
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 21 June 2021
Sec. Clinical Microbiology
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.685068
 Changrui Qian1,2,3†
Changrui Qian1,2,3† Hongmao Liu1,2,3†
Hongmao Liu1,2,3† Jiawei Cao2
Jiawei Cao2 Yongan Ji2,3
Yongan Ji2,3 Wei Lu2,3
Wei Lu2,3 Junwan Lu2,3
Junwan Lu2,3 Aifang Li4
Aifang Li4 Xinyi Zhu1,2,3
Xinyi Zhu1,2,3 Kai Shen2,3
Kai Shen2,3 Haili Xu1
Haili Xu1 Qianqian Chen2,3
Qianqian Chen2,3 Wangxiao Zhou2,3
Wangxiao Zhou2,3 Hongyun Lu5
Hongyun Lu5 Hailong Lin1,2,3
Hailong Lin1,2,3 Xueya Zhang1,2,3
Xueya Zhang1,2,3 Qiaoling Li1,2,3
Qiaoling Li1,2,3 Xi Lin2,3
Xi Lin2,3 Kewei Li2,3
Kewei Li2,3 Teng Xu6,7
Teng Xu6,7 Mei Zhu8*
Mei Zhu8* Qiyu Bao1,2,3*
Qiyu Bao1,2,3* Hailin Zhang1,3*
Hailin Zhang1,3*Florfenicol is widely used to control respiratory diseases and intestinal infections in food animals. However, there are increasing reports about florfenicol resistance of various clinical pathogens. floR is a key resistance gene that mediates resistance to florfenicol and could spread among different bacteria. Here, we investigated the prevalence of floR in 430 Pseudomonas aeruginosa isolates from human clinical samples and identified three types of floR genes (designated floR, floR-T1 and floR-T2) in these isolates, with floR-T1 the most prevalent (5.3%, 23/430). FloR-T2 was a novel floR variant identified in this study, and exhibited less identity with other FloR proteins than FloRv. Moreover, floR-T1 and floR-T2 identified in P. aeruginosa strain TL1285 were functionally active and located on multi-drug resistance region of a novel incomplete Tn4371-like integrative and conjugative elements (ICE) in the chromosome. The expression of the two floR variants could be induced by florfenicol or chloramphenicol. These results indicated that the two floR variants played an essential role in the host’s resistance to amphenicol and the spreading of these floR variants might be related with the Tn4371 family ICE.
Florfenicol is a fluorinated synthetic analog of thiamphenicol (Syriopoulou et al., 1981), mainly used to control respiratory tract diseases and enteric infections in food-producing animals (Zhao et al., 2016). However, due to inappropriate medication use, florfenicol resistance has become increasingly serious in veterinary medicine (Chang et al., 2014). Although florfenicol is not approved for use in humans, an increasing number of studies have reported dramatic and serious florfenicol resistance in various clinical strains, such as Pasteurella multocida, Salmonella, and Klebsiella pneumoniae (Lu et al., 2018; Ujvari et al., 2019; Zhan et al., 2019).
To date, seven florfenicol resistance genes (excluding variants), floR, fexA, fexB, cfr, optrA, pexA and estDL136, have been reported (Arcangioli et al., 1999; Schwarz et al., 2000; Kehrenberg and Schwarz, 2004; Schwarz et al., 2004; Lang et al., 2010; Liu et al., 2012; Tao et al., 2012; Wang et al., 2015). Among them, floR is one of the main florfenicol resistance genes in Gram-negative bacteria (He et al., 2015). Several variants of the floR gene, including pp-flo, cmlA-like, floRv and floSt, have been documented, and most of them encode 404 aa proteins. These floR variants are closely related to each other, and floRv from Stenotrophomonas maltophilia shares the lowest amino acid identity (88.4%-91.8%) with the others excluding pp-flo (He et al., 2015). The floR gene has been identified either on chromosomes or plasmids of various bacteria and has often been associated with mobile genetic elements and genomic islands (Lai et al., 2013; Gabida et al., 2015; da Silva et al., 2017).
Pseudomonas aeruginosa is an opportunistic pathogen that can cause numerous acute or chronic infections, and is notorious for its intrinsic and acquired resistance to numerous antibiotics (Breidenstein et al., 2011; Domalaon et al., 2018). Generally, P. aeruginosa chromosomes do not carry the floR gene. Although P. aeruginosa is clinically resistant to chloramphenicol (Morita et al., 2014), rifampicin-tobramycin conjugates could break the intrinsic resistance of P. aeruginosa to chloramphenicol in vitro and in vivo, making it suitable for clinical treatment (Idowu et al., 2019). However, the floR gene carried by this pathogen may cause this strategy to fail when chloramphenicol is used. The prevalence of the floR gene in P. aeruginosa hasn’t been previously investigated. In this study, we determined the prevalence of floR gene among 430 clinical P. aeruginosa isolates collected from Wenzhou, China in the years 2008-2009 and 2015-2017. The combination of whole-genome sequencing, genotyping and gene expression methods was used to characterize the floR variants. A novel Tn4371-like integrative and conjugative element (ICE) carrying floR-T1 and floR-T2 was identified, which indicated that the Tn4371-like ICE might play an important role in the dissemination of floR-T2.
A total of 430 clinical P. aeruginosa strains isolated from sputum, urine or blood samples of patients were collected from a teaching hospital of Wenzhou Medical University. Among these isolates, 200 strains were isolated during 2008-2009, and 230 strains were isolated in 2015-2017. The strains were identified using the Vitek-60 microorganism auto-analysis system (BioMerieux Corporate, Craponne, France).
Minimum inhibitory concentrations (MICs) of 17 antimicrobial agents were determined using an agar dilution method with Mueller-Hinton agar recommended by the Clinical and Laboratory Standards Institute (CLSI document M100-S27, 2017). Broad range concentrations of 0.125-1024 μg/mL were used for all the agents. MICs were interpreted according to CLSI breakpoints for P. aeruginosa.
Each purified isolate was incubated overnight in 5 ml of Luria-Bertani (LB) broth at 37°C for 16 hours, and genomic DNA was extracted using an AxyPrep Bacterial Genomic DNA Miniprep kit (Axygen Scientific, Union City, CA, USA). According to the time period of isolation, two mixed DNA collections consisting of equal amounts of genomic DNA of each strain were obtained. One collection (designated TL0809) contained the bacteria isolated from 2008-2009 and the other (designated TL151617) contained those isolated among 2015-2017. The library with an average insert size of 400 bp was prepared using NEBNext Ultra II DNA library preparation kit, and subsequently high-throughput sequenced by the Illumina Novaseq (paired-end run; 2×150 bp). In addition, a 10- to 20-kb insert library was obtained from the genomic DNA of P. aeruginosa TL1285 and sequenced by Pacific Bioscience RSII sequencers at Annoroad Gene Technology Co., Ltd. (Beijing, China).
