
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 18 May 2021
Sec. Molecular Bacterial Pathogenesis
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.661026
This article is part of the Research Topic RNA-mediated Regulation of Bacterial Pathogenesis View all 9 articles
 David D. Sarpong1,2,3
David D. Sarpong1,2,3 Erin R. Murphy2,3,4*
Erin R. Murphy2,3,4*The dynamic host environment presents a significant hurdle that pathogenic bacteria must overcome to survive and cause diseases. Consequently, these organisms have evolved molecular mechanisms to facilitate adaptation to environmental changes within the infected host. Small RNAs (sRNAs) have been implicated as critical regulators of numerous pathways and systems in pathogenic bacteria, including that of bacterial Toxin-Antitoxin (TA) systems. TA systems are typically composed of two factors, a stable toxin, and a labile antitoxin which functions to protect against the potentially deleterious activity of the associated toxin. Of the six classes of bacterial TA systems characterized to date, the toxin component is always a protein. Type I and Type III TA systems are unique in that the antitoxin in these systems is an RNA molecule, whereas the antitoxin in all other TA systems is a protein. Though hotly debated, the involvement of TA systems in bacterial physiology is recognized by several studies, with the Type II TA system being the most extensively studied to date. This review focuses on RNA-regulated TA systems, highlighting the role of Type I and Type III TA systems in several pathogenic bacteria.
At each point in the transmission and infection cycle, pathogenic bacteria are presented with various, and often extreme environmental conditions to which they must quickly adapt to survive. Their rapid response to environmental change allows bacteria to survive under the harshest of conditions ranging from nutritional stress, extreme temperatures, changing osmolarity, alterations in pH, and even threats of attacks from viral predators (bacteriophages) and the host immune response (Fernández et al., 2018). Pathogenic bacteria have evolved a variety of mechanisms by which to adapt to environmental changes, many of which involve protein factors that regulate the expression of a specific gene(s) in response to specific environmental signals (Brooks et al., 2011). An increasing number of studies, however, have revealed the vast potential of small RNAs (sRNAs) in controlling complex regulatory circuits connected to bacterial survival and/or virulence (reviewed in Plaza and José, 2020). One role that sRNAs play in controlling bacterial physiology and virulence is that of the anti-toxin component of Type I and Type III Toxin-Antitoxin (TA) systems (Brantl and Jahn, 2015; Holmqvist and Wagner, 2017).
TA systems are composed of two components: a stable toxin, and an unstable antitoxin which represses the synthesis or activity of the associated toxin (Harms et al., 2018). TA systems are categorized into six different types (Types I-VI) based on the nature and regulatory action of the antitoxin component (Page and Peti, 2016). Whereas the toxin of a TA system is always a protein, the antitoxin is either a protein (Types II, IV-VI) or an sRNA (Types I and III) (Unterholzner et al., 2013; Harms et al., 2018). Specifically, Type I and III TA systems involve sRNA antitoxins which either repress the translation (Type I) or activity (Type III) of the associated toxin protein. The Type II TA system is the most studied and involves a protein antitoxin which neutralizes the cognate toxin protein through protein-protein interactions. Antitoxin proteins of Type IV TA systems disrupt the activity of the associated toxin indirectly by competitively interfering with binding to their cellular targets. Type V antitoxin is an RNase that represses its cognate toxin at the level of transcription by cleaving the mRNA molecule on which the toxin is encoded. Finally, the newly discovered Type VI TA system involves an antitoxin protein which promotes toxin degradation by recruiting a protease to cleave it (Unterholzner et al., 2013; Harms et al., 2018; Song and Wood, 2020b).
Numerous studies have linked bacteria survival and pathogenesis to TA systems (Lobato-Márquez et al., 2015; Fernández-García et al., 2016; Lobato-Márquez et al., 2016a). Most of these studies focus on Type II TA systems (Korch et al., 2009; Zhu et al., 2009; Heaton et al., 2012; Wang et al., 2015). In this review, we highlight Type I and Type III TA systems and the role they play in the survival, pathogenicity, and persistence of select bacterial pathogens.
TA system toxins target important cellular components of the bacteria producing it, affecting vital physiological processes such as ATP synthesis, DNA replication, transcription, translation, and cell wall synthesis (Unterholzner et al., 2013; Harms et al., 2018; Jurėnas and Van Melderen, 2020). It is this activity that ultimately leads to toxin-associated bacterial growth inhibition or lysis.
Toxins from Type I TA systems are typically small (<60 aa) hydrophobic transmembrane proteins, (Fozo et al., 2008a). The toxic activity of these small proteins varies (Brielle et al., 2016), however the lethal effect of most Type I toxins is mediated by pore formation in the bacterial membrane, leading to disruption of membrane integrity, membrane depolarization, and subsequent ATP depletion (Wen and Fozo, 2014). This toxic mechanism is exemplified by the TisB, DinQ and HoK toxins of Escherichia coli (Wen and Fozo, 2014), as well as the SprG1, PepA1 and PepA2 toxins of S. aureus (Pinel-Marie et al., 2014; Germain-Amiot et al., 2019). Rather than disrupting the membrane potential, the BsrG toxin of B. subtilis and the AapA1 toxin of Helicobacter pylori target cell envelope synthesis directly, resulting in the formation of invaginations within the cell and ultimately cell lysis (Jahn et al., 2015; El Mortaji et al., 2020). Other Type I TA system toxins mediate their lethal effect via endoribonuclease activity (Kawano, 2012), endodeoxyribonuclease activity (Guo et al., 2014) or by promoting nucleoid condensation (Weaver, 2020). These toxic activities are exemplified by the toxin component of the E. coli SymE-SymR TA system, the E. coli RalR-RalA TA system, and the Enterococcus faecalis Fst-RNAII TA system, respectively (Kawano, 2012; Guo et al., 2014; Weaver, 2020). The activity, target(s) and function of select Type I and Type III toxin is summarized in Table 1.
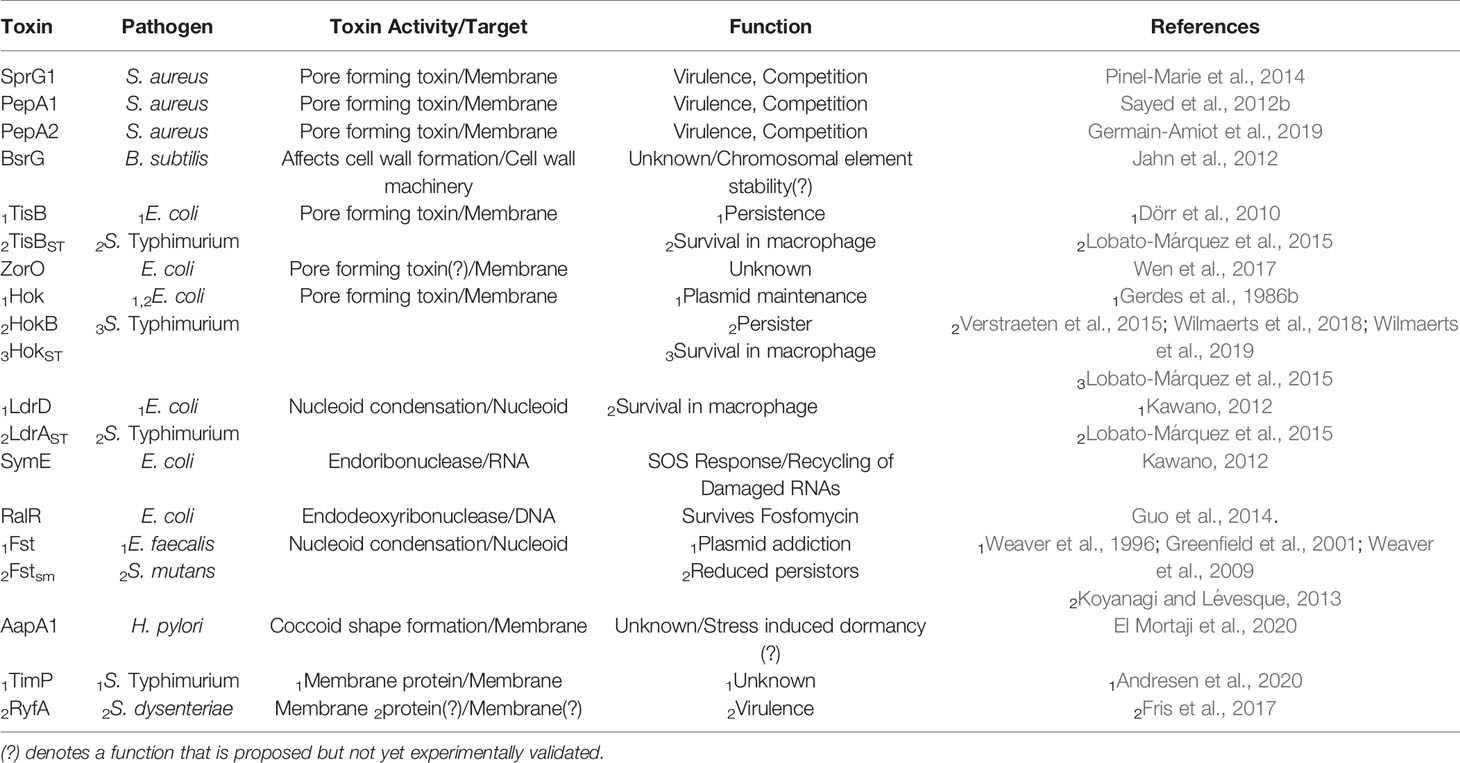
Table 1 Summary of the activity, target and function of select toxins produced by Type I and Type III TA systems in pathogenic bacteria.
The antitoxin within Type I TA systems is an sRNA that represses production of its cognate toxin through complementary base paring to the toxin encoding transcript, leading to toxin mRNA degradation, translation inhibition, or both (Wen and Fozo, 2014; Brantl and Jahn, 2015). The genes encoding each component of a Type I TA system are encoded within a single locus, and in many cases the antitoxin is encoded antisense (cis) to the toxin (Brielle et al., 2016). Some antitoxins, however, are encoded adjacent to, or partially overlapping with, the coding region of the toxin (trans) in either a divergent, convergent or parallel orientation (Fozo et al., 2008b). Type I TA systems of most Gram-positive bacteria have toxin and antitoxin genes arranged antisense to each other with partial overlap at their 3’ ends, resulting in complete complementarity between the corresponding areas of the antitoxin sRNA and toxin transcript (Masachis and Darfeuille, 2018; Brantl and Müller, 2019). Interestingly, the functional interaction between these antitoxin sRNAs and their target toxin transcript, however, does not always occur via these complementary sequences. For example, in Staphylococcus aureus sprA2/SprA2AS and sprA1/SprA1AS Type I TAs, though the 3’ ends of the toxin and antitoxin genes overlap, antitoxin-mediated regulation occurs via interactions between the 5’ end of the molecule (Sayed et al., 2012a; Germain-Amiot et al., 2019).
Functional regulation by the sRNA antitoxin is often mediated by binding between partially complementary nucleic acid sequences of the antitoxin sRNA and the target toxin transcript that results in occlusion of the Shine Dalgarno (SD) site on the toxin transcript, and thus inhibition of translation and/or degradation of the transcript (Figure 1) (Wen and Fozo, 2014). Alternatively, binding of a Type I antitoxin sRNA can occur at the 3’ end or elsewhere within the toxin transcript, generating an sRNA-mRNA heteroduplex that is cleaved by an RNase (Durand et al., 2012a; Wen and Fozo, 2014). Of note, the interaction between an sRNA antitoxin and target toxin transcript can lead to both translation inhibition and transcript degradation (Jahn et al., 2012; Brantl and Müller, 2019). Another mechanism by which Type I TA antitoxins regulate toxin production is to block a reading frame whose translation is coupled to that of the toxin gene (Figure 2) (Thisted et al., 1994). Finally, rather than preventing translation by binding directly to the SD of their target toxin mRNA, some Type I antitoxins achieve this end by binding to a ribosome standby site required for translation of the toxin (Figure 3) (Darfeuille et al., 2007).
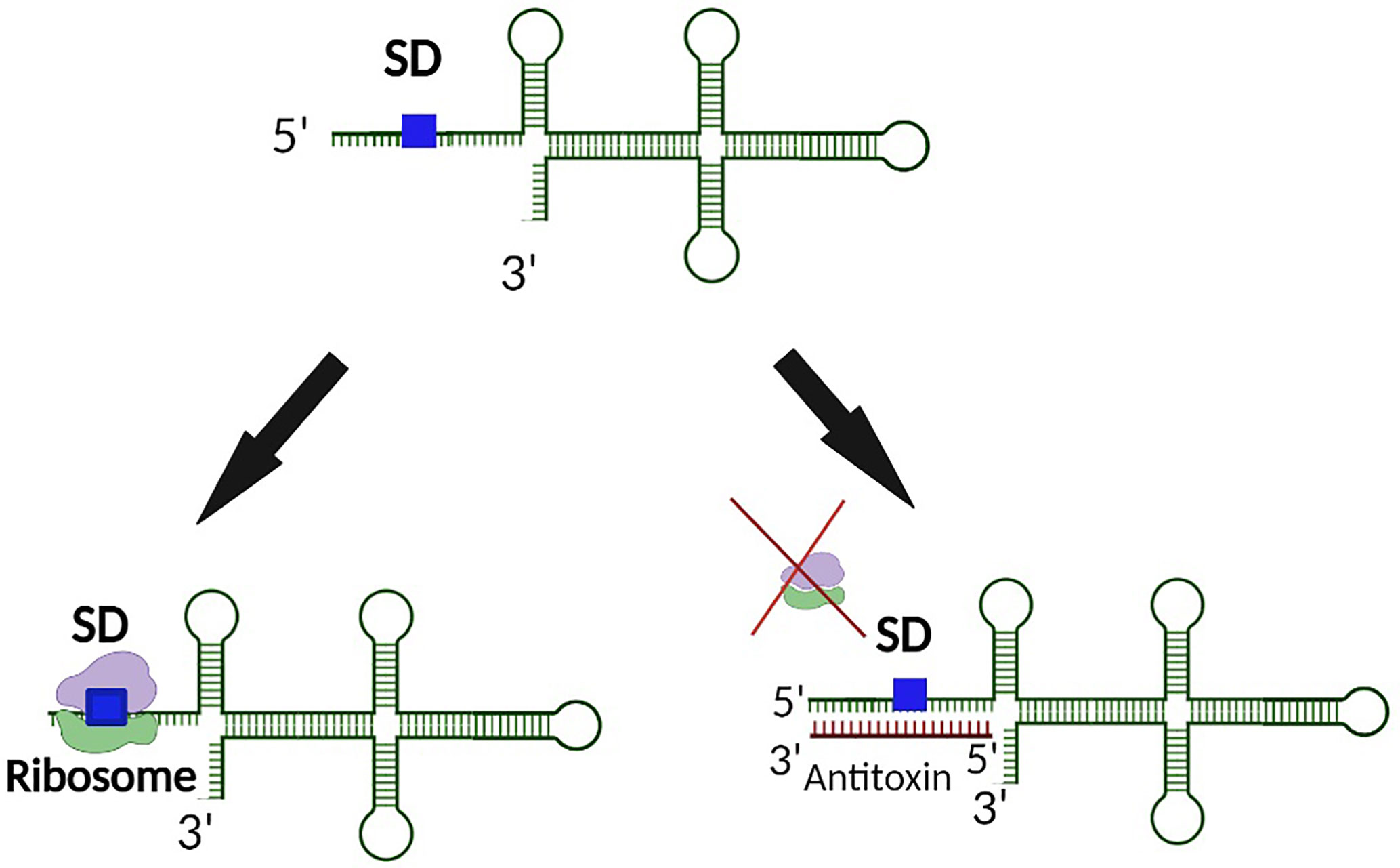
Figure 1 Binding of a Type I antitoxin occludes the SD site on the toxin transcript. In a typical mechanism of regulation, the antitoxin of a Type I TA systems binds the target toxin transcript such that binding of the ribosome to the SD site is physically blocked. As such, translation of the Type I toxin is prevented.
