
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 14 July 2021
Sec. Microbiome in Health and Disease
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.695515
This article is part of the Research TopicThe Pivotal Role of Oral Microbiota Dysbiosis and Microbiota-host Interactions in DiseasesView all 15 articles
 Shuang Li1†
Shuang Li1† Junping Zhu2†
Junping Zhu2† Bin Su1†
Bin Su1† Huanhuan Wei2
Huanhuan Wei2 Fei Chen2
Fei Chen2 Hongshan Liu2
Hongshan Liu2 Jiaqi Wei1
Jiaqi Wei1 Xiaodong Yang1
Xiaodong Yang1 Qiuyue Zhang1
Qiuyue Zhang1 Wei Xia1
Wei Xia1 Hao Wu1
Hao Wu1 Qiushui He2,3*
Qiushui He2,3* Tong Zhang1*
Tong Zhang1*Despite the antiretroviral therapy (ART), human immunodeficiency virus (HIV)-related oral disease remains a common problem for people living with HIV (PLWH). Evidence suggests that impairment of immune function in HIV infection might lead to the conversion of commensal bacteria to microorganisms with increased pathogenicity. However, limited information is available about alteration in oral microbiome in PLWH on ART. We performed a longitudinal comparative study on men who have sex with men (MSM) with acute HIV infection (n=15), MSM with chronic HIV infection (n=15), and HIV-uninfected MSM controls (n=15). Throat swabs were collected when these subjects were recruited (W0) and 12 weeks after ART treatment (W12) from the patients. Genomic DNAs were extracted and 16S rRNA gene sequencing was performed. Microbiome diversity was significantly decreased in patients with acute and chronic HIV infections compared with those in controls at the sampling time of W0 and the significant difference remained at W12. An increased abundance of unidentified Prevotellaceae was found in patients with acute and chronic HIV infections. Moreover, increased abundances of Prevotella in subjects with acute HIV infection and Streptococcus in subjects with chronic HIV infection were observed. In contrast, greater abundance in Lactobacillus, Rothia, Lautropia, and Bacteroides was found in controls. After effective ART, Bradyrhizobium was enriched in both acute and chronic HIV infections, whereas in controls, Lactobacillus, Rothia, Clostridia, Actinobacteria, and Ruminococcaceae were enriched. In addition, we found that lower CD4+ T-cell counts (<200 cells/mm3) were associated with lower relative abundances of Haemophilus, Actinomyces, unidentified Ruminococcaceae, and Rothia. This study has shown alteration in oral microbiome resulting from HIV infection and ART. The results obtained warrant further studies in a large number of subjects with different ethnics. It might contribute to improved oral health in HIV-infected individuals.
Human immunodeficiency virus (HIV) infection is characterized by rapid and substantial loss of CD4+ T cells that impairs host defense and increases the risk of opportunistic microbial infections. Worldwide, there are approximately 38 million people living with HIV infection (PLWH), with about 25.4 million receiving antiretroviral therapy (ART). Although PLWH with ART might achieve a stable virus suppression, several oral diseases, such as oropharyngeal candidiasis (OPC), oral hairy leukoplakia, periodontitis, oral warts, ulcers, herpes, and Kaposi’s sarcoma, are frequently reported (Goldberg et al., 2015; Heron and Elahi, 2017; El Howati and Tappuni, 2018).
It has been shown that impairment of immune function in HIV infection might lead to the conversion of commensal bacteria to microorganisms with increased pathogenicity and contributes to opportunistic infections (Saxena et al., 2016; Griffen et al., 2019). Several studies reported the changes of gut microbiota composition observed in HIV infection (Sun et al., 2016; Zhou et al., 2018; Rocafort et al., 2019), including the increase in Prevotella and decrease in Bacteroides (Dillon et al., 2014; Mutlu et al., 2014). Alterations in the gut microbiota will eventually lead to an imbalance between microbes and their metabolites and could result in HIV-associated immune activation and inflammation (Dinh et al., 2015; Zevin et al., 2016). In addition, various studies have demonstrated that microbial translocation is a cause of HIV-associated immune activation and inflammation (Brenchley et al., 2006; Marchetti et al., 2013; Koay et al., 2018). Early in HIV infection, the observed loss of Th17 cells results in impaired integrity of the mucosal epithelial barriers, leading to microbial translocation from the gut lumen into the systemic circulation (Brenchley et al., 2006; Mudd and Brenchley, 2016). Although a great number of studies have focused on the contribution of gut microbiota to HIV infection, a handful of studies have also characterized the oral microbiome in HIV-infected individuals (Fulcher, 2020). Li et al reported that in comparison to the HIV-negative individuals, PLWH had higher levels of total cultivable microbes in saliva, including oral streptococci, Streptococcus mutans, lactobacilli, and Candida (Li et al., 2014). Recently Annavajhala et al. have shown that oral microbiome communities likely contribute to systemic inflammation and immune activation in PLWH (Annavajhala et al., 2020). However, data are still limited about alterations in oral microbiome in HIV infection on ART.
Previous studies have also demonstrated that the abundance and diversity of oral microbiomes in PLWH were significantly different from those observed in healthy controls. Indeed after ART, the oral microbiome composition was not completely recovered, although it turned to be similar to that observed in healthy controls (Presti et al., 2018; Li et al., 2020). It is known that the composition and homeostasis of oral microbiome are affected by multiple factors, such as diet, medication, and host responses (Samaranayake and Matsubara, 2017). It has been shown that Prevotella-rich microbiomes in the gut are associated with MSM and with HIV infection status (Noguera-Julian et al., 2016; Armstrong et al., 2018). Therefore, HIV-uninfected MSM could serve as useful controls when the effect of HIV infection itself on the oral microbiomes is studied. Further, to explore the impact of ART treatment on oral microbiomes, longitudinal studies and serial samples are needed. In this longitudinal study, we aimed to compare and identify changes in the oral microbiome in MSM with acute and chronic HIV infection before and after ART. Paired throat swab samples were collected within an interval of 12 weeks.
Participants with acute HIV infection or chronic HIV infection (both were from MSM population) were recruited from Beijing Youan Hospital, Beijing, China from May to November 2019. All acute HIV-infected individuals (referred as A0, n=15) and chronic HIV-infected individuals (B0, n=15) had not initiated ART. Fifteen HIV-uninfected MSM (D, n=15) were selected and included as controls. Throat swabs were collected from controls and all HIV-infected ART-naive individuals at the time of recruitment. Thereafter, all acute HIV-infected individuals (A12, n=15) and chronic HIV-infected individuals (B12, n=15) received ART, and throat swabs were collected at 12 weeks of ART. Acute HIV infection was defined as a positive HIV RNA but with negative or indeterminate HIV antibody results. Participants who have used antibiotics, probiotics, and prebiotics within the previous 4 weeks were excluded. In addition, the patients accompanied by active opportunistic infection and HBV/HCV were also excluded. The demographic and clinical characteristics of the study subjects are shown in Table 1. This study received approval from the ethics committee of the Beijing Youan Hospital ([2018]025), and all participants provided written informed consents.
Altogether 73 throat swabs from PLWH (except two patients at 12 weeks of ART were lost to follow-up), and controls were collected. The clean throat swabs were used by the clinical nurses to collect oral samples from the posterior throat and tonsil areas. The swabs were used to repeatedly wipe the sampling site two times and quickly transported and stored at −20°C in the laboratory before polymerase chain reaction (PCR). Genomic DNAs were extracted from swabs using the QIAamp Fast DNA Stool Mini Kit (50) and amplified by PCR for the sequencing of 16S rRNA V4-V5 region. Sequencing libraries were generated using Illumina TruSeq DNA PCR-Free Library Preparation Kit (Illumina, USA). The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an Illumina NovaSeq platform and 250 bp paired-end reads were generated.
