- Department of Microbiology and Biochemistry, University of the Free State, Bloemfontein, South Africa
The yeast Candida albicans exhibits multiple morphologies dependent on environmental cues. Candida albicans biofilms are frequently polymicrobial, enabling interspecies interaction through proximity and contact. The interaction between C. albicans and the bacterium, Pseudomonas aeruginosa, is antagonistic in vitro, with P. aeruginosa repressing the yeast-to-hyphal switch in C. albicans. Previous transcriptional analysis of C. albicans in polymicrobial biofilms with P. aeruginosa revealed upregulation of genes involved in regulation of morphology and biofilm formation, including SET3, a component of the Set3/Hos2 histone deacetylase complex (Set3C). This prompted the question regarding the involvement of SET3 in the interaction between C. albicans and P. aeruginosa, both in vitro and in vivo. We found that SET3 may influence early biofilm formation by C. albicans and the interaction between C. albicans and P. aeruginosa. In addition, although deletion of SET3 did not alter the morphology of C. albicans in the presence of P. aeruginosa, it did cause a reduction in virulence in a Caenorhabditis elegans infection model, even in the presence of P. aeruginosa.
Introduction
Interkingdom interactions are ubiquitous in nature and can affect various aspects of the growth, antimicrobial resistance and virulence of species within a consortium (Peters et al., 2012; Stacy et al., 2016). In the opportunistic fungal pathogen, Candida albicans, these interactions are frequently encountered in polymicrobial associations formed with commensal microorganisms as well as pathobionts in humans (Morales and Hogan, 2010; Diaz et al., 2012; Neville et al., 2015). This is, in part, due to the ability to form biofilms on both abiotic and biotic surfaces (Polke et al., 2015).
Candida albicans exhibits polymorphism, with up to nine distinct phenotypes being formed (Noble et al., 2017). This includes the classical morphotypes - yeast, hyphae, pseudohyphae and chlamydospores - as well as non-classical phenotypes dependent on the expression of the white-opaque regulator, Wor1p (Lan et al., 2002; Pande et al., 2013; Tong et al., 2014; Noble et al., 2017). These different phenotypes show alterations in mode of growth, morphology, carbon source utilisation and virulence. Importantly, distinctive phenotypes also show differences in competitive fitness with resident or co-infecting bacteria and may alter the population dynamics of these bacteria (Pande et al., 2013; Fox et al., 2014).
Candida albicans is frequently co-isolated with the Gram-negative bacterium, Pseudomonas aeruginosa from the lungs of cystic fibrosis patients (Chotirmall et al., 2010; Leclair and Hogan, 2010; Haiko et al., 2019). In vitro, the interaction is characterised as antagonistic, with both species influencing each other (reviewed by Fourie and Pohl, 2019). The bacterium was found to lyse and kill hyphae of C. albicans through physical interaction (Hogan and Kolter, 2002; Brand et al., 2008; Bandara et al., 2010) and affects C. albicans biofilm formation and morphogenesis through various secreted factors and cell wall components (Kerr et al., 1999; Hogan et al., 2004; McAlester et al., 2008; Xu et al., 2008; Bandara et al., 2010; Holcombe et al., 2010; Reen et al., 2011; Bandara et al., 2013). This includes inhibition of morphogenesis from yeast to hyphal morphologies by phenazines, quorum sensing molecules, lipopolysaccharides and via sequestration of iron, and promotion of morphogenesis by peptidoglycan. These stimuli elicit their effects through various signalling pathways in C. albicans, including stimulation of morphogenesis through the mitogen activated protein (MAP) kinase signalling pathway and the cyclic adenosine monophosphate (cAMP)/protein kinase A (PKA), as well as repression by the transcriptional repressor, Tup1p (Shareck and Belhumeur, 2011). Therefore, multiple conflicting stimuli, occurring simultaneously, from co-incubation with P. aeruginosa may play a role to affect the morphology of C. albicans. Transcriptomic evaluation of C. albicans indicated the upregulation of SET3 [a component of the Set3/Hos2 histone deacetylase complex (Set3C)] in the presence of P. aeruginosa (Fourie et al., 2021). This led to the question if Set3C may influence the interaction between C. albicans and P. aeruginosa, by integrating various external stimuli to influence biofilm morphology. As the contribution of this gene in the interaction of C. albicans with P. aeruginosa has not been evaluated before, its role in in vitro polymicrobial biofilm formation and virulence in Caenorhabditis elegans was examined.
Materials and Methods
Strain Maintenance
Candida albicans strains were stored at -80°C with 15% glycerol. Yeast strains were revived and maintained on yeast malt (YM) agar (3 g l-1 malt extract, 3 g l-1 yeast extract, 5 g l-1 peptone, 10 g l-1 glucose, 16 g l-1 agar) at 30°C. Pseudomonas aeruginosa PAO1 was stored at -80°C with 25% glycerol and revived/maintained on Luria-Bertani (LB) agar (5 g l-1 yeast extract, 10 g l-1 tryptone, 10 g l-1 sodium chloride and 15 g l-1 agar).
Construction of Homozygous Deletion Mutants With CRISPR/Cas9
A published CRISPR-Cas9 system (Nguyen et al., 2017) was used for the construction of homozygous mutants for SET3 with minor modifications. This method entails the introduction of a homozygous double stranded break at the site of interest and modification of the sites of interest with donor DNA in the wild type SC5314 strain. A detailed description of the procedure can be found in Supplementary Text S1. Following homozygous deletion of SET3 yielding set3Δ/Δ, a complemented strain (set3Δ/Δ::SET3) was constructed by reintroduction of the wild-type gene by modified donor DNA.
