- 1Department of Infectious Diseases, Ningbo Medical Center Lihuili Hospital, Ningbo, China
- 2Department of Rheumatology, Ningbo Medical Center Lihuili Hospital, Ningbo, China
- 3Department of Respiratory Medicine, Ningbo Medical Center Lihuili Hospital, Ningbo, China
Background: There is a paucity of studies using clinical characteristics and whole-genome sequencing together to fully identify the risk factors of patients with Klebsiella pneumoniae (KP) bloodstream infection (BSI).
Methods: We retrospectively analyzed the clinical and microbiological characteristics of patients with KP BSI. Isolates were processed using Illumina NGS, and relevant bioinformatics analysis was conducted (multi-locus sequence typing, serotype, phylogenetic reconstruction, detection of antibiotic resistance, and virulence genes). A logistic regression model was used to evaluate the risk factors of hosts and causative KP isolates associated with 30-day mortality in patients infected with KP BSI.
Results: Of the 79 eligible patients, the 30-day mortality rate of patients with KP BSI was 30.4%. Multivariate analysis showed that host-associated factors (increased APACHE II score and septic shock) were strongly associated with increased 30-day mortality. For the pathogenic factors, carriage of iutA (OR, 1.46; 95% CI, 1.11–1.81, p = 0.002) or Kvar_1549 (OR, 1.31; 95% CI, 1.02–1.69, p = 0.043) was an independent risk factor, especially when accompanied by a multidrug-resistant phenotype. In addition, ST11-K64 hypervirulent carbapenem-resistant KP co-harbored acquired blaKPC-2 together with iutA (76.5%, 13/17) and Kvar_1549 (100%, 17/17) genes. Comparative genomic analysis showed that they were clustered together based on a phylogenetic tree, and more virulence genes were observed in the group of ST11-K64 strains compared with ST11-non-K64. The patients infected with ST11-K64 strains were associated with relatively high mortality (47.2%, 7/17).
Conclusion: The carriage of iutA and Kvar_1549 was seen to be an independent mortality risk factor in patients with KP BSI. The identification of hypervirulent and carbapenem-resistant KP strains associated with high mortality should prompt surveillance.
Introduction
Klebsiella pneumoniae (KP) is one of the most common bacterial pathogens that causes community or nosocomial acquired infections, such as pneumonia, urinary tract and surgical site infections, bloodstream infections (BSIs), and hepatobiliary infections (Paczosa and Mecsas, 2016). The management of infections due to KP has been complicated by the emergence of antimicrobial resistance and hypervirulent K. pneumoniae (HvKp). In particular, disappointing clinical outcomes have been observed in patients with KP BSIs when accompanied by hypervirulent phenotype or multidrug-resistant organisms. The drugs of β-lactams are recognized as the most common use antimicrobial agents due to their high efficiency and lower toxicity. However, carbapenem resistance in KP is a growing problem worldwide, and in China, as China Antimicrobial Surveillance Network had shown, resistance to carbapenems in KP has increased from 2.9% in 2005 to 25% in 2018 (Zheng et al., 2019; Hu et al., 2020). More importantly, carbapenem-resistant HvKp have emerged in clinical settings, although most KP of hypervirulent and antimicrobial-resistant populations was largely non-overlapping (Bialek-Davenet et al., 2014; Zhang et al., 2016; Gu et al., 2018). The prevalence of such organisms poses a significant challenge for clinicians worldwide and often leads to the failure of clinical treatment (Lin et al., 2010; Mohammad Ali Tabrizi et al., 2018). Previous studies have indicated that mortality rates of KP BSIs range from 16% to 40% depending on the conditions of patients and characteristics of bacterial isolates as well as administrations of appropriate antibiotic therapy (Shon et al., 2013; Kohler et al., 2017; Xu et al., 2017; Kim et al., 2019). For the bacterial pathogenic traits, evaluating the elements of virulence factors and antimicrobial susceptibility are crucial in the improvements of clinical practice for managing KP BSI patients.
Several classical traits have been associated with HvKp as compared to classical KP, such as a hypermucoviscous phenotype, predominance of K1 and K2 capsule type, and carriage of multiple virulence genes (e.g., rmpA, rmpA2, iroBCDN, iucABCD, and iutA) (Lee et al., 2017; Russo et al., 2018). The risk factors for HvKp infections in patients with KP BSIs have been widely studied (Li et al., 2018; Harada et al., 2019). Impacts of virulence factors on mortality of KP BSIs in human patients have not been fully revealed, although fatal outcomes of HvKp infections have been observed in in vivo mouse infection models (Lery et al., 2014; Russo et al., 2015).
Most previous studies have reported that severity of underlying disease, intensive care unit (ICU) stay at infection onset, infection with extended-spectrum β-lactamase (ESBL)-producing or carbapenem-resistant KP (CRKP), and delayed administrations of appropriate therapy are the common risk factors for mortality in patients with KP BSIs (Viale et al., 2013). Among the pathogenic factors, studies focusing on the virulence factors related to mortality in patients with KP BSI are scarce (Kim et al., 2019; Namikawa et al., 2019). Kim et al. indicated that carriage of the pks gene cluster was a relevant marker of early mortality using multivariate Cox hazards modeling (Kim et al., 2019). Another study of multivariate analysis showed that iutA was an independent predictor associated with increased 30-day mortality in patients with KP BSIs (Namikawa et al., 2019). Notably, the virulence genes detected in previous studies were assessed by PCR, which may not fully address the clinically important genes influencing KP pathogenicity and virulence. The increasing availability of bacterial whole-genome sequencing (WGS) allows for the practical identification of virulence factors in bacteria, including K. pneumoniae (Holt et al., 2015). WGS and bioinformatics analyses combined with clinical characteristics can fully identify associations between bacterial accessory genome and mortality in patients with BSIs.
The aims of our study were to (i) describe the molecular epidemiology of KP in BSI patients at a tertiary hospital and (ii) evaluate the clinical variables and genetic backgrounds associated with mortality in patients with KP BSIs.
Methods
Study Design
This retrospective cohort study included all patients with KP BSI at a 1200-bed tertiary teaching hospital between January 1, 2016, and December 31, 2019, in Zhejiang Province, China. BSI was defined as one or more positive blood cultures with concomitant symptoms of infection, according to established criteria (Garner et al., 1988). Only the first episode of BSI was included, and the remaining sequential isolates were discarded. The detailed inclusion criteria were as follows: (a) patients aged ≥18 years; (b) hospitalization with a complete clinical data set; (c) a blood culture positive for KP and the sample preserved in our laboratory; and (d) clinical manifestations of infection. Outpatients, patients with incomplete data, and those lacking KP isolates were excluded. This study was approved by the Ethics Committee of Lihuili Hospital, Ningbo Medical Center (no. KY2019PJ028).
Definitions and Patient Assessment
The probable infectious source was determined using the CDC/National Healthcare Safety Network surveillance definitions (Centers for Disease Control and Prevention). Corticosteroid therapy was defined as the administration of >20 mg/day prednisone (or its equivalent) for a period of ≥7 days. Antimicrobial drug exposure was defined as the use of antibiotics for more than 48 h within 90 days prior to the onset of BSI. Empirical antimicrobial treatment was used to treat suspected KP BSI without in vitro antimicrobial susceptibility information, while definitive antimicrobial treatment was revised based on the in vitro antimicrobial susceptibility results. Antimicrobial therapy was determined to be appropriate when the treatment regimen included antibiotics active against pathogens in vitro. The final outcome was determined as survival and all-cause death at 30 days after the date of BSI onset.
Data Collection
The demographic and clinical information of the enrolled patients, including age, sex, underlying disease, according to the age-adjusted Charlson Comorbidity Index (aCCI), possible sources of BSI, antibiotic regimen, patient outcomes, and other relevant information, were retrieved from the electronic medical records system (Charlson et al., 1994). Moreover, the Acute Physiology and Chronic Health Evaluation (APACHE) II and Pitt bacteremia scores calculated at the time of BSI onset were used to assess illness severity (Knaus et al., 1985).
Bacterial Isolates and Antimicrobial Resistance Susceptibility
The string test was performed with a standard bacteriologic loop to evaluate hypermucoviscosity, and the formation of viscous strings >5 mm in length was considered positive (Fang et al., 2004). The virulence genes iucA, iroB, peg-344, rmpA, and rmpA2 were assessed using PCR (Russo et al., 2018). The primers used in this study are listed in Supplementary Table S1.
The minimum inhibitory concentrations (MICs) of ceftriaxone (CRO), cefepime (FEP), cefoperazone-sulbactam (1:1) (CFZ/SBT), piperacillin-tazobactam (PIP/TZP), imipenem (IPM), meropenem (MEM), levofloxacin (LEV), fosfomycin (FOS), and amikacin (AMK) against the collected strains were determined using the agar dilution method, according to the Clinical and Laboratory Standards Institute (CLSI) (Clinical and Laboratory Standards Institute). For colistin (CST) and tigecycline (TGC), the broth microdilution method was used.
