
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 18 March 2021
Sec. Clinical Microbiology
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.646991
This article is part of the Research Topic Advances in Bacterial Quorum Sensing System and Quenching View all 6 articles
 Sujatha Subramoni1
Sujatha Subramoni1 Muhammad Zulfadhly Bin Mohammad Muzaki1,2
Muhammad Zulfadhly Bin Mohammad Muzaki1,2 Sean C. M. Booth1
Sean C. M. Booth1 Staffan Kjelleberg1,2
Staffan Kjelleberg1,2 Scott A. Rice1,2,3*
Scott A. Rice1,2,3*Bacterial biofilms are important medically, environmentally and industrially and there is a need to understand the processes that govern functional synergy and dynamics of species within biofilm communities. Here, we have used a model, mixed-species biofilm community comprised of Pseudomonas aeruginosa PAO1, Pseudomonas protegens Pf-5 and Klebsiella pneumoniae KP1. This biofilm community displays higher biomass and increased resilience to antimicrobial stress conditions such as sodium dodecyl sulfate and tobramycin, compared to monospecies biofilm populations. P. aeruginosa is present at low proportions in the community and yet, it plays a critical role in community function, suggesting it acts as a keystone species in this community. To determine the factors that regulate community composition, we focused on P. aeruginosa because of its pronounced impact on community structure and function. Specifically, we evaluated the role of the N-acyl homoserine lactone (AHL) dependent quorum sensing (QS) system of P. aeruginosa PAO1, which regulates group behaviors including biofilm formation and the production of effector molecules. We found that mixed species biofilms containing P. aeruginosa QS mutants had significantly altered proportions of K. pneumoniae and P. protegens populations compared to mixed species biofilms with the wild type P. aeruginosa. Similarly, inactivation of QS effector genes, e.g. rhlA and pvdR, also governed the relative species proportions. While the absence of QS did not alter the proportions of the two species in dual species biofilms of P. aeruginosa and K. pneumoniae, it resulted in significantly lower proportions of P. aeruginosa in dual species biofilms with P. protegens. These observations suggest that QS plays an important role in modulating community biofilm structure and physiology and affects interspecific interactions.
Bacterial biofilms are made up of surface-attached or suspended aggregates encased in self-produced extracellular polymeric substances (EPS). Biofilms in natural systems normally contain a rich mixture of species. In addition to the species level diversity, biofilms can also contain cells that are physiologically diverse due to various chemical gradients (e.g. oxygen, carbon source etc.) in different parts of the biofilm (exposed surface vs. interior) (Davey and O’Toole G, 2000). Biofilm communities have been shown to display a number of emergent properties, including enhanced stress and antimicrobial tolerance, increased biomass and reduced genetic variation relative to comparable monospecies biofilms (Flemming et al., 2016). Such effects are not observed for planktonic communities suggesting that these behaviors are biofilm specific. Biofilm formation may enable such responses due to the ability of species to optimize their spatial organization, e.g. to colocalize with collaborating species or to avoid competitors. The biofilm matrix plays a key role in these processes, enabling and stabilizing localization. The matrix further facilitates the establishment of gradients that result in for the formation of microniches that further allow for specialized growth, e.g. growth of anaerobes under otherwise oxygenic conditions (Flemming and Wingender, 2010). In this context, it is clear that microorganisms can modify their environment for optimal growth.
Interactions between individual species that contribute to the assembly and stability of a multi-species biofilm are still not well characterized. It is also clear that, just as observed in macroscale ecosystems, successionary processes are important for microbial communities. For example, in an oral biofilm, the initial colonizer tends to be Streptococcus sp., followed by the bridging species Fusobacterium nucleatum and finally by Porphyromonas gingivalis (Kommerein et al., 2018). Another example is the association of Aggregatibacter actinomycetemcomitans utilizing lactate produced by other oral bacteria such as Streptococcus gordonii to form biofilms of higher biomass (Brown and Whiteley, 2007). Thus, it is clear that bacteria undergo ecological processes that are similar to those described for higher organisms.
In this study, we investigated a 3-species biofilm community consisting of Pseudomonas aeruginosa PAO1, Pseudomonas protegens Pf-5 and Klebsiella pneumoniae KP1. These three species are reported to occur together as a community in the gut of Bombyx mori and in other environments along with various bacterial species (Chazal, 1995; Anand et al., 2010). Moreover, this mixed-species biofilm is highly reproducible in structure and viable cell counts under well-defined conditions, and therefore suitable for molecular studies into community dynamics (Jackson et al., 2001; Stoodley et al., 2001; Lee et al., 2014). Previously, we showed that this mixed-species community generated higher biomass and resistance to stress conditions, such as antibiotics (tobramycin), detergent sodium dodecyl sulfate (SDS) and predation (unpublished), than individual biofilms (Lee et al., 2014). Further, these effects were only observed when the community grew in a spatially organized biofilm and were not apparent for planktonic communities. Interestingly, P. aeruginosa, arguably one of the best studied model biofilm forming organisms, was the least abundant, between 1-10% of the entire community. When the percentage of P. aeruginosa was artificially increased by substituting the wild type strain with a more competitive, natural genetic variant, many of the emergent properties, e.g. stress resistance of the community, were lost. This would argue that, despite its low overall abundance, P. aeruginosa exerts a disproportionate effect on the rest of the community. Such species are frequently described as keystone organisms. We therefore wanted to explore the mechanisms by which this keystone organism controls community biofilm development.
One of the mechanisms used by P. aeruginosa PA01 to regulate biofilm formation, maturation and interspecies interactions is quorum sensing (QS). QS is a cell-cell communication system where bacteria produce and respond to signals to coordinate gene expression at the population level. There are four primary QS gene circuits reported for P. aeruginosa PAO1, the AHL-dependent Las and Rhl systems and the quinolone dependent PQS and IQS signaling systems (Lee et al., 2013; Lee and Zhang, 2015). The Las QS system, which produces and responds to 3-Oxododecanoyl)-L-homoserine lactone (3-oxo-C12-HSL), is the dominant system that can regulate both the Rhl and PQS systems (Gambello and Iglewski, 1991). The Rhl system produces and responds to N-butanoyl-L-homoserine lactone (C4-HSL) while the PQS system mainly produces and responds to 2-heptyl-3-hydroxy-4-quinolone (PQS) (Ochsner and Reiser, 1995; Pearson et al., 1995; Lee and Zhang, 2015). QS controls the expression of multiple effector molecules that have been shown to play important roles in surface colonization, nutrient acquisition and virulence for monospecies biofilms. Most studies of the role of QS in species interactions have focused on cross-species signaling. For example, it has been shown that signal production by Pantoea agglomerans or Erwinia toletana could rescue virulence in a signal deficient mutant of Pseudomonas savastanoi (Hosni et al., 2011). Similarly, it has been shown that P. aeruginosa and Burkholderia cepacia display signal cross talk in a mouse lung infection model (Riedel et al., 2001). While there are several reports of the role of QS in controlling expression of compounds with antimicrobial activity (Moons et al., 2006; Lesic et al., 2009; Abdel-Mawgoud et al., 2010), the overall role of QS as a regulator of interspecies interactions with mixed species biofilms is relatively poorly understood, especially in the context of QS regulated effectors.
Here, we report a role for P. aeruginosa PAO1 AHL QS in determining the composition of the three species biofilm community consisting also of P. protegens Pf-5 and K. pneumoniae KP1. We have generated isogenic, double-mutants of P. aeruginosa PAO1 in Las and Rhl systems and determined its effect on composition and structural properties of the 3-species biofilm.
Bacteria (Table 1) were grown either in M9 minimal medium (48 mM Na2HPO4; 22 mM KH2PO4; 9 mM NaCl; 19 mM NH4Cl; 2 mM MgSO4; 0.1 mM CaCl2; and 0.04% w/v glucose) supplemented with 0.2% w/v casamino acids (M9 Cas glu minimal medium), LB medium (Lysogeny Broth; 10 g l−1 NaCl; 10 g l−1 tryptone; 5 g l−1 yeast extract) or ABTC medium (A + B salts of Clark and Maaløe supplemented with 2.5 mg thiamine liter-1 and 10 mM citrate) (Clark and Maaløe, 1967). Antibiotics when required were used at the following concentrations unless otherwise stated: gentamicin 60 µg ml-1 (Gm; MP Biomedicals, Singapore), carbenicillin 200 µg ml−1 (Cb; MP Biomedicals, Singapore) and chloramphenicol 10 µg ml-1 (Cm; MP Biomedicals, Singapore). All P. aeruginosa, P. protegens and K. pneumoniae cultures were grown at room temperature (25 °C) for 24 h with shaking (200 rpm) and used for flow-cell cultivation. Overnight Escherichia coli cultures were grown at 37 °C with shaking (200 rpm). Overnight cultures of P. aeruginosa were also grown at 37 °C with shaking (200 rpm) for mutant generation and conjugation was carried out at 30 °C.
To generate P. aeruginosa PAO1 mutants, lasR, rhlI, and rhlR were amplified to include positions 500 bp and 1000 bp upstream and downstream, respectively, using Phusion polymerase (New England Biolabs, NEB, Ipswich, MA, USA) and cloned into the suicide vector pK18GT (Table 1). For lasI, 500 bp flanking the 5’ and 3’ ends of the gene were amplified in the same manner and cloned into the suicide vector pEX18ApGW (primers are indicated in Table S1). The PCR temperature cycling conditions for amplifying the upstream and downstream regions were as follows: initial denaturation for 30 s at 98°C, followed by 35 standard cycles: denaturation at 98°C for 10 s, primer annealing for 30 s at 69°C, and primer extension at 72°C for 15 s for 500 bp or 30 s for 1000 bp. Both cloning reactions were carried out using the Gibson Assembly Cloning kit (NEB, Beverly, MA, USA). The suicide plasmid constructs were verified by sequencing using vector specific primers pK18F and pK18R (Table S1) and mobilized into wild type P. aeruginosa PAO1 either by biparental or triparental mating using pRK600 as the helper plasmid. For biparental mating, the deletion constructs in the suicide plasmids were maintained in Escherichia coli S17-1 λpir. The single recombinants were selected on ABTC medium containing gentamicin (60 µg ml-1). Mutant colonies were selected on ABTC plates containing 15% sucrose and further checked for sensitivity to gentamicin, which would be indicative of double recombination. Finally, each putative mutant was tested by PCR to confirm the in-frame deletion by PCR amplification using primers specific to flanking region (Table S1) and Sanger sequencing (AITbiotech, Singapore). The lasIrhlI mutant was generated by deleting lasI in the rhlI mutant background while the lasRrhlR double mutant was generated by deleting lasR in the rhlR mutant background using the same strategy.
The eyfp gene was introduced as described previously (Choi and Schweizer, 2006). Briefly, overnight cultures were washed with 300 mM sucrose at room temperature to generate electrocompetent cells. These were simultaneously electroporated with 500 ng of pUC18T-mini-Tn7T-Gm-eyfp and pTNS1 and plated on LB agar containing gentamicin 100 µg ml-1. Positive colonies were verified by checking for the expression of eyfp by confocal laser scanning microscopy (CLSM) (LSM780, Carl Zeiss, Singapore) (excitation wavelength 514 nm and emission wavelength 527 nm).