Preparation of Cells for Mono- and Polymicrobial Biofilms
Preparation of C. albicans Cells for Monomicrobial Biofilms
Candida albicans SC5314 (wild type) was grown on YM agar for 24 h at 30°C and was inoculated into 10 mL yeast nitrogen base (YNB) broth (10 g l-1 glucose, 16 g l-1 YNB) and incubated at 30°C for 24 h. Cells were harvested (1878 x g, 5 minutes) and the supernatant removed. This was followed by washing the cells twice with phosphate buffered saline (PBS) (Oxoid, England). The cells were counted with a hemocytometer and diluted to 1 x 106 cells/mL in filter sterilized (0.22 μm nitrocellulose filter, Merck Millipore, Ireland) RPMI-1640 medium (Sigma-Aldrich, USA).
Preparation of C. albicans and P. aeruginosa Cells for Polymicrobial Biofilms
Pseudomonas aeruginosa PAO1 (wild type) was grown on LB plates for 24 h at 37°C. Cells were inoculated into 5 mL nutrient broth (1 g l-1 malt extract, 2 g l-1 yeast extract, 5 g l-1 peptone and 8 g l-1 sodium chloride) and incubated at 37°C for 24 h with shaking (150 rpm). These cells were washed (X3) and diluted to an optical density (OD600) of approximately 0.05 in RPMI-1640 medium containing 1 x 106 cells/mL C. albicans (prepared as described in previous section).
Quantification and Characterisation of Biofilm Formation
Biofilm Biomass of Mono- and Polymicrobial Biofilms
Cells were prepared as described above and 200 µL was dispensed into a 96-well plate (Corning Incorporated, Costar®, USA). The plate was incubated for 6h and 48h respectively at 37°C to allow the formation of biofilms. The crystal violet assay was performed on biofilms according to Jin and co-workers (2003) with minor modifications. Briefly, the supernatant from each well was removed and the biofilms were washed twice with sterile PBS. Biofilms were then left to air dry for 45 minutes and stained with 110 µL crystal violet (0.4% w/v; Merck, Germany) for 45 min (Jin et al., 2003). Biofilms were washed three times with 350 µL sterile H2O and de-stained with 200 µL 95% ethanol for 45 min. One hundred microliter of de-staining solution was then transferred to a clean 96-well plate and absorbance was measured at 595 nm. This experiment was performed in triplicate with four technical replicates per biological replicate.
Quantification of C. albicans and P. aeruginosa Colony Forming Units in Biofilms
Cells for mono- and polymicrobial biofilms were prepared as described above in flat-bottom 6 well culture plates (Corning Incorporated, USA) in 3 mL medium and incubated for 48h at 37°C to allow biofilm formation to take place. After incubation, biofilms were washed twice with sterile PBS, scraped off and suspended in PBS. Biofilms were then vortexed 3 times for 1 minute to remove adherent cells from one another (adapted from Fourie et al., 2017). For quantification of C. albicans, serially diluted cells were plated onto YM medium acidified with tartaric acid (final concentration 0.08%). For bacterial quantification, serially diluted cells were plated onto LB supplemented with 10 µg/mL amphotericin B (Sigma-Aldrich, USA) (Pires et al., 2013). Plates were incubated overnight, to allow formation of colonies, and counted. This experiment was performed in triplicate.
Morphology of Mono- and Polymicrobial Biofilms
Cells for mono- and polymicrobial biofilms were prepared as described above in flat-bottom 6 well culture plates in 3 mL medium (Fourie et al., 2017). After incubation for 24h at 37°C (to allow for mature biofilms without extensive killing by P. aeruginosa), supernatant was removed and approximately 5 mm rectangular sections of the wells were cut and placed in PBS. Cells were fixed overnight in 3% (v/v) glutardialdehyde (Merck, Germany) in phosphate buffer. This was followed by washing of biofilms with PBS and fixation with 1% osmium tetroxide (Merck, Germany) for 1h. Following fixation, biofilms were sequentially dehydrated with increasing concentration of ethanol (50% to 100%) and subjected to critical point drying (Samdri-795 Critical point dryer, Tousimis, USA). A gold layer was applied to the biofilms with a SEM coating system (Bio-Rad, UK) and examined using a JSM-7800F Extreme-resolution Analytical Field Emission SEM.
Infection of Caenorhabditis elegans by C. albicans and P. aeruginosa
Caenorhabditis elegans AU37 [glp-4(bn2) I; sek-1(km4 X] (RRID : WB-STRAIN : WBStrain00000261), obtained for the Caenorhabditis Genetic Centre (University of Minnesota), was used for all infections. The nematodes were propagated and maintained on Nematode Growth Medium (3 g l-1 NaCl, 2.5 g l-1 peptone, 5 μg ml-1 cholesterol, 1 mM CaCl2, 1 mM MgSO4, 25 mM KPO4, 20 g l-1 agar) with Escherichia coli OP50 as a food source at 15°C (Brenner, 1974).
For infection by C. albicans alone, C. albicans strains were inoculated in YPD broth overnight at 30°C. Overnight cells were diluted to an OD600 of 0.8 and 100 μL was plated onto brain-heart infusion (BHI)-agar plates and incubated overnight at 30°C. Synchronised L4-stage nematodes were washed with M9 buffer (3 g l-1 KH2PO4, 6 g l-1 Na2PO4 and 1 mM MgSO4) and added to plates with C. albicans. Nematodes were incubated with C. albicans for 4 hours at 25°C and washed three times with M9 buffer to remove non-ingested C. albicans cells. Nematodes were then added at approximately 60 per well in a 6 well plate (Corning Incorporated, USA) containing 2 mL 80% M9 buffer and 20% BHI broth, with 90 μg/mL kanamycin and incubated at 25°C. Nematodes were monitored daily and dead nematodes (non-motile after mechanical stimulation with sterile pipette tip or penetration of C. elegans cuticle by C. albicans hyphae) were counted and removed.
