
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 28 June 2021
Sec. Parasite and Host
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.654216
This article is part of the Research Topic Systems Biology of Hosts, Parasites and Vectors View all 11 articles
Malaria transmission relies on parasite-mosquito midgut interaction. The interactive proteins are hypothesized to be ideal targets to block malaria transmission to mosquitoes. We chose 76 genes that contain signal peptide-coding regions and are upregulated and highly abundant at sexual stages. Forty-six of these candidate genes (60%) were cloned and expressed using the baculovirus expression system in insect cells. Six of them, e.g., PF3D7_0303900, PF3D7_0406200 (Pfs16), PF3D7_1204400 (Pfs37), PF3D7_1214800, PF3D7_1239400, and PF3D7_1472800 were discovered to interact with blood-fed mosquito midgut lysate. Previous works showed that among these interactive proteins, knockout the orthologs of Pfs37 or Pfs16 in P. berghei reduced oocysts in mosquitoes. Here we further found that anti-Pfs16 polyclonal antibody significantly inhibited P. falciparum transmission to Anopheles gambiae. Investigating these candidate proteins will improve our understanding of malaria transmission and discover new targets to break malaria transmission.
Malaria is a deadly infectious disease spread by mosquitoes. It affects half of the world’s population and claims nearly a half-million lives annually. Although the global malaria cases have decreased since 2010, the ten most burdened countries in Africa still had 3.5 million more cases than the previous year (Who, 2018). The fast spread of insecticide-resistant mosquitoes partially causes this. Mosquitoes become resistant to the five major chemical pesticides, making traditional vector control methods by pesticides often fail to control malaria epidemics (Alout et al., 2017). Concerns also come from the artemisinin-resistant Plasmodium falciparum throughout Cambodia and parts of Thailand. The spread of drug-resistant parasites may cause devastating consequences in the future (Chookajorn, 2018; Conrad and Rosenthal, 2019). Malaria vaccines that protect humans from infecting would be ideal for controlling malaria epidemics. However, no malaria vaccines have been developed. The most promising malaria vaccine candidate is RTS,S. It is mainly composed of circumsporozoite protein fragments presented on lipoprotein particles (Duffy and Gorres, 2020). However, the efficacy of RTS,S in Phase III trial was less than 36% (Rts, 2014). These issues have prompted researchers to find new strategies to stop the spread of malaria.
Since Plasmodium-infected mosquitoes spread malaria, a vaccine or medicine that prevents parasites from infecting mosquitoes is an alternative for malaria control. The targets of transmission-blocking reagents may participate in parasite developmental processes such as differentiation of sexual forms of microgametes and macrogametes, zygote formation, ookinete invasion of mosquito midgut, and sporozoite infection (Acquah et al., 2019). Several antigens from parasites have been examined as targets to block malaria transmission. Antibodies against Pfs25 or members of s48/45 6-Cysteine family (Pfs230, Pfs48/45, and Pfs47) on the surface of gametocytes (MacDonald et al., 2016) and zygotes (Young et al., 2005) have been shown to inhibit malaria transmission. Some proteins targeted by TBV studies were summarized in a recent review (Bennink et al., 2016).
The interaction between parasites and the mosquito midgut is a crucial determinant of successful infections in a mosquito. Quite some studies have shown mosquito proteins are essential for the ookinete formation in the mosquito midgut and their penetration of the peritrophic matrix (PM) and midgut epithelial cells. Mosquito proteins AnAPN1 and FREP1 have been identified to play roles in malaria transmission. AnAPN1 localizes on the apical surfaces of Anopheles gambiae midguts (Blagborough and Sinden, 2009), and studies have shown that anti-AnAPN1 antibodies significantly inhibited malaria infection in mosquitoes (Dinglasan et al., 2007; Pritsch et al., 2016). FREP1 was identified by a direct association study of clinically circulating P. falciparum infection in wild An. gambiae mosquitoes (Li et al., 2013). FREP1, located at PM, interacts with parasites, thereby facilitating malaria transmission (Zhang et al., 2015). Antibodies targeting the FREP1 FBG domain inhibited transmission of P. falciparum and P. vivax to An. gambiae and An. dirus, respectively (Niu et al., 2017). CRISPR/Cas9-mediated gene knockout of FREP1 further elucidated the function of FREP1 as a host factor in mosquitoes (Dong et al., 2018).
Parasitic proteins involved in gametogenesis, fertilization of macro- and micro-gametes, zygote-to-ookinete transformation, and penetration of the midgut endothelium are also critical for malaria transmission (Bennink et al., 2016). Recently, protein Pfs47 (PF3D7_1346800) was reported to interact with AgP47Rec in the mosquito midguts to evade mosquito immunity (Molina-Cruz et al., 2020). Also, some soluble proteins secreted by the sexual stage parasite or membrane-associated proteins such as chitinase and von Willebrand factor A-domain-related protein (WARP) are also important to complete parasite transmission to mosquitoes (Li et al., 2004). More research is required to study parasitic proteins that interact with mosquito midguts to elucidate molecular mechanisms of malaria transmission and provide more targets for malaria control.
Besides targeted by transmission-blocking vaccines, these interactive proteins can also be targeted by small molecules to block malaria transmission. The fungal secondary metabolite P-orlandin from Aspergillus niger (Niu et al., 2015), asperaculane B from Aspergillus aculeatus (Niu et al., 2020b), and pulixin (Niu et al., 2021) that prevent FREP1 in mosquito midgut from binding to sexual stage Plasmodium falciparum inhibit malaria transmission to mosquitoes. A synthetic polysulfonated polymer limits malaria transmission by inhibiting the interaction between midgut chondroitin sulfate glycosaminoglycans and ookinetes (Mathias et al., 2013).
Therefore, this study focused on sexual stage parasitic proteins that interact with mosquito midguts. These proteins should be at the cytoplasmic membrane or secreted from the parasites. The identification of these proteins will help us to understand the molecular mechanisms of malaria transmission and provide targets for vaccines and drugs to control malaria transmission.
An. gambiae (G3 strain) obtained from BEI Resources was maintained in an insectary set at 28°C, 80% relative humidity, 12 hours (h) day/night cycle. Larvae were fed with grounded fish food, and adult mosquitoes were maintained with 8% sucrose solution. The commercial human (AB+, Oklahoma Blood Institute, Oklahoma City, OK) was washed with the same volume of RPMI-1640 three times by centrifugation (500xg for 5 minutes (m)). The human serum (O+, Interstate Blood Bank, Memphis, TN) was heat-inactivated at 56°C for 30 m. The red blood cells and serum were mixed at the ratio of 1:1 (v/v) to feed mosquitoes through a glass feeding device (Chemglass, Vineland, NJ) to lay eggs.
P. falciparum (NF54 strain) was obtained from BEI. The parasites were cultured with the RPMI-1640 (Gibco) complete medium that contains 4% fresh O+ human red blood cells (Oklahoma Blood Institute, Oklahoma City, OK), 12.5 µg/mL hypoxanthine, and 10% human AB+ serum (Interstate Blood Bank, Memphis, TN) in a candle jar at 37°C. The culture was initiated at 0.25-0.5% parasitemia, and the medium was replaced daily. The day 15-17 P. falciparum centrifugation at 500×g for 3 m) and resuspended in human AB+ serum/packed O+ red blood cells (1:1 by volume) and used for standard membrane feeding assays (SMFA) (Zhang et al., 2015).
The sequences of P. falciparum (3D7) proteins were downloaded from PlasmoDB. The protein sequences in fasta format were uploaded into TMHMM Server (v2.0) online (http://www.cbs.dtu.dk/services/TMHMM/). The output format parameter was “Extensive, no graphics”. The number of transmembrane regions in each protein was predicted with the TMHMM algorithm (Krogh et al., 2001).
The information of the signal peptide for each protein was obtained from PlasmoDB. The data of the gene expression of the published RNA-Seq data (Lopez-Barragan et al., 2011) at the different developmental stages of the parasites was downloaded from PlasmoDB. It is inside mosquito midguts, and the gametocytes become gametes that further form ookinetes. The interactions between midguts and gametocytes as well as between midguts and ookinetes are important. These genes are expected to be upregulated at the sexual stage, and their products are cytoplasmic membrane proteins or secretory proteins. Therefore, the candidate genes were selected according to the following three criteria: 1) the coding proteins containing signal peptides; 2) gene expression at sexual stages (median values of stage II gametocytes, stage V gametocytes, and ookinetes) were >5-fold higher than that at any asexual stages (maximum schizont); and 3) genes are abundantly expressed at the sexual stage with >60 RNA-Seq reads at any sexual stages (minimum value). The protein annotations, subcellular locations, and other features were obtained from the UniProt and PlasmoDB. We did not use the mass spectrometry proteomics data because it is challenging to separate cells in different stages (Swearingen and Lindner, 2018).
