
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 17 June 2021
Sec. Molecular Bacterial Pathogenesis
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.663933
 Cong Cheng1,2†
Cong Cheng1,2† Wangxiao Zhou2,3†
Wangxiao Zhou2,3† Xu Dong2,3
Xu Dong2,3 Peiyao Zhang2,3
Peiyao Zhang2,3 Kexin Zhou2,3
Kexin Zhou2,3 Danying Zhou2,3
Danying Zhou2,3 Changrui Qian2,3
Changrui Qian2,3 Xi Lin2,3
Xi Lin2,3 Peizhen Li2,3
Peizhen Li2,3 Kewei Li2,3
Kewei Li2,3 Qiyu Bao2,3
Qiyu Bao2,3 Teng Xu4*
Teng Xu4* Junwan Lu2,3*
Junwan Lu2,3* Jun Ying2,3*
Jun Ying2,3*Delftia tsuruhatensis has become an emerging pathogen in humans. There is scant information on the genomic characteristics of this microorganism. In this study, we determined the complete genome sequence of a clinical D. tsuruhatensis strain, TR1180, isolated from a sputum specimen of a female patient in China in 2019. Phylogenetic and average nucleotide identity analysis demonstrated that TR1180 is a member of D. tsuruhatensis. TR1180 exhibited resistance to β-lactam, aminoglycoside, tetracycline and sulphonamide antibiotics, but was susceptible to phenicols, fluoroquinolones and macrolides. Its genome is a single, circular chromosome measuring 6,711,018 bp in size. Whole-genome analysis identified 17 antibiotic resistance-related genes, which match the antimicrobial susceptibility profile of this strain, as well as 24 potential virulence factors and a number of metal resistance genes. Our data showed that Delftia possessed an open pan-genome and the genes in the core genome contributed to the pathogenicity and resistance of Delftia strains. Comparative genomics analysis of TR1180 with other publicly available genomes of Delftia showed diverse genomic features among these strains. D. tsuruhatensis TR1180 harbored a unique 38-kb genomic island flanked by a pair of 29-bp direct repeats with the insertion of a novel In4-like integron containing most of the specific antibiotic resistance genes within the genome. This study reports the findings of a fully sequenced genome from clinical D. tsuruhatensis, which provide researchers and clinicians with valuable insights into this uncommon species.
Delftia is a genus of opportunistic, Gram-negative, aerobic, motile, non-spore-forming and nonfermenting bacilli belonging to the family Comamonadaceae (Wen et al., 1999). Members of the genus were initially classified into the genus Comamonas until 1999, when Wen et al., reevaluated their taxonomy and proposed a new genus named Delftia based on phylogenetic and phenotypic evidence (Wen et al., 1999). This genus is grouped into six species: D. acidovorans (Wen et al., 1999), D. tsuruhatensis (Shigematsu et al., 2003), D. lacustris (Jørgensen et al., 2009), D. litopenaei (Chen et al., 2012), D. deserti (Li et al., 2015) and D. rhizosphaerae (Carro et al., 2017), which reside in soil, water, sludge and human microflora (Lipuma et al., 2015). In addition, D. acidovorans confers innate resistance to aminoglycosides (Lipuma et al., 2015), which are drugs used as an empirical treatment option for most Gram-negative infections.
D. tsuruhatensis was first reported in Japan by Shigematsu et al., in 2003 (Shigematsu et al., 2003). This species was isolated from activated sludge and identified as a terephthalate-assimilating bacterium. Distinguishing D. tsuruhatensis from closely related species, such as D. acidovorans, using commercial biochemical systems is challenging, thus 16S rRNA gene sequencing is recommended for accurate identification of D. tsuruhatensis isolates (Preiswerk et al., 2011). This species is mainly studied for environmental applications, since it can biodegrade organic pollutants, such as phenolic compounds and chlorobenzene (Jimenez et al., 2012; Ye et al., 2019). The species inhibits growth of various plant pathogens and thus is referred as a plant growth-promoting bacterium (Han et al., 2005). Moreover, some D. tsuruhatensis isolates have exhibited anti-biofilm activities (Singh et al., 2017) and resistance to heavy metals, such as zinc, lead and cadmium (Dorian et al., 2012; Lin et al., 2016).
Although D. tsuruhatensis has been rarely associated with human infections, the microorganism is a possible causative pathogen for life-threatening infections in immunocompromised patients and patients with port-related infections (Preiswerk et al., 2011; Tabak et al., 2013; Ranc et al., 2018). In early 2011, Preiswerk et al., reported a catheter-related infection caused by D. tsuruhatensis in a 53-year-old female from Switzerland (Preiswerk et al., 2011). The isolate was resistant to ampicillin, cefuroxime, cefalothin, gentamicin, tobramycin, amikacin and colistin but susceptible to amoxicillin-clavulanate, piperacillin-tazobactam, the third- and fourth-generation cephalosporins, fluoroquinolones and carbapenems. Similar resistance phenotypes were observed in a clinical D. tsuruhatensis strain collected from a 53-year-old woman with breast cancer (Tabak et al., 2013). However, a recent study by Ranc et al. on the antimicrobial susceptibility of D. tsuruhatensis isolated from bronchial samplings of a premature infant from France (Ranc et al., 2018), reported that the strain was categorized as resistant to amoxicillin–clavulanate (MIC >256 mg/L), which was suggestive of the potential evolution of drug resistance for D. tsuruhatensis strains; moreover, Ranc et al. found 11 healthcare-associated infections caused by D. tsuruhatensis strains through case investigations in the university hospitals in Marseille, France, during 2008-2015 and in the literature, which were mainly isolated from blood cultures (5/11 cases) and respiratory specimens (5/11), but also from a urine sample. And patients in whom D. tsuruhatensis had been isolated from blood cultures all had an intravascular device (Ranc et al., 2018).
Currently, there are seven genome sequences of D. tsuruhatensis available in the NCBI genome database, including a complete genome of D. tsuruhatensis CM13 (accession number CP017420) isolated from murine proximal colonic tissue. However, the specific genomic characteristics of D. tsuruhatensis CM13 and the drug resistance of these D. tsuruhatensis strains have not been reported. In this study, we analyzed the genomic features of the clinical D. tsuruhatensis strain TR1180 with resistance to multiple antibiotics to characterize drug/metal resistance, potential virulence factors and other functional annotations of this strain. We also performed extensive comparative genomics analysis of D. tsuruhatensis TR1180 with other sequenced Delftia strains to elucidate the genome divergence of these microorganisms at the genus level.
D. tsuruhatensis TR1180 wild-type strain was isolated in 2019 from the sputum of a 91-year-old female patient with respiratory failure at the Central Hospital of Lishui City, China. The strain was initially identified using the Vitek-60 microorganism autoanalysis system (BioMerieux Corporate, Craponne, France). Considering the challenges in discriminating closely related species in the genus of Delftia by commercial biochemical systems (Preiswerk et al., 2011), subsequent verification was carried out by phylogenetic analysis of D. tsuruhatensis TR1180 with other closely related species. A 16S RNA gene-based tree was generated by MEGA X (Kumar et al., 2018) using neighbor-joining method with 1,000 bootstrap replicates and a whole genome-based tree was constructed using the Type Strain Genome Server (TYGS) (Meier-Kolthoff and Göker, 2019). Further, the identity of the strain was verified by calculating the average nucleotide identity (ANI) values using OrthoANI (Lee et al., 2016).