Whole-Genome Sequencing and Bioinformatics Analysis
To characterize the genetic features of BSI isolates, WGS was performed using the Illumina HiSeq platform (Illumina, San Diego, CA, USA). Sequencing data were assembled using SPAdes v. 3.15.0. The antimicrobial resistance genes were identified using the resistance gene identifier through the Comprehensive Antibiotic Research Database. The virulence genes were annotated using the Diamond software through the Virulence Factors Database (VFDB). KP surface polysaccharide locus typing was annotated using Kaptive (Wyres et al., 2016). Multi-locus sequence typing (MLST) was performed using the MLST software (https://github.com/tseemann/mlst), which incorporates the components of the PubMLST database. The kSNP program based on the k-mer analysis was used to identify the core genomic single-nucleotide polymorphisms in the WGS data. Kchooser was used to evaluate the optimal value of k-mer before kSNP. The output file of the maximum likelihood tree was generated using the iTOL (https://itol.embl.de/). As the strains of R5 and R18 (Sequence Read Archive database with SRP141269) were regarded as carbapenem-resistant HvKp via bacteriological test, neutrophil killing assay, and Galleria mellonella infection model in a previous study, comparative genomic analysis on the phylogenies and virulence of ST11 KP was conducted using by kSNP and Roary (Yang et al., 2020). The phylogenies of the ST11 strains were performed by kSNP software based on the core genome SNPs. The Roary software was used to calculate the pan-genome (Page et al., 2015). The virulence factors were identified by blasting the VFDB database using Diamond (Buchfink et al., 2021).
Statistical Analysis
Analyses were conducted using the R software (version 4.0.2; http://www.R-project.org/). Continuous variables are expressed as the mean ± standard deviation (assessed using Student’s t-test) or as the median (range) (evaluated using the Wilcoxon rank-sum test) when the distribution was not normal. Categorical data were expressed as frequency distributions, and the chi-square test or Fisher’s exact test was used to determine the distribution when appropriate. The clinically important variables, antimicrobial resistance, and virulence determinants (p < 0.1) in univariate analysis were then selected into a logistic regression model for multivariate analysis to evaluate the risk factors for KP BSI mortality. Notably, the above variables were determined using the variance inflation factor (VIF) and the eigenmatrix method to detect multicollinearity before logistic regression analysis. Binary logistic regression (step: Akaike information criterion [AIC], direction: backward and forward) was used to identify independent predictors of 30-day mortality. The strength of associations was determined by calculating the odds ratio (OR) and 95% confidence intervals (CIs). Two-tailed tests were used to determine the statistical significance. The survival distribution function was estimated using the Kaplan–Meier product limit method. Nonparametric (log-rank and Wilcoxon) tests were used to compare survival functions between the groups. In all analyses, a p-value of ≤0.05 was considered significant.
Nucleotide Sequence Accession Numbers
The whole-genome sequences described in this paper have been deposited in the National Microbiology Data Center under the accession numbers NMDC60014864-NMDC60014942.
Results
Demographic and Clinical Characteristics
In total, 92 unique cases of KP BSI during the 4-year study period were identified. Of these, five patients were excluded because of missing KP isolates, and eight cases were excluded owing to early death. Finally, 79 patients (48 males and 31 females) with laboratory-confirmed KPs were enrolled. The clinical characteristics of KP BSI between non-survivors and survivors are summarized in Table 1 and Supplementary Table S2. The median age of all patients was 67 years (interquartile range, 59.0–77.5). Hepatobiliary disease (27.8%) was the most prevalent comorbidity, followed by diabetes mellitus (21.5%) and malignancy (15.2%). The median age-aCCI was 3 (IQR, 2–4). The most common probable source of infection was pneumonia (43.0%), followed by biliary tract infections (13.9%) and liver abscesses (12.7%). The median time to the onset of KP BSI was 8 days (IQR, 1–23.5). Almost half of the patients resided in the ICU within 90 days prior to the onset of BSI. The median APACHE II and Pitt bacteremia scores at the time of the initial blood culture were 17 (IQR, 13–26) and 2 (IQR, 1–6), respectively. In addition, the 30-day mortality rate of patients infected with KP BSI was 30.4%.
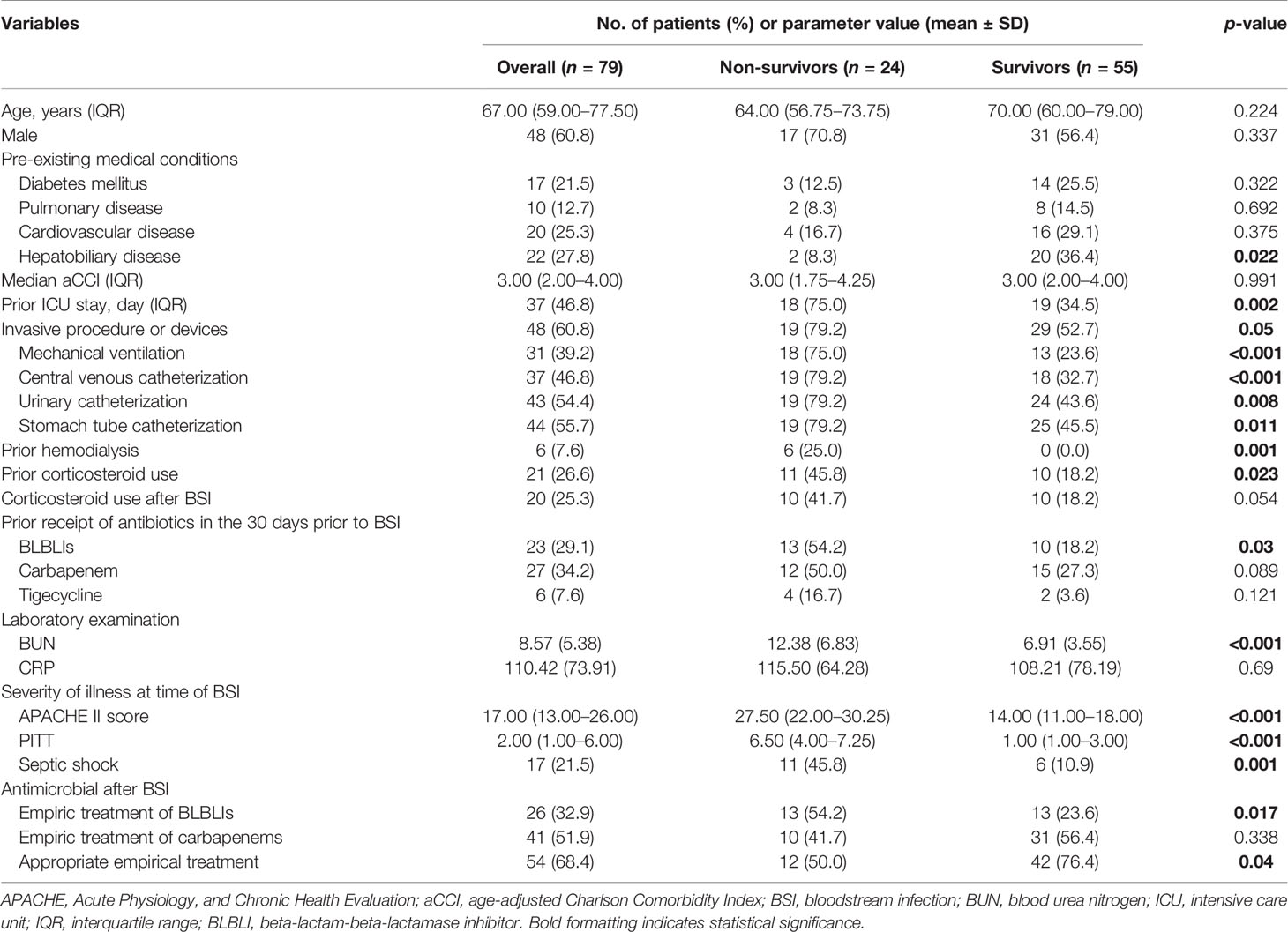
Table 1 Clinical characteristics and laboratory findings of non-survivor and survivor patients with K. pneumoniae bloodstream infection.
Bacterial Characteristics
The MIC results demonstrated that MDRKP and CRKP isolates, especially the latter, exhibited multidrug resistance to most classes of antimicrobial agents, except FOS, TGC, and CST (Figure 1 and Supplementary Table S3). CRKP was more likely to be observed in patients with prior admission to the ICU, and higher mortality was observed in the CRKP group (Figure 1 and Table 2). Regarding antimicrobial resistance genes, 114 resistance genes were detected in our study, including blaCTX-M, blaSHV, blaTEM, blaKPC-2, blaOXA-1, aac(3)-IId, qnr, tet, fosA5, sul, and aadA (Table 2 and Supplementary Table S4). Among them, blaSHV (89.9%, n = 71) and fosA6 (91.1%, n = 72) were the most commonly identified resistance genes in the tested isolates. The most prevalent blaCTX-M gene was blaCTX-M-65 (22.8%, n = 18), followed by blaCTX-M-14 (6.3%, n = 5), and blaCTX-M-15 (5.1%, n = 4). The blaKPC-2 gene, which is responsible for carbapenem resistance, was also observed in 24 cases.