For dual-infection by both C. albicans and P. aeruginosa, C. albicans was prepared on BHI-agar plates as described above. P. aeruginosa was inoculated in LB broth overnight and diluted to OD600 of 0.8. One hundred microliter of the P. aeruginosa suspension was plated on BHI-plates and incubated overnight at 37°C. Synchronised L4-stage nematodes were washed with M9 buffer and added to plates incubated with C. albicans for 2 hours at 25°C. Nematodes were washed three times with M9 buffer and added to BHI-plates with P. aeruginosa for 2 hours at 25°C. Nematodes were washed again with M9 buffer to remove non-ingested P. aeruginosa cells and placed in M9 buffer in 6 well plates (approximately 60 nematodes per well) and incubated at 25°C. Nematodes were monitored daily and dead worms (non-motile after mechanical stimulation with sterile pipette tip or penetration of C. elegans cuticle by C. albicans hyphae) were counted and removed. Nematodes incubated with E. coli OP50 prior to survival assay was used as a control. Infection was performed in triplicate for a total of 180 nematodes per strain of C. albicans. Survival metrics including Kaplan-Meier statistics, median survival time and log-rank test were performed with online application for survival analysis 2 (OASIS 2; Han et al., 2016).
Statistical Analysis
Differences between control and experimental, as well as between C. albicans strains were determined with t-test, performed with GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com) unless otherwise stated.
Results
Deletion of SET3 Influences Early Biofilm Formation and Interaction With P. aeruginosa
To evaluate the effect of the deletion of SET3 on C. albicans biofilm formation, biomass production during early biofilm development (6h) as well as after 48 hours (mature biofilms) in both mono- and polymicrobial biofilms were quantified using the crystal violet assay. In addition, the population dynamics in polymicrobial biofilms were studied using CFUs of C. albicans and P. aeruginosa, as presented in Figures 1, 2.
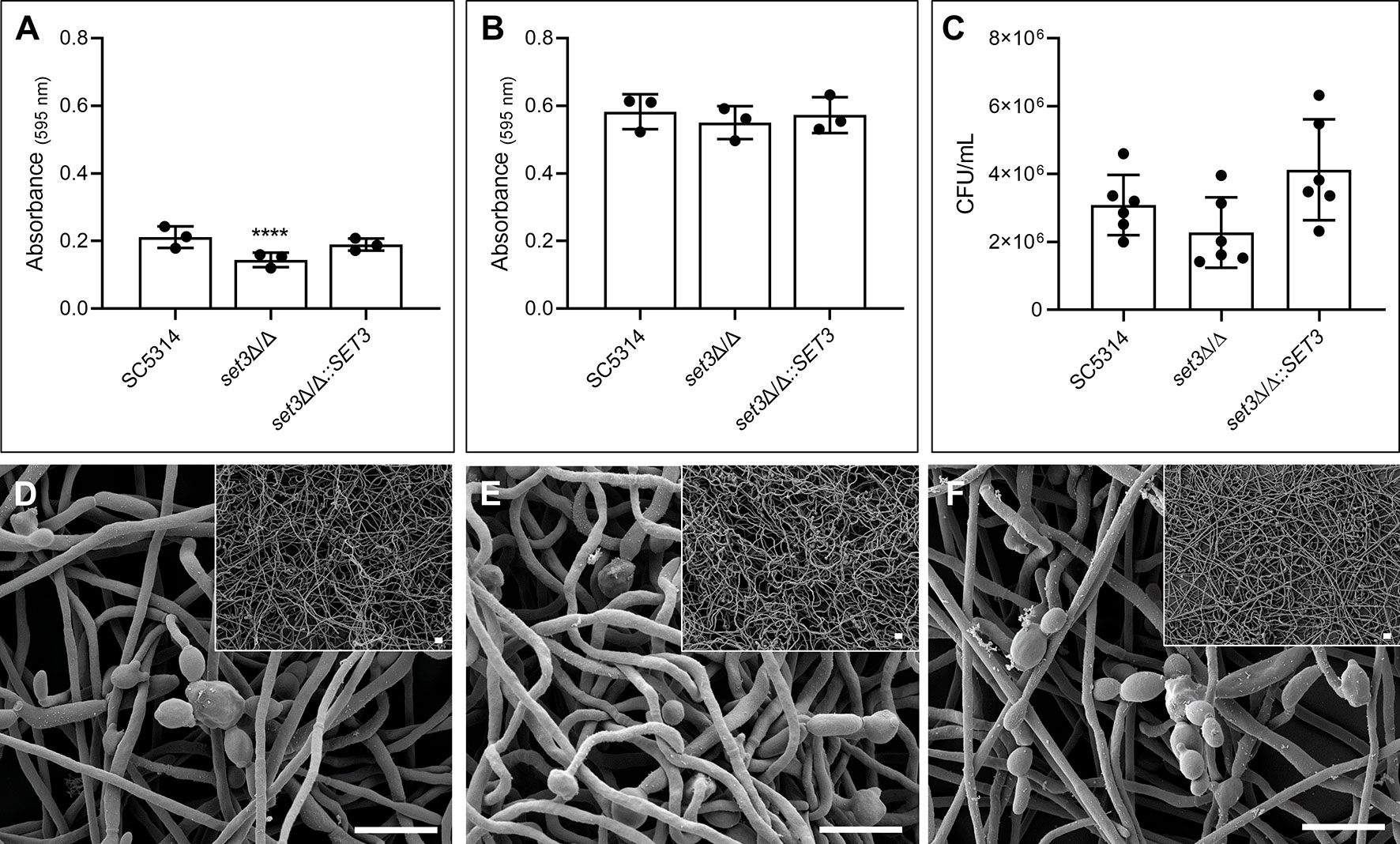
Figure 1 Effect of SET3 deletion on Candida albicans monomicrobial biofilms. Monomicrobial biofilm biomass after 6 h (A) and 48 h (B) of the homozygous mutant of SET3 (set3Δ/Δ) as well as the homozygous mutant with add-back of the wild-type gene (set3Δ/Δ::SET3). Colony forming units (CFU) of mature C. albicans biofilms are indicated in (C) as well as biofilm morphology of mature biofilms in (D) (SC5314), (E) (set3Δ/Δ) and (F) (set3Δ/Δ::SET3). Small panels on right corners indicate biofilms with lower magnification. Scale bars on all panels indicate 10 μm. *Significantly different from wild type (SC5314) (****P < 0.0001).
