- 1Centro Internacional de Entrenamiento e Investigaciones Médicas (CIDEIM), Cali, Colombia
- 2Universidad IcesiI, Cali, Colombia
- 3Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD, United States
- 4Center for Bioinformatics and Computational Biology, University of Maryland, College Park, MD, United States
- 5Pontificia Universidad Javeriana, Cali, Colombia
Early host-pathogen interactions drive the host response and shape the outcome of natural infections caused by intracellular microorganisms. These interactions involve a number of immune and non-immune cells and tissues, along with an assortment of host and pathogen-derived molecules. Our current knowledge has been predominantly derived from research on the relationships between the pathogens and the invaded host cell(s), limiting our understanding of how microbes elicit and modulate immunological responses at the organismal level. In this study, we explored the early host determinants of healing and non-healing responses in human cutaneous leishmaniasis (CL) caused by Leishmania (Viannia) panamensis. We performed a comparative transcriptomic profiling of peripheral blood mononuclear cells from healthy donors (PBMCs, n=3) exposed to promastigotes isolated from patients with chronic (CHR, n=3) or self-healing (SH, n=3) CL, and compared these to human macrophage responses. Transcriptomes of L. V. panamensis-infected PBMCs showed enrichment of functional gene categories derived from innate as well as adaptive immune cells signatures, demonstrating that Leishmania modulates adaptive immune cell functions as early as after 24h post interaction with PBMCs from previously unexposed healthy individuals. Among differentially expressed PBMC genes, four broad categories were commonly modulated by SH and CHR strains: cell cycle/proliferation/differentiation, metabolism of macromolecules, immune signaling and vesicle trafficking/transport; the first two were predominantly downregulated, and the latter upregulated in SH and CHR as compared to uninfected samples. Type I IFN signaling genes were uniquely up-regulated in PBMCs infected with CHR strains, while genes involved in the immunological synapse were uniquely downregulated in SH infections. Similarly, pro-inflammatory response genes were upregulated in isolated macrophages infected with CHR strains. Our data demonstrate that early responses during Leishmania infection extend beyond innate cell and/or phagocytic host cell functions, opening new frontiers in our understanding of the triggers and drivers of human CL.
Introduction
Clinical manifestations of human infections with Leishmania Viannia species (primarily L. V. braziliensis and L.V. panamensis) range from single self-healing skin lesions, to severe and chronic cutaneous or mucosal disease (Weigle et al., 1993; Murray et al., 2005). Contrary to infections where microorganisms inflict direct damage to the host via toxins or cellular injury, symptomatic dermal leishmaniasis (cutaneous, mucosal or muco-cutaneous) results from parasite-elicited immunopathology, and disease severity depends on the degree of immunopathology and concomitant immunoregulation (Murray et al., 2005; Rodriguez-Pinto et al., 2012). For these immunopathologically-driven infections, the early host-microbe interactions and the elicited innate immune responses, together with specific features of the anatomical sites and tissues affected, shape the adaptive responses that contribute to pathology and disease severity. However, the triggers and drivers of immunopathology of dermal leishmaniasis remain largely unknown.
Genotypic and phenotypic variability among Leishmania isolates of the same species has been long-recognized. Within the L. Viannia subgenus, a number of phenotypically discernable populations (zymodemes) co-circulate in endemic foci of transmission, with L. V braziliensis and L.V. panamensis being the two species with the highest heterogeneity (Saravia et al., 1998). Distinct host responses as well as intrinsic parasite phenotypes have been correlated with different zymodemes. Disease caused by L. V. braziliensis zymodeme 1.1 has been associated with a longer time of evolution in humans (Saravia et al., 1998), and L. V. panamensis zymodeme 2.3 has been identified as a population intrinsically resistant to antimony (Fernandez et al., 2014). Thus, parasite factors, as well as their interaction with the host may contribute to the distinct immunological responses unleashed upon infection with different strains of the same Leishmania species. Although the specific determinants of such responses are currently unknown, variations in virulence factors involved in modulating the early stages of the host-pathogen interaction [e.g. GP63 copy number or function (Gomez and Olivier, 2010)], could contribute to divergent clinical outcomes of disease.
Characterization of the early Leishmania-host interactions has been primarily conducted in the context of isolated phagocyte infections [macrophages, dendritic cells and neutrophils, and predominantly using murine or human cell lines (Cohen-Freue et al., 2007; Ovalle-Bracho et al., 2015)]. We have previously shown that L. V. panamensis strains isolated from patients with chronic cutaneous leishmaniasis (CL) induced significantly more expression of pro-inflammatory chemokines and cytokines in human macrophages (Navas et al., 2014), consistent with parasite-elicited hyperactivation of the host-cell inflammatory response driving disease severity. However, during natural vector-borne infections, the initial host-parasite interaction involves a multitude of other immune and non-immune cells, which constitute the cellular micro-environment to which, promastigotes are exposed after inoculation into the human host. Murine models of infection have provided evidence of the early participation of keratinocytes in susceptibility to L. major via induction of pro-inflammatory chemokines and cytokines (Ehrchen et al., 2010; Ronet et al., 2019). In humans, keratinocytes were proposed to participate in the development of post-kala azar dermal leishmaniasis, and in the pathologic changes of ulcerating skin lesions (Gasim et al., 1998; Tasew et al., 2010). The participation of non-immune and non-phagocytic immune cells in the early interactions of L. Viannia species and the host, and the contribution of those responses to development of symptomatic disease and in disease severity, are not completely understood.
Gene expression profiling of peripheral blood mononuclear cells (PBMCs) responses, both at the basal level and after antigenic recall, has been successfully implemented as a tool to identify immunological gene signatures of clinical and therapeutic outcomes in human infections. Illustrating this, transcriptional profiling of PBMCs has revealed gene signatures associated with immunological protection in human malaria vaccine studies (Moncunill et al., 2020), responsiveness to antiviral treatment in human hepatitis C infections (Orr et al., 2020), and long-lasting immune dysfunction in patients with chronic ebola infection (Wiedemann et al., 2020), among others. More recently, consistency between results from bulk RNA-seq and single cell sequencing has been demonstrated with PBMCs from COVID-19 patients (Arunachalam et al., 2020), supporting the use of bulk transcriptomics to explore immunological signatures of infectious diseases and disease severity.
Antigen-specific recall responses of PBMCs from CL patients has demonstrated that a mixed Th1/Th2 response, accompanied by immune deregulation, participates in the clinical manifestations and disease severity caused by L. Viannia (Carvalho et al., 1985; Saravia et al., 1989; Diaz et al., 2010). In this study, we examined how clinical strains of L.V. panamensis which cause different degrees of severity of CL in humans (chronic or self-healing CL), modulate the early responses of PBMCs within the first 24 hours of parasite-host interaction. We hypothesize that these initial PBMC-promastigote interactions contribute to define the nature and magnitude of activated or repressed host functions, ultimately determining the clinical outcome of infection. Identification of immunological signatures associated with development of more severe disease manifestations and triggered during the early phases of infection, will serve as a knowledge base for the development of prognostic tools, ultimately allowing timely intervention for these cases, known to be more refractory to first-line antileishmanial drugs (Amato et al., 2008; Maurer-Cecchini et al., 2009; Bamorovat et al., 2021).
Materials and Methods
Isolation of Total WBCs and PBMCs
Peripheral blood samples from healthy volunteers without history of CL and residing in an urban non-endemic city, were collected and processed to obtain total WBCs and PBMCs. WBCs were isolated by centrifugation at 400 g for 15 min at RT and cells were collected from the interface between the plasma and red blood cells. WBCs were incubated in RBC lysis buffer ([150 mM] NH4CL, [10 mM] KHCO3, [0.1 mM] Na2EDTA) for 5 min at RT, washed with PBS, and resuspended in RPMI supplemented with 10% FBS for subsequent procedures. PBMCs were obtained by centrifugation of PBS-diluted (1:1) blood samples over a Ficoll-Hypaque gradient (Sigma-Aldrich) following the manufacturer’s instructions.
Leishmania Strains
Clinical strains were obtained from the CIDEIM BioBank. Strains were originally isolated by needle aspirate of cutaneous lesions or from biopsies of mucosal lesions, propagated in Senekjie´s biphasic blood agar and immediately stored in liquid nitrogen until use. Strains were typed by immunoreactivity to monoclonal antibodies. L. V. panamensis strains MHOM/CO/11/5430 (5430chr), MHOM/CO/08/5433 (5433chr) and MHOM/CO/08/5397 (5397chr) were isolated from patients with chronic CL of > 6 months´ evolution. MHOM/CO/87/1320 (1320chr) and MHOM/CO/85/2504 (2504chr) were isolated from lesion biopsies of nasal mucosa from patients with >10 years of muco-cutaneous disease. Strains MHOM/CO/85/2272 (2272sh), MHOM/CO/85/2271 (2271sh), MHOM/CO/89/2189 (2189sh) and MHOM/CO/83/1022 (1022sh) were isolated from CL patients who clinically resolved disease in the absence of any treatment (self-healing CL).