Genome assembly of pooled DNA sequencing data was performed using megahit (Li et al., 2015), and contigs less than 400 bp were discarded. The complete genome of P. aeruginosa TL1285 was assembled using Canu (Koren et al., 2017) with long reads obtained from PacBio sequencing. Error correction of tentative complete circular sequence was performed using Pilon (Walker et al., 2014) with short read sets derived from Illumina sequencing. Open reading frames (ORFs) of pooled DNA sequences were predicted using Prodigal (Hyatt et al., 2010) with default parameters. Using the antibiotic resistance genes of the CARD (Jia et al., 2017) and ResFinder (Ea et al., 2012) databases as a query, a BLASTN search was performed against the two assembled sequences of the pooled DNA with thresholds of >70% nucleotide identity and >80% alignment coverage. Gene prediction and annotation of TL1285 were initially performed with RAST (Aziz et al., 2008) and then verified by BLASTP searches against the UniProtKB/Swiss-Prot (Boutet et al., 2016) and RefSeq (O’Leary et al., 2016) databases. Annotation of mobile genetic elements was carried out using online databases including ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and the Tn Number Registry (Roberts et al., 2008). Comparison of the TL1285 genome with the other six genomes was performed using BLAST Ring Image Generator (Alikhan et al., 2011). Gene organization diagrams were generated using R script and modified with Inkscape 1.0 (https://inkscape.org/en/).
Genomic DNA of each of the 430 isolates was screened for the floR gene using PCR with primers listed in Table 1. PCR amplification was carried out under the following conditions: initial denaturation for 10 min at 94°C; 35 cycles of denaturation (30 s at 94°C), annealing (30 s at 58°C) and extension (90 s at 72°C) and a final extension for 10 min at 72°C. The floR-T1 and floR-T2 gene sequences with promoter regions were amplified from P. aeruginosa TL1285 and cloned into pUCP24. Electroporation transformation was used to introduce the recombinant plasmids into P. aeruginosa PAO1 by Bio-rad MicroPulser with a voltage at 2.6 kv, resistance at 200 Ω and pulse time of 5 ms (Dennis and Sokol, 1995).
Quantitative reverse transcription PCR (qRT-PCR) was used to investigate the expression of the floR variants of TL1285 and transformants in the presence or absence of 2 mg/L florfenicol or chloramphenicol. In brief, RNA was extracted from 3 mL of LB broth culture (OD600 = 1) of P. aeruginosa TL1285 and the transformants using TRIzol Reagent (Invitrogen, USA) following the manufacturer’s instructions. RNA (1 μg) was used as the template for cDNA synthesis using HiScript II Reverse Transcriptase (Vazyme, Nanjing, China) following the manufacturer’s instructions. qRT-PCR was used to quantify the amount of floR-T1 and floR-T2 in cDNAs using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) following the manufacturer’s instructions with the qPCR primers (Table 1).
Inverse PCR using the primers beside the attL and attR sites could be utilized for the rapid identification of the extrachromosomal intermediate of Tn4371 (Ryan et al., 2009). PCR product was obtained only when integrative and conjugative element (ICE) was excised from the chromosome and circularized. Since no attL site was identified in TL1285, we designed two primers (P2 and P3) located beside the integrase genes as the forward primers. PCR amplification was carried out under the following conditions: an initial denaturation of 10 min at 94°C; 33 cycles of denaturation (94°C for 30 s), annealing (62°C for 30 s), and extension (72°C for 90 s); and a final extension step at 72°C for 10 min.
The complete chromosome sequence of the P. aeruginosa TL1285 (CP053390) has been submitted to NCBI GenBank.
This study uses strains obtained from a teaching hospital of Wenzhou Medical University. It did not require the study to be reviewed or approved by an ethics committee because individual patient data was not involved, and only anonymous clinical residual samples during routine hospital laboratory procedures were used in this study.
The MICs of florfenicol and chloramphenicol were determined for the 430 clinical P. aeruginosa isolates. It showed that 21 (4.88%) and 23 (5.35%) of the strains exhibited much higher resistance levels to florfenicol and chloramphenicol with the same MICs of ≥512 μg/mL for them (Figure 1). A total of 94.65% (407/430) of the strains were resistant to either florfenicol or chloramphenicol (or both), and only 5.35% (23/430) isolates were susceptible to both florfenicol and chloramphenicol with MIC ≤16 μg/mL.
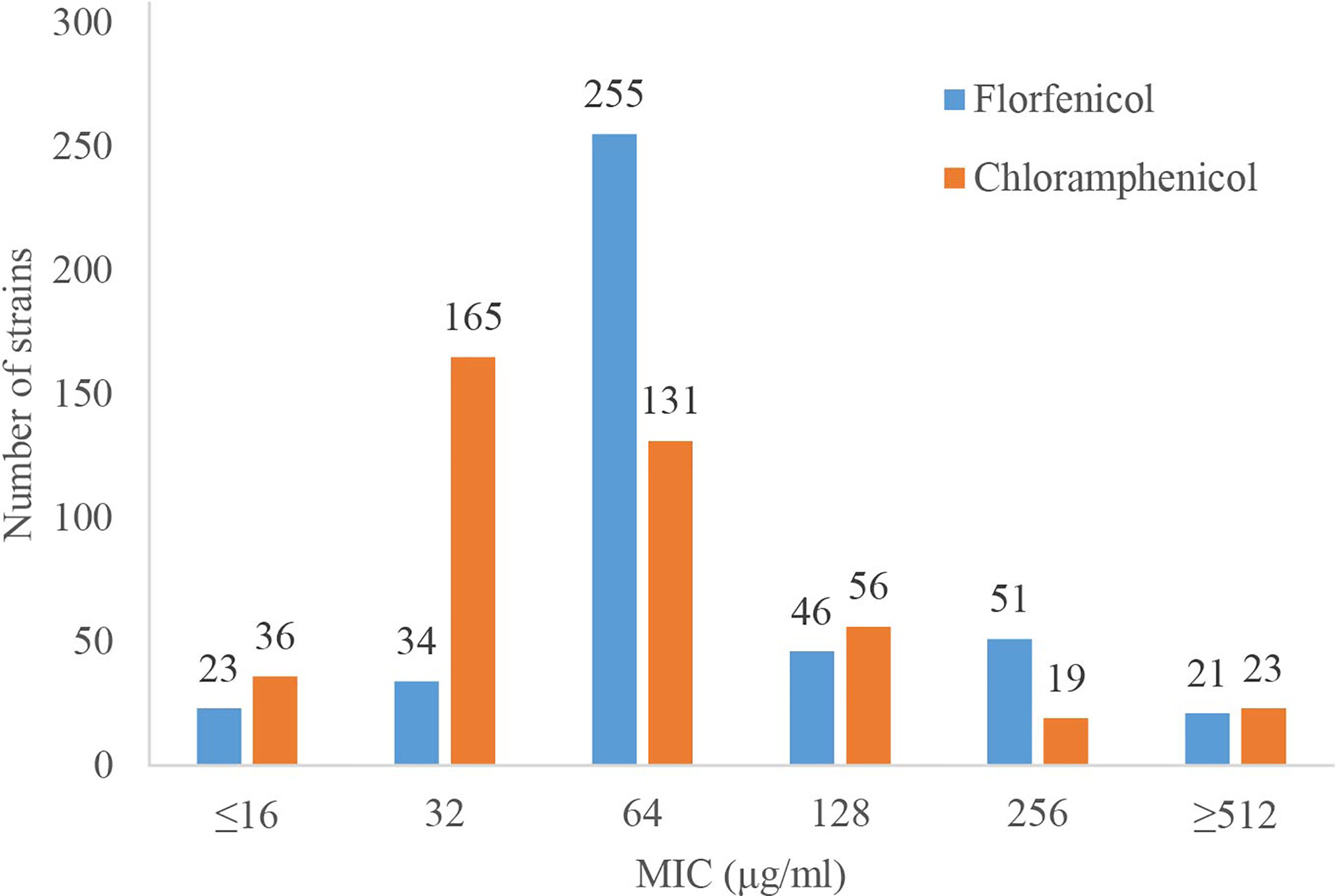
Figure 1 MIC results of the 430 clinical P. aeruginosa isolates against florfenicol and chloramphenicol.
