- Department of Microbiology, Immunology and Parasitology, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, Brazil
The development of genetic manipulation of Plasmodium falciparum in the 1980s was key to study malaria biology. Genetically modified parasites have been used to study several aspects of the disease, such as red blood cell invasion, drug resistance mechanisms, gametocyte development and mosquito transmission. However, biological and genetic differences between P. falciparum and the other human malaria parasites make P. falciparum a poor model to study different species. The lack of robust systems of long-term in vitro culture of P. vivax and the other human malaria parasites lagged the genetic manipulation of these species. Here we review the efforts to generate genetically modified non-falciparum human malaria parasites, in vivo and in vitro. Using in vivo models – infection of non-human primates such as rhesus macaques and saimiri monkeys – researchers were able to generate transgenic lines of P. knowlesi, P. cynomolgi, and P. vivax. The development of long-term in vitro culture of P. knowlesi in the 2000’s, using rhesus and human red blood cells, created a platform to genetically manipulate non-falciparum malaria parasites. Recently, the use of CRISPR/Cas9 technology to genome edit P. knowlesi provides another tool to non-falciparum malaria research, extending the possibilities and allowing researchers to study different aspects of the biology of these parasites and understand the differences between these species and P. falciparum.
Background
Bacterial transformation was discovered in 1955 (Hotchkiss, 1955) and since then scientists have been able to manipulate the genomes of several organisms using the same principle. The transformation mechanism itself was used first to manipulate bacterial genes (Stocker, 1963). In 1982, a group used electric pulses to transfer the herpes simplex thymidine kinase gene into mouse lyoma cells (Neumann et al., 1982). This was the first report of the use of electroporation to facilitate the incorporation of exogenous DNA by cells. The first protozoan parasite transfection was reported in 1987 when an exogenous plasmid was introduced in Trypanosoma brucei (Eid and Sollner-Webb, 1987). The first malaria parasite transfected was Plasmodium gallinaceum in 1993 (Goonewardene et al., 1993).
Since then, four of the six Plasmodium species that cause human malaria (Plasmodium falciparum, Plasmodium vivax, Plasmodium knowlesi, and Plasmodium cynomolgi) (Wu et al., 1995; Van Der Wel et al., 1997; Kocken et al., 1999; Barros et al., 2015) have been submitted to genetic transformation (van Dijk et al., 1995; Mota et al., 2001; Reece and Thompson, 2008). Different techniques were applied: integration of circular and linear DNA by homologous recombination (van Dijk et al., 1995; Crabb and Cowman, 1996; Mota et al., 2001; Kocken et al., 2002), Plasmodium Artificial Chromosomes (PACs) (Voorberg-van der Wel et al., 2013), and double-strand break based genome editing techniques, such as Zinc Finger Nucleases (ZFNs) (Straimer et al., 2012; Barros et al., 2015) and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 (Ghorbal et al., 2014; Lee and Fidock, 2014; Wagner et al., 2014; Zhang et al., 2014).
Given its impact on economy and global health, robust methods for genetic analysis are needed to study malaria parasites. However, research is still focused on P. falciparum, species responsible for the majority of human deaths. Research of the other human malaria parasites (P. vivax, P. knowlesi, P. cynomolgi, P. malariae and P. ovale) remains neglected (Carlton et al., 2011), including the development of transgenic parasites. In this paper we review all the published papers on genetic transformation of non-falciparum human malaria parasites, a field that should grow exponentially with the recent development of in vitro culture of P. knowlesi and P. cynomolgi (Kocken et al., 2002; Moon et al., 2013; Chua et al., 2019) combined with the use of genome editing techniques.
