
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 28 July 2021
Sec. Molecular Bacterial Pathogenesis
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.701625
Resistance to colistin, especially mobilized colistin resistance (mcr), is a serious threat to public health since it may catalyze a return of the “pre-antibiotic era”. Outer membrane vesicles (OMVs) play a role in antibiotic resistance in various ways. Currently, how OMVs participate in mcr-1-mediated colistin resistance has not been established. In this study, we showed that both OMVs from the mcr-1 negative and positive Escherichia coli (E. coli) strains conferred dose-dependent protection from colistin. However, OMVs from the mcr-1 positive strain conferred attenuated protection when compared to the OMVs of a mcr-1 negative strain at the same concentration. The attenuated protective effect of OMVs was related to the reduced ability to absorb colistin from the environment, thus promoting the killing of colistin sensitive E. coli strains. Lipid A modified with phosphoethanolamine was presented in the OMVs of the mcr-1 positive E. coli strain and resulted in decreased affinity to colistin and less protection. Meanwhile, E. coli strain carrying the mcr-1 gene packed more unmodified lipid A in OMVs and kept more phosphoethanolamine modified lipid A in the bacterial cells. Our study provides a first glimpse of the role of OMVs in mcr-1 -mediated colistin resistance.
Infections caused by multidrug-resistant (MDR) Gram-negative bacteria have led to the rebirth of the “last-line” antibiotic colistin. However, a gradual increase in colistin resistance has been reported in the past few years (Cai et al., 2012; Ah et al., 2014; Monaco et al., 2014; Bialvaei and Samadi Kafil, 2015; El-Sayed Ahmed et al., 2020), which is becoming a great challenge for clinical practice. Thus, understanding the mechanism of resistance to colistin and developing new therapeutic strategies against colistin-resistant superbugs are of great significance.
Colistin belongs to the amphipathic ‘detergent-like’ polypeptide antibiotics. The bactericidal action of colistin involves initial electrostatic binding to lipopolysaccharide (LPS) in the outer membrane of Gram-negative bacteria, with the resultant displacement of Ca2+ and Mg2+, followed by the insertion of the fatty acyl chain within the outer membrane leading to the leakage of intracellular contents (Falagas et al., 2005; Biswas et al., 2012; Cai et al., 2015). The most identified mechanism of resistance to colistin in Gram-negative bacteria involves modifications to the LPS which consists of the O antigen, the core oligosaccharide and the lipid A moiety (Nikaido, 2003). In E. coli, modification of lipid A with 4-amino-4-deoxy-L-arabinose, phosphoethanolamine (pEtN) and/or galactosamine results in a reduced negative charge of LPS and reduced interaction between colistin and the LPS (Breazeale et al., 2005; Chen and Groisman, 2013; Moffatt et al., 2019). While the genes involved in most of these modifications are chromosomally encoded, the first mobilized colistin resistance gene (mcr-1) conferring plasmid-mediated resistance was reported in 2016 and poses a great threat in colistin resistance dissemination globally (Liu et al., 2016; Ling et al., 2020). So far, 10 mcr genes (mcr-1–mcr-10) have been identified in Enterobacteriaceae (Wang et al., 2020). The MCR-1 enzyme is attached to the inner-membrane of Gram-negative bacteria (Gao et al., 2016) and belongs to the phosphoethanolamine (pEtN) transferase enzyme family. In E. coli, expression of the mcr-1 gene results in the addition of phosphoethanolamine to lipid A and resistance to colistin (Ling et al., 2020).
Outer membrane vesicles (OMVs) are spherical structures (20–300 nm) derived from the cell envelope of Gram-negative bacteria. OMVs have been found to mediate a variety of functions, including delivery of virulence factors (Bonnington and Kuehn, 2014), secretion of misfolded proteins to relieve cell envelope stress (McBroom and Kuehn, 2007), and resistance to antimicrobials (Chattopadhyay and Jagannadham, 2015). OMVs can help bacteria to fight against antimicrobials either by horizontally transferring antibiotic-resistant genes or directly absorbing the antibiotic from the environment (Schwechheimer and Kuehn, 2015). Previous research has shown that the addition of OMVs provided immediate resistance to colistin in E. coli and Pseudomonas syringae (Manning and Kuehn, 2011; Kulkarni et al., 2014; Kulkarni et al., 2015). However, the underlying molecular mechanisms have not been fully elucidated. More importantly, little is known about the role of OMVs in MCR-mediated colistin resistance. Therefore, this study set out to assess the role of OMVs in colistin resistance mediated by mcr-1 and to investigate the underlying molecular mechanisms.
In this work, OMVs isolated from E. coli strains carrying the mcr-1 gene, as well as control strains without the mcr-1 gene, were investigated for their ability to protect against colistin. The ability of OMVs to bind colistin was evaluated. The lipid A contents were characterized and analyzed to investigate the mechanisms of protection against colistin. Our data, for the first time, showed how OMVs were involved in mcr-1 related colistin resistance.
E. coli 08-85 was isolated from the blood of a patient and carried the mcr-1 gene on the plasmid (Liang et al., 2021). This clinical pathogenic strain and E. coli ATCC25922 selected as an mcr-1 negative control strain, were used in the study. In our previous work, E. coli K12 was constructed with mcr-1 which is transcriptionally fused to a constitutive promoter on the chromosome. This isogenic strain (E. coli 50434), as well as E. coli K12, was also used in the study. The transcriptional expression level of mcr-1 of E. coli 50434 was 102.6 ± 66.05 (compared with housekeeping genes cysG and hcaT) and resistant to colistin with the minimal inhibitory concentration (MIC) of 4 μg/ml (Qiao, 2019), while the MIC of colistin against E. coli K12 was 1 μg/ml. E. coli ATCC25922 and E. coli K12 were purchased from the American Type Culture Collection (ATCC, VA, USA), and all bacterial strains used in this study were stored in the Collection Center of Pathogen Microorganism of Chinese Academy of Medical Sciences (CAMS-CCPM-A, China). Strains used in this study are listed in Table 1.
All strains were cultivated in LB broth (Difco, NJ, USA) or grown on an LB agar plate. The log-phase culture was obtained by starting with a 1:1,000 dilution of overnight culture and growing to OD600nm of 0.8 with shaking at 220 rpm, 37°C.
OMVs were isolated from the log-phase culture of E. coli 25922, E. coli 08-85, E. coli K12, and E. coli 50434 according to the procedure published previously (Chutkan et al., 2013; Klimentová and Stulík, 2015). Briefly, 500 ml of LB broth was inoculated with 0.5 ml of overnight culture and incubated at 37°C with shaking at 220 rpm with or without colistin (2 μg/ml) until OD600nm reached 0.8. Cells were pelleted by centrifugation (4,700 rpm for 30 min), and the supernatant was filtered through a 0.22-μm membrane (Millipore, MA, USA). The filtrates were concentrated to 10 ml using an Amicon® stirred cell (Millipore, MA, USA) with the membrane of 100 kDa cutoff. The concentrated filtrates were ultracentrifuged (125,000 × g) for 1 h at 4°C. The pellets were resuspended in phosphate buffered saline (PBS).
The OMV preparations were checked for the presence of bacterial cells by plating the vesicle suspension on LB agar. The total protein concentration in the purified OMV preparations was examined by Bradford Coomassie assay (Thermo Fisher, MA, USA), and the OMV concentrations used in subsequent assays refer to this protein-based value.
The OMVs were resuspended in PBS and negatively stained with 2% phosphotungstic acid solution (pH 7.3). The OMVs were then fixed with 2.5% glutaraldehyde overnight at 4°C. Samples were washed, dehydrated, and placed on copper grids for examination by transmission electron microscope (JEOL, Japan).