The minimum inhibitory concentrations (MICs) were determined using the standard agar dilution method following the Clinical and Laboratory Standards Institute (CLSI) guidelines. Susceptibilities to each antimicrobial agent were determined according to the interpretive criteria for “other non-Enterobacteriaceae” and “Enterobacteriaceae” of CLSI (2019) guidelines. The resistance breakpoints for florfenicol (≥32 mg/L) and streptomycin (≥32 mg/L) were defined as reported in a previous study describing antimicrobial resistance (AMR) of commensal Escherichia coli (Wasyl et al., 2013) and the US Food and Drug Administration (FDA), respectively. E. coli ATCC 25922 was used as the reference strain for quality control.
Oxford Nanopore and Illumina HiSeq paired-end sequencing (400-bp insert sizes) of the D. tsuruhatensis TR1180 genome were performed. The long nanopore sequencing reads were initially assembled by Canu v1.8 (Koren et al., 2017), then the processed Illumina reads (adaptor trimming and quality filtering) were mapped onto the primary assembly to correct the consensus using BWA-0.7.17 and Pilon v1.23 (Li, 2013; Walker et al., 2014). Potential open reading frames (ORFs) were predicted using Prodigal v2.6.3 (Hyatt et al., 2010), and annotated using the Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi) program against UniProt/Swiss-Prot and non-redundant protein databases with an E-value cutoff of 1e-5. EggNOG-mapper v1.0.3 (Huerta-Cepas et al., 2017) was used to assign ORFs to Clusters of Orthologous Groups of proteins (COG) categories by searches against the eggNOG database (Huerta-Cepas et al., 2019). Gene Ontology (GO) and functional pathway annotations of genes were performed using InterProScan-5.36-75.0 and KAAS (Moriya et al., 2007), respectively. The AMR genes, virulence factors, two-component system (TCS) genes and metal resistance genes were predicted using the CARD (Jia et al., 2017), VFDB (Liu et al., 2019), P2CS (Ortet et al., 2015) and BacMet (Pal et al., 2014) with BLASTP (minimum 60% identity and 80% coverage), respectively. RNAmmer 1.2 (Lagesen et al., 2007), tRNAscan-SE 2.0 (Lowe and Chan, 2016) and Infernal 1.1.2 (Nawrocki et al., 2009) were used to identify rRNA, tRNA and noncoding RNA (ncRNA) genes, respectively. Detection of insertion sequences was performed using ISfinder (Siguier et al., 2006). Clustered regularly interspaced short palindromic repeat (CRISPR) arrays were detected using the CRISPRFinder (Grissa et al., 2007). The genome-wide comparison of orthologous genes was performed using GET_HOMOLOGUES (Contreras-Moreira and Vinuesa, 2013). Pan-genome analysis of Delftia based on the orthologous genes was performed by PanGP v1.0.1 (Zhao et al., 2014). The graphical map of the D. tsuruhatensis TR1180 genome, which was subsequently employed as the reference genome in comparative analysis, was constructed by BRIG v0.95 (Alikhan et al., 2011). The completeness of the genome assemblies was assessed using BUSCO v4.0.5 (Seppey et al., 2019). Comparison of nucleotide sequences was performed using BLASTN. Statistical analysis was performed using R software (https://www.r-project.org/). Other bioinformatics scripts were written using Python (https://www.python.org/) and Biopython (Cock et al., 2009).
A clinical D. tsuruhatensis strain designated TR1180 was isolated from the Central Hospital of Lishui City, China. Phylogenetic trees based on the 16S rRNA gene and whole-genome sequences showed that TR1180 belonged to the genus Delftia and exhibited the closest evolutionary relationship with D. tsuruhatensis NBRC 16741 (NZ_BCTO00000000) (Figure 1). Further, a search against the NCBI nucleotide database revealed that TR1180 shared the highest sequence similarity (at 99% identity and 91% coverage) with D. tsuruhatensis CM13. The ANI values between TR1180 and the above two D. tsuruhatensis strains were 98.09% (NBRC 16741) and 98.01% (CM13), respectively, which exceeded the threshold of 95-96% for species circumscription (Richter and Rosselló-Móra, 2009). Therefore, the study strain was grouped into the species D. tsuruhatensis.
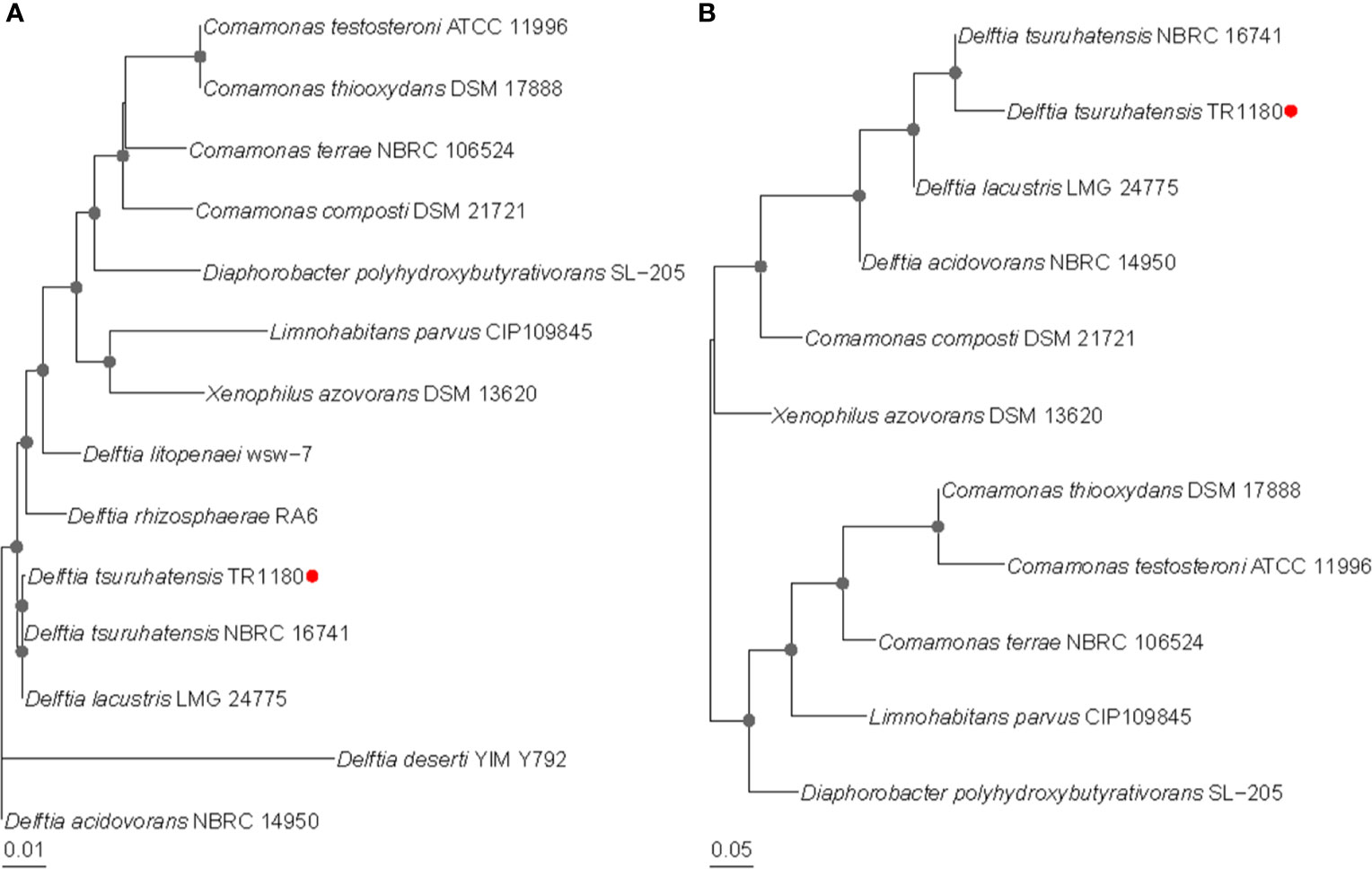
Figure 1 Phylogenetic relationships of D. tsuruhatensis TR1180 with other type strains of Delftia species and genera within the family Comamonadaceae. Phylogenetic trees were based on (A) 16S RNA gene sequences and (B) whole-genome sequences. Bootstrap values greater than 50% are indicated by gray dots at the nodes. D. tsuruhatensis TR1180 is highlighted with a red dot.