Plasmodium knowlesi
Historically, P. knowlesi has been extensively used as a model for malaria research because their natural and experimental hosts (Macaca fascicularis and Macaca mulatta, respectively) were easily available for research. However, the development of in vitro culture of P. falciparum and the restrictions to the use of non-human primates shifted the focus from P. knowlesi research in the 1970s (Butcher and Mitchell, 2016; Pasini et al., 2016). Transfection of P. knowlesi was first described in 1997. The authors developed a parasite line resistant to the antifolate pyrimethamine, using an episomal construct containing the Toxoplasma gondii dihydrofolate reductase-thymidylate synthase gene (tgdhfr-ts). The episomal plasmids also harbored expression control sequences originally from P. berghei and P. falciparum (Van Der Wel et al., 1997). Transgenic parasites were obtained in vivo, after the inoculation of electroporated parasitized red-blood cells in rhesus macaques. The results demonstrated that P. knowlesi parasites recognize expression control sequences from evolutionary distant malaria species, such as the rodent malaria P. berghei and the laverania malaria P. falciparum.
Plasmodium knowlesi was successfully adapted to long term in vitro culture using rhesus red-blood cells in 2001 (Kocken et al., 2002). Culture adapted parasites could be used later for rhesus infections, allowing studies in vitro and in vivo. Furthermore, parasites were transfected transiently or stably in vitro, using circular episomes or genomic integration of linearized DNA by single or double crossover (Kocken et al., 2002; Ozwara et al., 2003). The adaptation of Plasmodium knowlesi to culture using human red-blood cells in 2013 was a key advancement for non-falciparum malaria research (Moon et al., 2013). The need for expensive animal infections or rhesus blood was eliminated, facilitating parasite maintenance and allowing the use of selective agents potentially toxic to animal models. The authors also presented results of parasite transfections, showing fast and highly effective development of transgenic parasites, using GFP as reporter gene and human dhfr (hdhfr) as selective marker (Moon et al., 2013).
One of the major challenges for Plasmodium transfection have been the use of reliable selection markers. A limited number of drugs can be used as selective agents, especially in vivo, where pyrimethamine is the only drug available to select transgenic parasites in non-human primates. The first paper to address this issue was published in 2004 (Wel et al., 2004), when the authors evaluated the sensitivity of P. knowlesi to drugs previously used to select transgenic P. falciparum: Blasticidin S, G418 and WR99210. The genes that confer resistance to these drugs are blasticidin S deaminase (bsd), neomycin phosphotransferase II (neo) and human dihydrofolate reductase (hdhfr), respectively. The authors concluded that these genes can be used as positive selective markers for transfected parasites. However, drug concentrations of Blasticidin necessary to inhibit growth of P. knowlesi in vitro were 20-fold higher than the ones used to select transgenic P. falciparum, limiting the use of this drug (Wel et al., 2004). Several years later, in 2017, a study presented the antiplasmodial activity against P. knowlesi and P. falciparum of three compounds used for transgenic parasite selection in vitro: Blasticidin, the antifolate WR99210 and DSM-1. Results confirmed that P. knowlesi is naturally less sensitive to BSD than P. falciparum, and also showed that DSM-1 is required in a concentration at least 3-fold higher to inhibit P. knowlesi growth when compared to P. falciparum (van Schalkwyk et al., 2017).
Different methods can be used to genetic transform Plasmodium parasites, such as electroporation of parasitized red-blood cells or transformation by spontaneous plasmid uptake by P. falciparum after invasion of electroporated uninfected red-blood cells (Deitsch et al., 2001). This technique was applied to P. knowlesi in 2017, providing a convenient and unexpensive method to generate transgenic parasites. The study also showed that late-stage schizonts are mandatory for high electroporation efficiency of parasitized red-blood cells (Moraes Barros et al., 2017).