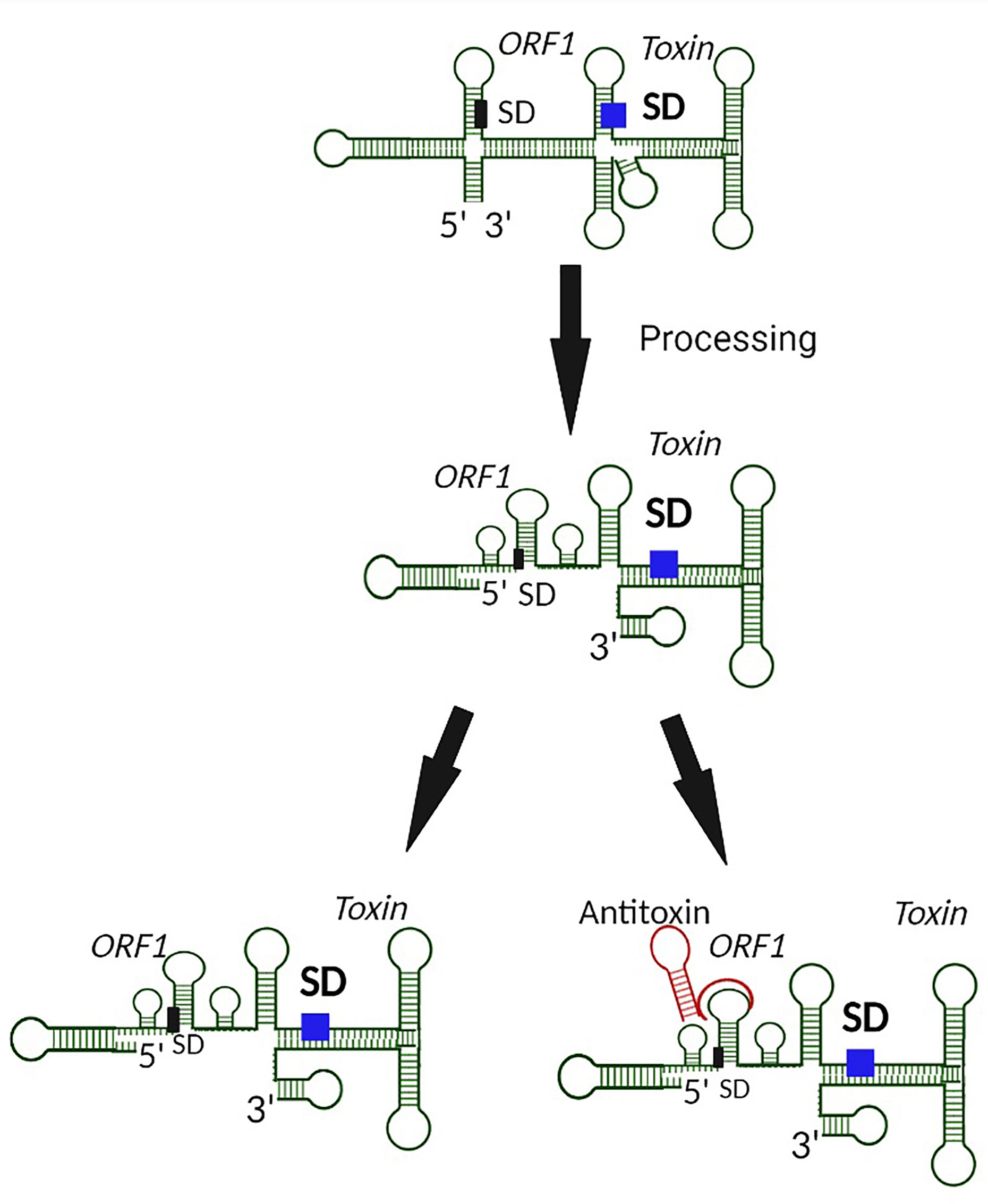
Figure 2 Prevention of Type I toxin production by blocking of an upstream coupled open reading frame by the antitoxin. In the case where translation of a toxin is coupled to translation of an upstream open reading frame (ORF1), binding of the antitoxin to the upstream SD prevents translation from both open reading frames, including from that encoded Type I toxin.
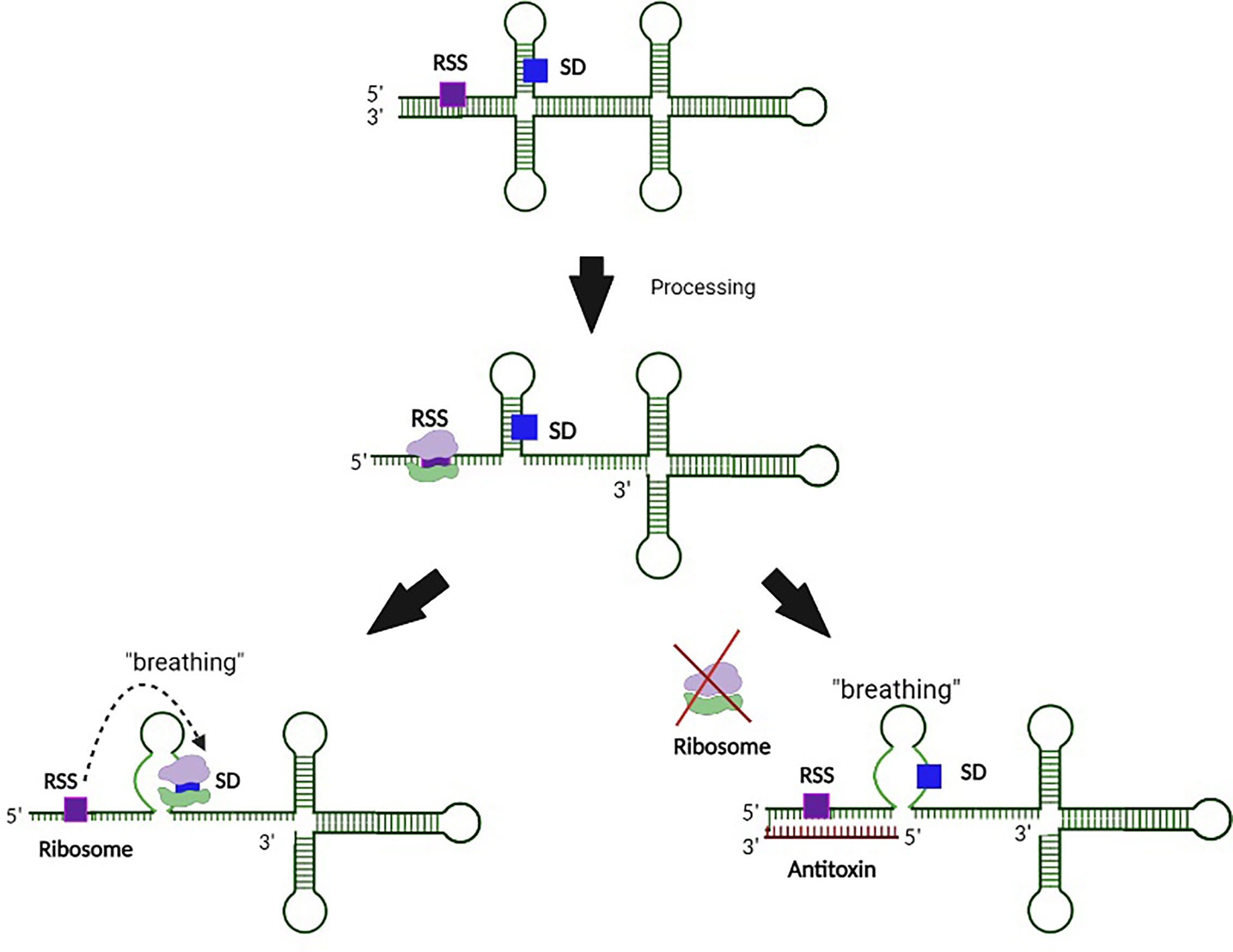
Figure 3 Regulation of Type I toxin production by binding of the antitoxin to an associated ribosomal standby site. In this example, processing of the toxin transcript exposes a ribosomal standby site (RSS) that facilitates binding of the ribosome to the SD site. Interaction between the antitoxin and RSS on the toxin transcript prevents ribosomal binding to this site, and thus subsequent binding to the SD. Such regulation prevents translation of the Type I toxin.
While many Type I sRNA antitoxins are encoded antisense to their target transcript, and thus have an extensive amount of perfect sequence complementarity with their cognate toxin transcript, those encoded in trans often display limited complementarity to their targets (Fozo et al., 2008b). Many bacterial sRNAs that share limited nucleic acid complementarity with their target mRNAs require a protein chaperone such as Hfq to facilitate regulation (Brennan and Link, 2007; Waters and Storz, 2009). Interestingly, the regulatory function of antitoxin sRNAs, even for those with limited nucleic acid complementary to their cognate toxin transcript, such as TisB/IstR-1, ShoB/OhsC, ZorO/OrzO in E. coli (Unoson and Wagner, 2008; Fozo, 2012; Wen et al., 2014) or BsrG/SR4, BsrE/SR5, yonT/SR6 (Jahn et al., 2012; Müller et al., 2016; Reif et al., 2018), does not require Hfq (Dambach et al., 2013). A notable exception to this rule is the sRNA antitoxin RalA of the E. coli RalR/RalA Type I TA system (Guo et al., 2014). Additionally, though the SR5 Type I antitoxin of B. subtilis is stabilized by Hfq, its regulatory activity can occur in its absence (Müller et al., 2016).
Premature synthesis of the toxin in the absence or low abundance of the corresponding antitoxin would be deleterious to bacterial survival. Hence additional steps of regulation are required to prevent such premature toxin production. To this end, secondary layers of toxin regulation which transcend the simplistic view of antitoxin regulation limited to complementary base-pairing between the toxin mRNA and antitoxin sRNA have evolved (Masachis and Darfeuille, 2018). In these instances, transcription of the toxin gene is uncoupled from its translation via mechanisms such as the sequestration of the ribosomal binding site within a hairpin structure in the 5’ untranslated region (Weaver et al., 2009; Durand et al., 2012a; Sayed et al., 2012a; Kristiansen et al., 2016; Müller et al., 2016; Wen et al., 2017; Masachis and Darfeuille, 2018). Translation of the toxin, and in some cases, interaction of the toxin with the antitoxin then requires processing of the transcript at either the 5’ or 3’ end by ribonucleases (Wen and Fozo, 2014; Masachis et al., 2019). Such processing results in a structural alteration of the toxin transcript which exposes the SD, thus facilitating translation and, potentially, interaction with the antitoxin (Figure 4). In another example, a sequence complementary to the Shine Dalgarno sequence (anti SD) is located within the 3’ end of the toxin transcript. Such complementarity results in the formation of a 5’-3’ long distance interaction (LDI) that inhibits translation by occluding the SD (Arnion et al., 2017; Masachis et al., 2019). Once again, processing of the transcript is required for binding of the transcript by the ribosome or antitoxin (Figure 5). Finally, in some cases the rate of toxin translation is reduced by the presence of a non-canonical start codon (Figure 6) (Durand et al., 2012b; Masachis and Darfeuille, 2018). Alternatively, the SD sequence on a toxin transcript may bind ‘too perfectly’ to the anti-SD of the 16S rRNA of the 30s ribosomal subunit leading to ribosomal pausing, slower release of the ribosome and thus, a reduced rate of translation (Figure 7) (Daou-Chabo et al., 2009; Durand et al., 2012a; Masachis and Darfeuille, 2018).
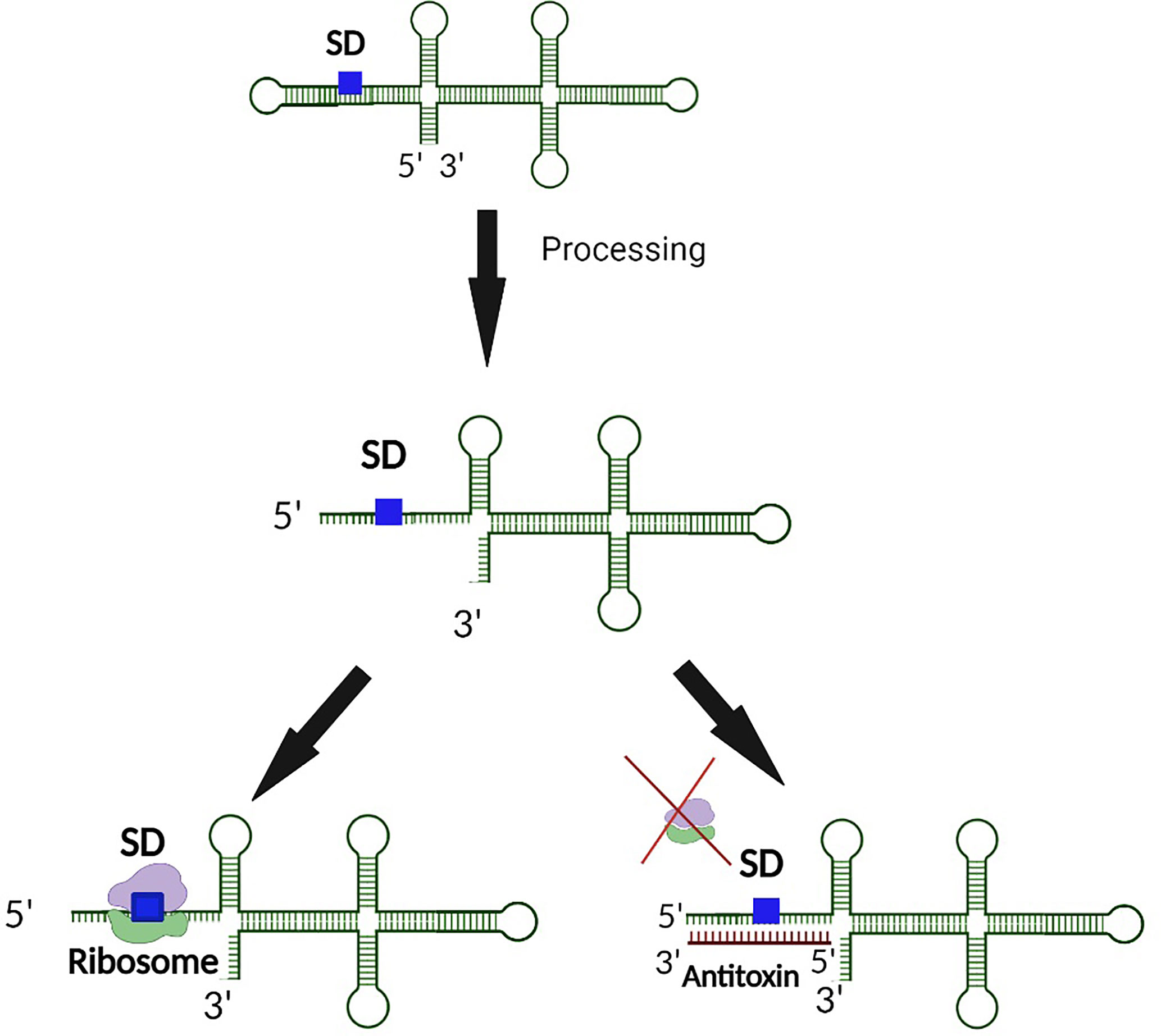
Figure 4 5’ end processing represents a second layer of regulation controlling the production of some Type I toxins. In the event an inhibitory secondary structure exists at the 5’ end of the toxin transcript, 5’ end processing exposes both the SD and antitoxin binding site. Upon exposure of the binding region, translation of the toxin is further regulated by an interaction between the antitoxin and the SD of the toxin transcript such that ribosomal binding is inhibited. Such dual regulation ensures minimal production of the Type I toxin when its activity would be deleterious to the bacterium.