To investigate the prevalence of the floR gene among clinical P. aeruginosa isolates, two mixed DNA collections TL0809 (containing 200 P. aeruginosa strains isolated from 2008-2009) and TL151617 (containing 230 P. aeruginosa strains isolated from 2015-2017) were sequenced. Using the floR gene (AF231986) as a reference, three types of floR variants (the reference floR, floR-T1 and floR-T2 in this study) were identified in the two mixed genomes, of which TL0809 contained all three and TL151617 contained only two floR variants (the reference floR and floR-T1) (Table 2). Other antimicrobial resistance genes (ARGs) identified in TL0809 and TL151617 were listed in Table S1.
The result of PCR amplification of the reference floR, floR-T1 and floR-T2 genes showed that the most prevalent variant was floR-T1, while floR-T2 was only identified in the isolates collected from 2008-2009 (Table 2). The positive rates were consistent with the abundance [expressed as ‘copy of ARG per copy of 16S-rRNA gene’ (Li et al., 2015)] of the corresponding genes in the pooled genomic DNA sequencing libraries. The positive rate of the floR-T1 gene in the strains collected from 2015-2017 (7.39%, 17/230) was higher compared with that from 2008-2009 (3.00%, 6/200).
Among all P. aeruginosa strains, only one strain named TL1285, isolated from a sputum sample in 2008, carried both floR-T1 and floR-T2. P. aeruginosa TL1285 was resistant to chloramphenicol, florfenicol and many other antibacterial agents (Table 3). The fragment containing floR-T1 or floR-T2 gene and its putative promoter region was amplified from TL1285 genomic DNA and subsequently cloned into pUCP24, and then transformed into E. coli DH5α and P. aeruginosa ΔPAO1 (P. aeruginosa PAO1 deleted of ampG), respectively. As a result, compared with the recipients (E. coli DH5α and P. aeruginosa ΔPAO1), the recombinants with the cloned floR-T1 (DH5α/pUCP24-floR-T1 and ΔPAO1/pUCP24-floR-T1) increased ≥4 folds of MIC levels to both chloramphenicol and florfenicol and the recombinants with the cloned floR-T2 (DH5α/pUCP24-floR-T2 and ΔPAO1/pUCP24-floR-T2) increased ≥8 folds of MIC levels to both chloramphenicol and florfenicol, respectively. The results indicated that the floR-T1 and floR-T2 genes of P. aeruginosa TL1285 were functionally active.
The expression of the two floR variants with or without florfenicol (or chloramphenicol) induction were detected (Figure 2). It revealed that the mRNA levels of floR-T2 in P. aeruginosa TL1285 and the corresponding transformants (DH5α/pUCP24-floR-T2 and ΔPAO1/pUCP24-floR-T2) were significantly increased, while the mRNA levels of floR-T1 in P. aeruginosa TL1285 and the transformants (DH5α/pUCP24-floR-T1 and ΔPAO1/pUCP24-floR-T1) were only slightly increased in the presence of florfenicol or chloramphenicol.
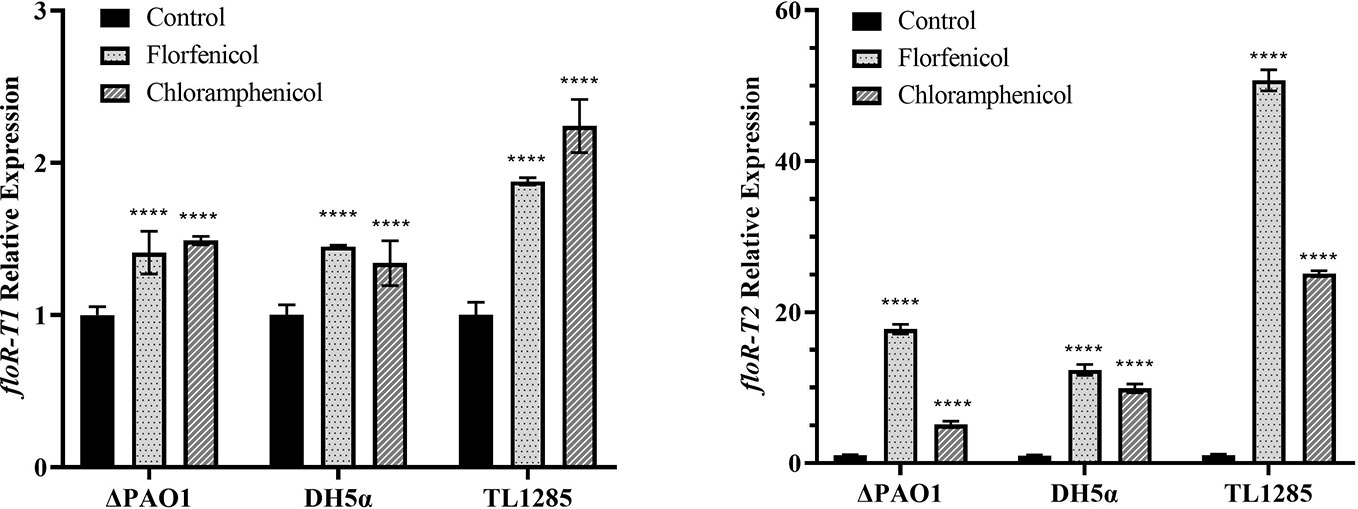
Figure 2 Expression of the floR variants in TL1285 and corresponding transformants with or without florfenicol or chloramphenicol induction. **** (P-value < 0.0001).
Using phylogenetic analysis, the amino acid identities of FloR-T1 and FloR-T2 with the known FloR proteins ranged from 90.80% to 100% and 86.10% to 88.90%, respectively (Figure S1). FloR-T1 was identical to the FloR protein (YP_001715371.1) identified in Acinetobacter baumannii, while FloR-T2 showed the highest identity (88.90%) with the FloR protein (YP_005351917.1) identified in Klebsiella pneumoniae.
The translational attenuator that consisted of a single pair inverted repeat (IR) sequence, and a short reading frame of 6-9 aa peptide was identified upstream of the floR variants (Figure 3). IR1 and IR2 can form a stable stem-loop structure blocking the resistance gene-associated ribosome binding site (RBS). The short peptides of floR-T1 and floR-T2 differ in three amino acids. The attenuator sequences of floR-T2 and floRv encode an identical peptide, although one synonymous variation (A>T) in their nucleotide sequences. The attenuators’ IR resulted also differently, and the stem-loop structures formed in distinct stable states. Among these variants, floR-T2 and floRv showed the most stable structure. However, the stable stem-loop structure of the attenuator sequence did not overlap with the RBS site of the floR gene.
