- 1State Key Laboratory of Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Research Units of Discovery of Unknown Bacteria and Function (2018RU010), Chinese Academy of Medical Sciences, Beijing, China
- 2Microbiology Department, Maanshan Center for Clinical Laboratory, Ma’anshan, China
- 3Microbiology Department, Maanshan Center for Disease Control and Prevention, Ma’anshan, China
- 4School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, NSW, Australia
Objectives: This prospective study was carried out to investigate molecular characteristics and antimicrobial susceptibility patterns of Citrobacter spp. from extraintestinal infections.
Methods: Forty-six clinical Citrobacter spp. isolates were isolated from hospital patients with extraintestinal infections and analyzed by multilocus sequence typing (MLST) using seven housekeeping genes. Antimicrobial susceptibility testing was performed by disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) recommendations. Adhesion and cytotoxicity to HEp-2 cells were assessed.
Results: The 46 clinical Citrobacter spp. isolates were typed into 38 sequence types (STs), 9 of which belonged to four clonal complexes (CCs). None of the isolates shared the same ST or CCs with isolates from other countries or from other parts of China. Over half of the isolates were multidrug-resistant (MDR), with 17/26 C. freundii, 5/6 C. braakii, and 3/14 C. koseri isolates being MDR. Moreover, four isolates were carbapenem resistant with resistance to imipenem or meropenem. Among eight quinolone resistant C. freundii, all had a mutation in codon 59 (Thr59Ile) in quinolone resistance determining region of the gyrA gene. Only a small proportion of the isolates were found to be highly cytotoxic and adhesive with no correlation to sample sources.
Conclusions: There was a diverse range of Citrobacter isolates causing extraintestinal infections and a high prevalence of MDR.
Introduction
Citrobacter spp. are facultative anaerobic Gram negative bacteria within the family Enterobacteriaceae. Citrobacter spp. have been associated with nosocomial infections involving the urinary tract, liver, biliary tract, peritoneum, intestines, bone, respiratory tract, endocardium, wounds, soft tissue, meninges, and the bloodstream (Khorasani et al., 2008; Kumar et al., 2013; Liu et al., 2018b). C. freundii is the most frequently isolated Citrobacter species from a range of infections (Khorasani et al., 2008) and has also caused small outbreaks in healthcare settings (Nada et al., 2004; Mohanty et al., 2007; Samonis et al., 2009; Bai et al., 2012). C. koseri can cause meningitis and brain abscesses in neonates and central nervous system (CNS) infections in head trauma, facial fractures, post neurosurgical procedures, or immunocompromised individuals (Vaz Marecos et al., 2012; Chao et al., 2013; Lechowicz et al., 2017; Reyes et al., 2017). C. braakii has been reported to cause bacteriemia (Lai et al., 2010; Hirai et al., 2016; Oyeka and Antony, 2017).
Emergence of multidrug-resistant (MDR) Citrobacter strains is an increasing concern (Khorasani et al., 2008). MDR C. freundii strains have been associated with a higher rate of in-hospital mortality compared to susceptible strains (Deal et al., 2007). MDR Citrobacter spp. with production of β-lactamase (Amp-C), broad-spectrum β-lactamase, extended-spectrum β-lactamase (ESBL), or even carbapenemase has been reported by several international surveillance programs (Wang et al., 2000; Mohanty et al., 2007; Zhang et al., 2008; Samonis et al., 2009; Kanamori et al., 2011; Lee et al., 2015; Liu et al., 2018b). It has been reported that 39–48% of C. freundii isolates were resistant to broad-spectrum cephalosporins (ceftriaxone, ceftazidime), piperacillin, and piperacillin/tazobactam (Khorasani et al., 2008). Moreover, a few studies have reported C. freundii harboring carbapenemases, particularly metallo-β-lactamases (MBLs) or Klebsiella pneumoniae carbapenemase (KPC) types (Weile et al., 2007; Protonotariou et al., 2008; Zhang et al., 2008; Gaibani et al., 2013; Schweizer et al., 2019; Gobeille Paré et al., 2020; Räisänen et al., 2021). Quinolone resistance determinant including qnr and aac(6’)-Ib-cr genes have been reported in Citrobacter spp. (Park et al., 2007; Zhang et al., 2012). Numerous qnrB alleles have been detected, and about 40 qnrB variants are located on the chromosome of Citrobacter spp., especially C. freundii (Jacoby et al., 2014, Liao et al., 2015). Fluoroquinolone resistance is associated with mutations in gyrA and parC genes (Minarini and Darini, 2012). Mutations in gyrA were found in fluoroquinolone resistant C. freundii isolates (Weigel et al., 1998; Minarini and Darini, 2012).
Studies on Citrobacter spp. from extraintestinal infections have been mostly focused on antibiotic resistance, and little is known about their genetic diversity and virulence properties. Citrobacter spp. can be isolated from fecal samples of healthy individuals and can also cause food-borne infections (Tassew et al., 2010; Bai et al., 2012; Ifeadike et al., 2012; Minarini and Darini, 2012; Liu et al., 2017; Liu et al., 2018a; Liu et al., 2020). The possible source of strains causing extraintestinal infections and their relationships to strains from other infections and other sources have not been well studied. In our previous studies, we analyzed Citrobacter isolates from diarrheal patients, foods, and environment in China (Bai et al., 2012; Liu et al., 2017; Liu et al., 2018a; Liu et al., 2020). We found high diversity of Citrobacter strains from these sources in sequence types (STs), antibiotic resistance profiles, and virulence properties (Liu et al., 2017; Liu et al., 2018a; Liu et al., 2020). In this study, we collected Citrobacter spp. isolates from extraintestinal infections of inpatients in Maanshan people's hospital, Anhui Province, China and examined these isolates by multilocus sequence typing (MLST), antibiotic resistance profiling, and in vitro virulence testing to obtain an insight into their genetic diversity, antibiotic resistance, and virulence.
Materials and Methods
Citrobacter Isolates
Forty-six Citrobacter spp. isolates were obtained from 26 urine, 15 sputum, 2 bile, 2 secretion, and 1 blood samples from 2014 to 2018 in Maanshan people hospital, Anhui Province, China. The 26 urine samples included 16 C. freundii, 1 C. braakii, and 9 C. koseri isolates; the 15 sputum samples contained 8 C. freundii, 3 C. braakii, and 4 C. koseri isolates; 2 bile samples contained C. freundii isolates; 2 secretion samples contained C. braakii isolates; 1 blood sample had C. koseri isolates. No other pathogens were isolated from these clinical specimens with the exception of a sputum sample where Citrobacter is the predominant pathogen. The identity of each isolate was determined using API 20E test strips (bioMérieux, La Balme les Grottes, France) at the time of isolation, and they were stored as glycerol stocks at -80°C. Bacteria were grown in Luria-Bertani (LB) broth or on LB and Mueller–Hinton agar plates (pH 7.4) at 37°C.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was carried out using the disk diffusion method according to CLSI recommendations (Clinical and Laboratory Standards Institute, 2016). We tested the following 20 antimicrobial agents: ampicillin (AMP, 10 μg), cefotaxime (CTX, 30 μg), ceftazidime (CAZ, 30 μg), cefepime (FEP, 30 μg), cefoxitin (FOX, 30 μg), imipenem (IMP, 10 μg), aztreonam (ATM, 30 μg), meropenem (MEM, 10 μg), nalidixic acid (NA, 30 μg), ciprofloxacin (CIP, 5 μg), levofloxacin (LEV, 5 μg), gentamicin (CN, 10 μg), amikacin (AK, 30 μg), streptomycin (S, 10 μg), kanamycin (K, 30 μg), tetracycline (TE, 30 μg), doxycycline (DO, 30 μg), chloramphenicol (C, 30 μg), trimethoprim/sulfamethoxazole (SXT, 25 μg), and azithromycin (AZM, 15 μg) (Oxoid, Hampshire, UK). Quality control was performed using the reference strain E. coli ATCC 25922. Results were used to classify isolates as being resistant or susceptible to a particular antibiotic comparing with the standard reference values (Clinical and Laboratory Standards Institute, 2016).
