E, M908L, T958M, K997R, F1076L, K1393N, and S1450L), P8L was significantly associated with the decreased IC50 to CQ, and G698S had a significant association with increased IC50 to CQ (P < 0.05; Figure 2). Based on the cutoff IC50 value of 220 nM used by others (Suwanarusk et al., 2007), two P. vivax isolates were categorized as CQR. P8L had a significant association with the decreased IC50 to AS, and G698S had a significant association with the increased IC50 to AS (P < 0.01; Figure 2). P8L and L845F had a significant association with the decreased IC50 to DHA, and G698S had a significant association with the increased IC50 to DHA (P < 0.01; Figure 2). L845F had a significant association with the decreased IC50 to PPQ (P < 0.01; Supplementary Figure S2). G520D and L845F had a significant association with the increased IC50 to PND (P < 0.05; Supplementary Figure S2). L845F also had a significant association with the increased IC50 to MFQ (P < 0.01; Supplementary Figure S2).
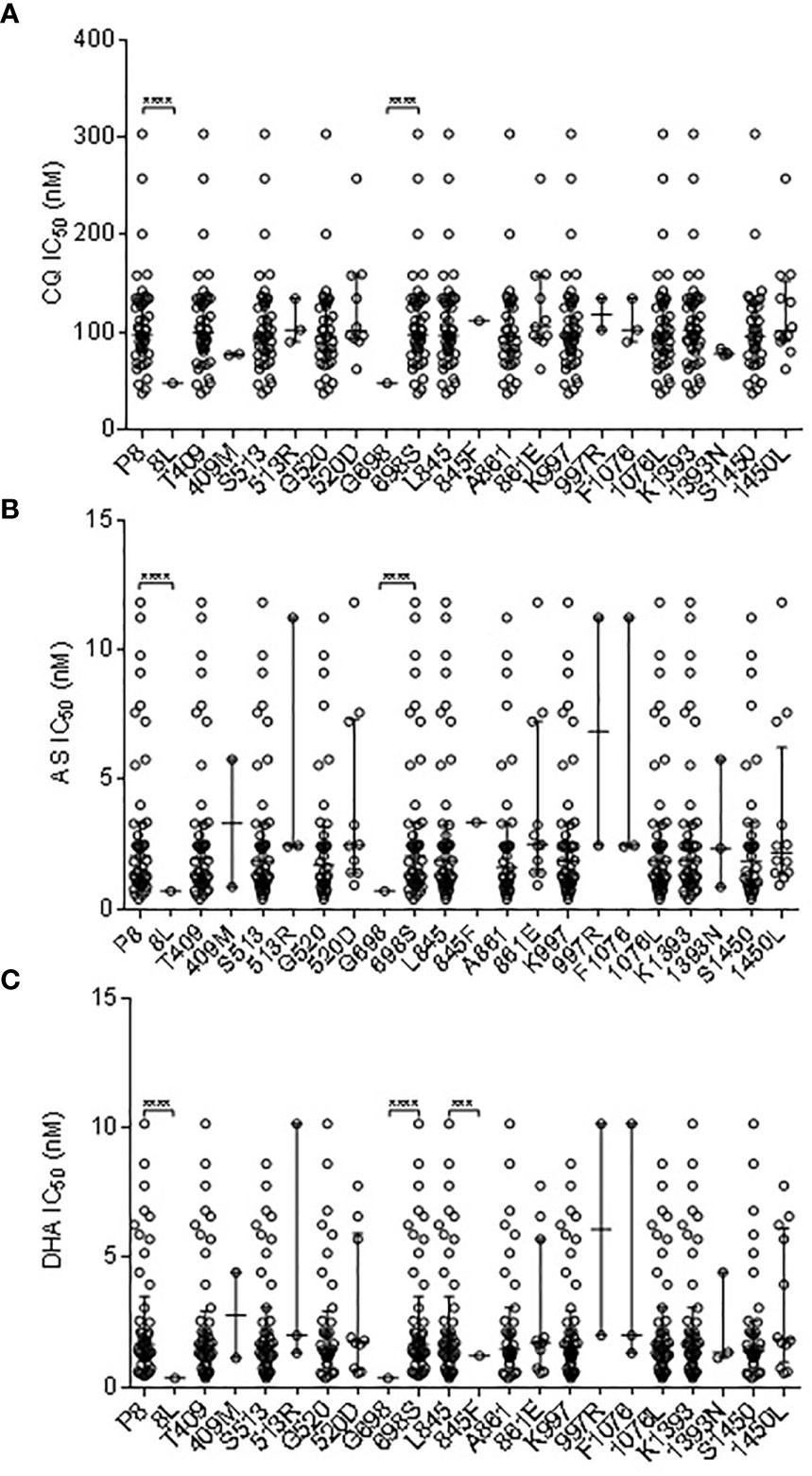
Figure 2 Correlation Association of SNPs in pvmdr1 with ex vivo susceptibilities to CQ (A), AS (B), and DHA (C). *** and **** represent indicate significant differences between the two alleles at P < 0.01 and <0.0001, respectively.
Of the three non-synonymous pvmrp1 mutations (Y1393D, G1419A, and V1478I), G1419A and V1478I had a significant association with the IC50 to CQ (P < 0.01; Figure 3). In addition, the G1419A and V1478I substitutions were associated with the decreased susceptibilities to AS and DHA (P < 0.01, Figure 3). G1419A was also associated with the decreased susceptibilities to PPQ, MFQ, and QN (P < 0.01; Figure 3).
Figure 3 Correlation Association of SNPs in pvmrp1 with ex vivo susceptibilities to CQ (A), AS (B), and DHA (C). **** represents indicate significant differences between the two alleles at P < 0.0001.