Genome editing of P. knowlesi using the CRISPR/Cas9 technology was achieved in 2019 (Mohring et al., 2019). The authors tested two different strategies to deliver the donor sequence for homologous recombination. Parasites were transfected with one plasmid harboring the genes encoding Cas9, the sgRNA, and the selective markers. The donor sequences were provided in a second plasmid or a linear DNA construct, obtained by PCR. Both approaches successfully generated transgenic parasites with high efficiency, with stable lines observed in 12 days. The use of linear PCR constructs as donor sequences is a fast and reliable method for gene tagging and knockout. The group also defined optimal parameters and limits to the CRISPR/Cas9 technique in P. knowlesi. Observing integration rate and parasite culture recovery period after drug pressure, results suggest that the optimal size for homology regions is between 200 bp and 800 bp. As for the distance between the homology regions and the double strand break site, integrations efficiency varies depending on the size of the homologous region - larger homologous regions allow more distant DSB sites. So larger constructs require larger homologous sequences for successful DSB repair. The authors concluded that the biggest bottleneck in the CRISPR/Cas9 system is the parasite’s homologous recombination mediated DNA repair system - as the majority of Cas9 induced DSBs are not successfully repaired (Mohring et al., 2019).
Recently, P. knowlesi parasites were genetic modified in vitro using circular plasmids containing P. vivax centromeric repeats. The presence of centromeric sequences of P. vivax provided higher stability to the episomes, allowing faster selection of transgenic parasites and the maintenance of the plasmid throughout the complete life cycle, in vitro and in vivo. The paper also presented for the first time that P. vivax regulatory sequences not recognized by P. falciparum are recognized by P. knowlesi, providing an in vitro model for P. vivax genetic studies. A transgenic line developed in this work expresses the reporter gene nanoluc and can be used for drug activity studies against blood stage and liver stage parasites and transmission blocking studies (Barros et al., 2021).
Plasmodium cynomolgi
Transfection of P. cynomolgi was reported for the first time in 1999, using parameters and DNA constructs similar to the ones used to transform P. knowlesi in 1997 (Kocken et al., 1999). After this first report, no paper was published presenting results of P. cynomolgi transformation until 2013, when this species was transformed using Plasmodium Artificial Chromosomes (PACs) (Voorberg-van der Wel et al., 2013).
In 2010, Iwanaga et al. (2010) identified and characterized the Plasmodium centromere and constructed a circular P. berghei artificial centromere (PAC) containing the new-found centromere sequences and also a linear P. berghei artificial centromere (L-PAC), containing the centromeric and telomeric repeats. Unlike transfections with circular episomal DNA, transfections with PAC are capable of stable segregation without drug selective pressure, remaining stable throughout the parasite’s life cycle (Iwanaga et al., 2010; Barale and Ménard, 2010). PACs containing P. cynomolgi centromeric repeats were designed in 2013 to genetic modify P. cynomolgi parasites in vivo, allowing the development of a transgenic line expressing reporter gene (GFP) throughout the parasite’s life cycle, including hypnozoites. This transgenic line can be used to analyze reactivation conditions and for high throughput drug screening against liver stage parasites (Voorberg-van der Wel et al., 2013). PACs containing the GFP gene under control of the heat shock 70 gene (pchsp70) promoter sequence were used to transform P. cynomolgi in 2020, allowing real time visualization of hypnozoite reactivation (Voorberg-van der Wel et al., 2020). In addition to that, a second reporter gene (mCherry) was added to the PAC, under transcriptional control of the schizont-specific promoter sequence of the liver-specific protein 2 (lisp2) gene, highlighting the parasites that had initiated schizogony (Voorberg-van der Wel et al., 2020). To this date, all P. cynomolgi transfections were performed in vivo, using macaque infections and pyrimethamine as selection agent (Kocken et al., 1999; Voorberg-van der Wel et al., 2013; Voorberg-van der Wel et al., 2020).