Antimicrobial susceptibility testing showed that D. tsuruhatensis TR1180 exhibited a multidrug-resistant (MDR) phenotype, including resistance to β-lactams (ampicillin and cefazolin), all tested aminoglycosides, tetracycline (tetracycline) and sulphonamide (trimethoprim/sulfamethoxazole) (Table 1). As mentioned earlier, to our knowledge, only three studies have reported the AMR profiles of clinical D. tsuruhatensis isolates (Preiswerk et al., 2011; Tabak et al., 2013; Ranc et al., 2018). Our work represented the first study that determined the MICs of phenicol, tetracycline, sulphonamide and macrolide antibiotics for clinical D. tsuruhatensis.
D. tsuruhatensis TR1180 genome was assembled into one circular chromosome of approximately 6.7 Mb in size with an average GC content of 66.52%, and there was no evidence of plasmids (Table 2 and Figure 2). The D. tsuruhatensis TR1180 genome harbored 6,009 protein-coding genes, including 17 antimicrobial genes [blaOXA-118, oqxB, dfrA16, aac(6’)-Ib3, aadA2, 2sul1, floR, tet(G) and other multidrug resistance genes], 24 putative virulence genes (mainly related to bacterial motility and adherence, iron acquisition and metabolism, and secretion system) (Table 3). In addition, the strain contained a series of predicted metal resistance genes (mercury, copper, arsenate, lead, chromate and other resistance) clustered or scattered on the chromosome (Table 3). The presence of drug-resistance genes in the TR1180 genome is in agreement with the antimicrobial susceptibility profile of the strain, except for its susceptibility to phenicols (MIC levels for chloramphenicol and florfenicol were 8 and 4 mg/L, respectively, Table 1). The susceptibility to phenicols implies that the floR gene in TR1180 may not be functional. Notably, more than half of the virulence genes (62.5%, 15/24) detected in the TR1180 genome are members of offensive virulence factors (Table 3). These genes include acpXL gene involved in lipopolysaccharide biosynthesis, which may have harmful effects on immunocompromised hosts. Analysis of TR1180 genome identified 15 rRNAs, 87 tRNAs and 14 ncRNAs (Table 2). Functional analysis using antiSMASH (Blin et al., 2019) identified a large number of gene loci responsible for the synthesis of the polyketide, nonribosomal peptide, terpene, bacteriocin and resorcinol. Resorcinol has been reported to participate in nematocidal and antimicrobial activity (Sultana et al., 2010; Calderón et al., 2014). The results of COG and GO annotation for TR1180 genome were shown in the Supplementary Figures S1A, B, respectively.
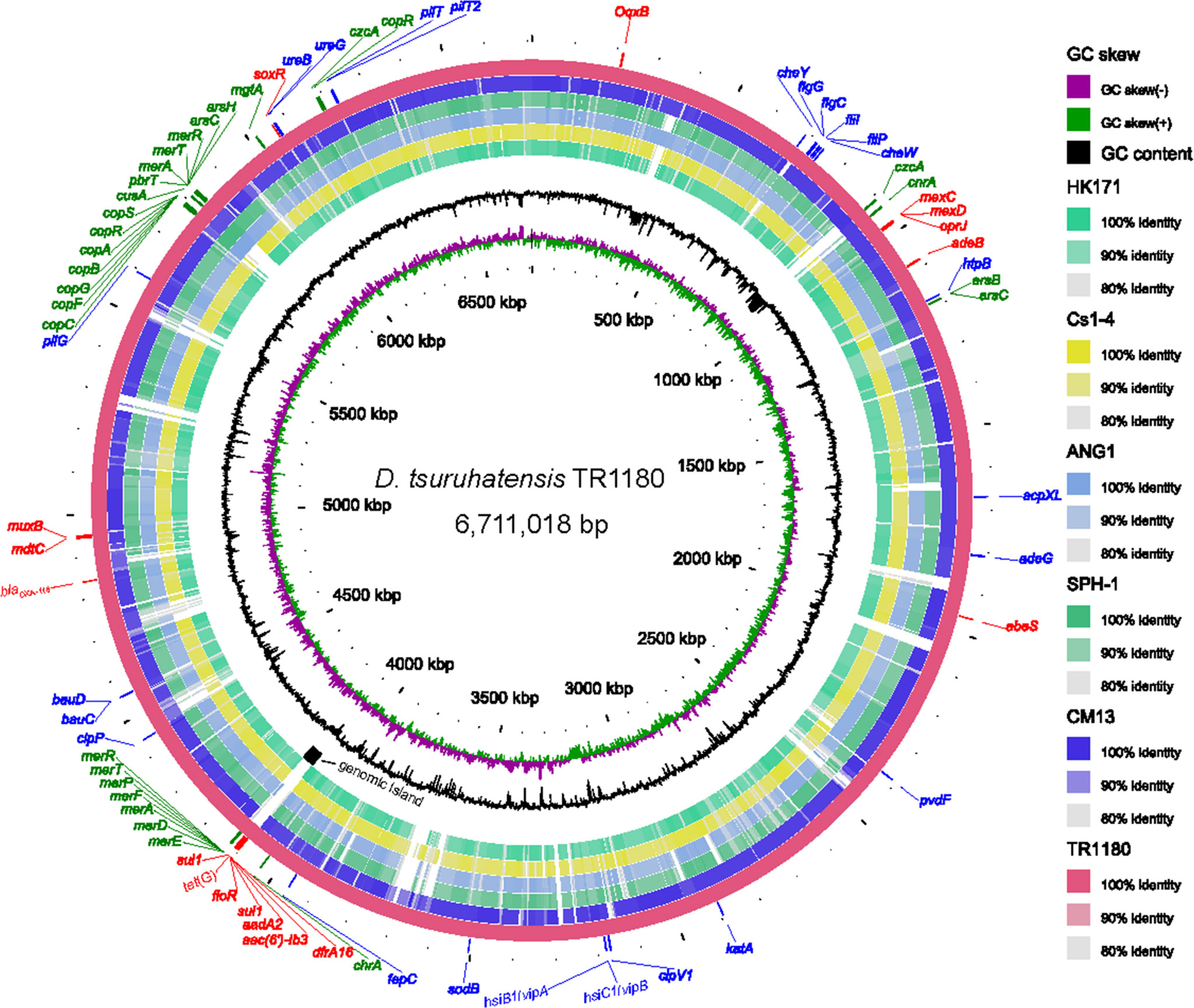
Figure 2 Circular representation of the D. tsuruhatensis TR1180 genome and comparative genomics analysis with other Delftia strains generated by BRIG. Counting from the outside toward the center: circle 1 shows AMR genes (colored in red), virulence genes (colored in blue) and metal resistance genes (colored in green) encoded on the TR1180 genome; circle 2 indicates the complete genome of TR1180 (as the reference genome); circles 3-7 refer to regions of D. tsuruhatensis CM13 (CP017420), D. acidovorans SPH-1 (CP000884), D. acidovorans ANG1 (CP019171), D. sp. Cs1-4 (CP002735) and D. sp. HK171 (CP018101) which have high sequence similarities (>80%) with D. tsuruhatensis TR1180, where empty regions indicate parts without similar hits between them; circle 8 displays the specific genomic island in TR1180; circles 9 and 10 represent GC content and GC skew of TR1180, respectively.