Total RNA was extracted from the P. falciparum (NF54) infected RBCs using RNAzol (Sigma-Aldrich, MO). The cDNA was synthesized using SuperScript First-Strand Synthesis System (Invitrogen, CA). The genes were amplified by PCR using DNA Engine Dyad Thermal Cycler (Bio-Rad, CA) with gene-specific primers (Table S1). The DNA fragments were purified by GeneJet PCR Purification Kit (Thermo Scientific) and cloned into the modified plasmid pFastBac1, which contains a 6xHis tag at the C-terminus of the multiple cloning site. The recombinant plasmid was transformed into DH5α competent cells. PCR-positive recombinant plasmids were confirmed by sequencing and then transformed into DH10Bac competent cells to obtain recombinant bacmids. The white colonies on culture plates were picked and confirmed by PCR. About 1 μg of recombinant bacmid in 100 μL of Grace’s Insect Medium, unsupplemented (Invitrogen) was mixed with 100 μL of Grace’s Insect Medium, unsupplemented (Gibco) containing 5 μL of cellfectin II, and incubated at room temperature (RT) for 15 m. The mixture was then added to 1.5 mL of Grace’s Insect Medium, unsupplemented containing 1×106 sf9 cells in a 6-well culture plate, and incubated for 4 h at 27°C. The supernatant was replaced with 2 mL of Grace’s Insect Medium (Gibco) with 10% FBS (Invitrogen) and cultured at 27°C for 3 days to produce recombinant baculoviruses. After amplification for 2-3 generations, 100 μL of the culture supernatant containing the recombinant virion was added to 2 mL of Express Five serum-free medium (Gibco) supplemented with 20 mM L-glutamine and 1 million High Five cells in the wells of a 6-well culture plate. After incubation at 27°C for three days, both supernatant and cells were harvested by centrifugation at 500×g for 5 m at 4°C. The cells were suspended in native cell lysis buffer (Invitrogen) and sonicated (pause for 30 s every 10 s) for 5 m on ice. Cell debris was removed by centrifugation (10,000 g for 5 m). The recombinant protein concentrations in culture medium and cell lysate were measured with anti-His monoclonal antibodies comparing to a standard protein concentration curve.
The 3-5-day old adult female mosquitoes were fed with fresh blood using a membrane feeding device. Starting 16 h post blood meal and lasting more than 4 h, ~100 midguts were dissected from blood-engorged mosquitoes. The blood was carefully removed from the midguts. The midguts were then placed in a 1.5 mL plastic tube containing lysis buffer (50 mM Tris-HCl, 0.15 mM NaCl, 0.2% Tween-20, pH7.8) and grounded with a micro pestle (Sigma-Aldrich). Subsequently, the tissues were lysed in native cell lysis buffer (Clontech) with ultrasonication for 10 m on ice with 30 s pulse and 10 s sonication. The debris was removed by centrifugation (10,000 g for 5 m) at 4°C. The protein concentration in the supernatant was measured by the Bradford method.
About 50 µL of 1 mg/mL midgut proteins were used to coat each well on a 96 well plate at 4°C overnight. The plate was then blocked with 100 µL of 2% BSA in PBS for 2 h at RT. About 100 ng of recombinant candidate proteins in 50 µl PBS were added to each well. Each corresponding recombinant protein was heat-inactivated for 15 m at 65°C as a control. The plates were incubated for 1 h at RT. Then, 50 µl anti-His mouse monoclonal antibody (Sigma, 1:1, 000 dilutions in blocking buffer) was added and incubated at RT for 1 h, followed by incubation with 50 µl of goat anti-mouse IgG-Alkaline Phosphatase conjugate (Sigma, 1:10, 000 dilutions in blocking buffer) for 1 h at RT. Finally, 50 µL of p-nitrophenyl phosphate (pNPP) (Sigma-Aldrich, MO) was added to develop the plate. The plate was then washed three times with PBST (1xPBS containing 0.2% Tween 20) between two incubations. The signal was detected with an Epoch Microplate Spectrophotometer at A405 nm (BioTek, Winooski, VT). Because one set of data from the same sample (midgut lysate) was used multiple times (46 proteins), statistical analysis was conducted by multiple t-test using the two-stage linear step-up procedure (Benjamini et al., 2006) with 1% of False Discovery Rate (Q) implemented in GraphPad Prism 8 (GraphPad Software, USA).
These six recombinant proteins were further verified by an alternative ELISA. About 10 unhomogenized midguts (collected 18 h after bloodmeal) in a 1.5 mL plastic tube were suspended in 50 µl PBS containing 100 ng of a recombinant candidate protein and incubated for 1 h at RT. The PBS buffer with chloramphenicol acetyltransferase (CAT), which was expressed with the baculovirus system in the High Five cells, was used as a blank control. Then, the midguts were collected through centrifugation (500 g for 5 m), washed with 100 µl PBS three times, and homogenized in 100 µl lysis buffer (50 mM Tris-HCl, 0.15 mM NaCl, 0.2% Tween-20, pH7.8) by a micro pestle (Sigma-Aldrich) and ultrasonication as described above. After removing the debris by centrifugation (10,000 g for 5 m) at 4°C, 50 µl of the supernatant was used to coat ELISA, followed by incubation with anti-His mouse monoclonal antibody, goat anti-mouse IgG-Alkaline Phosphatase conjugate, and pNPP as described above. The plate was washed three times with PBST (1xPBS containing 0.2% Tween 20) between two incubations. The signal was detected with an Epoch Microplate Spectrophotometer at A405 nm (BioTek, Winooski, VT). Statistical analysis was conducted by multiple t-test using the Two-stage linear step-up procedure (Benjamini et al., 2006) with 1% of False Discovery Rate (Q) implemented in GraphPad Prism 8 (GraphPad Software, USA).
E. coli-expression of the full-length mature Pfs16 protein and generation of the antibody was conducted by a company (Bosterbio, Pleasanton, CA). The polyclonal anti-Pfs16 antibody was generated in rabbits and purified with protein A/G immunogen affinity. The titer of the antibody was measured by coating 0.1 µg antigen to the wells in a 96-well plate and calculated by testing a series dilution of the antibody. To conduct antibody transmission-blocking assays, we suspended day 15-17 cultured P. falciparum with the fresh O+ type human blood to get the 0.2% final concentration of stage V gametocytes. Approximately 15 μL PBS containing 45 μg purified anti-Pfs16 rabbit polyclonal antibody was mixed with 285 μL infectious blood (final antibody concentration is 0.15 mg/ml), and an SMFA (Niu et al., 2020a) was conducted using 3-5 days old female naïve mosquitoes. After feeding for 30 m, the engorged mosquitoes were separated and maintained with 8% sugar in a BSL2 insectary (27°C, 12-h light/dark cycle, 80% humidity). Seven days after infection, midguts were dissected, stained with 0.1% mercurochrome, and examined using light microscopy to count oocysts. Equivalent amounts of irrelevant antibody or BSA were used as controls. Since the distribution of oocyst numbers does not follow the normal distribution, the results were analyzed with the nonparametric test, Wilcox-Mann-Whitney test, and the experiments were repeated three times. Also, 15 μL PBS containing 45 μg, 15 μg, 3 μg, 1 μg or 0 μg purified anti-Pfs16 rabbit polyclonal antibody were mixed with 210 μL infectious blood to conduct SMFA. Due to infection rates and mosquito mortality, >50 mosquitoes were used for infection.
Mosquito midguts were isolated from 3-5-day-old naïve mosquitoes. The mosquito midguts were also isolated from mosquitoes 18 hr post bloodmeal. The midguts were cut in half, and the content was rinsed with 200 μL 1xPBS three times. Midguts were incubated sequentially with the recombinant Pfs16 protein (experimental group) or CAT (control group) expressed in High Five cells culture supernatant (10 μg/mL) for 1 h at RT, the rabbit antibodies against Pfs16 protein in PBS (1μg/mL) for 1 h at RT, Alexa Fluor 594-conjugated goat against rabbit antibodies (ThermoFisher, 1:500 dilutions in PBS) for 1 h in the dark at RT. The sample midguts were washed with 1XPBS 5 min for 3 times between each incubation. Finally, the treated midguts were examined under a fluorescence microscope. All midguts were exposed with the same activated light intensity, and images were taken with the same exposure time. The fluorescence pixel in images was measured with Adobe Photoshop 2020.
