
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 19 August 2021
Sec. Parasite and Host
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.666469
This article is part of the Research TopicInfection and Control of Vector-Borne DiseasesView all 34 articles
 Jianhai Yin1
Jianhai Yin1 Frederick Yamba2
Frederick Yamba2 Canjun Zheng3
Canjun Zheng3 Shuisen Zhou1
Shuisen Zhou1 Samuel Juana Smith2*
Samuel Juana Smith2* Lili Wang4
Lili Wang4 Hongmei Li1
Hongmei Li1 Zhigui Xia1
Zhigui Xia1 Ning Xiao1*
Ning Xiao1*Vector control interventions including long-lasting insecticidal nets and indoor residual spraying are important for malaria control and elimination. And effectiveness of these interventions depends entirely on the high level of susceptibility of malaria vectors to insecticides. However, the insecticide resistance in majority of mosquito vector species across African countries is a serious threat to the success of vector control efforts with the extensive use of insecticides, while no data on insecticide resistance was reported from Sierra Leone in the past decade. In the present study, the polymerase chain reaction was applied for the identification of species of 757 dry adult female Anopheles gambiae mosquitoes reared from larvae collected from four districts in Sierra Leone during May and June 2018. And the mutations of kdr, rdl, ace-1 genes in An. gambiae were detected using SNaPshot and sequencing. As a result, one sample from Western Area Rural district belonged to Anopheles melas, and 748 An. gambiae were identified. Furthermore, the rdl mutations, kdr west mutations and ace-1 mutation were found. The overall frequency was 35.7%, 0.3%, 97.6% and 4.5% in A296G rdl, A296S rdl, kdrW and ace-1, respectively. The frequencies of A296G rdl mutation (P < 0.001), kdrW mutation (P = 0.001) and ace-1 mutation (P < 0.001) were unevenly distributed in four districts, respectively, while no statistical significance was found in A296S rdl mutation (P = 0.868). In addition, multiple resistance patterns were also found. In conclusion, multiple mutations involved in insecticide resistance in An. gambiae populations in Sierra Leone were detected in the kdrW, A296G rdl and ace-1 alleles in the present study. It is necessary to monitor vector susceptibility levels to insecticides used in this country, and update the insecticide resistance monitoring and management strategy.
Malaria is one of the most widespread infectious diseases globally, with a major burden causing the death of more than 400,000 people yearly in the past three years, most of them were reported in the WHO African Region and children aged under five years (WHO, 2018; WHO, 2019; WHO, 2020). Sierra Leone is an area of stable malaria endemicity. The high malaria disease burden in this country accounted for approximately 48% of outpatient morbidity and approximately 38% of mortality in children under five years according to the national Malaria Indicator Survey 2016 (National Malaria Control Programme (NMCP) et al., 2016).
Vector control is an essential component of malaria control and elimination. At present, long-lasting insecticidal nets and indoor residual spraying are still two core interventions for malaria vector control. The effectiveness of these interventions depends entirely on the high level of susceptibility of malaria vectors to the insecticides. However, the insecticide resistance in majority of mosquito vector species across African countries is a serious threat to the success of vector control efforts with the extensive use of insecticides (https://anopheles.irmapper.com/).
Target-site resistance is one of the most main mechanisms of insecticide resistance. Alterations in the target sites can reduce sensitivity of the target receptors to insecticide. Four main chemical classes (organochlorines, organophosphates, carbamates, and pyrethroids) are used in the vector control, and only pyrethroids are for use on nets. Moreover, mutations in the ace-1 gene encoding the target site of acetylcholinesterase (AChE) for organophosphate and carbamate insecticides (Weill et al., 2003; Weill et al., 2004; Fournier, 2005; Djogbenou et al., 2008), the rdl (resistance to dieldrin) gene (Ffrench-Constant, 1994; Du et al., 2005), and the amino acid sequence in the voltage-gated sodium channel referred to as knockdown resistance (kdr) (resistance to DDT and pyrethroids) (Martinez Torres et al., 1998; Ranson et al., 2000; Davies et al., 2007; Donnelly et al., 2009), have been found in Anopheles gambiae, which is a main malaria vector in Africa. For example, pyrethroid resistance is high in An. gambiae in West Africa including Benin, Burkina Faso, Ghana, Mali, Nigeria, etc. (Vincent and N’Guessan, 2013), while no data was reported from Sierra Leone in the past decade.