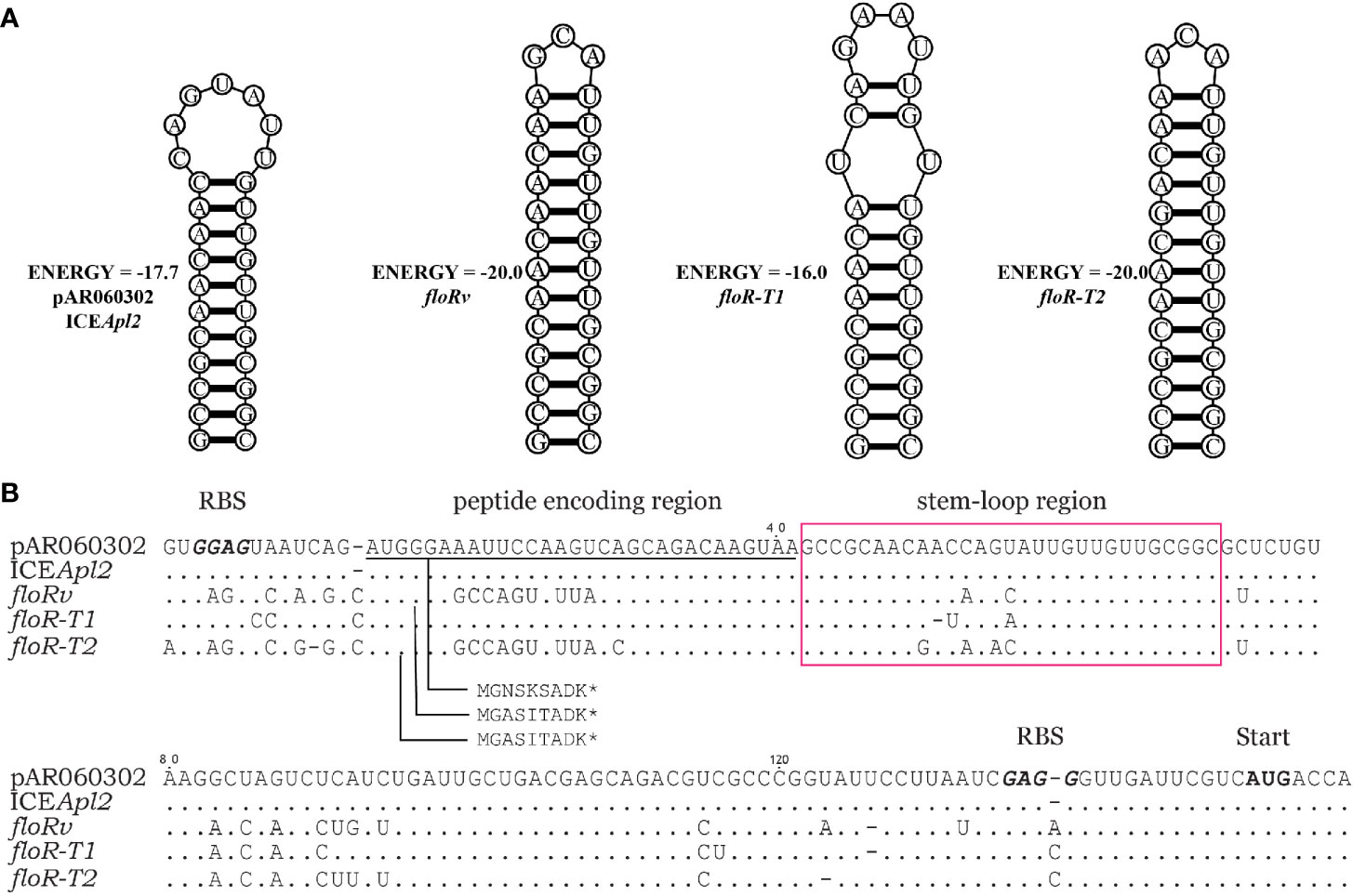
Figure 3 Comparison of sequences upstream of the floR variants. (A) The stable mRNA secondary structure of the floR variants formed by inverted repeat sequences boxed in (B). (B) The attenuator of the floR variants consists of a peptide-encoding region (underlined) and stem-loop region (boxed). The start codons and ribosome binding sites (RBS) of the short peptide and floR are labeled and displayed in bold type letters.
Whole genome sequencing (WGS) was performed for P. aeruginosa TL1285 carrying both floR-T1 and floR-T2, and only produced a circular 6,609,407 bp chromosome with an average GC content of 66.06% encoding 5,611 ORFs. Multiple ARGs, including resistance genes for β-lactams (blaOXA-50 and blaPDC-3), aminoglycosides (aadA5 and aac(3)-IIa), sulfonamides (sul1), tetracycline (tetG), chloramphenicol (catB7, floR-T1 and floR-T2) and fosfomycin (fosA), were identified in the P. aeruginosa TL1285 genome. The florfenicol-resistant genes floR-T1 and floR-T2 were embedded in an 86-kb Tn4371-like integrative and conjugative element (ICE) (Figure 4).
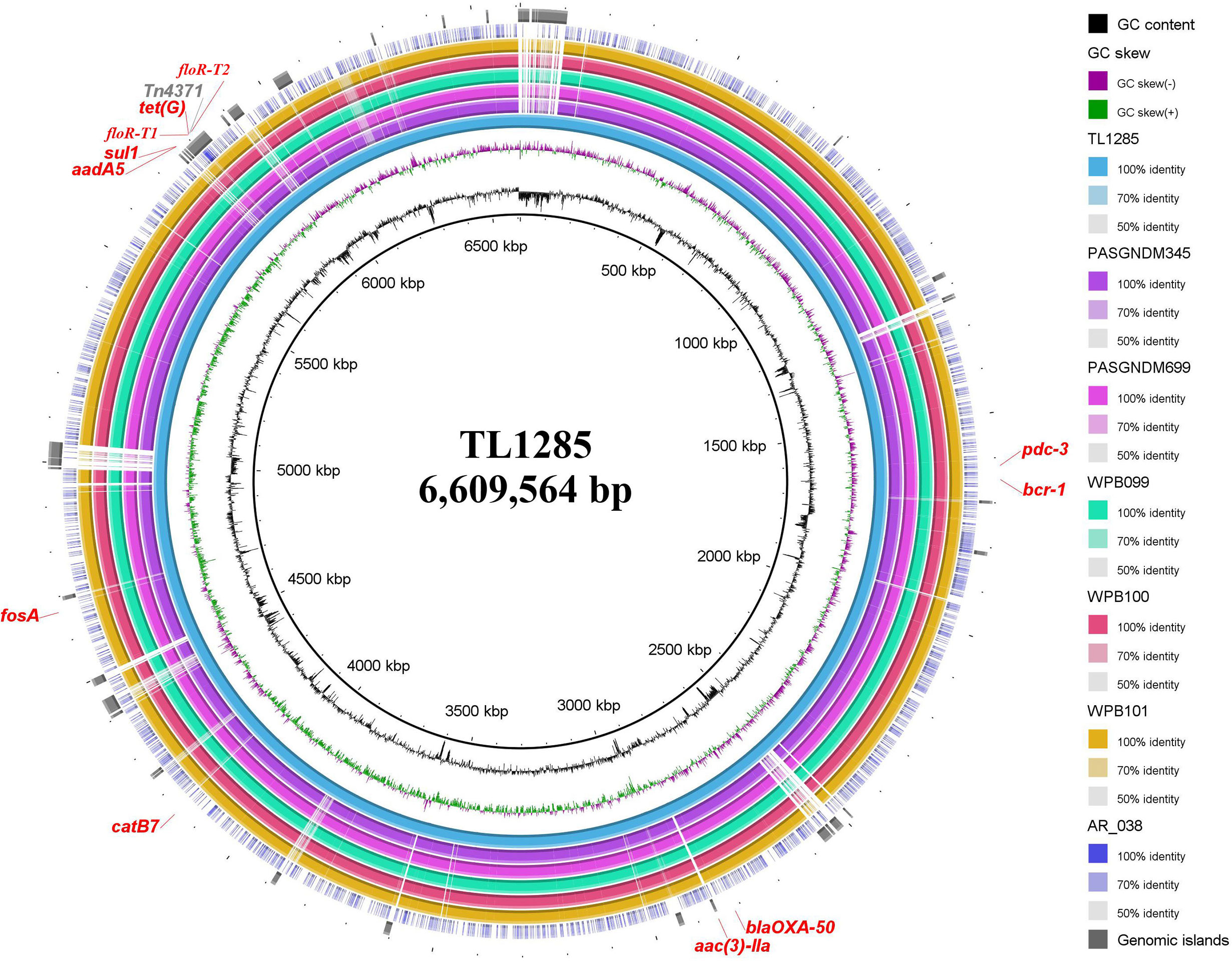
Figure 4 Sequence conservation among P. aeruginosa TL1285 and 6 other genomes carrying floR-T2. From innermost to outermost: Circle 1 shows the scale in kb; Circles 2 and 3 represent the GC content and GC skew maps of TL1285, respectively; Circle 4 represents the genome of TL1285; Circles 5-10 represent the homologous regions of PASGNDM345, PASGNDM699, WPB099, WPB100, WPB101 and AR_038 compared to those of TL1285, while the regions without similar hits between them were left blank; Circle 11 displays the genomic islands in TL1285; Circle 12 displays the antibiotic resistance genes in TL1285.