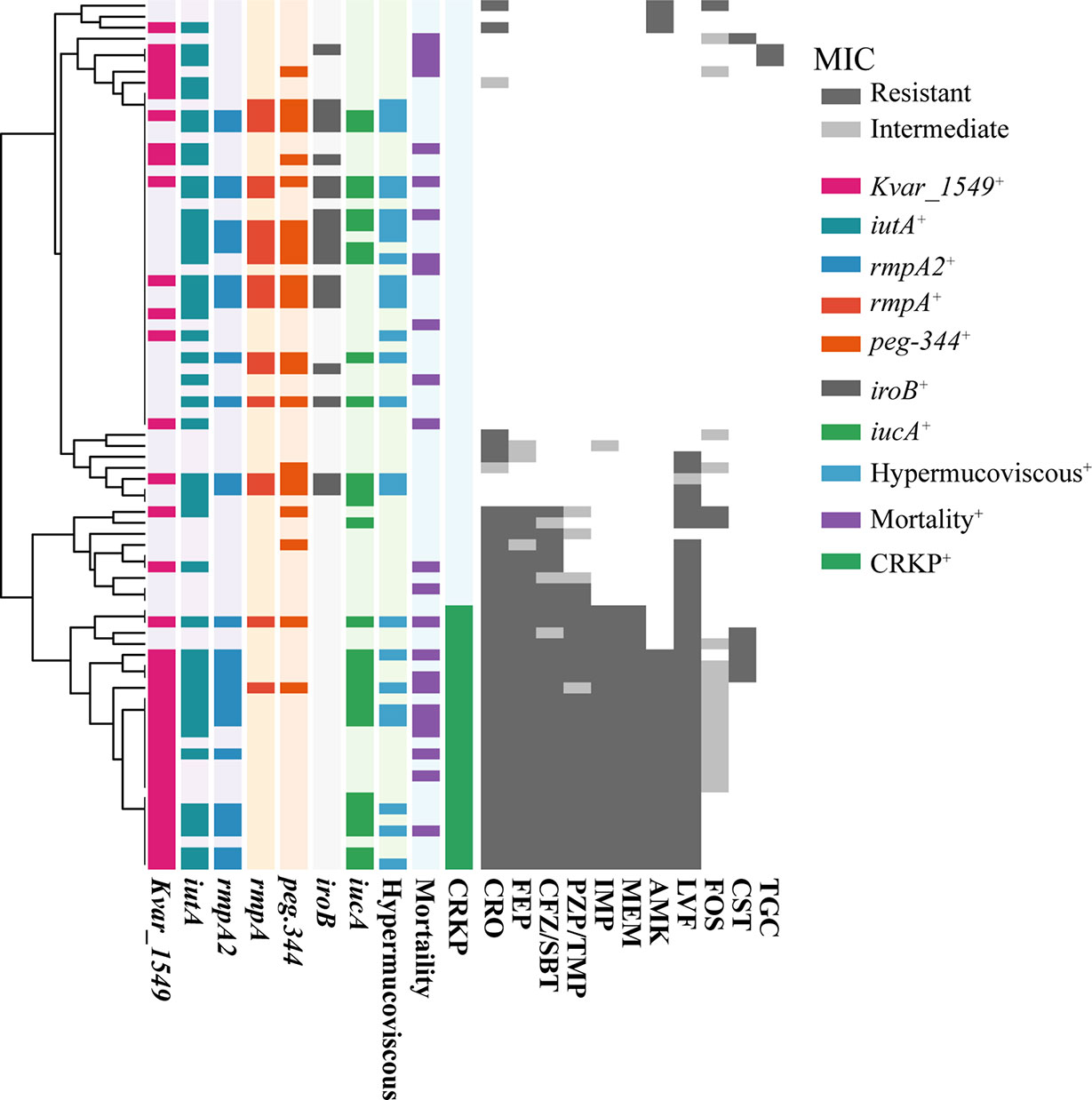
Figure 1 The MIC distribution of all antibiotics against K. pneumoniae isolated from bloodstream infection. MICs were using agar dilution methods for all antibiotics except for colistin and tigecycline, for which broth microdilution was used. Results were interpreted using the CLSI guidelines. Dark gray indicates resistance, light gray indicates mediation, and white indicates sensitivity. The tree on the left side was reconstructed by hierarchical clustering of MIC results using the Euclidean agglomeration method. The colored blocks in the middle represent the presence of virulence genes/phenotype, and white blocks represent absence. CRO, Ceftriaxone; FEP, Cefepime; CFZ/SBT, Ceftolozane-sulbactam; PZP/TMP, Piperacillin-tazobactam; IMP, Imipenem; MEM, Meropenem; AMK, Amikacin; LVF, Levofloxacin; FOS, Fosfomycin; CST, Colistin; TGC, Tigecycline. Mortality, 30-day mortality. CRKP, carbapenem-resistant K. pneumoniae.
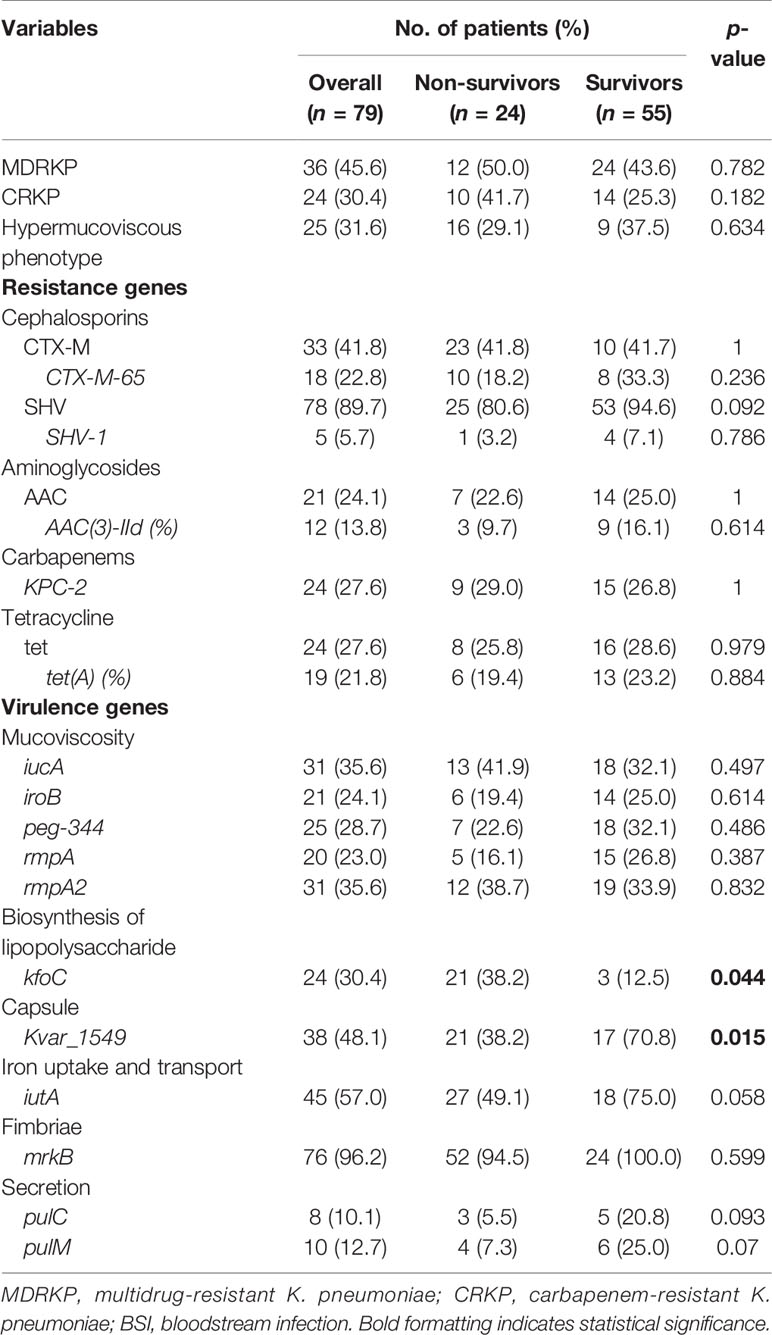
Table 2 Bacterial characteristics of the non-survivor and survivor patients with K. pneumoniae bloodstream infection.
