- 1Department of Pathogen Biology and Immunology, Kunming Medical University, Kunming, China
- 2Program in Public Health, College of Health Sciences, University of California at Irvine, Irvine, CA, United States
- 3Department of Pathology, Kunming Medical University, Kunming, China
- 4Department of Cell Biology and Medical Genetics, Kunming Medical University, Kunming, China
- 5Transfusion Medicine Research Department, Yunnan Kunming Blood Center, Kunming, China
- 6Animal Parasitic Disease Laboratory, Agricultural Research Service, US Department of Agriculture, Beltsville, MD, United States
Drug-resistant Plasmodium vivax malaria impedes efforts to control, eliminate, and ultimately eradicate malaria in Southeast Asia. P. vivax resistance to antifolate drugs derives from point mutations in specific parasite genes, including the dihydropteroate synthase (pvdhps), dihydrofolate reductase (pvdhfr), and GTP cyclohydrolase I (pvgch1) genes. This study aims to investigate the prevalence and spread of drug resistance markers in P. vivax populating the China-Myanmar border. Blood samples were collected from symptomatic patients with acute P. vivax infection. Samples with single-clone P. vivax infections were sequenced for pvdhps and pvdhfr genes and genotyped for 6 flanking microsatellite markers. Copy number variation in the pvgch1 gene was also examined. Polymorphisms were observed in six different codons of the pvdhps gene (382, 383, 512, 549, 553, and 571) and six different codons of the pvdhfr gene (13, 57, 58, 61, 99, 117) in two study sites. The quadruple mutant haplotypes 57I/L/58R/61M/117T of pvdhfr gene were the most common (comprising 76% of cases in Myitsone and 43.7% of case in Laiza). The double mutant haplotype 383G/553G of pvdhps gene was also prevalent at each site (40.8% and 31%). Microsatellites flanking the pvdhfr gene differentiated clinical samples from wild type and quadruple mutant genotypes (FST= 0.259-0.3036), as would be expected for a locus undergoing positive selection. The lack of copy number variation of pvgch1 suggests that SP-resistant P. vivax may harbor alternative mechanisms to secure sufficient folate.
Introduction
Plasmodium vivax, the most broadly distributed source of severe malarial morbidity and mortality, puts almost 40% of the world population at risk (Naing et al., 2014; Rahimi et al., 2014; WHO, 2020), resulting in cases that can be life threatening (Gupta et al., 2015; Gupta et al., 2016). Data from 2017 shows that about 3.3 billion people live in the P. vivax transmission areas, and about 1.5 billion people are at risk of exposure in foci of stable transmission (Battle et al., 2019). Southeast Asian countries shoulder the highest proportion of global burden, with about 7.3 million people infected with P. vivax in 2017, alone (Battle et al., 2019).
Asia is the focus of P. vivax transmission, and Myanmar is among the most severely affected in the region. As one of the most severe malaria burden countries from this area, Myanmar is a source of cases exported to neighboring countries such as mainland China, where low endemic transmission is otherwise leading to successful eradication. From 2010 to 2014, there were 1893 P. vivax malaria cases imported from Myanmar into China, far exceeding importation from any other Asian country (Zhou et al., 2016). Most imported cases are adult male Chinese laborers working in farming, construction, and mining (Zhou et al., 2016). Reported cases in Myanmar have risen steadily since 2012 (WHO, 2016).
Most cases imported to China occur in Yunnan province, the southwestern frontier which borders Myanmar to the west, and Laos and Vietnam to the south (Lai et al., 2019). In particular, Tengchong County (in Yunnan, China) reported the highest number of imported malaria cases from Myitsone, Myanmar from 2010 to 2014 (Wang et al., 2015). Previous studies identified boundary townships, where roads span the China-Myanmar Border, facilitate the spread of P. vivax (Hu et al., 2016; Xu et al., 2016).Therefore, the area merits significance for global malaria control.
The recommended first-line medication for vivax malaria patients in Myanmar is a radical cure with chloroquine-primaquine (CQ-PQ) (Htun et al., 2017). However, most local cases of malaria derive from Plasmodium falciparum (or from mixed infections with P. falciparum and P. vivax). Although the prevalence of mixed-species malaria infections was previously estimated at <2% in some surveys of Asian locales, therapeutic studies identify coinfection in over 30% of cases (Mayxay et al., 2004). The incidence of P. vivax infection after falciparum malaria treatment exceeds that which would be expected from entomological inoculation rates in Southeast Asia (Douglas et al., 2011). A survey from 2008 to 2012 in Yunnan found 17% of malaria cases were co-infections with P. vivax and P. falciparum, underscoring the difficulty in diagnosing low-level and co-infection cases (Zhou et al., 2014).
Given such high rates of coinfection, treatments meant to target chloroquine-resistant P. falciparum likely impose substantial selective pressure for resistance to sulfadoxine-pyrimethamine (SP) in the local P. vivax population, and may induce high-grade antifolate resistance in P. vivax. SP has been the recommended treatment for falciparum and other non-vivax malaria since the 1980s, replacing chloroquine (CQ) as the front-line anti-malaria treatment when large-scale CQ resistance developed in many countries; but growing drug resistance soon necessitated replacement of SP with artemisinin-based combination therapy (ACT) (Jiang et al., 2020). In addition to typical treatment proscribed by WHO guidelines (WHO, 2013), SP is also used for intermittent preventive treatment in infants (IPTi) and pregnant women (IPTp) in areas endemic for malaria.
Meanwhile, China developed its own strategy of malaria control: seasonal mass drug administration with pyrimethamine and primaquine. This approach attempts to treat carriers of P. vivax before seasonal transmission begins (Hsiang et al., 2013). These several uses of such antimalarials near the China-Myanmar Border necessitate continuous monitoring of antifolate resistance in P. vivax in this area. Given growing concerns of drug resistance, it is essential to monitor antifolate resistance in P. vivax in this area, which has undergone changes in transmission intensity and drug therapy.
Antifolate resistance derives, in part, from point mutations of enzymes used by the parasite to synthesize folate. These include dihydropteroate synthase (DHPS) and dihydrofolate reductase (DHFR). Barriers to continuous culture of P. vivax severely limit efforts to link pvdhfr and pvdhps genotypes with parasite responses to SP in vitro. Instead, investigators assay drug sensitivity by expressing P. vivax DHPS and DHFR in E. colior yeast (Auliff et al., 2010). In P. falciparum, SP resistance has also been attributed to amplification of GTP-cyclohydrolase (gch1), the first gene in the folate biosynthesis pathway (Nair et al., 2008); this phenomenon has not yet been demonstrated in P. vivax. Drug resistance was last surveyed from the vicinity (Nabang Town, Yingjiang County, Yunnan Province, and Kachin, Myanmar were surveyed in 2015 and 2017) (Wang et al., 2020; Zhao et al., 2020); sampling from Myitsone has not yet been reported.
Despite their likely contribution to emerging regional drug resistance, the presence and prevalence of mutations in genes of the folate biosynthesis pathways in P. vivax remain uncharacterized. This study aims to investigate the extent and spread of SP drug resistance alleles in P. vivax from local people in Laiza, Myanmar and from migrant Chinese laborers returning from Myitsone, Myanmar between 2012 and 2015.To do so, we sought signatures of selection by sequencing SP resistance-associated molecular markers and genotyped microsatellite markers around target genes.
