
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 24 June 2021
Sec. Molecular Bacterial Pathogenesis
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.694789
This article is part of the Research Topic Pseudomonas aeruginosa Pathogenesis: Virulence, Antibiotic Tolerance and Resistance, Stress Responses and Host-pathogen Interactions. View all 7 articles
Pseudomonas aeruginosa is a metabolically versatile opportunistic pathogen capable of infecting distinct niches of the human body, including skin wounds and the lungs of cystic fibrosis patients. Eradication of P. aeruginosa infection is becoming increasingly difficult due to the numerous resistance mechanisms it employs. Adaptive resistance is characterized by a transient state of decreased susceptibility to antibiotic therapy that is distinct from acquired or intrinsic resistance, can be triggered by various environmental stimuli and reverted by removal of the stimulus. Further, adaptive resistance is intrinsically linked to lifestyles such as swarming motility and biofilm formation, both of which are important in infections and lead to multi-drug adaptive resistance. Here, we demonstrated that NtrBC, the master of nitrogen control, had a selective role in host colonization and a substantial role in determining intrinsic resistance to ciprofloxacin. P. aeruginosa mutant strains (ΔntrB, ΔntrC and ΔntrBC) colonized the skin but not the respiratory tract of mice as well as WT and, unlike WT, could be reduced or eradicated from the skin by ciprofloxacin. We hypothesized that nutrient availability contributed to these phenomena and found that susceptibility to ciprofloxacin was impacted by nitrogen source in laboratory media. P. aeruginosa ΔntrB, ΔntrC and ΔntrBC also exhibited distinct host interactions, including modestly increased cytotoxicity toward human bronchial epithelial cells, reduced virulence factor production and 10-fold increased uptake by macrophages. These data might explain why NtrBC mutants were less adept at colonizing the upper respiratory tract of mice. Thus, NtrBC represents a link between nitrogen metabolism, adaptation and virulence of the pathogen P. aeruginosa, and could represent a target for eradication of recalcitrant infections in situ.
Pseudomonas aeruginosa is a highly adaptable opportunistic pathogen that is most commonly associated with severe, often fatal, nosocomial infections of the bloodstream (Tschudin-Sutter et al., 2018), urinary tract (Ferreiro et al., 2017) and upper respiratory tract of cystic fibrosis (CF) patients (Winstanley et al., 2016). Eradication of P. aeruginosa has become very difficult due to its capacity to resist conventional antibiotic therapy through various intrinsic, acquired and adaptive mechanisms (Pang et al., 2019). Adaptive resistance is particularly troubling since it is not genetically encoded and is induced by a wide array of environmental stimuli including antibiotic exposure, pH stress, anaerobiosis, and starvation (Breidenstein et al., 2011). Importantly, adaptive resistance is distinct from tolerance and persistence, which are characterized by metabolic dormancy that enable bacterial survival but not growth in the presence of antibiotics (Brauner et al., 2016). In contrast, adaptively multi-drug resistant cells actively divide and emerge during social lifestyles such as swarming motility and biofilm formation (Moradali et al., 2017; Coleman et al., 2020a).
Swarming is a complex adaptation involving rapid surface motility in which groups of cells collectively move using flagella and type IV pili to propel themselves over semi-solid agar in the presence of a weak nitrogen source, assisted by production of surface-wetting rhamnolipids (Yeung et al., 2009). Swarming is involved in bacterial dissemination from localized infection sites to distal organs (Coleman et al., 2020b) and is associated with increased resistance to most conventional antibiotics including ciprofloxacin, tobramycin and tetracycline (Coleman et al., 2020a). Similarly, surface-attached matrix-encapsulated communities of cells called biofilms exhibit up to 1000-fold higher (adaptive) multiple-antibiotic resistance than their planktonic counterparts (Olivares et al., 2020). Due to their ubiquity in nature, biofilms are often considered to represent the preferred growth state for bacteria in the environment and are implicated in more than 60% of all infections, often leading to chronic, recurrent infections (Flemming et al., 2016). The molecular mechanisms underlying swarming- and biofilm-mediated multi-drug resistance are not fully understood but are known to be multifactorial and largely independent of canonical resistance mechanisms (Lai et al., 2009; Ciofu and Tolker-Nielsen, 2019; Coleman et al., 2020a). A better understanding of these mechanisms is warranted for development of novel treatment strategies that target adaptive processes and dismantle bacterial virulence in vivo (Alford et al., 2019).
Adding to their complexity, swarming and biofilm lifestyles exhibit plasticity driven by environmental and genetic determinants (Kollaran et al., 2019; Thöming et al., 2020). For example, P. aeruginosa swarming is conserved but distinct on various nitrogen sources independent of growth differences (Alford et al., 2020). These nitrogen sources include nitrate, which is abundant in the lungs of CF patients (up to 400 μM; Line et al., 2014), as well as glutamate, which is also abundant in CF sputum (Palmer et al., 2005) and several other anatomical niches. Similarly, antibiotic susceptibility is dependent on central metabolism and can be potentiated or inhibited by addition of dicarboxylates such as fumarate or glyoxylate, respectively (Meylan et al., 2017; Hall et al., 2019). Tight regulation of gene expression through two-component system signaling allows P. aeruginosa to rapidly respond to environmental heterogeneity, contributing to its adaptability (Kollaran et al., 2019). NtrBC is a two-component system that regulates swarming motility, biofilm formation and nitrogen metabolism as well as invasiveness and virulence in a murine model of high-density infection (Alford et al., 2020). Since the transcriptional profile of NtrB and NtrC mutants demonstrated that genes involved in ciprofloxacin resistance were differentially expressed, we hypothesized they would be more susceptible in vitro. While confirming this data in vivo, we observed that, in the absence of ciprofloxacin, NtrBC mutants colonized the respiratory tracts, but not the skin, of mice to a lesser extent than WT. We hypothesized this effect was due to different availability of nitrogen in tissues and explored the impact of nitrogenous species on biofilm formation in vitro to confirm this. Furthermore, we demonstrated that NtrBC regulated host-directed cytotoxicity, virulence factor production and macrophage-mediated uptake, which might also contribute to differential colonization of host tissues.
Bacterial strains and plasmids used in this study are described in Table S1. Overnight cultures were routinely maintained in Luria-Bertani (LB) broth prepared according to the manufacturer’s specifications (Thermo Scientific). Overnight and sub-cultures were incubated for no longer than 18 h at 37°C with shaking (250 rpm). Modified forms of basal medium (BM2; containing 62 mM potassium phosphate buffer (pH = 7.0), 0.1% casamino acids (CAA), 2 mM MgSO4, 10 μM FeSO4, 20 mM glucose) were used for biofilm assays, kill curves and minimal inhibitory concentration (MIC) assays. For testing the influence of nitrogen on biofilm formation and ciprofloxacin killing, equimolar concentrations of nitrate , nitrite , glutamate (Glu) or urea replaced 0.1% casamino acids (CAA). Other media used in specific assays are described elsewhere. For plasmid selection in P. aeruginosa WT strains PA14 and LESB58, 50 μg/ml and 500 μg/ml gentamicin (Gm) was added to growth media. Bacterial growth was monitored by measuring optical density (OD600) with a spectrophotometer (Eppendorf, Missisauga, ON).
The Simian virus 40 (SV40)-transformed, immortalized human bronchial epithelial cell line 16HBE14o− (HBE) was used for the cytotoxicity assay. HBE cells were cultured in minimum essential medium with Earle’s Salts (MEM) supplemented with 10% fetal bovine serum (FBS, Gibco) and 2 mM L-glutamine (Gibco). The cell line was routinely cultured to 85 to 90% confluence in 100% humidity and 5% CO2 at 37°C and used between passages 9-15.
The human monocytic-like cell line THP-1 was obtained from the American Type Culture Collection (ATCC) and was routinely cultured in RPMI-1640 supplemented with 2 mM L-glutamine and 10% heat-inactivated FBS. Cells were differentiated into mature macrophages by stimulation with 100 ng/ml phorbol-12-myristate-13-acetate (PMA [P1585; Sigma]) for 48 h and then replaced with fresh medium without PMA for 24 h prior to the assay. The cell line was routinely cultured to 85 to 90% confluence in 100% humidity and 5% CO2 at 37°C.
Broth microdilution assays were performed according to the standard protocol outlined by the Clinical and Laboratory Standards Institute (CLSI, 2020) with minor modifications. Bacteria were seeded at ~105 CFU/ml in a 2-fold concentration gradient of antibiotic in Mueller-Hinton Broth (MHB) or BM2 with N-source, as indicated, at 200 μl/well in 96-well polystyrene flat bottom plates. Plates were incubated for 18 h at 37°C. MIC of antibiotics was determined as the lowest concentration that visibly inhibited bacterial growth.
Overnight cultures were diluted to a starting OD600 = 0.1 in 5 ml MHB. Cultures were grown to mid-log phase (OD600 = 0.4-0.6) at 37°C with aeration, and then treated with a high concentration of ciprofloxacin (25 μg/ml). Aliquots were taken every 30 or 60 min for up to 240 min following inoculation, diluted in PBS, pH 7.4 and plated on LB medium plates for bacterial enumeration.
Animal experiments were performed in accordance with the Canadian Council on Animal Care (CCAC) guidelines and were approved by the University of British Columbia Animal Care Committee (protocol A17-0253). Mice used in this study were outbred C57Bl/6 mice (female, aged 11-13 weeks) or CD-1 mice (female, aged 5-7 weeks). All animals were purchased from Charles River Laboratories, Inc. (Wilmington, MA). C57Bl/6 and CD-1 mice weighed 20 ± 5 g and 25 ± 5 g, respectively, at the time of experiment and were group housed in cohorts of 4-5 littermates exposed to the same bacterial strain. Littermates were randomly assigned to experimental groups. Standard animal husbandry protocols were employed.
Ciprofloxacin susceptibility of LESB58 WT and mutants was assessed in vivo using a nuanced subcutaneous abscess model, as previously described (Pletzer et al., 2017). Bacterial cultures were grown to an OD600 of 1.0 in LB, washed twice in sterile PBS and resuspended to give a final inoculum of ~5 x 107 CFU (in 50 μl). Bacteria were injected subcutaneously into the left dorsum of CD-1 mice and treated 1 h later with 50 μl of 0.2 mg/ml ciprofloxacin in endotoxin-free water. Abscesses were formed for 72 h, visible dermonecrosis was measured using a caliper at experimental endpoint and abscesses were harvested in PBS for bacterial enumeration on LB.