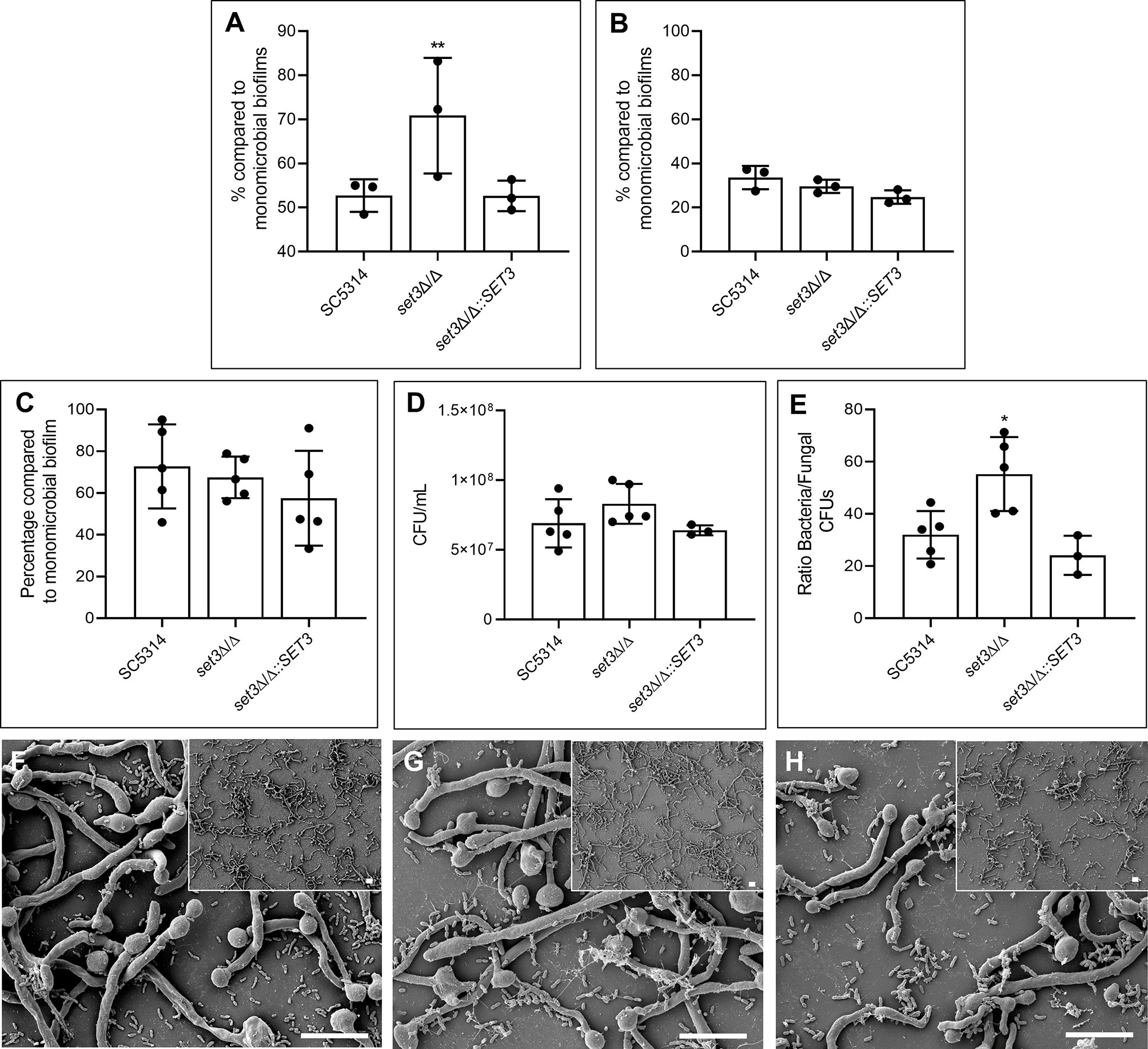
Figure 2 Effect of SET3 deletion on Candida albicans polymicrobial biofilms with Pseudomonas aeruginosa. Polymicrobial biofilm biomass after 6 h (A) and 48 h (B) of the homozygous mutant of SET3 (set3Δ/Δ) as well as the homozygous mutant with add-back of the wild-type gene (set3Δ/Δ::SET3) with P. aeruginosa. Colony forming units (CFU) of mature C. albicans biofilms are indicated in (C) and P. aeruginosa CFUs in (D, E) indicates the ratio of bacterial/fungal CFUs. (F) (SC5314), (G) (set3Δ/Δ) and (H) (set3Δ/Δ::SET3) indicates the morphology of mature polymicrobial biofilms. Small panels on right corners indicate biofilms with lower magnification. Scale bars on all panels indicate 10 μm. *Significantly different from wild type (SC5314) (*P < 0.05; **P < 0.005).