We selected P. falciparum proteins with signal peptides, and their gene expression was upregulated and abundantly present at the sexual stage as described in methods. Based on protein sequences from PlasmoDB, 1,079 P. falciparum proteins contain signal peptides. The RNA-Seq data of seven developmental stages of these genes (Lopez-Barragan et al., 2011) were downloaded from PlasmoDB. Differential gene expression analysis of RNA-seq data was performed using the R package, and a heatmap was generated. The result showed stage-specific gene expression patterns, with about half of them being upregulated at the sexual stage (Figures 1A, S1). Among them, 670 gene-encoded proteins contained one or more transmembrane regions (TM), and 413 contained signal peptides without TM. We selected the genes that were >5-fold upregulated at sexual stages and abundantly expressed (>60 reads per gene) at the sexual stages (gametocyte II, V, and ookinete). These criteria were selected because only sexual stage proteins are critical for parasites to infect mosquitoes, and abundantly expressed genes have fewer measurement errors and are relatively easier to study their mechanisms. Based on the selection criteria, 76 candidate proteins were obtained for further study (Figure 1B). Many important genes, such as CelTOS, GEST, and SOAP, which have been proved to be necessary for parasite invasion, were included in this list (Table 1). The expression data from another study (Pelle et al., 2015) were used to verify the expression of candidate genes. Except for gene PF3D7_1115100, the microarray expression profiles of 75 candidate genes were obtained. Results showed that >90% of candidate genes had expression signals greater than 4, confirming their abundance at the sexual stage (Figure S2A). Also, the microarray data confirmed that they were upregulated at the sexual stage comparing to the asexual stage (Figure S2B).
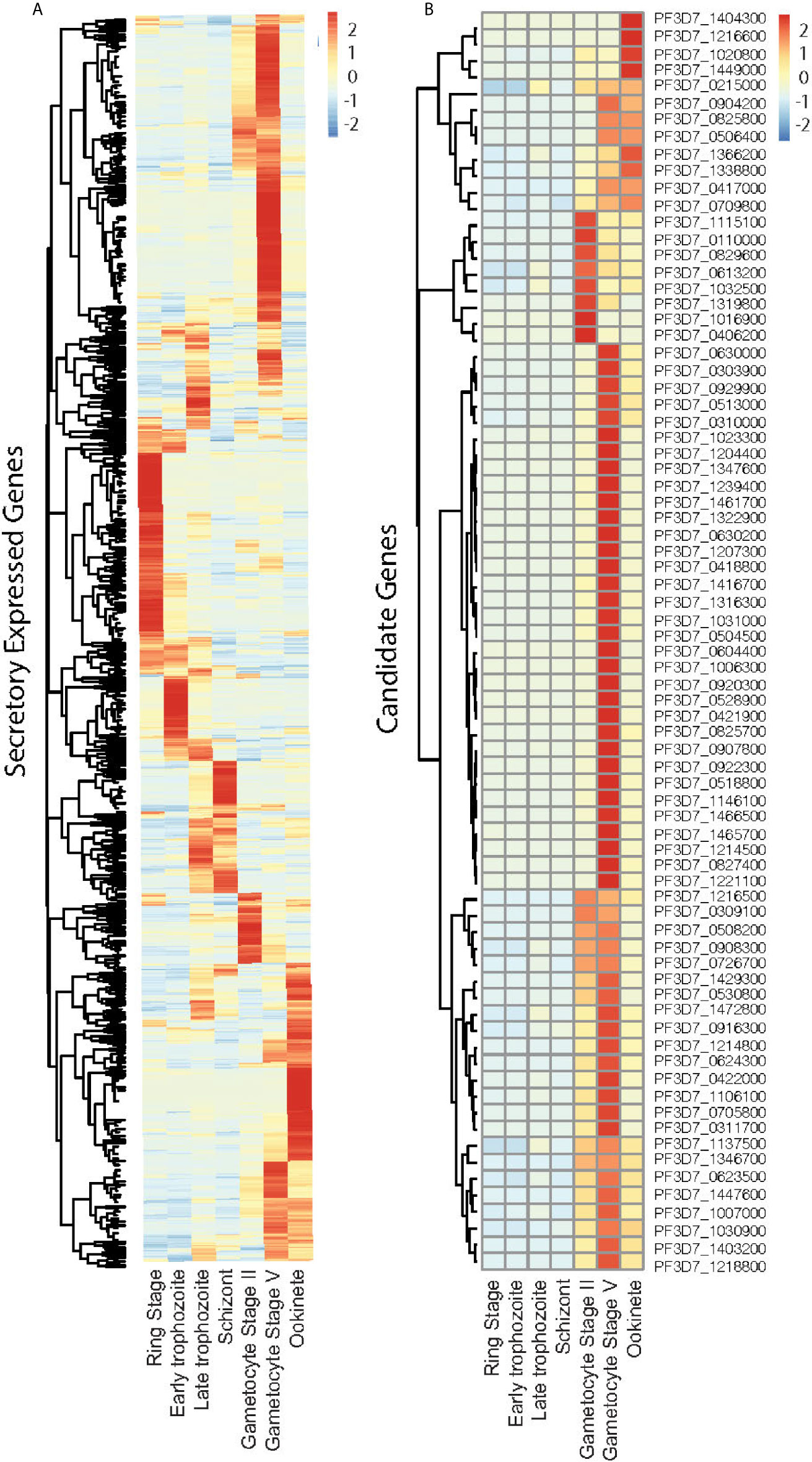
Figure 1 Expression profile of the Plasmodium falciparum genes containing signal peptide at different developmental stages. (A) The expression profiles of 1,079 parasitic proteins containing signal peptides using RNA-seq data (Lopez-Barragan et al., 2011). (B) The expression profiles of candidate genes.
Because not all membrane proteins have signal peptides, we also investigated the parasitic proteins that had TM while lacking signal peptides. About 949 P. falciparum proteins have TM and lack signal peptides (Table S2). However, none of them satisfied the expression criteria, e.g., >5-fold upregulated at sexual stages and abundantly expressed at the sexual stages (>60 reads per gene).
About 39 of the 76 candidate proteins contained TM. These 76 candidate proteins were localized in different subcellular compartments, although subcellular locations of 35% of these proteins (n=27) are still unknown (Figure 2). The rest were distributed in 9 different locations, including rhoptry neck, apicoplast, cell surface, crystalloid, cytoplasm, endoplasmic membrane, microneme, nucleus, and osmiophilic body. Among them, 18% of these proteins (n=14) are located on the apicoplast. The apicoplast is a specific organelle important for Plasmodium development. Also, a decent number of candidate proteins are located on the membrane (15%), which include parasitophorous vacuole membrane and cytomembrane.