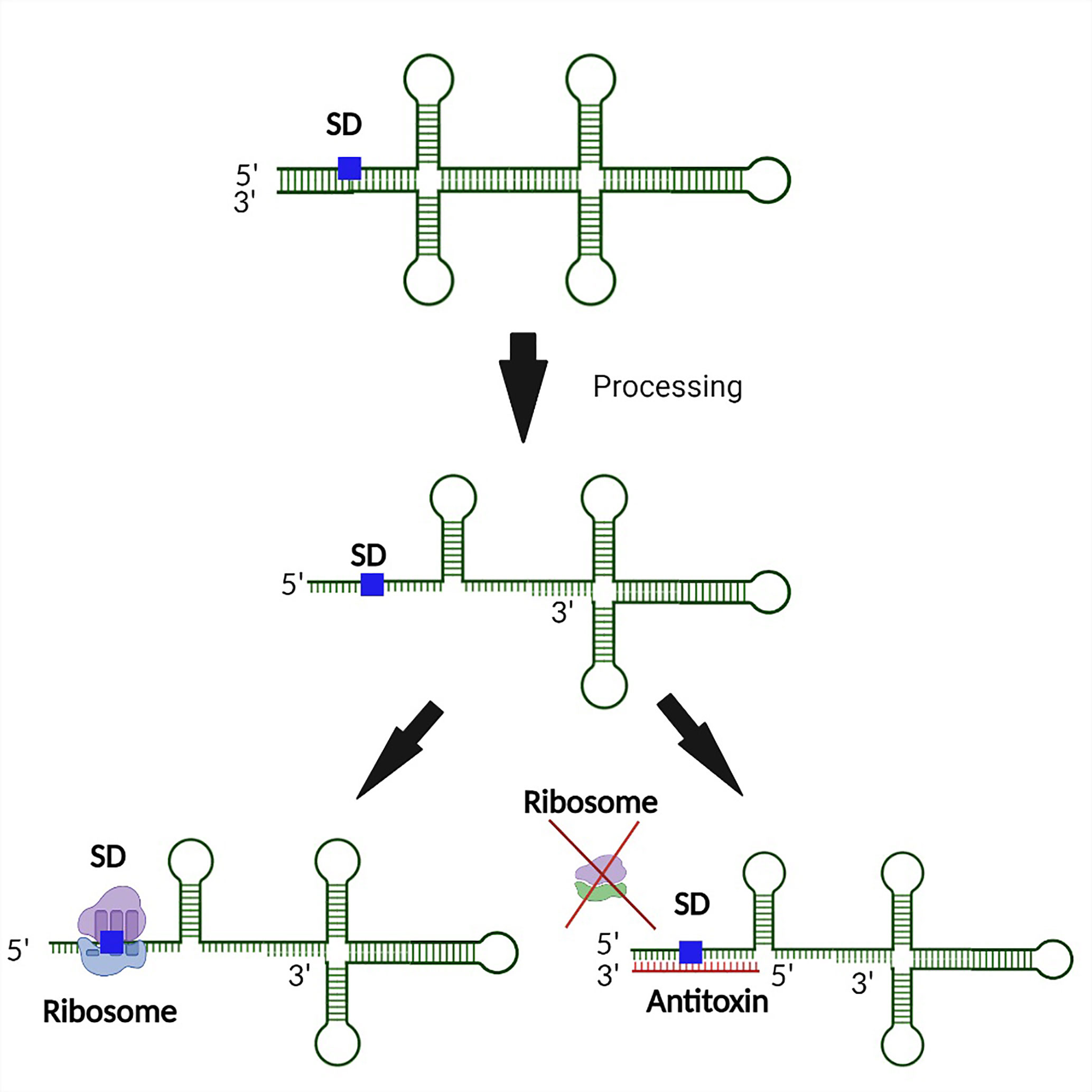
Figure 5 3’ end processing represents a second layer of regulation controlling the production of some Type I toxins. In the event an inhibitory secondary structure formed by binding between the 5’ and 3’ end of the toxin transcript, 5’ end processing is required to expose both the SD and antitoxin binding site. Upon exposure of the binding region, translation of the toxin is further regulated by an interaction between the antitoxin and the SD of the toxin transcript such that ribosomal binding is inhibited. Such dual regulation ensures minimal production of the Type I toxin when its activity would be deleterious to the bacterium.
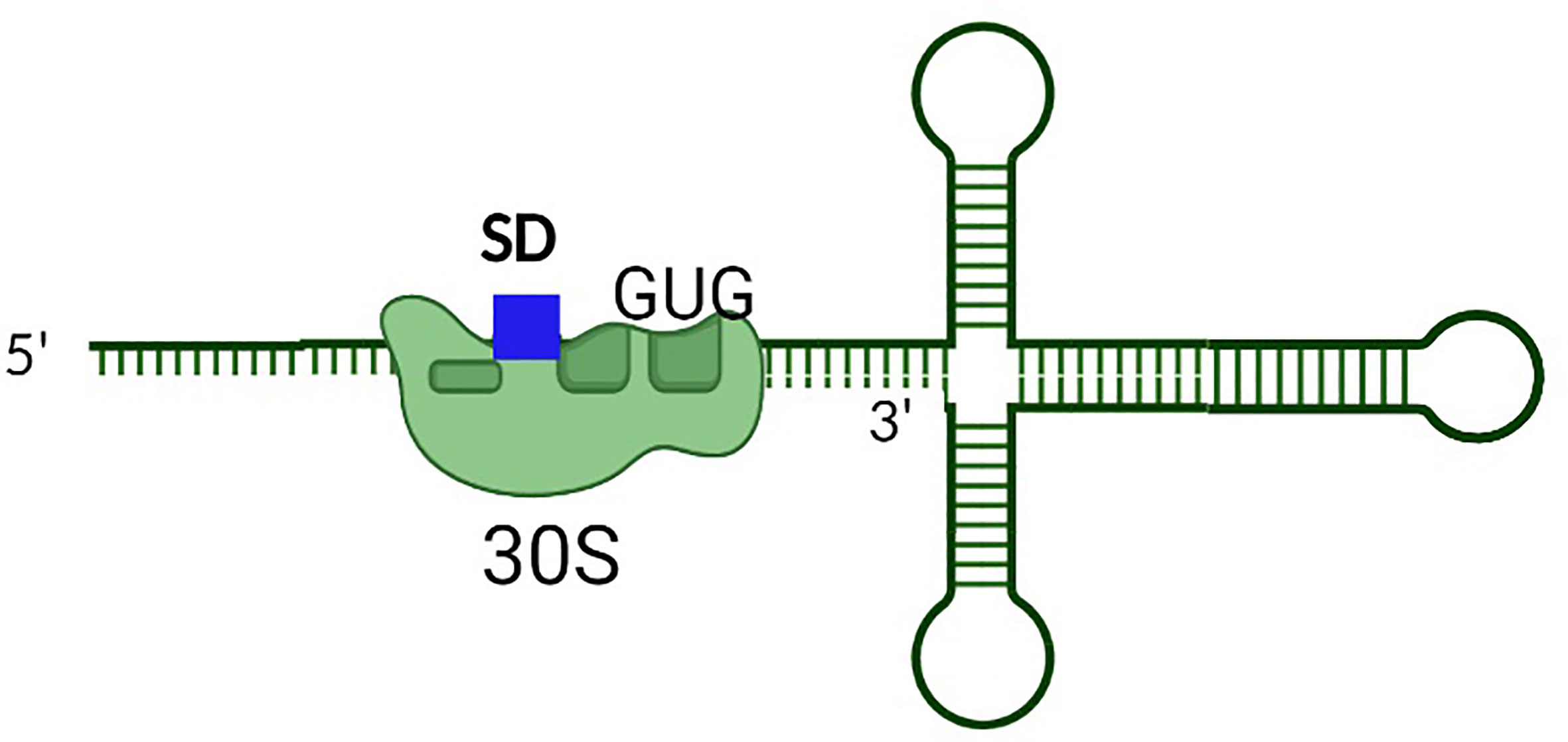
Figure 6 The presence of a non-AUG translational start codon is a secondary mechanism to limit the production of some Type I toxins. In this example, the presence of the non-AUG translational start codon results in partial inhibition of translation from the toxin transcript. Such regulation limits the production of Type I toxins when its activity would be deleterious to the bacterium.
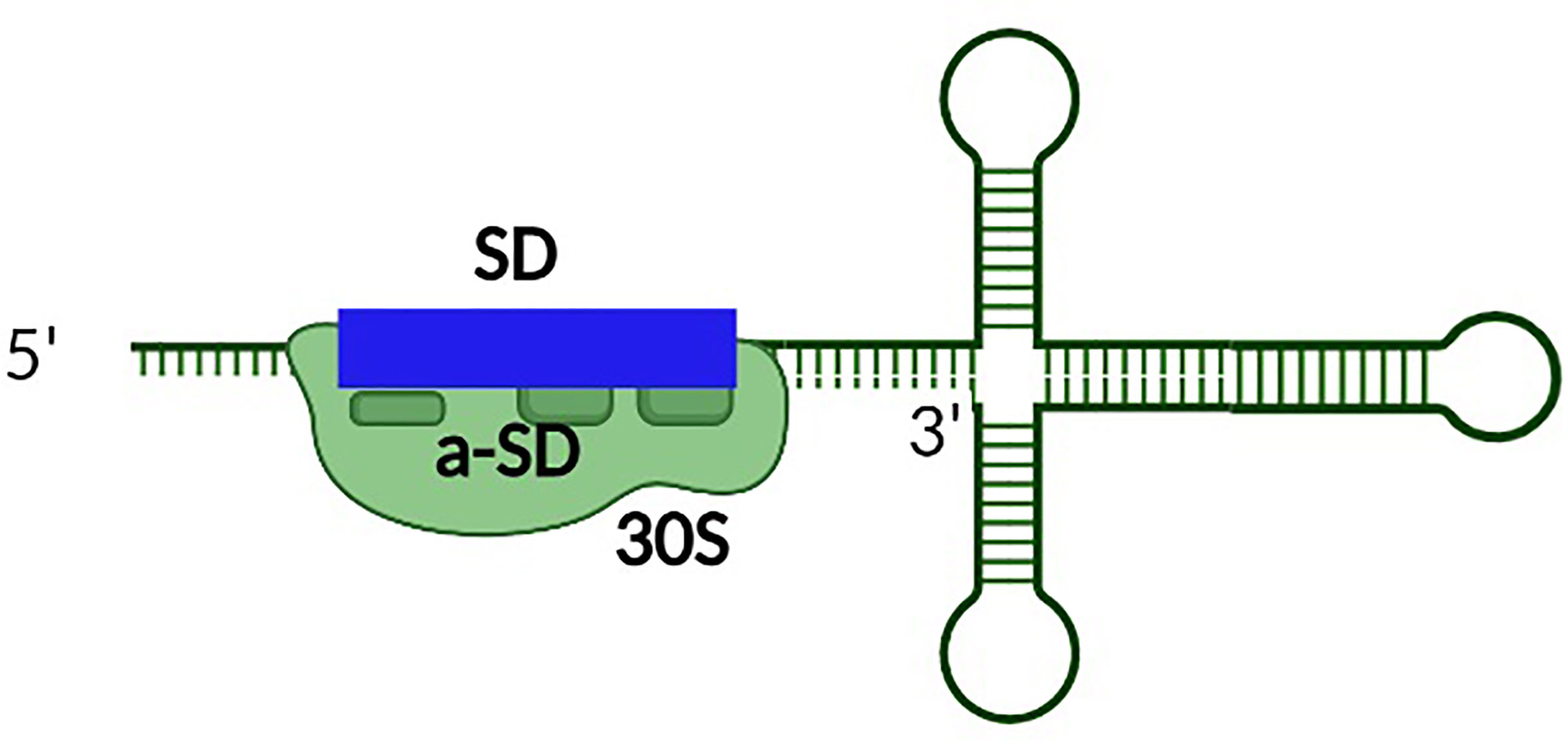
Figure 7 The presence of perfect complementarity between the SD site of the toxin transcript and the anti-SD sequence within the rRNA represses the production of some Type I toxins. Unusually tight binding between the SD site and anti-SD site within the rRNA of the ribosome reduces toxin production by inducing ribosomal pausing.
Originally discovered in 1983 and characterized as plasmid addiction modules, the increased recognition and characterization of TA systems on the chromosomes of bacteria and archaea has revealed their involvement in several other important processes (Gerdes et al., 1986a; Gerdes et al., 1986b; Thisted et al., 1994; Lee and Lee, 2016; Harms et al., 2018). Currently, bacterial Type I TA systems are hypothesized to function in numerous processes including plasmid maintenance, phage abortive infection, chromosome stabilization, persistence, and virulence (Gerdes, 2016; Kędzierska and Hayes, 2016; Sierra et al., 2019; Peltier et al., 2020). While it is clear that Type I TA systems play an important role in the physiology and virulence of many bacterial species, much is left to learn about these diverse systems (Wang and Wood, 2011; Germain-Amiot et al., 2019; Ma et al., 2019; Song and Wood, 2020a).