Figure 1A indicates a significant (P < 0.0001) reduction in biomass of early (6h) monomicrobial biofilms (approximately 33.6%) of set3Δ/Δ compared to the wild type. This was restored to wild type values with the re-introduction of the SET3 gene. Mature (48h) biofilms of set3Δ/Δ did not exhibit this reduction in biomass (Figure 1B), and although a slight reduction in set3Δ/Δ CFUs is seen after 48 hours (Figure 1C), this is not statistically significant. In addition, Figures 1D–F indicate that the mature monomicrobial biofilms of set3Δ/Δ is composed of a thicker layer of hyphal cells (Figure 1E), confirming previous observations by Nobile et al. (2014).
When set3Δ/Δ was exposed to P. aeruginosa in a polymicrobial biofilm (Figure 2), a significant increase in polymicrobial biofilm biomass, compared to the polymicrobial biofilms with wild type C. albicans, is seen after 6h (Figure 2A). This effect could be restored to the wild type phenotype with the re-introduction of SET3 and was lost during biofilm maturation (Figure 2B). Interestingly, although no statistically significant effect on C. albicans (Figure 2C) or P. aeruginosa (Figure 2D) CFUs is seen in the mature (48h) polymicrobial biofilms (due to the large variation between samples), a significant increase in the ratio of bacterial CFUs over C. albicans CFUs in polymicrobial biofilms of set3Δ/Δ is observed (Figure 2E). Deletion of SET3 did not prevent the inhibition of C. albicans morphogenesis by P. aeruginosa (Figures 2F–H). However, longer hyphae are present in set3Δ/Δ polymicrobial biofilms (Figure 2G) than in either the wild type biofilms (Figure 2F) or the complemented strain (Figure 2H).
SET3 Influences Virulence of Mono- and Polymicrobial Infection in C. elegans
Figure 3 indicates the percentage survival of C. elegans with survival statistics of C. elegans infected with C. albicans alone, or co-infected with P. aeruginosa. A significant (P < 0.0001) increase in survival of C. elegans infected with set3Δ/Δ compared to the wild type was found (Figure 3A). Virulence of set3Δ/Δ in C. elegans was restored through re-introduction of the wild-type gene (set3Δ/Δ::SET3). Notably, the deletion of SET3 did not influence the ability of C. albicans to form hyphae and pierce the cuticle of C. elegans. Similar to single-species infection, co-infection by C. albicans set3Δ/Δ and P. aeruginosa also exhibited decreased virulence compared to the co-infection with the wild type (Figure 3B), indicating that Set3C contributes to virulence of C. albicans in C. elegans, even in the presence of P. aeruginosa.
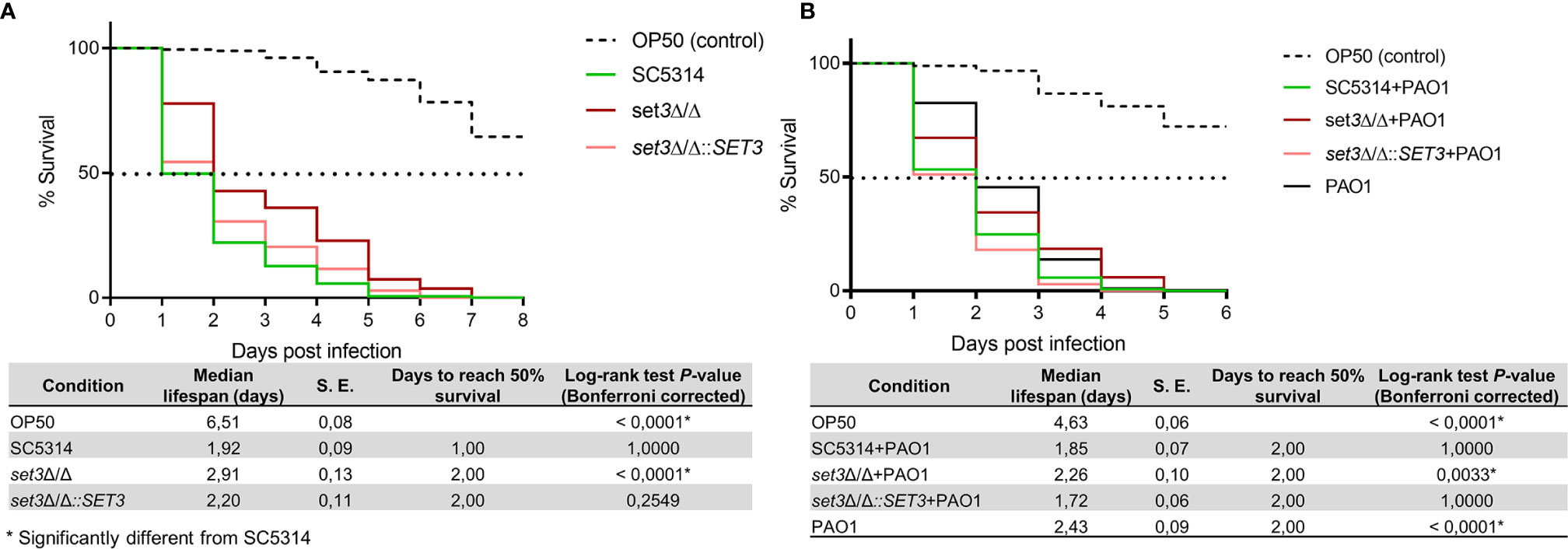
Figure 3 Survival of Caenorhabditis elegans infected with Candida albicans mutants and Pseudomonas aeruginosa. (A) - Percentage survival of C. elegans infected with C. albicans wild type (SC5314) or the homozygous mutant of SET3 (set3Δ/Δ) as well as the homozygous mutant with add-back of the wild-type gene (set3Δ/Δ::SET3). (B) – Percentage survival of C. elegans co-infected with C. albicans mutants and P. aeruginosa (PAO1). Controls consists of C. elegans allowed to feed on Escherichia coli OP50 (OP50). The tables represent median lifespan with standard error (S. E.) along with days to reach 50% mortality. P-values are included for the Log-rank test for overall differences in survival. *Significantly different from wild type C. albicans (SC5314).