Discussion
Drug resistance is a crucial issue for malaria control and prevention. In many malaria-endemic areas, CQ is still the drug of choice for the treatment of vivax malaria. CQR P. vivax was first reported from Papua New Guinea in 1989 (Rieckmann et al., 1989), followed by many P. vivax endemic areas, including the GMS (Cooper, 1994; Marlar et al., 1995; Singh, 2000; Bright et al., 2014; Shalini et al., 2014; Thanh et al., 2015; Baird, 2017). In the GMS, the high prevalence of mixed P. falciparum and P. vivax infections suggests that the extensive use of ACTs might have an exerted collateral selection pressure on the sympatric P. vivax populations. Here, we examined the ex vivo sensitivity of the P. vivax isolates collected from the China–Myanmar border area to seven commonly used antimalarial drugs. The overall result is that for most drugs, the IC50 values had a wide range, indicating the existence of parasites with reduced susceptibilities to commonly used antimalarials. For the correlation analysis of the antimalarial drugs, we did not find a significant correlation except for a weak correlation between DHA and CQ. This was consistent with our previous study, which found a weak correlation between CQ and AS (Li et al., 2020). Although this may not mean a similar resistance mechanism, it is noteworthy that these drugs are the most frequently used frontline treatment for P. vivax and P. falciparum, respectively, in the study area. Comparison with other ex vivo testing conducted in other endemic areas such as central China (Lu et al., 2011a), Thailand (Tjitra et al., 2008), South Korea (Chotivanich et al., 2009), Colombia (Fernández et al., 2014), and Indonesia (Pava et al., 2016), the CQ IC50 value for the China–Myanmar border isolates was lower than Thailand and Indonesia but higher than the IC50 value for parasites from the other regions. Due to the lack of a defined cutoff value for P. vivax CQ resistance, some researchers used 100 nM, while others used 220 nM as the cutoff IC50 value for CQ resistance (Suwanarusk et al., 2007; Barnadas et al., 2008). Using 220 nM as the cutoff value, we found that 4.3% (2/46) of the P. vivax isolates from the China-Myanmar border might be considered CQR, although this proportion was lower than our previous report (Li et al., 2020). Nevertheless, this result is consistent with our in vivo study on CQ efficacy in P. vivax patients in the same study area, where 2.6–5.2% of patients developed recurrent parasitemia within 28 days after CQ treatment, suggesting CQ resistance (Yuan et al., 2015; Xu et al., 2020). Regrettably, the clinical efficacy data of the parasite isolates used in this ex vivo study is lacking, so we cannot make further comparisons to obtain a more realistic resistance cutoff value for the parasites in this area. We also measured the ex vivo susceptibility of the parasite isolates to six other antimalarial drugs. For the artemisinin derivatives, the median IC50 values were relatively low, and did not correlate with drug susceptibilities to the quinoline drugs. This result is consistent with our previous report (Li et al., 2020), indicating that if P. vivax developed CQ resistance, ACT could be considered as an alternative for the treatment of CQR vivax malaria (Douglas et al., 2010; Baird, 2011). Compared with our previous study (Li et al., 2020), we found an upward trend of the IC50 values for PND between 2012 and 2016. While this may not indicate the emergence of PND resistance, this is coindental with the popular use of PND injections for malaria treatment in this region. For MFQ, the IC50 values were lower than those found in Thailand (Tjitra et al., 2008) and South Korea (Chotivanich et al., 2009), but close to Indonesia (Pava et al., 2016) and central China (Lu et al., 2011a), while the values were lower than those determined earlier at the same site (Supplementary Table S3) (Li et al., 2020). This appears consistent with that MFQ has not been used as the primary medicine for malaria in China and Myanmar.
There is no confirmed molecular marker for CQ resistance of P. vivax yet. Pvmdr1 is often used to monitor CQ resistance assuming similarity to the pfmdr1 gene, which mediates resistance to quinoline drugs in P. falciparum. In this study, sequencing of pvmdr1 identified 13 non-synonymous substitutions, of which, G698S was associated with a reduced ex vivo CQ, AS, and DHA susceptibility. In addition, we found that the G520D and L845F were associated with a reduced susceptibility to PND, and that the L845F was also associated with a reduced susceptibility to MFQ. However, Y976F was not found in our study. This finding is consistent with other parasite populations from China and Myanmar (Nyunt et al., 2017; Wang et al., 2020), although it is often prevalent in different endemic areas, including Indonesia, Thailand, Cambodia, India, Papua New Guinea, and Ethiopia (Brega et al., 2005; Suwanarusk et al., 2007; Suwanarusk et al., 2008; Lu et al., 2011b; Golassa et al., 2015; Schousboe et al., 2015; Chaorattanakawee et al., 2017).
Population genomic analyses of P. vivax populations in Southeast Asia and Oceania found evidence that the pvmrp1 gene is under a strong selection (Pearson et al., 2016; Benavente et al., 2017; Benavente et al., 2021). In South Asian populations, the pvmrp1 gene exhibited high frequencies of two non-synonymous substitutions (D1393Y and G1419A). Similarly, this study detected fixation of the D1393Y mutation and G1419A approaching fixation (97.83%). Furthermore, we found that G1419A in our samples was associated with a reduced sensitivity to AS, DHA, PPQ, MFQ, and QN. It is noteworthy that V1478I was associated with a reduced ex vivo CQ, AS, and DHA susceptibility.
