- 1Department of Clinical Laboratory, The First Affiliated Hospital of Nanchang University, Nanchang, China
- 2Department of Medical Microbiology, School of Medicine, Nanchang University, Nanchang, China
- 3Department of Preventive Medicine and Public Health, School of Public Health, Nanchang University, Nanchang, China
Background: The incidence of invasive candidiasis is increasing worldwide. However, the epidemiology, antifungal susceptibility, and virulence of Candida spp. in most hospitals remain unclear. This study aimed to evaluate invasive candidiasis in a tertiary care hospital in Nanchang City, China.
Methods: MALDI-TOF MS and 18S rDNA ITS sequencing were used to identify Candida strains. Randomly amplified polymorphic DNA analysis was used for molecular typing; biofilm production, caseinase, and hemolysin activities were used to evaluate virulence. The Sensititre™ YeastOne YO10 panel was used to examine antifungal susceptibility. Mutations in ERG11 and the hotspot regions of FKS1 of drug-resistant strains were sequenced to evaluate the possible mechanisms of antifungal resistance.
Results: We obtained 110 Candida strains, which included 40 Candida albicans (36.36%), 37 C. parapsilosis (33.64%), 21 C. tropicalis (19.09%), 9 C. glabrata (8.18%), 2 C. rugose (1.82%), and 1 C. haemulonii (0.91%) isolates. At a limiting point of 0.80, C. albicans isolates could be grouped into five clusters, C. parapsilosis and C. tropicalis isolates into seven clusters, and C. glabrata isolates into only one cluster comprising six strains by RAPD typing. Antifungal susceptibility testing revealed that the isolates showed the greatest overall resistance against fluconazole (6.36%), followed by voriconazole (4.55%). All C. albicans and C. parapsilosis isolates exhibited 100% susceptibility to echinocandins (i.e., anidulafungin, caspofungin, and micafungin), whereas one C. glabrata strain was resistant to echinocandins. The most common amino acid substitutions noted in our study was 132aa (Y132H, Y132F) in the azole-resistant strains. No missense mutation was identified in the hotpot regions of FKS1. Comparison of the selected virulence factors detectable in a laboratory environment, such as biofilm, caseinase, and hemolysin production, revealed that most Candida isolates were caseinase and hemolysin producers with a strong activity (Pz < 0.69). Furthermore, C. parapsilosis had greater total biofilm biomass (average Abs620 = 0.712) than C. albicans (average Abs620 = 0.214, p < 0.01) or C. tropicalis (average Abs620 = 0.450, p < 0.05), although all C. glabrata strains were either low- or no-biofilm producers. The virulence level of the isolates from different specimen sources or clusters showed no obvious correlation. Interesting, 75% of the C. albicans from cluster F demonstrated azole resistance, whereas two azole-resistant C. tropicalis strains belonged to the cluster Y.
Conclusion: This study provides vital information regarding the epidemiology, pathogenicity, and antifungal susceptibility of Candida spp. in patients admitted to Nanchang City Hospital.
Introduction
Fungal infections in humans are relatively common and range from common or mild superficial infections to life-threatening invasive infections. In recent years, the widespread use of broad-spectrum antibiotics, hormones, immunosuppressive agents, chemotherapy, and central venous catheters have led to an increase in the incidence of invasive fungal infections. More than 1.6 million people worldwide have been reported to suffer from serious fungal diseases that have a profound impact on patients and can even be fatal (Bongomin et al., 2017).
Infections caused by Candida spp. can be divided into superficial, cutaneous, mucosal, and invasive infections (deep and extensive). Up to date, Candida spp. have become the third most common cause of bloodstream infections, including candidemia (Wisplinghoff et al., 2004). Candida albicans is responsible for approximately 50% of all candidiasis, and the other species account for the remaining Candida infections. Specifically, infections caused by C. tropicalis, C. glabrata, C. parapsilosis, C. krusei, and C. auris have attracted significant scientific attention (Mba and Nweze, 2020). Invasive candidiasis has been attributed to 40%–50% mortality in intensive care units (ICUs) (De Rosa et al., 2009). Candidemia is currently the fourth most common nosocomial bloodstream infection in America (Wisplinghoff et al., 2004; Li et al., 2016). In Europe, the incidence of candidemia was estimated to be approximately 79 cases per day, of which 29 patients were estimated to have a fatal outcome on Day 30 (Koehler et al., 2019).
As the mortality rate in candidiasis is high, identifying the relationship among the various Candida species and assessing disease epidemiology, species distribution, and drug resistance to facilitate the control of nosocomial candidiasis is crucial. This study describes the results from an analysis of 110 Candida strains isolated from sterile body fluid samples obtained between 2014 and 2019 from patients in a tertiary hospital in Nanchang City. Candida spp. in the samples were identified, tested, and analyzed to evaluate the prevalence of Candida infection and determine virulence profiles.
Materials and Methods
Strains
This retrospective study was conducted between September 2014 and September 2019. Candida spp. were isolated from sterile fluid samples (i.e., blood, ascites, bile, secretions, and abscess) of patients admitted in the First Affiliated Hospital of Nanchang University, China. The ethics committee of the university hospital approved this study (approval no. 2014036).
Separation and Identification
The desired strains were isolated and purified on Sabouraud’s dextrose agar (SDA; 1% peptone, 4% dextrose, and 2% agar) plates and cultured at 30°C for 24 h. The first crucial step in our study is the correct identification of the Candida species. The strains were initially identified by MALDI-TOF mass spectrometry analyses using a Clin-ToF-II mass spectrometer (Bioyong, China). To ensure accuracy, 18S rDNA ITS sequencing was performed for further identification. Briefly, the contiguous ITS1-5.8S rDNA-ITS2 region was amplified with the universal fungal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Salehi et al., 2017). PCR reactions were performed using the Bio-Rad S1000 thermal cycler in 50 μL volumes containing 1 μL of extracted DNA, 47 μL of T3 Super PCR mix (TSINGKE, China), and 1 μL of each ITS1 and ITS4 primers. Amplification was performed with cycles of 3 min at 98°C for primary denaturation, followed by 35 cycles at 98°C (10 s), 55°C (10 s), and 72°C (20 s), along with a final extension at 72°C for 5 min. The PCR amplification products were sequenced by Tsingke Biological Company (Changsha, China). The sequences were analyzed using the NCBI BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Randomly Amplified Polymorphic DNA Analysis
DNA from individual strains was extracted as described by Jain et al. but with certain modifications (Jain et al., 2001). Briefly, overnight-cultured Candida strains were harvested via centrifugation at 12000 rpm after washing with sterile PBS, and the precipitate was resuspended in 100 μL STES buffer (0.2 M Tris-HCl, 0.5 M NaCl, 0.1% SDS, and 0.01 M EDTA, pH = 8.0) and 20 μL TE buffer (0.01 M Tris-HCl and 0.001 M EDTA, pH = 8.0). Acid-washed glass beads (Sigma-Aldrich, USA) and 120 μL phenol–chloroform–isoamyl alcohol mixture (25:24:1 v/v/v) were added to lyse cells and release DNA. After 10 min of vortexing, lysates were centrifuged at 12000 rpm and 4°C for 10 min. DNA from the aqueous phase was precipitated with 2 times the volume of absolute ethanol at −20°C for 15 min, centrifuged at 12000 rpm and 4°C for 10 min, dried, and resuspended in 50 μL TE buffer. The concentration of nucleic acids was measured using a NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific, USA).
Genomic DNA was amplified using previously described primers for five genes, namely, CD16AS (5′-CTCTTGAAACTGGGGAGACTTGA-3′), HP1247 (5′-AAGAGCCCGT-3′), ERIC-2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′), OPE-3 (5′-CCAGATGCAC-3′), and OPE-18 (5′-GGACTGCAGA-3′) (Paluchowska et al., 2014). The primers used were synthesized by the Tsingke Biological Technology Company. Randomly amplified polymorphic DNA (RAPD) reactions were performed in a final volume of 50 μL, which included 1 μL of the DNA template (approximately 50 ng), 1 μL of each primer (10 μM), and the T3 Super PCR mix. Amplification reactions were performed for 40 cycles using a Bio-Rad S1000 thermal cycler under the following conditions: initial denaturation at 98°C for 3 min, followed by denaturation at 98°C for 10 s, annealing at 36°C for 1 min, and extension at 72°C for 2 min, along with a final extension at 72°C for 5 min. The DNA fragments were separated by electrophoresis in a 1% agarose gel run at 100 V for 30 min. Fragment sizes were determined by comparison with a DL5000 DNA Marker (Vazyme, China), and bands were visualized in the ChemiDocXRS+ system (Bio-Rad, USA). Similarity analysis of RAPD patterns was performed using NTSYS 2.10 software.
Drug Sensitivity Test
In vitro susceptibility of Candida strains to nine antifungal drugs, namely, 5-flucytosine, anidulafungin, caspofungin, itraconazole, micafungin, posaconazole, voriconazole, amphotericin B, and fluconazole, was determined using the Sensititre™ YeastOne YO10 panel (Thermo Scientific, USA) based on manufacturer’s instructions. C. krusei ATCC 6258 and C. parapsilosis ATCC 22019 were used as controls, as per the Clinical and Laboratory Standards Institute guidelines. Plates were incubated at 35°C for 24 h, and minimum inhibitory concentrations (MICs) were interpreted according to documents M27-A3 and M27-S4 (Institut, C.a.L.S 2008; Institute, C.a.L.S 2012).