Material and Methods
Samples
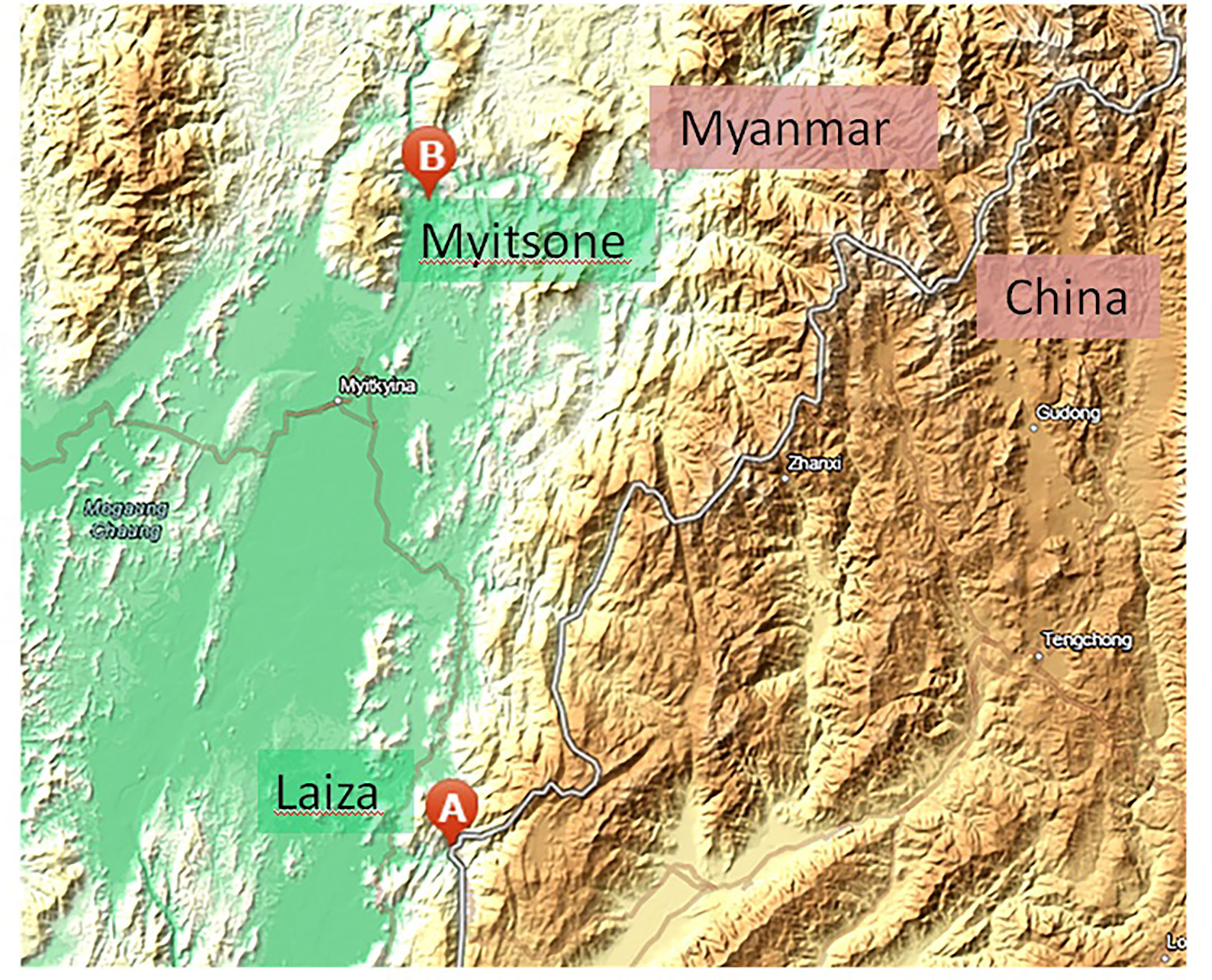
Between 2012 and 2015, finger-prick blood samples were collected from symptomatic patients with acute P. vivax infection. Infections were diagnosed by microscopy using Giemsa-stained thick and thin blood films at 13 local clinics around the Laiza township (N 24° 45’ 24”, E 97° 33’ 02”), Myanmar, and in returning migrant Chinese laborers from the Myitsone area (N 25° 41’ 23”, E 97° 31’ 04”), Myanmar (Figure 1). Patients who provided blood were well-informed by our study team and signed a consent form before sampling. The Institutional Review Board of Kunming Medical University, China, and the local Bureau of Health, Kachin State, Myanmar approved the sampling procedures.
Parasite DNA was extracted from each sample using the High Pure PCR Template Preparation Kit (Roche, Germany). Multiclonal infections were identified by genotyping the single-copy antigenic genes msp-3α and msp-3β as described previously (Khan et al., 2014); these were excluded from the ensuing analysis, leaving a total of 308 single-clone P. vivax isolates (101 from Myitsone, 207 from Laiza). A sample of fewer than 100 would suffice to achieve 80% power (with a type I error <.05) given reported mutation frequencies of in pvdhfr and pvdhps.
Pvdhfr and pvdhps Gene Sequencing, Pvgch1 Copy Number Quantification, and Microsatellite Genotyping
Parasite genes pvdhfr and pvdhps were amplified by nested PCR using Premix Taq Version 2.0 plus dye (TaKaRa Bio, Japan), according to the manufacturer’s instructions. Primers and PCR conditions were as described in Supplementary Table S1. PCR products were purified and sequenced by Sanger at SinoGenoMax (Kunming, China). For accuracy, sequences from the forward and reverse were examined by alignment using DNASTAR software (v.7.1). To avoid technical contamination, all PCR and sequencing assays include positive control (MRA-41 Sal I, gDNA, MR4/Bei resources) and negative controls (uninfected samples and water). Mutations were scored only when confirmed by each bidirectional sequencing read with high quality BioEdit (v.7.2.5) (Sarmah et al., 2017) was used to compare individual sequences with the reference sequences pvdhfr (PVX_089950) and pvdhps (PVX_123230). Nucleotide sequences were translated into amino acid sequences to examine mutant codons.
To determine copy number variation in pvgch1, PCR primers were designed (Supplementary Table S1) and quantitative real-time (qRT-PCR) performed using SuperRealPreMix Plus (SYBR Green) (TIANGEN, China) on the Stratagene Mx3000P PCR machine (Stratagene, CA, USA), according to the manufacturer’s instructions. The threshold cycle value (Ct value) of pvgch1 was estimated using pvaldolase the reference gene. In particular, we assessed, via real-time PCR, the proportion of mutant alleles of pvgch1 compared with the reference allele. This was calculated using Pfaffl method for each patient sample as amplification efficiency of the target and reference was not similar. Results were recorded as a ratio calculated from the mean Ct values for each gene in each sample. The pUC57 plasmid inserted with a single copy of pvgch1 or pvaldolase was purchased from Sangon Biotech (Shanghai, China) and used for amplification efficiency calibration.
Microsatellites flanking the pvdhfr gene were genotyped as previously reported (Ding et al., 2013). In brief, we analysed six microsatellite loci, with more than eight repeats, located proximate to the dhfr gene (-93 kb, -38 kb, -2.6 kb, +4.9 kb, +37 kb, and +94 kb) on chromosome 5 (GI: 157098056). Products were amplified by PCR using Premix TaqVersion 2.0 plus dye (TaKaRa Bio, Japan). Products labelled with Hex fluorophore were separated and detected on ABI 3730 capillary sequencer (Thermo Fisher Scientific, US) by Sangon Biotech (Shanghai, China). DNA fragment sizes were determined and visualized using Gene Marker software (v.1.5.).
Data Analysis
Population genetic parameters, pairwise linkage disequilibrium (LD), and tests of neutrality were estimated using DNASP software (v.5.0.) (Mardani et al., 2012). The copy number of pvgch1 in each sample was estimated by the Pfaffl method using the Ct value, rounded to the nearest integer (Raju et al., 2019).
The Chi-square test and Fisher’s exact test were employed to compare the independence of categorical variables, using GraphPad Prism 9.0.0 software. A statistical significance was set at the P-value of < 0.05. The length polymorphism of microsatellite loci and allele frequencies were calculated using GenAlEx software (v.6.5.) (He et al., 2018). Expected heterozygosity (He) was estimated for each locus based on allele frequencies following previously reported methods (Anderson et al., 2000). The sampling variance for Hewas estimated using ARLEQUIN software (v.3.5.), allowing estimation of genetic diversity indices and assessment of population differentiation between haplotypes. The Bottleneck program (Cornuet and Luikart, 1996) was used to investigate the influence of selection on allele distributions under the infinite alleles (IMM) and step wise mutation models (SMM). Wilcoxon tests were used to determine the significance of departures from He.