A positive string test was observed in one-third of the isolates (31.6%, n = 25), and most of them harbored at least one of the rmpA/rmpA2/iucA/iroB/peg-344 genes (5/5, n = 7; 4/5, n = 9; 3/5, n = 1; 2/5, n = 7; 1/5, n = 1; 0/5, n = 1) (Table 2 and Figure 1). The most prevalent virulence genes related to mucoviscosity were iucA (35.6%, n = 31) and rmpA2 (35.6%, n = 31), followed by peg-344 (28.7%, n = 25) and iroB (24.1%, n = 21) (Table 2). To better understand the genetic basis of KP strains, MLSTs and serotypes were further analyzed. For K1 and K2 isolates, hypermucoviscous phenotypes were frequently detected (78.6%, 11/14) while resistance genes were scarce. MLST results showed that ST11 was the most commonly identified ST (30.4%, n = 24), followed by ST25 (6.3%, n = 5) and ST23 (5.1%, n = 4) (Supplementary Table S5). The most prevalent capsular type in our strains was K64 (25.3%, n = 20), followed by K2 (10.1%, n = 8) and K1 (7.6%, n = 6). Notably, most of the K64 isolates co-carried genes for beta-lactamases (blaCTX-M, 90%, n = 18) and carbapenemases (blaKPC-2, 95%, n = 19). All the ST11-K64 strains (n = 17) were related to the acquisition of blaKPC-2 gene, and most co-harbored rmpA2 (76.5%, 13/17), iucA (70.6%, 12/17), iutA (76.5%, 13/17), and Kvar_1549 (100%, 17/17) genes. Furthermore, all the ST11-K64 strains in our study were clustered together with ST11-K64 strains R5 and R18, which were confirmed as carbapenem-resistant HvKp in a previous study. ST11-K64 and ST11-non-K64 strains appear to have evolved separately into two different branches. For virulence factors, more virulence genes, including rmpA, iutA, iucA, iucB, iucC, and iucD, were observed in ST11-K64 strains compared with ST11-non-K64 strains (Figure 2). The mortality rate of ST11-K64 strains was 47.2% (7/17) compared with 0% (0/6) of ST11-non-K64 strains.
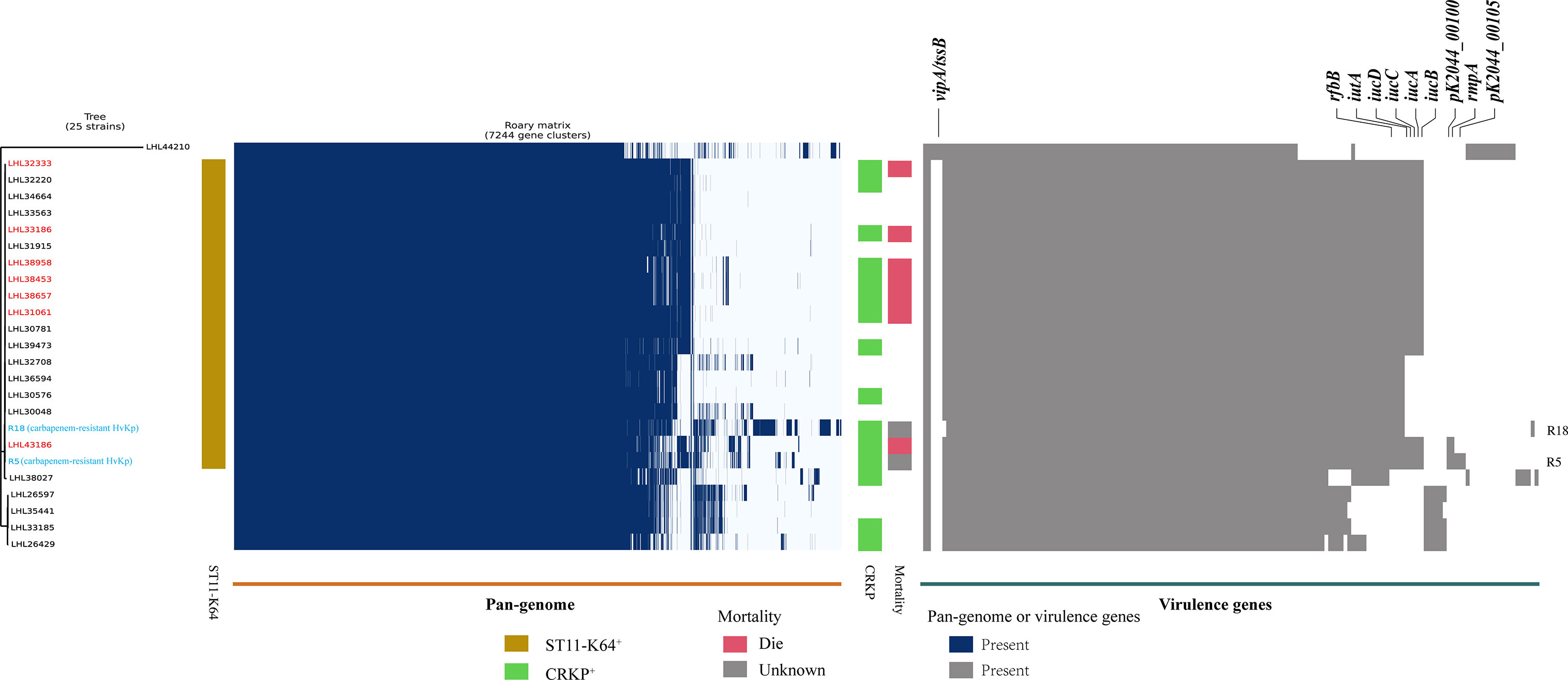
Figure 2 Phylogenetic analysis, pan-genome analyses, and virulence genes of ST11 K. pneumoniae strains. Phylogenetic analysis, pan-genome, and virulence genes are shown on the left, middle, and right, respectively. Phylogenetic analysis was performed by kSNP software based on the core genome SNPs. Strains are color-coded on the tree. Two carbapenem-resistant hypervirulent K. pneumoniae R5 and R18 (marked with blue) were selected from the NCBI SRA database as references. Patients with a fatal outcome were marked with red color. Each row of the heatmap indicated a strain, and each column represented a gene. Colored blocks represent the presence of factors, and white blocks represent absence. The mortality bar of R5 and R18 strains were marked with gray color as unknown outcomes.
The BLAST results identified 268 virulence genes in the tested KP, including biosynthesis of lipopolysaccharide, iron uptake and transport, capsule, fimbriae, and secretion (Supplementary Table S4). As shown in Figure 3, the heatmap of virulence traits showed that secretion-associated genes and genes related to iron uptake and transport and fimbriae were frequently detected in our strains. It should be noted that relatively more virulence genes in iron uptake and secretion were possessed by ST11-K64 strains than by some carbapenem-susceptible KP strains (Figure 3).
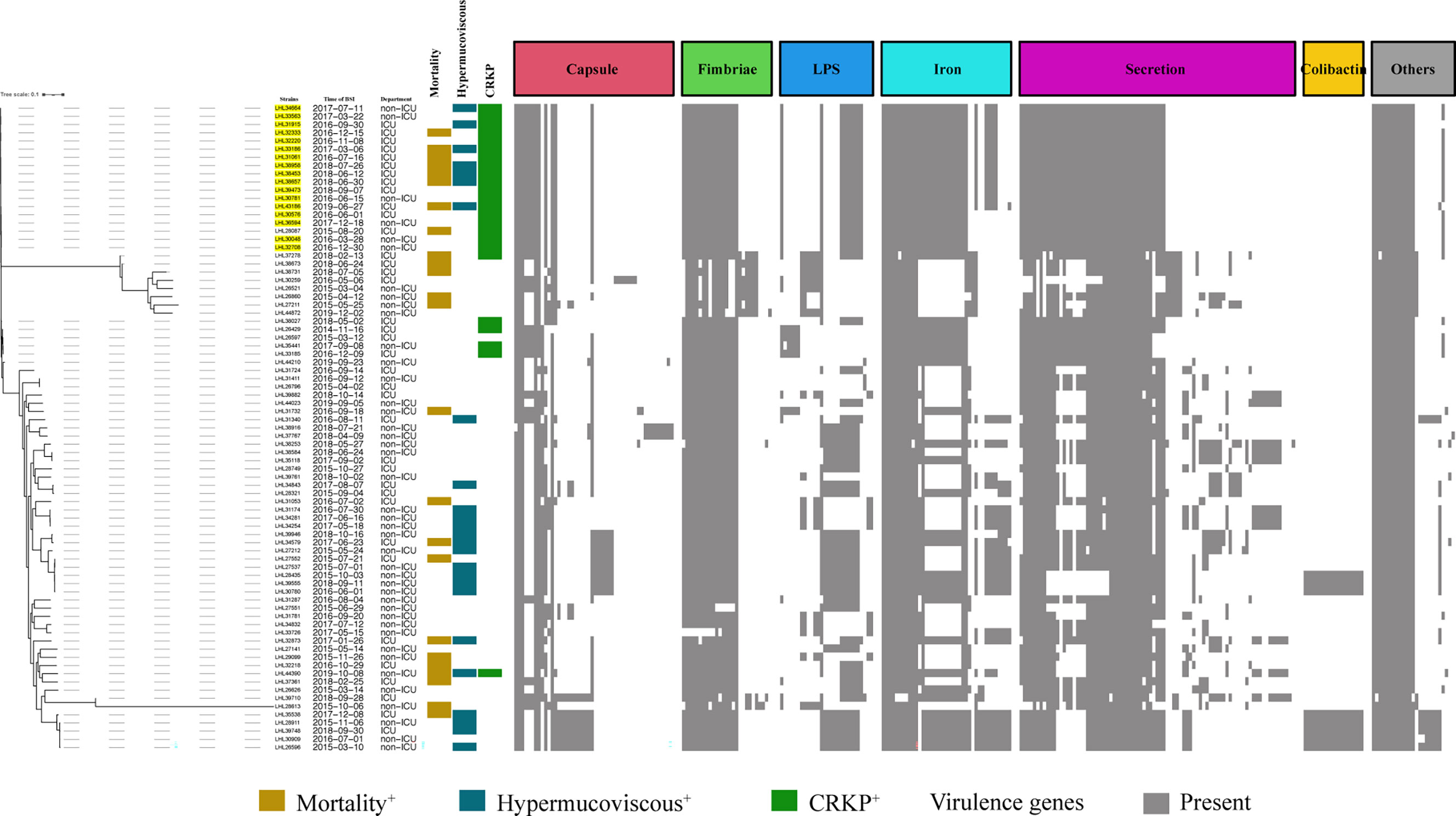
Figure 3 Evolutionary relationships, virulence genes, and distribution of K. pneumoniae strains. Evolutionary relationships and virulence genes are shown on the left and right, respectively. ST11-K64 strains are color-coded on the tree (yellow). Each row of the heatmap indicated a strain, and each column represented a virulence gene that belonged to the indicated functional clusters shown at the top. Colored blocks represent the presence of genes, and white blocks represent absence.