Sequencing of ERG11 and FKS1
The entire open reading frame (ORF) of ERG11 from azole-resistant strains were amplified and sequenced with specific primers (Supplementary Table S1). The hotspot 1 (HS1) and hotspot 2 (HS2) sequences of FKS1 were amplified using PCR with specific primers shown in Supplementary Table S1 based on their key role in echinocandin resistance. Conventional PCRs were performed in a 50 μL reaction mixture containing 47 μL of T3 Super PCR mix, 0.2 μM of each primer, and 100 ng of the extracted DNA. The amplification conditions were as follows: initial denaturation at 98°C for 3 min followed by 35 cycles of denaturation (98°C, 10 s), annealing (52°C, 20 s), and extension (72°C, 5 min), followed by a final extension of 72°C for 10 min. The PCR products were purified and sequenced by the Tsingke Biological Technology Company. The ERG11 sequences were aligned using BLAST and compared with the related published GenBank sequence for C. albicans SC5314 (Gene ID: 3641571), C. parapsilosis ATCC 22019 (Gene ID GQ302972.1), C. tropicalis ATCC 750 (Gene ID: M23673), C. glabrata CBS138 (Gene ID: XM 445876), and C. haemulonis (Gene ID: XM_025486744.1). The FKS1 sequences were compared with the reference sequences of C. tropicalis ATCC 750 (Gene ID: EU676168) and C. glabrata (Gene ID: XM_446406).
Caseinase Activity
Caseinase activity was measured by the single diffusion technique in SDA plates containing 1% casein (Ramesh et al., 2011). A standard inoculum (106 cells/mL) was prepared in saline solution from an overnight yeast culture for each isolate and 10 μL of this standard inoculum was plated. Plates were incubated at 37°C for 5 days. Three independent replicates were tested for each strain. Colony diameter (a) and colony diameter plus precipitation zone (b) were measured using a vernier caliper (Guanglu, China). Phospholipase index (defined as Pz = a/b, based on the study by Price et al) was used to evaluate the extent of hydrolytic enzyme production by various Candida species (Price et al., 1982). According to this definition, a lower Pz value indicates a greater enzymatic activity, which was scored into four categories as follows: Pz = 1.00 was defined as no enzymatic activity, Pz = 0.90–0.99 as weak enzymatic activity, Pz = 0.89–0.70 as moderate activity, and Pz < 0.69 as strong enzymatic activity (Price et al., 1982).
Hemolysin Activity
Hemolysin assay for Candida strains was measured using blood agar plates that were prepared by adding 6% human blood to 100 mL of SDA supplemented with 3% glucose (final concentration, wt/vol; pH = 5.6) (Neji et al., 2017). Standard inoculum (10 μL of 106 cells/mL) was spotted onto the blood plates, plates were incubated at 37°C for 5 days, and hemolytic activity was measured and evaluated using the method described by Price et al. (1982). The assay was performed in triplicate for each isolate.
Biofilm Formation
Biofilm production in Candida species was evaluated using a modified crystal violet assay described elsewhere (Silva et al., 2009; Mancera et al., 2015; Neji et al., 2017). Briefly, biofilm formation was spectrophotometrically determined wherein each well of a 96-well polystyrene plate was treated with 200 μL of 5% BSA (dissolved in sterilized PBS) at 4°C for 48 h and washed once with sterile PBS. Each experimental condition was designed such that five replicates were distributed in one column of the microplate. Sterility controls, i.e., without cell suspension, were used as a negative control to ensure no accidental contamination during treatment. Clinical isolates and the control strain Sc5314 were first cultured on SDA plates at 30°C for 24 h and then in 5 mL SDA medium overnight at 30°C with shaking. Cells were collected by centrifugation at 2458 g for 5 min, washed twice with sterilized PBS, and resuspended in spider medium (20 g/L nutrient broth, 20 g/L mannitol, and 4 g/L K2HPO4; pH 7.2). The turbidity of each strain suspension was spectrophotometrically adjusted to OD600 = 0.5; 200 μL of the cell suspension (OD600 = 0.5) was transferred to each well of the 96-well plates, which were incubated at 37°C for 90 min. Next, the strain suspension was removed, 200 μL of fresh spider medium was added, and the plates were incubated at 37°C for 48 h without shaking (static culture). Plates were washed twice with PBS to remove nonadherent cells after the adhesion stage and biofilms were fixed by incubation with 200 μL methanol for 30 min. Afterward, the plates were dried at room temperature and 200-μL 1% crystal violet (Solarbio, China) was added to each well and incubated for 30 min. The wells were then gently washed with water until the washing was colorless and incubated with 200 µL of acetic acid for 1 h to dissolve the biofilm. The absorbance of the obtained solution was read at 620 nm in a microtiter plate reader (Molecular Devices, USA). Absorbance values of the controls were subtracted from test values to eliminate background interference.
Statistical Analysis
Data were analyzed with SPSS (version 25.0) and GraphPad Prism (version 5.0). Unless otherwise mentioned, data are presented as mean ± standard deviation (SD). The normality of data distribution was evaluated using the D’ Agostino-Pearson omnibus normality test. Differences between two independent groups were performed with the nonparametric Mann–Whitney U test or the Kolmogorov–Smirnov Z test for the relative values. A p value of ≤0.05 was defined as statistically significant.
Results
Prevalence of Invasive Candida spp. Isolates
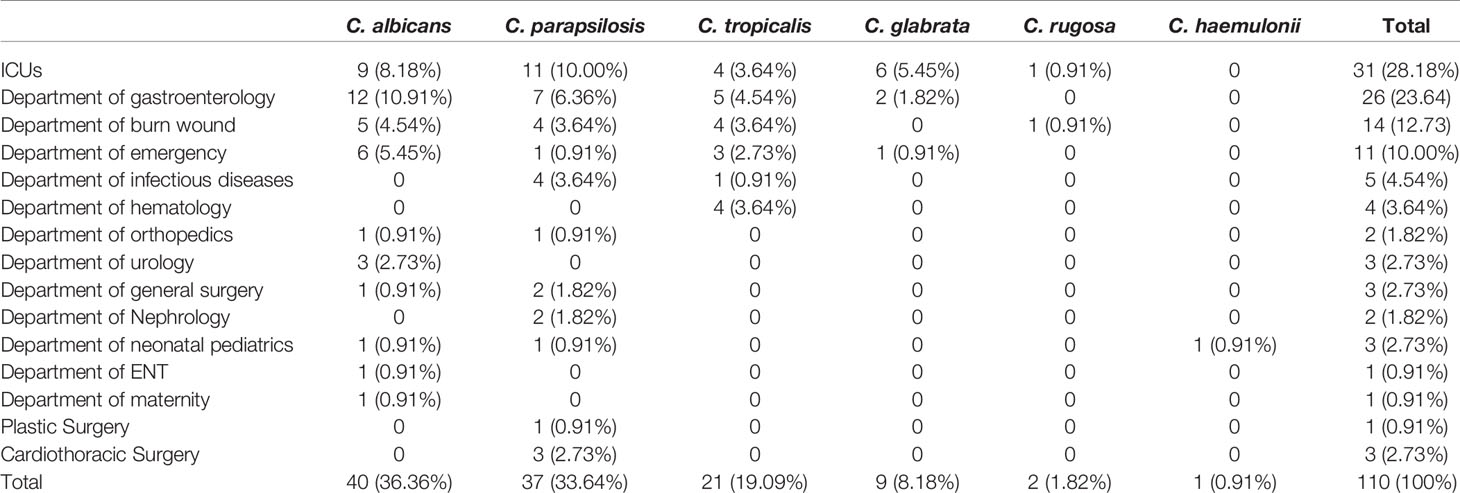
A total of 105 patients were enrolled in this study. The study population comprised 77 (73.3%) males and 28 (26.7%) females with a median age of 53 years (range 0–87 years). Although most isolates were obtained from patients hospitalized in the ICU (n = 31, 28.18%), 26 (23.6%) came from the department of gastroenterology, 14 (12.73%) from the burns unit, 11 (10.0%) from emergency services unit, 5 (4.54%) from infectious diseases unit, 4 (3.6%) from the hematology department, and 17 (15.5%) from other non-ICU departments (Table 1). Overall, 22.9% (24/105) of the patients died, mainly from acute pancreatitis, which accounted for approximately 37.5% of all mortality cases. Antifungal use (oral or injected) was reported for 42.9% (45/105) of the patients. In our study, the most common antifungal drug used was fluconazole (44.4%, 20/45), followed by voriconazole (40.0%, 18/45) and caspofungin (15.6%, 7/45).
A total of 110 clinical isolates (40 C. albicans, 37 C. parapsilosis, 21 C. tropicalis, 9 C. glabrata, 2 C. rugose, and 1 C. haemulonii isolates) were studied (Figure 1). Each isolate was obtained from different patients, with the following exceptions: C. albicans NCU_B040, NCU_B049, and NCU_B059, and C. tropicalis NCU_B047 and NCU_B050 strains, were acquired from the blood of one patient and three strains of C. glabrata (NCU_B103, NCU_O109, and NCU_O145) were isolated from different source sites of the same patient.