For fluoroquinolones resistant isolates, susceptibility testing to quinolones including nalidixic acid (NA), ciprofloxacin (CIP), norfloxacin (NOR), and levofloxacin (LEV) was carried out using the broth microdilution method according to CLSI recommendations, as previously described (Liu et al., 2017). Minimum inhibitory concentration (MIC) results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines. Quality control for MICs was performed using the reference E. coli ATCC 25922.
PCR Amplification and Sequencing
All the isolates were screened for qnrA, qnrB, qnrS, qnrC, qnrD, aac(6’)-Ib-cr, and qepA genes by PCR using previously published primers and protocols (Liu et al., 2020). All primers were synthesized by Shanghai Sangon Biological Engineering Technology and Services (Shanghai, China). Positive PCR products were confirmed by sequencing.
MultiLocus Sequence Typing
The seven housekeeping genes, including aspC, clpX, fadD, mdh, arcA, dnaG, and lysP, were typed by PCR using previously published primers and protocols (Liu et al., 2020). Alleles and STs were assigned using the MLST database (http://pubmlst.org/cfreundii/).
PHYLOViZ version 2.0 (Francisco et al., 2012; Luo et al., 2021), using the goeBURST algorithm, was used to calculate and visualize clonal complexes (CCs) between the STs of the isolates. MEGA X (Kumar et al., 2018) was used to construct phylogenetic trees using the neighbor-joining algorithm with the default parameters based on the concatenated sequences of the seven housekeeping genes. Bootstraps with 1000 replicates was performed to evaluate the robustness of the branches of the tree.
In Vitro Adhesion and Cytotoxicity Assays
In vitro adhesion to host cells was performed as previously described (Liu et al., 2017). An adhesion index (<1; >1 and <50; >50) describing the mean number of bacteria per HEp-2 after examination of 10 visual fields was determined (Liu et al., 2017). Infections were repeated three times in duplicate.
The lactate dehydrogenase (LDH) released by the HEp-2 cells was determined using the Cytotox 96 kit (Promega) according to the manufacturer’s instructions. The relative amount of cytotoxicity was expressed as previously described (Liu et al., 2017). All experiments were performed three times in duplicate.
Statistical Analysis
SPSS software version 13.0 (SPSS Inc., Chicago, IL, USA) was used to conduct all statistical comparisons. A nonparametric test (Mann–Whitney U-test) was employed to compare the different groups. Two-tailed p-value of 0.05 or less was considered to be statistically significant.
Results
Clinical Characteristics of the Patients
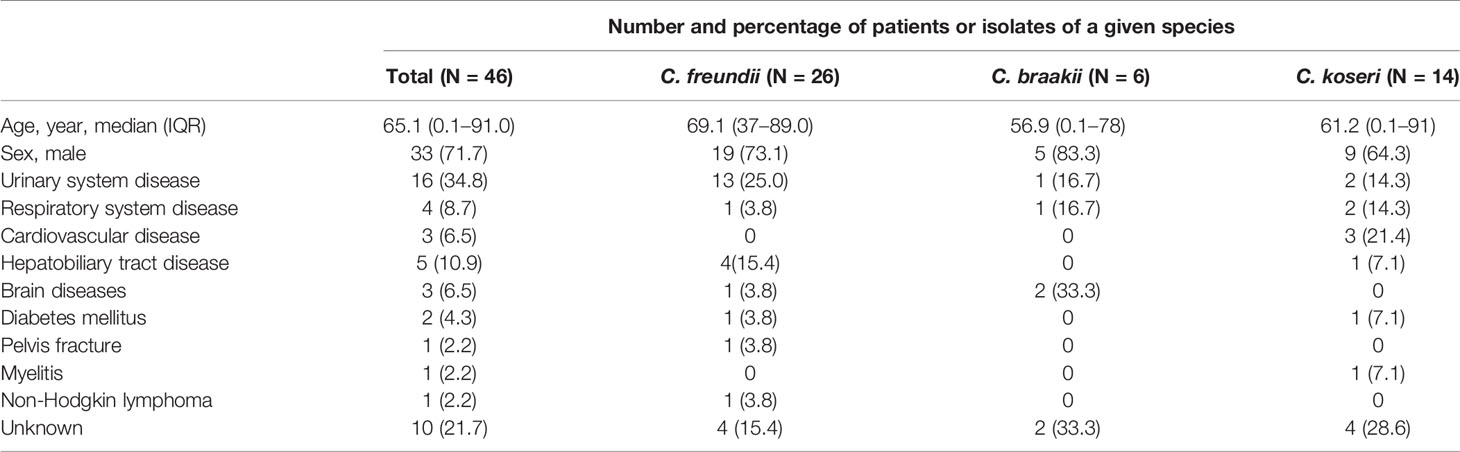
From September 2014 through August 2018, 46 cases of Citrobacter infections were identified and the distribution of isolates by year was presented in Figure 1. Among them, 26 (56.5%) cases had infections caused by C. freundii and 20 had non-C. freundii infections (6 of C. braakii and 14 of C. koseri). There was no clustering of cases or suspected outbreak during the study period. The median age was 65.1 years with a range of 0.1–91. Urinary system disease (16/46, 34.8%) was the common underlying disease among the patients (Table 1).
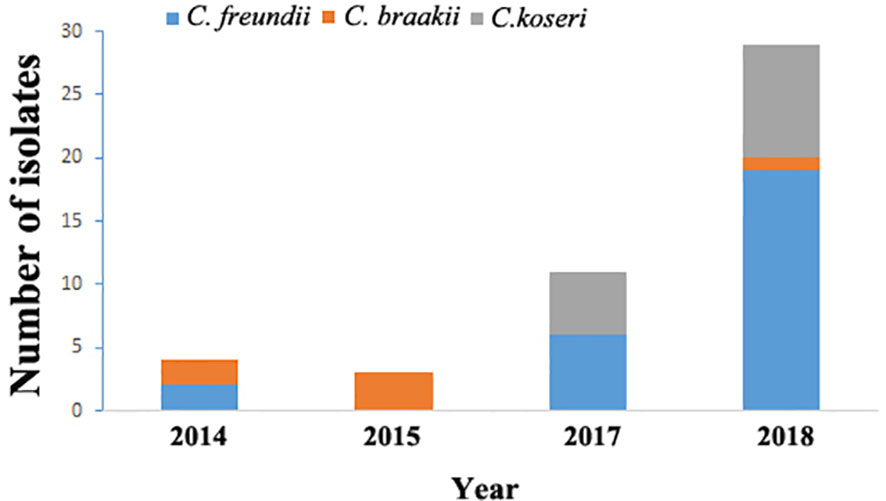
Figure 1 Distribution of Citrobacter isolates by year in Maanshan people’s hospital, Anhui Province, China from 2014 to 2018.
Multilocus Sequence Typing of Citrobacter Isolates
The 46 Citrobacter isolates including 26 C. freundii, 6 C. braakii, and 14 C. koseri isolates were divided into 38 STs, with the 26 C. freundii isolates dividing into 22 STs, the 6 C. braakii isolates into 4 STs, and the 14 C. koseri isolates into 12 STs (Table 2 and Figure 2). All of the 38 STs were novel STs in comparison to the STs in the public MLST database. Eight STs each contained two isolates with four C. freundii STs (ST434, ST441, ST451, and ST458), two C. braakii STs (ST435 and ST438), and two C. koseri STs (ST440 and ST455).