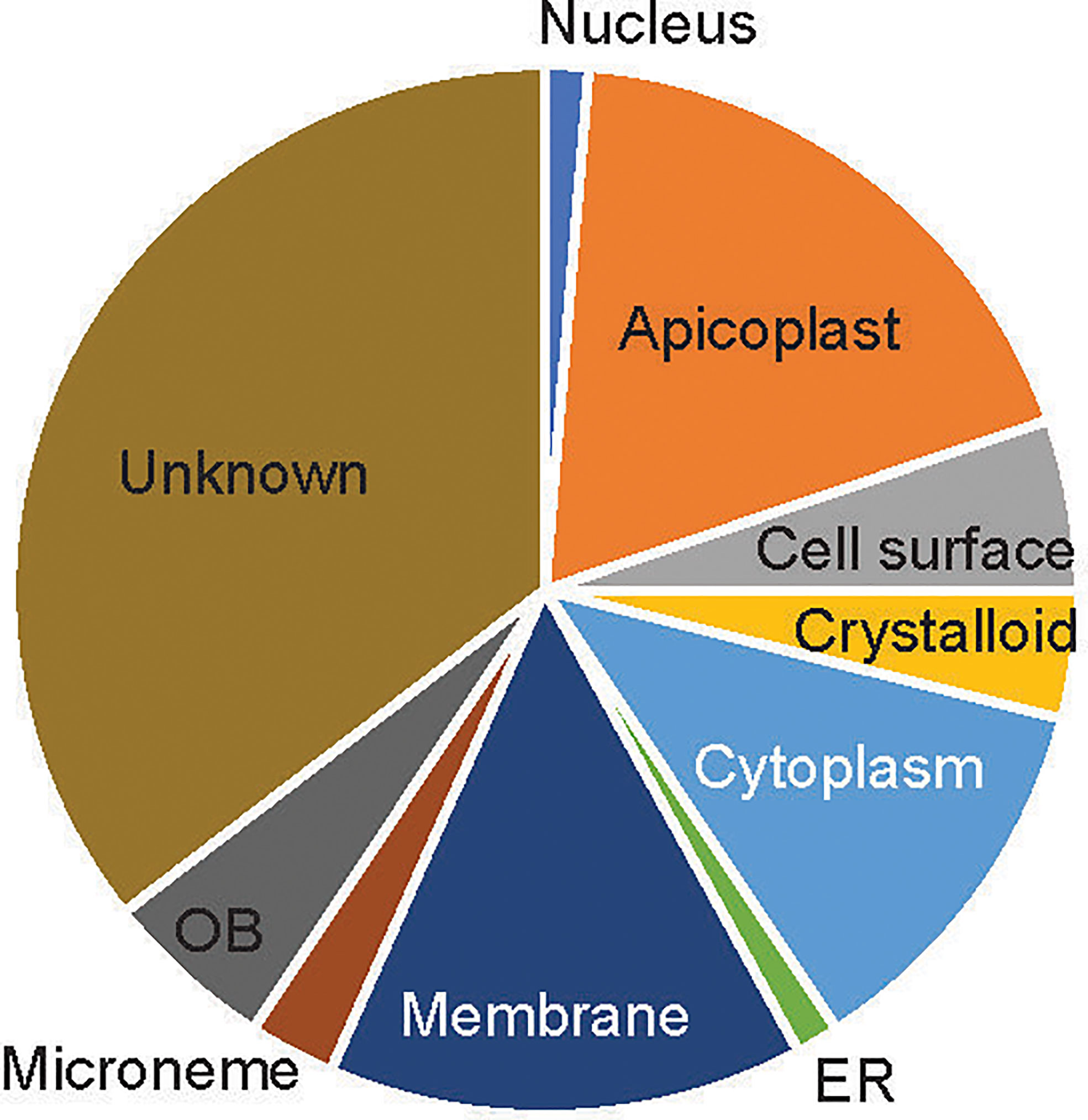
Figure 2 The subcellular location of the candidate proteins. ER, endoplasmic reticulum; OB, osmiophilic body.
Candidate genes were PCR-cloned and the recombinant proteins were expressed in High Five insect cells using the baculovirus expression system in the serum-free medium. A monoclonal antibody against the 6xHis tag at the C-terminus of recombinant proteins was used to quantify the recombinant protein concentration. We have finally cloned and expressed 46 genes successfully at ELISA detectable level for further analysis (Figure 3A).
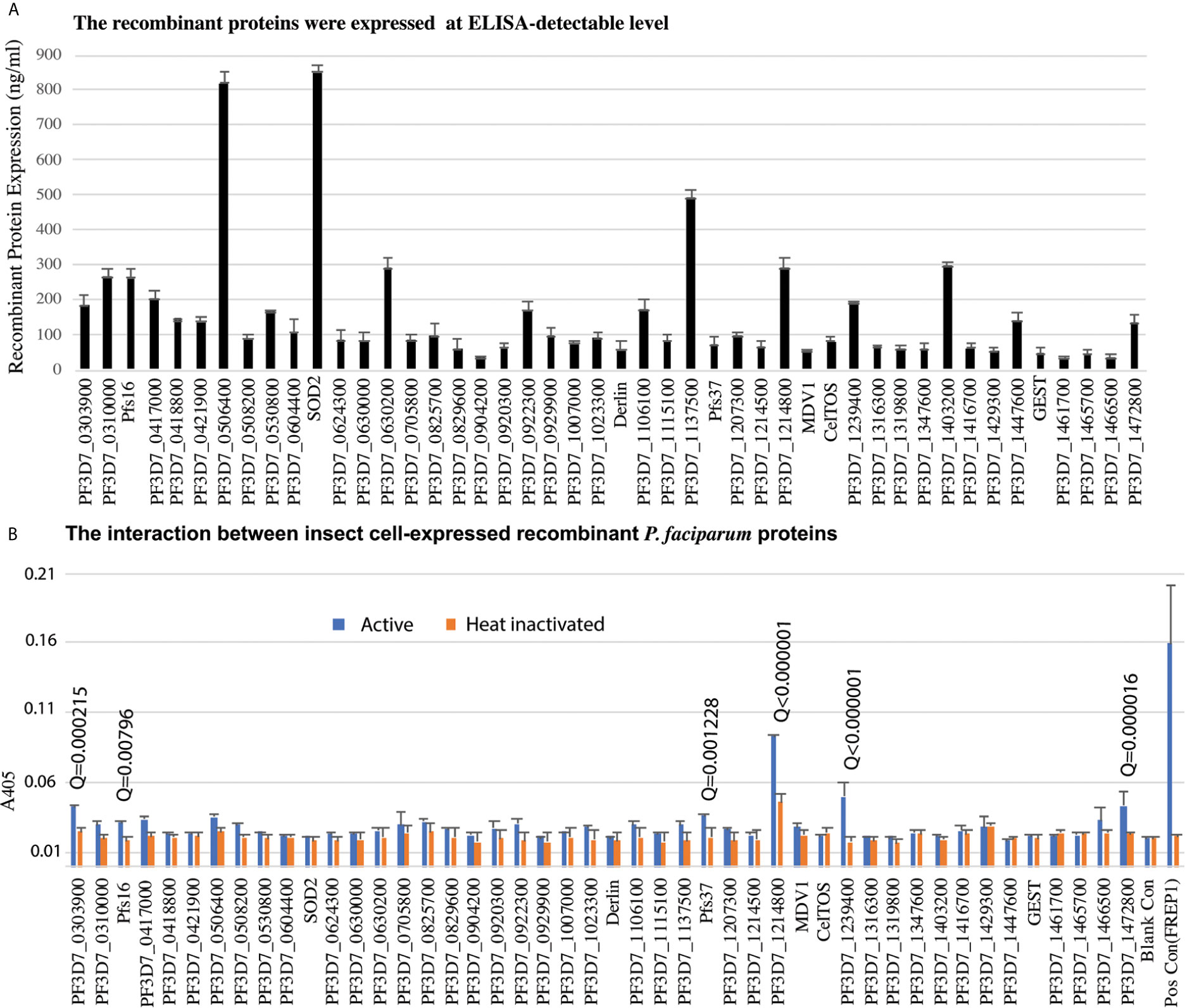
Figure 3 The insect-cell expressed recombinant parasitic proteins and their interaction with midguts. (A) The expression of candidate proteins in high five cells. Totally, forty-six genes were expressed in high five cells with a baculovirus expression system at a detectable level. Each sample was measured with triplicates. The expression experiments were conducted three times. (B) Interaction between recombinant proteins and midgut lysate. The concentration of recombinant proteins was normalized. Naïve recombinant proteins and heat-inactivated proteins were used as experimental groups and control groups with triplets. The false discovery rates (Q) were calculated by multiple t-test using the Two-stage linear step-up procedure (Benjamini et al., 2006). Those proteins with Q <1% were labeled with the Q values. The same experiments were repeated three times, and the results were similar.
To determine if P. falciparum candidate proteins interacted with An. gambiae midgut lysate, we performed ELISA. Mosquitoes were fed with blood, and their midguts, including peritrophic matrix, were isolated 15-21 h post blood meal. A 96-well 100 ng of recombinant protein was added into wells to determine the interaction between a candidate protein and the midgut lysate. The heat-inactivated proteins were used as the corresponding negative controls. Both the experimental group and control were done in triplicates per treatment. CAT, which was expressed with the baculovirus system in the High Five cells, was used as a blank control. The A405 of experimental and control wells were measured (Figure 3B). Since the same samples were used in 46 genes, the A405 values were analyzed with multiple t-test using the two-stage linear step-up procedure (Benjamini et al., 2006) with 1% of False Discovery Rate (Q). The result showed that six P. falciparum proteins (PF3D7_0303900, PF3D7_0406200 (Pfs16), PF3D7_1204400 (Pfs37), PF3D7_1214800, PF3D7_1239400, PF3D7_1472800) were retained by midgut lysates at a level significantly higher than the corresponding controls (Figure 3B, False Discovery Rate (Q) < 0.01).
These six recombinant proteins were further verified by an alternative ELISA using unhomogenized midguts to pull down the recombinant proteins following by homogenizing and detection. The results (Figure 4) showed the difference between each recombinant protein and the blank control was significant (Q < 0.01), which confirmed the interaction between the candidate proteins and midguts.