Moreover, strategies for insecticide resistance management in vector control must be implemented on the basis of local epidemiological and entomological data. Hence, bioassays using WHO defined diagnostic doses of insecticide, dose response bioassays, biochemical assays to detect activity of enzymes associated with insecticide resistance, and molecular assays to detect known resistance alleles, have been developed to facilitate the implementation of vector control interventions (Ranson et al., 2011; Vincent and N’Guessan, 2013). Among them, the molecular assays are very sensitive, and they can detect resistant alleles at low frequencies at an early stage of resistance development, which, therefore, provide an early warning of future resistance.
In addition, the World Health Organization (WHO) supported the Sierra Leone National Malaria Control Program (NMCP) to develop an Insecticide Resistance Monitoring and Management Program (IRMMP) 2017-2020 to conduct insecticide resistance monitoring in the four pilot districts - Bo, Bombali, Kono and Western Area Rural district, in order to maintain the effectiveness of existing insecticidal vector control interventions, adhering to the existing malaria strategic plan and linking with other specific implementation documents of the NMCP and the Ministry of Health and Sanitation (MOHS), Sierra Leone.
In this study, molecular detection of mutations in the three genes of ace-1, rdl and kdr were performed to understand the insecticide resistance of the female An. gambiae to the four classes of insecticides recommended by WHO for vector control especially the pyrethroids in Sierra Leone.
Adult female Anopheles gambiae mosquitoes were stored in the 1.5 ml Eppendorf tubes with cottons and silico gels (one mosquito per tube). They were reared from larvae, which were collected from natural breeding water bodies in Kayangba community from Bo district, Magbema community from Bombali district, Lebanon community from Kono district, and Bolima community from Western Area Rural district, respectively, during May and June 2018. And these four districts were selected into a four-year insecticide resistance monitoring and management program (2017-2020) which was supported by WHO to the National Malaria Control Program, Sierra Leone. Upon collection, larvae were kept in separate labelled buckets, transported to the insectary and maintained under optimal condition for mosquitoes rearing.
All adult females used for molecular characterization were morphologically identified as belonging to the An. gambiae complex. Genomic DNA was extracted from one mosquito respectively using the Qiagen DNA Mini Kit (Qiagen, Germany) according to the manufacturer’s instruction. The identification of single specimens of the An. gambiae complex was performed by a PCR (Scott et al., 1993).
Three pairs of primers to detect kdr west and kdr east mutation, rdl Ala296Gly and rdl Ala296Ser mutation, and ace-1 G119S mutation were used (Table 1) (Bass et al., 2007; Bass et al., 2015a; Bass et al., 2015b). Multiplex PCR was performed in a volume of 20 μl containing 1x GC-I buffer (Takara, Japan), 3.0 mM Mg2+ (Takara, Japan), 0.3 mM dNTP (Generay Biotech, Shanghai), 1 U Hot Star Taq polymerase (Qiagen, Germany), 1 µl of template DNA and 1 µl of primers (Sangon Biotech, Shanghai) (Table 1). Thermal cycler conditions were: 95°C for 2 min; 11 cycles of 94°C for 20 s, 65°C for 40 s, and 72°C for 1.5 min; 24 cycles of 94°C for 20 s, 59°C for 30 s, and 72°C for 1.5 min; and finally 2 min at 72°C. Multiplex PCR products were separated by electrophoresis on 2% agarose gels stained with ethidium bromide (Sangon Biotech, Shanghai). The remaining PCR products were purified with 5 units of shrimp alkaline phosphatase (SAP) (Promega, USA) and 2 units of exonuclease I (EXO I) (Epicentre, USA) at 37°C for 1 h and 75°C for 15 min, to remove excess dNTPs and primers, respectively.

Table 1 Multiplex PCR primers for detection of multiple insecticide resistance genes in An. gambiae.