It was necessary to excise the gentamicin marker to enable complementation with a gentamicin resistant vector. Therefore, the gentamicin cassette was removed from the eYFP tagged wild type and QS mutants lasRrhlR and lasIrhlI as previously described (Choi and Schweizer, 2006). Briefly, electrocompetent cells were prepared by washing with 300 mM sucrose solution and electroporated with 20-50 ng pFLP2 plasmid (Choi and Schweizer, 2006). The transformants were diluted 1000 fold and plated on LB+ Cb200 plates. Single colonies were checked for antibiotic susceptibility by patching on both LB+ Gm100 and LB+ Cb200 plates. Gentamicin sensitive colonies were checked for the presence of eYFP by CLSM (LSM780, Carl Zeiss, Singapore).
The full length lasR (1220 bp) and rhlR (1226 bp) genes, along with their endogenous promoter regions, were amplified from the P. aeruginosa wild type (Table 1). The PCR temperature cycling conditions for amplifying the full length genes were as follows: initial denaturation for 30 s at 98°C, followed by 35 standard cycles: denaturation at 98°C for 10 s, primer annealing for 30 s at 69°C, and primer extension at 72°C for 45 s. Both lasR and rhlR fragments were ligated together by overlap splice PCR to obtain a single PCR fragment of 2446 bp. This was done by setting up a PCR reaction containing both full length gene PCR products without any primer for 10 cycles (for overlap extension) followed by the standard PCR with primers RhlRHinF2 and LasREcoR2 for the next 30 cycles (Table S1). This was then cloned into pUCP22notI at the smaI site to generate pUCP22notI+R2. The presence of both genes was confirmed by sequencing using vector specific universal primers M13F and M13R (AITbiotech, Singapore). The gentamicin sensitive P. aeruginosa lasRrhlR mutant was then transformed with the pUCP22notI+R2 by electroporation (25 μF, 200 Ω and 2.5 kV cm−1) using a Gene Pulsar (Biorad, Hercules, CA, USA).
Swarming plates containing 0.5% w/v Bacto agar, 0.5% w/v peptone, 0.2% w/v yeast extract and 1% w/v glucose were prepared as previously described (Chang et al., 2014). Bacteria were diluted to 0.1 OD600 and 2 µl was inoculated at the center of the swarming agar plate. Plates were incubated for 16 h and the diameter of swarming zone was measured. Elastase activity was determined as previously described (Yin et al., 2015). Briefly, 100 µl of culture supernatant was added to 900 µl of Elastin-Congo Red (ECR) buffer (100 mM Tris, 1 mm CaCl2, pH 7.5) containing 20 mg ECR. This was then incubated overnight at 37˚C with shaking. The samples were then centrifuged (4000 g for 10 min at 4˚C) to remove insoluble ECR. The absorbance (OD495) of the supernatant was determined and sterile medium was included as a negative control. The change in absorbance was calculated to determine the change in elastase activity. Protease activity was determined by inoculating strains onto LB agar plates containing 1% skimmed milk. After incubation at 37˚C overnight, the diameter of clearance zones around the colonies, corresponding the protease activity was measured.
Flow-cell biofilms were established as previously described (Lee et al., 2014). Briefly, biofilms were cultivated in 3 channel flow cells having channel dimensions 40 mm x 4 mm x 1 mm. The flow cells were supplied with M9 cas glucose at the rate of 9 ml h-1 per channel. Each channel was injected with bacterial suspensions adjusted to approximately 1x108 cells ml-1. Mixed species biofilms were established by inoculating mixed cultures of P. aeruginosa PAO1, P. protegens Pf-5 and K. pneumoniae in the ratio of 5:5:1, respectively. Biofilms were imaged by CLSM (LSM780, Carl Zeiss, Singapore) as described (Lee et al., 2014). For each flow cell channel, 5 image stacks were acquired from the center of the channel at a distance of 5-10 mm from inlet to give a total of 15 image stacks per strain/genotype of mixed species biofilm. Three independent, replicate flow cells evaluated for each strain/combination. The image stacks were quantified using IMARIS (Bitplane AG, Belfast, UK). Statistical analyses were carried out by GraphPad Prism version 9 for windows, GraphPd software (La Jolla, California, USA, www.graphpad.com). Biofilm structure analysis was carried out with the COMSTAT 2.1 image analysis software package with default setting (Heydorn et al., 2000a; Heydorn et al., 2000b).
The growth of strains in M9 medium supplemented with casamino acids and glucose was monitored by inoculating in 96 well plates at an initial OD600 of 0.01 and incubating at room temperature for 24 h. Batch biofilm experiments were carried out as described previously (Schleheck et al., 2009). Briefly, overnight cultures of P. aeruginosa mutants or the wild type grown in M9 supplemented with casamino acids and glucose were adjusted to an OD600 of 0.01 and 1 ml of this suspension was distributed into wells of a 24 well plate. Biofilm attached to the wells of the plate was quantified by Crystal Violet (CV) assay. Briefly, the wells were washed once with 1.5 ml M9 medium to remove loosely attached cells. Staining was carried out with 1.75 ml CV solution (0.1% w/v). Unbound CV was removed by 3 washes with 2 ml M9 medium. Finally, 2 ml of absolute ethanol was used to extract bound CV by incubating on a rotary shaker 100 rpm for 10 min at RT after which time, the OD570 was determined for 200 µl of the ethanol extract using Tecan M200 Infinite microplate reader (Tecan Group Ltd., Männedorf, Switzerland).
In vitro competition experiments between P. aeruginosa strains and P. protegens were carried out in M9 cas glucose medium. Overnight cultures of P. aeruginosa QS regulated mutants and P. protegens Pf-5 were diluted 100 fold and 30 fold, respectively. The different dilutions were used to ensure similar CFUs (1X106 cfu ml-1) were present at the start of the experiment, based on comparisons of CFUs and OD (data not shown). Equal volumes of each of the P. aeruginosa mutants and P. protegens were mixed and incubated with shaking at 200 rpm at RT. Bacteria were enumerated at 0 and 24 h by dilution drop plating in duplicate. One set of plates at 37˚C allowed growth of P. aeruginosa strains, whereas incubation of duplicate plates at RT allowed growth of P. protegens. Bacteria were differentiated based on colony morphology. The fitness of each species was calculated as follows: Log[Fold change of mutant/fold change of wild type].
Cultures of P. aeruginosa strains grown at room temperature for 24 h were adjusted to OD600 = 0.1. A 500 μl aliquot of these bacterial suspensions were plated on LB agar plates. P. aeruginosa bacterial lawns were incubated for 2 h at room temperature. Cultures of P. protegens Pf-5 or K. pneumoniae KP-1 grown at room temperature for 24 h were adjusted to OD600 = 2 and 2 μl of each of these suspensions were spotted on the P. aeruginosa lawns. Plates were incubated for 2 to 3 d at room temperature. Growth inhibition zones were observed after 36-48 h. P. aeruginosa strains were screened for differences in the diameter of the growth inhibition zone surrounding the P. protegens Pf-5 or K. pneumoniae KP-1 colony in the middle on LB agar.
To determine the role of P. aeruginosa AHL quorum sensing (QS) in the 3 species biofilm community, a comparison of wild type P. aeruginosa relative to the QS mutants when present as part of the mixed species biofilms was carried out after 3 and 5 d (Figure 1). At day 3, the biovolumes per unit area of the mutant mixed-species biofilms (16.0 ± 3.0 µm3 µm-2 for lasIrhlI mixed-species and 16 ± 2.6 µm3 µm-2 for lasRrhlR mixed-species) were similar to when the wild type P. aeruginosa was present in the community. This trend was similar on day 5 and the wild type mixed species biofilm had biovolumes that were similar to the QS mutants (data not shown). While the overall biovolumes were similar, the proportions of K. pneumoniae and P. protegens were significantly different for the QS mutant mixed-species biofilms compared to when the wild type P. aeruginosa was present in the biofilm community (Figures 1A, B). Quantitative image analysis of the wild type mixed species revealed that it was composed of 74.8 ± 11.7% and 76.7 ± 6.9% K. pneumoniae, 6.3 ± 5% and 3.1 ± 2.8% P. aeruginosa and 18.8 ± 10.1% and 20.1 ± 7.9% P. protegens at days 3 and 5, respectively. In contrast, when the signal or receptor mutants of P. aeruginosa were present, the relative proportions of K. pneumoniae decreased, with a concomitant increase in P. protegens and a slight increase in P. aeruginosa for both time points. The community with the lasIrhlI mutant had lower proportions of K. pneumoniae and higher proportions P. protegens compared to the wild type community (Figures 1A, B). After 3 d, biofilms formed with the lasIrhlI mutant had 28 ± 13.5% K. pneumoniae, 17.6 ± 11.5% P. aeruginosa and 54.2 ± 21.8% P. protegens and at 5 d, these were 33.7 ± 13.5% K. pneumoniae, 5.5 ± 7.6% P. aeruginosa and 60.7 ± 20.7% P. protegens. The community with the lasRrhlR mutant also had similar changes in proportions of K. pneumoniae and P. protegens (Figures 1A, B). After 3 d, biofilms formed with the lasRrhlR mutant had 17.3 ± 15.7% K. pneumoniae, 9.7 ± 7.2% P. aeruginosa and 72.8 ± 20.5% P. protegens and at 5 d, these were 38.1 ± 11.6% K. pneumoniae, 2.9 ± 3.3% P. aeruginosa and 58.8 ± 12.2% P. protegens. These results show that the proportions of K. pneumoniae and P. protegens were affected by a functional QS system of P. aeruginosa, while there was no significant change in the relative abundance of P. aeruginosa.