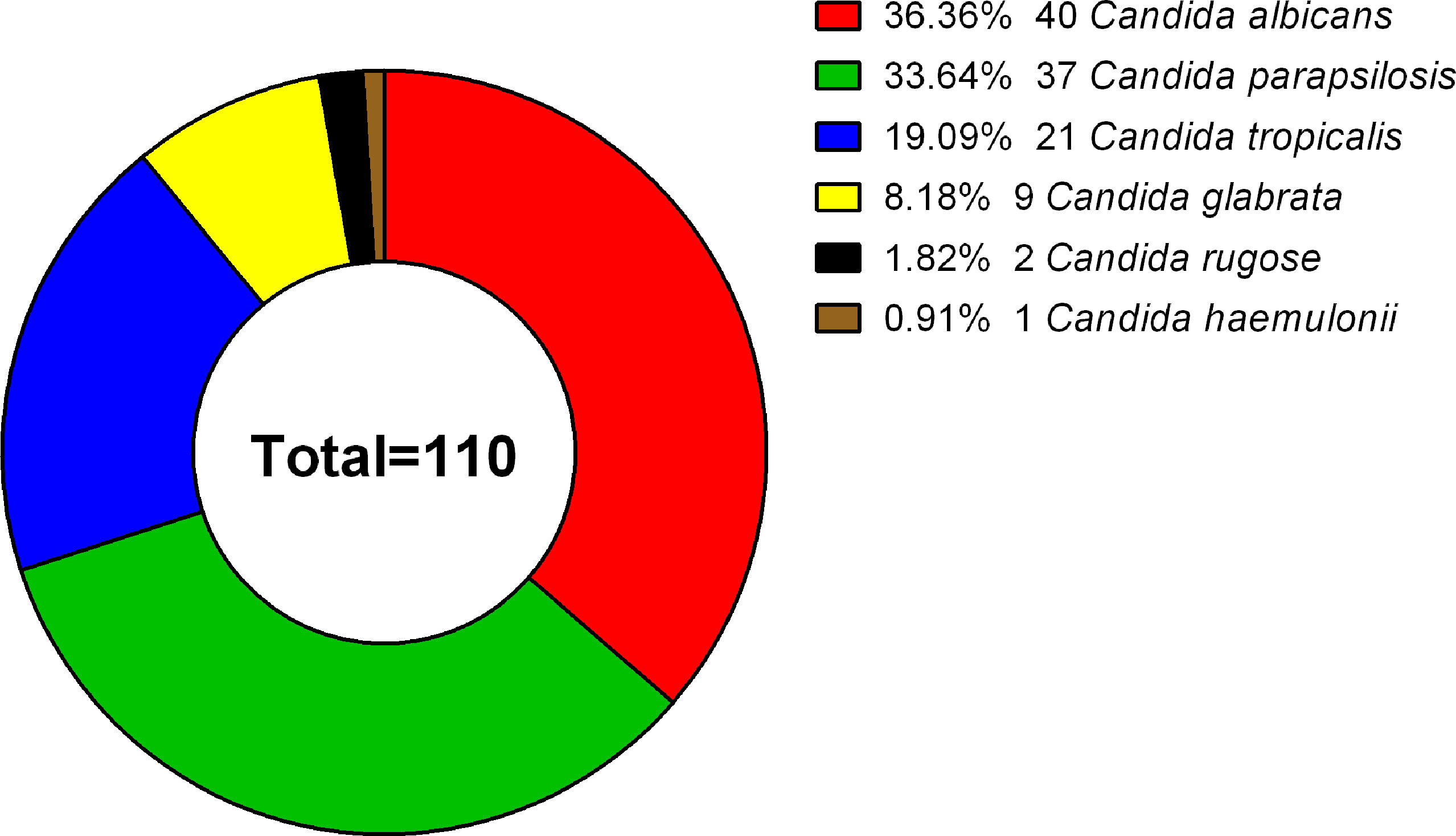
Figure 1 Species distribution of 110 clinical isolates. The 110 Candida strains analyzed included 40 C. albicans (36.36%), 37 C. parapsilosis (33.64%), 21 C. tropicalis (19.09%), 9 C. glabrata (8.18%), 2 C. rugose (1.82%), and 1 C. haemulonii (0.91%) isolate.
Sources of the 110 isolates (Table 2) were predominantly blood (n = 64, 58.18%), followed by catheter tips (n = 9, 8.18%) and drainage fluid (n = 17, 15.45%), and also included abscesses (1.82%), secretions (1.82%), and other sources (14.55%). Furthermore, as shown in Table 2 and Figure 2, C. albicans could be isolated from a wider range of specimens than other species. However, C. haemulonii and C. rugose were isolated only from blood.
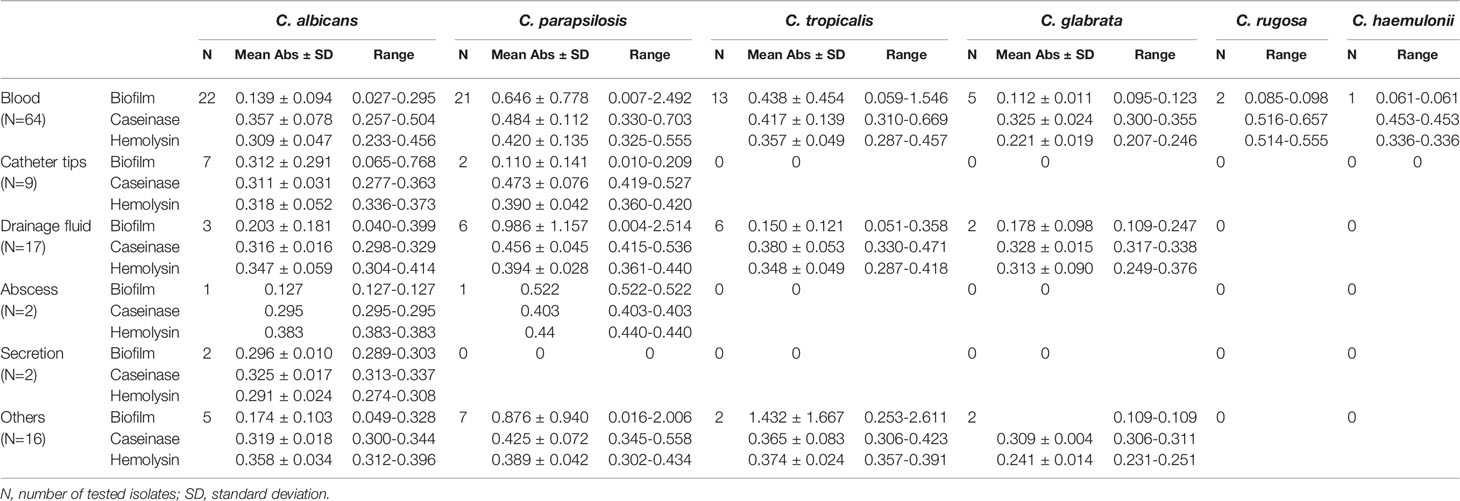
Table 2 Comparison of the virulence factors among Candida strains isolated from different sources (mean optical density ± SD).
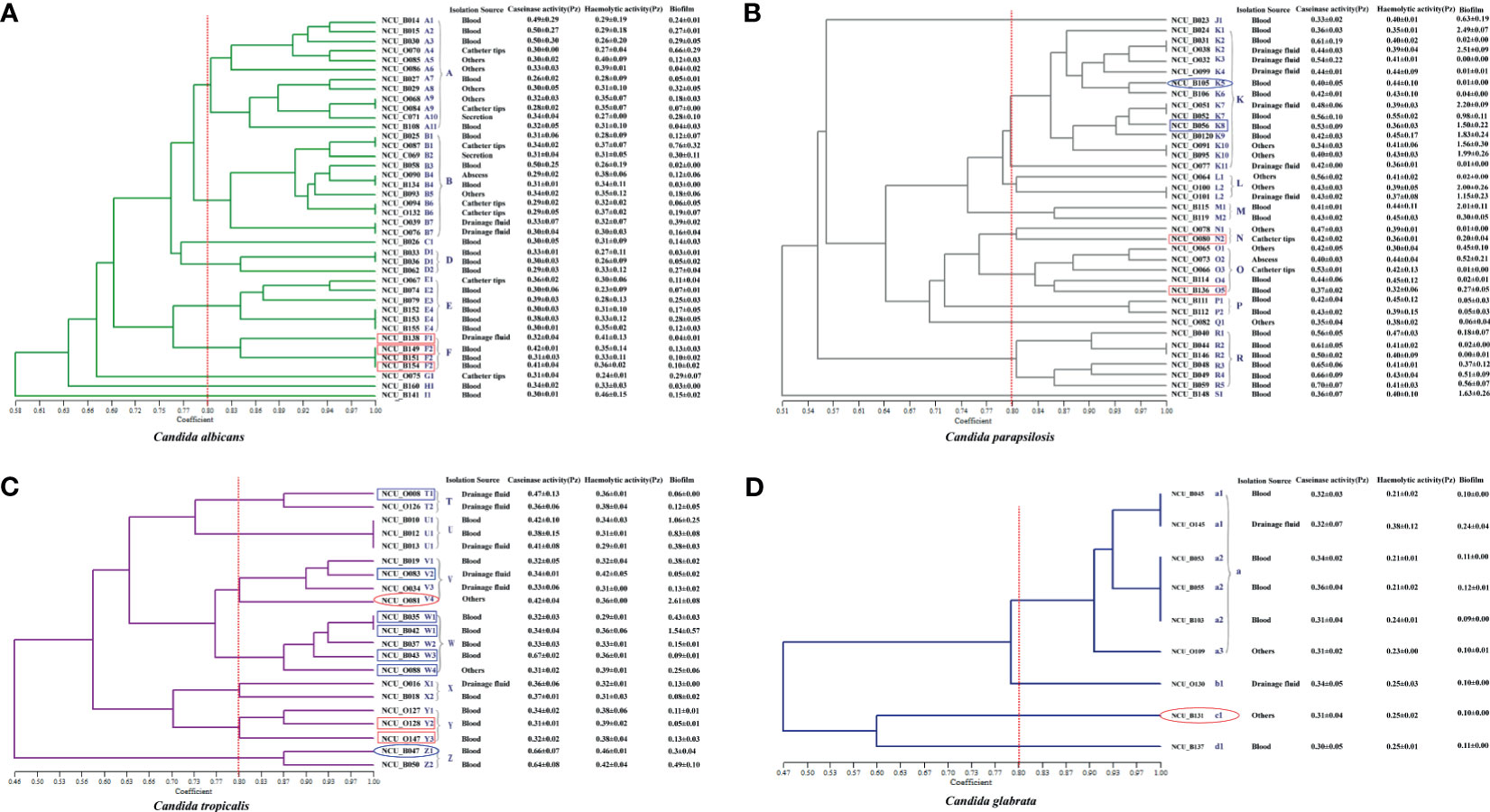
Figure 2 Dendrogram presenting the genetic relatedness of 107 Candida spp. based on the random amplified polymorphic DNA (RAPD) data. The virulence factors (i.e., caseinase, hemolysin, and biofilm), drug resistance, and specimen sources are indicated in the figure. The vertical line divides the strains based on the level of genetic similarity into related and unrelated strains. The red boxes indicate azole resistance (fluconazole and voriconazole), whereas the blue boxes indicate azole-susceptible (S) dose dependence (SDD). Red ellipses represent resistance to echinocandins, while the blue ellipses represent intermediate resistance to micafungin. (A) Dendrogram of 40 C. albicans isolates. (B) Dendrogram of 37 C. parapsilosis isolates. (C) Dendrogram of 21 C. tropicalis isolates. (D) Dendrogram of 9 C. glabrata isolates.
