
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 20 July 2021
Sec. Microbiome in Health and Disease
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.639667
 Lisa Vork1*†
Lisa Vork1*† John Penders2†
John Penders2† Jonna Jalanka3
Jonna Jalanka3 Svetlana Bojic4
Svetlana Bojic4 Sander M. J. van Kuijk5
Sander M. J. van Kuijk5 Anne Salonen3
Anne Salonen3 Willem M. de Vos3,6
Willem M. de Vos3,6 Mirjana Rajilic-Stojanovic4
Mirjana Rajilic-Stojanovic4 Zsa Zsa R. M. Weerts1
Zsa Zsa R. M. Weerts1 Ad A. M. Masclee1
Ad A. M. Masclee1 Marta Pozuelo7
Marta Pozuelo7 Chaysavanh Manichanh7,8*
Chaysavanh Manichanh7,8* Daisy M. A. E. Jonkers1 on behalf of COST Action GENIEUR/BM1106 Microbiome Working Group
Daisy M. A. E. Jonkers1 on behalf of COST Action GENIEUR/BM1106 Microbiome Working GroupIntroduction: Stool consistency has been associated with fecal microbial composition. Stool consistency often varies over time, in subjects with and without gastrointestinal disorders, raising the question whether variability in the microbial composition should be considered in microbiota studies. We evaluated within-subject day-to-day variability in stool consistency and the association with the fecal microbiota in irritable bowel syndrome (IBS) and healthy subjects, over seven days.
Methods: Twelve IBS patients and 12 healthy subjects collected fecal samples during seven consecutive days. Stool consistency was determined by the patient-reported Bristol Stool Scale (BSS) and fecal dry weight percentage. 16S rRNA V4 gene sequencing was performed and microbial richness (alpha diversity; Chao1 index, observed number of species, effective Shannon index) and microbial community structure (beta diversity; Bray-Curtis distance, generalized UniFrac, and taxa abundance on family level) were determined.
Results: Linear mixed-effects models showed significant associations between stool consistency and microbial richness, but no time effect. This implies that between-subject but not within-subject variation in microbiota over time can partially be explained by variation in stool consistency. Redundancy analysis showed a significant association between stool consistency and microbial community structure, but additional linear mixed-effects models did not demonstrate a time effect on this.
Conclusion: This study supports an association between stool consistency and fecal microbiota, but no effect of day-to-day fluctuations in stool consistency within seven days. This consolidates the importance of considering stool consistency in gut microbiota research, though confirms the validity of single fecal sampling to represent an individual’s microbiota at a given time point. NCT00775060.
Over the past decades, the gut microbiota has been studied extensively in the context of gastrointestinal (GI) function in health and disease. Stool consistency measured with the Bristol Stool Scale (BSS) or fecal moisture has been associated with gut microbiota diversity and composition (Tigchelaar et al., 2016; Vandeputte et al., 2016), and has been identified as important covariate of the microbiota composition in population-based studies (Falony et al., 2016; Vandeputte et al., 2017). This indicates the importance of considering stool consistency as a potential confounding factor in both cross-sectional and longitudinal intestinal microbiota analyses. However, stool consistency can fluctuate over time (Matto et al., 2005), possibly associated with temporal fluctuations in the microbiota composition. This raises the question whether a single fecal sample is representative for a subject’s microbial profile at a given time point and whether within-subject day-to-day variability should be considered in microbiota studies, by collecting repeated fecal samples.
While stool consistency can fluctuate from day to day in healthy subjects, fluctuations are often more pronounced in subjects with irritable bowel syndrome (IBS), a GI disorder characterized by abdominal pain and altered bowel habits. According to the Rome criteria, IBS is typically divided into four subtypes, often defined by BSS: diarrhea predominant (IBS-D), constipation predominant (IBS-C), a combination of both (IBS-M; mixed type), or unspecified in which both diarrhea and constipation are not predominantly present (IBS-U) (Longstreth et al., 2006). Multiple studies have shown changes in gut microbiota composition as well as functionality in IBS compared to healthy subjects (Salonen et al., 2010; Rajilic-Stojanovic et al., 2011; Jeffery et al., 2012). Some results point towards differences in microbiota between subgroups of IBS subjects (Malinen et al., 2005; Kassinen et al., 2007; Jeffery et al., 2012), though findings on specific bacterial taxa are not consistent. Additionally, in cross-sectional studies changes in microbial composition have been correlated to IBS symptom scores (Malinen et al., 2010; Rajilic-Stojanovic et al., 2011; Jeffery et al., 2012), which also generally vary over time.
So far, a few studies have focused on temporal (in)stability of the fecal microbiota and found a more unstable microbial composition in IBS patients compared to healthy volunteers over a period of months (Matto et al., 2005; Lyra et al., 2009; Durban et al., 2013). However, none have considered possible day-to-day variation and its association with stool consistency and symptoms within subjects over time. Since both stool consistency and GI symptoms can fluctuate from day to day, especially in IBS populations, a key question is whether a single fecal sample at one time point suffices or whether repeated sampling over a period of time is needed in order to take into account day-to-day variability in microbial composition.
Therefore, we aimed to evaluate within-subject day-to-day variability in stool consistency and gastrointestinal symptoms, and the association with the fecal microbiota composition in IBS and healthy subjects, over a seven-day course.
This study was embedded in the Maastricht IBS Cohort. The study protocol has been approved by the Maastricht University Medical Center+ (MUMC+) Committee of Ethics (METC 08-2-066) and was executed according to the revised Declaration of Helsinki (64th WMA General Assembly, Brazil 2013). The study has been registered in the US National Library of Medicine (http://www.clinicaltrials.gov, NCT00775060).
Between January 2015 and March 2016, IBS patients aged 18-75 years were recruited at the outpatient department of Gastroenterology-Hepatology of MUMC+. All subjects fulfilled the Rome III criteria and were assigned to IBS subtypes based on predominant bowel habits (Drossman, 2006). A history of abdominal surgery, except for uncomplicated appendectomy, cholecystectomy, or hysterectomy, was reason for exclusion. Additional investigations to exclude organic disease were performed if deemed necessary by a gastroenterologist.