To explore the genome divergence and bacterial evolution of the genus Delftia, we performed pan-genome analysis of all Delftia genomes publicly available in the NCBI genome database (genome completeness ≥95%, Supplementary Table S1). The pan-genome possessed 15,316 gene families and had not reached saturation based on its accumulation curve (Supplementary Figure S2A). According to the fitted-line model generated by PanGP (Zhao et al., 2014), the Delftia pan-genome was open and evolving, indicating species of this genus can colonize different environments and have various ways of exchanging genetic material (Medini et al., 2005). Among these gene families, a total of 2,905 families existed in all thirty-one Delftia genomes and hence represented the core genome (Figures S2B). The percentages of core genes of each Delftia strain ranged from 46.57% to 58.05% (Supplementary Figure S3), implying that the core genes comprised a large proportion of the total genes in each genome and hence played a key role in the stability and evolution of the genus Delftia. The unique genes (strain-specific genes) are acquired through horizontal gene transfer (HGT) among species to increase genetic diversity and accelerate genome innovation and evolution (Jain et al., 2003). In our study, the number of unique genes across different genomes exhibited a wide distribution, varying from 3 (0.05%) in D. acidovorans ANG1 to 1,025 (16.83%) in D. acidovorans B15 (Supplementary Figure S3). This finding suggests the diverse genetic evolution in different Delftia strains. Of the 146 strain-specific genes identified in TR1180 genome, 39.04% were annotated as hypothetical proteins. Therefore, further analysis should be performed to understand the specific functions of these hypothetical genes in TR1180.
COG annotations were assigned to the core, dispensable and unique genes of Delftia strains to better understand the functional differences among them (Figure 3A). The result showed that most of the genes in the core genome were involved in housekeeping functions, such as energy production and conversion (9.95%), translation, ribosomal structure and biogenesis (7.42%) as well as transport- and metabolism-related functions (43.02%), and were significantly higher in proportion (all P<0.01; chi-square test) than dispensable (7.27%, 3.21% and 34.68%) and unique genes (7.26%, 2.72% and 35.28%). Compared with core genes, dispensable and unique genes were enriched in functions poorly characterized (14.38% and 16.44%) and functions associated with survival of bacteria in different environments, such as cell envelope biogenesis (7.83% and 6.67%) and mobilome (7.46% and 8.99%). The bacteria cell envelope is a typical example of a complex multilayered structure, which in most bacteria is essential for protecting them from changing and often hostile environment (Silhavy et al., 2010). Substantial mobilome suggests frequent HGT events in Delftia genomes, which allow the bacteria to adapt to a new niche or to be more successful in its current niche (Perry and Wright, 2013). These findings support the notion that some dispensable and unique genes in Delftia genomes may also be related to niche adaptation.
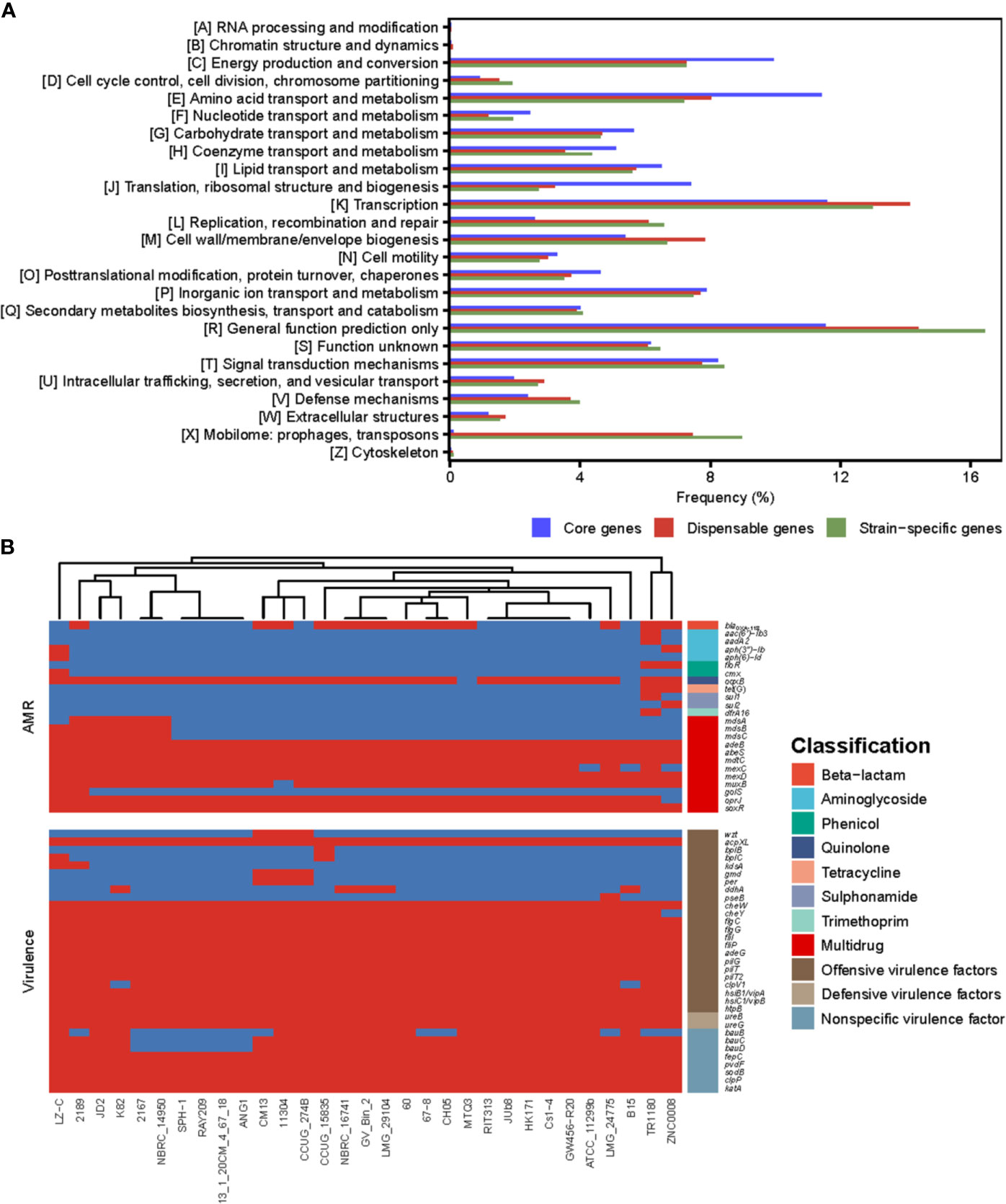
Figure 3 (A) The COG function classification comparison of core genes, dispensable genes and unique genes in the genus Delftia. (B) The resistome and virulome of 31 Delftia genomes.