Risk Factors for 30-Day Mortality in Patients With KP BSI
Of the 79 patients enrolled, 55 (69.6%) were classified as survivors and 24 (30.4%) were classified as non-survivors. In the univariate logistic analysis modeling by host factors, prior admission to the ICU, invasive procedures or devices, increased Pitt and APACHE II score at onset of BSI, corticosteroid use before or after BSI, and septic shock exhibited a significant association with increased 30-day mortality. Conversely, the adequacy of empirical antimicrobial treatment and empirical treatment with β-lactam-β-lactamase inhibitor (BLBLI) was associated with decreased 30-day mortality.
Regarding antimicrobial susceptibility, neither CRKP/MDRKP nor the hypermucoviscous phenotype showed a positive association with 30-day mortality. Accordingly, none of the resistance genes were significantly associated with increased 30-day mortality. For virulence traits, several virulence genes were associated with increased 30-day mortality. Notably, the tested strains carried pulC, and all of them co-harbored KP1_2101, A79E_1988, N559_2629, Kvar_1936, Kvar_0795, and Kvar_2690. Similar results were found for pulM and kfoc (Supplementary Table S4). A detailed univariate logistic analysis is shown in Supplementary Table S4.
Because of multicollinearity among the clinical and interconnected nature of the bacterial characteristics, the eigenmatrix method and VIF were conducted before the variables were selected for logistic modeling. Multivariate logistic modeling was performed using four separate sets of adjusted analyses. The variables of the host factors were included in all sets. The increased APACHE II score and septic shock were strongly associated with increased 30-day mortality in all four sets. For the virulence traits, iutA, Kvar_1549, pulM, and pulC were conducted using four separate sets because of multicollinearity among them. iutA (OR, 1.17; 95% CI, 0.97–1.37; p = 0.093), Kvar_1549 (OR, 1.16; 95% CI, 0.96–1.35; p = 0.102), pulM (OR, 1.31; 95% CI, 1.02–1.69; p = 0.038), and pulC (OR, 1.31; 95% CI, 0.99–1.73; p = 0.066) were independent predictors of the 30-day mortality of KP BSI (Table 3).
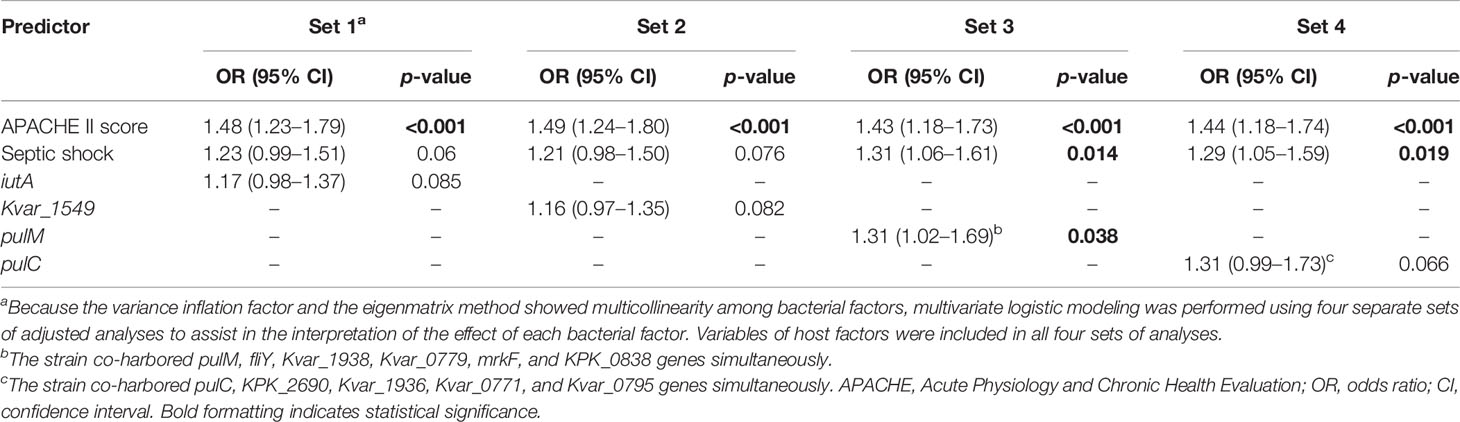
Table 3 Multivariate analysis of predictors associated with the 30-day mortality of K. pneumoniae bloodstream infection.
Risk Factors for 30-Day Mortality in Patients With MDRKP BSI
Subgroup analysis was performed according to the MDR phenotype. In total, 36 patients were enrolled, including 12 non-survivors and 24 survivors. The univariate logistic analysis indicated that increased Pitt and APACHE II scores at the onset of BSI, hypermucoviscous phenotype, mechanical ventilation, septic shock, iutA, rmpA2, and Kvar_1549 were associated with increased 30-day mortality (Supplementary Tables S6, S7). Multivariate analysis revealed that increased APACHE II score, septic shock, and iutA (OR, 1.46; 95% CI, 1.11–1.81, p = 0.002) in set 1 and APACHE II score, septic shock, hypermucoviscous phenotype (OR, 1.24; 95% CI, 0.92–1.68, p = 0.168), and Kvar_1549 (OR, 1.31; 95% CI, 1.02–1.69, p = 0.043) in set 2 increased the 30-day mortality of MDRKP BSI (Table 4).
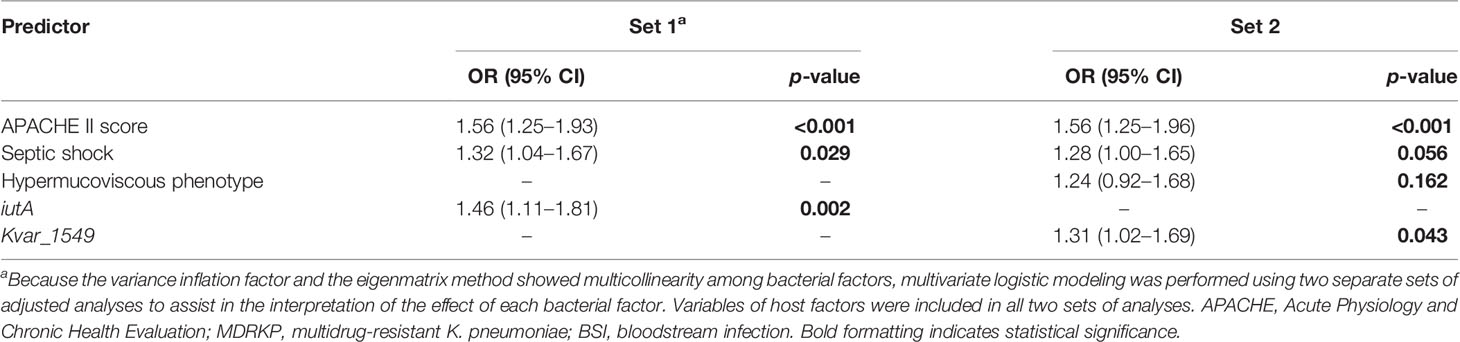
Table 4 Multivariate analysis of predictors associated with the 30-day mortality of MDRKP bloodstream infection.
The survival curves showed a significant impact on 30-day mortality for iutA (75.0% vs. 25.0%, p = 0.041) and Kvar_1549 (70.8% vs. 29.2%, p = 0.006) (Figures 4A, B). Similar results were observed in the subgroup of MDRKP BSI carrying iutA (83.3% vs. 16.7%, p = 0.002) or Kvar_1549 (91.7% vs. 8.3%, p = 0.017) genes (Figures 4C, D).