Our results indicate that P. vivax in the region was relatively resistant to the antifolate drug SP. Similar to P. falciparum, the resistance of P. vivax to antifolate drugs is related to mutations in the pvdhfr and pvdhps genes (Hawkins et al., 2007). Mutations at codons 50, 58, 117, and 173 of the pvdhfr gene confer resistance to pyrimethamine (Leartsakulpanich et al., 2002). Double mutations (S58R and S117N) were associated with a high level of resistance in P. vivax, whereas quadruple mutations (F57L/I, S58R, T61M, and S117T) were associated with SP treatment failure (Tjitra et al., 2002; Hastings et al., 2005). In this study, point mutations at codons 57, 58, 61, 99, and 117 of the pvdhfr gene were detected in 10.9–56.5% of the isolates. Here, the prevalence of the pvdhfr double or quadruple mutations was much lower than that found along the Thailand border and other areas of Myanmar (Nyunt et al., 2017; Tantiamornkul et al., 2018), but close to that from southern China (Miao et al., 2010). The most prevalent haplotype of the pvdhfr was FSTSS, consistent with our previous report (Zeng et al., 2021). Mutant tandem repeats are also suggested to be associated with P. vivax antifolate resistance, and the frequencies of Type II (H99S type) and Type III (deletion type) were 56.52 and 28.26%, respectively, as compared to Type II being the predominant mutation in isolates from central China (Huang et al., 2014). Nevertheless, the highest frequency of tandem repeat variants was for the wild type in southern Thailand and Xishuangbanna Prefecture, Yunnan Province, China (Huang et al., 2014; Noisang et al., 2019). In India and Cambodia, Type III (deletion type) was the most common (de Pécoulas et al., 2004; Prajapati et al., 2011). The prevalent haplotypes of the pvdhps were GAE and the wild type, which is consistent with our previous report (Zeng et al., 2021). Similar to pvdhfr, the pvdhps mutations, triple or quadruple mutation haplotypes were less common than those in southern Thailand and southern Myanmar (Tantiamornkul et al., 2018; Noisang et al., 2019), suggesting that the parasites from the China–Myanmar border regions were less resistant to SP than other sites of the GMS.
Conclusions
The ex vivo assays showed 4.3% of the P. vivax parasites in the China–Myanmar border areas as potential CQR isolates, which supports the results from in vivo CQ efficacy studies. While we identified correlations of the pvmdr1 G698S and pvmrp1 V1478I mutations with increased CQ IC50 values, further studies are required to validate the finding. We also found an increasing trend of PND IC50 values from 2012 to 2016, suggesting the emergence of PND resistance in the P. vivax population. Continuous in vivo and ex vivo studies to monitor the susceptibilities of the parasites to antimalarial drugs are needed to ensure the effective management of P. vivax cases and elucidate the mechanisms of resistance.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank (pvdhps gene accession numbers: MZ746778 - MZ746823, pvdhfr gene accession numbers: MZ746824 - MZ746869, pvmdr1 gene accession numbers: MZ746870 - MZ746915 and pvmrp1 gene accession numbers: MZ746916 - MZ746961).
Ethics Statement
The studies involving human participants were reviewed and approved by Kunming Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
WZe participated in the data collection, analysis, interpretation, and manuscript preparation. HZ, WZh, QY, XinL, XiaL, MD, XW, CL, ZX, and XC participated in the laboratory procedures, data collection, and analysis. ZY and LC contributed to the conception and design of the study. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Institute for Allergy and Infectious Diseases (U19 AI089672), The National Institute of Health (NIH), USA. ZY was funded by the National Science Foundation of China (31860604 and U1802286), Major science and technology projects of Yunnan Province (2018ZF0081), and International Science and Technology Cooperation-Yunnan International Science and Technology Cooperation Base (202003AE140004). XC and ZX were sponsored by the Yunnan Applied Basic Research Projects-Union Foundation (2018FE001-190 and 2019FE001-015, respectively). WZ was supported by the Education Department Fund of Yunnan Province (2019J1184).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank all the patients for volunteering to participate in the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.738075/full#supplementary-material