Age- and sex-matched healthy controls (HC) were recruited via public advertising and recruitment websites. A brief medical history was taken by a trained research physician to exclude the presence of any GI disorders or current GI symptoms. All participants gave written informed consent prior to inclusion.
A seven-day symptom diary was used to record daily symptom scores and bowel habits using BSS [Lewis and Heaton, 1997; U.S. Department of Health and Human Services FaDA and Center for Drug Evaluation and Research (CDER), 2012]. Symptoms (i.e. abdominal pain, bloating, fecal urgency, diarrhea, and constipation) were scored at the end of the day on an 11-point numeric rating scale [i.e. 0 (none) to 10 (severe) [U.S. Department of Health and Human Services FaDA and Center for Drug Evaluation and Research (CDER), 2012] and BSS for stool consistency was reported for every bowel movement. Additionally, medication use was reported every day and participants were instructed to maintain their habitual dietary habits during the study period.
The first fecal samples of each day, and one additional sample when subjects reported diarrhea, were collected. Subjects stored the samples at -20°C at home directly after collection. Following the seven-day study period, all samples were transported to MUMC+ on dry ice and stored at -80°C.
In addition to the BSS, the dry weight content of each fecal sample was determined, as a more objective measure of stool consistency. Therefore, an aliquot of 0.5 g (wet weight) was dried at 60°C in a vacuum dryer for 5 hours. Directly after drying, the samples were weighed again (dry weight). The percentage of dry weight was calculated as: [dry weight(g)/wet weight(g)] * 100.
Fecal microbiota profiling was achieved by next-generation sequencing of 16S rRNA V4-region gene amplicons. Detailed information on DNA extraction, sequencing, and data analysis can be found under Supporting Information – Supplementary Methods.
All statistical analyses were performed using QIIME version 1.9.1 and R version 3.4.2. Categorical patient characteristics are presented as proportions and differences between groups were tested using χ2 or Fisher’s exact test. Continuous characteristics are presented as mean and standard deviation (SD) or median with interquartile range in case of skewness. Differences between groups were tested using the independent t-test or the Mann-Whitney U test, depending on the normality of the distribution.
Alpha diversity data are expressed as Chao1 index, observed species, and effective Shannon index (exp[Shannon index]). To evaluate the within-subject variability in alpha diversity over time, inter-item (Pearson) correlations between the consecutively collected fecal samples, as well as intra-class correlations (ICC), were calculated for IBS and healthy subjects separately. Data from subjects that collected at least five consecutive samples were included in these analyses. The Bray-Curtis distance and generalized UniFrac (i.e. beta diversity) were used to quantify the (dis)similarity in microbial community structure between samples. Non-parametric tests were used for taxonomical abundance data. Principal coordinates analyses (PCoA) were performed on beta diversity indices to evaluate possible clustering of the microbial community structure. A Mann Whitney-U test was used to compare the average within-subject diversity distance between IBS and healthy subjects.
To evaluate the correlation between stool consistency and the microbiota, a constrained redundancy analysis (RDA) was carried out, using routines from R package “vegan”. The zeros from the count data (summarized on family level) were imputed using R package “zCompositions” and data were clr (centered log ratio) transformed. In addition, two-level mixed-effects linear regression models (level 1: consecutive stool samples; level 2: subjects) were used to examine the association between stool consistency and microbiota using all longitudinal measurements. Separate models were used for different measures of microbial diversity and composition, with alpha diversity indices and clr transformed taxonomical abundance data (family level), respectively, as the dependent variables. The two-way interaction term “stool consistency*time” and stool consistency were used as independent variables and a random intercept was chosen to correct for clustering of multiple measurements within each participant. A p-value of ≤0.05 was considered statistically significant.
In all analyses, stool consistency was primarily based on fecal dry weight percentage, and additional analyses were performed using BSS.
Twelve IBS patients and 12 healthy subjects were included. Demographics are shown in Table 1. In total, 71 and 78 samples were available for the IBS and healthy group, respectively.
The day-to-day variability in stool consistency, measured by fecal dry weight percentage, was found to be high in the IBS group (ICC: 0.223) and moderate (ICC: 0.622) in the healthy population. BSS scores showed high day-to-day variability in both groups (ICC for IBS: 0.397; ICC for HC: 0.276). This variability in stool consistency is illustrated in Supplementary Figure 1.
The predominant phyla in both healthy and IBS subjects were Firmicutes (average relative abundance of 79.5% and 81.9%, respectively), followed by Bacteroidetes, Actinobacteria, Verrucomicrobia, and Proteobacteria (Supplementary Figure 2). The relative abundances of these five predominant phylae are shown in Supplementary Figure 3. While evaluating differences between IBS and healthy subjects is beyond the scope of this study, the abundances are shown for both groups separately for better visibility.
For both IBS and healthy subjects, microbial richness showed high correlations between subsequent samples, demonstrating low within-subject variability from day to day. Inter-item correlations between different samples were all above 0.800 and a high degree of agreement in microbial richness was found between the different samples of one subject (single measure ICCs all above 0.893). Inter-item matrices and ICCs for Chao1 index are shown in Table 2. Similar results were found for observed species, and correlations were moderate for effective Shannon index (Supplementary Tables 1A, B).

Table 2 Inter-item (Pearson) correlations between Chao1-index of consecutive samples, for healthy subjects and IBS patients.
The microbial community structure clustered per individual, suggesting that the dissimilarity in microbiota between consecutive samples is larger between than within subjects, while no clear separation between IBS and healthy subjects was demonstrated (Figure 1A). Moreover, no significant difference was found in the average within-subject beta diversity between IBS and healthy subjects, indicating that the day-to-day variation in overall microbial community structure is similar between IBS and healthy subjects (Figure 1B). Comparable results were found for the generalized UniFrac as measure of beta diversity (Supplementary Figures 4A, B). In addition, in order to evaluate whether the dissimilarity in microbiota between different samples (i.e. within individual subjects) becomes larger over time, we illustrate the Bray Curtis Dissimilarity for each day (i.e. day 2 – 7) compared to day 1, separately for each subject in Supplementary Figure 5A (IBS patients) and Supplementary Figure 5B (healthy subjects). No apparent increase in beta diversity over the seven days is shown, indicating a rather stable microbial community structure per individual during the study period.