Results
Genetic Diversity of pvdhfr and pvdhps and Copy Number of pvgch1
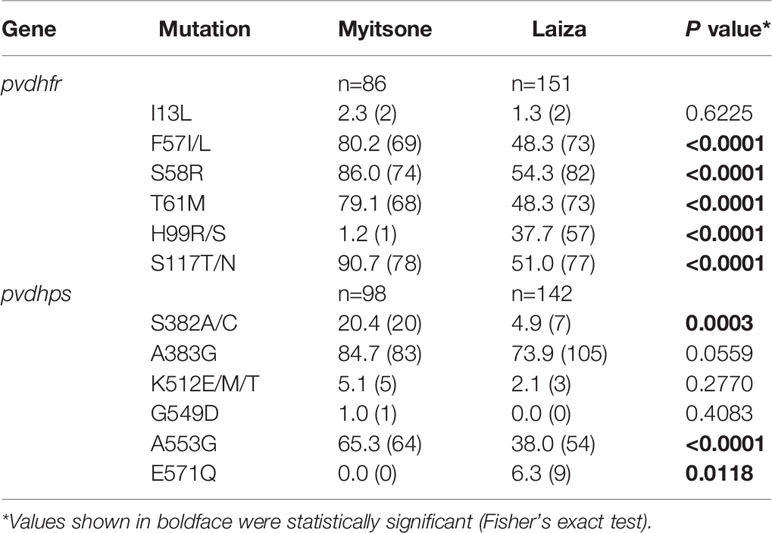
A total 237 (76.9%) samples for pvdhfr (86 of Laiza and 151 of Myitsone) and 240 (77.9%) samples for pvdhps (98 of Laiza and142 of Myitsone) were successfully sequenced and analysed (Table 1). The amplification products of pvdhfr and pvdhps were 632 bp (PVX_089950, nucleotide positions at -38 to 5941-767, amino acids positions at 1-198)) and 767 bp (PVX_123230, nucleotide positions at 1380-2146, amino acid positions at 349-604), respectively. No length polymorphism was detected in either gene. More variant sites haplotypes were observed in pvdhps (14, 16) than pvdhfr (10,14).
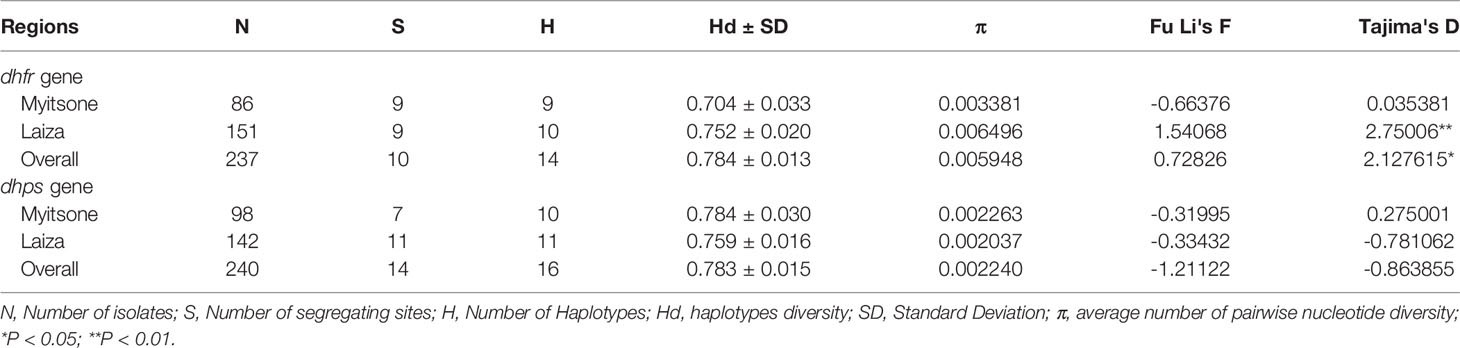
Table 1 Genetic diversity and tests of neutrality of the pvdhps and pvdhfr genes in P. vivax samples from the China/Myanmar border.
Nucleotide diversity in pvdhfr was almost twice as great in Laiza than in Myitsone (0.006496 v.s.0.003381); diversity was lower, and more regionally uniform, in pvdhps (0.002263 v.s.0.002037). For the Laiza population, neutrality tests showed significant positive values for Tajima’s D in the pvdhfr gene, indicating a dearth of singletons and consistent with balancing selection. However, for the pvdhps gene, we observed negative values for Fu & Li’s F and Tajima’s D, which indicates an excess of low-frequency alleles more consistent with population expansions or positive selection.
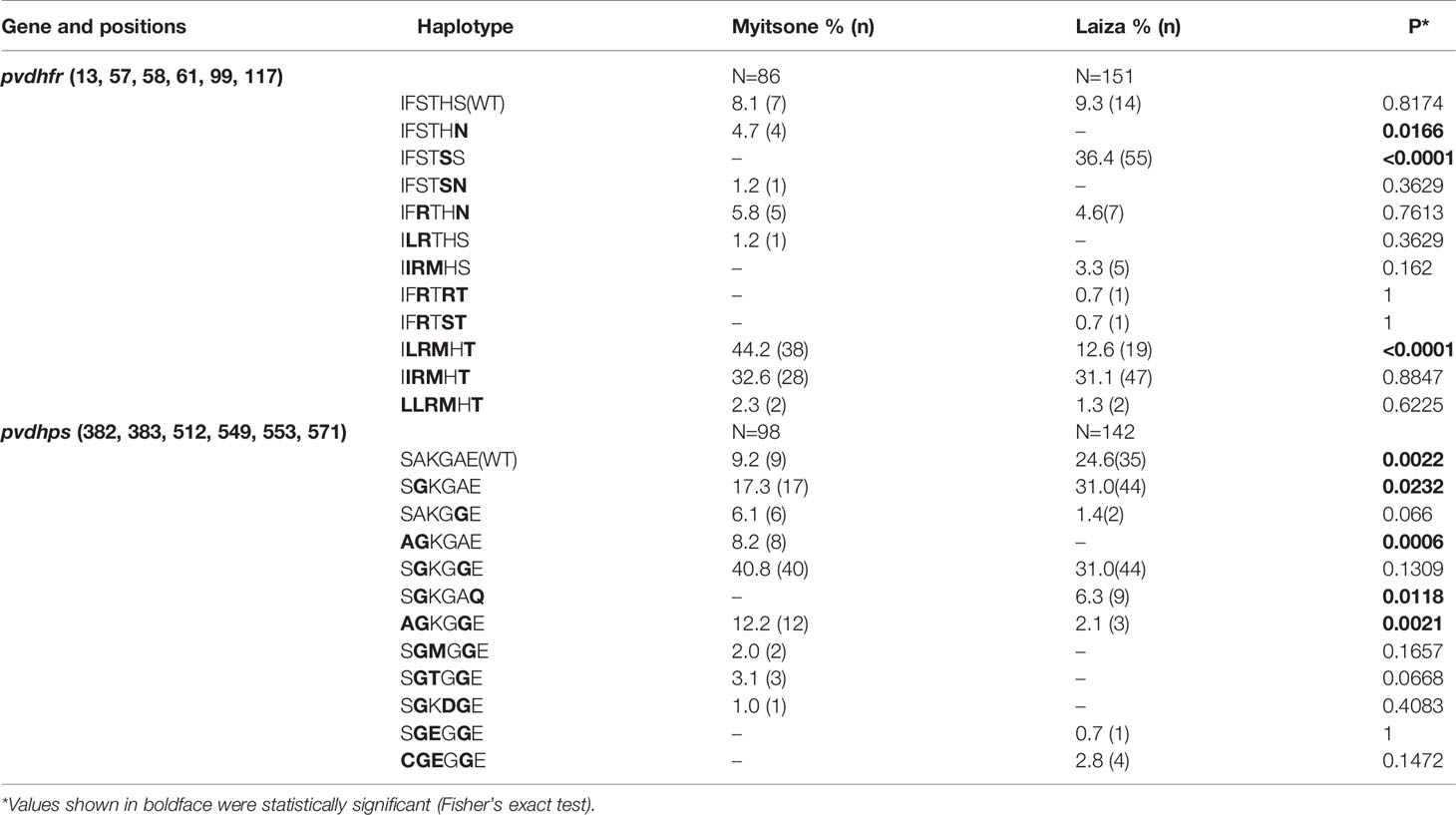
In the pvdhfr gene, Myitsone samples and Laiza samples contained mutations at six codons (13, 57, 58, 61, 99, 117)(Table 2). Compared to the pvdhfr reference sequence in Salvador I (PVX_089950), more than ninety percent of samples from each sampling locale had at least one codon change. Mutations in codons F57I/L, S58R, T61M, and S117T/N were highly prevalent in both populations. In the Myitsone samples, the F57I/L, S58R, T61M and S117T/N mutation in pvdhfr was significantly more frequent than in the Laiza samples; the H99R/S mutation in the Laiza samples was significantly more frequent than in the Myitsone samples. The quadruple mutant L57R58M61T117was observed significantly more frequent in Myitsone (44.2%) than in Laiza (12.6%) (Table 3). The quadruple mutant I57R58M61T117was observed in both areas (32.6% in Myitsone and 31.1% in Laiza). Genotypes R99 and S99 were only in Laiza (37.8%) (Table 2). Rare genotypes with triple mutations I57R58M61 (5 cases, 3.3%), R58R99T117 (1 case, 0.7%), and R58S99T117 (1 case, 0.7%) were also found in Laiza. The double mutant allele R58N117 existed in both areas, with prevalence of 5.8% and 4.6% in Myitsone and Laiza, respectively (Table 3).