References
Amaratunga, C., Sreng, S., Mao, S., Tullo, G. S., Anderson, J. M., Chuor, C. M., et al. (2014a). Chloroquine Remains Effective for Treating Plasmodium Vivax Malaria in Pursat Province, Western Cambodia. Antimicrob. Agents Chemother. 58, 6270–6272. doi: 10.1128/AAC.03026-14
Amaratunga, C., Witkowski, B., Khim, N., Menard, D., Fairhurst, R. M. (2014b). Artemisinin Resistance in Plasmodium Falciparum. Lancet Infect. Dis. 14, 449–450. doi: 10.1016/S1473-3099(14)70777-7
Ashley, E. A., Dhorda, M., Fairhurst, R. M., Amaratunga, C., Lim, P., Suon, S., et al. (2014). Spread of Artemisinin Resistance in Plasmodium Falciparum Malaria. N Engl. J. Med. 371, 411–423. doi: 10.1056/NEJMoa1314981
Baird, J. K. (2011). Resistance to Chloroquine Unhinges Vivax Malaria Therapeutics. Antimicrob. Agents Chemother. 55, 1827–1830. doi: 10.1128/AAC.01296-10
Baird, J. K. (2017). Asia-Pacific Malaria is Singular, Pervasive, Diverse and Invisible. Int. J. Parasitol 47, 371–377. doi: 10.1016/j.ijpara.2016.06.006
Baird, J. K., Basri, H., Purnomo Bangs, M. J., Subianto, B., Patchen, L. C., et al. (1991). Resistance to Chloroquine by Plasmodium Vivax in Irian Jaya, Indonesia. Am. J. Trop. Med. Hyg 44, 547–552. doi: 10.4269/ajtmh.1991.44.547
Barnadas, C., Ratsimbasoa, A., Tichit, M., Bouchier, C., Jahevitra, M., Picot, S., et al. (2008). Plasmodium Vivax Resistance to Chloroquine in Madagascar: Clinical Efficacy and Polymorphisms in Pvmdr1 and Pvcrt-O Genes. Antimicrob. Agents Chemother. 52, 4233–4240. doi: 10.1128/AAC.00578-08
Benavente, E. D., Manko, E., Phelan, J., Campos, M., Nolder, D., Fernandez, D., et al. (2021). Distinctive Genetic Structure and Selection Patterns in Plasmodium Vivax From South Asia and East Africa. Nat. Commun. 12, 3160. doi: 10.1038/s41467-021-23422-3
Benavente, E. D., Ward, Z., Chan, W., Mohareb, F. R., Sutherland, C. J., Roper, C., et al. (2017). Genomic Variation in Plasmodium Vivax Malaria Reveals Regions Under Selective Pressure. PloS One 12, e0177134. doi: 10.1371/journal.pone.0177134
Brega, S., Meslin, B., De Monbrison, F., Severini, C., Gradoni, L., Udomsangpetch, R., et al. (2005). Identification of the Plasmodium Vivax Mdr-Like Gene (Pvmdr1) and Analysis of Single-Nucleotide Polymorphisms Among Isolates From Different Areas of Endemicity. J. Infect. Dis. 191, 272–277. doi: 10.1086/426830
Bright, A. T., Manary, M. J., Tewhey, R., Arango, E. M., Wang, T., Schork, N. J., et al. (2014). A High Resolution Case Study of a Patient With Recurrent Plasmodium Vivax Infections Shows That Relapses Were Caused by Meiotic Siblings. PloS Negl. Trop. Dis. 8, e2882. doi: 10.1371/journal.pntd.0002882
Bruce, M. C., Galinski, M. R., Barnwell, J. W., Snounou, G., Day, K. P. (1999). Polymorphism at the Merozoite Surface Protein-3alpha Locus of Plasmodium Vivax: Global and Local Diversity. Am. J. Trop. Med. Hyg 61, 518–525. doi: 10.4269/ajtmh.1999.61.518
Chaorattanakawee, S., Lon, C., Chann, S., Thay, K. H., Kong, N., You, Y., et al. (2017). Measuring Ex Vivo Drug Susceptibility in Plasmodium Vivax Isolates From Cambodia. Malar J. 16, 392. doi: 10.1186/s12936-017-2034-2
Chehuan, Y. F., Costa, M. R., Costa, J. S., Alecrim, M. G., Nogueira, F., Silveira, H., et al. (2013). In Vitro Chloroquine Resistance for Plasmodium Vivax Isolates From the Western Brazilian Amazon. Malar J. 12, 226. doi: 10.1186/1475-2875-12-226
Chotivanich, K., Sattabongkot, J., Choi, Y. K., Park, J. S., Sritabal, J., Lim, C. S., et al. (2009). Antimalarial Drug Susceptibility of Plasmodium Vivax in the Republic of Korea. Am. J. Trop. Med. Hyg. 80, 902–904. doi: 10.4269/ajtmh.2009.80.902
Chu, C. S., Phyo, A. P., Lwin, K. M., Win, H. H., San, T., Aung, A. A., et al. (2018). Comparison of the Cumulative Efficacy and Safety of Chloroquine, Artesunate, and Chloroquine-Primaquine in Plasmodium Vivax Malaria. Clin. Infect. Dis. 67, 1543–1549. doi: 10.1093/cid/ciy319
Cooper, R. D. (1994). Studies of a Chloroquine-Resistant Strain of Plasmodium Vivax From Papua New Guinea in Aotus and Anopheles Farauti s.L. J. Parasitol 80, 789–795. doi: 10.2307/3283259
Cui, L., Yan, G., Sattabongkot, J., Cao, Y., Chen, B., Chen, X., et al. (2012). Malaria in the Greater Mekong Subregion: Heterogeneity and Complexity. Acta Trop. 121, 227–239. doi: 10.1016/j.actatropica.2011.02.016
De Pécoulas, P. E., Tahar, R., Ouatas, T., Mazabraud, A., Basco, L. K. (1998). Sequence Variations in the Plasmodium Vivax Dihydrofolate Reductase-Thymidylate Synthase Gene and Their Relationship With Pyrimethamine Resistance. Mol. Biochem. Parasitol 92, 265–273. doi: 10.1016/S0166-6851(97)00247-8