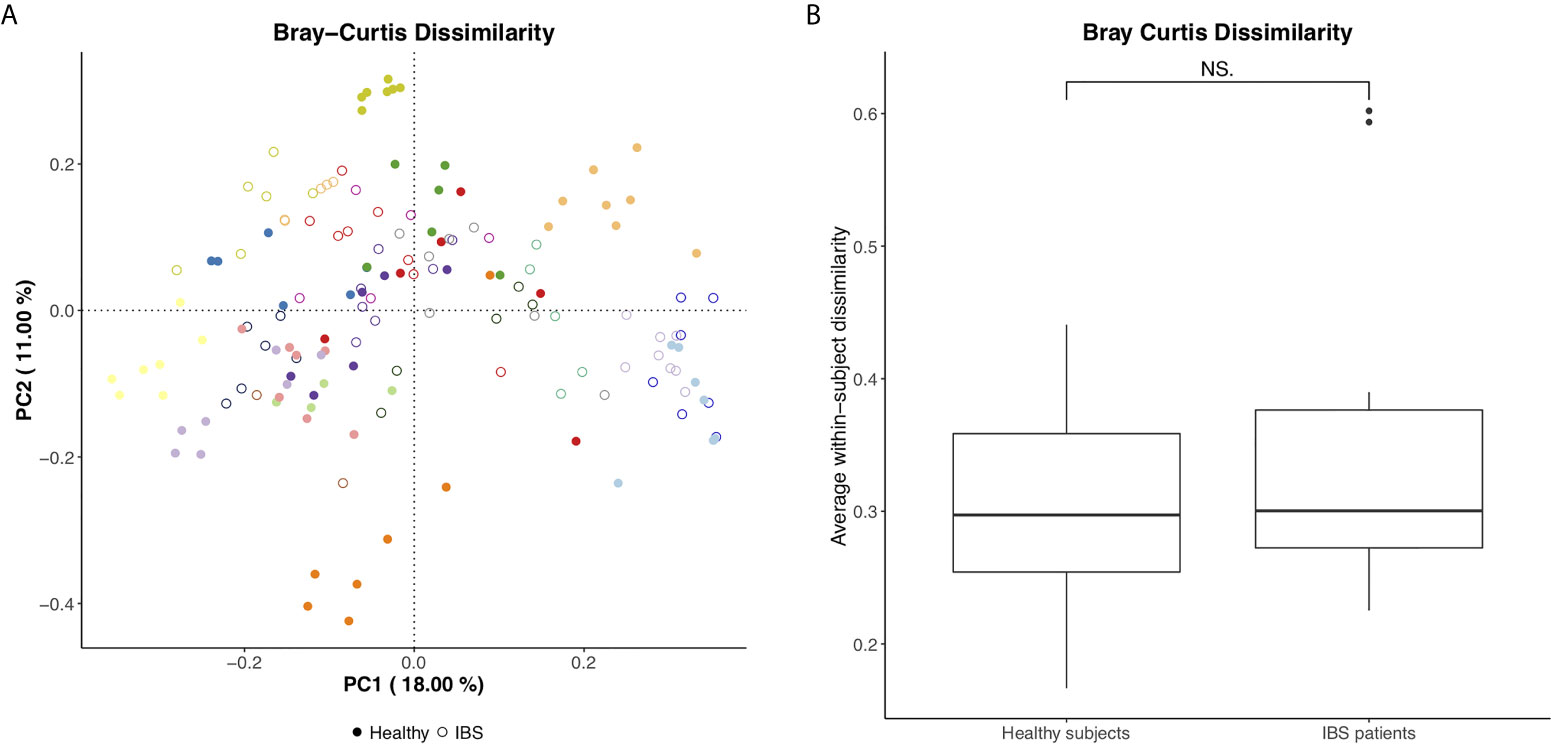
Figure 1 (A) Bray Curtis PCoA plot per individual (different colors) and for healthy subjects vs. IBS patients (figure annotations). (B) Average within-subject beta diversity (Bray Curtis Dissimilarity) for healthy subjects vs. IBS patients, significance tested using Mann Whitney U test, NS, not significant (p=0.71).
A linear mixed-effects model with the Chao1 index as dependent variable demonstrated no significant effect of the two-way interaction “fecal dry weight*time” on microbial richness (B: 0.030, SE: 0.072, p=0.676), indicating that the association between stool consistency and microbial richness was not different between subsequent samples. After removal of this term from the model, stool consistency was found to be a significant predictor of microbial richness (B: 1.231, SE: 0.200, p<0.001) (Table 3), showing an overall significant association between stool consistency and the microbiota, but independent of time. Subsequent linear mixed-effects models with observed species and the effective Shannon index as dependent variables, showed similar results (Supplementary Tables 2A, B). Models using BSS instead of fecal dry weight as a measure of stool consistency also showed similar results (Table 3, Supplementary Tables 2A, B).
A redundancy analysis, including data from both IBS and healthy subjects, showed a significant association between stool consistency and microbial composition on the family level, mainly driven by Christensenellaceae, Enterobacteriaceae, Verrucomicrobiaceae, Methanobacteriaceae, and Veillonellaceae (Figure 2). To correct for within-subject day-to-day variability in these associations between specific bacterial groups and stool consistency, linear mixed-effects models with the respective bacterial groups as the dependent variable were performed. For Christensenellaceae, Enterobacteriaceae, Verrucomicrobiaceae, and Methanobacteriaceae, stool consistency was indeed a significant predictor (i.e. higher abundance in firmer stools), but not for Veillonellaceae.