Infection
Promastigotes were grown at 25°C in Senekjie’s biphasic medium and passed for a maximum of two sub-passages into RPMI 1640 supplemented with 10% heat-inactivated FBS and 5 mg/ml hemin. Ten million WBCs or PBMCs were infected at a 10:1 parasite-to-monocyte ratio with human AB+ serum-opsonized stationary phase promastigotes for 24h. Selection of 24 hours post infection (hpi) as the time to characterize the early interaction of PBMCs and Leishmania, was based on prior transcriptomic data showing a strong modulation of gene expression at 4 hpi, and stabilization of these responses at 24 hpi, remaining essentially unaltered up to 72 hpi (Dillon et al., 2015; Fernandes et al., 2016).
Macrophage Samples
All macrophage samples used were those previously collected and characterized in a previous study from our group (Navas et al., 2014). Briefly, macrophages were differentiated from PBMCs by adherence to cell culture plasticware in serum-free RPMI for 2h, followed by culture for 7 days in RPMI supplemented with 20% FBS at 37°C and 5% CO2. Cells were infected for 24h at a 1:10 ratio as described above.
RNA Isolation and cDNA Synthesis
Total RNA was extracted from uninfected and infected cultured cells using Trizol (Invitrogen, USA) followed by RNA cleanup with RNeasy Mini Kit columns (Qiagen, USA). RNA integrity was assessed using an Agilent 2100 bioanalyzer. For RNA-seq, poly(A)-enriched cDNA libraries were generated using the Illumina TruSeq v2 sample preparation kit (San Diego, CA) and checked for quality and quantity using bioanalyzer and quantitative PCR. For qRT-PCR analyses, RNA was reverse transcribed with RT First Strand Kit (SABiosciences-Qiagen).
Gene Expression by qRT-PCR
Gene expression analysis of 84 inflammatory genes and receptors was conducted by qRT-PCR using Qiagen PCR Arrays (PAHS-077Z) on a BioRad® CFX-96 detection platform. Gene expression was normalized to a five gene panel composed of β2-microglobulin, hypoxanthine phosphoribosyltransferase-1, β-actin, GAPDH and ribosomal protein L13a. Data was analyzed using the ΔΔCt method and fold change calculated compared to uninfected cells and expressed as 2-ΔΔCt. Data was processed and analyzed on the RT² Profiler™ PCR Array Data Analysis online tool provided by the manufacturer.
RNA-Seq Data Generation, Preprocessing, and Quality Trimming
Paired-end reads (100 bp) were obtained using an Illumina HiSeq 1500. Trimmomatic (Bolger et al., 2014) was used to remove Illumina adapter sequences from reads and to trim bases off the start or the end of a read when the quality score fell below a threshold of 25. Sequence quality metrics were assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Mapping cDNA Fragments, Abundance Estimation, and Data Normalization
Reads were aligned against the human (hg38 revision 91), L.V. panamensis (TriTrypDb release 36), and L.V. braziliensis (release 26) genomes with TopHat (2.1.0) (Trapnell et al., 2012) using parameters to randomly place multi-matches (-g 1), using an existing set of splice junctions (-G), and using the more sensitive bowtie2 options (–b2-very-sensitive). The resulting accepted hits and mapped reads were sorted and indexed via SAMtools (Li et al., 2009) and passed to HTSeq (Anders et al., 2015) for generating count tables.
Global Data Assessment, Visualization and Differential Expression Analysis
Biological replicates and batch effects were assessed and visualized using the hpgltools (https://github.com/elsayed-lab/hpgltools) R package. The process included creating density plots, boxplots of depth, coefficient of variance, hierarchical clustering analyses based on Pearson’s correlation coefficient and Euclidean distance, variance partition analyses (Hoffman and Schadt, 2016), and principal component analyses before and after normalization. Several combinations of normalization and batch adjustment strategies were evaluated. The normalization methods tested included trimmed median of M-values, relative log expression, and quantile. These were combined with different batch evaluation strategies including: surrogate variable analysis (SVA) (Leek et al., 2012), ComBat (in sva), RUV (Risso et al., 2014), and batch factor removal via residuals (performed manually and via limma’s (Ritchie et al., 2015) removeBatchEffect function). Normalized data were visualized using log2 transformed counts per million reads following filtering to remove low counts (defined as any gene with fewer counts than twice the number of samples or when any sample had fewer than 2 counts).
Differential expression analyses were performed using a single pipeline which performed all pairwise comparisons using the Bioconductor packages: limma, edgeR (Robinson et al., 2010), DESeq2 (Love et al., 2014), EBSeq (Leng et al., 2013), and a statistically uninformed basic analysis. In each case (except EBSeq and the basic analysis), the surrogate variable estimates provided by SVA were used to adjust the statistical model in an attempt to address the batch/surrogate effects. The quality of each contrast was evaluated by the degree of agreement among methods, but the interpretations were primarily informed by the DESeq2 results. Detailed information on the analytical pipeline and scripts is available at (https://github.com/abelew/cideim_early_leukocyte).
Genes with significant changes in abundance (fold change ≥ |1.5| and false discovery rate adjusted P values ≤ 0.05) were passed to a few gene set enrichment methods including: GOseq (Young et al., 2010), clusterprofiler (Yu et al., 2012), topGO, GOstats (Falcon and Gentleman, 2007), gProfileR (Reimand et al., 2016) and gene set variation analysis (GSVA) (Hanzelmann et al., 2013). Gene ontology analyses were supplemented with manual data curation. GSVA was performed to produce an enrichment score for each gene set per sample. These scores were passed to limma to evaluate the difference in GSVA score distributions for each gene set in the samples. Limma results were then filtered according to log2 fold change, adjusted p-value, and maximum GSVA score mean. GSVA was conducted using the immunologic signature gene sets publicly available at GSEA|MSigDB (C7 collection) (Liberzon et al., 2011). Network analyses were done with STRING 11.0 (Szklarczyk et al., 2019).
Detection of Leishmania RNA Virus (LRV)
Total RNA was extracted from stationary-phase promastigotes using TRIzol (Invitrogen, USA), and cDNA was synthesized using a high-capacity cDNA reverse transcription kit (Applied Biosystems). LRV was detected by qRT-PCR using the primer sets described by (Zangger et al., 2013): Forward 5′-CTG ACT GGA CGG GGG GTA AT-3′ and Reverse 5′-CAA AAC ACT CCC TTA CGC-3′, derived from LRV1-4 genome sequences (GenBank accession number: NC_003601). As a quality control procedure for nucleic acids, a 372 bp fragment of the Leishmania β-tubulin gene was also amplified. Positive and negative controls for LRV were included in each run: L. V. guyanensis M5313 (WHI/BR/78/M5313 – LRV+) and L. V. panamensis (MHOM/CO/2002/3594 stably transfected with the luciferase reporter gene, L.p.LUC 001, LRV-).
Results and Discussion
Profiling of Inflammatory Mediators in WBCs and PBMCs Reveals a Similar Signature
The macrophage-Leishmania infection model has been consistently used as the preferred in vitro and ex vivo experimental system, because it portrays the interaction of the parasite and its primary host cell. However, recruitment of other cells to the inflammatory microenvironment generated during the vector blood meal, implies that the early interaction between the parasite and the host extends beyond the macrophage-Leishmania contact. Different experimental approaches and cell preparations can be used to explore these Leishmania-leukocyte interactions (e.g. whole blood, total white blood cells -buffy coat-, isolated PBMCs). To select the ex vivo cellular system for our experiments, we performed a comparative expression profiling of 84 immune response genes in total human white blood cells (WBCs) and PBMCs collected from healthy donors (n=3), which were then exposed to L.V. panamensis promastigotes for 24 hours. Eleven genes (ccl11, ccl16, ccl21, ccl8, crp, IL17a, il23r, il9, kng1, nos2 and sele) were not detected in either PBMCs or WBCs. Il22 was only detected in infected PBMCs from one donor, and ccl19 was not detected in WBCs, but was induced in infected PBMCs (Table S1). 56 genes were modulated by exposure to L. V. panamensis (fold change, FC ≥ |1.5|), and 39 were common in PBMCs and WBCs (28 upregulated and 11 downregulated). Modulation of il1r, il1rap and tlr1 was mainly found in WBCs, which is consistent with the predominant expression in granulocytes (Uhlen et al., 2015). The magnitude of modulation was consistently higher in PBMCs than WBCs, in line with higher transcriptional activity of mononuclear compared to polymorphonuclear cells (Ericson et al., 2014; Grassi et al., 2018) (Figure 1). The similarity of gene expression profiles supports the use of PBMCs as an informative cell population to explore the predominant immunological signatures elicited during the early stages of L.V. panamensis infections.