The OMV samples were serially diluted to make the final concentration within the measurement range in particle-free PBS (filtered through a 0.1-μm membrane). The measurement of OMV diameter was performed using ZetaView (Particle Metrix, Germany) following the instructions of the manufacturer.
For the protection of growth, an overnight culture of E. coli ATCC25922 or E. coli K12 was diluted 1:1,000 with LB broth and treated with colistin (2 μg/ml) alone or simultaneously with OMVs isolated from E. coli ATCC25922, E. coli 08-85, E. coli K12, or E. coli 50434 at the indicated concentrations (20, 2, or 0.2 μg/ml). A 350-μl aliquot was added to each well of a honeycomb plate (Oy Growth Curves Ab Ltd, Finland) in triplicate. The turbidity was read every 15 min using a Bioscreen C analyzer (Oy Growth Curves Ab Ltd, Finland). The mean optical density at each time point was calculated. The experiment was repeated independently twice.
For protection in the killing assay, 1 ml of log-phase culture (OD600nm = 0.8) of E. coli ATCC25922 or E. coli K12 was treated with 2 μg/ml colistin alone, or simultaneously with colistin and OMVs at the same concentration used in the Bioscreen assay. E. coli ATCC25922 or E. coli K12 alone was used as a positive control. Cultures were incubated at 37°C for 6 h and plated on LB agar to determine the colony-forming units (CFUs).
To determine if OMVs were able to bind to BC and quench its fluorescence, the fluorescence titration of OMVs was performed based on a previous assay (Wood et al., 2004). BC was diluted in 50 mM Tris-HCl (pH 7.4) to make a final concentration of 4 μM. The BC-OMV solutions were prepared by the addition of aliquots of stock solutions of OMVs into BC solutions, of which the final concentration of OMVs ranged from 0.1 to 4 μg/ml. A 100 μl of BC-OMV solution was added into a non-binding surface (NBS) black 96 well microplate (Corning, NY, USA) in triplicate. The fluorescence intensity was read with excitation at 580 nm and emission at 620 nm (Enspire 2300, PerkinElmer, MA, USA).
The displacement of OMV-bound BC by colistin was performed as previously described (French et al., 2019) with slight modifications. Briefly, 100 μl of 50 mM Tris-HCl (pH 7.4) buffer containing 4 μM BC, 2 μg/ml OMVs and serially diluted colistin (0.02 to 200 μg/ml) was added into NBS black 96 well microplate (Corning), and the fluorescence intensity was read.
Isolation of lipid A was performed according to the protocol of Henderson et al. (2013). The OMV pellets and bacterial cells were resuspended in 1.6 ml of PBS, and 2 ml of chloroform and 4 ml of methanol were added for a single-phase Bligh-Dyer (chloroform: methanol: water; 1:2:0.8 v/v). The samples were lysed for 20 min at room temperature, and pellets were collected (2,000 × g for 20 min) and washed with single phase Bligh-Dyer. The pellets were resuspended in 3.6 ml of mild acid hydrolysis buffer (50 mM sodium acetate, pH 4.5; 1% SDS), assisted by sonication at a constant duty cycle for 5 s at 25% output, and incubated at 95°C for 1 h. Lipid A was then extracted by adding 4 ml of chloroform and 4 ml of methanol for a two-phase Bligh-Dyer (chloroform:methanol:water, 2:2:1.8 v/v). The lower phase was collected, washed twice with neutral upper phase, and dried in a rotational vacuum centrifuge.
An U3000 system (ThermoFisher, MA, USA) coupled to a Triple TOF 5600 mass spectrometry (AB sciex, WA, USA) was employed in the study. The extracted lipid A was eluted using a gradient that consists of buffer A (MeOH/ACN/H2O, 1:1:1, 5 mM NH4OAC) and buffer B (MeOH/IPA, 1:4, 5 mM NH4OAC), which started from 10% buffer B for 1 min, 1–6 min to 60% buffer B, 6–18 min to 100% buffer B, then kept at 100% buffer B for 2 min, followed by equilibration with 10% B for 5 min at a flow rate of 0.5 ml/min. ZORBAX RRHD 300Å SB C8 (4.6 × 150 mm, 5 µm, Agilent, CA, USA) was maintained at 40°C. The extracted lipids were dissolved in dichloromethane-methanol (2:1, vol/vol) and infused into the ion source at 5 to 10 μl/min. Lipid A was analyzed in negative mode at 4,500 V with a mass range from 50 to 2,400 m/z. The relative quantitation of lipid A species was determined by calculating the peak area with MultiQuant 3.0. The percent composition was calculated as the peak area of each lipid A species divided by the sum of all lipid A species.
To quantitate OMV yield, log-phase culture (OD600nm = 0.8) of E. coli K12 and E. coli 50434 growing with or without colistin (2 μg/ml) was harvested for OMV isolation, and a portion of the culture was aliquoted and plated on LB agar to determine CFUs. The lipid content in OMV preparations was measured with the lipophilic dye FM4-64 (Molecular Probes, OR, USA) as described previously (McBroom et al., 2006). In brief, OMVs were incubated with FM4-64 (3.3 μg/ml) for 10 min at 37°C. The negative controls were set as OMVs alone and FM4-64 alone. The fluorescence intensity was measured using a PerkinElmer Enspire 2300 with excitation and emission at 506 nm and 750 nm respectively.
Statistical analysis was performed with GraphPad Prism 8 software. P-values were calculated by t-test, one-way ANOVA or two-way ANOVA followed by Tukey’s multiple comparisons test to analyze the differences between each pair of groups. P-values of <0.05 were considered significant. Experiments were performed in triplicate and repeated at least two times.
Our previous work identified a pathogenic E. coli strain (E. coli 08-85) carrying a mcr-1 gene (Liang et al., 2021). E. coli ATCC25922 was selected as a control strain that is colistin sensitive and mcr-1 negative. E. coli K12 and the isogenic E. coli 50434 (E. coli K12 with the mcr-1 gene) were also used throughout this study. OMVs of these strains were purified. Transmission electron microscopy analysis revealed that these E. coli strains released OMVs at the late log-phase of growth. The vesicles were spherical and ranged in diameter from 50 to 200 nm (Figures 1A, B). The nanoparticle tracking analysis (NTA) of the OMVs determined that the median diameters were 134.2 and 124.9 nm for OMVs isolated from E. coli 08-85 and E. coli K12 respectively (Figures 1C, D). The size distribution of the vesicles was in general agreement by both methods.
