- 1Department of Virology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland
- 2Department of Forensic Medicine, University of Helsinki, Helsinki, Finland
- 3Forensic Medicine Unit, Finnish Institute for Health and Welfare, Helsinki, Finland
- 4Department of Electronics, Telecommunications and Informatics, University of Aveiro, Aveiro, Portugal
- 5Institute of Electronics and Informatics Engineering of Aveiro, University of Aveiro, Aveiro, Portugal
The long-term impact of viruses residing in the human bone marrow (BM) remains unexplored. However, chronic inflammatory processes driven by single or multiple viruses could significantly alter hematopoiesis and immune function. We performed a systematic analysis of the DNAs of 38 viruses in the BM. We detected, by quantitative PCRs and next-generation sequencing, viral DNA in 88.9% of the samples, up to five viruses in one individual. Included were, among others, several herpesviruses, hepatitis B virus, Merkel cell polyomavirus and, unprecedentedly, human papillomavirus 31. Given the reactivation and/or oncogenic potential of these viruses, their repercussion on hematopoietic and malignant disorders calls for careful examination. Furthermore, the implications of persistent infections on the engraftment, regenerative capacity, and outcomes of bone marrow transplantation deserve in-depth evaluation.
Introduction
Hematopoiesis occurs in the bone marrow (BM) as a result of the crosstalk between, among others, mesenchymal and hematopoietic stem cells. Their interaction occurs in niches in which complex networks are built upon concerted cell-to-cell architectures. These compartments have been delineated in animal models at single-cell resolution (Tikhonova et al., 2019; Baccin et al., 2020), revealing a large diversity in the cellular components of arteriolar, endosteal, and sinusoidal niches. This heterogeneity gives rise to unique signaling patterns accounting for the multiple effector functions of this organ.
Alterations in these microenvironments, e.g. by viral infections, may result in multiple pathological conditions. Indeed, persistent viruses can perturb the homeostasis, either by cytotoxic damage of the niche´s structural integrity, transformation of the signaling repertoire, or interference with the cellular trafficking (Alexaki and Wigdahl, 2008; Pascutti et al., 2016). However, while the pathogenic effects of viruses in the BM are primarily known in the context of acute infection (Scadden et al., 1989; Rosenfeld and Young, 1991; Luppi et al., 2000) the full spectrum of viruses here persisting and the long-term repercussions on the marrow’s physiology and composition remain poorly understood.
Moreover, while most studies have focused on single viruses (Baranski et al., 1988; Cantin et al., 1994; Cassinotti et al., 1997; Eis-Hübinger et al., 2001; Heegaard et al., 2002; Khaiboullina et al., 2004; Lundqvist et al., 2005; Manning et al., 2007; Rollín et al., 2007; Corcioli et al., 2008; Tan et al., 2009; Garcia et al., 2011; Matsushita et al., 2013; Pambrun et al., 2014), a comprehensive analysis of the different types concomitantly persisting in the BM of an individual is lacking. This is however important, as their chronic effects, either concerted or combined, may contribute to BM dysfunction.
In the present study, we systematically investigated the genomic prevalence and copy numbers of altogether 38 clinically significant DNA viruses in the BM of 27 recently deceased individuals. Included were members of the Parvoviridae, Herpesviridae, Polyomaviridae, Hepadnaviridae, Anelloviridae, and Papillomaviridae families. To the best of our knowledge, this is the most comprehensive assessment of viral DNAs present in the BM to date.
Methods
Study Cohort
The study cohort consisted of 27 deceased individuals of Finnish origin. The age range of the study subjects was 36 to 85 years (mean 68) and the postmortem interval on average 8 days (Figure 1). None of the individuals had records of the use of immunosuppressants, neither of virus-related cancers or ongoing viral infections at the time of death, except for one (case number 6) with a history of facial shingles a few weeks earlier. The causes and manners of deaths are presented in Figure 1. The study was reviewed by the Ethics Committee of Helsinki and Uusimaa Hospital District (approval 164/13/03/00/2014).