We also reconstructed the Delftia metabolic pathways using the pan-genome and found, although core genes were only significantly enriched in ribosome and oxidative phosphorylation (both FDR<0.01; Fisher′s exact test), several of them were assigned into biofilm formation, beta-lactam resistance and two-component system (TCS) (Supplementary Table S2), which were closely associated with pathogenicity and resistance of Delftia strains. All Delftia genomes shared a similar virulence pattern as shown in Figure 3B, and 20 of 33 virulence factors identified in the pan-genome were significantly enriched in the core genome (P<0.01, Supplementary Table S3). Interestingly, although most of the AMR genes in these Delftia genomes exhibited multidrug resistance (mainly related to antibiotic efflux pumps), some of them conferring resistance to a specific class of antimicrobials were co-located on D. tsuruhatensis TR1180, D. sp. ZNC0008 and D. lacustris LZ-C (Figure 3B). This suggested the potential presence of HGT events or mutations among the three Delftia genomes. TCS, which is typically composed of a histidine kinase and a cognate response regulator in bacteria, was reported to be involved in the virulence and AMR responses of opportunistic bacterial pathogens (Stephenson and Hoch, 2002). In this study, we identified 157 TCS genes in the pan-genome and found 72 of them were significantly enriched in the core genome (P<0.01, Supplementary Table S3). These findings indicated that the core genome of Delftia spp. could contribute to pathogenicity and resistance of Delftia strains.
Comparative genomics analysis of the complete Delftia genomes showed that D. tsuruhatensis TR1180 harbored a unique 38-kb genomic island containing 49 ORFs (ranging from 4,095 to 4,133 kb in the genome map, Figures 2 and 4). The genomic island comprised a phage integrase-encoding gene, replication-associated genes (repAC) and partial elements of the conjugal transfer genes (trbL-trbK-trbJ-traJ-traK), but other essential components (such as the origin of transfer and relaxase gene) for an integrative and conjugative element were not identified (Wozniak and Waldor, 2010). This genomic island was integrated into the chromosome between orfA (encoding the ATP-binding protein) and guaA (encoding the glutamine-hydrolyzing GMP synthase). A pair of 29-bp direct repeats (DRs) with four nucleotide differences was identified by scanning its upstream and downstream sequences, implying the acquisition of the genomic island was possibly mediated by integrase through site-specific recombination. Further analysis demonstrated that the genomic island consisted of a backbone and several variable regions, including an In4-like integron and two truncated transposons Tn6050 and Tn5053, which conferred resistance to different classes of antibiotics and mercury (Figure 4 and Table 3).
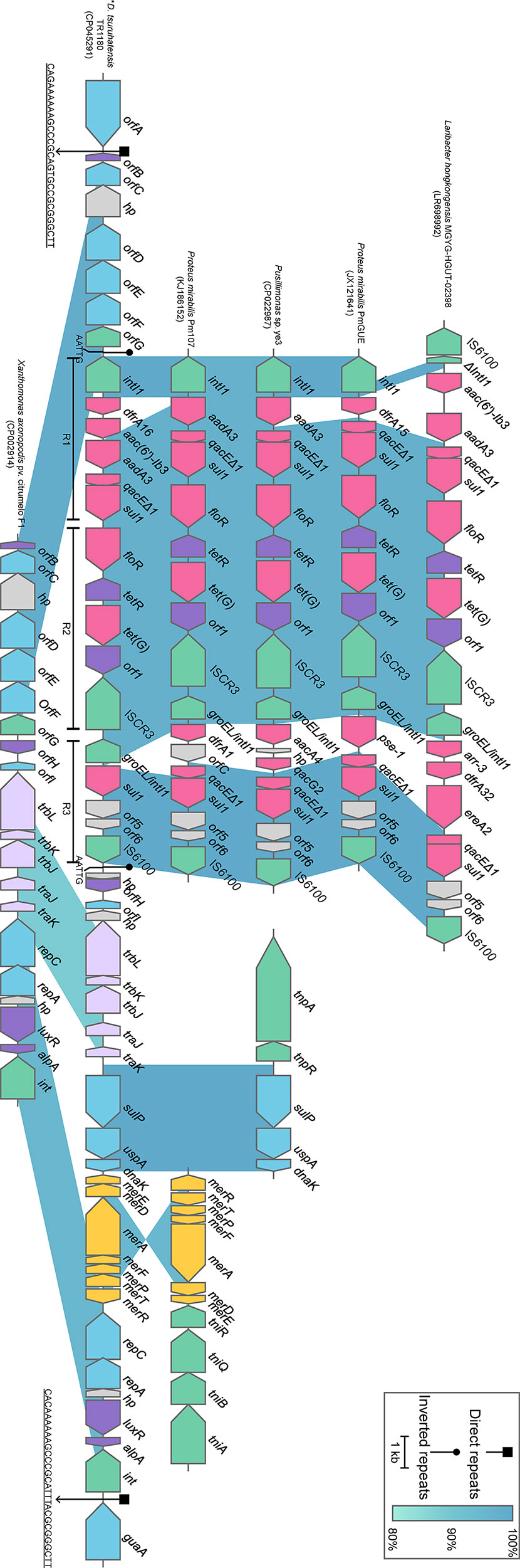
Figure 4 Schematic view of the genomic island integrated in D. tsuruhatensis TR1180 and linear comparison with related regions. IRL and IRR sequences flanking the In4-like integron of D. tsuruhatensis TR1180 are 5’-TGTCGTTTTCAGAAGACGGCTGCAC-3’, 5’-TGTCATTTTCAGAAGACGACTGCAC-3’ and 5’-GTGCAGTCGTCTTCTGAAAATGACA-3’, respectively. Genes are represented by arrowed boxes and colored based on gene function classification. Uncharacterized genes are illustrated as orfA and orfE, ATP-binding protein; orfB and orfH, transcriptional regulator; orfC, NADAR family protein; orfD, RES family NAD+ phosphorylase; orfF, sce7726 family protein; orfG, recombinase family protein; and orfI, type I toxin-antitoxin system ptaRNA1 family toxin. In4-like elements from Proteus mirabilis 09MAS2416, Salmonella enterica SC23 and Salmonella enterica subsp. enterica Z4 are identical to those of P. mirabilis Pm107 and hence are not displayed in the figure. D. tsuruhatensis TR1180 is indicated by an asterisk.