Molecular Phylogenetic Analysis of the Invasive Candida Spp. Isolates
To explore the diversity of the isolated Candida spp., RAPD analysis of the four major species of clinical isolates was performed with five random primers using the UPGMA method. C. rugose and C. haemulonii strains were not analyzed because of their low isolation rates. The number of patterns generated by C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata isolates with each primer changed between 3–10, 4–12, 3–10, and 6–10, respectively. All primers were used for the dendrogram construction.
We recognized 32 distinct RAPD profiles among the 40 C. albicans isolates analyzed. Figure 2A shows the cluster analysis of the electrophoretic bands of the C. albicans isolates, and the Sj value ranged from 0.58 to 1.00. Four strains presented unique patterns (C1, G1, H1, and I1) and were considered to be unrelated. At a limiting point of 0.80, 36 C. albicans isolates could be allocated into five clusters (A, B, and D–F). For C. parapsilosis, the primers produced up to 12 bands, and therefore, this set displayed the greatest discriminatory power. In their dendrogram, C. parapsilosis isolates could be grouped into seven clusters (K–P, and R) when 0.80 was assumed as the limiting value, whereas three isolates (NCU_B023, NCU_O082, and NCU_B148) were deemed unrelated (Figure 2B). Among the 21 C. tropicalis isolates, 18 RAPD profiles could be distinguished, and they were accommodated into seven clusters (T–Z) (Figure 2C). Six genotypes (a1– a3, b1, c1, and d1) were observed upon the RAPD analysis of the nine C. glabrata isolates. A total of 6 C. glabrata isolates with a similarity level >80% were allocated into the same cluster; of these, NCU_B103, NCU_O109, and NCU_O145 were different sites of the same patient and were grouped in the same cluster. Three genotypes (b1, c1, and d1) were represented by only one isolate each, and they were considered unrelated (Figure 2D).
Antifungal Susceptibility for the Invasive Candida spp. Isolates
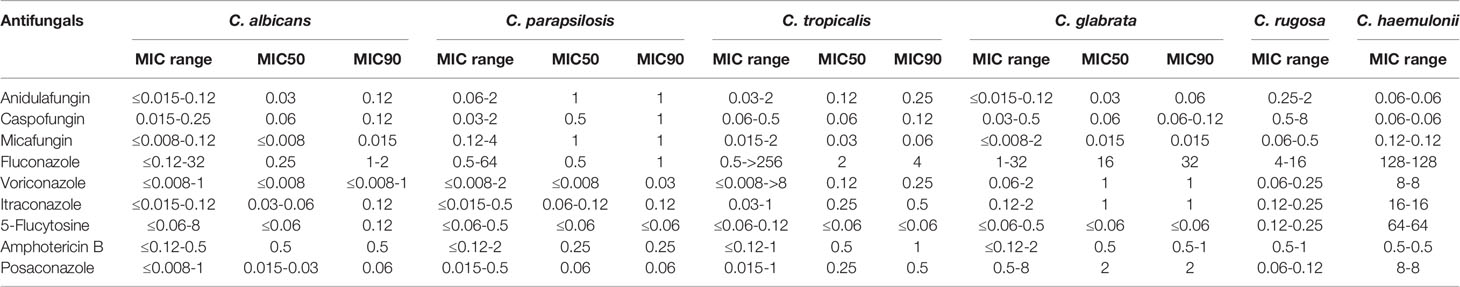
Table 3 shows the susceptibility profiles of the Candida species to the nine antifungal drugs tested. MICs were also determined for two rare yeast species (one C. haemulonii and two C. rugose isolates); however, fewer than five isolates were studied for these two Candida spp. Furthermore, drug-resistant strains of C. haemulonii and C. rugose could not be accurately determined because of a lack of validated clinical breakpoints. Nevertheless, on the basis of CLSI document M27-S4, we found that nine isolates were resistant to azoles. The MIC90 values for fluconazole were 1–2, 1, 4, and 32 μg/mL for C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata, respectively. MIC90 values for voriconazole, itraconazole, and posaconazole were in the range of ≤0.08–1, 0.12–1, and 0.06–2 μg/mL, respectively, for C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata, respectively. In addition, MIC for fluconazole ranged from 4 to 16 μg/mL for C. rugose isolates, whereas that for C. haemulonii was high at 128 μg/mL. MIC90 values of echinocandins (anidulafungin, caspofungin, and micafungin) against C. parapsilosis were higher than those for C. albicans, C. tropicalis, and C. glabrata.
Table 4 shows the overall drug susceptibility pattern of these four Candia species against the five antifungal drug types tested. All C. albicans and C. parapsilosis isolates exhibited 100% of susceptibility to echinocandins, whereas one strain of C. glabrata was resistant (Table 4). Overall resistance was greatest against fluconazole (6.36%), followed by voriconazole (4.55%). As far as species-specific antifungal resistance rates were concerned, C. tropicalis was resistant to almost all drugs tested (except for caspofungin), with the least resistance rate of 4.76%. Isolates of C. tropicalis were susceptible to fluconazole and voriconazole with sensitivity rates of 80.95% and 61.90%, respectively. It is worth noting that the current data were insufficient for distinguishing between drug-resistant and -sensitive strains of C. glabrata based on the CLSI document M27-S4 (Institute, 2012). One (11.11%) of the nine C. glabrata strains was resistant to echinocandins, and all C. glabrata isolates met the criteria for “susceptible-dose dependence (S-DD)” for fluconazole (Table 4).
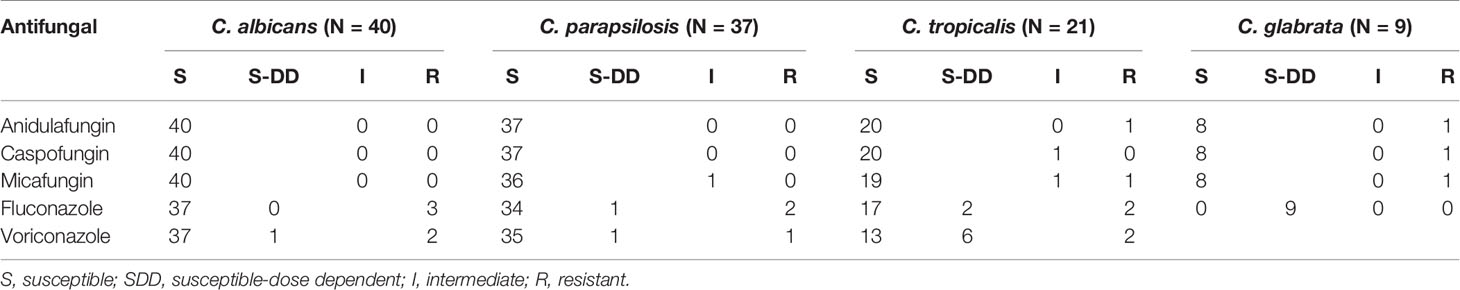
Table 4 Antifungal susceptibility testing results of four main Candida species against five antifungal drugs.
Mutations In ERG11 and the HS Regions of FKS1
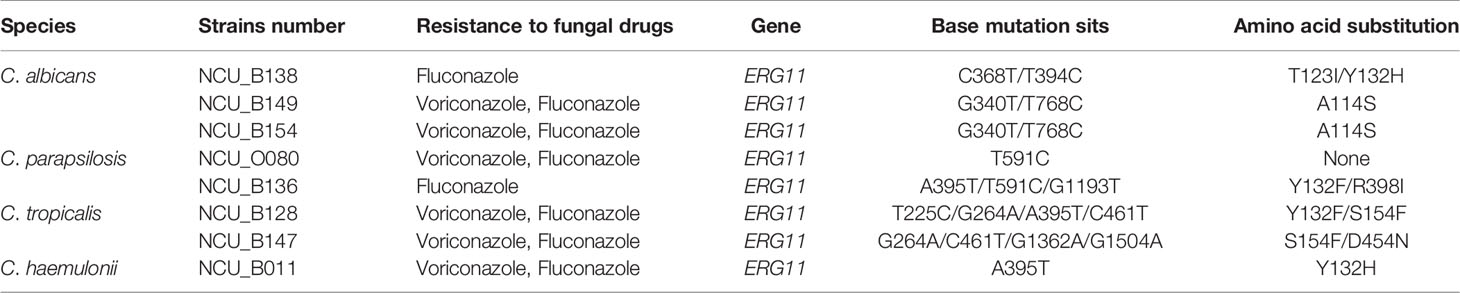
Comparison of the complete ORF of ERG11 of the eight isolates with that of the published wild-type sequences revealed 12 mutations, of which five were silent without any amino acid changes (data not shown). Seven missense mutations were detected in these seven strains. Of the seven distinct amino acid substitutions identified, three (i.e., A114S, Y132H, and Y132F) have been reported previously (Morio et al., 2010). One strains (NCU_O080) only showed a synonymous mutation. Mutations in ERG11 and the resultant amino acid changes are indicated in the Table 5. DNA sequencing of HS1 and HS2 of the drug target FKS1 that is known to confer echinocandin resistance was performed on one C. tropicalis isolate (NCU_O081) and one C. glabrata isolate (NCU_B131). However, no mutation was detected in the hotspot regions of FKS1 (Supplementary Table 2).
Virulence Evaluation of the Invasive Candida spp. Isolates
Several virulence factors are involved in the pathogenesis of candidiasis, and they allow the fungal cells to escape or evade host defense mechanisms. These include phenotypic switching, biofilm formation, and secretion of multiple hydrolases (Neji et al., 2017). We investigated three virulence factors, namely, biofilm, caseinase, and hemolysin.
Caseinase Activity
All 110 isolates were caseinase positive and 109 displayed strong activity. The mean Pz value in caseinase-positive C. albicans and C. parapsilosis isolates was 0.34 ± 0.06 and 0.47 ± 0.10, respectively, indicating a lower enzymatic activity in C. parapsilosis than that in C. albicans (Table 2 and Figure 2). Of the 37 C. parapsilosis isolates, 36 displayed strong enzymatic activity, whereas one had moderate activity. All C. tropicalis (mean Pz = 0.40 ± 0.11) and C. glabrata isolates (mean Pz = 0.32 ± 0.02) were strong caseinase producers. No appreciable difference was noted in the caseinase activity from any of the specimen sources (Figure 3).