Paired-end reads were assigned to samples based on the unique barcodes and merged by using FLASH (V1.2.7). The raw tags were obtained, and the quality filtering was performed under specific filtering conditions to obtain the high-quality clean tags according to the QIIME (V1.9.1, http://qiime.org/scripts/split_libraries_fastq.html) quality control process. Then the tags were compared with the reference database (Silva database, https://www.arb-silva.de/) using UCHIME algorithm (UCHIME algorithm, http://www.drive5.com/usearch/manual/uchime_algo.html) to detect chimera sequences. The chimera sequences were removed, and the effective tags were finally obtained. Operational taxonomic units (OTUs) were clustered by Uparse software (Uparse v7.0.1001), and representative sequence for each OTU was screened for further annotation. For each representative sequence, the Silva Database was used based on Mothur algorithm to annotate taxonomic information. Alpha diversity is applied in analyzing complexity of species diversity for a sample, including Observed-species, Chao1, Shannon, Simpson, ACE, Good-coverage, and PD whole tree. All these indices in our samples were calculated with QIIME (Version 1.7.0) and displayed with R software (Version 2.15.3). Beta diversity analysis was used to evaluate differences of samples in species complexity. Beta diversity on both weighted and unweighted unifrac was calculated by QIIME software (Version 1.9.1). Principal Coordinate Analysis (PCoA) analysis was displayed by WGCNA package, stat packages, and ggplot2 package in R software (Version 2.15.3).
Alpha diversity and beta diversity in oral microbiome among groups were tested by the Wilcoxon rank-sum test. To assess the differences in the microbial abundance between samples, significance test was conducted with some statistical analysis methods, including t-test and LEfSe. Mann-Whitney test and Kruskal Wallis test were used for comparing continuous variables. Two sides of p < 0.05 were considered statistically significant.
Samples were collected from 15 patients with acute HIV infection, 15 patients with chronic HIV infection, and 15 healthy controls. All participants (≥18 years old) were MSM, and all PLWH with acute and chronic HIV infection had not initiated ART. Throat swabs were collected from controls and HIV-infected patients at baseline and 12 weeks of ART. The baseline CD4+ T-cell count was 387 cells/mm3 in acute HIV infection and 376.2 cells/mm3 in chronic HIV infection. After ART for 12 weeks, all patients had increased CD4+ T-cell counts, with 397.4 cells/mm3 in acute HIV infection and 486.5 cells/mm3 in chronic HIV infection (Table 1).
Sequencing resulted in an average of 59,794 high-quality sequences after quality checks. Rarefaction curves and rank abundance showed a great sequencing depth and an even species distribution of the samples in our study (Supplementary Figures S1A, B). By clustering the sequences into OTUs with 97% similarity, an average of 683 OTUs were identified in controls. The corresponding numbers were 298 and 276 OTUs in the A0 and B0 groups, respectively. Following 12 weeks of ART, the corresponding numbers were 299 and 300 OTUs in the A12 and B12 groups. The following dominant phyla were observed in the oral microbiome: Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria, Actinobacteria, and Spirochaetes, etc (Figure 1A). These six dominant bacterial phyla accounted for more than 98% of the total oral microbiome. The most abundantly detected bacterial genus in the oral microbiome were Streptococcus, Neisseria, unidentified Prevotellaceae, with lower relative abundance of Veillonella, Haemophilus, Actinobacillus, Alloprevotella, and Fusobacterium in controls and the HIV-infected patients prior to ART. In addition, the abundance of Bradyrhizobium in both acute and chronic HIV infection groups was increased following 12 weeks of ART (Figure 1B).
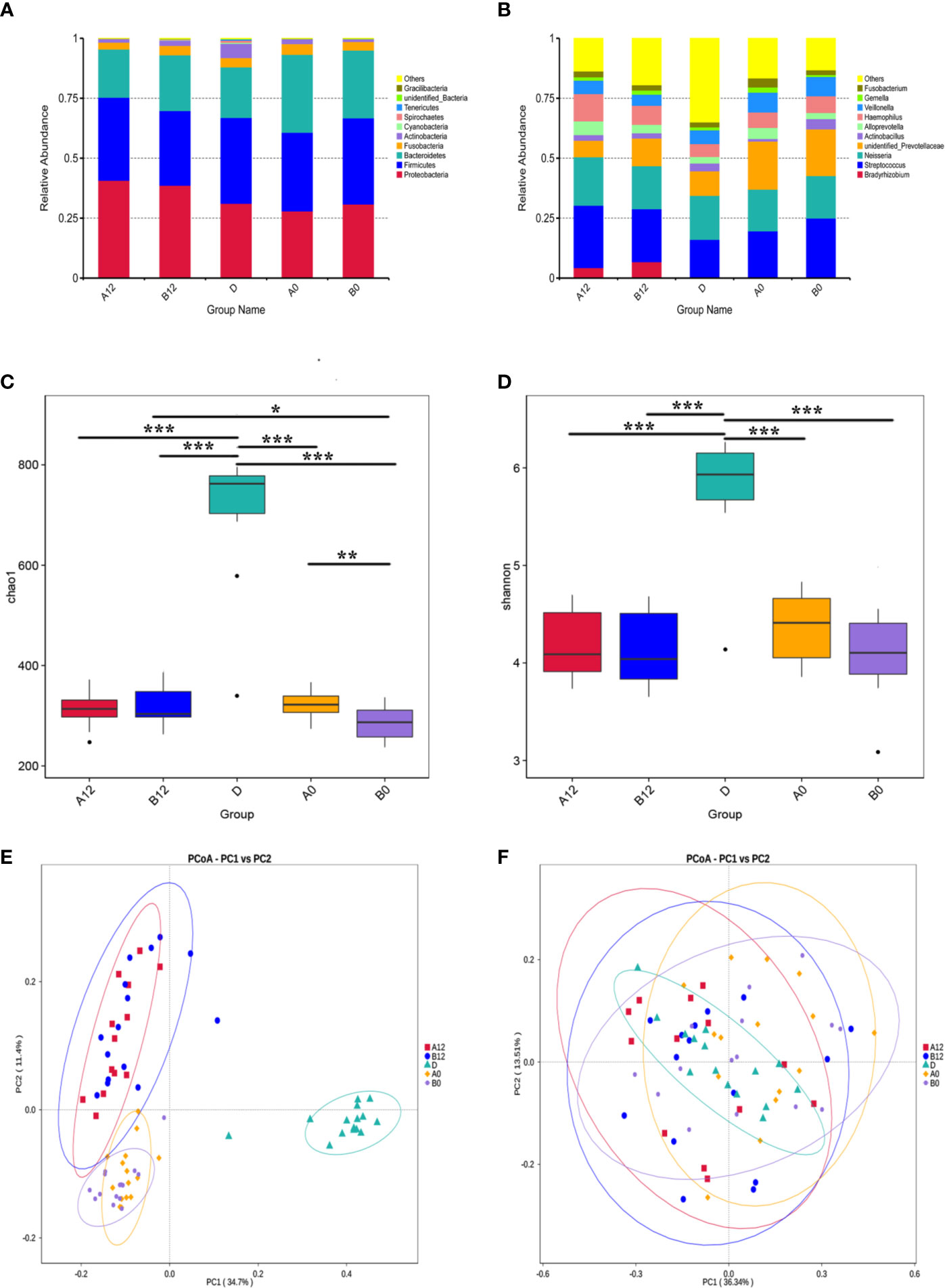
Figure 1 The composition, relative abundance, and diversity of oral microbiota in HIV-infected groups and controls (A) at the phylum level; (B) at the genus level; (C) Alpha diversity exemplified by the Chao1 index; (D) Alpha diversity exemplified by the Shannon index; (E) Beta diversity represented by Principal coordinate analysis (PcoA) of unweighted UniFrac distances; (F) Beta diversity represented by Principal coordinate analysis (PcoA) of weighted UniFrac distances. A0, people living with acute HIV infection at baseline; B0, people living with chronic HIV infection at baseline; D, HIV-uninfected controls; A12, people living with acute HIV infection after 12 weeks of ART; B12, people living with chronic HIV infection after 12 weeks of ART. *p < 0.05; **p < 0.01; ***p < 0.001.
We next used richness estimators (Chao) and diversity index (Shannon index) to compare microbial alpha diversity among different groups of samples. Compared with the control group D, the Chao1 index and Shannon index of oral microbiomes in both A0 group and B0 group were significantly decreased (Figures 1C, D) (all p<0.001). Although after 12 weeks of ART, the Chao1 index and Shannon index were still significantly lower in acute and chronic HIV-treated groups when compared with healthy controls (Figures 1C, D) (Supplementary Table S1) (all p<0.001). However, the Chao1 index of oral microbiomes in B12 group was significantly increased compared to B0 group (Figure 1C) (p<0.001). In addition, the Chao1 index in B0 group was significant decreased compared to A0 group (p<0.01), whereas the differences was not significant between the A12 and B12 (Figure 1C) (p=0.75). To identify the differences in microbial community composition of participants in these groups, the Principal Co-ordinates Analysis (PCoA) was used for beta diversity analysis. A conspicuous separation between HIV infection groups and control group was observed, beta diversity of oral microbiomes was significantly different between HIV-infected individuals and controls (Figures 1E, F) (p<0.001). There was no significant difference in beta diversity represented by PcoA of weighted UniFrac distances when comparing A0 group and B0 group (p=0.58), but there was a significant difference between groups A12 and B12 (p<0.001) (Figure 1F). Likewise, no significant difference in beta diversity of weighted UniFrac distances was observed between A0 and A12 groups (p=0.32), but there was a significant difference between groups B0 and B12 (Figure 1F) (p<0.001).