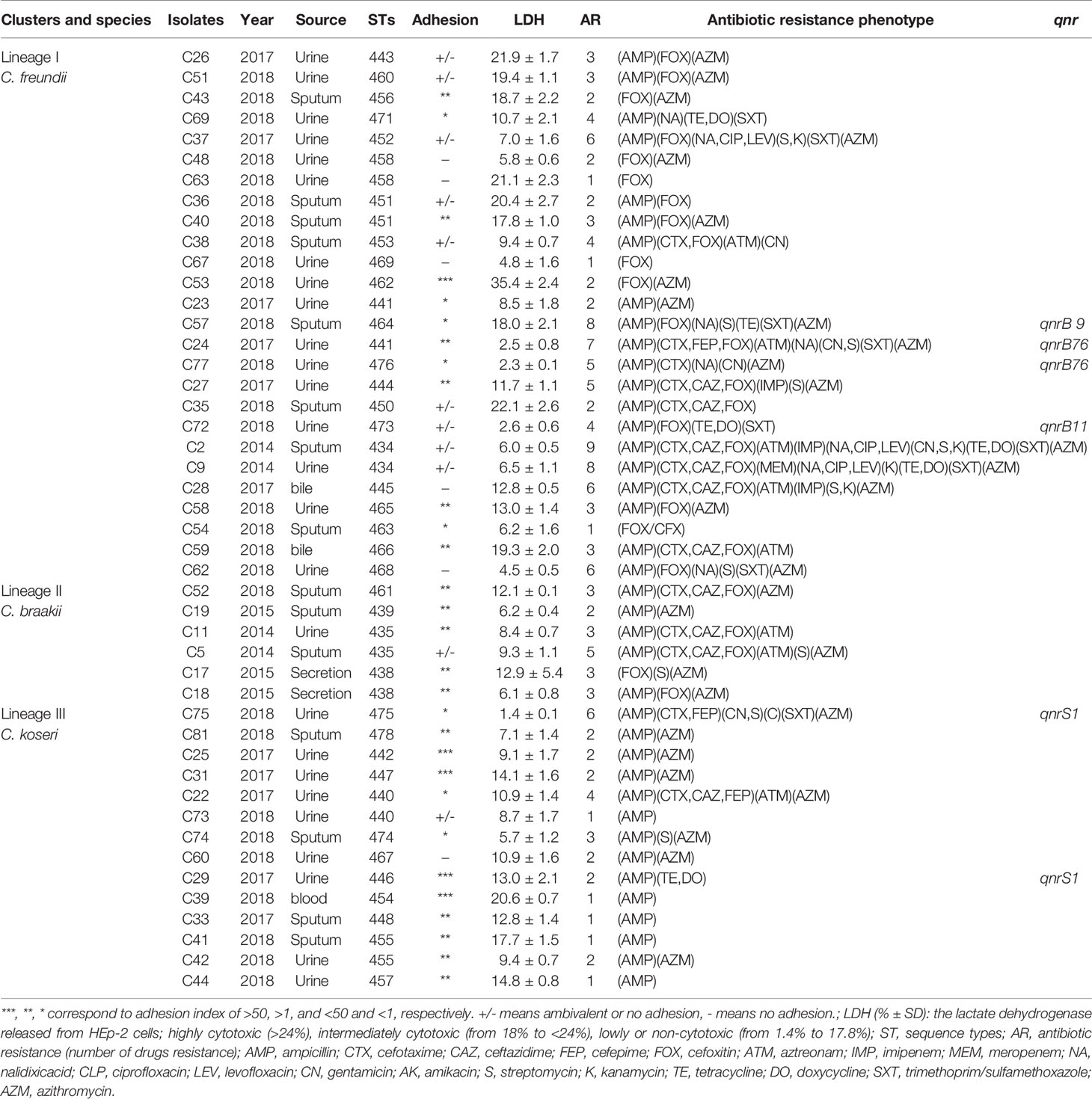
Table 2 Adherence, cytotoxicity, source, antibiotic resistance, genotypes, and antibiotic resistance phenotype of Citrobacter isolates.
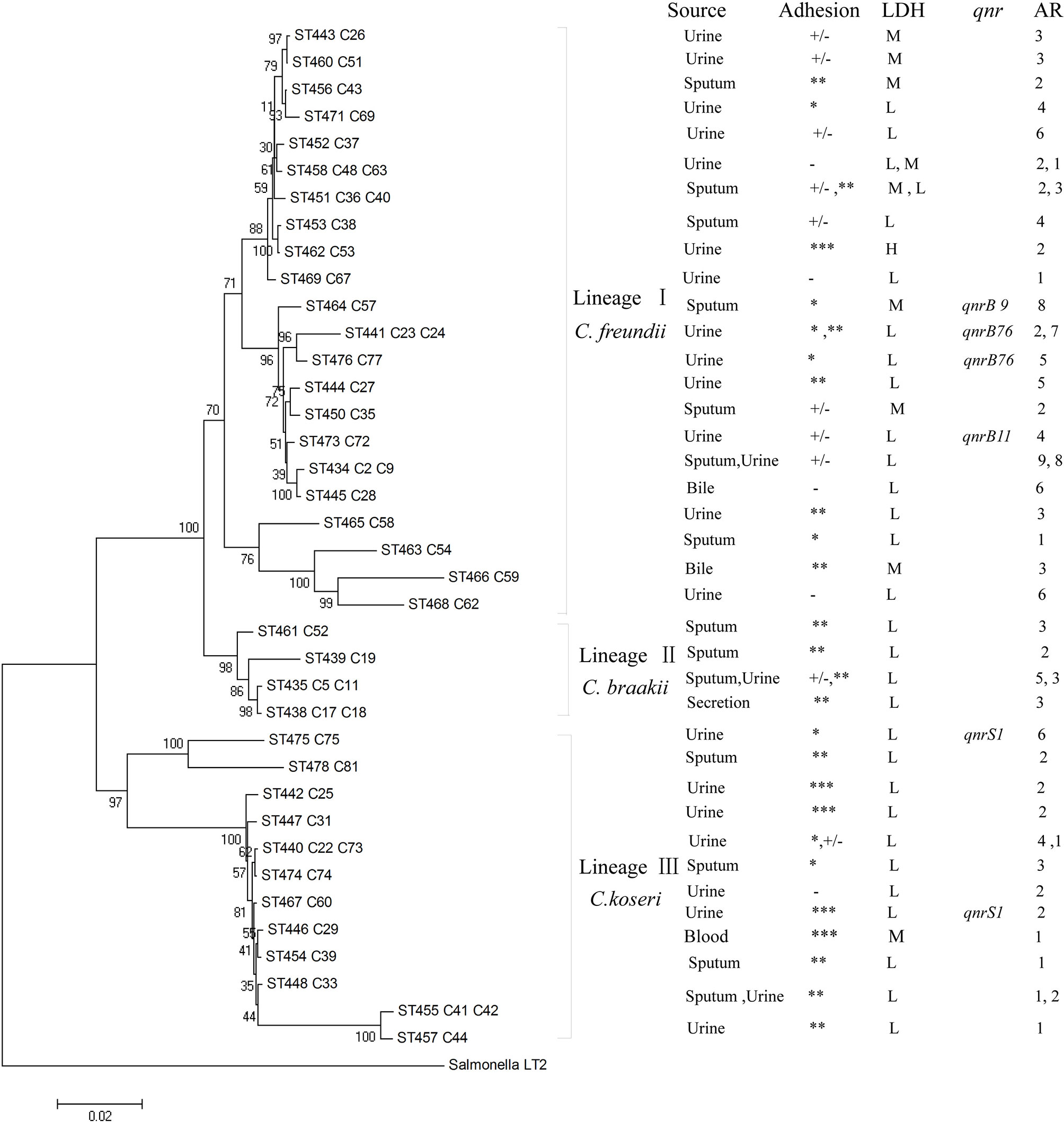
Figure 2 Phylogenetic relationships of the 46 Citrobacter isolates as determined by MLST data. The isolates were first marked with ST number followed by isolate name or names if more than one isolate. The presence of qnr genes, AR denoting antibiotic resistance with number of drugs resistant to, adhesion, LDH, and source among Citrobacter isolates were shown on the right. Note that for any STs with two isolates, properties for both were listed and separated by a comma. The tree was constructed using neighbor joining algorithm. ST and LDH indicate sequence types and lactate dehydrogenase, respectively. Cluster divisions are marked. Numbers on or near the nodes are bootstrap values from 1,000 replicates. Adhesion index: ***, >50; **, >1 and <50; *, <1; +/‐, ambivalent or no adhesion; ‐, no adhesion. Under LDH for cytotoxicity: H denotes highly cytotoxic if LDH values of >24%; M for intermediately cytotoxic if values from 18% to <24%; L for lowly or non-cytotoxic if LDH values from 1.4% to 17.8%. See Table 2 for actual values.