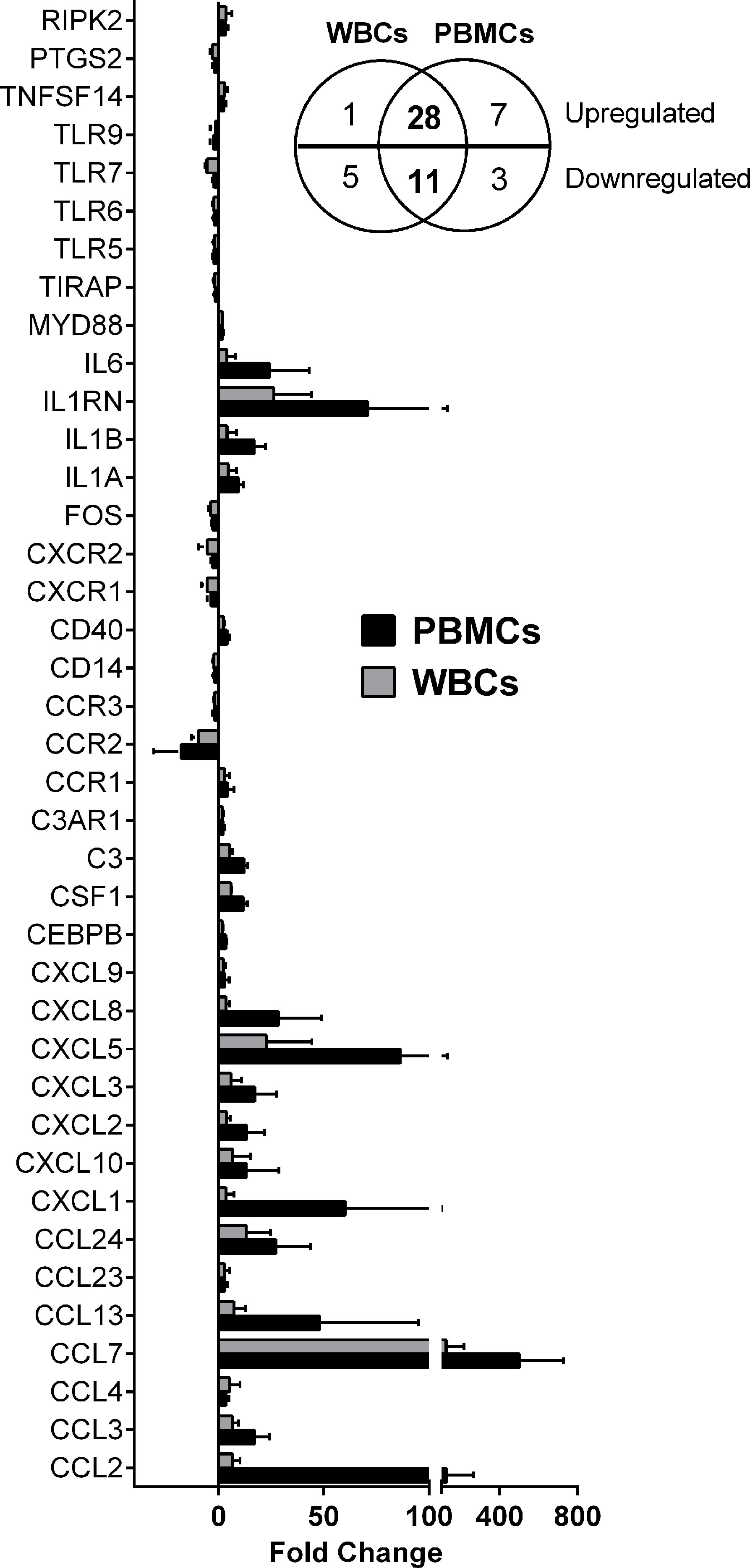
Figure 1 Differentially expressed genes in WBCs and PBMCs upon L. V. panamensis infection. Fold change gene expression (infected vs. uninfected) of genes modulated in WBCs and PBMCs from three healthy donors after ex vivo infection with L. V. panamensis for 24h. Insert: Venn diagram representing the number of upregulated and downregulated genes.
Exposure of PBMCs to L. Viannia Modulates Transcriptional Signatures of Lymphocyte Activation
PBMCs from three healthy donors from a non-endemic area, were exposed for 24h to stationary phase promastigotes isolated from patients with chronic (CHR, n=3) or self-healing (SH, n=3) CL. Leishmania virulence factors such as the major surface metalloprotease GP63, lipophosphoglycan (LPG), and the Leishmania RNA virus (LRV), contribute to disease severity in murine models of infection (Beverley and Turco, 1998; Gomez et al., 2009; Ives et al., 2011). However, a clear relationship of these and other virulence determinants in the clinical outcome of human infections remains elusive. We evaluated the presence of LRV in all clinical strains used in this study. All strains except for the positive control L. V. guyanensis M5313, which was LRV(+) with an avergae Ct value of 24,3.
Poly(A)-enriched cDNA libraries were constructed and 100 bp paired-end reads were generated (Table S2 and Figure S1). Each sample from infected cells consisted of a pool of mixed RNAs from parasites and human macrophages. Post-experimental authentication of Leishmania strains used in these experiments was carried out by SNP profiling of parasite-derived sequences. This confirmed that all strains used corresponded to L.V. panamensis, except for strain MHOM/CO/11/5430 which was identified as a mixed L. V. panamensis/L. V. braziliensis isolate.
A principal component analysis (PCA) showed that a significant amount of the variance observed in the host transcriptomic data resulted from either the infection status (infected vs. uninfected, Figure 2A) or, in the absence of uninfected samples, from inter-donor variability (Figure 2B). To account for these sources of variance, an adjusted statistical model was developed using SVA. A PCA of the SVA-adjusted data showed a partial separation of samples based on infection with CHR vs. SH strains (Figure 2C).
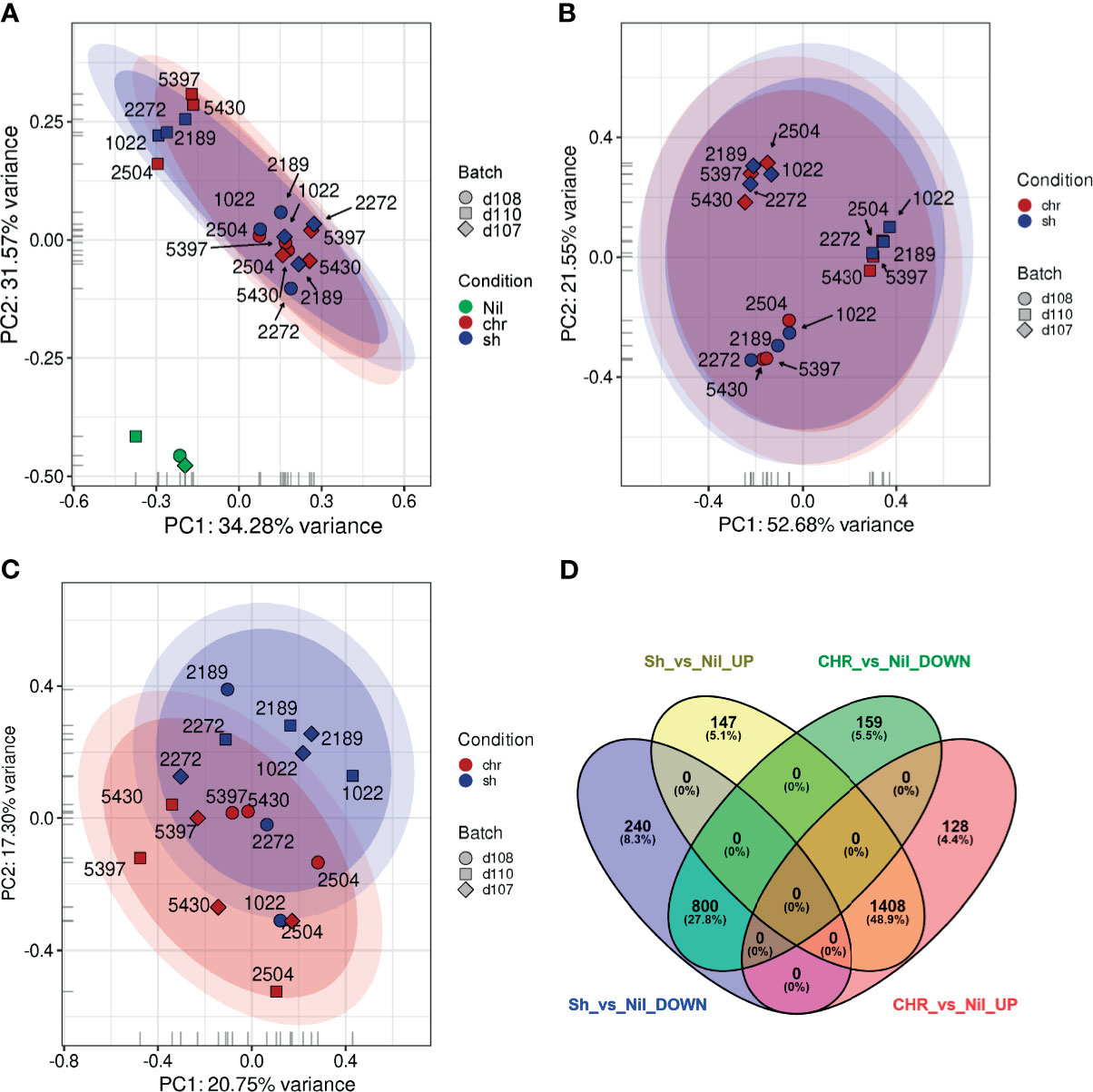
Figure 2 Principal Component Analysis plots and Differential Expression analyses of PBMCs exposed to SH and CHR strains. An initial PCA was performed (A) using log2, quantile, count per million (cpm), low-count-filtered data and displayed a strong clustering with respect to the three donors (represented as the different symbol shapes). (B) This donor-specific effect was made even clearer when the uninfected samples (green) were removed from the analysis. (C) When surrogate variable estimates from SVA were included, it became possible to visualize differences between the PBMCs exposed to CHR (red) and self-healing (blue) strains. (D) Venn diagrams showing the relationships between the overlaps of differentially expressed genes in SH vs. uninfected (Nil) and CHR vs. uninfected samples.