Sinusitis infections were performed as previously described (Alford et al., 2021). Briefly, bacterial subcultures of P. aeruginosa LESB58 WT and mutant strains were washed twice with sterile PBS and resuspended at an OD600 of 1.0. Bacteria were instilled (20 μl), dropwise, into the left naris of C57Bl/6 mice under anesthesia (2.5% isoflurane) at 106 CFU. At the experimental endpoint, mice were euthanized and nasal lavage as well as excision of lungs were performed for bacterial enumeration.
PA14 WT was examined for biofilm formation in the presence of different nitrogen (N)-sources using a high-throughput microtiter assay as described elsewhere (Fuente-Nunez et al., 2013). Overnight cultures were diluted to a starting OD600 = 0.1 in BM2 medium with 20 mM glucose and equimolar amounts of different N-sources and grown in polypropylene 96-well plates (Falcon). Following 18-24 h static incubation at 37°C, biomass was stained with 0.1% CV and dissolved in 70% ethanol. The OD595 was read using a BioTek SynergyH1 microplate reader (BioTek, Winooski, VT). Three independent experiments containing three biological replicates each were performed.
Pyoverdine was assessed as previously described (Imperi et al., 2009). Briefly, bacteria were incubated in Casamino acid medium (0.5% CAA, 0.1 mM MgSO4, 0.4% glucose, 7 mM potassium phosphate buffer, pH = 7.0) at 37°C (250 rpm). Turbid cultures were pelleted, and the supernatant was collected in a fresh microfuge tube. Five μl of supernatant was mixed with 995 μl 10 mM Tris-HCl (pH = 6.8). Fluorescence was measured at an excitation wavelength of 400 nm and emission 460 nm (Synergy H1 Microplate Reader, Biotek). Pyocyanin concentrations were determined spectrophotometrically after extraction with chloroform and 0.2 M HCl as described elsewhere (Essar et al., 1990). Absorbance at 520 nm was read (Synergy H1 Microplate Reader, Biotek). Elastase was determined by proteolysis of the Elastin-Congo red complex (Sigma) as described elsewhere (Ohman et al., 1980). Five hundred μl of supernatant from cultures grown for 16 h was collected, added to 10 mg/ml Elastin-Congo red in PBS (pH = 7.4) and incubated at 37°C (250 rpm) for 8 h. Absorbance of the aqueous fraction was examined at 495 nm (Synergy H1 Microplate Reader, Biotek).
Confluent HBE cells were washed once with PBS, pH 7.4 (Gibco), removed by trypsinization with 0.25% Trypsin-EDTA (Gibco) and counted. HBE cells were seeded at ~7.5 x 105 cells/well in 500 μl in a 24 well plate and grown again to confluency (2-3 days). Then, medium (MEM, 1% FBS, 2 mM L-glutamine) was refreshed and cells were rested for 1 h. Bacterial cultures grown to mid-log phase were pelleted, washed once with PBS and resuspended in medium. Bacterial cells were added to host cells at an MOI = 1, and co-cultures were maintained at 37°C with 5% CO2 for 12-16 h. HBE cell cytotoxicity was evaluated by measuring the release of lactate dehydrogenase (LDH) into the supernatant as previously described (Kumar et al., 2018).
Macrophage-mediated uptake of PA14 WT and mutants was performed as described (Yeung et al., 2014) with minor modifications. Briefly, cultures were grown to mid-log phase (OD600 = 0.4-0.6), then washed with RPMI-1640 and resuspended in 1 ml of the medium. Mature macrophages were seeded in 24 well plates at ~3.5 × 105 cells/well. Bacteria were added at an MOI of 10 and incubated for 1 h at 37°C since preliminary studies revealed that cytotoxicity was low at this point. Cells were washed with PBS and treated with 400 μg/ml gentamicin for 30 min at 37°C to remove residual bacteria from the well. Following treatment, macrophages were again washed, then left to rest for an additional 30-60 min or lysed with 0.1% Triton X-100. Macrophage lysate was plated onto LB agar for bacterial enumeration at 30, 60 or 90 min.
Statistics were performed using GraphPad Prism 8.0 (La Jolla, CA). P values were calculated as stated in the Figure captions. Statistical significance was established when P < 0.05.
The two-component regulatory system NtrBC was recently shown to impact on the invasiveness of P. aeruginosa in an in vivo model as well as rhamnolipid synthesis, swarming motility and biofilm formation (Alford et al., 2020). We assessed whether it also directly impacted on susceptibility to antibiotics in MHB, a standard medium for determining MIC (Table 1). The MIC of the fluoroquinolone ciprofloxacin was 8-fold lower for ΔntrB and ΔntrC strains and ≥16-fold lower for ΔntrBC. However, there were no significant differences in the MICs of tobramycin, chloramphenicol or tetracycline between P. aeruginosa PA14 strains.
We then tested whether NtrBC directly impacted on the rate of killing of P. aeruginosa PA14 by treating mid-log phase cultures with a high concentration (25 µg/ml) of ciprofloxacin in MHB (Figure 1). Deletion of the entire two-component system (ΔntrBC) resulted in significantly reduced viability by 2-log10 within 30 min of ciprofloxacin treatment and in most cases complete killing within 30-60 min of treatment, cf. WT that required 120 min for all bacteria to be killed. Deletion of just the response regulator (ΔntrB) or the sensor kinase (ΔntrC) from the chromosome of PA14 had no significant impact on the rate of killing. Consistent with this, N-source impacted on swarming-mediated resistance to ciprofloxacin (Figure S1 and Table S2) exhibited by P. aeruginosa.
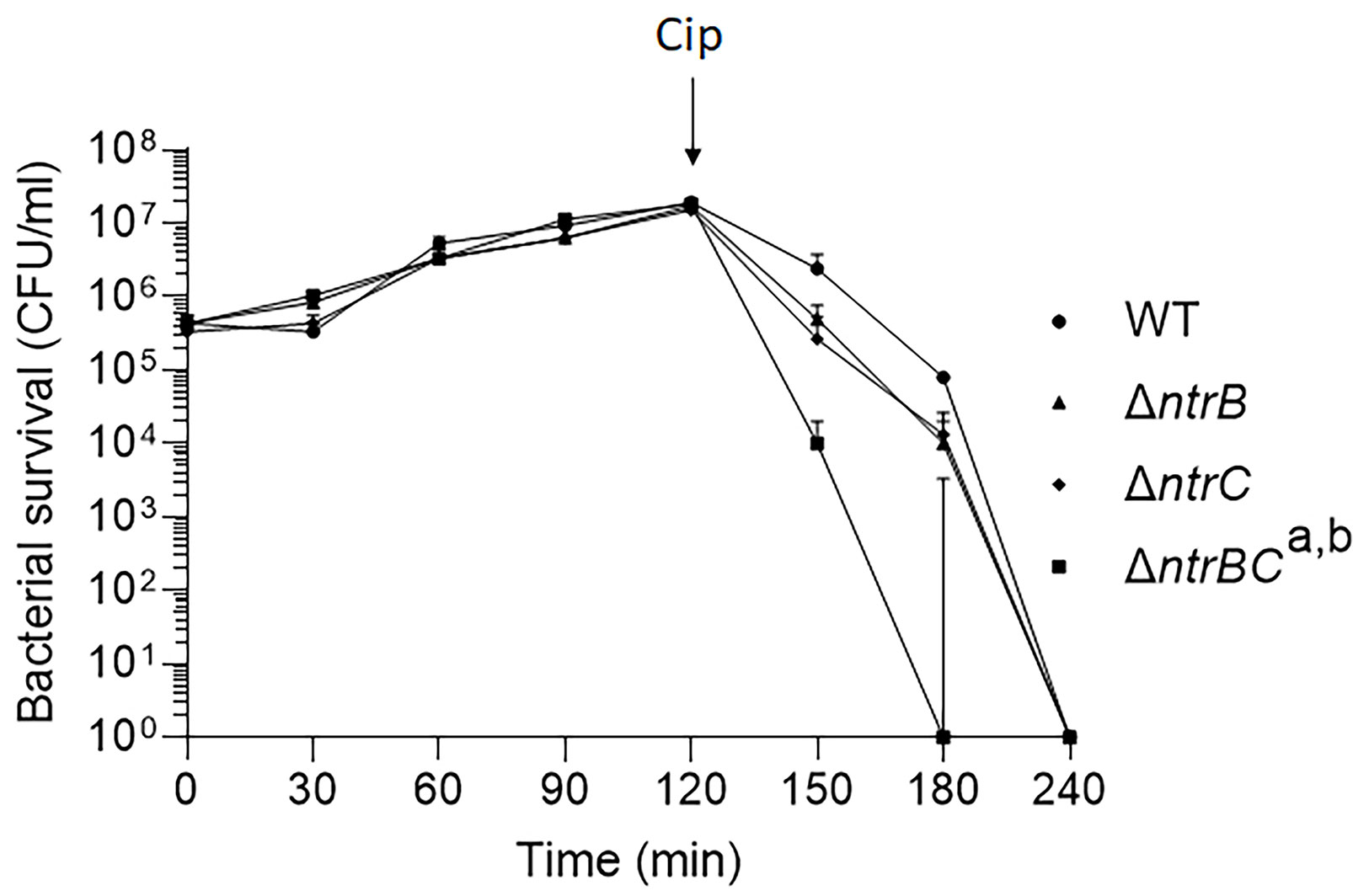
Figure 1 Mutation of the nitrogen regulatory two-component system, NtrBC, increased P. aeruginosa susceptibility to ciprofloxacin. Kill curves were performed in MHB after addition of 25 µg/ml ciprofloxacin at t = 120 min. Data are reported as mean ± standard error of the mean (SEM) from three independent experiments performed in duplicate. a,bP < 0.05 at t = 150 min and t = 180 min according to Kruskal Wallis nonparametric test followed by Dunn’s post-hoc analysis.
To explain the mechanism by which NtrBC influenced susceptibility to ciprofloxacin, we utilized published data (Alford et al., 2020) to examine the differential expression of genes in mutants belonging to the ciprofloxacin resistome (Table 2). Compared to WT, 17 of 287 ciprofloxacin resistome genes were downregulated in ΔntrB and/or ΔntrC strains.
The most downregulated ciprofloxacin resistome gene was lhpH, the product of which is implicated in 4-hydroxyproline catabolism, that was expressed 16.9- and 9.7-fold less in ΔntrB and ΔntrC when compared to WT, respectively. Other substantially downregulated resistome genes encoded a putative allophanate hydrolase subunit with a carboxytransferase domain (PA14_37310/PA2110), a flavohemoprotein (fhp), a threonine dehydratase biosynthetic protein (ilvA2).