The concatenated sequences of the seven housekeeping genes were used to construct a phylogenetic tree by the neighbor-joining algorithm to infer the relationship of the 46 Citrobacter isolates (Figure 2). Salmonella LT2 was used as the outgroup. The tree could be divided into three lineages corresponding to species divisions with high bootstrap support. Lineage I, II, and III contained C. freundii isolates, C. braakii isolates, and C. koseri isolates, respectively. Citrobacter isolates from urine, sputum, and secretion samples were distributed among the different lineages (Table 2 and Figure 2).
We further analyzed the 38 STs using the goeBURST algorithm to identify CCs. In this study, we defined CCs as clusters of STs differing by no more than one of the seven alleles to identify the most closely related STs. We computed CCs using our STs from this study and STs from the PubMLST database and identified 51 CCs including 4 CCs in this study and 47 CCs contained isolates from China and other countries (Figure 3A and Supplementary Table S1). The four CCs from this study (CC46–CC48) included nine STs from this study only and contained no isolates or STs from other Chinese studies or from other countries (Figure 3 and Supplementary Table S1). All isolates of CC46 and CC48 were from urine samples, while CC45 and CC47 contained two isolates with one from urine and one from sputum, respectively (Supplementary Figure S1).
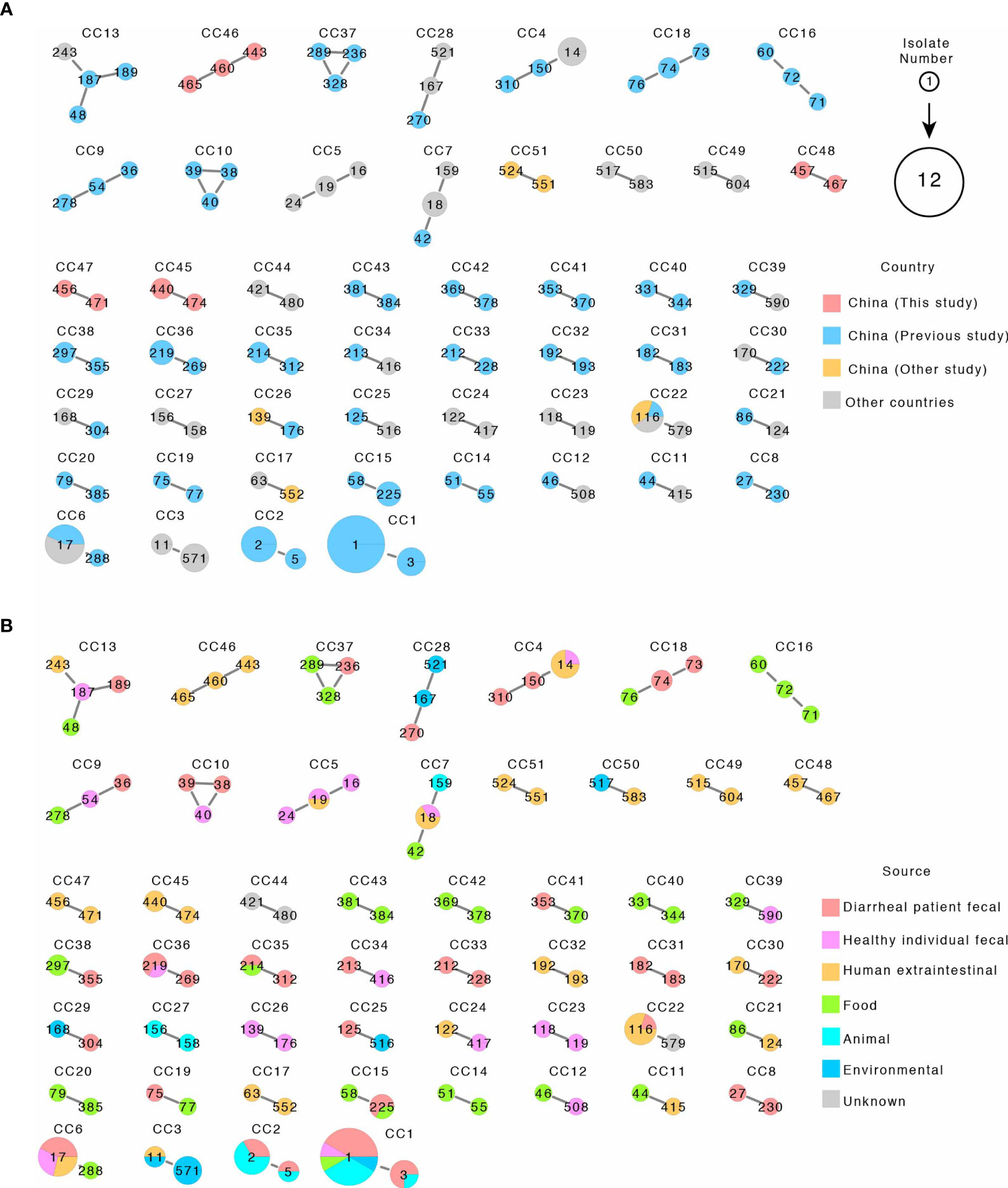
Figure 3 The 51 clonal complexes (CCs) by eBURST of 116 STs from this study and related STs from the public MLST database. (A, B) are the same CCs with A colored by country and B colored by source. Each circle represents an ST, and STs within CC are connected by solid line. ST numbers are marked inside the circle. Circle sizes are proportional to number of isolates. The pie chart within circles represented proportion of isolates from different country or sources as shown.
As shown in Figure 3A and Supplementary Table S1, some CCs were globally distributed and some CCs contained isolates from mixed sources including human clinical samples. CC6 contained two STs (ST288 and ST17). ST288 has one isolate from food, while ST17 has seven isolates with three isolated from diarrheal patients from China, two from healthy individual fecal samples in Latvia, and two from human extraintestinal samples in Poland. CC7 contained three STs (ST42, ST18, and ST159) and five isolates. The ST42 isolate was from food from China, the only ST159 isolate was from an animal source in Japan, while for the three ST18 isolates, two were from human extraintestinal samples in Poland and Spain, and one from healthy individual fecal samples in Greece. CC13 contained four STs each with one isolate which was found in China (three isolates) and USA, and the four isolates were separately from diarrheal patient fecal, healthy individual fecal, and human extraintestinal samples and food samples. CC4 contained three STs and six isolates, two isolates were from diarrheal patient fecal samples in China, and four isolates were from ST14 from other countries, including three human extraintestinal samples from Malaysia and one healthy individual fecal sample from Israel. CC22 contained two STs with five isolates and was found in China (three isolates) and other countries (two isolates from Thailand and one from The Netherlands) and with one from diarrheal patient fecal samples and four from human extraintestinal samples (blood, urine, and sputum) (Figure 3 and Supplementary Table S1).