Plasmodium vivax
The first transfection experiments of P. vivax were reported in 2005, transient transfection of parasites with episomal plasmids containing the luciferase reporter gene and P. falciparum regulatory sequences (Pfahler et al., 2006). No advancements were made in P. vivax transfection until 2015, when the genome of this species was manipulated for the first time, in vivo (Barros et al., 2015). Using the genome editing technique of Zinc-Finger Nucleases, that creates DNA double strand-breaks at specific sites, the authors were capable to insert point mutations in the P. vivax dhfr-ts gene, transforming parasites sensitive to pyrimethamine in resistant to this drug. This was the first report of use of genome editing techniques in non-falciparum Plasmodium, demonstrating the possibility of site-specific genome editing (Barros et al., 2015). The experiments were performed in vivo, using Saimiri boliviensis monkeys. The resistant/transfected parasites were selected for three weeks in animals treated with pyrimethamine in a method similar to the one used to select transgenic P. knowlesi (Van Der Wel et al., 1997) and P. cynomolgi in vivo, using rhesus macaques (Kocken et al., 1999; Voorberg-van der Wel et al., 2013; Voorberg-van der Wel et al., 2020).
Conclusions
Plasmodium vivax, P. malariae, P. ovale, P. knowlesi and P. cynomolgi are phylogenetically grouped together in the Primate malaria clade, evolutionary distant from P. falciparum, grouped with P. reichenowi and P. gabonii in the Laverania malaria clade (Loy et al., 2017). The large evolutionary distance between P. falciparum and the other human malaria parasites helps to explain the biological differences in the species, and makes mandatory specific genetic studies for non-falciparum human malaria species. Genetic manipulation is key to elucidate different aspects of these parasites’ biology, and these studies have been neglected for several years.
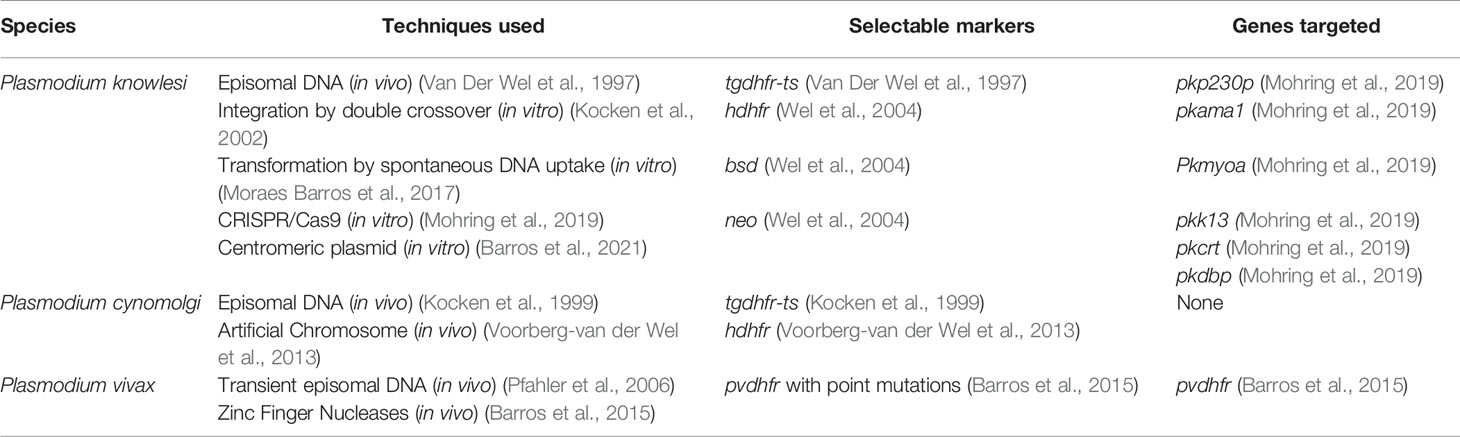
Transfection of non-falciparum human Plasmodium was reported for the first time in 1997, and since then only 12 papers were published describing new results using this technology. More than 20 years after the first transfection of P. knowlesi we have barely scratched the surface of what can be performed using these techniques. Little is known about specific processes of these parasites, such as red blood cell invasion, liver stage development and gametocyte biology. The first paper to use genetic modification to study specific features of non-falciparum human malaria parasites was published in 2019 when the authors used CRISPR/Cas9 to edit the duffy-binding protein (pkdbp-1) gene and were able to evaluate its role in red-blood cell invasion (Mohring et al., 2019). The Table 1 summarizes the techniques used, selectable markers and genes targeted.