Differences in the composition of oral microbiome between controls and PLWH were analyzed. At the genus level, we observed the increased abundance of unidentified Prevotellaceae in both A0 and B0 groups. The abundances of Prevotella in the A0 group and Streptococcus in the B0 group were significantly increased. In contrast, several groups of bacteria, such as Lactobacillus, Rothia, Lautropia, and Bacteroides were found in greater abundance in D group (Figures 2A, B).
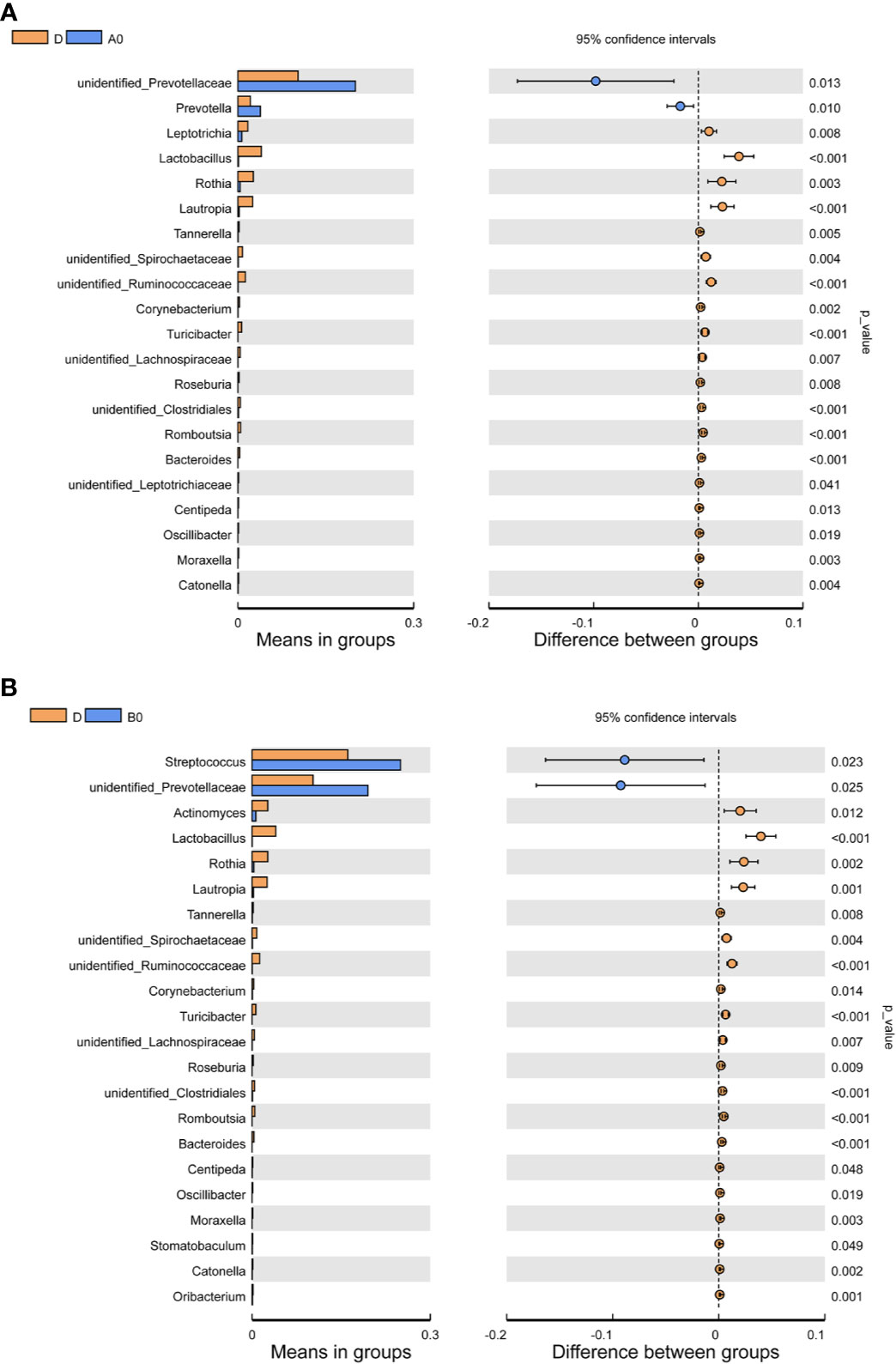
Figure 2 Comparisons of the relative abundance in oral microbiota at the genus level (A) Between A0 and D groups; (B) Between B0 and D groups. A0, people living with acute HIV infection at baseline; B0, people living with chronic HIV infection at baseline; D, HIV-uninfected controls; A12, people living with acute HIV infection after 12 weeks of ART; B12, people living with chronic HIV infection after 12 weeks of ART.
All of the PLWH included in this study received ART after the swab samples at baseline were taken. The second swabs were taken after 12 weeks of ART. LEfSe analyses showed that Bradyrhizobium was enriched in both A12 and B12 groups, whereas Clostridia, Actinobacteria, Lactobacillus, Ruminococcaceae, Rothia were enriched in the D group (Figures 3A–D).
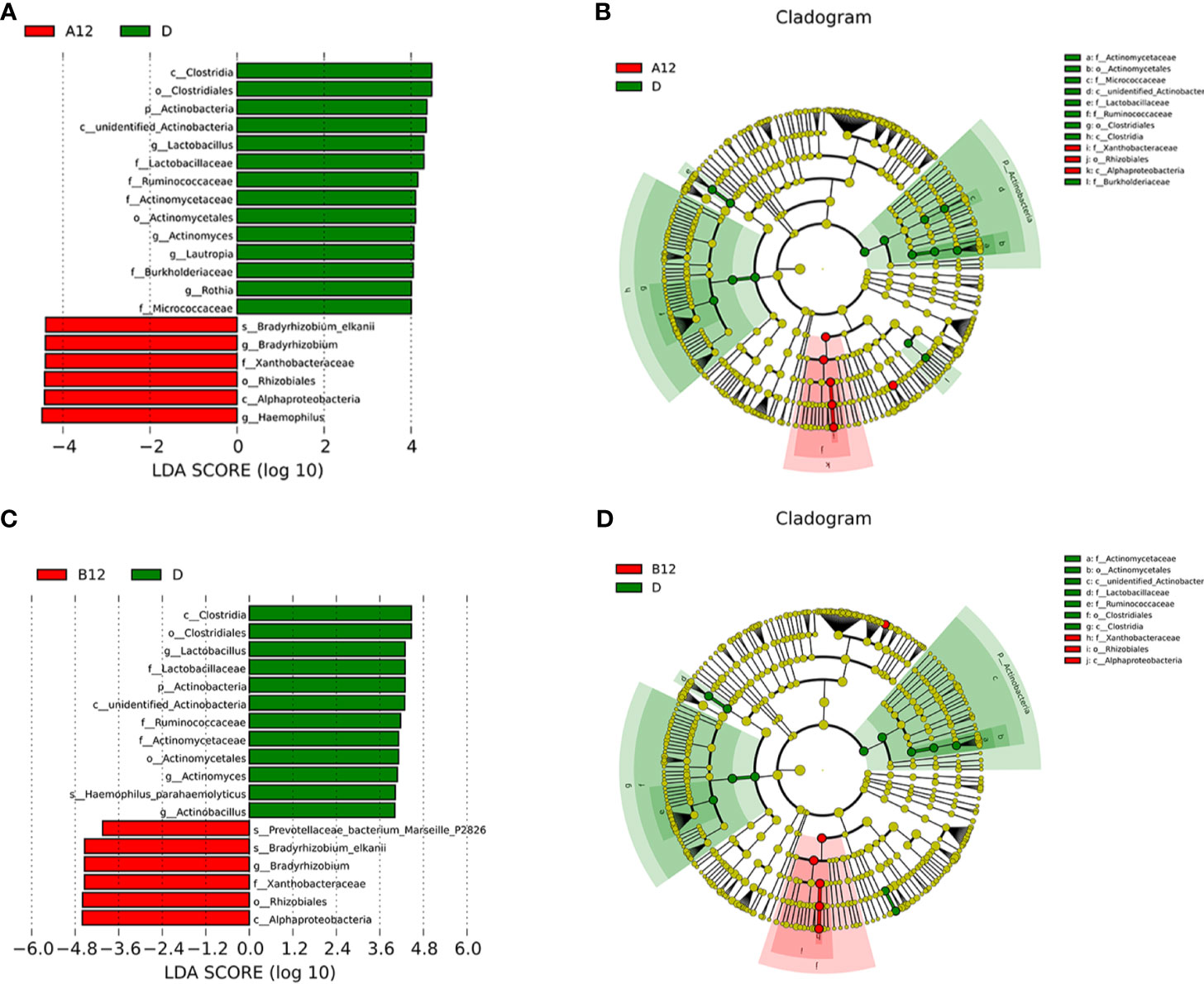
Figure 3 Linear discriminative analysis (LDA) effect size (LefSe) at the genus level. (A, B) shown between A12 and D groups and (C, D) between B12 and D groups. In (A, B), LDA scores for the significant taxa in D group are represented on the positive scale (green), and LDA-negative scores represent enriched taxa in A12 group (red); and in (C, D), LDA scores for the significant taxa in D group are represented on the positive scale (green), and LDA-negative scores represent enriched taxa in B12 group (red). A12, people living with acute HIV infection after 12 weeks of ART; B12, people living with chronic HIV infection after 12 weeks of ART; D, HIV-uninfected controls.