Antibiotic Resistance of the Citrobacter Isolates and Prevalence of Multidrug Resistance
The 46 Citrobacter isolates were tested for susceptibility to 19 antibiotics belonging to 9 antibiotic classes using the disk diffusion method according to CLSI recommendations. The C. freundii isolates had higher antibiotic resistance rate than C. braakii and C. koseri isolates, although the number of isolates was small (Table 3). Most of the 26 C. freundii isolates were resistant to one or more of the β-lactam antibiotics, especially to penicillins (76.9%), cephalosporins (3.8–88.5%), monobactams (19.2%), and carbapenems (3.8–11.5%) (Table 3). For the four carbapenem resistant C. freundii isolates, three were resistant to IMP and one to MEM. Over half of the Citrobacter isolates (25/46) were MDR and were isolated from different years and sources (Table 2). The C. freundii isolates from urine (11/16, 68.8%) showed higher rate of MDR than the C. freundii isolates from sputum samples with 4/8 (50%) being MDR (Table 2).
Prevalence of Fluoroquinolones Resistant Isolates
Among the 46 Citrobacter isolates, 8 C. freundii isolates were resistant to fluoroquinolones, all of which were resistant to NAL; 7 resistant to CIP and NOR; and 6 resistant to LEV (Table 4). Six fluoroquinolones resistant isolates were from urine and two from sputum, all of which were MDR. All of these eight NA-resistant isolates contained the mutation in codon 59 (Thr59Ile) in the gyrA gene. No mutation was found in the parC gene (Table 4).
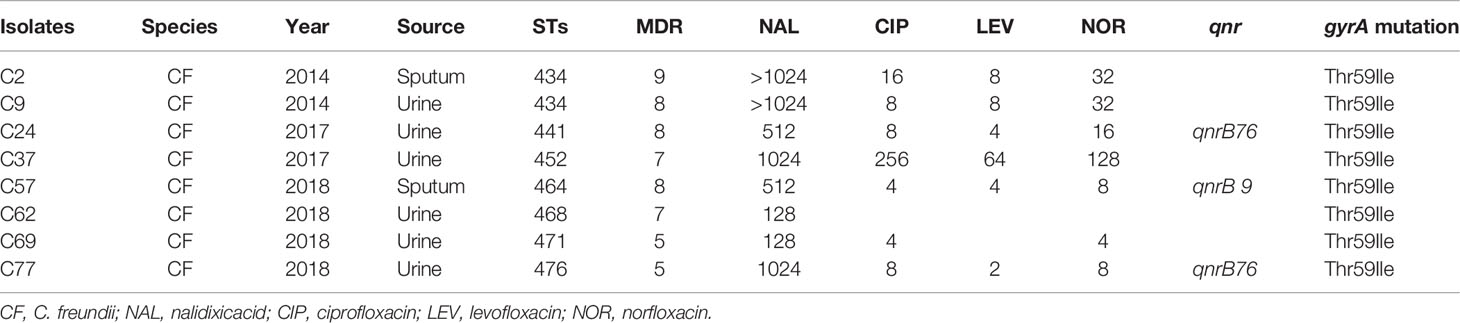
Table 4 Fluoroquinolone resistant values and alterations detected in the gyrA genes of Citrobacter isolates.
Prevalence of qnrB Genes
All the isolates were screened for qnrA, qnrB, qnrS, qnrC, qnrD, aac(6’)-Ib-cr, and qepA genes by PCR. One isolate was positive for qnrS, and four isolates were positive for qnrB (Table 2). PCR sequencing found that the qnrB alleles carried by the isolates were qnrB9 (one isolate), qnrB11 (one isolate), and qnrB76 (two isolates). Two qnrB76 harboring C. freundii isolates were all isolated from urine, all of which were resistant to NA, and were MDR. The qnrB9 harboring C. freundii isolate was isolated from sputum which was resistant to NA and was MDR.
Adherence and Cytotoxicity of Citrobacter Isolates
We tested the Citrobacter isolates for adhesion and cytotoxicity to HEp-2 cells in vitro as done previously (Bai et al., 2012; Liu et al., 2017; Liu et al., 2018a; Liu et al., 2020). Five isolates showed high adhesion, with an adhesion index greater than 50, four of which belonged C. koseri and one C. freundii. Other isolates showed no to intermediate adhesion. Only one isolate released LDH more than 24% and was classified as highly cytotoxic, and nine isolates released LDH from 18% to <24% and were classified as intermediately cytotoxic, while the remaining 36 isolates showed LDH release from 1.4% to 17.8% and were lowly or non-cytotoxic (Figures 4A, B and Table 2). When sample sources were considered, similar proportion of urine isolates (7/26, 26.7%) and sputum isolates (3/15, 20%) were highly/intermediately cytotoxic and/or highly adhesive (Table 2).
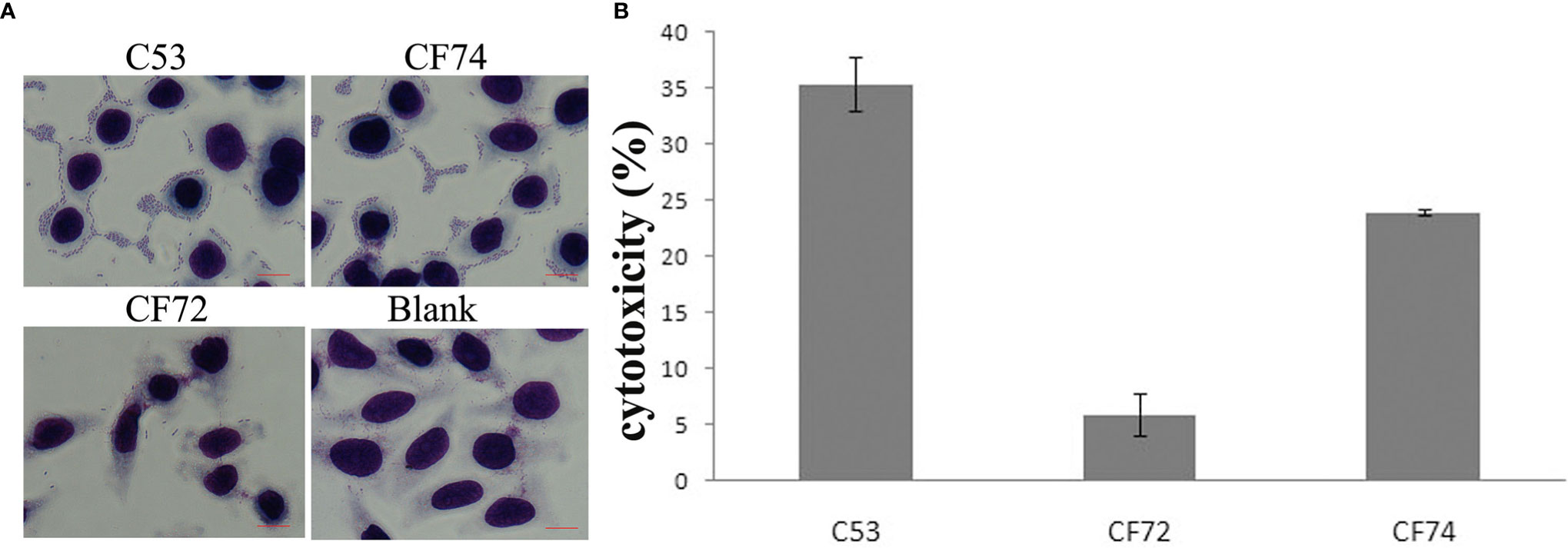
Figure 4 HEp-2 cell adhesion and cytotoxicity of Citrobacter isolates. (A) Light micrographs show the adherence patterns displayed by one strongly cytotoxic Citrobacter isolate (C53). Cells were stained with 1% Giemsa stain and examined under a light transmission microscope at a magnification of ×1,000. HEp-2 cell nuclei and bacteria were stained purple. C. freundii strain CF74 was used as highly adhesive control, C. freundii strain CF72 as lowly adhesive control, and Blank which had no bacteria as negative control. C53 displayed patterns of aggregative adherence to HEp-2 cells similar to CF74. Few bacteria were seen aggregated around HEp-2 cells in CF72. Bar: 10 µm. (B) Cytotoxicity of one highly cytotoxic Citrobacter isolate (C53) which was based on the LDH released from HEp-2 cells after exposure at 8 h. CF72 was used as lowly cytotoxic control strain and CF74 as highly cytotoxic strain. Y- axis is percentage of LDH released as measure of cytotoxicity.