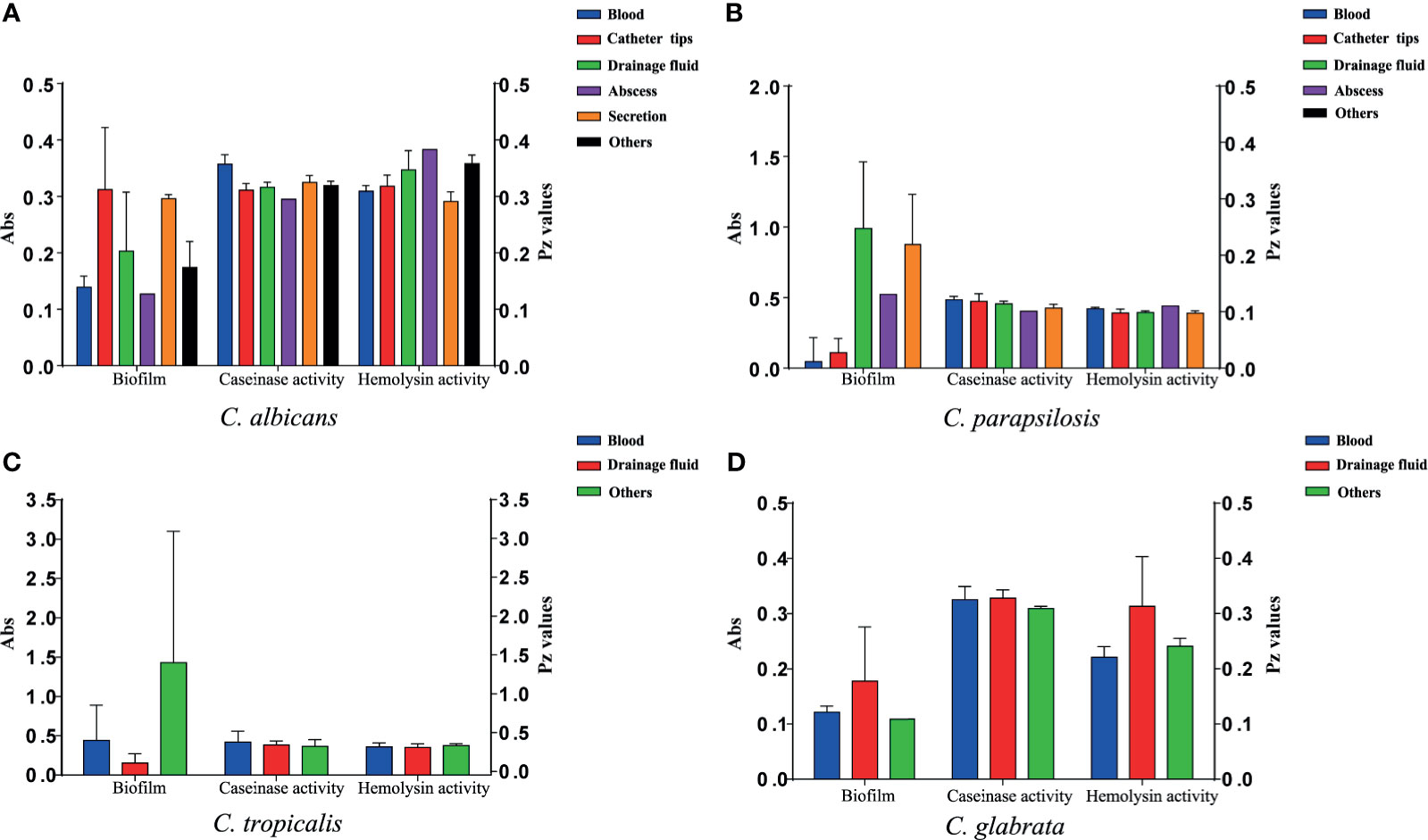
Figure 3 Virulence level (biofilm biomass, caseinase activity, and hemolysin activity) of the 107 Candida species isolates from different specimen sources. Y-axis (far left panel): biofilm biomass (positively correlated with virulence); Y-axis (far right panel): the Pz values of caseinase and hemolysin activities (negatively correlated with virulence). No significant differences were noted in these factors. (A) The virulence level of 40 C. albicans isolates from different specimen sources. (B) The virulence level of 37 C. parapsilosis isolates from different specimen sources. (C) The virulence level of 21 C. tropicalis isolates from different specimen sources. (D) The virulence level of 9 C. glabrata isolates from different specimen sources.
Hemolytic Activity
All of the tested isolates demonstrated strong hemolytic activity on human blood SDA plates with Pz values <0.69 (Figure 2). The mean Pz values for hemolytic activity were 0.32 ± 0.05, 0.41 ± 0.04, 0.36 ± 0.05, 0.25 ± 0.05, and 0.47 ± 0.10 for C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata, respectively (Table 2). No noticeable differences were detected in the hemolytic activity of the strains from different sources (Figure 3).
Biofilm Formation
Figure 2 and Table 2 shows the results of biofilm quantification. Although C. parapsilosis isolates produced greater total biomass (average Abs620 = 0.712) than did C. albicans (average Abs620 = 0.186, p < 0.05) or C. tropicalis isolates (average Abs620 = 0.450; Figure 4), one C. tropicalis isolate (NCU_O081) yielded the greatest biomass (Abs620 = 2.611 ± 0.087), and this was significantly higher than that of C. parapsilosis. By contrast, all C. glabrata strains were low biofilm producers with Abs620 values ranging between 0.095 and 0.247. No significant differences were found in the extent of biofilm formation among all C. glabrata isolates (p > 0.05). Among C. rugose and C. haemulonii, it was impossible to derive any conclusions because of the small sample size. Furthermore, strains isolated from drainage fluid generally produced more biofilm than those isolated from blood, except for C. tropicalis species. C. albicans isolates obtained from catheter tips were the highest biofilm producers, whereas, for C. parapsilosis, these were the isolates obtained from drainage fluids (Figure 3 and Table 2).
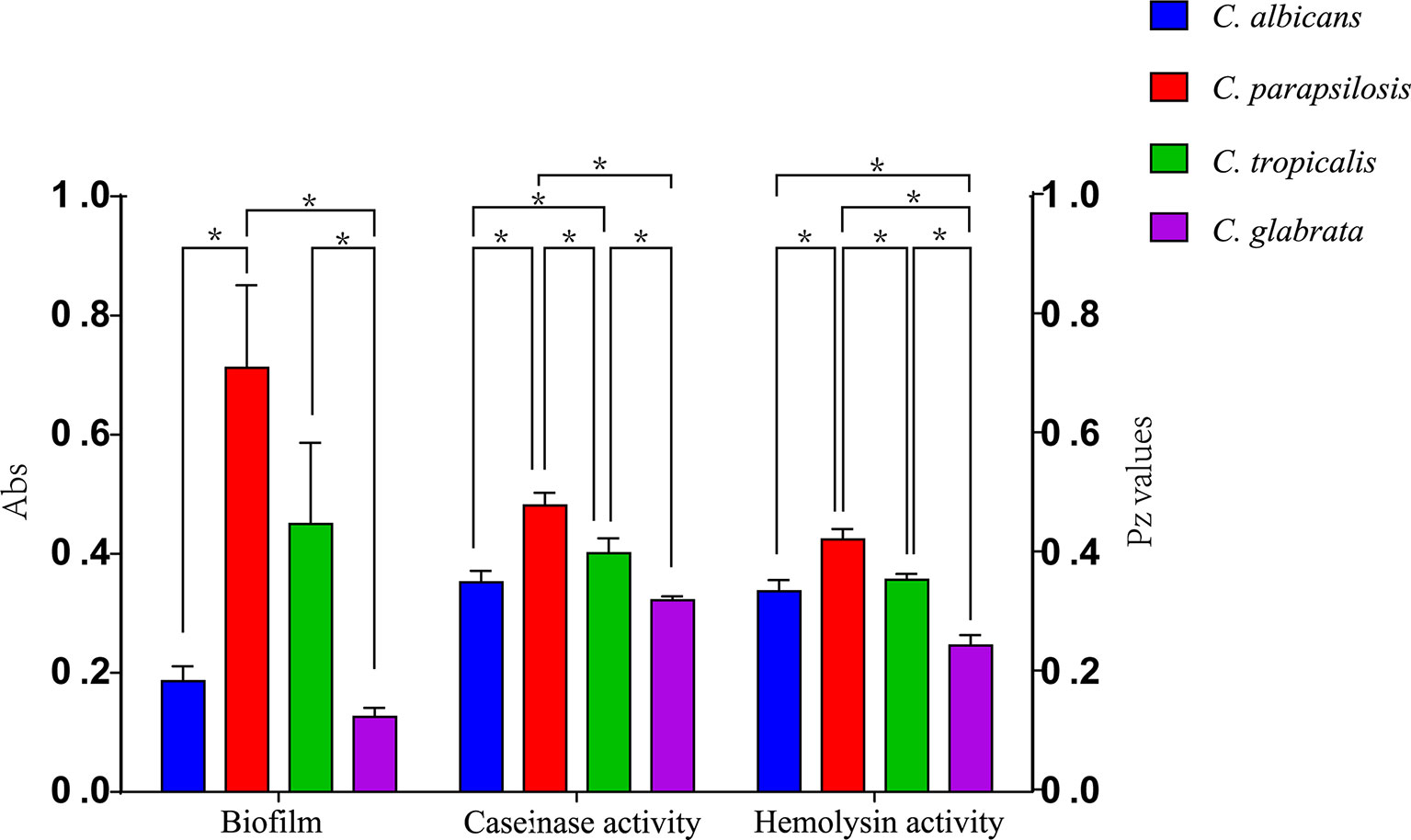
Figure 4 Comparison of the virulence levels (biofilm biomass, casein activity and hemolysin activity) of four Candida species. * indicates statistically significant (p < 0.05).