To track the epidemiological correlation between floR-T2 and genome islands, a BLASTN search was performed against the GenBank database using floR-T2 as a query. A total of five P. aeruginosa chromosomes, WPB099 (CP031878), WPB100 (CP031877), WPB101 (CP031876), PASGNDM345 (CP020703) and PASGNDM699 (CP020704), and one E. cloacae chromosome, AR_038 (CP030347), were found carrying floR-T2. Through MLST analysis, the five P. aeruginosa belonged to ST308, while TL1285 to ST316. Interestingly, these floR-T2-carrying strains came from different sources. WPB099, WPB100 and WPB101 were isolated from hospital wastewaters in Singapore, PASGNDM345 and PASGNDM699 from patient sputum in Singapore, while E. cloacae AR_038 and TL1285 were from patient sputum collected in United States and China, respectively. Whole genome alignment of the six P. aeruginosa strains revealed high identity, and their differences were mainly in some genomic islands (Figure 4). The Tn4371-like ICE carrying floR-T2 in TL1285 was also partially present in these five P. aeruginosa strains. Nevertheless, it should be noted that WPB099, WPB100 and WPB101 were not fully sequenced, and the floR-T2 gene was located on an approximately 10 kb separate segment, which means the precise genetic environments around floR-T2 could not be described.
Comparative analysis of the Tn4371-like ICE regions of six P. aeruginosa strains revealed that the plasmid maintenance system (repA, parA and parB) and conjugational transfer systems were conserved (Figure 5). The variable region between the traF and traR genes, which encoded a biphenyl catabolic bph gene cluster in Tn4371 (AJ536756), was different in these six P. aeruginosa isolates. The variable regions of WPB099, WPB100 and WPB101 were a 20-kb fragment encoding the oqxB32 gene, which confers resistance to quinolone. The variable regions of PASGNDM345 and PASGNDM699 shared high identity with those of WPB099, WPB100 and WPB101. The only difference was that in PASGNDM345 and PASGNDM699, a 13.7-kb fragment flanked by 695 bp direct repeats was inserted between czcD and lysR, which encode blaNDM-1, msr(E) and floR-T2 genes. The variable region of TL1285 was similar to those of PASGNDM345 and PASGNDM699, except that the blaNDM-1-hp-msr(E) genes of PASGNDM345 and PASGNDM699 were replaced by floR-T1-tetR-tetA-lysR in TL1285.
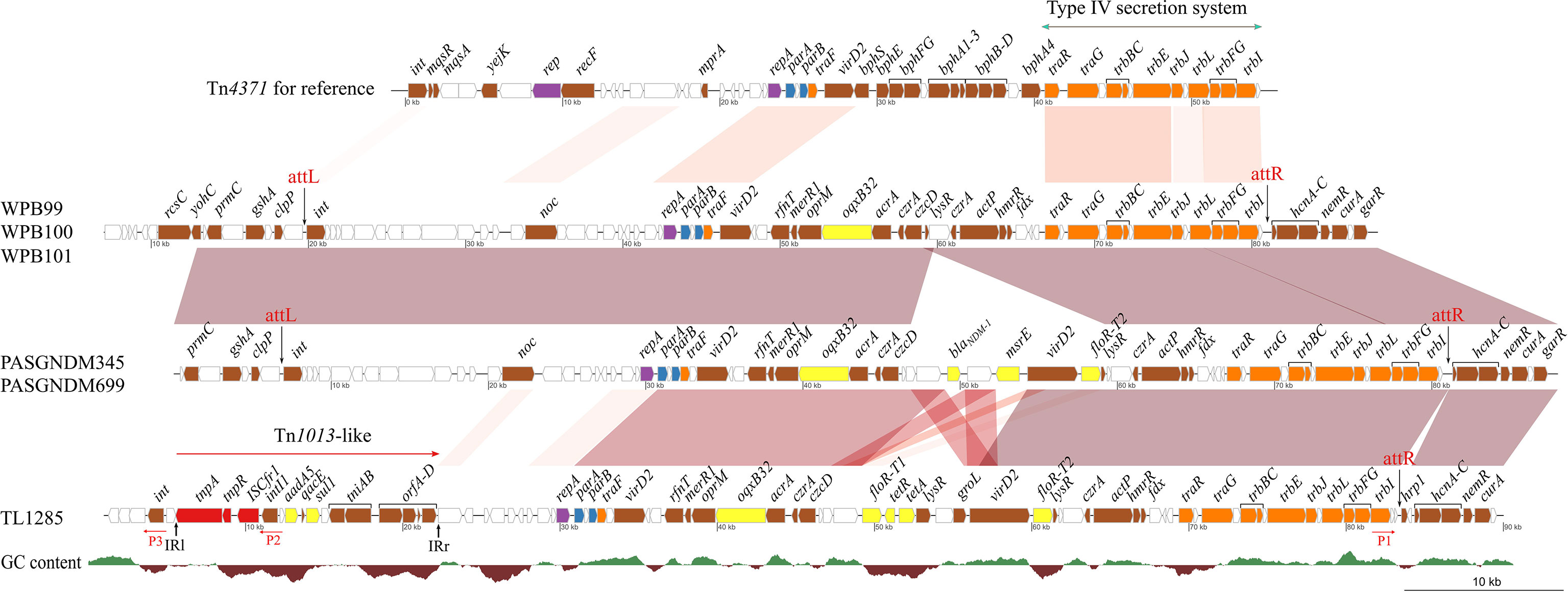
Figure 5 Comparative genomic analysis of the ICE region of TL1285 and 5 other floR-T2-carrying P. aeruginosa isolates. Genes with different functions are shown in different colors: red, transposable elements; yellow, drug resistance; orange, conjugational transfer; blue, plasmid maintenance; purple, replication; brown, genes with other functions; white, hypothetical proteins.