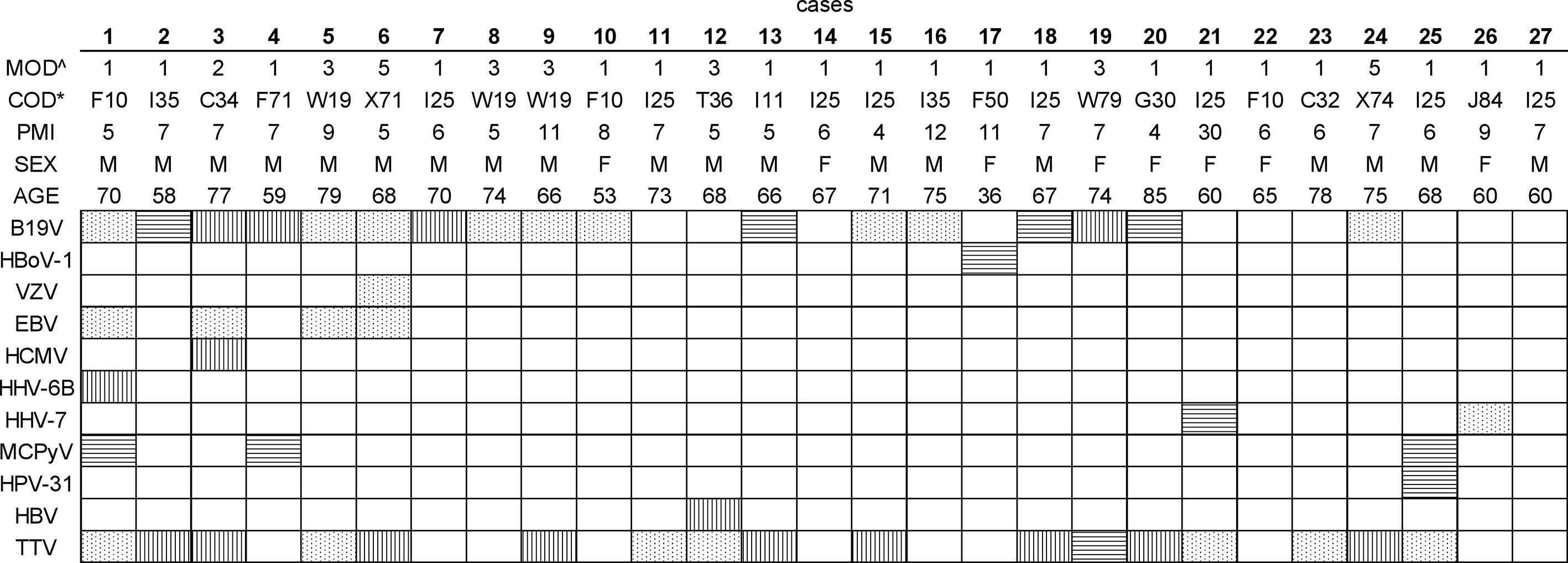
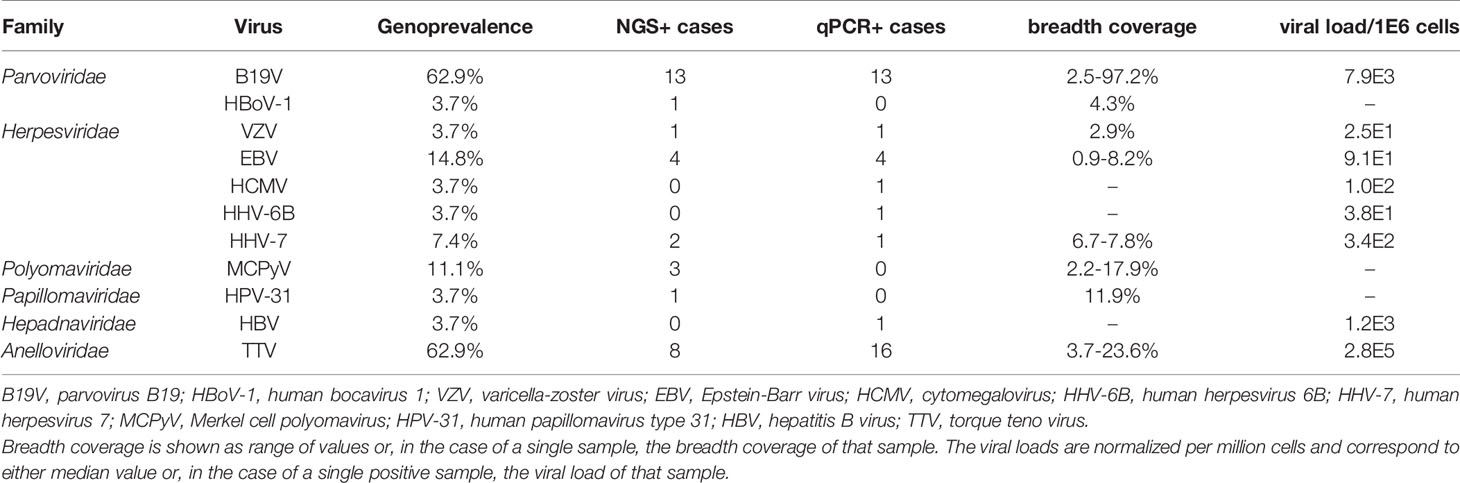
Figure 1 Viral DNA prevalences in bone marrow per study subject. The viral findings are presented as dots (NGS+, qPCR+), horizontal lines (NGS+, qPCR-), vertical lines (NGS-, qPCR+), and white (NGS-, qPCR-). ^The manner of death (MOD) is presented as 1=disease, 2=occupational disease, 3=injury, 5=suicide. *The cause of death (COD) is given according to the WHO ICD10 classification (https://icd.who.int/browse10/2019/en). Age is given in years. PMI, Post-mortem interval (given in days); B19V, parvovirus B19; HBoV-1, human bocavirus 1; VZV, varicella-zoster virus; EBV, Epstein-Barr virus; HCMV, cytomegalovirus; HHV-6B, human herpesvirus 6B; HHV-7, human herpesvirus 7; MCPyV, Merkel cell polyomavirus; HPV-31, human papillomavirus type 31; HBV, hepatitis B virus; TTV, torque teno virus.
Sample Processing
The BM was collected from the diaphysis of femoral bone and the DNA extracted with QIAamp DNA Mini Kit (Qiagen) as per manufacturer protocol. The quality and integrity of total DNA in each sample was confirmed with a Genomic DNA Reagent Kit in a LabChip GX Instrument.
To avoid contamination, strict precautions were taken throughout the experiments including the use of single-use disposable plastics and filter tips, the handling in hoods dedicated exclusively to nucleic acid work, and the use of negative controls through all steps. The reaction mixes, DNA extracts, plasmid controls, and amplification reactions, were each handled in separate rooms.
Virus Analysis
The viral DNAs were analyzed by targeted enrichment followed by sequencing in NovaSeq 6000 (Illumina), as well as by quantitative PCRs.