NtrBC was previously shown to be required for invasion in a murine abscess model of high-density infection (Alford et al., 2020). However, there were no differences in bacterial loads between P. aeruginosa LESB58 strains deleted for ntrB, ntrC or both. Since we observed that NtrBC is needed for resistance to ciprofloxacin in vitro, and ciprofloxacin was able to eradicate CF lung infection in a clinical trial (Hewer et al., 2020), we tested the ability of ciprofloxacin to kill these mutants in vivo (Figure 2). Treatment of WT with 0.1 mg ciprofloxacin did not significantly reduce abscess size when compared to untreated cells. Consistent with prior observations (Alford et al., 2020), deletion of ntrB, ntrC or both also failed to significantly reduce bacterial load compared to WT in the absence of treatment. However, mutants were more susceptible to ciprofloxacin in vivo. NtrBC mutants were reduced 4-log10 compared to WT following ciprofloxacin treatment.
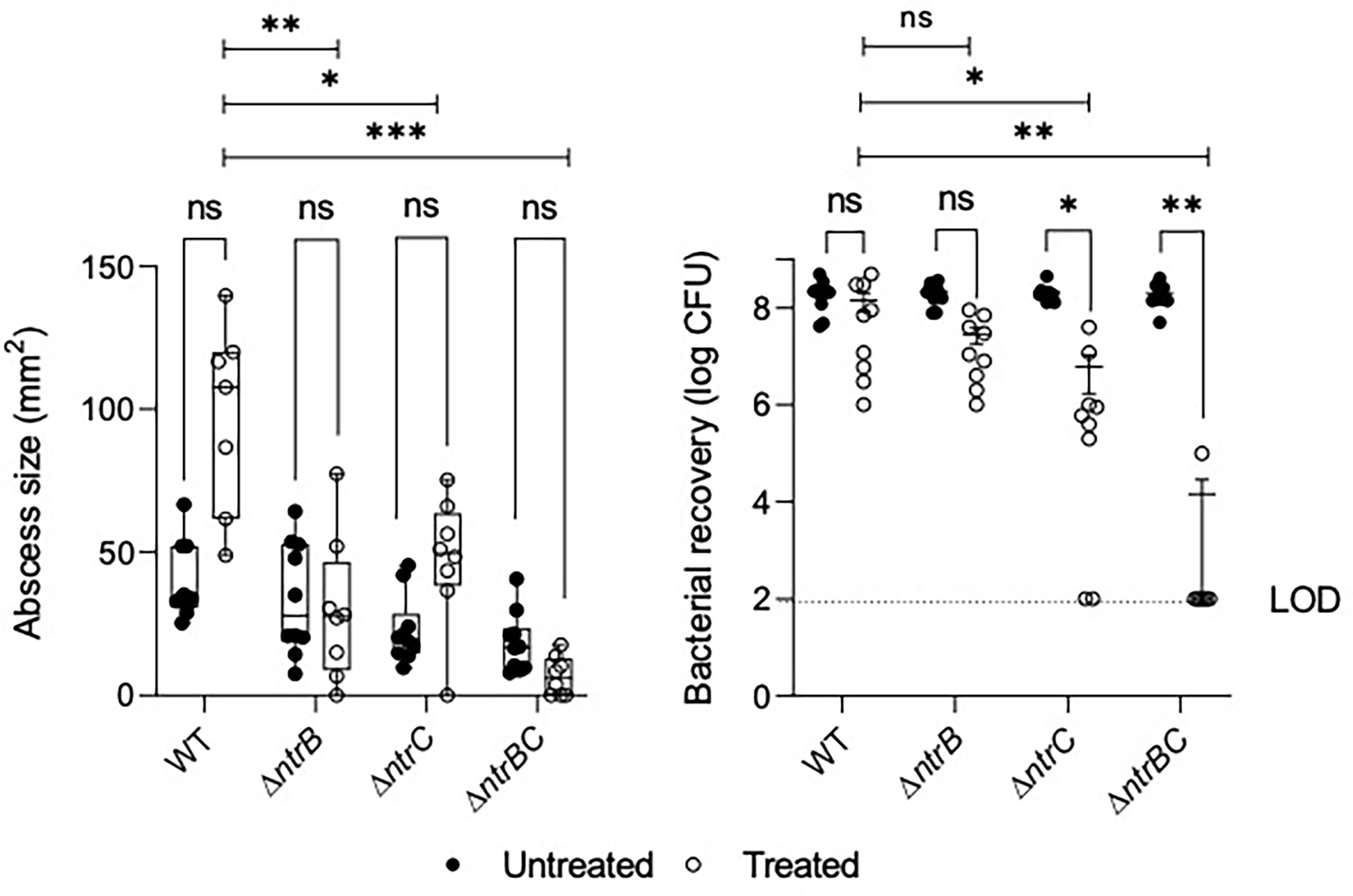
Figure 2 Mutation of the nitrogen regulatory two-component system, NtrBC, sensitized P. aeruginosa LESB58 to ciprofloxacin treatment in vivo. Abscesses formed by LESB58 ΔntrBC following ciprofloxacin treatment (0.1 mg) were significantly smaller and contained fewer bacteria than those formed by WT. Briefly, mice were subcutaneously injected 5 x 107 planktonic cells and treated 1 h later. After 72 h, abscesses were measured and harvested in phosphate buffered saline (PBS), homogenized and plated on LB for bacterial enumeration. Box and whiskers delineate interquartile range with geometric error from three independent experiments containing 2-3 biological replicates each (n = 8-10). Bacterial recovery data are reported as geometric mean ± standard deviation (SD). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significantly different compared to WT according to Two-Way ANOVA followed by Dunnett’s post-hoc analysis.
Ciprofloxacin treatment also reduced bacterial load of LESB58 deletion mutants when compared to untreated cells of the same strain. In contrast, abscesses formed by LESB58 ΔntrBC after ciprofloxacin treatment contained an average of only 1.4 x 104 CFU (with most mice being completely cured), even though the ΔntrBC deletion did not affect counts in the absence of ciprofloxacin treatment (2.0 x 108 CFU recovered) compared to WT. Abscesses formed by LESB58 ΔntrC exhibited a more moderate 30-fold decrease in bacterial load from 1.8 x 108 CFU to 6.1 x 106 CFU following ciprofloxacin treatment, while ciprofloxacin did not have a significant effect on LESB58 ΔntrB.
The abscess model described above involves inoculation with a high density inoculum to create a chronic infection. We wondered whether the lack of effect of ntrBC deletion on abscess size or bacterial numbers would also hold in an acute model with lower input bacterial inocula. To investigate this, P. aeruginosa LESB58 strains were tested for colonization and virulence in the upper respiratory tract of mice (Figure 3) using our recently described airway/sinus infection model (Alford et al., 2021). In this model we determined that ntrBC deletion had a very substantial and significant effect when compared to WT, with significantly fewer ΔntrB, ΔntrC or ΔntrBC isolated from the nasal cavity and lungs of mice, 72 h post-infection. Thus 96- to 298-fold fewer bacteria were isolated from the nasal cavity, while 8.0- to 10-fold less bacteria were found in the lungs of mice, infected with LESB58 ΔntrB or ΔntrC mutants. Moreover, the LESB58 ΔntrBC double mutant demonstrated an even more profound effect with 2,155-fold and 766-fold fewer bacteria isolated from the nasal cavity and lungs respectively, although the latter was not significant. Conversely, no significant differences were detected for bacterial load of the same strain between nasal cavity and lungs, suggesting that these differences were consistent in these different areas of the respiratory tract. Importantly, ntrBC mutation reduced P. aeruginosa LESB58 colonization of the respiratory tract to a far greater extent than was observed in the high-density skin abscess model. This effect could be reversed by complementation of the mutants with the cloned ntrBC coding region (Figure S2).
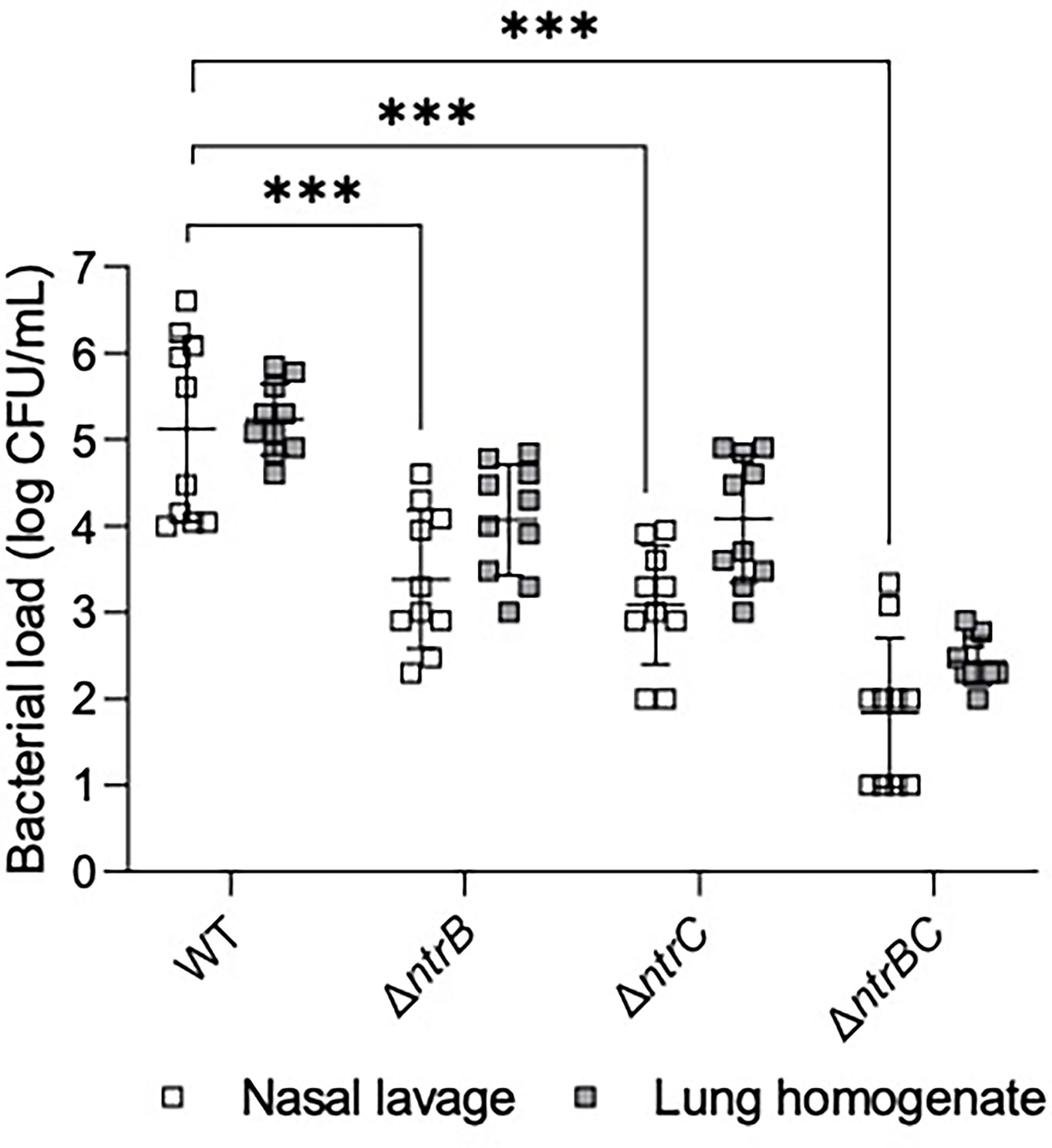
Figure 3 Mutation of the nitrogen regulatory two-component system, NtrBC, reduced bacterial load of P. aeruginosa in a murine model of sinusitis. Stationary-phase bacteria were inoculated dropwise in the left naris of C57Bl/6 mice (106 CFU). 72 h later mice were euthanized, and lung tissue or nasal lavage fluid was collected for bacterial enumeration following serial dilution. Significantly less bacteria were recovered from the lungs and nasal cavities of mice infected with P. aeruginosa ΔntrB, ΔntrC or ΔntrBC mutants. Data are presented as geometric mean ± standard deviation for three independent experiments containing 3-4 biological replicates each (n = 10). ***P < 0.001 according to Two-Way ANOVA followed by Dunnett’s post-hoc analysis.