De Pécoulas, P. E., Tahar, R., Yi, P., Thai, K. H., Basco, L. K. (2004). Genetic Variation of the Dihydrofolate Reductase Gene in Plasmodium Vivax in Snoul, Northeastern Cambodia. Acta Trop. 92, 1–6. doi: 10.1016/j.actatropica.2004.03.011
Djimdé, A., Doumbo, O. K., Steketee, R. W., Plowe, C. V. (2001). Application of a Molecular Marker for Surveillance of Chloroquine-Resistant Falciparum Malaria. Lancet 358, 890–891. doi: 10.1016/S0140-6736(01)06040-8
Dondorp, A. M., Nosten, F., Yi, P., Das, D., Phyo, A. P., Tarning, J., et al. (2009). Artemisinin Resistance in Plasmodium Falciparum Malaria. N Engl. J. Med. 361, 455–467. doi: 10.1056/NEJMoa0808859
Douglas, N. M., Anstey, N. M., Angus, B. J., Nosten, F., Price, R. N. (2010). Artemisinin Combination Therapy for Vivax Malaria. Lancet Infect. Dis. 10, 405–416. doi: 10.1016/S1473-3099(10)70079-7
Duraisingh, M. T., Jones, P., Sambou, I., Von Seidlein, L., Pinder, M., Warhurst, D. C. (2000). The Tyrosine-86 Allele of the Pfmdr1 Gene of Plasmodium Falciparum is Associated With Increased Sensitivity to the Anti-Malarials Mefloquine and Artemisinin. Mol. Biochem. Parasitol 108, 13–23. doi: 10.1016/S0166-6851(00)00201-2
Eldin De Pécoulas, P., Basco, L. K., Tahar, R., Ouatas, T., Mazabraud, A. (1998). Analysis of the Plasmodium Vivax Dihydrofolate Reductase-Thymidylate Synthase Gene Sequence. Gene 211, 177–185. doi: 10.1016/S0378-1119(98)00118-8
Fernández, D., Segura, C., Arboleda, M., Garavito, G., Blair, S., Pabón, A. (2014). In Vitro Susceptibility of Plasmodium Vivax to Antimalarials in Colombia. Antimicrob. Agents Chemother. 58, 6354–6359. doi: 10.1128/AAC.03191-14
Fidock, D. A., Nomura, T., Talley, A. K., Cooper, R. A., Dzekunov, S. M., Ferdig, M. T., et al. (2000). Mutations in the P. Falciparum Digestive Vacuole Transmembrane Protein PfCRT and Evidence for Their Role in Chloroquine Resistance. Mol. Cell 6, 861–871. doi: 10.1016/S1097-2765(05)00077-8
Geng, J., Malla, P., Zhang, J., Xu, S., Li, C., Zhao, Y., et al. (2019). Increasing Trends of Malaria in a Border Area of the Greater Mekong Subregion. Malar J. 18 (1), 309. doi: 10.1186/s12936-019-2924-6
Golassa, L., Erko, B., Baliraine, F. N., Aseffa, A., Swedberg, G. (2015). Polymorphisms in Chloroquine Resistance-Associated Genes in Plasmodium Vivax in Ethiopia. Malar J. 14, 164. doi: 10.1186/s12936-015-0625-3
Gomes, L. R., Almeida-De-Oliveira, N. K., De Lavigne, A. R., De Lima, S. R., De Pina-Costa, A., Brasil, P., et al. (2016). Plasmodium Vivax Mdr1 Genotypes in Isolates From Successfully Cured Patients Living in Endemic and non-Endemic Brazilian Areas. Malar J. 15, 96. doi: 10.1186/s12936-016-1141-9
Gupta, B., Xu, S., Wang, Z., Sun, L., Miao, J., Cui, L., et al. (2014). Plasmodium Falciparum Multidrug Resistance Protein 1 (Pfmrp1) Gene and its Association With In Vitro Drug Susceptibility of Parasite Isolates From North-East Myanmar. J. Antimicrob. Chemother. 69, 2110–2117. doi: 10.1093/jac/dku125
Guthmann, J. P., Pittet, A., Lesage, A., Imwong, M., Lindegardh, N., Min Lwin, M., et al. (2008). Plasmodium Vivax Resistance to Chloroquine in Dawei, Southern Myanmar. Trop. Med. Int. Health 13, 91–98. doi: 10.1111/j.1365-3156.2007.01978.x
Happi, C. T., Gbotosho, G. O., Folarin, O. A., Bolaji, O. M., Sowunmi, A., Kyle, D. E., et al. (2006). Association Between Mutations in Plasmodium Falciparum Chloroquine Resistance Transporter and P. Falciparum Multidrug Resistance 1 Genes and In Vivo Amodiaquine Resistance in P. Falciparum Malaria-Infected Children in Nigeria. Am. J. Trop. Med. Hyg 75, 155–161. doi: 10.4269/ajtmh.2006.75.155
Hastings, M. D., Maguire, J. D., Bangs, M. J., Zimmerman, P. A., Reeder, J. C., Baird, J. K., et al. (2005). Novel Plasmodium Vivax Dhfr Alleles From the Indonesian Archipelago and Papua New Guinea: Association With Pyrimethamine Resistance Determined by a Saccharomyces Cerevisiae Expression System. Antimicrob. Agents Chemother. 49, 733–740. doi: 10.1128/AAC.49.2.733-740.2005
Hawkins, V. N., Joshi, H., Rungsihirunrat, K., Na-Bangchang, K., Sibley, C. H. (2007). Antifolates can Have a Role in the Treatment of Plasmodium Vivax. Trends Parasitol 23, 213–222. doi: 10.1016/j.pt.2007.03.002
Holmgren, G., Hamrin, J., Svärd, J., Mårtensson, A., Gil, J. P., Björkman, A. (2007). Selection of Pfmdr1 Mutations After Amodiaquine Monotherapy and Amodiaquine Plus Artemisinin Combination Therapy in East Africa. Infect. Genet. Evol. 7, 562–569. doi: 10.1016/j.meegid.2007.03.005