The most well understood role of plasmid encoded Type I TA systems in bacteria is for plasmid addiction. This function ensures that the plasmids on which the TA system is encoded are preserved in a population of actively growing bacteria, through post-segregational killing of any daughter cell lacking the plasmid (Gerdes et al., 1986a; Thisted et al., 1994). Due to the relative stability of the toxin as compared to that of the antitoxin, daughter cells lacking the plasmid on which the TA system is encoded become exposed to the lethal effect of the stable toxin. The role of bacterial TA systems in plasmid addiction was originally characterized with the study of the E. coli Hok/Sok Type I TA system, a system encoded on the R1 plasmid (Gerdes et al., 1986b; Gerdes et al., 1986a; Thisted et al., 1994). A similar function was subsequently observed for the Fst/RNAII Type I TA system present on the paDI plasmid of E. faecalis (Weaver et al., 1996; Greenfield et al., 2001; Weaver et al., 2009).
Persister state formation is observed in many bacterial species. In this phenomenon, a heterogeneous population of phenotypically variant bacteria is formed in which metabolism and growth are arrested in a subpopulation, making them resistant to killing by antibiotics (Brauner et al., 2016). This metabolic halt is relieved when antibiotic is removed from the environment, and the persister cells propagate to reestablish the population (Michiels et al., 2016). This bet-hedging strategy may facilitate recurrent and chronic bacterial infections (Fauvart et al., 2011). Known to be controlled by a variety of transcriptional factors and global regulators, several studies now suggest a role for chromosomally encoded TA systems in persister cell formation (Wang and Wood, 2011; Gerdes, 2016; Page and Peti, 2016; Berghoff and Wagner, 2017). For instance, the SOS-dependent TisB/IstR Type I TA system of Bacillus subtilis has been shown to play a role in persistence, as induction of TisB led to increased numbers of ciprofloxacin persisters and a deletion of the entire locus abolished the phenotype (Dörr et al., 2010) This involves the SOS-induced cleavage of the IstRI antitoxin upon DNA damage caused by antibiotic, leaving the TisB toxin to damage the E. coli membrane, decrease bacterial transcription and translation, and by doing so reduce metabolism and increase antibiotic tolerance (Unoson and Wagner, 2008). Additionally, a recent report demonstrated that overexpression of a chromosomally encoded Hok homologue, HokB in E. coli strain BW25113, facilitates persister cell formation through pore induced-ATP leakage and loss of electrochemical potential (Verstraeten et al., 2015; Wilmaerts et al., 2018). Interestingly, for the first time, resuscitation of a Type I TA-induced persister state was demonstrated in a follow up study on HokB, and was shown to occur via two main processes (Wilmaerts et al., 2019). Firstly, the pores induced by HokB dimers are destabilized upon disassembly of the toxin by an DsbC oxidoreductase, and the resulting monomers degraded by the DegQ protease (Wilmaerts et al., 2019). Secondly, the ATP-depleted cells can be replenished through membrane repolarization by Complex I of the Electron Transport Chain (Wilmaerts et al., 2019). Adding to the Type I TA-induced persistence repertoire, the Type I AapAI toxin of H. pylori has been shown to transform the bacterium from a spiral to a coccoid in vitro, a phenotype which coincides with infections that are resistant to antibiotic-mediated killing. Finally, through an unknown mechanism, the RalR/RalA Type I TA system in E. coli was shown to confer protection to Fosfomycin, a broad-spectrum antibiotic (Guo et al., 2014). It is important to note however, that several studies have questioned the direct involvement of TA systems with persister cell formation. According to one hypothesis, the TA-induced persistence phenotype is primarily due to reduced metabolism and not to toxin mediated loss of ATP or ppGpp as some studies suggest (Pontes and Groisman, 2019). These issues, together with other ‘persistent persister misperceptions’ are reviewed in Kim and Wood (2016). Clearly, further studies are required to elucidate the exact molecular mechanisms of persister cell formation and the role that Type I TA systems play in these processes.
In a similar fashion to plasmid addiction, Type I TA systems have been suggested to play a role in stabilizing portions of the chromosome (Durand et al., 2012b, 1). Several B. subtilis Type I TAs are located near prophage regions within the chromosome and are predicted to protect these regions from being deleted (Durand et al., 2012b; Brantl and Müller, 2019). For example, the BsrG/SR4 and YonT/SR6 Type I TA systems in B. subtilis are both found on the SPβ prophage element on the chromosome and are suggested to play a role in its stabilization (Reif et al., 2018; Brantl and Müller, 2019). The SPβ prophage element contains genes which may aid B. subtilis in survival, including protection from UV light and from the SP10 bacteriophage (Wen and Fozo, 2014). Other B. subtilis Type I TA systems whose function might be linked to chromosome stabilization are the TxpA/RatA and BsrH/as-BsrH system. These systems are encoded within B. subtilis skin element, a chromosomal region crucial for sporulation (Silvaggi et al., 2005; Wen and Fozo, 2014). Finally, the proximity of Type I TA systems to CRISPR modules (Maikova et al., 2018)and prophage regions (Peltier et al., 2020) in C. difficile is suggestive of a role in the chromosome stabilization of the bacterium’s adaptive immunity against invading phages (Soutourina, 2019).
Recently, the direct or indirect impact of TA systems, especially Type II TA systems, on virulence of different pathogens has been supported by experimental analyses. The presence of TA systems on virulence-associated plasmids could contribute to virulence by ensuring maintenance of these plasmids, as studies of Type II TA systems have shown (Brendler et al., 2004; Lobato-Márquez et al., 2016b; McVicker and Tang, 2016). Additionally, several chromosomally encoded Type I TA systems have been shown to contribute to bacterial virulence in a variety of ways. The SprA1/SprA1AS and SprA2/SprA2AS Type I TA systems in S. aureus encode the toxins, PepA1 and PepA2 respectively, that differ slightly in function (Sayed et al., 2012a; Sayed et al., 2012b; Bronsard et al., 2017; Germain-Amiot et al., 2019). Whereas both toxins can target human polymorphonuclear neutrophils and erythrocytes, PepA1 is ten times more cytotoxic than PepA2 and is suggested to play a role in inter-bacterial competition (Germain-Amiot et al., 2019). Once secreted, both toxins can act together to destroy erythrocytes and competing bacteria. It is hypothesized that destruction of surrounding cells aids in the spread of the S. aureus within the host (Germain-Amiot et al., 2019). Another S. aureus Type I TA system which likely impacts virulence is the sprG1/sprF1 system (Riffaud et al., 2019, 1). Here, the toxin gene encodes two small toxins of unequal lengths which are secreted extracellularly. Once secreted both toxins can lyse human host erythrocytes, with the longer peptide doing so more efficiently. These toxins are also involved in interbacterial competition where, in this case, the shorter peptide is more effective (Riffaud et al., 2019). Additionally, the ability of a Salmonella Typhimurium Type I TA system homologous to the E. coli hok-sok, ldrA-rdl and tisB-istR systems to aid in the intracellular lifestyle of this pathogen, suggests a role in virulence (Lobato-Márquez et al., 2015). Interestingly, deleting the antitoxin of the E. faecalis Fst/RNAII Type I TA system, results in hypervirulence in mice and larvae models, increased resistance to oxidative and acid stress, and higher survival rates in macrophages as compared to the wild type strain (Michaux et al., 2014).
Type I TA systems have been suggested to defend bacteria from phage attack. This phenotype was first identified in E. coli where the Hok/Sok Type I TA system was shown to prevent the T4 phage from spreading throughout the bacteria population through a mechanism termed altruistic killing (Pecota and Wood, 1996). According to this hypothesis, E. coli inhibits T4 phage by downregulating the Sok antitoxin within a subpopulation of phage infected cells, thus releasing the Hok toxin to induce apoptosis (Pecota and Wood, 1996). Despite such evidence to support the role of TA systems in phage resistance via the killing of infected cells, the lack of direct evident to demonstrate Hok-induced killing somewhat weakens this hypothesis. A recent alternative hypothesis suggests that TA-mediated phage resistance results from a general reduction in metabolism caused by production of the toxin, rather than direct killing of the infected cell by the toxin (Song and Wood, 2018).
Aside from S. aureus, another pathogen of the ESKAPE group mentioned in this review is E. faecalis (Rice, 2008). Though the genus Enterococcus has approximately about forty species, the two mostly associated with human disease are E. faecium and E. faecalis (Van Tyne et al., 2013). Commensals in the gastrointestinal tract, these organisms can cause infection when carried to different sites (Kristich et al., 2014). Additionally, E. faecalis is associated with a number of nosocomial infections such as surgical wound infections, sepsis, urinary tract infections and endocarditis (Kau et al., 2005).
Though a number of TA systems have been described for E. faecalis, the only Type I TA system described in this pathogen is the par locus (Weaver, 2012). Discovered in 1993 on the pAD1 plasmid, E. faecalis par locus was the first Type I TA system described in a Gram-positive bacterium and is now a well-known plasmid addiction module (Weaver et al., 1993). The existence of additional copies of the par TA system on the E. faecalis chromosome, as well as that of Streptococcus mutans (reviewed in Weaver et al., 2009; Koyanagi and Lévesque, 2013), Lactobacillus casei (Weaver et al., 2009) and, Lactobacillus rhamnosus (Weaver et al., 2009; Folli et al., 2017; Maggi et al., 2019) suggests additional functions for this system (Kwong et al., 2010).
The par locus consists of two convergently encoded genes; RNAI which encodes for the Fst toxin, and RNAII, which encodes an antisense sRNA antitoxin that represses Fst translation. Both RNAI and RNAII share a common bidirectional terminator (Greenfield et al., 2001). Also, at the 5’ end of each toxin and antitoxin transcript is a direct repeat sequence, DRa and DRb respectively (Greenfield and Weaver, 2000; Greenfield et al., 2000; Greenfield et al., 2001). These direct repeats and the common bidirectional terminator sequence constitute the base pairing interaction regions between the antitoxin and the toxin transcript. The Fst toxin is the founding member of a large superfamily of toxins. A Fst-like toxin is typically a 33 amino acid protein, consisting of a transmembrane domain and a charged C-terminal tail, both of which are essential for toxin activity. Fst-like toxins cause nucleoid condensation within the pathogen, leading to septal displacement and disproportional division of DNA into daughter cells (Maggi et al., 2019). Göbl et al., 2010 also suggested that the lethal activity of the Fst toxin is mediated by insertion of the alpha helical hydrophobic domain into the bacterial membrane, an activity which affects membrane permeability.
S. mutans is a Gram-positive facultative anaerobe that, together with other streptococcal species, is associated with the human oral cavity, pharynx and intestine (Loesche, 1986). S. mutans is associated with dental caries in humans as well as infections of the muscle, skin, cardiovascular and nervous systems (Forssten et al., 2010).
The S. mutans chromosomally encoded Fst-like toxin, Fst-Sm, together with its associated cis-encoded sRNA antitoxin called srSm constitute the first reported functional Type I TA system in the genus Streptococcus (Koyanagi and Lévesque, 2013). A homologue of E. faecalis Fst toxin, Fst-Sm was an unannotated open reading frame found within IGR176 intergenic region of the S. mutans UA159 reference strain (Fozo et al., 2010). Convergently encoded at the 3’ end of fst-Sm is the antitoxin sRNA, srSm, which is suggested to block Fst-Sm expression by binding to a tandem repeat close to the translation initiation region of the toxin transcript. Fst-Sm is reported to encode for a small hydrophobic peptide, the overexpression of which results in growth inhibition of S. mutans. Counterintuitively, the ectopic expression of the entire Fst-Sm/srSm Type I TA locus reduced, rather than increased, the number of cell-wall tolerant persisters. Further experimental validation is required to fully understand the role that the Fts-Sm/srSm Type I TA system plays in the virulence and physiology of S. mutans.
Bacillus species are Gram-positive bacilli which, much like Clostridia, are known for their spore forming ability that enables them to survive harsh environmental conditions. Composed of approximately 200 members, Bacillus is a large genus consisting of both pathogenic and non-pathogenic species. B anthracis and B. cereus are associated with anthrax in livestock and food poisoning in humans, respectively. Of the more than a dozen Type I TA systems identified in B. subtilis, the TxpA/RatA (Silvaggi et al., 2005; Durand et al., 2012a), BsrG/SR4 (Jahn et al., 2012), bsrE/SR5 (Müller et al., 2016) and yonT/yoyJ/SR6 (Reif et al., 2018) systems are the most extensively characterized to date.
Through microarray work that sought to identify novel untranslated RNAs within the B. subtilis genome in 2005, the first Type I TA system in Bacillus, txpA/RatA was identified, and shown to be located on chromosomal skin element (Silvaggi et al., 2005). This consists of the 59 amino acid long toxin and a convergently encoded antitoxin, RatA. Unlike BsrG/SR4, binding of the RatA sRNA antitoxin to the TxpA toxin transcript does not induce translation inhibition (Silvaggi et al., 2005). Instead, the RatA sRNA antitoxin interacts with the toxin’s mRNA at the 3’ end via an extensive amount of nucleic acid complementarity forming an RNA duplex which is cleavable by RNAse III. Similar to BsrG/SR4, deletion of ratA antitoxin caused lysis of B. subtilis on agar plates, but only after 5 days of incubation (Silvaggi et al., 2005). Aside from RatA repression, another possible mechanism by which B. subtilis is protected from the toxin gene is through an unusually strong (11-12) base pairing between the TxpA SD sequence and the anti-SD 3’ end of 16S rRNA (Daou-Chabo et al., 2009; Durand et al., 2012a). This near perfect interaction could potentially lead to ribosomal pause and slow translation as reviewed above (Daou-Chabo et al., 2009; Durand et al., 2012a). TxpA/RatA, is distinct from BsrG/SR4 in that RNAse III is indispensable in TxpA repression unlike in BsrG, as rnc mutants (lacking RNAse III) showed lysis of B. subtilis, even in the presence of the RatA (Silvaggi et al., 2005; Durand et al., 2012a; Jahn et al., 2012; Brantl and Müller, 2019). Since its characterization, no work has been done on elucidating the exact mechanism of TxpA activity, though with its predicted N-terminal transmembrane domain, it is likely a membrane targeting toxin like many Toxins of Type I TA systems (Silvaggi et al., 2005; Fozo et al., 2008b)
The bsrG/sr4 loci encodes the first B. subtillis temperature dependent Type I TA system described (Jahn et al., 2012; Jahn et al., 2015). This locus consists of bsrG, a gene encoding a small hydrophobic peptide, and sr4, a gene encoding an sRNA antitoxin that functions to repress production of the BsrG toxin. Having a predicted transmembrane domain and charged C-terminus, BsrG was shown to localize to the B. subtilis membrane (Jahn et al., 2015). However, unlike many Type I TA toxins, BsrG does not perforate the cell membrane, but rather disturbs envelope biosynthesis which leads to cell membrane invagination, dislocation of the cell wall synthesis machinery, and ultimately cell lysis (Jahn et al., 2015). Cognate toxin repression by SR4 antitoxin is mediated by an extensive amount of nucleic acid complementarity to the toxin transcript. The BsrG/SR4 system is unique in that toxin repression is dependent on both translation inhibition and degradation of the toxin transcript. Upon SR4 binding to the 3’ end of the bsrG transcript, structural changes occur in the area around the SD that inhibit translation. Additionally, the mRNA-sRNA duplex attracts an RNase for cleavage. In the steady state, antitoxin abundance is higher than that of the toxin due to the antitoxin promoter having 6-10-fold higher strength. At 48°C, the half-life of the bsrG mRNA decreases, resulting in significantly lower levels of the toxin at that temperature (Jahn et al., 2012).