For the enrichment, a customized panel of biotinylated RNA oligonucleotides (Arbor Biosciences) was used. The viruses targeted were parvovirus B19 (B19V), human bocaviruses 1-4 (HBoV1-4), cutavirus, human papillomavirus (HPV) types 2, 6, 11, 16, 18, 21, and 45, human polyomaviruses (HPyV) 1-13, simian virus 40, human herpesviruses (HHV) 1-8, hepatitis B virus (HBV), torque teno virus (TTV) and variola virus (minor and major). A complete list of the reference strains used for the design of the RNA oligonucleotides, as well as the details of the NGS methods, are presented in (Toppinen et al., 2020). In brief, the sequencing libraries were prepared from sonicated total DNA (10-1000 ng) using the KAPA HyperPlus library preparation kit (Roche). The viral genomes were enriched with MyBaits v4 kit (Arbor Biosciences) using two consecutive rounds of hybridization on individual samples, following the manufacturer recommendations for low input DNA. The viral genomic sequences were reconstructed after removal of PCR duplicates, using a customized bioinformatics pipeline, TRACESPipe (Pratas et al., 2020). The adapter sequences were removed using Trimmomatic (Bolger et al., 2014), by explicitly removing content from an adapters’ list having a maximum mismatch that allowed a full match of 2. The palindrome and simple clip threshold were set at 30 and 10, respectively. The minimum quality score required to keep a base at the beginning and the end was fixed to 3. Low-quality data was filtered using a sliding window of 4 with an average quality of 15 and low complexity regions were flagged with GTO (Almeida et al., 2020). Reads shorter than 25 bases were discarded.
The consensus, as well as single sequences (when in low coverage), were confirmed by BLAST (NCBI).
To confirm the NGS findings, validated in-house quantitative PCRs were used for the detection of B19V (Toppinen et al., 2015), Merkel cell polyomavirus (MCPyV) (Goh et al., 2009), TTV (Toppinen et al., 2020), and the nine HHVs (Pyöriä et al., 2020). HBV and HPV-31 were tested with commercial kits (Hepatitis B Virus PCR Kit, GeneProof, Genesig; and Human papillomavirus 31 Standard kit, PrimerDesign, respectively) according to the manufacturer protocol. The human single-copy gene RNase P qPCR was utilized for normalization of the viral DNA copies to the cell number (Toppinen et al., 2015). See Supplementary Table 1 for further information on the qPCRs used.
Results
We conducted a systematic analysis of viral DNAs present in the bone marrow by two complementary approaches, NGS and qPCR.
We first analyzed the integrity of the total genomic DNA of each sample to control for bias in the prevalences and loads detected by qPCR. Based on the size distribution, the total DNA was of high quality and non-fragmented, except for one sample, taken 30 days post-mortem, exhibiting a wider range of fragment sizes (Figure 2).
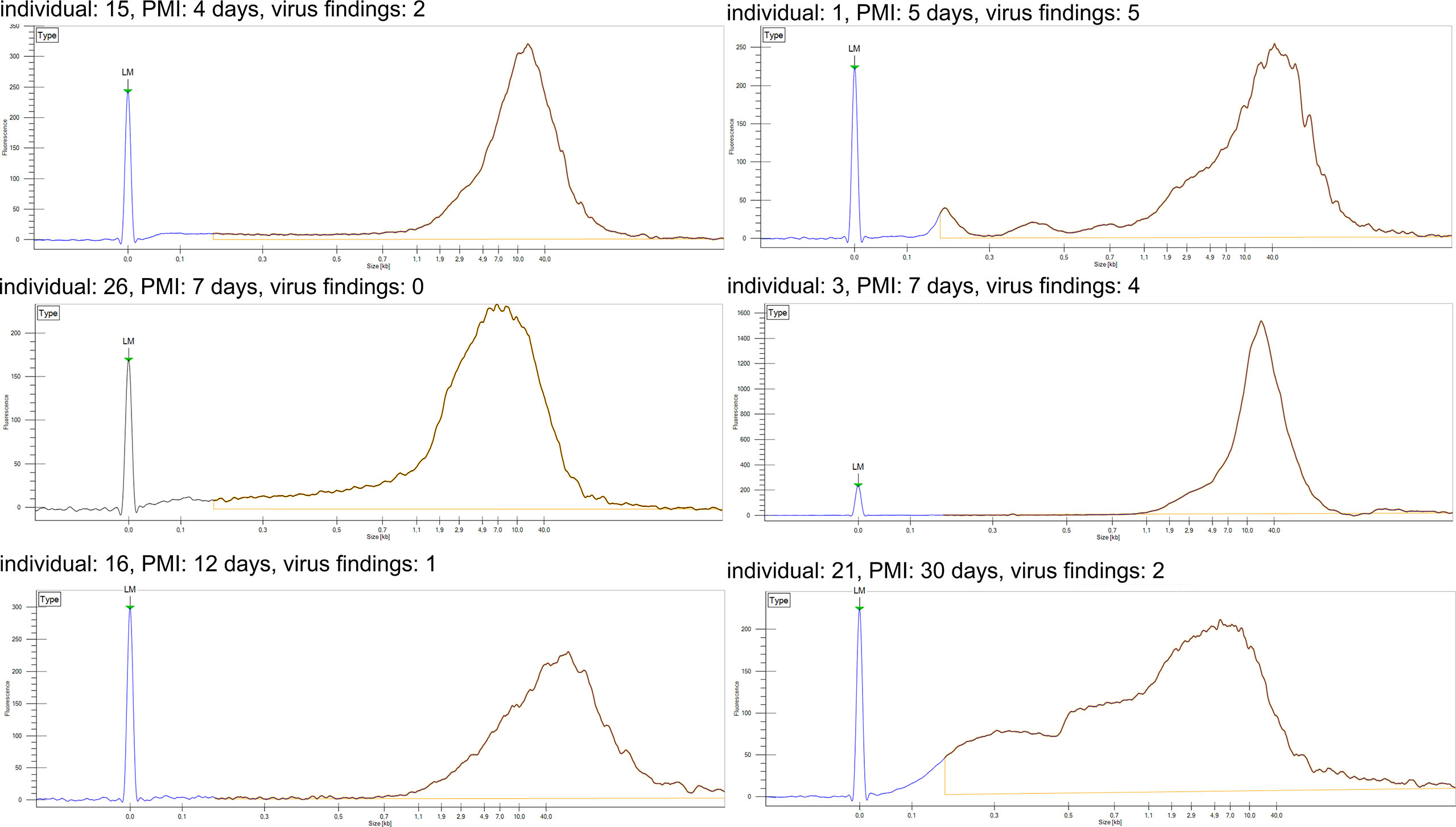
Figure 2 DNA fragment analysis of total DNA. Fragment length distribution of genomic DNA for six representative samples as established with LabChip GX. Individual number refers to Figure 1. LM, lower marker; PMI, post-mortem interval.