Discussion
In order to evaluate the possible role of SET3 in the interaction between C. albicans and P. aeruginosa, the impact of homozygous deletion of SET3 on biofilm formation of C. albicans was first evaluated. Using this approach, it was found that deletion of SET3 negatively influence early biofilm formation (Figure 1), but that this effect was lost in mature biofilms, which also showed robust formation of hyphae after 48h, similar to results reported previously (Hnisz et al., 2010). Thus, although binding of the Set3C correlates with gene expression during morphogenesis, deletion of SET3 leaves the expression of most genes unaffected (Hnisz et al., 2012) and may only transiently affect expression levels of key morphogenesis-related genes.
Upon evaluation of the effect of SET3 deletion on the interaction between C. albicans and P. aeruginosa, an increase in biomass of the set3Δ/Δ polymicrobial biofilm was seen at 6h (Figure 2), however this effect was also lost upon maturation of the biofilms (48h). It must be noted that the previous upregulation of SET3 in the presence of P. aeruginosa was also in 6h biofilms (Fourie et al., 2021), strengthening the finding that SET3 may modulate early biofilm formation, especially in the presence of P. aeruginosa. Interestingly, although the Set3C may provide additional regulation, deletion of SET3, a core component of the complex, is unable to prevent the inhibition of C. albicans hyphal formation by P. aeruginosa. However, longer hyphae were observed, which may be as a result of the hypersensitivity of the cAMP/PKA signaling pathway previously reported for set3Δ/Δ (Hnisz et al., 2010). This indicates that SET3 is required for the full wild type response of C. albicans to P. aeruginosa, mediating the morphological switch to the yeast morphology, which may allow dispersal and escape of C. albicans from the antagonistic effect of P. aeruginosa.
Caenorhabditis elegans is susceptible to C. albicans and P. aeruginosa (Tan et al., 1999; Pukkila-Worley et al., 2009) and is known to share a similar innate immune response with humans (Pukkila-Worley et al., 2011). The increased survival of nematodes infected with set3Δ/Δ (Figure 3) (even in the presence of P. aeruginosa) corroborates findings of reduced virulence of set3Δ/Δ as found previously in a murine model of systemic candidiasis (Hnisz et al., 2010) and further validates the use of this alternative infection model in the study of C. albicans virulence. Candida albicans primarily relies on hyphal formation to kill C. elegans in this assay (Pukkila-Worley et al., 2009). Notably, similar to the results in the murine model (Hnisz et al., 2010) in which hyperfilamentous set3Δ/Δ was less virulent, the deletion of SET3 also did not decrease the ability of C. albicans to form hyphae and pierce the cuticle of C. elegans. This confirms that the reduction in virulence is not due to a lack of hyphae, even in this simpler model. This adds further complexity to the role of the regulation of morphogenesis in virulence. In mammals, the importance of a yeast phase during certain stages of dissemination via the blood stream, as well as the timing of the yeast to hyphal switch is considered crucial for virulence. However, further research needs to be done to better understand the role of the yeast phase in the C. elegans model, where infection does not spread via dissemination. Due to the low homology of the Set3C to human or other histone deacetylases, it deserves further attention as a therapeutic target as it may not only affect the virulence of C. albicans during single species infection, but also during polymicrobial infection with P. aeruginosa.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
RF and CP conceptualised study and RF performed experiments. JA provided scholarly input regarding mutant construction and RF and CP co-wrote the manuscript. OS and OG provided resources and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Research foundation of South Africa [grant numbers 118543 and 115566 to CP and 121998 to OG].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.680732/full#supplementary-material
References
Bandara, H. M. H. N., Cheung, B. P. K., Watt, R. M., Jin, L. J., Samaranayake, L. P. (2013). Pseudomonas Aeruginosa Lipopolysaccharide Inhibits Candida albicans Hyphae Formation and Alters Gene Expression During Biofilm Development. Mol. Oral. Microb. 28, 54–69. doi: 10.1111/omi.12006
Bandara, H. M. H. N., Yau, J. Y. Y., Jin, L. J., Samaranayake, L. P. (2010). Pseudomonas aeruginosa Inhibits In-Vitro Candida Biofilm Development. BMC Microbiol. 10, 125. doi: 10.1186/1471-2180-10-125