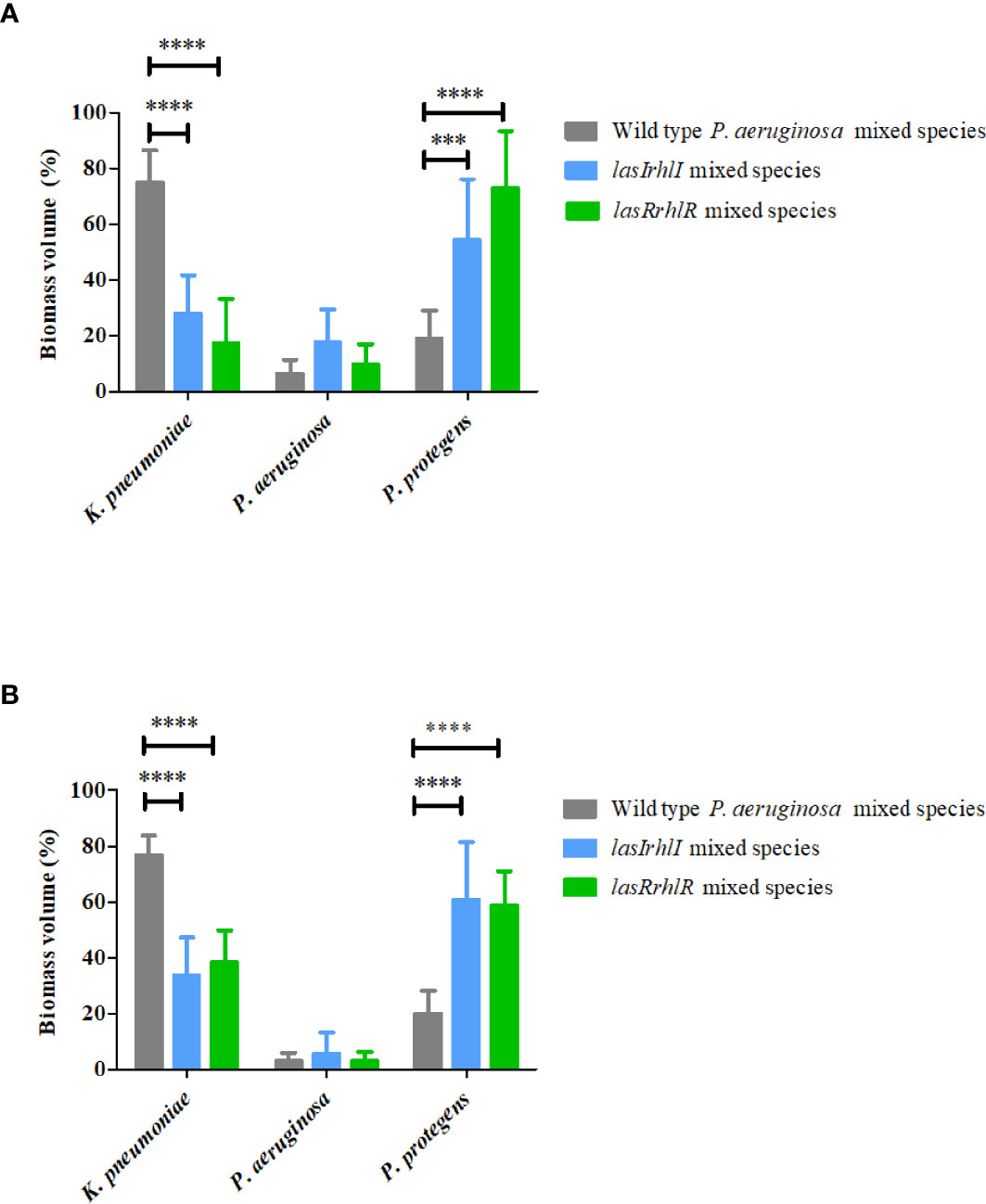
Figure 1 Proportions of the mixed species biofilm. The proportions of the three species in the mixed species biofilm containing either the P. aeruginosa wild type or the QS mutants (lasIrhlI or lasrRrhlR) after (A) 3 days and (B) 5 days of cultivation. Bar charts depict the percentages of K. pneumoniae, P. aeruginosa, and P. protegens in the wild type or QS mutant mixed species biofilm. The proportions were derived by cultivating biofilms under flow cell conditions followed by quantitative image analysis of two independent biological replicates. Error bars represent standard deviations (n = 2). *** and **** denote significant difference by two-way ANOVA followed by Tukey’s post-test (P < 0.001 and P < 0.0001).
Analyses of the images of wild type and QS mutant mixed species biofilms was also performed for five parameters, microcolony size, thickness, roughness coefficient, surface to biovolume ratio and substratum coverage (Table 2). At both 3 and 5 d of biofilm growth, the P. aeruginosa wild type mixed species community had a larger microcolony size for K. pneumoniae when compared to K. pneumoniae in the lasIrhlI and lasRrhlR mixed species biofilms. Similarly, the P. aeruginosa wild type mixed species had thicker K. pneumoniae and thinner P. protegens biofilm structures when compared to K. pneumoniae and P. protegens in the lasIrhlI and lasRrhlR mixed species biofilms. P. aeruginosa thickness was not significantly different between these biofilms. The roughness coefficient provides a measure of how much the thickness of the biofilm varies. Both at 3 and 5 days of biofilm growth, the P. aeruginosa wild type mixed species community had a less heterogeneous K. pneumoniae and more heterogeneous P. protegens biofilm structure when compared to K. pneumoniae and P. protegens in the lasIrhlI and lasRrhlR mixed species biofilms. The surface to biovolume ratio values supports the thickness measurements. Finally, both at 3 and 5 d of biofilm growth, the P. aeruginosa wild type mixed species biofilms had higher substratum coverage for K. pneumoniae and lower substratum coverage for P. protegens biofilm when compared to K. pneumoniae and P. protegens in the lasIrhlI and lasRrhlR mixed species biofilms.
We hypothesized that the effect of P. aeruginosa QS on community composition could be due to specific interactions with the other two species in the community. Therefore, dual-species biofilms of P. aeruginosa wild type with P. protegens and P. aeruginosa wild type with K. pneumoniae were cultivated separately and compared to biofilms where the wild type P. aeruginosa was substituted with the isogenic QS mutants (Figure 2). After 3 d, the proportions were 52.5 ± 17.8% wild type P. aeruginosa and 47.4 ± 17.8% K. pneumoniae. When either of the QS mutants were introduced, the proportions of K. pneumoniae increased slightly, but not significantly, with a similar reduction in the proportion of the mutants. When cultivated for 5 d, we again saw only a slight increase in K. pneumoniae when cultivated with the QS mutants compared to when the wild type was present. This shows that QS does not drive the interactions between P. aeruginosa and K. pneumoniae, as defined by their relative proportions, between these two species under these conditions (Figures 2A, B).
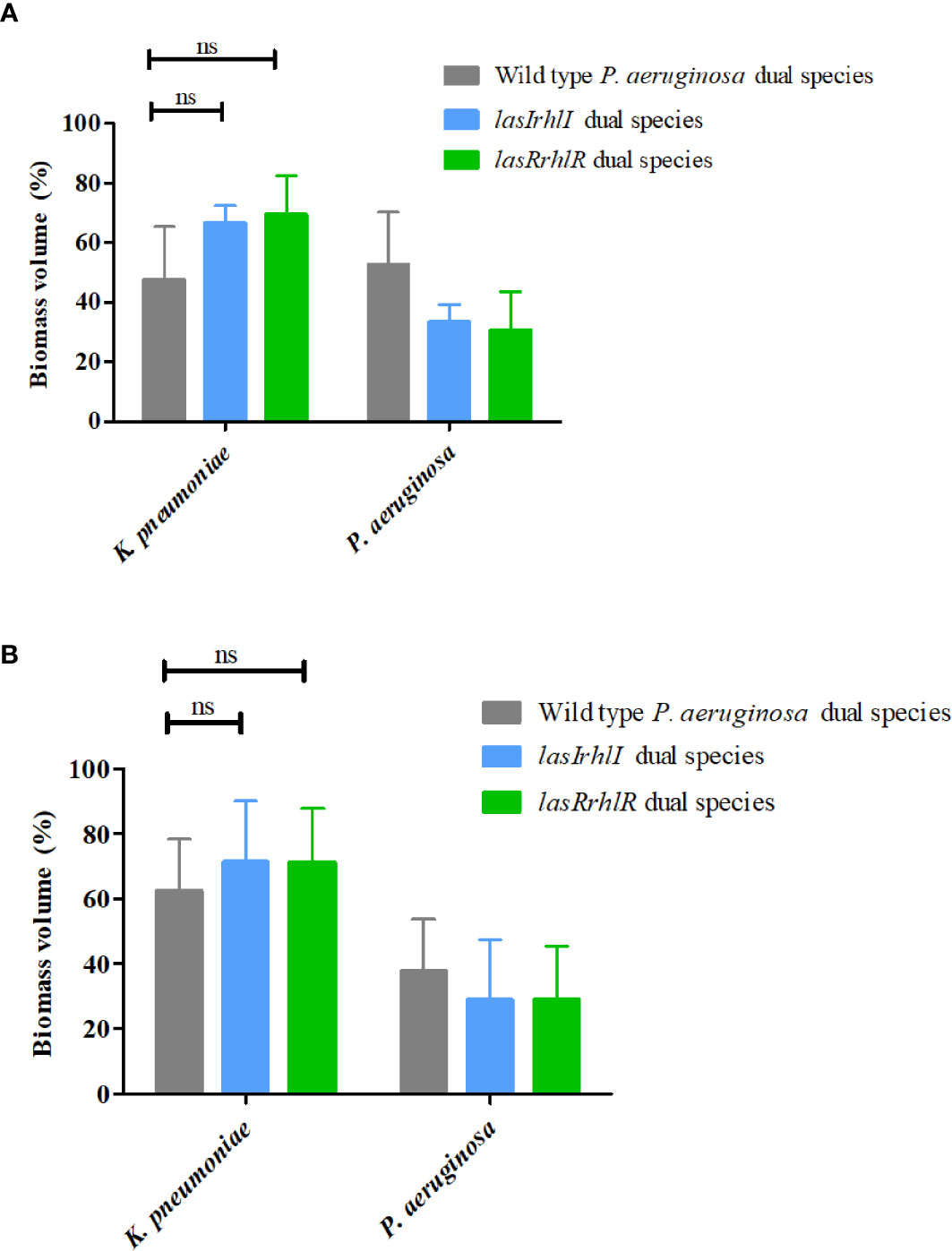
Figure 2 Proportions of K. pneumoniae-P. aeruginosa dual-species biofilm. The proportions of the two species in the dual species biofilm containing either the P. aeruginosa wild type or the QS mutants (lasIrhlI or lasrRrhlR) after (A) 3 days and (B) 5 days of cultivation. Bar charts depict the percentages of K. pneumoniae and P. aeruginosa in the wild type or QS mutant containing dual species biofilms. The proportions were derived by cultivating biofilms under flow cell conditions followed by quantitative image analysis of two independent biological replicates. Error bars represent standard deviations (n = 2). ns denotes there were no significant differences.
In contrast, deletion of QS in P. aeruginosa led to significant changes in the composition of dual species biofilms in the presence of P. protegens. When the wild type P. aeruginosa was present, it accounted for approximately 40% of the biofilm biomass with the remaining ~60% contributed by P. protegens at 3 and 4 d (Figures 3A, B). In contrast, on day 3 the proportions of the lasRrhlR and lasIrhlI mutants in the dual-species biofilms were significantly lower (13.5 ± 4.4% and 16.9 ± 4.3%, respectively), with the remaining 83-85% contributed by P. protegens. By day 4, the QS mutants were present at only 4-8%. Collectively, these results show that P. aeruginosa and P. protegens exhibit a competitive interaction that is controlled by the P. aeruginosa QS system.