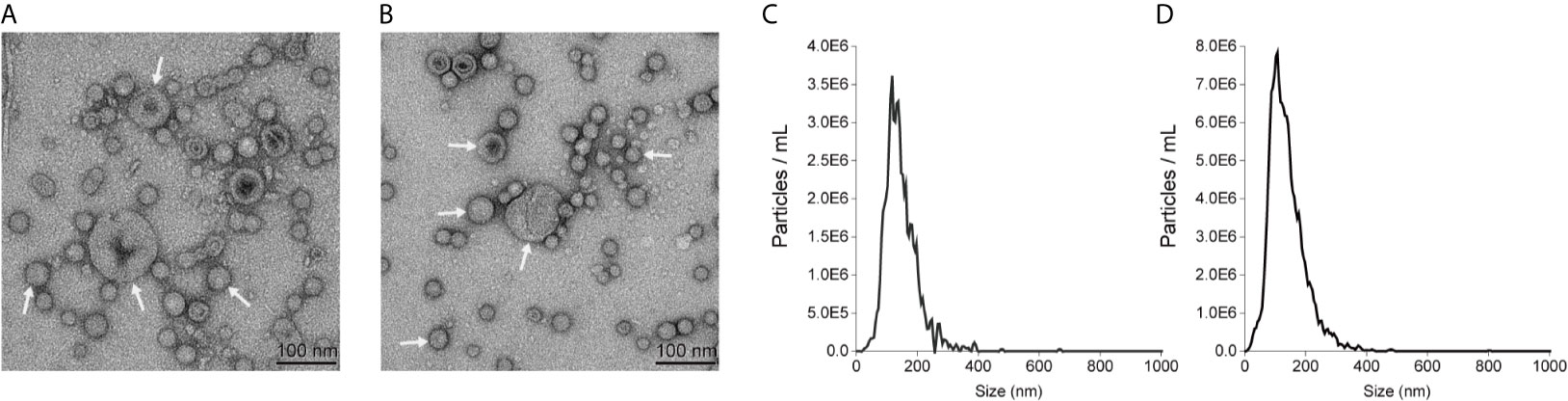
Figure 1 Representatives of transmission electron microscopy images of E coli 08-85 OMVs (A) and E coli K12 OMVs (B) recorded with negative staining with 2% phosphotungstic acid. The size distribution of OMVs of E. coli 08-85 (C) and E coli K12 OMVs (D) observed by nanoparticle tracking analysis. The OMVs were free of contaminating bacteria and the diameter ranged from 40 to 200 nm.
OMVs from clinical E. coli ATCC25922 and E. coli 08-85, as well as E. coli K12 and E. coli 50434, were purified, and the effect of OMVs on the bacterial growth and protection of killing from colistin was studied. The MICs of colistin were 1 μg/ml against E. coli ATCC25922 and E. coli K12, while the minimal bactericidal concentrations (MBCs) of colistin were 2 μg/ml against E. coli ATCC25922 and E. coli K12 (Table 1).
E. coli ATCC25922 or E. coli K12, incubated with serial concentrations of OMVs ranging from 0.2 to 20 μg/ml, was treated with 2× MIC colistin (2 μg/ml) to ensure that the bacteria did not grow at this concentration. As shown in Figures 2A, B, the bacterial cells of E. coli ATCC25922 or E. coli K12 in the absence of OMVs did not grow at all. OMVs protected E. coli ATCC25922 from growth inhibitory in a concentration-dependent manner, with the bacterial cells growing fastest with 20 μg/ml OMVs and slowest or no growth with 0.2 μg/ml OMVs (Figure 2A). Notably, OMVs from the mcr-1 positive strain E. coli 08-85 conferred less protection than OMVs isolated from E. coli ATCC25922 at the same concentration (Figure 2A). In the same way, E. coli K12 was protected by 20 μg/ml E. coli K12 OMVs, while the growth was totally inhibited within the first 12 h with 20 μg/ml E. coli 50434 OMVs (Figure 2B).
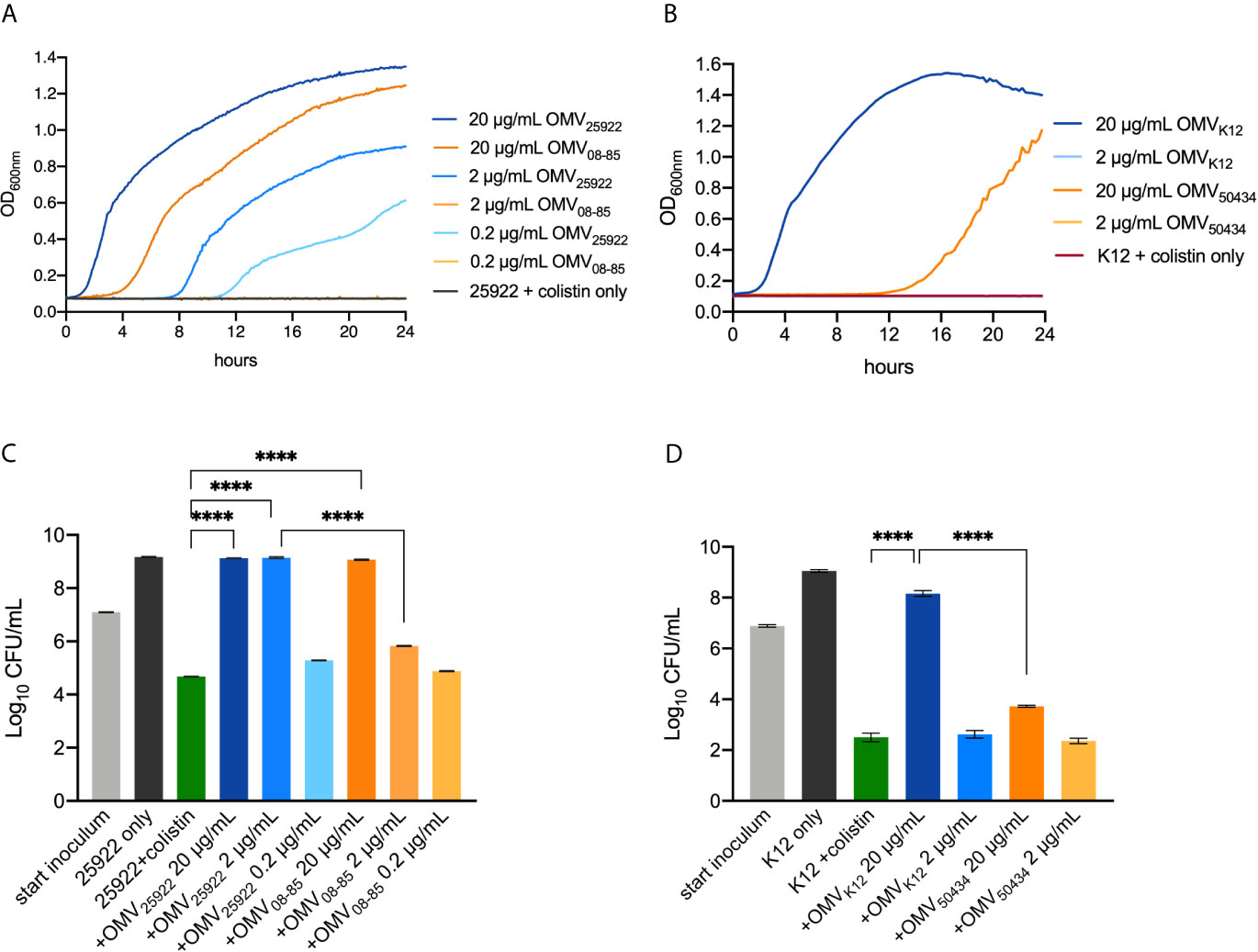
Figure 2 Growth curves (A, B) and killing protection assays (C, D) showed that OMVs of mcr-1 positive E coli strains provided attenuated protection from colistin. (A) A dilution (1:1,000) of overnight culture of E coli ATCC25922 (colistin-susceptible) co-incubated with 20, 2, or 0.2 μg/ml OMVs isolated from E. coli ATCC25922 (mcr-1 negative) or E. coli 08-85 (mcr-1 positive) was treated with 2 μg/ml colistin. (B) A 1:1,000 dilution of overnight culture of E. coli K12 (colistin-susceptible) co-incubated with 20 or 2 μg/ml OMVs isolated from E. coli K12 (mcr-1 negative) or E. coli 50434 (mcr-1 positive) was treated with 2 μg/ml colistin. The turbidity was read every 15 min for 24 h. E. coli ATCC25922 without OMVs was set as the negative control. The concentration of OMVs tested in the killing assay was the same as the growth curve assay. Log-phase culture (107 CFU/ml) of E. coli ATCC25922 (C) or E. coli K12 (D) was treated with colistin for 6 h, and the surviving bacterial cells were counted. The bacterial cells alone and bacterial cells with colistin alone were set as positive and negative controls respectively. Asterisks (*) denote statistical significance which was determined by one-way ANOVA followed by Turkey’s multiple-comparison test (****P < 0.0001, n = 3, bars represent mean ± s.e.m.).