We hypothesized that the influence of site of infection on NtrBC effects on colonization might reflect different N-sources at these sites. For example, it has been proposed that urea might be the predominant N-source in skin (Grice and Segre, 2011), while amino acids are the major N-source in the lung (Palmer et al., 2005). Thus, we examined the influence of N-source on biofilm formation (Figure 4), which is important for bacterial survival during chronic infection, and susceptibility of P. aeruginosa PA14 to ciprofloxacin killing in vitro.
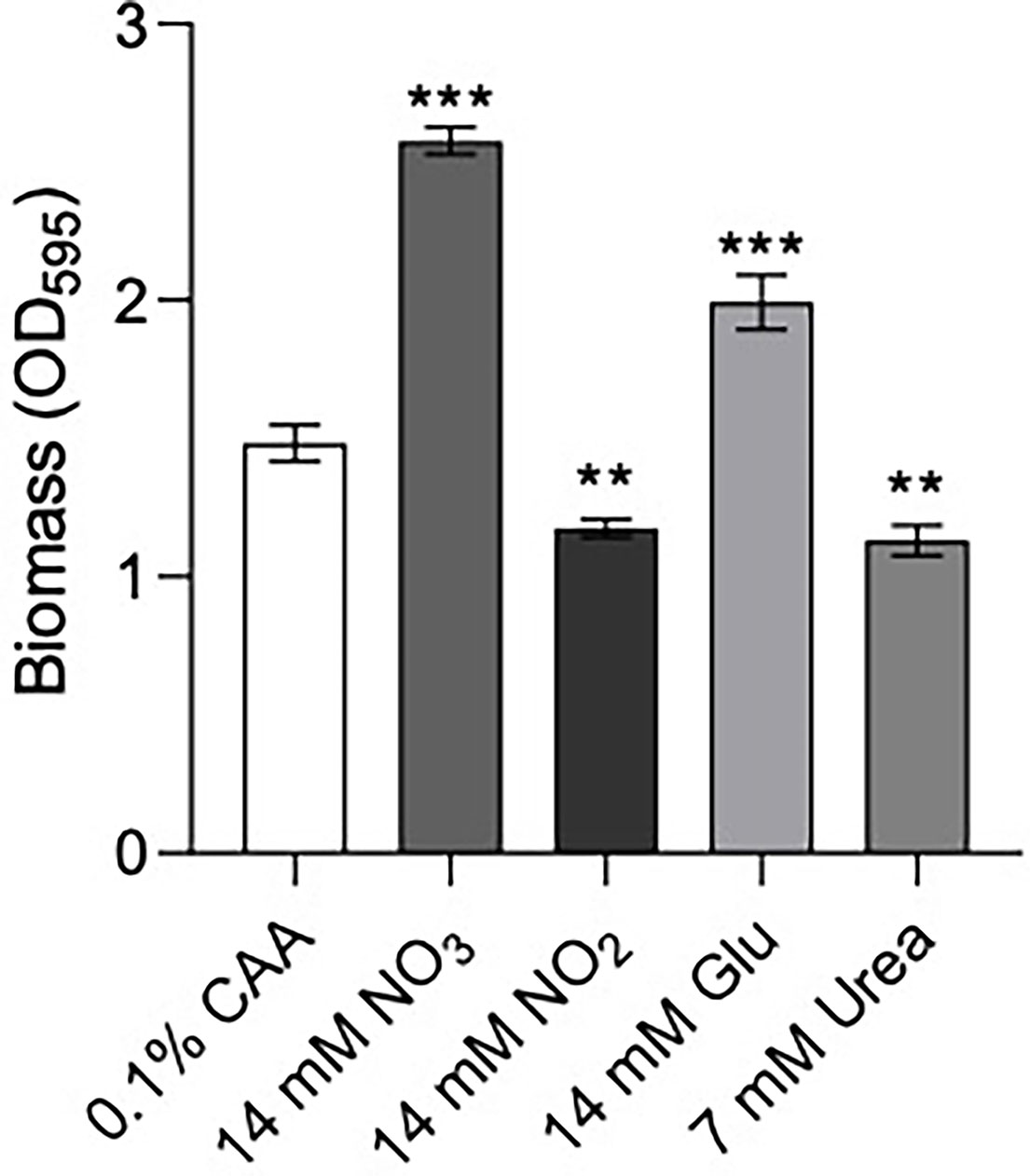
Figure 4 Biofilm formation of P. aeruginosa PA14 was influenced by nitrogen source. Biofilm was formed in BM2 medium containing casamino acids (CAA), nitrate , nitrite glutamate (Glu), or urea in equimolar amounts. 96-well polypropylene plates were inoculated with bacteria suspended in basal medium (BM2) supplemented with 0.4% glucose. 18-20 h later, biomass was stained with crystal violet and measured by scanning its OD595. Data reported as mean ± standard error of the mean (SEM) from three independent experiments containing 2-3 biological replicates each (n = 6-9). **P < 0.01, ***P < 0.001 according to Kruskal-Wallis nonparametric test followed by Dunn’s post-hoc analysis.
Previous data suggested that ΔntrBC but not ΔntrB, or ΔntrC alone had a substantial impact on biofilm formation (Alford et al., 2020). Therefore, we examined biofilm formation of P. aeruginosa PA14 in N-sources characteristic of different bodily niches, including casamino acids (CAA), nitrate , nitrite , glutamate (Glu) and urea (Figure 4). Relative to that in CAA, biofilm biomass staining (OD595) was significantly increased to 174% and 130% when and Glu replaced CAA in the growth medium. In contrast, when or Urea replaced CAA, OD595 was decreased to 79.2% and 76.3% respectively.
Since pyoverdine synthesis genes were expressed by 2.0- to 2.5-fold less in ΔntrB and ΔntrC strains compared to WT (Table 2 and Table S3), and because mutants were less capable of colonizing the respiratory tract, we measured production of virulence factors including pyoverdine, pyocyanin and elastase by PA14 strains grown overnight for 16 hours (Figure 5). Although there were no statistically significant differences between PA14 WT and ΔntrB, production of pyoverdine and pyocyanin in ΔntrC was significantly reduced by 38.2% and 31.7%, respectively. Production of all virulence factors was significantly and substantially reduced in the ΔntrBC double mutant, by 48.2%, 65.2% and 89.8% in pyoverdine, pyocyanin and elastase respectively. This coincided with the downregulated expression of 19 genes involved in the production of siderophores or phenazines in ΔntrB and/or ΔntrC (Table S3). Substantial reductions in virulence determinants might be expected to reduce the ability to colonize.
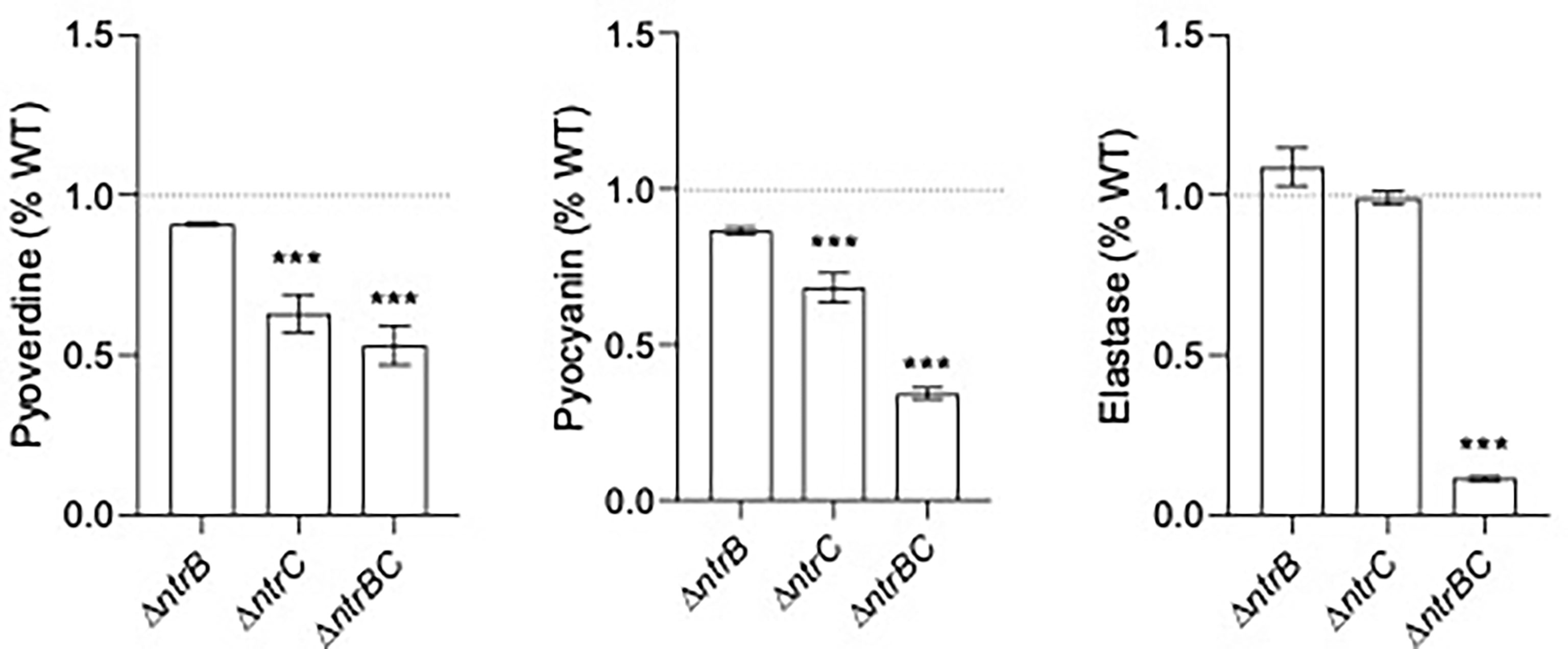
Figure 5 Mutation of the nitrogen regulatory two-component system, NtrBC, reduced production of virulence factors by P. aeruginosa. The supernatants of overnight cultures of similar density were assessed for pyoverdine (Pvd), pyocyanin (Pcn) and elastase (Las). Data were reported as mean ± standard error of the mean (SEM) from three independent experiments each containing three biological replicates (n = 9). ***P < 0.001 different from WT according to One-Way ANOVA followed by Dunn’s post-hoc analysis.