Moreover, Prevotella histicola, Prevotella melaninogenica, Bacteroidales, and unidentified Prevotellaceae were enriched in the A0 group, while Bradyrhizobium were enriched in the A12 group (Figures 4A, B). In the chronic HIV-infected patients, Prevotella histicola, Prevotella melaninogenica, Veillonellaceae, and unidentified Prevotellaceae were enriched in the B0 group, whereas the relative abundance of Bradyrhizobium in the B12 group was significantly higher (Figures 4C, D).
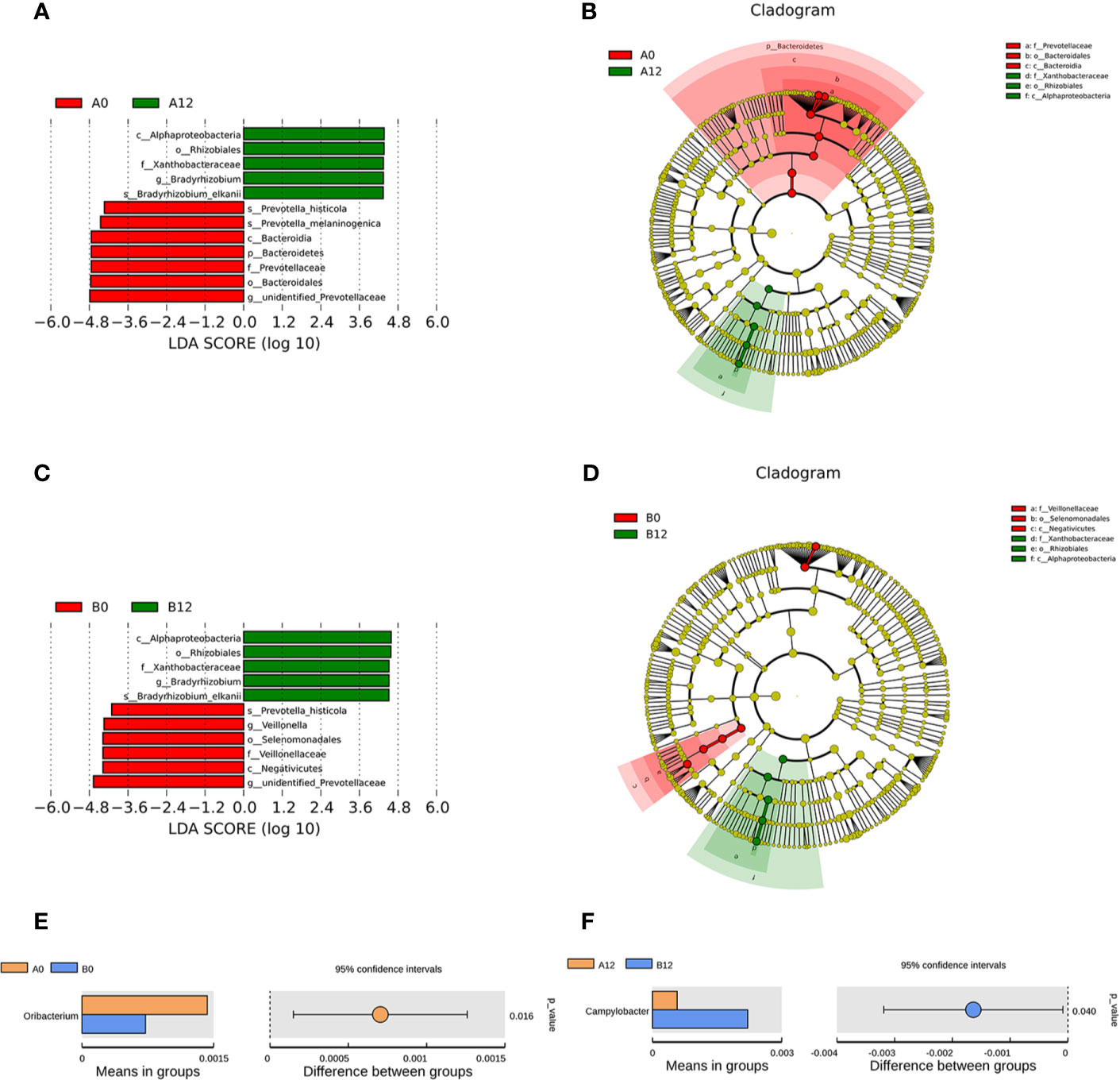
Figure 4 Linear discriminative analysis (LDA) effect size (LefSe) at the genus level. (A, B) shown between A0 and A12 groups and (C, D) between B0 and B12 groups. In (A, B), LDA scores for the significant taxa in A12 group are represented on the positive scale (green), and LDA-negative scores represent enriched taxa in A0 group (red); and in (C, D), LDA scores for the significant taxa in B12 group are represented on the positive scale (green), and LDA-negative scores represent enriched taxa in B0 group (red). Comparisons of the relative abundance in oral microbiota at the genus level (E) Between A0 and B0 groups; (F) Between A12 and B12 groups. A0, people living with acute HIV infection at baseline; B0, people living with chronic HIV infection at baseline; D, HIV-uninfected controls; A12, people living with acute HIV infection after 12 weeks of ART; B12, people living with chronic HIV infection after 12 weeks of ART.
Additionally, we also compared the composition of oral microbiome in people living with acute HIV infection and chronic HIV infection. Compared with A0 group, we noticed lower abundances of Oribacterium in the B0 group (Figure 4E). However, after 12 weeks of ART, we found that the abundance of Campylobacter in the B12 group was significantly higher than those in the A12 group (Figure 4F).
To study whether there is relationship between the alteration observed in oral microbiome and CD4+ T-cell count of patients, we performed analyses of these parameters. We found that the abundance of Haemophilus in patients with CD4<200 cells/mm3 was significantly decreased when comparing with HIV-uninfected controls and patients with CD4>200 cells/mm3. However, there was no significant difference between controls and patients with CD4>200 cells/mm3. In addition, the abundances of Actinomyces, unidentified Ruminococcaceae, and Rothia collected from both subjects with CD4 <200 cells/mm3 and subjects with CD4>200 cells/mm3 were significantly lower than those in HIV-uninfected subjects. Furthermore, lower CD4+ T-cell counts (<200 cells/mm3) were associated with lower relative abundances of Haemophilus, Actinomyces, unidentified Ruminococcaceae, and Rothia (Figures 5A–D).