Htun, M. W., Mon, N. C. N., Aye, K. M., Hlaing, C. M., Kyaw, M. P., Handayuni, I., et al. (2017). Chloroquine Efficacy for Plasmodium Vivax in Myanmar in Populations With High Genetic Diversity and Moderate Parasite Gene Flow. Am. J. Trop. Med. Hyg 16, 281. doi: 10.1186/s12936-017-1912-y
Huang, B., Huang, S., Su, X. Z., Tong, X., Yan, J., Li, H., et al. (2014). Molecular Surveillance of Pvdhfr, Pvdhps, and Pvmdr-1 Mutations in Plasmodium Vivax Isolates From Yunnan and Anhui Provinces of China. Malar J. 13, 346. doi: 10.1186/1475-2875-13-346
Imwong, M., Nguyen, T. N., Tripura, R., Peto, T. J., Lee, S. J., Lwin, K. M., et al. (2015a). The Epidemiology of Subclinical Malaria Infections in South-East Asia: Findings From Cross-Sectional Surveys in Thailand-Myanmar Border Areas, Cambodia, and Vietnam. Malar J. 14, 381. doi: 10.1186/s12936-015-0906-x
Imwong, M., Pukrittakayamee, S., Looareesuwan, S., Pasvol, G., Poirreiz, J., White, N. J., et al. (2001). Association of Genetic Mutations in Plasmodium Vivax Dhfr With Resistance to Sulfadoxine-Pyrimethamine: Geographical and Clinical Correlates. Antimicrob. Agents Chemother. 45, 3122–3127. doi: 10.1128/AAC.45.11.3122-3127.2001
Imwong, M., Pukrittayakamee, S., Rénia, L., Letourneur, F., Charlieu, J. P., Leartsakulpanich, U., et al. (2003). Novel Point Mutations in the Dihydrofolate Reductase Gene of Plasmodium Vivax: Evidence for Sequential Selection by Drug Pressure. Antimicrob. Agents Chemother. 47, 1514–1521. doi: 10.1128/AAC.47.5.1514-1521.2003
Korsinczky, M., Fischer, K., Chen, N., Baker, J., Rieckmann, K., Cheng, Q. (2004). Sulfadoxine Resistance in Plasmodium Vivax is Associated With a Specific Amino Acid in Dihydropteroate Synthase at the Putative Sulfadoxine-Binding Site. Antimicrob. Agents Chemother. 48, 2214–2222. doi: 10.1128/AAC.48.6.2214-2222.2004
Leartsakulpanich, U., Imwong, M., Pukrittayakamee, S., White, N. J., Snounou, G., Sirawaraporn, W., et al. (2002). Molecular Characterization of Dihydrofolate Reductase in Relation to Antifolate Resistance in Plasmodium Vivax. Mol. Biochem. Parasitol 119, 63–73. doi: 10.1016/S0166-6851(01)00402-9
Li, J., Tao, Z., Li, Q., Brashear, A., Wang, Y., Xia, H., et al. (2017). Further Evaluation of the NWF Filter for the Purification of Plasmodium Vivax-Infected Erythrocytes. Malar J. 16, 201. doi: 10.1186/s12936-017-1855-3
Li, J., Zhang, J., Li, Q., Hu, Y., Ruan, Y., Tao, Z., et al. (2020). Ex Vivo Susceptibilities of Plasmodium Vivax Isolates From the China-Myanmar Border to Antimalarial Drugs and Association With Polymorphisms in Pvmdr1 and Pvcrt-O Genes. PloS Negl. Trop. Dis. 14 (6), e0008255. doi: 10.1371/journal.pntd.0008255
Li, P., Zhao, Z., Wang, Y., Xing, H., Parker, D. M., Yang, Z., et al. (2014). Nested PCR Detection of Malaria Directly Using Blood Filter Paper Samples From Epidemiological Surveys. Malar J. 13, 175. doi: 10.1186/1475-2875-13-175
Lu, F., Gao, Q., Chotivanich, K., Xia, H., Cao, J., Udomsangpetch, R., et al. (2011a). In Vitro Anti-Malarial Drug Susceptibility of Temperate Plasmodium Vivax From Central China. Am. J. Trop. Med. Hyg. 85, 197–201. doi: 10.4269/ajtmh.2011.10-0070
Lu, F., Lim, C. S., Nam, D. H., Kim, K., Lin, K., Kim, T. S., et al. (2011b). Genetic Polymorphism in Pvmdr1 and Pvcrt-O Genes in Relation to In Vitro Drug Susceptibility of Plasmodium Vivax Isolates From Malaria-Endemic Countries. Acta Trop. 117, 69–75. doi: 10.1016/j.actatropica.2010.08.011
Lu, F., Wang, B., Cao, J., Sattabongkot, J., Zhou, H., Zhu, G., et al. (2012). Prevalence of Drug Resistance-Associated Gene Mutations in Plasmodium Vivax in Central China. Korean J. Parasitol. 50, 379–384. doi: 10.3347/kjp.2012.50.4.379
Marlar, T., Myat Phone, K., Aye Yu, S., Khaing Khaing, G., Ma, S., Myint, O. (1995). Development of Resistance to Chloroquine by Plasmodium Vivax in Myanmar. Trans. R Soc. Trop. Med. Hyg. 89, 307–308. doi: 10.1016/0035-9203(95)90556-1
Miao, M., Yang, Z., Cui, L., Ahlum, J., Huang, Y., Cui, L. (2010). Different Allele Prevalence in the Dihydrofolate Reductase and Dihydropteroate Synthase Genes in Plasmodium Vivax Populations From China. Am. J. Trop. Med. Hyg 83, 1206–1211. doi: 10.4269/ajtmh.2010.10-0259
Mint Lekweiry, K., Ould Mohamed Salem Boukhary, A., Gaillard, T., Wurtz, N., Bogreau, H., Hafid, J. E., et al. (2012). Molecular Surveillance of Drug-Resistant Plasmodium Vivax Using Pvdhfr, Pvdhps and Pvmdr1 Markers in Nouakchott, Mauritania. J. Antimicrob. Chemother. 67, 367–374. doi: 10.1093/jac/dkr464