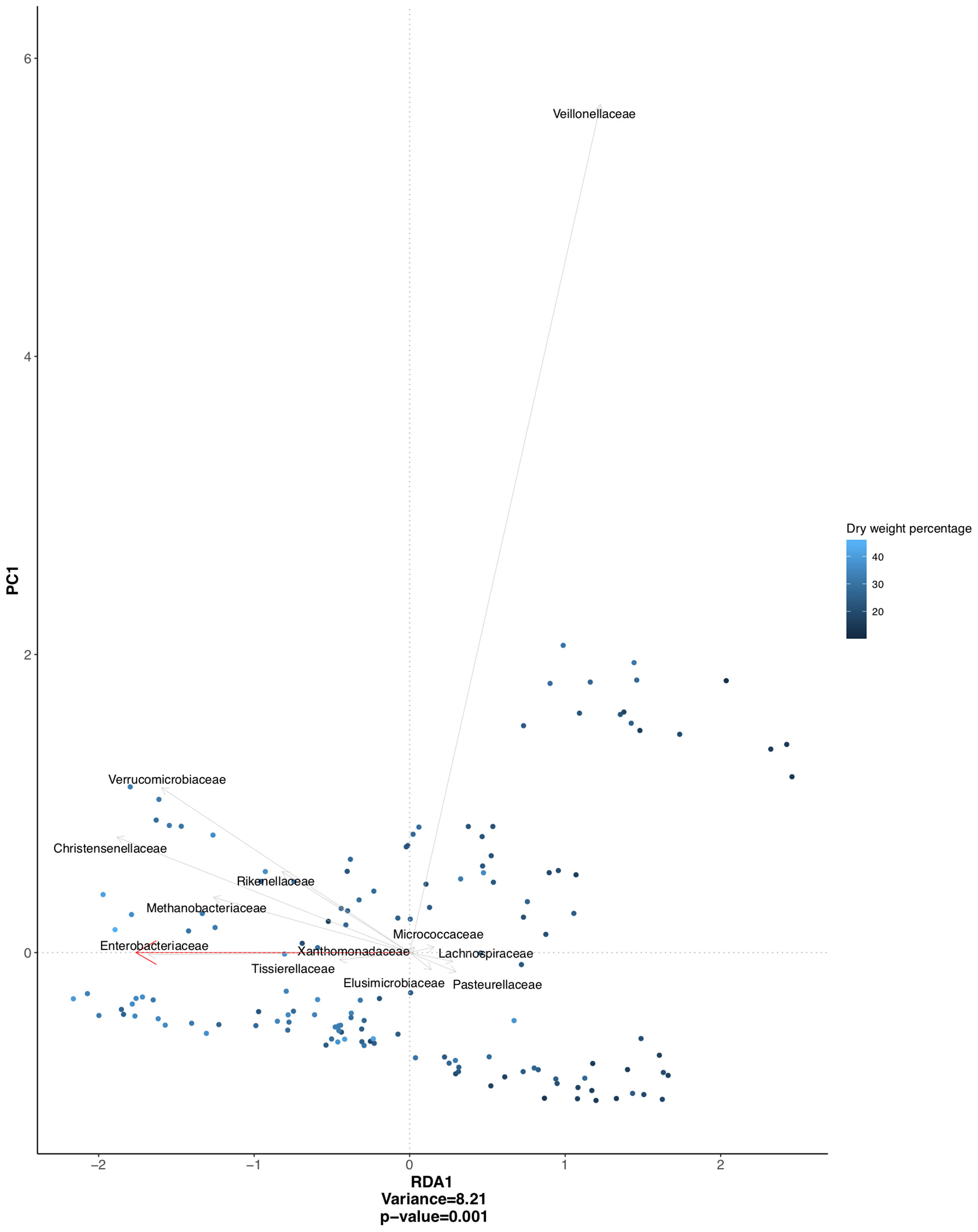
Figure 2 Redundancy analysis plot based on clr transformed abundancies, and constrained on stool consistency (dry weight percentage), with individual variation partialled-out. Significant association between stool consistency and microbial composition (p = 0.001), mainly driven by bacterial families depicted in the figure.
Exploratory analysis showed no significant association between the fecal microbiota (i.e. alpha and beta diversity) and abdominal pain and bloating (Figure 3), and can be found in Supporting Information – Supplementary Results.
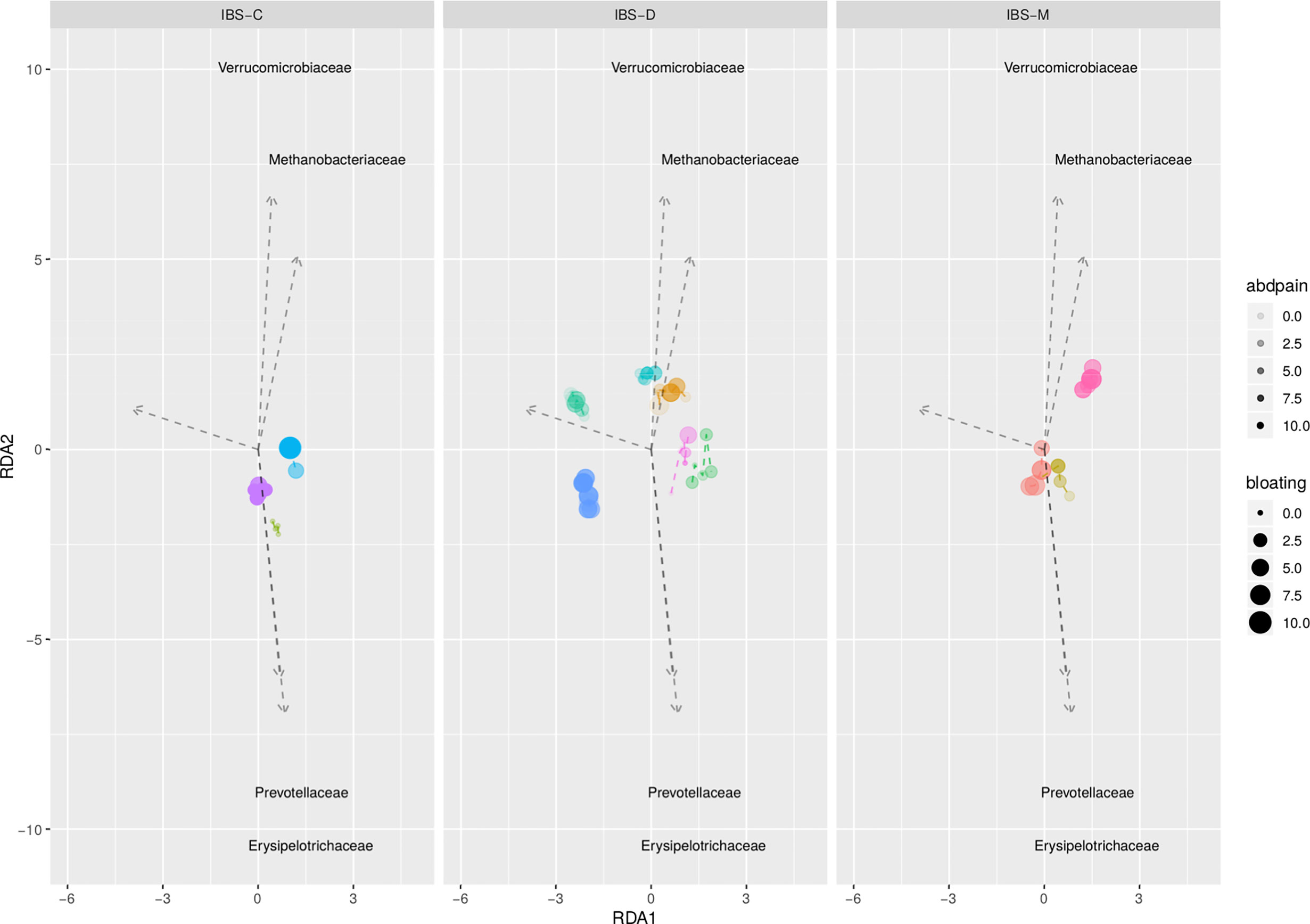
Figure 3 Redundancy analysis plot based on clr transformed abundancies, and constrained on individual, abdominal pain, and abdominal bloating. Each dot represents an individual sample; IBS patients are depicted by different colors. No significant association between both GI symptoms (p = 0.368 for abdominal pain; p = 0.521 for bloating) and microbial composition.
Previously described strong correlations between stool consistency and the fecal microbiota pointed towards the importance of stool consistency and/or gut transit time as confounding factors in microbiota analyses (Falony et al., 2016; Vandeputte et al., 2016; Vandeputte et al., 2017). Our study confirms this, though demonstrates that within-subject variation in the fecal microbiota composition between consecutive samples over seven days is limited in both IBS and healthy subjects, even in case of fluctuating stool consistency or GI symptoms.
Stool consistency is known to vary within individuals over time (Matto et al., 2005), suggesting that the fecal microbiota may also change over short time periods. Therefore, this study focused on the microbial stability from day to day and the association with stool consistency. Both IBS patients and healthy subjects were evaluated, since stool consistency is known to show high within-subject variability from day to day in IBS, but variability occurs in healthy subjects as well. We here demonstrate low within-subject day-to-day variability in fecal microbiota over seven consecutive days. Overall variability in the microbial community structure was mainly driven by between-subject variability, with rather small within-subject changes between consecutive samples. Although significant associations between stool consistency and microbial richness and composition were found, no significant effect of time on this association could be demonstrated. Altogether, this indicates that regardless of fluctuations in stool consistency, the variation in fecal microbiota composition within subjects over a one-week period, in both IBS patients and healthy subjects, is rather limited when compared to variations between subjects.