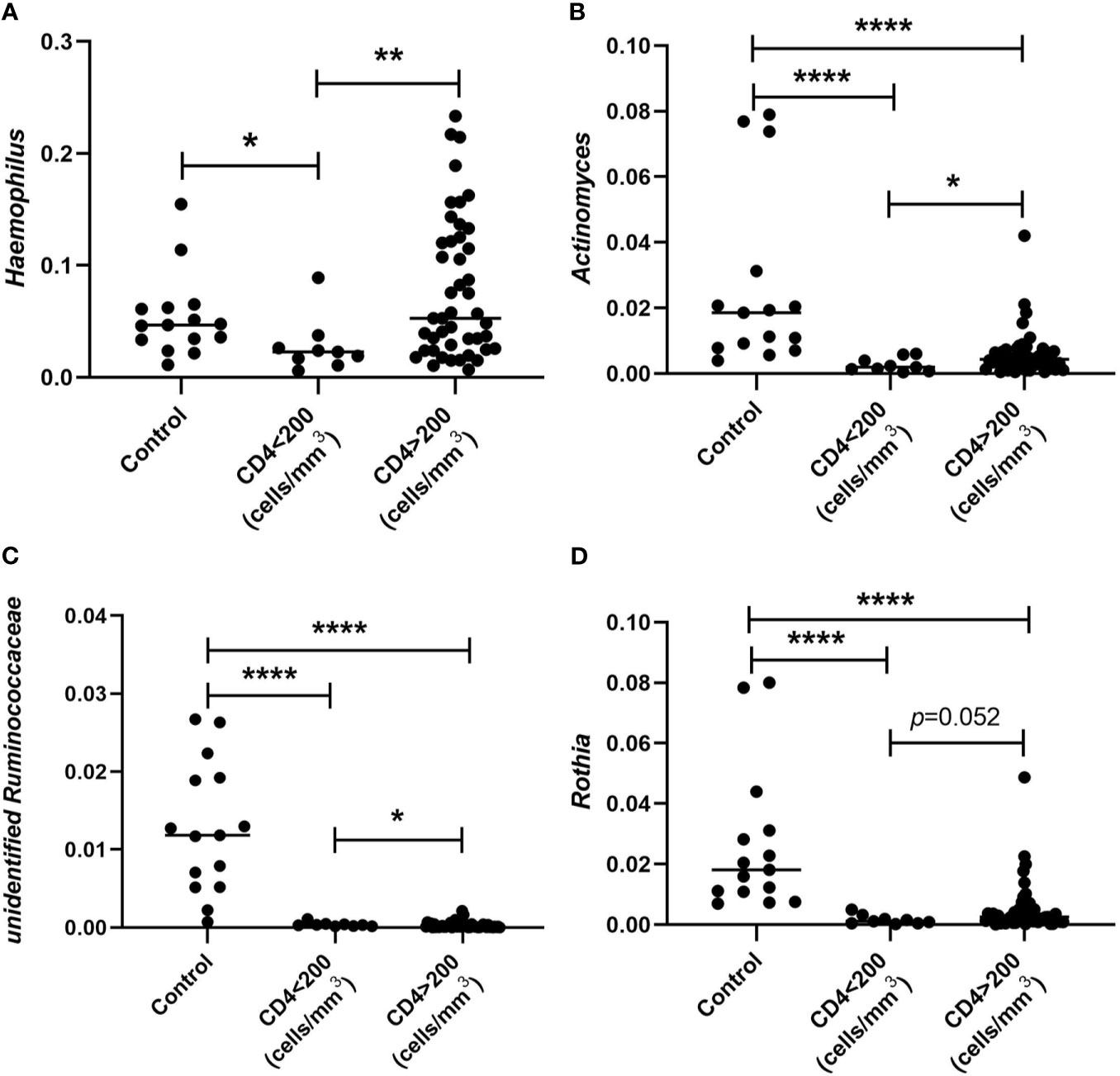
Figure 5 Comparison of relative abundances of Haemophilus (A), Actinomyces (B), unidentified Ruminococcaceae (C), and Rothia (D) collected from subjects whose CD4+ T cells were defined < and CD4>200 cells/mm3 and HIV-uninfected MSM controls. *p < 0.05; **p < 0.01; ****p < 0.0001.
According to the functional annotation, we used PICRUSt metagenome prediction to estimate the functional role of oral microbiome in PLWH and healthy controls. Compared with the D group, we noticed that pathways involved in cell growth and death, glycan biosynthesis, and metabolism increased in HIV-infected patients prior to ART (p<0.05). Moreover, replication and repair of DNA, genetic information processing, and translation-related pathways also increased in both A0 and B0 groups (p<0.05). In contrast, pathways related to xenobiotics biodegradation and metabolism, signal transduction, and cell motility (p<0.05) were decreased in both A0 and B0 groups (Figure 6 and Supplementary Figures S2A, B).
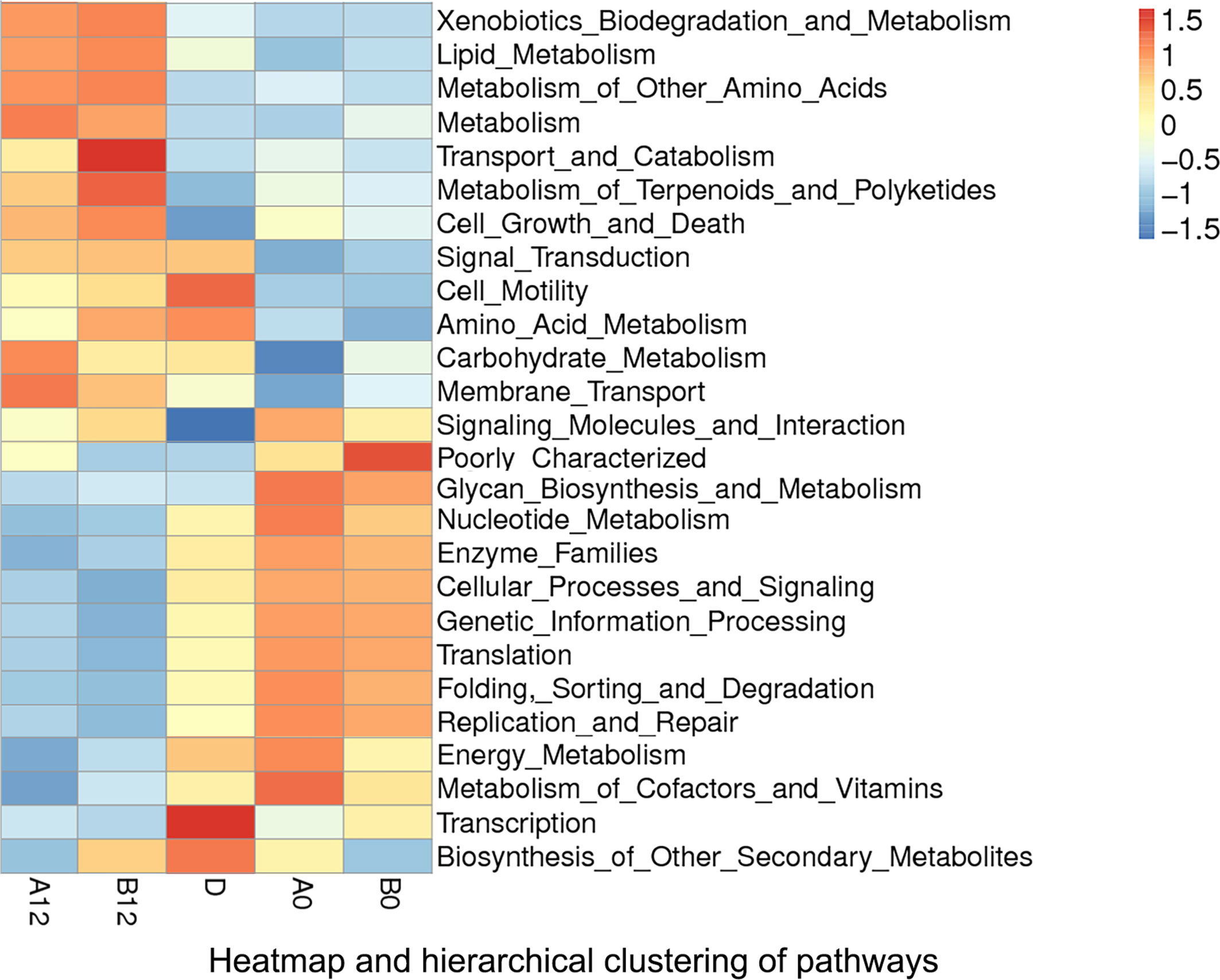
Figure 6 Heatmap and hierarchical clustering of pathways in A0, B0, D, A12 and B12 groups. A0, people living with acute HIV infection at baseline; B0, people living with chronic HIV infection at baseline; D, HIV-uninfected controls; A12, people living with acute HIV infection after 12 weeks of ART; B12, people living with chronic HIV infection after 12 weeks of ART.