Differential expression (DE) analysis of the transcriptomes from PBMCs exposed to SH strains against CHR strains yielded no statistically significant DE genes. Considering that most of the variance between samples was attributed to either donor variability or infection status, we conducted three additional analyses: 1) an independent DE analysis of infection with CHR vs. SH strains for each donor; 2) a grouped analysis of PBMCs from the three donors infected with SH strains against uninfected controls, and 3) a grouped analysis of PBMCs from the three donors infected with CHR strains against uninfected controls. For the independent donor analyses, transcriptomes of PBMCs from one donor did not identify statistically significant DE genes. For the second and third donors, only 3 and 28 DE genes were found respectively (Table S3). Grouped analysis of infected PBMCs against uninfected controls yielded over 2800 significantly DE genes in infections with either SH or CHR strains. Of these, 2208 (78.8%) were common to CHR and SH infections: 1408 upregulated and 800 downregulated (cut off values: p ≤ 0.05; log2FC ≥ |0.58|, i.e. fold change ≥ |1.5|), Figure 2D. Despite the statistical significance of this enrichment analysis, the biological information extracted from these GO categories is limited.
Among up- and down-regulated genes, the top 10 significantly enriched categories (ranked by % representation within the dataset) corresponded to broad categories related to immune functions, cell signaling, transcription, and cell proliferation, among others (Table S4).
To provide a more insightful biological interpretation of the effect of early exposure of PBMCs to L.V. panamensis, and to explore the potential contribution of specific cell types to the bulk transcriptomic profile of PBMCs, we conducted gene set variation analysis (GSVA) using the immunologic signature gene sets publicly available at GSEA|MSigDB (C7 collection) (Liberzon et al., 2011). 140 categories were significantly and differentially enriched in L.V. panamensis infected vs. uninfected PBMCs (Figure 3 and Table S5), independent of whether cells were exposed to CHR or SH strains. Interestingly, 37 gene sets were derived from experiments involving microbial agents, including infections with old world Leishmania species (L. donovani and L. major) and other intracellular microbes (Toxoplasma gondii and Mycobacterium tuberculosis). This potentially shows common early response mechanisms against intracellular pathogens.
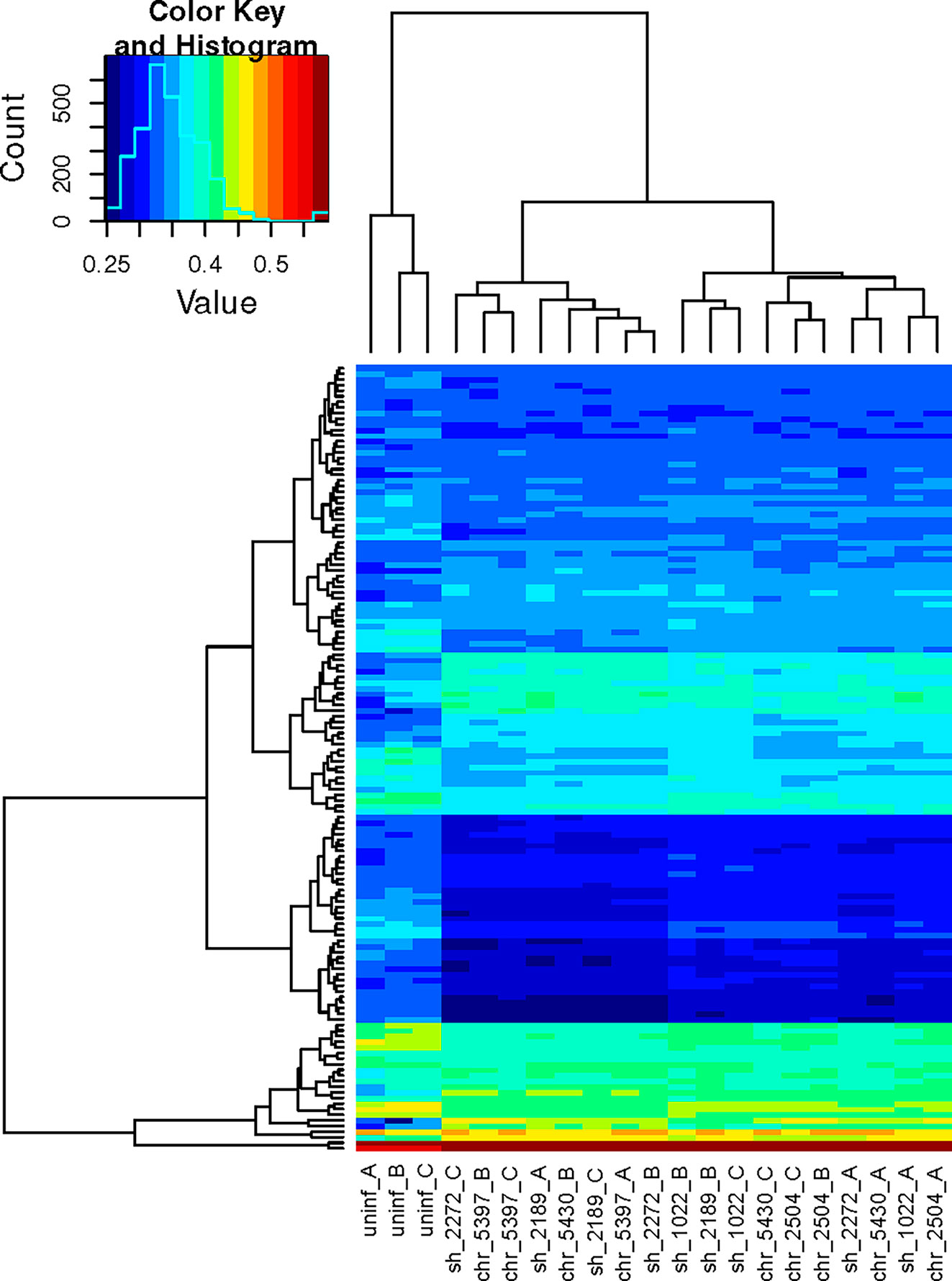
Figure 3 Hierarchical clustering heatmap of significant mSigDB c7 gene sets. GSVA was performed using the normalized expression data and the mSigDB c7 gene sets. The resulting scored gene sets were examined using limma and by de-novo comparisons of the mean group scores in order to extract the most significant gene sets. The remaining scores were passed to a hierarchical clustering algorithm and plotted. Each column corresponds to an independent sample, each row to a unique gene set. Columns 1- 3, uninfected PBMCs from three independent healthy donors (A–C). Columns 4-18, infected PBMCs. Sample codes correspond to the phenotype of the disease caused by the Leishmania strain (sh, self healing; chr, chronic), followed by the strain code and the PBMC donor.
Among enriched categories, 47 corresponded exclusively to innate cell signatures (monocytes, macrophages, NK cells, dendritic cells), while 56 were related to signatures derived from adaptive immune cells (CD8 and CD4 T cells, Th17, Tregs, T follicular cells, B cells) (Table S5). This indicates that as early as 24h after interaction, Leishmania modulates adaptive immune cell functions in PBMCs from previously unexposed healthy individuals, revealing a largely unexplored and exploitable dimension of the host-pathogen interactome in Leishmania infections. The mechanism(s) mediating this rapid activation, especially of naïve T cells, remains to be determined, but could occur via bystander (antigen-independent) activation of naïve or memory T cells (Kim and Shin, 2019; Lee et al., 2020), cross-reactivity of memory T cells, or potentially via antigen-presentation by circulating fibrocytes (Chesney et al., 1997; Grab et al., 2004). Interestingly, bystander activation of Lysteria-specific and LCMV-specific memory T-cells has been reported in chronic L. donovani infection of Lysteria-immune mice (Polley et al., 2005), and in acutely L. major infected mice, respectively (Crosby et al., 2014).
Exposure of PBMCs to SH and CHR Strains Differentially Modulate Genes Related to IFN Responses and the T Cell Synapse
We next analyzed the gene set differentially modulated by SH and CHR infections. A total of 387 genes were uniquely modulated in PBMCs after exposure to SH strains (240 down- and 157 up-regulated), and 287 were unique to infection with CHR strains (159 down- and 128 upregulated) (Table S6). Functional interpretation of DE genes was performed via GO enrichment, complemented by network analyses and manual curation. For infections with SH strains, no significantly enriched GO categories were found. For infections with CHR strains, a single GO category was enriched (FDR=0.00029): Type I IFN signaling (reactome pathway HSA-1606322) (Table S7).
Upon manual curation, 17 functional categories were defined (all containing ≥ 5 genes, except for “antimicrobial activity”). Four categories (metabolism of macromolecules, cell cycle/proliferation/differentiation, immune signaling and vesicle trafficking/transport) were common to infections with SH and CHR strains (Table S7). Genes involved in metabolism of macromolecules (lipid, glycan and nucleic acids metabolism) were found in both upregulated and downregulated gene lists, however, most of these genes were significantly downregulated in infected PBMCs. Rapid downregulation (at 4hpi) of fatty acid metabolism, amino acid catabolism and glycan degradation has been previously documented in murine and human macrophages infected with L. major (Dillon et al., 2015; Fernandes et al., 2016); here we show that this is maintained as long as 24hpi. This suggests that basic metabolic functions are stalled during the early time-points of Leishmania infection in both phagocytic host cells and other blood leukocytes, potentially revealing an early re-programming of basic metabolic functions towards a state of immune cell activation (O'Neill et al., 2016).