To understand the function of candidate proteins, we searched their orthologs in P. berghei and analyzed the P. berghei infectious phenotypes for their knockout in public databases RMgmDB (Khan et al., 2013; Gomes et al., 2015), PlasmoGEM (Bushell et al., 2017), and other works (Zhang et al., 2018). Results (Table 2) showed that three P. berghei genes were knocked out successfully, and two P. falciparum genes could be mutated by transposon insertions (Zhang et al., 2018). Disruption of orthologs of PF3D7_12148, PF3D7_1204400 (Pfs37) or PF3D7_0406200 (Pfs16) did not change P. berghei infection at asexual stage. Knockout of PBANKA_1430600, the ortholog of PF3D7_1214800, did not affect asexual stage parasites, and the effect on sexual stage parasites and transmission has not been reported yet. Knocking out the orthologs of Pfs37 or Pfs16 rendered Plasmodium parasites to generate fewer oocysts in mosquitoes. Pbg37, the ortholog of Pfs37, is a protein of 37 kDa with a signal peptide and multiple transmembrane domains and only expressed at sexual stages of parasites such as gametocytes, zygotes, and ookinetes. Knockout of Pbg37 led to reduced gametocyte production and significantly affected the further development of ookinetes from gametocytes. Notably, the anti-Pbg37 antiserum from the immunized mice significantly reduced the number of oocysts in the midgut, which indicates that it could be a target against malaria infection (Liu et al., 2018).
Next, we selected Pfs16 to study the antibody effects on malaria transmission. Pfs16 was found in the P. falciparum gametocyte parasitophorous vacuole membrane. Knockout of PBANKA_1003900 (Pbg16), the ortholog of Pfs16 in P. berghei, did not change Plasmodium asexual stage (Kongkasuriyachai et al., 2004) or formation of ookinetes (Deligianni et al., 2018). Disruption of Pbg16 reduced the number of oocysts in mosquitoes (Table 2) (Kongkasuriyachai et al., 2004). However, the amino acid sequences of Pfs16 and Pbg16 are largely different. Thus, we further examined the function of Pfs16 on parasite transmission using transmission-blocking assays.
The anti-Pfs16 polyclonal antibody was generated in rabbits immunized with E. coli-expressed Pfs16 as the antigen and further purified using protein A/G immune affinity column. The specificity of the anti-Pfs16 antibody was verified with Western blot analysis using the recombinant protein expressed in insect cells. The result (Figure 5A) showed two bands with the size of about 19kD and 16kD, which matched the precursor (with signal peptide) and mature forms of recombinant protein containing 6xHis and V5 tags. Therefore, a rabbit polyclonal antibody against Pfs16 could specifically recognize recombinant Pfs16. Next, we determined the inhibition of anti-Pfs16 polyclonal antibody against P. falciparum transmission to mosquitoes. The P. falciparum culture containing 0.2% stage V gametocytes and 0.15 mg/ml anti-Pfs16 antibody (titer = 103/mg) was used to infect An. gambiae (G3) using SMFA. The equal amounts of unrelated polyclonal antibody (anti-V5) or BSA were used to substitute anti-Pfs16 antibody as the control. The results showed that compared with the anti-V5 or BSA controls, the anti-Pfs16 Ab significantly reduced the number of oocysts by 50% and 57%, respectively (Figure 5B), supporting that Pfs16 is a potential target for malaria control. This experiment was repeated three times and obtained similar results. We also examined the different concentrations (0.2 mg/mL, 77 μg/mL, 20 μg/mL, 7.7 μg/mL and 0 μg/mL) of anti-Pfs16 on P. falciparum transmission to An. gambiae using SMFA. The results showed that the antibody inhibition rate decreased with the reduction of antibody concentration (Figure 5C). When antibody concentration was greater than 0.078 mg/mL, the inhibition against malaria transmission to mosquito was significant (p < 0.05).
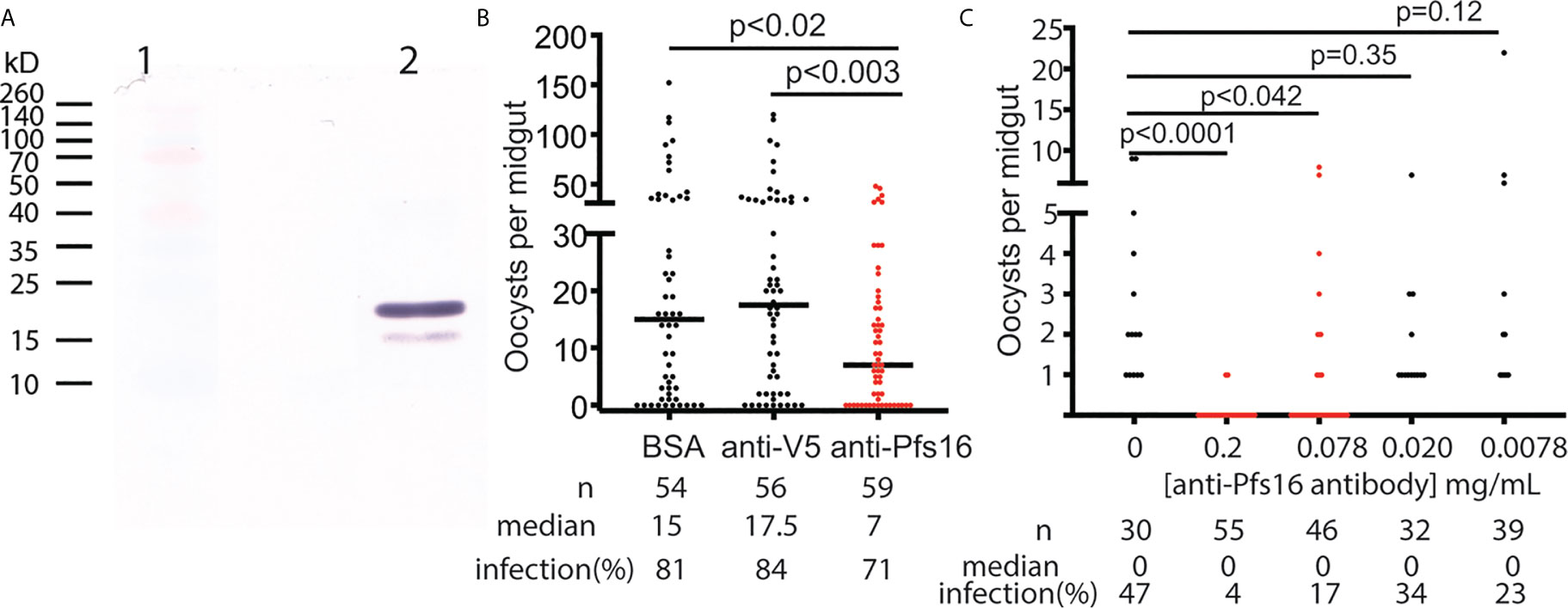
Figure 5 The functional analysis of Pfs16 on Plasmodium transmission to mosquitoes. (A) Western blot assay showed that anti-Pfs16 antibody specifically recognized the insect cell-expressed recombinant Pfs16 protein in lane 2. Two bands in lane 2 are the precursor and mature forms of Pfs16 with His- and V5-tag Lane 1 displayed standard protein markers with some color proteins. (B) Rabbit anti-Pfs16 polyclonal antibody inhibited P. falciparum transmission to An. gambiae by SMFA. The same concentration of BSA and an unrelated antibody (anti-V5) were used as the controls. The p-values between the experimental and the control groups were calculated using Wilcoxon-Mann-Whitney tests. The assays were repeated, and the results were similar. Horizontal bars are the median numbers of oocysts per mosquito midguts. (C) The antibody inhibition on malaria transmission was dose-dependent. This experiment was repeated twice, and the results were similar.
Finally, we examined the binding tissues of Pfs16. The naïve mosquito midgut contains endothelia and the basal lamina, while a peritrophic matrix was formed after a blood meal. The mosquito midguts were isolated from the naïve and blood-fed mosquitoes and incubated with insect cell-expressed recombinant Pfs16 protein or the non-specific insect cell-expressed protein (CAT) as the control. The rabbit polyclonal antibody against Pfs16 was used to detect the bound Pfs16. Results showed that the fluorescence from naïve mosquitoes incubated with CAT was the weakest (Figure 6A). Substitution of the CAT with Pfs16 protein resulted in a slighter increase of fluorescence intensity (Figure 6C); however, it was similar to the blood-fed mosquito midguts incubated with CAT and anti-Pfs16 (Figure 6B). The fluorescence was the highest for the blood-fed mosquito midguts incubation with Pfs16 and anti-Pfs16 (Figure 6D). The medians of fluorescence intensity of four treatments were shown in Figure 6E. Collective, these data suggest that Pfs16 protein interacted with the midgut peritrophic matrix.