Discussion
Invasive candidiasis is an emerging infection that is closely related to advances in medical technology, and it is widely recognized as a major cause of morbidity and mortality in high-risk groups such as immunosuppressed patients and those admitted to ICUs. At least 15 different species of Candida can cause human diseases, but a majority of the invasive infections are caused by five pathogens—C. albicans, C. parapsilosis, C. glabrata, C. tropicalis, and C. krusei (Pappas et al., 2018). Although C. albicans is currently the most common pathogen, non-albicans Candida spp. could collectively represent >50% of all bloodstream isolates in some regions (Pappas et al., 2018). A study of 141 clinical Candida specimens in Brazil reported that C. albicans is the most frequently isolated species (45.4%), followed by C. parapsilosis (28.4%), C. tropicalis (14.2%), and C. glabrata (1.4%) (Neufeld et al., 2015). A 6-year retrospective analysis of 351 patients with candidiasis showed that 48.1% of the candidemia episodes were due to C. albicans, followed by C. parapsilosis (25.1%) and C. glabrata (11.7%) (Ulu Kilic et al., 2017). In recent years, studies have described an increase in the incidence of C. parapsilosis infection (Ulu Kilic et al., 2017). We also show that C. albicans is the most prevalent disease-causing Candida spp., followed by C. parapsilosis, C. tropicalis, and C. glabrata, and that most isolates were obtained from patients hospitalized in ICUs.
RAPD analysis is a valuable tool for studying the genetic epidemiology of Candida infections, and multiple studies have confirmed its applicability and high discriminatory power for Candida genotyping at a local level (Paluchowska et al., 2014). Such accurate identification and molecular typing of Candida species provide valuable information for the prevention and control of nosocomial infections caused by these yeasts. Thus, the genetic diversity of 107 Candida species comprising C. albicans (n = 40), C. parapsilosis (n = 37), C. tropicalis (n = 21), and C. glabrata (n = 9), when an Sj value of 0.80 was defined as the limiting point, showed that C. parapsilosis and C. tropicalis isolates were more diverse than C. albicans isolates because of the greater range of Sj values in the dendrogram generated using the five primer pairs. The present study revealed two identical C. albicans isolates (NCU_B153 and NCU_B155) from the blood of two different patients admitted to ICU from different admission years. In addition, two C. glabrata isolates (NCU_B045 and NCU_B055) from different patients sharing the same ward showed similar genotype profiles. Notably, the aforementioned C. glabrata strains were isolated within the same month. These isolates may have come either from the patients, hospital personnel, medical devices, or environment, suggesting that Candida infection can spread exogenously.
Establishing antifungal susceptibility patterns among Candida species isolates from clinical specimens was an important aspect of our study. Fluconazole has remained the drug of choice for treating candidemia over several years now because of its efficacy against clinical Candida infection (Pappas et al., 2004; Autmizguine et al., 2018). However, the current treatment guidelines recommend the use of echinocandins as the first-line empirical treatment because of the increased resistance and treatment failure associated with the use of fluconazole (Bassetti et al., 2018; Zeng et al., 2019). Although we found that most isolates were sensitive to the echinocandins, one C. glabrata isolate was simultaneously resistant to three echinocandins and one C. tropicalis isolate was resistant to both anidulafungin and micafungin. Resistance to fluconazole (6.36%, 7/110) was high among the isolates tested here, and some strains displayed multiazole resistance, i.e., simultaneous resistance to both fluconazole and voriconazole among C. albicans, C. parapsilosis, and C. tropicalis isolates were 5% (2/40), 2.70% (1/37), and 9.52% (2/21), respectively. The appearance of multiazole-resistant strains could be associated with the high rates of azole usage in hospitals (36.2%, 38/105). The azole-resistant rates of patients with azole treatment to C. albicans, C. parapsilosis, C. tropicalis and C. glabrata were 30.0% (3/10), 25.0% (2/8), 25.0% (2/8), and 33.3% (1/3). Among these, 37.5% of the patients received long-course azole treatment for >15 days, which may be a risk factor for azole resistance. Although only two C. rugose strains and one C. haemulonii strain were isolated in our study, their high MICs against antifungal drugs could have been a cause for concern. In our study, the azole-resistance rate of C. albicans (7.5%, 3/10), C. parapsilosis (5.4%, 2/37), and C. tropicalis (9.5%, 2/21) were similar to those reported by Maubon et al. (Maubon et al., 2014). However, the resistance rate of C. albicans (7.5%) was significantly lower than that reported for a hospital in Southwest China (>20.0%) (Zeng et al., 2019). This discrepancy may be induced by the difference in the regional population, medical resources, and distribution of patient types.
DNA sequencing of the two drug target genes, ERG11 and FKS1, was performed using drug-resistant strains. The pharmacological target of azoles is the enzyme 14-α-demethylase (encoded by ERG11), which is an enzyme for ergosterol biosynthesis (Spettel et al., 2019). The mutation leading to amino acid substitution in the ERG11 sequence is one of the main mechanisms contributing to azole resistance in clinical isolates (Morio et al., 2010). As observed in previous studies, all ERG11 mutations in the present study also occurred at three diffuse hotspot regions: amino acids 105–165, 266–287, and 405–488 (Marichal et al., 1999). The most common amino acid substitution in our study was 132aa (50%, 4/8) in the azole-resistant strains. A single Y132H substitution has been shown to reduce susceptibility to voriconazole and fluconazole, with MIC increased by four and eight times, respectively (Wang et al., 2005). The Y132H substitution was detected in one C. albicans isolate (NCU_B138) and one C. haemulonii isolate (NCU_B011). The Y132F substitution was also detected in a C. parapsilosis isolate (NCU_B136) and a C. tropicalis isolate (NCU_B128). A previous study confirmed that the Y132H substitution interfered with the interaction between the heme center of the enzyme and fluconazole, thereby reducing the affinity of the target enzyme to fluconazole, leading to fluconazole resistance (Kelly et al., 1999). Hence, our data support the involvement of Y132F and Y132H in azole resistance and their potential use as predictive markers of azole resistance. FKS1 encode the β-1,3-glucan synthase, which is responsible for the synthesis of fungal cell wall and is the target enzyme of echinocandin (Sfeir et al., 2020). The most common cause of resistance is the serine-to-proline mutation in the HS1 region of FKS1 at the position 654 (Sfeir et al., 2020). No mutation was detected in the HS1 and HS2 regions of FKS1, although some mutations in other regions of this gene may have been missed because of sequencing only the HS1 and HS2 regions. Notably, the finding of amino acid substitutions in these isolates does not indicate that there may not be other coexisting mechanisms of antifungal resistance, such as the overexpression of ERG11, CDR1, and/or MDR1, although these points were not proved in our study.
Several virulence factors in Candida species are indispensable for their ability to cause disease, and these include extracellular hydrolytic enzymes, particularly proteases, phospholipases, and hemolysins, which facilitate food acquisition, adherence, colonization, invasion, diffusion, and escape from host immune responses (Abbes et al., 2017; Figueiredo-Carvalho et al., 2017). Accurate identification of putative virulence factors in Candida spp. is critical for predicting the response of antifungal drugs and detecting the emergence of strains with greater resistance. França et al. have reported that although 68% of patients with candidiasis had been prescribed antifungal therapy with a favorable susceptibility profile, the mortality rate was 56%, implying that other factors are involved in determining patient prognosis (França et al., 2008). Two such factors may be biofilm formation and the expression of virulence-associated factors. Biofilm formation is a major virulence factor (Mba and Nweze, 2020) because, when generated on host tissues and indwelling medical devices such as central venous catheters, they hinder drug diffusion and render infection eradication extremely difficult (Chen et al., 2020). We compared certain virulence factors among Candida species that are detectable in a laboratory environment, such as caseinase, hemolysin, and biofilm production, to assess the pathogenic potential of Candida species.
Proteases can degrade the host epithelial and mucosal barrier proteins such as albumin, collagen, and mucin. They also help Candida resist attacks by host immune system thorough the degradation of antibodies, complements, and cytokines (Borst and Fluit, 2003; Neji et al., 2017). Casein has also been widely reported as a substrate to evaluate the protease activity of pathogenic strains (Fernandes et al., 2012; Neji et al., 2017). Our results show that all the Candida spp. were caseinase producers with a strong enzymatic activity (99.09%). Furthermore, all C. glabrata strains showed a strong enzymatic activity and our results are consistent with findings reported by Abbes et al. who also tested C. glabrata isolates (Abbes et al., 2017). However, these results are in contrast to those described by Figueiredo-Carvalho et al. who reported no caseinase activity in 91 C. glabrata isolates (Figueiredo-Carvalho et al., 2017). Using casein as a substrate, all isolates of C. albicans, C. parapsilosis, C. tropicalis, C. haemulonii, and C. rugose were found to be proteinase positive. Lower caseinase activities were detected for C. parapsilosis than for C. albicans, C. tropicalis, and C. glabrata isolates with a statistically significant difference (Figure 4); these differences were independent of the specimen sources and genotypes (Figure 4; Supplementary Figure 1).
Hemolytic activity is another virulence factor exhibited by pathogenic microorganisms, which enables fungal pathogens to utilize hemoglobin as an iron source for growth in the iron-scarce host environment (Weissman et al., 2021). Previous studies have shown a close relationship between cellular iron and drug susceptibility in C. albicans (Prasad et al., 2006), and iron uptake mechanisms have also been reported to be necessary for the virulence of C. glabrata (Figueiredo-Carvalho et al., 2017). In our study, all Candida spp. showed strong hemolytic activity. Moreover, C. tropicalis exhibited greater hemolysin production than C. glabrata. We noted that the hemolytic activity of C. albicans isolates was higher than that of C. parapsilosis isolates; these findings are inconsistent with those of previous studies (Figure 4) (Chin et al., 2013; Neji et al., 2017).