Plasmodium knowlesi seems to be a formidable research model for members of the Primate Malarias clade. This species can be maintained in vitro, with a 24-hour assexual life cycle that provides rapid growth and fast generation of transgenic parasites. In vitro cultivated parasites can be used for blood stage, liver stage and transmission studies without the need of animal models (Grüring et al., 2014; Armistead et al., 2018). Furthermore, this parasite recognizes regulatory sequences such as transcriptional promoters and centromeric repeats of P. vivax, P. falciparum and P. berghei, allowing comparative genetic studies of different malaria species (Ozwara et al., 2003; Pasini et al., 2016; Barros et al., 2021).
The biggest bottleneck to non-falciparum malaria studies was the need for non-human primate infections and blood, but the development of long term in vitro culture of P. knowlesi using human red-blood cells changed the landscape (Moon et al., 2013). The recent development of P. cynomolgi in vitro culture is an exciting achievement (Chua et al., 2019), allowing specific studies of liver stage parasites, specially hipnozoites, that are not observed in P. falciparum and P. knowlesi, the other species that can be cultivated in vitro. Laboratories across the globe will also benefit from the success of new technologies that increase the efficiency of genetic transformation, such as artificial chromosomes and genome editing techniques. A second challenge that must be targeted for the expansion of Plasmodium transfection studies is the development of new selection markers, limited to antifolates. CRISPR derived techniques facilitate genetic knockout and knockdown and should be used to generate these transgenic lines. However, to improve the methods of genetic modification and create platforms of consecutive knockdowns of functional genes, new selection markers are necessary.
In the next few years, we hope that different genes and pathways will be the targets of genetic modification in non-falciparum malaria parasites, allowing a better comprehension of specific features of the biology of these parasites, leading to the development of better mechanisms of control and treatment of non-falciparum human malaria.
Author Contributions
RRMB has contributed designing and writing the manuscript. TBV and TPA have contributed writing this mini-review. All authors contributed and approved the submitted version.
Funding
This work has been supported by FAPESP grants JP 2018/06219-8 (RRMB), 2019/07223-1 (TPA), 2020/02303-4 (TBV) and CNPq 434011/2018-5 (RRMB).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Armistead, J. S., Moraes Barros, R. R., Gibson, T. J., Kite, W. A., Mershon, J. P., Lambert, L. E., et al. (2018). Infection of Mosquitoes From In Vitro Cultivated Plasmodium Knowlesi H Strain. Int. J. Parasitol. 48 (8), 601–610. doi: 10.1016/j.ijpara.2018.02.004
Barale, J. C., Ménard, R. (2010). Centromeric Plasmids and Artificial Chromosomes: New Kids on the Plasmodium Transfection Block. Cell Host Microbe 7 (3), 181–183. doi: 10.1016/j.chom.2010.03.002