For the pvdhps gene, nucleotide polymorphisms were observed at five different codons (382, 383, 512, 549, and 553) in samples from Myitsone and five codons (382, 383, 512, 553, and 571) from Laiza. As compared to the reference dhps sequence in Salvador I (PVX_123230), more than ninety percent and seventy percent of the samples had at least one codon change in Myitsone and Laiza samples, respectively. The most prevalent mutations were at codon A383G and A553G (Table 2). Similarly, these two codons also were more prevalent in the Myitsone population (84.7 and 65.3%, respectively) than in the Laiza population (74 and 38.1%, respectively). Mutation S382A also reached a high frequency (20.4%) in the Myitsone population. In the Myitsone samples the S382A/C and A553G mutation in pvdhps was significantly more frequent than in the Laiza samples; the E571Q mutation in the Laiza samples was significantly more frequent than in the Myitsone samples.
The parasites in Mytsome and Laiza each possessed certain unique pvdhps haplotypes (Table 3). For instance, parasites carrying the double mutations G383Q571 were identified only in Laiza, where they reached a sample prevalence of 6.3%. The double mutation A382G383 in pvdhps were 8.2% in Myitsone but not found in Laiza. Also, genotypes with triple mutations G383M512G553 (in 2 cases, 2%), G383T512G553 (in 3 cases, 3.1%), and G383D549G553 (in 1 case, 1%) were only found in Myitsone. However, the genotype with G383E512G553 were only found in 1 case (0.7%) in Laiza. Multiple mutations 382C/383G/512E/553G were found in 4 cases (3%) from Laiza. The distribution of major haplotypes of the pvdhfr and pvdhps genes in P. vivax from the two study areas are listed in Table 3.
Pairwise LD analysis revealed significant associations between several different nucleotide sites (as shown in Supplementary Tables S2, S3). The proportion of locus pairs showing significant LD was twice as high in Laiza (53.6%, 15/18 for pvdhfr; 50.9%, 28/55 for pvdhfr) than in Myitsone (25.0%, 7/28 for pvdhfr; 26.7%, 4/15 for pvdhfr). Only 7 of 101 samples in Myitsone had two copies of gch1; only 2 out of 101 had two gch1 copies in Laiza (Supplement Figure S1). There was no significant difference in the two regions (P >0.05, Fisher’s exact test).
Genetic Hitchhiking of Microsatellites Flanking the pvdhfr Gene
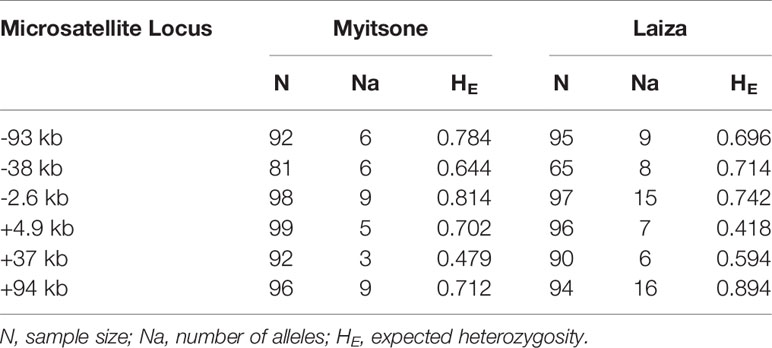
Polymorphisms of six microsatellite loci flanking pvdhfr (located at -93 kb, -38 kb, -2.6 kb, +4.9 kb, +37 kb, and +94 kb from the pvdhfr gene on chromosome 5) were examined in 100 samples from Laiza and 101 from Myitsone. Genotypes were obtained from 81 to 99 of the Myitone samples at these six loci, and from 65 to 97 of the Laiza samples (Table 4).
The mean number of observed alleles was higher in Laiza (10.2 ± 4.2) than in Myitsone (6.3 ± 2.3). A significant reduction in variability (compared to wild-type isolates) occurred across at the +4.9 kb region in Laiza samples and over a 7.5 kb region (spanning -2.6 to +4.9 kb) in Myitsone samples (P<0.01, Tukey’s multiple comparisons test) (Figures 2 and 3).
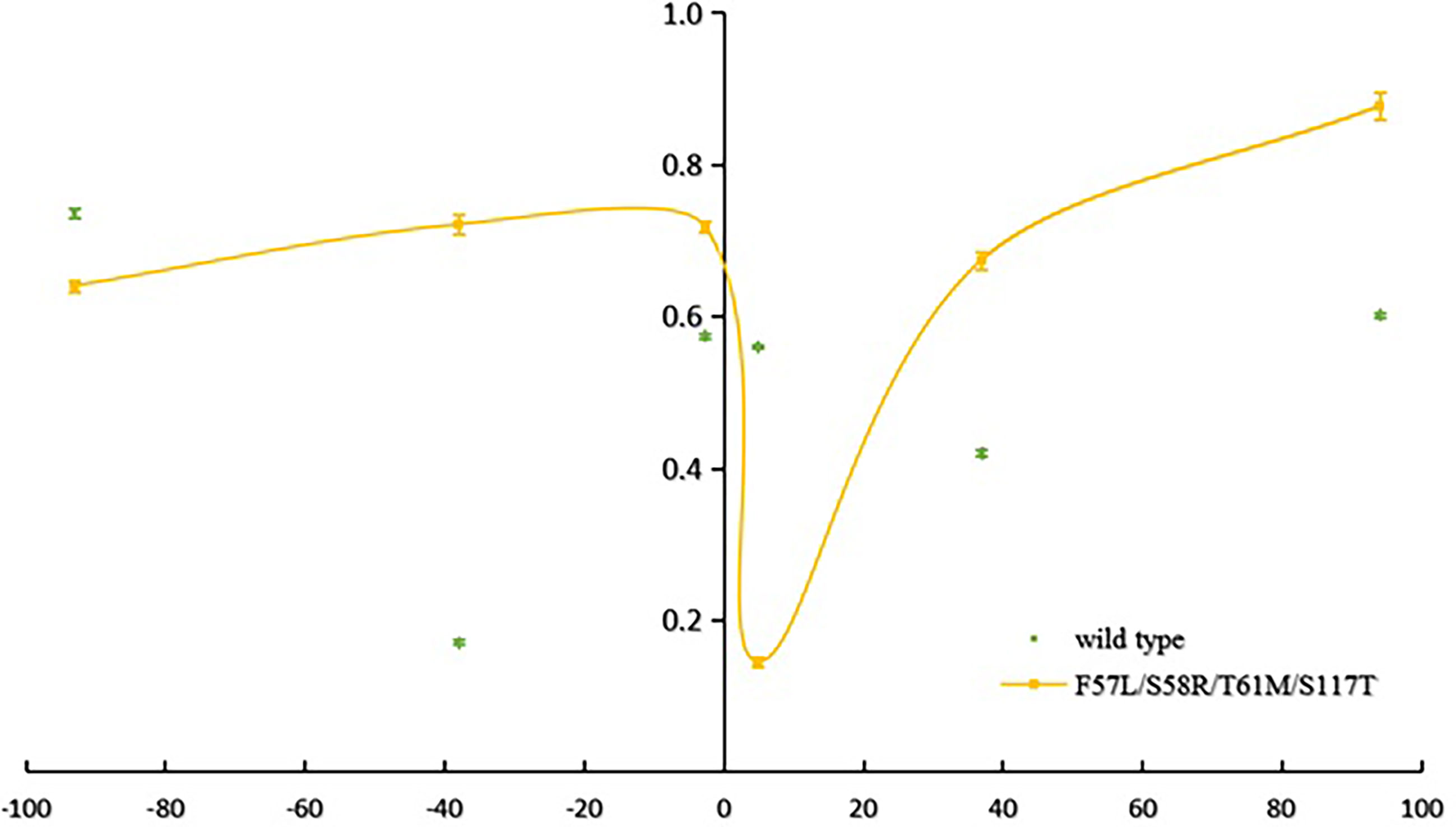
Figure 2 Reduced haplotype diversity proximate to mutant pvdhfr alleles in parasites sampled from Lazia (He, compared to wild type).