SNaPshot extension primers for detection of kdr west (L1014F) and kdr east (L1014S) mutations, A296G rdl (Ala-Gly) and A296S rdl (A296S) mutations, and G119S ace-1 (Gly-Ser) mutation were designed (Table 2) to anneal on the sense strand immediately adjacent to the mutation site. Each extension primer was synthesized with a different length of poly (dT) tail to allow separation of SNaPshot products on the basis of size. SNaPshot analysis was performed using an Applied Biosystems SNaPshot Multiplex Kit (Applied Biosystems Co., Ltd., USA). Extension reactions were performed in a volume of 10 μl containing 5 μl of SNaPshot Ready Multiplex Ready Reaction Mix, 1 μl of extension primer mix (Table 2) and 2 μl of ddH2o. Thermal cycler conditions were: 96°C for 1 min; 28 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 30 s. Each 10 μl of extension products were purified with 1 unit of SAP at 37°C for 1 h and 75°C for 15 min. Then, 0.5 μl of the purified extension products were mixed with 0.5 μl of Liz120 size standard, and 9 μl of Hi-Di™ formamide, and were denatured at 95°C for 5 minutes then sequenced using ABI3730XL. Analysis was performed using GeneMapper 4.1 (Applied Biosystems Co., Ltd., USA).

Table 2 SNaPshot extension primers for the detection of multiple insecticide resistance genes mutations in An. gambiae.
Data was entered in Microsoft Excel 2010 and analyzed using IBM SPSS Statistic 20. Frequency counts for mutations of insecticide resistance genes in mosquitoes from different districts were compared using Pearson Chi-Square test or Fisher’s exact test performed at 0.05 level of significance.
A total of 757 adult anopheline mosquitoes reared from larvae were analyzed in the present study. In detail, 271, 253, 112, and 121 mosquitoes were from Kayangba community, Magbema community, Lebanon community, and Bolima community, respectively.
Moreover, a total of 8 samples from Kayangba (1), Magbema (1), and Bolima (6) were detected as negative results of the An. gambiae complex by PCR (Scott et al., 1993), and one sample belonged to Anopheles melas, the remaining 748 samples were An. gambiae (Table 3).
A total of 748 adult An. gambiae were analyzed for presence of the rdl mutations, kdr mutations and G119S ace-1 mutation (Table 4). A296G rdl mutation, kdrW mutation and ace-1 mutation were identified at all 4 geographical sites, while A296S rdl mutation was only found in the samples from Kayangba and Magbema communities, and no kdrE mutation was found at all 4 sites. The overall mutation frequency was 35.7%, 0.3%, 0.0%, 97.6% and 4.5% in A296G rdl, A296S rdl, kdrE, kdrW and ace-1, respectively (Table 4). Moreover, the frequencies of A296G rdl mutation (χ2 = 377.148, df = 3, P < 0.001), kdrW mutation (Fisher’s exact test, P = 0.001) and ace-1 mutation (χ2 = 42.989, df = 3, P < 0.001) were significantly different among four districts, respectively, while no statistical significance was found in A296S rdl mutation (Fisher’s exact test, P = 0.868). In addition, there were 12 types of allelic combinations totally including the sites that test failed, and a simple homozygous resistant mutation of kdrW (319), or combined homozygous resistant mutation of A296G rdl (174), or combined heterozygous mutation of A296G rdl (154), were the three most common combinations (Table 5). And four samples are provided as examples to show the results of sequencing (Figure 1).
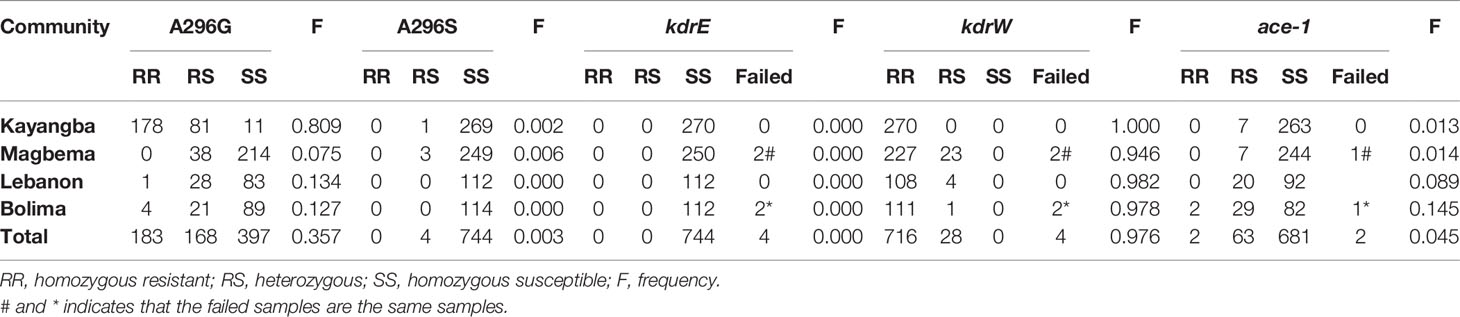
Table 4 Genotypes of multiple insecticide resistance genes and their mutation frequency in the An. gambiae.