We detected viral DNA in 88.9% of the samples, comprising altogether 11 distinct virus types (Table 1). The most prevalent genomes were of B19V and TTV each found in 62.9% of the individuals, followed by Epstein-Barr virus (EBV) in 14.8%, MCPyV in 11.1%, and human herpesvirus 7 (HHV-7) in 7.4%. Other viral genomes, identified in single cases, were of varicella-zoster virus (VZV), human cytomegalovirus (HCMV), human herpesvirus 6B (HHV-6B), HBoV-1, HPV-31, and HBV.
On average, we identified two virus types per individual, the most common co-detections being of B19V and TTV, in 44.4% of the BMs (Figure 1). A third of these double-positive marrows were also positive for EBV (i.e. 14.8% of all cases). The maximum number of viruses per individual was five, corresponding to a single case in which B19V, EBV, HHV-6B, MCPyV, and TTV were found.
When normalized to copies per million cells, the highest quantities were of B19V and TTV with a median of 7.9E3 and 2.8E5, respectively, to be followed by EBV with 9.1E1 copies. VZV, HCMV, HHV-6B, HHV-7 and HBV had respectively 2.5E1, 1.0E2, 3.8E1, 3.4E2 and 1.2E3 copies. In general, the quantities of most viral DNAs were very low, speaking against active replication.
Via in-solution hybridization, we captured and reconstructed 11 viral genomic sequences, with a minimum breadth coverage of 15% (Figure 3). Highest qualities were obtained for B19V (n=7, average breath coverage 88.2%; Genbank accession numbers MT988397-MT9888403), MCPyV (n=1, breath 17.9%; Supplementary Table 2), and TTV (n=3, average breath 19.8%; Supplementary Table 3). Remarkably, we found in one individual 14 unique reads (total length 936 nucleotides, Supplementary Table 4) matching to human papillomaviruses, with the highest similarity to HPV-31.
Figure 3 Coverage profiles of reconstructed viral DNA sequences. In the x axis is the breadth coverage (= reads covering the noted viral reference) and in the y axis the depth (number of reads covering a specific nucleotide/area). Represented are parvovirus B19, torque teno virus, Merkel cell polyomavirus, human papillomavirus type 31 and Epstein-Barr virus. Each profile is specified by the virus name, breadth coverage and the genomic sequence similarity to its closest match by NCBI-BLAST.
Moreover, albeit in low coverage, we manually and visually confirmed by BLAST each of the unique sequences (mean length 100 nucleotides) specific for the following viruses: VZV (1 individual, 103 reads), EBV (4 individuals, altogether 342 reads), HHV-7 (2 individuals, altogether 76 reads), MCPyV (2 additional findings, a total of 5 reads), TTV (6 additional findings, altogether 22 reads), B19V (5 additional findings, altogether 66 reads), and HBoV-1 (1 individual, 2 reads).
Because of our cohort’s advanced age and the sampling location, a higher proportion of mesenchymal cells are to be expected. We compared the sequences of the viruses found in the BM with those detected in the same individuals´ external surface of the femoral bone (Toppinen et al., 2020). We found a match in one or more individuals for EBV, HCMV, HHV-6B, MCPyV, B19V, TTV, and HBV. Having the reconstructed sequences of B19V the highest quality, we analyzed the nucleotide similarities between bone and BM and found they were nearly identical (with a maximum of four single nucleotide polymorphisms; Supplementary Table 5).
The reconstructed consensus sequences of B19V have been deposited in GenBank with accession numbers: MT988397-MT988403.
Discussion
A comprehensive analysis of the virus types residing in the human bone marrow is lacking. Yet, this knowledge is essential for assessment of the impact of viral DNA persistence on the marrow’s integrity and on the emergence of many pathological conditions. Since the BM homeostasis is founded on many precise regulatory networks, any alterations resulting from single or cumulative effects of multiple viruses can foreseeably have profound effects on its output and function.
We evaluated the prevalences of viral DNAs residing in the bone marrow and found 11 different virus types, in copy numbers compatible with latency.
Remarkably, among the viral genomes identified, we found HPV-31 in a male individual aged 68, who died of atherosclerotic heart disease. Since infections by this oncovirus are normally self-limited and restricted to epithelial surfaces, the detection of its DNA in BM is perplexing. Other HPV types have been reported in BM by Payne et al. (Payne et al., 1995; Payne et al., 1996), albeit in patients with multiple myeloma and monoclonal gammopathy of unknown significance. Our discovery in BM is supported by a parallel retrieval of the full-length genome of HPV-31 in the femoral bone of another individual (Toppinen et al., 2020). Unfortunately, we were unable to match the detection of this virus type in the bone and BMs of the subjects, possibly due to the very low copy numbers. Since neither individual had an HPV related clinical history, nor compatible pathological findings at the post-mortem evaluation, our discovery points to the possibility of bone and BM as novel sites of HPV-31 persistence and raise critical questions on its clinical significance (Flynn et al., 2010).