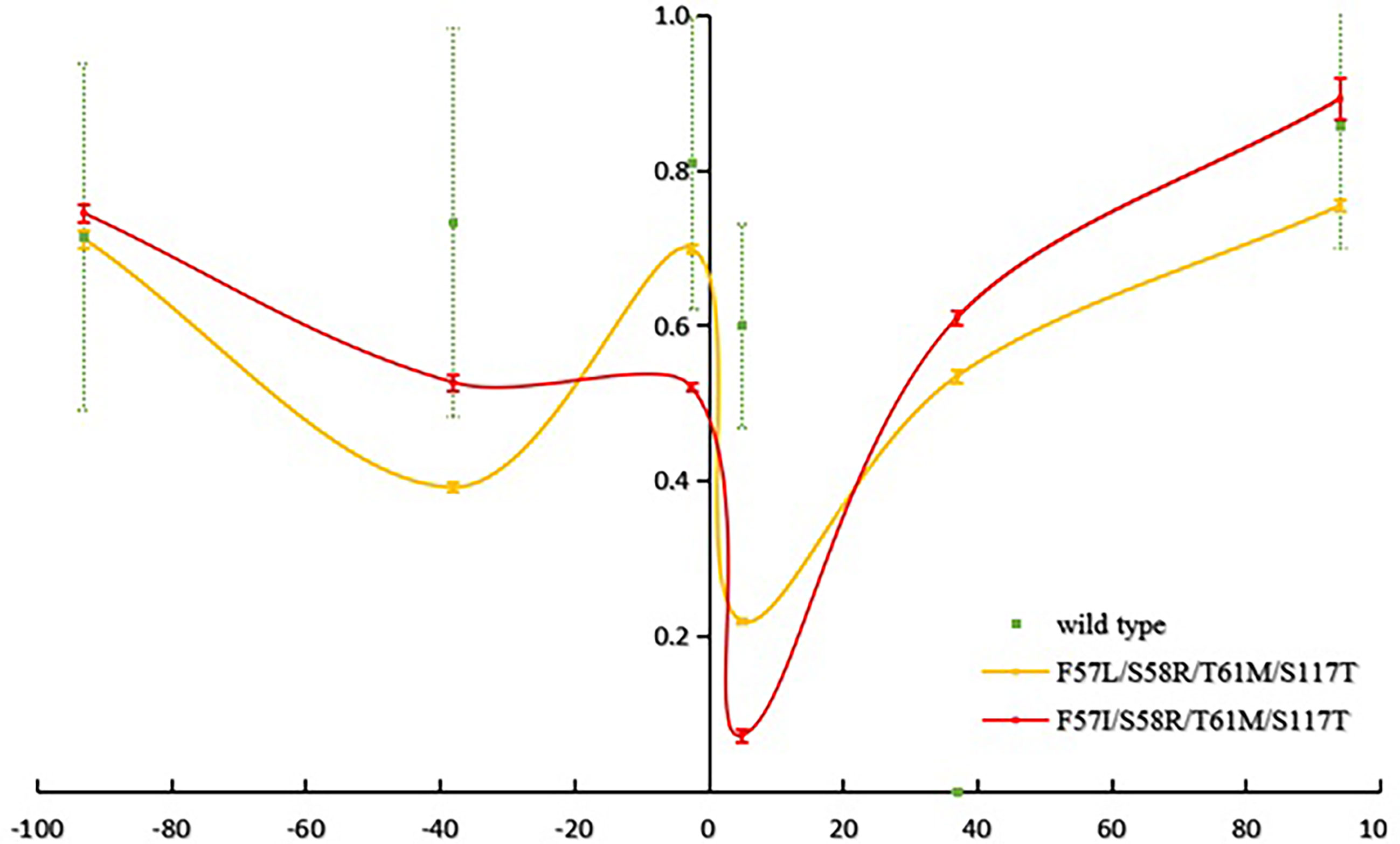
Figure 3 Reduced haplotype diversity proximate to mutant pvdhfr alleles in parasites sample from Myitsone (He, compared to wild type).
Tests using Bottleneck software, assuming the stepwise mutation model (SMM), indicated that parasites with the haplotype 57I/L-58R-61M-117T in the Myitsone population had significant heterozygote deficiency (P<0.05), suggestive of positive selection (Supplementary Table S4). However, significant heterozygosity excess (P<0.05) was found in parasites with genotype S58R-S117N in the Myitsone population, indicating that they could be suffering a bottleneck. There was no bottleneck effect detected in parasites with wild-type genotypes in either population when either an infinite allele (IMM) or stepwise mutation model (SMM) was assumed, suggesting parasite population under mutation-drift equilibrium. Significant genetic differentiation was observed between the wild type and the quadruple mutant genotypes 57I/L/58R/61M/117T (FST=0.25492 for 57I/L/58R/61M/117T, P<0.00, FST=0.30369 for 57I/L/58R/61M/117T, P<0.001) (as shown in Table 5).

Table 5 FST values for allele comparisons between different genotypes, determined using microsatellites around dhfr of P. vivax population in two regions of Myanmar.
Discussion
An increasing number of drug resistance markers in P. vivax have spread throughout Southeast Asia (Lu et al., 2010; Thongdee et al., 2013; Asih et al., 2015). These include polymorphisms at codon 373, 380, 382, 383, 384, 512, 553, 585, and 601 in the pvdhps gene, which are associated with resistance to sulfadoxine, and mutations at codon 15, 33, 50, 57, 58, 61, 64, 117, and 173 in the pvdhfr gene, contributing to treatment failure using pyrimethamine. We found polymorphisms in five different codons of the pvdhfr gene (13, 57, 58, 61, 117) in each locale; mutations in a sixth codon (99) were restricted to Laiza. These locales each had polymorphisms in codons 382, 383, 512, 553 of the pvdhps gene; observed polymorphisms in codon 549 were restricted to Myitsone, and in codon 571 were restricted to Laiza. These results were consistent with those reported previously in Southeast Asia (Lu et al., 2010; Thongdee et al., 2013; Asih et al., 2015).
Though most of the pvdhfr and pvdhps haplotypes identified in this study have been reported in previous studies, some new haplotypes were also detected. For example, 2 cases each from Laiza and Myitsone bore quintuple-mutant 13L/57I/58R/61M/117T pvdhfr allele. To the best of our knowledge, this is the first report of this haplotype in the China-Myanmar border region. The S117N mutation has been suggested to represent the first mutational step conferring drug-resistance (Brega et al., 2004). However, as shown in the Table 6, only 5 cases in Myitsone bore the single S117N mutation, and the quadruple pvdhfr 57L/58R/61M/117T allele was the most prevalent; this differs from Yangon, Myanmar, where the double 58R/117N allele was most common (Lu et al., 2010).