Since virulence factor production is a key determinant of P. aeruginosa pathogenesis (Alford et al., 2019, Strateva and Mitov, 2011), and NtrBC directly impacted on the levels of secreted virulence factors including cytotoxins like elastase (Figure 5) and rhamnolipids (Alford et al., 2020), we studied whether NtrBC modulated host-directed cytotoxicity in response to in vitro infection (Figure 6). A modest but non-significant increase in cytotoxicity of HBE cells was observed following infection with ΔntrB or ΔntrC (13.2% and 7.03% greater than WT, respectively) at an MOI = 1, and a modest but significant 21.5% increase in cytotoxicity was observed following infection with ΔntrBC.
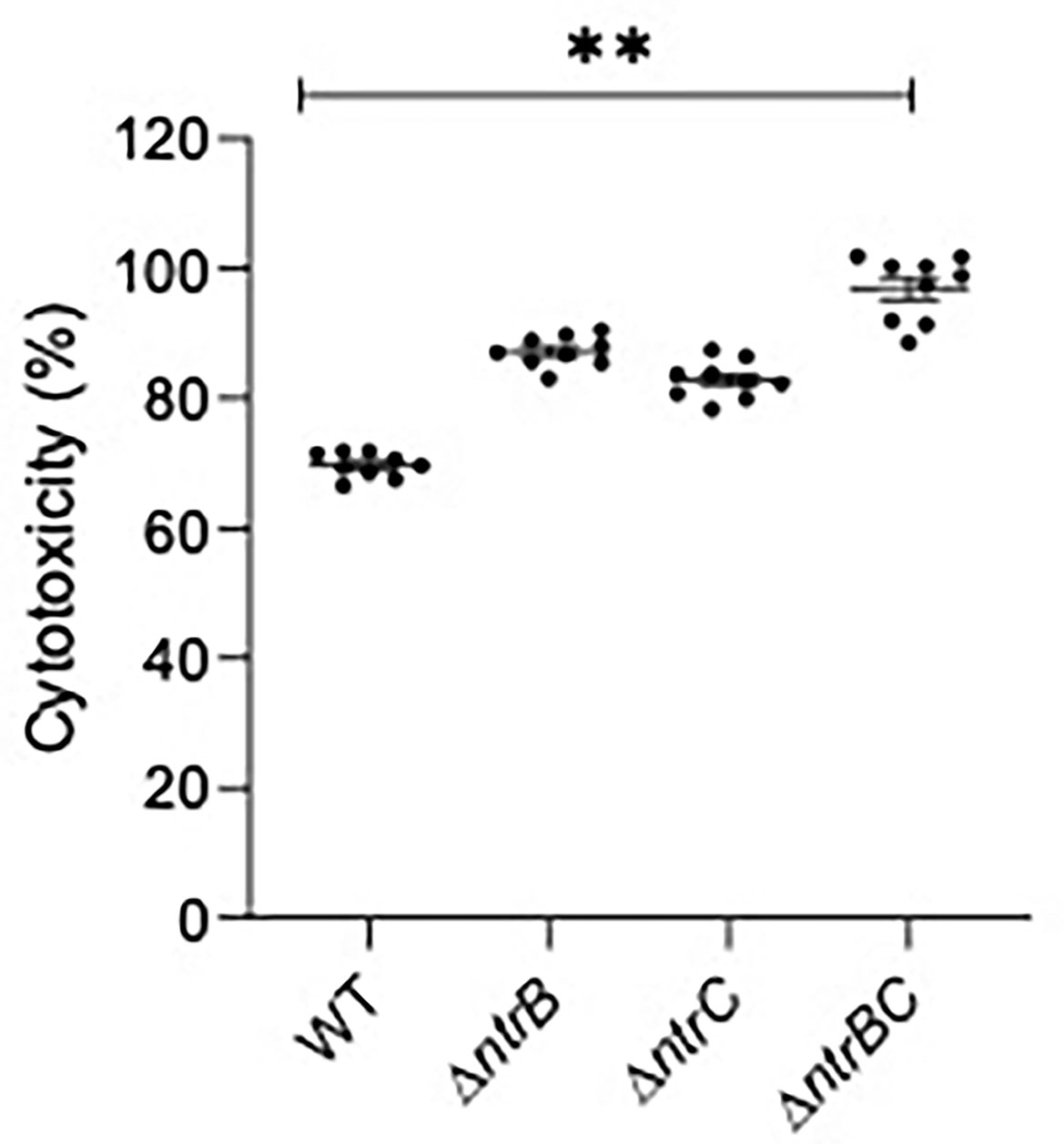
Figure 6 Mutation of the nitrogen regulatory two-component system, NtrBC, increased host-directed cytotoxicity of P. aeruginosa. Confluent human bronchial epithelial (HBE) cells (~7.5 x 104 cells/well) were treated with mid-log phase bacteria at a multiplicity of infection (MOI) = 1 and incubated at 37°C for 16-18 h. Cell-free supernatants were collected and host-directed cytotoxicity (%) was estimated by LDH release from cells relative to cells lysed with Triton X-100. Significantly more cytotoxicity was caused after infection with P. aeruginosa ΔntrBC mutants. Data are presented as mean ± standard error of the mean from four independent experiments containing three biological replicates each (n = 12). **P < 0.01 according to One-Way ANOVA followed by Dunn’s post-hoc analysis.
We next tested susceptibility to nonopsonic phagocytosis by examining macrophage-mediated uptake and clearance of pathogens at an MOI = 10 (Figure 7). Using a gentamicin-protection assay to record bacteria taken up by macrophages, lysates from infections with ΔntrB, ΔntrC or ΔntrBC contained 9.0- to 13.2-fold more bacteria than lysates from infections with WT at 30 min, indicating substantially greater uptake of these mutants. This coincided with significant upregulation of 38 genes important for macrophage uptake (Felgner et al., 2020) (Table S3). There were no statistically significant differences between mutants and WT at 90 min. Nonetheless, each strain was reduced, on average, by ~10-fold at the later time point, indicating similar rates of clearance. In contrast, pilot studies showed that neutrophil-mediated uptake of PA14 WT and mutants were not significantly different (Figure S3).
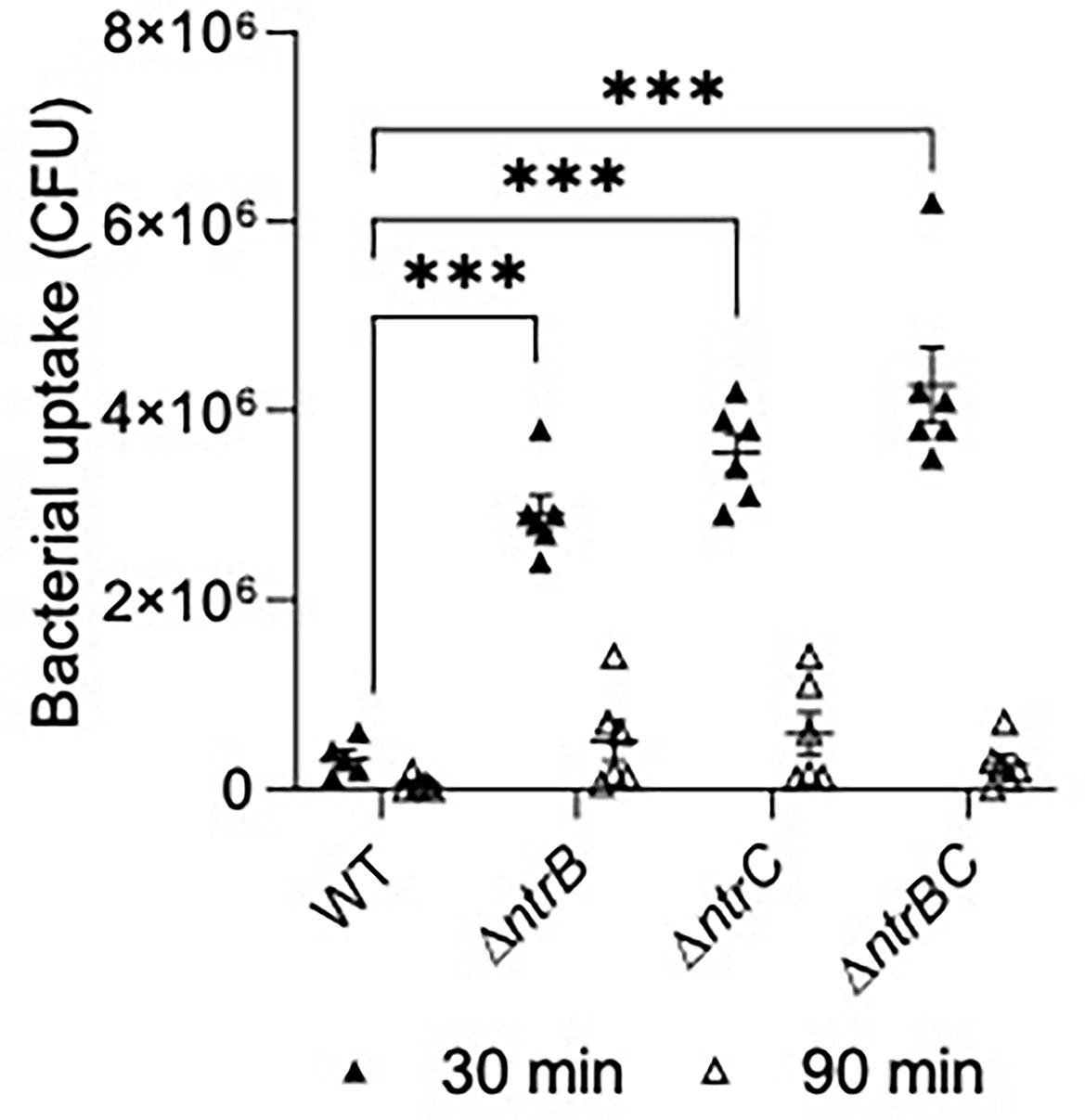
Figure 7 Mutation of the nitrogen regulatory two-component system, NtrBC, increased uptake of P. aeruginosa by macrophages. Confluent THP-1 monocytes (~3.0 x 105 cells/well) were differentiated and treated with mid-log phase bacteria at a multiplicity of infection (MOI) = 10 and incubated at 37°C for 30 min. Cells were washed, treated with gentamicin (500 µg/ml) for 30 min, and washed again to remove any residual bacteria in the supernatant. Cell lysates at t = 0, 60 min were plated for bacterial enumeration following serial dilution. Mutants were taken up and cleared from macrophages more efficiently than the WT. Data are presented as geometric mean ± standard deviation for three independent experiments containing 2 biological replicates each (n = 6). ns, not statistically different; ***P < 0.001 different according to Two-way ANOVA followed by Dunnett’s post-hoc analysis.