Several studies have established the importance of biofilm formation in clinical infections due to Candida strains (Nett and Andes, 2006; Akers et al., 2015). Estivill et al. found that biofilm formation was observed on implantable medical devices and all Candida strains tested (including C. albicans, C. parapsilosis, C. tropicalis, C. glabrata, and C. krusei) were able to form biofilms (Estivill et al., 2011). Our results correspond with those reported previously, especially that C. glabrata showed no biofilm production (Marak and Dhanashree, 2018). This is likely, in part, to be related to its intrinsic inability to form hyphae (Seidler et al., 2006). More C. albicans isolates produced biofilms than C. parapsilosis or C. tropicalis isolates, but C. rugose and C. haemulonii showed no biofilm production in our study. We also performed subgroup analyses by specimen source and found that isolates from drainage fluids exhibited greater biofilm formation than those originating from blood, except among C. tropicalis isolates.
The association between clusters and virulence factors was compared (Supplementary Figure 1). No significant difference was noted in the hemolytic and caseinase activities among the strains of different genotypes, although their biofilm production was found to vary with the different clusters. For instance, for C. parapsilosis, three clusters (K–M) showed higher biofilm formation ability than the others (Supplementary Figure 1B). A similar situation was also noted for C. tropicalis, although the difference failed to reach a statistical significance (Supplementary Figure 1C). We also compared the virulence factors of the isolates from different specimen sources, although no obvious correlation was recorded. Interestingly, a correlation was noted between the RAPD genotypes and antifungal resistance. Moreover, 75% of the C. albicans from cluster F demonstrated azole resistance, whereas only two azole-resistant C. tropicalis strains belonged to the cluster Y (Figure 2A). Moreover, 66.7% of the C. tropicalis strains, which were S-DD to azole drugs, were allocated into cluster W (Figure 2C). These findings together suggest that RAPD results allow the selection of drug-resistant isolates with diverse genetic backgrounds, which has significance in clinical medication and applications.
Conclusion
To summarize, this study is able to investigate the molecular relationship and genetic diversity among 110 clinical Candida isolates obtained from hospital inpatients admitted in the Nanchang region of China. Although these data are expected to help understand the epidemiology of Candida species in Nanchang City, our results also contribute toward a greater understanding of the pathogenicity of Candida species in patients with candidiasis and the susceptibility of Candida strains to the most commonly used antifungal drugs; the latter may also help prevent the occurrence and outbreak of Candida infections. Furthermore, our data provide experimental evidence for designing appropriate clinical monitoring and treatment strategies for candidiasis. Because of the rising incidence of Candida infections and their resistance to antifungal drugs, further studies are required to develop treatment strategies against Candida strains.
Data Availability Statement
The sequencing results of ERG11 and FKS1 in the study are deposited in the NCBI BankIt database under accession numbers MZ711431 to MZ711438 and MZ711439 to MZ711442, respectively.
Ethics Statement
The ethics committee of the First Affiliated Hospital of Nanchang University approved this study (approval no. 2014036).
Author Contributions
LZ, NH, QL, XY, and XH designed the study. JC, NH, HX, YZ,YH, and JT performed the experiments and analyses, and wrote the original draft. LZ and XH edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32060040 and 31760261), the Natural Science Foundation of Jiangxi Province (20202BABL216084, 20192ACBL21042, 20202BAB216045, 20192BBG70067, and 20204BCJL23054), the Science and Technology Research Project of Education Department of Jiangxi Province (GJJ180130), and the Major Science and Technology Project of Jiangxi Province (20181BBG70030).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We wish to thank Nanchang University Medical School, for providing the space and equipment necessary to conduct the present study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.721439/full#supplementary-material
Supplementary Figure 1 | Comparison of the virulence levels (biofilm biomass, casein activity, and hemolysin activity) of three Candida species isolates by genotypic relatedness. Y-axis (far left panel): biofilm biomass (positively correlated with virulence); Y-axis (far right panel): the Pz values of caseinase and hemolysin activities (negatively correlated with virulence). No significant differences can be noted in this figure. (A) The virulence level of 40 C. albicans isolates from different gene clusters; (B) The virulence level of 37 C. parapsilosis isolates from different gene clusters; (C) The virulence level of 21 C. tropicalis isolates from different gene clusters.
References
Abbes, S., Amouri, I., Trabelsi, H., Neji, S., Sellami, H., Rahmouni, F., et al. (2017). Analysis of Virulence Factors and In Vivo Biofilm-Forming Capacity of Yarrowia Lipolytica Isolated From Patients With Fungemia. Med. Mycology 55, 193–202. doi: 10.1093/mmy/myw028
Akers, K. S., Cardile, A. P., Wenke, J. C., Murray, C. K. (2015). Biofilm Formation by Clinical Isolates and its Relevance to Clinical Infections. Adv. Exp. Med. Biol. 830, 1–28. doi: 10.1007/978-3-319-11038-7_1
Autmizguine, J., Smith, P. B., Prather, K., Bendel, C., Natarajan, G., Bidegain, M., et al. (2018). Effect of Fluconazole Prophylaxis on Candida Fluconazole Susceptibility in Premature Infants. J. Antimicrobial. Chemother. 73, 3482–3487. doi: 10.1093/jac/dky353
Bassetti, M., Righi, E., Montravers, P., Cornely, O. A. (2018). What has Changed in the Treatment of Invasive Candidiasis? A Look at the Past 10 Years and Ahead. J. Antimicrobial. Chemother. 73, i14–i25. doi: 10.1093/jac/dkx445
Bongomin, F., Gago, S., Oladele, R., Denning, D. (2017). Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. J. Fungi 3, 57. doi: 10.3390/jof3040057
Borst, A., Fluit, A. C. (2003). High Levels of Hydrolytic Enzymes Secreted by Candida Albicans Isolates Involved in Respiratory Infections. J. Med. Microbiol. 52, 971–974. doi: 10.1099/jmm.0.05228-0
Chen, J., Liu, Q., Zeng, L., Huang, X. (2020). Protein Acetylation/Deacetylation: A Potential Strategy for Fungal Infection Control. Front. Microbiol. 11, 574736. doi: 10.3389/fmicb.2020.574736
Chin, V. K., Foong, K. J., Maha, A., Rusliza, B., Norhafizah, M., Ng, K. P., et al. (2013). Candida Albicans Isolates From a Malaysian Hospital Exhibit More Potent Phospholipase and Haemolysin Activities Than Non-Albicans Candida Isolates. Trop. Biomed 30, 654–662.
Clinical and Laboratory Standards Institut (2008). Reference method for broth dilution antifungal susceptibility testing of yeasts; third edition, M27-A3. Wayne, PA, USA.
Clinical and Laboratory Standards Institute (2012). Reference method for broth dilution antifungal susceptibility testing of yeasts; fourth informational supplement, M27–S4. Wayne, PA, USA.
De Rosa, F. G., Garazzino, S., Pasero, D., Di Perri, G., Ranieri, V. M. (2009). Invasive Candidiasis and Candidemia: New Guidelines. Minerva Anestesiologica 75, 453–458.
Desnos-Ollivier, M., Bretagne, S., Raoux, D., Hoinard, D., Dromer, F., Dannaoui, E. (2008). Mutations in the Fks1 Gene in Candida Albicans, C. Tropicalis and C. Krusei Correlate With Elevated Caspofungin MICs Uncovered in AM3 Medium Using the Method of the European Committee on Antibiotic Susceptibility Testing. Antimicrobial. Agents Chemother. 52, 3092–3098. doi: 10.1128/AAC.00088-08
Estivill, D., Arias, A., Torres-Lana, A., Carrillo-Muñoz, A. J., Arévalo, M. P. (2011). Biofilm Formation by Five Species of Candida on Three Clinical Materials. J. Microbiol. Methods 86, 238–242. doi: 10.1016/j.mimet.2011.05.019
Fernandes, E. G., Valério, H. M., Feltrin, T., van der Sand, S. T. (2012). Variability in the Production of Extracellular Enzymes by Entomopathogenic Fungi Grown on Different Substrates. Braz. J. Microbiol. [publication Braz. Soc. Microbiology] 43, 827–833. doi: 10.1590/S1517-83822012000200049
Figueiredo-Carvalho, M. H. G., Ramos, L. S., Barbedo, L. S., de Oliveira, J. C. A., Dos Santos, A. L. S., Almeida-Paes, R., et al. (2017). Relationship Between the Antifungal Susceptibility Profile and the Production of Virulence-Related Hydrolytic Enzymes in Brazilian Clinical Strains of Candida Glabrata. Mediators Inflamm. 2017, 8952878. doi: 10.1155/2017/8952878
França, J. C., Ribeiro, C. E., Queiroz-Telles, F. (2008). [Candidemia in a Brazilian Tertiary Care Hospital: Incidence, Frequency of Different Species, Risk Factors and Antifungal Susceptibility]. Rev. da Sociedade Bras. Medicina Trop. 41, 23–28. doi: 10.1590/s0037-86822008000100005