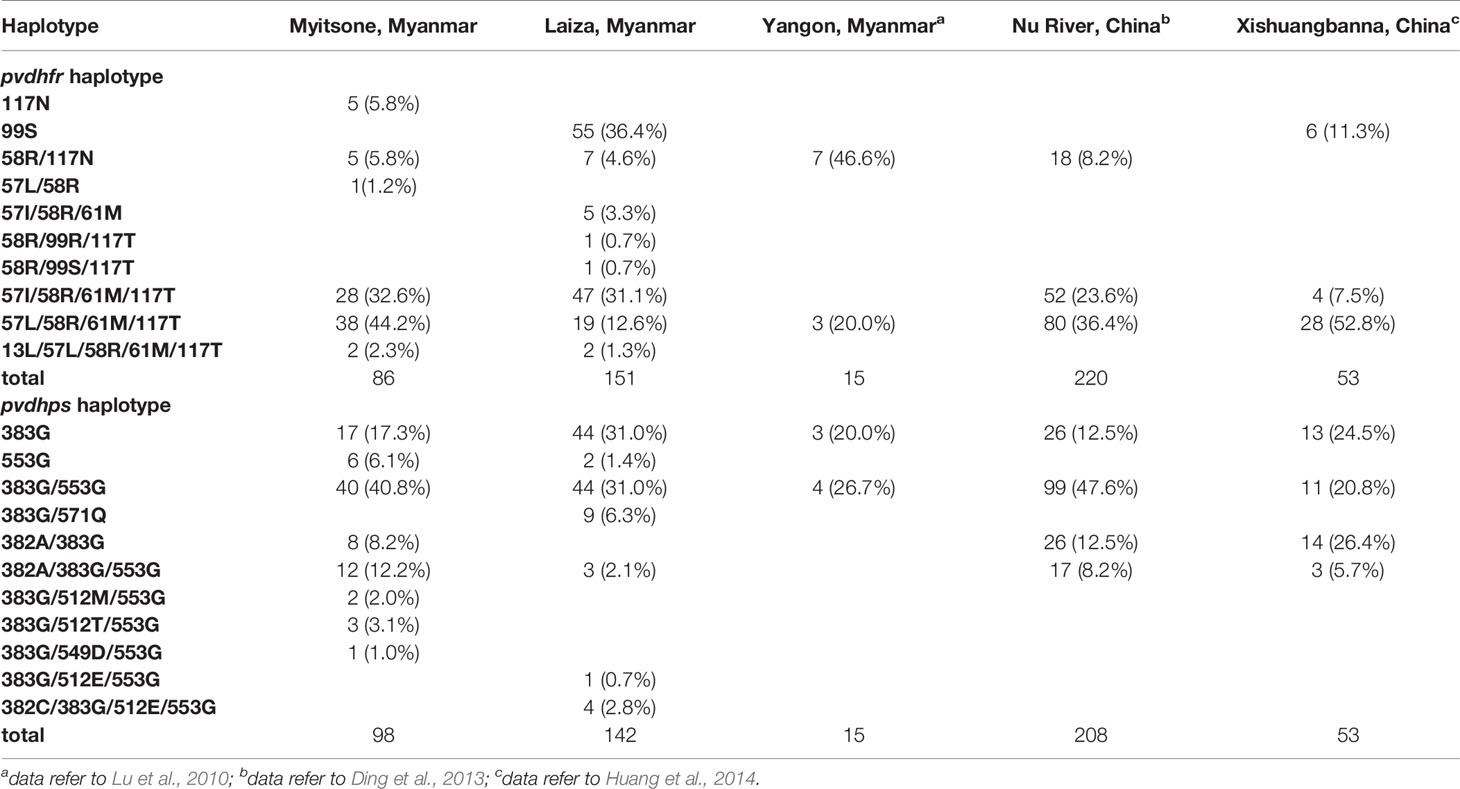
Table 6 Distribution of major haplotypes of the pvdhfr and pvdhps genes in P. vivax from different regions in Myanmar and Yunnan, China.
Studies in China and Myanmar have found the prevalence of pvdhfr and pvdhps drug-resistant mutations in P. vivax in regions of Yunnan province, China, including the Xishuangbanna prefecture and areas along the Nu River, and these studies identified parasites highly resistant to SP in subtropical China and Yangon, Myanmar from 2010 to 2014 (Lu et al., 2010; Ding et al., 2013; Huang et al., 2014). The quadruple-mutant 57L/58R/61M/117T allele was the most prevalent pvdhfr allele in two study sites in the Yunnan province. However, there were no mutations at positions 13 or 173 in the pvdhfr gene reported in parasites from the Xishuangbanna. An altered 173 allele was, however, found in 3.18% of samples analyzed from areas along the Nu River. In addition to this, Yunnan province also reported haplotypes with quintuple allelic variants. Our study profiled mutations in marker genes associated with SP resistance among the P. vivax from the China-Myanmar border. The quadruple-mutant 57I/58R/61M/117T allele was also prevalent in Myitsone, Myanmar, and in Yunnan, China (Ding et al., 2013; Huang et al., 2014) but absent from our survey of Laiza and from a previous study in Yangon, Myanmar (Lu et al., 2010).
Beneficial mutations reduce variation in physically proximate regions of the chromosome through “genetic hitchhiking” (McCollum et al., 2008; Artimovich et al., 2015). The polymorphism in six microsatellite loci flanking mutant pvdhfr (S58R-S117N and the quadruple mutant genotypes 57I/L/58R/61M/117T) was significantly less than that would be expected in wild type genetic backgrounds. The valley of reduced variation spanning pvdhfr is resembles that found for samples from Yunnan and central China (Ding et al., 2013). In each, an approximately 100 kb region suggests local genomic variability has been suppressed by a recent selective sweep. These results strengthen the evidence for a relationship between the specific mutant alleles and selection for drug drug-resistant phenotypes.
Furthermore, favorable alleles experience increased linkage disequilibrium (LD), at least until sufficient time has elapsed to enable recombination to distribute alleles randomly among haplotypes in the haploid blood stages of Plasmodium parasites. Therefore, the multiple mutations with S117T can serve as a marker for drug resistance at the China-Myanmar Border. Interestingly, 36.4% of Laiza samples bore the single H99S mutation, which has been reported from other studies (Lu et al., 2012; Huang et al., 2014; Wang et al., 2020).
Our results also elucidate the drug pressure exerted by the antifolatesulfadoxine. Previous study has focused on the A383G and A553G in pvdhps gene, which are directly related to sulfadoxine resistance (Korsinczky et al., 2004). All of the mutant cases from our study carried at least one of these two mutations. The most prevalent pvdhps haplotype we observed was the double mutant 383G/553G allele. There are additional mutant alleles found in Yunnan, including one non-synonymous mutation at codon 512T/M and triple mutant alleles at codons 372, 495, 561. We observed the triple mutant 382A/383G/553G allele and the double mutant 382A/383G allele in Myitsone, Myanmar, and Yunnan, China, but not in Laiza. Nor were these previously observed in Yangon, Myanmar (Lu et al., 2010). We observed the double mutant 383G/571Q only in nine Laiza cases. Besides the common triple mutant 382A/383G/553G allele described earlier, there are other types of triple mutant pvdhps alleles in Laiza and Myitsone. They included one case from Laiza with a 383G/512E/553G allele, one from Myitsone with a 383G/549D/553G allele, two from Myitsone with a 383G/512M/553G allele, and three from Myitsone with a 383G/512T/553G allele. Moreover, we observed the quadruple-mutant 382A/383G/512E/553G allele in four Myitsone cases.
SP combination therapy targets two enzymes in the folate-synthesis pathway, dihydropteroate synthase (DHPS) and dihydrofolate reductase (DHFR). The first enzyme in this pathway, GTP-cyclohydrolase 1 (GCH-1), might also respond to drug pressure. Previous work on P. falciparum (Kümpornsin et al., 2014) suggested a compensating effect, by copy-number variation (CNV) of gch-1, in parasites bearing resistance mutations in dhps and dhfr. However, such a relationship has not been established in P. vivax.Our study, the first to explore such a relationship in P. vivax, identified little evidence of copy number variation in pvgch1, suggesting that SP-resistant P. vivax may harbor alternative mechanisms to secure sufficient folate. Although microsatellite analysis showed that drug resistance genes experience selective pressure, pvgch1 copy number variation was only observed only in a small number of cases, suggesting that P. vivax population might not undergo pvgch1 compensation.
The first-line medication for vivax malaria patients in Myanmar is CQ-PQ. Currently, the molecular markers of primaquine resistance are still unknown. And no molecular markers are confirmed for chloroquine resistance in P. vivax, while pvmdr1 and pvcrt-o have often been queried since these two genes are involved in chloroquine resistance in P. falciparum, but the possible role of Pvcrt-o and pvmdr1 in chloroquine resistance is controversial. At this area, the pvmdr1 and pvcrt-o genes were reported (Li et al., 2020).The limit of detection of the PCR and sequencing assays used in the study may rule out the possibility of low-frequency mutation isolates. If we want to get the results, deep-sequence needed to be involved in further study.
Conclusion
We found that most P. vivax sampled from symptomatic patients at the China-Myanmar boarder harbor mutations in pvdhfr and pvdhps genes associated with drug resistance. In this region of intense transmission, which is the source of many cases of vivax malaria introduced to China, pvdhfr, a single mutation (S117T) serves as a marker for the presence of multiple mutations. All of the mutant cases from our study carried mutation A383G or A553G in this gene. Parasites throughout the studied region show genetic evidence of undergoing strong drug pressure. The lack of copy number variation of pvgch1 suggests that SP-resistant P. vivax may harbor alternative mechanisms to secure sufficient folate.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank (pvdhps gene accession numbers: MZ234760-MZ234999 and pvdhfr gene accession numbers: MZ235000-MZ235236).