In the killing protection assays (Figures 2C, D), the bacterial cells alone (black bars) and bacterial cells with colistin (2 μg/ml, green bars) were set as the positive and negative controls respectively. For clinical E. coli ATCC25922 (Figure 2C), both 20 μg/ml E. coli ATCC25922 OMVs and 20 μg/ml E. coli 08-85 OMVs provided protection from colistin treatment compared with the negative control. However, when incubated with 2 μg/ml OMVs, bacterial cells were completely protected with E. coli ATCC25922 OMVs, but not OMVs of E. coli 08-85. A 0.2 μg/ml OMVs of E. coli ATCC25922 or E. coli 08-85 was not able to protect E. coli ATCC25922 against colistin. As shown in Figure 2D, the colistin sensitive E. coli K12 was protected by 20 μg/ml E. coli K12 OMVs from colistin treatment compared with E. coli K12 without OMVs. However, the addition of 20 μg/ml E. coli 50434 OMVs conferred significantly attenuated protection against colistin. Neither 2 μg/ml E. coli K12 OMVs nor 2 μg/ml E. coli 50434 OMVs could protect E. coli K12 from colistin.
These results indicated that OMVs from the mcr-1 positive strain attenuated the protective effect when compared to the mcr-1 negative strain at the same concentration.
It has been reported that OMVs could serve as decoys that absorb antibiotics and provide protection for sensitive bacteria (Manning and Kuehn, 2011). To test whether the protection of OMVs observed in the protection assay was due to the absorption of colistin by OMVs, a fluorescent displacement assay using BODIPY TR cadaverine (BC), which binds to lipid A, was used according to a previously described assay (Wood et al., 2004). The fluorescence of BC is quenched by binding to lipid A phosphate groups and de-quenched by compounds displaying an affinity for lipid A. OMVs isolated from E. coli K12 and E. coli 50434 were used for the following studies. Strain E. coli 50434 was engineered from E. coli K12 with the mcr-1 gene introduced into the chromosome (Qiao, 2019). As shown in Figure 3A, the titration of OMVs against BC resulted in a concentration-dependent quenching of BC fluorescence. If colistin binds to the lipid A of OMVs similarly, it should displace the BC probe and lead to an increase of fluorescence intensity. Serial 10-fold dilutions of colistin were added to BC (4 μM) solution that was pre-incubated with 2 μg/ml OMVs of E. coli K12 or E. coli 50434. The fluorescence of BC increased while the concentration of colistin increased, which demonstrated that colistin binds to OMVs in a dose-dependent manner (Figure 3B).
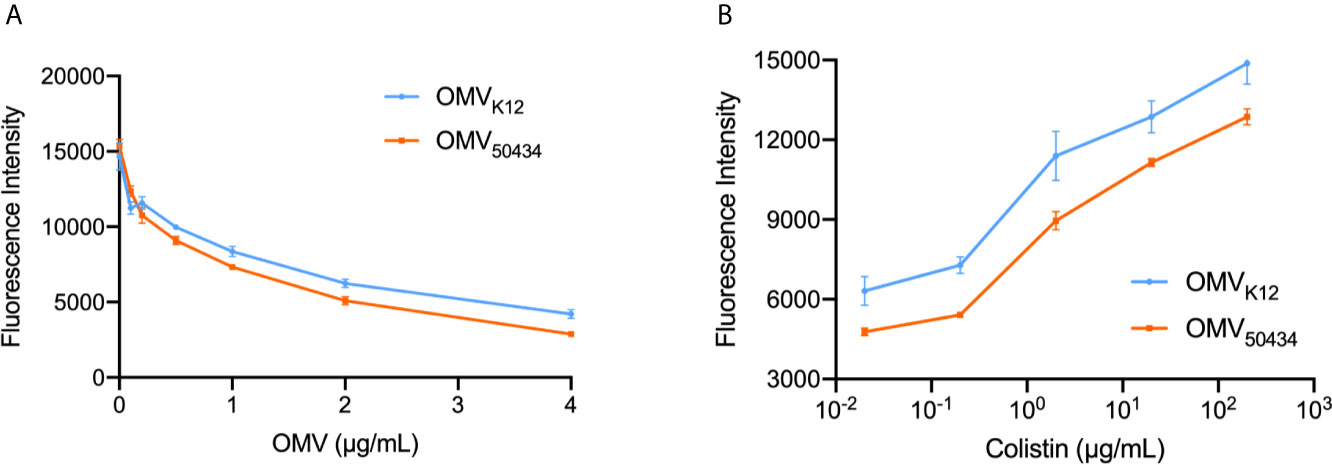
Figure 3 Titration of OMVs against BC (A) and displacement of OMV-bound BC by colistin (B). (A) BCs (4 μM) were titrated by OMVs isolated from E. coli K12 or E. coli 50434 (E. coli K12 with mcr-1) ranging from 0.1 to 4 μg/ml protein concentration. (B) Colistin (0.02–200 μg/ml) was successively added to the BC : OMV complex. Excitation: 580 nm; Emission: 620 nm. n = 3.
NPN uptake analysis was also performed to determine the interaction of OMVs and colistin. NPN is a lipophilic probe with low fluorescence quantum yield in an aqueous environment and becomes fluorescent when partitioned in a hydrophobic environment (Helander and Mattila-Sandholm, 2000). The binding of colistin to OMVs results in increased membrane permeability and increased fluorescence of NPN. As shown in Figure S1, the fluorescence of NPN increased while the concentration of colistin increased, which indicated that the colistin was absorbed by OMVs.
Notably, when incubated with the same concentration of colistin, there was more BC de-quenched by E. coli K12 OMVs than OMVs of E. coli 50434 (Figure 3B). Similarly, the uptake of NPN by OMVs of E. coli 50434 was less than that of E. coli K12 OMVs (Figure S1). These results suggested OMVs of E. coli K12 absorbed more colistin than OMVs of E. coli 50434 at the same concentration and could explain the attenuated protection provided by OMVs of mcr-1 positive E. coli strains. To assess if the difference in the ability to bind colistin was due to lipid A modification caused by the MCR-1 enzyme, the lipid A contents were then analyzed.
Lipid A of OMVs purified from E. coli K12 and E. coli 50434 was isolated and identified with LC-MS. Two species of unmodified lipid A, 1 or 4′-phosphate (Lipid A-P, m/z of 1716.2) and 1,4-bisphosphate (Lipid A-2P, m/z of 1796.2) were confirmed to be present in both E. coli K12 and E. coli 50434 OMVs. A single pEtN attached to these unmodified lipid A, lipid A-P-pEtN (m/z of 1839.2), and lipid A-2P-pEtN (m/z of 1919.2) was observed in the OMVs of E. coli 50434 (Figures 4A, B).