Downregulation of central elements of cell cycle progression and pro-growth signaling (MAPK and PI3K), and concomitant induction of genes involved in oxidative phosphorylation and intracellular vesicle trafficking, were also observed (Table S7). Downregulation of genes involved in cell cycle and metabolism of macromolecules, and induction of genes of the tricarboxylic acid (TCA) cycle, is consistent with a state of arrested cell cycle progression. High metabolic demands (provided by metabolism of macromolecules, and not the TCA cycle) are required during cell proliferation and differentiation, while oxidative phosphorylation is a major metabolic pathway in non-proliferative cells (O'Neill et al., 2016). These data further support that the early interaction of L. Viannia with PBMCs leads to reduced metabolic capacity, potentially skewing immune cells towards mechanisms of fast energy production to support rapid immune cell activation in otherwise metabolically “paused” cells (Alonso and Nungester, 1956; Newsholme et al., 1986).
Among the uniquely downregulated genes in PBMCs infected with SH strains were primary cilium proteins (Table S7). The primary cilium was long considered a vestigial organelle (Cassioli and Baldari, 2019), and re-discovered as a signaling hub (Nachury and Mick, 2019). Although hematopoietic cells do not have primary cilia, a more recent understanding of the T cell immunological synapse (IS) suggests participation of ciliary proteins in IS formation (Cassioli and Baldari, 2019). The assembly and function of the IS and the primary cilium depend on cytoskeleton dynamics and polarized vesicle trafficking, and evidence of strong modulation of these processes was found in the transcriptomic profiles of L. Viannia-infected PBMCs (Table S7). Within the first hour of interaction between T-cells and antigen presenting cells (APC), an actin-rich invasive pseudopodia emerges from T-cells and probes deep within the APC. The actin cap is then cleared to allow orientation of the microtubule organization center (MTOC) to the IS, resulting in a matured IS (Ueda et al., 2015). Interestingly, disruption of the MTOC does not impact the level of cytokine production in T cells; however, it does change the “directionality” of cytokine secretion so that it is no longer directed at the IS (Ueda et al., 2015). Dampened expression of molecules involved in primary cilia/IS formation during infection of PBMCs with SH strains suggests a cytokine micro-environment that may lead to subtler activation of APCs during interaction with T cells, potentially limiting excessive pro-inflammatory cytokine production that can result in uncontrolled inflammation and immunopathology (Scott and Novais, 2016).
Macrophage Functions Are Central Drivers of the Divergent Immune Responses Elicited Upon SH and CHR Infections
We explored the effect of SH and CHR infections in macrophages as one of the central APCs and principal host cells for Leishmania. Total transcriptomic profiles were obtained from primary macrophages infected for 24 h with SH and CHR strains [RNA samples used here were those previously reported in (Navas et al., 2014)]. PCA showed clustering of samples by disease phenotype (Figures 4A and S2). Macrophages infected with SH strains clustered together and close to uninfected cells, indicating minimal modulation of macrophage gene expression by SH strains, and supporting the controlled APC response that also emerged from SH-infected PBMC transcriptomes.
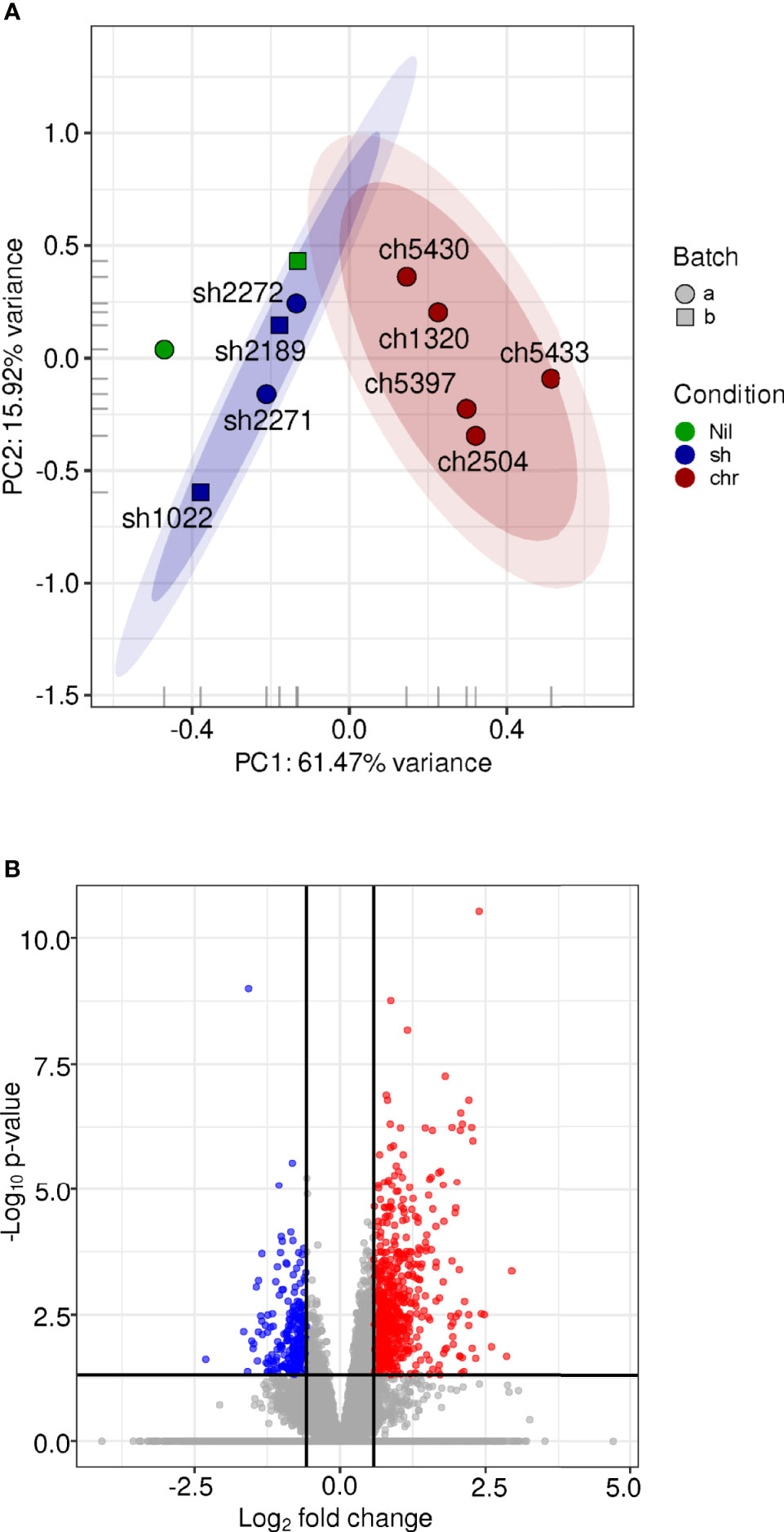
Figure 4 Gene expression profiles of human macrophages infected with SH and CHR L. V. panamensis strains CL. The (H) sapiens RNA-seq expression data for annotated coding genes after low-count filtering (7,019 genes remaining) and log2[quantile(cpm)] normalization was examined via PCA (A). Each glyph represents an experimental sample where the color represents the infection state, the shape represents the experimental batch. The 0.90, 0.95 confidence intervals are shown as shaded ovals. Differential expression analysis was performed using DESeq2, including experimental date (batch) in the statistical model. The volcano plot (B) shows the distribution of significance (-log adjusted P value) with respect to log2-fold change. Points colored in blue are deemed significantly downregulated in the macrophage samples infected with CHR vs. SH strains while those in red are significantly up-regulated.
Differential gene expression analysis of macrophages infected with CHR vs. SH strains identified 884 DE genes (Figure 4B and Table S8), representing 30% more DE genes compared to those in PBMCs. The macrophage DE gene profile consisted predominantly of upregulated inflammatory response genes in infections with CHR vs. SH strains [also previously reported in microarray and qRT-PCR datasets of L.V. panamensis infected macrophages (Ramirez et al., 2012; Navas et al., 2014)]. In infections with CHR strains, up-regulation of chemokines involved in monocyte (CCL2, CCL13 and CCR1) and CD4+-TH1 (CXCL10) activation and recruitment (Figure 5 and Table S8), as well as genes associated with TLR and IFN signaling (TLR4, CD14, TLR1, IFNGR1, IFNAR1, JAK2) were found. In addition, an upregulation of complement components including C3, C1QA, C1QC, CFD and CFP (complement factors D and properdin), ficolin (FCN1), CR1 and the anaphylatoxin receptor C5AR1 (and downstream signaling molecules Rap1b, Rap2a) was observed, which have been associated with a potent induction of inflammation and cell recruitment during acute and chronic inflammatory processes.