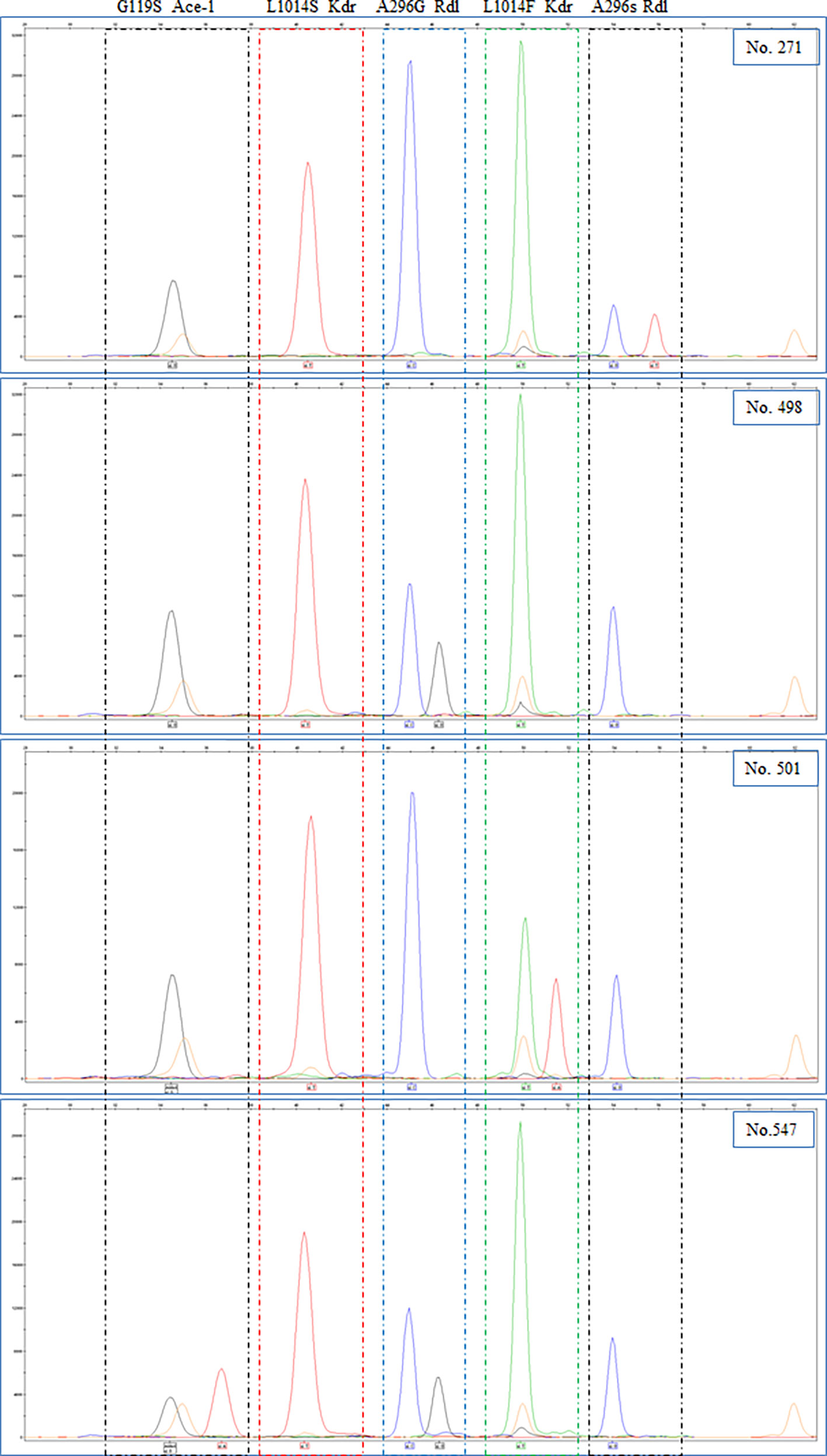
Figure 1 Electropherograms of genscan analysis of the SNaPshot reaction and alleles were indicated at the bottom. (Blue peak: G; Black peak: C; Green peak: A; Red peak: T; Orange peak: size standard).