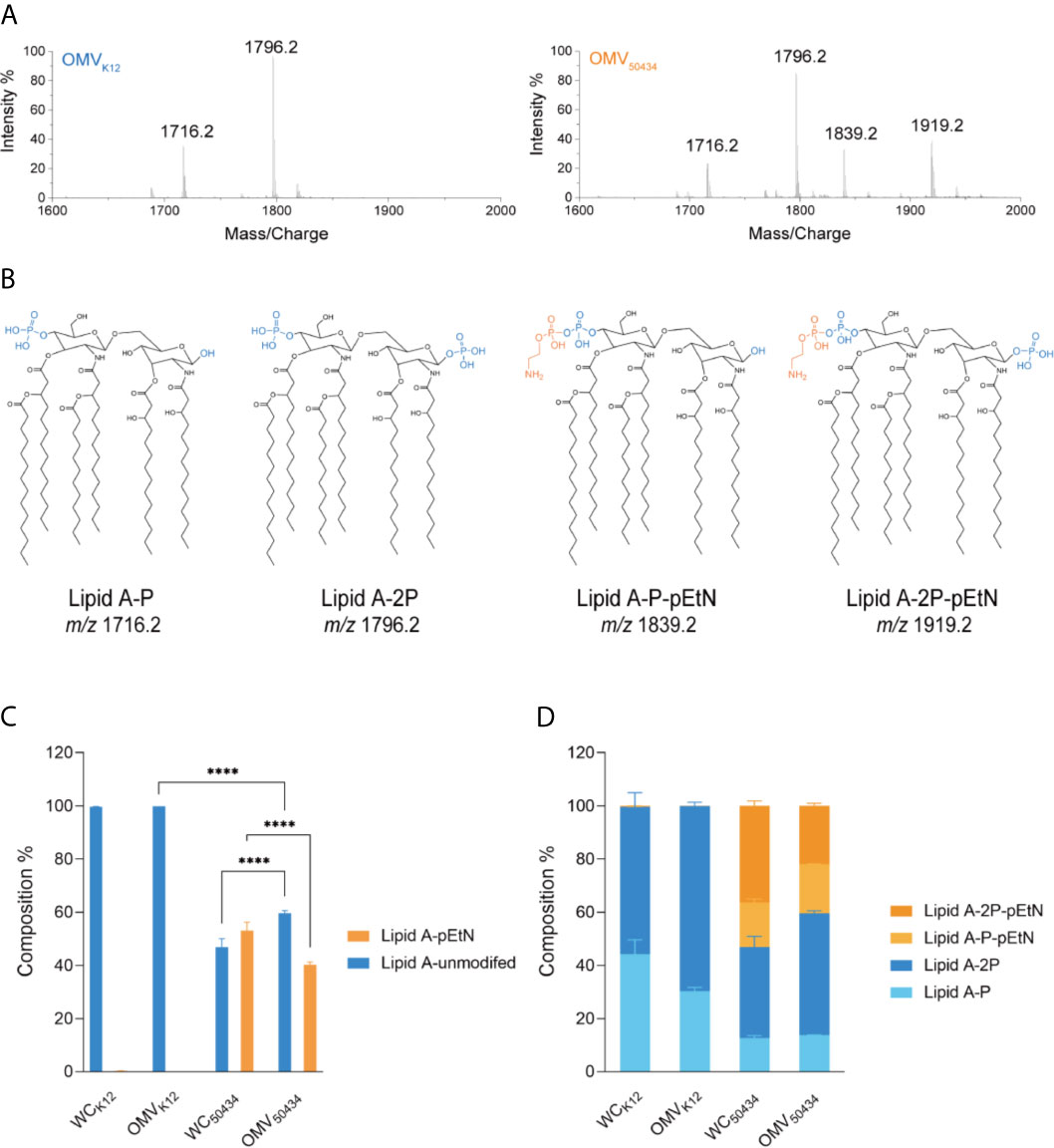
Figure 4 Characterization and relative quantitation of lipid A in OMVs and the producer whole cells (WCs) by LC-MS. (A) Representative mass spectra of lipid A from E. coli K12 OMVs (left) and E. coli 50434 OMVs (right). (B) Structures of lipid A species identified by mass spectrometry. Phosphate group, blue; pEtN, orange. (C, D) The composition of lipid A species was relatively quantitated of peak area with MultiQuant 3.0. Asterisks (*) denote statistical significance as determined by two-way ANOVA followed by Turkey’s multiple-comparison test (****P < 0.0001, n = 3).
The relative amounts of the lipid A species detected were calculated. The lipid A species with two phosphate groups were more abundant than that with only one phosphate group, whether modified with pEtN or not (Figure 4D). Interestingly, there was less pEtN-modified lipid A present in the OMVs compared with bacterial cells, with 46 and 60% in composition respectively (Figure 4C).
These results confirmed that the pEtN-modified lipid A was present in the OMVs of the mcr-1 positive E. coli strain. This could explain why OMVs of the mcr-1 positive strain conferred attenuated protection compared with OMVs of the mcr-1 negative strain at the same concentration. Interestingly, we noticed that the relative amounts of pEtN-modified and native lipid A between bacterial cells and OMVs of E. coli 50434 were not identical; the unmodified lipid A was more likely to be secreted into OMVs, and the pEtN modified lipid A was enriched in bacterial cells.
Finally, we investigated the impact of the mcr-1 gene on OMV production. The relative concentrations of OMVs from E. coli K12 or E. coli 50434 were determined by measuring lipid content via fluorescence from the lipophilic dye FM4-64 (McBroom et al., 2006). The OMV production was normalized by dividing by the number of CFU/ml, and the relative fold OMV production was calculated as these values were further divided by the OMV production of the control condition. Compared with E. coli K12, the mcr-1 positive E. coli 50434 released 2.5-fold more OMVs (Figure 5A). For strain E. coli 50434, growing with colistin pressure (2 μg/ml) resulted in a significant increase of OMV production (2.3-fold more) compared with E. coli 50434 growing without colistin (Figure 5B).
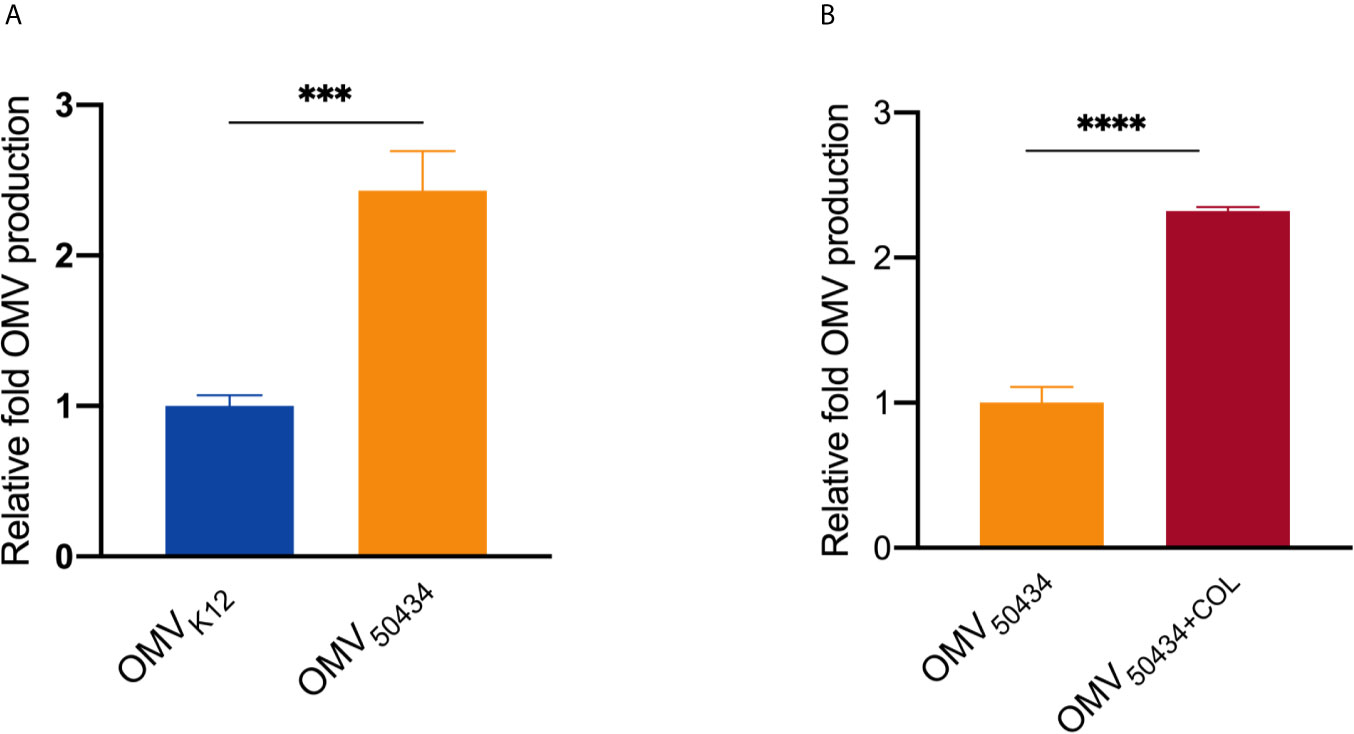
Figure 5 Relative quantitation of OMV production. OMV production was normalized to CFU counts and quantitated for lipid content by FM4-64. (A) The OMV production of E. coli K12 vs E. coli 50434 (E. coli K12 with mcr-1). (B) The OMV production of E. coli 50434 grown with or without colistin (COL, 2 μg/ml). Asterisks (*) denote statistical significance as determined by unpaired t-test (***P < 0.001, ****P < 0.0001, n = 3, bars represent mean ± s.e.m.)