The integrase genes (int) of PASGNDM345, PASGNDM699, WPB99, WPB100 and WPB101 were identical and shared 78% identity with that of Tn4371. However, no homologue of intTn4371 was found in TL1285 (Figure 5). Tn4371 family ICEs could be integrated into the genome through an 8-bp attB site, generating direct repeat attL and attR element chromosomal junctions (Merlin et al., 1999). In PASGNDM345, PASGNDM699, WPB99, WPB100 and WPB101, 8-bp repeats (5′-TTTTTTGT-3′) were identified in both extremities of the ICE region. However, in TL1285, only attR was found (Figure 5). The noc gene upstream of repA in TL1285 was truncated by a novel Tn402 family transposon. The transposon is formed by ISCfr1 and In2 carrying a single aadA5 cassette embedded downstream of the tnpR gene of Tn1013, and this Tn402 family transposon was surrounded by 37-bp imperfect inverted repeats (Figure S2).
Inverse PCR using primers P1, P2 and P3 (Figure 5) was performed to detect whether the ICE in TL1285 could generate a circular extrachromosomal form, but no positive result was observed. Taken together, we speculate that the ICE in TL1285 is an incomplete member of the Tn4371 family and may have lost the excising or integrating ability. The insertion of the Tn402 family transposon leads to the loss of the upstream sequence of the nucleoid occlusion protein coding gene noc, including the integrase gene int of ICE.
In this work, we found that among the 430 clinical P. aeruginosa isolates detected, most (94.65%, 407/430) of them were resistant to florfenicol or/and chloramphenicol. In fact, P. aeruginosa was intrinsically resistant to amphenicols, and the MICs to florfenicol and chloramphenicol for P. aeruginosa ATCC27853 were both 64 μg/mL (Fass and Barnishan, 1979). Active efflux and chloramphenicol acetyltransferase (CAT) encoded on P. aeruginosa chromosome are two major mechanisms of amphenicol resistance (Nitzan and Rushansky, 1981; Li et al., 1994), and different types of CAT determinant also contribute to amphenicol resistance (White et al., 1999). However, there were still 5.35% (23/430) isolates susceptible to amphenicol. The mutation and deletion of multidrug efflux system (such as MexA-MexB-OprK) and other resistance mechanisms might play a role in the loss of resistance to florfenicol or/and chloramphenicol of these bacteria.
Three floR variants (floR, floR-T1 and floR-T2) were identified in a number of clinical P. aeruginosa isolates, in which floR-T1 was the most prevalent variant and floR-T2 was a novel variant identified in this study. The positive rate of the floR-T1 gene in the strains collected from 2015-2017 (7.39%, 17/230) was similar to that of the clinical K. pneumoniae isolates collected from the same district during 2010-2014 (7.01%, 23/328) (Lu et al., 2018). The protein showing the highest identity (88.90%) with FloR-T2 was a FloR protein (YP_005351917.1) identified in Klebsiella pneumoniae. Currently, FloRv was the FloR variant with the lowest identity (88.40%-91.80%) to other previously reported FloR proteins (He et al., 2015). FloR-T2 exhibited less identity with other FloR proteins than FloRv. Furthermore, FloR-T2 was shown to be one of the most divergent members of the FloR family, followed by FloRv (Figure S1).
It was interesting to find that the expression levels of floR-T2 increased much more significantly in the host TL1285 or the recombinant than those of floR-T1 when induced by the amphenicols. Using transcriptome sequencing, Lang et al. found that the expression of the floR gene of the E. coli plasmid pAR060302 increased 8-fold under the induction of florfenicol (Lang et al., 2012). Yinghui et al. also reported that the mRNA levels of the floR gene encoded by ICEApl2 on chromosomes increased in the presence of chloramphenicol (Li et al., 2018). However, the modulation mechanisms of mRNA expression of floR variants remain unclear. As reported by Yinghui et al. (Li et al., 2018), we also identified the translational attenuator region upstream of the floR variants. In addition, we found that the peptide encoding region of floR-T1 was identical to those of pA060302 and ICEApl2 reported by Yinghui (Li et al., 2018) (Figure 3). It is known that the expression of chloramphenicol resistance genes, including catA, cmlA and fexA, could be induced by chloramphenicol, and this induction is mediated by translational attenuator structure at the post-transcriptional level (Stokes and Hall, 1991; Kehrenberg and Schwarz, 2004; Schwarz et al., 2004). However, considering that the stem-loop structure is distant to the RBS site of the floR gene, it is not clear whether this structure participates in the induced expression of the floR gene.
WGS result revealed that floR-T1 and floR-T2 of P. aeruginosa TL1285 were related with a novel Tn4371-like ICE. Tn4371 is a 55-kb ICE that can be integrated into the attB site (5’-TTTTCAT-3’) through a site-specific recombination process since the ends of the element can be detected covalently as a transfer intermediate (Merlin et al., 1999; Toussaint et al., 2003). The Tn4371-like ICEs are mosaic in structure and consist of Ti-RP4-like transfer systems, an integrase region, plasmid maintenance genes and accessory genes (Toussaint et al., 2003). Any ICE that encodes an integrase gene closely related to intTn4371 (>70% protein homology) and has similar maintenance and transfer genes could be considered as a member of the Tn4371 family (Ryan et al., 2009). The Tn4371-like ICEs carrying floR variants have been identified in the P. aeruginosa strains of different MLST types (such as ST308 and ST316) isolated from different samples of different countries. P. aeruginosa ST308 is a high-risk clone that can locally acquire resistance determinants from water-distribution system and was involved in a five-year outbreak in a French hospital between 2005 and 2010 (Jeanvoine et al., 2019). The variable region of these Tn4371-like ICEs also carried other ARGs like blaNDM-1, tetA and msr(E). These findings indicate that the Tn4371-like ICEs might have emerged as a potential vehicle to mediate the spread of drug resistance genes in P. aeruginosa isolates.
In this study, we determined the prevalence of floR among 430 clinical isolates of P. aeruginosa and characterized two floR variants, floR-T1 and floR-T2, in a P. aeruginosa strain TL1285. The floR-T1 gene was the most prevalent variant in clinical P. aeruginosa strains. The floR-T2 is a novel floR variant that showed less identities with the other FloR proteins than FloRv. The mRNA levels of the two floR variants could be induced by florfenicol and chloramphenicol and the expression level of floR-T2 was significantly higher than that of floR-T1. Inverted repeat sequences as well as stem-loop regions of the translational attenuators differed among the floR variants. The floR-T1 and floR-T2 of TL1285 were located on an incomplete novel Tn4371 family ICE, while floR-T2-carrying ICEs were also identified in other five P. aeruginosa genomes. These results indicate that Tn4371 family ICEs might be related with the dissemination of floR-T2 among P. aeruginosa strains.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
JL, HX, HYL, XL, KL, and HZ collected the strains. HML, WL, JC, XZhang, KS, and QL performed the experiments. QC, HLL, and XZhu analyzed the experimental results. CQ, WZ, and AL performed the bioinformatics analysis. CQ, TX, and QB co-led the writing of the manuscript. HZ, ZM, and QB designed the work. All authors contributed to the article and approved the submitted version.
This study was supported by the Science and Technology Project of Wenzhou City, China (Y20170205 and 2019Y0358), Science and Technology Project of Lishui City, China (2017GYX07), the National Natural Science Foundation of China (81973382, 81960381), the Natural Science Foundation of Zhejiang Province, China (LQ17H190001 and LY19C060002) and the Science & Technology Project of Inner Mongolia Autonomous Region, China (201802125).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflict of interest.