Discussion
In this study, we surveyed Citrobacter extraintestinal infections in a hospital and found that Citrobacter isolates were mostly isolated from urine with 54.3% of the 46 isolates, followed by sputum samples (30.4%). Our findings were similar to those previously reported in India (Mohanty et al., 2007) and the USA (Hodges et al., 1978).
The 46 isolates were separated into 38 STs. The 38 STs from this study were compared with 566 STs from the Citrobacer MLST database, including isolates from all countries and sources. No ST in this study was shared with isolates from the database. In our previous study, 11 STs were shared with isolates from the database from other countries or regions or from different sources, and isolates of the same ST may be widely present in human fecal, food, and human extra intestinal samples (Liu et al., 2018a). We further analyzed the STs by CCs to ascertain any sharing of CCs and to determine whether there are any widely distributed CCs. The four CCs found in this study did not share any ST or CCs with other countries or other Chinese isolates. Fifteen CCs were found to be globally distributed. Some CCs contained human clinical isolates, including from diarrheal patient fecal samples and human extra-intestinal samples, which suggests that some CCs are more likely to cause diseases in humans.
A key strength of this study was that the population diversity and relationships of the isolates were assessed by MLST. The use of a standardized MLST scheme allowed comparison of data from this study with local and international MLST data from different sources. The combined MLST data from isolates from this study, isolates from human fecal samples and food samples from our previous studies, and other international isolates have revealed that there was no prevalent strains or clones, unlike many other bacterial pathogens such as UTI causing MDR E. coli ST131 with global distribution (Petty et al., 2014). However it is much needed of more studies using MLST or genome sequencing to better understand the genetic diversity and virulence of Citrobacter populations.
Clinical Citrobacter spp. strains are often resistant to multiple classes of antibiotics (Leski et al., 2016). Infections by MDR Citrobacter strains have been associated with a higher rate of in-hospital mortality compared to susceptible strains (Leski et al., 2016). Similarly, our study found that 54.3% of the isolates from extraintestinal infections were MDR with resistance to penicillin (84.8%), cephalosporins (67.4%), and azithromycin (65.2%), but susceptible to carbapenems (87.0%).
Carbapenem resistant Enterobacteriaceae (CRE) has become a major public health threat that requires urgent attention (Ramsamy et al., 2020). Carbapenem-resistant Citrobacter spp. isolates have been reported due to the acquisition of worldwide disseminated carbapenemases, such as New Delhi Metallo-β-lactamase (NDM), VIM-1, OXA-48, KPC-2, and VIM-2 (Hammerum et al., 2016; Arana et al., 2017; Faccone et al., 2019; Schweizer et al., 2019; Gobeille Paré et al., 2020; Räisänen et al., 2021). blaNDM-1-positive C. freundii has been increasingly reported in China, India, Denmark, and South Africa (Yang et al., 2018) and VIM-1- and VIM-2-positive C. freundii have also been reported in Europe (Gaibani et al., 2013; Porres-Osante et al., 2014; Santos et al., 2017). In this study, four isolates were resistant to IMP or MEM. We did not determine the molecular mechanisms of carbapenem resistance of these four isolates which will be done in future studies. In our previous study, all isolates which were isolated from food, diarrheal patient fecal, healthy individual fecal, and environmental samples were susceptible to carbapenems (Liu et al., 2017; Liu et al., 2018a; Liu et al., 2020).
The prevalence of quinolone resistance and mutations of quinolone resistance genes varied among Citrobacter isolates. In our previous study, C. braakii had the highest proportion of quinolone resistant isolates (52.6%), followed by C. freundii with 23.7% (Liu et al., 2020). In this study, C. freundii had the highest proportion of quinolone resistant isolates. Citrobacter isolates with mutations in the quinolone resistance determining region of gyrA, including Thr83Ile and Asp87Asn, have shown reduced susceptibility to fluoroquinolones (Weigel et al., 1998; Minarini and Darini, 2012). In our previous studies (Liu et al., 2018a; Liu et al., 2020), four quinolone resistant C. freundii isolates had mutations in Thr59Ile, Gln111Arg, and Ile134Val. Twenty-seven quinolone resistant isolates carried mutations in Thr59Ile and one having three mutations in Thr59Ile, Gln111Arg, and Ile134Val. In this study, among eight quinolone resistant C. freundii isolates, all had the Thr59Ile mutation in the gyrA gene.
Cytotoxicity and adhesive ability in vitro were assessed for all isolates, which varied widely. Among the 50 isolates, only five and one were shown to be highly adhesive and highly cytotoxic, respectively. We did not find any association of cytotoxicity and adhesive ability with the source of the isolates (urine or sputum samples). Since all isolates were from clinical infections, it seems that in vitro cytotoxicity and adhesive ability of an isolate may not be indicative of their disease causing ability. However, the numbers of isolates were small and there were no patient data to determine whether any of these parameters is suggestive of more severe disease outcomes.
Conclusion
We analyzed 46 extraintestinal clinical Citrobacter isolates (26 C. freundii, 6 C. braakii, and 14 C. koseri isolates) from 2014 to 2018 in Maanshan people’s hospital of Anhui Province, China. The isolates showed high diversity with 38 STs, all of which were novel STs. Nine of the 38 STs belonged to four CCs, but no isolates or STs from this study shared the same CCs with isolates from other countries or other Chinese isolates reported. MDR was prevalent among the isolates causing extraintestinal infections at 54.3%, and four isolates (8.7%) were carbapenem resistant (IMP or MEM). All eight quinolone resistant C. freundii isolates carried the Thr59Ile mutation in the gyrA gene. Only a small proportion of the isolates were found to be highly cytotoxic and adhesive with no correlation to sample sources. This study has shed more light on the genetic diversity and antibiotic resistance of extraintestinal infection causing Citrobacter in China.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical Committee of the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, China (No. ICDC-2016007). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LL and JX designed the project. YW and LZ carried out the sampling work. HZ, MY and HS carried out the experiments. LL, RL, and DH analyzed data. LL and RL drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2019YFC1200505 and 2019YFC1200500) and grants from National Natural Science Foundation of China (No. 81301401).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.737636/full#supplementary-material
References
Arana, D. M., Ortega, A., González-Barberá, E., Lara, N., Bautista, V., Gómez-Ruíz, D., et al. (2017). Carbapenem-Resistant Citrobacter Spp. Isolated in Spain From 2013 to 2015 Produced a Variety of Carbapenemases Including VIM-1, OXA-48, KPC-2, NDM-1 and VIM-2. J. Antimicrob. Chemother. 72 (12), 3283–3287. doi: 10.1093/jac/dkx325
Bai, L., Xia, S., Lan, R., Liu, L., Ye, C., Wang, Y., et al. (2012). Isolation and Characterization of Cytotoxic, Aggregative Citrobacter Freundii. PloS One 7 (3), e33054. doi: 10.1371/journal.pone.0033054
Chao, C. T., Lee, S. Y., Yang, W. S., Chen, H. W., Fang, C. C., Yen, C. J., et al. (2013). Citrobacter Peritoneal Dialysis Peritonitis: Rare Occurrence With Poor Outcomes. Int. J. Med. Sci. 10 (9), 1092–1098. doi: 10.7150/ijms.6251
Clinical and Laboratory Standards Institute. (2016). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI Document M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute.