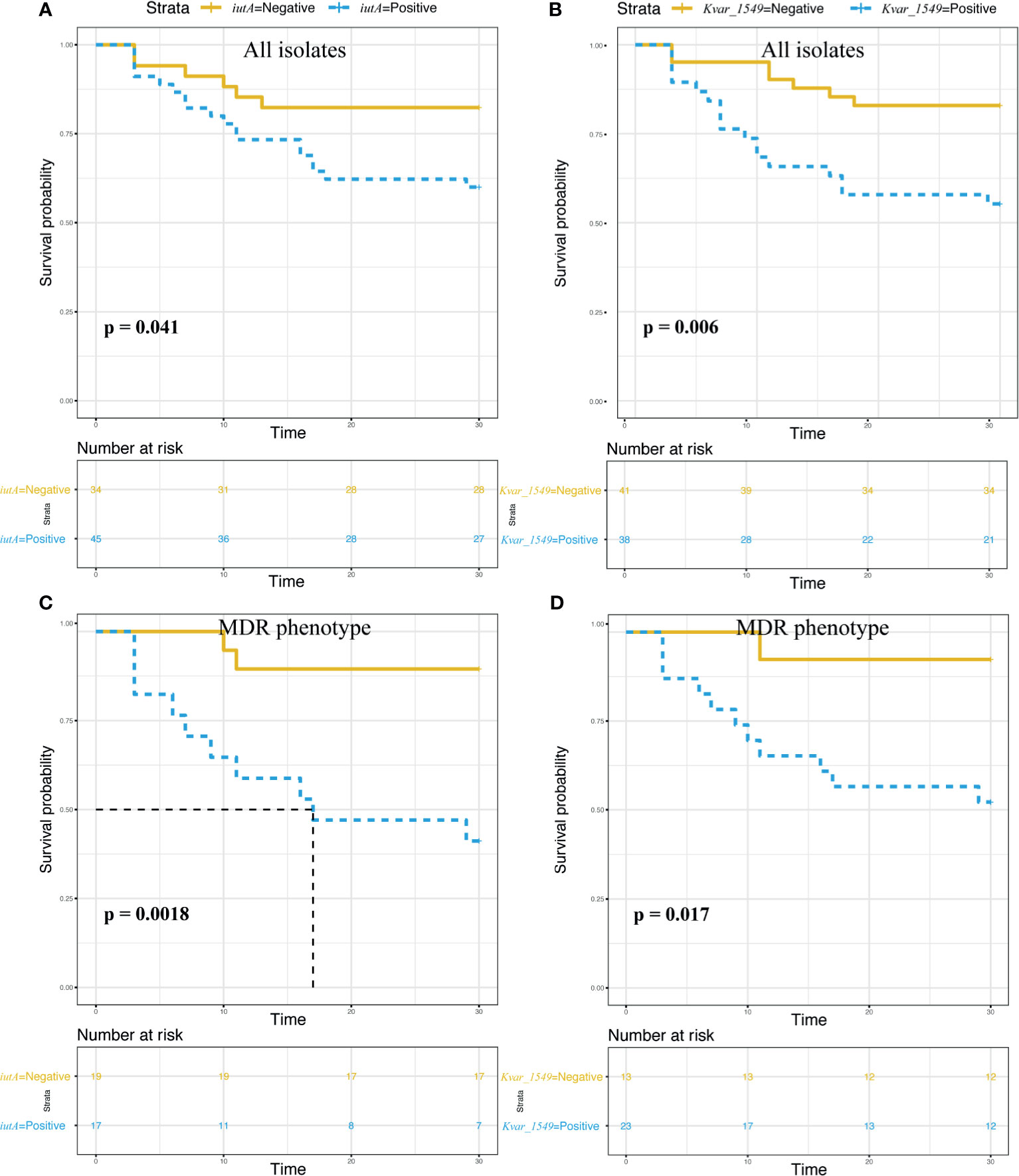
Figure 4 Survival analysis according to the carriage of the iutA and Kvar_1549 genes. Statistical significance was used by the log-rank test. (A, C) iutA gene in all the K. pneumoniae and MDR groups, respectively. (B, D) Kvar_1549 gene in all the K. pneumoniae and MDR groups, respectively.
Discussion
This study highlighted that iutA and Kvar_1549 were associated with increased 30-day mortality in KP BSI. Of note, higher mortality was observed when iutA and Kvar_1549 carriage by causative strains was accompanied by MDR. Some tested strains in the present study co-harbored resistance determinants and hypervirulence genes. Previous studies have indicated that hypervirulent and MDR strains evolved separately in different clonal groups, and MDRKP had lower virulence, whereas HvKp was sensitive to most antibiotics (Russo and Marr, 2019). The presence of either drug resistance or virulence genes in bacterial isolates may lead to treatment failure, severe life-threatening infections, and subsequent prolongation of the illness (Tumbarello et al., 2012; Viale et al., 2013; Russo and Marr, 2019). The integration of these factors together may further complicate clinical practice. MDR or carbapenem-resistant HvKp isolated from different clinical settings result in fatality and have begun to spread in China (Zhang et al., 2015; Gu et al., 2018; Huang et al., 2018; Zheng et al., 2020). Recently, a multi-center epidemiological and comparative genomic analysis indicated that the ST11-K64 hypervirulent CRKP, isolated from bacterial liver abscesses, was simultaneously MDR and hypervirulent using comprehensive test combinations including string test, human neutrophil killing assay, and G. mellonella infection model (Yang et al., 2020). We identified 17 ST11-K64 isolates, which were all related to the acquisition of blaKPC-2 gene, and most co-harbored rmpA2 (76.5%, 13/17), iucA (70.6%, 12/17), iutA (76.5%, 13/17), and Kvar_1549 (100%, 17/17) genes in our study. Comparative genome analysis showed that the ST11-K64 strains were phylogenetically closely related to carbapenem-resistant HvKp strains R5 and R18. More virulence factors were observed in the group of ST11-K64 carbapenem-resistant HvKp, which may account for its high mortality (47.2%, 7/17) rate in patients infected with this strain. In addition, these strains exhibited sustained survival during the 4-year study period, especially in the ICU department, indicating that they may chronically colonize the surrounding environment. This study provides clinical evidence of the hypervirulence of ST11-K64 carbapenem-resistant HvKp. Therefore, urgent control measures are required for its dissemination.
Capsule polysaccharides (CPS) are a major virulence factor in most isolates because of their ability to evade phagocytosis and complement-mediated killing and further inhibit complement activation of the host (Fang et al., 2004; Cozzone, 2005). Hypervirulence is associated with the overproduction of CPS, and in the absence of related virulence genes, its virulence is reduced or abolished (Favre-Bonte et al., 1999; Wyres et al., 2016). Shon et al. (2013) assumed that the high level of virulence was due to an increased expression of CPS combined with increased efficiency of iron acquisition and other traits. Kvar_1549, identified in our logistic model, was found to be associated with CPS. Increased mortality was observed in patients infected with these strains, with borderline differences in all KP and significant differences in the MDR phenotype. The protein encoded by Kvar_1549 is a type 2 phosphatidic acid phosphatase-like (PAP2) protein, which controls various physiological functions, such as CPS and lipopolysaccharide synthesis. However, to date, no cases of Kvar_1549 have been associated with increased 30-day mortality in patients with KP BSI. Zhao et al. (2019) indicated that LpxE, a PAP2 encoded protein, participates in multiple layers of biogenesis of the gram-negative bacterial envelope and increases antibiotic resistance and bacterial virulence. Thus, Kvar_1549 may be responsible for the formation of CPS, leading to enhanced virulence. Notably, the prevalence of Kvar_1549 in the tested strains was 48.1%. Continued studies regarding the role of Kvar_1549 should be performed using molecular analysis and applicable infection models because of its high prevalence in KP isolates.
iutA, the fifth gene of the aerobactin operon, encodes a specific outer membrane receptor protein for iron uptake. The ability to acquire iron is essential for bacterial growth and replication owing to its role as a cofactor for several enzymes, such as those involved in electron transport and amino acid and DNA biosynthesis (Wandersman and Stojiljkovic, 2000). This iron uptake system plays a crucial role in the progression of infection. Vargas et al. (2019) indicated that iutA promotes biofilm formation. Tang et al. (2010) showed that iutA is an independent pathogenicity factor for abscess formation. An in vitro study revealed that the expression of iutA was upregulated in pathogenic bacteria under iron-depleted conditions (Torres et al., 2012). Further tests revealed that the iutA mutant was outcompeted by the wild-type strain in the murine model and unable to persist in vivo (Torres et al., 2012). Similar findings were observed in a chicken infection model. An aerobactin-defective mutant of iutA was constructed and showed significantly decreased pathogenicity compared with the wild-type strain, as evidenced by the low extent of colonization in selected organs or being outcompeted in vivo (Gao et al., 2015). Moreover, a case–control clinical study found that iutA was an independent risk factor associated with 30-day mortality in patients with KP BSI. Consistent with previous results, we further determined that iutA carriage by causative strains was an independent risk factor for 30-day mortality in patients with KP BSI, and a higher mortality rate was observed when it was accompanied by MDR. We did not further analyze the risk factors of mortality in patients with CRKP BSI because of the limited number of cases. Notably, a fatal outcome was observed in patients with iutA-positive CRKP BSI (81.3%, 13/16). In addition, iutA was first found to be located on pColV plasmids, and subsequent studies revealed that it could also be chromosomally encoded in many strains (Warner et al., 1981; Lawlor and Payne, 1984; Mcdougall and Neilands, 1984). The emergence and dissemination of such hypervirulence plasmids, such as the virulence plasmid pLVPK that resulted in a fatal outbreak in ST11 CRKP (Gu et al., 2018), is concerning. Taken together, the iutA-related virulence factor plays a significant role in the pathogenesis of KP BSI and is a related marker associated with poor prognosis, especially in pathogenic strains accompanied by MDR and carbapenem-resistant phenotypes.