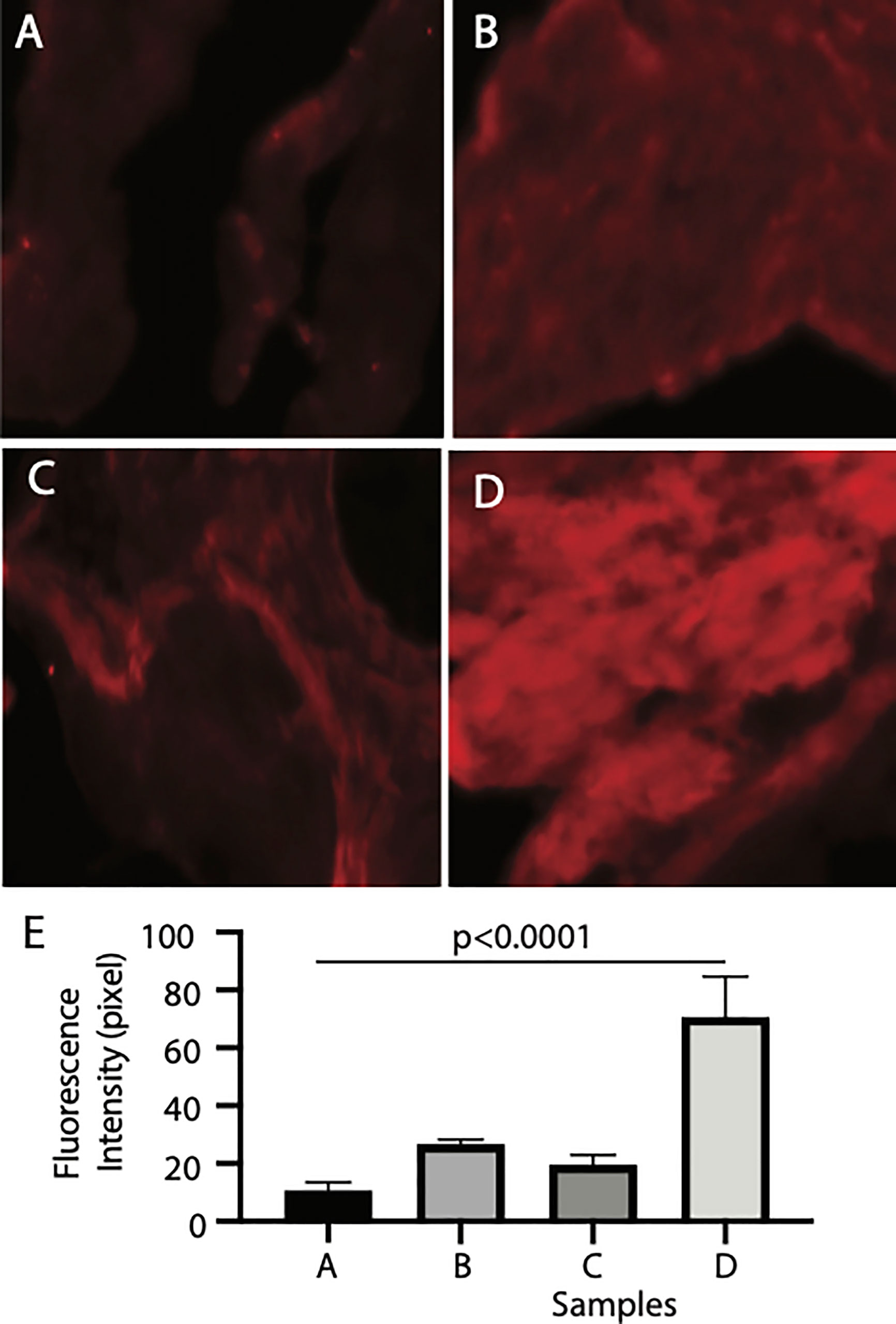
Figure 6 Recombinant Pfs16 protein bound to blood-fed mosquito midguts. (A) The naïve mosquito midguts were incubated with unrelated protein (CAT). (B) The blood-fed mosquito midguts incubated with un-related protein (CAT). (C) The naïve mosquito midguts incubated with Pfs16. (D) The blood-fed mosquito midguts incubated with Pfs16 protein. (E) The median fluorescence intensity of differently treated midguts. P. Calculated with one-way ANOVA.
Mosquito midgut is the first place for Plasmodium development into ookinetes and the subsequent invasion. The interaction between Plasmodium parasites and mosquito midgut plays a key role in malaria transmission (Zhang et al., 2015). However, the parasite-midgut interactome is not well-understood. This project investigated parasitic secreted proteins that interact with mosquito midguts.
About 76 parasitic proteins with signal peptides and abundant at the sexual stage of parasites were selected for investigation. Many of these candidate proteins reportedly play essential roles in malaria transmission. For instance, the CelTOS is essential for ookinetes to rupture the midgut epithelial cell membrane (Kariu et al., 2006), and GEST is the gamete egress and sporozoite traversal protein vital for gametes exit hosts in mosquito midguts (Stone et al., 2018). Our data showed that both CelTOS and GEST did not directly bind to the mosquito midguts, consistent with the reported mechanisms that CelTOS was secreted directly into endothelial cytoplasm (Kariu et al., 2006) and PEST ruptured gametocyte parasitophorous vacuole membrane (Talman et al., 2011).
The 76 candidate proteins locate at different subcellular locations, such as apicoplast, osmiophilic body, and microneme. Previous studies have shown that the abundant proteins in apicoplast have been often the targets to develop malaria drugs (Biddau and Sheiner, 2019). Micronemes play a critical role during the invasion of parasites in mosquitoes (Li et al., 2004). Three ookinete-secreted proteins, PgCHT1 chitinase, WARP, and CTRP from micronemes, can break mosquito midgut to facilitate malaria transmission. Antibodies against these proteins interfere with parasite invasion (Li et al., 2004). There are several membrane proteins e.g., Pfs25 (PF3D7_1031000), P28 (PF3D7_1030900), and P48/45 (PF3D7_1346700) that under investigation as TBV by communities.
In the meantime, our bioinformatics analysis might miss some genes if they lack signal peptides or their expression data were incomplete. For instance, mosquito AgP47Rec was recently reported to interact with parasitic protein PF3D7_0134800 (P47) (Molina-Cruz et al., 2020). It was missed in our candidates because the RNA-seq expression data lacks its expression at schizonts (Lopez-Barragan et al., 2011).
Out of the 76 candidate genes, 30 were not cloned into the plasmid due to incorrect annotation or were cloned into plasmids with low expression in insect cells. This is because some proteins such as SOAP rapturing insect cells and some membrane proteins are hard to clone and express in insect cells. Notably, forty-six genes (60.5%) were successfully cloned and highly expressed with a baculovirus expression system in High Five cells. By screening these proteins via ELISA analysis, we identified six P. falciparum proteins that interacted with the midgut of mosquitoes. The orthologs of these six P. falciparum genes were found in P. berghei. Knockout of Pbg37 and Pbg16, orthologs of Pfs37 and Pfs16, respectively, from P. berghei reduced Plasmodium oocysts in mosquitoes (Kongkasuriyachai et al., 2004; Liu et al., 2018). Disruption of the Pbg37 gene negatively impacted malaria transmission, which might be caused by attenuated gametocytes. Importantly, the anti-Pbg37 antiserum inhibited P. berghei transmission to mosquitoes (Liu et al., 2018), supporting Pfs37 is an excellent target to break malaria transmission.
Knockout of the Pbg16 gene did not change parasites at the sexual stage, and the formation of ookinetes was normal. The amino acid sequences between Pfs16 and Pbg16 are strikingly different, with only 35% Pfs16 was covered by Pbg16, and covered regions shared 26% identical amino acids. Therefore, we generated polyclonal antibodies to examine Pfs16 function in malaria transmission. Our data show that the ingested antibodies against Pfs16 significantly inhibited parasite transmission to mosquitoes. This result supports that Pfs16 is accessible to extracellular proteins such as antibodies. Pfs16 was observed as a gene at the onset of P. falciparum gametocytogenesis and often used as a marker for the production of the sexual stage of parasites (Bruce et al., 1994; Dechering et al., 1997; Lanfrancotti et al., 2007). Pfs16 seems not essential in the development of gametocytes and is not involved in the formation of zygote and ookinetes. However, it may play some role in the optimal production of sexual parasites. During the entire gametocyte maturation from stages I–V Pfs16 (Lobo et al., 1994; Dechering et al., 1997; Berry et al., 2009), it was detected to be localized on the parasitophorous vacuole membrane (Moelans et al., 1991; Baker et al., 1994; Eksi and Williamson, 2011). One publication reported that anti-Pfs16 antisera did not show any transmission-blocking activities. However, no data were presented in that paper (Moelans et al., 1995). Differently, our results showed that the purified anti-Pfs16 polyclonal antibody (0.15 mg/mL) significantly inhibited parasite transmission to mosquitoes.