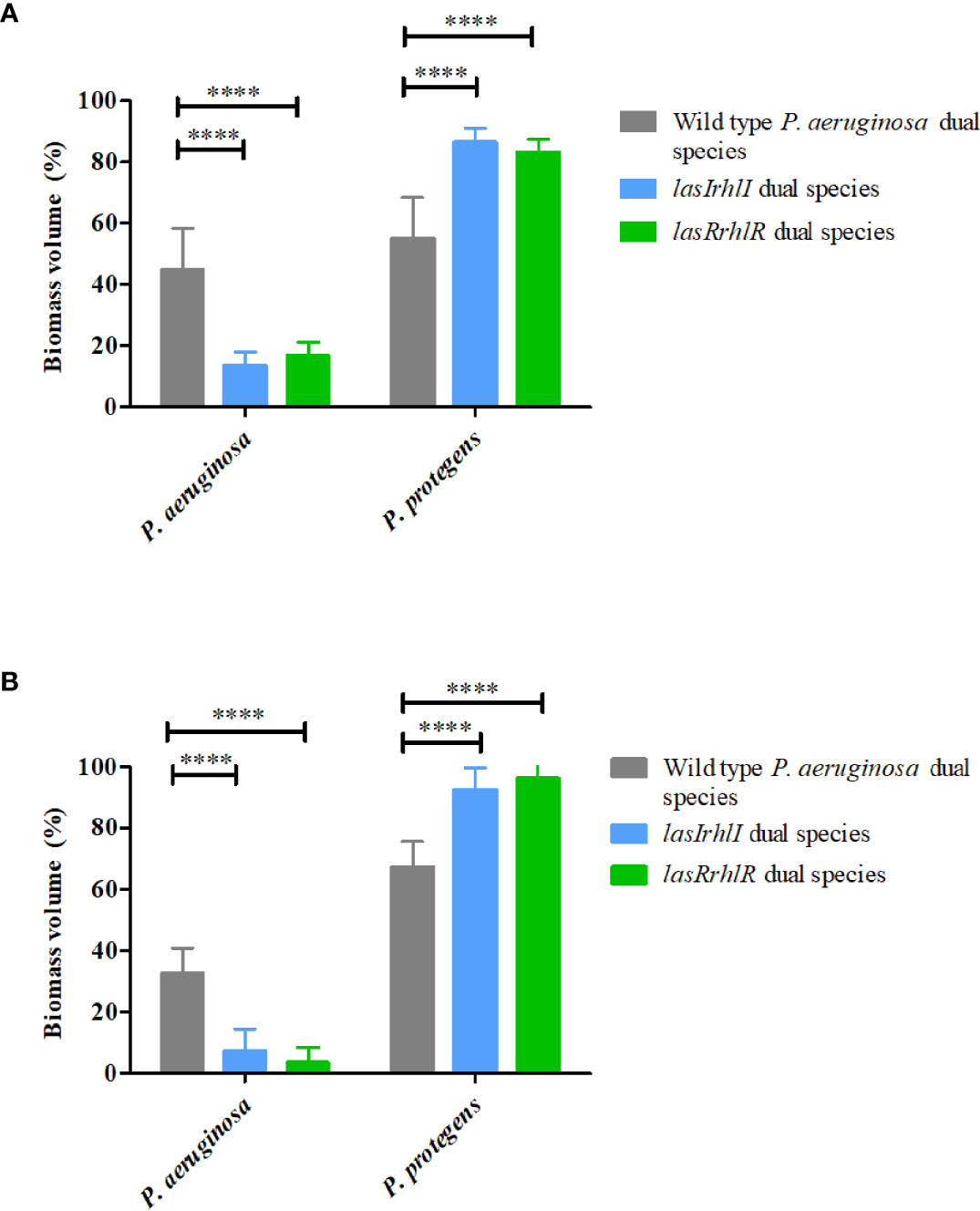
Figure 3 Proportions of P. protegens–P. aeruginosa dual-species biofilm. The proportions of the two species in the dual species biofilm containing either the P. aeruginosa wild type or the QS mutants (lasIrhlI or lasrRrhlR) after (A) 3 days and (B) 4 days of cultivation. Bar charts depict the percentages of P. protegens and P. aeruginosa in the wild type or QS mutant dual species biofilm. The proportions were derived by cultivating biofilms under flow cell conditions followed by quantitative image analysis of two independent biological replicates. Error bars represent standard deviations (n =2). **** denote significant difference by two-way ANOVA followed by Tukey’s post-test (P < 0.0001).
Complementation of the QS mutants showed full restoration of phenotypes such as swarming, elastase activity and protease activity to wild type levels (Figures S1A–C). Dual species biofilms were also formed with lasRrhlR mutant carrying the complementation vector, pUC22notI+R2, for P. aeruginosa-P. protegens dual species biofilms (Figure S2). After 3 d, the dual species biofilm containing the complemented lasRrhlR mutant had biovolumes of 24.4 ± 17.9% lasRrhlR/pUCP22notI+R2 and 75.5 ± 17.3% P. protegens, respectively. Therefore, the complemented lasRrhlR mutant formed dual species biofilms that were not significantly different from, but slightly lower than, when the wild type was present with P. protegens.
QS regulated genes in P. aeruginosa include those coding for secondary metabolites (phenazines, PQS), virulence factors, nutrient acquisition (elastase, protease, pyoverdine) and toxins (HCN, exotoxin) among others. It is possible that one or more of these provide fitness advantage to P. aeruginosa wild type in the presence of P. protegens. We therefore tested a range of mutants for their impact on monospecies biofilm formation and growth. These mutants were chosen as they represent some of the well-characterized genes of the Las, Rhl and Pqs regulon. Most of the mutants tested, including aprA, hcnB, lasB, phzA1, phzM, pqsA, pqsE, pvdR, rhlA and sdsA1, showed no significant difference in growth or monospecies biofilm formation (Figures S3 and S4). Only aprD showed a significant reduction in monospecies biofilm formation. There was also no difference in the fitness of these mutants relative to the wild type P. aeruginosa when grown in the presence of P. protegens during planktonic growth. (Figure S5).
In order to pinpoint which QS effector was involved in competitive interaction of P. aeruginosa with P. protegens and to screen a larger number of mutants, we carried out a growth inhibition assay on agar plates. Based on this screening, the lasIrhlI and lasRrhlR mutants showed an altered inhibition zone in the presence of P. protegens when compared to P. aeruginosa wild type. This sensitivity of the lasRrhlR mutant was abolished by providing the wild type copies of both genes in trans in the complemented strain (Figure 4). No such zone of inhibition was found when these mutants were tested in the presence of K. pneumoniae (Figure S6). Furthermore, our results also revealed that none of the mutants of QS effectors tested showed this altered inhibition zone in the presence of P. protegens (Figure S7).
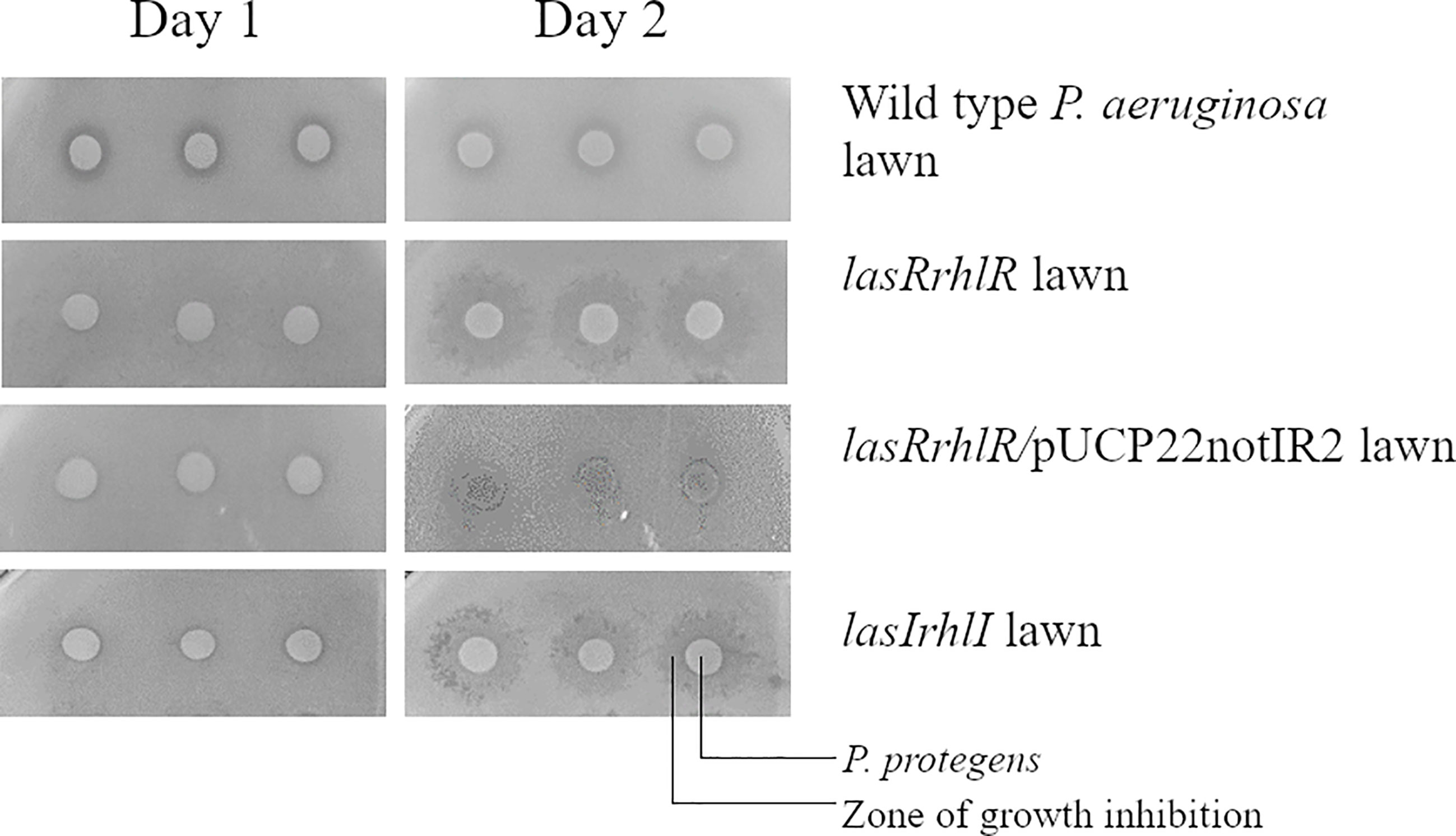
Figure 4 Pseudomonas aeruginosa mutants affected in competition with P. protegens. P. protegens was spotted on a lawn of P. aeruginosa wild type, QS mutants or complemented strains on LB agar plate. This panel shows the plate phenotype on day 1 and day 2 and the inhibition zone is observed from the second day.