The intact backbone region ranging from orfB (encoding the transcriptional regulator) to orfG (encoding the recombinase family protein) was found only in the chromosomes of Xanthomonas axonopodis pv. citrumelo F1 (with 98.60% nucleotide sequence identity, CP002914), which was reported as a citrus pathogen causing citrus bacterial spot disease (Jalan et al., 2011), and Ottowia sp. oral taxon 894 (with 88.44% identity, CP012073). Notably, most of the backbone genes downstream of the In4-like integron of TR1180 could also be identified in X. axonopodis pv. citrumelo F1 genome (with 95.47% identity) but not in Ottowia sp. genome, which implied that the genome islands of TR1180 and X. axonopodis pv. citrumelo F1 might have evolved from a common ancestor (Figure 4).
The 14-kb In4-like complex integron within the genomic island lacked the partial IS6100 element found in In4 and harbored different gene cassettes (In4 carrying an aacC1-orfE-aadA2-cmlA1 gene cassette array) (Partridge et al., 2001). The whole region was flanked by 25-bp imperfect inverted repeats (noted as IRL and IRR, representing terminal left and right inverted repeats, respectively) and further bracketed by 5-bp DRs (AATTG) at both ends. A BLAST search against the NCBI nucleotide database indicated that the overall structure of the In4-like element in TR1180 resembled that of In4-like elements found in seven bacterial chromosomes (>97% identity and >95% coverage) from three different orders (Neisseriales, Burkholderiales and Enterobacterales) (Figure 4). The similarity of In4-like elements among different strains implied that the lateral gene transfer of the In4-like integrons may happen among phylogenetically remote bacteria. Notably, the ISCR3-carrying In4-like element was initially described in the Salmonella genomic island 1 (SGI1) of S. enterica Typhymurium DT104 (Boyd et al., 2000; Boyd et al., 2002). Further analysis showed the In4-like elements of the aforementioned genomes were also located within the SGI1 except for those of TR1180, Laribacter hongkongensis MGYG-HGUT-02398 (LR698992) and Pusillimonas sp. ye3 (CP022987), which may have been transferred from SGI1. Moreover, alignment of the In4-like integron in TR1180 with that of the other seven sequences showed that the single dfrA16 was lost in L. hongkongensis MGYG-HGUT-02398. In Proteus mirabilis PmGUE (JX121641), the dfrA16-aac(6′)-Ib3-aadA2 gene cassette array was replaced by dfrA15 while the deletions of dfrA16-aac(6′)-Ib3 occurred in the other five sequences. To the best of our knowledge, the genomic architecture of a class I integron carrying the dfrA16-aac(6’)-Ib3-aadA2 gene array has not previously been reported. In addition, the class I integron downstream of the ISCR3 of TR1180 underwent loss of qacEΔ1 (quaternary ammonium resistance) at the 3’-CS and lacked the variable region in comparison with the retrieved sequences.
Tn6050 and Tn5053 were inserted into the genomic island region between the conjugative transfer and replication-related genes with loss of corresponding transposase-encoding genes, which may have occurred during the transposition process. The remnants of Tn6050 were similar (>99.87% sequence identity) to that of the prototype Tn6050 located on a megaplasmid from Cupriavidus metallidurans CH34 (Figure 4), which was isolated from the heavy metal-contaminated sludge in Belgium (Van Houdt et al., 2009). The remnants of Tn5053 shared a nucleotide sequence identity of 95.15% with the prototype Tn5053, as observed in the chromosome of a mercury mine isolate Xanthomonas sp. W17 (Kholodii et al., 1993), which was an 8.4-kb transposon containing a mer operon (merRTPFADE, mercury resistance) (Figure 4).
A combination of various MGEs (especially an In4-like integron carrying AMR genes) within the backbone sequence of this genomic island formed a large MDR region with a complex mosaic structure.
To our knowledge, this study reports novel whole-genomic features and antimicrobial susceptibility patterns of clinical D. tsuruhatensis with a fully sequenced genome. Comparative analysis of Delftia genomes provided a better understanding of genome divergence among strains of this genus. In addition, all D. tsuruhatensis TR1180 AMR genes were embedded in an In4-like integron, which was part of a genomic island, implying that these genes may have been acquired through HGT. Further research should explore the epidemiological characteristics of D. tsuruhatensis, which may provide useful information for the clinical control of this bacterium.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
QB, TX, JL, and JY designed the experiment. CC, WZ, XD, PZ, KZ, DZ, and CQ performed experiments. XL, PL, and KL contributed to analysis the experimental data. CC, JY, and QB wrote the manuscript. KL, QB, and TX critically revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Natural Science Foundation of Zhejiang Province (LY19C060002 and LY21C010004), the National Natural Science Foundation of China (81960381), and the Science & Technology Project of Inner Mongolia Autonomous Region, China (201802125).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.663933/full#supplementary-material
Alikhan, N. F., Petty, N. K., Zakour, N. L. B., Beatson, S. A. (2011). Blast Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC. Genomics 12, 402. doi: 10.1186/1471-2164-12-402
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic. Acids Rea. 47, W81–W87. doi: 10.1093/nar/gkz310