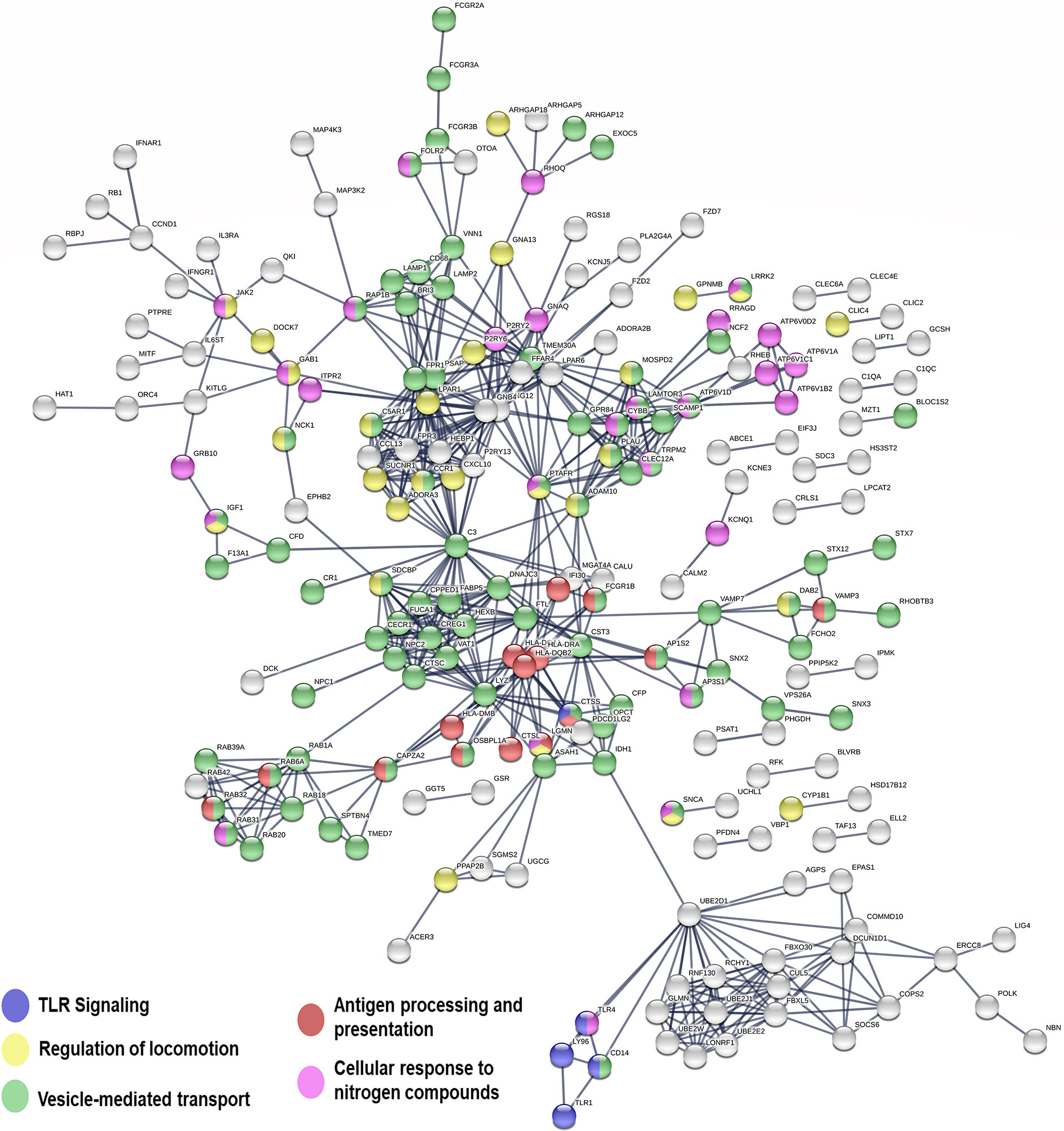
Figure 5 Network representation of genes significantly upregulated in macrophages infected with CHR vs. SH strains. Line thickness represents the strength of data support (displaying relationships > 0.9 confidence value). Interaction sources for network building in STRING included experiments, databases, co-expression, neighborhood, gene fusion and co-occurrence. Nodes not in networks are hidden.
Genes involved in opsonophagocytosis and vesicle mediated transport were also induced in CHR-infected macrophages relative to SH-infected cells (Figure 5 and Table S8). Among those were CR1, properdin and C3b, which have been associated with opsonophagocytosis of intracellular pathogens (Rosales and Uribe-Querol, 2017; Lu et al., 2018). Complement receptors participate in uptake of Leishmania by macrophages, and a direct relationship between permissiveness of macrophage infection, CR1 and CR3 activity, and chronic CL caused by L. V. panamensis has been reported (Robledo et al., 1994). Increased C3 gene expression has been demonstrated in lesion biopsies from CL patients unresponsive to antimonial treatment, potentially leading to enhanced recruitment of polymorphonuclear cells to the affected tissues, promoting immunopathology and a non-healing phenotype (Navas et al., 2020). Induction of multiple Fc gamma receptors (FcgR, including the high affinity receptor FcgR1 as well as low affinity receptors 2a, 2b, 3a and 3b), and a number of genes involved in vesicle-mediated transport (sortins, syntaxins), phagolysosomal acidification (Rab39A, Rab32, Rab20, Vamp7, lamp1, lamp2, V-ATPase subunits among others), and antigen processing and presentation (MHC-II molecules, lyzozyme, cathepsine L) was also observed in macrophages infected with CHR strains, supporting enhanced opsonophagocytosis (Lu et al., 2018) and intracellular parasite killing. Consistent with this hypothesis was the finding that parasite loads were lower in primary human macrophages infected with CHR compared to SH strains (Table S2, Figure S3).
Concluding Remarks
The exploration of transcriptomic signatures from both isolated human primary macrophages and PBMCs consistently provides evidence that pathology and severity of CL infection is determined, at least in part, by the immune response. Chronic CL is characterized by the induction of pro-inflammatory leukocyte responses, while benign, self-resolving CL, may be primarily marked by limited and controlled T-cell and APC activation. The early interaction of PBMCs and L. Viannia provides a more comprehensive view of additional mechanisms involved in the establishment of infection in the human host. Downregulation of genes involved in biosynthesis and metabolism of macromolecules, cell cycle progression and cellular proliferation, together with upregulation of functional categories related to glycolysis and oxidative phosphorylation, suggest that the parasite-host interplay of Leishmania and PBMCs drives cellular functions and energy production towards immunological activation rather than to promote metabolism of macromolecules. Whether this immunological activity is skewed towards a pro-inflammatory environment or a controlled adaptive response, emerges as a potential key aspect for defining the course of human CL. Results from this study instigate further explorations of the Leishmania-host interactions involving functional aspects of B and T lymphocytes in the early response to infection (within the first 24 hours of contact). Whether the functions modulated in these cells are dependent on APCs, are a direct effect elicited by the parasite, or are a bystander mechanism of adaptive cell activation, remains to be determined.
Data Availability Statement
All sequence data is publicly available in NCBI’s Short Read Archive (SRA) under bioproject PRJNA685835 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA685835).
Ethics Statement
This study was approved and monitored by the institutional review board for ethical conduct of research involving human subjects of the Centro Internacional de Entrenamiento e Investigaciones Médicas (CIDEIM) in accordance with national (Resolution 008430, República de Colombia, Ministry of Health, 1993) and international (Declaration of Helsinki and amendments, World Medical Association, Fortaleza, Brazil, October 2013) guidelines. All individuals voluntarily participated in the study and written informed consent was obtained from each participant.
Author Contributions
MG, MR-C, AN, and NE-S contributed to conception and design of the study. AM-V conducted patient recruitment and all clinical management of study participants. AN, MR-C, JM, AB, and LD performed the experiments. AB, MG, and NE-S performed the statistical analysis. MG wrote the first draft of the manuscript. AB, AN, JM, LD, TA, and NE-S wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded in part by the American Society of Tropical Medicine and Hygiene Gorgas Memorial Institute Research Award, and received support from NIAID/NIH U19AI129910 and Wellcome Trust 107595/Z/15/Z.
Author Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other agencies.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.687607/full#supplementary-material
References
Alonso, D., Nungester, W. J. (1956). Comparative Study of Host Resistance of Guinea Pigs and Rats: V. The Effect of Pneumococcal Products on Glycolysis and Oxygen Uptake by Polymorphonuclear Leucocytes. J. Infect. Dis. 99 (2), 174–181.
Amato, V. S., Tuon, F. F., Bacha, H. A., Neto, V. A., Nicodemo, A. C. (2008). Mucosal Leishmaniasis . Current Scenario and Prospects for Treatment. Acta Trop. 105 (1), 1–9. doi: 10.1016/j.actatropica.2007.08.003
Anders, S., Pyl, P. T., Huber, W. (2015). HTSeq–a Python Framework to Work With High-Throughput Sequencing Data. Bioinformatics 31 (2), 166–169. doi: 10.1093/bioinformatics/btu638
Arunachalam, P. S., Wimmers, F., Mok, C. K. P., Perera, R., Scott, M., Hagan, T., et al. (2020). Systems Biological Assessment of Immunity to Mild Versus Severe COVID-19 Infection in Humans. Science 369 (6508), 1210–1220. doi: 10.1126/science.abc6261
Bamorovat, M., Sharifi, I., Dabiri, S., Shamsi Meymandi, S., Karamoozian, A., et al. (2021). Major Risk Factors and Histopathological Profile of Treatment Failure, Relapse and Chronic Patients With Anthroponotic Cutaneous Leishmaniasis: A Prospective Case-Control Study on Treatment Outcome and Their Medical Importance. PloS Negl. Trop. Dis. 15 (1), e0009089. doi: 10.1371/journal.pntd.0009089
Beverley, S. M., Turco, S. J. (1998). Lipophosphoglycan (LPG) and the Identification of Virulence Genes in the Protozoan Parasite Leishmania. Trends Microbiol. 6 (1), 35–40. doi: 10.1016/S0966-842X(97)01180-3
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Carvalho, E. M., Johnson, W. D., Barreto, E., Marsden, P. D., Costa, J. L., Reed, S., et al. (1985). Cell Mediated Immunity in American Cutaneous and Mucosal Leishmaniasis. J. Immunol. 135 (6), 4144–4148.