Since flow-cell conditions used for cultivation of biofilms are very different from agar plates, we decided to evaluate the roles of a subset of these QS effectors in dual and 3-species biofilms. First, we tested mutants of various effector genes belonging to the Las, Rhl and Pqs systems in dual species biofilms with P. protegens. For the P. aeruginosa wild type and lasRrhlR communities the proportions of P. aeruginosa were 64.8 ± 17.7% and 8.4 ± 8.8%, respectively, whereas the proportions of P. protegens were 35.1 ± 17.3% and 91.5 ± 8.8%, respectively as expected. Although, the proportions of P. aeruginosa, 20.6 ± 10.5% and 13.6 ± 14.8%, respectively, were lower for the communities containing mutants of pvdR and rhlA compared to dual species biofilms with P. aeruginosa wild type, only rhlA was significantly different (Figure 5). The proportions of P. protegens was also much higher (79.3 ± 10.5% and 86.3 ± 14.8%) for these two mutant communities compared to wild type community. The proportions of P. aeruginosa and P. protegens in hcnB, lasB and pqsA communities were not significantly different when compared to the wild type community.
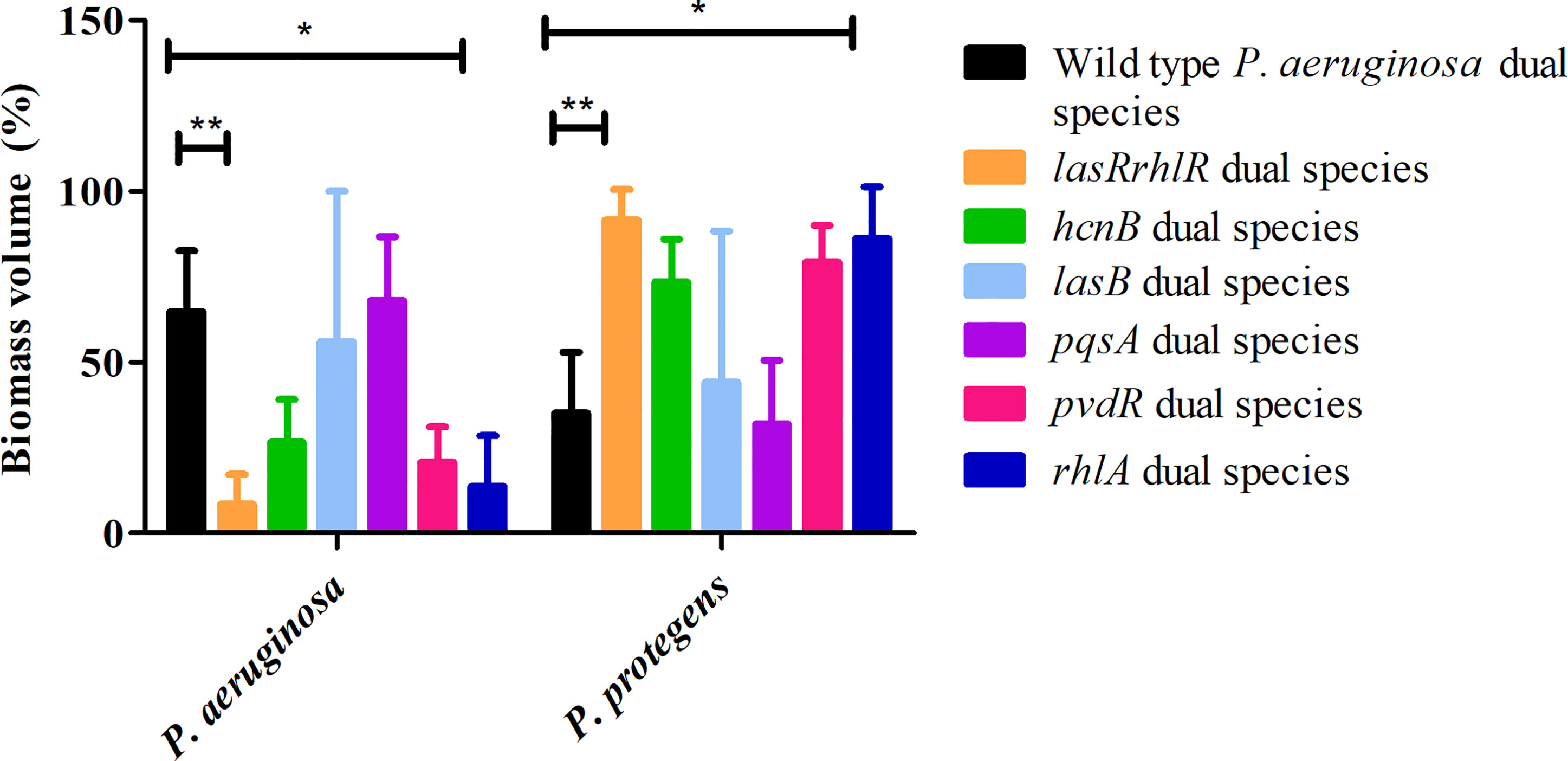
Figure 5 Proportions of P. protegens - P. aeruginosa dual species biofilm containing mutants of QS regulated effectors. The proportions of the two species in the mixed species biofilm containing either the P. aeruginosa wild type, the QS mutant lasRrhlR or the QS target mutants hcnB, lasB, pvdR, rhlA, or pqsA after 3 days of cultivation. Bar charts depict the percentages of P. aeruginosa and P. protegens in the wild type or the mutant dual species biofilms. The proportions were derived by cultivating biofilms under flow cell conditions followed by quantitative image analysis of two independent biological replicates. Error bars represent standard deviations (n = 2). * and ** denote significant difference by two-way ANOVA followed by Tukey’s post-test (P < 0.05; P < 0.01).
When tested in triple species biofilms, for the communities containing mutants of pvdR and rhlA, the proportions of K. pneumoniae, 39.1 ± 18.3% and 24.7 ± 17.4%, respectively, were lower compared to biofilms with the wild type P. aeruginosa but only rhlA was significantly different (Figure 6). The proportion of P. protegens was significantly different only for the pvdR mutant mixed community (47.6 ± 8.5%). For the lasRrhlR community the proportions of K. pneumoniae and P. protegens were 34.7 ± 15.5% and 48.8 ± 14.8%, respectively, as expected. Interestingly, the mixed species biofilm community formed with the rhlA mutant had a significantly higher proportion of P. aeruginosa (45.4 ± 27.1%) compared to the wild type. Furthermore, the mixed species biofilm community formed with the lasB mutant had a significantly higher proportion of K. pneumoniae (89.8 ± 4.5%), and significantly lower proportion of P. protegens (8 ± 2.9%), when compared to wild type P. aeruginosa containing biofilm community (70 ± 10.8% and 16.2 ± 11.3%, respectively) (Figure 6). The mixed species biofilm community formed with the hcnB and pqsA mutant did not differ significantly from wild type P. aeruginosa containing biofilm in its composition. These results suggest that QS regulated genes do not contribute equally to the community composition and that none have the same magnitude of effect as the lasrRrhlR or lasIrhlI mutants. Thus, the QS effectors may have a cumulative effect in determining community composition.
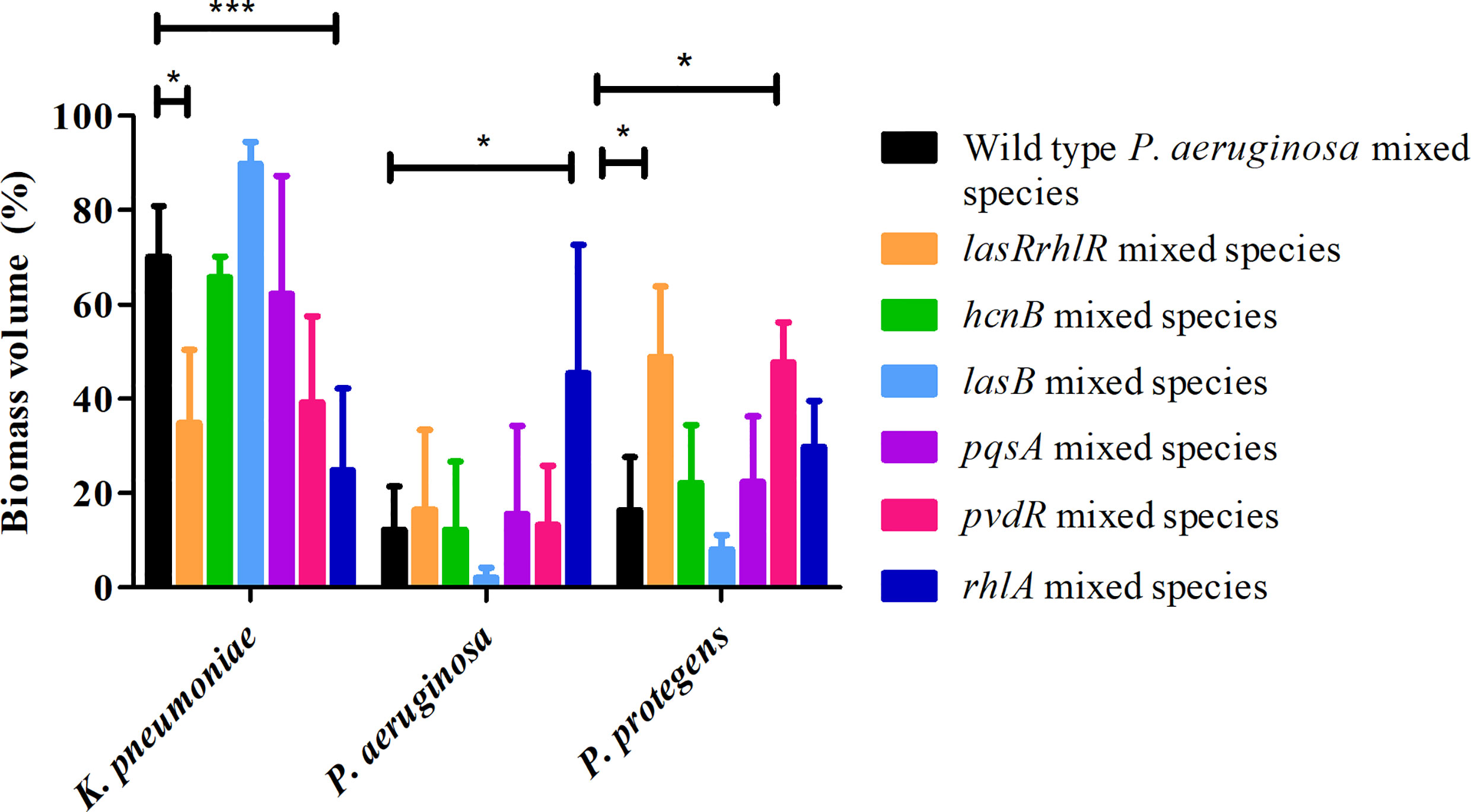
Figure 6 Proportions of mixed species biofilm containing mutants of QS regulated effectors. The proportions of the three species in the mixed species biofilm containing either the P. aeruginosa wild type, the QS mutant lasRrhlR or the QS target mutants hcnB, lasB, pvdR, rhlA, or pqsA after 3 days of cultivation. Bar charts depict the percentages of K. pneumoniae, P. aeruginosa and P. protegens in the wild type or the mutant mixed species biofilms. The proportions were derived by cultivating biofilms under flow cell conditions followed by quantitative image analysis of 2 independent biological replicates. Error bars represent standard deviations (n = 2). * and *** denote significant difference by two-way ANOVA followed by Tukey’s post-test (P < 0.05; P < 0.001).