Brand, A., Barnes, J. D., Mackenzie, K. S., Odds, F. C., Gow, N. A. R. (2008). Cell Wall Glycans and Soluble Factors Determine the Interaction Between the Hyphae of Candida albicans and Pseudomonas aeruginosa. FEMS Microbiol. Lett. 287, 48–55. doi: 10.1111/j.1574-6968.2008.01301.x
Brenner, S. (1974). The Genetics of Caenorhabditis elegans. Genetics 77, 71–94. doi: 10.1093/genetics/77.1.71
Chotirmall, S. H., O’Donoghue, E., Bennett, K., Gunaratnam, C., O’Neill, S. J., McElvaney, N. G. (2010). Sputum Candida albicans Presages FEV 1 Decline and Hospital-Treated Exacerbations in Cystic Fibrosis. Chest 138, 1186–1195. doi: 10.1378/chest.09-2996
Diaz, P. I., Xie, Z., Sobue, T., Thompson, A., Biyikoglu, B., Ricker, A., et al. (2012). Synergistic Interaction Between Candida albicans and Commensal Oral Streptococci in a Novel In Vitro Mucosal Model. Infect. Immun. 80, 620–632. doi: 10.1128/IAI.05896-11
Fourie, R., Cason, E. D., Albertyn, J., Pohl, C. H. (2021). Transcriptional Response oftoin a Polymicrobial Biofilm. G3 Genes Genomes Genet 11, jkab042. doi: 10.1093/g3journal/jkab042
Fourie, R., Ells, R., Kemp, G., Sebolai, O. M., Albertyn, J., Pohl, C. H. (2017). Pseudomonas aeruginosa Produces Aspirin Insensitive Eicosanoids and Contributes to the Eicosanoid Profile of Polymicrobial Biofilms With Candida albicans. Prostaglandins Leukot. Essent. Fatty Acids 117, 36–46. doi: 10.1016/j.plefa.2017.01.008
Fourie, R., Pohl, C. H. (2019). Beyond Antagonism: The Interaction Between Candida Species and Pseudomonas aeruginosa. J. Fungi 5, 34. doi: 10.3390/jof5020034
Fox, E. P., Cowley, E. S., Nobile, C. J., Hartooni, N., Newman, D. K., Johnson, A. D. (2014). Anaerobic Bacteria Grow Within Candida albicans Biofilms and Induce Biofilm Formation in Suspension Cultures. Curr. Biol. 24, 2411–2416. doi: 10.1016/j.cub.2014.08.057
Haiko, J., Saeedi, B., Bagger, G., Karpati, F., Özenci, V. (2019). Coexistence of Candida Species and Bacteria in Patients With Cystic Fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 38, 1071–1077. doi: 10.1007/s10096-019-03493-3
Han, S. K., Lee, D., Lee, H., Kim, D., Son, H. G., Yang, J. S., et al. (2016). OASIS 2: Online Application for Survival Analysis 2 With Features for the Analysis of Maximal Lifespan and Healthspan in Aging Research. Oncotarget 7, 56147–56152. doi: 10.18632/oncotarget.11269
Hnisz, D., Bardet, A. F., Nobile, C. J., Petryshyn, A., Glaser, W., Schöck, U., et al. (2012). A Histone Deacetylase Adjusts Transcription Kinetics at Coding Sequences During Candida albicans Morphogenesis. PloS Genet. 8, e1003118. doi: 10.1371/journal.pgen.1003118
Hnisz, D., Majer, O., Frohner, I. E., Komnenovic, V., Kuchler, K. (2010). The Set3/Hos2 Histone Deacetylase Complex Attenuates cAMP/PKA Signalling to Regulate Morphogenesis and Virulence of Candida albicans. PloS Pathog. 6, e1000889. doi: 10.1371/journal.ppat.1000889
Hogan, D. A., Kolter, R. (2002). Pseudomonas-Candida Interactions: An Ecological Role for Virulence Factors. Science 296, 2229–2232. doi: 10.1126/science.1070784
Hogan, D. A., Vik, Å., Kolter, R. (2004). A Pseudomonas aeruginosa Quorum-Sensing Molecule Influences Candida albicans Morphology. Mol. Microbiol. 54, 1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x
Holcombe, L. J., McAlester, G., Munro, C. A., Enjalbert, B., Brown, A. J. P., Gow, N. A. R., et al. (2010). Pseudomonas aeruginosa Secreted Factors Impair Biofilm Development in Candida albicans. Microbiology 156, 1476–1485. doi: 10.1099/mic.0.037549-0
Jin, Y., Yip, H. K., Samaranayake, Y. H., Yau, J. Y., Samaranayake, L. P. (2003). Biofilm-Forming Ability of Candida albicans is Unlikely to Contribute to High Levels of Oral Yeast Carriage in Cases of Human Immunodeficiency Virus Infection. J. Clin. Microbiol. 41, 2961–2967. doi: 10.1128/JCL.41.7.2961-2967.2003
Kerr, J. R., Taylor, G. W., Rutman, A., Høiby, N., Cole, P. J., Wilson, R. (1999). Pseudomonas Aeruginosa Pyocyanin and 1-Hydroxyphenazine Inhibit Fungal Growth. J. Clin. Pathol. 52, 385–387. doi: 10.1136/jcp.52.5.385
Lan, C.-Y., Newprot, G., Murillo, L. A., Jones, T., Scherer, S., Davis, R. W., et al. (2002). Metabolic Specialization Associated With Phenotypic Switching in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 99, 14907–14912. doi: 10.1073/pnas.232566499
Leclair, L. W., Hogan, D. A. (2010). Mixed Bacterial-Fungal Infections in the CF Respiratory Tract. Med. Mycol. 48, S125–S132. doi: 10.3109/13693786.2010.521522