Resistance to colistin, especially the recently reported plasmid-mediated colistin resistance, is a serious threat to global public health since the distribution of these plasmids among Gram-negative bacterial may be a further step towards the “pre-antibiotic era” (Bialvaei and Samadi Kafil, 2015). Thus, understanding the mechanism of colistin resistance is of great significance for developing new strategies against colistin-resistant bacteria. Previous reports have shown that OMVs mediated antibiotic resistance mainly in two ways: a) horizontally transferring antibiotic resistance gene (e.g. the extended spectrum βD;-lactamase gene blaCTX-M-15 (Bielaszewska et al., 2020), the New Delhi metallo-β-lactamase-1 gene blaNDM-1 (Chatterjee et al., 2017), and the β-lactamase gene blaOXA-24 (Rumbo et al., 2011)); b) reducing the antibiotic concentration in the environment either by directly absorbing it (e.g. membrane-active antibiotics) (Manning and Kuehn, 2011; Kulkarni et al., 2015) or packing antibiotic resistance enzymes in their lumen (e.g. β-lactamase) (Lee et al., 2013). So far, little is known about the role of OMVs in mcr-1 mediated colistin resistance. It is not surprising to speculate that OMVs may participate in the horizontal transfer of the mcr-1 gene which is carried on a plasmid. The mcr-1 gene was detected in the OMVs of E. coli 08-85 by dot blotting and PCR (Figure S2 and Figure S3A). Although there was no plasmid detected in E. coli 08-85 OMVs by pulsed-field gel electrophoresis (PFGE) digested with S1-nuclease (Figure S3B), we do not know whether the presence of the mcr-1 gene in E. coli 08-85 OMVs was associated with the plasmid or not since the PFGE method is not sensitive enough. However, the bacterial cells of E. coli ATCC25922 that survived the colistin treatment in the killing protection assay were collected, and the mcr-1 gene was not detected in any of them by PCR (Figure S4). Thus, it seems likely that the protective effect conferred by OMVs is not associated with the acquisition of the mcr-1 gene from E. coli 08-85 OMVs.
Manning and Kuehn (2011) and Kulkarni et al. (2015) showed that the addition of purified OMVs to the E. coli cultures provided substantial protection against antimicrobial peptides (AMPs). Moving a step forward on their research, we found that OMVs of a mcr-1 positive strain attenuated the effect of protection. Consistent with their study, we showed that OMVs, both from the mcr-1 positive and negative E. coli strains, conferred dose-dependent protection against colistin sensitive E. coli strains. However, the addition of 2 μg/ml E. coli ATCC25922 (mcr-1 negative) OMVs achieved 100% protection to the producer organism from colistin treatment, while 2 μg/ml E. coli 08-85 (mcr-1 positive) OMVs could not protect the colistin sensitive strain E. coli ATCC25922 (Figure 2C). We noticed that the protective effects of 2 μg/ml E. coli ATCC25922 OMVs and 20 μg/ml E. coli 08-85 OMVs in the growth curve assay were not exactly the same as in the killing protection assay. This could be due to the inoculum effect in which the initial density of cells affects the activity of many antibiotics including membrane-active peptides (Loffredo et al., 2021). The growth curve assay was started with a 1:1,000 dilution of the overnight culture (starting inoculum of 105 CFU/ml), while the log-phase culture was used as the starting inoculum which was 107 CFU/ml in the killing protection assay. Colistin was absorbed by OMVs (Figure 3), and there should be less free colistin left in the environment for killing bacterial cells after co-incubation with 20 μg/ml E. coli ATCC25922 OMVs than 2 μg/ml E. coli ATCC25922 OMVs. When incubated with 20 μg/ml E. coli ATCC25922 OMVs, the free colistin left might not be enough to either inhibit the growth or kill bacterial cells. However, when co-incubated with 2 μg/ml E. coli ATCC25922 OMVs or E. coli 08-85 OMVs, the free colistin left in the environment might be able to inhibit the growth of bacterial cells at a 105 CFU/ml inoculum, but might not be enough to kill an inoculum of 107 CFU/ml due to the inoculum effect. Also, we noticed that there were still 10,000 viable bacteria of colistin (2× MIC) treated E. coli ATCC25922 in the killing protection assays (Figure 2C). This could be due to the inoculum effect as well since the MBCs were determined with the inoculum of 105 CFU/ml (according to CLSI), while the starting inoculum was 107 CFU/ml in the killing protection assay. The attenuated protective effect was also observed in E. coli K12 and the isogenic strain E. coli 50434 when incubated with 20 μg/ml OMVs. LPSs are complex molecules that consist of O antigen, the core oligosaccharide and the lipid A moiety. The differences in genetic background between E. coli ATCC25922 and E. coli K12 might result in the different structure of LPS and the bacterial outer membrane, thus leading to the different binding affinity of OMVs to colistin (McInerney et al., 2016). The OMVs of E. coli ATCC25922 might have a better binding affinity to colistin than E. coli K12 OMVs. Therefore, 2 μg/ml of E. coli K12 OMVs might not be enough to protect from 2 μg/ml colistin due to the strain specificity, and this could be the reason for the relatively weaker protection provided by E. coli K12 OMVs compared with E. coli ATCC25922 OMVs, especially at 2 μg/ml.