In this work, we investigated the role of QS in interspecies interactions in a defined 3-species bacterial community during biofilm formation. We observed that the proportions of K. pneumoniae and P. protegens were dictated by the presence or absence of a functional P. aeruginosa QS system. We also show that there is no effect of deletion of the various QS genes on the relative fitness of the strains, further suggesting that it is the loss of QS function that drives the changes in community composition and structure. This effect was surprising as P. aeruginosa represents only 1-10% of the mature biofilm community, suggesting that even at a relatively low abundance, this species can significantly modulate community interactions. QS was shown to play a significant role in specifically modulating community composition between P. aeruginosa and P. protegens, with little to no effect on K. pneumoniae. We also showed that several downstream QS targets cumulatively mediate this competitive effect.
The complemented lasRrhlR mutant completely restored known QS regulated phenotypes, e.g. swarming, elastase activity and protease activity in in vitro assays. In addition, complementation also restored competitive advantage over P. protegens in growth assays on agar surface (Figure 4). While the relative abundances of the complemented strains were not significantly different from the wild type when grown in the mixed species biofilm, they were none the less slightly lower than for the wild type (Figure S2). The lower abundances achieved for the complemented strains might be due to higher and earlier expression of the QS genes from the multi-copy plasmids used for complementation. In addition to the amount of signal production, the level of signal receptor, LasR, production is known to be a critical factor in QS mediated gene expression linked to biofilm formation (Schuster et al., 2003; Rutherford and Bassler, 2012). This may in part be due to the expression of LasR and RhlR at late logarithmic and early stationary phase in planktonic cultures. Thus, given that the receptor genes were expressed on a multicopy plasmid, it is likely that the receptors were produced earlier in the complemented strain than normal. This could lead to the expression of QS regulated functions at a lower cell density by the complemented strain, which might consequently lead to its earlier detection by the competing bacterial species, P. protegens, likely leading to its removal and lower abundance. Therefore, optimal timing of expression of QS regulators LasR and RhlR might play an important role in the competition with P. protegens during early biofilm formation.
K. pneumoniae KP1 and P. protegens Pf-5 carry one and two luxR homologs, respectively, but they do not produce AHLs. It is possible that both K. pneumoniae and P. protegens in multispecies communities might sense and respond to foreign AHLs to regulate biofilm formation. This has previously been reported for multispecies biofilms formed by P. aeruginosa and Burkholderia cepacia in the lungs of CF patients (Van Delden and Iglewski, 1998; Huber et al., 2001; Riedel et al., 2001). Indeed, cross-species or cross-kingdom signal perception is the most commonly reported role of QS for communities. However, both of the P. aeruginosa lasRrhlR and lasIrhlI mutants show similar proportions in the 3-species community as do the individual QS effector mutants. This suggests that it is the P. aeruginosa QS regulon that mediates interaction with the other two species and not simply signal perception by those species through their LuxR homologs. However, it will be interesting to study whether AHLs directly influence biofilm formation by P. protegens and K. pneumoniae in future studies.
In dual species biofilms, there was no effect of QS on the relative abundance of either K. pneumoniae or P. aeruginosa (Figure 2) under the conditions used in this study. Also, the K. pneumoniae - P. protegens interaction appeared to be neutral, where both species were present in equal abundance (data not shown). However, QS provided a competitive advantage to P. aeruginosa when cultivated with P. protegens (Figure 3). Although there are several reports on the role of QS in laboratory dual species co-culture systems and it is predicted to influence more complex community dynamics, there is not much direct evidence for the latter, which has been shown here (Mazzola et al., 1992; Flemming and Wingender, 2010; Tan et al., 2014). Specifically, P. aeruginosa interaction with B. cepacia sp., A. tumefaciens, S. aureus and B. cenocepacia has been shown to be due to QS regulated antimicrobial production and AHL eavesdropping in planktonic cultures (Lewenza et al., 2002; An et al., 2006; Costello et al., 2014; Smalley et al., 2015). In Agrobacterium tumefaciens-P. aeruginosa dual species biofilms, the wild type P. aeruginosa completely blanketed A. tumefaciens and progressively excluded A. tumefaciens from the biofilm while a P. aeruginosa QS mutant did not. This was controlled by antimicrobial production and swarming (An et al., 2006). We propose that P. aeruginosa - P. protegens interactions in dual species biofilms is a two-way interaction involving functions from both bacteria which provide them competitive advantage and results in mutual exclusion. A number of QS regulated P. aeruginosa products such as rhamnolipids, hydrogen cyanide, pyocyanin and pyoverdine are known to be important for competition with other bacteria; these effects are likely to be multifactorial with some antimicrobials having a greater effect (Smalley et al., 2015). Similarly, P. protegens is also known to produce a cocktail of both antifungal and antibacterial secondary metabolites including 2,4-diacetylphloroglucinol, pyoluteorin, pyrrolnitrin, extracellular protease, hydrogen cyanide and siderophores pyochelin and pyoverdine (Paulsen et al., 2005). It has been proposed that the natural function of multi-drug resistance efflux pumps in P. aeruginosa could be to excrete a particular class of secondary metabolite (Poole, 1994). It is thus possible that QS regulated efflux systems also protects P. aeruginosa against one or more antimicrobials produced by P. protegens and future work will address these mechanisms.
Our results with QS and target mutants revealed that most of the mutants except aprD were proficient for biofilm formation in batch cultivation assays (Figure S4). Previous reports indicate that P. aeruginosa QS is required for biofilm formation. Specifically, the Las system has been shown to control progression from reversible to irreversible attachment whereas the Rhl system is responsible for biofilm maturation (Jaffar-Bandjee et al., 1995; Davies et al., 1998). However, in our experiments, monospecies biofilms, formed by lasIrhlI and lasRrhlR mutants under flow-cell conditions did not show any difference compared to P. aeruginosa wild type in terms of biovolume and thickness (Figure S8). Moreover, among the QS target mutants, pqsA, pqsE and rhlA have also shown to be defective for biofilm maturation (Yang et al., 2009; Darveau et al., 2012; Hajishengallis et al., 2012). The effects of Pqs system on biofilm structure was shown to be iron dependent. Although rhlA mutant has been shown to form flat biofilms lacking heterogeneity of wild type biofilms in the flow-cell system, in the same study it has been reported that in microtiter dish assays the rhlA mutant is more proficient at early colonization (up to 24 h) than is the wild type (Darveau et al., 2012). Active iron uptake has been shown to be important for P. aeruginosa biofilm architecture as pvdS and pvdA mutants form thin biofilms (Banin et al., 2005; Yang et al., 2009). However, pvdA and pvdE mutants form nearly normal biofilms in microtiter dish assays (Kang and Kirienko, 2017). Our experiments to test biofilm formation by QS target mutants was carried out under batch culture conditions. As described above, there are differences in the medium and carbon source/s, cultivation temperature and cultivation system between the previous studies with these mutants and the work we have presented here; this might account for the observed biofilm formation phenotypes. For example, Davies et al. (Davies et al., 1998) showed that the QS defect in biofilm formation by P. aeruginosa could be overcome by growing the mutant in complex (LB) rather than defined media.
In order to narrow down the processes involved in competition, we investigated interactions between P. aeruginosa and P. protegens in dual and 3-species biofilms where the wild type P. aeruginosa was substituted by mutants of several QS regulated genes, including rhlA and pvdR. rhlA mutant was significantly less competitive than wild type in dual species community with P. protegens (Figure 5). Interestingly, when wild type P. aeruginosa was substituted by the rhlA mutant in the 3-species community, there was a significant increase in the biomass of the rhlA mutant (Figure 6). Mutants in rhlA are defective for the production of the biosurfactant rhamnolipid which promotes motility and affects the architecture of monospecies P. aeruginosa biofilms (Davey et al., 2003). Rhamnolipids might affect both cell-cell and cell-surface interactions (Neu, 1996). In fact, in the wild type, when rhamnolipids are produced, they might prevent self-colonization as well as colonization by other species. This inhibition is lacking in the rhlA mutant community. Only pvdR showed trends of alteration of both K. pneumoniae and P. protegens similar to the QS mutants in 3-species biofilms, although in dual species pvdR community this difference was not significant. PvdR is involved pyoverdine transport and recycling in P. aeruginosa and its mutants accumulate pyoverdine in the periplasmic space and are unable to acquire ferric iron efficiently via pyoverdine pathway (Imperi et al., 2009). This suggests that optimal iron uptake and possibly use of heterologous siderophores might be important for maintaining the 3-species community composition as fluorescent Pseudomonads are known to produce multiple pyoverdines (Cornelis and Matthijs, 2002). Together, these results suggest that QS regulated effectors affect the community composition in different ways. Specifically, we found that reduction of K. pneumoniae biomass was not always linked to increase in P. protegens biomass in the 3-species biofilm. It is also interesting that QS did not alter the relationship between P. aeruginosa and K. pneumoniae despite the clear reduction in the latter when the QS mutants of P. aeruginosa are present in the three species community. Further work will be needed to elucidate how QS effectors operate in the three species community to control composition. Similarly, the mechanisms of competition mediated by secondary metabolites of P. protegens and their role in biofilms require further investigation and are beyond the scope of the present study.
The least abundant bacterium in this 3-species biofilm community, P. aeruginosa, determined the proportions of the other two species. This finding shows an effect of QS on non-clonal cells even when present in low abundance in a polymicrobial community. It has been similarly reported that Microbacterium oxydans, the least abundant species, determined the spatial organization of others in a four-species community (Liu et al., 2017). Additionally, P. gingivalis, which is present in relatively low abundance, is responsible for triggering changes in both amount and composition of oral commensal microbiota (Hajishengallis et al., 2011). These bacteria are examples of keystone species in a community (Hajishengallis and Lamont, 2016). Low abundant species in communities are often overlooked due to the limitation of omics approaches in delineating their role in these biofilms. Despite some of those limitations, it is none the less possible to identify potential keystone species in complex communities, such as wastewater systems, through network analysis. Indeed, such network analyses have been extended to functional genes within sludge communities to identify those genes, termed keystone genes, that play essential, linking roles, between community members and presumably sludge function (Roume et al., 2015). The role of low-abundance keystone pathogens that promote the formation and stabilization of dysbiotic and disease-inducing communities in the human gut have been reported (Hajishengallis et al., 2012). There are also keystone organisms that act as stabilizers that kill opportunistic pathogens but not the indigenous gut bacteria (Stecher et al., 2013). For example, it has been shown that P. aeruginosa senses peptidoglycan and diffusible signals produced by other bacteria to modify community composition and persistence in P. aeruginosa-dominated disease conditions (Twomey et al., 2012; Korgaonkar et al., 2013). The role played by low abundance bacteria producing these signals in shaping community-level changes would be consistent with their position as keystone bacteria. Our findings clearly demonstrate that a bacterium does not need to be the most abundant organism in the biofilm to be important for controlling the community structure and composition of bacterial species. In this way, P. aeruginosa serves as a keystone species in this biofilm community. The results presented here also demonstrate how a genetically amenable system can be well suited to test hypotheses about the roles and mechanisms of keystone species in mixed species communities.