McAlester, G., O’Gara, F., Morrissey, J. P. (2008). Signal-Mediated Interactions Between Pseudomonas aeruginosa and Candida albicans. J. Med. Microbiol. 57, 563–569. doi: 10.1099/jmm.0.47705-0
Morales, D. K., Hogan, D. A. (2010). Candida albicans Interactions With Bacteria in the Context of Human Health and Disease. PloS Pathog. 6, e1000886. doi: 10.1371/journal.ppat.1000886
Neville, B. A., d’Enfert, C., Bougnoux, M.-E. (2015). Candida albicans Commensalism in the Gastrointestinal Tract. FEMS Yeast Res. 15, fov081. doi: 10.1093/femsyr/fov081
Nguyen, N., Quail, M. M. F., Hernday, A. D. (2017). An Efficient, Rapid, and Recyclable System for CRISPR-Mediated Genome Editing in Candida albicans. mSphere 2, e00149–e00117. doi: 10.1128/mSphereDirect.00149-17
Nobile, C. J., Fox, E. P., Hartooni, N., Mitchell, K. F., Hnisz, D., Andes, D. R., et al. (2014). A Histone Deacetylase Complex Mediates Biofilm Dispersal and Drug Resistance in Candida albicans. mBio 5, e01201–e01214. doi: 10.1128/mBio.01201-14
Noble, S. M., Gianetti, B. A., Witchley, J. N. (2017). Candida albicans Cell-Type Switching and Functional Plasticity in the Mammalian Host. Nat. Rev. Microbiol. 15, 96–108. doi: 10.1038/nrmicro.2016.157
Pande, K., Chen, C., Noble, S. M. (2013). Passage Through the Mammalian Gut Triggers a Phenotypic Switch That Promotes Candida albicans Commensalism. Nat. Genet. 45, 1088–1091. doi: 10.1038/ng.2710
Peters, B. M., Jabra-Rizk, M. A., O’May, G. A., Costerton, J. W., Shirtliff, M. E. (2012). Polymicrobial Interactions: Impact on Pathogenesis and Human Disease. Clin. Microbiol. Rev. 25, 193–213. doi: 10.1128/CMR.00013-11
Pires, D. P., Silva, S., Almeida, C., Henriques, M., Anderson, E. M., Lam, J. S., et al. (2013). Evaluation of the Ability of C. albicans to Form Biofilm in the Presence of Phage-Resistant Phenotypes of P. Aeruginosa. Biofouling 29, 1169–1180. doi: 10.1080/08927014.2013.831842
Polke, M., Hube, B., Jacobsen, I. D. (2015). Candida Survival Strategies. Adv. Appl. Microbiol. 91, 139– 235. doi: 10.1016/bs.aambs.2014.12.002
Pukkila-Worley, R., Ausubel, F. M., Mylonakis, E. (2011). Candida albicans Infection of Caenorhabditis elegans Induces Antifungal Immune Defences. PloS Pathog. 7 (6), e1002074. doi: 10.1371/journal.ppat.1002074
Pukkila-Worley, R., Peleg, A. Y., Tampakakis, E., Mylonakis, E. (2009). Candida albicans Hyphal Formation and Virulence Assessed Using a Caenorhabditis elegans Infection Model. Eukaryot. Cell 11, 1750–1758. doi: 10.1128/EC.00163-09
Reen, F. J., Mooij, M. J., Holcombe, L. J., Mcsweeney, C. M., Mcglacken, G. P., Morrissey, J. P., et al. (2011). The Pseudomonas Quinolone Signal (PQS), and its Precursor HHQ, Modulate Interspecies and Interkingdom Behaviour. FEMS Microbiol. Ecol. 77, 413–428. doi: 10.1111/j.1574-6941.2011.01121.x
Shareck, J., Belhumeur, P. (2011). Modulation of Morphogenesis in Candida albicans by Various Small Molecules. Eukaryot. Cell 10, 1004–1012. doi: 10.1128/EC.05030-11
Stacy, A., McNally, L., Darch, S. E., Brown, S. P., Whiteley, M. (2016). The Biogeography of Polymicrobial Infection. Nat. Rev. Microbiol. 14, 93–105. doi: 10.1038/nrmicro.2015.8
Tan, M. W., Rahme, L. G., Sternberg, J. A., Tompkins, R. G., Ausubel, F. M. (1999). Pseudomonas aeruginosa Killing of Caenorhabditis elegans Used to Identify P. aeruginosa Virulence Factors. Proc. Natl. Acad. Sci. U. S. A. 96, 2408–2413. doi: 10.1073/pnas.96.5.2408
Tong, Y., Cao, C., Xie, J., Ni, J., Guan, G., Tao, L., et al. (2014). N-Acetylglucosamine-Induced White-to-Opaque Switching in Candida albicans is Independent of the Wor2 Transcription Factor. Fungal Genet. Biol. 62, 71–77. doi: 10.1016/j.fgb.2013.10.005
Keywords: biofilm, Candida albicans, Caenorhabditis elegans, Pseudomonas aeruginosa, SET3
Citation: Fourie R, Albertyn J, Sebolai O, Gcilitshana O and Pohl CH (2021) Candida albicans SET3 Plays a Role in Early Biofilm Formation, Interaction With Pseudomonas aeruginosa and Virulence in Caenorhabditis elegans. Front. Cell. Infect. Microbiol. 11:680732. doi: 10.3389/fcimb.2021.680732
Received: 15 March 2021; Accepted: 26 May 2021;
Published: 10 June 2021.
Edited by:
Scott Rice, Singapore Center for Environmental Life Sciences Engineering, SingaporeReviewed by:
Sarah Maddocks, Cardiff Metropolitan University, United KingdomKarishma S. Kaushik, Savitribai Phule Pune University, India
Copyright © 2021 Fourie, Albertyn, Sebolai, Gcilitshana and Pohl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carolina H. Pohl, UG9obENIQHVmcy5hYy56YQ==
 Ruan Fourie
Ruan Fourie Jacobus Albertyn
Jacobus Albertyn Olihile Sebolai
Olihile Sebolai Carolina H. Pohl
Carolina H. Pohl