NtrBC is a two-component system of P. aeruginosa that is essential for sensing environmental nitrogen levels and responding to nitrogen starvation through initiation of nitrate assimilation (Luque-Almagro et al., 2011). NtrC is known to activate a variety of other physiological processes, in part by increasing the binding affinity of the alternative sigma factor RpoN/σ54 to RNA polymerase, including histidine utilization (Naren and Zhang, 2021) and the stringent stress response (Brown et al., 2014). We previously showed that NtrBC impacts on P. aeruginosa adaptive phenotypes including biofilm formation and swarming motility as well as invasiveness and virulence in vivo (Alford et al., 2020), and that the effects of each of NtrB and NtrC appear to be additive for selected phenotypes. The results described here are distinct from those shown previously, since we demonstrated that NtrBC regulated colonization in a tissue-specific manner by affecting certain host-pathogen interactions and production of virulence factors. We also explored the role of nitrogen in adaptive phenotypes, including biofilm formation and swarming-mediated antibiotic resistance.
Adding to our knowledge of NtrBC as a global regulator, the results shown here indicated that activity of this two-component system directly impacted on ciprofloxacin resistance of P. aeruginosa (Figure 1 and Table 1) without influencing susceptibility to tobramycin, tetracycline or chloramphenicol. This was recapitulated in an abscess model of high-density infection (Figure 2). Ciprofloxacin is one of the most important antibiotics used for the treatment of CF lung infections of both children and adults (Andriole, 2005). However, ciprofloxacin resistance is on the rise, with 40% of CF isolates sampled in one study exhibiting resistance (Landman et al., 2007). Ciprofloxacin resistance is usually multifactorial, involving expulsion by the multidrug efflux-porin systems (Rehman et al., 2019), as well as target site mutation in the DNA gyrase (gyrAB) or topoisomerase (parCE) that are targeted by ciprofloxacin (Feng et al., 2019). We identified other, non-canonical effectors of ciprofloxacin resistance downstream of NtrBC that spanned multiple physiological categories (Table 1). Some of the most significantly dysregulated effectors included the intracellular protease PfpI (Fernández et al., 2012), which has been shown to affect swarming and biofilm formation as well as resistance, the acyl-coA ligase PA3924 (Zarzycki-Siek et al., 2013), which is needed for nutrient acquisition in the murine lung, flavohemoprotein fhp (Koskenkorva-Frank and Kallio, 2009), which is induced to protect against nitrosative stress, and catabolic protein LhpH (Li and Lu, 2016), which is implicated in use of the major non-proteinogenic amino acid in mammals, hydroxyproline. Clearly, NtrBC is a regulatory system at the intersection of ciprofloxacin resistance and metabolic processes of P. aeruginosa.
As a metabolically versatile organism with a large genome comprising numerous regulatory systems, P. aeruginosa is able to establish chronic, recurrent infections in various niches of the human body (Frimmersdorf et al., 2010). In the airways of CF patients, P. aeruginosa undergoes a broad metabolic rewiring (Bartell et al., 2017; La Rosa et al., 2019) to accommodate the needs of the cell according to diversity and availability of nutrients, including free amino acids and that are not detected in abundance in other bodily niches (Palmer et al., 2005). Thus, interference with metabolism of these nitrogen sources could inhibit colonization ability in a tissue-specific manner. In support, we showed that P. aeruginosa LESB58 ΔntrB and/or ΔntrC mutants, which are deficient for metabolism of nitrogenous species including (Alford et al., 2020) and Glu, but not urea, colonized the skin to a similar extent as WT in the absence of ciprofloxacin treatment, but did not colonize the respiratory tract of mice as well as WT (Figure 3). This effect could be mitigated by complementation of the ntrBC coding region back into mutants (Figure S2). This contributes to our understanding of the complex physiological processes assumed by bacteria during infection that impacts pathogenesis and antimicrobial intervention (Alford et al., 2019) and indicates that NtrBC is important for metabolic adaptation in the nasal cavity and lungs, but not the skin. This data also implicates NtrBC in recurrent CF infection, since P. aeruginosa may persist in the sinus cavity of patients, representing a reservoir where it can adapt and disseminate into the lower respiratory tract over time (Fothergill et al., 2014). As well, increased susceptibility to ciprofloxacin was conferred by the LESB58 ΔntrB and/or ΔntrC mutation in vivo (Figure 2), validating in vitro MIC results in physiologically relevant conditions, which is considered meaningful since bacterial susceptibility to antimicrobials is heavily dependent on environmental conditions.
Indeed, pathoadaptivity and metabolism are intrinsically associated, and nutritional cues in CF sputum impact on multicellular behaviors such as biofilm formation and swarming motility (Palmer et al., 2007). We previously showed that N-source impacted on swarming motility of P. aeruginosa (Alford et al., 2020) and that certain nitrogen-containing compounds, including ammonium and urea, did not support swarming motility as did CAA and . Here we provide further evidence showing that N-source impacts on biofilm formation (Figure 4), and that the effects on biofilm and swarming phenotypes are correlated, which taken together could partially explain the distinct tissue colonization patterns of P. aeruginosa LESB58 strains. More specifically, the nitrogen sources that promoted biofilm formation and swarming ( and the free amino acid Glu, relative to CAA) are those that are abundant in the respiratory tract but not the skin (Palmer et al., 2005; Grice and Segre, 2011). Further, ΔntrB and/or ΔntrC mutants do not grow well in these N-sources, nor do they undergo biofilm formation or swarming motility as well as WT in BM2 containing CAA as the N-source (Alford et al., 2020). Thus, there is the potential that other adaptive species are outcompeting ΔntrBC mutants for the respiratory tract. Furthermore, N-source influenced the ability of P. aeruginosa to swarm in the presence of a high concentration of ciprofloxacin (Figure S1 and Table S2). At ~8x its MIC, ciprofloxacin inhibited swarming of PA14 on BM2 containing CAA as the N-source. However, swarming was still observed in the presence of this concentration of ciprofloxacin following substitution of CAA for Glu and NO3-. Thus, PA14 exhibited swarming-associated resistance to ciprofloxacin in a nitrogen-dependent manner. This data might have implications for the development of metabolism-based therapeutics. For instance, compounds that antagonize and Glu metabolism may be explored as possible therapies to prevent P. aeruginosa adaptations in situ since provision of and Glu as the sole N-source promoted adaptive phenotypes across the board. This data shows the importance of considering experimental conditions and bacterial lifestyle when determining the activity of emerging antibacterial compounds (Alford et al., 2019). Further, swarming-mediated resistance of bacteria to several clinically important antibiotics was conserved across motile species including Salmonella enterica and Gram-positive Bacillus subtilis (Table S2). Thus, therapies targeting adaptive lifestyles of bacteria could be developed as broad-spectrum antimicrobial agents with potential for limiting bacterial dissemination in vivo (Coleman et al., 2020b).
Since genes implicated in production of pyoverdine (pvdD, pvdJ) were also significantly downregulated in ΔntrB and ΔntrC mutants (Tables 2 and S3), we assessed PA14 strains for their production of virulence factors (Figure 5). It was shown that strains defective for nitrogen metabolism produced less pyocyanin, pyoverdine and elastase. Although pathoadapted strains of P. aeruginosa isolated from chronic CF lung infections are characterized by reduced virulence overall, a recent study identified significant within-patient phenotypic diversity of isolates (O’Brien et al., 2017) characterized by co-existence of strains that under-produce or overproduce pyoverdine and elastase. This might be caused by different availability of nutrients in niches of the lung, since virulence factor production is associated with core metabolic activity of P. aeruginosa (Panayidou et al., 2020) and is inhibited following perturbation of metabolism during biofilm formation (Perinbam et al., 2020). These observations align with what we observed herein.
Though virulence factor production was downregulated, we noted significantly more cytotoxicity toward HBE cells by the PA14 ΔntrBC mutant strain than the WT (Figure 6). The first step in establishing P. aeruginosa infection of the airway is receptor-mediated binding to the apical surface of the epithelium, which depends on type IV pili and the type III secretion system (Bucior et al., 2014). Type IV pili are functional in the PA14 ΔntrB, ΔntrC and ΔntrBC strains since they twitched normally (Alford et al., 2020). Similarly, the type III secretion system is upregulated in mutants compared to WT (Table S3). Indeed, expression of the type III apparatus encoded by pscBCEP (Yang et al., 2007) is upregulated 3.0- to 7.5-fold in ΔntrB or ΔntrC when compared to WT (Alford et al., 2020), and expression of type III effectors encoded by pcr1 and pcr3 are upregulated 6.1-fold and 12-fold, respectively (Alford et al., 2020). However, since we observed that PA14 mutants were less virulent in the skin abscess model (i.e. caused less visible dermonecrosis even in the absence of ciprofloxacin), we were surprised by these results. Innate immunity mediated by phagocytic clearance by neutrophils and macrophages is key to the endogenous control of P. aeruginosa (Lovewell et al., 2014) and may also explain prior observations that ΔntrBC mutants was less invasive (Alford et al., 2020). Therefore, we studied whether uptake by macrophages was also affected by mutation of this regulatory system (Figure 7). Indeed ΔntrB, ΔntrC and ΔntrBC mutants were more susceptible to phagocytosis by mature macrophages, but demonstrated similar clearance rates (Figure 7) when compared to PA14 WT. In this regard it is worth noting that ntrBC mutants have functional flagella and swim and twitch normally (Alford et al., 2020) given that both flagella and pili are involved in non-opsonic phagocytosis by macrophages (Kelly et al., 1989; Lovewell et al., 2014). Production of type III toxins such as exoSTY has been shown to limit uptake of P. aeruginosa by macrophages (Hauser, 2009), and the expression of these is modestly downregulated in ntrBC mutants. Type III toxin (exoSTY) secretion by mutants should be assessed in future studies, particularly since genes encoding type III apparatus (pscBEP) and other effectors (pcr1 and pcr3) exhibited opposite expression.