Previous results on temporal stability of the microbial composition are contradictory. Particularly inter-individual differences have been demonstrated, but less is known about within-subject microbial variability (Jalanka-Tuovinen et al., 2011; Rajilic-Stojanovic et al., 2012). Instability in both IBS and healthy subjects has been demonstrated, with possibly a more instable microbiota in IBS patients, assessed over three to six months (Matto et al., 2005; Maukonen et al., 2006). Others found low variability between fecal microbial profiles in healthy subjects with intervals of 14 days to several months. However, even during periods of overall community stability, intra-individual dynamics of select microbial taxa could still be observed (Zoetendal et al., 1998; Zoetendal et al., 2001; Vanhoutte et al., 2004; Jalanka-Tuovinen et al., 2011; Rajilic-Stojanovic et al., 2012; Johnson et al., 2019). According to our results, such within-subject variation in microbial profiles is limited in both IBS and healthy subjects and not related to stool consistency over a seven-day period.
We used fecal dry weight percentage and patient-reported BSS scores as measures of stool consistency. From day to day, moderate-to-high levels of variability in both measures were found in both groups. This confirms that our data should be suitable to detect any day-to-day changes in the microbiota related to variability in stool consistency. Previous findings demonstrated that looser stools, according to BSS, were associated with lower species richness (Vandeputte et al., 2016). Our findings, both using fecal dry weight percentage and the BSS, confirm this association, but in addition indicate that there is no major within-subject shifting in microbial richness and composition from day to day, associated with short-term changes in stool consistency. Also, the previously demonstrated correlation between specific genera abundances and stool consistency was not dependent of time in our study. An increase in abundance of members of the Methanobacteriaceae and Verrucomicrobiaceae, and the Enterobacteriaceae and Christensenellaceae families in association with firmer stools has been described (Tigchelaar et al., 2016; Vandeputte et al., 2016; Vandeputte et al., 2017; Jalanka et al., 2019). However, linear mixed-effects models could not confirm an association between Veillonellaceae and stool consistency, suggesting that this association is mainly driven by between-subject differences and becomes less important when correcting for within-subject variation. In addition to these previous findings, we could not demonstrate important within-subject shifting in these bacterial groups in relation to fluctuations in stool consistency.
Since previous studies suggested a link between the microbiota and abdominal symptoms in IBS (Malinen et al., 2010; Rajilic-Stojanovic et al., 2011; Jeffery et al., 2012), and these can vary from day to day, we exploratory evaluated this association for abdominal pain and bloating. Our results indicate that there are no daily shifts in the gut microbiota related to daily individual symptom patterns.