Ethics Statement
The studies involving human participants were reviewed and approved by Kunming Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SF, SW, and WZ performed the statistical analysis and wrote the first draft of the manuscript. DZ, YH, YB, YR, YS, HZ, QY, XL, XC, YZ, CL, ZX, YW, FC, PS, and BR wrote sections of the manuscript. ZY contributed to conception and design of the study. ZY organized the database. All authors contributed to the article and approved the submitted version.
Funding
This study was under the support of the National Science Foundation of China (31860604 and U1802286), Major science and technology projects of Yunnan Province (2018ZF0081) and International Science and Technology Cooperation-Yunnan International Science and Technology Cooperation Base (202003AE140004. Furthermore, XC and YZ were under sponsoring from the Yunnan Applied Basic Research Projects-Union Foundation (2018FE001-190, 2015FB034, respectively). Also, WZ was under the support of the Education Department Fund of Yunnan Province (2019J1184). BR was supported by USDA Project Detection and Control of Foodborne Parasites for Food Safety (8042-32420-007-00D).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank all the patients for volunteering to participate in the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.683423/full#supplementary-material
References
Anderson, T. J., Haubold, B., Williams, J. T., Estrada-Franco, J. G., Richardson, L., Mollinedo, R., et al (2000). Microsatellite Markers Reveal a Spectrum of Population Structures in the Malaria Parasite Plasmodium Falciparum. Mol. Biol. Evol. 17, 1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247
Artimovich, E., Kapito-Tembo, A., Pensulo, P., Nyirenda, O., Brown, S., Joshi, S., et al (2015). The Effect of Local Variation in Malaria Transmission on the Prevalence of Sulfadoxine-Pyrimethamine Resistant Haplotypes and Selective Sweep Characteristics in Malawi. Malar. J. 14, 387. doi: 10.1186/s12936-015-0860-7
Asih, P. B., Marantina, S. S., Nababan, R., Lobo, N. F., Rozi, I. E., Sumarto, W., et al (2015). Distribution of Plasmodium Vivax Pvdhfr and Pvdhps Alleles and Their Association With Sulfadoxine-Pyrimethamine Treatment Outcomes in Indonesia. Malar. J. 14, 365. doi: 10.1186/s12936-015-0903-0
Auliff, A. M., Adams, J. H., O’neil, M. T., Cheng, Q. (2010). Defining the Role of Mutations in Plasmodium Vivax Dihydrofolate Reductase-Thymidylate Synthase Gene Using an Episomal Plasmodium Falciparum Transfection System. Antimicrob. Agents Chemother. 54, 3927–3932. doi: 10.1128/AAC.00628-10
Battle, K. E., Lucas, T. C. D., Nguyen, M., Howes, R. E., Nandi, A. K., Twohig, K. A., et al (2019). Mapping the Global Endemicity and Clinical Burden of Plasmodium Vivax 2000-17: A Spatial and Temporal Modelling Study. Lancet 394, 332–343. doi: 10.1016/S0140-6736(19)31096-7
Brega, S., De Monbrison, F., Severini, C., Udomsangpetch, R., Sutanto, I., Ruckert, P., et al (2004). Real-Time PCR for Dihydrofolate Reductase Gene Single-Nucleotide Polymorphisms in Plasmodium Vivax Isolates. Antimicrob. Agents Chemother. 48, 2581–2587. doi: 10.1128/AAC.48.7.2581-2587.2004
Cornuet, J. M., Luikart, G. (1996). Description and Power Analysis of Two Tests for Detecting Recent Population Bottlenecks From Allele Frequency Data. Genetics 144, 2001–2014.
Ding, S., Ye, R., Zhang, D., Sun, X., Zhou, H., Mccutchan, T. F., et al (2013). Anti-Folate Combination Therapies and Their Effect on the Development of Drug Resistance in Plasmodium Vivax. Sci. Rep. 3, 1008. doi: 10.1038/srep01008
Douglas, N. M., Nosten, F., Ashley, E. A., Phaiphun, L., Van Vugt, M., Singhasivanon, P., et al (2011). Plasmodium Vivax Recurrence Following Falciparum and Mixed Species Malaria: Risk Factors and Effect of Antimalarial Kinetics. Clin. Infect. Dis. 52, 612–620. doi: 10.1093/cid/ciq249
Gupta, H., Afsal, M. P., Shetty, S. M., Satyamoorthy, K., Umakanth, S. (2015). Plasmodium Vivax Infection Causes Acute Respiratory Distress Syndrome: A Case Report. J. Infect. Dev. Ctries. 9, 910–913. doi: 10.3855/jidc.6813
Gupta, H., Dhunputh, P., Bhatt, A. N., Satyamoorthy, K., Umakanth, S. (2016). Cerebral Malaria in a Man With Plasmodium Vivax Mono-Infection: A Case Report. Trop. Doct. 46, 241–245. doi: 10.1177/0049475515624857
He, Y., Jun, F., Jian-Hai, Y., Tian-Mu, C., Shui-Sen, Z. (2018). [Polymorphism Analysis of Microsatellite Marker Loci of Plasmodium Falciparum From Different Geographical Origins]. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi 30, 504–507. doi: 10.16250/j.32.1374.2017238
Hsiang, M. S., Hwang, J., Tao, A. R., Liu, Y., Bennett, A., Shanks, G. D., et al (2013). Mass Drug Administration for the Control and Elimination of Plasmodium Vivax Malaria: An Ecological Study From Jiangsu Province, China. Malar. J. 12, 383. doi: 10.1186/1475-2875-12-383
Htun, M. W., Mon, N. C. N., Aye, K. M., Hlaing, C. M., Kyaw, M. P., Handayuni, I., et al (2017). Chloroquine Efficacy for Plasmodium Vivax in Myanmar in Populations With High Genetic Diversity and Moderate Parasite Gene Flow. Malar. J. 16, 281. doi: 10.1186/s12936-017-1912-y
Huang, B., Huang, S., Su, X. Z., Tong, X., Yan, J., Li, H., et al (2014). Molecular Surveillance of Pvdhfr, Pvdhps, and Pvmdr-1 Mutations in Plasmodium Vivax Isolates From Yunnan and Anhui Provinces of China. Malar. J. 13, 346. doi: 10.1186/1475-2875-13-346
Hu, Y., Zhou, G., Ruan, Y., Lee, M. C., Xu, X., Deng, S., et al (2016). Seasonal Dynamics and Microgeographical Spatial Heterogeneity of Malaria Along the China-Myanmar Border. Acta Trop. 157, 12–19. doi: 10.1016/j.actatropica.2016.01.022
Jiang, T., Cheng, W., Yao, Y., Tan, H., Wu, K., Li, J. (2020). Molecular Surveillance of Anti-Malarial Resistance Pfdhfr and Pfdhps Polymorphisms in African and Southeast Asia Plasmodium Falciparum Imported Parasites to Wuhan, China. Malar. J. 19, 434. doi: 10.1186/s12936-020-03509-w
Khan, S. N., Khan, A., Khan, S., Ayaz, S., Attaullah, S., Khan, J., et al (2014). PCR/RFLP-Based Analysis of Genetically Distinct Plasmodium Vivax Population of Pvmsp-3α and Pvmsp-3β Genes in Pakistan. Malar. J. 13, 355. doi: 10.1186/1475-2875-13-355
Korsinczky, M., Fischer, K., Chen, N., Baker, J., Rieckmann, K., Cheng, Q. (2004). Sulfadoxine Resistance in Plasmodium Vivax is Associated With a Specific Amino Acid in Dihydropteroate Synthase at the Putative Sulfadoxine-Binding Site. Antimicrob. Agents Chemother. 48, 2214–2222. doi: 10.1128/AAC.48.6.2214-2222.2004
Kümpornsin, K., Kotanan, N., Chobson, P., Kochakarn, T., Jirawatcharadech, P., Jaru-Ampornpan, P., et al (2014). Biochemical and Functional Characterization of Plasmodium Falciparum GTP Cyclohydrolase I. Malar. J. 13, 150. doi: 10.1186/1475-2875-13-150