Another interesting finding was that of the genome of MCPyV, likewise of mucocutaneous tropism and oncogenic potential. We detected it in three individuals, two of whom were also positive in the bone. MCPyV DNA in BM was reported by Matsushita et al. (Matsushita et al., 2013), who found a similar prevalence (17%) in Japanese cadavers, but not by Dang et al. (2011) who surveyed also KI and WU polyomaviruses in 13 patients with different medical conditions.
Importantly, advanced stages of malignant transformation by both papillomaviruses and Merkel cell polyomavirus can present with manifestations in bone (Kamijo et al., 2002; Ley et al., 2014; Lee et al., 2018; Lewis et al., 2020). While normally considered metastatic, the contribution of the viruses already residing in these tissues should be assessed.
Our study is limited by the lack of distinction of the specific host-cell types. To date, the cellular reservoirs of most viruses in the BM are actually unknown, although in vitro a wide range of constituent and memory cells have been shown permissive for one or more (Chiara et al., 1996; Movassagh et al., 1996; Bahner et al., 1997; Babcock et al., 1998; Okamoto et al., 2000; Khaiboullina et al., 2004; Luppi et al., 2005; Sundin et al., 2006; Wu et al., 2006; Rollín et al., 2007; Smirnov et al., 2007; Alexaki and Wigdahl, 2008; Rong et al., 2008; Sundin et al., 2008; Ma et al., 2011; Bua et al., 2016; Furuta et al., 2017; Pyöriä et al., 2017).
When comparing the viral genome findings in the BM to those of the adjacent femoral bone, we found a match for EBV, HCMV, HHV-6B, MCPyV, B19V, TTV, and HBV in one or more individuals. This raises the possibility of infection taking place already at the precursor stage, whereafter the virus could be carried through the mature cell populations constituents of bone. Previously, stem cells have been proposed as renewable sources of B19V (Sundin et al., 2008), HCV (Ivantes et al., 2004), HIV (Alexaki and Wigdahl, 2008), and JCPyV (Chiara et al., 1996), thereby contributing to viral spread and disease progression. Taken together, our results call for further study on the implications of the resident viral genomes to the bone’s structure and metabolism, a topic until now unexplored.
Given the critical roles of BM in the support and regulation of hematopoiesis, lineage-specific differentiation, as well as homing and survival of memory cells (Visnjic et al., 2004; Bowers et al., 2015), our findings call for careful consideration of the clinical implications of viral persistence. Indeed, since many of the viruses detected can reactivate, special attention should be paid to their association with the development of lymphoproliferative and/or malignant disorders (Bartenhagen et al., 2017). Furthermore, the burden of persistent infections on the engraftment, regenerative capacity, and outcomes (Ivantes et al., 2004; Nombela-Arrieta and Isringhausen, 2017) of BM transplantation deserve in-depth evaluation.
In summary, our findings provide evidence of a wide spectrum of viral DNAs present in the human BM. In-depth studies are essential to determine their pathogenic potential and clinical significance.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Helsinki and Uusimaa Hospital District (approval 164/13/03/00/2014). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
MT, AS, KH, and MP designed the study. MT performed the experiments. MT, DP, and MP analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Finnish Medical Society (MP), the Jane and Aatos Erkko Foundation (KH), the Finnish Cultural Foundation (MT, MP), the Sigrid Jusélius Foundation (KH), the Life and Health Medical Foundation (AS, MP), the Magnus Ehrnrooth Foundation (AS, KH), Finnish Society of Sciences and Letters (AS, KH), the Kone Foundation (AS), the Research Funds of University of Helsinki and Helsinki University Hospital (KH), Finska Läkaresällskapet (KH, MP), Fundação para a Ciência e a Tecnologia, I.P., under the Scientific Employment Stimulus - Institutional Call - CI-CTTI-94-ARH/2019 (DP).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.657245/full#supplementary-material
References
Alexaki, A., Wigdahl, B. (2008). Hiv-1 Infection of Bone Marrow Hematopoietic Progenitor Cells and Their Role in Trafficking and Viral Dissemination. PloS Pathog. 4, e1000215. doi: 10.1371/journal.ppat.1000215
Almeida, J. R., Pinho, A. J., Oliveira, J. L., Fajarda, O., Pratas, D. (2020). Gto: A Toolkit to Unify Pipelines in Genomic and Proteomic Research. SoftwareX 12, 100535. doi: 10.1016/j.softx.2020.100535
Babcock, G. J., Decker, L. L., Volk, M., Thorley-Lawson, D. A. (1998). EBV Persistence in Memory B Cells In Vivo. Immunity 9, 395–404. doi: 10.1016/S1074-7613(00)80622-6
Baccin, C., Al-Sabah, J., Velten, L., Helbling, P. M., Grünschläger, F., Hernández-Malmierca, P., et al. (2020). Combined Single-Cell and Spatial Transcriptomics Reveal the Molecular, Cellular and Spatial Bone Marrow Niche Organization. Nat. Cell Biol. 22, 38–48. doi: 10.1038/s41556-019-0439-6
Bahner, I., Kearns, K., Coutinho, S., Leonard, E., Kohn, D. (1997). Infection of Human Marrow Stroma by Human Immunodeficiency Virus-1 (HIV-1) is Both Required and Sufficient for HIV-1-induced Hematopoietic Suppression In Vitro: Demonstration by Gene Modification of Primary Human Stroma. Blood 90, 1787–1798. doi: 10.1182/blood.V90.5.1787
Baranski, B., Armstrong, G., Truman, J. T., Quinnan, G. V., Straus, S. E., Young, N. S. (1988). Epstein-Barr Virus in the Bone Marrow of Patients With Aplastic Anemia. Ann. Internal Med. 109, 695–704. doi: 10.7326/0003-4819-109-9-695
Bartenhagen, C., Fischer, U., Korn, K., Pfister, S. M., Gombert, M., Chen, C., et al. (2017). Infection as a Cause of Childhood Leukemia: Virus Detection Employing Whole Genome Sequencing. Haematologica 102, e179–e183. doi: 10.3324/haematol.2016.155382
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bowers, M., Zhang, B., Ho, Y., Agarwal, P., Chen, C.-C., Bhatia, R. (2015). Osteoblast Ablation Reduces Normal Long-Term Hematopoietic Stem Cell Self-Renewal But Accelerates Leukemia Development. Blood 125, 2678–2688. doi: 10.1182/blood-2014-06
Bua, G., Manaresi, E., Bonvicini, F., Gallinella, G. (2016). Parvovirus B19 Replication and Expression in Differentiating Erythroid Progenitor Cells. PloS One 11, e0148547. doi: 10.1371/journal.pone.0148547
Cantin, E., Chen, J., Gaidulis, L., Valo, Z., McLaughlin-Taylor, E. (1994)Detection of Herpes Simplex Virus DNA Sequences in Human Blood and Bone Marrow Cells (Accessed April 26, 2019).