Of the three species used in this study, P. aeruginosa and K. pneumoniae are opportunistic human pathogens known to occur together in human-associated infections such as respiratory tract, chronic wound and urinary tract infections (Harmsen et al., 2010; Rezaei et al., 2011; Magill et al., 2014). Both bacteria adapt to airways and other biotic surfaces through biofilm formation which contributes to the success of these pathogens (Riquelme et al., 2018). These bacteria often occur with other bacteria in polymicrobial interactions associated with human disease and some of these interactions have been studied (Peters et al., 2012). P. protegens, predominantly studied as a plant-protecting bacterium, is not generally considered a human pathogen although bacteria of the P. fluorescens complex have been isolated in clinical samples from mouth, stomach and lungs (Paulsen et al., 2005; Scales et al., 2014). Colonization studies of roots and biofilm formation by P. protegens have been reported but interaction with other members of rhizosphere communities is less studied. Most of the community studies use a top-down approach to interrogate the properties of community dynamics and function. However, these studies have yielded less information regarding specific mechanisms, interactions and community assembly. Understanding the different 2-species interactions in our mixed-species community is an advantage for future studies with increasing community complexity made up of bacterial species having either clinical or environmental relevance.
In conclusion, P. aeruginosa QS is an important modulator of the composition and structure of the 3-species biofilm community. In this system, P. aeruginosa functions as a keystone species that modulates the relative abundances of the species within its local environment. Future studies need to focus on the mechanisms behind changes in the proportions of both K. pneumoniae and P. protegens in the 3-species biofilm community. Competitive interactions between two members of this community, P. aeruginosa and P. protegens, appear to be responsible for this phenotype. Therefore, further studies on P. aeruginosa QS regulated functions and P. protegens secondary metabolites might help us to understand principles of antagonistic and competitive interactions within a developing biofilm.
The original contributions presented in the study are publicly available. This data can be found here: https://doi.org/10.21979/N9/GV37YR.
SS, SK, and SR conceived and planned the experiments. SS and MM carried out the experiments. SS, SB, MM, SK, and SR contributed to the interpretation of the results. SS, SK, and SR wrote the manuscript. All authors contributed to discussions on the manuscript. All authors contributed to the article and approved the submitted version.
The authors would like to acknowledge the financial support from the National Research Foundation and Ministry of Education Singapore under its Research Centre of Excellence Program. The research was further supported by a grant from the Singapore Ministry of Education (MOE2019-T2-1-050).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.646991/full#supplementary-material
Supplementary Figure 1 | QS regulated phenotypes of P. aeruginosa wild type, lasRrhlR mutant and the complemented strain. (A) Swarming motility of indicated strains on 0.5% swarm agar. Plates were incubated at 37°C for 16 h. Bars represent average diameter of the swarm area. (B) Protease activity of indicated strains on 1% skimmed milk containing LB agar plates. Plates were incubated at 37°C for 16 h. Bars represent average diameter of the zone of clearance around the colonies. (C) Elastase activity of indicated strains measured by mixing 100 µl of culture supernatant with 900 µl of Elastin-Congo Red containing buffer and incubating overnight at 37°C. Elastase activity was calculated by measuring the absorbance (OD495) of the supernatant. Bars represent OD495 values obtained after subtracting the values of medium control from the sample values. The data are averages from three independent biological replicates. **** denotes significant difference by One-way ANOVA followed by Tukey’s post-test (P<0.0001). 1, wild type P. aeruginosa; 2, lasRrhlR; 3, lasRrhlR/pUCP22notI+R2.
Supplementary Figure 2 | Proportions of P. aeruginosa wild type, lasRrhlR mutant and the complemented strain in dual-species biofilms. The proportions of the two species in the dual-species biofilm containing either the P. aeruginosa wild type, the QS mutant lasRrhlR or the complemented strain after 3 d of cultivation. Bar charts depict the percentages of P. aeruginosa and P. protegens in the wild type, the mutant or the complemented dual-species biofilms. The proportions were derived by cultivating biofilms under flow cell conditions followed by quantitative image analysis of two independent biological replicates. Error bars represent standard deviations (n=2). * denotes significant difference by Two-way ANOVA followed by Tukey’s post-test (P<0.05).
Supplementary Figure 3 | Growth curves of wild type and QS effector mutants of P. aeruginosa in M9 minimal medium containing glucose and casamino acids. P. aeruginosa wild type and different QS effector mutants were incubated at room temperature in 96 well microtiter plates and absorbance (OD600) recorded at regular intervals. Data shown are average of three independent biological replicates.
Supplementary Figure 4 | Biofilm formation of wild type and QS effector mutants of P. aeruginosa in M9 minimal medium containing glucose and casamino acids. P. aeruginosa WT and different QS effector mutants were incubated at room temperature in 24 well microtiter plates and the amount of biofilm formed quantified as absorbance (OD570) after crystal violet staining. Data shown are average of 3 biological replicates.
Supplementary Figure 5 | Fitness assays for wild type and QS effector mutants of P. aeruginosa in the presence of P. protegens in planktonic cultures. Equal volumes of P. aeruginosa wild type or QS effector mutants were mixed with P. protegens and allowed to grow for 24 h at room temperature. Bacteria were enumerated at zero and 24 h. Fitness was plotted as Log[Fold change of mutant/fold change of wild type]. Data shown are average of three independent biological replicates.
Supplementary Figure 6 | Pseudomonas aeruginosa mutants are not affected in competition with K. pneumoniae. K. pneumoniae was spotted on a lawn of P. aeruginosa wild type, QS mutants or complemented strains on LB agar plate. This panel shows the plate phenotype on day1 and day2.
Supplementary Figure 7 | Pseudomonas aeruginosa QS target mutants are not affected in competition with P. protegens. P. protegens was spotted on a lawn of P. aeruginosa wild type or QS target mutants on LB agar plate. This panel shows the plate phenotype on day1 and day2.
Supplementary Figure 8 | Proportions of the P. aeruginosa monospecies biofilm. The biovolume per unit area in the monospecies biofilm containing either the P. aeruginosa wild type or the QS mutants (lasIrhlI or lasrRrhlR) after 3 d of cultivation. Bar charts depict the biovolume per unit area of P. aeruginosa wild type or QS mutant monospecies biofilm. The values were derived by cultivating biofilms under flow cell conditions followed by quantitative image analysis of two independent biological replicates.
Abdel-Mawgoud A. M., Lepine F., Deziel E. (2010). Rhamnolipids: diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 86 (5), 1323–1336. doi: 10.1007/s00253-010-2498-2
An D., Danhorn T., Fuqua C., Parsek M. R. (2006). Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl. Acad. Sci. U. S. A. 103 (10), 3828–3833. doi: 10.1073/pnas.0511323103
An S., Wu J., Zhang L. H. (2010). Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl. Environ. Microbiol. 76 (24), 8160–8173. doi: 10.1128/AEM.01233-10
Anand A. A., Vennison S. J., Sankar S. G., Prabhu D. I., Vasan P. T., Raghuraman T., et al. (2010). Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J. Insect Sci. 10:107. doi: 10.1673/031.010.10701
Banin E., Vasil M. L., Greenberg E. P. (2005). Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 102 (31), 11076–11081. doi: 10.1073/pnas.0504266102
Brown S. A., Whiteley M. (2007). A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J. Bacteriol 189 (17), 6407–6414. doi: 10.1128/JB.00554-07
Chang C. Y., Krishnan T., Wang H., Chen Y., Yin W. F., Chong Y. M., et al. (2014). Non-antibiotic quorum sensing inhibitors acting against N-acyl homoserine lactone synthase as druggable target. Sci. Rep. 4, 7245. doi: 10.1038/srep07245
Chazal P. M. (1995). Pollution of modern metalworking fluids containing biocides by pathogenic bacteria in France. Reexamination of chemical treatments accuracy. Eur. J. Epidemiol. 11 (1), 1–7. doi: 10.1007/BF01719939
Choi K. H., Schweizer H. P. (2005). An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30. doi: 10.1186/1471-2180-5-30
Choi K. H., Schweizer H. P. (2006). mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1 (1), 153–161. doi: 10.1038/nprot.2006.24
Choi K. H., Gaynor J. B., White K. G., Lopez C., Bosio C. M., Karkhoff-Schweizer R. R., et al. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2 (6), 443–448. doi: 10.1038/nmeth765
Clark D. J., Maaløe O. (1967). DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23 (1), 99 – 112. doi: 10.1016/S0022-2836(67)80070-6
Cornelis P., Matthijs S. (2002). Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ. Microbiol. 4 (12), 787–798. doi: 10.1046/j.1462-2920.2002.00369.x
Costello A., Reen F. J., O’Gara F., Callaghan M., McClean S. (2014). Inhibition of co-colonizing cystic fibrosis-associated pathogens by Pseudomonas aeruginosa and Burkholderia multivorans. Microbiology (Reading) 160 (Pt 7), 1474–1487. doi: 10.1099/mic.0.074203-0
Darveau R. P., Hajishengallis G., Curtis M. A. (2012). Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 91 (9), 816–820. doi: 10.1177/0022034512453589
Davey M. E., O’Toole G A. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64 (4), 847–867. doi: 10.1128/mmbr.64.4.847-867.2000
Davey M. E., Caiazza N. C., O’Toole G. A. (2003). Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol 185 (3), 1027–1036. doi: 10.1128/jb.185.3.1027-1036.2003
Davies D. G., Parsek M. R., Pearson J. P., Iglewski B. H., Costerton J. W., Greenberg E. P. (1998). The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280 (5361), 295–298. doi: 10.1126/science.280.5361.295
Flemming H. C., Wingender J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8 (9), 623–633. doi: 10.1038/nrmicro2415
Flemming H. C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14 (9), 563–575. doi: 10.1038/nrmicro.2016.94
Gambello M. J., Iglewski B. H. (1991). Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol 173 (9), 3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991