Barros, R. R. M., Straimer, J., Sa, J. M., Salzman, R. E., Melendez-Muniz, V. A., Mu, J., et al. (2015). Editing the Plasmodium Vivax Genome, Using Zinc-Finger Nucleases. J. Infect. Dis. 211 (1), 125–129. doi: 10.1093/infdis/jiu423
Barros, R. R. M., Thawnashom, K., Gibson, T. J., Armistead, J. S., Caleon, R. L., Kaneko, M., et al. (2021). Activity of Plasmodium Vivax Promoter Elements in Plasmodium Knowlesi, and a Centromere-Containing Plasmid That Expresses NanoLuc Throughout the Parasite Life Cycle. Malar J. 20 (1), 247. doi: 10.1186/s12936-021-03773-4
Butcher, G. A., Mitchell, G. H. (2016). The Role of Plasmodium Knowlesi in the History of Malaria Research. Parasitology 145, 1–12. doi: 10.1017/S0031182016001888
Carlton, J. M., Sina, B. J., Adams, J. H. (2011). Why Is Plasmodium Vivax a Neglected Tropical Disease? PloS Negl. Trop. Dis. 5 (6), e1160. doi: 10.1371/journal.pntd.0001160
Chua, A. C. Y., Ong, J. J. Y., Malleret, B., Suwanarusk, R., Kosaisavee, V., Zeeman, A. M., et al. (2019). Robust Continuous In Vitro Culture of the Plasmodium Cynomolgi Erythrocytic Stages. Nat. Commun. 10 (1), 3635. doi: 10.1038/s41467-019-11332-4
Crabb, B. S., Cowman, A. F. (1996). Characterization of Promoters and Stable Transfection by Homologous and Nonhomologous Recombination in Plasmodium Falciparum. Proc. Natl. Acad. Sci. U. S. A. 93 (14), 7289–7294. doi: 10.1073/pnas.93.14.7289
Deitsch, K. W., Driskill, C. L., Wellems, T. E. (2001). Transformation of Malaria Parasites by the Spontaneous Uptake and Expression of DNA From Human Erythrocytes. Nucleic Acids Res 29, 850–853. doi: 10.1093/nar/29.3.850
Eid, J., Sollner-Webb, B. (1987). Efficient Introduction of Plasmid DNA Into Trypanosoma Brucei and Transcription of a Transfected Chimeric Gene. Proc. Natl. Acad. Sci. U. S. A. 84, 7812–7816. doi: 10.1073/pnas.84.22.7812
Ghorbal, M., Gorman, M., MacPherson, C. R., Martins, R. M., Scherf, A., Lopez-Rubio, J. J. (2014). Genome Editing in the Human Malaria Parasite Plasmodium Falciparum Using the CRISPR-Cas9 System. Nat. Biotechnol. 32 (8), 819–821. doi: 10.1038/nbt.2925
Goonewardene, R., Daily, J., Kaslow, D., Sullivan, T. J., Duffy, P., Carter, R., et al. (1993). Transfection of the Malaria Parasite and Expression of Firefly Luciferase. Proc. Natl. Acad. Sci. U. S. A. 90 (11), 5234–5236. doi: 10.1073/pnas.90.11.5234
Grüring, C., Moon, R. W., Lim, C., Holder, A. A., Blackman, M. J., Duraisingh, M. T. (2014). Human Red Blood Cell-Adapted Plasmodium Knowlesi Parasites: A New Model System for Malaria Research. Cell Microbiol. 16 (5), 612–620. doi: 10.1111/cmi.12275
Hotchkiss, R. D. (1955). Bacterial Transformation. J. Cell. Physiol. Suppl. 45 (Suppl. 2), 1–22. doi: 10.1002/jcp.1030450503
Iwanaga, S., Khan, S. M., Kaneko, I., Christodoulou, Z., Newbold, C., Yuda, M., et al. (2010). Functional Identification of the Plasmodium Centromere and Generation of a Plasmodium Artificial Chromosome. Cell Host Microbe 7 (3), 245–255. doi: 10.1016/j.chom.2010.02.010