The txpA/RatA Type I TA system, located on B. subtilis chromosomal skin element, was the first Type I TA system described in this species (Silvaggi et al., 2005; Durand et al., 2012a.) The RatA sRNA antitoxin interacts with the 3’ end of the toxin encoding txpA transcript via an extensive amount of nucleic acid complementarity, forming an RNA duplex which is cleavable by RNAse III.
A multi-stress responsive module BsrE/SR5 is another B. subtilis Type I TA and is located on the P6 prophage on the B. subtilis chromosome (Irnov et al., 2010; Durand et al., 2012b; Müller et al., 2016). One major characteristic of this module is the susceptibility of the toxin and antitoxin transcripts to RNAse degradation by a variety of ribonucleases, in response to different kinds of stress. Under O2, acidic and iron stress, the antitoxin SR5 is degraded by endoribonuclease J1. On the other hand, J1 degradation of bsrE transcript occurs in the presence of temperature shock and alkaline stress. Both transcripts however are rapidly degraded by endoribonuclease Y under ethanol stress. Whereas the antitoxin promoter strength is 10-fold more than that of the toxin, the toxin transcript is also about 10-fold more stable than its cognate antitoxin. The antitoxin transcript is cleaved by the exonuclease PnP prior to binding to the target toxin transcript.
S. aureus colonizes the skin and mucous membranes of approximately a third of the world’s population (Williams, 1963; Otto, 2010). Though it exists on human skins and nares as normal flora, S. aureus can cause a variety of infections upon entry into the bloodstream or deeper tissues of the body. S. aureus is the etiologic agent of several human diseases ranging from relatively mild skin and soft tissue infections, to life threatening infections such as necrotizing fasciitis, empyema, meningitis, and sepsis (Kobayashi et al., 2015). S. aureus is armed with several virulence factors that aid in survival within their human host (Otto, 2010), including five Type I and three Type III TA systems (Schuster and Bertram, 2016a; Harms et al., 2018; Habib et al., 2020).A homologue of E. faecalis Fst/RNAII, SprA1-SprA1AS was the first Type I TA system discovered in S. aureus. This TA system is encoded within the SaPIn3 pathogenicity island of the Newman strain (Pichon and Felden, 2005; Sayed et al., 2012a; Schuster and Bertram, 2016b; Bronsard et al., 2017; Habib et al., 2020). Subsequent analyses revealed a homologous locus termed SprA2/SprA2As (Germain-Amiot et al., 2019). This second system is located within the same S. aureus pathogenicity island, and shares 75% sequence similarity with the previously characterized SprA1/SprA1AS TA system (Germain-Amiot et al., 2019). As with Type I TA systems in other Gram-positive bacteria, genes encoding the toxin and antitoxin components of the SprA/SprAAS system are encoded antisense to each other and overlap at their 3’ ends, leading to perfect complementarity between these portions of the toxin mRNA and the antitoxin sRNA (Sayed et al., 2012a; Germain-Amiot et al., 2019). Experimental characterization of the SprA2/SprA2AS system revealed however, that the functional interaction between the antitoxin sRNA and the toxin mRNA occurs via the 5’ end of each molecule. This interaction abolishes toxin translation by blocking the ribosomal binding site and impeding ribosome loading (Habib et al., 2020).
Despite the high level of similarity, cross-regulation experiments demonstrated that between the sprA1/SprA1AS and sprA2/SprA2AS systems, antitoxin regulation is specific, each antitoxin only regulating production of its cognate toxin (Germain-Amiot et al., 2019).
The sprA1/SprA1AS and sprA2/SprA2AS system both encode for short toxic peptides, PepA1 and PepA2 (Sayed et al., 2012b; Germain-Amiot et al., 2019). While both PepA1 and PepA2 have been shown to be cytotoxic against human neutrophils, (Sayed et al., 2012a; Sayed et al., 2012b; Germain-Amiot et al., 2019), functional differences between these proteins have been identified. Firstly, unlike PepA2, PepA1 has been shown to play a role in interbacterial competition. Also, whereas osmotic shock and stringent conditions trigger the production of PepA2, PepA1 production is stimulated by acidic and oxidative stress. Such findings suggest differential production and different roles of these two Type I TA systems in the physiology and virulence of S. aureus (Germain-Amiot et al., 2019).
Another S. aureus Type I TA system, sprG1/SprF1, was discovered on the ΦSa3 PI mobile genetic element (Riffaud et al., 2019). This Type I TA locus is a homologue of the TxpA-RatA system in B. subtilis (Fozo et al., 2010) An additional 3 copies of this system, designated sprG2-4/SprF2-SprF4, were later identified on the S. aureus core genome (Riffaud et al., 2019). The toxin gene sprG1 encodes two peptides of unequal lengths utilizing different translational start sites on the transcript (Pinel-Marie et al., 2014; Wen and Fozo, 2014; Schuster and Bertram, 2016b). In addition to being self-lethal to S. aureus, these pore forming toxins have been shown to be capable of lysing both human red blood cells and competing bacterial species (Pinel-Marie et al., 2014). sprG1 shares complementarity at its 3’ end with SprF1, a relatively unstable sRNA antitoxin whose constitutive expression represses the production of the SprG1 toxin. Toxins from the sprG2-SprF2, sprG3-SprF3, sprG4-SprF4 TA systems also mediate bacteriostatic action to different extents in S. aureus, with a significant level of cross talk amongst antitoxin-mediated regulation (Riffaud et al., 2019; Riffaud et al., 2020).
H. pylori is a Gram-negative spiral bacterium that colonizes fifty percent of the human population. H. pylori is the first direct link between bacteria and cancer (gastric cancer), as well as the major cause of other gastric diseases such as peptic ulcer and gastritis (Kusters et al., 2006). With the site of infection being the human stomach, H. pylori possesses diverse molecular mechanisms that aid in survival within that environment (Wroblewski et al., 2010).
H. pylori carries six chromosomally encoded TA systems, each encoding a small toxin designated AapA1-6 and a cis encoded antisense sRNA, designated IsoA1-6. These systems were identified by the analysis of transcriptomic data, and were found to be expressed at high levels during the exponential phase of growth (Sharma et al., 2010). Of these TA systems, two (AapA1/IsoA1 and AapA3/IsoA3) were predicted to be members of the Type I TA family due to the arrangement of the genes within the loci, and the fact that production of the toxin is inhibited by the cognate antisense RNA (Sharma et al., 2010; Arnion et al., 2017). Further studies in H. pylori confirmed that the aapA1/isoA1 locus encodes for a typical Type I TA system, with the translation of the AapA1 peptide leading to reduced growth and lysis of the bacterium (Arnion et al., 2017). Interestingly, the full length AapA1 mRNA is translationally inactive upon transcription, as the SD is sequestered by a long-distance interaction (LDI) between the 5’ and 3’ ends of the transcript (Arnion et al., 2017). This structure gives the full-length transcript increased stability, not only preventing translation but also preventing interaction with the antitoxin. Processing of the aapA transcript at the 3’ end, induces structural alterations within the 5’ UTR that result in exposure of the SD sequence and translation of the peptide. This 3’ processing also allows for the IsaA1 antitoxin to interact with the aapA1 transcript through two kissing loop interactions, forming an RNA duplex that is subsequently cleavable by RNAse III (Arnion et al., 2017).
In addition to the LDI mediated prevention of toxin production, subsequent analyses of the H. pylori AapA3/IsoA3 TA system demonstrated that premature toxin translation is prevented by the formation of transiently stable stem loop structures within the 5’ UTR and the toxin open reading frame that is capable of sequestering the SD sequence within the growing transcript (Masachis et al., 2019). Such a mechanism prevents ribosomal loading and toxin translation before the 3’ end of the transcript is generated by progression of the RNA polymerase.
C. difficile is the major cause of antibiotic resistant, hospital acquired diarrhea (Heinlen and Ballard, 2010). Resistance to stressful conditions, and the ability to reestablish infection within treated hosts is due largely to spore formation by this pathogen. Additionally, TA system may contribute to the pathogen’s survival under adverse conditions (Vedantam et al., 2012).
Compared to those of Type II TA systems, Type I TA systems are relatively difficult to identify (Lobato-Márquez et al., 2015; Coray et al., 2017). Homology searches using previously characterized Type I TA systems from other bacteria did not identify many candidate Type I TAs in C. difficile (Soutourina, 2019). However, RNAseq-based promoter mapping and subsequent in silico analyses identified 251 putative sRNAs in C. difficile (Soutourina et al., 2013). This finding facilitated further data mining in the search for transcripts antisense to, or overlapping with genes that encode for small proteins of unknown function (Maikova et al., 2018). From this, six TA candidates were identified and three of them, CD2517.1-RCd8, CD2907.1-RCd9, and CD0956.2-RCd10, were studied further (Maikova et al., 2018). The identified putative toxins are predicted to be 50 amino acid long transmembrane proteins with conserved charged amino acids at the C-terminus. Production of these toxin proteins arrest C. difficile growth, a phenotype which is inhibited by cognate sRNA antitoxins with high specificity (Maikova et al., 2018). Interestingly, a close association of these TA systems to CRSPR sequences is observed, suggesting a possible role in maintenance of these important regions of the bacterial chromosomal (Maikova et al., 2018).
E coli is a Gram-negative rod-shaped bacterium. Though a major constituent of normal intestinal flora, more than six pathotypes have been noted to cause gastrointestinal and urinary tract infections. E. coli is one of the most extensively studied bacteria for Type I TA systems. In total, approximately 26 Type 1 TA systems have been identified in both pathogenic and non-pathogenic strains of E. coli (Fozo et al., 2010). Selected systems will be highlighted here.
Apart from the E. faecalis Fst/RNAII system, the early days of bacteria TA research saw the discovery of the plasmid encoded Hok/Sok Type I TA system within E. coli, now one of the most studied Type I TA system in this species (Gerdes and Wagner, 2007; Yang and Walsh, 2017). A plasmid addiction module, Hok/Sok is involved in maintaining the R1 plasmid through post segregational killing, a phenomenon largely adapted to explain the roles of plasmid encoded TA systems within bacteria (Gerdes et al., 1986b; Gerdes et al., 1986a). The Hok protein is a transmembrane protein that has been shown to target the bacterial membrane, destroying its integrity through the formation of tiny anion selective pores that result in the depletion of ATP and the formation of so called ‘ghost cells’ (Gerdes et al., 1986b; Peng et al., 2019). The Sok antitoxin is an sRNA that is involved in the indirect inhibition of Hok translation, by blocking an open reading frame, mok, that overlaps with the hok sequence, whose translation is coupled to hok translation. Importantly, for either mok translation or Sok inhibitory binding to occur, the full length hok mRNA must first undergo 3’ end processing to release the transcript from a fold back inhibition mediated by binding between the 5’ and 3’ ends of the transcript (Thisted et al., 1994).
Like the Hok/Sok system is the Ldr/Rdl putative Type I TA system (Kawano, 2012). This system is composed of a short LdrD toxin whose production in E. coli is lethal to the organism and whose production is regulated by the sRNA antitoxin, RdlD. Much like the Hok/Sok/Mok arrangement, the ldrD toxin transcript has a small open reading frame, ldrX within its long 5’ UTR which overlaps with the translation initiation region of the toxin gene. Hence translation of the toxin gene is coupled to that of the small open reading frame ldrX. RdlD inhibits the production of LdrD by blocking the translation of ldrX (Kawano et al., 2002; Kawano, 2012).
Another E. coli Type I TA system is the SymE/SymR system (Kawano et al., 2007). This system is composed of an endonuclease toxin, SymE, and a sRNA antitoxin, SymR. Unlike most Type I antitoxins, SymR is unique in having a higher stability than its toxin counterpart. At steady state, symE expression is tightly repressed by three different processes. Firstly, the symE gene is transcriptionally regulated by the LexA repressor. Second, following transcription, the SymR antitoxin prevents translation from the symE transcript through complementary base pairing in the 5’ UTR of the toxin transcript. Finally, Lon protease acts post-translationally to degrade the SymE toxin (Kawano, 2012). This multilayer repression of the toxin likely protects the bacteria from toxin production until it is needed, such as when the bacterium experiences DNA damage. Under such circumstances, production of the SymR antitoxin is repressed by the SOS response, allowing toxin production to proceed. Once produced, SymE inhibits growth by degrading DNA and decreasing protein synthesis (Kawano et al., 2007, Kawano, 2012).
Another well studied Type I TA system in E. coli is TisB/IstR system. This is another SOS-induced Type I TA toxin like SymE/SymR (Kawano et al., 2007). As an SOS-induced toxin gene, tisB is repressed in the absence of stress by the master regulator, LexA. In addition, TisB production is repressed by a trans encoded sRNA antitoxin, IstR. Unique among Type I TA systems, tisB expression is inhibited by a structure in its 5’ UTR that occludes a ribosome standby site (RSS) on the transcript. Like most Type I TA toxin transcripts, a processing event is required to render tisB translatable. This processed transcript, however, can also be bound by the antitoxin IstR-1, to generate a RNA duplex that is cleaved by RNAse III (Vogel et al., 2004). In a IstR-1 deleted strain, TisB is translated and causes growth inhibition due to membrane depolarization.
Present on the chromosome of E. coli O157:H7 are two nearly identical Type I TA systems designated zorO/orzO and zorP/orzP (Wen and Fozo, 2013). Of these two systems, zorO/orzo has been studied in more detail (Wen and Fozo, 2013). Increased production of ZorO is toxic to E. coli, a phenotype that can be reversed by the coproduction of the OrzO antitoxin in a manner dependent upon RNase III. In addition to repression by ZorO, the full-length ZorO 5’UTR is sufficient to inhibit its own translation by forming an inhibitory structure that sequesters the SD (Wen et al., 2017). OrzP-dependent repression of zorO is mediated by nucleic acid complementarity between the sRNA antitoxin and the target toxin transcript. Interestingly, despite nucleic acid sequence similarity between the two antitoxins, repression of zorO is achieved only by the OrzO antitoxin and not by neighboring OrzP molecule.