This study has analyzed the species composition in the An. gambiae complex and the mutations of the multiple insecticide resistance genes in An. gambiae in Sierra Leone, 2018. Adult female mosquitoes were reared from larvae, which were collected from several identified water bodies in four pilot districts for insecticide resistance monitoring in Sierra Leone. And only An. gambiae (99.9%, 748/749) and An. melas (0.1%, 1/749) were characterized in the present study, which is consistent with Sierra Leone’s records on the species distribution of An. gambiae complex (National Malaria Control Programme, 2015).
Key to the success of malaria control and elimination is a strategic plan on malaria vector control informed by comprehensive monitoring and evaluation of insecticide resistance, this is becoming more important as insecticide resistance increases and spreads across Africa, but there was absence of insecticide resistance monitoring and entomological work in general in the presence of large scale use of insecticides for malaria control in Sierra Leone. Therefore, it is necessary to carry out work in this area to promote the malaria control and elimination in this country.
Currently most resistance monitoring is dependent on bioassays which detect resistance under defined insecticide concentrations and exposure times recommended by WHO, but these methods require large numbers of alive mosquitoes and lack sensitivity (Ranson et al., 2011; Vincent and N’Guessan, 2013). And several alternative methods for detecting resistance have been developed, especially molecular tests based on DNA providing an early warning of emergence of the resistance are available for target sites with high sensitivity, although they are just a complement rather than a substitute for bioassays. And PCR or an intentional mismatched primer - PCR (IMP - PCR) followed by agarose gel electrophresis, or real - time TaqMan assays have typically been applied to the characterization of allelic mutations of the single insecticide resistance genes, respectively (Bass et al., 2010; Donnelly et al., 2016; Mavridis et al., 2018). Although reliable, these are time consuming with large numbers of samples for detection of multiple resistance mutations. The SNaPshot method used in the present study offers a specific, sensitive, inexpensive and rapid alternative to the simultaneous screening for multiple mutations and subsequent confirmation by sequencing (Quintans et al., 2004; Alina et al., 2005; Fondevila et al., 2017). This method has been extensively used in the blood group typing (Palacajornsuk et al., 2009; Latini et al., 2014; Chen et al., 2019) and common mutations in genes related to some cancers (Filippini et al., 2007; Hurst et al., 2009; Fariña Sarasqueta et al., 2011).
With regard to the frequency of mutations, a high mutation frequency of kdrW (97.6%) was found totally and most of them were homozygous (Table 4), and distributed in all four pilot sites. It supports the findings that pyrethroid resistance is predominant in An. gambiae s.s. in West Africa (Vincent and N’Guessan, 2013). Moreover, the kdrW mutation combined with one to two mutations at other three sites (A296G rdl, A296S rdl, ace-1) were found (Table 5), it revealed for the first time in Sierra Leone to our knowledge that multiple resistance patterns existed. However, standardized bioassays are still needed to test the resistance phenotype in individuals and combined with the molecular tests, to understand the real resistance status in this country. For example, there was a report that An. gambiae s.l. with the L1014F mutation were still susceptible to pyrethroids in Guinea Bissau (Dabire et al., 2008). Meanwhile, the failure to detect allelic mutations by molecular tests also cannot be interpreted as an absence of resistance in a population, and it reminds us that the detection system used in the present study should be further optimized. In addition, resistance mutations in different genes of A296G rdl, kdrW and ace-1 were unevenly distributed in four pilot sites (Table 4). The mutation in A296G rdl (80.9%) was more frequent in samples from Kayangba community than other three communities. And no mutation was detected in A296S rdl in samples from Lebanon and Bolima communities, and very few samples were heterozygous individuals in the other two sites. Furthermore, the mutation in ace-1 was more frequent in samples from Bolima community than other three communities.