Kocken, C. H. M., Ozwara, H., van der Wel, A., Beetsma, A. L., Mwenda, J. M., Thomas, A. W. (2002). Plasmodium Knowlesi Provides a Rapid In Vitro and In Vivo Transfection System That Enables Double-Crossover Gene Knockout Studies. Infect. Immun. 70 (2), 655–660. doi: 10.1128/IAI.70.2.655-660.2002
Kocken, C. H. M., van der Wel, A., Thomas, A. W. (1999). Plasmodium Cynomolgi: Transfection of Blood-Stage Parasites Using Heterologous DNA Constructs. Exp. Parasitol 93 (1), 58–60. doi: 10.1006/expr.1999.4430
Lee, M. C. S., Fidock, D. A. (2014). CRISPR-Mediated Genome Editing of Plasmodium Falciparum Malaria Parasites. Genome Med. 6 (8), 63. doi: 10.1186/s13073-014-0063-9
Loy, D. E., Liu, W., Li, Y., Learn, G. H., Plenderleith, L. J., Sundararaman, S. A., et al. (2017). Out of Africa: Origins and Evolution of the Human Malaria Parasites Plasmodium Falciparum and Plasmodium Vivax. Int. J. Parasitol 47, 87–97. doi: 10.1016/j.ijpara.2016.05.008
Mohring, F., Hart, M. N., Rawlinson, T. A., Henrici, R., Charleston, J. A., Diez Benavente, E., et al. (2019). Rapid and Iterative Genome Editing in the Malaria Parasite Plasmodium Knowlesi Provides New Tools for P. Vivax Research. Elife 8, 1–29. doi: 10.7554/eLife.45829
Moon, R. W., Hall, J., Rangkuti, F., Ho, Y. S., Almond, N., Mitchell, G. H., et al. (2013). Adaptation of the Genetically Tractable Malaria Pathogen Plasmodium Knowlesi to Continuous Culture in Human Erythrocytes. Proc. Natl. Acad. Sci. 110 (2), 531–536. doi: 10.1073/pnas.1216457110
Moraes Barros, R. R., Gibson, T. J., Kite, W. A., Sá, J. M., Wellems, T. E. (2017). Comparison of Two Methods for Transformation of Plasmodium Knowlesi: Direct Schizont Electroporation and Spontaneous Plasmid Uptake From Plasmid-Loaded Red Blood Cells. Mol. Biochem. Parasitol 218, 16–22. doi: 10.1016/j.molbiopara.2017.10.001
Mota, M. M., Thathy, V., Nussenzweig, R. S., Nussenzweig, V. (2001). Gene Targeting in the Rodent Malaria Parasite Plasmodium Yoelii. Mol. Biochem. Parasitol. 113 (2), 271–278. doi: 10.1016/s0166-6851(01)00228-6
Neumann, E., Schaefer-Ridder, M., Wang, Y., Hofschneider, P. H. (1982). Gene Transfer Into Mouse Lyoma Cells by Electroporation in High Electric Fields. EMBO J 1, 841–845. doi: 10.1002/j.1460-2075.1982.tb01257.x
Ozwara, H., van der Wel, A., Kocken, C. H. M., Thomas, A. W. (2003). Heterologous Promoter Activity in Stable and Transient Plasmodium Knowlesi Transgenes. Mol. Biochem. Parasitol 130 (1), 61–64. doi: 10.1016/S0166-6851(03)00141-5
Pasini, E. M., Zeeman, A. M., Voorberg-van der Wel, A., Kocken, C. H. M. (2016). Plasmodium Knowlesi: A Relevant, Versatile Experimental Malaria Model. Parasitology 145, 1–15. doi: 10.1017/S0031182016002286
Pfahler, J. M., Galinski, M. R., Barnwell, J. W., Lanzer, M. (2006). Transient Transfection of Plasmodium Vivax Blood Stage Parasites. Mol. Biochem. Parasitol 149 (1), 99–101. doi: 10.1016/j.molbiopara.2006.03.018
Reece, S. E., Thompson, J. (2008). Transformation of the Rodent Malaria Parasite Plasmodium Chabaudi and Generation of a Stable Fluorescent Line PcGFPCON. Malar. J. 7, 183.