After 12 weeks of ART, comparison of pathways between HIV-infected groups and healthy controls showed that ART could partly reverse the changes of pathways in the acute and chronic HIV infection, but pathways related to metabolism, cell growth and death were still significantly increased (p<0.05), whereas transcription was significantly decreased in both A12 and B12 groups (p<0.05) (Figure 6 and Supplementary Figures S2C, D).
Our present study clearly showed that the OTUs identified in oral microbiome in people living with acute or chronic HIV infection were significantly lower compared with those in controls, and the number of OTUs in patients did not return to normal level even after 12 weeks of ART. Moreover, the alpha diversity of oral microbiomes in the patients was decreased significantly. In addition, beta diversity was also found to be different between patients and controls. Our results were in line with an earlier finding in which a decrease in microbial diversity in HIV-infected individuals was observed (Li et al., 2014). The decreased alpha diversity may be attributed to the increased proportions of opportunistic microbes as a result of immunocompromised state of patients (Li et al., 2014). Recently, Jiménez-Hernández et al. reported an increase of diversity parameters in salivary microbiota in HIV-infected individuals, mainly in those viremic ART-naive patients (Jimenez-Hernandez et al., 2019). It is conceivable that the impaired immune function resulting from HIV infection can disrupt the normally constituted commensal oral bacterial colonization, and the elevated viremia in untreated PLWH is associated with significantly higher proportions of potentially pathogenic Veillonella, Prevotella, Megasphaera, and Campylobacter species than in healthy controls (Dang et al., 2012).
In this study, the Chao1 index and Shannon index of oral microbiomes in both A0 group and B0 group were significantly decreased when compared with HIV-uninfected controls, and the significant difference remained after 12 weeks of ART. We also found that before the ART, the Chao1 index in chronic HIV-infected patients B0 group was significantly lower than that in the acute HIV-infected individuals A0 group, whereas after 12 weeks of the ART, the differences was not significant. Further, after 12 weeks of ART, the Chao1 index in B12 group significantly increased compared with the B0 group. These results suggested that the richness and diversity of oral microbiomes decreased with the progression of HIV infection. Although such reduction can be improved by ART, the oral microbiota dysbiosis cannot be fully restored after the 12-week ART in PLWH (Saxena et al., 2016; Presti et al., 2018). Moreover, there was no significant difference observed in beta diversity when comparing A0 group with B0 group, but a significant difference was noticed between groups A12 and B12. Likewise, no significant difference in beta diversity was found between A0 and A12 groups, whereas there was a significant difference between groups B0 and B12. These results further indicated that the dysbiosis in oral microbiome in people living with acute HIV infection could be partially restored if the ART is early initiated. Recent studies have also revealed that ART initiated in acute HIV infection can limit reservoir size and mitigate systemic chronic immune activation (Cheret et al., 2015; Hey-Cunningham et al., 2015). Altogether, these findings suggested that ART initiated during acute HIV infection might play an important role in protecting commensal bacterial colonization and preventing the occurrence of HIV-related oral diseases.
Alterations in oral microbiota have been noted in individuals with HIV. However, the mechanisms remain unclear and the compromised mucosal immunity might contribute to the dysbiosis of the oral microbiota in PLWH (Heron and Elahi, 2017). It is well documented that Th17 cells are depleted in HIV infection (Cote et al., 2019), however, other studies have shown that oral-resident Th17 cells can play an important role in controlling oral fungal colonization (Conti et al., 2014; Kirchner and LeibundGut-Landmann, 2021). Additionally, salivary components, such as salivary IgA, lactoferrin, defensins, and epithelial cell-mediated cytokines, are altered in PLWH, resulting in the occurrence of frequent oral infections (Muller et al., 1992). A recent study reported that the changes of salivary microbiome in HIV infection and found that Streptococcus was enriched in HIV-infected individuals, whereas the richness of Neisseria was high in healthy controls (Li et al., 2020). In present study, we observed that the abundance of unidentified Prevotellaceae in both groups of patients with acute and chronic HIV infection was increased as well as the abundances of Prevotella in acute HIV individuals and Streptococcus in chronic HIV individuals.
Interestingly, the changes observed in the oral microbiomes were similar to those in the gut microbiomes in HIV-infected patients. One possible explanation is that oral microbiome could indirectly impact gut bacteria by eating and dispersing (Schmidt et al., 2019). The gut microbes might also in turn affect the oral microbiome through microbial translocation or systemic immune regulation. In addition, it has been shown that Prevotella exhibit an increased ability to induce inflammatory mediators, such as IL-6, IL-8, and tumor necrosis factor-α (TNF-α), when compared with strict commensal oral bacteria (Larsen, 2017). The Prevotella can also promote periodontitis by driving the recruitment of neutrophil via Th17 immune responses (Uriarte et al., 2016). Another study also found that Streptococcus was enriched in AIDS patients with periodontitis (Zhang et al., 2015). Moreover, a recent study revealed a significant enrichment of Streptococcus in the saliva of HIV-infected individuals with high sCD14 levels, which might contribute to HIV-associated immune activation (Annavajhala et al., 2020). In contrast, several groups of bacteria, such as Lactobacillus, Rothia, Lautropia, and Bacteroides, were found in greater abundance in healthy controls. It has been demonstrated that Lactobacillus species can produce various antimicrobial factors including hydrogen peroxide, acetic acid, lactic acid, and bacteriocins (Spinler et al., 2008). Salari et al. also demonstrated in an in vitro study that the lactobacilli have antifungal effects on different oral Candida species isolated from HIV/AIDS patients (Salari and Ghasemi Nejad Almani, 2020). It therefore seems that HIV-induced oral microbiota dysbiosis might be characterized by an increased abundance of bacteria that are potentially inflammatory or pathogenic and a decreased abundance of bacteria that are anti-inflammatory or protective.
Although the similar oral microbiome composition has been reported between the HIV-infected individuals undergoing ART and healthy controls, there are significant differences (Li et al., 2020). In the longitudinal study, we collected samples from acute and chronic HIV-infected MSM at baseline and 12 weeks of ART. We found that Bradyrhizobium was enriched in both acute and chronic HIV-infected individuals after 12 weeks of ART, whereas Clostridia, Actinobacteria, Lactobacillus, Ruminococcaceae, and Rothia were enriched in controls. Yang et al. reported that Bradyrhizobium was enriched in the proximal gut of PLWH (Yang et al., 2016). They also found that Lactobacillus species might have a co-avoidant relationship with Bradyrhizobium pachyrhizi in the duodenum. However, little is known about the mechanisms of Bradyrhizobium colonization in PLWH, and the exact effects of ART on Bradyrhizobium colonization remain to be shown. Furthermore, we found that the abundances of potentially pathogenic bacteria, such as Prevotella histicola, Prevotella melaninogenica, and Veillonellaceae, were decreased when comparing samples collected at baseline with those collected following 12 weeks of ART in PLWH. Our data indicated that ART can partially reverse the effects of HIV on the oral bacteriome, whereas the commensal bacteria with “protective capacity in the oral cavity have been not fully recovered (Moyes et al., 2016).
After 12 weeks of ART, comparing the composition of oral microbiome in people living with acute HIV infection with that of chronic HIV infection, we found that the abundance of Campylobacter in the chronic HIV-infected individuals was significantly higher than those in acute HIV-infected individuals. Campylobacter spp are gram-negative bacteria with predominant enteric pathogenicity (Manfredi et al., 2002). Molina et al. showed that Campylobacter can cause acute diarrhea in PLWH (Molina et al., 1995). In untreated PLWH, Campylobacter species in the lingual microbiome was associated with high-level viremia (Dang et al., 2012). However, another study showed that ART increased the risk for recovering Campylobacter species in saliva of HIV-positive women (Navazesh et al., 2005).
Of note, we also observed that alterations in the oral microbiome are associated with CD4+ T-cell count in patients. The abundances of Haemophilus, Actinomyces, unidentified Ruminococcaceae, and Rothia were significantly decreased in subjects with lower CD4+ T-cell counts (<200 cells/mm3) when comparing with subjects with CD4>200 cells/mm3. It is known that Haemophilus bacteria can implicate in various opportunistic infections. A previous study reported that Haemophilus parainfluenzae was significantly associated with HIV-positive individuals, and it was positively correlated with CD4+ T cell counts within the HIV-positive group (Kistler et al., 2015). However, a recent study found that the genus Haemophilus was correlated negatively with CD4+ T cell count (Li et al., 2020). The possible reasons for the different results might include the different study subjects, samples, the small sample sizes, and other possible factors not involved in the analysis.