Previous studies also showed rapid changes of microbial composition over the course of several days after dietary changes, especially on animal-based diet (David et al., 2014). This demonstrates that the gut microbial community can change rapidly and underlines the importance of considering time when evaluating potential confounders. In the current study, participants were not allowed to introduce major dietary changes, and we do not expect diet to be an important confounder in our results.
This is the first study to examine a short-term within-subject association between stool consistency and the gut microbiota, using repeated fecal sampling over one week. Both IBS and healthy subjects were evaluated in order to capture highly fluctuating as well as more stable day-to-day patterns of stool consistency, but this study was not designed to draw any conclusions on differences in the microbial composition between IBS and healthy subjects. Fecal dry weight percentage was used as an objective measure of stool consistency, which might have an advantage over the commonly used Bristol Stool Scale, since the latter was developed as a surrogate marker of whole-gut transit time and is subject to inter-individual differences in interpretation (Lewis and Heaton, 1997). A more detailed recommendation on the use of fecal dry weight percentage to correct for stool consistency in future (microbiota) studies was recently published elsewhere (Vork et al., 2019). Nevertheless, results were similar for BSS scores and fecal dry weight percentage, confirming that our results are suitable to make comparisons with previous studies. The results on the day-to-day stability of microbial richness were slightly less pronounced for the effective Shannon index when compared to Chao1 index and observed species. The Shannon index is a measure of biodiversity, taking into account the evenness next to the richness, and is therefore more affected by shifts in abundance of microbial taxa. Nevertheless, we could not demonstrate a significant effect of time on the association between stool consistency and microbial richness for any of the three measures.
A sample size of 24 subjects might be relatively small, but the use of repeated measures increases statistical power (Schuster et al., 2020). Furthermore, the performed analyses focus on within-subject rather than on between-subject differences. The female predominance in our study population reflects the sex distribution in IBS, which might question the generalizability to the general population. Previous studies, however, demonstrated no important differences in microbial composition between men and women (Arumugam et al., 2011; Falony et al., 2016).
In conclusion, this study supports an association between stool consistency and the fecal microbiota, but the overall microbial composition was not significantly related to day-to-day fluctuations in stool consistency. This consolidates the importance of considering stool consistency in gut microbiota research, but on the other hand, confirms the validity of the use of single fecal sampling for between-subject comparisons in the context of cross-sectional studies. Likewise, this indicates that in longitudinal studies evaluating within-subject microbial stability over longer periods (i.e. months or years) one fecal sample per time point should suffice. Concluding, a single fecal sample appears sufficient to gain a reliable picture of an individual’s microbiota within a short time period, even within subjects with large fluctuations in stool consistency.
Beate Niesler Chair COST-Action GENIEUR.
Department of Human Molecular Genetics, Institute of Human Genetics, Heidelberg University Hospital, Heidelberg, Germany.
nCounter Core Facility, Institute of Human Genetics, Heidelberg University Hospital, Heidelberg, Germany.
Interdisciplinary Center for Neurosciences (IZN), Heidelberg University, Heidelberg, Germany.
Gerard Clarke Department of Psychiatry and Alimentary Pharmabiotic Centre, University College Cork, Cork, Ireland.
Paul Enck Department of Psychosomatic Medicine and Psychotherapy, University Hospital Tuebingen, T̈übingen, Germany
Natasa Golic Laboratory for Molecular Microbiology, Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Belgrade, Serbia.
Kurt Hanevik Department of Clinical Science, University of Bergen, Bergen, Norway.
Fuad A. Iraqi Department of Clinical Microbiology and Immunology, Sackler Faculty of Medicine, Tel-Aviv University, Tel-Aviv, Israel.
Elena Philippou Department of Life and Health Sciences, University of Nicosia, Cyprus.
Division of Diabetes and Nutritional Sciences, King’s College London, London, UK.
Jeroen Raes Department of Microbiology and Immunology, KU Leuven, Leuven, Belgium.
Robin C. Spiller Nottingham Digestive Diseases Biomedical Research Unit, University of Nottingham, Queens Medical Centre, Nottingham, UK.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA682378.
The studies involving human participants were reviewed and approved by Medisch-etische toetsingscommissie azM/UM. The patients/participants provided their written informed consent to participate in this study.
Study concept and design: JP, WV, and DJ. Collection study materials: LV and ZW. Data analysis: LV, JJ, SB, SK, AS, MR-S, MP, and CM. Manuscript writing: LV, JP, and DJ. Constructive review of manuscript: JJ, SB, AS, WV, MR-S, ZW, AM, MP, and CM. All authors contributed to the article and approved the submitted version.
This manuscript results in part from collaboration and network activities promoted under the frame of the international network GENIEUR (Genes in Irritable Bowel Syndrome Research Network Europe), which has been funded by the COST program (BM1106, www.GENIEUR.eu) and is currently supported by the European Society of Neurogastroenterology and Motility (ESNM, www.ESNM.eu).
Part of the work of JP is financed by the Joint Programming Initiative A healthy diet for a healthy life (HDHL) Joint Action Intestinal Microbiomics (project number 50–52905–98–599). WV was partially supported by the SIAM Gravitation Grant 024.002.002 and Spinoza Award of the Netherlands Organization for Scientific Research. MR-S performed consultation services for Hemofarm AD, Serbia. ZW was supported to attend a scientific meeting by Will Pharma S.A. AM has received a ZonMw, The Netherlands Organization for Health Research and Development, health care efficiency grant to evaluate efficacy of peppermint oil in IBS, has received an unrestricted research grant from Will Pharma S.A., and received research funding from Allergan and Grünenthal on IBS topics. AM has given scientific advice to Bayer and Kyowa Kirin related to IBS and constipation, and received funding from Pentax Europe GmBH. Part of the work of CM is supported by the Instituto de Salud Carlos III, grant PI/17/00614 co-financed by the European Regional Development Fund (ERDF). Part of the work of DJ is financed by Grant Top Knowledge Institute (Well on Wheat), the Carbokinietics program as part of the NWO-CCC Partnership program and H2020 Nr. 848228/DISCOvERIE.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.639667/full#supplementary-material
Supplementary Table 1A | Inter-item correlations (Pearson correlations) between observed species of consecutive samples, for healthy subjects and IBS patients separately.