High frequencies of kdrW alleles and uneven frequencies of A296G rdl and ace-1 in An. gambiae populations in Sierra Leone detected in the present study prompts the need for close vector monitoring of susceptibility levels to insecticides used in this country. It provides important information to the Sierra Leone Malaria Control Programme for development of insecticide resistance monitoring and adjusting the strategy to implement in this country.
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding authors.
The project was informed to the study sites, and consent was obtained from the local authorities.
JY, SS, and NX conceived the study. FY and SS conducted sample collection and preparation. JY and FY performed the laboratory works. JY performed the result analysis. JY, CZ, SS, LW, and NX contributed to the study design. JY, SZ, HL, ZX, and NX contributed to experimental materials. JY drafted and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Science and Technology Major Program of China (No. 2018ZX10101002–002), Technical Reserve Programme for Global Tropical Diseases Prevention and Control (No. 131031104000160004), the Fifth Round of Three-Year Public Health Action Plan of Shanghai (No. GWV-10.1-XK13), and Sierra Leone-China Second Phase of the Fixed Biological Safety Laboratory Technical Cooperation Project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We appreciate the Consultant Medical Entomologists, staffs from National Malaria Control Program, Directorate for Environmental Health and Sanitation, Onchocerciasis Control Program, and District Health Management Teams for their wonderful work in the sample collection and other works.
Alina, D., Yolanda, V., John, P. N. (2005). Multiplexed SNP Genotyping Using Primer Single - Base Extension (SBE) and Microsphere Arrays. Curr. Protoc. Cytom. 13, 13–14. doi: 10.1002/0471142956.cy1304s34
Bass, C., Nikou, D., Donnelly, M. J., Williamson, M. S., Ranson, H., Ball, A., et al. (2007). Detection of Knockdown Resistance (Kdr) Mutations in Anopheles Gambiae: A Comparison of Two New High-Throughput Assays With Existing Methods. Malar. J. 6, 111. doi: 10.1186/1475-2875-6-111
Bass, C., Nikou, D., Vontas, J., Donnelly, M. J., Williamson, M. S., Field, L. M. (2010). The Vector Population Monitoring Tool (VPMT): High-Throughput DNA-Based Diagnostics for the Monitoring of Mosquito Vector Populations. Malar. Res. Treat. 2010, 190434. doi: 10.4061/2010/190434
Bass, C., Williamson, M. S., Vontas, J., Ranson, H., Donnelly, M. J., Field, L. M. (2015a). “Insensitive Acetylcholinesterase (Iache) Assay,” in Methods in Anopheles Research, 2015 edition. chapter 8.5.1.4. BEI Resources.
Bass, C., Williamson, M. S., Vontas, J., Ranson, H., Donnelly, M. J., Field, L. M. (2015b). “Resistance to Dieldrin (Rdl) Assay,” in Methods in Anopheles Research, 2015 edition. chapter 8.5.1.5. BEI Resources.
Chen, D. P., Wen, Y. H., Lu, J. J., Tseng, C. P., Wang, W. T. (2019). Rapid Rare ABO Blood Typing Using a Single PCR Based on a Multiplex SNaPshot Reaction. J. Formos. Med. Assoc. 118, 395–400. doi: 10.1016/j.jfma.2018.06.014
Dabire, K. R., Diabate, A., Agostinho, F., Alves, F., Manga, L., Faye, O., et al. (2008). Distribution of the Members of Anopheles Gambiae and Pyrethroid Knock-Down Resistance Gene (Kdr) in Guinea-Bissau, West Africa. Bull. Soc Pathol. Exot. 101 (2), 119–123.