Cassinotti, P., Burtonboy, G., Fopp, M., Siegl, G. (1997)Evidence for Persistence of Human Parvovirus B19 DNA in Bone Marrow (Accessed September 2, 2019).
Chiara, M., Monaco, G., Atwood, W. J., Gravell, M., Tornatore, C. S., Major, E. O. (1996). Jc Virus Infection of Hematopoietic Progenitor Cells, Primary B Lymphocytes and Tonsillar Stromal Cells: Implications for Viral Latency. J. Virol. 70, 7004–7012. doi: 10.1128/JVI.70.10.7004-7012.1996
Corcioli, F., Zakrzewska, K., Rinieri, A., Fanci, R., Innocenti, M., Civinini, R., et al. (2008). Tissue Persistence of Parvovirus B19 Genotypes in Asymptomatic Persons. J. Med. Virol. 80, 2005–2011. doi: 10.1002/jmv
Dang, X., Bialasiewicz, S., Nissen, M. D., Sloots, T. P., Koralnik, I. J., Tan, C. S. (2011). Infrequent Detection of KI, WU and MC Polyomaviruses in Immunosuppressed Individuals With or Without Progressive Multifocal Leukoencephalopathy. PloS One 6, e16736. doi: 10.1371/journal.pone.0016736
Eis-Hübinger, A. M., Reber, U., Abdul-Nour, T., Glatzel, U., Lauschke, H., Pütz, U. (2001). Evidence for Persistence of Parvovirus B19 DNA in Livers of Adults. J. Med. Virol. 65, 395–401. doi: 10.1002/jmv.2047
Flynn, J. M., Andritsos, L., Lucas, D., Byrd, J. C. (2010). Second Malignancies in B-cell Chronic Lymphocytic Leukaemia: Possible Association With Human Papilloma Virus. Br. J. Haematol. 149, 388–390. doi: 10.1111/j.1365-2141.2010.08110.x
Furuta, R., Yasunaga, J., Miura, M., Sugata, K., Saito, A., Akari, H., et al. (2017). Human T-cell Leukemia Virus Type 1 Infects Multiple Lineage Hematopoietic Cells In Vivo. PloS Pathog. 13, e1006722. doi: 10.1371/journal.ppat.1006722
Garcia, S., Kleine Neto, W., da Costa, A. C., Sanabani, S. S., Mendrone, A., Pereira, J., et al. (2011). Parvovirus Among Patients With Cytopenia of Unknown Origin in Brazil: A Case-Control Study. J. Clin. Microbiol. 49, 1578–1580. doi: 10.1128/JCM.00077-11
Goh, S., Lindau, C., Tiveljung-Lindell, A., Allander, T. (2009). Merkel Cell Polyomavirus in Respiratory Tract Secretions. Emerg. Infect. Dis. 15, 489–491. doi: 10.3201/eid1503.081206
Heegaard, E. D., Petersen, B. L., Heilmann, C. J., Hornsleth, A. (2002). Prevalence of Parvovirus B19 and Parvovirus V9 DNA and Antibodies in Paired Bone Marrow and Serum Samples From Healthy Individuals. J. Clin. Microbiol. 40, 933–936. doi: 10.1128/jcm.40.3.933-936.2002
Ivantes, C. A. P., Amarante, H., Ioshii, S. O., Pasquini, R. (2004). Hepatitis C Virus in Long-Term Bone Marrow Transplant Survivors. Bone Marrow Transplant. 33, 1181–1185. doi: 10.1038/sj.bmt.1704519
Kamijo, A., Koshino, T., Hirakawa, K., Saito, T. (2002). Merkel Cell Carcinoma With Bone Metastasis: A Case Report. J. Orthop. Sci. 7, 574–577. doi: 10.1007/s007760200102
Khaiboullina, S. F., Maciejewski, J. P., Crapnell, K., Spallone, P. A., Dean Stock, A., Pari, G. S., et al. (2004). Human Cytomegalovirus Persists in Myeloid Progenitors and is Passed to the Myeloid Progeny in a Latent Form. Br. J. Haematol. 126, 410–417. doi: 10.1111/j.1365-2141.2004.05056.x
Lee, K., Newbold, K., Thway, K., Weller, A. (2018). Bone Marrow Failure in HPV-Associated Oropharyngeal Squamous Cell Carcinoma. J. Otolaryngol.-ENT Res. 10, 302. doi: 10.15406/joentr.2018.10.00302
Lewis, C. W., Qazi, J., Hippe, D. S., Lachance, K., Thomas, H., Cook, M. M., et al. (2020). Patterns of Distant Metastases in 215 Merkel Cell Carcinoma Patients: Implications for Prognosis and Surveillance. Cancer Med. 9, 1374–1382. doi: 10.1002/cam4.2781