Supplementary Table 1B | Inter-item correlations (Pearson correlations) between effective Shannon index of consecutive samples, for healthy subjects and IBS patients separately.
Supplementary Table 2A | Results from linear mixed-effects models (with random intercept, fixed slopes, and scaled identity covariance structure). Regression coefficient indicates the direction and strength of the association between the predictor and dependent variable.
Supplementary Table 2B | Results from linear mixed-effects models (with random intercept, fixed slopes, and scaled identity covariance structure). Regression coefficient indicates the direction and strength of the association between the predictor and dependent variable.
Supplementary Figure 1 | Stool consistency for each sample and separately for all subjects (i.e. each color represents a subject), measured by fecal dry weight percentage (A) and Bristol Stool Scale (B).
Supplementary Figure 2 | Overview of the microbial composition of IBS and healthy subjects based on relative abundances on the phylum level.
Supplementary Figure 3 | Relative abundances for healthy subjects vs. IBS patients, significance tested using Mann Whitney U test. ** p < 0.01, NS, Not significant. (A) Firmicutes, (B) Bacteroidetes, (C) Actinobacteria, (D) Verrucomicrobia, (E) Proteobacteria.
Supplementary Figure 4 | (A) Generalized UniFrac PCoA plot per individual (different colors) and for healthy subjects vs. IBS patients (see figure annotations). (B) Average within-subject beta diversity (generalized UniFrac) for healthy subjects vs. IBS patients, significance tested using Mann Whitney U test, p = 0.80.
Supplementary Figure 5 | Bray Curtis Dissimilarity for days 2-7 compared to day 1 for IBS patients (A) and healthy subjects (B). Different colors representing individual subjects; black and dashed line representing the mean Bray Curtis Dissimilarity.
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the Human Gut Microbiome. Nature 473 (7346), 174–180. doi: 10.1038/nature09944
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 505 (7484), 559–563. doi: 10.1038/nature12820
Drossman, D. A. (2006). The Functional Gastrointestinal Disorders and the Rome III Process. Gastroenterology 130 (5), 1377–1390. doi: 10.1053/j.gastro.2006.03.008
Durban, A., Abellan, J. J., Jimenez-Hernandez, N., Artacho, A., Garrigues, V., Ortiz, V., et al. (2013). Instability of the Faecal Microbiota in Diarrhoea-Predominant Irritable Bowel Syndrome. FEMS Microbiol. Ecol. 86 (3), 581–589. doi: 10.1111/1574-6941.12184
Falony, G., Joossens, M., Vieira-Silva, S., Wang, J., Darzi, Y., Faust, K., et al. (2016). Population-Level Analysis of Gut Microbiome Variation. Science 352 (6285), 560–564. doi: 10.1126/science.aad3503
Jalanka, J., Major, G., Murray, K., Singh, G., Nowak, A., Kurtz, C., et al. (2019). The Effect of Psyllium Husk on Intestinal Microbiota in Constipated Patients and Healthy Controls. Int. J. Mol. Sci. 20 (2), 433. doi: 10.3390/ijms20020433
Jalanka-Tuovinen, J., Salonen, A., Nikkila, J., Immonen, O., Kekkonen, R., Lahti, L., et al. (2011). Intestinal Microbiota in Healthy Adults: Temporal Analysis Reveals Individual and Common Core and Relation to Intestinal Symptoms. PloS One 6 (7), e23035. doi: 10.1371/journal.pone.0023035
Jeffery, I. B., O’Toole, P. W., Ohman, L., Claesson, M. J., Deane, J., Quigley, E. M., et al. (2012). An Irritable Bowel Syndrome Subtype Defined by Species-Specific Alterations in Faecal Microbiota. Gut 61 (7), 997–1006. doi: 10.1136/gutjnl-2011-301501
Jeffery, I. B., Quigley, E. M., Ohman, L., Simren, M., O’Toole, P. W. (2012). The Microbiota Link to Irritable Bowel Syndrome: An Emerging Story. Gut Microbes 3 (6), 572–576. doi: 10.4161/gmic.21772
Johnson, A. J., Vangay, P., Al-Ghalith, G. A., Hillmann, B. M., Ward, T. L., Shields-Cutler, R. R., et al. (2019). Daily Sampling Reveals Personalized Diet-Microbiome Associations in Humans. Cell Host Microbe 25 (6), 789–802.e5. doi: 10.1016/j.chom.2019.05.005
Kassinen, A., Krogius-Kurikka, L., Makivuokko, H., Rinttila, T., Paulin, L., Corander, J., et al. (2007). The Fecal Microbiota of Irritable Bowel Syndrome Patients Differs Significantly From That of Healthy Subjects. Gastroenterology 133 (1), 24–33. doi: 10.1053/j.gastro.2007.04.005
Lewis, S. J., Heaton, K. W. (1997). Stool Form Scale as a Useful Guide to Intestinal Transit Time. Scand. J. Gastroenterol. 32 (9), 920–924. doi: 10.3109/00365529709011203
Longstreth, G. F., Thompson, W. G., Chey, W. D., Houghton, L. A., Mearin, F., Spiller, R. C. (2006). Functional Bowel Disorders. Gastroenterology 130 (5), 1480–1491. doi: 10.1053/j.gastro.2005.11.061
Lyra, A., Rinttila, T., Nikkila, J., Krogius-Kurikka, L., Kajander, K., Malinen, E., et al. (2009). Diarrhoea-Predominant Irritable Bowel Syndrome Distinguishable by 16S rRNA Gene Phylotype Quantification. World J. Gastroenterol. 15 (47), 5936–5945. doi: 10.3748/wjg.15.5936