Jain, P., Khan, Z. K., Bhattacharya, E., Ranade, S. A. (2001). Variation in Random Amplified Polymorphic DNA (RAPD) Profiles Specific to Fluconazole-Resistant and -Sensitive Strains of Candida Albicans. Diagn. Microbiol. Infect. Dis. 41, 113–119. doi: 10.1016/s0732-8893(01)00292-9
Kelly, S. L., Lamb, D. C., Kelly, D. E. (1999). Y132H Substitution in Candida Albicans Sterol 14alpha-Demethylase Confers Fluconazole Resistance by Preventing Binding to Haem. FEMS Microbiol. Lett. 180, 171–175. doi: 10.1111/j.1574-6968.1999.tb08792.x
Koehler, P., Stecher, M., Cornely, O. A., Koehler, D., Vehreschild, M. J. G. T., Bohlius, J., et al. (2019). Morbidity and Mortality of Candidaemia in Europe: An Epidemiologic Meta-Analysis. Clin. Microbiol. Infect. 25, 1200–1212. doi: 10.1016/j.cmi.2019.04.024
Li, Y., Du, M., Chen, L. A., Liu, Y., Liang, Z. (2016). Nosocomial Bloodstream Infection Due to Candida Spp. In China: Species Distribution, Clinical Features, and Outcomes. Mycopathologia 181, 485–495. doi: 10.1007/s11046-016-9997-3
Mancera, E., Porman, A. M., Cuomo, C. A., Bennett, R. J., Johnson, A. D. (2015). Finding a Missing Gene: EFG1 Regulates Morphogenesis in Candida Tropicalis. G3 (Bethesda Md.) 5, 849–856. doi: 10.1534/g3.115.017566
Marak, M. B., Dhanashree, B. (2018). Antifungal Susceptibility and Biofilm Production of Candida Spp. Isolated From Clinical Samples. Int. J. Microbiol. 2018, 7495218. doi: 10.1155/2018/7495218
Marichal, P., Koymans, L., Willemsens, S., Bellens, D., Verhasselt, P., Luyten, W., et al. (1999). Contribution of Mutations in the Cytochrome P450 14alpha-Demethylase (Erg11p, Cyp51p) to Azole Resistance in Candida Albicans. Microbiol. (Reading) 145 (Pt 10), 2701–2713. doi: 10.1099/00221287-145-10-2701
Maubon, D., Garnaud, C., Calandra, T., Sanglard, D., Cornet, M. (2014). Resistance of Candida Spp. To Antifungal Drugs in the ICU: Where Are We Now? Intensive Care Med. 40, 1241–1255. doi: 10.1007/s00134-014-3404-7
Mba, I. E., Nweze, E. I. (2020). Mechanism of Candida Pathogenesis: Revisiting the Vital Drivers. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 39, 1797–1819. doi: 10.1007/s10096-020-03912-w
Morio, F., Loge, C., Besse, B., Hennequin, C., Le Pape, P. (2010). Screening for Amino Acid Substitutions in the Candida Albicans Erg11 Protein of Azole-Susceptible and Azole-Resistant Clinical Isolates: New Substitutions and a Review of the Literature. Diagn. Microbiol. Infect. Dis. 66, 373–384. doi: 10.1016/j.diagmicrobio.2009.11.006
Neji, S., Hadrich, I., Trabelsi, H., Abbes, S., Cheikhrouhou, F., Sellami, H., et al. (2017). Virulence Factors, Antifungal Susceptibility and Molecular Mechanisms of Azole Resistance Among Candida Parapsilosis Complex Isolates Recovered From Clinical Specimens. J. Biomed. Sci. 24, 67. doi: 10.1186/s12929-017-0376-2
Nett, J., Andes, D. (2006). Candida Albicans Biofilm Development, Modeling a Host-Pathogen Interaction. Curr. Opin. Microbiol. 9, 340–345. doi: 10.1016/j.mib.2006.06.007
Neufeld, P. M., Melhem Mde, S., Szeszs, M. W., Ribeiro, M. D., Amorim Ede, L., da Silva, M., et al. (2015). Nosocomial Candidiasis in Rio De Janeiro State: Distribution and Fluconazole Susceptibility Profile. Braz. J. Microbiol. [publication Braz. Soc. Microbiology] 46, 477–484. doi: 10.1590/s1517-838246220120023
Paluchowska, P., Tokarczyk, M., Bogusz, B., Skiba, I., Budak, A. (2014). Molecular Epidemiology of Candida Albicans and Candida Glabrata Strains Isolated From Intensive Care Unit Patients in Poland. Memorias do Instituto Oswaldo Cruz 109, 436–441. doi: 10.1590/0074-0276140099
Pappas, P. G., Lionakis, M. S., Arendrup, M. C., Ostrosky-Zeichner, L., Kullberg, B. J. (2018). Invasive Candidiasis. Nat. Rev. Dis. Primers 4, 18026. doi: 10.1038/nrdp.2018.26
Pappas, P. G., Rex, J. H., Sobel, J. D., Filler, S. G., Dismukes, W. E., Walsh, T. J., et al. (2004). Guidelines for Treatment of Candidiasis. Clin. Infect. Dis. 38, 161–189. doi: 10.1086/380796
Prasad, T., Chandra, A., Mukhopadhyay, C. K., Prasad, R. (2006). Unexpected Link Between Iron and Drug Resistance of Candida Spp.: Iron Depletion Enhances Membrane Fluidity and Drug Diffusion, Leading to Drug-Susceptible Cells. Antimicrobial Agents Chemother. 50, 3597–3606. doi: 10.1128/aac.00653-06
Price, M. F., Wilkinson, I. D., Gentry, L. O. (1982). Plate Method for Detection of Phospholipase Activity in Candida Albicans. Sabouraudia 20, 7–14. doi: 10.1080/00362178285380031
Ramesh, N., Priyadharsini, M., Sumathi, C. S., Balasubramanian, V., Hemapriya, J., Kannan, R. (2011). Virulence Factors and Anti Fungal Sensitivity Pattern of Candida Sp. Isolated From HIV and TB Patients. Indian J. Microbiol. 51, 273–278. doi: 10.1007/s12088-011-0177-3
Salehi, F., Esmaeili, M., Mohammadi, R. (2017). Isolation of Candida Species From Gastroesophageal Lesions Among Pediatrics in Isfahan, Iran: Identification and Antifungal Susceptibility Testing of Clinical Isolates by E-Test. Adv. BioMed. Res. 6, 103. doi: 10.4103/2277-9175.213662
Seidler, M., Salvenmoser, S., Müller, F. M. (2006). In Vitro Effects of Micafungin Against Candida Biofilms on Polystyrene and Central Venous Catheter Sections. Int. J. Antimicrobial. Agents 28, 568–573. doi: 10.1016/j.ijantimicag.2006.07.024
Sfeir, M. M., Jiménez-Ortigosa, C., Gamaletsou, M. N., Schuetz, A. N., Soave, R., Van Besien, K., et al. (2020). Breakthrough Bloodstream Infections Caused by Echinocandin-Resistant: An Emerging Threat to Immunocompromised Patients With Hematological Malignancies. J. Fungi (Basel) 6, 20. doi: 10.3390/jof6010020
Silva, S., Henriques, M., Martins, A., Oliveira, R., Williams, D., Azeredo, J. (2009). Biofilms of non-Candida Albicans Candida Species: Quantification, Structure and Matrix Composition. Med. Mycology 47, 681–689. doi: 10.3109/13693780802549594
Spettel, K., Barousch, W., Makristathis, A., Zeller, I., Nehr, M., Selitsch, B., et al. (2019). Analysis of Antifungal Resistance Genes in Candida Albicans and Candida Glabrata Using Next Generation Sequencing. PloS One 14, e0210397. doi: 10.1371/journal.pone.0210397
Ulu Kilic, A., Alp, E., Cevahir, F., Ture, Z., Yozgat, N. (2017). Epidemiology and Cost Implications of Candidemia, a 6-Year Analysis From a Developing Country. Mycoses 60, 198–203. doi: 10.1111/myc.12582
Vatanshenassan, M., Arastehfar, A., Boekhout, T., Berman, J., Lass-Flörl, C., Sparbier, K., et al. (2019). Anidulafungin Susceptibility Testing of Candida Glabrata Isolates From Blood Cultures by the MALDI Biotyper Antibiotic (Antifungal) Susceptibility Test Rapid Assay. Antimicrobial. Agents Chemother. 63(9). doi: 10.1128/AAC.00554-19
Wang, Y.-b., Wang, H., Guo, H.-y., Zhao, Y.-z., Luo, S.-q. (2005). Analysis of ERG11 Gene Mutation in Candida Albicans. Di Yi Jun Yi Da Xue Xue Bao 25, 1390–1393.
Weissman, Z., Pinsky, M., Donegan, R. K., Reddi, A. R., Kornitzer, D. (2021). Using Genetically Encoded Heme Sensors to Probe the Mechanisms of Heme Uptake and Homeostasis in Candida Albicans. Cell. Microbiol. 23, e13282. doi: 10.1111/cmi.13282
Wisplinghoff, H., Bischoff, T., Tallent, S. M., Seifert, H., Wenzel, R. P., Edmond, M. B. (2004). Nosocomial Bloodstream Infections in US Hospitals: Analysis of 24,179 Cases From a Prospective Nationwide Surveillance Study. Clin. Infect. Dis. 39, 309–317. doi: 10.1086/421946
Zeng, Z.-R., Tian, G., Ding, Y.-H., Yang, K., Liu, J.-B., Deng, J. (2019). Surveillance Study of the Prevalence, Species Distribution, Antifungal Susceptibility, Risk Factors and Mortality of Invasive Candidiasis in a Tertiary Teaching Hospital in Southwest China. BMC Infect. Dis. 19, 939. doi: 10.1186/s12879-019-4588-9
Keywords: Candida species, RAPD, caseinase, hemolysin, biofilm, antifungal susceptibility
Citation: Chen J, Hu N, Xu H, Liu Q, Yu X, Zhang Y, Huang Y, Tan J, Huang X and Zeng L (2021) Molecular Epidemiology, Antifungal Susceptibility, and Virulence Evaluation of Candida Isolates Causing Invasive Infection in a Tertiary Care Teaching Hospital. Front. Cell. Infect. Microbiol. 11:721439. doi: 10.3389/fcimb.2021.721439
Received: 07 June 2021; Accepted: 09 August 2021;
Published: 15 September 2021.
Edited by:
Min Chen, Shanghai Changzheng Hospital, ChinaReviewed by:
Mohammad Javad Najafzadeh, Mashhad University of Medical Sciences, IranFarnaz Daneshnia, Hackensack University Medical Center, United States
Copyright © 2021 Chen, Hu, Xu, Liu, Yu, Zhang, Huang, Tan, Huang and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaotian Huang, eHRodWFuZ0BuY3UuZWR1LmNu; Lingbing Zeng, bGluZ2JpbmdfemVuZ0AxNjMuY29t
†These authors have contributed equally to this work
 Junzhu Chen
Junzhu Chen Niya Hu
Niya Hu Hongzhi Xu
Hongzhi Xu Qiong Liu
Qiong Liu Xiaomin Yu
Xiaomin Yu Yuping Zhang
Yuping Zhang Yongcheng Huang
Yongcheng Huang Junjun Tan2
Junjun Tan2 Xiaotian Huang
Xiaotian Huang Lingbing Zeng
Lingbing Zeng