Boyd, D., Cloeckaert, A., Chaslus-Dancla, E., Mulvey, M. R. (2002). Characterization of Variant Salmonella Genomic Island 1 Multidrug Resistance Regions From Serovars Typhimurium DT104 and Agona. Antimicrob. Agents. Chemother. 46, 1714–1722. doi: 10.1128/AAC.46.6.1714-1722.2002
Boyd, D. A., Peters, G. A., Ng, L. K., Mulvey, M. R. (2000). Partial Characterization of a Genomic Island Associated With the Multidrug Resistance Region of Salmonella Enterica Typhymurium Dt104. FEMS. Microbiol. Lett. 189, 285–291. doi: 10.1111/j.1574-6968.2000.tb09245.x
Calderón, C. E., Vicente, A., Cazorla, F. M. (2014). Role of 2-Hexyl, 5-Propyl Resorcinol Production by Pseudomonas Chlororaphis PCL1606 in the Multitrophic Interactions in the Avocado Rhizosphere During the Biocontrol Process. FEMS. Microbiol. Ecol. 89, 20–31. doi: 10.1111/1574-6941.12319
Carro, L., Mulas, R., Pastor-Bueis, R., Blanco, D., Terron, A., Gonzalez- Andres, F., et al. (2017). Delftia Rhizosphaerae Sp. Nov. Isolated From the Rhizosphere of Cistus Ladanifer. Int. J. Syst. Evol. Microbiol. 67, 1957–1960. doi: 10.1099/ijsem.0.001892
Chen, W. M., Lin, Y. S., Sheu, D. S., an Sheu, S. Y. (2012). Delftia Litopenaei Sp. Nov., a Poly-β-Hydroxybutyrate-Accumulating Bacterium Isolated From a Freshwater Shrimp Culture Pond. Int. J. Syst. Evol. Microbiol. 62, 2315–2321. doi: 10.1099/ijs.0.037507-0
Cock, P. J., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: Freely Available Python Tools for Computational Molecular Biology and Bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Contreras-Moreira, B., Vinuesa, P. (2013). GET_HOMOLOGUES, a Versatile Software Package for Scalable and Robust Microbial Pangenome Analysis. Appl. Environ. Microbiol. 79, 7696–7701. doi: 10.1128/AEM.02411-13
Dorian, A. B. H., Landy, I. R. B., Enrique, D. P., Luis, F. L. (2012). Zinc and Lead Biosorption by Delftia Tsuruhatensis: A Bacterial Strain Resistant to Metals Isolated From Mine Tailings. J. Water Res. Protect. 4, 207–212. doi: 10.4236/jwarp.2012.44023
Grissa, I., Vergnaud, G., Pourcel, C. (2007). Crisprfinder: A Web Tool to Identify Clustered Regularly Interspaced Short Palindromic Repeats. Nucleic. Acids Res. 35, W52–W57. doi: 10.1093/nar/gkm360
Han, J., Sun, L., Dong, X., Cai, Z., Sun, X., Yang, H., et al. (2005). Characterization of a Novel Plant Growth-Promoting Bacteria Strain Delftia Tsuruhatensis HR4 Both as a Diazotroph and a Potential Biocontrol Agent Against Various Plant Pathogens. Syst. Appl. Microbiol. 28, 66–76. doi: 10.1016/j.syapm.2004.09.003
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., Mering, C. (2017). Fast Genome-Wide Functional Annotation Through Orthology Assignment by Eggnog-Mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernandez-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic. Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Hyatt, D., Chen, G. L., LoCascio, P. F., Land, M., Larimer, F. W., Hauser, L. J. (2010). Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC. Bioinf. 11, 119. doi: 10.1186/1471-2105-11-119
Jørgensen, N. O., Brandt, K. K., Nybroe, O., Hansen, M. (2009). Delftia Lacustris Sp. Nov., a Peptidoglycan-Degrading Bacterium From Fresh Water, and Emended Description of Delftia Tsuruhatensis as a Peptidoglycan-Degrading Bacterium. Int. J. Syst. Evol. Microbiol. 59, 2195–2199. doi: 10.1099/ijs.0.008375-0
Jain, R., Rivera, M. C., Moore, J. E., Lake, J. A. (2003). Horizontal Gene Transfer Accelerates Genome Innovation and Evolution. Mol. Biol. Evol. 20, 1598–1602. doi: 10.1093/molbev/msg154
Jalan, N., Aritua, V., Kumar, D., Yu, F., Jones, J. B., Graham, J. H., et al. (2011). Comparative Genomic Analysis of Xanthomonas Axonopodis Pv. Citrumelo F1, Which Causes Citrus Bacterial Spot Disease, and Related Strains Provides Insights Into Virulence and Host Specificit. J. Bacteriol. 193, 6342–6357. doi: 10.1128/JB.05777-11
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K., et al. (2017). CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic. Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jimenez, B. J., Rivas, P. R., Lopez, J. G., Pesciaroli, C., Barghini, P., Fenice, M. (2012). Immobilization of Delftia Tsuruhatensis in Macro-Porous Cellulose and Biodegradation of Phenolic Compounds in Repeated Batch Process. J. Biotechnol. 157, 148–153. doi: 10.1016/j.jbiotec.2011.09.026
Kholodii, G. Y., Yurieva, O. V., Lomovskaya, O. L., Gorlenko, Z., Mindlin, S. Z., Nikiforov, V. G. (1993). Tn5053, a Mercury Resistance Transposon With Integron’s Ends. J. Mol. Biol. 230, 1103–1107. doi: 10.1006/jmbi.1993.1228
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., Phillippy, A. M. (2017). Canu: Scalable and Accurate Long-Read Assembly Via Adaptive K-Mer Weighting and Repeat Separation. Genome. Res. 27, 722–736. doi: 10.1101/gr.215087.116
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). Mega X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., Ussery, D. W. (2007). Rnammer: Consistent and Rapid Annotation of Ribosomal RNA Genes. Nucleic. Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lee, I., Kim, Y. O., Park, S. C., Chun, J. (2016). OrthoANI: An Improved Algorithm and Software for Calculating Average Nucleotide Identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Li, H. (2013). Aligning Sequence Reads, Clone Sequences and Assembly Contigs With BWA-MEM. arXiv preprint arXiv:1303.3997.