The authors would like to acknowledge all study participants and individuals who contributed to the study. This manuscript has been released as a pre-print at Research Square, (Qian et al., 2020).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.685068/full#supplementary-material
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., Beatson, S. A. (2011). Blast Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402
Arcangioli, M. A., Leroy-Sétrin, S., Martel, J. L., Chaslus-Dancla, E. (1999). A New Chloramphenicol and Florfenicol Resistance Gene Flanked by Two Integron Structures in Salmonella Typhimurium DT104. FEMS Microbiol. Lett. 174 (2), 327–332. doi: 10.1111/j.1574-6968.1999.tb13586.x
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genomics 9 (1), 75–70. doi: 10.1186/1471-2164-9-75
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt Knowledgebase: How to Use the Entry View. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Breidenstein, E. B. M., de la Fuente-Núñez, C., Hancock, R. E. W. (2011). Pseudomonas Aeruginosa: All Roads Lead to Resistance. Trends Microbiol. 19 (8), 419–426. doi: 10.1016/j.tim.2011.04.005
Chang, S.-K., Lo, D.-Y., Wei, H.-W., Kuo, H.-C. (2014). Antimicrobial Resistance of Escherichia Coli Isolates From Canine Urinary Tract Infections. J. Vet. Med. Sci. 77 (1), 59–65. doi: 10.1292/jvms.13-0281
da Silva, G. C., Rossi, C. C., Santana, M. F., Langford, P. R., Bossé, J. T., Bazzolli, D. M. S. (2017). p518, a Small Flor Plasmid From a South American Isolate of Actinobacillus Pleuropneumoniae. Vet. Microbiol. 204, 129–132. doi: 10.1016/j.vetmic.2017.04.019
Dennis, J. J., Sokol, P. A. (1995). Electrotransformation of Pseudomonas. Methods Mol. Biol. 47, 125–133. doi: 10.1385/0-89603-310-4:125
Domalaon, R., Idowu, T., Zhanel, G. G., Schweizer, F. (2018). Antibiotic Hybrids: The Next Generation of Agents and Adjuvants Against Gram-Negative Pathogens? Clin. Microbiol. Rev. 31 (2). doi: 10.1128/CMR.00077-17
Ea, Z., Henrik, H., Salvatore, C., Martin, V., Simon, R., Ole, L., et al. (2012). Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 67 (11), 11. doi: 10.1093/jac/dks261
Fass, R. J., Barnishan, J. (1979). Minimal Inhibitory Concentrations of 34 Antimicrobial Agents for Control Strains Escherichia Coli ATCC 25922 and Pseudomonas Aeruginosa ATCC 27853. Antimicrob. Agents Chemother. 16 (5), 622–624. doi: 10.1128/aac.16.5.622
Gabida, M., Gombe, N. T., Chemhuru, M., Takundwa, L., Bangure, D., Tshimanga, M. (2015). Foodborne Illness Among Factory Workers, Gweru, Zimbabwe, 2012: A Retrospective Cohort Study. BMC Res. Notes 8 (1), 493. doi: 10.1186/s13104-015-1512-2
He, T., Shen, J., Schwarz, S., Wu, C., Wang, Y. (2015). Characterization of a Genomic Island in Stenotrophomonas Maltophilia That Carries a Novel Flor Gene Variant. J. Antimicrob. Chemother. 70 (4), 1031–1036. doi: 10.1093/jac/dku491
Hyatt, D., Chen, G.-L., LoCascio, P. F., Land, M. L., Larimer, F. W., Hauser, L. J. (2010). Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinformatics 11 (1), 119–110. doi: 10.1186/1471-2105-11-119
Idowu, T., Arthur, G., Zhanel, G. G., Schweizer, F. (2019). Heterodimeric Rifampicin-Tobramycin Conjugates Break Intrinsic Resistance of Pseudomonas Aeruginosa to Doxycycline and Chloramphenicol In Vitro and in a Galleria Mellonella In Vivo Model. Eur. J. Med. Chem. 174, 16–32. doi: 10.1016/j.ejmech.2019.04.034
Jeanvoine, A., Meunier, A., Puja, H., Bertrand, X., Valot, B., Hocquet, D. (2019). Contamination of a Hospital Plumbing System by Persister Cells of a Copper-Tolerant High-Risk Clone of Pseudomonas Aeruginosa. Water Res. 157, 579–586. doi: 10.1016/j.watres.2019.04.011
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 45 (D1), D566–DD73. doi: 10.1093/nar/gkw1004
Kehrenberg, C., Schwarz, S. (2004). fexA, a Novel Staphylococcus Lentus Gene Encoding Resistance to Florfenicol and Chloramphenicol. Antimicrob. Agents Chemother. 48 (2), 615–618. doi: 10.1128/AAC.48.2.615-618.2004
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., Phillippy, A. M. (2017). Canu: Scalable and Accurate Long-Read Assembly Via Adaptive K-Mer Weighting and Repeat Separation. Genome Res. 27 (5), 722–736. doi: 10.1101/gr.215087.116
Lai, J., Wang, Y., Shen, J., Li, R., Han, J., Foley, S. L., et al. (2013). Unique Class 1 Integron and Multiple Resistance Genes Co-Located on IncHI2 Plasmid Is Associated With the Emerging Multidrug Resistance of Salmonella Indiana Isolated From Chicken in China. Foodborne Pathog. Dis. 10 (7), 581–588. doi: 10.1089/fpd.2012.1455
Lang, K. S., Anderson, J. M., Schwarz, S., Williamson, L., Handelsman, J., Singer, R. S. (2010). Novel Florfenicol and Chloramphenicol Resistance Gene Discovered in Alaskan Soil by Using Functional Metagenomics. Appl. Environ. Microbiol. 76 (15), 5321–5326. doi: 10.1128/AEM.00323-10
Lang, K. S., Danzeisen, J. L., Xu, W., Johnson, T. J. J. A., Microbiology, E. (2012). Transcriptome Mapping of pAR060302, a blaCMY-2-Positive Broad-Host-Range IncA/C Plasmid. Appl. Environ. Microbiol. 78 (9), 3379–3386. doi: 10.1128/AEM.07199-11
Li, Y., Li, Y., Fernandez Crespo, R., Leanse, L. G., Langford, P. R., Bosse, J. T. (2018). Characterization of the Actinobacillus Pleuropneumoniae SXT-related Integrative and Conjugative Element ICEApl2 and Analysis of the Encoded FloR Protein: Hydrophobic Residues in Transmembrane Domains Contribute Dynamically to Florfenicol and Chloramphenicol Efflux. J. Antimicrob. Chemother. 73 (1), 57–65. doi: 10.1093/jac/dkx342
Li, D., Liu, C. M., Luo, R., Kunihiko, S., Tak-Wah, L. J. B. (2015). MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly Via Succinct De Bruijn Graph. Bioinformatics 31 (10), 16740–1676. doi: 10.1093/bioinformatics/btv033
Li, X. Z., Livermore, D. M., Nikaido, H. (1994). Role of Efflux Pump(s) in Intrinsic Resistance of Pseudomonas Aeruginosa: Resistance to Tetracycline, Chloramphenicol, and Norfloxacin. Antimicrob. Agents Chemother. 38 (8), 1732–1741. doi: 10.1128/aac.38.8.1732