This study has several limitations. The retrospective nature of the study and the small sample size are intrinsic study limitations. In addition, the amount of missing patient data may have resulted in bias. Second, data from a single center limited generalizability to other geographical areas or institutions. Finally, although all the virulence genes presented in the tested strains were evaluated in our study, the expression of related genes was not determined in in vitro and in vivo experiments, such as knockout, qRT-PCR, and infection animal models.
In summary, this study identified that iutA or Kvar_1549 gene was associated with poor prognosis, especially in MDR phenotypes of KP BSI. Moreover, the long-term colonization and dissemination of ST11-K64 carbapenem-resistant HvKP isolates resulted in increased difficulties and challenges in the treatment of infections. Therefore, clinicians should carefully manage patients with BSI caused by these difficult-to-treat strains. Further studies are needed to elucidate the pathogenic mechanisms and transmission dynamics of HvKP and carbapenem-resistant HvKP.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: the National Microbiology Data Center (https://nmdc.cn/en) under the accession numbers NMDC60014864-NMDC60014942.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Lihuili Hospital, Ningbo Medical Center (no. KY2019PJ028). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
XW, CH, and HW conceptualized and planned the work that led to the manuscript. QS and SS collected the clinical and MIC data. CH and XW analyzed the data. XW and CH drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Ningbo (Grant number 2019A610232).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Editage for their assistance in language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.688989/full#supplementary-material
References
Bialek-Davenet, S., Criscuolo, A., Ailloud, F., Passet, V., Jones, L., Delannoy-Vieillard, A. S., et al. (2014). Genomic Definition of Hypervirulent and Multidrug-Resistant Klebsiella Pneumoniae Clonal Groups. Emerg. Infect. Dis. 20, 1812–1820. doi: 10.3201/eid2011.140206
Buchfink, B., Reuter, K., Drost, H. G. (2021). Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 18, 366–368. doi: 10.1038/s41592-021-01101-x
Centers for Disease Control and Prevention. CDC/NHSN Surveillance Definitions for Specific Types of Infections. Available at: http://www.cdc.gov/nhsn/PDFs/pscManual/17pscNosInfDef_current.pdf (Accessed 15 January 2014).
Charlson, M., Szatrowski, T. P., Peterson, J., Gold, J. (1994). Validation of a Combined Comorbidity Index. J. Clin. Epidemiol. 47, 1245–1251. doi: 10.1016/0895-4356(94)90129-5
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Available at: http://www.clsi.org/ (Accessed January 21, 2020).
Cozzone, A. J. (2005). Role of Protein Phosphorylation on Serine/Threonine and Tyrosine in the Virulence of Bacterial Pathogens. J. Mol. Microbiol. Biotechnol. 9, 198–213. doi: 10.1159/000089648
Fang, C. T., Chuang, Y. P., Shun, C. T., Chang, S. C., Wang, J. T. (2004). A Novel Virulence Gene in Klebsiella Pneumoniae Strains Causing Primary Liver Abscess and Septic Metastatic Complications. J. Exp. Med. 199, 697–705. doi: 10.1084/jem.20030857
Favre-Bonte, S., Licht, T. R., Forestier, C., Krogfelt, K. A. (1999). Klebsiella Pneumoniae Capsule Expression Is Necessary for Colonization of Large Intestines of Streptomycin-Treated Mice. Infect. Immun. 67, 6152–6156. doi: 10.1128/IAI.67.11.6152-6156.1999
Gao, Q. Q., Jia, X. X., Wang, X. B., Xiong, L. P., Gao, S., Liu, X. F. (2015). The Avian Pathogenic Escherichia Coli O2 Strain E058 Carrying the Defined Aerobactin-Defective iucD or Iucdiuta Mutation Is Less Virulent in the Chicken. Infect. Genet. Evol. 30, 267–277. doi: 10.1016/j.meegid.2014.12.038
Garner, J. S., Jarvis, W. R., Emori, T. G., Horan, T. C., Hughes, J. M. (1988). Cdc Definitions for Nosocomial Infections 1988. Am. J. Infect. Control 16, 128–140. doi: 10.1016/0196-6553(88)90053-3
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A Fatal Outbreak of ST11 Carbapenem-Resistant Hypervirulent Klebsiella Pneumoniae in a Chinese Hospital: A Molecular Epidemiological Study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Harada, S., Aoki, K., Yamamoto, S., Ishii, Y., Sekiya, N., Kurai, H., et al. (2019). Clinical and Molecular Characteristics of Klebsiella Pneumoniae Isolates Causing Bloodstream Infections in Japan: Occurrence of Hypervirulent Infections in Health Care. J. Clin. Microbiol. 57 (11), e01206-19. doi: 10.1128/JCM.01206-19
Holt, K. E., Wertheim, H., Zadoks, R. N., Baker, S., Whitehouse, C. A., Dance, D., et al. (2015). Genomic Analysis of Diversity, Population Structure, Virulence, and Antimicrobial Resistance in Klebsiella Pneumoniae, an Urgent Threat to Public Health. Proc. Natl. Acad. Sci. U. S. A. 112, E3574–E3581. doi: 10.1073/pnas.1501049112
Huang, Y. H., Chou, S. H., Liang, S. W., Ni, C. E., Lin, Y. T., Huang, Y. W., et al. (2018). Emergence of an XDR and Carbapenemase-Producing Hypervirulent Klebsiella Pneumoniae Strain in Taiwan. J. Antimicrobial Chemother. 73, 2039–2046. doi: 10.1093/jac/dky164
Hu, F., Guo, Y., Zhu, D., Wang, F., Jiang, X., Fu, Y., et al. (2020). CHINET Surveillance of Bacterial Resistance in China: 2018 Report. Chin. J. Infect. Chemother. 20, 1–10. doi: 10.16718/j.1009-7708.2020.01.001
Kim, D., Park, B. Y., Choi, M. H., Yoon, E. J., Lee, H., Lee, K. J., et al. (2019). Antimicrobial Resistance and Virulence Factors of Klebsiella Pneumoniae Affecting 30 Day Mortality in Patients With Bloodstream Infection. J. Antimicrob. Chemother. 74, 190–199. doi: 10.1093/jac/dky397
Knaus, W. A., Draper, E. A., Wagner, D. P., Zimmerman, J. E. (1985). Apache-Ii - A Severity of Disease Classification-System. Crit. Care Med. 13, 818–829. doi: 10.1097/00003246-198510000-00009
Kohler, P. P., Volling, C., Green, K., Uleryk, E. M., Shah, P. S., Mcgeer, A. (2017). Carbapenem Resistance, Initial Antibiotic Therapy, and Mortality in Klebsiella Pneumoniae Bacteremia: A Systematic Review and Meta-Analysis. Infect. Control Hosp. Epidemiol. 38, 1319–1328. doi: 10.1017/ice.2017.197
Lawlor, K. M., Payne, S. M. (1984). Aerobactin Genes in Shigella Spp. J. Bacteriol. 160, 266–272. doi: 10.1128/jb.160.1.266-272.1984
Lee, C. R., Lee, J. H., Park, K. S., Jeon, J. H., Kim, Y. B., Cha, C. J., et al. (2017). Antimicrobial Resistance of Hypervirulent Klebsiella Pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell Infect. Microbiol. 7, 483. doi: 10.3389/fcimb.2017.00483
Lery, L. M., Frangeul, L., Tomas, A., Passet, V., Almeida, A. S., Bialek-Davenet, S., et al. (2014). Comparative Analysis of Klebsiella Pneumoniae Genomes Identifies a Phospholipase D Family Protein as a Novel Virulence Factor. BMC Biol. 12, 41. doi: 10.1186/1741-7007-12-41
Lin, Y. T., Jeng, Y. Y., Chen, T. L., Fung, C. P. (2010). Bacteremic Community-Acquired Pneumonia Due to Klebsiella Pneumoniae: Clinical and Microbiological Characteristics in Taiwan 2001-2008. BMC Infect. Dis. 10, 307. doi: 10.1186/1471-2334-10-307