MCR-1 catalyzes the transfer of pEtN to lipid A and decreases colistin affinity. Thus, we wondered if the protection of OMVs observed in the protection assay was due to the absorption of colistin by OMVs. If that was the case, the attenuated protection of OMVs from the mcr-1 positive strain could be related to the reduced ability to absorb colistin which is caused by the modification of lipid A. To address this, we first determined the ability of OMVs to bind to colistin. Since E. coli ATCC25922 and clinical isolate E. coli 08-85 are two strains with different genetic backgrounds, E. coli K12 and E. coli K12 recombineered with the mcr-1 gene (E. coli 50434) were used for the molecular mechanism studies. To avoid the interference of the potential horizontal transfer of mcr-1 genes, strain E. coli 50434 was engineered with the mcr-1 gene introduced into the chromosome. We confirmed that colistin was absorbed by OMVs by both the BC replacement (Figure 3) and NPN uptake assays (Figure S1). In both methods, E. coli 50434 OMVs showed less ability to bind to colistin, and this is consistent with the attenuated protection of mcr-1 positive strain OMVs exhibited in Figure 2.
We confirmed that E. coli 50434 OMVs contained pEtN-modified lipid A molecules, while lipid A in E. coli K12 OMVs was unmodified. This explained the observation of the attenuated protective effect and binding affinity to colistin of OMVs from the mcr-1 positive strain compared to the negative strain (Figures 2, 3). For the mcr-1 positive strain, when comparing the composition of lipid A species between E. coli 50434 OMVs and its producer cells, we found that they packed more unmodified lipid A in OMVs and retained more pEtN modified lipid A in the outer membrane of bacterial cells. Both the colistin-sensitive strain and themselves could benefit from this arrangement as they release OMVs with more unmodified lipid A for better protection for sensitive strains and left more pEtN-modified lipid A for themselves to combat against colistin. Several investigations have shown that OMV biogenesis is a deliberate process with cargo selection (Ayalew et al., 2013; Bonnington and Kuehn, 2014). Our results provided further support to this phenomenon by adding evidence of adjustment of lipid A species in OMVs of the mcr-1 positive E. coli strain for combating colistin and causing and spreading resistance to colistin. Nevertheless, it should be noted that the E. coli strain with mcr-1 released more OMVs than E. coli K12 and two-fold more under colistin pressure. The mechanism of increased OMV production of the mcr-1 positive strain is unclear and needs further investigation. Our work, for the first time, sheds light on how mcr-1 affected the lipid contents of OMVs and added new insights into the research field of OMVs and antibiotic resistance.
Our study found that OMVs from the mcr-1 positive strain conferred attenuated protection against colistin when compared to OMVs from the mcr-1 negative strain. This could be due to the presence of pEtN-modified lipid A in the OMVs of mcr-1 positive E. coli strain, which resulted in the decreased ability to absorb colistin in the environment.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
YZ and XL conceived and designed the study. XL, YZ, LS, XW, XH, and TN carried out the experiments and analyzed the data. CL and XYa revised the manuscript. XYo is the director of this work and responsible for the general supervision of the study. All authors contributed to the article and approved the submitted version.
This research was funded by the National Natural Science Foundation of China (81803413), the Peking Union Medical College Youth Fund (PUMC) (3332018093), the National Mega-project for Innovative Drugs (2019ZX09721001), the CAMS Initiative for Innovative Medicine (2016-I2M-3- 014).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.701625/full#supplementary-material
Ah, Y.-M., Kim, A.-J., Lee, J.-Y. (2014). Colistin Resistance in Klebsiella Pneumoniae. Int. J. Antimicrob. Agents 44 (1), 8–15. doi: 10.1016/j.ijantimicag.2014.02.016
Ayalew, S., Confer, A. W., Shrestha, B., Wilson, A. E., Montelongo, M. (2013). Proteomic Analysis and Immunogenicity of Mannheimia Haemolytica Vesicles. Clin. Vaccine Immunol. 20 (2), 191–196. doi: 10.1128/CVI.00622-12
Bialvaei, A. Z., Samadi Kafil, H. (2015). Colistin, Mechanisms and Prevalence of Resistance. Curr. Med. Res. Opin. 31 (4), 707–721. doi: 10.1185/03007995.2015.1018989
Bielaszewska, M., Daniel, O., Karch, H., Mellmann, A. (2020). Dissemination of the Bla CTX-M-15 Gene Among Enterobacteriaceae via Outer Membrane Vesicles. J. Antimicrob. Chemother. 75 (9), 2442–2451. doi: 10.1093/jac/dkaa214
Biswas, S., Brunel, J.-M., Dubus, J.-C., Reynaud-Gaubert, M., Rolain, J.-M. (2012). Colistin: an Update on the Antibiotic of the 21st Century. Expert Rev. Anti-infect. Ther. 10 (8), 917–934. doi: 10.1586/eri.12.78
Bonnington, K., Kuehn, M. (2014). Protein Selection and Export via Outer Membrane Vesicles. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 1843 (8), 1612–1619. doi: 10.1016/j.bbamcr.2013.12.011
Breazeale, S. D., Ribeiro, A. A., McClerren, A. L., Raetz, C. R. (2005). A Formyltransferase Required for Polymyxin Resistance in Escherichia Coli and the Modification of Lipid A With 4-Amino-4-Deoxy-L-Arabinose: Identification and Function of UDP-4-Deoxy-4-Formamido-L-Arabinose. J. Biol. Chem. 280 (14), 14154–14167. doi: 10.1074/jbc.M414265200
Cai, Y., Chai, D., Wang, R., Liang, B., Bai, N. (2012). Colistin Resistance of Acinetobacter Baumannii: Clinical Reports, Mechanisms and Antimicrobial Strategies. J. Antimicrob. Chemother. 67 (7), 1607–1615. doi: 10.1093/jac/dks084
Cai, Y., Lee, W., Kwa, A. L. (2015). Polymyxin B Versus Colistin: an Update. Expert Rev. Anti-infect. Ther. 13 (12), 1481–1497. doi: 10.1586/14787210.2015.1093933
Chatterjee, S., Mondal, A., Mitra, S., Basu, S. (2017). Acinetobacter Baumannii Transfers the blaNDM-1 Gene via Outer Membrane Vesicles. J. Antimicrob. Chemother. 72 (8), 2201–2207. doi: 10.1093/jac/dkx131
Chattopadhyay, M. K., Jagannadham, M. V. (2015). Vesicles-Mediated Resistance to Antibiotics in Bacteria. Front. Microbiol. 6, 758. doi: 10.3389/fmicb.2015.00758
Chen, H. D., Groisman, E. A. (2013). The Biology of the PmrA/PmrB Two-Component System: the Major Regulator of Lipopolysaccharide Modifications. Annu. Rev. Microbiol. 67, 83–112. doi: 10.1146/annurev-micro-092412-155751
Chutkan, H., MacDonald, I., Manning, A., Kuehn, M. J. (2013). “Quantitative and Qualitative Preparations of Bacterial Outer Membrane Vesicles,” in Bacterial Cell Surfaces (New York, USA: Springer), 259–272.