Malinen, E., Krogius-Kurikka, L., Lyra, A., Nikkila, J., Jaaskelainen, A., Rinttila, T., et al. (2010). Association of Symptoms With Gastrointestinal Microbiota in Irritable Bowel Syndrome. World J. Gastroenterol. 16 (36), 4532–4540. doi: 10.3748/wjg.v16.i36.4532
Malinen, E., Rinttila, T., Kajander, K., Matto, J., Kassinen, A., Krogius, L., et al. (2005). Analysis of the Fecal Microbiota of Irritable Bowel Syndrome Patients and Healthy Controls With Real-Time PCR. Am. J. Gastroenterol. 100 (2), 373–382. doi: 10.1111/j.1572-0241.2005.40312.x
Matto, J., Maunuksela, L., Kajander, K., Palva, A., Korpela, R., Kassinen, A., et al. (2005). Composition and Temporal Stability of Gastrointestinal Microbiota in Irritable Bowel Syndrome–a Longitudinal Study in IBS and Control Subjects. FEMS Immunol. Med. Microbiol. 43 (2), 213–222. doi: 10.1016/j.femsim.2004.08.009
Maukonen, J., Satokari, R., Matto, J., Soderlund, H., Mattila-Sandholm, T., Saarela, M. (2006). Prevalence and Temporal Stability of Selected Clostridial Groups in Irritable Bowel Syndrome in Relation to Predominant Faecal Bacteria. J. Med. Microbiol. 55 (Pt 5), 625–633. doi: 10.1099/jmm.0.46134-0
Rajilic-Stojanovic, M., Biagi, E., Heilig, H. G., Kajander, K., Kekkonen, R. A., Tims, S., et al. (2011). Global and Deep Molecular Analysis of Microbiota Signatures in Fecal Samples From Patients With Irritable Bowel Syndrome. Gastroenterology 141 (5), 1792–1801. doi: 10.1053/j.gastro.2011.07.043
Rajilic-Stojanovic, M., Heilig, H. G., Tims, S., Zoetendal, E. G., de Vos, W. M. (2012). Long-Term Monitoring of the Human Intestinal Microbiota Composition. Environ. Microbiol. 15 (4), 1146–1159. doi: 10.1111/1462-2920
Salonen, A., de Vos, W. M., Palva, A. (2010). Gastrointestinal Microbiota in Irritable Bowel Syndrome: Present State and Perspectives. Microbiology 156 (Pt 11), 3205–3215. doi: 10.1099/mic.0.043257-0
Schuster, R., Schreyer, M. L., Kaiser, T., Berger, T., Klein, J. P., Moritz, S., et al. (2020). Effects of Intense Assessment on Statistical Power in Randomized Controlled Trials: Simulation Study on Depression. Internet Interv. 20, 100313. doi: 10.1016/j.invent.2020.100313
Tigchelaar, E. F., Bonder, M. J., Jankipersadsing, S. A., Fu, J., Wijmenga, C., Zhernakova, A. (2016). Gut Microbiota Composition Associated With Stool Consistency. Gut 65 (3), 540–542. doi: 10.1136/gutjnl-2015-310328
U.S. Department of Health and Human Services FaDA, Center for Drug Evaluation and Research (CDER) (2012). “Guidance for Industry Irritable Bowel Syndrome”, in Clinical Evaluation of Drugs for Treatment.
Vandeputte, D., Falony, G., Vieira-Silva, S., Tito, R. Y., Joossens, M., Raes, J. (2016). Stool Consistency Is Strongly Associated With Gut Microbiota Richness and Composition, Enterotypes and Bacterial Growth Rates. Gut 65 (1), 57–62. doi: 10.1136/gutjnl-2015-309618
Vandeputte, D., Kathagen, G., D’Hoe, K., Vieira-Silva, S., Valles-Colomer, M., Sabino, J., et al. (2017). Quantitative Microbiome Profiling Links Gut Community Variation to Microbial Load. Nature 551 (7681), 507–511. doi: 10.1038/nature24460
Vanhoutte, T., Huys, G., Brandt, E., Swings, J. (2004). Temporal Stability Analysis of the Microbiota in Human Feces by Denaturing Gradient Gel Electrophoresis Using Universal and Group-Specific 16S rRNA Gene Primers. FEMS Microbiol. Ecol. 48 (3), 437–446. doi: 10.1016/j.femsec.2004.03.001
Vork, L., Wilms, E., Penders, J., Jonkers, D. (2019). Stool Consistency: Looking Beyond the Bristol Stool Form Scale. J. Neurogastroenterol. Motil. 25 (4), 625. doi: 10.5056/jnm19086
Zoetendal, E. G., Akkermans, A. D., Akkermans - van Vliet, W. M., de Visser, A. G. M., de Vos, W. M. (2001). The Host Genotype Affects the Bacterial Community in the Human Gastrointestinal Tract. Microb. Ecol. Health Dis. 13, 129–134. doi: 10.3402/mehd.v13i3.8013
Keywords: fecal microbiota, intestinal microbiome, stool consistency, irritable bowel syndrome, adult
Citation: Vork L, Penders J, Jalanka J, Bojic S, van Kuijk SMJ, Salonen A, de Vos WM, Rajilic-Stojanovic M, Weerts ZZRM, Masclee AAM, Pozuelo M, Manichanh C and Jonkers DMAE (2021) Does Day-to-Day Variability in Stool Consistency Link to the Fecal Microbiota Composition? Front. Cell. Infect. Microbiol. 11:639667. doi: 10.3389/fcimb.2021.639667
Received: 01 February 2021; Accepted: 11 June 2021;
Published: 20 July 2021.
Edited by:
Marloes Dekker Nitert, The University of Queensland, AustraliaReviewed by:
Muriel Derrien, Nutricia Research (France), FranceCopyright © 2021 Vork, Penders, Jalanka, Bojic, van Kuijk, Salonen, de Vos, Rajilic-Stojanovic, Weerts, Masclee, Pozuelo, Manichanh and Jonkers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisa Vork, bC52b3JrQG1hYXN0cmljaHR1bml2ZXJzaXR5Lm5s; Chaysavanh Manichanh, Y21hbmljaGFAZ21haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.