Davies, T. G., Field, L. M., Usherwood, P. N., Williamson, M. S. (2007). DDT, Pyrethrins, Pyrethroids and Insect Sodium Channels. IUBMB Life 59 (3), 151–162. doi: 10.1080/15216540701352042
Djogbenou, L., Chandre, F., Berthomieu, A., Dabire, R., Koffi, A., Alout, H., et al. (2008). Evidence of Introgression of the Ace-1(R) Mutation and of the Ace-1 Duplication in West African Anopheles Gambiae s. s. PloS One 3 (5), e2172. doi: 10.1371/journal.pone.0002172
Donnelly, M. J., Corbel, V., Weetman, D., Wilding, C. S., Williamson, M. S., Black, W. (2009). Does Kdr Genotype Predict Insecticide-Resistance Phenotype in Mosquitoes? Trends Parasitol. 25 (5), 213–219. doi: 10.1016/j.pt.2009.02.007
Donnelly, M. J., Isaacs, A. T., Weetman, D. (2016). Identification, Validation, and Application of Molecular Diagnostics for Insecticide Resistance in Malaria Vectors. Trends Parasitol. 32, 197–206. doi: 10.1016/j.pt.2015.12.001
Du, W., Awolola, T. S., Howell, P., Koekemoer, L. L., Brooke, B. D., Benedict, M. Q., et al. (2005). Independent Mutations in the Rdl Locus Confer Dieldrin Resistance to Anopheles Gambiae and An. Arabiensis. Insect. Mol. Biol. 14 (2), 179–183. doi: 10.1111/j.1365-2583.2005.00544.x
Fariña Sarasqueta, A., Moerland, E., de Bruyne, H., de Graaf, H., Vrancken, T., van Lijnschoten, G., et al. (2011). SNaPshot and StripAssay as Valuable Alternatives to Direct Sequencing for KRAS Mutation Detection in Colon Cancer Routine Diagnostics. J. Mol. Diagn. 13, 199–205. doi: 10.1016/j.jmoldx.2010.10.006
Ffrench-Constant, R. H. (1994). The Molecular and Population Genetics of Cyclodiene Insecticide Resistance. Insect. Biochem. Mol. Biol. 24 (4), 335–345. doi: 10.1016/0965-1748(94)90026-4
Filippini, S., Blanco, A., Fernández-Marmiesse, A., Álvarez-Iglesias, V., Ruíz-Ponte, C., Carracedo, Á., et al. (2007). Multiplex SNaPshot for Detection of BRCA1/2 Common Mutations in Spanish and Spanish Related Breast/Ovarian Cancer Families. BMC Med. Genet. 8, 40. doi: 10.1186/1471-2350-8-40
Fondevila, M., Børsting, C., Phillips, C., de la Puente, M., Consortium, E. N., Carracedo, A., et al. (2017). Forensic SNP Genotyping With SNaPshot: Technical Considerations for the Development and Optimization of Multiplexed SNP Assays. Forensic Sci. Rev. 29, 57–76.
Fournier, D. (2005). Mutations of Acetylcholinesterase Which Confer Insecticide Resistance in Insect Populations. Chem. Biol. Interact. 157-158, 257–261. doi: 10.1016/j.cbi.2005.10.040
Hurst, C. D., Zuiverloon, T. C., Hafner, C., Zwarthoff, E. C., Knowles, M. A. (2009). A SNaPshot Assay for the Rapid and Simple Detection of Four Common Hotspot Codon Mutations in the PIK3CA Gene. BMC Res. Notes 2, 66. doi: 10.1186/1756-0500-2-66
Latini, F. R., Gazito, D., Arnoni, C. P., Muniz, J. G., de Medeiros Person, R., Carvalho, F. O., et al. (2014). A New Strategy to Identify Rare Blood Donors: Single Polymerase Chain Reaction Multiplex SNaPshot Reaction for Detection of 16 Blood Group Alleles. Blood Transfus. 12 (Suppl 1), s256–s263. doi: 10.2450/2013.0242-12
Martinez Torres, D., Chandre, F., Williamson, M. S., Darriet, F., Berge, J. B., Devonshire, A. L., et al. (1998). Molecular Characterization of Pyrethroid Knockdown Resistance (Kdr) in the Major Malaria Vector Anopheles Gambiae s.s. Insect. Mol. Biol. 7 (2), 179–184. doi: 10.1046/j.1365-2583.1998.72062.x
Mavridis, K., Wipf, N., Müller, P., Traoré, M. M., Muller, G., Vontas, J. (2018). Detection and Monitoring of Insecticide Resistance Mutations in Anopheles Gambiae: Individual vs Pooled Specimens. Genes 9, 479. doi: 10.3390/genes9100479
National Malaria Control Programme (2015). Sierra Leone Malaria Control Strategic Plan (2016-2020). 40–42.