S. enterica serovar Typhimurium is a member of the Enterobacteriaceae family and has been implicated in a variety of food-borne illnesses. Three E. coli Type I TA homologues found in Salmonella enterica serovar Typhimurium, hok-sokST, ldrA-rdlAST, tisB-istRST (described above) have been shown to play a role in the intracellular lifestyle of the pathogen (Lobato-Márquez et al., 2015). As functional Type I TA toxins, overproduction of HokST, LdrAST or TisBST inhibits S. Typhimurium growth (Lobato-Márquez et al., 2015). S. Typhimurium is known to inhibit its proliferation to survive within fibroblasts (Cano et al., 2001). Consistent with their growth inhibition phenotype, genes encoding these three toxins were shown to be upregulated, in bacteria isolated from fibroblasts. More importantly, deletion of the toxin genes resulted in limited intracellular survival within fibroblasts, based on survival assays, indicating the significance of the toxin genes in survival within such intracellular environments (Lobato-Márquez et al., 2015). Relevance of a TA system in survival of S. Typhimurium is not new, as Type II TA modules have previously been suggested in S. Typhimurium persistence within macrophages (Stårsta et al., 2020). However, this study highlights for the first time that Type I TAs are equally important in facilitating adaptation of S. Typhimurium to the intracellular environment.
Recently, a new Type I TA system has been proposed in Salmonella, termed TimR/TimP (Andresen et al., 2020). TimP transcript was originally identified in E. coli, through a transcriptomic study as a RNA of unknown function, RyfA, and no conserved open reading frame (ORF) was identified for it in that study (Wassarman et al., 2001; Andresen et al., 2020). Subsequently, conservation within different genomic contexts, and expression of RyfA were observed in other members of the Enterobacteriaceae family (Hershberg et al., 2003; Andresen et al., 2020), and relevance in virulence and biofilm formation observed for RyfA expression in Shigella (Fris et al., 2017) and ocular pathogenic E. coli respectively (Ranjith et al., 2019). However, the work done in S. enterica serovar Typhimurium showed the production of small toxic membrane peptide, TimP from previously termed ryfA sRNA. Translation of this toxin was shown to be repressed by a divergently encoded sRNA, TimR. When overexpressed, TimP damages the membrane, leading to growth inhibition of Salmonella (Andresen et al., 2020).
Like Type I TA systems, Type III TA systems have a protein toxin and an sRNA antitoxin. Unlike Type I TA systems, however, the Type III sRNA antitoxin functions to protect against the lethal activity of the toxin by directly binding the toxin itself. The locus encoding each Type III TA system has a single promoter which constitutively drives the transcription of a polycistronic transcript encoding for both the toxin protein and antitoxin sRNA. The antitoxin gene is encoded upstream that of the toxin, with a Rho-independent transcriptional terminator separating them. This Rho-independent terminator is suggested to control the sRNA to toxin mRNA stoichiometry, allowing excess of antitoxins via infrequent read-throughs. The antitoxin transcript is 200-nucleotide long when initially transcribed, and it is characterized by the presence of nearly identical tandemly arranged repeat sequences. The number of repeat sequences is specific for each Type III TA system described. Each repetitive sequence is 36 nucleotides long and can be separated by cleavage by the toxin which is an endonuclease. Following cleavage, the antitoxin fragments form a complex with the endonuclease toxin that functionally, inactivates the toxin. Such inactivation prevents the toxin from cleaving other essential cellular targets (Blower et al., 2012; Brantl and Jahn, 2015).
Research on Type III TA systems is limited, with just one such system identified in a pathogenic bacterium to date; the ToxI/ToxN system in the plant pathogen, Pectobacterium atroseptica. (Short et al., 2018). Here, the plasmid encoded Type III TA locus consists of the ToxN toxin and the ToxI antitoxin.
While the best studied Type III TA system is the ToxI/ToxN system in the plant pathogen, Pectobacterium atroseptica (Short et al., 2018), other type III TA systems have been studied based on similarity to this system. In Lactobacillus lactis, the plasmid encoded AbiQ/AntiQ TA system is similar in genetic context; having a long antitoxin transcript at the 5’ end of a polycistronic transcript separated from the gene encoding the endonuclease toxin by a weak terminator (Samson et al., 2013). Two other Type III systems, TenpN/TenpI and CptN/CptI, have been predicted based on homology to the amino acid sequence of ToxN. Together with the well characterized ToxI/ToxN module, these make up three families of Type III TA systems (Blower et al., 2012).
Despite the recent discoveries of new Type I TA systems in bacteria, the number of RNA-regulated TA systems identified and verified to date still trails behind that of the Type II TA system. This could be due in part to the relatively small size of Type I toxin proteins, and to the fact that regulatory sRNAs can be difficult to identify by sequence analyses. Additionally, the toxins produced by Type I TA systems do not share extensive conservation with each other but rather only share the characteristic transmembrane domain and a C-terminus made of aromatic amino acids. This results in many Type I TA systems having sequence divergence across different bacteria. In fact, Fozo et al. (2010) observed the difficulty in predicting Type I TA systems using the default parameters of the NCBI homology search platforms alone. However, by employing a computational approach involving an exhaustive TBLASTN and PSI-BLAST across approximately 800 sequenced genomes, the group demonstrated that, contrary to what was previously reported, Type I TA systems are not narrowly distributed within bacteria by horizontal transfer, but rather are widely distributed across different bacteria (Fozo et al., 2010). More recently this approach was used to identify 5 Type I TA homologs in Salmonella Typhimurium strain SL1344 (Lobato-Márquez et al., 2015). Additionally, the study increased the known number of Type I TA systems by discovering new loci using search parameters based on other known Type I TAs, including location of the short open reading frames in the intergenic region, the presence of a transmembrane domain and a bulky C-terminus residue, and the proximity of the putative antitoxin sRNA (Fozo et al., 2010).
The identification of new Type III TA systems, on the other hand, has highly relied on homology searches of structural motifs across different bacteria, leading to the discovery of new putative Type III TA homologues though many lack experimental validation to date. Future investigations will no doubt, provide additional information about the distribution and function of Type III TA systems in pathogenic bacteria (Blower et al., 2012).
A recent finding suggests the existence of a new class of TA systems, one in which both the toxin and antitoxin are sRNA molecules. The inaugural member of this new class of TA systems is the SdsR/RyeA in E. coli (Choi et al., 2018). Here, the RyeA toxin targets and represses expression of the tolC and mutS genes in an Hfq-dependent manner, leading to cell lysis of the bacterium. Interestingly, a previously characterized E. coli sRNA has recently been shown to encode for a small peptide in Salmonella and is now suggested to be a part of a Type I TA pair (Andresen et al., 2020). Together, these findings therefore present the interesting possibility of a Type I TA toxin having regulatory function as both an sRNA and a protein. Similarly, an interesting question was raised in a recent review about the possibility of Type I and III sRNA antitoxins having additional regulatory activity within bacteria, aside from toxin repression. Dual functionality of components of bacterial TA systems is clearly worthy of further investigating (Riffaud et al., 2020).
Different studies assign different functions to various Type I TA systems. Deletion experiments, overexpression experiments and identifying the conditions in which the toxins are expressed provide clues about the putative role(s) of a TA system. However, a number of these findings are lacking in their universality and are not fully confirmed. A possible explanation for the difficulty in functional characterization of the sRNA-regulated TA systems is the reliance on overexpression experiments, which might not represent the physiological levels of the molecules in the bacterium (Masachis et al., 2019).
In conclusion, the RNA regulated TA systems provide additional evidence to the importance of regulatory RNAs in controlling bacteria gene expression. As with all sRNA-mediated regulation, the involvement of a sRNA antitoxin in a TA system could provide an advantage over a protein counterpart. The extra energy investment required for the production of a protein antitoxin is relieved if that antitoxin is instead, an sRNA molecule (Fozo et al., 2008b). There is no doubt that with further investigation the recognized significance of the role that RNA regulated TA systems play in the physiology and virulence of pathogenic bacteria will continue to grow as will the knowledge of the molecular mechanism underlying their activity.
DS wrote the first draft which was contributed to, and edited, by EM. All authors contributed to the article and approved the submitted version.
NIH (R15AI103887-01A1) - supported studies related to this paper. Ohio University Heritage College of Osteopathic Medicine: Will help cover the publication costs. Infectious and Tropical Disease Institute, Ohio University: Will help cover the publication costs. Molecular and Cellular Biology Program, Ohio University: Will help cover the publication costs.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge Ohio University, Ohio University Heritage College of Osteopathic Medicine, the Tropical and Infectious Disease Institute at Ohio University, the Ohio University Molecular and Cellular Biology Program, and NIH NIAID (R15AI103887-01A1) for support of this work. Figures were created with biorender.com.
Andresen, L., Martínez-Burgo, Y., Zangelin, J. N., Rizvanovic, A., Holmqvist, E. (2020). The Small Toxic Salmonella Protein TimP Targets the Cytoplasmic Membrane and Is Repressed by the Small RNA Timr. mBio 11, 1–16. doi: 10.1128/mBio.01659-20
Arnion, H., Korkut, D. N., Masachis Gelo, S., Chabas, S., Reignier, J., Iost, I., et al. (2017). Mechanistic Insights Into Type I Toxin Antitoxin Systems in Helicobacter pylori: The Importance of mRNA Folding in Controlling Toxin Expression. Nucleic Acids Res. 45 (8), 4782–4795. doi: 10.1093/nar/gkw1343
Berghoff, B. A., Wagner, E. G. H. (2017). RNA-Based Regulation in Type I Toxin–Antitoxin Systems and its Implication for Bacterial Persistence. Curr. Genet. 63, 1011–1016. doi: 10.1007/s00294-017-0710-y
Blower, T. R., Short, F. L., Rao, F., Mizuguchi, K., Pei, X. Y., Fineran, P. C., et al. (2012). Identification and Classification of Bacterial Type III Toxin-Antitoxin Systems Encoded in Chromosomal and Plasmid Genomes. Nucleic Acids Res. 40, 6158–6173. doi: 10.1093/nar/gks231
Brantl, S., Jahn, N. (2015). sRNAs in Bacterial Type I and Type III Toxin-Antitoxin Systems. FEMS Microbiol. Rev. 39, 413–427. doi: 10.1093/femsre/fuv003
Brantl, S., Müller, P. (2019). Toxin–Antitoxin Systems in Bacillus Subtilis. Toxins (Basel) 11, 1–18. doi: 10.3390/toxins11050262
Brauner, A., Fridman, O., Gefen, O., Balaban, N. Q. (2016). Distinguishing Between Resistance, Tolerance and Persistence to Antibiotic Treatment. Nat. Rev. Microbiol. 14, 320–330. doi: 10.1038/nrmicro.2016.34
Brendler, T., Reaves, L., Austin, S. (2004). Interplay Between Plasmid Partition and Postsegregational Killing Systems. J. Bacteriol. 186, 2504–2507. doi: 10.1128/JB.186.8.2504-2507.2004
Brennan, R. G., Link, T. M. (2007). Hfq Structure, Function and Ligand Binding. Curr. Opin. Microbiol. 10, 125–133. doi: 10.1016/j.mib.2007.03.015
Brielle, R., Pinel-Marie, M.-L., Felden, B. (2016). Linking Bacterial Type I Toxins With Their Actions. Curr. Opin. Microbiol. 30, 114–121. doi: 10.1016/j.mib.2016.01.009
Bronsard, J., Pascreau, G., Sassi, M., Mauro, T., Augagneur, Y., Felden, B. (2017). sRNA and Cis-Antisense sRNA Identification in Staphylococcus Aureus Highlights an Unusual sRNA Gene Cluster With One Encoding a Secreted Peptide. Sci. Rep. 7, 4565. doi: 10.1038/s41598-017-04786-3
Brooks, A. N., Turkarslan, S., Beer, K. D., Lo, F. Y., Baliga, N. S. (2011). Adaptation of Cells to New Environments. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 544–561. doi: 10.1002/wsbm.136
Cano, D. A., Martínez-Moya, M., Pucciarelli, M. G., Groisman, E. A., Casadesús, J., García-Del Portillo, F. (2001). Salmonella Enterica Serovar Typhimurium Response Involved in Attenuation of Pathogen Intracellular Proliferation. Infect. Immun. 69, 6463–6474. doi: 10.1128/IAI.69.10.6463-6474.2001
Choi, J. S., Kim, W., Suk, S., Park, H., Bak, G., Yoon, J., et al. (2018). SdsR, Acts as a Novel Type of Toxin in Escherichia Coli. RNA Biol. 15, 1319–1335. doi: 10.1080/15476286.2018.1532252
Coray, D. S., Wheeler, N. E., Heinemann, J. A., Gardner, P. P. (2017). Why So Narrow: Distribution of Anti-Sense Regulated, Type I Toxin-Antitoxin Systems Compared With Type II and Type III Systems. RNA Biol. 14, 275–280. doi: 10.1080/15476286.2016.1272747
Dambach, M., Irnov, I., Winkler, W. C. (2013). Association of RNAs With Bacillus Subtilis Hfq. PloS One 8, e55156. doi: 10.1371/journal.pone.0055156
Daou-Chabo, R., Mathy, N., Bénard, L., Condon, C. (2009). Ribosomes Initiating Translation of the Hbs mRNA Protect it From 5′-to-3′ Exoribonucleolytic Degradation by RNase J1. Mol. Microbiol. 71, 1538–1550. doi: 10.1111/j.1365-2958.2009.06620.x
Darfeuille, F., Unoson, C., Vogel, J., Wagner, E. G. H. (2007). An Antisense RNA Inhibits Translation by Competing With Standby Ribosomes. Mol. Cell 26, 381–392. doi: 10.1016/j.molcel.2007.04.003
Dörr, T., Vulić, M., Lewis, K. (2010). Ciprofloxacin Causes Persister Formation by Inducing the TisB Toxin in Escherichia Coli. PloS Biol. 8, e1000317. doi: 10.1371/journal.pbio.1000317
Durand, S., Gilet, L., Condon, C. (2012a). The Essential Function of B. Subtilis RNase Iii Is to Silence Foreign Toxin Genes. PloS Genet. 8, e1003181. doi: 10.1371/journal.pgen.1003181
Durand, S., Jahn, N., Condon, C., Brantl, S. (2012b). Type I Toxin-Antitoxin Systems in Bacillus Subtilis. RNA Biol. 9, 1491–1497. doi: 10.4161/rna.22358
El Mortaji, L., Tejada-Arranz, A., Rifflet, A., Boneca, I. G., Pehau-Arnaudet, G., Radicella, J. P., et al. (2020). A Peptide of a Type I Toxin–Antitoxin System Induces Helicobacter Pylori Morphological Transformation From Spiral Shape to Coccoids. Proc. Natl. Acad. Sci. U.S.A. 117, 31398–31409. doi: 10.1073/pnas.2016195117
Fauvart, M., De Groote, V. N., Michiels, J. (2011). Role of Persister Cells in Chronic Infections: Clinical Relevance and Perspectives on Anti-Persister Therapies. J. Med. Microbiol. 60, 699–709. doi: 10.1099/jmm.0.030932-0