Deal, E. N., Micek, S. T., Ritchie, D. J., Reichley, R. M., Dunne, W. M., Jr., Kollef, M. H. (2007). Predictors of in-Hospital Mortality for Bloodstream Infections Caused by Enterobacter Species or Citrobacter Freundii. Pharmacotherapy 27 (2), 191–199. doi: 10.1592/phco.27.2.191
Faccone, D., Albornoz, E., Tijet, N., Biondi, E., Gomez, S., Pasterán, F., et al. (2019). Characterization of a Multidrug Resistant Citrobacter Amalonaticus Clinical Isolate Harboring Bla(NDM-1) and Mcr-1.5 Genes. Infect. Genet. Evol. 67, 51–54. doi: 10.1016/j.meegid.2018.10.020
Francisco, A. P., Vaz, C., Monteiro, P. T., Melo-Cristino, J., Ramirez, M., Carriço, J. A. (2012). PHYLOViZ: Phylogenetic Inference and Data Visualization for Sequence Based Typing Methods. BMC Bioinformatics 13, 87. doi: 10.1186/1471-2105-13-87
Gaibani, P., Ambretti, S., Farruggia, P., Bua, G., Berlingeri, A., Tamburini, M. V., et al. (2013). Outbreak of Citrobacter Freundii Carrying VIM-1 in an Italian Hospital, Identified During the Carbapenemases Screening Actions, June 2012. Int. J. Infect. Dis. 17 (9), e714–e717. doi: 10.1016/j.ijid.2013.02.007
Gobeille Paré, S., Mataseje, L. F., Ruest, A., Boyd, D. A., Lefebvre, B., Trépanier, P., et al. (2020). Arrival of the Rare Carbapenemase OXA-204 in Canada Causing a Multispecies Outbreak Over 3 Years. J. Antimicrob. Chemother. 75 (10), 2787–2796. doi: 10.1093/jac/dkaa279
Hammerum, A. M., Hansen, F., Nielsen, H. L., Jakobsen, L., Stegger, M., Andersen, P. S., et al. (2016). Use of WGS Data for Investigation of a Long-Term NDM-1-Producing Citrobacter Freundii Outbreak and Secondary In Vivo Spread of blaNDM-1 to Escherichia Coli, Klebsiella Pneumoniae and Klebsiella Oxytoca. J. Antimicrob. Chemother. 71 (11), 3117–3124. doi: 10.1093/jac/dkw289
Hirai, J., Uechi, K., Hagihara, M., Sakanashi, D., Kinjo, T., Haranaga, S., et al. (2016). Bacteremia Due to Citrobacter Braakii: A Case Report and Literature Review. J. Infect. Chemother. 22 (12), 819–821. doi: 10.1016/j.jiac.2016.07.003
Hodges, G. R., Degener, C. E., Barnes, W. G. (1978). Clinical Significance of Citrobacter Isolates. Am. J. Clin. Pathol. 70 (1), 37–40. doi: 10.1093/ajcp/70.1.37
Ifeadike, C. O., Ironkwe, O. C., Adogu, P. O., Nnebue, C. C., Emelumadu, O. F., Nwabueze, S. A., et al. (2012). Prevalence and Pattern of Bacteria and Intestinal Parasites Among Food Handlers in the Federal Capital Territory of Nigeria. Niger. Med. J. 53 (3), 166–171. doi: 10.4103/0300-1652.104389
Jacoby, G. A., Strahilevitz, J., Hooper, D. C. (2014). Plasmid Mediated Quinolone Resistance. Microbiol. Spectr. 2, PLAS-0006-2013. doi: 10.1128/microbiolspec.PLAS-0006-2013
Kanamori, H., Yano, H., Hirakata, Y., Endo, S., Arai, K., Ogawa, M., et al. (2011). High Prevalence of Extended-Spectrum β-Lactamases and Qnr Determinants in Citrobacter Species From Japan: Dissemination of CTX-M-2. J. Antimicrob. Chemother. 66 (10), 2255–2262. doi: 10.1093/jac/dkr283
Khorasani, G., Salehifar, E., Eslami, G. (2008). Profile of Microorganisms and Antimicrobial Resistance at a Tertiary Care Referral Burn Centre in Iran: Emergence of Citrobacter Freundii as a Common Microorganism. Burns 34 (7), 947–952. doi: 10.1016/j.burns.2007.12.008
Kumar, P., Ghosh, S., Rath, D., Gadpayle, A. K. (2013). Multidrug Resistant Citrobacter: An Unusual Cause of Liver Abscess. BMJ Case Rep. 2013 (apr22 1). doi: 10.1136/bcr-2013-008714
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Lai, C. C., Tan, C. K., Lin, S. H., Liu, W. L., Liao, C. H., Huang, Y. T., et al. (2010). Bacteraemia Caused by Non-Freundii, Non-Koseri Citrobacter Species in Taiwan. J. Hosp. Infect. 76 (4), 332–335. doi: 10.1016/j.jhin.2010.06.006
Lechowicz, M., Dąbek, K., Majewska, U., Bekesińska-Figatowska, M., Borszewska-Kornacka, M. K., Bokiniec, R. (2017). Multiple Brain Abscesses Caused by Citrobacter Koseri in a Preterm Neonate - Case Report. Pol. J. Radiol. 82, 837–841. doi: 10.12659/pjr.903276
Lee, C. H., Lee, Y. T., Kung, C. H., Ku, W. W., Kuo, S. C., Chen, T. L., et al. (2015). Risk Factors of Community-Onset Urinary Tract Infections Caused by Plasmid-Mediated AmpC β-Lactamase-Producing Enterobacteriaceae. J. Microbiol. Immunol. Infect. 48 (3), 269–275. doi: 10.1016/j.jmii.2013.08.010
Leski, T. A., Taitt, C. R., Bangura, U., Stockelman, M. G., Ansumana, R., Cooper, W. H., 3rd, et al. (2016). High Prevalence of Multidrug Resistant Enterobacteriaceae Isolated From Outpatient Urine Samples But Not the Hospital Environment in Bo, Sierra Leone. BMC Infect. Dis. 16, 167. doi: 10.1186/s12879-016-1495-1
Liao, X., Fang, L., Li, L., Sun, J., Li, X., Chen, M., et al. (2015). Characterization of Chromosomal qnrB and ampC alleles in Citrobacter freundii Isolates From Different Origins. Infect. Genet. Evol. 35, 214–220. doi: 10.1016/j.meegid.2015.07.011
Liu, L., Chen, D., Liu, L., Lan, R., Hao, S., Jin, W., et al. (2018a). Genetic Diversity, Multidrug Resistance, and Virulence of Citrobacter Freundii From Diarrheal Patients and Healthy Individuals. Front. Cell Infect. Microbiol. 8, 233. doi: 10.3389/fcimb.2018.00233
Liu, L., Lan, R., Liu, L., Wang, Y., Zhang, Y., Wang, Y., et al. (2017). Antimicrobial Resistance and Cytotoxicity of Citrobacter Spp. In Maanshan Anhui Province, China. Front. Microbiol. 8, 1357. doi: 10.3389/fmicb.2017.01357
Liu, L., Qin, L., Hao, S., Lan, R., Xu, B., Guo, Y., et al. (2020). Lineage, Antimicrobial Resistance and Virulence of Citrobacter Spp. Pathogens 9 (3), 195. doi: 10.3390/pathogens9030195
Liu, L. H., Wang, N. Y., Wu, A. Y., Lin, C. C., Lee, C. M., Liu, C. P. (2018b). Citrobacter Freundii Bacteremia: Risk Factors of Mortality and Prevalence of Resistance Genes. J. Microbiol. Immunol. Infect. 51 (4), 565–572. doi: 10.1016/j.jmii.2016.08.016
Luo, Y., Wang, H., Liang, J., Qian, H., Ye, J., Chen, L., et al. (2021). Population Structure and Multidrug Resistance of Non-O1/Non-O139 Vibrio Cholerae in Freshwater Rivers in Zhejiang, China. Microb. Ecol. doi: 10.1007/s00248-020-01645-z. Online ahead of print.