Successful parasite invasion of mosquitoes begins with parasites’ interaction with mosquito midguts. Through bioinformatics analysis followed by protein interaction assays, we identified six midgut-binding parasitic proteins. Moreover, we showed that antibodies against Pfs16 inhibited malaria transmission. Further investigation of these proteins will improve our understanding of the molecular mechanisms of malaria transmission and provide targets to break malaria transmission.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
GN and YC conducted experiments, interpreted the data, and wrote the manuscript draft. XW conducted bioinformatics and statistical analysis. YK expressed Pfs16 proteins in insect cells. JL conceived concepts, designed the project, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work is supported by NIAID (No. 1R01AI125657) and NSF Career Award (No. 1453287). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.654216/full#supplementary-material
Supplementary Table 1 | Specific primers for candidate genes.
Supplementary Table 2 | The expression data of genes that contain transmembrane domains and lack signal peptides.
Supplementary Figure 1 | The heatmap of 1,079 parasitic proteins containing signal peptides.
Supplementary Figure 2 | The expression of parasitic proteins based on microarray showing the abundance of candidate genes at sexual stages.
Acquah, F. K., Adjah, J., Williamson, K. C., Amoah, L. E. (2019). Transmission-Blocking Vaccines: Old Friends and New Prospects. Infect. Immun. 87, e00775–18. doi: 10.1128/IAI.00775-18
Alout, H., Roche, B., Dabire, R. K., Cohuet, A. (2017). Consequences of Insecticide Resistance on Malaria Transmission. PLoS Pathog. 13, e1006499. doi: 10.1371/journal.ppat.1006499
Baker, D. A., Daramola, O., McCrossan, M. V., Harmer, J., Targett, G. A. (1994). Subcellular Localization of Pfs16, A Plasmodium Falciparum Gametocyte Antigen. Parasitology 108 (Pt 2), 129–137. doi: 10.1017/s0031182000068219
Benjamini, Y., Krieger, A. M., Yekutieli, D. (2006). Adaptive Linear Step-Up Procedures That Control the False Discovery Rate. Biometrika 93, 17. doi: 10.1093/biomet/93.3.491
Bennink, S., Kiesow, M. J., Pradel, G. (2016). The Development of Malaria Parasites In the Mosquito Midgut. Cell Microbiol. 18, 905–918. doi: 10.1111/cmi.12604
Berry, A., Deymier, C., Sertorio, M., Witkowski, B., Benoit-Vical, F. (2009). Pfs 16 Pivotal Role in Plasmodium Falciparum Gametocytogenesis: A Potential Antiplasmodial Drug Target. Exp. Parasitol. 121, 189–192. doi: 10.1016/j.exppara.2008.10.010
Biddau, M., Sheiner, L. (2019). Targeting The Apicoplast in Malaria. Biochem. Soc. Trans. 47 (4), 973–983. doi: 10.1042/BST20170563
Blagborough, A. M., Sinden, R. E. (2009). Plasmodium Berghei Hap2 Induces Strong Malaria Transmission-Blocking Immunity in Vivo and In Vitro. Vaccine 27, 5187–5194. doi: 10.1016/j.vaccine.2009.06.069
Bruce, M. C., Carter, R. N., Nakamura, K., Aikawa, M., Carter, R. (1994). Cellular Location and Temporal Expression of the Plasmodium Falciparum Sexual Stage Antigen Pfs16. Mol. Biochem. Parasitol. 65, 11–22. doi: 10.1016/0166-6851(94)90111-2
Bushell, E., Gomes, A. R., Sanderson, T., Anar, B., Girling, G., Herd, C., et al. (2017). Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes. Cell 170, 260–272.e268. doi: 10.1016/j.cell.2017.06.030
Chookajorn, T. (2018). How To Combat Emerging Artemisinin Resistance: Lessons From “The Three Little Pigs”. PLoS Pathog. 14, e1006923. doi: 10.1371/journal.ppat.1006923
Conrad, M. D., Rosenthal, P. J. (2019). Antimalarial Drug Resistance in Africa: The Calm Before the Storm? Lancet Infect. Dis. 10 (4), e3380e351. doi: 10.1016/S1473-3099(19)30261-0
Dechering, K. J., Thompson, J., Dodemont, H. J., Eling, W., Konings, R. N. (1997). Developmentally Regulated Expression of pfs16, a Marker for Sexual Differentiation of the Human Malaria Parasite Plasmodium Falciparum. Mol. Biochem. Parasitol. 89, 235–244. doi: 10.1016/s0166-6851(97)00123-0
Deligianni, E., Andreadaki, M., Koutsouris, K., Siden-Kiamos, I. (2018). Sequence and Functional Divergence of Gametocyte-Specific Parasitophorous Vacuole Membrane Proteins in Plasmodium Parasites. Mol. Biochem. Parasitol. 220, 15–18. doi: 10.1016/j.molbiopara.2018.01.002
Dinglasan, R. R., Kalume, D. E., Kanzok, S. M., Ghosh, A. K., Muratova, O., Pandey, A., et al. (2007). Disruption of Plasmodium Falciparum Development by Antibodies Against a Conserved Mosquito Midgut Antigen. Proc. Natl. Acad. Sci. U. S. A. 104, 13461–13466. doi: 10.1073/pnas.0702239104
Dong, Y., Simoes, M. L., Marois, E., Dimopoulos, G. (2018). CRISPR/Cas9 -Mediated Gene Knockout of Anopheles Gambiae FREP1 Suppresses Malaria Parasite Infection. PLoS Pathog. 14, e1006898. doi: 10.1371/journal.ppat.1006898
Duffy, P. E., Gorres, J. P. (2020). Malaria Vaccines Since 2000: Progress, Priorities, Products. NPJ Vaccines 5, 48. doi: 10.1038/s41541-020-0196-3
Eksi, S., Williamson, K. C. (2011). Protein Targeting to the Parasitophorous Vacuole Membrane of Plasmodium Falciparum. Eukaryot. Cell 10, 744–752. doi: 10.1128/EC.00008-11
Gomes, A. R., Bushell, E., Schwach, F., Girling, G., Anar, B., Quail, M. A., et al. (2015). A Genome-Scale Vector Resource Enables High-Throughput Reverse Genetic Screening in a Malaria Parasite. Cell Host Microbe 17, 404–413. doi: 10.1016/j.chom.2015.01.014
Kariu, T., Ishino, T., Yano, K., Chinzei, Y., Yuda, M. (2006). CelTOS, a Novel Malarial Protein That Mediates Transmission to Mosquito and Vertebrate Hosts. Mol. Microbiol. 59, 1369–1379. doi: 10.1111/j.1365-2958.2005.05024.x
Khan, S. M., Kroeze, H., Franke-Fayard, B., Janse, C. J. (2013). Standardization in Generating and Reporting Genetically Modified Rodent Malaria Parasites: The RMgmDB Database Methods. Mol. Biol. 923, 139–150. doi: 10.1007/978-1-62703-026-7_9
Kongkasuriyachai, D., Fujioka, H., Kumar, N. (2004). Functional Analysis of Plasmodium Falciparum Parasitophorous Vacuole Membrane Protein (Pfs16) During Gametocytogenesis and Gametogenesis by Targeted Gene Disruption. Mol. Biochem. Parasit. 133, 275–285. doi: 10.1016/j.molbiopara.2003.10.014
Krogh, A., Larsson, B., von Heijne, G., Sonnhammer, E. L. (2001). Predicting Transmembrane Protein Topology With a Hidden Markov Model: Application to Complete Genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Lanfrancotti, A., Bertuccini, L., Silvestrini, F., Alano, P. (2007). Plasmodium Falciparum: mRNA Co-Expression and Protein Co-Localisation of Two Gene Products Upregulated in Early Gametocytes. Exp. Parasitol. 116, 497–503. doi: 10.1016/j.exppara.2007.01.021