One of the limitations of this study was the small sample size. Clearly, studies with large sample size are required. Other factors, such as age, might also affect the results of our study. In addition, previous studies have indicated that oral fungal colonization is altered in HIV-infected individuals (Mukherjee et al., 2014; Hager and Ghannoum, 2018). Although the occurrence of OPC in HIV infection has significantly declined since the introduction of ART, it remains a common opportunistic infection in AIDS diseases (Thompson et al., 2010; Patil et al., 2018). The present study was focused on the oral bacterial community in HIV infection, future studies should also address how the oral mycobiome shifts in the setting of HIV infection and ART initiation.
In conclusion, this longitudinal study has shown important alterations in oral microbiome resulting from HIV infection as well as among MSM with acute and chronic HIV infection before and after ART. The findings might contribute to improved oral health in HIV-infected individuals and provide some clues to exploring the association of the oral and the intestinal microbiome during the disease courses.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA PRJNA739016.
The studies involving human participants were reviewed and approved by the Ethics Committee of the Beijing Youan Hospital ([2018]025). The patients/participants provided their written informed consent to participate in this study.
All authors made a substantial, direct, and intellectual contribution to the work, including the study design, subject recruitment, sample collection, laboratory experiments, data analysis, and manuscript drafting. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (82072271, 81772165, and 81974303), the National 13th Five-Year Grand Program on Key Infectious Disease Control (2017ZX10202101-004-001, 2017ZX10202102-005-003), Beijing Natural Science Foundation (7172016), the NSFC-NIH Biomedical collaborative research program (81761128001), the Beijing Key Laboratory for HIV/AIDS Research (BZ0089), and Beijing Natural Science Foundation and Handian Innovation Joint Project (19L2043).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank all of the staff from Beijing Youan Hospital, Capital Medical University, and Novogene who worked on this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.695515/full#supplementary-material
Annavajhala, M. K., Khan, S. D., Sullivan, S. B., Shah, J., Pass, L., Kister, K., et al. (2020). Oral and Gut Microbial Diversity and Immune Regulation in Patients with HIV on Antiretroviral Therapy. mSphere 5 (1), e00798-19. doi: 10.1128/mSphere.00798-19
Armstrong, A. J. S., Shaffer, M., Nusbacher, N. M., Griesmer, C., Fiorillo, S., Schneider, J. M., et al. (2018). An Exploration of Prevotella-Rich Microbiomes in HIV and Men Who Have Sex With Men. Microbiome 6, 198. doi: 10.1186/s40168-018-0580-7
Brenchley, J. M., Price, D. A., Schacker, T. W., Asher, T. E., Silvestri, G., Rao, S., et al. (2006). Microbial Translocation is a Cause of Systemic Immune Activation in Chronic HIV Infection. Nat. Med. 12, 1365–1371. doi: 10.1038/nm1511
Cheret, A., Bacchus-Souffan, C., Avettand-Fenoel, V., Melard, A., Nembot, G., Blanc, C., et al. (2015). Combined ART Started During Acute HIV Infection Protects Central Memory CD4+ T Cells and can Induce Remission. J. Antimicrob. Chemother. 70, 2108–2120. doi: 10.1093/jac/dkv084
Conti, H. R., Peterson, A. C., Brane, L., Huppler, A. R., Hernandez-Santos, N., Whibley, N., et al. (2014). Oral-Resident Natural Th17 Cells and Gammadelta T Cells Control Opportunistic Candida Albicans Infections. J. Exp. Med. 211, 2075–2084. doi: 10.1084/jem.20130877
Cote, S. C., Stilla, A., Burke Schinkel, S. C., Berthoud, T. K., Angel, J. B. (2019). IL-7-Induced Proliferation of Peripheral Th17 Cells Is Impaired in HAART-Controlled HIV Infection. AIDS 33, 985–991. doi: 10.1097/QAD.0000000000002164
Dang, A. T., Cotton, S., Sankaran-Walters, S., Li, C. S., Lee, C. Y., Dandekar, S., et al. (2012). Evidence of an Increased Pathogenic Footprint in the Lingual Microbiome of Untreated HIV Infected Patients. BMC Microbiol. 12:153. doi: 10.1186/1471-2180-12-153
Dillon, S. M., Lee, E. J., Kotter, C. V., Austin, G. L., Dong, Z., Hecht, D. K., et al. (2014). An Altered Intestinal Mucosal Microbiome in HIV-1 Infection Is Associated With Mucosal and Systemic Immune Activation and Endotoxemia. Mucosal Immunol. 7, 983–994. doi: 10.1038/mi.2013.116
Dinh, D. M., Volpe, G. E., Duffalo, C., Bhalchandra, S., Tai, A. K., Kane, A. V., et al. (2015). Intestinal Microbiota, Microbial Translocation, and Systemic Inflammation in Chronic HIV Infection. J. Infect. Dis. 211, 19–27. doi: 10.1093/infdis/jiu409
El Howati, A., Tappuni, A. (2018). Systematic Review of the Changing Pattern of the Oral Manifestations of HIV. J. Investig. Clin. Dent. 9, e12351. doi: 10.1111/jicd.12351
Fulcher, J. A. (2020). Is the Oral Microbiome Important in HIV-Associated Inflammation? mSphere 5 (1), e00034-20. doi: 10.1128/mSphere.00034-20
Goldberg, B. E., Mongodin, E. F., Jones, C. E., Chung, M., Fraser, C. M., Tate, A., et al. (2015). The Oral Bacterial Communities of Children With Well-Controlled HIV Infection and Without HIV Infection. PloS One 10, e0131615. doi: 10.1371/journal.pone.0131615
Griffen, A. L., Thompson, Z. A., Beall, C. J., Lilly, E. A., Granada, C., Treas, K. D., et al. (2019). Significant Effect of HIV/HAART on Oral Microbiota Using Multivariate Analysis. Sci. Rep. 9, 19946. doi: 10.1038/s41598-019-55703-9
Hager, C. L., Ghannoum, M. A. (2018). The Mycobiome in HIV. Curr. Opin. HIV AIDS 13, 69–72. doi: 10.1097/COH.0000000000000432
Heron, S. E., Elahi, S. (2017). HIV Infection and Compromised Mucosal Immunity: Oral Manifestations and Systemic Inflammation. Front. Immunol. 8:241. doi: 10.3389/fimmu.2017.00241
Hey-Cunningham, W. J., Murray, J. M., Natarajan, V., Amin, J., Moore, C. L., Emery, S., et al. (2015). Early Antiretroviral Therapy With Raltegravir Generates Sustained Reductions in HIV Reservoirs But Not Lower T-Cell Activation Levels. AIDS 29, 911–919. doi: 10.1097/QAD.0000000000000625
Jimenez-Hernandez, N., Serrano-Villar, S., Domingo, A., Pons, X., Artacho, A., Estrada, V., et al. (2019). Modulation of Saliva Microbiota through Prebiotic Intervention in HIV-Infected Individuals. Nutrients 11 (6), 1346. doi: 10.3390/nu11061346
Kirchner, F. R., LeibundGut-Landmann, S. (2021). Tissue-Resident Memory Th17 Cells Maintain Stable Fungal Commensalism in the Oral Mucosa. Mucosal Immunol. 14, 455–467. doi: 10.1038/s41385-020-0327-1
Kistler, J. O., Arirachakaran, P., Poovorawan, Y., Dahlen, G., Wade, W. G. (2015). The Oral Microbiome in Human Immunodeficiency Virus (HIV)-Positive Individuals. J. Med. Microbiol. 64, 1094–1101. doi: 10.1099/jmm.0.000128
Koay, W. L. A., Siems, L. V., Persaud, D. (2018). The Microbiome and HIV Persistence: Implications for Viral Remission and Cure. Curr. Opin. HIV AIDS 13, 61–68. doi: 10.1097/COH.0000000000000434
Larsen, J. M. (2017). The Immune Response to Prevotella Bacteria in Chronic Inflammatory Disease. Immunology 151, 363–374. doi: 10.1111/imm.12760
Li, J., Chang, S., Guo, H., Ji, Y., Jiang, H., Ruan, L., et al. (2020). Altered Salivary Microbiome in the Early Stage of HIV Infections among Young Chinese Men Who Have Sex with Men (MSM). Pathogens 9 (1), 960. doi: 10.3390/pathogens9110960
Li, Y., Saxena, D., Chen, Z., Liu, G., Abrams, W. R., Phelan, J. A., et al. (2014). HIV Infection and Microbial Diversity in Saliva. J. Clin. Microbiol. 52, 1400–1411. doi: 10.1128/JCM.02954-13
Manfredi, R., Calza, L., Chiodo, F. (2002). Enteric and Disseminated Campylobacter Species Infection During HIV Disease: A Persisting But Significantly Modified Association in the HAART Era. Am. J. Gastroenterol. 97, 510–511. doi: 10.1111/j.1572-0241.2002.05522.x
Marchetti, G., Tincati, C., Silvestri, G. (2013). Microbial Translocation in the Pathogenesis of HIV Infection and AIDS. Clin. Microbiol. Rev. 26, 2–18. doi: 10.1128/CMR.00050-12
Molina, J., Casin, I., Hausfater, P., Giretti, E., Welker, Y., Decazes, J., et al. (1995). Campylobacter Infections in HIV-Infected Patients: Clinical and Bacteriological Features. AIDS 9, 881–885. doi: 10.1097/00002030-199508000-00008
Moyes, D. L., Saxena, D., John, M. D., Malamud, D. (2016). The Gut and Oral Microbiome in HIV Disease: A Workshop Report. Oral. Dis. 22 Suppl 1, 166–170. doi: 10.1111/odi.12415
Mudd, J. C., Brenchley, J. M. (2016). Gut Mucosal Barrier Dysfunction, Microbial Dysbiosis, and Their Role in HIV-1 Disease Progression. J. Infect. Dis. 214 Suppl 2, S58–S66. doi: 10.1093/infdis/jiw258
Mukherjee, P. K., Chandra, J., Retuerto, M., Sikaroodi, M., Brown, R. E., Jurevic, R., et al. (2014). Oral Mycobiome Analysis of HIV-Infected Patients: Identification of Pichia as an Antagonist of Opportunistic Fungi. PloS Pathog. 10, e1003996. doi: 10.1371/journal.ppat.1003996
Muller, F., Holberg-Petersen, M., Rollag, H., Degre, M., Brandtzaeg, P., Froland, S. S. (1992). Nonspecific Oral Immunity in Individuals With HIV Infection. J. Acquir. Immune Defic. Syndr. (1988) 5 (1), 46–51.