National Malaria Control Programme (NMCP) [Sierra Leone]Statistics Sierra LeoneUniversity of Sierra LeoneCatholic Relief ServicesICF. (2016). Sierra Leone Malaria Indicator Survey. Freetown, Sierra Leone: NMCP, SSL, CRS, and ICF.
Palacajornsuk, P., Halter, C., Isakova, V., Tarnawski, M., Farmar, J., Reid, M. E., et al. (2009). Detection of Blood Group Genes Using Multiplex SNaPshot Method. Transfusion 49, 740–749. doi: 10.1111/j.1537-2995.2008.02053.x
Quintans, B., Alvarez-Iglesias, V., Salas, A., Phillips, C., Lareu, M. V., Carracedo, A. (2004). Typing of Mitochondrial DNA Coding Region SNPs of Forensic and Anthropological Interest Using SNaPshot Minisequencing. Forensic Sci. Int. 140, 241e57. doi: 10.1016/j.forsciint.2003.12.005
Ranson, H., Jensen, B., Vulule, J. M., Wang, X., Hemingway, J., Collins, F. H. (2000). Identification of a Point Mutation in the Voltage-Gated Sodium Channel Gene of Kenyan Anopheles Gambiae Associated With Resistance to DDT and Pyrethroids. Insect. Mol. Biol. 9 (5), 491–497. doi: 10.1046/j.1365-2583.2000.00209.x
Ranson, H., N’Guessan, R., Lines, J., Moiroux, N., Nkuni, Z., Corbel, V. (2011). Pyrethroid Resistance in African Anopheline Mosquitoes: What Are the Implications for Malaria Control? Trends Parasitol. 27 (2), 91–98. doi: 10.1016/j.pt.2010.08.004
Scott, J. A., Brogdon, W. G., Collins, F. H. (1993). Identification of Single Specimens of the Anopheles Gambiae Complex by the Polymerase Chain Reaction. Am. J. Trop. Med. Hyg. 49, 520–529. doi: 10.4269/ajtmh.1993.49.520
Vincent, C., N’Guessan, R. (2013). Distribution, Mechanism, Impact and Management of Insecticide Resistance in Malaria Vectors: A Pragmatic Review. IntechOpen: Anopheles Mosq.: New Insights Malaria Vectors, 579–633. doi: 10.5772/56117
Weill, M., Lutfalla, G., Mogensen, K., Chandre, F., Berthomieu, A., Berticat, C., et al. (2003). Comparative Genomics: Insecticide Resistance in Mosquito Vectors. Nature 423 (6936), 136–137. doi: 10.1038/423136b
Weill, M., Malcolm, C., Chandre, F., Mogensen, K., Berthomieu, A., Marquine, M., et al. (2004). The Unique Mutation in Ace-1 Giving High Insecticide Resistance Is Easily Detectable in Mosquito Vectors. Insect. Mol. Biol. 13 (1), 1–7. doi: 10.1111/j.1365-2583.2004.00452.x
Keywords: Multiple insecticide resistance, Anopheles gambiae, kdr, SNaPshot, Sierra Leone, rdl, ace-1
Citation: Yin J, Yamba F, Zheng C, Zhou S, Smith SJ, Wang L, Li H, Xia Z and Xiao N (2021) Molecular Detection of Insecticide Resistance Mutations in Anopheles gambiae from Sierra Leone Using Multiplex SNaPshot and Sequencing. Front. Cell. Infect. Microbiol. 11:666469. doi: 10.3389/fcimb.2021.666469
Received: 10 February 2021; Accepted: 05 August 2021;
Published: 19 August 2021.
Edited by:
Xiaojun Chen, Nanjing Medical University, ChinaReviewed by:
Isabel Mauricio, New University of Lisbon, PortugalCopyright © 2021 Yin, Yamba, Zheng, Zhou, Smith, Wang, Li, Xia and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ning Xiao, eGlhb25pbmdAbmlwZC5jaGluYWNkYy5jbg==; Samuel Juana Smith, c2FtdWVsanVhbmFAeWFob28uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.