Li, F. W., Templeton, T. J., Popov, V., Comer, J. E., Tsuboi, T., Torii, M., et al. (2004). Plasmodium Ookinete-Secreted Proteins Secreted Through a Common Micronemal Pathway are Targets of Blocking Malaria Transmission. J. Biol. Chem. 279, 26635–26644. doi: 10.1074/jbc.M401385200
Liu, F., Li, L., Zheng, W. Q., He, Y. W., Wang, Y. R., Zhu, X. T., et al. (2018). Characterization of Plasmodium Berghei Pbg37 as Both a Pre- and Postfertilization Antigen With Transmission-Blocking Potential. Infect. Immun. 86, e00785–17. doi: 10.1128/IAI.00785-17
Li, J., Wang, X., Zhang, G., Githure, J. I., Yan, G., James, A. A. (2013). Genome-Block Expression-Assisted Association Studies Discover Malaria Resistance Genes in Anopheles Gambiae. Proc. Natl. Acad. Sci. U. S. A. 110, 20675–20680. doi: 10.1073/pnas.1321024110
Lobo, C. A., Konings, R. N., Kumar, N. (1994). Expression of Early Gametocyte-Stage Antigens Pfg27 and Pfs16 in Synchronized Gametocytes and non-Gametocyte Producing Clones of Plasmodium Falciparum. Mol. Biochem. Parasitol. 68, 151–154. doi: 10.1016/0166-6851(94)00155-3
Lopez-Barragan, M. J., Lemieux, J., Quinones, M., Williamson, K. C., Molina-Cruz, A., Cui, K., et al. (2011). Directional Gene Expression and Antisense Transcripts in Sexual and Asexual Stages of Plasmodium Falciparum. BMC Genomics 12, 587. doi: 10.1186/1471-2164-12-587
MacDonald, N. J., Nguyen, V., Shimp, R., Reiter, K., Herrera, R., Burkhardt, M., et al. (2016). Structural and Immunological Characterization of Recombinant 6-Cysteine Domains of the Plasmodium Falciparum Sexual Stage Protein Pfs230. J. Biol. Chem. 291, 19913–19922. doi: 10.1074/jbc.M116.732305
Mathias, D. K., Pastrana-Mena, R., Ranucci, E., Tao, D., Ferruti, P., Ortega, C., et al. (2013). A Small Molecule Glycosaminoglycan Mimetic Blocks Plasmodium Invasion of the Mosquito Midgut. PLoS Pathog. 9, e1003757. doi: 10.1371/journal.ppat.1003757
Moelans, I. I., Cohen, J., Marchand, M., Molitor, C., de Wilde, P., van Pelt, J. F., et al. (1995). Induction of Plasmodium Falciparum Sporozoite-Neutralizing Antibodies Upon Vaccination With Recombinant Pfs16 Vaccinia Virus and/or Recombinant Pfs16 Protein Produced in Yeast. Mol. Biochem. Parasitol. 72, 179–192. doi: 10.1016/0166-6851(95)00072-9
Moelans, I. I., Klaassen, C. H., Kaslow, D. C., Konings, R. N., Schoenmakers, J. G. (1991). Minimal Variation in Pfs16, a Novel Protein Located in the Membrane of Gametes and Sporozoites of Plasmodium Falciparum. Mol. Biochem. Parasitol. 46, 311–313. doi: 10.1016/0166-6851(91)90056-c
Molina-Cruz, A., Canepa, G. E., Alves, E. S. T. L., Williams, A. E., Nagyal, S., Yenkoidiok-Douti, L., et al. (2020). Plasmodium Falciparum Evades Immunity of Anopheline Mosquitoes by Interacting With a Pfs47 Midgut Receptor. Proc. Natl. Acad. Sci. U. S. A. 117, 2597–2605. doi: 10.1073/pnas.1917042117
Niu, G., Annamalai, T., Wang, X., Li, S., Munga, S., Niu, G. M., et al. (2020a). A Diverse Global Fungal Library for Drug Discovery. PeerJ 8, e10392. doi: 10.7717/peerj.10392
Niu, G., Franca, C., Zhang, G., Roobsoong, W., Nguitragool, W., Wang, X., et al. (2017). The Fibrinogen-Like Domain of FREP1 Protein Is a Broad-Spectrum Malaria Transmission-Blocking Vaccine Antigen. J. Biol. Chem. 292, 11960–11969. doi: 10.1074/jbc.M116.773564
Niu, G., Hao, Y., Wang, X., Gao, J. M., Li, J. (2020b). Fungal Metabolite Asperaculane B Inhibits Malaria Infection and Transmission. Molecules 25, 3018. doi: 10.3390/molecules25133018
Niu, G., Wang, X., Hao, Y., Kandel, S., Niu, G. M., Raptis, R. G., et al. (2021). A Novel Fungal Metabolite Inhibits Plasmodium Falciparum Transmission and Infection. Parasit. Vectors 14, 177. doi: 10.1186/s13071-021-04677-7
Niu, G., Wang, B., Zhang, G., King, J. B., Cichewicz, R. H., Li, J. (2015). Targeting Mosquito FREP1 With a Fungal Metabolite Blocks Malaria Transmission. Sci. Rep. 5, 14694. doi: 10.1038/srep14694
Pelle, K. G., Oh, K., Buchholz, K., Narasimhan, V., Joice, R., Milner, D. A., et al. (2015). Transcriptional Profiling Defines Dynamics of Parasite Tissue Sequestration During Malaria Infection. Genome Med. 7, 19. doi: 10.1186/s13073-015-0133-7
Pritsch, M., Ben-Khaled, N., Chaloupka, M., Kobold, S., Berens-Riha, N., Peter, A., et al. (2016). Comparison of Intranasal Outer Membrane Vesicles with Cholera Toxin and Injected MF59C.1 as Adjuvants for Malaria Transmission Blocking Antigens AnAPN1 and Pfs48/45. J. Immunol. Res., 2016, 3576028. doi: 10.1155/2016/3576028
Rts SCTP (2014). Efficacy and Safety of the RTS,S/AS01 Malaria Vaccine During 18 Months After Vaccination: A Phase 3 Randomized, Controlled Trial in Children and Young Infants at 11 African Sites. PLoS Med. 11, e1001685. doi: 10.1371/journal.pmed.1001685
Stone, W. J. R., Campo, J. J., Ouedraogo, A. L., Meerstein-Kessel, L., Morlais, I., Da, D., et al. (2018). Unravelling the Immune Signature of Plasmodium Falciparum Transmission-Reducing Immunity. Nat. Commun. 9, 558. doi: 10.1038/s41467-017-02646-2
Swearingen, K. E., Lindner, S. E. (2018). Plasmodium Parasites Viewed Through Proteomics. Trends Parasitol. 34, 945–960. doi: 10.1016/j.pt.2018.08.003
Talman, A. M., Lacroix, C., Marques, S. R., Blagborough, A. M., Carzaniga, R., Menard, R., et al. (2011). PbGEST Mediates Malaria Transmission to Both Mosquito and Vertebrate Host. Mol. Microbiol. 82, 462–474. doi: 10.1111/j.1365-2958.2011.07823.x
Young, J. A., Fivelman, Q. L., Blair, P. L., de la Vega, P., Le Roch, K. G., Zhou, Y., et al. (2005). The Plasmodium Falciparum Sexual Development Transcriptome: A Microarray Analysis Using Ontology-Based Pattern Identification. Mol. Biochem. Parasitol. 143, 67–79. doi: 10.1016/j.molbiopara.2005.05.007
Zhang, G., Niu, G., Franca, C. M., Dong, Y., Wang, X., Butler, N. S., et al. (2015). Anopheles Midgut Frep1 Mediates Plasmodium Invasion. J. Biol. Chem. 290, 16490–16501. doi: 10.1074/jbc.M114.623165
Keywords: Plasmodium, malaria transmission, sexual stage, parasite-mosquito interaction, Pfs16, mosquito midgut invasion
Citation: Niu G, Cui Y, Wang X, Keleta Y and Li J (2021) Studies of the Parasite-Midgut Interaction Reveal Plasmodium Proteins Important for Malaria Transmission to Mosquitoes. Front. Cell. Infect. Microbiol. 11:654216. doi: 10.3389/fcimb.2021.654216
Received: 15 January 2021; Accepted: 10 May 2021;
Published: 28 June 2021.
Edited by:
Luiz Gustavo Gardinassi, Instituto de Patologia Tropical e Saúde Pública, Universidade Federal de Goiás (IPTSP – UFG), BrazilReviewed by:
Nazzy Pakpour, California State University, East Bay, United StatesCopyright © 2021 Niu, Cui, Wang, Keleta and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Li, bGlqQGZpdS5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.