Fernández-García, L., Blasco, L., Lopez, M., Bou, G., García-Contreras, R., Wood, T., et al. (2016). Toxin-Antitoxin Systems in Clinical Pathogens. Toxins (Basel) 8, 1–23. doi: 10.3390/toxins8070227
Fernández, L., Rodríguez, A., García, P. (2018). Phage or Foe: An Insight Into the Impact of Viral Predation on Microbial Communities. ISME J. 12, 1171–1179. doi: 10.1038/s41396-018-0049-5
Folli, C., Levante, A., Percudani, R., Amidani, D., Bottazzi, S., Ferrari, A., et al. (2017). Toward the Identification of a Type I Toxin-Antitoxin System in the Plasmid DNA of Dairy Lactobacillus Rhamnosus. Sci. Rep. 7, 12051. doi: 10.1038/s41598-017-12218-5
Forssten, S. D., Björklund, M., Ouwehand, A. C. (2010). Streptococcus Mutans, Caries and Simulation Models. Nutrients 2, 290–298. doi: 10.3390/nu2030290
Fozo, E. M. (2012). New Type I Toxin-Antitoxin Families From “Wild” and Laboratory Strains of E. Coli: Ibs-Sib, ShoB-OhsC and Zor-Orz. RNA Biol. 9, 1504–1512. doi: 10.4161/rna.22568
Fozo, E. M., Hemm, M. R., Storz, G. (2008a). Small Toxic Proteins and the Antisense Rnas That Repress Them. Microbiol. Mol. Biol. Rev. 72, 579–589. doi: 10.1128/MMBR.00025-08
Fozo, E. M., Hemm, M. R., Storz, G. (2008b). Small Toxic Proteins and the Antisense RNAs That Repress Them. Microbiol. Mol. Biol. Rev. 72, 579–589. doi: 10.1128/MMBR.00025-08
Fozo, E. M., Makarova, K. S., Shabalina, S. A., Yutin, N., Koonin, E. V., Storz, G. (2010). Abundance of Type I Toxin-Antitoxin Systems in Bacteria: Searches for New Candidates and Discovery of Novel Families. Nucleic Acids Res. 38, 3743–3759. doi: 10.1093/nar/gkq054
Fris, M. E., Broach, W. H., Klim, S. E., Coschigano, P. W., Carroll, R. K., Caswell, C. C., et al. (2017). Sibling Srna RyfA1 Influences Shigella Dysenteriae Pathogenesis. Genes 8:50. doi: 10.3390/genes8020050
Gerdes, K. (2016). Hypothesis: Type I Toxin–Antitoxin Genes Enter the Persistence Field—a Feedback Mechanism Explaining Membrane Homoeostasis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 371, 1–9. doi: 10.1098/rstb.2016.0189
Gerdes, K., Bech, F. W., Jørgensen, S. T., Løbner-Olesen, A., Rasmussen, P. B., Atlung, T., et al. (1986a). Mechanism of Postsegregational Killing by the Hok Gene Product of the Parb System of Plasmid R1 and its Homology With the Relf Gene Product of the E. Coli Relb Operon. EMBO J. 5, 2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x
Gerdes, K., Rasmussen, P. B., Molin, S. (1986b). Unique Type of Plasmid Maintenance Function: Postsegregational Killing of Plasmid-Free Cells. Proc. Natl. Acad. Sci. U.S.A. 83, 3116–3120. doi: 10.1073/pnas.83.10.3116
Gerdes, K., Wagner, E. G. H. (2007). RNA Antitoxins. Curr. Opin. Microbiol. 10, 117–124. doi: 10.1016/j.mib.2007.03.003
Germain-Amiot, N., Augagneur, Y., Camberlein, E., Nicolas, I., Lecureur, V., Rouillon, A., et al. (2019). A Novel Staphylococcus Aureus Cis–Trans Type I Toxin–Antitoxin Module With Dual Effects on Bacteria and Host Cells. Nucleic Acids Res. 47, 1759–1773. doi: 10.1093/nar/gky1257
Göbl, C., Kosol, S., Stockner, T., Rückert, H. M., Zangger, K. (2010). Solution Structure and Membrane Binding of the Toxin Fst of the Par Addiction Module. Biochemistry 49, 6567–6575. doi: 10.1021/bi1005128
Greenfield, T. J., Ehli, E., Kirshenmann, T., Franch, T., Gerdes, K., Weaver, K. E. (2000). The Antisense RNA of the Par Locus of pAD1 Regulates the Expression of a 33-Amino-Acid Toxic Peptide by an Unusual Mechanism. Mol. Microbiol. 37, 652–660. doi: 10.1046/j.1365-2958.2000.02035.x
Greenfield, T. J., Franch, T., Gerdes, K., Weaver, K. E. (2001). Antisense RNA Regulation of the Par Post-Segregational Killing System: Structural Analysis and Mechanism of Binding of the Antisense RNA, RNAII and its Target, RNAI. Mol. Microbiol. 42, 527–537. doi: 10.1046/j.1365-2958.2001.02663.x
Greenfield, T. J., Weaver, K. E. (2000). Antisense RNA Regulation of the pAD1 Par Post-Segregational Killing System Requires Interaction at the 5′ and 3′ Ends of the Rnas. Mol. Microbiol. 37, 661–670. doi: 10.1046/j.1365-2958.2000.02034.x
Guo, Y., Quiroga, C., Chen, Q., McAnulty, M. J., Benedik, M. J., Wood, T. K., et al. (2014). Ralr (a DNase) and RalA (a Small RNA) Form a Type I Toxin–Antitoxin System in Escherichia Coli. Nucleic Acids Res. 42, 6448–6462. doi: 10.1093/nar/gku279
Habib, G., Zhu, J., Sun, B. (2020). A Novel Type I Toxin-Antitoxin System Modulates Persister Cell Formation in Staphylococcus Aureus. Int. J. Med. Microbiol. 310, 151400. doi: 10.1016/j.ijmm.2020.151400
Harms, A., Brodersen, D. E., Mitarai, N., Gerdes, K. (2018). Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 70, 768–784. doi: 10.1016/j.molcel.2018.01.003
Heaton, B. E., Herrou, J., Blackwell, A. E., Wysocki, V. H., Crosson, S. (2012). Molecular Structure and Function of the Novel BrnT/BrnA Toxin-Antitoxin System of Brucella Abortus. J. Biol. Chem. 287, 12098–12110. doi: 10.1074/jbc.M111.332163
Heinlen, L., Ballard, J. D. (2010). Clostridium Difficile Infection. Am. J. Med. Sci. 340, 247–252. doi: 10.1097/MAJ.0b013e3181e939d8
Hershberg, R., Altuvia, S., Margalit, H. (2003). Survey AND Summary: A Survey of Small RNA-encoding Genes in Escherichia Coli. Nucleic Acids Res. 31, 1813–1820. doi: 10.1093/nar/gkg297
Holmqvist, E., Wagner, E. G. H. (2017). Impact of Bacterial sRNAs in Stress Responses. Biochem. Soc. Trans. 45, 1203–1212. doi: 10.1042/BST20160363
Irnov, I., Sharma, C. M., Vogel, J., Winkler, W. C. (2010). Identification of Regulatory RNAs in Bacillus Subtilis. Nucleic Acids Res. 38, 6637–6651. doi: 10.1093/nar/gkq454
Jahn, N., Brantl, S., Strahl, H. (2015). Against the Mainstream: The Membrane-Associated Type I Toxin BsrG From Bacillus Subtilis Interferes With Cell Envelope Biosynthesis Without Increasing Membrane Permeability. Mol. Microbiol. 98, 651–666. doi: 10.1111/mmi.13146
Jahn, N., Preis, H., Wiedemann, C., Brantl, S. (2012). BsrG/SR4 From Bacillus Subtilis–the First Temperature-Dependent Type I Toxin-Antitoxin System. Mol. Microbiol. 83, 579–598. doi: 10.1111/j.1365-2958.2011.07952.x
Jurėnas, D., Van Melderen, L. (2020). The Variety in the Common Theme of Translation Inhibition by Type Ii Toxin–Antitoxin Systems. Front. Genet. 11:262. doi: 10.3389/fgene.2020.00262
Kędzierska, B., Hayes, F. (2016). Emerging Roles of Toxin-Antitoxin Modules in Bacterial Pathogenesis. Molecules 21, 1–25. doi: 10.3390/molecules21060790
Kau, A. L., Martin, S. M., Lyon, W., Hayes, E., Caparon, M. G., Hultgren, S. J. (2005). Enterococcus Faecalis Tropism for the Kidneys in the Urinary Tract of C57BL/6J Mice. Infect. Immun. 73, 2461–2468. doi: 10.1128/IAI.73.4.2461-2468.2005
Kawano, M. (2012). Divergently Overlapping Cis-Encoded Antisense RNA Regulating Toxin-Antitoxin Systems From E. Coli: Hok/Sok, Ldr/Rdl, Syme/Symr. RNA Biol. 9, 1520–1527. doi: 10.4161/rna.22757
Kawano, M., Oshima, T., Kasai, H., Mori, H. (2002). Molecular Characterization of Long Direct Repeat (LDR) Sequences Expressing a Stable mRNA Encoding for a 35-Amino-Acid Cell-Killing Peptide and a Cis-Encoded Small Antisense RNA in Escherichia Coli. Mol. Microbiol. 45, 333–349. doi: 10.1046/j.1365-2958.2002.03042.x
Kawano, M., Aravind, L., Storz, G. (2007). An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 64 (3), 738–754.
Kim, J.-S., Wood, T. K. (2016). Persistent Persister Misperceptions. Front. Microbiol. 7:2134. doi: 10.3389/fmicb.2016.02134
Kobayashi, S. D., Malachowa, N., DeLeo, F. R. (2015). Pathogenesis of Staphylococcus Aureus Abscesses. Am. J. Pathol. 185, 1518–1527. doi: 10.1016/j.ajpath.2014.11.030
Korch, S. B., Contreras, H., Clark-Curtiss, J. E. (2009). Three Mycobacterium Tuberculosis Rel Toxin-Antitoxin Modules Inhibit Mycobacterial Growth and are Expressed in Infected Human Macrophages. J. Bacteriol. 191, 1618–1630. doi: 10.1128/JB.01318-08
Koyanagi, S., Lévesque, C. M. (2013). Characterization of a Streptococcus Mutans Intergenic Region Containing a Small Toxic Peptide and Its Cis-Encoded Antisense Small RNA Antitoxin. PloS One 8, e54291. doi: 10.1371/journal.pone.0054291
Kristiansen, K. I., Weel-Sneve, R., Booth, J. A., Bjørås, M. (2016). Mutually Exclusive RNA Secondary Structures Regulate Translation Initiation of DinQ in Escherichia Coli. RNA 22, 1739–1749. doi: 10.1261/rna.058461.116
Kristich, C. J., Rice, L. B., Arias, C. A. (2014)Enterococcal Infection—Treatment and Antibiotic Resistance. In: Enterococci: From Commensals to Leading Causes of Drug Resistant Infection (Boston: Massachusetts Eye and Ear Infirmary). Available at: http://www.ncbi.nlm.nih.gov/books/NBK190420/ (Accessed January 29, 2021).
Kusters, J. G., van Vliet, A. H. M., Kuipers, E. J. (2006). Pathogenesis of Helicobacter Pylori Infection. Clin. Microbiol. Rev. 19, 449–490. doi: 10.1128/CMR.00054-05
Kwong, S. M., Jensen, S. O., Firth, N. (2010). Prevalence of Fst-like Toxin-Antitoxin Systems. Microbiol. (Reading) 156, 975–977. doi: 10.1099/mic.0.038323-0
Lee, K.-Y., Lee, B.-J. (2016). Structure, Biology, and Therapeutic Application of Toxin–Antitoxin Systems in Pathogenic Bacteria. Toxins 8:305. doi: 10.3390/toxins8100305
Lobato-Márquez, D., Díaz-Orejas, R., García-del Portillo, F. (2016a). Toxin-Antitoxins and Bacterial Virulence. FEMS Microbiol. Rev. 40, 592–609. doi: 10.1093/femsre/fuw022
Lobato-Márquez, D., Molina-García, L., Moreno-Córdoba, I., García-del Portillo, F., Díaz-Orejas, R. (2016b). Stabilization of the Virulence Plasmid pSLT of Salmonella Typhimurium by Three Maintenance Systems and Its Evaluation by Using a New Stability Test. Front. Mol. Biosci. 3:66. doi: 10.3389/fmolb.2016.00066
Lobato-Márquez, D., Moreno-Córdoba, I., Figueroa, V., Díaz-Orejas, R., García-del Portillo, F. (2015). Distinct Type I and Type II Toxin-Antitoxin Modules Control Salmonella Lifestyle Inside Eukaryotic Cells. Sci. Rep. 5, 9374. doi: 10.1038/srep09374
Loesche, W. J. (1986). Role of Streptococcus Mutans in Human Dental Decay. Microbiol. Rev. 50, 353–380. doi: 10.1128/MR.50.4.353-380.1986
Maggi, S., Yabre, K., Ferrari, A., Lazzi, C., Kawano, M., Rivetti, C., et al. (2019). Functional Characterization of the Type I Toxin Lpt From Lactobacillus Rhamnosus by Fluorescence and Atomic Force Microscopy. Sci. Rep. 9, 15208. doi: 10.1038/s41598-019-51523-z
Maikova, A., Peltier, J., Boudry, P., Hajnsdorf, E., Kint, N., Monot, M., et al. (2018). Discovery of New Type I Toxin–Antitoxin Systems Adjacent to CRISPR Arrays in Clostridium Difficile. Nucleic Acids Res. 46, 4733–4751. doi: 10.1093/nar/gky124
Ma, D., Mandell, J. B., Donegan, N. P., Cheung, A. L., Ma, W., Rothenberger, S., et al. (2019). The Toxin-Antitoxin Mazef Drives Staphylococcus Aureus Biofilm Formation, Antibiotic Tolerance, and Chronic Infection. mBio 10, 1–15. doi: 10.1128/mBio.01658-19
Masachis, S., Darfeuille, F. (2018). Type I Toxin-Antitoxin Systems: Regulating Toxin Expression Via Shine-Dalgarno Sequence Sequestration and Small Rna Binding. Microbiol. Spectr. 6, 1–18. doi: 10.1128/microbiolspec.RWR-0030-2018
Masachis, S., Tourasse, N. J., Lays, C., Faucher, M., Chabas, S., Iost, I., et al. (2019). A Genetic Selection Reveals Functional Metastable Structures Embedded in a Toxin-Encoding Mrna. eLife 8, e47549. doi: 10.7554/eLife.47549
McVicker, G., Tang, C. M. (2016). Deletion of Toxin–Antitoxin Systems in the Evolution of Shigella Sonnei as a Host-Adapted Pathogen. Nat. Microbiol. 2, 1–8. doi: 10.1038/nmicrobiol.2016.204
Michaux, C., Hartke, A., Martini, C., Reiss, S., Albrecht, D., Budin-Verneuil, A., et al. (2014). Involvement of Enterococcus Faecalis Small Rnas in Stress Response and Virulence. Infect. Immun. 82, 3599. doi: 10.1128/IAI.01900-14
Michiels, J. E., Van den Bergh, B., Verstraeten, N., Michiels, J. (2016). Molecular Mechanisms and Clinical Implications of Bacterial Persistence. Drug Resist. Upd. 29, 76–89. doi: 10.1016/j.drup.2016.10.002
Müller, P., Jahn, N., Ring, C., Maiwald, C., Neubert, R., Meißner, C., et al. (2016). A Multistress Responsive Type I Toxin-Antitoxin System: bsrE/SR5 From the B. Subtilis Chromosome. RNA Biol. 13, 511–523. doi: 10.1080/15476286.2016.1156288
Otto, M. (2010). Staphylococcus Colonization of the Skin and Antimicrobial Peptides. Expert Rev. Dermatol. 5, 183–195. doi: 10.1586/edm.10.6
Page, R., Peti, W. (2016). Toxin-Antitoxin Systems in Bacterial Growth Arrest and Persistence. Nat. Chem. Biol. 12, 208–214. doi: 10.1038/nchembio.2044
Pecota, D. C., Wood, T. K. (1996). Exclusion of T4 Phage by the Hok/Sok Killer Locus From Plasmid R1. J. Bacteriol. 178, 2044–2050. doi: 10.1128/jb.178.7.2044-2050.1996
Peltier, J., Hamiot, A., Garneau, J. R., Boudry, P., Maikova, A., Hajnsdorf, E., et al. (2020). Type I Toxin-Antitoxin Systems Contribute to the Maintenance of Mobile Genetic Elements in Clostridioides Difficile. Commun. Biol. 3, 1–13. doi: 10.1038/s42003-020-01448-5
Peng, J., Triplett, L. R., Schachterle, J. K., Sundin, G. W. (2019). Chromosomally Encoded Hok-Sok Toxin-Antitoxin System in the Fire Blight Pathogen Erwinia Amylovora: Identification and Functional Characterization. Appl. Environ. Microbiol. 85, 1–16. doi: 10.1128/AEM.00724-19
Pichon, C., Felden, B. (2005). Small RNA Genes Expressed From Staphylococcus Aureus Genomic and Pathogenicity Islands With Specific Expression Among Pathogenic Strains. Proc. Natl. Acad. Sci. U.S.A. 102, 14249–14254. doi: 10.1073/pnas.0503838102
Pinel-Marie, M.-L., Brielle, R., Felden, B. (2014). Dual Toxic-Peptide-Coding Staphylococcus Aureus RNA Under Antisense Regulation Targets Host Cells and Bacterial Rivals Unequally. Cell Rep. 7, 424–435. doi: 10.1016/j.celrep.2014.03.012
Plaza, G., José, J. (2020). Small RNAs as Fundamental Players in the Transference of Information During Bacterial Infectious Diseases. Front. Mol. Biosci. 7, 101. doi: 10.3389/fmolb.2020.00101
Pontes, M. H., Groisman, E. A. (2019). Slow Growth Dictates non-Heritable Antibiotic Resistance in Salmonella Enterica. Sci. Signal 12, 1–10. doi: 10.1126/scisignal.aax3938
Ranjith, K., Ramchiary, J., Prakash, J. S. S., Arunasri, K., Sharma, S., Shivaji, S. (2019). Gene Targets in Ocular Pathogenic Escherichia Coli for Mitigation of Biofilm Formation to Overcome Antibiotic Resistance. Front. Microbiol. 10, 1308. doi: 10.3389/fmicb.2019.01308
Reif, C., Löser, C., Brantl, S. (2018). Bacillus Subtilis Type I Antitoxin SR6 Promotes Degradation of Toxin Yont mRNA and Is Required to Prevent Toxic Yoyj Overexpression. Toxins (Basel) 10, 1–18. doi: 10.3390/toxins10020074
Rice, L. B. (2008). Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No Eskape. J. Infect. Dis. 197, 1079–1081. doi: 10.1086/533452
Riffaud, C., Pinel-Marie, M.-L., Felden, B. (2020). Cross-Regulations Between Bacterial Toxin–Antitoxin Systems: Evidence of an Interconnected Regulatory Network? Trends Microbiol. 28, 851–866. doi: 10.1016/j.tim.2020.05.016
Riffaud, C., Pinel-Marie, M.-L., Pascreau, G., Felden, B. (2019). Functionality and Cross-Regulation of the Four SprG/SprF Type I Toxin–Antitoxin Systems in Staphylococcus Aureus. Nucleic Acids Res. 47, 1740–1758. doi: 10.1093/nar/gky1256
Samson, J. E., Belanger, M., Moineau, S. (2013). Effect of the abortive infection mechanism and type III toxin/antitoxin system AbiQ on the lytic cycle of Lactococcus lactis phages. J. Bacteriol. 195 (17), 3947–3956.