Despite extensive studies on the physiology of P. aeruginosa, we have only a limited understanding of the relationship between core metabolism (including nitrogen assimilation) and adaptive resistance or virulence, particularly in pre-clinical disease models. This study shows a clear role for nitrogen metabolism and NtrBC regulation in ciprofloxacin susceptibility and virulence attributes of P. aeruginosa and suggests NtrBC as a potential target for the development of future metabolism-directed or virulence-attenuating therapeutics. Further studies should be performed to elucidate the interacting partners of NtrB and NtrC that could be contributing to their apparent dose-dependent influence on certain phenotypes and to predict compensatory mechanisms that could be triggered by inhibition of NtrB and NtrC in situ.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by the Canadian Council on Animal Care (CCAC) guidelines and were approved by the University of British Columbia Animal Care Committee (protocol A17-0253).
MA was responsible for investigation, validation and visualization of data, formal analysis, writing (drafting and editing) and project administration. BB, AA, and KY-C were responsible for experiments and writing (editing). RH was responsible for funding acquisition, provision of resources and supervision, project administration and writing (editing). All authors contributed to the article and approved the submitted version.
We gratefully acknowledge funding to RH from the Canadian Institutes for Health Research grant FDN-154287 and Michael Smith Foundation for Health Research grant 17774. RH holds a Canada Research Chair in Health and Genomics and a UBC Killam Professorship. MA holds a UBC Killam Doctoral Scholarship, Four-Year Fellowship and CIHR Vanier Graduate Scholarship. AA also holds a UBC Killam Doctoral Scholarship while K-YC holds a Michael Smith Foundation for Health Research postdoctoral fellowship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.694789/full#supplementary-material
Alford, M. A., Baghela, A., Yeung, A. T. Y., Pletzer, D., Hancock, R. E. W. (2020). NtrBC Regulates Invasiveness and Virulence of Pseudomonas aeruginosa During High-Density Infection. Front. Microbiol. 11, 773. doi: 10.3389/fmicb.2020.00773
Alford, M. A., Choi, K. Y. G., Trimble, M. J., Masoudi, H., Kalsi, P., Pletzer, D., et al. (2021). Model of Sinusitis Infection for Screening Antimicrobial and Immunomodulatory Therapies. Front. Cell. Infect. Microbiol. 11, 621081. doi: 10.3389/fcimb.2021.621081
Alford, M. A., Pletzer, D., Hancock, R. E. W. (2019). Dismantling the Bacterial Virulence Program. Microb. Biotechnol. 12 (3), 409–413. doi: 10.1111/1751-7915.13388
Andriole, V. T. (2005). The Quinolones: Past, Present, and Future. Clin. Infect. Dis. 41 (2), S113–S119. doi: 10.1086/428051
Bartell, J. A., Blazier, A. S., Yen, P., Thørgensen, J. C., Jelsbak, L., Goldberg, J. B., et al. (2017). Reconstruction of the Metabolic Network of Pseudomonas aeruginosa to Interrogate Virulence Factor Synthesis. Nat. Commun. 8:14631. doi: 10.1038/ncomms14631
Brauner, A., Fridman, O., Gefen, O., Balaban, N. Q. (2016). Distinguishing Between Resistance, Tolerance and Persistence to Antibiotic Treatment. Nat. Rev. Microbiol. 14, 320–330. doi: 10.1038/nrmicro.2016.34
Brazas, M. D., Breidenstein, E. B. M., Overhage, J., Hancock, R. E. W. (2007). Role of Lon, An ATP-Dependent Protease Homolog, in Resistace of Pseudomonas Aeruginosa to Ciprofloxacin. Antimicrob. Agents Chemother. 51 (12), 4276–4283. doi: 10.1128/AAC.00830-07
Breidenstein, E. B. M., Fuente-Nuñez, C., Hancock, R. E. W. (2011). Pseudomonas Aeruginosa: All Roads Lead to Resistance. Trends Microbiol. 19 (8), 419–426. doi: 10.1016/j.tim.2011.04.005
Breidenstein, E. B. M., Khaira, B. K., Wiegand, I., Overhage, J., Hancock, R. E. W. (2008). Complex Ciprofloxacin Resistome Revealed by Screening a Pseudomonas Aeruginosa Mutant Library for Altered Susceptibility. Antimicrob. Agents Chemother. 52 (12), 4486–4491. doi: 10.1128/AAC.00222-08
Brown, D. R., Barton, G., Pan, Z., Buck, M., Wigneshweraraj, S. (2014). Nitrogen Stress Response and Stringent Response are Coupled in Escherichia Coli. Nat. Commun. 5, 4115. doi: 10.1038/ncomms5115
Bucior, I., Tran, C., Engel, J. (2014). Assessing Pseudomonas Virulence Using Host Cells. Methods Mol. Biol. 1149, 741–755. doi: 10.1007/978-1-4939-0473-0_57
Ciofu, O., Tolker-Nielsen, T. (2019). Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents - How P. Aeruginosa Can Escape Antibiotics. Front. Microbiol. 10, 913. doi: 10.3389/fmicb/2019.00913
Clinical and Laboratory Standards Institute (CLSI) (2020). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed, Wayne, PA: Clinical and Laboratory Standards Institute. ISBN: 978-1-68440-067-6, CLSI supplement M100.
Coleman, S. R., Blimkie, T., Falsafi, R., Hancock, R. E. W. (2020a). Multi-Drug Adaptive Resistance of Pseudomonas aeruginosa Swarming Cells. Antimicrob. Agents Chemother. 64 (3), e01999–e01919. doi: 10.1128/AAC.01999-19
Coleman, S. R., Pletzer, D., Hancock, R. E. W. (2020b). Contribution of Swarming Motility to Dissemination in a Pseudomonas aeruginosa Murine Skin Abscess Infection Model. J. Infect. Dis. jiaa778. doi: 10.1093/infdis/jiaa778
Dötsch, A., Becker, T., Pommerenke, C., Magnowska, Z., Jänsch, L., Häussler, S. (2009). Genomewide Identification of Genetic Determinants of Antimicrobial Drug Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53 (6), 2522–2531. doi: 10.1128/AAC.00035-09
Essar, D. W., Eberly, L., Hadero, A., Crawford, I. P. (1990). Identification and Characterization of Genes for a Second Anthranilate Synthase in Pseudomonas aeruginosa: Interchangeability of the Two Anthranilate Synthases and Evolutionary Implications. J. Bacteriol. 172, 884–900. doi: 10.1128/jb.172.2.884-900.1990
Fajardo, A., Martínez-Martín, N., Mercadillo, M., Galán, J. C., Ghysels, B., Matthijs, S., et al. (2008). The Neglected Instrinsic Resistome of Bacterial Pathogens. PLoS One 3 (2), e1619. doi: 10.1371/journal.pone.0001619
Felgner, S., Preusse, M., Beutling, U., Stahnke, S., Pawar, V., Rohde, M., et al. (2020). Host-Induced Spermidine Production in Motile Pseudomonas aeruginosa Triggers Phagocytic Uptake. eLife 9, e55744. doi: 10.7554/eLife.55744
Feng, X., Zhang, Z., Li, X., Song, Y., Kang, J., Yin, D., et al. (2019). Mutations in gyrB Play an Important Role in Ciprofloxacin-Resistant Pseudomonas aeruginosa. Infect. Drug Resist. 12, 261–272. doi: 10.2147/idr.s182272
Fernández, L., Breidenstein, E. B. M., Song, D., Hancock, R. E. W. (2012). Role of Intracellular Proteases in the Antibiotic Resistance, Motility and Biofilm Formation of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56 (2), 1128–1132. doi: 10.1128/AAC.05336-11
Ferreiro, J. L. L., Otero, J. Á., González, L. G., Lamazares, L. N., Blanco, A. A., Sajurjo, J. R. B., et al. (2017). Pseudomonas aeruginosa Urinary Tract Infections in Hospitalized Patients: Mortality and Prognostic Factors. PLoS One 12 (5), e0178178. doi: 10.1371/journal.pone.0178178
Flemming, H., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., Kjelleberg, S. (2016). Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Fothergill, J. L., Neill, D. R., Loman, N., Winstanley, C., Kadioglu, A. (2014). Pseudomonas aeruginosa Adaptation in the Nasopharyngeal Reservoir Leads to Migration and Persistence in the Lungs. Nat. Commun. 5, 4780. doi: 10.1038/ncomms5780
Frimmersdorf, E., Horatzek, S., Pelnikevich, A., Wiehlmann, L., Schomburg, D. (2010). How Pseudomonas aeruginosa Adapts to Various Environments: A Metabolomic Approach. Environ. Microbiol. 12, 1734–1747. doi: 10.1111/j.1462-2920.2010.02253.x
Fuente-Nunez, C., Reffuveille, F., Fernandez, L., Hancock, R. E. W. (2013). Bacterial Biofilm Development as a Multicellular Adaptation: Antibiotic Resistance and New Therapeutic Strategies. Curr. Opin. Microbiol. 16 (5), 580–589. doi: 10.1016/j.mib.2013.06.013
Grice, E. A., Segre, J. A. (2011). The Skin Microbiome. Nat. Rev. Microbiol. 9 (4), 244–253. doi: 10.1038/nrmicro2537
Hall, C. W., Farkas, E., Zhang, L., Mah, T. F. (2019). Potentiation of Aminoglycoside Lethality by C4-dicarboxylates Requires RpoN in Antibiotic-Tolerant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63 (10), e01313–e01319. doi: 10.1128/AAC.01313-19
Hauser, A. R. (2009). The Type III Secretion System of Pseudomonas aeruginosa: Infection by Injection. Nat. Rev. Microbiol. 7 (9), 654–665. doi: 10.1038/nrmicro2199