Minarini, L. A., Darini, A. L. (2012). Mutations in the Quinolone Resistance-Determining Regions of gyrA and parC in Enterobacteriaceae Isolates From Brazil. Braz. J. Microbiol. 43 (4), 1309–1314. doi: 10.1590/s1517-838220120004000010
Mohanty, S., Singhal, R., Sood, S., Dhawan, B., Kapil, A., Das, B. K. (2007). Citrobacter Infections in a Tertiary Care Hospital in Northern India. J. Infect. 54 (1), 58–64. doi: 10.1016/j.jinf.2006.01.015
Nada, T., Baba, H., Kawamura, K., Ohkura, T., Torii, K., Ohta, M. (2004). A Small Outbreak of Third Generation Cephem-Resistant Citrobacter Freundii Infection on a Surgical Ward. Jpn. J. Infect. Dis. 57 (4), 181–182.
Oyeka, M., Antony, S. (2017). Citrobacter Braakii Bacteremia: Case Report and Review of the Literature. Infect. Disord. Drug Targets 17 (1), 59–63. doi: 10.2174/1871526516666161005155847
Park, Y. J., Yu, J. K., Lee, S., Oh, E. J., Woo, G. J. (2007). Prevalence and Diversity of qnr Alleles in AmpC-Producing Enterobacter cloacae, Enterobacter Aerogenes, Citrobacter Freundii and Serratia Marcescens: A Multicentre Study from Korea. J. Antimicrob. Chemother. 60, 868–871. doi: 10.1093/jac/dkm266
Petty, N. K., Ben Zakour, N. L., Stanton-Cook, M., Skippington, E., Totsika, M., Forde, B. M., et al. (2014). Global Dissemination of a Multidrug Resistant Escherichia Coli Clone. Proc. Natl. Acad. Sci. U.S.A. 111 (15), 5694–5699. doi: 10.1073/pnas.1322678111
Porres-Osante, N., Estepa, V., Seral, C., Rojo-Bezares, B., Salvo, S., Algarate, S., et al. (2014). First Description of a blaVIM-2-Carrying Citrobacter Freundii Isolate in Spain. Antimicrob. Agents Chemother. 58 (10), 6331–6332. doi: 10.1128/aac.03168-14
Protonotariou, E., Tsalidou, M., Vitti, D., Kalogeridis, A., Sofianou, D. (2008). First Identification of VIM-1-Producing Citrobacter Freundii in Greece. Int. J. Antimicrob. Agents 32 (5), 460–461. doi: 10.1016/j.ijantimicag.2008.05.008
Räisänen, K., Sarvikivi, E., Arifulla, D., Pietikäinen, R., Forsblom-Helander, B., Tarkka, E., et al. (2021). Three Clusters of Carbapenemase-Producing Citrobacter Freundii in Finland 2016-20. J. Antimicrob. Chemother. dkab209. doi: 10.1093/jac/dkab209. Online ahead of print.
Ramsamy, Y., Mlisana, K. P., Amoako, D. G., Allam, M., Ismail, A., Singh, R., et al. (2020). Pathogenomic Analysis of a Novel Extensively Drug-Resistant Citrobacter Freundii Isolate Carrying a Bla(NDM-1) Carbapenemase in South Africa. Pathogens 9 (2), 89. doi: 10.3390/pathogens9020089
Reyes, F., Singh, N., Anjuman-Khurram, N., Lee, J., Chow, L. (2017). Strongyloides Hyperinfection Syndrome Causing Fatal Meningitis and Septicemia by Citrobacter Koseri. IDCases 10, 102–104. doi: 10.1016/j.idcr.2017.09.005
Samonis, G., Karageorgopoulos, D. E., Kofteridis, D. P., Matthaiou, D. K., Sidiropoulou, V., Maraki, S., et al. (2009). Citrobacter Infections in a General Hospital: Characteristics and Outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 28 (1), 61–68. doi: 10.1007/s10096-008-0598-z
Santos, C., Ramalheira, E., Da Silva, G., Mendo, S. (2017). Genetically Unrelated Multidrug- and Carbapenem-Resistant Citrobacter Freundii Detected in Outpatients Admitted to a Portuguese Hospital. J. Glob. Antimicrob. Resist. 8, 18–22. doi: 10.1016/j.jgar.2016.09.010
Schweizer, C., Bischoff, P., Bender, J., Kola, A., Gastmeier, P., Hummel, M., et al. (2019). Plasmid-Mediated Transmission of KPC-2 Carbapenemase in Enterobacteriaceae in Critically Ill Patients. Front. Microbiol. 10, 276. doi: 10.3389/fmicb.2019.00276
Tassew, H., Abdissa, A., Beyene, G., Gebre-Selassie, S. (2010). Microbial Flora and Food Borne Pathogens on Minced Meat and Their Susceptibility to Antimicrobial Agents. Ethiop. J. Health Sci. 20 (3), 137–143. doi: 10.4314/ejhs.v20i3.69442
Vaz Marecos, C., Ferreira, M., Ferreira, M. M., Barroso, M. R. (2012). Sepsis, Meningitis and Cerebral Abscesses Caused by Citrobacter Koseri. BMJ Case Rep. 2012, bcr1020114941. doi: 10.1136/bcr.10.2011.4941
Wang, J. T., Chang, S. C., Chen, Y. C., Luh, K. T. (2000). Comparison of Antimicrobial Susceptibility of Citrobacter Freundii Isolates in Two Different Time Periods. J. Microbiol. Immunol. Infect. 33 (4), 258–262.
Weigel, L. M., Steward, C. D., Tenover, F. C. (1998). gyrA Mutations Associated With Fluoroquinolone Resistance in Eight Species of Enterobacteriaceae. Antimicrob. Agents Chemother. 42 (10), 2661–2667. doi: 10.1128/aac.42.10.2661
Weile, J., Rahmig, H., Gfröer, S., Schroeppel, K., Knabbe, C., Susa, M. (2007). First Detection of a VIM-1 Metallo-Beta-Lactamase in a Carbapenem-Resistant Citrobacter Freundii Clinical Isolate in an Acute Hospital in Germany. Scand. J. Infect. Dis. 39 (3), 264–266. doi: 10.1080/00365540600868388
Yang, L., Li, P., Liang, B., Hu, X., Li, J., Xie, J., et al. (2018). Multidrug-Resistant Citrobacter Freundii ST139 Co-Producing NDM-1 and CMY-152 From China. Sci. Rep. 8 (1), 10653. doi: 10.1038/s41598-018-28879-9
Zhang, R., Ichijo, T., Huang, Y. L., Cai, J. C., Zhou, H. W., Yamaguchi, N., et al. (2012). High Prevalence of qnr and aac(6’)-Ib-cr Genes in Both Water-Borne Environmental Bacteria and Clinical Isolates of Citrobacter freundii in China. Microb. Environ. 27, 158–163. doi: 10.1264/jsme2.ME11308
Keywords: Citrobacter spp., sequence types, multidrug resistance, gyrA, cytotoxicity
Citation: Liu L, Zhang L, Zhou H, Yuan M, Hu D, Wang Y, Sun H, Xu J and Lan R (2021) Antimicrobial Resistance and Molecular Characterization of Citrobacter spp. Causing Extraintestinal Infections. Front. Cell. Infect. Microbiol. 11:737636. doi: 10.3389/fcimb.2021.737636
Received: 07 July 2021; Accepted: 03 August 2021;
Published: 27 August 2021.
Edited by:
Krisztina M. Papp-Wallace, Louis Stokes Cleveland VA Medical Center, United StatesReviewed by:
Rezvan Moniri, Kashan University of Medical Sciences, IranLeili Shokoohizadeh, Hamadan University of Medical Sciences, Iran
Copyright © 2021 Liu, Zhang, Zhou, Yuan, Hu, Wang, Sun, Xu and Lan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liyun Liu, bGl1bGl5dW5AaWNkYy5jbg==; Ruiting Lan, ci5sYW5AdW5zdy5lZHUuYXU=
†These authors have contributed equally to this work
 Liyun Liu
Liyun Liu Ling Zhang2†
Ling Zhang2† Min Yuan
Min Yuan Ruiting Lan
Ruiting Lan