Sayed, N., Jousselin, A., Felden, B. (2012a). A Cis -Antisense RNA Acts in Trans in Staphylococcus Aureus to Control Translation of a Human Cytolytic Peptide. Nat. Struct. Mol. Biol. 19, 105–112. doi: 10.1038/nsmb.2193
Sayed, N., Nonin-Lecomte, S., Réty, S., Felden, B. (2012b). Functional and Structural Insights of a Staphylococcus Aureus Apoptotic-Like Membrane Peptide From a Toxin-Antitoxin Module. J. Biol. Chem. 287, 43454–43463. doi: 10.1074/jbc.M112.402693
Schuster, C., Bertram, R. (2016a). Toxin-Antitoxin Systems of Staphylococcus Aureus. Toxins 8:140. doi: 10.3390/toxins8050140
Schuster, C. F., Bertram, R. (2016b). Toxin-Antitoxin Systems of Staphylococcus Aureus. Toxins 8:140. doi: 10.3390/toxins8050140
Sharma, C. M., Hoffmann, S., Darfeuille, F., Reignier, J., Findeiss, S., Sittka, A., et al. (2010). The Primary Transcriptome of the Major Human Pathogen Helicobacter Pylori. Nature 464, 250–255. doi: 10.1038/nature08756
Short, F. L., Akusobi, C., Broadhurst, W. R., Salmond, G. P. C. (2018). The Bacterial Type III Toxin-Antitoxin System, ToxIN, is a Dynamic protein-RNA Complex With Stability-Dependent Antiviral Abortive Infection Activity. Sci. Rep. 8, 1013. doi: 10.1038/s41598-017-18696-x
Sierra, R., Viollier, P., Renzoni, A. (2019). Linking Toxin-Antitoxin Systems With Phenotypes: A Staphylococcus Aureus Viewpoint. Biochim. Biophys. Acta (BBA) - Gene Regul. Mech. 1862, 742–751. doi: 10.1016/j.bbagrm.2018.07.009
Silvaggi, J. M., Perkins, J. B., Losick, R. (2005). Small Untranslated Rna Antitoxin in Bacillus Subtilis. J. Bacteriol. 187, 6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005
Song, S., Wood, T. K. (2018). Post-Segregational Killing and Phage Inhibition Are Not Mediated by Cell Death Through Toxin/Antitoxin Systems. Front. Microbiol. 9:814. doi: 10.3389/fmicb.2018.00814
Song, S., Wood, T. K. (2020a). A Primary Physiological Role of Toxin/Antitoxin Systems Is Phage Inhibition. Front. Microbiol. 1, 895. doi: 10.3389/fmicb.2020.01895
Song, S., Wood, T. K. (2020b). Toxin/Antitoxin System Paradigms: Toxins Bound to Antitoxins are Not Likely Activated by Preferential Antitoxin Degradation. Adv. Biosyst. 4, 1900290. doi: 10.1002/adbi.201900290
Soutourina, O. (2019). Type I Toxin-Antitoxin Systems in Clostridia. Toxins (Basel) 11, 1–16. doi: 10.3390/toxins11050253
Soutourina, O. A., Monot, M., Boudry, P., Saujet, L., Pichon, C., Sismeiro, O., et al. (2013). Genome-Wide Identification of Regulatory RNAs in the Human Pathogen Clostridium Difficile. PloS Genet. 9, e1003493. doi: 10.1371/journal.pgen.1003493
Stårsta, M., Hammarlöf, D. L., Wäneskog, M., Schlegel, S., Xu, F., Gynnå, A. H., et al. (2020). RHS-Elements Function as Type II Toxin-Antitoxin Modules That Regulate Intra-Macrophage Replication of Salmonella Typhimurium. PloS Genet. 16, e1008607. doi: 10.1371/journal.pgen.1008607
Thisted, T., Sørensen, N. S., Wagner, E. G., Gerdes, K. (1994). Mechanism of Post-Segregational Killing: Sok Antisense RNA Interacts With Hok mRNA Via its 5’-End Single-Stranded Leader and Competes With the 3’-End of Hok mRNA for Binding to the Mok Translational Initiation Region. EMBO J. 13, 1960–1968. doi: 10.1002/j.1460-2075.1994.tb06465.x
Unoson, C., Wagner, E. G. H. (2008). A Small SOS-induced Toxin is Targeted Against the Inner Membrane in Escherichia Coli. Mol. Microbiol. 70, 258–270. doi: 10.1111/j.1365-2958.2008.06416.x
Unterholzner, S. J., Poppenberger, B., Rozhon, W. (2013). Toxin–Antitoxin Systems. Mob. Genet. Elements 3, e26219. doi: 10.4161/mge.26219
Van Tyne, D., Martin, M. J., Gilmore, M. S. (2013). Structure, Function, and Biology of the Enterococcus Faecalis Cytolysin. Toxins (Basel) 5, 895–911. doi: 10.3390/toxins5050895
Vedantam, G., Clark, A., Chu, M., McQuade, R., Mallozzi, M., Viswanathan, V. K. (2012). Clostridium Difficile Infection. Gut Microbes 3, 121–134. doi: 10.4161/gmic.19399
Verstraeten, N., Knapen, W. J., Kint, C. I., Liebens, V., Van den Bergh, B., Dewachter, L., et al. (2015). Obg and Membrane Depolarization Are Part of a Microbial Bet-Hedging Strategy That Leads to Antibiotic Tolerance. Mol. Cell 59, 9–21. doi: 10.1016/j.molcel.2015.05.011
Vogel, J., Argaman, L., Wagner, E. G., Altuvia, S. (2004). The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 14 (24), 2271–2276.
Wang, Y., Wang, H., Hay, A. J., Zhong, Z., Zhu, J., Kan, B. (2015). Functional RelBE-Family Toxin-Antitoxin Pairs Affect Biofilm Maturation and Intestine Colonization in Vibrio Cholerae. PloS One 10, e0135696. doi: 10.1371/journal.pone.0135696
Wang, X., Wood, T. K. (2011). Toxin-Antitoxin Systems Influence Biofilm and Persister Cell Formation and the General Stress Response. Appl. Environ. Microbiol. 77, 5577–5583. doi: 10.1128/AEM.05068-11
Wassarman, K. M., Repoila, F., Rosenow, C., Storz, G., Gottesman, S. (2001). Identification of Novel Small RNAs Using Comparative Genomics and Microarrays. Genes Dev. 15, 1637–1651. doi: 10.1101/gad.901001
Waters, L. S., Storz, G. (2009). Regulatory RNAs in Bacteria. Cell 136, 615–628. doi: 10.1016/j.cell.2009.01.043
Weaver, K. E. (2012). The Par Toxin-Antitoxin System From Enterococcus Faecalis Plasmid pAD1 and its Chromosomal Homologs. RNA Biol. 9, 1498–1503. doi: 10.4161/rna.22311
Weaver, K. (2020). The Fst/Ldr Family of Type I Ta System Toxins: Potential Roles in Stress Response, Metabolism and Pathogenesis. Toxins (Basel) 12, 1–12. doi: 10.3390/toxins12080474
Weaver, K. E., Clewell, D. B., An, F. (1993). Identification, Characterization, and Nucleotide Sequence of a Region of Enterococcus Faecalis Pheromone-Responsive Plasmid pAD1 Capable of Autonomous Replication. J. Bacteriol. 175, 1900–1909. doi: 10.1128/JB.175.7.1900-1909.1993
Weaver, K. E., Jensen, K. D., Colwell, A., Sriram, S. (1996). Functional Analysis of the Enterococcus Faecalis Plasmid pAD1-encoded Stability Determinant Par. Mol. Microbiol. 20, 53–63. doi: 10.1111/j.1365-2958.1996.tb02488.x
Weaver, K. E., Reddy, S. G., Brinkman, C. L., Patel, S., Bayles, K. W., Endres, J. L. (2009). Identification and Characterization of a Family of Toxin–Antitoxin Systems Related to the Enterococcus Faecalis Plasmid pAD1 Par Addiction Module. Microbiol. (Reading) 155, 2930–2940. doi: 10.1099/mic.0.030932-0
Wen, J., Fozo, E. (2014). Srna Antitoxins: More Than One Way to Repress a Toxin. Toxins 6, 2310–2335. doi: 10.3390/toxins6082310
Wen, J., Harp, J. R., Fozo, E. M. (2017). The 5′ UTR of the Type I Toxin ZorO can Both Inhibit and Enhance Translation. Nucleic Acids Res. 45, 4006–4020. doi: 10.1093/nar/gkw1172
Wen, J., Won, D., Fozo, E. M. (2014). The ZorO-OrzO Type I Toxin-Antitoxin Locus: Repression by the OrzO Antitoxin. Nucleic Acids Res. 42, 1930–1946. doi: 10.1093/nar/gkt1018
Williams, R. E. O. (1963). Healthy Carriage of Staphylococcus Aureus: Its Prevalence and Importance1. Bacteriol. Rev. 27, 56–71. doi: 10.1128/BR.27.1.56-71.1963
Wilmaerts, D., Bayoumi, M., Dewachter, L., Knapen, W., Mika, J. T., Hofkens, J., et al. (2018). The Persistence-Inducing Toxin HokB Forms Dynamic Pores That Cause ATP Leakage. mBio 9, 1–12. doi: 10.1128/mBio.00744-18
Wilmaerts, D., Dewachter, L., De Loose, P.-J., Bollen, C., Verstraeten, N., Michiels, J. (2019). Hokb Monomerization and Membrane Repolarization Control Persister Awakening. Mol. Cell 75, 1031–1042.e4. doi: 10.1016/j.molcel.2019.06.015
Wroblewski, L. E., Peek, R. M., Wilson, K. T. (2010). Helicobacter Pylori and Gastric Cancer: Factors That Modulate Disease Risk. Clin. Microbiol. Rev. 23, 713–739. doi: 10.1128/CMR.00011-10
Yang, Q. E., Walsh, T. R. (2017). Toxin–Antitoxin Systems and Their Role in Disseminating and Maintaining Antimicrobial Resistance. FEMS Microbiol. Rev. 41, 343–353. doi: 10.1093/femsre/fux006
Keywords: sRNA, pathogen, bacteria, RNA, toxin-antitoxin (TA)
Citation: Sarpong DD and Murphy ER (2021) RNA Regulated Toxin-Antitoxin Systems in Pathogenic Bacteria. Front. Cell. Infect. Microbiol. 11:661026. doi: 10.3389/fcimb.2021.661026
Received: 30 January 2021; Accepted: 29 April 2021;
Published: 18 May 2021.
Edited by:
John S. Gunn, The Research Institute at Nationwide Children's Hospital, United StatesReviewed by:
Clayton Caswell, Virginia Tech, United StatesCopyright © 2021 Sarpong and Murphy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erin R. Murphy, bXVycGh5ZUBvaGlvLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.