Hajishengallis G., Lamont R. J. (2016). Dancing with the Stars: How Choreographed Bacterial Interactions Dictate Nososymbiocity and Give Rise to Keystone Pathogens, Accessory Pathogens, and Pathobionts. Trends Microbiol. 24 (6), 477–489. doi: 10.1016/j.tim.2016.02.010
Hajishengallis G., Liang S., Payne M. A., Hashim A., Jotwani R., Eskan M. A., et al. (2011). Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10 (5), 497–506. doi: 10.1016/j.chom.2011.10.006
Hajishengallis G., Darveau R. P., Curtis M. A. (2012). The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10 (10), 717–725. doi: 10.1038/nrmicro2873
Harmsen M., Yang L., Pamp S. J., Tolker-Nielsen T. (2010). An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 59 (3), 253–268. doi: 10.1111/j.1574-695X.2010.00690.x
Heydorn A., Ersboll B. K., Hentzer M., Parsek M. R., Givskov M., Molin S. (2000a). Experimental reproducibility in flow-chamber biofilms. Microbiology (Reading) 146 ( Pt 10), 2409–2415. doi: 10.1099/00221287-146-10-2409
Heydorn A., Nielsen A. T., Hentzer M., Sternberg C., Givskov M., Ersboll B. K., et al. (2000b). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology (Reading) 146 ( Pt 10), 2395–2407. doi: 10.1099/00221287-146-10-2395
Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212 (1), 77–86. doi: 10.1016/s0378-1119(98)00130-9
Hosni T., Moretti C., Devescovi G., Suarez-Moreno Z. R., Fatmi M. B., Guarnaccia C., et al. (2011). Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. ISME J. 5 (12), 1857–1870. doi: 10.1038/ismej.2011.65
Huber B., Riedel K., Hentzer M., Heydorn A., Gotschlich A., Givskov M., et al. (2001). The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147 (Pt 9), 2517–2528. doi: 10.1099/00221287-147-9-2517
Imperi F., Tiburzi F., Visca P. (2009). Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 106 (48), 20440–20445. doi: 10.1073/pnas.0908760106
Jackson G., Beyenal H., Rees W. M., Lewandowski Z. (2001). Growing reproducible biofilms with respect to structure and viable cell counts. J. Microbiol. Methods 47 (1), 1–10. doi: 10.1016/s0167-7012(01)00280-9
Jaffar-Bandjee M. C., Lazdunski A., Bally M., Carrere J., Chazalette J. P., Galabert C. (1995). Production of elastase, exotoxin A, and alkaline protease in sputa during pulmonary exacerbation of cystic fibrosis in patients chronically infected by Pseudomonas aeruginosa. J. Clin. Microbiol. 33 (4), 924–929. doi: 10.1128/JCM.33.4.924-929.1995
Kang D., Kirienko N. V. (2017). High-Throughput Genetic Screen Reveals that Early Attachment and Biofilm Formation Are Necessary for Full Pyoverdine Production by Pseudomonas aeruginosa. Front. Microbiol. 8, 1707. doi: 10.3389/fmicb.2017.01707
Kessler B., de Lorenzo V., Timmis K. N. (1992). A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233 (1-2), 293–301. doi: 10.1007/BF00587591
Kommerein N., Doll K., Stumpp N. S., Stiesch M. (2018). Development and characterization of an oral multispecies biofilm implant flow chamber model. PloS One 13 (5), e0196967. doi: 10.1371/journal.pone.0196967
Korgaonkar A., Trivedi U., Rumbaugh K. P., Whiteley M. (2013). Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. U. S. A. 110 (3), 1059–1064. doi: 10.1073/pnas.1214550110
Lee J., Zhang L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6 (1), 26–41. doi: 10.1007/s13238-014-0100-x
Lee J., Wu J., Deng Y., Wang J., Wang C., Wang J., et al. (2013). A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 9 (5), 339–343. doi: 10.1038/nchembio.1225
Lee K. W., Periasamy S., Mukherjee M., Xie C., Kjelleberg S., Rice S. A. (2014). Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 8 (4), 894–907. doi: 10.1038/ismej.2013.194
Lesic B., Starkey M., He J., Hazan R., Rahme L. G. (2009). Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 155 (Pt 9), 2845–2855. doi: 10.1099/mic.0.029082-0
Lewenza S., Visser M. B., Sokol P. A. (2002). Interspecies communication between Burkholderia cepacia and Pseudomonas aeruginosa. Can. J. Microbiol. 48 (8), 707–716. doi: 10.1139/w02-068
Lim C. K., Hassan K. A., Penesyan A., Loper J. E., Paulsen I. T. (2013). The effect of zinc limitation on the transcriptome of Pseudomonas protegens Pf-5. Environ. Microbiol. 15 (3), 702–715. doi: 10.1111/j.1462-2920.2012.02849.x
Liu W., Russel J., Roder H. L., Madsen J. S., Burmolle M., Sorensen S. J. (2017). Low-abundant species facilitates specific spatial organization that promotes multispecies biofilm formation. Environ. Microbiol. 19 (7), 2893–2905. doi: 10.1111/1462-2920.13816
Magill S. S., Edwards J. R., Bamberg W., Beldavs Z. G., Dumyati G., Kainer M. A., et al. (2014). Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 370 (13), 1198–1208. doi: 10.1056/NEJMoa1306801
Mazzola M., Cook R. J., Thomashow L. S., Weller D. M., Pierson L. S. (1992). Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 58 (8), 2616–2624. doi: 10.1128/AEM.58.8.2616-2624.1992
Moons P., Van Houdt R., Aertsen A., Vanoirbeek K., Engelborghs Y., Michiels C. W. (2006). Role of Quorum Sensing and Antimicrobial Component Production by Serratia plymuthica in Formation of Biofilms, Including Mixed Biofilms with Escherichia coli. Appl. Environ. Microbiol. 72 (11), 7294–7300. doi: 10.1128/AEM.01708-06
Neu T. R. (1996). Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 60 (1), 151–166. doi: 10.1128/MR.60.1.151-166.1996
Ochsner U. A., Reiser J. (1995). Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 92 (14), 6424–6428. doi: 10.1073/pnas.92.14.6424
Pamp S. J., Tolker-Nielsen T. (2007). Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol 189 (6), 2531–2539. doi: 10.1128/JB.01515-06
Paulsen I. T., Press C. M., Ravel J., Kobayashi D. Y., Myers G. S., Mavrodi D. V., et al. (2005). Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23 (7), 873–878. doi: 10.1038/nbt1110
Pearson J. P., Passador L., Iglewski B. H., Greenberg E. P. (1995). A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 92 (5), 1490–1494. doi: 10.1073/pnas.92.5.1490
Peters B. M., Jabra-Rizk M. A., O’May G. A., Costerton J. W., Shirtliff M. E. (2012). Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 25 (1), 193–213. doi: 10.1128/CMR.00013-11
Poole K. (1994). Bacterial multidrug resistance–emphasis on efflux mechanisms and Pseudomonas aeruginosa. J. Antimicrob. Chemother. 34 (4), 453–456. doi: 10.1093/jac/34.4.453
Rahim R., Ochsner U. A., Olvera C., Graninger M., Messner P., Lam J. S., et al. (2001). Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 40 (3), 708–718. doi: 10.1046/j.1365-2958.2001.02420.x
Ramette A., Frapolli M., Fischer-Le Saux M., Gruffaz C., Meyer J. M., Defago G., et al. (2011). Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 34 (3), 180–188. doi: 10.1016/j.syapm.2010.10.005
Rezaei E., Safari H., Naderinasab M., Aliakbarian H. (2011). Common pathogens in burn wound and changes in their drug sensitivity. Burns 37 (5), 805–807. doi: 10.1016/j.burns.2011.01.019
Riedel K., Hentzer M., Geisenberger O., Huber B., Steidle A., Wu H., et al. (2001). N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147 (Pt 12), 3249–3262. doi: 10.1099/00221287-147-12-3249
Riquelme S. A., Ahn D., Prince A. (2018). Pseudomonas aeruginosa and Klebsiella pneumoniae Adaptation to Innate Immune Clearance Mechanisms in the Lung. J. Innate Immun. 10 (5-6), 442–454. doi: 10.1159/000487515
Roume H., Heintz-Buschart A., Muller E. E. L., May P., Satagopam V. P., Laczny C. C., et al. (2015). Comparative integrated omics: identification of key functionalities in microbial community-wide metabolic networks. NPJ Biofilms Microbiomes 1 (1), 15007. doi: 10.1038/npjbiofilms.2015.7
Rutherford S. T., Bassler B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2 (11), a012427. doi: 10.1101/cshperspect.a012427
Scales B. S., Dickson R. P., LiPuma J. J., Huffnagle G. B. (2014). Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clin. Microbiol. Rev. 27 (4), 927–948. doi: 10.1128/CMR.00044-14
Schleheck D., Barraud N., Klebensberger J., Webb J. S., McDougald D., Rice S. A., et al. (2009). Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PloS One 4 (5), e5513. doi: 10.1371/journal.pone.0005513
Schuster M., Lostroh C. P., Ogi T., Greenberg E. P. (2003). Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol 185 (7), 2066–2079. doi: 10.1128/jb.185.7.2066-2079.2003
Simon R., Priefer U., Pühler A. (1983). A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Biotechnology 1 (9), 784–791. doi: 10.1038/nbt1183-784
Smalley N. E., An D., Parsek M. R., Chandler J. R., Dandekar A. A. (2015). Quorum Sensing Protects Pseudomonas aeruginosa against Cheating by Other Species in a Laboratory Coculture Model. J. Bacteriol 197 (19), 3154–3159. doi: 10.1128/JB.00482-15
Stecher B., Maier L., Hardt W. D. (2013). ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 11 (4), 277–284. doi: 10.1038/nrmicro2989
Stoodley P., Wilson S., Hall-Stoodley L., Boyle J. D., Lappin-Scott H. M., Costerton J. W. (2001). Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol. 67 (12), 5608–5613. doi: 10.1128/AEM.67.12.5608-5613.2001
Tan C. H., Koh K. S., Xie C., Tay M., Zhou Y., Williams R., et al. (2014). The role of quorum sensing signalling in EPS production and the assembly of a sludge community into aerobic granules. ISME J. 8 (6), 1186–1197. doi: 10.1038/ismej.2013.240
Twomey K. B., O’Connell O. J., McCarthy Y., Dow J. M., O’Toole G. A., Plant B. J., et al. (2012). Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J. 6 (5), 939–950. doi: 10.1038/ismej.2011.167
Van Delden C., Iglewski B. H. (1998). Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4 (4), 551–560. doi: 10.3201/eid0404.980405
West S. E., Schweizer H. P., Dall C., Sample A. K., Runyen-Janecky L. J. (1994). Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148 (1), 81–86. doi: 10.1016/0378-1119(94)90237-2
Yang L., Nilsson M., Gjermansen M., Givskov M., Tolker-Nielsen T. (2009). Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol. Microbiol. 74 (6), 1380–1392. doi: 10.1111/j.1365-2958.2009.06934.x
Keywords: quorum sensing, species interactions, multispecies, biofilms, Pseudomonas aeruginosa, Pseudomonas protegens, Klebsiella pneumoniae
Citation: Subramoni S, Muzaki MZBM, Booth SCM, Kjelleberg S and Rice SA (2021) N-Acyl Homoserine Lactone-Mediated Quorum Sensing Regulates Species Interactions in Multispecies Biofilm Communities. Front. Cell. Infect. Microbiol. 11:646991. doi: 10.3389/fcimb.2021.646991
Received: 28 December 2020; Accepted: 25 February 2021;
Published: 18 March 2021.
Edited by:
Mingkai Li, Fourth Military Medical University, ChinaReviewed by:
William Schwan, University of Wisconsin–La Crosse, United StatesCopyright © 2021 Subramoni, Muzaki, Booth, Kjelleberg and Rice. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Scott A. Rice, cnNjb3R0QG50dS5lZHUuc2c=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.