Ley, J., Wildes, T., El-Mofty, S., Adkins, D. (2014). Metastasis Occurring Eleven Years After Diagnosis of Human Papilloma Virus-Related Oropharyngeal Squamous Cell Carcinoma. Ecancermedicalscience 8, 480. doi: 10.3332/ecancer.2014.480
Lundqvist, A., Isa, A., Tolfvenstam, T., Kvist, G., Broliden, K. (2005). High Frequency of Parvovirus B19 DNA in Bone Marrow Samples From Rheumatic Patients. J. Clin. Virol. 33, 71–74. doi: 10.1016/J.JCV.2004.11.011
Luppi, M., Barozzi, P., Schulz, T. F., Setti, G., Staskus, K., Trovato, R., et al. (2000). Bone Marrow Failure Associated With Human Herpesvirus 8 Infection After Transplantation. New Engl. J. Med. 343, 1378–1385. doi: 10.1056/NEJM200011093431905
Luppi, M., Trovato, R., Barozzi, P., Gibellini, F., Potenza, L., Riva, G., et al. (2005). Suppressive Effect of Human herpesvirus-8/Kaposi Sarcoma-Associated Herpesvirus on In Vitro Colony Formation of Hematopoietic Progenitor Cells. Leukemia Res. 29, 951–953. doi: 10.1016/j.leukres.2005.01.012
Manning, A., Willey, S. J., Bell, J. E., Simmonds, P. (2007). Comparison of Tissue Distribution, Persistence and Molecular Epidemiology of Parvovirus B19 and Novel Human Parvoviruses PARV4 and Human Bocavirus. J. Infect. Dis. 195, 1345–1352. doi: 10.1086/513280
Matsushita, M., Kuwamoto, S., Iwasaki, T., Higaki-Mori, H., Yashima, S., Kato, M., et al. (2013). Detection of Merkel Cell Polyomavirus in the Human Tissues From 41 Japanese Autopsy Cases Using Polymerase Chain Reaction. Intervirology 56, 1–5. doi: 10.1159/000338620
Ma, R., Xing, Q., Shao, L., Wang, D., Hao, Q., Li, X., et al. (2011). Hepatitis B Virus Infection and Replication in Human Bone Marrow Mesenchymal Stem Cells. Virol. J. 8, 486. doi: 10.1186/1743-422X-8-486
Movassagh, M., Gozlan, J., Senechal, B., Baillou, C., Petit, J. C., Lemoine, F. M. (1996). Direct Infection of CD34+ Progenitor Cells by Human Cytomegalovirus: Evidence for Inhibition of Hematopoiesis and Viral Replication. Blood 88, 1277–1283. doi: 10.1182/blood.v88.4.1277.bloodjournal8841277
Nombela-Arrieta, C., Isringhausen, S. (2017). The Role of the Bone Marrow Stromal Compartment in the Hematopoietic Response to Microbial Infections. Front. Immunol. 7, 689. doi: 10.3389/fimmu.2016.00689
Okamoto, H., Nishizawa, T., Tawara, A., Takahashi, M., Kishimoto, J., Sai, T., et al. (2000). TT Virus mRNAs Detected in the Bone Marrow Cells From an Infected Individual. Biochem. Biophys. Res. Commun. 279, 700–707. doi: 10.1006/bbrc.2000.4012
Pambrun, E., Mengelle, C., Fillola, G., Laharrague, P., Esposito, L., Cardeau-Desangles, I., et al. (2014). An Association Between BK Virus Replication in Bone Marrow and Cytopenia in Kidney-Transplant Recipients. J. Transplant. 2014, 252914. doi: 10.1155/2014/252914
Pascutti, M. F., Erkelens, M. N., Nolte, M. A. (2016). Impact of Viral Infections on Hematopoiesis: From Beneficial to Detrimental Effects on Bone Marrow Output. Front. Immunol. 7, 364. doi: 10.3389/fimmu.2016.00364
Payne, D. A., Chan, T. S., Ostermeyer Schoaib, B., Patten, B. M., Tyring, S. K. (1996). Human Papillomavirus is Associated With Monoclonal Gammopathies of Unknown Significance. J. Biomed. Sci. 3, 319–322. doi: 10.1007/BF02257961
Payne, D. A., Chan, T. S., Patten, B., Tyring, S. K. (1995). Extrachromosomal Human Papillomavirus (HPV) in Multiple Myeloma and Monoclonal Gammopathy of Unknown Significance (MGUS) Patients. Anticancer Res. 15, 2213–2215.