Lin, X., Mou, R., Cao, Z., Xu, P., Wu, X., Zhu, Z., et al. (2016). Characterization of Cadmium-Resistant Bacteria and Their Potential for Reducing Accumulation of Cadmium in Rice Grains. Sci. Total. Environ. 569, 97–104. doi: 10.1016/j.scitotenv.2016.06.121
Lipuma, J. J., Currie, B. J., Peacock, S. J., Vandamme, P. A. (2015). Burkholderia, Stenotrophomonas, Ralstonia, Cupriavidus, Pandoraea, Brevundimonas, Comamonas, Delftia, and Acidovorax. Manual Clin. Microbiol. Eleventh Edition: Am. Soc. Microbiol. p, 791–812. doi: 10.1128/9781555817381.ch43
Liu, B., Zheng, D., Jin, Q., Chen, L., Yang, J. (2019). VFDB 2019: A Comparative Pathogenomic Platform With an Interactive Web Interface. Nucleic. Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Li, C. T., Yan, Z. F., Chu, X., Hussain, F., Xian, W. D., Yunus, Z., et al. (2015). Delftia Deserti Sp. Nov., Isolated From a Desert Soil Sample. Antonie. Van. Leeuwenhoek. 107, 1445–1450. doi: 10.1007/s10482-015-0440-4
Lowe, T. M., Chan, P. P. (2016). Trnascan-SE On-line: Integrating Search and Context for Analysis of Transfer RNA Genes. Nucleic. Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Medini, D., Donati, C., Tettelin, H., Masignani, V., Rappuoli, R. (2005). The Microbial Pan-Genome. Curr. Opin. Genet. Dev. 15, 589–594. doi: 10.1016/j.gde.2005.09.006
Meier-Kolthoff, J. P., Göker, M. (2019). TYGS is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat. Commun. 10, 1–10. doi: 10.1038/s41467-019-10210-3
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., Kanehisa, M. (2007). KAAS: An Automatic Genome Annotation and Pathway Reconstruction Server. Nucleic. Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Nawrocki, E. P., Kolbe, D. L., Eddy, S. R. (2009). Infernal 1.0: Inference of RNA Alignments. Bioinformatics 25, 1335–1337. doi: 10.1093/bioinformatics/btp157
Ortet, P., Whitworth, D. E., Santaella, C., Achouak, W., Barakat, M. (2015). P2CS: Updates of the Prokaryotic Two-Component Systems Database. Nucleic. Acids Res. 43, D536–D541. doi: 10.1093/nar/gku968
Pal, C., Bengtsson-Palme, J., Rensing, C., Kristiansson, E., Larsson, D. J. (2014). BacMet: Antibacterial Biocide and Metal Resistance Genes Database. Nucleic. Acids Res. 42, D737–D743. doi: 10.1093/nar/gkt1252
Partridge, S. R., Recchia, G. D., Stokes, H. W., Hall, R. M. (2001). Family of Class 1 Integrons Related to In4 From Tn1696. Antimicrob. Agents. Chemother. 45, 3014–3020. doi: 10.1128/AAC.45.11.3014-3020.2001
Perry, J., Wright, G. (2013). The Antibiotic Resistance “Mobilome”: Searching for the Link Between Environment and Clinic. Front. Microbiol. 4, 138. doi: 10.3389/fmicb.2013.00138
Preiswerk, B., Ullrich, S., Speich, R., Bloemberg, G. V., Hombach, M. (2011). Human Infection With Delftia Tsuruhatensis Isolated From a Central Venous Catheter. J. Med. Microbiol. 60, 246–248. doi: 10.1099/jmm.0.021238-0
Ranc, A., Dubourg, G., Fournier, P. E., Raoult, D., Fenollar, F. (2018). Delftia Tsuruhatensis, an Emergent Opportunistic Healthcare-Associated Pathogen. Emerg. Infect. Dis. 24, 594. doi: 10.3201/eid2403.160939
Richter, M., Rosselló-Móra, R. (2009). Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Seppey, M., Manni, M., Zdobnov, E. M. (2019). Busco: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. 1962, 227–245. doi: 10.1007/978-1-4939-9173-0_14
Shigematsu, T., Yumihara, K., Ueda, Y., Numaguchi, M., Morimura, S., Kida, K. (2003). Delftia Tsuruhatensis Sp. Nov., a Terephthalate-Assimilating Bacterium Isolated From Activated Sludge. Inter. J. Syst. Evol. Microbiol. 53, 1479–1483. doi: 10.1099/ijs.0.02285-0
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). Isfinder: The Reference Centre for Bacterial Insertion Sequences. Nucleic. Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Silhavy, T. J., Kahne, D., Walker, S. (2010). The Bacterial Cell Envelope. Cold. Spring. Harb. Perspect. Biol. 2, a000414. doi: 10.1101/cshperspect.a000414
Singh, V. K., Mishra, A., Jha, B. (2017). Anti-Quorum Sensing and Anti-Biofilm Activity of Delftia Tsuruhatensis Extract by Attenuating the Quorum Sensing-Controlled Virulence Factor Production in Pseudomonas Aeruginosa. Front. Cell. Infect. Microbiol. 7, 337. doi: 10.3389/fcimb.2017.00337
Stephenson, K., Hoch, J. A. (2002). Virulence-and Antibiotic Resistance-Associated Two-Component Signal Transduction Systems of Gram-positive Pathogenic Bacteria as Targets for Antimicrobial Therapy. Pharmacol. Ther. 93, 293–305. doi: 10.1016/S0163-7258(02)00198-5
Sultana, N., Akhter, M., Khatoon, Z. (2010). Nematicidal Natural Products From the Aerial Parts of Rubus Niveus. Nat. Prod. Res. 24, 407–415. doi: 10.1080/14786410802696429
Tabak, O., Mete, B., Aydin, S., Mandel, N. M., Otlu, B., Ozaras, R., et al. (2013). Port-Related Delftia Tsuruhatensis Bacteremia in a Patient With Breast Cancer. New. Microbiol. 36, 199–201.
Van Houdt, R., Monchy, S., Leys, N., Mergeay, M. (2009). New Mobile Genetic Elements in Cupriavidus Metallidurans CH34, Their Possible Roles and Occurrence in Other Bacteria. Antonie. Van. Leeuwenhoek. 96, 205–226. doi: 10.1007/s10482-009-9345-4
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PloS One 9, e112963. doi: 10.1371/journal.pone.0112963
Wasyl, D., Hoszowski, A., Zając, M., Szulowski, K. (2013). Antimicrobial Resistance in Commensal Escherichia Coli Isolated From Animals at Slaughter. Front. Microbiol 4, 221. doi: 10.3389/fmicb.2013.00221
Wen, A., Fegan, M., Hayward, C., Chakraborty, S., Sly, L. I. (1999). Phylogenetic Relationships Among Members of the Comamonadaceae, and Description of Delftia Acidovorans (Den Dooren De Jong 1926 and Tamaoka Et Al. 1987) Gen. Nov., Comb. Nov. Int. J. Syst. Bacteriol. 49, 567–576. doi: 10.1099/00207713-49-2-567
Wozniak, R. A., Waldor, M. K. (2010). Integrative and Conjugative Elements: Mosaic Mobile Genetic Elements Enabling Dynamic Lateral Gene Flow. Nat. Rev. Microbiol. 8, 552–563. doi: 10.1038/nrmicro2382
Ye, J. X., Lin, T. H., Hu, J. T., Poudel, R., Cheng, Z. W., Zhang, S. H., et al. (2019). Enhancing Chlorobenzene Biodegradation by Delftia Tsuruhatensis Using a Water-Silicone Oil Biphasic System. Int. J. Environ. Res. Public. Health 16, 1629. doi: 10.3390/ijerph16091629
Keywords: Delftia tsuruhatensis, antimicrobial susceptibility profile, pan-genome, whole-genome analysis, comparative genomics analysis, In4-like integron
Citation: Cheng C, Zhou W, Dong X, Zhang P, Zhou K, Zhou D, Qian C, Lin X, Li P, Li K, Bao Q, Xu T, Lu J and Ying J (2021) Genomic Analysis of Delftia tsuruhatensis Strain TR1180 Isolated From A Patient From China With In4-Like Integron-Associated Antimicrobial Resistance. Front. Cell. Infect. Microbiol. 11:663933. doi: 10.3389/fcimb.2021.663933
Received: 04 February 2021; Accepted: 31 May 2021;
Published: 17 June 2021.
Edited by:
Ghassan M. Matar, American University of Beirut, LebanonReviewed by:
Jessica Kajfasz, University of Florida, United StatesCopyright © 2021 Cheng, Zhou, Dong, Zhang, Zhou, Zhou, Qian, Lin, Li, Li, Bao, Xu, Lu and Ying. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Ying, eWluZ2p1bkB3bXUuZWR1LmNu; Junwan Lu, bHVqdW53YW4zMDQwNDE0NzRAcXEuY29t; Teng Xu, eHV0ZW5nQHdtdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.