Hewer, S. C. L., Smyth, A. R., Brown, M., Jones, A. P., Hickey, H., Kenna, D., et al. (2020). Intravenous Versus Oral Antibiotics for Eradication of Pseudomonas aeruginosa in Cystic Fibrosis (TORPEDO-CF): A Randomised Controlled Trial. Lancet Respir. Med. 8 (10), 975–986. doi: 10.1016/S2213-2600(20)30331-3
Imperi, F., Tiburzi, F., Visca, P. (2009). Molecular Basis of Pyoverdine Siderophore Recycling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 106 (48), 20440–20445. doi: 10.1073/pnas.0908760106
Kelly, N. M., Kluftinger, J. L., Pasloske, B. L., Paranchych, W., Hancock, R. E. W. (1989). Pseudomonas aeruginosa Pili as Ligands for Nonopsonic Phagocytosis by Fibronectin-Stimulated Macrophages. Infect. Immun. 57 (12), 3841–3845. doi: 10.1128/IAI.57.12.3841-3845.1989
Kollaran, A. M., Joge, S., Kotian, H. S., Badal, D., Prakash, D., Mishra, A., et al. (2019). Context-Specific Requirement of Forty-Four Two-Component Loci in Pseudomonas aeruginosa Swarming. iScience 13, 305–317. doi: 10.1016/j.isci.2019.02.028
Koskenkorva-Frank, T. S., Kallio, P. T. (2009). Induction of Pseudomonas aeruginosa Fhp and fhpR by Reactive Oxygen Species. Can. J. Microbiol. 55 (6), 657–663. doi: 10.1139/w09-024
Kumar, P., Nagarajan, A., Uchil, P. D. (2018). Analysis of Cell Viability by the Lactate Dehydrogenase Assay. Cold Spring Harb. Protoc. 6. doi: 10.1101/pdb.prot095497
Lai, A., Tremblay, J., Déziel, E. (2009). Swarming Motility: A Multicellular Behaviour Conferring Antimicrobial Resistance. Environ. Microbiol. 11 (1), 126–136. doi: 10.1111/j.1462-2920.2008.01747.x
Landman, D., Bratu, S., Kochar, S., Panwar, M., Trehan, M., Doymaz, M., et al. (2007). Evolution of Antimicrobial Resistance Among Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae in Brooklyn, NY. J. Antimicrob. Chemother. 60, 78–82. doi: 10.1093/jac/dkm129
La Rosa, R., Johansen, H. K., Molin, S. (2019). Adapting to the Airways: Metabolic Requirements of Pseudomonas aeruginosa During the Infection of Cystic Fibrosis Patients. Metabolites 9 (10):234. doi: 10.3390/metabo9100234
Li, G., Lu, C. D. (2016). Molecular Characterization of LhpR in Control of Hydroxyproline Catabolism and Transport in Pseudomonas aeruginosa PAO1. Microbiology (Read) 162 (7), 1232–1242. doi: 10.1099/mic.0.000300
Line, L., Alhede, M., Kolpen, M., Kühl, M., Ciofu, O., Bjarnsholt, T., et al. (2014). Physiological Levels of Nitrate Support Anoxic Growth by Denitrification of Pseudomonas aeruginosa at Growth Rates Reported in Cystic Fibrosis Lungs and Sputum. Front. Microbiol. 5, 554. doi: 10.3389/fmicb.2014.00554
Lovewell, R. R., Patankar, Y. R., Berwin, B. (2014). Mechanisms of Phagocytosis and Host Clearance of Pseudomonas aeruginosa. Am. J. Physiol. Lung Cell Mol. Physiol. 306 (7), L591–L603. doi: 10.1152/ajplung.00335.2013
Luque-Almagro, V. M., Gates, A. J., Moreno-Vivian, C., Ferguson, S. J., Richardson, D. J., Roldan, M. D. (2011). Bacterial Nitrate Assimilation: Gene Distribution and Regulation. Biochem. Soc. Trans. 39, 1838–1843. doi: 10.1042/BST20110688
Meylan, S., Porter, C. B. M., Yang, J. H., Belenky, P., Gutierrez, A., Lobritz, M. A., et al. (2017). Carbon Sources Tune Antibiotic Susceptibility in Pseudomonas aeruginosa via Tricarboxylic Acid Cycle Control. Cell Chem. Biol. 24, 195–206. doi: 10.1016/j.chembiol.2016.12.015
Moradali, M. F., Ghods, S., Rehm, B. H. A. (2017). Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival and Persistence. Front. Cell. Infect. Microbiol. 7, 39. doi: 10.3389/fcimb.2017.00039
Naren, N., Zhang, X.-X. (2021). Role of a Local Transcription Factor in Governing Cellular Carbon/Nitrogen Homeostasis in Pseudomonas fluorescens. Nucleic Acids Res. 49 (6), 3204–3216. doi: 10.1093/nar/gkab091
O’Brien, S., Williams, D., Fothergill, J. L., Paterson, S., Winstanley, C., Brockhurst, M. A. (2017). High Virulence Sub-Populations in Pseudomonas aeruginosa Long-Term Cystic Fibrosis Airway Infections. BMC Microbiol. 17 (1), 30. doi: 10.1186/s12866-017-0941-6
Ohman, D. E., Cryz, S. J., Iglewski, B. H. (1980). Isolation and Characterization of Pseudomonas aeruginosa PAO Mutant That Produces Altered Elastase. J. Bacteriol. 142, 836–842. doi: 10.1128/jb.142.3.836-842.1980
Olivares, E., Badel-Berchoux, S., Provot, C., Prévost, G., Bernardi, T., Jehl, F. (2020). Clinical Impact of Antibiotics for the Treatment of Pseudomonas aeruginosa Biofilm Infections. Front. Microbiol. 10, 2894. doi: 10.3389/fmicb.2019.02894
Palmer, K. L., Aye, L. M., Whiteley, M. (2007). Nutritional Cues Control Pseudomonas aeruginosa Behavior in Cystic Fibrosis Sputum 189, 22, 8079–8087. doi: 10.1128/JB.01138-07
Palmer, K. L., Mashburn, L. M., Singh, P. K., Whiteley, M. (2005). Cystic Fibrosis Sputum Supports Growth and Cues Key Aspects of Pseudomonas aeruginosa Physiology. J. Bacteriol. 187 (15), 5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005
Panayidou, S., Georgiades, K., Christofi, T., Tamana, S., Promponas, V. J., Apidianakis, Y. (2020). Pseudomonas aeruginosa Core Metabolism Exerts a Widespread Growth-Independent Control on Virulence. Sci. Rep. 10, 9505. doi: 10.1038/s41598-020-66194-4
Pang, Z., Raudonis, R., Glick, B. R., Lin, T., Cheng, Z. (2019). Antibiotic Resistance in Pseudomonas aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 37 (1), 177–192. doi: 10.1016/j.biotechadv.2018.11.013
Perinbam, K., Chacko, J. V., Kannan, A., Digman, M. A., Siryaporn, A. (2020). A Shift in Central Metabolism Accompanies Virulence Activation in Pseudomonas aeruginosa. mBio 11 (2), e02730–e02718. doi: 10.1128/mBio.02730-18
Pletzer, D., Mansour, S. C., Wuerth, K., Rahanjam, N., Hancock, R. E. W. (2017). New Mouse Model for Chronic Infections by Gram-Negative Bacteria Enabling the Study of Anti-Infective Efficacy and Host-Microbe Interactions. mBio 8 (1), e00140-17. doi: 10.1128/mBio.00140-17
Rehman, A., Patrick, W. M., Lamont, I. L. (2019). Mechanisms of Ciprofloxacin Resistance in Pseudomonas aeruginosa: New Approaches to an Old Problem. J. Med. Microbiol. 68, 1–10. doi: 10.1099/jmm.0.000873
Strateva, T., Mitov, I. (2011). Contribution of an Arsenal of Virulence Factors to Pathogenesis of Pseudomonas aeruginosa. Ann. Microbiol. 61, 717–732. doi: 10.1007/s13213-011-0273-y
Thöming, J. G., Tomasch, J., Preusse, M., Koska, M., Grahl, N., Pohl, S., et al. (2020). Parallel Evolutionary Paths to Produce More Than One Pseudomonas aeruginosa Biofilm Phenotype. NPJ Biofilms Microbiomes 6, 2. doi: 10.1038/s41522-019-0113-6
Tschudin-Sutter, S., Fosse, N., Frei, R., Widmer, A. F. (2018). Combination Therapy for Treatment of Pseudomonas aeruginosa Bloodstream Infections. PLoS One 13 (9), e0203295. doi: 10.1371/journal.pone.0203295
Winstanley, C., O’Brien, S., Brockhurst, M. A. (2016). Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends Microbiol. 24 (5), 327–337. doi: 10.1016/j.tim.2016.01.008
Yang, H., Shan, Z., Kim, J., Wu, W., Li, W., Zeng, L., et al. (2007). Regulatory Role of PopN and Its Interacting Partners in Type III Secretion of Pseudomonas aeruginosa. J. Bacteriol. 189 (7), 2599–2609. doi: 10.1128/JB.01680-06
Yeung, A. T. Y., Janot, L., Pena, O. M., Neidig, A., Kukavica-Ibrulj, I., Hilchie, A., et al. (2014). Requirement of the Pseudomonas aeruginosa CbrA Sensor Kinase for Full Virulence in a Murine Acute Lung Infection Model. Infect. Immun. 82 (3), 1256–1267. doi: 10.1128/IAI.01527-13
Yeung, A. T. Y., Torfs, E. C. W., Jamshidi, F., Bains, M., Wiegand, I., Hancock, R. E. W., et al. (2009). Swarming of Pseudomonas aeruginosa is Controlled by a Broad Spectrum of Transcriptional Regulators, Including MetR. J. Bacteriol. 191 (18), 5592–5602. doi: 10.1128/JB.00157-09
Keywords: nitrogen metabolism, virulence, abscess, respiratory infection, antibiotic resistance
Citation: Alford MA, Baquir B, An A, Choi K-YG and Hancock REW (2021) NtrBC Selectively Regulates Host-Pathogen Interactions, Virulence, and Ciprofloxacin Susceptibility of Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 11:694789. doi: 10.3389/fcimb.2021.694789
Received: 13 April 2021; Accepted: 03 June 2021;
Published: 24 June 2021.
Edited by:
Ruggero La Rosa, Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain), DenmarkReviewed by:
Jose L. Martinez, Consejo Superior de Investigaciones Científicas (CSIC), SpainCopyright © 2021 Alford, Baquir, An, Choi and Hancock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert E. W. Hancock, Ym9iQGhhbmNvY2tsYWIuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.