Li, J., Ren, J., Wang, W., Wang, G., Gu, G., Wu, X., et al. (2018). Risk Factors and Clinical Outcomes of Hypervirulent Klebsiella Pneumoniae Induced Bloodstream Infections. Eur. J. Clin. Microbiol. Infect. Dis. 37, 679–689. doi: 10.1007/s10096-017-3160-z
Mcdougall, S., Neilands, J. B. (1984). Plasmid-Coded and Chromosome-Coded Aerobactin Synthesis in Enteric Bacteria - Insertion Sequences Flank Operon in Plasmid-Mediated Systems. J. Bacteriol. 159, 300–305. doi: 10.1128/jb.159.1.300-305.1984
Mohammad Ali Tabrizi, A., Badmasti, F., Shahcheraghi, F., Azizi, O. (2018). Outbreak of Hypervirulent Klebsiella Pneumoniae Harbouring blaVIM-2 Among Mechanically-Ventilated Drug-Poisoning Patients With High Mortality Rate in Iran. J. Glob. Antimicrob. Resist. 15, 93–98. doi: 10.1016/j.jgar.2018.06.020
Namikawa, H., Niki, M., Niki, M., Yamada, K., Nakaie, K., Sakiyama, A., et al. (2019). Clinical and Virulence Factors Related to the 30-Day Mortality of Klebsiella Pneumoniae Bacteremia at a Tertiary Hospital: A Case-Control Study. Eur. J. Clin. Microbiol. Infect. Dis. 38, 2291–2297. doi: 10.1007/s10096-019-03676-y
Paczosa, M. K., Mecsas, J. (2016). Klebsiella Pneumoniae: Going on the Offense With a Strong Defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Russo, T. A., Marr, C. M. (2019). Hypervirulent Klebsiella Pneumoniae. Clin. Microbiol. Rev. 32 (3), e00001-19. doi: 10.1128/CMR.00001-19
Russo, T. A., Olson, R., Fang, C. T., Stoesser, N., Miller, M., Macdonald, U., et al. (2018). Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella Pneumoniae From Classical K-Pneumoniae. J. Clin. Microbiol. 56 (9), e00776-18. doi: 10.1128/JCM.00776-18
Russo, T. A., Olson, R., Macdonald, U., Beanan, J., Davidson, B. A. (2015). Aerobactin, But Not Yersiniabactin, Salmochelin, or Enterobactin, Enables the Growth/Survival of Hypervirulent (Hypermucoviscous) Klebsiella Pneumoniae Ex Vivo and In Vivo. Infect. Immun. 83, 3325–3333. doi: 10.1128/IAI.00430-15
Shon, A. S., Bajwa, R. P. S., Russo, T. A. (2013). Hypervirulent (Hypermucoviscous) Klebsiella Pneumoniae A New and Dangerous Breed. Virulence 4, 107–118. doi: 10.4161/viru.22718
Tang, H. L., Chiang, M. K., Liou, W. J., Chen, Y. T., Peng, H. L., Chiou, C. S., et al. (2010). Correlation Between Klebsiella Pneumoniae Carrying pLVPK-Derived Loci and Abscess Formation. Eur. J. Clin. Microbiol. Infect. Dis. 29, 689–698. doi: 10.1007/s10096-010-0915-1
Torres, A. G., Cieza, R. J., Rojas-Lopez, M., Blumentritt, C. A., Souza, C. S., Johnston, R. K., et al. (2012). In Vivo Bioluminescence Imaging of Escherichia Coli O104:H4 and Role of Aerobactin During Colonization of a Mouse Model of Infection. BMC Microbiol. 12, 112. doi: 10.1186/1471-2180-12-112
Tumbarello, M., Viale, P., Viscoli, C., Trecarichi, E. M., Tumietto, F., Marchese, A., et al. (2012). Predictors of Mortality in Bloodstream Infections Caused by Klebsiella Pneumoniae Carbapenemase-Producing K. Pneumoniae: Importance of Combination Therapy. Clin. Infect. Dis. 55, 943–950. doi: 10.1093/cid/cis588
Vargas, J. M., Mochi, M. P. M., Nunez, J. M., Caceres, M., Mochi, S., Moreno, R. D., et al. (2019). Virulence Factors and Clinical Patterns of Multiple-Clone Hypermucoviscous KPC-2 Producing K. Pneumoniae. Heliyon 5 (6), e01829. doi: 10.1016/j.heliyon.2019.e01829
Viale, P., Giannella, M., Lewis, R., Trecarichi, E. M., Petrosillo, N., Tumbarello, M. (2013). Predictors of Mortality in Multidrug-Resistant Klebsiella Pneumoniae Bloodstream Infections. Expert Rev. Anti Infect. Ther. 11, 1053–1063. doi: 10.1586/14787210.2013.836057
Wandersman, C., Stojiljkovic, I. (2000). Bacterial Heme Sources: The Role of Heme, Hemoprotein Receptors and Hemophores. Curr. Opin. Microbiol. 3, 215–220. doi: 10.1016/S1369-5274(00)00078-3
Warner, P. J., Williams, P. H., Bindereif, A., Neilands, J. B. (1981). ColV Plasmid-Specific Aerobactin Synthesis by Invasive Strains of Escherichia Coli. Infect. Immun. 33, 540–545. doi: 10.1128/iai.33.2.540-545.1981
Wyres, K. L., Wick, R. R., Gorrie, C., Jenney, A., Follador, R., Thomson, N. R., et al. (2016). Identification of Klebsiella Capsule Synthesis Loci From Whole Genome Data. Microb. Genom. 2, e000102. doi: 10.1099/mgen.0.000102
Xu, L., Sun, X., Ma, X. (2017). Systematic Review and Meta-Analysis of Mortality of Patients Infected With Carbapenem-Resistant Klebsiella Pneumoniae. Ann. Clin. Microbiol. Antimicrob. 16, 18. doi: 10.1186/s12941-017-0191-3
Yang, Q., Jia, X., Zhou, M., Zhang, H., Yang, W., Kudinha, T., et al. (2020). Emergence of ST11-K47 and ST11-K64 Hypervirulent Carbapenem-Resistant Klebsiella Pneumoniae in Bacterial Liver Abscesses From China: A Molecular, Biological, and Epidemiological Study. Emerg. Microbes Infect. 9, 320–331. doi: 10.1080/22221751.2020.1721334
Zhang, R., Lin, D., Chan, E. W., Gu, D., Chen, G. X., Chen, S. (2016). Emergence of Carbapenem-Resistant Serotype K1 Hypervirulent Klebsiella Pneumoniae Strains in China. Antimicrob. Agents Chemother. 60, 709–711. doi: 10.1128/AAC.02173-15
Zhang, Y., Zeng, J., Liu, W., Zhao, F., Hu, Z., Zhao, C., et al. (2015). Emergence of a Hypervirulent Carbapenem-Resistant Klebsiella Pneumoniae Isolate From Clinical Infections in China. J. Infect. 71, 553–560. doi: 10.1016/j.jinf.2015.07.010
Zhao, J. S., An, J. S., Hwang, D., Wu, Q. L., Wang, S., Gillespie, R. A., et al. (2019). The Lipid A 1-Phosphatase, LpxE, Functionally Connects Multiple Layers of Bacterial Envelope Biogenesis. Mbio 10 (3), e00886-19. doi: 10.1128/mBio.00886-19
Zheng, S., Li, P., Zhang Zhengliang, P. H. (2019). CHINET Surveilance of Carbapenem-Resistant Gram-Negative Bacteria in China From 2005 to 2017. J. Clin. Emerg. (China) 20, 45–49. doi: 10.13201/j.issn.1009-5918.2019.01.008
Zheng, B., Xu, H., Lv, T., Guo, L., Xiao, Y., Huang, C., et al. (2020). Stool Samples of Acute Diarrhea Inpatients as a Reservoir of ST11 Hypervirulent KPC-2-Producing Klebsiella Pneumoniae. mSystems 5 (3), e00498-20. doi: 10.1128/mSystems.00498-20
Keywords: Klebsiella pneumoniae, bloodstream infection (BSI), mortality, hypervirulent, carbapenem-resistant, risk factors
Citation: Wu X, Shi Q, Shen S, Huang C and Wu H (2021) Clinical and Bacterial Characteristics of Klebsiella pneumoniae Affecting 30-Day Mortality in Patients With Bloodstream Infection. Front. Cell. Infect. Microbiol. 11:688989. doi: 10.3389/fcimb.2021.688989
Received: 31 March 2021; Accepted: 13 August 2021;
Published: 16 September 2021.
Edited by:
Krisztina M. Papp-Wallace, Louis Stokes Cleveland VA Medical Center, United StatesReviewed by:
Maria Fernanda Mojica, Case Western Reserve University, United StatesShuguang Li, Peking University People’s Hospital, China
Ru Yue Tian, Fudan University, China
Copyright © 2021 Wu, Shi, Shen, Huang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Huang, ZHJoY2hlbkAxNjMuY29t; Hongcheng Wu, ZG9jdG9yd3UxOTY3QDEyNi5jb20=
 Xingbing Wu1
Xingbing Wu1 Chen Huang
Chen Huang Hongcheng Wu
Hongcheng Wu