Pratas, D., Toppinen, M., Pyöriä, L., Hedman, K., Sajantila, A., Perdomo, M. F. (2020). A Hybrid Pipeline for Reconstruction and Analysis of Viral Genomes At Multi-Organ Level. GigaScience 9, giaa086. doi: 10.1093/gigascience/giaa086
Pyöriä, L., Jokinen, M., Toppinen, M., Salminen, H., Vuorinen, T., Hukkanen, V., et al. (2020). Herq-9 Is a New Multiplex PCR for Differentiation and Quantification of All Nine Human Herpesviruses. mSphere 5, e00265–e00220. doi: 10.1128/mSphere.00265-20
Pyöriä, L., Toppinen, M., Mäntylä, E., Hedman, L., Aaltonen, L.-M., Vihinen-Ranta, M., et al. (2017). Extinct Type of Human Parvovirus B19 Persists in Tonsillar B Cells. Nat. Commun. 8, 14930. doi: 10.1038/ncomms14930
Rollín, R., Álvarez-Lafuente, R., Marco, F., Jover, J. A., Hernández-García, C., Rodríguez-Navas, C., et al. (2007). Human Parvovirus B19, Varicella Zoster Virus and Human Herpesvirus-6 in Mesenchymal Stem Cells of Patients With Osteoarthritis: Analysis With Quantitative Real-Time Polymerase Chain Reaction. Osteoarthr. Cartil. 15, 475–478. doi: 10.1016/j.joca.2006.11.007
Rong, Q., Zhang, L., Su, E., Li, J., Li, J., Liu, Z., et al. (2008). Bone Marrow-Derived Mesenchymal Stem Cells are Capable of Mediating Hepatitis B Virus Infection in Injured Tissues. J. Viral Hepat. 15, 607–614. doi: 10.1111/j.1365-2893.2008.00978.x
Rosenfeld, S. J., Young, N. S. (1991). Viruses and Bone Marrow Failure. Blood Rev. 5, 71–77. doi: 10.1016/0268-960X(91)90037-D
Scadden, D., Zon, L., Groopman, J. (1989). Pathophysiology and Management of HIV-associated Hematologic Disorders. Blood 74, 1455–1463. doi: 10.1182/blood.v74.5.1455.1455
Smirnov, S. V., Harbacheuski, R., Lewis-Antes, A., Zhu, H., Rameshwar, P., Kotenko, S. V. (2007). Bone-Marrow-Derived Mesenchymal Stem Cells as a Target for Cytomegalovirus Infection: Implications for Hematopoiesis, Self-Renewal and Differentiation Potential. Virology 360, 6–16. doi: 10.1016/j.virol.2006.09.017
Sundin, M., Lindblom, A., Örvell, C., Barrett, A. J., Sundberg, B., Watz, E., et al. (2008). Persistence of Human Parvovirus B19 in Multipotent Mesenchymal Stromal Cells Expressing the Erythrocyte P Antigen: Implications for Transplantation. Biol. Blood Marrow Transplant. 14, 1172–1179. doi: 10.1016/j.bbmt.2008.08.003
Sundin, M., Rasmusson, I., Sundberg, B., Ringdén, R., Le Blanc, K. (2006). Mesenchymal Stem Cells are Susceptible to Human Herpesviruses, But Viral DNA Cannot be Detected in the Healthy Seropositive Individual. Bone Marrow Transplant. 37, 1051–1059. doi: 10.1038/sj.bmt.1705368
Tan, C. S., Dezube, B. J., Bhargava, P., Autissier, P., Wüthrich, C., Miller, J., et al. (2009). Detection of JC Virus DNA and Proteins in the Bone Marrow of HIV-Positive and HIV-Negative Patients: Implications for Viral Latency and Neurotropic Transformation. J. Infect. Dis. 199, 881–888. doi: 10.1086/597117
Tikhonova, A. N., Dolgalev, I., Hu, H., Sivaraj, K. K., Hoxha, E., Cuesta-Domínguez, Á., et al. (2019). The Bone Marrow Microenvironment At Single-Cell Resolution. Nature 569, 222–228. doi: 10.1038/s41586-019-1104-8
Toppinen, M., Norja, P., Aaltonen, L. M., Wessberg, S., Hedman, L., Söderlund-Venermo, M., et al. (2015). A New Quantitative PCR for Human Parvovirus B19 Genotypes. J. Virol. Methods 218, 40–45. doi: 10.1016/j.jviromet.2015.03.006
Toppinen, M., Pratas, D., Väisänen, E., Söderlund-Venermo, M., Hedman, K., Perdomo, M. F., et al. (2020). The Landscape of Persistent Human DNA Viruses in Femoral Bone. Forensic Sci. Int. Genet. 48, 102353. doi: 10.1016/j.fsigen.2020.102353
Visnjic, D., Kalajzic, Z., Rowe, D. W., Katavic, V., Lorenzo, J., Aguila, H. L. (2004). Hematopoiesis is Severely Altered in Mice With an Induced Osteoblast Deficiency. Blood 103, 3258–3264. doi: 10.1182/blood-2003-11-4011
Keywords: human bone marrow, DNA viruses, human papillomavirus 31, transplantation, hematopoietic disorders, virome
Citation: Toppinen M, Sajantila A, Pratas D, Hedman K and Perdomo MF (2021) The Human Bone Marrow Is Host to the DNAs of Several Viruses. Front. Cell. Infect. Microbiol. 11:657245. doi: 10.3389/fcimb.2021.657245
Received: 22 January 2021; Accepted: 06 April 2021;
Published: 22 April 2021.
Edited by:
Georges Michel Verjans, Erasmus Medical Center, NetherlandsReviewed by:
Ugo Moens, Arctic University of Norway, NorwayEstanislao Nistal-Villan, San Pablo CEU University, Spain
Richard Thompson, University of Cincinnati, United States
Copyright © 2021 Toppinen, Sajantila, Pratas, Hedman and Perdomo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria F. Perdomo, bWFyaWEucGVyZG9tb0BoZWxzaW5raS5maQ==
 Mari Toppinen
Mari Toppinen Antti Sajantila
Antti Sajantila Diogo Pratas
Diogo Pratas Klaus Hedman
Klaus Hedman Maria F. Perdomo1*
Maria F. Perdomo1*