El-Sayed Ahmed, M.A.E.-G., Zhong, L.-L., Shen, C., Yang, Y., Doi, Y., Tian, G.-B. (2020). Colistin and its Role in the Era of Antibiotic Resistance: an Extended Review (2000–2019). Emerging. Microbes Infect. 9 (1), 868–885. doi: 10.1080/22221751.2020.1754133
Falagas, M. E., Kasiakou, S. K., Saravolatz, L. D. (2005). Colistin: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. 40 (9), 1333–1341. doi: 10.1086/429323
French, S., Farha, M., Ellis, M. J., Sameer, Z., Côté, J.-P., Cotroneo, N., et al. (2019). Potentiation of Antibiotics Against Gram-Negative Bacteria by Polymyxin B Analogue SPR741 From Unique Perturbation of the Outer Membrane. ACS Infect. Dis. 6 (6), 1405–1412. doi: 10.1021/acsinfecdis.9b00159
Gao, R., Hu, Y., Li, Z., Sun, J., Wang, Q., Lin, J., et al. (2016). Dissemination and Mechanism for the MCR-1 Colistin Resistance. PloS Pathog. 12 (11), e1005957. doi: 10.1371/journal.ppat.1005957
Helander, I., Mattila-Sandholm, T. (2000). Fluorometric Assessment of Gram-Negative Bacterial Permeabilization. J. Appl. Microbiol. 88 (2), 213–219. doi: 10.1046/j.1365-2672.2000.00971.x
Henderson, J. C., O'Brien, J. P., Brodbelt, J. S., Trent, M. S. (2013). Isolation and Chemical Characterization of Lipid A From Gram-Negative Bacteria. JoVE (J. Visual. Experiments). 79), e50623. doi: 10.3791/50623
Klimentová, J., Stulík, J. (2015). Methods of Isolation and Purification of Outer Membrane Vesicles From Gram-Negative Bacteria. Microbiol. Res. 170, 1–9. doi: 10.1016/j.micres.2014.09.006
Kulkarni, H. M., Nagaraj, R., Jagannadham, M. V. (2015). Protective Role of E. Coli Outer Membrane Vesicles Against Antibiotics. Microbiol. Res. 181, 1–7. doi: 10.1016/j.micres.2015.07.008
Kulkarni, H. M., Swamy, C. V., Jagannadham, M. V. (2014). Molecular Characterization and Functional Analysis of Outer Membrane Vesicles From the Antarctic Bacterium Pseudomonas Syringae Suggest a Possible Response to Environmental Conditions. J. Proteome Res. 13 (3), 1345–1358. doi: 10.1021/pr4009223
Lee, J., Lee, E.-Y., Kim, S.-H., Kim, D.-K., Park, K.-S., Kim, K. P., et al. (2013). Staphylococcus Aureus Extracellular Vesicles Carry Biologically Active β-Lactamase. Antimicrob. Agents Chemother. 57 (6), 2589–2595. doi: 10.1128/AAC.00522-12
Liang, Z., Pang, J., Hu, X., Nie, T., Lu, X., Li, X., et al. (2021). Low Prevalence of Mcr-1 Among Clinical Enterobacteriaceae Isolates and Co-Transfer of Mcr-1 and Bla NDM-1 From Separate Donors. Microbial. Drug Resist. 27 (4), 476–484. doi: 10.1089/mdr.2020.0212
Ling, Z., Yin, W., Shen, Z., Wang, Y., Shen, J., Walsh, T. R. (2020). Epidemiology of Mobile Colistin Resistance Genes Mcr-1 to Mcr-9. J. Antimicrob. Chemother. 75 (11), 3087–3095. doi: 10.1093/jac/dkaa205
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: a Microbiological and Molecular Biological Study. Lancet Infect. Dis. 16 (2), 161–168. doi: 10.1016/S1473-3099(15)00424-7
Loffredo, M. R., Savini, F., Bobone, S., Casciaro, B., Franzyk, H., Mangoni, M. L., et al. (2021). Inoculum Effect of Antimicrobial Peptides. Proc. Natl. Acad. Sci. 118 (21), e2014364118. doi: 10.1073/pnas.2014364118
Manning, A. J., Kuehn, M. J. (2011). Contribution of Bacterial Outer Membrane Vesicles to Innate Bacterial Defense. BMC Microbiol. 11 (1), 1–15. doi: 10.1186/1471-2180-11-258
McBroom, A. J., Johnson, A. P., Vemulapalli, S., Kuehn, M. J. (2006). Outer Membrane Vesicle Production by Escherichia Coli is Independent of Membrane Instability. J. Bacteriol. 188 (15), 5385–5392. doi: 10.1128/JB.00498-06
McBroom, A. J., Kuehn, M. J. (2007). Release of Outer Membrane Vesicles by Gram-Negative Bacteria is a Novel Envelope Stress Response. Mol. Microbiol. 63 (2), 545–558. doi: 10.1111/j.1365-2958.2006.05522.x
McInerney, M. P., Roberts, K. D., Thompson, P. E., Li, J., Nation, R. L., Velkov, T., et al. (2016). Quantitation of Polymyxin–Lipopolysaccharide Interactions Using an Image-Based Fluorescent Probe. J. Pharm. Sci. 105 (2), 1006–1010. doi: 10.1016/j.xphs.2015.10.028
Moffatt, J. H., Harper, M., Boyce, J. D. (2019). Mechanisms of Polymyxin Resistance. Adv. Exp. Med. Biol. 1145, 55–71. doi: 10.1007/978-3-030-16373-0_5
Monaco, M., Giani, T., Raffone, M., Arena, F., Garcia-Fernandez, A., Pollini, S., et al. (2014). Colistin Resistance Superimposed to Endemic Carbapenem-Resistant Klebsiella Pneumoniae: a Rapidly Evolving Problem in Italy, November 2013 to April 2014. Eurosurveillance 19 (42), 20939. doi: 10.2807/1560-7917.ES2014.19.42.20939
Nikaido, H. (2003). Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 67 (4), 593–656. doi: 10.1128/MMBR.67.4.593-656.2003
Qiao, H. (2019). Effects of Different Levels of Mcr-1 Expression on Escherichia Coli and Study on the Mechanism of Carbapenem Heteroresistance in Pseudomonas Aeruginosa. [Master’s Thesis] (Beijing, China: Chinese Academy of Medical Sciences and Peking Union Medical College).
Rumbo, C., Fernández-Moreira, E., Merino, M., Poza, M., Mendez, J. A., Soares, N. C., et al. (2011). Horizontal Transfer of the OXA-24 Carbapenemase Gene via Outer Membrane Vesicles: a New Mechanism of Dissemination of Carbapenem Resistance Genes in Acinetobacter Baumannii. Antimicrob. Agents Chemother. 55 (7), 3084–3090. doi: 10.1128/AAC.00929-10
Schwechheimer, C., Kuehn, M. J. (2015). Outer-Membrane Vesicles From Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 13 (10), 605–619. doi: 10.1038/nrmicro3525
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., Zong, Z. (2020). Identification of Novel Mobile Colistin Resistance Gene Mcr-10. Emerging. Microbes Infect. 9 (1), 508–516. doi: 10.1080/22221751.2020.1732231
Wood, S. J., Miller, K. A., David, S. A. (2004). Anti-Endotoxin Agents. 1. Development of a Fluorescent Probe Displacement Method Optimized for the Rapid Identification of Lipopolysaccharide-Binding Agents. Combinatorial. Chem. High Throughput. Screening. 7 (3), 239–249. doi: 10.2174/1386207043328832
Keywords: outer membrane vesicles, mcr-1, colistin resistance, lipid A, protection
Citation: Li X, Sun L, Li C, Yang X, Wang X, Hu X, Nie T, Zhang Y and You X (2021) The Attenuated Protective Effect of Outer Membrane Vesicles Produced by a mcr-1 Positive Strain on Colistin Sensitive Escherichia coli. Front. Cell. Infect. Microbiol. 11:701625. doi: 10.3389/fcimb.2021.701625
Received: 28 April 2021; Accepted: 09 July 2021;
Published: 28 July 2021.
Edited by:
Jaipalreddy Panga, Institute for Systems Biology (ISB), United StatesReviewed by:
Michael John Calcutt, University of Missouri, United StatesCopyright © 2021 Li, Sun, Li, Yang, Wang, Hu, Nie, Zhang and You. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youwen Zhang, eW91d2VuemhhbmdAaW1iLmNhbXMuY24=; Xuefu You, eHVlZnV5b3VAaW1iLnB1bWMuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.