Müller, I. B., Hyde, J. E. (2013). Folate Metabolism in Human Malaria Parasites–75 Years on. Mol. Biochem. Parasitol 188, 63–77. doi: 10.1016/j.molbiopara.2013.02.008
Myat Phone, K., Myint, O., Myint, L., Thaw, Z., Kyin Hla, A., Nwe Nwe, Y. (1993). Emergence of Chloroquine-Resistant Plasmodium Vivax in Myanmar (Burma). Trans. R Soc. Trop. Med. Hyg. 87 (6), 687. doi: 10.1016/0035-9203(93)90294-z
Nawaz, F., Nsobya, S. L., Kiggundu, M., Joloba, M., Rosenthal, P. J. (2009). Selection of Parasites With Diminished Drug Susceptibility by Amodiaquine-Containing Antimalarial Regimens in Uganda. J. Infect. Dis. 200, 1650–1657. doi: 10.1086/647988
Noedl, H., Se, Y., Schaecher, K., Smith, B. L., Socheat, D., Fukuda, M. M. (2008). Evidence of Artemisinin-Resistant Malaria in Western Cambodia. N Engl. J. Med. 359, 2619–2620. doi: 10.1056/NEJMc0805011
Noisang, C., Prosser, C., Meyer, W., Chemoh, W., Ellis, J., Sawangjaroen, N., et al. (2019). Molecular Detection of Drug Resistant Malaria in Southern Thailand. Malar J. 18, 275. doi: 10.1186/s12936-019-2903-y
Nomura, T., Carlton, J. M., Baird, J. K., Del Portillo, H. A., Fryauff, D. J., Rathore, D., et al. (2001). Evidence for Different Mechanisms of Chloroquine Resistance in 2 Plasmodium Species That Cause Human Malaria. J. Infect. Dis. 183, 1653–1661. doi: 10.1086/320707
Nyunt, M. H., Han, J. H., Wang, B., Aye, K. M., Aye, K. H., Lee, S. K., et al. (2017). Clinical and Molecular Surveillance of Drug Resistant Vivax Malaria in Myanma-2016). Malar J. 16, 117. doi: 10.1186/s12936-017-1770-7
Pava, Z., Handayuni, I., Wirjanata, G., To, S., Trianty, L., Noviyanti, R., et al. (2016). Expression of Plasmodium Vivax Crt-O Is Related to Parasite Stage But Not Ex Vivo Chloroquine Susceptibility. Antimicrob. Agents Chemother. 60, 361–367. doi: 10.1128/AAC.02207-15
Pearson, R. D., Amato, R., Auburn, S., Miotto, O., Almagro-Garcia, J., Amaratunga, C., et al. (2016). Genomic Analysis of Local Variation and Recent Evolution in Plasmodium Vivax. Nat. Genet. 48, 959–964. doi: 10.1038/ng.3599
Prajapati, S. K., Joshi, H., Dev, V., Dua, V. K. (2011). Molecular Epidemiology of Plasmodium Vivax Anti-Folate Resistance in India. Malar J. 10, 102. doi: 10.1186/1475-2875-10-102
Price, R. N., Uhlemann, A. C., Brockman, A., Mcgready, R., Ashley, E., Phaipun, L., et al. (2004). Mefloquine Resistance in Plasmodium Falciparum and Increased Pfmdr1 Gene Copy Number. Lancet 364, 438–447. doi: 10.1016/S0140-6736(04)16767-6
Price, R. N., Von Seidlein, L., Valecha, N., Nosten, F., Baird, J. K., White, N. J. (2014). Global Extent of Chloroquine-Resistant Plasmodium Vivax: A Systematic Review and Meta-Analysis. Lancet Infect. Dis. 14, 982–991. doi: 10.1016/S1473-3099(14)70855-2
Rieckmann, K. H., Davis, D. R., Hutton, D. C. (1989). Plasmodium Vivax Resistance to Chloroquine? Lancet 2, 1183–1184. doi: 10.1016/S0140-6736(89)91792-3
Rijken, M. J., Boel, M. E., Russell, B., Imwong, M., Leimanis, M. L., Phyo, A. P., et al. (2011). Chloroquine Resistant Vivax Malaria in a Pregnant Woman on the Western Border of Thailand. Malar J. 10, 113. doi: 10.1186/1475-2875-10-113
Russell, B. M., Udomsangpetch, R., Rieckmann, K. H., Kotecka, B. M., Coleman, R. E., Sattabongkot, J. (2003). Simple In Vitro Assay for Determining the Sensitivity of Plasmodium Vivax Isolates From Fresh Human Blood to Antimalarials in Areas Where P. Vivax Is Endemic. Antimicrob. Agents Chemother. 47, 170–173. doi: 10.1128/AAC.47.1.170-173.2003
Sá, J. M., Nomura, T., Neves, J., Baird, J. K., Wellems, T. E., Del Portillo, H. A. (2005). Plasmodium Vivax: Allele Variants of the Mdr1 Gene do Not Associate With Chloroquine Resistance Among Isolates From Brazil, Papua, and Monkey-Adapted Strains. Exp. Parasitol 109, 256–259. doi: 10.1016/j.exppara.2004.12.005
Schousboe, M. L., Ranjitkar, S., Rajakaruna, R. S., Amerasinghe, P. H., Morales, F., Pearce, R., et al. (2015). Multiple Origins of Mutations in the Mdr1 Gene–A Putative Marker of Chloroquine Resistance in P. Vivax. PloS Negl. Trop. Dis. 9, e0004196. doi: 10.1371/journal.pntd.0004196
Shalini, S., Chaudhuri, S., Sutton, P. L., Mishra, N., Srivastava, N., David, J. K., et al. (2014). Chloroquine Efficacy Studies Confirm Drug Susceptibility of Plasmodium Vivax in Chennai, India. Malar J. 13, 129. doi: 10.1186/1475-2875-13-129
Singh, R. K. (2000). Emergence of Chloroquine-Resistant Vivax Malaria in South Bihar (India). Trans. R Soc. Trop. Med. Hyg. 94 (3), 327.