Lai, S., Sun, J., Ruktanonchai, N. W., Zhou, S., Yu, J., Routledge, I., et al (2019). Changing Epidemiology and Challenges of Malaria in China Towards Elimination. Malar. J. 18, 107. doi: 10.1186/s12936-019-2736-8
Li, J., Zhang, J., Li, Q., Hu, Y., Ruan, Y., Tao, Z., et al (2020). Ex Vivo Susceptibilities of Plasmodium Vivax Isolates From the China-Myanmar Border to Antimalarial Drugs and Association With Polymorphisms in Pvmdr1 and Pvcrt-o Genes. PLoS Negl. Trop. Dis. 14, e0008255. doi: 10.1371/journal.pntd.0008255
Lu, F., Lim, C. S., Nam, D. H., Kim, K., Lin, K., Kim, T. S., et al (2010). Mutations in the Antifolate-Resistance-Associated Genes Dihydrofolate Reductase and Dihydropteroate Synthase in Plasmodium Vivax Isolates From Malaria-Endemic Countries. Am. J. Trop. Med. Hyg. 83, 474–479. doi: 10.4269/ajtmh.2010.10-0004
Lu, F., Wang, B., Cao, J., Sattabongkot, J., Zhou, H., Zhu, G., et al (2012). Prevalence of Drug Resistance-Associated Gene Mutations in Plasmodium Vivax in Central China. Korean J. Parasitol. 50, 379–384. doi: 10.3347/kjp.2012.50.4.379
Mardani, A., Keshavarz, H., Heidari, A., Hajjaran, H., Raeisi, A., Khorramizadeh, M. R. (2012). Genetic Diversity and Natural Selection at the Domain I of Apical Membrane Antigen-1 (AMA-1) of Plasmodium Falciparum in Isolates From Iran. Exp. Parasitol. 130, 456–462. doi: 10.1016/j.exppara.2012.01.006
Mayxay, M., Pukrittayakamee, S., Newton, P. N., White, N. J. (2004). Mixed-Species Malaria Infections in Humans. Trends Parasitol. 20, 233–240. doi: 10.1016/j.pt.2004.03.006
McCollum, A. M., Basco, L. K., Tahar, R., Udhayakumar, V., Escalante, A. A. (2008). Hitchhiking and Selective Sweeps of Plasmodium Falciparum Sulfadoxine and Pyrimethamine Resistance Alleles in a Population From Central Africa. Antimicrob. Agents Chemother. 52, 4089–4097. doi: 10.1128/AAC.00623-08
Naing, C., Whittaker, M. A., Nyunt Wai, V., Mak, J. W. (2014). Is Plasmodium Vivax Malaria a Severe Malaria?: A Systematic Review and Meta-Analysis. PLoS Negl. Trop. Dis. 8, e3071. doi: 10.1371/journal.pntd.0003071
Nair, S., Miller, B., Barends, M., Jaidee, A., Patel, J., Mayxay, M., et al (2008). Adaptive Copy Number Evolution in Malaria Parasites. PLoS Genet. 4, e1000243. doi: 10.1371/journal.pgen.1000243
Rahimi, B. A., Thakkinstian, A., White, N. J., Sirivichayakul, C., Dondorp, A. M., Chokejindachai, W. (2014). Severe Vivax Malaria: A Systematic Review and Meta-Analysis of Clinical Studies Since 1900. Malar. J. 13, 481. doi: 10.1186/1475-2875-13-481
Raju, L. S., Kamath, S., Shetty, M. C., Satpathi, S., Mohanty, A. K., Ghosh, S. K., et al (2019). Genome Mining-Based Identification of Identical Multirepeat Sequences in Plasmodium Falciparum Genome for Highly Sensitive Real-Time Quantitative PCR Assay and Its Application in Malaria Diagnosis. J. Mol. Diagn. 21, 824–838. doi: 10.1016/j.jmoldx.2019.04.004
Sarmah, N. P., Sarma, K., Bhattacharyya, D. R., Sultan, A. A., Bansal, D., Singh, N., et al (2017). Antifolate Drug Resistance: Novel Mutations and Haplotype Distribution in Dhps and Dhfr From Northeast India. J. Biosci. 42, 531–535. doi: 10.1007/s12038-017-9706-5
Thongdee, P., Kuesap, J., Rungsihirunrat, K., Tippawangkosol, P., Mungthin, M., Na-Bangchang, K. (2013). Distribution of Dihydrofolate Reductase (Dhfr) and Dihydropteroate Synthase (Dhps) Mutant Alleles in Plasmodium Vivax Isolates From Thailand. Acta Trop. 128, 137–143. doi: 10.1016/j.actatropica.2013.07.005
Wang, D., Li, S., Cheng, Z., Xiao, N., Cotter, C., Hwang, J., et al (2015). Transmission Risk From Imported Plasmodium Vivax Malaria in the China-Myanmar Border Region. Emerg. Infect. Dis. 21, 1861–1864. doi: 10.3201/eid2110.150679
Wang, X., Ruan, W., Zhou, S., Feng, X., Yan, H., Huang, F. (2020). Prevalence of Molecular Markers Associated With Drug Resistance of Plasmodium Vivax Isolates in Western Yunnan Province, China. BMC Infect. Dis. 20, 307. doi: 10.1186/s12879-020-05032-4
World Health Organization (2013). WHO Policy Brief for the Implementation of Intermittent Preventive Treatment of Malaria in Pregnancy Using Sulfadoxine-Pyrimethamine (IPTp-SP) (Geneva: World Health Organization). Available at: https://www.afro.who.int/pt/node/6507 (Accessed 13 May, 2013).
World Health Organization (2016). World Malaria Report 2016 (Geneva: World Health Organization). Available at: https://www.who.int/malaria/publications/world-malaria-report-2016/report/en/ (Accessed 13 December, 2016).
World Health Organization (2020). World Malaria Report 2020 (Geneva: World Health Organization). Available at: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2020 (Accessed 30 November, 2020).
Xu, X., Zhou, G., Wang, Y., Hu, Y., Ruan, Y., Fan, Q., et al. (2016). Microgeographic Heterogeneity of Border Malaria During Elimination Phase, Yunnan Province, China 2011-2013. Emerg. Infect. Dis. 22, 1363–1370. doi: 10.3201/eid2208.150390
Zhao, Y., Wang, L., Soe, M. T., Aung, P. L., Wei, H., Liu, Z., et al. (2020). Molecular Surveillance for Drug Resistance Markers in Plasmodium Vivax Isolates From Symptomatic and Asymptomatic Infections at the China-Myanmar Border. Malar. J. 19, 281. doi: 10.1186/s12936-020-03354-x
Zhou, X., Huang, J. L., Njuabe, M. T., Li, S. G., Chen, J. H., Zhou, X. N. (2014). A Molecular Survey of Febrile Cases in Malaria-Endemic Areas Along China-Myanmar Border in Yunnan Province, People’s Republic of China. Parasite 21, 27. doi: 10.1051/parasite/2014030
Keywords: Plasmodium vivax, sulfadoxine-pyrimethamine, dihydropteroate synthase gene, dihydrofolate reductase gene, GTP cyclohydrolase I gene
Citation: Zeng W, Wang S, Feng S, Zhong D, Hu Y, Bai Y, Ruan Y, Si Y, Zhao H, Yang Q, Li X, Chen X, Zhang Y, Li C, Xiang Z, Wu Y, Chen F, Su P, Rosenthal BM and Yang Z (2021) Polymorphism of Antifolate Drug Resistance in Plasmodium vivax From Local Residents and Migrant Workers Returned From the China-Myanmar Border. Front. Cell. Infect. Microbiol. 11:683423. doi: 10.3389/fcimb.2021.683423
Received: 21 March 2021; Accepted: 03 June 2021;
Published: 24 June 2021.
Edited by:
Jun Cao, Jiangsu Institute of Parasitic Diseases (JIPD), ChinaReviewed by:
Himanshu Gupta, London School of Hygiene and Tropical Medicine, United KingdomLemu Golassa, Addis Ababa University, Ethiopia
Copyright © 2021 Zeng, Wang, Feng, Zhong, Hu, Bai, Ruan, Si, Zhao, Yang, Li, Chen, Zhang, Li, Xiang, Wu, Chen, Su, Rosenthal and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoqing Yang, emhhb3Fpbmd5OTJAaG90bWFpbC5jb20=
†These authors have contributed equally to this work
 Weilin Zeng
Weilin Zeng Siqi Wang1†
Siqi Wang1† Shi Feng
Shi Feng Hui Zhao
Hui Zhao Zheng Xiang
Zheng Xiang Benjamin M. Rosenthal
Benjamin M. Rosenthal