Cassioli, C., Baldari, C. T. (2019). A Ciliary View of the Immunological Synapse. Cells 8 (8), 1–24. doi: 10.3390/cells8080789
Chesney, J., Bacher, M., Bender, A., Bucala, R.. (1997). The Peripheral Blood Fibrocyte is a Potent Antigen-Presenting Cell Capable of Priming Naive T Cells in Situ. Proc. Natl. Acad. Sci. U. S. A. 94 (12), 6307–6312. doi: 10.1073/pnas.94.12.6307
Cohen-Freue, G., Holzer, T. R., Forney, J. D., McMaster, W. R. (2007). Global Gene Expression in Leishmania. Int. J. Parasitol. 37 (10), 1077–1086. doi: 10.1016/j.ijpara.2007.04.011
Crosby, E. J., Goldschmidt, M. H., Wherry, E. J., Scott, P. (2014). Engagement of NKG2D on Bystander Memory CD8 T Cells Promotes Increased Immunopathology Following Leishmania Major Infection. PloS Pathog. 10 (2), e1003970. doi: 10.1371/journal.ppat.1003970
Diaz, Y. R., Rojas, R., Valderrama, L., Saravia, N. G. (2010). T-Bet, GATA-3, and Foxp3 Expression and Th1/Th2 Cytokine Production in the Clinical Outcome of Human Infection With Leishmania (Viannia) Species. J. Infect. Dis. 202 (3), 406–415. doi: 10.1086/653829
Dillon, L. A., Suresh, R., Okrah, K., Corrada Bravo, H., Mosser, D. M., El-Sayed, N. M. (2015). Simultaneous Transcriptional Profiling of Leishmania Major and Its Murine Macrophage Host Cell Reveals Insights Into Host-Pathogen Interactions. BMC Genomics 16, 1108. doi: 10.1186/s12864-015-2237-2
Ehrchen, J. M., Roebrock, K., Foell, D., Nippe, N., von Stebut, E., Weiss, J. M., et al. (2010). Keratinocytes Determine Th1 Immunity During Early Experimental Leishmaniasis. PloS Pathog. 6 (4), e1000871. doi: 10.1371/journal.ppat.1000871
Ericson, J. A., Duffau, P., Yasuda, K., Ortiz-Lopez, A., Rothamel, K., Rifkin, I. R., et al. (2014). Gene Expression During the Generation and Activation of Mouse Neutrophils: Implication of Novel Functional and Regulatory Pathways. PloS One 9 (10), e108553. doi: 10.1371/journal.pone.0108553
Falcon, S., Gentleman, R. (2007). Using GOstats to Test Gene Lists for GO Term Association. Bioinformatics 23 (2), 257–258. doi: 10.1093/bioinformatics/btl567
Fernandes, M. C., Dillon, L. A., Belew, A. T., Bravo, H. C., Mosser, D. M., El-Sayed, N. M. (2016). Dual Transcriptome Profiling of Leishmania-Infected Human Macrophages Reveals Distinct Reprogramming Signatures. mBio 7 (3). doi: 10.1128/mBio.00027-16
Fernandez, O. L., Diaz-Toro, Y., Ovalle, C., Valderrama, L., Muvdi, S., Rodriguez, I., et al. (2014). Miltefosine and Antimonial Drug Susceptibility of Leishmania Viannia Species and Populations in Regions of High Transmission in Colombia. PloS Negl. Trop. Dis. 8 (5), e2871. doi: 10.1371/journal.pntd.0002871
Gasim, S., Elhassan, A. M., Khalil, E. A., Ismail, A., Kadaru, A. M., Kharazmi, A., et al. (1998). High Levels of Plasma IL-10 and Expression of IL-10 by Keratinocytes During Visceral Leishmaniasis Predict Subsequent Development of Post-Kala-Azar Dermal Leishmaniasis. Clin. Exp. Immunol. 111 (1), 64–69. doi: 10.1046/j.1365-2249.1998.00468.x
Gomez, M. A., Contreras, I., Halle, M., Tremblay, M. L., McMaster, R. W., Olivier, M. (2009). Leishmania GP63 Alters Host Signaling Through Cleavage-Activated Protein Tyrosine Phosphatases. Sci. Signal. 2 (90), ra58. doi: 10.1126/scisignal.2000213
Gomez, M. A., Olivier, M. (2010). Proteases and Phosphatases During Leishmania-Macrophage Interaction: Paving the Road for Pathogenesis. Virulence 1 (4), 314–318. doi: 10.4161/viru.1.4.12194
Grab, D. J., Salem, M. L., Dumler, J. S., Bucala, R. (2004). A Role for the Peripheral Blood Fibrocyte in Leishmaniasis? Trends Parasitol. 20 (1), 12. doi: 10.1016/j.pt.2003.10.012
Grassi, L., Pourfarzad, F., Ullrich, S., Merkel, A., Were, F., Carrillo-de-Santa-Pau, E., et al. (2018). Dynamics of Transcription Regulation in Human Bone Marrow Myeloid Differentiation to Mature Blood Neutrophils. Cell Rep. 24 (10), 2784–2794. doi: 10.1016/j.celrep.2018.08.018
Hanzelmann, S., Castelo, R., Guinney, J. (2013). GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinf. 14, 7. doi: 10.1186/1471-2105-14-7
Hoffman, G. E., Schadt, E. E. (2016). Variancepartition: Interpreting Drivers of Variation in Complex Gene Expression Studies. BMC Bioinf. 17 (1), 483. doi: 10.1186/s12859-016-1323-z
Ives, A., Ronet, C., Prevel, F., Ruzzante, G., Fuertes-Marraco, S., Schutz, F., et al. (2011). Leishmania RNA Virus Controls the Severity of Mucocutaneous Leishmaniasis. Science 331 (6018), 775–778. doi: 10.1126/science.1199326
Kim, T. S., Shin, E. C. (2019). The Activation of Bystander CD8(+) T Cells and Their Roles in Viral Infection. Exp. Mol. Med. 51 (12), 1–9. doi: 10.1038/s12276-019-0316-1
Lee, H. G., Cho, M. Z., Choi, J. M. (2020). Bystander CD4(+) T Cells: Crossroads Between Innate and Adaptive Immunity. Exp. Mol. Med. 52 (8), 1255–1263. doi: 10.1038/s12276-020-00486-7
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E., Storey, J. D. (2012). The Sva Package for Removing Batch Effects and Other Unwanted Variation in High-Throughput Experiments. Bioinformatics 28 (6), 882–883. doi: 10.1093/bioinformatics/bts034
Leng, N., Dawson, J. A., Thomson, J. A., Ruotti, V., Rissman, A. I., Smits, B. M., et al. (2013). EBSeq: An Empirical Bayes Hierarchical Model for Inference in RNA-Seq Experiments. Bioinformatics 29 (8), 1035–1043. doi: 10.1093/bioinformatics/btt087
Liberzon, A., Subramanian, A., Pinchback, R., Thorvaldsdottir, H., Tamayo, P., Mesirov, J. P. (2011). Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 27 (12), 1739–1740. doi: 10.1093/bioinformatics/btr260
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map Format and SAMtools. Bioinformatics 25 (16), 2078–2079. doi: 10.1093/bioinformatics/btp352
Love, M. I., Huber, W., Anders, S. (2014). Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data With Deseq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Lu, J., Mold, C., Du Clos, T. W., Sun, P. D. (2018). Pentraxins and Fc Receptor-Mediated Immune Responses. Front. Immunol. 9, 2607. doi: 10.3389/fimmu.2018.02607
Maurer-Cecchini, A., Decuypere, S., Chappuis, F., Alexandrenne, C., De Doncker, S., Boelaert, M., et al. (2009). Immunological Determinants of Clinical Outcome in Peruvian Patients With Tegumentary Leishmaniasis Treated With Pentavalent Antimonials. Infect. Immun. 77 (5), 2022–2029. doi: 10.1128/IAI.01513-08
Moncunill, G., Scholzen, A., Mpina, M., Nhabomba, A., Hounkpatin, A. B., Osaba, L., et al. (2020). Antigen-Stimulated PBMC Transcriptional Protective Signatures for Malaria Immunization. Sci. Transl. Med. 12 (543). doi: 10.1126/scitranslmed.aay8924
Murray, H. W., et al. (2005). Advances in Leishmaniasis. Lancet 366 (9496), 1561–1577. doi: 10.1016/S0140-6736(05)67629-5
Nachury, M. V., Mick, D. U. (2019). Establishing and Regulating the Composition of Cilia for Signal Transduction. Nat. Rev. Mol. Cell Biol. 20 (7), 389–405. doi: 10.1038/s41580-019-0116-4
Navas, A., Vargas, D. A., Freudzon, M., McMahon-Pratt, D., Saravia, N. G., et al. (2014). Chronicity of Dermal Leishmaniasis Caused by Leishmania Panamensis Is Associated With Parasite-Mediated Induction of Chemokine Gene Expression. Infect. Immun. 82 (7), 2872–2880. doi: 10.1128/IAI.01133-13
Navas, A., Fernandez, O., Gallego-Marin, C., Castro, M. D. M., Rosales-Chilama, M., Murillo, J., et al. (2020). Profiles of Local and Systemic Inflammation in the Outcome of Treatment of Human Cutaneous Leishmaniasis Caused by Leishmania (Viannia). Infect. Immun. 88 (3). doi: 10.1128/IAI.00764-19
Newsholme, P., Curi, R., Gordon, S., Newsholme, E. A. (1986). Metabolism of Glucose, Glutamine, Long-Chain Fatty Acids and Ketone Bodies by Murine Macrophages. Biochem. J. 239 (1), 121–125. doi: 10.1042/bj2390121
O'Neill, L. A., Kishton, R. J., Rathmell, J. (2016). A Guide to Immunometabolism for Immunologists. Nat. Rev. Immunol. 16 (9), 553–565. doi: 10.1038/nri.2016.70
Oliveros, J. C. Venny. An Interactive Tool for Comparing Lists With Venn's Diagrams. (2007-2015). Available at: https://bioinfogp.cnb.csic.es/tools/venny/index.html.