Snounou, G., Viriyakosol, S., Zhu, X. P., Jarra, W., Pinheiro, L., Do Rosario, V. E., et al. (1993). High Sensitivity of Detection of Human Malaria Parasites by the Use of Nested Polymerase Chain Reaction. Mol. Biochem. Parasitol 61, 315–320. doi: 10.1016/0166-6851(93)90077-B
Suwanarusk, R., Chavchich, M., Russell, B., Jaidee, A., Chalfein, F., Barends, M., et al. (2008). Amplification of Pvmdr1 Associated With Multidrug-Resistant Plasmodium Vivax. J. Infect. Dis. 198, 1558–1564. doi: 10.1086/592451
Suwanarusk, R., Russell, B., Chavchich, M., Chalfein, F., Kenangalem, E., Kosaisavee, V., et al. (2007). Chloroquine Resistant Plasmodium Vivax: In Vitro Characterisation and Association With Molecular Polymorphisms. PloS One 2, e1089. doi: 10.1371/journal.pone.0001089
Tantiamornkul, K., Pumpaibool, T., Piriyapongsa, J., Culleton, R., Lek-Uthai, U. (2018). The Prevalence of Molecular Markers of Drug Resistance in Plasmodium Vivax From the Border Regions of Thailand in 2008 and 2014. Int. J. Parasitol Drugs Drug Resist. 8, 229–237. doi: 10.1016/j.ijpddr.2018.04.003
Tao, Z. Y., Xia, H., Cao, J., Gao, Q. (2011). Development and Evaluation of a Prototype non-Woven Fabric Filter for Purification of Malaria-Infected Blood. Malar J. 10, 251. doi: 10.1186/1475-2875-10-251
Thanh, P. V., Hong, N. V., Van, N. V., Louisa, M., Baird, K., Xa, N. X., et al. (2015). Confirmed Plasmodium Vivax Resistance to Chloroquine in Central Vietnam. Antimicrob. Agents Chemother. 59, 7411–7419. doi: 10.1128/AAC.00791-15
Tjitra, E., Anstey, N. M., Sugiarto, P., Warikar, N., Kenangalem, E., Karyana, M., et al. (2008). Multidrug-Resistant Plasmodium Vivax Associated With Severe and Fatal Malaria: A Prospective Study in Papua, Indonesia. PloS Med. 5, e128. doi: 10.1371/journal.pmed.0050128
Tjitra, E., Baker, J., Suprianto, S., Cheng, Q., Anstey, N. M. (2002). Therapeutic Efficacies of Artesunate-Sulfadoxine-Pyrimethamine and Chloroquine-Sulfadoxine-Pyrimethamine in Vivax Malaria Pilot Studies: Relationship to Plasmodium Vivax Dhfr Mutations. Antimicrob. Agents Chemother. 46, 3947–3953. doi: 10.1128/AAC.46.12.3947-3953.2002
Veiga, M. I., Ferreira, P. E., Jörnhagen, L., Malmberg, M., Kone, A., Schmidt, B. A., et al. (2011). Novel Polymorphisms in Plasmodium Falciparum ABC Transporter Genes are Associated With Major ACT Antimalarial Drug Resistance. PloS One 6, e20212. doi: 10.1371/journal.pone.0020212
Wang, X., Ruan, W., Zhou, S., Feng, X., Yan, H., Huang, F. (2020). Prevalence of Molecular Markers Associated With Drug Resistance of Plasmodium Vivax Isolates in Western Yunnan Province, China. BMC Infect. Dis. 20, 307. doi: 10.1186/s12879-020-05032-4
Wellems, T. E., Plowe, C. V. (2001). Chloroquine-Resistant Malaria. J. Infect. Dis. 184, 770–776. doi: 10.1086/322858
Who (1967). Chemotherapy of Malaria: Report of the WHO Scientific Group. WHO Technical Report Series No. 375. Available at: https://apps.who.int/iris/bitstream/handle/10665/40671/WHO_TRS_375.pdf.
Who (2015). Strategy for Malaria Elimination in the Greater Mekong Subregion, (2015–2030) (Geneva: World Health Organization).
Xu, S., Zeng, W., Ngassa Mbenda, H. G., Liu, H., Chen, X., Xiang, Z., et al. (2020). Efficacy of Directly-Observed Chloroquine-Primaquine Treatment for Uncomplicated Acute Plasmodium Vivax Malaria in Northeast Myanmar: A Prospective Open-Label Efficacy Trial. Travel Med. Infect. Dis. 36, 101499. doi: 10.1016/j.tmaid.2019.101499
Yuan, L., Wang, Y., Parker, D. M., Gupta, B., Yang, Z., Liu, H., et al. (2015). Therapeutic Responses of Plasmodium Vivax Malaria to Chloroquine and Primaquine Treatment in Northeastern Myanmar. Antimicrob. Agents Chemother. 59, 1230–1235. doi: 10.1128/AAC.04270-14
Keywords: Plasmodium vivax, drug sensitivity, multidrug resistance-1 gene, multidrug resistance protein 1 gene, China–Myanmar border
Citation: Zeng W, Zhao H, Zhao W, Yang Q, Li X, Li X, Duan M, Wang X, Li C, Xiang Z, Chen X, Cui L and Yang Z (2021) Molecular Surveillance and Ex Vivo Drug Susceptibilities of Plasmodium vivax Isolates From the China–Myanmar Border. Front. Cell. Infect. Microbiol. 11:738075. doi: 10.3389/fcimb.2021.738075
Received: 08 July 2021; Accepted: 06 August 2021;
Published: 01 November 2021.
Edited by:
Gaoqian Feng, Burnet Institute, AustraliaReviewed by:
Cindy Chu, Mahidol Oxford Tropical Medicine Research Unit (MORU), ThailandHargobinder Kaur, Yale University, United States
Copyright © 2021 Zeng, Zhao, Zhao, Yang, Li, Li, Duan, Wang, Li, Xiang, Chen, Cui and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoqing Yang, emhhb3Fpbmd5OTJAaG90bWFpbC5jb20=; Liwang Cui, bGl3YW5nY3VpQHVzZi5lZHU=
 Weilin Zeng
Weilin Zeng Hui Zhao
Hui Zhao Wei Zhao1
Wei Zhao1 Zheng Xiang
Zheng Xiang