Liu, H., Wang, Y., Wu, C., Schwarz, S., Shen, Z., Jeon, B., et al. (2012). A Novel Phenicol Exporter Gene, fexB, Found in Enterococci of Animal Origin. J. Antimicrob. Chemother. 67 (2), 322–325. doi: 10.1093/jac/dkr481
Li, B., Yang, Y., Ma, L., Ju, F., Guo, F., Tiedje, J. M., et al. (2015). Metagenomic and Network Analysis Reveal Wide Distribution and Co-Occurrence of Environmental Antibiotic Resistance Genes. ISME J. 9 (11), 2490–2502. doi: 10.1038/ismej.2015.59
Lu, J., Zhang, J., Xu, L., Liu, Y., Li, P., Zhu, T., et al. (2018). Spread of the Florfenicol Resistance Flor Gene Among Clinical Klebsiella Pneumoniae Isolates in China. Antimicrob. Resist. Infect. Control 7, 127. doi: 10.1186/s13756-018-0415-0
Merlin, C., Springael, D., Toussaint, A. (1999). Tn4371: A Modular Structure Encoding a Phage-Like Integrase, a Pseudomonas-like Catabolic Pathway, and RP4/Ti-like Transfer Functions. Plasmid 41 (1), 40–54. doi: 10.1006/plas.1998.1375
Morita, Y., Tomida, J., Kawamura, Y. (2014). Responses of Pseudomonas Aeruginosa to Antimicrobials. Front. Microbiol. 4, 422. doi: 10.3389/fmicb.2013.00422
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., Correia, A. (2009). INTEGRALL: A Database and Search Engine for Integrons, Integrases and Gene Cassettes. Bioinformatics 25 (8), 1096–1098. doi: 10.1093/bioinformatics/btp105
Nitzan, Y., Rushansky, N. M. (1981). Chloramphenicol Acetyltransferase fromPseudomonas Aeruginosa—A New Variant of the Enzyme. Curr. Microbiol. 5 (5), 261–265. doi: 10.1007/BF01567915
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 44 (D1), D733–D745. doi: 10.1093/nar/gkv1189
Qian, C., Liu, H., Lu, W., Cao, J., Lu, J., Li, A., et al. (2020). Identification of Flor Variants Associated With Tn4371-Like Integrative and Conjugative Elements in Human Clinical Pseudomonas Aeruginosa Isolates. Res. Sq. doi: 10.21203/rs.3.rs-67840/v1
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised Nomenclature for Transposable Genetic Elements. Plasmid 60 (3), 167–173. doi: 10.1016/j.plasmid.2008.08.001
Ryan, M. P., Pembroke, J. T., Adley, C. C. (2009). Novel Tn4371-ICE Like Element in Ralstonia Pickettii and Genome Mining for Comparative Elements. BMC Microbiol. 9, 242. doi: 10.1186/1471-2180-9-242
Schwarz, S., Kehrenberg, C., Doublet, B., Cloeckaert, A. (2004). Molecular Basis of Bacterial Resistance to Chloramphenicol and Florfenicol. FEMS Microbiol. Rev. 28 (5), 519–542. doi: 10.1016/j.femsre.2004.04.001
Schwarz, S., Werckenthin, C., Kehrenberg, C. (2000). Identification of a Plasmid-Borne Chloramphenicol-Florfenicol Resistance Gene in Staphylococcus Sciuri. Antimicrob. Agents Chemother. 44 (9), 2530–2533. doi: 10.1128/AAC.44.9.2530-2533.2000
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: The Reference Centre for Bacterial Insertion Sequences. Nucleic Acids Res. 34 (Database issue), D32–D36. doi: 10.1093/nar/gkj014
Stokes, H., Hall, R. (1991). Sequence Analysis of the Inducible Chloramphenicol Resistance Determinant in the Tn1696 Integron Suggests Regulation. Plasmid 26, 10–19. doi: 10.1016/0147-619X(91)90032-R
Syriopoulou, V. P., Harding, A. L., Goldmann, D. A., Smith, A. L. (1981). In Vitro Antibacterial Activity of Fluorinated Analogs of Chloramphenicol and Thiamphenicol. Antimicrob. Agents Chemother. 19 (2), 294–297. doi: 10.1128/aac.19.2.294
Tao, W., Lee, M. H., Wu, J., Kim, N. H., Kim, J.-C., Chung, E., et al. (2012). Inactivation of Chloramphenicol and Florfenicol by a Novel Chloramphenicol Hydrolase. Appl. Environ. Microbiol. 78 (17), 6295–6301. doi: 10.1128/AEM.01154-12
Toussaint, A., Merlin, C., Monchy, S., Benotmane, M. A., Leplae, R., Mergeay, M., et al. (2003). The Biphenyl- and 4-Chlorobiphenyl-Catabolic Transposon Tn4371, a Member of a New Family of Genomic Islands Related to IncP and Ti Plasmids. Appl. Environ. Microbiol. 69 (8), 4837–4845. doi: 10.1128/aem.69.8.4837-4845.2003
Ujvari, B., Weiczner, R., Deim, Z., Terhes, G., Urban, E., Toth, A. R., et al. (2019). Characterization of Pasteurella Multocida Strains Isolated From Human Infections. Comp. Immunol. Microbiol. Infect. Dis. 63, 37–43. doi: 10.1016/j.cimid.2018.12.008
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PloS One 9 (11), e112963. doi: 10.1371/journal.pone.0112963
Wang, Y., Lv, Y., Cai, J., Schwarz, S., Cui, L., Hu, Z., et al. (2015). A Novel Gene, optrA, That Confers Transferable Resistance to Oxazolidinones and Phenicols and its Presence in Enterococcus Faecalis and Enterococcus Faecium of Human and Animal Origin. J. Antimicrob. Chemother. 70 (8), 2182–2190. doi: 10.1093/jac/dkv116
White, P. A., Stokes, H. W., Bunny, K. L., Hall, R. M. (1999). Characterisation of a Chloramphenicol Acetyltransferase Determinant Found in the Chromosome of Pseudomonas Aeruginosa. FEMS Microbiol. Lett. 175 (1), 27–35. doi: 10.1111/j.1574-6968.1999.tb13598.x%JFEMSMicrobiologyLetters
Zhan, Z., Xu, X., Shen, H., Gao, Y., Zeng, F., Qu, X., et al. (2019). Rapid Emergence of Florfenicol-Resistant Invasive Non-Typhoidal Salmonella in China: A Potential Threat to Public Health. Am. J. Trop. Med. Hyg. 101 (6), 1282–1285. doi: 10.4269/ajtmh.19-0403
Keywords: floR, florfenicol resistance, Pseudomonas aeruginosa, Tn4371, integrative and conjugative elements
Citation: Qian C, Liu H, Cao J, Ji Y, Lu W, Lu J, Li A, Zhu X, Shen K, Xu H, Chen Q, Zhou W, Lu H, Lin H, Zhang X, Li Q, Lin X, Li K, Xu T, Zhu M, Bao Q and Zhang H (2021) Identification of floR Variants Associated With a Novel Tn4371-Like Integrative and Conjugative Element in Clinical Pseudomonas aeruginosa Isolates. Front. Cell. Infect. Microbiol. 11:685068. doi: 10.3389/fcimb.2021.685068
Received: 24 March 2021; Accepted: 01 June 2021;
Published: 21 June 2021.
Edited by:
Costas C. Papagiannitsis, University of Thessaly, GreeceReviewed by:
Tamara Salloum, Lebanese American University, LebanonCopyright © 2021 Qian, Liu, Cao, Ji, Lu, Lu, Li, Zhu, Shen, Xu, Chen, Zhou, Lu, Lin, Zhang, Li, Lin, Li, Xu, Zhu, Bao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei Zhu, emh1bWVpX2RAMTYzLmNvbQ==; Qiyu Bao, YmFvcXlAZ2Vub21pY3MuY24=; Hailin Zhang, emhsd3o5N0Bob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.