Mutlu, E. A., Keshavarzian, A., Losurdo, J., Swanson, G., Siewe, B., Forsyth, C., et al. (2014). A Compositional Look at the Human Gastrointestinal Microbiome and Immune Activation Parameters in HIV Infected Subjects. PloS Pathog. 10, e1003829. doi: 10.1371/journal.ppat.1003829
Navazesh, M., Mulligan, R., Pogoda, J., Greenspan, D., Alves, M., Phelan, J., et al. (2005). The Effect of HAART on Salivary Microbiota in the Women’s Interagency HIV Study (WIHS). Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. Endod. 100, 701–708. doi: 10.1016/j.tripleo.2004.10.011
Noguera-Julian, M., Rocafort, M., Guillen, Y., Rivera, J., Casadella, M., Nowak, P., et al. (2016). Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine 5, 135–146. doi: 10.1016/j.ebiom.2016.01.032
Patil, S., Majumdar, B., Sarode, S. C., Sarode, G. S., Awan, K. H. (2018). Oropharyngeal Candidosis in HIV-Infected Patients-An Update. Front. Microbiol. 9:980. doi: 10.3389/fmicb.2018.00980
Presti, R. M., Handley, S. A., Droit, L., Ghannoum, M., Jacobson, M., Shiboski, C. H., et al. (2018). Alterations in the Oral Microbiome in HIV-Infected Participants After Antiretroviral Therapy Administration Are Influenced by Immune Status. AIDS 32, 1279–1287. doi: 10.1097/QAD.0000000000001811
Rocafort, M., Noguera-Julian, M., Rivera, J., Pastor, L., Guillen, Y., Langhorst, J., et al. (2019). Evolution of the Gut Microbiome Following Acute HIV-1 Infection. Microbiome 7, 73. doi: 10.1186/s40168-019-0687-5
Salari, S., Ghasemi Nejad Almani, P. (2020). Antifungal Effects of Lactobacillus Acidophilus and Lactobacillus Plantarum Against Different Oral Candida Species Isolated From HIV/AIDS Patients: An In Vitro Study. J. Oral. Microbiol. 12:1769386. doi: 10.1080/20002297.2020.1769386
Samaranayake, L., Matsubara, V. H. (2017). Normal Oral Flora and the Oral Ecosystem. Dent. Clin. North Am. 61, 199–215. doi: 10.1016/j.cden.2016.11.002
Saxena, D., Li, Y., Devota, A., Pushalkar, S., Abrams, W., Barber, C., et al. (2016). Modulation of the Orodigestive Tract Microbiome in HIV-Infected Patients. Oral. Dis. 22 Suppl 1, 73–78. doi: 10.1111/odi.12392
Schmidt, T. S., Hayward, M. R., Coelho, L. P., Li, S. S., Costea, P. I., Voigt, A. Y., et al. (2019). Extensive Transmission of Microbes Along the Gastrointestinal Tract. Elife 8, e42693. doi: 10.7554/eLife.42693
Spinler, J. K., Taweechotipatr, M., Rognerud, C. L., Ou, C. N., Tumwasorn, S., Versalovic, J. (2008). Human-Derived Probiotic Lactobacillus Reuteri Demonstrate Antimicrobial Activities Targeting Diverse Enteric Bacterial Pathogens. Anaerobe 14, 166–171. doi: 10.1016/j.anaerobe.2008.02.001
Sun, Y., Ma, Y., Lin, P., Tang, Y. W., Yang, L., Shen, Y., et al. (2016). Fecal Bacterial Microbiome Diversity in Chronic HIV-Infected Patients in China. Emerg. Microbes Infect. 5, e31. doi: 10.1038/emi.2016.25
Thompson, G. R., 3rd, Patel, P. K., Kirkpatrick, W. R., Westbrook, S. D., Berg, D., Erlandsen, J., et al. (2010). Oropharyngeal Candidiasis in the Era of Antiretroviral Therapy. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. Endod. 109, 488–495. doi: 10.1016/j.tripleo.2009.11.026
Uriarte, S. M., Edmisson, J. S., Jimenez-Flores, E. (2016). Human Neutrophils and Oral Microbiota: A Constant Tug-of-War Between a Harmonious and a Discordant Coexistence. Immunol. Rev. 273, 282–298. doi: 10.1111/imr.12451
Yang, L., Poles, M. A., Fisch, G. S., Ma, Y., Nossa, C., Phelan, J. A., et al. (2016). HIV-Induced Immunosuppression Is Associated With Colonization of the Proximal Gut by Environmental Bacteria. AIDS 30, 19–29. doi: 10.1097/QAD.0000000000000935
Zevin, A. S., McKinnon, L., Burgener, A., Klatt, N. R. (2016). Microbial Translocation and Microbiome Dysbiosis in HIV-Associated Immune Activation. Curr. Opin. HIV AIDS 11, 182–190. doi: 10.1097/COH.0000000000000234
Zhang, F., He, S., Jin, J., Dong, G., Wu, H. (2015). Exploring Salivary Microbiota in AIDS Patients With Different Periodontal Statuses Using 454 GS-FLX Titanium Pyrosequencing. Front. Cell Infect. Microbiol. 5:55. doi: 10.3389/fcimb.2015.00055
Keywords: human immunodeficiency virus, oral microbiome, 16S rRNA sequencing, antiretroviral therapy, men who have sex with men
Citation: Li S, Zhu J, Su B, Wei H, Chen F, Liu H, Wei J, Yang X, Zhang Q, Xia W, Wu H, He Q and Zhang T (2021) Alteration in Oral Microbiome Among Men Who Have Sex With Men With Acute and Chronic HIV Infection on Antiretroviral Therapy. Front. Cell. Infect. Microbiol. 11:695515. doi: 10.3389/fcimb.2021.695515
Received: 15 April 2021; Accepted: 24 June 2021;
Published: 14 July 2021.
Edited by:
Yulong Niu, Sichuan University, ChinaReviewed by:
Pengfan Zhang, Max Planck Institute for Plant Breeding Research, GermanyCopyright © 2021 Li, Zhu, Su, Wei, Chen, Liu, Wei, Yang, Zhang, Xia, Wu, He and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tong Zhang, enRfZG9jQGNjbXUuZWR1LmNu; Qiushui He, cWl1aGVAdXR1LmZp
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.