Orr, C., Xu, W., Masur, H., Kottilil, S., Meissner, E. G. (2020). Peripheral Blood Correlates of Virologic Relapse After Sofosbuvir and Ribavirin Treatment of Genotype-1 HCV Infection. BMC Infect. Dis. 20 (1), 929. doi: 10.1186/s12879-020-05657-5
Ovalle-Bracho, C., Franco-Munoz, C., Londono-Barbosa, D., Restrepo-Montoya, D., Clavijo-Ramirez, C. (2015). Changes in Macrophage Gene Expression Associated With Leishmania (Viannia) Braziliensis Infection. PloS One 10 (6), e0128934. doi: 10.1371/journal.pone.0128934
Polley, R., Sanos, S. L., Prickett, S., Haque, A., Kaye, P. M. (2005). Chronic Leishmania Donovani Infection Promotes Bystander CD8+-T-Cell Expansion and Heterologous Immunity. Infect. Immun. 73 (12), 7996–8001. doi: 10.1128/IAI.73.12.7996-8001.2005
Ramirez, C., Diaz-Toro, Y., Tellez, J., Castilho, T. M., Rojas, R., Ettinger, N. A., et al. (2012). Human Macrophage Response to L. (Viannia) Panamensis: Microarray Evidence for an Early Inflammatory Response. PloS Negl. Trop. Dis. 6 (10), e1866. doi: 10.1371/journal.pntd.0001866
Reimand, J., Arak, T., Adler, P., Kolberg, L., Reisberg, S., Peterson, H., et al. (2016). G:Profiler-A Web Server for Functional Interpretation of Gene Lists (2016 Update). Nucleic Acids Res. 44 (W1), W83–W89. doi: 10.1093/nar/gkw199
Risso, D., Ngai, J., Speed, T. P., Dudoit, S. (2014). Normalization of RNA-Seq Data Using Factor Analysis of Control Genes or Samples. Nat. Biotechnol. 32 (9), 896–902. doi: 10.1038/nbt.2931
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 43 (7), e47. doi: 10.1093/nar/gkv007
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26 (1), 139–140. doi: 10.1093/bioinformatics/btp616
Robledo, S., Wozencraft, A., Valencia, A. Z., Saravia, N. (1994). Human Monocyte Infection by Leishmania (Viannia) Panamensis. Role of Complement Receptors and Correlation of Susceptibility In Vitro With Clinical Phenotype. J. Immunol. 152 (3), 1265–1276.
Rodriguez-Pinto, D., Navas, A., Blanco, V. M., Ramirez, L., Garcerant, D., Cruz, A., et al. (2012). Regulatory T Cells in the Pathogenesis and Healing of Chronic Human Dermal Leishmaniasis Caused by Leishmania (Viannia) Species. PloS Negl. Trop. Dis. 6 (4), e1627. doi: 10.1371/journal.pntd.0001627
Ronet, C., Passelli, K., Charmoy, M., Scarpellino, L., Myburgh, E., Hauyon La Torre, Y., et al. (2019). TLR2 Signaling in Skin Nonhematopoietic Cells Induces Early Neutrophil Recruitment in Response to Leishmania Major Infection. J. Invest. Dermatol. 139 (6), 1318–1328. doi: 10.1016/j.jid.2018.12.012
Rosales, C., Uribe-Querol, E. (2017). Phagocytosis: A Fundamental Process in Immunity. BioMed. Res. Int 2017, 9042851. doi: 10.1155/2017/9042851
Saravia, N. G., Valderrama, L., Labrada, M., Holguin, A. F., Navas, C., Palma, G., et al. (1989). The Relationship of Leishmania Braziliensis Subspecies and Immune Response to Disease Expression in New World Leishmaniasis. J. Infect. Dis. 159 (4), 725–735. doi: 10.1093/infdis/159.4.725
Saravia, N. G., Segura, I., Holguin, A. F., Santrich, C., Valderrama, L., Ocampo, C. (1998). Epidemiologic, Genetic, and Clinical Associations Among Phenotypically Distinct Populations of Leishmania (Viannia) in Colombia. Am. J. Trop. Med. Hyg. 59 (1), 86–94. doi: 10.4269/ajtmh.1998.59.86
Scott, P., Novais, F. O. (2016). Cutaneous Leishmaniasis: Immune Responses in Protection and Pathogenesis. Nat. Rev. Immunol. 16 (9), 581–592. doi: 10.1038/nri.2016.72
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING V11: Protein-Protein Association Networks With Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 47 (D1), D607–D613. doi: 10.1093/nar/gky1131
Tasew, G., Nylen, S., Lieke, T., Lemu, B., Meless, H., Ruffin, N., et al. (2010). Systemic FasL and TRAIL Neutralisation Reduce Leishmaniasis Induced Skin Ulceration. PloS Negl. Trop. Dis. 4 (10), e844. doi: 10.1371/journal.pntd.0000844
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, R., et al. (2012). Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments With TopHat and Cufflinks. Nat. Protoc. 7 (3), 562–578. doi: 10.1038/nprot.2012.016
Ueda, H., Zhou, J., Xie, J., Davis, M. M. (2015). Distinct Roles of Cytoskeletal Components in Immunological Synapse Formation and Directed Secretion. J. Immunol. 195 (9), 4117–4125. doi: 10.4049/jimmunol.1402175
Uhlen, M., Fagerberg, L., Hallstrom, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Tissue-Based Map of the Human Proteome. Science 347 (6220), 1260419. doi: 10.1126/science.1260419
Weigle, K. A., Santrich, C., Martinez, F., Valderrama, L., Saravia, N. G. (1993). Epidemiology of Cutaneous Leishmaniasis in Colombia: A Longitudinal Study of the Natural History, Prevalence, and Incidence of Infection and Clinical Manifestations. J. Infect. Dis. 168 (3), 699–708. doi: 10.1093/infdis/168.3.699
Wiedemann, A., Foucat, E., Hocini, H., Lefebvre, C., Hejblum, B. P., Durand, M., et al. (2020). Long-Lasting Severe Immune Dysfunction in Ebola Virus Disease Survivors. Nat. Commun. 11 (1), 3730. doi: 10.1038/s41467-020-17489-7
Young, M. D., Wakefield, M. J., Smyth, G. K., Oshlack, A. (2010). Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 11 (2), R14. doi: 10.1186/gb-2010-11-2-r14
Yu, G., Wang, L. G., Han, Y., He, Q. Y. (2012). Clusterprofiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS 16 (5), 284–287. doi: 10.1089/omi.2011.0118
Keywords: Leishmania Viannia, PBMCs, leukocytes, host-pathogen interactions, RNA-Seq
Citation: Gomez MA, Belew AT, Navas A, Rosales-Chilama M, Murillo J, Dillon LAL, Alexander TA, Martinez-Valencia A and El-Sayed NM (2021) Early Leukocyte Responses in Ex-Vivo Models of Healing and Non-Healing Human Leishmania (Viannia) panamensis Infections. Front. Cell. Infect. Microbiol. 11:687607. doi: 10.3389/fcimb.2021.687607
Received: 29 March 2021; Accepted: 16 August 2021;
Published: 07 September 2021.
Edited by:
Wander Pavanelli, State University of Londrina, BrazilReviewed by:
Izabela Marques Dourado Bastos, University of Brasilia, BrazilWalderez Ornelas Dutra, Federal University of Minas Gerais, Brazil
Maria Fernanda Laranjeira-Silva, University of São Paulo, Brazil
Copyright © 2021 Gomez, Belew, Navas, Rosales-Chilama, Murillo, Dillon, Alexander, Martinez-Valencia and El-Sayed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Adelaida Gomez, bWdvbWV6QGNpZGVpbS5vcmcuY28=; Najib M. El-Sayed, ZWxzYXllZEB1bWQuZWR1
 Maria Adelaida Gomez
Maria Adelaida Gomez Ashton Trey Belew
Ashton Trey Belew Adriana Navas
Adriana Navas Mariana Rosales-Chilama
Mariana Rosales-Chilama Julieth Murillo
Julieth Murillo Laura A. L. Dillon3,4
Laura A. L. Dillon3,4 Najib M. El-Sayed
Najib M. El-Sayed