Stocker, B. A. (1963). Transformation of Bacillus Subtilis To Motility and Prototrophy. J. Bacteriol. 86, 797–804. doi: 10.1128/jb.86.4.797-804.1963
Straimer, J., Lee, M. C. S., Lee, A. H., Zeitler, B., Williams, A. E., Pearl, J. R., et al. (2012). Site-Specific Genome Editing in Plasmodium Falciparum. Using Engineered Zinc-Finger Nucleases. Nat. Methods 9 (10), 993–998. doi: 10.1038/nmeth.2143
Van Der Wel, A. M., Tomás, A. M., Kocken, C. H. M., Malhotra, P., Janse, C. J., Waters, A. P., et al. (1997). Transfection of the Primate Malaria Parasite Plasmodium Knowlesi Using Entirely Heterologous Constructs. J. Exp. Med. 185 (8), 1499–1503. doi: 10.1084/jem.185.8.1499
van Dijk, M. R., Waters, A. P., Janse, C. J. (1995). Stable Transfection of Malaria Parasite Blood Stages. Science 268 (5215), 1358–1362. doi: 10.1084/jem.185.8.1499.
van Schalkwyk, D. A., Moon, R. W., Blasco, B., Sutherland, C. J. (2017). Comparison of the Susceptibility of Plasmodium Knowlesi and Plasmodium Falciparum to Antimalarial Agents. J. Antimicrob. Chemother. 72, 3051–3058. doi: 10.1093/jac/dkx279
Voorberg-van der Wel, A. M., Zeeman, A. M., Nieuwenhuis, I. G., van der Werff, N. M., Klooster, E. J., Klop, O., et al. (2020). A Dual Fluorescent Plasmodium Cynomolgi Reporter Line Reveals In Vitro Malaria Hypnozoite Reactivation. Commun. Biol. 1, 1–9. doi: 10.1038/s42003-019-0737-3
Voorberg-van der Wel, A., Zeeman, A. M., van Amsterdam, S. M., van den Berg, A., Klooster, E. J., Iwanaga, S., et al. (2013). Transgenic Fluorescent Plasmodium Cynomolgi Liver Stages Enable Live Imaging and Purification of Malaria Hypnozoite-Forms. PloS One 8 (1), e54888. doi: 10.1371/journal.pone.0054888
Wagner, J. C., Platt, R. J., Goldfless, S. J., Zhang, F., Niles, J. C. (2014). Efficient CRISPR-Cas9-Mediated Genome Editing in Plasmodium Falciparum. Nat. Methods 11 (9), 915–918. doi: 10.1038/nmeth.3063
Wel, A. V. D., Kocken, C. H. M., Pronk, T. C., Franke-Fayard, B., Thomas, A. W. (2004). New Selectable Markers and Single Crossover Integration for the Highly Versatile Plasmodium Knowlesi Transfection System. Mol. Biochem. Parasitol 134 (1), 97–104. doi: 10.1016/j.molbiopara.2003.10.019
Wu, Y., Sifri, C. D., Lei, H. H., Su, X. Z., Wellems, T. E. (1995). Transfection of Plasmodium Falciparum Within Human Red Blood Cells. Proc. Natl. Acad. Sci. U. S. A. 92 (4), 973–977. doi: 10.1073/pnas.92.4.973
Keywords: transfection, reporter genes, drug selection, Plasmodium knowlesi, Plasmodium vivax, Plasmodium cynomolgi
Citation: Vieira TB, Astro TP and Moraes Barros RRd (2021) Genetic Manipulation of Non-Falciparum Human Malaria Parasites. Front. Cell. Infect. Microbiol. 11:680460. doi: 10.3389/fcimb.2021.680460
Received: 14 March 2021; Accepted: 11 August 2021;
Published: 30 August 2021.
Edited by:
Bruce Malcolm Russell, University of Otago, New ZealandReviewed by:
Hassan Hakimi, Nagasaki University, JapanShaojun Long, China Agricultural University, China
Copyright © 2021 Vieira, Astro and Moraes Barros. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Rudge de Moraes Barros, bW9yYWVzLmJhcnJvc0B1bmlmZXNwLmJy
 Taís Baruel Vieira
Taís Baruel Vieira Thafne Plastina Astro
Thafne Plastina Astro Roberto Rudge de Moraes Barros
Roberto Rudge de Moraes Barros